SUMMARY
In this hypothesis article, we explore the origin of the eukaryotic nucleus. In doing so, we first look afresh at the nature of this defining feature of the eukaryotic cell and its core functions—emphasizing the utility of seeing the eukaryotic nucleoplasm and cytoplasm as distinct regions of a common compartment. We then discuss recent progress in understanding the evolution of the eukaryotic cell from archaeal and bacterial ancestors, focusing on phylogenetic and experimental data which have revealed that many eukaryotic machines with nuclear activities have archaeal counterparts. In addition, we review the literature describing the cell biology of representatives of the TACK and Asgardarchaeaota - the closest known living archaeal relatives of eukaryotes. Finally, bringing these strands together, we propose a model for the archaeal origin of the nucleus that explains much of the current data, including predictions that can be used to put the model to the test.
KEYWORDS: nucleus, evolution, archaea, eukaryogenesis, inside-out model, theory, asgard archaea, nucleolus, origins, topology
INTRODUCTION
The nucleus is the defining feature of eukaryotic cells. All eukaryotes have one and its presence marks them out as different from the vast array of bacteria and archaea (Fig. 1). While the average bacterial or archaeal cell tends to be structurally simple [with notable exceptions (1–4)], all eukaryotic cells possess an elaborate maze of internal membranes that extends outwards from the nuclear envelope. Sitting at the heart of the cell, the nuclear compartment acts as a safe haven for the genome. Beginning with transcription, splicing, and mRNA export, the directed flow of this genetically encoded information propagates out from the nucleus to the cell periphery (Fig. 1 and 2). While all eukaryotes share this dynamic organization, it is not clear how it arose during evolution. In an attempt to answer this question, in this article we bring together knowledge about nuclear structure and function, phylogenetic data, and recent cell biological studies in archaea. As we will see, the synthesis of these disparate strands of information suggests an archaeal origin for many nuclear activities, and leads us to propose a possible path for the gradual emergence of a separate nucleoplasm and cytoplasm early on during eukaryogenesis.
Fig 1.
These diagrams compare the spatial organization of gene expression in a schematic eukaryotic and bacterial cell. The images emphasize that, while transcription and translation are separate in eukaryotic cells, they are coupled in many bacteria. A key question posed by this article is whether or not transcription and translation are strongly coupled or partially uncoupled to enable the local translation of some transcripts in close archaeal relatives of eukaryotes.
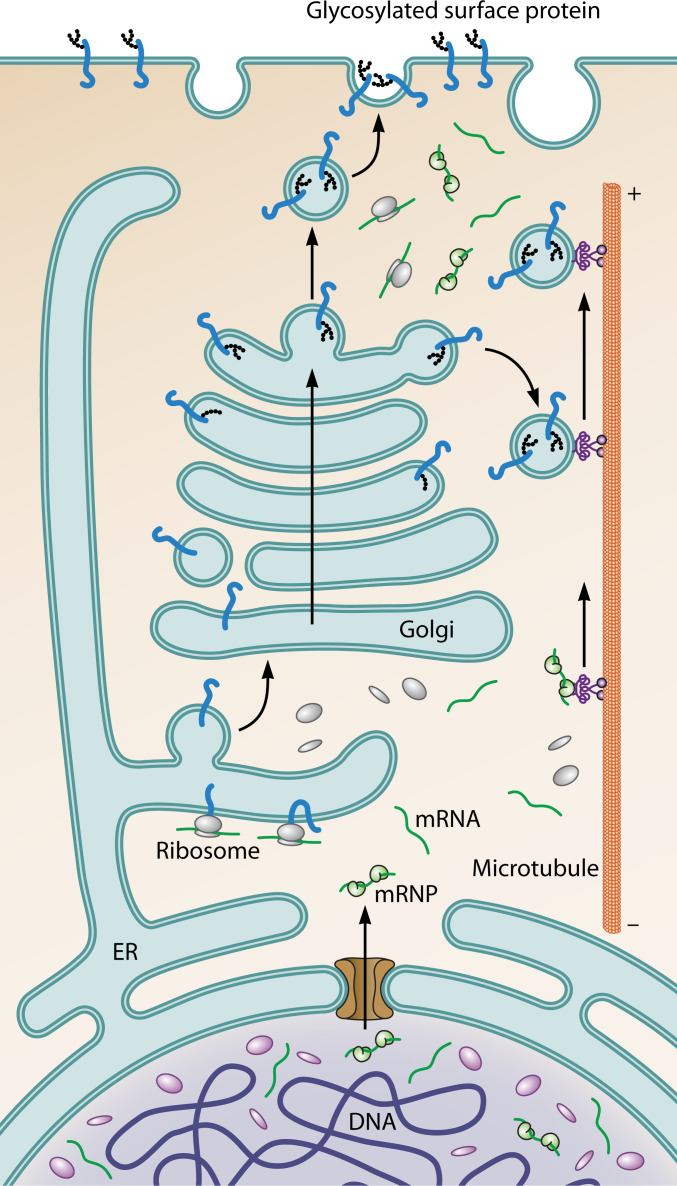
Fig 2.
Diagram shows the path of genetically encoded information in eukaryotic cells as it moves out of the nucleus (bottom) toward the cell periphery (top). DNA is transcribed in the nucleus. The RNAs generated are then processed and exported through nuclear pores into the cytoplasm. While many messenger RNAs (mRNAs) are rapidly translated upon entering the cytoplasm, others remain inaccessible to ribosomes as the result of RNA-binding proteins. Some of these are trafficked along microtubules in an inactive state, enabling local protein synthesis, e.g., at the tips of axons. In parallel, proteins carrying signal peptides, which are translated by ribosomes situated at the surface of the rough endoplasmic reticulum (ER), move through the ER and Golgi, where they are modified by glycosylation, packaged into vesicles, and trafficked out to the cell periphery along microtubules.
It should be noted that, while this article builds on ideas put forward in the inside-out model of eukaryogenesis (5), the model we propose here is distinct and more delimited in scope than the original model in that it does not attempt to explain the evolution of the eukaryotic cell as a whole. As a result, it is possible for elements of this model of nuclear evolution to be wrong without this invalidating the inside-out model. Conversely, certain elements of the inside-out model of eukaryogenesis could be erroneous without this having an impact on the validity of the model of the evolution of the nucleus presented here. Because this article focuses on the nucleus, it does not include a comprehensive overview of the alternative models put forward to explain eukaryogenesis. While these are covered in brief in this article (see later), we point readers interested in a detailed discussion of the topic to some excellent recent reviews on the subject (6–11).
NUCLEAR ARCHITECTURE
To begin, we discuss the nature of the eukaryotic nucleus, distinct nuclear sub-compartments, the nuclear envelope, and the pores that connect the nucleoplasm and cytoplasm.
The nuclear-cytoplasm divide
Because the nucleus appears to be enveloped by two lipid bilayers when imaged in cross-section using electron microscopy, it is commonly said to possess a bounding “double membrane.” This view is inaccurate though because the inner and outer nuclear membranes are physically continuous (Fig. 2 and 3). The two bilayers are, in fact, connected to one another via numerous highly curved membrane connections or pores (Fig. 3). As a result, lipids are rapidly exchanged between the inner nuclear envelope and the endoplasmic reticulum (ER), without the need for protein channels like those required to transport lipids between the ER and the plasma membrane and mitochondria (12). Furthermore, the lumen of the ER and the space that lies between the inner and outer nuclear envelope forms a single continuous fluid-filled network that defines the nuclear boundary and extends throughout the eukaryotic cell (Fig. 2). In addition, because the nuclear envelope contains numerous pores, the nucleoplasm and cytoplasm should not be viewed as distinct compartments separated by a double membrane, but rather as sub-domains or discrete phases of a single compartment between which small molecules freely diffuse. In most eukaryotic cells, this mixing of the nucleoplasm and cytoplasm is accentuated at every mitosis following the loss of nuclear pores (13). The continual exchange of nuclear material with the cytoplasm marks the nucleus out as very different from the other membrane-bound compartments within the eukaryotic cell. The contrast is clear when we consider non-nuclear organelles, such as endosomes, lysosomes, mitochondria, and peroxisomes, which tend to have bounding membranes that function as physical barriers across which hydrophilic molecules cannot move unaided. As a result, each of these organelles is able to establish a unique internal milieu that is both suited to its function and profoundly different from the surrounding cytoplasm. In the case of the lumen of the lysosome, for example, this includes a low internal pH [generated by active transport via pumps like the AV-type ATP synthase (14)], a high calcium concentration, an oxidizing rather than reducing environment, and limited pools of nucleotides (15). Because of this, physical insults that perforate the bounding membrane of any one of these organelles can have catastrophic consequences for cell function and often serve as triggers warning cells of pathogen invasion (16).
Fig 3.
(A) Diagram shows the structure of the eukaryotic nucleus, and the endoplasmic reticulum to which it is connected. The nucleus is studded with nuclear pore complexes (NPCs). These NPCs sit at sites of high membrane curvature where the inner and outer nuclear membranes meet, and function as gated channels through which material can move between the nucleoplasm and cytoplasm. (B) Image shows a single eightfold symmetric nuclear pore complex inserted into the membrane viewed from one side (with kind permission of Agnieszka Obarska and Martin Beck). (C) New nuclear pores are inserted into the nuclear envelope via two processes: (i) insertion into gaps in the nuclear envelope as it reforms at mitotic exit and (ii) via interphase insertion. The diagram (adapted from Otsuke and Ellenberg, 2016, and based on electron microscopy data) shows a proposed path for interphase nuclear pore insertion from the inside out.
Sub-regionalization of the nucleus
At the same time, the nucleus is not structurally or biochemically uniform (17). Like the cytoplasm, the nucleoplasm possesses functionally distinct domains (18). It contains a nucleolus (Fig. 3A), one of the first organelles to be described by cell biologists, where rRNAs are processed and assembled into pre-ribosomes (19). Nuclei also contain near spherical Cajal bodies, sites of RNP processing, and maturation, where telomerase and spliceosomal machinery are concentrated (17). DNA replication (20) and repair (21) activities tend to be clustered in discrete foci in the nucleus. Transcribed and silenced chromatin have distinct nuclear locations (22). Individual chromosomes have been reported to occupy their own territories (23). And when perturbed, nuclei accumulate local stress granules (24) and, when infected, viral assembly factories (25). Because these nuclear structures are not bound by membranes, they are likely established and maintained by weak multivalent interactions between proteins and/or nucleic acids. In some cases, these structures resemble vinegar droplets in olive oil, distinct liquid phases that some authors have termed “biomolecular condensates” (26). Thus, even though nuclei remain largely free of internal membranes during both interphase and mitosis (27), the nucleus is home to an ensemble of local sub-structures where different cellular components can be concentrated as and when required (26).
NPCs: gate-keepers controlling traffic between the nucleoplasm and cytoplasm
The exchange of material between the nucleus and cytoplasm is regulated by huge rivet-like nuclear pore complexes (NPCs) (28) (Fig. 3B). These NPCs physically support the highly curved membrane at the sites at which inner and outer nuclear membranes meet, and are among the largest and most complex molecular assemblies in all of biology. NPCs are constructed from a set of ~500 proteins in yeast and ~1,000 in humans, with a total mass of 50 or 120 megadaltons, respectively. The pore’s central scaffold is composed of two 8-fold symmetric rings stacked on top of one another (one facing the cytoplasm, the other the nucleoplasm). These symmetric rings act as a physical support for more peripheral proteins that sit at the cytoplasmic and/or nuclear face of the pore (29). NPCs are anchored within the curved portion of the membrane by a small number of proteins that are physically embedded in the membrane. The NPC scaffold surrounds a central aqueous hole that has an internal diameter of ~50 nm (30), which is enormous by cell standards. Thus, while chromatin, small vesicles, and most viral capsids cannot move between the nucleoplasm and cytoplasm, NPCs are large enough to allow the passage of some of the largest multicomponent molecular complexes present in cells, such as 25 nm diameter pre-ribosomes. The transit of large proteins and protein complexes through the NPC is regulated by a central plug of disordered peptides rich in phenylalanine/glycine (FG) repeats (28). By engaging in multiple low-affinity interactions, these FG repeats generate a dynamic mesh that acts as a selectivity filter to limit the passage of macromolecules in a manner that depends on their size (31). As a result, while small water-soluble molecules diffuse freely through the NPC between nuclear and cytoplasmic compartments, molecules or complexes greater than ~30 kDa in size, such as large proteins, RNPs, pre-ribosomes, and mature tRNAs, only cross the nuclear/cytoplasm compartment boundary if recognized by importins or exportins. These carrier proteins alter the biophysical properties of the FG repeats that form the sieve at the center of NPCs to facilitate the passage of cargo to which they are bound (32). Flux across the nuclear envelope has been estimated to be ≈1 million macromolecules/second (32).
The association of cargo proteins with importins and exportins depends on the presence of specific Nuclear Localization Sequences (NLS) (33) and Nuclear Export Sequences, respectively, and is regulated by the nucleotide status of an associated small GTPase called Ran [reviewed in reference (34)]. Ran is loaded with GTP in the nucleus (by a chromatin-bound guanine nucleotide exchange factor or GEF) but switches to its GDP form in the cytoplasm as a result of the local stimulation of Ran’s GTPase activity by cytoplasmic GTPase activating proteins (GAPs). The physical separation of GEFs and GAPs sets up a gradient in Ran-GTP/Ran-GDP concentration across the nuclear/cytoplasmic compartment boundary that cells use to power the net, directed movement of large complexes either into or out of the nucleus, depending on whether the cargo is bound by importins (released by Ran-GTP) or exportins (released by Ran-GDP). It is important to note, however, that this Ran shuttle doesn’t require a membrane. All that is needed for Ran to polarize cellular space is the physical separation of GEFs and GAPs. In fact, Ran is used in exactly this way to polarize a single aqueous environment (35) following the loss of the nuclear/cytoplasmic compartment barrier in oocytes (36), as well as during mitosis in fertilized eggs and somatic cells (34, 37). In these instances, the Ran-GTP/GDP gradient is used as a measure of distance (38, 39) to induce the nucleation of microtubules (40–42) and the remodeling of the cell cortex in the vicinity of RanGEF-bound chromatin (43–45). More controversially, Ran, importin, and a small number of nuclear pore proteins have also been implicated in the regulation of traffic across the diffusion barrier at the base of cilia (46), enabling, among other things, the local translation of cilia-targeted mRNAs. This suggests the possibility that some of the machinery used to govern the passage of molecules across the nuclear pore may have been re-deployed to regulate traffic into and out of cilia (5, 47).
Ran is not the only system controlling traffic across the nuclear pore. The Ntf2 system (32, 48) operates in parallel to Ran in many eukaryotes to support the unidirectional movement of mRNAs and pre-ribosomes from the nucleoplasm to the cytoplasm (49), which is one of the most important activities of the NPC (32, 48). In this process, Ntf2 homologs help processed mRNAs that are part of large protein-RNA complexes, called mRNPs (ribonucleoprotein particles containing mRNA), to cross the disordered mesh of FG repeats (50). Following export, Ntf2 proteins are then removed from these mRNPs by a family of ATP-dependent DEAD-box helicases that sit at the cytosolic face of the nucleus (51). This renders the export process irreversible. A similar logic applies to mRNAs packaged as part of inactive mRNPs that are trafficked to peripheral regions of the eukaryotic cell, like the tips of axons and cilia. Again, this type of regulation doesn’t require a compartment boundary. In these cases, mRNPs are silent when trafficked, but are disassembled when required by DEAD-box helicases located at the cell periphery, enabling ribosome binding and the initiation of protein synthesis at the appropriate time and place (52) (Fig. 1 and 3).
NPC assembly and disassembly
Although NPCs are some of the most stable structures in the cell (53), they are removed from the nuclear envelope at every division. The extent, timing, and location of mitotic nuclear pore disassembly vary widely across eukaryotes depending on the extent to which nuclear division is accompanied by a loss of the nuclear/cytoplasmic compartment boundary (13). The loss of NPCs opens up transient holes in the mitotic nuclear barrier. In an “open” mitosis, say in a human cell, the entire set of NPCs are disassembled, leading to extensive mixing of the mitotic nucleoplasm and cytoplasm. By contrast, in a dividing fission yeast nucleus (54), a classical example of “closed” mitosis in which the nucleo-cytoplasmic diffusion barrier is retained during the process of nuclear division, NPCs are only disassembled at the center of the late anaphase nuclear bridge. This local loss of NPCs facilitates remodeling of the nuclear envelope and ER at the center of the bridge, allowing nuclear division to go to completion, yielding two daughter nuclei. Whatever the mode of division, to counter the loss of NPCs induced by rounds of mitosis and their dilution during interphase nuclear envelope growth new NPCs must be continually inserted into the eukaryotic nuclear envelope (Fig. 3C). NPC insertion occurs via two distinct pathways (55, 56). First, nucleoporin sub-complexes are recruited into gaps left in the bounding nuclear envelope when the nuclear-cytoplasmic compartment boundary is re-established at mitotic exit (55). During this process, ESCRT-III (endosomal sorting complexes required for transport -III) proteins repair errors and seal holes that remain (57). Second, during interphase, pores are inserted into growing nuclei in discrete steps from the inside-out (5), via a process that has only recently been studied in any detail (55, 56) (Fig. 3C). This insertion begins with the out-folding of the inner nuclear envelope. Nascent nuclear pore components then accumulate at the neck of small membrane nuclear blebs, stabilizing their structure. The fusion of the bulging inner membrane with the overlying outer nuclear envelope then completes the process, generating a membrane-lined NPC that connects the nucleoplasm and cytoplasm. Finally, in some cells, like oocytes, which undergo extensive periods of growth, nuclear growth is fueled by the insertion of cytoplasmic membrane sheets termed annulate lamellae that contain pre-assembled pores (58). In many eukaryotes, AAA-ATPase Torsin helps power the membrane remodeling that accompanies NPC insertion (59). While capturing snapshots of putative intermediates by electron microscopy has proven hard in typical eukaryotic cells imaged under physiological conditions, similar structures have been observed by electron microscopy under perturbed conditions across a wide range of eukaryotes (60, 61), implying that this process of pore insertion may be a generic and ancient one, as suggested by the inside-out model of eukaryogenesis (5).
WHAT IS THE NUCLEUS FOR?
Having reviewed nuclear architecture and the machinery used to control the exchange of material between the nucleus and the cytoplasm, we now discuss what might be considered the main functions of the nucleus. We can point to six, more or less distinct roles of the nucleus.
First, the eukaryotic nucleus provides a well-defined space in which to house and organize the genome. In eukaryotes, whose genomes (from 10 Mb to >100,000 Mb) tend to be much larger than those found in prokaryotes (0.5–10 Mb), this requires extensive genome compaction, which is made possible by a large ensemble of abundant DNA-binding proteins.
Second, the eukaryotic nucleus protects the DNA from physical insults. In mechanically soft cells, like animal cells and Dictyostelium, the nuclear envelope is supported by an underlying lamina, analogous to the skull under our scalps, which prevents external physical forces from disrupting the information processing activities going on within the nucleus. Under moderate pressure, mechanical signaling in the nuclear envelope triggers an increased cortical contractility, which likely further insulates the cell from external forces (62, 63). If the mechanical resistance of the lamina is overcome, the resulting rupture of the nuclear/cytoplasmic barrier allows antiviral defense systems concentrated in the cytoplasm, such as TREX-1, to leak into the nucleus, inducing DNA damage (64).
Third, the nucleus likely provides a chemically distinct and more reducing environment in which to house the genome. This is achieved, in part, through the exclusion of metabolically active organelles like mitochondria. This has the effect of limiting the exposure of DNA to genotoxic free radicals generated by respiration, perhaps helping to keep mutation rates within manageable levels.
Fourth, the eukaryotic nuclear envelope provides a physical boundary that separates transcription and translation. This distinguishes the core steps in eukaryotic gene expression from those described for most bacteria and some archaea (65, 66)—where the translation of genes frequently starts before transcription has finished [see reference (67) for an exception] (Fig. 1). In eukaryotes, the physical uncoupling of these processes facilitates the processing of newly transcribed RNAs through the addition of a 5′ cap, a polyA tail, and intron removal (68) - something that takes time. Quality control machinery ensures that mRNAs are only exported to the cytoplasm once processing is complete (49). The spatial separation of transcription and translation also relies on the confinement of active ribosomes to the cytoplasm, through a carefully choreographed process of rRNA processing and ribosome assembly. This begins in the nucleolus where rRNAs are transcribed, processed, and modified (69). [Note that rRNAs and tRNAs are processed via the removal of insertions and through modifications in bacteria and archaea as well as in eukaryotes (70)]. Pre-ribosomes assembled within the eukaryotic nucleus are kept in an inactive state by proteins such as eIF6 (71). Inactive pre-ribosomes are then exported from the nucleus through NPCs so that the final steps of ribosomal assembly and activation can be completed in the cytoplasm, where mature ribosomal subunits encounter fully processed mRNAs - their substrates. In combination, these two activities, intranuclear mRNA processing and extranuclear ribosome assembly, function to spatially and temporally uncouple transcription from translation in all extant eukaryotic cells. This has been suggested to augment the ability of eukaryotes to regulate gene expression, for example, through alternative splicing (72).
Fifth, the nucleus functions as a coherent aqueous membrane-free space in which genomes composed of multiple chromosomes can be aligned and segregated. Since the forced association of mitotic chromatin with membranes has been shown to interfere with chromosome segregation (27), the transient loss of chromatin-nuclear envelope binding may be a functionally important part of the mitotic program.
Finally, the partial separation of the nucleus and cytoplasm likely acts as a barrier to the replication of viruses entering the cell from the environment by enabling DNA nucleases to police the cytoplasm (73), and by sequestering away host machinery capable of replicating genomic DNA within the nucleus.
THE ORIGINS OF THE EUKARYOTIC ENDOMEMBRANE SYSTEM
Having discussed the structure and function of the nucleus, in the following section we discuss the challenges of determining when the nucleus first emerged during evolution. In doing so, we look at recent phylogenomic data which suggest that eukaryotic cells likely arose as the result of a merger between an alphaproteobacterial cell and an asgardarchaeotal cell (Fig. 4). We then briefly discuss the different types of models that have been proposed to explain the available data.
Fig 4.
Schematic view of the tree of life with Bacteria in green, Archaea in purple, and eukaryotes (i.e. Eukaryota) in pink. Labels on the tree mark the Last Universal Common Ancestor (LUCA), the Last Archaeal Common Ancestor (LACA), the Last Bacterial Common Ancestor (LBCA), and the Last Eukaryotic Common Ancestor (LECA). As depicted in the diagram, LECA is hypothesized to be derived from the merging of at least two partners: an alphaproteobacterial symbiont and an asgardarchaeotal host.
The absence of evidence
While the nucleus plays an indispensable role in the life of all eukaryotic cells, its origin and the timing of its emergence relative to other eukaryotic traits remain unclear. The challenge of establishing a precise order of events that led to the formation of a eukaryotic cell with a nucleus and a complex endomembrane system, in part, reflects the lack of “simple eukaryotes,” i.e., direct descendants of intermediates along the path toward the formation of today’s complex eukaryotes that have remained relatively unchanged over long periods of evolutionary time. Diplomonads like Giardia were once thought to be early diverging “primitive” eukaryotes of this type. This is because Giardia cells lack canonical mitochondria and appear to have a simplified endomembrane system with a single internal membrane (including a continuous inner and outer nuclear envelope, ER, Golgi, and endocytic/lysosomal compartment) (74). More recently, however, Giardia cells have been shown to possess mitochondrial-like organelles, referred to as mitosomes (75, 76). Thus, if aspects of Giardia cell biology appear simple, this simplicity is likely a consequence of evolutionary streamlining—as has been shown to be the case for other microbial eukaryotes, including in the oxymonad Monocercomonoides sp., which has lost mitochondria altogether (77). Thus, as far as we know, all extant eukaryotic lineages emerged from a complex Last Eukaryotic Common Ancestor (LECA) (78, 79) that had all the hallmarks of a modern eukaryote, via an explosive evolutionary radiation sometime between about 1 and 2.3 billion years ago (80–83) (Fig. 4 and 5). The absence of architecturally simple eukaryotes situated as sister to a clade of more complex eukaryotes sets up a kind of evolutionary event horizon beyond which we cannot see. The search for the origin of the eukaryotic nucleus, therefore, requires a journey deeper into our prokaryotic past.
Fig 5.
Schematic tree of life with Bacteria in green, Archaea in purple, and Eukaryota in pink—emerging from the merger of an alphaproteobacterial symbiont with an asgardarchaeotal host. Horizontal gene transfer, which can complicate the phylogenetic analysis, is indicated by interconnecting lines. The light-shaded boxes indicate enzymes of likely archaeal (purple), bacterial (green), or unknown (gray) origin. Question marks indicate putative origins that are less clear.
An overview of theories of eukaryogenesis
It has long been postulated that the complex intracellular architecture of eukaryotes arose via endosymbiosis, when one or more prokaryotic cells took up residence inside another cell [reviewed in reference (84)]. In line with this hypothesis, early phylogenetic studies showed that the eukaryotic genome is composite in nature, including a strong alphaproteobacterial signal (85–89), together with a strong archaeal signal (90–95). These bacterial and archaeal ancestors of eukaryotes appear to have made complementary contributions to the biology of the eukaryotic cell. As an example of this, the core mitochondrial machinery (e.g., mitochondrial rRNA, ribosomal proteins, together with many of the enzymes involved in electron transport) appears to be of alphaproteobacterial origin (96–100) (Fig. 5). Conversely, the core information processing machinery, including that involved in DNA replication (101–104), chromatin structure (105–109), transcription (110–115), translation (116–122), and RNA processing (123, 124), along with cell division (125–127) (Fig. 5), appears to be of archaeal origin. These data are consistent with the possibility that an alphaproteobacterial symbiont that gave rise to mitochondria was taken up by an archaeal host cell.
The discovery of the Lokiarchaeota (now Lokiarchaeia (128)) and other members of the Asgard archaea (now Asgardarchaeota (128)) in metagenomic assemblies from environmental samples (129–133) significantly strengthened the case for eukaryotes having emerged from an archaeal host. This is because phylogenetic analyses showed that Asgardarchaeota are the closest living archaeal relatives of eukaryotes (Fig. 4). Furthermore, different Asgardarchaeota genomes were found to encode a wide range of so-called eukaryotic signature proteins (ESPs)—proteins whose homologs were previously thought to be specific to eukaryotes (129–131, 133). Strikingly, these include cytoskeletal actin and several actin regulators (134, 135), the ESCRT membrane remodeling machinery, ubiquitin, and its associated ligases and de-ubiquitinases (136), together with numerous small GTPases—most notably the Rags (129–131, 137, 138) (Fig. 5). While some of these proteins have homologs among members of the TACK archaea and Euryarchaeota (78, 94, 139), these proteins are more widely distributed across the various Asgardarchaeota, where they often appear to be part of cellular machinery that is more similar in terms of complexity and component parts to the corresponding eukaryotic machinery (131, 133, 140). Many of these genes were likely present in the last common ancestor of Asgardarchaeota and eukaryotes, and they have likely played important roles in the evolution of cellular complexity during eukaryogenesis (129–131, 133).
While these findings support the hypothesis that eukaryotes emerged from a partnership between at least one alphaproteobacterial ancestor (85), which gave rise to the mitochondria, and an asgardarchaeotal partner (78) (see Fig. 4 and 5), the debate about eukaryotic origins is far from settled. The reasons for this are severalfold. First, families of genes assigned to the last eukaryotic common ancestor (LECA) include representatives with diverse or unresolved prokaryotic origins (96–100, 141). This has led some to postulate three or more partners contributing to the emergence of eukaryotes (8). The difficulties in assessing such claims have been confounded by the fact that all alphaproteobacterial (142), asgardarchaeotal (130, 143, 144), and eukaryotic genomes (145–147) have been shaped by horizontal gene transfer events throughout their evolutionary history (see Fig. 5). Furthermore, eukaryotic genomes are characterized by a large set of proteins and domains absent from prokaryotes, whose origins remain unknown (148, 149). Finally, the current phylogenetic data provide few insights as to the origins of organelles, including the nucleus, and have done little to reveal the relative timing of organelle acquisition. Because of these uncertainties, the current data are compatible with aspects of different models put forward in an attempt to explain the origins of the eukaryotic cell (see Box 1 and Box 2 together with Fig. 6 and 7).
Box 1. Brief overview of select models proposed as explanations for the origin of the nucleus.
Models of eukaryogenesis can be divided into those that imagine the nucleus having been acquired via symbiosis and those that imagine it having arisen by the remodeling of an ancestral cell. We refer readers to comprehensive reviews (6–8, 11, 78, 90) for an in-depth discussion. Note that all models have to contend with: (i) the phylogenetic data; (ii) the acquisition of alphaproteobacterial-derived mitochondria with two membranes; (iii) the acquisition or gradual evolution of a nucleus; (iv) eukaryotic cell organization (the presence of a continuous nuclear envelope and ER, and a topologically separate plasma membrane); and (v) the fact that, while much of the core protein machinery underlying eukaryotic cell organization appears to have its origins in archaea, eukaryotic membranes are largely constructed from bacterial-type lipids, which host both bacterial-derived respiratory chains (e.g., in mitochondria) as well as important transmembrane proteins of archaeal origin (e.g., the translocon) (Fig. 5).
Endosymbiotic nuclear origin:
Tripartite hydrogen and sulfur-transfer-based model (syntrophy). The syntrophy model proposes that the nucleus derives from an endosymbiotic archaeon that was engulfed by a sulfate-reducing deltaproteobacterial host cell, which also internalized a facultatively aerobic sulfide-oxidizing alphaproteobacterial cell that gave rise to mitochondria (8). The model’s strength is that it can explain why eukaryotic cells possess bacterial rather than archaeal-type lipid membranes. A weakness of the model is the lack of clarity regarding how requisite changes in membrane organization occurred. First, it is not clear how the archaeon entered the cytoplasm of the bacterial host. Second, under the model, the bacterial host formed membranes around this central archaeal cell via the ingathering of membranes as in classical outside-in models. As explained in Fig. 7, this requires extensive membrane remodeling both to seal invaginating membranes and to form a nuclear compartment that contains pores and is continuous with the endoplasmic reticulum. Third, the model proposes that the archaeal membrane was then somehow lost. This would have induced the mixing of deltaproteobacterial and archaeal ribosomal machinery within in a single compartment, likely compromising translation. In this regard, it is worth noting that eukaryotic ribosomes seem to be derived from archaeal, alphaproteobacterial, and, in the case of plastid-containing eukaryotes, cyanobacterial homologs, without strong indications of the existence of an additional prokaryotic source. Finally, the model proposes that one of the two outer membranes of the deltaproteobacterial host was lost and the ER internalized. The problem here is that this could not have been achieved in any simple way. Remove the outer membrane, and the ER lumen now becomes continuous with the environment. Remove the inner membrane, and the ER is lost and, more problematically, the intra-membrane luminal space fuses with the cytoplasm.
Viral origin hypothesis. This model proposes that the nucleus arose from a DNA virus replication factory in an archaeal cell, which also hosted the alphaproteobacterial endosymbiont (150, 151). A key feature of this model is its focus on explaining evolution of the separation of transcription and translation, given that both archaea and bacteria tend to lack the machinery that generates and reads the 5′ m7G cap characteristic of eukaryotic mRNAs (152). A significant problem with this model is that the double-membrane compartment that viruses generate to allow RNA processing arises via the remodeling of internal membranes (153, 154). Thus, the model assumes a pre-existing dynamic endomembrane system - whose origin all other models of eukaryogenesis aim to resolve. It is also not clear where traces of this virus are to be found in eukaryotic genomes (155).
Box 2. Autogenous nuclear origin:
Outside-in model: This model proposes that the nucleus arose as cytoplasmic membranes within a host cell, usually imagined to be an archaeon, were recruited around DNA in a manner resembling nuclear reformation at the exit from an open mitosis. Such internal membranes would, in turn, have arisen via internalization of the plasma membrane (Fig. 7) via a process akin to the initial steps of endocytosis/phagocytosis (11). Note that extensive membrane fusion is required to generate a single membrane-bound nuclear compartment that is connected to the rest of the cell via pores and to generate a single ER network (Fig. 7). At the same time, multiple membrane fission events are required to topologically separate the plasma membrane from the internal membranes (Fig. 7).
Mitochondrial-derived membrane model: Under this model, the nucleus arose from the extrusion of membranes from the outer surface of the mitochondria (156) via the coalescence of internal membrane vesicles—similar to that envisaged by outside-in models. The strength of the model is its ability to explain why the membranes of eukaryotic cells appear similar to those of alphaproteobacteria. The model also takes inspiration from the observation that vesicles have been seen budding off the outer membranes of gram-negative bacteria. However, the model doesn’t explain many specifics of eukaryotic cell architecture. For example, while vesicular trafficking of this type is used by eukaryotic cells to move lipids between organelles like the ER, Golgi, and plasma membrane, it is not used to move lipids to or from the mitochondria as proposed (12). Instead, the movement of lipids between mitochondria and the ER in eukaryotes is mediated by protein channels at membrane contact sites. In addition, it is hard to understand how nuclear pores arose under this model.
Inside-out model: Under the inside-out model, the first step in eukaryogenesis was the gradual emergence of distinct nuclear and cytoplasmic domains within a common compartment. This was initiated when simple archaeal cells that housed their genomes in a cell body began to interact with bacterial partners in their environment via protrusions (5) (Fig. 6 and 7). Under the model, the sites at which these protrusions emerged from the cell body gave rise to nuclear pores, which became elaborated to control directional traffic between the nascent nucleoplasm and cytoplasm (Fig. 6, step 3). After the acquisition of membrane remodeling activities and the growth of protrusions, a topologically separate plasma membrane was formed via a membrane scission event (Fig. 6, step 4). The model also envisages protrusion-mediated contact between syntropic partners led to a gradual process of symbiotic integration, as bacterial genes for lipid biosynthesis were incorporated into the archaeal host genome (5). This model has the advantage of requiring very few membrane remodeling steps to generate a cell that is topologically equivalent to the eukaryotic cell. The model also proved prescient in predicting the existence of archaeal cells that lacked internal compartments but which might interact with partners via protrusions (Fig. 8). A perceived weakness of this model is that it requires a switch from archaeal to bacterial lipid membranes, which under the model were acquired during the long partnership between the bacterial cell and its archaeal host. While this remains an active area of research, recent findings have shown that bacteria with mixed membranes exist in nature (157) and that Escherichia coli cells with an engineered hybrid heterochiral membrane are viable (158). In addition, it is clear that the eukaryotic counterparts of archaeal proteins that once resided in archaeal membranes, like Sec61, made the transition during eukaryogenesis and now perform their function in eukaryotic membranes. Taken together, these data suggest that a transition period with a mixed membrane may not have posed a significant barrier to eukaryogenesis.
Fig 6.
Diagram depicts the stepwise evolution of the eukaryotic cell as imagined under the inside-out model (5). The cell in step 1 resembles a TACK archaeal cell. It possesses a nucleolar-like domain where rRNAs are assembled into ribosomes, a single bounding membrane, and a complete surface S-layer. The cell in step 2 has protrusions whose close contacts with bacterial partners (red) are facilitated by reduced S-layer coverage. The internal space within protrusions acts as a nascent cytoplasmic compartment, which is separated from the cell body by protrusion necks, where a region of high-membrane curvature is stabilized by multimeric proteins that bind to the membrane from the cytoplasmic side. These structures also function to confine the genome to the cell body. In step 3, the separation of a nascent nucleoplasm and cytoplasm is enhanced by the duplication of the machinery at protrusion necks (brown in the inset), and by the onset of directional, energy-dependent trafficking across this nuclear/cytoplasmic boundary. As a result, RNAs are only translated upon entry into the cytoplasm (indicated in green in inset). The increased curvature induced by duplication of the machinery at the neck of protrusions, together with the emergence of proteins that encourage the self-association of membranes, force the membrane to fold back over the cell body, effectively insulating the cell body from the chemical and physical environment. The partial fusion of protrusions leads to the formation of a more continuous cytoplasm. Proto-mitochondria reside in the spaces in between neighboring cytoplasmic compartments which are topologically equivalent to the lumen of the endoplasmic reticulum. Small black arrows indicate the flow of genetic information out from the center in steps 2 and 3. In the final step, step 4, the formation of a plasma membrane by a process of self-engulfment [as a single cytoplasmic bleb wraps around the whole and undergoes a single membrane scission event (see also Fig. 7)], yields a cell with a structure similar to that of a eukaryotic cell, with a topologically separate ER and plasma membrane, a continuous nuclear envelope-endoplasmic reticulum, and trapped vertically-inherited mitochondria, which later enter the cytoplasm. For an in-depth description of the entire process, see the original inside-out model.
Fig 7.
Diagram shows an intermediate step in the process of eukaryogenesis expected under an outside-in model (left) and an inside-out model (right). Note that under the outside-in model, left, in order to generate a cell topologically similar to a eukaryotic cell (see Fig. 6, step 4), multiple membrane remodeling events are required to fuse initially separate endoplasmic reticulum (ER)-like compartments to generate a single lumenal network that is continuous with the nuclear envelope, and to generate a nuclear compartment that is connected to the cytoplasm via pores. In addition, additional membrane remodeling events are required to cut all the connections linking the nascent NE-ER compartment to the outer membrane to generate a topologically separate plasma membrane. By contrast, under the inside-out model shown in the diagram on the right, at this stage cells already possess a nascent nuclear compartment, a continuous ER and NE, and nuclear pores, but lack a single continuous cytoplasm and a plasma membrane. A topologically separate plasma membrane can be generated by a process of “auto-phagocytosis” whereby one protrusion extends around the whole and undergoes a single scission event.
Models of eukaryogenesis (see Box 1 and Box 2) exist on a continuum, but can be broadly divided into “mito early” scenarios, in which the acquisition of the alphaproteobacterial endosymbiont represents the first step on the path to the eukaryotic cell, and “mito-late” scenarios, which imagine mitochondria being acquired through a process akin to phagocytosis relatively late in the process of eukaryogenesis by a complex proto-eukaryotic host cell that already possessed a nucleus (78, 160). In addition, models can be grouped by topology into “outside-in” versions, which propose that the nucleus was formed de novo following the coalescence of membranes around the DNA in a manner resembling nuclear reformation following exit from an open mitosis (Fig. 7); models that imagine a third partner in the form of a virus (150, 151) or another cell (8) giving rise to the nucleus; and the “inside-out” model (5), in which the nucleus and ER are envisioned as having arisen from the original bounding membrane of an archaeal cell (Fig. 6 and 7).
Nuclear origins in light of cell biological features of archaea
The existence of very different models that all claim to explain the evolution of the various cellular features of eukaryotes might lead one to conclude that the origin of the nucleus is unlikely to be resolved anytime soon. However, by focusing on the eukaryotic membrane system and membrane trafficking, the plethora of models obscures the long-appreciated fact that many of the core functions of the nucleus are executed by proteins that have their origins in archaea (Fig. 5). In fact, numerous studies over the past decades have characterized molecular machines in archaea that have close, if more complex, counterparts in eukaryotes, many of which function within the nucleus (101–127). These parallels are most evident from molecular cell biology studies carried out using Sulfolobus cells—currently the most experimentally tractable member of the TACK (94) archaea - a sister group of the Asgardarchaeota and Eukarya (78). This type of analyses has shown that Sulfolobus cells use close counterparts of the machinery found in eukaryotes to carry out many of the core information processing steps that are central to life including: DNA replication (102, 161), DNA-dependent RNA transcription (113), aspects of RNA processing (162), rRNA modification, ribosome assembly (163), messenger RNA processing (164), translation (120), protein secretion, protein degradation (165), and protein glycosylation (166). In addition, as they grow and divide (167, 168), Sulfolobus cells pass through discrete G1, S, G2, and Division phases, which resemble phases of the eukaryotic cell cycle. Since Sulfolobus cells lack obvious homologs of the cell cycle clock (CDK/Cyclins), it is not yet known how they might regulate orderly passage through the cell cycle. However, Sulfolobus cells express proteins involved in the regulation of transcription that possess a Cyclin-box fold and whose expression oscillates across the cycle (169). In addition, proteasome-mediated degradation has been implicated in resetting the Sulfolobus cell cycle, just as it has been in eukaryotic cells (170). Furthermore, Sulfolobus cells initiate S-phase (167) via the near synchronous firing of multiple replication origins using counterparts of the machinery used in eukaryotes, which include homologs of ORC/Cdc6, Cdc45, MCM, and GINS (168). Like eukaryotes, Sulfolobus cells also organize their genome into domains using SMC proteins (171) and express chromatin organizing proteins that, like histones [which are present in the vast majority of archaea, including other TACK archaea, but have been lost from Sulfolobales (106, 108, 172, 173)], are subject to regulation by acetylation (113)—including a chromatin protein Alba that has homologs in eukaryotes (174). Thus, the parallels between the core information processing machinery present in TACK family archaea and the corresponding nuclear machinery present in eukaryotes are striking.
Despite the presence of homologs of proteins that function in the eukaryotic nucleus, there is no sign of TACK archaeal cells having anything like a nuclear envelope. With a few notable exceptions, including Ignococcus, which has two membranes and no S-layer (1, 175), most TACK archaeal cells described thus far have a single compartment bounded by a plasma membrane. These data could be used to argue that these archaeal relatives cannot tell us much about the origins of the nucleus. However, there is a way out of this conundrum. The difficulties of imagining how an archaeal cell might have given rise to a cell with a nucleus and cytoplasm come, in part, from the tendency to look at the nucleus as a distinct organelle that is separated from the cytoplasm by a double membrane. When the cytoplasm and nucleoplasm are viewed as sub-domains of a common compartment, as discussed above, it is easy to see the nucleus as having arisen via many small stepwise changes in the organization of a simple archaeal cell that resulted in the separation of information storage and processing activities (found in the modern eukaryotic nucleus) from protein synthesis and metabolism (activities usually confined to the eukaryotic cytoplasm). This idea plays a central role in the inside-out model of eukaryogenesis (5).
Under the inside-out model (5), the nucleus is suggested to have its origin in a simple archaeal cell (Fig. 6, step 1) that develops protrusions (Fig. 6, step 2). This change in cellular organization establishes spatially separate domains equivalent to a nascent nucleoplasm, where the DNA is housed, and a proto-cytoplasm, i.e., the main site of protein synthesis, metabolism, and contact with the environment. In such a cell, machinery would likely be required to keep the genome out of protrusions, and to stabilize the sites of high local membrane curvature at junctions connecting protrusions to the cell body by preventing scission by the membrane remodeling ESCRT-III machinery, which induces membrane remodeling at topologically similar structures in both archaea and eukaryotes (136). Under the inside-out model, this would be achieved by the elaboration of the machinery at the interface connecting the cell body and protrusions. This then set the stage for the directional traffic of large macromolecules (Fig. 6, step 3), by helping to prevent chromatin and intermediates in the information processing pipeline (like immature mRNAs and partially assembled ribosomes) from entering the nascent cytoplasm, i.e., protrusions. In this sense, the machinery at the necks of protrusions would come to resemble the nuclear pore complex and the diffusion barrier at the base of cilia in modern eukaryotes (5).
If this view of early eukaryotic cell evolution is correct, the genomes of the Asgardarchaeota might be expected to hold clues to the origins of eukaryotic cell organization. However, while asgardarchaeal genomes encode numerous homologs of proteins involved in eukaryotic information processing, cytoskeletal proteins, membrane remodeling complexes, and regulators, the dynamic organization of a cell is not something that can be deduced from genomic information alone (176). A genome encodes the machinery required for cell growth and division rather than explicitly encoding cell biological features like cell size, structure, or numbers of membrane-bound compartments. As a case in point, there is currently no simple way to analyze the genomes of Sulfolobus acidocaldarius and Ignicoccus hospitalis cells and say how many membrane compartments the two related archaea possess (one and two, respectively). Thus, the availability of high-quality genomes of Lokiarchaeia and other Asgardarchaota is insufficient to determine their cell biology (176). To do so, one must observe cells under a microscope.
This only became possible in 2020, once a representative of the Lokiarchaeia—Candidatus Prometheoarchaeum syntrophicum (159)—had been cultivated. This feat was 12 years in the making, and was followed by the description of an enrichment culture of a second member of the Lokiarchaeia, Candidatus Lokiarchaeum ossiferum in 2023 (177). Strikingly, the Lokiarchaeia enriched in the two studies closely resemble one another when imaged using electron microscopy. Both studies described small, micron-sized cells with long finger-like protrusions emanating from a spherical cell body (159, 177) (Fig. 8), through which they were seen contacting other cells in the culture (Fig. 8A). Significantly, the Lokiarchaeia in both studies also lacked both internal membranes and anything resembling a separate internal nuclear compartment. These observations led to the proposition that Lokiarchaeia use protrusions to make contact with syntropic partners through which they can exchange metabolites (159), as was suggested as a first step toward eukaryogenesis under the inside-out model (5). Furthermore, in line with this suggestion, when grown on organic compounds like amino acids, the growth of both cultivated representatives of Lokiarchaeia was found to depend on partner organisms to which they can transfer electrons in the form of hydrogen and formate—consistent with predictions from genomic analyses (178). This type of syntrophic lifestyle has been hypothesized as a potential driving force leading to intricate interactions of the prokaryotic ancestors of eukaryotes (5, 8, 178–180).
Fig 8.
This figure, kindly reproduced with permission of the authors from Rodrigues-Oliveira et al. (159), shows electron microscopic images of Lokiarchaeia cells. (A) An SEM shows a Candidatus Lokiarchaeum ossiferum cell (left) making contact with a possible syntropic partner via protrusions. (B) A zoomed in CryoEM image shows the highly curved neck that separates the Lokiarchaeum cell body from its protrusions, and the electron dense layer that underlies it. (C) and (D) show Candidatus Lokiarchaeum ossiferum cells in which actin filaments (ochre), ribosomes (gray), and the bounding membrane have been imaged using CryoEM and highlighted in different colors.
While it is tempting to speculate, based on these data, that the asgardarchaeotal ancestor of eukaryotes was capable of syntrophic growth, it is important to note that Lokiarchaeia are only distantly related to the Heimdallarcheia, including the Hodarchaeales, which currently appear to comprise the lineages most closely related to eukaryotes, and which have more diverse metabolic repertoires than Lokiarchaeia (131, 143, 178, 181). Thus, gaining a more comprehensive view of the metabolic and cellular diversity of the Asgardarchaeota will require the enrichment, cultivation and imaging of additional taxa.
Possible archaeal origins of nuclear functions
Given the archaeal origins of a large proportion of the information processing machinery present in eukaryotic cells, cell biological studies looking at the structure of Lokiarchaeia and TACK archaea, like Sulfolobus, provide a useful starting point for thinking about the origin of the nucleus. Specifically, they clarify ways in which the regionalization of cellular space could have emerged - leading to the spatial separation of information processing events from translation and metabolism.
Several pieces of evidence already point to the possibility of some archaea having an ordered internal space despite lacking distinct membrane-bound compartments. First, the genome in several TACK archaea appears physically confined to a small portion of the available cytoplasm, rather than being spread throughout the cytoplasm as it often is in bacteria. In Sulfolobus acidocaldarius, where this process has been studied using live cell imaging, the genome appears spread along a portion of the membrane in interphase (182). Then, as cells prepare to divide, the genome appears to detach from the membrane as it compacts, leading to the formation of two discrete and separated DNA masses, each of which is partitioned into one of the two daughter cells at cytokinesis (182). This is not specific to S. acidocaldarius. The nucleioids of other Sulfolobales species as well as Nitrosopumulus maritimus, a member of the Thaumarchaeota, have been reported to undergo similar changes in organization across the cell cycle (183). Superficially, at least, this process resembles mitotic chromosome condensation in eukaryotes. Even more strikingly, in fluorescent images of Lokiarchaeia (177), the genome appears confined to the cell body and absent from protrusions—just as was envisaged for an early intermediate on the path to eukaryogenesis under the inside-out model (Fig. 6). Conversely, while a few actin filaments were observed in the central body of these Lokiarchaeia using cryogenic-electron microscopy (cryo-EM), the majority of the signal generated using a fluorescently labeled actin antibody was seen in protrusions (177). These data suggest that Lokiarchaeia may possess a spatially distinct proto-nucleoplasm and proto-cytoplasm.
In eukaryotic cells, the spatial separation of cellular functions is enforced and amplified by diffusion barriers (at the nuclear pore and at the base of cilia), gradients in GTPase activity, protein and RNA trafficking via cytoskeletal elements, local translation, and the use of peptide localization tags, such as nuclear localization signals. Therefore, it is important to determine whether similar factors operate in archaea. As we discuss below, the published data already suggest that archaeal cells possess simple versions of all five types of regulatory systems.
Taking each in turn:
Diffusion barriers
Asgardarchaeota have membrane connections at the base of protrusions that are similar in width (~100 nm) to the size of nuclear pores in eukaryotes (177). In cryo-EM images, the membrane at the funnel-shaped necks of these protrusions appears to be associated with cytoplasmic protein density, potentially pointing to the existence of membrane-associated proteins that provide the highly curved membrane with structural stability (Fig. 8B). This is one of the main functions of NPCs in eukaryotes (184). At the same time, associated proteins could function to limit the free diffusion of large macro-molecular complexes, such as chromatinized DNA or RNPs, between the cell body and protrusions. Furthermore, machinery at the neck of protrusions may limit transmembrane proteins to different parts of the continuous bounding cell membrane—as occurs in eukaryotes in the partitioning of proteins between the inner and outer nuclear membrane, and between the rough and smooth ER.
GTPase gradients
While other Bacteria and Archaea, including TACK archaea and Euryarchaeota, possess a number of small GTPases (138, 185), Lokiarchaeia and other Asgardarchaeota possess them in abundance (129, 130, 137, 138). It is not clear why this might be. Thus far, there is little evidence that these archaeal small GTPases bind membranes or are modified by the covalent attachment of lipid moieties [with the exception of a few with transmembrane domains (176)]. Nevertheless, it is possible that some of these GTPases function like Ran to organize intracellular space in conjunction with spatially separate GEFs and GAPs. Given that GTPases can act as diffusible switches, they may also enable regionalization of the single cytoplasmic space and/or the single bounding membrane.
Cytoskeletal trafficking of protein and RNAs
Lokiarchaeia and other Asgardarchaeota possess close homologs of eukaryotic Actin and Tubulin (130, 134, 135). In Lokiarchaeia, while actin filaments have been seen extending into protrusions (159, 177), it is not yet known if they help to generate and/or stabilize protrusions. In the available images, the actin filaments in Lokiarchaeia don’t appear highly organized or polarized with respect to one another or the membrane (Fig. 8C). Nevertheless, in vitro, these filaments appear to be dynamic (134, 135). This suggests the possibility that the growth, shrinkage, and treadmilling of such dynamic cytoskeletal polymers in cells that lack clear homologs of actin-dependent molecular motors might generate flows that stir the cytoplasm and/or direct the transport of material associated with filament ends (186, 187). This type of “cytomotive” behavior is used by eukaryotic microtubules to power the movement of chromosomes during mitosis. In cells with long protrusions where the diffusion of large complexes, such as mRNPs, to and from protrusion tips is likely to be very slow (188), such assisted diffusion may be critical.
Local translation
Many archaea possess counterparts of ribosome processing machinery present in eukaryotes that are absent from bacteria (120). These factors include SnoRNAs (189), homologs of Fibrillarin, and several Fibrillarin-associated proteins that act as rRNA methylases, all of which are key components of the eukaryotic nucleolus (163, 190). This suggests the possibility that such proteins may function to ensure the spatial separation of active and inactive ribosomes in archaea. If these proteins and RNAs accumulate in spatially segregated domains, this may explain the physical separation of DNA and rRNA observed in Lokiarchaeia as imaged by Avci et al. (191). This type of regionalization could be enhanced by other RNA-processing enzymes. Archaeal cells (and many bacteria) possess a host of RNA-processing enzymes (192) and RNA-binding proteins that could function in this way. These include homologs of the enzymes that cleave the 3′ of RNAs, and DDX helicases, which induce local mRNA unwinding and translation in eukaryotes (32, 48). Furthermore, some archaeal possess homologs of Nmd3 (32, 48, 120) and TIF6/SBDS, which function to keep pre-ribosomes inactive in the nucleus in eukaryotes (193, 194). It should be noted here that, although archaea possess type-II self-splicing introns (195), and RNA insertions [predominantly in rRNAs and tRNAs (70, 196)], there is no evidence of archaea possessing eukaryotic-like introns within protein-coding RNAs (195). Thus, for the moment, the evolutionary origins of the eukaryotic spliceosome remain unresolved (197, 198). Taken together, however, these data suggest the possibility that some archaea may be able to separate transcription and RNA processing from the site of translation, using a simplified version of the process taking place in eukaryotes.
Short protein localization tags
NLS sequences in eukaryotic proteins typically consist of short stretches of basic amino acids or a mixture of basic and surface-exposed hydrophobic amino acids. Similar sequences are present in ribosomal proteins in most archaeal groups (199), including DPANN and Euryarchaeota [all of which are thought to lack anything resembling a nuclear compartment], where they likely function as rRNA-binding motifs. NLSs are also present in subunits of the proteasome in eukaryotic cells and in some archaea (200). These data suggest the possibility that NLSs may have originated in archaea as low-affinity nucleic acid binding motifs that concentrate proteins in parts of the cell rich in rRNA or DNA even if these cells lack a spatially distinct nucleus. Such sequences could have then been redeployed to direct nuclear/cytoplasmic traffic with the advent of a complex nuclear pore. By the same argument, other small motifs used to direct different types of cellular traffic in eukaryotic cells may have counterparts in archaea that function to locally concentrate specific proteins in the context of a single compartment.
AN UPDATED MODEL FOR AN INSIDE-OUT ORIGIN OF THE EUKARYOTIC NUCLEUS
Bringing these arguments and data together enables us to propose a stepwise path for the evolution of different nuclear functions that builds upon the inside-out model of eukaryogenesis. This is detailed below and depicted in Fig. 6 and 7.
A: The separation of transcription and translation
The starting point for the model is a structurally simple early archaeal ancestor of eukaryotes that lacks internal membranes, but which possesses machinery that enables the partial physical separation of transcription and translation for some transcripts. We propose that this separation is achieved via several processes working in tandem. First, rRNA processing and modifications [including Fibrillarin-dependent methylation (163)] are spatially confined to sites of rRNA transcription through the establishment of a phase-separated region of the cytoplasm that acts like a primitive nucleolus (201). Second, the delayed completion of ribosome assembly ensures that a subset of ribosomes remains inactive until meeting an RNA substrate at a relevant site in a cell [as occurs in eukaryotes for secreted proteins as a result of SRP14-mediated inhibition of translation (202)]. Third, newly transcribed and processed RNAs (203) are rapidly assembled into inactive RNPs, preventing their association with active ribosomes until they reach the correct cellular site. As a result, the translation of some RNAs is contingent on RNP disassembly driven by the action of specific local RNA helicases (204).
B: The separation of a nascent cytoplasm from a nascent nucleoplasm
In a more recent common ancestor of Asgardarchaeota and eukaryotes, this partial separation of transcription from translation is augmented by the spatial confinement of these processes to the cell body and protrusions. This is facilitated by machinery localized at the necks of protrusions (Fig. 6, step 2). Protein assemblies, like those visible in the electron dense material underlying curved membranes in electron micrographs of lokiarchaeial cells (177) (Fig. 8), function to prevent the movement of chromatin into metabolically active protrusions. Later in the process of eukaryogenesis, the elaboration of machinery at protrusion necks acts to limit the free diffusion of large macromolecules (e.g., cytoskeletal polymers and RNPs). Together, this machinery facilitates the spatial differentiation of the cell into a metabolically inactive central domain (a proto-nucleoplasm), in which nucleic acids are stored and processed, and peripheral protrusions (a proto-cytoplasm), where active metabolism occurs and where physical contact with symbionts is established—enabling the efficient transfer of electrons and/or substrates between partners (159). Over time, the evolution of machinery accentuating local differences in the accumulation of proteins in different parts of the cell enhances regional specialization of the single bounding membrane and cytoplasm.
In cells with long thin protrusions, diffusion is slow. This both limits the cell’s ability to coordinate distant processes (205, 206) (Fig. 6, step 2) and facilitates the separation of activities, like metabolism and information storage, which might otherwise interfere with one another. In such a diffusion-limited system, we imagine cells using dynamic cytoskeletal filaments that bind and hydrolyze NTPs, like those present in Lokiarchaea, to regulate the flow of genetically encoded information between regions, by coupling the unidirectional transport of large macromolecular complexes to discrete phases of filament growth or shrinkage and/or by stirring the cytoplasm. Note that the distance over which such cytomotive filaments (207) operate depends on their persistence length, which is determined by the structure of the polymer (208), so that any increase in cell size will need to be accompanied by an increase in filament stiffness, e.g. via filament bundling or the formation of microtubules. Such cytomotive forces will be especially important for directing the traffic of large, slowly diffusing particles, like pre-ribosomes or RNPs to the tips of protrusions (note a 25-kDa protein is encoded for by an RNA of ~200 kDa). Over time, we imagine enzymes emerging to bias these movements. For example, homologs of the rRNA remodeling complex Midasin (209)—an ATPase involved in the activation of translation (210, 211) and related to eukaryotic Dynein (212, 213)—may aid this process, setting the stage for the later emergence of ATP-hydrolyzing cytoskeletal molecular motors.
C: Formation of a double nuclear envelope
At the next stage of eukaryogenesis (Fig. 6, step 3), we imagine protrusions expanding and folding back onto the surface of the cell body to generate a double nuclear envelope. This change in membrane organization would have been aided by the evolution of complementary surface adhesive proteins, perhaps similar to the glycosylated SUN and KASH proteins (48), which in eukaryotes function to hold inner and outer nuclear membranes together at a fixed distance away from one another (5), and by the duplication of the machinery supporting the curved membrane at protrusion necks (Fig. 6, step 3). This is depicted in the model as a switch from a half-pore with eightfold rotational symmetry to a full nuclear pore with an additional axis of symmetry. This change in cellular organization, in which the peripheral proto-cytoplasm and membrane wrap the proto-nucleus, physically isolates the cell body housing the genome from the external environment (5). The augmented pore can then also participate in the regulation of bi-directional nuclear-cytoplasmic transport, perhaps by coupling movement across the neck of protrusions to a Ran-GTP’ase-like system, regulated by a chromatin-localized GEF working in concert with diffusible or membrane-associated GAPs, using importin/exportin and FG repeat proteins. In this case, GTP hydrolysis could power the directional transport of material into or out of the nascent nuclear compartment, where the genome resides. Together, these innovations would help enforce a more complete separation of transcription, ribosomal assembly, and RNA processing, from active protein synthesis. This sets the stage for the evolution of splicing, perhaps using components derived from Asgardarchaeota, Alphaprotebacteria, and other Bacteria (195). As such cells divide, we envision the partial opening of the curved necks at the base of protrusions [equivalent to a semi-open mitosis (13)] facilitating the mixing of proto-cytoplasm and proto-nucleoplasm to aid the fair partitoning of the full complement of cellular material between daughter cells.
The role of symbiosis in the origins of nuclear functions
Having outlined a possible scheme above for the stepwise evolution of a nuclear-like compartment, it is worth emphasizing that our focus here on the archaeal host and earliest stages of eukaryogenesis deliberately leaves aside the role of the symbiotic partnership with the alpha-proteobacterial cell that gave rise to the mitochondria, which was covered in detail in the original inside-out model (5). It is clear, however, that the mitochondrial ancestor contributed to the selective pressure driving the emergence of certain cellular features and played a critical role in the evolution of eukaryotic cell organization (6–8, 79, 91) (Fig. 5). For instance, the transfer of genes from bacterial partners to the archaeal host nucleus (181), together with horizontal gene transfer from other bacteria (8, 142, 214), likely contributed to the expansion of the nuclear genome, to changes in translation required to prevent crosstalk between the host and mitochondrial ribosome assembly pathways, to the emergence of introns, and to the acquisition of new metabolic functions. This latter process included the synthesis of fatty acids and sterols (215, 216), leading to a switch in membrane composition that is likely to have facilitated the acquisition of dynamic membrane trafficking (5). The advantage of putting these aspects of the inside-out model aside in this discussion of the origin of the nucleus is that it allows us to propose a framework that can be explored by experiment through the study of existing cultures of TACK archaea and Asgardarchaeota. Thus, while this extension of the original inside-out model is speculative, it suggests a simple series of steps that could have led to the emergence of a proto-eukaryotic cell with a functioning nucleus and nuclear envelope in a way that does not necessarily depend on the prior emergence of functioning internal mitochondria and a sophisticated vesicle trafficking system.
Experimentally testable questions raised by the model
In order to test the validity of this model for the origin of the nucleus, it is important to improve the cell biological analysis of TACK and Asgardarchaeota though studies exploring regionalization of archaea cells and their bounding membranes. This should include experiments to determine:
How are protein-coding transcripts in TACK and Asgardarchaeota processed prior to translation (192)?
Is there a temporal delay and/or physical separation between transcription and translation in some TACK and Asgardarchaeota?
Are pre-ribosomes assembled in a nucleolus-like structure in members of the TACK and Asgardarchaeota (201), and do these cells possess mechanisms to ensure that ribosomes become activated at specific sites far from their assembly?
Do different subsets of RNAs accumulate in the cell body and protrusions of Lokiarchaeia prior to translation? Do Lokiarchaeia express transcripts whose translation is inhibited by RNA-binding proteins before being locally activated by RNA helicases?
Is genomic DNA excluded from Lokiarchaeia protrusions? If so, how?
Do proteins at the curved base of Lokiarchaeia protrusions serve as barriers to the diffusion of large protein and/or RNA complexes? Do these proteins also block the ESCRT-III mediated scission of protrusion necks?
Does the cytomotive activity of cytoskeleton polymers in Asgardarchaeota aid facilitated transport in diffusion-limited protrusions?
At division, do structures at the base of protrusions in Lokiarchaeia open up to enable the mixing of different domains as an aid to division symmetry?
Do short peptide tags, including NLS sequences, aid the local accumulation of proteins at specific sites within the cytoplasm or at specific membrane domains in Lokiarchaeia?
Do small GTPases help establish distinct domains of activity across lokiarchaeial cells?
Key challenges faced by the model
With the caveat that extant representatives derived from the asgardarchaeotal sister-lineage of eukaryotes have evolved for as much time as the eukaryotic lineage, future experiments on archaea could turn up data that would be hard to reconcile with the model of nuclear evolution put forward here. Such data might also favour alternative models of eukaryogenesis by, for example, supporting the idea that vesicle trafficking evolved prior to the separation of distinct nuclear and cytoplasmic activities. These could include the following:
The demonstration of direct coupling between transcription and translation of all protein-coding transcripts in the closest archaeal relatives of eukaryotes.
Failure to demonstrate that Asgardarchaeota are able to establish and maintain locally distinct domains where specific RNAs, proteins, and localization tags accumulate.
The identification of close archaeal relatives of eukaryotes that possess a complex and dynamic internal membrane organization [emanating from the cell surface (as shown in Fig. 7, left) or the mitochondria], but which are not able to physically separate the genome and early information processing events (transcription and ribosome assembly) from protein synthesis and metabolism.
The identification of close archaeal relatives of eukaryotes that use coatamer-like assemblies to regulate vesicle trafficking but which lack anything resembling a scaffolded nuclear pore.
The identification of close archaeal relatives of eukaryotes that cannot physically separate early information processing events from protein synthesis and metabolism, but which are capable of phagocytosis.
The identification of symbiotic consortia consisting of bacterial cells that act as hosts to intracellular archaeal symbionts related to the asgardarchaeotal ancestor of eukaryotes—as envisioned in the syntropy model.
The identification of close archaeal relatives of eukaryotes that possess viral assembly factories that resemble nuclei.
CONCLUSION
In this hypothesis article, we propose that our closest living archaeal relatives, TACK and Asgardarchaeota, hold clues to the early stages in the origin of the nucleus. This model builds on decades of molecular and cell biological evidence from studies using TACK archaea like Sulfolobus (217), and on a very limited number of cell biological studies in Asgardarchaeota, which together suggest that, prior to the association of a mitochondrial symbiont, archaeal ancestors of eukaryotes already possessed a nucleolar-like domain and a cytoplasmic-like compartment in the form of protrusions that exclude DNA (177). These data support the inside-out model (5) and imply that archaea can differentiate a single continuous cytoplasm and membrane into distinct genome-storage and metabolically active domains—just as eukaryotic cells do through the establishment of a nuclear-cytoplasmic compartment boundary. Bringing these data together, we propose that an array of biochemical functions that are associated with eukaryotic nuclei first arose archaea. These were then elaborated during eukaryogenesis, alongside changes in cell shape, to give rise to the nucleus. Under this model, the nucleus has its origin in archaea.
ACKNOWLEDGMENTS
We would like to thank many wonderful colleagues for their constructive criticism and for their many fresh insights in reading one or more of the many drafts of this manuscript. First, we would like to thank David Baum, co-author of the original inside-out model, for his insightful feedback. In addition, we thank Martin Beck, Simon Bullock, Gautam Dey, Maria Hondele, Jan Löwe, Lori Passmore, Berend Snel, Finn Werner, Tom Williams, Carlos Santana Molina, and the anonymous reviewers for critically reading the manuscript. We thank Konstantinos Alexandrou at the LMB Vislab and Patrick Lane for help with Figures; Agnieszka Obarska and Martin Beck for the image in Fig. 3B; and Thiago Rodrigues-Oliveira, Martin Pilhofer, and Christa Schleper for the images in Fig. 8. We thank the Life Sciences-Moore-Simons foundation (735929LPI) and a Gordon and Betty Moore Foundation’s Symbiosis in Aquatic Systems Initiative (GBMF9346) for supporting our collaborative work together.
In addition, B.B. received generous support from the MRC-LMB, the Wellcome Trust (203276/Z/16/Z), and the VW Foundation (94933). Finally, A.S. is thankful for funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 947317, ASymbEL) and from the Gordon and Betty Moore Foundation through grant GBMF9741.
B.B. formulated the plan for the article and updated the inside-out model to emphasize its implications for the origin of the eukaryotic nucleus. B.B. and A.S. co-wrote the manuscript, generated the figures (supported by professional artists), and completed the revisions.
AFTER EPUB
[An error in the Acknowledgments section of the version of this article published on 29 November was corrected in the current version, posted on 20 December.]
Contributor Information
Buzz Baum, Email: bbaum@mrc-lmb.cam.ac.uk.
Amy K. Schmid, Duke University, Durham, North Carolina, USA
REFERENCES
- 1. Heimerl T, Flechsler J, Pickl C, Heinz V, Salecker B, Zweck J, Wanner G, Geimer S, Samson RY, Bell SD, Huber H, Wirth R, Wurch L, Podar M, Rachel R. 2017. A complex endomembrane system in the archaeon Ignicoccus hospitalis tapped by Nanoarchaeum equitans. Front Microbiol 8:1072. doi: 10.3389/fmicb.2017.01072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greening C, Lithgow T. 2020. Formation and function of bacterial organelles. Nat Rev Microbiol 18:677–689. doi: 10.1038/s41579-020-0413-0 [DOI] [PubMed] [Google Scholar]
- 3. Volland J-M, Gonzalez-Rizzo S, Gros O, Tyml T, Ivanova N, Schulz F, Goudeau D, Elisabeth NH, Nath N, Udwary D, Malmstrom RR, Guidi-Rontani C, Bolte-Kluge S, Davies KM, Jean MR, Mansot J-L, Mouncey NJ, Angert ER, Woyke T, Date SV. 2022. A centimeter-long bacterium with DNA contained in metabolically active, membrane-bound organelles. Science 376:1453–1458. doi: 10.1126/science.abb3634 [DOI] [PubMed] [Google Scholar]
- 4. Katayama T, Nobu MK, Kusada H, Meng XY, Hosogi N, Uematsu K, Yoshioka H, Kamagata Y, Tamaki H. 2020. Isolation of a member of the candidate phylum 'Atribacteria' reveals a unique cell membrane structure. Nat Commun 11:6381. doi: 10.1038/s41467-020-20149-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baum DA, Baum B. 2014. An inside-out origin for the eukaryotic cell. BMC Biol 12:76. doi: 10.1186/s12915-014-0076-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin WF, Garg S, Zimorski V. 2015. Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond B Biol Sci 370:20140330. doi: 10.1098/rstb.2014.0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guy L, Saw JH, Ettema TJG. 2014. The archaeal legacy of eukaryotes: a phylogenomic perspective. Cold Spring Harb Perspect Biol 6:a016022. doi: 10.1101/cshperspect.a016022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. López-García P, Moreira D. 2020. The Syntrophy hypothesis for the origin of eukaryotes revisited. Nat Microbiol 5:655–667. doi: 10.1038/s41564-020-0710-4 [DOI] [PubMed] [Google Scholar]
- 9. Koonin EV. 2015. Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: eukaryogenesis just made easier? Philos Trans R Soc Lond B Biol Sci 370:20140333. doi: 10.1098/rstb.2014.0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baum B, Baum DA. 2020. The merger that made us. BMC biology 18:72. doi: 10.1186/s12915-020-00806-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baum DA. 2015. A comparison of autogenous theories for the origin of eukaryotic cells. Am J Bot 102:1954–1965. doi: 10.3732/ajb.1500196 [DOI] [PubMed] [Google Scholar]
- 12. Reinisch KM, Prinz WA. 2021. Mechanisms of nonvesicular lipid transport. J Cell Biol 220:e202012058. doi: 10.1083/jcb.202012058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dey G, Baum B. 2021. Nuclear envelope remodelling during mitosis. Curr Opin Cell Biol 70:67–74. doi: 10.1016/j.ceb.2020.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mulkidjanian AY, Makarova KS, Galperin MY, Koonin EV. 2007. Inventing the dynamo machine: the evolution of the F-type and V-type ATPases. Nat Rev Microbiol 5:892–899. doi: 10.1038/nrmicro1767 [DOI] [PubMed] [Google Scholar]
- 15. Csala M, Bánhegyi G, Benedetti A. 2006. Endoplasmic reticulum: a metabolic compartment. FEBS Lett 580:2160–2165. doi: 10.1016/j.febslet.2006.03.050 [DOI] [PubMed] [Google Scholar]
- 16. Thurston TLM, Wandel MP, von Muhlinen N, Foeglein A, Randow F. 2012. Galectin 8 targets damaged Vesicles for Autophagy to defend cells against bacterial invasion. Nature 482:414–418. doi: 10.1038/nature10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu L, Brangwynne CP. 2015. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol 34:23–30. doi: 10.1016/j.ceb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sawyer IA, Sturgill D, Dundr M. 2019. Membraneless nuclear organelles and the search for phases within phases. Wiley Interdiscip Rev RNA 10:e1514. doi: 10.1002/wrna.1514 [DOI] [PubMed] [Google Scholar]
- 19. Brown DD, Gurdon JB. 1964. Absence of ribosomal RNA synthesis in the anucleolate mutant of xenopus laevis. Proc Natl Acad Sci U S A 51:139–146. doi: 10.1073/pnas.51.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Natsume T, Tanaka TU. 2010. Spatial regulation and organization of DNA replication within the nucleus. Chromosome Res 18:7–17. doi: 10.1007/s10577-009-9088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, Gottesman ME, Gautier J. 2018. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559:61–66. doi: 10.1038/s41586-018-0237-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Briand N, Collas P. 2020. Lamina-associated domains: peripheral matters and internal affairs. Genome Biol 21:85. doi: 10.1186/s13059-020-02003-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cremer T, Cremer C. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2:292–301. doi: 10.1038/35066075 [DOI] [PubMed] [Google Scholar]
- 24. McCluggage F, Fox AH. 2021. Paraspeckle nuclear condensates: global sensors of cell stress? Bioessays 43:e2000245. doi: 10.1002/bies.202000245 [DOI] [PubMed] [Google Scholar]
- 25. Caragliano E, Bonazza S, Frascaroli G, Tang J, Soh TK, Grünewald K, Bosse JB, Brune W. 2022. Human cytomegalovirus forms phase-separated compartments at viral genomes to facilitate viral replication. Cell Rep 38:110469. doi: 10.1016/j.celrep.2022.110469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. 2021. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol 22:165–182. doi: 10.1038/s41580-020-0272-6 [DOI] [PubMed] [Google Scholar]
- 27. Schlaitz A-L, Thompson J, Wong CCL, Yates JR, Heald R. 2013. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell 26:315–323. doi: 10.1016/j.devcel.2013.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hampoelz B, Andres-Pons A, Kastritis P, Beck M. 2019. Structure and assembly of the nuclear pore complex. Annu Rev Biophys 48:515–536. doi: 10.1146/annurev-biophys-052118-115308 [DOI] [PubMed] [Google Scholar]
- 29. Obado SO, Brillantes M, Uryu K, Zhang W, Ketaren NE, Chait BT, Field MC, Rout MP. 2016. Interactome mapping reveals the evolutionary history of the nuclear pore complex. PLoS Biol 14:e1002365. doi: 10.1371/journal.pbio.1002365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zimmerli CE, Allegretti M, Rantos V, Goetz SK, Obarska-Kosinska A, Zagoriy I, Halavatyi A, Hummer G, Mahamid J, Kosinski J, Beck M. 2021. Nuclear pores dilate and constrict in cellulo. Science 374:eabd9776. doi: 10.1126/science.abd9776 [DOI] [PubMed] [Google Scholar]
- 31. Lim RYH, Huang N-P, Köser J, Deng J, Lau KHA, Schwarz-Herion K, Fahrenkrog B, Aebi U. 2006. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc Natl Acad Sci U S A 103:9512–9517. doi: 10.1073/pnas.0603521103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Güttler T, Görlich D. 2011. Ran-dependent nuclear export mediators: a structural perspective. EMBO J 30:3457–3474. doi: 10.1038/emboj.2011.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu J, Wu T, Zhang B, Liu S, Song W, Qiao J, Ruan H. 2021. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun Signal 19:60. doi: 10.1186/s12964-021-00741-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cavazza T, Vernos I. 2015. The RanGTP pathway: from nucleo-cytoplasmic transport to spindle assembly and beyond. Front Cell Dev Biol 3:82. doi: 10.3389/fcell.2015.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalab P, Heald R. 2008. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci 121:1577–1586. doi: 10.1242/jcs.005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaláb P, Solc P, Motlík J. 2011. The role of RanGTP gradient in vertebrate oocyte maturation. Results Probl Cell Differ 53:235–267. doi: 10.1007/978-3-642-19065-0_12 [DOI] [PubMed] [Google Scholar]
- 37. Ozugergin I, Piekny A. 2021. Complementary functions for the Ran gradient during division. Small GTPases 12:177–187. doi: 10.1080/21541248.2020.1725371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kalab P, Weis K, Heald R. 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295:2452–2456. doi: 10.1126/science.1068798 [DOI] [PubMed] [Google Scholar]
- 39. Kaláb P, Pralle A, Isacoff EY, Heald R, Weis K. 2006. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440:697–701. doi: 10.1038/nature04589 [DOI] [PubMed] [Google Scholar]
- 40. Silverman-Gavrila RV, Hales KG, Wilde A. 2008. Anillin-mediated targeting of peanut to pseudocleavage furrows is regulated by the GTPase Ran. Mol Biol Cell 19:3735–3744. doi: 10.1091/mbc.e08-01-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilde A, Zheng Y. 1999. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284:1359–1362. doi: 10.1126/science.284.5418.1359 [DOI] [PubMed] [Google Scholar]
- 42. Ohba T, Nakamura M, Nishitani H, Nishimoto T. 1999. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science 284:1356–1358. doi: 10.1126/science.284.5418.1356 [DOI] [PubMed] [Google Scholar]
- 43. Dimitracopoulos A, Srivastava P, Chaigne A, Win Z, Shlomovitz R, Lancaster OM, Le Berre M, Piel M, Franze K, Salbreux G, Baum B. 2020. Mechanochemical crosstalk produces cell-intrinsic patterning of the cortex to orient the mitotic spindle. Curr Biol 30:3687–3696. doi: 10.1016/j.cub.2020.06.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rodrigues NTL, Lekomtsev S, Jananji S, Kriston-Vizi J, Hickson GRX, Baum B. 2015. Kinetochore-localized PP1-Sds22 couples chromosome segregation to polar relaxation. Nature 524:489–492. doi: 10.1038/nature14496 [DOI] [PubMed] [Google Scholar]
- 45. Kiyomitsu T, Cheeseman IM. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol 14:311–317. doi: 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fan S, Whiteman EL, Hurd TW, McIntyre JC, Dishinger JF, Liu CJ, Martens JR, Verhey KJ, Sajjan U, Margolis B. 2011. Induction of Ran GTP drives ciliogenesis. Mol Biol Cell 22:4539–4548. doi: 10.1091/mbc.E11-03-0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang S, Dougherty LL, Avasthi P. 2021. Separable roles for RanGTP in nuclear and ciliary trafficking of a kinesin-2 subunit. J Biol Chem 296:100117. doi: 10.1074/jbc.RA119.010936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mans BJ, Anantharaman V, Aravind L, Koonin EV. 2004. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3:1612–1637. doi: 10.4161/cc.3.12.1316 [DOI] [PubMed] [Google Scholar]
- 49. De Magistris P. 2021. The great escape: mRNA export through the nuclear pore complex. Int J Mol Sci 22:11767. doi: 10.3390/ijms222111767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Derrer CP, Mancini R, Vallotton P, Huet S, Weis K, Dultz E. 2019. The RNA export factor Mex67 functions as a mobile nucleoporin. J Cell Biol 218:3967–3976. doi: 10.1083/jcb.201909028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stewart M. 2007. Ratcheting mRNA out of the nucleus. Mol Cell 25:327–330. doi: 10.1016/j.molcel.2007.01.016 [DOI] [PubMed] [Google Scholar]
- 52. Jao L-E, Akef A, Wente SR. 2017. A role for Gle1, a regulator of DEAD-box RNA helicases, at centrosomes and basal bodies. Mol Biol Cell 28:120–127. doi: 10.1091/mbc.E16-09-0675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. D’Angelo MA, Raices M, Panowski SH, Hetzer MW. 2009. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136:284–295. doi: 10.1016/j.cell.2008.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dey G, Culley S, Curran S, Schmidt U, Henriques R, Kukulski W, Baum B. 2020. Closed mitosis requires local disassembly of the nuclear envelope. Nature 585:119–123. doi: 10.1038/s41586-020-2648-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Otsuka S, Tempkin JOB, Zhang W, Politi AZ, Rybina A, Hossain MJ, Kueblbeck M, Callegari A, Koch B, Morero NR, Sali A, Ellenberg J. 2023. A quantitative map of nuclear pore assembly reveals two distinct mechanisms. Nature 613:575–581. doi: 10.1038/s41586-022-05528-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Otsuka S, Bui KH, Schorb M, Hossain MJ, Politi AZ, Koch B, Eltsov M, Beck M, Ellenberg J. 2016. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. Elife 5:e19071. doi: 10.7554/eLife.19071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Webster BM, Colombi P, Jäger J, Lusk CP. 2014. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159:388–401. doi: 10.1016/j.cell.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hampoelz B, Mackmull M-T, Machado P, Ronchi P, Bui KH, Schieber N, Santarella-Mellwig R, Necakov A, Andrés-Pons A, Philippe JM, Lecuit T, Schwab Y, Beck M. 2016. Pre-assembled nuclear pores insert into the nuclear envelope during early development. Cell 166:664–678. doi: 10.1016/j.cell.2016.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chase AR, Laudermilch E, Schlieker C. 2017. Torsin ATPases: harnessing dynamic instability for function. Front Mol Biosci 4:29. doi: 10.3389/fmolb.2017.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rampello AJ, Laudermilch E, Vishnoi N, Prophet SM, Shao L, Zhao C, Lusk CP, Schlieker C. 2020. Torsin ATPase deficiency leads to defects in nuclear pore biogenesis and sequestration of MLF2. J Cell Biol 219:e201910185. doi: 10.1083/jcb.201910185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Panagaki D, Croft JT, Keuenhof K, Larsson Berglund L, Andersson S, Kohler V, Büttner S, Tamás MJ, Nyström T, Neutze R, Höög JL. 2021. Nuclear envelope budding is a response to cellular stress. Proc Natl Acad Sci U S A 118:e2020997118. doi: 10.1073/pnas.2020997118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lomakin AJ, Cattin CJ, Cuvelier D, Alraies Z, Molina M, Nader GPF, Srivastava N, Sáez PJ, Garcia-Arcos JM, Zhitnyak IY, Bhargava A, Driscoll MK, Welf ES, Fiolka R, Petrie RJ, De Silva NS, González-Granado JM, Manel N, Lennon-Duménil AM, Müller DJ, Piel M. 2020. The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science 370:eaba2894. doi: 10.1126/science.aba2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Venturini V, Pezzano F, Català Castro F, Häkkinen H-M, Jiménez-Delgado S, Colomer-Rosell M, Marro M, Tolosa-Ramon Q, Paz-López S, Valverde MA, Weghuber J, Loza-Alvarez P, Krieg M, Wieser S, Ruprecht V. 2020. The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science 370:eaba2644. doi: 10.1126/science.aba2644 [DOI] [PubMed] [Google Scholar]
- 64. Nader G de F, Agüera-Gonzalez S, Routet F, Gratia M, Maurin M, Cancila V, Cadart C, Palamidessi A, Ramos RN, San Roman M, Gentili M, Yamada A, Williart A, Lodillinsky C, Lagoutte E, Villard C, Viovy J-L, Tripodo C, Galon J, Scita G, Manel N, Chavrier P, Piel M. 2021. Compromised nuclear envelope integrity drives TREX1-dependent DNA damage and tumor cell invasion. Cell 184:5230–5246. doi: 10.1016/j.cell.2021.08.035 [DOI] [PubMed] [Google Scholar]
- 65. French SL, Santangelo TJ, Beyer AL, Reeve JN. 2007. Transcription and translation are coupled in Archaea. Mol Biol Evol 24:893–895. doi: 10.1093/molbev/msm007 [DOI] [PubMed] [Google Scholar]
- 66. Weixlbaumer A, Grünberger F, Werner F, Grohmann D. 2021. Coupling of transcription and translation in Archaea: cues from the bacterial world. Front Microbiol 12:661827. doi: 10.3389/fmicb.2021.661827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Johnson GE, Lalanne J-B, Peters ML, Li G-W. 2020. Functionally uncoupled transcription-translation in Bacillus subtilis. Nature 585:124–128. doi: 10.1038/s41586-020-2638-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rogozin IB, Carmel L, Csuros M, Koonin EV. 2012. Origin and evolution of spliceosomal introns. Biol Direct 7:11. doi: 10.1186/1745-6150-7-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pederson T. 2011. The nucleolus. Cold Spring Harb Perspect Biol 3:a000638–a000638. doi: 10.1101/cshperspect.a000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. 2011. Evolution of introns in the archaeal world. Proc Natl Acad Sci U S A 108:4782–4787. doi: 10.1073/pnas.1100862108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jaako P, Faille A, Tan S, Wong CC, Escudero-Urquijo N, Castro-Hartmann P, Wright P, Hilcenko C, Adams DJ, Warren AJ. 2022. eIF6 rebinding dynamically couples ribosome maturation and translationon. Nat Commun 13:1562. doi: 10.1038/s41467-022-29214-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gehring NH, Roignant J-Y. 2021. Anything but ordinary - emerging splicing mechanisms in eukaryotic gene regulation. Trends Genet 37:355–372. doi: 10.1016/j.tig.2020.10.008 [DOI] [PubMed] [Google Scholar]
- 73. Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479–480:2–25. doi: 10.1016/j.virol.2015.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Abodeely M, DuBois KN, Hehl A, Stefanic S, Sajid M, DeSouza W, Attias M, Engel JC, Hsieh I, Fetter RD, McKerrow JH. 2009. A contiguous compartment functions as endoplasmic reticulum and endosome/lysosome in Giardia lamblia. Eukaryot Cell 8:1665–1676. doi: 10.1128/EC.00123-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Roger AJ, Svärd SG, Tovar J, Clark CG, Smith MW, Gillin FD, Sogin ML. 1998. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc Natl Acad Sci U S A 95:229–234. doi: 10.1073/pnas.95.1.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tovar J, León-Avila G, Sánchez LB, Sutak R, Tachezy J, van der Giezen M, Hernández M, Müller M, Lucocq JM. 2003. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature 426:172–176. doi: 10.1038/nature01945 [DOI] [PubMed] [Google Scholar]
- 77. Karnkowska A, Vacek V, Zubáčová Z, Treitli SC, Petrželková R, Eme L, Novák L, Žárský V, Barlow LD, Herman EK, Soukal P, Hroudová M, Doležal P, Stairs CW, Roger AJ, Eliáš M, Dacks JB, Vlček Č, Hampl V. 2016. A eukaryote without a mitochondrial organelle. Curr Biol 26:1274–1284. doi: 10.1016/j.cub.2016.03.053 [DOI] [PubMed] [Google Scholar]
- 78. Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG. 2018. Archaea and the origin of eukaryotes. Nat Rev Microbiol 16:120. doi: 10.1038/nrmicro.2017.154 [DOI] [PubMed] [Google Scholar]
- 79. Koumandou VL, Wickstead B, Ginger ML, van der Giezen M, Dacks JB, Field MC. 2013. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol 48:373–396. doi: 10.3109/10409238.2013.821444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Eme L, Sharpe SC, Brown MW, Roger AJ. 2014. On the age of eukaryotes: evaluating evidence from fossils and molecular clocks. Cold Spring Harb Perspect Biol 6:a016139–a016139. doi: 10.1101/cshperspect.a016139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Betts HC, Puttick MN, Clark JW, Williams TA, Donoghue PCJ, Pisani D. 2018. Integrated genomic and fossil evidence illuminates life's early evolution and eukaryote origin. Nat Ecol Evol 2:1556–1562. doi: 10.1038/s41559-018-0644-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Strassert JFH, Irisarri I, Williams TA, Burki F. 2021. A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids. Nat Commun 12:1879. doi: 10.1038/s41467-021-22044-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mahendrarajah TA, Moody ERR, Schrempf D, Szánthó LL, Dombrowski N, Davín AA, Pisani D, Donoghue PCJ, Szöllősi GJ, Williams TA, Spang A. 2023. ATP synthase evolution on a cross-braced dated tree of life. bioRxiv. doi: 10.1101/2023.04.11.536006 [DOI] [PMC free article] [PubMed]
- 84. López-García P, Moreira D. 2015. Open questions on the origin of eukaryotes. Trends Ecol Evol 30:697–708. doi: 10.1016/j.tree.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Roger AJ, Muñoz-Gómez SA, Kamikawa R. 2017. The origin and diversification of mitochondria. Curr Biol 27:R1177–R1192. doi: 10.1016/j.cub.2017.09.015 [DOI] [PubMed] [Google Scholar]
- 86. Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG. 2018. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557:101–105. doi: 10.1038/s41586-018-0059-5 [DOI] [PubMed] [Google Scholar]
- 87. Fan L, Wu D, Goremykin V, Xiao J, Xu Y, Garg S, Zhang C, Martin WF, Zhu R. 2020. Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within Alphaproteobacteria. Nat Ecol Evol 4:1213–1219. doi: 10.1038/s41559-020-1239-x [DOI] [PubMed] [Google Scholar]
- 88. Muñoz-Gómez SA, Susko E, Williamson K, Eme L, Slamovits CH, Moreira D, López-García P, Roger AJ. 2022. Site-and-branch-heterogeneous analyses of an expanded dataset favour mitochondria as sister to known aphaproteobacteria. Nat Ecol Evol 6:253–262. doi: 10.1038/s41559-021-01638-2 [DOI] [PubMed] [Google Scholar]
- 89. Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG. 2022. Phylogenetic affiliation of mitochondria with Alpha-II and Rickettsiales is an artefact. Nat Ecol Evol 6:1829–1831. doi: 10.1038/s41559-022-01871-3 [DOI] [PubMed] [Google Scholar]
- 90. Koonin EV, Yutin N. 2014. The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb Perspect Biol 6:a016188. doi: 10.1101/cshperspect.a016188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lake JA, Henderson E, Oakes M, Clark MW. 1984. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci U S A 81:3786–3790. doi: 10.1073/pnas.81.12.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Foster PG, Cox CJ, Embley TM. 2009. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Philos Trans R Soc Lond B Biol Sci 364:2197–2207. doi: 10.1098/rstb.2009.0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Raymann K, Brochier-Armanet C, Gribaldo S. 2015. The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci U S A 112:6670–6675. doi: 10.1073/pnas.1420858112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Guy L, Ettema TJG. 2011. The archaeal 'TACK' superphylum and the origin of eukaryotes. Trends Microbiol 19:580–587. doi: 10.1016/j.tim.2011.09.002 [DOI] [PubMed] [Google Scholar]
- 95. Williams TA, Foster PG, Cox CJ, Embley TM. 2013. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504:231–236. doi: 10.1038/nature12779 [DOI] [PubMed] [Google Scholar]
- 96. Pittis AA, Gabaldon T. 2016. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 531:101–104. doi: 10.1038/nature16941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rochette NC, Brochier-Armanet C, Gouy M. 2014. Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes. Mol Biol Evol 31:832–845. doi: 10.1093/molbev/mst272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Thiergart T, Landan G, Schenk M, Dagan T, Martin WF. 2012. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol 4:466–485. doi: 10.1093/gbe/evs018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Alvarez-Ponce D, Lopez P, Bapteste E, McInerney JO. 2013. Gene similarity networks provide tools for understanding eukaryote origins and evolution. Proc Natl Acad Sci U S A 110:E1594–E1603. doi: 10.1073/pnas.1211371110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Vosseberg J, van Hooff JJE, Marcet-Houben M, van Vlimmeren A, van Wijk LM, Gabaldón T, Snel B. 2021. Timing the origin of eukaryotic cellular complexity with ancient duplications. Nat Ecol Evol 5:92–100. doi: 10.1038/s41559-020-01320-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Grabowski B, Kelman Z. 2003. Archeal DNA replication: eukaryal proteins in a bacterial context. Annu Rev Microbiol 57:487–516. doi: 10.1146/annurev.micro.57.030502.090709 [DOI] [PubMed] [Google Scholar]
- 102. Barry ER, Bell SD. 2006. DNA replication in the archaea. Microbiol Mol Biol Rev 70:876–887. doi: 10.1128/MMBR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Greci MD, Bell SD. 2020. Archaeal DNA replication. Annu Rev Microbiol 74:65–80. doi: 10.1146/annurev-micro-020518-115443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Makarova KS, Koonin EV. 2013. Archaeology of eukaryotic DNA replication. Cold Spring Harb Perspect Biol 5:a012963. doi: 10.1101/cshperspect.a012963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sandman K, Krzycki JA, Dobrinski B, Lurz R, Reeve JN. 1990. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc Natl Acad Sci U S A 87:5788–5791. doi: 10.1073/pnas.87.15.5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Grayling RA, Sandman K, Reeve JN. 1996. Histones and chromatin structure in hyperthermophilic Archaea. FEMS Microbiol Rev 18:203–213. doi: 10.1111/j.1574-6976.1996.tb00237.x [DOI] [PubMed] [Google Scholar]
- 107. Mattiroli F, Bhattacharyya S, Dyer PN, White AE, Sandman K, Burkhart BW, Byrne KR, Lee T, Ahn NG, Santangelo TJ, Reeve JN, Luger K. 2017. Structure of histone-based chromatin in Archaea. Science 357:609–612. doi: 10.1126/science.aaj1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Grau-Bové X, Navarrete C, Chiva C, Pribasnig T, Antó M, Torruella G, Galindo LJ, Lang BF, Moreira D, López-Garcia P, Ruiz-Trillo I, Schleper C, Sabidó E, Sebé-Pedrós A. 2022. A phylogenetic and proteomic reconstruction of eukaryotic chromatin evolution. Nat Ecol Evol 6:1007–1023. doi: 10.1038/s41559-022-01771-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Laursen SP, Bowerman S, Luger K. 2021. Archaea: the final frontier of chromatin. J Mol Biol 433:166791. doi: 10.1016/j.jmb.2020.166791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Huet J, Schnabel R, Sentenac A, Zillig W. 1983. Archaebacteria and eukaryotes possess DNA-dependent RNA polymerases of a common type. EMBO J 2:1291–1294. doi: 10.1002/j.1460-2075.1983.tb01583.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Langer D, Hain J, Thuriaux P, Zillig W. 1995. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci U S A 92:5768–5772. doi: 10.1073/pnas.92.13.5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kwapisz M, Beckouët F, Thuriaux P. 2008. Early evolution of eukaryotic DNA-dependent RNA polymerases. Trends Genet 24:211–215. doi: 10.1016/j.tig.2008.02.002 [DOI] [PubMed] [Google Scholar]
- 113. Werner F, Grohmann D. 2011. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 9:85–98. doi: 10.1038/nrmicro2507 [DOI] [PubMed] [Google Scholar]
- 114. Kramm K, Endesfelder U, Grohmann D. 2019. A single-molecule view of archaeal transcription. J Mol Biol 431:4116–4131. doi: 10.1016/j.jmb.2019.06.009 [DOI] [PubMed] [Google Scholar]
- 115. Blombach F, Matelska D, Fouqueau T, Cackett G, Werner F. 2019. Key concepts and challenges in archaeal transcription. J Mol Biol 431:4184–4201. doi: 10.1016/j.jmb.2019.06.020 [DOI] [PubMed] [Google Scholar]
- 116. Henderson E, Oakes M, Clark MW, Lake JA, Matheson AT, Zillig W. 1984. A new ribosome structure. Science 225:510–512. doi: 10.1126/science.6429855 [DOI] [PubMed] [Google Scholar]
- 117. Lecompte O, Ripp R, Thierry J-C, Moras D, Poch O. 2002. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res 30:5382–5390. doi: 10.1093/nar/gkf693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yutin N, Puigbò P, Koonin EV, Wolf YI. 2012. Phylogenomics of prokaryotic ribosomal proteins. PLoS One 7:e36972. doi: 10.1371/journal.pone.0036972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Schmitt E, Coureux P-D, Kazan R, Bourgeois G, Lazennec-Schurdevin C, Mechulam Y. 2020. Recent advances in archaeal translation initiation. Front Microbiol 11:584152. doi: 10.3389/fmicb.2020.584152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Londei P, Ferreira-Cerca S. 2021. Ribosome biogenesis in archaea. Front Microbiol 12:686977. doi: 10.3389/fmicb.2021.686977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kisly I, Tamm T. 2023. Archaea/eukaryote-specific ribosomal proteins - guardians of a complex structure. Comput Struct Biotechnol J 21:1249–1261. doi: 10.1016/j.csbj.2023.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Jüttner M, Ferreira-Cerca S. 2022. Looking through the lens of the ribosome biogenesis evolutionary history: possible implications for archaeal phylogeny and eukaryogenesis. Mol Biol Evol 39:msac054. doi: 10.1093/molbev/msac054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. 2003. An exosome-like complex in sulfolobus solfataricus. EMBO Rep 4:889–893. doi: 10.1038/sj.embor.embor929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gauernack AS, Lassek C, Hou L, Dzieciolowski J, Evguenieva-Hackenberg E, Klug G. 2017. Nop5 interacts with the archaeal RNA exosome. FEBS Lett 591:4039–4048. doi: 10.1002/1873-3468.12915 [DOI] [PubMed] [Google Scholar]
- 125. Bernander R, Poplawski A. 1997. Cell cycle characteristics of thermophilic archaea. J Bacteriol 179:4963–4969. doi: 10.1128/jb.179.16.4963-4969.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lindås A-C, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. 2008. A unique cell division machinery in the archaea. Proc Natl Acad Sci U S A 105:18942–18946. doi: 10.1073/pnas.0809467105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Samson RY, Obita T, Freund SM, Williams RL, Bell SD. 2008. A role for the ESCRT system in cell division in archaea. Science 322:1710–1713. doi: 10.1126/science.1165322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Rinke C, Chuvochina M, Mussig AJ, Chaumeil P-A, Davín AA, Waite DW, Whitman WB, Parks DH, Hugenholtz P. 2021. A standardized Archaeal Taxonomy for the genome Taxonomy database. Nature microbiology 6:946–959. doi: 10.1038/s41564-021-00918-8 [DOI] [PubMed] [Google Scholar]
- 129. Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521:173–179. doi: 10.1038/nature14447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJG. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541:353–358. doi: 10.1038/nature21031 [DOI] [PubMed] [Google Scholar]
- 131. Liu Y, Makarova KS, Huang WC, Wolf YI, Nikolskaya AN, Zhang X, Cai M, Zhang CJ, Xu W, Luo Z, Cheng L, Koonin EV, Li M. 2021. Expanded diversity of asgard archaea and their relationships with eukaryotes. Nature 593:553–557. doi: 10.1038/s41586-021-03494-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Williams TA, Cox CJ, Foster PG, Szöllősi GJ, Embley TM. 2020. Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol 4:1568–1568. doi: 10.1038/s41559-020-01347-2 [DOI] [PubMed] [Google Scholar]
- 133. Eme L, Tamarit D, Caceres EF, Stairs CW, De Anda V, Schön ME, Seitz KW, Dombrowski N, Lewis WH, Homa F, Saw JH, Lombard J, Nunoura T, Li W-J, Hua Z-S, Chen L-X, Banfield JF, John ES, Reysenbach A-L, Stott MB, Schramm A, Kjeldsen KU, Teske AP, Baker BJ, Ettema TJG. 2023. Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes. Nature 618:992–999. doi: 10.1038/s41586-023-06186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Akıl C, Robinson RC. 2018. Genomes of asgard archaea encode profilins that regulate actin. Nature 562:439–443. doi: 10.1038/s41586-018-0548-6 [DOI] [PubMed] [Google Scholar]
- 135. Akıl C, Tran LT, Orhant-Prioux M, Baskaran Y, Manser E, Blanchoin L, Robinson RC. 2020. Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from asgard archaea. Proc Natl Acad Sci U S A 117:19904–19913. doi: 10.1073/pnas.2009167117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hatano T, Palani S, Papatziamou D, Salzer R, Souza DP, Tamarit D, Makwana M, Potter A, Haig A, Xu W, Townsend D, Rochester D, Bellini D, Hussain HMA, Ettema TJG, Löwe J, Baum B, Robinson NP, Balasubramanian M. 2022. Asgard archaea shed light on the evolutionary origins of the eukaryotic ubiquitin-ESCRT machinery. Nat Commun 13:3398. doi: 10.1038/s41467-022-30656-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Klinger CM, Spang A, Dacks JB, Ettema TJG. 2016. Tracing the archaeal origins of eukaryotic membrane-trafficking system building blocks. Mol Biol Evol 33:1528–1541. doi: 10.1093/molbev/msw034 [DOI] [PubMed] [Google Scholar]
- 138. Surkont J, Pereira-Leal JB. 2016. Are there rab GTPases in archaea? Mol Biol Evol 33:1833–1842. doi: 10.1093/molbev/msw061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Spang A, Caceres EF, Ettema TJG. 2017. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science 357:eaaf3883. doi: 10.1126/science.aaf3883 [DOI] [PubMed] [Google Scholar]
- 140. Spang A, Mahendrarajah TA, Offre P, Stairs CW. 2022. Evolving perspective on the origin and diversification of cellular life and the virosphere. Genome Biol Evol 14:evac034. doi: 10.1093/gbe/evac034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Stairs CW, Dharamshi JE, Tamarit D, Eme L, Jørgensen SL, Spang A, Ettema TJG. 2020. Chlamydial contribution to anaerobic metabolism during eukaryotic evolution. Sci Adv 6:eabb7258. doi: 10.1126/sciadv.abb7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ku C, Nelson-Sathi S, Roettger M, Sousa FL, Lockhart PJ, Bryant D, Hazkani-Covo E, McInerney JO, Landan G, Martin WF. 2015. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524:427–432. doi: 10.1038/nature14963 [DOI] [PubMed] [Google Scholar]
- 143. Wu F, Speth DR, Philosof A, Crémière A, Narayanan A, Barco RA, Connon SA, Amend JP, Antoshechkin IA, Orphan VJ. 2022. Unique mobile elements and scalable gene flow at the prokaryote-eukaryote boundary revealed by circularized asgard archaea genomes. Nat Microbiol 7:200–212. doi: 10.1038/s41564-021-01039-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Filée J, Becker HF, Mellottee L, Eddine RZ, Li Z, Yin W, Lambry JC, Liebl U, Myllykallio H. 2023. Bacterial origins of thymidylate metabolism in asgard archaea and eukarya. Nat Commun 14:838. doi: 10.1038/s41467-023-36487-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Archibald JM. 2015. Evolution: gene transfer in complex cells. Nature 524:423–424. doi: 10.1038/nature15205 [DOI] [PubMed] [Google Scholar]
- 146. Leger MM, Eme L, Stairs CW, Roger AJ. 2018. Demystifying eukaryote lateral gene transfer. Bioessays 40. doi: 10.1002/bies.201700115 [DOI] [PubMed] [Google Scholar]
- 147. Irwin NAT, Pittis AA, Richards TA, Keeling PJ. 2022. Systematic evaluation of horizontal gene transfer between eukaryotes and viruses. Nat Microbiol 7:327–336. doi: 10.1038/s41564-021-01026-3 [DOI] [PubMed] [Google Scholar]
- 148. Hirt RP, Alsmark C, Embley TM. 2015. Lateral gene transfers and the origins of the eukaryote proteome: a view from microbial parasites. Curr Opin Microbiol 23:155–162. doi: 10.1016/j.mib.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Sibbald SJ, Eme L, Archibald JM, Roger AJ. 2020. Lateral gene transfer mechanisms and pan-genomes in eukaryotes. Trends Parasitol 36:927–941. doi: 10.1016/j.pt.2020.07.014 [DOI] [PubMed] [Google Scholar]
- 150. Takemura M. 2020. Medusavirus ancestor in a proto-eukaryotic cell: updating the hypothesis for the viral origin of the nucleus. Front Microbiol 11:571831. doi: 10.3389/fmicb.2020.571831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bell PJL. 2022. Eukaryogenesis: the rise of an emergent superorganism. Front Microbiol 13:858064. doi: 10.3389/fmicb.2022.858064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Doamekpor SK, Sharma S, Kiledjian M, Tong L. 2022. Recent insights into noncanonical 5' capping and decapping of RNA. J Biol Chem 298:102171. doi: 10.1016/j.jbc.2022.102171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. de Castro IF, Volonté L, Risco C. 2013. Virus factories: biogenesis and structural design. Cell Microbiol 15:24–34. doi: 10.1111/cmi.12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Snijder EJ, Limpens RWAL, de Wilde AH, de Jong AWM, Zevenhoven-Dobbe JC, Maier HJ, Faas FFGA, Koster AJ, Bárcena M, Cimarelli A. 2020. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol 18:e3000715. doi: 10.1371/journal.pbio.3000715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Krupovic M, Dolja VV, Koonin EV. 2023. The virome of the last eukaryotic common ancestor and eukaryogenesis. Nat Microbiol 8:1008–1017. doi: 10.1038/s41564-023-01378-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Gould SB, Garg SG, Martin WF. 2016. Bacterial vesicle secretion and the evolutionary origin of the eukaryotic endomembrane system. Trends Microbiol 24:525–534. doi: 10.1016/j.tim.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 157. Villanueva L, von Meijenfeldt FAB, Westbye AB, Yadav S, Hopmans EC, Dutilh BE, Damsté JSS. 2021. Bridging the membrane lipid divide: bacteria of the FCB group superphylum have the potential to synthesize archaeal ether lipids. ISME J 15:168–182. doi: 10.1038/s41396-020-00772-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Caforio A, Siliakus MF, Exterkate M, Jain S, Jumde VR, Andringa RLH, Kengen SWM, Minnaard AJ, Driessen AJM, van der Oost J. 2018. Converting Escherichia coli into an archaebacterium with a hybrid heterochiral membrane. Proc Natl Acad Sci U S A 115:3704–3709. doi: 10.1073/pnas.1721604115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, Matsui Y, Miyazaki M, Murata K, Saito Y, Sakai S, Song C, Tasumi E, Yamanaka Y, Yamaguchi T, Kamagata Y, Tamaki H, Takai K. 2020. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature 577:519–525. doi: 10.1038/s41586-019-1916-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Ettema TJG. 2016. Evolution: mitochondria in the second act. Nature 531:39–40. doi: 10.1038/nature16876 [DOI] [PubMed] [Google Scholar]
- 161. Makarova KS, Krupovic M, Koonin EV. 2014. Evolution of replicative DNA polymerases in archaea and their contributions to the eukaryotic replication machinery. Front Microbiol 5:354. doi: 10.3389/fmicb.2014.00354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Phung DK, Etienne C, Batista M, Langendijk-Genevaux P, Moalic Y, Laurent S, Liuu S, Morales V, Jebbar M, Fichant G, Bouvier M, Flament D, Clouet-d’Orval B. 2020. RNA processing machineries in archaea: the 5'-3' exoribonuclease aRNase J of the β-CASP family is engaged specifically with the helicase ASH-Ski2 and the 3'-5' exoribonucleolytic RNA exosome machinery. Nucleic Acids Res 48:3832–3847. doi: 10.1093/nar/gkaa052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Rodriguez-Corona U, Sobol M, Rodriguez-Zapata LC, Hozak P, Castano E. 2015. Fibrillarin from archaea to human. Biol Cell 107:159–174. doi: 10.1111/boc.201400077 [DOI] [PubMed] [Google Scholar]
- 164. Evguenieva-Hackenberg E, Hou L, Glaeser S, Klug G. 2014. Structure and function of the archaeal exosome. Wiley Interdiscip Rev RNA 5:623–635. doi: 10.1002/wrna.1234 [DOI] [PubMed] [Google Scholar]
- 165. Maupin-Furlow JA. 2013. Archaeal proteasomes and sampylation. Subcell Biochem 66:297–327. doi: 10.1007/978-94-007-5940-4_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Albers SV, Meyer BH. 2011. The archaeal cell envelope. Nat Rev Microbiol 9:414–426. doi: 10.1038/nrmicro2576 [DOI] [PubMed] [Google Scholar]
- 167. Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. 2004. Identification of two origins of replication in the single chromosome of the archaeon sulfolobus solfataricus. Cell 116:25–38. doi: 10.1016/s0092-8674(03)01034-1 [DOI] [PubMed] [Google Scholar]
- 168. Xu Y, Gristwood T, Hodgson B, Trinidad JC, Albers SV, Bell SD. 2016. Archaeal orthologs of Cdc45 and GINS form a stable complex that stimulates the helicase activity of MCM. Proc Natl Acad Sci U S A 113:13390–13395. doi: 10.1073/pnas.1613825113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Noble ME, Endicott JA, Brown NR, Johnson LN. 1997. The cyclin box fold: protein recognition in cell-cycle and transcription control. Trends Biochem Sci 22:482–487. doi: 10.1016/s0968-0004(97)01144-4 [DOI] [PubMed] [Google Scholar]
- 170. Tarrason Risa G, Hurtig F, Bray S, Hafner AE, Harker-Kirschneck L, Faull P, Davis C, Papatziamou D, Mutavchiev DR, Fan C, Meneguello L, Arashiro Pulschen A, Dey G, Culley S, Kilkenny M, Souza DP, Pellegrini L, de Bruin RAM, Henriques R, Snijders AP, Šarić A, Lindås A-C, Robinson NP, Baum B. 2020. The proteasome controls ESCRT-III-mediated cell division in an archaeon. Science 369:eaaz2532. doi: 10.1126/science.aaz2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Takemata N, Samson RY, Bell SD. 2019. Physical and functional compartmentalization of archaeal chromosomes. Cell 179:165–179. doi: 10.1016/j.cell.2019.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Henneman B, van Emmerik C, van Ingen H, Dame RT. 2018. Structure and function of archaeal histones. PLoS Genet 14:e1007582. doi: 10.1371/journal.pgen.1007582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Hocher A, Borrel G, Fadhlaoui K, Brugère JF, Gribaldo S, Warnecke T. 2022. Growth temperature and chromatinization in archaea. Nat Microbiol 7:1932–1942. doi: 10.1038/s41564-022-01245-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Goyal M, Banerjee C, Nag S, Bandyopadhyay U. 2016. The alba protein family: structure and function. Biochim Biophys Acta 1864:570–583. doi: 10.1016/j.bbapap.2016.02.015 [DOI] [PubMed] [Google Scholar]
- 175. Junglas B, Briegel A, Burghardt T, Walther P, Wirth R, Huber H, Rachel R. 2008. Ignicoccus hospitalis and nanoarchaeum equitans: ultrastructure, cell-cell interaction, and 3D reconstruction from serial sections of freeze-substituted cells and by electron cryotomography. Arch Microbiol 190:395–408. doi: 10.1007/s00203-008-0402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Dey G, Thattai M, Baum B. 2016. On the archaeal origins of eukaryotes and the challenges of inferring phenotype from genotype. Trends Cell Biol 26:476–485. doi: 10.1016/j.tcb.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo RI, Xu J, Rittmann SK-MR, Klingl A, Pilhofer M, Schleper C. 2023. Actin cytoskeleton and complex cell architecture in an asgard archaeon. Nature 613:332–339. doi: 10.1038/s41586-022-05550-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Spang A, Stairs CW, Dombrowski N, Eme L, Lombard J, Caceres EF, Greening C, Baker BJ, Ettema TJG. 2019. Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of asgard archaeal metabolism. Nat Microbiol 4:1138–1148. doi: 10.1038/s41564-019-0406-9 [DOI] [PubMed] [Google Scholar]
- 179. Martin W, Müller M. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392:37–41. doi: 10.1038/32096 [DOI] [PubMed] [Google Scholar]
- 180. Moreira D, López-García P. 1998. Symbiosis between methanogenic archaea and delta-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J Mol Evol 47:517–530. doi: 10.1007/PL00006408 [DOI] [PubMed] [Google Scholar]
- 181. Bulzu P-A, Andrei A-Ş, Salcher MM, Mehrshad M, Inoue K, Kandori H, Beja O, Ghai R, Banciu HL. 2019. Casting light on asgardarchaeota metabolism in a sunlit microoxic niche. Nat Microbiol 4:1129–1137. doi: 10.1038/s41564-019-0404-y [DOI] [PubMed] [Google Scholar]
- 182. Pulschen AA, Mutavchiev DR, Culley S, Sebastian KN, Roubinet J, Roubinet M, Risa GT, van Wolferen M, Roubinet C, Schmidt U, Dey G, Albers S-V, Henriques R, Baum B. 2020. Live imaging of a hyperthermophilic archaeon reveals distinct roles for two ESCRT-III homologs in ensuring a robust and symmetric division. Curr Biol 30:2852–2859. doi: 10.1016/j.cub.2020.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Pelve EA, Lindås A-C, Martens-Habbena W, de la Torre JR, Stahl DA, Bernander R. 2011. Cdv-based cell division and cell cycle organization in the thaumarchaeon nitrosopumilus maritimus. Mol Microbiol 82:555–566. doi: 10.1111/j.1365-2958.2011.07834.x [DOI] [PubMed] [Google Scholar]
- 184. Beck M, Hurt E. 2017. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18:73–89. doi: 10.1038/nrm.2016.147 [DOI] [PubMed] [Google Scholar]
- 185. Mills J, Gebhard LJ, Schubotz F, Shevchenko A, Speth DR, Liao Y, Duggin IG, Marchfelder A, Erdmann S. 2023. Extracellular vesicles of euryarchaeida: precursor to eukaryotic membrane trafficking. bioRxiv. doi: 10.1101/2023.03.03.530948:2023.03.03.530948 [DOI] [Google Scholar]
- 186. Baumann SJ, Grawenhoff J, Rodrigues EC, Speroni S, Gili M, Komissarov A, Maurer SP. 2022. APC couples neuronal mRNAs to multiple kinesins, EB1, and shrinking microtubule ends for bidirectional mRNA motility. Proc Natl Acad Sci U S A 119:e2211536119. doi: 10.1073/pnas.2211536119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Mili S, Moissoglu K, Macara IG. 2008. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453:115–119. doi: 10.1038/nature06888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Luby-Phelps K, Taylor DL, Lanni F. 1986. Probing the structure of cytoplasm. J Cell Biol 102:2015–2022. doi: 10.1083/jcb.102.6.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP. 2000. Homologs of small nucleolar RNAs in archaea. Science 288:517–522. doi: 10.1126/science.288.5465.517 [DOI] [PubMed] [Google Scholar]
- 190. Ye K, Jia R, Lin J, Ju M, Peng J, Xu A, Zhang L. 2009. Structural organization of box C/D RNA-guided RNA methyltransferase. Proc Natl Acad Sci U S A 106:13808–13813. doi: 10.1073/pnas.0905128106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Avcı B, Brandt J, Nachmias D, Elia N, Albertsen M, Ettema TJG, Schramm A, Kjeldsen KU. 2022. Spatial separation of ribosomes and DNA in asgard archaeal cells. ISME J 16:606–610. doi: 10.1038/s41396-021-01098-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Clouet-d’Orval B, Batista M, Bouvier M, Quentin Y, Fichant G, Marchfelder A, Maier LK. 2018. Insights into RNA-processing pathways and associated RNA-degrading enzymes in archaea. FEMS Microbiol Rev 42:579–613. doi: 10.1093/femsre/fuy016 [DOI] [PubMed] [Google Scholar]
- 193. Basu U, Si K, Warner JR, Maitra U. 2001. The saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol 21:1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Lo Gullo G, De Santis ML, Paiardini A, Rosignoli S, Romagnoli A, La Teana A, Londei P, Benelli D. 2021. The archaeal elongation factor EF-2 induces the release of aIF6 from 50s ribosomal subunit. Front Microbiol 12:631297. doi: 10.3389/fmicb.2021.631297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Vosseberg J, Stolker D, von der Dunk SHA, Snel B, Arkhipova I. 2023. Integrating phylogenetics with intron positions illuminates the origin of the complex spliceosome. Mol Biol Evol 40:msad011. doi: 10.1093/molbev/msad011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Yokobori S, Itoh T, Yoshinari S, Nomura N, Sako Y, Yamagishi A, Oshima T, Kita K, Watanabe Y. 2009. Gain and loss of an Intron in a protein-coding gene in archaea: the case of an archaeal RNA pseudouridine synthase gene. BMC Evol Biol 9:198. doi: 10.1186/1471-2148-9-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Penny D, Hoeppner MP, Poole AM, Jeffares DC. 2009. An overview of the introns-first theory. J Mol Evol 69:527–540. doi: 10.1007/s00239-009-9279-5 [DOI] [PubMed] [Google Scholar]
- 198. Koonin EV. 2006. The origin of Introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate?. Biol Direct 1:22. doi: 10.1186/1745-6150-1-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Melnikov S, Kwok H-S, Manakongtreecheep K, van den Elzen A, Thoreen CC, Söll D. 2020. Archaeal ribosomal proteins possess nuclear localization signal-type motifs: implications for the origin of the cell nucleus. Mol Biol Evol 37:124–133. doi: 10.1093/molbev/msz207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Nederlof PM, Wang HR, Baumeister W. 1995. Nuclear localization signals of human and thermoplasma proteasomal alpha subunits are functional in vitro. Proc Natl Acad Sci U S A 92:12060–12064. doi: 10.1073/pnas.92.26.12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Islas-Morales PF, Cárdenas A, Mosqueira MJ, Jiménez-García LF, Voolstra CR. 2023. Ultrastructural and proteomic evidence for the presence of a putative nucleolus in an archaeon. Front Microbiol 14:1075071. doi: 10.3389/fmicb.2023.1075071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Lakkaraju AKK, Mary C, Scherrer A, Johnson AE, Strub K. 2008. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 133:440–451. doi: 10.1016/j.cell.2008.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Yue L, Li J, Zhang B, Qi L, Li Z, Zhao F, Li L, Zheng X, Dong X. 2020. The conserved ribonuclease aCPSF1 triggers genome-wide transcription termination of archaea via a 3'-end cleavage mode. Nucleic Acids Res 48:9589–9605. doi: 10.1093/nar/gkaa702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Linder P, Jankowsky E. 2011. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12:505–516. doi: 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- 205. Bressloff PC, Newby JM. 2013. Stochastic models of intracellular transport. Rev Mod Phys 85:135–196. doi: 10.1103/RevModPhys.85.135 [DOI] [Google Scholar]
- 206. Theriot JA. 2013. Why are bacteria different from eukaryotes? BMC Biol 11:119. doi: 10.1186/1741-7007-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Löwe J, Amos LA. 2009. Evolution of cytomotive filaments: the cytoskeleton from prokaryotes to eukaryotes. Int J Biochem Cell Biol 41:323–329. doi: 10.1016/j.biocel.2008.08.010 [DOI] [PubMed] [Google Scholar]
- 208. Gittes F, Mickey B, Nettleton J, Howard J. 1993. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol 120:923–934. doi: 10.1083/jcb.120.4.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Böttcher B, Hurt E. 2009. Mechanochemical removal of ribosome biogenesis factors from nascent 60s ribosomal subunits. Cell 138:911–922. doi: 10.1016/j.cell.2009.06.045 [DOI] [PubMed] [Google Scholar]
- 210. Schmidt H, Carter AP. 2016. Review: structure and mechanism of the dynein motor ATPase. Biopolymers 105:557–567. doi: 10.1002/bip.22856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Iyer LM, Leipe DD, Koonin EV, Aravind L. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol 146:11–31. doi: 10.1016/j.jsb.2003.10.010 [DOI] [PubMed] [Google Scholar]
- 212. Han G, Liu B, Zhang J, Zuo W, Morris NR, Xiang X. 2001. The aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr Biol 11:719–724. doi: 10.1016/s0960-9822(01)00200-7 [DOI] [PubMed] [Google Scholar]
- 213. Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. 2006. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat Cell Biol 8:264–270. doi: 10.1038/ncb1370 [DOI] [PubMed] [Google Scholar]
- 214. Santana-Molina C, Rivas-Marin E, Rojas AM, Devos DP. 2020. Origin and evolution of polycyclic triterpene synthesis. Mol Biol Evol 37:1925–1941. doi: 10.1093/molbev/msaa054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Hoshino Y, Gaucher EA. 2021. Evolution of bacterial steroid biosynthesis and its impact on eukaryogenesis. Proc Natl Acad Sci U S A 118:e2101276118. doi: 10.1073/pnas.2101276118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Lombard J, López-García P, Moreira D. 2012. The early evolution of lipid membranes and the three domains of life. Nat Rev Microbiol 10:507–515. doi: 10.1038/nrmicro2815 [DOI] [PubMed] [Google Scholar]
- 217. van Wolferen M, Pulschen AA, Baum B, Gribaldo S, Albers S-V. 2022. The cell biology of archaea. Nat Microbiol 7:1744–1755. doi: 10.1038/s41564-022-01215-8 [DOI] [PMC free article] [PubMed] [Google Scholar]