SUMMARY
Methicillin-resistant Staphylococcus aureus (MRSA) is a leading cause of severe and often fatal infections. MRSA epidemics have occurred in waves, whereby a previously successful lineage has been replaced by a more fit and better adapted lineage. Selection pressures in both hospital and community settings are not uniform across the globe, which has resulted in geographically distinct epidemiology. This review focuses on the mechanisms that trigger the establishment and maintenance of current, dominant MRSA lineages across the globe. While the important role of antibiotic resistance will be mentioned throughout, factors which influence the capacity of S. aureus to colonize and cause disease within a host will be the primary focus of this review. We show that while MRSA possesses a diverse arsenal of toxins including alpha-toxin, the success of a lineage involves more than just producing toxins that damage the host. Success is often attributed to the acquisition or loss of genetic elements involved in colonization and niche adaptation such as the arginine catabolic mobile element, as well as the activity of regulatory systems, and shift metabolism accordingly (e.g., the accessory genome regulator, agr). Understanding exactly how specific MRSA clones cause prolonged epidemics may reveal targets for therapies, whereby both core (e.g., the alpha toxin) and acquired virulence factors (e.g., the Panton-Valentine leukocidin) may be nullified using anti-virulence strategies.
KEYWORDS: methicillin-resistant Staphylococcus aureus, virulence, toxins, superantigens, metabolism, gene regulation, mobile genetic elements
INTRODUCTION
Staphylococcus aureus remains a threat to public health despite collective efforts designed to mitigate its impact upon healthcare systems, and the community. S. aureus is a commensal of the skin and mucosal surface of about 30% of the human population (1–3). However, when opportunity presents, it is capable of causing a range of infections affecting virtually all of the body’s tissues including the skin, soft tissue, blood, bone, heart, and lungs (4–13).
Compounding the problem of S. aureus infection is the emergence of antibiotic-resistant strains (14–19). Methicillin-resistant S. aureus (MRSA) infections have been a clinical challenge for decades, and compared with infections due to methicillin-susceptible S. aureus (MSSA), morbidity and mortality rates are typically higher, which is associated with increased length of hospital stay and associated economic expenses (20–24). The deployment of promising anti-staphylococcal agents has been met with the rapid emergence of resistance, including against last-line agents such as linezolid, daptomycin, and anti-MRSA cephalosporins (18, 25–30). Vancomycin resistance in MRSA remains rare, but the rise of vancomycin-intermediate S. aureus strains is concerning (31, 32).
To define the global epidemiology of MRSA, strains are grouped into lineages based on shared molecular characteristics. The success and spread of MRSA lineages are not uniform, with some remaining geographically restricted and others capable of causing global pandemics (33, 34). The regional distribution, frequency, and persistence of dominant MRSA lineages are multifactorial and involve a range of pathogenic factors. In addition to developing resistance to antibiotics, hospital disinfectants, and other toxic compounds, a selection of surface proteins may be present in a given lineage to mediate human infection, including adherence and immune interaction, as well as a range of lytic proteins that contribute to host tissue damage (35–44). Pathogenic factors are often carried on mobile genetic elements that originate in other non-pathogenic species, which serve as a genetic reservoir for the adaptive evolution of MRSA (45, 46). Additionally, subtle genetic changes such as single nucleotide polymorphisms (SNPs), small insertions/deletions (InDels), and genome rearrangements can emerge under selection pressure. This genetic evolution can affect bacterial clone survival and success within a given host through its impact on metabolism and/or virulence gene expression (47–50). The purpose of this review is to summarize current knowledge of the pathogenic mechanisms that contribute to the successes of current epidemic and pandemic MRSA lineages from around the world.
EMERGENCE OF MRSA
Penicillin-resistant S. aureus
Mortality rates associated with staphylococcal bloodstream infections in the pre-antibiotic era exceeded 80% (51). In 1941, the outlook for patients with S. aureus bacteremia dramatically changed following the introduction of the β-lactam antibiotic, penicillin. β-Lactams bind covalently to penicillin-binding protein transpeptidases (PBPs), inhibiting the final cross-linking reaction in the synthesis of peptidoglycan, which is a critical component of the bacterial cell wall (52). However, penicillin resistance became prevalent soon after the clinical introduction of β-lactams, and by 1948, close to 60% of isolates were penicillin resistant (53). Penicillin resistance was subsequently attributed to the production of β-lactamase, an enzyme that hydrolyzes the β-lactam ring of penicillin resulting in drug deactivation (54–57). The bla gene, coding for β-lactamase, is typically carried on plasmids that facilitate horizontal gene transfer (HGT) between bacterial strains and species (54–57).
Methicillin-resistant S. aureus
In 1961, the semi-synthetic β-lactam methicillin was introduced, which was resistant to the action of β-lactamases that hydrolyzed penicillin. However, shortly after its clinical introduction, MRSA isolates were reported in the United Kingdom (58), and by the late 1960s, MRSA infections had been described in Australia, the United States (US), Switzerland, Denmark, France, India, and Japan (59–62). MRSA evolved from MSSA via the acquisition of a mobile genetic element known as the staphylococcal cassette chromosome mec (SCCmec) (63). The mec gene complex contains a structural gene (mecA, mecB, mecC, or mecD), which encodes a specific penicillin-binding protein (PBP2a, also known as PBP2′), as well as regulatory elements mecI and mecRI that control mec gene expression (34, 63–65). PBP2a has transpeptidase activity and harbors lower affinity for β-lactams when compared to native PBPs (66, 67). In concert with the transglycosylase activity of PBP2, PBP2a can restore peptidoglycan synthesis and generate high-level β-lactam resistance (68). It was thought that MRSA was restricted to healthcare settings until the early 1980s, when cases of community-acquired MRSA (CA-MRSA) infection were first reported (69–71).
Community-acquired S. aureus
CA-MRSA and hospital-associated MRSA (HA-MRSA) were traditionally classified by epidemiological definitions. The term “community-acquired” was applied loosely, as the acquisition of infection was often unclear, and many reported CA-MRSA cases that were associated with healthcare risk factors. The term was soon replaced by “community-associated MRSA” and the US Centers for Disease Control and Prevention (CDC) introduced a case definition to distinguish CA-MRSA from healthcare-associated infections (72). An infection was classified as CA-MRSA if it was diagnosed in an outpatient setting or less than 48 hours after hospital admission (72). In addition, the patient would have none of the following healthcare risk factors: history of hospitalization, surgery, dialysis, and residence in a long-term care facility within the previous year of MRSA culture date; permanent indwelling catheter or percutaneous device; or previous isolation of MRSA (72). It is clear that the distinction between CA-MRSA and HA-MRSA has become increasingly blurred as both continue to cause infections and outbreaks in hospital and community settings (72, 73).
Livestock-associated MRSA
MRSA is also an important veterinary and zoonotic pathogen present in a broad range of animal species, including pigs, cattle, horses, rabbits, poultry, dogs, and cats (74–80). The emergence of livestock-associated MRSA (LA-MRSA) in animals is often associated with an increase of LA-MRSA colonization in humans, particularly in farm workers (81, 82). LA-MRSA is capable of host adaptation to animals, and the selection pressure of antibiotics used in animal husbandry has raised concerns of multidrug-resistant LA-MRSA as a reservoir for human MRSA infections (83–85). Control measures for the evolution of antibiotic resistance, virulence, and transmission of LA-MRSA are required to reduce the impact of emerging LA-MRSA lineages on public health.
In addition to mecA, mecC is another genetic determinant that was first identified in human and bovine MRSA isolates from Denmark and the UK in 2011 (86). The origin of mecC remains unknown. Interestingly, Larsen et al. recently reported that particular MRSA lineages carrying mecC emerged in European hedgehogs prior to the clinical use of antibiotics (87). One speculative theory of the mecC appearance in these MRSA lineages was likely driven by co-evolution, in which S. aureus adapted to hedgehog dermatophyte Trichophyton erinacei that naturally produces two β-lactams (87).
DEFINING MRSA LINEAGES
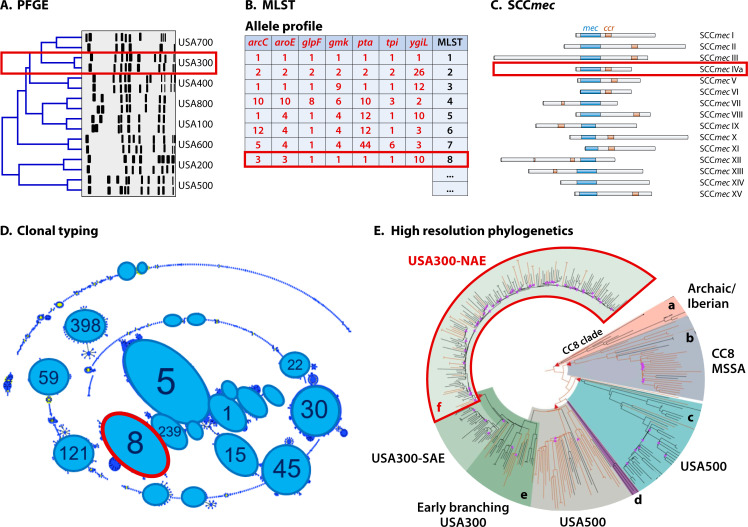
Several well-established techniques have been used to genetically categorize MRSA strains in order to describe population structure, and these methods have been discussed in a recent review (34). Here, the focus is on relevant techniques that will provide context when dominant lineages are subsequently described, including pulsed-field gel electrophoresis (PFGE), multi-locus sequence typing (MLST), SCCmec typing, and whole-genome sequencing (WGS) (Fig. 1). For other typing methods such as spa typing, readers are referred to other reviews (34). Of note, the techniques are not mutually exclusive; often, multiple techniques can be used in combination to define a lineage, and two different techniques can generate definitions that can be used interchangeably.
Fig 1.
Defining MRSA lineages. (A) Pulsed-field gel electrophoresis (PFGE) was previously the standard method to discriminate S. aureus lineages and is based on comparing DNA patterns following restriction enzyme digestion [adapted from reference (88)]. (B) Characterization of the relatedness between S. aureus strains was advanced by comparing the sequence of seven housekeeping genes, which is known as multi-locus sequence typing (MLST). (C) As all MRSA strains carry the staphylococcal cassette chromosome mec (SCCmec), identifying sequence and structural similarities of SCCmec between the isolates provides another dimension for lineage definition [based on reference (34)]. (D) Sequence types of MRSA strains defined by MLST can be grouped into clonal complexes (CCs) to infer evolutionary descent across MRSA lineages [adapted from reference (89) with permission of the publisher). (E) Whole-genome sequencing is increasingly being used for phylogenetic analysis to trace the evolution and transmission of successful MRSA clones at high resolution. In the example shown, phylogenetic analysis of 348 genomes illustrates the relationship structure of clinically important CC8 groups. These groups include (A) CC8a Archaic/Iberian, (B) CC8b MSSA clade, (C) CC8c USA500, (D) CC8d CMRSA-9, (E) CC8e (USA500, EB-USA300, USA300-SAE), and (F) CC8f USA300-NAE. Throughout the figure, red boxes indicate how these technologies can be integrated to define a lineage. Here, we have highlighted MRSA-ST8-IVa (USA300-NAE) [adapted from reference (90)].
Pulsed-field gel electrophoresis
Traditionally, PFGE has been a standard method for typing MRSA. This method involves in-gel digestion of chromosomal DNA with a restriction endonuclease, typically SmaI. The DNA fragments are resolved by gel electrophoresis using an instrument that switches current directions based on a predetermined pattern. The relatedness of S. aureus isolates can subsequently be determined by comparing the DNA restriction patterns (91, 92) (Fig. 1A). PFGE was formerly the standard method used by the CDC for bacterial strain typing (91). MRSA strains were classified into pulsed-field types (PFTs), for example, PFT USA100 (88). In the early 2000s, PFGE played an important role in identifying USA300 as a dominant CA-MRSA lineage in the US (88, 93). However, PFGE is time consuming and requires costly reagents and specialized equipment, which are its major limitations (94–97). Furthermore, there is insufficient resolution for bands dissimilar in size by <5% and inconsistent interpretation of PFGE bands between different facilities (94–97). Consequently and due to the advent of newer methods for defining MRSA lineages, PFGE is a less commonly used approach nowadays.
Multi-locus sequence typing
MLST is the most widely recognized typing technique for describing MRSA populations. MLST involves sequencing internal fragments of seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and ygiL) that are present in all strains of S. aureus (Fig. 1B). The strain of interest is assigned a sequence type (ST) based on the allele profiles of the seven genes via an online database (https://pubmlst.org/saureus/) (98). MLST has the advantage of comparing strains between laboratories and provides the opportunity to explore patterns of evolutionary descent using the algorithm, eBURST, which arranges isolates of similar STs into groups known as clonal complexes (CC) and facilitates the prediction of founder STs (99) (Fig. 1D). STs that differ at only one of the seven MLST loci are grouped together, and the ST with the largest number strains in the group is determined as the founding ST using the principle of parsimony (99). This principle based on the simplest explanation, which is initial diversification of a strain from the founder would differ at only one of the seven alleles (99). For example, CC8 harbors genetically similar STs including ST8 and ST247, whereby ST247 most likely originated from ST8. Conversely, ST93 is a singleton and does not cluster with other STs.
SCCmec typing
The SCCmec element, which harbors the mec gene facilitating methicillin resistance, can also be used to describe MRSA epidemiology. SCCmec is heterogeneous and ranges in size from 21 to 72.5 kb (100, 101). SCCmec is comprised of three basic elements: the mec gene complex, the ccr gene complex, and the joining region (J region) (63). The ccr gene complex of SCCmec contains different kinds of recombinases (ccrA, ccrB, or ccrC) that are responsible for the excision and integration of the SCCmec element into the chromosome at a specific site (attSCC) at the 3′ end of the rRNA methyltransferase gene orfX/rlmH (63, 65, 102–104). SCCmec also harbors open reading frames in addition to mec and ccr gene complexes, including additional antibiotic and heavy metal resistance genes (65, 102, 105). It is the combination of these elements (mec, ccr complex, and additional genes) that allow for the classification of MRSA into various SCCmec types and subtypes (34, 106). To date, there are 15 SCCmec types (I to XV) that have been discovered, with subtypes (e.g., IVa and IVb) categorized by variations in the linking region between the mec and ccr elements (34, 100, 107, 108) (Fig. 1C). Although multiplex PCR is a commonly used method for SCCmec typing (109, 110), web-based tools such as SCCmecFinder have been develop to determine SCCmec types using whole-genome sequencing data (111). MLST is often used in combination with SCCmec typing to define a lineage. For instance, PFGE type (pulsotype) USA300 belongs to ST8 and harbors a SCCmec type IVa element and can be described as ST8-IVa.
Whole-genome sequencing
The application of WGS to strain typing has risen dramatically due to the advances in sequencing technology coupled with significant reductions in cost (112). At the time of writing, 31,252 whole genomes of S. aureus had been deposited at the National Center for Biotechnology Information, of which 1,279 were classed as “complete.” Based on these data, the S. aureus genome is on average 2.84 Mbp in length, has a guanine-cytosine (GC) content of 32.8%, and has 2,728 predicted coding sequences (CDS). Phylogenomics of S. aureus is based on divergence of SNPs between genomes (113). Difference between genomes within the same CC is up to 3,000 SNPs (113). In contrast, a difference of more than 15,000 SNPs can be found when comparing genomes from different CCs (113).
Recent studies have demonstrated the promise of routine WGS of S. aureus isolates for epidemiological surveillance and identification of high-risk clones based on clonal relatedness, abundance, virulence, and antimicrobial resistance properties inferred from WGS data (114). WGS together with phylogenomic analyses have also been utilized in the clinical setting to define outbreaks, characterize transmission, and exclude unrelated cases (115–118). WGS enables the expansion of traditional MLST (seven genes) to core genome MLST (cgMLST) that includes 1,861 gene loci (119, 120). Leopoid et al. show that cgMLST identified MRSA transmission events that were unsuspected during epidemiological investigation using spa typing, showing the precision and discriminatory power of cgMLST (119). An example is the delineation of clonal relationships between previously indistinguishable MRSA ST398 isolates (n = 66) using SeqSphere+ software to process WGS for cgMLST, which differentiated the isolates by between 3 and 78 alleles (121). Whole-genome MLST (wgMLST) is an extension of cgMLST and uses both the core and accessory genomes for analysis, potentially providing higher resolution than cgMLST (120). A consistent naming system for clones or lineages defined using cgMLST and wgMLST will be required for sharing data and reproducibility in the future (120).
The identification of genome-wide SNPs across MRSA isolates is a common method to define the genetic relatedness. Unlike cgMLST and wgMLST that require a reference genome, SNP calling can be performed with or without a reference genome (120). Using high-resolution SNPs analysis, chains of transmission that were originally unsuspected were uncovered during MRSA outbreaks in neonatal intensive care units (115, 122). Defining genetic relatedness using a consensus SNP threshold is crucial for the interpretation of outbreak and infection control management. Recently, Coll et al. have suggested guidelines for determining MRSA transmission based on genetic differences between strains measured as SNPs (123). If the differences are greater than 25 whole-genome SNPs or 15 core-genome SNPs, it suggests that MRSA transmission within the past 6 months is unlikely (123).
The wealth of information garnered from WGS facilitates the in silico prediction of antibiotic resistance profiles (114). One approach involves establishing databases of antibiotic resistance determinants based on existing literature (114, 124). This is followed by cross-referencing the genome of an inquiry sequence against these databases to identify the presence of genes or mutations associated with antibiotic resistance (114, 124). Another method employs genome-wide association studies to pinpoint specific genetic variations in antibiotic-resistant strains in comparison to susceptible strains (125). These identified genetic variations can then be utilized to predict antibiotic resistance in genomes of unknown strains (126). However, the potential limitation lies in our incomplete understanding of the genetic basis of antibiotic resistance when using WGS-based prediction method (126). Therefore, it is advisable to incorporate traditional culture-based antimicrobial testing as a quality control measure to validate phenotypic resistance predictions obtained through these approaches (126).
WGS is poised to replace other typing methods and become the new gold standard for epidemiological surveillance (114, 118). It assists us in precisely defining lineages and, through comparative genomics, identifying genes that may be crucial for patho-adaptation (Fig. 1E). Importantly, the current key challenge lies in the need for universal bioinformatic tools that can seamlessly integrate biological and clinical data with WGS data in a timely manner. The development of WGS bioinformatic pipelines, such as EpiSeq and BacPipe, will help overcome the hurdle of data analysis and promote the routine use of WGS in monitoring infection transmissions in hospitals and public health (127, 128).
FACTORS CONTRIBUTING TO MRSA CLONAL EXPANSION
Virulence factors
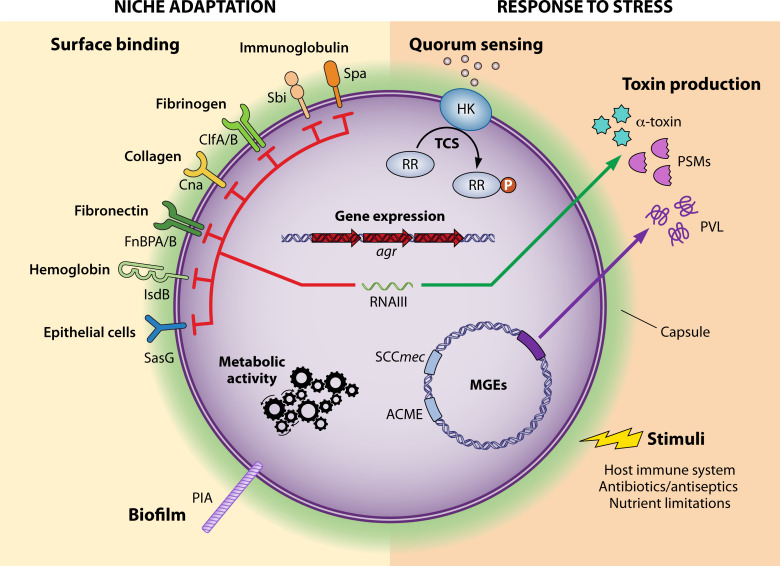
S. aureus is a versatile pathogen that, when interacting with a host, can be either colonizing, persistent, or disease causing (129). Here, we will define virulence as “the relative capacity of a microbe to cause damage in a host” (130), and we will thus define any microbial component that contributes to virulence, by facilitating colonization, persistence, immune evasion, or damage to the host, as a virulence factor (Fig. 2). The success of a lineage can thus be determined by the acquisition of a virulence factor on a mobile genetic element (e.g., the sasX gene facilitating colonization) or the specific regulation of an intrinsic factor (for example, the accessory gene regulator, Agr). When possible, we will highlight instances where the contribution of a virulence factor to pathogenesis was confirmed using the molecular postulates, including gene deletion, complementation, and overexpression studies.
Fig 2.
Virulence factors in Staphylococcus aureus. The factors are encoded in the genome or acquired via horizontal gene transfer and can be grouped into two categories: establishing niches and responding to stress. Control of these factors is intertwined via two-component regulatory systems (TCS) and the accessory genome regulator (agr). ACME, arginine catabolic mobile element; ClfA/B, clumping factor A/B; Cna, collagen adhesin; FnBPA/B, fibronectin-binding protein A/B; HK, histidine kinase; IsdB, iron-regulated surface determinant B; MGEs, mobile genetic elements; PIA, polysaccharide intercellular adhesin; PSMs, phenol soluble modulins; PVL, Panton-Valentine leucocidin; RR, response regulator; SasG, S. aureus surface protein G; SCCmec, staphylococcal cassette chromosome mec; Sbi, immunoglobulin-binding protein; Spa, staphylococcal protein A.
Cell envelope
The S. aureus cell envelope is at the forefront of the dynamic interaction between host and pathogen during colonization. The cell envelope is mainly composed of capsular polysaccharides, peptidoglycans, wall teichoic acids (WTAs), lipoteichoic acids (LTAs), and surface proteins. The role of cell envelope components in S. aureus pathogenesis is multifaceted. For example, polysaccharide encapsulation of S. aureus promotes colonization on mucosal surfaces and interferes with opsonophagocytosis to facilitate bacterial persistence in human blood [for detailed review, see reference (131)]. Thus, the serotype and extent of capsular polysaccharide produced by a lineage are likely to contribute to its success.
Surface-associated proteins are important for adherence to host tissue, a critical factor in colonization and the initiation of MRSA infection. These proteins facilitate binding to host extracellular matrices (ECM), including collagen, fibrinogen, fibronectin, elastin, and bone sialoprotein (132–136). Based on structural and functional analyses, surface proteins are categorized into five distinct groups: microbial surface component recognizing adhesive matrix molecules (MSCRAMMs), near iron transporter motif family, three-helical bundle, G5-E repeat family, and structurally uncharacterized proteins (35). MSCRAMMs are the largest class of surface proteins and have important roles in adhesion to ECM and immune evasion (136). MSCRAMMs include clumping factor A (ClfA) and B (ClfB); collagen adhesin (Cna); fibronectin-binding proteins A (FnBPA) and B (FnBPB); and serine-aspartate repeat proteins C (SdrC), D (SdrD), and E (SdrE) (35). The full repertoire of surface proteins varies among strains, and many surface proteins have multiple roles in virulence, with functional redundancy between these surface proteins (35, 137).
Once infection is established, MRSA surface proteins play an integral role in disturbing the host immune system. Protein A (SpA) is a three-helical bundle surface protein that binds human immunoglobulin Fc fragment to inhibit opsonophagocytosis and B-cell receptors to induce B-cell apoptosis (138–142). Inflammatory responses of epithelial cells can also be manipulated by SpA via activating tumor-necrosis factor receptor 1 (TNFR1) and type I interferon (IFN) signaling to promote the pathogenesis of staphylococcal pneumonia (143–145). Sbi is an additional immunoglobulin-binding protein that is released and is capable of inactivating the host complement pathway via interacting with complement component C3 (146, 147).
In addition to facilitating colonization and immune evasion, cell envelope components, peptidoglycan, WTA, and LTA, are major bacterial factors to induce cytokine release and systemic inflammation in the host, which can lead to sepsis, septic shock, and multiple-organ failure if infection is not rapidly controlled (148–151). In MRSA, the alteration of peptidoglycan linkage caused by PBP2a, the product of mecA, results in the release of peptidoglycan and the induction of exacerbated inflammation in the host (152). Recent findings also show that specific MRSA strains with the capacity to cause more severe skin abscess in a mouse infection model are closely correlated with higher content of WTA (153). Taken together, coordination of the cell envelope components is critical for MRSA to establish a niche, to evade the host immune system, and to cause disease. The intricacies of a given lineage’s outer surface is likely to play a role in defining its clinical impact.
Pore-forming toxins
MRSA expresses multiple extracellular toxins and enzymes that facilitate tissue dissemination and host cell lysis during the course of infection (154). One major group is the β-barrel pore-forming toxins, including α-toxin (Hla) and the bicomponent leukocidins (Luk) (155, 156). Hla inserts into the plasma membrane of host cells to form a monomeric heptamer complex, leading to the uncontrolled flux of ions and water followed by membrane damage and cell lysis (157–159). The binding affinity of Hla to cells contributes to the sensitivity of different cell types or host species to Hla (160). Hla reduces dendritic cell accumulation in skin during infection, leading to the suppression of antigen-specific T cell responses (161). A disintegrin and metalloprotease 10 (ADAM10) has been identified as a high-affinity binding receptor for Hla on cells to cleave the adherence junction protein E-cadherin, contributing to lethal infection and skin infection in mice (162–164). The important role of Hla in virulence has been confirmed in mouse lung and skin infection models (165–169). Deletion of hla resulted in smaller skin lesion in the model while complementation of hla in the mutant restored the abscess size at the same level as wild type (165). Vaccination with a non-cytolytic mutant form of Hla (HlaH35L) or targeting of ADAM10 was validated as an effective strategy to reduce the severity of S. aureus infections (165–169). Interestingly, a recent study indicates that Hla induces specialized pro-resolving mediators in human M2-like macrophages to resolve infectious inflammation (170), highlighting the multifaceted functions of Hla during host-pathogen interactions.
The bicomponent leukocidins are comprised of the paired fast-eluting F-subunit and the slow-eluting S-subunit, which are so-called based on the rate of migration in liquid chromatography (171, 172). So far, five leukocidins related to human infections have been characterized: LukSF-PV [originally known as Panton-Valentine leukocidin (PVL)], LukED, γ-hemolysins AB and CB (HlgAB and HlgCB), and LukAB (also known as LukGH) (44, 155, 156). The S-subunit of the leukocidins recognizes and binds with high affinity to a target protein receptor on the host cell membrane, which causes the recruitment of the F-subunit to the cell surface. Dimerization of F-S subunits leads to oligomer formation and the assembly of an octameric β-barrel pore that spans the host cell membrane bilayer, resulting in cell lysis (44, 155, 156). Each of these leukocidins target different cell receptors and have host species specificity for toxin binding. Further details of these bicomponent leukocidins are provided in other reviews (44, 155, 156, 173).
Phenol-soluble modulins
Phenol-soluble modulins (PSMs) are a family of secreted short peptides with α-helical and amphipathic physicochemical properties (42, 174). PSMs are grouped into the smaller α-type (PSMα1 to PSMα4, δ-toxin, and PSM-mec) and the larger β-type (PSMβ1 and PSMβ2) (174, 175). The psmα and psmβ operons encode four PSMα peptides (PSMα1 to PSMα4) and two PSMβ peptides (PSMβ1 and PSMβ2), respectively (42, 174). The δ-toxin is encoded by hld within the agr effector, RNAIII, while PSM-mec is found in SCCmec types II, III, and VIII (174, 176, 177). All S. aureus lineages contain a single highly conserved allele of the psmα operon, while psmβ2 is absent in some lineages (178).
PSMs are able to attract, activate, and lyse human neutrophils and have demonstrated significant contribution to the virulence of CA-MRSA strains in murine bacteremia and skin infection models using PSMα deletion strains (42). Complementation of PSMα in the deletion strain completely restored lytic activities against human neutrophils (42). PSMα3 causes the most profound effects on human neutrophils and inflammatory response compared with other PSMs (42). The cytotoxicity of PSMα3 to human cells is highly related to self-associating amyloid-like PSMα3 fibrils, which are formed by stacking amphipathic α helices perpendicular to the fibril axis (179). The δ-toxin is a strong inducer of mast cell degranulation to release immunoglobulin-E, which mediates allergic skin diseases such as atopic dermatitis (AD) (180). Replication within the host cell cytoplasm as an intracellular pathogen is another strategy for MRSA to evade the host immune system, and PSMα is important for the escape of MRSA from the phagosome to grow intracellularly (181). All PSMs mediate structural biofilm formation and detachment processes, which was demonstrated by the dissemination of S. aureus cells in a murine catheter infection model (182). PSMs are critical to form fibrils that have amyloid-like properties to stabilize S. aureus biofilms, and PSMα1 and PSMα4 assemble these fibrils through joining steric zipper interfaces of β-sheets (183, 184).
Superantigens
S. aureus superantigens are secreted virulence factors that disrupt the host adaptive immunity by stimulating T cell hyper-activation and, therefore, contribute to toxic shock syndrome, pneumonia, and sepsis (38, 40, 185, 186). The family of S. aureus superantigens contain toxic shock syndrome toxin-1 (TSST-1) and staphylococcal enterotoxins (SEs) and enterotoxin-like (SEls) proteins (186). SEs have demonstrated emetic activity in monkeys; however, TSST-1 and SEls have no proven emetic activity in the non-human primate model despite these toxins being structurally similar to SEs (187). Genes encoding enterotoxins are often found as a cluster of genes in an operon present on a variety of different mobile genetic elements (188). For example, the enterotoxin gene clusters (egc) containing seg, sei, sem, sen, and seo are found in a genomic island vSAβ (189). Enterotoxin genes sea, sep, sek, and seq form an immune evasion gene cluster (IEC), which is present in a prophage φSa3int (190, 191). Staphylococcal enterotoxins B (SEB) and C (SEC) promote pathogenic production of interferon γ (IFN-γ) to facilitate bacterial colonization in the liver, contributing to MRSA bloodstream infections in mice expressing human MHC class II (192). The findings of SEB and SEC in liver colonization were validated by successfully complementing the genes in trans to restore their functions (192). This excessive IFN-γ from CD4+ T cells induced by SEB and SEC allows MRSA to replicate efficiently within macrophages (192). In addition to superantigen activity, recent data indicate that SEC has antiangiogenic effects to inhibit branching microvessel formation and the expression of angiogenesis mediator, contributing to MRSA endocarditis (193). The critical roles of SEC and staphylococcal enterotoxin-like X (SElX) for MRSA are shown in rabbit infection models and were associated with infective endocarditis, sepsis, acute kidney injury, and necrotizing pneumonia (194, 195). Loss of SEC or SElX in MRSA led to attenuated virulence, while complementation or mutation repair restored disease production (194, 195).
Other secreted enzymes
In addition to exotoxins, MRSA produces other excreted enzymes including lipases, phospholipases, proteases, esterases, and hyaluronidases that contribute to host tissue invasion, immune evasion, and pathogenesis (196, 197). Secreted factors including formyl peptide receptor-like 1 inhibitor, chemotaxis inhibitory protein of S. aureus (CHIPS), and staphylococcal complement inhibitor (SCIN) contribute to immune evasion by inhibiting neutrophil chemotaxis and complement activation (154, 198–200). CHIPS is encoded by the gene chp, and CHIPS specifically binds to human neutrophils to inhibit calcium mobilization induced by formylated peptides and complement activation C5a (199). A secreted lipase of MRSA, glycerol ester hydrolase (Geh), hydrolyzes host-derived lipoprotein particles to utilize liberated free fatty acids for the bacterial membrane phospholipid biosynthesis as an adaptive strategy (201). Geh is also capable of inhibiting activation of the innate immune system via ester hydrolysis of MRSA lipoproteins, which is a major pathogen-associated molecular pattern recognized by Toll-like receptor 2 of host immune cells (197). Phosphatidylinositol (PI)-specific phospholipase C releases glycosyl-PI-linked proteins from the host cell membrane and contributes to the survival of MRSA in human blood and neutrophils (202). IEC also contains genes responsible for evasion of the human immune response, including the staphylokinase sak, chp, and scn encoding SCIN (190, 191).
Shaping virulence in MRSA
Regulation
The response to stress both inside and outside of a mammalian host has a fundamental role in the success of MRSA (203). These responses are coordinated by a complex and finely tuned regulatory network controlling virulence and metabolism to adapt to different environments. A bioinformatic analysis predicted MRSA to code for 135 transcription factors; a large number of which are yet to be experimentally characterized (204). Three well-studied systems pertaining to virulence are the two-component regulatory systems (TCRSs), the staphylococcal accessory regulator (Sar) nucleic acid-binding protein family, and alternative sigma factors (205–207). Of these, the most well-characterized and central virulence regulator is Agr, which will be a key focus of this discussion. The functioning of these regulatory systems is further influenced by central metabolism and is dependent on numerous environmental factors.
Two-component regulatory systems
TCRSs are critical mediators of signal transduction in prokaryotes. In their simplest form, they consist of a sensor histidine kinase that responds to an environmental signal by autophosphorylating a response regulator (208, 209). The activated response regulator then binds specifically to target DNA sequences resulting in a transcriptional response (208). Most strains of S. aureus have 16 TCRSs, while MRSA strains harbor an additional TCRS within SCCmec that mediates methicillin resistance (210, 211). AgrCA is one of the best characterized staphylococcal TCRSs. The agr locus encodes two transcripts, RNAII and RNAIII, which are controlled by two divergent promoters, P2 and P3, respectively (205). RNAII contains agrC and agrA, which encode the histidine kinase and response regulator, respectively, as well as agrB and agrD (205, 212, 213). AgrD is the precursor of the agr autoinducing peptide (AIP), and AgrB catalyzes the formation of an AIP biosynthesis intermediate, the AgrD (1–32) thiolactone (214). Maturation of AIP is mediated by the protease regulator MroQ in agr specificity groups I and II (215, 216). AgrC binds AIP resulting in phosphorylation of AgrA, which drives the transcription of RNAIII (217). In addition to coding for the δ-toxin, RNAIII is the effector of the agr system, regulating the expression of numerous extracellular toxins and enzymes including α-toxin, PSMs, PVL, enterotoxins, TSST-1, exfoliative toxin and serine proteases, as well as surface-associated virulence factors including SpA, FnBPA, FnBPB, and the capsule (42, 217–221). In addition to AgrCA, numerous TCRSs are confirmed regulators of virulence as determined by gene deletion followed by complementation in trans, including SaeRS, LytRS, GraRS, VraRS, SsrAB, ArlRS, and WalKR (222–228).
The association between dysfunctional agr and reduced vancomycin susceptibility in MRSA is of interest and has been reported in several studies (229–231). However, other researchers found no significant difference of agr dysfunction between high- (≥2 µg/mL) and low-vancomycin minimum inhibitory concentration (MIC) (≤1.0 µg/mL) groups (232). Furthermore, Butterfield et al. showed that there was no association between agr dysfunction and vancomycin-intermediate S. aureus (MIC ≥4 µg/mL) (233). In line with this discrepancy, agr dysfunction did not correlate with vancomycin treatment failure of MRSA in a rabbit infective endocarditis (IE) model (234). The relationship between agr and reduced vancomycin susceptibility is further complicated by mutations in walKR during vancomycin exposure (235, 236). Rao et al. recently found that the WalK(S221P) mutation that is responsible for a vancomycin-intermediate phenotype failed to activate WalR to bind the promoter of the agr system, leading to silenced Agr gene expression and attenuated virulence (235). This study showcases how gene expression is moderated by this type of regulatory crosstalk and can influence colonization, persistence, and disease-causing behaviors (235).
Sar transcriptional regulators
Sar transcriptional regulators are classed based on their homology to the SarA prototype. The sar locus consists of three overlapping transcripts driven from three promoters (P1, P2, and P3), each of which contain the sarA gene (237). SarA binds to promoter regions termed Sar boxes (238), which directly enhances the expression of hemolysins and surface proteins, including SpA, FnBPA, FnBPB, and Cna (239–242). In addition, SarA binds to both P2 and P3 of the agr locus, regulating the production of RNAII and RNAIII (243). As such, SarA regulates virulence factor expression in both agr-dependent and agr-independent manners (240, 243), further highlighting the complex and intertwined nature of S. aureus virulence regulation. The role of SarA-like homologs and repressor of toxins (Rot) in the regulation of virulence factor expression has also been described (244–250).
Alternative sigma factors
Sigma factors bind to RNA polymerase providing gene target specificity during the process of transcription initiation. The primary sigma factor, σ70, is responsible for the expression of housekeeping genes typical of exponential growth, while alternative sigma factors take over in response to adverse conditions. Sigma factor B (σB) influences the expression of 200–250 genes with various functions, many of which relate to virulence (251, 252). SigB activity has been shown to influence virulence gene expression independently and in concert with other regulatory systems including SarA and Agr (253). The S. aureus genome codes for an additional alternative sigma factor, σH; however, its contribution to pathogenesis is currently unclear (254).
Influence of metabolism
S. aureus virulence is influenced by nutrient composition in the environment, as well as the activity of seemingly unrelated metabolic pathways (255). The contribution of carbohydrate and amino acid metabolism has been appreciated for many years, whereas the role of nucleotide metabolism and lipid biosynthesis is only beginning to be delineated.
S. aureus responds to changes in carbohydrate (glucose, fructose, and glycerol) accessibility via carbon catabolite repression regulators, CcpA and CcpE, each of which have been shown to influence virulence gene expression (256, 257). In modeled diabetic infections, MRSA acquires excess glucose via two glucose transporters to significantly enhance the production of Hla, leading to worse skin infection outcomes (258). Deletion of CcpA results in reduced Hla production, attenuated virulence in the murine diabetic model, and decreased level of bacterial cellular ATP (259). Additionally, pH shifts resulting from glucose catabolism have been shown to inhibit Agr function and downstream virulence gene transcription (260). More recently, the central metabolite, pyruvate, which is a key nutrient in the human host, has been shown to induce the expression of leukocidins and increase the virulence of CA-MRSA by inactivating key TCRSs (AgrCA, SaeRS, and ArlRS) (261). Fatty acid metabolism regulated by NADH-dependent respiration is sensed by SaeRS in S. aureus (262). Deficiency of NADH dehydrogenase NdhC impairs S. aureus biofilm formation, Hla production, and bacterial colonization in a murine model of systemic infection (262). Complementation of ndhC in the ndhC mutant restored biofilm formation and Hla production (262).
The relationship between metabolism and virulence is finely tuned by moonlighting or multitasking regulators. Branched-chain amino acids (BCAAs; isoleucine, leucine and valine) are vital nutrients for the growth of S. aureus, as they are essential to the biosynthesis of proteins and membrane branched-chain fatty acids (263–265). Depletion of BCAAs is sensed by the transcriptional regulator CodY to lift its repression of the operon for BCAA biosynthesis (266, 267). In addition to amino acid metabolism, CodY is also a global repressor of virulence and mediates the expression of hla, the agr system, and saeRS (266, 268–270). Deletion of codY in MRSA strains was shown to impact virulence in mouse models of necrotizing pneumonia, skin infection, and bacteremia (270, 271). However, deletion of codY in a MSSA strain had no effect on host survival and bacterial burden in the same murine bacteremia model (272), suggesting that more studies are required to investigate the role of codY in virulence.
The intricate relationship between metabolism and virulence is further illustrated by the link between the nucleic acid biosynthesis pathway and virulence control in MRSA (273, 274). PurR is the master negative regulator of de novo purine biosynthesis in bacteria (273–276). Inactivation of PurR in murine infection models leads to a greater amount of secreted leukocidins and Hla and up-regulation of FnBPs, resulting in hypervirulence independent of enhanced purine production (273, 274). The role of PurR in virulence is further confirmed by complementation of PurR, which reverses toxin secretion and hypervirulence phenotypes (273). Interestingly, in a comparative study using clinical MRSA bacteremia isolates from the same clonal complexes, the expression of purine synthesis genes is higher in isolates that were persistent in patients longer than 6 days, compared to the isolates that were resolved within 4 days after therapy (277). These studies suggest that purine biosynthesis may have an important role in MRSA persistence in vivo.
Lipid metabolism is important as S. aureus cell membranes are involved in crucial cellular processes including stress response, antimicrobial resistance, and virulence (278). Host antimicrobial peptides target S. aureus cell membranes composed by phospholipids, mainly anionic phosphatidylglycerol (PG) (278). Aminoacylation of PG with an L-lysine group to form lysyl-phosphatidylglycerol (L-PG) is a defense strategy mediated by multiple peptide resistance factor (MprF) in S. aureus (279). Depletion of MprF leads to hyper susceptibility to neutrophil killing and attenuated virulence in animal models (279, 280). Zheng et al. recently showed that secretion of virulence factors LukAB on cell surface and to extracellular milieu depends on L-PG and LTA biosynthesis, which are controlled by MprF and YpfP, respectively (281). Stimulation of host fatty acids induces the expression of the S. aureus type VII secretion system genes to export virulence factors during infection (282). This process is mediated by S. aureus fatty acid kinase (Fak) pathway to incorporate extracellular fatty acids into bacterial membranes (282, 283). Depletion of the kinase FakA compromises α-hemolysin production, enhances proteases SspAB and aureolysin secretions, and increases resistance to host antimicrobial peptides (284–286). The role of FakA in virulence appears to be tissue specific, as deletion of fakA enhances S. aureus pathogenesis in a murine skin infection model but reduces the virulence in a mouse model of S. aureus bacteremia (282, 286).
Core genome diversity and mutations
Genes that are present in all MRSA strains not only constitute a core component of approximately 75% of the genome and are principally responsible for essential cellular functions but also contribute to virulence (287). Natural variations within the core genome can have remarkable effects upon gene expression and protein function and contribute to the evolution of successful clones under various selection pressures (195, 288). These variations include SNPs, InDels, repeat variations, and operon arrangements (287).
SNPs can result in nonsense mutations that introduce premature stop codons, producing pseudogenes without function. A classic example is the hla pseudogene of CC30, which eliminates α-toxin production for this lineage (48). SNPs can result in non-synonymous amino acid substitutions that alter protein function (289). An SNP causing a non-synonymous amino acid substitution within the regulator of the pyrimidine biosynthetic operon (PyrR K126I) resulted in upregulation of the operon and promoted colonization and transmission for a dominant subclone of USA300 (47). SNPs outside of coding sequences can also influence virulence gene expression. Polymorphism in the promoter region upstream of hla is a genetic marker for hyper α-toxin production for strains of S. aureus isolated from bovine mastitis (289, 290).
Recent evidence supports adaptive shaping of MRSA pathogenicity via phase variation. The function of agr can be influenced by a multitude of mechanisms including nonsense mutations, non-synonymous mutations, frameshift mutations, poly(A) tract alterations, and inversion duplication mutations (291–296). Reactivation of agr function can be triggered by host-mediated stress such as phagocytosis, resulting in functional reversion of the shutdown mutations (291). Ramond et al. recently showed that loss of agr function is associated with a proinflammatory response in the lung, contributing to the long-term colonization of S. aureus in young cystic fibrosis patients (297). In a Japanese study, S. aureus isolates from infants who did not develop AD had increased frequency of agr mutations, compared with the isolates from infants who later developed AD (298). More studies are needed to elucidate the contribution of agr in the evolution of S. aureus during colonization in different host tissues.
Diversity in operon arrangements can extend variation within core genomes and impact upon bacterial competition and host immune responses (299–301). Again, using agr as an example, the operon contains an internal variable region ranging from the C terminus of AgrB to the N terminus of AgrC and spanning AgrD (302, 303). This variation results in four distinct agr groups with divergent capacity of AIP to activate or inhibit quorum sensing between strains carrying a different agr system (300, 302, 304).
A second example of operon arrangement/composition influencing pathogenicity is the cap operon, coding for capsular polysaccharide. Most clinical MRSA isolates express capsular polysaccharides serotype 5 (CP5) or 8 (CP8), which consist of the same repeating element of trisaccharide with a difference only in the linkages between the sugars and O acetylation positions (131, 305–307). The gene clusters responsible for CP5 and CP8 biosynthesis contain 12 essentially identical genes and four type-specific genes (cap5HIJK and cap8HIJK), which display low sequence similarity (308). The capsular serotype is highly associated with strain lineage as most of CC5 and CC8 strains are CP5 while CC30 strains are CP8 (306, 309, 310). In a murine infection model, CP5 was associated with better bacterial survival compared with CP8 in a bacteremia model, indicating that the difference between CP5 and CP8 likely contributes to the relative virulence of serotype 5 and 8 MRSA in vivo (311). CP5-specific monoclonal antibodies were shown to protect mice from bacteremia caused by serotype 5 strains (312–314); however, CP8-specific monoclonal antibodies failed to protect against serotype 8 staphylococcal infections in mice and was associated with a high amount of CP8 release from serotype 8 strains, which hindered the development of CP8 vaccines or antibodies for passive immunotherapy (314).
The accessory genome
Much of the genetic material that exists outside of the core genome is present on various distinct elements that are either mobile [termed mobile genetic elements (MGE)] or were once likely mobile but have since become fixed within the genome [termed genomic islands (GI)]. The presence and arrangement of these elements play a crucial role in shaping S. aureus lineages.
Genomic islands
Staphylococcal GIs are stably maintained within the chromosome; however, they present evidence of historic mobility including incomplete integration machinery (315). S. aureus GIs do not contain core/essential genes, but they typically harbor genes that contribute to virulence and/or niche adaptation. Often, multiple virulence genes with highly similar sequences appear in series of variable length and composition (316–318). The complement of these genes differs between lineages but is highly conserved within them (316, 318), suggesting a key role for GIs in lineage-specific successes.
S. aureus genomes typically contain two major GIs: vSAα and vSAβ (315, 319). Multiple staphylococcal superantigen-like genes (ssl, also referred to as staphylococcal enterotoxin-like, set) and lipoprotein genes (lpl) are located on vSAα, each of which appear in extended series (320). vSAβ typically carries a serine protease-like (spl) gene cluster and an egc and often harbors a lantibiotic/bacteriocin biosynthesis operon (bsa), a hyaluronate lyase precursor gene (hysA), and genes coding for a bicomponent leucocidin (lukED) (316, 320).
Mobile genetic elements
S. aureus contains many MGEs that can move between and across species, including bacteriophages, pathogenicity islands, staphylococcal cassette chromosomes (SCC), insertion sequences, transposons, and plasmids (321). MGEs can provide genes that contribute to both virulence and antibiotic resistance. We will focus our discussion on the specific contribution of MGEs to virulence. MGEs related to antibiotic resistance are discussed in another review (322).
Bacteriophages
Temperate bacteriophages of the Siphoviridae family are frequently integrated in S. aureus genomes. Siphoviridae have highly organized genomes that are approximately 40 kb and arranged in functional modules that facilitate lysogeny/integration, DNA replication, transcriptional regulation, packaging, head proteins, tail proteins, and lysis (323). A useful classification scheme is centered upon the sequence of the integrase (int). Here, the majority of prophages cluster within seven major groups (φSa1int–φSa7int) (191).
Many important virulence genes are carried by temperate phages. The most common prophage is φSa3int, which is present in approximately 75% of S. aureus genomes (191). φSa3int harbors the IEC that contains various combinations of sek, seq, sea, sak, scn, and chp (190). Interestingly, φSa3int further modulates S. aureus virulence as its typical site of insertion results in inactivation of hlb (324). φSa3int integration is strongly associated with human nasal colonization isolates and is less frequent in isolates from acute infection, suggesting that it is an important mediator of the switch from commensal to pathogen (191). Other factors that can affect nasal colonization include nasal microbiome, the specific composition of which can either promote or inhibit S. aureus persistence (325). φSa2int is the second most common prophage and is the major carrier of lukFS-PV, which codes for Panton-Valentine leucocidin (191, 326). Thus, like PVL, the presence of φSa2int is strongly associated with necrotizing pneumonia and skin and soft tissue infections (SSTIs) in humans (327, 328). Less frequently detected phage groups φSa7int and φSa1int have been shown to harbor the virulence genes coding for staphylokinase (sak) and exfoliative toxin A (eta), respectively (191). The recently identified virulence gene sasX, which codes for the cell wall-anchored virulence determinant SasX, is present on an atypically large (127 kb) staphylococcal φSPβ-like prophage (329). The prophage was a marker for an epidemic lineage of MRSA (ST239) that spreads through Chinese hospitals in the 2000s (329), reaffirming the important contribution of MGEs to S. aureus clonal expansion (330).
S. aureus pathogenicity islands (SaPIs)
SaPIs share similarities with bacteriophages, including a modular genetic architecture with conserved regions for integration, regulation, and replication (331). However, they do not encode the machinery that facilitates HGT but instead can be mobilized by hijacking the capsid of so-called helper phages (332). Toxin genes are commonly present as accessory genes in SaPIs, including TSST and a host of superantigens (i.e., seb, sec, sek, sel, sep, and seq) (332, 333).
Additional elements
Plasmids are more commonly involved in the horizontal transfer of antibiotic resistance genes; however, some code for virulence factors (334). For example, plasmids can contain various combinations of enterotoxin genes (including sea, seb, sed, seg, sej, sep, ser, ses, and set) (334–338), exfoliative toxin B (339, 340), and an ica-like locus that may contribute to biofilm formation (341). As is the case for plasmids, the SCCmec element is more commonly associated with resistance to antibiotics and heavy metals; however, certain SCCmec variants contain a PSM (termed PSMmec) (342), and non-mec SCC elements can code for capsule genes (SCCcap) (343).
Animal host adaptation
Comparative analyses of S. aureus isolates from various sources, including humans and animals, have revealed an additional role for MGEs in host-specific adaptation (344). As mentioned above, the hlb converting phage φSa3int carries a set of human innate immunomodulatory genes (the IEC), and this element is infrequently related to animal-associated lineages (345, 346). For some avian-adapted MRSA, φSa3int is replaced by an alternative hlb converting phage, φAvβ, which is not associated with human isolates and harbors genes predicted to be involved in avian niche-specific adaptation including an ornithine cyclodeaminase and a putative protease (80, 345). SaPIs have also been shown to harbor host-specific adaptive genes including SaPIbov2, which codes for the Bap adhesion protein that was shown to contribute to persistence in a bovine intramammary gland infection model (347). Additionally, differential coagulation capacities of ruminant associated S. aureus (348) have been attributed to the SaPI encoded von Willebrand factor binding homologs that have livestock blood clotting specificities (349, 350).
Gene reservoirs
S. aureus is a frequent colonizer of the human skin and shares this niche with a multitude of commensals, including the clinically important coagulase-negative staphylococci (CoNS). Due to their close physical proximity and genetic relatedness, genetic material can be exchanged within and between staphylococci via HGT. Multiple MGEs that have contributed to the success of specific S. aureus lineages appear to have originated in the CoNS, suggesting that these species may act as a reservoir for pathoadaptive genes (45).
The archetypal S. aureus MGE, SCCmec, appears to have originated in the commensal species Staphylococcus sciuri. Some taxonomists have suggested the reclassification of S. sciuri into a novel genus known as Mammaliicoccus (351). The mecA gene homolog of M. sciuri shares high sequence identity (80%–99%) with mecA of contemporary MRSA (352, 353). Importantly, mecA from M. sciuri confers resistance to β-lactams upon introduction into S. aureus (354, 355). Additional genes present on prototypical MRSA SCCmec elements have been identified in Staphylococcus vitulinus, Staphylococcus fleurettii, and M. sciuri, suggesting that these three early branching species each contributed to the modular assembly of SCCmec (356). Homologs of psm-mec, which codes for a toxin that contributes to sepsis for Staphylococcus epidermidis and acts at the interface between virulence and antibiotic resistance in MRSA (342, 357), also appear to have its origins in the M. sciuri group (356).
Multiple lines of evidence support that S. epidermidis is a source for the introduction of SCCmec into S. aureus (45). SCCmec elements are widespread in S. epidermidis, and they share high sequence identity with those found in MRSA lineages. Of these, SCCmec type IV particularly seems to have appeared in S. epidermidis earlier than in S. aureus (45, 358). In vivo, the conversion of an MSSA to MRSA in a patient undergoing antibiotic therapy was attributed to the horizontal acquisition of SCCmec from a co-colonizing S. epidermidis (359, 360). However, this particular genetic exchange could not be recapitulated in the laboratory (360).
In addition to the acquisition of SCCmec from CoNS, select lineages have benefited from additional elements that have provided an adaptive advantage. These include the arginine catabolic mobile element (ACME), which was assembled in S. epidermidis and transferred to MRSA USA300 (361), and the φSPβ-like prophage that harbors the sesI homolog sasX, which contributed to the spread of ST239 in Asia (362). Each of these elements will be discussed in more detail in the section describing USA300.
EVOLUTION OF SUCCESSFUL MRSA LINEAGES IN THE CONTEXT OF VIRULENCE
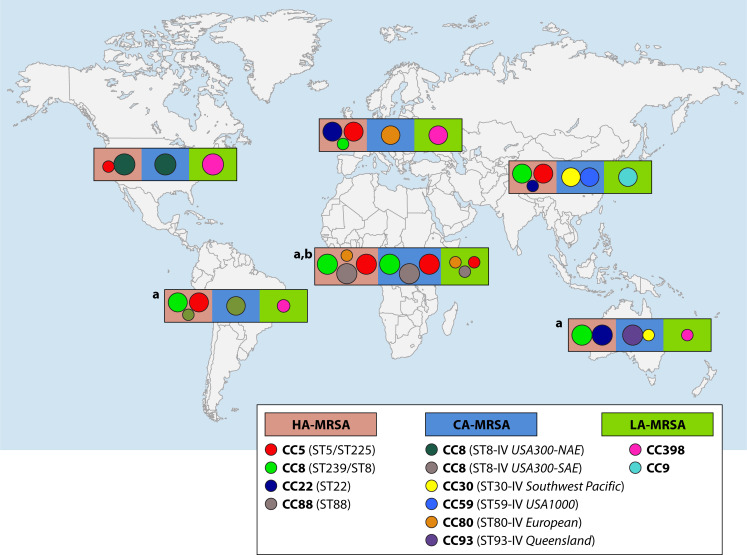
The emergence and spread of MRSA across the globe have resulted in distinct clones circulating in different settings and regions. While some clonal types are disseminated, others are restricted to specific geographical locations (Fig. 3). In this section, we will discuss the virulence attributes of current dominant lineages typically associated with HA-MRSA, CA-MRSA, and LA-MRSA from across the globe. The details of the representative strains used to characterize the pathogenicity of dominant lineages are also summarized in Table 1. For each lineage, we will discuss its origins and definitions, epidemiology in humans and/or animals, virulence in animal models, and the current known molecular mechanisms contributing to its success.
Fig 3.
Distribution of current dominant MRSA lineages. The major lineages of HA-MRSA, CA-MRSA, and LA-MRSA reported in each continent or region are shown. (a) Data on LA-MRSA from Africa, Latina America, and Australia were retrieved from single reports due the paucity of available data and, therefore, should not be considered as predominant LA-MRSA lineages in these regions. (b) Although there is no clear distinction between HA- and CA-MRSA clones reported from the African continent, the results should be interpreted with caution since they may reflect the lack of epidemiological data.
TABLE 1.
Representative Staphylococcus aureus strains of common clonal complexes used in virulence studies
| Clone name | Synonymsa | Representative | NARSAb | ATCCc | Region | Accession (reference) | Virulence studies |
|---|---|---|---|---|---|---|---|
| CC1d | |||||||
| ST1-IV | USA400; Canada-7e | MW2 | NRS123 | BAA-1707 | USA |
NC_003923 (315) |
(167, 301, 363–378) |
| CC5 | |||||||
| ST5-II/ST225-II | New York Japan; Rhine Hesse; USA100; UK-3f; Canadian-2 | N315 | NRS70 | Japan |
NC_002745 (210) |
(301, 315, 377, 379, 380) | |
| 04-02981 | Germany |
CP001844 (381) |
|||||
| ST5-IV | Pediatric clone; USA800 | 1045 | NRS387 | USA | SRR1014708 | ||
| ST105 | JH1 | USA |
NC_009632 (382) |
||||
| ST228-I | South German Epidemic; Italian clone | 16035 | Switzerland |
NC_020533 (383) |
|||
| CC8 | |||||||
| ST8-IV | USA500 | 2395 | USA |
CP007499 (384) |
(384, 385) | ||
| BD02-25 | (364, 386) | ||||||
| 95938 | NRS385 | ||||||
| ST8-IVa | USA300; USA300-NAE; Canadian-10; WA-12g | FPR3757 | BAA-1556 | USA |
CP000255 (387) |
(369, 377) | |
| LAC | USA | (47, 167, 363, 365–368, 371, 373, 374, 376, 378, 388–393) | |||||
| SF8300 | USA | (364, 378, 386) | |||||
| USA300-BKV | USA | PRJNA497094 | (47) | ||||
| ST8-IVc | USA300-SAE | CA12 | South America |
CP007672.1 (394) |
|||
| ST72-IVa | USA700 | CN1 | Korea |
NC_022226 (395) |
(364) | ||
| ST239-III | Hungarian, Portuguese, Brazilian, Czech of Vienna clone | TW2 | UK |
NC_017331 (396) |
(372) | ||
| UK-1, UK-4, UK-7, or UK-11; Canadian-3 or Canadian-6. | JKD6008 | New Zealand |
NC_017341 (397) |
(398) | |||
| T0131 | China |
CP002643.1 (399) |
|||||
| GV69 | Brazil |
CP009681.1 (400) |
|||||
| ST247-I | Iberian; North German Epidemic; UK-5, UK-8, UK-17 | HPV107 | BAA-44 | Portugal | SAMN06320797 (90) |
(386) | |
| ST250-I | Early; ancestral | COL | NRS100 | UK |
CP000046 (320) |
(202, 363, 366, 374, 386) | |
| CC9 | |||||||
| ST9-XII | ZY462471 | China | GCA_015070865 (401) |
(401) | |||
| CC22 | |||||||
| ST22-IV | UK-15; Canadian-8; Barnim Epidemic | H-EMRSA-15 | Denmark |
CP007659.1 (402) |
(372) | ||
| CC30 | |||||||
| ST36-II | UK-16; USA200; Canadian-4 | MRSA252 | BAA-1720 | UK |
BX571856 (403) |
(367, 370, 372, 374, 404) | |
| ST30-IV | Southwest Pacific; USA1100 | TCH60 | NR-10129 | USA | CP002110.1 | (202) | |
| CC45 | |||||||
| ST45-II | USA600; Canadian-1 | CA-347 | NRS648 | USA | NC_021554 (405) | (406) | |
| ST45-IV | Berlin Epidemic; WA-75 | ||||||
| CC59 | |||||||
| ST59-IV | USA1000 | AIS2006061 | NRS483 | USA | |||
| ST59-VT | Taiwan clone | SA957 | Taiwan |
CP003603.1 (407) |
(371, 407–409) | ||
| CC80 | |||||||
| ST80-IV | European CA-MRSA clone | 11819-97 | Denmark |
CP003194.1 (410) |
|||
| ST93 | |||||||
| ST93-IV | Queensland clone | JKD6159 | Australia |
NC_017338 (411) |
(372, 391, 412–414) | ||
| CC398 | |||||||
| ST398-V/VT | S0385 | NR-28983 | Netherlands |
AM990992.1 (415) |
(416–420) | ||
Based on guidelines in reference (33).
NRSA, Network on Antimicrobial Resistance in Staphylococcus aureus.
ATCC, American Type Culture Collection.
CC, clonal complex.
Canadian-, Canadian-MRSA-.
UK-, UK-EMRSA-.
WA-, WA-MRSA-.
CC5
CC5 is a widespread clonal complex, which comprises a large number of different pandemic HA-MRSA clones worldwide (Fig. 3). Although ST5 is the dominant and presumed ancestor of CC5, this lineage comprises many other epidemic clonal types mainly spread within Europe (33), including ST225-II in Central Europe (381), ST125-IV/VI mainly in Spain (421), and ST228-I (South German/Italian clone) mostly in Germany, Hungary, Austria, and Italy (33). ST5-II (also called USA100) was reported as the main clone in New York hospitals in the late 1990s (422). Subsequently, it was also reported in Japanese hospitals and designated as the “New York/Japan clone” (423). ST5-II remained the predominant HA-MRSA clone all over the US during the following 15 years (88) and is still the major HA-MRSA clone in Japan and in other countries in Eastern Asia (424). On the other hand, ST5-IV (also called USA800), which was initially detected among pediatric isolates and referred to as the “Pediatric clone” (425), has also achieved pandemic spread and clinical relevance, including in the African continent (426).
Our current understanding of the virulence mechanisms of CC5 is relatively limited compared with the knowledge of CC8 lineages. Clinical MRSA CC5 isolates were able to cause mortality at similar levels as other major lineages in rabbit endocarditis, murine sepsis, and Galleria mellonella infection models (363, 427, 428). G. mellonella is a non-mammalian model for studying the pathogenesis of S. aureus infections (429). However, contributions of specific virulence factors during CC5 infections have yet to be fully investigated in vivo. Gerlach et al. recently showed that CC5 MRSA clones altered cell glycosylation to evade host immunity by abrogating IgG response in vivo, which compromised neutrophil phagocytosis of CC5 strains (301). This immune evasion is mediated by TarP encoded in φSaint3, which is an alternative WTA glycosyltransferase transferring N-acetylglucosamine to a different hydroxyl group of the WTA ribitol-phosphate than the standard enzyme TarS (301). In addition to immune evasion, CC5 MRSA isolates from patients with bacteremia formed stronger bonds with fibronectin compared with CC45 counterparts, which likely promote binding to target tissues (430).
Phylogenomic analyses and phenotypic studies of clinical isolates provide hints of molecular mechanisms behind the success of MRSA CC5. Clinical CC5 isolates from various infection sites exhibit strong hemolysis of rabbit erythrocytes and strong biofilm formation compared with CC30 (USA200), CC8 (USA300), CC1 (USA400), and CC45 (USA600) isolates in the US, showing the toxicity and virulence of this lineage (363). This is corroborated by a high prevalence of virulence factors in CC5, including IEC in phage φSaint3, egc, and lukED on genomic island υSaβ (363, 431, 432). Among these virulence factors, the TSST-1 gene was strongly associated with lethal infections in a Chinese hospital (433), and staphylococcal enterotoxin P (Sep) was a significant predictor of bacteremia in hospitalized patients colonized with MRSA (434). Sep has been shown to disrupt the immune response by inducing proliferation of human lymphocytes and cytokine production of human T cells (435). Interestingly, clonal expansion of CC5 across the Americas was preceded by convergent loss of sep and gains of resistance to fluoroquinolone, macrolide, and lincosamide antibiotics, suggesting that more antibiotic-resistant and less virulent MRSA CC5 clones are more likely to spread geographically (431). However, a new local variant within CC5, ST764, emerged and disseminated endemically via acquiring new virulence determinants, ACME and SaPInn54 (436, 437).
CC8
CC8 is common in the community and in hospital settings, particularly in the US. CC8 encompasses numerous lineages of historic and contemporary importance including the notorious USA300 (ST8-IVa), which will be of particular focus in this review, as well as the closely related USA500 (also mainly ST8-IV), Archaic (ST250-I), and Iberian (ST247-I) clones. ST239 shares sequence similarity with CC8 lineages and will be discussed in the following section.
USA300 (ST8-IVa)
The clonal lineage USA300 emerged in the 1990s and rapidly became the dominant CA-MRSA strain in the US (438–440) (Fig. 3). USA300 was originally defined by its PFGE profile (88). The emergence and spread of USA300 coincided with the increased use of WGS as a diagnostic and epidemiological tool. Comparative genomic analyses revealed several prototypical molecular markers that were then used to define the lineage including MLST ST8, SCCmecIVa, genes coding for PVL, as well as either an ACME or copper and mercury resistance (COMER) element, which differentiates the North American epidemic (USA300-NAE) and South American epidemic (USA-300-SAE or USA300-LV) sublineages, respectively (288, 387, 394).
Given the success of USA300, a number of studies compared the virulence of representative strains with isolates from non-USA300 lineages using animal infection models (364, 386). In a landmark study, Li et al. showed that USA300 and the closely related lineage USA500 had enhanced virulence when compared to others related to CC8, including the archaic clone (ST250-I), the Iberian clone (ST247-I), and the Brazilian/Portuguese clone (ST239-III), based on mortality and abscess size in murine bacteremia and skin models, respectively (386). In a subsequent study, the same group showed that CC8 representatives USA300 and USA500, as well as ST80, were more virulent compared with isolates from diverse clonal complexes including CC5 (USA100), CC30 (USA200, USA1100), CC1 (USA400), CC59 (USA1000), and ST72. USA300 produced larger abscesses in a rabbit skin infection model, and this correlated with enhanced host immune markers of infection including leukocyte infiltration and cytokine levels (IL-8 and TNF-α) (364). In a rodent pneumonia model, USA300 produced more severe disease based on mortality and lung tissue pathology compared to an alternative CA-MRSA lineage, USA400 (365). Despite a limited capacity to form robust cardiac vegetations, USA300 isolates were highly lethal in rabbit models of infective endocarditis (363, 366). Taken together, USA300 represents a highly virulent and transmissible clonal lineage, and the molecular characteristics driving these traits are under close scrutiny.
Virulence of USA300 is commonly attributed to the virulence regulator Agr, as evidenced by deletion of the agr locus resulting in reduced abscess size and less dermonecrosis for USA300 in a murine subcutaneous infection model (367). USA300 displays striking agr-dependent expression of α-toxin, PSMs and PVL (367). Additionally, virulence was attenuated in agr mutants, as well as hla and psm deletion mutants, in rabbit infection models, thus supporting the role of these genes in USA300 pathogenesis (388). In contrast, despite the strong epidemiological association between the enigmatic PVL and S. aureus SSTIs (328), deletion of PVL had no impact on pathogenicity in a murine skin infection model (388) or murine models of pneumonia and bacteremia (167, 368, 389, 441). The lack of a pathogenic contribution for PVL in animal models has been attributed to host specificity of the toxin, whereby PVL is lytic toward human neutrophils, but its effect against murine neutrophils is benign (369). Interestingly, the lytic effects of PVL were most pronounced in the presence of an additional virulence factor, PSMα3 (369). Additionally, the lytic effects of USA300 culture supernatants toward human cells were neutralized by an anti-PVL monoclonal antibody. In a separate study using an ex vivo human skin model, PVL was toxic, albeit to a lesser extent when compared to α-toxin (442). More recently, virulence attenuation was observed for PVL deletion mutants in SSTI and pneumonia models when using humanized mice expressing PVL-sensitive receptors (390, 443). Deletion of PVL reduces bacterial burden in lung tissues in the humanized mice and improves clearance of PVL-deficient cells, while complementation of PVL restores these phenotypes (390). Together, there may be a role for PVL in human disease; however, it is likely to be less pronounced compared to other toxins and may be dependent on the activity of other virulence factors (444).
The exact reason for the distinct agr regulation profile of USA300 is not completely defined. One possible explanation is the presence of SCCmecIV. Unlike SCCmecII, SCCmecIII, and SCCmecVIII, which are associated with HA-MRSA lineages, SCCmecIV does not code for the psm-mec locus. The psm-mec transcription product binds to agrA mRNA, which inhibits its translation (370). Deletion of psm-mec from select HA-MRSA strain backgrounds increased AgrA production and enhanced virulence in murine models of skin infection and sepsis (370). Conversely, introduction of psm-mec into USA300 reduced the expression of AgrA (370), suggesting that its absence may facilitate high agr activity for this lineage. In a similar vein, the mecA gene itself has been shown to reduce the virulence of MRSA lineages harboring SCCmec types II, III, and VIII, which may represent a general explanation for the reduced toxicity of HA-MRSA when compared to MSSA (404). In contrast, strains harboring SCCmecIV had lower levels of oxacillin resistance and expressed less PBP2a, which correlated with high toxicity similar to that of MSSA (404). In addition, SCCmecIV is not associated with the in vitro and in vivo fitness costs described for other SCCmec types (445, 446). Together, SCCmec type and its relationship with agr functionality may explain a selective advantage for SCCmecIV harboring lineages; however, given the distribution of SCCmecIV among additional CA-MRSA lineages, it does not specifically explain the success of USA300.
Unlike SCCmecIVa, the prototypical ACME is rarely detected in non-USA300 isolates making it an attractive potential explanation for the success of USA300-NAE (447, 448). In addition, despite the close proximity of the elements, when SCCmec is infrequently lost from USA300, ACME is retained, suggesting that it provides an appreciable selective advantage (90). However, ACME’s contribution to acute virulence in S. aureus is unclear. While reduced fitness was attributed to ACME deletion in a rabbit model of bacteremia (445), a subsequent study using murine infection models found that deletion of ACME had no appreciable impact upon virulence endpoints (449). Here, the absence of ACME did not impact upon mortality, organ bacterial density, or lung pathology in a necrotizing pneumonia model or skin dermonecrosis in an SSTI model (449). Taken together, there is some evidence to suggest that ACME may improve in vivo fitness, but it does not enhance the severity of invasive S. aureus disease in animals.
USA300 acquired ACME from S. epidermidis, a predominant member of the human skin microflora (387, 450). This horizontal gene transfer event coincided with the rapid emergence of SSTIs caused by USA300 and displacement of other dominant clonal types causing SSTIs, suggesting that ACME likely contributed to improved colonization and/or transmission as opposed to enhanced acute virulence. Indeed, the skin provides an inhospitable environment for bacterial pathogens and in order to colonize it, S. aureus has to overcome low pH as well as innate and adaptive immune responses (451). ACME harbors several genetic systems that support this hypothesis: an auxiliary arginine deiminase pathway coded for by the arcACME operon, a spermidine (Spd)/spermine (Spm) acetyltransferase (speG), and the copper resistance locus copXL. Each system mediates subtle metabolic adaptations that improve survival in conditions relevant to human skin.
The ArcACME system facilitates acid tolerance for USA300 at pH levels associated with the skin (pH ~5.0) (452). Arc converts arginine to ornithine and concomitantly generates ammonia and ATP. The core S. aureus genome codes for an intrinsic Arc; however, this system functions in anoxic conditions not typical of the skin (453). In contrast, arcACME is constitutively active, and ArcACME-mediated ammonification effectively neutralizes physiologically relevant acid levels (452). However, circumventing skin pH via arginine deamination presents an additional obstacle; excessive ornithine is converted by the host to polyamines such as spermine and spermidine (452). Polyamines are present at high levels during inflammation and wound healing; they synergize with antibiotics and are toxic toward non-USA300 S. aureus (454–456). For USA300, polyamines can be mitigated by the function of ACME encoded SpeG (457). The ΔACME and ΔspeG mutants are susceptible to polyamines, while introduction of speG in trans in these mutants recovers the resistance to polyamines (457). However, the exact mechanisms underscoring this detoxification remain unclear. In addition to facilitating polyamide resistance, speG has recently been shown to provide additional benefits, including improved adherence, biofilm formation, and resistance to keratinocyte-mediated killing (361). Together, there appears to be a strong selective advantage for ACME elements harboring speG, whereby Arc and SpeG are both physically and functionally linked within the ACME, and SpeG works to detoxify a byproduct of Arc activity as well as provide tolerance toward naturally occurring polyamines present in human tissues.
ACME is not present in USA300-SAE. In its place, USA300-SAE has acquired the distinct COMER element. The COMER and ACME regions share two genes, copX and copL. Phylogenetic analysis of the orthologs revealed that the two major USA300 subtypes acquired the genes from other staphylococcal species independently (394, 458), further highlighting the important role of skin commensals as reservoirs for genes involved in USA300 adaptation. The copXL locus is involved in copper resistance (458). While copper is an important cofactor, elevated levels are toxic for bacteria, and it is exploited by the innate immune system for its antibacterial properties, particularly by macrophages patrolling the skin and respiratory tract (458–460). The S. aureus core genome codes for an intrinsic copper efflux system mediated by the P1B-1-type ATPase copper efflux transporter CopA and the copper chaperone protein CopZ (461). However, acquisition of copXL has been shown to confer copper hyper-resistance for USA300. CopX is a P1B-3-type ATPase efflux transporter which extrudes copper with high efficiency, and it is postulated that CopL may sequester copper and interact with the CopX and CopA transporters (458). Copper hyper-resistance for copXL harboring strains has been shown to promote survival in macrophages, suggesting that it may enhance USA300 fitness by circumventing innate immunity (458).
Despite its rapid spread over the past 25 years, there is some evidence that USA300 may be in decline in some regions of the US (462). This is thought to be due in part to improved clinical practices focused on hand hygiene, environmental disinfection, and decolonization strategies. Alarmingly, Copin et al. have recently reported the clonal expansion of a USA300 sublineage, Brooklyn variant (USA300-BKV), which was driven by the acquisition of resistance genes for topical antimicrobials chlorhexidine and mupirocin, leading the authors to postulate that the spread was attributable to excessive clinical intervention (47). In addition to resistance, the USA300-BKV sublineage has adaptive genome alterations that has enhanced its pathogenicity. First, USA300-BKV has a mutation in the regulator of pyrimidine biosynthesis, which resulted in a subtle metabolic shift that improved the fitness of the lineage in a murine colonization and transmission model (47). Second, the lineage harbors a mosaic phage that contributed to abscess formation in a murine SSTI model (47). This continued adaptation toward enhanced virulence and antibiotic resistance suggests that we are a long way from seeing the end of USA300.
USA500 (ST8-IV)
USA500 is an additional pulsotype describing a group of MRSA isolates from CC8 that are closely related to USA300 and commonly cause invasive infections in North America (88, 463). It was originally thought that USA500 was the direct progenitor of USA300 (386); however, multiple comprehensive phylogenetic studies have suggested that the two lineages most likely arose independently from a common ancestor (384, 463–465). Although USA500 and USA300 emerged in clinics at similar times, the prevalence of USA500 decreased as USA300 proliferated to become the most dominant MRSA clonal type in the US (464–467).
Phylogenetic analysis reveals unique features of USA500 compared with USA300 (463). Insertions of the mobile element IS256 are prevalent in USA500 strains, but no USA300 strains have IS256 (463). Unlike USA300 strains, ACME (or COMER) elements, speG, and PVL toxins are uncommon in the USA500 lineage (463). USA500 contains a frameshift mutation in adsA encoding adenosine synthase, resulting in a truncated protein of 131 amino acids instead of full-length 773 amino acids (463). The cell wall-anchored protein AdsA generates deoxyadenosine and deoxyguanosine extracellularly to modulate host immune responses (468, 469). AdsA is important for S. aureus to escape phagocytic clearance in blood, induce cell death of macrophages, and contribute to bacterial survival in organs in a murine infection model (468, 469). However, it is unclear how this adsA mutation in USA500 affects virulence and transmission. A recent study has found that deletion of adsA in MRSA induces potent inflammatory cytokines release in mice and promotes protective T cell responses against re-infection (470). More studies are required to examine how USA300 outcompetes USA500, including if USA500 elicits enhanced inflammatory and T cell responses as compared to USA300.
USA500 can cause severe invasive infections associated with high morbidity and mortality and has caused outbreaks in both community and hospital settings (463, 466). The severity of disease caused by USA500 in humans has also been modeled in laboratory animals whereby USA500 showed a similarly high virulence potential when compared to that of USA300 (364, 386). In agreement with these findings, USA500 expresses high levels of toxic exoproducts including α-toxin (384, 386). At least for a subset of USA500 isolates, hypervirulence could be attributed to the acquisition and insertion of the mobile element IS256 in the promoter of rot, which codes for the Rot (384). This event resulted in derepression of toxin production for USA500 and subsequently increased virulence in a murine infection model. Additionally, IS256 has also occasionally integrated in the agr locus suggesting that the movement of this element can result in both increased or decreased toxin production (384). However, this study was limited to strains from one area (New York). By analyzing a larger, more diverse collection of USA500 isolates, Frisch et al. found that 76 of the included 539 isolates harbored at least one copy of IS256 and that these isolates typically clustered in one of three clades within USA500 (463). This suggests that IS256-mediated toxin regulation cannot entirely explain the hypervirulence of USA500.
Taken together, USA500 and USA300 are very closely related, and each have high virulence potential. Of note, USA500 harbors the same SCCmec element as USA300, suggesting that it may also have virulence benefits when compared to HA-MRSA. Nevertheless, it is clear that virulence per se is not the sole determining factor for an MRSA lineage and that more subtle changes in physiology that result in improved colonization and transmission likely contribute to the comparative success of USA300.
CC9
MRSA CC9 clones were first isolated from pig farms in China in 2008 and have become the predominant LA-MRSA clonal lineage in Asia (471, 472) (Fig. 3). In the US, it is concerning that colonization of the CC9 lineage has emerged among pigs raised in industrial hog operations and persons who work or live close to hog operations (473, 474). Recent genomic analyses of 191 ST9 strains collected from 12 countries suggest that ST9 emerged as a human MSSA approximately two centuries ago (475). The loss of the immune evasion cluster genes (scn, chp, and sak) for human infections and the acquisition of animal-specific virulence factor SaPIbov4-like element-encoding vwb in ST9 support the evolution of host shift from human to animals (475).
Phylogenetic analysis of S. aureus isolates from different sources has indicated that pigs are an important reservoir for CC9 transmission to human and bovine hosts (476). Close contact is a likely risk factor for transmission given that the carriage rate of MRSA CC9 for pig farm workers was significantly higher than that of the general population (477–480). However, the pathogenicity of CC9 in humans remains largely unknown. Chen et al. recently identified eight MRSA CC9 isolates after screening 3,328 clinical MRSA isolates from a national database (481). The majority of these CC9 isolates carried SCCmecXII and were associated with lethal bacteremia and osteomyelitis, indicating that CC9 strains can be pathogenic to humans (481). Jin et al. recently reported a highly virulent ST-SCCmecXII isolate with IEC-carrying βC-φ, and phylogenetic analysis indicated that this strain is likely evolved from an MSSA predecessor rather than LA-MRSA ST9 (401).
Virulence factors of MRSA CC9 have not been fully investigated in animal models; only vertical perinatal transmission from sow to newborn piglets was shown in a swine model (482). Nevertheless, WGS analysis has shown that MRSA CC9 has acquired many MGEs harboring functional antimicrobial resistance and virulence genes, possibly due to the use of antimicrobials in industrial animal food productions (476, 479, 483, 484). Taken together, MRSA CC9 presents a potential risk as a zoonotic pathogen, and multidrug resistance is of particular concern.
CC22
CC22 clones cause the majority of HA-MRSA infections worldwide, particularly ST22-IV, which is a highly epidemic MRSA (EMRSA) clone designated as EMRSA-15 (Fig. 3) (33). It is considered the most rapidly expanding and persistent HA-MRSA clone in Europe (485). First isolated in the United Kingdom in the mid-1980s, EMRSA-15 quickly became endemic in UK hospitals and subsequently disseminated throughout Europe and beyond, namely, to Australia, Asia, and the Middle East (33, 486). Currently, EMRSA-15 is the main HA-MRSA clone in Australia (56%) and Singapore (424, 487). Using phylogenomic analyses, Holden et al. (488) showed that the current pandemic ST22-IV clone evolved from a healthcare-associated EMRSA-15 subclone spread in England during the mid-1980s (488).
The success of CC22 is likely related to antibiotic resistance, virulence, and stress tolerance. The presence of a fluoroquinolone resistance trait in CC22 provided a selective advantage, while there was extensive use of this class of antibiotic in the early 1990s (488). CC22 MRSA strains show greater fitness, strong biofilm formation, and superior capacity to survive desiccation, antiseptics, oxidative stress, heat, and pH changes compared with other competing clonal types, including CC5 (ST228-I), CC30 (ST36-II), and CC8 (ST239-III) (489–492). Animal models of infection have also been used to assess the virulence of CC22 (489, 493, 494). In a mouse model of acute lung infection, CC22 exhibited higher virulence, resulting in lethal infections and causing a higher bacterial load in the lung, which in turn led to severe inflammation, as compared to CC5 (ST228-I) (489). ST228-I was the predominant clone replaced by CC22 (489). Further comparisons with CC5 (ST228-I) revealed stronger α-hemolysin activity and β-hemolysin production, as well as an active agr, which partly explained the enhanced virulence of CC22 (491).
Naturally occurring variations in the agr system in CC22 strains can directly impact on virulence, suggesting that it is a virulence switch for this lineage (494). For example, amino acid substitution Y223C in AgrC, caused by an SNP in argC, led to the destabilization of the AgrA-AgrC interactions. This caused differential regulation of virulence genes, leading to a switch from a cytotoxic to a colonizing phenotype and less severe skin tissue damage in a murine skin infection model (494). This phenotypic switch likely promotes the emergence of new variants for the ongoing persistence of CC22. However, given that this observation is based on a limited number of isolates, further comprehensive studies are required to investigate the role of the agr system in CC22 virulence.
Despite our current understanding of CC22 virulence factors informed by experimental animal studies, it remains to be fully elucidated if these virulence factors are important in human infections. A recent study extensively investigated the role of bacterial factors in determining disease outcome of S. aureus bacteremia in humans (495). Recker et al. quantitatively phenotyped cytolytic activity and biofilm formation of a collection of 135 sequenced clinical CC22 isolates from patients with bacteremia. The researchers utilized a machine-learning framework to analyze the pooled data of bacterial phenotype and genotype together with clinical metadata (495). Elevated cytolytic toxicity in combination with low levels of biofilm formation was predictive of an increased risk of mortality in infections caused by CC22 strains (495). A virulence factor for S. aureus, CapA, was identified to be predictive of mortality within the CC22 collections in their model (495). The capA gene encodes a dual-function phosphodiesterase/kinase activator and forms a protein complex with CapB to positively control multiple enzymatic checkpoints of capsule biosynthesis (496). Expression of capsular polysaccharide is important for protection from host immune responses and is a determinant of virulence in a mouse bacteremia model (497, 498). A naturally occurring SNP in capA was identified in CC22 isolates from patients who survived their bacteremia compared with isolates from patients who died. This SNP led to amino acid substitution P146S in CapA and was associated with reduced capsule production, a lack of reactivity to the antisera and susceptibility to human neutrophil killing compared with wild-type CapA (495).
Taken together, antibiotic resistance and high tolerance to environmental stress contribute to the success of CC22. Further genomic and phenotypic analysis of clinical CC22 isolates combined with clinical records will improve our understanding of factors contributing to bacterial virulence and pathogenesis in humans. This knowledge will potentially improve management of infectious diseases caused by this lineage.
CC30
S. aureus isolates belonging to CC30 have been causing bacterial epidemics for close to 70 years in both hospital and community settings (48, 499). The first lineage of CC30 to become prominent was the penicillin-resistant, methicillin-sensitive clone known as phage-type 80/81, which emerged in Australia, Europe, and North America in the early 1950s (500–502). The decline of phage-type 80/81 coincided with the clinical introduction of penicillinase-resistant β-lactams (i.e., methicillin) in 1961. Two CC30 MRSA lineages largely replaced phage-type 80/81: the hospital-acquired ST36-II clone known as EMRSA-16 in the UK (503), which is closely related to USA200 in the US (88), and the community-acquired ST30-IV PVL+ lineage known as the Southwest Pacific Clone (SWP) (499).
EMRSA-16/USA200 (ST36-II)
For the purpose of this review, we will refer to EMRSA-16, USA200, and closely related CC30 MSSA isolates as contemporary CC30 hospital isolates (cCC30), as previously described (48). cCC30 were a predominant source of hospital-acquired infections, particularly in the UK throughout the 1990s (503, 504), but they have more recently been displaced in hospital settings by CC22 (505). Nevertheless, in humans, cCC30 MRSA is associated with persistent bacteremia and hematogenous complications such as IE (506–511). In a rabbit infection model, cCC30 has a propensity to cause IE rather than lethality (366), which correlates with what has been observed clinically. When compared with the other closely related CC30 clones, phage-type 80/81 and SWP, cCC30 infections were less lethal in both bacteremia and pneumonia models (48). cCC30 isolates taken from patients with persistent bacteremia were more resistant to host innate immune defenses and displayed enhanced adherence to host cells in vitro, compared with isolates causing resolving bacteremia (from CC8). However, this did not correlate with any differences in acute virulence endpoints in a rabbit IE model. Nevertheless, in the same IE model, the cCC30 strain was more resistant to vancomycin therapy evidenced by greater cardiac vegetations after treatment, highlighting the potential for enhanced persistence of this lineage (511).
WGS has been crucial for the molecular differentiation of CC30 and has revealed a number of genetic contributors to explain the pathogenicity profile of each sublineage (512). Numerous mechanisms have been identified that appear to direct cCC30 away from acute bloodstream infection and toward persistence and niche adaptation, including gene mutations that directly alter toxicity and virulence gene expression, acquisition of MGEs, amplification of insertion sequence (IS) elements, and altered metabolism (513). cCC30 harbors a mutation that generates a premature stop codon in hla, which abolishes the production of functional α-toxin and has been shown to directly contribute to reduced virulence in bacteremia models (48). cCC30 also possesses an SNP mutation in agrC, which results in a non-synonymous amino acid change (G55R) that reduces RNAIII transcription. However, the impact of this specific mutation to virulence in animal models was less pronounced when compared to that of hla (48, 514). Additionally, CC30 codes for a unique variant of PSMα3, which is typically the most proinflammatory example of these cytolytic peptides (42). The non-synonymous mutation to psmα3 in CC30 isolates (PSMα3N22Y) reduces the potential for the peptide to stimulate neutrophil chemotaxis and reduces its contribution to cytotoxicity when compared to classical non-CC30 PSMα3 (514). In a murine bacteremia model, the CC30-specific PSMα3N22Y contributed to hematogenous seeding, as determined by kidney abscess formation. No enhanced virulence profile was observed using an SSTI model, suggesting that the contribution of PSMα3N22Y to virulence is infection setting specific, which may explain the association between CC30 and hematogenous infections in humans. Of note, PSMα3N22Y is present in both historic and contemporary CC30 strains, so it cannot explain the particular virulence profile of cCC30 (514). However, it is possible that reduced toxicity mediated by the combination of hla and agr mutations and PSMα3N22Y may synergistically contribute to persistence in humans by circumventing the host immune response.
In addition to causing persistent bacteremia and infections associated with hematogenous spread, CC30 is largely responsible for cases of toxic shock syndrome (TSS) (515, 516). In CC30, the gene coding for TSST-1 (tst) is carried on SaPI2, which is commonly harbored by both MSSA and MRSA strains (516, 517). However, TSS is more commonly caused by CC30 MSSA, and this correlates with increased production of TSST-1 compared to tst-positive CC30 MRSA, as demonstrated in vitro (515). Reduced production of TSST-1 for tst-positive MRSA has been associated with a non-synonymous SNP in ccpA that resulted in an amino acid change (T87I) in the catabolite control protein CcpA, which has been shown to directly influence the expression of tst (518). While the association was strong [33/39 tst-positive MRSA compared with 0/23 tst-positive MSSA (518)], the direct contribution for CcpA T87I to TSST-1 production or TSS is yet to be confirmed using allelic replacement. This possibly represents another example of the evolution of cCC30 MRSA away from acute toxicity; however, the benefits of reduced TSST-1 production remain to be elucidated.
SWP clone/USA1100 (ST30-IV)
The SWP clone is a pandemic lineage of CA-MRSA that has been identified from numerous locations across the globe (519). It was originally thought that SWP was a direct descendant of phage-type 80/81 (499); however, multiple reports using high-resolution molecular differentiation facilitated by genome sequencing revealed that the clone, along with cCC30, arose from a common ancestor (48, 517). Nevertheless, SWP and phage-type 80/81 share high toxin production profiles and are highly virulent in murine sepsis and pneumonia (520). Compared with cCC30 and CA-MRSA clones such as USA300, the lineage-specific molecular contributors to SWP virulence are not well characterized. One study assessed the contribution of PVL to cytotoxicity toward human osteoblasts and found it to be negligible (521). Given its known contribution to severe infection and its global distribution, more work is warranted to address the pathogenicity of the SWP clone.
ST239
One of the more successful global clones of HA-MRSA is ST239 (Fig. 3). The lineage emerged as the result of a major recombination event involving ST8 and ST30 (522). Recent genomic analyses indicate that ST239 originated between 1920 and 1945, predating the clinical use of methicillin in 1959 (523). ST239 has evolved toward antibiotic resistance and virulence, with the cost of lower competitive fitness compared with ST8 and ST30 (523). Comparing with ST8, ST22, and ST93, ST239 and ST36 isolates were less cytotoxic toward monocyte-macrophage THP-1 cells (524). Epidemiological studies have revealed that ST239 MRSA isolates cluster into regional clades, which indicates local expansion and is associated with only limited and sporadic intercontinental spread of evolved representatives (525, 526). In line with this, regional ST239 MRSA sublineages often have distinct pathogenicity profiles.
A contributing factor for the success of ST239-III in many Asian hospitals is the presence of sasX, which codes for an LPXTG motif surface-anchored protein (329, 396). The gene is typically found on ϕSPβ-like prophages and shares sequence similarity with sesI from S. epidermidis (396, 527). A comparative analysis of global ST239 revealed that the majority of isolates from the “Asian clade” harbored sasX and, conversely, few non-Asian representatives possessed the gene (528). The rate of sasX-positive MRSA in Chinese hospitals increased between 2003 and 2011, and the same increases were not observed in the community, therefore suggesting that sasX is specifically linked to hospital-acquired infections (329). However, the adaptive benefit of sasX is likely dependent on regional selection pressures, as imported sasX-positive ST239-III strains were unable to persist in Japanese hospitals (529).
Cell surface-bound SasX promotes adhesion to human nasal epithelial cells, which has epidemiological significance as nasal carriage is linked with infection (530). Deletion of sasX reduces biofilm formation, bacterial survival in human blood, and lysis of human neutrophils, whereas complementation of sasX restores these phenotypes (329). SasX also contributes to immune evasion and virulence in animal skin and lung infection models (329), suggesting that it may represent an attractive anti-virulence target. Indeed, passive and active immunization strategies using the SasX protein effectively reduced nasal colonization in mice and reduced the severity of disease in animal infection models (531).
When compared to MSSA ST398, ST239 MRSA has an enhanced capacity for nasal colonization in mice and is less virulent in a septic murine infection model (532). At the proteomic level, the production of AgrCA was lower in ST239 MRSA, and this correlated with enhanced production of surface-related proteins under its repressional control, including SpA, FnbpA, ClfA, IsaA, IsaB, LtaS, SsaA, and Cna (533). Most notably, expression of SpA contributed to the impressive nasal colonization of ST239 MRSA. Additionally, SpA contributed to long-term tissue damage in a persistent murine renal abscess model (532). Taken together with sasX acquisition, ST239-III strains from China have a characteristic cell surface decoration, which facilitates efficient colonization and persistence in hospital environments.
In Australian hospitals, ST239-III has persisted for many decades. Two distinct clades have been identified, each of which have undergone convergent adaptation toward the hospital environment, manifested by temporal increases in antibiotic resistance and virulence attenuation (525). Reduced susceptibility of MRSA ST239-III to antibiotics, including vancomycin, teicoplanin, and daptomycin, has been observed over time in Australia (525). SNPs in the walKR locus are significantly associated with increased vancomycin MICs (525). Reduced virulence for late isolates was also observed, and this correlated with agr dysfunction. Despite the loss of acute virulence traits in a murine model of septicemia, the adapted isolates maintained the capacity to persist in the kidney, suggesting that the adapted Australian ST239-III clades favor persistence as opposed to acute pathogenicity (525).
CC59
CC59 is an epidemic lineage of CA-MRSA in the Asia-Pacific region (Fig. 3) with carriage and infection commonly observed in children (534–537). CC59 is also becoming part of HA-MRSA in the Asia-Pacific region (538, 539). Phylogenomic analysis of global CC59 strains shows that two distinct major clades emerged in the US and in East Asia/Taiwan between 1960 and 1970, followed by dissemination to Europe and Australia separately (540). Of note, CC59 strains from East Asia/Taiwan contain a greater number of antibiotic resistance genes compared with the USA CC59 strains (540).
Recent population studies of S. aureus infections in China show that CC59 is replacing CC8 (ST239-III) and CC5 (ST5-II), indicating the success of this lineage in the region (541, 542). In murine infection models, ST59 CA-MRSA isolates caused significantly larger skin abscesses and more pronounced lung damage compared with HA-MRSA ST5 and ST239 isolates from the same Chinese hospital (543). ST59 also displays higher growth rate and better competitive advantage compared to ST239 but increased susceptibility to rifampicin and fluoroquinolones (541). Regarding virulence, PVL appears not to be essential for the success of CC59 given that PVL is not the common feature of CC59 USA and East Asia/Taiwan clones (543, 544). Genetic association studies identified a possible link between chp and enhanced virulence potential of MRSA ST59 when compared with MRSA ST239 isolates (545). This association was validated in the lysis assay of human erythrocytes using chp knockout MRSA ST59 mutants (545).
The virulence of ST59 MRSA is correlated with increased secretion of PSMα and δ-toxin compared with ST5 and ST239 MRSA isolates (543). Consistently, deletion of hla, psmα, and agr significantly compromised the virulence and pathogenesis of CC59 CA-MRSA in skin, lung, and blood infections, suggesting that these virulence factors play an important role in CC59 MRSA infections (543). Analysis of toxin genes also indicated that CC59 MRSA isolates from Chinese pediatric patients with bloodstream infections contain a specific toxin gene profile, that is, seb-sek-seq (546, 547). Recently, Bae et al. identified that Seb has a significant role in the virulence of ST59 MRSA isolates, as deletion of seb reduced the cytokine storm and increased host survival in a murine systemic infection model (548). The contribution of sek and seq in the pathogenesis of CC59 MRSA remains to be determined.
Genomic comparison further indicates that the East Asia/Taiwan CC59 clade is composed of a “Taiwan clone” [PVL positive, SCCmec V(5C2&5)] and an “Asian-Pacific clone” (PVL negative, SCCmec IV) (106, 407, 535). The Taiwan clone is frequently isolated from patients with severe disease, while the Asian-Pacific clone is a common colonizer of healthy children (407). The Taiwan clone induced more severe infections with a higher mortality rate in comparison with an Asia-Pacific clone in a murine bacteremia model (407). Loss of sak in prophage φSaint3 contributes to virulence in the Taiwan clone, as complementation of sak expression was shown to reduce the level of virulence in murine skin infections and bacteremia models (407, 549). The G10S variant of δ-toxin is also a characteristic of CC59, which leads to reduced chemotaxis and lysis of human neutrophils (371).
CC80
CC80 (ST80-IV) emerges as an important CA-MRSA lineage in Europe in late 1990s (550–552). MRSA ST80 was first reported in a Greek hospital and was the dominant CA-MRSA strain to cause SSTIs in Denmark (550, 551). Since then, MRSA CC80 has been prevalent in Europe, the Middle East, and North Africa (553–555). It is likely that this lineage was imported into Europe as many infections caused by MRSA ST80 in Scandinavia were related to travels to the Middle East and Africa (556, 557). Phylogenetic analyses of genomes from global MSSA and MRSA CC80 isolates indicate that a PVL-positive MSSA from sub-Saharan Africa is most likely to be the ancestor of the European epidemic ST80-IV (558). During this evolution, ST80-IV also acquired a plasmid conferring resistance to fusidic acid, which is a commonly used antibiotic to treat skin infections (558).
The success of MRSA ST80 is potentially attributed to specific properties of this lineage (558, 559). In addition to antibiotic resistance, MRSA ST80 harbors a specific non-synonymous SNP in agrC that distinguishes this strain from its MSSA ST80 ancestor (558). This SNP results in L184I amino acid substitution within the sensor domain of the AgrC receptor, where the AIP binds (558, 560, 561). Moreover, ST80-IV induces lower cytokine production (TNF-α, IL-1b, IL-6, IL-8, IL-10, IFN-γ, and IL-2) by monocytes compared with the response induced by ST30-IV, ST225-II, ST239-III, and ST5-IV (559). These properties possibly promote persistent colonization of MRSA ST80 by evading host immune responses and increasing its fitness.
PVL, epidermal differentiation inhibitor B (EdinB), and the exfoliative toxin D (EtD) have frequently been associated with MRSA-ST80 (562). EdinB targets host Rho GTPase (563), and deletion of edinB in MRSA-ST80 reduced the occurrence of bacteremia in mice with pneumonia, suggesting that EdinB facilitates bacterial dissemination in tissues (564). Furthermore, EtD induced skin exfoliation with the destruction of cell-to-cell adhesion in mice (565). These virulence factors might play a pathogenic role during MRSA ST80 infections.
ST93
In Australia, the dominant CA-MRSA clone is ST93 and is colloquially termed the “Queensland or QLD clone” (566) (Fig. 3). ST93 MRSA is typically SCCmec type IVa (2B) and PVL positive and is an MLST singleton as determined by eBURST, suggesting distant molecular relationships with other global lineages (567, 568). Recent studies indicate that ST93-MRSA-IV originated from MSSA strains in remote indigenous communities of northwestern Australia, emerged as MRSA and spread along the Australian East Coast in the early 2000s, followed by spreading overseas to New Zealand, the UK, and Papua New Guinea (569, 570).
ST93 commonly causes SSTIs in humans, as well as additional severe clinical manifestations such as necrotizing pneumonia (571, 572). Accordingly, the common MRSA ST93 reference strain (JKD6159) has shown high virulence potential in animal models, producing larger lesions even when compared to other notorious CA-MRSA lineages such as USA300 in mice skin infection models (391, 412). In support of these findings, enhanced virulence was associated with hyperproduction of α-toxin, whereby deletion of hla from JKD6159 reduced the severity of disease (413). In contrast, deletion of the psmα locus or PVL had little impact on virulence endpoints, further highlighting the key role of α-toxin for the pathogenicity of MRSA ST93 in this model (413). While high expression of α-toxin is a hallmark feature of ST93, some naturally occurring clinical isolates were shown to produce low levels of exotoxins as a result of convergent mutations affecting the agr locus, and this resulted in virulence attenuation (414). Toxin production for MRSA ST93 was also shown to be potentiated by the activity of an AraC/XylS family regulator, AryK (413).
Recently, MRSA ST93 isolates with human origin have been detected in pigs and farm workers in Australia. Transmission was followed by livestock adaptation in many cases including the loss of φSaint3 and its characteristic human invasion gene cluster (~70%), absence of PVL (~30%), and increased antibiotic resistance (573). Thus, ST93 is a well-established CA-MRSA threat and an emerging occupational risk for piggery workers in Australia (574).
ST398
LA-ST398
ST398, originally identified as PFGE non-typeable MRSA, is the major clone of CC398 which dominates LA-MRSA strains isolated from animals in North America, Europe, and Asia (Fig. 3) (34, 575). In particular, strains belonging to CC398 frequently colonize pigs and other livestock hosts as well as people exposed to pigs and pig farmer’s households (575–577). The origin of ST398 in farmed animals is not completely clear. Phylogenetic analyses of MRSA and MSSA from animals and humans across 19 countries and four continents indicate that it is likely that ST398 LA-MRSA originated in humans as MSSA and then jumped to livestock and companion animals followed by host adaptive evolution (74, 345, 578). Of note, a recent study has also shown that human-adapted ST398 MSSA can acquire an SCCmec-V class D to become CA-MRSA ST398, which is different from SCCmec-V class C present in most of MRSA ST398 (579). This lineage is capable of adapting to hosts and environments via obtaining or losing MGEs, raising concerns of MRSA ST398 as a potential zoonotic pathogen (84, 345, 580).
In light of the threat of MRSA ST398, animal models have been developed to examine the colonization, transmission, and virulence factors of this lineage in the hope to develop control strategies (482, 579, 581–586). MRSA has often been detected in the air and housing environments of pig barns, suggesting that transmission of LA-MRSA can occur via the environment (587–589). Colonization of MRSA ST398 in pigs via inhalation was demonstrated in a swine model when piglets were exposed to MRSA in an aerosol chamber (581). Additionally, MRSA ST398 was able to co-colonize piglet skin or the nose in the presence of coagulase-negative staphylococci or MSSA, suggesting that this lineage is able to adapt to the microbial ecology of farmed animals (584, 585). This may provide a reservoir for genes that can be important for adaptation in different animals, as has been seen for human MRSA USA300.
The spread of LA-MRSA by vertical perinatal transmission was observed when vaginal inoculation of a sow with LA-MRSA led to persistent colonization in all newborn piglets (482). In a study carried out in two intensive pig production systems in the US, S. aureus was prevalent among vaginal swabs (39.6%) from sows, and 91.1% of pigs had S. aureus in at least one site of nares, tonsils, skin, or rectum (590). Transmission of MRSA ST398 between pigs was also evident as naïve pigs were colonized after exposure to pigs orally inoculated with MRSA ST398 (586). Together, these studies provide insights underlying the success of colonizing MRSA ST398 in farmed animals.
Comparative genomic analyses have shown the characteristics that differentiate human- and animal-adapted ST398 strains. Human ST398 MSSA isolate harbors φSaint3, which contains IEC and is associated with enhanced adhesion ability to human skin keratinocytes and keratin (416). Host adaptive evolution of ST398 from human to livestock involves loss of φSaint3 and IEC and acquisition of resistance to antibiotics, including tetracycline and methicillin (84, 345). However, it is likely that LA ST398 still maintains the capacity to at least temperately colonize humans. In a study involving healthy human volunteers inoculated with a mixture of bovine MSSA ST398 and human MSSA ST931 into the nose, ST398 was able to compete with the human strain and survived in the nose for 21 days (591). Acquisition of prophages was also suggested to increase adhesion expression of LA-MRSA ST398 and virulence in a rat IE model (592). It remains rare but possible that human-to-human transmission of LA-MRSA ST398 can occur, given that dissemination between caretaker and patient in a hospital setting has been reported (593). LA-MRSA ST398 is constantly evolving and is able to re-acquire IEC for the readaptation to human host (580).
LA-MRSA ST398 strains display the lowest content of virulence genes compared with ST5, ST15, ST22, CC30, CC97, CC130, and CC151 (594). The lack of accessory virulence genes and no specific virulence markers in LA-MRSA ST398 suggest that the genetic background of this lineage is unique compared with other major MRSA lineages from humans (594). Recent analysis of superantigens gene distribution reveals that CC398 isolates have only selw but no other superantigens (595). SELW is required for a CC398 clinical isolate to induce Vβ-specific T cell proliferation, and SELW contributes to bacterial load in liver by a CC398 isolate in a murine bloodstream infection model (595). Complementation of selw in Δselw mutant in trans leads to more severe disease outcomes in the model compared with wild type (595). Despite the absence of many perceived virulence factors, LA-MRSA CC398 has retained its capacity to cause infections in humans who have close contact with livestock and companion animals (593, 596, 597).
The pathogenicity of LA-MRSA CC398 strains from industrial hog operation workers was assessed in a mouse SSTI model, which showed that larger lesion sizes and reduced IL-1β expression could be induced by LA-MRSA CC398 compared with an MRSA USA300 strain, SF8300 (598). Comparing ST398 MRSA isolates from pigs with those from humans, Schmidt et al. showed that human ST398 MRSA isolates have better cell adhesion and stronger lysis activity toward human neutrophils (599). This stronger cytolytic activity of human MRSA ST398 is likely due to the regulation of toxin expression and exportation as no pivotal difference in virulence factors was detected between LA-MRSA ST398 and human MRSA ST398 (599).
Human-adapted ST398
In addition to transmission from animals to humans, ST398 MRSA strains can also evolve from human-adapted ST398 MSSA and cause severe infections (579). A recent study shows that severe and fatal human infections in a Chinese community were caused by human-adapted ST398 MRSA, which acquired a low level of methicillin resistance while maintaining high virulence as ST398 MSSA (579). In both murine lung and skin infection models, the ST398 CA-MRSA isolates showed more severe disease than HA-MRSA clones (ST5 and ST239) and LA-MRSA ST398 strain S0385. These human-adapted ST398 MRSA clones harbor a class D SCCmec-V element compared with class C SCCmec-V from LA-MRSA ST398 clones and have a lower oxacillin MIC (4 µg/mL) compared with HA-MRSA clones’ MIC (128 µg/mL) (579). Oxacillin MIC 4 µg/mL is the susceptibility breakpoint to define MRSA. This study shows that distinct ST398 MRSA lineages emerged by multiple recent SCCmec uptake events from ST398 MSSA ancestors, indicating that the development of novel and highly virulent MRSA lineages may be a frequently occurring scenario.
At the molecular level, community-acquired ST398 MRSA isolates from China exhibited higher expression levels of ESAT-6 secretion system (ESS) genes compared with ST239, which is the predominant hospital-associated lineage MRSA in China (582). ESS is important for the virulence of the ST398 isolates collected from humans in China (582). The disruption of ESS by deletion of the structural component, essB, was shown to significantly reduce resistance to neutrophil killing and decrease virulence in murine skin and blood infection models (582). Neutrophil killing was restored after complementation of essB in the ΔessB mutant (582). The ESS-secreted protein, EsxX, is highly conserved only in the ST398 MRSA isolates from humans in China (600). EsxX promotes neutrophil lysis, evades neutrophil killing, and contributes to virulence in murine infection models (600). Comparative proteomic analysis of ST398 and ST239 MRSA lineages illustrates that ST398 has a higher expression of the Agr system (AgrA and AgrC), which enhances ESS and Agr interactive factors (PhoP, SrrB, WalK, SarX, SigB, and ClpP) (533, 582). High production of α-toxin and a highly functional agr system has also been associated with fatal pneumonia caused by ST398 MRSA isolates in Brazil (417).
Characterization of 35 prophages found in the collection of 76 ST398 MRSA bacteremia isolates showed that these prophages harbor many genes that are associated with virulence or immune evasion (601). An increasing prevalence of poly-lysogeny in ST398 bloodstream infection isolates over time was identified, therefore highlighting that this lineage may increase its capacity to spread in humans by continuously acquiring virulence and/or antibiotic resistance genes via HGT (601). ST398 MRSA expresses S. aureus Toll/IL-1 receptor (TIR)-like protein 1 (SaTlp1) and S. aureus TIR-like protein 2 (SaTlp2), which contain domains similar to TIR (583). SaTlp1 and SaTlp2 enhance activation of the transcription factor NF-κb and downstream pro-inflammatory cytokines and immune effectors (583).
Taken together, ST398 represents a successful lineage for its ability to constantly evolve, adapt, and readapt to changing host environments. Surveillance of antimicrobial resistance and factors contributing to transmission and virulence remains a priority for monitoring this lineage. Future studies are required to develop control measures to mitigate the threat of MRSA ST398 as a zoonotic pathogen important to public health.
CONCLUDING REMARKS
Although the rate of MRSA infections has stabilized or even decreased in many countries, it is still a significant concern for society and a threat to global economies. MRSA infections remain severe and difficult to treat, especially with the emergence of strains resistant to last-line antibiotics. As S. aureus continues to evolve and to surprise, now is not the time for complacency.
In the current review, we have summarized a large body of work from the last 25 years that has defined the specific character of successful MRSA lineages. WGS and comparative genomics have played a major role in our understanding of MRSA pathobiology. Here, numerous core and lineage-specific virulence factors have been identified and discussed. Factors that contributed to MRSA infections, including toxins, immune evasion proteins, and agr, have been investigated to characterize their contribution in the success of a lineage. Recent studies have also shed light on how MRSA pathogenicity and metabolism are closely regulated. The success of USA300 lineage is attributed to virulence regulation and metabolic adaptation, whereas it is less clear how ST80-IV is persistent among European countries. Further investigations on host factors for MRSA colonization and pathogenesis are likely to improve our understanding of the success of a specific MRSA lineage. Together, these findings will identify interesting potential targets for future intervention. Neutralization of toxins, inactivation of the central virulence regulator Agr, inhibition of the transfer of MGEs, and microbiome-mediated exclusion of MRSA colonization are likely to be important strategies to prevent continued global MRSA pandemics.
Biographies

Jhih-Hang Jiang is a molecular microbiologist with special interests on bacterial pathogens. He obtained a Ph.D in the laboratory of Richard Strugnell (the University of Melbourne, Australia), where he investigated how pathogenic Neisseria species manipulate host cell death. His postdoctoral training was undertaken in the laboratory of Anton Peleg (Monash University, Melbourne, Australia), where he investigated antimicrobial resistance and host immune evasion in Staphylococcus aureus. He currently leads Staphylococcal Translational Research Program in the Department of Infectious Diseases, Monash University. His program is to develop new therapeutic strategies to treat MRSA.

David R. Cameron is a molecular microbiologist focused on understanding staphylococcal pathogenesis and developing new approaches for treatment. He obtained a Ph.D. in the laboratory of Anton Peleg (Monash University, Melbourne, Australia), where he worked to define the relationship between antibiotic resistance and virulence for Staphylococcus aureus. His postdoctoral training was undertaken in the laboratory of Kim Lewis (Northeastern University, Boston, USA), where he explored the clinical importance of antibiotic-tolerant persister cells. He arrived in Switzerland in 2018 and worked closely with Yok-Ai Que at the University Hospital of Bern to assess bacteriophage therapy for the treatment of staphylococcal infections using translational animal models. Currently, he is a Senior Scientist at Debiopharm International S.A. and functions as the lead microbiologist on each of the company’s anti-infective programs.

Cara Nethercott completed her PhD under the supervision of Prof. Anton Peleg in the Department of Microbiology, Monash University, Australia. Her PhD research involved characterizing Staphylococcus aureus virulence factors and rapid techniques for detecting antibiotic resistance. Her research background and interests include infectious diseases, immunology and antibiotic resistance. She is now a Medical Writer at Oxford PharmaGenesis Pty Ltd, where she writes peer-reviewed publications on clinical trials and develops medical education for healthcare professionals.

Marta Aires-de-Sousa is a molecular microbiologist focused on understanding the molecular epidemiology and antimicrobial resistance mechanisms in Staphylococcus aureus and Enterobacteriaceae. She obtained a Ph.D. in the laboratory of Hermínia de Lencastre (Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Portugal), where she explored the molecular epidemiology of methicillin resistant S. aureus under a One-Health approach, among humans, animals and the environment. More recently she is involved in both surveillance of antimicrobial resistance and analysis of the molecular genetics of antimicrobial resistance in Enterobacteriaceae. She is a Professor of Microbiology and the chairman of the Direction Board at the Higher School of Health of the Portuguese Red Cross, Lisbon, Portugal.

Anton Peleg is a Professor of Infectious Diseases and Microbiology, Australian National Health and Medical Research Council Practitioner Fellow and Director of the Department of Infectious Diseases at The Alfred Hospital and Monash University, Melbourne, VIC, Australia. He is Theme Leader for Infection and Immunity at Monash Academic Health Research and Translational Centre, and has recently been elected as a Fellow of the Australian Academy of Health and Medical Sciences. He completed his infectious diseases clinical training in Australia in 2005 and then went to the USA for four years and worked at the Harvard-affiliated hospitals; Beth Israel Deaconess Medical Center and Massachusetts General Hospital. He completed a Masters of Public Health at Harvard School of Public Health, and also completed a PhD in Infectious Diseases and Microbiology with a focus on antimicrobial resistance (AMR) and microbial genomics in 2010. His research interests are in hospital acquired infections, AMR and novel solutions, bacterial genomics, mechanisms of pathogenesis and infections in immunocompromised hosts, and he has received numerous national and international awards for his advanced research and contribution to Infectious Diseases and Microbiology.
Contributor Information
Anton Y. Peleg, Email: anton.peleg@monash.edu.
Graeme N. Forrest, Rush University, Chicago, Illinois, USA
REFERENCES
- 1. Sollid JUE, Furberg AS, Hanssen AM, Johannessen M. 2014. Staphylococcus aureus: determinants of human carriage. Infect Genet Evol 21:531–541. doi: 10.1016/j.meegid.2013.03.020 [DOI] [PubMed] [Google Scholar]
- 2. Wertheim HFL, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5:751–762. doi: 10.1016/S1473-3099(05)70295-4 [DOI] [PubMed] [Google Scholar]
- 3. Krismer B, Weidenmaier C, Zipperer A, Peschel A. 2017. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat Rev Microbiol 15:675–687. doi: 10.1038/nrmicro.2017.104 [DOI] [PubMed] [Google Scholar]
- 4. Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 5. Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Esposito S, Noviello S, Leone S. 2016. Epidemiology and microbiology of skin and soft tissue infections. Curr Opin Infect Dis 29:109–115. doi: 10.1097/QCO.0000000000000239 [DOI] [PubMed] [Google Scholar]
- 7. Lew DP, Waldvogel FA. 2004. Osteomyelitis. Lancet 364:369–379. doi: 10.1016/S0140-6736(04)16727-5 [DOI] [PubMed] [Google Scholar]
- 8. Mylonakis E, Calderwood SB. 2001. Infective endocarditis in adults. N Engl J Med 345:1318–1330. doi: 10.1056/NEJMra010082 [DOI] [PubMed] [Google Scholar]
- 9. Rubinstein E, Kollef MH, Nathwani D. 2008. Pneumonia caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis 46 Suppl 5:S378–85. doi: 10.1086/533594 [DOI] [PubMed] [Google Scholar]
- 10. Hatlen TJ, Miller LG. 2021. Staphylococcal skin and soft tissue infections. Infect Dis Clin North Am 35:81–105. doi: 10.1016/j.idc.2020.10.003 [DOI] [PubMed] [Google Scholar]
- 11. Masters EA, Ricciardi BF, Bentley KL de M, Moriarty TF, Schwarz EM, Muthukrishnan G. 2022. Skeletal infections: microbial pathogenesis, immunity and clinical management. Nat Rev Microbiol 20:385–400. doi: 10.1038/s41579-022-00686-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Talha KM, DeSimone DC, Sohail MR, Baddour LM. 2020. Pathogen influence on epidemiology, diagnostic evaluation and management of infective endocarditis. Heart 106:1878–1882. doi: 10.1136/heartjnl-2020-317034 [DOI] [PubMed] [Google Scholar]
- 13. Pivard M, Moreau K, Vandenesch F. 2021. Staphylococcus aureus arsenal to conquer the lower respiratory tract. mSphere 6:e00059-21. doi: 10.1128/mSphere.00059-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi: 10.1038/nrmicro2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sakoulas G, Moellering RC. 2008. Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin Infect Dis 46 Suppl 5:S360–367. doi: 10.1086/533592 [DOI] [PubMed] [Google Scholar]
- 16. Lowy FD. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest 111:1265–1273. doi: 10.1172/JCI18535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giulieri SG, Tong SYC, Williamson DA. 2020. Using genomics to understand meticillin- and vancomycin-resistant Staphylococcus aureus infections. Microb Genom 6:e000324. doi: 10.1099/mgen.0.000324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nguyen AH, Hood KS, Mileykovskaya E, Miller WR, Tran TT. 2022. Bacterial cell membranes and their role in daptomycin resistance: a review. Front Mol Biosci 9:1035574. doi: 10.3389/fmolb.2022.1035574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, Holland TL, Fowler VG. 2019. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 17:203–218. doi: 10.1038/s41579-018-0147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cameron DR, Howden BP, Peleg AY. 2011. The interface between antibiotic resistance and virulence in Staphylococcus aureus and its impact upon clinical outcomes. Clin Infect Dis 53:576–582. doi: 10.1093/cid/cir473 [DOI] [PubMed] [Google Scholar]
- 21. Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis 36:53–59. doi: 10.1086/345476 [DOI] [PubMed] [Google Scholar]
- 22. Engemann JJ, Carmeli Y, Cosgrove SE, Fowler VG, Bronstein MZ, Trivette SL, Briggs JP, Sexton DJ, Kaye KS. 2003. Adverse clinical and economic outcomes attributable to methicillin resistance among patients with Staphylococcus aureus surgical site infection. Clin Infect Dis 36:592–598. doi: 10.1086/367653 [DOI] [PubMed] [Google Scholar]
- 23. Miyakis S, Brentnall S, Masso M, Reynolds G, Byrne MK, Newton P, Crawford S, Fish J, Nicholas B, Hill T, van Oijen AM, Wollongong Antimicrobial Resistance Research Alliance (WARRA) and One Health Understanding Through Bacterial Resistance to Antibiotics Knowledge (OUTBREAK) Consortium . 2022. Key predictors and burden of meticillin-resistant Staphylococcus aureus infection in comparison with meticillin-susceptible S. aureus infection in an Australian hospital setting. J Hosp Infect 129:41–48. doi: 10.1016/j.jhin.2022.07.004 [DOI] [PubMed] [Google Scholar]
- 24. Klein EY, Jiang W, Mojica N, Tseng KK, McNeill R, Cosgrove SE, Perl TM. 2019. National costs associated with methicillin-susceptible and methicillin-resistant Staphylococcus aureus hospitalizations in the United States, 2010-2014. Clin Infect Dis 68:22–28. doi: 10.1093/cid/ciy399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsiodras S, Gold HS, Sakoulas G, Eliopoulos GM, Wennersten C, Venkataraman L, Moellering RC, Ferraro MJ. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207–208. doi: 10.1016/S0140-6736(01)05410-1 [DOI] [PubMed] [Google Scholar]
- 26. Belousoff MJ, Eyal Z, Radjainia M, Ahmed T, Bamert RS, Matzov D, Bashan A, Zimmerman E, Mishra S, Cameron D, Elmlund H, Peleg AY, Bhushan S, Lithgow T, Yonath A. 2017. Structural basis for linezolid binding site rearrangement in the Staphylococcus aureus ribosome. mBio 8:e00395-17. doi: 10.1128/mBio.00395-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fowler VG, Boucher HW, Corey GR, Abrutyn E, Karchmer AW, Rupp ME, Levine DP, Chambers HF, Tally FP, Vigliani GA, et al. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med 355:653–665. doi: 10.1056/NEJMoa053783 [DOI] [PubMed] [Google Scholar]
- 28. Peleg AY, Miyakis S, Ward DV, Earl AM, Rubio A, Cameron DR, Pillai S, Moellering RC, Eliopoulos GM. 2012. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 7:e28316. doi: 10.1371/journal.pone.0028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones RN, Mendes RE, Sader HS. 2010. Ceftaroline activity against pathogens associated with complicated skin and skin structure infections: results from an international surveillance study. J Antimicrob Chemother 65 Suppl 4:iv17–iv31. doi: 10.1093/jac/dkq252 [DOI] [PubMed] [Google Scholar]
- 30. McNeil JC, Sommer LM, Vallejo JG, Hulten KG, Kaplan SL, Flores AR. 2022. Reduced ceftaroline susceptibility among invasive MRSA infections in children: a clinical and genomic investigation. Antimicrob Agents Chemother 66:e0074522. doi: 10.1128/aac.00745-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, Shah S, Rudrik JT, Pupp GR, Brown WJ, Cardo D, Fridkin SK, Vancomycin-Resistant Staphylococcus aureus Investigative Team . 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med 348:1342–1347. doi: 10.1056/NEJMoa025025 [DOI] [PubMed] [Google Scholar]
- 32. Sujatha S, Praharaj I. 2012. Glycopeptide resistance in Gram-positive cocci: a review. Interdiscip Perspect Infect Dis 2012:781679. doi: 10.1155/2012/781679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O’Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan H-L, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lakhundi S, Zhang K. 2018. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31:e00020-18. doi: 10.1128/CMR.00020-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Foster TJ, Geoghegan JA, Ganesh VK, Höök M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. doi: 10.1038/nrmicro3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fleury OM, McAleer MA, Feuillie C, Formosa-Dague C, Sansevere E, Bennett DE, Towell AM, McLean WHI, Kezic S, Robinson DA, Fallon PG, Foster TJ, Dufrêne YF, Irvine AD, Geoghegan JA. 2017. Clumping factor B promotes adherence of Staphylococcus aureus to corneocytes in atopic dermatitis. Infect Immun 85:e00994-16. doi: 10.1128/IAI.00994-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lower SK, Lamlertthon S, Casillas-Ituarte NN, Lins RD, Yongsunthon R, Taylor ES, DiBartola AC, Edmonson C, McIntyre LM, Reller LB, Que YA, Ros R, Lower BH, Fowler VG. 2011. Polymorphisms in fibronectin binding protein A of Staphylococcus aureus are associated with infection of cardiovascular devices. Proc Natl Acad Sci USA 108:18372–18377. doi: 10.1073/pnas.1109071108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dinges MM, Orwin PM, Schlievert PM. 2000. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 13:16–34, doi: 10.1128/CMR.13.1.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ladhani S, Joannou CL, Lochrie DP, Evans RW, Poston SM. 1999. Clinical, microbial, and biochemical aspects of the exfoliative toxins causing staphylococcal scalded-skin syndrome. Clin Microbiol Rev 12:224–242. doi: 10.1128/CMR.12.2.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Todd J, Fishaut M, Kapral F, Welch T. 1978. Toxic-shock syndrome associated with phage-group-I staphylococci. Lancet 2:1116–1118. doi: 10.1016/s0140-6736(78)92274-2 [DOI] [PubMed] [Google Scholar]
- 41. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29:1128–1132. doi: 10.1086/313461 [DOI] [PubMed] [Google Scholar]
- 42. Wang R, Braughton KR, Kretschmer D, Bach T-HL, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13:1510–1514. doi: 10.1038/nm1656 [DOI] [PubMed] [Google Scholar]
- 43. Berry KA, Verhoef MTA, Leonard AC, Cox G. 2022. Staphylococcus aureus adhesion to the host. Ann N Y Acad Sci 1515:75–96. doi: 10.1111/nyas.14807 [DOI] [PubMed] [Google Scholar]
- 44. Spaan AN, van Strijp JAG, Torres VJ. 2017. Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat Rev Microbiol 15:435–447. doi: 10.1038/nrmicro.2017.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Otto M. 2013. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays 35:4–11. doi: 10.1002/bies.201200112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miragaia M. 2018. Factors contributing to the evolution of mecA-mediated β-lactam resistance in staphylococci: update and new insights from whole genome sequencing (WGS). Front Microbiol 9:2723. doi: 10.3389/fmicb.2018.02723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Copin R, Sause WE, Fulmer Y, Balasubramanian D, Dyzenhaus S, Ahmed JM, Kumar K, Lees J, Stachel A, Fisher JC, Drlica K, Phillips M, Weiser JN, Planet PJ, Uhlemann A-C, Altman DR, Sebra R, van Bakel H, Lighter J, Torres VJ, Shopsin B. 2019. Sequential evolution of virulence and resistance during clonal spread of community-acquired methicillin-resistant Staphylococcus aureus Proc Natl Acad Sci USA 116:1745–1754. doi: 10.1073/pnas.1814265116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. DeLeo FR, Kennedy AD, Chen L, Bubeck Wardenburg J, Kobayashi SD, Mathema B, Braughton KR, Whitney AR, Villaruz AE, Martens CA, Porcella SF, McGavin MJ, Otto M, Musser JM, Kreiswirth BN. 2011. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc Natl Acad Sci USA 108:18091–18096. doi: 10.1073/pnas.1111084108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guérillot R, Kostoulias X, Donovan L, Li L, Carter GP, Hachani A, Vandelannoote K, Giulieri S, Monk IR, Kunimoto M, Starrs L, Burgio G, Seemann T, Peleg AY, Stinear TP, Howden BP. 2019. Unstable chromosome rearrangements in Staphylococcus aureus cause phenotype switching associated with persistent infections. Proc Natl Acad Sci USA 116:20135–20140. doi: 10.1073/pnas.1904861116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jiang JH, Bhuiyan MS, Shen HH, Cameron DR, Rupasinghe TWT, Wu CM, Le Brun AP, Kostoulias X, Domene C, Fulcher AJ, McConville MJ, Howden BP, Lieschke GJ, Peleg AY. 2019. Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc Natl Acad Sci USA 116:3722–3727. doi: 10.1073/pnas.1812066116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Skinner D, Keefer CS. 1941. Significance of bacteremia caused by Staphylococcus aureus: a study of one hundred and twenty-two cases and a review of the literature concerned with experimental infection in animals. Arch Intern Med 68:851–875. doi: 10.1001/archinte.1941.00200110003001 [DOI] [Google Scholar]
- 52. Tipper DJ, Strominger JL. 1965. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci USA 54:1133–1141. doi: 10.1073/pnas.54.4.1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barber M, Rozwadowska-Dowzenko M. 1948. Infection by penicillin-resistant staphylococci. Lancet 2:641–644. doi: 10.1016/s0140-6736(48)92166-7 [DOI] [PubMed] [Google Scholar]
- 54. Bondi A, Dietz CC. 1945. Penicillin resistant staphylococci. Proc Soc Exp Biol Med 60:55–58. doi: 10.3181/00379727-60-15089 [DOI] [PubMed] [Google Scholar]
- 55. Abraham EP, Chain E. 1988. An enzyme from bacteria able to destroy penicillin. Rev Infect Dis 10:677–678. [PubMed] [Google Scholar]
- 56. Kirby WM. 1944. Extraction of a highly potent penicillin inactivator from penicillin resistant staphylococci. Science 99:452–453. doi: 10.1126/science.99.2579.452 [DOI] [PubMed] [Google Scholar]
- 57. Medeiros AA. 1997. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin Infect Dis 24 Suppl 1:S19–45. doi: 10.1093/clinids/24.supplement_1.s19 [DOI] [PubMed] [Google Scholar]
- 58. Rolinson GN. 1961. Celbenin” - resistant staphylococci. BMJ 1:125–126. doi: 10.1136/bmj.1.5219.125 [DOI] [Google Scholar]
- 59. Rountree PM, Beard MA. 1968. Hospital strains of Staphylococcus aureus, with particular reference to methicillin-resistant strains. Med J Aust 2:1163–1168. doi: 10.5694/j.1326-5377.1968.tb83502.x [DOI] [PubMed] [Google Scholar]
- 60. Barrett FF, McGehee RF, Finland M. 1968. Methicillin-resistant Staphylococcus aureus at Boston city hospital . N Engl J Med 279:441–448. doi: 10.1056/NEJM196808292790901 [DOI] [PubMed] [Google Scholar]
- 61. Rosendal K, Bülow P, Bentzon MW, Eriksen KR. 1976. Staphylococcus aureus strains isolated in Danish hospitals from January 1st. Acta Pathol Microbiol Scand B 84B:359–368. doi: 10.1111/j.1699-0463.1976.tb01953.x [DOI] [PubMed] [Google Scholar]
- 62. PAL SC, RAY BG. 1964. Methicillin-resistant staphylococci. J Indian Med Assoc 42:512–517. [PubMed] [Google Scholar]
- 63. Katayama Y, Ito T, Hiramatsu K. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 44:1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Matsuhashi M, Song MD, Ishino F, Wachi M, Doi M, Inoue M, Ubukata K, Yamashita N, Konno M. 1986. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J Bacteriol 167:975–980. doi: 10.1128/jb.167.3.975-980.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ito T, Katayama Y, Hiramatsu K. 1999. Cloning and nucleotide sequence determination of the entire MEC DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother 43:1449–1458. doi: 10.1128/AAC.43.6.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hartman BJ, Tomasz A. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol 158:513–516. doi: 10.1128/jb.158.2.513-516.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pinho Mariana G., Filipe SR, de Lencastre H, Tomasz A. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus . J Bacteriol 183:6525–6531. doi: 10.1128/JB.183.22.6525-6531.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pinho M G, de Lencastre H, Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci USA 98:10886–10891. doi: 10.1073/pnas.191260798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Centers for Disease Control & Prevention . 1981. Community-acquired methicillin-resistant Staphylococcus aureus infections - Michigan. Morb Mortal Wkly Rep 30:185–187. [PubMed] [Google Scholar]
- 70. Saravolatz LD, Markowitz N, Arking L, Pohlod D, Fisher E. 1982. Methicillin-resistant Staphylococcus aureus. Epidemiologic observations during a community-acquired outbreak. Ann Intern Med 96:11–16. doi: 10.7326/0003-4819-96-1-11 [DOI] [PubMed] [Google Scholar]
- 71. Levine DP, Cushing RD, Jui J, Brown WJ. 1982. Community-acquired methicillin-resistant Staphylococcus aureus endocarditis in the detroit medical center. Ann Intern Med 97:330–338. doi: 10.7326/0003-4819-97-3-330 [DOI] [PubMed] [Google Scholar]
- 72. David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. doi: 10.1128/CMR.00081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bal AM, Coombs GW, Holden MTG, Lindsay JA, Nimmo GR, Tattevin P, Skov RL. 2016. Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: blurring of the traditional definitions. J Glob Antimicrob Resist 6:95–101. doi: 10.1016/j.jgar.2016.04.004 [DOI] [PubMed] [Google Scholar]
- 74. Weese JS. 2010. Methicillin-resistant Staphylococcus aureus in animals. ILAR J 51:233–244. doi: 10.1093/ilar.51.3.233 [DOI] [PubMed] [Google Scholar]
- 75. Aires-de-Sousa M. 2017. Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin Microbiol Infect 23:373–380. doi: 10.1016/j.cmi.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 76. Cuny C, Witte W. 2017. MRSA in equine hospitals and its significance for infections in humans. Vet Microbiol 200:59–64. doi: 10.1016/j.vetmic.2016.01.013 [DOI] [PubMed] [Google Scholar]
- 77. Agnoletti F, Mazzolini E, Bacchin C, Bano L, Berto G, Rigoli R, Muffato G, Coato P, Tonon E, Drigo I. 2014. First reporting of methicillin-resistant Staphylococcus aureus (MRSA) ST398 in an industrial rabbit holding and in farm-related people. Vet Microbiol 170:172–177. doi: 10.1016/j.vetmic.2014.01.035 [DOI] [PubMed] [Google Scholar]
- 78. Kaspar U, von Lützau A, Schlattmann A, Roesler U, Köck R, Becker K. 2018. Zoonotic multidrug-resistant microorganisms among small companion animals in Germany. PLoS One 13:e0208364. doi: 10.1371/journal.pone.0208364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bortolaia V, Espinosa-Gongora C, Guardabassi L. 2016. Human health risks associated with antimicrobial-resistant enterococci and Staphylococcus aureus on poultry meat. Clin Microbiol Infect 22:130–140. doi: 10.1016/j.cmi.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 80. Lowder BV, Guinane CM, Ben Zakour NL, Weinert LA, Conway-Morris A, Cartwright RA, Simpson AJ, Rambaut A, Nübel U, Fitzgerald JR. 2009. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc Natl Acad Sci USA 106:19545–19550. doi: 10.1073/pnas.0909285106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, Voss A, Kluytmans J. 2007. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis 13:1834–1839. doi: 10.3201/eid1312.070384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Graveland H, Wagenaar JA, Bergs K, Heesterbeek H, Heederik D. 2011. Persistence of livestock associated MRSA CC398 in humans is dependent on intensity of animal contact. PLoS One 6:e16830. doi: 10.1371/journal.pone.0016830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kadlec K, Fessler AT, Hauschild T, Schwarz S. 2012. Novel and uncommon antimicrobial resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect 18:745–755. doi: 10.1111/j.1469-0691.2012.03842.x [DOI] [PubMed] [Google Scholar]
- 84. Sieber RN, Skov RL, Nielsen J, Schulz J, Price LB, Aarestrup FM, Larsen AR, Stegger M, Larsen J. 2018. Drivers and dynamics of methicillin-resistant livestock-associated Staphylococcus aureus. mBio 9:e02142-18. doi: 10.1128/mBio.02142-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pantosti A. 2012. Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front Microbiol 3:127. doi: 10.3389/fmicb.2012.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. García-Álvarez L, Holden MTG, Lindsay H, Webb CR, Brown DFJ, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RLR, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. 2011. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis 11:595–603. doi: 10.1016/S1473-3099(11)70126-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Larsen J, Raisen CL, Ba X, Sadgrove NJ, Padilla-González GF, Simmonds MSJ, Loncaric I, Kerschner H, Apfalter P, Hartl R, et al. 2022. Emergence of methicillin resistance predates the clinical use of antibiotics. Nature 602:135–141. doi: 10.1038/s41586-021-04265-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rao Q, Shang W, Hu X, Rao X. 2015. Staphylococcus aureus ST121: a globally disseminated hypervirulent clone. J Med Microbiol 64:1462–1473. doi: 10.1099/jmm.0.000185 [DOI] [PubMed] [Google Scholar]
- 90. Bowers JR, Driebe EM, Albrecht V, McDougal LK, Granade M, Roe CC, Lemmer D, Rasheed JK, Engelthaler DM, Keim P, Limbago BM, Fey PD. 2018. Improved subtyping of Staphylococcus aureus clonal complex 8 strains based on whole-genome phylogenetic analysis. mSphere 3:e00464-17. doi: 10.1128/mSphere.00464-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Parizad EG, Parizad EG, Valizadeh A. 2016. The application of pulsed field gel electrophoresis in clinical studies. J Clin Diagn Res 10:DE01–DE04. doi: 10.7860/JCDR/2016/15718.7043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kazakova SV, Hageman JC, Matava M, Srinivasan A, Phelan L, Garfinkel B, Boo T, McAllister S, Anderson J, Jensen B, Dodson D, Lonsway D, McDougal LK, Arduino M, Fraser VJ, Killgore G, Tenover FC, Cody S, Jernigan DB. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med 352:468–475. doi: 10.1056/NEJMoa042859 [DOI] [PubMed] [Google Scholar]
- 94. Weller TM. 2000. Methicillin-resistant Staphylococcus aureus typing methods: which should be the international standard J Hosp Infect 44:160–172. doi: 10.1053/jhin.1999.0701 [DOI] [PubMed] [Google Scholar]
- 95. Goering RV. 2010. Pulsed field gel electrophoresis: a review of application and interpretation in the molecular epidemiology of infectious disease. Infect Genet Evol 10:866–875. doi: 10.1016/j.meegid.2010.07.023 [DOI] [PubMed] [Google Scholar]
- 96. Cookson BD, Aparicio P, Deplano A, Struelens M, Goering R, Marples R. 1996. Inter-centre comparison of pulsed-field gel electrophoresis for the typing of methicillin-resistant Staphylococcus aureus. J Med Microbiol 44:179–184. doi: 10.1099/00222615-44-3-179 [DOI] [PubMed] [Google Scholar]
- 97. van Belkum A, van Leeuwen W, Kaufmann ME, Cookson B, Forey F, Etienne J, Goering R, Tenover F, Steward C, O’Brien F, Grubb W, Tassios P, Legakis N, Morvan A, El Solh N, de Ryck R, Struelens M, Salmenlinna S, Vuopio-Varkila J, Kooistra M, Talens A, Witte W, Verbrugh H. 1998. Assessment of resolution and Intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J Clin Microbiol 36:1653–1659. doi: 10.1128/JCM.36.6.1653-1659.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Boyle-Vavra S, Daum RS, Hiramatsu K. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother 46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wilson LK, Coombs GW, Christiansen K, Grubb WB, O’Brien FG. 2016. Characterization of a novel staphylococcal cassette chromosome composite island from community-associated MRSA isolated in aged care facilities in Western Australia. J Antimicrob Chemother 71:3372–3375. doi: 10.1093/jac/dkw317 [DOI] [PubMed] [Google Scholar]
- 102. Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. 2004. Novel type V staphylococcal cassette chromosome MEC driven by a novel cassette chromosome recombinase, ccrC . Antimicrob Agents Chemother 48:2637–2651. doi: 10.1128/AAC.48.7.2637-2651.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Boundy S, Safo MK, Wang L, Musayev FN, O’Farrell HC, Rife JP, Archer GL. 2013. Characterization of the Staphylococcus aureus rRNA methyltransferase encoded by orfX, the gene containing the staphylococcal chromosome cassette mec (SCCmec) insertion site. J Biol Chem 288:132–140. doi: 10.1074/jbc.M112.385138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hiramatsu K, Cui L, Kuroda M, Ito T. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol 9:486–493. doi: 10.1016/s0966-842x(01)02175-8 [DOI] [PubMed] [Google Scholar]
- 106. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) . 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. doi: 10.1128/AAC.00579-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Urushibara N, Aung MS, Kawaguchiya M, Kobayashi N. 2020. Novel staphylococcal cassette chromosome mec (SCCmec) type XIV (5A) and a truncated SCCmec element in SCC composite Islands carrying speG in ST5 MRSA in Japan. J Antimicrob Chemother 75:46–50. doi: 10.1093/jac/dkz406 [DOI] [PubMed] [Google Scholar]
- 108. Wang W, Hu Y, Baker M, Dottorini T, Li H, Dong Y, Bai Y, Fanning S, Li F. 2022. Novel SCCmec type XV (7A) and two pseudo-SCCmec variants in foodborne MRSA in China. J Antimicrob Chemother 77:903–909. doi: 10.1093/jac/dkab500 [DOI] [PubMed] [Google Scholar]
- 109. Milheiriço C, Oliveira DC, de Lencastre H. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: 'SCCmec IV Multiplex J Antimicrob Chemother 60:42–48. doi: 10.1093/jac/dkm112 [DOI] [PubMed] [Google Scholar]
- 110. Zhang K, McClure JA, Conly JM. 2012. Enhanced multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. Mol Cell Probes 26:218–221. doi: 10.1016/j.mcp.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 111. Kaya H, Hasman H, Larsen J, Stegger M, Johannesen TB, Allesøe RL, Lemvigh CK, Aarestrup FM, Lund O, Larsen AR, Limbago BM. 2018. Sccmecfinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3. doi: 10.1128/mSphere.00612-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Didelot X, Bowden R, Wilson DJ, Peto TEA, Crook DW. 2012. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13:601–612. doi: 10.1038/nrg3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Planet PJ, Narechania A, Chen L, Mathema B, Boundy S, Archer G, Kreiswirth B. 2017. Architecture of a species: phylogenomics of Staphylococcus aureus. Trends Microbiol 25:153–166. doi: 10.1016/j.tim.2016.09.009 [DOI] [PubMed] [Google Scholar]
- 114. Aanensen DM, Feil EJ, Holden MTG, Dordel J, Yeats CA, Fedosejev A, Goater R, Castillo-Ramírez S, Corander J, Colijn C, Chlebowicz MA, Schouls L, Heck M, Pluister G, Ruimy R, Kahlmeter G, Åhman J, Matuschek E, Friedrich AW, Parkhill J, Bentley SD, Spratt BG, Grundmann H, European SRL Working Group . 2016. Whole-genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. mBio 7:e00444-16. doi: 10.1128/mBio.00444-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Köser CU, Holden MTG, Ellington MJ, Cartwright EJP, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Miller RM, Price JR, Batty EM, Didelot X, Wyllie D, Golubchik T, Crook DW, Paul J, Peto TEA, Wilson DJ, Cule M, Ip CLC, Day NPJ, Moore CE, Bowden R, Llewelyn MJ. 2014. Healthcare-associated outbreak of meticillin-resistant Staphylococcus aureus bacteraemia: role of a cryptic variant of an epidemic clone. J Hosp Infect 86:83–89. doi: 10.1016/j.jhin.2013.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Price JR, Golubchik T, Cole K, Wilson DJ, Crook DW, Thwaites GE, Bowden R, Walker AS, Peto TEA, Paul J, Llewelyn MJ. 2014. Whole-genome sequencing shows that patient-to-patient transmission rarely accounts for acquisition of Staphylococcus aureus in an intensive care unit. Clin Infect Dis 58:609–618. doi: 10.1093/cid/cit807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Azarian T, Cook RL, Johnson JA, Guzman N, McCarter YS, Gomez N, Rathore MH, Morris JG, Salemi M. 2015. Whole-genome sequencing for outbreak investigations of methicillin-resistant Staphylococcus aureus in the neonatal intensive care unit: time for routine practice. Infect Control Hosp Epidemiol 36:777–785. doi: 10.1017/ice.2015.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Simar SR, Hanson BM, Arias CA. 2021. Techniques in bacterial strain typing: past, present, and future. Curr Opin Infect Dis 34:339–345. doi: 10.1097/QCO.0000000000000743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mellmann A, Bletz S, Böking T, Kipp F, Becker K, Schultes A, Prior K, Harmsen D. 2016. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol 54:2874–2881. doi: 10.1128/JCM.00790-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Harris SR, Cartwright EJP, Török ME, Holden MTG, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 13:130–136. doi: 10.1016/S1473-3099(12)70268-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Coll F, Raven KE, Knight GM, Blane B, Harrison EM, Leek D, Enoch DA, Brown NM, Parkhill J, Peacock SJ. 2020. Definition of a genetic relatedness cutoff to exclude recent transmission of meticillin-resistant Staphylococcus aureus: a genomic epidemiology analysis. Lancet Microbe 1:e328–e335. doi: 10.1016/S2666-5247(20)30149-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TEA, Crook DW, Walker AS, Golubchik T, Carroll KC. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191. doi: 10.1128/JCM.03117-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Alam MT, Petit RA, Crispell EK, Thornton TA, Conneely KN, Jiang Y, Satola SW, Read TD. 2014. Dissecting vancomycin-intermediate resistance in Staphylococcus aureus using genome-wide association. Genome Biol Evol 6:1174–1185. doi: 10.1093/gbe/evu092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Su M, Satola SW, Read TD. 2019. Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 57:e01405-18. doi: 10.1128/JCM.01405-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Durand G, Javerliat F, Bes M, Veyrieras J-B, Guigon G, Mugnier N, Schicklin S, Kaneko G, Santiago-Allexant E, Bouchiat C, Martins-Simões P, Laurent F, Van Belkum A, Vandenesch F, Tristan A. 2018. Routine whole-genome sequencing for outbreak investigations of Staphylococcus aureus in a national reference center. Front Microbiol 9:511. doi: 10.3389/fmicb.2018.00511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Xavier BB, Mysara M, Bolzan M, Ribeiro-Gonçalves B, Alako BTF, Harrison P, Lammens C, Kumar-Singh S, Goossens H, Carriço JA, Cochrane G, Malhotra-Kumar S. 2020. Bacpipe: a rapid, user-friendly whole-genome sequencing pipeline for clinical diagnostic bacteriology. iScience 23:100769. doi: 10.1016/j.isci.2019.100769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Howden BP, Giulieri SG, Wong Fok Lung T, Baines SL, Sharkey LK, Lee JYH, Hachani A, Monk IR, Stinear TP. 2023. Staphylococcus aureus host interactions and adaptation. Nat Rev Microbiol 21:380–395. doi: 10.1038/s41579-023-00852-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Casadevall A, Pirofski LA. 1999. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun 67:3703–3713. doi: 10.1128/IAI.67.8.3703-3713.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. O’Riordan K, Lee JC. 2004. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev 17:218–234. doi: 10.1128/CMR.17.1.218-234.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. McDevitt D, Francois P, Vaudaux P, Foster TJ. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol 11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x [DOI] [PubMed] [Google Scholar]
- 133. Jönsson K, Signäs C, Müller HP, Lindberg M. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. the complete nucleotide sequence and characterization of the second gene. Eur J Biochem 202:1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x [DOI] [PubMed] [Google Scholar]
- 134. Switalski LM, Speziale P, Höök M. 1989. Isolation and characterization of a putative collagen receptor from Staphylococcus aureus strain cowan 1. J Biol Chem 264:21080–21086. [PubMed] [Google Scholar]
- 135. Tung H s, Guss B, Hellman U, Persson L, Rubin K, Rydén C. 2000. A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal SDR family. Biochem J 345 Pt 3:611–619. [PMC free article] [PubMed] [Google Scholar]
- 136. Patti JM, Allen BL, McGavin MJ, Höök M. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol 48:585–617. doi: 10.1146/annurev.mi.48.100194.003101 [DOI] [PubMed] [Google Scholar]
- 137. McCarthy AJ, Lindsay JA. 2010. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol 10:173. doi: 10.1186/1471-2180-10-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Deisenhofer J. 1981. Crystallographic refinement and atomic models of a human FC fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry 20:2361–2370. [PubMed] [Google Scholar]
- 139. Cedergren L, Andersson R, Jansson B, Uhlén M, Nilsson B. 1993. Mutational analysis of the interaction between staphylococcal protein A and human Igg1. Protein Eng 6:441–448. doi: 10.1093/protein/6.4.441 [DOI] [PubMed] [Google Scholar]
- 140. Silverman GJ, Goodyear CS. 2006. Confounding B-cell defences: lessons from a staphylococcal superantigen. Nat Rev Immunol 6:465–475. doi: 10.1038/nri1853 [DOI] [PubMed] [Google Scholar]
- 141. Graille M, Stura EA, Corper AL, Sutton BJ, Taussig MJ, Charbonnier JB, Silverman GJ. 2000. Crystal structure of a Staphylococcus aureus protein A domain complexed with the fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci USA 97:5399–5404. doi: 10.1073/pnas.97.10.5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ulloa-Morales AJ, Goodyear CS, Silverman GJ. 2018. Essential domain-dependent roles within soluble IgG for In vivo superantigen properties of staphylococcal protein A: resolving the B-cell superantigen paradox. Front Immunol 9:2011. doi: 10.3389/fimmu.2018.02011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Gómez MI, Lee A, Reddy B, Muir A, Soong G, Pitt A, Cheung A, Prince A. 2004. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat Med 10:842–848. doi: 10.1038/nm1079 [DOI] [PubMed] [Google Scholar]
- 144. Gómez MI, O’Seaghdha M, Magargee M, Foster TJ, Prince AS. 2006. Staphylococcus aureus protein A activates TNFR1 signaling through conserved IgG binding domains. J Biol Chem 281:20190–20196. doi: 10.1074/jbc.M601956200 [DOI] [PubMed] [Google Scholar]
- 145. Martin FJ, Gomez MI, Wetzel DM, Memmi G, O’Seaghdha M, Soong G, Schindler C, Prince A. 2009. Staphylococcus aureus activates type I IFN signaling in mice and humans through the Xr repeated sequences of protein A. J Clin Invest 119:1931–1939. doi: 10.1172/jci35879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Burman JD, Leung E, Atkins KL, O’Seaghdha MN, Lango L, Bernadó P, Bagby S, Svergun DI, Foster TJ, Isenman DE, van den Elsen JMH. 2008. Interaction of human complement with Sbi, a staphylococcal immunoglobulin-binding protein: indications of a novel mechanism of complement evasion by Staphylococcus aureus. J Biol Chem 283:17579–17593. doi: 10.1074/jbc.M800265200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Upadhyay A, Burman JD, Clark EA, Leung E, Isenman DE, van den Elsen JMH, Bagby S. 2008. Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi. J Biol Chem 283:22113–22120. doi: 10.1074/jbc.M802636200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kengatharan KM, De Kimpe S, Robson C, Foster SJ, Thiemermann C. 1998. Mechanism of gram-positive shock: identification of peptidoglycan and lipoteichoic acid moieties essential in the induction of nitric oxide synthase, shock, and multiple organ failure. J Exp Med 188:305–315. doi: 10.1084/jem.188.2.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. De Kimpe SJ, Kengatharan M, Thiemermann C, Vane JR. 1995. The cell wall components peptidoglycan and lipoteichoic acid from Staphylococcus aureus act in synergy to cause shock and multiple organ failure. Proc Natl Acad Sci U.S.A 92:10359–10363. doi: 10.1073/pnas.92.22.10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Wang JE, Jørgensen PF, Almlöf M, Thiemermann C, Foster SJ, Aasen AO, Solberg R. 2000. Peptidoglycan and lipoteichoic acid from Staphylococcus aureus induce tumor necrosis factor alpha, interleukin 6 (IL-6), and IL-10 production in both T cells and monocytes in a human whole blood model. Infect Immun 68:3965–3970. doi: 10.1128/IAI.68.7.3965-3970.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Myhre AE, Stuestøl JF, Dahle MK, Øverland G, Thiemermann C, Foster SJ, Lilleaasen P, Aasen AO, Wang JE. 2004. Organ injury and cytokine release caused by peptidoglycan are dependent on the structural integrity of the glycan chain. Infect Immun 72:1311–1317. doi: 10.1128/IAI.72.3.1311-1317.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Müller S, Wolf AJ, Iliev ID, Berg BL, Underhill DM, Liu GY. 2015. Poorly cross-linked peptidoglycan in MRSA due to mecA induction activates the inflammasome and exacerbates immunopathology. Cell Host Microbe 18:604–612. doi: 10.1016/j.chom.2015.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Wanner S, Schade J, Keinhörster D, Weller N, George SE, Kull L, Bauer J, Grau T, Winstel V, Stoy H, Kretschmer D, Kolata J, Wolz C, Bröker BM, Weidenmaier C. 2017. Wall teichoic acids mediate increased virulence in Staphylococcus aureus. Nat Microbiol 2:17048. doi: 10.1038/nmicrobiol.2017.48 [DOI] [PubMed] [Google Scholar]
- 154. Powers ME, Bubeck Wardenburg J. 2014. Igniting the fire: Staphylococcus aureus virulence factors in the pathogenesis of sepsis. PLoS Pathog 10:e1003871. doi: 10.1371/journal.ppat.1003871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Seilie ES, Bubeck Wardenburg J. 2017. Staphylococcus aureus pore-forming toxins: the interface of pathogen and host complexity. Semin Cell Dev Biol 72:101–116. doi: 10.1016/j.semcdb.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Alonzo F, Torres VJ. 2014. The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol Mol Biol Rev 78:199–230. doi: 10.1128/MMBR.00055-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. 1996. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274:1859–1866. doi: 10.1126/science.274.5294.1859 [DOI] [PubMed] [Google Scholar]
- 158. von Hoven G, Qin Q, Neukirch C, Husmann M, Hellmann N. 2019. Staphylococcus aureus alpha-toxin: small pore, large consequences. Biol Chem 400:1261–1276. doi: 10.1515/hsz-2018-0472 [DOI] [PubMed] [Google Scholar]
- 159. Gouaux E. 1998. Alpha-hemolysin from Staphylococcus aureus: an archetype of beta-barrel, channel-forming toxins. J Struct Biol 121:110–122. doi: 10.1006/jsbi.1998.3959 [DOI] [PubMed] [Google Scholar]
- 160. Hildebrand A, Pohl M, Bhakdi S. 1991. Staphylococcus aureus alpha-toxin. Dual mechanism of binding to target cells. J Biol Chem 266:17195–17200. doi: 10.1016/S0021-9258(19)47358-4 [DOI] [PubMed] [Google Scholar]
- 161. Lee B, Olaniyi R, Kwiecinski JM, Wardenburg JB. 2020. Staphylococcus aureus toxin suppresses antigen-specific T cell responses. J Clin Invest 130:1122–1127. doi: 10.1172/JCI130728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Wilke GA, Bubeck Wardenburg J. 2010. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus alpha-hemolysin-mediated cellular injury. Proc Natl Acad Sci USA 107:13473–13478. doi: 10.1073/pnas.1001815107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Inoshima I, Inoshima N, Wilke GA, Powers ME, Frank KM, Wang Y, Bubeck Wardenburg J. 2011. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat Med 17:1310–1314. doi: 10.1038/nm.2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Inoshima N, Wang Y, Bubeck Wardenburg J. 2012. Genetic requirement for ADAM10 in severe Staphylococcus aureus skin infection. J Invest Dermatol 132:1513–1516. doi: 10.1038/jid.2011.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Kennedy AD, Bubeck Wardenburg J, Gardner DJ, Long D, Whitney AR, Braughton KR, Schneewind O, DeLeo FR. 2010. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J Infect Dis 202:1050–1058. doi: 10.1086/656043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Bubeck Wardenburg J, Patel RJ, Schneewind O. 2007. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun 75:1040–1044. doi: 10.1128/IAI.01313-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O. 2007. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med 13:1405–1406. doi: 10.1038/nm1207-1405 [DOI] [PubMed] [Google Scholar]
- 168. Bubeck Wardenburg J, Schneewind O. 2008. Vaccine protection against Staphylococcus aureus pneumonia. J Exp Med 205:287–294. doi: 10.1084/jem.20072208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Sampedro GR, DeDent AC, Becker REN, Berube BJ, Gebhardt MJ, Cao H, Bubeck Wardenburg J. 2014. Targeting Staphylococcus aureus alpha-toxin as a novel approach to reduce severity of recurrent skin and soft-tissue infections. J Infect Dis 210:1012–1018. doi: 10.1093/infdis/jiu223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Jordan PM, Gerstmeier J, Pace S, Bilancia R, Rao Z, Börner F, Miek L, Gutiérrez-Gutiérrez Ó, Arakandy V, Rossi A, Ialenti A, González-Estévez C, Löffler B, Tuchscherr L, Serhan CN, Werz O. 2020. Staphylococcus aureus-derived alpha-hemolysin evokes generation of specialized pro-resolving mediators promoting inflammation resolution. Cell Rep 33:108247. doi: 10.1016/j.celrep.2020.108247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Woodin AM. 1960. Purification of the two components of leucocidin from Staphylococcus aureus. Biochem J 75:158–165. doi: 10.1042/bj0750158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wooodin AM1959. Fractionation of a leucocidin from Staphylococcus aureus. Biochem J 73:225–237. doi: 10.1042/bj0730225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Tam K, Torres VJ. 2019. Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol Spectr 7:16. doi: 10.1128/microbiolspec.GPP3-0039-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Peschel A, Otto M. 2013. Phenol-soluble modulins and staphylococcal infection. Nat Rev Microbiol 11:667–673. doi: 10.1038/nrmicro3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Otto M. 2014. Phenol-soluble modulins. Int J Med Microbiol 304:164–169. doi: 10.1016/j.ijmm.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Queck SY, Khan BA, Wang R, Bach T-HL, Kretschmer D, Chen L, Kreiswirth BN, Peschel A, Deleo FR, Otto M. 2009. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog 5:e1000533. doi: 10.1371/journal.ppat.1000533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Chatterjee SS, Joo H-S, Duong AC, Dieringer TD, Tan VY, Song Y, Fischer ER, Cheung GYC, Li M, Otto M. 2013. Essential Staphylococcus aureus toxin export system. Nat Med 19:364–367. doi: 10.1038/nm.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. McCarthy AJ, Lindsay JA. 2013. Staphylococcus aureus innate immune evasion is lineage-specific: a bioinfomatics study. Infect Genet Evol 19:7–14. doi: 10.1016/j.meegid.2013.06.012 [DOI] [PubMed] [Google Scholar]
- 179. Tayeb-Fligelman E, Tabachnikov O, Moshe A, Goldshmidt-Tran O, Sawaya MR, Coquelle N, Colletier JP, Landau M. 2017. The cytotoxic Staphylococcus aureus PSMalpha3 reveals a cross-alpha amyloid-like fibril. Science 355:831–833. doi: 10.1126/science.aaf4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Muñoz-Planillo R, Hasegawa M, Villaruz AE, Cheung GYC, McGavin MJ, Travers JB, Otto M, Inohara N, Núñez G. 2013. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature 503:397–401. doi: 10.1038/nature12655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Grosz M, Kolter J, Paprotka K, Winkler A-C, Schäfer D, Chatterjee SS, Geiger T, Wolz C, Ohlsen K, Otto M, Rudel T, Sinha B, Fraunholz M. 2014. Cytoplasmic replication of Staphylococcus aureus upon phagosomal escape triggered by phenol-soluble modulin alpha. Cell Microbiol 16:451–465. doi: 10.1111/cmi.12233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Periasamy S, Joo H-S, Duong AC, Bach T-HL, Tan VY, Chatterjee SS, Cheung GYC, Otto M. 2012. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci USA 109:1281–1286. doi: 10.1073/pnas.1115006109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Salinas N, Colletier JP, Moshe A, Landau M. 2018. Extreme amyloid polymorphism in Staphylococcus aureus virulent PSMalpha peptides. Nat Commun 9:3512. doi: 10.1038/s41467-018-05490-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. 2012. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog 8:e1002744. doi: 10.1371/journal.ppat.1002744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Bergdoll MS, Crass BA, Reiser RF, Robbins RN, Davis JP. 1981. A new staphylococcal enterotoxin, enterotoxin F, associated with toxic-shock-syndrome Staphylococcus aureus isolates. Lancet 1:1017–1021. doi: 10.1016/s0140-6736(81)92186-3 [DOI] [PubMed] [Google Scholar]
- 186. Fraser JD, Proft T. 2008. The bacterial superantigen and superantigen-like proteins. Immunol Rev 225:226–243. doi: 10.1111/j.1600-065X.2008.00681.x [DOI] [PubMed] [Google Scholar]
- 187. Lina G, Bohach GA, Nair SP, Hiramatsu K, Jouvin-Marche E, Mariuzza R, Staphylococcal S. 2004. Standard nomenclature for the superantigens expressed by Staphylococcus. J Infect Dis 189:2334–2336. doi: 10.1086/420852 [DOI] [PubMed] [Google Scholar]
- 188. Fisher EL, Otto M, Cheung GYC. 2018. Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front Microbiol 9:436. doi: 10.3389/fmicb.2018.00436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, Etienne J, Vandenesch F, Bonneville M, Lina G. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol 166:669–677. doi: 10.4049/jimmunol.166.1.669 [DOI] [PubMed] [Google Scholar]
- 190. van Wamel WJB, Rooijakkers SHM, Ruyken M, van Kessel KPM, van Strijp JAG. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J Bacteriol 188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, Bröker BM, Doskar J, Wolz C. 2009. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol 191:3462–3468. doi: 10.1128/JB.01804-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Tuffs SW, Goncheva MI, Xu SX, Craig HC, Kasper KJ, Choi J, Flannagan RS, Kerfoot SM, Heinrichs DE, McCormick JK. 2022. Superantigens promote Staphylococcus aureus bloodstream infection by eliciting pathogenic interferon-gamma production. Proc Natl Acad Sci USA 119:e2115987119. doi: 10.1073/pnas.2115987119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Kinney KJ, Tang SS, Wu X-J, Tran PM, Bharadwaj NS, Gibson-Corley KN, Forsythe AN, Kulhankova K, Gumperz JE, Salgado-Pabón W. 2022. SEC is an antiangiogenic virulence factor that promotes Staphylococcus aureus endocarditis independent of superantigen activity. Sci Adv 8:eabo1072. doi: 10.1126/sciadv.abo1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Salgado-Pabón W, Breshears L, Spaulding AR, Merriman JA, Stach CS, Horswill AR, Peterson ML, Schlievert PM. 2013. Superantigens are critical for Staphylococcus aureus infective endocarditis, sepsis, and acute kidney injury. mBio 4:e00494-13. doi: 10.1128/mBio.00494-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Wilson GJ, Seo KS, Cartwright RA, Connelley T, Chuang-Smith ON, Merriman JA, Guinane CM, Park JY, Bohach GA, Schlievert PM, Morrison WI, Fitzgerald JR, DeLeo FR. 2011. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog 7:e1002271. doi: 10.1371/journal.ppat.1002271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Gordon RJ, Lowy FD. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46 Suppl 5:S350–9. doi: 10.1086/533591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Chen X, Alonzo F III. 2019. Bacterial lipolysis of immune-activating ligands promotes evasion of innate defenses. Proc Natl Acad Sci U.S.A 116:3764–3773. doi: 10.1073/pnas.1817248116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Prat C, Bestebroer J, de Haas CJC, van Strijp JAG, van Kessel KPM. 2006. A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J Immunol 177:8017–8026. doi: 10.4049/jimmunol.177.11.8017 [DOI] [PubMed] [Google Scholar]
- 199. de Haas CJC, Veldkamp KE, Peschel A, Weerkamp F, Van Wamel WJB, Heezius ECJM, Poppelier MJJG, Van Kessel KPM, van Strijp JAG. 2004. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J Exp Med 199:687–695. doi: 10.1084/jem.20031636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Rooijakkers SHM, Ruyken M, Roos A, Daha MR, Presanis JS, Sim RB, van Wamel WJB, van Kessel KPM, van Strijp JAG. 2005. Immune evasion by a staphylococcal complement inhibitor that acts on C3 Convertases. Nat Immunol 6:920–927. doi: 10.1038/ni1235 [DOI] [PubMed] [Google Scholar]
- 201. Delekta PC, Shook JC, Lydic TA, Mulks MH, Hammer ND. 2018. Staphylococcus aureus utilizes host-derived lipoprotein particles as sources of fatty acids. J Bacteriol 200:e00728-17. doi: 10.1128/JB.00728-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. White MJ, Boyd JM, Horswill AR, Nauseef WM. 2014. Phosphatidylinositol-specific phospholipase C contributes to survival of Staphylococcus aureus USA300 in human blood and neutrophils. Infect Immun 82:1559–1571. doi: 10.1128/IAI.01168-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Clements MO, Foster SJ. 1999. Stress resistance in Staphylococcus aureus. Trends Microbiol 7:458–462. doi: 10.1016/s0966-842x(99)01607-8 [DOI] [PubMed] [Google Scholar]
- 204. Ibarra JA, Pérez-Rueda E, Carroll RK, Shaw LN. 2013. Global analysis of transcriptional regulators in Staphylococcus aureus. BMC Genomics 14:126. doi: 10.1186/1471-2164-14-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640 [DOI] [PubMed] [Google Scholar]
- 206. Liu Q, Yeo WS, Bae T. 2016. The saers two-component system of Staphylococcus aureus. Genes 7:81. doi: 10.3390/genes7100081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Cheung AL, Nishina KA, Trotonda MP, Tamber S. 2008. The sara protein family of Staphylococcus aureus. Int J Biochem Cell Biol 40:355–361. doi: 10.1016/j.biocel.2007.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- 209. George Cisar EA, Geisinger E, Muir TW, Novick RP. 2009. Symmetric signalling within asymmetric dimers of the Staphylococcus aureus receptor histidine kinase AgrC. Mol Microbiol 74:44–57. doi: 10.1111/j.1365-2958.2009.06849.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2 [DOI] [PubMed] [Google Scholar]
- 211. Haag AF, Bagnoli F. 2017. The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr Top Microbiol Immunol 409:145–198. doi: 10.1007/82_2015_5019 [DOI] [PubMed] [Google Scholar]
- 212. Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. 1995. The AGR P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet 248:446–458. doi: 10.1007/BF02191645 [DOI] [PubMed] [Google Scholar]
- 213. Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (AGR) in Staphylococcus aureus. J Bacteriol 170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Wang B, Zhao A, Novick RP, Muir TW. 2015. Key driving forces in the biosynthesis of autoinducing peptides required for staphylococcal virulence. Proc Natl Acad Sci USA 112:10679–10684. doi: 10.1073/pnas.1506030112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Zhao A, Bodine SP, Xie Q, Wang B, Ram G, Novick RP, Muir TW. 2022. Reconstitution of the S. aureus AGR quorum sensing pathway reveals a direct role for the integral membrane protease MroQ in pheromone biosynthesis. Proc Natl Acad Sci USA 119:e2202661119. doi: 10.1073/pnas.2202661119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Stock MR, Fang L, Johnson KR, Cosgriff C, Teoh WP, Alonzo F. 2022. Characterization of MroQ-dependent maturation and export of the Staphylococcus aureus accessory gene regulatory system autoinducing peptide. Infect Immun 90:e0026322. doi: 10.1128/iai.00263-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Ji G, Beavis RC, Novick RP. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA 92:12055–12059. doi: 10.1073/pnas.92.26.12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator rot by an antisense mechanism. Genes Dev 21:1353–1366. doi: 10.1101/gad.423507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gruss A, Novick RP. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol Gen Genet 202:58–61. doi: 10.1007/BF00330517 [DOI] [PubMed] [Google Scholar]
- 221. Luong T, Sau S, Gomez M, Lee JC, Lee CY. 2002. Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA. Infect Immun 70:444–450. doi: 10.1128/IAI.70.2.444-450.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Dubrac S, Msadek T. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J Bacteriol 186:1175–1181. doi: 10.1128/JB.186.4.1175-1181.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Liang X, Yu C, Sun J, Liu H, Landwehr C, Holmes D, Ji Y. 2006. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect Immun 74:4655–4665. doi: 10.1128/IAI.00322-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Fournier B, Klier A, Rapoport G. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol 41:247–261. doi: 10.1046/j.1365-2958.2001.02515.x [DOI] [PubMed] [Google Scholar]
- 225. Yarwood JM, McCormick JK, Schlievert PM. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol 183:1113–1123. doi: 10.1128/JB.183.4.1113-1123.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Kraus D, Herbert S, Kristian SA, Khosravi A, Nizet V, Götz F, Peschel A. 2008. The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol 8:85. doi: 10.1186/1471-2180-8-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Gardete S, Kim C, Hartmann BM, Mwangi M, Roux CM, Dunman PM, Chambers HF, Tomasz A. 2012. Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLoS Pathog 8:e1002505. doi: 10.1371/journal.ppat.1002505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Yang SJ, Xiong YQ, Yeaman MR, Bayles KW, Abdelhady W, Bayer AS. 2013. Role of the LytSR two-component regulatory system in adaptation to cationic antimicrobial peptides in Staphylococcus aureus. Antimicrob Agents Chemother 57:3875–3882. doi: 10.1128/AAC.00412-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Howden BP, Davies JK, Johnson PDR, Stinear TP, Grayson ML. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 23:99–139. doi: 10.1128/CMR.00042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Chong YP, Kim ES, Park SJ, Park KH, Kim T, Kim MN, Kim SH, Lee SO, Choi SH, Woo JH, Jeong JY, Kim YS. 2013. Accessory gene regulator (agr) dysfunction in Staphylococcus aureus bloodstream isolates from South Korean patients. Antimicrob Agents Chemother 57:1509–1512. doi: 10.1128/AAC.01260-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Chen CJ, Lin MH, Shu JC, Lu JJ. 2014. Reduced susceptibility to vancomycin in Isogenic Staphylococcus aureus strains of sequence type 59: tracking evolution and identifying mutations by whole-genome sequencing. J Antimicrob Chemother 69:349–354. doi: 10.1093/jac/dkt395 [DOI] [PubMed] [Google Scholar]
- 232. Park SY, Oh IH, Lee HJ, Ihm CG, Son JS, Lee MS, Kim MN. 2013. Impact of reduced vancomycin MIC on clinical outcomes of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob Agents Chemother 57:5536–5542. doi: 10.1128/AAC.01137-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Butterfield JM, Tsuji BT, Brown J, Ashley ED, Hardy D, Brown K, Forrest A, Lodise TP. 2011. Predictors of AGR dysfunction in methicillin-resistant Staphylococcus aureus (MRSA) isolates among patients with MRSA bloodstream infections. Antimicrob Agents Chemother 55:5433–5437. doi: 10.1128/AAC.00407-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Seidl K, Chen L, Bayer AS, Hady WA, Kreiswirth BN, Xiong YQ. 2011. Relationship of AGR expression and function with virulence and vancomycin treatment outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 55:5631–5639. doi: 10.1128/AAC.05251-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Rao Y, Peng H, Shang W, Hu Z, Yang Y, Tan L, Li M, Zhou R, Rao X. 2022. A vancomycin resistance-associated walk(S221P) mutation attenuates the virulence of vancomycin-intermediate Staphylococcus aureus. J Adv Res 40:167–178. doi: 10.1016/j.jare.2021.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. McEvoy CRE, Tsuji B, Gao W, Seemann T, Porter JL, Doig K, Ngo D, Howden BP, Stinear TP. 2013. Decreased vancomycin susceptibility in Staphylococcus aureus caused by IS256 tempering of WalkR expression. Antimicrob Agents Chemother 57:3240–3249. doi: 10.1128/AAC.00279-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Bayer MG, Heinrichs JH, Cheung AL. 1996. The molecular architecture of the SAR locus in Staphylococcus aureus. J Bacteriol 178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Chien Y, Manna AC, Projan SJ, Cheung AL. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for SAR-dependent gene regulation. J Biol Chem 274:37169–37176. doi: 10.1074/jbc.274.52.37169 [DOI] [PubMed] [Google Scholar]
- 239. Cheung AL, Koomey JM, Butler CA, Projan SJ, Fischetti VA. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (SAR) distinct from agr. Proc Natl Acad Sci USA 89:6462–6466. doi: 10.1073/pnas.89.14.6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Wolz C, Pöhlmann-Dietze P, Steinhuber A, Chien YT, Manna A, van Wamel W, Cheung A. 2000. Agr-independent regulation of fibronectin-binding protein(S) by the regulatory locus SAR in Staphylococcus aureus. Mol Microbiol 36:230–243. doi: 10.1046/j.1365-2958.2000.01853.x [DOI] [PubMed] [Google Scholar]
- 241. Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Blevins JS, Gillaspy AF, Rechtin TM, Hurlburt BK, Smeltzer MS. 1999. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol Microbiol 33:317–326. doi: 10.1046/j.1365-2958.1999.01475.x [DOI] [PubMed] [Google Scholar]
- 243. Cheung AL, Bayer MG, Heinrichs JH. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J Bacteriol 179:3963–3971. doi: 10.1128/jb.179.12.3963-3971.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Manna A, Cheung AL. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect Immun 69:885–896. doi: 10.1128/IAI.69.2.885-896.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. 2004. Regulation of virulence determinants In vitro and In vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol 40:1–9. doi: 10.1016/S0928-8244(03)00309-2 [DOI] [PubMed] [Google Scholar]
- 246. Kaito C, Morishita D, Matsumoto Y, Kurokawa K, Sekimizu K. 2006. Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus. Mol Microbiol 62:1601–1617. doi: 10.1111/j.1365-2958.2006.05480.x [DOI] [PubMed] [Google Scholar]
- 247. Manna AC, Ingavale SS, Maloney M, van Wamel W, Cheung AL. 2004. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J Bacteriol 186:5267–5280. doi: 10.1128/JB.186.16.5267-5280.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Saïd-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. 2003. Global regulation of Staphylococcus aureus genes by rot. J Bacteriol 185:610–619. doi: 10.1128/JB.185.2.610-619.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Tegmark K, Karlsson A, Arvidson S. 2000. Identification and characterization of Sarh1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol 37:398–409. doi: 10.1046/j.1365-2958.2000.02003.x [DOI] [PubMed] [Google Scholar]
- 250. Truong-Bolduc QC, Zhang X, Hooper DC. 2003. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J Bacteriol 185:3127–3138. doi: 10.1128/JB.185.10.3127-3138.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Guldimann C, Boor KJ, Wiedmann M, Guariglia-Oropeza V. 2016. Resilience in the face of uncertainty: sigma factor B fine-tunes gene expression to support homeostasis in gram-positive bacteria. Appl Environ Microbiol 82:4456–4469. doi: 10.1128/AEM.00714-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bächi B, Projan S. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J Bacteriol 186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Bischoff M, Entenza JM, Giachino P. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J Bacteriol 183:5171–5179. doi: 10.1128/JB.183.17.5171-5179.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Jenul C, Horswill AR. 2018. Regulation of Staphylococcus aureus virulence. Microbiol Spectr 6:29. doi: 10.1128/9781683670131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Somerville GA, Proctor RA. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol Mol Biol Rev 73:233–248. doi: 10.1128/MMBR.00005-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, Harris L, Berger-Bächi B, Bischoff M. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother 50:1183–1194. doi: 10.1128/AAC.50.4.1183-1194.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Hartmann T, Zhang B, Baronian G, Schulthess B, Homerova D, Grubmüller S, Kutzner E, Gaupp R, Bertram R, Powers R, Eisenreich W, Kormanec J, Herrmann M, Molle V, Somerville GA, Bischoff M. 2013. Catabolite control protein E (CcpE) is a Lysr-type transcriptional regulator of tricarboxylic acid cycle activity in Staphylococcus aureus. J Biol Chem 288:36116–36128. doi: 10.1074/jbc.M113.516302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Thurlow LR, Stephens AC, Hurley KE, Richardson AR. 2020. Lack of nutritional immunity in diabetic skin infections promotes Staphylococcus aureus virulence. Sci Adv 6:eabc5569. doi: 10.1126/sciadv.abc5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Stephens AC, Thurlow LR, Richardson AR. 2022. Mechanisms behind the indirect impact of metabolic regulators on virulence factor production in Staphylococcus aureus. Microbiol Spectr 10:e0206322. doi: 10.1128/spectrum.02063-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Regassa LB, Novick RP, Betley MJ. 1992. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect Immun 60:3381–3388. doi: 10.1128/iai.60.8.3381-3388.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Harper L, Balasubramanian D, Ohneck EA, Sause WE, Chapman J, Mejia-Sosa B, Lhakhang T, Heguy A, Tsirigos A, Ueberheide B, Boyd JM, Lun DS, Torres VJ, Richardson AR, Dunman P. 2018. Staphylococcus aureus responds to the central metabolite pyruvate to regulate virulence. mBio 9:e02272-17. doi: 10.1128/mBio.02272-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Schurig-Briccio LA, Parraga Solorzano PK, Lencina AM, Radin JN, Chen GY, Sauer JD, Kehl-Fie TE, Gennis RB. 2020. Role of respiratory NADH oxidation in the regulation of Staphylococcus aureus virulence. EMBO Rep 21:e45832. doi: 10.15252/embr.201845832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Onoue Y, Mori M. 1997. Amino acid requirements for the growth and enterotoxin production by Staphylococcus aureus in chemically defined media. Int J Food Microbiol 36:77–82. doi: 10.1016/s0168-1605(97)01250-6 [DOI] [PubMed] [Google Scholar]
- 264. Lincoln RA, Leigh JA, Jones NC. 1995. The amino acid requirements of Staphylococcus aureus isolated from cases of bovine mastitis. Vet Microbiol 45:275–279. doi: 10.1016/0378-1135(95)00041-8 [DOI] [PubMed] [Google Scholar]
- 265. Kaiser J.C, Sen S, Sinha A, Wilkinson BJ, Heinrichs DE. 2016. The role of two branched-chain amino acid transporters in Staphylococcus aureus growth, membrane fatty acid composition and virulence. Mol Microbiol 102:850–864. doi: 10.1111/mmi.13495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Waters NR, Samuels DJ, Behera RK, Livny J, Rhee KY, Sadykov MR, Brinsmade SR. 2016. A spectrum of cody activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol Microbiol 101:495–514. doi: 10.1111/mmi.13404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Kaiser JC, King AN, Grigg JC, Sheldon JR, Edgell DR, Murphy MEP, Brinsmade SR, Heinrichs DE. 2018. Repression of branched-chain amino acid synthesis in Staphylococcus aureus is mediated by isoleucine via cody, and by a leucine-rich attenuator peptide. PLoS Genet 14:e1007159. doi: 10.1371/journal.pgen.1007159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. 2008. Staphylococcus aureus cody negatively regulates virulence gene expression. J Bacteriol 190:2257–2265. doi: 10.1128/JB.01545-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. 2009. Cody in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 191:2953–2963. doi: 10.1128/JB.01492-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Montgomery CP, Boyle-Vavra S, Roux A, Ebine K, Sonenshein AL, Daum RS. 2012. Cody deletion enhances In vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect Immun 80:2382–2389. doi: 10.1128/IAI.06172-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Rom JS, Atwood DN, Beenken KE, Meeker DG, Loughran AJ, Spencer HJ, Lantz TL, Smeltzer MS. 2017. Impact of Staphylococcus aureus regulatory mutations that modulate biofilm formation in the USA300 strain LAC on virulence in a murine bacteremia model. Virulence 8:1776–1790. doi: 10.1080/21505594.2017.1373926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Rom JS, Beenken KE, Ramirez AM, Walker CM, Echols EJ, Smeltzer MS. 2021. Limiting protease production plays a key role in the pathogenesis of the divergent clinical isolates of Staphylococcus aureus LAC and UAMS-1. Virulence 12:584–600. doi: 10.1080/21505594.2021.1879550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Sause WE, Balasubramanian D, Irnov I, Copin R, Sullivan MJ, Sommerfield A, Chan R, Dhabaria A, Askenazi M, Ueberheide B, Shopsin B, van Bakel H, Torres VJ. 2019. The purine biosynthesis regulator purr moonlights as a virulence regulator in Staphylococcus aureus Proc Natl Acad Sci USA 116:13563–13572. doi: 10.1073/pnas.1904280116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Goncheva MI, Flannagan RS, Sterling BE, Laakso HA, Friedrich NC, Kaiser JC, Watson DW, Wilson CH, Sheldon JR, McGavin MJ, Kiser PK, Heinrichs DE. 2019. Stress-induced inactivation of the Staphylococcus aureus purine biosynthesis repressor leads to hypervirulence. Nat Commun 10:775. doi: 10.1038/s41467-019-08724-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Weng M, Nagy PL, Zalkin H. 1995. Identification of the bacillus subtilis pur operon repressor. Proc Natl Acad Sci USA 92:7455–7459. doi: 10.1073/pnas.92.16.7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Cho B-K, Federowicz SA, Embree M, Park Y-S, Kim D, Palsson BØ. 2011. The purr regulon in Escherichia coli K-12 Mg1655. Nucleic Acids Res 39:6456–6464. doi: 10.1093/nar/gkr307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Li L, Abdelhady W, Donegan NP, Seidl K, Cheung A, Zhou YF, Yeaman MR, Bayer AS, Xiong YQ. 2018. Role of purine biosynthesis in persistent methicillin-resistant Staphylococcus aureus infection. J Infect Dis 218:1367–1377. doi: 10.1093/infdis/jiy340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Kuhn S, Slavetinsky CJ, Peschel A. 2015. Synthesis and function of phospholipids in Staphylococcus aureus. Int J Med Microbiol 305:196–202. doi: 10.1016/j.ijmm.2014.12.016 [DOI] [PubMed] [Google Scholar]
- 279. Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KP, van Strijp JA. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MPRF is based on modification of membrane lipids with l-lysine. J Exp Med 193:1067–1076. doi: 10.1084/jem.193.9.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Cameron DR, Mortin LI, Rubio A, Mylonakis E, Moellering RC, Eliopoulos GM, Peleg AY. 2015. Impact of daptomycin resistance on Staphylococcus aureus virulence. Virulence 6:127–131. doi: 10.1080/21505594.2015.1011532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Zheng X, Marsman G, Lacey KA, Chapman JR, Goosmann C, Ueberheide BM, Torres VJ. 2021. The cell envelope of Staphylococcus aureus selectively controls the sorting of virulence factors. Nat Commun 12:6193. doi: 10.1038/s41467-021-26517-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Lopez MS, Tan IS, Yan D, Kang J, McCreary M, Modrusan Z, Austin CD, Xu M, Brown EJ. 2017. Host-derived fatty acids activate type VII secretion in Staphylococcus aureus Proc Natl Acad Sci USA 114:11223–11228. doi: 10.1073/pnas.1700627114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Parsons JB, Broussard TC, Bose JL, Rosch JW, Jackson P, Subramanian C, Rock CO. 2014. Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc Natl Acad Sci USA 111:10532–10537. doi: 10.1073/pnas.1408797111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Li M, Rigby K, Lai Y, Nair V, Peschel A, Schittek B, Otto M. 2009. Staphylococcus aureus mutant screen reveals interaction of the human antimicrobial peptide dermcidin with membrane phospholipids. Antimicrob Agents Chemother 53:4200–4210. doi: 10.1128/AAC.00428-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Bose JL, Daly SM, Hall PR, Bayles KW. 2014. Identification of the Staphylococcus aureus vfrAB operon, a novel virulence factor regulatory locus. Infect Immun 82:1813–1822. doi: 10.1128/IAI.01655-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Ridder MJ, Daly SM, Triplett KD, Seawell NA, Hall PR, Bose JL. 2020. Staphylococcus aureus fatty acid kinase faka modulates pathogenesis during skin infection via proteases. Infect Immun 88:e00163-20. doi: 10.1128/IAI.00163-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Lindsay JA, Holden MTG. 2006. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct Integr Genomics 6:186–201. doi: 10.1007/s10142-005-0019-7 [DOI] [PubMed] [Google Scholar]
- 288. Strauß L, Stegger M, Akpaka PE, Alabi A, Breurec S, Coombs G, Egyir B, Larsen AR, Laurent F, Monecke S, Peters G, Skov R, Strommenger B, Vandenesch F, Schaumburg F, Mellmann A. 2017. Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. Proc Natl Acad Sci USA 114:E10596–E10604. doi: 10.1073/pnas.1702472114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Liang X, Hall JW, Yang J, Yan M, Doll K, Bey R, Ji Y, Aziz RK. 2011. Identification of single nucleotide polymorphisms associated with hyperproduction of alpha-toxin in Staphylococcus aureus. PLoS One 6:e18428. doi: 10.1371/journal.pone.0018428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Hall JW, Ji Y. 2012. Identification of predominant SNPs as a novel method for genotyping bovine Staphylococcus aureus isolates. Virulence 3:98–102. doi: 10.4161/viru.3.1.18724 [DOI] [PubMed] [Google Scholar]
- 291. Gor V, Takemura AJ, Nishitani M, Higashide M, Medrano Romero V, Ohniwa RL, Morikawa K. 2019. Finding of AGR phase variants in Staphylococcus aureus. mBio 10:e00796-19. doi: 10.1128/mBio.00796-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Somerville GA, Beres SB, Fitzgerald JR, DeLeo FR, Cole RL, Hoff JS, Musser JM. 2002. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence . J Bacteriol 184:1430–1437. doi: 10.1128/JB.184.5.1430-1437.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Shopsin B, Eaton C, Wasserman GA, Mathema B, Adhikari RP, Agolory S, Altman DR, Holzman RS, Kreiswirth BN, Novick RP. 2010. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J Infect Dis 202:1593–1599. doi: 10.1086/656915 [DOI] [PubMed] [Google Scholar]
- 294. Traber K, Novick R. 2006. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate delta- and alpha-haemolysins. Mol Microbiol 59:1519–1530. doi: 10.1111/j.1365-2958.2006.04986.x [DOI] [PubMed] [Google Scholar]
- 295. Adhikari RP, Arvidson S, Novick RP. 2007. A nonsense mutation in agrA accounts for the defect in agr expression and the avirulence of Staphylococcus aureus 8325-4 traP::Kan. Infect Immun 75:4534–4540. doi: 10.1128/IAI.00679-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Raghuram V, Alexander AM, Loo HQ, Petit RA, Goldberg JB, Read TD, Sharma G, Hasan N. 2022. Species-wide phylogenomics of the Staphylococcus aureus agr operon revealed convergent evolution of frameshift mutations. Microbiol Spectr 10:e0133421. doi: 10.1128/spectrum.01334-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Ramond E, Lepissier A, Ding X, Bouvier C, Tan X, Euphrasie D, Monbernard P, Dupuis M, Saubaméa B, Nemazanyy I, Nassif X, Ferroni A, Sermet-Gaudelus I, Charbit A, Coureuil M, Jamet A. 2022. Lung-adapted Staphylococcus aureus isolates with dysfunctional agr system trigger a proinflammatory response. J Infect Dis 226:1276–1285. doi: 10.1093/infdis/jiac191 [DOI] [PubMed] [Google Scholar]
- 298. Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, Kusuya Y, Shoji T, Takada S, Nakagawa S, Oguma R, et al. 2020. Staphylococcus agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med 12. doi: 10.1126/scitranslmed.aay4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Fischetti VA, Thakker M, Park J-S, Carey V, Lee JC. 1998. Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model . Infect Immun 66:5183–5189. doi: 10.1128/IAI.66.11.5183-5189.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Ji G, Beavis R, Novick RP. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030. doi: 10.1126/science.276.5321.2027 [DOI] [PubMed] [Google Scholar]
- 301. Gerlach D, Guo Y, De Castro C, Kim S-H, Schlatterer K, Xu F-F, Pereira C, Seeberger PH, Ali S, Codée J, Sirisarn W, Schulte B, Wolz C, Larsen J, Molinaro A, Lee BL, Xia G, Stehle T, Peschel A. 2018. Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity. Nature 563:705–709. doi: 10.1038/s41586-018-0730-x [DOI] [PubMed] [Google Scholar]
- 302. Dufour P, Jarraud S, Vandenesch F, Greenland T, Novick RP, Bes M, Etienne J, Lina G. 2002. High genetic variability of the agr locus in Staphylococcus species. J Bacteriol 184:1180–1186. doi: 10.1128/jb.184.4.1180-1186.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Wright JS, Traber KE, Corrigan R, Benson SA, Musser JM, Novick RP. 2005. The agr radiation: an early event in the evolution of staphylococci. J Bacteriol 187:5585–5594. doi: 10.1128/JB.187.16.5585-5594.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Jarraud S, Lyon GJ, Figueiredo AM, Lina G, Vandenesch F, Etienne J, Muir TW, Novick RP. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol 182:6517–6522. doi: 10.1128/JB.182.22.6517-6522.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Fournier JM, Vann WF, Karakawa WW. 1984. Purification and characterization of Staphylococcus aureus type 8 capsular polysaccharide. Infect Immun 45:87–93. doi: 10.1128/iai.45.1.87-93.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Mohamed N, Timofeyeva Y, Jamrozy D, Rojas E, Hao L, Silmon de Monerri NC, Hawkins J, Singh G, Cai B, Liberator P, Sebastian S, Donald RGK, Scully IL, Jones CH, Creech CB, Thomsen I, Parkhill J, Peacock SJ, Jansen KU, Holden MTG, Anderson AS, Rohde H. 2019. Molecular epidemiology and expression of capsular polysaccharides in Staphylococcus aureus clinical isolates in the United States. PLoS One 14:e0208356. doi: 10.1371/journal.pone.0208356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Moreau M, Richards JC, Fournier JM, Byrd RA, Karakawa WW, Vann WF. 1990. Structure of the type 5 capsular polysaccharide of Staphylococcus aureus. Carbohydr Res 201:285–297. doi: 10.1016/0008-6215(90)84244-o [DOI] [PubMed] [Google Scholar]
- 308. Sau S, Bhasin N, Wann ER, Lee JC, Foster TJ, Lee CY. 1997. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology (Reading) 143 ( Pt 7):2395–2405. doi: 10.1099/00221287-143-7-2395 [DOI] [PubMed] [Google Scholar]
- 309. Murphy E, Lin SL, Nunez L, Andrew L, Fink PS, Dilts DA, Hoiseth SK, Jansen KU, Anderson AS. 2011. Challenges for the evaluation of Staphylococcus aureus protein based vaccines: monitoring antigenic diversity. Hum Vaccin 7 Suppl:51–59. doi: 10.4161/hv.7.0.14562 [DOI] [PubMed] [Google Scholar]
- 310. Nanra JS, Timofeyeva Y, Buitrago SM, Sellman BR, Dilts DA, Fink P, Nunez L, Hagen M, Matsuka YV, Mininni T, Zhu D, Pavliak V, Green BA, Jansen KU, Anderson AS. 2009. Heterogeneous in vivo expression of clumping factor A and capsular polysaccharide by Staphylococcus aureus: implications for vaccine design. Vaccine 27:3276–3280. doi: 10.1016/j.vaccine.2009.01.062 [DOI] [PubMed] [Google Scholar]
- 311. Watts A, Ke D, Wang Q, Pillay A, Nicholson-Weller A, Lee JC. 2005. Staphylococcus aureus strains that express serotype 5 or serotype 8 capsular polysaccharides differ in virulence. Infect Immun 73:3502–3511. doi: 10.1128/IAI.73.6.3502-3511.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Park S, Gerber S, Lee JC, Camilli A. 2014. Antibodies to Staphylococcus aureus serotype 8 capsular polysaccharide react with and protect against serotype 5 and 8 isolates. Infect Immun 82:5049–5055. doi: 10.1128/IAI.02373-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Wacker M, Wang L, Kowarik M, Dowd M, Lipowsky G, Faridmoayer A, Shields K, Park S, Alaimo C, Kelley KA, Braun M, Quebatte J, Gambillara V, Carranza P, Steffen M, Lee JC. 2014. Prevention of Staphylococcus aureus infections by glycoprotein vaccines synthesized in Escherichia coli. J Infect Dis 209:1551–1561. doi: 10.1093/infdis/jit800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Liu B, Park S, Thompson CD, Li X, Lee JC. 2017. Antibodies to Staphylococcus aureus capsular polysaccharides 5 and 8 perform similarly in vitro but are functionally distinct in vivo. Virulence 8:859–874. doi: 10.1080/21505594.2016.1270494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. The Lancet 359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5 [DOI] [PubMed] [Google Scholar]
- 316. Klaui AJ, Boss R, Graber HU. 2019. Characterization and comparative analysis of the Staphylococcus aureus genomic island vSabeta: an in silico approach. J Bacteriol 201:e00777–18. doi: 10.1128/JB.00777-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Lindsay JA. 2019. Staphylococci: evolving genomes. Microbiol Spectr 7. doi: 10.1128/microbiolspec.GPP3-0071-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Sung JM-L, Lloyd DH, Lindsay JA. 2008. Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology (Reading) 154:1949–1959. doi: 10.1099/mic.0.2007/015289-0 [DOI] [PubMed] [Google Scholar]
- 319. Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 190:300–310. doi: 10.1128/JB.01000-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Gill SR, Fouts DE, Archer GL, Mongodin EF, DeBoy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, et al. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Malachowa N, DeLeo FR. 2010. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci 67:3057–3071. doi: 10.1007/s00018-010-0389-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Brüssow H, Desiere F. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol Microbiol 39:213–222. doi: 10.1046/j.1365-2958.2001.02228.x [DOI] [PubMed] [Google Scholar]
- 324. Coleman DC, Sullivan DJ, Russell RJ, Arbuthnott JP, Carey BF, Pomeroy HM. 1989. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J Gen Microbiol 135:1679–1697. doi: 10.1099/00221287-135-6-1679 [DOI] [PubMed] [Google Scholar]
- 325. Laux C, Peschel A, Krismer B. 2019. Staphylococcus aureus colonization of the human nose and interaction with other microbiome members. Microbiol Spectr 7. doi: 10.1128/microbiolspec.GPP3-0029-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Kaneko J, Kimura T, Narita S, Tomita T, Kamio Y. 1998. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage phiPVL carrying Panton-Valentine leukocidin genes. Gene 215:57–67. doi: 10.1016/s0378-1119(98)00278-9 [DOI] [PubMed] [Google Scholar]
- 327. Gillet Y, Issartel B, Vanhems P, Fournet J-C, Lina G, Bes M, Vandenesch F, Piémont Y, Brousse N, Floret D, Etienne J. 2002. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753–759. doi: 10.1016/S0140-6736(02)07877-7 [DOI] [PubMed] [Google Scholar]
- 328. Shallcross LJ, Fragaszy E, Johnson AM, Hayward AC. 2013. The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: a systematic review and meta-analysis. Lancet Infect Dis 13:43–54. doi: 10.1016/S1473-3099(12)70238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Li M, Du X, Villaruz AE, Diep BA, Wang D, Song Y, Tian Y, Hu J, Yu F, Lu Y, Otto M. 2012. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat Med 18:816–819. doi: 10.1038/nm.2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Ingmer H, Gerlach D, Wolz C. 2019. Temperate phages of Staphylococcus aureus. Microbiol Spectr 7:1. doi: 10.1128/microbiolspec.GPP3-0058-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Novick RP, Christie GE, Penadés JR. 2010. The phage-related chromosomal islands of gram-positive bacteria. Nat Rev Microbiol 8:541–551. doi: 10.1038/nrmicro2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol 29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x [DOI] [PubMed] [Google Scholar]
- 333. Novick RP. 2003. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 49:93–105. doi: 10.1016/s0147-619x(02)00157-9 [DOI] [PubMed] [Google Scholar]
- 334. McCarthy AJ, Lindsay JA. 2012. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol 12:104. doi: 10.1186/1471-2180-12-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Zhang S, Iandolo JJ, Stewart GC. 1998. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol Lett 168:227–233. doi: 10.1111/j.1574-6968.1998.tb13278.x [DOI] [PubMed] [Google Scholar]
- 336. Bayles KW, Iandolo JJ. 1989. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J Bacteriol 171:4799–4806. doi: 10.1128/jb.171.9.4799-4806.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Omoe K, Hu DL, Takahashi-Omoe H, Nakane A, Shinagawa K. 2003. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect Immun 71:6088–6094. doi: 10.1128/IAI.71.10.6088-6094.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Shalita Z, Hertman I, Sarid S. 1977. Isolation and characterization of a plasmid involved with enterotoxin B production in Staphylococcus aureus. J Bacteriol 129:317–325. doi: 10.1128/jb.129.1.317-325.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Jackson MP, Iandolo JJ. 1986. Cloning and expression of the exfoliative toxin B gene from Staphylococcus aureus. J Bacteriol 166:574–580. doi: 10.1128/jb.166.2.574-580.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Yamaguchi T, Hayashi T, Takami H, Ohnishi M, Murata T, Nakayama K, Asakawa K, Ohara M, Komatsuzawa H, Sugai M. 2001. Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect Immun 69:7760–7771. doi: 10.1128/IAI.69.12.7760-7771.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Feßler AT, Zhao Q, Schoenfelder S, Kadlec K, Brenner Michael G, Wang Y, Ziebuhr W, Shen J, Schwarz S. 2017. Complete sequence of a plasmid from a bovine methicillin-resistant Staphylococcus aureus harbouring a novel ica-like gene cluster in addition to antimicrobial and heavy metal resistance genes. Vet Microbiol 200:95–100. doi: 10.1016/j.vetmic.2016.07.010 [DOI] [PubMed] [Google Scholar]
- 342. Qin L, McCausland JW, Cheung GYC, Otto M. 2016. PSM-mec-a virulence determinant that connects transcriptional regulation, virulence, and antibiotic resistance in staphylococci. Front Microbiol 7:1293. doi: 10.3389/fmicb.2016.01293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Luong TT, Ouyang S, Bush K, Lee CY. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J Bacteriol 184:3623–3629. doi: 10.1128/JB.184.13.3623-3629.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Guinane CM, Ben Zakour NL, Tormo-Mas MA, Weinert LA, Lowder BV, Cartwright RA, Smyth DS, Smyth CJ, Lindsay JA, Gould KA, Witney A, Hinds J, Bollback JP, Rambaut A, Penadés JR, Fitzgerald JR. 2010. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol Evol 2:454–466. doi: 10.1093/gbe/evq031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, et al. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. doi: 10.1128/mBio.00305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Mrochen DM, Grumann D, Schulz D, Gumz J, Trübe P, Pritchett-Corning K, Johnson S, Nicklas W, Kirsch P, Martelet K, Brandt J van den, Berg S, Bröker BM, Wiles S, Holtfreter S. 2018. Global spread of mouse-adapted Staphylococcus aureus lineages CC1, CC15, and CC88 among mouse breeding facilities. Int J Med Microbiol 308:598–606. doi: 10.1016/j.ijmm.2017.11.006 [DOI] [PubMed] [Google Scholar]
- 347. Ubeda C, Tormo MA, Cucarella C, Trotonda P, Foster TJ, Lasa I, Penadés JR. 2003. Sip, an Integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol Microbiol 49:193–210. doi: 10.1046/j.1365-2958.2003.03577.x [DOI] [PubMed] [Google Scholar]
- 348. Devriese LA. 1984. A simplified system for biotyping Staphylococcus aureus strains isolated from animal species. J Appl Bacteriol 56:215–220. doi: 10.1111/j.1365-2672.1984.tb01341.x [DOI] [PubMed] [Google Scholar]
- 349. Viana D, Blanco J, Tormo-Más MA, Selva L, Guinane CM, Baselga R, Corpa JM, Lasa I, Novick RP, Fitzgerald JR, Penadés JR. 2010. Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol Microbiol 77:1583–1594. doi: 10.1111/j.1365-2958.2010.07312.x [DOI] [PubMed] [Google Scholar]
- 350. Novick RP. 2019. Pathogenicity islands and their role in staphylococcal biology. Microbiol Spectr 7:21. doi: 10.1128/microbiolspec.GPP3-0062-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Madhaiyan M, Wirth JS, Saravanan VS. 2020. Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int J Syst Evol Microbiol 70:5926–5936. doi: 10.1099/ijsem.0.004498 [DOI] [PubMed] [Google Scholar]
- 352. Couto I, de Lencastre H, Severina E, Kloos W, Webster JA, Hubner RJ, Sanches IS, Tomasz A. 1996. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb Drug Resist 2:377–391. doi: 10.1089/mdr.1996.2.377 [DOI] [PubMed] [Google Scholar]
- 353. Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. 2010. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother 54:4352–4359. doi: 10.1128/AAC.00356-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Wu SW, de Lencastre H, Tomasz A. 2001. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J Bacteriol 183:2417–2424. doi: 10.1128/JB.183.8.2417-2424.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355. Antignac A, Tomasz A. 2009. Reconstruction of the phenotypes of methicillin-resistant Staphylococcus aureus by replacement of the Staphylococcal cassette chromosome mec with a plasmid-borne copy of Staphylococcus sciuri pbpD gene. Antimicrob Agents Chemother 53:435–441. doi: 10.1128/AAC.01099-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356. Rolo J, Worning P, Nielsen JB, Bowden R, Bouchami O, Damborg P, Guardabassi L, Perreten V, Tomasz A, Westh H, Lencastre H, Miragaia M. 2017. Evolutionary origin of the staphylococcal cassette chromosome mec (SCCmec). Antimicrob Agents Chemother 61:e02302-16. doi: 10.1128/AAC.02302-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357. Qin L, Da F, Fisher EL, Tan DCS, Nguyen TH, Fu C-L, Tan VY, McCausland JW, Sturdevant DE, Joo H-S, Queck SY, Cheung GYC, Otto M, Grundling A. 2017. Toxin mediates sepsis caused by methicillin-resistant Staphylococcus epidermidis. PLoS Pathog 13:e1006153. doi: 10.1371/journal.ppat.1006153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Wisplinghoff H, Rosato AE, Enright MC, Noto M, Craig W, Archer GL. 2003. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob Agents Chemother 47:3574–3579. doi: 10.1128/AAC.47.11.3574-3579.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Wielders CL, Vriens MR, Brisse S, de Graaf-Miltenburg LA, Troelstra A, Fleer A, Schmitz FJ, Verhoef J, Fluit AC. 2001. In-vivo transfer of mecA DNA to Staphylococcus aureus. Lancet 357:1674–1675. doi: 10.1016/s0140-6736(00)04832-7 [DOI] [PubMed] [Google Scholar]
- 360. Bloemendaal ALA, Brouwer EC, Fluit AC. 2010. Methicillin resistance transfer from Staphylocccus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS One 5:e11841. doi: 10.1371/journal.pone.0011841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Planet PJ, LaRussa SJ, Dana A, Smith H, Xu A, Ryan C, Uhlemann AC, Boundy S, Goldberg J, Narechania A, Kulkarni R, Ratner AJ, Geoghegan JA, Kolokotronis SO, Prince A. 2013. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio 4:e00889-13. doi: 10.1128/mBio.00889-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Otto M. 2013. How colonization factors are linked to outbreaks of methicillin-resistant Staphylococcus aureus: the roles of SasX and ACME. Biomol Concepts 4:533–537. doi: 10.1515/bmc-2013-0025 [DOI] [PubMed] [Google Scholar]
- 363. King JM, Kulhankova K, Stach CS, Vu BG, Salgado-Pabón W. 2016. Phenotypes and virulence among Staphylococcus aureus USA100, USA200, USA300, USA400, and USA600 clonal lineages. mSphere 1:e00071-16. doi: 10.1128/mSphere.00071-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Li M, Cheung GYC, Hu J, Wang D, Joo H-S, Deleo FR, Otto M. 2010. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis 202:1866–1876. doi: 10.1086/657419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus Pulsotypes USA300 and Usa400 in a rat model of pneumonia. J Infect Dis 198:561–570. doi: 10.1086/590157 [DOI] [PubMed] [Google Scholar]
- 366. Spaulding AR, Satterwhite EA, Lin YC, Chuang-Smith ON, Frank KL, Merriman JA, Schaefers MM, Yarwood JM, Peterson ML, Schlievert PM. 2012. Comparison of Staphylococcus aureus strains for ability to cause infective endocarditis and lethal sepsis in rabbits. Front Cell Infect Microbiol 2:18. doi: 10.3389/fcimb.2012.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Cheung GYC, Wang R, Khan BA, Sturdevant DE, Otto M. 2011. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun 79:1927–1935. doi: 10.1128/IAI.00046-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, Long RD, Dorward DW, Gardner DJ, Lina G, Kreiswirth BN, DeLeo FR. 2006. Is panton-valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease J Infect Dis 194:1761–1770. doi: 10.1086/509506 [DOI] [PubMed] [Google Scholar]
- 369. Hongo I, Baba T, Oishi K, Morimoto Y, Ito T, Hiramatsu K. 2009. Phenol-soluble modulin alpha 3 enhances the human neutrophil lysis mediated by panton-valentine leukocidin. J Infect Dis 200:715–723. doi: 10.1086/605332 [DOI] [PubMed] [Google Scholar]
- 370. Kaito C, Saito Y, Ikuo M, Omae Y, Mao H, Nagano G, Fujiyuki T, Numata S, Han X, Obata K, Hasegawa S, Yamaguchi H, Inokuchi K, Ito T, Hiramatsu K, Sekimizu K. 2013. Mobile genetic element SCCmec-encoded PSM-MEC RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog 9:e1003269. doi: 10.1371/journal.ppat.1003269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371. Cheung GYC, Yeh AJ, Kretschmer D, Duong AC, Tuffuor K, Fu C-L, Joo H-S, Diep BA, Li M, Nakamura Y, Nunez G, Peschel A, Otto M. 2015. Functional characteristics of the Staphylococcus aureus Delta-toxin allelic variant G10S. Sci Rep 5:18023. doi: 10.1038/srep18023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Lacoma A, Edwards AM, Young BC, Domínguez J, Prat C, Laabei M. 2019. Cigarette smoke exposure redirects Staphylococcus aureus to a virulence profile associated with persistent infection. Sci Rep 9:10798. doi: 10.1038/s41598-019-47258-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Burlak C, Hammer CH, Robinson MA, Whitney AR, McGavin MJ, Kreiswirth BN, Deleo FR. 2007. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol 9:1172–1190. doi: 10.1111/j.1462-5822.2006.00858.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Saïd-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol 175:3907–3919. doi: 10.4049/jimmunol.175.6.3907 [DOI] [PubMed] [Google Scholar]
- 375. Guimarães MA, Ramundo MS, Américo MA, de Mattos MC, Souza RR, Ramos-Júnior ES, Coelho LR, Morrot A, Melo PA, Fracalanzza SEL, Ferreira FA, Figueiredo AMS. 2015. A comparison of virulence patterns and in vivo fitness between hospital- and community-acquired methicillin-resistant Staphylococcus aureus related to the USA400 clone. Eur J Clin Microbiol Infect Dis 34:497–509. doi: 10.1007/s10096-014-2253-1 [DOI] [PubMed] [Google Scholar]
- 376. Spentzas T, Kudumula R, Acuna C, Talati AJ, Ingram KC, Savorgnan F, Meals EA, English BK. 2011. Role of bacterial components in macrophage activation by the LAC and Mw2 strains of community-associated, methicillin-resistant Staphylococcus aureus. Cell Immunol 269:46–53. doi: 10.1016/j.cellimm.2011.03.009 [DOI] [PubMed] [Google Scholar]
- 377. Kaito C, Saito Y, Nagano G, Ikuo M, Omae Y, Hanada Y, Han X, Kuwahara-Arai K, Hishinuma T, Baba T, Ito T, Hiramatsu K, Sekimizu K. 2011. Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog 7:e1001267. doi: 10.1371/journal.ppat.1001267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378. Diep BA, Palazzolo-Ballance AM, Tattevin P, Basuino L, Braughton KR, Whitney AR, Chen L, Kreiswirth BN, Otto M, DeLeo FR, Chambers HF. 2008. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS One 3:e3198. doi: 10.1371/journal.pone.0003198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379. Chatterjee SS, Chen L, Joo H-S, Cheung GYC, Kreiswirth BN, Otto M. 2011. Distribution and regulation of the mobile genetic element-encoded phenol-soluble modulin psm-mec in methicillin-resistant Staphylococcus aureus. PLoS One 6:e28781. doi: 10.1371/journal.pone.0028781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380. Monecke S, Ehricht R, Slickers P, Wiese N, Jonas D. 2009. Intra-strain variability of methicillin-resistant Staphylococcus aureus strains ST228-MRSA-I and ST5-MRSA-II. Eur J Clin Microbiol Infect Dis 28:1383–1390. doi: 10.1007/s10096-009-0796-3 [DOI] [PubMed] [Google Scholar]
- 381. Nübel U, Dordel J, Kurt K, Strommenger B, Westh H, Shukla SK, Zemlicková H, Leblois R, Wirth T, Jombart T, Balloux F, Witte W. 2010. A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog 6:e1000855. doi: 10.1371/journal.ppat.1000855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382. Mwangi MM, Wu SW, Zhou Y, Sieradzki K, de Lencastre H, Richardson P, Bruce D, Rubin E, Myers E, Siggia ED, Tomasz A. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci USA 104:9451–9456. doi: 10.1073/pnas.0609839104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383. Vogel V, Falquet L, Calderon-Copete SP, Basset P, Blanc DS. 2012. Short term evolution of a highly transmissible methicillin-resistant Staphylococcus aureus clone (ST228) in a tertiary care hospital. PLoS One 7:e38969. doi: 10.1371/journal.pone.0038969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384. Benson MA, Ohneck EA, Ryan C, Alonzo F III, Smith H, Narechania A, Kolokotronis S, Satola SW, Uhlemann A, Sebra R, Deikus G, Shopsin B, Planet PJ, Torres VJ. 2014. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element . Molecular Microbiology 93:664–681. doi: 10.1111/mmi.12682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385. Alonzo III F, Benson MA, Chen J, Novick RP, Shopsin B, Torres VJ. 2012. Staphylococcus aureus leucocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo . Molecular Microbiology 83:423–435. doi: 10.1111/j.1365-2958.2011.07942.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386. Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA 106:5883–5888. doi: 10.1073/pnas.0900743106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 388. Kobayashi SD, Malachowa N, Whitney AR, Braughton KR, Gardner DJ, Long D, Bubeck Wardenburg J, Schneewind O, Otto M, Deleo FR. 2011. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect Dis 204:937–941. doi: 10.1093/infdis/jir441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Bubeck Wardenburg J, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo FR. 2008. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J Infect Dis 198:1166–1170. doi: 10.1086/592053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390. Prince A, Wang H, Kitur K, Parker D. 2017. Humanized mice exhibit increased susceptibility to Staphylococcus aureus pneumonia. J Infect Dis 215:1386–1395. doi: 10.1093/infdis/jiw425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391. Tong SYC, Sharma-Kuinkel BK, Thaden JT, Whitney AR, Yang S-J, Mishra NN, Rude T, Lilliebridge RA, Selim MA, Ahn SH, Holt DC, Giffard PM, Bayer AS, Deleo FR, Fowler VG. 2013. Virulence of endemic nonpigmented northern Australian Staphylococcus aureus clone (Clonal complex 75, S. argenteus) is not augmented by staphyloxanthin. J Infect Dis 208:520–527. doi: 10.1093/infdis/jit173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392. Loughran AJ, Gaddy D, Beenken KE, Meeker DG, Morello R, Zhao H, Byrum SD, Tackett AJ, Cassat JE, Smeltzer MS. 2016. Impact of sarA and phenol-soluble modulins on the pathogenesis of osteomyelitis in diverse clinical isolates of Staphylococcus aureus. Infect Immun 84:2586–2594. doi: 10.1128/IAI.00152-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Howden BP, McEvoy CRE, Allen DL, Chua K, Gao W, Harrison PF, Bell J, Coombs G, Bennett-Wood V, Porter JL, Robins-Browne R, Davies JK, Seemann T, Stinear TP. 2011. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog 7:e1002359. doi: 10.1371/journal.ppat.1002359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394. Planet PJ, Diaz L, Kolokotronis SO, Narechania A, Reyes J, Xing G, Rincon S, Smith H, Panesso D, Ryan C, Smith DP, Guzman M, Zurita J, Sebra R, Deikus G, Nolan RL, Tenover FC, Weinstock GM, Robinson DA, Arias CA. 2015. Parallel epidemics of community-associated methicillin-resistant Staphylococcus aureus USA300 infection in North and South America. J Infect Dis 212:1874–1882. doi: 10.1093/infdis/jiv320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395. Chen Y, Chatterjee SS, Porcella SF, Yu Y-S, Otto M, Li M. 2013. Complete genome sequence of a Panton-Valentine leukocidin-negative community-associated methicillin-resistant Staphylococcus aureus strain of sequence type 72 from Korea. PLoS One 8:e72803. doi: 10.1371/journal.pone.0072803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396. Holden MTG, Lindsay JA, Corton C, Quail MA, Cockfield JD, Pathak S, Batra R, Parkhill J, Bentley SD, Edgeworth JD. 2010. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW). J Bacteriol 192:888–892. doi: 10.1128/JB.01255-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397. Howden BP, Seemann T, Harrison PF, McEvoy CR, Stanton J-AL, Rand CJ, Mason CW, Jensen SO, Firth N, Davies JK, Johnson PDR, Stinear TP. 2010. Complete genome sequence of Staphylococcus aureus strain JKD6008, an ST239 clone of methicillin-resistant Staphylococcus aureus with intermediate-level vancomycin resistance. J Bacteriol 192:5848–5849. doi: 10.1128/JB.00951-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398. Cameron DR, Lin YH, Trouillet-Assant S, Tafani V, Kostoulias X, Mouhtouris E, Skinner N, Visvanathan K, Baines SL, Howden B, Monk IR, Laurent F, Stinear TP, Howden BP, Peleg AY. 2017. Vancomycin-intermediate Staphylococcus aureus isolates are attenuated for virulence when compared with susceptible progenitors. Clin Microbiol Infect 23:767–773. doi: 10.1016/j.cmi.2017.03.027 [DOI] [PubMed] [Google Scholar]
- 399. Li Y, Cao B, Zhang Y, Zhou J, Yang B, Wang L. 2011. Complete genome sequence of Staphylococcus aureus T0131, an ST239-MRSA-SCCmec type III clone isolated in China. J Bacteriol 193:3411–3412. doi: 10.1128/JB.05135-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400. Botelho AMN, Costa MOC, Beltrame CO, Ferreira FA, Côrtes MF, Bandeira PT, Lima NCB, Souza RC, Almeida LGP, Vasconcelos ATR, Nicolás MF, Figueiredo AMS. 2016. Complete genome sequence of an agr-dysfunctional variant of the ST239 lineage of the methicillin-resistant Staphylococcus aureus strain GV69 from Brazil. Stand Genomic Sci 11:34. doi: 10.1186/s40793-016-0154-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401. Jin Y, Yu X, Chen Y, Chen W, Shen P, Luo Q, Zhang S, Kong X, Zheng B, Xiao Y. 2020. Characterization of highly virulent community-associated methicillin-resistant Staphylococcus aureus ST9-SCCmec XII causing bloodstream infection in China. Emerg Microbes Infect 9:2526–2535. doi: 10.1080/22221751.2020.1848354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402. Sabirova JS, Xavier BB, Hernalsteens J-P, De Greve H, Ieven M, Goossens H, Malhotra-Kumar S. 2014. Complete genome sequences of two prolific biofilm-forming Staphylococcus aureus isolates belonging to USA300 and EMRSA-15 clonal lineages. Genome Announc 2:e00610-14. doi: 10.1128/genomeA.00610-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. Holden MTG, Feil EJ, Lindsay JA, Peacock SJ, Day NPJ, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, et al. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA 101:9786–9791. doi: 10.1073/pnas.0402521101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404. Rudkin JK, Edwards AM, Bowden MG, Brown EL, Pozzi C, Waters EM, Chan WC, Williams P, O’Gara JP, Massey RC. 2012. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J Infect Dis 205:798–806. doi: 10.1093/infdis/jir845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405. Stegger M, Driebe EM, Roe C, Lemmer D, Bowers JR, Engelthaler DM, Keim P, Andersen PS. 2013. Genome sequence of Staphylococcus aureus strain CA-347, a USA600 methicillin-resistant isolate. Genome Announc 1:e00517-13. doi: 10.1128/genomeA.00517-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406. Beenken KE, Mrak LN, Griffin LM, Zielinska AK, Shaw LN, Rice KC, Horswill AR, Bayles KW, Smeltzer MS. 2010. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One 5:e10790. doi: 10.1371/journal.pone.0010790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407. Chen C-J, Unger C, Hoffmann W, Lindsay JA, Huang Y-C, Götz F. 2013. Characterization and comparison of 2 distinct epidemic community-associated methicillin-resistant Staphylococcus aureus clones of ST59 lineage. PLoS One 8:e63210. doi: 10.1371/journal.pone.0063210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408. Morvan C, Halpern D, Kénanian G, Hays C, Anba-Mondoloni J, Brinster S, Kennedy S, Trieu-Cuot P, Poyart C, Lamberet G, Gloux K, Gruss A. 2016. Environmental fatty acids enable emergence of infectious Staphylococcus aureus resistant to FASII-targeted antimicrobials. Nat Commun 7:12944. doi: 10.1038/ncomms12944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409. Bosi E, Monk JM, Aziz RK, Fondi M, Nizet V, Palsson BØ. 2016. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc Natl Acad Sci USA 113:E3801–E3809. doi: 10.1073/pnas.1523199113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410. Stegger M, Price LB, Larsen AR, Gillece JD, Waters AE, Skov R, Andersen PS. 2012. Genome sequence of Staphylococcus aureus strain 11819-97, an ST80-IV European community-acquired methicillin-resistant isolate. J Bacteriol 194:1625–1626. doi: 10.1128/JB.06653-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411. Chua K, Seemann T, Harrison PF, Davies JK, Coutts SJ, Chen H, Haring V, Moore R, Howden BP, Stinear TP. 2010. Complete genome sequence of Staphylococcus aureus strain JKD6159, a unique Australian clone of ST93-IV community methicillin-resistant Staphylococcus aureus. J Bacteriol 192:5556–5557. doi: 10.1128/JB.00878-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412. Chua KYL, Seemann T, Harrison PF, Monagle S, Korman TM, Johnson PDR, Coombs GW, Howden BO, Davies JK, Howden BP, Stinear TP. 2011. The dominant Australian community-acquired methicillin-resistant Staphylococcus aureus clone ST93-IV [2B] is highly virulent and genetically distinct. PLoS One 6:e25887. doi: 10.1371/journal.pone.0025887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413. Chua KYL, Monk IR, Lin Y-H, Seemann T, Tuck KL, Porter JL, Stepnell J, Coombs GW, Davies JK, Stinear TP, Howden BP. 2014. Hyperexpression of alpha-hemolysin explains enhanced virulence of sequence type 93 community-associated methicillin-resistant Staphylococcus aureus. BMC Microbiol 14:31. doi: 10.1186/1471-2180-14-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414. Stinear TP, Holt KE, Chua K, Stepnell J, Tuck KL, Coombs G, Harrison PF, Seemann T, Howden BP. 2014. Adaptive change inferred from genomic population analysis of the ST93 epidemic clone of community-associated methicillin-resistant Staphylococcus aureus. Genome Biol Evol 6:366–378. doi: 10.1093/gbe/evu022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415. Schijffelen MJ, Boel CHE, van Strijp JAG, Fluit AC. 2010. Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics 11:376. doi: 10.1186/1471-2164-11-376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416. Uhlemann AC, Porcella SF, Trivedi S, Sullivan SB, Hafer C, Kennedy AD, Barbian KD, McCarthy AJ, Street C, Hirschberg DL, Lipkin WI, Lindsay JA, DeLeo FR, Lowy FD. 2012. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. mBio 3:e00027-12. doi: 10.1128/mBio.00027-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417. Bonesso MF, Yeh AJ, Villaruz AE, Joo H-S, McCausland J, Fortaleza CMCB, Cavalcante RS, Sobrinho MT, Ronchi CF, Cheung GYC, Cunha MLRS, Otto M. 2016. Key role of alpha-toxin in fatal pneumonia caused by Staphylococcus aureus sequence type 398. Am J Respir Crit Care Med 193:217–220. doi: 10.1164/rccm.201506-1225LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418. McCarthy AJ, Loeffler A, Witney AA, Gould KA, Lloyd DH, Lindsay JA. 2014. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol Evol 6:2697–2708. doi: 10.1093/gbe/evu214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419. Christiansen MT, Kaas RS, Chaudhuri RR, Holmes MA, Hasman H, Aarestrup FM. 2014. Genome-wide high-throughput screening to investigate essential genes involved in methicillin-resistant Staphylococcus aureus sequence type 398 survival. PLoS One 9:e89018. doi: 10.1371/journal.pone.0089018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420. Warne B, Harkins CP, Harris SR, Vatsiou A, Stanley-Wall N, Parkhill J, Peacock SJ, Palmer T, Holden MTG. 2016. The Ess/type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genomics 17:222. doi: 10.1186/s12864-016-2426-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421. Ariza-Miguel J, Hernández M, Fernández-Natal I, Rodríguez-Lázaro D. 2014. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in a university hospital in northwestern Spain. Int Microbiol 17:149–157. doi: 10.2436/20.1501.01.217 [DOI] [PubMed] [Google Scholar]
- 422. Roberts RB, de Lencastre A, Eisner W, Severina EP, Shopsin B, Kreiswirth BN, Tomasz A, MRSA Collaborative Study Group . 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J Infect Dis 178:164–171. doi: 10.1086/515610 [DOI] [PubMed] [Google Scholar]
- 423. Aires de Sousa M, de Lencastre H, Santos Sanches I, Kikuchi K, Totsuka K, Tomasz A. 2000. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus isolates widely spread in hospitals in New York city and in a hospital in Tokyo, Japan. Microb Drug Resist 6:253–258. doi: 10.1089/mdr.2000.6.253 [DOI] [PubMed] [Google Scholar]
- 424. Chen CJ, Huang YC. 2014. New epidemiology of Staphylococcus aureus infection in Asia. Clin Microbiol Infect 20:605–623. doi: 10.1111/1469-0691.12705 [DOI] [PubMed] [Google Scholar]
- 425. Sá-Leão R, Santos Sanches I, Dias D, Peres I, Barros RM, de Lencastre H. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain J Clin Microbiol 37:1913–1920. doi: 10.1128/JCM.37.6.1913-1920.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426. Abdulgader SM, Shittu AO, Nicol MP, Kaba M. 2015. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol 6:348. doi: 10.3389/fmicb.2015.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427. Karauzum H, Ferry T, de Bentzmann S, Lina G, Bes M, Vandenesch F, Schmaler M, Berger-Bächi B, Etienne J, Landmann R. 2008. Comparison of adhesion and virulence of two predominant hospital-acquired methicillin-resistant Staphylococcus aureus clones and clonal methicillin-susceptible S. aureus isolates. Infect Immun 76:5133–5138. doi: 10.1128/IAI.01697-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428. Pérez-Montarelo D, Viedma E, Murcia M, Muñoz-Gallego I, Larrosa N, Brañas P, Fernández-Hidalgo N, Gavaldà J, Almirante B, Chaves F. 2017. Pathogenic characteristics of Staphylococcus aureus endovascular infection isolates from different clonal complexes. Front Microbiol 8:917. doi: 10.3389/fmicb.2017.00917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429. Peleg AY, Monga D, Pillai S, Mylonakis E, Moellering RC, Eliopoulos GM. 2009. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J Infect Dis 199:532–536. doi: 10.1086/596511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430. Xiong YQ, Sharma-Kuinkel BK, Casillas-Ituarte NN, Fowler VG, Rude T, DiBartola AC, Lins RD, Abdel-Hady W, Lower SK, Bayer AS, Camilli A. 2015. Endovascular infections caused by methicillin-resistant Staphylococcus aureus are linked to clonal complex-specific alterations in binding and invasion domains of fibronectin-binding protein A as well as the occurrence of fnbB. Infect Immun 83:4772–4780. doi: 10.1128/IAI.01074-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431. Challagundla L, Reyes J, Rafiqullah I, Sordelli DO, Echaniz-Aviles G, Velazquez-Meza ME, Castillo-Ramírez S, Fittipaldi N, Feldgarden M, Chapman SB, Calderwood MS, Carvajal LP, Rincon S, Hanson B, Planet PJ, Arias CA, Diaz L, Robinson DA. 2018. Phylogenomic classification and the evolution of clonal complex 5 methicillin-resistant Staphylococcus aureus in the Western hemisphere. Front Microbiol 9:1901. doi: 10.3389/fmicb.2018.01901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432. He C, Xu S, Zhao H, Hu F, Xu X, Jin S, Yang H, Gong F, Liu Q. 2018. Leukotoxin and pyrogenic toxin superantigen gene backgrounds in bloodstream and wound Staphylococcus aureus isolates from Eastern region of China. BMC Infect Dis 18:395. doi: 10.1186/s12879-018-3297-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433. Wang M, Zheng Y, Mediavilla J, Chen L, Kreiswirth B, Song Y, Yang R, Du H. 2017. Hospital dissemination of TST-1-positive Clonal complex 5 (CC5) methicillin-resistant Staphylococcus aureus. Front Cell Infect Microbiol 7:101. doi: 10.3389/fcimb.2017.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434. Calderwood MS, Desjardins CA, Sakoulas G, Nicol R, Dubois A, Delaney ML, Kleinman K, Cosimi LA, Feldgarden M, Onderdonk AB, Birren BW, Platt R, Huang SS, Program CDCPE. 2014. Staphylococcal enterotoxin P predicts bacteremia in hospitalized patients colonized with methicillin-resistant Staphylococcus aureus . J Infect Dis 209:571–577. doi: 10.1093/infdis/jit501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435. Omoe K, Imanishi K, Hu DL, Kato H, Fugane Y, Abe Y, Hamaoka S, Watanabe Y, Nakane A, Uchiyama T, Shinagawa K. 2005. Characterization of novel staphylococcal enterotoxin-like toxin type P. Infect Immun 73:5540–5546. doi: 10.1128/IAI.73.9.5540-5546.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436. Aung MS, Kawaguchiya M, Urushibara N, Sumi A, Ito M, Kudo K, Morimoto S, Hosoya S, Kobayashi N. 2017. Molecular characterization of methicillin-resistant Staphylococcus aureus from outpatients in northern Japan: increasing tendency of ST5/ST764 MRSA-IIa with arginine catabolic mobile element. Microb Drug Resist 23:616–625. doi: 10.1089/mdr.2016.0176 [DOI] [PubMed] [Google Scholar]
- 437. Takano T, Hung WC, Shibuya M, Higuchi W, Iwao Y, Nishiyama A, Reva I, Khokhlova OE, Yabe S, Ozaki K, Takano M, Yamamoto T. 2013. A new local variant (ST764) of the globally disseminated ST5 lineage of hospital-associated Methicillin-resistant Staphylococcus aureus (MRSA) carrying the virulence determinants of community-associated MRSA. Antimicrob Agents Chemother 57:1589–1595. doi: 10.1128/AAC.01147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438. King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. 2006. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med 144:309–317. doi: 10.7326/0003-4819-144-5-200603070-00005 [DOI] [PubMed] [Google Scholar]
- 439. Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA, Group EMINS. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med 355:666–674. doi: 10.1056/NEJMoa055356 [DOI] [PubMed] [Google Scholar]
- 440. Diekema DJ, Richter SS, Heilmann KP, Dohrn CL, Riahi F, Tendolkar S, McDanel JS, Doern GV. 2014. Continued emergence of USA300 Methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect Control Hosp Epidemiol 35:285–292. doi: 10.1086/675283 [DOI] [PubMed] [Google Scholar]
- 441. Villaruz AE, Bubeck Wardenburg J, Khan BA, Whitney AR, Sturdevant DE, Gardner DJ, DeLeo FR, Otto M. 2009. A point mutation in the agr locus rather than expression of the panton-valentine leukocidin caused previously reported phenotypes in Staphylococcus aureus pneumonia and gene regulation. J Infect Dis 200:724–734. doi: 10.1086/604728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442. Olaniyi RO, Pancotto L, Grimaldi L, Bagnoli F. 2018. Deciphering the pathological role of staphylococcal alpha-toxin and panton-valentine leukocidin using a novel ex vivo human skin model. Front Immunol 9:951. doi: 10.3389/fimmu.2018.00951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443. Tseng CW, Biancotti JC, Berg BL, Gate D, Kolar SL, Müller S, Rodriguez MD, Rezai-Zadeh K, Fan X, Beenhouwer DO, Town T, Liu GY. 2015. Increased susceptibility of humanized NSG mice to panton-valentine leukocidin and Staphylococcus aureus skin infection. PLoS Pathog 11:e1005292. doi: 10.1371/journal.ppat.1005292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444. Thurlow LR, Joshi GS, Richardson AR. 2012. Virulence strategies of the dominant USA300 lineage of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). FEMS Immunol Med Microbiol 65:5–22. doi: 10.1111/j.1574-695X.2012.00937.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445. Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, des Etages S-A, Jones A, Palazzolo-Ballance AM, Perdreau-Remington F, Sensabaugh GF, DeLeo FR, Chambers HF. 2008. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of Methicillin-resistant Staphylococcus aureus. J Infect Dis 197:1523–1530. doi: 10.1086/587907 [DOI] [PubMed] [Google Scholar]
- 446. Ender M, McCallum N, Adhikari R, Berger-Bächi B. 2004. Fitness cost of SCCmec and Methicillin resistance levels in Staphylococcus aureus. Antimicrob Agents Chemother 48:2295–2297. doi: 10.1128/AAC.48.6.2295-2297.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447. Goering RV, McDougal LK, Fosheim GE, Bonnstetter KK, Wolter DJ, Tenover FC. 2007. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J Clin Microbiol 45:1981–1984. doi: 10.1128/JCM.00273-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448. Shore AC, Rossney AS, Brennan OM, Kinnevey PM, Humphreys H, Sullivan DJ, Goering RV, Ehricht R, Monecke S, Coleman DC. 2011. Characterization of a novel arginine catabolic mobile element (ACME) and staphylococcal chromosomal cassette mec composite island with significant homology to Staphylococcus epidermidis ACME type II in Methicillin-resistant Staphylococcus aureus genotype ST22-MRSA-IV. Antimicrob Agents Chemother 55:1896–1905. doi: 10.1128/AAC.01756-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449. Montgomery CP, Boyle-Vavra S, Daum RS. 2009. The arginine catabolic mobile element is not associated with enhanced virulence in experimental invasive disease caused by the community-associated methicillin-resistant Staphylococcus aureus USA300 genetic background . Infect Immun 77:2650–2656. doi: 10.1128/IAI.00256-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450. Barbier F, Lebeaux D, Hernandez D, Delannoy A-S, Caro V, François P, Schrenzel J, Ruppé E, Gaillard K, Wolff M, Brisse S, Andremont A, Ruimy R. 2011. High prevalence of the arginine catabolic mobile element in carriage isolates of methicillin-resistant staphylococcus epidermidis. J Antimicrob Chemother 66:29–36. doi: 10.1093/jac/dkq410 [DOI] [PubMed] [Google Scholar]
- 451. Miller LS, Cho JS. 2011. Immunity against Staphylococcus aureus cutaneous infections. Nat Rev Immunol 11:505–518. doi: 10.1038/nri3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452. Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107. doi: 10.1016/j.chom.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453. Makhlin J, Kofman T, Borovok I, Kohler C, Engelmann S, Cohen G, Aharonowitz Y. 2007. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J Bacteriol 189:5976–5986. doi: 10.1128/JB.00592-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454. El Baze P, Milano G, Verrando P, Renée N, Ortonne JP. 1983. Polyamine levels in normal human skin. a comparative study of pure epidermis, pure dermis, and suction blister fluid. Arch Dermatol Res 275:218–221. doi: 10.1007/BF00416663 [DOI] [PubMed] [Google Scholar]
- 455. Seiler N, Atanassov CL. 1994. The natural polyamines and the immune system. Prog Drug Res 43:87–141. doi: 10.1007/978-3-0348-7156-3_4 [DOI] [PubMed] [Google Scholar]
- 456. Kwon DH, Lu CD. 2007. Polyamine effects on antibiotic susceptibility in bacteria. Antimicrob Agents Chemother 51:2070–2077. doi: 10.1128/AAC.01472-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457. Joshi GS, Spontak JS, Klapper DG, Richardson AR. 2011. Arginine catabolic mobile element encoded SpeG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous Polyamines. Mol Microbiol 82:9–20. doi: 10.1111/j.1365-2958.2011.07809.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458. Purves J, Thomas J, Riboldi GP, Zapotoczna M, Tarrant E, Andrew PW, Londoño A, Planet PJ, Geoghegan JA, Waldron KJ, Morrissey JA. 2018. A horizontally gene transferred copper resistance locus confers hyper-resistance to antibacterial copper toxicity and enables survival of community acquired methicillin resistant Staphylococcus aureus USA300 in macrophages. Environ Microbiol 20:1576–1589. doi: 10.1111/1462-2920.14088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459. Djoko KY, Ong CY, Walker MJ, McEwan AG. 2015. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem 290:18954–18961. doi: 10.1074/jbc.R115.647099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460. White C, Lee J, Kambe T, Fritsche K, Petris MJ. 2009. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem 284:33949–33956. doi: 10.1074/jbc.M109.070201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461. Sitthisak S, Knutsson L, Webb JW, Jayaswal RK. 2007. Molecular characterization of the copper transport system in Staphylococcus aureus. Microbiology (Reading) 153:4274–4283. doi: 10.1099/mic.0.2007/009860-0 [DOI] [PubMed] [Google Scholar]
- 462. Planet PJ. 2017. Life after USA300: the rise and fall of a superbug. J Infect Dis 215:S71–S77. doi: 10.1093/infdis/jiw444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463. Frisch MB, Castillo-Ramírez S, Petit RA, Farley MM, Ray SM, Albrecht VS, Limbago BM, Hernandez J, See I, Satola SW, Read TD. 2018. Invasive methicillin-resistant Staphylococcus aureus USA500 strains from the U.S. emerging infections program constitute three geographically distinct lineages. mSphere 3:e00571-17. doi: 10.1128/mSphere.00571-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464. Boyle-Vavra S, Li X, Alam MT, Read TD, Sieth J, Cywes-Bentley C, Dobbins G, David MZ, Kumar N, Eells SJ, Miller LG, Boxrud DJ, Chambers HF, Lynfield R, Lee JC, Daum RS. 2015. USA300 and USA500 clonal lineages of Staphylococcus aureus do not produce a capsular polysaccharide due to conserved mutations in the CAP5 locus. mBio 6:e02585-14. doi: 10.1128/mBio.02585-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465. Jamrozy DM, Harris SR, Mohamed N, Peacock SJ, Tan CY, Parkhill J, Anderson AS, Holden MTG. 2016. Pan-genomic perspective on the evolution of the Staphylococcus aureus USA300 epidemic. Microbial Genomics 2. doi: 10.1099/mgen.0.000058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466. Seybold U, Kourbatova EV, Johnson JG, Halvosa SJ, Wang YF, King MD, Ray SM, Blumberg HM. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis 42:647–656. doi: 10.1086/499815 [DOI] [PubMed] [Google Scholar]
- 467. Nelson MU, Bizzarro MJ, Baltimore RS, Dembry LM, Gallagher PG. 2015. Clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit in the decade following implementation of an active detection and isolation program. J Clin Microbiol 53:2492–2501. doi: 10.1128/JCM.00470-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468. Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. 2009. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J Exp Med 206:2417–2427. doi: 10.1084/jem.20090097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469. Tantawy E, Schwermann N, Ostermeier T, Garbe A, Bähre H, Vital M, Winstel V. 2022. Staphylococcus aureus multiplexes death-effector deoxyribonucleosides to neutralize phagocytes. Front Immunol 13:847171. doi: 10.3389/fimmu.2022.847171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470. Deng J, Zhang B-Z, Chu H, Wang X-L, Wang Y, Gong H-R, Li R, Yang D, Li C, Dou Y, Gao P, Cai J-P, Jin M, Du Q, Chan JF-W, Kao RY-T, Yuen K-Y, Huang J-D. 2021. Adenosine synthase A contributes to recurrent Staphylococcus aureus infection by dampening protective immunity. EBioMedicine 70:103505. doi: 10.1016/j.ebiom.2021.103505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471. Wagenaar JA, Yue H, Pritchard J, Broekhuizen-Stins M, Huijsdens X, Mevius DJ, Bosch T, Van Duijkeren E. 2009. Unexpected sequence types in livestock associated methicillin-resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Vet Microbiol 139:405–409. doi: 10.1016/j.vetmic.2009.06.014 [DOI] [PubMed] [Google Scholar]
- 472. Chuang YY, Huang YC. 2015. Livestock-associated meticillin-resistant Staphylococcus aureus in Asia: an emerging issue Int J Antimicrob Agents 45:334–340. doi: 10.1016/j.ijantimicag.2014.12.007 [DOI] [PubMed] [Google Scholar]
- 473. Rinsky JL, Nadimpalli M, Wing S, Hall D, Baron D, Price LB, Larsen J, Stegger M, Stewart J, Heaney CD, Cloeckaert A. 2013. Livestock-associated methicillin and multidrug resistant Staphylococcus aureus is present among industrial, not antibiotic-free livestock operation workers in North Carolina. PLoS One 8:e67641. doi: 10.1371/journal.pone.0067641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474. Nadimpalli M, Rinsky JL, Wing S, Hall D, Stewart J, Larsen J, Nachman KE, Love DC, Pierce E, Pisanic N, Strelitz J, Harduar-Morano L, Heaney CD. 2015. Persistence of livestock-associated antibiotic-resistant Staphylococcus aureus among industrial hog operation workers in North Carolina over 14 days. Occup Environ Med 72:90–99. doi: 10.1136/oemed-2014-102095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475. Yu F, Cienfuegos-Gallet AV, Cunningham MH, Jin Y, Wang B, Kreiswirth BN, Chen L, Cleary DW. 2021. Molecular evolution and adaptation of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) sequence type 9. mSystems 6. doi: 10.1128/mSystems.00492-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476. Zhou W, Li X, Osmundson T, Shi L, Ren J, Yan H. 2018. WGS analysis of ST9-MRSA-XII isolates from live pigs in China provides insights into transmission among porcine, human and bovine hosts. J Antimicrob Chemother 73:2652–2661. doi: 10.1093/jac/dky245 [DOI] [PubMed] [Google Scholar]
- 477. Fang H-W, Chiang P-H, Huang Y-C, de Lencastre H. 2014. Livestock-associated Methicillin-resistant Staphylococcus aureus ST9 in pigs and related personnel in Taiwan. PLoS One 9:e88826. doi: 10.1371/journal.pone.0088826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478. Wang JT, Liao CH, Fang CT, Chie WC, Lai MS, Lauderdale TL, Lee WS, Huang JH, Chang SC. 2009. Prevalence of and risk factors for Colonization by Methicillin-resistant Staphylococcus aureus among adults in community settings in Taiwan. J Clin Microbiol 47:2957–2963. doi: 10.1128/JCM.00853-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479. Ye X, Wang X, Fan Y, Peng Y, Li L, Li S, Huang J, Yao Z, Chen S. 2016. Genotypic and phenotypic markers of livestock-associated methicillin-resistant Staphylococcus aureus CC9 in humans. Appl Environ Microbiol 82:3892–3899. doi: 10.1128/AEM.00091-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480. Randad PR, Larsen J, Kaya H, Pisanic N, Ordak C, Price LB, Aziz M, Nadimpalli ML, Rhodes S, Stewart JR, Love DC, Mohr D, Davis MF, Miller LS, Hall D, Carroll KC, Perl TM, Heaney CD. 2021. Transmission of antimicrobial-resistant Staphylococcus aureus clonal complex 9 between pigs and humans, United States . Emerg Infect Dis 27:740–748. doi: 10.3201/eid2703.191775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481. Chen C-J, Lauderdale T-LY, Lu C-T, Chuang Y-Y, Yang C-C, Wu T-S, Lee C-Y, Lu M-C, Ko W-C, Huang Y-C. 2018. Clinical and molecular features of MDR livestock-associated MRSA ST9 with staphylococcal cassette chromosome mecXII in humans. J Antimicrob Chemother 73:33–40. doi: 10.1093/jac/dkx357 [DOI] [PubMed] [Google Scholar]
- 482. Moodley A, Latronico F, Guardabassi L. 2011. Experimental colonization of pigs with methicillin-resistant Staphylococcus aureus (MRSA): insights into the colonization and transmission of livestock-associated MRSA. Epidemiol Infect 139:1594–1600. doi: 10.1017/S0950268810002888 [DOI] [PubMed] [Google Scholar]
- 483. Li SM, Zhou YF, Li L, Fang LX, Duan JH, Liu FR, Liang HQ, Wu YT, Gu WQ, Liao XP, Sun J, Xiong YQ, Liu YH. 2018. Characterization of the multi-drug resistance gene CFR in methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from animals and humans in China. Front Microbiol 9:2925. doi: 10.3389/fmicb.2018.02925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484. Jiang N, Wyres KL, Li J, Feßler AT, Krüger H, Wang Y, Holt KE, Schwarz S, Wu C. 2021. Evolution and genomic insight into methicillin-resistant Staphylococcus aureus ST9 in China. J Antimicrob Chemother 76:1703–1711. doi: 10.1093/jac/dkab106 [DOI] [PubMed] [Google Scholar]
- 485. Grundmann H, Schouls LM, Aanensen DM, Pluister GN, Tami A, Chlebowicz M, Glasner C, Sabat AJ, Weist K, Heuer O, Friedrich AW, ESGoME M, European Staphylococcal Reference Laboratory Working G. 2014. The dynamic changes of dominant clones of Staphylococcus aureus causing bloodstream infections in the European region: results of a second structured survey. Euro Surveill 19:20987. doi: 10.2807/1560-7917.es2014.19.49.20987 [DOI] [PubMed] [Google Scholar]
- 486. Udo EE, Boswihi SS, Al-Sweih N. 2016. High prevalence of toxic shock syndrome toxin-producing epidemic methicillin-resistant Staphylococcus aureus 15 (EMRSA-15) strains in Kuwait hospitals. New Microbes New Infect 12:24–30. doi: 10.1016/j.nmni.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487. Coombs GW, Pearson JC, Nimmo GR, Collignon PJ, Bell JM, McLaws M-L, Christiansen KJ, Turnidge JD, Australian Group on Antimicrobial Resistance . 2013. Antimicrobial susceptibility of Staphylococcus aureus and molecular epidemiology of meticillin-resistant S. aureus isolated from Australian hospital inpatients: report from the Australian group on antimicrobial resistance 2011 Staphylococcus aureus surveillance programme. J Glob Antimicrob Resist 1:149–156. doi: 10.1016/j.jgar.2013.04.005 [DOI] [PubMed] [Google Scholar]
- 488. Holden MTG, Hsu L-Y, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, et al. 2013. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res 23:653–664. doi: 10.1101/gr.147710.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489. Baldan R, Testa F, Lorè NI, Bragonzi A, Cichero P, Ossi C, Biancardi A, Nizzero P, Moro M, Cirillo DM. 2012. Factors contributing to epidemic MRSA clones replacement in a hospital setting. PLoS One 7:e43153. doi: 10.1371/journal.pone.0043153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490. Knight GM, Budd EL, Whitney L, Thornley A, Al-Ghusein H, Planche T, Lindsay JA. 2012. Shift in dominant hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) clones over time. J Antimicrob Chemother 67:2514–2522. doi: 10.1093/jac/dks245 [DOI] [PubMed] [Google Scholar]
- 491. Baldan R, Rancoita PMV, Di Serio C, Mazzotti M, Cichero P, Ossi C, Biancardi A, Nizzero P, Saracco A, Scarpellini P, Cirillo DM. 2015. Epidemic MRSA clone ST22-IV is more resistant to multiple Host- and environment-related stresses compared with ST228-I. J Antimicrob Chemother 70:757–765. doi: 10.1093/jac/dku467 [DOI] [PubMed] [Google Scholar]
- 492. Hughes C, Ferguson J. 2017. Phenotypic chlorhexidine and triclosan susceptibility in clinical Staphylococcus aureus isolates in Australia. Pathology 49:633–637. doi: 10.1016/j.pathol.2017.05.008 [DOI] [PubMed] [Google Scholar]
- 493. Boakes E, Marbach H, Lynham S, Ward M, Edgeworth JD, Otter JA. 2016. Comparative analysis of phenol-soluble modulin production and galleria mellonella killing by community-associated and healthcare-associated meticillin-resistant Staphylococcus aureus strains. J Med Microbiol 65:1429–1433. doi: 10.1099/jmm.0.000379 [DOI] [PubMed] [Google Scholar]
- 494. Mairpady Shambat S, Siemens N, Monk IR, Mohan DB, Mukundan S, Krishnan KC, Prabhakara S, Snäll J, Kearns A, Vandenesch F, Svensson M, Kotb M, Gopal B, Arakere G, Norrby-Teglund A. 2016. A point mutation in AgrC determines cytotoxic or colonizing properties associated with phenotypic variants of ST22 MRSA strains. Sci Rep 6:31360. doi: 10.1038/srep31360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495. Recker M, Laabei M, Toleman MS, Reuter S, Saunderson RB, Blane B, Török ME, Ouadi K, Stevens E, Yokoyama M, Steventon J, Thompson L, Milne G, Bayliss S, Bacon L, Peacock SJ, Massey RC. 2017. Clonal differences in Staphylococcus aureus bacteraemia-associated mortality. Nat Microbiol 2:1381–1388. doi: 10.1038/s41564-017-0001-x [DOI] [PubMed] [Google Scholar]
- 496. Rausch M, Deisinger JP, Ulm H, Müller A, Li W, Hardt P, Wang X, Li X, Sylvester M, Engeser M, Vollmer W, Müller CE, Sahl HG, Lee JC, Schneider T. 2019. Coordination of capsule assembly and cell wall biosynthesis in Staphylococcus aureus. Nat Commun 10:1404. doi: 10.1038/s41467-019-09356-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497. Tzianabos AO, Wang JY, Lee JC. 2001. Structural rationale for the modulation of abscess formation by Staphylococcus aureus capsular polysaccharides. Proc Natl Acad Sci USA 98:9365–9370. doi: 10.1073/pnas.161175598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498. Nilsson IM, Lee JC, Bremell T, Rydén C, Tarkowski A. 1997. The role of staphylococcal polysaccharide microcapsule expression in septicemia and septic arthritis. Infect Immun 65:4216–4221. doi: 10.1128/iai.65.10.4216-4221.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499. Robinson DA, Kearns AM, Holmes A, Morrison D, Grundmann H, Edwards G, O’Brien FG, Tenover FC, McDougal LK, Monk AB, Enright MC. 2005. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired Meticillin-resistant clone. Lancet 365:1256–1258. doi: 10.1016/S0140-6736(05)74814-5 [DOI] [PubMed] [Google Scholar]
- 500. Rountree PM, Beard MA. 1958. Further observations on infection with phage type 80 staphylococci in Australia. Med J Aust 45:789–795. [PubMed] [Google Scholar]
- 501. Bynoe ET, Elder RH, Comtois RD. 1956. Phage-typing and antibiotic-resistance of staphylococci isolated in a general hospital. Can J Microbiol 2:346–358. doi: 10.1139/m56-041 [DOI] [PubMed] [Google Scholar]
- 502. Gillespie WA, Alder VG. 1957. Control of an outbreak of staphylococcal infection in a hospital. Lancet 272:632–634. doi: 10.1016/s0140-6736(57)91091-7 [DOI] [PubMed] [Google Scholar]
- 503. Cox RA, Conquest C, Mallaghan C, Marples RR. 1995. A major outbreak of methicillin-resistant staphylococcus aureus caused by a new phage-type (EMRSA-16). J Hosp Infect 29:87–106. doi: 10.1016/0195-6701(95)90191-4 [DOI] [PubMed] [Google Scholar]
- 504. Johnson AP. 2001. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European antimicrobial resistance surveillance system (EARSS). J Antimicrob Chemother 48:143–144. doi: 10.1093/jac/48.1.143 [DOI] [PubMed] [Google Scholar]
- 505. Ellington MJ, Hope R, Livermore DM, Kearns AM, Henderson K, Cookson BD, Pearson A, Johnson AP. 2010. Decline of EMRSA-16 amongst methicillin-resistant Staphylococcus aureus causing bacteraemias in the UK between 2001 and 2007. J Antimicrob Chemother 65:446–448. doi: 10.1093/jac/dkp448 [DOI] [PubMed] [Google Scholar]
- 506. Sharma-Kuinkel BK, Mongodin EF, Myers JR, Vore KL, Canfield GS, Fraser CM, Rude TH, Fowler VG, Gill SR. 2015. Potential influence of Staphylococcus aureus clonal complex 30 genotype and transcriptome on hematogenous infections. Open Forum Infect Dis 2:ofv093. doi: 10.1093/ofid/ofv093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507. Rasigade J-P, Leclère A, Alla F, Tessier A, Bes M, Lechiche C, Vernet-Garnier V, Laouénan C, Vandenesch F, Leport C, The AEPEI Study Group . 2018. Staphylococcus aureus CC30 lineage and absence of SED,J,R-harboring plasmid predict embolism in infective endocarditis. Front Cell Infect Microbiol 8:187. doi: 10.3389/fcimb.2018.00187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508. Fowler VG, Nelson CL, McIntyre LM, Kreiswirth BN, Monk A, Archer GL, Federspiel J, Naidich S, Remortel B, Rude T, Brown P, Reller LB, Corey GR, Gill SR. 2007. Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. J Infect Dis 196:738–747. doi: 10.1086/520088 [DOI] [PubMed] [Google Scholar]
- 509. Nienaber JJC, Sharma Kuinkel BK, Clarke-Pearson M, Lamlertthon S, Park L, Rude TH, Barriere S, Woods CW, Chu VH, Marín M, Bukovski S, Garcia P, Corey GR, Korman T, Doco-Lecompte T, Murdoch DR, Reller LB, Fowler VG, International Collaboration on Endocarditis-Microbiology Investigators . 2011. Methicillin-susceptible Staphylococcus aureus Endocarditis isolates are associated with clonal complex 30 genotype and a distinct repertoire of enterotoxins and adhesins. J Infect Dis 204:704–713. doi: 10.1093/infdis/jir389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510. Ferreira-González I, Fernández-Hidalgo N, Ribera A. 2018. “'A pragmatic approach for mortality prediction after surgery in infective endocarditis' - author's reply”. Clin Microbiol Infect 24:1354. doi: 10.1016/j.cmi.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 511. Xiong YQ, Fowler VG, Yeaman MR, Perdreau-Remington F, Kreiswirth BN, Bayer AS. 2009. Phenotypic and Genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia In vitro and in an experimental endocarditis model. J Infect Dis 199:201–208. doi: 10.1086/595738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512. Di Gregorio S, Haim MS, Vielma Vallenilla J, Cohen V, Rago L, Gulone L, Aanensen DM, Argimón S, Mollerach M, Fey PD. 2021. Genomic epidemiology of CC30 Methicillin-resistant Staphylococcus aureus strains from Argentina reveals four major clades with distinctive genetic features. mSphere 6. doi: 10.1128/mSphere.01297-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513. McGavin MJ, Arsic B, Nickerson NN. 2012. Evolutionary blueprint for host- and niche-adaptation in Staphylococcus aureus clonal complex CC30. Front Cell Infect Microbiol 2:48. doi: 10.3389/fcimb.2012.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514. Cheung GYC, Kretschmer D, Duong AC, Yeh AJ, Ho TV, Chen Y, Joo H-S, Kreiswirth BN, Peschel A, Otto M. 2014. Production of an attenuated phenol-soluble modulin variant unique to the MRSA clonal complex 30 increases severity of bloodstream infection. PLoS Pathog 10:e1004298. doi: 10.1371/journal.ppat.1004298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515. Sharma H, Smith D, Turner CE, Game L, Pichon B, Hope R, Hill R, Kearns A, Sriskandan S. 2018. Clinical and molecular epidemiology of staphylococcal toxic shock syndrome in the United Kingdom. Emerg Infect Dis 24:258–266. doi: 10.3201/eid2402.170606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516. Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM. 2001. Evolutionary genomics of Staphylococcus aureus: Insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci U S A 98:8821–8826. doi: 10.1073/pnas.161098098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517. McAdam PR, Templeton KE, Edwards GF, Holden MTG, Feil EJ, Aanensen DM, Bargawi HJA, Spratt BG, Bentley SD, Parkhill J, Enright MC, Holmes A, Girvan EK, Godfrey PA, Feldgarden M, Kearns AM, Rambaut A, Robinson DA, Fitzgerald JR. 2012. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA 109:9107–9112. doi: 10.1073/pnas.1202869109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518. Seidl K, Bischoff M, Berger-Bächi B. 2008. Ccpa mediates the Catabolite repression of TST in Staphylococcus aureus. Infect Immun 76:5093–5099. doi: 10.1128/IAI.00724-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519. Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. 2012. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr Opin Microbiol 15:588–595. doi: 10.1016/j.mib.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 520. DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521. Saadatian-Elahi M, Tristan A, Laurent F, Rasigade J-P, Bouchiat C, Ranc A-G, Lina G, Dauwalder O, Etienne J, Bes M, Vandenesch F. 2013. Basic rules of hygiene protect health care and lab workers from nasal colonization by Staphylococcus aureus: an international cross-sectional study. PLoS One 8:e82851. doi: 10.1371/journal.pone.0082851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522. Robinson DA, Enright MC. 2004. Evolution of Staphylococcus aureus by large Chromosomal replacements. J Bacteriol 186:1060–1064. doi: 10.1128/JB.186.4.1060-1064.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523. Gill JL, Hedge J, Wilson DJ, MacLean RC. 2021. Evolutionary processes driving the rise and fall of Staphylococcus aureus ST239, a dominant hybrid pathogen. mBio 12:e0216821. doi: 10.1128/mBio.02168-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524. Laabei M, Peacock SJ, Blane B, Baines SL, Howden BP, Stinear TP, Massey RC. 2021. Significant variability exists in the cytotoxicity of global methicillin-resistant Staphylococcus aureus lineages. Microbiology (Reading) 167:001119. doi: 10.1099/mic.0.001119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525. Baines SL, Holt KE, Schultz MB, Seemann T, Howden BO, Jensen SO, van Hal SJ, Coombs GW, Firth N, Powell DR, Stinear TP, Howden BP. 2015. Convergent adaptation in the dominant global hospital clone ST239 of Methicillin-resistant Staphylococcus aureus. mBio 6:e00080. doi: 10.1128/mBio.00080-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526. Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527. Söderquist B, Andersson M, Nilsson M, Nilsdotter-Augustinsson Å, Persson L, Friberg Ö, Jacobsson S. 2009. Staphylococcus epidermidis surface protein I (SesI): a marker of the invasive capacity of S. epidermidis? J Med Microbiol 58:1395–1397. doi: 10.1099/jmm.0.008771-0 [DOI] [PubMed] [Google Scholar]
- 528. Botelho AMN, Cerqueira E Costa MO, Moustafa AM, Beltrame CO, Ferreira FA, Côrtes MF, Costa BSS, Silva DNS, Bandeira PT, Lima NCB, Souza RC, de Almeida LGP, Vasconcelos ATR, Narechania A, Ryan C, O’Brien K, Kolokotronis S-O, Planet PJ, Nicolás MF, Figueiredo AMS. 2019. Local diversification of methicillin-resistant Staphylococcus aureus ST239 in South America after its rapid worldwide dissemination. Front Microbiol 10:82. doi: 10.3389/fmicb.2019.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529. Nakaminami H, Ito T, Han X, Ito A, Matsuo M, Uehara Y, Baba T, Hiramatsu K, Noguchi N. 2017. First report of sasX-positive methicillin-resistant Staphylococcus aureus in Japan. FEMS Microbiol Lett 364. doi: 10.1093/femsle/fnx171 [DOI] [PubMed] [Google Scholar]
- 530. von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia . N Engl J Med 344:11–16. doi: 10.1056/NEJM200101043440102 [DOI] [PubMed] [Google Scholar]
- 531. Liu Q, Du X, Hong X, Li T, Zheng B, He L, Wang Y, Otto M, Li M. 2015. Targeting surface protein SasX by active and passive vaccination to reduce Staphylococcus aureus Colonization and infection. Infect Immun 83:2168–2174. doi: 10.1128/IAI.02951-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532. Hong X, Qin J, Li T, Dai Y, Wang Y, Liu Q, He L, Lu H, Gao Q, Lin Y, Li M. 2016. Staphylococcal protein A promotes colonization and immune evasion of the epidemic healthcare-associated MRSA ST239. Front Microbiol 7:951. doi: 10.3389/fmicb.2016.00951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533. He L, Meng H, Liu Q, Hu M, Wang Y, Chen X, Liu X, Li M. 2018. Distinct virulent network between healthcare- and community-associated Staphylococcus aureus based on Proteomic analysis. Clin Proteomics 15:2. doi: 10.1186/s12014-017-9178-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534. Chuang YY, Huang YC. 2013. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis 13:698–708. doi: 10.1016/S1473-3099(13)70136-1 [DOI] [PubMed] [Google Scholar]
- 535. Huang YC, Chen CJ. 2011. Community-associated meticillin-resistant Staphylococcus aureus in children in Taiwan, 2000S. Int J Antimicrob Agents 38:2–8. doi: 10.1016/j.ijantimicag.2011.01.011 [DOI] [PubMed] [Google Scholar]
- 536. Qiao Y, Dong F, Song W, Wang L, Yang Y, Shen X. 2013. Hospital- and community-associated methicillin-resistant Staphylococcus aureus: a 6-year surveillance study of invasive infections in Chinese children. Acta Paediatr 102:1081–1086. doi: 10.1111/apa.12386 [DOI] [PubMed] [Google Scholar]
- 537. Coombs GW, Monecke S, Ehricht R, Slickers P, Pearson JC, Tan HL, Christiansen KJ, O’Brien FG. 2010. Differentiation of clonal complex 59 community-associated methicillin-resistant Staphylococcus aureus in Western Australia. Antimicrob Agents Chemother 54:1914–1921. doi: 10.1128/AAC.01287-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538. Chen YJ, Liu KL, Chen CJ, Huang YC. 2015. Comparative molecular characteristics of community-associated and healthcare-associated methicillin-resistant Staphylococcus aureus isolates from adult patients in northern Taiwan. Medicine 94:e1961. doi: 10.1097/MD.0000000000001961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539. Hu L, Li Y, Lu Y, Klena JD, Qiu Y, Lin Y, Jiang M, Shi X, Chen L, Liu X, Ma H, Cheng J, Wu S, Kan B, Hu Q. 2016. Clinical characteristics, virulence factors and molecular typing of methicillin-resistant Staphylococcus aureus infections in Shenzhen city, China. Epidemiol Infect 144:3037–3045. doi: 10.1017/S0950268816001552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540. Ward MJ, Goncheva M, Richardson E, McAdam PR, Raftis E, Kearns A, Daum RS, David MZ, Lauderdale TL, Edwards GF, Nimmo GR, Coombs GW, Huijsdens X, Woolhouse MEJ, Fitzgerald JR. 2016. Identification of source and sink populations for the emergence and global spread of the East-Asia clone of community-associated MRSA. Genome Biol 17:160. doi: 10.1186/s13059-016-1022-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541. Li S, Sun S, Yang C, Chen H, Yin Y, Li H, Zhao C, Wang H. 2018. The changing pattern of population structure of Staphylococcus aureus from bacteremia in China from 2013 to 2016: ST239-030-MRSA replaced by ST59-T437. Front Microbiol 9:332. doi: 10.3389/fmicb.2018.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542. Chen Y, Sun L, Wu D, Wang H, Ji S, Yu Y. 2018. Using core-genome multilocus sequence typing to monitor the changing epidemiology of methicillin-resistant Staphylococcus aureus in a teaching hospital. Clin Infect Dis 67:S241–S248. doi: 10.1093/cid/ciy644 [DOI] [PubMed] [Google Scholar]
- 543. Li M, Dai Y, Zhu Y, Fu CL, Tan VY, Wang Y, Wang X, Hong X, Liu Q, Li T, Qin J, Ma X, Fang J, Otto M. 2016. Virulence determinants associated with the Asian community-associated methicillin-resistant Staphylococcus aureus lineage ST59. Sci Rep 6:27899. doi: 10.1038/srep27899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544. Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. 2006. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J Infect Dis 193:1495–1503. doi: 10.1086/503777 [DOI] [PubMed] [Google Scholar]
- 545. Chen H, Yin Y, van Dorp L, Shaw LP, Gao H, Acman M, Yuan J, Chen F, Sun S, Wang X, Li S, Zhang Y, Farrer RA, Wang H, Balloux F. 2021. Drivers of methicillin-resistant Staphylococcus aureus (MRSA) lineage replacement in China. Genome Med 13. doi: 10.1186/s13073-021-00992-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546. Wang X, Liu Q, Zhang H, Li X, Huang W, Fu Q, Li M. 2018. Molecular characteristics of community-associated Staphylococcus aureus isolates from pediatric patients with bloodstream infections between 2012 and 2017 in Shanghai, China. Front. Microbiol 9:1211. doi: 10.3389/fmicb.2018.01211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547. Wu D, Li X, Yang Y, Zheng Y, Wang C, Deng L, Liu L, Li C, Shang Y, Zhao C, Yu S, Shen X. 2011. Superantigen gene profiles and presence of exfoliative toxin genes in community-acquired meticillin-resistant Staphylococcus aureus isolated from Chinese children. J Med Microbiol 60:35–45. doi: 10.1099/jmm.0.023465-0 [DOI] [PubMed] [Google Scholar]
- 548. Bae JS, Da F, Liu R, He L, Lv H, Fisher EL, Rajagopalan G, Li M, Cheung GYC, Otto M. 2021. Contribution of staphylococcal enterotoxin B to Staphylococcus aureus systemic infection. J Infect Dis 223:1766–1775. doi: 10.1093/infdis/jiaa584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549. Feng Y, Chen HL, Chen CJ, Chen CL, Chiu CH. 2017. Genome comparisons of two Taiwanese community-associated methicillin-resistant Staphylococcus aureus ST59 clones support the multi-origin theory of CA-MRSA. Infect Genet Evol 54:60–65. doi: 10.1016/j.meegid.2017.06.018 [DOI] [PubMed] [Google Scholar]
- 550. Aires de Sousa M, Bartzavali C, Spiliopoulou I, Sanches IS, Crisóstomo MI, de Lencastre H. 2003. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in patras, greece. J Clin Microbiol 41:2027–2032. doi: 10.1128/JCM.41.5.2027-2032.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551. Faria NA, Oliveira DC, Westh H, Monnet DL, Larsen AR, Skov R, de Lencastre H. 2005. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J Clin Microbiol 43:1836–1842. doi: 10.1128/JCM.43.4.1836-1842.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552. Dufour P, Gillet Y, Bes M, Lina G, Vandenesch F, Floret D, Etienne J, Richet H. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces panton-valentine leukocidin. Clin Infect Dis 35:819–824. doi: 10.1086/342576 [DOI] [PubMed] [Google Scholar]
- 553. Alkharsah KR, Rehman S, Alkhamis F, Alnimr A, Diab A, Al-Ali AK. 2018. Comparative and molecular analysis of MRSA isolates from infection sites and carrier colonization sites. Ann Clin Microbiol Antimicrob 17:7. doi: 10.1186/s12941-018-0260-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554. Witte W, Strommenger B, Cuny C, Heuck D, Nuebel U. 2007. Methicillin-resistant Staphylococcus aureus containing the panton-valentine leucocidin gene in Germany in 2005 and 2006. J Antimicrob Chemother 60:1258–1263. doi: 10.1093/jac/dkm384 [DOI] [PubMed] [Google Scholar]
- 555. Antri K, Rouzic N, Dauwalder O, Boubekri I, Bes M, Lina G, Vandenesch F, Tazir M, Ramdani-Bouguessa N, Etienne J. 2011. High prevalence of Methicillin-resistant Staphylococcus aureus clone ST80-IV in hospital and community settings in Algiers. Clin Microbiol Infect 17:526–532. doi: 10.1111/j.1469-0691.2010.03273.x [DOI] [PubMed] [Google Scholar]
- 556. Larsen AR, Böcher S, Stegger M, Goering R, Pallesen LV, Skov R. 2008. Epidemiology of European community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV strains isolated in Denmark from 1993 to 2004. J Clin Microbiol 46:62–68. doi: 10.1128/JCM.01381-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557. Larsson A-K, Gustafsson E, Johansson PJH, Odenholt I, Petersson AC, Melander E. 2014. Epidemiology of MRSA in southern Sweden: strong relation to foreign country of origin, health care abroad and foreign travel. Eur J Clin Microbiol Infect Dis 33:61–68. doi: 10.1007/s10096-013-1929-2 [DOI] [PubMed] [Google Scholar]
- 558. Stegger M, Wirth T, Andersen PS, Skov RL, De Grassi A, Simões PM, Tristan A, Petersen A, Aziz M, Kiil K, Cirković I, Udo EE, del Campo R, Vuopio-Varkila J, Ahmad N, Tokajian S, Peters G, Schaumburg F, Olsson-Liljequist B, Givskov M, Driebe EE, Vigh HE, Shittu A, Ramdani-Bougessa N, Rasigade J-P, Price LB, Vandenesch F, Larsen AR, Laurent F. 2014. Origin and evolution of European Community-acquired methicillin-resistant Staphylococcus aureus. mBio 5:e01044-14. doi: 10.1128/mBio.01044-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559. Kolonitsiou F, Papadimitriou-Olivgeris M, Spiliopoulou A, Drougka E, Jelastopulu E, Anastassiou ED, Spiliopoulou I. 2018. Methicillin-resistant Staphylococcus aureus ST80 induce lower cytokine production by monocytes as compared to other sequence types. Front Microbiol 9:3310. doi: 10.3389/fmicb.2018.03310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560. Jensen RO, Winzer K, Clarke SR, Chan WC, Williams P. 2008. Differential recognition of Staphylococcus aureus Quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. Journal of Molecular Biology 381:300–309. doi: 10.1016/j.jmb.2008.06.018 [DOI] [PubMed] [Google Scholar]
- 561. Chen LC, Tsou LT, Chen FJ. 2009. Ligand-receptor recognition for activation of quorum sensing in Staphylococcus aureus. J Microbiol 47:572–581. doi: 10.1007/s12275-009-0004-2 [DOI] [PubMed] [Google Scholar]
- 562. Mairi A, Touati A, Lavigne JP. 2020. Methicillin-resistant Staphylococcus aureus ST80 clone: a systematic review. Toxins (Basel) 12:119. doi: 10.3390/toxins12020119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563. Sugai M, Chen CH, Wu HC. 1992. Bacterial ADP-ribosyltransferase with a substrate specificity of the rho protein disassembles the golgi apparatus in vero cells and mimics the action of brefeldin A. Proc Natl Acad Sci USA 89:8903–8907. doi: 10.1073/pnas.89.19.8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564. Courjon J, Munro P, Benito Y, Visvikis O, Bouchiat C, Boyer L, Doye A, Lepidi H, Ghigo E, Lavigne JP, Vandenesch F, Lemichez E. 2015. EDIN-B promotes the translocation of Staphylococcus aureus to the bloodstream in the course of pneumonia. Toxins (Basel) 7:4131–4142. doi: 10.3390/toxins7104131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565. Yamaguchi T, Nishifuji K, Sasaki M, Fudaba Y, Aepfelbacher M, Takata T, Ohara M, Komatsuzawa H, Amagai M, Sugai M. 2002. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect Immun 70:5835–5845. doi: 10.1128/IAI.70.10.5835-5845.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566. Coombs GW, Daley DA, Lee YT, Pang S, Antimicrobial R. 2018. Australian group on antimicrobial resistance (AGAR) Australian Staphylococcus aureus sepsis outcome programme (ASSOP)
- 567. Coombs Geoffrey W, Goering RV, Chua KYL, Monecke S, Howden BP, Stinear TP, Ehricht R, O’Brien FG, Christiansen KJ. 2012. The molecular epidemiology of the highly virulent ST93 Australian community Staphylococcus aureus strain. PLoS One 7:e43037. doi: 10.1371/journal.pone.0043037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568. Munckhof WJ, Schooneveldt J, Coombs GW, Hoare J, Nimmo GR. 2003. Emergence of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) infection in Queensland, Australia. Int J Infect Dis 7:259–264. doi: 10.1016/s1201-9712(03)90104-4 [DOI] [PubMed] [Google Scholar]
- 569. van Hal SJ, Steinig EJ, Andersson P, Holden MTG, Harris SR, Nimmo GR, Williamson DA, Heffernan H, Ritchie SR, Kearns AM, Ellington MJ, Dickson E, de Lencastre H, Coombs GW, Bentley SD, Parkhill J, Holt DC, Giffard PM, Tong SYC. 2018. Global scale dissemination of ST93: a divergent Staphylococcus aureus epidemic lineage that has recently emerged from remote northern Australia. Front. Microbiol 9:1453. doi: 10.3389/fmicb.2018.01453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570. Steinig E, Aglua I, Duchene S, Meehan MT, Yoannes M, Firth C, Jaworski J, Drekore J, Urakoko B, Poka H, Wurr C, Ebos E, Nangen D, Müller E, Mulvey P, Jackson C, Blomfeldt A, Aamot HV, Laman M, Manning L, Earls M, Coleman DC, Greenhill A, Ford R, Stegger M, Syed MA, Jamil B, Monecke S, Ehricht R, Smith S, Pomat W, Horwood P, Tong SYC, McBryde E. 2022. Phylodynamic signatures in the emergence of community-associated MRSA. Proc Natl Acad Sci USA 119:e2204993119. doi: 10.1073/pnas.2204993119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571. Peleg AY, Munckhof WJ. 2004. Fatal necrotising pneumonia due to community-acquired methicillin-resistant Staphylococcus aureus (MRSA). Med J Aust 181:228–229. doi: 10.5694/j.1326-5377.2004.tb06247.x [DOI] [PubMed] [Google Scholar]
- 572. Coombs GW, Nimmo GR, Pearson JC, Christiansen KJ, Bell JM, Collignon PJ, McLaws ML, Antimicrobial R. 2009. Prevalence of MRSA strains among Staphylococcus aureus isolated from outpatients, 2006. Commun Dis Intell Q Rep 33:10–20. [PubMed] [Google Scholar]
- 573. Sahibzada S, Abraham S, Coombs GW, Pang S, Hernández-Jover M, Jordan D, Heller J. 2017. Transmission of highly virulent community-associated MRSA ST93 and livestock-associated MRSA St398 between humans and pigs in Australia. Sci Rep 7:5273. doi: 10.1038/s41598-017-04789-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574. Sahibzada Shafi, Hernández-Jover M, Jordan D, Thomson PC, Heller J. 2018. Emergence of highly prevalent CA-MRSA St93 as an occupational risk in people working on a pig farm in Australia. PLoS One 13:e0195510. doi: 10.1371/journal.pone.0195510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575. Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis 11:1965–1966. doi: 10.3201/eid1112.050428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576. Baba K, Ishihara K, Ozawa M, Tamura Y, Asai T. 2010. Isolation of meticillin-resistant Staphylococcus aureus (MRSA) from swine in Japan. Int J Antimicrob Agents 36:352–354. doi: 10.1016/j.ijantimicag.2010.06.040 [DOI] [PubMed] [Google Scholar]
- 577. Smith TC, Male MJ, Harper AL, Kroeger JS, Tinkler GP, Moritz ED, Capuano AW, Herwaldt LA, Diekema DJ. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One 4:e4258. doi: 10.1371/journal.pone.0004258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578. Vanderhaeghen W, Hermans K, Haesebrouck F, Butaye P. 2010. Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol Infect 138:606–625. doi: 10.1017/S0950268809991567 [DOI] [PubMed] [Google Scholar]
- 579. He L, Zheng HX, Wang Y, Le KY, Liu Q, Shang J, Dai Y, Meng H, Wang X, Li T, Gao Q, Qin J, Lu H, Otto M, Li M. 2018. Detection and analysis of methicillin-resistant human-adapted sequence type 398 allows insight into community-associated methicillin-resistant Staphylococcus aureus evolution. Genome Med 10:5. doi: 10.1186/s13073-018-0514-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580. Cuny C, Abdelbary M, Layer F, Werner G, Witte W. 2015. Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet Microbiol 177:219–223. doi: 10.1016/j.vetmic.2015.02.031 [DOI] [PubMed] [Google Scholar]
- 581. Rosen K, Roesler U, Merle R, Friese A. 2018. Persistent and transient airborne MRSA colonization of piglets in a newly established animal model. Front Microbiol 9:1542. doi: 10.3389/fmicb.2018.01542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582. Wang Y, Hu M, Liu Q, Qin J, Dai Y, He L, Li T, Zheng B, Zhou F, Yu K, Fang J, Liu X, Otto M, Li M. 2016. Role of the ESAT-6 secretion system in virulence of the emerging community-associated Staphylococcus aureus lineage ST398. Sci Rep 6:25163. doi: 10.1038/srep25163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583. Patterson NJ, Günther J, Gibson AJ, Offord V, Coffey TJ, Splitter G, Monk I, Seyfert H-M, Werling D. 2014. Two TIR-like domain containing proteins in a newly emerging zoonotic Staphylococcus aureus strain sequence type 398 are potential virulence factors by Impacting on the host innate immune response. Front Microbiol 5:662. doi: 10.3389/fmicb.2014.00662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584. Verstappen KM, Duim B, van Nes A, Snijders S, van Wamel WJB, Wagenaar JA. 2014. Experimental nasal colonization of piglets with methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Vet Microbiol 174:483–488. doi: 10.1016/j.vetmic.2014.09.019 [DOI] [PubMed] [Google Scholar]
- 585. Giotis ES, Loeffler A, Knight-Jones T, Lloyd DH. 2012. Development of a skin colonization model in gnotobiotic piglets for the study of the microbial ecology of meticillin-resistant Staphylococcus aureus ST398. J Appl Microbiol 113:992–1000. doi: 10.1111/j.1365-2672.2012.05397.x [DOI] [PubMed] [Google Scholar]
- 586. Broens EM, Graat EAM, van de Giessen AW, Broekhuizen-Stins MJ, de Jong MCM. 2012. Quantification of transmission of livestock-associated methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol 155:381–388. doi: 10.1016/j.vetmic.2011.09.010 [DOI] [PubMed] [Google Scholar]
- 587. Schulz J, Friese A, Klees S, Tenhagen BA, Fetsch A, Rösler U, Hartung J. 2012. Longitudinal study of the contamination of air and of soil surfaces in the vicinity of pig barns by livestock-associated methicillin-resistant Staphylococcus aureus. Appl Environ Microbiol 78:5666–5671. doi: 10.1128/AEM.00550-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588. Bos MEH, Verstappen KM, van Cleef BAGL, Dohmen W, Dorado-García A, Graveland H, Duim B, Wagenaar JA, Kluytmans JAJW, Heederik DJJ. 2016. Transmission through air as a possible route of exposure for MRSA. J Expo Sci Environ Epidemiol 26:263–269. doi: 10.1038/jes.2014.85 [DOI] [PubMed] [Google Scholar]
- 589. Friese A, Schulz J, Hoehle L, Fetsch A, Tenhagen BA, Hartung J, Roesler U. 2012. Occurrence of MRSA in air and housing environment of pig barns. Vet Microbiol 158:129–135. doi: 10.1016/j.vetmic.2012.01.019 [DOI] [PubMed] [Google Scholar]
- 590. Linhares LL, Yang M, Sreevatsan S, Munoz-Zanzi CA, Torremorell M, Davies PR. 2015. The effect of anatomic site and age on detection of Staphylococcus aureus in pigs. J Vet Diagn Invest 27:55–60. doi: 10.1177/1040638714559598 [DOI] [PubMed] [Google Scholar]
- 591. Slingerland BCGC, Tavakol M, McCarthy AJ, Lindsay JA, Snijders SV, Wagenaar JA, van Belkum A, Vos MC, Verbrugh HA, van Wamel WJB. 2012. Survival of Staphylococcus aureus ST398 in the human nose after artificial inoculation. PLoS One 7:e48896. doi: 10.1371/journal.pone.0048896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592. Laumay F, Corvaglia A-R, Diene SM, Girard M, Oechslin F, van der Mee-Marquet N, Entenza JM, François P. 2019. Temperate prophages increase bacterial adhesin expression and virulence in an experimental model of endocarditis due to Staphylococcus aureus from the CC398 lineage. Front Microbiol 10:742. doi: 10.3389/fmicb.2019.00742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593. Wulf MW, Markestein A, van der Linden FT, Voss A, Klaassen C, Verduin CM. 2008. First outbreak of Methicillin-resistant Staphylococcus aureus St398 in a dutch hospital. Euro Surveill 13:8051. [PubMed] [Google Scholar]
- 594. Jamrozy DM, Fielder MD, Butaye P, Coldham NG. 2012. Comparative genotypic and phenotypic characterisation of methicillin-resistant Staphylococcus aureus ST398 isolated from animals and humans. PLoS One 7:e40458. doi: 10.1371/journal.pone.0040458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595. Vrieling M, Tuffs SW, Yebra G, van Smoorenburg MY, Alves J, Pickering AC, Park JY, Park N, Heinrichs DE, Benedictus L, Connelley T, Seo KS, McCormick JK, Fitzgerald JR. 2020. Population analysis of Staphylococcus aureus reveals a cryptic, highly prevalent superantigen selw that contributes to the pathogenesis of bacteremia. mBio 11:e02082-20. doi: 10.1128/mBio.02082-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596. Krziwanek K, Metz-Gercek S, Mittermayer H. 2009. Methicillin-resistant Staphylococcus aureus ST398 from human patients, upper Austria. Emerg Infect Dis 15:766–769. doi: 10.3201/eid1505.080326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597. Pan A, Battisti A, Zoncada A, Bernieri F, Boldini M, Franco A, Giorgi M, Iurescia M, Lorenzotti S, Martinotti M, Monaci M, Pantosti A. 2009. Community-acquired methicillin-resistant Staphylococcus aureus ST398 infection, Italy. Emerg Infect Dis 15:845–847. doi: 10.3201/eid1505.081417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598. Randad PR, Dillen CA, Ortines RV, Mohr D, Aziz M, Price LB, Kaya H, Larsen J, Carroll KC, Smith TC, Miller LS, Heaney CD. 2019. Comparison of livestock-associated and community-associated Staphylococcus aureus pathogenicity in a mouse model of skin and soft tissue infection. Sci Rep 9:12811. doi: 10.1038/s41598-019-46940-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599. Schmidt T, Zündorf J, Grüger T, Brandenburg K, Reiners A-L, Zinserling J, Witte W, Schnitzler N. 2013. Phenotyping of Staphylococcus aureus reveals a new virulent ST398 lineage. Clin Microbiol Infect 19:279–285. doi: 10.1111/j.1469-0691.2012.03775.x [DOI] [PubMed] [Google Scholar]
- 600. Dai Y, Wang Y, Liu Q, Gao Q, Lu H, Meng H, Qin J, Hu M, Li M. 2017. A novel ESAT-6 secretion system-secreted protein ESXX of community-associated Staphylococcus aureus lineage ST398 contributes to immune evasion and virulence. Front Microbiol 8:819. doi: 10.3389/fmicb.2017.00819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601. Diene SM, Corvaglia AR, François P, van der Mee-Marquet N, Regional Infection Control Group of the Centre Region . 2017. Prophages and adaptation of Staphylococcus aureus ST398 to the human clinic. BMC Genomics 18:133. doi: 10.1186/s12864-017-3516-x [DOI] [PMC free article] [PubMed] [Google Scholar]