Abstract
Epithelial-mesenchymal transition (EMT) contributes to normal tissue patterning and carcinoma invasiveness. We show that transforming growth factor (TGF)-β/activin members, but not bone morphogenetic protein (BMP) members, can induce EMT in normal human and mouse epithelial cells. EMT correlates with the ability of these ligands to induce growth arrest. Ectopic expression of all type I receptors of the TGF-β superfamily establishes that TGF-β but not BMP pathways can elicit EMT. Ectopic Smad2 or Smad3 together with Smad4 enhanced, whereas dominant-negative forms of Smad2, Smad3, or Smad4, and wild-type inhibitory Smad7, blocked TGF-β–induced EMT. Transcriptomic analysis of EMT kinetics identified novel TGF-β target genes with ligand-specific responses. Using a TGF-β type I receptor that cannot activate Smads nor induce EMT, we found that Smad signaling is critical for regulation of all tested gene targets during EMT. One such gene, Id2, whose expression is repressed by TGF-β1 but induced by BMP-7 is critical for regulation of at least one important myoepithelial marker, α-smooth muscle actin, during EMT. Thus, based on ligand-specific responsiveness and evolutionary conservation of the gene expression patterns, we begin deciphering a genetic network downstream of TGF-β and predict functional links to the control of cell proliferation and EMT.
INTRODUCTION
The TGF-β superfamily of secreted polypeptides has diverse roles during metazoan embryonic development and adult tissue homeostasis (Piek et al., 1999a; Siegel and Massagué, 2003). TGF-β ligands signal through complexes of type II and type I receptor serine/threonine kinases, which induce Smad and alternative signaling pathways (Shi and Massagué, 2003). Receptor-regulated Smads (R-Smads) are phosphorylated by type I receptors and oligomerize with Smad4 (CoSmad). In the nucleus, R-Smads and Smad4 regulate gene transcription. Inhibitory Smads (I-Smads), Smad6 and Smad7, are negative signal regulators that block R-Smad phosphorylation and induce type I receptor dephosphorylation and/or proteasomal degradation (Shi and Massagué, 2003).
The superfamily is divided into the TGF-β/activin/nodal branch, hereafter abbreviated as TGF-β, and the BMP/growth and differentiation factor branch, hereafter referred to as BMP, based on their downstream R-Smad pathways (Miyazawa et al., 2002). TGF-β activates Smad2 and Smad3, whereas BMP activates Smad1, Smad5, and Smad8. These pathways show cell type specificity but many coexist in one cell type (Chang et al., 2002). Currently, no systematic studies of multiple TGF-β pathways in distinct cell types have yet been performed. This approach is important for the understanding of major physiological responses to these factors, i.e., cell proliferation, apoptosis or differentiation (ten Dijke et al., 2002; Siegel and Massagué, 2003).
Epithelial-mesenchymal transition (EMT) is the process by which an epithelial cell becomes a more motile mesenchymal cell (Condeelis and Segall, 2003; Grünert et al., 2003). EMT operates during early embryonic cell layer movements and later during organogenesis (Tosh and Slack, 2002). During EMT, cell-cell junctions are altered, cells lose epithelial polarity, express the mesenchymal markers vimentin and α-smooth muscle actin (α-SMA), and the resulting reorganization of the actin cytoskeleton supports cell migration. EMT is also important during tumor progression by stimulating invasiveness and metastasis (Condeelis and Segall, 2003; Grünert et al., 2003). In vivo verification of EMT is hard to assess due to the transient and reversible nature of the process and due to the lack of analytical tools that distinguish carcinoma cells undergoing EMT from neighboring stromal fibroblasts.
During embryonic development and in cell culture systems, EMT is triggered by various growth factors, fibroblast growth factor and hepatocyte growth factor/scatter factor being the most extensively studied (Thiery, 2002). TGF-β acts as a tumor suppressor by inhibiting epithelial cell growth, but it also promotes tumor progression in vivo (Siegel and Massagué, 2003; Tang et al., 2003). For example, TGF-β induces differentiation of squamous carcinoma into invasive spindle cell carcinoma in vivo and Smad2 plays critical roles in this process (Cui et al., 1996; Portella et al., 1998; Oft et al., 2002). TGF-β stimulates EMT in vitro in Namru murine mammary gland (NMuMG) and mouse proximal tubule kidney epithelial cells, in immortalized human keratinocytes HaCaT, as well as, in human colonic and hepatocellular carcinomas (Miettinen et al., 1994; Zavadil et al., 2001; Bates and Mercurio, 2003; Xu et al., 2003; Brown et al., 2004). In NMuMG cells, TGF-β–mediated EMT involves the TGF-β type I and type II receptor complexes and Smad3 (Piek et al., 1999b). In addition, signaling inputs of the p38 mitogen-activated protein kinase (Bhowmick et al., 2001b; Bakin et al., 2002; Yu et al., 2002), the phosphoinositide-3′-kinase-Akt pathway (Bakin et al., 2000), and the RhoA GTPase (Bhowmick et al., 2001a) contribute to EMT of NMuMG cells in response to TGF-β. Thus, EMT in response to TGF-β involves multiple intracellular effectors and the role of the Smad pathway in this cellular response has been disputed repeatedly (Bhowmick et al., 2001a, 2001b; Bakin et al., 2002; Thiery, 2002; Yu et al., 2002; Grünert et al., 2003). This is in contrast to the process of epithelial growth suppression by TGF-β where the central role of Smads is firmly established (ten Dijke et al., 2002; Siegel and Massagué, 2003). It is therefore important to systematically analyze the role of Smads during EMT. There is also a growing interest in identifying gene targets that execute EMT, in order to effectively intervene against this process, in the context of invasive/metastatic carcinomas.
In the present study, we investigated the effects of various TGF-β superfamily members on the EMT process of several normal mouse and human epithelial cell types. Based on our previous report on the role of the type I receptor kinase ALK-5 in EMT (Piek et al., 1999b), we analyzed for the first time the role of all type I receptor kinases and Smads of the superfamily. Using a cDNA microarray screen, we uncovered several new candidate genes possibly involved in TGF-β-mediated EMT. We therefore describe an extensive dissection of the signaling and genetic network that supports EMT.
MATERIALS AND METHODS
Cell Culture, Adenoviruses, Reagents, and Ligands
Human HaCaT keratinocytes, primary normal human mammary epithelial cells (HMEC), normal human epidermal keratinocytes (NHEK) of adult origin, and murine mammary epithelial NMuMG cells have been described previously (Kowanetz et al., 2004). NMe cells, a clonal epithelial derivative of NMuMG were provided by K. Vershueren (Leuven; Comijn et al., 2001). Immortalized normal human mammary epithelial cells, MCF-10A were obtained from ATCC and immortalized human lung epithelial cells, HPL1 (clones A and D) were donated by T. Takahashi (Nagoya; Masuda et al., 1997).
Adenoviruses expressing: a) the control protein β-galactosidase (Ad-LacZ), the C-terminally hemaglutinin (HA)-tagged constitutively active ALK-1(QD), -2(QD), -3(QD), -6(QD), and ALK-4(TD), -5(TD), -7(TD) receptors, the HA-tagged kinase-inactive ALK1–7(KR) receptors, the N-terminally Flag-tagged Smad1–7 proteins, the C-terminally deleted form of Smad4 (Smad4 515-ter), and the dominant-negative Smad3 (Smad3(D407E)) were donated by K. Miyazono (Tokyo; Fujii et al., 1999); b) N-terminally Flag-tagged constitutively active (T204D) and L45 loop-mutated ALK-5 (caALK-5mL45, here referred to as ALK5(TD)mL45) was provided by V. Kaartinen (Los Angeles; Dudas et al., 2004); c) the CAGA9-MLP-Luciferase promoter-reporter module was from S. Dooley (Aachen; Dooley et al., 2001); and d) GFP was based on the bicistronic Adeasy viral system from B. Vogelstein (Baltimore; He et al., 1998). Adenoviruses were amplified and titrated as previously described (Piek et al., 1999b).
The small-molecular-weight inhibitor SB431542 was a gift from N. J. Laping (GlaxoSmithKline Pharmaceuticals) and has been shown to inhibit selectively the ALK-4/-5/-7 protein kinase activity (Inman et al., 2002).
Recombinant mature TGF-β1 was purchased from PeproTech EC (London, United Kingdom) or provided by N. Ferrara (Genentech, San Francisco, CA), and recombinant mature TGF-β2 and -β3 were provided by R&D Systems (Minneapolis, MN), and recombinant mature BMP-7 was from K. Sampath (Curis, Cambridge, MA). Recombinant mature activin-A was donated by Y. Eto (Ajinomoto, Tokyo, Japan).
Transient Cell Infections and Gene Reporter Assays
Adenoviral transient infection of cells, using the multiplicities of infection (MOIs) specified in the figure legends or tables, were performed as previously described (Piek et al., 1999b). Luciferase reporter assays were performed with the enhanced luciferase assay kit from BD PharMingen (San Diego, CA), according to the protocol of the manufacturer. A Smad3/Smad4-specific luciferase reporter, CAGA9-Luc was used. This reporter contains 9 repeats of the Smad-binding element found in the plasminogen activator inhibitor 1 promoter and is an excellent read-out system for testing Smad3/Smad4 activation (Dennler et al., 1998; Dooley et al., 2001).
Transient and Stable Transfections
NMuMG or NMe cells were transiently transfected with siRNAs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol and as described (Kowanetz et al., 2004). The two mouse Idb2-specific siRNAs used were described recently (Kowanetz et al., 2004). Twenty-four hours after transfection, cells were stimulated with BMP-7 for the times indicated in the figures and before extraction of total proteins or total RNA. For the establishment of stable NMuMG clones expressing inducible wild-type Smad2 or the dominant-negative form of Smad2 (Smad2SA), N-terminally Flag-tagged wild-type Smad2 and Smad2SA were provided subcloned into the inducible vector pMEP4 (Invitrogen) by S. Souchelnytskyi (Uppsala; Souchelnytskyi et al., 1997). Empty vector pMEP4, wild-type Smad2 or Smad2SA were transfected into NMuMG cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Two days after transfection, cells were subcultured in the presence of 400 μg/ml hygromycin-B (Calbiochem, La Jolla, CA), and pools of antibiotic-resistant cells or individual cell clones were derived. For induction of transfected Smad2 and immunofluorescence or Western blot assays, cells growing continuously in the presence of hygromycin-B were cultured in the presence of 10 μM CdCl2 (Sigma, St. Louis, MO) for 24 h, at which point vehicle or TGF-β1 was added for another 24–48 h in the presence of hygromycin-B and inducer.
Immunoblotting and Immunofluorescence Microscopy
Total proteins from infected and/or stimulated NMuMG cells were extracted and subjected to SDS-PAGE and analyzed by Western blotting as described previously (Kurisaki et al., 2003). Mouse monoclonal anti-Flag (M2 and M5), mouse anti-α-smooth muscle actin (A2547) and mouse anti-β-tubulin (T8535) antibodies were purchased from Sigma-Aldrich, mouse monoclonal anti-hemagglutinin (HA, 12CA5) from Roche (Nutley, NJ), mouse monoclonal anti-E-cadherin (C20820) from BD Transduction Laboratories (Lexington, KY), rat monoclonal anti-ZO-1 (MAB1520) from Chemicon International (Temecula, CA), and mouse monoclonal anti-Smad1/2/3 (H2) and rabbit polyclonal anti-Id2 (C-20) from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-phospho-Smad3 antibody was a gift from Michael Reiss (New Brunswick). Rabbit polyclonal anti-phospho-Smad1 and anti-phospho-Smad2 antibodies were produced in house. Secondary anti-mouse-IgG and anti-rabbit-IgG coupled to horseradish peroxidase (HRP) were from Amersham Biosciences (Piscataway, NJ). The enhanced chemiluminescence detection system was purchased from Santa Cruz Biotechnology.
For morphological analysis, cell monolayers were stimulated with TGF-β1/2/3, BMP-7, or activin-A, and/or infected with adenoviruses as indicated in the figure legends. Fixed cell preparations were stained with tetramethyl-rhodamine isothiocyanate (TRITC)-labeled phalloidin (Sigma-Aldrich) or processed for immunofluorescence microscopy as described (Kowanetz et al., 2004). For immunofluorescence, primary antibodies were those listed above, and fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse-IgG antibody was from DAKO (Carpinteria, CA) and TRITC-conjugated goat anti-rat-IgG antibody was from Jackson ImmunoResearch Laboratories (West Grove, PA).
Thymidine Incorporation Assays
Cells were cultured, stimulated with growth factors, and labeled metabolically as described before (Kowanetz et al., 2004). The data are plotted as average values with SEs of triplicate repeats per independent experiment. Each independent experiment was repeated at least twice.
cDNA Microarray Analysis
The cDNA microarray analysis was performed with RNAs isolated from NMuMG cells cultured in the presence of 10% fetal bovine serum for 2, 8, or 36 h in the presence or absence of TGF-β1 (10 ng/ml) and from three independent cell cultures. Total RNA extraction and cDNA probe labeling was performed as described (Kowanetz et al., 2004). Equal amount of labeled cDNA probes per pair was hybridized to cDNA microarray chips (Mver1.1.1) from the Sanger/LICR/CRUK Consortium (http://www.sanger.ac.uk/Projects/Microarrays/for details and hybridization protocols). This microarray chip contained 11,500 unique single-stranded cDNA elements of 1.5-kb average length spotted on glass, which represent roughly 8850 unique mouse genes. Hybridizations were carried out in quadruplicate or quintuplicate, using the RNAs from the three independent cultures and including the dye swap control. Microarray scanning, image analysis, and primary spot intensity statistical analysis was performed as described (Kowanetz et al., 2004). Regulated genes were selected based on the average ratio value ≥1.7 for up-regulated genes and ≤0.57 for down-regulated genes. In addition, regulated genes had to be expressed at least on three arrays out of four (or on four arrays out of five, for the 8- and 36-h time points) and with a t test value for the ratios within replicates corresponding to p < 0.05. Statistically significant genes were clustered based on their expression values using the K-means statistical algorithm that is incorporated into the GeneSpring 6.0 data mining software (Silicon Genetics, Redwood City, CA). For all time points, we considered as 0-h time point the condition of duplicate cell cultures treated in an identical manner as the TGF-β1–treated samples, with the only difference being that we replaced TGF-β1 with vehicle. Functional classification of the highly regulated genes was performed manually based on exhaustive searches in PubMed (http://www.ncbi.nlm.nih.gov/PubMed/). All files containing the raw data of the microarray analysis have been deposited to ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) at the European Bioinformatics Institute (Hinxton) and are publicly available with Acc. No. E-MEXP-139.
Semiquantitative RT-PCR and Quantitative Real-time RT-PCR
Total RNA from NMuMG cells was extracted and analyzed as previously described (Kowanetz et al., 2004), using specific primers designed according to sequences available in the databanks or published by other authors (Supplementary Table 1). Primers for mouse glyceraldehyde-3′-phosphate dehydrogenase (Gapdh) were used to ascertain that an equivalent amount of cDNA was synthesized. Controls where reverse transcriptase was omitted (-RT) and where cDNAs were replaced with water were performed in order to demonstrate the specificity of the reactions.
DNase RQI-digested RNA from HaCaT and HMEC cells was reverse-transcribed as described (Kurisaki et al., 2003). PCR was performed in a total volume of 25 μl with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 1 μl (24 ng) of cDNA, and a 200 nM concentration of each primer (Supplementary Table 2). Primers were designed with the computer program Primer Express (Applied Biosystems), using parameters recommended by the manufacturer. Reactions were carried out in an ABI-prism 7000 sequence detector (Applied Biosystems) in triplicate, using the following conditions: an initial denaturation step consisted of 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Levels of gene expression in each sample were determined with the comparative Ct method (after validation assays for each gene primer set), using GAPDH gene as an endogenous control. For each condition, the ground condition (-TGF-β1) was set as 1 and expression data are presented as bar graphs of average expression values plus SD.
RESULTS
EMT Correlates with Growth Arrest Induced by TGF-β in Normal Human and Mouse Epithelial Cells
We investigated the potencies of different members of the TGF-β superfamily (TGF-β1, -β2, -β3, activin-A or BMP-7) to induce EMT and growth inhibition in epithelial cells of different origins. Primary human mammary epithelial cells (HMEC), normal human and mouse mammary epithelial cells (MCF-10A and NMuMG), immortalized normal lung epithelial cells (HPL1), primary normal human epidermal keratinocytes (NHEK), and immortalized normal human keratinocytes (HaCaT) were analyzed (Figure 1A). TGF-β1, -β2, and -β3 induced characteristic features of EMT in the mammary and lung cells: formation of actin stress fibers (Figure 1A), down-regulation and delocalization of E-cadherin and elongated (spindle-like) cell morphology (unpublished results). This effect was less pronounced in normal keratinocytes NHEK and HaCaT (Figure 1A). Signs of cell scattering were weakly obvious after prolonged stimulation of keratinocytes for several days (unpublished results). Activin-A did not induce EMT in mammary cells or keratinocytes, but caused scattering and spindle-like morphology of HPL1 cells (Figure 1A). Finally, BMP-7 did not induce EMT or scattering of any cell type tested (Figure 1A). BMP-7 induced thin stress fibers and led to increased cell volume but preserved the cuboidal architecture of HMEC and HPL1 cells.
Figure 1.
TGF-β induces EMT and growth inhibition in human and mouse normal epithelial cells. (A) Actin cytoskeleton direct fluorescence microscopy of HMEC, MCF-10A, NMuMG, HPL1 (clone D), NHEK, and HaCaT cells treated with vehicle (control), TGF-β1 (2.5 ng/ml), TGF-β2 (2.5 ng/ml), TGF-β3 (2.5 ng/ml), activin-A (10 ng/ml), or BMP-7 (150 ng/ml) for 36 h. Bars represent 10 μm and the magnification power of each series of photomicrographs is shown above the bars. (B) Dose-response growth inhibition assays performed in HMEC, NMuMG, NHEK, and HaCaT cells in response to TGF-β1 (black bars) and BMP-7 (gray bars). The percentage of radiolabeled thymidine incorporated after TGF-β1 or BMP-7 treatment is plotted relative to the level of vehicle-treated cells (set as 100%; open bars). Error bars represent SEs of the mean derived from triplicate determinations per experiment.
We also analyzed whether the six cell types were growth-inhibited because this is a common response of epithelial cells to TGF-β (ten Dijke et al., 2002). Cells were treated with different doses of TGF-β1 or BMP-7, and growth inhibition was estimated by incorporation of [3H]thymidine (Figure 1B and unpublished results). All the epithelial cells tested were growth-inhibited by TGF-β1, whereas BMP-7 had a weak effect even at high doses, as expected (Kowanetz et al., 2004). We conclude that normal keratinocytes exhibit strong growth suppression but relatively weaker scattering/EMT responses to TGF-β, whereas normal mammary and lung epithelial cells show both growth inhibition and EMT in relatively good correlation. Furthermore, ligands (such as TGF-β1) that induce potent EMT also elicit potent growth suppression, whereas ligands (such as BMP7) that induce weak growth arrest are essentially incapable of promoting EMT in normal epithelial cells.
Only type I Receptors of the TGF-β Branch Are Able to Drive EMT in NMuMG Cells
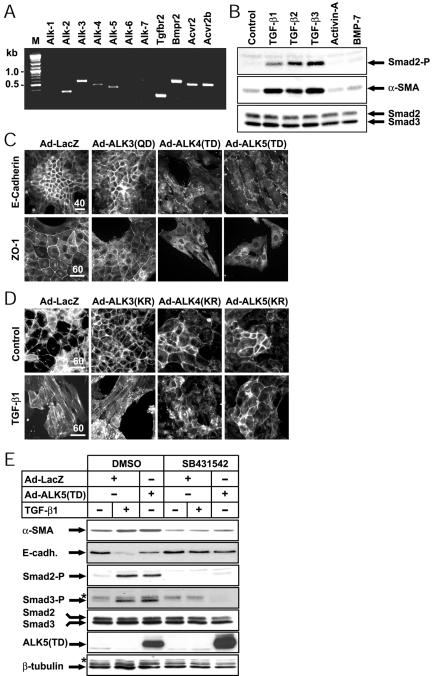
The differential responsiveness of epithelial cells to various TGF-β members might reflect the repertoire of receptors expressed by those cells or the differential activation of intracellular pathways downstream of the receptors. We investigated these two possibilities in NMuMG cells, because these cells exhibit the ligand-specific EMT response and thus represent a suitable in vitro model (Figure 1A). We analyzed by RT-PCR the expression of all seven TGF-β superfamily type I receptors, also known as activin receptor-like kinases (ALK-1 to -7), and four type II receptors (Figure 2A). NMuMG cells express Alk-5 (TGF-β receptor type I), Alk-2 (activin receptor type IA), and Tgfbr2 (TGF-β receptor type II) mRNA. This is in accordance with previous receptor-ligand cross-linking experiments (Miettinen et al., 1994; Piek et al., 1999b). However, NMuMG cells did not express detectable Alk-1 (endothelial TGF-β type I receptor) mRNA. Although NMuMG cells did not undergo EMT in the presence of BMP-7 or activin-A, they did express at high levels Alk-3 (BMP receptor type IA), and Bmpr2 (BMP receptor type II) mRNA. However, they did not express detectable Alk-6 (BMP receptor type IB) mRNA. They also expressed high levels of Acvr2 (activin receptor type IIA) and Acvr2b (activin receptor type IIB), lower mRNA levels of Alk-4 (activin receptor type IB) but did not express any Alk-7 (activin receptor type IC) mRNA. Thus, the inability of activin-A or BMP-7 to induce robust EMT in NMuMG cells (Figure 1A) cannot be explained by the lack of expression of receptors for these ligands.
Figure 2.
All three type I receptors of the TGF-β branch are capable of eliciting EMT. (A) Expression of TGF-β superfamily type I and type II receptors in NMuMG cells. The expression of genes encoding the ALK and type II receptors was analyzed by RT-PCR. Lane M shows markers of molecular size (in kbp, kb). (B) Immunoblot analysis of phospho-Smad2 (Smad2-P) and α-SMA levels in NMuMG cells. Cells were stimulated with the indicated ligands for 36 h, under the same conditions as in Figure 1A, and total cell extracts were analyzed with antibodies for the indicated proteins (arrows). (C) Effect of constitutively active ALK-3, -4, and -5 receptors on EMT. NMuMG cells were infected with control adenovirus LacZ (Ad-LacZ; MOI 100) or adenoviruses (MOI 100) expressing constitutively active forms of the BMP, activin, or TGF-β type I receptors, and the localization of E-cadherin and ZO-1 was assessed 36 h later. (D) NMuMG cells were infected either with the control adenovirus (Ad-LacZ, MOI 200) or with the adenoviruses expressing the dominant-negative versions of the indicated type I receptors (Ad-ALK(KR), MOI 200) and were treated with vehicle (Control) or TGF-β1 (2.5 ng/ml) for 36 h. Direct fluorescence microphotographs of the actin cytoskeleton are shown. Bars, (C) and (D) 10 μm; the magnification power of each series of photomicrographs is shown above the bars. (E) NMe cells were infected with Ad-LacZ (MOI 100) or Ad-ALK-5(TD) (MOI 100) and then left unstimulated or stimulated with TGF-β1 (5 ng/ml), in the presence of vehicle (dimethylsulfoxide) or SB431542 inhibitor (10 μM), and the EMT process was analyzed 48 h later. The levels of endogenous α-SMA, E-cadherin (E-cadh.), phosphorylated Smad2 (Smad2-P) and Smad3 (Smad3-P), total Smad2 and Smad3, exogenous ALK5(TD), and β-tubulin were determined by immunoblotting. Relevant protein bands (arrows) and background bands (asterisk) are marked.
To confirm the relative inability of activin-A in causing EMT of NMuMG cells, we measured the levels of phosphorylated Smad2, which is an immediate intracellular effector of the pathway downstream the type I receptors (Figure 2B). TGF-β1–3 induced potent phospho-Smad2 levels within 30 min of stimulation (unpublished observations) that were sustained up to 36 h, at which point cells exhibit full EMT. In contrast, activin-A showed the same background response as vehicle or the negative control used, BMP-7. The Smad2 phosphorylation level reflects directly the EMT process as verified by immunoblot analysis of an established marker of EMT, α-SMA (Figure 2B). Thus, the relative failure of activin-A in inducing EMT in NMuMG cells correlates with the poor activation of the Smad pathway, and it must reflect a defective signaling component proximal to the receptors or a very low level of functional receptor.
We then tried to test whether ectopic expression of activin and BMP type I receptors in NMuMG cells could rescue the observed inability of these ligands to induce EMT. We used an adenoviral system to transduce transiently all 7 type I receptors of the superfamily (in a constitutively active mutant form). EMT was followed by immunofluorescence microscopy of the adherens junction marker E-cadherin and the tight junction marker ZO-1 (Figure 2C and Supplementary Figure 1A). The exogenous type I receptors were expressed in >95% of the infected cells (Supplementary Figure 1B), at high levels (Supplementary Figure 1C). Staining of LacZ-transduced cells demonstrated that infection per se did not alter epithelial differentiation. Overexpression of constitutively active receptors of the BMP branch did not affect the epithelial phenotype (Figure 2C for ALK-3(QD) and Supplementary Figure 1A for ALK-1(QD), -2(QD), and -6(QD)). In contrast, constitutively active activin, TGF-β and nodal type I receptors ALK-4(TD), -5(TD), -7(TD) induced a clear EMT (Figure 2C and Supplementary Figure 1A). The constitutively active type I receptors were signaling properly because they induced endogenous Smad1 (BMP receptors) or Smad2 (TGF-β receptors) phosphorylation even at the time (36–48 h) at which EMT is fully manifested (Supplementary Figure 1C). Thus, among all type I receptors of the TGF-β superfamily, only those of the TGF-β branch were able to induce EMT in NMuMG cells. The activin type I receptor (ALK-4) is therefore sufficient to elicit EMT of NMuMG cells under conditions of ectopic expression.
To test the possibility that endogenous ALK-4 could also contribute to EMT, we overexpressed in NMuMG cells dominant-negative versions of all type I receptors of the TGF-β superfamily and stimulated cells with TGF-β1 (Figure 2D and Supplementary Figure 2A). Dominant negative ALK-1 to -7(KR) have a lysine to arginine mutation in the kinase active site, which obliterates their enzymatic activity. Overexpression of ALK-3(KR) receptor (Figure 2D) or ALK-1(KR), -2(KR), -6(KR), and -7(KR) (Supplementary Figure 2A) did not block EMT induced by TGF-β1. Only ALK-5(KR), and to a lower extent ALK-4(KR), were able to inhibit EMT induced by TGF-β1 (Figure 2D). The exogenous type I receptors were distributed in >95% of the infected cells (Supplementary Figure 2B) and were expressed at high levels (Supplementary Figure 2C). Moreover, as a specificity control for the dominant-negative activity of the ectopic receptors, we showed that ALK-5(KR), but neither ALK-2(KR) nor ALK-3(KR), were able to block Smad2 phosphorylation induced by TGF-β1 (Supplementary Figure 2D). Thus, all the above experiments establish that in addition to ALK-5, also ALK-4 and ALK-7 are capable of inducing EMT in NMuMG cells; however, the latter receptor is not expressed at all in NMuMG cells.
We confirmed the above data on type I receptor specificity by using a low-molecular-weight inhibitor (SB431542), which selectively inactivates the ALK-4, -5, and -7 receptors (Inman et al., 2002). NMuMG and their clonal derivatives, NMe cells were treated with TGF-β1 or infected with the constitutively active ALK-5 adenovirus, in the presence or absence of the SB431542 compound. TGF-β1 induced α-SMA expression and robustly down-regulated E-cadherin levels, and SB431542 blocked these effects (Figure 2E). We found the same results of SB431542 at the morphological level (Supplementary Figure 3). These experiments also indicated that TGF-β is secreted by NMuMG cells and acts in an autocrine and/or paracrine manner because after treating control cells with the inhibitor, we observed a stronger epithelial phenotype with more polarized cuboidal morphology (Figure 2E and Supplementary Figure 3). We confirmed that SB431542 could indeed block the activity of the endogenous and exogenous ALK-5 receptors, as it abolished the basal and TGF-β1– or ALK-5(TD)–mediated phosphorylation of Smad2 and Smad3 in NMuMG cells (Figure 2E). Thus, the combined experiments with constitutively active type I receptors and SB431542 suggest that EMT of NMuMG cells can be mediated by the TGF-β branch of pathways in the superfamily.
The Endogenous Smad Pathway Plays Critical Roles in TGF-β–induced EMT
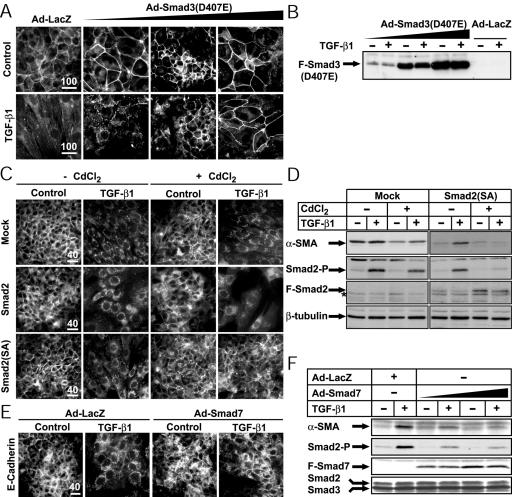
The only signaling effectors known so far to be shared by ALK-4, -5, and -7 receptors are Smad2 and Smad3 (Miyazawa et al., 2002). We previously provided evidence that Smad3, when overexpressed together with ALK-5(TD), could enhance synergistically the EMT response of NMuMG cells (Piek et al., 1999b). However, those experiments did not establish whether endogenous Smad pathways are indeed implicated in the EMT response, a question of importance, as described in the Introduction. Overexpression of a dominant-negative Smad3(D407E) (Goto et al., 1998) reverted the TGF-β1 effect in a dose-dependent manner and fully blocked EMT at the highest dose of adenovirus tested (Figure 3, A and B, and Supplementary Figure 4A). As a control of the potency of dominant-negative Smad3, we found that Smad3(D407E) partially blocked the dose-dependent growth inhibition of NMuMG cells by TGF-β1 (Supplementary Figure 4B). Smad3(D407E) also interfered with the activation of endogenous Smads in NMuMG cells, because it blocked dose-dependently the inducibility of a Smad3/Smad4-specific luciferase reporter, CAGA9-Luc, by TGF-β1 (Supplementary Figure 4C). Similar inhibitory effects were measured in stably transfected NMuMG cells with a dominant-negative Smad2 mutant (Smad2SA) whose two C-terminal serines were replaced with alanines (Figure 3, C and D, and Supplementary Figure 4D). Empty-vector and wild-type Smad2 transfected clones served as controls. In mixed pools of transfected NMuMG cells and in two distinct clones per transfectant, we consistently observed only minor enhancement of morphological EMT by wild-type Smad2 and efficient blocking of the response by Smad2SA (Figure 3C and Supplementary Figure 4E). Furthermore, induction of Smad2SA effectively blocked α-SMA levels in response to TGF-β1 (Figure 3D). As a control, Smad2SA was shown to block effectively endogenous phospho-Smad2 but not as potently phospho-Smad3 activation by TGF-β1 (Figure 3D and Supplementary Figure 4F).
Figure 3.
Endogenous Smad pathways mediate EMT in NMuMG cells. (A) NMuMG cells were infected either with Ad-LacZ (MOI 300) or with increasing doses of Ad-Smad3(D407E) (triangle: MOI 20, 100, 300) and were treated with vehicle (Control) or TGF-β1 (2.5 ng/ml) for 36 h. Localization of ZO-1 was visualized by immunofluorescence. (B) Flag-tagged Smad3(D407E) levels measured by immunoblotting using extracts from cells treated as in A. (C) E-cadherin immunofluorescence microscopy of individual NMuMG stable clones transfected with empty vector pMEP4 (Mock), pMEP4-Smad2, and pMEP4-Smad2(SA). Transfected cells were induced (+) or not (-) with CdCl2 and subsequently treated with vehicle (Control) or 5 ng/ml TGF-β1 for 36 h. (D) Immunoblot analysis of endogenous α-SMA in pools of NMuMG cells stably transfected with empty vector (Mock) or Smad2(SA) and treated under identical conditions as in C. Control immunoblots of endogenous phospho-Smad2 (Smad2-P), exogenous Flag-tagged Smad2 (F-Smad2), and endogenous β-tubulin, which serves as loading reference are also shown. (E) Immunofluorescence microscopy of E-cadherin in NMuMG cells infected with Ad-LacZ (MOI 100) or with Ad-Smad7 (MOI 100) and treated with vehicle (Control) or TGF-β1 (5 ng/ml) for 36 h. Bars, (A, C, and E) 10 μm; the magnification power of each series of photomicrographs is shown above the bars. (F) Smad7 blocks α-SMA protein synthesis induced by TGF-β1. NMuMG cells were infected with Ad-LacZ (MOI 100) or increasing doses of Ad-Smad7 (triangle: MOI 30, 100), and then treated with vehicle (-) or with 5 ng/ml TGF-β1 (+) for 48 h. The levels of α-SMA, phosphorylated Smad2 (Smad2-P), exogenous Flag-Smad7 (F-Smad7), and total Smad2 and Smad3 were determined by immunoblotting. In B, D, and F the relevant protein bands are marked with arrows, and an asterisk marks background bands.
We obtained similar but weaker effects using a C-terminally deleted Smad4(515-ter) (Supplementary Figure 5, A and B). Smad4(515-ter) is found in pancreatic tumors and acts as a dominant-negative protein, which is verified here by its effects on TGF-β–induced growth inhibition (Supplementary Figure 5D) and CAGA9-Luc reporter activity (Supplementary Figure 5E). The weaker effect of Smad4(515-ter) compared with Smad3(D407E) or Smad2SA is due to the low expression level of the former protein (Supplementary Figure 5C), which results from its documented unstable nature (Maurice et al., 2001).
We also tested the inhibitory Smads, Smad6, and Smad7, which block endogenous R-Smad activation by type I receptors and can also mediate receptor degradation (Shi and Massagué, 2003). Smad7 overexpression inhibited completely EMT induced by TGF-β1 (Figure 3E and Supplementary Figure 6A), as well as the changes induced by ALK-4(TD), ALK-5(TD), and ALK-7(TD) (Supplementary Figure 7A). Moreover, Smad7 blocked in a dose-dependent manner α-SMA induction (Figure 3F). As controls, we showed that Smad7 fully blocked NMuMG growth inhibition by TGF-β1 (Supplementary Figure 6B) and diminished both the basal and TGF-β1,-induced phosphorylation of Smad2 (Figure 3F). In contrast, Smad6, which mainly blocks the BMP signaling pathways, was not able to block EMT mediated either by TGF-β1 or by constitutively active type I receptors ALK-4, -5, -7 (Supplementary Figure 7A), supporting the observed Smad effector specificity during EMT.
As a last test of the signaling specificity among TGF-β superfamily pathways that are capable to elicit EMT in NMuMG cells, we ectopically expressed either each R-Smad alone, or together with Smad4, in the presence or absence of constitutively active type I receptors (these data are summarized in Supplementary Figure 7A). We used ALK-4/-5/-7(TD) receptors for the activation of Smad2 and Smad3 proteins, and ALK-1/-2/-3/-6(QD) receptors for Smad1 and Smad5. None of the R-Smads when expressed alone transiently via the adenovirus system were able to drive EMT. However, Smad3, and more weakly Smad2, in combination with Smad4 could induce EMT as previously described (Piek et al., 1999b). In addition, only Smad2 and Smad3 (together with Smad4) were able to efficiently synergize with a low dose of each of the three type I receptors, ALK-4(TD), -5(TD), and -7(TD). In contrast, none of the BMP-specific Smad1 or Smad5 could support similar effects on EMT when coexpressed with several constitutively active type I receptors. We verified the synergistic contribution of Smads together with constitutively active type I receptors on EMT by monitoring the expression levels of ZO-1, E-cadherin, and α-SMA. We present only limited data for the effect of Smad3 alone or in combination with Smad4 and in the absence or presence of increasing MOI of the ALK-4(TD) receptor (Supplementary Figure 7B). The combined data on the three protein markers recapitulate rather faithfully the microscopic analysis of EMT in NMuMG cells (Supplementary Figure 7A).
All the above data showed that interfering with the endogenous Smads using dominant-negative Smad mutants and inhibitory Smads resulted in inhibition of EMT induced by TGF-β1 in NMuMG cells. Overexpressing Smad2 and Smad3 could also lead to EMT, whereas the BMP-specific Smad1 and Smad5 could not. We conclude that all three Smad proteins of the TGF-β pathway, Smad2, Smad3, and Smad4, are involved in EMT.
Transcriptome Analysis of TGF-β1–mediated EMT in NMuMG Cells
To understand the mechanisms by which TGF-β1 alters cellular differentiation and leads to EMT in mammary epithelial cells, we used a transcriptomic approach. We focused on the early (2 h) and intermediate (8 h) transcriptional events triggered by TGF-β1 (and leading to EMT) in NMuMG cells. Besides, in order to define (possibly new) markers of EMT, we analyzed genes differentially expressed in NMuMG cells showing clear EMT after long stimulation with TGF-β1 (36 h). Of ∼9000 unique genes represented on the mouse cDNA chip of 11,500 cDNA elements used, we found 344 independent genes regulated by TGF-β1, among which 205 (60%) were up-regulated and 139 (40%) were down-regulated (Figure 4A; Supplementary Tables 3 and 4). Many of the regulated genes (38%) do not have any annotation or known function. Among the regulated genes with clear annotation (213 genes out of 344), we identified 28 previously studied TGF-β gene targets and 22 additional genes that are shared with recent microarray screens of the TGF-β response in various cell types. The rest 163 gene targets of the pathway have not been previously reported. We clustered the regulated genes according to their expression profile (K-means method) and inside each cluster we classified the genes according to their functional annotation (Figure 4, B and C; Supplementary Tables 3 and 4). It is likely that genes that represent markers of EMT belong to clusters 3 and 4, whereas genes that induce EMT are in clusters 1 and 2. This is based on the kinetics of phenotypic change induced by TGF-β, because no obvious morphological change, alteration in cell-cell adhesion, or tight junction integrity can be observed in NMuMG cells earlier than 8–12 h poststimulation.
Figure 4.
Summary and clustering of highly regulated genes by TGF-β1 in NMuMG cells. (A) Venn diagrams of gene expression itemized in three ways, up-regulated genes (left), down-regulated genes (middle), and total regulated genes (right). The three time points (2, 8, and 36 h) are indicated outside each circle, and gene numbers are shown within each section. (B and C) Clustering of highly regulated genes. Up-regulated (B) and down-regulated (C) genes upon TGF-β1 stimulation were clustered according to their expression profile using the K-means algorithm. Graphs represent the average values of normalized ratios of all genes per cluster plotted with their SEs (ratios of gene expression have no unit). The number of genes belonging to each cluster is indicated.
Among the annotated up-regulated genes, 18% encode functions linked to the extracellular matrix/cell adhesion/cytoskeleton and are regulated mostly at the late time point. This is in agreement with the EMT process as fibroblastic cells start to synthesize an extracellular matrix and increase their migratory properties. Many cytoskeletal genes are also in agreement with the observed dramatic reorganization of the cytoskeleton. We could also observe changes that support the well-established model of TGF-β auto-induction (van Obberghen-Schilling et al., 1988). Accordingly, this group includes genes such as the up-regulated Ltbp1, Cd44, Itgb5, Bmp1, and Tgfbr1 that link directly to processes of TGF-β activation and signaling. Approximately 34% of the annotated down-regulated genes encode functions linked to metabolism/transport/folding and are regulated between 8 and 36 h (clusters 3 and 4). A substantial proportion of these (15 genes) encode functions linked to redox regulation and hypoxia regulatory factors. Genes encoding proteins linked to cell cycle/transcription/apoptosis/development corresponded roughly to 9% of the annotated regulated genes, 63% of them being up-regulated and 37% down-regulated, and were enriched in the corresponding immediate-early clusters 1 and 2. In addition to the known transcriptional program that underlies the growth inhibitory response of epithelial cells to TGF-β (Siegel and Massagué, 2003), we identified a new set of strong candidate genes for the cytostatic program (up-regulated Hrasls3, N4wbp4/Pmepa1, Cks-2, Drp, Gas1, Cfh, and down-regulated Igfbp5, Cxcl1, Ccnd2, Pold1), and few new regulators of the apoptotic response (up-regulated Bag3, Capn2, Tgfb1i4/Tsc-22, Pea15). This functional group also includes several transcription factors, some of which may possibly regulate genes of the EMT program, including the architectural transcription factor Hmga2, the hypoxia-induced factor Hif1a, the Krüppel factor Klf4/Gklf and the Ets family member Ehf. Ongoing studies aim at analyzing the hypothesis that these proteins are directly involved in EMT.
Functional Validation of TGF-β Target Genes Based on Pathway Specificity, Smad Requirement, and Contribution to EMT
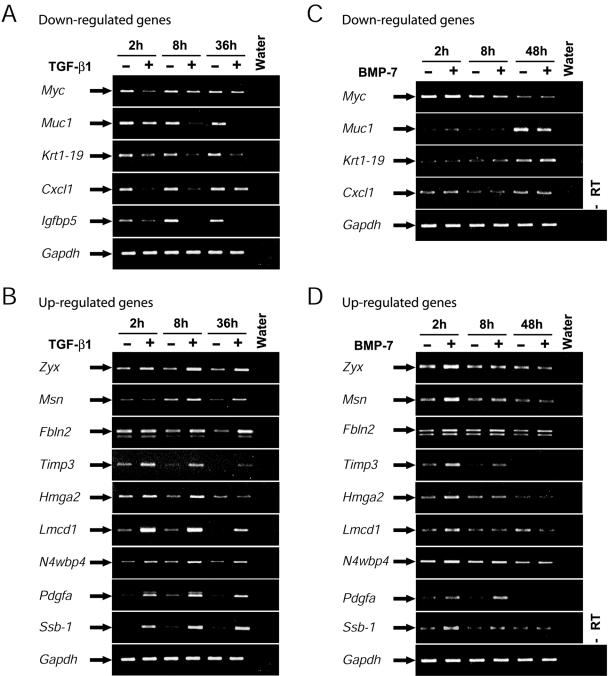
To validate the microarray experiment, we chose a set of highly regulated genes (Table 1) based on several criteria: 1) high intensity of gene expression (Lmcd1, Ssb-1, Pdgfa, N4wbp4/Pmepa1, Hmga2, and Cxcl1), 2) known TGF-β responsive genes (Tgif, Idb2, Idb3, Myc, and Timp3), 3) known markers of epithelial cells (Krt1–19), and 4) late and possibly novel markers of EMT based on our literature screen (Zyx, Msn, Muc1, Igfbp5, and Fbln2). We assessed the expression of these genes with RT-PCR (Figures 5, A and B, and 7A; unpublished results for Tgif; and Kowanetz et al. (2004) for Idb3). In general, we found a very strong concordance between microarray values of gene expression and levels measured by semiquantitative RT-PCR for all the genes tested.
Table 1.
Selected genes chosen for functional validation
| Genes/Hours | 2 h | 8 h | 36 h | Cluster | ||||
|---|---|---|---|---|---|---|---|---|
| Up-regulated genes (ratio ≥ 1,7, t test p < 0.05) | ||||||||
| Lmcd1 | 6.0 | 8.8 | 3.4 | 2 | ||||
| Ssb-1 | 4.2 | 5.0 | 2.2 | 2 | ||||
| Pdgfa | 3.3 | 4.9 | 3.0 | 2 | ||||
| N4wbp4 | 3.3 | 4.5 | 2.5 | 2 | ||||
| Hmga2 | 1.9 | 3.3 | 0.8 | 2 | ||||
| Tgif | 1.9 | 2.5 | 1.3 | 2 | ||||
| Zyx | 1.6 | 2.4 | 1.9 | 2 | ||||
| Timp3 | 1.5 | 2.5 | 2.2 | 3 | ||||
| Msn | 1.1 | 2.1 | 2.2 | 3 | ||||
| Fbln2 | 1.4 | 2.3 | 3.6 | 4 | ||||
| Down-regulated genes (ratio ≤ 0,57, t test p < 0.05) | ||||||||
| Igfbp5 | 0.68 | 0.16 | 0.39 | 3 | ||||
| Muc1 | 0.86 | 0.45 | 0.18 | 3 | ||||
| Krt1-19 | 0.91 | 0.58 | 0.44 | 3 | ||||
| Idb3 | 0.93 | 0.20 | 0.41 | 3 | ||||
| Idb2 | 0.60 | 0.16 | 0.58 | 2 | ||||
| Cxcl1 | 0.32 | 0.32 | 0.75 | 2 | ||||
| Myc | 0.54 | 0.82 | 0.74 | 1 | ||||
Short list of genes regulated by TGF-β1. Gene names use the Ensembl nomenclature and values represent average fold-change of the mRNA levels estimated by microarray analysis (bold values: significance above or below the threshold levels; regular values: not statistically significant changes). The cluster to which genes have been classified is indicated in the last column and refers to the graphs of Figure 4, B and C.
Figure 5.
Differential response of selected gene targets to TGF-β1 versus BMP-7. Semiquantitative RT-PCR analysis showing the mRNA steady-state levels of 14 selected genes. NMuMG cells were treated (+) or not (-) with 5 ng/ml TGF-β1 for 2, 8, or 36 h (A and B) or cells were treated with 300 ng/ml BMP-7 for 2, 8, or 48 h (C and D). Expression of down-regulated (A) and up-regulated (B) genes is shown. The classification to down-regulated (C) and up-regulated (D) genes in the BMP-7 experiments is based on gene responsiveness to TGF-β1. Gapdh serves as a control of equivalent cDNA synthesis, and water and minus RT controls show the specificity of the reactions.
Figure 7.

Id2 inhibits BMP-7 from regulating α-SMA expression. (A and B) Semiquantitative RT-PCR analysis showing the mRNA steady state levels of Idb2 and control Gapdh. NMuMG cells were treated with vehicle (-), 5 ng/ml TGF-β1 for 2, 8, and 36 h (A) or 300 ng/ml BMP-7 (+) for 2, 8, and 48 h (B). Water and minus RT controls show the specificity of the reactions. (C) Knockdown of Id2 allows BMP-7 to cause EMT and enhance α-SMA protein synthesis in a time-dependent manner. NMe clonal cells were transfected with control (siRNA-luc) or specific (siRNA-Id2) and then treated with vehicle (0) or with 300 ng/ml BMP-7 for 8 or 36 h. The levels of endogenous α-SMA, Id2, and β-tubulin were determined by immunoblotting.
Because we have shown that BMP-7 or BMP type I receptors fail to induce EMT (Figures 1 and 2), we hypothesized that genes coregulated by both TGF-β1 and BMP-7 might not be involved in the EMT process. We therefore compared the expression of the same set of genes in the presence of BMP-7 (Figure 5, C and D). BMP-7 did not affect the expression levels of any of the down-regulated genes tested (Figure 5C). The same was true for all the up-regulated genes tested except Timp3 and Pdgfa, whose expression was not induced by BMP-7 at the late time point (48 h), a characteristic difference from the TGF-β1 response (Figure 5C). Furthermore, some of the up-regulated genes showed a transient induction by BMP-7 at 2 h, which was considerably weaker and not sustained later compared with the TGF-β1 response. This may reflect a transient and physiologically irrelevant response to the acutely high dose of BMP-7 (150 or 300 ng/ml) used in most of these experiments. This was done in order to ensure maximal responsiveness by BMP-7. These experiments allow us to conclude that the differential response of genes to TGF-β versus BMP may prove useful as a means of validation of candidate targets that play key roles in EMT.
To rapidly validate the involvement of R-Smad activation downstream of TGF-β1 in regulation of the selected genes, we used a double mutant type I receptor that exhibits constitutively active kinase function and lacks critical amino acids in its L45 loop, the docking site for R-Smads. This receptor (ALK5(TD)mL45) is completely inactive in terms of phosphorylating R-Smads (Figure 6A), but otherwise can elicit downstream signaling via Smad-independent pathways (Yu et al., 2002; Itoh et al., 2003). The ALK5(TD)mL45 adenoviral vector was equally ineffective in inducing EMT of NMuMG cells (unpublished results), or α-SMA levels (Figure 6A), as the previously published retroviral vectors carrying the same or equivalent mutations in the L45 loop (Yu et al., 2002; Itoh et al., 2003). This insufficiency correlated tightly with the failure of this mutant to induce phospho-Smad2 levels. ALK5(TD)mL45 receptor rendered NMuMG cells more polarized, mimicking the effect of the ALK-4/-5/-7 kinase inhibitor described in Figure 2E. Indeed, using α-SMA as a quantitative marker of EMT, we showed that increasing MOI of the ALK5(TD)mL45 adenovirus led to a gradual decrease of background α-SMA levels, thus enhancing the evidence that low level of constitutive endogenous TGF-β signaling operates in NMuMG cells (Figure 6B). These experiments indicate that the ALK5(TD)mL45 receptor can act as a dominant-negative mutant against endogenous (autocrine) TGF-β receptor signaling.
Figure 6.
Role of the L45 loop of the TGF-β type I receptor in EMT and gene regulation. (A) ALK5(TD)mL45 fails to induce α-SMA protein synthesis. NMuMG cells were infected with Ad-LacZ (MOI 100) or increasing doses of Ad-ALK5(TD) or Ad-ALK5(TD)mL45 (triangles: MOI 50, 100), or alternatively they were treated with vehicle (-) or with 5 ng/ml TGF-β1 (+) for 48 h. The levels of endogenous α-SMA, phosphorylated Smad2 (Smad2-P), and Smad2 and Smad3 and exogenous ALK5(TD) receptors were determined by immunoblotting. The two different receptors are detected using anti-HA and anti-Flag immunoblotting as shown. (B) ALK5(TD)mL45 reduces α-SMA protein synthesis. NMuMG cells were infected with Ad-LacZ (MOI 100) or increasing doses of Ad-ALK5(TD)mL45 (triangle: MOI 2, 5, 20, 50, 100) for 48 h. The levels of endogenous α-SMA, exogenous ALK5(TD)mL45 receptor and total Smad2 and Smad3 were determined by immunoblotting. In A and B relevant protein bands (arrows) and background bands (asterisk) are marked. (C) Semiquantitative RT-PCR analysis showing the mRNA steady state levels of nine selected genes from Table 1. NMuMG cells were infected with Ad-LacZ, Ad-ALK5(TD), or Ad-ALK5(TD)mL45 (MOI 100) and then treated with vehicle (-) or with 5 ng/ml TGF-β1 (+) for 36 h. Gapdh serves as a control of equivalent cDNA synthesis, and water and minus RT controls show the specificity of the reactions.
Next, we measured mRNA expression of the selected genes of Table 1 in NMuMG cells stimulated with TGF-β1 (as control), cells infected with Ad-ALK5(TD) or with Ad-ALK5(TD)mL45 (Figure 6C). Although ALK5(TD) gave the same response as TGF-β1, the ALK5(TD)mL45 receptor gave essentially background response, similar to vehicle. This is clearly shown for Muc1, Krt1–19, Msn, Fbln2, Hmga2, Lmcd1, N4wbp4/Pmepa1, Pdgfa, and Ssb1 (Figure 6C). The levels of Hmga2 do not show any regulation by the positive control ALK5(TD) as this gene does not respond after 36 h post-TGF-β1 treatment (Figure 5B), and adenoviral transduction of ALK5(TD) mimics a long-term stimulation (>36 h) with TGF-β1. Thus, all of the selected genes appear to require an intact L45 loop in the TGF-β type I receptor in order to be properly regulated, suggesting a role of R-Smad activation in their regulation.
One of the target genes identified in this screen, which also showed a distinct mode of regulation by TGF-β1 versus BMP-7 is the inhibitor of differentiation Idb2 that encodes for the known protein Id2 (Table 1). We have recently established the functional role of Id2 (and Id3) in regulation of EMT mediated by TGF-β (Kowanetz et al., 2004). Id2 (and Id3) must be down-regulated by TGF-β in order for EMT to occur, whereas BMP fails to induce EMT because of the robust induction of the levels of Id proteins. Figure 7, A and B, shows the patterns of Idb2 gene expression in response to TGF-β1 and BMP-7 in NMuMG cells. This pattern is conserved in many epithelial cell types including HMEC, NHEK, and HaCaT cells and is also faithfully reflected at the protein level (Figure 7C and Kowanetz et al., 2004). To functionally link one of the EMT markers used throughout this study, α-SMA, with the Smad-regulated Idb2, we analyzed NMuMG or clonal NMe cells transiently transfected with Id2-specific siRNAs that could down-regulate endogenous Id2 by 75% and stimulated the cells with BMP-7 (Figure 7C). Under conditions of Id2 knockdown, BMP-7 is capable of inducing significant α-SMA levels, in a time-dependent manner, much like TGF-β1 under normal conditions. This is in full agreement with the fact that partial knockdown of Id2 is sufficient to allow BMP-7 to elicit EMT (Kowanetz et al., 2004). We conclude that Id2 could play a primary role in regulating some of the genes identified in our primary transcriptomic screen, and we are in the process of testing this hypothesis using more complex cDNA microarray analyses.
Evolutionary Conservation of Gene Expression Patterns Downstream of TGF-β
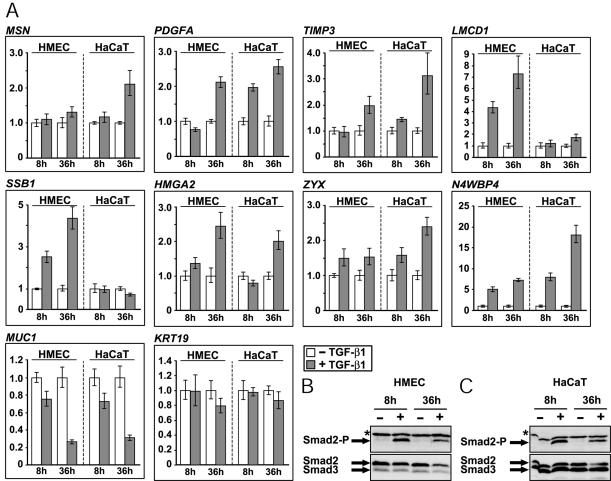
In the first part of this study we established that TGF-β can elicit EMT in several epithelial cell types of both mouse and human origin (Figure 1A). Two common characteristics of these cell types were their relatively normal phenotype (as opposed to carcinoma cells) and their ability to be growth inhibited by TGF-β (Figure 1B). We reasoned that an additional criterion for validation of the gene targets obtained through our microarray screen should be evolutionary conservation of the expression patterns, at least between mouse and human epithelial cells that exhibit robust EMT responses. We therefore validated the selected gene list in two human cell types, HMEC and HaCaT cells by monitoring gene expression at 8 and 36 h post-TGF-β1 treatment (Figure 8A), because higher degree of regulation was obtained in these two time points in NMuMG cells (Figure 5). HMEC respond to TGF-β1 with robust EMT, whereas HaCaT could only exhibit weak scattering and cytoskeletal responses (Figure 1A). To control for the ability of TGF-β1 to signal in these two cell types, we measured and confirmed the proper induction of phospho-Smad2 levels at comparable levels between the two human cell types (Figure 8, B and C).
Figure 8.
Patterns of gene regulation by TGF-β1. (A) Expression patterns of 10 selected genes from those tested in Figure 5. Quantitative real-time RT-PCR analysis performed with HMEC and HaCaT cells stimulated (gray bars) or not (white bars) with TGF-β1 (5 ng/ml) for 8 and 36 h. Normalized values of relative gene expression (arbitrary units) are plotted in bar graphs as described in Materials and Methods. Smad2 phosphorylation status in HMEC (B) and HaCaT (C) cells in response to TGF-β1 under the identical conditions used for the RT-PCR analysis. Immunoblots for phospho-Smad2 (Smad2-P) and total Smad2 and Smad3 are shown. The relevant protein bands are marked with arrows and an asterisk marks background bands.
This analysis revealed several genes that showed a unique regulatory pattern in mammary epithelial cells, which is conserved among mice and humans, and is not observed in human keratinocytes. A few genes with a common pattern among mammary cells and keratinocytes were also observed. Of 14 tested genes, the patterns of gene regulation that were observed are summarized in Table 2. We therefore suggest that Muc1, Fbln2, Hmga2, Lmcd1, Ssb1, Zyx, and Msn are all newly identified targets of the TGF-β pathway whose expression patterns across species, and in response to various ligands of the superfamily, suggests that they might play functional roles in the process of EMT. We currently test this prediction experimentally for these seven genes. The comparative approach of gene expression we have undertaken, so far has revealed significant gene targets and our future aim is to exploit these targets in order to establish the process of EMT in normal human epithelial cells at the molecular level.
Table 2.
Expression patterns of 14 selected target genes
| TGF-β response
|
BMP response
|
||||
|---|---|---|---|---|---|
| Gene | Regulation | NMuMG | HMEC | HaCaT | NMuMG |
| Msn | Up | M; L | L | L | NR |
| Pdgfa | Up | E; M; L | L | M; L | E; M |
| Timp3 | Up | E; M; L | L | M; L | E; M |
| Lmcd1 | Up | E; M; L | M; L | L | NR |
| Ssb-1 | Up | E; M; L | M; L | NR | NR |
| Hmga2 | Up | E; M | M; L | L | NR |
| Zyx | Up | E; M; L | M; L | M; L | NR |
| Fbln2 | Up | M; L | –* | L* | NR |
| N4wbp4 | Up | E; M; L | M; L | M; L | NR |
| Muc1 | Down | M; L | M; L | M; L | NR |
| Myc | Down | E; M; L | n.d. | E; M; L* | NR |
| Krt1-19 | Down | E; M; L | NR | NR | NR |
| Cxcl1 | Down | E; M | n.d. | n.d. | NR |
| Igfbp5 | Down | E; M; L | n.d. | n.d. | NR |
Tabulation of the expression patterns of all 14 gene targets analyzed in this study. The mouse gene name, the mode of regulation, and the gene response to TGF-β1 or BMP-7 are listed as follows: 2 h (early, E), 8 h (middle, M), 36/48 h (late, L), no response (NR), undetectable expression (–), not determined (n.d.). An asterisk star (*) indicates unpublished data.
DISCUSSION
The role of epithelial plasticity in normal embryogenesis and in carcinoma invasiveness and metastasis is a topic of intense current research (Condeelis and Segall, 2003; Grünert et al., 2003). This emphasizes the need for identification of factors that mediate cancer metastasis. EMT characterizes changes in epithelial plasticity and probably represents a transitory stage of carcinoma cell evolution toward aggressive malignancy (Radisky et al., 2001; Thiery, 2002; Grünert et al., 2003). Changes in epithelial plasticity during carcinoma progression are controlled by epigenetic, environmental, and cell communication mechanisms (Radisky et al., 2001). In this context, secreted growth factors are of paramount importance, and the TGF-β superfamily includes many members that contribute to regulation of epithelial plasticity and growth (ten Dijke et al., 2002; Siegel and Massagué, 2003). One major task in the analysis of the TGF-β pathway is therefore the molecular dissection of components that contribute to processes such as cell cycle arrest or EMT.
Here we provide a comprehensive analysis of most components of the TGF-β superfamily signaling pathways using appropriate model cell systems and furthermore identify a group of novel gene targets of this pathway that potentially link basic TGF-β biology to epithelial cell invasiveness. A major conclusion is that EMT is a physiological response of both normal mouse and human epithelial cells to TGF-β. We observed a more robust scattering and depolarization in mammary and lung epithelial cells than in keratinocytes (Figure 1A), which may reflect the developmental history of each tissue type. Despite this, all cell types tested here, as well as several others such as kidney, prostate or lens epithelial cells, undergo EMT (reviewed in Gotzmann et al., 2004). The observations in normal keratinocytes are in agreement with another report, which analyzed HaCaT cell EMT in response to TGF-β1 (Zavadil et al., 2001). Malignant keratinocytes expressing Ras oncogenes exhibit stronger EMT and architectural depolarization, which is mediated by TGF-β and appears more similar to the response described here for normal mammary cells (Portella et al., 1998). The data also suggest that TGF-β is sufficient to elicit a transient but fully developed EMT response in normal epithelial cells without the need for predisposing oncogenic transformation as documented for the Ras oncogene (Janda et al., 2002; Oft et al., 2002; Grünert et al., 2003). We also demonstrate that the in vitro EMT response of normal epithelial cells is conserved between mice and humans. Our data suggest that NMuMG cells represent a faithful in vitro cell model of EMT despite the fact that these cells carry no autonomous tumorigenic potential.
Although the role of TGF-β on modulation of epithelial plasticity and tumor progression has become a major point of current research (ten Dijke et al., 2002; Siegel and Massagué, 2003), the contribution of the other members of the TGF-β superfamily to such processes remains poorly studied. Using adenovirus-mediated expression of all type I receptors of the superfamily we could delineate for the first time that only signaling pathways that belong to the TGF-β/activin/nodal branch can induce EMT (Figure 2C, Supplementary Figure 1). The low-molecular-weight inhibitor of the three receptor kinases ALK-4, -5, -7, gave evidence for constitutive autocrine TGF-β signaling that maintains low level gene expression in NMuMG cells (Figure 2E). Although all three ALK-4(TD), -5(TD), and -7(TD) receptors induced EMT in NMuMG cells, only ALK-5(KR) (and partially ALK-4(KR)) were able to block TGF-β1-induced EMT (Figure 2D). This reflects the fact that all three constitutively active receptors can activate robust Smad2/Smad3 signaling, whereas only ALK-5(KR) can efficiently interfere with the function of the endogenous TGF-β receptor complex. Because overexpressed activin type I receptor (ALK-4) is able to elicit EMT, we conclude that the failure of activin-A to induce EMT and Smad2 phosphorylation in NMuMG cells may reflect a suboptimal level of ALK-4 on the cell surface in this cell type.
A corollary from the comprehensive receptor analysis was that specific R-Smads, i.e., Smad2 and Smad3, might be involved in the process of EMT. Indeed, we previously reported a role for Smad3 downstream of TGF-β in NMuMG cell responses (Piek et al., 1999b). We extended our analysis to essentially all Smad components of the superfamily, Smad1–7, with the sole exception of Smad8 for technical reasons (Figure 3, Supplementary Figures 4–7). The new results strongly implicate the endogenous Smads of the TGF-β branch in signal transduction that orchestrates the EMT response. However, a previous report that used stably transfected NMuMG clones with wild-type Smad7 or a different dominant negative Smad3 mutant than Smad3(D407E), failed to observe effects on EMT in response to TGF-β (Bhowmick et al., 2001a). In our hands it has been impossible to obtain stable clones of epithelial cells that express wild-type or mutant Smads constitutively (unpublished results). We either had to rely on inducible expression systems (Figure 3C) or on transient adenoviral infections (Figure 3A). Thus, it is possible that NMuMG clones selected for survival in the continuous presence of high levels of exogenous mutant Smad3 or Smad7 might adapt by exhibiting differential responses to TGF-β. The data described here are in good agreement with several recent reports on the role of Smad4 in mammary EMT and on the ability of transcription factors YY1 and c-ski, which interact and inactivate Smad protein function, to block specifically EMT of NMuMG cells (Kurisaki et al., 2003; Li et al., 2003; Takeda et al., 2004). The dominant-negative Smad2 and Smad3 mutants used unfortunately have not allowed us to pinpoint possible differential effects between the two R-Smads of the TGF-β pathway. This is in part due to the fact that these mutants interfere at least partially with both Smad2 and Smad3 activation by TGF-β receptors. This has been previously established for mutant Smad3(D407E) (Goto et al., 1998) but not for mutant Smad2SA. In the stable clones expressing Smad2SA we could measure robust phospho-Smad3 levels induced by TGF-β1 (Supplementary Figure 4F). However, this finding does not necessarily exclude the possibility that the mutant interferes with other critical signaling effects, e.g., proper receptor-Smad endocytosis and trafficking. The fact that ectopic expression of Smad2 (or Smad3) alone could not elicit robust EMT in NMuMG cells (Figure 3C, Supplementary Figure 7) suggests that the Smad pathway as a whole is needed for this response, in addition to the cooperating non-Smad effectors.
The evidence supporting participation of Smads in the establishment of EMT by TGF-β is also corroborated by the analysis of the ALK-5 receptor with mutations in its L45 loop (Figure 6), a result in full agreement with previous reports that examined the role of the L45 loop of ALK-5 on EMT of NMuMG cells (Yu et al., 2002; Itoh et al., 2003). Despite this, other evidence has supported the role of alternative signaling pathways in the same phenotypic response (Bakin et al., 2000, 2002; Bhowmick et al., 2001a, 2001b; Yu et al., 2002). Our evidence suggests that modulation of epithelial plasticity may be a more complex phenomenon than previously appreciated, in which both Smad and non-Smad effectors cooperate in order to execute a complete program of EMT. This would be consistent with the fact that EMT incorporates an extensive program of changes in gene expression (see below), part of which at least depends on Smad transcriptional inputs. Another part of this program may require the non-Smad signaling pathways, and a challenge for the immediate future remains to establish the points of cross-talk between the various effectors.
A remaining question concerned the types and numbers of genes regulated by TGF-β during the course of EMT. Transcriptomic analysis gave a rather rich outcome of regulated genes (Table 1, Figure 4, and Supplementary Tables 3 and 4). This was anticipated based on recent transcriptomic screens of the TGF-β pathway in various cell types (Perou et al., 1999; Akiyoshi et al., 2001; Verrecchia et al., 2001; Zavadil et al., 2001; Ota et al., 2002; Chambers et al., 2003; Coyle et al., 2003; Jechlinger et al., 2003; Kang et al., 2003a; Xie et al., 2003; Kowanetz et al., 2004). On the other hand, the gene list identified here shares relatively small similarities (only 22 common genes in contrast to 163 unique genes) with other transcriptomic analyses of the TGF-β pathway, including one recently reported in the same cell line (Xie et al., 2003).
In our effort to define genes whose expression links to the EMT process, analyzing both early and late gene responses was instrumental. Thus, we were able to define gene clusters based strictly on expression values, which include genes with kinetics that suit with the onset and establishment of EMT in NMuMG cells (clusters 3 and 4, Figure 4, B and C). Combining this information with systematic screens of PubMed and cross-referencing with microarray data from studies of breast cancer, epithelial invasiveness, and metastasis (Clark et al., 2000; Ryu et al., 2001; van't Veer et al., 2002; Jechlinger et al., 2003; Kang et al., 2003b), we selected a group of genes that possibly links to the EMT process. A number of these genes, such as Myh9, Cfh, Hif1a, Pdgfa, Ext1, Col6a1, Fn1, Sdc1, Gas1, Igfbp5, Iqgap1, Idb2, Cks-2, Vamp8, and Pold1 have already been enlisted as poor prognosis markers for breast cancer or invasion and metastasis markers, but most of them have not been previously recognized as TGF-β responsive genes. Thus, we conclude that the NMuMG in vitro system, although more distant from mammary carcinoma cells undergoing EMT in vivo, represents a useful tool for the identification of candidate genes of EMT. Future work will focus on the actual in vivo validation of the novel TGF-β target genes.
To evaluate functionally some of the genes identified in this screen, we made use of the results on signaling specificity obtained in the first part of this study. We postulated that genes that are differentially regulated by TGF-β1 versus BMP-7 should be relevant targets that characterize the EMT process and can be used for future in vivo validation. Furthermore the newly established role of endogenous Smads on mammary EMT suggested that relevant gene targets might require an intact Smad pathway for proper regulation. Finally, we reasoned that critical TGF-β targets for regulation of EMT should exhibit conservation of their expression pattern among species. In agreement with our postulates, all genes tested showed a strict requirement for R-Smad activation by the TGF-β type I receptor (Figure 6). In addition, most of the genes tested were not regulated by BMP-7 (Figure 5), or, as is the case of the transcriptional regulators of the Id family, they exhibited the inverse response to BMP-7. Analysis of functional inactivation of Id2 using siRNA technology established a causal relationship between BMP-7 signaling and proper regulation of the myoepithelial marker α-SMA (Figure 7). This is in perfect agreement with the model we have recently established, based on which, arrest of epithelial proliferation and induction of EMT require the down-regulation of Id2 (and Id3) by TGF-β. On the other hand, BMPs that induce high levels of Id2 (or Id3) in epithelial cells fail to exhibit robust growth inhibition or EMT because of the inhibitory role Id proteins play in these processes (Kowanetz et al., 2004). Examination of the pattern of regulation of the selected genes in HMEC and HaCaT cells (Figure 8, Table 2) revealed that a number of the selected genes showed cell type specificity and evolutionary conservation in terms of their expression and mode of regulation.
In conclusion, we show that epithelial-mesenchymal transition is a multigenic cellular response to TGF-β superfamily members. Among such members, TGF-βs, activins and possibly other ligands such as nodals, which have the common feature of activating Smad2 and Smad3, appear to be competent to elicit EMT, whereas other members cannot. The combined signaling specificity and transcriptomic analysis provides a first degree of molecular dissection of novel gene functions that link and orchestrate the differentiative programs underlying epithelial plasticity.
Supplementary Material
Acknowledgments
We thank J. Zavadil, Y-C. Yang, E. Böttinger, S. White, and E. Hunter for advice and training on cDNA microarray analysis; S. Dooley, Y. Eto, N. Ferrara, N. Fusenig, V. Kaartinen, N. J. Laping, F. D. Miller, K. Miyazono, M. Reiss, K. Sampath, S. Souchelnytskyi, T. Takahashi, and B. Vogelstein for reagents; and Irene Apostolou for technical assistance. The microarray consortium is funded by the Wellcome Trust, Cancer Research UK, and the Ludwig Institute of Cancer Research. We thank the staff of the Sanger Institute Microarray Facility http://www.sanger.ac.uk/Projects/Microarrays/for supplying arrays, lab protocols, and technical advice (David Vetrie, Cordelia Langford, Adam Whittaker, Neil Sutton) and Quantarray/GeneSpring datafiles and databases relating to array elements (Kate Rice, Rob Andrews, Adam Butler, Harish Chudasama). The mouse cDNA clone collection was from the National Institute on Aging (Bethesda, MD). cDNA clone resequencing was performed by Team 56 at the Sanger Institute. This research was funded in part by grants from the Human Frontier Science Program and the Swedish Cancer Society (Project number 4855-B03-01XAC to A.M.) and post-doctoral fellowships from the French Association pour la Recherche sur le Cancer and the Swedish Cancer Society (Project number 4812-B03-01VAA to U.V.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-08-0658) on February 2, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Akiyoshi, S., Ishii, M., Nemoto, N., Kawabata, M., Aburatani, H., and Miyazono, K. (2001). Targets of transcriptional regulation by transforming growth factor-β: expression profile analysis using oligonucleotide arrays. Jpn. J. Cancer Res. 92, 257-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin, A. V., Rinehart, C., Tomlinson, A. K., and Arteaga, C. L. (2002). p38 mitogen-activated protein kinase is required for TGFβ-mediated fibroblastic transdifferentiation and cell migration. J. Cell Sci. 115, 3193-3206. [DOI] [PubMed] [Google Scholar]
- Bakin, A. V., Tomlinson, A. K., Bhowmick, N. A., Moses, H. L., and Arteaga, C. L. (2000). Phosphatidylinositol 3-kinase function is required for transforming growth factor β-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 275, 36803-36810. [DOI] [PubMed] [Google Scholar]
- Bates, R. C., and Mercurio, A. M. (2003). Tumor necrosis factor-α stimulates the epithelial-to-mesenchymal transition of human colonic organoids. Mol. Biol. Cell 14, 1790-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick, N. A., Ghiassi, M., Bakin, A., Aakre, M., Lundquist, C. A., Engel, M. E., Arteaga, C. L., and Moses, H. L. (2001a). Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell 12, 27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick, N. A., Zent, R., Ghiassi, M., McDonnell, M., and Moses, H. L. (2001b). Integrin β1 signaling is necessary for transforming growth factor-β activation of p38MAPK and epithelial plasticity. J. Biol. Chem. 276, 46707-46713. [DOI] [PubMed] [Google Scholar]
- Brown, K. A., Aakre, M. E., Gorska, A. E., Price, J. O., Eltom, S. E., Pietenpol, J. A., and Moses, H. L. (2004). Induction by transforming growth factor-β1 of epithelial to mesenchymal transition is a rare event in vitro. Breast Cancer Res. 6, R215-R231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers, R. C., Leoni, P., Kaminski, N., Laurent, G. J., and Heller, R. A. (2003). Global expression profiling of fibroblast responses to transforming growth factor-β1 reveals the induction of inhibitor of differentiation-1 and provides evidence of smooth muscle cell phenotypic switching. Am. J. Pathol. 162, 533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H., Brown, C. W., and Matzuk, M. M. (2002). Genetic analysis of the mammalian transforming growth factor-β superfamily. Endocr. Rev. 23, 787-823. [DOI] [PubMed] [Google Scholar]
- Clark, E. A., Golub, T. R., Lander, E. S., and Hynes, R. O. (2000). Genomic analysis of metastasis reveals an essential role for RhoC. Nature 406, 532-535. [DOI] [PubMed] [Google Scholar]
- Comijn, J., Berx, G., Vermassen, P., Verschueren, K., van Grunsven, L., Bruyneel, E., Mareel, M., Huylebroeck, D., and van Roy, F. (2001). The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 7, 1267-1278. [DOI] [PubMed] [Google Scholar]
- Condeelis, J., and Segall, J. E. (2003). Intravital imaging of cell movement in tumours. Nat. Rev. Cancer 3, 921-930. [DOI] [PubMed] [Google Scholar]
- Coyle, B., Freathy, C., Gant, T. W., Roberts, R. A., and Cain, K. (2003). Characterization of the transforming growth factor-β 1-induced apoptotic transcriptome in FaO hepatoma cells. J. Biol. Chem. 278, 5920-5928. [DOI] [PubMed] [Google Scholar]
- Cui, W., Fowlis, D. J., Bryson, S., Duffie, E., Ireland, H., Balmain, A., and Akhurst, R. J. (1996). TGFβ1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell 86, 531-542. [DOI] [PubMed] [Google Scholar]
- Dennler, S., Itoh, S., Vivien, D., ten Dijke, P., Huet, S., and Gauthier, J.-M. (1998). Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17, 3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley, S., Delvoux, B., Streckert, M., Bonzel, L., Stopa, M., ten Dijke, P., and Gressner, A. M. (2001). Transforming growth factor β signal transduction in hepatic stellate cells via Smad2/3 phosphorylation, a pathway that is abrogated during in vitro progression to myofibroblasts. TGFβ signal transduction during transdifferentiation of hepatic stellate cells. FEBS Lett. 502, 4-10. [DOI] [PubMed] [Google Scholar]
- Dudas, M., Nagy, A., Laping, N. J., Moustakas, A., and Kaartinen, V. (2004). Tgf-beta3-induced palatal fusion is mediated by Alk-5/Smad pathway. Dev. Biol. 266, 96-108. [DOI] [PubMed] [Google Scholar]
- Fujii, M., Takeda, K., Imamura, T., Aoki, H., Sampath, T. K., Enomoto, S., Kawabata, M., Kato, M., Ichijo, H., and Miyazono, K. (1999). Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol. Biol. Cell 10, 3801-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, D., Yagi, K., Inoue, H., Iwamoto, I., Kawabata, M., Miyazono, K., and Kato, M. (1998). A single missense mutant of Smad3 inhibits activation of both Smad2 and Smad3, and has a dominant negative effect on TGF-β signals. FEBS Lett. 430, 201-204. [DOI] [PubMed] [Google Scholar]
- Gotzmann, J., Mikula, M., Eger, A., Schulte-Hermann, R., Foisner, R., Beug, H., and Mikulits, W. (2004). Molecular aspects of epithelial cell plasticity: implications for local tumor invasion and metastasis. Mutat. Res. 566, 9-20. [DOI] [PubMed] [Google Scholar]
- Grünert, S., Jechlinger, M., and Beug, H. (2003). Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat. Rev. Mol. Cell. Biol. 4, 657-665. [DOI] [PubMed] [Google Scholar]
- He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W., and Vogelstein, B. (1998). A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman, G. J., Nicolas, F. J., Callahan, J. F., Harling, J. D., Gaster, L. M., Reith, A. D., Laping, N. J., and Hill, C. S. (2002). SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 62, 65-74. [DOI] [PubMed] [Google Scholar]
- Itoh, S., Thorikay, M., Kowanetz, M., Moustakas, A., Itoh, F., Heldin, C.-H., and ten Dijke, P. (2003). Elucidation of Smad requirement in transforming growth factor-β type I receptor-induced responses. J. Biol. Chem. 278, 3751-3761. [DOI] [PubMed] [Google Scholar]
- Janda, E., Lehmann, K., Killisch, I., Jechlinger, M., Herzig, M., Downward, J., Beug, H., and Grünert, S. (2002). Ras and TGFβ cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J. Cell Biol. 156, 299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jechlinger, M., Grunert, S., Tamir, I. H., Janda, E., Ludemann, S., Waerner, T., Seither, P., Weith, A., Beug, H., and Kraut, N. (2003). Expression profiling of epithelial plasticity in tumor progression. Oncogene 22, 7155-7169. [DOI] [PubMed] [Google Scholar]
- Kang, Y., Chen, C. R., and Massagué, J. (2003a). A self-enabling TGFβ response coupled to stress signaling. Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell 11, 915-926. [DOI] [PubMed] [Google Scholar]
- Kang, Y., Siegel, P. M., Shu, W., Drobnjak, M., Kakonen, S. M., Cordon-Cardo, C., Guise, T. A., and Massagué, J. (2003b). A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537-549. [DOI] [PubMed] [Google Scholar]
- Kowanetz, M., Valcourt, U., Bergström, R., Heldin, C.-H., and Moustakas, A. (2004). Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor β and bone morphogenetic protein. Mol. Cell. Biol. 24, 4241-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisaki, K., Kurisaki, A., Valcourt, U., Terentiev, A. A., Pardali, K., ten Dijke, P., Heldin, C.-H., Ericsson, J., and Moustakas, A. (2003). Nuclear factor YY1 inhibits transforming growth factor β- and bone morphogenetic protein-induced cell differentiation. Mol. Cell. Biol. 23, 4494-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Qiao, W., Chen, L., Xu, X., Yang, X., Li, D., Li, C., Brodie, S. G., Meguid, M. M., Hennighausen, L., and Deng, C.-X. (2003). Squamous cell carcinoma and mammary abscess formation through squamous metaplasia in Smad4/Dpc4 conditional knockout mice. Development 130, 6143-6153. [DOI] [PubMed] [Google Scholar]
- Masuda, A., Kondo, M., Saito, T., Yatabe, Y., Kobayashi, T., Okamoto, M., Suyama, M., and Takahashi, T. (1997). Establishment of human peripheral lung epithelial cell lines (HPL1) retaining differentiated characteristics and responsiveness to epidermal growth factor, hepatocyte growth factor, and transforming growth factor β1. Cancer Res. 57, 4898-4904. [PubMed] [Google Scholar]
- Maurice, D., Pierreux, C. E., Howell, M., Wilentz, R. E., Owen, M. J., and Hill, C. S. (2001). Loss of Smad4 function in pancreatic tumors: C-terminal truncation leads to decreased stability. J. Biol. Chem. 276, 43175-43181. [DOI] [PubMed] [Google Scholar]
- Miettinen, P. J., Ebner, R., Lopez, A. R., and Derynck, R. (1994). TGF-β induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 127, 2021-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa, K., Shinozaki, M., Hara, T., Furuya, T., and Miyazono, K. (2002). Two major Smad pathways in TGF-β superfamily signalling. Genes Cells 7, 1191-1204. [DOI] [PubMed] [Google Scholar]
- Oft, M., Akhurst, R. J., and Balmain, A. (2002). Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat. Cell Biol. 4, 487-494. [DOI] [PubMed] [Google Scholar]
- Ota, T., Fujii, M., Sugizaki, T., Ishii, M., Miyazawa, K., Aburatani, H., and Miyazono, K. (2002). Targets of transcriptional regulation by two distinct type I receptors for transforming growth factor-β in human umbilical vein endothelial cells. J. Cell. Physiol. 193, 299-318. [DOI] [PubMed] [Google Scholar]
- Perou, C. M. et al. (1999). Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl. Acad. Sci. USA 96, 9212-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek, E., Heldin, C.-H., and ten Dijke, P. (1999a). Specificity, diversity, and regulation in TGF-β superfamily signaling. FASEB J. 13, 2105-2124. [PubMed] [Google Scholar]
- Piek, E., Moustakas, A., Kurisaki, A., Heldin, C.-H., and ten Dijke, P. (1999b). TGF-β type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J. Cell Sci. 112, 4557-4568. [DOI] [PubMed] [Google Scholar]
- Portella, G., Cumming, S. A., Liddell, J., Cui, W., Ireland, H., Akhurst, R. J., and Balmain, A. (1998). Transforming growth factor β is essential for spindle cell conversion of mouse skin carcinoma in vivo: implications for tumor invasion. Cell Growth Differ. 9, 393-404. [PubMed] [Google Scholar]
- Radisky, D., Hagios, C., and Bissell, M. J. (2001). Tumors are unique organs defined by abnormal signaling and context. Semin. Cancer Biol. 11, 87-95. [DOI] [PubMed] [Google Scholar]
- Ryu, B., Jones, J., Hollingsworth, M. A., Hruban, R. H., and Kern, S. E. (2001). Invasion-specific genes in malignancy: serial analysis of gene expression comparisons of primary and passaged cancers. Cancer Res. 61, 1833-1838. [PubMed] [Google Scholar]
- Shi, Y., and Massagué, J. (2003). Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685-700. [DOI] [PubMed] [Google Scholar]
- Siegel, P. M., and Massagué, J. (2003). Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer 3, 807-820. [DOI] [PubMed] [Google Scholar]
- Souchelnytskyi, S., Tamaki, K., Engström, U., Wernstedt, C., ten Dijke, P., and Heldin, C.-H. (1997). Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J. Biol. Chem. 272, 28107-28115. [DOI] [PubMed] [Google Scholar]
- Takeda, M., Mizuide, M., Oka, M., Watabe, T., Inoue, H., Suzuki, H., Fujita, T., Imamura, T., Miyazono, K., and Miyazawa, K. (2004). Interaction with Smad4 is indispensable for suppression of BMP signaling by c-Ski. Mol. Biol. Cell 15, 963-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, B., Vu, M., Booker, T., Santner, S. J., Miller, F. R., Anver, M. R., and Wakefield, L. M. (2003). TGF-β switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J. Clin. Invest. 112, 1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dijke, P., Goumans, M.-J., Itoh, F., and Itoh, S. (2002). Regulation of cell proliferation by Smad proteins. J. Cell. Physiol. 191, 1-16. [DOI] [PubMed] [Google Scholar]
- Thiery, J.-P. (2002). Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442-454. [DOI] [PubMed] [Google Scholar]
- Tosh, D., and Slack, J. M. (2002). How cells change their phenotype. Nat. Rev. Mol. Cell. Biol. 3, 187-194. [DOI] [PubMed] [Google Scholar]
- van Obberghen-Schilling, E., Roche, N. S., Flanders, K. C., Sporn, M. B., and Roberts, A. B. (1988). Transforming growth factor β 1 positively regulates its own expression in normal and transformed cells. J. Biol. Chem. 263, 7741-7746. [PubMed] [Google Scholar]
- van't Veer, L. J. et al. (2002). Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530-536. [DOI] [PubMed] [Google Scholar]
- Verrecchia, F., Chu, M. L., and Mauviel, A. (2001). Identification of novel TGF-β/Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J. Biol. Chem. 276, 17058-17062. [DOI] [PubMed] [Google Scholar]
- Xie, L., Law, B. K., Aakre, M. E., Edgerton, M., Shyr, Y., Bhowmick, N. A., and Moses, H. L. (2003). Transforming growth factor β-regulated gene expression in a mouse mammary gland epithelial cell line. Breast Cancer Res. 5, R187-R198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z., Shen, M. X., Ma, D. Z., Wang, L. Y., and Zha, X. L. (2003). TGF-β1-promoted epithelial-to-mesenchymal transformation and cell adhesion contribute to TGF-β1-enhanced cell migration in SMMC-7721 cells. Cell Res. 13, 343-350. [DOI] [PubMed] [Google Scholar]
- Yu, L., Hebert, M. C., and Zhang, Y. E. (2002). TGF-β receptor-activated p38 MAP kinase mediates Smad-independent TGF-β responses. EMBO J. 21, 3749-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil, J., Bitzer, M., Liang, D., Yang, Y. C., Massimi, A., Kneitz, S., Piek, E., and Böttinger, E. P. (2001). Genetic programs of epithelial cell plasticity directed by transforming growth factor-β. Proc. Natl. Acad. Sci. USA 98, 6686-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.