Abstract
Site-specific exchange of genetic information is mediated by DNA recombinases, such as FLP or Cre, and has become a valuable tool in modern molecular biology. The so far low number of suitable recombinating enzymes has driven current research activities towards alteration of catalytic properties, such as thermostability or recognition sequences. However, identification and analysis of new mutants requires sensitive in vitro activity assays, which traditionally are based on gel electrophoresis. Here, we describe the development of a new sensitive DNA recombination assay based on dual-color fluorescence cross-correlation spectroscopy (DC-FCCS), which works in homogenous solution and does not require any separation step such as electrophoresis. The assay was validated with unlabeled FLP recombinase and different fluorescently labeled DNA substrates containing the FLP recognition target (FRT). This strategy fulfills all requirements for possible application in high throughput screening and engineering of new site-specific DNA recombinases starting from the FLP-FRT system, and is easily adjustable to other systems like Cre/loxP.
INTRODUCTION
In nature, exchange of genetic information plays an important role for phenotype diversification and evolution. Recombination of two DNA double strands can be mediated in a highly site-specific way by FLP recombinase from yeast or other members of the integrase family, such as phage λ Int or the phage P1 Cre protein. For the enzyme-catalyzed DNA exchange reaction, no sequence homology is required except the recognition sequence. The FLP recognition target (FRT) is composed of three 13 bp symmetry elements a, b and c each representing a binding site for one FLP monomer. For recombination, the symmetry element c is not necessary. Elements a and b are flanking an 8 bp asymmetrical core region in opposite directions giving the FRT site a total length of 34 bp. The recognition site of Cre-recombinase is of similar structure and called loxP. The reaction mechanisms and biochemical properties of FLP and Cre recombinases have been well elucidated during the last two decades (1–4). The unique property of very accurate recombination at a specific, naturally rarely occurring sequence has turned both the FLP-FRT and the Cre-loxP systems into very valuable tools in genetic engineering (5). Especially for in vivo applications, enzymatic recombination has gained strong importance for excision, inversion, integration and exchange of genetic elements. Conditional knock-out mouse model systems employ site-specific recombination as well as transgenic plant systems (6–8).
Unfortunately, there is only limited number of site-specific recombinases known from nature, and therefore few recombination sites are available. It would be of enormous scientific and therapeutical importance to have more or—in ideal case—virtually any site available for specific recombination/exchange of genetic parts. Current scientific research addresses this problem using mutagenesis and directed evolution of recombinases in order to change the specificity of existing enzymes (9–13). Identification and characterization of newly engineered recombinase mutants is usually done by conventional DNA recombination assays, which in most cases are dependent on separation steps, such as electrophoresis, and therefore are time-consuming and cumbersome (14–17).
Here we describe, as a novel approach, the use of dual-color fluorescence cross-correlation spectroscopy (DC-FCCS) to follow site-specific DNA recombination in solution. Applying this single-molecule sensitive technique, we are able to monitor and quantify recombination events in homogenous solution, without any need for separation steps. A short introduction to DC-FCCS can be found in Material and Methods section. For a more detailed theoretical and experimental background, as well as other biochemical applications of DC-FCCS, please refer to previous published articles which give a comprehensive overview of this biophysical technique (18–21). The recombination assay presented here fulfills all requirements for the rapid characterization of new recombinases.
MATERIALS AND METHODS
Preparation and storage of active FLP-protein
For expression in E.coli, the gene of the FLP-recombinase [EC 2.7.7.-] from yeast was completely codon-optimized. For each of the 424 amino acids, optimal triplets were chosen according to the E.coli codon-frequency table from the GCG-Wisconsin package. The FLP gene was assembled from 64 overlapping synthetic oligonucleotides custom-synthesized by Eurogentec, Seraing, Belgium [for exact sequences, see (22)] using the protocol from Stemmer et al. (23). The 1280 bp product was cloned with the TOPO-blunt cloning kit (Invitrogen, Karlsruhe, Germany) and sequenced. Subsequent cloning in the expression vector pET24a(+) (Novagen, Bad Schwalbach/Ts., Germany) was done by PCR using primers with BamHI and SalI sites yielding the FLP construct with C-terminal His-Tag fusion. E.coli strain BL21(DE3) was used for expression. A culture of 4.8 l LB medium supplemented with 30 μg/ml kanamycin was grown at 37°C with an OD (600 nm) of ∼0.7, and then cooled down to 23°C. Expression was induced with 1 mM IPTG over 2 h at 23°C before cooling on ice. Cells were harvested by centrifugation, washed once with 50 mM Tris–HCl, pH 7.4 at 4°C, 150 mM NaCl, 10 mM imidazol and resuspended in 60 ml of the same buffer. Before lysis, 2400 U DNaseI and 1 mM PMSF were added. Cell disruption was done in a french pressure cell and the lysate was centrifuged 45 min at 43 000 g and applied on a 1 ml HiTrap-chelating column (Amersham, Freiburg, Germany). FPLC-purification was performed with step-wise elution at 20, 40, 80, 120 and 200 mM imidazole in the lysis buffer containing 10% (v/v) glycerol. His-tagged FLP protein was in the 200 mM fraction in which the concentration of NaCl was raised to 400 mM by dropwise adding 1/19 vol of a 5 M NaCl stock solution. Removal of residual DNA was achieved via a second purification step using a 1 ml Q-Sepharose anion-exchange column. This column was equilibrated with 50 mM Tris–HCl, pH 7.4 at 4°C, 10% (v/v) glycerol, 400 mM NaCl, 200 mM imidazol, and the FLP sample from the first step was applied. The flow-through fraction contained pure FLP and was concentrated and simultaneously dialyzed against 50 mM Tris–HCl, pH 7.4 at 4°C, 10% (v/v) glycerol, 1 M NaCl, 1 mM EDTA and 2 mM DTT using ultra thimbles (Schleicher & Schuell, Dassel, Germany). Storage of the FLP protein was done in small aliquots at −80°C and a conventional gel-shift assay was performed to ensure activity of the FLP preparation.
DNA substrates
The DNA substrate for intramolecular recombination was prepared by PCR and had a length of 368 bp. Primers contained the FLP-recognition targets in equal orientation (bold), and were purchased from IBA, Göttingen, Germany with synthetic fluorophores attached on different positions. Upper primer: Ev3FLP1-Cy5: 5′-Cy5-AT GCCGGCCAGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCCCCGCGAAATTAATACGACTCACTATAGGG-3′; Lower Primer: Ev3FLP2-int50-RG: 5′-TGCCGGTACTGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCCTCC(T-internal Rhodamine Green)TTCAGCAAAAAACCCCTCAAGACCC-3′. The PCR product was gel-purified from agarose using the NucleoSpin Extract Kit (Macherey & Nagel, Düren, Germany) and finally precipitated with ethanol/acetate.
Assay conditions
Recombination reactions were performed with 20 nM double-labeled DNA-substrate in FLP-buffer [50 mM Tris–HCl, pH 7.4, 10 mM MgCl2, 0.05% (w/v) Pluronic-F127 (Sigma P-2443)]. For the native PAGE, 4 μM FLP was added in a total volume of 30 μl. Samples were allowed to react for ∼2 h at room temperature before addition of 1.5 μl proteinase K (Stratagene, 10 mg/ml stock solution in 10 mM CaCl2). After digestion of ∼1 h, the samples were purified using the Nucleotide Removal Kit (Qiagen) and analyzed on a 8% native PAGE. The gel was scanned with the Typhoon-Instrument (Amersham) using filter sets for Rhodamine green and Cy5 fluorescence. After scanning, the gel was stained with ethidium bromide in order to visualize the non-fluorescent marker lane and estimate the sizes of the bands.
For DC-FCCS measurements, 20 nM substrate was incubated with different amounts of purified FLP protein in FLP-buffer as above. All DC-FCCS reactions were performed in 8-well LabTek chambered coverglass (NalgeNunc) at room temperature with a total volume of 20 μl. Direct after mixing, initial DC-FCCS measurement was recorded. Samples were incubated and digested with Proteinase K as described above. After digestion the samples were measured again with DC-FCCS.
Dual-color fluorescence cross-correlation spectroscopy
In DC-FCCS, two superimposed laser lines of different wavelength are focused by a single microscope objective in order to generate overlapping focal volume elements. In this way, two spectrally distinct dyes can be simultaneously excited and their fluorescence emission detected via the same objective in two separate channels, i and j. Both intensity traces are subjected to autocorrelation analysis. The additionally calculated cross-correlation function G(τ) is obtained by correlating intensity fluctuations of both channels with each other:
| 1 |
From the logarithmic plot and curve fitting of Gij(τ), the number of double-labeled species can be obtained and used to calculate absolute concentrations when the effective volume element Veff is known from preceding calibration measurements. Further theoretical background of DC-FCCS is described elsewhere (18,21).
Optical setup, measurements and data analysis
All DC-FCCS measurements of this work were carried out using the ConfoCor2 instrument (Zeiss, Germany). For excitation of the Rhodamine green and Cy5 fluorophores, we used the 488 nm laser line of the argon-ion laser and the He–Ne 633 nm laser line, respectively. Correlation curves were averaged from 20 measurements of 10 s each, exported and processed in Origin 7.0 (OriginLab, Northhampton, MA, USA). Nonlinear-least-square fit routines with standard single-component models for auto- and cross-correlation analysis were used to obtain parameters such as amplitude, G(0) and diffusion time, τd.
The remaining substrate concentration after recombination reaction (and proteinase K digestion) was calculated using the following equation:
| 2 |
where G(0)CC designates the cross-correlation amplitude and G(0)AC means the lower of the two autocorrelation amplitudes (which is most often the curve derived from the red channel, as the volume element from the red excitation light is larger due to wavelength-dependent diffraction).
RESULTS AND DISCUSSION
FLP protein
As FLP protein from yeast is produced with extremely low yields upon heterologous expression in E.coli (24,25), we performed a complete codon optimization of the FLP gene and removed about 40 rare codons. This step drastically improved expression levels and enabled us to purify reasonable amounts of active his-tagged FLP protein from low culture volumes. It is known that the commercially available FLP plasmid pOG44, whose sequence served as a template for the codon optimization, carries a point mutation (26). The resulting FLP-F70L mutant is slightly thermolabile at 37°C, but exhibits the same activity compared to wild-type FLP when reacting at room temperature, where our experiments were performed. In literature, contradictory data can be found concerning the effect of ions on recombination, but it is a consensus, that divalent ions are required for intermolecular DNA exchange reaction (24,25,27,28). We therefore included Mg2+ in the buffer. From previous DC-FCCS experiments, we also experienced that adding small amounts of detergent helps to avoid depletion of the fluorophores from the solution, probably by preventing adsorption of dye-labeled substrates to the glass surface. With FLP protein, we tried 8 different detergents [Triton X-100, Pluronic F-127, Nonidet-P40, Brij-35, Span 80, Tween-20, Tween-80, Polybutylen/ethylen-Polyethylenoxyd-Block-Copolymer (PBE/PEO, gift from K. Landfester, University of Ulm, Germany)] and found that six of them completely inhibited recombination activity, even when used at concentrations as low as 0.05%. Only Pluronic F-127 and PBE/PEO did not interfere with FLP activity.
Assay principles for detection of DNA recombination using DC-FCCS
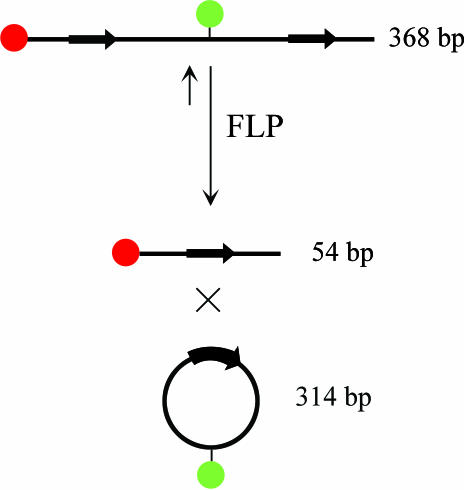
The DC-FCCS technique is suitable for sensitive detection of the relative amount of double-labeled molecules versus single-labeled species. From that, in principle all biochemical reactions can be monitored where connections between two spectrally distinct fluorophores are formed or broken. As the site-specific DNA recombination mechanism involves a transient strand break with a subsequent ligation step, the resulting repositioning of the markers can efficiently be detected with DC-FCCS. Principally both, inter- and intramolecular recombination can be followed via the rearrangement of appropriately positioned fluorescent labels. In the case of intermolecular reaction, the concentration of double-labeled molecules will decrease or increase during the reaction course, depending on the used educts. In contrast, intramolecular recombination can only be measured via decreasing cross-correlation amplitudes from diminishing double-labeled educts (Figure 1).
Figure 1.
Principle of observation of intramolecular recombination using a double-labeled substrate. Arrows represent FLP-recognition-targets (FRTs) and their orientation. To distribute labels on different product molecules during the excision reaction, it is important that in the educt one label is positioned outside the FRTs, while the other needs to be positioned between the FRTs.
As FLP is known to bind very tight to its recognition target (29,30), it is necessary to digest the reactions with proteinase K in order to release the DNA from the DNA–protein complexes. This digestion step is also common in conventional assay approaches and allows for analysis of the free DNA.
Measurement results for intramolecular recombination
At low nanomolar concentrations required for DC-FCCS, the intramolecular recombination reaction is favored. The substrate we used for intramolecular recombination studies was easily prepared by PCR and purified. Both FRT-sites had the same direction, leading to excision of the enclosed part on recombination with FLP. For separation of the dyes during recombination, it is therefore important that on the linear substrate one dye is positioned outside of and the other fluorophore in between the FRT-sites (Figure 1). Electrophoretic analysis with subsequent fluorescent scanning verified that the system used here indeed delivers two different-colored and different-sized products upon FLP-mediated recombination (Figure 2). From the gel-image, the double-labeled educt (yellow) can clearly be distinguished from the small 54 bp linear (red) and 314 bp circular (green) recombination products.
Figure 2.
Native PAGE to visualize the recombination reaction. Approximately 150 ng (20 nM in 30 μl) of the 368 bp educt was incubated with 4 μM FLP in FLP buffer either without BSA or supplemented with 100 μg/ml BSA. After reaction samples were proteinase K digested, purified, analyzed on 8% PAGE and scanned for fluorescence of Cy5 and Rhodamine green. While the negative controls without FLP (2 lanes on the right-hand side) only showed the double-labeled educt (yellow color), in the FLP-containing reactions (2 lanes of the left-hand side) two new bands appeared corresponding to the expected recombination products: 314 bp circle (green) and 54 bp linear fragment (red). Owing to its topological state, the band of the circular green-labeled DNA runs above the 368 bp linear band.
The main topic of this work was the development of a DC-FCCS assay for detection of recombination in homogenous solution. Typical DC-FCCS measurements of intramolecular FLP-mediated recombination are end-point measurements and were performed as follows: directly after setting up the reaction, an initial DC-FCCS measurement was recorded and then incubated at room temperature to allow for FLP recombination. Then proteinase K was added and the chamber was incubated at 37°C to complete digestion. Finally, a DC-FCCS measurement was performed again, and from the difference between final and initial measurements, the remaining substrate concentration was calculated (Equation 2).
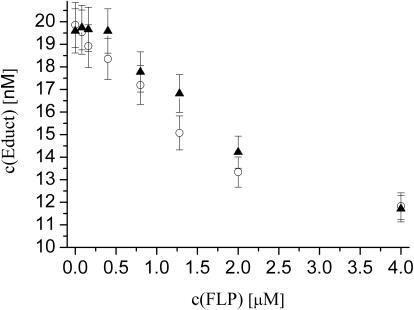
For in vitro assays, it has been reported that FLP needs to be used in stochiometric amounts or even in higher excess to its binding sites (FRT-half-sites), rather than in catalytic amounts (25,28). The maximum concentration we used here was 4 μM FLP per 20 nM substrate (containing two FRT-sites with two FRT-half-sites each), corresponding to a 50-fold ratio FLP:FRT-half-site. Using this ratio, we observed a maximal decrease in substrate concentration down to ∼60% of the initial value after recombination and digestion with proteinase K (Figure 3). It is not completely understood, why an excess of FLP is necessary, however Meyer-Leon et al. (25) excluded the possibility that partial inactive FLP is the reason, because they used a substrate-affinity purification procedure and claimed their FLP preparation to be 100% active. We also performed a series of different ratios of FLP:FRT-halfsites. In Figure 4, the remaining substrate concentration after the recombination reaction is plotted against the employed FLP concentration. We found that different buffer compositions (±BSA) have influence on the amount of FLP needed.
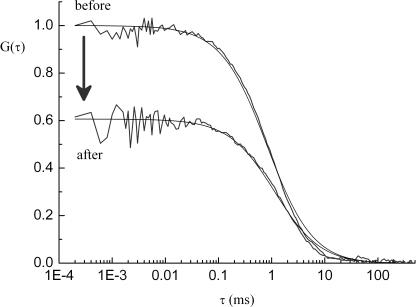
Figure 3.
Normalized measured and fitted cross-correlation curves of intramolecular recombination. The decrease in G(0) between measurements taken before and after the excision reaction directly reflects the reduced relative amount of double-labeled molecules versus single-labeled species. c(FLP) = 4 μM; c(Educt) = 20 nM.
Figure 4.
Equilibrium substrate concentrations depend on the amount of FLP used and on buffer composition. The equilibrium concentration is equal to the concentration of remaining double-labeled molecules after recombination/proteinase K digestion and was obtained from DC-FCCS measurements according to Equation 2. Standard 5% error bars are shown. The concentration of FLP is plotted on the x-axis. Filled triangles, without BSA; Open circles, with 100 μg/ml BSA.
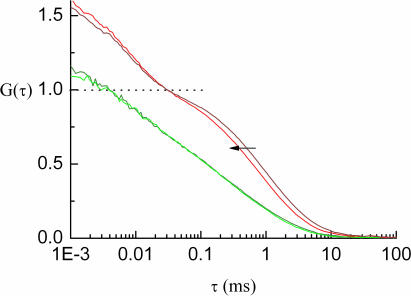
To further verify our results, we also evaluated the autocorrelation data. From Figure 5, it can be confirmed that the green-labeled part of the 368 bp substrate is excised and nearly does not change its diffusion behavior during recombination. The green-labeled recombination product is 314 bp in length, while the red-labeled product is only 54 bp long (Figure 1). Autocorrelation curves obtained before and after recombination clearly show a decrease of diffusion times (increase in mobility) of the red-labeled species, but not of the green molecules. These results, as well as negative controls without added FLP, excluded the possibility that any other DNA cleaving agent (e.g. from DNase contaminations of the proteinase K) could be responsible for the observed decrease in cross-correlation amplitudes.
Figure 5.
Only the red-labeled molecules show a reduced diffusion time after recombination. In this graph, autocorrelation curves are plotted for the red and green channel, both before and after recombination reaction. Note that for each channel, the curves are normalized to a particle number of 1, and therefore intersect at G(τ) = 1, which is indicated by a dashed line. However, the triplet-induced fraction of the decay curves varies between measurements before and after recombination.
Intermolecular recombination
Monitoring intermolecular recombination with DC-FCCS would have the advantage of simple substrate generation. Annealing of two labeled oligonucleotides might be sufficient to generate the substrate and would allow for fast and flexible alteration of the FRT.
However, there are several considerations when testing intermolecular recombination with DC-FCCS. First, compared to intramolecular recombination, the expectable signal range of an intermolecular reaction is much lower, because of several possible intermolecular reaction pathways. Two differently labeled reaction partners may not only react with each other, but also react with themselves, not contributing to the signal. Second, at low concentrations, intermolecular reactions are generally disfavored. For FLP we found that, at low concentrations, intermolecular recombination is strongly dependent on the substrate length. In the nanomolar range, no recombination was detected with linear substrates up to a few hundred bp length, while 2.7 kb long substrates performed well at the same concentrations (data not shown). We cannot give a logical explanation for this observation, but speculate that these findings have to do with the fact that an excess of FLP is needed for recombination (25,28). If up to 50 FLP monomers per FRT-half-site (binding site) are needed for efficient recombination, the protein presumably occupies also free DNA stretches other than its 13 bp recognition site within the FRT. It is not totally clear and has not yet been reported in literature, why and how this FLP characteristic contributes to formation of the holliday intermediate and finally recombination. However, it might lead to failure of recombination, if the substrate molecules are too short to accommodate all FLP monomers.
Sensitivity and detection limit
The DC-FCCS FLP recombination assay in this work was developed using 20 nM substrate in 20 μl total volume per sample. This reaction condition is typically used for in vitro recombination assays and was chosen here to assure proper function of the new assay. However, considerably less sample can be processed by DC-FCCS in two ways: reducing the volume or reducing the concentration. Regarding the volume, it should be noted that the ConfoCor2 instrument has a focal volume element of ∼0.15–0.2 fl. Besides technical limitations in liquid handling and evaporation, volumes down to nanoliters are applicable. Reduction of concentration is possible ∼10-fold when using the ConfoCor2 instrument with its small focal volume. Using different instruments with larger focal volume elements, further reduction in concentration can be achieved. However, it remains always to decide, whether extremely low concentrations still make sense in enzymatic reactions.
For accurate characterization and comparison of newly isolated FLP mutants, most often the detection limit is more important than sensitivity. The question is: how little recombination activity can be reliably discriminated? The answer is given by the quality of the measured data, and in turn the quality and reliability of its mathematical fit. As all parameters finally are obtained from the fitting of the curves, it is obvious that better smoother data curves yield data within a smaller error range. As DC-FCCS is dealing with correlation of fluorescent time traces, it becomes clear that longer measurement times are required for more accurate discrimination of small effects in recombination activity, whereas shorter measurement times can be used for rapid qualitative yes-or-no decisions, e.g. for screening purposes. Here, we averaged curves from 20× 10 s measurements and were able to distinguish 19 nM remaining substrate from 20 nM initial substrate (Figure 4), which corresponds to 5% difference.
Outlook: possible application of the assay in enzyme-engineering
The site-specific DNA recombination assay as presented in this paper is ideally suited for rapid characterization of recombinase functions and specificities. The use of DC-FCCS for this purpose has several advantages over other conventional methods. The main benefit of the technique used here arises from its ability to drastically minimize measurement times. This is achieved in two ways: first, working in homogenous solution, time-consuming handling and separation steps are completely eliminated. Second, the measurement times per sample are low and even have the potential to be further reduced down to the sub-second range by employing alternative evaluating algorithms other than correlation analysis, e.g. coincidence analysis (31).
Other than that, DC-FCCS exhibits single-molecule sensitivity and therefore helps reducing sample volume (low- to sub-microliter range) and concentration (nanomolar range). This in turn saves valuable compounds during screening. As for all fluorescent techniques, labeling is necessary and is here achieved by generating a PCR product with custom-labeled DNA primers.
The biochemical part of the new assay has been designed to be as flexible as possible concerning the DNA site to be investigated. Virtually, any site can easily be incorporated via the PCR primers. This strategy allows for quick change of the recognition site and enables fast negative and positive specificity-screening of recombinase variants.
CONCLUSIONS
We have demonstrated here a novel approach based on DC-FCCS to monitor site-specific recombination of DNA. From both, inter- and intramolecular recombination processes, the intramolecular recombination is clearly favored for measurements in DC-FCCS. We could clearly demonstrate a decrease of substrate amount after recombination using substrate concentrations as low as 20 nM and an excess of FLP. Although the separation-free detection technique used here would ideally be suited for recording of real-time recombination kinetics (32), this is unfortunately impossible with the FLP/FRT system. Due to the fact that FLP binds very tight to its target, protease K digestion is necessary in our assay (as well as in other traditional assays), permitting only end-point measurements.
This concept was verified using the FLP/FRT system, but due to easily interchangeable recognition-sites, we suggest that adaptation to other site-specific DNA recombinase systems, e.g. the Cre/loxP system can be achieved.
Taken together, the versatile and quick DNA recombination assay developed in this work builds the base not only for biochemical characterizations of different site-specific recombinases and chimeras thereof, but also for screening purposes, e.g. in directed molecular enzyme evolution.
Acknowledgments
The experiments were carried out in the BioFuture group Experimental Biophysics at the Max-Planck-Institute for Biophysical Chemistry, Göttingen, Germany. The authors would like to thank Dr Guido Böse (MPI Göttingen), Dr Andre Koltermann and Dr Ulrich Kettling (both DIREVO Biotech, Cologne, Germany) for helpful discussions. The excellent technical assistance of Karin Birkenfeld, Inge Grüneberg and Sylvia Löbermann is gratefully acknowledged. We thank Carl Zeiss (Jena, Germany) for collaborations on the ConfoCor2 instrument. Financial support was provided by the German Ministry of Education and Research (BMBF). Funding to pay the OpenAccess publication charges for this article was provided by the Saxony Ministry of Science and Arts, Germany.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chen Y., Rice P.A. New insight into site-specific recombination from Flp recombinase-DNA structures. Annu. Rev. Biophys. Biomol. Str. 2003;32:135–159. doi: 10.1146/annurev.biophys.32.110601.141732. [DOI] [PubMed] [Google Scholar]
- 2.Shaikh A.C., Sadowski P.D. Chimeras of the flp and cre recombinases: tests of the mode of cleavage by flp and cre. J. Mol. Biol. 2000;302:27–48. doi: 10.1006/jmbi.2000.3967. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y., Narendra U., Iype L.E., Cox M.M., Rice P.A. Crystal structure of a flp recombinase-holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell. 2000;6:885–897. [PubMed] [Google Scholar]
- 4.Sadowski P.D. The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 1995;51:53–91. [PubMed] [Google Scholar]
- 5.Lewandoski M. Conditional control of gene expression in the mouse. Nature Rev. Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 6.Luo H., Kausch A.P. Application of FLP/FRT site-specific DNA recombination system in plants. Genet. Eng. (New York) 2002;24:1–16. doi: 10.1007/978-1-4615-0721-5_1. [DOI] [PubMed] [Google Scholar]
- 7.O'Gorman S., Fox D.T., Wahl G.M. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 8.Koch K.S., Aoki T., Wang Y., Atkinson A.E., Gleiberman A.S., Glebov O.K., Leffert H.L. Site-specific integration of targeted dna into animal cell genomes. Gene. 2000;249:135–144. doi: 10.1016/s0378-1119(00)00153-0. [DOI] [PubMed] [Google Scholar]
- 9.Buchholz F., Angrand P.O., Stewart A.F. Improved properties of flp recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 10.Santoro S.W., Schultz P.G. Directed evolution of the site specificity of Cre recombinase. Proc. Natl Acad. Sci. USA. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voziyanov Y., Konieczka J.H., Stewart A.F., Jayaram M. Stepwise manipulation of DNA specificity in Flp recombinase: progressively adapting Flp to individual and combinatorial mutations in its target site. J. Mol. Biol. 2003;326:65–76. doi: 10.1016/s0022-2836(02)01364-5. [DOI] [PubMed] [Google Scholar]
- 12.Rufer A.W., Sauer B. Non-contact positions impose site selectivity on Cre recombinase. Nucleic Acids Res. 2002;30:2764–2771. doi: 10.1093/nar/gkf399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchholz F., Stewart A.F. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 14.Snaith M.R., Kilby N.J., Murray J.A.H. An Escherichia coli system for assay of flp site-specific recombination on substrate plasmids. Gene. 1996;180:225–227. doi: 10.1016/s0378-1119(96)00449-0. [DOI] [PubMed] [Google Scholar]
- 15.Buchholz F., Angrand P.O., Stewart A.F. A simple assay to determine the functionality of cre or flp recombination targets in genomic manipulation constructs. Nucleic Acids Res. 1996;24:3118–3119. doi: 10.1093/nar/24.15.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panigrahi G., Zhao B.P., Krepinsky J.J., Sadowski P.D. Toward a mechanism-based fluorescent assay for site-specific recombinases and topoisomerases—assay design and syntheses of fluorescent substrates. J. Am. Chem. Soc. 1996;118:12004–12011. [Google Scholar]
- 17.Seibler J., Bode J. Double-reciprocal crossover mediated by flp-recombinase—a concept and an assay. Biochemistry. 1997;36:1740–1747. doi: 10.1021/bi962443e. [DOI] [PubMed] [Google Scholar]
- 18.Schwille P., Meyeralmes F.J., Rigler R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 1997;72:1878–1886. doi: 10.1016/S0006-3495(97)78833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwille P. Cross-correlation analysis in FCS. In: Rigler R., Elson E., editors. Fluorescence Correlation Spectroscopy—Theory and Applications. Heidelberg: Springer-Verlag; 2000. pp. 360–378. [Google Scholar]
- 20.Medina M.A., Schwille P. Fluorescence correlation spectroscopy for the detection and study of single molecules in Biology. Bioessays. 2002;24:758–764. doi: 10.1002/bies.10118. [DOI] [PubMed] [Google Scholar]
- 21.Haustein E., Schwille P. Ultrasensitive investigations of biological systems by fluorescence correlation spectroscopy. Methods. 2003;29:153–166. doi: 10.1016/s1046-2023(02)00306-7. [DOI] [PubMed] [Google Scholar]
- 22.Jahnz M. 2003. Evolutive Biotechnologie und Fluoreszenz-Korrelations-Spektroskopie: Systeme zur Änderung enzymatischer Substratspezifität und Auffindung neuer Wirkstoffe. Thesis/Dissertation, University of Hannover, Germany. [Google Scholar]
- 23.Stemmer W.P.C., Crameri A., Ha K.D., Brennan T.M., Heyneker H.L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 24.Babineau D., Vetter D., Andrews B.J., Gronostajski R.M., Proteau G.A., Beatty L.G., Sadowski P.D. The FLP protein of the 2-micron plasmid of yeast. Purification of the protein from Escherichia coli cells expressing the cloned FLP gene. J. Biol. Chem. 1985;260:12313–12319. [PubMed] [Google Scholar]
- 25.Meyer-Leon L., Gates C.A., Attwood J.M., Wood E.A., Cox M.M. Purification of the FLP site-specific recombinase by affinity chromatography and re-examination of basic properties of the system. Nucleic Acids Res. 1987;15:6469–6488. doi: 10.1093/nar/15.16.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchholz F., Ringrose L., Angrand P.O., Rossi F., Stewart A.F. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 1996;24:4256–4262. doi: 10.1093/nar/24.21.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gronostajski R.M., Sadowski P.D. The FLP protein of the 2-micron plasmid of yeast. Inter- and intramolecular reactions. J. Biol. Chem. 1985;260:12328–12335. [PubMed] [Google Scholar]
- 28.Ringrose L., Lounnas V., Ehrlich L., Buchholz F., Wade R., Stewart A.F. Comparative kinetic analysis of FLP and CRE recombinases—mathematical models for DNA binding and recombination. J. Mol. Biol. 1998;284:363–384. doi: 10.1006/jmbi.1998.2149. [DOI] [PubMed] [Google Scholar]
- 29.Waite L.L., Cox M.M. A protein dissociation step limits turnover in FLP recombinase-mediated site-specific recombination. J. Biol. Chem. 1995;270:23409–23414. doi: 10.1074/jbc.270.40.23409. [DOI] [PubMed] [Google Scholar]
- 30.Gates C.A., Cox M.M. FLP recombinase is an enzyme. Proc. Natl Acad. Sci. USA. 1988;85:4628–4632. doi: 10.1073/pnas.85.13.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winkler T., Kettling U., Koltermann A., Eigen M. Confocal fluorescence coincidence analysis: an approach to ultra high-throughput screening. Proc. Natl Acad. Sci. USA. 1999;96:1375–1378. doi: 10.1073/pnas.96.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kettling U., Koltermann A., Schwille P., Eigen M. Real-time enzyme kinetics monitored by dual-color fluorescence cross-correlation spectroscopy. Proc. Natl Acad. Sci. USA. 1998;95:1416–1420. doi: 10.1073/pnas.95.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]