Summary
Mycobacterium tuberculosis (Mtb) causes 1.6 million deaths annually. Active tuberculosis correlates with a neutrophil-driven type I interferon (IFN) signature, but the cellular mechanisms underlying tuberculosis pathogenesis remain poorly understood. We found interstitial macrophages (IMs) and plasmacytoid dendritic cells (pDCs) are dominant producers of type I IFN during Mtb infection in mice and non-human primates, and pDCs localize near human Mtb granulomas. Depletion of pDCs reduces Mtb burdens, implicating pDCs in tuberculosis pathogenesis. During IFN-driven disease, we observe abundant DNA-containing neutrophil extracellular traps (NETs) described to activate pDCs. Cell type-specific disruption of the type I IFN receptor suggests IFNs act on IMs to inhibit Mtb control. Single cell RNA-seq indicates type I IFN-responsive cells are defective in their response to IFNγ, a cytokine critical for Mtb control. We propose pDC-derived type I IFNs act on IMs to permit bacterial replication, driving further neutrophil recruitment, and active tuberculosis disease.
In Brief
Induction of an anti-viral type I interferon response strongly correlates with progression to active tuberculosis disease in humans. Using a tuberculosis-susceptible mouse model, two cell types were identified as the major producers of type I interferon and one as a critical sensor, thereby revealing mechanisms underlying type I interferon-driven susceptibility to tuberculosis.
Graphical Abstract
Introduction
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis disease, caused 1.6 million deaths in 20211. Treatment requires a 4–6 month course of antibiotics, or up to 2 years for increasingly prevalent multi-drug resistant strains. Moreover, the only approved vaccine for Mtb has variable or no efficacy in adults2. The pathophysiology of tuberculosis remains poorly understood. The mouse model has been used to discover most of the host factors known to control tuberculosis in humans, including tumor necrosis (TNF) factor and interferon-γ3–5. Nevertheless, mouse models have been criticized for poorly recapitulating key aspects of human disease6.
In humans, active tuberculosis disease is reproducibly associated with the induction of type I interferons (IFNs)7–9, a family of cytokines that includes IFNβ and multiple IFNα isoforms. Type I IFNs signal via the type I IFN receptor (IFNAR) to elicit an anti-viral response that overlaps but is insufficient to recapitulate the protective anti-Mtb response elicited by IFNγ. Evidence that type I IFNs exacerbate tuberculosis in humans comes from observations that viral infections are associated with worse Mtb infection outcomes. For example, influenza infection correlates with an increased risk of death among pulmonary tuberculosis patients, and infants with cytomegalovirus have an increased risk of tuberculosis disease10–12. A causal role for type I IFNs in driving human tuberculosis is also supported by the finding that a partial loss-of-function mutation in IFNAR is associated with Mtb resistance in humans13. How type I IFNs drive Mtb progression in humans is poorly understood, but one mechanism may involve antagonism of the critical, protective IFNγ response during infection14,15.
Mice are able to model the virus-induced susceptibility to Mtb seen in humans and have established causality between type I IFN and loss of Mtb control. Chronic lymphocytic choriomeningitis virus (LCMV), acute LCMV, pneumonia virus of mice, and influenza A virus all exacerbate tuberculosis infection16–20. However, co-infection studies make it challenging to determine the cellular mechanism behind Mtb susceptibility, as perturbations such as type I IFN receptor blockade simultaneously impact viral and bacterial control. Therefore, an ideal platform to study type I IFN-driven Mtb susceptibility would be a mouse model where Mtb infection is itself sufficient to elicit the exacerbated type I IFN response observed in humans with active TB. However, C57BL/6 (B6) mice, the most used model for Mtb infection, generate a weak type I IFN response in response to Mtb infection; indeed, Ifnar1 deletion from unmanipulated B6 mice does not consistently impact Mtb lung burden21–24. Investigators have circumvented this issue by intranasal injection of B6 mice with polyI:C, an IFN-inducing viral mimic8,25,26. Such studies demonstrate a convincing causal link between type I IFNs and Mtb susceptibility in mice, and have shown that a major detrimental effect of type I IFNs is to impair interleukin-1-dependent control of Mtb26,27. Despite these advances, it has been challenging to decipher the cellular mechanisms underlying the detrimental effects of type I IFN on bacterial control, and in particular, the cells required to produce or respond to type I IFNs to mediate Mtb-susceptibility remain unknown.
Unlike B6 mice, C3H and 129 mice exhibit a type I interferon-driven susceptibility to Mtb28–30, but there are limited genetic tools in these mouse strains, making mechanistic studies difficult. We recently discovered that congenic B6 mice with the ‘super susceptibility to tuberculosis 1’ region from C3H mice (i.e., B6.Sst1S mice)29,30 are more susceptible to Mtb infection (compared to isogenic B6 mice) due to their strong type I IFN response. Ifnar1 deletion fully rescues the enhanced susceptibility of B6.Sst1S mice at early timepoints and increases survival27. We further identified Sp140 as the gene within the Sst1 genetic interval that controls Mtb susceptibility, and confirmed that the early Mtb susceptibility of Sp140−/− mice is also rescued by Ifnar1 deletion31. Our Sp140−/− mice were generated on a pure C57BL/6J background, enabling the use of existing genetic tools to dissect tuberculosis pathogenesis.
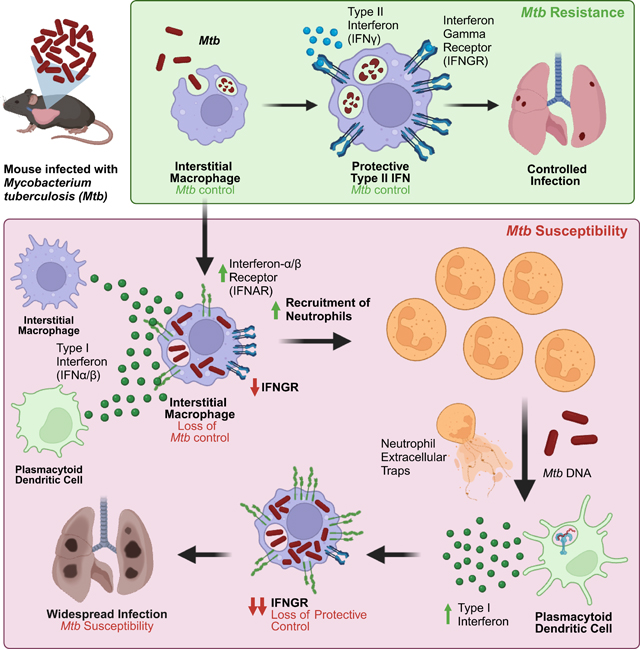
In this study, we leveraged Sp140−/− mice to determine the cellular mechanisms of type I IFN-driven Mtb susceptibility. Single cell RNA-sequencing (scRNA-seq) identified interstitial macrophages (IMs) as a major type I IFN producer. A sensitive genetic reporter of type I IFN production corroborated the scRNA-seq findings, and also revealed that plasmacytoid dendritic cells (pDCs) are an additional source of type I IFN during Mtb infection. Type I IFN production by pDCs drives disease since pDC depletion rescued the enhanced susceptibility of Sp140−/− mice. Loss of bacterial control in Sp140−/− mice leads to neutrophil influx and abundant production of DNA-rich neutrophil extracellular traps (NETs), ligands described to promote type I IFN production by pDCs32,33. We developed transcriptional signatures that distinguish the response elicited by type I IFN from that elicited by IFNγ. Application of these signatures to our scRNA-seq data indicated that Mtb-infected type I IFN-responsive IMs in the lungs have impaired IFNγ responses15,34,35. Cell-type specific deletion of Ifnar1 validated that type I IFN confers susceptibility by acting on IMs. Our findings suggest a model of tuberculosis pathogenesis in which type I IFNs drive an initial loss of bacterial control, possibly by impairing IFNγ responses, that in turn initiates a positive feedback loop of NET production and type I IFN expression by pDCs, leading to uncontrolled bacterial replication and active tuberculosis disease.
Results
Myeloid cells harbor Mtb in Sp140−/− mice
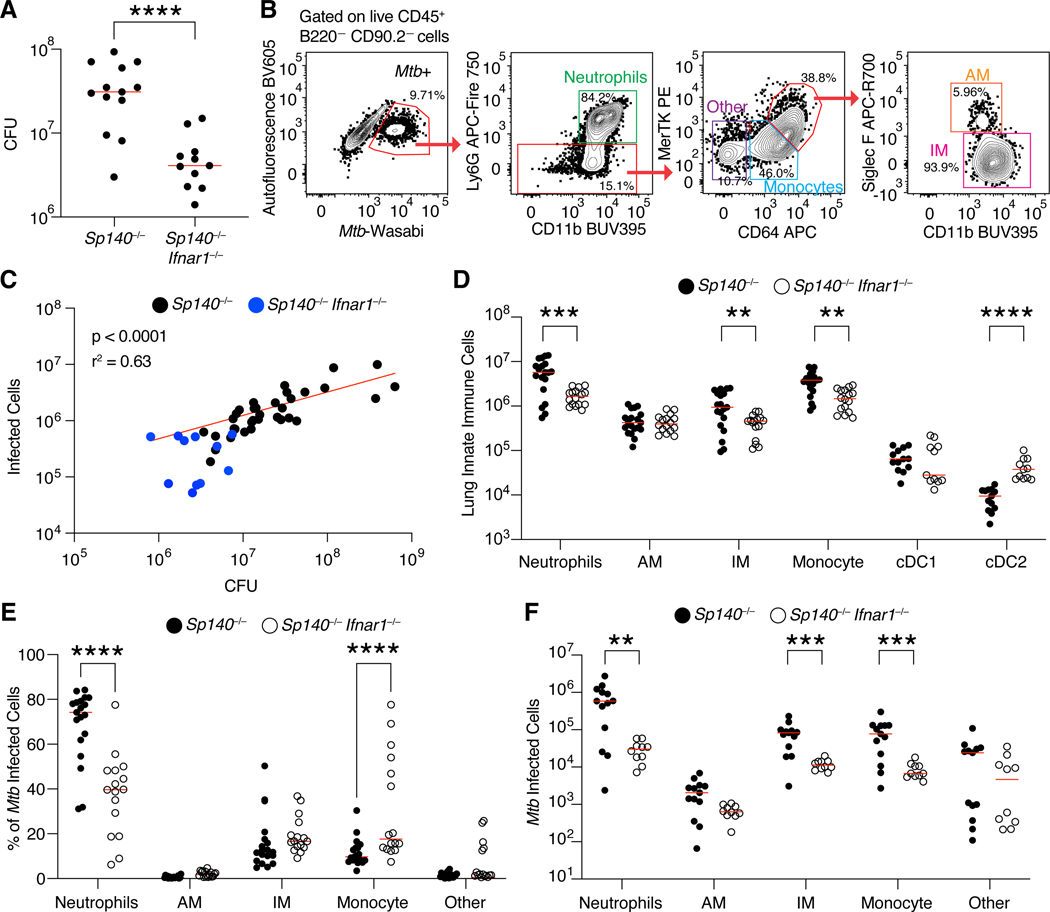
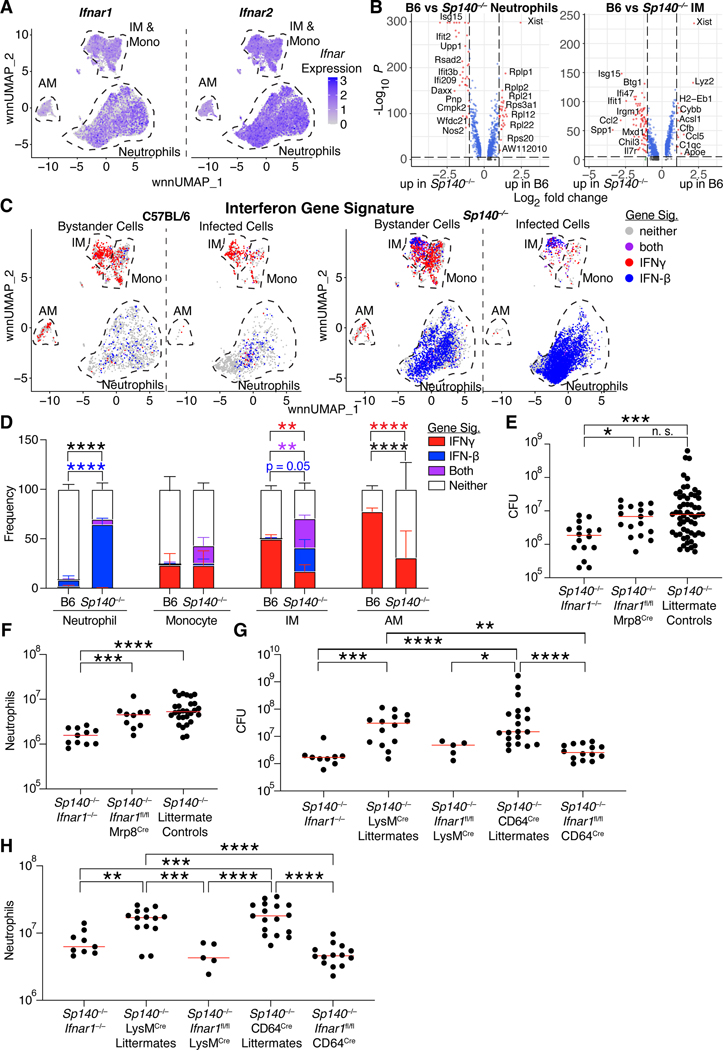
As we previously demonstrated, the susceptibility of Sp140−/− mice to Mtb is driven by type I IFN, and mirrors the correlation between type I IFN and active tuberculosis disease seen in humans. Genetic or antibody-mediated depletion of IFNAR fully rescues the enhanced susceptibility of Sp140−/− animals (Fig. 1A)31. To better characterize the immune response of Sp140−/− mice to Mtb, we infected mice with Mtb expressing the fluorescent protein Wasabi (Mtb-Wasabi)36. With this approach, we identified Mtb-harboring cells in mouse lungs 25 days post-infection (Fig. 1B). Flow cytometry reliably reported overall lung bacterial burdens as the number of Mtb-infected cells detected by flow cytometry correlated with lung Mtb CFU (R2 = 0.63; Fig. 1C). Infected lungs from Sp140−/− animals contained significantly more neutrophils, IMs, and monocytes as compared to Sp140−/− Ifnar1−/− mice, with no difference in the number of AMs and a reduction in the number of cDC2 (Fig. 1D, Supplementary Fig. 1). Over 90% of infected cells were myeloid cells (Fig. 1B, 1E), in line with previous reports37,38. Consistent with their higher abundance in infected Sp140−/− lungs, neutrophils comprised a considerably larger percentage and absolute number of the Mtb-infected cells in Sp140−/− mice compared to Sp140–/–Ifnar1−/− animals (Fig. 1E, 1F). There were also more infected IMs and monocytes in Sp140−/− mice compared to Sp140−/− Ifnar1−/− animals, in line with the overall increase in these immune populations in the lungs of infected Sp140−/− mice (Fig. 1D, 1F). Importantly, we did not observe substantial alterations in the immune compartment of uninfected Sp140−/− mice, implying grossly normal hematopoietic development in these mice (Supplemental Fig. 2A-C) in contrast to prior suggestions39–41. However, the exact mechanisms causing the infection-induced differences in myeloid cells from Sp140−/− mice was unclear, and thus required more in-depth profiling of the myeloid compartment.
Figure 1. Myeloid cells are the dominant Mtb harboring cells in Sp140−/− and Sp140−/− Ifnar1−/− mice.
(A) Colony forming units (CFU) of Mtb in the lungs of Sp140−/− (n = 13) and Sp140−/− Ifnar1−/− (n = 11) mice. (B) Representative flow cytometry plots of an Mtb-infected Sp140−/− mouse lung gated on live CD45+B220−CD90.2− cells to identify Mtb-infected cells, subset into neutrophils (green; Ly6G+ CD11b+), other cells (purple; Ly6G−CD64−MerTK−), monocytes (blue; Ly6G−CD64+MerTKlow), alveolar macrophages (AMs; orange; Ly6G− CD64+MerTKhighSiglec F+), and interstitial macrophages (IMs; pink; Ly6G−CD64+MerTKhighSiglec F−). (C) Correlation between infected cell numbers identified by flow cytometry of total lung digests to CFU from the same infected lung (black; Sp140−/− and blue; Sp140−/−Ifnar1−/− combined; n = 45). Red line indicates a nonlinear regression. (D) Number of innate immune cells by cell type in Mtb-infected Sp140−/− (n = 19; closed circles) and Sp140−/−Ifnar1−/− lungs (n = 16; open circles). (E) Frequency and (F) number of immune cell populations of Mtb-infected cells in Sp140−/− (n = 12–19; closed circles) and Sp140−/− Ifnar1−/− mice (n = 10–15; open circles). Lungs were analyzed 24–26 days after Mtb infection. The bars in (A), (D), (E), and (F) represent the median. Pooled data from two or three independent experiments are shown. A linear regression performed on log transformed data was used to calculate significance and R2 for (C). An unpaired t test was used to determine significance for (A), a two-way ANOVA with Sidak’s multiple comparisons test was used to calculate significance for (D), (E), and (F). **p < 0.01, ***p < 0.001, ****p < 0.0001.
Macrophages and neutrophils exhibit a variety of activation states during Mtb infection
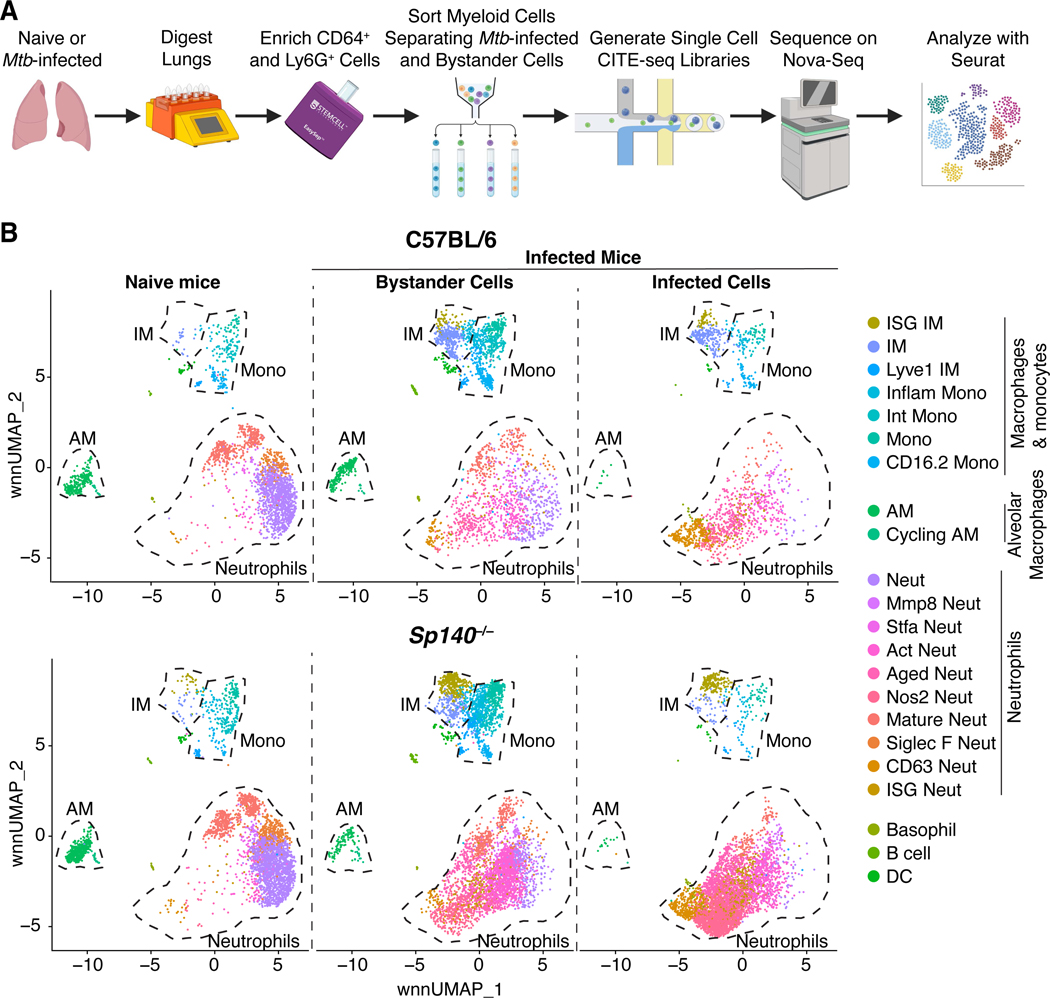
To further characterize the Mtb-infected myeloid cells in B6 and Sp140−/− mice, we performed scRNA-seq on myeloid cells from Mtb-infected or uninfected lungs 25 days after infection. For this experiment, CD64+ and Ly6G+ cells were magnetically enriched, sort purified, and processed for library generation with the 10X Genomics platform (Fig. 2A). Infected and uninfected (bystander) cells from Mtb-infected mice were sorted and barcoded separately. mRNA transcripts and protein expression for select lineage markers were simultaneously measured by CITE-seq, allowing for Weighted Nearest Neighbor (WNN) analysis to cluster cells on mRNA and protein expression and WNN uniform manifold approximation and projection (wnnUMAP) reductions for data visualization (Supplementary Fig. 1)42–46. The resulting dataset consists of 6,604 B6 and 13,668 Sp140−/− cells, almost exclusively consisting of myeloid cells (Fig. 2B). Each cluster is represented in the two datasets, however the proportions of some clusters are altered between genotypes. Most notably, the ratio of IFN stimulated gene (ISG)+ IM to ISG– IM was higher in the Sp140−/− mice, as expected from the exacerbated type I IFN response in this strain (Fig. 2B). The largest changes in composition were seen when comparing cells from naïve lungs to bystander and Mtb-infected cells from Mtb-infected lungs (Fig. 2B). For example, AMs are relatively abundant in naïve lungs but are rare among the Mtb-infected cells 25 days post-infection, as also seen by flow cytometry (Fig. 1F, 2B). B6 and Sp140−/− myeloid cells from uninfected mice were highly transcriptionally similar (Supplemental Fig. 2D-E), confirming that the type I IFN-driven changes in B6 and Sp140−/− mice occur after Mtb infection.
Figure 2. Single cell RNA-sequencing analysis of B6 and Sp140−/− myeloid cells from Mtb-infected and naïve lungs.
CITE-seq was used to integrate transcriptomic and protein expression of single cells, as detailed in Supplementary Fig. 1. (A) Model of the processing steps involved in generating the scRNA-seq dataset. (B) Unbiased clustering of myeloid cells in B6 and Sp140−/− Mtb-infected and naïve lungs (n = 10 lungs; n= 20,272 cells) distinguishing cells from naïve mice, the bystander cells from Mtb-infected mice, and the Mtb-infected cells from Mtb-infected mice. Lungs were analyzed 25 days after Mtb-Wasabi infection.
Bystander pDCs and IMs are the primary sources of type I IFN during Mtb infection
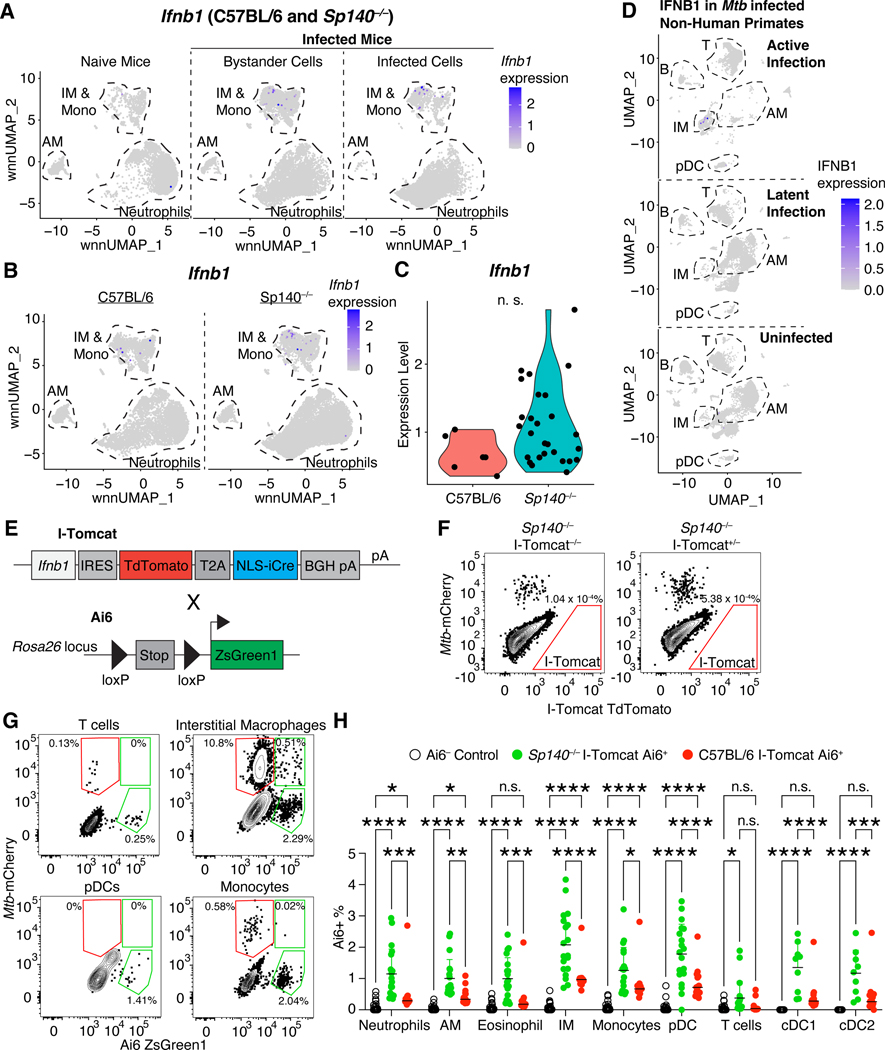
To determine the cellular mechanism of type I IFN-driven Mtb susceptibility, we first sought to identify the type I IFN-producing cells. In general, our scRNA-seq analysis revealed that very few cells were Ifnb1 positive, which may reflect a lack of sensitivity of scRNA-seq, and/or the transient and stochastic expression pattern of this gene (Fig. 3A)47–50. Mtb infection resulted in increased expression of Ifnb1 in infected and bystander mononuclear phagocytes, with a slight bias towards Ifnb1 production by IMs compared to monocytes, and no production by AMs (Fig. 3A). While there was no major difference in the cell types producing Ifnb1 between B6 and Sp140−/− cells, a greater number and frequency of Sp140−/− cells expressed Ifnb1 (Fig. 3B). Additionally, Ifnb1-expressing cells in Sp140−/− mice trended towards a higher per cell expression of Ifnb1 than B6 cells (Fig. 3C). A prior scRNA-seq study of Mtb-infected and naïve lungs from non-human primates largely mirrors our findings in mice51. Our analyses of these data indicate that IMs were also the dominant IFNB1-expressing cells in non-human primates with active tuberculosis, and IMs did not express IFNB1 in naïve or latently infected lungs (Fig. 3D). These results suggest that mice faithfully recapitulate the Mtb-induced type I IFN production seen in non-human primates.
Figure 3. Bystander pDCs, IMs, and monocytes are the primary IFN-β producers in mice and non-human primates.
(A) Ifnb1 expression in myeloid cells from naïve mice, bystander myeloid cells from infected mice, and Mtb-infected myeloid cells from infected mice (B6 and Sp140−/− combined). (B) Ifnb1 expression in myeloid cells (combined infected and bystander) from B6 and Sp140−/− mice. (C) Ifnb1 expression in B6 and Sp140−/− cells that express Ifnb1. (D) Analysis of GSE149758 scRNA-seq data from Esaulova E., et al. 2021 depicting IFNB1 expression in cells from non-human primates with active Mtb infection, latent Mtb infection, or that are uninfected. (E) Schematic representation of the genetic structure of I-Tomcat mice and Ai6 mice. (F) Representative flow cytometry plot of TdTomato expression in immune cells. (G) Representative flow cytometry plots of ZsGreen expression and Mtb-mCherry detection in T cells, IMs, pDCs, and monocytes. (H) Frequency of Ai6 expressing cells in lung immune cells from Ai6− control (n = 34; open circles), Sp140−/− I-Tomcat Ai6 (n = 19; green circles), and I-Tomcat Ai6 (n = 15; red circles) mice. The bars in (H) represent the median. Pooled data from four independent experiments are shown in (H). Lungs were analyzed 25 days after Mtb infection. Statistical significance in (C) was calculated by non-parametric Wilcoxon rank sum test with Bonferroni correction and in (H) by one-way ANOVA with Tukey’s multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The type I IFN producers identified in the scRNA-seq datasets were validated using an Ifnb1 genetic reporter called I-Tomcat mice, which express TdTomato and Cre downstream of Ifnb1 (Fig. 3E, Supplementary Fig. 3). While TdTomato expression was sufficient to identify Ifnb1 expression by bone marrow-derived macrophages following in vitro stimulation with poly I:C (Supplementary Fig. 3), TdTomato+ cells were not detected 25 days after Mtb infection (Fig. 3F). TdTomato detection was not improved in I-Tomcat homozygous mice, examining an earlier timepoint of 19 days post-infection, or by gating on specific immune populations such as IMs (Supplementary Fig. 3). Even though type I IFN drives Mtb susceptibility in Sp140−/− mice, it is unclear when type I IFN production occurs (Fig. 1A). Type I IFN production may be an early and/or transient event, which would be missed by analyzing a single timepoint. To address this issue, we crossed I-Tomcat mice with the Ai6 Cre reporter mice (I-Tomcat Ai6) on B6 and Sp140−/− backgrounds (Fig. 3E)52. In these mice, any cell that has ever expressed Ifnb1 will constitutively express ZsGreen. Mtb-infected I-Tomcat Ai6 mice contained reporter-positive myeloid cells and low background was detected among cell populations that are not expected to express Ifnb1 (e.g., ~0.1% of T cells were Ai6+) (Fig. 3G). Consistent with the scRNA-seq analysis, IMs and monocytes were the primary Ifnb1-expressing cells in B6 and Sp140−/− mice (Fig. 3H, Supplementary Fig. 3). Interestingly, Sp140−/− mice exhibited elevated Ai6+ expression frequency in all cell types, suggesting SP140 broadly modulates the sensitivity for inducing Ifnb1 expression (Fig. 3H). In addition to corroborating the scRNA-seq data, the I-Tomcat mice also identified pDCs as a major type I IFN-producing cell population. Lung pDCs are very rare CD64–/Ly6G– cells and were therefore not represented in our scRNA-seq dataset. However, despite their scarcity, pDCs are extremely robust producers of type I IFNs on a per cell basis53.
While we expected IMs to be a major type I IFN-producing population given the scRNA-seq results, we were surprised that the majority of the Ai6+ IMs were Mtb– and most Mtb+ IMs were Ai6– (Fig. 3G). These results suggest that direct infection of IMs is neither required nor sufficient for IFN-β production. To examine this phenomenon in greater detail, we performed confocal microscopy and histo-cytometry analysis of Mtb-infected I-Tomcat Ai6 and Sp140−/− I-Tomcat Ai6 lungs54,55. While lesions of diseased tissue were clearly identifiable in I-Tomcat Ai6 mice, the size and myeloid cell influx into the diseased tissue were greatly exacerbated in Sp140−/− I-Tomcat Ai6 (Fig. 4A). Additionally, Ai6 expressing cells were identifiable throughout the lungs, with an increased propensity to localize in diseased rather than healthy tissue (Fig. 4A–4C). Within diseased tissue, Ai6 expressing cells were primarily located near Mtb harboring cells in I-Tomcat Ai6 and Sp140−/− I-Tomcat Ai6 lungs (Fig. 4B). Similar to the flow cytometry results, SIRPɑ+ cells, which are primarily macrophages in Mtb-infected lungs as they are ~100 fold more abundant than SIRPɑ expressing cDC2s, were a major Ai6 expressing cell population (Fig. 1D, 4B, 4D). The SIRPɑ+ macrophages expressed Ai6 at a higher frequency than CD4+ T cells in the diseased tissue but not healthy tissue (Fig. 3H, 4B, 4D). Direct infection by Mtb did not appear to be a major driver of IFN-β expression, as ~2–3% of infected macrophages were Ai6+ and ~12–15% of Ai6+ cells were Mtb-infected, in line with the frequencies seen in IMs by flow cytometry (Fig. 3H, 4E). These results suggest that IM localization to Mtb rich regions provides the activating signals required for IFN-β expression, while direct infection of IMs is not required for IFN-β expression.
Figure 4. Cells producing IFN-β are enriched in diseased tissue, but only a minority harbor Mtb.
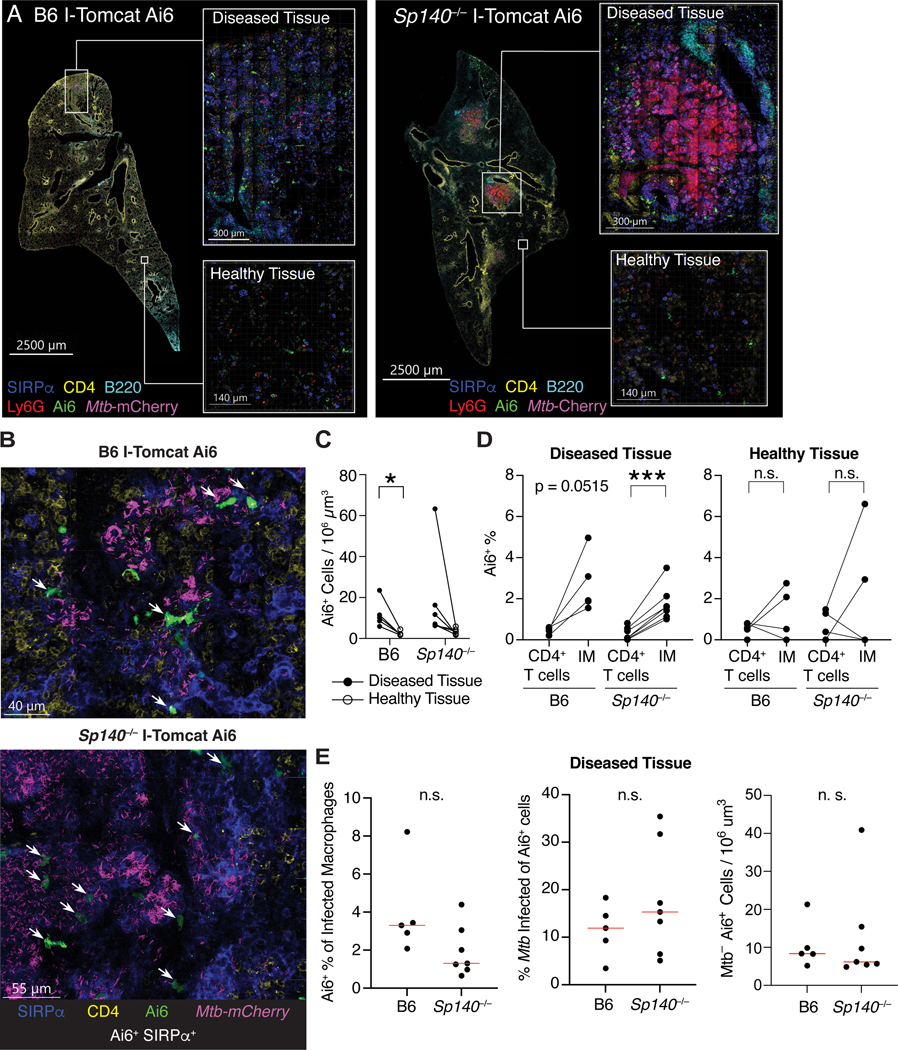
(A) Representative images of Mtb-infected I-Tomcat Ai6 and Sp140−/− I-Tomcat Ai6 lung sections stained for SIRPɑ (dark blue), CD4 (yellow), B220 (teal), Ly6G (red), Ai6 (green), and Mtb-expressed mCherry (magenta). Inset images depict higher magnification of diseased and healthy tissue for both genotypes. (B) Representative images of Ai6+ cell localization near Mtb in the diseased portions of I-Tomcat Ai6 and Sp140−/− I-Tomcat Ai6 lungs. Sections were stained with SIRPɑ (dark blue), CD4 (yellow), Ai6 (green), and Mtb-expressed mCherry (magenta). White arrows indicate cells co-expressing Ai6 and SIRPɑ. (C) Number of Ai6+ cells per 106 um3 in diseased (closed circle) and healthy tissue (open circle) from B6 (n = 5) and Sp140−/− (n = 7) Mtb-infected mouse lungs. (D) Image quantification of the frequency of Ai6 expression in CD4+ T cells and SIRPɑ+ IMs in the diseased and healthy tissue of B6 I-Tomcat Ai6 (n = 5) and Sp140−/− I-Tomcat Ai6 (n = 7) lungs. (E) Image quantification of the frequency of Ai6 expression among Mtb-infected macrophages, frequency of Mtb infection among Ai6+ cells, and number of uninfected Ai6+ cells for B6 I-Tomcat Ai6 (n = 5) and Sp140−/− I-Tomcat Ai6 (n = 7) lungs. All samples were analyzed 25 days after Mtb infection. Pooled data from two independent experiments are shown in (C), (D), and (E). Statistical significance in (C), (D) and (E) was calculated with a paired or unpaired t test. *p < 0.05, ***p < 0.001.
pDCs contribute significantly to the susceptibility of Sp140−/− animals to Mtb
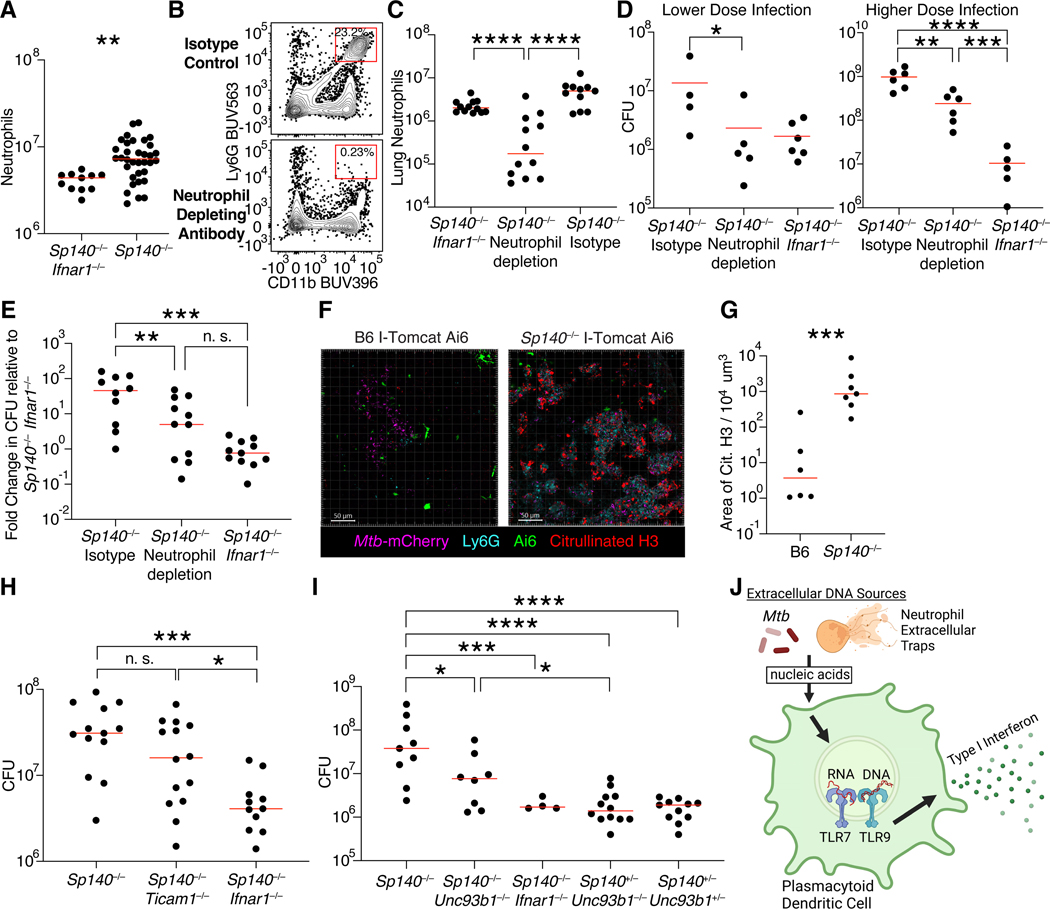
While pDCs have a well-established role in anti-viral immunity in the lung, limited work has assessed their contribution during Mtb infection56,57. pDCs may have been previously overlooked because of their scarcity in the lung. Indeed, we observe only ~20,000 pDCs in the lungs of naïve B6 and Sp140−/− mice, but this number increases 10-fold following Mtb infection and is modestly but significantly higher in Mtb-infected Sp140−/− than B6 mice (Fig. 5A). Despite their scarcity, pDCs can have major effects due to the extremely high levels of interferons produced per cell53. Consistent with a role for pDCs during Mtb infection in mice, Khader and colleagues described the presence of pDCs in lungs of non-human primates with active pulmonary TB51. However, the lack of genetic tools in non-human primates precluded functional studies of pDCs during TB. Therefore, we assessed whether pDCs affect Mtb control using our experimentally tractable mouse model and an anti-BST2 antibody58–60. This strategy efficiently depleted pDCs and resulted in a partial rescue of Mtb control in Sp140−/− mice (Fig. 5B, 5C). However, BST2 is known to be upregulated by cells other than pDCs in inflammatory environments; thus, antibody depletion could have been protective against Mtb by depleting non-pDC cells59. We therefore also tested the contribution of pDCs by using a genetic pDC depletion strategy by crossing Sp140−/− mice with mice expressing the diphtheria toxin receptor (DTR) downstream of the human BDCA2 promoter (pDC-DTR)61. We used Sp140+/− pDC-DTR littermates as wild-type controls since a single copy of Sp140 is sufficient to restore Mtb control. DT administration efficiently ablated pDCs in Sp140−/− and Sp140+/− mice, with the depletion specifically affecting pDCs (Fig. 5D, Supplementary Fig. 4). pDC-DTR depletion of pDCs fully rescued bacterial control in Sp140−/− mice, while depletion in Sp140-sufficient animals did not affect lung bacterial burden, as expected (Fig. 5E). Additionally, pDC-DTR depletion of pDCs in Sp140−/− mice reduced expression of type I IFN stimulated genes to the level of Sp140-sufficient animals, while rescuing expression of the type II IFN stimulated gene H2-Ab1 (Fig. 5F, 5G). These results demonstrate a substantial contribution of pDCs in limiting Mtb control in animals with a hyper type I IFN response.
Figure 5. pDC depletion reduces Mtb burdens in Sp140−/− mice, and pDCs are present in the lymphocytic cuff surrounding granulomas in Mtb-infected human lymph nodes and lungs.
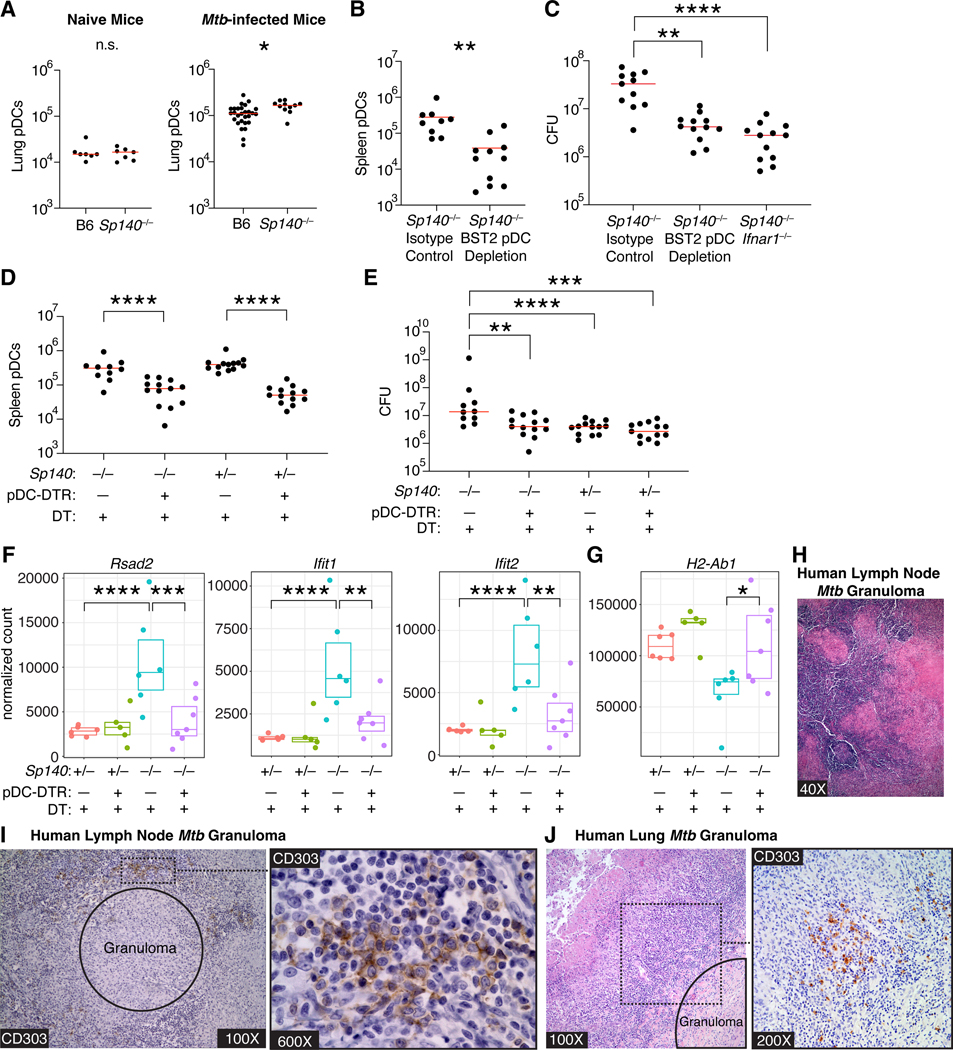
(A) Number of lung pDCs in B6 and Sp140−/− mice in naïve (n = 7) and Mtb-infected mice (n = 11–28). (B) Number of splenic pDCs and (C) bacterial burden in Sp140−/− mice that received isotype or pDC depleting anti-BST2 antibody from days 12–24 post-infection (n = 9–12). (D) Number of splenic pDCs, (E) bacterial burden, (F) lung expression of Rsad2, Ifit1, and Ifit2 as representative type I IFN-stimulated genes, and (G) lung expression of H2-Ab1 as a representative type II IFN-stimulated gene in Sp140−/− pDC-DTR mice or Sp140−/− mice controls that received DT from days 12 to 24 after infection (n = 10–13 for (D) and (E); n = 5–7 for (F) and (G)). (H) Representative hematoxylin and eosin or (I) anti-CD303 (brown) and hematoxylin staining on serial sections of Mtb-infected human lymph nodes (n = 8). (J) Representative hematoxylin and eosin and anti-CD303 (brown) and hematoxylin staining on serial sections of Mtb-infected human lung samples (n = 8). Mouse lungs were harvested 25 days post-infection. The bars in (A), (B), (C), (D), and (E) represent the median. Pooled data from two independent experiments are shown in (A), (B), (C), (D), and (E). Statistical significance was calculated by one-way ANOVA with Tukey’s multiple comparison test for (C), (D), and (E) and by an unpaired t test for (A) and (B). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
We next sought to understand the role of lung pDCs during human Mtb infection by analyzing human lung and lymph node biopsies taken from Mtb culture-positive patients for the presence of pDCs near Mtb granulomas (Fig. 5H). Based on CD303 and CD123 staining, pDCs localized to the lymphocytic cuff surrounding Mtb granulomas in human lungs and lymph nodes (Fig. 5I, 5I, Supplementary Fig. 4). Of the 8 patient samples analyzed, 5 lung samples and 7 lymph node samples had pDCs in the same 400× field as an Mtb granuloma (Supplementary Table 1). The majority of the pDCs in the lung samples were distributed as individual cells, while lymph node pDCs were primarily grouped together in clusters of over 20 cells or scattered individually (Supplementary Table 1). These results demonstrate that pDCs, which are overall a rare cell type, cluster near Mtb-infected cells in granulomas in human lung and lymph nodes during infection. These results, along with our results in mice and previous studies in non-human primates51, implicate pDCs as a plausible source of type I IFN that drives active tuberculosis in humans.
We sought to understand why pDCs contribute to the susceptibility of Sp140−/− but not Sp140-sufficient animals. pDCs are not directly infected and typically produce type I IFNs after recognition of extracellular-derived nucleic acid ligands. Thus, we examined the availability of ligands that might potentially activate pDCs to produce type I IFNs. We focused on DNA-rich neutrophil extracellular traps (NETs) as a potential pDC-activating ligand because extracellular DNA can activate pDCs via TLR9, and NETs have been described as a stimulus for type I interferon production by pDCs in mice and humans in the context of autoimmunity32,33,62. Additionally, another Mtb susceptible mouse model with a hyper type I interferon response identified the presence of NETs in the lungs of susceptible mice and humans with active Mtb disease63. We found a 5-fold enrichment in lung neutrophils in Mtb-infected Sp140−/− relative to Sp140−/− Ifnar1−/− mice (Fig. 1D, 6A). Additionally, neutrophil depletion partially rescued the susceptibility of Sp140−/− mice at both lower and higher bacterial burdens (Fig. 6B-E). Given the importance of neutrophils, we assessed NET production in Sp140−/− and B6 mice by staining for citrullinated H3 in the lungs of Mtb-infected mice (Fig. 6F). Sp140−/− mice had over a 100-fold increase in NET staining as compared to B6 animals, indicating that the lungs of Sp140−/− mice harbor substantially more ligand to activate type I interferon production by pDCs as compared to wild-type hosts (Fig. 6G). As NETs are a source of nucleic acids, we hypothesized that pDCs would sense the NETs through endosomal TLRs. In line with this prediction, deletion of Unc93b1, which is an essential chaperone required for TLR7 and TLR9 function, partially rescued the Mtb susceptibility of Sp140−/− mice (Fig. 6H). By contrast, deletion of Ticam1, which encodes for TRIF, an adapter molecule critical for type I IFN production downstream of TLR3 and TLR4, had no effect on bacterial control in Sp140−/− mice (Fig. 6I). Together, these data suggest a model in which extracellular nucleic acid, potentially from NETs, is sensed by endosomal TLRs triggering pDC production of type I IFNs (Fig. 6J).
Figure 6. Role of neutrophils, neutrophil extracellular traps (NETs) and endosomal TLRs during type I IFN-driven Mtb pathogenesis.
(A) Quantification of lung neutrophils in Mtb-infected Sp140−/− (n = 34) and Sp140−/− Ifnar1−/− (n = 11) mice. (B) Representative flow cytometry plot and (C) quantification of lung neutrophils (n = 10–11) as well as (D) bacterial burden after lower (n = 4–6) or higher dose (n = 5–6) infection and (E) combined normalized bacterial burden (n = 10–11) in Sp140−/− mice that received isotype or neutrophil depleting anti-Ly6G clone 1A8 antibody from days 12 to 24 post-infection. (F) Representative images and (G) quantification of NET production based on citrullinated H3 staining in the diseased portions of I-Tomcat Ai6 and Sp140−/− I-Tomcat Ai6 lungs. Sections were stained with citrullinated H3 (red), Ai6 (green), Ly6G (teal), and Mtb-expressed mCherry (magenta) (n = 6–7). (H) Lung bacterial burden in Mtb-infected Sp140−/− (n = 13), Sp140−/− Ticam1−/− (Ticam1 encodes TRIF; n = 14), and Sp140−/− Ifnar1−/− mice (n = 11). (I) Lung bacterial burden in Mtb-infected Sp140−/− (n = 9), Sp140−/− Unc93b1−/− (n = 8), Sp140−/− Ifnar1−/− (n = 4), Sp140+/− Unc93b1−/− mice (n = 12), and Sp140+/− Unc93b1+/− mice (n = 11). (J) Model of potential extracellular DNA sources stimulating pDC production of type I IFNs through endosomal TLR signaling. The bars in (A), (C), (D), (E), (G), (H), and (I) represent the median. Lungs were analyzed 25 days after Mtb infection. Statistical significance was calculated by one-way ANOVA with Tukey’s multiple comparison test for (E), (H), and (I), by one-tailed unpaired t test for (D), and by two-tailed unpaired t test for (A) and (G). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Neutrophils and IMs are the major sensors of type I IFNs during Mtb infection
Having identified pDCs, IMs, and monocytes as the main cells producing type I IFN during Mtb infection, we next sought to identify the cells responding to type I IFN. As expected, IFNAR was uniformly expressed by all lung myeloid cells, and was therefore not informative for identifying IFN responsive cells (Fig. 7A)64. However, comparing differentially expressed genes in B6 and Sp140−/− neutrophils and IMs showed a clear induction of IFN stimulated genes in cells from Sp140−/− over B6 animals (Fig. 7B). A major complication is that many genes induced by type I IFN are also induced by type II IFN (IFNγ), and most existing studies do not distinguish the two. Therefore, we sought to develop type I IFN-specific and IFNγ-specific transcriptional signatures. RNA-sequencing analysis of human macrophages and mouse bone marrow-derived macrophages stimulated with IFNγ, IFN-β, TNF, transforming growth factor-β, or nothing were used to define cytokine-induced genes (Supplementary Fig. 5, 6)65. Similar gene families were preferentially upregulated by type I or II IFNs in human and mouse macrophages. Genes included in the signatures for type I or II IFN were not strongly induced by a panel of other cytokines (Supplementary Fig. 5). We next applied the mouse gene signatures to our mouse lung myeloid scRNA-seq dataset. The strength of signature expression in naïve mice was used as the threshold for classifying cells as responding to IFNγ or type I IFN (Supplementary Fig. 6). As expected, naïve mice had very few cells responding to either cytokine, while bystander and Mtb-infected cells responded strongly to type I and/or II IFNs (Fig. 7C, Supplementary Fig. 6). Interestingly, the type I IFN response was limited to IMs and neutrophils, even though monocytes and AMs were responsive to IFNγ. Potentially, differences in the localization of these cells could explain their differences in cytokine responsiveness. As expected, Mtb-infected neutrophils and IMs from Sp140−/− mice exhibited a significant increase in type I IFN signaling relative to cells from B6 lungs (Fig. 7D). Consistent with considerable prior work demonstrating that type I IFNs impair responsiveness to IFNγ15,34,35,66, the Sp140−/− mice harbored a distinct population of infected IMs that exhibited the signature of type I IFN-responsiveness but lacked the signature of IFNγ responsiveness (note the distinct population of blue IMs among the Mtb-infected cells in Fig. 7C, 7D). The reduction in IFNγ signaling in Mtb-infected IMs in Sp140−/− mice correlated with reduced IFNγ receptor 1 expression on these cells in Sp140−/− relative to B6 mice (Supplementary Fig. 7). This reduced IFNγ receptor 1 expression also correlated with increased type I IFN stimulated genes and reduced type II IFN stimulated genes (Supplementary Fig. 7). Since IFNγ is critical for Mtb control, these results suggest that type I IFN impairs Mtb control at least in part by opening a niche of susceptible IMs that fail to respond to IFNγ.
Figure 7. Macrophage recognition of type I IFN drives Mtb susceptibility of Sp140−/− mice.
(A) Ifnar1 and Ifnar2 mRNA expression in innate immune cells from Mtb-infected lungs (B6 and Sp140−/− combined). (B) Differentially expressed genes comparing B6 and Sp140−/− neutrophils and IMs, with higher log fold change indicating greater expression in B6. (C) Bystander, and Mtb-infected lung myeloid cells from B6 and Sp140−/− mice classified by their responsiveness to IFNγ (red), type I IFN (blue), both (purple), or neither (grey). (D) Graph of neutrophils, monocytes, IMs, and AMs frequencies from Mtb-infected B6 (n = 3) and Sp140−/− (n = 3) lungs that are responsive to IFNγ (red), type I IFN (blue), both (purple), or neither (white). (E) Lung bacterial burden and (F) neutrophils in Mtb-infected Sp140−/− Ifnar1−/− (n = 16), Sp140−/− Ifnar1fl/fl Mrp8cre (n = 18), and Sp140−/− littermate control (n = 57) mice. (G) Lung bacterial burden and (H) neutrophils in Mtb-infected Sp140−/− Ifnar1−/− (n = 9), Sp140−/− LysMcre littermate control (n = 14), Sp140−/− Ifnar1fl/fl LysMcre (n = 5), Sp140−/− CD64cre littermate control (n = 20), and Sp140−/− Ifnar1fl/fl CD64cre (n = 14) mice. The bars in (E-H) represent the median. Lungs were analyzed 24–26 days after Mtb infection. Pooled data from two-three independent experiments are shown. Statistical significance in (B) was calculated by non-parametric Wilcoxon rank sum test with Bonferroni correction, by one-way ANOVA with Tukey’s multiple comparison test for (E-H), and by two-way ANOVA with Tukey’s multiple comparisons test in (D). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
As neutrophils and IMs were the primary sensors of type I IFN, these cell types were tested as potential mediators of type I IFN-driven susceptibility in Sp140−/− mice. Neutrophil-specific deletion of Ifnar1 was insufficient to rescue the Mtb susceptibility or the increase in lung neutrophils exhibited by Sp140−/− mice (Fig. 7E, 7F). Conversely, deletion of Ifnar1 expression on myeloid cells with LysMCre Ifnar1fl/fl mice, or specifically in macrophages with CD64Cre Ifnar1fl/fl mice, rescued bacterial control and reduced lung neutrophil numbers to the same extent as global Ifnar1 deletion (Fig. 7G, 7H). Taken together, these results are consistent with a model in which type I IFNs act on IMs to inhibit IFNγ signaling in these cells, thereby reducing their ability to restrict Mtb growth.
Discussion
The dominant gene signature identified in humans with active tuberculosis disease is a type I IFN signature7,8,67,68. Type I IFNs are critical for effective anti-viral immunity67 but impair Mtb control in human and animal models7,8,26,27,27,28. For example, viral co-infections or chemical interferon inducers exacerbate Mtb disease in mice16–20. Interestingly, although type I IFN and IFNγ induce a highly overlapping set of target genes (Supplementary Fig. 5), type I IFNs promote Mtb disease while IFNγ potently protects against mycobacterial infections in mice5 and humans8,69–72. Indeed, type I IFNs can exacerbate bacterial infections in mice and humans by directly antagonizing IFNγ signaling15,34,35. The underlying mechanism is poorly understood but may in part be due to downregulation of the IFNγ receptor34,35. Type I IFNs can also impair IL-1 signaling, an additional pathway critical for Mtb control, through the induction of IL-1 receptor antagonist and eicosanoid imbalance24,26,27,73. Thus, the antagonism of protective IFNγ and IL-1 responses by type I IFNs may be a key driver of progression to active tuberculosis. Therefore, we sought a genetically tractable model of Mtb infection to establish the cellular mechanisms by which type I IFN drives Mtb susceptibility.
The commonly used B6 mouse model does not exhibit a strong type I IFN response after Mtb infection23,27,31. Consistent with the modest type I IFN response of B6 mice, Ifnar1 deletion on the B6 background does not reliably impact survival or lung bacterial burden after Mtb infection21–24,27. Therefore, we sought a different mouse model that recapitulated two key aspects of human disease: the hyper type I IFN response, and the accompanying neutrophilic inflammation7,74,75. Previously, we identified B6.Sst1S mice as a mouse model that exhibits type I IFN-driven susceptibility to Mtb infection27. We then demonstrated that the absence of Sp140 in B6.Sst1S mice explains their susceptibility to Mtb31. SP140 is a member of the Speckled Protein family of epigenetic readers and is widely expressed in leukocytes76. Sp140-deficient macrophages have been hypothesized to exhibit inherent defects in bacterial control causing increased susceptibility to dextran sulfate sodium-induced colitis39,40. However, an inherent defect in bacterial control is not evident during Mtb infection, as Sp140−/− mice lacking Ifnar1 restrict Mtb as well as B6 animals 25 days after infection31, a time point at which macrophages are critical for Mtb control38. This result suggests that the early susceptibility of Sp140−/− mice is due to their strong type I IFN response rather than an inherent defect in bacterial killing by macrophages. In addition to their hyper type I IFN response, Sp140−/− mice have more lung neutrophils after Mtb infection than Sp140−/−Ifnar1−/− animals (Fig. 1D, 7B, 7D). Therefore, Sp140−/− mice recapitulate the fundamental type I IFN and neutrophilic character of human active Mtb disease, and can serve as a platform for understanding the cellular mechanism of type I IFN-driven Mtb susceptibility.
Sp140−/− mice provide an ideal model of the aberrant type I IFN response, as they are on a pure B6 genetic background, and do not require repeated administration of TLR agonists, viral co-infection, or other perturbations of the innate immune system for type I IFN production16,17,25,26,63. Other groups have also modeled the type I IFN response by infecting B6 mice with a lineage 2 clinical Mtb strain, such as HN87877,78. However, Ifnar1 deletion had no impact on survival or bacterial control at early time points in B6 mice infected with HN878, unlike Sp140−/− mice infected with Mtb Erdman31,79,80. Thus, we believe the Sp140−/− mouse model recapitulates the hyper type I IFN response exhibited by humans, and provides a tool to study the mechanistic basis of the aberrant type I IFN response.
We used Sp140−/− mice to identify the type I IFN producers and responders that mediate Mtb disease. Flow cytometry and imaging of I-Tomcat Ai6 Ifnb1 reporter mice identified IMs and pDCs as the major IFN-β producers during Mtb infection (Figure 3, 4). Imaging provided insight into why these cells expressed type I IFN, as the frequency of type I IFN-expressing macrophages was enriched relative to CD4+ T cells in diseased tissue but not in healthy tissue. This result suggests that proximity to Mtb dictates access to activating signals required to induce IFN-β expression by macrophages. However, most IFN-β expressing IMs were not infected with Mtb, and most infected IMs were not Ifnb1 or reporter positive, indicating direct infection is insufficient and may not be the main driver of type I IFN expression in vivo. In vitro studies have shown that bone marrow-derived macrophages infected with Mtb induce type I IFN via the cytosolic DNA-sensing cGAS-STING pathway. However, this pathway does not appear to play a major role in vivo27,81–84. Instead, our results suggest that uninfected bystander cells responding to extracellular ligands may be the primary producers of type I IFN during Mtb infection. Consistent with this hypothesis, we found that mice lacking Unc93b1, a chaperone required for TLRs that sense extracellular nucleic acids, exhibit enhanced control of Mtb (Figure 6I). Of note, pDCs, which we found to be important type I IFN producers during Mtb infection, are robust producers of type I IFN after TLR7/9 sensing of exogenous nucleic acids58,85,86.
Expression of type I IFNs by pDCs during Mtb infection was particularly noteworthy as limited work exists on the effect of pDCs on Mtb control. Production of type I IFNs by pDCs is important for control of viral infections56,57,61. pDCs also demonstrate a protective function against bacterial infections such as Citrobacter rodentium, Chlamydia pneumoniae, and Klebsiella pneumoniae87–90. However, the contribution of pDCs during Mtb infection remains unclear. Blood pDC numbers were reduced in Mtb-infected humans, but lung pDC numbers or function were not assessed91,92. In non-human primates, active pulmonary Mtb correlated with pDC influx and IFN-responsive macrophages into the lungs of rhesus macaques51. The granulomas of Mtb-infected cynomolgus macaques also contained pDCs, but the pDC frequency did not correlate with bacterial burdens in the granulomas93. A major issue in the studies using NHPs or humans is the difficulty in depleting or otherwise functionally assessing the role of pDCs. To address this limitation, we generated Sp140−/− pDC-DTR mice59,61. Using these mice, we found that pDCs contribute significantly to the susceptibility of Sp140−/− mice (Figure 5). Additionally, we identified pDCs in the lymphocytic cuff surrounding Mtb granulomas in human lungs and lymph nodes. Together, these results suggest that type I IFN produced by pDCs drives tuberculosis disease in mice, and is likely conserved across non-human primates and humans.
While pDC depletion rescued Sp140−/− mice, it had no impact on bacterial burden in B6 animals. This result was expected given that very few myeloid cells in B6 mice expressed a type I IFN signature and Ifnar1 deficiency also has only modest effects in the B6 background23,27. As pDCs are present in B6 and Sp140−/− mice, we speculated that the difference in pDC type I IFN production in these mouse strains could be due in part to differences in the availability of activating ligands. As seen in another Mtb-susceptible mouse model63, and in Mtb-infected human lungs63, Sp140−/− mice had a significant enrichment in NET production compared to B6 mice. NETs are DNA-rich products of neutrophils and may act as ligands for TLR9 on the pDCs, as described in mouse and human autoimmunity32,33,62,94. In support of this hypothesis, neutrophil depletion partially rescued the susceptibility of Sp140−/− mice. Given that NET formation was also identified in Mtb granulomas in human lung sections63, pDC sensing of NETs may contribute to the type I IFN response detected in humans with active Mtb disease.
Having defined the cells producing type I IFNs in vivo after Mtb infection, we then sought to identify the type I IFN responders. To do this, we first developed transcriptional signatures that distinguish the response to type I IFN from the closely related response to IFNγ. Applying these signatures to our scRNA-seq data, we identified neutrophils and IMs as type I IFN sensors. Both IMs and neutrophils harbor Mtb, so the effect of type I IFN could be on either or both cell types. In a GM-CSF blockade model of type I IFN-driven Mtb susceptibility, neutrophil-specific deletion of Ifnar1 rescued bacterial control63. By contrast, we were unable to detect any rescue of Sp140−/− mice when neutrophils lacked Ifnar1 (Figure 7E-F). Instead we found that deletion of Ifnar1 on macrophages rescued Sp140−/− mouse bacterial control (Figure 7G-H). GM-CSF is critical for maintaining lung alveolar macrophages and enhances the responsiveness of lung monocytes and macrophages to infections, including Mtb infection95–98. Therefore, it is possible that impairing lung macrophages by GM-CSF blockade shifted the impact of type I IFN on Mtb control from macrophages to neutrophils. Our scRNA-seq dataset only contains myeloid cells and therefore cannot address the contribution of type I IFN signaling in other cell types. However, it is likely that type I IFN largely acts through myeloid cells to drive Mtb susceptibility, as macrophage-specific deletion of Ifnar1 rescued Mtb control to a similar extent as global Ifnar1 deficiency. These results suggest that during a GM-CSF sufficient response, type I IFN signaling in macrophages reduces their ability to restrict Mtb. The mechanism by which type I IFNs impair Mtb clearance by IMs remains unknown. However, we observed that infected IMs in Sp140−/− mice express lower levels of IFNγ receptor and IFNγ target genes, and instead primarily exhibited a type I IFN signature, consistent with the exacerbated type I IFN response in these mice (Supplementary Fig. 7). The transcriptional response of Sp140−/− IMs contrasted dramatically with that of infected IMs in B6 mice, which control Mtb and which exhibited a uniform signature of responsiveness to IFNγ (Figure 7). Given the essential role of IFNγ in controlling Mtb in mice and humans, our results suggest that one detrimental effect of type I IFNs is inhibition of IFNγ signaling in infected macrophages14,34,35.
Taken together, our results identify the cell types that produce and respond to type I IFNs during IFN-driven tuberculosis disease. We propose that in addition to its previously described role in inhibition of IL-1 signaling, type I IFNs also impair responsiveness to IFNγ, leading to an initial loss of bacterial control. Bacterial replication then leads to neutrophil influx and NET production within the diseased tissue. DNA-rich NETs may be one source of extracellular-derived ligands sensed by endosomal TLRs in pDCs, though cellular RNA or bacterial DNA may be additional sources. Activation of pDCs results in very robust per-cell production of type I IFNs, which we propose acts in a positive feedback loop to further antagonize IFNγ signaling and reduce the ability of IMs to restrict Mtb growth. Given the correlations between our results and findings in rhesus macaques and humans with active Mtb, we believe that our proposed mechanism of type I IFN-driven loss of Mtb control is conserved across species. These findings open the door for the development of therapies targeting NET production or pDC function as host-directed strategies for treating active Mtb infection.
Limitations of the Study
Although we see a strong correlation between NET formation and type I IFN-driven Mtb susceptibility, the current study does not directly test the contribution of NETs in this response. Additionally, the present study provides data suggesting that type I IFN signaling correlates with a loss of type II IFN signaling in IMs during Mtb infection, but does not directly examine whether Mtb-harboring IMs in Sp140−/− mice are unable to control Mtb infection because of a lack of response to IFNγ. We also limited our studies to ~25 days post-infection, which is an early time point for Mtb infection. It is possible that the cellular sources and targets of type I IFN shift to other cell types at later time points in the infection.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Russell Vance (rvance@berkeley.edu).
Materials availability
Materials used in this study will be provided upon request and available upon publication.
Data and code availability
Raw and processed bulk RNA- and single cell RNA-sequencing data is deposited in the NCBI Gene Expression Omnibus: GSE216023, GSE232827, GSE232922. This paper also analyzes existing, publicly available data, for which the accession numbers are listed in the key resources table.
Code for bulk RNA- and scRNA-sequencing analysis is available on Github: https://github.com/dmitrikotov/Sp140-Type-I-Inteferon.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| TruStain FcX PLUS (anti-mouse CD16/32) clone S17011E | BioLegend | Cat # 156604; RRID:AB_2783138 |
| BUV496 anti-mouse CD45 clone 30-F11 | BD Biosciences | Cat # 749889; RRID:AB_2874129 |
| APC anti-mouse CD64 clone X54–5/7.1 | BioLegend | Cat # 139306; RRID:AB_1121939 1 |
| BV480 anti-mouse B220 clone RA3–6B2 | BD Biosciences | Cat # 565631; RRID:AB_2739311 |
| BV480 anti-mouse CD90.2 clone 53–2.1 | BD Biosciences | Cat # 566082; RRID:AB_2739494 |
| APC-Fire 750 anti-mouse Ly6G clone 1A8 | BioLegend | Cat # 127652; RRID:AB_2616733 |
| BUV395 anti-mouse CD11b clone M1/70 | BD Biosciences | Cat # 563553; RRID:AB_2738276 |
| BUV737 anti-mouse CD11c clone HL3 | BD Biosciences | Cat # 612796; RRID:AB_2870123 |
| APC-R700 anti-mouse Siglec F clone E50–2440 | BD Biosciences | Cat # 565183; RRID:AB_2739097 |
| PE anti-mouse MerTK clone DS5MMER | Thermo Fisher Scientific | Cat # 12–5751-82; RRID:AB_2572623 |
| Super Bright 645 anti-mouse MHC II clone M5/114.15.2 |
Thermo Fisher Scientific |
Cat # 64–5321-82; RRID:AB_2662402 |
| BV421 anti-mouse PD-L1 clone MIH5 | BD Biosciences | Cat # 564716; RRID:AB_2738911 |
| BV711 anti-mouse Ly6C clone HK1.4 | BioLegend | Cat # 128037; RRID:AB_2562630 |
| PE anti-mouse IFNAR-1 clone MAR1–5A3 | BioLegend | Cat # 127311; RRID:AB_1134011 |
| PE-Cy7 anti-mouse MerTK clone DS5MMER | Thermo Fisher Scientific | Cat # 25–5751-82; RRID:AB_2573466 |
| APC-eFluor 780 anti-mouse CD11b clone M1/70 | Thermo Fisher Scientific | Cat # 47–0112-82; RRID:AB_1603193 |
| BUV395 anti-mouse CCRL2 clone BZ2E3 | BD Biosciences | Cat # 743689; RRID:AB_2741676 |
| BUV563 anti-mouse Ly6G clone 1A8 | BD Biosciences | Cat # 612921; RRID:AB_2870206 |
| Percp-Cy5.5 anti-mouse B220 clone RA3–6B2 | BioLegend | Cat # 103235; RRID:AB_893356 |
| BV421 anti-mouse Siglec H clone 440c | BD Biosciences | Cat # 566581; RRID:AB_2739747 |
| BV480 anti-mouse CD19 clone 1D3 | BD Biosciences | Cat # 566167; RRID:AB_2739564 |
| BV605 anti-mouse MHC II clone M5/114.15.2 | BD Biosciences | Cat # 563413; RRID:AB_2738190 |
| BV785 anti-mouse Ly6C clone HK1.4 | BioLegend | Cat # 128041; RRID:AB_2565852 |
| BV605 anti-mouse CD4 clone GK1.5 | BioLegend | Cat # 100451; RRID:AB_2564591 |
| BUV805 anti-mouse CD8ɑ clone 53–6.7 | BD Biosciences | Cat # 612898; RRID:AB_2870186 |
| PE-Cy7 anti-mouse PDCA-1 clone eBio927 | Thermo Fisher Scientific | Cat # 25–3172-80; RRID:AB_2573439 |
| PE-Cy7 anti-mouse CD63 clone NVG-2 | BioLegend | Cat # 143909 |
| Percp-eFluor 710 anti-mouse iNOS clone CXNFT | Thermo Fisher Scientific | Cat # 46–5920-82; RRID:AB_2688059 |
| BV785 anti-mouse CD206 clone C068C2 | BioLegend | Cat # 141729; RRID:AB_2565823 |
| Anti-mouse BST2 clone 927 | BioXCell | Cat # BE0311; RRID:AB_2736991 |
| Rat IgG2b isotype antibody clone LTF-2 | BioXCell | Cat # BE0090; RRID:AB_1107780 |
| Anti-mouse Ly6G clone 1A8 | BioXCell | Cat # BE0075–1; RRID:AB_1107721 |
| Rat IgG2a isotype antibody clone 2A3 | BioXCell | Cat # BE0089; RRID:AB_1107769 |
| Anti-human CD123 clone 6h6 | Thermo Fisher Scientific | Cat # 14–1239-82; RRID:AB_467453 |
| Anti-human CD303 clone 124B3.13 | Dendritics | Cat # DDX0043; RRID:AB_1149764 |
| BV421 anti-mouse SIRP𝘢 clone P84 | BD Biosciences | Cat # 740071; RRID:AB_2739835 |
| Pacific Blue anti-mouse B220 clone RA3–6B2 | BioLegend | Cat # 103227; RRID:AB_492876 |
| eF506 anti-mouse CD4 clone RM4–5 | Thermo Fisher Scientific | Cat # 69–0042-80; RRID:AB_2637458 |
| AF647 anti-mouse Ly6G clone 1A8 | BioLegend | Cat # 127609; RRID:AB_1134162 |
| BV421 anti-mouse Ly6G clone 1A8 | BioLegend | Cat # 127628; RRID:AB_2562567 |
| Rabbit polyclonal anti-citrullinated histone-H3 (citrulline R2, R8, R17) |
Abcam | Cat # ab5103; RRID:AB_304752 |
| AF488 donkey anti-rabbit secondary clone Poly4064 | BioLegend | Cat # 406416; RRID:AB_2563203 |
| APC anti-mouse Ly6G clone 1A8 | BioLegend | Cat # 127614; RRID:AB_2227348 |
| TotalSeq-A anti-mouse Ly6G clone 1A8 | BioLegend | Cat # 127655; RRID:AB_2749962 |
| TotalSeq-A anti-mouse Ly6C clone HK1.4 | BioLegend | Cat # 128047; RRID:AB_2749961 |
| TotalSeq-A anti-mouse CD44 clone IM7 | BioLegend | Cat # 103045; RRID:AB_2734154 |
| TotalSeq-A anti-mouse CD169 clone 3D6.112 | BioLegend | Cat # 142425; RRID:AB_2783106 |
| TotalSeq-A anti-mouse CD274 clone MIH6 | BioLegend | Cat # 153604; RRID:AB_2783125 |
| TotalSeq-A anti-mouse Siglec F clone S17007L | BioLegend | Cat # 155513; RRID:AB_2832540 |
| TotalSeq-A anti-mouse CSF1R clone AFS98 | BioLegend | Cat # 135533; RRID:AB_2734198 |
| TotalSeq-A anti-mouse CD11b clone M1/70 | BioLegend | Cat # 101265; RRID:AB_2734152 |
| TotalSeq-A anti-mouse CD86 clone GL-1 | BioLegend | Cat # 105047; RRID:AB_2750348 |
| TotalSeq-A anti-mouse MHC II clone M5/114.15.2 | BioLegend | Cat # 107653; RRID:AB_2750505 |
| TotalSeq-A anti-mouse CX3CR1 clone SA011F11 | BioLegend | Cat # 149041; RRID:AB_2783121 |
| TotalSeq-A anti-mouse Hashtag 1 | BioLegend | Cat # 155801; RRID:AB_2750032 |
| TotalSeq-A anti-mouse Hashtag 2 | BioLegend | Cat # 155803; RRID:AB_2750033 |
| TotalSeq-A anti-mouse Hashtag 3 | BioLegend | Cat # 155805; RRID:AB_2750034 |
| TotalSeq-A anti-mouse Hashtag 4 | BioLegend | Cat # 155807; RRID:AB_2750035 |
| TotalSeq-A anti-mouse Hashtag 5 | BioLegend | Cat # 155809; RRID:AB_2750036 |
| TotalSeq-A anti-mouse Hashtag 6 | BioLegend | Cat # 155811; RRID:AB_2750037 |
| PE anti-mouse B220 clone RA3–6B2 | Tonbo Biosciences | Cat # 50–0452-U100; RRID:AB_2621764 |
| PE anti-mouse CD90.2 clone 30-H12 | Tonbo Biosciences | Cat # 50–0903-U025; RRID:AB_2940772 |
| BV785 anti-mouse CD45.2 clone 104 | BioLegend | Cat # 109839; RRID:AB_2562604 |
| Pacific Blue anti-mouse B220 clone RA3–6B2 | BioLegend | Cat # 103230; RRID:AB_492877 |
| Pacific Blue anti-mouse CD90.2 clone 53–2.1 | BioLegend | Cat # 140305; RRID:AB_1064533 5 |
| PE anti-mouse F4/80 clone BM8 | Thermo Fisher Scientific |
Cat # 12–4801-80; RRID:AB_465922 |
| PE anti-mouse CXCL9 clone MIG-2F5.5 | BioLegend | Cat # 515603; RRID:AB_2245490 |
| Bacterial and virus strains | ||
| Bacteria: Mtb strain Erdman | A gift from Sarah Stanley, University of California, Berkeley |
N/A |
| Bacteria: Mtb-Wasabi | This study | N/A |
| Bacteria: Mtb-mCherry | This study | N/A |
| Biological samples | ||
| Human lung and lymph node samples | Surgical pathology archives of Emory University Hospital |
N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ghost Dye Violet 510 | Tonbo Biosciences | Cat # 13–0870-T500 |
| Super Bright Complete Staining Buffer | Thermo Fisher Scientific | Cat # SB-4401–75 |
| True-Stain Monocyte Blocker | BioLegend | Cat # 426102 |
| Cytofix/cytoperm | BD biosciences | Cat # 554722 |
| AccuCheck Counting Beads | Invitrogen | Cat # PCB100 |
| Vector Laboratories Hematoxylin and Eosin Stain Kit | Thermo Fisher Scientific | Cat # NC1470670 |
| Diphtheria toxin | Millipore Sigma | Cat # D0564–1MG |
| DreamTaq Green PCR Master Mix (2X) | Thermo Fisher Scientific | Cat # K1082 |
| Middlebrook 7H9 Broth (Dehydrated) | Thermo Fisher Scientific | Cat # R454012 |
| BBL seven H11 agar base | BD biosciences | Cat # 212203 |
| Middlebrook OADC | Thermo Fisher Scientific | Cat # b12351 |
| Hygromycin B Gold | Invivogen | Cat # ant-hg-1 |
| Kanamycin | Millipore Sigma | Cat # K4000–5G |
| Liberase TM | Roche | Cat # 5401127001 |
| Dnase I | Roche | Cat # 11284932001 |
| Newborn Calf Serum | Thermo Fisher Scientific | Cat # 26010074 |
| Trizol LS | Thermo Fisher Scientific | Cat # 10296010 |
| M-CSF | Vance lab | N/A |
| IFN-β | BioLegend | Cat # 581302 |
| IFNg | Peprotech | Cat # 315–05 |
| Tumor necrosis factor | Peprotech | Cat # 315–01A |
| Transforming growth factor-β | BioLegend | Cat # 763102 |
| TRK lysis buffer | Omega Bio-Tek | Cat # PR021 |
| 2-mercaptoethanol | Thermo Fisher Scientific | Cat # 21985023 |
| Ultracomp eBeads Plus | Thermo Fisher Scientific | Cat # 01–3333-42 |
| Sytox Blue Dead Cell Stain | Invitrogen | Cat # S11348 |
| RNaseOUT Recombinant Ribonuclease Inhibitor | Invitrogen | Cat # 10777019 |
| Critical commercial assays | ||
| RNeasy Micro | Qiagen | Cat # 74004 |
| E.Z.N.A Total RNA Kit I | Omega Bio-Tek | Cat # R6834–02 |
| EasySep APC Positive Selection Kit II | StemCell Technologies | Cat # 17681 |
| Chromium Single Cell 3’ Reagent Kit v3.1 chemistry | 10X Genomics | Cat # 1000268 |
| Deposited data | ||
| scRNA-seq of Mtb-infected non-human primates | Esaulova et al.51 | GEO: GSE149758 |
| Bulk RNA-seq of cytokine stimulated human macrophages | Nilsson et al.65 | GEO: GSE20251 |
| Bulk RNA-seq of cytokine stimulated mouse bone marrow-derived macrophages | This study | GEO: GSE232827 |
| Bulk RNA-seq of Mtb-infected lungs from pDC depleted Sp140+/− and Sp140−/− mice | This study | GEO: GSE232922 |
| scRNA-seq of myeloid cells from naïve and Mtb-infected B6 and Sp140−/− mice | This study | GEO: GSE216023 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Cat # 000664; RRID:IMSR_JAX:0 00664 |
| Mouse: Ifnar1−/−: B6.129S2-Ifnar1tm1Agt/Mmjax | The Jackson Laboratory | Cat # 032045-JAX RRID:MMRRC_03 2045-JAX |
| Mouse: Ai6: B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J | The Jackson Laboratory | Cat # 007906 RRID:IMSR_JAX:0 07906 |
| Mouse: Ifnar1fl: B6(Cg)-Ifnar1tm1.1Ees/J | The Jackson Laboratory | Cat # 028256 RRID:IMSR_JAX:0 28256 |
| Mouse: Mrp8Cre: B6.Cg-Tg(S100A8-cre,-EGFP)1Ilw/J | The Jackson Laboratory | Cat # 021614 RRID:IMSR_JAX:0 21614 |
| Mouse: LysMCre: B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | Cat # 004781 RRID:IMSR_JAX:0 04781 |
| Mouse: C57BL/6J-Ticam1Lps2/J (Ticam1−/−) | The Jackson Laboratory | Cat # 005037 RRID:IMSR_JAX:0 05037 |
| Mouse: C57BL/6N-Unc93b1tm1(KOMP)Vlcg/Mmucd (Unc93b1−/−) | MMRRC, KOMP repository, Regeneron Pharmaceuticals |
Cat # 050296-UCD RRID:MMRRC_05 0296-UCD |
| Mouse: I-Tomcat: Ifnb1-Tomato-Cre-pA Terminator | This study | N/A |
| Mouse: FLPer: B6N.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/J | The Jackson Laboratory | Cat # 016226 RRID:IMSR_JAX:0 16226 |
| Mouse: CD64Cre: B6-Fcgr1tm2Ciphe | Scott et al.106 | N/A |
| Mouse: pDC-DTR: B6-Tg(CLEC4C-HBEGF)956Cln/J | The Jackson Laboratory | Cat # 014176; RRID:IMSR_JAX:0 14176 |
| Mouse: Sp140−/− | Ji et al.31 | N/A |
| Recombinant DNA | ||
| pTEC15 | Takaki et al.36 | Addgene plasmid # 30174 |
| pMSP12::mCherry | A gift from Lalita Ramakrishnan, University of Cambridge |
Addgene plasmid # 30167 |
| Software and algorithms | ||
| Chrysalis | Kotov et al.55 | https://github.com/Histo-cytometry/Chrysalis |
| Generate Compensation Matrix | Kotov et al. 55 | https://github.com/Histo-cytometry/Chrysalis |
| Imaris version 9.9.1 | Bitplane | N/A |
| FlowJo version 10 | BD Biosciences | N/A |
| Trimmomatic v.0.36 | Bolger et al.109 | https://github.com/usadellab/Trimmomatic |
| STAR aligner v.2.5.2b | Dobin et al.110 | https://github.com/alexdobin/STAR |
| CellRanger version 4.0.0 | 10X Genomics | N/A |
| CITE-Seq-Count v.1.4.3 | Roelli et al.113 | https://github.com/Hoohm/CITE-seq-Count |
| R version 3.16 | R Development Core Team114 | http://www.r-project.org/ |
| RStudio “Cherry Blossom” Release | Posit | https://posit.co/products/open-source/rstudio/ |
| DESeq2 v.1.38.3 | Love et al.111 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Seurat v.4.1.1 | Hao et al.42 | https://satijalab.org/seurat/index.html |
| EnhancedVolcano v1.16.0 | Blighe et al.116 | http://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html |
| Tidyverse v2.0.0 | Wickham et al.115 | https://www.tidyverse.org/ |
| UCell v2.2.0 | Andreatta et al.117 | http://www.bioconductor.org/packages/release/bioc/html/UCell.html |
| Adobe Illustrator | Adobe.com | N/A |
| Prism | GraphPad | N/A |
| Other | ||
| 4 laser SH-800 cell sorter | Sony | N/A |
| LSM 880 laser scanning confocal microscope | Zeiss | N/A |
| 5 laser LSRFortessa analyzer | BD Biosciences | N/A |
| 5 laser Aurora analyzer | Cytek | N/A |
| GentleMACS | Miltenyi Biotec | N/A |
| Chromium controller | 10X Genomics | N/A |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals
Mice were maintained under specific pathogen-free conditions and housed at 23°C with a 12 hour light-dark cycle in accordance with the regulatory standards of the University of California Berkeley Institutional Animal Care and Use Committee. All mice were sex- and age-matched and were 6–12 weeks old at the start of infections. Male and female mice were used in all experiments. Littermate controls were used when possible, as indicated in the figure legends. B6, B6.129S2-Ifnar1tm1Agt/Mmjax (Ifnar1−/−)99, B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J (Ai6)52, B6(Cg)-Ifnar1tm1.1Ees/J (Ifnar1fl)100, B6.Cg-Tg(S100A8-cre,-EGFP)1Ilw/J (Mrp8Cre)101, B6.129P2-Lyz2tm1(cre)Ifo/J (LysMCre)102, C57BL/6J-Ticam1Lps2/J (Ticam1−/−)103, and B6N.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/J (FLPer)104 mice were purchased from Jackson Laboratories. C57BL/6N-Unc93b1tm1(KOMP)Vlcg/Mmucd (Unc93b1−/−) mice were obtained from the Mutant Mouse Resource and Research Center (MMRRC) at the University of California, Davis, was donated to the MMRRC by the KOMP repository at University of California, Davis, originated from David Valenzuela of Regeneron Pharmaceuticals105, and were provided by Gregory Barton at the University of California, Berkeley. Ifnb1-Tomato-Cre-pA Terminator (I-Tomcat) mice were generated by Daniel Stetson at the University of Washington as described below. B6-Fcgr1tm2Ciphe (CD64Cre)106 mice were generated by Bernard Malissen at Centre d’Immunologie de Marseille-Luminy and provided by Yasmine Belkaid at the National Institutes of Health. B6-Tg(CLEC4C-HBEGF)956Cln/J (pDC-DTR)61 mice were provided by Adam Lacy-Hulbert at the Benaroya Research Institute. Sp140−/− mice were previously generated in-house31. Sp140−/− Ifnar1−/− mice were generated by crossing Sp140−/− mice with Ifnar1−/− mice in-house. I-Tomcat Ai6 mice were generated by crossing I-Tomcat mice with Ai6 mice, while Sp140−/− I-Tomcat Ai6 mice were the result of crossing I-Tomcat mice with Sp140−/− and Ai6 mice in-house. Sp140−/− mice were crossed in-house with pDC-DTR mice to generate Sp140−/− pDC-DTR mice. Sp140−/− Ifnar1fl LysMcre, Sp140−/− Ifnar1fl Mrp8Cre, and Sp140−/− Ifnar1fl CD64Cre mice were generated by crossing Sp140−/− mice with Ifnar1fl and LysMCre or Mrp8Cre or CD64Cre mice in-house.
Bacterial strains
Mtb strain Erdman was a gift from Sarah Stanley at the University of California, Berkeley. Frozen aliquoted stocks were produced after passing the strain in vivo to ensure virulence. Mtb expressing Wasabi (Mtb-Wasabi) and Mtb-mCherry were generated using Mtb that had been passaged 2 or fewer times in vitro. For these fluorescent strains, Mtb was grown in Middlebrook 7H9 liquid medium supplemented with 10% albumin-dextrose-saline, 0.4% glycerol, and 0.05% Tween-80 for 5 days at 37°C. The cells were pelleted and washed in 10% glycerol to remove salt. The bacteria were then electroporated with 1 μg DNA using a 2 mm electroporation cuvette and the following settings: 2500 volts, 1000 Ohms, 25 μF. The pTEC15 plasmid36 (Addgene plasmid # 30174), which expresses Wasabi under the control of the Mycobacterium Strong Promoter, was electroporated into Mtb to generate Mtb-Wasabi36. The pMSP12::mCherry plasmid (a gift from Lalita Ramakrishnan, University of Cambridge; Addgene plasmid # 30167), which expresses mCherry under the control of the Mycobacterium Strong Promoter, was electroporated into Mtb to generate Mtb-mCherry. Following electroporation, bacteria were grown on 7H11 plates supplemented with 10% oleic acid, albumin, dextrose, and catalase, 0.5% glycerol, and either 200 μg / mL Hygromycin for Mtb-Wasabi or 50 μg / mL Kanamycin for Mtb-mCherry for 3–4 weeks at 37°C. Individual colonies where then propagated in 10 mL inkwell flask cultures using 7H9 medium supplemented with 10% albumin-dextrose-saline, 0.4% glycerol, 0.05% Tween-80, and either Hygromycin for Mtb-Wasabi or Kanamycin for Mtb-mCherry for 7 days at 37°C. The inkwell cultures were expanded into a 100 mL culture using the same 7H9 supplemented media with antibiotics and cultured for 4–5 days at 37°C. Once the bacteria were in log phase, the culture was filtered with a 5 μm syringe filter and frozen in 1 mL aliquots in 10% glycerol.
METHOD DETAILS
Generation of the I-TOMCAT IFNβ reporter mice
Targeting of C57BL6/J embryonic stem (ES) cells and generation of chimeric mice was performed by Biocytogen. A construct was targeted immediately downstream of the endogenous Ifnb stop codon using Cas9 and a gRNA targeting the genomic site TGCAACCACCACTCATTCTGAGG; the underlined sequence represents the protospacer adjacent motif (PAM). The construct included an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES), coding sequence for the TdTomato red fluorescent protein, a picornavirus T2A “self-cleaving” peptide, a nuclear localization sequence (NLS)-containing Cre recombinase, and a bovine growth hormone (BGH) polyadenylation (pA) sequence that bypasses the endogenous polyadenylation site in the 3’ untranslated region of the Ifnb gene.
After the BGH pA sequence, the construct contained a FRT site-flanked phosphoglycerate kinase (PGK)-neomycin resistance cassette. The insertion cassette was flanked by ~2 kilobase homology arms on either side, and a Diptheria Toxin A (DTA) gene at the 3’ end to select against random insertions. Successful targeting of ES cells was confirmed by PCR. After germline transmission, the knockin mice were confirmed by sequencing and then bred to FLPer mice104 to remove the FRT-flanked neo cassette.
Mtb infections
For infection, a frozen aliquot of Mtb-Wasabi or Mtb-mCherry was diluted in distilled H2O, and 9 mL of diluted culture was loaded into the nebulizer of a inhalation exposure system (Glas-Col, Terre Haute, IN) to deliver ~20–100 bacteria per mouse as determined by measuring CFU in lungs 1 day post-infection.
Tissue Processing for CFU and Flow cytometry
Mice were harvested at various days post-infection (as described in figure legends) to measure CFUs by plating and innate immune populations by flow cytometry. All lung lobes were harvested into a gentleMACS C tube (Miltenyi Biotec) containing 3 mL of RPMI media with 70 μg / mL of Liberase TM (Roche) and 30 μg / mL of Dnase I (Roche). Samples were processed into chunks using the lung_01 setting on the gentleMACS (Miltenyi Biotec) and incubated for 30 minutes at 37°C. Tissue was then homogenized into a single cell suspension by running the samples on the lung_02 setting on the gentleMACS. The digestion was quenched by adding 2 mL of PBS with 20% Newborn Calf Serum (Thermo Fisher Scientific) and filtered through 70 μm SmartStrainers (Miltenyi Biotec).
For measuring plasmacytoid dendritic cell numbers, spleens were harvested into a 12 well plate with 1 mL of PBS with 2% Newborn Calf Serum and 0.05% sodium azide in each well. The spleens were sandwiched between 100 uM mesh filters and mashed into a single cell suspension with the back of a syringe plunger. The single cell suspensions were filtered through 70 μm SmartStrainers (Miltenyi Biotec).
Measuring Bacterial Burden
To measure CFU, 50 μL was taken from each single cell suspension and then serially diluted in phosphate-buffered saline (PBS) with 0.05% Tween-80. Serial dilutions were plated on 7H11 plates supplemented with 10% oleic acid, albumin, dextrose, and catalase and 0.5% glycerol. Colonies were counted after 3 weeks.
Flow Cytometry
For flow cytometry, lung single cell suspensions were pelleted and resuspended in 500 μL of PBS with 2% Newborn Calf Serum and 0.05% Sodium azide and 100–150 μL were stained with antibodies for analysis. Spleen single cell suspensions were pelleted and resuspended in 5 mL of PBS with 2% Newborn Calf Serum and 0.05% Sodium azide, of which 50 μL were stained with antibodies. Single cell suspensions were stained for 45 minutes to an hour at room temperature with the following antibodies: TruStain FcX PLUS (S17011E, BioLegend), BUV496-labeled CD45 (30-F11, BD Biosciences), APC-labeled CD64 (X54–5/7.1, BioLegend), BV480-labeled B220 (RA3–6B2, BD Biosciences), BV480-labeled CD90.2 (53–2.1, BD Biosciences), APC-Fire 750-labeled Ly6G (1A8, BioLegend), BUV395-labeled CD11b (M1/70, BD Biosciences), BUV737-labeled CD11c (HL3, BD Biosciences), APC-R700-labeled Siglec F (E50–2440, BD Biosciences), PE-labeled MerTK (DS5MMER, Thermo Fisher Scientific), Super Bright 645-labeled MHC II (M5/114.15.2, Thermo Fisher Scientific), BV421-labeled PD-L1 (MIH5, BD Biosciences), BV711-labeled Ly6C (HK1.4, BioLegend), PE-labeled IFNAR-1 (MAR1–5A3, BioLegend), PE-Cy7-labeled MerTK (DS5MMER, Thermo Fisher Scientific), APC-eFluor 780-labeled CD11b (M1/70, Thermo Fisher Scientific), BUV395-labeled CCRL2 (BZ2E3, BD Biosciences), BUV563-labeled Ly6G (1A8, BD Biosciences), Percp-Cy5.5-labeled B220 (RA3–6B2, BioLegend), BV421-labeled Siglec H (440c, BD Biosciences), BV480-labeled CD19 (1D3, BD Biosciences), BV605-labeled MHC II (M5/114.15.2, BioLegend), BV785-labeled Ly6C (HK1.4, BioLegend), BV605-labeled CD4 (GK1.5, BioLegend), BUV805-labeled CD8ɑ (53–6.7, BD Biosciences), and PE-Cy7-labeled PDCA-1 (eBio927, Thermo Fisher Scientific). All samples also received fixable viability dye (Ghost Dye Violet 510; Tonbo Biosciences), Super Bright Complete Staining Buffer (Thermo Fisher Scientific), and True-Stain Monocyte Blocker (BioLegend) at the same time as the antibodies. Stained samples were fixed with cytofix/cytoperm (BD biosciences) for 20 minutes at room temperature before samples were removed from the BSL3. For intracellular staining, the fixed samples were stained with the following antibodies: PE-Cy7-labeled CD63 (NVG-2, BioLegend), Percp-eFluor 710-labeled iNOS (CXNFT, Thermo Fisher Scientific), BV785-labeled CD206 (C068C2, BioLegend). Cell numbers were calculated by adding fluorescent AccuCheck Counting Beads (Invitrogen) to each sample. Cells were then analyzed on a Fortessa (BD Biosciences) or an Aurora (Cytek) flow cytometer. Data were analyzed with Flowjo version 10 (BD Biosciences).
Immune cell depletion
Injections for cell depletions were started 12 days after Mtb infection and continued every other day until the mice were harvested 25 days post-infection. Antibody depletion of pDCs was performed by administering 200 μg of anti-BST2 (927, BioXCell) or rat IgG2b isotype control antibody (LTF-2, BioXCell) in 200 μL PBS via intraperitoneal injection59. Genetic depletion involved administering 100 ng diphtheria toxin (Millipore Sigma) in 100 μL PBS via intraperitoneal injection into Sp140−/− pDC-DTR mice and littermate controls61. Antibody depletion of neutrophils was performed by administering 200 μg of anti-Ly6G (1A8, BioXCell) or rat IgG2a isotype control antibody (2A3, BioXCell) in 200 μL PBS via intraperitoneal injection.
Immunostaining human lymph nodes and lungs
Human lung and lymph node samples were acquired from the surgical pathology archives of Emory University Hospital with appropriate institutional approval. 8 lung samples and 8 lymph node samples were analyzed. Each sample had been previously culture verified for Mycobacterium tuberculosis infection. The samples were formalin-fixed and paraffin-embedded. Sections were cut and stained with anti-CD123 (6h6, Thermo Fisher Scientific), anti-CD303 (124B3.13, Dendritics), or hematoxylin and eosin107,108. Primary antibodies were detected by immunoperoxidase staining with the LSAB+ System and a standard DAB reaction following manufacturer’s instructions (DakoCytomation). Sections were counterstained with hematoxlyin prior to mounting and microscopy. pDCs were assessed in multiple 400× fields for each section to calculate the frequency of samples containing pDCs and the clustering of pDCs within each sample, defined as either singe cells, loose clusters of 5–20 cells, or tight clusters of more than 20 cells.
Confocal microscopy
Confocal Microscopy was performed using a Zeiss LSM 880 laser scanning confocal microscope (Zeiss) equipped with two photomultiplier detectors, a 34-channel GaASP spectral detector system, and a 2-channel AiryScan detector as well as 405, 458, 488, 514, 561, 594, and 633 lasers. 20 μm paraformaldehyde fixed lung sections from Mtb-mCherry infected I-Tomcat Ai6 and Sp140−/− I-Tomcat Ai6 mice were stained at 4°C overnight with BV421-labeled SIRPα (P84, BD Biosciences), Pacific Blue–labeled B220 (RA3–6B2, BioLegend), eF506-labeled CD4 (RM4–5, BioLegend), and AF647-labeled Ly6G (1A8, BioLegend). Sections detecting the presence of Neutrophil extracellular traps (NETs) were stained with BV421-labeled Ly6G (1A8, BioLegend) and rabbit polyclonal anti-citrullinated histone-H3 (citrulline R2, R8, R17; Abcam), stained with AF488 donkey anti-rabbit secondary (Poly4064, BioLegend). Stained sections were inspected with a 5× air objective to find representative lesions and distal sites and then imaged using a 63× oil immersion objective lens with a numerical aperture of 1.4. For each infected lung, one Mtb-heavy lesion image and one distal site image was taken consisting of 20 μm z-stacks acquired at a 1.5 μm step size. For representative NET images, 4×4 tiled images were captured without a z-stack. Additionally, the Zeiss LSM 880 microscope was used to image single color-stained Ultracomp eBeads Plus (Thermo Fisher Scientific) for generating a compensation matrix.
Image processing and histo-cytometry analysis
Image analysis was performed using Chrysalis software55. Briefly, a compensation matrix was generated by automatic image-based spectral measurements on single color-stained controls in ImageJ by using Generate Compensation Matrix script. This compensation matrix was used to perform linear unmixing on three-dimensional images with Chrysalis. Chrysalis was also used for further image processing, including rescaling data and generating new channels by performing mathematical operations using existing channels. For histo-cytometry analysis, Imaris 9.9.1 (Bitplane) was used for surface creation to digitally identify cells in images based on protein expression54. Statistics for the identified cells were exported from Imaris and then imported into FlowJo version 10 (BD Biosciences) for quantitative image analysis.
Bulk RNA-seq sample preparation and analysis
RNA-seq of Mtb-infected Sp140-sufficient and -deficient mouse lungs genetically depleted of pDCs was performed on 20% of each lung single cell suspension, prepared as described for CFU and flow cytometry analysis. Single cell suspensions were preserved in Trizol LS (Thermo Fisher Scientific) and stored at −80°C. The samples were thawed at room temperature for 5 minutes, then 200 μL of chloroform (Thermo Fisher Scientific) was added per 0.75 mL of Trizol LS to each sample and the samples were removed from the BSL3. Samples were centrifuged in Phasemaker tubes (Thermo Fisher Scientific) to isolate total RNA, which was then purified following the RNeasy Micro (Qiagen) protocol starting at the ethanol addition step. Library preparation, sequencing, and read alignment to the mouse genome was performed by Azenta Life Sciences. Libraries were prepared from total RNA using an Illumina kit for Poly(A) selection. Samples were sequenced on an Illumina HiSeq sequencer with paired 150 bp reads to a depth of 20–30 million reads per sample. Sequence reads were trimmed of adapter sequences and low quality nucleotides with Trimmomatic v.0.36109 and then mapped to the Mus musculus GRCm38 reference genome with STAR aligner v.2.5.2b110. The raw counts were used as input for DESeq2111 analysis of differential gene expression.
RNA-seq of cytokine stimulated bone marrow-derived macrophages was performed by differentiating bone marrow from B6 mice in DMEM supplemented with 10% FBS, PenStrep, Glutamin, HEPES, and M-CSF for 7 days then reseeding the cells in a 6-well plate and incubating for 5 days at 37°C and 5% CO2. The macrophages were left untreated or stimulated with 10 ng / mL of IFN-β (BioLegend), IFNγ (Abcam), TNF (Peprotech), or transforming growth factor-β (BioLegend) for 6 hours. Cells were lysed with TRK lysis buffer (Omega Bio-Tek) and 2-mercaptoethanol (Thermo Fisher Scientific) followed by total RNA isolation using the E.Z.N.A Total RNA Kit I (Omega Bio-Tek). The library preparation, sequencing, and read alignment to the mouse genome was performed by Azenta Life Sciences as described for the Mtb-infected mouse lung samples. Raw counts were used as input for analysis with DESeq2111.
Sorting Immune Cells for scRNA-seq analysis
3 B6 and 3 Sp140−/− mice were infected with less than 100 CFU of Mtb-Wasabi bacteria. Lungs from infected animals as well as 2 naïve control mice per genotype were harvested 25 days post-infection and processed as described for CFU and flow cytometry analysis. Single cell suspensions were resuspended in PBS with 2% Newborn Calf Serum and stained with TruStain FcX PLUS (S17011E, BioLegend), APC-labeled Ly6G (1A8, BioLegend), APC-labeled CD64 (X54–5/7.1, BioLegend), and TotalSeq-A-labeled Ly6G (1A8, BioLegend) anti-mouse antibodies on ice for 30 minutes. Myeloid cells were then magnetically enriched using an EasySep APC Positive Selection Kit II (StemCell Technologies) and MojoSort Magnets (BioLegend). All enriched samples were stained with the following panel of TotalSeq-A-labeled anti-mouse antibodies to detect protein expression in the scRNA-seq dataset: Ly6C (HK1.4, BioLegend), CD44 (IM7, BioLegend), CD169 (3D6.112, BioLegend), CD274 (MIH6, BioLegend), Siglec F (S17007L, BioLegend), CSF1R (AFS98, BioLegend), CD11b (M1/70, BioLegend), CD86 (GL-1, BioLegend), MHC II (M5/114.15.2, BioLegend), and CX3CR1 (SA011F11, BioLegend). Each sample was also stained with a unique anti-mouse TotalSeq-A Hashtag antibody (1–6; BioLegend) to allow up to 6 populations to be multiplexed in a single lane on a 10X Genomics Chromium Next GEM Chip112. Enriched cells from infected mice were also stained with PE-labeled B220 (RA3–6B2, Tonbo), PE-labeled CD90.2 (30-H12, Tonbo), and BV785-labeled CD45.2 (104, BioLegend) anti-mouse antibodies. Enriched cells from naïve mice were stained with Pacific Blue-labeled B220 (RA3–6B2, BioLegend), Pacific Blue-labeled CD90.2 (53–2.1, BioLegend), PE-labeled F4/80 (BM8, Thermo Fisher Scientific), and BV785-labeled CD45.2 (104, BioLegend) anti-mouse antibodies. All post-enrichment antibody staining was performed on ice for 45 minutes in the presence of True-Stain Monocyte Blocker (BioLegend). Following staining, cells were resuspended in PBS with 2% Newborn Calf Serum and Sytox Blue Dead Cell Stain (Thermo Fisher Scientific). Cells were sort purified using a 100 μm microfluidic sorting chip in a 4 laser SH-800 cell sorter (Sony) on the purity setting. The isolated populations from infected lungs were Mtb-infected cells and bystander myeloid cells. Macrophages and a mixture of neutrophils and monocytes were isolated from the naïve lungs and the macrophages were combined with neutrophil/monocyte mixture at a 1:2 ratio for better macrophage representation in the resulting dataset.
Single cell RNA: Library generation and sequencing
The scRNA-sequencing libraries were generated using the v3.1 chemistry Chromium Single Cell 3’ Reagent Kit (10X Genomics) largely following the manufacturer protocol with the following minor modifications. Cells were loaded into 3 different lanes on a Chromium Next GEM Chip. Lane 1 was loaded with Mtb-infected cells from all 3 B6 and 3 Sp140−/− lungs. Lane 2 was loaded with bystander myeloid cells from the 3 infected B6 lungs as well as the myeloid cell mixture from the 2 naïve B6 lungs. Lane 3 was loaded with bystander myeloid cells from the 3 infected Sp140−/− lungs as well as the myeloid cell mixture from the 2 naïve Sp140−/− lungs. All 3 lanes of the Chromium Next GEM Chip were super-loaded with 29000 cells with a target of 14800 single cells per lane, as hashtag barcoding allows for a lower effective multiplet rate due to the ability to identify most of the multiplets (https://satijalab.org/costpercell/) 112. 0.5 U/μL RNaseOUT Recombinant Ribonuclease Inhibitor (Invitrogen) was added to single cell RT master mix during the loading step and 1 μL of ADT and HTO additive primers (0.2 μM stock) were added during the cDNA amplification, as recommended by the CITE-seq and Cell Hashing Protocol (https://cite-seq.com/protocols/)46. Following cDNA, ADT, and HTO purification, samples were decontaminated by 2 rounds of centrifugation through 0.2 μM filter microcentrifuge tubes and then removed from the BSL3. Library preparations were completed outside of the BSL3 following the 10X Genomics protocol for the cDNA and the CITE-seq and Cell Hashing Protocol for the ADT and HTO libraries. Quality control of the libraries was performed with a Fragment Analyzer (Agilent). The mRNA, ADT, and HTO libraries were pooled at the following proportions: 85% mRNA, 9% ADT, and 6% HTO. Libraries were sequenced on a NovaSeq 6000 (Illumina) using two lanes of a S1 flow cell and the following cycles read 1 (28 cycles), i7 index (10 cycles), i5 index (10 cycles), read 2 (90 cycles).
ScRNA-seq: data processing
Raw sequencing reads for the mRNA libraries were processed into raw count matrices with CellRanger version 4.0.0 (10X Genomics). The ADT and HTO libraries were processed into raw count matrices with CITE-Seq-Count version 1.4.3 (https://hoohm.github.io/CITE-seq-Count/) 113. The raw counts for mRNA, ADT, and HTO were analyzed in R114 via the RStudio integrated development environment with Seurat v4.1.142 using default settings for normalizing the data, finding variable features, and scaling the data. HTO demultiplexing was performed with the HTODemux function. Data was filtered to only include single cells with between 200 and 4500 genes and less than 5% mitochondrial reads. The resulting datasets were integrated together using 30 dimensions for the FindIntegrationAnchors function and 30 dimensions for the IntegrateData function. The data was then scaled and analyzed by PCA with 30 principal components followed by UMAP analysis with 30 dimensions. Clustering was performed by using 30 dimensions with the FindNeighbors function and a resolution of 0.8 for the FindClusters function.
To improve resolution for clustering innate immune cells, weighted nearest neighbor analysis was used to combine the protein data (ADTs) and the mRNA data when clustering cells. For this analysis, variable ADT features were identified and then normalized using centered log ratio transformation and a margin of 2. The normalized ADT data was then scaled and analyzed by PCA. The ADT and mRNA data was then combined with the FindMultiModalNeighbors function using 30 dimensions for the mRNA and 10 for the protein. The resulting dataset was analyzed by UMAP and clusters were identified with the FindClusters function using algorithm 3 and a resolution of 1.544. Tidyverse115, EnhancedVolcano116, and various Seurat functions were used for plotting the scRNA-seq data.
Type I IFN and IFNγ Gene Signature Analysis
For generating the type I IFN and IFNγ gene signatures, we utilized a published RNA-seq dataset (GEO: GSE20251) of primary human macrophages that were unstimulated or stimulated with 10 ng/mL of TNF, IFNγ, IFN-β, transforming growth factor-β, or other ligands for 24 hours and then processed for RNA-sequencing65. We also generated an RNA-seq dataset mouse bone marrow-derived macrophages left unstimulated or stimulated with 10 ng/mL of TNF, IFNγ, IFN-β, or transforming growth factor-β for 6 hours. For both datasets, counts were normalized by DESeq2’s median of ratios method and biological replicates were averaged to generate average normalized counts for each cytokine condition. The average normalized counts were used to calculate a max to second-max ratio for the 4 cytokine conditions to determine gene specificity. Next, gene expression was compared across all stimulation conditions and filtered to only include genes that were induced relative to the untreated cells by a log2 fold change of 1 by IFNγ or IFN-β and to exclude genes induced by a log2 fold change of 1.5 by TNF or transforming growth factor-β. Using this filtered gene list, a ratio was calculated of the log2 fold change following IFNγ stimulation to the log2 fold change following IFN-β stimulation. For the human dataset, IFN-β specific genes were defined as those having a fold change ratio < 0, a log2 fold change upon IFN-β stimulation > 1, an average normalized count following IFN-β stimulation > 2000 and a max to second-max ratio > 2.5. Human IFNγ specific genes were defined as those with a fold change ratio > 1.5, a log2 fold change upon IFNγ stimulation > 2, an average normalized count following IFNγ stimulation > 1000 and a max to second-max ratio > 2.5. Mouse macrophage IFN-β specific genes were defined as those with a fold change ratio < 0.66, a log2 fold change upon IFN-β stimulation > 4, an average normalized count following IFN-β stimulation > 4000 and a max to second-max ratio > 3. Mouse IFNγ specific genes were defined as those that had a fold change ratio > 1.5, a log2 fold change upon IFNγ stimulation > 2, an average normalized count following IFNγ stimulation > 500 and a max to second-max ratio > 3.
Human IFN-β and IFNγ gene signatures were validated by examining their induction following human macrophage stimulation with a variety of cytokines, such as IL-4, IL-6 and IL-10, using the published RNA-seq dataset (GEO: GSE20251). The specificity of the mouse macrophage IFNγ signature was validated by stimulating bone marrow-derived macrophages for >16 hours with 10 ng/mL of IFNγ, IFN-β, or nothing and then examining CXCL9 expression by flow cytometric analysis of intracellular staining with PE-labeled anti-mouse CXCL9 antibody (MIG-2F5.5; BioLegend). The mouse gene signatures were then used to score cells in the mouse myeloid scRNA-seq dataset based on their gene expression with the UCell R package117.
QUANTIFICATION AND STATISTICAL ANALYSIS
Group sizes were informed by the results of preliminary experiments and power calculations. The number of animals in each figure is indicated in the legends as n = x mice per group. Statistical significance was determined using Prism (GraphPad) software for unpaired one-tailed or two-tailed Student t test when comparing two populations, one-way or two-way ANOVA tests with Tukey’s or Sidak’s multiple comparisons test when comparing multiple groups. Prism (GraphPad) was also used to calculate linear correlations and R2. R was used to calculate statistical significance for bulk RNA- and scRNA-sequencing datasets using the Wald test with multiple testing correction by the Benjamini and Hochberg method and the Wilcoxon Rank-Sum test with Bonferroni correction, respectively. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s. = not significant. See figure legends for more information on statistical tests.
Supplementary Material
Supplementary Figure 1. Identifying innate immune cell populations in Mtb-infected lungs by flow cytometry and scRNA-seq. Related to Figure 1 and 2. (A) Gating strategy for identifying neutrophils, eosinophils, monocytes, DCs, AMs, IMs, conventional type 1 DCs (cDC1), conventional type 2 DCs (cDC2) and Mtb-infected cells. Very few Mtb+ cells were detected among the lymphoid and non-hematopoietic-derived cells. (B) Cell clustering by mRNA expression, protein expression, or combined mRNA and protein expression (B6 and Sp140−/− combined). (C) Cell clustering by biological replicate in naïve and infected B6 and Sp140−/− mice. (D) Protein and mRNA expression of lineage-defining markers used for cluster classification. Markers used for annotating (E) neutrophil and (F) monocyte and macrophage clusters based on maturity, activation status, and specific gene expression. The data represents a total of 10 mice (n = 3 for the infected lung samples and n = 2 for the naïve lung samples from B6 and Sp140−/− mice).
Supplementary Figure 2. Naïve Sp140−/− mice exhibit minimally altered immune cell numbers by flow cytometry and scRNA-seq. Related to Figure 2. Comparison of innate and adaptive immune cell numbers between B6 (n = 7; closed circles) and Sp140−/− (n = 8; open circles) (A) spleen, (B) lung, (C) and thymus. (D) Clustering of myeloid cells from naïve lungs of B6 (n = 2) and Sp140−/− (n = 2) mice. (E) Differentially expressed genes between B6 and Sp140−/− AMs, IMs, monocytes, and neutrophils. Greater fold change indicates higher expression in B6 relative to Sp140−/−. The bars in (A), (B), and (C) represent the median. Pooled data from two independent experiments are shown in (A), (B), and (C). Statistical significance was calculated by multiple unpaired t tests in (A), (B), and (C), and by Wilcoxon Rank-Sum test with Bonferroni correction in (B). *p < 0.05.
Supplementary Figure 3. I-Tomcat cells express TdTomato following in vitro stimulation, but the TdTomato signal is undetectable in vivo during Mtb infection. Using I-Tomcat Ai6 mice, pDC, IMs, and monocytes are the major type I interferon producing cells following Mtb infection in B6 and Sp140−/− mice. Related to Figure 3. (A) We targeted the mouse Ifnb locus immediately downstream of the ORF and upstream of the endogenous polyadenylation (pA) site using CRISPR in C57BL/6 ES cells, inserting a reporter cassette containing the indicated elements. Targeted mice were then bred to FLPer mice to excise the Neo cassette.(B) Representative flow cytometry histogram of TdTomato expression by I-Tomcat bone marrow-derived macrophages that were unstimulated (grey) or stimulated with poly I:C (red line). (C) Representative flow cytometry plot of TdTomato expression in immune cells in Mtb-infected lungs of Sp140−/− I-Tomcat−/−, Sp140−/− I-Tomcat+/−, and Sp140−/− I-Tomcat+/+ mice 25 days after infection. (D) Representative flow cytometry plot of TdTomato expression in AM, pDCs, monocytes, and IM from Mtb-infected lungs of Sp140−/− I-Tomcat+/+ mice 19 days post-infection or (E) 25 days post-infection. Frequency of Ai6 expression by immune cell type in the lungs of Mtb-infected (F) Sp140−/− I-Tomcat Ai6 and (G) I-Tomcat Ai6 mice. The bars in (E) and (F) represent the median. Lungs were analyzed 25 days after Mtb infection. Pooled data from two independent experiments. Statistical significance was calculated by one-way ANOVA with Tukey’s multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Figure 4. pDC-DTR mice specifically deplete pDCs without affecting major lung immune cell populations, and CD123 also identifies pDCs in Mtb-infected human lymph node samples. Related to Figure 5. (A) Representative flow cytometry plot of splenic pDCs in Sp140+/− pDC-DTR mice treated with PBS or DT from days 12 to 24 after Mtb infection. (B) RNA-sequencing validation of Sp140 expression and pDC depletion efficiency based on Siglech expression in Sp140+/− pDC-DTR mice treated with PBS or DT from days 12 to 24 after Mtb infection. (C) Number of various immune cell populations in Mtb-infected lungs of Sp140+/− pDC-DTR (n = 13; filled circles) and Sp140+/− mice (n = 13; open circles). Mice received DT from days 12 to 24 post-infection. (D) anti-CD123 (brown) and hematoxylin staining on Mtb-infected human lymph nodes. Mouse lungs and spleens were harvested 25 days after infection. Pooled data from two independent experiments are shown in (B) and (C). The bars in (C) represent the median. Statistical significance in (B) was calculated by the Wald test with multiple testing correction using the Benjamini and Hochberg method and in (C) by multiple unpaired t tests. *p < 0.05, ***p < 0.001, ****p < 0.0001.
Supplementary Figure 5. Generating gene signatures for identifying IFNγ and type I IFN responding cells. Related to Figure 7. (A) Plots depicting the log2 fold change of genes upregulated in human macrophages or mouse bone marrow-derived macrophages following stimulation with IFNγ or IFN-β. Dots colored red indicate genes used for the IFNγ signaling gene signature while blue dots indicate those used for the gene signature for type I IFN responsiveness. Representative genes induced preferentially by IFNγ or IFN-β are labeled. (B) Number of genes upregulated in human macrophages following stimulation with each indicated cytokine and (C) Number of type I or II IFN gene signature genes induced after cytokine stimulation. (D) Representative flow cytometry histogram of CXCL9 expression by mouse bone marrow-derived macrophages that were untreated (grey), IFN-β stimulated (blue), or IFNγ stimulated (red).
Supplementary Figure 6. Applying gene signatures for identifying IFNγ and type I IFN responding cells to the myeloid scRNA-seq dataset. Related to Figure 7. (A) List of genes from the cytokine stimulated mouse macrophages that were used for the gene signature for IFNγ or type I IFN responsiveness. (B) IFNγ gene signature or (C) type I IFN gene signature expression visualized by wnnUMAP plots of naïve, bystander, or Mtb-infected lung myeloid cells from B6 and Sp140−/− mice. (D) IFNγ and type I IFN gene signature expression on neutrophils, monocytes, IM, and AM from naïve B6 and Sp140−/− mouse lungs. The red line indicates the 99% cutoff used to classify cells as type I or II IFN responders. (E) Comparison of naïve, bystander, and Mtb-infected cells classified as cells that responded to type I IFN (blue), type II IFN (red), both (purple), or neither (grey) in lungs (B6 and Sp140−/− combined).
Supplementary Figure 7. IFNγ receptor and IFNγ responsive genes are expressed at lower levels in Mtb-infected IMs in Sp140−/− relative to B6 mice. Related to Figure 7. (A) Ifngr1 and Ifngr2 expression on bystander and Mtb-infected cells from B6 and Sp140−/− mice. (B) Expression of Ifngr1 and Ifngr2, (C) representative type I IFN stimulated genes, including Isg15, Ifit1, and Oasl1, and (D) representative type II IFN stimulated genes, including H2-Ab1 mRNA, MHC II protein, and Cxcl9 mRNA on bystander and Mtb-infected IM and ISG+ IM from B6 and Sp140−/− mice. (E) Gene ontology term enrichment of the Hallmark Interferon-a Response and the REACTOME Translation terms on Mtb-infected IM and ISG+ IM from B6 and Sp140−/− mice. Statistical significance in (B), (C), and (D) was calculated by non-parametric Wilcoxon rank sum test with Bonferroni correction. ***p < 0.001, ****p < 0.0001.
Highlights.
Sp140-deficient mice model type I interferon-driven tuberculosis disease seen in humans
Plasmacytoid dendritic cells and interstitial macrophages produce IFN during tuberculosis
Plasmacytoid dendritic cell depletion rescues tuberculosis susceptibility
Type I interferons act on infected macrophages to impair bacterial control
Acknowledgments
We thank members of the Vance, Barton, Stanley, and Cox laboratories for advice and discussions; M. Colonna for advice on pDC depletion; B. Malissen, Y. Belkaid, and A. Lacy-Hulbert for mouse lines; P. Dietzen, R. Chavez, and J. Morales for technical assistance; A. Valeros, H. Nolla, and K. Heydari of the UC Berkeley Cancer Research Laboratory Flow Cytometry facility for assistance with flow cytometry; H. Aaron and F. Ives of the UC Berkeley Cancer Research Laboratory Molecular Imaging Center (RRID:SCR_017852) for assistance with confocal microscopy (supported by the Helen Wills Neuroscience Institute). Model figures were created with BioRender.com. D.I.K. was supported by an NIH Postdoctoral Fellowship (F32HL158250) and S.A.F. by a Postdoctoral Fellowship from EMBO (ALTF 617–2021) and the Swiss National Science Foundation (P500PB_206801). R.E.V. is an Investigator of the Howard Hughes Medical Institute and is funded by NIH grants AI075039, AI063302 and AI155634.
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Declaration of Interests
R.E.V. consults for Ventus Therapeutics, Tempest Therapeutics, and X-biotix Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.WHO (2021). Global tuberculosis report 2021. https://www.who.int/publications-detail-redirect/9789240037021. [Google Scholar]
- 2.Katelaris AL, Jackson C, Southern J, Gupta RK, Drobniewski F, Lalvani A, Lipman M, Mangtani P, and Abubakar I. (2020). Effectiveness of BCG Vaccination Against Mycobacterium tuberculosis Infection in Adults: A Cross-sectional Analysis of a UK-Based Cohort. J. Infect. Dis 221, 146–155. 10.1093/infdis/jiz430. [DOI] [PubMed] [Google Scholar]
- 3.Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, and Orme IM (1993). Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med 178, 2243–2247. 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, Schreiber R, Mak TW, and Bloom BR (1995). Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity 2, 561–572. 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 5.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, and Bloom BR (1993). An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med 178, 2249–2254. 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell DG, Barry CE, and Flynn JL (2010). Tuberculosis: what we don’t know can, and does, hurt us. Science 328, 852–856. 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry MPR, Graham CM, McNab FW, Xu Z, Bloch SAA, Oni T, Wilkinson KA, Banchereau R., Skinner J., Wilkinson RJ., et al. (2010). An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977. 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreira-Teixeira L, Mayer-Barber K, Sher A, and O’Garra A. (2018). Type I interferons in tuberculosis: Foe and occasionally friend. J. Exp. Med 215, 1273–1285. 10.1084/jem.20180325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scriba TJ, Penn-Nicholson A, Shankar S, Hraha T, Thompson EG, Sterling D, Nemes E, Darboe F, Suliman S, Amon LM, et al. (2017). Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLOS Pathog. 13, e1006687. 10.1371/journal.ppat.1006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez L, Nicol MP, Wedderburn CJ, Stadler A, Botha M, Workman L, le Roux DM, and Zar HJ (2021). Cytomegalovirus acquisition in infancy and the risk of tuberculosis disease in childhood: a longitudinal birth cohort study in Cape Town, South Africa. Lancet Glob. Health 9, e1740–e1749. 10.1016/S2214-109X(21)00407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller J, Tanner R, Matsumiya M, Snowden MA, Landry B, Satti I, Harris SA, O’Shea MK, Stockdale L, Marsay L, et al. (2019). Cytomegalovirus infection is a risk factor for tuberculosis disease in infants. JCI Insight 4, 130090. 10.1172/jci.insight.130090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walaza S, Tempia S, Dawood H, Variava E, Moyes J, Cohen AL, Wolter N, Groome M, von Mollendorf C, Kahn K, et al. (2015). Influenza virus infection is associated with increased risk of death amongst patients hospitalized with confirmed pulmonary tuberculosis in South Africa, 2010–2011. BMC Infect. Dis 15, 26. 10.1186/s12879-015-0746-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang G, deWeerd NA, Stifter SA, Liu L, Zhou B, Wang W, Zhou Y, Ying B, Hu X, Matthews AY, et al. (2018). A proline deletion in IFNAR1 impairs IFN-signaling and underlies increased resistance to tuberculosis in humans. Nat. Commun 9, 85. 10.1038/s41467-017-02611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singhal A, Jaiswal A, Arora VK, and Prasad HK (2007). Modulation of Gamma Interferon Receptor 1 by Mycobacterium tuberculosis: a Potential Immune Response Evasive Mechanism. Infect. Immun 75, 2500–2510. 10.1128/IAI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teles RMB, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, Komisopoulou E, Kelly-Scumpia K, Chun R, Iyer SS, et al. (2013). Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science 339, 1448–1453. 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang TG, Kwon KW, Kim K, Lee I, Kim MJ, Ha S-J, and Shin SJ (2022). Viral coinfection promotes tuberculosis immunopathogenesis by type I IFN signaling-dependent impediment of Th1 cell pulmonary influx. Nat. Commun 13, 3155. 10.1038/s41467-022-30914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redford PS., Mayer-Barber KD., McNab FW., Stavropoulos E., Wack A., Sher A., and O’Garra A. (2014). Influenza A Virus Impairs Control of Mycobacterium tuberculosis Coinfection Through a Type I Interferon Receptor–Dependent Pathway. J. Infect. Dis 209, 270–274. 10.1093/infdis/jit424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volkert M, Pierce C, Horsfall FL, and Dubos RJ (1947). THE ENHANCING EFFECT OF CONCURRENT INFECTION WITH PNEUMOTROPIC VIRUSES ON PULMONARY TUBERCULOSIS IN MICE. J. Exp. Med 86, 203–214. 10.1084/jem.86.3.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, Snell LM, Guo M, Boukhaled G, Macleod BL, Li M, Tullius MV, Guidos CJ, Tsao M-S, Divangahi M, et al. (2021). Early innate and adaptive immune perturbations determine long-term severity of chronic virus and Mycobacterium tuberculosis coinfection. Immunity 54, 526–541.e7. 10.1016/j.immuni.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ring S, Eggers L, Behrends J, Wutkowski A, Schwudke D, Kröger A, Hierweger AM, Hölscher C, Gabriel G, and Schneider BE Blocking IL-10 receptor signaling ameliorates Mycobacterium tuberculosis infection during influenza-induced exacerbation. JCI Insight 4, e126533. 10.1172/jci.insight.126533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNab FW, Ewbank J, Rajsbaum R, Stavropoulos E, Martirosyan A, Redford PS, Wu X, Graham CM, Saraiva M, Tsichlis P, et al. (2013). TPL-2-ERK1/2 signaling promotes host resistance against intracellular bacterial infection by negative regulation of type I IFN production. J. Immunol. Baltim. Md 1950 191, 1732–1743. 10.4049/jimmunol.1300146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreira-Teixeira L, Redford PS, Stavropoulos E, Ghilardi N, Maynard CL, Weaver CT, Freitas do Rosário AP, Wu X, Langhorne J, and O’Garra A. (2017). T Cell-Derived IL-10 Impairs Host Resistance to Mycobacterium tuberculosis Infection. J. Immunol. Baltim. Md 1950 199, 613–623. 10.4049/jimmunol.1601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanley SA, Johndrow JE, Manzanillo P, and Cox JS (2007). The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. Baltim. Md 1950 178, 3143–3152. 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 24.Mayer-Barber KD, Andrade BB, Barber DL, Hieny S, Feng CG, Caspar P, Oland S, Gordon S, and Sher A. (2011). Innate and Adaptive Interferons Suppress IL-1α and IL-1β Production by Distinct Pulmonary Myeloid Subsets during Mycobacterium tuberculosis Infection. Immunity 35, 1023–1034. 10.1016/j.immuni.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antonelli LRV, Gigliotti Rothfuchs A, Gonçalves R, Roffê E, Cheever AW, Bafica A, Salazar AM, Feng CG, and Sher A. (2010). Intranasal Poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogen-permissive monocyte/macrophage population. J. Clin. Invest 120, 1674–1682. 10.1172/JCI40817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL, Gonzales J, Derrick SC, Shi R, Kumar NP, Wei W, et al. (2014). Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature 511, 99–103. 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji DX., Yamashiro LH., Chen KJ., Mukaida N., Kramnik I., Darwin KH., and Vance RE. (2019). Type I interferon-driven susceptibility to Mycobacterium tuberculosis is mediated by IL-1Ra. Nat. Microbiol 4, 2128–2135. 10.1038/s41564-019-0578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorhoi A, Yeremeev V, Nouailles G, Weiner J, Jörg S, Heinemann E, Oberbeck-Müller D, Knaul JK, Vogelzang A, Reece ST, et al. (2014). Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur. J. Immunol 44, 2380–2393. 10.1002/eji.201344219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan H, Yan B-S, Rojas M, Shebzukhov YV, Zhou H, Kobzik L, Higgins DE, Daly MJ, Bloom BR, and Kramnik I. (2005). Ipr1 gene mediates innate immunity to tuberculosis. Nature 434, 767–772. 10.1038/nature03419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichugin AV, Yan B-S, Sloutsky A, Kobzik L, and Kramnik I. (2009). Dominant role of the sst1 locus in pathogenesis of necrotizing lung granulomas during chronic tuberculosis infection and reactivation in genetically resistant hosts. Am. J. Pathol 174, 2190–2201. 10.2353/ajpath.2009.081075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji DX, Witt KC, Kotov DI, Margolis SR, Louie A, Chevée V, Chen KJ, Gaidt MM, Dhaliwal HS, Lee AY, et al. (2021). Role of the transcriptional regulator SP140 in resistance to bacterial infections via repression of type I interferons. eLife 10, e67290. 10.7554/eLife.67290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Romo GS, Caielli S, Vega B, Connolly J, Allantaz F, Xu Z, Punaro M, Baisch J, Guiducci C, Coffman RL, et al. (2011). Netting Neutrophils Are Major Inducers of Type I IFN Production in Pediatric Systemic Lupus Erythematosus. Sci. Transl. Med 3, 73ra20–73ra20. 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J, Meller S, Chamilos G, Sebasigari R, Riccieri V, et al. (2011). Neutrophils Activate Plasmacytoid Dendritic Cells by Releasing Self-DNA–Peptide Complexes in Systemic Lupus Erythematosus. Sci. Transl. Med 3, 73ra19–73ra19. 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eshleman EM, Delgado C, Kearney SJ, Friedman RS, and Lenz LL (2017). Down regulation of macrophage IFNGR1 exacerbates systemic L. monocytogenes infection. PLOS Pathog. 13, e1006388. 10.1371/journal.ppat.1006388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rayamajhi M, Humann J, Penheiter K, Andreasen K, and Lenz LL (2010). Induction of IFN-αβ enables Listeria monocytogenes to suppress macrophage activation by IFN-γ. J. Exp. Med 207, 327–337. 10.1084/jem.20091746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takaki K, Davis JM, Winglee K, and Ramakrishnan L. (2013). Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat. Protoc 8, 1114–1124. 10.1038/nprot.2013.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen SB., Gern BH., Delahaye JL., Adams KN., Plumlee CR., Winkler JK., Sherman DR., Gerner MY., and Urdah KB. (2018). Alveolar Macrophages Provide an Early Mycobacterium tuberculosis Niche and Initiate Dissemination. Cell Host Microbe 24, 439–446.e4. 10.1016/j.chom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang L, Nazarova EV, Tan S, Liu Y, and Russell DG (2018). Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J. Exp. Med 215, 1135–1152. 10.1084/jem.20172020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amatullah H, Fraschilla I, Digumarthi S, Huang J, Adiliaghdam F, Bonilla G, Wong LP, Rivard M-E, Beauchamp C, Mercier V, et al. (2022). Epigenetic reader SP140 loss of function drives Crohn’s disease due to uncontrolled macrophage topoisomerases. Cell 185, 3232–3247.e18. 10.1016/j.cell.2022.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fraschilla I, Amatullah H, Rahman R-U, and Jeffrey KL (2022). Immune chromatin reader SP140 regulates microbiota and risk for inflammatory bowel disease. Cell Host Microbe, S1931–3128(22)00417–6. 10.1016/j.chom.2022.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehta S, Cronkite DA, Basavappa M, Saunders TL, Adiliaghdam F, Amatullah H, Morrison SA, Pagan JD, Anthony RM, Tonnerre P, et al. (2017). Maintenance of macrophage transcriptional programs and intestinal homeostasis by epigenetic reader SP140. Sci. Immunol 2, eaag3160. 10.1126/sciimmunol.aag3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schyns J, Bai Q, Ruscitti C, Radermecker C, De Schepper S, Chakarov S, Farnir F, Pirottin D, Ginhoux F, Boeckxstaens G, et al. (2019). Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat. Commun 10, 3964. 10.1038/s41467-019-11843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2019). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44. 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 45.Pisu D, Huang L, Narang V, Theriault M, Lê-Bury G, Lee B, Lakudzala AE, Mzinza DT, Mhango DV, Mitini-Nkhoma SC, et al. (2021). Single cell analysis of M. tuberculosis phenotype and macrophage lineages in the infected lung. J. Exp. Med 218, e20210615. 10.1084/jem.20210615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, and Smibert P. (2017). Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868. 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu J, Sealfon SC, Hayot F, Jayaprakash C, Kumar M, Pendleton AC, Ganee A, Fernandez-Sesma A, Moran TM, and Wetmur JG (2007). Chromosome-specific and noisy IFNB1 transcription in individual virus-infected human primary dendritic cells. Nucleic Acids Res. 35, 5232–5241. 10.1093/nar/gkm557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheu S., Dresing P., and Locksley RM. (2008). Visualization of IFNbeta production by plasmacytoid versus conventional dendritic cells under specific stimulation conditions in vivo. Proc. Natl. Acad. Sci. U. S. A 105, 20416–20421. 10.1073/pnas.0808537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomsen EA, Andersen S, Marqvorsen MHS, Skipper KA, Paludan SR, and Mikkelsen JG (2021). Single-Cell Monitoring of Activated Innate Immune Signaling by a d2eGFP-Based Reporter Mimicking Time-Restricted Activation of IFNB1 Expression. Front. Cell. Infect. Microbiol 11, 784762. 10.3389/fcimb.2021.784762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao M, Zhang J, Phatnani H, Scheu S, and Maniatis T. (2012). Stochastic expression of the interferon-β gene. PLoS Biol. 10, e1001249. 10.1371/journal.pbio.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Esaulova E, Das S, Singh DK, Choreño-Parra JA, Swain A, Arthur L, Rangel-Moreno J, Ahmed M, Singh B, Gupta A, et al. (2021). The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell Host Microbe 29, 165–178.e8. 10.1016/j.chom.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fitzgerald-Bocarsly P, Dai J, and Singh S. (2008). Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev. 19, 3–19. 10.1016/j.cytogfr.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, and Germain RN (2012). Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376. 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kotov DI, Pengo T, Mitchell JS, Gastinger MJ, and Jenkins MK (2019). Chrysalis: A New Method for High-Throughput Histo-Cytometry Analysis of Images and Movies. J. Immunol 202, 300–308. 10.4049/jimmunol.1801202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cervantes-Barragan L, Lewis KL, Firner S, Thiel V, Hugues S, Reith W, Ludewig B, and Reizis B. (2012). Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc. Natl. Acad. Sci 109, 3012–3017. 10.1073/pnas.1117359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smit JJ, Rudd BD, and Lukacs NW (2006). Plasmacytoid dendritic cells inhibit pulmonary immunopathology and promote clearance of respiratory syncytial virus. J. Exp. Med 203, 1153–1159. 10.1084/jem.20052359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asselin-Paturel C, Brizard G, Pin J-J, Brière F, and Trinchieri G. (2003). Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody. J. Immunol. Baltim. Md 1950 171, 6466–6477. 10.4049/jimmunol.171.12.6466. [DOI] [PubMed] [Google Scholar]
- 59.Blasius AL., Giurisato E., Cella M., Schreiber RD., Shaw AS., and Colonna M. (2006). Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J. Immunol. Baltim. Md 1950 177, 3260–3265. 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- 60.Krug A, French AR, Barchet W, Fischer JAA, Dzionek A, Pingel JT, Orihuela MM, Akira S, Yokoyama WM, and Colonna M. (2004). TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity 21, 107–119. 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 61.Swiecki M, Gilfillan S, Vermi W, Wang Y, and Colonna M. (2010). Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity 33, 955–966. 10.1016/j.immuni.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cervantes-Luevano KE, Caronni N, Castiello MC, Fontana E, Piperno GM, Naseem A, Uva P, Bosticardo M, Marcovecchio GE, Notarangelo LD, et al. (2018). Neutrophils drive type I interferon production and autoantibodies in patients with Wiskott-Aldrich syndrome. J. Allergy Clin. Immunol 142, 1605–1617.e4. 10.1016/j.jaci.2017.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moreira-Teixeira L, Stimpson PJ, Stavropoulos E, Hadebe S, Chakravarty P, Ioannou M, Aramburu IV, Herbert E, Priestnall SL, Suarez-Bonnet A, et al. (2020). Type I IFN exacerbates disease in tuberculosis-susceptible mice by inducing neutrophil-mediated lung inflammation and NETosis. Nat. Commun 11, 5566. 10.1038/s41467-020-19412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Weerd NA, and Nguyen T. (2012). The interferons and their receptors—distribution and regulation. Immunol. Cell Biol 90, 483–491. 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nilsson A, Peters JM, Meimetis N, Bryson B, and Lauffenburger DA (2022). Artificial neural networks enable genome-scale simulations of intracellular signaling. Nat. Commun 13, 3069. 10.1038/s41467-022-30684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crisler WJ, and Lenz LL (2018). Crosstalk between type I and II interferons in regulation of myeloid cell responses during bacterial infection. Curr. Opin. Immunol 54, 35–41. 10.1016/j.coi.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McNab F, Mayer-Barber K, Sher A, Wack A, and O’Garra A. (2015). Type I interferons in infectious disease. Nat. Rev. Immunol 15, 87–103. 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ottenhoff THM, Dass RH, Yang N, Zhang MM, Wong HEE, Sahiratmadja E, Khor CC, Alisjahbana B, Crevel R. van, Marzuki S, et al. (2012). Genome-Wide Expression Profiling Identifies Type 1 Interferon Response Pathways in Active Tuberculosis. PLOS ONE 7, e45839. 10.1371/journal.pone.0045839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levin M, Newport MJ, D’Souza S, Kalabalikis P, Brown IN, Lenicker HM, Agius PV, Davies EG, Thrasher A, and Klein N. (1995). Familial disseminated atypical mycobacterial infection in childhood: a human mycobacterial susceptibility gene? Lancet Lond. Engl 345, 79–83. 10.1016/s0140-6736(95)90059-4. [DOI] [PubMed] [Google Scholar]
- 70.Newport MJ., Huxley CM., Huston S., Hawrylowicz CM., Oostra BA., Williamson R., and Levin M. (1996). A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N. Engl. J. Med 335, 1941–1949. 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 71.Jouanguy E, Altare F, Lamhamedi S, Revy P, Emile J-F, Newport M, Levin M, Blanche S, Seboun E, Fischer A, et al. (1996). Interferon-γ –Receptor Deficiency in an Infant with Fatal Bacille Calmette–Guérin Infection. N. Engl. J. Med 335, 1956–1962. 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 72.Casanova J-L, and Abel L. (2022). From rare disorders of immunity to common determinants of infection: Following the mechanistic thread. Cell 185, 3086–3103. 10.1016/j.cell.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bohrer AC, Tocheny C, Assmann M, Ganusov VV, and Mayer-Barber KD (2018). Cutting Edge: IL-1R1 Mediates Host Resistance to Mycobacterium tuberculosis by Trans-Protection of Infected Cells. J. Immunol. Baltim. Md 1950 201, 1645–1650. 10.4049/jimmunol.1800438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lowe DM, Bandara AK, Packe GE, Barker RD, Wilkinson RJ, Griffiths CJ, and Martineau AR (2013). Neutrophilia independently predicts death in tuberculosis. Eur. Respir. J 42, 1752–1757. 10.1183/09031936.00140913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panteleev AV, Nikitina IY, Burmistrova IA, Kosmiadi GA, Radaeva TV, Amansahedov RB, Sadikov PV, Serdyuk YV, Larionova EE, Bagdasarian TR, et al. (2017). Severe Tuberculosis in Humans Correlates Best with Neutrophil Abundance and Lymphocyte Deficiency and Does Not Correlate with Antigen-Specific CD4 T-Cell Response. Front. Immunol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fraschilla I, and Jeffrey KL (2020). The Speckled Protein (SP) Family: Immunity’s Chromatin Readers. Trends Immunol. 41, 572–585. 10.1016/j.it.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Akter S, Chauhan KS, Dunlap MD, Choreño-Parra JA, Lu L, Esaulova E, Zúñiga J, Artyomov MN, Kaushal D, and Khader SA (2022). Mycobacterium tuberculosis infection drives a type I IFN signature in lung lymphocytes. Cell Rep. 39, 110983. 10.1016/j.celrep.2022.110983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE, Freedman VH, and Kaplan G. (2001). Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha /beta. Proc. Natl. Acad. Sci. U. S. A 98, 5752–5757. 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Howard NC, Marin ND, Ahmed M, Rosa BA, Martin J, Bambouskova M, Sergushichev A, Loginicheva E, Kurepina N, Rangel-Moreno J, et al. (2018). Mycobacterium tuberculosis carrying a rifampicin drug resistance mutation reprograms macrophage metabolism through cell wall lipid changes. Nat. Microbiol 3, 1099–1108. 10.1038/s41564-018-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ordway D., Henao-Tamayo M., Harton M., Palanisamy G., Troudt J., Shanley C., Basaraba RJ., and Orme IM. (2007). The Hypervirulent Mycobacterium tuberculosis Strain HN878 Induces a Potent TH1 Response followed by Rapid Down-Regulation. J. Immunol 179, 522–531. 10.4049/jimmunol.179.1.522. [DOI] [PubMed] [Google Scholar]
- 81.Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J, Schmid-Burgk JL, Schmidt T, Hornung V, Cole ST, et al. (2015). Mycobacterium tuberculosis Differentially Activates cGAS- and Inflammasome-Dependent Intracellular Immune Responses through ESX-1. Cell Host Microbe 17, 799–810. 10.1016/j.chom.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 82.Collins AC, Cai H, Li T, Franco LH, Li X-D, Nair VR, Scharn CR, Stamm CE, Levine B, Chen ZJ, et al. (2015). Cyclic GMP-AMP Synthase (cGAS) Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell Host Microbe 17, 820–828. 10.1016/j.chom.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, Vance RE, Stallings CL, Virgin HW, and Cox JS (2015). The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe 17, 811–819. 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yamashiro LH, Wilson SC, Morrison HM, Karalis V, Chung J-YJ, Chen KJ, Bateup HS, Szpara ML, Lee AY, Cox JS, et al. (2020). Interferon-independent STING signaling promotes resistance to HSV-1 in vivo. Nat. Commun 11, 3382. 10.1038/s41467-020-17156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Piersma SJ, Poursine-Laurent J, Yang L, Barber GN, Parikh BA, and Yokoyama WM (2020). Virus infection is controlled by hematopoietic and stromal cell sensing of murine cytomegalovirus through STING. eLife 9, e56882. 10.7554/eLife.56882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zucchini N, Bessou G, Traub S, Robbins SH, Uematsu S, Akira S, Alexopoulou L, and Dalod M. (2008). Cutting Edge: Overlapping Functions of TLR7 and TLR9 for Innate Defense against a Herpesvirus Infection1. J. Immunol 180, 5799–5803. 10.4049/jimmunol.180.9.5799. [DOI] [PubMed] [Google Scholar]
- 87.Crother TR, Ma J, Jupelli M, Chiba N, Chen S, Slepenkin A, Alsabeh R, Peterson E, Shimada K, and Arditi M. (2012). Plasmacytoid Dendritic Cells Play a Role for Effective Innate Immune Responses during Chlamydia pneumoniae Infection in Mice. PLOS ONE 7, e48655. 10.1371/journal.pone.0048655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lippitsch A, Baal N, Chukovetskyi Y, Cunningham S, Michel G, Dietert K, Gurtner C, Gruber AD, Bein G, and Hackstein H. (2019). Plasmacytoid dendritic cell depletion modifies FoxP3+ T cell homeostasis and the clinical course of bacterial pneumonia in mice. J. Leukoc. Biol 106, 977–985. 10.1002/JLB.3AB0119-014RR. [DOI] [PubMed] [Google Scholar]
- 89.Rahman T, Brown AS, Hartland EL, van Driel IR, and Fung KY (2019). Plasmacytoid Dendritic Cells Provide Protection Against Bacterial-Induced Colitis. Front. Immunol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu V., Smith AA., You H., Nguye TA., Ferguson R., Taylor M., Park JE., Llontop P., Youngman KR., and Abramson T. (2016). Plasmacytoid dendritic cell-derived IFNα modulates Th17 differentiation during early Bordetella pertussis infection in mice. Mucosal Immunol. 9, 777–786. 10.1038/mi.2015.101. [DOI] [PubMed] [Google Scholar]
- 91.Lichtner M, Rossi R, Mengoni F, Vignoli S, Colacchia B, Massetti AP, Kamga I, Hosmalin A, Vullo V, and Mastroianni CM (2006). Circulating dendritic cells and interferon-α production in patients with tuberculosis: correlation with clinical outcome and treatment response. Clin. Exp. Immunol 143, 329–337. 10.1111/j.1365-2249.2005.02994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu Y-B, Xiao D-Q, Liang K-D, Zhang J-A, Wang W-D, Yu S-Y, Zheng B-Y, Gao Y-C, Dai Y-C, Jia Y, et al. (2017). Profiling dendritic cell subsets in the patients with active pulmonary tuberculosis. Mol. Immunol 91, 86–96. 10.1016/j.molimm.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 93.Gideon HP, Hughes TK, Tzouanas CN, Wadsworth MH, Tu AA, Gierahn TM, Peters JM, Hopkins FF, Wei J-R, Kummerlowe C, et al. (2022). Multimodal profiling of lung granulomas in macaques reveals cellular correlates of tuberculosis control. Immunity 55, 827–846.e10. 10.1016/j.immuni.2022.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caielli S, Athale S, Domic B, Murat E, Chandra M, Banchereau R, Baisch J, Phelps K, Clayton S, Gong M, et al. (2016). Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J. Exp. Med 213, 697–713. 10.1084/jem.20151876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bryson BD, Rosebrock TR, Tafesse FG, Itoh CY, Nibasumba A, Babunovic GH, Corleis B, Martin C, Keegan C, Andrade P, et al. (2019). Heterogeneous GM-CSF signaling in macrophages is associated with control of Mycobacterium tuberculosis. Nat. Commun 10, 2329. 10.1038/s41467-019-10065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gschwend J, Sherman SPM, Ridder F, Feng X, Liang H-E, Locksley RM, Becher B, and Schneider C. (2021). Alveolar macrophages rely on GM-CSF from alveolar epithelial type 2 cells before and after birth. J. Exp. Med 218, e20210745. 10.1084/jem.20210745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu X, Boyer MA, Holmgren AM, and Shin S. (2020). Legionella-Infected Macrophages Engage the Alveolar Epithelium to Metabolically Reprogram Myeloid Cells and Promote Antibacterial Inflammation. Cell Host Microbe 28, 683–698.e6. 10.1016/j.chom.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rothchild AC, Jayaraman P, Nunes-Alves C, and Behar SM (2014). iNKT Cell Production of GM-CSF Controls Mycobacterium tuberculosis. PLOS Pathog. 10, e1003805. 10.1371/journal.ppat.1003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, and Aguet M. (1994). Functional role of type I and type II interferons in antiviral defense. Science 264, 1918–1921. 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 100.Prigg JR., Hoy TR., Dobrine E., Capecch MR., Schmid EE., and Meissne N. (2015). Type I IFNs Act upon Hematopoietic Progenitors To Protect and Maintain Hematopoiesis during Pneumocystis Lung Infection in Mice. J. Immunol. 195, 5347–5357. 10.4049/jimmunol.1501553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Passegué E, Wagner EF, and Weissman IL (2004). JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 119, 431–443. 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 102.Clausen BE, Burkhardt C, Reith W, Renkawitz R, and Förster I. (1999). Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277. 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 103.Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Lin P, Mann N, Mudd S, et al. (2003). Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature 424, 743–748. 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 104.Farley FW, Soriano P, Steffen LS, and Dymecki SM (2000). Widespread recombinase expression using FLPeR (flipper) mice. Genes. N. Y. N 2000 28, 106–110. [PubMed] [Google Scholar]
- 105.Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. (2003). High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotechnol 21, 652–659. 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 106.Scott CL, T’Jonck W, Martens L, Todorov H, Sichien D, Soen B, Bonnardel J, Prijck SD, Vandamme N, Cannoodt R, et al. (2018). The Transcription Factor ZEB2 Is Required to Maintain the Tissue-Specific Identities of Macrophages. Immunity 49, 312–325.e5. 10.1016/j.immuni.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jaye DL, Geigerman CM, Herling M, Eastburn K, Waller EK, and Jones D. (2006). Expression of the plasmacytoid dendritic cell marker BDCA-2 supports a spectrum of maturation among CD4+ CD56+ hematodermic neoplasms. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 19, 1555–1562. 10.1038/modpathol.3800679. [DOI] [PubMed] [Google Scholar]
- 108.Jaye DL, Iqbal J, Fujita N, Geigerman CM, Li S, Karanam S, Fu K, Weisenburger DD, Chan WC, Moreno CS, et al. (2007). The BCL6-associated transcriptional co-repressor, MTA3, is selectively expressed by germinal centre B cells and lymphomas of putative germinal centre derivation. J. Pathol 213, 106–115. 10.1002/path.2199. [DOI] [PubMed] [Google Scholar]
- 109.Bolger AM, Lohse M, and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Love MI., Huber W., and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM, Smibert P, and Satija R. (2018). Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224. 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roelli P, bbimber, Flynn B, santiagorevale, and Gui G. (2019). Hoohm/CITE-seq-Count: 1.4.2. 10.5281/zenodo.2590196. [DOI] [Google Scholar]
- 114.R Core Team (2021). R: A language and environment for statistical computing. [Google Scholar]
- 115.Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, et al. (2019). Welcome to the Tidyverse. J. Open Source Softw 4, 1686. 10.21105/joss.01686. [DOI] [Google Scholar]
- 116.Blighe K, Rana S, and Lewis M. (2023). EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. [Google Scholar]
- 117.Andreatta M, and Carmona SJ (2021). UCell: Robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J 19, 3796–3798. 10.1016/j.csbj.2021.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Identifying innate immune cell populations in Mtb-infected lungs by flow cytometry and scRNA-seq. Related to Figure 1 and 2. (A) Gating strategy for identifying neutrophils, eosinophils, monocytes, DCs, AMs, IMs, conventional type 1 DCs (cDC1), conventional type 2 DCs (cDC2) and Mtb-infected cells. Very few Mtb+ cells were detected among the lymphoid and non-hematopoietic-derived cells. (B) Cell clustering by mRNA expression, protein expression, or combined mRNA and protein expression (B6 and Sp140−/− combined). (C) Cell clustering by biological replicate in naïve and infected B6 and Sp140−/− mice. (D) Protein and mRNA expression of lineage-defining markers used for cluster classification. Markers used for annotating (E) neutrophil and (F) monocyte and macrophage clusters based on maturity, activation status, and specific gene expression. The data represents a total of 10 mice (n = 3 for the infected lung samples and n = 2 for the naïve lung samples from B6 and Sp140−/− mice).
Supplementary Figure 2. Naïve Sp140−/− mice exhibit minimally altered immune cell numbers by flow cytometry and scRNA-seq. Related to Figure 2. Comparison of innate and adaptive immune cell numbers between B6 (n = 7; closed circles) and Sp140−/− (n = 8; open circles) (A) spleen, (B) lung, (C) and thymus. (D) Clustering of myeloid cells from naïve lungs of B6 (n = 2) and Sp140−/− (n = 2) mice. (E) Differentially expressed genes between B6 and Sp140−/− AMs, IMs, monocytes, and neutrophils. Greater fold change indicates higher expression in B6 relative to Sp140−/−. The bars in (A), (B), and (C) represent the median. Pooled data from two independent experiments are shown in (A), (B), and (C). Statistical significance was calculated by multiple unpaired t tests in (A), (B), and (C), and by Wilcoxon Rank-Sum test with Bonferroni correction in (B). *p < 0.05.
Supplementary Figure 3. I-Tomcat cells express TdTomato following in vitro stimulation, but the TdTomato signal is undetectable in vivo during Mtb infection. Using I-Tomcat Ai6 mice, pDC, IMs, and monocytes are the major type I interferon producing cells following Mtb infection in B6 and Sp140−/− mice. Related to Figure 3. (A) We targeted the mouse Ifnb locus immediately downstream of the ORF and upstream of the endogenous polyadenylation (pA) site using CRISPR in C57BL/6 ES cells, inserting a reporter cassette containing the indicated elements. Targeted mice were then bred to FLPer mice to excise the Neo cassette.(B) Representative flow cytometry histogram of TdTomato expression by I-Tomcat bone marrow-derived macrophages that were unstimulated (grey) or stimulated with poly I:C (red line). (C) Representative flow cytometry plot of TdTomato expression in immune cells in Mtb-infected lungs of Sp140−/− I-Tomcat−/−, Sp140−/− I-Tomcat+/−, and Sp140−/− I-Tomcat+/+ mice 25 days after infection. (D) Representative flow cytometry plot of TdTomato expression in AM, pDCs, monocytes, and IM from Mtb-infected lungs of Sp140−/− I-Tomcat+/+ mice 19 days post-infection or (E) 25 days post-infection. Frequency of Ai6 expression by immune cell type in the lungs of Mtb-infected (F) Sp140−/− I-Tomcat Ai6 and (G) I-Tomcat Ai6 mice. The bars in (E) and (F) represent the median. Lungs were analyzed 25 days after Mtb infection. Pooled data from two independent experiments. Statistical significance was calculated by one-way ANOVA with Tukey’s multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Figure 4. pDC-DTR mice specifically deplete pDCs without affecting major lung immune cell populations, and CD123 also identifies pDCs in Mtb-infected human lymph node samples. Related to Figure 5. (A) Representative flow cytometry plot of splenic pDCs in Sp140+/− pDC-DTR mice treated with PBS or DT from days 12 to 24 after Mtb infection. (B) RNA-sequencing validation of Sp140 expression and pDC depletion efficiency based on Siglech expression in Sp140+/− pDC-DTR mice treated with PBS or DT from days 12 to 24 after Mtb infection. (C) Number of various immune cell populations in Mtb-infected lungs of Sp140+/− pDC-DTR (n = 13; filled circles) and Sp140+/− mice (n = 13; open circles). Mice received DT from days 12 to 24 post-infection. (D) anti-CD123 (brown) and hematoxylin staining on Mtb-infected human lymph nodes. Mouse lungs and spleens were harvested 25 days after infection. Pooled data from two independent experiments are shown in (B) and (C). The bars in (C) represent the median. Statistical significance in (B) was calculated by the Wald test with multiple testing correction using the Benjamini and Hochberg method and in (C) by multiple unpaired t tests. *p < 0.05, ***p < 0.001, ****p < 0.0001.
Supplementary Figure 5. Generating gene signatures for identifying IFNγ and type I IFN responding cells. Related to Figure 7. (A) Plots depicting the log2 fold change of genes upregulated in human macrophages or mouse bone marrow-derived macrophages following stimulation with IFNγ or IFN-β. Dots colored red indicate genes used for the IFNγ signaling gene signature while blue dots indicate those used for the gene signature for type I IFN responsiveness. Representative genes induced preferentially by IFNγ or IFN-β are labeled. (B) Number of genes upregulated in human macrophages following stimulation with each indicated cytokine and (C) Number of type I or II IFN gene signature genes induced after cytokine stimulation. (D) Representative flow cytometry histogram of CXCL9 expression by mouse bone marrow-derived macrophages that were untreated (grey), IFN-β stimulated (blue), or IFNγ stimulated (red).
Supplementary Figure 6. Applying gene signatures for identifying IFNγ and type I IFN responding cells to the myeloid scRNA-seq dataset. Related to Figure 7. (A) List of genes from the cytokine stimulated mouse macrophages that were used for the gene signature for IFNγ or type I IFN responsiveness. (B) IFNγ gene signature or (C) type I IFN gene signature expression visualized by wnnUMAP plots of naïve, bystander, or Mtb-infected lung myeloid cells from B6 and Sp140−/− mice. (D) IFNγ and type I IFN gene signature expression on neutrophils, monocytes, IM, and AM from naïve B6 and Sp140−/− mouse lungs. The red line indicates the 99% cutoff used to classify cells as type I or II IFN responders. (E) Comparison of naïve, bystander, and Mtb-infected cells classified as cells that responded to type I IFN (blue), type II IFN (red), both (purple), or neither (grey) in lungs (B6 and Sp140−/− combined).
Supplementary Figure 7. IFNγ receptor and IFNγ responsive genes are expressed at lower levels in Mtb-infected IMs in Sp140−/− relative to B6 mice. Related to Figure 7. (A) Ifngr1 and Ifngr2 expression on bystander and Mtb-infected cells from B6 and Sp140−/− mice. (B) Expression of Ifngr1 and Ifngr2, (C) representative type I IFN stimulated genes, including Isg15, Ifit1, and Oasl1, and (D) representative type II IFN stimulated genes, including H2-Ab1 mRNA, MHC II protein, and Cxcl9 mRNA on bystander and Mtb-infected IM and ISG+ IM from B6 and Sp140−/− mice. (E) Gene ontology term enrichment of the Hallmark Interferon-a Response and the REACTOME Translation terms on Mtb-infected IM and ISG+ IM from B6 and Sp140−/− mice. Statistical significance in (B), (C), and (D) was calculated by non-parametric Wilcoxon rank sum test with Bonferroni correction. ***p < 0.001, ****p < 0.0001.
Data Availability Statement
Raw and processed bulk RNA- and single cell RNA-sequencing data is deposited in the NCBI Gene Expression Omnibus: GSE216023, GSE232827, GSE232922. This paper also analyzes existing, publicly available data, for which the accession numbers are listed in the key resources table.
Code for bulk RNA- and scRNA-sequencing analysis is available on Github: https://github.com/dmitrikotov/Sp140-Type-I-Inteferon.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| TruStain FcX PLUS (anti-mouse CD16/32) clone S17011E | BioLegend | Cat # 156604; RRID:AB_2783138 |
| BUV496 anti-mouse CD45 clone 30-F11 | BD Biosciences | Cat # 749889; RRID:AB_2874129 |
| APC anti-mouse CD64 clone X54–5/7.1 | BioLegend | Cat # 139306; RRID:AB_1121939 1 |
| BV480 anti-mouse B220 clone RA3–6B2 | BD Biosciences | Cat # 565631; RRID:AB_2739311 |
| BV480 anti-mouse CD90.2 clone 53–2.1 | BD Biosciences | Cat # 566082; RRID:AB_2739494 |
| APC-Fire 750 anti-mouse Ly6G clone 1A8 | BioLegend | Cat # 127652; RRID:AB_2616733 |
| BUV395 anti-mouse CD11b clone M1/70 | BD Biosciences | Cat # 563553; RRID:AB_2738276 |
| BUV737 anti-mouse CD11c clone HL3 | BD Biosciences | Cat # 612796; RRID:AB_2870123 |
| APC-R700 anti-mouse Siglec F clone E50–2440 | BD Biosciences | Cat # 565183; RRID:AB_2739097 |
| PE anti-mouse MerTK clone DS5MMER | Thermo Fisher Scientific | Cat # 12–5751-82; RRID:AB_2572623 |
| Super Bright 645 anti-mouse MHC II clone M5/114.15.2 |
Thermo Fisher Scientific |
Cat # 64–5321-82; RRID:AB_2662402 |
| BV421 anti-mouse PD-L1 clone MIH5 | BD Biosciences | Cat # 564716; RRID:AB_2738911 |
| BV711 anti-mouse Ly6C clone HK1.4 | BioLegend | Cat # 128037; RRID:AB_2562630 |
| PE anti-mouse IFNAR-1 clone MAR1–5A3 | BioLegend | Cat # 127311; RRID:AB_1134011 |
| PE-Cy7 anti-mouse MerTK clone DS5MMER | Thermo Fisher Scientific | Cat # 25–5751-82; RRID:AB_2573466 |
| APC-eFluor 780 anti-mouse CD11b clone M1/70 | Thermo Fisher Scientific | Cat # 47–0112-82; RRID:AB_1603193 |
| BUV395 anti-mouse CCRL2 clone BZ2E3 | BD Biosciences | Cat # 743689; RRID:AB_2741676 |
| BUV563 anti-mouse Ly6G clone 1A8 | BD Biosciences | Cat # 612921; RRID:AB_2870206 |
| Percp-Cy5.5 anti-mouse B220 clone RA3–6B2 | BioLegend | Cat # 103235; RRID:AB_893356 |
| BV421 anti-mouse Siglec H clone 440c | BD Biosciences | Cat # 566581; RRID:AB_2739747 |
| BV480 anti-mouse CD19 clone 1D3 | BD Biosciences | Cat # 566167; RRID:AB_2739564 |
| BV605 anti-mouse MHC II clone M5/114.15.2 | BD Biosciences | Cat # 563413; RRID:AB_2738190 |
| BV785 anti-mouse Ly6C clone HK1.4 | BioLegend | Cat # 128041; RRID:AB_2565852 |
| BV605 anti-mouse CD4 clone GK1.5 | BioLegend | Cat # 100451; RRID:AB_2564591 |
| BUV805 anti-mouse CD8ɑ clone 53–6.7 | BD Biosciences | Cat # 612898; RRID:AB_2870186 |
| PE-Cy7 anti-mouse PDCA-1 clone eBio927 | Thermo Fisher Scientific | Cat # 25–3172-80; RRID:AB_2573439 |
| PE-Cy7 anti-mouse CD63 clone NVG-2 | BioLegend | Cat # 143909 |
| Percp-eFluor 710 anti-mouse iNOS clone CXNFT | Thermo Fisher Scientific | Cat # 46–5920-82; RRID:AB_2688059 |
| BV785 anti-mouse CD206 clone C068C2 | BioLegend | Cat # 141729; RRID:AB_2565823 |
| Anti-mouse BST2 clone 927 | BioXCell | Cat # BE0311; RRID:AB_2736991 |
| Rat IgG2b isotype antibody clone LTF-2 | BioXCell | Cat # BE0090; RRID:AB_1107780 |
| Anti-mouse Ly6G clone 1A8 | BioXCell | Cat # BE0075–1; RRID:AB_1107721 |
| Rat IgG2a isotype antibody clone 2A3 | BioXCell | Cat # BE0089; RRID:AB_1107769 |
| Anti-human CD123 clone 6h6 | Thermo Fisher Scientific | Cat # 14–1239-82; RRID:AB_467453 |
| Anti-human CD303 clone 124B3.13 | Dendritics | Cat # DDX0043; RRID:AB_1149764 |
| BV421 anti-mouse SIRP𝘢 clone P84 | BD Biosciences | Cat # 740071; RRID:AB_2739835 |
| Pacific Blue anti-mouse B220 clone RA3–6B2 | BioLegend | Cat # 103227; RRID:AB_492876 |
| eF506 anti-mouse CD4 clone RM4–5 | Thermo Fisher Scientific | Cat # 69–0042-80; RRID:AB_2637458 |
| AF647 anti-mouse Ly6G clone 1A8 | BioLegend | Cat # 127609; RRID:AB_1134162 |
| BV421 anti-mouse Ly6G clone 1A8 | BioLegend | Cat # 127628; RRID:AB_2562567 |
| Rabbit polyclonal anti-citrullinated histone-H3 (citrulline R2, R8, R17) |
Abcam | Cat # ab5103; RRID:AB_304752 |
| AF488 donkey anti-rabbit secondary clone Poly4064 | BioLegend | Cat # 406416; RRID:AB_2563203 |
| APC anti-mouse Ly6G clone 1A8 | BioLegend | Cat # 127614; RRID:AB_2227348 |
| TotalSeq-A anti-mouse Ly6G clone 1A8 | BioLegend | Cat # 127655; RRID:AB_2749962 |
| TotalSeq-A anti-mouse Ly6C clone HK1.4 | BioLegend | Cat # 128047; RRID:AB_2749961 |
| TotalSeq-A anti-mouse CD44 clone IM7 | BioLegend | Cat # 103045; RRID:AB_2734154 |
| TotalSeq-A anti-mouse CD169 clone 3D6.112 | BioLegend | Cat # 142425; RRID:AB_2783106 |
| TotalSeq-A anti-mouse CD274 clone MIH6 | BioLegend | Cat # 153604; RRID:AB_2783125 |
| TotalSeq-A anti-mouse Siglec F clone S17007L | BioLegend | Cat # 155513; RRID:AB_2832540 |
| TotalSeq-A anti-mouse CSF1R clone AFS98 | BioLegend | Cat # 135533; RRID:AB_2734198 |
| TotalSeq-A anti-mouse CD11b clone M1/70 | BioLegend | Cat # 101265; RRID:AB_2734152 |
| TotalSeq-A anti-mouse CD86 clone GL-1 | BioLegend | Cat # 105047; RRID:AB_2750348 |
| TotalSeq-A anti-mouse MHC II clone M5/114.15.2 | BioLegend | Cat # 107653; RRID:AB_2750505 |
| TotalSeq-A anti-mouse CX3CR1 clone SA011F11 | BioLegend | Cat # 149041; RRID:AB_2783121 |
| TotalSeq-A anti-mouse Hashtag 1 | BioLegend | Cat # 155801; RRID:AB_2750032 |
| TotalSeq-A anti-mouse Hashtag 2 | BioLegend | Cat # 155803; RRID:AB_2750033 |
| TotalSeq-A anti-mouse Hashtag 3 | BioLegend | Cat # 155805; RRID:AB_2750034 |
| TotalSeq-A anti-mouse Hashtag 4 | BioLegend | Cat # 155807; RRID:AB_2750035 |
| TotalSeq-A anti-mouse Hashtag 5 | BioLegend | Cat # 155809; RRID:AB_2750036 |
| TotalSeq-A anti-mouse Hashtag 6 | BioLegend | Cat # 155811; RRID:AB_2750037 |
| PE anti-mouse B220 clone RA3–6B2 | Tonbo Biosciences | Cat # 50–0452-U100; RRID:AB_2621764 |
| PE anti-mouse CD90.2 clone 30-H12 | Tonbo Biosciences | Cat # 50–0903-U025; RRID:AB_2940772 |
| BV785 anti-mouse CD45.2 clone 104 | BioLegend | Cat # 109839; RRID:AB_2562604 |
| Pacific Blue anti-mouse B220 clone RA3–6B2 | BioLegend | Cat # 103230; RRID:AB_492877 |
| Pacific Blue anti-mouse CD90.2 clone 53–2.1 | BioLegend | Cat # 140305; RRID:AB_1064533 5 |
| PE anti-mouse F4/80 clone BM8 | Thermo Fisher Scientific |
Cat # 12–4801-80; RRID:AB_465922 |
| PE anti-mouse CXCL9 clone MIG-2F5.5 | BioLegend | Cat # 515603; RRID:AB_2245490 |
| Bacterial and virus strains | ||
| Bacteria: Mtb strain Erdman | A gift from Sarah Stanley, University of California, Berkeley |
N/A |
| Bacteria: Mtb-Wasabi | This study | N/A |
| Bacteria: Mtb-mCherry | This study | N/A |
| Biological samples | ||
| Human lung and lymph node samples | Surgical pathology archives of Emory University Hospital |
N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ghost Dye Violet 510 | Tonbo Biosciences | Cat # 13–0870-T500 |
| Super Bright Complete Staining Buffer | Thermo Fisher Scientific | Cat # SB-4401–75 |
| True-Stain Monocyte Blocker | BioLegend | Cat # 426102 |
| Cytofix/cytoperm | BD biosciences | Cat # 554722 |
| AccuCheck Counting Beads | Invitrogen | Cat # PCB100 |
| Vector Laboratories Hematoxylin and Eosin Stain Kit | Thermo Fisher Scientific | Cat # NC1470670 |
| Diphtheria toxin | Millipore Sigma | Cat # D0564–1MG |
| DreamTaq Green PCR Master Mix (2X) | Thermo Fisher Scientific | Cat # K1082 |
| Middlebrook 7H9 Broth (Dehydrated) | Thermo Fisher Scientific | Cat # R454012 |
| BBL seven H11 agar base | BD biosciences | Cat # 212203 |
| Middlebrook OADC | Thermo Fisher Scientific | Cat # b12351 |
| Hygromycin B Gold | Invivogen | Cat # ant-hg-1 |
| Kanamycin | Millipore Sigma | Cat # K4000–5G |
| Liberase TM | Roche | Cat # 5401127001 |
| Dnase I | Roche | Cat # 11284932001 |
| Newborn Calf Serum | Thermo Fisher Scientific | Cat # 26010074 |
| Trizol LS | Thermo Fisher Scientific | Cat # 10296010 |
| M-CSF | Vance lab | N/A |
| IFN-β | BioLegend | Cat # 581302 |
| IFNg | Peprotech | Cat # 315–05 |
| Tumor necrosis factor | Peprotech | Cat # 315–01A |
| Transforming growth factor-β | BioLegend | Cat # 763102 |
| TRK lysis buffer | Omega Bio-Tek | Cat # PR021 |
| 2-mercaptoethanol | Thermo Fisher Scientific | Cat # 21985023 |
| Ultracomp eBeads Plus | Thermo Fisher Scientific | Cat # 01–3333-42 |
| Sytox Blue Dead Cell Stain | Invitrogen | Cat # S11348 |
| RNaseOUT Recombinant Ribonuclease Inhibitor | Invitrogen | Cat # 10777019 |
| Critical commercial assays | ||
| RNeasy Micro | Qiagen | Cat # 74004 |
| E.Z.N.A Total RNA Kit I | Omega Bio-Tek | Cat # R6834–02 |
| EasySep APC Positive Selection Kit II | StemCell Technologies | Cat # 17681 |
| Chromium Single Cell 3’ Reagent Kit v3.1 chemistry | 10X Genomics | Cat # 1000268 |
| Deposited data | ||
| scRNA-seq of Mtb-infected non-human primates | Esaulova et al.51 | GEO: GSE149758 |
| Bulk RNA-seq of cytokine stimulated human macrophages | Nilsson et al.65 | GEO: GSE20251 |
| Bulk RNA-seq of cytokine stimulated mouse bone marrow-derived macrophages | This study | GEO: GSE232827 |
| Bulk RNA-seq of Mtb-infected lungs from pDC depleted Sp140+/− and Sp140−/− mice | This study | GEO: GSE232922 |
| scRNA-seq of myeloid cells from naïve and Mtb-infected B6 and Sp140−/− mice | This study | GEO: GSE216023 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Cat # 000664; RRID:IMSR_JAX:0 00664 |
| Mouse: Ifnar1−/−: B6.129S2-Ifnar1tm1Agt/Mmjax | The Jackson Laboratory | Cat # 032045-JAX RRID:MMRRC_03 2045-JAX |
| Mouse: Ai6: B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J | The Jackson Laboratory | Cat # 007906 RRID:IMSR_JAX:0 07906 |
| Mouse: Ifnar1fl: B6(Cg)-Ifnar1tm1.1Ees/J | The Jackson Laboratory | Cat # 028256 RRID:IMSR_JAX:0 28256 |
| Mouse: Mrp8Cre: B6.Cg-Tg(S100A8-cre,-EGFP)1Ilw/J | The Jackson Laboratory | Cat # 021614 RRID:IMSR_JAX:0 21614 |
| Mouse: LysMCre: B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | Cat # 004781 RRID:IMSR_JAX:0 04781 |
| Mouse: C57BL/6J-Ticam1Lps2/J (Ticam1−/−) | The Jackson Laboratory | Cat # 005037 RRID:IMSR_JAX:0 05037 |
| Mouse: C57BL/6N-Unc93b1tm1(KOMP)Vlcg/Mmucd (Unc93b1−/−) | MMRRC, KOMP repository, Regeneron Pharmaceuticals |
Cat # 050296-UCD RRID:MMRRC_05 0296-UCD |
| Mouse: I-Tomcat: Ifnb1-Tomato-Cre-pA Terminator | This study | N/A |
| Mouse: FLPer: B6N.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/J | The Jackson Laboratory | Cat # 016226 RRID:IMSR_JAX:0 16226 |
| Mouse: CD64Cre: B6-Fcgr1tm2Ciphe | Scott et al.106 | N/A |
| Mouse: pDC-DTR: B6-Tg(CLEC4C-HBEGF)956Cln/J | The Jackson Laboratory | Cat # 014176; RRID:IMSR_JAX:0 14176 |
| Mouse: Sp140−/− | Ji et al.31 | N/A |
| Recombinant DNA | ||
| pTEC15 | Takaki et al.36 | Addgene plasmid # 30174 |
| pMSP12::mCherry | A gift from Lalita Ramakrishnan, University of Cambridge |
Addgene plasmid # 30167 |
| Software and algorithms | ||
| Chrysalis | Kotov et al.55 | https://github.com/Histo-cytometry/Chrysalis |
| Generate Compensation Matrix | Kotov et al. 55 | https://github.com/Histo-cytometry/Chrysalis |
| Imaris version 9.9.1 | Bitplane | N/A |
| FlowJo version 10 | BD Biosciences | N/A |
| Trimmomatic v.0.36 | Bolger et al.109 | https://github.com/usadellab/Trimmomatic |
| STAR aligner v.2.5.2b | Dobin et al.110 | https://github.com/alexdobin/STAR |
| CellRanger version 4.0.0 | 10X Genomics | N/A |
| CITE-Seq-Count v.1.4.3 | Roelli et al.113 | https://github.com/Hoohm/CITE-seq-Count |
| R version 3.16 | R Development Core Team114 | http://www.r-project.org/ |
| RStudio “Cherry Blossom” Release | Posit | https://posit.co/products/open-source/rstudio/ |
| DESeq2 v.1.38.3 | Love et al.111 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Seurat v.4.1.1 | Hao et al.42 | https://satijalab.org/seurat/index.html |
| EnhancedVolcano v1.16.0 | Blighe et al.116 | http://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html |
| Tidyverse v2.0.0 | Wickham et al.115 | https://www.tidyverse.org/ |
| UCell v2.2.0 | Andreatta et al.117 | http://www.bioconductor.org/packages/release/bioc/html/UCell.html |
| Adobe Illustrator | Adobe.com | N/A |
| Prism | GraphPad | N/A |
| Other | ||
| 4 laser SH-800 cell sorter | Sony | N/A |
| LSM 880 laser scanning confocal microscope | Zeiss | N/A |
| 5 laser LSRFortessa analyzer | BD Biosciences | N/A |
| 5 laser Aurora analyzer | Cytek | N/A |
| GentleMACS | Miltenyi Biotec | N/A |
| Chromium controller | 10X Genomics | N/A |