Abstract
Sigma 54 associates with bacterial core RNA polymerase and converts it into an enhancer-responsive enzyme. Deletion of the N-terminal 40 amino acids is known to result in loss of the ability to respond to enhancer binding proteins. In this work PCR mutagenesis and genetic screens were used to identify a small patch, from amino acids 33 to 37, that is required for proper response to activator in vivo. Site-directed single point mutants within this segment were constructed and studied. Two of these were defective in responding to the enhancer binding protein in vitro. The mutants could still direct the polymerase to bind to DNA and initiate transient melting. However, they failed in directing activator-dependent formation of a heparin-stable open complex. Thus, amino acid region 33 to 37 includes critical activation response determinants. This region overlaps the larger leucine patch negative-control region, suggesting that anti-inhibition and positive activation are closely coupled events.
Sigma 54 is an obligatory factor in transcription from bacterial promoters that rely on enhancer elements. It associates with the common core RNA polymerase and directs the polymerase to promoters containing appropriate recognition sequences near −12 and −24 (14, 16). The holoenzyme bound to these elements remains inactive until signaled by an activator protein bound to a remote enhancer sequence (3, 17, 20–22). The activation event is the energy-dependent melting of a previously unmelted DNA segment bound by the holoenzyme (1, 20, 21). Once the open promoter complex is formed, the template strand may be read and transcription can proceed.
This reliance on enhancers and on energy for DNA melting is characteristic of eukaryotic RNA polymerase II mechanisms. It is uncharacteristic of typical bacterial transcription mechanisms that use the common sigma 70 family of proteins. Thus, sigma 54 is thought to cause the prokaryotic RNA polymerase to adopt a mechanism that is a hybrid in the sense that it has both eukaryotic and prokaryotic properties.
The amino acid sequence of sigma 54 is not similar to that of any other protein (13, 14), except perhaps for a tiny homologous segment that contributes to RNA polymerase binding (26, 28). The functional domain structure of the protein is complex, but it is thought to contain three primary domains: a C-terminal portion needed for binding DNA, a central portion needed to bind the polymerase, and an N-terminal portion needed for proper regulation of activation (4, 7, 9, 10, 22, 32). When the N-terminal 40 amino acids are deleted, sigma 54 can still bind RNA polymerase and direct it to DNA. However, the bound holoenzyme fails to respond to enhancer protein in that it fails to form a stable open complex that can initiate transcription. If extreme solution conditions that trigger transient DNA melting are used, the holoenzyme with an N-terminally deleted sigma 54 can produce transcript (30), showing that its catalytic activity is intact. This transcript is unusual in that it results from heparin-sensitive transcription. Its production is not enhanced by the addition of activator, implying that the N terminus contains essential activation response determinants.
The N-terminal 40 amino acids have the unusual composition of 40% leucines and glutamines. Site-directed mutagenesis has implicated some of the leucines in protein function (9). Multiple leucine substitutions can alter several properties of the holoenzyme. These include a loss of function, a reduction in ability to melt the DNA, and a reduction in protection of the −12 region of the promoter. The roles of individual leucines in these various processes have not been firmly established.
It is known, however, that a subset of these leucines, including four between amino acids 25 and 31, have a role in keeping unregulated transcription in check. Certain “bypass” mutations in this leucine patch allow some leaky transcription to occur in vivo. These mutants also mimic one of the in vitro properties just described for N-terminally deleted sigma: they allow some transcription to occur in the absence of enhancer protein (25, 31). As with the N-terminal deletion mutants, this in vitro transcription is unusual in being sensitive to heparin and in requiring solution conditions that favor DNA melting.
These leucine patch bypass substitution mutants differ in vitro from the deletion mutant in one important aspect. Deletions destroy the ability to respond to activator, whereas the leucine point substitutions do not. That is, activator can stimulate transcription from the point mutants and can cause the transcription to be heparin resistant (30). Thus, it appears that the N terminus contains additional determinants outside of this leucine patch that are required for the response to activator.
The determinants within the N-terminal region that are needed for the activation response are not known. The aim of this study was to begin to identify these determinants. The initial approach was to create a library of N-terminal mutants and screen it to identify specific amino acids that might be important. Candidate residues were then altered by site-directed mutagenesis and tested for function in vivo. Mutants that showed a defect were then purified and studied further in vitro. The results led to the identification of a small activation response region between amino acids 33 and 37. The properties of mutants in this region assist in understanding how sigma 54 converts the polymerase into an enhancer-responsive enzyme.
MATERIALS AND METHODS
Strains, plasmids, and mutagenesis.
The plasmid pAS54 was derived from expression plasmid pJF5401 (6, 23). The two differ only in that a proline substitution of unknown origin at position 13 was corrected by replacement with the wild-type leucine. The plasmid carries the Escherichia coli sigma 54 gene and the gene for ampicillin resistance. E. coli YMC109, lacking a wild-type chromosomal copy of the sigma 54 gene and containing a glnAp2-lacZ fusion on the chromosome, was used as the host. A functional copy of sigma 54 is required to get blue colonies on W-arginine (W-Arg) minimal medium plates (although very small white colonies can grow in the absence of sigma 54).
A library of mutations covering the N terminus of sigma 54 was created via PCR methods essentially as described previously (12, 26). The miscopying by Taq polymerase is stimulated by increasing the Mg2+ ion concentration to 6 mM and introducing Mn2+ to the PCR. Titrations indicated that 0.3 to 0.5 mM Mn2+ gave a small but detectable amount of PCR product. This PCR product was assumed to have the highest level of errors. In brief, pAS54 was linearized by restriction at the unique EcoRI site just after the C terminus of the gene. Two primers 24 bases long flanking the start site of the gene and 3′ to the unique AlwNI site at the codon for amino acid 39 were used for amplification. The PCR was performed for 25 cycles, and the 100-μl reaction mixture contained the following: 5 mM MgCl2, 0.3 to 0.5 mM Mn2+, 1× magnesium-free Taq polymerase buffer (Promega), linearized pAS54 (25 ng), 100 ng of each primer, 1 mM deoxynucleoside triphosphates (dNTPs), and 0.5 μl of Taq DNA polymerase (Promega).
The PCR product was phenol extracted, passed over a G50 spin column, and digested with AlwNI and NdeI. There are two AlwNI sites on the plasmid, so it was necessary to get a partially digested plasmid. Ten minutes of digestion with 1 U of AlwNI accomplished this. The partially digested plasmid was run on a 1% agarose gel overnight at 20 V, and the linearized plasmid was excised and purified. It was then further digested with NdeI and treated with shrimp alkaline phosphatase, and the 5.2-kb band was purified. The digested PCR product and plasmid were then ligated, transformed via electroporation into YMC109, and plated on Luria-Bertani (LB) plates containing appropriate antibiotics. These colonies were screened as described below.
Site-directed mutants were made by using the Pfu mutagenesis kit (Stratagene) with two complementary 24-nucleotide primers containing the new sequence. Approximately two-thirds of the colonies picked were the correct mutant.
Functional screen.
Transformants from the library in YMC109 cells were grown on LB plates at 30°C. Colonies were then picked and transferred onto W-Arg–X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates (500 ml of W-Arg medium contains 5.25 g of K2HPO4, 2.25 g of KH2PO4, 7.5 g of agar, 10 ml of 20% glucose, 1.0 ml of thiamine [10.0 mg/ml], 0.215 ml of 1 M MgSO4, 0.5 g of l-arginine, and appropriate antibiotics) and grown overnight at 30°C. The next day the plates were transferred to 43°C and checked every hour to observe the change of color to blue. Colonies with wild-type sigma 54 show a clear blue color after approximately 2 h. After 6 h, all of the colonies begin to turn blue, and subsequently, the screen becomes unreliable.
Protein purification.
Sigma 54 was purified as described previously (29–31). In brief, HB101 cells were transformed with expression vector pAS54 and grown in 250 ml of Luria broth medium at 30°C. When the optical density at 600 nm reached 0.8, the culture was shifted to 43°C and the cells were grown for another 3 to 4 h to induce expression. The harvested cells were resuspended in 12 ml of buffer S (10 mM Tris HCl [pH 8.0], 200 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 5% glycerol), and disrupted twice in a French press. The lysate was spun at 1,200 × g for 40 min at 4°C, and the pellet was collected and resuspended in buffer S with 4 M guanidium HCl and 0.1% Nonidet P-40 (nonionic detergent). Sonication was used to help dissolve the pellet, which was then dialyzed, first against buffer S with 1 M guanidium HCl and then against buffer S. After each dialysis the undissolved material was discarded. The dialysate was loaded onto a Q-Sepharose column (1.5 by 15 cm), washed with 10 column volumes of buffer S, and eluted with a 100-ml linear gradient of 0.2 to 0.8 M KCl in buffer S. The protein elutes near fraction 12 to 16, and the fractions with the highest purity were pooled and precipitated with 70% saturated ammonium sulfate (0.436 g/ml at 4°C). The pellet was dissolved in 1 ml of buffer S and dialyzed, first against buffer S and then against buffer S with 40% glycerol. The concentration of sigma 54 protein was determined by A260 absorption (1 A260 unit = 1.26 mg/ml) and checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis against known protein markers. The proteins purified from inclusion bodies by this one-column purification are >90% pure. The mutant proteins were grown in YMC109 cells (a strain lacking sigma 54) and purified as described above.
Standard purification was used for NtrC (15). RNA core polymerase was obtained from Epicentre Technologies, Madison, Wis.
In vitro experiments.
Standard in vitro transcriptions were done as described previously (30). In brief, 20 μl of reaction mixture contains 1× HEPES buffer (50 mM HEPES [pH 7.9], 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 50 μg of bovine serum albumin/ml, and 3.5% (wt/vol) polyethylene glycol), 5 nM pTH8 DNA, 100 nM sigma 54, 36 nM core polymerase (1 U; Epicentre Technologies), 10 mM carbamyl phosphate (Sigma), 4.5 mM ATP, and 100 nM NtrC unless otherwise stated. Reaction mixtures were assembled on ice and preincubated for 20 min at 37°C. Heparin was then added with NTPs (0.5 mM each) and incubated for an additional 10 min at 37°C. The NTPs included 4 μCi of α-32P-labeled UTP (50 μM). The reaction was stopped with urea-saturated formamide dye, loaded directly onto a 6% polyacrylamide-urea gel, and analyzed with a PhosphorImager (Molecular Dynamics). Variations, in which heparin challenge was not used, are described in Results.
Bandshift incubations were done as described previously (28). The conditions were as described above for transcription, with the following differences: NtrC, ATP, and carbamyl phosphate were absent; 1 nM annealed probe replaced pTH7; and 6 ng of dI-dC was present. The reaction mixtures were assembled on ice, incubated at 37°C for 20 min, and loaded directly on a cold 5% polyacrylamide gel in 0.5× Tris-borate-EDTA. The gel was run at 300 V for 25 min until the xylene cyanol dye came to the bottom, dried, and analyzed as described above.
RESULTS
Screening libraries created by random mutagenesis.
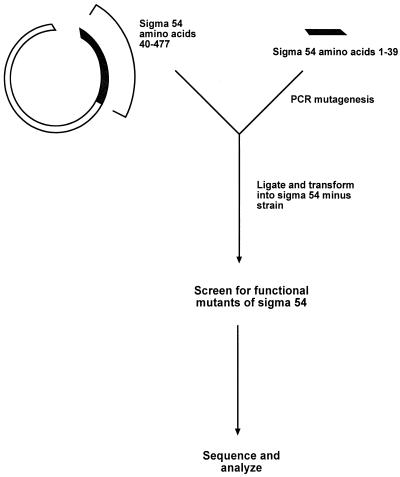
The N-terminal region of sigma 54 was targeted for mutagenesis in order to find determinants needed for activation. Error-prone PCR-copying procedures were used to create a library of mutants covering the N-terminal region prior to amino acid 40. The 150-bp PCR product was ligated in frame to the remainder of the wild-type sigma 54 gene contained on a recipient plasmid (Fig. 1). A library was created by transformation into E. coli YMC109. This strain lacks a chromosomal copy of sigma 54. Thus, the sole gene copy is provided from the plasmid containing potential N-terminal mutants. The host cell chromosome contains a fusion between the sigma 54-dependent glnAp2 promoter and lacZ, allowing color screening for expression (10). The plates contained W-Arg medium, which induces glnAp2 expression. Thus, functional and nonfunctional forms of sigma 54 may be distinguished by color, with only functional colonies producing a blue color on X-Gal indicator plates.
FIG. 1.
Outline of the PCR mutagenesis protocol used to create the N-terminal library.
In initial experiments a library with a low frequency of mutation was made and screened. White colonies were identified as nonfunctional, and the N termini of the sigma 54 genes from these colonies were sequenced. Almost all changes were found to be either frame shifts or stop codons. Thus, the information content from this approach was too low to proceed efficiently. In order to identify important amino acids, we turned to the study of a library with a high frequency of mutation, as described previously for analysis of other regions of this protein (7, 26).
The object of this approach is to sort N-terminal amino acids into groups that either can or cannot easily tolerate changes and still retain function. This should identify amino acids of potential importance. Approximately 1,700 colonies were screened. Of these, 30% retained a dark-blue color, indicating that the sigma 54 protein retained function. The plasmids were isolated and retransformed into YMC109 to confirm that the blue phenotype followed sigma 54 on the plasmid DNA. The plasmid-based sigma 54 N termini were sequenced and found to contain on average two to three changes in the N-terminal region. Selected examples of nonfunctional colonies were sequenced and found to contain many more mutations on average.
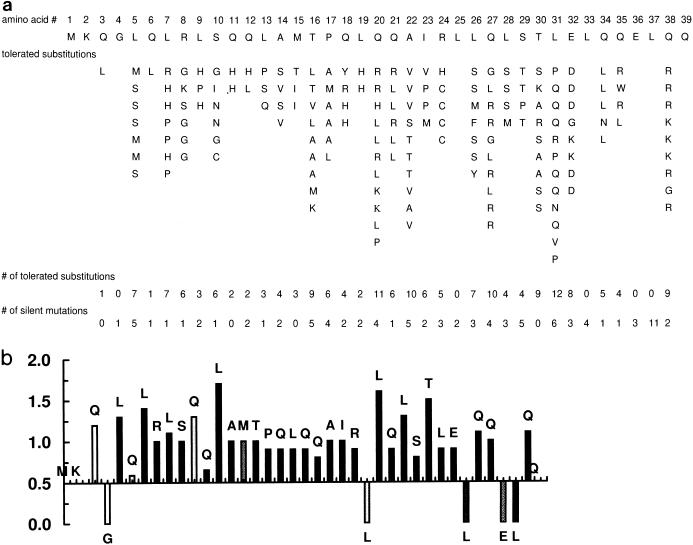
The collection of mutations from plasmids isolated from dark-blue colonies defines changes that are tolerated without loss of function. These tolerated changes are shown in Fig. 2a. The data show that most amino acids can be substituted without destroying sigma function. The screen does not give information about amino acids 1 to 4 and 39, as their proximity to the primers used for PCR amplification and restriction cleavage apparently eliminates or severely restricts their ability to be changed. Of the remaining 34 amino acids, only 4 appear to be resistant to change: L37, E36, L33, and L25. As these four amino acids are rarely mutated in sigmas that retain function, they are the prime candidates for residues with important functions.
FIG. 2.
(a) Summary of changes that are tolerated without loss of function. Sequence changes found in functional forms of sigma 54 are shown in columns beneath the wild-type sequence of the N-terminal region. The total number of tolerated amino acid substitutions and the total number of silent mutations at each position are shown at the bottom. (b) Evaluation of significance of observed pattern of tolerated changes. The fraction of nucleotide changes that led to tolerated amino acid changes was calculated for each codon. This was compared to the theoretical fraction of substitutions expected from random mutagenesis. The ratio of actual fraction/theoretical fraction is shown and is expected to be unity for positions that are not critical for function. A ratio of less than unity indicates that nucleotide changes were introduced but were not tolerated when they led to a changed amino acid. The darkness of the shading of the bars indicates the total number of changes observed, with black bars indicating at least five changes and lighter bars having progressively fewer changes.
This interpretation must be considered preliminary for several reasons. Most importantly, in order for it to be validated, the lack of tolerated mutations in these positions should not be due to a general resistance of the particular DNA sequence to being mutated in the PCR protocol. To assess this, we collected the silent mutations that accumulated in this same library (Fig. 2a, bottom). Each of these four amino acids accumulated silent mutations, showing that the codons were subject to mutation. The number of silent changes varied from a low of 2 to a high of 11. We extended this analysis to the whole data set to see if other positions of importance might have been overlooked. This would be indicated if their codons accumulated an abnormal ratio of silent to nonsilent mutations. The analysis requires that each codon be considered independently.
For each amino acid the codon was used to determine how many point nucleotide substitutions could result in a changed amino acid and how many could produce a silent change. For example, the theoretical fraction of nucleotide substitutions giving a changed amino acid for leucine codon CTG is five-ninths, whereas the theoretical fraction giving a change for methionine codon ATG is nine-ninths. We compared these fractions expected from random mutagenesis to the actual fractions observed at each amino acid in the experimental analysis. As an extreme example, 7 of 8 nucleotide changes at leucine 7 resulted in an amino acid change, whereas 0 of 11 nucleotide changes at leucine 37 resulted in an amino acid change. Both leucines should have five-ninths of the changed nucleotides lead to a new amino acid. For leucine 7, the actual and theoretical patterns match closely, suggesting that changes there were random and had no influence on whether the protein passed the screen for function. By contrast, although leucine 37 was also heavily mutated, none of the mutations passed the screen for function. Thus, L37 has a very high probability of being important for function whereas L7 does not.
Figure 2b systematizes this analysis and presents it at every position. The ratio of the actual fraction to the theoretical fraction of nucleotide changes giving a changed amino acid was determined for each position. A ratio of 1 indicates that all changes detected are acceptable, as all appeared to be functional. This suggests that the amino acid is not critical for function. As the ratio gets smaller, it indicates that many changes did not pass the screen for function and thus did not appear in the functional library. These amino acids have a higher probability of being important for function.
The data show that almost all amino acids have ratios of close to 1. The exceptions are the four amino acids already discussed (which have ratios of 0). Only glutamines 6 and 12 deviate even slightly from the expected distribution of mutations, and these deviations are small. The data give no indication that any positions are underrepresented in the library of functional mutants, except for the four positions at which no changes were observed. These four candidate amino acids are not all associated with the same level of confidence, based on the total number of changes observed. At one extreme is L37, a very strong candidate with 11 silent changes, and at the other extreme is L25, with only 2 silent changes.
Site-directed mutagenesis of candidate amino acids.
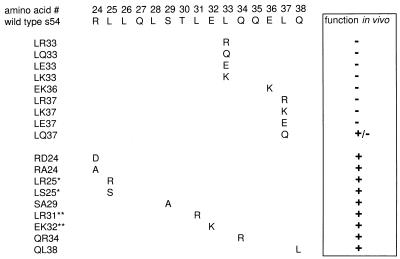
Next we used site-directed mutagenesis (or existing point mutants) to assess the role of single changes at these four positions. Other mutants were also made for the purposes of comparison. All four candidate amino acids were changed to positively charged amino acids as a common example of an extreme change. The resulting sigmas were assayed for function in vivo with the same screen developed for the library analysis.
Figure 3 shows that LR33, LR37, and EK36 are all nonfunctional, as predicted from the preceding analysis. By contrast LR25 retains function; recall that this was the candidate with the lowest confidence level. The data show that a total of nine single changes in L37, E36, and L33 were associated with a loss of function. By contrast amino acid L25 could be changed to either K or S and still retain function. We infer that a small patch between amino acids 33 and 37 contains residues of primary functional importance.
FIG. 3.
Functionality of sigmas made by site-directed mutagenesis. Substitutions are indicated below the wild-type sequence. The results of a plate test for function are shown at the right (+, functional; −, nonfunctional). ∗, mutants from previous work (25); ∗∗, mutants from the library.
There are three glutamine residues within and adjacent to this 33-to-37 region. The analysis does not suggest that they are important for function. However, we noticed that Q34 and Q38 sustained different substitutions even though they are associated with the same codon. Among the nine changes at glutamine 38, none were to leucine, whereas glutamine 34 was primarily changed to leucine. We made mutant QL38 to see if this would lead to loss of function, but the data showed that it did not (Fig. 3). Similarly, we made QR34 to test whether the kinds of changes associated with position 38 would have an effect at position 34. The data show that the change does not lead to loss of function (Fig. 3). Note that this substitution of a positively charged amino acid at position 34 is without effect in this assay in contrast to the loss of function when similar changes are made in the nearby residues 33, 36, and 37. We also made mutations in positions 24 and 29 because of the lack of certain expected amino acid substitutions at those positions, but the mutant proteins remained functional.
Several other changes in nearby residues also retained function. Substitution EK32 is without effect, whereas the data described above showed that EK36 was nonfunctional. A leucine-to-arginine change in position 31 (LR31) retains function, whereas the very similar nearby mutant LR33 does not. These and the above-mentioned comparisons emphasize that the effects on amino acids 33, 36, and 37 are strongly position specific; similar substitutions at the nearby positions 31, 32, 34, and 38 do not interfere with function.
In vitro transcription.
Three sigmas containing point mutations that lead to loss of function in vivo were purified and tested in vitro. The induction and purification was additional confirmation that the in vivo defects were not due to lack of expression. Prior in vitro transcription experiments with point mutants have failed to show significant reductions in function, even when transcription in vivo showed some defect. By contrast the effects of more radical mutations, such as the small N-terminal deletion Sal58 (amino acids 18 to 31 deleted), were easily detectable (30). Sal58 polymerase was used as a negative comparison in these studies.
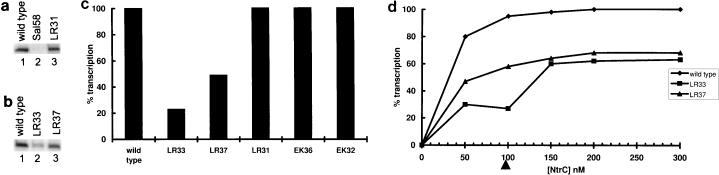
Figure 4a shows transcription in vitro from the down mutant Sal58 and from mutant LR31, which passed the in vivo screen for function. The transcription was done with a glnAp2 promoter template in the presence of the required activator NtrC. The protocol used was the standard one-round heparin challenge assay (30). The results are consistent with expectations: LR31 transcribes at a wild-type level, much higher than that of Sal58.
FIG. 4.
In vitro transcription at the glnAp2 promoter with the standard heparin challenge protocol. (a) Results with control sigmas. (b) Results with point mutants in residues deemed important. (c) Summary of results from repeated experiments. (d) Transcription changes as a function of added activator (the arrow indicates 100 nM, above which NtrC can begin to activate by the nonphysiological mechanism from solution).
The properties of down mutants LR33 and LR37 were compared to the similar but functional mutant LR31. The results of in vitro transcription are shown in Fig. 4a and b. The data show that LR33 is strongly defective in in vitro transcription. LR37 is defective, but apparently to a lesser extent. These results are consistent with the down phenotype of LR33 and LR37.
The experiments were repeated numerous times to enhance the reliability of the quantitation. Figure 4c shows the average of 14 experiments for LR33 and LR37 and the average of 6 experiments for other mutants. LR37 is down twofold in transcription, and the stronger defect in LR33 is approximately fourfold. These defects were not cured by increasing the concentration of sigma 54 (not shown).
We were unable to detect a defect in this transcription assay for mutant EK36, even though it was nonfunctional in vivo. The sigma 54 and NtrC concentrations were varied to see if these changes would reveal a defect in EK36, but they did not. We also assayed the rate of open-complex formation of EK36, but it was normal. EK36 is an expressed protein in vivo, so the defect is not due to lack of protein. Possible reasons for the lack of function in vivo by EK36 will be discussed below.
The two mutants defective in vitro, LR33 and LR37, were studied with regard to their response to activator. The concentration of activator was varied, and each was used in transcription reactions in parallel with wild-type holoenzyme. Below 100 nM the activator is known to stimulate transcription of wild-type sigma 54 holoenzyme by the physiological mechanism of binding DNA and looping to polymerase. Over this range (Fig. 4d) wild-type transcription always exceeds that of LR37, which in turn exceeds LR33. At concentrations higher than 100 nM (Fig. 4d), NtrC can begin to activate by the nonphysiological mechanism from solution (18). This mechanism appears to help LR33 somewhat, but neither it nor LR37 reaches the wild-type level over the experimentally accessible range of concentrations. This result differs from that of other N-terminal region mutants, such as the prototype HRS456 bypass mutant, which reaches the wild-type level of activation (29). We also did kinetic studies to learn if providing longer binding times could cure the defects, but this did not happen (not shown).
These heparin challenge assays measure transcription from fully formed open complexes, which are the only ones that can resist inactivation by heparin. We showed previously that the pathway to full open complexes proceeds through two experimentally detectable intermediate complexes (25, 29). The first step of the mechanism takes a closed complex to a heparin-sensitive partially open complex. The second step carries the heparin-sensitive complex to a fully heparin-resistant open complex. Both steps have been shown to require NtrC and ATP. We wished to learn if LR33 and LR37 were defective in any of these individual steps.
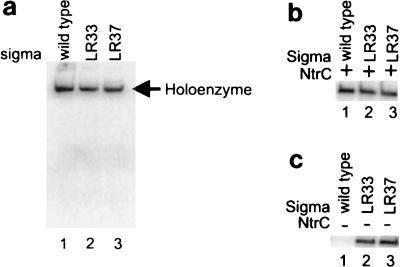
We tested whether LR33 and LR37 were able to direct closed-complex formation by using holoenzymes in a band shift assay without NtrC and with a nifH promoter probe (28). Figure 5a shows that LR33, which is strongly defective in transcription, forms a closed complex at the same level as the wild type. LR37 is slightly defective, perhaps up to the 20% level. The ability of these mutant holoenzymes to bind DNA was confirmed by DNase I footprinting (data not shown). The results show that the mutant sigmas can bind polymerase and DNA to form closed complexes but do not rule out lowering of binding affinity.
FIG. 5.
Characterization of properties of mutant sigmas. Band shift experiments of mutant holoenzymes with the nifH probe (a) and in vitro transcription without heparin challenge (b) and without activator (c) are shown.
As LR33 and LR37 are not obviously defective in forming a closed complex, we assessed whether they could form a heparin-sensitive partially open complex. This assay has been established previously with wild-type sigma 54 and various mutants (30). The experiment begins with the addition of NtrC and ATP to allow complexes to form. It then diverges from the standard in vitro heparin challenge assay, such as that described above. In this modified assay, initiating nucleotides are added to allow any partially open complexes to initiate. If heparin-sensitive complexes are present, they will be driven to initiate under these conditions (30). At this point in the protocol the addition of heparin is ineffective and simply limits the initiated complexes to a single transcript.
The results show that the mutants are not defective when this protocol is used (Fig. 5b). This is clearly different from the result observed when complexes are first challenged with heparin before nucleotides are added (Fig. 4b). The comparison shows that the mutants are defective in converting a heparin-sensitive complex to a heparin-resistant complex.
Mutants LR33 and LR37 are directly adjacent to the “leucine patch” that plays a critical role in preventing the holoenzyme from opening the DNA prior to activation. Mutations within this patch of leucines can lead to holoenzymes that transcribe in the absence of activator proteins (25, 31). In vitro, this transcription can be enhanced by specialized bypass conditions primarily involving preincubation with initiating nucleotides. When bypass conditions are used, both LR33 and LR37 produce transcripts whereas the wild-type does not (Fig. 5c), nor does mutant EK36 (not shown). Most likely, mutants LR33 and LR37 were not picked up in our prior enhancer bypass screen because they are not functional in vivo. Thus, L33 and L37, which are required for the activator-dependent full opening of the DNA, also play a role in keeping the DNA closed prior to activation.
DISCUSSION
Prior to this work it was known that the N-terminal 40 residues of sigma 54 were involved in activation, but little was known about the importance of individual amino acids (30). There had been no systematic study of this issue; information was based on the properties of deletions or multiple-substitution mutants. In this work we introduced mutations randomly into this region and screened for function. Analysis of the data led to proposals as to which amino acids were important for function and which were not.
The results showed that most of the amino acids could be changed in multiple ways and still retain function. Analysis indicated that loss of function was likely to be associated with changes in three amino acids within a small patch between residues 33 and 37. Amino acids L33, E36, and L37 appeared to be the most important. This was confirmed directly by creating site-directed mutants in these positions and observing a loss of function. Very similar changes were made in several nearby amino acids, and these retained function. Thus, most amino-terminal residues assayed by substitution did not show defects. It might require specific multiple mutations outside the 33-to-37 region for a loss of function to be observed. Moreover, the type of substitution is probably important, as substitution of serine at position 37 was not deleterious (9) and substitution of glutamine within this region had a modest effect (see Results). It is also possible that the other amino acids within the N terminus might be important for promoter-activator combinations that differed from the one used as the basis for the genetic screen.
Sigma proteins with inactivating changes in each of these positions were purified and studied in vitro. Changes in L33 and L37 caused defects in transcription. A change in E36 did not lead to lower transcription in vitro (discussed further below), as has been observed for some multiple point mutations that were defective in vivo (unpublished data).
L33 and L37 mutants were studied by using more detailed assays to attempt to identify the step at which the transcription defect occurred. Changes at either leucine did not strongly interfere with the binding of polymerase to the promoter or with the formation of a heparin-sensitive transcription complex. However, the formation of a heparin-resistant complex was defective. Prior studies have associated this formation with the use of ATP and activator to form fully stable open complexes (25, 29). Thus, the data indicate that L33 and L37 have specific roles in the enhancer- and energy-dependent melting of the DNA. Recently, we determined that formation of heparin-resistant complexes involves a novel fork-binding activity of sigma 54 (8). Preliminary experiments suggest that mutations in L33 and L37 can strongly inhibit this activity. Thus, the residues may be important in creating or maintaining the fork-binding activity of sigma.
Both leucine 33 and leucine 37 are seen to be very highly conserved when sigma 54 genes from different organisms are compared (2). This suggests that they may play a common role in all organisms. By contrast glutamate 36, which was important in vivo but did not show its defect in vitro, is not conserved. Possibly E36 has a role in steps that are not assayed here, such as reinitiation (27). Alternatively, it may be required for the assembly of transcription complexes in vivo using sets of proteins that are not used in the particular in vitro assay system. It is interesting to note that E36 becomes protease sensitive during initiation (5). Other amino acids near the 33-to-37 patch, such as glutamate 32, are strongly conserved, and their functions remain to be established.
This patch is predominantly leucines and glutamines, and it is within the N-terminal region that has a high (40%) content of these two residues. Some of the residues in this patch have been changed as part of multiple leucine and glutamine mutations in prior studies (9, 10). L33 is part of a heptad hydrophobic repeat that has been previously identified as being important for activation and −12-region recognition, but in vitro studies were not done (9, 10). The 33-to-37 patch is also covered by deletions that were known to eliminate the response to activation (30). The glutamines near and within this patch could be changed without loss of function, consistent with the modest defects associated with multiple glutamine mutations (10). Overall, the identification of L33 and L37 as important residues is consistent with prior studies.
The 33-to-37 region is directly adjacent to a region between amino acids 25 and 31 that is associated with a different critical property of sigma 54. The 25-to-31 region contains leucines that have roles in locking sigma 54 holoenzyme in a closed-complex form prior to activation (25, 31). Mutating leucines in this patch allows some unactivated transcription to occur. However, NtrC and ATP can fully stimulate when leucines between 25 and 31 are mutated, in contrast to the properties of mutations at L33 and L37. The above-mentioned data show that this locking determinant probably extends beyond position 31 to include the small 33-to-37 region identified in the present study. Figure 6 shows the overlap between the locking leucine patch and the activation response determinants uncovered here. The existing data indicate that the entire 25-to-37 region helps keep the polymerase inactive initially. After signal transduction the phosphorylated NtrC would loop (24) to the inactive polymerase and use determinants that include the 33-to-37 subregion to complete the activation process. The 33-to-37 subregion need not contact the activator directly but could be downstream in the response pathway, perhaps, as discussed above, being defective in maintaining an activator-induced DNA fork junction binding activity (8).
FIG. 6.
Overlap between inhibitory leucine patch and activation response determinants. The region involved in positive activation (defective) is boxed, and the region required to keep melting in check prior to activation (bypass) is underlined.
Thus, at this stage of investigation it is clear that residues between amino acids 25 and 37 are involved in multiple aspects of activation. The entire region keeps the polymerase from melting the DNA inappropriately. The 33-to-37 subregion is then required in the activation response pathway; the inhibition is first overcome, and eventually the melting reaction is fully completed. One can speculate that NtrC makes contact with sigma (11) to disorganize an inhibitory structure and drive conformational changes (5) that lead eventually to stable melting. The subregion would not be expected to contain all of the required response determinants (19), nor need it control the response to all sigma 54-dependent activators. Other elements within the sigma 54 response pathways are likely to be uncovered.
ACKNOWLEDGMENTS
We thank Yi Song for sequencing and Shomi Sanjabi and Kristine Hetzel for help in making and testing mutants.
The work was supported by NIH grant GM35754.
REFERENCES
- 1.Austin S, Dixon R. The prokaryotic enhancer binding protein NtrC has an ATPase activity which is phosphorylation and DNA dependent. EMBO J. 1992;11:2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brun Y V, Shapiro L. A temporally controlled sigma-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 1992;6:2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- 3.Buck M, Cannon W. Activator-independent formation of a closed complex between sigma 54-holoenzyme and nifH and nifU promoters of Klebsiella pneumoniae. Mol Microbiol. 1992;6:1625–1630. doi: 10.1111/j.1365-2958.1992.tb00887.x. [DOI] [PubMed] [Google Scholar]
- 4.Cannon W V, Chaney M K, Wang X, Buck M. Two domains within sigmaN (sigma54) cooperate for DNA binding. Proc Natl Acad Sci USA. 1997;94:5006–5011. doi: 10.1073/pnas.94.10.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casaz P, Buck M. Probing the assembly of transcription initiation complexes through changes in sigmaN protease sensitivity. Proc Natl Acad Sci USA. 1997;94:12145–12150. doi: 10.1073/pnas.94.22.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng J, Goss T J, Bender R A, Ninfa A J. Activation of transcription initiation from the nac promoter of Klebsiella pneumoniae. J Bacteriol. 1995;177:5523–5534. doi: 10.1128/jb.177.19.5523-5534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Y, Gralla J D. DNA-binding determinants of sigma 54 as deduced from libraries of mutations. J Bacteriol. 1997;179:1239–1245. doi: 10.1128/jb.179.4.1239-1245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo, Y., and J. D. Gralla. Promoter opening using a novel DNA fork junction binding activity. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 9.Hsieh M, Gralla J D. Analysis of the N-terminal leucine heptad and hexad repeats of sigma 54. J Mol Biol. 1994;239:15–24. doi: 10.1006/jmbi.1994.1347. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh M, Tintut Y, Gralla J D. Functional roles for the glutamines within the glutamine-rich region of the transcription factor sigma 54. J Biol Chem. 1994;269:373–378. [PubMed] [Google Scholar]
- 11.Lee J H, Hoover T R. Protein crosslinking studies suggest that Rhizobium meliloti C4-dicarboxylic acid transport protein D, a sigma 54-dependent transcriptional activator, interacts with sigma 54 and the beta subunit of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:9702–9706. doi: 10.1073/pnas.92.21.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 13.Lonetto M, Gribskov M, Gross C A. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1993;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (sigma N) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 15.Moore J B, Shiau S P, Reitzer L J. Alterations of highly conserved residues in the regulatory domain of nitrogen regulator I (NtrC) of Escherichia coli. J Bacteriol. 1993;175:2692–2701. doi: 10.1128/jb.175.9.2692-2701.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morett E, Buck M. In vitro studies on the interactions of DNA polymerase-ς54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters. The role of NifA in the formation of an open promoter complex. J Mol Biol. 1989;210:65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- 17.Ninfa A J, Reitzer L J, Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 18.North A K, Kustu S. Mutant forms of the enhancer-binding protein NtrC can activate transcription from solution. J Mol Biol. 1997;267:17–36. doi: 10.1006/jmbi.1996.0838. [DOI] [PubMed] [Google Scholar]
- 19.Oguiza J A, Buck M. DNA-binding domain mutants of sigma-N defective between closed and stable open promoter complex formation. Mol Microbiol. 1997;26:655–664. doi: 10.1046/j.1365-2958.1997.5861954.x. [DOI] [PubMed] [Google Scholar]
- 20.Popham D L, Szeto D, Keener J, Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 21.Sasse-Dwight S, Gralla J D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasse-Dwight S, Gralla J D. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor sigma 54. Cell. 1990;62:945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- 23.Schauder B, Blocker H, Frank R, McCarthy J E. Inducible expression vectors incorporating the Escherichia coli atpE translational initiation region. Gene. 1987;52:279–283. doi: 10.1016/0378-1119(87)90054-0. [DOI] [PubMed] [Google Scholar]
- 24.Su W, Porter S, Kustu S, Echols H. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Syed A, Gralla J D. Isolation and properties of enhancer-bypass mutants of sigma 54. Mol Microbiol. 1997;23:987–995. doi: 10.1046/j.1365-2958.1997.2851651.x. [DOI] [PubMed] [Google Scholar]
- 26.Tintut Y, Gralla J D. PCR mutagenesis identifies a polymerase-binding sequence of sigma 54 that includes a sigma 70 homology region. J Bacteriol. 1995;177:5818–5825. doi: 10.1128/jb.177.20.5818-5825.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tintut Y, Wang J T, Gralla J D. A novel bacterial transcription cycle involving sigma(54) Genes Dev. 1995;9:2305–2313. doi: 10.1101/gad.9.18.2305. [DOI] [PubMed] [Google Scholar]
- 28.Tintut Y, Wong C, Jiang Y, Hsieh M, Gralla J D. RNA polymerase binding using a strongly acidic hydrophobic-repeat region of sigma 54. Proc Natl Acad Sci USA. 1994;91:2120–2124. doi: 10.1073/pnas.91.6.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J T, Gralla J D. The transcription initiation pathway of sigma 54 mutants that bypass the enhancer protein requirement: implications for the mechanism of activation. J Biol Chem. 1996;271:32707–32713. doi: 10.1074/jbc.271.51.32707. [DOI] [PubMed] [Google Scholar]
- 30.Wang J T, Syed A, Gralla J D. Multiple pathways to bypass the enhancer requirement of sigma 54 RNA polymerase: roles for DNA and protein determinants. Proc Natl Acad Sci USA. 1997;94:9538–9543. doi: 10.1073/pnas.94.18.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J T, Syed A, Hsieh M, Gralla J D. Converting Escherichia coli RNA polymerase into an enhancer-responsive enzyme: role of an NH2-terminal leucine patch in sigma 54. Science. 1995;270:992–994. doi: 10.1126/science.270.5238.992. [DOI] [PubMed] [Google Scholar]
- 32.Wong C, Tintut Y, Gralla J D. The domain structure of sigma 54 as determined by analysis of a set of deletion mutants. J Mol Biol. 1994;236:81–90. doi: 10.1006/jmbi.1994.1120. [DOI] [PubMed] [Google Scholar]