Abstract
The human microbiome, an intricate ecological network, has garnered significant attention due to its potential implications in oncogenesis. This paper delves into the multifaceted relationships between the microbiome, its metabolites, and cancer development, emphasizing the human intestinal tract as the primary microbial habitat. Highlighting the potential causative associations between microbial disturbances and cancer progression, we underscore the role of specific bacterial strains in various cancers, such as stomach and colorectal cancer. Traditional causality assessment methods, like randomized controlled trials (RCTs), have limitations. Therefore, we advocate using Mendelian Randomization (MR) as a powerful alternative to study causal relationships, leveraging genetic variants as instrumental variables. With the proliferation of genome-wide association studies, MR harnesses genetic variations to infer causality, which is especially beneficial when addressing confounders like diet and lifestyle that can skew microbial research. We systematically review MR’s application in understanding the microbiome-cancer nexus, emphasizing its strengths and challenges. While MR offers a unique perspective on causality, it faces hurdles like horizontal pleiotropy and weak instrumental variable bias. Integrating MR with multi-omics data, encompassing genomics, transcriptomics, proteomics, and metabolomics, holds promise for future research, potentially heralding groundbreaking discoveries in microbiology and genetics. This comprehensive review underscores the critical role of the human microbiome in oncogenesis and champions MR as an indispensable tool for advancing our understanding in this domain.
Keywords: Microbiome, cancer, Mendelian randomization
Introduction
The human microbiome is an intricate ecological network, teeming with various microorganisms encompassing bacteria, viruses, fungi, and protozoa. These microorganisms have colonized numerous sites within our bodies, such as the skin, oral cavity, vagina, and most prominently, the gastrointestinal tract [1]. Astoundingly, the human intestinal tract, serving as the epicenter of this microbial diversity, is a sanctuary to approximately 40 trillion bacteria. The combined genetic repertoire of these microorganisms eclipses that of the human genome, being about 150 times more extensive [2]. Current scientific endeavors indicate that the intricate interactions between the microbiome and the metabolites they produce profoundly influence the tumor microenvironment [3]. An imbalance or disruption in this microbiome can wreak havoc on cellular signaling pathways, ignite localized inflammation, and undermine the epithelial barrier’s function, potentially accelerating cancer progression [4,5]. One illustrative example is stomach cancer, which is believed to have a close association with the bacterium Helicobacter pylori. The potential interplay between Helicobacter pylori infections and alterations in the intestinal microbiota composition could be instrumental in the genesis of gastric carcinoma.
Furthermore, there has been a significant uptick in the presence of certain bacteria such as Escherichia coli, enterotoxin-producing Enterotoxin fragile-like bacilli, anaerobic digestive Streptococci, and Enterococcus faecalis in colorectal cancer tissues [6,7]. Additionally, an overabundance of Clostridium perfringens has been implicated in aiding tumor invasion and fostering metastatic proliferation. In breast cancer contexts, shifts in the microbiota composition could play a pivotal role in hormone regulation, potentially influencing the disease’s etiology [8]. One notable player in this arena is Lactobacillus johnsonii, an Enterococcus genus member, which has been observed to exert a profound influence on breast cancer progression, primarily by promoting elevated estrogen levels [9].
The pursuit of unraveling the causal links between microorganisms and cancer holds paramount significance for advancing pathologic research and refining cancer prevention and treatment strategies. The randomized controlled trial (RCT) is traditionally the gold standard for deducing causality [10]. Yet, the practicalities of RCTs, including substantial human, material, and financial outlays and ethical considerations, delineate its limitations in clinical research scenarios. An alternative avenue to shed light on the nexus between exposure and disease is the observational study [11-13]. Notably, various cancers, such as colorectal, lung, breast, and pancreatic cancers, have been associated with the microbiome through observational studies [9,14,15]. However, these traditional observational investigations often grapple with inherent constraints, including confounding at the local level and the looming specter of reverse causality.
Mendelian randomization (MR), drawing inspiration from Gregor Mendel’s foundational laws of heredity, leverages genetic variants with robust correlations to exposure factors as instrumental variables (IVs) [16,17]. MR resembles RCTs in its approach and can navigate the pitfalls of unmeasured confounders, thereby mitigating biases inherent to observational studies [18]. Moreover, MR’s versatility enables it to probe exposures that are unfeasible to randomize in traditional RCTs. The comparison of RCTs, observational studies, and MR analysis is in Figure 1. The burgeoning domain of genome-wide association studies (GWAS) has further bolstered MR’s potential, transforming genetic variation into a formidable arsenal for causal inference. Traditional observational studies often find themselves in a dilemma when trying to discern the causal relationship between microbial composition and disease risk, especially given that shifts in microbial composition might be intertwined with other disease risk factors, such as dietary and lifestyle habits. MR rises to this challenge by meticulously selecting genetic variants associated with microbial composition as IVs, offering a more refined estimation of the causal relationship between microbial composition and disease risks [16,19]. This review aims to distill insights from the application of MR in understanding the intricate relationship between the microbiome and cancers, laying a theoretical foundation to fathom the deep-seated influence of the microbiome in systemic malignancies.
Figure 1.
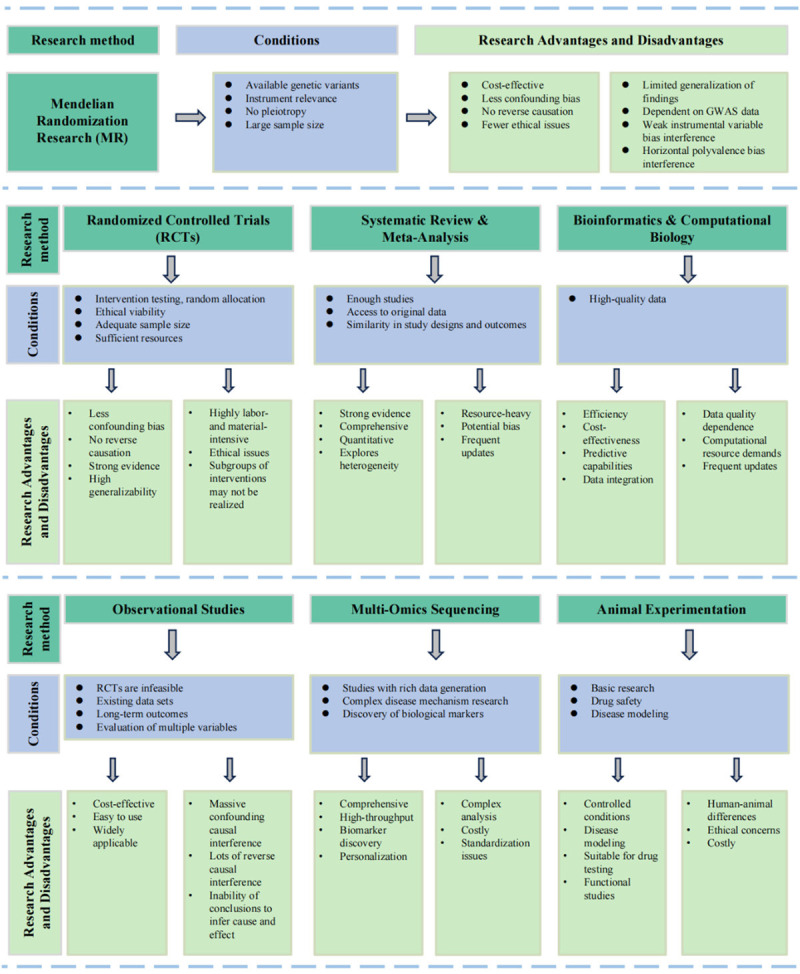
Advantages, disadvantages, and application conditions of six research methodologies: Mendelian randomization, randomized controlled trials, systematic reviews and meta-analyses, bioinformatics and computational biology, observational studies, multi-omics sequencing, and animal models.
Bidirectional causal relationship between microbiome and cancer
Microbial imbalances, commonly called dysbiosis, have been intrinsically linked to the onset and progression of cancer [20-22]. The interplay between cancer and the microbiome encompasses a range of complex interactions, including the association of specific microbes with cancer development and alterations in microbial composition within cancer patients. About 20% of human cancers may be linked to microbes [23], with specific types like Helicobacter pylori, Fusobacterium nucleatum, Escherichia coli, Bacteroides fragilis, and Porphyromonas gingivalis associated with the development of malignancies such as colorectal, gastric, and pancreatic cancer. In patients with colorectal cancer (CRC), for instance, the composition of the gut microbiome differs significantly from that of healthy individuals, and the tumor microbiome in CRC tissue also shows distinct variations from non-cancerous tissue. Additionally, the discovery of bacteria within many tumors, such as large microbial communities in the pancreatic tissue of patients with Pancreatic Ductal Adenocarcinoma (PDAC), challenges the notion that tumors are sterile environments. These microbes inhabit tumors and contribute to carcinogenesis through pathogenic products that cause chronic inflammation and subsequent damage. Specific virulence factors of bacteria, such as those produced by the microbes mentioned above, are implicated in various cancers, influencing the pathogenesis and progression of these diseases.
Furthermore, the microbiome plays a crucial role in shaping the host’s immune response to tumors and significantly affects the response to cancer treatments, particularly in therapies like immune checkpoint blockade. Recent research further emphasizes this by linking microbial cytotoxins and probiotic strains to cancer cell death, highlighting their potential as targeted tools in cancer therapy [2,14,24,25]. Interestingly, microorganisms can migrate to various bodily regions irrespective of their origins, potentially influencing tumorigenic processes [26]. Beyond the direct implications of pathogenic organisms in cancer genesis, it’s crucial to underscore the role of commensal bacterial communities. Through the induction of symbiotic imbalances, these communities can influence cancer genesis via several underlying pathways [27-30].
Metabolomics, which stands at the crossroads of environmental factors, metabolic molecules, host genes, and diseases, has emerged as a focal point of attention for researchers aiming to dissect tumor development [31-33]. Several metabolites, including short-chain fatty acids (SCFAs), amino acids, vitamins, bile acids, and toxins, are intricately linked with microbial actions [34,35]. For instance, SCFAs, pivotal metabolites synthesized by gut microbes, profoundly affect intestinal function and overall metabolism [36,37]. Studies have illuminated that augmenting the abundance of SCFA-producing strains can potentially impede the progression of colorectal cancer. A notable mention is an isovaleric acid (IVA), a specific SCFA that has been intertwined with colorectal cancer [38]. IVA is known to trigger the upregulation of proteins, thereby elevating levels of 5-hydroxytryptophan (5-HT), which, in turn, can exert direct influence over tumor cells, enhancing their self-renewal capacity and, consequently, increasing susceptibility to intestinal cancer [39].
Moreover, inevitable microbial byproducts have been identified to influence T cell activities potentially. A case in point is isotretinoin, which is known to bolster mitochondrial functionality while promoting the production of regulatory T cells [40]. Another metabolite, oxodeoxycholic acid, has been associated with a heightened risk for stomach cancer. Furthermore, specific bacterial strains can initiate intracellular signaling cascades, producing reactive oxygen species (ROS) and triggering immune responses [41]. Some bacteria also release formate, which can activate cancer-related signaling pathways. Notably, trimethylamine oxide (TMAO) in the intestinal milieu has been identified to enhance the anticancer properties of CD8+ T cells. In addition, compounds like gallic acid have demonstrated regulatory effects on immune cells, potentially improving the efficacy of cancer treatments.
The clinical significance of cancer’s impact on the microbiome is multifaceted and complex. Studies have indicated that the microbiome, particularly the gut microbiome, is instrumental in the evolution and advancement of cancer as well as the reaction to its treatment. Factors such as inflammation, tumor development, and cancer therapy can significantly alter a person’s microbiome. In particular, altered metabolism, a hallmark of cancer, is influenced by the gut microbiota, affecting human health and cancer therapy. Cancer may modify the microbiome by inducing tissue inflammation, altering host immune responses, or directly affecting the gut environment. Cancer treatments like chemotherapy, radiotherapy, immunotherapy, and targeted therapies not only affect cancer cells but also alter the microbial composition of patients, potentially leading to dysbiosis, increased infection risks, and even influencing treatment responses. Analyzing a patient’s microbial composition clinically allows for more precise treatment choices. At the same time, measures to maintain or restore a healthy microbiome, such as using probiotics or prebiotics, can reduce gastrointestinal discomfort caused by chemotherapy.
Moreover, monitoring changes in the microbiome of the gut or other body parts can aid in the timely detection of treatment or side effects. Appropriate dietary and lifestyle adjustments can also help maintain a healthy microbiome. Overall, understanding how cancer affects the microbiome and leveraging this knowledge to refine therapeutic strategies, such as adjusting anti-cancer treatments to minimize the destruction of beneficial microbes, is vital for enhancing treatment effectiveness and reducing side effects.
Overview of methodologies microbiome-cancer research: advantages, limitations, and practical applications
Randomized controlled trial
As the gold standard in research design, RCTs minimize bias with random assignment and controlled conditions, making them practical for testing new drugs or therapies [42]. RCTs have a rigorous approach to reducing bias through randomly allocating subjects into experimental and control groups, paired with stringent control over experimental conditions. This robust framework is particularly adept at assessing the efficacy and safety of new pharmaceuticals and therapeutic strategies. RCTs play a crucial role in microbial research related to cancer. They are the cornerstone for evaluating the effectiveness of antimicrobials or clinical testing of anti-cancer drugs [43-45], providing high-quality evidence while minimizing bias. Such trials are fundamental in addressing antimicrobial resistance, which presents a significant challenge in cancer care. However, while RCTs are excellent for assessing immediate outcomes of interventions against oncogenic microbes, they may not fully capture the extended interplay between the microbiome and cancer progression. Here, observational studies complement RCTs by providing long-term data on the evolution of microbial communities throughout cancer development, thereby offering a more comprehensive view of the interactions that influence carcinogenesis.
Observational studies
Observational studies provide critical insights into the dynamics of real-world settings, encompassing expansive populations and the long-term consequences of variable and uncontrollable factors. Observational studies in microbial research have significantly advanced our understanding of the complex interactions between the microbiome and various health outcomes, including cancer [23]. They generate longitudinal data that elucidate the evolution of microbial communities in concert with disease progression or changes in the tumor microenvironment [46,47]. Such studies have unveiled potential causal linkages by establishing associations between specific microorganisms or variations in microbial diversity and distinct disease states. Moreover, observational studies have been fundamental in elucidating the microbiome’s influence on the efficacy of pharmacological treatments and immunotherapeutic approaches, as well as in identifying biomarkers critical for diagnosing and monitoring disease trajectories. The insights derived from these studies are paramount for developing hypotheses that can subsequently be tested within the confines of more rigorously controlled experimental designs, such as RCTs. Nonetheless, the intrinsic nature of observational studies, which are devoid of the randomization characteristic of RCTs, renders them more vulnerable to the effects of confounding variables [48]. These confounding factors can veil the authentic interrelations between variables, thereby necessitating the application of advanced statistical methodologies to differentiate actual effects from those that are merely coincidental [49].
Systematic reviews and meta-analyses
In microbial research, systematic reviews and meta-analyses have precipitated considerable advancements by synthesizing extant data to elucidate the complex roles of the microbiome in human health and various pathologies. These systematic analyses have illuminated distinct microbial signatures associated with various diseases, including neoplasms and immune dysfunctions [50], through comparative assessments of microbiota compositions. Furthermore, these analytical approaches have underscored the representational imbalances in microbiome research, notably within African cohorts [51], thereby identifying pivotal research lacunae that necessitate further scholarly inquiry. In aggregate, these methodological endeavors bolster the statistical robustness required to extract authentic correlations and inform subsequent investigative directions by amalgamating results from smaller studies that may be subject to bias, thus reinforcing the pertinence and practicality of findings within the microbiome research milieu [52]. However, it is imperative to acknowledge that these methodologies are not impervious to publication bias and must meticulously consider the heterogeneity in study design and quality during data amalgamation.
Multi-omics sequencing
Multi-omics sequencing approaches have significantly propelled microbiome and cancer research, offering nuanced insights into the complex interplay between microbial communities and cancer development and progression. Integrating genomics, epigenomics, transcriptomics, proteomics, and metabolomics has illuminated the functional consequences of genomic alterations and the associations between mutations and downstream signaling pathways in cancer. This technique has facilitated the identification of distinct molecular patterns, biomarkers for disease diagnosis and prognosis, and the potential for personalized treatment strategies based on the interactions between a patient’s microbiome and their response to therapy [53]. Moreover, multi-omics has been particularly impactful in tumor microbiome research, providing a comprehensive toolbox for researchers to dissect the intricate relationships between the tumor microenvironment and microbial inhabitants [54]. In essence, multi-omics analyses facilitate the clustering of biological samples into meaningful groups, providing a deeper understanding of prognostic and predictive phenotypes. They play a pivotal role in dissecting cellular responses to therapy and assist in translational research by offering integrative models that bridge the gap between benchtop research and clinical application [55]. However, these methods are costly, require intensive data analysis, and necessitate extensive expertise in biology and bioinformatics for data interpretation.
Animal models
Animal models are essential in microbial oncology, providing a means to study cancer’s complex mechanisms and the microbiome’s impact on its development. Researchers can utilize models such as mice, zebrafish, and fruit flies to investigate genetic and environmental contributions to cancer influenced by microbial interactions [56,57]. These models are crucial for assessing the efficacy and safety of new cancer treatments and understanding the microbiome’s effect on drug metabolism and immune response. They also allow for examining tumor microenvironments to understand microbial effects on cancer progression. Animal models thus remain fundamental for translating research from lab to clinic, driving forward the fight against cancer. Nonetheless, biological differences between animal models and humans restrict the findings’ direct applicability, and ethical concerns for animal welfare are growing.
Bioinformatics and computational biology
Recent developments in computational biology and bioinformatics have significantly increased our awareness of the microbiome’s influence on cancer. Bioinformatics is crucial to microbiome research, enabling the analysis of how microbial changes correlate with cancer development and treatment outcomes. This field supports multi-omic studies to elucidate the microbiome’s impact on cancer at the molecular level. New computational frameworks shed light on the microbiota’s role in cancer development [58]. This demonstrates the innovative methods addressing the complexities of microbiome data analysis. Machine learning, part of computational biology, excels in microbiome studies, especially in selecting features, identifying biomarkers, and predicting disease and treatment outcomes [59]. These techniques can decode complex microbiome patterns, potentially revealing disease conditions or therapeutic responses. Clinical bioinformatics employs these methods to aid in diagnosing, treating, preventing, and managing diseases like cancer. Bioinformatics tools are vital for identifying biomarkers crucial to diagnosing and monitoring conditions and crafting personalized treatments. For example, big data analytics and machine learning studies reveal the gut microbiome’s complex role in cancer [60]. While bioinformatics and computational biology have propelled research forward, they encounter critical challenges, such as the complexity of managing and interpreting extensive datasets like 16S rRNA and metagenomics [61]. Furthermore, the difficulty in integrating data from different studies and the lack of necessary computational infrastructure hinder effective collaboration and slow the pace of scientific discovery.
Mendelian randomization
MR utilizes genetic variants for randomization, addressing the confounding biases common in observational studies, and applies where direct experimentation is impractical or unethical. This genetic epidemiology tool offers a complementary approach to the methods discussed, providing a quasi-experimental design to infer causality, which is particularly invaluable when traditional experimental studies are not feasible. However, MR necessitates robust statistical tools and in-depth genetic knowledge, as interpretations may be complex due to genetic heterogeneity and pleiotropic pathways. The MR will be explored in depth later. Figure 1 summarizes the strengths, weaknesses, and application conditions of all research methods mentioned above.
Mendelian randomization: principles and applications
MR is a method for assessing causal relationships in epidemiology and genetics using genetic variants as instrumental variables (IVs). To ensure the credibility of MR studies, the single-nucleotide polymorphisms (SNPs) chosen as IVs must satisfy three core assumptions: relevance, independence, and exclusion restriction. These dictate a strong IV-exposure association, the absence of confounder associations, and a direct influence on the outcome through the exposure, respectively [62]. The “relevance assumption” mandates a robust association between the instrumental variable and the exposure under study. The “independence assumption” dictates that the chosen SNPs should be devoid of associations with potential confounders that might skew the relationship between the exposure and the outcome. Finally, the “exclusion restriction assumption” stipulates that SNPs should influence the outcome solely via the exposure of interest, without any alternative pathways intervening [62]. Figure 2 shows the MR model and three key assumptions. The validity of these assumptions in MR can be compromised by issues such as pleiotropy, where genetic variants influence more than one trait, potentially violating the exclusion restriction assumption. Heterogeneity tests can address this by identifying and excluding variants with pleiotropic effects. Robust MR methods like median and mode-based approaches represent the true causal effect by the median or mode of estimates across variants despite potential instrument invalidity. The Mendelian randomization-Egger (MR-Egger) method estimates causal effects, assuming instrument strength is unaffected by direct outcomes, thus accommodating invalid instruments. Despite their advancement, these methods depend on inherently untestable assumptions about the heterogeneity and distribution of pleiotropic effects. Therefore, critically assessing these assumptions and the robustness of the employed data and methods is crucial.
Figure 2.
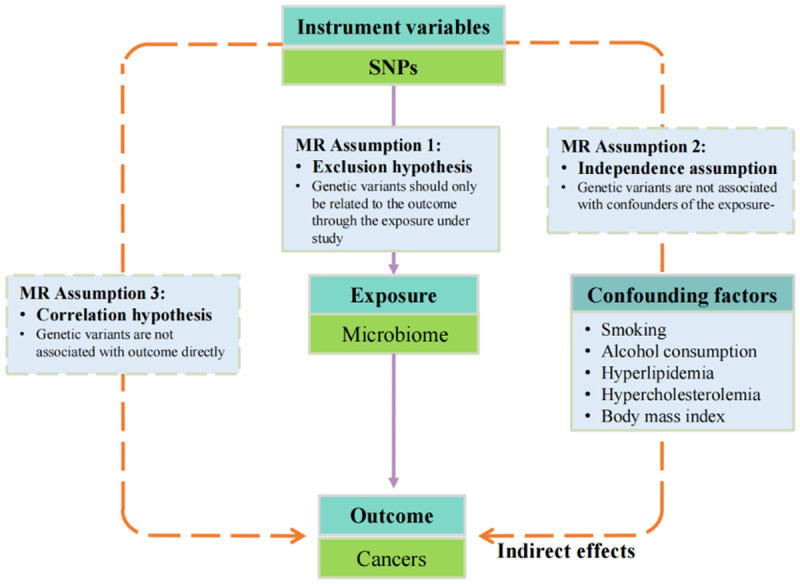
Mendelian randomization model and three key assumptions of a Mendelian randomization analysis. MR, Mendelian randomization.
Several classifications of MR have emerged, including independent-sample MR, two-sample MR, multivariate MR, bidirectional MR, and network MR [63]. In extensive biobanks, one-sample MR is being increasingly utilized to investigate causal links, as genomic associations with diverse exposures and outcomes become more prevalent. One-sample MR, which differs from two-sample MR, derives gene-exposure and gene-outcome association estimates from the same set of individuals. However, this approach may encounter challenges in scenarios with confounding variables. Consequently, researchers have employed an array of techniques, including both fixed and random effects meta-analyses, weighted median estimators, weighted mode estimators, and MR-Egger regression. These methods are applicable to both single-sample and two-sample MR, and their efficacy has been evaluated across various contexts, encompassing the existence of actual causal effects, the extent of confounding factors, and the nature of polymorphism.
In instances where confounders create significant correlations, methods designed for two-sample MR, when applied in one-sample MR settings, generally show similar performance to their use in two-sample MR scenarios. However, MR-Egger may introduce biases that mirror the direction and intensity of the confounding factors. Despite this, studies indicate that, within large biobanks, two-sample MR techniques can be securely implemented in single-sample MR analyses, except for MR-Egger. This particular method is advised against in single-sample MR unless the correlations caused by confounding are substantially reduced or there is a high variability in the strength of the instruments. This suite of findings offers vital methodological insights for probing causality in intricate biomedical datasets.
In a typical two-sample MR study, researchers embark on a journey encompassing five crucial stages:
(1) Study Typology and Genetic Instrumentation: The purpose of MR analysis is to quantify and test the causal effect of one trait on another by utilizing genetic variants as IVs. The first step necessitates the meticulous curation of genetic variants strongly associated with the exposure under investigation. This often involves delving into extensive biobanks or meta-analytic databases.
(2) Instrumental Variable Screening: Post selection, these IVs undergo rigorous validation checks to ensure their adherence to MR’s foundational principles, thus bolstering the study’s statistical power. In order for an IV to be considered genuine, there needs to be a clear connection between the genetic variant and the exposure. Additionally, any relationship between the variant and the outcome should be influenced by the exposure. Direct correlations between the genetic variant and the result or a confounding factor are prohibited.
(3) Causal Inference and Interpretation: Combine the selected genetic tools with outcome data, using various MR models such as MR-Egger, weighted median methods, etc., to ensure consistency and reliability of results. Analyze potential biological mechanisms, interpreting how genetic variants might influence the outcome through environmental exposure.
(4) Statistical Paradigms for Effect Estimation: Estimating causal effects predominantly harnesses a suite of statistical methods, from Wald ratios to two-stage least squares and inverse variance weighting. For pooled data studies, the analytical toolbox expands to include techniques such as weighted median methods and MR-Egger regression.
(5) Sensitivity Analyses: An integral component, sensitivity analysis aims to reinforce the credibility of the study’s findings. Conduct various methods (like leave-one-out, MR-Egger regression) to detect and adjust for potential biases, such as genetic pleiotropy or heterogeneity in the outcomes. Test the robustness of the results to ensure the inferred causal relationships are not influenced by specific analytical strategies or assumptions.
A variety of statistical methodologies have been harnessed, ranging from the inverse variance weighted (IVW) method to the Mendelian randomization-Egger (MR-Egger) method, and include techniques like the Weighted median (WM) method, Simple mode, and the Weighted mode method [64]. While the IVW method often emerges as the primary technique in MR studies, the other methods serve as supplementary tools [17]. For instance, MR-Egger is tailored to detect and adjust for pleiotropy - a situation where a single genetic variant impacts multiple traits. Despite its ability to yield unbiased estimates even when faced with invalid instruments, MR-Egger demands larger sample sizes and typically has diminished statistical power [65]. On the other hand, the weighted median approach is often reserved for scenarios with substantial sample sizes and aids in deriving weighted median estimates by prioritizing causal effect estimates for individual SNPs.
Logistic regression analysis is essential in MR studies with binary outcomes as it helps identify associations between exposures and outcomes and compute ratio estimates. The study by Park et al. [66] was analyzed by logistic regression analysis in the MR study for short or long sleep duration and CKD. Although not as frequently discussed, factor analysis can uncover latent variables that influence observed variables and may act as instruments or confounders in MR. The role of factor analysis in MR studies is equally important yet often understated. This method reveals latent variables, unobserved factors affecting multiple observed variables. In MR, latent variables provide insights as potential instruments or confounders. Identifying and accounting for these hidden factors allows researchers to refine models to reflect the genetic complexity affecting health outcomes more accurately. Factor analysis further enriches MR’s statistical landscape by clarifying the complex genetic architecture of traits. It enables the exploration of genetic variations affecting multiple phenotypes, which is invaluable for understanding SNPs’ pleiotropic effects. Meta-analysis can also be used in MR to combine results from separate epidemiological studies using a small but select group of genetic variants [67]. Such analytical depth is crucial for validating MR’s instrumental variables, ensuring they fulfill the rigorous assumptions necessary for credible causal inference. Consequently, integrating logistic regression and factor analysis into MR studies bolsters the precision and reliability of findings, advancing genetic epidemiology.
Advantages and limitations of Mendelian randomization in microbiological research
Thanks to its distinctive advantages, MR has carved a niche for itself in microbiological research. One of the foremost benefits of MR is its adeptness in circumventing the intricate web of confounders - variables such as diet, lifestyle, and medication - which often muddy the waters in microbial research [68-73]. By leveraging instrumental variables, MR effectively delineates causal impacts, rendering clarity to the analyses. Secondly, MR capitalizes on the inherent randomness of genetic variations, thereby allowing researchers to ascertain definitive causal links [16]. This is particularly invaluable when probing the intricate interplay between microbial ecosystems and prevalent health conditions, such as obesity, diabetes, and cardiovascular diseases.
Furthermore, MR offers a buffer against the perennial challenge of reverse causality. By anchoring its analyses on immutable genetic variables, MR enhances the veracity of causal interpretations, offering more reliable insights. Another notable advantage lies in the realm of ethical considerations. MR can harness data from pre-existing genetic databases, eliminating the need for interventions that might be deemed ethically questionable, such as microbial transplantation. Lastly, as the scientific community witnesses rapid strides in genome sequencing technologies and a concurrent expansion in microbiome datasets, the precision and feasibility of MR are poised to witness significant enhancements.
MR offers a distinctive edge in the investigation of causality. It’s imperative to remain vigilant of its inherent limitations. A primary concern arises from the issue of horizontal pleiotropy, which can potentially disrupt the assumptions of independence and exclusivity. There’s also the looming specter of weak instrumental variable bias, which can undermine the association assumption.
Several external factors further compound these challenges. LD, population heterogeneity, and the Beavis effect can subtly influence the foundational assumptions of MR [74]. Moreover, pinpointing the specific mechanisms underlying disease development remains an intricate endeavor [75]. While extensively studied, the relationships between genes and diseases are yet to be fully deciphered [76]. Furthermore, the conclusions drawn from GWAS can sometimes be hemmed in by population-specific nuances, limiting their broader applicability. Achieving the statistical firepower required in MR studies often requires larger sample sizes [77]. Another nuance to consider is that MR predominantly zeroes in on the long-term ramifications of risk factors on outcomes. This focus can make it challenging to parse out the causal effects at specific junctures of disease progression. As a result, while MR stands as a robust tool in causality assessment, it’s crucial to bolster its findings with corroborative evidence from high-quality RCTs, ensuring a holistic and rigorous approach to understanding complex biological relationships.
Mendelian randomization illuminating microbiome-cancer causality
To gain a holistic understanding of the gut microbiome’s role in cancer etiology, an exhaustive literature search was conducted, spanning publications from 2017 to 2023, sourced from the esteemed PubMed database. The search incorporated terms such as “Mendelian randomization”, “microbiota”, and “cancer” and their synonymous counterparts. After filtering out unrelated studies, 12 research pieces, all focusing on the microbiome, emerged as pivotal. Of these, several studies embarked on a broader trajectory, scrutinizing diverse cancers. Zhu et al. embarked on an extensive exploration, leveraging data from the IEU Open GWAS project, to comprehend the causal nexus between gut microbiota and eight distinct cancer types, including breast, colorectal, and prostate. Their findings unveiled 11 unambiguous causal links, particularly emphasizing the genetic predisposition of the gut microbiota, especially within the genus Bifidobacterium, and their impact on diverse cancer types [78]. In another analytical endeavor, researchers probed into the relationships between specific gut microbiota and five prevalent cancers, inclusive of their subtypes. Their observations underscored a direct association between heightened levels of the genus Sellimonas and an increased predisposition to estrogen receptor-positive breast cancer. Conversely, an elevated concentration of the Alphaproteobacteria class was associated with diminished prostate cancer risk [79]. This nexus was further elucidated by Su et al. [80] and Xie et al. [81], who cast a wider net, encompassing a myriad of gastrointestinal cancers.
Colorectal cancer stood out, being the focal point of eight discerning studies [78-80,82-86]. Ni et al. [86] highlighted the protective role of Blautia in thwarting the onset of colorectal cancer. Conversely, Li et al. [82] discerned an augmented risk associated with Bacteroides and pinpointed protective attributes in microbes such as Faecalibacterium, Blautia, and Ruminococcus. Xie et al. [84] further bolstered the evidence base, suggesting a potential link between Bacteroides and enhanced susceptibility to colorectal and stomach cancers. Hatcher C’s [85] study presented an array of microorganisms, including Fusobacterium and Peptostreptococcus, as potential risk enhancers for colorectal cancer.
In addition to colorectal cancer, other cancers have also been addressed. In a study conducted by Zhou et al. [87], a correlation was established between Weissella and Oscillospira with a decreased susceptibility to lung cancer. Conversely, Yang et al. [88] found a positive association between Prevotella and Veillonella and an increased incidence of Barrett’s esophagus while also noting a potential protective effect of Lactobacillus. Ma et al. [89] has identified that the families Ruminococcaceae, Porphyromonadaceae, and Bacteroidetes are associated with reduced susceptibility to liver cancer. Bowdon et al. [90] combined meta-analyses of different exposure contrast from 50 studies with MR analysis, indicating an altered vaginal microbiome (RR=1.59 (95% CI=1.40-1.81)), is supported by strong and highly suggestive evidence for an association with HPV persistence, CIN or cervical cancer.
In conclusion, the findings above provide substantial evidence for the causal involvement of gut microbiota in the development of cancer while presenting promising avenues for further research on the mechanisms and clinical implications of microbiota-related cancer causes. Results are summarized in Table 1 and Figure 3.
Table 1.
Summary of current research on microbiome causality in relation to multiple cancers
| Author, year | Exposure/Outcomes | Outcomes/Exposure | Direction | Causality |
|---|---|---|---|---|
| Long Y, 2023 | Gut microbiota | Eight cancer types (BC, LC, CRC, PCA, GC, Head and neck cancer, Endometrial cancer, OC) | Bidirectional | Actinobacteria, Bifidobacteriaceae, higher risk of BC; Ruminococcaceae, Bifidobacteriaceae, lower risk of BC; Actinobacteria, Tyzzerella3, Lactobacillales, higher risk of LC; Burkholderiales, lower risk of LC; Tyzzerella3, lower risk of CRC; Ruminococcustorquesgroup, Verrucomicrobiae, Desulfovibrionales, higher risk of CRC; Ruminococcustorquesgroup, Verrucomicrobiales, Terrisporobacter, lower risk of PCA; AlphaproteobacteriaM, higher risk of PCA; Peptostreptococcaceae, higher risk of GC; Gastranaerophilales, Actinobacteria, lower risk of head and neck cancer; Gammaproteobacteria, lower risk of endometrial cancer; Ruminiclostridium 6, higher risk of OC |
| Wei Z, 2023 | Gut microbiota | Five cancers and their subtypes (BC, LC, PCA, Endometrial cancer, OC) | One-way | Bifidobacterium: lower risk of BC; Lactobacillus: lower risk of BC and endometrial cancer |
| Bacteroides & Ruminococcus: higher risk of BC and LC; Prevotella: higher risk of BC and endometrial cancer; Faecalibacterium, Roseburia, Blautia, Akkermansia & Eubacterium: lower risk of LC; Alphaproteobacteria: lower risk of PCA; Christensenellaceae, Streptococcaceae, Peptostreptococcaceae: higher risk of OC | ||||
| Ma J, 2023 | Gut microbiota | Liver cancer | One-way | Ruminococcaceae, Porphyromonadaceae & Bacteroidetes: lower risk of liver cancer |
| Ni JJ, 2022 | Gut microbiota | CRC | One-way | Blautia: lower risk of CRC |
| Li W, 2023 | Gut microbiota | CRC | Bidirectional | Bacteroides: higher risk; Faecalibacterium, Blautia & Ruminococcus: lower risk |
| Xie N, 2021 | Gut microbiota | CRC, GC, EC, liver cancer, and PC | Bidirectional | Bacteroides: higher risk of CRC and GC; Faecalibacterium: lower risk of CRC and GC; Blautia & Ruminococcus: lower risk of CRC; Prevotella: higher risk of EC |
| Su Q, 2023 | Gut microbiota | CRC, GC, EC, PC, liver cancer, gallbladder cancer | Bidirectional | Bifidobacterium: lower risk of CRC; Faecalibacterium: lower risk of GC; Ruminococcus: higher risk of GC; Lactobacillus: lower risk of EC; Prevotella: higher risk of PC; Bacteroides: higher risk of liver cancer; Clostridium: higher risk of gallbladder cancer |
| Hatcher, 2023 | Gut microbiota | CRC | One-way | Fusobacterium, Peptostreptococcus & Parvimonas: higher risk |
| Zhou H, 2020 | Gut microbiota | LC | One-way | Weissella & Oscillospira: lower risk |
| Yang Z, 2022 | Gut microbiota | Barrett’s esophagus | One-way | Prevotella & Veillonella: higher risk; Lactobacillus: lower barrett’s esophagus risk |
| Li H, 2023 | Gut microbiota | CRC | One-way | Bacteroides: higher risk of left-sided colon cancer and stage III CRC |
| Faecalibacterium: lower risk of right-sided colon cancer and stage I CRC | ||||
| Fusobacterium: higher risk of rectal cancer and stage IV CRC | ||||
| Hong W, 2023 | Gut microbiota | BC | One-way | Bifidobacterium: lower risk of ER-positive BC; Lactobacillus: lower risk of ER-negative BC; Prevotella: higher risk of ER-positive BC |
BC, breast cancer; CRC, Colorectal cancer; LC, Lung cancer; GC, Gastric cancer; EC, Esophageal cancer; PC, Pancreatic cancer; PCA, Prostate cancer; OC, Ovarian cancer.
Figure 3.
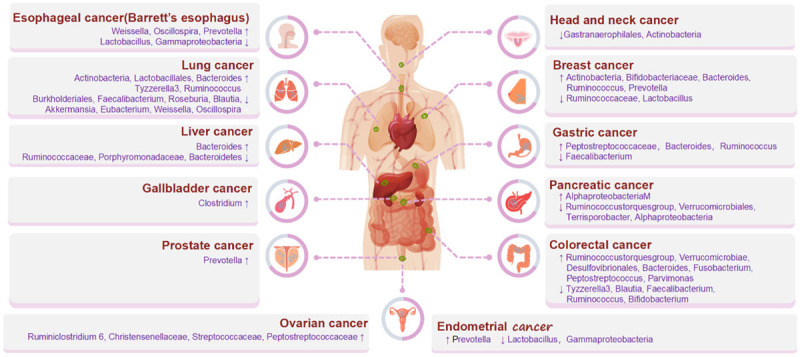
Summary of current research on microbiome causality in relation to multiple cancers.
Mendelian randomization illuminating cancer-microbiome causality
In addition to microbes promoting tumorigenesis and progression, cancer may modify microbial communities’ composition. Research has revealed that various forms of cancer, along with tumors that possess distinct clinicopathologic characteristics such as cancer stage, grade, and receptor status, display notable variations in the relative prevalence of microbial communities [2]. The gut microbiota in individuals diagnosed with colon cancer frequently exhibits substantial alterations in comparison to those in healthy persons. As an illustration, specific pathogenic bacteria, such as Fusobacterium nucleatum, tend to exhibit higher levels of prevalence among individuals diagnosed with colon cancer [15]. Sagarika et al. [91] breast cancer subtypes exhibit distinct microbiome compositions, with estrogen receptor-positive tumors displaying the highest microbial diversity and triple-negative tumors exhibiting the lowest. Different microbial profiles enable the discrimination of various breast cancer subtypes.
Four MR studies explored the bidirectional correlation between cancer and microbes. Zhu et al. [78] also employed a methodology wherein cancer was considered the independent variable (exposure) and the gut microbiota as the dependent variable (outcome). The authors observed a significant association between lung adenocarcinoma and the genus Tyzzerella 3 (P=1.02×10-2, IVW), indicating a bidirectional causal relationship between these entities. Li et al. [82] suggest a significant association between genetic susceptibility to colorectal adenomas and the heightened prevalence of the class Gammaproteobacteria and the family Enterobacteriaceae by the reverse MR analysis. Su et al. [80] indicated significant links between genetic susceptibility to digestive system cancers and the relative prevalence of certain bacterial species. In the study on gut microbiota and digestive system cancers, Xie et al. [81] identified specific cancers can regulate the relative abundance of particular strains of gut microbiota by the reverse MR.
In summation, the body of evidence presented underscores the pivotal role of cancer in gut microbiota, offering a fertile ground for further exploration into the underlying mechanisms and potential therapeutic interventions.
MR studies of microbiome, metabolites, and cancers
A comprehensive study encompassed a sample size of 3432 individuals from China [92]. The study employed bidirectional MR methods to examine the causal associations between the gut microbiome and blood metabolites. This work used a hierarchical clustering approach to analyze the associations between 12 microbial features and eight blood metabolites. The analysis revealed 17 causal linkages from the gut microbiota to the blood metabolites. The clustering process resulted in the formation of two distinct groups. One cohort observed reduced plasma triglyceride and alanine levels by manipulating gut microbial taxa or functional modules.
In contrast, another cohort observed a drop in 5-methyltetrahydrofuran or progesterone levels by manipulating gut microbial characteristics while monitoring increased serum uric acid or plasma glutamate levels. Alistipes negatively affected blood triglycerides, reduced the risk of hepatocellular carcinoma (P=0.045), and increased the risk of colorectal cancer (P=0.047). Escherichia coli increased the risk of hepatocellular carcinoma (P=0.04). Similarly, Salmonella enterica increased the risk of prostate cancer, and Pseudomonadales increased the risk of gastric (P=0.008), esophageal (P=0.027), and biliary tract (P=0.034) cancers. Streptococcus parasanguinis had a positive effect on colorectal cancer. These results illustrate the potential significance of the gut microbe-blood metabolite relationship in understanding and preventing cancer. The research suggested the value of human genetic data in determining the importance of gut microbial characteristics for additional mechanical and their involvement in cancers [92].
Utilizing bidirectional MR and a substantial sample size sourced from China, this study investigated the complex interconnections between blood metabolites and the gut microbiome. The findings from this study spotlighted the transformative potential of harnessing the synergies between microbiota, metabolites, and malignancies, promising a new era of insights in cancer biology.
Mendelian randomization in the age of multi-omics: a confluence of opportunities
MR emerges as a pivotal instrument in causal assessment, seamlessly integrating with novel technological advancements in microbiological research. A salient application of MR lies in its capability to harness the power of multi-omics, a holistic approach that delves deep into various tiers of a biological system. From the meticulous examination of DNA sequences in genomics to probing mRNA expression in transcriptomics, to the deep-dive scrutiny of protein dynamics in proteomics, and finally, to the expansive analysis of cellular metabolites in metabolomics, multi-omics stands as a comprehensive beacon in biomedical research [93]. Auwerx et al. [94] present a novel multivariate MR methodology that combines findings from GWAS with information on genetic variations influencing transcript levels or metabolite composition. The variants, as mentioned above, known as eQTL (expressed quantitative trait loci) and mQTL (metabolite QTL), can be accessed from various demographic cohorts, enabling researchers to leverage the considerable wealth of existing information [95]. The causal links between transcripts and metabolites were established by employing overlapping mQTL and eQTL as IVs. Determining causal effects between metabolites and phenotypes of interest is subsequently conducted utilizing mQTL and genetic variations found in GWAS. Subsequently, statistical computations were performed on the three entities, employing metabolism-related variation as an instrumental variable to establish a causal link between transcript, metabolite, and trait. This analysis aims to ascertain the extent to which transcription, either directly or through unidentified mediators, influences the trait and the extent to which fluctuation in metabolite levels mediates the association between transcription and the trait [96].
The pioneering work of Xu et al. [97] introduced the utilization of multi-omics data within the framework of multivariate MR. Their study was designed to unravel potential causal pathways and molecular mechanisms linking osteoporosis (OS) genes to Crohn’s disease (CD). The primary data sources encompassed both blood and intestinal tissues. A meticulous meta-analysis was executed on the intestinal transcriptome to identify genes exhibiting differential expression in Crohn’s disease patients. By adopting the Summary-based MR approach, the research integrated GWAS data of CD with blood eQTL and DNA mQTL. Notably, incorporating gut eQTL and fecal microbial mBQTL in this study offered novel insights into potential interactions between host OS genes and the gastrointestinal microbiome. Two additional MR techniques were incorporated for sensitivity analyses to ensure robustness, addressing potential heterogeneity. The validity of the findings was further underscored by their partial replication in distinct multi-omics datasets.
Subsequently, the research by Darci-Maher et al. [98] amalgamated Mendelian randomization with RNA-seq sequencing techniques, focusing on adipose and liver tissues. Their primary objective was to illuminate the implications of variations in blood triglyceride levels for non-alcoholic fatty liver disease (NAFLD).
In a distinct avenue of exploration, Klerk et al. [99] embarked on an investigative journey to discern fluctuations in long non-coding RNAs (lncRNAs) and messenger RNAs (mRNAs) within the blood profiles of individuals afflicted with type 2 diabetes mellitus. Leveraging the two-sample MR methodology, the researchers probed the genetic determinants modulating mRNA expression. Moreover, they endeavored to delineate the potential causative roles of both lncRNAs and mRNAs in diabetes-associated phenotypes, particularly emphasizing lipid metabolism and anthropometric determinants [100].
The integration of this multi-faceted data unfolds a treasure trove of biological insights. It bestows researchers with a panoramic view of biological processes, disease mechanisms, and the nuances of therapeutic responses [101,102]. For instance, genetic anomalies linked to a particular disease might ripple through the biological hierarchy, influencing gene expression, protein functionality, and metabolite levels [103]. Through MR, it becomes feasible to discern the intricate web of causal associations between these biomarkers and specific health conditions or diseases.
Moreover, the confluence of multi-omics data promises to unveil novel biomarkers, pivotal for timely disease detection, prognostic evaluations, and tailoring therapeutic interventions [104]. In the burgeoning arena of personalized medicine, this integrated data becomes instrumental in identifying patients more receptive to specific treatments, thus heralding a new era of bespoke medical interventions [105,106]. Yet, as with all grand endeavors, challenges abound. Multi-omics data’s sheer volume and intricacy demand innovative statistical methodologies and sophisticated computational tools. Furthermore, reconciling data from varied sour-ces or platforms necessitates vigilant data harmonization to forestall potential inconsistencies or biases. It becomes paramount to institute meticulous data preprocessing and standardization regimes.
Future prospects in Mendelian randomization research
As the field of microbiomics evolves towards a paradigm emphasizing quality over quantity and functional understanding over mere structural characterization, the integration of novel bioinformatics tools becomes imperative for surmounting existing challenges and enhancing the efficacy of MR methodologies. This paradigm shift necessitates the adoption of sophisticated sequencing technologies, enabling the acquisition of intricate multi-omics data. Concurrently, there is a growing need for the development of refined data analysis techniques and computational models tailored for such complex datasets.
A notable advancement in this realm is the integration of machine learning and deep learning approaches. These methodologies are instrumental in discerning intricate patterns and interrelations within multi-omics data. The increasing relevance of Artificial Intelligence (AI) in bioinformatics, particularly in the context of microbiome research, cannot be overstated. AI and machine learning algorithms are pivotal in processing and analyzing large-scale bioinformatics data with enhanced efficiency. Such technological advancements facilitate researchers in identifying key patterns and associations within microbiome datasets, thereby enabling more accurate predictions and inferences.
In the analysis of microbiome data, traditional machine learning models like linear regression, random forests, and support vector machines have demonstrated efficacy and have been applied in various studies, including those predicting host ecological imbalances [107]. Furthermore, fully connected neural network architectures have shown a higher degree of classification accuracy in predicting host phenotypes from raw macrogenomic data compared to traditional methodologies. Advanced deep learning methods, such as Convolutional Neural Networks (CNNs), as evidenced in applications like TaxNN and PopPhy-CNN, highlight the superiority of these approaches in tasks involving prediction of host phenotypes [108].
Another significant trajectory in microbiome research is integrated metagenomics analysis, encompassing Metabolomics, Proteomics, and Genomics. This integrative approach allows researchers to construct a holistic view of the microbiome’s structure and function. Such multidimensional analyses are crucial for understanding the dynamics of microbial interactions and adaptability in varying environments. Utilizing network analysis tools facilitates the elucidation of complex interactions both among microbiome constituents and between microbes and their hosts, thereby deepening our comprehension of the microbiome’s influence on host health and disease states [109].
For instance, Recurrent Neural Networks (RNNs) have been effectively employed for detecting dependencies and dynamic patterns in time-series data, with models based on long-term tracking data, such as three-year infant allergy phenotype studies, outperforming traditional models [109,110]. Additionally, autoencoders are increasingly being used for data dimensionality reduction, exemplified by DeepMicro’s neural network, which employs a variant of multilayer autoencoder architecture, unveiling how diverse underlying information can enhance the prediction accuracy for conditions like Irritable Bowel Syndrome and Type 2 Diabetes.
In conclusion, the incorporation of cloud computing and big data technologies marks a transformative phase in microbiome research. These technologies are capable of managing and analyzing vast microbiome datasets, significantly improving data processing efficiency and scalability. They also foster enhanced data sharing and collaborative efforts, thereby propelling the pace of scientific discoveries in this domain.
Conclusions
In summation, MR offers a robust scaffold, bridging the realms of microbiology and oncogenesis. While challenges persist, the relentless march of technological progress in biomedical research promises a horizon replete with opportunities and transformative breakthroughs.
Acknowledgements
We acknowledge all the participating authors for their contributions to this article. In addition, we appreciate that this research was supported by the National Natural Science Foundation of China (No. 82072097), the CAMS Innovation Fund for Medical Sciences (GIFMS, 2021-I2M-1-014), and the Breast Cancer Single Disease Diagnosis and Treatment Capacity Enhancement Project (RXDBZ-2022-11).
Disclosure of conflict of interest
None.
Abbreviations
- MR
Mendelian randomization
- RCT
randomized controlled trial
- GWAS
genome-wide association analysis
- GWAS
genome-wide association studies
- SCFAs
short-chain fatty acids
- 5-HT
5-hydroxytryptophan
- ROS
oxygen species
- TMAO
trimethylamine oxide
- SNPs
single-nucleotide polymorphisms
- IVW
inverse variance weighted
- WM
weighted median
- LD
Linkage Disequilibrium
References
- 1.Gunjur A. Cancer and the microbiome. Lancet Oncol. 2020;21:888. doi: 10.1016/S1470-2045(20)30351-X. [DOI] [PubMed] [Google Scholar]
- 2.Knippel RJ, Drewes JL, Sears CL. The cancer microbiome: recent highlights and knowledge gaps. Cancer Discov. 2021;11:2378–2395. doi: 10.1158/2159-8290.CD-21-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xavier JB, Young VB, Skufca J, Ginty F, Testerman T, Pearson AT, Macklin P, Mitchell A, Shmulevich I, Xie L, Caporaso JG, Crandall KA, Simone NL, Godoy-Vitorino F, Griffin TJ, Whiteson KL, Gustafson HH, Slade DJ, Schmidt TM, Walther-Antonio MRS, Korem T, Webb-Robertson BM, Styczynski MP, Johnson WE, Jobin C, Ridlon JM, Koh AY, Yu M, Kelly L, Wargo JA. The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends Cancer. 2020;6:192–204. doi: 10.1016/j.trecan.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 5.Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. Br J Cancer. 2016;114:237–242. doi: 10.1038/bjc.2015.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158:322–340. doi: 10.1053/j.gastro.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong C, Liang L, Liu G, Du L, Yang Y, Liu J, Shi D, Li X, Ma Y. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut. 2023;72:1129–1142. doi: 10.1136/gutjnl-2022-327156. [DOI] [PubMed] [Google Scholar]
- 8.Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, Li H, Bai H, Liu X, Zhang Y, Wang C, Guo Y, Li N, Cai S. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. 2022;185:1356–1372. e1326. doi: 10.1016/j.cell.2022.02.027. [DOI] [PubMed] [Google Scholar]
- 9.Papakonstantinou A, Nuciforo P, Borrell M, Zamora E, Pimentel I, Saura C, Oliveira M. The conundrum of breast cancer and microbiome - a comprehensive review of the current evidence. Cancer Treat Rev. 2022;111:102470. doi: 10.1016/j.ctrv.2022.102470. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, Robinson MR, McGrath JJ, Visscher PM, Wray NR, Yang J. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernán MA, Robins JM. Instruments for causal inference: an epidemiologist’s dream? Epidemiology. 2006;17:360–372. doi: 10.1097/01.ede.0000222409.00878.37. [DOI] [PubMed] [Google Scholar]
- 12.Fu R, Kim SJ. Inferring causality from observational studies: the role of instrumental variable analysis. Kidney Int. 2021;99:1303–1308. doi: 10.1016/j.kint.2021.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Lousdal ML. An introduction to instrumental variable assumptions, validation and estimation. Emerg Themes Epidemiol. 2018;15:1. doi: 10.1186/s12982-018-0069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong-Rolle A, Wei HK, Zhao C, Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell. 2021;12:426–435. doi: 10.1007/s13238-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saus E, Iraola-Guzmán S, Willis JR, Brunet-Vega A, Gabaldón T. Microbiome and colorectal cancer: roles in carcinogenesis and clinical potential. Mol Aspects Med. 2019;69:93–106. doi: 10.1016/j.mam.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekula P, Del Greco M F, Pattaro C, Köttgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. 2016;27:3253–3265. doi: 10.1681/ASN.2016010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26:2333–2355. doi: 10.1177/0962280215597579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ference BA, Holmes MV, Smith GD. Using Mendelian randomization to improve the design of randomized trials. Cold Spring Harb Perspect Med. 2021;11:a040980. doi: 10.1101/cshperspect.a040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23:R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohtani N. Microbiome and cancer. Semin Immunopathol. 2015;37:65–72. doi: 10.1007/s00281-014-0457-1. [DOI] [PubMed] [Google Scholar]
- 21.Zyoud SH, Al-Jabi SW, Amer R, Shakhshir M, Shahwan M, Jairoun AA, Akkawi M, Abu Taha A. Global research trends on the links between the gut microbiome and cancer: a visualization analysis. J Transl Med. 2022;20:83. doi: 10.1186/s12967-022-03293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahman MM, Islam MR, Shohag S, Ahasan MT, Sarkar N, Khan H, Hasan AM, Cavalu S, Rauf A. Microbiome in cancer: role in carcinogenesis and impact in therapeutic strategies. Biomed Pharmacother. 2022;149:112898. doi: 10.1016/j.biopha.2022.112898. [DOI] [PubMed] [Google Scholar]
- 23.Doocey CM, Finn K, Murphy C, Guinane CM. The impact of the human microbiome in tumorigenesis, cancer progression, and biotherapeutic development. BMC Microbiol. 2022;22:53. doi: 10.1186/s12866-022-02465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science. 2021;371:eabc4552. doi: 10.1126/science.abc4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matson V, Chervin CS, Gajewski TF. Cancer and the microbiome-influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology. 2021;160:600–613. doi: 10.1053/j.gastro.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elinav E, Garrett WS, Trinchieri G, Wargo J. The cancer microbiome. Nat Rev Cancer. 2019;19:371–376. doi: 10.1038/s41568-019-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Deng Y, Chu Q, Zhang P. Gut microbiome and cancer immunotherapy. Cancer Lett. 2019;447:41–47. doi: 10.1016/j.canlet.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 28.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25:377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 29.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33:570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajagopala SV, Vashee S, Oldfield LM, Suzuki Y, Venter JC, Telenti A, Nelson KE. The human microbiome and cancer. Cancer Prev Res (Phila) 2017;10:226–234. doi: 10.1158/1940-6207.CAPR-16-0249. [DOI] [PubMed] [Google Scholar]
- 31.Naik PP, Panigrahi S, Parida R, Praharaj PP, Bhol CS, Patil S, Manjunath N, Ghosh D, Patra SK, Bhutia SK. Metabostemness in cancer: linking metaboloepigenetics and mitophagy in remodeling cancer stem cells. Stem Cell Rev Rep. 2022;18:198–213. doi: 10.1007/s12015-021-10216-9. [DOI] [PubMed] [Google Scholar]
- 32.Moindjie H, Rodrigues-Ferreira S, Nahmias C. Mitochondrial metabolism in carcinogenesis and cancer therapy. Cancers (Basel) 2021;13:3311. doi: 10.3390/cancers13133311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M, Soga T, Pollard PJ. Oncometabolites: linking altered metabolism with cancer. J Clin Invest. 2013;123:3652–3658. doi: 10.1172/JCI67228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomes SD, Oliveira CS, Azevedo-Silva J, Casanova MR, Barreto J, Pereira H, Chaves SR, Rodrigues LR, Casal M, Côrte-Real M, Baltazar F, Preto A. The role of diet related short-chain fatty acids in colorectal cancer metabolism and survival: prevention and therapeutic implications. Curr Med Chem. 2020;27:4087–4108. doi: 10.2174/0929867325666180530102050. [DOI] [PubMed] [Google Scholar]
- 35.Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 36.Wang G, Yu Y, Wang YZ, Wang JJ, Guan R, Sun Y, Shi F, Gao J, Fu XL. Role of SCFAs in gut microbiome and glycolysis for colorectal cancer therapy. J Cell Physiol. 2019;234:17023–17049. doi: 10.1002/jcp.28436. [DOI] [PubMed] [Google Scholar]
- 37.Yang HJ, Kim JH. Role of microbiome and its metabolite, short chain fatty acid in prostate cancer. Investig Clin Urol. 2023;64:3–12. doi: 10.4111/icu.20220370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu P, Wang Y, Yang G, Zhang Q, Meng L, Xin Y, Jiang X. The role of short-chain fatty acids in intestinal barrier function, inflammation, oxidative stress, and colonic carcinogenesis. Pharmacol Res. 2021;165:105420. doi: 10.1016/j.phrs.2021.105420. [DOI] [PubMed] [Google Scholar]
- 39.Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Becker E, Bengs S, Aluri S, Opitz L, Atrott K, Stanzel C, Castro PAR, Rogler G, Frey-Wagner I. Doxycycline, metronidazole and isotretinoin: do they modify microRNA/mRNA expression profiles and function in murine T-cells? Sci Rep. 2016;6:37082. doi: 10.1038/srep37082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hassabo AG, Nada AA, Ibrahim HM, Abou-Zeid NY. Impregnation of silver nanoparticles into polysaccharide substrates and their properties. Carbohydr Polym. 2015;122:343–350. doi: 10.1016/j.carbpol.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Zabor EC, Kaizer AM, Hobbs BP. Randomized controlled trials. Chest. 2020;158:S79–S87. doi: 10.1016/j.chest.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou L, Qiu W, Wang J, Zhao A, Zhou C, Sun T, Xiong Z, Cao P, Shen W, Chen J, Lai X, Zhao LH, Wu Y, Li M, Qiu F, Yu Y, Xu ZZ, Zhou H, Jia W, Liao Y, Retnakaran R, Krewski D, Wen SW, Clemente JC, Chen T, Xie RH, He Y. Effects of vaginal microbiota transfer on the neurodevelopment and microbiome of cesarean-born infants: a blinded randomized controlled trial. Cell Host Microbe. 2023;31:1232–1247. e1235. doi: 10.1016/j.chom.2023.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Alemu BK, Azeze GG, Wu L, Lau SL, Wang CC, Wang Y. Effects of maternal probiotic supplementation on breast milk microbiome and infant gut microbiome and health: a systematic review and meta-analysis of randomized controlled trials. Am J Obstet Gynecol MFM. 2023;5:101148. doi: 10.1016/j.ajogmf.2023.101148. [DOI] [PubMed] [Google Scholar]
- 45.Rinott E, Meir AY, Tsaban G, Zelicha H, Kaplan A, Knights D, Tuohy K, Scholz MU, Koren O, Stampfer MJ, Wang DD, Shai I, Youngster I. The effects of the Green-Mediterranean diet on cardiometabolic health are linked to gut microbiome modifications: a randomized controlled trial. Genome Med. 2022;14:29. doi: 10.1186/s13073-022-01015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Tekle G, Garrett WS. Bacteria in cancer initiation, promotion and progression. Nat Rev Cancer. 2023;23:600–618. doi: 10.1038/s41568-023-00594-2. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Zhang Y. Intratumor microbiome in cancer progression: current developments, challenges and future trends. Biomark Res. 2022;10:37. doi: 10.1186/s40364-022-00381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D’Onofrio BM, Sjölander A, Lahey BB, Lichtenstein P, Öberg AS. Accounting for confounding in observational studies. Annu Rev Clin Psychol. 2020;16:25–48. doi: 10.1146/annurev-clinpsy-032816-045030. [DOI] [PubMed] [Google Scholar]
- 49.Skrivankova VW, Richmond RC, Woolf BAR, Davies NM, Swanson SA, VanderWeele TJ, Timpson NJ, Higgins JPT, Dimou N, Langenberg C, Loder EW, Golub RM, Egger M, Davey Smith G, Richards JB. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375:n2233. doi: 10.1136/bmj.n2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Islam MZ, Tran M, Xu T, Tierney BT, Patel C, Kostic AD. Reproducible and opposing gut microbiome signatures distinguish autoimmune diseases and cancers: a systematic review and meta-analysis. Microbiome. 2022;10:218. doi: 10.1186/s40168-022-01373-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allali I, Abotsi RE, Tow LA, Thabane L, Zar HJ, Mulder NM, Nicol MP. Human microbiota research in Africa: a systematic review reveals gaps and priorities for future research. Microbiome. 2021;9:241. doi: 10.1186/s40168-021-01195-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lampeter T, Love C, Tang TT, Marella AS, Lee HY, Oganyan A, Moffat D, Kareem A, Rusling M, Massmann A, Orr M, Bongiorno C, Yuan LL. Risk of bias assessment tool for systematic review and meta-analysis of the gut microbiome. Gut Microbiome. 2023;4:e13. doi: 10.1017/gmb.2023.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang N, Kandalai S, Zhou X, Hossain F, Zheng Q. Applying multi-omics toward tumor microbiome research. iMeta. 2023;2:e73. doi: 10.1002/imt2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Menyhárt O, Győrffy B. Multi-omics approaches in cancer research with applications in tumor subtyping, prognosis, and diagnosis. Comput Struct Biotechnol J. 2021;19:949–960. doi: 10.1016/j.csbj.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arıkan M, Muth T. Integrated multi-omics analyses of microbial communities: a review of the current state and future directions. Mol Omics. 2023;19:607–623. doi: 10.1039/d3mo00089c. [DOI] [PubMed] [Google Scholar]
- 56.Morais LH, Schreiber HL 4th, Mazmanian SK. The gut microbiota-brain axis in behaviour and brain disorders. Nat Rev Microbiol. 2021;19:241–255. doi: 10.1038/s41579-020-00460-0. [DOI] [PubMed] [Google Scholar]
- 57.Hajishengallis G. Illuminating the oral microbiome and its host interactions: animal models of disease. FEMS Microbiol Rev. 2023;47:fuad018. doi: 10.1093/femsre/fuad018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu Y, Ling Z, Arabnia H, Deng Y. Current trend and development in bioinformatics research. BMC Bioinformatics. 2020;21(Suppl 9):538. doi: 10.1186/s12859-020-03874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marcos-Zambrano LJ, Karaduzovic-Hadziabdic K, Loncar Turukalo T, Przymus P, Trajkovik V, Aasmets O, Berland M, Gruca A, Hasic J, Hron K, Klammsteiner T, Kolev M, Lahti L, Lopes MB, Moreno V, Naskinova I, Org E, Paciência I, Papoutsoglou G, Shigdel R, Stres B, Vilne B, Yousef M, Zdravevski E, Tsamardinos I, Carrillo de Santa Pau E, Claesson MJ, Moreno-Indias I, Truu J. Applications of machine learning in human microbiome studies: a review on feature selection, biomarker identification, disease prediction and treatment. Front Microbiol. 2021;12:634511. doi: 10.3389/fmicb.2021.634511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cammarota G, Ianiro G, Ahern A, Carbone C, Temko A, Claesson MJ, Gasbarrini A, Tortora G. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat Rev Gastroenterol Hepatol. 2020;17:635–648. doi: 10.1038/s41575-020-0327-3. [DOI] [PubMed] [Google Scholar]
- 61.Bharti R, Grimm DG. Current challenges and best-practice protocols for microbiome analysis. Brief Bioinform. 2021;22:178–193. doi: 10.1093/bib/bbz155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng XT, Xia LY, Zhang YG, Li S, Leng WD, Kwong JS. Periodontal disease and incident lung cancer risk: a meta-analysis of cohort studies. J Periodontol. 2016;87:1158–1164. doi: 10.1902/jop.2016.150597. [DOI] [PubMed] [Google Scholar]
- 63.Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park S, Lee S, Kim Y, Lee Y, Kang MW, Kim K, Kim YC, Han SS, Lee H, Lee JP, Joo KW, Lim CS, Kim YS, Kim DK. Short or long sleep duration and CKD: a mendelian randomization study. J Am Soc Nephrol. 2020;31:2937–2947. doi: 10.1681/ASN.2020050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: a review. Res Synth Methods. 2019;10:486–496. doi: 10.1002/jrsm.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meng C, Deng P, Miao R, Tang H, Li Y, Wang J, Wu J, Wang W, Liu S, Xia J, Lu Y. Gut microbiome and risk of ischaemic stroke: a comprehensive Mendelian randomization study. Eur J Prev Cardiol. 2023;30:613–620. doi: 10.1093/eurjpc/zwad052. [DOI] [PubMed] [Google Scholar]
- 69.Gagnon E, Mitchell PL, Manikpurage HD, Abner E, Taba N, Esko T, Ghodsian N, Thériault S, Mathieu P, Arsenault BJ. Impact of the gut microbiota and associated metabolites on cardiometabolic traits, chronic diseases and human longevity: a Mendelian randomization study. J Transl Med. 2023;21:60. doi: 10.1186/s12967-022-03799-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Zhang X, Chen D, Lu J, Gong Q, Fang J, Jiang J. Causal associations between gut microbiome and cardiovascular disease: a Mendelian randomization study. Front Cardiovasc Med. 2022;9:971376. doi: 10.3389/fcvm.2022.971376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo Q, Hu Y, Chen X, Luo Y, Chen J, Wang H. Effects of gut microbiota and metabolites on heart failure and its risk factors: a two-sample Mendelian randomization study. Front Nutr. 2022;9:899746. doi: 10.3389/fnut.2022.899746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Q, Ni JJ, Han BX, Yan SS, Wei XT, Feng GJ, Zhang H, Zhang L, Li B, Pei YF. Causal relationship between gut microbiota and autoimmune diseases: a two-sample Mendelian randomization study. Front Immunol. 2022;12:746998. doi: 10.3389/fimmu.2021.746998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ni JJ, Xu Q, Yan SS, Han BX, Zhang H, Wei XT, Feng GJ, Zhao M, Pei YF, Zhang L. Gut microbiota and psychiatric disorders: a two-sample Mendelian randomization study. Front Microbiol. 2022;12:737197. doi: 10.3389/fmicb.2021.737197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burgess S, Thompson SG CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40:755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 75.Evans DM, Davey Smith G. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genomics Hum Genet. 2015;16:327–350. doi: 10.1146/annurev-genom-090314-050016. [DOI] [PubMed] [Google Scholar]
- 76.Taylor AE, Davies NM, Ware JJ, VanderWeele T, Smith GD, Munafò MR. Mendelian randomization in health research: using appropriate genetic variants and avoiding biased estimates. Econ Hum Biol. 2014;13:99–106. doi: 10.1016/j.ehb.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 78.Long Y, Tang L, Zhou Y, Zhao S, Zhu H. Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study. BMC Med. 2023;21:66. doi: 10.1186/s12916-023-02761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei Z, Yang B, Tang T, Xiao Z, Ye F, Li X, Wu S, Huang JG, Jiang S. Gut microbiota and risk of five common cancers: a univariable and multivariable Mendelian randomization study. Cancer Med. 2023;12:10393–10405. doi: 10.1002/cam4.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Su Q, Jin C, Bo Z, Yang Y, Wang J, Wang J, Zhou J, Chen Y, Zeng H, Chen G, Wang Y. Association between gut microbiota and gastrointestinal cancer: a two-sample bi-directional Mendelian randomization study. Front Microbiol. 2023;14:1181328. doi: 10.3389/fmicb.2023.1181328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xie N, Wang Z, Shu Q, Liang X, Wang J, Wu K, Nie Y, Shi Y, Fan D, Wu J. Association between gut microbiota and digestive system cancers: a bidirectional two-sample Mendelian randomization study. Nutrients. 2023;15:2937. doi: 10.3390/nu15132937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li W, Zhou X, Yuan S, Wang L, Yu L, Sun J, Chen J, Xiao Q, Wan Z, Zheng JS, Zhang CX, Larsson SC, Farrington SM, Law P, Houlston RS, Tomlinson I, Ding KF, Dunlop MG, Theodoratou E, Li X. Exploring the complex relationship between gut microbiota and risk of colorectal neoplasia using bidirectional Mendelian randomization analysis. Cancer Epidemiol Biomarkers Prev. 2023;32:809–817. doi: 10.1158/1055-9965.EPI-22-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H, Sheng D, Jin C, Zhao G, Zhang L. Identifying and ranking causal microbial biomarkers for colorectal cancer at different cancer subsites and stages: a Mendelian randomization study. Front Oncol. 2023;13:1224705. doi: 10.3389/fonc.2023.1224705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xie Y, Xie F, Zhou X, Zhang L, Yang B, Huang J, Wang F, Yan H, Zeng L, Zhang L, Zhou F. Microbiota in tumors: from understanding to application. Adv Sci (Weinh) 2022;9:e2200470. doi: 10.1002/advs.202200470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hatcher C, Richenberg G, Waterson S, Nguyen LH, Joshi AD, Carreras-Torres R, Moreno V, Chan AT, Gunter M, Lin Y, Qu C, Song M, Casey G, Figueiredo JC, Gruber SB, Hampe J, Hampel H, Jenkins MA, Keku TO, Peters U, Tangen CM, Wu AH, Hughes DA, Rühlemann MC, Raes J, Timpson NJ, Wade KH. Application of Mendelian randomization to explore the causal role of the human gut microbiome in colorectal cancer. Sci Rep. 2023;13:5968. doi: 10.1038/s41598-023-31840-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ni JJ, Li XS, Zhang H, Xu Q, Wei XT, Feng GJ, Zhao M, Zhang ZJ, Zhang L, Shen GH, Li B. Mendelian randomization study of causal link from gut microbiota to colorectal cancer. BMC Cancer. 2022;22:1371. doi: 10.1186/s12885-022-10483-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou H, Liu J, Shen J, Fang W, Zhang L. Gut microbiota and lung cancer: a Mendelian randomization study. JTO Clin Res Rep. 2020;1:100042. doi: 10.1016/j.jtocrr.2020.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Z, Yu R, Deng W, Wang W. Roles of 21 genera of human gut microbiota in Barrett’s esophagus risk: a Mendelian randomization study. Front Genet. 2022;13:894900. doi: 10.3389/fgene.2022.894900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ma J, Li J, Jin C, Yang J, Zheng C, Chen K, Xie Y, Yang Y, Bo Z, Wang J, Su Q, Wang J, Chen G, Wang Y. Association of gut microbiome and primary liver cancer: a two-sample Mendelian randomization and case-control study. Liver Int. 2023;43:221–233. doi: 10.1111/liv.15466. [DOI] [PubMed] [Google Scholar]
- 90.Bowden SJ, Doulgeraki T, Bouras E, Markozannes G, Athanasiou A, Grout-Smith H, Kechagias KS, Ellis LB, Zuber V, Chadeau-Hyam M, Flanagan JM, Tsilidis KK, Kalliala I, Kyrgiou M. Risk factors for human papillomavirus infection, cervical intraepithelial neoplasia and cervical cancer: an umbrella review and follow-up Mendelian randomisation studies. BMC Med. 2023;21:274. doi: 10.1186/s12916-023-02965-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Banerjee S, Wei Z, Tian T, Bose D, Shih NNC, Feldman MD, Khoury T, De Michele A, Robertson ES. Prognostic correlations with the microbiome of breast cancer subtypes. Cell Death Dis. 2021;12:831. doi: 10.1038/s41419-021-04092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu X, Tong X, Zou Y, Lin X, Zhao H, Tian L, Jie Z, Wang Q, Zhang Z, Lu H, Xiao L, Qiu X, Zi J, Wang R, Xu X, Yang H, Wang J, Zong Y, Liu W, Hou Y, Zhu S, Jia H, Zhang T. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat Genet. 2022;54:52–61. doi: 10.1038/s41588-021-00968-y. [DOI] [PubMed] [Google Scholar]
- 93.Wörheide MA, Krumsiek J, Kastenmüller G, Arnold M. Multi-omics integration in biomedical research - a metabolomics-centric review. Anal Chim Acta. 2021;1141:144–162. doi: 10.1016/j.aca.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Auwerx C, Sadler MC, Woh T, Reymond A, Kutalik Z, Porcu E. Exploiting the mediating role of the metabolome to unravel transcript-to-phenotype associations. Elife. 2023;12:e81097. doi: 10.7554/eLife.81097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weith M, Beyer A. The next step in Mendelian randomization. Elife. 2023;12:e86416. doi: 10.7554/eLife.86416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu S, Li X, Zhang S, Qi C, Zhang Z, Ma R, Xiang L, Chen L, Zhu Y, Tang C, Bourgonje AR, Li M, He Y, Zeng Z, Hu S, Feng R, Chen M. Oxidative stress gene expression, DNA methylation, and gut microbiota interaction trigger Crohn’s disease: a multi-omics Mendelian randomization study. BMC Med. 2023;21:179. doi: 10.1186/s12916-023-02878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Darci-Maher N, Alvarez M, Arasu UT, Selvarajan I, Lee SHT, Pan DZ, Miao Z, Das SS, Kaminska D, Örd T, Benhammou JN, Wabitsch M, Pisegna JR, Männistö V, Pietiläinen KH, Laakso M, Sinsheimer JS, Kaikkonen MU, Pihlajamäki J, Pajukanta P. Cross-tissue omics analysis discovers ten adipose genes encoding secreted proteins in obesity-related non-alcoholic fatty liver disease. EBioMedicine. 2023;92:104620. doi: 10.1016/j.ebiom.2023.104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Klerk JA, Beulens JWJ, Mei H, Bijkerk R, van Zonneveld AJ, Koivula RW, Elders PJM, ’t Hart LM, Slieker RC. Altered blood gene expression in the obesity-related type 2 diabetes cluster may be causally involved in lipid metabolism: a Mendelian randomisation study. Diabetologia. 2023;66:1057–1070. doi: 10.1007/s00125-023-05886-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rappoport N, Shamir R. Multi-omic and multi-view clustering algorithms: review and cancer benchmark. Nucleic Acids Res. 2018;46:10546–10562. doi: 10.1093/nar/gky889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zeng D, Ye Z, Shen R, Yu G, Wu J, Xiong Y, Zhou R, Qiu W, Huang N, Sun L, Li X, Bin J, Liao Y, Shi M, Liao W. IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front Immunol. 2021;12:687975. doi: 10.3389/fimmu.2021.687975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Poirion OB, Jing Z, Chaudhary K, Huang S, Garmire LX. DeepProg: an ensemble of deep-learning and machine-learning models for prognosis prediction using multi-omics data. Genome Med. 2021;13:112. doi: 10.1186/s13073-021-00930-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kopczynski D, Coman C, Zahedi RP, Lorenz K, Sickmann A, Ahrends R. Multi-OMICS: a critical technical perspective on integrative lipidomics approaches. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:808–811. doi: 10.1016/j.bbalip.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 104.Olivier M, Asmis R, Hawkins GA, Howard TD, Cox LA. The need for multi-omics biomarker signatures in precision medicine. Int J Mol Sci. 2019;20:4781. doi: 10.3390/ijms20194781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wishart D. Metabolomics and the multi-omics view of cancer. Metabolites. 2022;12:154. doi: 10.3390/metabo12020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Akhoundova D, Rubin MA. Clinical application of advanced multi-omics tumor profiling: shaping precision oncology of the future. Cancer Cell. 2022;40:920–938. doi: 10.1016/j.ccell.2022.08.011. [DOI] [PubMed] [Google Scholar]
- 107.Carter AR, Sanderson E, Hammerton G, Richmond RC, Davey Smith G, Heron J, Taylor AE, Davies NM, Howe LD. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur J Epidemiol. 2021;36:465–478. doi: 10.1007/s10654-021-00757-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hernández Medina R, Kutuzova S, Nielsen KN, Johansen J, Hansen LH, Nielsen M, Rasmussen S. Machine learning and deep learning applications in microbiome research. ISME Commun. 2022;2:98. doi: 10.1038/s43705-022-00182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lo C, Marculescu R. MetaNN: accurate classification of host phenotypes from metagenomic data using neural networks. BMC Bioinformatics. 2019;20(Suppl 12):314. doi: 10.1186/s12859-019-2833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sharma D, Xu W. phyLoSTM: a novel deep learning model on disease prediction from longitudinal microbiome data. Bioinformatics. 2021;37:3707–3714. doi: 10.1093/bioinformatics/btab482. [DOI] [PubMed] [Google Scholar]


