Abstract
Triple-negative breast cancer (TNBC) is an aggressive disease characterized by remarkable intratumor heterogeneity (ITH), which poses therapeutic challenges. However, the clinical relevance and key determinant of ITH in TNBC are poorly understood. Here, we comprehensively characterized ITH levels using multi-omics data across our center’s cohort (n = 260), The Cancer Genome Atlas cohort (n = 134), and four immunotherapy-treated cohorts (n = 109). Our results revealed that high ITH was associated with poor patient survival and immunotherapy resistance. Importantly, we identified zinc finger protein 689 (ZNF689) deficiency as a crucial determinant of ITH formation. Mechanistically, the ZNF689–TRIM28 complex was found to directly bind to the promoter of long interspersed element-1 (LINE-1), inducing H3K9me3-mediated transcriptional silencing. ZNF689 deficiency reactivated LINE-1 retrotransposition to exacerbate genomic instability, which fostered ITH. Single-cell RNA sequencing, spatially resolved transcriptomics and flow cytometry analysis confirmed that ZNF689 deficiency-induced ITH inhibited antigen presentation and T-cell activation, conferring immunotherapy resistance. Pharmacological inhibition of LINE-1 significantly reduced ITH, enhanced antitumor immunity, and eventually sensitized ZNF689-deficient tumors to immunotherapy in vivo. Consistently, ZNF689 expression positively correlated with favorable prognosis and immunotherapy response in clinical samples. Altogether, our study uncovers a previously unrecognized mechanism underlying ZNF689 deficiency-induced ITH and suggests LINE-1 inhibition combined with immunotherapy as a novel treatment strategy for TNBC.
Subject terms: Breast cancer, Tumour heterogeneity, Cancer immunotherapy
Introduction
Triple-negative breast cancer (TNBC) is a highly heterogeneous disease that has long been considered a major challenge due to its aggressive behavior and poor prognosis.1 An important factor in the failure of TNBC treatment is intratumor heterogeneity (ITH), which refers to the concept that a single tumor comprises many different subclones of cells.2 These subclones exhibit distinct genotypic, phenotypic, biological, and morphological patterns, presenting a degree of ITH that enables tumor progression and resistance to treatment.3 Therefore, directly blocking ITH may improve outcomes in patients with TNBC.
Immunotherapy has shown remarkable clinical benefits for several solid tumor types, including TNBC.4,5 However, the benefits were not durable and were limited to a small subset of patients with TNBC. Recent clinical trial reports state that the anti-programmed cell death (PD)-1 antibody pembrolizumab had an objective response rate (ORR) of 21.4%, which dropped to 5.7% in the second or later lines of therapy.6,7 ITH has been reported as the key obstacle to the success of immunotherapy.8,9 Increased ITH could lead to immunoediting and immune evasion, potentially diminishing immune response.10 Therefore, blocking ITH may serve as a promising therapeutic strategy to overcome immunotherapy resistance in TNBC.
ITH is a highly complex and multifactorial phenomenon.11 Genomic instability, an aberrant state that allows cancer cells to accumulate genetic alterations, is regarded as a major cause of genetic ITH.2,9,12 In addition, phenotypic and microenvironmental heterogeneity, as well as epigenetic, transcriptional, metabolic and secretory heterogeneity, also contribute to the broader concept of ITH.8,13 TNBC is generally considered a type of breast cancer featuring a high level of ITH, and this includes a notable amount of genetic ITH.14 However, the key determinants and underlying mechanisms of genetic ITH and other forms of ITH in TNBC remain to be fully elucidated.
Conventionally, genetic ITH is evaluated using multi-region sampling15 or single-cell analysis,13 which is difficult to scale up for studies of hundreds of tumors. Next-generation sequencing data derived from even single regions can be analyzed to uncover subclonal populations. This approach has been shown to provide sufficient resolution for a window into a tumor’s overall level of genetic ITH and enables the study of a larger tumor sample set.14,16,17 Recently, based on both copy number alterations and tumor-specific mutations, the bioinformatics tool PyClone has been proven to be a simple, quantitative, and generally applicable way to evaluate genetic ITH.18 In addition, histologic ITH can be assessed using the different morphologies of tumor clones on hematoxylin and eosin (H&E) images.19,20 However, due to the lack of large-scale cohorts with multi-omics data in TNBC, a comprehensive landscape of ITH has not been explored.
Based on this background, our study aimed to extensively characterize the degrees of both genetic ITH and histologic ITH — two key components within the vast spectrum of ITH — in TNBC. To achieve this, we utilized multi-omics data derived from the Fudan University Shanghai Cancer Center (FUSCC) and The Cancer Genome Atlas (TCGA) datasets. We undertook a discovery effort to identify the important determinants and underlying mechanisms of ITH in TNBC, and provided a potential therapeutic strategy by which blocking ITH would suppress cancer progression and improve patient survival.
Results
High ITH reduces patient survival and confers immunotherapy resistance in TNBC
We leveraged multi-omics data across our center’s cohort (FUSCC cohort, n = 260) and TCGA cohort (n = 134) to comprehensively characterize the ITH of TNBC, including whole-exome sequencing (WES) results, somatic copy number variation (SCNV) data, transcriptomic RNA sequencing (RNA-seq) data, H&E images, and clinical data (Fig. 1a). To infer genetic ITH, we used PyClone to estimate the number of subclones per tumor. To quantify histologic ITH, we measured the size and staining intensity of every nucleus detected in the tumor’s H&E-stained images using the established image analysis software CellProfile.21 In the FUSCC cohort, patients were divided into high and low genetic ITH groups using 3 subclones as the cut-off level (Supplementary information, Fig. S1a). Patients were also classified into high and low histologic ITH groups using the median value of 0.5 as the cut-off level. A strong correlation was observed between genetic ITH and histologic ITH in the FUSCC cohort (Supplementary information, Fig. S1b, c), suggesting that histologic ITH could be used to discriminate the genetic ITH level. Moreover, we characterized the genetic ITH and histologic ITH of TNBC within the TCGA cohort, and our findings were in line with the results from our cohort (Supplementary information, Fig. S1d–f).
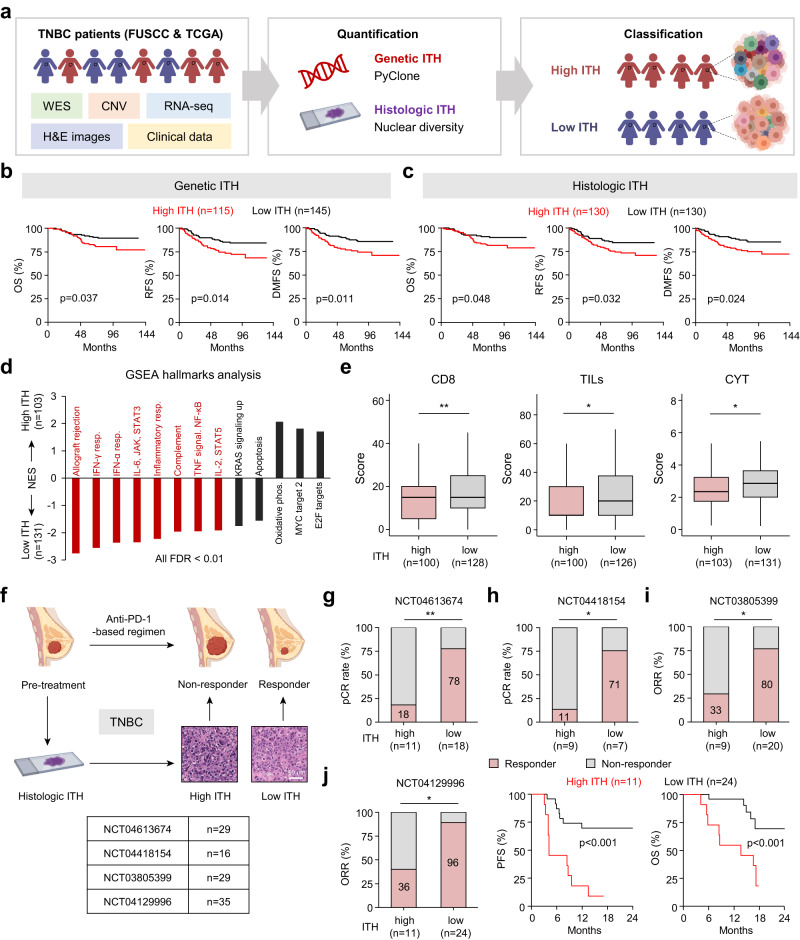
Fig. 1. High ITH reduces patient survival and confers immunotherapy resistance in TNBC.
a Flow chart of ITH analysis in TNBC. b, c Kaplan–Meier analyses of the OS, RFS and DMFS of FUSCC patients grouped according to genetic ITH (b) and histologic ITH (c). d Normalized enrichment score (NES) for hallmark gene sets correlated with genetic ITH levels in the FUSCC cohort. Red indicates immune gene sets. FDR false discovery rate, phos. phosphorylation, resp. response. e The CD8, TILs, and CYT scores of patients in the FUSCC cohort with different levels of genetic ITH. f Schematic overview of the analytical workflow. Four clinical trials were used to document the relationship between pretreatment ITH and anti-PD-1 responses in TNBC. Scale bar, 60 μm. g–i Analysis of the responses in patients with different levels of histologic ITH in NCT04613674 (g), NCT04418154 (h) and NCT03805399 (i). j Analysis of the response, PFS and OS in patients with different levels of histologic ITH in NCT04129996. P values were determined using log-rank tests (b, c, j), Wilcoxon tests (e), and Fisher’s exact tests (g–j). *P < 0.05, **P < 0.01.
To investigate the clinical relevance of ITH in TNBC, we performed Kaplan–Meier survival analysis. Our findings suggested that increased genetic or histologic ITH correlated with lower overall survival (OS), recurrence-free survival (RFS), and distant metastasis-free survival (DMFS) in the FUSCC cohort (Fig. 1b, c), and with reduced OS and disease-specific survival in the TCGA cohort (Supplementary information, Fig. S1g). In the multivariate analyses, both high genetic ITH and high histologic ITH were found to be independent poor prognostic factors (Supplementary information, Fig. S1h, i). However, no associations were found between ITH levels and the clinicopathological characteristics of TNBC patients in the two cohorts (Supplementary information, Tables S1 and S2).
To determine the downstream pathways of ITH in TNBC, we analyzed the transcriptomic differences from FUSCC cohorts. Gene set enrichment analysis (GSEA) results revealed anti-correlation of high genetic ITH with canonical immune signatures, such as IFN responses and IL-6, JAK, and STAT3 pathways (Fig. 1d). Moreover, high ITH was negatively correlated with the CD8 score, tumor-infiltrating lymphocyte (TIL) score, and cytolytic (CYT) score (Fig. 1e), indicating that high ITH may result in immune-excluded tumors. Previous studies have demonstrated that ITH has a major impact on the efficacy of immunotherapy.8,22,23 Therefore, we explored the impact of ITH (evaluated by H&E section-derived histologic ITH) on the antitumor immune response in four anti-PD-1-based clinical trials (n = 109) in TNBC (Fig. 1f). Analyses of the neoadjuvant trials NCT04613674 and NCT04418154, which included 29 and 16 patients’ H&E images, respectively, showed a significant decrease in the pathological complete response (pCR) rates in high-ITH tumors (Fig. 1g, h). In the third cohort (NCT03805399), we examined 29 patients’ H&E images, and observed a lower ORR in high-ITH tumors (Fig. 1i). In the last cohort (NCT04129996), we assessed 35 patients’ H&E images, and a lower ORR and shorter progression-free survival (PFS) and OS were noted for patients with high-ITH tumors (Fig. 1j). Collectively, our findings demonstrate that high ITH reduces patient survival and confers immunotherapy resistance in TNBC.
ZNF689 deficiency promotes ITH in TNBC
To explore the key determinant of ITH in TNBC, we analyzed copy number alterations, somatic mutations, and transcriptomes using data from our cohort. Since high genetic ITH or histologic ITH tumors exhibited SCNVs and mutation landscapes similar to those of low-ITH tumors (Supplementary information, Fig. S2a–d), we focused on transcriptomic analysis. After obtaining the differentially expressed genes between high-ITH and low-ITH groups at genetic and histologic levels separately (Supplementary information, Fig. S2e, f), we found that there were 6 common differentially expressed genes (MUC19, PGC, DHRS2, ZNF689, TDRD12, and C20orf114) (Fig. 2a; Supplementary information, Fig. S2g, h).
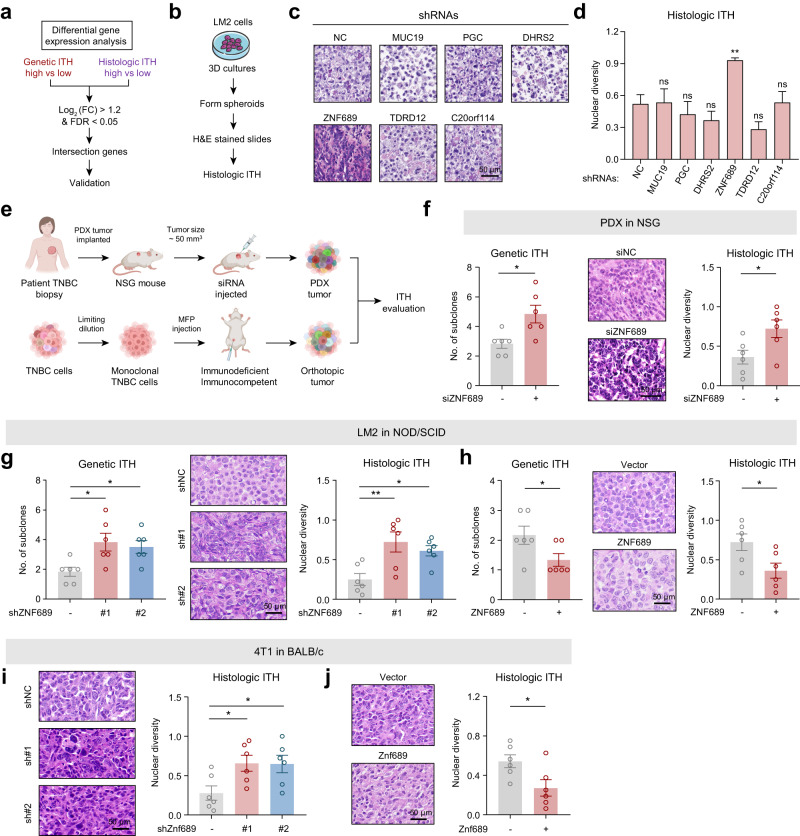
Fig. 2. ZNF689 deficiency promotes ITH in TNBC.
a Flowchart of the screening process. FC fold change; FDR false discovery rate. b Protocols of the 3D tumor sphere assay to evaluate the histologic ITH of LM2 cells. c Representative images of H&E-stained LM2 spheroids. d Histologic ITH analysis of LM2 spheroids. e Graphic illustration of different mouse models to examine the role of ZNF689 in ITH. f The genetic ITH and histologic ITH of TNBC PDX tumors from NSG mice (n = 6) after intratumoral administration with either siNC or siZNF689. g The genetic ITH and histologic ITH of orthotopic tumors generated by inoculating NOD/SCID mice (n = 6) in the MFPs with shNC or shZNF689 LM2 cells. h The genetic ITH and histologic ITH of orthotopic tumors generated by inoculating NOD/SCID mice (n = 6) in the MFPs with vector- or ZNF689-overexpressing LM2 cells. i Histologic ITH of orthotopic tumors generated by injecting shNC or shZnf689 4T1 cells into the MFPs of BALB/c mice (n = 6). j Histologic ITH of orthotopic tumors generated by injecting vector- or Znf689-overexpressing 4T1 cells into the MFPs of BALB/c mice (n = 6). Scale bars, 50 μm. P values were determined using one-way ANOVA (d, g, i) and two-tailed unpaired Student’s t-tests (f, h, j). ns not significant; *P < 0.05, **P < 0.01.
Next, we designed a 3D tumor sphere assay to evaluate histologic ITH in 9 TNBC cell lines (Supplementary information, Fig. S3a, b). We chose the LM2 cell line, which has low genetic and histologic ITH (Supplementary information, Fig. S3c), to experimentally identify the most pivotal gene inducing TNBC ITH (Fig. 2b). Examination of H&E-stained images of LM2 spheroids revealed that knockdown of ZNF689 using short hairpin RNAs (shRNAs) markedly increased histologic ITH (Fig. 2c, d; Supplementary information, Fig. S3d). Specifically, the high genetic and histologic ITH groups exhibited a 3-fold and 2.6-fold average reduction in ZNF689 expression, respectively, compared to the low ITH groups. Furthermore, we observed an inverse relationship between ZNF689 protein expression and the extent of ITH in TNBC cell lines (Supplementary information, Fig. S3e, f). In alignment with this, low ZNF689 expression in tumors was correlated with elevated genetic and histologic ITH levels in both our cohort and the TCGA cohort (Supplementary information, Fig. S3g). Collectively, these findings indicate that ZNF689 may serve as a pivotal factor in modulating ITH in TNBC, warranting more in-depth experimental exploration.
To investigate whether ZNF689 could influence TNBC ITH in vivo, we designed a series of experiments using both immunodeficient and immunocompetent mice (Fig. 2e). (1) We selected a TNBC patient-derived xenograft (PDX) specimen with low genetic ITH (evidenced by two subclones) and low histologic ITH, alongside heightened ZNF689 expression (Supplementary information, Fig. S4a, b). Following the establishment of this PDX model, we conducted intratumoral injections of siRNAs directed against ZNF689. Notably, the depletion of ZNF689 significantly increased the genetic ITH and histologic ITH levels (Fig. 2f). The PDX tumors treated with siZNF689 also showed faster growth rates than the control group (Supplementary information, Fig. S4c, d). (2) LM2 cells stably expressing shNC and shZNF689 were subcutaneously injected into the mammary fat pads (MFPs) of NOD/SCID mice. We found that ZNF689 knockdown markedly increased the genetic and histologic ITH levels in immunodeficient mouse xenografts (Fig. 2g). Furthermore, the xenograft tumors expressing shZNF689 grew faster than those expressing shNC (Supplementary information, Fig. S4e, f). (3) LM2 cells stably expressing vector and ZNF689 were subcutaneously injected into the MFPs of NOD/SCID mice. We observed that ZNF689 overexpression restricted both genetic ITH and histologic ITH in LM2 tumor xenografts (Fig. 2h). Moreover, ZNF689-overexpressing xenograft tumors grew more slowly (Supplementary information, Fig. S4g, h). (4) The shZnf689- or Znf689-overexpressing 4T1 cells were subcutaneously injected into the MFPs of BALB/c mice. We found that Znf689 deficiency resulted in significantly higher histologic ITH in 4T1 syngeneic grafts (Fig. 2i). In contrast, Znf689 overexpression resulted in a lower degree of histologic ITH (Fig. 2j). In addition, Znf689 knockdown significantly promoted tumor growth, while Znf689 overexpression had the opposite effects (Supplementary information, Fig. S4i–l). Together, these results reveal that ZNF689 deficiency promotes ITH in TNBC both in vitro and in vivo.
ZNF689 represses LINE-1 retrotransposition via the TRIM28 complex
To dissect the underlying mechanism of how ZNF689 deficiency could promote ITH in TNBC, we applied stable isotope labeling with amino acids in cell culture (SILAC)-based quantitative proteomics to screen ZNF689-interacting proteins (Supplementary information, Fig. S5a). Among the top 7 potential interactors of ZNF689 ranked by Log2 (ratio H/L) (Supplementary information, Fig. S5b), we selected the top-ranked protein TRIM28, a universal cofactor for Krüppel-associated box domain zinc finger protein (KRAB-ZFP) transcription factors, for further binding validation. Co-immunoprecipitation (co-IP) experiments with endogenous ZNF689 and TRIM28 followed by western blotting demonstrated that these two proteins could interact at endogenous levels in LM2, Hs578T, and HEK293T cells (Fig. 3a; Supplementary information, Fig. S5c). In vitro pull-down assays using recombinant ZNF689 and TRIM28 proteins further suggested that the interaction was likely to be direct (Fig. 3b). Immunofluorescence (IF) showed that ZNF689 colocalized with TRIM28 in the nucleus (Supplementary information, Fig. S5d). We also mapped out the interaction between the N-terminus of the KRAB domain of ZNF689 and the RING domain, B-boxes and coiled-coil (RBCC) domain of TRIM28 (Supplementary information, Fig. S5e–g), which are the critical regions mediating the binding of KRAB-ZFPs and TRIM28.24
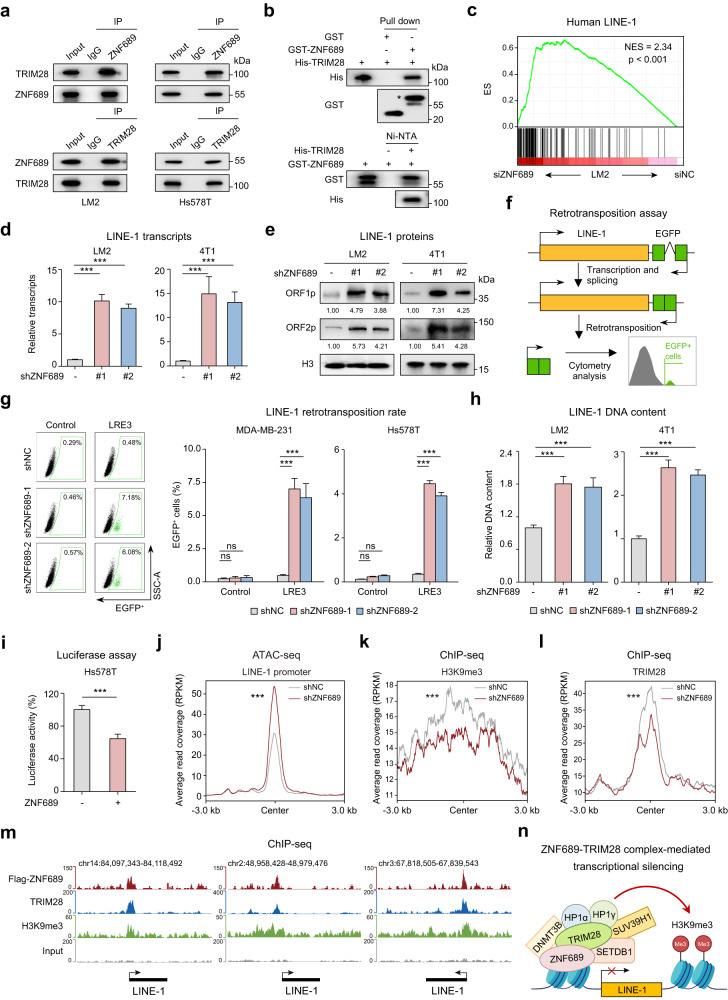
Fig. 3. ZNF689 represses LINE-1 retrotransposition via TRIM28 complex-mediated transcriptional silencing.
a Co-IP experiments of endogenous ZNF689 and TRIM28 followed by western blotting. b In vitro pull-down assays of GST-tagged ZNF689 and His-tagged TRIM28 recombinant proteins followed by western blotting. A single asterisk represents the specific band. c Upregulation of the human LINE-1 gene signature in siZNF689-treated LM2 cells by GSEA. d RT-qPCR analysis of LINE-1 (ORF2) transcript levels in shNC and shZNF689 cells. e Western blot analysis of LINE-1 ORF1p and ORF2p proteins in shNC and shZNF689 cells. f Schematic illustration of the LINE-1 retrotransposition reporter assay. g Representative flow cytometry graphs are shown for cells harboring the LRE3-EGFP retrotransposition reporter or the retrotransposition-deficient JM111 control (left). Quantification of de novo retrotransposition events (EGFP-positive cells) in shNC and shZNF689 cells (right). h RT-qPCR analysis of the relative LINE-1 (5′-UTR for LM2; ORF2 for 4T1) genomic DNA content in shNC and shZNF689 cells. i Dual-luciferase reporter assay detecting the activity of the LINE-1 promoter in ZNF689-overexpressing cells. j ATAC-seq peak signals affected by ZNF689 knockdown at the full-length LINE-1 promoter in LM2 cells. k, l Metaplots showing changes in H3K9me3 (k) and TRIM28 (l) ChIP-seq signals upon ZNF689 knockdown in the full-length LINE-1 promoter. m ChIP-seq tracks showing the binding patterns of ZNF689, TRIM28, H3K9me3 and input at full-length LINE-1. n Schematic diagram showing that ZNF689 represses LINE-1 retrotransposition via TRIM28 complex-mediated transcriptional silencing. P values were determined using one-way ANOVA (d, g, h), two-tailed unpaired Student’s t-tests (i) and Wilcoxon tests (j–l). ns not significant; ***P < 0.001.
The primary role of KRAB-ZNFs is to silence repetitive elements (REs) by recruiting the transcriptional regulator TRIM28 and the associated mediators of histone H3 Lys9 trimethylation (H3K9me3)-dependent heterochromatin formation and DNA methylation.25,26 Since REs are major players in genetic variability and genome evolution,27 we speculated that ZNF689 might regulate REs to repress TNBC ITH. To study whether any of the known REs were regulated upon ZNF689 depletion, we performed RNA-seq of siNC- and siZNF689-treated LM2 cells. The data were analyzed using RepEnrich software to quantify RE expression. Among the REs with changed expression upon ZNF689 depletion, LINE, one of the most abundant REs in human genomes,28 showed significantly increased expression (Supplementary information, Fig. S6a). GSEA also revealed marked upregulation of the human LINE-1 gene signature in siZNF689-treated cells (Fig. 3c). To validate the transcriptomic analyses, we performed real-time quantitative polymerase chain reaction (RT-qPCR) and western blotting to quantify LINE-1. We observed that ZNF689 deficiency significantly upregulated LINE-1 mRNAs and their encoding proteins ORF1p and ORF2p in human (LM2 and Hs578T) and murine (4T1 and AT3) TNBC cell lines (Fig. 3d, e; Supplementary information, Fig. S6b, c). To examine whether the retrotransposition activity of LINE-1 is increased in the absence of ZNF689, we conducted retrotransposition analysis in ZNF689-knockdown cells (MDA-MB-231 and Hs578T) with an engineered LINE-1 reporter system (Fig. 3f). Notably, a higher retrotransposition frequency of LINE-1 was observed in ZNF689-knockdown cells than in control cells (Fig. 3g). We also detected an elevated genomic DNA content of LINE-1 in ZNF689-knockdown human and murine TNBC cell lines with RT-qPCR (Fig. 3h; Supplementary information, Fig. S6d). LINE-1 transcription is driven by an internal promoter within the LINE-1 5′-untranslated region (UTR).29 To test whether ZNF689 directly regulates LINE-1 transcription, we cloned the human LINE-1 5′-UTR into a luciferase reporter plasmid. ZNF689 overexpression strongly repressed luciferase activity in Hs578T and HEK293T cells (Fig. 3i; Supplementary information, Fig. S6e). Assay for transposase-accessible chromatin using sequencing (ATAC-seq) supported the notion that ZNF689 deficiency led to derepression and increased chromatin accessibility of the full-length LINE-1 promoter (Fig. 3j; Supplementary information, Fig. S6f). Using a chromatin immunoprecipitation-qPCR (ChIP-qPCR) assay, we revealed that ZNF689 was specifically enriched at the 5′-UTR of LINE-1 loci in LM2 cells (Supplementary information, Fig. S6g). Analysis of ChIP-seq data confirmed ZNF689 binding to LINE-1 promoters (Supplementary information, Fig. S6h, i). Accordingly, LINE-1 shows significant overexpression in high-ITH TNBC tumors (Supplementary information, Fig. S6j).
TRIM28 acts as a scaffold for a silencing complex that comprises the histone methyltransferase SETDB1, the nucleosome remodeling and deacetylation (NuRD) complex, heterochromatin protein 1 (HP1), and DNA methyltransferases.24 By performing co-IP assays, we identified SETDB1, DNMT3B, HP1α, HP1γ, and SUV39H1 as additional ZNF689-interacting proteins (Supplementary information, Fig. S6k). The promoter of LINE-1 is selectively enriched in histone variant H3.3 as well as the histone mark H3K9me3, which are essential for maintaining the repressed state of LINE-1.30 Therefore, we hypothesized that ZNF689 might recruit these enzymes to the LINE-1 promoter to enhance H3K9me3-mediated transcriptional silencing. ChIP-seq analysis revealed a decrease in H3K9me3 modifications at the full-length LINE-1 promoter upon ZNF689 knockdown (Fig. 3k). Subsequent assays showed attenuated TRIM28 recruitment to this locus in the absence of ZNF689 (Fig. 3l). These data support the coordinated binding of ZNF689 and TRIM28 to mediate H3K9me3 modifications at LINE-1 promoters (Fig. 3m). Moreover, ZNF689 knockdown did not significantly alter TRIM28 or SETDB1 protein levels (Supplementary information, Fig. S6l). Taken together, these results suggest that ZNF689 represses LINE-1 retrotransposition via TRIM28 complex-mediated transcriptional silencing (Fig. 3n).
ZNF689 deficiency-induced LINE-1 retrotransposition exacerbates genomic instability and promotes ITH
LINE-1 retrotransposition is a major source of genomic instability, which manifests as increased double-strand DNA breaks and chromosomal instability (CIN).31–33 Therefore, we hypothesized that ZNF689 deficiency might lead to increased genomic instability by derepressing LINE-1 retrotransposition to promote ITH. First, we examined the phosphorylated histone H2AX (γH2AX), a marker of DNA double-strand breaks used to assess genomic instability.34 IF assays (Fig. 4a) and western blotting (Supplementary information, Fig. S7a) showed that ZNF689 knockdown enhanced γH2AX level, consistent with the result in ZNF689-knockdown orthotopic LM2 tumors (Fig. 4b). We next explored the potential role of ZNF689 deficiency in CIN. Metaphase chromosome spreading assays revealed a marked increase in chromosomal aberrations in ZNF689-knockdown cells (Fig. 4c). Furthermore, low ZNF689 expression in our cohort significantly correlated with elevated CIN gene signature scores (Supplementary information, Fig. S7b). TNBC tumors with reduced ZNF689 expression also had higher aneuploidy scores (Supplementary information, Fig. S7c). Histological examination of H&E sections also confirmed a significant correlation between ZNF689 deficiency and increased chromosomal missegregation during anaphase (Fig. 4d). These results reveal that ZNF689 deficiency led to increased genomic instability in TNBC.
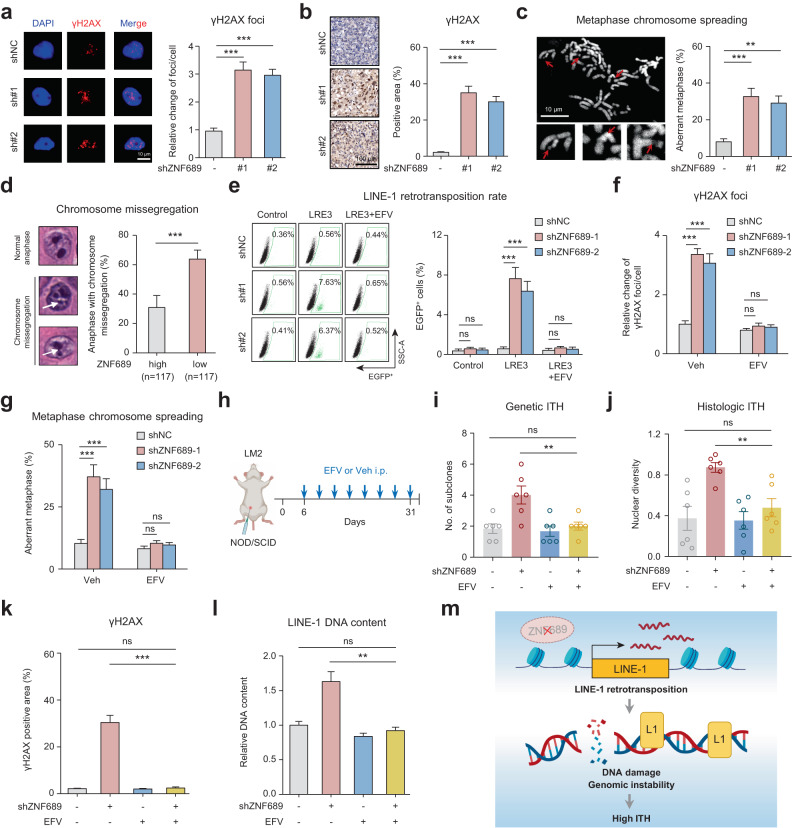
Fig. 4. ZNF689 deficiency-induced LINE-1 retrotransposition exacerbates genomic instability and promotes ITH.
a Representative IF images and quantification of γH2AX foci in shNC and shZNF689 LM2 cells (n = 30). Scale bar, 10 μm. b Representative IHC images and quantification of γH2AX in orthotopic shNC and shZNF689 LM2 tumors (n = 6). Scale bar, 100 μm. c Representative images and quantification of chromosome aberrations (red arrows) in shNC and shZNF689 LM2 cells at passage 20 (n = 50). Scale bar, 10 μm. d Representative images and quantification of chromosome missegregation errors (white arrows) in diagnostic H&E samples in the FUSCC cohort. e Representative flow cytometry graphs and quantification of de novo retrotransposition events (EGFP-positive cells) in shNC and shZNF689 MDA-MB-231 cells treated with EFV (20 μM). f Quantification of γH2AX foci in shNC and shZNF689 LM2 cells treated with EFV (20 μM) (n = 30). g Quantification of chromosome aberrations in shNC and shZNF689 LM2 cells at passage 20 treated with EFV (20 μM) (n = 50). h Schematic diagram of the EFV treatment regimen. After inoculation of shNC or shZNF689 LM2 cells into the MFP, the NOD/SCID mice were randomly divided into groups and treated daily with Veh or EFV via i.p. injection (n = 6 mice/group). i, j The genetic ITH (i) and histologic ITH (j) of orthotopic LM2 tumors from mice in h (n = 6). k Quantification of γH2AX in orthotopic tumors from mice in h (n = 6). l RT-qPCR analysis of LINE-1 (5′-UTR) DNA content of tumors dissected from mice in h (n = 6). m Schematic diagram showing that ZNF689 deficiency exacerbates genomic instability to promote ITH by reactivating LINE-1 retrotransposition. P values were determined using one-way ANOVA (a–c, e–g, i–l) and two-tailed unpaired Student’s t-tests (d). ns not significant; **P < 0.01, ***P < 0.001.
Reverse transcriptase inhibitors such as efavirenz (EFV) can effectively block the enzymatic activity of endogenous reverse transcriptase and are regarded as potential specific inhibitors for LINE-1 retrotransposition.35–38 We observed that EFV effectively mitigated LINE-1 retrotransposition and genomic DNA content alterations induced by ZNF689 knockdown (Fig. 4e; Supplementary information, Fig. S7d, e). Subsequent IF assays (Fig. 4f) and western blotting (Supplementary information, Fig. S7f) revealed that EFV treatment attenuated the ZNF689 knockdown-induced upregulation of γH2AX. EFV also reduced chromosomal aberrations in ZNF689-depleted cells (Fig. 4g). Importantly, EFV did not significantly affect DNA damage, cell apoptosis, or proliferation in TNBC cells (Supplementary information, Fig. S7g–i). These results suggest that ZNF689 deficiency exacerbates genomic instability via LINE-1 retrotransposition.
Next, we investigated whether ZNF689 deficiency-induced ITH could be reversed by inhibition of LINE-1 retrotransposition in vivo. We constructed the MFP xenograft model with shNC or shZNF689 LM2 cells. After one week, NOD/SCID mice were randomly divided into groups and treated daily with the vehicle (Veh) control or EFV via intraperitoneal (i.p.) injection (Fig. 4h). The results showed that EFV completely abrogated ZNF689 knockdown-induced tumor growth (Supplementary information, Fig. S7j). Notably, EFV significantly decreased the genetic ITH (Fig. 4i) and histologic ITH (Fig. 4j) in ZNF689-knockdown tumors, suggesting that the ITH-promoting effect of ZNF689 deficiency is LINE-1 retrotransposition dependent. Immunohistochemistry (IHC) analysis of orthotopic tumors showed that EFV effectively reduced γH2AX-positive area in ZNF689-deficient tumors (Fig. 4k). In addition, EFV significantly downregulated the DNA content of LINE-1 in ZNF689-deficient tumors (Fig. 4l). In sum, the results support that ZNF689 deficiency exacerbates genomic instability to promote ITH by derepressing LINE-1 retrotransposition (Fig. 4m).
ZNF689 deficiency-induced high ITH impairs antigen presentation and T-cell activation
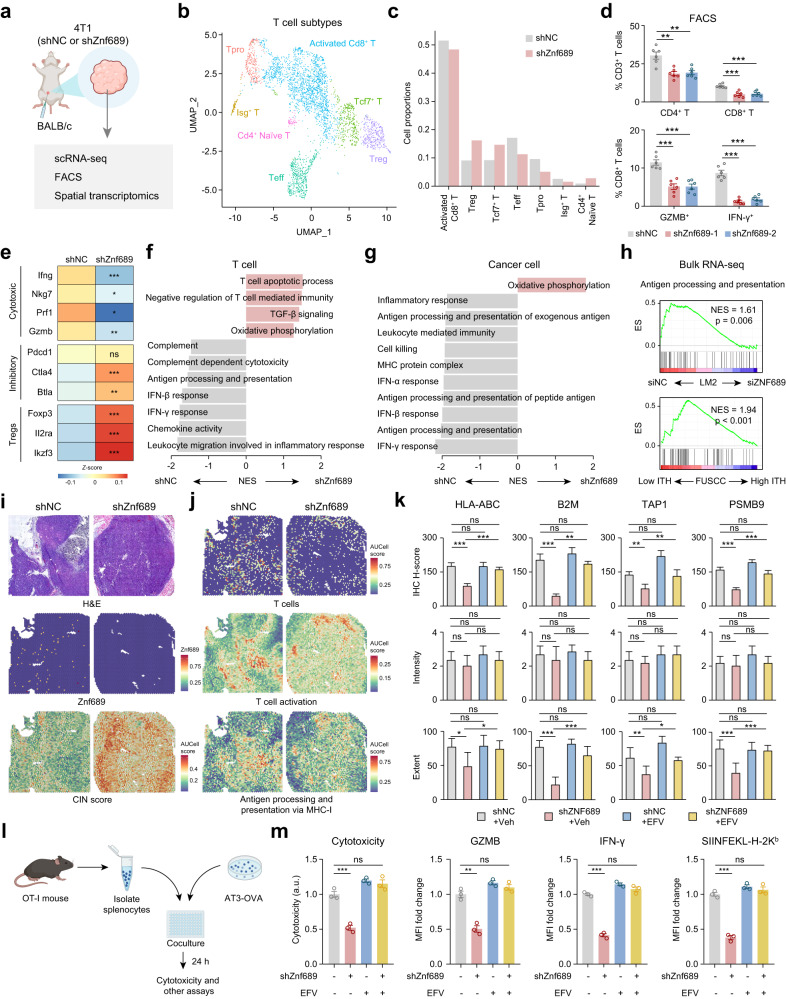
We revealed that high ITH conferred immunotherapy resistance in TNBC (Fig. 1f–j). To directly evaluate tumor microenvironment remodeling under ITH induction, we performed single-cell RNA sequencing (scRNA-seq) to characterize changes in the transcriptome of cells harvested from shZnf689 or shNC 4T1 syngeneic grafts (Fig. 5a). Finally, we collected 10,913 cells from the shZnf689 group and 14,788 cells from the shNC group. After integrating the transcriptomic data from all acquired cells, uniform manifold approximation and projection (UMAP) visualization showed six cell types, including cancer cells, myeloid cells, B cells, T cells, cancer-associated fibroblasts (CAFs) and endothelial cells (Supplementary information, Fig. S8a, b). We confirmed that cancer cells were copy number-unstable, while other cell populations were copy number-stable (Supplementary information, Fig. S8c). Notably, the proportion of T cells in Znf689-deficient tumor cells was decreased (Supplementary information, Fig. S8d). To accurately define the T cells, we subclustered T cells into seven subtypes: activated CD8+ T cells, regulatory T cells (Treg), Tcf7+ T cells, effector T cells (Teff), proliferating T cells (Tpro), Isg+ T cells and CD4+ naïve T cells (Fig. 5b; Supplementary information, Fig. S8e). Activated CD8+ T cells, Teff, Tpro and Isg+ T cells were decreased, while Tregs, Tcf7+ T cells and CD4+ naïve T cells were increased in the Znf689-deficient group (Fig. 5c). This finding was also validated by flow cytometric analysis of immune cell profiling of 4T1 tumors from BALB/c mice, in which ZNF689 knockdown decreased tumor CD4+ T-cell infiltration, CD8+ T-cell infiltration, and the percentages of granzyme B+ (GZMB+) and IFN-γ+ CD8+ T cells (Fig. 5d; Supplementary information, Fig. S8f). Conversely, a significant increase in the percentages of CD4+ T cells and CD8+ T cells among CD3+ T cells and GZMB+ and IFN-γ+ cells among CD8+ T cells was observed in 4T1 tumors with Znf689 overexpression (Supplementary information, Fig. S8g). Similarly, scRNA-seq data showed that T cells in Znf689-deficient tumors expressed decreased levels of cytotoxic T cell markers (Ifng, Nkg7, Prf1 and Gzmb) but increased levels of inhibitory T cell (Ctla4 and Btla) and Treg (Foxp3, II2ra and Ikzf3) markers (Fig. 5e).
Fig. 5. ZNF689 deficiency-induced ITH impairs antigen presentation and T-cell activation.
a shNC and shZnf689 4T1 cells were subcutaneously injected into the MFPs of BALB/c mice. Then tumors were used for scRNA-seq, fluorescence activated cell sorting (FACS) and spatial transcriptomics. b UMAP plot of reclassification of intratumoral T cells. c The distribution of T-cell subtypes in shNC and shZnf689 tumors. d Flow cytometry analysis of the percentages of CD4+ T cells, CD8+ T cells, GZMB+ CD8+ T cells, and IFN-γ+ CD8+ T cells in tumors (n = 6). e Heatmap showing the expression of marker genes for cytotoxic T cells, inhibitory T cells and Tregs in T cells of shNC and shZnf689 tumors. f, g Pathways downregulated in T cells (f) and cancer cells (g) in shZnf689 tumors. h Downregulation of antigen processing and presentation in siZNF689-treated LM2 cells and high genetic ITH tumors from the FUSCC cohort by GSEA. i H&E-stained tissue images, Znf689 expression and signature scores of CIN in the spots of shNC and shZnf689 tumors. j Signature scores of T cells, T-cell activation, and antigen processing and presentation via MHC-I in the spots of shNC and shZnf689 tumors. k IHC H-scores, intensity, and extent for HLA-ABC, B2M, TAP1, and PSMB9 protein expression in tumors from mice in Fig. 4h. l Schematic diagram of the in vitro tumor-immune cell coculture assay. m Cytotoxicity analysis of culture medium at 24 h after tumor cell and OT-I CD8+ T-cell coculture. Flow cytometry analysis was used to determine the expression of GZMB and IFN-γ in CD8+ T cells, and OVA (SIINFEKL–H-2Kb) presentation in AT3-OVA tumor cells at 24 h after tumor cell and OT-I splenocyte coculture. P values were determined using one-way ANOVA (d, k, m) and two-tailed unpaired Student’s t-tests (e). ns not significant; *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we investigated the detailed mechanisms by which ZNF689 deficiency-induced ITH regulates T-cell-mediated antitumor immunity. Pathway analyses of T cells and cancer cells from scRNA-seq revealed downregulation of IFN responses and antigen processing and presentation (APP) in the Znf689-deficient group (Fig. 5f, g). Bulk RNA-seq confirmed that the APP pathway was significantly lost in siZNF689-treated LM2 cells and high-ITH tumors (Fig. 5h). RT-qPCR analysis validated that ZNF689 knockdown resulted in lower mRNA levels of major histocompatibility complex class I (MHC-I) APP-associated genes in LM2 and 4T1 cells (Supplementary information, Fig. S9a, b), suggesting that ZNF689 is involved in antigen presentation regulation. To reveal whether ZNF689 deficiency affected the spatial distribution of single cells and gene expression, we performed spatially resolved transcriptomics on 4T1 tumors from BALB/c mice. In line with the findings illustrated before, the Znf689-deficient tumor showed visible and strong activities by the CIN score (Fig. 5i). As shown in Fig. 5j and Supplementary information, Fig. S9c, d, the signature scores of T cells (including CD4+ and CD8+ T cells), T-cell activation, and APP via MHC-I were lower and heterogeneous in the spots of ZNF689-deficient tumor. These signatures were mainly observed in the peritumor region of the ZNF689-deficient tumor, indicating a more immune-excluded microenvironment. Moreover, IHC analysis of HLA-ABC, B2M, TAP1, and PSMB9 in orthotopic LM2 tumors indicated that the downregulation of those MHC-I APP-associated markers upon ZNF689 knockdown was mainly due to the reduced percentages of stained cells, not the staining intensity (Fig. 5k; Supplementary information, Fig. S9e), suggesting that ITH also induced the spatial heterogeneity of MHC-I APP-associated protein expression. However, treatment with the LINE-1 inhibitor EFV significantly abrogated the increase in the spatial heterogeneity of MHC-I APP-associated protein expression caused by ZNF689 knockdown in orthotopic LM2 tumors (Fig. 5k; Supplementary information, Fig. S9e).
To further elucidate the specific mechanisms through which ZNF689 regulates MHC-I APP-associated genes, analysis of our Flag-ZNF689 ChIP-seq data revealed that ZNF689 did not directly bind to the promoters of these key genes (Supplementary information, Fig. S10a). Conversely, ATAC-seq showed reduced chromatin accessibility for these genes upon ZNF689 knockdown (Supplementary information, Fig. S10b), suggesting a role of ZNF689 in the observed gene expression decline. RT-qPCR results further demonstrated that EFV counteracted the APP-associated gene downregulation induced by ZNF689 depletion (Supplementary information, Fig. S10c), consistent with the hypothesis that this suppression is linked to elevated LINE-1 retrotransposition in the absence of ZNF689.
To assess whether ZNF689 deficiency restricts CD8+ T-cell cytotoxic activities due to impaired MHC-I expression, we cultured mouse ovalbumin (OVA)-specific CD8+ T cells (OT-I) with OVA-expressing shNC and shZnf689 AT3 tumor cells (Fig. 5l). Znf689 deficiency in tumor cells resulted in a decrease in CD8+ T-cell cytotoxic activities and CD8+ T-cell activation, as indicated by lower GZMB, IFN-γ and CD137 expression (Fig. 5m; Supplementary information, Fig. S10d, e). These data correspond to reduced presentation of SIINFEKL (OVA peptide)–H-2Kb complex in shZnf689 AT3 tumor cells compared to shNC cells (Fig. 5m). However, when the tumor-immune cell coculture system was treated with the LINE-1 inhibitor EFV, we found that antigen presentation of tumor cells and T-cell activation were rescued (Fig. 5m; Supplementary information, Fig. S10d, e). Additionally, we ruled out the direct effects of EFV on CD8+ T cells (Supplementary information, Fig. S11a–f). Together, these data suggest that ZNF689 deficiency-induced ITH reduces antigen presentation and inhibits T-cell infiltration and activation, thereby contributing to immune escape and immunotherapy resistance in TNBC.
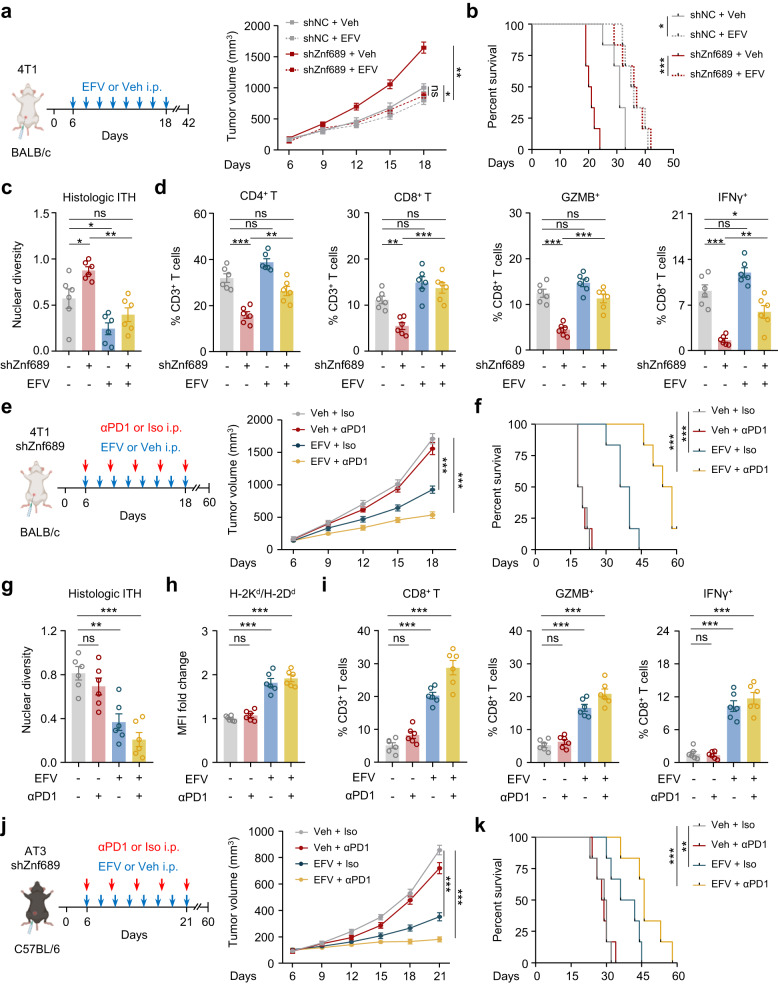
LINE-1 inhibition sensitizes ZNF689 deficiency-induced high-ITH tumors to immunotherapy
Given the importance of LINE-1 inhibition in regulating ITH and antigen presentation, we further explored whether the LINE-1 inhibitor EFV could reverse the immunosuppressive tumor microenvironment. We then implanted 4T1 cells into the MFPs of BALB/c mice. EFV significantly inhibited tumor growth (Fig. 6a) and prolonged survival time (Fig. 6b) without affecting mouse body weight or food consumption (Supplementary information, Fig. S12a, b) in the Znf689-deficient group. EFV effectively reduced histologic ITH in Znf689-deficient tumors (Fig. 6c). Using flow cytometry, we found that EFV-treated Znf689-deficient tumors had more CD4+ T cells, CD8+ T cells, GZMB+ and IFN-γ+ CD8+ T cells than Veh controls (Fig. 6d). These observations suggest a potential synergistic antitumor effect of combining LINE-1 inhibition and immunotherapy. Therefore, we performed orthotopic MFP transplantation of Znf689-knockdown 4T1 cells in BALB/c mice to investigate whether targeting LINE-1 could sensitize high-ITH tumors to anti-PD-1 treatment. While the anti-PD-1 response exhibited minimal variation between the shNC and shZnf689 groups (Supplementary information, Fig. S12c), the combined treatment demonstrated a synergistic antitumor effect in the shZnf689 groups when assessed by tumor volume in comparison to the control (Fig. 6e). In addition, EFV together with anti-PD-1 treatment dramatically prolonged mouse survival (Fig. 6f). Orthotopic 4T1 tumors showed that combination therapy significantly decreased histologic ITH in ZNF689-deficient tumors (Fig. 6g). Furthermore, we detected microenvironment remodeling in mice treated with combination therapy; the related changes included significant increases in H2d MHC-I alloantigen levels (Fig. 6h), the infiltration of CD4+ T cells (Supplementary information, Fig. S12d) and CD8+ T cells, and the percentages of GZMB+ and IFN-γ+ CD8+ T cells (Fig. 6i).
Fig. 6. LINE-1 inhibition sensitizes ZNF689 deficiency-induced high-ITH tumors to immunotherapy in TNBC.
a Schematic diagram for the establishment of the orthotopic 4T1 syngeneic tumor model in BALB/c mice and the treatment schedule for the LINE-1 inhibitor EFV. Tumor growth curves are shown (n = 6 mice/group). b Kaplan–Meier survival curves for the mice in a. Survival data were obtained from another independent experiment. c Histologic ITH of orthotopic tumors from mice in a (n = 6). d Primary tumors from mice in a were harvested for flow cytometry to determine the percentages of CD4+ and CD8+ T cells among CD3+ T cells and GZMB+ and IFN-γ+ cells among CD8+ T cells (n = 6). e Schematic diagram for the establishment of the orthotopic shZnf689 4T1 syngeneic tumor model in BALB/c mice and the treatment schedule for PD-1 antibody and LINE-1 inhibitor EFV. Tumor growth curves are shown (n = 6 mice/group). f Kaplan–Meier survival curves for the mice in e. Survival data were obtained from another independent experiment. g The histologic ITH of orthotopic tumors from mice in e (n = 6). h Quantitative estimate of MHC-I levels on the surface of 4T1 tumors from mice in e (n = 6). MFI mean fluorescence intensity. i Primary tumors from mice in e were harvested for flow cytometry to determine the percentages of CD8+ T cells among CD3+ T cells and GZMB+ and IFN-γ+ cells among CD8+ T cells (n = 6). j Schematic diagram for the establishment of the orthotopic shZnf689 AT3 syngeneic tumor model in C57BL/6 mice and the treatment schedule for PD-1 antibody and LINE-1 inhibitor EFV. Tumor growth curves are shown (n = 6 mice/group). k Kaplan–Meier survival curves for the mice in j. Survival data were obtained from another independent experiment. P values were determined using two-way ANOVA (a, e, j), log-rank tests (b, f, k) and one-way ANOVA (c, d, g–i). ns not significant; *P < 0.05, **P < 0.01, ***P < 0.001.
To exclude a cell line- or mouse strain-specific effect, we established the AT3 breast cancer model by injecting AT3 cells orthotopically into the MFPs of C57BL/6 mice. Again, we observed delayed tumor growth (Fig. 6j) and prolonged survival (Fig. 6k) in the Znf689-deficient group treated with the LINE-1 inhibitor EFV in combination with anti-PD-1. Moreover, combination therapy significantly decreased histologic ITH, enhanced antigen presentation, and orchestrated the infiltration and activation of CD8+ T cells in ZNF689-deficient tumors (Supplementary information, Fig. S12e, f). Collectively, these data suggest that pharmacological inhibition of LINE-1 in combination with immunotherapy induces potent antitumor immunity by promoting the recruitment and activation of CD8+ T cells in the tumor microenvironment via blocking ITH and enhancing antigen presentation.
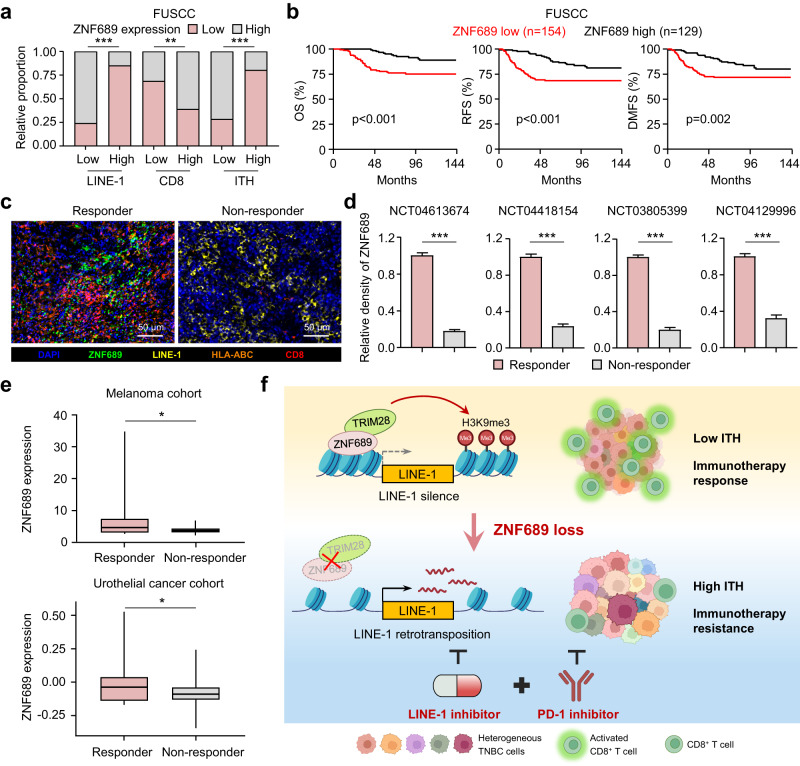
ZNF689 expression positively correlates with a favorable prognosis and immunotherapy response in TNBC
To further validate our preclinical findings in human TNBC samples, we investigated the expression levels of ZNF689, LINE-1, and CD8 in a cohort of TNBC patients by IHC analysis of a tissue microarray (n = 283). Consistent with our findings, the IHC results showed that ZNF689 expression was negatively correlated with LINE-1 ORF1p level but positively correlated with CD8+ T-cell infiltration in TNBC tissues (Fig. 7a; Supplementary information, Fig. S13a). Furthermore, our analysis revealed a negative correlation between ZNF689 expression and histologic ITH levels based on H&E staining (Fig. 7a), suggesting that ZNF689 is a negative factor associated with ITH in TNBC. For the survival analysis, we observed that low ZNF689 levels indicated a significantly worse prognosis in TNBC patients than corresponding high ZNF689 expression, as evidenced by shorter OS, RFS, and DMFS (Fig. 7b). The results from the public database (Kaplan-Meier Plotter) also showed that low ZNF689 expression was correlated with poor patient outcomes in basal-like breast cancer (Supplementary information, Fig. S13b).
Fig. 7. ZNF689 expression positively correlates with a favorable prognosis and immunotherapy response in TNBC.
a Analysis of the correlation of ZNF689 expression with LINE-1 ORF1p, CD8, and histologic ITH in TNBC tissues (n = 283). b Kaplan–Meier analysis of OS, RFS, and DMFS in TNBC patients from FUSCC grouped according to the expression of ZNF689. c Multiplex IF staining in one representative responder and one non-responder treated with anti-PD-1-based immunotherapy. Scale bars, 50 µm. d Quantification of ZNF689 expression in responders and non-responders in four trials. e Analysis of ZNF689 mRNA levels in all responder versus non-responder patients in the melanoma cohort (GSE91061) and urothelial cancer cohort (GSE176307). f Illustration of the proposed working model. P values were determined using Pearson’s χ2 test (a), log-rank test (b), two-tailed unpaired Student’s t-test (d), and one-tailed Student’s t-test (e). *P < 0.05, **P < 0.01, ***P < 0.001.
Moreover, we further assessed the protein expression levels of ZNF689, LINE-1, HLA-ABC and CD8 by multicolor IF on tumors from four TNBC trials treated with anti-PD-1 therapy (n = 100). The IF staining results showed a positive relationship between ZNF689, HLA-ABC and CD8 expression, and a negative relationship between ZNF689 and LINE-1 ORF1p expression in anti-PD-1 therapy responders (Fig. 7c; Supplementary information, Fig. S13c). Importantly, we found that high ZNF689 expression in tumors correlated with a positive response to immunotherapy (Fig. 7d). These results were consistent with our in vitro and in vivo studies. To assess whether these observations also apply to other cancer types, we examined the melanoma cohort (GSE91061) and urothelial cancer cohort (GSE176307) treated with anti-PD-1 therapy and observed higher levels of ZNF689 in responders (Fig. 7e). Similar to the effect of ZNF689 loss, we confirmed that the multi-subclone PDX model exhibited heightened sensitivity to EFV in comparison to single subclone PDX model (Supplementary information, Fig. S14a–c). Supporting our findings, the patient-derived tumor fragment (PDTF) model demonstrated that combining EFV and anti-PD-1 therapy for high-ITH PDTFs with multiple subclones induced a rapid reprogramming of tumor microenvironment to foster T-cell accumulation and effector activity (Supplementary information, Fig. S14d–f), which may be promising for precision treatment of high-ITH patients. Overall, these findings demonstrate that ZNF689 expression positively correlates with patient outcomes and immunotherapy response in TNBC.
Discussion
In the present study, we uncovered the underlying mechanism of ITH in TNBC. Our data revealed ITH as an important determinant of TNBC patient survival and the response to immunotherapy. Remarkably, we found that ZNF689 deficiency promotes ITH through TRIM28 complex-mediated transcriptional derepression of LINE-1 retrotransposition. ZNF689 deficiency-induced ITH impaired antigen presentation and CD8+ T-cell infiltration. LINE-1 inhibition using a reverse transcriptase inhibitor reduced ITH, thereby potentiating T-cell activation and boosting antitumor immune responses. Therefore, we suggest that blocking ITH with LINE-1 inhibitors can sensitize highly heterogeneous tumors to immunotherapy (Fig. 7f).
Although ITH has significant clinical relevance in many tumor types, assessing heterogeneity in human tissue samples in large-scale cohorts remains a major challenge.11 Here, we comprehensively characterized ITH at both the genetic and histologic levels in TNBC tumors from the FUSCC and TCGA cohorts. The bioinformatics tool PyClone was used to detect subclones, which inferred genetic ITH; H&E images were used to detect nuclear diversity, which defined histologic ITH. We confirmed that the degree of ITH in TNBC was highly variable. In addition, our study found that histologic ITH was associated with genetic ITH, suggesting that H&E-stained tumor sections are clinically applicable to estimate the degree of ITH in TNBC. Therefore, we combined data of patient survival and immunotherapy response to document a relationship between ITH and clinical value in TNBC. Consistent with observations in other cancer types,22,39–41 TNBC patients with high ITH had significantly worse survival than those with low ITH. The results from four anti-PD-1 clinical trials revealed that high ITH conferred immunotherapy resistance in patients with TNBC. These observations highlighted the importance of using ITH as a biomarker for assessing the success of immunotherapy.
Research on the exact dominant factors and underlying mechanisms of ITH remains rudimentary. Our work identified ZNF689 as a core protein controlling the levels of ITH in TNBC. ZNF689 is a C2H2-type zinc finger transcription factor,42–44 yet its expression, regulation, and function in breast cancer have not been elucidated. We demonstrated that ZNF689 deficiency contributes to ITH in vitro and in vivo. Mechanistically, we provided the first evidence that ZNF689 represses LINE-1 retrotransposition by TRIM28 complex-mediated transcriptional silencing. Specifically, ZNF689 directly binds to the promoter of LINE-1 and recruits the TRIM28 complex to promote H3K9me3-mediated epigenetic silencing. LINE-1 retrotransposons are the only autonomously mobile transposons in humans, occupying 17% of the human genome.28 The mobility of LINE-1 is largely repressed in somatic tissues but is derepressed in many cancers.27 As such, LINE-1 has been considered mostly deleterious since its activity can lead to insertional mutagenesis, chromosomal rearrangements, and genome instability, which contribute to cancer development.45 Emerging evidence indicates that genomic instability is the major cause of genetic heterogeneity in cancer.2,12 We present detailed evidence that the LINE-1 retrotransposon is an important source of ITH that exerts its effects by exacerbating genomic instability, suggesting the potential of blocking LINE-1 activity to limit tumor heterogeneity. LINE-1 retrotransposition can be efficiently controlled with reverse transcriptase inhibitors,35–38,46 which are frequently used to treat viral diseases. These drugs, including EFV, have demonstrated anticancer effects by inhibiting the proliferation, invasion, and metastasis of several malignant cancers.35,37,47–49 Our study verified that the reverse transcriptase inhibitor EFV profoundly restricted genetic and histologic ITH and reduced tumor growth in ZNF689-deficient tumors. Therefore, these findings provide new insights into how ZNF689 functionally modulates LINE-1 retrotransposition to restrain ITH in TNBC.
An important finding of our study is that high ITH not only accelerates tumor proliferation but also concurrently dampens immune response in TNBC. The growth promotion in high-ITH tumors can be largely attributed to the dual effects of conferred plasticity in evolving tumors and a weakened immune response. Previous studies revealed that high-ITH tumors effectively evade immune attack through complex mechanisms, such as neoantigen silencing,50 impaired antigen presentation,51,52 and impaired IFN-γ sensing pathways.53 Moreover, clones with subclonal antigens may undergo “dilution” within vastly heterogeneous tumors.22 We demonstrated that ZNF689 deficiency impairs antigen presentation by downregulating key MHC-I genes through LINE-1 retrotransposition, likely leading to decreased chromatin accessibility for these genes. Previous research has indicated that LINE-1 activation can disrupt promoter function and decrease chromatin accessibility upon insertion,54 while also serving as insulators that affect promoter–enhancer interactions.55–58 Additionally, LINE-1 retrotransposition can cause chromosomal alterations and structural variations,45,59,60 which in turn can reshape chromatin’s 3D structure, impacting its accessibility.61–65 Considering that ZNF689 does not directly bind to antigen presentation-related genes but LINE-1 significantly influences genome structure, epigenome, and transcription,66,67 it is plausible that ZNF689 deficiency-induced downregulation of antigen presentation-related genes is a consequence of LINE-1 retrotransposition. This could thereby lead to diminished immune infiltration and weakened cytotoxic activity of CD8+ T cells. However, the reciprocal influence between ITH and antitumor immunity is complex;10 while immune pressure might favor low-ITH tumors, immunotherapy could reduce tumor diversity.68 Hence, fully understanding the relationship of ITH and immune response remains a challenge.
The majority of patients with TNBC do not receive long-term benefits from immunotherapy.1 As discussed above, ITH serves as a mechanism enabling tumors to evade immune surveillance and conferring tumor resistance to immunotherapy, thus constituting a promising therapeutic target. We demonstrated that targeting ITH by LINE-1 inhibition could transform the tumor microenvironment to be favorable in TNBC, manifested by enhanced antigen presentation and CD8+ T-cell infiltration. Similar results reported that LINE-1 retrotransposition negatively correlated with immune activation, especially the infiltration of CD8+ cytotoxic T cells.37,69,70 While existing research indicates that LINE-1 reactivation could enhance tumor immunogenicity,71,72 our research in TNBC reveals a more intricate relationship between LINE-1 retrotransposition and immunogenicity. We propose that the decline in immunogenicity following the loss of ZNF689, and the subsequent activation of LINE-1, results from an interplay between the specific attributes of TNBC and the selective influence of ZNF689 on LINE-1. Further study showed that LINE-1 inhibition can improve the antitumor immune environment in lung carcinoma models.37 Consistently, our study confirmed that targeting the ITH source by LINE-1 inhibition sensitizes ZNF689-deficient tumors to anti-PD-1 therapy and, in combination with this treatment, effectively limits ITH and tumor growth. Therefore, our research advocates exploring LINE-1 inhibitors as potential immunosensitizers to enhance immunotherapy in TNBC. This approach is currently being tested in a proof-of-concept clinical trial (NCT05076682) for metastatic TNBC patients resistant to immunotherapy, highlighting the promise of this novel therapeutic strategy.
We acknowledge two limitations in our study. First, our models may not fully replicate human tumor ITH, which is constrained by the necessity of shorter study durations due to animal welfare considerations. Second, the intricate effects of ZNF689 deficiency-induced LINE-1 retrotransposition on chromatin accessibility within antigen presentation-related genes remain to be elucidated.
In summary, our study elucidates the underlying mechanisms and uncovers a therapeutic strategy by which targeting LINE-1 renders high-ITH tumors eradicable by synergizing with immunotherapy, providing the basis for the combinatorial use of LINE-1 inhibitors and immunotherapy for TNBC precision therapy.
Materials and methods
Datasets
Our study included two datasets. The primary dataset was the multi-omics TNBC dataset from FUSCC. Detailed information on the biospecimen collection, generation of expression profiles, WES data, SCNV data and H&E digital whole-slide images were described in our previous study.73,74 Finally, our cohort included 260 patients with corresponding multi-omics data and H&E images. All tissue samples were obtained after approval of the research by the FUSCC Ethics Committee, and each patient provided written informed consent for data and tissue use.
The second dataset comprised TNBC cases from the TCGA. We obtained multi-omics TNBC data from the TCGA portal (https://tcga-data.nci.nih.gov/tcga/). The diagnostic H&E images were downloaded from the Genomic Data Commons portal (https://portal.gdc.cancer.gov/). Finally, the TCGA cohort consisted of 134 patients with multi-omics data and H&E images.
Clinical studies and human samples
For the neoadjuvant trials (NCT04613674 and NCT04418154), we obtained 26 and 16 H&E digital whole-slide images, respectively, from early-stage TNBC patients treated with a PD-1 inhibitor-based regimen in FUSCC. The therapeutic response was evaluated according to the pCR rate using the definition of ypT0/Tis ypN0 (i.e., no invasive residual in breast or nodes; noninvasive breast residuals allowed) at the time of definitive surgery determined by experienced pathologists. The FUTURE (NCT03805399)75 and FUTURE-C-PLUS (NCT04129996)76 trial cohorts included 29 and 35 H&E digital whole-slide images from TNBC patients treated with a PD-1 inhibitor-based regimen in FUSCC. Clinical responses were evaluated based on radiologic assessments of tumor sizes by the investigators according to the Response Evaluation Criteria in Solid Tumors (RECIST, v1.1)77 for the FUTURE and FUTURE-C-PLUS trials. For multiplex IF staining, 100 formalin-fixed paraffin-embedded slides were collected before the patients received therapy from the aforementioned four trials. The histologic ITH of these four trials was calculated by incorporating the FUSCC TNBC cohort. Furthermore, a tissue microarray composed of TNBC samples was obtained from FUSCC (n = 283). All tissue samples were derived from untreated patients. Acquisition of all clinical samples was approved by the FUSCC Ethics Committee, and signed informed consent was obtained from each patient. This study conforms to the Declaration of Helsinki.78
Data for the GSE91061 (51 melanoma patients who had anti-PD-1 therapy with RNA-seq)79 and GSE176307 (33 metastatic urothelial cancer patients who had anti-PD-1 therapy with RNA-seq)80 cohorts were retrieved from https://www.ncbi.nlm.nih.gov/geo/.
Quantification of genetic ITH
Genetic ITH was determined using PyClone (v0.13.1)18 with default options to estimate the number of subclonal clusters within each tumor sample. Allele counts used for the PyClone input were extracted from the GATK output MAF files, whereas copy number input data were generated by ASCAT (v2.5.2) from probe-level data of OncoScan array processed by Chromosome Analysis Suite software (v4.1), or WES allele counts generated by alleleCounter (v4.0.1).
Quantification of histologic ITH
Utilizing CellProfiler software, we identified nuclei within representative H&E images for each tumor sample. The detailed procedure has been described previously.19 For each specific image, referenced as I, we meticulously extracted a suite of features that encompassed the multifaceted nuclear characteristics. This extraction honed in on the mean radius of the nuclei, shedding light on the morphological variations. Additionally, to gauge the disparities in staining intensity, both median and mean intensities were captured. The ensuing nuclear diversity intrinsic to an image, articulated as , is computed through the median absolute deviation (MAD) of these features:
Upon collating the images for a distinct tumor sample, denoted as T, we derived the median nuclear diversity across these images, offering an exhaustive depiction of that tumor’s nuclear heterogeneity. This process culminated in the formation of nuclear diversity ranks, , for every tumor. These ranks emerged by arranging the tumor samples in tandem with their respective diversity attributes:
Given the plausible impact of tumor purity on our metrics, we delved into a linear regression analysis. This strategic move aimed to discern the correlation between nuclear diversity ranks and tumor purity — the latter being quantified as the percentage of tumor nuclei discerned from histopathological assessment. A subsequent normalization of these ranks adeptly countered potential biases or distortions springing from tumor purity nuances (Supplementary information, Fig. S15).
Rounding off our methodological framework, the conclusive histologic ITH metric for every tumor, denominated as , was extrapolated from the median of the aforementioned nuclear diversity ranks:
Cell lines
Briefly, MDA-MB-231, Hs578T, BT549, HCC1143, HCC1806, MDA-MB-468, MDA-MB-453, BT20, 4T1, AT3 and HEK293T cells were obtained from the American Type Culture Collection. LM2 cells, a high lung metastatic subline of MDA-MB-231, were kindly provided by Guohong Hu (Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences, Shanghai, China). All cell lines were cultured under standard conditions. Cells were regularly checked for mycoplasma contamination and authenticated. For this study, cells were grown under limiting dilution conditions to obtain monoclonal status to decrease the ITH levels at baseline.
Plasmid construction, transfection, and lentiviral shRNA vectors
To generate ZNF689 and TRIM28 overexpression plasmids, ZNF689 (NM_138447.3) and TRIM28 (NM_005762.3) cDNAs were amplified by PCR and cloned into pCDH-CMV-MCS-EF1-Puro (System Biosciences) and pcDNA6/Myc-HisB (Invitrogen), respectively. ZNF689 and TRIM28 truncation mutants were cloned into pcDNA3.1 (Invitrogen) and pcDNA6/Myc-HisB, respectively.
Plasmid transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer’s instructions. Lentiviruses were generated using the pLKO.1 vector and packaging plasmids (psPAX2 and pMD2.G) in the HEK293T cell line. Annealing oligonucleotides targeting MUC19, PGC, DHRS2, ZNF689, TDRD12, and C20orf114 were synthesized (Sangon Biotech) and cloned into the pLKO.1-Puro vector (Supplementary information, Table S3). The supernatants were collected and filtered through a 0.45 μm syringe filter. Target cells were infected with lentiviruses and subjected to selection with 1–2 μg/mL puromycin for 4 days. The knockdown efficiencies and specificities of all the shRNAs were validated by RT-qPCR or western blotting. siRNAs were purchased from RiboBio (Supplementary information, Table S4). siRNA transfection was performed using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific). The silencing efficiency was assessed via RT-qPCR after 48 h of transfection.
DNA and RNA analyses
Total RNA was purified using the RNeasy Mini Kit (QIAGEN) and reverse transcribed into cDNA using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa). Genomic DNA was then purified with a DNA extraction kit (QIAGEN), and PCR was carried out with PrimeSTAR polymerase. RT-qPCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme) in a 7900HT Fast RealTime PCR System (Applied Biosystems). The primer sequences are shown in Supplementary information, Tables S5 and S6.
Western blotting and co-IP
Western blotting analysis was performed using a standard protocol.71 ImageJ was used to quantify the relative expression of proteins, which is presented as the ratio of test protein integrated density to internal control integrated density. Detailed information on the antibodies used in this study is provided in Supplementary information, Table S7. For co-IP, the cells were lysed in RIPA lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, supplemented with protease inhibitor cocktail). The lysates were incubated with the indicated antibodies overnight at 4 °C. The protein–antibody complexes were subjected to precipitation with Dynabeads Protein A beads (Invitrogen) for 2 h, washed five times in lysis buffer, and eluted with SDS-PAGE loading buffer by boiling for 5 min.
IF and multiplex IF
For IF analyses, cells grown on glass coverslips were fixed in 4% paraformaldehyde, permeabilized in 0.2% Triton X-100, and blocked in 10% normal goat serum in phosphate-buffered saline (PBS). Cells were incubated with primary antibodies overnight at 4 °C, washed three times in PBS, and then detected using the appropriate fluorescent secondary antibody. DNA staining was performed using ProLong Gold Antifade Mountant with DAPI (Invitrogen). Microscopic analyses were performed using a Leica SP5 confocal laser scanning microscope (Leica Microsystems). For multiplex IF, a five-color multiplex fluorescent immunohistochemical staining kit (Absin) was used following the manufacturer’s manual. Antibody information is listed in Supplementary information, Table S7. The stained slides were scanned using a Vectra Polaris System (PerkinElmer) and analyzed using QuPath software (v0.3.2).
IHC
Paraffin-embedded tissue sections were deparaffinized at 60 °C for 20 min, cleared in xylene, and rehydrated in a graded alcohol series. For H&E staining, the slides were stained with Mayer’s hematoxylin (Sigma-Aldrich), blued with 0.1% sodium bicarbonate, and counterstained with eosin Y solution (Sigma-Aldrich). IHC staining was performed with anti-ZNF689 (1:100), anti-LINE-1 ORF1p (1:200), anti-CD8 (1:500), anti-γH2AX (1:100), anti-HLA-ABC (1:500), anti-B2M (1:500), anti-TAP1 (1:500), and anti-PAMB9 (1:500) primary antibodies. Images visible under an Olympus BX43 microscope were acquired. Interpretation of the IHC results was performed by two independent pathologists who were blinded to the clinicopathological information. The expression of ZNF689 and LINE-1 ORF1p was scored and quantified as previously described.81 For CD8, the amount of staining (0–3) yielded a 4-point immunoreactivity score that ranged from 0 (no staining) to 3 (extensive, strong staining). Low and high expressions were defined according to the median immunoreactivity score. For HLA-ABC, B2M, TAP1 and PAMB9 expression, sections were scored by pathologists for intensity (0–3+) and extent (0–100%) of staining by light microscopy. The intensity and extent were multiplied to assign each tumor an H-score (range of 0–300).82
3D tumor sphere assay and histological analyses
Briefly, 1 × 106 cells expressing the indicated shRNAs were plated into 6-well ultralow attachment plates (Corning) to allow the cells to form spheroids. The culture medium was replaced once every three days, and spheroids were cultured for up to 14 days. Spheroids were collected and washed with PBS, fixed in 4% paraformaldehyde for 2 h, embedded in 2% HistoGel in 4% paraformaldehyde, and finally embedded in paraffin. Later, spheroids were cut into 3 μm sections and stained with H&E.
Animal studies
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of FUSCC. Five- to six-week-old female NSG mice, NOD/SCID mice, BALB/c mice and C57BL/6 mice were obtained from Shanghai Jihui Laboratory Animal Care Co., Ltd. OT-I mice were purchased from Shanghai Model Organisms Center, Inc.
For the PDX models, tumors sourced from TNBC patients were promptly implanted into the MFPs of NSG mice and then expanded. Upon reaching a volume of ~50 mm3, treatments were initiated. PDX1 tumors received intratumoral injections of either cholesterol-modified ZNF689 siRNA or control siRNA (RiboBio, 5 nmol/kg) in diluted water every 4 days over a 28-day period. For PDX2 and PDX3, tumors were treated daily with either Veh or EFV (S4685, Selleck), and delivered at 30 mg/kg/day via i.p. injections for 21 days.
For orthotopic injection of NOD/SCID mice, 1 × 106 LM2 cells with or without ZNF689 knockdown or overexpression were injected into the MFPs of the mice. For EFV drug treatment, mice inoculated with LM2 cells with or without ZNF689 knockdown were subjected daily to i.p. injection with either Veh or EFV (30 mg/kg/day). For orthotopic injection of BALB/c mice, 1 × 105 4T1 cells with or without ZNF689 knockdown or overexpression were injected into the MFPs of the mice. For orthotopic injection of C57BL/6 mice, 1 × 105 AT3 cells with ZNF689 knockdown were injected into the MFPs of the mice. For combination treatments, BALB/c mice and C57BL/6 mice were randomly divided into four different treatment groups after the tumor was palpable. Veh or EFV (30 mg/kg/day) was administered in combination with isotype rat IgG2a (BP0089, BioXcell) or anti-PD-1 (BP0273, BioXcell) (100 µg per mouse, every 3 days) antibodies via i.p. injection. Tumor size was measured twice or thrice weekly using a caliper. Tumor volume in mm3 was calculated using the formula: tumor volume = 0.5 × L × W2, where L is the longest dimension and W is the perpendicular dimension. Mouse survival was monitored with tumor volume exceeding 2000 mm3, weight loss > 20%, and decreasing behavioral conditions considered endpoints.
WES of mouse tumors
DNA was extracted from tumors derived from PDX and LM2 xenografts utilizing the DNeasy kit (QIAGEN). Whole-exome library was constructed using the Agilent SureSelect Human All Exon V6 Kit (Agilent Technologies). According to the manufacturer’s instructions, the index-coded samples were clustered on a cBot Cluster Generation System using a HiSeq PE Cluster Kit (Illumina). After cluster generation, the DNA libraries were sequenced on the Illumina HiSeq platform (Illumina) and 150 bp paired-end reads were generated.
Flow cytometry
For in vitro analysis, the cells were washed with PBS and dissociated from the plates with Accutase (Gibco) for 5–10 min at 37 °C to generate single-cell suspensions. For in vivo studies, tumors were excised postmortem and enzymatically digested using a mixture of 0.5 mg/mL collagenase type I (Sigma-Aldrich), 1 mg/mL dispase (Roche), and 1 mg/mL hyaluronidase (Sigma-Aldrich) with antibiotics for 30 min at 37 °C. The dissociated cells were passed through a 40 µm filter to collect single-cell suspensions. Single-cell suspensions were washed twice in flow staining buffer and incubated with the appropriate flow antibodies at 4 °C for 30 min in the dark. For intracellular staining of mouse GZMB and IFN-γ, cells were stimulated with Leukocyte Activation Cocktail (BD Biosciences) for 6 h and then subjected to surface and intracellular staining using a Cytofix/Cytoperm Soln Kit (BD Biosciences). A live/dead stain was used to discriminate between viable and dead cells. All antibodies used for flow cytometry are listed in Supplementary information, Table S7. Data were analyzed with CytExpert software (Beckman Coulter). The gating strategy is shown in Supplementary information, Fig. S8f.
SILAC labeling and mass spectrometry
For SILAC experiments, empty vector or Flag-ZNF689 LM2 cells were grown in DMEM supplemented with 10% FBS and either heavy form 13C615N4-arginine (Arg10) and 13C614N2-lysine (Lys6) or light form 12C614N4-arginine (Arg0) and 12C614N2-lysine (Lys0). The cells were grown for more than six generations before being harvested to achieve > 97% labeling efficiency. For mass spectrometry, the gel-containing samples were decolorized and washed to make them transparent and then freeze-dried. The disulfide bonds of the samples were reduced by dithiothreitol and alkylation before enzymatic hydrolysis. Then, the peptide fraction was extracted and dried in a vacuum. The samples were desalted, and the supernatant was added to the sample flask for mass spectrometry (Q Exactive) detection. The MaxQuant search database was searched. The results of the proteomics analysis are provided in Supplementary information, Table S8.
RNA-seq and data analysis
Total RNA samples were prepared from LM2 cells using the RNeasy kit (QIAGEN), and the following RNA preparation and sequencing procedures were performed as described elsewhere.81 GSEA was performed using the GSEA v4.2 desktop application. For analysis of RE expression from RNA-seq data, reads were mapped to the Homo sapiens genome (hg19) using Bowtie 2 and assigned to REs using RepEnrich2 with the recommended parameters (https://github.com/nerettilab/RepEnrich2).83 The RE annotation file was provided by RepEnrich2. The resulting counts for REs were analyzed by the edgeR package to obtain counts per million reads values. GSEA was performed using the LINE-1 signature annotated by RepeatMasker open-4.0.5.
Retrotransposition reporter assay
We used an EGFP reporter assay to measure retrotransposition.84 We transfected 2 × 105 MDA-MB-231 and Hs578T cells with 2 µg LINE-1 reporter plasmids (MT525, JM111) or 2 µg EGFP plasmid and selected them with 1 μg/mL puromycin for 12 days. The cells were trypsinized and resuspended in cytometry buffer (HBSS, no phenol red, 1% FBS, 1 mM EDTA) at a concentration of 1 × 106 cells/mL and then subjected to flow cytometry. Singlets were gated on SSC-A/SSC-H and FSC-A/FSC-H, and EGFP thresholds were set such that untransfected cells showed 0.1% EGFP+ cells. We normalized the percentage of EGFP+ cells in the experimental groups to the percentage of EGFP+ cells in the EGFP-transfected control groups.
ChIP and ChIP-seq
ChIP assays were conducted as described previously.85 Briefly, cells were crosslinked with 1% formaldehyde for 10 min at room temperature. Chromatin was fragmented to 150–800 bp by sonication. ChIP was carried out using either control IgG or the Flag (F3165, Sigma-Aldrich) primary antibody. Following washing and reverse cross-linking steps, the quantity of eluted DNA was determined via qPCR. All utilized primer sequences are provided in Supplementary information, Table S9.
For ChIP-seq, 150–800 bp chromatin fragments were immunoprecipitated with protein A/G beads conjugated to specific antibodies: anti-Flag (F3165, Sigma-Aldrich), anti-H3K9me3 (ab8898, Abcam), and anti-TRIM28 (ab10483, Abcam). After reverse crosslinking, both ChIP and input DNA were sequenced on the Illumina NextSeq 500 in paired-end 2 × 150 mode. Raw reads underwent quality refinement with Fastp (v0.20.0), trimming sequencing adapters, short reads (< 30 bp), and inferior sequences. These curated reads were mapped to the human genome (hg19) via Bowtie2 (v2.2.6). Read count normalization was performed using deepTools (v3.3.2), and peaks were identified with MACS2 under specific parameters. Annotation was performed with ChIPseeker and UCSC hg19 r1_repeat_rmsk for LINE-1, while distribution analysis over LINE-1 and genes utilized deepTools 3.3.2 computeMatrix, segmenting gene models into 20 bins, including ±3 kb surrounding regions.
Immune and tumor cell coculture assay
AT3-OVA tumor cells were plated in 6-well plates for in vitro coculture in the DMEM/F12 listed above. Once the cells reached 50%–75% confluency, OT-I splenocytes or CD8+ T cells were added to the tumor cells at a 1:10 ratio (tumor cell: immune cell). After 24 h of coculture, the cells and culture medium were collected for further experiments. To assess tumor cell viability after CD8+ T-cell coculture, we used the lactate dehydrogenase cytotoxicity assay (Yeasen) to measure the T-cell killing of AT3-OVA tumor cells according to the manufacturer’s instructions. Furthermore, IFN-γ levels were quantified using a Mouse IFN-γ ELISA Kit (Lianke) following the manufacturer’s guidelines.
scRNA-seq
Single-cell suspensions were generated using the Mouse Tumor Dissociation Kit (Miltenyi) according to the manufacturer’s protocol. The cell suspension was loaded into the 10× Genomics Chromium platform, and sequencing libraries were constructed with reagents from a Chromium Single Cell 3′ Reagent Kit v3 (10× Genomics) according to the manufacturer’s instructions. Sequencing was performed with the Illumina sequencing platform (NovaSeq) according to the manufacturer’s instructions (Illumina). Raw reads were demultiplexed and mapped to the reference genome (mm10, GRCm38) using 10× Genomics Cell Ranger. The cellranger aggr pipeline was used to normalize and merge two samples into one.
We used the R package Seurat (v4.3.0) to perform downstream analysis. All cells expressing < 200 or > 7500 genes were removed, as well as cells that contained > 20% mitochondrial counts. The anchor-based canonical correlation analysis (CCA) method in the Seurat package was performed for dataset integration and batch effect correction after normalization. Then, the integrated data were adopted for dimensional reduction, clustering, and data visualization using default parameters. Cell clusters were annotated based on canonical cell markers from published literature86–89 and databases including CellMarker90 and PanglaoDB.91 The R package fgsea (v1.24.0) was used to perform GSEA based on hallmark, REACTOME and GO gene sets from MSigDB. Copy number instability was assessed with the R package inferCNV.92
Spatial transcriptomics
Fresh tissues were concurrently frozen and embedded in optical cutting tissue (OCT) compound in liquid nitrogen. Tissue blocks were cut into 10 μm sections and processed using the Visium Spatial Gene Expression Slide & Reagent kit (10× Genomics) according to the manufacturer’s instructions. First, the Visium Spatial Tissue Optimization Slide & Reagent kit (10× Genomics) was used to optimize permeabilization conditions for the tissue. Sections were stained with H&E and imaged, followed by tissue permeabilization. Then, a reverse transcription experiment was conducted, and sequencing libraries were prepared following the manufacturer’s protocol. Sequencing was performed with a Novaseq PE150 platform according to the manufacturer’s instructions (Illumina) at an average depth of 300 million read pairs per sample. Spots were manually annotated by an experienced pathologist.
Reads were demultiplexed and mapped to the reference genome (mm10, GRCm38) using Space Ranger software v1.2.0 (10× Genomics). The filtered gene-spot matrix and the fiducial-aligned low-resolution image were loaded into the R package Seurat (v4.3.0) for all subsequent data normalization, dimensional reduction and data visualization with default parameters. Data normalization was performed on independent tissue sections using the SCTransform function in Seurat.93 Signature scores were calculated by the R package AUCell (v1.20.2).94
Statistics
The statistical details and methods are indicated in the figure legends or supplementary information. The survival curves were constructed according to the Kaplan–Meier method and compared with a log-rank test. Patients without events or death were censored at the last follow-up. Multivariate analyses were performed by Cox proportional hazards regression, and hazard ratios and 95% confidence intervals were reported. Boxplot limits indicate the minimum and maximum, and the boxplot centerline indicates the median. Statistical significance tests, including one- or two-tailed unpaired and paired Student’s t-test, one- or two-way ANOVA, Spearman correlation test, Wilcoxon test, Pearson’s χ2 test, permutation test and Fisher’s exact test were performed using R or GraphPad Prism software, as denoted in each analysis. P < 0.05 was considered to indicate a statistically significant difference. The data are presented as the mean ± SEM for a minimum of three independent experiments unless otherwise indicated.
Other methods are described in detail in Supplementary information, Data S1.
Supplementary information
Acknowledgements
We thank Hong-Jie Shen, Wen-Qi Xu and Jin Wang from the Shanghai Key Laboratory of Medical Epigenetics, Institutes of Biomedical Sciences, Fudan University, for their expert guidance on the regulatory mechanism of ZNF689 on LINE-1. This work was supported by the National Key R&D Program of China (2020YFA0112304), the National Natural Science Foundation of China (82272822, 81922048, 82341003, 82002799 and 92159301), the Natural Science Foundation of Shanghai (22ZR1479200 and 23ZR1411800), the Shanghai Key Laboratory of Breast Cancer (12DZ2260100), the SHDC Municipal Project for Developing Emerging and Frontier Technology in Shanghai Hospitals (SHDC12021103) and the Youth Talent Program of Shanghai Health Commission (2022YQ012).
Author contributions
Y.-Z.J., Z.-M.S. and G.-H.D. conceived the research project. Z.-M.S. and Y.-Z.J. secured funding and supervised the study. L.-P.G. designed and performed most experiments, analyzed the data, and wrote the manuscript. X.J. designed the experiments and supervised the study. D.M., Z.-Y.W., C.-L.L. and X.-Y.L performed the bioinformatics analyses. C.-Z.Z, S.Z. and T.-J.Y. assisted with flow cytometry and histology imaging. Y.-Z.J., L.-P.G., X.J., D.M., Z.-Y.W. and S.Z. discussed the results and manuscript. All authors have read and approved the manuscript.
Data availability
Sequence data have been deposited in the NCBI Gene Expression Omnibus (OncoScan array; GEO: GSE118527) and Sequence Read Archive (WES and RNA-seq; SRA: SRP157974). Other data that support the findings of this study are presented in the main text and in the online Supplementary information. Sequencing coverage for our datasets is detailed in Supplementary information, Table S10, while quality control metrics are provided in Supplementary information, Table S11. Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Li-Ping Ge, Xi Jin, Ding Ma, Zi-Yu Wang.
Contributor Information
Gen-Hong Di, Email: genhongdi@163.com.
Zhi-Ming Shao, Email: zhimingshao@fudan.edu.cn.
Yi-Zhou Jiang, Email: yizhoujiang@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-023-00909-w.
References
- 1.Bianchini G, De Angelis C, Licata L, Gianni L. Treatment landscape of triple-negative breast cancer — expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2022;19:91–113. doi: 10.1038/s41571-021-00565-2. [DOI] [PubMed] [Google Scholar]
- 2.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Vitale I, et al. Mutational and antigenic landscape in tumor progression and cancer immunotherapy. Trends Cell Biol. 2019;29:396–416. doi: 10.1016/j.tcb.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Schmid P, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 5.Cortes J, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020;396:1817–1828. doi: 10.1016/S0140-6736(20)32531-9. [DOI] [PubMed] [Google Scholar]
- 6.Adams S, et al. Pembrolizumab monotherapy for previously untreated, PD-L1-positive, metastatic triple-negative breast cancer: cohort B of the phase II KEYNOTE-086 study. Ann. Oncol. 2019;30:405–411. doi: 10.1093/annonc/mdy518. [DOI] [PubMed] [Google Scholar]
- 7.Winer EP, et al. Pembrolizumab versus investigator-choice chemotherapy for metastatic triple-negative breast cancer (KEYNOTE-119): a randomised, open-label, phase 3 trial. Lancet Oncol. 2021;22:499–511. doi: 10.1016/S1470-2045(20)30754-3. [DOI] [PubMed] [Google Scholar]
- 8.Vitale I, Shema E, Loi S, Galluzzi L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 2021;27:212–224. doi: 10.1038/s41591-021-01233-9. [DOI] [PubMed] [Google Scholar]
- 9.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 10.Wolf Y, Samuels Y. Intratumor heterogeneity and anti-tumor immunity shape one another bidirectionally. Clin. Cancer Res. 2022;28:2994–3001. doi: 10.1158/1078-0432.CCR-21-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marusyk A, Janiszewska M, Polyak K. Intratumor heterogeneity: the Rosetta stone of therapy resistance. Cancer Cell. 2020;37:471–484. doi: 10.1016/j.ccell.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 13.Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 2018;20:1349–1360. doi: 10.1038/s41556-018-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGranahan N, et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 2015;7:283ra254. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dentro SC, et al. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell. 2021;184:2239–2254.e39. doi: 10.1016/j.cell.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth A, et al. PyClone: statistical inference of clonal population structure in cancer. Nat. Methods. 2014;11:396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. Patient-derived xenograft models in cancer therapy: technologies and applications. Signal Transduct. Target. Ther. 2023;8:160. doi: 10.1038/s41392-023-01419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf Y, et al. UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell. 2019;179:219–235.e21. doi: 10.1016/j.cell.2019.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czerwińska P, Mazurek S, Wiznerowicz M. The complexity of TRIM28 contribution to cancer. J. Biomed. Sci. 2017;24:63. doi: 10.1186/s12929-017-0374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs FMJ, et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imbeault M, Helleboid P-Y, Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017;543:550–554. doi: 10.1038/nature21683. [DOI] [PubMed] [Google Scholar]
- 27.Payer LM, Burns KH. Transposable elements in human genetic disease. Nat. Rev. Genet. 2019;20:760–772. doi: 10.1038/s41576-019-0165-8. [DOI] [PubMed] [Google Scholar]
- 28.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Meter M, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elsässer SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belancio VP, Deininger PL, Roy-Engel AM. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu Z, et al. Silencing of LINE-1 retrotransposons is a selective dependency of myeloid leukemia. Nat. Genet. 2021;53:672–682. doi: 10.1038/s41588-021-00829-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKerrow W, et al. LINE-1 expression in cancer correlates with p53 mutation, copy number alteration, and S phase checkpoint. Proc. Natl. Acad. Sci. USA. 2022;119:e2115999119. doi: 10.1073/pnas.2115999119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, et al. Gut stem cell necroptosis by genome instability triggers bowel inflammation. Nature. 2020;580:386–390. doi: 10.1038/s41586-020-2127-x. [DOI] [PubMed] [Google Scholar]
- 35.Patnala R, et al. Inhibition of LINE-1 retrotransposon-encoded reverse transcriptase modulates the expression of cell differentiation genes in breast cancer cells. Breast Cancer Res. Treat. 2014;143:239–253. doi: 10.1007/s10549-013-2812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon M, et al. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 2019;29:871–885.e5. doi: 10.1016/j.cmet.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang R, et al. LINE-1 retrotransposition promotes the development and progression of lung squamous cell carcinoma by disrupting the tumor-suppressor gene FGGY. Cancer Res. 2019;79:4453–4465. doi: 10.1158/0008-5472.CAN-19-0076. [DOI] [PubMed] [Google Scholar]
- 38.Bi S, et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell. 2020;11:483–504. doi: 10.1007/s13238-020-00728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mroz EA, et al. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12:e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma D, Jiang Y-Z, Liu X-Y, Liu Y-R, Shao Z-M. Clinical and molecular relevance of mutant-allele tumor heterogeneity in breast cancer. Breast Cancer Res. Treat. 2017;162:39–48. doi: 10.1007/s10549-017-4113-z. [DOI] [PubMed] [Google Scholar]
- 41.Mazor T, Pankov A, Song JS, Costello JF. Intratumoral heterogeneity of the epigenome. Cancer Cell. 2016;29:440–451. doi: 10.1016/j.ccell.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva FP, Hamamoto R, Furukawa Y, Nakamura Y. TIPUH1 encodes a novel KRAB zinc-finger protein highly expressed in human hepatocellular carcinomas. Oncogene. 2006;25:5063–5070. doi: 10.1038/sj.onc.1209517. [DOI] [PubMed] [Google Scholar]
- 43.Shigematsu S, et al. ZNF689 suppresses apoptosis of hepatocellular carcinoma cells through the down-regulation of Bcl-2 family members. Exp. Cell Res. 2011;317:1851–1859. doi: 10.1016/j.yexcr.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Zeng H, et al. MicroRNA-339 inhibits human hepatocellular carcinoma proliferation and invasion via targeting ZNF689. Drug Des. Devel. Ther. 2019;13:435–445. doi: 10.2147/DDDT.S186352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez-Martin B, et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat. Genet. 2020;52:306–319. doi: 10.1038/s41588-019-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai L, Huang Q, Boeke JD. Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem. 2011;12:18. doi: 10.1186/1471-2091-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellisai C, et al. Reverse transcriptase inhibitors promote the remodelling of nuclear architecture and induce autophagy in prostate cancer cells. Cancer Lett. 2020;478:133–145. doi: 10.1016/j.canlet.2020.02.029. [DOI] [PubMed] [Google Scholar]
- 48.Sciamanna I, et al. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene. 2005;24:3923–3931. doi: 10.1038/sj.onc.1208562. [DOI] [PubMed] [Google Scholar]
- 49.Rajurkar M, et al. Reverse transcriptase inhibition disrupts repeat element life cycle in colorectal cancer. Cancer Discov. 2022;12:1462–1481. doi: 10.1158/2159-8290.CD-21-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenthal R, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567:479–485. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sade-Feldman M, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 2017;8:1136. doi: 10.1038/s41467-017-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Golkaram M, et al. Spatiotemporal evolution of the clear cell renal cell carcinoma microenvironment links intra-tumoral heterogeneity to immune escape. Genome Med. 2022;14:143. doi: 10.1186/s13073-022-01146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaretsky JM, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl J. Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goubert C, Zevallos NA, Feschotte C. Contribution of unfixed transposable element insertions to human regulatory variation. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2020;375:20190331. doi: 10.1098/rstb.2019.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lunyak VV, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 56.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, et al. MIR retrotransposon sequences provide insulators to the human genome. Proc. Natl. Acad. Sci. USA. 2015;112:E4428–E4437. doi: 10.1073/pnas.1507253112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cournac A, Koszul R, Mozziconacci J. The 3D folding of metazoan genomes correlates with the association of similar repetitive elements. Nucleic Acids Res. 2016;44:245–255. doi: 10.1093/nar/gkv1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Helman E, et al. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24:1053–1063. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doucet-O’Hare TT, et al. LINE-1 expression and retrotransposition in Barrett’s esophagus and esophageal carcinoma. Proc. Natl. Acad. Sci. USA. 2015;112:E4894–E4900. doi: 10.1073/pnas.1502474112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spielmann M, Lupiáñez DG, Mundlos S. Structural variation in the 3D genome. Nat. Rev. Genet. 2018;19:453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- 63.Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018;19:789–800. doi: 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019;20:207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 65.Dubois F, Sidiropoulos N, Weischenfeldt J, Beroukhim R. Structural variations in cancer and the 3D genome. Nat. Rev. Cancer. 2022;22:533–546. doi: 10.1038/s41568-022-00488-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lawson HA, Liang Y, Wang T. Transposable elements in mammalian chromatin organization. Nat. Rev. Genet. 2023;24:712–723. doi: 10.1038/s41576-023-00609-6. [DOI] [PubMed] [Google Scholar]
- 67.Fueyo R, Judd J, Feschotte C, Wysocka J. Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell Biol. 2022;23:481–497. doi: 10.1038/s41580-022-00457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milo I, et al. The immune system profoundly restricts intratumor genetic heterogeneity. Sci. Immunol. 2018;3:eaat1435. doi: 10.1126/sciimmunol.aat1435. [DOI] [PubMed] [Google Scholar]
- 69.Jung H, Choi JK, Lee EA. Immune signatures correlate with L1 retrotransposition in gastrointestinal cancers. Genome Res. 2018;28:1136–1146. doi: 10.1101/gr.231837.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Solovyov A, et al. Global cancer transcriptome quantifies repeat element polarization between immunotherapy responsive and T cell suppressive classes. Cell Rep. 2018;23:512–521. doi: 10.1016/j.celrep.2018.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen JZ, et al. FBXO44 promotes DNA replication-coupled repetitive element silencing in cancer cells. Cell. 2021;184:352–369.e23. doi: 10.1016/j.cell.2020.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Griffin GK, et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature. 2021;595:309–314. doi: 10.1038/s41586-021-03520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang YZ, et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell. 2019;35:428–440.e5. doi: 10.1016/j.ccell.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 74.Zhao, S. et al. Deep learning framework for comprehensive molecular and prognostic stratifications of triple-negative breast cancer. Fundamental Res. 10.1016/j.fmre.2022.06.008 (2022). [DOI] [PMC free article] [PubMed]
- 75.Jiang YZ, et al. Molecular subtyping and genomic profiling expand precision medicine in refractory metastatic triple-negative breast cancer: the FUTURE trial. Cell Res. 2021;31:178–186. doi: 10.1038/s41422-020-0375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen L, et al. Famitinib with camrelizumab and nab-paclitaxel for advanced immunomodulatory triple-negative breast cancer (FUTURE-C-PLUS): an open-label, single-arm, phase 2 trial. Clin. Cancer Res. 2022;28:2807–2817. doi: 10.1158/1078-0432.CCR-21-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 78.World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 79.Riaz N, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rose TL, et al. Fibroblast growth factor receptor 3 alterations and response to immune checkpoint inhibition in metastatic urothelial cancer: a real world experience. Br. J. Cancer. 2021;125:1251–1260. doi: 10.1038/s41416-021-01488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu T-J, et al. PDSS1-mediated activation of CAMK2A-STAT3 signaling promotes metastasis in triple-negative breast cancer. Cancer Res. 2021;81:5491–5505. doi: 10.1158/0008-5472.CAN-21-0747. [DOI] [PubMed] [Google Scholar]
- 82.Pietanza MC, et al. Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J. Clin. Oncol. 2018;36:2386–2394. doi: 10.1200/JCO.2018.77.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Criscione SW, Zhang Y, Thompson W, Sedivy JM, Neretti N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics. 2014;15:583. doi: 10.1186/1471-2164-15-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu W, et al. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature. 2021;591:317–321. doi: 10.1038/s41586-021-03210-1. [DOI] [PubMed] [Google Scholar]
- 86.Gubin MM, et al. High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell. 2018;175:1014–1030.e19. doi: 10.1016/j.cell.2018.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zilionis R, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–1334.e10. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hollern DP, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 2019;179:1191–1206.e21. doi: 10.1016/j.cell.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cortellino S, et al. Fasting renders immunotherapy effective against low-immunogenic breast cancer while reducing side effects. Cell Rep. 2022;40:111256. doi: 10.1016/j.celrep.2022.111256. [DOI] [PubMed] [Google Scholar]
- 90.Zhang X, et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47:D721–D728. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Franzén O, Gan LM, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database. 2019;2019:baz046. doi: 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Puram SV, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited in the NCBI Gene Expression Omnibus (OncoScan array; GEO: GSE118527) and Sequence Read Archive (WES and RNA-seq; SRA: SRP157974). Other data that support the findings of this study are presented in the main text and in the online Supplementary information. Sequencing coverage for our datasets is detailed in Supplementary information, Table S10, while quality control metrics are provided in Supplementary information, Table S11. Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors.