Abstract
Human microorganisms, including bacteria, fungi, and viruses, play key roles in several physiological and pathological processes. Some studies discovered that tumour tissues once considered sterile actually host a variety of microorganisms, which have been confirmed to be closely related to oncogenesis. The concept of intratumoural microbiota was subsequently proposed. Microbiota could colonise tumour tissues through mucosal destruction, adjacent tissue migration, and hematogenic invasion and affect the biological behaviour of tumours as an important part of the tumour microenvironment. Mechanistic studies have demonstrated that intratumoural microbiota potentially promote the initiation and progression of tumours by inducing genomic instability and mutations, affecting epigenetic modifications, promoting inflammation response, avoiding immune destruction, regulating metabolism, and activating invasion and metastasis. Since more comprehensive and profound insights about intratumoral microbiota are continuously emerging, new methods for the early diagnosis and prognostic assessment of cancer patients have been under examination. In addition, interventions based on intratumoural microbiota show great potential to open a new chapter in antitumour therapy, especially immunotherapy, although there are some inevitable challenges. Here, we aim to provide an extensive review of the concept, development history, potential sources, heterogeneity, and carcinogenic mechanisms of intratumoural microorganisms, explore the potential role of microorganisms in tumour prognosis, and discuss current antitumour treatment regimens that target intratumoural microorganisms and the research prospects and limitations in this field.
Subject terms: Tumour immunology, Cancer
Introduction
Approximately 38 trillion microorganisms are found in the human microbiota, including bacteria, fungi, and viruses, and their number roughly equals that of human cells.1,2 These microorganisms have previously been found in open cavities and organs such as the gut, skin, mouth, and vagina.3 However, with the breakthroughs of technology, the tissues and organs once considered sterile, including the lung, breast, liver, pancreas, prostate, and kidney, have also demonstrated to harbour low-biomass microbial communities, which leads to further research in related fields.4,5 In particular, the concept of intratumoural microbiota present in tumour tissues is proposed,6 and such microorganisms have been found in at least 33 major cancer types.7–9 The intratumoural microbiota is an integral part of the tumour microenvironment (TME), mainly in cancer and immune cells.7,10–12 Intratumoural microorganisms can significantly change the biology of different cell compartments, affecting the occurrence, development and metastasis of tumours and antitumour immunity.7,13
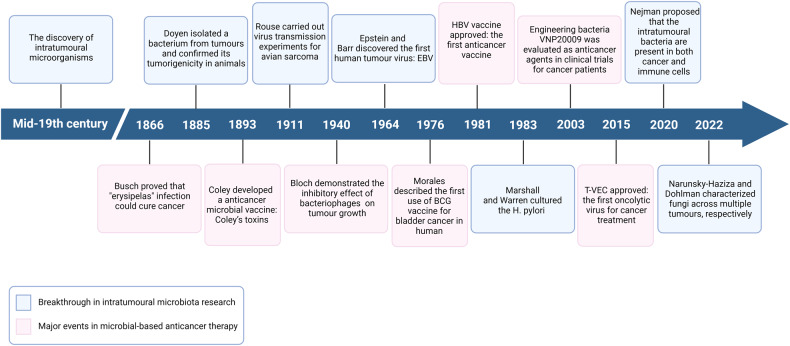
So far, many important discoveries about intratumoural microbiota have been reported (Fig. 1). As early as in the mid-19th century, microbiologists discovered the presence of a variety of microorganisms in tumours. In 1885, Doyen isolated a bacterium, Micrococcus neoformans, from different tumours and confirmed its tumorigenicity in animals.14 However, due to the limited sterile conditions available at that time, this findings were questioned. In the early 20th century, Rouse discovered that avian sarcoma can be transmitted in animals through filtered cell-free tumour extracts which suggested that causative agent in the tumour extracts, roussarcoma virus, could induce cancer, thereby establishing viruses as the causal agents of cancer for the first time.15 In 1964, Epstein and Barr discovered the first human virus particle in Burkitt’s lymphoma—Epstein-Barr virus (EBV).16 Since then, evidence that viral infections cause cancer in humans began to emerge. In 1983, Marshall and Warren cultured H. pylori and further established its role in gastric cancer aetiology.17,18 In 2020, the most rigorous and comprehensive survey of bacteria in seven human tumour samples by Nejman et al. revealed that different cancer types involve different bacterial species.7 Subsequently, Narunsky-Haziza and Dohlman separately characterised fungi in human cancer specimens from multiple tumour types to further explore the role of intratumoural fungi in cancer diagnosis and prognosis.11,12
Fig. 1.
Milestone events of intratumoural microbiota. The key findings on intratumoural microbiota and the major achievements of microbial-based anticancer therapy were reviewed retrospectively. Created with BioRender.com
Meanwhile, cancer treatment methods based on microbial intervention have also been developed. In 1866, Busch purposefully infected cancer patients with erysipelas and found that the tumours of the patients subsided.19 In 1893, Coley invented an anticancer drug microbial vaccine—Coley toxin— which has been observed to alleviate advanced cancers in some cases.20 As similar approaches became more widely researched, numerous anticancer vaccines emerged in the following decades.21 In addition, since Bloch demonstrated the interaction of bacteriophages with malignant cells and their ability to inhibit tumour growth in 1940,22 scientists have also started paying attention to the potential of these dynamic viral entities to treat cancer. In 1981, the hepatitis B virus (HBV) vaccine was approved as the first anticancer vaccine, saving millions of potential victims from developing hepatocellular carcinoma (HCC).21 By now, far beyond natural microbes, scientists have shifted their sights to modifying bacteria and viruses to fight cancer.
Based on the need for a comprehensive and in-depth understanding of the current research progress in intratumoural microbiota, this review summarises the characteristics and emerging functions of intratumoural microorganisms in various tumours as well as their effects on cancer development and antitumour immunity. We also discuss prospective applications of intratumoural microbiota in cancer prognosis and therapy. These findings complement the results of previous studies on intratumoural microorganisms and may provide potential theoretical support for future research on intratumoural microbiota-targeted cancer treatments.
Characteristics of intratumoural microbiota
Colonisation of the tumour by microorganisms
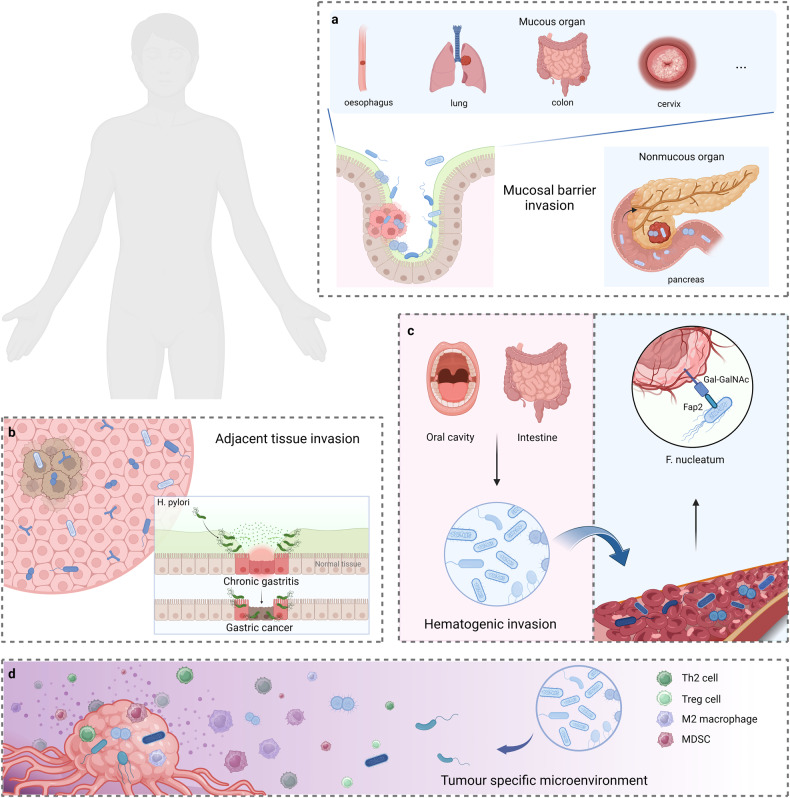
There are three possible origins of intratumoural microorganisms (Fig. 2). The first is mucosal barrier invasion, in which microorganisms colonising the mucosa may invade the tumour through the damaged mucosa.23,24 Tjalsma et al. proposed a bacterial driver–passenger model in which “driver” bacteria such as the genus Bacteroides and the family Enterobacteriaceae colonise the intestine and drive tumorigenesis. With microenvironment change, driving bacteria are gradually replaced by “passenger” bacteria, including opportunistic pathogens and commensal or probiotic bacteria, which further affect tumour progression.25 Many intratumoural microbiota have been found to colonise mucosal organs, such as the oesophagus, lung, colon, and cervix. Moreover, the intratumoural microbiota in non-mucosal organs, such as the pancreas, has also been found to translocate from the intestinal tract with impaired mucosal barriers and into the pancreas through the pancreatic duct, thereby reshaping the TME and increasing susceptibility to microbial translocation.26 The second is adjacent tissue invasion, where the microbiome communities between the tumour and adjacent normal tissue share many similarities.7,27,28 Moreover, many studies have found that viral infections29 and specific bacterially mediated chronic inflammation,30 such as Helicobacter pylori (H. pylori) and gastritis, can eventually evolve into tumours. Nonetheless, the origin of microorganisms in normal tissues of most organs remains unclear, and these microbes may also disseminate from the tumour site; therefore, further research is required to confirm this hypothesis. Finally, the third is hematogenic invasion, in which microorganisms from the oral cavity, intestine, and other potential locations may be carried to the tumour locations and colonise the tumour through destroyed blood vessels.31 Abed et al. reported that intravenously injected Fusobacterium nucleatum (F. nucleatum) interacts with the host polysaccharide D-galactose-β(1–3)-N-acetyl-D-galactosamine (Gal-GalNAc) in a lectin Fap2-dependent manner to localise to mouse tumour tissues, indicating that fusobacteria reached colon adenocarcinomas via a hematogenous pathway.32 A similar result was found in mouse mammary tumours.33 However, intestinal localisation of these tumours does not fully summarise the complex histological features of human colorectal adenomas. Furthermore, specific microenvironments in tumours may enhance microbial colonisation, such as immunosuppressive, hypoxic, and metabolic nutrient-enriched environments.34 However, these conjectures must be confirmed through holistic metagenomic sequencing and genetic identification.
Fig. 2.
The potential origins of intratumoural microbiota. a Mucosal barrier invasion. Microorganisms may invade the tumour through the damaged mucosa. b Adjacent tissue invasion. the microbiome community between the tumour and adjacent normal tissue share many similarities. c Hematogenic invasion. microorganisms from the oral cavity, intestine, and other potential locations may be carried to the tumour locations and colonise the tumour through destroyed blood vessels. d Attraction of tumour specific microenvironment. immunosuppressive, hypoxic, and metabolic nutrient-enriched environments in tumours may enhance microbial colonisation. Created with BioRender.com
Diversity of intratumoural microbiota
The structure and abundance of the intratumoural microbial population vary substantially across different types, subtypes, and stages of cancer. Here, we have summarised the intratumoural microbiota in a few cancers to understand their role in cancer progression (Table 1).
Table 1.
Characterization of the intratumoural microbiota in various cancers
| Cancer types | Microbiome compositions | Quantitative dynamics | Function |
|---|---|---|---|
| Lung cancer | Genus Modestobacter36 | Increase | — |
|
Genus Propionibacterium, genus Enterobacteriaceae36 |
Dcrease | — | |
| Genus Blastomyces11 | Increase | — | |
|
Class Agaricomycetes, genus Aspergillus12 |
Increase | — | |
| Genus Acidovorax37 | Increase | Related to tumours with high TP53 mutation; | |
|
Genus Klebsiella, genus Anaerococcus37 |
Increase | — | |
|
Genus Acinetobacter, genus Brevundimonas, genus Propionibacterium38 |
Increase | — | |
| Phylum Cyanobacteria39 | Increase | — | |
|
Genus Veillonella, genus Megasphaera40 |
Increase | As the diagnostic biomarker of tumour; | |
|
Family Coriobacteriaceae, genus Pasteurella201 |
— | Related to the number of CD8 + T cells and M2 macrophages; | |
| Species Nontypeable Haemophilus influenzae188 | Increase | Released IL-17C and recruited the neutrophils; | |
| Liver cancer | Hepatitis B virus43,130,179,203 | Increase |
Integrated viral genome into the host chromosome; induced m6A modification of RNA Recruited Treg cells; |
| Hepatitis C virus43,204 | Increase | Recruited Treg cells; | |
| Species Helicobacter pylori48–50 | Increase | — | |
| Order Gammaproteobacteria53 | Increase | — | |
|
Family Streptococcaceae, genus Lactococcus53 |
Increase | As the hallmark groups of cirrhosis hepatocellular carcinoma; | |
| Family Enterobacteriaceae54 | Increase | — | |
|
Family Caulobacteraceae, family Rickettsiaceae54 |
Decrease | — | |
| Species Paraburkholderia fungorum55 | Decrease | Related to antitumor activity; | |
| Colorectal cancer | Species Enterotoxigenic Bacteroides fragilis56,140,186 | Increase |
Secreted carcinogenic toxins; initiated pro-inflammatory signalling cascade; |
| Genus Fusobacterium57–59,182 | Increase | Promoted the polarization of M2-like macrophages; | |
|
Genus Lactococcus, genus Bacteroides, genus Prevotella, genus Streptococcus59 |
Increase | — | |
|
Genus Pseudomonas, genus Escherichia-Shigella59 |
Decrease | — | |
| Species Fusobacterium nucleatum60,141,166–168,174,190,199,210,211,242,248 | Increase |
Related to advanced-stage tumour; induced histone modification; upregulated DNA methyltransferases; inhibited autophagic process; activated β-catenin signalling; aggregated tumour-infiltrating myeloid cells; activated TIGIT and CEACAM1 receptors expressed on immune cells; induced EMT; upregulated ICAM1 and promoted the adhesion of cancer cells to endothelial cells; |
|
|
Genus Bifidobacterium, genus Romboutsia62 |
Increase | Related to left-sided colon cancers; | |
|
Genus Haemophilus, genus Veillonella62 |
Increase | Related to right-sided colon cancers; | |
|
Phylum Ascomycota Class Malasseziomycetes65 |
Increase | — | |
|
Class Saccharomycetes, class Pneumocystidomycetes65 |
Dcrease | — | |
| Species Escherichia coli140,249 | Increase |
Secreted carcinogenic toxins; related to metastasis; |
|
| Species Enteropathogenic Escherichia coli144 | Increase | Disrupted mechanisms of DNA mismatch repair; | |
| Species Hungatella hathewayi168 | — | Upregulated DNA methyltransferases; | |
| Genus Akkermansia194 | — | Increased IL-17 production and B cell infiltration; | |
| Genus Candida11 | Increase | Involved in the downregulation of genes mediating cellular adhesion; | |
| Gastric cancer | Species Helicobacter pylori68,69,76,145,161,162,212–214 | Increase |
Related to early-stage of tumours; disrupted DNA mismatch repair mechanisms; inducted abnormal DNA methylation; activated CEACAM1 on immune cells; upregulated PD-L1 expression; |
|
Genus Prevotella, genus Streptococcus, genus Veillonella, genus Haemophilus genus Neisseria73 |
Increase | — | |
| Genus Helicobacter73,227 | Decrease | Metabolic regulation; | |
| Genus Lactobacillus74,227 | Increase |
Related to tumour progression; metabolic regulation; |
|
|
Phylum Nitrospirae, family Lachnospiraceae, genus Escherichia-Shigella, species Burkholderia fungorum74 |
Increase | Related to tumour progression; | |
|
Genus Oceanobacter, genus Methylobacterium, genus Syntrophomonas75 |
Increase | — | |
|
Species Propionibacterium acnes, species Prevotella copri76 |
Increase | Related to early-stage of tumours; | |
|
Phylum TM7, genus Porphyromonas, genus Neisseria species Streptococcus sinensis77 |
Decrease | Related to benign gastric disease; | |
|
Family Lachnospiraceae, species Lactobacillus coleohominis77 |
Increase | Related to malignant gastric disease; | |
| Epstein-Barr virus70–72 | Increase | Related to DNA hypermethylation; | |
|
Order Clostridium, family Comamonadaceae, genus Moryella, genus Vibro, genus Paludibacter, genus Agrobacterium30 |
Increase | Monitor the risk of gastric cancer development; | |
|
Species Kytococcus sedentarius, species Actinomyces oris163 |
Increase | Inducted abnormal DNA methylation; | |
| Breast cancer |
Genus Pseudomonas, genus Proteus81 |
Increase | — |
| Species Methylobacterium radiotolerans82,83 | Increase | — | |
| Species Sphingomonas yanoikuyae82 | Decrease | — | |
| Genus Methylobacterium84 | Decrease | — | |
| Genus Cladosporium12 | Increase | — | |
| Genus Lactobacillus85,225 | Increase |
Metabolic regulation; enhanced the resistance of tumour cells to flow shear stress; |
|
|
Genus Streptococcus, genus Staphylococcus85 |
Increase | Enhanced the resistance of tumour cells to flow shear stress; | |
| Family Streptococcaceae88 | Increase | — | |
| Genus Bosea88 | Increase | Related to tumour progression; | |
|
Species Escherichia coli, species Staphylococcus epidermis143 |
Increase | Caused double-stranded DNA breaks; | |
|
Genus Fusobacterium, genus Atopobium, genus Hydrogenophaga, genus Gluconacetobacter225 |
Increase | Metabolic regulation; | |
| Species Enterotoxigenic Bacteroides fragilis246 | Increase | Enhanced stemness potential and metastatic progression; | |
| Species Fusobacterium nucleatum33,252 | Increase | Related to metastasis; | |
| Pancreatic cancer | Phylum Proteobacteria91 | Increase | — |
| Species Helicobacter pylori92,93 | Increase | Related to activation of molecular pathways for tumour growth and progression; | |
| Genus Pseudomonas91,94 | Increase | Related to carcinogenesis; | |
| Genus Elizabethkingia91 | Increase | — | |
|
Genus Acinetobacter, genus Sphingopyxis94 |
Increase | Related to carcinogenesis; | |
| Genus Malassezia95 | Increase | — | |
| Oral cancer | Genus Fusobacterium101,112 | Increase | — |
| Species Fusobacterium nucleatum142,229 | Increase |
Caused double-stranded DNA breaks; Promoted GLUT1 upregulation and lactic acid accumulation; |
|
| Species Porphyromonas gingivalis102–105,154,155,244 | Increase |
Related to the formation of DNA adducts or the inhibition of DNA repair enzymes; Related to the expression of EMT-related transcription factors; |
|
| Genus Prevotella106 | Icrease | — | |
| Species Prevotella intermedia107 | Increase | Related to carcinogenesis; | |
| Species Treponema denticola108,109 | Increase | — | |
| Species Streptococcus anginosus110,111,155 | Increase | Related to the formation of DNA adducts or the inhibition of DNA repair enzymes; | |
| Genus Streptococcus112 | Decrease | — | |
|
Species Pseudomonas aeruginosa, Campylobacter sp. Oral taxon 44113 |
Increase | — | |
| Human papillomavirus98 | Increase | — | |
| Epstein-Barr virus99 | Increase | — | |
| Herpes Simplex Virus Type 1100 | Increase | — | |
|
Species Candida albicans, species Candida etchellsii, species Hannaella luteola-like114 |
Increase | — | |
| Head-and-neck squamous cell carcinomas | Genus Parvimonas116 | Increase | — |
| Genus Actinomyces116 | Decrease | — | |
| Human papillomavirus types 16117,134 | Increase | Inhibited the cGas-STING pathway; | |
| Oesophageal cancer | Species Campylobacter conisus189 | Increase | Upregulated PRRs and aggregated IFI16 inflammasome; |
| Nasopharyngeal carcinoma |
Genus Corynebacterium, genus Staphylococcus28 |
Increase | — |
| Epstein-Barr virus119,217 | Increase | Downregulated IDO expression; | |
| Ovarian carcinoma |
Phylum Aquificae, phylum Planctomycetes120 |
Increase | — |
| Phylum Crenarchaeota120 | Decrease | — | |
| Human papillomavirus types 16, 18, and 45121 | Related to advanced-stage tumours; | ||
| Endometrial cancer |
Genus Bacteroides, genus Faecalibacterium122 |
Increase | — |
|
Genus Staphylococcus, genus Blautia, genus Parabacteroides122 |
Decrease | — | |
| Cervical cancer |
Genus Gardnerella, genus Prevotella, genus Streptococcus, genus Atopobium226 |
Increase | Metabolic regulation; |
| Human papillomavirus131,205,208,209 | Increase |
Integrated viral genome into the host chromosome; recruited Treg cells; inhibited cytotoxic T and NK cell activation; upregulated PD-L1 expression; |
|
| Prostatic cancer | Species Cutibacterium acnes123,124 | Increase | — |
| Species Staphylococcus aureus202 | Increase | Recruited Treg cells; | |
| Bladder cancer |
Species Escherichia coli, species Butyrate-producing bacteria SM4/1, a species of Oscillatoria243 |
Increase | Related to the expression of EMT-related genes; |
| Pituitary neuroendocrine tumour |
Order Clostridiales, family Fusobacteriaceae, family Tissierellaceae, family Aerococcaceae, family Corynebacteriaceae, family S24-7, F16125 |
Increase | Related to different clinical phenotypes of tumour; |
| Burkitt’s lymphoma | Epstein-Barr virus126 | Increase | — |
| Hodgkin’s lymphoma | Epstein-Barr virus126 | Increase | — |
| NK cell and T cell lymphomas | Epstein-Barr virus126 | Increase | — |
| Human T-lymphotropic virus type 1139 | Increase | Inhibited DNA repair pathway. | |
| Lymphocytic leukaemia | Human endogenous retroviruses118 | Increase | — |
Lung cancer
As the mucosa is in primary contact with the external environment, the lungs are exposed to microbes and environmental factors and harbour diverse microbes.35 In lung cancer tissues, the prevalence of Modestobacter was higher than that in adjacent normal tissues, while the prevalence of Propionibacterium and Enterobacteriaceae was lower.36 Dohlman et al. found that abundant Blastomyces existed in lung tumours.11 In addition, current smokers had higher intratumoural fungal diversity and abundance of Aspergillus and Agaricomycetes than never smokers.12 The intratumoural microbiota is also related to the histological subtype and tumour stage of lung cancer. Compared to lung adenocarcinoma, lung squamous cell carcinoma microbiota is more diverse. The relative abundances of Acidovorax, Klebsiella, and Anaerococcus in squamous cell carcinoma were elevated, especially in patients with tumour protein p53 (TP53) mutations, where Acidovorax was more abundant.37 In adenocarcinomas, Acinetobacter, Brevundimonas, and Propionibacterium were more abundant.38 Furthermore, Apopa et al. discovered that the phylum Cyanobacteria was more enriched in adenocarcinoma.39 Additionally, it has been found that the genera Veillonella and Megasphaera have a high area under the curve for predicting lung cancer.40
A few studies have aimed to describe the intratumoural micropopulation associated with lung cancer. Most of the samples used in these studies were based on indirect specimens of bronchoalveolar lavage fluid (BALF), sputum, and airway brushing tissue, which could be problematic because the upper and lower respiratory tracts have distinct microbial populations that might result in cross-contamination.41 Therefore, surgical specimens would provide a more precise evaluation of microorganisms in lung cancer.
Liver cancer
Primary liver cancer (PLC) is a malignancy that originates from malignant hepatocellular tumours and precursors such as HCC, the main type of PLC.42 PLC generally progresses from chronic liver diseases, such as viral hepatitis, as ~56% of liver cancers are associated with HBV and 20% with the hepatitis C virus (HCV).43 The interaction between microbiota and PLC has been intensively investigated because of the gut-liver axis.44–47 However, studying microorganisms in liver tumours remains limited. H. pylori and similar species have been identified in patients with HCC liver tissues.48–50 Although in vitro studies have suggested the potential connection between H. pylori and liver cancer development, no evidence has been obtained showing that this species directly contributes to tumorigenesis.51,52 Huang et al. found microbes present in hepatocytes and erythrocytes, and the abundance of Gammaproteobacteria in cancerous tissues was considerably higher than that in normal tissues. In particular, compared with that in non-cirrhosis HCC, Streptococcaceae and Lactococcus were significantly increased in cirrhosis HCC, suggesting that they could be used as marker taxa for cirrhosis HCC.53 Qu et al. demonstrated that the abundance of Enterobacteriaceae was substantially higher in the HCC group, whereas the abundances of Caulobacteraceae and Rickettsiaceae were substantially lower in the combined hepatocellular carcinoma and intrahepatic cholangiocarcinoma (cHCC‐ICC) group.54 Moreover, the abundance of Paraburkholderia fungorum in ICC was higher in paraneocancerous tissues than in cancerous tissues, and the value was inversely correlated with carbohydrate antigen 199 (CA199) levels. The results of in vitro and in vivo experiments further indicated that the fungus had antitumour activity.55 Currently, studies on the intratumoural microorganisms of HCC are limited, and further investigation is needed to identify the significance of other microbiota in HCC.
Colorectal cancer
Fusobacterium and enterotoxigenic Bacteroides fragilis (ETBF) have been demonstrated to be carcinogenic factors in the advancement of colorectal cancer (CRC).56–58 Unlike normal tissues, Lactococcus, Bacteroides, Fusobacterium, Prevotella, and Streptococcus were more abundant in tumour tissues. Pseudomonas and Escherichia-Shigella were substantially enriched in adjacent normal tissues compared to those in tumour tissues.59 Yamamoto et al. also discovered a varied abundance of F. nucleatum in the progression of CRC, with the highest prevalence in stage III/IV tumours.60 However, another study reported that F. nucleatum was not detected in most CRC samples and that there were no substantial differences in the microbiota between tumours and adjacent tissues.61 This may be due to differences in patient selection and techniques used among the studies. In addition, distinctions between left and right colon cancers were detected, where the loadings of Bifidobacterium and Romboutsia were higher in left-sided colon cancers (LSCCs), whereas Haemophilus and Veillonella were higher in right-sided colon cancers (RSCCs).62 This may partially explain the difference in the biological subtypes of CRC between the LSCCs and RSCCs, with increased levels of the “microsatellite unstable/immune” consensus molecular subtype (CMS)1 and the “metabolic” CMS3 identified in RSCCs.63 Regarding fungi, Ascomycota, Glomeromycota, and Basidiomycota were identified as the predominant phyla between adenomas and adjacent tissues,64 and the abundance of Ascomycota was higher in patients with CRC, with an increase in Malasseziomycetes and a decrease in Saccharomycetes and Pneumocystidomycetes.65
Several studies on the CRC microbiome utilised faecal samples due to the simple and non-invasive sample collection procedure. However, intestinal mucosal tissue samples are more suitable for assessing the physiopathology of CRC, and distinct microbiome patterns in mucosal and faecal samples have also been reported.66,67 The composition of CRC-related micropopulations has not yet been unified and requires further research.
Gastric cancer
H. pylori is the most predominant microorganism detected in gastric cancer (GC) and has been implicated in promoting premalignant lesions that can ultimately progress to GC.68,69 EBV was found within malignant epithelial cells in 9% of GC.70 EBV-GC may be the most common type of EBV-associated cancer, highly correlated with cyclin-dependent kinase inhibitor 2 A (CDKN2A) promoter hypermethylation.71,72 Shao et al. found that the microbial diversity in GC cells was considerably greater than that in benign stomach lesions. Specifically, the genera Prevotella, Streptococcus, Veillonella, Haemophilus, and Neisseria were more abundant, whereas Helicobacter was less abundant.73 Moreover, GC tissues have a high load of potentially carcinoma-promoting bacteria, including Lactobacillus, Escherichia-Shigella, Lachnospiraceae, Nitrospirae, and Burkholderia fungorum.74 Peng et al. observed that the Oceanobacter, Methylobacterium, and Syntrophomonas genera were enriched in tumour tissue, and the intratumoural Methylobacterium was considerably correlated with a poor prognosis in GC patients.75 Recent research has demonstrated that as GC progresses, the abundance of H. pylori may gradually reduce, and the microbiota diversity may also change. In the early-stages of GC, the numbers of H. pylori, Propionibacterium acnes, and Prevotella copri were higher than that in non-cancer patients.76 In addition, the microbiome of patients at different histological stages, from gastritis to precancerous lesions to stomach cancer, also showed changes. The abundance of TM7, Porphyromonas, Neisseria, and Streptococcus sinensis decreased with disease progression, while that of Lactobacillus coleohominis and Lachnospiraceae were reversed.77 In addition, a longitudinal prospective study identified a total of six microbial taxonomic features, namely the Moryella genus, Vibro genus, Comamonadaceae family, Paludibacter genus, Agrobacterium genus, and Clostridium order, at baseline that could be used to indicate the risk of future GC development.30
Breast cancer
Breast cancer (BC) is the most widespread malignancy among women.78 The relationship between the microbiota and carcinogenesis has been evaluated to determine its function in the initiation and progression of BC.79,80 Breast tumours have the greatest bacterial diversity and abundance among all tumours.7 Tzeng et al. discovered that the genera Pseudomonas and Proteus were highly enriched in BC tissues.81 Xuan et al. demonstrated that Methylobacterium radiotolerans was increased in tumour tissue, while Sphingomonas yanoikuyae was increased in normal tissue. Moreover, the tumour stage was inversely associated with total bacterial load at the tumour site, which may provide clues for BC diagnosis.82 Another study also revealed an increased abundance of Methylobacterium radiotolerans in the tumour sentinel lymph nodes.83 However, Wang et al. demonstrated a decreased abundance of Methylobacterium in BC tissues.84 Narunsky-Haziza et al. identified that the Cladosporium genus was increased in the BC of patients ≥50 years old.12 The unique intratumoural microbiota present in BC tissues, mainly comprising Lactobacillus, Streptococcus, and Staphylococcus, is a potential factor contributing to tumour metastasis.85 Multiple investigations have found that the microbial community varies according to subtype.86,87 The family Streptococcaceae is more abundant in the triple-negative BC subtype, and the abundance of the genus Bosea increases with the progression of the tumour.88
Pancreatic cancer
Pancreatic cancer (PC) is a malignant cancer characterised by a dismal prognosis; pancreatic ductal adenocarcinoma (PDAC) accounts for most of PC.89 Recent research has revealed the existence of bacteria in the pancreas; PC tissues contain a larger proportion of bacteria than normal pancreatic tissues.90 Proteobacteria are the most prevalent intratumoural microorganisms in PDAC, similar to the normal duodenal microbial composition.91 Studies have suggested that H. pylori colonised PC cells and was associated with the activation of molecular pathways for tumour initiation and development.92,93 However, the subspecies of Helicobacter found in pancreatic and gastroduodenal tissues were different, suggesting that Helicobacter in the pancreas may not have migrated from the gastroduodenum.92 Moreover, the microbes in PDAC are completely distinct from those in normal pancreatic tissues, especially regarding Pseudomonas and Elizabethkingia, which were highly abundant in tumours.91 Basal-like tumours, a highly aggressive PDAC subtype, were found to have distinctive intratumoural microbiota, and increased abundance of Acinetobacter, Pseudomonas, and Sphingopyxis was strongly associated with carcinogenesis.94 In addition, the fungal population of PDAC tumours was found to be significantly enriched with Malassezia.95
Oral cancer
The oral microbiome hosts >750 common oral species.96 The normal oral microbiome mainly comprises aerobes; the percentage of anaerobes increases with the development of oral cancer (OC). Oral squamous cell carcinoma (OSCC) constitutes 90% of epithelial malignancies in the oral cavity.97 Human papillomavirus (HPV) has been recognised as a potential contributor to OSCC, with HPV type 16 being the most significant subtype. Approximately 25–35% of OSCCs are attributed to HPV infection.98 Other oncogenic viruses, such as EBV and Herpes Simplex Virus Type 1 (HSV-1), have also been demonstrated to be associated with OSCC.99,100 F. nucleatum is the natural flora present in oral mucosa that has been implicated in the development of oral malignancies. Nagy et al. observed a higher abundance of Fusobacterium in OSCC tissue than in normal mucosal tissue.101 Porphyromonas gingivalis (P. gingivalis) is another independent and critical risk factor for OC.102,103 Katz et al. detected a significant enrichment of P. gingivalis in gingival squamous cell carcinoma.104 Similarly, Chang et al. revealed that the level of P. gingivalis in OSCC tissue was higher than that in normal tissue; they also found a positive correlation between P. gingivalis infection and advanced-stage, poor differentiation, and lymph node metastasis among OSCC patients.105 Another study revealed that Prevotella were enriched in OSCC tissue.106 In particular, Zhang et al. found that the abundance of Prevotella intermedia was significantly increased in OSCC; functional prediction further suggested that the bacteria were associated with carcinogenesis.107 Moreover, studies have shown that Treponema denticola is closely correlated with OSCC and oropharyngeal squamous cell carcinoma (OPSCC).108,109 The aerobic bacterium Streptococcus has also been found to be associated with OC. Sasaki et al. showed that the levels of Streptococcus anginosus (S. anginosus) were elevated in patients with OC.110 Rai et al. reported similar results.111 Some studies, however, have reported contrasting results. Su et al. found that Fusobacterium was substantially enriched at tumour sites, while Streptococcus showed the opposite results.112 Besides, Pseudomonas aeruginosa and Campylobacter sp. Oral taxon 44 were also abundant in OSCC.113 Moreover, Perera et al. found that Candida albicans, Candida etchellsii, and a Hannaella luteola-like species were relatively abundant in OSCC.114
Additionally, the oral microbiota changed with the progress of OC. Yang et al. demonstrated that as cancer progressed, the prevalence of Fusobacterium increased, while that of Streptococcus, Haemophilus, Porphyromonas, and Actinomyces decreased.115
Other cancers
In addition to the cancers reported above, multiple studies have also shown the presence of microbiota in other tumours. In an investigation of head-and-neck squamous cell carcinomas (HNSCCs), Actinomyces was significantly reduced, while Parvimonas was elevated relative to that in normal tissues.116 Another study demonstrated that HPV 16 was detected in HNSCCs,117 and a significant exclusivity of HPV and driver mutations in TP53, CDKN2A, and telomerase reverse transcriptase was exhibited in the tumour.118 In addition to EBV,119 a recent study found that some bacteria, mainly Corynebacterium and Staphylococcus, existed in nasopharyngeal carcinoma (NPC) tumour tissues, and the total intratumoural bacterial load was negatively correlated with prognosis.28 Among other reproductive system tumours, the microbiota in ovarian cancer tissue consisted of increased Aquificae and Planctomycetes abundance and decreased Crenarchaeota abundance.120 Compared to normal adjacent tissues, ovarian carcinoma contained a higher proportion of HPVs, and high-risk HPV types 16, 18, and 45 were significantly correlated with advanced-stage tumours.121 Bacteroides and Faecalibacterium were particularly associated with endometrial cancer, while Staphylococcus, Blautia, and Parabacteroides were more associated with benign uterine disease.122 Certain bacterial species, especially Cutibacterium acnes, can persist in prostatic tissue specimens.123,124 Intracranial tumours, such as glioblastomas7 and pituitary neuroendocrine tumours (PitNETs),125 were found to contain intratumoural microorganisms, and the abundance of microorganisms in different subtypes of the PitNETs was also different, which Fusobacteriaceae, Tissierellaceae, and Aerococcaceae were substantially enriched in adrenocorticotropic hormone-secreting PitNET (ACTH-PitNET) tissues and Corynebacteriaceae, S24-7, Aerococcaceae, Clostridiales, and F16 were more enriched in growth hormone-secreting PitNET (GH-PitNET) tissues. Moreover, EBV was present in the blood system’s tumour cells, such as Burkitt’s lymphoma, Hodgkin’s lymphoma, and some natural killer (NK) and T cell lymphomas.126 Human endogenous retroviruses (HERVs) have been found in chronic lymphocytic leukaemia, where ERV1 was strongly expressed.118 However, the relationships between intratumoural microbiota and other types of cancers have not been thoroughly studied, and further research is required.
Role of intratumoural microbiota in the development of cancer
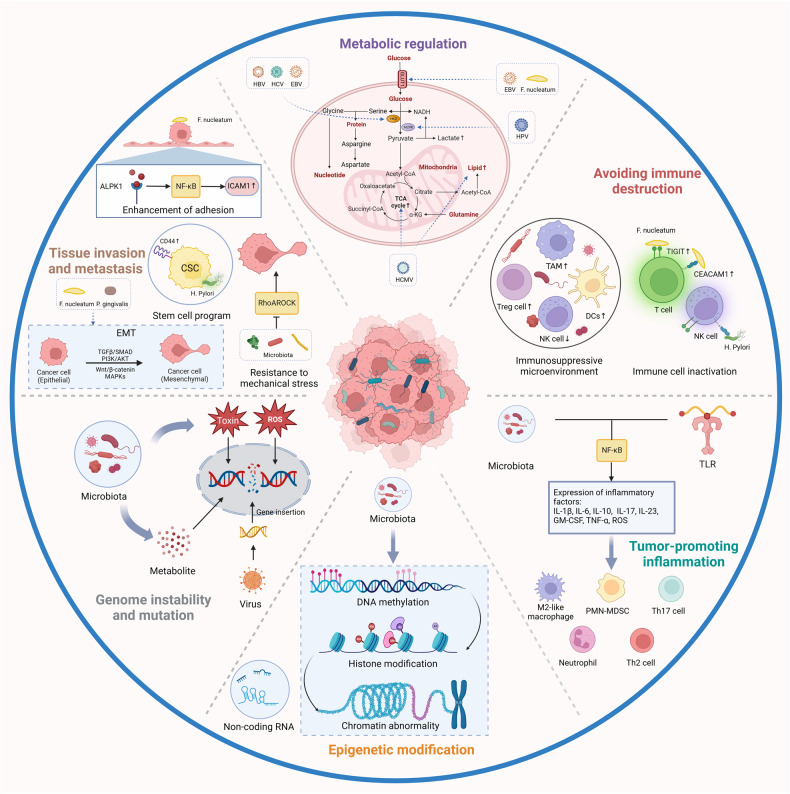
Cancer cells influence disease progression by maintaining proliferation, evading growth inhibition, resisting cell death, enabling replicative immortality, inducing angiogenesis, and activating invasion and metastasis.127,128 Although the potential role of the microbiome in the initiation and advancement of cancer remains elusive, it could be associated with modulating the most relevant tumour-promoting functions between malignant and non-malignant cells. Understanding these mechanisms is crucial for cancer prediction and treatment (Fig. 3).
Fig. 3.
The role of intratumoural microbiota in the development of cancer. The potential effects of the microbiome on the cancer remain elusive. Six major mechanisms have been proposed to explain how the intratumoural microbiota influence the initiation and advancement of cancer, including genome instability and mutation, epigenetic modification, chronic inflammation, immune evasion, metabolic regulation, activation of invasion and metastasis. TAM, tumour-associated macrophage; HK2, hexokinase 2; TCA, tricarboxylic acid. Created with BioRender.com
Genome instability and mutation
The induction of genomic instability and mutation is one of the carcinogenic mechanisms of the microbiome. More than 10% of human malignancies are primarily caused by oncoviruses.129 Various studies have suggested that oncoviruses cause cancer by integrating the viral genome into the host chromosome and triggering genetic mutations, such as HPVs in cervical, head-and-neck, and several other cancers, and HBV in liver cancer.130–132 Integrated deoxyribonucleic acid (DNA) also leads to viral oncoprotein production, which modulates host signalling pathways and alters genes’ and ribonucleic acid (RNA) expression. In mouse models, the HPV E7 oncoprotein directly inhibits the cyclic guanosine monophosphate (MP)-adenosine MP synthase (cGas)-stimulator of interferon genes (STING) pathway and specifically reduces the expression of genes encoding type I interferon and pro-inflammatory factors, thereby driving immune escape in multiple HPV-related tumours.133,134 Moreover, EBV and Kaposi sarcoma-associated herpesvirus (KSHV) oncoproteins can upregulate oncogenic cellular proteins and microRNAs in mouse, downregulate tumour suppressors, and trigger signalling pathways, such as the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway, to drive the proliferation and transformation of B cells and endothelial cells.135–138 Human T-lymphotropic virus type 1 (HTLV-1), the retrovirus that triggers adult T cell leukaemia, inhibits the DNA repair pathway through the HTLV-1 Tax protein, leading to genome instability and the accumulation of carcinogenic mutations.139
Certain carcinogenic bacteria, such as pks+ Escherichia coli (E. coli) and ETBF, encode and secrete carcinogenic toxins that induce DNA damage, which results in faster tumour onset and greater mortality.140 Moreover, FadA, a key adhesin secreted by F. nucleatum, promotes E-cadherin/β-catenin activation to upregulate checkpoint kinase 2 (CHK2), causing DNA damage in mouse CRC cells.141 F. nucleatum infection promotes OSCC by causing Ku70/p53 pathway-dependent DNA double-strand breaks (DSBs).142 E. coli and Staphylococcus epidermis isolated from breast tumour tissue cause DSBs in HeLa cells.143 In addition, EspF-expressing enteropathogenic E. coli and H. pylori may interfere with DNA mismatch repair mechanisms, thereby aggravating genome instability and promoting tumorigenesis.144,145 However, further studies are required to identify the direct involvement of bacterial virulence factors (VirFs) in cancer induction. The fundamental role of these VirFs could be to induce DNA damage response and subsequently activate the immune system, which results in pro-inflammatory outcomes and forms a microenvironment conducive to cancer.146 Microbial activities can also elicit the generation of reactive oxygen species (ROS), hydrogen sulphide, and superoxide dismutase.147–149 ETBF toxin increases the expression of spermine oxidase (SMO) in HT29/c1 and T84 colonic epithelial cells, triggering the SMO-dependent production of ROS and activation of γ-H2A, which causes DNA damage.56
In addition, many microbial metabolites affect tumour development by promoting DNA damage. BC tissues harbour increased levels of β-glucuronidase, which has been identified as a carcinogenic enzyme.150,151 β-glucuronidase may release reactive intermediates from 2-amino-3-methylimidazo[4,5-f]quinoline to cause DNA damage in rats.152 S. anginosus and P. gingivalis can convert ethanol into acetaldehyde, resulting in the formation of DNA adducts or the inhibition of DNA repair enzymes, which might cause DNA damage and oral carcinogenesis.153–155
Epigenetic modification
Epigenetic pathways also play an essential part in oncogenesis by aberrantly silencing tumour suppressor genes (TSGs) and activating oncogenes.156 According to reports, bacterial infections survive, replicate, and evade destruction by the host immune system by regulating the host epigenome.157–160 Induction of abnormal DNA methylation is the main pathway for H. pylori infection to induce gastric adenocarcinoma.161,162 Other gastric microbiota, such as Kytococcus sedentarius and Actinomyces oris, are also involved in this mechanism, which promotes the occurrence of gastric adenocarcinoma and metastasis and affects its prognosis.163 A study reported that the significantly rich microorganisms in the high-cell subtype thyroid cancer patients were associated with higher tumour suppressor gene methylation.164 A recent study compared intratumoural microbiome and DNA methylation profiles of HCC tissue and normal liver tissue, and correlation analysis showed that 10 metabolome-related microbiome groups were closely related to 25 methylation-related differentially expressed genes.165 The abundance of intratumoural F. nucleatum was linked to increased infiltration of macrophages and promoter CpG island hypermethylation of CDKN2A in CRC patients.166 However, the molecular mechanism underlying the host epigenetic changes induced by intratumoural microbiota has not yet been fully characterised. On the one hand, some studies have reported that microorganisms directly regulate host epigenetic modifications. Liu et al. reported that in vivo and in vitro H. pylori infection promoted guanine nucleotide-binding protein subunits β-44 (GNB4) demethylation by activating NF-κB to upregulate TET1, inducing the carcinogenic pathway.161 Besides, F. nucleatum upregulates the transcription of long non-coding RNA (lncRNA) enolase1-intronic transcript 1 (ENO1-IT1) through transcription factor SP1. Elevated ENO1-IT instructs KAT7 histone acetyltransferase to change the histone decorator pattern on its target genes, enhancing CRC glycolysis and tumorigenesis.167 F. nucleatum and Hungatella hathewayi could mediate TSG promoter hypermethylation by upregulating DNA methyltransferases in CRC.168 On the other hand, it was reported that microorganisms synthesised and metabolised abundant compounds that serve as epigenetic substrates and cofactors or regulate epigenetic enzymes, indirectly affecting host epigenetic modifications. For example, folate and other B vitamins (B2, B12) are the primary substrates for DNA and histone methylation.169 Microorganism-derived short-chain fatty acids (SCFAs) trigger genomic epigenetic changes by affecting the activities of histone acetylase and histone deacetylase.170,171 A recent study found that specific bacteria produce methionine in lung cancer patients,172 and they behave as the main methyl donors for nucleic acid and protein methylation, making epigenetic reprogramming of host cells possible.173 Moreover, microorganisms may activate other pathways that indirectly alter the epigenetics of host cells. Koi et al. found that chronic F. nucleatum infection produces ROS, which causes DNA damage and triggers MSH2/MSH6-dependent repair, resulting in DNA hypermethylation.174 Previous reviews have described the changes in various non-coding RNAs induced by bacterial infection and their role in modifying chromatin structure.175–177 However, few studies have focused on intracellular microorganisms in this area. Epigenetic regulation crosstalk between the virus and the host also occurs. Pietropaolo et al. reviewed the research progress on seven viruses causing cancer through epigenetic changes in host cells.178 Another review summarised several mechanisms by which HBV induces hepatocarcinogenesis by inducing m6A modification of RNA, including virus replication, immune escape, and carcinogenesis, indicating a complex interaction between the microbiome and host.179
In conclusion, multiple studies have demonstrated that intratumoural microorganisms (mainly viruses and bacteria) could directly or indirectly regulate host epigenetic modifications, including DNA modification, histone modification, RNA modification, and non-coding RNA. However, the molecular mechanisms underlying the host epigenetic changes induced by intratumoural microorganisms must be further explored.
Chronic inflammation
Persistent chronic inflammation is correlated with the advancement of most cancer types.180 The intratumoural microbiome can activate inflammatory signalling pathways and cascades by interacting with pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), in the TME.
TLRs are expressed on a variety of immune cells, including macrophages, dendritic cells, B cells, certain types of T cells, as well as non-immune cells like fibroblasts and epithelial cells.181 Fusobacterium recognised by TLR4 enhanced the interleukin (IL)-6/phospho-signal transducer and activator of transcription 3 (p-STAT3)/c-MYC signalling pathway, resulting in M2-like macrophage polarisation and mouse CRC progression.182 Pushalkar et al. showed that the PDAC microbiota inhibits the type 1 T-helper (Th1) polarisation of cluster of differentiation (CD)4 + T cells and M1-like macrophage differentiation by activating TLR, thus generating a tolerant immune programme.91 Moreover, in a mouse colitis-associated tumorigenic model, Yang et al. found that microbial-derived lipopolysaccharide /TLR4 mediates the chemokine-dependent recruitment of monocyte-like macrophages to promote IL-1β production, further promoting Th17 cell expansion. This process may enhance intestinal permeability and allow the excessive release of microbial products, thus promoting M2-like macrophage differentiation, generating a positive feedback loop to attract immunosuppressive cells and forming a tolerogenic microenvironment.183,184 Another mouse model showed that microbial products elicited by tumorous epithelial barrier disruption activate tumour-infiltrating inflammatory dendritic cells (DCs), thereby inducing γδT17 cells polarisation, which can release a lot of IL-17, IL-8, granulocyte-macrophage colony-stimulating factor, tumour necrosis factor alpha (TNF-α), and other cytokines to promote inflammation. These cytokines further attract the accumulation of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs), thus transforming the inflammatory microenvironment into an immunosuppressor microenvironment and promoting the progression of CRC.185 Moreover, ETBF toxin triggers a STAT3–NF-κB-dependent pro-inflammatory signalling cascade to release cytokines like IL-17 and IL-23, which has been demonstrated to attract pro-tumoural myeloid cells and promote distal colon tumorigenesis.186 Park et al. demonstrated that F. nucleatum and Aggregatibacter actinomycetemcomitans activate TLR2 and TLR4 and downstream NF-κB in bone marrow-derived macrophages to stimulate IL-6 production in mouse.187 Nontypeable Haemophilus influenzae can stimulate mouse epithelial cells to release IL-17C, recruit neutrophils into inflamed tissues, and promote lung tumour growth.188 In addition, macrophage infection with Campylobacter conisus (C. conisus) leads to the upregulation of PRRs and aggregation of interferon-inducible protein 16 inflammasome, which may be a related pro-inflammatory mechanism underlying the ability of C. conisus to cause oesophageal cancer.189 Moreover, F. nucleatum-mediated inhibition of autophagy in colon cancer epithelial cells promotes ROS accumulation, triggering the production of pro-inflammatory cytokines, such as IL-8, IL-1β, and TNF-α.190
In addition, microbes can also induce macropinocytosis, an endocytic activity that cells use for antigen capture and delivery, to activate inflammation.191 A mouse study found that the Wnt pathway activates macrophage proliferation and stimulates macrophage uptake of bacteria and their products into the colon. However, transcriptional targets for macropinocytosis activation by the Wnt pathway remain unknown; some potential candidates for Wnt-dependent transcription factors involve RAB5, PDK1, and PAK1.192,193 The crosstalk between intratumoural microorganisms and polymorphonuclear neutrophils (PMNS) can also promote cancer development. The loss of neutrophils was found to promote the enrichment of Akkermansia in a mouse model of CRC. The intratumoural bacteria can boost IL-17 production and intratumoural B cell infiltration, thereby promoting tumour growth and cancer progression.194
The inflammatory reaction is accompanied by the immune response. Stress and tissue damage from microbial infection recruit immune cells to promote inflammation and further activate various pro-tumoural inflammatory factors.195 The chronic inflammatory microenvironment eventually transforms into an immunosuppressive microenvironment to promote tumour progression and inhibit antitumour immunity.196 Moreover, inflammatory cells at sites of infection can also produce ROS to induce DNA damage. The latter can also amplify inflammatory responses, leading to increased DNA damage.197 Therefore, crosstalk between various pathogenic mechanisms may contribute to cancer development.
Immune evasion
The interaction between microorganisms and their host is essential for sustaining immune homoeostasis.198 Intratumoural microbes can evade the immune response and affect tumorigenesis by promoting an immunosuppressive microenvironment and immune cell inactivation.
F. nucleatum regulates the tumour immune microenvironment by selectively aggregating tumour-infiltrating myeloid cells, including CD11b+ myeloid cells, MDSCs, tumour-associated macrophages, classical myeloid DCs, and CD103+ regulatory DCs, thereby potentiating tumorigenesis.199 Besides this, commensal bacteria triggered Myd88-dependent IL-1β and IL-23 production, driving the activation of Vγ6 + Vδ1 + γδ T cells and the subsequent release of IL-17 and other cytotoxic effectors to promote an inflammatory immunosuppressive environment and lung tumour cell proliferation in mouse.200 Another mouse study revealed that Pasteurella was positively correlated with cytotoxic CD8+ tumour-infiltrating lymphocytes (TILs) and negatively correlated with M2-like macrophages, whereas Coriobacteriaceae was positively associated with M2-like macrophages and negatively associated with CD8+ cells. All these immune responses influence the initiation and development of lung tumours.201 Certain microorganisms, such as Staphylococcus aureus, HBV, and HCV, potentially promote the progression of prostate and liver cancers by promoting the immunosuppression mediated by T regulatory cells (Tregs).202–204 Moreover, a large number of Tregs have been observed in HPV-induced cervical lesions, and the level of Tregs is associated with the severity of the disease, suggesting that Tregs may be involved in the interference of anti-HPV immune response.205 Furthermore, in both in vivo and in vitro trials, the fungal community of PDAC tissue stimulated the expression of cancer-cell-specific IL-33, which leads to the recruitment and activation of Th2 cells and innate lymphoid cells 2, consequently promoting tumour progression.206
As noted previously, intratumoural microbiota can also evade immune responses by promoting the inactivation of immune cells. In a mouse model, the PC microbiome promotes suppressive M1 macrophage differentiation via differentially activating selective TLR to induce T cell anergy.91 Elevated lung SCFA levels from lower airway anaerobic bacteria may hinder the production of interferon gamma (IFN-γ) by CD4+ and CD8 + T cells and cause effector T cell depletion, which promotes tumour growth.207 HPV downregulates the antigen-presenting pathway through its gene expression programme to inhibit cytotoxic T and NK cell activation, thereby increasing virus replication and transmission and promoting malignant transformation in human cervical cancer cells.208 Another in vitro study showed that HPV E7 in cervical cancer directly upregulated intratumoural surface programmed death-ligand 1 (PD-L1) and inhibited cytotoxic T cell function.209 Moreover, F. nucleatum protein could bind and activate T Cell immunoreceptor with immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domains (TIGIT) and carcinoembryonic antigen cell adhesion molecule 1 (CEACAM1) receptors express on human NK cells and other lymphocytes, inhibiting antitumour immune cell function in CRC.210,211 Similarly, the HopQ outer-membrane adhesin of H. pylori interacts with CEACAM1 to inactivate immune cells and mediate the migration of VirF cytotoxin-associated gene A (CagA) into host cells and production of IL-8, thereby promoting GC progression.212,213 In addition, CagA can stimulate PD-L1 expression in gastric epithelial cells, creating premalignant lesions progressing to GC.214 Further studies have shown that the process might be mediated by the Sonic Hedgehog signalling pathway.215 EBV infection of B cells and NPC cell lines can induce the downregulation of indoleamine 2, 3-dioxygenase (IDO), resulting in T cell surveillance inactivation.216,217
However, many studies have demonstrated that intratumoural microbiota can mediate immune activation and produce antitumour immunity. HPV was associated with massive infiltration of IFN-γ + CD8 + T cells and IL-17 + CD8 + T cells in HNSCC, which might play a key role in the significantly better response to immunotherapy in HPV-positive patients.218 The intestinal microbe Bifidobacterium has been shown to preferentially colonise in tumour sites and enhance STING/IFN-I signalling in tumour-infiltrating DCs, thereby promoting T cell-dependent antitumour responses in mouse.31 Another mouse model of CRC showed that colonisation with Helicobacter hepaticus correlates with increased infiltration of CD11c+ myeloid cells, T and B cells, leading to reduced tumour burden.219 Moreover, Lactobacillus plantarum-derived indole-3-lactic acid increased IL-12a production of DCs by promoting H3K27ac binding in the IL-12a enhancer region, therefore initiating the CD8 + T cell immunity against tumour growth.220 Another mouse experiment found that intratumoural Lactobacillus reuteri (Lr) releases dietary tryptophan catabolite indole-3-aldehyde to promote IFN-γ-producing CD8 + T cells, thereby enhancing immune checkpoint inhibitors (ICIs).221 The Clostridiales-derived metabolite trimethylamine N-oxide activates the endoplasmic reticulum stress kinase PERK, which causes gasdermin E-mediated pyroptosis in tumour cells and promotes CD8 + T cell-mediated antitumour immunity in BC in vivo.222 The metabolite inosine produced by the bacterium Bifidobacterium pseudolongum in a mouse model of CRC induces the expression of Th1-regulating genes in CD4 + T cells, thus promoting antitumour immunotherapy.223
Overall, microbiota and their derived metabolites are also potential therapeutic targets for supporting immunotherapy, and their effects are context-dependent and require further clarification prior to clinical translation.
Metabolic regulation
Alterations in human metabolism caused by the microbiome may lead to various metabolic diseases and cancers.224 Hieken et al. reported that microbiota in benign breast disease tissue was associated with increased cysteine and methionine metabolism, glycosyltransferases, and fatty acid biosynthesis. By comparison, microbiota in BC tissue, including Fusobacterium, Atopobium, Hydrogenophaga, Gluconacetobacter, and Lactobacillus, reduce inositol phosphate metabolism.225 In cervical cancer, non-Lactobacillus dominated communities, comprising Gardnerella, Prevotella, Streptococcus, and Atopobium, affected amino acid and nucleotide metabolism.226 Dai et al. discovered that the relative level of carbohydrates, carbohydrate conjugates, amino acids, glycerophospholipids, and nucleosides in gastric tumour tissues was higher than that in non-tumour tissues through untargeted metabolomic analysis; subsequent analysis indicated that Helicobacter and Lactobacillus exhibited negative and positive correlations, respectively, with the majority of differential metabolites in the amino acids, carbohydrates, nucleosides, nucleotides, and glycerophospholipids classes.227 Yost et al. showed that the microbiota in patients with OSCC was associated with the upregulation of the activities of iron ion transport-related enzymes, tryptophanase, glutamate dehydrogenase, starch synthase, and superoxide dismutase.228 Moreover, Sun et al. found that F. nucleatum promoted glucose transporter 1 (GLUT1) upregulation and lactic acid accumulation by triggering GalNAc-Autophagy-TBC1D5 signalling, leading to OSCC progression.229 In vitro experiments showed that the expression of Merkel Cell Polyomavirus (MCPyV) oncoprotein can increase the expression of glycolytic genes, including the monocarboxylate lactate transporter SLC16A1 (MCT1), and induce aerobic glycolysis, which is typically characteristic of malignant and rapidly proliferating tumour cells.230 KSHV infection has been demonstrated to induce stable glycolysis via increasing HIF1α expression and activating the PI3K/Akt/mTOR signalling pathway.231–233 Metabolic flux experiments confirmed that human cytomegalovirus (HCMV) markedly upregulated the glycolytic pathway, tricarboxylic acid cycle, and fatty acid biosynthesis pathway in infected cells.234 In addition, HPV E7 protein interacts with and accumulates the dimeric form of M2 type pyruvate kinase (M2PK), a low-activity form that has been found to be upregulated in multiple tumours.235 Other cancer-causing viruses, such as EBV, HBV, and HCV, promote cancer by targeting transcription factors, oncogenes, and tumour suppressors to regulate metabolic enzymes and signalling pathways.236 However, many non-carcinogenic viral infections have similar metabolic alterations. This suggests that metabolic changes induced by tumour viruses may not be sufficient to cause carcinogenic effects. Nevertheless, in the microenvironment of hypoxia, inflammation and immunosuppression, the presence of pro-tumoural metabolism may potentially promote virus-induced cancerisation.
Activating invasion and metastasis
Intratumoural microorganisms can alter the internal features of oncocytes and their external microenvironment to promote cancer metastasis.237 On the one hand, they directly modulate cancer cells to cope with an unfavourable environment.238,239
The epithelial-mesenchymal transition (EMT) process imparts the transition of carcinoma cells with a metastatic mesenchymal phenotype via the TGFβ/SMAD, PI3K/AKT, Wnt/β-catenin, and MAPKs signalling pathways, which drives the invasion and spread of carcinoma cells.240 Multiple investigations have demonstrated the association between microorganisms and EMT.241 A novel virulence protein of F. nucleatum, Fn-Dps, induces EMT by upregulating chemokines CCL2/CCL7, thus promoting the invasion and metastasis of mouse CRC cells.242 Li et al. discovered that E. coli, Butyrate-producing bacteria SM4/1, and a species of Oscillatoria were correlated with the production of EMT-related genes in bladder cancer, including E-cadherin, vimentin, SNAI2, SNAI3, and TWIST1.243 In an in vitro study, P. gingivalis was found to increase the expression of the EMT-associated transcription factors Slug (SNAI2), Snail, and Zeb1 as well as the levels of phosphorylated glycogen synthase kinase-3 beta, an important EMT regulator, thereby promoting the migration of oral epithelial cells.244 The presence of Candida in advanced metastatic colon tumours might play a role in the downregulation of genes mediating cellular adhesion, including PTK2B, CDKN2C, and NET1, thereby leading to metastasis.11 The microbiota can affect EMT and enhance the expression of cancer stem cell (CSC) markers. Bessède et al. reported that infection with H. Pylori CagA-positive strain triggers EMT-like changes and high expression of CD44 in vitro, a known gastric CSC marker, leading to an enhanced ability of cells to migrate, invade, and form tumour spheres.245 In a mouse experiment, ETBF toxin induces downstream β-catenin nuclear localisation by cleavage of E-cadherin in BC and subsequently enhances stemness potential, tumour growth, and metastatic progression.246 Staphylococcus, Lactobacillus, and Streptococcus were highly abundant in a mouse spontaneous breast tumour model and could restrain the RhoAROCK signalling pathway to remodel the actin cell skeleton, thereby improving the resistance of tumour cells to flow shear stress (FSS) and promoting tumour metastasis.85 The adhesion of tumour cells to endothelial cells in the bloodstream is another key stage of invasion and metastasis.247 F. nucleatum mediated a novel PRR, ALPK1, that triggered the NF-κB signalling pathway and upregulated ICAM1, thereby boosting CRC cell adhesion to endothelial cells and metastasis.248
Further, the intratumoural microbiota forms a microenvironment conducive to cancer metastasis. In a mouse model of CRC, tumour-resident E. coli disrupted the gut vascular barrier through the VirF, promoting the spread of bacteria to the liver and the recruitment of metastatic cells.249 Furthermore, F. nucleatum colonises BC and suppresses the accumulation of tumour-infiltrating T cells through the abundant Gal-GalNAc on tumour cells, thereby facilitating tumour metastasis in mouse.33 Many studies have shown that extracellular vesicles (EVs) trigger pro-inflammatory signalling and activate immunosuppression by modulating communication between cancer cells and their surrounding microenvironment as well as distal organ cells, thereby establishing a pre-metastatic niche for tumour metastasis.250,251 A recent work indicated that F. nucleatum-derived EVs in BC tissue enhance tumour cell migration and invasion via TLR4.252 However, few studies have investigated whether intratumoural microbiota-derived EVs play a key role in promoting metastasis or fully described the mechanisms involved, which may be a worthy direction for future research.
Prognostic role of intratumoural microbiota
Microbiota composition differs significantly between tumours and healthy tissues, as well as among distinct tumour stages, gene mutations, and distant tumour metastasis, making it a potential prognostic tool.8 As our understanding of the influence of intratumoural microorganisms on tumorigenesis deepens, applying these profiles in precision oncology becomes more likely. Here, we briefly summarise the application of various intratumoural microbiota to predict cancer survival and therapeutic efficacy (Table 2).
Table 2.
Prognostic role of intratumoural microbiota
| Cancer types | Pathological type | Stage | Treatment | Microorganism | Outcomes | Survival |
|---|---|---|---|---|---|---|
| Oesophageal cancer |
Squamous cell carcinoma, adenocarcinoma, and others |
— | Neoadjuvant chemotherapy/ chemoradiotherapy, surgery | Species Fusobacterium nucleatum253 | Risk factor | OS |
| Squamous cell carcinoma | All stages | Neoadjuvant chemotherapy/ chemoradiotherapy, surgery | Species Fusobacterium nucleatum269 | Risk factor | PFS | |
| Lung cancer | Non-small cell lung cancer | Stage II | Surgery |
Order Actinomycetales, order Pseudomonadales261 |
Risk factor | DFS |
| Non-small cell lung cancer | Stage III, IV | Surgery/ biopsy, targeted therapy/ chemotherapy | Speices Haemophilus parainfluenzae262 | Risk factor | OS | |
|
Species Serratia marcescens, species Acinetobacter jungii, species Streptococcus constellation262 |
Protective factor | OS | ||||
| Non-small cell lung cancer | All stages | — |
Genus Thermus, Genus Legionella263 |
Risk factor | — | |
| Non-small cell lung cancer | Stage IIIB, IV | Chemotherapy, immunotherapy | Species Helicobacter pylori274 | Risk factor | OS, PFS | |
| Liver cancer | Hepatocellular carcinoma, intrahepatic cholangiocarcinoma, combined HCC and iCCA | — | Surgery | Family Pseudomonadaceae54 | Protective factor | OS |
| Colorectal cancer | — | All stages | Surgery | Species Fusobacterium nucleatum254 | Risk factor | OS |
| — | All stages | Surgery | Genus Fusobacterium, genus Granulicatella, genus Gemella260 | Protective factor | DFS | |
| — | All stages | Biopsy | Species Porphyromonas gingivalis265 | Risk factor | OS | |
| — | Stage IV | — | Genus Candida11 | Protective factor | — | |
| Genus Saccharomyces11 | Risk factor | — | ||||
| Gastric Cancer | — | All stages | Surgery | Species Fusobacterium nucleatum255 | Risk factor | OS |
| — | All stages | Surgery, chemotherapy | Species Helicobacter pylori270 | Protective factor | OS | |
| — | — | Surgery | Species Helicobacter pylori271 | Protective factor | OS | |
| — | Stage II, III | Surgery, chemotherapy | Species Helicobacter pylori272 | Protective factor | OS, DFS | |
| Adenocarcinoma, squamous carcinoma, carcinoid carcinoma | Stage III, IV | Immunotherapy | Species Helicobacter pylori275 | Risk factor | PFS | |
| Pancreatic cancer | Pancreatic ductal adenocarcinoma | All stages | Surgery, chemotherapy | Genus Fusobacterium257 | Risk factor | OS |
| Pancreatic ductal adenocarcinoma | Stage I, II | Surgery, neoadjuvant chemoradiotherapy |
Genus Pseudoxanthomonas, genus Streptomyces, genus Saccharopolyspora, genus Bacillus clausii264 |
Protective factor | OS | |
| Melanoma | — | — | Immunotherapy | Genus Clostridium7 | Protective factor | — |
| Species Gardnerella vaginalis7 | Risk factor | — | ||||
| — | Stage III, IV | Immunotherapy | Species Helicobacter pylori273 | Risk factor | OS | |
| — | — | Immunotherapy | Genus Cladosporium12 | Risk factor | — | |
| Oral cancer | Oral squamous cell carcinoma | All stages | Surgery | Species Fusobacterium nucleatum258 | Protective factor | OS |
| Anal Squamous Cell Carcinoma | — | All stages | Radiotherapy/chemoradiotherapy, surgery | Species Fusobacterium nucleatum259 | Protective factor | OS |
| Vulvar squamous cell carcinoma | — | Stage I, II, and III | Surgery, chemotherapy/ radiotherapy |
Species Fusobacterium nucleatum, species Pseudomonas aeruginosa256 |
Risk factor | PFS |
| Ovarian cancer | — | — | Surgery | Genus Phaeosphaeriaceae12 | Risk factor | PFS |
| Kidney cancer | — | — | — | Human endogenous retrovirus118 | Risk factor | OS |
The abundance of F. nucleatum is correlated with the short survival of many cancers.253–255 F. nucleatum and Pseudomonas aeruginosa have been identified as tumour-promoting bacteria linked to poor outcomes in vulvar squamous cell carcinoma patients.256 The colonisation of Fusobacterium species in patients with PDAC is also associated with short survival.257 However, in some cases, F. nucleatum was unexpectedly found to be positively correlated with survival, such as in OSCC258 and anal squamous cell carcinoma.259 Moreover, Alexander et al. recently found that a cluster of microbiota that included Fusobacterium, Granulicatella, and Gemella independently predicted higher disease-free survival (DFS) in patients after CRC resection,260 despite previous studies linking high F. nucleatum abundance to poor prognosis of CRC.254 This paradox may be due to the enhanced immune response triggered by an increased abundance of the microbial cluster, potentially leading to persistent immune memory against CRC post-surgery and vigilant monitoring for recurrence and metastasis. Therefore, the contribution of microorganisms to prognosis should be considered in the context of different organ tumours and treatment modalities.
In addition to F. nucleatum, other microbiota associations with prognosis have been identified. For instance, a higher burden of the Actinomycetales and Pseudomonadales orders was related to lower DFS in stage II non-small cell lung cancer (NSCLC) tumour tissue.261 Analysis of the microbiota in first-line treatment of NSCLC samples revealed that the prevalence of certain bacteria, including Haemophilus parainfluenzae, Serratia marcescens, Acinetobacter jungii, and Streptococcus constellation, effectively predicted 2-year survival.262 Moreover, Thermus is more prevalent in advanced-stage patients, whereas Legionella is more abundant in metastatic patients.263 Conversely, the abundance of Pseudomonadaceae, which have antitumour effects, was reduced in tumour tissues and linearly related to the prognosis of PLC patients.54 Riquelme et al. discovered that the intratumoural microbiota, including Pseudoxanthomonas, Streptomyces, Saccharopolyspora, and Bacillus clausii, could be utilised as an ideal combination to predict the long-term survival of PC.264 Patients with P. gingivalis infection were found to exhibit substantial reduction in cancer-specific survival in CRC.265 The Candida-to-Saccharomyces ratio was positively correlated with the stage of CRC.11 In ovarian cancer, patients with intratumoural Phaeosphaeriaceae, or related Phaeosphaeria genus, had substantially decreased progression-free survival (PFS).12 In addition, high HERV expression in kidney cancer was associated with poorer survival.118 A recent prospective analysis demonstrated the prognostic significance of the measurable microbiome in soft tissue sarcoma, especially intratumoural viral microbiome, which is associated with higher NK cell infiltration and improved clinical outcomes.266 Beyond the individual roles of certain microorganisms, microbial clustering or combinations demonstrate unique prognostic value. Sun et al. identified two hepatotypes based on the microbial profile clustering, representing independent prognostic factors in patients with resected HCC.267 Song et al. developed a microbiome-related score model based on 27 microorganisms that acted as an independent prognostic factor for HCC patients.268
Specific intracellular microorganisms have also been related to immunotherapy efficacy across various cancers. A higher abundance of F. nucleatum is correlated with the reduced efficacy of neoadjuvant chemotherapy in oesophageal squamous cell carcinoma. Targeting this bacterium with antibiotics may potentially enhance the curative effect in oesophageal squamous cell carcinoma patients.269 In patients with metastatic melanoma receiving immunotherapy, Clostridium is enriched in tumours that responded to ICIs, while Gardnerella vaginalis is enriched in the tumours of non-responders.7 Interestingly, previous research suggested that H. pylori infection is correlated with a favourable outcome in GC patients.270–272 However, recent studies have indicated that the presence of H. pylori may affect the efficacy of ICIs in melanoma,273 NSCLC,274 and advanced GC,275 introducing a new paradox regarding the varied effects of the same microorganism on cancer occurrence, prognosis, and treatment efficacy. Fungi also play a role in immunotherapy response in metastatic melanoma, with the Cladosporium genus significantly enriched in non-responders.12 Therefore, further understanding of whether intracellular microbiota could interfere with drug efficacy is essential as well as exploring whether targeted microbiota therapy represents an effective strategy to improve tumour efficacy and patient prognosis.
Prospective microbiome studies have aided in cancer diagnosis and prognosis. Although the microbiome as a prognostic factor requires further research and validation, if its predictive and prognostic capabilities are confirmed, this biomarker could significantly advance the goal of precision medicine for patients with cancer. Unlike intestinal and blood microbiota, intratumoural microbiota is in close proximity to tumour cells, allowing for a more accurate reflection of the actual state of the tumour. However, given that obtaining tissue samples are invasive and difficult, many current studies often use alternative specimens, such as sputum, BALF, and airway brushing tissue, to screen for lung cancer; however, they may introduce certain errors. With the rapid advances in technology and bioinformatics approaches, combining microbiome-based blood diagnostics and imaging evidence of their spatial distribution may improve cancer detection and prognosis. Furthermore, integrating multiple-omics data through artificial intelligence with other emerging sampling techniques, such as ingestible capsules for microbiome sampling, will enable further precision diagnosis and prognostic strategies.276–278
Nevertheless, challenges still exist regarding environmental pollution, low microbial biomass, and antibiotic perturbation. Moreover, most research has focused on advanced tumours, highlighting the need for future work to focus on detecting precancerous lesions or early-stage tumours to achieve early diagnosis and treatment.
Applications of intratumoural microbiota in cancer therapy
Rapid identification and treatment achieve better outcomes in patients with cancer; treatment strategies vary depending on the type of cancer. Different cancer subtypes, mutation states, and degrees of invasion need specific treatment for optimal efficacy, making individualised treatment particularly important.279 Some specific microorganisms have been reported as direct factors responsible for the development of cancer. Certain medications have been used clinically to eradicate microorganisms to prevent cancer as much as possible and to assist in the treatment of cancer; these include the use of quadruple antibiotics to prevent and treat H. pylori-derived GC,280–282 direct antiviral drugs (DAAs) against HCV,283,284 as well as vaccination against HPV and HBV to prevent cervical, head-and-neck, and liver cancer.285,286 Due to the significance of the microbiome in carcinogenesis, targeting intratumoural pathogenic microbiota may be beneficial for precise cancer therapy and prevention of recurrence.
Antibiotics
Gamma-Proteobacteria present in PDAC produces the bacterial enzyme cytidine deaminase (CDD) to metabolise the chemotherapy drug gemcitabine (2’, 2’-difluorodeoxycytidine) into its ineffective form (2’, 2’-difluorodeoxyuridine), thereby reducing its efficacy and leading to drug resistance in PC.90 F. nucleatum is abundant in CRC chemotherapy patients’ tissues, reduces CRC apoptosis, and causes CRC resistance to Oxaliplatin and 5-fluorouracil.287 Multiple retrospective investigations have found that antibiotic therapy improves cancer patients’ survival (Fig. 4a). For example, patients treated with gemcitabine-based and 5-FU-based chemotherapy respectively, were found to have significantly prolonged PFS after antibiotic therapy.288 In addition, metronidazole reduced the amount of Fusobacterium, reproduction of carcinoma cells, and development of tumours in a mouse model of CRC.289 A recent study found that aerosolised and oral absorbable antibiotics can reduce mouse mammary tumour growth, and it also showed that paclitaxel treatment in combination with ampicillin improved the chemotherapeutic efficacy in BC.290 Another mouse study indicated that antibiotics could counteract the metastatic effects of F. nucleatum in BC.33
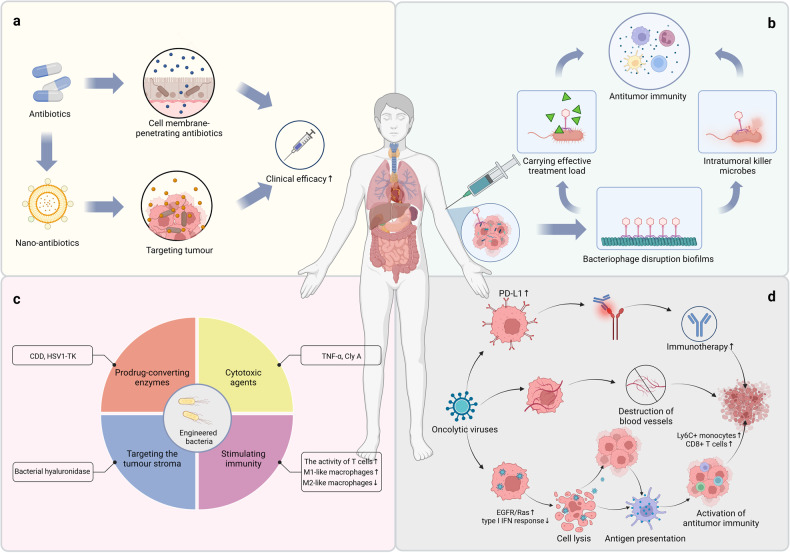
Fig. 4.
Application of intratumoural microbiota in cancer therapy. a Antibiotics. Cell membrane-penetrating antibiotics and nano-antibiotics can target intratumoural microorganisms, thereby improving clinical drug efficacy. b Bacteriophages. Bacteriophages can precisely target and eliminate harmful microbes within tumours and provide an effective therapeutic load to attract antitumour immune cells to the attack site by being modified into programmable bacterial assassins. c Engineered bacteria. Engineered bacteria can exert significant antitumour effects through a variety of payload delivery and effector systems, such as the production of prodrug-converting enzymes, expression of controlled cytotoxic agents, stimulation of immune responses, and targeting of tumour stroma. d Oncolytic viruses. Oncolytic viruses can exploit dysregulated signalling pathways to cause cell lysis and death or damage blood vessels to reduce tumour cell growth. Oncolytic viruses can also increase PD-L1 expression in tumours and immune cells, resulting in more sensitive targets for anti-PD-L1 immunotherapy. Created with BioRender.com
However, the systemic administration of antibiotics can not only eliminate the tumour microbiota but also disturb the gut microbiome, which further affects the efficacy of antitumour therapy. As the intestinal microbiota plays a crucial role in tumour development, the consequences of antibiotic use via different administration routes can be complicated.85 A study demonstrated that exposure to antibiotics shortly before or after the start of ICI could lead to the disruption of microbiota, which may be detrimental to ICI efficacy.291 Moreover, Jing et al. found that antibiotic treatment was linked with an increased risk of immune-related adverse events among cancer patients receiving anti-programmed cell death protein 1 (PD-1)/PD-L1 therapies.292 Stein-Thoeringer et al. recently demonstrated that treatment with wide-spectrum antibiotics before CD19-targeted chimeric antigen receptor (CAR)-T cell therapy is associated with adverse outcomes.293 Additionally, Laura et al.’s study indicated that gut dysbiosis induced by the wide-spectrum antibiotic cocktail treatment in mouse epithelial ovarian cancer could promote tumour growth and suppress cisplatin sensitivity.294 Although the advantages and disadvantages of implementing antibiotics in cancer treatment are still difficult to clarify due to their intricate effects on gut microbiota, clearance of the tumour-infiltrating microbiome (TIM) is a key point in cancer therapy. Thus, further exploration is required into precisely targeting TIM without disrupting the gut microbiome. Administration of the cell membrane-penetrating antibiotics, such as doxycycline, can be a potential choice.85 Gao et al. developed metronidazole-fluorouridine nanoparticles (MTI-FDU) in a hydrophilic solution, and they achieved TIM targeting through increasing penetrability and retention effect in solid tumours.295 Further in-depth analysis of how to efficiently eliminate the pro-tumoural TIM and keep a balanced microbiota system will provide more insights into cancer therapies.
Bacteriophages
The dysbiotic influence of antibiotics on microorganisms, bacterial resistance, and the impenetrability of biofilms have made it challenging to accurately ablate the microbiome in tumours using antibiotics.296–299 However, bacteriophages can precisely target and eliminate harmful microbes within tumours,300 and several Fusobacterium-targeted phages were shown to effectively invade bacteria within tumours (Fig. 4b). Researchers are now utilising synthetic biology techniques to modify phages into programmable bacterial assassins capable of delivering an effective therapeutic load to attract antitumour immune cells to the attack site.301 For example, azide-modified phages targeting F. nucleatum have been studied to covalently bind irinotecan nanoparticles and improve their delivery to mouse colon tumours.302 Although many studies have reported the safety of bacteriophages because they only reproduce in bacteria, they have been shown to induce host inflammation and immune responses.303,304 Therefore, determining the type, dosage, and mode of administration of bacteriophages is essential.305
Specificity, one of the advantages of bacteriophages as a customised fungicide, instead limits the therapeutic efficacy of bacteriophages by restricting their ability to kill only a specific type of bacteria. In this case, therapeutic bacteriophages must be specially selected and produced according to the different bacterial strains of different patients. Expanding the bactericidal range of bacteriophages is the focus of current research. Of those, cocktail therapy with a mixture of multiple bacteriophages is currently considered the best bacteriophage regimen.306 Bacteriophages are more suitable as a complement for antibiotics or a last resort when other effective therapies are not available; however, they are not a complete substitute for antibiotics.307
Engineered bacteria
Given the advent of synthetic biology, multiple bacteria have been modified to have lower toxicity and higher capability to accurately target tumour tissues.308 These tumour-targeting bacteria play a significant antitumour role via a variety of payload delivery and effector systems, such as the production of prodrug-converting enzymes, expression of controlled cytotoxic agents, stimulation of the immune response, and targeting the tumour stroma (Fig. 4c).309
Some bacteria produce prodrug-converting enzymes that enhance drug efficacy, including CDD. A study discovered that intratumoural injection of attenuated Salmonella typhimurium (S. typhimurium) strains VNP20009 expressing E. coli CDD significantly increased intratumoural 5-FU levels in cancer patients.310 The Bifidobacterium infantis-mediated prodrug enzyme delivery system of herpes simplex virus type I thymidine kinase/ganciclovir (HSV1-TK/GCV) significantly reduced bladder cancer progression in a rat model.311 Moreover, tumour-targeting bacteria can be engineered to express controlled cytotoxic drugs with antitumour effects. Utilising tumour-targeting systems with the non-pathogenic E. coli strain, MG1655 induced targeted production of TNF-α in mouse tumours, which demonstrated the tumour-suppressive effect.312 In two other mouse studies, the expression of cytolysin A (Cly A) in E. coli or S. typhimurium was regulated by inducible or constitutive promoters, achieving localisation to tumour tissue.313,314 Engineered bacteria can enhance antitumour activity by stimulating immune responses. A probiotic strain of E. coli Nissle 1917 was engineered to target mouse tumours and transform the accumulated metabolic waste product, ammonia, in the TME into L-arginine, which increased T cell activity and enhanced the efficacy of immunotherapy.315 Moreover, researchers designed an attenuated S. typhimurium strain that released Vibrio trauma flagellum B (FlaB) in the tumour tissue of the mouse and caused M1-like macrophages to increase and M2-like macrophages to decrease through TLR4 signalling.316 In addition, Listeria harbouring-galactosylceramide could enhance the activity of NKT cells and further reduce metastasis in a mouse breast tumour model.317 By taking advantage of bacterial enzymes targeting the tumour stroma, engineered bacteria can also be applied to treat solid tumours that are difficult to target using conventional therapies. Ebelt et al. engineered an attenuated tumour-targeting strain of S. typhimurium secreting functional bacterial hyaluronidase that degraded the deposition of human hyaluronic acid and significantly enhanced penetration of chemotherapeutic agents in orthotopic human PDAC mouse models.318 Moreover, many natural bacteria have antitumour effects, such as Staphylococcus epidermidis, which is capable of producing 6-N-hydroxy-aminopurine, a molecule that inhibits DNA polymerase activity to selectively inhibit the proliferation and development of skin tumours in mouse.319 Modifying these natural bacteria to target tumour inhibition will be a new research prospect.
Bacteria-based tumour therapy could provide multiple opportunities, but most research is still in the preclinical phase. This may be due to underlying differences between model organisms and humans in their genetic backgrounds or disease heterogeneity. Moreover, safety issues cannot be ignored. First, as the pharmacokinetics and dose responses of live bacteria differ from the norm, it poses a challenge in determining the optimal initial dose and schedule of administration. Secondly, engineered bacterial therapy involves transforming tumour foci into locally destructive tumour infections that, if not managed properly, can result in life-threatening infections or sepsis. Third, it is essential to prevent the contamination of the adjacent medical environment by living bacteria.
Oncolytic viruses
Oncolytic virus (OV) therapy involves intratumoural injection of genetically modified and attenuated viruses to selectively target cancer cells. OVs utilise the susceptibility of tumour cells to viral infection and exploit dysregulated signalling pathways to cause cell lysis and death. The antigens secreted from tumour cell lysis are taken up by antigen-presenting cells to further stimulate antitumour immunity. Moreover, OV can damage blood vessels, thus reducing tumour cell growth (Fig. 4d).320
Both in vivo and in vitro studies have demonstrated that the oncolytic poxvirus JX-594 replicates within and attacks tumour cells of various types of cancer by activating the EGFR/Ras signalling pathway and deactivating the type I IFN response pathway.321 Moreover, the arenavirus lymphocytic choriomeningitis virus replicates within tumour cells in multiple mouse and human cancers, inducing antitumour immunity by enlisting IFN-producing Ly6C+ monocytes and increasing the abundance of tumour-specific CD8 + T cells.322 Chen et al. found that variant OVs generating IL-23 increased the level of antitumour factors and infiltration of activated T cells in the TME, thereby converting the immunosuppressive phenotype and eliciting antitumour immunity in multiple mouse tumour models.323 In addition, OVs enhance PD-L1 expression in tumours and immune cells, resulting in more sensitive anti-PD-L1 immunotherapy targets. Coxsackievirus A21 combined with pembrolizumab partially increased the number of PD-L1+ tumour cells and exhibited well tolerance in a clinical trial.324 This indicates that the combination of OVs and ICIs may enhance antitumour efficacy while limiting toxicity, compared with that of monotherapy.325 Thus far, four OVs have been approved for cancer treatment worldwide, among which Talimogene laherparepvec (T-VEC) for melanoma is the first widely approved and recognised OV.326
However, the clinical transformation of OVs remains challenging. Combining OVs and radiotherapy, chemotherapy, or immunotherapy generally achieves a better therapeutic effect than the modalities alone. As replicable live viruses, OVs have received considerable attention in terms of safety. It is necessary to carefully examine the administration method, dosage, volume, time, and drug preservation method. Although intratumoural injection is the main administration route of the OVs,326 growing evidence has supported the possibility of intravenous injection.327 This possibility raises new problems; for example, OVs may be recognised and eliminated by the immune system, which would significantly decrease their efficacy. Therefore, new delivery methods for OVs have been in development, such as polymer coating or cell vectors.328–330 In addition, no reliable predictive biomarkers are available to identify which patients are candidates for OV treatment. Song et al. found that matrix remoulding associated 8 (MXRA8), which is widely and highly expressed in multiple solid tumours, is the receptor and therapeutic biomarker of oncolytic virus M1 (OVM). Further research showed that the tumour selectivity of OVM mainly hinges upon the combination of MXRA8 and zinc-finger antiviral protein.331 More similar studies are needed to provide precision medicine guidance for OV treatment.
Microbial intervention in the context of immunotherapy
Cancer immunotherapy has emerged as a significant means of combating cancer, mainly including immune checkpoint therapy represented by CTLA-4 and PD-1 and adoptive T cell therapy represented by CAR-T therapy.332 Based on the established role of host microbiota in modulating immunotherapy responses, many studies have begun to attempt to manipulate the microbiome for therapeutic purposes; faecal microflora transplantation (FMT), probiotics, and antibiotics are examples of this approach.
Vetizou et al. revealed the correlation between Bacteroides fragilis (B. fragilis) and the efficacy of CTLA-4 blocking in mouse models and melanoma patients. Oral gavage (OG) with B. fragilis and FMT from responding humans to mouse altered the immunotherapy response of germ-free, non-responding mouse.333 In addition, through FMT from JAX mouse that were effective for anti-PD-L1 therapy to TAC mouse, Sivan et al. showed that PD-L1 antibody levels were enhanced to inhibit melanoma in TAC mouse and further identified Bifidobacterium to be the responsible microbe.334 Other studies have reported similar results that microbial intervention boosts immunotherapy.223,335
These results indicate the benefits of microbial-based interventions in the context of immunotherapy; nevertheless, further studies in patients with different cancer types are warranted. Given the feasibility of stool collection from human donors, the use of FMT in immunotherapy is undoubtedly a promising area. Clinical trials have shown that FMT can benefit melanoma patients.336,337 In addition, immunotherapy combined with probiotic administration is another powerful research direction, which is more targeted to combine one or multiple microorganisms into a single formula.338 It is worth noting that there are few studies on whether bacteriophages affect immunotherapy responses in cancer patients. A study has shown that a bacteriophage-specific T cell epitope is correlated with anti-PD-1 therapy response in mouse.339 Accordingly, screening for differential bacteriophages from patients with different immune response states and combined immunotherapy through an approach similar to probiotic supplementation may help maximise benefits.
However, integrating animal model data into complex human tumours and antitumour immunotherapy is challenging. As mentioned earlier, broad-spectrum antibiotics disrupt the gut microbiota and impair immunotherapy efficacy and long-term patient survival. How to use specific antibiotics to remove immunosuppressive microbes is a key question. In addition, studies have found that non-targeted use of commercially available probiotics may not be beneficial to immunotherapy efficacy and may cause immunotherapy-related autoimmune events.340–342 Therefore, additional research is required to identify the underlying mechanisms of these microbial interventions and to develop personalised beneficial probiotics for patients with different living environments and dietary habits.343 Meanwhile, in order to avoid the risk of infection in cancer patients with weakened immune systems, beneficial substances produced by microorganisms or prebiotics can be used instead of probiotics.344
In addition, most current studies are based on gut microbes, and there are few studies on the role of tumour microbes in immunotherapy efficacy. It remains unknown whether there is a crosstalk between the gut microbes and the intratumoural microbes and whether changing the gut microbes will affect the intratumoural microbes and the characteristics of the host immune microenvironment; these aspects need to be explored. The study by Riquelme et al. may provide a way forward.264 Using human-mouse FMT from short- and long-term survival or control donors, they found that gut microbes can differentially regulate pancreatic tumour microbiome, thereby affecting tumour growth and tumour immune infiltration. Moreover, a recent study showed that OG probiotic Lr translocated from the gut and persistently colonised the tumours in melanoma mice, promoting the response of CD8 + T cells in TME and enhancing the therapeutic effect of anti-PD-L1.221 Thus, these microbial interventions may function through the gut-tumour microbiome crosstalk or a suitable niche provided by the TME, and direct intratumoural targeting may further enhance immunotherapy efficacy in the future. In addition, the research progress of CAR-T therapy for solid tumours is limited, so there are few studies on the influence of intratumoural microorganisms on CAR-T therapy. It is not yet known whether microorganisms can be a new breakthrough for CAR-T treatment of solid tumours.
Conclusion and outstanding questions
The microbiota has received widespread attention as a critical component in tumours. TME microbiota that modulates tumour development and affects cancer therapy is drawing increasing attention. Multiple tumours are thought to harbour distinct microbial communities, and the intratumoural microbiota may facilitate cancer development through various mechanisms, such as genomic instability and mutation, epigenetic modification, tumour-promoting inflammation, immune evasion, metabolic regulation, and activation of invasion and metastasis. Understanding this complex relationship between microbes and tumours could provide valuable insights into potential and existing cancer treatment options. Many bacteria that have been modified to be less virulent and accurately target tumour tissues have shown significant antitumour effects in preclinical studies. Moreover, microbial combinations with chemotherapy/immunotherapy may reduce drug resistance and improve anticancer efficacy.
Several studies have reported contradictory conclusions when describing the relationship between microorganisms and tumours.60,61,110–112 Therefore, standardised protocols for the study of intratumoural microorganisms should be developed and generally adopted. In addition, most studies are microbe-based cross-sectional studies that could not determine a causal relationship between intratumoural microorganisms and tumorigenesis. Although many studies have suggested that intratumoural microbes are the cause of cancer, we cannot deny the fact that microbial changes are the result of the development of certain cancers. Nevertheless, it is meaningful to use longitudinal prospective studies with large sample sizes and further explore the effects of the intratumoural microbiota on cancer cells of different phenotypes as well as immune cells. Recently, Bullman proposed that cancer cells and intratumoural microbiota may have a mutualistic relationship; that is, they both need to evade the immune system, and both have the capability to migrate or spread to new permitted niches.345 Therefore, despite the complex crosstalk between microbiota and tumours, it may be possible to treat tumours by modifying or manipulating the intratumoural microbiota.
The study of intratumoural microbiota in tumorigenesis and progression is just the beginning. Sophisticated animal models are needed in the future to trace tumour cells affected by microorganisms to provide more preclinical evidence. Interdisciplinary approaches are also necessary to quantitatively understand the relationship between microbes in tumours and tumour formation and development. Future work should also focus on the relationship between intratumoural microbes and other clinical factors known to be linked to the risk of various cancers, such as inflammatory and metabolic markers, to determine whether these factors have an additional effect on the composition of intratumoural microbiota. In addition, linking tissue microbial differences between healthy and high-risk individuals with cancer development might contribute to establishing more effective cancer prevention and diagnosis methods; however, the effort is complicated by the ethical and accessibility challenges associated with normal human tissue. In terms of antitumour therapy, studies linking identified microbial signatures to tumour response regulation could identify new targets for clinical intervention. Exploring combination therapy strategies based on microbial intervention to improve the clinical treatment effect is another promising research direction.
Acknowledgements
We are grateful to Dr. Mengfei Guo (Wuhan Union Hospital), Dr. Min Zhang (Second Affiliated Hospital of Zhejiang University School of Medicine), and Prof. Weimin Wang (Tongji Medical College) for many useful discussions through the formation of this work. We would like to thank Editage (www.editage.cn) for English language editing. Funding: This work was supported by the National Natural Science Foundation of China (No.82330003; No.82070099; No. 82102496; No. 82001988); National Science and Technology Major Project of the Ministry of Science and Technology of China (2022YFF1203300); Major Project of the Department of Science and Technology of Hubei (2022BCA016) and Hubei Key Laboratory of Biological Targeted Therapy (2022swbx009).
Author contributions
Y.C., H.X. and X.T. conceived and drafted the manuscript, generated the figures and summarized the tables; C.S. and Y.M. discussed the details of the manuscript; D.M., Z.L. and M.Z. collected the references; S.W. and Y.J. reviewed and edited the manuscript; All authors have read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yaqi Cao, Hui Xia, Xueyun Tan.
Contributor Information
Sufei Wang, Email: wangsufei0@163.com.
Yang Jin, Email: whuhjy@126.com.
References
- 1.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azevedo MM, Pina-Vaz C, Baltazar F. Microbes and cancer: friends or faux? Int J. Mol. Sci. 2020;21:3115. doi: 10.3390/ijms21093115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong-Rolle A, Wei HK, Zhao C, Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Prot. Cell. 2021;12:426–435. doi: 10.1007/s13238-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gagliani N, Hu B, Huber S, Elinav E, Flavell RA. The fire within: microbes inflame tumors. Cell. 2014;157:776–783. doi: 10.1016/j.cell.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Nejman D, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poore GD, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Kalaora S, et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature. 2021;592:138–143. doi: 10.1038/s41586-021-03368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galeano Niño JL, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 2022;611:810–817. doi: 10.1038/s41586-022-05435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dohlman AB, et al. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell. 2022;185:3807–3822. doi: 10.1016/j.cell.2022.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narunsky-Haziza L, et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. 2022;185:3789–3806. doi: 10.1016/j.cell.2022.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elinav E, Garrett WS, Trinchieri G, Wargo J. The cancer microbiome. Nat. Rev. Cancer. 2019;19:371–376. doi: 10.1038/s41568-019-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudgeon LS, Dunkley EV. The micrococcus neoformans: its cultural characters and pathogenicity and the results of the estimation of the opsonic and agglutinative properties of the serum of patients suffering from malignant disease on this organism and on the Staphylococcus Albus. J. Hyg. Lond. 1907;7:13–21. doi: 10.1017/S002217240003309X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rous P. A sarcoma of the fowl transmissible by an agent separable from the tumor cells. J. Exp. Med. 1911;13:397–411. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt’s Lymphoma. Lancet. 1964;1:702–703. doi: 10.1016/S0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- 17.Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1:1273–1275. [PubMed] [Google Scholar]
- 18.Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR, Isaacson PG. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet. 1991;338:1175–1176. doi: 10.1016/0140-6736(91)92035-Z. [DOI] [PubMed] [Google Scholar]
- 19.Felgner S, Kocijancic D, Frahm M, Weiss S. Bacteria in cancer therapy: renaissance of an old concept. Int J. Microbiol. 2016;2016:8451728. doi: 10.1155/2016/8451728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiemann B, Starnes CO. Coley’s toxins, tumor necrosis factor and cancer research: a historical perspective. Pharm. Ther. 1994;64:529–564. doi: 10.1016/0163-7258(94)90023-X. [DOI] [PubMed] [Google Scholar]
- 21.Mohammed S, Bakshi N, Chaudri N, Akhter J, Akhtar M. Cancer vaccines: past, present, and future. Adv. Anat. Pathol. 2016;23:180–191. doi: 10.1097/PAP.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 22.Budynek P, Dabrowska K, Skaradziński G, Górski A. Bacteriophages and cancer. Arch. Microbiol. 2010;192:315–320. doi: 10.1007/s00203-010-0559-7. [DOI] [PubMed] [Google Scholar]
- 23.González-Sánchez P, DeNicola GM. The microbiome(s) and cancer: know thy neighbor(s) J. Pathol. 2021;254:332–343. doi: 10.1002/path.5661. [DOI] [PubMed] [Google Scholar]
- 24.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 26.Leinwand J, Miller G. Regulation and modulation of antitumor immunity in pancreatic cancer. Nat. Immunol. 2020;21:1152–1159. doi: 10.1038/s41590-020-0761-y. [DOI] [PubMed] [Google Scholar]
- 27.Desai S, et al. Fusobacterium nucleatum is associated with inflammation and poor survival in early-stage HPV-negative tongue cancer. NAR Cancer. 2022;4:zcac006. doi: 10.1093/narcan/zcac006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiao H, et al. Association of intratumoral microbiota with prognosis in patients with Nasopharyngeal Carcinoma from 2 hospitals in China. JAMA Oncol. 2022;8:1301–1309. doi: 10.1001/jamaoncol.2022.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin D, Gutkind JS. Human tumor-associated viruses and new insights into the molecular mechanisms of cancer. Oncogene. 2008;27:S31–42. doi: 10.1038/onc.2009.351. [DOI] [PubMed] [Google Scholar]
- 30.Png CW, et al. Mucosal microbiome associates with progression to gastric cancer. Theranostics. 2022;12:48–58. doi: 10.7150/thno.65302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J. Exp. Med. 2020;217:e20192282. doi: 10.1084/jem.20192282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abed J, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host. Microb. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parhi L, et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 2020;11:3259. doi: 10.1038/s41467-020-16967-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker SP, Tangney M, Claesson MJ. Sequence-based characterization of intratumoral bacteria-a guide to best practice. Front Oncol. 2020;10:179. doi: 10.3389/fonc.2020.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charlson ES, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao Q, et al. Differential flora in the microenvironment of lung tumor and paired adjacent normal tissues. Carcinogenesis. 2020;41:1094–1103. doi: 10.1093/carcin/bgaa044. [DOI] [PubMed] [Google Scholar]
- 37.Greathouse KL, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomes S, et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci. Rep. 2019;9:12838. doi: 10.1038/s41598-019-49195-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apopa PL, et al. PARP1 is up-regulated in non-small cell lung cancer tissues in the presence of the Cyanobacterial toxin microcystin. Front. Microbiol. 2018;9:1757. doi: 10.3389/fmicb.2018.01757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. 2016;102:89–95. doi: 10.1016/j.lungcan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Mao Q, et al. Interplay between the lung microbiome and lung cancer. Cancer Lett. 2018;415:40–48. doi: 10.1016/j.canlet.2017.11.036. [DOI] [PubMed] [Google Scholar]
- 42.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 43.Maucort-Boulch D, de Martel C, Franceschi S, Plummer M. Fraction and incidence of liver cancer attributable to hepatitis B and C viruses worldwide. Int J. Cancer. 2018;142:2471–2477. doi: 10.1002/ijc.31280. [DOI] [PubMed] [Google Scholar]
- 44.Kho ZY, Lal SK. The human gut microbiome - A potential controller of wellness and disease. Front. Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Cao ZM, Zhang LL, Li JM, Lv WL. The role of gut microbiota in some liver diseases: from an immunological perspective. Front. Immunol. 2022;13:923599. doi: 10.3389/fimmu.2022.923599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macpherson AJ, Heikenwalder M, Ganal-Vonarburg SC. The liver at the nexus of host-microbial interactions. Cell Host. Microb. 2016;20:561–571. doi: 10.1016/j.chom.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 47.Kanmani P, Suganya K, Kim H. The gut microbiota: how does it influence the development and progression of liver diseases. Biomedicines. 2020;8:501. doi: 10.3390/biomedicines8110501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pellicano R, Ménard A, Rizzetto M, Mégraud F. Helicobacter species and liver diseases: association or causation? Lancet Infect. Dis. 2008;8:254–260. doi: 10.1016/S1473-3099(08)70066-5. [DOI] [PubMed] [Google Scholar]
- 49.Tian XF, et al. Procuration and identification of bacteria in paraffin-embedded liver tissues of hepatocellular carcinoma by laser-assisted microdissection technique. Apmis. 2008;116:10–15. doi: 10.1111/j.1600-0463.2008.00739.x. [DOI] [PubMed] [Google Scholar]
- 50.Testerman TL, Morris J. Beyond the stomach: an updated view of Helicobacter pylori pathogenesis, diagnosis, and treatment. World J. Gastroenterol. 2014;20:12781–12808. doi: 10.3748/wjg.v20.i36.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Comparative proteome analysis of untreated and Helicobacter pylori-treated HepG2. World J. Gastroenterol. 2005;11:3485–3489. doi: 10.3748/wjg.v11.i22.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, et al. Ubiquitin specific peptidase 5 mediates Histidine-rich protein Hpn induced cell apoptosis in hepatocellular carcinoma through P14-P53 signaling. Proteomics. 2017;17:1600350. doi: 10.1002/pmic.201600350. [DOI] [PubMed] [Google Scholar]
- 53.Huang JH, et al. The intratumoral bacterial metataxonomic signature of hepatocellular carcinoma. Microbiol. Spectr. 2022;10:e0098322. doi: 10.1128/spectrum.00983-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qu D, et al. Intratumoral microbiome of human primary liver cancer. Hepatol. Commun. 2022;6:1741–1752. doi: 10.1002/hep4.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chai X, et al. Intratumor microbiome features reveal antitumor potentials of intrahepatic cholangiocarcinoma. Gut. Microbes. 2023;15:2156255. doi: 10.1080/19490976.2022.2156255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goodwin AC, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl Acad. Sci. USA. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Z, Chen J, Yao H, Hu H. Fusobacterium and colorectal cancer. Front. Oncol. 2018;8:371. doi: 10.3389/fonc.2018.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kostic AD, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto S, et al. Heterogeneous distribution of Fusobacterium nucleatum in the progression of colorectal cancer. J. Gastroenterol. Hepatol. 2021;36:1869–1876. doi: 10.1111/jgh.15361. [DOI] [PubMed] [Google Scholar]
- 61.Repass J. Replication study: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Elife. 2018;7:e25801. doi: 10.7554/eLife.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kneis B, et al. Colon cancer microbiome landscaping: differences in right- and left-sided colon cancer and a tumor microbiome-Ileal microbiome association. Int J. Mol. Sci. 2023;24:3265. doi: 10.3390/ijms24043265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee MS, Menter DG, Kopetz S. Right versus left colon cancer biology: integrating the consensus molecular subtypes. J. Natl Compr. Cancer Netw. 2017;15:411–419. doi: 10.6004/jnccn.2017.0038. [DOI] [PubMed] [Google Scholar]
- 64.Luan C, et al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci. Rep. 2015;5:7980. doi: 10.1038/srep07980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coker OO, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2019;68:654–662. doi: 10.1136/gutjnl-2018-317178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng Q, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 67.Wirbel J, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019;25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai H, et al. Helicobacter pylori blood biomarker for gastric cancer risk in East Asia. Int J. Epidemiol. 2016;45:774–781. doi: 10.1093/ije/dyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Correa P, Houghton J. Carcinogenesis of helicobacter pylori. Gastroenterology. 2007;133:659–672. doi: 10.1053/j.gastro.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 70.Murphy G, Pfeiffer R, Camargo MC, Rabkin CS. Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology. 2009;137:824–833. doi: 10.1053/j.gastro.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee JH, et al. Clinicopathological and molecular characteristics of Epstein-Barr virus-associated gastric carcinoma: a meta-analysis. J. Gastroenterol. Hepatol. 2009;24:354–365. doi: 10.1111/j.1440-1746.2009.05775.x. [DOI] [PubMed] [Google Scholar]
- 72.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shao D, et al. Microbial characterization of esophageal squamous cell carcinoma and gastric cardia adenocarcinoma from a high-risk region of China. Cancer. 2019;125:3993–4002. doi: 10.1002/cncr.32403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang L, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur. J. Gastroenterol. Hepatol. 2016;28:261–266. doi: 10.1097/MEG.0000000000000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peng R, et al. Gastric microbiome alterations are associated with decreased CD8+ tissue-resident memory T cells in the tumor microenvironment of gastric cancer. Cancer Immunol. Res. 2022;10:1224–1240. doi: 10.1158/2326-6066.CIR-22-0107. [DOI] [PubMed] [Google Scholar]
- 76.Gunathilake MN, et al. Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Sci. Rep. 2019;9:13589. doi: 10.1038/s41598-019-50054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 2014;4:4202. doi: 10.1038/srep04202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coughlin SS. Epidemiology of breast cancer in women. Adv. Exp. Med. Biol. 2019;1152:9–29. doi: 10.1007/978-3-030-20301-6_2. [DOI] [PubMed] [Google Scholar]
- 79.Alpuim Costa D, et al. Human microbiota and breast cancer-is there any relevant link? A literature review and new horizons toward personalised medicine. Front. Microbiol. 2021;12:584332. doi: 10.3389/fmicb.2021.584332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urbaniak C, et al. Microbiota of human breast tissue. Appl. Environ. Microbiol. 2014;80:3007–3014. doi: 10.1128/AEM.00242-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tzeng A, et al. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. 2021;13:60. doi: 10.1186/s13073-021-00874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xuan C, et al. Microbial dysbiosis is associated with human breast cancer. PLoS One. 2014;9:e83744. doi: 10.1371/journal.pone.0083744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yazdi HR, et al. Evaluation of Methylobacterium radiotolerance and Sphyngomonas yanoikoaie in Sentinel Lymph Nodes of breast cancer cases. Asian Pac. J. Cancer Prev. 2016;17:279–285. doi: 10.7314/APJCP.2016.17.S3.279. [DOI] [PubMed] [Google Scholar]
- 84.Wang H, et al. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget. 2017;8:88122–88138. doi: 10.18632/oncotarget.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu A, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. 2022;185:1356–1372. doi: 10.1016/j.cell.2022.02.027. [DOI] [PubMed] [Google Scholar]
- 86.Parida S, Siddharth S, Xia Y, Sharma D. Concomitant analyses of intratumoral microbiota and genomic features reveal distinct racial differences in breast cancer. NPJ Breast Cancer. 2023;9:4. doi: 10.1038/s41523-023-00505-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith A, et al. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Sci. Rep. 2019;9:11940. doi: 10.1038/s41598-019-48348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thyagarajan S, et al. Comparative analysis of racial differences in breast tumor microbiome. Sci. Rep. 2020;10:14116. doi: 10.1038/s41598-020-71102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 90.Geller LT, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pushalkar S, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8:403–416. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nilsson HO, Stenram U, Ihse I, Wadstrom T. Helicobacter species ribosomal DNA in the pancreas, stomach and duodenum of pancreatic cancer patients. World J. Gastroenterol. 2006;12:3038–3043. doi: 10.3748/wjg.v12.i19.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takayama S, Takahashi H, Matsuo Y, Okada Y, Manabe T. Effects of Helicobacter pylori infection on human pancreatic cancer cell line. Hepato. Gastroenterol. 2007;54:2387–2391. [PubMed] [Google Scholar]
- 94.Guo W, et al. Tumor microbiome contributes to an aggressive phenotype in the basal-like subtype of pancreatic cancer. Commun. Biol. 2021;4:1019. doi: 10.1038/s42003-021-02557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aykut B, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574:264–267. doi: 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tuominen H, Rautava J. Oral microbiota and cancer development. Pathobiology. 2021;88:116–126. doi: 10.1159/000510979. [DOI] [PubMed] [Google Scholar]
- 97.Tandon P, Dadhich A, Saluja H, Bawane S, Sachdeva S. The prevalence of squamous cell carcinoma in different sites of oral cavity at our rural health care centre in Loni, Maharashtra - a retrospective 10-year study. Contemp. Oncol. 2017;21:178–183. doi: 10.5114/wo.2017.68628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bussu F, et al. HPV infection in squamous cell carcinomas arising from different mucosal sites of the head and neck region. Is p16 immunohistochemistry a reliable surrogate marker? Br. J. Cancer. 2013;108:1157–1162. doi: 10.1038/bjc.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guidry JT, Birdwell CE, Scott RS. Epstein-Barr virus in the pathogenesis of oral cancers. Oral. Dis. 2018;24:497–508. doi: 10.1111/odi.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jain M. Assesment of correlation of herpes simplex virus-1 with oral cancer and precancer- a comparative study. J. Clin. Diagn. Res. 2016;10:ZC14–ZC17. doi: 10.7860/JCDR/2016/18593.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nagy KN, Sonkodi I, Szöke I, Nagy E, Newman HN. The microflora associated with human oral carcinomas. Oral. Oncol. 1998;34:304–308. doi: 10.1016/S1368-8375(98)80012-2. [DOI] [PubMed] [Google Scholar]
- 102.Ahn J, Segers S, Hayes RB. Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis. 2012;33:1055–1058. doi: 10.1093/carcin/bgs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Geng F, et al. Persistent exposure to porphyromonas gingivalis promotes proliferative and invasion capabilities, and tumorigenic properties of human immortalized oral epithelial cells. Front. Cell Infect. Microbiol. 2017;7:57. doi: 10.3389/fcimb.2017.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int. J. Oral. Sci. 2011;3:209–215. doi: 10.4248/IJOS11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang C, et al. The prevalence rate of periodontal pathogens and its association with oral squamous cell carcinoma. Appl. Microbiol. Biotechnol. 2019;103:1393–1404. doi: 10.1007/s00253-018-9475-6. [DOI] [PubMed] [Google Scholar]
- 106.Sarkar P, et al. Dysbiosis of oral microbiota during oral squamous cell carcinoma development. Front. Oncol. 2021;11:614448. doi: 10.3389/fonc.2021.614448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang L, Liu Y, Zheng HJ, Zhang CP. The oral microbiota may have influence on oral cancer. Front. Cell Infect. Microbiol. 2019;9:476. doi: 10.3389/fcimb.2019.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nieminen MT, et al. Treponema denticola chymotrypsin-like proteinase may contribute to orodigestive carcinogenesis through immunomodulation. Br. J. Cancer. 2018;118:428–434. doi: 10.1038/bjc.2017.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kylmä AK, et al. Treponema denticola chymotrypsin-like protease as associated with HPV-negative oropharyngeal squamous cell carcinoma. Br. J. Cancer. 2018;119:89–95. doi: 10.1038/s41416-018-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sasaki M, et al. Streptococcus anginosus infection in oral cancer and its infection route. Oral. Dis. 2005;11:151–156. doi: 10.1111/j.1601-0825.2005.01051.x. [DOI] [PubMed] [Google Scholar]
- 111.Rai AK, et al. Dysbiosis of salivary microbiome and cytokines influence oral squamous cell carcinoma through inflammation. Arch. Microbiol. 2021;203:137–152. doi: 10.1007/s00203-020-02011-w. [DOI] [PubMed] [Google Scholar]
- 112.Su SC, et al. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis. 2021;42:127–135. doi: 10.1093/carcin/bgaa062. [DOI] [PubMed] [Google Scholar]
- 113.Al-Hebshi NN, et al. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci. Rep. 2017;7:1834. doi: 10.1038/s41598-017-02079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Perera M, et al. A dysbiotic mycobiome dominated by Candida albicans is identified within oral squamous-cell carcinomas. J. Oral. Microbiol. 2017;9:1385369. doi: 10.1080/20002297.2017.1385369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang CY, et al. Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front. Microbiol. 2018;9:862. doi: 10.3389/fmicb.2018.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang H, et al. Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas. Genome Med. 2017;9:14. doi: 10.1186/s13073-017-0405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mork J, et al. Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2001;344:1125–1131. doi: 10.1056/NEJM200104123441503. [DOI] [PubMed] [Google Scholar]
- 118.Zapatka M, et al. The landscape of viral associations in human cancers. Nat. Genet. 2020;52:320–330. doi: 10.1038/s41588-019-0558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wolf H, zur Hausen H, Becker V. EB viral genomes in epithelial nasopharyngeal carcinoma cells. Nat. N. Biol. 1973;244:245–247. doi: 10.1038/newbio244245a0. [DOI] [PubMed] [Google Scholar]
- 120.Wang Q, et al. The differential distribution of bacteria between cancerous and noncancerous ovarian tissues in situ. J. Ovarian. Res. 2020;13:8. doi: 10.1186/s13048-019-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Banerjee S, et al. The ovarian cancer oncobiome. Oncotarget. 2017;8:36225–36245. doi: 10.18632/oncotarget.16717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Walther-António MR, et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016;8:122. doi: 10.1186/s13073-016-0368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen Y, Wei J. Identification of pathogen signatures in prostate cancer using RNA-seq. PLoS One. 2015;10:e0128955. doi: 10.1371/journal.pone.0128955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kustrimovic N, Bombelli R, Baci D, Mortara L. Microbiome and prostate cancer: a novel target for prevention and treatment. Int J. Mol. Sci. 2023;24:1511. doi: 10.3390/ijms24021511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ye L, et al. Evidence for an intra-tumoral microbiome in pituitary neuroendocrine tumors with different clinical phenotypes. J. Neuro.Oncol. 2023;163:133–142. doi: 10.1007/s11060-023-04318-2. [DOI] [PubMed] [Google Scholar]
- 126.Young LS, Yap LF, Murray PG. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat. Rev. Cancer. 2016;16:789–802. doi: 10.1038/nrc.2016.92. [DOI] [PubMed] [Google Scholar]
- 127.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 128.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 129.Khan G, Hashim MJ. Global burden of deaths from Epstein-Barr virus attributable malignancies 1990-2010. Infect. Agent Cancer. 2014;9:38. doi: 10.1186/1750-9378-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jiang Z, et al. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 2012;22:593–601. doi: 10.1101/gr.133926.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hu Z, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat. Genet. 2015;47:158–163. doi: 10.1038/ng.3178. [DOI] [PubMed] [Google Scholar]
- 132.Krump NA, You J. Molecular mechanisms of viral oncogenesis in humans. Nat. Rev. Microbiol. 2018;16:684–698. doi: 10.1038/s41579-018-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lau L, Gray EE, Brunette RL, Stetson DB. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350:568–571. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- 134.Luo X, et al. HPV16 drives cancer immune escape via NLRX1-mediated degradation of STING. J. Clin. Invest. 2020;130:1635–1652. doi: 10.1172/JCI129497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jones T, et al. Viral cyclin promotes KSHV-induced cellular transformation and tumorigenesis by overriding contact inhibition. Cell Cycle. 2014;13:845–858. doi: 10.4161/cc.27758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ballon G, Chen K, Perez R, Tam W, Cesarman E. Kaposi sarcoma herpesvirus (KSHV) vFLIP oncoprotein induces B cell transdifferentiation and tumorigenesis in mice. J. Clin. Invest. 2011;121:1141–1153. doi: 10.1172/JCI44417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang LW, Jiang S, Gewurz BE. Epstein-Barr Virus LMP1-Mediated Oncogenicity. J. Virol. 2017;91:e01718–01716. doi: 10.1128/JVI.01718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Allday MJ, Bazot Q, White RE. The EBNA3 family: Two oncoproteins and a tumour suppressor that are central to the biology of EBV in B cells. Curr. Top. Microbiol. Immunol. 2015;391:61–117. doi: 10.1007/978-3-319-22834-1_3. [DOI] [PubMed] [Google Scholar]
- 139.Giam CZ, Semmes OJ. HTLV-1 infection and adult T-Cell leukemia/lymphoma-A tale of two proteins: tax and HBZ. Viruses. 2016;8:161. doi: 10.3390/v8060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dejea CM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Guo P, et al. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J. Exp. Clin. Cancer Res. 2020;39:202. doi: 10.1186/s13046-020-01677-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Geng F, Zhang Y, Lu Z, Zhang S, Pan Y. Fusobacterium nucleatum caused DNA damage and promoted cell proliferation by the Ku70/p53 pathway in oral cancer cells. DNA Cell Biol. 2020;39:144–151. doi: 10.1089/dna.2019.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Urbaniak C, et al. The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 2016;82:5039–5048. doi: 10.1128/AEM.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Maddocks OD, Scanlon KM, Donnenberg MS. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. mBio. 2013;4:e00152–00113. doi: 10.1128/mBio.00152-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Santos JC, et al. Helicobacter pylori infection modulates the expression of miRNAs associated with DNA mismatch repair pathway. Mol. Carcinog. 2017;56:1372–1379. doi: 10.1002/mc.22590. [DOI] [PubMed] [Google Scholar]
- 146.Martin OCB, Frisan T. Bacterial genotoxin-induced DNA damage and modulation of the host immune microenvironment. Toxins Basel. 2020;12:63. doi: 10.3390/toxins12020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. ROS and the DNA damage response in cancer. Redox Biol. 2019;25:101084. doi: 10.1016/j.redox.2018.101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huycke MM, Gaskins HR. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp. Biol. Med. 2004;229:586–597. doi: 10.1177/153537020422900702. [DOI] [PubMed] [Google Scholar]
- 149.Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012;3:448. doi: 10.3389/fphys.2012.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chan AA, et al. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 2016;6:28061. doi: 10.1038/srep28061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim DH, Jin YH. Intestinal bacterial beta-glucuronidase activity of patients with colon cancer. Arch. Pharm. Res. 2001;24:564–567. doi: 10.1007/BF02975166. [DOI] [PubMed] [Google Scholar]
- 152.Humblot C, et al. Beta-glucuronidase in human intestinal microbiota is necessary for the colonic genotoxicity of the food-borne carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline in rats. Carcinogenesis. 2007;28:2419–2425. doi: 10.1093/carcin/bgm170. [DOI] [PubMed] [Google Scholar]
- 153.Mizumoto A, et al. Molecular mechanisms of acetaldehyde-mediated carcinogenesis in squamous epithelium. Int J. Mol. Sci. 2017;18:1943. doi: 10.3390/ijms18091943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Olsen I, Yilmaz Ö. Possible role of Porphyromonas gingivalis in orodigestive cancers. J. Oral. Microbiol. 2019;11:1563410. doi: 10.1080/20002297.2018.1563410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang WL, et al. Who is who in oral cancer? Exp. Cell Res. 2019;384:111634. doi: 10.1016/j.yexcr.2019.111634. [DOI] [PubMed] [Google Scholar]
- 156.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 157.Hamon MA, Cossart P. Histone modifications and chromatin remodeling during bacterial infections. Cell Host. Microb. 2008;4:100–109. doi: 10.1016/j.chom.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 158.Bierne H, Hamon M, Cossart P. Epigenetics and bacterial infections. Cold Spring Harb. Perspect. Med. 2012;2:a010272. doi: 10.1101/cshperspect.a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rolando M, Gomez-Valero L, Buchrieser C. Bacterial remodelling of the host epigenome: functional role and evolution of effectors methylating host histones. Cell Microbiol. 2015;17:1098–1107. doi: 10.1111/cmi.12463. [DOI] [PubMed] [Google Scholar]
- 160.Arbibe L. Immune subversion by chromatin manipulation: a ‘new face’ of host-bacterial pathogen interaction. Cell Microbiol. 2008;10:1582–1590. doi: 10.1111/j.1462-5822.2008.01170.x. [DOI] [PubMed] [Google Scholar]
- 161.Liu D, et al. Helicobacter pylori-induced aberrant demethylation and expression of GNB4 promotes gastric carcinogenesis via the Hippo-YAP1 pathway. BMC Med. 2023;21:134. doi: 10.1186/s12916-023-02842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Niwa T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–1440. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 163.Yue K, et al. Bidirectional mediation effects between intratumoral microbiome and host DNA methylation changes contribute to stomach adenocarcinoma. Microbiol. Spectr. 2023;11:e0090423. doi: 10.1128/spectrum.00904-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gnanasekar A, et al. The intratumor microbiome predicts prognosis across gender and subtypes in papillary thyroid carcinoma. Comput. Struct. Biotechnol. J. 2021;19:1986–1997. doi: 10.1016/j.csbj.2021.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xue C, et al. Intratumoral bacteria interact with metabolites and genetic alterations in hepatocellular carcinoma. Signal Transduct. Target Ther. 2022;7:335. doi: 10.1038/s41392-022-01159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Park HE, Kim JH, Cho NY, Lee HS, Kang GH. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchow. Arch. 2017;471:329–336. doi: 10.1007/s00428-017-2171-6. [DOI] [PubMed] [Google Scholar]
- 167.Hong J, et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2021;70:2123–2137. doi: 10.1136/gutjnl-2020-322780. [DOI] [PubMed] [Google Scholar]
- 168.Xia X, et al. Bacteria pathogens drive host colonic epithelial cell promoter hypermethylation of tumor suppressor genes in colorectal cancer. Microbiome. 2020;8:108. doi: 10.1186/s40168-020-00847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Woo V, Alenghat T. Epigenetic regulation by gut microbiota. Gut. Microbes. 2022;14:2022407. doi: 10.1080/19490976.2021.2022407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.van der Hee B, Wells JM. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. 2021;29:700–712. doi: 10.1016/j.tim.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 171.Fawad JA, et al. Histone deacetylase inhibition by Gut microbe-generated short-chain fatty acids entrains intestinal epithelial circadian rhythms. Gastroenterology. 2022;163:1377–1390. doi: 10.1053/j.gastro.2022.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vega, A.A. et al. Methionine-producing tumor micro(be) environment fuels growth of solid tumors. Cell Oncol.10.1007/s13402-023-00832-7 (2023). [DOI] [PMC free article] [PubMed]
- 173.Clare CE, Brassington AH, Kwong WY, Sinclair KD. One-carbon metabolism: linking nutritional biochemistry to epigenetic programming of long-term development. Annu. Rev. Anim. Biosci. 2019;7:263–287. doi: 10.1146/annurev-animal-020518-115206. [DOI] [PubMed] [Google Scholar]
- 174.Koi M, Okita Y, Carethers JM. Fusobacterium nucleatum infection in colorectal cancer: linking inflammation, DNA mismatch repair and genetic and epigenetic alterations. J. Anus Rectum Colon. 2018;2:37–46. doi: 10.23922/jarc.2017-055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Duval M, Cossart P, Lebreton A. Mammalian microRNAs and long noncoding RNAs in the host-bacterial pathogen crosstalk. Semin. Cell Dev. Biol. 2017;65:11–19. doi: 10.1016/j.semcdb.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zur Bruegge J, Einspanier R, Sharbati S. A long journey ahead: long non-coding RNAs in bacterial infections. Front. Cell Infect. Microbiol. 2017;7:95. doi: 10.3389/fcimb.2017.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Aguilar C, Mano M, Eulalio A. MicroRNAs at the host-bacteria interface: host defense or bacterial offense. Trends Microbiol. 2019;27:206–218. doi: 10.1016/j.tim.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 178.Pietropaolo V, Prezioso C, Moens U. Role of virus-induced host cell epigenetic changes in cancer. Int J. Mol. Sci. 2021;22:8346. doi: 10.3390/ijms22158346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kostyusheva A, Brezgin S, Glebe D, Kostyushev D, Chulanov V. Host-cell interactions in HBV infection and pathogenesis: the emerging role of m6A modification. Emerg. Microbe. Infect. 2021;10:2264–2275. doi: 10.1080/22221751.2021.2006580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhao H, et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct. Target Ther. 2021;6:2426–2471. doi: 10.1038/s41392-021-00658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 182.Chen T, et al. Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol. Immunother. 2018;67:1635–1646. doi: 10.1007/s00262-018-2233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yang Y, et al. Cross-talk between the gut microbiota and monocyte-like macrophages mediates an inflammatory response to promote colitis-associated tumourigenesis. Gut. 2020;70:1495–1506. doi: 10.1136/gutjnl-2020-320777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wu P, et al. γδT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity. 2014;40:785–800. doi: 10.1016/j.immuni.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Grivennikov SI, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491:254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chung L, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host. Microb. 2018;23:203–214. doi: 10.1016/j.chom.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Park SR, et al. Diverse Toll-like receptors mediate cytokine production by Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans in macrophages. Infect. Immun. 2014;82:1914–1920. doi: 10.1128/IAI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jungnickel C, et al. IL-17C mediates the recruitment of tumor-associated neutrophils and lung tumor growth. Oncogene. 2017;36:4182–4190. doi: 10.1038/onc.2017.28. [DOI] [PubMed] [Google Scholar]
- 189.Kaakoush NO, et al. Transcriptomic and proteomic analyses reveal key innate immune signatures in the host response to the gastrointestinal pathogen Campylobacter concisus. Infect. Immun. 2015;83:832–845. doi: 10.1128/IAI.03012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tang B, et al. Fusobacterium nucleatum-induced impairment of autophagic flux enhances the expression of proinflammatory cytokines via ROS in Caco-2 Cells. PLoS One. 2016;11:e0165701. doi: 10.1371/journal.pone.0165701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Redelman-Sidi G, Iyer G, Solit DB, Glickman MS. Oncogenic activation of Pak1-dependent pathway of macropinocytosis determines BCG entry into bladder cancer cells. Cancer Res. 2013;73:1156–1167. doi: 10.1158/0008-5472.CAN-12-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Redelman-Sidi G, et al. The canonical Wnt pathway drives macropinocytosis in cancer. Cancer Res. 2018;78:4658–4670. doi: 10.1158/0008-5472.CAN-17-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 194.Triner D, et al. Neutrophils restrict tumor-associated microbiota to reduce growth and invasion of colon tumors in mice. Gastroenterology. 2019;156:1467–1482. doi: 10.1053/j.gastro.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kuraishy A, Karin M, Grivennikov SI. Tumor promotion via injury- and death-induced inflammation. Immunity. 2011;35:467–477. doi: 10.1016/j.immuni.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shalapour S, Karin M. Pas de Deux: control of anti-tumor immunity by cancer-associated inflammation. Immunity. 2019;51:15–26. doi: 10.1016/j.immuni.2019.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kidane D, et al. Interplay between DNA repair and inflammation, and the link to cancer. Crit. Rev. Biochem. Mol. Biol. 2014;49:116–139. doi: 10.3109/10409238.2013.875514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Borowsky J, et al. Association of Fusobacterium nucleatum with specific T-cell subsets in the colorectal carcinoma microenvironment. Clin. Cancer Res. 2021;27:2816–2826. doi: 10.1158/1078-0432.CCR-20-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jin C, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176:998–1013. doi: 10.1016/j.cell.2018.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zheng L, et al. Microbiome related cytotoxically active CD8+ TIL are inversely associated with lung cancer development. Front. Oncol. 2020;10:531131. doi: 10.3389/fonc.2020.531131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ma J, et al. Influence of intratumor microbiome on clinical outcome and immune processes in prostate cancer. Cancers. 2020;12:2524. doi: 10.3390/cancers12092524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gao Y, et al. Intratumoral stem-like CCR4+ regulatory T cells orchestrate the immunosuppressive microenvironment in HCC associated with hepatitis B. J. Hepatol. 2022;76:148–159. doi: 10.1016/j.jhep.2021.08.029. [DOI] [PubMed] [Google Scholar]
- 204.Ouaguia L, et al. Hepatitis C virus improves human tregs suppressive function and promotes their recruitment to the liver. Cells. 2019;8:1296. doi: 10.3390/cells8101296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Adurthi S, et al. Regulatory T cells in a spectrum of HPV-induced cervical lesions: cervicitis, cervical intraepithelial neoplasia and squamous cell carcinoma. Am. J. Reprod. Immunol. 2008;60:55–65. doi: 10.1111/j.1600-0897.2008.00590.x. [DOI] [PubMed] [Google Scholar]
- 206.Alam A, et al. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. 2022;40:153–167. doi: 10.1016/j.ccell.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Segal LN, et al. Anaerobic bacterial fermentation products increase tuberculosis risk in antiretroviral-drug-treated HIV patients. Cell Host. Microb. 2017;21:530–537. doi: 10.1016/j.chom.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hemmat N, Baghi HB. Human papillomavirus E5 protein, the undercover culprit of tumorigenesis. Infect. Agent Cancer. 2018;13:31. doi: 10.1186/s13027-018-0208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Liu C, et al. Increased expression of PD‑L1 by the human papillomavirus 16 E7 oncoprotein inhibits anticancer immunity. Mol. Med. Rep. 2017;15:1063–1070. doi: 10.3892/mmr.2017.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gur C, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gur C, et al. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology. 2019;8:e1581531. doi: 10.1080/2162402X.2019.1581531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Javaheri A, et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat. Microbiol. 2016;2:16189. doi: 10.1038/nmicrobiol.2016.189. [DOI] [PubMed] [Google Scholar]
- 213.Gur C, et al. The Helicobacter pylori HopQ outermembrane protein inhibits immune cell activities. Oncoimmunology. 2019;8:e1553487. doi: 10.1080/2162402X.2018.1553487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wu YY, et al. Increased programmed death-ligand-1 expression in human gastric epithelial cells in Helicobacter pylori infection. Clin. Exp. Immunol. 2010;161:551–559. doi: 10.1111/j.1365-2249.2010.04217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Holokai L, et al. Increased programmed death-ligand 1 is an early epithelial cell response to helicobacter pylori infection. PLoS Pathog. 2019;15:e1007468. doi: 10.1371/journal.ppat.1007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Song H, et al. IDO metabolite produced by EBV-transformed B cells inhibits surface expression of NKG2D in NK cells via the c-Jun N-terminal kinase (JNK) pathway. Immunol. Lett. 2011;136:187–193. doi: 10.1016/j.imlet.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 217.Liu P, et al. Expression of indoleamine 2,3-dioxygenase in nasopharyngeal carcinoma impairs the cytolytic function of peripheral blood lymphocytes. BMC Cancer. 2009;9:416. doi: 10.1186/1471-2407-9-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Partlová S, et al. Distinct patterns of intratumoral immune cell infiltrates in patients with HPV-associated compared to non-virally induced head and neck squamous cell carcinoma. Oncoimmunology. 2015;4:e965570. doi: 10.4161/21624011.2014.965570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Overacre-Delgoffe AE, et al. Microbiota-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. Immunity. 2021;54:2812–2824. doi: 10.1016/j.immuni.2021.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhang Q, et al. Lactobacillus plantarum-derived indole-3-lactic acid ameliorates colorectal tumorigenesis via epigenetic regulation of CD8(+) T cell immunity. Cell Metab. 2023;35:943–960. doi: 10.1016/j.cmet.2023.04.015. [DOI] [PubMed] [Google Scholar]
- 221.Bender MJ, et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell. 2023;186:1846–1862. doi: 10.1016/j.cell.2023.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang H, et al. The microbial metabolite trimethylamine N-oxide promotes antitumor immunity in triple-negative breast cancer. Cell Metab. 2022;34:581–594. doi: 10.1016/j.cmet.2022.02.010. [DOI] [PubMed] [Google Scholar]
- 223.Mager LF, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369:1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 224.Thomas S, et al. The host microbiome regulates and maintains human health: a primer and perspective for non-microbiologists. Cancer Res. 2017;77:1783–1812. doi: 10.1158/0008-5472.CAN-16-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hieken TJ, et al. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci. Rep. 2016;6:30751. doi: 10.1038/srep30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ilhan ZE, et al. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine. 2019;44:675–690. doi: 10.1016/j.ebiom.2019.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Dai D, et al. Interactions between gastric microbiota and metabolites in gastric cancer. Cell Death Dis. 2021;12:1104. doi: 10.1038/s41419-021-04396-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Yost S, et al. Increased virulence of the oral microbiome in oral squamous cell carcinoma revealed by metatranscriptome analyses. Int J. Oral. Sci. 2018;10:32. doi: 10.1038/s41368-018-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sun J, et al. F. nucleatum facilitates oral squamous cell carcinoma progression via GLUT1-driven lactate production. EBioMedicine. 2023;88:104444. doi: 10.1016/j.ebiom.2023.104444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Berrios C, et al. Merkel cell polyomavirus small T antigen promotes pro-glycolytic metabolic perturbations required for transformation. PLoS Pathog. 2016;12:e1006020. doi: 10.1371/journal.ppat.1006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Carroll PA, Kenerson HL, Yeung RS, Lagunoff M. Latent Kaposi’s sarcoma-associated herpesvirus infection of endothelial cells activates hypoxia-induced factors. J. Virol. 2006;80:10802–10812. doi: 10.1128/JVI.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Singh RK, Lang F, Pei Y, Jha HC, Robertson ES. Metabolic reprogramming of Kaposi’s sarcoma associated herpes virus infected B-cells in hypoxia. PLoS Pathog. 2018;14:e1007062. doi: 10.1371/journal.ppat.1007062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bhatt AP, Damania B. AKTivation of PI3K/AKT/mTOR signaling pathway by KSHV. Front. Immunol. 2012;3:401. doi: 10.3389/fimmu.2012.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Munger J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zwerschke W, et al. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proc. Natl Acad. Sci. USA. 1999;96:1291–1296. doi: 10.1073/pnas.96.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lévy P, Bartosch B. Metabolic reprogramming: a hallmark of viral oncogenesis. Oncogene. 2016;35:4155–4164. doi: 10.1038/onc.2015.479. [DOI] [PubMed] [Google Scholar]
- 237.Fu A, Yao B, Dong T, Cai S. Emerging roles of intratumor microbiota in cancer metastasis. Trends Cell Biol. 2023;33:583–593. doi: 10.1016/j.tcb.2022.11.007. [DOI] [PubMed] [Google Scholar]
- 238.Welch DR, Hurst DR. Defining the hallmarks of metastasis. Cancer Res. 2019;79:3011–3027. doi: 10.1158/0008-5472.CAN-19-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct. Target Ther. 2020;5:28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 241.Vergara D, et al. The cancer microbiota: EMT and inflammation as shared molecular mechanisms associated with plasticity and progression. J. Oncol. 2019;2019:1253727. doi: 10.1155/2019/1253727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wu Y, et al. Fn-Dps, a novel virulence factor of Fusobacterium nucleatum, disrupts erythrocytes and promotes metastasis in colorectal cancer. PLoS Pathog. 2023;19:e1011096. doi: 10.1371/journal.ppat.1011096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Li WT, et al. The bladder microbiome is associated with epithelial-mesenchymal transition in muscle invasive urothelial bladder carcinoma. Cancers. 2021;13:3649. doi: 10.3390/cancers13153649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lee J, et al. Human primary epithelial cells acquire an epithelial-mesenchymal-transition phenotype during long-term infection by the oral opportunistic pathogen, porphyromonas gingivalis. Front. Cell Infect. Microbiol. 2017;7:493. doi: 10.3389/fcimb.2017.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Bessède E, et al. Helicobacter pylori generates cells with cancer stem cell properties via epithelial-mesenchymal transition-like changes. Oncogene. 2014;33:4123–4131. doi: 10.1038/onc.2013.380. [DOI] [PubMed] [Google Scholar]
- 246.Parida S, et al. A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and β-Catenin axes. Cancer Discov. 2021;11:1138–1157. doi: 10.1158/2159-8290.CD-20-0537. [DOI] [PubMed] [Google Scholar]
- 247.Zhong X, Rescorla FJ. Cell surface adhesion molecules and adhesion-initiated signaling: understanding of anoikis resistance mechanisms and therapeutic opportunities. Cell Signal. 2012;24:393–401. doi: 10.1016/j.cellsig.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 248.Zhang Y, et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-κB/ICAM1 axis. Gut. Microbe. 2022;14:2038852. doi: 10.1080/19490976.2022.2038852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Bertocchi A, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell. 2021;39:708–724. doi: 10.1016/j.ccell.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 250.Shang A, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol. Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Channon LM, et al. Small extracellular vesicles (exosomes) and their cargo in pancreatic cancer: key roles in the hallmarks of cancer. Biochim. Biophys. Acta. Rev. Cancer. 2022;1877:188728. doi: 10.1016/j.bbcan.2022.188728. [DOI] [PubMed] [Google Scholar]
- 252.Li G, et al. Fusobacterium nucleatum-derived small extracellular vesicles facilitate tumor growth and metastasis via TLR4 in breast cancer. BMC Cancer. 2023;23:473. doi: 10.1186/s12885-023-10844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Yamamura K, et al. Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin. Cancer Res. 2016;22:5574–5581. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- 254.Mima K, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hsieh YY, Kuo WL, Hsu WT, Tung SY, Li C. Fusobacterium nucleatum-induced tumor mutation burden predicts poor survival of gastric cancer patients. Cancers. 2022;15:269. doi: 10.3390/cancers15010269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Rustetska N, et al. The intratumour microbiota and neutrophilic inflammation in squamous cell vulvar carcinoma microenvironment. J. Transl. Med. 2023;21:285. doi: 10.1186/s12967-023-04113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Mitsuhashi K, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209–7220. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Neuzillet C, et al. Prognostic value of intratumoral Fusobacterium nucleatum and association with immune-related gene expression in oral squamous cell carcinoma patients. Sci. Rep. 2021;11:7870. doi: 10.1038/s41598-021-86816-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hilmi M, et al. Prognostic value of Fusobacterium nucleatum after abdominoperineal resection for anal squamous cell carcinoma. Cancers. 2022;14:1606. doi: 10.3390/cancers14071606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Alexander JL, et al. Pathobionts in the tumour microbiota predict survival following resection for colorectal cancer. Microbiome. 2023;11:100. doi: 10.1186/s40168-023-01518-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Peters BA, et al. The lung microbiome, peripheral gene expression, and recurrence-free survival after resection of stage II non-small cell lung cancer. Genome Med. 2022;14:121. doi: 10.1186/s13073-022-01126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zhang M, et al. Intratumoral microbiota impacts the first-line treatment efficacy and survival in non-small cell lung cancer patients free of lung infection. J. Health. Eng. 2022;2022:5466853. doi: 10.1155/2022/5466853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Yu G, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17:163. doi: 10.1186/s13059-016-1021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Riquelme E, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178:795–806. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kerdreux M, et al. Porphyromonas gingivalis in colorectal cancer and its association to patient prognosis. J. Cancer. 2023;14:1479–1485. doi: 10.7150/jca.83395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Perry LM, et al. Human soft tissue sarcomas harbor an intratumoral viral microbiome which is linked with natural killer cell infiltrate and prognosis. J. Immunother. Cancer. 2023;11:e004285. doi: 10.1136/jitc-2021-004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sun L, et al. Intratumoural microbiome can predict the prognosis of hepatocellular carcinoma after surgery. Clin. Transl. Med. 2023;13:e1331. doi: 10.1002/ctm2.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Song Y, et al. Identification of a brand intratumor microbiome signature for predicting prognosis of hepatocellular carcinoma. J. Cancer Res Clin. Oncol. 2023;149:11319–11332. doi: 10.1007/s00432-023-04962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yamamura K, et al. Intratumoral Fusobacterium nucleatum levels predict therapeutic response to neoadjuvant chemotherapy in esophageal squamous cell carcinoma. Clin. Cancer Res. 2019;25:6170–6179. doi: 10.1158/1078-0432.CCR-19-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Kang SY, et al. Helicobacter pylori infection as an independent prognostic factor for locally advanced gastric cancer patients treated with adjuvant chemotherapy after curative resection. Int J. Cancer. 2012;130:948–958. doi: 10.1002/ijc.26081. [DOI] [PubMed] [Google Scholar]
- 271.Marrelli D, et al. Negative Helicobacter pylori status is associated with poor prognosis in patients with gastric cancer. Cancer. 2009;115:2071–2080. doi: 10.1002/cncr.24253. [DOI] [PubMed] [Google Scholar]
- 272.Nishizuka SS, et al. Helicobacter pylori infection is associated with favorable outcome in advanced gastric cancer patients treated with S-1 adjuvant chemotherapy. J. Surg. Oncol. 2018;117:947–956. doi: 10.1002/jso.24977. [DOI] [PubMed] [Google Scholar]
- 273.Tonneau M, et al. Helicobacter pylori serology is associated with worse overall survival in patients with melanoma treated with immune checkpoint inhibitors. Oncoimmunology. 2022;11:2096535. doi: 10.1080/2162402X.2022.2096535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Oster P, et al. Helicobacter pylori infection has a detrimental impact on the efficacy of cancer immunotherapies. Gut. 2022;71:457–466. doi: 10.1136/gutjnl-2020-323392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Che H, et al. Association of Helicobacter pylori infection with survival outcomes in advanced gastric cancer patients treated with immune checkpoint inhibitors. BMC Cancer. 2022;22:904. doi: 10.1186/s12885-022-10004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Cammarota G, et al. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 2020;17:635–648. doi: 10.1038/s41575-020-0327-3. [DOI] [PubMed] [Google Scholar]
- 277.Karlsson FH, Nookaew I, Petranovic D, Nielsen J. Prospects for systems biology and modeling of the gut microbiome. Trends Biotechnol. 2011;29:251–258. doi: 10.1016/j.tibtech.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 278.Ding Z, et al. Novel scheme for non-invasive gut bioinformation acquisition with a magnetically controlled sampling capsule endoscope. Gut. 2021;70:2297–2306. doi: 10.1136/gutjnl-2020-322465. [DOI] [PubMed] [Google Scholar]
- 279.Jackson SE, Chester JD. Personalised cancer medicine. Int J. Cancer. 2015;137:262–266. doi: 10.1002/ijc.28940. [DOI] [PubMed] [Google Scholar]
- 280.Fallone CA, et al. The toronto consensus for the treatment of helicobacter pylori infection in adults. Gastroenterology. 2016;151:51–69.e14. doi: 10.1053/j.gastro.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 281.Choi IJ, et al. Family history of gastric cancer and helicobacter pylori treatment. N. Engl. J. Med. 2020;382:427–436. doi: 10.1056/NEJMoa1909666. [DOI] [PubMed] [Google Scholar]
- 282.Stolte M, et al. Helicobacter and gastric MALT lymphoma. Gut. 2002;50:III19–24. doi: 10.1136/gut.50.suppl_3.iii19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Roche B, Coilly A, Duclos-Vallee JC, Samuel D. The impact of treatment of hepatitis C with DAAs on the occurrence of HCC. Liver Int. 2018;38:139–145. doi: 10.1111/liv.13659. [DOI] [PubMed] [Google Scholar]
- 284.Calvaruso V, et al. Incidence of hepatocellular carcinoma in patients with HCV-associated cirrhosis treated with direct-acting antiviral agents. Gastroenterology. 2018;155:411–421.e414. doi: 10.1053/j.gastro.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 285.Lowy DR, Schiller JT. Preventing cancer and other diseases caused by human papillomavirus infection: 2017 Lasker-DeBakey clinical research award. Jama. 2017;318:901–902. doi: 10.1001/jama.2017.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.McAleer WJ, et al. Human hepatitis B vaccine from recombinant yeast. Nature. 1984;307:178–180. doi: 10.1038/307178a0. [DOI] [PubMed] [Google Scholar]
- 287.Yu T, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–563.e516. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Mohindroo C, et al. Antibiotic use influences outcomes in advanced pancreatic adenocarcinoma patients. Cancer Med. 2021;10:5041–5050. doi: 10.1002/cam4.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Bullman S, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Bernardo G, et al. Reduction of Staphylococcus epidermidis in the mammary tumor microbiota induces antitumor immunity and decreases breast cancer aggressiveness. Cancer Lett. 2023;555:216041. doi: 10.1016/j.canlet.2022.216041. [DOI] [PubMed] [Google Scholar]
- 291.Lurienne L, et al. NSCLC immunotherapy efficacy and antibiotic use: a systematic review and meta-analysis. J. Thorac. Oncol. 2020;15:1147–1159. doi: 10.1016/j.jtho.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 292.Jing Y, et al. Association of antibiotic treatment with immune-related adverse events in patients with cancer receiving immunotherapy. J. Immunother. Cancer. 2022;10:e003779. doi: 10.1136/jitc-2021-003779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Stein-Thoeringer CK, et al. A non-antibiotic-disrupted gut microbiome is associated with clinical responses to CD19-CAR-T cell cancer immunotherapy. Nat. Med. 2023;29:906–916. doi: 10.1038/s41591-023-02234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Chambers LM, et al. Disruption of the gut microbiota confers cisplatin resistance in epithelial ovarian cancer. Cancer Res. 2022;82:4654–4669. doi: 10.1158/0008-5472.CAN-22-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Gao C, et al. Synergistic target of intratumoral microbiome and tumor by metronidazole-fluorouridine nanoparticles. ACS Nano. 2023;17:7335–7351. doi: 10.1021/acsnano.2c11305. [DOI] [PubMed] [Google Scholar]
- 296.Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016;4:10. doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Boursi B, Mamtani R, Haynes K, Yang YX. Recurrent antibiotic exposure may promote cancer formation-another step in understanding the role of the human microbiota? Eur. J. Cancer. 2015;51:2655–2664. doi: 10.1016/j.ejca.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Dicks LMT, Mikkelsen LS, Brandsborg E, Marcotte H. Clostridium difficile, the difficult “Kloster” fuelled by antibiotics. Curr. Microbiol. 2019;76:774–782. doi: 10.1007/s00284-018-1543-8. [DOI] [PubMed] [Google Scholar]
- 299.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J. Antimicrob. Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 300.Kabwe M, Dashper S, Bachrach G, Tucci J. Bacteriophage manipulation of the microbiome associated with tumour microenvironments-can this improve cancer therapeutic response? FEMS Microbiol. Rev. 2021;45:fuab017. doi: 10.1093/femsre/fuab017. [DOI] [PubMed] [Google Scholar]
- 301.Dolgin E. Fighting cancer with microbes. Nature. 2020;577:S16–S18. doi: 10.1038/d41586-020-00199-x. [DOI] [PubMed] [Google Scholar]
- 302.Zheng DW, et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 2019;3:717–728. doi: 10.1038/s41551-019-0423-2. [DOI] [PubMed] [Google Scholar]
- 303.Gogokhia L, et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host. Microb. 2019;25:285–299. doi: 10.1016/j.chom.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Sinha A, et al. Transplantation of bacteriophages from ulcerative colitis patients shifts the gut bacteriome and exacerbates the severity of DSS colitis. Microbiome. 2022;10:105. doi: 10.1186/s40168-022-01275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Malik DJ, et al. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv. Colloid Inter. Sci. 2017;249:100–133. doi: 10.1016/j.cis.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 306.Hatfull GF, Dedrick RM, Schooley RT. Phage therapy for antibiotic-resistant bacterial infections. Annu Rev. Med. 2022;73:197–211. doi: 10.1146/annurev-med-080219-122208. [DOI] [PubMed] [Google Scholar]
- 307.Chang RYK, Nang SC, Chan HK, Li J. Novel antimicrobial agents for combating antibiotic-resistant bacteria. Adv. Drug Deliv. Rev. 2022;187:114378. doi: 10.1016/j.addr.2022.114378. [DOI] [PubMed] [Google Scholar]
- 308.Riglar DT, Silver PA. Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 2018;16:214–225. doi: 10.1038/nrmicro.2017.172. [DOI] [PubMed] [Google Scholar]
- 309.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer. 2010;10:785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Nemunaitis J, et al. Pilot trial of genetically modified, attenuated Salmonella expressing the E. coli cytosine deaminase gene in refractory cancer patients. Cancer Gene. Ther. 2003;10:737–744. doi: 10.1038/sj.cgt.7700634. [DOI] [PubMed] [Google Scholar]
- 311.Tang W, He Y, Zhou S, Ma Y, Liu G. A novel Bifidobacterium infantis-mediated TK/GCV suicide gene therapy system exhibits antitumor activity in a rat model of bladder cancer. J. Exp. Clin. Cancer Res. 2009;28:155. doi: 10.1186/1756-9966-28-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Murphy C, Rettedal E, Lehouritis P, Devoy C, Tangney M. Intratumoural production of TNFα by bacteria mediates cancer therapy. PLoS One. 2017;12:e0180034. doi: 10.1371/journal.pone.0180034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Jiang SN, et al. Inhibition of tumor growth and metastasis by a combination of Escherichia coli-mediated cytolytic therapy and radiotherapy. Mol. Ther. 2010;18:635–642. doi: 10.1038/mt.2009.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Jiang SN, et al. Engineering of bacteria for the visualization of targeted delivery of a cytolytic anticancer agent. Mol. Ther. 2013;21:1985–1995. doi: 10.1038/mt.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Canale FP, et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature. 2021;598:662–666. doi: 10.1038/s41586-021-04003-2. [DOI] [PubMed] [Google Scholar]
- 316.Zheng JH, et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 2017;9:eaak9537. doi: 10.1126/scitranslmed.aak9537. [DOI] [PubMed] [Google Scholar]
- 317.Singh M, et al. Direct incorporation of the NKT-cell activator α-galactosylceramide into a recombinant Listeria monocytogenes improves breast cancer vaccine efficacy. Br. J. Cancer. 2014;111:1945–1954. doi: 10.1038/bjc.2014.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Ebelt ND, Zuniga E, Passi KB, Sobocinski LJ, Manuel ER. Hyaluronidase-expressing Salmonella effectively targets tumor-associated hyaluronic acid in pancreatic ductal adenocarcinoma. Mol. Cancer Ther. 2020;19:706–716. doi: 10.1158/1535-7163.MCT-19-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Nakatsuji T, et al. A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci. Adv. 2018;4:eaao4502. doi: 10.1126/sciadv.aao4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Fukuhara H, Ino Y, Todo T. Oncolytic virus therapy: a new era of cancer treatment at dawn. Cancer Sci. 2016;107:1373–1379. doi: 10.1111/cas.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Parato KA, et al. The oncolytic poxvirus JX-594 selectively replicates in and destroys cancer cells driven by genetic pathways commonly activated in cancers. Mol. Ther. 2012;20:749–758. doi: 10.1038/mt.2011.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kalkavan H, et al. Spatiotemporally restricted arenavirus replication induces immune surveillance and type I interferon-dependent tumour regression. Nat. Commun. 2017;8:14447. doi: 10.1038/ncomms14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Chen L, et al. Intratumoral expression of interleukin 23 variants using oncolytic vaccinia virus elicit potent antitumor effects on multiple tumor models via tumor microenvironment modulation. Theranostics. 2021;11:6668–6681. doi: 10.7150/thno.56494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Boyero L, et al. Primary and acquired resistance to immunotherapy in lung cancer: unveiling the mechanisms underlying of immune checkpoint blockade therapy. Cancers Basel. 2020;12:3729. doi: 10.3390/cancers12123729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Liu Z, Ravindranathan R, Kalinski P, Guo ZS, Bartlett DL. Rational combination of oncolytic vaccinia virus and PD-L1 blockade works synergistically to enhance therapeutic efficacy. Nat. Commun. 2017;8:14754. doi: 10.1038/ncomms14754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Shalhout SZ, Miller DM, Emerick KS, Kaufman HL. Therapy with oncolytic viruses: progress and challenges. Nat. Rev. Clin. Oncol. 2023;20:160–177. doi: 10.1038/s41571-022-00719-w. [DOI] [PubMed] [Google Scholar]
- 327.Liu Y, et al. Intravenous injection of the oncolytic virus M1 awakens antitumor T cells and overcomes resistance to checkpoint blockade. Cell Death Dis. 2020;11:1062. doi: 10.1038/s41419-020-03285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Nosaki K, et al. A novel, polymer-coated oncolytic measles virus overcomes immune suppression and induces robust antitumor activity. Mol. Ther. Oncol. 2016;3:16022. doi: 10.1038/mto.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Draganov DD, et al. Delivery of oncolytic vaccinia virus by matched allogeneic stem cells overcomes critical innate and adaptive immune barriers. J. Transl. Med. 2019;17:100. doi: 10.1186/s12967-019-1829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Minev BR, et al. First-in-human study of TK-positive oncolytic vaccinia virus delivered by adipose stromal vascular fraction cells. J. Transl. Med. 2019;17:271. doi: 10.1186/s12967-019-2011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Song D, et al. Identification of the receptor of oncolytic virus M1 as a therapeutic predictor for multiple solid tumors. Sign. Transduct. Target Ther. 2022;7:100. doi: 10.1038/s41392-022-00921-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Vétizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Routy B, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 336.Davar D, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Baruch EN, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 338.Dizman N, et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: a randomized phase 1 trial. Nat. Med. 2022;28:704–712. doi: 10.1038/s41591-022-01694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Fluckiger A, et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science. 2020;369:936–942. doi: 10.1126/science.aax0701. [DOI] [PubMed] [Google Scholar]
- 340.Spencer CN, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science. 2021;374:1632–1640. doi: 10.1126/science.aaz7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Zegarra-Ruiz DF, et al. A diet-sensitive commensal lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host. Microb. 2019;25:113–127. doi: 10.1016/j.chom.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Pandey SP, et al. Tet2 deficiency drives liver microbiome dysbiosis triggering Tc1 cell autoimmune hepatitis. Cell Host. Microb. 2022;30:1003–1019. doi: 10.1016/j.chom.2022.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Behnsen J, Deriu E, Sassone-Corsi M, Raffatellu M. Probiotics: properties, examples, and specific applications. Cold Spring Harb. Perspect. Med. 2013;3:a010074. doi: 10.1101/cshperspect.a010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat. Med. 2019;25:716–729. doi: 10.1038/s41591-019-0439-x. [DOI] [PubMed] [Google Scholar]
- 345.Bullman S. The intratumoral microbiota: from microniches to single cells. Cell. 2023;186:1532–1534. doi: 10.1016/j.cell.2023.03.012. [DOI] [PubMed] [Google Scholar]