Abstract
The timing of mammalian diversification in relation to the Cretaceous-Paleogene (KPg) mass extinction continues to be a subject of substantial debate. Previous studies have either focused on limited taxonomic samples with available whole-genome data or relied on short sequence alignments coupled with extensive species samples. In the present study, we improved an existing dataset from the landmark study of Meredith et al. (2011) by filling in missing fragments and further generated another dataset containing 120 taxa and 98 exonic markers. Using these two datasets, we then constructed phylogenies for extant mammalian families, providing improved resolution of many conflicting relationships. Moreover, the timetrees generated, which were calibrated using appropriate molecular clock models and multiple fossil records, indicated that the interordinal diversification of placental mammals initiated before the Late Cretaceous period. Additionally, intraordinal diversification of both extant placental and marsupial lineages accelerated after the KPg boundary, supporting the hypothesis that the availability of numerous vacant ecological niches subsequent to the mass extinction event facilitated rapid diversification. Thus, our results support a scenario of placental radiation characterized by both basal cladogenesis and active interordinal divergences spanning from the Late Cretaceous into the Paleogene.
Keywords: Mammals, Phylogeny, Diversification, Evolutionary model
INTRODUCTION
Considerable efforts have been made to decipher higher-level relationships among extant mammalian orders in recent decades (Hu et al., 2012; Liu et al., 2017; McCormack et al., 2012; Meredith et al., 2011; Springer et al., 2003; Tarver et al., 2016), resulting in the categorization of placental mammals into four superordinal groups: Laurasiatheria, Euarchontoglires, Xenarthra, and Afrotheria. Recent advances in genomic resources have enabled extensive investigation of several interordinal controversies regarding the positions of Scandentia (tree shrews), Perissodactyla (horses, rhinoceroses), Artiodactyla (cows, dolphins), Carnivora (dogs, cats), and Chiroptera (bats) (Doronina et al., 2017a; Lv et al., 2021; Murphy et al., 2021; Zhou et al., 2012). Similarly, phylogenetic histories at the genus level have also been intensively studied (Guo et al., 2021; He et al., 2018; Missoup et al., 2018; Wildman et al., 2009). Nonetheless, family-level phylogenetic relationships within the placental mammalian tree have received less attention and remain inadequately resolved. In their landmark study, Meredith et al. (2011) examined relationships and divergence times among all living mammalian families utilizing a molecular supermatrix containing 26 gene fragments from 164 mammals and five outgroups, yielding the most comprehensive family-level mammalian tree to date. They further posited that interordinal diversification occurred after the Cretaceous Terrestrial Revolution (KTR), whereas intraordinal diversification occurred across the Cretaceous-Paleogene (KPg) boundary. Nevertheless, their supermatrix exhibited some limitations due to the occurrence of nonrandom gaps and missing data—for example, the family Calomyscidae was represented by only four gene fragments—which may have introduced biases. Emerling et al. (2015) and Foley et al. (2016) later provided updated species-level supermatrices based on the work of Meredith et al. (2011) but did not rectify the data gaps. Upham et al. (2019) constructed the most expansive species-level phylogeny for extant mammals to date, employing “backbone-and-patch” tree methodology, similar to the supertree approach, but did not extend their study to detailing low-support phylogenetic relationships, such as the patch clade of Emballonuroidea and Vespertilionoidea. More recently, Álvarez-Carretero et al. (2022) created a species-level mammalian timetree by integrating phylogenomic data across 13 subtree partitions, although each subtree partition alignment exhibited more than 45% missing nuclear sites. In parallel, Foley et al. (2023) employed 241 placental genomes to estimate phylogenetic relationships and divergence time, albeit with limited representation of mammalian families.
In addition to resolving phylogenetic controversies, determining divergence times among all mammalian families is crucial to ascertain whether placental radiation occurred before or after the KPg boundary, which marks the extinction of nonavian dinosaurs and many other Mesozoic vertebrates (Archibald & Deutschman, 2001; dos Reis et al., 2012, 2014; Grossnickle & Newham, 2016; Song et al., 2012; Springer et al., 2003; Wible et al., 2007). For many mammalian species, the KPg mass extinction event created ecological and evolutionary opportunities that were previously unattainable. However, recent research has suggested that mammalian diversification did not occur uniformly or universally across the KPg boundary, but rather was limited to certain placental orders, including Artiodactyla and Chiroptera (Bininda-Emonds et al., 2007). Consequently, three models have been proposed to characterize the evolutionary history of placental crown groups (Archibald & Deutschman, 2001; Springer et al., 2019): (i) explosive model, which contends that placental origins and diversification transpired after the KPg mass extinction (Phillips, 2016); (ii) short fuse model, which posits that both interordinal and intraordinal cladogenesis occurred before the KPg boundary, indicating deeper temporal roots (Bininda-Emonds et al., 2007; Puttick et al., 2016); and (iii) long fuse model, which proposes that placental origins took place the Late Cretaceous and that interordinal diversification primarily occurred before the KPg boundary, while intraordinal diversification primarily occurred after the boundary (Foley et al., 2016; Springer et al., 2003). More recently, Liu et al. (2017) proposed the trans-KPg model, which posits that interordinal diversification began in the Late Cretaceous and continued into the early Cenozoic without interruption by the KPg boundary, while intraordinal diversification followed rapidly during the Paleogene. Additionally, the explosive radiation of angiosperms (Dilcher, 2000) and insects (Magallón et al., 2015) during the Cretaceous Terrestrial Revolution (KTR, 125 to 80 million years ago, Ma) raises questions regarding whether similar radiation events occurred at the mammalian order level. As a result, mammalian diversification patterns and phylogenetics at the order level remain inadequately understood. To resolve these uncertainties, taxonomic sampling at the family level is considered sufficient. Hence, in the current study, we aimed to construct a robust phylogeny at the family level for extant mammals by generating new multi-sequence alignments for a majority of mammalian families, thereby facilitating a more comprehensive understanding of their evolutionary history and patterns of radiation.
MATERIALS AND METHODS
Taxonomic sampling and dataset construction
All animal care and research protocols were approved by the Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Sciences, China (approval No. IOZ-IACUC-2021-129). For the purposes of this study, we generated two distinct datasets for mammalian phylogenetic analysis. Dataset A comprised 162 mammalian lineages, including 141 single species and 21 chimerics, and five nonmammalian outgroups: Danio rerio (zebrafish), Xenopus tropicalis (African clawed frog), Anolis carolinensis (green anole), Gallus gallus (chicken), and Taeniopygia guttata (house finch). The primary data were downloaded from the National Center for Biotechnology Information (NCBI) database according to the GenBank accession numbers cited in Meredith et al. (2011) and consisted of 21 exonic segments (A2AB, ADORA3, ADRB2, ApoB, ATP7A, BCHE, BNDF, BRCA1, BRCA2, CNR1, DMP1, EDG1, ENAM, GHR, IRBP, PNOC, RAG1, RAG2, TTN, TYR1, and vWF) and five noncoding regions (APP, BMI1, CREM, FBN1, and PLCB4). To fill the gaps in the primary data, orthologous sequences were extracted from available genome sequences using local BLAST v2.10.0+ (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/~LATEST/) with the parameter “-evalue 1e-5” and human DNA sequences serving as the query. For each family represented in the dataset, a single species was deemed sufficient for sequence retrieval. Custom Perl scripts (m6_file.pl and m6_get_fa.pl) were used to extract the best hit sequence from the BLAST output files. The remaining gaps were filled by retrieving 115 DNA sequences from available genomes and six sequences from the NCBI nucleotide database (Supplementary Table S1). Dataset B included 120 mammalian species, 14 of which were subjected to genomic DNA extraction using QIAGEN DNeasy Blood & Tissue Kit (Cat. 69504, Germany). Species identification was conducted following Ivanova et al. (2012) using the DNA barcoding primer sets dgLCO-1490/dgHCO-2198 (Meyer, 2003). Sample information is provided in Supplementary Table S2. Nondestructive blood sampling was conducted for Nycticebus sp. (slow loris) and Paguma larvata (masked palm civet), while the remaining 12 muscle samples were obtained from deceased or dying individuals. All sampling protocols, including nondestructive sampling, abided by the relevant ethical and legal regulations and guidelines. Dataset B contained 98 exonic markers, including 32 newly developed primers in this research and 66 primers developed by Zhou et al. (2012). Details on the primer sequences, annealing temperatures for polymerase chain reaction (PCR) amplification, and PCR conditions are provided in Supplementary Table S3. Finally, 9 474 exonic fragments were retrieved from 106 genomic datasets using localized BLAST searches and custom Perl scripts. Additionally, 964 exonic sequences from 14 species listed in Supplementary Table S2 were submitted to GenBank (accession numbers OR503106–OR504069). The custom Perl scripts and sequence alignments are available in the Dryad dataset (DOI: https://doi.org/10.5061/dryad.crjdfn36w).
Data comparison
We initially downloaded dataset information, including sequence IDs, from the supplementary files provided by Meredith et al. (2011), Upham et al. (2019), and Álvarez-Carretero et al. (2022). Because the dataset in Álvarez-Carretero et al. (2022) lacked corresponding family and order information for each taxon, we used NCBItaxa in the Python package “ete3” (Huerta-Cepas et al., 2016) for family and order identification, with manual determination for those taxa not found in NCBItaxa. Subsequently, we developed custom Perl scripts to obtain information on nuclear gene fragment gaps in each dataset, with a gene fragment considered present in a family if at least one species within that family had the corresponding gene ID information. Furthermore, as gene fragment lengths varied among species, we adopted the gene fragment length of humans (Homo sapiens) before alignment. Although Álvarez-Carretero et al. (2022) provided the gene Ensembl IDs, they did not specify fragment lengths; thus, we employed the maximum length across all transcripts for these genes. Gap percentages were then calculated by dividing the number of gaps in the alignment by the number of taxa times the number of gene fragments. Finally, the comparison of gene fragment gap data was visualized using the R package “ComplexHeatmap” (Gu et al., 2016).
Alignment simulation
An increase in gene fragment gaps in alignments is often correlated with an increase in missing sites, assuming the length of each gene sequence is the same or extremely similar. To investigate the effects of missing sites on phylogenetic inference, using a maximum-likelihood (ML) tree as an example, we simulated sequence alignments from the OrthoMaM database (Scornavacca et al., 2019). We selected autosomal orthologous nuclear markers based on the following parameters: “relative evolution rate: 0–1.3; %GC: 40.0–60.0; α shape (Γ distribution): 1.0–5.3 and genes present in at least one or more species”. A total of 108 filtered coding sequences met these criteria and were concatenated (Supplementary Data). Subsequently, we generated concatenated alignments for 100 species, retaining one representative species at the genus level to infer the optimal ML tree using IQ-TREE v2.2.0 (Minh et al., 2020) under the substitution model “GTR+G”.
In recent phylogenetic studies, datasets have commonly consisted of either relatively short sequence lengths and hundreds of species or very large lengths (millions of base pairs (bp) but limited to several dozen taxa. Accordingly, we simulated sequence alignments with variable numbers of taxa (20, 60, and 100) and sequence lengths (1k, 10k, and 100k). Subtrees containing 20 and 60 taxa were extracted from the optimal ML topology of 100 taxa to serve as the simulated original tree. Sequence alignments without missing sites were simulated using IQ-TREE v2.2.0 (Minh et al., 2020) under the substitution model “GTR+G” and original trees containing 20, 60, and 100 taxa, with alignment length set with the parameter “-length 1k/10k/100k”. Subsequently, random deletions were applied to 20%, 40%, and 80% of alignment matrices using a custom Python script. All simulated alignments were then used to infer the ML tree with the consistent simulated substitution model and 1 000 bootstrap (BS) replicates. IQ-TREE v2.2.0 (Minh et al., 2020) was used to evaluate the Robinson-Foulds (RF) distance between the simulated topology and the original tree. A Python script was used to compute the mean BS value of the optimal ML tree. Finally, RF distances and mean BS values were visualized using the R package “ggplot2” (Wickham et al., 2016).
Sequence alignment
The DNA sequences from Dataset A, used to fill the gaps in the alignments of Meredith et al. (2011), are shown in Supplementary Table S3. These sequences were aligned to the original alignments in Dataset A using MAFFT v7.471 (Katoh et al., 2002), preserving the lengths of the original alignments. In Dataset B, the sequencing base peak profiles were verified prior to alignment using Chromas software (http://technelysium.com.au/wp/chromas/). The multiple sequence alignments (MSAs) of the DNA coding sequences in Dataset B were realigned using OMM_MACSE v10.01 (https://github.com/ranwez/MACSE_V2_PIPELINES) with default settings (Delsuc & Ranwez, 2020). The OMM_MACSE pipeline encompassed a series of operations, including translation of nucleotide sequences into amino acids, detection of frameshifts (using MACSE v2.03) (Ranwez et al., 2018), alignment of amino acid sequences (using MAFFT v7.471) (Katoh et al., 2002), filtering of resulting alignments (using HmmCleaner v1.8) (Di Franco et al., 2019), and post-filtering (using MACSE v2.03) (Ranwez et al., 2018).
Model selection
The heterogeneity in substitution rates across various coding/noncoding sequences, genes, and codon positions can impact both optimal substitution model selection and phylogenetic tree inference. For nucleotide alignments in Dataset A, the following partitioning strategies were employed: ① concatenation; ② coding+noncoding; ③ gene; ④ codon1/2+codon3+noncoding; ⑤ gene+codon1/2+codon3+noncoding; ⑥ optimized partition determined by PartitionFinder2 (Lanfear et al., 2017) according to corrected Akaike information criterion (AICc). For amino acid alignments in Dataset A, the following strategies were used: ① concatenation; ② gene; ③ optimized partition determined by PartitionFinder2 according to AICc. For nucleotide alignments in Dataset B, the following partitioning strategies were used: ① concatenation; ② gene; ③ codon1/2+codon3; ④ gene+codon1/2+codon3; ⑤ optimized partition determined by PartitionFinder2 according to AICc. For amino acid alignments in Dataset B, the following strategies were applied: ① concatenation; ② gene; ③ optimized partition determined by PartitionFinder2 according to AICc. The best-fit substitution models of nucleotides and amino acids were chosen using IQ-TREE v2.2.0 (Minh et al., 2020), as suggested by Bayesian information criterion. The model selected included both a proportion of invariant sites and a gamma distribution to account for rate heterogeneity. If the model suggested by ModelFinder was not supported in either RAxML-ng v1.0.3 (Kozlov et al., 2019) or MrBayes v3.2.7a (Ronquist et al., 2012), a suboptimal model was selected among the models available.
Phylogenetic tree analyses
Bayesian phylogenetic analyses were carried out using MrBayes v3.2.7a (Ronquist et al., 2012). Two independent Markov chain Monte Carlo (MCMC) sets were run, each with three heated chains and one cold chain and 50 000 000 and 10 000 000 generations for nucleotide and amino acid alignments, respectively. Trees were separately sampled for 5 000 and 1 000 generations, and convergence was confirmed with Tracer v1.7.2 (https://github.com/beast-dev/tracer/releases/tag/v1.7.2). RAxML-ng v1.0.3 (Kozlov et al., 2019) was used for ML analyses and run for 500 BS replicates while searching for the best-scoring ML tree. The difference in gene-wise log-likelihood scores (△GLS) was calculated, known as partitioned-likelihood support (Lee & Hugall, 2003) among hypothetical topologies using the equation used in Shen et al. (2017). The obtained △GLS values were plotted using the “ggplot2” package in R (Wickham et al., 2016). Approximately unbiased (AU) tests (Shimodaira, 2002) were conducted in Consel v0.1 (Shimodaira & Hasegawa, 2001) to evaluate the likelihood ratio difference test between the best ML tree and alternative topologies. The Shimodaira-Hasegawa (SH) test (Shimodaira & Hasegawa, 1999) was performed to measure the congruence between single-gene trees. Site-wise log-likelihood values for the hypothetical topologies were estimated with the substitution model “GTR+GAMMA” (Yang, 1994) using RAxML v8.2.12 (option -f G) (Stamatakis, 2014).
Guanine-cytosine (GC)-rich bias
The GC content of each gene alignment was determined using PhyKIT v1.5.1 (Steenwyk et al., 2021). Due to the presence of degenerate bases at the third codon position, which could potentially introduce bias into GC content calculations, we specifically gauged the GC content at the third codon position for exons, named GC3. Average GC content in human genomes ranges from 35% to 60% across 100 kb fragments (International Human Genome Sequencing Consortium, 2001). Here, alignments exhibiting a GC content exceeding 60% were categorized as GC-rich genes.
Species tree analyses
Coalescence-based species trees were generated with ASTRAL v5.7.3 (Zhang et al., 2017). Support for each node was evaluated using local posterior probabilities assigned by calculating the proportion of gene trees containing each node after collapsing the gene tree nodes with SH-like test scores <80. Moreover, the quartet frequencies were checked for controversial branching using DiscoVista (Discordance Visualization Tool) (Sayyari et al., 2018).
Molecular clock test
Molecular substitution was evaluated under a strict clock, relaxed clock with independent-rate (IR) and autocorrelated-rate (AR) models using the “mcmc3r” package in R (dos Reis et al., 2018). The MCMCTree program in PAML v4.9j (Yang, 2007) was used to calculate Bayes factors (BF) and posterior model probabilities assuming equal prior model probabilities. Marginal likelihoods were calculated by sampling trees from power-posteriors using the stepping-stone method, with β values selected using mcmc3r::make.beta (n=8, a=5, methods=“step-stones”). Due to the computational intensity of marginal likelihood calculations for large sequence alignments, shorter MCMC chains were run, and more frequent sampling was conducted using nsample=10 000 000 and samplefreq=2 with 100 BS replicates to calculate the standard error (SE) following the stationary block BS method (Politis & Romano, 1994). Four datasets were tested but convergence could not be achieved for nucleotide alignments of Dataset A, even after more than one month on our server with a 2.30 GHz Intel (R) Xeon (R) Gold 5218 CPU. The IR and AR models were further compared by plotting mean divergence time of the internal nodes against the width of 95% highest posterior density (HPD).
Molecular dating analyses
Molecular dating with calibration nodes from the fossil record was performed using the MCMCTree program. The best optimized tree topologies were used as the input trees for MCMCTree analyses with Datasets A and B. All timetree analyses were performed using the IR and AR models under soft-bounded constraints. Analyses were run for 100 000 generations after a burn-in of 10 000 generations. Chains were sampled every 100 generations. Each run was conducted with both evolutionary rate models (IR and AR) at least twice to check for convergence using Tracer v1.7.2. To test fossil effects, tree topologies and sequence alignments were conducted without fossil calibrations.
Lineage-through-time (LTT) analysis
The accumulation of extant mammalian families was examined using LTT plots. Diversification-rate, pure-birth, and birth-death scenarios were applied to fit the LTT model (Etienne et al., 2016). The R packages RPANDA v2.2 (Morlon et al., 2016) and Laser v2.3 (Rabosky, 2006) were used to describe the diversification rate-constant (RC) and rate-variable (RV) models using likelihood tests. The AICc scores of the best-fitting RC and RV models were obtained following the birth-death likelihood tests (Rabosky, 2006). Laser v2.3 included two RC models identical to those in RPANDA v2.2, along with four distinct RV models. To further confirm whether lineage accumulation showed continuous growth in marsupials around the KPg boundary, LTT and diversification rate shifts were analyzed using our timetree as well as those in Duchêne et al. (2018) and Upham et al. (2019).
Branch-specific diversification rate estimation
To ascertain the heterogeneity of speciation rates across different clades, three approaches were used to mitigate potential bias associated with any individual method. Initially, Bayesian analysis of macroevolutionary mixtures (BAMM) was used to identify distinct diversification models across the phylogeny. BAMM employs reversible-jump MCMC to transition among rate frequencies configurations derived from various prior models, including constant rate, birth-death, pure-birth, and modeling evolutionary diversification using stepwise AIC (MEDUSA)-like model. The RV model is considered optimal compared to the RC model, contingent upon the observed results. Accordingly, an RV MEDUSA-like model was adapted to describe the evolutionary scenario. The BAMM sample priors were inferred from setBAMMpriors, and a no-data run was conducted to assess estimates based only on prior information. Each analysis was run for 200 million generations with sampling every 20 000 generations. BAMM outputs were analyzed using BAMMtools v2.1.10 (Rabosky et al., 2014). Multiple runs were conducted to check for convergence using Tracer v1.7.2. The effective sample size (ESS) in each estimation exceeded 200. The first 10% of samples were discarded as burn-in. Subsequently, branch-specific diversification rates were evaluated under the conditional birth-death-shift process via RevBayes v1.0 (Höhna et al., 2016). Priors for both speciation and extinction rates followed a lognormal distribution, containing six and 20 rate categories, respectively. Two independent MCMC chains were executed in RevBayes, each with 10 000 generations, and a burn-in of 1 000 generations. Convergence within and between MCMC chains was confirmed using Tracer. Lastly, the fit_ClaDS function (Maliet et al., 2019) in the RPANDA package was employed, using a Bayesian method to estimate the branch-specific diversification rates in a phylogeny context, which accounts for changes in diversification rates, from multiple small shifts to occasional major shifts (Maliet et al., 2019). Inference of branch-specific speciation rates and the hyper parameters was conducted under a constant turnover rate (ClaDS2) over a span of MCMC 5 000 step.
RESULTS
Improvement in dataset matrices by filling gaps and expanding alignment length
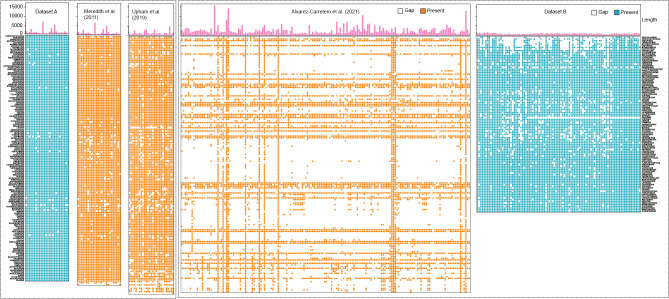
As both complete alignment and dense sampling are crucial for estimating phylogeny and divergence time, we generated two datasets in this study (Figure 1). Dataset A, derived from Meredith et al. (2011), comprised an extensively augmented alignment of 26 nuclear gene fragments (35 063 bp), containing 21 protein-coding genes, plus five noncoding fragments (Supplementary Table S1) from 162 mammalian lineages within 147 families (accounting for approximately 95%–99% of all extant mammalian families) (Duff & Lawson, 2004), as well as five nonmammalian outgroups. Meredith et al. (2011) initially reported 74 species with missing gene fragments. However, Dataset A added corresponding sequences for 30 of these species, reducing the proportion of missing genes from 6.3% in the previous study to 1.8% in this study, markedly lower than the missing rate (>12%) reported in Upham et al. (2019) (Figure 1).
Figure 1.
Comparison of datasets between this and previous studies
Top panel is length of gene fragments corresponding to each column in bottom panel. Left and right annotations are family name (ordered by phylogenetic relationships). Left four panels follow left family name annotation and right panels follow right family name annotation. Gap information of gene fragments from Dataset A, Meredith et al. (2011), Upham et al. (2019), Álvarez-Carretero et al. (2022), and Dataset B. Blue and yellow indicate present gene fragments in this and other studies, respectively, white indicates missing gene fragments.
Álvarez-Carretero et al. (2022) performed a multisequence alignment involving 168 nuclear genes from 2 939 species. Notably, the majority of families included only fragmentary gene sequences, with the exception of Hominidae and Muridae, which contained complete genes (Figure 1). As such, Dataset B was generated, consisting of an alignment of 98 nuclear gene fragments (67 080 bp, Supplementary Table S2) from 120 mammalian species (including species in Supplementary Table S3) representing 105 families. The proportion of missing nuclear gene fragments at the family level in Dataset B was 11.8%, substantially lower than the 70.1% missing rate reported in Álvarez-Carretero et al. (2022). Dataset B additionally incorporated sequence alignments for the Tachyglossidae and Calomyscidae families, absent from Dataset A. Reduced proportions of missing sequences in Datasets A and B were expected to enhance the accuracy of phylogenetic topologies and BS support. This expectation was supported by our simulations, which indicated that decreased rates of missing data and/or increased alignment lengths, assuming a constant number of taxa, tended to diminish deviations in accurate phylogenetic topologies and improve BS support (Supplementary Figure S1).
New phylogenies resolved several relationships
Phylogenetic relationships were reconstructed using both datasets based on Bayesian inference (BI) and ML approaches. Given that concatenation strategies under a single substitution model may not adequately represent the substitution rate heterogeneity evolutionary history, multiple partition strategies were employed, including noncoding+coding regions, genes, codon positions and PartitionFinder. AICc selected GTR+F+I+G as the optimal model for most DNA partitions in all partitioning strategies.
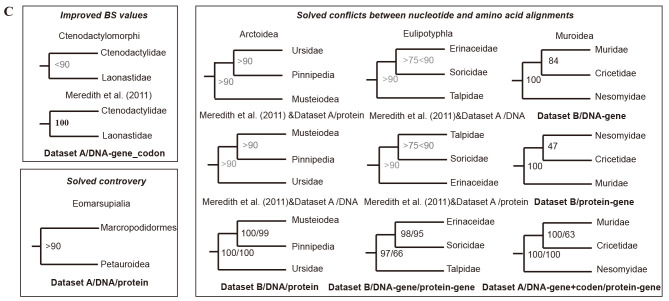
The resulting phylogenies revealed robust support, with approximately 88% of the internal nodes exhibiting BS support ≥90% and Bayesian posterior probabilities (BPP) ≥0.95 (Figure 2A, B). In these phylogenetic reconstructions, the Theria clade bifurcated into marsupials and placentals. Within marsupials, paraphyletic Ameridelphia was rooted prior to monophyletic Australidelphia, while placentals were divided into Atlantogenata (Afrotheria+Xenarthra) and Boreoeutheria (Euarchontoglires+Laurasiatheria). In both datasets, over 94% of internal nodes demonstrated concordance between the BI and ML trees, regardless of partitioning strategies or sequence types (DNA or protein) (Figure 2A, B), strengthening some relationships and resolving certain controversial relationships. For example, BS support for the sister groups Laonastidae and Ctenodactylidae increased to 100% in our study compared to less than 90% in Meredith et al. (2011) and Gatesy et al. (2017). The taxonomic organization of the marsupial suborder Phalangeriformes, comprising two superfamilies: Phalangeroidea (Burramuidae+Phalangeridae) and Petauroidea (Pseudocheiridae+Petauridae+Tarsipedidae+Acrobatidae), was previously based on morphological traits related to the auditory region bone, e.g., fusion of the ectotympanic bone with other bones of the skull (Springer & Woodburne, 1989), with the recent identification of orthologous short interspersed element (SINE) markers attesting to its monophyly (Doronina et al., 2022). However, molecular sequence data have challenged the monophyly of Phalangeriformes, favoring a closer affinity between Macropodiformes and Phalangeroidea (Beck, 2008; Meredith et al., 2009; Phillips & Pratt, 2008) or Petauroidea (May-Collado et al., 2015; Mitchell et al., 2014), with the latter relationship supported by recent research of 1 550 exonic loci (Duchêne et al., 2018). Consistent with these molecular insights, Dataset A also favored the latter grouping, with more than 90% BS support, higher than the support levels reported in previous studies (Figure 2C).
Figure 2.
Phylogenetic trees and estimated divergence times of extant mammalian families
A: Phylogenetic timetree of 162 mammals, rooted with five vertebrate outgroups (chicken, zebra finch, green anole, frog, and zebrafish) generated with Dataset A. B: Phylogenetic timetree of 120 mammals, rooted with one Monotremata outgroup (platypus) generated with Dataset B. All nodes were strongly supported (BS≥90% and Bayesian posterior probabilities (BPP)≥0.95) except for nodes denoted by solid blue circles (consistent between DNA and amino acid trees, with BS≥70<90, BPP≥0.90) or solid red circles (conflict between DNA and amino acid trees under different partitioning strategies). Strongly supported nodes showing disagreement between the two datasets are indicated with solid black circles. Fossil calibration points are tagged with bone cartoon icons. Dashed line corresponds to KPg boundary. Long branches are truncated with double slashes. C: Improved phylogenetic relationships among families compared to previous studies or different datasets with consistent topology. Meredith et al. (2011) & Dataset A/protein mean topologies between Meredith et al. (2011) and Dataset A/protein are consistent, and BS values sourced from the Dataset A/protein.
The high degree of consistency in phylogenetic relationships between the two alignments was considered reliable as BS support exceeded 90% for the consistent nodes. For example, analyses of Dataset A and data from Meredith et al. (2011) showed inconsistent positions for Ursidae within Caniformia. However, nucleotide and protein alignments of Dataset B positioned Ursidae as the basal-most lineage of Arctoidea, with identical BS support for the DNA and protein alignments (BSdata B/DNA=BSdata B/protein=100%), regardless of partitioning strategies (Figure 2C). This placement is supported by retrophylogenomic evidence (Doronina et al., 2015) and mitogenomic data (Arnason et al., 2007) but contradicts the reported sister relationship of Ursidae and Pinnipedia (Yu et al., 2011). Similar conflict occurred for the placement of Soricidae within Eulipotyphla. In the ML tree generated from Dataset B, Erinaceidae, not Talpidae, was strongly supported as the sister group of Soricidae (BSdata B/DNA-gene=98%; BSdata B/protein-gene=95%) (Figure 2C), as substantiated by nuclear and mitochondrial gene analyses (Brace et al., 2016; Narita et al., 2001; Sato et al., 2019) and dental characteristics (Archer et al., 2011). In contrast, nucleotide alignment of Dataset B only moderately supported Cricetidae as sister to Muridae (BSdata B/DNA-gene=84%), while protein alignment weakly supported Cricetidae as sister to Nesomyidae (BSdata B/protein-gene=47%). The ML tree generated from Dataset A, which involved dense sampling, yielded concordant evidence grouping Cricetidae with Muridae (BSdata A/DNA-gene+codon=100%; BSdata A/protein-gene=63%) (Figure 2C), as verified by multiple mitochondrial and nuclear genes (Blanga-Kanfi et al., 2009; Steppan et al., 2004; Steppan & Schenk, 2017) and thousands of ultraconserved elements (UCEs) (Swanson et al., 2019).
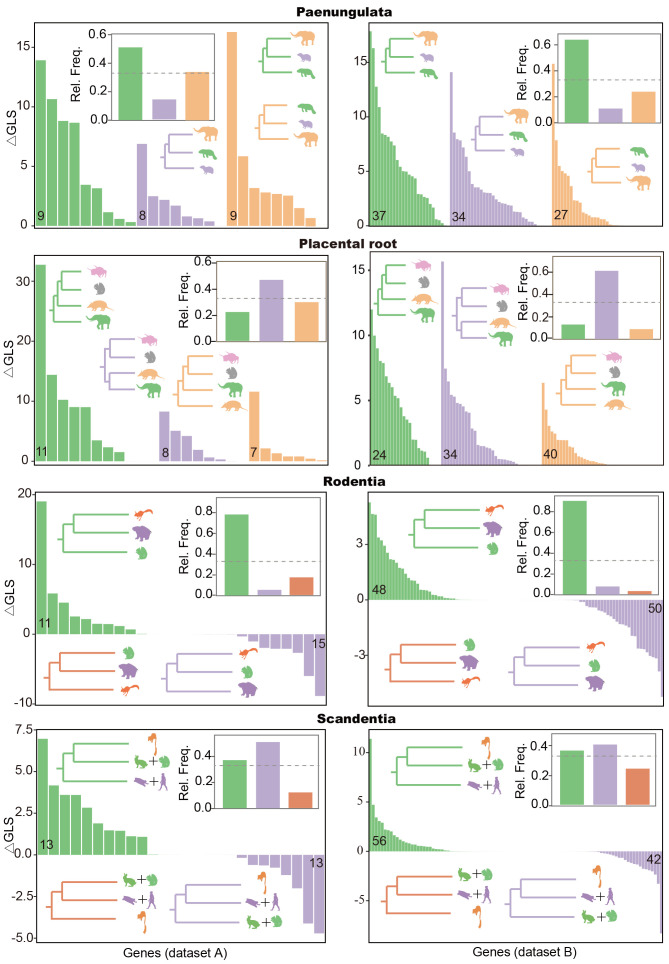
Exploration of sequence and tree heterogeneity for conflicting topologies
We adopted two methods to estimate the reliability of optimal topologies in long-standing phylogenetic relationships, including placental root, phylogenetic position of Scandentia, interordinal relationships within Laurasiatheria and Paenungulata, and intraordinal group surrounding the sister group of Sciuromorpha in Rodentia (Figure 3; Supplementary Tables S4–S7). First, recent research has suggested that a small subset of genes in a large data matrix may yield extreme log-likelihood scores that bias specific topologies, thereby obscuring the unbiased phylogenetic signals of the remaining genes (Shen et al., 2017). Based on △GLS values between contentious branches, our results revealed no pronounced bias towards any specific branch (Figure 3) and no substantial difference in the number of genes supporting conflicting topologies (Figure 3). Second, the AU (Shimodaira, 2002) and SH tests (Shimodaira & Hasegawa, 1999) applied to nucleotide alignments demonstrated no significant differences across various branch arrangements (Supplementary Tables S4, S6). However, when the two tests were applied to amino acid alignments, they consistently rejected the sister-group relationships between Artiodactyla and Ferae (Carnivora+Pholidota) (Pdata A/protein-SH<0.05, SH probability of amino acid alignment A; Pdata A/protein-AU<0.05; Supplementary Table S5) and between Perissodactyla and Ferae (Pdata B/protein-SH<0.01; Pdata B/protein-AU<0.001; Supplementary Table S7) in Laurasiatheria. In our previous work, we used 97 orthologous genes and 1 608 exons to resolve this puzzle with reliable topology, (Chiroptera, ((Perissodactyla, Artiodactyla), Ferae)) (Zhou et al., 2012), as corroborated by datasets containing 12 931 genes (Jebb et al., 2020) and 3 787 UCEs (Esselstyn et al., 2017). This topology currently represents the prevailing phylogenetic relationship at the genomic level data, although 3 638 intron sequences displayed a different phylogenetic topology: ((Perissodactyla, Chiroptera), (Ferae, Artiodactyla)) (Chen et al., 2017).
Figure 3.
Comparison of gene-wise log-likelihood scores and relative frequencies of gene tree quartets for four phylogenetic groups with conflicting topologies
Four phylogenetic groups are shown (Paenungulata, Placental Root, Rodentia and Scandentia). Relative frequency inferred from species tree (inset) based on gene alignments from Datasets A (left) and B (right). Conflicting topologies are shown in different colors, supported by△GLS histogram (y axis) from different genes (x axis) and relative frequencies inferred from species tree. Numbers of genes supporting the topology are indicated at the bottom of histogram.
Recent phylogenomic studies have reported that base composition of markers carries unambiguous phylogenetic signals, but GC-content rich genes are biased phylogenetic tree reconstructions (Betancur-R et al., 2013; Collins et al., 2005; Nabholz et al., 2011; Rodríguez-Ezpeleta et al., 2007; Romiguier et al., 2013a, 2013b). The ML tree exclusive of GC-rich genes (Supplementary Table S8) showed no differences in topologies (Supplementary Table S9), indicating the GC-rich regions introduced no bias in this study.
Incomplete lineage sorting (ILS), a process whereby different loci yield topologically distinct phylogenies, is a major source of ambiguous relationships. To address this, we applied coalescent-based “species tree” methods (Edwards, 2009), known to perform well in situations involving conflict caused by ILS. While Meredith et al. (2011) and Song et al. (2012) stated that 26 gene fragments either introduce bias or are insufficient for resolving the phylogeny of Eutherian mammals using coalescence-based methods, Gatesy et al. (2017) argued that the observed limitations may be attributable to simplistic DNA substitution models rather than insufficient fragments. In that respect, our coalescence-based results from Datasets A and B were still reliable. Moreover, the phylogenetic relationships between the “species tree” and ML tree were highly consistent, even when resolving conflicting relationships, including those within Laurasiatheria and Paenungulata, the placental root, and the sister group of Sciuromorpha in Rodentia (Figure 3). Here, we focused on the relative frequency of gene trees, rather than “species trees”, to analyze ambiguous relationships and avoid potential bias associated with the “species tree” method. First, over 45% of gene tree quartets in ASTRAL analyses of the two datasets supported Proboscidea and Hyracoidea as a sister group, as corroborated by multiple nuclear and mitochondrial gene fragments and thousands of UCEs (Esselstyn et al., 2017; Meredith et al., 2011; Shimodaira, 2002). However, the AU and SH tests applied to the amino acid alignment of Dataset B contradicted the monophyly of Tethytheria (Proboscidea plus Sirenia) (Pdata B/protein-SH<0.05; Pdata B/protein-AU<0.05, Supplementary Table S7), as previously proposed by various studies based on nuclear and mitochondrial sequences, transposon insertion/deletion matrices, fossil records, and morphological characters (Beck et al., 2014; Lavergne et al., 1996; Murata et al., 2003; Nishihara et al., 2005; Puttick & Thomas, 2015; Romiguier et al., 2013a). Second, although the AU tests applied to the amino acid alignment of Dataset B opposed the Atlantogenata rooting hypothesis (Pdata B/protein-AU<0.05; Supplementary Table S7), 40% of gene tree quartets supported Atlantogenata as the root of placental mammals (Figure 3), consistent with many genomic data studies (Esselstyn et al., 2017; Liu et al., 2017; Song et al., 2012; Tarver et al., 2016). Third, more than 70% of gene tree quartets placed Sciuromorpha at the basal position of Rodentia, followed by divergence between the Hystricomorpha and mouse-related clades (Figure 3). This topology coincides with phylogenomic analyses based on exons (Feijoo & Parada, 2017; Scornavacca & Galtier, 2017) and transposons (Churakov et al., 2010; Doronina et al., 2017b) but conflicts with the proposed sister relationship between Sciuromorpha and Hystricomorpha (Montgelard et al., 2008; Swanson et al., 2019). Finally, with respect to the position of Scandentia, although the relative frequency of gene tree quartets indicated Scandentia and Primatomorpha as sister groups over the other two topologies, the evidence was not highly compelling (Figure 3). Based on 3 787 UCEs, Esselstyn et al. (2017) also supported Scandentia as a sister group to Primatomorpha but failed to reject a sister relationship between Scandentia and Glires. Moreover, Liu et al. (2017) constructed a species tree using 4 388 genes, suggesting a sister group between Scandentia and Glires. However, this phylogenetic affinity was challenged by Lin et al. (2014), who posited that the perceived relationship could be attributed to an accelerated evolutionary rate in these two groups. Consequently, Scandentia is more likely to be a sister group of Primatomorpha than to Glires. In summary, these findings indicate that ILS is a critical determinant in existing deep-level incongruences in the phylogeny of placental mammals.
Placental diversification initiated in Late Cretaceous
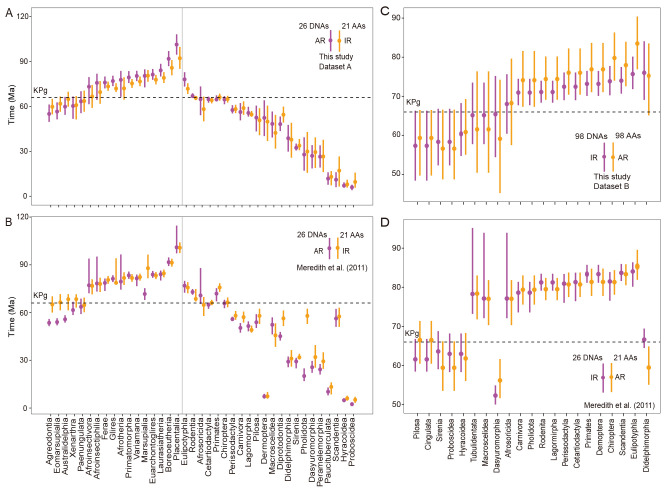
We conducted dating analyses using MCMCTree (Yang, 2007), incorporating 76 and 43 selected fossil calibrations for Datasets A and B, respectively (Supplementary Table S10). Inaccurate application of molecular clocks may introduce bias to dating estimates (dos Reis et al., 2015, 2016). Thus, we employed two strategies to determine the most appropriate molecular clock for estimating divergence time. Bayesian model selection using thermodynamic integration indicated that the AR model exhibited the highest marginal likelihood for both the amino acid alignment of Dataset A and nucleotide alignment of Dataset B (posterior model probability=1.00), while the IR model exhibited the highest marginal likelihood for the amino acid alignment of Dataset B (posterior model probability=0.98) (Supplementary Table S11). Given the computational burden of marginal likelihood calculations, particularly for the nucleotide alignment of Dataset A, we plotted the mean divergence time of internodes against the 95% HPD width for this dataset. The divergence time plot showed that the IR model was positively correlated with finite-sites from Dataset A nucleotide alignment (Supplementary Figure S2). The divergence times for most interordinal and intraordinal lineages determined from nucleotide alignment exhibited narrower 95% HPD intervals compared to the estimates derived from amino acid alignment (Supplementary Figure S2). Therefore, the IR model was preferable to the AR model in divergence time estimation for Dataset A nucleotide alignment and Dataset B amino acid alignment, whereas the AR model was preferable over the IR model for Dataset A amino acid alignment and Dataset B nucleotide alignment.
Compared with the timetree reported in Meredith et al. (2011) (Figure 4), the divergence time of the nucleotide alignment of Dataset A was more ancient (Supplementary Table S12), while the amino acid alignment estimation was relatively younger (Supplementary Table S13). This may have resulted from the efforts of filling the gaps in Meredith et al. (2011) and/or updated fossil calibrations (Supplementary Table S14). Our dating analyses revealed that placental radiation was incompatible with current placental radiation models. The split of Theria occurred approximately 163 Ma, close to the recent species-level mammalian tree (Upham et al., 2019). The diversification of placentals was dated to 101.4 Ma (95% HPD, 94.6–108.2 Ma) for the DNA alignment (Supplementary Table S12) and 92.2 Ma (95% HPD, 85.1–99.8 Ma) (Supplementary Table S13) for the protein alignment of Dataset A. These estimates correspond with, or even predate, the Late Cretaceous (66.0–100.5 Ma), consistent with the dates reported in Bininda-Emonds et al. (2007) (mean: 98.5 Ma, 95% HPD: 93.2–108.1 Ma) and Upham et al. (2019) (mean: 91.8 Ma, 95% HPD: 77.4–105.0), but older than the trans-KPg (mean: 75.2 Ma) (Liu et al., 2017) and explosive models (mean: 77.2 Ma) (Phillips, 2016) and Álvarez-Carretero et al. (2022) (mean: 80.3 Ma) (Table 1; Supplementary Figure S3). In parallel, the most recent interordinal split, between Sirenia and Proboscidea, was estimated at approximately 56.6 Ma (95% HPD, 48.8–66.6 Ma), which was not accounted for by either the long or short fuse models. Consequently, our results suggested that placental diversification may have commenced during, or even prior to, the Late Cretaceous, with subsequent interordinal lineage divergences extending to the Paleocene (56–66 Ma) and more than 40% (10/21) of intraordinal lineages crossing the KPg boundary (Figure 4A, C), a model we termed the long trans-KPg model.
Figure 4.
Divergence times of extant mammals
A, C: Mean values (solid dots) and 95% HPD intervals (solid lines) for interordinal (left part of gray vertical line) and intraordinal (right part of gray vertical line) divergence times, derived from DNA sequences (purple solid dots and lines) and protein alignments (orange solid dots and lines) of Datasets A and B, respectively. B, D: Intraordinal divergence times as reported by Meredith et al. (2011) from corresponding sequences of A and C, respectively. 26 DNAs and 98 DNAs represented DNA sequences, 21 AAs and 98 AAs represented protein alignments. AR represented autocorrelated clock and IR represented the independent clock. Dotted line indicates KPg boundary.
Table 1. Divergence time (Ma) differences between this and other studies.
| Taxon | This study | Bininda-Emonds et al., 2007 | Phillip, 2016 | Upham et al., 2019 | Álvarez-Carretero et al., 2022 | ||||||||
| Mean | Mean | Difference | Mean | Difference | Mean | Difference | Mean | Difference | |||||
| Difference represented the mean value of our study minus other studies. | |||||||||||||
| Placentalia | 101.4 | 98.5 | 2.9 | 77.2 | 24.2 | 91.8 | 9.6 | 80.3 | 21.1 | ||||
| Xenarthra | 60.3 | 70.5 | −10.2 | 46.8 | 13.5 | 67.4 | −7.1 | 63.6 | −3.3 | ||||
| Afrotheria | 77.8 | 90.4 | −12.6 | 57.7 | 20.1 | 80.0 | −2.2 | 70.8 | 7.0 | ||||
| Afrosoricida | 64.9 | 82.4 | −17.5 | 51.4 | 13.5 | 70.4 | −5.5 | 60.0 | 4.9 | ||||
| Macroscelidea | 48.4 | 47.5 | 0.9 | 38.7 | 9.7 | 59.0 | −10.6 | 49.8 | −1.4 | ||||
| Paenungulata | 63.4 | 75.8 | −12.4 | 45.2 | 18.2 | 54.0 | 9.4 | 60.3 | 3.1 | ||||
| Hyracoidea | 7.1 | 18.6 | −11.5 | 3.1 | 4.0 | 10.0 | −2.9 | 13.14 | −6.04 | ||||
| Proboscidea | 6.0 | 19.5 | −13.5 | 1.3 | 4.7 | 10.1 | −4.1 | 5.44 | 0.56 | ||||
| Sirenia | 32.4 | 52.2 | −19.8 | 5.7 | 26.7 | 14.3 | 18.1 | 34.3 | −1.9 | ||||
| Boreoeutheria | 91.8 | 96.1 | −4.3 | 72.3 | 19.5 | 83.5 | 8.3 | 75.1 | 16.7 | ||||
| Laurasiatheria | 84.1 | 87.8 | −3.7 | 64.6 | 19.5 | 75.1 | 9.0 | 73.8 | 10.3 | ||||
| Eulipotyphla | 78.1 | 82.5 | −4.4 | 59.1 | 19.0 | 74.3 | 3.8 | 70.1 | 8.0 | ||||
| Chiroptera | 64.7 | 71.2 | −6.5 | 50.3 | 14.4 | 57.1 | 7.6 | 57.9 | 6.8 | ||||
| Perissodactyla | 57.6 | 55.8 | 1.8 | 27.9 | 29.7 | 38.7 | 18.9 | 59.5 | −1.9 | ||||
| Pholidota | 52.6 | 19.1 | 33.5 | 15.3 | 37.3 | 27.1 | 25.5 | 34.8 | 17.8 | ||||
| Carnivora | 56.3 | 63.4 | −7.1 | 36.7 | 19.6 | 40.2 | 16.1 | 51.7 | 4.6 | ||||
| Artiodactyla | 64.9 | 70.7 | −5.8 | 45.4 | 19.5 | 58.4 | 6.5 | 55.7 | 9.2 | ||||
| Euarchontoglires | 81.3 | 91.8 | −10.5 | 67.6 | 13.7 | 77.0 | 4.3 | 69.8 | 11.5 | ||||
| Primatomorpha | 79.4 | 88.5 | −9.1 | 66.9 | 12.5 | / | / | 66.5 | 12.9 | ||||
| Primates | 64.7 | 84.5 | −19.8 | 59.9 | 4.8 | 67.1 | −2.4 | 63.1 | 1.6 | ||||
| Dermoptera | 52.4 | 15.0 | 37.4 | 5.5 | 46.9 | 9.6 | 42.8 | 20.5 | 31.9 | ||||
| Scandentia | 11.1 | 31.7 | −20.6 | 44.3 | −33.2 | 52.8 | −41.7 | 58.1 | −47.0 | ||||
| Glires | 77.0 | 88.9 | −11.9 | 65.8 | 11.2 | 72.4 | 4.6 | 66.2 | 10.8 | ||||
| Rodentia | 67.0 | 82.8 | −15.8 | 60.4 | 6.6 | 67.9 | −0.9 | 60.8 | 6.2 | ||||
| Lagomorpha | 55.7 | 64.3 | −8.6 | 41.4 | 14.3 | 50.9 | 4.8 | 48.3 | 7.4 | ||||
| Sum | (P=4.34e-03) | −149.1 | (P=1.94e-05) | 389.9 | (P=1.12e-02) | 111.9 | (P=5.17e-04) | 130.82 | |||||
Progressive radiation of extant mammals after KPg boundary
We examined the dynamics of lineage accumulation through time in the mammalian timetree by fitting the RC and RV models to mammalian diversification. The likelihood tests using RPANDA (Foley et al., 2016) and Laser (Rabosky, 2006) both demonstrated that the best RC model of extant mammalian diversification was pure birth, and that the RV models were superior to the RC models for fitting mammalian lineage accumulation throughout evolutionary history (Table 2; Supplementary Table S15).
Table 2. Likelihood tests of RC and RV models of mammalian diversification from R packages.
| R package | laser | RPANDA | |||||
| Model | Parameter | LH | AIC | LH | AICc | ||
| LH, log-likelihood; AIC, Akaike information criterion; AICc, corrected Akaike information criterion; PB, pure birth (Yule) model with constant speciation rate; BD, birth-death model with constant speciation and extinction rate; yule2rate, pure birth model, assumes clade has diversified under net diversification rate r1 until some time point st, at which point the diversification rate shifts to a new rate r2; yule3rate assumes a diversification rate r1 until some time point st1 shifts to a new rate r2, then until another time point st2 shifts to a new rate r2; B-Lin, pure birth with linear variation of speciation rate; D-Lin, birth-death model with linear variation of extinction rate. −: Not available, | |||||||
| RC model | PB | −114.036 | 230.071 | −773.751 | 1 549.528 | ||
| BD | −111.036 | 232.071 | −773.752 | 1 551.578 | |||
| RV model | yule2rate | −78.601 | 163.202 | − | − | ||
| yule3rate | −69.722 | 149.444 | − | − | |||
| B-Lin_D | − | − | −752.352 | 1 510.855 | |||
| B_D-Lin | − | − | −741.540 | 1 489.232 | |||
Our results further showed that lineage accumulation progressively increased from 100 Ma to 20 Ma for extant mammals, marsupials, and placentals (Supplementary Figures S4, S5). Only one order, Eulipotyphla, had a 95% HPD that was entirely restricted to the Cretaceous, with the remaining intraordinal diversification occurring later or across the KPg. To further confirm whether lineage accumulation showed continuous growth around the KPg boundary, we analyzed lineages through time and diversification rate shifts using our timetree as well as those from Duchêne et al. (2018) and Upham et al. (2019) (Table 2; Supplementary Figure S5B). The significantly negative Gamma statistic rejected the constant diversification rate through time hypothesis and indicated a concentration of nodes toward the root of tree, suggesting a decreasing diversification rate over time for extant family-level mammals, marsupials, and placentals (r<0 and P <0.05; Table 3). Additionally, branch-specific diversification rate shifts appeared to be randomly distributed and discordance shifts were disordered on the mammalian tree (Supplementary Figure S6), thereby excluding the possibility that specific taxonomic groups exerted a disproportionate influence on overall patterns of diversification. Therefore, the legacy of KTR boosted mammalian diversification and was not interrupted by the KPg boundary in living placentals and marsupials. Thus, it is reasonable to speculate that numerous vacant niches after the KPg catastrophe contributed to extant mammalian radiation, especially intraordinal diversification (Meredith et al., 2011). However, we cannot rule out the benefits of angiosperm radiation (Liu et al., 2017) and mammalian adaptation to environmental challenges (Upham et al., 2019).
Table 3. Test for deviations from constant diversification rate through time using γ statistic.
DISCUSSION
Our study resolved the phylogenetic relationships among the vast majority of extant mammalian families, including specific nodes that have been the subject of long-standing debate. Resolution was achieved using multiple partitioning strategies employed in the ML and BI phylogenetic reconstructions, as well as the relative frequency of gene tree quartets. Our results corroborated the division of Theria into marsupials and placentals. Furthmore, within marsupials, Ameridelphia was paraphyletic and gave rise to a monophyletic Australidelphia, whereas, within placentals, the Atlantogenata and Boreoeutheria clades were distinctly delineated. No evidence was found that extreme log-likelihood values from a limited number of genes or GC-rich regions led to biased results. Rather, ILS was identified as a crucial factor contributing to the existing deep-level incongruences found in the phylogeny of placental mammals, refuting the suggestion that ILS is the minor determinant of existing phylogenetic conflicts in mammals (Scornavacca & Galtier, 2017).
The proper translation of fossil data into calibration points is essential for the reliability of divergence dating estimations. Here, the estimated divergence times for the calibration internodes in the phylogenetic tree largely coincided with known fossil records (Supplementary Figures S7–S13), with a few exceptions. Notably, we identified several ghost lineages, inferred to exist but with no fossil record, in three placental groups, i.e., Anthropoidea, Octodontoidea, and Loridae+Galagidae (Supplementary Table S16). The divergence times for Anthropoidea, estimated using the IR clock applied to DNA and amino acid alignments of Dataset B, were 6.4–27.5 Ma and 6.9–26.7 Ma, respectively (Supplementary Figures S11–S14 and Table S16). However, these estimates conflict with the youngest known fossil of the Anthropoidea genus Branisella, dated at 28.3 Ma (Kay et al., 1998). Comparable discrepancies were observed for Octodontoidea and Loridae+Galagidae. We also identified two zombie lineages, which extended beyond known fossil records. Specifically, the divergence times of Primates and Haplorrhini, estimated using the AR clock applied to nucleotide alignment of Dataset A, were approximately 67.5–71.7 Ma and 62.2–67.5 Ma, respectively (Supplementary Figure S8 and Table S16). These estimates exceed the maximum known fossil ages for these groups: 66 Ma for Primates, based on early Paleocene fossils of plesiadapids, paromomyids, and carpolestids (Springer et al., 2017); and 59.2 Ma for Haplorrhini based on the potential late Paleocene discovery of the stem anthropoid genus Altiatlasius (Springer et al., 2017).
Additionally, the mean divergence time estimated for the most common ancestor of Cetacea was 31.8 Ma, according to Dataset A (Supplementary Figures S7–S10). This is more recent than the oldest known crown cetacean fossil, the stem mysticete Mystacodon selenensis, dated at around 36.4 Ma (Lambert et al., 2017). The mean divergence time estimated from the amino acid alignment of Dataset B did not precede the oldest known fossil time for crown cetaceans. The maximum mean divergence time for the Cetacea split was 37.1 Ma, aligning more closely with the estimates of McGowen et al. (2020) and predating those of Chen et al. (2011) and Zhou et al. (2011). To assess the influence of calibration points falling outside the 95% HPD interval, we conducted additional dating without those points. Results showed that the recalibrations had no obvious effect on mean estimated divergence times (Pdata A-DNA=0.89; Pdata A-protein=0.02; Supplementary Tables S17, S18), although they did potentially impact the 95% HPD of divergence times (Pdata A-DNA=0.27; Pdata A-protein=4.46e-05; Supplementary Tables S17, S18). Consequently, both zombie and ghost lineages should be carefully compared to the fossil record in divergence time estimation analyses.
Our timetree offered a comprehensive dating estimation for mammalian radiation, with extensive coverage of extant families and approximately 100 molecular markers. However, our divergence dating differed from previous molecular clock-based estimates (P<0.05, Table 1). Long-standing discrepancies surround the onset time of placental radiation and whether interordinal and intraordinal diversification occurred before or after the KPg boundary. Neither the morphological-based short fuse model (Archibald & Deutschman, 2001), which posits interordinal radiation prior to the KPg, nor the molecular-based explosive and soft explosive models (Archibald & Deutschman, 2001), which posit that intraordinal radiation occurred post-KPg, were consistent with placental radiation patterns identified in our study. Our findings were similar with the trans-KPg model (Liu et al., 2017), i. e. interordinal and intordinal radiations across the KPg boundary, but showed more ancient divergent onset time than trans-KPg model (Liu et al., 2017). Specially, the diversification of placental orders began approaching (the mean value of 92.2 Ma estimated from protein sequences) and even earlier the Late Cretaceous (the mean value of 101.4 Ma estimated from nucleotide sequences). Our model of placental radiation was constructed using carefully aligned DNA and protein alignments and an appropriately selected molecular clock. However, we noted that certain lineages, such as the aforementioned zombie and ghost lineages, may lie outside the scope of fossil calibration. Gatesy & Springer (2017) suggested that the presence of zombie lineages may compromise the reliability of timetree estimates. Whether the disparities in our results are due to limitations in clock methodology or inconsistencies in the fossil record remains a subject for future investigation.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
SCIENTIFIC FIELD SURVEY PERMISSION INFORMATION
The field-collected Taphozous melanopogon, Aselliscus stoliczkanus, Chaerephon plicatus, and Hipposideros larvatus samples used in this study are not from endangered species and are not included in the “List of Protected Animals in China”. No specific permissions were required for sampling activities.
Acknowledgments
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
X.M.Z. conceived and supervised the project. J.D., Z.H.L., L.W., X.X.Z., Z.Z., Y.Y., M.L. P.C.W., X.W., and M.L. performed data curation. G.M.L., Q.P., P.F.Z., W.Q.L., C.Y.H., and Y.C.D. conducted hypothesis testing, phylogenetic analyses, and divergence time estimation. G.M.L. and X.M.Z. wrote the manuscript with input from all authors. All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank Shao-Ying Liu, Wen-Hua Yu, Song Li, Chen-Ling Zhang, Alice Hughes, Xing Chen, Yan-Nan Chen, Zhi-Qian Yu, Cheng-Ming Huang, Hui-Zhong Fan, and Shijiazhuang Zoo (Shijiazhuang City, China) for sample collection.
Funding Statement
This work was supported by the National Key Research and Development Projects of the Ministry of Science and Technology of China (2021YFC2301300), National Natural Science Foundation of China (82050002, 32070528, 32100335, 32000287), and Beijing Natural Sciences Foundation for Distinguished Young Scholars (JQ19022)
DATA AVAILABILITY
The generated sequences were submitted to GenBank (accession numbers OR503106–OR504069). Custom Perl scripts and alignments are available at Dryad (DOI: 10.5061/dryad.crjdfn36w).
References
- Álvarez-Carretero S, Tamuri AU, Battini M, et al A species-level timeline of mammal evolution integrating phylogenomic data. Nature. 2022;602(7896):263–267. doi: 10.1038/s41586-021-04341-1. [DOI] [PubMed] [Google Scholar]
- Archer M, Beck R, Gott M, et al Australia's first fossil marsupial mole (Notoryctemorphia) resolves controversies about their evolution and palaeoenvironmental origins. Proceedings of the Royal Society B:Biological Sciences. 2011;278(1711):1498–1506. doi: 10.1098/rspb.2010.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JD, Deutschman DH Quantitative analysis of the timing of the origin and diversification of extant placental orders. Journal of Mammalian Evolution. 2001;8(2):107–124. doi: 10.1023/A:1011317930838. [DOI] [Google Scholar]
- Arnason U, Gullberg A, Janke A, et al Mitogenomic analyses of caniform relationships. Molecular Phylogenetics and Evolution. 2007;45(3):863–874. doi: 10.1016/j.ympev.2007.06.019. [DOI] [PubMed] [Google Scholar]
- Beck RMD A dated phylogeny of marsupials using a molecular supermatrix and multiple fossil constraints. Journal of Mammalogy. 2008;89(1):175–189. doi: 10.1644/06-MAMM-A-437.1. [DOI] [Google Scholar]
- Beck RMD, Travouillon KJ, Aplin KP, et al The osteology and systematics of the enigmatic australian oligo-miocene metatherian Yalkaparidon (Yalkaparidontidae; Yalkaparidontia;? Australidelphia; Marsupialia) Journal of Mammalian Evolution. 2014;21(2):127–172. doi: 10.1007/s10914-013-9236-3. [DOI] [Google Scholar]
- Betancur-R R, Li CH, Munroe TA, et al Addressing gene tree discordance and non-stationarity to resolve a multi-locus phylogeny of the flatfishes (Teleostei: Pleuronectiformes) Systematic Biology. 2013;62(5):763–785. doi: 10.1093/sysbio/syt039. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Cardillo M, Jones KE, et al The delayed rise of present-day mammals. Nature. 2007;446(7135):507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Blanga-Kanfi S, Miranda H, Penn O, et al Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evolutionary Biology. 2009;9(1):71. doi: 10.1186/1471-2148-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brace S, Thomas JA, Dalén L, et al Evolutionary history of the Nesophontidae, the last unplaced recent mammal family. Molecular Biology and Evolution. 2016;33(12):3095–3103. doi: 10.1093/molbev/msw186. [DOI] [PubMed] [Google Scholar]
- Chen MY, Liang D, Zhang P Phylogenomic resolution of the phylogeny of Laurasiatherian mammals: exploring phylogenetic signals within coding and noncoding sequences. Genome Biology and Evolution. 2017;9(8):1998–2012. doi: 10.1093/gbe/evx147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Xu SX, Zhou KY, et al Whale phylogeny and rapid radiation events revealed using novel retroposed elements and their flanking sequences. BMC Evolutionary Biology. 2011;11(1):314. doi: 10.1186/1471-2148-11-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G, Sadasivuni MK, Rosenbloom KR, et al Rodent evolution: back to the root. Molecular Biology and Evolution. 2010;27(6):1315–1326. doi: 10.1093/molbev/msq019. [DOI] [PubMed] [Google Scholar]
- Collins TM, Fedrigo O, Naylor GJ Choosing the best genes for the job: the case for stationary genes in genome-scale phylogenetics. Systematic Biology. 2005;54(3):493–500. doi: 10.1080/10635150590947339. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Ranwez V. 2020. Accurate alignment of (meta)barcoding data sets using MACSE. In: Scornavacca C, Delsuc F, Galtier N. Phylogenetics in the Genomic Era. No Commercial Publisher, 2.3: 1–2.3: 31.
- Di Franco A, Poujol R, Baurain D, et al Evaluating the usefulness of alignment filtering methods to reduce the impact of errors on evolutionary inferences. BMC Evolutionary Biology. 2019;19(1):21. doi: 10.1186/s12862-019-1350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilcher D Toward a new synthesis: major evolutionary trends in the angiosperm fossil record. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(13):7030–7036. doi: 10.1073/pnas.97.13.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina L, Churakov G, Kuritzin A, et al Speciation network in Laurasiatheria: retrophylogenomic signals. Genome Research. 2017a;27(6):997–1003. doi: 10.1101/gr.210948.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina L, Churakov G, Shi JJ, et al Exploring massive incomplete lineage sorting in arctoids (Laurasiatheria, Carnivora) Molecular Biology and Evolution. 2015;32(12):3194–3204. doi: 10.1093/molbev/msv188. [DOI] [PubMed] [Google Scholar]
- Doronina L, Feigin CY, Schmitz J Reunion of Australasian possums by shared SINE insertions. Systematic Biology. 2022;71(5):1045–1053. doi: 10.1093/sysbio/syac025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina L, Matzke A, Churakov G, et al The beaver’s phylogenetic lineage illuminated by retroposon reads. Scientific Reports. 2017b;7(1):43562. doi: 10.1038/srep43562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Donoghue PCJ, Yang ZH Neither phylogenomic nor palaeontological data support a Palaeogene origin of placental mammals. Biology Letters. 2014;10(1):20131003. doi: 10.1098/rsbl.2013.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Donoghue PCJ, Yang ZH Bayesian molecular clock dating of species divergences in the genomics era. Nature Reviews Genetics. 2016;17(2):71–80. doi: 10.1038/nrg.2015.8. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Inoue J, Hasegawa M, et al Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proceedings of the Royal Society B: Biological Sciences. 2012;279(1742):3491–3500. doi: 10.1098/rspb.2012.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Thawornwattana Y, Angelis K, et al Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Current Biology. 2015;25(22):2939–2950. doi: 10.1016/j.cub.2015.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis MD, Gunnell GF, Barba-Montoya J, et al Using phylogenomic data to explore the effects of relaxed clocks and calibration strategies on divergence time estimation: primates as a test case. Systematic Biology. 2018;67(4):594–615. doi: 10.1093/sysbio/syy001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne DA, Bragg JG, Duchêne S, et al Analysis of phylogenomic tree space resolves relationships among marsupial families. Systematic Biology. 2018;67(3):400–412. doi: 10.1093/sysbio/syx076. [DOI] [PubMed] [Google Scholar]
- Duff A, Lawson A. 2004. Mammals of the World. A Checklist. London: A and C Black.
- Edwards SV Is a new and general theory of molecular systematics emerging? Evolution. 2009;63(1):1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- Emerling CA, Huynh HT, Nguyen MA, et al Spectral shifts of mammalian ultraviolet-sensitive pigments (short wavelength-sensitive opsin 1) are associated with eye length and photic niche evolution. Proceedings of the Royal Society B:Biological Sciences. 2015;282(1819):20151817. doi: 10.1098/rspb.2015.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esselstyn JA, Oliveros CH, Swanson MT, et al Investigating difficult nodes in the placental mammal tree with expanded taxon sampling and thousands of ultraconserved elements. Genome Biology and Evolution. 2017;9(9):2308–2321. doi: 10.1093/gbe/evx168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne RS, Pigot AL, Phillimore AB How reliably can we infer diversity-dependent diversification from phylogenies? Methods in Ecology and Evolution. 2016;7(9):1092–1099. doi: 10.1111/2041-210X.12565. [DOI] [Google Scholar]
- Feijoo M, Parada A Macrosystematics of eutherian mammals combining HTS data to expand taxon coverage. Molecular Phylogenetics and Evolution. 2017;113:76–83. doi: 10.1016/j.ympev.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Foley NM, Mason VC, Harris AJ, et al A genomic timescale for placental mammal evolution. Science. 2023;380(6643):eabl8189. doi: 10.1126/science.abl8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley NM, Springer MS, Teeling EC Mammal madness: is the mammal tree of life not yet resolved? Philosophical Transactions of the Royal Society B: Biological Sciences. 2016;371(1699):20150140. doi: 10.1098/rstb.2015.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatesy J, Meredith RW, Janecka JE, et al Resolution of a concatenation/coalescence kerfuffle: partitioned coalescence support and a robust family-level tree for Mammalia. Cladistics. 2017;33(3):295–332. doi: 10.1111/cla.12170. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Springer MS Phylogenomic red flags: homology errors and zombie lineages in the evolutionary diversification of placental mammals. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(45):E9431–E9432. doi: 10.1073/pnas.1715318114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossnickle DM, Newham E Therian mammals experience an ecomorphological radiation during the Late Cretaceous and selective extinction at the K-Pg boundary. Proceedings of the Royal Society B:Biological Sciences. 2016;283(1832):20160256. doi: 10.1098/rspb.2016.0256. [DOI] [Google Scholar]
- Gu ZG, Eils R, Schlesner M Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Guo YT, Zhang J, Xu DM, et al Phylogenomic relationships and molecular convergences to subterranean life in rodent family Spalacidae. Zoological Research. 2021;42(5):671–674. doi: 10.24272/j.issn.2095-8137.2021.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Chen X, Chen P, et al A new genus of Asiatic short-tailed shrew (Soricidae, Eulipotyphla) based on molecular and morphological comparisons. Zoological Research. 2018;39(5):321–334. doi: 10.24272/j.issn.2095-8137.2018.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhna S, Landis MJ, Heath TA, et al RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Systematic Biology. 2016;65(4):726–736. doi: 10.1093/sysbio/syw021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JY, Zhang YP, Yu L Summary of Laurasiatheria (Mammalia) phylogeny. Zoological Research. 2012;33(E5-6):65–74. doi: 10.3724/SP.J.1141.2012.E05-06E65. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Molecular Biology and Evolution. 2016;33(6):1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Ivanova NV, Clare EL, Borisenko AV. 2012. DNA barcoding in mammals. In: Kress WJ, Erickson DL. DNA Barcodes. Totowa: Humana Press, 153–182.
- Jebb D, Huang ZX, Pippel M, et al Six reference-quality genomes reveal evolution of bat adaptations. Nature. 2020;583(7817):578–584. doi: 10.1038/s41586-020-2486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma KI, et al MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay RF, Macfadden BJ, Madden RH, et al Revised age of the Salla beds, Bolivia, and its bearing on the age of the Deseadan South American Land Mammal “Age”. Journal of Vertebrate Paleontology. 1998;18(1):189–199. doi: 10.1080/02724634.1998.10011043. [DOI] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, et al RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21):4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert O, Martínez-Cáceres M, Bianucci G, et al Earliest mysticete from the Late Eocene of Peru sheds new light on the origin of baleen whales. Current Biology. 2017;27(10):1535–1541.e2. doi: 10.1016/j.cub.2017.04.026. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, et al PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution. 2017;34(3):772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- Lavergne A, Douzery E, Stichler T, et al Interordinal mammalian relationships: evidence for paenungulate monophyly is provided by complete mitochondrial 12S rRNA sequences. Molecular Phylogenetics and Evolution. 1996;6(2):245–258. doi: 10.1006/mpev.1996.0074. [DOI] [PubMed] [Google Scholar]
- Lee MSY, Hugall AF Partitioned likelihood support and the evaluation of data set conflict. Systematic Biology. 2003;52(1):15–22. doi: 10.1080/10635150390132650. [DOI] [PubMed] [Google Scholar]
- Lin JN, Chen GF, Gu L, et al Phylogenetic affinity of tree shrews to Glires is attributed to fast evolution rate. Molecular Phylogenetics and Evolution. 2014;71:193–200. doi: 10.1016/j.ympev.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang J, Rheindt FE, et al Genomic evidence reveals a radiation of placental mammals uninterrupted by the KPg boundary. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(35):E7282–E7290. doi: 10.1073/pnas.1616744114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Hu JY, Hu YW, et al Diverse phylogenomic datasets uncover a concordant scenario of Laurasiatherian interordinal relationships. Molecular Phylogenetics and Evolution. 2021;157:107065. doi: 10.1016/j.ympev.2020.107065. [DOI] [PubMed] [Google Scholar]
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes L, et al A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist. 2015;207(2):437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- Maliet O, Hartig F, Morlon H A model with many small shifts for estimating species-specific diversification rates. Nature Ecology & Evolution. 2019;3(7):1086–1092. doi: 10.1038/s41559-019-0908-0. [DOI] [PubMed] [Google Scholar]
- May-Collado LJ, Kilpatrick CW, Agnarsson I Mammals from ‘down under’: a multi-gene species-level phylogeny of marsupial mammals (Mammalia, Metatheria) PeerJ. 2015;3:e805. doi: 10.7717/peerj.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JE, Faircloth BC, Crawford NG, et al Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis. Genome Research. 2012;22(4):746–754. doi: 10.1101/gr.125864.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowen MR, Tsagkogeorga G, Álvarez-Carretero S, et al Phylogenomic resolution of the cetacean tree of life using target sequence capture. Systematic Biology. 2020;69(3):479–501. doi: 10.1093/sysbio/syz068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Janečka JE, Gatesy J, et al Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- Meredith RW, Westerman M, Springer MS A phylogeny of Diprotodontia (Marsupialia) based on sequences for five nuclear genes. Molecular Phylogenetics and Evolution. 2009;51(3):554–571. doi: 10.1016/j.ympev.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Meyer CP Molecular systematics of cowries (Gastropoda: Cypraeidae) and diversification patterns in the tropics. Biological Journal of the Linnean Society. 2003;79(3):401–459. doi: 10.1046/j.1095-8312.2003.00197.x. [DOI] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, et al IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missoup AD, Yemchui GD, Denys C, et al Molecular phylogenetic analyses indicate paraphyly of the genus Hybomys (Rodentia: Muridae): taxonomic implications. Journal of Zoological Systematics and Evolutionary Research. 2018;56(3):444–452. doi: 10.1111/jzs.12213. [DOI] [Google Scholar]
- Mitchell KJ, Pratt RC, Watson LN, et al Molecular phylogeny, biogeography, and habitat preference evolution of marsupials. Molecular Biology and Evolution. 2014;31(9):2322–2330. doi: 10.1093/molbev/msu176. [DOI] [PubMed] [Google Scholar]
- Montgelard C, Forty E, Arnal V, et al Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evolutionary Biology. 2008;8(1):321. doi: 10.1186/1471-2148-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlon H, Lewitus E, Condamine FL, et al RPANDA: an R package for macroevolutionary analyses on phylogenetic trees. Methods in Ecology and Evolution. 2016;7(5):589–597. doi: 10.1111/2041-210X.12526. [DOI] [Google Scholar]
- Murata Y, Nikaido M, Sasaki T, et al Afrotherian phylogeny as inferred from complete mitochondrial genomes. Molecular Phylogenetics and Evolution. 2003;28(2):253–260. doi: 10.1016/S1055-7903(03)00035-6. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Foley NM, Bredemeyer KR, et al Phylogenomics and the genetic architecture of the placental mammal Radiation. Annual Review of Animal Biosciences. 2021;9:29–53. doi: 10.1146/annurev-animal-061220-023149. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Künstner A, Wang R, et al Dynamic evolution of base composition: causes and consequences in avian phylogenomics. Molecular Biology and Evolution. 2011;28(8):2197–2210. doi: 10.1093/molbev/msr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita Y, Oda SI, Takenaka O, et al Phylogenetic position of Eulipotyphla inferred from the cDNA sequences of pepsinogens A and C. Molecular Phylogenetics and Evolution. 2001;21(1):32–42. doi: 10.1006/mpev.2001.0996. [DOI] [PubMed] [Google Scholar]
- Nishihara H, Satta Y, Nikaido M, et al A retroposon analysis of Afrotherian phylogeny. Molecular Biology and Evolution. 2005;22(9):1823–1833. doi: 10.1093/molbev/msi179. [DOI] [PubMed] [Google Scholar]
- Phillips MJ Geomolecular dating and the origin of placental mammals. Systematic Biology. 2016;65(3):546–557. doi: 10.1093/sysbio/syv115. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Pratt RC Family-level relationships among the Australasian marsupial “herbivores” (Diprotodontia: Koala, wombats, kangaroos and possums) Molecular Phylogenetics and Evolution. 2008;46(2):594–605. doi: 10.1016/j.ympev.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Politis DN, Romano JP The stationary bootstrap. Journal of the American Statistical Association. 1994;89(428):1303–1313. doi: 10.1080/01621459.1994.10476870. [DOI] [PubMed] [Google Scholar]
- Puttick MN, Thomas GH Fossils and living taxa agree on patterns of body mass evolution: a case study with Afrotheria. Proceedings of the Royal Society B: Biological Sciences. 2015;282(1821):20152023. doi: 10.1098/rspb.2015.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttick MN, Thomas GH, Benton MJ Dating placentalia: morphological clocks fail to close the molecular fossil gap. Evolution. 2016;70(4):873–886. doi: 10.1111/evo.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL Likelihood methods for detecting temporal shifts in diversification rates. Evolution. 2006;60(6):1152–1164. [PubMed] [Google Scholar]
- Rabosky DL, Grundler M, Anderson C, et al BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods in Ecology and Evolution. 2014;5(7):701–707. doi: 10.1111/2041-210X.12199. [DOI] [Google Scholar]
- Ranwez V, Douzery EJP, Cambon C, et al MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Molecular Biology and Evolution. 2018;35(10):2582–2584. doi: 10.1093/molbev/msy159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Roure B, et al Detecting and overcoming systematic errors in genome-scale phylogenies. Systematic Biology. 2007;56(3):389–399. doi: 10.1080/10635150701397643. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Delsuc F, et al Less is more in mammalian phylogenomics: AT-rich genes minimize tree conflicts and unravel the root of placental mammals. Molecular Biology and Evolution. 2013a;30(9):2134–2144. doi: 10.1093/molbev/mst116. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJP, et al Genomic evidence for large, long-lived ancestors to placental mammals. Molecular Biology and Evolution. 2013b;30(1):5–13. doi: 10.1093/molbev/mss211. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato JJ, Bradford TM, Armstrong KN, et al Post K-Pg diversification of the mammalian order Eulipotyphla as suggested by phylogenomic analyses of ultra-conserved elements. Molecular Phylogenetics and Evolution. 2019;141:106605. doi: 10.1016/j.ympev.2019.106605. [DOI] [PubMed] [Google Scholar]
- Sayyari E, Whitfield JB, Mirarab S DiscoVista: interpretable visualizations of gene tree discordance. Molecular Phylogenetics and Evolution. 2018;122:110–115. doi: 10.1016/j.ympev.2018.01.019. [DOI] [PubMed] [Google Scholar]
- Shen XX, Hittinger CT, Rokas A Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nature Ecology & Evolution. 2017;1(5):0126. doi: 10.1038/s41559-017-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution. 1999;16(8):1114. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- Shimodaira H, Hasegawa M CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17(12):1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Shimodaira H An approximately unbiased test of phylogenetic tree selection. Systematic Biology. 2002;51(3):492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Song S, Liu L, Edwards SV, et al Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(37):14942–14947. doi: 10.1073/pnas.1211733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Emerling CA, Meredith RW, et al Waking the undead: implications of a soft explosive model for the timing of placental mammal diversification. Molecular Phylogenetics and Evolution. 2017;106:86–102. doi: 10.1016/j.ympev.2016.09.017. [DOI] [PubMed] [Google Scholar]
- Springer MS, Foley NM, Brady PL, et al Evolutionary models for the diversification of placental mammals across the KPg boundary. Frontiers in Genetics. 2019;10:1241. doi: 10.3389/fgene.2019.01241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Murphy WJ, Eizirik E, et al Placental mammal diversification and the Cretaceous–Tertiary boundary. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(3):1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Woodburne MO The distribution of some basicranial characters within the Marsupialia and a phylogeny of the Phalangeriformes. Journal of Vertebrate Paleontology. 1989;9(2):210–221. doi: 10.1080/02724634.1989.10011755. [DOI] [Google Scholar]
- Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Buida III TJ, Labella AL, et al PhyKIT: a broadly applicable UNIX shell toolkit for processing and analyzing phylogenomic data. Bioinformatics. 2021;37(16):2325–2331. doi: 10.1093/bioinformatics/btab096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppan SJ, Adkins RM, Anderson J Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Systematic Biology. 2004;53(4):533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Steppan SJ, Schenk JJ Muroid rodent phylogenetics: 900-species tree reveals increasing diversification rates. PLoS One. 2017;12(8):e0183070. doi: 10.1371/journal.pone.0183070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MT, Oliveros CH, Esselstyn JA A phylogenomic rodent tree reveals the repeated evolution of masseter architectures. Proceedings of the Royal Society B: Biological Sciences. 2019;286(1902):20190672. doi: 10.1098/rspb.2019.0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarver JE, dos Reis M, Mirarab S, et al The interrelationships of placental mammals and the limits of phylogenetic inference. Genome Biology and Evolution. 2016;8(2):330–344. doi: 10.1093/gbe/evv261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upham NS, Esselstyn JA, Jetz W Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biology. 2019;17(12):e3000494. doi: 10.1371/journal.pbio.3000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wible JR, Rougier GW, Novacek MJ, et al Cretaceous eutherians and Laurasian origin for placental mammals near the K/T boundary. Nature. 2007;447(7147):1003–1006. doi: 10.1038/nature05854. [DOI] [PubMed] [Google Scholar]
- Wickham H, Chang W, Wickham MH Package ‘ggplot2’. Create Elegant Data Visualisations Using the Grammar of Graphics. Version. 2016;2(1):1–189. [Google Scholar]
- Wildman DE, Jameson NM, Opazo JC, et al A fully resolved genus level phylogeny of neotropical primates (Platyrrhini) Molecular Phylogenetics and Evolution. 2009;53(3):694–702. doi: 10.1016/j.ympev.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Yang ZH Estimating the pattern of nucleotide substitution. Journal of Molecular Evolution. 1994;39(1):105–111. doi: 10.1007/BF00178256. [DOI] [PubMed] [Google Scholar]
- Yang ZH PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yu L, Luan PT, Jin W, et al Phylogenetic utility of nuclear introns in interfamilial relationships of Caniformia (order Carnivora) Systematic Biology. 2011;60(2):175–187. doi: 10.1093/sysbio/syq090. [DOI] [PubMed] [Google Scholar]
- Zhang C, Sayyari E, Mirarab S. 2017. ASTRAL-III: increased scalability and impacts of contracting low support branches. In: Proceedings of the 15th International Workshop on RECOMB International Workshop on Comparative Genomics. Barcelona, Spain: Springer, 53–75.
- Zhou XM, Xu SX, Xu JX, et al Phylogenomic analysis resolves the interordinal relationships and rapid diversification of the Laurasiatherian mammals. Systematic Biology. 2012;61(1):150. doi: 10.1093/sysbio/syr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XM, Xu SX, Yang YX, et al Phylogenomic analyses and improved resolution of Cetartiodactyla. Molecular Phylogenetics and Evolution. 2011;61(2):255–264. doi: 10.1016/j.ympev.2011.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
The generated sequences were submitted to GenBank (accession numbers OR503106–OR504069). Custom Perl scripts and alignments are available at Dryad (DOI: 10.5061/dryad.crjdfn36w).