Abstract
Background
We profiled circulating plasma metabolites to identify systemic biochemical changes in clinical and biomarker-assisted diagnosis of Alzheimer’s disease (AD).
Methods
We used an untargeted approach with liquid chromatography coupled to high-resolution mass spectrometry to measure small molecule plasma metabolites from 150 clinically diagnosed AD patients and 567 age-matched healthy elderly of Caribbean Hispanic ancestry. Plasma biomarkers of AD were measured including P-tau181, Aβ40, Aβ42, total-tau, neurofilament light chain (NfL) and glial fibrillary acidic protein (GFAP). Association of individual and co-abundant modules of metabolites were tested with clinical diagnosis of AD, as well as biologically-defined AD pathological process based on P-tau181 and other biomarker levels.
Results
Over 6000 metabolomic features were measured with high accuracy. First principal component (PC) of lysophosphatidylcholines (lysoPC) that bind to or interact with docosahexaenoic acid (DHA), eicosapentaenoic acid (EPA) and arachidonic acid (AHA) was associated with decreased risk of AD (OR = 0.91 [0.89–0.96], p = 2e-04). Association was restricted to individuals without an APOE ε4 allele (OR = 0.89 [0.84–0.94], p = 8.7e-05). Among individuals carrying at least one APOE ε4 allele, PC4 of lysoPCs moderately increased risk of AD (OR = 1.37 [1.16–1.6], p = 1e-04). Essential amino acids including tyrosine metabolism pathways were enriched among metabolites associated with P-tau181 levels and heparan and keratan sulfate degradation pathways were associated with Aβ42/Aβ40 ratio.
Conclusions
Unbiased metabolic profiling can identify critical metabolites and pathways associated with β-amyloid and phosphotau pathology. We also observed an APOE-ε4 dependent association of lysoPCs with AD and biologically based diagnostic criteria may aid in the identification of unique pathogenic mechanisms.
Summary
Untargeted metabolomics identifies Lysophosphatidylcholines association with plasma Ptau181 levels and biological Alzheimer’s Disease in an APOE-ε4 dependent manner.
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder characterized by cognitive and memory decline, affecting millions of individuals worldwide. Despite extensive research, the underlying pathogenic mechanisms of AD have not been fully revealed, hindering the development of effective therapeutic strategies. However, recent advancements in high-throughput omics technologies have provided a powerful platform to explore the complex molecular landscape of AD1.
Mass spectrometry-based metabolic profiling, a.k.a. metabolomics, offers a comprehensive analysis of small molecules involved in cellular metabolism. It provides a unique opportunity to unravel metabolic alterations associated with disease pathogenesis, thus contributing to a better understanding of AD at the molecular level2–5. Untargeted metabolomics allows the unbiased profiling of the entire metabolome, including both known and unknown metabolites.
Metabolomic studies in AD have revealed a range of altered metabolic signaling. Several studies have demonstrated dysregulation of energy metabolism pathways in AD6–8. These alterations involve changes in glucose metabolism9, including reduced glycolysis10–14 and impaired mitochondrial function15–17, and decreased levels of metabolites such as glucose, lactate, and pyruvate18. Alterations in the tricarboxylic acid (TCA) cycle intermediates, have also been observed. Studies have reported lower levels of phosphatidylcholine, phosphatidylethanolamine, and sphingomyelins in AD19–22, suggesting disruptions in membrane integrity and signaling pathways, altered cholesterol metabolism has also been implicated. Other studies have uncovered alterations in amino acid metabolism in AD23. Reduced levels of certain amino acids, such as tryptophan24, phenylalanine25,26, tyrosine27,28, and branched-chain amino acids (valine, leucine, isoleucine)3,29–38 may reflect disruptions in neurotransmitter synthesis, neuroinflammation, and protein homeostasis. Studies have also shown alterations in the levels of neurotransmitters such as acetylcholine, glutamate, and γ-aminobutyric acid (GABA) in AD patients39–43. These changes may contribute to cognitive dysfunction and synaptic alterations in the disease.
Several groups have reported elevated levels of reactive oxygen species (ROS) and oxidative damage markers, along with alterations in antioxidant metabolites and enzymes, have been observed in whole blood and brains of AD patients. These findings suggest a role for altered redox status in AD pathogenesis44–47.
Given that metabolomics is the omics layer closest to the phenotype, it has the potential to uncover critical insights into the disease risk and progression, and potentially uncover therapeutic targets. By integrating metabolomics data with clinical diagnosis and plasma biomarker levels of AD, we aim to identify metabolic networks underlying the disease. In this study, we investigated the association between metabolites and clinical and biomarker assisted diagnosis of AD to detect early and mid-stage metabolic changes in disease.
Methods
Participants.
The Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA) has been recruiting individuals with suspected sporadic and familial AD and healthy controls similar in age through advertisements in local newspapers and radio stations, and through clinical referrals in the Dominican Republic and in the Washington Heights neighborhood of New York City48. Participants in this study provided informed consent under protocols approved by the Columbia University Irving Medical Center Institutional Review Board, and the National Health Bioethics Committee of the Dominican Republic (CONABIOS). They underwent a medical and neurological history and detailed examinations, neuropsychological testing, and collection of blood for plasma and DNA processing. CSF were performed in a subgroup of participants. The clinical diagnosis of Alzheimer’s disease (AD) was based on NIA-AA criteria49. All clinical diagnoses were determined in a consensus conference attended by a neurologist, a neuropsychologist, and an internist with expertise in dementia and geriatrics. Briefly, individuals with clinical AD must have a history of progressive cognitive decline in the absence of other brain disorders and objective evidence of a decline memory and in at least two other cognitive domains such as verbal fluency or executive function. Healthy controls showed no evidence of cognitive decline. For the analyses in this manuscript, only biological samples and data from individuals recruited between January 1, 2018, and April 30, 2022, were considered.
Sample collection
Blood was collected in K2EDTA tubes by standard venipuncture and transported to a laboratory for centrifugation, preparation of plasma, and storage at −80°C within 2 hours of collection. CSF was obtained by standard aseptic technique, distributed into aliquots of 400 μL each in polypropylene tubes, frozen, and stored at −80°C48.
Plasma and CSF metabolomics data generation
Plasma and CSF metabolites were extracted using acetonitrile and the extracts were injected in triplicate on two chromatographic columns: a hydrophilic interaction column (HILIC) under positive ionization (HILIC+)50 and a C18 column under negative ionization (C18-)51 coupled to a Thermo Orbitrap HFX Q-Exactive mass spectrometer, scanning for molecules within 85–1250 kDa. This produced three technical replicates per sample per column. The untargeted mass spectral data were processed through a computational pipeline that leverages open source feature detection and peak alignment software, apLCMS52 and xMSanalyzer53. The feature tables were generated containing information on the mass-to-charge (m/z) ratio, retention time, and median summarized abundance/intensity of each ion for each sample. Correction for batch effects was performed using ComBat, which uses an empirical Bayesian framework to adjust for known batches in which the samples were run54. Each of these ions are referred to as metabolic features. For the analysis, metabolic features detected in at least 70% of the samples were retained, leaving 3253 features from the HILIC + column and 3628 features from the C18- column for plasma samples and 4460 features from the HILIC + column and 4501 features from the C18- column for CSF samples. Zero-intensity values were considered below the detection limit of the instrument and were imputed with half the minimum intensity observed for each metabolic feature. The intensity of each metabolic feature was log10 transformed, quantile normalized, and auto-scaled for normalization and standardization.
Metabolite annotation
Annotations were made using an internal library and by matching to the Human Metabolome Database (HMDB) using the R package xMSannotator (version 1.3.2)53. This uses a multistage clustering algorithm method to determine metabolic pathway associations, intensity profiles, retention time, mass defect, and isotope/adduct patterns to assign putative annotations to metabolic features. When a feature had multiple matches, we used the following rules to assign an annotation: first, we screened features based on the confidence score assigned by xMSannotator, and the annotation with the highest score was used. Second, if all annotations had the same score, we chose the annotation with the lowest difference in expected and observed mass (delta parts per million (ppm)). Finally, if all features had the same score and delta ppm, we indicated the identity as “multiple matches” since we couldn’t decipher a unique putative annotation. If a feature did not match any database entries, it was denoted as “unknown” (33% from HILIC + column and 40% from C18 – column). The confidence in annotation was based on criteria defined by Schymanski et al55, where level 1 corresponds to a confirmed structure identified through MS/MS and/or comparison to an authentic standard; level 2 to a probable structure identified through spectral matches to a database; level 3 to a putative identification with a speculative structure; level 4 to an unequivocal molecular formula but with insufficient evidence to propose a structure; and level 5 to an exact mass but not enough information to assign a formula.
Blood based biomarker analyses
The methods have been previously described in detail48. Briefly, the plasma biomarkers assays were performed in duplicate using the SIMOA HD-X platform. Neurology 3-Plex A kits were used to determine levels of Aβ42, Aβ40, and T-tau, the Advantage V2 kit for P-tau181, and the Neurology 2-Plex B for GFAP and NfL. Ratio of Aβ42/Aβ40 was also calculated.
Biomarker positive for AD
Based on previous analysis48, P-tau-181 plasma level < 2.33 considered biomarker status negative and ≥ 2.33 considered biomarker status positive. We use biomarker positive and biological AD interchangeably in the manuscript.
Statistical analysis approach
We used two approaches to find circulating metabolic features associated with outcomes of interest: 1) a metabolome-wide association study (MWAS) framework with correction for multiple comparisons by controlling the false discovery rate (FDR) at 5%, and 2) a co-abundance analysis to find modules of metabolic features associated with outcomes, providing a means of unsupervised dimensionality reduction based on correlation between the metabolic features. Both analyses were conducted separately for data from each column. All analyses were conducted in R (version 3.6.3).
Metabolome-wide association study (MWAS)
MWAS was conducted using multiple linear models, adjusted for age and sex. The analyses were conducted separately for data from each column. We corrected for multiple comparisons using an FDR of 5% and q-values were estimated using the Benjamini-Hochberg (BH) method.
Metabolite Co-abundance analysis:
Co-abundance modular analysis was conducted using weighted gene correlation network analysis56 using the WGCNA R package (version 1.69). Using normalized intensity values for each metabolic feature from each sample, we first constructed a metabolic feature co-abundance network using pairwise Pearson correlations between each metabolic feature. We used a soft threshold of 4 for the HILIC + data and 3 for the C18 – data, chosen based on saturation of the R2 at 0.9. This correlation network, where the nodes are metabolic features and edges are the scaled correlation coefficients, was used to create the topological overlap matrix (TOM), which provides a measure of similarity between a given pair of metabolic features in the network. This similarity matrix was used to create a dendrogram to assign metabolic features into modules based on their co-abundance pattern. We used the following parameters: minimum module size of 30, merge cutHeight of 0.25, an unsigned network, and a reassign threshold of 0. After network and dendrogram construction, modules were defined using the moduleEigengenes function in WGCNA. The module eigengene is a quantitative representation of a module derived from a principal component analysis (PCA) as the first PC, conducted using only those metabolic features that were part of the module. Association analyses were conducted to find modules associated with outcomes in linear regression models, adjusted for age and sex. We used the Bonferroni method to correct for multiple comparisons.
Subgroup comparisons:
Subgroup comparisons were conducted using logistic regression and multinomial logistic regression using the R package nnet (version 7.3–12). We created three different models to compare the six different subgroups: a) BM+/Cases, BM+/Controls and BM−/cases vs BM−/Controls using healthy participants with biomarker status negative (BM-) as the reference (i.e BM−/controls), we compared metabolic features with differential levels in participants with i) biomarker status positive and a clinical diagnosis of AD (BM+/Cases), ii) biomarker status negative and a clinical diagnosis of AD (BM−/Cases) and, iii) biomarker positive with no clinical diagnosis of AD (BM+/Controls) ; b) BM+/Case and BM+/Control vs BM−/Control− metabolic features with differential levels among BM+/control, BM+/case, compared to BM−/control; and c) BM+/case compared to BM+/control.
Pathway analysis.
To determine the biological relevance of the metabolic features associated with AD and biomarkers, we conducted pathway analysis using the “functional analysis” module in MetaboAnalyst (version 5.0, ref)57, a web-based interface for comprehensive metabolomic data analysis. We used the MWAS results from both columns and applied a nominal p-value cut-off of 0.01 to determine metabolic pathway enrichment using the mummichog algorithm and the human MFN reference database58. We present results for pathways with a Fisher’s exact test p-value < 0.3.
Chemical class enrichment.
This approach was used to determine the different chemical classes represented by metabolic feature members of WGCNA modules significantly associated with outcomes. The main chemical classes enriched was determined using the Enrichment Analysis module in MetaboAnalyst using the HMDBIDs for features with an annotation confidence score < 3.
Construction of lysophosphatidylcholine (lysoPCs) components and stratified analysis by APOE-ε4 status
Based on findings described below we performed principal component analyses using all features that were annotated as lysoPCs from both columns (44 from HILIC + and 13 from C18-). Since PCs 1–5 explained ~ 60% of the variance in the data, we used the first five PCs in logistic regression models to find the association with clinical diagnosis of AD and biomarker positive status, adjusted for age and sex. We tested for the presence of an interaction between the combinations of the lysoPCs and the presence of at least one APOE-ε4 allele and performed a stratified analysis since there was a significant interaction term between APOE-ε4 allele and PC4 (p-value for interaction = 0.0058).
Correlation between plasma and CSF metabolites
Among people with both plasma and CSF metabolomics data available (n = 113), plasma metabolites with level 1 confidence that were significantly associated with any outcome were tested for their correlation with the same metabolite identified in CSF using spearman correlation.
Association between lysoPCs and brain pathology in the ROSMAP cohort
To provide external validation of our findings with lysoPCs, we obtained data from the ROSMAP cohort and examined associations between lysoPCs and brain pathology. We identified metabolites identified as lysoPCs and computed principal components as described above. Amongst the 42 PCs generated, we tested association of the top five with amyloid, tangles, total global pathology, clinical and pathological diagnosis of Alzheimer’s disease (AD).
Results
Study participants
717 participants were included in the study of whom 150 (20.9%) were diagnosed with clinical AD and 567 were cognitively unimpaired controls (Table 1). The study population had a mean age of 69.6 years (standard deviation (SD) = 7.6), the individuals with clinical AD were slightly older, with a mean age of 73.2 (SD = 8.3), compared to controls who had a mean age of 68.6 (SD = 7.2). Two-thirds of the group were women (65%) and this proportion was similar among AD patients (67%) and controls (65%). A third of the study group had at least one APOE-ε4 allele (38%) and this proportion was only marginally higher in AD (43%) compared to controls (36%). Among AD, 58% were biomarker positive, while 29% of controls were biomarker positive. The mean levels of most plasma-based AD biomarkers were higher in AD than in controls, including P-tau181 (3.02 pg/mL (SD = 1.7) in AD and 2.13 pg/mL (SD = 1) in healthy controls), NfL pg/mL (26.4 (SD = 20.5) in AD and 17.3 pg/mL (SD = 19.2) in healthy controls), and GFAP pg/mL (219 (163) in AD and 140 pg/mL (96) in healthy controls). The mean ratio of Αβ42/Αβ40 was nominally lower in AD cases (0.049 (SD = 0.01)) compared to healthy controls (0.053 (SD = 0.03)). A subset of the study population, n = 113, also had CSF metabolomic data generated (S Table 1). Among them, 35 were clinically diagnosed with AD and 78 were controls. We also obtained postmortem brain tissue metabolomic data from a subset of participants from the ROSMAP cohort, n = 110 (S Table 2). Of them, 71 were diagnosed with AD and had brain pathology information available.
Table 1.
Characteristics of the study population.
| control (N = 567) | AD (N = 150) | All (N = 717) | |
|---|---|---|---|
| Age at diagnosis or last visit (years) | |||
| Mean (SD) | 68.6 (7.17) | 73.2 (8.26) | 69.6 (7.64) |
| Sex | |||
| Men | 200 (35.3%) | 50 (33.3%) | 250 (34.9%) |
| Women | 367 (64.7%) | 100 (66.7%) | 467 (65.1%) |
| Plasma pTau181 cut-off | |||
| < 2.33 (Biomarker −) | 376 (66.3%) | 57 (38.0%) | 433 (60.4%) |
| ≥ 2.33 (Biomarker +) | 163 (28.7%) | 87 (58.0%) | 250 (34.9%) |
| Missing | 28 (4.9%) | 6 (4.0%) | 34 (4.7%) |
| APOE\varvecϵ4 allele | |||
| ϵ4 allele absent | 358 (63.1%) | 86 (57.3%) | 444 (61.9%) |
| At least 1ϵ4 allele | 20 6 (36.3%) | 64 (42.7%) | 270 (37.7%) |
| Missing | 3 (0.5%) | 0 (0%) | 3 (0.4%) |
| Plasma pTau181 | |||
| Mean (SD) | 2.13 (1.00) | 3.02 (1.67) | 2.32 (1.23) |
| Missing | 28 (4.9%) | 6 (4.0%) | 34 (4.7%) |
| Plasma NfL | |||
| Mean (SD) | 17.3 (19.2) | 26.4 (20.5) | 19.2 (19.8) |
| Missing | 28 (4.9%) | 6 (4.0%) | 34 (4.7%) |
| Plasma GFAP | |||
| Mean (SD) | 140 (95.9) | 219 (163) | 157 (118) |
| Missing | 28 (4.9%) | 6 (4.0%) | 34 (4.7%) |
| Plasma A\varvecβ42/A\varvecβ40 ratio | |||
| Mean (SD) | 0.053 (0.031) | 0.049 (0.011) | 0.053 (0.028) |
| Missing | 33 (5.8%) | 7 (4.7%) | 40 (5.6%) |
Metabolome wide association study
We identified 6445 and 5827 metabolic features in the HILIC + and C18- columns respectively. Restricting to metabolic features seen in at least 70% of the group, 3253 and 3628 features were filtered for further analysis. 669 features were associated with at least one phenotype (clinical diagnosis of AD, P-tau181 biomarker positive for AD or biological AD, plasma levels of Αβ42/Αβ 40 ratio, NfL, P-tau181 and GFAP). Of those, 174 features were annotated with level 1 to level 3 confidence based on Schymanski scale (Fig. 1, Table 2, supplementary Table 1).
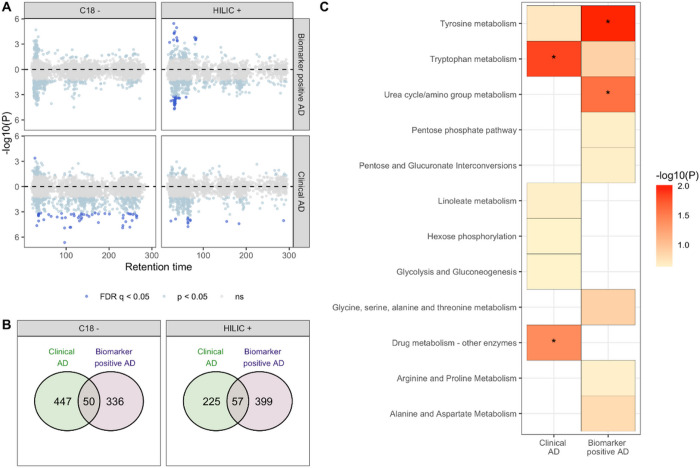
Figure 1: Metabolic features and pathways associated with clinical AD and with biomarker positive status.
In A, a modified Miami plot shows features with positive beta values above the zero line and those with negative beta values below the zero line. The dark blue points indicate features with FDR q-value < 0.05 for data obtained for each column (C18 and HILIC). In B, the overlap in features associated with clinical AD and biomarker positive status at nominal p < 0.05 (light blue and dark blue points) for each column. In C, the metabolic pathways, with Fisher’s exact test p < 0.3, enriched by features nominally associated with the clinical AD and biomarker positive status. An asterisk indicates pathway that were significantly enriched (p < 0.05).
Table 2. Metabolic features associated with outcomes investigated using a metabolome-wide association study framework.
m/z: mass-to-charge ratio, Time: Retention time, ID score: confidence in annotation based on Schymanski scale (1 being the highest and 5 the lowest), ESI: electrospray ionization, Delta ppm: mass difference in parts per million, ρCSF: correlation coefficient for metabolite measured in CSF (this was performed in a subset of participants, n = 113), Std: identify confirmed using chemical standard. See supplemental table for complete list of features associated at FDR of 5%.
| Outcome | m/z | Time (s) | Annotation | ID score | ESI | Delta ppm | Adduct | β | FDR q-value | ρ CSF | Pathway |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical AD | 203.0827 | 35.5 | Tryptophan | 1 | - | 0.49 | Std | −0.334 | 0.045 | 0.24 | Tryptophan metabolism |
| 133.0143 | 27.3 | Malic Acid | 1 | - | 0.38 | Std | 0.341 | 0.045 | TCA cycle, gluconeogenesis | ||
| 342.2649 | 109.1 | Dodecanoylcarnitine | 3 | - | 0.23 | M-H | −0.386 | 0.009 | Fatty acid oxidation | ||
| Biomarker/ pTau181 positive | 114.0663 | 33.5 | Creatinine | 1 | + | 0.96 | Std | 0.390 | 0.021 | 0.32 | Urea cycle/amino group metabolism |
| 524.3703 | 32.2 | LysoPC (18:0) | 1 | + | 1.47 | Std | −0.326 | 0.021 | Glycerophospholipid metabolism | ||
| 176.1033 | 82.9 | Citrulline | 1 | + | 1.87 | Std | 0.337 | 0.025 | 0.34 | Arginine and proline metabolism | |
| pTau181 | 203.1038 | 27.1 | Valyl-Serine | 1 | - | 0.34 | Std | −0.245 | 2.9E-04 | Glycine, serine, and threonine metabolism | |
| 114.0663 | 33.5 | Creatinine | 1 | + | 0.96 | Std | 0.251 | 6.2E-04 | 0.32 | Urea cycle/amino group metabolism | |
| 176.1033 | 82.9 | Citrulline | 1 | + | 1.87 | Std | 0.199 | 0.003 | 0.34 | Arginine and proline metabolism | |
| Aβ42/Aβ40 | 213.1497 | 242.2 | 3-Oxododecanoic acid | 3 | - | 0.38 | M-H | −0.011 | 4.0E-23 | Fatty acid metabolism | |
| 88.9881 | 236.8 | Oxalic acid | 3 | - | 0.79 | M-H | −0.006 | 5.4E-06 | Glyoxylate and dicarboxylate metabolism | ||
| 540.4982 | 22.9 | Hexacosanoyl carnitine | 3 | + | 0.81 | M + H | −0.006 | 8.1E-06 | Fatty acid oxidation | ||
| NfL | 195.051 | 33.4 | Gluconate | 1 | - | 0.15 | Std | 5.822 | 4.6E-11 | 0.52 | Pentose phosphate pathway |
| 160.0615 | 36.7 | 2-Aminoadipic acid/ N-methylglutamic acid | 1 | - | 0.12 | Std | 5.305 | 1.5E-09 | 0.16 | Lysine/Glutamate metabolism | |
| 149.0455 | 30.1 | Arabinose/lyxose/ribose/ xylose | 1 | - | 0.34 | Std | 5.030 | 2.9E-08 | 0.33 | Pentose phosphate pathway | |
| GFAP | 718.2797 | 178.4 | 5-Methyltetrahydropteroyltri glutamate | 3 | + | 0.85 | M + H | −17.369 | 0.032 | Methionine metabolism (gut bacteria) | |
| 522.3552 | 32.9 | LysoPC (18:1) | 3 | + | 0.42 | M + H | 16.776 | 0.044 | Glycerophospholipid metabolism | ||
| 405.3727 | 24.3 | Dihydroxycholestane | 3 | + | 0.02 | M + H | −16.646 | 0.044 | Bile acid biosynthesis |
We identified 107 metabolic features nominally associated (p < 0.05) with both clinical AD and biomarker positive status for AD. Metabolites associated with being biomarker positive for AD were enriched in tyrosine and urea cycle/amino acid metabolism pathways. Dodecanoylcarnitine (adj p = 0.009) and Ramipril (adj p = 0.02) were the top analytes associated with clinical AD while lysoPC(18:0) (adj p = 0.02) and creatinine (adj p = 0.02) were associated with biological AD. 151 metabolites with a level 1–3 confidence score for annotation were associated (adj p < 0.05) with at least one measured plasma biomarker (Fig. 2, Supplementary Table 1). The strongest association observed was 3-oxododecanoic acid with Αβ42/Αβ40 ratio (adj p = 4.03E-23). Oxalic acid and hexacosanoyl carnitine were also strongly associated with Αβ42/Αβ40 ratio. Metabolic features associated with Αβ42/Αβ40 ratio were enriched in heparan sulfate, chondroitin sulfate and keratan sulfate degradation processes. Increased sulfation and heparan sulfate proteoglycan degradation have been widely reported in AD related neuritic plaques previously.
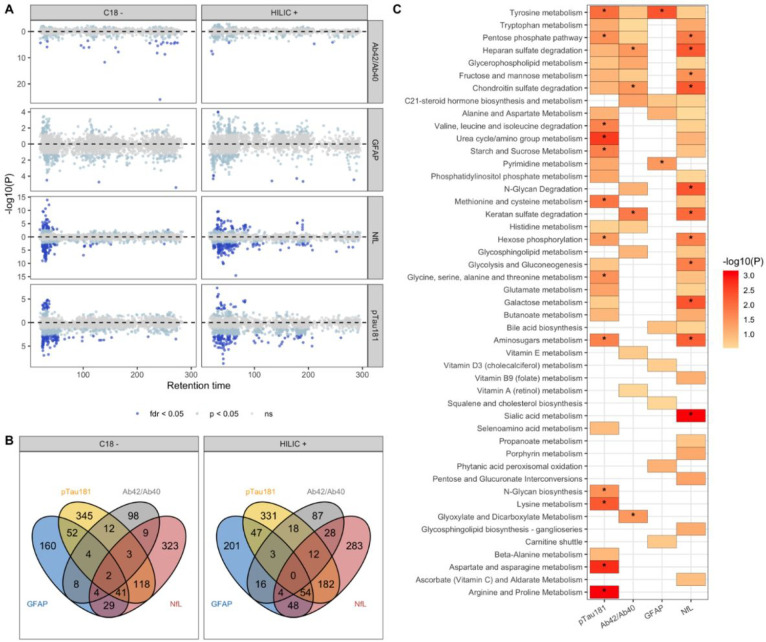
Figure 2: Metabolic features and pathways associated with plasma-based biomarkers of AD.
In A, a modified Miami plot shows features with positive beta values above the zero line and those with negative beta values below the zero line. The dark blue points indicate features with FDR q-value < 0.05 for data obtained for each column (C18 and HILIC). In B, the overlap in features associated with the four biomarkers at nominal p < 0.05 (light blue and dark blue points) for each column. In C, the metabolic pathways, with Fisher’s exact test p < 0.3, enriched by features nominally associated with the biomarkers. An asterisk indicates pathways that were significantly enriched (p < 0.05). 42/40: ratio of A 42 to A 40 measured in plasma, GFAP: glial fibrillary acidic protein, NfL: neurofilament light chain, pTau181: tau phosphorylated at threonine-181.
Valyl serine, creatinine and citrulline were among the 76 well annotated metabolic features associated with plasma levels of P-tau181. Several lysophosphatidylcholines (lysoPCs) including lysoPC(22:6), lysoPC(18:0) and lysoPC(20:4) were inversely associated after multiple testing correction with plasma P-tau181 levels (Supplementary Table 1). Metabolic features associated with P-tau181 levels were enriched in several essential amino acid metabolism pathways including tyrosine, arginine and proline metabolism, valine, leucine, and isoleucine degradation, and aminosugars, starch and sucrose metabolism. Lysine metabolism, aspartate and asparagine metabolism and arginine and proline metabolism were enriched only among P-tau181 associated metabolites.
406 metabolic features were associated with NfL levels in plasma, of which 107 were annotated with Level 1–3 confidence (Fig. 2, Supplementary Table 1). Gluconate (adj p = 4.56E-11) and Arabinose (adj p = 2.93E-08) in the pentose pathway and 2-Aminoadipic Acid (adj p = 1.53e-09) in Lysine/Glutamate metabolism were the top metabolites associated with NfL. Metabolic features associated with NfL shared common pathways with those associated with Αβ42/Αβ40 ratio including heparan sulfate, chondroitin sulfate and keratan sulfate degradation. Pentose phosphate, aminosugars metabolism and hexose phosphorylation pathways were shared by features associated with both NfL and P-tau181. Sialic acid metabolism was observed only amongst NfL associated metabolites.
Increased creatinine levels were associated with biological AD, increased amounts of plasma P-Tau181 and NfL. Several lysoPCs were observed to decrease in biological AD, and with increased amounts of plasma NfL and P-tau181 levels. Only lysoPC(18:1) was found to increase with increased levels of GFAP, although it was observed to decrease with increased P-tau181 and NfL levels indicating a time dependent abundance depending on the disease stage.
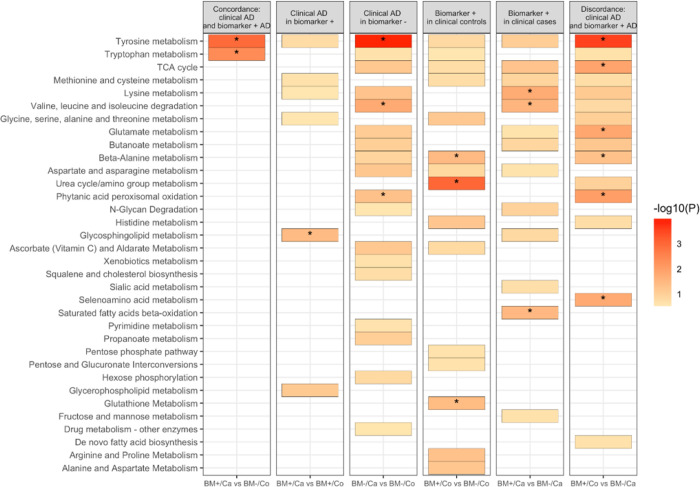
Metabolites in patients with discordant biological and clinical status of AD.
First, we compared healthy participants with no negative biomarker diagnosis (BM-/Control) with the other three groups (BM+/Case, BM+/Control and BM−/Case) (Fig. 3, Supplementary Figure S7). LysoPCs, in particular, lysoPC 22:6 (DHA) and lysoPC 20:5 (EPA) are reduced in clinical AD and biomarker positive patients. They are depleted the most in patients with both biological and clinical diagnosis of AD. Similarly, creatinine is increased in BM + and clinical AD patients and is highest in patients with elevated Ptau-181. Tyrosine metabolism is enriched amongst metabolites that are elevated in patients with either clinical or biological AD. Glycosphingolipid metabolism is altered only in patients with only clinical diagnosis of AD and urea cycle and amino group metabolism is altered in biological AD patients.
Figure 3: Subgroup analysis based on clinical diagnosis and biomarker positive status.
Results from multinomial and regular logistic regression were used to determine metabolic pathways enriched. A colored box indicates an enriched pathway with Fisher’s exact test p-value < 0.3 while an asterisk indicates statistically significant enrichment (p < 0.05).
Co-abundance analysis of metabolites.
We clustered co-abundant metabolic features using WGCNA independently on metabolic features detected in the HILIC and C18 columns. WGCNA identified 18 and 15 co-abundant metabolic color-coded modules in HILIC and C18 columns respectively with at least 30 metabolic features (Supplementary Figure S2). We then tested association of each module with clinical and biological AD and levels of the plasma biomarkers (Fig. 4A). Purple module was negatively associated with biological AD (adj p = 9e-05) while black module was associated with P-tau181 levels (adj p = 3e-04). Salmon and greenyellow modules were associated both with biological AD and P-tau181 levels. Enrichment analysis of the metabolites co-abundant in the purple module found that fatty amides (adj p = 5e-03), glycerophosphocholines (adj p = 5e-03) and sphingoid bases (adj p = 5e-03) were over-represented in the module (Fig. 4B). Glycerophosphocholines (adj p = 3e-22) were also significantly enriched in the greenyellow module, while amino acids and peptides were the top group over-represented in the black (adj p = 1.31e-16) and salmon modules (9.22E-07).
Figure 4: Results from co-expression analysis using data from the HILIC column.
In A, the volcano plot shows metabolic modules significantly associated with clinical AD, biomarker positive status and AD biomarkers using Bonferroni adjusted p-value. In B, the chemical classes enriched by module member metabolic features present at a proportion of at least 6.5%. In C, module hub members of the purple module with KME > 0.6 and associated with biomarker positive status at FDR q-value < 0.05. In D, module hub members of the salmon module with KME > 0.6 and associated with biomarker positive status at FDR q-value < 0.05. In E, module hub members of the greenyellow module with KME > 0.6 and associated with biomarker positive status at FDR q-value < 0.05. In F, module hub members of the yellow module with KME > 0.6 and associated with clinical AD at FDR q-value < 0.05.
We then identified the hub metabolites that are most connected to other metabolites in the purple, salmon and greenyellow modules. Interestingly, 12 lysoPCs were hub metabolites in the purple module and all of them were more abundant in biomarker negative participants compared to individuals who were biomarker positive and defined to have biological AD (Fig. 4C). Phosphatidylcholines (PC) and lysoPCs were also the hub metabolites in the greenyellow module and (Fig. 4E, Supplementary table 2) and were also more abundant in healthy participants compared to biomarker positive patients. Creatinine was the most connected metabolite in the salmon module and as previously described, was increased in AD patients compared to controls.
LysoPCs association with AD biomarkers
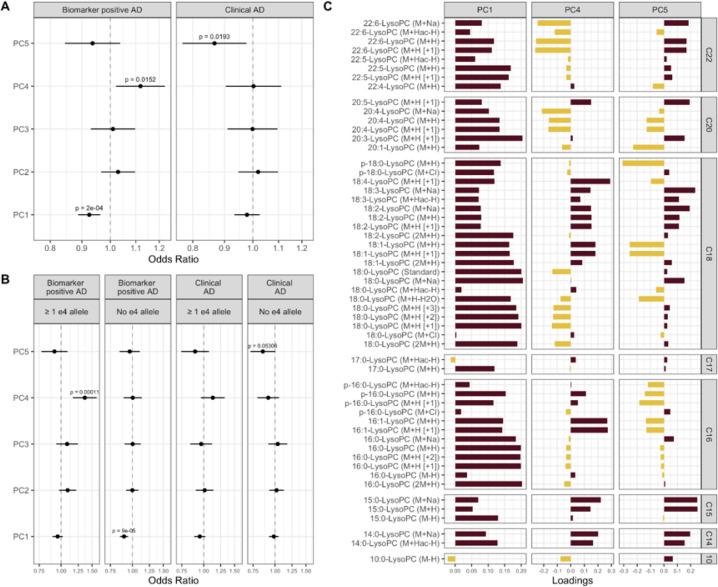
Both MWAS and WGCNA detected lysoPCs to be significantly associated with biological AD, P-tau181 and NFL levels. Thus, we tested joint association of all lysoPCs with clinical and biological AD by constructing lysoPC principal components. We constructed PCs for the 55 lysoPCs detected by HILIC and C18 columns and found that the first five PCs explained ~ 60% of the variance (Supplementary Figure S3). We tested association of the first five PCs together in a regression model adjust for age and sex (Fig. 5A). PC1 and PC4 were associated with biological AD whereas PC5 was associated with clinical diagnosis of AD. PC1 and PC5 were protective while PC4 increased risk of biological AD. Further stratifying participants by presence of absence of APOE ε4 allele (Fig. 5B), we found that PC1 and PC5 were protective of biological AD and clinical AD respectively, only in APOE ε4 non-carriers, whereas risk conferred by PC4 was restricted to APOE ε4 carriers. We investigated the loadings of the lysoPCs on PCs 1,4 and 5 and particularly focused on lysoPCs that have poly unsaturated fatty acids (PUFAs) at the sn-1 and sn-2 positions (Fig. 5C, Supplementary Fig. 5B). LysoPCs that carry eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA) and arachidonic acid (AHA) had positive loading on PC1 and hence decreased in biological AD, particularly in APOE ε4 non-carriers. Both DHA and AHA had negative loadings but EPA had positive loadings on PC4 which increased risk of biological AD. We also tested the correlation between PCs 1, 4 and 5 with circulating PUFAs in plasma (Supplementary Figure S5) and CSF. PC1 was positively correlated with circulating levels of EPA and AHA, and positively correlated with CSF levels of AHA. PC4 was negatively correlated with most measured plasma PUFAs and negatively correlated with CSF levels of DHA. PC5 was positively correlated with plasma linolenic acid, EPA and DHA, negatively correlated with plasma AHA, and positively correlated with CSF DHA. We also found that lysoPCs that carry EPA and DHA were positively correlated with the CSF levels of their respective PUFAs, while this correlation for AHA was negligible (Supplementary Figure S5). This indicates that lysoPCs, in conjunction with APOE, might play a role in AD biology since they transport long chain PUFAs into the central nervous system59.
Figure 5: Lysophosphatidylcholines (LysoPCs) associated with clinical AD and biomarker positive status.
In A, the odds ratio (point) and confidence interval (whiskers) of PC1 – 5 in relation to biomarker positive status and clinical AD. In B, the results from analysis stratified by APOE-ε4 allele status. In C, the loadings of lysoPCs on the three PCs (PC1, PC4, and PC5) significantly associated with biomarkers positive status or clinical AD.
LysoPC analysis in the ROSMAP cohort
To determine the generalizability of our results we examined the association between lysoPCs and AD pathology in the ROSMAP cohort. We used the metabolomics data derived from 110 brain samples in the ROSMAP cohort to test association of lysoPCs and phosphatidylcholines (PCs) with pathological definition of AD, amyloid burden, tangle density, and global pathology. First, we detected 14 LysoPCs and 13 PCs in the ROSMAP cohort. We constructed principal components from the LysoPCs and tested association with AD pathology (Supplementary Figure S6). We identified that PC3 is positively associated with increased tangle density, global pathology and a pathological diagnosis of AD. Of the lysoPCs carrying PUFAs, we only detected AHA in the ROSMAP cohort. AHA is increased in tangles, global pathology and pathological definition of AD. Three phosphatidylcholines were negatively associated with amyloid burden and tau tangles implying that lysoPCs and PCs are reduced in post-mortem AD brains. These findings are consistent with plasma lysoPCs observation in the EFIGA cohort.
Discussion
We investigated the association of metabolites with plasma biomarkers and clinical diagnoses in a cohort of Caribbean Hispanics to identify metabolic pathways associated with hallmarks of AD pathology. Two of the most notable findings were that metabolite profiles differed when a clinical diagnosis was used versus a validated plasma biomarker-based diagnosis and that lysoPCs, which have been reported in recent studies in AD59,60, were identified in our unbiased approach in a Hispanic population.
LysoPCs were associated with both quantitative levels of plasma P-tau181 and biological AD (defined by P-tau181). Co-abundance analysis revealed P-tau181 association of metabolic modules that harbor several lysoPCs as hub metabolites, suggesting a critical role in disease pathogenesis. Several studies have observed lower levels of lysoPCs in the brains, CSF and plasma of AD patients61–72. These changes often involve lysoPC species, particularly those that bind to anti-inflammatory PUFAs being decreased in patients with AD. Some lysoPC species have been implicated in promoting neurotoxicity and inflammation73–75. They can induce oxidative stress, impair mitochondrial function, and activate immune cells, leading to neuronal damage and death. LysoPCs are also involved in dysregulation of lipid metabolism these disturbances. The breakdown of phosphatidylcholine, a major lipid component of cell membranes, can generate lysoPCs. Disruptions in enzymes involved in this process, such as phospholipase A2 (PLA2), have been observed in AD and may contribute to altered lysoPC levels76–78.
We observed a differential effect of lysoPCs within APOE ε4 carriers and non-carriers. The risk conferred by lysoPCs was restricted to APOE ε4 carriers, while the protective effects were significant within APOE ε4 non-carriers. We previously showed significant differences in metabolic profiles in a small multi-ethnic AD cohort and these differences remained when the analysis was restricted to APOE ε4 carriers79. APOE ε4 carriers tend to exhibit higher levels of specific lysoPC species in CSF, plasma, and brain tissue compared to non-carriers80–83. Elevated levels of certain lysoPCs in APOE ε4 carriers have been linked to increased Aβ deposition, tau phosphorylation, and neuroinflammation. Distinct patterns of lysoPC alterations have been observed in APOE ε4 carriers compared to non-carriers.
We also found essential amino acids metabolism (tryptophan and tyrosine) were associated with clinical and biological diagnosis of AD. Urea cycle/amino group metabolism was associated with only the biological diagnosis of AD. Tyrosine is an essential amino acid and plays a crucial role in the synthesis of catecholamines. Limited research has focused on measuring tyrosine levels specifically in the AD patient brains but administering tyrosine orally can enhance memory and cognitive function84. Tryptophan is an essential amino acid and a precursor for the synthesis of serotonin, a neurotransmitter involved in mood regulation and cognition. Alterations in tryptophan metabolism may impact serotonin availability in the brain and contribute to AD pathophysiology, particularly (Aβ) pathology. Aβ accumulation can disrupt tryptophan metabolism, leading to altered levels of tryptophan and its metabolites. Conversely, tryptophan metabolites, such as kynurenic acid, can affect Aβ aggregation and clearance, potentially influencing disease progression. Interestingly Tryptophan levels in plasma were associated with clinical diagnosis of AD and were also mildly correlated with CSF levels (correlation = 0.24, Table 2).
Heparan sulfate, chondroitin sulfate and keratan sulfate degradation processes were associated with Aβ42/40 ratio. Heparan sulfate, chondroitin sulfate, and keratan sulfate are types of glycosaminoglycans (GAGs) or sulfated carbohydrates that are found in the extracellular matrix of cells. GAGs have been reported in accumulation and clearance of in Aβ. Heparan sulfate proteoglycans (HSPGs) are a type of protein with heparan sulfate chains that interact with Aβ and can contribute to the formation of amyloid plaques. Chondroitin sulfate proteoglycans (CSPGs) and HSPGs have been implicated in the regulation of Aβ clearance. These sulfated glycosaminoglycans can interact with various proteins involved in the clearance of Aβ, including neprilysin and insulin-degrading enzyme. Disruption of the balance between Aβ production and clearance, partly mediated by GAGs, may contribute to the accumulation of Aβ in AD. GAGs can interact with various inflammatory molecules, including cytokines and chemokines, and modulate neuroinflammatory processes in AD. Chondroitin sulfate and heparan sulfate chains present on proteoglycans can act as binding sites for inflammatory molecules, contributing to the activation of immune cells and the generation of a pro-inflammatory environment in the brain.
Taken together these results suggest that understanding metabolic heterogeneity in AD pathogenesis may enable identification of biological mechanisms for specific subgroups with the disease and that it is essential to combine biochemical analysis with biomarkers of disease. Specifically, identification of metabolic pathways associated with plasma biomarkers might indicate biological mechanisms underlying AD pathology at different stages of the disease. We observed common metabolic pathways perturbed in clinical AD and elevated Aβ42/40 ratio, indicating that these metabolites might be involved in both amyloidogenic and later (clinical) stages of the disease. Similarly, distinct set of metabolites were observed in association with elevated P-tau181 and NfL levels, suggesting processes that might be involved both in neurofibrillary change and neurodegeneration. However, more investigation specifically with longitudinal measures of biomarkers and metabolic assessments are needed to disentangle the metabolic cascades in different stages of disease progression. Finally, this study demonstrates the ability of high-resolution mass spectrometry-based untargeted metabolomics to reveal biochemical differences in participants with differential plasma biomarker profiles and to identify metabolic perturbations in different stages of the disease. This has the potential to open up a new era of biochemically-based discovery in AD.
Acknowledgements
EFIGA study is supported by NIA grants R56AG063908, R01AG067501 and RF1AG015473. We acknowledge the services of CEDIMAT for collaborating with sample collection and processing in the EFIGA cohort.
The metabolomics core that generated the metabolomics data for the project is supported by the National Center for Advancing Translational Sciences grant-5UL1TR001873.
Vrinda Kalia a, Dolly Reyes-Dumeyer b,c, Saurabh Dubey a, Renu Nandakumar a, Annie J. Lee b,c, Rafael Lantigua d, Martin Medrano e, Diones Rivera f, Lawrence S. Honig b,c,g, Richard Mayeux b,c,g,h, Gary W. Miller+ a,h, Badri N. Vardarajan+ b,c,g
Footnotes
Declarations
Competing interests: The authors do not have any conflict of interest with the research presented in this investigation.
Additional Declarations: There is NO Competing Interest.
Contributor Information
Badri Vardarajan, Columbia University.
Vrinda Kalia, Columbia University Mailman School of Public Health.
Dolly Reyes-Dumeyer, Columbia University Irving Medical Center.
Saurabh Dubey, Columbia University.
Renu Nandakumar, Columbia University.
Annie Lee, Center for Translational & Computational Neuroimmunology.
Rafael Lantigua, Columbia University.
Martin Medrano, Pontificia Universidad Catolica Madre y Masestra.
Diones Rivera, CEDIMAT.
Lawrence Honig, Columbia University.
Richard Mayeux, Columbia University.
Gary Miller, Columbia University.
Data and materials availability:
All intermediate and final results are included in the main text or the supplementary materials of the manuscript. The raw metabolomics and biomarker will be shared with qualified investigators using the request form available here https://cumc.co1.qualtrics.com/jfe/form/SV_dmck0uV3A91pmzb
References
- 1.Badhwar A et al. (2020) A multiomics approach to heterogeneity in Alzheimer’s disease: focused review and roadmap. Brain 143:1315–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson CH, Ivanisevic J, Siuzdak G (2016) Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 17:451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horgusluoglu E et al. (2022) Integrative metabolomics-genomics approach reveals key metabolic pathways and regulators of Alzheimer’s disease. Alzheimers Dement 18:1260–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkins JM, Trushina E (2017) Application of Metabolomics in Alzheimer’s Disease. Front Neurol 8:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huo Z et al. (2020) Brain and blood metabolome for Alzheimer’s dementia: findings from a targeted metabolomics analysis. Neurobiol Aging 86:123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu L, Jin J, Xu Y, Zhu X (2022) Aberrant Energy Metabolism in Alzheimer’s Disease. J Transl Int Med 10:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minhas PS et al. (2021) Restoring metabolism of myeloid cells reverses cognitive decline in ageing. Nature 590:122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartke A, Brannan S, Hascup E, Hascup K, Darcy J (2021) Energy Metabolism and Aging. World J Mens Health 39:222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunnane SC et al. (2020) Brain energy rescue: an emerging therapeutic concept for neurodegenerative disorders of ageing. Nat Rev Drug Discov 19:609– 633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoyer S, Nitsch R, Oesterreich K (1991) Predominant abnormality in cerebral glucose utilization in late-onset dementia of the Alzheimer type: a cross-sectional comparison against advanced late-onset and incipient early-onset cases. J Neural Transm Park Dis Dement Sect 3:1–14 [DOI] [PubMed] [Google Scholar]
- 11.An Y et al. (2018) Evidence for brain glucose dysregulation in Alzheimer’s disease. Alzheimers Dement 14:318–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madeira C et al. (2015) d-serine levels in Alzheimer’s disease: implications for novel biomarker development. Transl Psychiatry 5:e561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, Zhang X, Zhao L (2018) Human ApoE Isoforms Differentially Modulate Brain Glucose and Ketone Body Metabolism: Implications for Alzheimer’s Disease Risk Reduction and Early Intervention. J Neurosci 38:6665–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto K et al. (2004) Possible role of D-serine in the pathophysiology of Alzheimer’s disease. Prog Neuropsychopharmacol Biol Psychiatry 28:385–388 [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Zhao F, Ma X, Perry G, Zhu X (2020) Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol Neurodegener 15:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quntanilla RA, Tapia-Monsalves C (2020) The Role of Mitochondrial Impairment in Alzheimer s Disease Neurodegeneration: The Tau Connection. Curr Neuropharmacol 18:1076–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jadiya P et al. (2019) Impaired mitochondrial calcium efflux contributes to disease progression in models of Alzheimer’s disease. Nat Commun 10:3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Alshakhshir N, Zhao L (2021) Glycolytic Metabolism, Brain Resilience, and Alzheimer’s Disease. Front Neurosci 15:662242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosicek M, Hecimovic S (2013) Phospholipids and Alzheimer’s disease: alterations, mechanisms and potential biomarkers. Int J Mol Sci 14:1310–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiley L et al. (2014) Evidence of altered phosphatidylcholine metabolism in Alzheimer’s disease. Neurobiol Aging 35:271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin F (2023) Lipid metabolism and Alzheimer’s disease: clinical evidence, mechanistic link and therapeutic promise. FEBS J 290:1420–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patrick RP (2019) Role of phosphatidylcholine-DHA in preventing APOE4-associated Alzheimer’s disease. FASEB J 33:1554–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffin JW, Bradshaw PC (2017) Amino Acid Catabolism in Alzheimer’s Disease Brain: Friend or Foe? Oxid Med Cell Longev 5472792 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savonije K, Weaver DF (2023) The Role of Tryptophan Metabolism in Alzheimer’s Disease. Brain Sci 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P et al. (2021) Phenylalanine Metabolism Is Dysregulated in Human Hippocampus with Alzheimer’s Disease Related Pathological Changes. J Alzheimers Dis 83:609–622 [DOI] [PubMed] [Google Scholar]
- 26.Whiley L et al. (2021) Metabolic phenotyping reveals a reduction in the bioavailability of serotonin and kynurenine pathway metabolites in both the urine and serum of individuals living with Alzheimer’s disease. Alzheimers Res Ther 13:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCann A et al. (2021) Serum tyrosine is associated with better cognition in Lewy body dementia. Brain Res 1765:147481. [DOI] [PubMed] [Google Scholar]
- 28.Hajjar I, Liu C, Jones DP, Uppal K (2020) Untargeted metabolomics reveal dysregulations in sugar, methionine, and tyrosine pathways in the prodromal state of AD. Alzheimers Dement (Amst) 12:e12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basun H et al. (1990) Amino acid concentrations in cerebrospinal fluid and plasma in Alzheimer’s disease and healthy control subjects. J Neural Transm Park Dis Dement Sect 2:295–304 [DOI] [PubMed] [Google Scholar]
- 30.Fonteh AN, Harrington RJ, Tsai A, Liao P, Harrington MG (2007) Free amino acid and dipeptide changes in the body fluids from Alzheimer’s disease subjects. Amino Acids 32:213–224 [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Dominguez R, Garcia-Barrera T, Gomez-Ariza JL (2015) Metabolite profiling for the identification of altered metabolic pathways in Alzheimer’s disease. J Pharm Biomed Anal 107:75–81 [DOI] [PubMed] [Google Scholar]
- 32.Ibanez C et al. (2012) Toward a predictive model of Alzheimer’s disease progression using capillary electrophoresis-mass spectrometry metabolomics. Anal Chem 84:8532–8540 [DOI] [PubMed] [Google Scholar]
- 33.Larsson SC, Markus HS (2017) Branched-chain amino acids and Alzheimer’s disease: a Mendelian randomization analysis. Sci Rep 7:13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H et al. (2018) Defect of branched-chain amino acid metabolism promotes the development of Alzheimer’s disease by targeting the mTOR signaling. Biosci Rep 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz HH et al. (2016) Increased susceptibility to metabolic dysregulation in a mouse model of Alzheimer’s disease is associated with impaired hypothalamic insulin signaling and elevated BCAA levels. Alzheimers Dement 12:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toledo JB et al. (2017) Metabolic network failures in Alzheimer’s disease: A biochemical road map. Alzheimers Dement 13:965–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trushina E, Dutta T, Persson XM, Mielke MM, Petersen RC (2013) Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS ONE 8:e63644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tynkkynen J et al. (2018) Association of branched-chain amino acids and other circulating metabolites with risk of incident dementia and Alzheimer’s disease: A prospective study in eight cohorts. Alzheimers Dement 14:723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kandimalla R, Reddy PH (2017) Therapeutics of Neurotransmitters in Alzheimer’s Disease. J Alzheimers Dis 57:1049–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francis PT (2005) The interplay of neurotransmitters in Alzheimer’s disease. CNS Spectr 10:6–9 [DOI] [PubMed] [Google Scholar]
- 41.Yang Z, Zou Y, Wang L (2023) Neurotransmitters in Prevention and Treatment of Alzheimer’s Disease. Int J Mol Sci 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y et al. (2016) Implications of GABAergic Neurotransmission in Alzheimer’s Disease. Front Aging Neurosci 8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y et al. (2012) Neurotransmitter receptors and cognitive dysfunction in Alzheimer’s disease and Parkinson’s disease. Prog Neurobiol 97:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez de Toda I, Miguelez L, Vida C, Carro E (2019) De la Fuente, M. Altered Redox State in Whole Blood Cells from Patients with Mild Cognitive Impairment and Alzheimer’s Disease. J Alzheimers Dis 71:153–163 [DOI] [PubMed] [Google Scholar]
- 45.Holubiec MI, Gellert M, Hanschmann EM (2022) Redox signaling and metabolism in Alzheimer’s disease. Front Aging Neurosci 14:1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang WJ, Zhang X, Chen WW (2016) Role of oxidative stress in Alzheimer’s disease. Biomed Rep 4:519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butterfield DA, Halliwell B (2019) Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat Rev Neurosci 20:148–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Honig LS et al. (2023) Evaluation of Plasma Biomarkers for A/T/N Classification of Alzheimer Disease Among Adults of Caribbean Hispanic Ethnicity. JAMA Netw Open 6:e238214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKhann GM et al. (2011) The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 7:263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buszewski B, Noga S (2012) Hydrophilic interaction liquid chromatography (HILIC)--a powerful separation technique. Anal Bioanal Chem 402:231–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada T et al. (2013) Supercritical fluid chromatography/Orbitrap mass spectrometry based lipidomics platform coupled with automated lipid identification software for accurate lipid profiling. J Chromatogr A 1301:237–242 [DOI] [PubMed] [Google Scholar]
- 52.Yu T, Park Y, Johnson JM, Jones DP (2009) apLCMS–adaptive processing of high-resolution LC/MS data. Bioinformatics 25:1930–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uppal K, Walker DI, Jones DP, xMSannotator (2017) An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal Chem 89:1063–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28:882–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schymanski EL et al. (2014) Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol 48:2097–2098 [DOI] [PubMed] [Google Scholar]
- 56.Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia J, Psychogios N, Young N, Wishart DS (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res 37:W652–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li S et al. (2013) Predicting network activity from high throughput metabolomics. PLoS Comput Biol 9:e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Semba RD, Perspective (2020) The Potential Role of Circulating Lysophosphatidylcholine in Neuroprotection against Alzheimer Disease. Adv Nutr 11:760–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Law SH et al. (2019) An Updated Review of Lysophosphatidylcholine Metabolism in Human Diseases. Int J Mol Sci 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmerler D et al. (2012) Targeted metabolomics for discrimination of systemic inflammatory disorders in critically ill patients. J Lipid Res 53:1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mulder C et al. (2003) Decreased lysophosphatidylcholine/phosphatidylcholine ratio in cerebrospinal fluid in Alzheimer’s disease. J Neural Transm (Vienna) 110:949–955 [DOI] [PubMed] [Google Scholar]
- 63.Fan L et al. (2012) Identification of metabolic biomarkers to diagnose epithelial ovarian cancer using a UPLC/QTOF/MS platform. Acta Oncol 51:473–479 [DOI] [PubMed] [Google Scholar]
- 64.Chan RB et al. (2012) Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J Biol Chem 287:2678–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grimm MO et al. (2011) From brain to food: analysis of phosphatidylcholins, lyso-phosphatidylcholins and phosphatidylcholin-plasmalogens derivates in Alzheimer’s disease human post mortem brains and mice model via mass spectrometry. J Chromatogr A 1218:7713–7722 [DOI] [PubMed] [Google Scholar]
- 66.Villamil-Ortiz JG et al. (2018) Differential Pattern of Phospholipid Profile in the Temporal Cortex from E280A-Familiar and Sporadic Alzheimer’s Disease Brains. J Alzheimers Dis 61:209–219 [DOI] [PubMed] [Google Scholar]
- 67.Ross BM, Moszczynska A, Erlich J, Kish SJ (1998) Phospholipid-metabolizing enzymes in Alzheimer’s disease: increased lysophospholipid acyltransferase activity and decreased phospholipase A2 activity. J Neurochem 70:786–793 [DOI] [PubMed] [Google Scholar]
- 68.Li NJ et al. (2010) Plasma metabolic profiling of Alzheimer’s disease by liquid chromatography/mass spectrometry. Clin Biochem 43:992–997 [DOI] [PubMed] [Google Scholar]
- 69.Liu Y, Li N, Zhou L, Li Q, Li W (2014) Plasma metabolic profiling of mild cognitive impairment and Alzheimer’s disease using liquid chromatography/mass spectrometry. Cent Nerv Syst Agents Med Chem 14:113–120 [DOI] [PubMed] [Google Scholar]
- 70.Cui Y et al. (2014) Lysophosphatidylcholine and amide as metabolites for detecting alzheimer disease using ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry-based metabonomics. J Neuropathol Exp Neurol 73:954–963 [DOI] [PubMed] [Google Scholar]
- 71.Gonzalez-Dominguez R, Garcia-Barrera T, Gomez-Ariza JL (2014) Combination of metabolomic and phospholipid-profiling approaches for the study of Alzheimer’s disease. J Proteom 104:37–47 [DOI] [PubMed] [Google Scholar]
- 72.Casanova R et al. (2016) Blood metabolite markers of preclinical Alzheimer’s disease in two longitudinally followed cohorts of older individuals. Alzheimers Dement 12:815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inose Y, Kato Y, Kitagawa K, Uchiyama S, Shibata N (2015) Activated microglia in ischemic stroke penumbra upregulate MCP-1 and CCR2 expression in response to lysophosphatidylcholine derived from adjacent neurons and astrocytes. Neuropathology 35:209–223 [DOI] [PubMed] [Google Scholar]
- 74.Sheikh AM, Michikawa M, Kim SU, Nagai A (2015) Lysophosphatidylcholine increases the neurotoxicity of Alzheimer’s amyloid beta1–42 peptide: role of oligomer formation. Neuroscience 292:159–169 [DOI] [PubMed] [Google Scholar]
- 75.Knuplez E, Marsche G (2020) An Updated Review of Pro- and Anti-Inflammatory Properties of Plasma Lysophosphatidylcholines in the Vascular System. Int J Mol Sci 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gattaz WF, Maras A, Cairns NJ, Levy R, Forstl H (1995) Decreased phospholipase A2 activity in Alzheimer brains. Biol Psychiatry 37:13–17 [DOI] [PubMed] [Google Scholar]
- 77.Sanchez-Mejia RO et al. (2008) Phospholipase A2 reduction ameliorates cognitive deficits in a mouse model of Alzheimer’s disease. Nat Neurosci 11:1311–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanchez-Mejia RO, Mucke L (2010) Phospholipase A2 and arachidonic acid in Alzheimer’s disease. Biochim Biophys Acta 1801:784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vardarajan B et al. (2020) Differences in plasma metabolites related to Alzheimer’s disease, APOE epsilon4 status, and ethnicity. Alzheimers Dement (N Y) 6:e12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coughlan G et al. (2021) APOE epsilon4 alters associations between docosahexaenoic acid and preclinical markers of Alzheimer’s disease. Brain Commun 3:fcab085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim WL et al. (2013) Effects of a high-fat, high-cholesterol diet on brain lipid profiles in apolipoprotein E epsilon3 and epsilon4 knock-in mice. Neurobiol Aging 34:2217–2224 [DOI] [PubMed] [Google Scholar]
- 82.Kariv-Inbal Z et al. (2012) The isoform-specific pathological effects of apoE4 in vivo are prevented by a fish oil (DHA) diet and are modified by cholesterol. J Alzheimers Dis 28:667–683 [DOI] [PubMed] [Google Scholar]
- 83.Miranda AM et al. (2022) Effects of APOE4 allelic dosage on lipidomic signatures in the entorhinal cortex of aged mice. Transl Psychiatry 12:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van de Rest O, van der Zwaluw NL, de Groot LC (2013) Literature review on the role of dietary protein and amino acids in cognitive functioning and cognitive decline. Amino Acids 45:1035–1045 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All intermediate and final results are included in the main text or the supplementary materials of the manuscript. The raw metabolomics and biomarker will be shared with qualified investigators using the request form available here https://cumc.co1.qualtrics.com/jfe/form/SV_dmck0uV3A91pmzb