ABSTRACT
Butyrate, a physiologically active molecule, can be synthesized through metabolic interactions among colonic microorganisms. Previously, in a fermenting trial of human fecal microbiota, we observed that the butyrogenic effect positively correlated with the increasing Bifidobacterium population and an unidentified Megasphaera species. Therefore, we hypothesized that a cross-feeding phenomenon exists between Bifidobacterium and Megasphaera, where Megasphaera is the butyrate producer, and its growth relies on the metabolites generated by Bifidobacterium. To validate this hypothesis, three bacterial species (B. longum, B. pseudocatenulatum, and M. indica) were isolated from fecal cultures fermenting hydrolyzed xylan; pairwise cocultures were conducted between the Bifidobacterium and M. indica isolates; the microbial interactions were determined based on bacterial genome information, cell growth, substrate consumption, metabolite quantification, and metatranscriptomics. The results indicated that two Bifidobacterium isolates contained distinct gene clusters for xylan utilization and expressed varying substrate preferences. In contrast, M. indica alone scarcely grew on the xylose-based substrates. The growth of M. indica was significantly elevated by coculturing it with bifidobacteria, while the two Bifidobacterium species responded differently in the kinetics of cell growth and substrate consumption. Coculturing led to the depletion of lactate and increased the formation of butyrate. An RNA-seq analysis further revealed the upregulation of M. indica genes involved in the lactate utilization and butyrate formation pathways. We concluded that lactate generated by Bifidobacterium through catabolizing xylose fueled the growth of M. indica and triggered the synthesis of butyrate. Our findings demonstrated a novel cross-feeding mechanism to generate butyrate in the human colon.
IMPORTANCE
Butyrate is an important short-chain fatty acid that is produced in the human colon through microbial fermentation. Although many butyrate-producing bacteria exhibit a limited capacity to degrade nondigestible food materials, butyrate can be formed through cross-feeding microbial metabolites, such as acetate or lactate. Previously, the literature has explicated the butyrate-forming links between Bifidobacterium and Faecalibacterium prausnitzii and between Bifidobacterium and Eubacterium rectale. In this study, we provided an alternative butyrate synthetic pathway through the interaction between Bifidobacterium and Megasphaera indica. M. indica is a species named in 2014 and is indigenous to the human intestinal tract. Scientific studies explaining the function of M. indica in the human colon are still limited. Our results show that M. indica proliferated based on the lactate generated by bifidobacteria and produced butyrate as its end metabolic product. The pathways identified here may contribute to understanding butyrate formation in the gut microbiota.
KEYWORDS: cross-feeding, lactate, butyrate, Bifidobacterium, Megasphaera
INTRODUCTION
Butyrate, a four-carbon organic acid, plays an important role in regulating physiological responses and maintaining energy balances in the human body. Generated in the intestinal lumen, butyrate is uptaken by epithelial cells through diffusion or active transportation using specific channels, such as monocarboxylate transporters (1). As the primary energy source of colonocytes, butyrate is metabolized in the mitochondria through beta-oxidation and accounts for nearly 70% of the energy produced in intestinal cells (2, 3). The oxidation of butyrate in colonocytes depletes intracellular oxygen and preserves the anaerobic environment in the colon for strict anaerobes (4). In addition to cellular energy conversion, butyrate also serves as a signaling molecule and an epigenetic regulator (5). Butyrate molecules are recognized by cell surface G protein-coupled receptors (GPRs), including GPR41, GPR43, and GPR109A, which trigger signaling cascades responsible for the anti-inflammatory effect, T cell proliferation, and hormone secretion (6). Depending on the cellular concentration, butyrate molecules inhibit histone deacetylase (HDAC) activity, resulting in the hyperacetylation of histones, and they have been associated with antitumor functions and the apoptosis of colorectal cancer cells (7).
Microbial-derived butyrate is produced through the fermentation of undigested food materials in the colonic environment. Metabolic pathways involved in butyrate formation bridge the catabolism of carbohydrates, fatty acids, or amino acids through pyruvate and acetyl-CoA molecules (8). Generally, two molecules of acetyl-CoA condense into one molecule of butyryl-CoA via a four-step synthetic pathway, forming intermediate metabolites, including acetoacetyl-CoA, β-hydroxybutyryl-CoA, and crotonyl-CoA. These conversions are catalyzed by enzymes, such as acetyl-CoA C-acetyltransferase (EC 2.3.1.9), 3-hydroxybutyryl-CoA dehydrogenase (EC 1.1.1.157), enoyl-CoA hydratase (EC 4.2.1.17), and butyryl-CoA dehydrogenase (EC 1.3.8.1), in a stepwise manner. The final conversion of butyryl-CoA to butyrate is accomplished through the route of either phosphate butyryltransferase (EC 2.3.1.19) and butyrate kinase (EC 2.7.2.7) or butyryl-CoA:acetate CoA-transferase (EC 2.8.3.8) (9). In addition to the starting molecules from catabolizing carbohydrates, butyrate can be synthesized from L-lysine, L-glutamine, succinate, or lactate (10). Lactate, a three-carbon organic acid, is converted to pyruvate via L- or D-lactate dehydrogenases (EC 1.1.1.27 or EC 1.1.1.28) and is then involved in the butyrate synthesis steps (11).
The most known butyrate-producing bacteria that function in the human colon are affiliated with the phylum Bacillota (synonym Firmicutes). Bacterial genera involved in producing butyrate include Anaerostipes, Butyrivibrio, Coprococcus, Eubacterium, and Ruminococcus, which are classified under the family Lachnospiraceae, and Butyricicoccus, Faecalibacterium, Roseburia, and Suboligogranum, which are classified under the family Rumicococcaceae (6, 12). At the species level, Faecalibacterium prausnitzii and Eubacterium rectale (Agathobacter rectalis) are two dominant species that have been increasingly researched. Both F. prausnitzii and E. rectale utilize the butyryl-CoA acetate CoA-transferase route and account for 8%–10% of the human fecal microbiota that contributes substantially to colonic butyrate synthesis (11, 13). In addition, members of the genus Roseburia, including R. intestinalis, R. faecis, R. hominis, and R. inulinivorans, are also butyrate producers. Each Roseburia species accounts for 0.9%–5.0% of the human fecal microbiota (14). In particular, species of Roseburia contain a wide range of carbohydrate-active enzymes (CAZymes) that can be utilized to directly consume nondigestible polysaccharides, such as fructan, pectin, and xylan (15). Besides colonic bacillota, the genera Butyricimonas, Porphyromonas, Odoribacter, and Alistipes, affiliated with the phylum Bacteroidota (synonym Bacteroidetes), have also been reported to be involved in butyrate production (12).
Despite the notion that butyrate formation is considered beneficial to health, many colonic butyrate-producing bacteria are not able to catabolize nondigested food materials directly (16). Mechanisms of metabolic cross-feeding effects have been proposed to explain butyrate synthesis by connecting the primary degraders of food materials and butyrate producers (17, 18). Bifidobacterium species generally encompass a reservoir of CAZymes and serve as primary degraders of nondigestible carbohydrates, including polysaccharides and oligosaccharides, in the gut microbiota. Through the fructose-6-phosphate phosphoketolase pathway, bifidobacteria metabolize internalized sugars into acetates and lactates that can be further consumed as nutrients by the butyrate-producing population (19, 20). It has been reported that F. prausnitzii grows poorly in culture media containing starch or xylan substrates. However, it metabolizes acetates and enhances the production of butyrate when it is cocultured with Bifidobacterium adolescentis (21, 22).
In our previous study investigating the impact of xylan chain length on the human gut microbiota, we observed a highly correlated relationship between the dominant populations of Bifidobacterium and Megasphaera in the fecal cultures that fermented fragmented beechwood xylan (BWX; Fig. S1). Species of Bifidobacterium (OTU2 B. pseudocatenulatum and OTU13 B. longum) and Megasphaera (OTU10 M. unidentified) were simultaneously enriched when fermenting short-chain BWX. All three species (OTU2, OTU10, and OTU13) were significantly positively correlated with butyrate formation (P < 0.05) (23). The genus Megasphaera consists of gram-negative anaerobic cocci that are classified under the family Veillonellaceae and phylum Bacillota. To date, a total of 14 validated Megasphaera species have been isolated from various sources. Among them, M. elsdenii, a ruminal bacterium that is commonly found in cattle and sheep, has been investigated in multiple probiotic applications to relieve ruminal acidosis, which is attributed to the characteristic of lactate utilization. In addition, the species M. indica, M. butyrica, M. hominis, and M. massiliensis were isolated from human samples. Physiological tests and comparative genome analyses revealed that type strains of M. indica (DSM 25563) and M. elsdenni (DSM 20460) shared catabolic pathways exemplified by lactate utilization and volatile fatty acid production, while these two species can be distinguished based on genetic similarity and protein sequence analysis results (24, 25).
Based on the aforementioned observations and evidence, we hypothesized that a metabolic cross-feeding phenomenon exists between Bifidobacterium and Megasphaera species, which contributes to the degradation of BWX substrates and the formation of butyrate. To validate this hypothesis, two Bifidobacterium species (OTU2 and OTU13) and one unidentified Megasphaera species (OTU10) were isolated from the fecal cultures fermenting short-chain BWX. Microbial strains were characterized based on the full-length 16S rRNA gene sequence and whole genome sequencing results. Isolates of Bifidobacterium and Megasphaera were cocultured in BWX- or xylose-containing media to illustrate the kinetics of bacterial growth, substrate consumption, and metabolite production. Bacterial gene expression levels under coculture conditions were further revealed using RNA sequencing. Our results provide insight into the metabolic interactions between Bifidobacterium and Megasphaera that may take place in the human colonic environment.
RESULTS
Identification of bacterial isolates
Three bacterial isolates were obtained from fecal cultures fermenting xylan. To identify and classify the bacterial isolates, phylogenetic analyses based on the pangenome and 16S rRNA sequences were both conducted (Fig. S2). The phylogenetic tree of the genus Bifidobacterium demonstrated that OTU2 and OTU13 isolates fell into the clusters of B. pseudocatenulatum and B. longum, respectively (Figs. S2A, C, and D). The OTU2 genome isolates presented an average nucleotide identity (ANI) value of 98.17% with the B. pseudocatenulatum (JCM 1200) genome. The OTU13 genome isolates presented an ANI value of 98.83% with the B. longum (JCM 1217) genome (Table S1). The above results confirmed that OTU2 was a B. pseudocatenulatum strain and that OTU13 was a B. longum strain. These two isolates are denoted as B. pseudocatenulatum_BWX (BP) and B. longum_BWX (BL) below.
On the other hand, the codon tree of the genus Megasphaera was constructed among 23 Megasphaera genomes. As shown in Fig. S2B, OTU10 clustered with Megasphaera sp. NM10 and Megasphaera sp. BL7 in the same subclade with high nodal support numbers. These three genome samples were close but separated from the clade containing the species M. elsdenii. Strains of Megasphaera sp. NM10 and Megasphaera sp. BL7 were isolated from human feces, and their 16S rRNA gene sequences showed high similarity (>99%) with M. elsdenii JCM 1772T. However, based on the DNA-DNA hybridization results and phenotypic analysis, these two strains were proposed to be a new species, namely, Megasphaera indica (24, 25). The result of the 16S rRNA sequence analysis further supported the findings that OTU10 clustered with valid M. indica strains (Fig. S2E). Additionally, the OTU10 genome presented an ANI value of 99.09% with Megasphaera sp. NM10, which represented M. indica, and only presented an ANI value of 90.89% with M. elsdenii (Table S1). As reported, two strains displaying an ANI value of >95% are considered to belong to the same species (26). Collectively, the above results suggested that the isolated OTU10 was an M. indica strain, which is denoted as M. indica_BWX (MI) below.
Genomic characteristics of three bacterial isolates
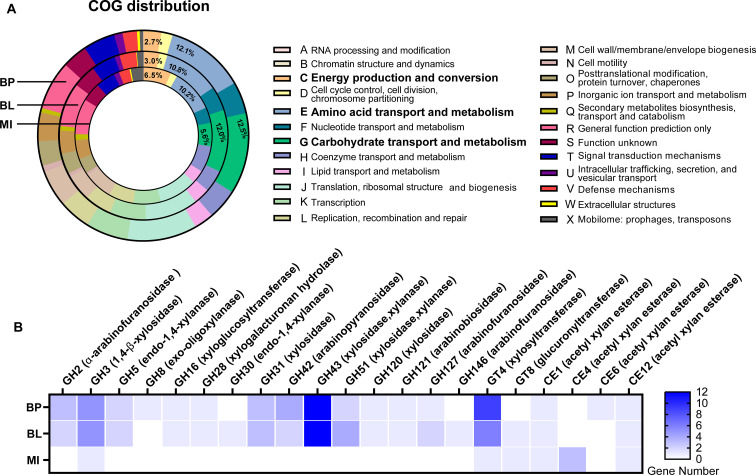
To depict the genetic potential of bacterial isolates, the genome sequences were annotated and functionally characterized (Fig. 1). Based on the COG (Clusters of Orthologous Genes) classification, B. pseudocatenulatum_BWX and B. longum_BWX possessed 12.5% and 12.0% of the genes involved in carbohydrate transport and metabolism, respectively. In contrast, the M. indica_BWX genome only contained 5.6% of the genes that are involved in carbohydrate utilization, whereas the proportion of COG-identified genes associated with energy production and conversion in the M. indica_BWX genome (6.5%) was more than twice as high as that observed in the B. pseudocatenulatum_BWX (2.7%) and B. longum_BWX (3.0%) genomes (Fig. 1A). In agreement with the COG identification results, B. pseudocatenulatum_BWX and B. longum_BWX possessed identical numbers of CAZyme genes covering families of glycoside hydrolase (GH), glycosyl transferase (GT), carbohydrate-binding module (CBM), and carbohydrate esterase (CE) (Fig. S3A). With respect to the GH family, GH13, which is the most prevalent CAZyme gene across both bifidobacterial genomes, is responsible for the hydrolysis of alpha-glucosidic linkages (Fig. S3B). GH43, the second most abundant family present in the bifidobacterial genomes, contains endo-1,4-β-xylanase, β-xylosidase, and α-l-arabinofuranosidase enzymes that are responsible for degrading xylo-oligosaccharides and/or xylo-polysaccharides. Other CAZyme families, including GH2, GH3, GH5, GH31, GH42, GH51, GH120, GH121, GH127, GT4, CE1, and CE12, were detected in both bifidobacterial genomes, while some CAZymes were uniquely present in B. pseudocatenulatum_BWX or B. longum_BWX. For instance, GH8 and CE6 were present primarily in the B. pseudocatenulatum_BWX genome, whereas GH30, GH146, and GT8 were only found in the B. longum_BWX genome, suggesting that the two bifidobacterial strains employed different mechanisms to degrade xylan-derived fibers (Fig. 1B). Despite the limited number of CAZyme genes that were characterized from the M. indica_BWX genome, these genes covered a broad range of GT and CE activities (Fig. S3C). Several M. indica_BWX genes exemplified by GH3, GT4, GT8, CE1, CE4, and CE12 were related to the xylanolytic function (Fig. 1B). The Kyoto Encyclopedia of Genes and Genomes (KEGG)-based analysis further revealed that the M. indica_BWX genome possessed multiple pathways for amino acid and fatty acid metabolism. The presence of the butyryl-CoA:acetate CoA transferase (EC 2.8.3.8) gene suggests its potential role in butyrate production. The findings from the genome characterization validate the difference in genetic traits between Bifidobacterium and Megasphaera. In particular, the former exhibited a broader spectrum in carbohydrate utilization, whereas the latter possessed the capability of butyrate formation.
Fig 1.
Functional classification of the protein-coding sequences in three bacterial isolates. (A) The proportion of genomes identified for each COG (Clusters of Orthologous Genes) category. The value indicated represents the percentage of total gene numbers in each genome. (B) The number of genes matched to a CAZyme family responsible for xylan utilization. BP, Bifidobacterium pseudocatenulatum_BWX; BL, Bifidobacterium longum_BWX; MI, Megasphaera indica_BWX; CBM, carbohydrate-binding module; CE, carbohydrate esterase; GH, glycoside hydrolase; GT, glycosyl transferase; AA, auxiliary activity family.
Growth kinetics of bacterial isolates
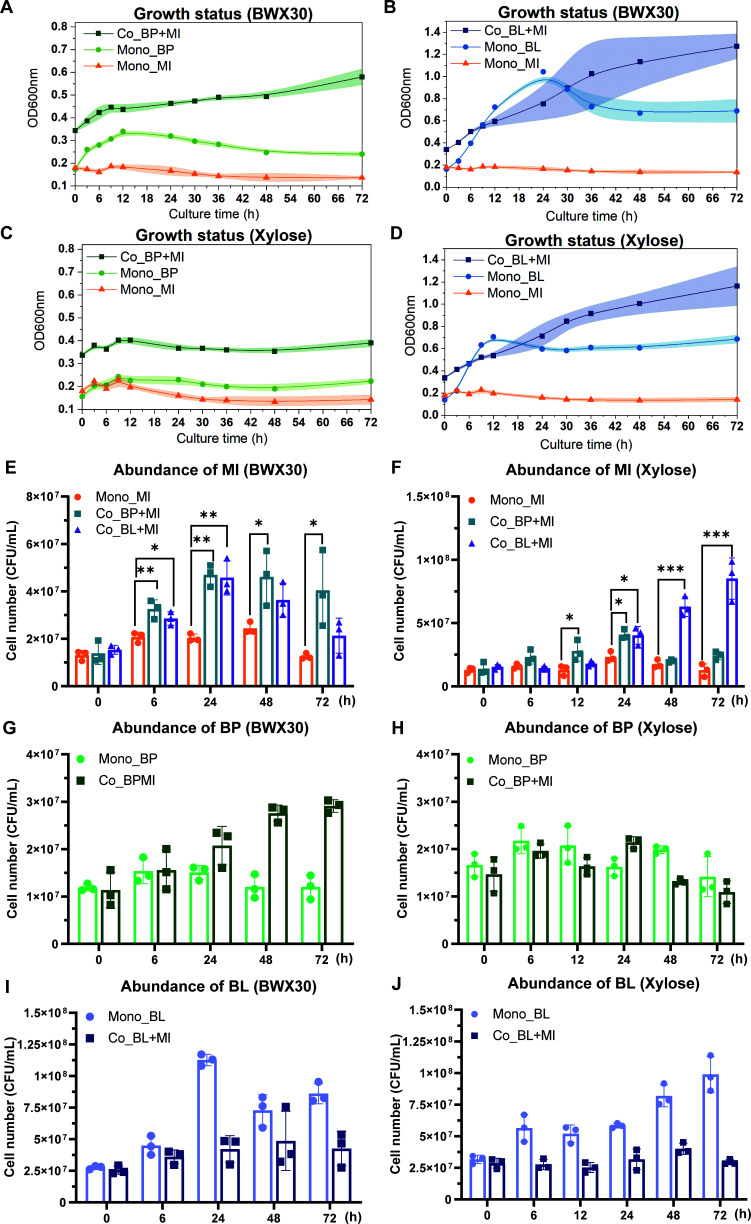
Phenotypic characterization of the growth of three isolated bacteria was investigated under monoculture and coculture conditions on hydrolyzed beechwood xylan (BWX30)- and xylose-based media. As shown in Fig. 2A through D, the three isolates exhibited distinct growth kinetics in monoculture fermentation. B. pseudocatenulatum_BWX presented a faster growth rate on BWX30 in comparison to xylose monomers. In contrast, B. longum_BWX displayed good utilization of both BWX30 and xylose. The cell density (OD600) of B. longum_BWX peaked at 12 h on xylose media, whereas the peak cell density was delayed to 24 h on BWX30 media. As for M. indica_BWX, it exhibited a poor ability to utilize either BWX30 or xylose. Interestingly, cocultures of M. indica_BWX with one of the bifidobacterial strains led to a stable increase in the overall cell densities after 24 h of fermentation. To determine the absolute abundance of each isolate in culture, the quantitative real-time PCR (qPCR) method was used to quantify the cell concentration. As shown in Fig. 2E and F, the presence of bifidobacteria significantly boosted the growth of M. indica_BWX on either BWX30- or xylose-based media (P < 0.05). In contrast, B. pseudocatenulatum_BWX and B. longum_BWX responded distinctively to the presence of M. indica_BWX. B. pseudocatenulatum_BWX displayed continuous growth throughout 72 h of coculture fermentation and reached a higher cell density than its monoculture on BWX30 (Fig. 2G), while B. longum_BWX did not reach the same level of cell density in coculture as it did in monoculture on either BWX30- or xylose-based media (Fig. 2I and J). Collectively, these results indicated that the growth of M. indica_BWX benefited from coculturing with the bifidobacterial strains, and they also revealed the impact of coculture on the growth of B. pseudocatenulatum_BWX and B. longum_BWX.
Fig 2.
Growth of three bacterial isolates on BWX30 and xylose substrates. (A through D) Growth kinetics were recorded using time-series measurements of OD600. Each point on the growth curve represents the mean, and the shaded regions represent a 95% confidence interval of three biological replicates. (E through J) The absolute abundance of each strain in monoculture and coculture was quantified by qPCR. The range bars in the bar charts show the standard deviation of three biological replicates. Statistical differences in the abundance of MI at each time point between monoculture and coculture were analyzed by a Student t-test (*P < 0.005, **P < 0.01, and ***P < 0.001). Mono, monoculture; Co, coculture; BP, Bifidobacterium pseudocatenulatum_BWX; BL, Bifidobacterium longum_BWX; MI, Megasphaera indica_BWX.
Substrate consumption during fermentation
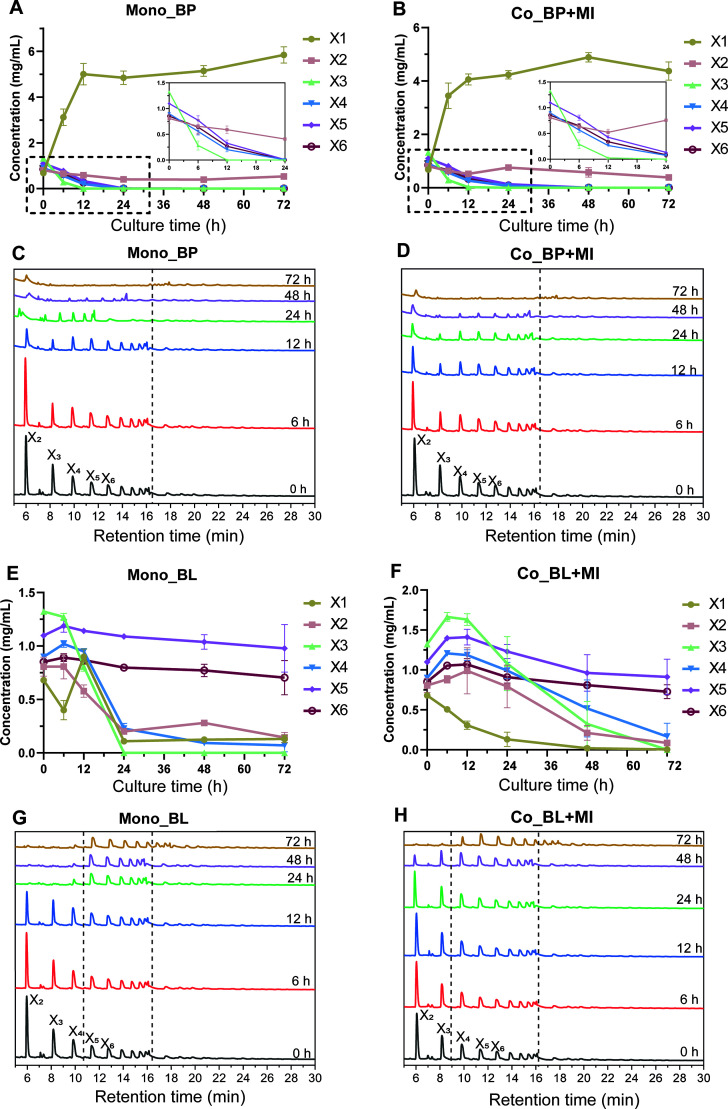
Total xylose consumption and concentrations of xylose oligomers were monitored during fermentation. In the xylose-based media, significant depletion of xylose monomers was only observed in cultures inoculated with B. longum_BWX (Fig. S4A). The depletion of total xylose substrates in the BWX30-based media is shown in Fig. S4B. B. longum_BWX displayed a better capability of utilizing BWX30 than B. pseudocatenulatum_BWX in monocultures, as the consumption of individual xylo-oligosaccharides was demonstrated in Fig. 3. Continuous degradation of X2-X10 oligomers and an accumulation of xylose monomers were observed in cultures inoculated with B. pseudocatenulatum_BWX (Fig. 3A and C). Alternatively, only short oligomers (X2-X4) were rapidly consumed in cultures inoculated with B. longum_BWX; concentrations of oligomers with longer chain lengths (X5-X10) remained unchanged throughout 72 h of fermentation (Fig. 3E and G). With respect to M. indica_BWX cultures, neither xylose monomer nor xylo-oligosaccharide substrates were consumed during monoculture (Figs. S4C and D). The presence of M. indica_BWX in the cocultures with B. longum_BWX led to delayed utilization of xylo-oligosaccharides, while it did not influence the BWX30 degradation pattern when coculturing with B. pseudocatenulatum_BWX (Fig. 3B through D and 3F through H).
Fig 3.
Substrate utilization during monoculture and coculture fermentation. (A and B; E and F) The concentration of xylose and xylo-oligosaccharides in the medium with BWX30 as the carbohydrate source. (C and D; G and H) Profile of the oligomers in the medium with BWX30 as the carbohydrate source. X1, xylose; X2, xylobiose; X3, xylotriose; X4, xylotetraose; X5, xylopentaose; X6, xylohexaose. The range bars in line charts denote the standard deviation of three biological replicates.
Metabolite production
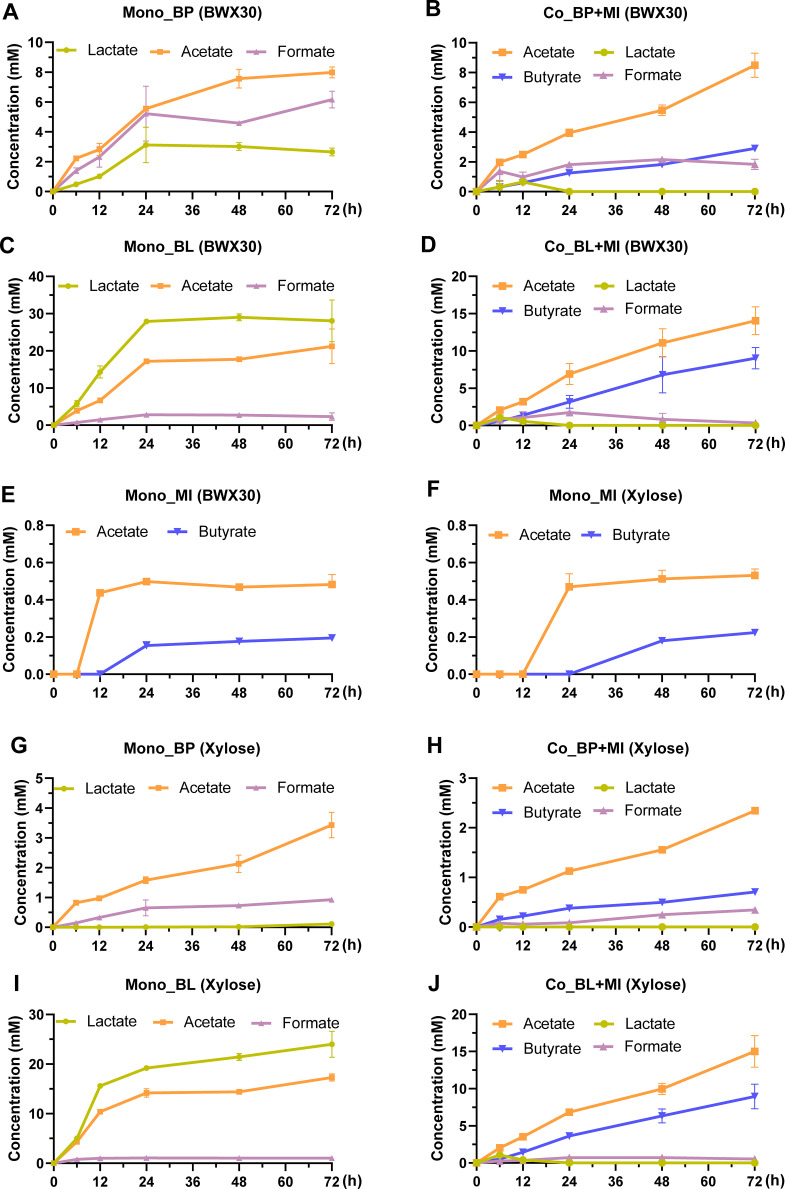
To explore the possible cross-feeding phenomenon between M. indica_BWX and bifidobacterial strains, concentrations of short-chain fatty acids in the monoculture and coculture fermentations of three isolates were quantified (Fig. 4). The primary metabolites of B. pseudocatenulatum_BWX and B. longum_BWX grown on BWX30 and xylose were lactate, acetate, and formate. The two bifidobacterial strains exhibited dissimilar production ratios of organic acids. B. pseudocatenulatum_BWX produced a greater amount of acetate and formate than lactate, whereas B. longum_BWX had a higher yield of lactate than acetate and formate (Fig. 4A and C). M. indica_BWX only generated low amounts of acetate and butyrate in the BWX30- and xylose-based media (Fig. 4E and F). Considerable alterations in the metabolic profiles were detected by coculturing M. indica_BWX with bifidobacteria, as lactate molecules were depleted in the media, while butyrate concentrations rose throughout fermentation (Fig. 4B, D, H, and J).
Fig 4.
Metabolite production during monoculture and coculture fermentation in a medium with either BWX30 (A through E) or xylose (F through J) as the sole carbohydrate source. The range bars denote the standard deviation of three biological replicates.
RNA-seq analysis
Transcriptomic analyses were conducted to clarify the underlying interactions during the coculture of M. indica_BWX with B. pseudocatenulatum_BWX or B. longum_BWX. Sixteen differentially expressed genes (DEGs) that were involved in lactate metabolism were identified by comparing the gene expression levels of M. indica_BWX in coculture versus monoculture stages (Fig. 5A). Out of the 16 selected DEGs, 11 genes were upregulated and encoded a series of enzymes, including lactate permease (lctP), an influx protein that imports lactate into cells, lactate racemase (larA), which is responsible for the interconversion of D-lactate and L-lactate, FAD-binding-4, a family of proteins comprising D-lactate dehydrogenase that require FAD as a cofactor in the conversion of D-lactate to pyruvate, and lactate utilization proteins (lutB and lutC), which play a role similar to L-lactate hydrogenase (ldh) and D-lactate hydrogenase (ddh). It is noteworthy that a greater number of genes encoding ddh were upregulated in comparison to those encoding ldh. Collectively, these elevated-expressed genes confirmed the capability of M. indica_BWX to utilize lactate.
Fig 5.
Differentially expressed genes that are involved in lactate utilization (A) and butyrate production (B) between coculture and monoculture fermentations of MI on the BWX30 substrate at 12 h. The log2 fold change values are plotted on the x-axis, and gene IDs are indicated on the y-axis. The red bars indicate the genes that are significantly upregulated, while the blue bars indicate the genes that are significantly downregulated. The gray bars indicate genes that are not significantly differentially expressed. The analysis was performed using RNA-seq data and a false discovery rate (FDR) cutoff of 0.05. (C) The predicted cross-feeding mechanism between Bifidobacterium species and Megaspharea indica. BP, Bifidobacterium pseudocatenulatum_BWX; BL, Bifidobacterium longum_BWX; MI, Megasphaera indica_BWX.
To decipher the pathway that M. indica_BWX employed to produce butyrate, 21 DEGs involved in the butyrate-producing pathway were identified (Fig. 5B). Genes encoding acetyl-CoA acetyltransferase (EC 2.3.1.9), 3-hydroxy butyryl-CoA dehydrogenase (EC 1.1.1.157), enoyl-CoA hydratase (EC 4.2.1.17), and butyryl-CoA dehydrogenase (EC 1.3.8.1) were significantly upregulated. A substantial increase in the expression of two genes that function as electron-transferring flavoproteins (ETFs) was detected. ETFA and ETFB proteins were reported to mediate the reduction of crotonyl-CoA in anaerobic bacteria for energy conservation. In addition, five genes encoding butyryl-CoA transferases (EC 2.8.3.8) that might contribute to the conversion of butyryl-CoA to butyrate were identified. Nevertheless, only one out of five genes was significantly upregulated. Collectively, the expression of M. indica_BWX genes that are involved in both lactate metabolism and butyrate production was influenced by the presence of B. pseudocatenulatum_BWX and B. longum_BWX during coculture fermentation, suggesting a lactate-mediated, cross-feeding interaction between the butyrate producer Megasphaera indica and Bifidobacterium species.
DISCUSSION
In this study, a bottom-up approach combined with bacterial genomic information and RNA-seq techniques was adopted to validate the metabolic interchange between Bifidobacterium sp. and Megasphaera indica. We hypothesized that Bifidobacterium was the primary degrader of xylan-derived fibers that catabolizes carbohydrate substrates into acetate and lactate, whereas Megasphaera indica served as the lactate-utilizing bacteria and was responsible for butyrate production in the microbial community. The phenomenon of cross-feeding between Bifidobacterium and other gut anaerobes to generate butyrate in the colonic ecosystem has been explored previously. Duncan et al. (16) isolated the butyrate-producing species Eubacterium hallii and Anaerostipes caccae from human fecal samples. E. hallii and A. caccae took up lactate molecules generated by Bifidobacterium adolescentis during coculture. Conversely, Roseburia hominis (DSM 16839) generated butyrate without consuming the available lactate (17). More recently, Rios-Covian et al. (22) demonstrated the cross-feeding interaction between Faecalibacterium prausnitzii and B. adolescentis. F. prausnitzii was not efficient in utilizing fructooligosaccharide (FOS P95). However, butyrate production was elevated when bifidobacteria-derived acetate was present. Rivière et al. (27) presented a case of mutual cross-feeding on arabinoxylan oligosaccharides (AXOS) between Bifidobacterium longum (strain NCC2705) and Eubacterium rectale (ATCC 33656). Interestingly, both bacterial strains were involved in the degradation of AXOS. The arabinose substitute of AXOS was consumed by B. longum, while E. rectale metabolized the bifidobacterial derived acetate into butyrate and hydrolyzed the AXOS backbone, which benefited the growth of bifidobacteria.
To explain the possible mechanisms of the cross-feeding between colonic bifidobacteria and butyrate-producing bacteria, Rivière et al. (20) categorized the mechanisms into three types. For the first type, both bifidobacterial and butyrate-producing strains were capable of utilizing substrates, such as polyfructose, and the acetate and lactate derived from bifidobacterial metabolism became the co-substrates of the butyrate-producing strains to enhance butyrate formation. As for the second type, only the bifidobacterial strain consumed carbohydrate substrates, whereas the growth of the butyrate-producing strain was dependent on the substrate breakdown products, lactate or acetate, which were generated by the bifidobacterial strain. The last cross-feeding type was settled based on a mutual cross-beneficial effect. Both the bifidobacterial and butyrate-producing strains utilized substrates, such as AXOS, whereas the growth of bifidobacteria was enhanced by consuming the carbohydrate breakdown products released by the extracellular degradation of the substrate that was triggered by the butyrate-producing strain.
M. indica, proposed as a novel species of the genus Megasphaera in 2014, is a human gut commensal. Presently, there are limited reports illustrating its function in the intestinal environment. Most recently, Nagara et al. (28) conducted a correlation analysis on a human trial feeding granular starch. A significant correlation between M. indica and B. adolescentis was reported. As M. indica was reclassified from the species M. elsdenii, which represented M. indica’s closest relative in the ruminant system, the two species shared certain traits but were distinct from others. Lanjekar et al. (24) compared the physiological and biochemical characteristics of M. indica and M. elsdenii. Two M. indica strains (NM10 and BL7) were similar to the M. elsdenii type strain (DSM20460) in morphology and their capability to catabolize DL-lactate, D-glucose, D-fructose, and maltose but differed in their capability to catabolize D-galactose and D-mannose and in their bacterial cell wall fatty acid profiles. Shetty et al. (25) conducted genomic analyses on M. indica (NM10 and BL7) and M. elsdenii DSM20460. The NM10 and BL7 strains were highly similar in their protein sequences and the distribution of CAZyme families in comparison to DSM20460. Nevertheless, the polysaccharide lyase (PL) family was not found in the genomes of all three strains. Shetty et al. (25) hypothesized that M. indica may play a comparable role in the human gut as M. elsdenii in the ruminant that targets end fermentation products, especially lactate. Consistent with previous findings, we found that the genome of the isolated M. indica_BWX strain contained several families of CAZymes, whereas the strain possessed a limited capability to utilize xylan-derived carbohydrates both genotypically and phenotypically.
We demonstrated that lactate tended to accumulate in monoculture conditions when inoculated with bifidobacteria. However, when the bifidobacterial strains were cocultured with M. indica, lactate was completely consumed, and the cell density of M. indica increased. The expression of lctP, larA, and associated FAD-binding oxidoreductase/transferase type 4 family genes in the M. indica_BWX genome significantly increased at 12 h of coculture fermentation. Hino and Kuroda (29) revealed that the enzyme activity of the lactate racemase of the lactate-utilizing bacteria (M. elsdenii) was only detected when the cells were fed lactate, suggesting that the M. indica_BWX strain did uptake the L-lactate liberated by bifidobacteria from the extracellular environment. Regarding the conversion of lactate to pyruvate, it is notable that the expression of the ddh and lactate utilization protein (lutB and lutC) genes was much higher than the expression of the ldh gene in M. indica. Although it is unclear whether the lutB and lutC proteins acted on the D-lactate or L-lactate substrates, the higher expression of ddh genes indicated that the M. indica_BWX strain preferred to utilize D-lactate, which corresponded with the characteristics of other Megasphaera strains (30).
Lactate metabolism by Megasphaera sp. leads to the production of volatile fatty acids, while the metabolite profiles often vary depending on the strain. In one metabolic route, lactate molecules can be catalyzed by lactate dehydrogenase to form pyruvate and connect with the acetate and butyrate formation pathways. Alternatively, lactate molecules can also be converted into propionate via the acrylate pathway by forming the essential intermediate metabolites lactyl-CoA and propionyl-CoA. Studies show that M. elsdenii contains both pathways converting either lactate to butyrate or lactate to propionate. Marounek et al. (31) evaluated organic acids produced from four M. elsdenii strains. They found that there were differences among the strains regarding organic acid production. Two strains (LC1 and AW106) converted lactate into acetate and propionate; one strain (J1) converted lactate into acetate, propionate, and butyrate; and the final strain (L8) converted lactate into acetate, propionate, butyrate, and valerate. Prabhu et al. (32) observed that the metabolite profile of the M. elsdenii ATCC17753 strain shifted from acetate and propionate to acetate and butyrate when the culture conditions changed from lactate-based batch fermentation to a carbon-limited steady state. They theorized that when the lactate concentration was low, the driving force of lactyl-CoA formation was too low to drive propionate generation. In this study, the M. indica_BWX strain also encompassed both the propionate and butyrate formation pathways, while only acetate and butyrate were detected during culture. This phenomenon may be explained by Prabhu et al.’s theory that the lactate concentration is constantly low during coculture, which triggers the production of butyrate.
Regarding the steps of butyrate formation, the M. indica_BWX strain encompassed two pathways (the acetyl-CoA pathway and the 4-aminobutyrate pathway) to synthesize the crucial intermediate crotonyl-CoA. Nevertheless, the acetyl-CoA pathway was postulated to be the dominant route because the available amino acids (glutamate, arginine, and proline) that can be converted to 4-aminobutyrate were in low concentrations in the fermentation media. The conversion of crotonyl-CoA to butyryl-CoA required the catalysis of butyryl-CoA dehydrogenase (EC 1.3.8.1), which corresponded to the elevated expression of multiple genes encoding butyryl-CoA dehydrogenase. Increased gene expression levels were also detected for two electron-transferring flavoproteins (ETFA and ETFB) that were expected to form a butyryl-CoA dehydrogenase/ETF complex. This complex is confirmed to commonly exist in butyrate-producing bacteria and plays a crucial role in butyrate synthesis (33). As for the final step of butyrate formation, we demonstrated that the M. indica_BWX strain converted butyryl-CoA to butyrate via butyryl-CoA:acetate CoA-transferase (EC 2.8.3.8) instead of using the phosphate butyryl transferase-butyrate kinase pathway. Even though multiple genes that encoded butyryl-CoA:acetate CoA-transferase were located, the expression was only elevated in one. A possible explanation might be that the RNA samples were collected at 12 h of fermentation, which was earlier than the occasion when butyrate concentrations were substantially raised in the bacterial culture. Conversely, the conversion step of butyryl-CoA to butyrate can also be accomplished by other CoA transferases, such as acetate CoA transferases and propionate CoA transferases, which have been reported to replace the butyrate CoA transferase function to produce butyrate (34).
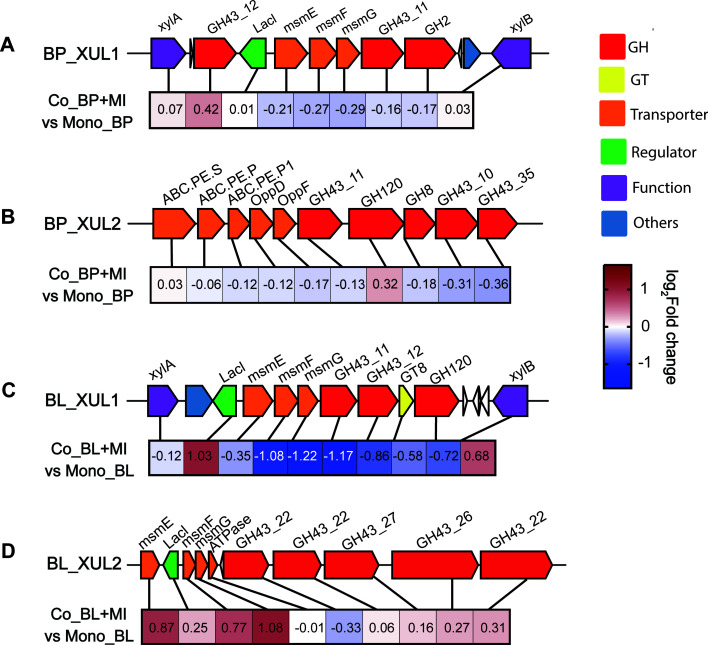
To explain the difference between the two bifidobacterial strains in BWX substrate degradation and cell growth, four gene clusters related to xylan utilization were identified based on genome information. As shown in Fig. 6, B. pseudocatenulatum_BWX was equipped with gene clusters of BP_XUL1 and BP_XUL2. The BP_XUL2 cluster was composed of a peptide ABC (ATP-binding cassette) transporter and several families of glycoside hydrolase families, including GH8, GH43_10, GH43_11, and GH43_35. GH8 is a family containing reducing-end xylose-releasing exo-oligoxylanase (EC 3.2.1.156) that targets long-chain xylo-oligosaccharides and generates the products xylotriose (X3) and xylotetraose (X4) (35, 36). Regarding GH43, multiple enzymes of this family have been characterized as possessing bi-functionality properties, containing both β-xylosidase and α-arabinofuranosidase activities (37). In particular, several GH43 members, identified as β-xylosidases, present preferences for the nonreducing end of short xylo-oligosaccharides (X2-X6) (38, 39). Therefore, the decomposition of the BWX30 substrate by B. pseudocatenulatum_BWX might be attributed to the synergistic effects of the GH8 and GH43 enzymes, by which GH8 acted on the reducing end of the long-chain xylan while GH43 acted on the nonreducing end of the short-chain oligosaccharides. In addition, the B. pseudocatenulatum_XUL1 cluster was found to consist of multiple-sugar metabolism (msm) operons and genes encoding xylose isomerase (xylA), xylose kinase (xylB), GH43 families (GH43_11 and GH43_12), and the GH2 (α-arabinofuranosidase) family. The msm operon was composed of a Lac repressor (LacI), a sugar-binding protein (msmE), and two membrane proteins (msmF and msmG) and was predicted to operate as an ABC-type xylo-oligosaccharide transporter (40). Interestingly, we found that the above gene clusters were arranged similarly to the arabinoxylan-hydrolysate (AXH) utilization systems identified in another B. pseudocatenulatum strain, a result that further supported the conclusion that the ability to consume xylo-oligosaccharide is a common feature shared within the species B. pseudocatenulatum (41). However, unlike the strain reported by Watanabe et al. (41), B. pseudocatenulatum_BWX lacked GH10 enzymes (endo-1,4-β-xylanase), which might affect its xylan degradation capability and cell growth.
Fig 6.
Schematic representation of xylan utilization loci (XUL) in the genomes of (A and B) BP and (C and D) BL. Heatmap of the log2 fold changes of the corresponding XUL genes between coculture and monoculture fermentations on BWX30 at 12 h. BP, Bifidobacterium pseudocatenulatum_BWX; BL, Bifidobacterium longum_BWX; MI, Megasphaera indica_BWX.
As for the B. longum_BWX strain, we identified a B. longum_XUL1 gene cluster that presented a similar structure to B. pseudocatenulatum_XUL1 (Fig. 6). The B. longum_BWX_XUL1 cluster contained the GT8 and GH120 genes that were not present in B. pseudocatenulatum_XUL1. Transferases of the GT8 family catalyze the removal of glucuronic acid (GlcA) and methyl glucuronic acid (MeGlcA) from xylan (42). Hydrolases of the GH120 family preferably facilitate the hydrolysis of X2-X4 molecules (43). Therefore, the B. longum_XUL1 gene cluster might contribute to the assimilation of short-chain oligosaccharides. Even though we did identify the B. longum_XUL2 cluster that contained the msm oligosaccharide transporter and a tandem of five GH43 families, the B. longum_BWX strain presented limitations in consuming substrates larger than X4. Additionally, there are multiple genes encoding monosaccharide transport systems in the B. longum_BWX genome, which might help explain its capability for utilizing xylose monomers. Conversely, the absence of xylose transport systems could be a possible explanation for the inability of the B. pseudocatenulatum_BWX strain to consume xylose effectively (Fig. S5).
Although the isolated M. indica_BWX strain was demonstrated in this study to be beneficial in both cell growth and metabolite production by coculturing it with Bifidobacterium species, the responses of the Bifidobacterium strains varied in the presence of M. indica. B. pseudocatenulatum_BWX exhibited a faster growth rate and reached a higher cell density when it was cocultured on BWX substrates. However, the substrate consumption rate of B. longum_BWX decreased from the beginning of coculture, leading to a reduced cell density. This suggests a mutually beneficial interaction between B. pseudocatenulatum and M. indica, unlike the commensal or possibly parasitic interaction between B. longum and M. indica. Given the weak competitive advantage of Megasphaera over Bifidobacterium in carbohydrate utilization, the differential impact of M. indica on two bifidobacterial strains may be associated with competition or cooperation to acquire noncarbohydrate nutrients, such as amino acids, minerals, or vitamins. Additionally, the growth of Bifidobacterium could be influenced by environmental factors, including medium acidity, oxygen levels, microbial toxins, and growth space (44). To reveal the physiological response of Bifidobacterium-Megaspaera interactions, an in-depth analysis of gene expression at different growth stages is needed in future research.
In conclusion, during the coculture of Bifidobacterium sp. and M. indica, we found that pure cultures of M. indica did not proliferate in the fermentation media containing BWX30, whereas the cell density of M. indica significantly increased when cocultured with one of the two bifidobacterial strains. Lactate induced the elevated expression of lactate permease, racemase, and D-lactate dehydrogenase genes and triggered the formation of butyrate by M. indica. These results validated our hypothesis. Furthermore, we propose that the cross-feeding phenomenon that occurred between Bifidobacterium species and M. indica can be classified as the second type of interaction that was proposed by Rivière et al. (20). The Bifidobacterium strains, B. longum and B. pseudocatenulatum, were the only xylan substrate consumers, whereas M. indica was the butyrate-producing strain that utilized the lactate generated by bifidobacterial metabolism.
MATERIALS AND METHODS
Substrate preparation, reagents, and chemicals
The fragmented beechwood xylan (BWX30), used as a substrate in bacterial cultures, was prepared using the autohydrolysis method as previously reported (23). Briefly, 1.5 g of BWX powder (BWX, Megazyme, Wicklow, Ireland) was mixed with 28.5 mL of deionized water to reach a solid content of 5% (wt/wt). The slurry (30 g) was loaded into a custom-fabricated pipe reactor (50 mL capacity, Swagelok, Chicago Fluid System Technologies, Chicago, IL, USA) that was 5.5 inches in tubular length (SS-T16-S-083–6ME; 1-inch OD × 0.083-inch wall thickness) and was adapted with two 316 stainless steel caps (SS-1610-C) at both ends. The reactors were manually sealed, immersed in a preheated fluidized sand bath (IFB-51 Industrial Fluidized Bath, Techne Inc., Burlington, NJ, USA), and heated to the destinated reaction temperature (175°C). The autohydrolysis reaction was carried out at 175°C for 30 min and stopped by chilling the reactors under flowing tap water. The collected hydrolyzates were decolorized by incubating them with 5% (wt/vol) Amberlite IR96 ion exchange resins for 24 h (50 rpm, 25°C). After the resins were removed, the resulting solutions were freeze-dried to form BWX30. All the reagents and chemicals used in this study, except those stated otherwise, were of bioreagent grade and procured from Merck or Fisher Scientific.
Isolation and identification of the Bifidobacterium and Megasphaera species
The bacterial strains were isolated from fecal culture stocks that had been fermenting in BWX30 for 24 h. Stock cultures were previously cryopreserved at −80°C. The isolation procedure was conducted in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI, USA) that was operated under a controlled atmosphere containing nitrogen (91%), carbon dioxide (5%), and hydrogen (4%). The selection of Bifidobacterium species was performed using modified de-Mann Rogosa Sharpe (MRS, Oxoid, Thermal Fisher Scientific) agar plates supplemented with 0.05% (wt/vol) L-cysteine-HCl and 0.005% (wt/vol) lithium mupirocin (69732, Merck). The selection of Megasphaera species was conducted using lactate-containing agar plates, as reported by Sato et al. (45). The composition of the selective agar plates was as follows (for 1 L): 2.0 g trypticase peptone, 0.5 g yeast extract, 0.5 g L-cysteine-HCl monohydrate, 0.6 g Na2CO3, 20 mL 0.05% hemin solution, 0.5 mL 1.0% resazurin solution, 3.25 mL DL-lactic acid, 0.11 g KH2PO4, 0.11 g K2HPO4, 0.23 g NaCl, 11 g (NH4)2SO4, 11 mg CaCl2, 24 mg MgSO4 heptahydrate, and 16 g agar. After incubating at 37°C for 24–48 h, microbial colonies were randomly picked from the agar plates. The streaking procedure was repeated three times to obtain pure cultures.
For the taxonomic identification of the isolated strains, bacterial genomic DNA was extracted using an alkaline lysis method that was described previously in D’hoe et al. (46) with minor modifications. Isolates were inoculated in 5 mL of MRS broth containing 0.05% (wt/vol) L-cysteine-HCl and were incubated at 37°C and 150 rpm for 24 h. The cells were then harvested by centrifugation at 16,200 × g (13,000 rpm) for 10 min and were then washed using 1 mL of phosphate-buffered saline (PBS, pH 7.2). Cell suspensions were centrifuged, and supernatants were removed by pipetting. Washed cells were resuspended in 1 mL of PBS and diluted to 20 mL in volume. One hundred microliters of the cell suspension was mixed with an equal volume of 0.2 M NaOH in a 2-mL Eppendorf tube. After heating at 90°C for 10 min to break the cell wall, 1,600 µL of 0.04 M Tris-HCl (pH 7.5, Fisher Scientific) was added for neutralization. A full-length 16S rRNA gene was amplified by PCR using the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′- TACGGYTACCTTGTTACGACTT-3′). PCR products were separated by electrophoresis on a 1% agarose gel. The DNA bands at around 1.5 kb were further purified using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) and were subjected to Sanger sequencing served by BioBasic Asia Pacific Pte Ltd. (Singapore). The resulting 16S rRNA gene sequences were aligned to the reference sequences in the NCBI database using BLAST.
Whole genome sequencing of the three bacterial isolates
Bacterial cells (OTU2, OTU10, and OTU13) cultured in MRS broth were harvested at 24 h after inoculation. Genomic DNA was extracted from cells using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Whole genome sequencing was conducted at NovogeneAIT Genomics Singapore Pte Ltd. (Singapore). Library preparation was performed using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs). Genomic DNA was first fragmented into 350-bp fragments, which were then end polished (A-tailing), ligated with adapters, and amplified by PCR. Subsequently, DNA fragments were sequenced on an Illumina NovaSeq 6000 PE150 platform. The original image data were transformed into sequencing reads by base calling in CASAVA and then were processed with the quality control treatment through fastp (v0.23.1) to obtain clean reads. The clean reads were subjected to a de novo assembly using SOAPdenovo (v2.04), SPAdes (v3.10.0), and ABySS software (v1.3.7). Assembled sequences were integrated using the CISA software (v1.3) and further processed with gap-closing and fragment-filtering steps. The resulting contigs were then submitted for genome component prediction and gene function annotation. Bacterial coding gene prediction was conducted using the GeneMarkS method (47). Functional annotations were performed using BLAST (E value ≤ 1e-5) against the COG and KEGG databases. The CAZyme annotation was performed using a diamond search against the CAZy pre-annotated CAZyme sequence database and an HMMER search against the dbCAN HMM (hidden Markov model) database (48).
Phylogenetic analysis
Phylogenetic analyses were conducted using the genome analysis tool provided by the Bacterial and Viral Bioinformatics Resource Center (BV-BRC, https://www.bv-brc.org) (49). First, the assembled contigs of the three bacterial isolates were annotated using the RAST tool kit (RASTtk, BV-BRC) (50). The codon tree was then constructed using the Bacterial Genome Tree service. Briefly, the phylogenetic comparison was performed between three bacterial isolates and their reference strains using PATtyFams based on the BV-BRC global Protein Families (PGFams) database (51). In particular, the OTU2 and OTU13 genome isolates were compared with 98 other Bifidobacterium strains (96 reference strains and two validated published strains), and the OTU10 isolate genome was compared with another 22 Megasphaera strain genomes (14 type strains and eight validated published strains). A specified number of amino acid and nucleotide sequences were selected for alignment using MUSCLE (52) and the Codon_align function of BioPython (53), respectively. One hundred bootstrap replicates were set for assessing the branch support. The corresponding phylogenomic tree was visualized with iTOL (Interactive Tree of Life, https://itol.embl.de). In addition, the 16S rRNA sequences of three isolates were analyzed on the Protologger webserver (www.protologger.de) by comparing them with those validly published species according to the DSMZ nomenclature list (54). The pairwise average nucleotide identity values between the genomes of three bacterial isolates and their closely related type strains were acquired using the Protologger webserver (www.protologger.de) and the OrthoANI tool (https://www.ezbiocloud.net/tools/ani) (54, 55).
Coculture of the Bifidobacterium and Megasphaera isolates
Cultivations were conducted in Hungate-type culture tubes at 37°C in an anaerobic chamber. To prepare the starting cultures, all the isolated strains were passaged in MRS broth containing 0.05% (wt/vol) L-cysteine-HCl three times to the mid-logarithmic phase. The cells were harvested by centrifugation at 12,857 × g (10,000 rpm) for 5 min, and the cell pellets were resuspended in carbohydrate-free fortified buffer supplemented with the vitamin and amino acid (FBVA) medium (56). The composition of the FBVA medium is listed in Table S2. Monoculture fermentation was conducted by inoculating 20 µL of the seed culture in 9.98 mL of FBVA medium containing 1% (wt/vol) BWX30 or xylose, reaching an initial OD600 ranging from 0.10 to 0.15. Coculture fermentations were conducted between the Bifidobacterium strains (OTU2 and OTU13) and the Megasphaera strain (OTU10). An equal volume (20 µL) of the seed cultures was inoculated into 9.96 mL of FBVA medium containing 1% (wt/vol) BWX30 or xylose. All the fermentation trials were conducted in triplicate. Cultures were incubated at 37°C and 150 rpm for 72 h. Growth kinetics were monitored by recording the OD600 readings using a DiluPhotometer (Implen, Munich, Germany). Meanwhile, 1 mL of bacterial culture was specifically sampled at 0, 6, 12, 24, 48, and 72 h of fermentation for cell number, microbial metabolite, and residual carbohydrate analyses. An illustration of the sampling time points is presented in Fig. S6.
Quantification of the bacterial numbers in culture using qPCR
Cells from the aliquoted samples were collected by centrifugation at 12,857 × g (10,000 rpm) for 10 min. Bacterial DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The extracted DNA was used as the template for the qPCRs. Primer pairs that targeted the Bifidobacterium cells were cited in Rivière et al. (57). Primers that targeted Megasphaera cells were designed in-house using Primer 3 (https://primer3.org/) based on the 16S rRNA sequence obtained from Sanger sequencing (Table 1). Designed Megasphaera primers were validated by checking PCR products on 1% agarose gels. The qPCRs were performed using a Bio-Rad CFX 96 system (Bio-Rad Laboratories, Hercules, CA, USA). Each qPCR (10 µL) was composed of 1 µL of the genomic DNA template, 5 µL of PowerUp SYBR Green Master Mix (Applied Biosystems, USA), 0.3 µL of each reverse and forward primer at its optimal concentration, and 3.4 µL of sterile DI water. Reactions without genomic DNA loading were used as negative controls. All the PCR conditions were run in triplicate under the following settings: incubation at 50°C for 2 min and initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 10 seconds and at 60°C for 30 seconds. The melting curve was generated at the end of each reaction to check for primer-dimer artifacts.
TABLE 1.
Primers used for quantitative real-time PCR (qPCR)
| Species | Primer pairs | Sequence (5′−3′) | Annealing temperature (°C) |
|---|---|---|---|
| Bifidobacterium pseudocatenulatum | BiCATg-1 | F: CGGATGCTCCGACTCCT | 60 |
| BiCATg-2 | R: CGAAGGCTTGCTCCCGAT | ||
| Bifidobacterium longum | BlonF | F: CAGTTGATCGCATGGTCTT | 60 |
| BlonR | R: TACCCGTCGAAGCCAC | ||
| Megasphaera indica | 0917_MegaF | F: TACGGGACGAATGGTACGACG | 60 |
| 0917_MegaR | R: CCCCGCACTTTTAAGACCGAC |
The bacterial cell concentration in each sample was determined using the absolute quantification method. To prepare the calibration curves, each isolate was grown anaerobically in MRS broth at 37°C for 12 h. The cells were harvested and diluted with the FBVA medium to reach OD600 readings from 1.0 to 1.5. One aliquot of diluted cells was used for DNA extraction using the DNeasy Blood and Tissue Kit. The extracted bacterial DNA was serially diluted and used as templates in the qPCR to establish the standard curves (Cq versus DNA concentration). Meanwhile, another aliquot of diluted cells was serially diluted (10−3–10−7) and plated on MRS agar plates. Plates were incubated anaerobically at 37°C for 48 h. The bacterial counts were determined as CFU per milliliter (CFU/mL). Standard curves were reestablished using a normalized DNA concentration (bacterial counts, CFU/mL). The abundance of each strain (CFU/mL) in the unknown samples was determined based on the standard curve of each specific strain.
Monitoring substrate consumption during fermentation
The total xylose in each culture sample was determined using Bial’s method with minor modifications (58). Bial’s reagent was prepared by dissolving orcinol (3 mg/mL) and ferric chloride (FeCl3, 0.6%, wt/vol) in concentrated hydrochloric acid (HCl, 12.2 M). Twenty microliters of cell-free supernatants was diluted 100 times and mixed with Bial’s reagent in equal volume. The resulting solutions were heated at 100°C for 10 min to form colorants. The OD670 readings were acquired using an ultraviolet-visible spectrophotometer (UV2450, Shimadzu, Kyoto, Japan). Xylose concentrations in each sample were determined by the calibration curves prepared by standards with known xylose concentrations.
Individual xylose oligomers were analyzed using high-performance anion exchange chromatography fitted with an electrochemical detector (HPAEC-ECD, Knauer, Berlin, Germany). Culture supernatants (4 µL) were loaded onto a CarboPac PA100 analytical column (4 × 250 mm) and eluted with the mobile phase made up of solution A (0.1 M NaOH) and solution B (1 M NaOAc with 0.1 M NaOH) at a flow rate of 1 mL/min following the gradient setting at the following settings: 0–35 min, 85% A and 15% B; 35–45 min, 50% A and 50% B; and 45–50 min, 100% A and 0% B; and a final washing step was carried out by running 100% A for 10 min. Quantification of the xylose oligomers with a degree of polymerization smaller than six was based on the standard curves that were prepared by mixed standard solutions composed of xylobiose (X2), xylotriose (X3), xylotetraose (X4), xylopentaose (X5), and xylohexaose (X6) that were purchased from Megazyme (Wicklow, Ireland).
Quantification of bacterial metabolites
The concentration of the short-chain fatty acids (acetate, propionate, and butyrate) was determined by gas chromatography. Briefly, the samples were centrifuged at 16,200 × g (13,000 rpm) for 10 min. The resulting supernatant was mixed with the internal standard solution (5% phosphoric acid containing 50 mM 4-methyvaleric acid) at a ratio of 4:1 (vol/vol). Samples were then analyzed using a gas chromatography system (GC, 6890N, Agilent Technologies, USA) that was equipped with a flame ionization detector and a fused silica column (Nukol Capillary GC Column, 30 m × 0.25-mm inner diameter × 0.25-µm film thickness). Other metabolites, including lactate, formate, ethanol, and free xylose, were quantitatively analyzed using a high-performance liquid chromatography-refractive index detector system (HPLC-RID, Agilent). Briefly, the cell-free supernatants obtained by microfiltration (0.22 µm) were loaded onto a Bio-Rad Aminex HPX-87H column (Bio-Rad, Hercules, CA, USA), with 0.005 M H2SO4 as the mobile phase. The quantification of lactate, formate, ethanol, and xylose was based on the calibration curves prepared using standards with known concentrations.
RNA isolation and RNA-seq analysis
Bacterial cultures grown on BWX30 that were harvested at 12 h in triplicate were preserved in the RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany). Cell pellets were immediately frozen at −80°C after removing the liquid portion by centrifugation at 16,200 × g (13,000 rpm) for 10 min. For RNA extraction, cell pellets were thawed at room temperature and mixed with 200 µL of lysis buffer (30 mM Tris-HCl, 1 mM EDTA, and 15 mg/mL lysozyme) and 15 µL of Puregene Proteinase K (Qiagen, Hilden, Germany) at 37°C for 1 h using a Thermomixer Comfort (300 rpm, Eppendorf, Hamburg, Germany). Bacterial RNA was then purified using the RNeasy mini kit (Qiagen, Hilden, Germany), during which RNase-free DNase (Qiagen, Hilden, Germany) was applied to the column to remove residual genomic DNA. All the steps were performed following the manufacturer’s instructions. The extracted RNA samples were quantitatively and qualitatively assessed using the Fragment Analyzer 5400 (Agilent, USA), and samples with integrity numbers ranging from 9 to 10 were subjected to the sequencing service provided by Novogene AIT Genomics Singapore Pte Ltd. Initially, rRNA molecules were removed using the Illumina Ribo-Zero Plus rRNA Depletion Kit, and the sequencing libraries were prepared using the NEBNext Ultra RNA Library Prep Kit. RNA sequencing was conducted on an Illumina NovaSeq 6000 sequencer with a PE150 strategy. Low-quality reads or reads with adapters were filtered using fastp (v0.23.1). The qualified reads were aligned against the combined reference genomes of OTU2, OTU10, and OTU13 using Bowtie2 (v2.3.4.3). The genomic data, including sequences (FASTA) and annotation files (GFF), were obtained from whole genome sequencing as described in the previous section. RNA-seq reads were assembled using Rockhopper (v2.0.3) to predict novel genes (59), and the novel transcripts were aligned to sequences in the NCBI NR database using Blastx (E value < 1e-5). Gene expression levels were quantified using featureCounts (v1.5.0-p3) and were normalized to fragments per kilobase of transcript per million mapped reads (FPKM). The differential gene expression analysis was performed using DESeq2 (R package v1.20.0) with an adjusted P value of <0.05 to identify differentially expressed genes. The enrichment of the differentially expressed genes in KEGG pathways and gene ontology (GO) function was assessed using clusterProfiler (R package v3.8.1).
ACKNOWLEDGMENTS
The authors would like to thank Professor Stefan Wuertz and Professor Kunn Hadinoto from Nanyang Technological University for their constructive comments. This work was conducted using the institutional funds provided by Nanyang Technological University and National Taiwan University.
Contributor Information
Ming-Hsu Chen, Email: minghsuchen@ntu.edu.tw.
Arpita Bose, Washington University in St. Louis, St. Louis, Missouri, USA.
DATA AVAILABILITY
Sequences of 16S rRNA genes of the three bacterial isolates (BL, BP, and MI) were submitted to GenBank under accession numbers of OR083355, OR083356, and OR084128, respectively. Raw reads (in FASTQ format) from the whole genome sequencing of bacterial isolates and the transcriptomic analyses were deposited in the NCBI Sequence Read Archive under the BioProject PRJNA979082, PRJNA979114, PRJNA979144, and PRJNA979349.
SUPPLEMENTAL MATERIAL
One supplementary file is available online.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.01019-23.
Supplementary tables and figures.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Hodgkinson K, El Abbar F, Dobranowski P, Manoogian J, Butcher J, Figeys D, Mack D, Stintzi A. 2023. Butyrate’s role in human health and the current progress towards its clinical application to treat gastrointestinal disease. Clin Nutr 42:61–75. doi: 10.1016/j.clnu.2022.10.024 [DOI] [PubMed] [Google Scholar]
- 2. Liu H, Wang J, He T, Becker S, Zhang G, Li D, Ma X. 2018. Butyrate: a double-edged sword for health? Adv Nutr 9:21–29. doi: 10.1093/advances/nmx009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Macfarlane GT, Macfarlane S. 2011. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol 45:S120–S127. doi: 10.1097/MCG.0b013e31822fecfe [DOI] [PubMed] [Google Scholar]
- 4. Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, Ehrentraut SF, Pickel C, Kuhn KA, Lanis JM, Nguyen V, Taylor CT, Colgan SP. 2015. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17:662–671. doi: 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gasaly N, de Vos P, Hermoso MA. 2021. Impact of bacterial metabolites on gut barrier function and host immunity: a focus on bacterial metabolism and its relevance for intestinal inflammation. Front Immunol 12:658354. doi: 10.3389/fimmu.2021.658354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Louis P, Hold GL, Flint HJ. 2014. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12:661–672. doi: 10.1038/nrmicro3344 [DOI] [PubMed] [Google Scholar]
- 7. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. 2016. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165:1332–1345. doi: 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 8. Shi L, Tu BP. 2015. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol 33:125–131. doi: 10.1016/j.ceb.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x [DOI] [PubMed] [Google Scholar]
- 10. Vital M, Howe AC, Tiedje JM. 2014. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 5:e00889. doi: 10.1128/mBio.00889-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Louis P, Flint HJ. 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19:29–41. doi: 10.1111/1462-2920.13589 [DOI] [PubMed] [Google Scholar]
- 12. Gasaly N, Hermoso MA, Gotteland M. 2021. Butyrate and the fine-tuning of colonic homeostasis: implication for inflammatory bowel diseases. Int J Mol Sci 22:3061. doi: 10.3390/ijms22063061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. 2002. Acetate utilization and Butyryl coenzyme A (CoA): acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol 68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh V, Lee G, Son H, Koh H, Kim ES, Unno T, Shin J-H. 2022. “Butyrate producers,“the sentinel of gut”: their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics”. Front Microbiol 13:1103836. doi: 10.3389/fmicb.2022.1103836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O Sheridan P, Martin JC, Lawley TD, Browne HP, Harris HMB, Bernalier-Donadille A, Duncan SH, O’Toole PW, P Scott K, J Flint H. 2016. Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microb Genom 2:e000043. doi: 10.1099/mgen.0.000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duncan SH, Louis P, Flint HJ. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Vuyst L, Leroy F. 2011. Cross-feeding between Bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol 149:73–80. doi: 10.1016/j.ijfoodmicro.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 19. Pokusaeva K, Fitzgerald GF, van Sinderen D. 2011. Carbohydrate metabolism in Bifidobacteria. Genes Nutr 6:285–306. doi: 10.1007/s12263-010-0206-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. 2016. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front Microbiol 7:979. doi: 10.3389/fmicb.2016.00979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJM, Garcia-Gil LJ, Flint HJ. 2012. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol 78:420–428. doi: 10.1128/AEM.06858-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG. 2015. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett 362:fnv176. doi: 10.1093/femsle/fnv176 [DOI] [PubMed] [Google Scholar]
- 23. Zhao S, Dien BS, Lindemann SR, Chen M-H. 2021. Controlling autohydrolysis conditions to produce xylan-derived fibers that modulate gut microbiota responses and metabolic outputs. Carbohydr Polym 271:118418. doi: 10.1016/j.carbpol.2021.118418 [DOI] [PubMed] [Google Scholar]
- 24. Lanjekar VB, Marathe NP, Ramana VV, Shouche YS, Ranade DR. 2014. Megasphaera indica sp. nov., an obligate anaerobic bacteria isolated from human faeces. Int J Syst Evol Microbiol 64:2250–2256. doi: 10.1099/ijs.0.059816-0 [DOI] [PubMed] [Google Scholar]
- 25. Shetty SA, Marathe NP, Lanjekar V, Ranade D, Shouche YS. 2013. Comparative genome analysis of Megasphaera sp. reveals niche specialization and its potential role in the human gut. PLoS One 8:e79353. doi: 10.1371/journal.pone.0079353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richter M, Rosselló-Móra R. 2009. Shifting the Genomic gold standard for the Prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rivière A, Gagnon M, Weckx S, Roy D, De Vuyst L. 2015. Mutual cross-feeding interactions between Bifidobacterium longum subsp. Longum NCC2705 and Eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan oligosaccharides. Appl Environ Microbiol 81:7767–7781. doi: 10.1128/AEM.02089-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagara Y, Fujii D, Takada T, Sato-Yamazaki M, Odani T, Oishi K. 2022. Selective induction of human gut-associated acetogenic/butyrogenic microbiota based on specific microbial colonization of indigestible starch granules. ISME J 16:1502–1511. doi: 10.1038/s41396-022-01196-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hino T, Kuroda S. 1993. Presence of lactate dehydrogenase and lactate racemase in Megasphaera elsdenii grown on glucose or lactate. Appl Environ Microbiol 59:255–259. doi: 10.1128/aem.59.1.255-259.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang X, Su Y, Zhu W. 2016. Fermentation characteristics of Megasphaera elsdenii J6 derived from pig feces on different lactate isomers. J Integr Agric 15:1575–1583. doi: 10.1016/S2095-3119(15)61236-9 [DOI] [Google Scholar]
- 31. Marounek M, Fliegrova K, Bartos S. 1989. Metabolism and some characteristics of ruminal strains of Megasphaera elsdenii. Appl Environ Microbiol 55:1570–1573. doi: 10.1128/aem.55.6.1570-1573.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prabhu R, Altman E, Eiteman MA. 2012. Lactate and acrylate metabolism by Megasphaera elsdenii under batch and steady-state conditions. Appl Environ Microbiol 78:8564–8570. doi: 10.1128/AEM.02443-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anand S, Kaur H, Mande SS. 2016. Comparative In silico analysis of butyrate production pathways in gut commensals and pathogens. Front Microbiol 7:1945. doi: 10.3389/fmicb.2016.01945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eeckhaut V, Van Immerseel F, Croubels S, De Baere S, Haesebrouck F, Ducatelle R, Louis P, Vandamme P. 2011. Butyrate production in phylogenetically diverse Firmicutes isolated from the chicken caecum. Microb Biotechnol 4:503–512. doi: 10.1111/j.1751-7915.2010.00244.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Collins T, Gerday C, Feller G. 2005. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev 29:3–23. doi: 10.1016/j.femsre.2004.06.005 [DOI] [PubMed] [Google Scholar]
- 36. Fowler CA, Hemsworth GR, Cuskin F, Hart S, Turkenburg J, Gilbert HJ, Walton PH, Davies GJ. 2018. Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae. Acta Crystallogr D Struct Biol 74:946–955. doi: 10.1107/S2059798318009737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morais MAB, Coines J, Domingues MN, Pirolla RAS, Tonoli CCC, Santos CR, Correa JBL, Gozzo FC, Rovira C, Murakami MT. 2021. Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations. Nat Commun 12:367. doi: 10.1038/s41467-020-20620-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang XJ, Wang L, Wang S, Chen ZL, Li YH. 2021. Contributions and characteristics of two bifunctional GH43 β-xylosidase /α-L-arabinofuranosidases with different structures on the xylan degradation of Paenibacillus physcomitrellae strain XB. Microbiol Res 253:126886. doi: 10.1016/j.micres.2021.126886 [DOI] [PubMed] [Google Scholar]
- 39. Rohman A, Dijkstra BW, Puspaningsih NNT. 2019. β-xylosidases: structural diversity, catalytic mechanism, and inhibition by monosaccharides. Int J Mol Sci 20:5524. doi: 10.3390/ijms20225524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ejby M, Fredslund F, Vujicic-Zagar A, Svensson B, Slotboom DJ, Abou Hachem M. 2013. Structural basis for arabinoxylo-oligosaccharide capture by the probiotic Bifidobacterium animalis subsp. lactis Bl-04. Mol Microbiol 90:1100–1112. doi: 10.1111/mmi.12419 [DOI] [PubMed] [Google Scholar]
- 41. Watanabe Y, Saito Y, Hara T, Tsukuda N, Aiyama-Suzuki Y, Tanigawa-Yahagi K, Kurakawa T, Moriyama-Ohara K, Matsumoto S, Matsuki T. 2021. Xylan utilisation promotes adaptation of Bifidobacterium pseudocatenulatum to the human gastrointestinal tract. ISME Commun 1:62. doi: 10.1038/s43705-021-00066-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee C, Teng Q, Zhong R, Ye ZH. 2012. Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol 53:1204–1216. doi: 10.1093/pcp/pcs064 [DOI] [PubMed] [Google Scholar]
- 43. Lagaert S, Pollet A, Delcour JA, Lavigne R, Courtin CM, Volckaert G. 2011. Characterization of two β-xylosidases from Bifidobacterium adolescentis and their contribution to the hydrolysis of prebiotic xylooligosaccharides. Appl Microbiol Biotechnol 92:1179–1185. doi: 10.1007/s00253-011-3396-y [DOI] [PubMed] [Google Scholar]
- 44. Coyte KZ, Rakoff-Nahoum S. 2019. Understanding competition and cooperation within the mammalian gut microbiome. Curr Biol 29:R538–R544. doi: 10.1016/j.cub.2019.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sato T, Matsumoto K, Okumura T, Yokoi W, Naito E, Yoshida Y, Nomoto K, Ito M, Sawada H. 2008. Isolation of lactate-utilizing butyrate-producing bacteria from human feces and In vivo administration of Anaerostipes caccae strain L2 and galacto-oligosaccharides in a rat model. FEMS Microbiol Ecol 66:528–536. doi: 10.1111/j.1574-6941.2008.00528.x [DOI] [PubMed] [Google Scholar]
- 46. D’hoe K, Vet S, Faust K, Moens F, Falony G, Gonze D, Lloréns-Rico V, Gelens L, Danckaert J, De Vuyst L, Raes J. 2018. Integrated culturing, modeling and transcriptomics uncovers complex interactions and emergent behavior in a three-species synthetic gut community. Elife 7:e37090. doi: 10.7554/eLife.37090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Besemer J, Lomsadze A, Borodovsky M. 2001. Genemarks: a self-training method for prediction of gene starts in microbial genomes. implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. doi: 10.1093/nar/29.12.2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. doi: 10.1093/nar/gky418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Olson RD, Assaf R, Brettin T, Conrad N, Cucinell C, Davis JJ, Dempsey DM, Dickerman A, Dietrich EM, Kenyon RW, et al. 2023. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res 51:D678–D689. doi: 10.1093/nar/gkac1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davis JJ, Gerdes S, Olsen GJ, Olson R, Pusch GD, Shukla M, Vonstein V, Wattam AR, Yoo H. 2016. PATtyFams: protein families for the microbial genomes in the PATRIC database. Front Microbiol 7:118. doi: 10.3389/fmicb.2016.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. 2009. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hitch TCA, Riedel T, Oren A, Overmann J, Lawley TD, Clavel T. 2021. Automated analysis of genomic sequences facilitates high-throughput and comprehensive description of bacteria. ISME Commun 1:16. doi: 10.1038/s43705-021-00017-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J. 2017. A large-scale evaluation of Algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. doi: 10.1007/s10482-017-0844-4 [DOI] [PubMed] [Google Scholar]
- 56. Yao T, Chen M-H, Lindemann SR. 2020. Structurally complex carbohydrates maintain diversity in gut-derived microbial consortia under high dilution pressure. FEMS Microbiol Ecol 96:fiaa158. doi: 10.1093/femsec/fiaa158 [DOI] [PubMed] [Google Scholar]
- 57. Rivière A, Selak M, Geirnaert A, Van den Abbeele P, De Vuyst L. 2018. Complementary mechanisms for degradation of inulin-type fructans and arabinoxylan oligosaccharides among bifidobacterial strains suggest bacterial cooperation. Appl Environ Microbiol 84:e02893-17. doi: 10.1128/AEM.02893-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pham PJ, Hernandez R, French WT, Estill BG, Mondala AH. 2011. A spectrophotometric method for quantitative determination of xylose in fermentation medium. Biomass Bioener 35:2814–2821. doi: 10.1016/j.biombioe.2011.03.006 [DOI] [Google Scholar]
- 59. McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables and figures.
Data Availability Statement
Sequences of 16S rRNA genes of the three bacterial isolates (BL, BP, and MI) were submitted to GenBank under accession numbers of OR083355, OR083356, and OR084128, respectively. Raw reads (in FASTQ format) from the whole genome sequencing of bacterial isolates and the transcriptomic analyses were deposited in the NCBI Sequence Read Archive under the BioProject PRJNA979082, PRJNA979114, PRJNA979144, and PRJNA979349.