Abstract
Cdc7/Dbf4 is a protein kinase that is required for the initiation of DNA replication in eukaryotes. Recent work has provided new clues to the role that Cdc7/Dbf4 plays in this process. A range of other observations suggest that Cdc7/Dbf4 also plays another, less well characterized, role in checkpoint function and in the maintenance of genomic integrity. In this review we attempt to bring together new information to explain how Cdc7/Dbf4 may perform these two distinct functions.
The function of Cdc7/Dbf4 during origin activation
The activation of eukaryotic replication origins can be divided into two stages. The first stage comprises the sequential assembly onto replication origins of the origin recognition complex (ORC), Cdc6 and Mcm2–7 (the MCM/P1 proteins). This assembly takes place during late mitosis and G1, and results in the origin becoming ‘licensed’ for DNA replication (Figure 1). In the second stage, licensed origins fire at different times during S phase to each initiate a pair of replication forks. This stage requires the action of two kinases: cyclin-dependent kinases (CDKs) and Cdc7/Dbf4.
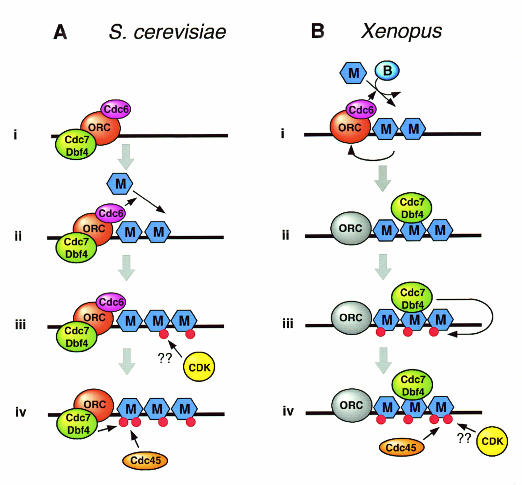
Fig. 1. Comparison of proposed Cdc7/Dbf4 function in S. cerevisiae and Xenopus. (A) Events occurring at an S. cerevisiae replication origin, taken from Pasero et al. (1999), Nougarede et al. (2000) and Zou et al. (2000). ‘M’ refers to the Mcm(2–7) heterohexamer. (i) Cdc6 and Cdc7/Dbf4 bind independently to ORC at a replication origin. (ii) Cdc6-dependent loading of Mcm(2–7) onto origin. (iii) CDK-dependent step, possibly phosphorylation of Mcm(2–7), that must occur prior to Cdc7/Dbf4 function. (iv) Cdc7/Dbf4 function [most likely Mcm(2–7) phosphorylation] leads to assembly of Cdc45 onto the origin. (B) Events occurring at a Xenopus replication origin, taken from Jares and Blow (2000). ‘M’ refers to the Mcm(2–7) heterohexamer, and ‘B’ refers to RLF-B component of the replication licensing system. (i) The loading of Mcm(2–7) proteins onto DNA (origin licensing), which depends on the function of ORC, Cdc6 and RLF-B, causes displacement of Cdc6 and destabilization of ORC. (ii) Cdc7/Dbf4 recruitment to the origin requires the Mcm(2–7) but not ORC or Cdc6. (iii) Cdc7/Dbf4 phosphorylation of Mcm(2–7) can occur in the absence of CDKs. (iv) Loading of Cdc45 onto the origin depends on both Cdc7 and CDK activity.
Cdc7 is a serine/threonine kinase, conserved from yeast to humans (reviewed in Johnston et al., 1999; Sclafani, 2000). It is activated by binding a regulatory protein called Dbf4 (Jackson et al., 1993). Although Cdc7 protein levels are approximately constant throughout the cell cycle, Cdc7 kinase activity peaks at the G1/S transition (Jackson et al., 1993; Yoon et al., 1993). This cyclic control of activity reflects changes in the abundance of Dbf4 (Brown and Kelly, 1999; Cheng et al., 1999; Jiang et al., 1999; Oshiro et al., 1999; Takeda et al., 1999), which is degraded in late mitosis and early G1 by the anaphase promoting complex (Cheng et al., 1999; Weinreich and Stillman, 1999; Ferreira et al., 2000). Cdc7 is required early in S phase for the initiation of early-firing origins as well as late in S phase for the initiation of late-firing origins (Bousset and Diffley, 1998; Donaldson et al., 1998). This means that instead of being a generalized trigger of S phase, Cdc7/Dbf4 is required for initiation to occur at each replication origin. Consistent with this proposal, Cdc7 and Dbf4 have been shown to associate with chromatin (Pasero et al., 1999; Weinreich and Stillman, 1999; Jares and Blow, 2000), whilst Dbf4 has been shown to localize to a replication origin using a one-hybrid assay (Dowell et al., 1994).
In Saccharomyces cerevisiae, Dbf4 binds to chromatin at the G1/S transition and remains attached during S phase, whilst Cdc7 appears to remain chromatin-associated throughout the cell cycle (Weinreich and Stillman, 1999). The association of Dbf4 with chromatin depends on the presence of ORC, but does not require Cdc6 or Mcm(2–7) (Figure 1A; Pasero et al., 1999). In the Xenopus cell-free system, Cdc7 also binds to chromatin during G1 and S phase, but its association with chromatin appears different from yeast. The association of Xenopus Cdc7 with chromatin depends on the presence of Mcm(2–7) (i.e. licensed origins), but does not require the continued presence of XORC or XCdc6 once they have fulfilled their essential role in licensing (Figure 1B; Jares and Blow, 2000). These results suggest that yeast and Xenopus may localize Cdc7/Dbf4 to replication origins differently.
The association of Cdc7/Dbf4 with replication origins suggests that their substrates may also be origin-bound. Several lines of evidence indicate that Mcm(2–7) may be the important targets of Cdc7/Dbf4. An allele of MCM5 (mcm5-bob1) is able to completely bypass the requirement for Cdc7 and Dbf4 (Hardy et al., 1997). Moreover, Mcm(2–7) physically interact with Cdc7/Dbf4 and Mcm2 is an excellent substrate both in vivo and in vitro (Lei et al., 1997; Sato et al., 1997; Brown and Kelly, 1998; Jiang et al., 1999; Roberts et al., 1999; Jares and Blow, 2000). Although Mcm(2–7) appear to be the most likely physiological substrates, other proteins including DNA polymerase α, Cdc45, Orc4, geminin and SV40 T antigen have recently been shown to be phosphorylated by Cdc7/Db4 in vitro (Weinreich and Stillman, 1999; Masai et al., 2000; Nougarede et al., 2000). The consequence of the phosphorylation of Mcm(2–7) by Cdc7 is currently unclear. Contrary to a previous report (Zou and Stillman, 1998), Cdc7/Dbf4 function is required in combination with CDKs for the binding of Cdc45 to origins in both yeast and Xenopus (Figure 1; Jares and Blow, 2000; Zou and Stillman, 2000). Direct interaction between Cdc45 and Mcm(2–7) has been described (Hopwood and Dalton, 1996; Zou and Stillman, 1998; Roberts et al., 1999), and it is therefore tempting to speculate that Cdc45 may bind directly to Mcm(2–7) once they have been phosphorylated by Cdc7/Dbf4 (Figure 1). After the Cdc7 execution point, or in mcm5-bob1 cells arrested in G1, structural changes at the yeast replication origin ARS1 are observed that may reflect DNA unwinding of the B2 domain (Geraghty et al., 2000). This B2 domain is thought to be involved in DNA unwinding and has recently been shown to be required for the stable association of Mcm2 with DNA (Zou and Stillman, 2000). These results would be consistent with Cdc7/Dbf4-dependent phosphorylation of the Mcm(2–7) complex allowing it to promote DNA unwinding in concert with Cdc45 (Walter and Newport, 2000).
Since both Cdc7/Dbf4 and CDKs are required for Cdc45 loading and origin activation, this raises the question of whether the kinases must act in a specific order. Recent studies in yeast and Xenopus have come to different conclusions. Reciprocal shift experiments in yeast have suggested that Cdc7/Dbf4 can perform its function only after CDK activation (Figure 1A; Nougarede et al., 2000). A potential biochemical explanation for this result is provided by in vitro experiments showing that phosphorylation of Mcm(2–7) by CDKs modestly stimulates their subsequent phosphorylation by Cdc7/Dbf4 (Masai et al., 2000). In contrast, experiments in Xenopus egg extracts suggest that Cdc7 is able to bind chromatin and fulfil its essential replicative function in the absence of CDK activity (Figure 1B; Jares and Blow, 2000). This discrepancy may represent a basic difference between the different systems. Alternatively, Cdc7/Dbf4 activity might be sufficiently high in the Xenopus system that although it normally functions after CDKs, an absolute requirement for prior phosphorylation of substrate proteins by CDKs is not evident. However, more work is needed to fully understand the function of Cdc7/Dbf4 and CDKs in origin activation.
Cdc7/Dbf4 and cell cycle checkpoints
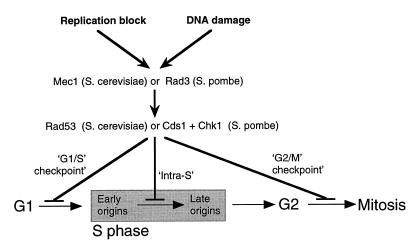
Checkpoints are surveillance mechanisms that prevent one cell cycle stage from starting if a previous cell cycle stage has not been successfully completed. Checkpoints can be considered signal transduction cascades with three components: sensors to detect incomplete or aberrant cell cycle events, transducers of the checkpoint signal, and targets that are modified by the transducers to cause cell cycle arrest (Figure 2; Elledge, 1996). At least two types of aberrant event are monitored during S phase: failure of ongoing replication forks and DNA damage. The signal transducers involve two types of protein kinase. The first type consists of members of the PI kinase superfamily, termed Mec1 in S. cerevisiae and Rad3 in Schizosaccharomyces pombe. These kinases act upstream of a second group of kinases: Rad53 in S. cerevisiae, and Cds1 plus Chk1 in S. pombe. As a consequence of checkpoint activation in S phase, further origin initiation is suppressed (‘intra-S checkpoint’) and progression into mitosis is blocked (‘G2/M checkpoint’). Evidence suggests that Cds1, but not Chk1, participates in the intra-S checkpoint (Elledge, 1996).
Fig. 2. Schematic overview of checkpoint functions. Two triggers of the checkpoint machinery (incomplete replication and DNA damage) lead to activation of the checkpoint kinases. Depending on where the cell happens to be in the cell cycle, this can cause cell cycle arrest via either the ‘G1/S’ checkpoint, the ‘intra-S’ checkpoint or the ‘G2/M’ checkpoint.
Early work showed that cdc7 mutants were hypersensitive to DNA-damaging agents, suggesting that it might play a role in the response to DNA damage (reviewed by Sclafani, 2000). One direct route by which Cdc7/Dbf4 might affect the checkpoint machinery is by being essential for the initiation of replication forks: if no replication forks are present in the cell, then failure of their progression cannot generate a checkpoint signal. This effect may explain why some CDC7/DBF4 mutants progress into mitosis despite having completely unreplicated DNA (Toyn et al., 1995; Tavormina et al., 1997; James et al., 1999). However, this is not sufficient to explain all the defects of CDC7 or DBF4 mutants. In both S. pombe and S. cerevisiae, CDC7/DBF4 mutants show enhanced sensitivity to hydroxyurea, which blocks DNA replication by depleting dNTP pools. Mutants of the S. pombe DBF4 homologue (dfp1/him1) that were able to support cell cycle progression showed sensitivity to hydroxyurea, similar to a cds1 mutant (Takeda et al., 1999). Schizosaccharomyces pombe dbf4 mutants also showed a slowed growth recovery following DNA damage. On release of S. pombe CDC7 (hsk1) mutants from hydroxyurea arrest at the non-permissive temperature, the mutants completed DNA synthesis properly but then underwent an abnormal mitosis (Snaith et al., 2000). Similarly, deletion of CDC7 in mcm5-bob1 mutants caused hypersensitivity to hydroxyurea (Weinreich and Stillman, 1999). Despite a severe effect on long-term growth of Δcdc7 mcm5-bob1 cells, hydroxyurea treatment did not cause an immediate drop in viability of these cells, indicating that the G2/M checkpoint was intact.
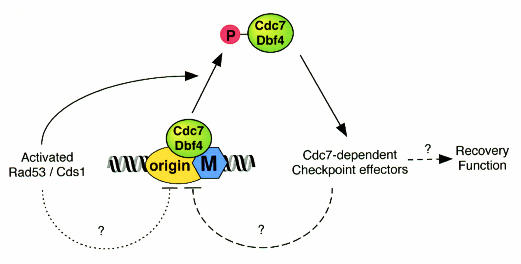
These results all suggest that Cdc7/Dbf4 plays a role in maintaining genome integrity in addition to being required for the initiation of replication forks (Figure 3). One possible explanation for this might be that Cdc7/Dbf4 plays a role in recovery from replication fork arrest. In S. pombe, although Cds1 is not required to block mitosis following treatment of cells with hydroxyurea, Cds1 deleted cells rapidly lose viability after hydroxyurea treatment (Murakami and Okayama, 1995; Boddy et al., 1998). This function of Cds1, termed the recovery function, is proposed to be part of a surveillance mechanism that somehow protects genome integrity when replication is blocked or DNA is damaged during S phase. This recovery function may act by preventing further initiation, by protecting existing replication forks from collapse or by re-activating replication forks once damage has been repaired.
Fig. 3. Model for proposed role of Cdc7/Dbf4 in the intra-S checkpoint. As a consequence of Rad53/Cds1 activation, Cdc7/Dbf4 becomes hyperphosphorylated and is displaced from chromatin. Should its initiation substrates, such as Mcm(2–7), become dephosphorylated, this by itself is likely to block further origins from initiating. To explain why the intra-S checkpoint would be intact in cdc7ts, mcm5-bob1 cells, two additional pathways are proposed (dashed lines). First, Rad53/Cds1 could have an additional repressive effect on replication origins, not mediated by Cdc7/Dbf4 (dotted line). Secondly, displaced Cdc7/Dbf4 could have an additional function in repressing the initiation of replication (dashed line). The latter pathway could also provide the signal for other Cdc7/Dbf4-dependent ‘recovery functions’.
The intra-S checkpoint: is Cdc7 a target or a transducer?
Another possible role of Cdc7/Dbf4 is as a target of the intra-S checkpoint. Treatment of cells with hydroxyurea prevents the association of Cdc45 with late origins in wild type but not in rad53 mutant yeast cells (Aparicio et al., 1999). Since Cdc45 loading onto origins is dependent on Cdc7/Dbf4 (Jares and Blow, 2000; Zou and Stillman, 2000), the effect of hydroxyurea could potentially be mediated by inhibition of Cdc7/Dbf4. Several recent reports support this possibility. Genetic and physical interactions between Cdc7 and Rad53 have been described in yeast (Dohrmann et al., 1999). Schizosaccharomyces pombe Cdc7/Hsk1 undergoes Cds1-dependent phosphorylation in response to hydroxyurea, and is a direct substrate of purified Cds1 in vitro (Snaith et al., 2000). Dbf4 also becomes phosphorylated after hydroxyurea treatment, and this phosphorylation is Rad53/Cds1-dependent (Brown and Kelly, 1999; Takeda et al., 1999; Weinreich and Stillman, 1999). This phosphorylation reduces the Cdc7/Dbf4 kinase activity (Weinreich and Stillman, 1999). Furthermore, there is a Rad53-dependent removal of Dbf4 from chromatin following hydroxyurea treatment (Pasero et al., 1999). All these results suggest that inhibition of Cdc7/Dbf4 could mediate the intra-S checkpoint and block late origin firing. One prediction of this might be that the intra-S checkpoint is defective in mcm5-bob1 cells where origin firing is independent of Cdc7/Dbf4 activity. However, unpublished experiments from Diffley and colleagues (K. Bousset and J.F. Diffley, personal communication) show that late origin firing is still blocked in cdc7ts, mcm5-bob1 cells treated with hydroxyurea. This result could be reconciled with checkpoint regulation of Cdc7/Dbf4 in two possible ways. One possibility would be that Rad53 could block late-firing origins by multiple redundant pathways, targeting other factors such as CDK, Cdc45 or RPA in addition to Cdc7/Dbf4 (Figure 3, dotted line). Indeed, a recent paper has suggested negative regulation of Cdc28 by checkpoint kinases that correlates with the presence of an unphosphorylated DNA polymerase α subunit (Pellicioli et al., 1999). Moreover, RPA undergoes a Mec1-dependent phosphorylation and is required for Cdc45 loading (Brush et al., 1996; Zou and Stillman, 2000). Another possibility would be that Cdc7/Dbf4, as discussed above, has multiple distinct roles. One role, where it induces the initiation of DNA replication, would be bypassed in an mcm5-bob1 mutant, whilst another, where it transduces checkpoint signals and blocks late origin firing, is still functional in cdc7ts, mcm5-bob1 cells (Figure 3, dashed lines). In this case, the checkpoint-dependent phosphorylation of Cdc7/Dbf4 complex might release the complex to function in some other aspect of checkpoint function. These two models are not mutually exclusive (Figure 3). The story of Cdc7/Dbf4 function is far from complete, but promises to give important insights into the way that the eukaryotic cell cycle responds to potentially lethal perturbations.
Acknowledgments
Acknowledgements
This work was supported by the Cancer Research Campaign. P.J. is the recipient of a post-doctoral fellowship from Ministerio de Educacion y Cultura of Spain. A.D. is a Royal Society University Research Fellow.
References
- Aparicio O.M., Stout, A.M. and Bell, S.P. (1999) Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl Acad. Sci. USA, 96, 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M.N., Furnari, B., Mondesert, O. and Russell, P. (1998) Replication checkpoint enforced by kinases Cds1 and Chk1. Science, 280, 909–912. [DOI] [PubMed] [Google Scholar]
- Bousset K. and Diffley, J.F. (1998) The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev., 12, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.W. and Kelly, T.J. (1998) Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J. Biol. Chem., 273, 22083–22090. [DOI] [PubMed] [Google Scholar]
- Brown G.W. and Kelly, T.J. (1999) Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase. Proc. Natl Acad. Sci. USA, 96, 8443–8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush G.S., Morrow, D.M., Hieter, P. and Kelly, T.J. (1996) The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc. Natl Acad. Sci. USA, 93, 15075–15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Collyer, T. and Hardy, C.F. (1999) Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol., 19, 4270–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann P.R., Oshiro, G., Tecklenburg, M. and Sclafani, R.A. (1999) RAD53 regulates DBF4 independently of checkpoint function in Saccharomyces cerevisiae. Genetics, 151, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A.D., Fangman, W.L. and Brewer, B.J. (1998) Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev., 12, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell S.J., Romanowski, P. and Diffley, J.F. (1994) Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science, 265, 1243–1246. [DOI] [PubMed] [Google Scholar]
- Elledge S.J. (1996) Cell cycle checkpoints: preventing an identity crisis. Science, 274, 1664–1672. [DOI] [PubMed] [Google Scholar]
- Ferreira M.F., Santocanale, C., Drury, L.S. and Diffley, J.F. (2000) Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol. Cell. Biol., 20, 242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty D.S., Ding, M., Heintz, N.H. and Pederson, D.S. (2000) Premature structural changes at replication origins in a yeast MCM mutant. J. Biol. Chem., 275, 18011–18021. [DOI] [PubMed] [Google Scholar]
- Hardy C.F., Dryga, O., Seematter, S., Pahl, P.M. and Sclafani, R.A. (1997) mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc. Natl Acad. Sci. USA, 94, 3151–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood B. and Dalton, S. (1996) Cdc45p assembles into a complex with Cdc46p/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc. Natl Acad. Sci. USA, 93, 12309–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Pahl, P.M., Harrison, K., Rosamond, J. and Sclafani, R.A. (1993) Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol. Cell. Biol., 13, 2899–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S.W. et al. (1999) nimO, an Aspergillus gene related to budding yeast Dbf4, is required for DNA synthesis and mitotic checkpoint control. J. Cell Sci., 112, 1313–1324. [DOI] [PubMed] [Google Scholar]
- Jares P. and Blow, J.J. (2000) Xenopus Cdc7 function is dependent on licensing but not on XORC, XCdc6 or CDK activity and is required for XCdc45 loading. Genes Dev., 14, 1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Jiang W., McDonald, D., Hope, T.J. and Hunter, T. (1999) Mammalian Cdc7–Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J., 18, 5703–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L.H., Masai, H. and Sugino, A. (1999) First the CDKs, now the DDKs. Trends Cell Biol., 9, 249–252. [DOI] [PubMed] [Google Scholar]
- Lei M., Kawasaki, Y., Young, M.R., Kihara, M., Sugino, A. and Tye, B.K. (1997) Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev., 11, 3365–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Matsui, E., You, Z., Ishimi, Y., Tamai, K. and Arai, K. (2000) Human Cdc7-related kinase complex: in vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a critical threonine residue of Cdc7 by Cdks. J. Biol. Chem. 275, 29042–29052. [DOI] [PubMed] [Google Scholar]
- Murakami H. and Okayama, H. (1995) A kinase from fission yeast responsible for blocking mitosis in S phase. Nature, 374, 817–819. [DOI] [PubMed] [Google Scholar]
- Nougarede R., Della Seta, F., Zarzov, P. and Schwob, E. (2000) Hierarchy of S-phase-promoting factors: yeast Dbf4-cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol., 20, 3795–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro G., Owens, J.C., Shellman, Y., Sclafani, R.A. and Li, J.J. (1999) Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability. Mol. Cell. Biol., 19, 4888–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P., Duncker, B.P., Schwob, E. and Gasser, S.M. (1999) A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev., 13, 2159–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lucca, C., Liberi, G., Marini, F., Lopes, M., Plevani, P., Romano, A., Di Fiore, P.P. and Foiani, M. (1999) Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J., 18, 6561–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B.T., Ying, C.Y., Gautier, J. and Maller, J.L. (1999) DNA replication in vertebrates requires a homolog of the Cdc7 protein kinase. Proc. Natl Acad. Sci. USA, 96, 2800–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Arai, K. and Masai, H. (1997) Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J., 16, 4340–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani R.A. (2000) Cdc7p-Dbf4p becomes famous in the cell cycle. J. Cell Sci., 113, 2111–2117. [DOI] [PubMed] [Google Scholar]
- Snaith H.A., Brown, G.W. and Forsburg, S.L. (2000) S. pombe hsk1+ is a Cds1p target required for genome integrity. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Ogino, K., Matsui, E., Cho, M.K., Kumagai, H., Miyake, T., Arai, K. and Masai, H. (1999) A fission yeast gene, him1(+)/dfp1(+), encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol. Cell. Biol., 19, 5535–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina P.A., Wang, Y.C. and Burke, D.J. (1997) Differential requirements for DNA replication in the activation of mitotic checkpoints in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 3315–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn J.H., Johnson, A.L. and Johnston, L.H. (1995) Segregation of unreplicated chromosomes in Saccharomyces cerevisiae reveals a novel G1/M-phase checkpoint. Mol. Cell. Biol., 15, 5312–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J. and Newport, J. (2000) Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol. Cell, 5, 617–627. [DOI] [PubMed] [Google Scholar]
- Weinreich M. and Stillman, B. (1999) Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J., 18, 5334–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.J., Loo, S. and Campbell, J.L. (1993) Regulation of Saccharomyces cerevisiae CDC7 function during the cell cycle. Mol. Biol. Cell, 4, 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. and Stillman, B. (1998) Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science, 280, 593–596. [DOI] [PubMed] [Google Scholar]
- Zou L. and Stillman, B. (2000) Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol., 20, 3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]