Abstract
Synthesis of monoselenophosphate, the selenium donor required for the synthesis of selenocysteine (Sec) is catalyzed by the enzyme selenophosphate synthetase (SPS), first described in Escherichia coli. SPS homologs were identified in archaea, mammals and Drosophila. In the latter, however, an amino acid replacement is present within the catalytic domain and lacks selenide-dependent SPS activity. We describe the identification of a novel Drosophila homolog, Dsps2. The open reading frame of Dsps2 mRNA is interrupted by an UGA stop codon. The 3′UTR contains a mammalian-like Sec insertion sequence which causes translational readthrough in both transfected Drosophila cells and transgenic embryos. Thus, like vertebrates, Drosophila contains two SPS enzymes one with and one without Sec in its catalytic domain. Our data indicate further that the selenoprotein biosynthesis machinery is conserved between mammals and fly, promoting the use of Drosophila as a genetic tool to identify components and mechanistic features of the synthesis pathway.
INTRODUCTION
Selenium is a trace element present in a number of prokaryotic and eukaryotic proteins in the form of the amino acid selenocysteine (Sec) (Low and Berry, 1996; Stadtman, 1996). Biosynthesis of selenoproteins rests on the production of the selenium donor compound monoselenophosphate (MSP) from selenide and ATP, a reaction catalyzed by the enzyme selenophosphate synthetase (SPS). SPS was initially identified as the product of the gene selD, one of four essential selenoprotein synthesis genes (selA–D) of Escherichia coli (Leinfelder et al., 1990). MSP is used to synthesize Sec from seryl-tRNA (Forchhammer and Böck, 1991). tRNASec, the selC gene product of E. coli (Leinfelder et al., 1988; Forchhammer and Böck, 1991), and eukaryotic homologs (Park et al., 1997) recognize UGA stop codons and cause incorporation of Sec into the elongating polypeptide. This process involves protein–RNA interactions mediated by the selB gene product, a Sec-specific elongation factor which interacts with tRNASec and a stem loop element, termed ‘Sec insertion sequence’ (SECIS) (Zinoni et al., 1990; Berry et al., 1991). In prokaryotes, SECIS elements reside immediately downstream of the UGA (Zinoni et al., 1990), whereas in archaea and eukaryotes SECIS elements reside in the 3′untranslated regions (3′UTRs) of the selenoprotein-coding mRNA (Berry et al., 1991; Wilting et al., 1997).
Selenoprotein synthesis per se is conserved in evolution (Low and Berry, 1996; Stadtman, 1996). As in E. coli, selenoprotein synthesis was shown to be essential in mammals, since targeted disruption of the mouse selenocysteine tRNA gene caused early embryonic lethality (Bosl et al., 1997). selD homologs have been identified in mammals, referred to as SPS1 and SPS2, respectively (Guimaraes, 1996). Furthermore, invertebrates have been demonstrated to synthesize selenoproteins. In case of thioredoxin reductase of Caenorhabditis elegans incorporation of Sec involves a UGA codon (Buettner et al., 1999; Gladyshev et al., 1999). Its alternative decoding is mediated by a functional 3′UTR-contained SECIS element, which differs by a G replacing the conserved single-stranded A in the mammalian SECIS element (Buettner et al., 1999). In addition, a selD homolog of Drosophila was identified. However, its gene product, which corresponds to mammalian SPS1 (Persson et al., 1997; Alsina et al., 1999), lacks selenide-dependent SPS activity due to an arginine substitution of the critical Cys (or Sec) residue in the catalytic domain of the enzyme when expressed in E. coli (Persson et al., 1997). None the less, disruption of the gene resulted in the reduction of selenoprotein synthesis, providing circumstantial evidence that the gene indeed does play a role in selenoprotein synthesis (Alsina et al., 1999).
Here we describe a novel selD homolog of Drosophila, termed Dsps2. The results show that in contrast to another invertebrate, C. elegans, the fly contains two Dsps genes of the same kind as observed in mammals. Its Dsps2 transcript contains a UGA stop codon in the catalytic center of the enzyme, but readthrough activity is provided by a mammalian-like SECIS element which functions in both transfected cultured cells and transgenic embryos. Furthermore, mammalian SECIS elements mediate readthrough activity in Drosophila cells, indicating that the components for SECIS-dependent selenoprotein biosynthesis are functionally conserved between mammals and Drosophila.
RESULTS AND DISCUSSION
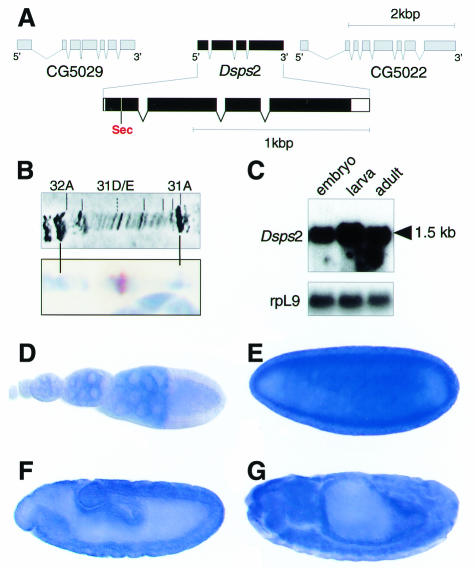
The structure of the Dsps2 gene, as revealed by sequence comparison of cDNA and corresponding genomic DNA, is shown in Figure 1A. Dsps2 contains four exons, spanning a genomic region of ∼2 kb (Figure 1A). The locus is located on the left arm of the second chromosome in section 31D/E (Figure 1B) and codes for a single transcript of ∼1.5 kb (Figure 1C).
Fig. 1. Structure, location and expression of the Drosophila sps2 gene. (A) Genomic organization of Dsps2 showing that the gene is composed of four exons; coding sequences (black bars) and untranslated regions (open bars) are indicated. Note the position of the UGA in position 24 of the deduced protein. (B) In situ hybridization of Dsps2 cDNA to polytene chromosome showing a signal in section 31D/E (see text). (C) Northern blot showing a single transcript in poly(A)+ RNA of embryos, larvae and adults; rpL9 is a control for similar RNA loading. (D–G) In situ hybridization to ovaries (D), blastoderm (E), gastrula (F) and a germ band retracted embryo (G) showing that maternal Dsps2 transcripts are expressed in nurse cells (D) and that zygotic expression occurs in a spatially restricted pattern (see text). Orientation: anterior to the left, lateral view, dorsal is top (E–G). For staging see Campos-Ortega and Hartenstein (1985).
Whole-mount in situ hybridization of ovaries and embryos shows that Dsps2 transcripts are expressed during all stages of oogenesis in nurse cells (Figure 1D). They accumulate in the oocyte and remain present at high levels up to the blastoderm stage (Figure 1E). Subsequently, the maternally derived transcripts decrease (not shown) and become replaced by zygotically expressed transcripts first detected in the midgut anlagen (Figure 1F). During subsequent stages, transcripts become enriched in a variety of tissues and organs including gut and nervous system (Figure 1G).
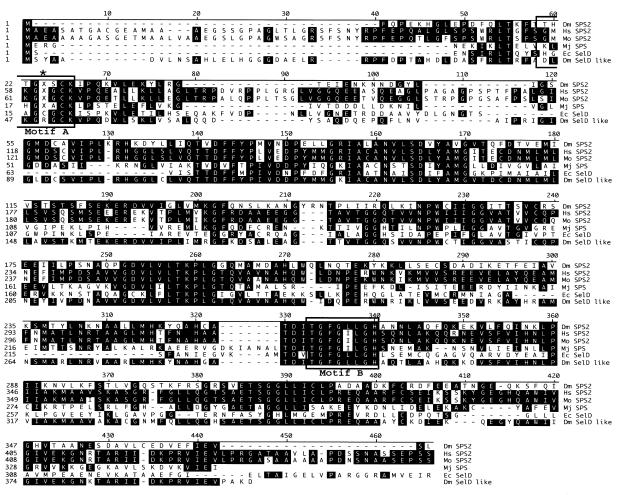
The 1324 bp Dsps2 cDNA includes a 48bp 5′UTR, a 1110 bp open reading frame (ORF) and a 166 bp 3′UTR including a conventional polyadenylation signal (DDBJ/EMBL/GenBank accession No. AJ278068). It codes for a 370 amino acid polypeptide with a high degree of overall similarity to the SPSs of mouse, human, Methanococus jannaschii and E. coli (Figure 2). DSPS2 contains a conserved ATP/GTP binding site required for ATP hydrolysis during monoselenophosphate production (Veres et al., 1994). The residue corresponding to Cys-17 in the catalytic domain of E. coli SelD is encoded by a UGA codon as observed in the mammalian class 2 SPSs (Figure 2). This observation implies that as in mammals, DSPS2 is a selenoprotein (Guimaraes, 1996) and its translation involves alternative decoding of the UGA stop codon in a SECIS element-dependent manner.
Fig. 2. Sequence comparison of DSPS2 and the known homologs. Alignment showing the Drosophila (Dm SPS2), human (Hs SPS2), mouse (Mo SPS2), M. jannaschii (Mj SPS), and E. coli (Ec SelD) SPS proteins and Drosophila SelD like protein (Dm SelD like). Position corresponding to Cys-17 of E. coli SelD is indicated by an asterisk, X denotes the Sec residue. Boxes: catalytic center (Motif A) and the ATP/GTP binding site (Motif B).
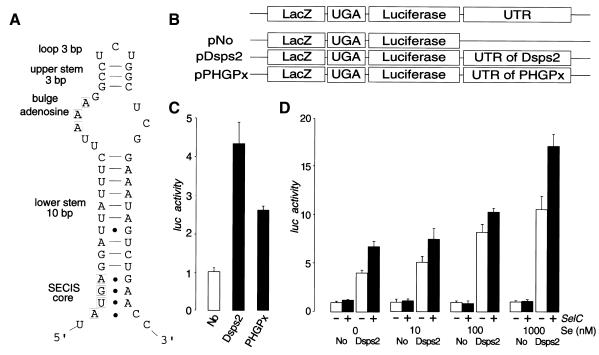
Eukaryotic SECIS elements can be grouped into two distinct classes (Grundner-Culemann et al., 1999). Both contain the SECIS core AUGA motif, a stem and adenosine loop as well as the critical GA or AA pairs on the 3′ side of the stem, opposite AUGA, forming non-Watson–Crick G–A, A–G tandem base pairs (Walczak et al., 1996; Martin et al., 1998; Grundner-Culemann et al., 1999). The predicted secondary structure of Dsps2 mRNA suggests that the 3′UTR can potentially fold into a form II SECIS element (Figure 3A).
Fig. 3. 3′UTR-dependent readthrough of UGA in Drosophila tissue culture cells. (A) Predicted secondary structure of the 3′UTR of Dsps2 showing a form II SECIS element [see text and details in Grundner-Culemann et al. (1999)]. (B) Diagrams of transfection constructs for experiment. (C) Readthrough activity as determined for different reporter genes [for nomenclature see (B)] containing different 3′UTRs in transiently transfected Schneider cells. Luciferase activities were normalized versus β-galactosidase activity and ‘Luc activity’ is represented as a fold-increase over the baseline value which is given by No-UTR. (‘Luc activity’ mean values of six independent transfection experiments and standard deviation are indicated). Note higher Dsps2 3′UTR-dependent readthrough activity as compared to SECIS elements of mammalian glutathione peroxidase (pPHGPx). (D) Effect of selenium (adding sodium selenite to the medium; see Methods) or SelC (by overexpressing co-transfected DselC as indicated by a +; see Methods) on Dsps2 3′UTR-dependent readthrough activity.
To demonstrate SECIS function of the Dsps2 3′UTR, we used a reporter gene construct in which the ORFs of bacterial LacZ and the luciferase genes were fused via an UGA stop codon (‘LacZ/TGA/luc reporter’). We transfected cultured Drosophila Schneider cells with DNA of LacZ/TGA/luc reporter genes (see Methods) with or without the Dsps2 3′UTR (pDsps2, pNo; Figure 3B). The results indicate that the 3′UTR of Dsps2 resulted in translational readthrough activity as shown by luciferase activity (Figure 3C) as previously demonstrated in response to the mammalian SECIS-element (Kollmus et al., 1996). Addition of limiting components of the selenoprotein biosynthesis pathway such as selenium and/or overexpressed tRNASec caused an increase of the SECIS-dependent readthrough activity (Figure 3D), indicating that the 3′UTR of Dsps2 acts in a Sec-specific manner.
Next, we asked whether mammalian SECIS elements can also increase readthrough activity in Drosophila cells. For this, we used LacZ/TGA/luc reporter genes, which contained functionally characterized SECIS-elements of pig heart phospholipid hydroperoxide glutathione peroxidase mRNA (PHGPx) (Methods; Figure 3B). Figure 3C shows that readthrough activity is increased by SECIS-elements of pig PHGPx, indicating that the components of selenoprotein biosynthesis are functionally conserved between mammals and fly. The same result was obtained with the SECIS-containing 3′UTR of rat 5′ deiodinase mRNA (data not shown). The reduced readthrough efficiency mediated by the mammalian SECIS elements might be due to minor incompatibilities between the mammalian and fly selenoprotein biosynthesis systems or reflect a distance effect between the UGA and the SECIS element, an aspect not addressed by our study.
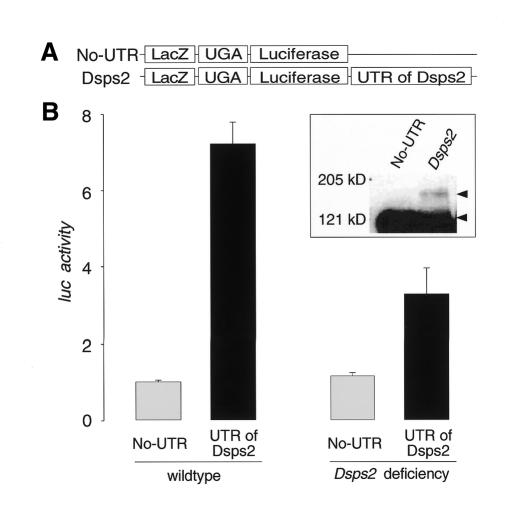
To demonstrate that the Dsps2 3′UTR mediates SECIS function in a living organism, we expressed LacZ/TGA/luc reporter genes with or without the Dsps2 3′UTR in transgenic embryos using the GAL4/UAS system (Brand and Perrimon, 1993) (Figure 4A). Embryos expressing a Dsps2 3′UTR-containing reporter gene showed significantly higher luciferase activities than those lacking the Dsps2 3′UTR (Figure 4B). In parallel to the enzymatic assays, embryonic homogenates were examined by immunostaining of western blots showing that the UGA stop codon is indeed suppressed in a Dsps2 3′UTR-dependent manner and that a β-galactosidase/luciferase fusion protein is produced (Figure 4B; inset). Furthermore, the Dsps2 3′UTR-dependent readthrough activity is reduced more than 2-fold in embryos homozygous for a Dsps2 deficiency, which derived from heterozygous Dsps2 deficiency females (Figure 4B). Residual DSPS2 in these embryos can be attributed to maternal Dsps2 activity (see Figure 1D, E). In the absence of distinct Dsps2, this activity could not be removed since additional essential genes are uncovered by the smallest available Dsps2 deficiency. Alternatively, part of the residual activity could also be contributed by Dsps1, provided that it carries SPS activity in vivo as suggested by Alsina et al. (1999).
Fig. 4. SECIS element-dependent readthrough of the UGA stop codon in Drosophila embryos. (A) Schematic representation of the reporter genes used to construct transgenic embryos (see Methods). (B) Readthrough activity as determined from extracts of wildtype (left side) or Dsps2 deficiency mutant embryos (right side; see text; readthrough activity was determined as described in the legend of Figure 3). Relative activities represent mean values from six independent collections of embryos; standard deviations are indicated. Inset: western blot indicating that embryonic extracts contain a 3′UTR-dependent β-galactosidase/luciferase fusion protein as revealed by anti-β-galactosidase-antibody staining. The fusion protein and UGA-terminated β-galactosidase are indicated by arrowheads.
The results reported here establish that: (i) Drosophila possesses the two vertebrate-type isoenzymes of SPSs; (ii) the SECIS element of Dsps2 mRNA functions both in transfected cells and transgenic embryos; (iii) the translational readthrough can be stimulated by addition of critical components of both Sec synthesis and Sec incorporation; and (iv) transgene-derived DSPS2 of the expected size can be immunostained on western blots (our unpublished result), suggesting that Drosophila SPS2 is a selenoprotein. This conclusion is supported by the observation that its homologs in Haemophilus influenzae, the archaeon, M. jannaschii, mouse and human carry a UGA-encoded Sec corresponding to Cys-17 in the activity center of SPS (Guimaraes et al., 1996).
The genomes of C. elegans and Drosophila melanogaster are sequenced (The C. elegans Sequencing Consortium, 1998; Adams et al., 2000). Whole genome database searches for selD homologs in the two species (www.sanger.ac.uk/Projects/C_elegans/ and www.fruitfly.org/), which represent model systems for unraveling molecular pathways by functional genetics, revealed only one selD homolog in C. elegans which contains a Cys in the critical position of the enzyme. The conservation of two isoforms of a key enzyme of selenoprotein biosynthesis, DSPS1 (Persson et al., 1997; Alsina et al., 1999) and DSPS2, in Drosophila and the readthrough activity obtained in response to the mammalian SECIS elements in Drosophila cell culture make it possible to use fly genetics to elucidate the precise function of each of the isoforms and to identify yet unknown components of the selenoprotein biosynthesis pathway more easily than in the vertebrate system.
METHODS
Cloning, sequencing and molecular procedures. Cloning of the Dsps2 cDNA was initiated with an expressed sequence tag (EST) obtained from the Berkeley Drosophila Genome Project (BDGP). Two of the longest cDNA clones obtained from a poly(A)+ RNA-derived library of 2–18 h old Drosophila embryos (Stratagene) were sequenced. Sequence comparison and secondary structure prediction were performed using the MegAlign program (DNASTAR Inc.) and the RNA-fold program of the GCG package (Genetic Computer Group), respectively. Database searches were done with both the Drosophila and C. elegans genomic sequences (www.fruitfly.org/ and www.sanger.ac.uk/Projects/C_elegans/).
Vectors pBPLUGA, and pBPHGPx3U (Kollmus et al., 1996) were obtained from Dr McCarthy. The Dsps2-3′UTR fragment was PCR-amplified (primers: 5′-AGA TCT AAT TAA TCA TTC AAC TTA TGA G-3′ and 5′- GGT ACC ATC GAC AAA CGA TTT TAT TAT-3′) and subcloned into BglI–KpnI sites of pBPLUGA (pBselD). Reporter gene fragments (LacZ/luciferase; with or without 3′UTRs) of pBPLUGA, pBPHGPx3U, and pBselD were subcloned into the EcoRV sites of pAc5.1 C (Invitrogen; pNo, pPHGPx and pDsps2, see Figure 3) and into the NotI–KpnI sites of pUAST (Brand and Perrimon, 1993; pUAS-No-UTR and pUAS-Dsps2, see Figure 4). DselC DNA was PCR-amplified (primers: 5′-GGT AAC CTT ACT CTT AAA ATT CCC AAG TGC TTA-3′ and 5′-GCG GCC GCC AGT TGC CTA GAT CGG GAG ACT CTG–3′) and subcloned into KpnI–NotI sites of the pAc5.1 C (pSelC). Northern blots and whole-mount in situ hybridization of preparations and chromosomes are described (Persson et al., 1997; Vorbrüggen et al., 2000).
Transgenic flies and cell transfections. Transgenic flies were generated by P-element-mediated germline transformation using pUAS-No-UTR and pUAS-Dsps2 DNA (Rubin and Spradling, 1983). Independent transformants were crossed with the actin-Gal4 driver (Bloomington Stock Center) to examine luciferase and LacZ activities in 15–24 h old wildtype and Df(2L)J27 mutant embryos which lack the Dsps2 gene (www.fruitfly.org).
Schneider Cells (Schneider, 1972) were transfected with 10 µg of plasmids (pNo, pPHGPx or pDsps2) and 5 µg of pBluescript (Niessing et al., 1999). For overexpression of DselC, 5 µg of pSelC instead of pBluescript were co-transfected with pNo or pDsps2. Twenty-four hours after transfection, fresh medium or medium with various concentrations of sodium selenite (Sigma) were added. Cells were harvested for enzymatic assay 48 h after transfection.
Enzymatic assay and western blot analysis. Cell lysates and homogenates of embryos were prepared in ‘Reporter Lysis Buffer’ (Promega). LacZ activities (and the LacZ reference; Sigma) were determined from 100 µl samples diluted with 500 µl assay buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgCl2, 12.5 mM β-mercaptoethanol) and 100 µl substrate (4 mg/ml o-nitrophenyl β-d-galactopyranoside in 60 mM Na2HPO4, 40 mM NaH2PO4). Reactions were stopped with 500 µl 1M NaCO3. LacZ activity was determined at 405 nm. Luciferase activity was measured in a PharMingen luminometer after injection of 100 µl Luciferase Assay Reagent (Promega).
Proteins were separated by SDS–PAGE. Western blots were stained with anti-β-galactosidase rabbit serum (Cappel; final dilution of 1:1000) and horseradish peroxidase-conjugated anti-rabbit IgG (Sigma) (secondary antibodies, dilution: 1:10000) assayed by SuperSignal west Dura Extended Duration substrate (PIERCE).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr McCarthy for providing constructs. We also thank G. Dowe, H. Taubert and S. Fellert for excellent technical support. We are grateful to our colleagues in the laboratory, especially R.P. Kühnlein and D. Niessing for helpful contributions, and A. Böck for criticism on the manuscript and encouragement. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SSP selenoprotein).
REFERENCES
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Alsina B., Corominas, M., Berry, M.J., Baguna, J. and Serras, F. (1999) Disruption of selenoprotein biosynthesis affects cell proliferation in the imaginal discs and brain of Drosophila melanogaster. J. Cell Sci., 112, 2875–2884. [DOI] [PubMed] [Google Scholar]
- Berry M.J., Banu, L., Chen, Y.Y., Mandel, S.J., Kieffer, J.D., Harney, J.W. and Larsen, P.R. (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature, 353, 273–276. [DOI] [PubMed] [Google Scholar]
- Bosl M.R., Takaku, K., Oshima, M., Nishimura, S. and Taketo, M.M. (1997) Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc. Natl Acad. Sci. USA, 94, 5531–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H. and Perrimon, N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Buettner C., Harney, J.W. and Berry, M.J. (1999) The Caenorhabditis elegans homologue of thioredoxin reductase contains a selenocysteine insertion sequence (SECIS) element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J. Biol. Chem., 274, 21598–21602. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J.A. and Hartenstein, V. (1985) The Embryonic Development of Drosophila melanogaster, Springer Verlag, Heidelberg, Germany.
- C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- Forchhammer K. and Böck, A. (1991) Selenocysteine synthase from Escherichia coli. Analysis of the reaction sequence. J. Biol. Chem., 266, 6324–6328. [PubMed] [Google Scholar]
- Gladyshev V.N., Krause, M., Xu, X.M., Korotkov, K.V., Kryukov, G.V., Sun, Q.A., Lee, B.J., Wootton, J.C. and Hatfield, D.L. (1999) Selenocysteine-containing thioredoxin reductase in C. elegans. Biochem. Biophys. Res. Commun., 259, 244–249. [DOI] [PubMed] [Google Scholar]
- Grundner-Culemann E., Martin, G.W., III, Harney, J.W. and Berry, M.J. (1999) Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA, 5, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes M.J. et al. (1996) Identification of a novel selD homolog from eukaryotes, bacteria, and archaea: is there an autoregulatory mechanism in selenocysteine metabolism? Proc. Natl Acad. Sci. USA, 93, 15086–15091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmus H., Flohe, L. and McCarthy, J.E. (1996) Analysis of eukaryotic mRNA structures directing cotranslational incorporation of selenocysteine. Nucleic Acids Res., 24, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinfelder W., Zehelein, E., Mandrand-Berthelot, M.A. and Böck, A. (1988) Gene for a novel tRNA species that accepts l-serine and cotranslationally inserts selenocysteine. Nature, 331, 723–725. [DOI] [PubMed] [Google Scholar]
- Leinfelder W., Forchhammer, K., Veprek, B., Zehelein, E. and Böck, A. (1990) In vitro synthesis of selenocysteinyl-tRNA(UCA) from seryl-tRNA(UCA): involvement and characterization of the selD gene product. Proc. Natl Acad. Sci. USA, 87, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low S.C. and Berry, M.J. (1996) Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci., 21, 203–208. [PubMed] [Google Scholar]
- Martin G.W., III, Harney, J.W. and Berry, M.J. (1998) Functionality of mutations at conserved nucleotides in eukaryotic SECIS elements is determined by the identity of a single nonconserved nucleotide. RNA, 4, 65–73. [PMC free article] [PubMed] [Google Scholar]
- Niessing D., Dostatni, N., Jäckle, H. and Rivera-Pomar, R. (1999) Sequence interval within the PEST motif of Bicoid is important for translational repression of caudal mRNA in the anterior region of the Drosophila embryo. EMBO J., 18, 1966–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.I., Park, J.M., Chittum, H.S., Yang, E.S., Carlson, B.A., Lee, B.J. and Hatfield, D.L. (1997) Selenocysteine tRNAs as central components of selenoprotein biosynthesis in eukaryotes. Biomed. Environ. Sci., 10, 116–124. [PubMed] [Google Scholar]
- Persson B.C., Böck, A., Jäckle, H. and Vörbruggen, G. (1997) SelD homolog from Drosophila lacking selenide-dependent monoselenophosphate synthetase activity. J. Mol. Biol., 274, 174–180. [DOI] [PubMed] [Google Scholar]
- Rubin G.M. and Spradling, A.C. (1983) Vectors for P-element-mediated gene transfer in Drosophila. Nucleic Acids Res., 11, 6341–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. (1972) Cell lines derived from late embryonic stages of Drosophila melanogaster. Embryol. Exp. Morphol., 27, 352–365. [PubMed] [Google Scholar]
- Stadtman T.C. (1996) Selenocysteine. Annu. Rev. Biochem., 65, 83–100. [DOI] [PubMed] [Google Scholar]
- Veres Z., Kim, I.Y., Scholtz, T.Z. and Stadtman, T.C. (1994) Selenophosphate synthetase. Enzyme properties and catalytic reaction. J. Biol. Chem., 269, 10597–10603. [PubMed] [Google Scholar]
- Vorbrüggen G., Önel, S. and Jäckle, H. (2000) Restricted expression and subnuclear localization of the Drosophila gene Dnop5, a member of the Nop/Sik family of the conserved rRNA processing factors. Mech. Dev., 90, 305–308. [DOI] [PubMed] [Google Scholar]
- Walczak R., Westhof, E., Carbon, P. and Krol, A. (1996) A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA, 2, 367–379. [PMC free article] [PubMed] [Google Scholar]
- Wilting R., Schorling, S., Persson, B.C. and Böck, A. (1997) Selenoprotein synthesis in archaea: identification of a mRNA element of Methanococcus jannaschii probably directing selenocysteine insertion. J. Mol. Biol., 266, 637–641. [DOI] [PubMed] [Google Scholar]
- Zinoni F., Heider, J. and Böck, A. (1990) Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc. Natl Acad. Sci. USA, 87, 4660–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]