Abstract
The incidence rate of colorectal cancer (CRC) in younger adults has been rising in developed countries. This trend may be attributed to environmental exposures as a result of lifestyle changes. Many of the lifestyle factors that promote CRC can also affect the gut microbiome, which may be associated with CRC risks. The role of the microbiome in the ongoing rise of early-onset CRC is unknown. Here, we aimed to investigate age-related differences in the gut microbiome of CRC patients and healthy individuals by examining both the fecal and tumor microbiomes. We utilized the publicly accessible data on fecal shotgun metagenomics from CuratedMetagenomeData and The Cancer Genome Atlas (TCGA) via the GDC Data Portal. Comparison of 701 CRC and 693 controls revealed that microbial features were age dependent, with a significant difference in species enrichment between early-onset (<50 years) and late-onset (>65 years) CRC patients. Analysis of the tumor-associated microbiome in a separate dataset of 85 CRC patients verified age-specific differences in taxon abundance between early- and late-onset CRC patients. Finally, using host gene expression data, we found a stronger microbe-host interaction in early- vs. late-onset CRCs. Altogether, these findings indicate that microbial features were age-dependent with stronger microbial-host interactions at the tumor site in early-onset CRCs, suggesting a direct role of microbes in tumorigenesis via interaction with cancer-related pathways in this age-group.
Keywords: Colorectal cancer, Microbiome, Bioinformatics
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second most deadly cancer globally. While the risk of CRC increases with age, screening programs have decreased the incidence rate among individuals aged 50 and older. However, CRC incidence has been rising among younger adults (<50 years) in high-income and developed countries (1). While genetic factors account for a small portion of cases even among this early-onset group, most CRCs are sporadic and thought to be linked to lifestyle (environmental) factors (2). Increased risk of early-onset colorectal cancer may be attributable to a number of variables, including those related to lifestyle and environmental exposures that may alter the gut microbiome (3). In addition, the clinical presentation, pathology, and molecular profile of early-onset CRC are different from those of late-onset CRC, indicating a distinctive disease biology (4). Accumulating evidence suggests that microorganisms that reside in the gut (intestinal microbiome) can mediate the contribution of lifestyle factors to colon carcinogenesis, a role that is particularly plausible considering that many of the established CRC risk factors affect the gut microbiome (5). Recently, polymorphic microbiomes have been proposed as a new dimension in the hallmarks of cancer with significant implications for both protective as well as detrimental impacts on cancer development and progression (6). Individuals with CRC have been shown to have an altered gut microbiome showing a disruption of the natural intestinal microbial community, characterized by loss of protective functions of the microbiome or an increase in cancer promoting ones (7). Among the microbiome are bacterial species that influence several biological processes in part via their metabolites. For example, our group and others demonstrated that loss of beneficial metabolites (i.e., short chain fatty acids/SCFA) that are involved in barrier function can increase inflammation and intestinal leakiness, which may affect the progression of CRC (8). Some other bacteria, such as Firmicutes, Bacteroidetes, enterotoxigenic Bacteroides fragilis, and the oral anaerobe Fusobacterium nucleatum, which are more prevalent in CRC, may play pathogenic roles in cancer formation (9). On the other hand, aging, the main CRC risk factor, is associated with gut microbiome changes by itself (10). Since the age-dependent divergent CRC epidemiology could be linked to lifestyle exposures that often impact the intestinal microbiome, we have tested the hypothesis that CRC-associated microbiomes may differ by age and that these differences may affect CRC-related pathways.
Materials and Methods
CuratedMetagenomeData
We used publicly available fecal shotgun metagenomics of patients with colorectal cancer from CuratedMetagenomeData (11). A total of 11 studies were identified in which subjects were diagnosed with CRC. All these studies also incorporated control groups for comparative analysis. During a screening visit, participants were categorized as having CRC based on biopsy results. Conversely, participants were classified as normal if they displayed no signs of CRC or adenomatous polyps. CuratedMetagenomeData have been uniformly processed for taxonomy profiling using MetaPhlAn3. The taxonomy profiles of the fecal microbiome from all studies (11 studies) that included CRC samples and their comparative control samples were retrieved. All of these microbiome data were compiled uniformly and were gathered from studies previously conducted by various institutions in nine countries. Although data from all the different studies have been uniformly processed, the number of total taxa reads varied per study (supplementary Table 1). Data regarding the DNA extraction methods were only available for 57.7% of the studies. Briefly, three different kits were used to isolate DNA from the stool samples, as follows: Gnome (28.8%), Qiagen (171.3), MoBio (11.6%, supplementary Table 2). Each research study included males and females of varying ages as well as a comparable number of CRC patients and control subjects, shown in Table 1. We excluded samples whose disease status was unknown, leaving 1394 samples that were included in this study. None of the subjects had current antibiotic use at the time of extraction. Diagnosis of CRC was established through the examination of tissue samples obtained via tissue biopsy analysis.
Table 1:
Subject Demographics
| control (N=693) |
CRC (N=701) |
Overall (N=1394) |
|
|---|---|---|---|
| Study name (country) | |||
| FengQ_2015 (AUT) | 61 (8.8%) | 46 (6.6%) | 107 (7.7%) |
| GuptaA_2019 (IND) | 30 (4.3%) | 30 (4.3%) | 60 (4.3%) |
| HanniganGD_2017 (USA&CAN) | 28 (4.0%) | 27 (3.9%) | 55 (3.9%) |
| ThomasAM_2018a (ITA) | 24 (3.5%) | 29 (4.1%) | 53 (3.8%) |
| ThomasAM_2018b (ITA) | 27 (3.9%) | 32 (4.6%) | 59 (4.2%) |
| ThomasAM_2019_c (JPN) | 40 (5.8%) | 40 (5.7%) | 80 (5.7%) |
| VogtmannE_2016 (USA) | 52 (7.5%) | 52 (7.4%) | 104 (7.5%) |
| WirbelJ_2018 ((DEU) | 65 (9.4%) | 60 (8.6%) | 125 (9.0%) |
| YachidaS_2019 (JPN) | 251 (36.2%) | 258 (36.8%) | 509 (36.5%) |
| YuJ_2015 (CHN) | 54 (7.8%) | 74 (10.6%) | 128 (9.2%) |
| ZellerG_2014 (FRA) | 61 (8.8%) | 53 (7.6%) | 114 (8.2%) |
| Age | |||
| Mean (SD) | 60.2 (12.3) | 63.3 (11.0) | 61.8 (11.7) |
| Median [Min, Max] | 63.0 [21.0, 84.0] | 64.0 [28.0, 90.0] | 64.0 [21.0, 90.0] |
| Age category | |||
| <50 | 125 (18.0%) | 82 (11.7%) | 207 (14.8%) |
| 50-65 | 277 (40.0%) | 278 (39.7%) | 555 (39.8%) |
| >65 | 291 (42.0%) | 341 (48.6%) | 632 (45.3%) |
| Gender | |||
| female | 310 (44.7%) | 257 (36.7%) | 567 (40.7%) |
| male | 383 (55.3%) | 444 (63.3%) | 827 (59.3%) |
| BMI | |||
| Mean (SD) | 24.1 (3.73) | 24.3 (4.31) | 24.2 (4.03) |
| Median [Min, Max] | 23.5 [16.4, 39.3] | 23.8 [13.3, 57.5] | 23.6 [13.3, 57.5] |
| Missing | 5 (0.7%) | 9 (1.3%) | 14 (1.0%) |
Age classification
We categorized age into three groups according to the clinical relevance for CRC screening: under 50 years, 50–65 years, and over 65 years. We chose the age cutoff of 50 years because it is the recommended age to initiate CRC screening among average-risk individuals. The age cutoff of 65 was selected based on the NIH’s definition of older adults.
Statistical analysis
Data analysis was conducted in R version 4.0.3 (RRID: SCR_000432, RRID:SCR_001905). MetaPhlAn 3.0 outputs of microbial relative abundance from selected studies were retrieved from CuratedMetagenomeData (11) and converted to Phyloseq objects (RRID:SCR_013080). The data reflects the available data from the CuratedMetagenomeData snapshotDate(): 2023-04-24. On the basis of subject characteristics, microbiome dissimilarity was evaluated in order to identify all of the factors influencing changes in our data. Using a phyloseq object, taxa were filtered based on abundance of >0.01% of the total data. The data were transformed using the Hellinger transformation “hell”, and redundancy analysis was then calculated using the rda function of the vegan package (RRID:SCR_011950) based on the study condition (CRC or control), age, country, gender, BMI, disease status, and DNA extraction method. Later, the envfit function was employed to build a model to investigate the microbiome-altering factors.
Microbiome dissimilarity (β diversity) was assessed based on disease status on the Bray-Curtis dissimilarity matrix at the species level using permutational ANOVA (PERMANOVA) using the adonis2 function of the vegan package. The analysis was repeated while adjusting for age, country, BMI, gender, and DNA extraction methods. Relative abundances were centered log-ratio (CLR) transformed, and then we used principal component analysis to examine beta-diversity ordination with the Aitchison distance, using R’s microbiome package (RRID: SCR_024699). After that, the "RDA" configuration from the Phyloseq package was used with the "ordinate" function to get the ordination from the transformed principal component. To investigate the different enrichment features of the microbiome between control and CRC, a multivariable linear regression analysis was performed using the MaAslin2 package (RRID: SCR_023241) at the species level. The significant features were selected based on criteria for significance, which included a p-value < 0.05, a q-value to control the False-Discovery Rate of < 0.25, and a log2 Fold Change of +/− 1.25. The association of patient age with the CRC microbiome was further assessed to discover species enrichment in early- vs. late-onset CRC. We divided the data into two subsets based on ages categories of early- (<50 years) and late (>65 years).
Tumor-associated microbiome and host’s pathway genes
TCGA-COAD clinical and RNA-seq data
To assess the interaction between the gut microbiome and the host’s gene pathways, the Cancer Genome Atlas (RRID:SCR_003193) Colon Adenocarcinoma (TCGA-COAD) patients’ sample IDs and clinical data were downloaded from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/) (12). We included patients with a diagnosis of CRC who were older than 18 years old. Patients were grouped into two age groups: early-onset (defined as <50 years, n = 15) and late-onset (defined as >65 years, n = 70). Only primary tumor samples were collected for analyses. Raw gene counts (HTSeq) were downloaded from the GDC portal (RRID:SCR_014514) and normalized using DESeq2 (RRID: SCR_015687). DESeq2 was also used to detect differential gene expression between two age groups.
De-contaminated and curated TCMA microbiota dataset
Corrected microbial abundance information of TCGA-COAD patients was downloaded from The Cancer Microbiome Atlas (TCMA) database (13). Briefly, TCMA detects metagenomics signals from the TCGA whole-genome sequencing data and de-contaminants from the tumor tissues according to their relative prevalence in tissue and blood. Microbial compositions must be grouped and averaged sample-wise (sample-level datasets). Counts are normalized using reads-per-million (RPM) by default. Subsequently, matched samples and cases were collapsed using the mean relative abundance (for case-level averaging, tissue and blood samples are averaged separately). Finally, a CLR transform is used to remove the positivity constraint and map the microbial compositions to a normalized distribution. The TCMA-COAD dataset’s Phyloseq object was processed in R. Taxa with a mean relative abundance of more than 1% in at least 5% of samples were retained for downstream analysis. Differential abundance analysis between two age groups was performed using the Wilcoxon rank sum test on CLR-transformed data.
Integrated analysis of interactions between host pathway genes expressions and taxa abundances
To investigate the possible interaction between patients’ microbiomes and matched hosts’ gene expression, we collected well-annotated pathways and pathway gene sets from the KEGG database (RRID: SCR_018145). To balance out the differences in sample numbers between the two age groups, 15 samples from the late-onset group were randomly picked without replacement, and Spearman’s rho correlation was computed. This step was repeated 1000 times (i.e., 1000 taxa-gene correlation matrices) and Spearman’s rho scores were converted into z-scores. Z-scores were finally averaged to get a single metric per KEGG pathway showing overall correlation between taxa abundance and host gene expressions. This average was converted back to the rho scores. Spearman correlation was used for this analysis, as it performs better with normalized counts (gene expression) as well as compositional data (microbiome relative abundance) compared to Pearson correlation. Correlations were visualized using the CorrPlot (RRID:SCR_023081) and iGraph (RRID:SCR_019225) packages.
From this bacteria-gene correlation matrix for each KEGG pathway, the number of correlated (positively or negatively, ∣rho∣ > 0.4) pathway genes was first counted. If the proportion of these disturbed genes in a given KEGG pathway was over 25%, we assumed the host pathway was strongly correlated with or even affected by the bacteria.
Ethic Statement
The institutional review board (IRB) at Rush University Medical Center granted a waiver to the study because it was considered Non-Human Subject Research (NHSR) due to the use of only de-identified publicly available data.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials.
Results
Microbiome Diversity Analysis Between Colorectal Cancer and Control Cohorts.
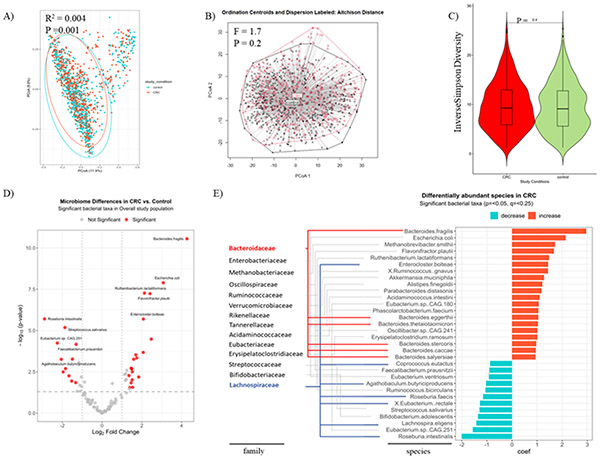
To gain insights into the overall compositional differences of the intestinal microbial communities between CRC and control samples, we compared the fecal microbiome compositions of 1394 individuals including 701 CRC patients and 693 control subjects. There was a difference in the fecal microbiome composition (β diversity) at the species level across disease statuses (control vs. CRC, R2 = 0.004, p = 0.001. Figure 1A). Despite this statistically significant difference, the effect size (R2) is notably small, indicating the observed differences account for a minor portion of the overall variance. Dispersion test and Aitchison distances after centered log-ratio (CLR) normalization of relative abundance of each taxa verified that difference in β diversity was non-significant (F: 1.7, p=0.2, Figure 1B), as is represented by a considerable overlap seen when plotting the ordinations using Non-Metric Multidimensional Scaling (NMDS) (Figure 1A) or Aitchison Distances (Figure 1B). The ordination figures continued to show extensive overlapping between study conditions when we plotted them by each study name or country (Supplementary Figure 1A, 1B).
Figure 1. Fecal microbiome compositions and differentially abundant species between CRC and control subjects.
(A) Variations in fecal microbiome composition (β diversity) across disease statuses assessed on Bray-Curtis dissimilarity matrix and tested using PERMANOVA. (B) Aitchison plot and distances from centroids based on disease statuses. (C) Alpha diversity Inverse Simpson index of the gut microbiome community across disease status.
A volcano plot showing varying enrichment of statistically significant bacterial taxa (red dots) in the CRC compared to control subjects (D) and a bar plot showing the coefficient change of significant taxa of species level in CRCs compared to controls using a multivariable linear model by disease status adjusting for country, BMI, gender, DNA extraction methods of the total 11 cohorts from the CuratedMetagenomeData (E). The analysis was done under the species level, however, both species and family taxonomy ranks are shown in (E). Both Bacteroidaceae (blue lines) and Lachnospiraceae (red lines) stand out for exhibiting the greatest numbers of species enrichment, with 6 and 7 species, respectively. The criteria for significance included a p-value < 0.05, a q-value to control the False-Discovery Rate of < 0.25, and a log2 Fold Change of +/− 1.25.
To unmask the potential role of any other factors that may be influencing microbial variation, we examined the contribution of all the available factors in the datasets, using a multivariate analysis. The tested covariates showed a statistically significant spatial effect on the microbial diversity (Supplementary Figure 1C) with country having the strongest effect, as was reported previously (14). Therefore, we corrected our subsequent analyses for all these factors. Age showed a significant effect on the microbiome composition in all 1394 subjects across the disease states (p = 0.001) as well as in CRC subjects (p = 0.002), independently of all the other covariates.
We then conducted within-group diversity indices (so called α diversity) between CRC and control conditions. Commonly used indices including richness (Chao1), evenness (Inverse Simpson), and coverage (diversity coverage) metrics showed comparable results between the two conditions (Figure 1C, Supplementary Figure 2). The α diversity results, combined with those obtained from the β diversity analyses, suggest that large microbial variations in our series which is composed of data from diverse populations is prohibitive of finding a distinct CRC-associated bacteria that could be less abundant. Next, we conducted the rarity-rare abundance that examines the diversity of less common species in the community and their distribution between the groups. This showed a significant distinction between the CRC and controls with the CRCs having a higher rate of rare taxa (p=0.002, Supplementary Figure 2C) suggesting that less abundant species rather than overall microbial diversity maybe associated with CRC.
Microbial Species Known to be Associated with CRC Were Enriched In The Cancer Group.
To examine potential difference of each taxon in CRC vs. control subjects, we conducted a species enrichment analysis. A multivariate regression was employed to account for all the available covariates including country, BMI, gender, DNA extraction method, and age. We identified a total of 32 taxa that exhibited statistically significant differences in abundance between the CRC and control subjects. The analysis revealed heightened levels of several previously reported CRC-associated bacterial species, including but not limited to Bacteroides fragilis (15), Escherichia coli (16), Methanobrevibacter smithii (17), Flavonifractor plautii (18) and Ruthenibacterium lactatiformans (19). Additionally, the analysis revealed reduced levels of beneficial species such as Roseburia intestinalis (20), Eubacterium.sp CAG 251 (21), Lachnospira eligens (22), Bifidobacterium adolescentis (23), and Streptococcus salivarius (24) among others (Figure 1D, 1E). These findings underscore the relevance of these microbial species in CRC in this large cohort and are consistent with prior research.
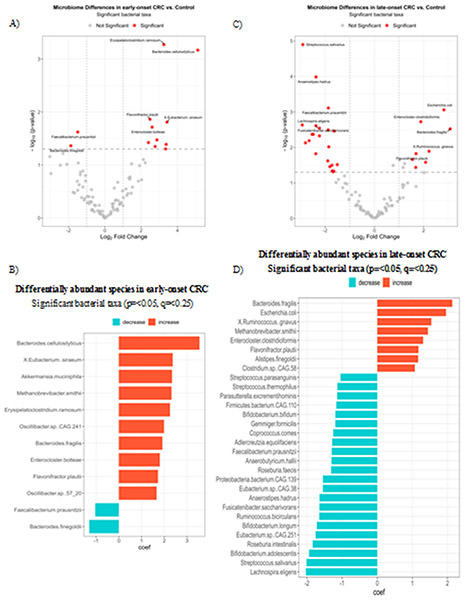
CRC-associated Microbial Species Differed by Age
We then aimed to explore potential distinctions in microbial species associated with early vs. late-onset CRC in our dataset by comparing the CRC-associated microbiome differences in early- (<50 years) separately from the late-onset (>65 years) CRCs. The age cutoffs are based on the definition of early vs. older adult onset CRC, respectively (3). Further, exclusion of the middle-aged adults which could have overlapping features with either group, would allow us to better distinguish the CRC-associated bacterial communities of younger from older age demographics. We found that species enrichment varied by age across disease statuses (early-onset Figure 2A, 2B, late-onset Figure 2C, 2D), corrected for all other covariates. The fecal microbiome of early-onset CRCs was characterized by an increase in known CRC promoting bacteria such as (Akkermansia muciniphila, Bacteroides fragilis, Methanobrevibacter smithii) as well as an enrichment in less commonly reported species (comprising ~2.5% of total taxa) such as Bacteroides cellulolyticus, Eubacterium siraeum, Erysipelatochlostridium ramosum, Oscillibbacter sp. CAG.241 and Enterocloster bolteae (Figure 2B). In the late-onset CRCs, the microbiome not only was enriched with CRC promoting bacteria but more frequently showed a decrease in several protective species, including SCFA-producing bacteria (Lachnospira eligens, Streptococcus salivarius, Bifidobacterium adolescents, Roseburia intestinalis, Eubacterium sp. CAG 251 etc. A full list is shown in Figure 2D).
Figure 2. Differentially abundant species in early- and late-onset CRC vs. Control.
A volcano plot showing differentially abundant species (red dots) in early-onset CRC vs. control subjects of respective age group of <50 years (A) and a bar plot showing the coefficient change of significant taxa of species-level using a multivariable linear model by disease status adjusting for country, BMI, gender, DNA extraction methods of early-onset CRC (B). A volcano plot showing differentially abundant species (red dots) in late-onset CRC vs. subjects of respective age group of >65 years (C). A bar plot showing the coefficient change of significant taxa of species-level using a multivariable linear model by disease status adjusting for country, BMI, gender, DNA extraction methods of late-onset CRC vs. control subjects (D). The criteria for significance included a p-value < 0.05, a q-value to control the False-Discovery Rate of < 0.25, and a log2 Fold Change of +/− 1.25.
Early-onset CRCs Show a Strong Microbial-host Interaction At the Tumor Site.
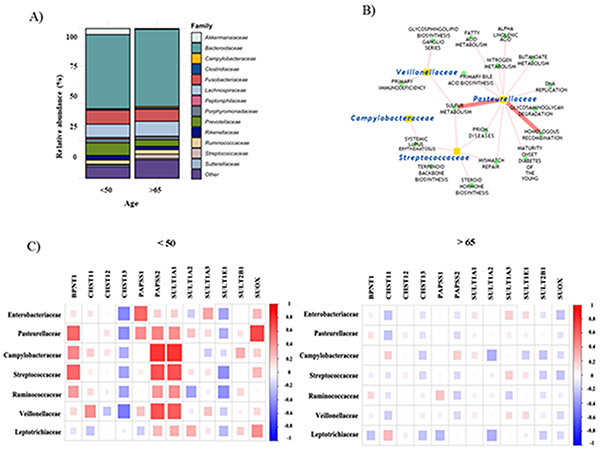
Next, we focused on the mucosal bacteria as the likely group of bacteria that are directly implicated in CRC tumorigenesis (25). We used the COAD dataset from The Cancer Genome Atlas (TCGA) via the GDC Data Portal (13), and interrogated the tumor-associated microbiome of early- (<50 years; mean age 44 ± 5.8 years) and late-onset (>65 years; 75 ± 5.7 years) CRCs. We found key similarities in the tumor-associated microbiome by age; Bacteroidaceae, Fusobacteriaceae, Lachnospiraceae, and Prevotellaceae were among the most abundant families in both early- and late-onset CRCs (Figure 3A). However, several bacterial families, which have been implicated in CRC, were differentially abundant between the two groups (Table 2). Of note, these observed differentially abundant tumor associated bacteria were among the CRC associated bacteria (Figure 1E).
Figure 3. Early-onset CRCs show distinct tumor-associated microbiomes.
(A) Relative taxa abundances of family-level in tumor-associated microbiomes in early- vs. late-onset CRC. (B) Network plot depicting strong correlations between microbial abundance and activity of KEGG pathways. (C) Correlation plots depicting correlations between tumor-associated taxa relative abundances and expression of genes in sulfur metabolism pathway in early- vs. late-onset CRC.
Table 2:
Differentially abundant bacterial families in tumor-associated microbiomes of early- versus late-onset patients and their CRC associations. CLR, centered-log ratio.
| Taxon | <50 Average CLR (SD) |
>65 Average CLR (SD) |
P-value | CRC association (PMID) |
|---|---|---|---|---|
| Enterobacteriaceae | −5.88 (8.20) | −0.11 (9.34) | 0.04 | 31582724 |
| Ruminococcaceae | −19.92 (33.11) | −0.71 (31.15) | 0.04 | 31582724 |
| Leptotrichiaceae | −7.22 (8.92) | 1.45 (13.95) | 0.02 | 24450771 |
| Campylobacteraceae | −5.92 (8.52) | 1.45 (9.44) | < 0.01 | 23733170 |
| Pasteurellaceae | −14.31 (12.84) | 1.21 (16.75) | < 0.01 | 31582724 |
| Streptococcaceae | −22.06 (21.71) | 3.44 (28.54) | < 0.01 | 29666615 |
| Veillonellaceae | −11.00 (10.30) | 1.03 (14.62) | < 0.01 | 34551683 |
Using the host gene counts, we calculated pathway enrichment scores according to the KEGG database,. Then we assessed the correlations between the age-dependent differentially abundant taxa and pathway enrichment scores. The correlation coefficients were significantly different between the age groups in most pathways (103 of 186 pathways, 55.3%, p < 0.05, Supplementary Table 3), suggesting a differential microbe-host association by age.
Using the percent of genes per pathway that showed strong correlations with the differentially abundant bacteria taxa (absolute coefficient > 0.4), we discovered that Early-onset CRCs had a significantly higher percentage of microbial-host pathway correlation (Supplementary Table 3). Interestingly, several of the pathways with the highest correlations were associated with CRC (Figure 3B). For example, the sulfur metabolism pathway demonstrated the strongest correlation between microbial and host pathways in young versus old (Figure 3C). Additionally, patients with CRC under 50 showed stronger microbial association with several DNA repair pathways (Supplementary Table 3).
Discussion
Given the increase in CRC incidence among younger adults (<50 years) and the accompanying lifestyle changes that are known to alter the intestinal microbiome, we examined if the microbiomes associated with CRC vary according to age.
Consistent with the association of the gut microbiome with CRC (26), our analysis of fecal shotgun metagenomics from CuratedMetagenomeData and The Cancer Genome Atlas (TCGA) showed that CRC had a statistically significant effect on the microbial composition. However, this effect was small due to a remarkable overlap of microbial features between the CRC and controls This could be explained by an overall large microbial variation in this series and presence of potential confounders that are known to affect intestinal microbiota, as was confirmed by our multivariate analysis. Of note, country showed the highest effect on the microbial variation in this dataset of 11 studies from diverse geographical regions. Indeed, geographical regions underlie the epidemiological variations in CRCs across populations, likely due to differences in environmental exposures such as dietary intake that affect the disease risk as well as the microbiome (27, 28) .
Adjustment of our data for the identified confounders revealed a significant effect of age on the microbial composition of the whole series in general and among the CRCs. The CRC microbiome in both age groups were characterized by an increase in CRC-promoting microbes, whereas only late-onset CRC showed a decrease in beneficial bacteria. These findings suggest that while early-onset CRC is directly associated with an increase in CRC promoting bacteria, late-onset CRCs could additionally be linked to age-related decreases in protective bacteria such as SCFA-producing microbes (29).
Since the rise in early-onset CRC is associated with changes in lifestyle exposures (30), which is reported to vary among age groups, the age-dependent CRC associated microbiome in our study may be in part explained by differential exposures to lifestyle factors across different age cohorts. While we did not have access to data on these factors, this notion is supported by the reported changes in the exposure to these CRC-associated lifestyle factors such as diet, physical activity, alcohol consumption, and disruptions in circadian rhythms in the younger populations (31). Age by itself can affect the microbiome (32, 33), which could be particularly relevant among late-onset CRCs that showed reductions in protective bacteria in our series.
Our analysis involves both fecal and mucosal microbiome. While consistent bacterial profiles are reported between fecal and biopsy samples in CRCs, mucosa-associated microbiota, given its proximity to the epithelium, is thought to have a profound impact on the mucosal homeostasis and probably tumorigenesis (34, 35). Our analysis of the tumor-associated bacteria showed that despite key similarities in the CRC-associated microbiome by age, several bacterial families that have been associated with CRC in our series (Figures 3A, 3B) were differentially abundant between the two groups. Implication of these tumor associated bacteria (e.g., increased levels of Bacteroidaceae and reduced levels of Lachnospiraceae, Figure 1E) have already been shown in CRC patients (36, 37, 38).
Accumulative data suggest that microbial signaling affects the host in part via changes in the gene expression of the tissue (24). Our analysis of the biological processes coupled with bacterial changes in early and late-onset CRCs revealed a differing microbe-host relationship between the two age groups. Early-onset CRCs showed stronger microbial-host pathway correlation terms. Interestingly, several of the highest correlation scores were among CRC-related pathways such as those related to the sulfur metabolism pathway. Sulfur source is exclusively from diet. Hydrogen sulfide (H2S), a bacterial product of meat metabolism, is involved in apoptosis and endoplasmic reticulum stress. At higher concentrations, H2S can cause DNA damage as well as fragmentation of the colonic mucosal barrier, leading to subsequent inflammation and carcinogenesis (39). Early-onset tumors also demonstrated stronger microbial correlation with several DNA repair pathways that are heavily involved in early-onset CRC, which is consistent with the emerging role of microbiota in genomic instability in CRC (5).
We are aware of the limitations of our study. Several factors that affect the risk of CRC and the microbiome such as dietary intake, alcohol consumption, and physical activity were not reported in the dataset. However, we adjusted our analyses for all the available confounding covariates including geographical area (country), which could represent differential lifestyle exposures among these populations. We had no access to the time of specimen collection post-diagnosis, the information on the use of medications or supplements at the time of sampling, the storage conditions, and the extent and the location of the tumor. All these elements may potentially act as confounding variables. For our tumor-associated bacteria, we used a well characterized though relatively small cohort of early-onset CRC. While this can restrict the scope of our findings at the tissue level, focusing on samples from one cohort eliminated the anticipated heterogeneity among different studies and enabled us to verify involvement of several CRC-associated bacteria at the tumor site. Exercising further statistical caution by employment of permutation analysis and FDR revealed novel age-dependent microbiome-host interactions that need to be verified in larger cohorts.
In summary, our findings suggest a distinct intestinal bacterial population in early- versus late-onset CRC. The CRC microbiome was characterized by an increase in CRC-promoting microbes overall, and a decrease in beneficial bacteria more among late-onset CRC. Age-dependent microbial differences also occurred at the tumor site. Strong interactions of tumor microbes with several cancer-related pathways among early-onset CRC supports a link between CRC in younger patients and the intestinal bacterial species, which are regulated by environmental exposures. This is in line with the accumulative observations that the uptrend of CRCs among the younger population coincides with changes to several key aspects of our environment, such as sedentary lifestyle, obesity, diet, and circadian disruption.
Supplementary Material
Prevention Relevance.
Early-onset colorectal can cer (CRC) is on the rise, presumably due to changes in environmental exposures. Lifestyle changes may contribute to CRC via alterations in gut microbes. Here, we show that microbial association with CRC is age-dependent, and microbe interactions with tumor pathways are stronger in young versus older CRCs.
Acknowledgements
We thank Dr. Shirin Moossavi for her help with the initial analysis of the older versions of the datasets and Dr. Kwangmin Choi for his help in data processing and data analysis.
F. Bishehsari was supported by NIH AA025387, CA279487 and CA277110. M. Alsayid was supported by the National Center For Advancing Translational Sciences of the National Institutes of Health under Award Number TL1TR002388. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- BMI
body mass index
- COAD
Colon Adenocarcinoma
- CRC
colorectal cancer
- DNA
deoxyribonucleic acid
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- TCGA
The Cancer Genome Atlas
Footnotes
Disclosures: The authors declare no potential conflicts of interest.
References
- 1.Lee JK, Merchant SA, Jensen CD, Murphy CC, Udaltsova N, Corley DA. Rising Early-onset Colorectal Cancer Incidence Is Not an Artifact of Increased Screening Colonoscopy Use in a Large, Diverse Healthcare System. Gastroenterology. 2022;162(1):325–7.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huxley RR, Ansary-Moghaddam A, Clifton P, Czernichow S, Parr CL, Woodward M. The impact of dietary and lifestyle risk factors on risk of colorectal cancer: a quantitative overview of the epidemiological evidence. Int J Cancer. 2009;125(1):171–80. [DOI] [PubMed] [Google Scholar]
- 3.Sinicrope FA. Increasing Incidence of Early-Onset Colorectal Cancer. The New England journal of medicine. 2022;386(16):1547–58. [DOI] [PubMed] [Google Scholar]
- 4.Yeo H, Betel D, Abelson JS, Zheng XE, Yantiss R, Shah MA. Early-onset Colorectal Cancer is Distinct From Traditional Colorectal Cancer. Clinical colorectal cancer. 2017;16(4):293–9.e6. [DOI] [PubMed] [Google Scholar]
- 5.Saus E, Iraola-Guzmán S, Willis JR, Brunet-Vega A, Gabaldón T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol Aspects Med. 2019;69:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer discovery. 2022;12(1):31–46. [DOI] [PubMed] [Google Scholar]
- 7.Yusuf F, Adewiah S, Syam AF, Fatchiyah F, editors. Altered profile of gut microbiota and the level short chain fatty acids in colorectal cancer patients. Journal of Physics: Conference Series; 2019: IOP Publishing. [Google Scholar]
- 8.Bishehsari F, Engen PA, Preite NZ, Tuncil YE, Naqib A, Shaikh M, et al. Dietary Fiber Treatment Corrects the Composition of Gut Microbiota, Promotes SCFA Production, and Suppresses Colon Carcinogenesis. Genes. 2018;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66(4):633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh TS, Das M, Jeffery IB, O'Toole PW. Adjusting for age improves identification of gut microbiome alterations in multiple diseases. Elife. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasolli E, Schiffer L, Manghi P, Renson A, Obenchain V, Truong DT, et al. Accessible, curated metagenomic data through ExperimentHub. Nat Methods. 2017;14(11):1023–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a Shared Vision for Cancer Genomic Data. The New England journal of medicine. 2016;375(12):1109–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohlman AB, Arguijo Mendoza D, Ding S, Gao M, Dressman H, Iliev ID, et al. The cancer microbiome atlas: a pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell host & microbe. 2021;29(2):281–98.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta VK, Paul S, Dutta C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front Microbiol. 2017;8:1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PloS one. 2015;10(3):e0119462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. 2020;580(7802):269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaci N, Borrel G, Tottey W, O'Toole PW, Brugere JF. Archaea and the human gut: new beginning of an old story. World journal of gastroenterology. 2014;20(43):16062–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Du L, Shi D, Kong C, Liu J, Liu G, et al. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nature Communications. 2021;12(1):6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trivieri N, Pracella R, Cariglia MG, Panebianco C, Parrella P, Visioli A, et al. BRAF(V600E) mutation impinges on gut microbial markers defining novel biomarkers for serrated colorectal cancer effective therapies. J Exp Clin Cancer Res. 2020;39(1):285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie K, Ma K, Luo W, Shen Z, Yang Z, Xiao M, et al. Roseburia intestinalis: A Beneficial Gut Organism From the Discoveries in Genus and Species. Front Cell Infect Microbiol. 2021;11:757718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee A, Lordan C, Ross RP, Cotter PD. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes. 2020;12(1):1802866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021;27(2):321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan L, Qi Y, Qu S, Chen X, Li A, Hendi M, et al. B. adolescentis ameliorates chronic colitis by regulating Treg/Th2 response and gut microbiota remodeling. Gut Microbes. 2021;13(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaci G, Goudercourt D, Dennin V, Pot B, Dore J, Ehrlich SD, et al. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol. 2014;80(3):928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS one. 2012;7(6):e39743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rebersek M. Gut microbiome and its role in colorectal cancer. BMC Cancer. 2021;21(1):1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moossavi S, Bishehsari F. Microbes: possible link between modern lifestyle transition and the rise of metabolic syndrome. Obes Rev. 2019;20(3):407–19. [DOI] [PubMed] [Google Scholar]
- 28.Bishehsari F, Mahdavinia M, Vacca M, Malekzadeh R, Mariani-Costantini R. Epidemiological transition of colorectal cancer in developing countries: environmental factors, molecular pathways, and opportunities for prevention. World journal of gastroenterology. 2014;20(20):6055–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, et al. Gut Microbiota and Extreme Longevity. Current biology : CB. 2016;26(11):1480–5. [DOI] [PubMed] [Google Scholar]
- 30.Stoffel EM, Murphy CC. Epidemiology and Mechanisms of the Increasing Incidence of Colon and Rectal Cancers in Young Adults. Gastroenterology. 2020;158(2):341–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ugai T, Sasamoto N, Lee HY, Ando M, Song M, Tamimi RM, et al. Is early-onset cancer an emerging global epidemic? Current evidence and future implications. Nat Rev Clin Oncol. 2022;19(10):656–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh TS, Shanahan F, O'Toole PW. The gut microbiome as a modulator of healthy ageing. Nature reviews Gastroenterology & hepatology. 2022;19(9):565–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragonnaud E, Biragyn A. Gut microbiota as the key controllers of "healthy" aging of elderly people. Immun Ageing. 2021;18(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rezasoltani S, Dabiri H, Asadzadeh-Aghdaei H, Sepahi AA, Modarressi MH, Nazemalhosseini-Mojarad E. The gut microflora assay in patients with colorectal cancer: in feces or tissue samples? Iran J Microbiol. 2019;11(1):1–6. [PMC free article] [PubMed] [Google Scholar]
- 35.Juge N. Relationship between mucosa-associated gut microbiota and human diseases. Biochem Soc Trans. 2022;50(5):1225–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Dong W, Zhao J, Wu J, Xia J, Xie S, et al. Gut microbiota profiling variated during colorectal cancer development in mouse. BMC Genomics. 2022;23(Suppl 4):848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Artemev A, Naik S, Pougno A, Honnavar P, Shanbhag NM. The Association of Microbiome Dysbiosis With Colorectal Cancer. Cureus. 2022;14(2):e22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Yu D, Wu D, Gao X, Shao F, Zhao M, et al. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell host & microbe. 2023;31(3):418–32 e8. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen LH, Cao Y, Hur J, Mehta RS, Sikavi DR, Wang Y, et al. The Sulfur Microbial Diet Is Associated With Increased Risk of Early-Onset Colorectal Cancer Precursors. Gastroenterology. 2021;161(5):1423–32.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials.