Abstract
The large ribosomal subunit protein Rpl10p is required for subunit joining and 60S export in yeast. We have recently shown that Rpl10p as well as the cytoplasmic GTPase Lsg1p are required for releasing the 60S nuclear export adapter Nmd3p from subunits in the cytoplasm. Here, we more directly address the order of Nmd3p and Rpl10p recruitment to the subunit. We show that Nmd3p can bind subunits in the absence of Rpl10p. In addition, we examined the basis of the previously reported dominant negative growth phenotype caused by overexpression of C-terminally truncated Rpl10p and found that these Rpl10p fragments are not incorporated into subunits in the nucleus but instead sequester the WD-repeat protein Sqt1p. Sqt1p is an Rpl10p binding protein that is proposed to facilitate loading of Rpl10p into the 60S subunit. Although Sqt1p normally only transiently binds 60S subunits, the levels of Sqt1p that can be coimmunoprecipitated by the 60S-associated GTPase Lsg1p are significantly increased by a dominant mutation in the Walker A motif of Lsg1p. This mutant Lsg1 protein also leads to increased levels of Sqt1p in complexes that are coimmunoprecipitated with Nmd3p. Furthermore, the dominant LSG1 mutant also traps a mutant Rpl10 protein that does not normally bind stably to the subunit. These results support the idea that Sqt1p loads Rpl10p onto the Nmd3p-bound subunit after export to the cytoplasm and that Rpl10p loading involves the GTPase Lsg1p.
In eukaryotic cells, ribosomes are assembled in the nucleolus and are transported across the nuclear membrane through nuclear pore complexes to the cytoplasm, where they are activated for translation. Many rRNA processing events must coincide with the assembly of proteins onto nascent ribosomal subunits during their biogenesis (25, 34, 35). The nascent 60S particle is preassembled in the nucleolus from locally transcribed and processed rRNAs and ribosomal proteins imported from the cytoplasm. The assembly events are highly organized and require over 150 trans-acting factors, many of which reside solely in the nucleolus. Much work has been done in elucidating the various intermediate forms of the pre-60S particle during its maturation in the nucleolus and nucleoplasm (references 7, 8, and 34 and references therein). Most of the assembly factors are removed from the preassembled complex before it leaves the nucleolus, and the complement of trans-acting factors is further reduced in the nucleoplasm prior to export (2, 29).
Nuclear export of the large subunit requires the adapter protein Nmd3p, which provides the nuclear export signal (NES) for the subunit (9, 15). 60S export also depends on the export receptor Crm1p, which recognizes the leucine-rich NES of Nmd3p (9, 15). This export pathway is conserved in humans, where it has also been suggested that Nmd3p binds to 60S subunits in the nucleolus and may be required for their release into the nucleoplasm (17, 32, 33).
NMD3 functionally interacts with the ribosomal protein RPL10. The interaction of NMD3 and RPL10 was first observed in a genetic screen for spontaneous suppressors of an rpl10 temperature-sensitive (ts) (rpl10[G161D]) mutant (20). Subsequently, high-copy NMD3 was found to suppress an rpl10 (rpl10[F85S]) mutant (37). Nmd3p and Rpl10p are reported to interact in vitro (9); however, these two factors do not interact through two-hybrid analysis (20). The in vitro binding and the genetic interaction have led to the plausible model that Rpl10p provides the binding site for Nmd3p on the 60S subunit, thereby recruiting the export adapter to the subunit in the nucleus. Once in the cytoplasm, release of Nmd3p from the subunit occurs prior to subunit joining during translation initiation, as it is not found on translating polysomes (13). Conversely, Rpl10p is required for subunit joining and translation (4, 6). We have recently shown that release of Nmd3p from 60S subunits after export to the cytoplasm requires the cytoplasmic GTPase Lsg1p and functional Rpl10p. We found that nmd3 mutants, identified as suppressors of rpl10[G161D], were also able to suppress lsg1 mutants. Furthermore, these mutant Nmd3 proteins recycled to the nucleus more efficiently and displayed weakened binding to 60S subunits. These results have led us to propose that the genetic interaction between NMD3 and RPL10 reflects their interplay during release of Nmd3p in the cytoplasm rather than during recruitment of Nmd3p to the subunit in the nucleus. However, this work did not directly address the order of Nmd3p and Rpl10p recruitment to nascent 60S subunits.
Rpl10p also interacts with the essential WD-repeat protein Sqt1p. SQT1 was identified as a high-copy suppressor of dominant negative truncated RPL10 mutants (5). Multiple WD-repeat proteins have been found to be associated with pre-60S particles, and it has been suggested that they act as molecular scaffolds to allow binding of other trans-acting factors that may have a more direct role in 60S formation (10). Along these lines, Sqt1p may facilitate loading of Rpl10p into 60S subunits (5), yet it exhibits only transient interaction with 60S through sucrose gradient sedimentation. It is likely that Sqt1p function is similar to that of another WD-repeat protein, Rrb1p, that is proposed to be a chaperone for free Rpl3p prior to or during its loading onto pre-60S subunits in the nucleolus (16).
Here, we further examine the physical and genetic relationships between Rpl10p, Sqt1p, and Nmd3p. We also examine the effect of dominant negative LSG1 mutants on Sqt1p and Rpl10p association with 60S subunits. Our results indicate that the loading of Rpl10p into subunits occurs in the cytoplasm after export by Nmd3p. Furthermore, our results suggest that the GTPase Lsg1p is involved in this process. Thus, the release of the export adapter may be triggered by loading Rpl10p.
MATERIALS AND METHODS
Yeast strains and media.
All strains were grown at 30°C unless otherwise indicated in rich medium (yeast extract-peptone) or dropout medium (synthetic complete [SC]) (18) containing either 2% glucose, 1% galactose, or 1% raffinose as the carbon source as described previously. The following strains were made for use in this study. A sqt1::KanMX4 heterozygous diploid (Research Genetics) was transformed with pAJ336 (GAL10::SQT1 LEU2-CEN), sporulated, and dissected to obtain strain AJY1433 (MATα sqt1::KanMX4 met15Δ10 leu2Δ0 ura3Δ0 his3Δ1 pAJ336). AJY1605 and AJY1640 were made by transforming AJY1433 with pAJ1062 (SQT1-myc URA3-CEN) and pAJ1065 (GAL10::SQT1 URA3-CEN), respectively, to replace pAJ336. AJY1961 was made by PCR amplifying 3xHA::KanMX6 from pFA6a-3HA-KanMX6 (27) using the 5′ oligonucleotide AJO718 (GAAAACAACATCAGAGAATTCCCAGAATACTTTGCTGCTCAAGCTCGGATCCCCGGGTTAATTAA) and the 3′ oligonucleotide AJO719 (TAATAAACTAGAATTTAAATCAAAAAAATTTCTCTTTTAAGTTAGGAATTCGAGCTCGTTTAAAC) and transforming the resulting product into wild-type W303. Transformants were selected on YPDG418 plates, and a clone containing hemagglutinin (HA)-tagged Rpl10p was verified by PCR (Table 1).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303 | MATaleu2-3,112 his3-11 ura3-1 trp1-1 ade2-1 can1-100 SSD1-d | J. Warner |
| CH1305 | MATaade2 ade3 leu2 lys2-801 ura3-52 | 24 |
| DEH221+ | MATα qsr1Δ1::HIS3 ade2-1 trp1-1 leu2-3,112 ura3-1 can1-100 (pDEGQ2) | 5 |
| AJY272 | MATaade2 ade3 leu2 lys2-801 ura3-52 NMD3-13cmyc::KanMX6 | 14 |
| AJY1433 | MATα sqt1::KanMX4 met15Δ10 leu2Δ0 ura3Δ0 his3Δ1 (pAJ336) | This study |
| AJY1605 | MATα sqt1::KanMX4 met15Δ10 leu2Δ0 ura3Δ0 his3Δ1 (pAJ1062) | This study |
| AJY1640 | MATα sqt1::KanMX4 met15Δ10 leu2Δ0 ura3Δ0 his3Δ1 (pAJ1065) | This study |
| AJY1961 | MATaRPL10-3xHA::KanMX6 leu2-3,112 his3-11 ura3-1 trp1-1 ade2-1 can1-100 SSD1-d | This study |
Construction of plasmids.
Unless otherwise noted, myc denotes 13 tandem copies of the c-myc epitope. The 5′ oligonucleotide AJO491 (GTGCCATGGCTAGAAGACCAGCT) and the 3′ oligonucleotide AJO493 (GGTTTAATTAAAGCTTGAGCAGCAAAGTA), AJO494 (GGTTTAATTAAAGCTTCTCTCTTCTTCAA), or AJO513 (GGTTTAATTAAAGCTTCAGAAGACAATTG) were used in PCR with CH1305 genomic DNA as template to amplify full-length RPL10, RPL10N187, or RPL10N64, respectively. The products were digested with NcoI and PacI and ligated into the same sites of pAJ544 (GAL10::NMD3-myc) to make pAJ792 (GAL10::RPL10-myc), pAJ793 (GAL10::RPL10N187-myc), and pAJ795 (GAL10::RPL10N64-myc), respectively. pAJ1056 (GAL10::RPL10[amino acids 26 to 187]-myc) was made essentially the same way as pAJ793, except the 5′ oligonucleotide used was AJO556 (GTGCCATGGTTCCAGACTCCAAGATC). pAJ794 (GAL10::RPL10C43-myc) was made the same way as pAJ792, except the 5′ oligonucleotide used was AJO492 (GTGCCATGGGTCCAGAATACTTGAAGAAG). pAJ796 (GAL10::RPL10-GFP), pAJ797 (GAL10::RPL10N187-GFP), pAJ798 (GAL10::RPL10C43-GFP), and pAJ799 (GAL10::RPL10N64-GFP) were all made by replacing the PacI-Bsm1 fragments of pAJ792, pAJ793, pAJ794, and pAJ795, respectively, with the PacI-BsmI green fluorescent protein (GFP)-containing fragment from pAJ582. pAJ1100 (GAL10::RPL10N187-GFP) was made by ligating a BamHI-PstI fragment from pAJ797 into the same sites of YEp352. Single myc-tagged SQT1 was cut from pSQTMYC1 (5) with BamHI and ligated into the same site of pRS416 to make pAJ1062 (SQT1-myc). GAL::SQT1, as a BamHI fragment from pAJ336 (GAL10::SQT1), was ligated into BamHI-digested pRS416 to make pAJ1065 (GAL10::SQT1). SQT1 was amplified by PCR with the 5′ oligonucleotide AJO521 (CGCGGATCCGAAATTCCCACTCGCCGC) and the 3′ oligonucleotide AJO522 (GAGAAGGATCCCCAAAGAAC), digested with KpnI, treated with T4 polymerase, and then digested with BamHI for ligation into BamHI- and SmaI-digested pAJ368 (GAL10::NMD3Δ100) (15) to make pAJ1063 (SQT1-myc). Randomly mutagenized loss-of-function and temperature-sensitive mutants of SQT1 were made by pooling 40 separate 20-cycle PCRs using Taq polymerase (GeneChoice) with the 5′ oligonucleotide AJO620 (ATGGAACCTCAAGAAGAG), the 3′ oligonucleotide AJO621 (CCTCGAACACCAGAGATA), and CH1305 genomic DNA as template. These products were cotransformed into AJY1605 along with MscI-gapped pAJ1063 for in vivo homologous recombination. Cotransformants were selected on SC-leu glucose plates and replica plated to 5-fluoroorotic acid plates to identify temperature-sensitive or constitutive-slow-growth mutants. Plasmids were recovered from selected mutants and transformed into Escherichia coli to obtain pAJ1264 (sqt1[T356A]-myc), pAJ1265 (sqt1[E40G/I370T]-myc), pAJ1266 (sqt1[N56I]-myc), and pAJ1252 (sqt1[Q193S/L194V]-myc ts). pAJ1340 (GAL10::LSG1-3xHA) and pAJ1341 (GAL10::LSG1[K349T]-3xHA) were constructed by replacing the c-myc epitopes (PacI-NheI) in pAJ879 and pAJ1109 (12), respectively, with three tandem copies of the HA epitope tag from pFA6a-3HA-KanMX6 as a restriction fragment (Table 2) (27).
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant marker(s) | Source or reference |
|---|---|---|
| pDEGQ2 | URA3-CEN (GAL1-10UAS-QSR1) | 5 |
| pDEGQ64 | URA3-CEN (GAL1-10UAS-qsr1-64Δ) | 5 |
| pDEGQ187 | URA3-CEN (GAL1-10UAS-qsr1-187Δ) | 5 |
| pAJ369 | URA3-CEN (GAL10-RPL25-GFP) | 15 |
| pAJ538 | LEU2-CEN (NMD3-myc) | 15 |
| pAJ582 | LEU2-CEN (NMD3-GFP) | 12 |
| pAJ754 | LEU2-CEN (NMD3AAA-GFP) | 12 |
| pAJ755 | URA3-CEN (NMD3-GFP) | 12 |
| pAJ792 | LEU2-CEN (GAL10::RPL10-myc) | This study |
| pAJ793 | LEU2-CEN (GAL10::RPL10N187-myc) | This study |
| pAJ794 | LEU2-CEN (GAL10::RPL10C43-myc) | This study |
| pAJ795 | LEU2-CEN (GAL10::RPL10N64-myc) | This study |
| pAJ796 | LEU2-CEN (GAL10::RPL10-GFP) | This study |
| pAJ797 | LEU2-CEN (GAL10::RPL10N187-GFP) | This study |
| pAJ798 | LEU2-CEN (GAL10::RPL10C43-GFP) | This study |
| pAJ799 | LEU2-CEN (GAL10::RPL10N64-GFP) | This study |
| pAJ907 | LEU2-CEN (RPL25-eGFP) | 12 |
| pAJ908 | URA3-CEN (RPL25-eGFP) | 12 |
| pAJ1056 | LEU2-CEN (GAL10::RPL10 [amino acids 26 to 187] myc) | This study |
| pAJ1062 | LEU2-CEN (GAL10::SQT1-myc) | This study |
| pAJ1063 | LEU2-CEN (SQT1-myc) | This study |
| pAJ1065 | URA3-CEN (GAL10::SQT1) | This study |
| pAJ1100 | URA3-2μm (GAL10::RPL10N187-GFP) | This study |
| pAJ1107 | LEU2-CEN (GAL10::LSG1-myc) | 12 |
| pAJ1108 | LEU2-CEN (GAL10::LSG1[K349T]) | 12 |
| pAJ1252 | LEU2-CEN (sqt1[Q193S/L194V]-myc ts) | This study |
| pAJ1264 | LEU2-CEN (sqt1[T356A]-myc) | This study |
| pAJ1265 | LEU2-CEN (sqt1[E40G/I370T]-myc) | This study |
| pAJ1266 | LEU2-CEN (sqt1[N561]-myc) | This study |
| pAJ1340 | LEU2-CEN (GAL10::LSG1-3xHA) | This study |
| pAJ1341 | LEU2-CEN (GAL10::LSG1[K349T]-3xHA) | This study |
Indirect immunofluorescence.
Indirect immunofluorescence was performed as described previously (15). Antibodies used were monoclonal α-c-myc (9E10) for the primary antibody (1:1,000 dilution; Covance) and Cy2-conjugated α-mouse antibody (1:300 dilution; Jackson IRL) for the secondary antibody. Fluorescence was visualized on a Zeiss Axiophot microscope fitted with a ×100 objective lens and a Princeton Electronics Micro-MAX charge-coupled device camera controlled with the IPLab Spectrum P software package from Signal Analytics Corp or on a Nikon E800 microscope fitted with a ×100 objective and a Diagnostic Instruments SPOT II camera controlled with SPOT software. Images were prepared by using Adobe Photoshop 5.0.
Immunoprecipitations.
For immunoprecipitations (IPs) of myc-tagged Rpl10p fragments, 100-ml cultures were grown to an optical density at 600 nm (OD600) of ∼0.3 in SC-leu raffinose and induced for 3 h by the addition of galactose. All subsequent steps were carried out on ice or at 4°C. Cells were harvested, washed in lysis buffer (10 mM Tris [pH 7.6], 40 mM NaCl, 10% glycerol, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, and 1 μg/ml each of leupeptins and pepstatin A), and repelleted. The cells were resuspended in 1 volume of lysis buffer, and extracts were made by glass bead lysis (vortexing five times for 50 s with 1-min intervals on ice). Insoluble material was pelleted at 15,000 × g for 10 min at 4°C. α-c-myc (9E10) antibody (1.5 μl) was added to equal OD260 units of sample supernatants and rocked for 1 h at 4°C. Thirty microliters of bovine serum albumin-blocked protein A agarose beads (Invitrogen) were then added, and rocking was continued for an additional 1 h. Beads were washed three times with lysis buffer and eluted in 50 μl of 1× Laemmli sample buffer without β-mercaptoethanol. For the IPs using α-GFP, cultures were grown as described in the figure legend, and IP steps were carried out as described above, except 5 μl of α-GFP antibody (Santa Cruz Biotech) was used in place of α-c-myc. For IP of Nmd3-myc from the free 60S sucrose gradient fraction (see Fig. 7), half of the fraction was diluted with an equal volume of IP buffer (10 mM Tris [pH 7.6], 50 mM KCl, 10 mM MgCl2, and protease inhibitors), and further steps were carried out as described above using 1.5 μl of α-c-myc. For IPs (see Fig. 8), cultures were grown as described in the figure legend, and IP steps were carried as described above for Fig. 1A using IP buffer supplemented with 1 mM MgCl2 and 5 μl of α-HA (see Fig. 8A) or 1.5 μl of α-c-myc (see Fig. 8B and C). All samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting analysis as described in the respective figure legends.
FIG. 7.
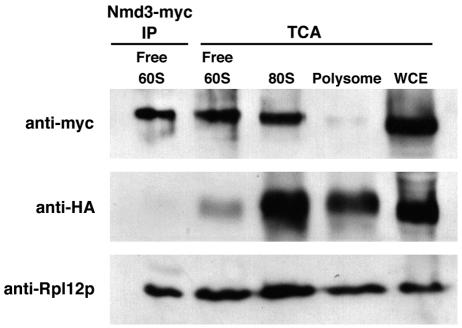
Nmd3p can bind to subunits in the absence of Rpl10p. Extracts were prepared from AJY1961 (RPL10-3xHA) containing pAJ538 (NMD3-myc) and resolved on a 7 to 47% sucrose gradient as described in the text. Immunoprecipitation of Nmd3-myc was performed using α-c-myc (9E10; Covance) from the free 60S gradient fraction. 60S subunits coimmunoprecipitating with Nmd3p and the level of Rpl10-HA in these subunits were monitored by Western blotting for the ribosomal protein Rpl12p and Rpl10-HA, respectively. The relative levels of Nmd3p-myc, Rpl10-HA, and Rpl12p in the free 60S, 80S, and polysomal fractions were analyzed by Western blotting of proteins precipitated from the respective fractions with trichloroacetic acid (TCA).
FIG. 8.
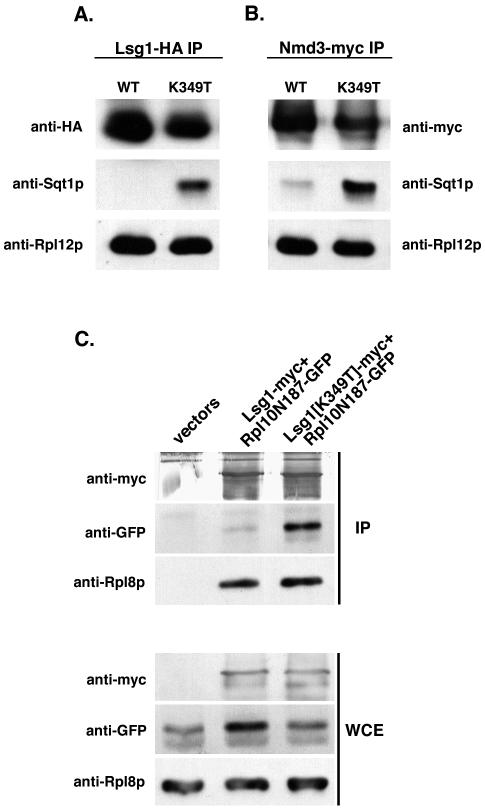
Sqt1p and Rpl10N187p accumulate on 60S subunits containing Lsg1(K349T). CH1305 and AJY272 (NMD3-myc) cells containing either pAJ1340 (GAL10::LSG1-HA) or pAJ1341 (GAL10:: LSG1[K349T]-HA) were grown to saturation and diluted to an OD600 of ∼0.15 in raffinose-containing medium. After incubation for 5 h, Lsg1p-HA or Lsg1(K349T)-HA was induced for 3.5 h by the addition of galactose. Extracts were prepared and immunoprecipitations carried out using α-HA (Covance) (A) or α-c-myc (9E10; Covance) (B) as described in the text. Immunoprecipitated samples were run on an SDS-10% PAGE gel followed by Western blotting for Lsg1-HA, Nmd3-myc, Sqt1p, and Rpl12p as a 60S binding control. (C) Cultures of CH1305 containing empty vectors pRS425 and pRS426 or pAJ1100 (GAL10::RPL10N187-GFP) with either pAJ1107 (GAL10::LSG1-myc) or pAJ1108 (GAL10::LSG1[K349T]-myc) were grown to saturation and diluted to an OD600 of ∼0.15 in raffinose-containing medium. After incubation for 6 h, Lsg1p and Rpl10N187p were coinduced for 3 h by the addition of galactose. Extracts were prepared and immunoprecipitations carried out using α-c-myc as described in the text. Immunoprecipitated samples (IP), as well as whole-cell extracts (WCE), were run on an SDS-12% PAGE gel followed by Western blotting for Lsg1-myc, Rpl10N187-GFP, or Rpl8p as a 60S reporter.
FIG. 1.
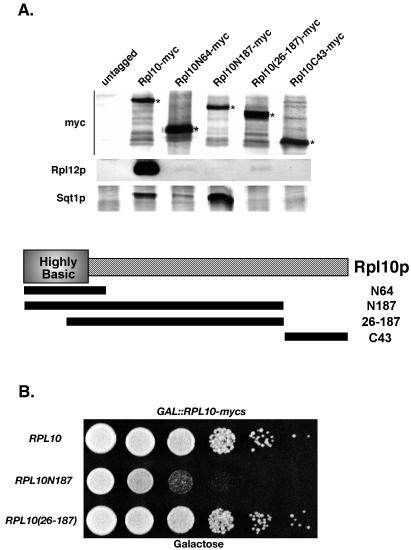
Coimmunoprecipitation of proteins associated with Rpl10p dominant negative fragments and relief of the dominant negative growth phenotype by deletion mapping. (A) Extracts were prepared from strain W303 (wild type) transformed with pAJ792 (GAL10::RPL10-myc), pAJ793 (GAL10::RPL10N187-myc), pAJ794 (GAL10::RPL10C43-myc), pAJ795 (GAL10::RPL10N64-myc), pAJ1056 (GAL10::RPL10[amino acids 25 to 187]-myc), or pAJ24 (empty vector) and immunoprecipitated as described in the text. The proteins associated with the affinity-purified complexes were separated on an SDS-12% PAGE gel and transferred to nitrocellulose for Western blotting with α-c-myc, α-Rpl12p (J. Ballesta) as a ribosomal marker, or α-Sqt1p (B. Trumpower). Asterisks indicate positions of various myc-tagged Rpl10p alleles. A schematic diagram of Rpl10p is shown in conjunction with graphic representations of the truncated mutant proteins used in this work. As noted, the first 40 amino acids of Rpl10p are highly basic. (B) 10× serial dilutions of W303 containing pAJ792, pAJ793, or pAJ1056 were spotted onto an SC-leu galactose plate and incubated for 3 days at 30°C.
GFP fusion protein localization.
GFP visualization was adapted from a method described previously (31). Culture conditions are given in corresponding figure legends. Cultured cells were fixed with a 1:9 volume of 37% formaldehyde for 40 min. The cells were washed in 0.1 M potassium phosphate, pH 6.6, and resuspended in Ksorb buffer (0.1 M potassium phosphate [pH 6.6], 1.2 M sorbitol). Triton X-100 (0.05%) was added to permeabilize cells for 4 min, and 1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) was added to stain nuclei. After 2 min, cells were washed twice with phosphate-buffered saline and visualized as described for indirect immunofluorescence.
Polysome analysis.
For sucrose density gradients, cultures were treated and collected as indicated in the corresponding figure legends. All of the following steps were carried out on ice or at 4°C. Culture pellets were washed with 2 ml of polysome lysis buffer (10 mM Tris-HCl [pH 7.6], 100 mM KCl, 10 mM MgCl2, 6 mM β-mercaptoethanol, 150 μg/ml cycloheximide, 1 mM phenylmethylsulfonyl fluoride, and 1 μg/ml each of leupeptins and pepstatin A). Cells were pelleted, resuspended in 1 volume of the same buffer, and broken open by glass bead lysis (vortexing four times for 30 s with 1-min intervals on ice). Insoluble material was pelleted by centrifugation at 15,000 × g for 10 min. Nine OD260 units of supernatant were loaded onto continuous 7 to 47% sucrose gradients in polysome lysis buffer without protease inhibitors. After a 2.5-h spin at 40,000 rpm in a Beckman SW40 rotor, gradient fractions were collected and precipitated with 10% trichloroacetic acid. After resuspension in 50 μl of 1× Laemmli buffer, fractions were run on SDS-PAGE gels and Western blotting was performed as for IPs with antibodies indicated in figure legends.
Growth assays.
For growth comparisons, saturated cultures were serially diluted and plated onto the appropriate medium as indicated in the corresponding figure legends.
RESULTS
The ribosomal protein Rpl10p shows genetic (9, 20, 37) and physical (9) interaction with Nmd3p, the export adapter that provides the NES for export of nascent 60S subunits from the nucleus in yeast. rpl10 mutants are defective for nuclear export of 60S subunits (9), and C-terminal truncations of Rpl10p inhibit cell growth when overexpressed in wild-type cells (5). We tested the model that these dominant negative Rpl10p fragments competed with wild-type Rpl10p for binding to the subunit in the nucleus (9), thus preventing recruitment of the export adapter Nmd3p.
Truncated Rpl10 proteins are not incorporated into subunits.
Full-length and truncated Rpl10p fragments were expressed from a galactose-inducible (GAL10) promoter in vivo as fusions to an oligomeric myc epitope and could be immunoprecipitated from extracts (Fig. 1A). Overexpression of the myc-tagged fusion proteins, as well as GFP-tagged fusion proteins, impaired cell growth to a degree similar to that observed with untagged proteins (5; data not shown). 60S subunits, monitored by Western blotting for the ribosomal protein Rpl12p, were efficiently coimmunoprecipitated with full-length Rpl10-myc but not with Rpl10N187-myc (containing amino acids 1 to 187), Rpl10N64-myc (containing amino acids 1 to 64), or the C-terminal 43-amino-acid fragment Rpl10C43-myc. These results indicate that only the full-length protein was stably incorporated into subunits. Thus, the dominant negative effect of overexpressing these truncated proteins does not appear to arise from their incorporation into subunits, thereby blocking subsequent assembly events.
Dominant negative fragments of Rpl10p sequester Sqt1p.
Previous work has shown that Rpl10p loads into the nascent 60S subunit late in the biogenesis pathway (26) and is one of several exchangeable proteins (36). The loading of Rpl10p into 60S subunits is thought to require the WD-repeat protein Sqt1p (5), as Rpl10p binds to Sqt1p in a two-hybrid assay and repression of Sqt1p expression leads to free 60S subunits lacking Rpl10p.
In our attempts to coimmunoprecipitate 60S subunits with truncated Rpl10 proteins, we noticed the copurification of a protein of approximately 47 kDa that was highly enriched in the Rpl10N187p immunoprecipitated sample and present in the Rpl10N64p sample but not in the immunoprecipitate obtained with the C-terminal fragment Rpl10C43p. Based on its apparent size and the reported interaction of Sqt1p with Rpl10p (5), the immunoprecipitates were tested by Western blotting for the presence of Sqt1p. Indeed, the 47-kDa protein strongly cross-reacted with anti-Sqt1p antisera (Fig. 1A). This result raised the possibility that the dominant negative effects of the C-terminally truncated Rpl10p proteins are caused by sequestering Sqt1p.
In order to test this idea, we asked whether disrupting the interaction of Rpl10N187p with Sqt1p would relieve its dominant negative effect. We reasoned that the interaction between these two proteins is mediated in part by electrostatic interactions between the highly acidic N terminus of Sqt1p and the basic N terminus of Rpl10p (Fig. 2A and 1A, respectively). We found that removal of the first 25 amino acids of Rpl10N187-myc eliminated its binding to Sqt1p (Fig. 1A) and relieved its dominant negative effect (Fig. 1B). Since this double truncation mutant was expressed at levels similar to or even higher than the single C-terminal deletion (Rpl10N187-myc) (data not shown), these results support the idea that the RPL10N187 dominant negative phenotype is due to sequestering Sqt1p, thereby effectively preventing its association with endogenous Rpl10p. This is consistent with the finding that overexpression of Sqt1p suppresses the slow-growth phenotype caused by Rpl10N187p (5). In this work, we found that Rpl10N187p appears to bind more Sqt1p than did Rpl10N64p (Fig. 1A), even though expression of either fragment is dominant negative. However, both Rpl10p truncations are suppressed by high-copy SQT1, suggesting that Sqt1p is depleted by both Rpl10p truncations. The dominant negative effect of Rpl10N64p is likely due to its higher level of expression compared to Rpl10N187p (Fig. 1A), resulting in efficient sequestration of Sqt1p.
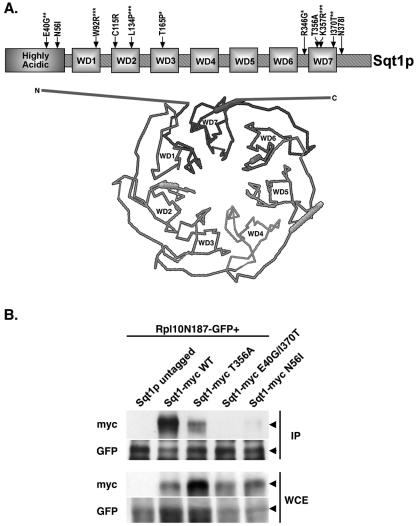
FIG. 2.
Mutations in Sqt1p lead to loss of interaction with an Rpl10p dominant negative fragment. (A) Linear schematic and predicted structure for Sqt1p. Arrows indicate mutations in the SQT1 sequence that lead to a decrease in Rpl10p binding. Asterisks indicate multiple mutations that occur in the same allele. All sequencing of wild-type and mutant SQT1 alleles revealed sequence differences (nucleotides C791 and C928) compared to the published sequence (A791 and T928, respectively). These represent polymorphisms in strain CH1305 genomic sequence. As noted, the first 60 amino acids of Sqt1p are highly acidic. Predicted structural representation of Sqt1p based on the crystal structure of TUP1 (www.sbg.bio.ic.ac.uk/∼3dpssm [21]). Sqt1p is a WD-repeat protein with seven well-predicted blades (WD1 to WD7) whose two termini are structurally adjacent. The N and C termini of Sqt1p are depicted here as unmodeled extensions from core structure. (B) Cultures of W303 containing pAJ1100 (GAL10::L10N187-GFP) with either pAJ1063 (Sqt1-myc WT), pAJ1264 (sqt1[T356A]-myc), pAJ1265 (sqt1[E40G/I370T]-myc), or pAJ1266 (sqt1[N56I]-myc) were grown to saturation in SC-leu-ura medium containing raffinose. Cultures were diluted to an OD600 of ∼0.15, incubated for 4 h, and induced with galactose for 3 h. Extracts were made and immunoprecipitations performed with α-GFP as described in the text. An extract without myc-labeled Sqt1p was used as the control (lane 1).
Sqt1p binding to Rpl10p is important for its function.
We randomly mutagenized SQT1 by PCR and identified recessive lethal loss-of-function mutants in order to correlate function with biochemical activity. We found that sqt1 loss-of-function mutants have reduced affinity for Rpl10N187p (Fig. 2B). Sqt1p is a WD-repeat protein with seven well-predicted blades. It is predicted to fold into a disk shape, typical of WD-repeat proteins, with an acidic amino-terminal extension. Based on threading the Sqt1p sequence onto the crystal structure of TUP1 from Saccharomyces cerevisiae (www.sbg.bio.ic.ac.uk/∼3dpssm [21]) (Fig. 2A), the two termini of Sqt1p are close together and on the same surface of the protein. Sequence analysis revealed that the sqt1 mutants contained point mutations that mapped to the N and C termini and repeats 1 to 3 and 7, all potentially on one face of the protein (Fig. 2A) and all of which disrupted binding to Rpl10N187-GFP (Fig. 2B and data not shown). The mutant proteins were expressed at levels similar to or greater than wild-type protein (Fig. 2B), indicating that protein stability was not significantly affected.
Cellular localization of C-terminally truncated Rpl10 proteins.
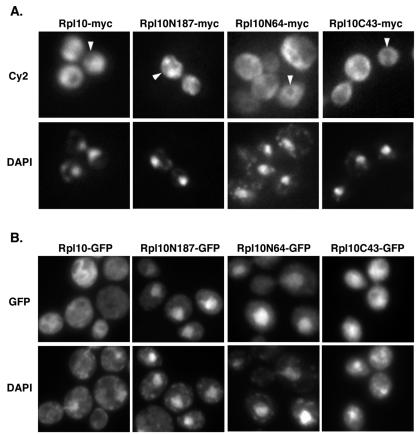
We next examined the localization of full-length and truncated forms of Rpl10p by indirect immunofluorescence. The first 64 N-terminal amino acids of Rpl10p are highly basic and have been shown to localize GFP to the nucleus, suggesting the presence of a nuclear localization signal (NLS) (9). Because the amino terminus of Rpl10p is predicted to be buried in the interface of Rpl10p and the 60S subunit (1), the NLS would be masked in the assembled subunit. We found that none of the myc-tagged proteins, including Rpl10N64p, accumulated in the nucleus after 3 h of expression (Fig. 3A), even though myc-tagged Rpl10N64p and Rpl10N187p were strongly dominant negative. To confirm the results reported with GFP-tagged Rpl10N64p (9), we examined the localization of GFP-tagged proteins. As previously reported, we found that GFP-tagged Rpl10N64p was predominantly nuclear (Fig. 3B). Additionally, both GFP-tagged Rpl10N187p and Rpl10C43p showed significant nuclear accumulation, while full-length Rpl10p was cytoplasmic. We have observed that the fusion of GFP to other proteins yields a nuclear bias in their distribution as well (see Discussion).
FIG. 3.
Localization of Rpl10p fragments. Cultures of strain W303 (wild type) transformed with either (A) pAJ792 (GAL10::RPL10-myc), pAJ793 (GAL10::RPL10N187-myc), pAJ794 (GAL10::RPL10C43-myc), or pAJ795 (GAL10::RPL10N64-myc) or (B) pAJ796 (GAL10::RPL10-GFP), pAJ797 (GAL10::RPL10N187-GFP), pAJ798 (GAL10::RPL10C43-GFP), or pAJ799 (GAL10::RPL10N64-GFP) were grown to saturation in SC-leu raffinose medium and diluted to an OD600 of ∼0.15 in fresh medium. After 4 h at 30°C, galactose was added and the cultures were grown for an additional 3 h. Cells were fixed and treated for microscopy as described in the text. Arrowheads in panel A point to nuclei.
Expression of RPL10N187 or repression of SQT1 blocks ribosome export and Nmd3p recycling.
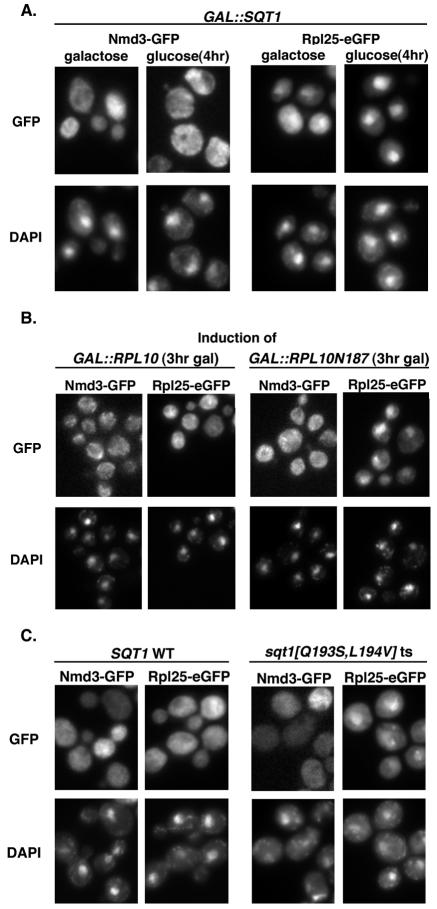
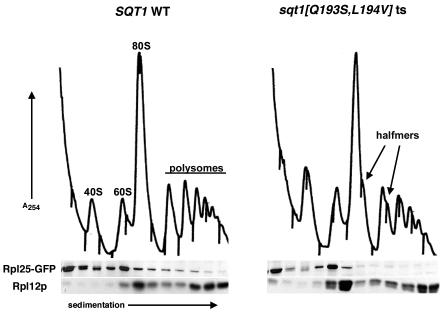
We have recently shown that release of the nuclear export adapter for the 60S subunit, Nmd3p, in the cytoplasm requires Rpl10p and the GTPase Lsg1p (12). A failure to recycle Nmd3p to the nucleus, in turn, leads to a block in 60S subunit export. If Sqt1p facilitates Rpl10p loading into the subunit, sqt1 mutants or truncated Rpl10 proteins that titrate out Sqt1p should show a similar effect on 60S export and Nmd3p recycling. When SQT1 was repressed or RPL10N187 was expressed, the 60S reporter Rpl25-eGFP accumulated in the nucleus (Fig. 4A and B, respectively). Nmd3p, on the other hand, remained cytoplasmic (Fig. 4A and B; also see below). Similar results were observed in temperature-sensitive sqt1 mutants (Fig. 4C), as well as in rpl10 mutants and cells repressed for RPL10 expression (12). The nuclear accumulation of Rpl25-eGFP suggests that the nascent subunits are blocked for export. To confirm that Rpl25-eGFP was incorporated into 60S subunits but that these subunits were not efficiently incorporated into the translationally active pool of ribosomes, we examined the sedimentation of Rpl25-GFP in an sqt1 ts mutant. Cells were grown at semipermissive temperature, followed by induction of Rpl25-GFP expression. Extracts were prepared and analyzed by sucrose gradient velocity sedimentation. Under these conditions, wild-type cells showed Rpl25-GFP present in 60S, 80S, and polysome fractions. In contrast, in the sqt1 ts mutant Rpl25-GFP was incorporated into free 60S, but its incorporation into polysomes was markedly reduced (Fig. 5).
FIG. 4.
Inhibition of SQT1 expression or function and overexpression of RPL10N187 lead to defects in ribosome export without Nmd3p nuclear entrapment. (A) Cultures of AJY1640 (GAL::SQT1) containing pAJ582 (NMD3-GFP) or pAJ907 (RPL25-eGFP) were grown to saturation in SC-leu medium containing galactose. Cultures were diluted to an OD600 of ∼0.15 and split into two aliquots. NMD3-GFP cultures were incubated for 4 h prior to addition of glucose to one set of aliquots, while glucose was immediately added to one set of RPL25-eGFP aliquots during recovery from lag phase. After 4 h, cells were fixed and DAPI stained according to the text. (B) W303 containing either pDEGQ2 (GAL1-10::RPL10) or pDEGQ187 (GAL1-10::RPL10N187), each with either pAJ582 (NMD3-GFP) or pAJ907 (RPL25-eGFP), were grown to saturation in SC-leu-ura medium containing raffinose. NMD3-GFP cultures were diluted to an OD600 of ∼0.15 and incubated for 4 h before the addition of galactose to induce the expression of wild-type and truncated RPL10 for 3 h. RPL25-eGFP cultures were also diluted to an OD600 of ∼0.15, but galactose was immediately added to induce wild-type and truncated RPL10 for 3 h during recovery from lag phase. Cells were treated and visualized as described for panel A. (C) Cultures of AJY1605 with either pAJ1063 (SQT1-myc) or pAJ1252 (sqt1[Q193S/L194V]-myc ts) replacing pAJ1062 and each with either pAJ908 (RPL25-eGFP) or pAJ755 (NMD3-GFP) were grown to saturation in SC-leu-ura medium. Cultures were diluted to an OD600 of ∼0.1 into fresh medium and incubated at 25°C for 5 h prior to shifting to 37°C for 2 h. Cells were fixed and DAPI stained as described in the text.
FIG. 5.
Rpl25-eGFP is incorporated into nascent subunits in an sqt1 ts mutant. Cultures of AJY1605 with either pAJ1063 (SQT1-myc) or pAJ1252 (sqt1[Q193S/L194V]-myc ts) replacing pAJ1062 and each with pAJ369 (GAL10::RPL25-GFP) were grown to saturation in SC-leu-ura raffinose. Cultures were diluted to an OD600 of ∼0.15 into fresh medium and incubated at 25°C. At an OD600 of ∼0.3, RPL25-GFP expression was induced with the addition of galactose. After 1 h, cycloheximide (150 μg/ml final concentration) was added to each culture, and cells were collected and analyzed by sucrose gradient fractionation as described in the text. Fractions were run on SDS-12% PAGE gels and transferred to nitrocellulose for Western blotting with α-GFP (Rpl25-GFP) or α-Rpl12p.
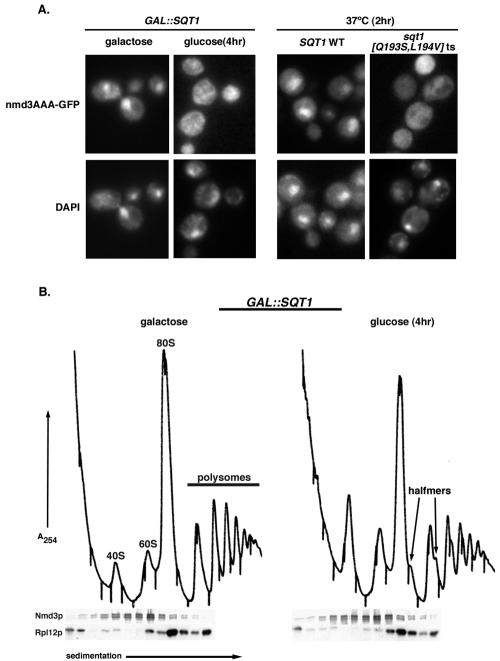
To determine if the cytoplasmic localization of Nmd3-GFP was caused by a failure of Nmd3-GFP to recycle to the nucleus in the absence of functional Sqt1p, we utilized an NMD3 mutant (NMD3AAA). This mutant contains three point mutations within its NES (I493A, L497A, and L500A) that significantly impair its nuclear export, resulting in a predominantly nuclear localization in wild-type cells (12). Nmd3AAA-GFP was redistributed to the cytoplasm when either SQT1 expression was repressed or an sqt1 ts mutant strain was cultured at nonpermissive temperature (Fig. 6A), similar to what was observed with rpl10 ts mutants or after repression of RPL10 (12). The Nmd3 protein in extracts prepared from SQT1-repressed cells cosedimented with free 60S subunits in sucrose gradients, indicating that it remained associated with 60S subunits and that cytoplasmic retention was due to a defect in Nmd3p release from subunits rather than an import defect for free Nmd3p (Fig. 6B).
FIG. 6.
Nmd3p fails to shuttle in the absence of functional SQT1. AJY1640 (GAL10::SQT1) or AJY1605 with either pAJ1063 (SQT1-myc) or pAJ1252 (sqt1[Q193S/L194V]-myc ts) as the sole copies of SQT1 were transformed with pAJ754 (NMD3AAA-GFP) and were grown to saturation in either galactose- or glucose-containing dropout medium, respectively. Cultures of cells expressing Nmd3AAA-GFP in either AJY1640 or AJY1605 were handled as described for Nmd3-GFP in legends to Fig. 4A or C, respectively. (B) Saturated culture of AJY1604 was diluted to an OD600 of ∼0.1 into two aliquots of fresh medium and incubated for 5 h prior to the addition of glucose and incubation for an additional 4 h. Cycloheximide (150 μg/ml final concentration) was added to each culture, and cells were collected. Extracts were analyzed by sucrose gradient fractionation as described in the text. Fractions were run on SDS-12% PAGE gels and transferred to nitrocellulose for Western blotting with α-Nmd3p or α-Rpl12p.
Nmd3p can bind to 60S subunits independently of Rpl10p.
It has been proposed previously that Rpl10p recruits Nmd3p to the nascent 60S subunit in the nucleus for nuclear export (9). However, we have shown that Nmd3p remains associated with 60S subunits in the cytoplasm and cannot shuttle to the nucleus when RPL10 transcription is repressed or in rpl10 mutants (12). This block in Nmd3p recycling to the nucleus accounts for the nuclear accumulation of nascent 60S subunits, as the export defect can be bypassed by high-copy expression of NMD3 (12). Thus, Nmd3p can direct export of subunits independently of Rpl10p. To test more directly if Nmd3p can bind subunits lacking Rpl10p, we asked if subunits immunoprecipitated with Nmd3p were substoichiometric for Rpl10p. To this end, we inserted three tandem copies of the HA epitope into the 3′ end of genomic RPL10. Extracts prepared from these cells coexpressing c-myc-tagged Nmd3p were fractionated by sucrose gradient sedimentation, and Nmd3p was immunoprecipitated from the free 60S fraction. We then compared the amount of Rpl10-HA to a second 60S subunit protein, Rpl12p, in the immunoprecipitated sample, in the total pool of free 60S subunits, in intact 80S subunits, and in a polysomal fraction. The subunits coimmunoprecipitated with Nmd3p contained a lower relative amount of Rpl10-HA than the total pool of free 60S subunits and were strikingly deficient for Rpl10-HA when compared to joined subunits in the 80S peak or translating subunits in polysomes (Fig. 7). Thus, Nmd3p is capable of binding to 60S subunits in the absence of stoichiometric levels of Rpl10p. Taken together, these results are more consistent with a model in which Rpl10p loads after Nmd3p in the biogenesis pathway prior to subunit incorporation into the translational pool. The substoichiometric level of Rpl10p in the free 60S species is surprising and could be due to the contribution of nuclear or cytoplasmic subunits not yet loaded with Rpl10p.
A dominant mutation in the Walker A motif of the GTPase Lsg1p traps Sqt1p on 60S subunits.
Nmd3p can bind subunits independently of Rpl10p (see above), and high-copy Nmd3p can bypass the export defect of RPL10-repressed cells (12), indicating that Rpl10p is not directly required for 60S export. Nevertheless, Nmd3p requires Rpl10p for its release from subunits in the cytoplasm. Release of Nmd3p from cytoplasmic 60S subunits also requires the cytoplasmic GTPase Lsg1p (12). Because Rpl10p appears to be required after Nmd3p, and rpl10 and lsg1 mutants have similar effects on Nmd3p release, we considered the possibility that Lsg1p may be involved in loading Rpl10p into the subunit. We reasoned that a dominant negative LSG1 mutant that blocks the release of Nmd3p (12) might also trap Sqt1p on the subunit during Rpl10p loading. To test this prediction, we used the dominant LSG1(K349T) allele (12), which contains a mutation in the lysine of the Walker A motif that is essential for GTP binding. Extracts were prepared from cells expressing wild-type LSG1 or LSG1(K349T). The Lsg1 proteins were immunoprecipitated, and the copurification of 60S subunits and Sqt1p was monitored by Western blotting. Figure 8A shows that, whereas only a low level of Sqt1p copurified with wild-type Lsg1p, significantly higher levels were associated with Lsg1(K349T)p. The enrichment of Sqt1p on 60S subunits by mutant Lsg1p suggests that Sqt1p loads Rpl10p in the presence of Lsg1p and that release of Sqt1p requires the GTPase function of Lsg1p.
To determine whether Sqt1p was enriched by mutant Lsg1p in the presence of Nmd3p, we repeated this experiment in cells expressing epitope-tagged Nmd3p. Indeed, when the immunoprecipitation was carried out against Nmd3p, the presence of Lsg1(K349T)p in cells led to increased levels of Sqt1p that copurified with Nmd3p (Fig. 8B). These results suggest that Sqt1p loads Rpl10p onto the Nmd3p-bound subunit in the presence of Lsg1p.
A mutant Rpl10 protein that is not stably incorporated into 60S subunits can be trapped on the subunit by dominant LSG1 mutants.
As we showed above, the C-terminally truncated fragment of Rpl10p, Rpl10N187p, does not stably bind to the 60S subunit, and its expression is deleterious to cells because of its affinity for Sqt1p. The absence of the Rpl10N187p fragment from subunits could be due to a complete failure to interact with the subunit, or it could be due to a failure in releasing from Sqt1p, leading to aborted loading into the subunit. Remarkably, Rpl10N187p was also efficiently coimmunoprecipitated with Lsg1(K349T)p (Fig. 8C, lane 3). Rpl10N187p was not found at appreciable levels in complexes immunoprecipitated with wild-type Lsg1p (Fig. 8C, lane 2) or in free or translating 60S subunits (Fig. 1A and data not shown). The presence of Rpl10N187p in the mutant Lsg1p complex shows that this Rpl10p fragment is capable of interacting with the subunit, presumably while complexed with Sqt1p. Its absence from subunits in wild-type cells suggests that it either fails to be accommodated into its binding site and is released from the subunit still bound to Sqt1p or that subunits containing the Rpl10p fragment are unstable and rapidly degraded. However, the accumulation of Rpl10N187p complexed with Sqt1p (Fig. 1A) supports the former possibility, that the mutant protein is not accommodated into the subunit and is released while still bound to Sqt1p. We believe that the entrapment of Rpl10N187p and Sqt1p by dominant negative Lsg1p is akin to stabilization of a transient assembly intermediate and provides physical evidence that Lsg1p is directly involved in loading Rpl10p into the 60S subunit in the cytoplasm.
DISCUSSION
Rpl10p is an integral component of the large ribosomal subunit, positioned between the central protuberance and the GTPase stalk, and makes contacts with 25S and 5S rRNAs (30). Eukaryotic Rpl10 proteins contain a C-terminal extension of approximately 50 amino acids. Removal of this C terminus from yeast Rpl10p yields a protein that is comparable in size and predicted structure to its prokaryotic counterparts and can be modeled into the 15A cryo-electron microscopy map of the yeast ribosome (30) using the crystal structure from Haloarcula marismortui (1, 22). Consequently, it seemed reasonable that truncated Rpl10p, lacking only the C-terminal 43 amino acids, would be incorporated into subunits. This truncation and smaller N-terminal fragments of Rpl10p inhibit cell growth when expressed in yeast (5), which could be explained if their incorporation into subunits in the nucleus blocked subsequent assembly or export events. We found, however, that these Rpl10p fragments did not assemble into 60S subunits, indicating that their dominant negative effect comes from interactions separate from the subunit. These truncated Rpl10 proteins were dominant negative regardless of whether they localized to the nucleus or cytoplasm, consistent with the idea that they act independently of subunit assembly in the nucleus.
Sqt1p-Rpl10p interaction.
In addition to its interactions within the 60S subunit, Rpl10p physically interacts with the WD-repeat protein Sqt1p that has been suggested to act as a chaperone for loading Rpl10p into subunits (5). In our attempts to test whether or not Rpl10p fragments were incorporated into 60S subunits, we found that these fragments bind, perhaps irreversibly, to Sqt1p. Our results show that the dominant negative effect of rpl10 truncations is due to titration of Sqt1p and not a direct effect on subunit assembly. Sqt1p is predicted to fold into a disk-shaped structure, typical of WD-repeat proteins, with a highly acidic amino-terminal extension (amino acids 1 to 50 have a calculated pI of 3.9). This amino terminus could bind electrostatically to the highly basic amino-terminal 40 amino acids of Rpl10p. Indeed, the deletion of 25 amino acids from the N terminus of Rpl10p prevented its binding to Sqt1p and, at the same time, relieved the dominant negative effect of a C-terminal truncation. Interestingly, this highly basic amino terminus of Rpl10p was also reported to contain an NLS. Although we have shown that this domain does not localize to the nucleus when tagged with the c-myc epitope, it is possible that Sqt1p binding helps mask the NLS activity of this peptide, thereby preventing Rpl10p nuclear entry. Because of the instability of Rpl10p in the absence of Sqt1p, we have not been able to test this idea.
SQT1 was originally identified as a high-copy suppressor of the dominant negative Rpl10p fragments (5). Based on the observation that free 60S subunits were depleted of Rpl10p when SQT1 was repressed, it was proposed that Sqt1p acts as a chaperone for Rpl10p (5). To determine whether Sqt1p stabilizes free Rpl10p, we tried to measure the half-life of Rpl10p in sqt1 temperature-sensitive mutants. However, we were not able to detect expression of Rpl10p in these cells at restrictive temperature (J. B. Hedges and A. W. Johnson, unpublished), suggesting that nascent Rpl10p is highly unstable in the absence of functional Sqt1p. This role for Sqt1p as a chaperone for free Rpl10p may be similar to the role of another WD-repeat protein, Rrb1p, in Rpl3p assembly into the large subunit (16). Additional WD-repeat proteins are required for ribosome synthesis as well (10), but their specific functions have not yet been identified.
Nmd3p binds to 60S subunits prior to Rpl10p.
Rpl10p is one of the last ribosomal proteins to be loaded into the 60S subunit (26). It has been thought that Rpl10p binds to the pre-60S subunit in the nucleus to recruit Nmd3p (9). However, our results are more consistent with a model in which Nmd3p loads in the nucleus and Rpl10p loads subsequently in the cytoplasm (Fig. 9). The idea that Rpl10p recruited Nmd3p to pre-60S subunits in the nucleus was based on several lines of evidence. Some rpl10 mutants are suppressed by high-copy NMD3 (37) or by dominant mutations in NMD3 (23), suggestive of physical interaction between Nmd3p and Rpl10p. Indeed, Rpl10p and Nmd3p were reported to copurify when coexpressed in E. coli (9). GFP fused to the N-terminal 64 amino acids of Rpl10p localizes to the nucleus, suggesting an NLS activity in this region of Rpl10p (9). Furthermore, Rpl10-GFP accumulates in the nucleus in dis3 mutant cells defective in 3′ RNA processing by the exosome (9), and RPL10 shows genetic interaction with RSA1, encoding a nonessential nuclear factor important for normal Rpl10p levels on free 60S subunits (25). Rpl10p has also been identified in mass spectrometric analysis of affinity-purified, nuclear pre-60S particles (29).
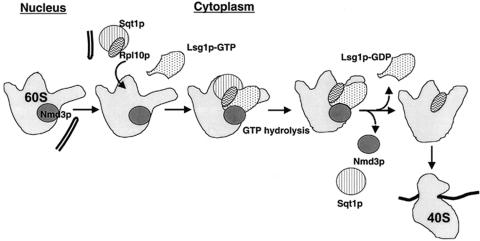
FIG. 9.
Rpl10p loads into 60S subunits after they are exported into the cytoplasm by Nmd3p. We propose here that the 60S export adapter, Nmd3p, binds to nascent subunits in the nucleus in the absence of Rpl10p. Following subunit export, Rpl10p binds to 60S in the cytoplasm, assisted by its chaperone, Sqt1p. The cytoplasmic GTPase Lsg1p (presumably in its GTP-bound form) also binds to subunits at this time, although the precise order of binding is currently unknown. GTP hydrolysis on Lsg1p may drive a conformational change in 60S that culminates in the accommodation of Rpl10p into the subunit and the release of Nmd3p and Rpl10p's chaperone, Sqt1p. The incorporation of Rpl10p into 60S subunits in this manner, coupled with the release of Nmd3p, may be a requisite step for activating subunits for translation initiation.
Are there plausible alternative explanations for these observations? We have recently shown that rpl10 mutants block release of Nmd3p from subunits in the cytoplasm, thereby preventing the recycling of Nmd3p to the nucleus (12). High-copy NMD3 bypasses this defect, and the dominant Nmd3 proteins that suppress Rpl10p mutants have reduced affinity for 60S subunits in vitro, allowing them to recycle to the nucleus more efficiently. Thus, the genetic interaction between these two proteins is due to functional interaction in the cytoplasm and not recruitment in the nucleus. Physical interaction between Nmd3p and Rpl10p does not distinguish between interactions in the nucleus and those in the cytoplasm. The putative NLS in Rpl10p (within amino acids 1 to 64) did not localize to the nucleus when fused to a c-myc epitope, even though this protein retained its dominant negative effect of sequestering Sqt1p. Although Rpl10p has been identified in mass spectrometric analysis of nuclear pre-60S particles, its identification in relatively early particles (for example, Nsa3p-containing particles [29]) seems contrary to Rpl10p being one of the last proteins loaded. Possibly, the Rpl10p identified in these particles results from cytoplasmic contamination during cell lysis. We cannot explain why Rpl10-GFP accumulates in dis3 cells. However, the low Rpl10 levels in rsa1 mutants could easily be explained if 60S export was slow in rsa1 mutants, resulting in nuclear accumulation of pre-60S subunits lacking Rpl10p.
Additional results support the idea that Rpl10p loads in the cytoplasm. Rpl10p does not accumulate in the nucleus of yeast cells treated with leptomycin B (LMB) (unpublished data), an inhibitor of Crm1p, the export receptor for 60S subunits (9, 15). This is in contrast to nascent 60S subunits and Nmd3p, which are readily trapped under these conditions (9, 15). Rpl10p also does not accumulate in cells expressing Nmd3Δ100 (unpublished data), a truncated form of Nmd3p lacking its NES that inhibits 60S export (15). Similarly, functional Sqt1-myc does not accumulate in nuclei of cells treated with LMB or expressing Nmd3Δ100p. Furthermore, the block in 60S export that arises when RPL10 transcription is repressed can be alleviated by increasing the levels of Nmd3p (12), indicating that Rpl10p is not directly required for subunit export. Moreover, human Rpl10p (QM) is reported to load in the cytoplasm (28).
GFP fusions show a nuclear bias in yeast.
We found that fusing GFP to truncated Rpl10 proteins resulted in their nuclear accumulation, whereas the same truncations tagged with an oligomeric c-myc epitope were cytoplasmic. To understand this disparity in localization, we compared the localization of native Nmd3p with Nmd3p that was tagged with GFP or with myc. Here again, we found that the GFP-tagged protein in live and fixed cells showed greater nuclear accumulation than both myc-tagged and untagged proteins (data not shown). A similar nuclear bias has been observed for signal recognition particle proteins in yeast when tagged with small epitopes versus GFP (3, 11). This difference in localization is probably not due to inhibition of import by the myc epitope, because myc-tagged mutant Nmd3 proteins readily accumulate in the nucleus (15). We have also observed that free GFP accumulates in the nucleus of LMB-sensitive yeast when treated with LMB (G. Kallstrom and A. W. Johnson, unpublished), suggesting that GFP itself may contribute to the nuclear accumulation of some fusion proteins. These results suggest that GFP can influence the distribution of proteins in yeast, particularly when assayed after treatment with LMB. Therefore, a degree of caution is merited when interpreting such localization data.
Rpl10p is loaded onto the subunit by Sqt1 in the presence of Nmd3p and the cytoplasmic GTPase Lsg1p.
We have recently shown that both Rpl10p and the cytoplasmic GTPase Lsg1p are required for release of Nmd3p from subunits in the cytoplasm, allowing for its recycling to the nucleus for subsequent rounds of 60S subunit export. We proposed that the correct assembly of Rpl10p into the subunit is required for Lsg1p to stimulate release of Nmd3p through a conformational change in the subunit. We further suggested that such a conformational change could at the same time be required for stabilization of Rpl10p in the subunit. Here we show that dominant mutations in LSG1 trap Sqt1p on the subunit. Because Sqt1p forms a complex with free Rpl10p that appears to be necessary for its stability prior to its assembly into the subunit, its entrapment by mutant Lsg1p is strong evidence that Rpl10p loading by Sqt1p takes place in the presence of the cytoplasmic GTPase Lsg1p.
Rpl10p lacking its C-terminal 43 amino acids does not assemble stably into the 60S subunit and remains bound to Sqt1p. Thus, the C terminus of Rpl10p may be necessary for its release from Sqt1p as it assembles into the subunit or it may be needed for Rpl10p to bind to the subunit either by adopting a conformation appropriate for 60S binding or by initiating the loading onto the subunit. In the cryo-electron microscopy map of yeast ribosomes, only amino acids 4 to 168 could be threaded onto the H. marismortui structure due to the fact that the C terminus of eukaryotic Rpl10 proteins (53 amino acids in yeast Rpl10p) is not conserved in archaea. However, an unassigned mass was observed in the expected position for the C terminus of Rpl10p and adjacent to helix 38 of 25S rRNA (30). This mass may in part represent the C terminus of yeast Rpl10p and would indicate that it makes direct contact with the 60S subunit. Although deletion of the C terminus of Rpl10p prevented its stable association with the subunit, this truncated Rpl10p, as well as Sqt1p, could be trapped on the subunit by a dominant mutant in the GTP binding loop of Lsg1p. Thus, the C-terminal extension is not required for initial binding but rather for release of Rpl10p from Sqt1p as it loads onto the subunit. During normal assembly of the large subunit, the release of Sqt1p and Nmd3p may be driven by an Lsg1p-dependent conformational change in the 60S subunit that accommodates the correct assembly of Rpl10p into its binding site (Fig. 9) (12). Considering the relatively small amounts of Rpl10p in Nmd3p-bound and free subunits, loading of Rpl10p may, in fact, require the release of Nmd3p.
Rpl10p exchange.
Rpl10p was identified many years ago as one of three exchangeable proteins on the 60S subunit (36). We have provided evidence previously that Lsg1p as well as Nmd3p bind to recycling mature subunits in addition to nascent subunits (14, 19). Thus, loading of Rpl10p by Sqt1p coupled with Lsg1p-dependent release of Nmd3p could provide a means of assessing the levels of free 60S subunits available for subunit joining and finely regulate the availability of free Nmd3p to recycle to the nucleus for 60S export. We are currently testing whether Lsg1p is required for Rpl10p exchange.
Acknowledgments
We thank Juan Ballesta for α-L12 antibody and Jonathan Warner for strain W303. We are also grateful to Bernard Trumpower for providing strain DEH221+, as well as plasmids pDEGQ2, pDEGQ64, and pDEGQ187 and α-Sqt1p antibody.
This work was supported by NIH grant GM53655 to A.W.J.
REFERENCES
- 1.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 3.Ciufo, L. F., and J. D. Brown. 2000. Nuclear export of yeast signal recognition particle lacking Srp54p by the Xpo1p/Crm1p NES-dependent pathway. Curr. Biol. 10:1256-1264. [DOI] [PubMed] [Google Scholar]
- 4.Dick, F. A., D. P. Eisinger, and B. L. Trumpower. 1997. Exchangeability of Qsr1p, a large ribosomal subunit protein required for subunit joining, suggests a novel translational regulatory mechanism. FEBS Lett. 419:1-3. [DOI] [PubMed] [Google Scholar]
- 5.Eisinger, D. P., F. A. Dick, E. Denke, and B. L. Trumpower. 1997. SQT1, which encodes an essential WD domain protein of Saccharomyces cerevisiae, suppresses dominant-negative mutations of the ribosomal protein gene QSR1. Mol. Cell. Biol. 17:5146-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisinger, D. P., F. A. Dick, and B. L. Trumpower. 1997. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol. Cell. Biol. 17:5136-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 8.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 9.Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 11.Grosshans, H., K. Deinert, E. Hurt, and G. Simos. 2001. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J. Cell Biol. 153:745-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedges, J., M. West, and A. W. Johnson. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho, J. H., and A. W. Johnson. 1999. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2389-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho, J. H., G. Kallstrom, and A. W. Johnson. 2000. Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA 6:1625-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho, J. H., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iouk, T. L., J. D. Aitchison, S. Maguire, and R. W. Wozniak. 2001. Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome biosynthesis. Mol. Cell. Biol. 21:1260-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, A. W., E. Lund, and J. Dahlberg. 2002. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27:580-585. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Kallstrom, G., J. Hedges, and A. W. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karl, T., K. Onder, R. Kodzius, A. Pichova, H. Wimmer, A. Thür, H. Hundsberger, M. Loffler, T. Klade, A. Beyer, M. Breitenbach, and L. Koller. 1999. GRC5 and NMD3 function in translational control of gene expression and interact genetically. Curr. Genet. 34:419-429. [DOI] [PubMed] [Google Scholar]
- 21.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 22.Klein, D. J., P. B. Moore, and T. A. Steitz. 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340:141-177. [DOI] [PubMed] [Google Scholar]
- 23.Koller, H. T., T. Klade, A. Ellinger, and M. Breitenbach. 1996. The yeast growth control gene GRC5 is highly homologous to the mammalian putative tumor suppressor gene QM. Yeast 12:53-65. [DOI] [PubMed] [Google Scholar]
- 24.Kranz, J. E., and C. Holm. 1990. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc. Natl. Acad. Sci. USA 87:6629-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kressler, D., P. Linder, and J. de la Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruiswijk, T., R. J. Planta, and J. M. Krop. 1978. The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta 517:378-389. [DOI] [PubMed] [Google Scholar]
- 27.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen, Y. H., A. A. Mills, and E. J. Stanbridge. 1998. Assembly of the QM protein onto the 60S ribosomal subunit occurs in the cytoplasm. J. Cell. Biochem. 68:281-285. [PubMed] [Google Scholar]
- 29.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spahn, C. M., R. Beckmann, N. Eswar, P. A. Penczek, A. Sali, G. Blobel, and J. Frank. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell 107:373-386. [DOI] [PubMed] [Google Scholar]
- 31.Stage-Zimmermann, T., U. Schmidt, and P. A. Silver. 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11:3777-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas, F., and U. Kutay. 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 116:2409-2419. [DOI] [PubMed] [Google Scholar]
- 33.Trotta, C. R., E. Lund, L. Kahan, A. W. Johnson, and J. E. Dahlberg. 2003. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 22:2841-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 35.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 36.Zinker, S., and J. R. Warner. 1976. The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins. J. Biol. Chem. 251:1799-1807. [PubMed] [Google Scholar]
- 37.Zuk, D., J. P. Belk, and A. Jacobson. 1999. Temperature-sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover. Genetics 153:35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]