Abstract
The dynamic interplay between the immunome and microbiome in reproductive health is a complex and rapidly advancing research field, holding tremendously vast possibilities for the development of reproductive medicine. This immunome–microbiome relationship influences the innate and adaptive immune responses, thereby affecting the onset and progression of reproductive disorders. However, the mechanisms governing these interactions remain elusive and require innovative approaches to gather more understanding. This comprehensive review examines the current knowledge on reproductive microbiomes across various parts of female reproductive tract, with special consideration of bidirectional interactions between microbiomes and the immune system. Additionally, it explores innate and adaptive immunity, focusing on immunoglobulin (Ig) A and IgM antibodies, their regulation, self-antigen tolerance mechanisms, and their roles in immune homeostasis. This review also highlights ongoing technological innovations in microbiota research, emphasizing the need for standardized detection and analysis methods. For instance, we evaluate the clinical utility of innovative technologies such as Phage ImmunoPrecipitation Sequencing (PhIP-Seq) and Microbial Flow Cytometry coupled to Next-Generation Sequencing (mFLOW-Seq). Despite ongoing advancements, we emphasize the need for further exploration in this field, as a deeper understanding of immunome–microbiome interactions holds promise for innovative diagnostic and therapeutic strategies for reproductive health, like infertility treatment and management of pregnancy.
Keywords: immunome, microbiome, reproductive health, phage immunoprecipitation sequencing, microbial flow cytometry, next-generation sequencing, diagnostic tools, therapeutic interventions, interactions
Overview of the Female Reproductive Tract Microbiota
The human body is known to host trillions of microorganisms, forming our microbiome, a complex ecosystem that significantly affects various aspects of health and disease. 1 2 This so-called microbiome-dominated world is often referred to as a “second genome.” 3 Gut microbiome has been extensively studied, along with the findings emphasizing the role of the female reproductive tract (FRT) microbiota in reproductive health. 4 5 6 The FRT microbiota comprises bacteria, fungi, viruses, archaea, and protozoa and is collectively referred to as the reproductive tract (RT) microbiota. FRT accounts for ∼9% of the total bacterial burden in the body. 7 Microorganisms produce biologically active substances interacting with endocrine, immune, nervous, metabolic, and reproductive systems, contributing to healthy development by maintaining homeostasis. Changes in the microbiota composition, known as dysbiosis, have been suggested to affect metabolism, immunity, gene expression, epigenetics, and disease risk by altering metabolic enzymes, hormones, and disease risks. 8
The term FRT refers to both the upper reproductive tract (URT), which includes the endocervix, endometrium, uterus, fallopian tubes, ovaries, peritoneal fluid, and placenta, and the lower reproductive tract (LRT), consisting of the ectocervix, vagina, and vulva (see Fig. 1 ). Previous studies have revised the notion of sterility in URT, revealing the presence of microorganisms in locations such as the ovarian follicles, fallopian tubes, uterus, and placenta. 9 10 11 12 13 14 15 16 Each segment of the FRT contains its own microbiome composition, which can vary based on factors such as age, presence of pregnancy, physiological state, lifestyle, and environmental elements. Microorganisms play an essential role in reproductive well-being by protecting against infections, encouraging fertility, and supporting an uncomplicated pregnancy. However, changes in FRT microbiome are suggested to be associated with reproductive disorders, including infertility, endometriosis, recurrent pregnancy loss, and preterm birth. 9 17 18 19 20 Therefore, understanding the interactions between microbiome and reproductive health is crucial for designing personalized treatments. 8 17
Fig. 1.
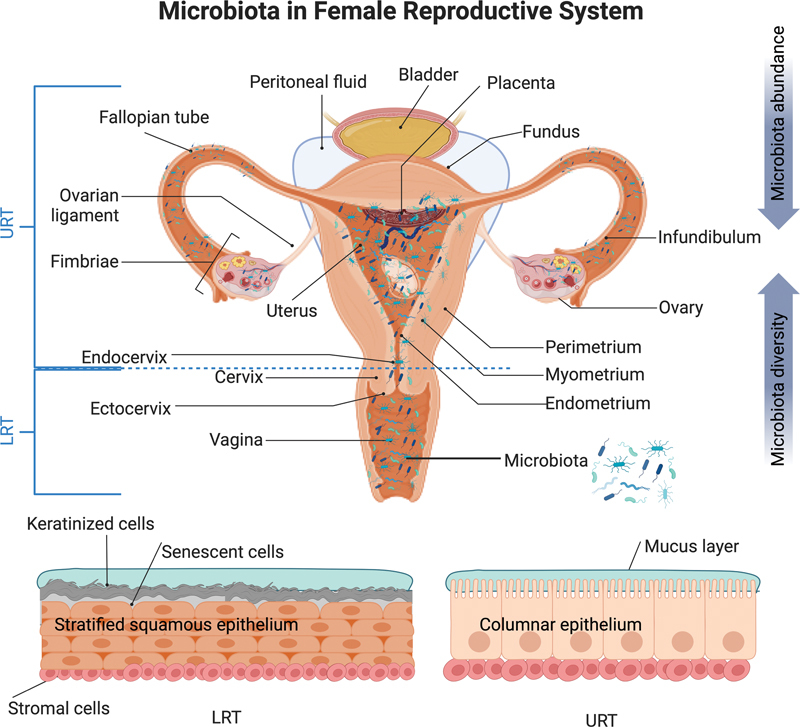
Distribution of microbiota in the female reproductive tract (FRT). This figure illustrates the microbial composition in the FRT, which is divided into upper and lower reproductive tracts (URT and LRT). The LRT includes the vagina and ectocervix, both lined with a stratified squamous epithelium. This epithelium originates from the basement membrane and culminates in fully keratinized senescent cells. The URTs comprise the endocervix, uterus with endometrium, fallopian tubes, ovaries, peritoneal fluid, and placenta and features a monolayer columnar epithelium. In particular, the transition from stratified squamous epithelium to single columnar epithelium occurs in the “transformation zone” between the ectocervix and endocervix. Microbial abundance was generally higher in the LRT than in the URT, whereas microbial diversity increases from the outermost to the innermost regions. This figure has been adapted from BioRender.com (2023). Source: https://app.Biorender.com .
The Role of the Microbiota in Reproductive Health
The microbiome plays an integral role in shaping reproductive health by intricately balancing microbial communities within the female reproductive organs. Notably, the vaginal microbiome, characterized by diverse bacteria, fungi, and viruses, is paramount for vaginal health and infection prevention. 11 21 Previous research has identified five “community state types (CSTs)” within the vaginal microbiome, primarily characterized by the prevalence of Lactobacillus -dominant species. More specifically, four out of five CSTs demonstrated dominance by distinct lactobacilli, while CST-IV was characterized by the prevalence of bacteria associated with bacterial vaginosis (BV). 22 23 Factors like hormonal fluctuations, sexual activity, and contraceptive use exert an influence on the vaginal microbiome. 24 25 26 Lactic acid production by Lactobacillus sp. plays a central role in neutralizing pathogens, including Chlamydia trachomatis , Escherichia coli , and herpes simplex virus 1 (HIV-1). 27 28 29 Dysbiosis leads to BV, linked to adverse outcomes like sexually transmitted infections (STIs), infertility, and pregnancy complications. 9 30 31 32 33 34 35 Variations in the vaginal microbiome are evident in conditions such as polycystic ovary syndrome (PCOS) and during pregnancy, predisposing to adverse pregnancy outcomes. 28 36 37 38
The cervix connects uterus and vagina, and its microbiota is essential for female reproduction. Despite being sparsely studied, the cervical microbiome includes Lactobacillus spp., Gardnerella vaginalis , and various bacteria. 39 40 41 42 43 However, it is important to note that sampling methods can impact the results, as they may introduce potential contamination from the vaginal microbiome. Pervious research established connections between cervical microbiota, cervical health, and outcomes such as preterm birth, cervical ripening, and labor. 44 45 Certain cervical bacteria can strengthen the immune response against human papillomavirus, potentially offering effective protection against infection. 46 Microbiota of endometrium is known to have lower biomass and greater diversity than vaginal and cervical tissues as a result of factors such as blood flow and increased pH. 28 47 48 49 Lactobacillus sp., prevalent in the vagina, exhibit variations in abundance in the endometrium. 47 50 51 52 Studies have suggested that Lactobacillus -dominated endometrial microbiota may improve reproductive outcomes; however, conflicting reports have been published. 49 53 54 55 56 57 58 Imbalances in endometrial microbiota are linked to various reproductive issues, including repeated implantation failure (RIF), pregnancy loss, endometriosis, endometritis, endometrial polyps, hyperplasia, and endometrial cancer (EC). 49 59 60 61 62 63 For instance, Fusobacterium infection of the endometrium causes inflammation, leading to conditions such as endometriosis. 19
In contrast to earlier assumptions that fallopian tubes (FT) are sterile, recent research has demonstrated the presence of host specific microbiome, including Lactobacillus, Staphylococcus, and Enterococcus sp. 9 64 Additionally, variations in bacterial composition may exist between the right and left FTs. Staphylococcus spp. was more abundant on the right, while Lactobacillus spp., Enterococcus spp., and Prevotella spp. were more prevalent on the left. 9 64 65 Reproductive-aged women exhibit overlapping bacterial profiles between FTs and endometrium, emphasizing shared microbial fingerprints. 64 66 Concerns have been expressed regarding G. vaginalis biofilms migrating from the vagina to fallopian tubes and disrupting pregnancy. 67 However, lactobacilli produce biosurfactants that counteract pathogenic biofilm formation. 68 Further investigations should explore the impact of FT microbiota on reproductive health.
Ovaries, once thought to be completely sterile, harbor distinct microbiome associated with various medical conditions. 15 69 Proteobacteria , Firmicutes , and Bacteroidetes predominate in the ovarian environment, with specific bacteria associated with ovarian cancer. 70 71 The source of the ovarian microbiota remains debated, with possible migration through the vaginal passageway or bloodstream. 72 73 Unfortunately, the ovarian microbiota has not been studied as extensively as other reproductive organs, and the focus of these studies has been given more on certain specific health issues and diseases rather than those of healthy and fertile state, emphasizing the need for further exploration and understanding. The placenta has traditionally been considered sterile. However, certain studies have revealed its richness in microbial composition. Nevertheless, its exploration poses challenges due to the low biomass involved. 74 75 76 77 Contrarily, some recent studies have revealed that healthy placenta has no microbiome. Almost all bacterial signals were related either to acquisition of bacteria during labor and delivery or contamination of laboratory reagents with bacterial DNA. 75 78 79 80 81 82 However, considering the conflicting results of previous studies, new and better-planned research is still necessary, as the placental microbiota provides a new frontier that may affect the health outcomes of pregnancy and newborns. Understanding the intricate interactions between reproductive organs and their microbiota is crucial for advancing women's and children's health.
Importance of the Immune System in Female Reproductive Health
The immune system is an intricate network composed of innate and adaptive components that work together to respond effectively and overcome various health threats. Immune cells provide defense mechanisms against potential pathogens, while tolerating self-antigens to avoid harmful immune reactions. 83 Simultaneously, the microbiota of FRT interacts with immune cells in an unpredictable environment to modulate immune responses and maintain balance. 84 However, our knowledge of the interaction between microbiota and immune cells in RT remains incomplete, necessitating further investigation. The acquisition of diverse microbiota coincides with the development of the immune system, suggesting a co-evolutionary relationship and creating a symbiotic partnership. 85 A notable component of this symbiotic relationship is the role of maternal secretory (s) immunoglobulin (Ig) A (sIgA) in reproductive health. sIgA plays an essential role in restricting immune activation and inhibiting microbial attachment by binding to nutritional and microbial antigens, contributing to healthier host–microbe interactions in infant intestines. 86 FRT maintains its balance through a complex network comprising epithelial defenses, natural killer cells (NKC), macrophages, dendritic cells (DCs), and T lymphocytes. Epithelial defenses include physical barriers, commensal bacteria with IgA antibodies, and antimicrobial peptides. 87 88 These commensal bacteria provide complex protection, known as colonization resistance.
It is suggested that even subtle disruptions in the delicate relationship between the microbiota and immune system can dramatically affect reproductive health, potentially leading to infertility, miscarriage, or premature birth. 17 88 89 Such imbalances may result in dysbiosis, inflammation, and various reproductive disorders. 17 90 Recent advances in our understanding of immune system dynamics have shed light on their intricate relationship with microbiomes, prompting further research on reproductive health. For example, Fusobacterium infections cause an innate immune response, activate transforming growth factor-β (TGF-β) signaling pathways, and promote the transformation of endometrial fibroblasts to myofibroblasts, ultimately contributing to endometriotic lesions. 19 This study established, for the first time, a solid link between the microbiome, immune dysfunction, and endometriosis development. 19 Similarly, women whose vaginal microbiome predominantly contained Lactobacillus crispatus showed higher IgA levels, suggesting increased immune protection. 32 91 Moreover, contradictory immune regulation and inflammation have been associated with reproductive/pregnancy complications, such as preeclampsia, fetal growth restriction (FGR), gestational diabetes, and maternal weight gain. 92 93 94 Therefore, understanding the intricate interactions between immune function and the microbiome is critical in unraveling the biological mechanisms that can enhance fertility and contribute to more effective disease treatments.
Rationale for Studying the Microbiota–Immunome Interaction
This comprehensive review examines the current knowledge of microbiome–immunome interactions in reproductive health using available technologies. We will evaluate the existing literature to understand better how microbiota affects and regulates the immune system in the RT. Understanding this interaction of microbiota–immunome interactions in the context of reproductive health is crucial to unravel the mechanisms underlying disease etiology, identify biomarkers, develop targeted therapies, and implement preventive strategies. Recent advances in multi-omics technologies, such as metagenomics, 16S rRNA sequencing, Phage ImmunoPrecipitation Sequencing (PhIP-Seq), and Microbial Flow Cytometry coupled to Next-Generation Sequencing (mFLOW-Seq), have allowed for a more comprehensive analysis of microbiota–immunome interactions. This provides valuable insight into personalized interventions and improved reproductive health outcomes, while creating opportunities to protect or restore reproductive health outcomes. Integrating knowledge of microbiota–immunome interactions with advanced technologies has great potential to improve our understanding of reproductive outcomes and enable personalized interventions.
Immunome–Microbiota Interplay in Female Reproductive Health
The FRT engages in intricate interactions with the external environment, involving resident immune cells and microbiota. 95 96 97 98 Pattern recognition receptors (PRRs), including Dectin-1, toll-like receptors (TLRs), and nucleotide-binding oligomerization domain (NOD)-like receptors, detect commensal bacteria in both URTs and LRTs environments. 96 99 100 101 102 After exposure to microbes, these receptors activate signaling pathways that result in secretion of proinflammatory cytokines, such as interleukins (IL)-1β, IL-6, IL-8, and tumor necrosis factor-α (TNF-α). This further triggers the activation and mobilization of various immune cell types, including NK cells, macrophages, CD4+ T cells, CD8+ cytotoxic T cells, and B lymphocytes (see Fig. 2 ). 102 103 104 105 106 107 Genetic variants in PRR genes, including TLR4, TLR9, IL-1R2, and TNF-α, can influence women's responses to microbiological challenges and pregnancy outcome. 104 PRRs are present in both the squamous epithelial cells lining the vagina and the columnar cells in the upper FRT. 95 96 (see Fig. 3 ). Commensal FRT bacteria contribute to an effective antimicrobial barrier by producing antimicrobial peptides (AMPs) and mucins, which reinforce tight junctions. 84 108 These bacteria also influence immune responses in similar ways as intestinal bacteria by impacting CD4+ T cell differentiation as well as pro- and anti-inflammatory responses. 109 110 Regulatory T (Treg) cells, which are essential for commensal bacterial tolerance, also possess anti-inflammatory properties. 111 112 113 Increased endocervical Tregs have been linked to reduced proinflammatory cytokine and CD4+ T cell counts. 114
Fig. 2.
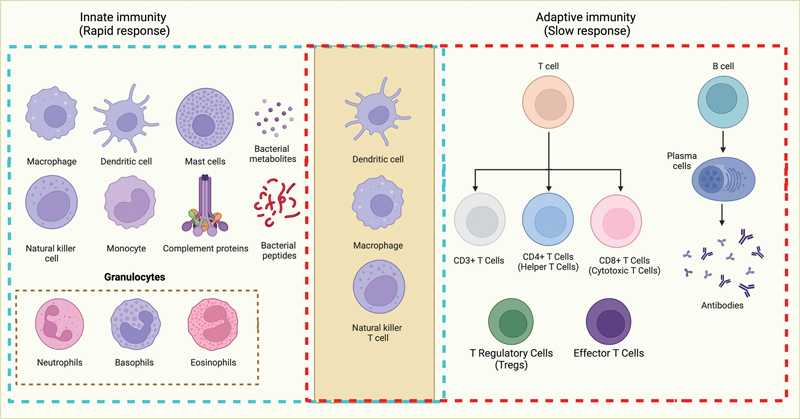
Schematic representation of innate and adaptive immune systems. The immune system comprises two main components: innate and adaptive immunity. Innate immunity, depicted on the left, provides a rapid, nonspecific initial defense against various pathogens, and includes cell types like macrophages, dendritic cells, granulocytes, natural killer cells, and mast cells. The adaptive immune system, shown on the right, offers highly specific protection and involves T and B lymphocytes, with CD8+ T cells (cytotoxic T-lymphocytes) and CD4+ T cells (helper T cells) among its key components. In the middle, the activation of T and B cells is predominantly orchestrated through the stimulation of dendritic cells, macrophages, and natural killer T cells, underscoring the central role of innate immunity in modulating adaptive immune responses. Figure created using BioRender.com (2023). Source: https://app.Biorender.com .
Fig. 3.
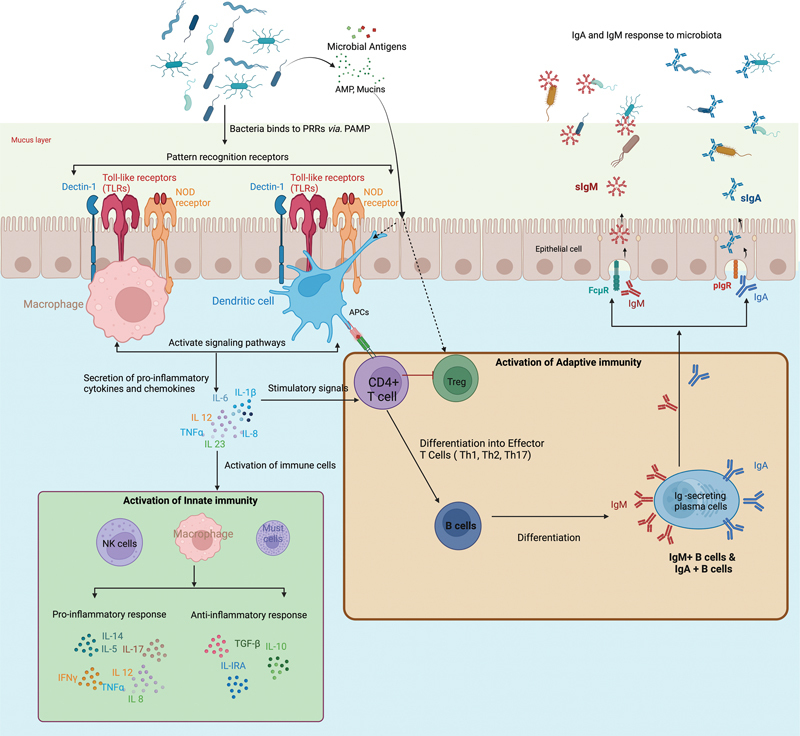
Interactions between microbiota in the reproductive and immune systems. The figure illustrates the mechanisms that govern the generation of IgA and IgM within the reproductive system and how these immunoglobulins, in turn, influence the microbiota. Microbiome-derived factors interact with pattern recognition receptors (PRR), including TLR, Dectin-1, and NOD ligands, while also affecting local and systemic immunity through metabolites. PRRs can directly bind to specific pathogen-associated molecular patterns (PAMPs) on the bacterial surface. Following this interaction, PRRs initiate intracellular signaling through macrophages and dendritic cells, inducing the production of cytokines and chemokines. These molecules, in turn, activate innate immunity through various innate immune cells, producing both pro- and anti-inflammatory responses for microbiota maintenance. The activation of CD4+ T cells by bacteria is crucial in initiating adaptive immune responses. This process involves the presentation of bacterial antigens to CD4+ T cells by antigen-presenting cells (APCs). CD4+ T cells, recognizing the antigens, are co-stimulated by cytokines produced by dendritic cells. Activated CD4 T cells differentiate into various effector cell types, such as regulatory T cells (Tregs), follicular helper T cells (Tfh), and T-helper 17 cells (Th17). Depending on the cytokine microenvironment, cytokines participate in antigen recognition and immune response regulation. These interactions facilitate B cell differentiation into plasma cells, which produce IgA and IgM, thereby contributing to the production of secretory IgA (sIgA) and secretory IgM (sIgM) via pIgR and FcμR receptors, consequently maintaining microbiota homeostasis. In addition, antimicrobial peptides (AMPs) and mucins play multifaceted roles in immunity, including direct antimicrobial defense, modulation of immune responses, influence on the generation of immunoglobulins (IgA and IgM), and contribution to the maintenance of immune tolerance and homeostasis. This figure was adapted from studies on immune interactions of the gut microbiota and was created using BioRender.com (2023). Source: https://app.Biorender.com .
Antigen-presenting cells (APC), such as DCs, can contribute to Treg selection through the TGF-β and retinoic acid (RA) signaling pathways. 115 116 Furthermore, microbiota can activate TLR4 at lower FRT to further support immune tolerance. 89 Immune–microbiota interactions involve leukocyte subsets, plasma cells, interleukin regulation, and antibodies of various classes (IgA, IgM, and IgG). 104 Defense mechanisms include epithelial cells, AMPs, mucous layers, IgA antibodies, commensal bacteria, and immune cells, such as NK cells, macrophages, DCs, and T lymphocytes. 88 99 117 Immune balance protects against pathogens while assuring fetal antigen tolerance during pregnancy. 99 118 Estrogen and progesterone steroid hormones affect immunity and bacterial growth, altering the bacterial composition and susceptibility to infection in FRT. 89 119 Recent research has highlighted the interrelation between microbiomes and adaptive immune systems. Antibodies such as IgA and IgM play a vital role in RT health. 111 120 IgA prevents adhesion of pathogens to epithelial cells, whereas IgM removes them. 121 122 Together, these interactions maintain reproductive health and require further research to understand them fully.
Immunity plays an integral role in reproductive health, especially during gestation, involving complex immune reactions. The orchestrated collaboration between innate and adaptive immunity is vital for maintaining immune homeostasis and defending against potential threats (see Fig. 2 ). This collaboration encompasses various components, such as IgA antibodies, decidual natural killer (dNK) cells, decidual macrophages (DM), DCs, and adaptive immune cells. 99 123 124 125 126 Immune responses vary during implantation, placental support, and labor. 99 127 Trophoblast–bacteria interactions activate interferon-β (IFN-β) and interferon-stimulating genes (ISG), helping maintain immune homeostasis. 128 Tregs provide immunity and tolerance; however, their exact role remains unknown. 127 The FRT has distinct anatomical layers. The URT is lined with a single-layered columnar epithelium, serving as a vital barrier between the external and internal environments. In contrast, the LRT is characterized by a stratified squamous, non-keratinized epithelium, offering enhanced protection. 89 117 Estrogen plays an essential role in modulating antimicrobial molecules and cytokine production for fertility, including sperm survival. 102 129 Commensal bacteria influence immune responses through immune cells and mechanisms that vary according to menstrual cycle phase. 126 Tissue-resident NK and Treg cells maintain immunity and tolerance. They are vital for angiogenesis, trophoblast migration, and immune tolerance. 102 126 Commensal bacteria found in the endometrium have been shown to regulate immune responses via NK cells, T-cell subsets, and cytokines. 130 131 132 The levels of pro-inflammatory cytokines and antimicrobial peptides increase during hormonal fluctuations. 122 129 Cytokine production plays a critical role in response to BV. 133 TLRs in mucosal cells detect molecular patterns associated with pathogens (PAMPs). 100 134 However, prolonged cytokine production can weaken the epithelial barrier and increase susceptibility to infection. 105
Interaction between Immunoglobulin A and Microbiota
The interplay between IgA and microbiota is a complex and finely tuned process within the human body, having significant implications for health and disease. 135 As the predominant class of antibodies found in mucosal tissues, IgA plays an integral role in maintaining relationships with various microbial communities on mucosal surfaces such as those found within the digestive, respiratory, and also in RT. 91 104 136 Additionally, IgA antibodies modulate immune defense, pathogen defense, and maintenance of mucosal homeostasis. 136 137 Mucosal immunity encompasses both tolerance for benign microbes and antigens as well as robust protection against pathogens. Mucosal IgA acts as a guardian with several essential functions, including improving microbial diversity, countering the effects of toxins and viruses, impeding the colonization of pathogenic bacteria, removing undesirable particles, and facilitating antigen sampling. 138
Furthermore, the interaction between IgA and the microbiota has far-reaching consequences for immune modulation, facilitating immune exclusion, pathogen defense, and maintenance of mucosal homeostasis. Humans have five classes of immunoglobulins: IgM, IgG, IgA, IgD, and IgE. IgA is the primary antibody class within mucosal tissues, with two subtypes, IgA1 and IgA2, identified by their unique glycosylation patterns. 136 IgA1 is a widely distributed variant found in the bloodstream, and its functional role remains relatively unknown. On the contrary, IgA2 is secreted mainly in mucosal regions. 138 Previous research has suggested that IgA1 and IgA2 antibodies broadly react with commensal bacteria found in the small intestine. Remarkably, these antibodies exhibited impressive cross-reactivity by binding to multiple targets simultaneously, even at the clonal level. However, it remains unclear about the selection process and specificity of antibodies independent from microbiota. 139 140 B cells producing IgA2 have become widely recognized for their resistance to bacteria-produced proteases compared with IgA1. 141
IgA plays an essential role in coating and encasing the commensal microbiota within the digestive tract, known as immune inclusion. Furthermore, immune exclusion is integral to the protection against enteric pathogens by blocking access to the intestinal epithelium. 121 142 IgA deficiency can arise due to B cell dysfunction, an imbalance in cytokines (e.g., IL-4, IL-6, IL-7, IL-10, IL-21, and TGF-β), and compromised survival of plasma cells producing IgA antibodies. To compensate for IgA deficiency, IgM and/or IgG production often increases to regulate intestinal immunity and prevent infection. 143 144 IgA interacts with multiple receptors on immune cells and serves various effector functions. When IgA binds to the polymeric immunoglobulin receptor (pIgR), it initiates the production of secretory IgA (sIgA) (see Fig. 3 ). sIgA is highly glycosylated, resistant to degradation, and effectively inhibits pathogen adhesion. 145
In particular, commensal-specific antibodies predominantly target microbial capsular polysaccharides and lipopolysaccharides through T cell-independent mechanisms, whereas T cell-dependent high-affinity IgA binding sites are used against more immunogenic and potentially harmful commensals. 91 141 IgA exerts its influence on microbiota by selectively coating antigens with antibodies. Furthermore, its interaction with immune cell Fc receptors such as Fc fragment of IgA receptor (FcαRI) has regulatory effects on their response mechanisms. The expression of FcαRI in myeloid cells results in a range of cellular responses when cross-linked with IgA immune complexes. 137 Opportunistic bacteria such as Staphylococcus aureus and Streptococcus have developed strategies to avoid IgA-mediated elimination by interfering with FcαRI binding. 137 146 Dysregulation of the IgA–FcαRI interaction contributes to autoimmune and inflammatory conditions. The reciprocal relationship between microbiota and IgA levels adds another layer of complexity, suggesting a potential feedback loop in which microbiota influences IgA production. Simultaneously, IgA can alter the composition of the microbiota, suggesting dynamic crosstalk between immune responses and microbial colonization. Hence, comprehending this interplay holds therapeutic promise for microbiota-related diseases.
Women with L. crispatus -dominated vaginal microbiota had higher IgA-coated bacterial levels than those without L. crispatus . L. crispatus predominates the healthy vaginal microbiota, whereas IgG dominates the vaginal mucosa, and its levels fluctuate throughout the reproductive cycle. 91 Furthermore, cervical immunoglobulin levels of IgA and IgG showed significant variations during ovulation. Factors including elevated estradiol and hemoglobin levels, menstrual cycle phases, advanced age, oral contraceptive use, and pregnancy have all been associated with increased levels of IgA and IgG antibodies. 147
In contrast, previous studies have demonstrated that lower estradiol levels during ovulation are strongly correlated with reduced cervical immunoglobulin levels. 147 However, little is known about the role of IgA, IgM, and IgG antibodies in FRT, with an inadequate understanding of their interactions with the microbiota. Furthermore, the composition of reproductive microbiota bound to IgA or IgM remains unknown. Unleashing IgA for therapeutic use requires in-depth knowledge of its complex interactions with microbiota. Addressing these complex interactions could open doors for therapies targeting inflammation, infectious agents, and even cancer, by modulating multiple roles.
Interaction between Immunoglobulin M and Microbiota
The interaction between immunoglobulin M (IgM) antibodies and human microbiota is fundamental to the innate immune response. IgM is an ancient and highly conserved immunoglobulin isotype found in all species from zebra fish to humans. It serves multiple functions, including acting as the first line of defense against infections, supporting immune cell functions, and regulating tissue homeostasis. 148 149 IgM is distinct from other immunoglobulins because of its ability to exist in both natural (nIgM) and antigen-induced forms, such as membrane-bound IgM-type B cell receptors (BCR) or secreted IgM. 150 nIgM is continuously produced by B1 cells and exhibits polyreactivity, which allows it to interact with autoantigens and pathogens. Its functions include removing apoptotic cells and debris, promoting B cell survival, forming lymphoid tissue architecture, and initiating immune responses. 151 152 153 Pathogen-induced IgM is produced by both B-1 and conventional B-2 cells in response to pathogens, strengthening early passive IgM-mediated defenses and regulating IgG production. Secreted IgM (sIgM) interacts with cells through various receptors and is essential for neutralizing microbes, T cell recognition, B cell isotype switching, and antigen transport to lymphoid tissues. 148 150 154
The IgM pentameric structure made up of five interconnected IgM monomers by the J-chain has a higher valency than other immunoglobulins, leading to superior agglutination efficacy, up to 10,000 times that of IgG. This structure allows IgM to bind effectively to multiple antigenic sites, which is known as avidity binding. IgM operates systemically and in the mucosal environment, thereby providing versatility. Unlike IgA, which works mainly on mucosal surfaces, IgM circulates systemically, allowing it to perform immune surveillance and strengthen defenses against threats. 138 153 155 In addition to the broad binding capabilities of circulating polyreactive nIgM, other immune IgM clones also display specific antigen affinity. 153 nIgM antibodies react with the conserved epitopes found in both microbes and self-antigens. Natural IgM production begins with self-antigen interactions. 156 IgM plays a key role in immune surveillance, recognition of threats, and the early detection of harmful microorganisms. 154 IgM is vital for the diversity and stability of microbes in mucosal tissues, particularly in the intestines and respiratory tract, where plasma cells secreting IgM interact with microbiota. 157 158
IgM antibodies bind to the Fc fragment of IgM receptor (FcμR) and pIgR, facilitating the transport of soluble secretory IgM across the mucosal epithelium (see Fig. 3 ). This prevents pathogens from attaching to epithelial cells and destroys the pathogens. 159 160 161 162 For example, IgA and IgM from HIV-1-positive women inhibit HIV-1 transport across the mucosal surfaces of the cervical–vaginal tissues. 161 163 IgM receptor FcαμR internalizes IgM-coated microorganisms similar to pIgR, which binds to IgM and IgA. FcμR is the only IgM-specific receptor in mammals that exclusively binds to IgM. The mechanisms underlying the regulation of sIgM, including its development, control, and function, are not yet fully understood. Previous studies have suggested that FcμR facilitates direct interaction of sIgM with B and T cells. 159 Selective and secreted IgM deficiencies are prone to viral, bacterial, and protozoal infections, and have a higher risk of allergies, inflammation, and autoimmune disorders. 156 The protective functions of IgG and IgM antibodies often involve opsonization and complement recruitment, whereas activation of the classical complement pathway of IgA is limited. 159 164
IgM tends to localize in the endometrial epithelia, unlike IgG, suggesting active transport of serum-derived polymeric IgM through the endometrial glands. 146 Estrogen increases total IgG and IgM production in the human peripheral blood mononuclear cells (PBMCs) of healthy individuals. 165 In contrast, women infected with Neisseria gonorrhoeae exhibited a much higher IgM response. 166 B cells found within the FRT are spread throughout but are concentrated primarily within the vagina, ectocervix, endocervix, and fallopian tubes, although small numbers also produce IgA, IgG, and IgM antibodies that exist primarily within these areas, but may also appear within the endometrium and ovary tissues. The endocervix, fallopian tubes, uterus, and ectocervical glands contain single-layer epithelial cells that express pIgR, which allows the selective transport of sIgA. 167 By contrast, the vagina and ectocervix, which contain multilayered cells lacking pIgR, do not transport pIgA or IgM. However, vaginal subepithelial connective tissue contains plasma cells that are positive for IgA and J chains, suggesting that locally produced immunoglobulins are transported from the blood to the cervical fluid. 168 Cervical shortening, a precursor of spontaneous preterm birth, is associated with Lactobacillus iners and elevated levels of IgM, C3b, C5, C5a, and IL-6. 169 The endocervix contains IgA+ and IgM+ plasma cells that regulate humoral immune response. 170 In cases of IgA deficiency, compensatory IgM responses targeting commensal bacteria arise. This phenomenon has been observed in both mice and humans, highlighting the adaptive nature of the immune system. 144 157 171 172 173
Mucosal antibody responses are heightened in the context of inflammatory bowel disease (IBD), including Crohn's disease and ulcerative colitis. This hyperactivity results from a complex interplay among host genetics, environmental factors, and microbiota composition. 174 Plasma IgM+ and IgG+ cells accumulate in the inflamed intestine, possibly worsening inflammation; however, their role in pathology is not fully understood. 175 176 The interaction of IgM with microbiota extends beyond its classical role in systemic immunity. IgM actively shapes the microbial landscape and contribute to immune surveillance, diversity maintenance, and pathogen protection. Understanding these interactions may lead to new treatments for microbiota-related health issues. Further research may uncover IgM and microbial interactions in reproductive health.
Future Directions and Advanced Technologies
The field of microbiota–immunome interactions in reproductive health is rapidly evolving and several future directions and therapeutic implications are emerging. These advances will enhance our understanding of the complex interactions between the microbiota, the immune system, and reproductive disorders and to create targeted interventions. This section highlights key areas for future research as well as possible diagnostic and therapeutic implications. However, host–microbiota interactions are dynamic, and their mechanisms remain poorly understood. Evidence has shown that reproductive microbiome dynamics play a pivotal role in reproductive health. It is critical to gain an in-depth understanding of their function and impact on host immunity responses. The precise mechanisms by which host antibodies recognize and respond to bacteria remain unclear. Understanding this interaction between the microbiota and immune system remains integral to understanding these interactions.
The current methods used to construct personalized profiles of IgM, IgA, and IgG antibodies that bind to endogenous microbiota require technological improvement before the knowledge can be used to provide tailored treatments more efficiently for reproductive diseases. Antibody-based assays have long been indispensable in the diagnosis of infectious diseases. IgM, IgG, IgA, and IgE antibodies offer additional information that aids in the diagnosis and understanding of the immunity against reinfection. These techniques, including Western blotting, immunofluorescence assays, and enzyme-linked immunosorbent assays, form the backbone of clinical diagnostics and research laboratories. However, limitations in throughput and multiplexing have necessitated the development of high-throughput multiplexable assays, including bead- and chip-based assays. These advanced assays have addressed some of these limitations. However, they can be inefficient because they are slow, incapable of simultaneously analyzing multiple pathogens, and require the expression and purification of target antigens, which may not be achievable for certain pathogen proteins. 177 178
The introduction of next-generation sequencing (NGS) has provided an innovative opportunity with far-reaching results for microbiome–immunome studies. NGS holds great promise for revolutionizing diagnostics by directly detecting microbial nucleic acids and deepening our understanding of host–microbe relationships. However, the limitations of DNA-based microbiome studies, as 16S and metagenome shotgun sequencing, pose significant challenges to its implementation and standardization, particularly in capturing the dynamics of live microbial functions. To bridge this gap, ongoing technological advancements are indispensable to unravel the intricate complexities of microbiota–immunome interactions in reproductive health. Innovative, cutting-edge methodologies, such as mFLOW-Seq and PhIP-Seq, offer powerful ways of studying the RT microbiota and immune cell dynamics at high resolution. These technologies enable high-throughput comprehensive profiling of microbial communities and antibody profiles, offering invaluable insights into new antibody detection modalities and helping researchers to discover biomarkers for various diseases.
Furthermore, the technologies mentioned above empower investigations into physical characteristics, interactions, and functional roles, shedding light on the complex network of interactions between the microbiota and immune system in the context of reproductive health. As this field continues to evolve, it holds promise for the advancement of disease diagnostics and treatment, and ultimately contributes to improved reproductive health outcomes.
Microbial Flow Cytometry coupled to NGS (mFLOW-Seq)
Flow cytometry, known for its high-throughput cell analysis capabilities, is a versatile tool for examining various cell types in research and clinical settings. Microbial flow cytometry (mFLOW) offers additional benefits in investigating microbes found in both environmental and clinical samples. It facilitates the analysis of physical properties and metabolic states associated with microbes, including size, shape, and granularity, using fluorescence markers. The utility of mFLOW-Seq has been expanded by incorporating host antibodies to assess humoral immune responses to specific microbes or microbial groups. This innovative approach uses FACS and NGS to sort and detect microbes that contain specific antibodies (IgA, IgM, and IgG) from complex communities. Fusing microbial flow cytometry with NGS (mFLOW-Seq) provides an effective means of deciphering intricate interactions between the immune system and endogenous microbial communities. This technology offers novel insights into commensal–host interactions in mucosal and systemic environments. mFLOW-Seq provides quantitative information on microbial communities. This approach discriminates microbial subcommunities based on the immunoglobulin-bound microbiota, cell size, and DNA content. mFLOW-Seq has enabled researchers to isolate microbiota targeted by systemic antibodies across various samples (see Fig. 4 ).
Fig. 4.
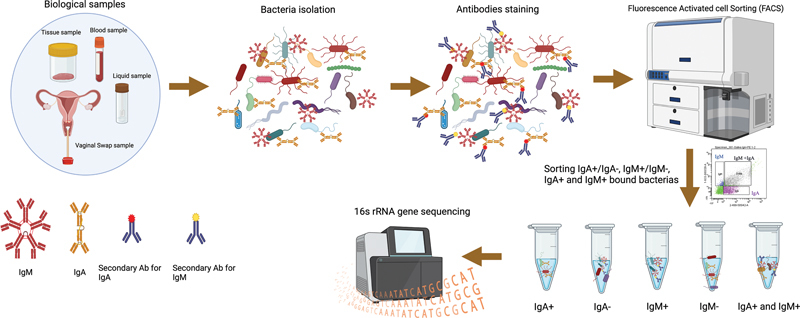
Schematic of mFLOW-Seq for identifying microbiota targeted by host antibodies in biological samples. The process involves several key steps. Isolation of bacteria: bacteria are extracted from the biological samples. Incubation with antibodies: The isolated bacteria are then incubated with fluorescently labeled secondary antibodies, specifically anti-IgA, anti-IgM, and anti-IgG. FACS: employed to segregate the bacterial populations into two groups: IgA +/− , IgM +/− , and IgG +/− according to antibodies bound to bacteria. Microbiota analysis: the sorted bacterial populations are subjected to 16S rRNA gene/metagenomic sequencing. This sequencing helps identify which microbes are enriched in the Ig fraction, revealing microbiota and immunome interactions. This figure was created using BioRender.com (2023). Source: https://app.Biorender.com .
Initially, the bacteria extracted from the samples are stained with fluorescently labeled secondary antibodies specific for IgA, IgG, or IgM. Subsequently, fluorescence-activated cell sorting (FACS) segregates bacteria into distinct populations based on their binding characteristics: IgA + , IgG + , IgM + , IgA − , IgG − , and IgM − . These separate populations are subjected to 16S rRNA gene sequencing to identify microbes that are enriched in each fraction. 16S rRNA gene sequencing is a standard method for microbiome analysis. It targets specific regions of bacterial 16S ribosomal RNA (rRNA) genes with nine hypervariable regions (V1–V9) with significant sequence diversity among bacterial species. Microbiome studies often incorporate multiple regions like V3–V4 to improve bacterial taxon identification, offering insights into bacterial and archaeal community composition at the genus level. In contrast, metagenome sequencing targets all genomic DNA in a sample, enabling the study of bacteria, archaea, DNA viruses, and other microorganisms in complex microbial communities. It offers a comprehensive view, allowing for species-level identification and broader detection of microorganisms. 179 180 This process revealed that IgA, IgG, IgM, and unbound fractions selectively bind to specific microbes, providing valuable information on antibody–microbe interactions 181 182 183 184 185 (see Fig. 4 ). This method excludes lysed bacteria and its fragments and concentrates solely on intact bacteria, thus offering both high sensitivity and a broad dynamic range. Remarkably, it operates effectively with minimal amounts of immunoglobulin, which makes it suitable for scenarios with low bacterial target densities, allowing for the analysis of immune responses, even against challenging bacterial species.
The previously established mFLOW-Seq technique has helped to quantify antibody binding specifically to live bacterial surface epitopes. mFLOW has been applied to fecal, intestinal, and various bodily fluids, such as serum, plasma, intestinal/bronchial/vaginal lavage, saliva, and breast milk bacterial samples. 186 187 188 189 Previous studies using mFLOW-Seq in pediatric selective immunoglobulin A (IgA) deficiency (SIgAD) patients found that mucosal and systemic antibodies target the same commensal microbes. The absence of secretory IgA results in abnormal systemic exposures and immune responses to commensal microbes, increasing the risk of immune dysregulation and symptomatic disease. IgA prevents microbial translocation and related immune dysregulation, leading to milder symptoms in SIgAD patients. 190 Furthermore, a study underscores commensal bacteria's impact on serum IgA levels, emphasizing T cell-dependent systemic responses. It links microbiota composition to serum IgA concentrations and identifies protective roles against sepsis when using mFLOW-Seq for microbiota analysis. 191 192
Although NGS, a groundbreaking method for exploring microbial communities, has a limitation in distinguishing between live and deceased cells, as it extracts DNA from all materials indiscriminately. 52 In contrast, mFLOW-Seq holds promise in this regard by utilizing FACS to differentiate between live and dead microbial members before NGS sequencing. 193 Several studies have used high-throughput FACS combined with 16S rRNA sequencing to investigate the interactions between immunoglobulin IgA, IgM, IgG, and microbiota. 143 172 191 194 195 196 197 However, a significant gap remains in the field of reproductive immuno-microbiota analysis, with only one study using the mFLOW-Seq method for vaginal swab analysis of IgA and IgG bound microbiota populations. 91 This underscores the need for further exploration in this specific area.
Therefore, this technique enables monitoring microbial community changes over time due to host environmental changes and disease progression. Further research is required to investigate whether antibodies binding to endogenous bacteria can predict the risk of infection (i.e., facilitating or preventing the transition from commensals to pathogens). In the move toward clinical use of mFLOW-Seq, further research is vital to fully grasp the significance of microbe-specific serum levels of IgA, IgM, and IgG in immunoglobulin replacement therapies. Immunoglobulin replacement therapy delivers IgA, IgM, and IgG to individuals with immunodeficiencies. Tailoring these antibody levels to specific pathogens is crucial in improving therapy effectiveness, minimizing over-treatment risks, and optimizing treatment for individual immunodeficiencies. Understanding the potential of microbe-specific serum levels of IgA, IgM, and IgG in immunoglobulin replacement therapies is crucial. Although antibody responses typically take several days to develop, limiting their utility during the early phases of infection, it may be beneficial to test whether specific antibody preparations can bind to endogenous microbiota to prevent invasive infections. It can potentially revolutionize our understanding of host–microbiota relationships in various health contexts, including reproductive health. These findings may pave the way for innovative diagnostic and therapeutic approaches. Still, further research and clinical investigations are required to fully unlock the potential of mFLOW-Seq in microbiome-related research and clinical applications for personalized reproductive healthcare.
Phage ImmunoPrecipitation Sequencing (PhIP-Seq)
PhIP-Seq has emerged as a powerful seroepidemiology tool that offers a promising avenue for investigating various aspects of human health including reproductive health. This cutting-edge technology combines T7 phage display with NGS, enabling the exploration of complex interactions between human microbiota and the immune system. PhIP-Seq is a versatile method that simultaneously profiles the antibody responses to various antigens. In the context of reproductive health, where the dynamic interaction between the microbiota and host immune system is of immense importance, this opens a unique avenue for investigation. In addition, their ability to detect antibody responses to new or less-explored RT pathogens is indispensable. PhIP-Seq, a relatively recent antibody detection technology, offers the ability to simultaneously investigate numerous antigens, thereby facilitating identification of microbial gene epitopes that interact with the host immune system. This technology sheds light on the intricate interactions between the microbiome and host immune system, uncovering the specific epitopes that drive these complex relationships. It also has the potential to identify biomarkers for various diseases, including those of an immune origin. The opportunity to investigate the diversity and distribution of microbiota within different organs and to understand how distinct antibody classes respond to pathogens is of significant value. Furthermore, understanding how the reproductive microbiome influences host immunity and the development of gynecological diseases offers promising avenues for research.
PhIP-Seq technology, based on phage-displayed synthetic oligo libraries representing the microbiome, offers a comprehensive approach to investigate the dynamics of the microbiome and its intricate association with host antibody responses. This versatile technology accommodates various sample and antibody isotypes including IgM, IgG, IgA, IgD, and IgE. This procedure can be adapted to construct libraries spanning different biological domains. Briefly, the protocol involves the design of peptides that tile across the proteins. Subsequently, these peptides are synthesized into overlapping oligo libraries and cloned into T7 bacteriophage display vectors. This streamlined approach is versatile and provides a straightforward method for crafting customized libraries. Typically, these libraries rely on protein reference sequences from public databases, with overlapping peptides ranging from 56 to 90 amino acids (see Fig. 5 ). This inclusivity allows the incorporation of various known strains and infectious pathogens, making it applicable to reproductive health scenarios. Furthermore, the ability of PhIP-Seq to propagate phage libraries provides a consistent source of antigenic material to study immune responses. 198 199 200 Phage display and immunoprecipitation are employed to assess antibody binding to all peptides in parallel, and DNA sequencing is used to determine the relative abundance of the immunoprecipitated microbial population. 201 The article by Mohan et al provides a comprehensive protocol for constructing a PhIP-Seq peptide library designed for the in-depth analysis of serum antibodies. 202
Fig. 5.
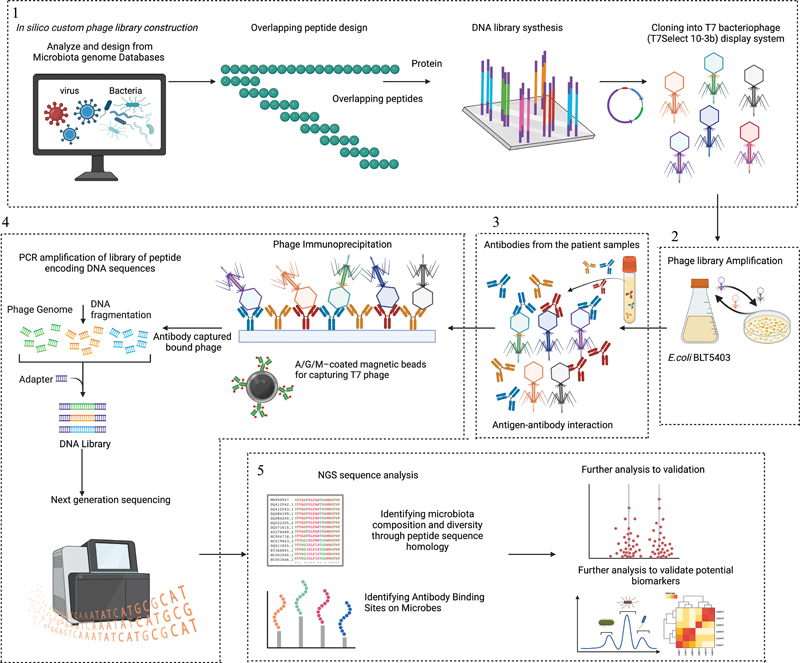
Overview of PhIP-Seq methodology. The PhIP-Seq methodology involves four key steps. (1) Phage library construction: Downloading or designing a protein database and utilizing bioinformatics tools to generate overlapping peptide sequences from microbiome database proteins. Synthesis of an oligonucleotide library encoding the peptide sequences. PCR-amplify the oligonucleotide library with adapters for cloning into the T7Select 10–3b mid-copy phage display system. (2) Propagation of phage library: The phage library amplified using Escherichia coli BLT 5403 to ensure library diversity and sufficient phage clones for subsequent experiments and sequencing. (3) Patient antibody–antigen interaction: Patient samples containing antibodies are incubated with the amplified phage library for specific interactions. (4) Phage immunoprecipitation and data analysis: Patient samples and their antigens are used in phage immunoprecipitation reactions to capture antibodies and their bound phages on beads coated with proteins A, M, and G. DNA recovered from the immunoprecipitated phage. IgA, IgM, and IgG contents in each sample quantified using ELISA for antibody input normalization. Amplify the library of peptide-encoding DNA sequences directly from the immunoprecipitate. PCR-amplified DNA, sample-specific barcodes, and sequencing adapters for NGS. Pool-barcoded amplicons for sequencing using NGS. (5) Data analysis and validation: Multiplexed data are aligned to reference sequences to create a count matrix. Statistical analysis identifies peptide enrichments, facilitating project-specific investigations, such as identifying common autoantigens, validating microbe-specific epitopes, and identifying potential biomarkers. This methodology provides a comprehensive approach for analyzing antibody epitopes and their association with microbiota. Figure adapted from BioRender.com (2023). Source: https://app.Biorender.com .
PhIP-Seq libraries have been meticulously designed to span many biological domains, including human proteome, allergens, protein toxins, microbiota, viruses, and gut bacteria. In 2011, Larman et al introduced a groundbreaking method that combined PhIP-Seq with a synthetic human proteome library. This innovative approach aims to identify autoantibodies and unravel the autoantigens linked to autoimmune diseases, particularly in individuals with paraneoplastic syndromes, type 1 diabetes, multiple sclerosis, and rheumatoid arthritis. 201 203 204 Subsequent studies with human virome and human proteome phage display libraries have focused on dissecting antiviral antibody profiles, 205 unraveling immune responses in membrane nephropathy, 206 antibody responses to environmental toxins and virulence factors, 207 and focusing on specific viral families such as anelloviruses. 208 In another groundbreaking study, PhIP-Seq was used to construct a human gut–bacterial microbiome library, examining serum antibody responses in 997 healthy individuals. Surprisingly, these responses are diverse and exceptionally stable over time, thereby introducing the concept of an “immunological fingerprint.” 209 Further research involving 1,003 nonallergic individuals explored the response of IgG antibodies to food and environmental antigens. They used an extensive library of bacterial and viral peptides displayed on phages covering various food and environmental allergens from multiple databases. 210 Previous studies have combined the human gut microbiome with an allergic phage library to examine human antibody repertoires in 1,443 individuals. These studies considered genetic, environmental, and intrinsic factors; assessed antibody responses in people with IBD such as Crohn's disease and ulcerative colitis; and compared them with a control group. 211 212
However, despite these advantages, PhIP-Seq has several limitations. It cannot identify autoantibodies that recognize post-translationally modified proteins (i.e., glycosylation isoforms); lack information on post-translational modifications, like sugar moieties; and misses the three-dimensional structure of antigens. It is also limited to linear epitopes and excludes non-protein immunogenic molecules such as lipids and glycans. 211 Research exploring the interactions between human microbiota and the immune system in reproductive health remains limited. There is a compelling opportunity to leverage PhIP-Seq to develop comprehensive microbiome libraries that are specific to RT. This would allow an in-depth study of the dynamic interactions between the microbiota and host immune system. This development has the potential for widespread commercial applications, particularly in the development of diagnostic tools and therapies related to microbiome–host interactions, and facilitates the RT probiotic strain studies. Personalized antibody profiles and assessment of antibody responses against microbiome antigens can be utilized for risk assessment, pathogen detection, and disease monitoring, thus presenting extensive healthcare and commercial potential. PhIP-Seq has immense potential to improve our understanding of reproductive health by providing a sophisticated means of scrutinizing human microbiota and immune interactions. Its adaptability, high-throughput capabilities, and precise data analysis make it an invaluable asset for reproductive health research. Given the profound impact of microbial communities and immune responses on reproductive health, PhIP-Seq has emerged as a cutting-edge tool to elucidate this intricate interplay. Together with other molecular techniques, harnessing the advantages of PhIP-Seq in reproductive health investigations promises breakthroughs in diagnosing, preventing, and managing reproductive health issues.
Conclusions
Microbiota–immunome interactions in reproductive health represent a complex and promising field of research with significant potential. This review covers various relevant sections that provide insights into these relationships, technological developments, diagnostic and therapeutic possibilities. The field of reproductive microbiota, particularly of that of upper RTs, requires extensive exploration to reveal their composition and effects on women's reproductive health. Moreover, the current knowledge regarding the microbiome in specific locations within the FRT, such as the ovaries, endometrium, and placenta, is limited and primarily obtained from women with preexisting conditions. Further research is essential to understand the microbiome in healthy nonpregnant and pregnant women. As a result, distinguishing between the normal microbiome and their potential causative agents of infections remains a challenge. To address this gap, further investigations should be conducted regarding the controversial endometrial/uterine, ovaries, fallopian tube, and placental resident microbiome. It is crucial to implement strictly controlled studies on live microbial cells to avoid contamination during sample processing, ensuring the reliability of results.
At the same time, the human microbiota has an immense effect on the immune system, shaping the adaptive and innate responses that are essential to overall health. Investigating how IgA and IgM regulate these responses is a continuous pursuit. Differentiating between healthy and pathological microbiota–immunome interactions would be critical. Acquiring knowledge on how antibodies identify and respond to bacteria is crucial for gaining insights into microbiota–immunome interactions. Still, manipulation of antibodies for diagnostic and therapeutic use requires further investigation. Standardized methods are essential to understand their roles in clinical disorders and provide the foundation for large-scale personalized medicine and diagnostic trials. Technologies such as mFLOW-Seq and PhIP-Seq provide exceptional insights into microbiota–immune interactions, including the epitopes that drive these interactions in FRT and larger in the entire organism. Current insights into microbiota–immunome interactions come from DNA-based microbiome studies, which may not fully capture the functional aspects of live microbes and their dynamics. To address this critical gap, future research should shift from microbiome to microbiota studies and use techniques such as mFLOW-Seq and PhIP-Seq, capable of assessing live microbes, for a more comprehensive understanding and more robust conclusions.
This comprehensive review highlights the potential to expand our knowledge of the composition and diversity of the microbiota within RT, which offers promise for personalized medicine for reproductive disorders and improved outcomes. These insights are helpful in predicting and preventing infectious diseases and autoimmune disorders that affect reproductive health. Innovative technologies such as mFLOW-Seq and PhIP-Seq are promising for revolutionizing disease diagnosis, monitoring, and offering truly personalized medical interventions. Technological innovations go beyond reproductive health and have become invaluable tools for studying microbiota–immunome interactions in various reproductive contexts. This undertaking provides the basis for new diagnostic and therapeutic avenues for reproductive healthcare.
Acknowledgments
This work has been supported by the Estonian Research Council (grant PRG1076), Horizon 2020 innovation grant (ERIN, grant no. EU952516), Enterprise Estonia (grant no EU48695), Ministry of Education and Research (KOGU-HUMB), and the Horizon Europe (NESTOR, grant no. 101120075).
Conflict of Interest All authors declare that they have no competing interests.
Authors' Contribution
All authors helped to conceive the topic for the review. P.L. prepared the initial draft of the manuscript; all authors edited the manuscript and confirmed the last version of the manuscript.
References
- 1.Wang B, Yao M, Lv L, Ling Z, Li L. The human microbiota in health and disease. Engineering. 2017;3(01):71–82. [Google Scholar]
- 2.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(08):e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brüls T, Weissenbach J.The human metagenome: our other genome? Hum Mol Genet 201120(R2):R142–R148. [DOI] [PubMed] [Google Scholar]
- 4.Donders G GG, Bellen G, Ruban K S. Abnormal vaginal microbiome is associated with severity of localized provoked vulvodynia. Role of aerobic vaginitis and Candida in the pathogenesis of vulvodynia. Eur J Clin Microbiol Infect Dis. 2018;37(09):1679–1685. doi: 10.1007/s10096-018-3299-2. [DOI] [PubMed] [Google Scholar]
- 5.Onderdonk A B, Delaney M L, Fichorova R N. The human microbiome during bacterial vaginosis. Clin Microbiol Rev. 2016;29(02):223–238. doi: 10.1128/CMR.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobel J D. Bacterial vaginosis. Annu Rev Med. 2000;51(01):349–356. doi: 10.1146/annurev.med.51.1.349. [DOI] [PubMed] [Google Scholar]
- 7.Human Microbiome Project Consortium . Huttenhower C, Gevers D, Knight R et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang N, Chen L, Yi K, Zhang B, Li C, Zhou X. The effects of microbiota on reproductive health: a review. Crit Rev Food Sci Nutr. 2022:1–22. doi: 10.1080/10408398.2022.2117784. [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Song X, Wei W et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun. 2017;8(01):875. doi: 10.1038/s41467-017-00901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowe M, Veerus L, Trosvik P, Buckling A, Pizzari T. The reproductive microbiome: an emerging driver of sexual selection, sexual conflict, mating systems, and reproductive isolation. Trends Ecol Evol. 2020;35(03):220–234. doi: 10.1016/j.tree.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Feng T, Liu Y. Microorganisms in the reproductive system and probiotic's regulatory effects on reproductive health. Comput Struct Biotechnol J. 2022;20:1541–1553. doi: 10.1016/j.csbj.2022.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoenmakers S, Steegers-Theunissen R, Faas M. The matter of the reproductive microbiome. Obstet Med. 2019;12(03):107–115. doi: 10.1177/1753495X18775899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomaiuolo R, Veneruso I, Cariati F, D'Argenio V. Microbiota and human reproduction: the case of female infertility. High Throughput. 2020;9(02):12. doi: 10.3390/ht9020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quaranta G, Sanguinetti M, Masucci L. Fecal microbiota transplantation: a potential tool for treatment of human female reproductive tract diseases. Front Immunol. 2019;10:2653. doi: 10.3389/fimmu.2019.02653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miles S M, Hardy B L, Merrell D SS. Investigation of the microbiota of the reproductive tract in women undergoing a total hysterectomy and bilateral salpingo-oopherectomy. Fertil Steril. 2017;107(03):813–8200. doi: 10.1016/j.fertnstert.2016.11.028. [DOI] [PubMed] [Google Scholar]
- 16.Moreno I, Simon C. Deciphering the effect of reproductive tract microbiota on human reproduction. Reprod Med Biol. 2018;18(01):40–50. doi: 10.1002/rmb2.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharya K, Dutta S, Sengupta P, Bagchi S. Reproductive tract microbiome and therapeutics of infertility. Middle East Fertil Soc J. 2023;28(01):11. [Google Scholar]
- 18.Garcia-Grau I, Perez-Villaroya D, Bau D et al. Taxonomical and functional assessment of the endometrial microbiota in a context of recurrent reproductive failure: A case report. Pathogens. 2019;8(04):4–6. doi: 10.3390/pathogens8040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muraoka A, Suzuki M, Hamaguchi T et al. Fusobacterium infection facilitates the development of endometriosis through the phenotypic transition of endometrial fibroblasts . Sci Transl Med. 2023;15(700):eadd1531. doi: 10.1126/scitranslmed.add1531. [DOI] [PubMed] [Google Scholar]
- 20.Payne M S, Newnham J P, Doherty D A et al. A specific bacterial DNA signature in the vagina of Australian women in midpregnancy predicts high risk of spontaneous preterm birth (the Predict1000 study) Am J Obstet Gynecol. 2021;224(02):2060–2.06E25. doi: 10.1016/j.ajog.2020.08.034. [DOI] [PubMed] [Google Scholar]
- 21.France M T, Ma B, Gajer P et al. VALENCIA: a nearest centroid classification method for vaginal microbial communities based on composition. Microbiome. 2020;8(01):166. doi: 10.1186/s40168-020-00934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Britto A MA, Siqueira J D, Curty G et al. Microbiome analysis of Brazilian women cervix reveals specific bacterial abundance correlation to RIG-like receptor gene expression. Front Immunol. 2023;14:1.14795E6. doi: 10.3389/fimmu.2023.1147950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravel J, Gajer P, Abdo Zet al. Vaginal microbiome of reproductive-age women Proc Natl Acad Sci U S A 2011108(Suppl 1, Suppl 1):4680–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cocomazzi G, De Stefani S, Del Pup L et al. The impact of the female genital microbiota on the outcome of assisted reproduction treatments. Microorganisms. 2023;11(06):1443. doi: 10.3390/microorganisms11061443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehtoranta L, Ala-Jaakkola R, Laitila A, Maukonen J. Healthy vaginal microbiota and influence of probiotics across the female life span. Front Microbiol. 2022;13:819958. doi: 10.3389/fmicb.2022.819958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mändar R, Punab M, Borovkova N et al. Complementary seminovaginal microbiome in couples. Res Microbiol. 2015;166(05):440–447. doi: 10.1016/j.resmic.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 27.O'Hanlon D E, Moench T R, Cone R A. Vaginal pH and microbicidal lactic acid when lactobacilli dominate the microbiota. PLoS ONE. 2013;8(11):e80074. doi: 10.1371/journal.pone.0080074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Younes J A, Lievens E, Hummelen R, van der Westen R, Reid G, Petrova M I. Women and their microbes: the unexpected friendship. Trends Microbiol. 2018;26(01):16–32. doi: 10.1016/j.tim.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Delgado-Diaz D J, Tyssen D, Hayward J A, Gugasyan R, Hearps A C, Tachedjian G. Distinct immune responses elicited from cervicovaginal epithelial cells by lactic acid and short chain fatty acids associated with optimal and non-optimal vaginal microbiota. Front Cell Infect Microbiol. 2020;9:446. doi: 10.3389/fcimb.2019.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adapen C, Réot L, Menu E. Role of the human vaginal microbiota in the regulation of inflammation and sexually transmitted infection acquisition: contribution of the non-human primate model to a better understanding? Front Reprod Health. 2022;4:992176. doi: 10.3389/frph.2022.992176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coleman J S, Gaydos C A. Molecular diagnosis of bacterial vaginosis: an update. J Clin Microbiol. 2018;56(09) doi: 10.1128/JCM.00342-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koort K, Sõsa K, Türk S et al. Lactobacillus crispatus-dominated vaginal microbiome and Acinetobacter-dominated seminal microbiome support beneficial ART outcome. Acta Obstet Gynecol Scand. 2023;102(07):921–934. doi: 10.1111/aogs.14598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar L, Dwivedi M, Jain N et al. The female reproductive tract microbiota: friends and foe. Life (Basel) 2023;13(06):1313. doi: 10.3390/life13061313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravel J, Moreno I, Simón C. Bacterial vaginosis and its association with infertility, endometritis, and pelvic inflammatory disease. Am J Obstet Gynecol. 2021;224(03):251–257. doi: 10.1016/j.ajog.2020.10.019. [DOI] [PubMed] [Google Scholar]
- 35.Štšepetova J, Baranova J, Simm J et al. The complex microbiome from native semen to embryo culture environment in human in vitro fertilization procedure. Reprod Biol Endocrinol. 2020;18(01):3. doi: 10.1186/s12958-019-0562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan C, Hong F, Xin G, Duan S, Deng X, Xu Y. Alterations in the vaginal microbiota of patients with preterm premature rupture of membranes. Front Cell Infect Microbiol. 2022;12:858732. doi: 10.3389/fcimb.2022.858732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fettweis J M, Serrano M G, Brooks J P et al. The vaginal microbiome and preterm birth. Nat Med. 2019;25(06):1012–1021. doi: 10.1038/s41591-019-0450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen A TC, Le Nguyen N T, Hoang T TA et al. Aerobic vaginitis in the third trimester and its impact on pregnancy outcomes. BMC Pregnancy Childbirth. 2022;22(01):432. doi: 10.1186/s12884-022-04761-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barczyński B, Frąszczak K, Grywalska E, Kotarski J, Korona-Głowniak I. Vaginal and cervical microbiota composition in patients with endometrial cancer. Int J Mol Sci. 2023;24(09):8266. doi: 10.3390/ijms24098266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bednarska-Czerwińska A, Morawiec E, Zmarzły N et al. Dynamics of microbiome changes in the endometrium and uterine cervix during embryo implantation: a comparative analysis. Med Sci Monit. 2023;29:e941289. doi: 10.12659/MSM.941289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong M, Dong Y, Bai Jet al. Interactions between microbiota and cervical epithelial, immune, and mucus barrier Front Cell Infect Microbiol 202313(February):1.124591E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang J, Li M, Zhang L et al. Analysis of the microbiota composition in the genital tract of infertile patients with chronic endometritis or endometrial polyps. Front Cell Infect Microbiol. 2023;13:1.12564E6. doi: 10.3389/fcimb.2023.1125640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tuominen H, Rautava S, Syrjänen S, Collado M C, Rautava J. HPV infection and bacterial microbiota in the placenta, uterine cervix and oral mucosa. Sci Rep. 2018;8(01):9787. doi: 10.1038/s41598-018-27980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Payne M S, Ireland D J, Watts R et al. Ureaplasma parvum genotype, combined vaginal colonisation with Candida albicans, and spontaneous preterm birth in an Australian cohort of pregnant women. BMC Pregnancy Childbirth. 2016;16(01):312. doi: 10.1186/s12884-016-1110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parnell L A, Briggs C M, Mysorekar I U. Maternal microbiomes in preterm birth: recent progress and analytical pipelines. Semin Perinatol. 2017;41(07):392–400. doi: 10.1053/j.semperi.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anahtar M N, Gootenberg D B, Mitchell C M, Kwon D S. Cervicovaginal microbiota and reproductive health: the virtue of simplicity. Cell Host Microbe. 2018;23(02):159–168. doi: 10.1016/j.chom.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell C M, Haick A, Nkwopara E et al. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am J Obstet Gynecol. 2015;212(05):6110–6.11E11. doi: 10.1016/j.ajog.2014.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koedooder R, Mackens S, Budding A et al. Identification and evaluation of the microbiome in the female and male reproductive tracts. Hum Reprod Update. 2019;25(03):298–325. doi: 10.1093/humupd/dmy048. [DOI] [PubMed] [Google Scholar]
- 49.Moreno I, Codoñer F M, Vilella F et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol. 2016;215(06):684–703. doi: 10.1016/j.ajog.2016.09.075. [DOI] [PubMed] [Google Scholar]
- 50.Molina N M, Sola-Leyva A, Saez-Lara M J et al. New opportunities for endometrial health by modifying uterine microbial composition: present or future? Biomolecules. 2020;10(04):593. doi: 10.3390/biom10040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molina N M, Sola-Leyva A, Haahr T et al. Analysing endometrial microbiome: methodological considerations and recommendations for good practice. Hum Reprod. 2021;36(04):859–879. doi: 10.1093/humrep/deab009. [DOI] [PubMed] [Google Scholar]
- 52.Sola-Leyva A, Andrés-León E, Molina N M et al. Mapping the entire functionally active endometrial microbiota. Hum Reprod. 2021;36(04):1021–1031. doi: 10.1093/humrep/deaa372. [DOI] [PubMed] [Google Scholar]
- 53.Franasiak J M, Werner M D, Juneau C R et al. Endometrial microbiome at the time of embryo transfer: next-generation sequencing of the 16S ribosomal subunit. J Assist Reprod Genet. 2016;33(01):129–136. doi: 10.1007/s10815-015-0614-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moreno I, Garcia-Grau I, Perez-Villaroya D et al. Endometrial microbiota composition is associated with reproductive outcome in infertile patients. Microbiome. 2022;10(01):1. doi: 10.1186/s40168-021-01184-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diaz-Martínez M DC, Bernabeu A, Lledó B et al. Impact of the vaginal and endometrial microbiome pattern on assisted reproduction outcomes. J Clin Med. 2021;10(18):4063. doi: 10.3390/jcm10184063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wee B A, Thomas M, Sweeney E L et al. A retrospective pilot study to determine whether the reproductive tract microbiota differs between women with a history of infertility and fertile women. Aust N Z J Obstet Gynaecol. 2018;58(03):341–348. doi: 10.1111/ajo.12754. [DOI] [PubMed] [Google Scholar]
- 57.Hiraoka T, Osuga Y, Hirota Y. Current perspectives on endometrial receptivity: A comprehensive overview of etiology and treatment. J Obstet Gynaecol Res. 2023;49(10):2397–2409. doi: 10.1111/jog.15759. [DOI] [PubMed] [Google Scholar]
- 58.Lüll K, Saare M, Peters M et al. Differences in microbial profile of endometrial fluid and tissue samples in women with in vitro fertilization failure are driven by Lactobacillus abundance. Acta Obstet Gynecol Scand. 2022;101(02):212–220. doi: 10.1111/aogs.14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riganelli L, Iebba V, Piccioni M et al. Structural variations of vaginal and endometrial microbiota: hints on female infertility. Front Cell Infect Microbiol. 2020;10:350. doi: 10.3389/fcimb.2020.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ojosnegros S, Seriola A, Godeau A L, Veiga A. Embryo implantation in the laboratory: an update on current techniques. Hum Reprod Update. 2021;27(03):501–530. doi: 10.1093/humupd/dmaa054. [DOI] [PubMed] [Google Scholar]
- 61.Silpe J E, Balskus E P. Deciphering human microbiota-host chemical interactions. ACS Cent Sci. 2021;7(01):20–29. doi: 10.1021/acscentsci.0c01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belizário J E, Napolitano M.Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches Front Microbiol 20156(Oct):1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purcell R V, Pearson J, Aitchison A, Dixon L, Frizelle F A, Keenan J I. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017;12(02):e0171602. doi: 10.1371/journal.pone.0171602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pelzer E S, Willner D, Buttini M, Hafner L M, Theodoropoulos C, Huygens F. The fallopian tube microbiome: implications for reproductive health. Oncotarget. 2018;9(30):21541–21551. doi: 10.18632/oncotarget.25059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peric A, Weiss J, Vulliemoz N, Baud D, Stojanov M. Bacterial colonization of the female upper genital tract. Int J Mol Sci. 2019;20(14):3405. doi: 10.3390/ijms20143405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Canha-Gouveia A, Pérez-Prieto I, Rodríguez C M et al. The female upper reproductive tract harbors endogenous microbial profiles. Front Endocrinol. 2023;14:1.09605E6. doi: 10.3389/fendo.2023.1096050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mondal A S, Sharma R, Trivedi N. Bacterial vaginosis: a state of microbial dysbiosis. Med Microecology. 2023;16:100082. [Google Scholar]
- 68.Wasfi R, Abd El-Rahman O A, Zafer M M, Ashour H M. Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J Cell Mol Med. 2018;22(03):1972–1983. doi: 10.1111/jcmm.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Banerjee S, Tian T, Wei Z et al. The ovarian cancer oncobiome. Oncotarget. 2017;8(22):36225–36245. doi: 10.18632/oncotarget.16717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, Zheng Y, Chen X et al. 2bRAD-M reveals the difference in microbial distribution between cancerous and benign ovarian tissues. Front Microbiol. 2023;14:1.231354E6. doi: 10.3389/fmicb.2023.1231354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou B, Sun C, Huang J et al. The biodiversity composition of microbiome in ovarian carcinoma patients. Sci Rep. 2019;9(01):1691. doi: 10.1038/s41598-018-38031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Païssé S, Valle C, Servant F et al. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion. 2016;56(05):1138–1147. doi: 10.1111/trf.13477. [DOI] [PubMed] [Google Scholar]
- 73.Wang Q, Zhao L, Han L et al. The differential distribution of bacteria between cancerous and noncancerous ovarian tissues in situ. J Ovarian Res. 2020;13(01):8. doi: 10.1186/s13048-019-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aagaard K, Ma J, Antony K M, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237):237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kennedy K M, de Goffau M C, Perez-Muñoz M E et al. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature. 2023;613(7945):639–649. doi: 10.1038/s41586-022-05546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perez-Muñoz M E, Arrieta M C, Ramer-Tait A E, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5(01):48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prince A L, Ma J, Kannan P S et al. The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am J Obstet Gynecol. 2016;214(05):6270–6.27E18. doi: 10.1016/j.ajog.2016.01.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Goffau M C, Lager S, Sovio U et al. Human placenta has no microbiome but can contain potential pathogens. Nature. 2019;572(7769):329–334. doi: 10.1038/s41586-019-1451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Farooqi H MU, Kim K H, Kausar F, Muhammad J, Bukhari H, Choi K H. Frequency and molecular characterization of Staphylococcus aureus from placenta of mothers with term and preterm deliveries . Life (Basel) 2022;12(02):257. doi: 10.3390/life12020257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuperman A A, Zimmerman A, Hamadia S et al. Deep microbial analysis of multiple placentas shows no evidence for a placental microbiome. BJOG. 2020;127(02):159–169. doi: 10.1111/1471-0528.15896. [DOI] [PubMed] [Google Scholar]
- 81.Parhi L, Abed J, Shhadeh A et al. Placental colonization by Fusobacterium nucleatum is mediated by binding of the Fap2 lectin to placentally displayed Gal-GalNAc. Cell Rep. 2022;38(12):110537. doi: 10.1016/j.celrep.2022.110537. [DOI] [PubMed] [Google Scholar]
- 82.Sterpu I, Fransson E, Hugerth L W et al. No evidence for a placental microbiome in human pregnancies at term. Am J Obstet Gynecol. 2021;224(03):2960–2.96E25. doi: 10.1016/j.ajog.2020.08.103. [DOI] [PubMed] [Google Scholar]
- 83.Belkaid Y, Hand T W. Role of the microbiota in immunity and inflammation. Cell. 2014;157(01):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hou K, Wu Z X, Chen X Y et al. Microbiota in health and diseases. Signal Transduct Target Ther. 2022;7(01):135. doi: 10.1038/s41392-022-00974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee Y K, Mazmanian S K. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330(6012):1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Donald K, Petersen C, Turvey S E, Finlay B B, Azad M B. Secretory IgA: linking microbes, maternal health, and infant health through human milk. Cell Host Microbe. 2022;30(05):650–659. doi: 10.1016/j.chom.2022.02.005. [DOI] [PubMed] [Google Scholar]
- 87.Bonney E A. Immune regulation in pregnancy: a matter of perspective? Obstet Gynecol Clin North Am. 2016;43(04):679–698. doi: 10.1016/j.ogc.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Al-Nasiry S, Ambrosino E, Schlaepfer M et al. The interplay between reproductive tract microbiota and immunological system in human reproduction. Front Immunol. 2020;11:378. doi: 10.3389/fimmu.2020.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gholiof M, Adamson-De Luca E, Wessels J M. The female reproductive tract microbiotas, inflammation, and gynecological conditions. Front Reprod Health. 2022;4:963752. doi: 10.3389/frph.2022.963752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Integrative HMP (iHMP) Research Network Consortium . Proctor L M, Creasy H H, Fettweis J M et al. The Integrative Human Microbiome Project. Nature. 2019;569(7758):641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Breedveld A C, Schuster H J, van Houdt R et al. Enhanced IgA coating of bacteria in women with Lactobacillus crispatus-dominated vaginal microbiota. Microbiome. 2022;10(01):15. doi: 10.1186/s40168-021-01198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jin J, Gao L, Zou X et al. Gut dysbiosis promotes preeclampsia by regulating macrophages and trophoblasts. Circ Res. 2022;131(06):492–506. doi: 10.1161/CIRCRESAHA.122.320771. [DOI] [PubMed] [Google Scholar]
- 93.Tao Z, Chen Y, He F et al. Alterations in the gut microbiome and metabolisms in pregnancies with fetal growth restriction. Microbiol Spectr. 2023;11(03) doi: 10.1128/spectrum.00076-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hasain Z, Mokhtar N M, Kamaruddin N A et al. Gut microbiota and gestational diabetes mellitus: a review of host-gut microbiota interactions and their therapeutic potential. Front Cell Infect Microbiol. 2020;10:188. doi: 10.3389/fcimb.2020.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mitchell C, Marrazzo J. Bacterial vaginosis and the cervicovaginal immune response. Am J Reprod Immunol. 2014;71(06):555–563. doi: 10.1111/aji.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Horne A W, Stock S J, King A E. Innate immunity and disorders of the female reproductive tract. Reproduction. 2008;135(06):739–749. doi: 10.1530/REP-07-0564. [DOI] [PubMed] [Google Scholar]
- 97.Witkin S S, Linhares I M, Giraldo P. Bacterial flora of the female genital tract: function and immune regulation. Best Pract Res Clin Obstet Gynaecol. 2007;21(03):347–354. doi: 10.1016/j.bpobgyn.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 98.Anahtar M N, Byrne E H, Doherty K E et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42(05):965–976. doi: 10.1016/j.immuni.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mor G, Aldo P, Alvero A B. The unique immunological and microbial aspects of pregnancy. Nat Rev Immunol. 2017;17(08):469–482. doi: 10.1038/nri.2017.64. [DOI] [PubMed] [Google Scholar]
- 100.Nasu K, Narahara H. Pattern recognition via the toll-like receptor system in the human female genital tract. Mediators Inflamm. 2010;2010:976024. doi: 10.1155/2010/976024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kogut M H, Lee A, Santin E. Microbiome and pathogen interaction with the immune system. Poult Sci. 2020;99(04):1906–1913. doi: 10.1016/j.psj.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Agostinis C, Mangogna A, Bossi F, Ricci G, Kishore U, Bulla R.Uterine immunity and microbiota: a shifting paradigm Front Immunol 201910(OCT):2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Atay S, Gercel-Taylor C, Suttles J, Mor G, Taylor D D. Trophoblast-derived exosomes mediate monocyte recruitment and differentiation. Am J Reprod Immunol. 2011;65(01):65–77. doi: 10.1111/j.1600-0897.2010.00880.x. [DOI] [PubMed] [Google Scholar]
- 104.Villa P, Cipolla C, D'Ippolito S et al. The interplay between immune system and microbiota in gynecological diseases: a narrative review. Eur Rev Med Pharmacol Sci. 2020;24(10):5676–5690. doi: 10.26355/eurrev_202005_21359. [DOI] [PubMed] [Google Scholar]
- 105.Arnold K B, Burgener A, Birse K et al. Increased levels of inflammatory cytokines in the female reproductive tract are associated with altered expression of proteases, mucosal barrier proteins, and an influx of HIV-susceptible target cells. Mucosal Immunol. 2016;9(01):194–205. doi: 10.1038/mi.2015.51. [DOI] [PubMed] [Google Scholar]
- 106.He Y, Fu L, Li Y et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8 + T cell immunity . Cell Metab. 2021;33(05):988–1.0E10. doi: 10.1016/j.cmet.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 107.Carvalho F A, Aitken J D, Vijay-Kumar M, Gewirtz A T. Toll-like receptor-gut microbiota interactions: perturb at your own risk! Annu Rev Physiol. 2012;74(01):177–198. doi: 10.1146/annurev-physiol-020911-153330. [DOI] [PubMed] [Google Scholar]
- 108.Baker J M, Chase D M, Herbst-Kralovetz M M.Uterine microbiota: residents, tourists, or invaders? Front Immunol 20189(MAR):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maslowski K M, Vieira A T, Ng A et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ruff W E, Greiling T M, Kriegel M A. Host-microbiota interactions in immune-mediated diseases. Nat Rev Microbiol. 2020;18(09):521–538. doi: 10.1038/s41579-020-0367-2. [DOI] [PubMed] [Google Scholar]
- 111.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(06):492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mu Q, Tavella V J, Luo X M. Role of Lactobacillus reuteri in human health and diseases Front Microbiol 20189(APR):757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maloy K J, Powrie F. Regulatory T cells in the control of immune pathology. Nat Immunol. 2001;2(09):816–822. doi: 10.1038/ni0901-816. [DOI] [PubMed] [Google Scholar]
- 114.Ssemaganda A, Cholette F, Perner M et al. Endocervical regulatory T cells are associated with decreased genital inflammation and lower HIV target cell abundance. Front Immunol. 2021;12:726472. doi: 10.3389/fimmu.2021.726472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Coombes J L, Siddiqui K RR, Arancibia-Cárcamo C V et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J Exp Med. 2007;204(08):1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Duluc D, Gannevat J, Anguiano E et al. Functional diversity of human vaginal APC subsets in directing T-cell responses. Mucosal Immunol. 2013;6(03):626–638. doi: 10.1038/mi.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Monin L, Whettlock E M, Male V. Immune responses in the human female reproductive tract. Immunology. 2020;160(02):106–115. doi: 10.1111/imm.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Frew L, Stock S J. Antimicrobial peptides and pregnancy. Reproduction. 2011;141(06):725–735. doi: 10.1530/REP-10-0537. [DOI] [PubMed] [Google Scholar]
- 119.Flores-Mireles A L, Walker J N, Caparon M, Hultgren S J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(05):269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sterlin D, Fadlallah J, Slack E, Gorochov G. The antibody/microbiota interface in health and disease. Mucosal Immunol. 2020;13(01):3–11. doi: 10.1038/s41385-019-0192-y. [DOI] [PubMed] [Google Scholar]
- 121.Chen K, Magri G, Grasset E K, Cerutti A. Rethinking mucosal antibody responses: IgM, IgG and IgD join IgA. Nat Rev Immunol. 2020;20(07):427–441. doi: 10.1038/s41577-019-0261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Torcia M G. Interplay among vaginal microbiome, immune response and sexually transmitted viral infections. Int J Mol Sci. 2019;20(02):266. doi: 10.3390/ijms20020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bulmer J N, Williams P J, Lash G E.Immune cells in the placental bed Int J Dev Biol 201054(2-3):281–294. [DOI] [PubMed] [Google Scholar]
- 124.Abelius M S, Janefjord C, Ernerudh J et al. The placental immune milieu is characterized by a Th2- and anti-inflammatory transcription profile, regardless of maternal allergy, and associates with neonatal immunity. Am J Reprod Immunol. 2015;73(05):445–459. doi: 10.1111/aji.12350. [DOI] [PubMed] [Google Scholar]
- 125.Muzzio D O, Soldati R, Ehrhardt J et al. B cell development undergoes profound modifications and adaptations during pregnancy in mice. Biol Reprod. 2014;91(05):115. doi: 10.1095/biolreprod.114.122366. [DOI] [PubMed] [Google Scholar]
- 126.Lee S K, Kim C J, Kim D J, Kang J H. Immune cells in the female reproductive tract. Immune Netw. 2015;15(01):16–26. doi: 10.4110/in.2015.15.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Y, Liu Z, Sun H. Fetal-maternal interactions during pregnancy: a ‘three-in-one’ perspective. Front Immunol. 2023;14:1.19843E6. doi: 10.3389/fimmu.2023.1198430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ding J, Maxwell A, Adzibolosu N et al. Mechanisms of immune regulation by the placenta: role of type I interferon and interferon-stimulated genes signaling during pregnancy. Immunol Rev. 2022;308(01):9–24. doi: 10.1111/imr.13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wira C R, Fahey J V, Rodriguez-Garcia M, Shen Z, Patel M V. Regulation of mucosal immunity in the female reproductive tract: the role of sex hormones in immune protection against sexually transmitted pathogens. Am J Reprod Immunol. 2014;72(02):236–258. doi: 10.1111/aji.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nagamatsu T, Schust D J. The immunomodulatory roles of macrophages at the maternal-fetal interface. Reprod Sci. 2010;17(03):209–218. doi: 10.1177/1933719109349962. [DOI] [PubMed] [Google Scholar]
- 131.Harding C V, Ramachandra L, Wick M J. Interaction of bacteria with antigen presenting cells: influences on antigen presentation and antibacterial immunity. Curr Opin Immunol. 2003;15(01):112–119. doi: 10.1016/s0952-7915(02)00008-0. [DOI] [PubMed] [Google Scholar]
- 132.Barnea E R, Kirk D, Todorova K, McElhinney J, Hayrabedyan S, Fernández N. PIF direct immune regulation: blocks mitogen-activated PBMCs proliferation, promotes TH2/TH1 bias, independent of Ca(2+) Immunobiology. 2015;220(07):865–875. doi: 10.1016/j.imbio.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 133.Cauci S. Vaginal immunity in bacterial vaginosis. Curr Infect Dis Rep. 2004;6(06):450–456. doi: 10.1007/s11908-004-0064-8. [DOI] [PubMed] [Google Scholar]
- 134.Wira C R, Fahey J V, Sentman C L, Pioli P A, Shen L. Innate and adaptive immunity in female genital tract: cellular responses and interactions. Immunol Rev. 2005;206(01):306–335. doi: 10.1111/j.0105-2896.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 135.Wines B D, Hogarth P M. IgA receptors in health and disease. Tissue Antigens. 2006;68(02):103–114. doi: 10.1111/j.1399-0039.2006.00613.x. [DOI] [PubMed] [Google Scholar]
- 136.Takeuchi T, Ohno H. IgA in human health and diseases: potential regulator of commensal microbiota. Front Immunol. 2022;13:1.02433E6. doi: 10.3389/fimmu.2022.1024330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Breedveld A, van Egmond M.IgA and FcαRI: pathological roles and therapeutic opportunities Front Immunol 201910(Mar):553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Macpherson A J, Yilmaz B, Limenitakis J P, Ganal-Vonarburg S C. IgA function in relation to the intestinal microbiota. Annu Rev Immunol. 2018;36:359–381. doi: 10.1146/annurev-immunol-042617-053238. [DOI] [PubMed] [Google Scholar]
- 139.Okai S, Usui F, Yokota S et al. High-affinity monoclonal IgA regulates gut microbiota and prevents colitis in mice. Nat Microbiol. 2016;1(09):16103. doi: 10.1038/nmicrobiol.2016.103. [DOI] [PubMed] [Google Scholar]
- 140.Bunker J J, Erickson S A, Flynn T M et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science. 2017;358(6361):eaan6619. doi: 10.1126/science.aan6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sterlin D, Fadlallah J, Adams O et al. Human IgA binds a diverse array of commensal bacteria. J Exp Med. 2020;217(03):e20181635. doi: 10.1084/jem.20181635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mantis N J, Rol N, Corthésy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4(06):603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Catanzaro J R, Strauss J D, Bielecka A et al. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci Rep. 2019;9(01):13574. doi: 10.1038/s41598-019-49923-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fadlallah J, El Kafsi H, Sterlin D et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med. 2018;10(439):eaan1217. doi: 10.1126/scitranslmed.aan1217. [DOI] [PubMed] [Google Scholar]
- 145.Johansen F E, Kaetzel C S. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 2011;4(06):598–602. doi: 10.1038/mi.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ramsland P A, Willoughby N, Trist H M et al. Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1 . Proc Natl Acad Sci U S A. 2007;104(38):15051–15056. doi: 10.1073/pnas.0706028104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Safaeian M, Falk R T, Rodriguez A C et al. Factors associated with fluctuations in IgA and IgG levels at the cervix during the menstrual cycle. J Infect Dis. 2009;199(03):455–463. doi: 10.1086/596060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jones K, Savulescu A F, Brombacher F, Hadebe S. Immunoglobulin M in health and diseases: How far have we come and what next? Front Immunol. 2020;11:595535. doi: 10.3389/fimmu.2020.595535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Flajnik M F, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11(01):47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Blandino R, Baumgarth N. Secreted IgM: new tricks for an old molecule. J Leukoc Biol. 2019;106(05):1021–1034. doi: 10.1002/JLB.3RI0519-161R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Litvack M L, Post M, Palaniyar N. IgM promotes the clearance of small particles and apoptotic microparticles by macrophages. PLoS One. 2011;6(03):e17223. doi: 10.1371/journal.pone.0017223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ehrenstein M R, Notley C A. The importance of natural IgM: scavenger, protector and regulator. Nat Rev Immunol. 2010;10(11):778–786. doi: 10.1038/nri2849. [DOI] [PubMed] [Google Scholar]
- 153.Racine R, Winslow G M. IgM in microbial infections: taken for granted? Immunol Lett. 2009;125(02):79–85. doi: 10.1016/j.imlet.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Grönwall C, Vas J, Silverman G J.Protective roles of natural IgM antibodies Front Immunol 20123(Apr):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cooper N R, Nemerow G R, Mayes J T.Methods to detect and quantitate complement activation Springer Semin Immunopathol 19836(2-3):195–212. [DOI] [PubMed] [Google Scholar]
- 156.Gupta S, Gupta A.Selective IgM deficiency-an underestimated primary immunodeficiency Front Immunol 20178(SEP):1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Magri G, Comerma L, Pybus M et al. Human secretory IgM emerges from plasma cells clonally related to gut memory b cells and targets highly diverse commensals. Immunity. 2017;47(01):118–1.34E10. doi: 10.1016/j.immuni.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Macpherson A J, Gatto D, Sainsbury E, Harriman G R, Hengartner H, Zinkernagel R M. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288(5474):2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 159.Li Y, Shen H, Zhang R et al. Immunoglobulin M perception by FcμR. Nature. 2023;615(7954):907–912. doi: 10.1038/s41586-023-05835-w. [DOI] [PubMed] [Google Scholar]
- 160.Shibuya A, Sakamoto N, Shimizu Y et al. Fc α/μ receptor mediates endocytosis of IgM-coated microbes. Nat Immunol. 2000;1(05):441–446. doi: 10.1038/80886. [DOI] [PubMed] [Google Scholar]
- 161.Devito C, Ellegård R, Falkeborn T et al. Human IgM monoclonal antibodies block HIV-transmission to immune cells in cervico-vaginal tissues and across polarized epithelial cells in vitro. Sci Rep. 2018;8(01):10180. doi: 10.1038/s41598-018-28242-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kirkland D, Benson A, Mirpuri J et al. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity. 2012;36(02):228–238. doi: 10.1016/j.immuni.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Devito C, Broliden K, Kaul R et al. Mucosal and plasma IgA from HIV-1-exposed uninfected individuals inhibit HIV-1 transcytosis across human epithelial cells. J Immunol. 2000;165(09):5170–5176. doi: 10.4049/jimmunol.165.9.5170. [DOI] [PubMed] [Google Scholar]
- 164.Dunkelberger J R, Song W C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20(01):34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 165.Kanda N, Tamaki K.Estrogen enhances immunoglobulin production by human PBMCs J Allergy Clin Immunol 1999103(2, Pt 1):282–288. [DOI] [PubMed] [Google Scholar]
- 166.So N SY, Ostrowski M A, Gray-Owen S D. Vigorous response of human innate functioning IgM memory B cells upon infection by Neisseria gonorrhoeae. J Immunol. 2012;188(08):4008–4022. doi: 10.4049/jimmunol.1100718. [DOI] [PubMed] [Google Scholar]
- 167.Rodriguez-Garcia M, Patel M V, Shen Z, Wira C R. The impact of aging on innate and adaptive immunity in the human female genital tract. Aging Cell. 2021;20(05):e13361. doi: 10.1111/acel.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mestecky J, Fultz P N. Mucosal immune system of the human genital tract. J Infect Dis. 1999;179 03:S470–S474. doi: 10.1086/314806. [DOI] [PubMed] [Google Scholar]
- 169.Chan D, Bennett P R, Lee Y S et al. Microbial-driven preterm labour involves crosstalk between the innate and adaptive immune response. Nat Commun. 2022;13(01):975. doi: 10.1038/s41467-022-28620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pudney J, Quayle A J, Anderson D J. Immunological microenvironments in the human vagina and cervix: mediators of cellular immunity are concentrated in the cervical transformation zone. Biol Reprod. 2005;73(06):1253–1263. doi: 10.1095/biolreprod.105.043133. [DOI] [PubMed] [Google Scholar]
- 171.Klemola T. Immunohistochemical findings in the intestine of IgA-deficient persons: number of intraepithelial T lymphocytes is increased. J Pediatr Gastroenterol Nutr. 1988;7(04):537–543. doi: 10.1097/00005176-198807000-00010. [DOI] [PubMed] [Google Scholar]
- 172.Bunker J J, Flynn T M, Koval J C et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43(03):541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bunker J J, Bendelac A. IgA responses to microbiota. Immunity. 2018;49(02):211–224. doi: 10.1016/j.immuni.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dalal S R, Chang E B. The microbial basis of inflammatory bowel diseases. J Clin Invest. 2014;124(10):4190–4196. doi: 10.1172/JCI72330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kanai T, Kawamura T, Dohi T et al. TH1/TH2-mediated colitis induced by adoptive transfer of CD4+CD45RB high T lymphocytes into nude mice. Inflamm Bowel Dis. 2006;12(02):89–99. doi: 10.1097/01.MIB.0000197237.21387.mL. [DOI] [PubMed] [Google Scholar]
- 176.Uo M, Hisamatsu T, Miyoshi J et al. Mucosal CXCR4+ IgG plasma cells contribute to the pathogenesis of human ulcerative colitis through FcγR-mediated CD14 macrophage activation. Gut. 2013;62(12):1734–1744. doi: 10.1136/gutjnl-2012-303063. [DOI] [PubMed] [Google Scholar]
- 177.Chan Y, Fornace K, Wu L et al. Determining seropositivity—a review of approaches to define population seroprevalence when using multiplex bead assays to assess burden of tropical diseases. PLoS Negl Trop Dis. 2021;15(06):e0009457. doi: 10.1371/journal.pntd.0009457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dowlatshahi S, Shabani E, Abdekhodaie M J. Serological assays and host antibody detection in coronavirus-related disease diagnosis. Arch Virol. 2021;166(03):715–731. doi: 10.1007/s00705-020-04874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Durazzi F, Sala C, Castellani G, Manfreda G, Remondini D, De Cesare A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci Rep. 2021;11(01):3030. doi: 10.1038/s41598-021-82726-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.López-Aladid R, Fernández-Barat L, Alcaraz-Serrano V et al. Determining the most accurate 16S rRNA hypervariable region for taxonomic identification from respiratory samples. Sci Rep. 2023;13(01):3974. doi: 10.1038/s41598-023-30764-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Brown E L, Essigmann H T, Hoffman K L et al. Impact of diabetes on the gut and salivary IgA microbiomes. Infect Immun. 2020;88(12):1–12. doi: 10.1128/IAI.00301-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jackson M A, Pearson C, Ilott N E et al. Accurate identification and quantification of commensal microbiota bound by host immunoglobulins. Microbiome. 2021;9(01):33. doi: 10.1186/s40168-020-00992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Morton J T, Marotz C, Washburne A et al. Establishing microbial composition measurement standards with reference frames. Nat Commun. 2019;10(01):2719. doi: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Palm N W, de Zoete M R, Cullen T W et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158(05):1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Silverman M A, Green J L. Insight into host-microbe interactions using microbial flow cytometry coupled to next-generation sequencing. J Pediatric Infect Dis Soc. 2021;10 04:S106–S111. doi: 10.1093/jpids/piab092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bos N A, Bun J CAM, Popma S H et al. Monoclonal immunoglobulin A derived from peritoneal B cells is encoded by both germ line and somatically mutated VH genes and is reactive with commensal bacteria. Infect Immun. 1996;64(02):616–623. doi: 10.1128/iai.64.2.616-623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kroese F GM, de Waard R, Bos N A. B-1 cells and their reactivity with the murine intestinal microflora. Semin Immunol. 1996;8(01):11–18. doi: 10.1006/smim.1996.0003. [DOI] [PubMed] [Google Scholar]
- 188.van der Waaij L A, Mesander G, Limburg P C, van der Waaij D. Direct flow cytometry of anaerobic bacteria in human feces. Cytometry. 1994;16(03):270–279. doi: 10.1002/cyto.990160312. [DOI] [PubMed] [Google Scholar]
- 189.Moor K, Fadlallah J, Toska A et al. Analysis of bacterial-surface-specific antibodies in body fluids using bacterial flow cytometry. Nat Protoc. 2016;11(08):1531–1553. doi: 10.1038/nprot.2016.091. [DOI] [PubMed] [Google Scholar]
- 190.Conrey P E, Denu L, O'Boyle K C et al. IgA deficiency destabilizes homeostasis toward intestinal microbes and increases systemic immune dysregulation. Sci Immunol. 2023;8(83):eade2335. doi: 10.1126/sciimmunol.ade2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wilmore J R, Gaudette B T, Gomez Atria D et al. Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe. 2018;23(03):302–311000. doi: 10.1016/j.chom.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kan B, Razzaghian H R, Lavoie P M. An immunological perspective on neonatal sepsis. Trends Mol Med. 2016;22(04):290–302. doi: 10.1016/j.molmed.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Emerson J B, Adams R I, Román C MB et al. Schrödinger's microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome. 2017;5(01):86. doi: 10.1186/s40168-017-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lambrecht J, Schattenberg F, Harms H, Mueller S. Characterizing microbiome dynamics - flow cytometry based workflows from pure cultures to natural communities. J Vis Exp. 2018;2018(137):58033. doi: 10.3791/58033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Koch M A, Reiner G L, Lugo K A et al. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell. 2016;165(04):827–841. doi: 10.1016/j.cell.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Doron I, Leonardi I, Li X V et al. Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell. 2021;184(04):1017–1.031E17. doi: 10.1016/j.cell.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kau A L, Planer J D, Liu J et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7(276):276ra24. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tiu C K, Zhu F, Wang L F, de Alwis R. Phage ImmunoPrecipitation Sequencing (PhIP-Seq): the promise of high throughput serology. Pathogens. 2022;11(05):568. doi: 10.3390/pathogens11050568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lingasamy P, Tobi A, Haugas M et al. Bi-specific tenascin-C and fibronectin targeted peptide for solid tumor delivery. Biomaterials. 2019;219:119373. doi: 10.1016/j.biomaterials.2019.119373. [DOI] [PubMed] [Google Scholar]
- 200.Lingasamy P.Development of Multitargeted Tumor Penetrating PeptidesUniversity of Tartu;2020http://hdl.handle.net/10062/70738
- 201.Larman H B, Zhao Z, Laserson U et al. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol. 2011;29(06):535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mohan D, Wansley D L, Sie B M et al. PhIP-Seq characterization of serum antibodies using oligonucleotide-encoded peptidomes. Nat Protoc. 2018;13(09):1958–1978. doi: 10.1038/s41596-018-0025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Larman H B, Laserson U, Querol L et al. PhIP-Seq characterization of autoantibodies from patients with multiple sclerosis, type 1 diabetes and rheumatoid arthritis. J Autoimmun. 2013;43:1–9. doi: 10.1016/j.jaut.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Román-Meléndez G D, Monaco D R, Montagne J M et al. Citrullination of a phage-displayed human peptidome library reveals the fine specificities of rheumatoid arthritis-associated autoantibodies. EBioMedicine. 2021;71:103506. doi: 10.1016/j.ebiom.2021.103506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Xu G J, Kula T, Xu Q et al. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348(6239):aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cantarelli C, Jarque M, Angeletti A et al. A comprehensive phenotypic and functional immune analysis unravels circulating anti-phospholipase A2 receptor antibody secreting cells in membranous nephropathy patients. Kidney Int Rep. 2020;5(10):1764–1776. doi: 10.1016/j.ekir.2020.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Angkeow J W, Monaco D R, Chen A et al. Phage display of environmental protein toxins and virulence factors reveals the prevalence, persistence, and genetics of antibody responses. Immunity. 2022;55(06):1051–1.066E7. doi: 10.1016/j.immuni.2022.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Venkataraman T, Swaminathan H, Arze C A et al. Comprehensive profiling of antibody responses to the human anellome using programmable phage display. Cell Rep. 2022;41(12):111754. doi: 10.1016/j.celrep.2022.111754. [DOI] [PubMed] [Google Scholar]
- 209.Vogl T, Klompus S, Leviatan S et al. Population-wide diversity and stability of serum antibody epitope repertoires against human microbiota. Nat Med. 2021;27(08):1442–1450. doi: 10.1038/s41591-021-01409-3. [DOI] [PubMed] [Google Scholar]
- 210.Leviatan S, Vogl T, Klompus S, Kalka I N, Weinberger A, Segal E. Allergenic food protein consumption is associated with systemic IgG antibody responses in non-allergic individuals. Immunity. 2022;55(12):2454–2.469E9. doi: 10.1016/j.immuni.2022.11.004. [DOI] [PubMed] [Google Scholar]
- 211.Lifelines Cohort Study . Andreu-Sánchez S, Bourgonje A R, Vogl T et al. Phage display sequencing reveals that genetic, environmental, and intrinsic factors influence variation of human antibody epitope repertoire. Immunity. 2023;56(06):1376–1.392E11. doi: 10.1016/j.immuni.2023.04.003. [DOI] [PubMed] [Google Scholar]
- 212.Bourgonje A R, Andreu-Sánchez S, Vogl T et al. Phage-display immunoprecipitation sequencing of the antibody epitope repertoire in inflammatory bowel disease reveals distinct antibody signatures. Immunity. 2023;56(06):1393–1.409E9. doi: 10.1016/j.immuni.2023.04.017. [DOI] [PubMed] [Google Scholar]


