Abstract
Multicellular microbial communities are the predominant form of existence for microorganisms in nature. As one of the most primitive social organisms, Myxococcus xanthus has been an ideal model bacterium for studying intercellular interaction and multicellular organization. Through previous genetic and EM studies, various extracellular appendages and matrix components have been found to be involved in the social behavior of M. xanthus, but none of them was directly visualized and analyzed under native conditions. Here, we used atomic force microscopy (AFM) imaging and in vivo force spectroscopy to characterize these cellular structures under native conditions. AFM imaging revealed morphological details on the extracellular ultrastructures at an unprecedented resolution, and in vivo force spectroscopy of live cells in fluid allowed us to nanomechanically characterize extracellular polymeric substances. The findings provide the basis for AFM as a useful tool for investigating microbial-surface ultrastructures and nanomechanical properties under native conditions.
Keywords: force spectroscopy, nonomechanics, bacteria, swarm
Bacteria live mainly as single- or multispecies multicellular communities (1, 2), usually assuming the form of surface-associated cell assemblages, or biofilms (3-5). Intercellular communication and concerted multicellular activities, through which the cells can differentiate and produce spatially organized structures, are common within microbial communities (refs. 6-8 and reviewed in ref. 2). An important component of these structures is the matrix, formed by extracellular polymeric substances, which plays a critical role in collective motility. Extensive cell-cell and cell-matrix interaction is essential to the building and maintaining of the microcommunities, and these interactions have been revealed in detail with scanning EM (SEM) (e.g., refs. 9 and 10). However, despite the high resolution that EM can achieve, the extensive sample processing involved limits its application in visualization under physiological conditions.
One of the best-studied social bacteria, Myxococcus xanthus, has been an excellent model system for investigating the intercellular interaction and multicellular organizations in microbial communities. When nutrients are available, M. xanthus forms swarms and cooperatively pools extracellular digestive enzymes to prey on other bacteria (11, 12). When the food supply runs low, cells initiate a complex developmental program that leads to the formation of dome-like structures called fruiting bodies, inside which individual cells undergo sporulation (13).
A key characteristic in both M. xanthus vegetative swarming and developmental aggregation is large-scale coordinated motion, which is realized through social (S) gliding motility (14, 15). Genetic and behavioral analyses revealed that S motility requires at least three major cellular components: type IV pili (TFP), extracellular fibril material, and LPS O antigen (16-18). The role of LPS O antigen in S motility remains elusive (18), but both TFP and fibril material have been studied in greater detail. M. xanthus TFP were seen under EM as polar filaments measuring 5-7 nm in diameter and 4-10 μm in length (19). Previous studies have revealed that TFP-dependent surface motilities are achieved through TFP extension and attachment to an external substrate followed by retraction, which pulls the cells forward (20-22). Extracellular fibril material has been visualized under SEM as 30- to 50-nm-thick intercellular fibers (23) that form a mesh-like network surrounding the cell body (24). Because mutants lacking fibril material are unable to agglutinate in liquid suspension, it was hypothesized that cohesion is the major role of fibril material (23). Biochemical analysis suggested that fibril material is composed of approximately equal amounts of protein and carbohydrate (25). A recent study indicated that, in addition to mere cohesion, the amine-containing polysaccharide of the fibril material may function to trigger pilus retraction (26), providing another perspective on the role of fibril material in S motility.
Despite extensive genetic and functional studies, neither TFP nor fibril material had been imaged under native conditions. Although the social behaviors of M. xanthus have been investigated in detail, its cellular organization in a social group had never been revealed under physiological conditions at high resolution. In this study, we used atomic force microscopy (AFM) (59) to visualize the cellular ultrastructures as well as social swarming groups of M. xanthus, revealing their morphological details under native conditions at an unprecedented resolution. By imaging a panel of M. xanthus mutants (Table 1), we were able to correlate nanoscale morphology and nanomechanical characterization of the cell wall properties with the mutants' genetic backgrounds, demonstrating the quantifiable phenotypic differences among the M. xanthus mutants. Through AFM imaging and in vivo force spectroscopy, we were also able to perform nanomechanical characterization of the cell surface and extracellular polymeric substances on live M. xanthus cells. These results revealed the elasticity of the M. xanthus surface and the mechanical nature of the adhesion mediated by fibril material; the latter provided nanomechanical evidence for the role of extracellular polysaccharides in cellular cohesion and the social behaviors of M. xanthus.
Table 1. Characteristics of the five M. xanthus strains used in this study.
| Genotype (strain) | Mutant characteristics | Pili | Fibril | LPS O antigen | S motility | Slime |
|---|---|---|---|---|---|---|
| Wild type (dk 1622) | • | • | • | • | • | |
| pilA (dk 10407) | Missing pili | ○ | • | • | ○ | • |
| dif (sw504) | Missing fibril material | ▪ | ○ | • | ○ | • |
| stk (dk3088) | Fibril overproduction | • | ▪ | • | ▾ | • |
| wzt wzm wbgA LPS O antigen− (hk1324) | Missing LPS O antigen | • | • | ○ | ○ | • |
•, Present; ○, not present; ▪, these cell types display an excess of the specified extracellular material; ▾, this cell type exhibits increased production of cell-surface fibrils, cell cohesion, and group S motility.
Materials and Methods
Bacterial Strains and Growth Conditions. The M. xanthus strains used were DK1622 (wild type) (19), DK10407 (pilA, pilus-) (strain supplied by D. Kaiser, Stanford University School of Medicine, Stanford, CA), SW504 (ΔdifA, fibril-) (27), DK3088 (sglA1 stk, fibril++) (28), and HK1324 (Δwzt wzm wbgA (Ω Kanr), LPS O antigen-) (18). The strains were grown at 32°C in CYE medium (10 g/liter casitone, 5 g/liter yeast extract, and 8 mM MgSO4 in 10 mM Mops buffer, pH 7.6) on a rotary shaker at 225 rpm.
Cell Immobilization. For imaging in air, logarithmic-phase M. xanthus cells were collected by centrifugation at 6,000 × g for 5 min, washed in Mops buffer (10 mM Mops/4 mM MgSO4, pH 7.6), and resuspended to 1 × 107 cells per ml in the same buffer. A 12-well Cel-Line glass slide (Erie Scientific, Portsmouth, NH) was cleaned with 75% ethanol and wiped dry with lens paper. Resuspended M. xanthus cells (20 μl) were added to one well for 30 min, and excess liquid was removed with filter paper. The slide was then air-dried before imaging. To image cell groups, cells were resuspended to 1 × 108 cells per ml and prepared as above; a wash step was added after the 30-min incubation to remove the unattached cells.
For imaging in fluid, samples were prepared as described in ref. 29, with slight modifications. An 18-× 18-mm coverslip was cleaned with 75% ethanol. A drop of 1% polyethyleneimine (PEI) (Mr 1,200) dissolved in deionized water was placed on one side of the glass and allowed to adsorb for 3 h, after which the drop was decanted, and the coverslip was rinsed in water and air-dried. Logarithmic-phase M. xanthus cells were collected by centrifugation at 6,000 × g for 5 min, washed with PBS (pH 7.4), and resuspended to 109 cells per ml. For fixing, the cells were stirred in 2.5% (vol/vol) glutaraldehyde for 2 h at 4°C, rinsed, and resuspended in 1 mM Tris (pH 7.5). One drop of the cell suspension (with or without fixing) was placed on a PEI-coated coverslip and placed in a CentriVap concentrator (Labconco, Kansas City, MO) (without spinning) for 20-30 min at 35°C to evaporate excess water without drying the cells. The coverslip was then adhered to a Petri dish and submerged in deionized water for AFM imaging. After imaging, cell viability was verified by streaking the M. xanthus cells on a CYE agar plate and checking growth after 24 h.
AFM. All imaging in air and fluid was carried out with a Nanoscope IV Bioscope (Veeco Digital Instruments, Santa Barbara, CA). Oxide-sharpened cantilevers (OTR4, Olympus, Tokyo) with spring constants of 0.02 N/m and a tip radius of <10 nm were used in contact mode. Fluid imaging and mechanical measurements were performed at room temperature. All force measurements were recorded at a pulling rate of 1 Hz. “Height” and “deflection” images were simultaneously recorded. Deflection images do not represent the true topography of the sample; however, they consistently revealed a higher sensitivity to small surface features and yielded images with greater detail (30). Images presented in this study are deflection images, but the quantitative measurements of cell structures were taken from the height data on the same sample.
Results
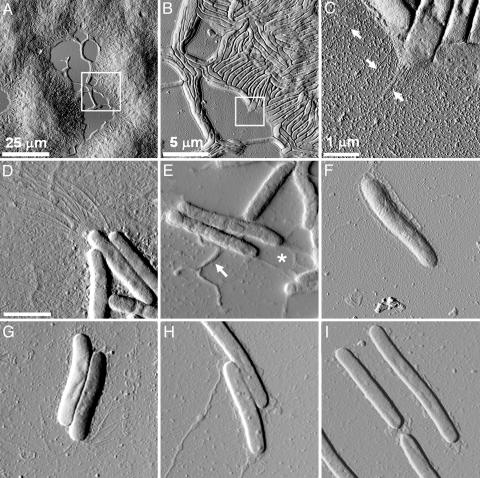
M. xanthus Cell Organization in a Social Group. Actively growing wild-type M. xanthus cells (DK1622) were dotted at various concentrations on glass slides and allowed to settle for 30 min (see Materials and Methods for details). The samples were washed several times to remove the unattached cells, air dried, and imaged under AFM in contact mode. As shown in Fig. 1A, attached cells were seen as large mound-like cell packs, and aligned individual cells were found connecting different “mounds.” When we zoomed in on the edge of a cell mound, where cells were in a single layer, the cell alignment and organization were clearly seen (Fig. 1B). Individual cells measured ≈5 μm long (sometimes as long as 10 μm) and ≈800 nm wide. When a higher-resolution image was taken on the edge of the cell group, pili were clearly seen at the cell poles (arrows, Fig. 1C).
Fig. 1.
AFM deflection-mode images of large social groups and individual M. xanthus cells in air. (A) A 100-μm2 scan of large mounds of wild-type cells. (B) A higher-resolution scan of the marked area in A reveals cellular ordering along the edge of the social group in domains of ≈10 cells. (C) A further, higher-resolution scan of the area marked in B reveals the presence of pili at the cell poles. (D-I) Individual cells of M. xanthus mutants. (Scale bar, 2 μm.) (D) Wild-type DK1622, showing polar pili. (E) Wild-type DK1622 cells displaying slime-like substances (*) and extruding blebs (arrow). (F) pilA mutant DK10407, showing the absence of pili at the cell pole. (G) dif mutant SW504, showing the presence of long pili that bend toward the cell body. (H) stk mutant DK3088, displaying an excess of extracellular substances in the form of filaments with variable diameters from 15 to 65 nm. (I) The LPS O antigen mutant HK1324.
Cell-Surface Ultrastructures. Wild type and S-motility mutants were imaged in air to examine their cellular ultrastructures. A lower concentration of actively growing M. xanthus cells was dotted directly from liquid culture onto glass slides. As described in Table 1, many S-motility mutants are defective in certain extracellular structures. The pilA mutant lacks pili, the dif mutant lacks fibril material and has been reported to be overpiliated (26), the stk mutant overproduces fibril material, and the LPS O antigen mutant lacks LPS O antigen (a component of the outer cell wall). To examine the effects of these genetic mutations on nanoscale surface morphology under native conditions, we harvested the cells from actively growing cultures, air-dried, and imaged directly with AFM (see Materials and Methods).
Polar TFP. As shown in Fig. 1D, TFP filaments measuring ≈4-6 μm in length and 5-8 nm in diameter could be clearly seen at the cell poles of wild-type strain DK1622, whereas the filaments were missing from the pilA mutant (Fig. 1F). The filaments extended from the cell pole and spread out from the long axis of the cell, with a slight curve in all filaments. This native pili morphology is rather different from that seen in EM micrographs, where pili appeared as randomly arranged fibers (19). It is interesting to note that cells adjacent to each other often have pili at the same end (Fig. 1D). Because little sample preparation was involved for AFM imaging, this observation suggested that cells in close proximity to each other may coordinate their pili-shooting direction.
Overpiliation has been reported in the dif mutants that lack extracellular fibril material (26). When a dif mutant, SW504 (ΔdifA), was examined under AFM, the cells did display longer pili at cell poles (Fig. 1G). Notably, the pili morphology in the dif mutant also appeared to be different from that of the wild-type cells, with the majority bending backwards toward the cell body instead of shooting forward as in the wild type (Fig. 1G). It has been reported that pili in dif mutants fail to retract, leading to the overpiliation phenotype and S-motility defect (26). The pili morphology observed here might be a result of the pili overextension and may correlate with the defective pili function in the S motility in dif mutants.
Extracellular polysaccharide matrix. The dif mutants lack extracellular fibril material. SEM observations had originally defined “fibrils” as a matrix material consisting of branching extensions ≈30 nm in diameter that surrounds the wild-type cells (23, 24). Later studies revealed that these fibrils likely form a mesh of extracellular polymeric substances over the entire cell body (25, 31). When wild-type M. xanthus cells were examined by using AFM, no filamentous structures were seen on the cell body (Fig. 1 D and E). Nevertheless, several structural features could be noticed on the AFM images. (i) The cell surfaces appeared rather “rough,” as shown in Fig. 1D, and (ii) slime-like structures were often seen covering or extending from the cell body, as shown in Fig. 1E. The dif mutant, however, appeared much “smoother” (Fig. 1G). To quantify the visual differences, a roughness analysis of the cellular surfaces was carried out. As detailed in Supporting Text, which is published as supporting information on the PNAS web site, we define roughness (R) as the standard deviation of the height values (h) away from the mean height (h0) of a given scan line over the cell surface (see Fig. 4, which is published as supporting information on the PNAS web site). The Rrms for the wild-type and pilA cells were determined to be 4.30 ± 1.09 nm and 3.43 ± 0.91 nm, respectively. However, the dif mutant displayed much less roughness (2.54 ± 0.77 nm).
The M. xanthus stk mutants are known to have constitutively high polysaccharide production and a higher-than-normal level of fibril material (28, 32). When this mutant (DK3088) was examined under AFM, an excessive amount of slime-like substances were seen on the cell surface, and long filamentous structures were often found extending from the cell body (Fig. 1H). The stk mutants were known to exhibit a variety of properties including the clumping of cells during growth in liquid culture, rapid agglutination, and the formation of colonies in which cells adhere tightly to each other and the agar surface (28). The striking amount of extracellular substances observed under physiological conditions provided the structural basis for these phenotypes. The surface roughness of the stk mutant was also measured and averaged 7.16 ± 2.74 nm, significantly higher than that of wild-type cells (4.30 ± 1.09 nm).
Another important type of extracellular polysaccharide in M. xanthus is LPS. M. xanthus LPS is typical of Gram-negative bacteria and consists of lipid A, which forms the outer leaflet of the outer membrane bilayer; core, which is a chain of carbohydrates attached to lipid A; and O antigen, which contains a variable number of repeating oligosaccharide units and extends outward from the core (33). Genetic studies showed that the wzm wzt wbgA genes in the sasA locus of M. xanthus encode LPS O antigen biogenesis proteins, and the LPS O antigen mutant (HK1324, Δwzm wzt wbgA) was defective in S motility (18). When this mutant was imaged with AFM, it exhibited a relatively “clean” cell surface (Fig. 1I) as compared with wild type. The cell-surface roughness of the mutant was quantified and averaged 4.05 ± 1.42 nm, comparable to that of wild-type cell surface. This finding is expected because LPS O antigen mutants exhibit a wild-type level of extracellular fibril material (18), which presumably masks the cell wall and contributes to a wild-type-like cell-surface roughness.
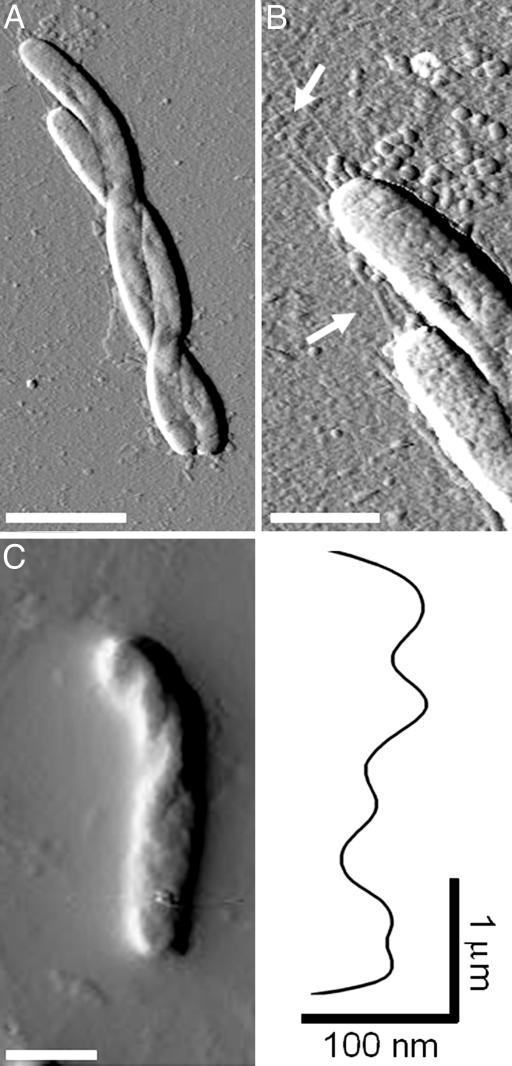
Morphology of Gliding Cells. Aside from the ultrastructure imaging, some unique cell morphology was also observed. As shown in Fig. 2A, M. xanthus cells were occasionally seen curling their cell bodies and twisting about each other, leading to a braid-like morphology. Bending and flexing of the cell body was often seen in M. xanthus cells and was believed to be a means for directional change during gliding (34). The formation observed here suggested that adjacent cells can somehow coordinate their flexing to twist around each other. Interestingly, when examined at a higher resolution, extending pili could be seen at the same cell pole of both twisted cells (Fig. 2B), demonstrating again that pili on adjacent cells tend to shoot from the same cell ends.
Fig. 2.
The twisting morphology of M. xanthus cells. (A) Two cells twisted around each other. (Scale bar, 2.5 μm.) (B) A higher-resolution image of the cells in A, showing pili at the same cell poles of both cells. (Scale bar, 1 μmin both B and C.) (C) Helical twists in a single cell and a height profile of the long axis of the cell body, revealing distinct 25- to 200-nm bumps separated by ≈500-nm “valleys.”
Another noteworthy observation involves individual cell morphology. Earlier studies using shock-freezing and SEM revealed motility-associated surface patterns in gliding M. xanthus (35). The cells appeared to be “rotated” along the long axis of the cell, and the rotation patterns were lost in sodium-azide-treated cells, suggesting that the rotation was associated with active gliding (35). Under AFM, similar patterns were observed (Fig. 2C). Individual cells were seen to be twisted along their long axis, with a helical surface fold running through the cell body. A height profile of the twisted cell body revealed ≈25- to 100-nm “peaks” separated by ≈500-nm “valleys” on the helical patterns along the cell axis (Fig. 2C), comparable to the patterns obtained by Lunsdorf et al. (35) (600- to 1,000-nm separations). When the cells were treated with 0.02% sodium azide for 10 min, no twisting was seen in the entire sample (data not shown), demonstrating that these morphological changes were correlated with active gliding, rather than representing a drying artifact.
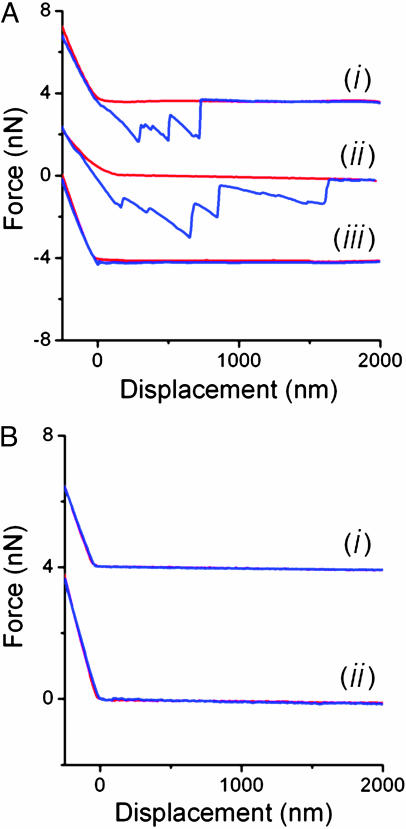
Local Elasticity of Live M. xanthus Cells in Fluid. The ability of M. xanthus to flex and twist its cell body suggests the high level of flexibility in live M. xanthus cells. Because of its force-spectroscopy capacity, AFM allows the investigation of cell local flexibility or elasticity that is unattainable with any other imaging tools. Wild-type M. xanthus cells were immobilized on a glass coverslip, submerged in fluid (see Materials and Methods), and probed with AFM. In contrast to imaging in air, the scanning of the AFM tip in fluid caused slight movements of the cell bodies that compromised image quality (see Fig. 5, which is published as supporting information on the PNAS web site). However, these cells were stable enough to be isolated and used for the determination of the local cell-wall elasticity or “stiffness” (Young's modulus, E) by measuring force curves (Fig. 3A and Supporting Text) on the cell (36). As detailed in Supporting Text, E can be determined by converting force-displacement curves (Fig. 3A) into force-indentation curves obtained on stable areas of cell surfaces (Fig. 6, which is published as supporting information on the PNAS web site). Based on these curves, wild-type cells were found to have a local Young's modulus of 0.25 ± 0.18 million pascals (MPa) (see Supporting Text for calculation details). After glutaraldehyde fixation (see Materials and Methods), wild-type cells displayed an E of 1.34 ± 0.66 MPa, demonstrating a significant change in cell-surface elasticity upon glutaraldehyde treatment.
Fig. 3.
Force-displacement curves. (A) Force curves measured on living wild-type (curve i), stk (curve ii), and glutaraldehyde-fixed wild-type M. xanthus (curve iii) cells (the curves are shifted 4 nN for clarity). Force-displacement curves were recorded as “approach” (red) and “retraction” (blue) curves. During the approach curve, the force acting on the AFM cantilever was recorded as a function of the displacement of the piezoelectric crystal, which moves the cantilever toward the sample. The force acting on the cantilever remains at zero as long as the AFM cantilever is not in contact with a surface, and the force increases monotonically after contact. (B) Force curves measured on a bare portion of the substrate before (curve i) and after (curve ii) the tip was used for force-spectroscopy measurements on the living cells.
Although measurements of bacterial turgor pressure and local spring constants have been reported in refs. 37-39, very little is known about the Young's modulus of bacterial cell walls, and the AFM measurement of E on a living bacterium in aqueous conditions has not been previously reported. Studies using “bacteria threads” have estimated E on “wet” Bacillus subtilis at ≈30 MPa (40), 100 times higher that the values we obtained with AFM on unfixed M. xanthus in aqueous conditions. AFM has been used to measure the Young's modulus on yeast cells (averaging ≈1 MPa) (36, 41) and mammalian cells, which have highly variable and spatially dependent E, and usually falls in the 1- to 200-kPa range (42-45). The E value we obtained on wild-type M. xanthus cells therefore reflects the relative stiffness of the M. xanthus cell wall as compared with other organisms.
In Vivo Force Spectroscopy of Extracellular Fibrils on the M. xanthus Cell Surface. Contact-mediated cell-cell interactions are an important aspect of the social behavior of M. xanthus and are facilitated by extracellular fibril material that exists on the surface of the cells (17, 23, 24). The force-spectroscopy capacity of AFM allows us to investigate the nanomechanical properties of the M. xanthus surface adhesive molecules.
Wild-type cells were immobilized in liquid as described above, and force-displacement curves were measured on the cell by lowering the tip and pressing it against the cell surface with a force of ≈10 nN. Upon tip retraction, a sequence of rupture events occurred at distances of 1-3 μm, presumably arising from the breakage of multiple adhesions between the AFM tip and the cell-surface substances (Fig. 3A). The cantilever displacement always returned to its initial zero position after the series of rupture events. Similar retraction curves have been reported for other adhesive polymers (DNA, proteins, and polysaccharides) (46-49), suggesting that the adhesive substances on the M. xanthus cell surface were extracellular polymeric molecules. Wild-type cells fixed with glutaraldehyde, which cross-links the extracellular polymers (see Materials and Methods), displayed retraction curves consistent with small or absent adhesion events (Fig. 3A, curve iii), thus confirming that it is extracellular polymer that adheres to AFM tips. Force curves were also measured on bare substrate, before and after measurement on the cell, confirming that there was no contamination on the AFM tip that may have caused nonspecific adhesion between the tip and the cell surface (Fig. 3B). Therefore, we ascribe the adhesion events to the stretching of extracellular polymeric substances with the AFM tip.
The average adhesion force of each rupture event is ≈2.5 nN, significantly greater than forces obtained on most other microbial surfaces (≈20-900 pN) (50, 51), suggesting the high degree of surface adhesiveness in this social bacterium. The stk mutants were examined under the same conditions and exhibited a greater number of major adhesions (one to nine events) compared with the wild type (one to three events). However, both cell types exhibit a similar average adhesion force (Fig. 3A), indicating that the fibril material from both strains is similar in its chemical adhesion properties. More than 100 measurements were made on each cell type for the force-spectroscopy study. The adhesion events on the stk mutant occurred after the tip was retracted more than 1 μm, whereas wild-type adhesion events usually ceased after ≈1 μm of tip retraction (Fig. 3A). Although the measured retraction length is partly determined by where the tip contacts the fibril, stk cells still display a 50-80% longer retraction length than do the wild type, indicating that the fibril-material molecules on the stk mutant are longer, in general, than those of the wild-type cells. These observations are consistent with the fibril-overproducing phenotype of the stk mutant and the morphological features observed by using AFM (Fig. 1H).
These in vivo systems are complex and very different from the idealized systems presented in previous polysaccharide force-spectroscopy studies (52, 53). In an artificial system, the concentration of polysaccharide on a surface can be precisely controlled so that only one polysaccharide filament can be pulled for each force-spectroscopy measurement. On a cell surface, however, this precision is impossible to achieve. Fibril material forms a complex mesh-like structure that has not been precisely determined, but the transient and local contact between an AFM tip and a cell surface is a reasonable model for examining the adhesive property of the M. xanthus cell surface.
Discussion
In this study, we achieved high-resolution AFM imaging of the social bacterium M. xanthus, revealing not only the cell coordination and organization in a social gliding group (Fig. 1 A-C) but also the morphological details of extracellular ultrastructures (TFP and extracellular substances) (Fig. 1 D and E) under native conditions and at a resolution comparable to EM. Three noteworthy observations were made during imaging. (i) Pili, the S-motility motor, were seen on the same pole of a group of active cells (considering the minimal sample preparation before imaging) (Fig. 1D), revealing that pili on adjacent gliding cells may coordinate shooting direction as a means to concert S motility. (ii) Roughness analysis on a variety of mutants with altered extracellular polysaccharide production was performed and clearly correlated the changes in surface topology with the genetic defects, allowing quantitative morphological characterization of the extracellular polysaccharide mutants. (iii) Individual M. xanthus cells with helical surface patterns obviously related to gliding were seen (Fig. 2), confirming the earlier discoveries made through SEM (35) and suggesting that gliding motility is correlated with helical twisting along the long axis of the cell body.
In addition to high-resolution imaging, the mechanical properties and surface features of M. xanthus cells were investigated with AFM in aqueous conditions. The local modulus of elasticity (Young's modulus, E), which averaged 0.25 MPa, was determined for live M. xanthus cells. This E value falls between that of yeast cells (≈1 MPa) (36, 41) and mammalian cells (≈1-200 kPa), measured with AFM (42-45). AFM measurements are innately local and deal with mechanical properties of the cell at the nanoscale. Therefore, the AFM results indicate the mechanical properties of the samples at local scales. The relatively small value of E for M. xanthus provided mechanical evidence for the often observed flexing of the cell body (Fig. 2) (34) and for the helical twisting observed on gliding cells (Fig. 2) (35). When the mechanism of adventurous (A) motility was investigated, it was proposed that slime-hydration extrusion powers cellular movement (54). The flexible cell surface will, therefore, allow such processes to take place and accommodates this hypothesis.
The unique ability of AFM to operate in fluid enables us to perform in vivo force spectroscopy on M. xanthus cell-surface molecules. Single-molecule force spectroscopy (SMFS) has been widely used on well defined monolayers of structurally characterized molecules (48, 55, 56). However, except for a few reports (57, 58), little SMFS study has been carried out on living systems, which display much higher morphological heterogeneity. Here, we reported SMFS analysis of the extracellular substances on wild-type M. xanthus cells that allowed us to estimate the strength of the adhesion mediated by fibril material and provided physical and quantitative evidence for the role of these extracellular polysaccharides in cellular cohesion and the social behaviors of M. xanthus.
As the predominant form of existence for microorganisms in nature, the microbial communities and biofilms have not been studied under native conditions at resolutions comparable to EM. Results from this study demonstrated the capacity of AFM in studying the social bacteria M. xanthus and serve as a good example for further studies on other microbial communities. The minimal sample preparation involved in AFM allows direct high-resolution imaging on individual cells as well as on microbial communities under native conditions, and the force sensitivity of AFM enables nanomechanical study of cell-surface properties, including cellular elasticity, the nature of adhesive molecules, adhesion forces, etc. The combination of these capacities makes AFM a powerful tool for investigating the morphological and nanomechanical details of microbial communities and biofilms that will greatly enhance our understanding of the biological nature of the microbial world.
Supplementary Material
Acknowledgments
W.S. and Y.L. acknowledge partial support from National Institutes of Health Grant GM54666. A.E.P. and J.K.G. acknowledge partial support from the Institute for Cell Mimetic Space Exploration, a National Aeronautics and Space Administration University Research Engineering Technology Institute.
Author contributions: A.E.P., Y.L., W.S., and J.K.G. designed research; A.E.P. and Y.L. performed research; A.E.P. and Y.L. analyzed data; A.E.P., Y.L., and W.S. wrote the paper; and W.S. and J.K.G. provided funding.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AFM, atomic force microscopy; S motility, social motility; SEM, scanning EM; TFP, type IV pili.
References
- 1.Shapiro, J. A. & Dworkin, M. (1997) Bacteria as Multicellular Organisms (Oxford Univ. Press, New York).
- 2.Shapiro, J. A. (1998) Annu. Rev. Microbiol. 52, 81-104. [DOI] [PubMed] [Google Scholar]
- 3.Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R. & Lappin-Scott, H. M. (1995) Annu. Rev. Microbiol. 49, 711-745. [DOI] [PubMed] [Google Scholar]
- 4.O'Toole, G., Kaplan, H. B. & Kolter, R. (2000) Annu. Rev. Microbiol. 54, 49-79. [DOI] [PubMed] [Google Scholar]
- 5.Davey, M. E. & O'Toole, G. A. (2000) Microbiol. Mol. Biol. Rev. 64, 847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray, K. M. (1997) Trends Microbiol. 5, 184-188. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser, D. & Losick, R. (1993) Cell 73, 873-885. [DOI] [PubMed] [Google Scholar]
- 8.Wirth, R., Muscholl, A. & Wanner, G. (1996) Trends Microbiol. 4, 96-103. [DOI] [PubMed] [Google Scholar]
- 9.Shapiro, J. A. (1985) J. Bacteriol. 164, 1171-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro, J. A. (1987) J. Bacteriol. 169, 142-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg, E., Keller, K. H. & Dworkin, M. (1977) J. Bacteriol. 129, 770-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reichenbach, H. (1984) in The Myxobacteria, ed. Rosenberg, E. (Springer, New York), pp. 1-50.
- 13.White, D. (1993) in Myxobacteria II, eds. Dworkin, M. & Kaiser, D. (Am. Soc. Microbiol., Washington, DC), pp. 307-333.
- 14.McBride, M. J., Hartzell, P. & Zusman, D. (1993) in Myxobacteria II, eds. Dworkin, M. & Kaiser, D. (Am. Soc. Microbiol., Washington, DC), pp. 285-305.
- 15.Hodgkin, J. & Kaiser, D. (1979) Mol. Gen. Genet. 171, 177-191. [Google Scholar]
- 16.Wu, S. S. & Kaiser, D. (1995) Mol. Microbiol. 18, 547-558. [DOI] [PubMed] [Google Scholar]
- 17.Shimkets, L. J. (1986) J. Bacteriol. 166, 842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowden, M. G. & Kaplan, H. B. (1998) Mol. Microbiol. 30, 275-284. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser, D. (1979) Proc. Natl. Acad. Sci. USA 76, 5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merz, A. J., So, M. & Sheetz, M. P. (2000) Nature 407, 98-102. [DOI] [PubMed] [Google Scholar]
- 21.Sun, H., Zusman, D. R. & Shi, W. (2000) Curr. Biol. 10, 1143-1146. [DOI] [PubMed] [Google Scholar]
- 22.Skerker, J. M. & Berg, H. C. (2001) Proc. Natl. Acad. Sci. USA 98, 6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnold, J. W. & Shimkets, L. J. (1988) J. Bacteriol. 170, 5771-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behmlander, R. M. & Dworkin, M. (1991) J. Bacteriol. 173, 7810-7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behmlander, R. M. & Dworkin, M. (1994) J. Bacteriol. 176, 6295-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y., Sun, H., Ma, X., Lu, A., Lux, R., Zusman, D. & Shi, W. (2003) Proc. Natl. Acad. Sci. USA 100, 5443-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, Z., Geng, Y., Xu, D., Kaplan, H. B. & Shi, W. (1998) Mol. Microbiol. 30, 1123-1130. [DOI] [PubMed] [Google Scholar]
- 28.Dana, J. R. & Shimkets, L. J. (1993) J. Bacteriol. 175, 3636-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Razatos, A., Ong, Y. L., Sharma, M. M. & Georgiou, G. (1998) Proc. Natl. Acad. Sci. USA 95, 11059-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paige, M. F., Rainey, J. K. & Goh, M. C. (1998) Biophys. J. 74, 3211-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merroun, M. L., Ben Chekroun, K., Arias, J. M. & Gonzalez-Munoz, M. T. (2003) Chemosphere 52, 113-120. [DOI] [PubMed] [Google Scholar]
- 32.Kim, S. H., Ramaswamy, S. & Downard, J. (1999) J. Bacteriol. 181, 1496-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fink, J. M. & Zissler, J. F. (1989) J. Bacteriol. 171, 2028-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spormann, A. M. (1999) Microbiol. Mol. Biol. Rev. 63, 621-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lunsdorf, H. & Schairer, H. U. (2001) Microbiology 147, 939-947. [DOI] [PubMed] [Google Scholar]
- 36.Touhami, A., Nysten, B. & Dufrene, Y. F. (2003) Langmuir 19, 4539-4543. [Google Scholar]
- 37.Arnoldi, M., Fritz, M., Bauerlein, E., Radmacher, M., Sackmann, E. & Boulbitch, A. (2000) Phys. Rev. E 62, 1034-1044. [DOI] [PubMed] [Google Scholar]
- 38.Yao, X., Walter, J., Burke, S., Stewart, S., Jericho, M. H., Pink, D., Hunter, R. & Beveridge, T. J. (2002) Colloids Surf. B 23, 213-230. [Google Scholar]
- 39.Velegol, S. B. & Logan, B. E. (2002) Langmuir 18, 5256-5262. [Google Scholar]
- 40.Thwaites, J. J. & Surana, U. C. (1991) J. Bacteriol. 173, 197-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelling, A. E., Sehati, S., Gralla, E. B., Valentine, J. S. & Gimzewski, J. K. (2004) Science 305, 1147-1150. [DOI] [PubMed] [Google Scholar]
- 42.Almqvist, N., Bhatia, R., Primbs, G., Desai, N., Banerjee, S. & Lal, R. (2004) Biophys. J. 86, 1753-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rotsch, C., Jacobson, K. & Radmacher, M. (1999) Proc. Natl. Acad. Sci. USA 96, 921-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radmacher, M., Fritz, M., Kacher, C. M., Cleveland, J. P. & Hansma, P. K. (1996) Biophys. J. 70, 556-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofmann, U. G., Rotsch, C., Parak, W. J. & Radmacher, M. (1997) J. Struct. Biol. 119, 84-91. [DOI] [PubMed] [Google Scholar]
- 46.van der Aa, B. C., Michel, R. M., Asther, M., Zamora, M. T., Rouxhet, P. G. & Dufrene, Y. F. (2001) Langmuir 17, 3116-3119. [Google Scholar]
- 47.Rief, M., Clausen-Schaumann, H. & Gaub, H. E. (1999) Nat. Struct. Biol. 6, 346-349. [DOI] [PubMed] [Google Scholar]
- 48.Rief, M., Oesterhelt, F., Heymann, B. & Gaub, H. E. (1997) Science 275, 1295-1297. [DOI] [PubMed] [Google Scholar]
- 49.Fisher, T. E., Marszalek, P. E. & Fernandez, J. M. (2000) Nat. Struct. Biol. 7, 719-724. [DOI] [PubMed] [Google Scholar]
- 50.van der Aa, B. C., Asther, M. & Dufrene, Y. F. (2002) Colloids Surf. B 24, 277-284. [Google Scholar]
- 51.van der Aa, B. C. & Dufrene, Y. F. (2002) Colloids Surf. B 23, 173-182. [Google Scholar]
- 52.Marszalek, P. E., Pang, Y. P., Li, H., El Yazal, J., Oberhauser, A. F. & Fernandez, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 7894-7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marszalek, P. E., Li, H., Oberhauser, A. F. & Fernandez, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 4278-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolgemuth, C., Hoiczyk, E., Kaiser, D. & Oster, G. (2002) Curr. Biol. 12, 369-377. [DOI] [PubMed] [Google Scholar]
- 55.Oesterhelt, F., Oesterhelt, D., Pfeiffer, M., Engel, A., Gaub, H. E. & Muller, D. J. (2000) Science 288, 143-146. [DOI] [PubMed] [Google Scholar]
- 56.Sattin, B. D., Pelling, A. E. & Goh, M. C. (2004) Nucleic Acids Res. 32, 4876-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, B. L., Schaffer, T. E., Viani, M., Thompson, J. B., Frederick, N. A., Kindt, J., Belcher, A., Stucky, G. D., Morse, D. E. & Hansma, P. K. (1999) Nature 399, 761-763. [Google Scholar]
- 58.Rong, W., Pelling, A. E., Ryan, A., Friedlander, S. K. & Gimzewski, J. K. (2004) Nano Lett. 4, 2287-2292. [Google Scholar]
- 59.Binning, G., Quate, C. F. & Gerber, C. (1986) Phys. Rev. Lett. 56, 930-933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.