ABSTRACT
Acinetobacter baumannii, an important pathogen known for its widespread antibiotic resistance, has been the focus of extensive research within its genus, primarily involving clinical isolates. Consequently, data on environmental A. baumannii and other Acinetobacter species remain limited. Here, we utilized Illumina and Nanopore sequencing to analyze the genomes of 10 Acinetobacter isolates representing 6 different species sourced from aquatic environments in South Australia. All 10 isolates were phylogenetically distinct compared to clinical and other non-clinical Acinetobacter strains, often tens of thousands of single-nucleotide polymorphisms from their nearest neighbors. Despite the genetic divergence, we identified pdif modules (sections of mobilized DNA) carrying clinically important antimicrobial resistance genes in species other than A. baumannii, including carbapenemase oxa58, tetracycline resistance gene tet(39), and macrolide resistance genes msr(E)-mph(E). These pdif modules were located on plasmids with high sequence identity to those circulating in globally distributed A. baumannii ST1 and ST2 clones. The environmental A. baumannii isolate characterized here (SAAb472; ST350) did not possess any native plasmids; however, it could capture two clinically important plasmids (pRAY and pACICU2) with high transfer frequencies. Furthermore, A. baumannii SAAb472 possessed virulence genes and a capsular polysaccharide type analogous to clinical strains. Our findings highlight the potential for environmental Acinetobacter species to acquire and disseminate clinically important antimicrobial resistance genes, underscoring the need for further research into the ecology and evolution of this important genus.
IMPORTANCE
Antimicrobial resistance (AMR) is a global threat to human, animal, and environmental health. Studying AMR in environmental bacteria is crucial to understand the emergence and dissemination of resistance genes and pathogens, and to identify potential reservoirs and transmission routes. This study provides novel insights into the genomic diversity and AMR potential of environmental Acinetobacter species. By comparing the genomes of aquatic Acinetobacter isolates with clinical and non-clinical strains, we revealed that they are highly divergent yet carry pdif modules that encode resistance to antibiotics commonly used in clinical settings. We also demonstrated that an environmental A. baumannii isolate can acquire clinically relevant plasmids and carries virulence factors similar to those of hospital-associated strains. These findings suggest that environmental Acinetobacter species may serve as reservoirs and vectors of clinically important genes. Consequently, further research is warranted to comprehensively understand the ecology and evolution of this genus.
KEYWORDS: Acinetobacter baumannii, Acinetobacter towneri, Acinetobacter gerneri, Acinetobacter johnsonii, Acinetobacter chinensis, antibiotic resistance, environmental, virulence, mobile genetic elements, plasmid
INTRODUCTION
Acinetobacter is a genus of Gram-negative bacteria comprising over 85 species with different ecological niches and clinical impacts. Some Acinetobacter species are benign and ubiquitous in nature (e.g., found in soil, water, and animals), while others are pathogenic and frequently found in clinical settings (1–5). Acinetobacter baumannii is an opportunistic pathogen that is prevalent in healthcare settings (6, 7), causing a range of nosocomial infections, including pneumonia, as well as bloodstream, urinary tract, and wound infections (8). A major challenge in treating A. baumannii infections arises from its remarkable ability to acquire resistance to multiple antibiotics, facilitated primarily by horizontal transfer of antibiotic resistance genes via mobile genetic elements (MGEs), such as transposons and plasmids (7, 9–12). Due to its clinical relevance and high levels of antimicrobial resistance, A. baumannii has been the subject of extensive research in the last two decades (8, 13).
Pathogenic Acinetobacter species have several virulence genes that aid in evading host immune system responses and increase survival and spread throughout its host. For example, the ompA gene encodes a key antigenic factor (OmpA) that enables immune evasion (14, 15). Other factors, such as iron acquisition systems and serum/complement resistance, also facilitate in vivo survival (14, 15). Moreover, Acinetobacter spp. produce a variety of complex carbohydrate structures on their cell surface, such as capsular polysaccharide (CPS), lipooligosaccharide (LOS), and/or lipopolysaccharide with an O-antigen covalently attached to the outer core (OC) moiety of the LOS (16). These structures are important virulence factors for Gram-negative bacteria. In Acinetobacter, surface polysaccharides have been studied most extensively in A. baumannii, where it has been established that the species produces CPS and LOS (16). The genes that direct the synthesis of CPS and the OC component of the LOS are diverse and can vary even between closely related strains belonging to the same sequence type (ST). Typing these genes is considered an important primary step in strain characterization (11, 16, 17).
The environment is an important yet understudied reservoir of resistant bacteria, despite the knowledge that prominent extended-spectrum β-lactamases, quinolone resistance genes, and carbapenemases originated from marine and soil bacterium and subsequently entered clinical isolates through plasmids (18–20). Given the wide environmental spread of Acinetobacter strains, there is a growing consensus that One Health issues—which recognize that humans, animals, and the environment are interconnected—must be addressed through a comprehensive, integrated research approach (21, 22). This involves studying strains of Acinetobacter that have been isolated in clinical settings alongside those found in the natural environment to better understand the complex relationships and genetic exchange events between and within each niche (22). However, to date, the primary focus of comparative genomics research has been on hospital-acquired A. baumannii strains. As a result, the evolution, genetic structure, virulence determinants, antimicrobial resistance genes, and their associated MGEs present in environmental strains, particularly those not belonging to the baumannii species, are poorly understood and remain largely unexplored.
In this study, we performed genomic analyses on 10 Acinetobacter isolates recovered from South Australian aquatic samples. We show that, while the isolates were genetically unrelated to clinical strains, they share common antibiotic resistance pdif modules. This study provides new evidence that environmental strains might act as reservoirs for some of the clinically significant antibiotic resistance genes.
MATERIALS AND METHODS
Sample collection and isolation of Acinetobacter species
Influent samples were collected from three Australian wastewater treatment plants in 2019, hereafter referred to as wastewater treatment plants (WWTPs) A, C, and D. Characteristics, influent quality, operating conditions, and flow schematics have been described previously (23, 24). Briefly, isolates SAAt364, SAAt401, and SAAt388 originated from WWTP A, which serves approximately 150,000 inhabitants and receives primarily domestic and commercial sewage (PE organic load 120,000; sewage flow 17.3 ± 1.1 mL/day). Isolate SAAg309 was retrieved from WWTP C, which serves approximately 700,000 inhabitants (PE organic load 1,150,000; sewage flow 174 ± 15 mL/day) and receives a large industrial/commercial component, including some meat-processing trade waste, as well as residential and hospital sources. Isolates SAAj643, SAAc573, and SAAc652 originated from WWTP D, a rural wastewater treatment plant that serves 5,000 inhabitants and treats around 1.2 mL/day, primarily from households and seasonal meat-processing facilities. Raw wastewater for all WWTPs is classified as having low-to-medium organic strength. Isolates were also retrieved from a shallow artificial lake (SAAs470, SAAb472, SAAs474) fed by recycled water and storm water. All water samples (~10 L) were collected in triplicate, stored on ice, and processed within 2–3 hours post collection. All samples were plated, in triplicate, on L agar and Oxoid Brilliance CRE agar plates (ThermoFisher Scientific, Australia) (for initial screening of carbapenem-resistant strains) after 10-fold serial dilutions, using 500 µL from two to three consecutive dilutions. All cultures were incubated at 25°C, 37°C, and 44°C for 24 hours. Single colonies growing on CRE agar were picked up and streaked on plates counting agar (PCA) (ThermoFisher Scientific, Australia). PCA cultures were then incubated at 37°C for 18–24 hours. Acinetobacter isolates were initially identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) using Bruker Daltonics, operated in linear positive mode under MALDI Biotyper 3.0 real-time classification v3.1. Sample spectra were identified against an MSP database (5989 MSP entries). Identification scores of 2.300–3.000 indicated highly probable species identification, scores of 2.000–2.299 indicated secure genus identification and probable species identification, scores of 1.700–1.999 indicated probable genus identification, and a score of ≤1.699 indicated that the identification is not reliable. Isolates were stored in glycerol (40% vol/vol) at −80°C.
Antibiotic resistance profile
The resistance patterns of 23 antibiotics, including meropenem (10 µg), imipenem (10 µg), ampicillin (25 µg), cefotaxime (30 µg), ceftriaxone (30 µg), ceftazidime (30 µg), ampicillin/sulbactam (10/10 µg), tobramycin (10 µg), gentamicin (10 µg), spectinomycin (25 µg), netilmicin (30 µg), kanamycin (30 µg), amikacin (30 µg), neomycin (30 µg), streptomycin (25 µg), sulfamethoxazole (100 µg), rifampicin (30 µg), trimethoprim (5 µg), nalidixic acid (30 µg), ciprofloxacin (5 µg), florfenicol (30 µg), chloramphenicol (30 µg), and tetracycline (30 µg) were determined using the standard disc diffusion and standard microbroth dilution methods, which were previously described (25). In addition, minimal inhibitory concentrations (MIC) for eight antibiotics (cefotaxime, ceftazidime, meropenem, imipenem, amikacin, kanamycin, gentamicin, and colistin) belonging to four important families (beta-lactams, carbapenems, aminoglycosides, and polymyxin) were determined using Etest (bioMerieux, Durham, NC, USA) according to the manufacturer’s instructions (https://www.biomerieux-diagnostics.com/etestr). The resistance patterns were analyzed based on the guidelines of CLSI (Clinical & Laboratory Standards Institute) for Acinetobacter spp. and the calibrated dichotomous sensitivity disc diffusion test (http://cdstest.net) in cases where a CLSI breakpoint was not available. This was applicable for antibiotics such as netilmicin, streptomycin, spectinomycin, sulfamethoxazole, nalidixic acid, and rifampicin.
Plasmid transfer assays
Transfer studies were performed using a rifampicin mutant of A. baumannii SAAb472 (named SAAb472rif), which was made in this study as previously described, as a recipient. A. baumannii ACICU, which carries the conjugative plasmid pACICU2 (GenBank accession number CP031382) was used as donor in conjugation assays (26). Conjugation assays were conducted using the traditional mating assay at 37°C on agar upon an overnight culture of donor and recipient cells as previously described (27). Briefly, equal amounts of overnight cultures of the donor (ACICU) and recipient (SAAb472rif) were mixed and incubated on an L-agar plate overnight. Cells were re-suspended and diluted in 0.9% saline, and transconjugants were selected by plating on MHA plates containing rifampicin (100 mg/L) and kanamycin (100 mg/L). Transfer frequency (transconjugants/donor) was the average of three determinations. Potential transconjugants were purified and checked for growth on L-agar-containing kanamycin (20 mg/L) and tobramycin (10 mg/L), to which the donor was resistant and the recipient susceptible. Transformation assays were done using electroporation as we previously described (28), and plasmid DNA purified from A. baumannii D36 (GenBank accession number CP012952), which carries the small non-conjugative plasmid pRAY (GenBank accession number CP012954) (28, 29). Transformation frequency was calculated as transformants per microgram plasmid DNA.
Whole-genome sequencing, genome assembly, quality control, and annotation
Short-read sequencing was performed using the Illumina Nextseq500 platform, and reads were assembled using Shovill v1.1 (https://github.com/tseemann/shovill) with --trim option and the default SPAdes assembler (30). Resulting draft genomes were QCed using assembly-stats v1.0.1 (https://github.com/sanger-pathogens/assembly-stats). Long-read sequencing was performed using the Nanopore GridION platform, and consensus long-read assemblies were achieved using Trycycler v0.5.4 (31) in conjunction with Flye v2.9.2 (32), miniasm/minipolish v0.1.3 (33), and Raven v1.8.1 (34) assemblers. To create accurate hybrid assemblies, consensus long-read assemblies were polished with short reads using Polypolish v0.5.0 (35) followed by POLCA (36). All genomes were annotated using the Prokka pipeline v1.14.6 (37) with the --compliant and --addgenes options.
Sequence analysis and screening for antibiotic resistance and virulence genes
Resistance genes and insertion sequences were annotated manually using ResFinder ( http://genepi.food.dtu.dk/resfinder) and ISFinder (https://isfinder.biotoul.fr), respectively. Sequence types were determined using the Institut Pasteur Multi-Locus Sequence Typing (MLST) scheme using the mlst v2.22.1 (https://github.com/tseemann/mlst). The sequence of a set of genes known to be associated with virulence in A. baumannii (38) was used to screen the genomes. Kaptive v. 2.0.5 (17) was initially used to detect genes for the surface polysaccharides, capsular polysaccharide (K). and the outer core component of the lipooligosaccharide. Command-line searches utilized the available A. baumannii K locus (KL) (39) and OC locus (OCL) (40) reference sequence databases, which include 241 KL and 22 OCL, respectively. The minimum identity cut-off parameter for tBLASTn gene searches conducted by Kaptive was set to 60%. Protein-coding regions were characterized using BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE = Proteins) and Pfam (pfam.xfam.org) searches. Standalone BLAST was used to further characterize the structure of plasmids. The SnapGene v6.0.5 software was used to manually annotate regions of interest and draw figures to scale using the Illustrator v26.2.1 program. EasyFig v. 2.2.5 (41) was used to generate KL and OCL sequence comparisons.
Phylogenetic analysis
Core genome alignments (Block Mapping and Gathering with Entropy and recombination filtered) were produced using Panaroo v1.3.2 (42) with --clean-mode strict and -a core options and fed into IQ-Tree2 v2.2.0.3 (43) to produce Maximum Likelihood phylogenetic trees using -m MFP (best fit determined by ModelFinder) and -B 1000 (bootstrap replicates) options. All trees were visualized using Interactive Tree of Life (iTOL) v5 (44). For comparative analyses, additional genomes (n = 351) were downloaded from National Center for Biotechnology Information (NCBI) databases (April 2023) using NCBI Datasets v14.18.0 (https://github.com/ncbi/datasets). For additional taxonomic classifications, average nucleotide identity BLAST (ANIb) (45) with a >95% cutoff and in silico DNA-DNA hybridizations (DDH) (46) with a >70% cutoff (formula 2) were performed for species delimitation using NCBI representative strains as references. Pairwise single-nucleotide polymorphisms (SNPs) were determined using snp-dists v0.8.2 (github.com/tseemann/snp-dists) SNP matrix heatmaps were produced in RStudio v4.0.5 using pheatmap and ape packages. Pangenomes were calculated using Panaroo and visualized using Phandango v1.3.0 (47). Genome-wide association studies (GWASs) were conducted using scoary v1.6.16 (48) with --no_pairwise options, in conjunction with a gene presence/absence binary matrix produced by Panaroo.
RESULTS
Phylogenetic analysis reveals significant genetic variability among environmental isolates
Ten Acinetobacter isolates were recovered from aquatic environments in 2019 in South Australia, including influent wastewater (IW) and a lake (Table 1). Initially identified using MALDI-TOF MS, the isolates were classified as A. baumannii (n = 1), Acinetobacter towneri (n = 3), Acinetobacter johnsonii (n = 1), and Acinetobacter spp. (n = 5). However, while MALDI-TOF MS is widely used for bacterial identification, environmental isolates, especially those from complex ecosystems like wastewater, can exhibit significant genetic variation leading to incorrect speciation or ambiguous results (49). Thus, to confirm MALDI-TOF speciation, a maximum likelihood core genome phylogeny was constructed using 80 representative genomes for Acinetobacter species available on NCBI (Fig. S1; Supplementary Data Set 1). Consequently, three of the five isolates that could not be identified beyond the genus level by MALDI-TOF MS were assigned a species: Acinetobacter gerneri (n = 1; ANIb = 97.04%; DDH = 80.20%) and Acinetobacter chinensis (n = 2; ANIb = 96.57% and 97.02%; DDH = 74.10% and 76.50%). The remaining two isolates, SAAs470 and SAAs474, formed their own distinct phylogenetic branch and likely represent a novel Acinetobacter species.
TABLE 1.
Properties of the Acinetobacter strains analyzed in this study
| Number | Species | Source | Antibiotic resistance profilea | Acquired antibiotic resistance genes | GyrA83 | ParC80 | No. of plasmids | GenBank acc. no. |
|---|---|---|---|---|---|---|---|---|
| SAAb472 | A. baumannii | Lake | AMP, SPT, STR, TMP, FFC, CHL | aadA1-pm, ampC158 | S | S | 0 | CP127906 |
| SAAg309 | A. gerneri | IW | AMP, SAM, SPT, STR, TOB, GEN, SUL, RIF, NAL, FFC, TET | oxa308, aph(3′)-IXa, tet39 | Fc | S | (5)b | JASVDU000000000 |
| SAAt364 | A. towneri | IW | CTX, CAZ, IMI, MEM, TET, CHL, SPT, STR, TMP, NAL | oxa58, tet39, msr-mph(E), cmlB1 | Yd | Y | (2)b | JASVDV000000000 |
| SAAt401 | A. towneri | IW | TMP, NAL, TET | tet39 | Y | F | 4 | CP127892 |
| SAAt388 | A. towneri | IW | CTX, CAZ, SPT, STR, NAL | msr-mph(E) | Y | F | (3)b | JASVDW000000000 |
| SAAs470 | Acinetobacter sp. | Lake | AMP | –e | S | S | 12 | CP127893 |
| SAAs474 | Acinetobacter sp. | Lake | – | – | S | S | 12 | CP127915 |
| SAAj643 | A. johnsonii | IW | CAZ, SPT, TMP, NAL | oxa211 | F | S | (4) | JASVDX000000000 |
| SAAc573 | A. chinensis | IW | AMP, CTX, CAZ, SAM, SPT, STR, TMP, TET | msr-mph(E), tet39 | S | S | 5 | CP127923 |
| SAAc652 | A. chinensis | IW | NA | – | F | S | (0) | JASVDY000000000 |
IPM, imipenem; MEM, meropenem; AMP, ampicillin; CTX, cefotaxime; CAZ, ceftazidime; SAM, ampicillin/sulbactam; GEN, gentamicin; TOB, tobramycin; SPT, spectinomycin; STR, streptomycin; SUL, sulfamethoxazole; RIF, rifampicin; TMP, trimethoprim; FFC, florfenicol; CHL, chloramphenicol; TET, tetracycline; NAL, nalidixic acid.
Numbers in brackets indicate the number of plasmids predicted.
Phenylalanine.
Tyrosine.
Dashes indicate the ebsence of resistance phenotypes or antibiotic resistance gene(s).
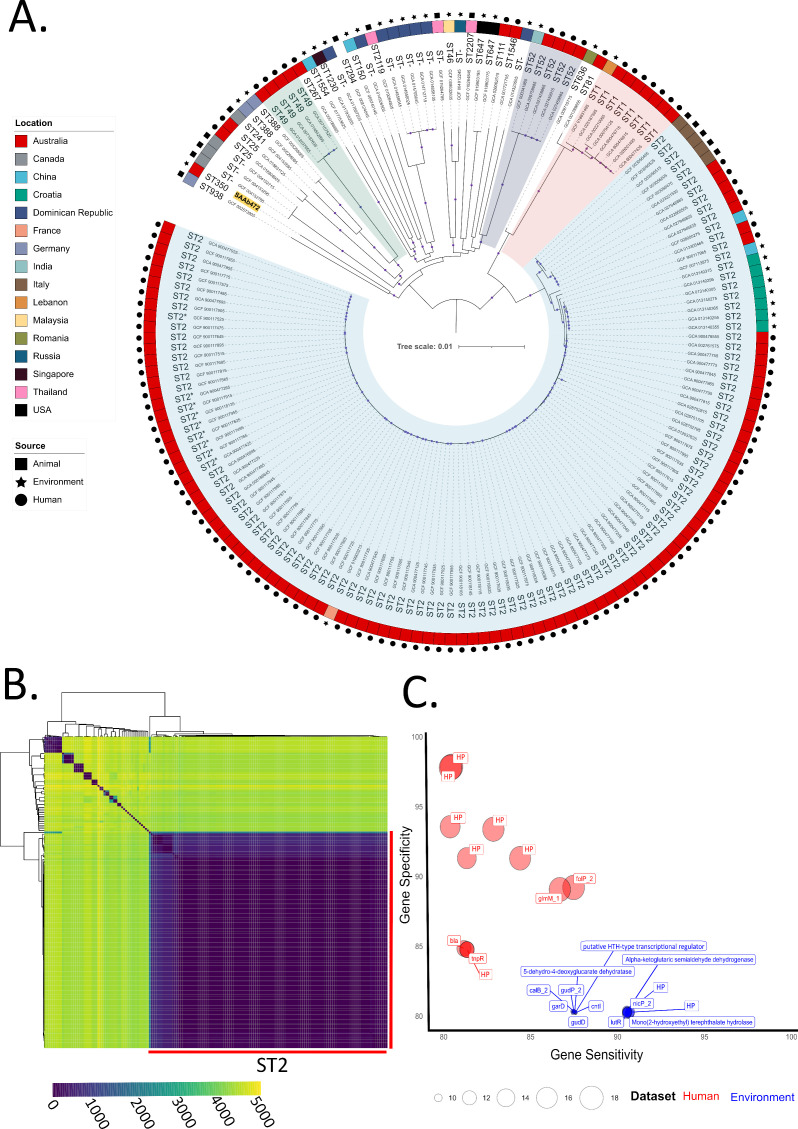
Further phylogenetic trees and SNP heatmaps were built to determine the genetic relatedness of our environmental isolates to Acinetobacter genomes previously deposited in public databases. These analyses were not performed for A. chinensis and A. gerneri due to the limited number of genomes currently available (excluding isolates from this study: A. chinensis = 2, A. gerneri = 1). For A. baumannii, assemblies available for clinical isolates from Australia (n = 122) and global non-clinical isolates (n = 48) were used to construct a maximum likelihood core genome phylogenetic tree (Fig. 1A; genome metadata provided in Supplementary Data Set 1). The analysis demonstrated a highly diverse A. baumannii population consisting of 26 STs, sharing 2,595 core genes of a total of 10,901 genes (24% of the pangenome), and an average SNP pairwise distance of 23,572 SNPs (Fig. 1B). The A. baumannii isolate from this study, SAAb472 obtained from a lake, belonged to ST350 and formed its own distinct branch in the phylogenetic tree. ST350 is a rare sequence type with only one representative on the PubMLST website (strain DV35). This strain, recovered from food in Switzerland in 2013, is a non-clinical isolate, suggesting that ST350 may be distributed in the environment across wide geographical regions. Interestingly, SAAb472 was found to be 43,916 SNPs from its nearest neighbor, A. baumannii strain GaenseEi-1 (GenBank accession: GCF_002573905) isolated from goose eggshells in Germany 2016.
Fig 1.
Genetic relatedness of A. baumannii clinical, environmental, and animal-sourced isolates. (A) Maximum likelihood phylogenetic tree of 171 A. baumannii genomes built using a core genome alignment (2,846,563 bp length). The colored outer ring denotes isolate geographic location, and isolate sources are marked as either a circle (human), rectangle (animal), or star (environmental). Tree branches belonging to STs with more than three isolates are highlighted—ST2 isolates in blue, ST1 in red, ST52 in purple, and ST49 in green. Bootstrap values of >0.95 marked by dots on branches. Isolate from this study is highlighted in yellow. (B) Heatmap illustrating pairwise SNP distances between A. baumannii isolates. (C) Bubble chart depicting highly sensitive and specific genes identified clinical isolates (shown in red) and environmental isolates (blue). The size of each bubble relates P-values and ranges from 2.31E−10 to 9.58E−19. HP, hypothetical protein.
While the environmental isolates generally displayed higher genetic heterogeneity than human-sourced isolates, even within the clinically significant ST2 population, the average pairwise SNPs distance was relatively high at 1,170 SNPs (range 0–8,525 SNPs). Interestingly, there were two distinct clades of Australian clinical ST2 strains. One clade consisted of 90 isolates with an average distance of 70 SNPs, while the other clade consisted of 5 isolates that were more closely related to 5 ST2 isolates from companion animals in Italy than to the larger clade of Australian clinical isolates (2,258 vs 4,528 SNPs). Also of note was an environmental ST2 strain (GCF_019903215) isolated from a river in France, which averaged 120 SNPs from the clonal clade of Australian clinical isolates. These observations are consistent with the ability of A. baumannii to spread and adapt to different environments (50). Nevertheless, GWASs did identify genes that were either significantly (P < 0.01) more or less represented in clinical and environmental isolates (Supplementary Data 2), including genes with >80% specificity and >80% sensitivity (Fig. 1C). These highly specific and sensitive genes in environmental isolates included those involved in D-glucarate and aldaric acid catabolic processes (5-dehydro-4-deoxyglucarate dehydratase, galactarate dehydratase garD, glucarate dehydratase gudD, and glucarate transporter gudP), and mono(2-hydroxyethyl) terephthalate hydrolase, shown to be involved in the degradation and metabolization of PET plastic for use as a carbon source (51). The genes that were highly sensitive and specific in clinical isolates mostly encoded hypothetical proteins (8 of 11 identified); however, beta-lactamase blaTEM and transposon Tn3 resolvase tnpR were found to be more characteristic of clinical isolates (Fig. 1C).
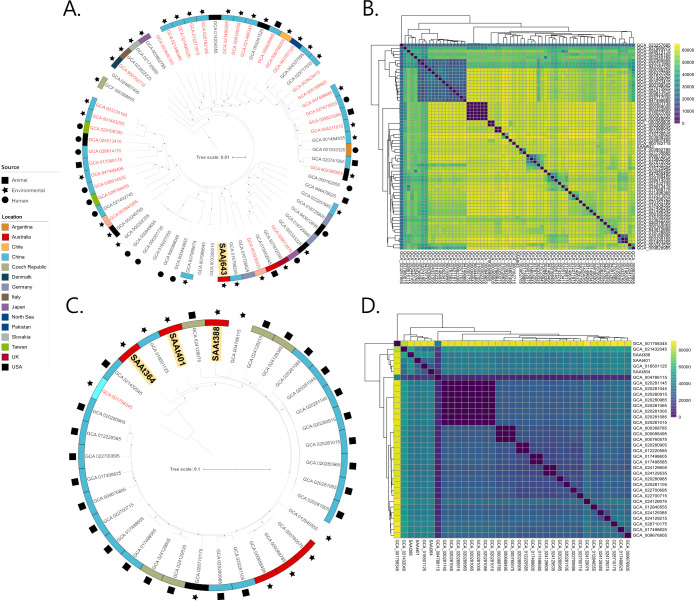
To study the evolutionary context of the A. johnsonii isolate, SAAj643 (wastewater) and the three A. towneri isolates, SAAt364, SSAt388, and SAAt401 (all wastewater), all assemblies available on NCBI for each respective species, were downloaded (A. johnsonii = 67; A. towneri = 31; genome metadata in Supplementary Data Set 1) and used to build phylogenetic trees (Fig. 2A and C). A. johnsonii SAAj643 formed its own distinct branch and was 50,729 SNPs from its closest neighbor—A. johnsonii strain new_MAG-226 (GCA_016790205) isolated from bioreactor sludge in China. On a cautionary note, A. johnsonii isolates only shared 294 core genes, and 31 (47%) publicly available genomes currently annotated as A. johnsonii fall below the ANI threshold for the species. Incorrect taxonomic assignments in the genus Acinetobacter are a recognized issue (52), and here, we highlight that misclassification may be prominent in A. johnsonii, though choice of NCBI representative strains may play a role in low ANIs. Regarding A. towneri, there are currently no human-sourced assemblies available on NCBI, and most isolates have originated from animal sources (pig and cattle) from China (n = 19; 59%). Like A. baumannii, A. towneri isolates were diverse averaging a pairwise distance of 30,445 SNPs (Fig. 2D). The wastewater isolates SAAt364, SSAt388, and SAAt401 each formed their own branches in the phylogenetic tree and had an average genetic distance of 27,660 SNPs between them.
Fig 2.
Genetic relatedness of A. johnsonii and A. towneri isolates. (A) Maximum likelihood phylogenetic tree of 52 A. johnsonii genomes built using a core genome alignment (1,948,641 bp length). Isolate from this study in bold and highlighted in yellow. Isolates shown in red mark those that fall under the ANI threshold for an A. johnsonii conclusive identification. (B) Heatmap illustrating pairwise SNP distances between A. johnsonii isolates. (C) Maximum likelihood phylogenetic tree of 34 A. towneri genomes built using a core genome alignment (1,758,026 bp length). The three A. towneri isolates from this study are in bold and highlighted in yellow. The isolate shown in red fell under the ANI threshold for an A. towneri conclusive identification. (D) Heatmap illustrating pairwise SNP distances between A. towneri isolates.
Antibiotic resistance and genetic context of resistance genes
Except marginal ampicillin resistance, the Acinetobacter sp. strains SAAs470 and SAAs474 recovered in a lake were susceptible to all other antibiotics tested, and consistent with this, they did not contain any acquired antibiotic resistance genes (i.e., known to be acquired via mobile genetic elements). However, in all other strains, analysis of the antibiotic resistance profiles indicated resistance to several clinically significant antibiotics, including resistance to carbapenems in A. towneri SAAt364 (Table 1); tetracycline resistance in A. gerneri SAAg309, A. towneri SAAt364, A. towneri SAAt401, and A. chinesis SAAc573 (Table 1); and aminoglycoside resistance in A. baumannii SAAb472 and A. gerneri SAAg309 (Table 1). MICs also revealed high levels of resistance to eight antibiotics in several strains consistent with the resistance genes they carry (see details in Table S1). However, few resistance phenotypes could not be explained by the presence or absence of horizontally acquired antibiotic resistance genes. For instance, four strains, SAAt364, SAAt388, SAAc573, and SAAc652 were resistant to ceftazidime and cefotaxime, which is often due to the presence of the insertion sequence ISAba1 upstream of the intrinsic ampC gene (53). However, none of these strains contained a copy of the IS in vicinity of their intrinsic ampC gene. It is likely that those novel intrinsic ampC genes have contributed to the phenotypes observed. Notably, these four strains (SAAt364, SAAt388, SAAc573, and SAAc652) were also highly resistant to colistin (Table S1), which is a last-resort antibiotic. Colistin resistance often occurs due to changes (often mutations) in several key genes (e.g., prmA prmB) or prmB) that cause the bacterium to alter the sugar moieties, resulting in the colistin being unable to bind to its target (7). Moreover, it could be due to changes in two-component systems that control the production and modification of LOS, namely in PhoPQ, PmrAB, BaeSR, or StkSR (7). Among these, several specific amino acid substitutions in PrmA (I13M and P102H) and PrmB (A227V, P233S, T235N, A262P, and Q270P) have been shown to be predominant changes driving colistin resistance in A. baumannii (54, 55). Here, it was found that SAAt364 and SAAt388 include the PrmA P102H, and PrMB T13V, and A262L substitutions. SAAc573 and SAAc652 also had PrMB T13V and A262I substitutions. Colistin resistance could also result from the acquisition of the mcr gene via a mobile genetic element (7). However, no mcr gene was detected in any of these strains, suggesting that the PrmA and PrmB amino acid substitutions found here are likely associated with the observed colistin resistance phenotypes.
Importantly, analysis of the genomes indicated that A. towneri SAAt364 carries a copy of the oxa58 carbapenem resistance gene (Table 1). The oxa58 gene is one of the most widespread carbapenem resistance genes found in clinical A. baumannii strains belonging to the major successful global clones such as ST1, ST2, ST25, etc. (10, 56). This gene is often located on R3-type plasmids surrounded by several complete and partial copies of ISAba2 and ISAba3. Recently, oxa58 and its companion insertion sequences were shown to be part of a pdif module (57). pdif modules are frequently found in Acinetobacter plasmids. They are made of pairs of Xer recombination sites called plasmid-dif (pdif) sites that flank a gene or genes (often resistance genes, toxin-antitoxin genes, etc.) (2, 57). Here, analysis of the draft genome of A. towneri SAAt364 showed that oxa58 is in a pdif module like those previously described (57). Analysis of the contig (containing oxa58) also suggested that the pdif module carrying the oxa58 carbapenem resistance gene is on an R3-type plasmid closely related to pMAD (with an addition of the oxa58 pdif module) in A. baumannii MAD (AY665723). A. baumannii MAD is a clinical strain recovered in Toulouse, France, in 2003 (58).
The tet39 tetracycline resistance gene was found in A. gerneri SAAg309, A. chinensis SAAc573, and A. towneri strains SAAt364 and SAAt401 (see the genetic structure of pSAAt401 in Fig. S2). In addition, the msr-mph(E) macrolide resistance genes were detected in A. towneri strains SAAt364 and SAAt388 and A. chinensis SAAc573.
Finally, six isolates (SAAg309, SAAt364, SAAt401, SAAt388, SAAj643, and SAAc652) showed resistance to nalidixic acid. Resistance to fluoroquinolones, including nalidixic acid, arises from amino acid substitutions at position 83 of the GyrA protein (DNA gyrase) and position 80 of the ParC protein (topoisomerase IV), resulting from point mutations in the gyrA and parC genes (59, 60). Here, we found these substitutions in all six nalidixic acid-resistant isolates consistent with their phenotypes (Table 1).
Among the complete genomes in this study (n = 5), A. baumannii SAAb472 did not contain plasmids. However, others, including A. towneri SAAt401, A. chinensis SAAc573, and the two Acinetobacter sp. strains (SAAs470 and SAAs474), carry several diverse novel plasmids, ranging in size from ~2 to over 150 kb (Table 2).
TABLE 2.
Properties of complete plasmids sequenced in this study
| Species/strain | Plasmid | Plasmid size (bp) | Rep | Mob | Resistance gene(s) | Important function | GenBank acc. no. |
|---|---|---|---|---|---|---|---|
| A. towneri SAAt401 | p1SAAt401 | 11,566 | R3-Tnew1 | MobQ | – | HigAB TAa | CP127888 |
| p2SAAt401 | 13,655 | – | – | tet39 | BrnAB TA | CP127889 | |
| p3SAAt401 | 26,382 | R3-Tnew2 | – | – | DNA methylase/invertase, YadV fimbrial pr | CP127891 | |
| p4SAAt401 | 153,370 | R3-T45 | MobP | – | BrnAB, DinJ/YafQ, HigAB TA, UvrABC, IcfB ligase, PhaAC acetyltransferase, BicA bicarbonate transporter, ChoD cholesterol oxidase, Blh beta-lactamase hydrolase, Dtd3 tRNA metabolism | CP127890 | |
| Acinetobacter sp. SAAs470b | p1SAAs470 | 2,120 | R3-Tnew3 | – | – | – | CP127905 |
| p2SAAs470 | 3,330 | R3-Tnew4 | MobV | – | – | CP127904 | |
| p3SAAs470 | 4,948 | R3-Tnew5 | MobQ | – | – | CP127903 | |
| p4SAAs470 | 5,105 | R3-Tnew6 | MobA | – | Phd/YefM TA | CP127901 | |
| p5SAAs470 | 7,689 | R3-Tnew7 | MobQ | – | RelB/DinJ TA, RutB peroxyureidoacrylate/ureidoacrylate amidohydrolase | CP127898 | |
| p6SAAs470 | 8,183 | R3-Tnew8 | – | – | Selenium=binding protein, cadmium, cobalt, and zinc/H(+)-K(+) antiporter | CP127900 | |
| p7SAAs470 | 8,767 | R3-T2 | – | – | DinJ/YafQ TA, zinc chelation protein | CP127899 | |
| p8SAAs470 | 9,125 | R3-T33 | – | – | HigAB TA | CP127902 | |
| p9SAAs470 | 9,830 | R3-Tnew9 | – | – | DNA adenine methylase | CP127896 | |
| p10SAAs470 | 11,846 | R3-Tnew10 | – | – | Mval/Bcnl restriction endonuclease, DNA cytosine methyltransferase | CP127895 | |
| p11SAAs470 | 12,273 | R3-Tnew11 | – | – | PemIK and HigBA TA, HdeA acid stress chaperone | CP127897 | |
| p12SAAs470 | 135,520 | R3-Tnew12 | MobP | bcr3, smvA, fsrc | Phd/YefM TA, sorbitol dehydrogenase, TraL conjugative pr, fimbria/pilus protein, BtuD vitamin B12 import p | CP127894 | |
| A. chinensis SAAc573 | p1SAAc573 | 2,341 | R3-Tnew13 | – | – | – | CP127922 |
| p2SAAc573 | 5,774 | R3-Tnew14 | MobQ | – | TrbLJ conjugative transfer pr | CP127924 | |
| p3SAAc573 | 8,141 | R3-Tnew15 | – | – | Export chaperone SecB | CP127925 | |
| p4SAAc573 | 10,750 | R3-T77 | – | tet39, msr-mph(E) | HigBA TA, putative ABC transporter YbiT | CP127920 | |
| p5SAAc573 | 11,013 | – | – | – | EsiB secretory immunoglobulin A-binding Pr | CP127921 |
TA, toxin/antitoxin.
The Acinetobacter sp. strain SAAs474 also includes 12 plasmids p1SAAs474–p12SAAs474 (CP127907–CP127919) identical to those found in Acinetobacter sp. SAAs470 shown in this table.
bcr3, bicyclomycin resistance gene; smvA, viologen (herbicide) resistance gene; fsr, fosmidomycin resistance gene.
Notably, all acquired antibiotic resistance genes were found in pdif modules located in different plasmid types (Table 2). pdif modules are frequently found in Acinetobacter plasmids (7, 28, 56, 61). Here, the analysis of the surrounding regions of the oxa58, tet39, and msr-mph(E) genes also indicated they are located within pdif modules. pdif modules containing the oxa58, tet39, and msr-mph(E) genes are widespread in strains recovered in clinical samples (56, 57, 62), and in this study, we report their presence in a set of environmental isolates, indicating that these resistance genes, or the plasmids they are associated with, are able to move between different Acinetobacter spp. or onto diverse plasmids.
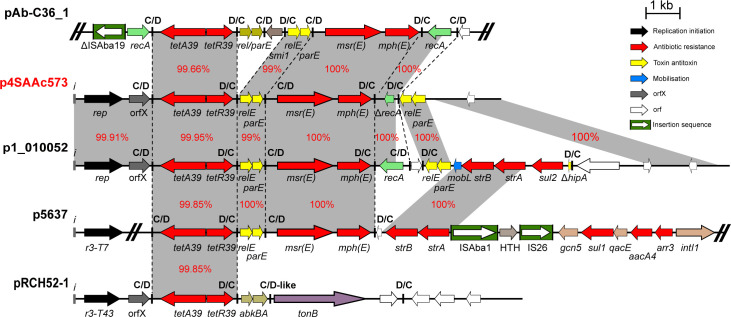
We also analyzed the genetic context of several plasmids that carry the tet39 and msr-mph(E) resistance genes and found several closely related plasmids (with at least 95% identity and 90% coverage) and of both environmental and clinical origins. This indicates that these plasmids can potentially be exchanged between clinical and environmental strains. As an example, analysis of p4AAc573 indicated that it is closely related to multiple plasmids from environmental and clinical strains (Fig. 3), particularly p1_010052 (carried by Acinetobacter sp. WCHAc010052 recovered in sewage in China in 2015; CP032138), p5637 (carried by A. baumannii 2016GDAB1 recovered in a bronchoalveolar lavage in China in 2016; CP065052), and pRCH52-1 (KT346360.1), which was also recovered in a clinical sample in Brisbane, Australia prior to 2012.
Fig 3.
Genetic structure of p2SAAc573 compared to pAb-C36_1, p1_010052, p5637, and pRCH52-1. Filled arrows indicate the orientation and extent of genes. Resistance genes are colored red, and the filled boxes colored green indicate ISAto1. Black arrows are putative replication initiation genes, and toxin/antitoxin genes are yellow. Vertical black lines indicate pdif sites. Scale bar is shown.
In addition to the closely related plasmids, the tet39, an msr-mph(E) pdif module found here, was also identified in several unrelated plasmids with clinical origins (Fig. 3; Fig. S2), suggesting that apart from plasmids, pdif modules can also be readily exchanged between different species and between strains with environmental and clinical origins.
A. baumannii SAAb472 can capture clinically important plasmids
A. baumannii plasmids play a crucial role in facilitating the spread of clinically significant antibiotic resistance genes. However, there remains a paucity of information concerning the ability of environmental strains to capture these clinically relevant plasmids. Hence, we sought to investigate whether plasmids of clinical importance could be transferred to A. baumannii strain SAAb472 (isolated from a lake in South Australia). Two plasmids were used in transfer assays, (i) pRAY (GenBank accession number CP012954.1), a 6-kb non-conjugative plasmid harboring the aadB tobramycin resistance gene (28), and (ii) pACICU2 (GenBank accession number CP031382), a 70-kb conjugative plasmid carrying the aphA6 amikacin resistance gene (63, 64). Both plasmids are widely recognized and prevalent across various A. baumannii STs and clinical settings in diverse geographical regions (28, 61, 63, 64). Given that pRAY is non-conjugative, transformation was used to move it into SAAb472. Using electroporation, pRAY was successfully transferred to SAAb472 with a high transfer frequency of 2.99 × 107 CFU/µg. While electroporation was used, the ability of pRAY to replicate and be maintained in SAAb472 suggests its potential uptake by naturally competent Acinetobacter strains (65, 66) (including baumannii and non-baumannii), which is consistent with its widespread dissemination. Similarly, pACICU2 was also successfully transferred to SAAb472rif (a rifampicin mutant of SAAb472 generated in this study) through conjugation assays, with a high frequency of 2.1 × 10−3.
Environmental strains encode various virulence functions
To understand variations in virulence potential within the South Australian environmental strains, we explored genes that have previously been identified as crucial for virulence in clinical isolates of A. baumannii. This included a set of genes encoding various functions such as biofilm formation, outer membrane proteins, capsular surface polysaccharides, regulatory proteins, and siderophores (38).
All genes, previously shown to be associated with virulence in A. baumannii (2), were identified in the A. baumannii isolate, signifying that it has the potential to cause infection (Table 3). All genes, encoding regulatory proteins (bmfRS, gigA, and gacA/S) were identified across all genomes studied (Table 3). The ompA outer membrane gene was absent in A. towneri strains SAAt364 and SAAt388 as well as the two Acinetobacter sp. strains (SAAs470 and SAAs474); however, the smpA and blc genes encoding outer membrane proteins were present in all species (Table 3). In contrast, the bap gene (associated with biofilm formation) and the bauA gene (associated with siderophore production) were absent in all isolates except for A. baumannii SAAb472 (Table 3).
TABLE 3.
Distribution of virulence functions encoded by South Australian aquatic strains
| Virulence determinant | Locus_id (ABUW_)a |
SAAb472 | SAAt364 | SAAt388 | SAAt401 | SAAs470 | SAAs474 | SAAg309 | SAAc652 | SAAc573 | SAAj643 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Outer membrane | |||||||||||
| Blc | 1048 | + | + | + | + | + | + | + | + | + | + |
| OmpA | 2571 | + | - | - | + | - | - | + | + | + | + |
| SmpA | 3034 | + | + | + | + | + | + | + | + | + | + |
| Biofilm formation | |||||||||||
| CsuA | 1488 | + | - | - | - | - | - | - | - | - | - |
| CsuB | 1489 | + | - | - | - | - | - | - | - | - | - |
| CsuC | 1490 | + | - | - | - | - | - | - | - | - | - |
| PgaA | 1557 | + | + | - | + | - | - | - | - | - | + |
| PgaB | 1558 | + | + | - | +b | - | - | - | +b | + | + |
| PgaC | 1559 | + | + | - | + | + | + | + | + | + | + |
| PgaD | 1560 | + | + | - | + | - | - | - | - | - | + |
| Bapb | 0885 | + | - | - | - | - | - | - | - | - | - |
| Capsule biosynthesis | |||||||||||
| Ptk | 3833 | + | + | + | + | + | + | + | + | + | - |
| Wzb | 3832 | + | + | + | + | + | + | + | + | + | - |
| GalUc | 3819 | + | + | + | + | + | + | - | + | + | + |
| Siderophore | |||||||||||
| RhbC_3 | 2189 | + | + | + | + | + | + | - | - | - | - |
| IucD | 2188 | + | - | + | - | - | - | - | - | - | - |
| EmrB | 2187 | + | - | - | - | + | + | - | - | - | + |
| RhbC_2 | 2186 | + | - | - | - | - | - | - | - | - | - |
| RhbC_1 | 2185 | + | - | - | - | - | - | - | - | - | - |
| 2Fe-2S | 2184 | + | - | - | - | - | - | - | - | - | - |
| RraA | 2183 | + | - | - | - | - | - | - | - | - | - |
| BauA | 1177 | + | - | - | - | + | + | + | + | + | + |
| Regulatory proteins | |||||||||||
| BmfR | 3181 | + | + | + | + | + | + | + | + | + | + |
| BmfS | 3180 | + | + | + | + | + | + | + | + | + | + |
| GigA | 3260 | + | + | + | + | + | + | + | + | + | + |
| GacA/S | 3504 | + | + | + | + | + | + | + | + | + | + |
Genome locus ids based on the genome sequence of A. baumannii strain AB5075-UW (GenBank accession number CP008706.1).
Interrupted.
Also involved in various cellular functions including stress response, biofilm formation, and interaction with host cells.
Together, several important virulence determinants were identified across our genomes, with a high degree of variation likely reflecting inter-species differences. Further functional studies are required to validate the roles of virulence determinants identified in our environmental strains, as well as to explore potential additional determinants in non-baumannii species. Given that most genomes (9 out of 10) returned hits for common CPS genes, we therefore studied the genetic context and variations within the CPS loci (see below).
Surface polysaccharide loci
In A. baumannii, CPS genes are clustered at the K locus (KL) between conserved fkpA and lldP genes, whereas genes for the OC of the LOS are at the OC locus (OCL) flanked by ilvE and aspS. While typing schemes are not currently available for other Acinetobacter spp., the K locus of a small number of studied strains has been shown to follow a similar arrangement. Therefore, the 10 Acinetobacter spp. genomes were screened against the available A. baumannii reference sequence databases of 241 KL and 22 OCL to identify common CPS and OC genes to locate the putative positions of polysaccharide loci for further analyses.
Only A. baumannii SAAb472 returned a significant match to a KL and OCL reference in the databases. The specific sequence at the K locus was identified as the KL121 CPS biosynthesis gene cluster (100% coverage, 97.93% DNA identity), previously described as carrying psa genes for the synthesis of a complex nonulosonic acid known as pseudaminic acid. KL121 has been detected among a range of A. baumannii isolates from clinical (blood, wound, and respiratory) or environmental (white stork and goose eggshells) samples obtained from either USA, Poland, Germany, Brazil, or Nigeria. At the OC locus, SAAb472 carries the OCL1 locus type (100% coverage, 98.42% DNA identity) previously found in ~75% of publicly available A. baumannii genome assemblies and common among members of the GC1 and GC2 clonal complexes (40).
Significant matches across the entire length of the KL or OCL were not obtained for any of the other Acinetobacter genomes. However, a common module of wza-wzb-wzc CPS export genes (61%–81% translated aa identity) was detected in eight of the nine genomes, identifying the contig(s) and possible location of the K locus. In all eight genomes, this module was found adjacent to other genes encoding products predicted to be involved in polysaccharide biosynthesis. While wza-wzb-wzc genes were not found in the SAAj643 genome, other genes associated with CPS synthesis were identified together in a single contig, suggesting that this strain may produce a complex polysaccharide but use different machineries to export it to the cell surface. Each genome was found to harbor a different locus sequence, though the overall genetic organization of the identified loci (shown in Fig. S3) resembles that of the A. baumannii K locus. Each locus included one or more modules of genes for different sugars, all of which have previously been identified in A. baumannii KL.
The putative OC locus in the non-baumannii genomes was identified via the finding of genes related to OCL genes from A. baumannii. A novel locus sequence was present in all genomes, though the two Acinetobacter spp. carried the same locus type (Fig. S4). As for A. baumannii, all loci identified consisted of one to two modules of genes for sugar biosynthesis, as well as acetyl-/acyltransferase gene(s) and/or several glycosyltransferase genes. Most included rmlA, rmlB, rmlC, and rmlD genes for the synthesis of L-rhamnose or rmlA/rmlB for a related sugar. However, like the K loci, very little sequence similarity was observed between the strains.
DISCUSSION
Environmental bacteria are ubiquitous and play important roles in biogeochemical cycles and ecological interactions. However, they are often intrinsically resistant to a range of antibiotics, and constant selective pressures facilitate the acquisition of additional antibiotic resistance genes through horizontal gene transfer (HGT). Thus, studying environmental bacteria in the context of AMR is crucial to understand the mechanisms, dynamics, and drivers of AMR emergence and spread, as well as to identify potential sources and reservoirs of AMR genes. The Acinetobacter genus includes a set of extremely diverse species, most of which are non-pathogenic and frequently found in the environment (2–5). Among them, A. baumannii has become a notorious opportunistic pathogen, mainly associated with antibiotic-resistant nosocomial infections (7). Moreover, it has been shown that non-clinical A. baumannii strains play an important role in posing a risk to human health (67). Non-baumannii species have also been identified in clinical samples. For instance, A. johnsonii has been detected in various environmental samples, such as aquatic samples, animals, food samples, and samples taken from human skin (68). Moreover, A. johnsonii has been demonstrated to cause severe human infections and carry clinically significant antibiotic resistance genes, including those conferring resistance to carbapenems indicating its clinical significance (68). Here, through a genomic analysis of 10 environmental Acinetobacter isolates from South Australia, we found that, despite being phylogenetically very distinct, several Acinetobacter species shared AMR-associated MGEs previously only associated with clinical A. baumannii strains.
Previous studies have highlighted the high genetic diversity of Acinetobacter populations (69, 70). However, the presence of antibiotic resistance genes in the environmental isolates underscores the significance of disseminating resistance determinants in the environment. It is known that the presence of residual antibiotics and other pharmaceutical substances stimulates the bacterial SOS system and promotes hypermutation and HGT (71). These substances are released into rivers and streams during manufacture and when antibiotics are metabolized. These residues also accumulate in municipal wastewater and urban runoff, creating selection pressure in the environment that fosters antibiotic resistance. These processes, in turn, increase the risk of antibiotic resistance acquisition and evolution (72). Additionally, the coexistence of diverse antibiotic resistance genes within a diverse population of pathogens and environmental bacteria, along with elevated rates of HGT, creates a conducive environment where new resistance gene arrangements or mutations can emerge due to these selection pressures. The identification of diverse plasmids harboring antibiotic resistance genes, including those associated with carbapenem resistance, suggests the mobility and adaptability of resistance elements across various strains. The presence of similar plasmids in both clinical and environmental strains indicates the potential for cross-species transfer, which has implications for public health interventions and strategies to combat the spread of antibiotic resistance. pdif sites are significant as they are associated with the movement of many genes, including antibiotic and heavy metal resistance genes. pdif modules carrying carbapenem, tetracycline, and macrolide resistance genes (e.g., tet39, mph-msr(E), oxa58, and oxa24) are widespread in major global clones of A. baumannii (28, 57). Here, the identification of the tet39, mph-msr(E), and oxa58 pdif modules that are often associated with plasmids circulating in globally distributed A. baumannii clones (e.g., ST1, ST2, ST25, etc.) is of particular significance. This observation raises concerns about the potential exchange of resistance genes between environmental and clinical strains, further emphasizing the need for holistic One Health approaches in studying the evolution and transmission of antibiotic resistance in the genus.
The successful transfer of clinically significant plasmids to the environmental A. baumannii strain SAAb472 highlights the adaptability of environmental strains to acquire plasmids of clinical importance. This combined with the identification of virulence determinants within the environmental isolates, particularly the A. baumannii strain SAAb472, underscores the potential of the environmental strains to cause infections in clinical settings.
The diversity in surface polysaccharide loci observed in this study reflects the complexity of virulence factor expression within Acinetobacter species. While some genes were shared among strains, the high degree of variation within species emphasizes the need for further investigation to understand the functional implications of these variations in terms of host interactions and environmental adaptations. The fact that CPS loci in other species are arranged in the same general genetic organization as sequences at the A. baumannii K locus may assist with recombination of the central region that determines the structural type driving CPS diversity. However, the KL and most OCL identified in this study were found at different locations in the genome in other species.
In this study, environmental Australian Acinetobacter strains were examined, providing valuable insights into genetic diversity, antibiotic resistance, plasmid dynamics, and virulence determinants. The findings not only shed light on the potential role of these strains as reservoirs for antibiotic resistance genes but also emphasize the interconnectedness of environmental and clinical strains. The identification of clinically significant genes within environmental strains underscores the potential risk of gene transfer between different niches, emphasizing the importance of ongoing research into the dynamics of antibiotic resistance and virulence factors in Acinetobacter populations. These insights contribute significantly to our understanding of the complex interactions between bacteria, the environment, and human health. They also provide a solid foundation for future studies aimed at mitigating the spread of antibiotic resistance.
ACKNOWLEDGMENTS
This research was supported by the Australian Institute for Microbiology and Infection, University of Technology Sydney, Data Generation Grant, and the Australian Centre for Genomic Epidemiological Microbiology (Ausgem), a collaborative research partnership between the New South Wales Department of Primary Industries and the University of Technology Sydney. J.K. is supported by an Australian Research Council Future Fellowship (FT230100400). M.H. is supported by an Australian Research Council DECRA fellowship (DE200100111).
M.H. conceptualization. L.T., V.M.J., J.K., M.H. methodology, investigation, formal analysis. B.D. resources. V.M.J., M.H. writing – original draft. V.M.J., J.K., B.D., S.D., M.H., methodology, writing – review & editing.
Contributor Information
Mehrad Hamidian, Email: mehrad.hamidian@uts.edu.au.
Christopher A. Elkins, Centers for Disease Control and Prevention, Atlanta, USA
DATA AVAILABILITY
The complete genome, plasmid sequences, and short-read data of all strains reported in this study have been deposited in the GenBank/EMBL/DDBJ database and are publicly available under the BioProject accession number PRJNA949389.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.01654-23.
Scoary GWAS raw output.
Genomes' metadata.
Minimal inhibitory concentrations (MICs).
Maximum-likelihood phylogenetic tree using 80 Acinetobacter representative species and ten South Australian environmental isolates.
Genetic structure of p2SAAt401 compared to pSSA3-1 and pRCH52-1.
Genetic arrangement of capsular polysaccharide biosynthesis loci identified in the genomes studied here.
Genetic arrangement of loci for synthesis of the outer core of the lipooligosaccharide/lipopolysaccharide for genomes studied here.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Koong J, Johnson C, Rafei R, Hamze M, Myers GSA, Kenyon JJ, Lopatkin AJ, Hamidian M. 2021. Phylogenomics of two ST1 antibiotic-susceptible non-clinical Acinetobacter baumannii strains reveals multiple lineages and complex evolutionary history in global clone 1. Microb Genom 7:000705. doi: 10.1099/mgen.0.000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prity FT, Tobin LA, Maharajan R, Paulsen IT, Cain AK, Hamidian M. 2023. The evolutionary tale of eight novel plasmids in a colistin-resistant environmental Acinetobacter baumannii isolate. Microb Genom 9:001010. doi: 10.1099/mgen.0.001010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Furlan JPR, de Almeida OGG, De Martinis ECP, Stehling EG. 2019. Characterization of an environmental multidrug-resistant Acinetobacter seifertii and comparative genomic analysis reveals co-occurrence of antimicrobial resistance and metal tolerance determinants. Front Microbiol 10:2151. doi: 10.3389/fmicb.2019.02151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dekić S, Klobučar G, Ivanković T, Zanella D, Vucić M, Bourdineaud J-P, Hrenović J. 2018. Emerging human pathogen Acinetobacter baumannii in the natural aquatic environment: a public health risk. Int J Environ Health Res 28:315–322. doi: 10.1080/09603123.2018.1472746 [DOI] [PubMed] [Google Scholar]
- 5. Adewoyin MA, Okoh AI. 2018. The natural environment as a reservoir of pathogenic and non-pathogenic Acinetobacter species. Rev Environ Health 33:265–272. doi: 10.1515/reveh-2017-0034 [DOI] [PubMed] [Google Scholar]
- 6. Doughari HJ, Ndakidemi PA, Human IS, Benade S. 2011. The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes Environ 26:101–112. doi: 10.1264/jsme2.me10179 [DOI] [PubMed] [Google Scholar]
- 7. Cain AK, Hamidian M. 2023. Portrait of a killer: uncovering resistance mechanisms and global spread of Acinetobacter baumannii. PLoS Pathog 19:e1011520. doi: 10.1371/journal.ppat.1011520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hamidian M, Hall RM. 2018. The AbaR antibiotic resistance Islands found in Acinetobacter baumannii global clone 1 - structure, origin and evolution. Drug Resist Updat 41:26–39. doi: 10.1016/j.drup.2018.10.003 [DOI] [PubMed] [Google Scholar]
- 10. Hamidian M, Nigro SJ. 2019. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom 5:e000306. doi: 10.1099/mgen.0.000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holt K, Kenyon JJ, Hamidian M, Schultz MB, Pickard DJ, Dougan G, Hall R. 2016. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb Genom 2:e000052. doi: 10.1099/mgen.0.000052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 13. Roca I, Espinal P, Vila-Farrés X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front Microbiol 3:148. doi: 10.3389/fmicb.2012.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. doi: 10.1038/nrmicro.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. doi: 10.1128/CMR.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kenyon JJ, Hall RM. 2013. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 8:e62160. doi: 10.1371/journal.pone.0062160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wyres KL, Cahill SM, Holt KE, Hall RM, Kenyon JJ. 2020. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb Genom 6:e000339. doi: 10.1099/mgen.0.000339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poirel L, Rodriguez-Martinez J-M, Mammeri H, Liard A, Nordmann P. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob Agents Chemother 49:3523–3525. doi: 10.1128/AAC.49.8.3523-3525.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poirel L, Vuillemin X, Kieffer N, Mueller L, Descombes M-C, Nordmann P. 2019. Identification of FosA8, a plasmid-encoded fosfomycin resistance determinant from Escherichia coli, and its origin in Leclercia adecarboxylata. Antimicrob Agents Chemother 63:01403–01419. doi: 10.1128/AAC.01403-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sekizuka T, Matsui M, Yamane K, Takeuchi F, Ohnishi M, Hishinuma A, Arakawa Y, Kuroda M. 2011. Complete sequencing of the blaNDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS ONE 6:e25334. doi: 10.1371/journal.pone.0025334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Castillo-Ramírez S. 2022. Zoonotic Acinetobacter baumannii: the need for genomic epidemiology in a one health context. Lancet Microbe 3:e895–e896. doi: 10.1016/S2666-5247(22)00255-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hernández-González IL, Castillo-Ramírez S. 2020. Antibiotic-resistant Acinetobacter baumannii is a one health problem. Lancet Microbe 1:e279. doi: 10.1016/S2666-5247(20)30167-1 [DOI] [PubMed] [Google Scholar]
- 23. Drigo B, Brunetti G, Aleer SC, Bell JM, Short MD, Vasileiadis S, Turnidge J, Monis P, Cunliffe D, Donner E. 2021. Inactivation, removal, and regrowth potential of opportunistic pathogens and antimicrobial resistance genes in recycled water systems. Water Res 201:117324. doi: 10.1016/j.watres.2021.117324 [DOI] [PubMed] [Google Scholar]
- 24. Hem S, Wyrsch ER, Drigo B, Baker DJ, Charles IG, Donner E, Jarocki VM, Djordjevic SP. 2022. Genomic analysis of carbapenem-resistant Comamonas in water matrices: implications for public health and wastewater treatments. Appl Environ Microbiol 88:e0064622. doi: 10.1128/aem.00646-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- 26. Hamidian M, Holt KE, Pickard D, Dougan G, Hall RM. 2014. A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J Antimicrob Chemother 69:955–958. doi: 10.1093/jac/dkt454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamidian M, Ambrose SJ, Hall RM. 2016. A large conjugative Acinetobacter baumannii plasmid carrying the sul2 sulphonamide and strAB streptomycin resistance genes. Plasmid 87–88:43–50. doi: 10.1016/j.plasmid.2016.09.001 [DOI] [PubMed] [Google Scholar]
- 28. Hamidian M, Hall RM. 2018. Genetic structure of four plasmids found in Acinetobacter baumannii isolate D36 belonging to lineage 2 of global clone 1. PLoS One 13:e0204357. doi: 10.1371/journal.pone.0204357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamidian M, Nigro SJ, Hall RM. 2012. Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J Antimicrob Chemother 67:2833–2836. doi: 10.1093/jac/dks318 [DOI] [PubMed] [Google Scholar]
- 30. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wick RR, Judd LM, Cerdeira LT, Hawkey J, Méric G, Vezina B, Wyres KL, Holt KE. 2021. Trycycler: consensus long-read assemblies for bacterial genomes. Genome Biol 22:266. doi: 10.1186/s13059-021-02483-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kolmogorov M, Bickhart DM, Behsaz B, Gurevich A, Rayko M, Shin SB, Kuhn K, Yuan J, Polevikov E, Smith TPL, Pevzner PA. 2020. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat Methods 17:1103–1110. doi: 10.1038/s41592-020-00971-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32:2103–2110. doi: 10.1093/bioinformatics/btw152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vaser R, Šikić M. 2021. Time- and memory-efficient genome assembly with Raven. Nat Comput Sci 1:332–336. doi: 10.1038/s43588-021-00073-4 [DOI] [PubMed] [Google Scholar]
- 35. Wick RR, Holt KE. 2022. Polypolish: short-read polishing of long-read bacterial genome assemblies. PLOS Comput Biol 18:e1009802. doi: 10.1371/journal.pcbi.1009802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zimin AV, Salzberg SL. 2020. The genome polishing tool POLCA makes fast and accurate corrections in genome assemblies. PLOS Comput Biol 16:e1007981. doi: 10.1371/journal.pcbi.1007981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 38. Hamidian M, Maharjan RP, Farrugia DN, Delgado NN, Dinh H, Short FL, Kostoulias X, Peleg AY, Paulsen IT, Cain AK. 2022. Genomic and phenotypic analyses of diverse non-clinical Acinetobacter baumannii strains reveals strain-specific virulence and resistance capacity. Microb Genom 8:000765. doi: 10.1099/mgen.0.000765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cahill SM, Hall RM, Kenyon JJ. 2022. An update to the database for Acinetobacter baumannii capsular polysaccharide locus typing extends the extensive and diverse repertoire of genes found at and outside the K locus. Microb Genom 8:mgen000878. doi: 10.1099/mgen.0.000878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sorbello BM, Cahill SM, Kenyon JJ. 2023. Identification of further variation at the lipooligosaccharide outer core locus in Acinetobacter baumannii genomes and extension of the OCL reference sequence database for Kaptive. Microb Genom 9:001042. doi: 10.1099/mgen.0.001042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, Gladstone RA, Lo S, Beaudoin C, Floto RA, Frost SDW, Corander J, Bentley SD, Parkhill J. 2020. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol 21:180. doi: 10.1186/s13059-020-02090-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Letunic I, Bork P. 2021. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M. 2022. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50:D801–D807. doi: 10.1093/nar/gkab902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. 2017. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34:292–293. doi: 10.1093/bioinformatics/btx610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:262. doi: 10.1186/s13059-016-1132-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barreiro JR, Gonçalves JL, Braga PAC, Dibbern AG, Eberlin MN, Veiga Dos Santos M. 2017. Non-culture-based identification of mastitis-causing bacteria by MALDI-TOF mass spectrometry. J Dairy Sci 100:2928–2934. doi: 10.3168/jds.2016-11741 [DOI] [PubMed] [Google Scholar]
- 50. Monem S, Furmanek-Blaszk B, Łupkowska A, Kuczyńska-Wiśnik D, Stojowska-Swędrzyńska K, Laskowska E. 2020. Mechanisms protecting Acinetobacter baumannii against multiple stresses triggered by the host immune response, antibiotics and outside-host environment. Int J Mol Sci 21:5498. doi: 10.3390/ijms21155498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Palm GJ, Reisky L, Böttcher D, Müller H, Michels EAP, Walczak MC, Berndt L, Weiss MS, Bornscheuer UT, Weber G. 2019. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat Commun 10:1717. doi: 10.1038/s41467-019-09326-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mateo-Estrada V, Graña-Miraglia L, López-Leal G, Castillo-Ramírez S. 2019. Phylogenomics reveals clear cases of misclassification and genus-wide phylogenetic markers for Acinetobacter. Genome Biol Evol 11:2531–2541. doi: 10.1093/gbe/evz178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hamidian M, Hall RM. 2013. ISAba1 targets a specific position upstream of the intrinsic ampC gene of Acinetobacter baumannii leading to cephalosporin resistance. J Antimicrob Chemother 68:2682–2683. doi: 10.1093/jac/dkt233 [DOI] [PubMed] [Google Scholar]
- 54. Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634. doi: 10.1128/AAC.00284-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sun B, Liu H, Jiang Y, Shao L, Yang S, Chen D. 2020. New mutations involved in colistin resistance in Acinetobacter baumannii. mSphere 5:e00895-19. doi: 10.1128/mSphere.00895-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jones NI, Harmer CJ, Hamidian M, Hall RM. 2022. Evolution of Acinetobacter baumannii plasmids carrying the oxa58 carbapenemase resistance gene via plasmid fusion, IS26-mediated events and dif module shuffling. Plasmid 121:102628. doi: 10.1016/j.plasmid.2022.102628 [DOI] [PubMed] [Google Scholar]
- 57. Blackwell GA, Hall RM. 2017. The tet39 determinant and the msrE-mphE genes in Acinetobacter plasmids are each part of discrete modules flanked by inversely oriented pdif (XerC-XerD) sites. Antimicrob Agents Chemother 61:e00780-17. doi: 10.1128/AAC.00780-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Poirel L, Marqué S, Héritier C, Segonds C, Chabanon G, Nordmann P. 2005. OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob Agents Chemother 49:202–208. doi: 10.1128/AAC.49.1.202-208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vila J, Ruiz J, Goñi P, Jimenez de Anta T. 1997. Quinolone-resistance mutations in the topoisomerase IV parC gene of Acinetobacter baumannii. J Antimicrob Chemother 39:757–762. doi: 10.1093/jac/39.6.757 [DOI] [PubMed] [Google Scholar]
- 60. Vila J, Ruiz J, Goñi P, Marcos A, Jimenez de Anta T. 1995. Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 39:1201–1203. doi: 10.1128/AAC.39.5.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lam MMC, Koong J, Holt KE, Hall RM, Hamidian M. 2023. Detection and typing of plasmids in Acinetobacter baumannii using rep genes encoding replication initiation proteins. Microbiol Spectr 11:e0247822. doi: 10.1128/spectrum.02478-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hamidian M, Holt KE, Pickard D, Hall RM. 2016. A small Acinetobacter plasmid carrying the tet39 tetracycline resistance determinant. J Antimicrob Chemother 71:269–271. doi: 10.1093/jac/dkv293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hamidian M, Wick RR, Hartstein RM, Judd LM, Holt KE, Hall RM. 2019. Insights from the revised complete genome sequences of Acinetobacter baumannii strains AB307-0294 and ACICU belonging to global clones 1 and 2. Microb Genom 5:e000298. doi: 10.1099/mgen.0.000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hamidian M, Hall RM. 2014. pACICU2 is a conjugative plasmid of Acinetobacter carrying the aminoglycoside resistance transposon TnaphA6. J Antimicrob Chemother 69:1146–1148. doi: 10.1093/jac/dkt488 [DOI] [PubMed] [Google Scholar]
- 65. Ramirez MS, Don M, Merkier AK, Bistué AJS, Zorreguieta A, Centrón D, Tolmasky ME. 2010. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol 48:1488–1490. doi: 10.1128/JCM.01264-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Santala S, Santala V. 2021. Acinetobacter baylyi ADP1-naturally competent for synthetic biology. Essays Biochem 65:309–318. doi: 10.1042/EBC20200136 [DOI] [PubMed] [Google Scholar]
- 67. Castillo-Ramírez S. 2023. The importance of Acinetobacter baumannii from non-human sources. Lancet Microbe 4:e761–e762. doi: 10.1016/S2666-5247(23)00246-X [DOI] [PubMed] [Google Scholar]
- 68. Castillo-Ramírez S, Mateo-Estrada V, Gonzalez-Rocha G, Opazo-Capurro A. 2020. Phylogeographical analyses and antibiotic resistance genes of Acinetobacter johnsonii highlight its clinical relevance. mSphere 5:e00581-20. doi: 10.1128/mSphere.00581-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Antunes LCS, Visca P, Towner KJ. 2014. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis 71:292–301. doi: 10.1111/2049-632X.12125 [DOI] [PubMed] [Google Scholar]
- 71. Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74. doi: 10.1038/nature02241 [DOI] [PubMed] [Google Scholar]
- 72. Wengenroth L, Berglund F, Blaak H, Chifiriuc MC, Flach C-F, Pircalabioru GG, Larsson DGJ, Marutescu L, van Passel MWJ, Popa M, Radon K, de Roda Husman AM, Rodríguez-Molina D, Weinmann T, Wieser A, Schmitt H. 2021. Antibiotic resistance in wastewater treatment plants and transmission risks for employees and residents: the concept of the AWARE study. Antibiotics (Basel) 10:478. doi: 10.3390/antibiotics10050478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scoary GWAS raw output.
Genomes' metadata.
Minimal inhibitory concentrations (MICs).
Maximum-likelihood phylogenetic tree using 80 Acinetobacter representative species and ten South Australian environmental isolates.
Genetic structure of p2SAAt401 compared to pSSA3-1 and pRCH52-1.
Genetic arrangement of capsular polysaccharide biosynthesis loci identified in the genomes studied here.
Genetic arrangement of loci for synthesis of the outer core of the lipooligosaccharide/lipopolysaccharide for genomes studied here.
Data Availability Statement
The complete genome, plasmid sequences, and short-read data of all strains reported in this study have been deposited in the GenBank/EMBL/DDBJ database and are publicly available under the BioProject accession number PRJNA949389.