Abstract
The p21 protein, a cyclin-dependent kinase (CDK) inhibitor, is capable of binding to both cyclin-CDK and the proliferating cell nuclear antigen (PCNA). Through its binding to PCNA, p21 can regulate the function of PCNA differentially in replication and repair. To gain an understanding of the precise mechanism by which p21 affects PCNA function, we have designed a new assay for replication factor C (RFC)-catalyzed loading of PCNA onto DNA, a method that utilizes a primer-template DNA attached to agarose beads via biotin-streptavidin. Using this assay, we showed that RFC remains transiently associated with PCNA on the DNA after the loading reaction. Addition of p21 did not inhibit RFC-dependent PCNA loading; rather, p21 formed a stable complex with PCNA on the DNA. In contrast, the formation of a p21-PCNA complex on the DNA resulted in the displacement of RFC from the DNA. The nonhydrolyzable analogs of ATP, adenosine-5′-O-(3-thiotriphosphate) (ATPγS) and adenyl-imidodiphosphate, each stabilized the primer recognition complex containing RFC and PCNA in the absence of p21. RFC in the ATPγS-activated complex was no longer displaced from the DNA by p21. We propose that p21 stimulates the dissociation of the RFC from the PCNA-DNA complex in a process that requires ATP hydrolysis and then inhibits subsequent PCNA-dependent events in DNA replication. The data suggest that the conformation of RFC in the primer recognition complex might change on hydrolysis of ATP. We also suggest that the p21-PCNA complex that remains attached to DNA might function to tether cyclin-CDK complexes to specific regions of the genome.
The p21 protein, also known as Waf1, Cip1, or Sdi1, is an inhibitor of cyclin-dependent kinase (CDK) and directly binds to both cyclin and CDK kinase (for reviews, see references 59 and 60). The expression of p21, among other stimulators, can be induced by the tumor suppressor gene product p53 in response to DNA damage, so that cells can arrest in G1 phase of the cell cycle by p21-dependent inhibition of the cyclin-CDKs that are required for the G1 progression or the G1-S transition (16, 18). This model for cell cycle checkpoint function involving p21 is supported by previous studies, including some involving gene knockout mice (6, 14). On the other hand, p21 can also be induced by serum or individual growth factors in a p53-independent manner (20, 46); hence, increased levels of p21 are present during G1 phase after the cells have been released from serum deprivation (42, 45). With respect to the p53-independent induction, it has been shown that p21 might function as an assembly factor for certain pairs of cyclin-CDK complexes (30, 37). Furthermore, p21 is also thought to be involved in other cellular functions, including control of apoptosis, differentiation, signal transduction, and senescence (5, 17, 29, 51, 61, 65).
Based on amino acid sequence similarity, the p21 protein belongs to a family of CDK inhibitors which also includes p27 (Kip1) and p57 (Kip2) (for a review, see reference 60). Among these, only p21 can bind directly to the proliferating-cell nuclear antigen (PCNA) (22, 72), an essential DNA replication and repair factor (for a review, see reference 34). Previous biochemical studies have shown that p21 exists as part of a quaternary complex with cyclin, CDK, and PCNA (77). Interestingly, the quaternary complex was seen in normal cells but not in many transformed cells (78), suggesting that it might play an important role in maintaining the integrity of the genome. Recent studies also suggest that this quaternary complex is a target for the human papillomavirus type 16 (HPV-16) E7 oncoprotein. In addition to the retinoblastoma protein, E7 binds directly to p21 in a region that overlaps with the PCNA binding region, and the E7 protein can reverse the inhibitory effect of p21 on DNA replication (25). Moreover, binding of E7 to p21 that is complexed with cyclin-CDK results in reactivation of the kinase activity (25, 33).
Through a direct interaction between p21 and PCNA, p21 inhibits simian virus 40 DNA replication and PCNA-dependent DNA synthesis by DNA polymerase δ in vitro (22, 72). A p21-derived peptide that also binds to PCNA has a similar inhibitory effect on DNA replication in vitro (26). In contrast to its effect on DNA replication, p21 has no detectable effect on nucleotide excision repair using cell extracts, even though the repair reaction is absolutely dependent on PCNA (40, 62). Consistent with the biochemical data, p21 has also been shown to colocalize with PCNA in the nuclei of UV-induced, DNA-damaged G1 cells (39). Interestingly, both p21 and PCNA became resistant to detergent extraction from nuclei upon the occurrence of DNA damage (39). This similarity of behavior of p21 and PCNA in response to DNA damage suggests that these proteins might function in concert during a certain step of the DNA repair reactions.
The structure of p21 has been extensively characterized. Mutational analyses of p21 have revealed distinct regions that are responsible for binding of cyclin, CDK, and PCNA (10, 12, 13, 23, 27, 43, 44, 47, 48, 75). The cyclin-CDK binding region is located near the N terminus of p21, while the region necessary for PCNA binding is near the C terminus. A second cyclin binding region, however, has been found in the C-terminal region that overlaps with the PCNA binding region (13). A crystal structure of a complex containing cyclin A-CDK2 and a peptide derived from the N-terminal region of p27 (Kip1), whose sequence is similar to that of the N-terminal region of p21, has been determined (58). However, it is not yet clear how the C-terminal region of p21 also influences the interaction between p21 and cyclin-CDK. Intriguingly, it has been shown that the HPV-16 E7 oncoprotein, but not the nononcogenic HPV-6 E7 protein, binds to the C-terminal region of p21 and reactivates the kinase activity of p21-bound cyclin E-CDK2 without displacing p21, indicating that the C-terminal region of p21 may affect the interaction between p21 and cyclin-CDK (25). Another structural study has shown that although free p21 does not seem to have a stable, high-order structure, p21 adopts an ordered and stable conformation upon its binding to cyclin-CDK, indicating that the protein has considerable structural flexibility (36).
The crystal structure of PCNA complexed with a peptide derived from the p21 C-terminal region has been also determined (28); the peptide binds to the interdomain connecting loop of PCNA, suggesting that p21 might block the interaction between PCNA and another protein(s) that requires this particular loop for binding. Related to this mutually exclusive binding, it has been shown that a complex containing PCNA and FEN1, a 5′-3′ exo- and endonuclease required for DNA replication and repair, can be disrupted by p21 (11).
PCNA functions as a processivity factor for DNA polymerase δ, and possibly DNA polymerase ɛ, during DNA replication (1, 4, 56, 66, 74). PCNA exists as a homotrimer, forming a donutlike ring shape (35), and can be loaded onto DNA catalytically by replication factor C (RFC) in an ATP-dependent manner (9, 70). During this process, RFC binds preferentially to the DNA at a primer-template junction and thus forms a primer recognition complex with PCNA (8, 9, 38, 70). PCNA then interacts with DNA polymerase δ, forming a processive polymerase holoenzyme. This series of events is thought to be required for initiation of leading-strand DNA synthesis, as well as for synthesis of each Okazaki fragment on the lagging strand during DNA replication (67, 73).
Although p21, as mentioned above, can interfere with the function of PCNA by directly binding to it, it is not known how p21 affects PCNA function, leading to inhibition of replication but not repair. To begin to address this issue, we have designed a new assay for RFC-catalyzed loading of PCNA onto DNA, a method which has allowed us to investigate the formation of the primer recognition complex in great detail. This assay utilizes a primer-template DNA fixed onto agarose beads through biotin-streptavidin binding. We have shown that PCNA can be efficiently loaded onto the DNA even if the DNA is fixed to beads. Furthermore, we have found that p21 does not inhibit the loading of PCNA; rather, it can form a stable complex with PCNA on the DNA. RFC, however, is displaced from the DNA upon formation of the p21-PCNA complex. Interestingly, the nonhydrolyzable analogs of ATP, adenosine-5′-O-(3-thiophosphate) (ATPγS) and adenyl-imidodiphosphate (AMP-PNP), each stabilize the primer recognition complex, and RFC in the ATPγS-activated complex is no longer displaced by p21. Hence, these results not only suggest a mechanism of inhibition of DNA replication by p21 but also provide some insight into the function of RFC upon ATP hydrolysis during the assembly of a primer recognition complex.
MATERIALS AND METHODS
Materials.
ATPγS and AMP-PNP were obtained from Boehringer Mannheim. Homopolymer poly(dA) (average length, 540 nucleotides) and oligomer dT12–18 were from Pharmacia. Streptavidin-agarose was purchased from Sigma. The antibodies used were an anti-RFC p140 monoclonal antibody (no. 6 [7]), an anti-PCNA monoclonal antibody (PC10 [76]), a rabbit anti-p21 antiserum (79), and an anti-RPA p70 monoclonal antibody (p70-9 [15]).
Proteins.
RFC (second phosphocellulose fraction [68]), PCNA (21), and histidine-tagged p21 (His-p21 [72]) were prepared as previously described. The Escherichia coli expression plasmid for human replication protein A (RPA) was kindly provided by M. Wold (University of Iowa), and recombinant RPA was purified as described elsewhere (31).
DNA primer-templates.
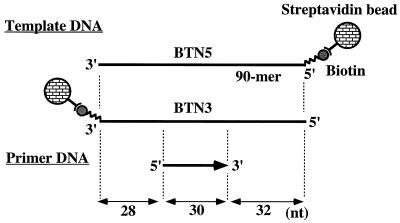
The 90-mer oligonucleotide, biotin labeled at the 5′ or 3′ end (BTN5 and BTN3, respectively [see Fig. 1]), was synthesized with an Applied Biosystems model 394 DNA synthesizer, using either biotin-cyanoethyl phosphoramidite at the 5′ end or a biotin-linked, controlled pore glass bead at the 3′ end (Peninsula Laboratories, Inc.), and purified by denaturing 6% polyacrylamide gel electrophoresis. The sequence of BTN5 and BTN3 corresponded to nucleotide positions 4833 and 4922 of the bacteriophage M13 mp18 single-stranded DNA. The sequence of the 30-mer primer (Fig. 1) was complementary to nucleotide positions 4865 to 4894 of the M13 mp18 DNA. To anneal the 30-mer primer to the biotin template, they were mixed at a 2:1 (primer/template) molar ratio in 100 mM NaCl–10 mM Tris-HCl (pH 8.0)–1 mM EDTA, heated at 95°C for 3 min, and slowly cooled down to 37°C.
FIG. 1.
The design of biotin-labeled templates and a primer for PCNA loading. The two biotin-labeled 90-mer DNA templates, BTN5 and BTN3, are identical in nucleotide sequence but differ in the end to which a biotin moiety is attached (5′ and 3′ ends for BTN5 and BTN3, respectively). The location of a primer and the length (in nucleotides [nt]) of each DNA segment are shown.
Nicked circular pSVO10 plasmid (55) was prepared by a brief treatment of the supercoiled plasmid with DNase I and then purified by phenol-chloroform extraction and ethanol precipitation. Under these conditions, more than 90% of the plasmid was converted to nicked circular DNA.
PCNA loading assay with biotin-labeled DNA.
The assay was performed as described below unless otherwise noted. Ten microliters of streptavidin-agarose beads was first preincubated with 1 pmol of biotin-labeled DNA template-primer in 20 μl of buffer A (40 mM HEPES-KOH [pH 8.0], 8 mM MgCl2, 0.1 mg of bovine serum albumin per ml, and 1 mM dithiothreitol) with 0.12 M NaCl for 30 min at room temperature. The DNA-coated beads were washed with 100 μl of 0.12 M NaCl in buffer A three times and resuspended in 20 μl of the same buffer. The DNA-coated beads were then preincubated with 2 pmol of RPA in the same buffer containing 0.12 M NaCl for 5 min at room temperature, and PCNA loading was started by further adding 1 mM ATP, RFC, and PCNA as indicated in the figures. The final reaction volume was adjusted to 100 μl with buffer A containing 0.12 M NaCl. The reaction mixture was incubated for 30 min at room temperature with occasional agitation. After incubation, the mixture was briefly chilled on ice and the beads were then washed with 400 μl of ice-cold buffer A containing 0.12 M NaCl three times. Finally, the beads were boiled for 3 min in a sodium dodecyl sulfate (SDS) loading buffer, and the proteins recovered were analyzed by SDS–10% polyacrylamide gel electrophoresis and Western blotting with the appropriate antibodies. The detection for Western blotting was done with the ECL Western blotting detection reagent (Amersham) or SuperSignal chemiluminescent substrate (Pierce). To reprobe with a different antibody, the blots were stripped by incubation in 62.5 mM Tris-HCl (pH 6.8)–2% SDS–0.1 M β-mercaptoethanol for 30 min at 50°C.
When p21 was tested in the loading reaction with biotin-labeled DNA, 0.2% Nonidet P-40 (NP-40) was included in the reaction mixture (see Results). When the effect of p21 on the stability of the PCNA-RFC-DNA complex was examined, the complexes bound to beads were washed as described above, resuspended in 100 μl of buffer A containing 0.12 M NaCl, 0.2% NP-40, and 5 μM poly(dA)-oligo(dT) (19:1, in nucleotides), and incubated with or without p21 under the conditions indicated in the figure legends. The preincubation of PCNA with p21 in the experiment described below (see Fig. 6) was done in buffer B (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 10% glycerol, 0.01% NP-40, 12% sucrose, and 1 mM dithiothreitol) containing 0.12 M NaCl.
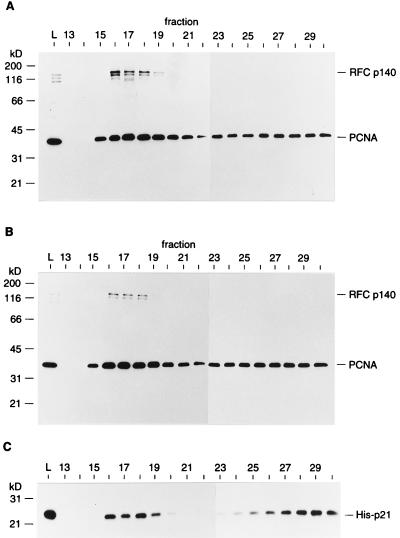
FIG. 6.
PCNA loaded onto nicked circular DNA remains associated with p21. The loading reaction was carried out with 0.3 pmol of nicked circular pSVO10 DNA, 6 pmol of RFC, and 2.2 pmol of PCNA trimer in the absence (A) or presence (B) of 22 pmol of His-p21. Under these conditions, the void volume was to around fraction 17. The proteins in each fraction were precipitated with trichloroacetic acid, separated in an SDS–12.5% polyacrylamide gel, and subjected to Western blotting with a mixture of anti-RFC p140 and anti-PCNA antibodies. (C) The blot shown in panel B was reprobed with anti-p21 antibody. Lane L represents 1/20 of the protein used for the loading reaction. The positions of molecular mass markers (in kilodaltons) are shown on the left.
Western blotting signals were quantitated by using an IS-1000 (version 1.97) digital imaging system (Alpha Innotech Corporation).
Gel filtration assay.
Nicked circular pSVO10 DNA (0.3 pmol) was incubated for 10 min at 37°C with RFC, PCNA, and 1 mM ATP, in the presence or absence of His-p21 (see Fig. 7), in a 100-μl reaction volume containing buffer A with 0.12 M NaCl. Then, the mixture was transferred onto ice and immediately applied onto a 5-ml (0.55- by 21-cm) Bio-Gel A1.5m (Bio-Rad) column equilibrated with buffer A with 0.12 M NaCl. Thirty-five fractions (170 μl each) were collected at 4°C. The proteins in each fraction were precipitated with 20% trichloroacetic acid and 0.08% sodium deoxycholate and analyzed by SDS–12.5% polyacrylamide gel electrophoresis and Western blotting as described above.
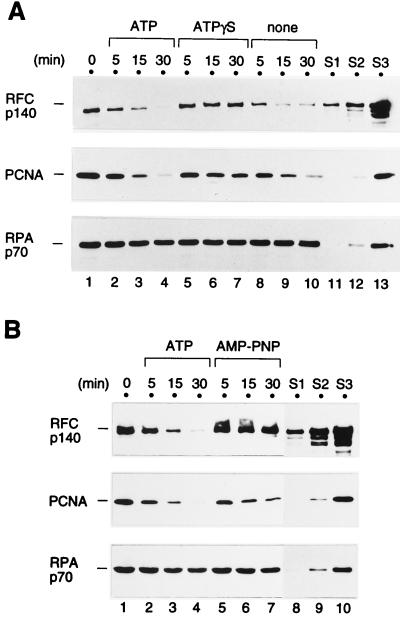
FIG. 7.
Stability of the primer recognition complex in the presence of ATP or one of its nonhydrolyzable analogs. The primer recognition complex was first formed on the primed BTN3 template DNA by incubating the DNA-coated beads with 2 pmol of RPA, 2.2 pmol of RFC, 2 pmol of PCNA trimer, and 1 mM ATP for 20 min at room temperature. The beads were washed with the buffer without nucleotide as described in Materials and Methods (lanes 1, 0 min [A and B]) and then resuspended in buffer A containing 0.12 M NaCl, 5 μM poly(dA)-oligo(dT), and 1 mM ATP (lanes 2 to 4 [A and B]), ATPγS (lanes 5 to 7 [A]), AMP-PNP (lanes 5 to 7 [B]), or no nucleotide (lanes 8 to 10 [A]), after which they were incubated further for the indicated period of time at room temperature. Finally, the beads were washed and the proteins that bound to them were analyzed by Western blotting. All of the reactions in both panels were processed at the same time, and the proteins were run on the same SDS-polyacrylamide gel. The standards (S1 to S3) are as described in the legend to Fig. 2.
RESULTS
PCNA loading on primed template DNA fixed on beads.
To examine the effect of the p21 protein on the formation of a PCNA-RFC-DNA complex on the primer-template, we designed a new assay for RFC-catalyzed loading of PCNA onto DNA, a method which allowed us to analyze multiple reactions relatively quickly without the use of any cross-linking reagent. This assay utilized a 90-mer DNA template that was primed and labeled with biotin at its 5′ or 3′ end (Fig. 1). As described in Materials and Methods, the biotin-labeled, primed template DNA was fixed onto streptavidin-agarose beads prior to the incubation with proteins. The primer was located in the middle of the template DNA, leaving about 30 nucleotides of single-stranded DNA at either end so that RPA, the single-stranded DNA binding protein, could bind to either side of the primer.
After the protein complex was formed on the DNA-coated beads, the unbound proteins were washed away, and the proteins remaining on the beads were subjected to Western blot analysis. Alternatively, the protein-bound beads were immediately processed further by resuspending them in another buffer. Because the washing process took approximately 30 min under these conditions, very transient DNA-protein interactions should not have been detected by this assay. Instead of capturing the preformed protein-DNA complexes directly, the biotin-primed template was fixed to streptavidin beads because it was possible that the recovery of protein-DNA complexes might vary, depending on the state of the complex formed.
The efficiency of the binding of biotin-labeled DNA to streptavidin-agarose beads was examined by using 32P-end-labeled biotin-DNA templates (BTN3 and BTN5). We determined that approximately 50 to 60% of input biotin-labeled DNA bound to the streptavidin beads under these conditions, irrespective of the end to which the biotin was attached (data not shown). Nonspecific binding of the biotin-labeled DNA to agarose beads was negligible under the same conditions (approximately 0.4% of input DNA bound to Sepharose CL-4B).
Requirement for RPA in PCNA loading.
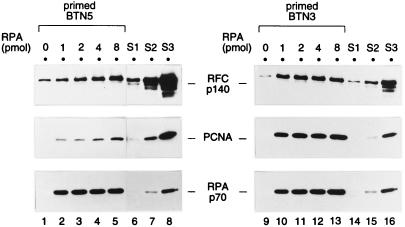
Previous studies of the formation of the primer recognition complex, using a gel shift assay (70), have shown that RPA can prevent RFC from binding to the single-stranded DNA region, thereby facilitating specific recognition of a primer-template junction by RFC (i.e., a junction between a single-stranded DNA template and the 3′ end of the duplex DNA section). Thus, first we examined whether RPA would be required in this new assay. Increasing amounts of RPA were tested in the presence of ATP with either primed BTN5 or primed BTN3 template DNA (Fig. 2). Both PCNA and RFC could be recovered with the DNA-coated beads. Importantly, the recovery of PCNA was observed only when RPA was present, indicating that RPA was required for PCNA loading in this assay. In the case of the primed BTN3 template DNA, the recovery of PCNA was saturated with 2 pmol of RPA (lane 11), which appeared to be a stoichiometric amount if one took into consideration that about 0.6 pmol of the primed template DNA had been fixed to the beads and that there were two binding sites for RPA in each primed template. There was a notable difference, however, between primed BTN5 and primed BTN3 in the efficiency of recovery of both RFC and PCNA, BTN3 being clearly the better substrate. This difference was not due to a different efficiency of binding of the biotin-labeled DNA to streptavidin-agarose beads (data not shown). This conclusion was also supported by the fact that RPA was recovered to a similar extent with both primed templates (Fig. 2, bottom panels). This template preference was further addressed, as described below. On the basis of this experiment, 2 pmol of RPA was used with the primed BTN3 template for most of the reactions described below.
FIG. 2.
RPA is required for PCNA loading onto a primer-template fixed on agarose beads. The loading reactions (100-μl reaction volumes) were carried out in the presence of 1 mM ATP with increasing amounts of RPA (0 to 8 pmol), 1 pmol of primed template BTN5 (lanes 1 to 5) or primed template BTN3 (lanes 9 to 13), 2 pmol of PCNA trimer, and 2.2 pmol of RFC. After incubation for 30 min at room temperature, the beads were washed at 0°C with buffer A containing 0.2 mM ATP. The proteins remaining on the beads, along with protein standards, were subjected to Western blotting. The protein standards were as follows: 0.2, 0.4, and 0.8 pmol of RFC; and 0.02, 0.05, and 0.2 pmol each of PCNA trimer and RPA in standards S1, S2, and S3, respectively. Note that the blots shown on the left (lanes 1 to 8), probed with anti-RFC p140 and anti-PCNA antibodies, were exposed longer than those on the right (lanes 9 to 16). Each set of samples (lanes 1 to 8 or 9 to 16) was run on the same SDS-polyacrylamide gel.
Requirement for ATP and primer in PCNA loading.
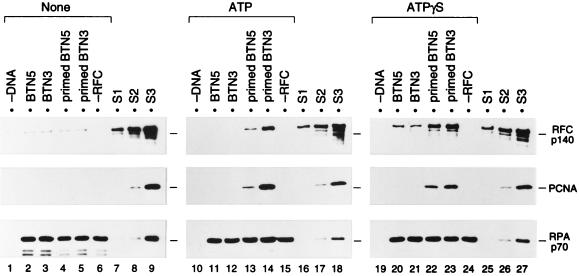
To verify that the recovery of both RFC and PCNA with the DNA-coated beads truly reflected ATP-dependent formation of the primer recognition complex, the requirement for ATP and primer was investigated. In the experiments shown in Fig. 3, constant amounts of both RFC and PCNA were incubated with primed or unprimed template DNA (BTN3 or BTN5)-coated beads in the absence or presence of ATP. Nonhydrolyzable analogs of ATP, ATPγS and AMP-PNP, were also tested (Fig. 3, right panels, and data not shown). In the absence of nucleotide, RPA bound to the DNA but neither RFC nor PCNA was recovered (lanes 1 to 5). The faint bands of RFC seen across the lanes are probably due to nonspecific binding of RFC to the agarose beads. In contrast, in the presence of ATP or ATPγS, both PCNA and RFC were recovered with either BTN3 or BTN5 template DNA only when a primer was present (lanes 13, 14, 22, and 23). PCNA was not recovered without RFC (lanes 15 and 24). Therefore, these results indicated that even when the primed template DNA was fixed on agarose beads, RFC could catalyze the loading of PCNA onto DNA, forming a primer-recognition complex in the presence of ATP or ATPγS. Such a complex was not observed when AMP-PNP was used instead of ATP (data not shown).
FIG. 3.
ATP- and primer-specific formation of a primer recognition complex on DNA fixed onto agarose beads. The loading reaction was carried out in the absence of nucleotide (lanes 1 to 6) or in the presence (lanes 10 to 15) of ATP or ATPγS (lanes 19 to 24), using 1 pmol each of BTN5 without primer (lanes 2, 11, and 20), BTN3 without primer (lanes 3, 12, and 21), primed BTN5 (lanes 4, 13, and 22), or primed BTN3 (lanes 5, 14, and 23), along with 2 pmol of RPA, 2.2 pmol of RFC, and 20 pmol of PCNA trimer. Reactions without DNA (lanes 1, 10, and 19) or RFC (lanes 6, 15, and 24) were also performed. The beads, after incubation for 30 min at room temperature, were washed at 0°C with buffer A containing no nucleotide (lanes 1 to 6), 0.2 mM ATP (lanes 10 to 15), or 0.2 mM ATPγS (lanes 19 to 24). The bound proteins and standards were analyzed as described in the legend to Fig. 2. Each set of samples (lanes 1 to 9, 10 to 18, or 19 to 27) was run on the same SDS-polyacrylamide gel. Note that although 10-fold more PCNA was used here as well as for the study shown in Fig. 4, the efficiency of loading of PCNA was essentially the same with either amount of PCNA. S1, S2, and S3, standards (see legend to Fig. 2).
The amount of PCNA recovered with primed BTN3 template DNA in the presence of ATP was clearly more than 0.2 pmol of trimer (compare to the 0.2 pmol of PCNA standard shown in Fig. 3, lane 18, middle panel). Therefore, since the streptavidin-agarose beads bound approximately 60% of the input DNA (equivalent to 0.6 pmol), a significant fraction of the primed template fixed on the beads must have been occupied by PCNA, assuming that only one PCNA trimer could be loaded per primed template DNA. Based on quantitative immunoblotting with highly purified RFC (data not shown), the RFC standard in lanes 8, 17, and 26 was equivalent to 0.4 pmol of RFC, indicating that the primer recognition complex was formed stoichiometrically on the DNA-coated beads (compare lanes 14 and 17).
Among the proteins used in the reactions described above, only RFC was partially purified (second phosphocellulose fraction [68]). However, PCNA could be loaded onto primed BTN3 to a similar extent even when highly purified RFC (glycerol gradient fraction [68]) was used (data not shown).
The results above are consistent with previous results obtained by using a gel shift assay with a hairpin DNA substrate (70). However, in contrast with the previous experiments (70), we have failed to detect the formation of an RFC-DNA complex without PCNA by this assay using DNA fixed to beads in the presence of ATP (data not shown). In the presence of ATPγS, a primer-independent complex of RFC with single-stranded DNA was observed (Fig. 3, lanes 20 and 21), suggesting that RFC may bind nonproductively to single-stranded DNA and then be released by hydrolysis of ATP. This nonproductive binding, however, could not facilitate loading of PCNA (lanes 20 and 21).
One unique feature revealed by this assay was that the efficiency of PCNA loading varied depending on the end of the template to which the biotin was attached. As shown in Fig. 2 and 3, primed BTN3 template was a better primer-template DNA than primed BTN5, whereas similar amounts of RPA bound to both primed templates. The same result was obtained when another primer located at the 3′ end of the template was used (data not shown). Interestingly, however, when ATPγS was used instead of ATP, both PCNA and RFC were recovered to relatively similar extents with both templates (Fig. 3, lanes 22 and 23). This could be due partly to stabilization of the primer recognition complex by ATPγS (see below for details). One possible reason for the more efficient recovery of PCNA with primed BTN3 is that the streptavidin-agarose beads attached to the 3′ end of the template might have prevented the loaded PCNA from sliding off the 3′ end of the DNA. This could have resulted in an increased half-life of the PCNA-DNA complex.
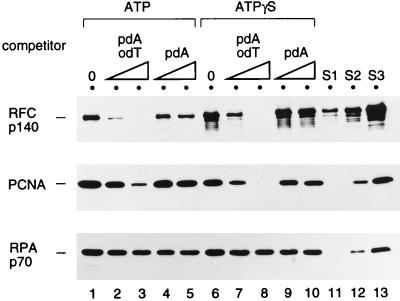
A competition experiment was also carried out to further verify that the RFC-catalyzed loading of PCNA was primer specific. In the loading reaction in the presence of ATP or ATPγS, different amounts of oligo(dT)-poly(dA) (primer-template) or poly(dA) (single-stranded DNA) were added. The amount of single-stranded poly(dA) was not sufficient to titrate the excess RPA in the reaction (Fig. 4). In contrast, oligo(dT)-poly(dA), but not poly(dA) alone, blocked formation of the RFC-PCNA-DNA complex, supporting the notion that the formation of the complex is primer specific. Considering the fact that 500 pmol of oligo(dT)-poly(dA) contains 1.6 pmol of oligo(dT) primer (lanes 3 and 8) and that approximately 0.6 pmol of biotin-labeled, primed template was detected on the beads, PCNA loading was completely inhibited by the addition of only a threefold molar excess of the primer-template competitor. Based on this result, oligo(dT)-poly(dA) was used as a trap to prevent the reformation of RFC-PCNA-DNA complexes on the DNA-coated beads in some of the experiments described below.
FIG. 4.
PCNA loading onto immobilized DNA is inhibited by soluble primed template DNA but not single-stranded DNA. A primed BTN3 template DNA fixed to the beads was first preincubated with 2 pmol of RPA for 5 min at room temperature and then mixed with 2.2 pmol of RFC, 20 pmol of PCNA trimer, and homopolymeric competitor DNA in the presence of 1 mM ATP (lanes 1 to 5) or ATPγS (lanes 6 to 10) for 30 min at room temperature. The beads were then washed with buffer A containing 0.2 mM ATP (lanes 1 to 5) or 0.2 mM ATPγS (lanes 6 to 10). The bound proteins were analyzed by Western blotting along with standards (S1 to S3) as described in the legend to Fig. 2. The competitor DNAs used were as follows: none (lanes 1 and 6), 250 pmol (in nucleotides) (lanes 2 and 7) or 500 pmol (lanes 3 and 8) of poly(dA)-oligo(dT) (pdA odT), and 250 pmol (lanes 4 and 9) or 500 pmol (lanes 5 and 10) of poly(dA) (pdA).
Effect of p21 on the loading of PCNA.
To examine the effect of p21 on the loading of PCNA, p21 was preincubated with PCNA and then the mixture was added to the loading reaction. p21 was added in a 6-fold (Fig. 5, lane 2) or a 17-fold (lane 3) molar excess over the amount of PCNA trimer. Thus, it is highly likely that almost all of the PCNA molecules were associated with p21 (72). Furthermore, a 10-fold molar excess of p21 over PCNA trimer fully inhibited RFC- and PCNA-dependent DNA synthesis by DNA polymerase δ on an RPA-coated, primed M13 DNA (data not shown).
FIG. 5.
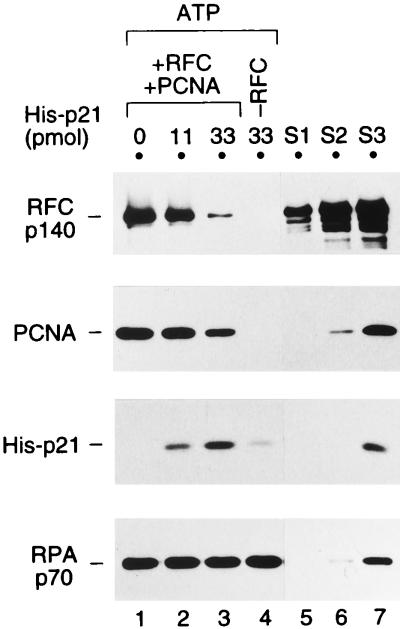
p21 does not inhibit PCNA loading; rather, it forms a complex with loaded PCNA. A loading reaction with 1 pmol of primed BTN3 template DNA and (except for lane 4) 2.2 pmol of RFC was carried out in the presence of 1 mM ATP as described in Materials and Methods, except that 2 pmol of PCNA trimer was preincubated with 0 (lane 1), 11 (lane 2), or 33 (lanes 3 and 4) pmol of His-p21 in buffer B containing 0.12 M NaCl for 20 min on ice. This mixture was then added into the PCNA loading reaction mixture supplemented with 0.25% NP-40. After incubation for 30 min at room temperature, the beads were washed with buffer A containing 0.2 mM ATP. The bound proteins were analyzed by Western blotting along with standards as described in the legend to Fig. 2. The reactions in lane 4 contained no RFC. For the standards, 0.06, 0.15, and 0.6 pmol of His-p21 were included in S1, S2, and S3, respectively, in addition to RFC, PCNA, and RPA as described in the legend to Fig. 2.
As shown in Fig. 5, when a sixfold molar excess of p21 was added to the loading reaction in the presence of ATP (lane 2), the formation of the primer recognition complex was hardly affected; more than 0.2 pmol of PCNA trimer was loaded (lanes 1 and 2; also compare to the 0.2-pmol PCNA standard in lane 7). Even when a 17-fold molar excess of p21 was added (lane 3), a significant amount of PCNA (approximately 0.1 pmol of trimer) was loaded (lane 3; compare to PCNA standards in lanes 6 and 7). However, the amount of RFC recovered was notably reduced upon addition of a 17-fold molar excess of p21 (compare lanes 1 and 3). This result suggests that p21 does not inhibit the loading of PCNA, even when p21 is present in a large excess, but p21 affects the stability of the formed primer recognition complex. Interestingly, p21 was also recovered with the beads (lanes 2 and 3). When RFC, PCNA, or RFC and PCNA were left out (lane 4 and data not shown), only faint bands of p21 were seen, probably due to its aggregation or nonspecific binding to the DNA or streptavidin-agarose. These data indicated that p21 remained associated with PCNA even after PCNA had been loaded onto the template DNA. Note that in order to reduce nonspecific binding of p21 to the beads, it was necessary to include 0.2 to 0.25% NP-40 in the loading reaction mixture. However, we have confirmed that neither RFC- and PCNA-dependent DNA synthesis nor its inhibition by p21 was affected in the presence of 0.25% NP-40 (data not shown).
In addition to the loading assay using the biotin-labeled template DNA, a gel filtration assay was also used to examine the effect of p21 on PCNA loading. In this assay, a nicked circular DNA was incubated with RFC and PCNA in the presence of ATP and then the mixture was directly applied to a gel filtration column. Under these conditions, free DNA as well as protein-DNA complexes, such as PCNA loaded onto nicked circular DNA, were large enough to be excluded from the gel filtration matrix, whereas unbound proteins were in the included volume (data not shown).
In the absence of p21, during the loading reaction a significant portion of input PCNA was recovered in the void-volume fractions (Fig. 6A, fractions 15 to 19). The recovery of PCNA in the void-volume fractions was ATP dependent (data not shown). Moreover, no PCNA was seen in the void-volume fractions when a supercoiled form of plasmid was used (data not shown). Consistent with the results from the loading assay described above, even when a 10-fold molar excess of p21 over PCNA trimer was present during the reaction, the loading of PCNA was hardly affected (Fig. 6B). Furthermore, some portion of input p21 was also recovered in the void-volume fractions (Fig. 6C). Since neither p21 nor PCNA was detected in the voided fractions when the DNA was linearized by BamHI digestion immediately after the loading reaction with p21 (data not shown), these results demonstrated that p21 could form a complex with the PCNA loaded on the DNA. It should be noted, however, that we could not conclude that RFC specifically bound to the nick under these conditions, because RFC was recovered in the void volume even when supercoiled DNA was used (data not shown). The same results were obtained by using the gel filtration assay with an RPA-coated, singly primed M13mp18 DNA; namely, even in the presence of a 24-fold molar excess of p21 over PCNA trimer, the loading of PCNA was not affected. Furthermore, both p21 and PCNA were recovered in the voided fractions in an RFC- and ATP-dependent manner (data not shown).
Taken together, the results of both PCNA loading assays demonstrated that p21 could not inhibit RFC-catalyzed loading of PCNA; rather, it formed a complx with the PCNA loaded onto the DNA.
Stability of the primer recognition complex.
The decreased recovery of RFC in the presence of p21 shown in Fig. 5 would imply that p21 might accelerate the dissociation of the primer recognition complex. Thus, we next wanted to examine the effect of p21 specifically on the preformed primer recognition complex.
In the experiment shown in Fig. 7, the stability of the preformed primer recognition complex was examined in the absence of p21. For this experiment, the complex was first formed on the primed BTN3 template in the absence of p21 and then the beads were extensively washed to remove unbound RFC and PCNA (Fig. 7, 0 min). Subsequently, the beads, on which the primer recognition complex remained, were resuspended in buffer containing a trap DNA and incubated further for various lengths of time at room temperature.
In the presence of ATP or in the absence of any nucleotide, the primer recognition complex gradually dissociated during the second incubation (Fig. 7A, lanes 1 to 4 and 8 to 10; Fig. 7B, lanes 1 to 4). Quantitation of the amount of bound RFC revealed that the half-life of the RFC-DNA complex was approximately 5 min under these conditions. In stark contrast, in the presence of ATPγS or AMP-PNP, RFC in the primer recognition complex was significantly more stable, since the majority of RFC remained on the DNA-coated beads even after 30 min (Fig. 7, lanes 5 to 7). This result indicated that even after the primer recognition complex was formed, these nonhydrolyzable analogs could bind to RFC and affect its stability on the DNA. As for PCNA, a slight stabilization was seen in the presence of the nonhydrolyzable analogs of ATP (Fig. 7A, middle panel, lanes 5 to 7).
Effect of p21 on a preformed primer recognition complex.
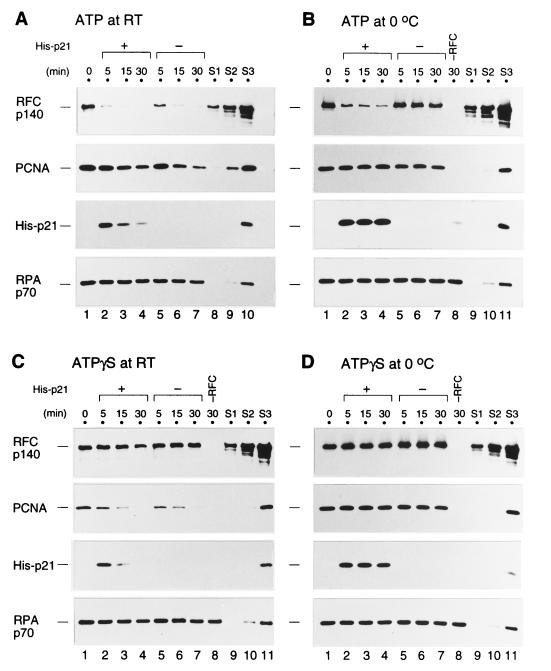
Next, a similar stability assay was performed in the presence of p21 (Fig. 8). In this experiment, the stability at 0°C and at room temperature was examined. As shown in Fig. 8A, no obvious effect of p21 on the stability of the loaded PCNA was detected in the presence of ATP at room temperature. In contrast, p21 did affect the stability of RFC, since slightly more RFC remained on the DNA-coated beads in the absence of p21, but the rate of RFC release was too rapid to easily observe this effect. Therefore, the rate of RFC release was slowed down by performing reactions at 0°C. In the absence of p21, the primer recognition complex was significantly stabilized by lowering the temperature of the second incubation, even in the presence of ATP (Fig. 8B, lanes 5 to 7). In the presence of p21, however, the majority of RFC was displaced from the DNA at 0°C (Fig. 8B, lanes 2 to 4). On the other hand, the majority of PCNA still remained on the DNA-coated beads throughout the incubation at 0°C (Fig. 8B, lanes 1 to 7), and p21 also remained bound to the PCNA (Fig. 8A and B, lanes 2 to 4). Only a faint band of p21 was seen without RFC (Fig. 8B, lane 8). These results indicated that the binding of p21 to the loaded PCNA stimulated the displacement of RFC from the DNA and consequently led to the disruption of the primer recognition complex, while the p21-PCNA complex remained on the DNA.
FIG. 8.
p21 facilitates the dissociation of RFC from an ATP-activated primer recognition complex but not from an ATPγS-activated complex. The complex was first formed in the absence of p21 on the primed BTN3 template DNA-coated beads as described in the legend to Fig. 7. The beads were then washed (0 min; lanes 1) and resuspended in buffer A containing 0.2% NP-40, 5 μM poly(dA)-oligo(dT), and either 1 mM ATP (A and B) or 1 mM ATPγS (C and D); then they were incubated further with 10 pmol of His-p21 (+) (lanes 2 to 4) or without His-p21 (−) (lanes 5 to 7) for the indicated time periods at room temperature (A and C) or 0°C (B and D). Finally, the beads were washed and the bound proteins were analyzed by Western blotting along with standards (S1 to S3) as described in the legend to Fig. 5. The loading reaction in lane 8 of panels B, C, and D contained no RFC.
The displacement of RFC by the addition of p21 was also seen without addition of ATP in the second incubation mixture (data not shown). Thus, additional ATP binding appears not to be required for the displacement of RFC by p21.
Interestingly, the primer recognition complex was no longer disrupted by p21 when ATPγS was used instead of ATP during the incubation with p21, either at room temperature or 0°C (Fig. 8C and D). Both PCNA and the PCNA-p21 complex were gradually displaced at room temperature, irrespective of the presence of p21 (Fig. 8C). At 0°C and with ATPγS, the primer recognition complex was stable, even in the presence of p21. Although we don’t know how ATPγS stabilizes a preformed primer recognition complex which has already hydrolyzed ATP during PCNA loading, the results shown here suggest that the state of the primer recognition complex stabilized by ATPγS would be different from that stabilized by lowering the temperature, based on the different response of RFC to the presence of p21 in the complex on the DNA.
DISCUSSION
Mechanism of inhibition of DNA replication by p21.
Our biochemical analysis of the loading of PCNA revealed that p21 did not immediately inhibit RFC-catalyzed loading of PCNA onto DNA. This means that the possible transient opening of the PCNA trimer, which is thought to be a prerequisite for its loading, is not affected, even if PCNA is complexed with p21 before loading. In addition, our results argue that the p21-PCNA complex in solution is also efficiently recognized by RFC. However, we cannot exclude the possibility that p21 transiently dissociates from the PCNA trimer during the loading of PCNA. Gibbs et al. (26) showed that adding p21 or a peptide derived from p21 that is capable of binding to PCNA inhibited the loading of PCNA onto nicked circular DNA by only 50%. Although their experiment did not address how much RFC remained on the DNA, the 50% inhibition they observed could be due to the accelerated dissociation of RFC from DNA upon the formation of the p21-PCNA complex on DNA, as shown in this study.
Both PCNA loading assays, i.e., the biotin-labeled DNA beads method and the gel filtration assay, have revealed that p21 can form a complex with PCNA loaded on the DNA. This is consistent with the crystal structure of the PCNA-p21 peptide complex, showing that the trimeric ring of PCNA and its central hole for the interaction with DNA are maintained even in the p21 peptide-bound form (28). The p21-PCNA complex on the DNA could be formed by either loading of preformed p21-PCNA complex or association of p21 with preloaded PCNA trimer. A quaternary complex consisting of p21, PCNA, RFC, and DNA may exist transiently in the presence of ATP, but subsequently RFC is displaced from the DNA, leaving the p21-PCNA complex on the DNA. PCNA stimulates the RFC ATPase activity and thus stimulates RFC dissociation (reference 69 and data described herein), and the present results suggest that the PCNA-p21 complex is a more effective activator of RFC dissociation from the DNA.
Although the role of RFC in the assembly of the DNA polymerase holoenzyme after PCNA loading is not yet known, it is conceivable that after PCNA is bound, the RFC-PCNA acts synergistically to load DNA polymerase and then remains part of the polymerase complex throughout DNA synthesis, as suggested previously (71). This is also supported by the observation by Pan et al. that one of the small subunits of RFC, p40, physically interacts with polymerase δ (49). In this regard, it will be interesting to see, using this newly designed assay, whether RFC would be displaced from DNA upon association of PCNA with polymerase δ or upon initiation of DNA synthesis.
It has been shown that p21 inhibits PCNA-dependent DNA synthesis by polymerase δ under conditions where RFC is not required (72). Thus, it is also likely that some function of PCNA itself is impaired by the formation of the p21-PCNA complex on DNA. The crystal structure of PCNA complexed with the p21 peptide has revealed that the peptide interacts with the interdomain connecting loop (amino acids 119 to 127) of PCNA that is located on its surface (28). In addition, Roos et al. have shown that a monoclonal antibody that specifically recognizes the interdomain connecting loop can inhibit PCNA-dependent DNA replication (57). Furthermore, a mutational analysis of PCNA by Fukuda et al. has shown that some of the alanine substitution mutations of PCNA which lead to impaired stimulation of DNA polymerase δ are located in the same loop (24). Therefore, it is likely that the interdomain connecting loop serves to interact with other replication proteins, such as DNA polymerase δ and FEN1 (11, 41), and that p21 blocks these interactions in a competitive manner. Taken together, these data indicate that the inhibition of DNA replication by p21 is initiated by the displacement of RFC from the DNA and further enhanced by the impaired association of PCNA with DNA polymerase δ.
We have shown previously that p21 does not inhibit PCNA-dependent nucleotide excision repair in vitro or in vivo (39, 40). However, biochemical data obtained by other groups have suggested that p21 does indeed inhibit nucleotide excision repair in vitro and that the assembly of polymerase δ holoenzyme may not be inhibited by p21 (50, 54). We expect that this discrepancy will be clarified by further investigation of the mechanism of polymerase δ holoenzyme assembly and determination of how RFC and PCNA are involved in DNA repair. What is clear is that p21 is localized with PCNA to sites of DNA repair in vivo (39), and thus it is unlikely that p21 inhibits DNA repair in vivo. We suggest that either PCNA-dependent polymerase processivity is altered by p21, a different DNA polymerase (perhaps polymerase ɛ) is involved in PCNA-dependent repair, or, alternatively, PCNA loading during nucleotide excision repair does not depend on RFC.
Mechanism of formation of the primer recognition complex.
PCNA functions as a processivity factor for DNA polymerase δ. However, the process through which PCNA is engaged in the DNA replication machinery is complex and includes energy-driven, dynamic transitions, e.g., ATP-dependent loading of PCNA, formation of a primer recognition complex, and association with polymerase δ. The new assay described here has allowed us to analyze some of these steps in detail, looking at each protein component, such as PCNA, RFC, RPA, or p21.
The PCNA loading assay revealed that the preformed primer recognition complex can be stabilized with ATPγS or AMP-PNP. It is highly likely that upon binding of these ATP analogs to RFC in the primer recognition complex, the conformation of RFC is altered or stabilized, leading to increased stability of the entire complex. This could be due, for instance, to an altered DNA binding activity of RFC when it is present in the ATP- or ATPγS-activated state. The possibility of a difference in conformation of the complex or state of DNA binding of RFC in the presence of ATP or ATPγS was further substantiated by the differential sensitivity of RFC to p21; p21 could displace RFC in the ATP-activated complex but not in the presence of ATPγS. In a previous analysis of the PCNA-RFC-DNA complex by a DNase I footprinting assay, protection of DNA by the RFC-PCNA complex was detected in the presence of ATPγS, but not ATP, while the formation of similar complexes was detected by a gel shift assay with either ATPγS or ATP (70). This ATPγS-specific DNase I protection could reflect differences between RFC in an ATP- and an ATPγS-activated primer recognition complex, as seen in this study.
It is not clear whether RFC in the complex can continuously hydrolyze ATP. It is possible that once ATP is hydrolyzed, turnover of RFC occurs. Regarding this aspect, previous studies of the bacteriophage T4 gp44/62 proteins, a functional equivalent of RFC, demonstrated that the addition of aluminum tetrafluoride (AlF4), a phosphate analog, resulted in the formation of a gp44/62-ADP-AlF4 complex in a stable transition state during the loading of gp45 onto DNA (gp45 is a functional homolog of PCNA). This indicates that the ADP-bound gp44/62 proteins may exist transiently but long enough for it to associate with AlF4 (3). Furthermore, a processive T4 DNA polymerase complex failed to form once the gp44/62-ADP-AlF4 complex was formed (3). This suggests that turnover of gp44/62 is necessary for the assembly of a processive DNA polymerase complex.
Based on these observations, it is reasonable to assume that immediately after ATP is hydrolyzed by RFC during the loading of PCNA, an ADP-bound RFC exists in a transition state. This ADP-bound RFC may be displaced from the 3′ end of the primer but still remain bound to DNA, and it may be required for the subsequent assembly of the polymerase holoenzyme. Our PCNA loading assay has indicated that this ADP-bound RFC on the DNA is unstable at room temperature. In contrast, it is stable at 0°C but sensitive to p21, so that RFC can completely dissociate from the DNA upon formation of the p21-PCNA complex. However, upon addition of ATPγS or AMP-PNP into the ADP-bound RFC on the DNA, an exchange of nucleotides at the same ATP binding site or an additional binding of the ATP analog to one of the potential sites present in another RFC subunit might occur. This ATPγS-activated RFC, which has been shown to be stable and resistant to p21, might structurally mimic the gp44/62-ADP-AlF4 complex. Thus, it will be interesting to see if the ATPγS-activated complex can associate with DNA polymerase δ.
We have also shown that AMP-PNP can stabilize the preformed RFC-PCNA complex, but in the presence of this ATP analog, neither binding of RFC to the primer template DNA nor loading of PCNA was detected under the conditions described here (data not shown). This also supports the notion that the conformation of RFC itself can be altered following assembly of the primer recognition complex, so that an AMP-PNP-reactivated RFC binds to DNA with a higher affinity while AMP-PNP-bound free RFC may not tightly bind to the DNA. However, at present, it is not clear whether free RFC actually binds AMP-PNP.
We expect that the new assay presented here will facilitate studies designed to gain a more precise understanding of how turnover of ATP as well as RFC occurs during the assembly of the primer recognition complex and the subsequent formation of a processive DNA polymerase complex. In addition, since cDNAs for all five subunits of RFC have been cloned (for a review, see reference 32) and recombinant RFC has been reconstituted by expression in baculovirus-infected insect cells (9, 19, 53), it will be possible to test how RFC mutants, such as those deficient in ATP binding or hydrolysis, function in polymerase assembly.
It is intriguing that there was a notable difference between the 5′- and 3′-biotin-DNA templates in the amounts of both RFC and PCNA recovered in the complex. The 3′-biotin-DNA template is a better template than the 5′-labeled one. It can be reasonably speculated that the presence of streptavidin-agarose at the 3′ end prevents the PCNA trimer from sliding off the end of the DNA. This different efficiency is, however, not due to the different efficiency of PCNA loading itself, because with either primed template, both RFC and PCNA were recovered to relatively similar extents when ATPγS was used instead of ATP (Fig. 3). In this situation, once the primer recognition complex is formed, the complex is stabilized further by ATPγS. This stabilization could account for the similar recoveries of both RFC and PCNA on the two templates in the presence of ATPγS. However, it remains to be determined whether RFC can actually hydrolyze ATPγS.
In vivo functions of p21-PCNA interaction.
Beamish et al. (2) have reported that delayed progression through S phase of the cell cycle occurs in response to DNA damage by γ-irradiation in wild-type cells but not in ataxia telangiectasia-derived cells, in which the ATM gene that is required for induction of p21 and p53 expression has been mutated. This suggests that downstream of ATM function there might exist some mechanism(s) to control progression through S phase. Moreover, Paulovich and Hartwell (52) also reported a slow rate of S-phase progression in response to DNA damage in the yeast Saccharomyces cerevisiae, in which this slowing was dependent on the MEC1 and RAD53 genes. Thus, although it is not known whether p21 can regulate ongoing DNA replication in vivo, it would be interesting to see if the p21-PCNA interaction is involved in slowing of the progression of S phase. One attractive but speculative possibility is that the displacement of RFC by p21 is a signal for the activation of a cell cycle checkpoint and that this signal results in slow progression through S phase or arrest at the G2-M transition of the cell cycle. It is therefore of interest that Rfc5p, one of the RFC subunits in S. cerevisiae, has been shown genetically to be involved in the S-phase checkpoint (63), although the precise biochemical mechanism for this checkpoint has not been determined. Another possibility is that during DNA replication, p21 binds to PCNA within the replication machinery only under specific circumstances—for example, when the DNA polymerase is stalled at a damaged site (64). If this is the case, the assembly of a p21-PCNA complex and subsequent displacement of RFC might trigger an arrest in DNA replication to allow DNA repair pathways to gain access to the DNA repair lesion.
Since PCNA is loaded onto the DNA and can remain bound after dissociation of the clamp loader protein (RFC) and, possibly, DNA polymerase, it has the potential to be a landing pad for other proteins. Since p21 can bind to PCNA and the cyclin-CDKs simultaneously (77), the DNA-bound p21-PCNA complex could tether the cyclin-CDKs to the DNA at critical sites in the genome and thereby coordinate cyclin-CDK-dependent processes at these sites. This would represent a novel mechanism for targeting a protein kinase to regions of the genome. Interestingly, the p21-PCNA-cyclin-CDK quaternary complex is disrupted in many tumor cells (78), suggesting that this complex plays a role in maintaining genome integrity.
ACKNOWLEDGMENTS
We thank Gregory J. Hannon and David Beach for rabbit anti-p21 antiserum and Spencer Teplin for synthesis of the biotin-labeled oligonucleotides. We are also grateful to Alain Verreault and Viola Ellison for reading the manuscript and to Mike Ockler, Jim Duffy, and Phil Renna for preparation of the figures.
This research was supported by a grant from the National Cancer Institute (PO1CA13106).
REFERENCES
- 1.Bauer G A, Burgers P M J. The yeast analog of mammalian cyclin/proliferating-cell nuclear antigen interacts with mammalian DNA polymerase δ. Proc Natl Acad Sci USA. 1988;85:7506–7510. doi: 10.1073/pnas.85.20.7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beamish H, Williams R, Chen P, Lavin M F. Defect in multiple cell cycle checkpoints in ataxia-telangiectasia postirradiation. J Biol Chem. 1996;271:20486–20493. doi: 10.1074/jbc.271.34.20486. [DOI] [PubMed] [Google Scholar]
- 3.Berdis A J, Benkovic S J. Mechanism of bacteriophage T4 DNA holoenzyme assembly: the 44/62 protein acts as a molecular motor. Biochemistry. 1997;36:2733–2743. doi: 10.1021/bi962139l. [DOI] [PubMed] [Google Scholar]
- 4.Bravo R, Frank R, Blundell P A, MacDonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase δ. Nature. 1987;326:515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- 5.Brown J P, Wei W, Sedivy J M. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 6.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 7.Bunz F, Kobayashi R, Stillman B. cDNAs encoding the large subunit of human replicator factor C. Proc Natl Acad Sci USA. 1993;90:11014–11018. doi: 10.1073/pnas.90.23.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgers P M J. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases δ and ɛ. J Biol Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- 9.Cai J, Uhlmann F, Gibbs E, Flores-Rozas H, Lee C-G, Phillips B, Finkelstein J, Yao N, O’Donnell M, Hurwitz J. Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc Natl Acad Sci USA. 1996;93:12896–12901. doi: 10.1073/pnas.93.23.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen I-T, Akamatau M, Smith M L, Lung F-D T, Duba D, Roller P P, Fornace A J, Jr, O’Connor P M. Characterization of p21Cip1/Waf1 peptide domains required for cyclin E/Cdk2 and PCNA interaction. Oncogene. 1996;12:595–607. [PubMed] [Google Scholar]
- 11.Chen J, Chen S, Saha P, Dutta A. p21Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex. Proc Natl Acad Sci USA. 1996;93:11597–11602. doi: 10.1073/pnas.93.21.11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Jackson P K, Kirschner M W, Dutta A. Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature. 1995;374:386–388. doi: 10.1038/374386a0. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 15.Din S, Brill S, Fairman M P, Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 1990;4:968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- 16.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 17.El-Deiry W S, Harper J W, O’Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, Wiman K G, Mercer W E, Kastan M B, Kohn K W, Elledge S J, Kinzler K W, Vogelstein B. WAF1/CIP1 in induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 18.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 19.Ellison V, Stillman B. Reconstitution of recombinant human replication factor C (RFC) and identification of an RFC subcomplex possessing DNA-dependent ATPase activity. J Biol Chem. 1998;273:5979–5987. doi: 10.1074/jbc.273.10.5979. [DOI] [PubMed] [Google Scholar]
- 20.Fan Z, Lu Y, Wu X, DeBlasio A, Koff A, Mendelsohn J. Prolonged induction of p21Cip1/WAF1/CDK2/PCNA complex by epidermal growth factor receptor activation mediates ligand-induced A431 cell growth inhibition. J Cell Biol. 1995;131:235–242. doi: 10.1083/jcb.131.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flores-Rozas H, Kelman Z, Dean F B, Pan Z-Q, Harper J W, Elledge S J, O’Donnell M, Hurwitz J. Cdk-interacting protein 1 directly binds with proliferating cell nuclear antigen and inhibits DNA replication catalyzed by the DNA polymerase δ holoenzyme. Proc Natl Acad Sci USA. 1994;91:8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fotedar R, Fitzgerald P, Rousselle T, Cannella D, Doree M, Messier H, Fotedar A. p21 contains independent binding sites for cyclin and cdk2: both sites are required to inhibit cdk2 kinase activity. Oncogene. 1996;12:2155–2164. [PubMed] [Google Scholar]
- 24.Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtsuka E, Tsurimoto T. Structure-function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. J Biol Chem. 1995;270:22527–22534. doi: 10.1074/jbc.270.38.22527. [DOI] [PubMed] [Google Scholar]
- 25.Funk J O, Waga S, Harry J B, Espling E, Stillman B, Galloway D A. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbs E, Kelman Z, Gulbis J M, O’Donnell M, Kuriyan J, Burgers P M J, Hurwitz J. The influence of the proliferating cell nuclear antigen-interacting domain of p21CIP1 on DNA synthesis catalyzed by the human and Saccharomyces cerevisiae polymerase δ holoenzymes. J Biol Chem. 1997;272:2373–2381. doi: 10.1074/jbc.272.4.2373. [DOI] [PubMed] [Google Scholar]
- 27.Goubin F, Ducommun B. Identification of binding domains on the p21Cip1 cyclin-dependent kinase inhibitor. Oncogene. 1995;10:2281–2287. [PubMed] [Google Scholar]
- 28.Gulbis J M, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 29.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 30.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tasi L, Zhang P, Dobrorolski S, Bai C, Connell-Crowley L, Swindell E, Fox M P, Wei N. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henricksen L A, Umbricht C B, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 32.Hübscher U, Maga G, Podust V N. DNA replication accessory proteins. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 525–543. [Google Scholar]
- 33.Jones D L, Alani R M, Münger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelman Z. PCNA: structure, functions and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- 35.Krishna T S R, Kong X-P, Gary S, Burgers P M, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 36.Kriwacki R W, Hengst L, Tennant L, Reed S I, Wright P E. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci USA. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 38.Lee S-H, Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase δ, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci USA. 1990;87:5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li R, Hannon G J, Beach D, Stillman B. Subcellular distribution of p21 and PCNA in normal and repair-deficient cells following DNA damage. Curr Biol. 1996;6:189–199. doi: 10.1016/s0960-9822(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 40.Li R, Waga S, Hanon G, Beach D, Stillman B. Differential effects by the p21 CDK inhibitor on PCNA dependent DNA replication and DNA repair. Nature. 1994;371:534–537. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Li J, Harrington J, Lieber M R, Burgers P M J. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Jenkins C W, Nichols M A, Xiong Y. Cell cycle expression and p53 regulation of the cyclin-dependent kinase inhibitor p21. Oncogene. 1994;9:2261–2268. [PubMed] [Google Scholar]
- 43.Lin J, Reichner C, Wu X, Levine A J. Analysis of wild-type and mutant p21WAF-1 gene activities. Mol Cell Biol. 1996;16:1786–1793. doi: 10.1128/mcb.16.4.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo Y, Hurwitz J, Massague J. Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature. 1995;375:159–161. doi: 10.1038/375159a0. [DOI] [PubMed] [Google Scholar]
- 45.Macleod J F, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 46.Michieli P, Chedid M, Lin D, Pierce J H, Mercer W E, Givol D. Induction of WAF1/CIP1 by a p53-independent pathway. Cancer Res. 1994;54:3391–3395. [PubMed] [Google Scholar]
- 47.Nakanishi M, Robetorye R S, Adami G R, Pereira-Smith O M, Smith J R. Identification of the active region of the DNA synthesis inhibitory gene p21Sdi1/CIP1/WAF1. EMBO J. 1995;14:555–563. doi: 10.1002/j.1460-2075.1995.tb07031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakanishi M, Robetorye R S, Pereira-Smith O M, Smith J R. The C-terminal region of p21SDI1/WAF1/CIP1 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J Biol Chem. 1995;270:17060–17063. doi: 10.1074/jbc.270.29.17060. [DOI] [PubMed] [Google Scholar]
- 49.Pan Z-Q, Chen M, Hurwitz J. The subunits of activator 1 (replication factor C) carry out multiple functions essential for proliferating-cell nuclear antigen-dependent DNA synthesis. Proc Natl Acad Sci USA. 1993;90:6–10. doi: 10.1073/pnas.90.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan Z-Q, Reardon J T, Li L, Flores-Rozas H, Legerski R, Sancar A, Hurwitz J. Inhibition of nucleotide excision repair by the cyclin-dependent kinase inhibitor p21. J Biol Chem. 1995;270:22008–22016. doi: 10.1074/jbc.270.37.22008. [DOI] [PubMed] [Google Scholar]
- 51.Parker S B, Eichele G, Zhang P, Rawls A, Sands A T, Bradley A, Olson E N, Harper J W, Elledge S J. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 52.Paulovich A G, Hartwell L H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 53.Podust V N, Fanning E. Assembly of functional replication factor C expressed using recombinant baculoviruses. J Biol Chem. 1997;272:6303–6310. doi: 10.1074/jbc.272.10.6303. [DOI] [PubMed] [Google Scholar]
- 54.Podust V N, Podust L M, Goubin F, Ducommun B, Hübscher U. Mechanism of inhibition of proliferating cell nuclear antigen-dependent DNA synthesis by the cyclin-dependent kinase inhibitor p21. Biochemistry. 1995;34:8869–8875. doi: 10.1021/bi00027a039. [DOI] [PubMed] [Google Scholar]
- 55.Prelich G, Stillman B. Coordinated leading and lagging strand synthesis during SV40 DNA replication in vitro requires PCNA. Cell. 1988;53:117–126. doi: 10.1016/0092-8674(88)90493-x. [DOI] [PubMed] [Google Scholar]
- 56.Prelich G, Tan C K, Kostura M, Mathews M B, So A G, Downey K M, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase δ auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 57.Roos G, Jiang Y, Landberg G, Nielsen N H, Zhang P, Lee M Y W T. Determination of the epitope of an inhibitory antibody to proliferating cell nuclear antigen. Exp Cell Res. 1996;226:208–213. doi: 10.1006/excr.1996.0220. [DOI] [PubMed] [Google Scholar]
- 58.Russo A A, Jeffrey P D, Patten A K, Massague J, Pavletich N P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 59.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 60.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 61.Shim J, Lee H, Park J, Kim H, Choi E-J. A non-enzymatic p21 protein inhibitor of stress-activated protein kinases. Nature. 1996;381:804–807. doi: 10.1038/381804a0. [DOI] [PubMed] [Google Scholar]
- 62.Shivji M K K, Grey S J, Strausfeld U P, Wood R D, Blow J J. Cip1 inhibits DNA replication but not PCNA-dependent nucleotide excision-repair. Curr Biol. 1994;4:1062–1068. doi: 10.1016/s0960-9822(00)00244-x. [DOI] [PubMed] [Google Scholar]
- 63.Sugimoto K, Shimomura T, Hashimoto K, Araki H, Sugino A, Matsumoto K. Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc Natl Acad Sci USA. 1996;93:7048–7052. doi: 10.1073/pnas.93.14.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svoboda D L, Vox J-M H. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc Natl Acad Sci USA. 1995;92:11975–11979. doi: 10.1073/pnas.92.26.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tahara H, Sato E, Noda A, Ide T. Increase in expression level of p21sdi1/cip1/waf1 with increasing division age in both normal and SV40-transformed human fibroblasts. Oncogene. 1995;10:835–840. [PubMed] [Google Scholar]
- 66.Tan C K, Castillo C, So A G, Downey K M. An auxiliary protein for DNA polymerase-δ from fetal calf thymus. J Biol Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 67.Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two DNA polymerase complexes at the SV40 DNA replication origin. Nature. 1990;346:534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- 68.Tsurimoto T, Stillman B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol Cell Biol. 1989;9:609–619. doi: 10.1128/mcb.9.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsurimoto T, Stillman B. Functions of replication factor C and proliferating cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci USA. 1990;87:1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory factors. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 71.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase α and δ during initiation of leading and lagging strand synthesis. J Biol Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
- 72.Waga S, Hannon G J, Beach D, Stillman B. The p21 cyclin dependent kinase inhibitor directly controls DNA replication via interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 73.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 74.Waga, S., and B. Stillman. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem., in press. [DOI] [PubMed]
- 75.Warbrick E, Lane D P, Glover D M, Cox L S. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- 76.Waseem N H, Lane D P. Monoclonal antibody analysis of the proliferating cell nuclear antigen (PCNA). Structural conservation and the detection of a nucleolar form. J Cell Sci. 1990;96:121–129. doi: 10.1242/jcs.96.1.121. [DOI] [PubMed] [Google Scholar]
- 77.Xiong Y, Zhang H, Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- 78.Xiong Y, Zhang H, Beach D. Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev. 1993;7:1572–1583. doi: 10.1101/gad.7.8.1572. [DOI] [PubMed] [Google Scholar]
- 79.Zhang H, Hannon G J, Beach D. p21-containing cyclin kinases exist in both active and inactive states. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]