Abstract
Accurate diagnosis of chronic obstructive pulmonary disease (COPD) and exacerbations by metabolic biomarkers enables individualized treatment. Advanced metabolic detection platforms rely on designed materials. Here, we design mesoporous PdPt alloys to characterize metabolic fingerprints for diagnosing COPD and exacerbations. As a result, the optimized PdPt alloys enable the acquisition of metabolic fingerprints within seconds, requiring only 0.5 μL of native plasma by laser desorption/ionization mass spectrometry owing to the enhanced electric field, photothermal conversion, and photocurrent response. Machine learning decodes metabolic profiles acquired from 431 individuals, achieving a precise diagnosis of COPD with an area under the curve (AUC) of 0.904 and an accurate distinction between stable COPD and acute exacerbations of COPD (AECOPD) with an AUC of 0.951. Notably, eight metabolic biomarkers identified accurately discriminate AECOPD from stable COPD while providing valuable information on disease progress. Our platform will offer an advanced nanoplatform for the management of COPD, complementing standard clinical techniques.
Short abstract
Plasmonic alloys enhanced metabolic fingerprints in plasma due to photoinduced charge/energy transfer and a size-exclusion effect, achieving a precise diagnosis of COPD and exacerbations.
Introduction
Chronic obstructive pulmonary disease (COPD) is a prevalent condition characterized by persistent respiratory symptoms and airflow limitation and impacts approximately 300 million people globally.1,2 It is a major cause of morbidity and mortality worldwide, resulting in a global public health problem with mortality of about 3.2 million individuals annually.2−4 Advanced diagnostic methods could facilitate the development of subsequent treatment plans in time, improving the health outcomes of patients.5,6 However, COPD is a systemic condition, and spirometry, which is a commonly used clinical method for COPD diagnosis through the assessment of lung function alone, is still limited by its poor accuracy, accessibility, and patient compliance.7,8 Moreover, this method fails to provide information on the disease’s progress. In particular, there are no objective clinical tools available for the diagnosis of COPD exacerbations, which are the main factors of hospitalizations and mortality in COPD.1,8,9 Therefore, a reliable and noninvasive method, realized through robust analytical platforms for the diagnosis of COPD and exacerbations, is highly required.
Biomarkers allow the characterization of disease progression at the molecular level through noninvasive techniques,10,11 holding promise for the diagnosis of COPD and exacerbations.9,12,13 Recently, various gene (e.g., circulating miRNA)14,15 and protein biomarkers (e.g., C-reactive protein)16,17 have been reported for the diagnosis or evaluation of COPD, but their performance is suboptimal for clinical use due to their poor accuracy.13 Compared with genes and proteins, metabolites function as immediate indicators of biochemical activity and exhibit a close correlation with the COPD phenotype.6,13 Mass spectrometry (MS), specifically laser desorption/ionization (LDI) MS, has emerged as a robust analytical instrument for the high-throughput and sensitive detection of various metabolites.18−24 However, metabolic analysis is often impeded by the inherent challenges of concentration and purification, given the low concentration of metabolites and high complexity of samples in clinical specimens.10,25−27
Matrix materials play crucial roles in analyte detection, deciding the performance of LDI MS.28−31 In particular, noble metals are superior candidates for enhancing LDI efficiency, concerning the surface plasmon resonance and hot carriers generated under laser irradiation.32,33 However, most of the current matrixes focus on monometallic nanoparticles (e.g., Au, Ag, Pt, and Pd)34,35 or their composites with other materials,36−38 exhibiting insufficient sensitivity in LDI MS for clinical application. Additionally, there are very few studies investigating the relationship between particle size and selective LDI. Furthermore, nanoparticles with porous structures enable the enrichment of small metabolites and the exclusion of proteins, achieving selective ionization of metabolites.22,39 Therefore, bimetallic alloys with synergistic effects and mesoporous structure promise to alleviate the constraints of monometallic/metal-composite and nonporous matrixes and advance metabolite analysis toward precise diagnosis.
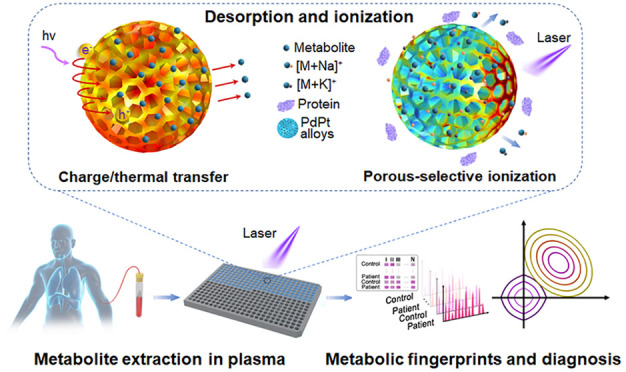
In this study, we constructed mesoporous PdPt-assisted LDI MS for metabolite profiling from plasma (Figure 1a), following the diagnosis of COPD and exacerbations and biomarker discovery via machine learning (Figure 1b). The excellent LDI performance of PdPt alloys was attributed to the enhanced electric field, photothermal conversion, photocurrent response, and mesoporous structure with size exclusion effect. The optimized PdPt alloys facilitated rapid metabolite profiling within seconds, requiring only 0.5 μL of native plasma without tedious pretreatment. By integration with machine learning, we achieved a precise diagnosis of COPD with an area under the curve (AUC) of 0.904 in the discovery cohort and an AUC of 0.955 in the validation cohort. Notably, our platform enables an accurate distinction between stable COPD (SCOPD) and acute exacerbations of COPD (AECOPD), with an AUC of 0.951 in the discovery cohort and an AUC of 0.976 in the validation cohort. Furthermore, eight identified metabolic biomarkers revealed a distinct signature for the discrimination of SCOPD and AECOPD. Our work provides an advanced nanoplatform for the precise diagnosis of COPD and exacerbations and valuable information on disease progress.
Figure 1.
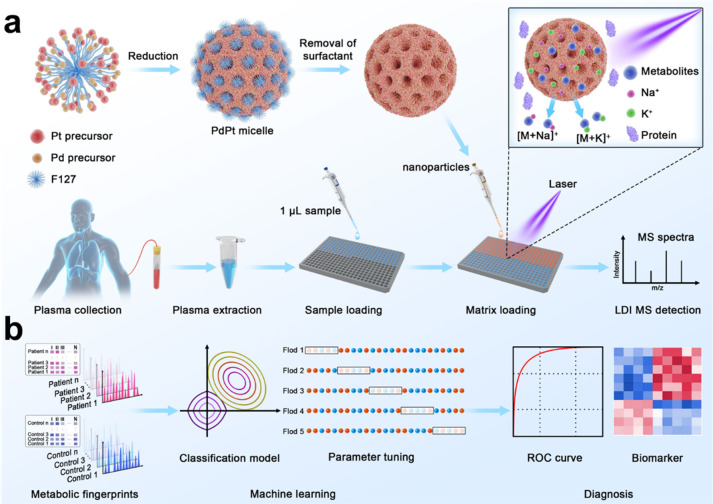
Schematics for extraction of plasma metabolic fingerprints toward the diagnosis of chronic obstructive pulmonary disease (COPD) and exacerbations. (a) Experimental procedure for the extraction of plasma metabolic fingerprints via laser desorption/ionization mass spectrometry (LDI MS) assisted by PdPt alloys. Mesoporous PdPt alloys were synthesized by utilizing F127 surfactant as a pore-directing template. One microliter of plasma extract (0.5 μL of native plasma) was used to obtain signals of metabolites with cation adducts (Na+/K+). (b) Schematic diagram for machine learning of metabolic fingerprints for diagnosis of COPD and exacerbations.
Results and Discussion
Synthesis and Characterization of Mesoporous PdPt Alloys
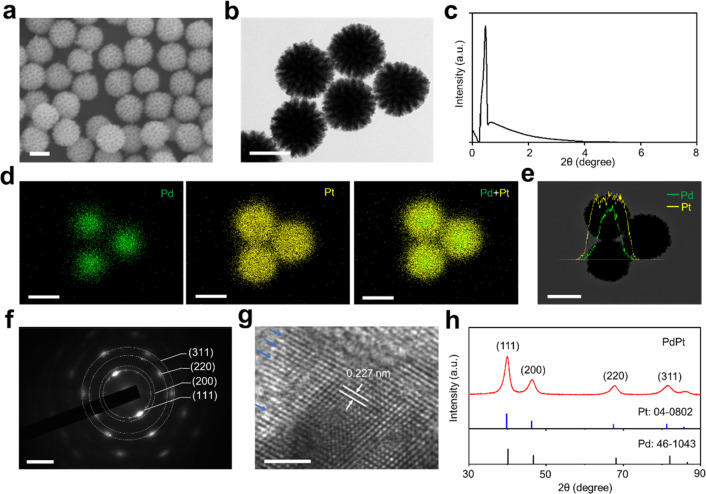
We successfully synthesized mesoporous PdPt spheres through a facile and modified surfactant-directing method.22 Briefly, the bimetallic alloys were obtained by the ascorbic acid-triggered reduction of Na2PdCl4 and H2PtCl6 in an aqueous solution, followed by the removal of F127 serving as pore-directing agents. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) images (Figure 2a,b) showed that the alloys were fairly uniform and possessed well-defined mesoporous structures with a pore size of approximately 20 nm. This was further characterized by low-angle X-ray diffraction (XRD) (Figure 2c) that showed a clear, sharp peak at 2θ = 0.46° (d = 19.0 nm), demonstrating the formation of a periodic mesoporous structure. The nitrogen adsorption–desorption isotherm of PdPt alloys exhibited a typical type-IV curve (Figure S1a) with a pronounced capillary condensation phenomenon at the relative pressure (P/P0) of 0.7–0.9, further evidencing the existence of a mesoporous structure.40,41 The specific surface area was ∼23.29 m2 g–1 as counted by the Brunauer–Emmett–Teller (BET) model. The pore size was ∼19.6 nm as calculated by the pore-size distribution curve (Figure S1b) based on the Barret–Joyner–Halenda (BJH) mode, which is almost consistent with the low-angle X-ray diffraction analysis. In particular, the mesopores on the alloy surface exhibited concavity toward the center of the particles, which was crucial to trapping small molecules while excluding larger nucleic acids and proteins in complex biofluids.22,42
Figure 2.
Characterization of PdPt alloys. Electron micrograph images of PdPt alloys, including (a) scanning electron microscopy (SEM) and (b) transmission electron microscopy (TEM). (c) Low-angle X-ray diffraction (XRD) pattern of PdPt with a sharp peak at 2θ = 0.46°. (d) Elemental mapping analysis and (e) line-scan energy-dispersive X-ray (EDX) results of PdPt alloys, with Pd in green and Pt in yellow, respectively. (f) Selected-area electron diffraction (SAED) pattern and (g) high-resolution TEM (HRTEM) of PdPt alloys. The arrows in (g) show the unsaturated Pt atoms. (h) Wide-angle XRD pattern of PdPt alloys, standard Pd (JCPDS: 46-1043) and Pt (JCPDS: 04-0802). Scale bar: (a, b, d, e) 100 nm for SEM, TEM, and mapping results, (f) 5/nm, and (g) 2 nm for HRTEM.
Furthermore, elemental mapping and line scan profiles were investigated to reveal the distributions of Pd and Pt in the particles. As shown in Figure 2d, elemental mapping showed a pronounced concentration of Pd within the core region of the particles, while Pt was found to be uniformly distributed across the entirety of the spherical structure. This was further revealed by the line scan profiles (Figure 2e). The selected-area electron diffraction (SAED) pattern (Figure 2f) and high-resolution TEM (Figure 2g) with clear lattice fringes demonstrated the high degree of crystallization of the bimetallic alloys.22,43 The crystal structure was evidenced by the wide-angle XRD (Figure 2h), yielding (111), (200), (220), and (311) diffraction peaks of face-centered-cubic (fcc) crystal structure assignable to Pd (JCPDS: 46-1043) and Pt (JCPDS: 04-0802).43,44 X-ray photoelectron spectroscopy (XPS) (Figure S2) showed doublet peaks of Pt0 4f7/2 and Pt0 4f5/2 as well as peaks of Pd 3f5/2 and Pd 3f3/2, respectively, evidencing the presence of metallic state Pt and Pd on the surface of the alloys.43,45 The surface Pt/Pd mole ratio of porous PdPt was calculated to be 6.3, demonstrating that the surface of PdPt is abundant in Pt, consistent with the elemental mapping result (Figure 2d,e). Notably, the PdPt alloys demonstrated an fcc crystal structure with an elevated density of surface atoms with low coordination, possessing high surface energy.46,47 In particular, the unsaturated Pt atomic structures (with some atomic edge and kink sites as shown in Figure 2g) on the surface can provide sufficient active sites and enhance analyte adsorption and charge transfer to analytes,43,48 which will be beneficial for the following LDI process enhancement.
Optimization of Mesoporous PdPt Alloys
LDI enhancement of nanomaterials was proven to be closely correlated with their elemental compositions and surface structures by numerous studies.22,49 However, rarely have reports demonstrated the effects of nanomaterial size on the LDI process. Therefore, we synthesized mesoporous PdPt nanoparticles with controlled particle size by changing the concentration of HCl (3 M (250 μL)/6 M (250 μL)/12 M (250 μL)/12 M (500 μL) in HCl aqueous solution, denoted as PdPt-1/2/3/4). As the HCl concentration increased, we observed increased particle size with an average size of approximately 90/150/200/240 nm for PdPt-1/2/3/4 alloys (Figure 3a–d and Figure S3). Energy-dispersive X-ray (EDX) analysis revealed similar Pd and Pt contents for PdPt-1/2/3/4 alloys (Figure S4 and Table S1), which was helpful in investigating the effect of particle size on the LDI process without the influence of elemental composition. The zeta potential demonstrated that the surface of PdPt-1/2/3/4 was negatively charged and the charge increased from −44.07 ± 0.65 mV to −22.77 ± 0.60 mV as the particle size increased (Figure S5 and Table S2), which is helpful for cation adduction and matrix use in LDI MS.22,49,50 All PdPt nanospheres exhibited strong absorption in the ultraviolet (UV) range (Figure S6). Considering the wavelength (355 nm) of the Nd:YAG laser used for LDI MS, mesoporous PdPt alloys may allow efficient laser energy transfer to analytes.22,51
Figure 3.
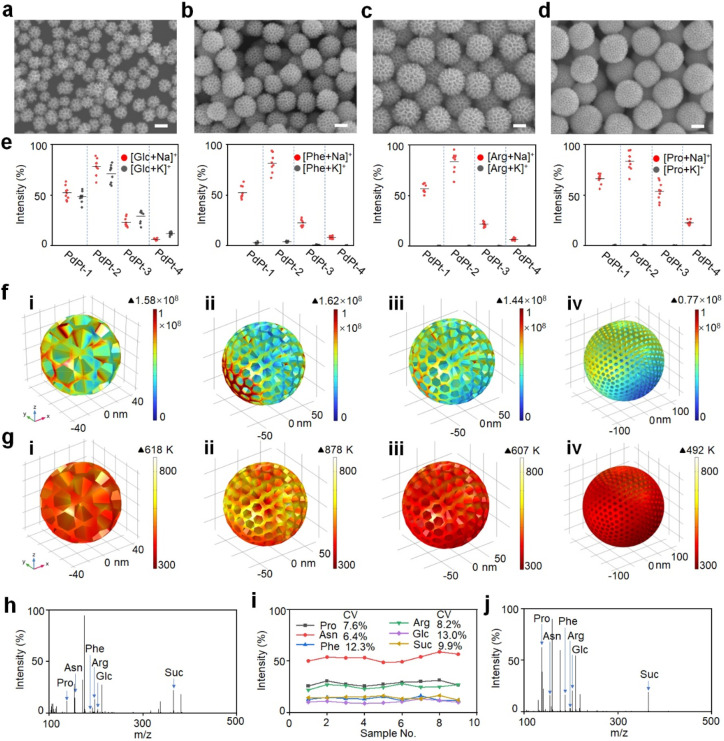
Optimization of PdPt alloys for metabolite detection. SEM images of (a) PdPt-1, (b) PdPt-2, (c) PdPt-3, and (d) PdPt-4 alloys with different particle sizes. (e) Intensities of sodium- and potassium-adducted signals of glucose (Glc), phenylalanine (Phe), arginine (Arg), and proline (Pro) detected by PdPt-1/2/3/4 alloy-assisted LDI MS. The error bars were determined as the standard deviation (s.d.) of nine measurements. Contour plots of (f) electric field amplitudes and (g) thermal field distribution shown on the color scale for (i) PdPt-1, (ii) PdPt-2, (iii) PdPt-3, and (iv) PdPt-4 alloys, for a 355 nm laser beam with polarization along the Y axis. Laser light was introduced along the Z-axis. The electric field amplitudes and thermal field distribution were counted by the finite element method. (h) Typical mass spectrometry of different metabolites (Pro, asparagine (Asn), Phe, Arg, Glc and sucrose (Suc)) in water solution, detected by PdPt-2 alloys assisted LDI MS. (i) Coefficient of variance (CV) distribution of different metabolites (Pro, Asn, Phe, Arg, Glc and Suc) analyzed by PdPt-2 alloys assisted LDI MS. The results come from nine independent experiments. (j) Typical mass spectrometry of different metabolites (Pro, Asn, Phe, Arg, Glc, and Suc) in a mixture solution of 0.5 M NaCl (salt tolerance detection) and 5 mg mL–1 bovine serum albumin (protein endurance detection), detected by PdPt-2 alloy-assisted LDI MS. Scale bar: (a, b, c, d) 100 nm.
To select the optimized materials for the following biofluids detection, a thorough LDI MS performance evaluation of the PdPt nanoparticles was conducted for glucose (Glc), phenylalanine (Phe), arginine (Arg), and proline (Pro) analysis. As a result, PdPt-2 presented significantly elevated signal intensities (p < 0.05) for different small molecules, superior to PdPt-1/3/4 alloys (Figure 3e). In particular, PdPt-2 alloys still exhibited higher signal intensity (p < 0.05) of glucose compared with that of other materials for plasma detection (Figure S7), indicating PdPt-2 alloys to be the matrix for the ideal detection of metabolites in clinical samples. Furthermore, various kinds of metabolites (30 small molecules) including nonpolar amino acids, polar amino acids, carbohydrates, alkaloids, nucleotides, fatty acids, and organic acids were used to comprehensively evaluate the LDI performance of PdPt-2 alloys. Consequently, PdPt alloys showed preferred performance for polar amino acids and carbohydrates (Figure S8) because metabolites with hydrophilic chemical groups (such as hydroxyl groups) can be desorbed and ionized on the surface of PdPt alloys with homogeneous cocrystallization.52,53 The superior LDI performance of PdPt-2 alloys was assigned to the enhancement of the electric field for desorption/ionization induced by light, photothermal conversion for desorption triggered by heat, and a photocurrent response for the inhibition of electron–hole recombination.
To demonstrate the electric field enhancement, a three-dimensional finite element simulation was conducted to model the electric/thermal field distribution when exposed to a 355 nm wavelength laser matched with the laser source of LDI MS. As a result, PdPt-2 offered the greatest electric field (shown as E2) of 1.62 × 108 (Figure 3f), a maximum ∼2.1-fold higher over PdPt-1/3/4 due to its unique particle size. Notably, porous bimetallic alloys displayed a superior electric field enhancement of 1.3–2.4-fold compared to that of porous/nonporous monometallic particles (Figure S9a), attributed to the facile charge transfer among multiple elements and porous structure.22,54 Specifically, a PdPt alloy with noble metal characteristics may generate hot carriers and strong optical near-field effects under UV (355 nm) illumination.32,55 Even though the resonance wavelengths of both Pd and Pt solid nanoparticles are not 355 nm, the PdPt alloy can generate a strong plasmonic effect under UV illumination due to the special alloy properties and porous structure. The alloy structure may lead to interactions between Pd and Pt, creating surface-isolated exciton coupling effects and altering the electromagnetic response of the single material.56,57 Furthermore, the nanopores act as resonators, concentrating the incoming electromagnetic field and generating high-density hotspots, despite the fact that the plasmons are not in resonance under short-wavelength irradiation.54,58 Thus, the surface adsorbates on the alloys are expected to generate an enhanced MS signal due to effective desorption/ionization induced by hot carriers that are triggered by near-field enhancement.22,59
For the photothermal conversion, a three-dimensional finite element simulation was first carried out to study the temperature of PdPt alloys under a 355 nm wavelength. The highest temperature reached 878 K for PdPt-2, higher than for PdPt-1/3/4 bimetallic alloys and monometallic particles (Figure 3g and Figure S9b), owing to its distinct particle size and lower thermal conductivity compared to that of monometals.60 The bimetallic alloys with lower thermal conductivity could minimize the diffusion of thermal energy and thus were heated to a higher temperature under the same laser irradiation fluence,60,61 leading to higher photothermal efficiency and the efficient desorption of nearby analytes compared to monometallic particles. The rapid relaxation processes of hot carriers produced by metals can cause local heating, resulting from the plasmon resonances, to boost light absorption in the surface area of the metal, consequently enhancing the conversion of absorbed light energy.62,63 In addition, the light absorption of a nonplanar (like porous) metal surface is more efficient than that of planar structures that reflect most of the incident light.62 Notably, the high temperature facilitating the partial melting of materials induced surface structural changes at high fluence areas (Figure S10), improving the thermal desorption and phase transition processes while facilitating the desorption/ionization of analytes.22,55,64 Although there are no significant differences in the pore diameter between PdPt-2 and PdPt-1/3 alloys, significant differences exist between PdPt-2 and PdPt-4 alloys (Figure S11a). We further compared the electric field and photothermal conversion of PdPt-2 and PdPt-4 alloys without significant differences in pore size. The PdPt-4 alloy model with a 20 nm pore size was built for COMSOL simulation, still exhibiting a lower maximum electric field (shown as E2) of 0.85 × 108 and a temperature of 513 K compared to those of PdPt-2 alloys (Figure S11b,c). For the photocurrent response, the PdPt-2 alloy displayed the strongest photocurrent intensity compared to PdPt-1/3/4 alloys (Figure S12), indicating a reduced rate of electron–hole recombination.51 Considering the distinctive optical, electric, and thermal characteristics, the PdPt-2 alloys offer notable advantages in the highly sensitive detection of metabolites.
To further demonstrate the advantage of the PdPt-2 matrix over traditional organic matrixes (α-cyano-4-hydroxycinnamic acid (CHCA) and 2,5-dihydroxybenzoic acid (DHB)), we compared the detection reproducibility in a mixture of six metabolites, including Pro, asparagine (Asn), Phe, Arg, Glc, and sucrose (Suc). Typical mass spectrometry of different metabolites is shown in Figure 3h. The intensity coefficients of variation (CVs) for six molecules were 7.6%, 6.4%, 12.3%, 8.2%, 13.0%, and 9.9%, respectively (Figure 3i), superior to CHCA and DHB with CV values of over 25.0% (Figure S13a,b). The excellent reproducibility of the PdPt-2 alloy benefited from the more homogeneous nanoparticle–analyte cocrystals, compared to the random sample crystallization with organic matrixes (Figure S13c–e).18,33 The limits of detection (LOD) for metabolites by PdPt alloys and organic matrixes were further studied by changing the molecule concentration from 100 to 0.1 μg mL–1. The regression equation exhibited excellent linear correlation results with R2 > 0.980 for each metabolite by PdPt alloy-assisted LDI MS analysis (Figure S14). The nanoplatform demonstrated high sensitivity detection for metabolites, with a LOD of as low as 0.3 pmol (Table S3). In contrast, organic CHCA and DHB matrixes showed poor linear correlation results with R2 = 0.697–0.975 (Figure S15). In particular, CHCA cannot detect the signals of Glc and Suc, and DHB cannot obtain the signals of Pro, Phe, Glc, and Suc, even at a high concentration of 100 μg mL–1 (Figure S15 and Table S3). Therefore, PdPt alloys outperform the organic matrix and exhibit lower limits of detection for small metabolite detection.
In light of metabolic abundance and sample complexity influencing the MS analysis, tedious pretreatment is essential for concentrating and segregating metabolites from complex biofluids.65 Nanoparticle with porous/crevices structure can concentrate metabolites in the nanopores for in situ macromolecule exclusion in biofluids, attaining a targeted LDI process of small metabolites.22,66 Consequently, the PdPt alloy with a porous structure realized the targeted and sensitive detection of metabolites (Figure 3j) in the presence of a high salt concentration (0.5 M NaCl) and protein content (5 mg mL–1 bovine serum albumin (BSA)). To further prove the size-selective effect in porous PdPt, glucose and BSA were used as model molecules for metabolites and protein, respectively, to observe the carbon element distribution in the alloy-analytes hybrids. The element mapping analysis revealed that the metabolites were captured by pores in alloy-metabolite hybrids, whereas such enrichment was not observed in the alloy-protein composites (Figure S16). Notably, we can observe a large number of small-molecule signal peaks in the detection of plasma, serum, and urine by PdPt-2 alloy-assisted LDI MS, outperforming CHCA and DHB as the matrix (Figure S17). Therefore, PdPt-2 alloys with optimized structure enabled the sensitive and selective detection of metabolites in complex biofluids toward clinical sample analysis.
Enhanced Metabolic Fingerprints for COPD Diagnosis
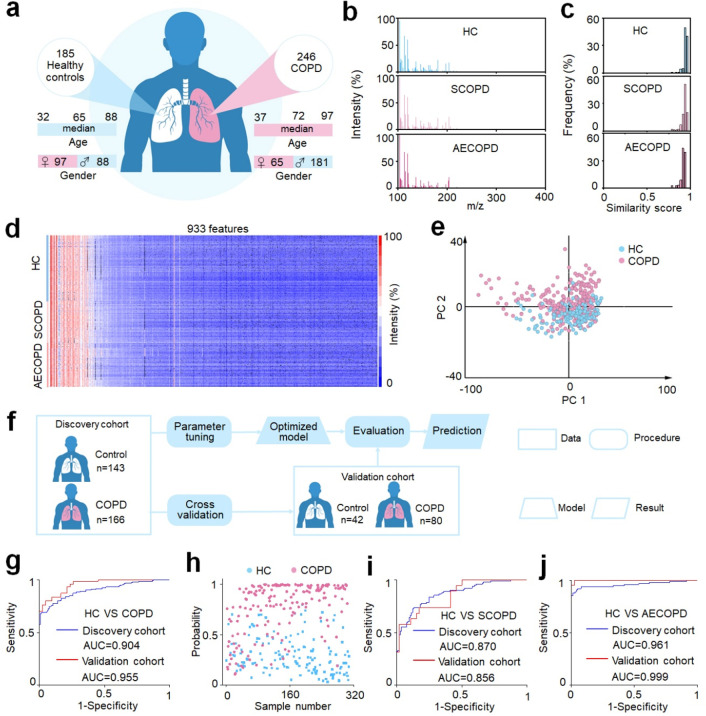
To uncover the specific metabolic signatures of COPD, we collected 431 plasma samples (Figure 4a) including 185 healthy controls and 246 COPD patients (122 SCOPD and 124 AECOPD individuals) for PdPt-2 alloy-assisted LDI MS analysis. The healthy controls were verified by spirometry with normal lung function (FEV1/FVC > 70%), and all patients were diagnosed with persistent airflow limitation based on the clinic criteria for the diagnosis of COPD.67,68 In this research, the test sample and the PdPt alloy suspension were prepared layer-by-layer (see details of Experimental Procedures in the Supporting Information) due to the high ionization efficiency.22,69 In contrast, the LDI performance showed a significant decrease (p < 0.05, Figure S18) if the alloy suspension was positioned ahead of the plasma sample or if the alloys and plasma were premixed. The metabolic fingerprints of plasma (Figure 4b) were successfully recorded in seconds in the low mass range (100–400 Da), with simple sample pretreatment and minimal sample consumption (0.5 μL of native plasma). Traditionally, liquid chromatography (LC) MS and nuclear magnetic resonance (NMR) are common methods for metabolite analysis, requiring tedious sample pretreatment (∼hours) and large sample consumption (∼milliliters).65,70 In contrast, PdPt-2 alloy-assisted LDI MS exhibited simple sample pretreatment (only protein precipitation required), improved detection speed, and minimized sample usage due to the enhanced sensitivity through optical/electrical/photothermal characteristics and in situ enrichment by the porous structure. In particular, over 95% of samples exhibited high similar scores over 0.85 for mass spectra in both the control and patient groups (Figure 4c), demonstrating the reliability of plasma metabolic fingerprints for further diagnostic applications. As a result, we obtained 933 m/z signals from the plasma via data processing (refer to the Experimental Procedures) and built the heat map composed of the metabolic data matrix of controls and patients (Figure 4d). However, we cannot see a significant difference between groups from the heat map. Furthermore, unsupervised analysis of the principle component analysis (PCA) showed a certain degree of separation without enough clarity between healthy controls and COPD patients (Figure 4e), indicating the imperative for developing a cutting-edge machine-learning method to find the distinct metabolic phenotype of COPD.
Figure 4.
Machine learning of metabolic fingerprints for COPD diagnosis. (a) Demographic characteristics of 431 clinical specimens, including age and gender information on 185 healthy controls (HC) and 246 COPD patients (122 stable COPD (SCOPD) patients and 124 acute exacerbations of COPD (AECOPD) patients). (b) Typical mass spectra of plasma extracts from HC, SCOPD, and AECOPD samples with m/z ranging from 100 to 400, using 0.5 μL of native plasma. (c) The frequency distribution of similarity scores was computed for HC, SCOPD, and AECOPD groups. (d) Metabolic fingerprints were extracted from raw mass spectra of 185 healthy controls and 246 COPD patients, each containing 933 m/z features. (e) The unsupervised principal component analysis (PCA) showed a certain degree of discrimination between 185 healthy controls and 246 COPD patients. (f) Workflow for the diagnosis of COPD by machine learning. The discovery cohort comprised 309 samples (143/166, HC/COPD) used for parameter tuning and model construction. The optimized model was evaluated using an independent validation cohort with 122 subjects (42/80, HC/COPD). No statistically significant differences in age and gender between HC and COPD in the discovery cohort (p > 0.05). (g) The receiver operator characteristic (ROC) curve differentiates HC from COPD for the discovery (blue) and validation (red) cohorts. (h) Scatter diagram for HC and COPD from the discovery cohort. A probability of close to 1 implied a high level of certainty in the model that the sample belonged to class 1 (patient). In contrast, a probability close to 0 indicated a model inclination toward classifying the sample as class 0 (healthy control).22,92 ROC curves differentiate (i) HC from SCOPD and (j) HC from AECOPD for the discovery (blue) and validation (red) cohorts.
To further reveal the unique metabolic signature of COPD, sparse learning was performed for metabolic data analysis. Initially, we grouped the 431 plasma samples into a discovery cohort (143 healthy controls and 166 COPD patients) for parameter tuning and model optimization and an independent validation cohort (42 healthy controls and 80 COPD patients) for model evaluation and prediction (Figure 4f). No statistically significant differences in age and gender were observed between the two groups in the discovery cohort (p > 0.05, Table S4). Subsequently, we carried out the power analysis on a preliminary study of 12 samples (6/6, healthy control/COPD patient) to count the minimum required sample size for detecting a statistically significant difference (Figure S19). As a result, a predicted power of 0.92 was obtained at a false discovery rate (FDR) of 0.1 with a sample number of 48 (24/24, healthy control/COPD patient), evidencing the statistics of machine learning at a confidence level. A receiver operating characteristic (ROC) curve was graphed to illustrate the performance of sparse learning for discriminating COPD from healthy controls, producing an averaged area under the curve (AUC) of 0.904 with a 95% confidence interval (CI) of 0.869–0.939 (with a sensitivity of 0.801, specificity of 0.853, and accuracy of 0.825) for the discovery cohort (Figure 4g). Furthermore, we confirmed that there was no overfitting for the sparse learning model via a permutation test (Figure S20a, 1000 times and p < 0.001). Similarly, a consistent performance generated by the optimized model for the independent validation cohort was obtained, showing an AUC of 0.955 with 95% CI of 0.923–0.988 (with a sensitivity of 0.838, specificity of 0.833, and accuracy of 0.836). Notably, the possibility of each subject (in the discovery cohort) being diagnosed as a patient by the classification model was plotted as the scatter diagram that exhibited a clear separation of these two groups, indicating metabolic alteration accompanied by disease occurrence (Figure 4h). These results evidenced the advanced diagnostic power toward COPD by metabolic fingerprints through the PdPt alloy-assisted LDI MS analysis.
To search for significant metabolic biomarkers for COPD diagnosis, we screened top-ranking 4 m/z features based on the frequency, p value, abundance, and AUC of a feature produced by the sparse learning for discovery cohort analysis (Figure S21a). The features were glucose, lactic acid, uric acid, and malondialdehyde (Table S5), confirmed by the human metabolome database (HMDB) (http://www.hmdb.ca/), accurate MS measurement on Fourier transform ion-cyclotron resonance mass spectrometry (FT-ICR-MS), and identification by ultraperformance liquid chromatography-MS (UHPLC-MS) analysis (<5 ppm). In particular, glucose was down-regulated, and lactic acid, uric acid, and malondialdehyde were up-regulated in COPD patients (Figure S21b–e), constructing a classification model with an AUC of 0.779 (CI: 0.727–0.831) for COPD diagnosis in the discovery cohort (Figure S21f). Globally, the prevalence of COPD in males was about double that in females mainly due to common tobacco smoking among men,1,71 a similar phenomenon found in this work (Table S4). However, in the discovery cohort, the p value of gender was 0.43, indicating no significant difference in gender between healthy controls and the COPD patients. Therefore, differences in the number of males and females did not affect diagnostic modeling and biomarker discovery for COPD. Additionally, we compared the metabolic differences between males and females. However, there were no significant metabolic differences (p > 0.05) between males and females (Figure S22) for the four top-ranking significant metabolic biomarkers (including glucose, lactic acid, uric acid, and malondialdehyde) in COPD diagnosis, further demonstrating that the diagnostic model was not affected by gender.
To comprehensively evaluate the performance of our platform in COPD diagnosis, for both stable and acute exacerbations, sparse learning was conducted for the discrimination of healthy controls and SCOPD as well as healthy controls and AECOPD, respectively. No significant differences existed in age and gender for both discovery cohorts (p > 0.05, Tables S6 and Table S7). Consequently, we obtained an AUC of 0.870 (with 95% CI of 0.823–0.971) in the discovery cohort (105 healthy controls and 103 SCOPD patients) and an AUC of 0.856 (with 95% CI of 0.760–0.952) in the validation cohort (46 healthy controls and 19 SCOPD patients; Table S6) for the discrimination of healthy controls and SCOPD (Figure 4i). Meanwhile, we got an AUC of 0.961 (with 95% CI of 0.932–0.991) in the discovery cohort (105 healthy controls and 102 AECOPD patients) and an AUC of 0.999 (with 95% CI of 0.995–1) in the validation cohort (46 healthy controls and 22 AECOPD patients; Table S7) for the distinction between healthy controls and AECOPD (Figure 4j). These results further demonstrated that the constructed PdPt alloy-assisted LDI MS platform was capable of achieving a precise diagnosis of COPD.
Clinically, the diagnosis of COPD is confirmed by the existence of consistent airflow limitation, as evaluated by postbronchodilator spirometry.1,67 However, this method generally underestimates the prevalence of COPD in younger patients and overestimates it in older patients.72 In particular, this method presents poor patient compliance for people with underlying diseases and acute exacerbations. Currently, there is a deficiency in reliable genes or protein biomarkers to assess the diagnosis of COPD.15,17,73 In contrast, metabolite biomarkers function as direct indicators of biochemistry activity, closely correlating the phenotype of disease.74 Here, we developed PdPt alloy-assisted LDI MS for the effective extraction of metabolic fingerprints in plasma due to the unique physiochemical property and porous structure. Notably, the AUC for the discrimination of healthy controls and COPD patients is beyond 0.90, demonstrating the dependability and application potential of metabolic fingerprints for COPD diagnosis.
Biomarker Discovery for AECOPD Diagnosis
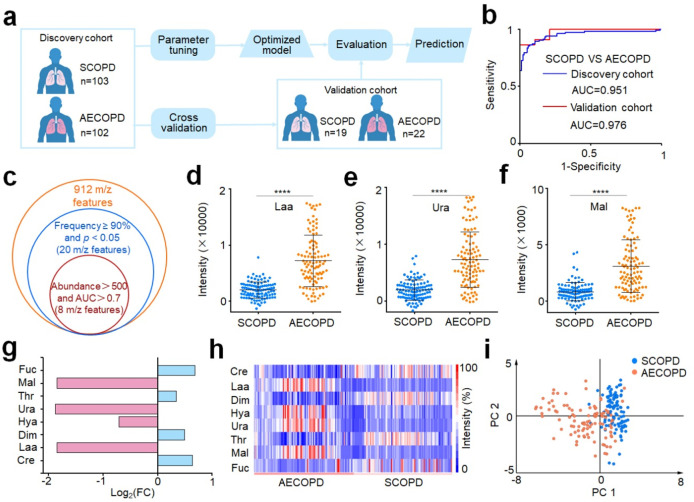
AECOPD features acute worsening of respiratory symptoms and is the primary cause of mortality from COPD,68,75 indicating that early diagnosis is important for patient survival and prognosis. To construct powerful tools for the diagnosis and assessment of AECOPD, we randomly grouped the 246 (122 SCOPD and 124 AECOPD patients) plasma samples into a discovery cohort (103 SCOPD and 102 AECOPD patients) for parameter tuning and model optimization and an independent validation cohort (19 SCOPD and 22 AECOPD patients) for model evaluation and prediction (Figure 5a). No statistically significant differences in age and gender were observed between the two groups in the discovery cohort (p > 0.05, Table S8). After obtaining the metabolic fingerprints of all plasma samples by PdPt alloy-assisted LDI MS, we extracted 912 m/z metabolic signals in plasma via data processing. Unsupervised PCA based on all of these signals was plotted to demonstrate the difference between SCOPD and AECOPD, yielding an unclear separation between the two groups (Figure S23a). We further performed sparse learning for the metabolic data analysis, achieving a precise diagnosis of AECOPD with an AUC of 0.951 (with 95% CI of 0.920–0.982, sensitivity of 0.852, specificity of 0.942, and accuracy of 0.898) in the discovery cohort (Figure 5b). Furthermore, we confirmed that there was no overfitting for the sparse learning model via a permutation test (Figure S20b, 1000 times and p < 0.001). Additionally, based on the optimized model, a comparable AUC of 0.976 (with 95% CI of 0.939-1, a sensitivity of 1, a specificity of 0.789, and an accuracy of 0.902) was obtained in the independent validation cohort (Figure 5b). The above results prove that our platform can realize the precise diagnosis of AECOPD, showing great potential in the management of AECOPD. SCOPD and AECOPD represent different stages of COPD’s disease course with different clinical features and treatment needs.68,76 In particular, AECOPD is a progressive stage of stable COPD (SCOPD), with a high rate of hospitalization and mortality. Distinguishing between them helps to tailor individualized treatment regimens more precisely, improving survival and prognosis.77 Furthermore, the onset of AECOPD accompanies the changes in biomarkers in the blood, and discriminating SCOPD and AECOPD will help to develop significant metabolic biomarkers for the early diagnosis and assessment of AECOPD progression.1,78
Figure 5.
Machine learning of metabolic fingerprints for AECOPD diagnosis and biomarker discovery. (a) Workflow for the diagnosis of AECOPD by machine learning. The discovery cohort included 205 samples (103/102, SCOPD/AECOPD) for parameter tuning and model construction. The optimized model was tested in an independent validation cohort with 41 individuals (19/22, COPD/AECOPD). No statistically significant differences in age and gender between SCOPD and AECOPD in the discovery cohort (p > 0.05). (b) ROC curves differentiate SCOPD from AECOPD for the discovery (blue) and validation (red) cohorts. (c) Venn diagram of 8 m/z features screened as the metabolic signature panel with frequency ≥90%, p < 0.05, abundance >500, and AUC of single feature >0.7. Scatter diagram of three key differential features for SCOPD and AECOPD, including (d) lactic acid (Laa), (e) uric acid (Ura), and (f) malondialdehyde (Mal). **** is represented by p < 0.0001. (g) Fold change of four up-regulated metabolites (Laa, Ura, Mal, and 3-hydroxybutyric acid (Hya) with magenta color) and four down-regulated metabolites (creatine (Cre), dimethylglycine (Dim), threonine (Thr), and fucose (Fuc) with cyan color) in AECOPD patients compared with SCOPD. (h) The heat map of the discovery cohort, including SCOPD and AECOPD patients, is constructed by eight metabolic biomarkers as potential signatures for AECOPD diagnosis. (i) PCA analysis showed a clear discrimination between SCOPD and AECOD patients based on eight metabolic biomarkers.
In clinical situations, there are no reliable biomarkers with enough sensitivity and specificity to predict AECOPD onset. To search for the distinct metabolic biomarkers between SCOPD and AECOPD, we screened top-ranking 8 m/z features based on the frequency, p value, abundance, and AUC of a single feature produced by the sparse learning for discovery cohort analysis (Figure 5c). These features were confirmed for eight metabolites according to the HMDB (http://www.hmdb.ca/), accurate MS measurement on FT-ICR-MS, and identification by UHPLC-MS analysis (<5 ppm). These biomarkers included creatine (Cre), lactic acid (Laa), dimethylglycine (Dim), 3-hydroxybutyric acid (Hya), uric acid (Ura), threonine (Thr), malondialdehyde (Mal), and fucose (Fuc). The scatter diagram demonstrated that four metabolites (Laa, Hya, Ura, and Mal) were up-regulated and four metabolites (Cre, Dim, Thr, and Fuc) were down-regulated in AECOPD compared with COPD (Figure 5d–f and Figure S24). This was further validated by the fold-change map, in which Laa, Hya, Ura, and Mal showed relatively high fold changes (Figure 5g). The heat map established with the eight-biomarker panel presented a noticeable distinction between SCOPD and AECOPD, offering proof of the promising metabolic assessment of AECOPD (Figure 5h). PCA of these eight metabolites (Figure 5i) displayed enhanced separation of SCOPD and AECOPD, compared with that of all 912 m/z features (Figure S23a). Notably, the diagnostic performance of the combination of eight biomarkers outperformed any single biomarker (Table S9), affording an enhanced AUC of 0.904 (95% CI of 0.859–0.950) for the discovery cohort and an AUC of 0.955 (95% CI of 0.874–1) for the validation cohort (Figure S23b). These results demonstrate that the eight screening metabolic biomarkers can accurately distinguish SCOPD from AECOPD, providing promising biomarkers for the diagnosis of COPD exacerbations and the assessment of COPD progress.
AECOPD often requires emergency department care and is also the primary cause of death from COPD.1,67 Ideal biomarkers facilitate the evaluation of the risk of exacerbation for individuals with stable COPD. Generally, C-reactive protein (CRP) is used for the diagnosis of AECOPD or the prediction of the frequency and severity of exacerbations.9,79 However, CRP is neither sufficiently sensitive nor specific, showing an AUC of 0.73 for the AECOPD diagnosis.80 In addition, fibrinogen and the leukocyte count are frequently used for the evaluation of risk in exacerbations, exhibiting poorer diagnostic results than the CRP biomarker.79,80 In contrast, the AUCs of the eight metabolic biomarkers identified for the discrimination SCOPD and AECOPD in this work, except for fucose, exceeded the AUC of the CRP biomarker (Table S9). Notably, the AUC of the eight metabolic biomarker panel is greater than 0.9, much better than for the CRP biomarker. Besides, the clinical diagnostic methods for discriminating SCOPD from AECOPD are time-consuming (the detection of multiple indicators, including respiratory rate, heart rate, oxygen saturation level, C-CRP levels, etc., will take several hours to days).77 In contrast, our nanoplatform offers a fast (within 1 h after getting the blood) tool for the diagnosis of AECOPD, facilitating the early diagnosis of AECOPD.
Biomarkers can not only be used for the construction of diagnostic models but also provide available information on the disease’s progress. For example, lactic acid is produced in large amounts in AECOPD patients due to their poor ability to breathe, which leads to the limitation of oxygen and enhancement of glycolysis.81,82 A serum uric acid level that increases significantly during hypoxia is associated with a higher risk of AECOPD and hospitalizations.83 Malondialdehyde concentrations are also elevated in AECOPD, requiring hospitalizations compared with stable COPD.84 Creatine and dimethylglycine commonly showed a decrease in blood due to the oxidation of creatine oxidation for AECOPD patients.85,86 3-Hydroxybutyric acid was increased, which may be attributed to the energy metabolism shift from carbohydrates to lipid utilization and amino acid metabolism in AECOPD.87 Threonine was found to be a potential biomarker for AECOPD diagnosis.88,89 Fucose may be the degradation product of the fucosylation glycoprotein, which was a biomarker of COPD.90 These metabolic biomarkers will provide powerful tools for the assessment and management of COPD. Furthermore, eight metabolic biomarkers were used with pathway enrichment analysis to explore biological relevance. The results demonstrated that the glycine, serine, and threonine metabolism were the most significantly altered metabolic pathways associated with COPD exacerbation (Figure S25, Table S10). The reason for amino acid metabolism dysfunction may arise from systemic inflammation and impaired energy metabolism in skeletal muscles.86,91
Conclusions
We constructed mesoporous PdPt alloys to reveal unique metabolic signatures for the diagnosis of COPD and exacerbations. The optimized PdPt alloys exhibited superior performance in metabolite detection, attributed to the enhanced electric field, robust photothermal conversion, and strengthened photocurrent response. The platform achieved precise diagnosis of COPD and exacerbations, with simple sample pretreatment, minimal sample consumption, and high speed. It is noteworthy that we screened distinct metabolic biomarkers for the diagnosis of COPD exacerbations, revealing the metabolic signatures in COPD progress. Although there are still limitations in the study, such as the fact that more samples should be collected from the multicenter to further validate the reliability of our platform and further study needs to be conducted to explore the mechanism of how metabolic biomarkers contribute to the AECOPD onset and progression, we believe that our platform would provide a noninvasive and robust tool to advance metabolic analysis for disease diagnosis and prognosis.
Acknowledgments
The authors gratefully acknowledge financial support from projects 2022YFE0103500, 2021YFF0703500, 2020YFC2003700, and 2022YFA0806200 by the National Key R&D Program of China, project BX2021182 by the China Postdoctoral Science Foundation, project 2022XYJG0001-03-0 by the Shanghai Jiao Tong University Inner Mongolia Research Institute, projects 81971771, 82070045, and 82270040 by the NSFC, and project 2021-01-07-00-02-E00083 by Shanghai Institutions of Higher Learning and the Innovative Research Team of High-Level Local Universities in Shanghai (SHSMU-ZDCX20210700). This work was also sponsored by the Innovation Group Project of Shanghai Municipal Health Commission (2019CXJQ03, 2022JC013, and 202240361), the Innovation Research Plan by the Shanghai Municipal Education Commission (ZXWF082101), the National Research Center for Translational Medicine Shanghai (TMSK-2021-124 and NRCTM(SH)-2021-06), the Medical-Engineering Joint Funds of Shanghai Jiao Tong University (YG2021ZD09, YG2022QN107, and YG2023ZD08), the Science and Technology Commission of Shanghai Municipality (20DZ2261200, 20DZ2254400, and 21DZ2200600), the Shanghai Municipal Key Clinical Specialty (shslczdzk02201), and the Shanghai Municipal Science and Technology Major Project (ZD2021CY001).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.3c01201.
Detailed chemicals, reagents, experimental procedures (preparation of mesoporous PdPt nanoparticles, material characterization, theoretical simulation, study cohort, LDI MS detection, machine-learning analysis, FT-ICR-MS analysis, UHPLC-MS analysis, statistical analysis), and supplementary figures and tables (PDF)
Author Contributions
Conceptualization: K.Q., J.Z., Y.S., and H.S. Methodology: H.S., S.Y., Z.Z., Y.S., S.C., and L.G. Investigation: L.Y., C.C., D.H., X.W., X.M., P.H., and D.S. Visualization: H.S. and J.Z. Supervision: Q.K., Y.S., J.Z., and D.S. Writing-original draft: H.S. Writing-review and editing: H.S., J.Z., Q.K., Y.S., S.Y., and Z.Z.
Author Contributions
△ H.S. and Y.S. contributed equally to this work.
The authors declare the following competing financial interest(s): The authors declare competing financial interest. The authors have filed patents for both the technology and the use of the technology to detect bio-samples.
Supplementary Material
References
- Christenson S. A.; Smith B. M.; Bafadhel M.; Putcha N. Chronic obstructive pulmonary disease. Lancet 2022, 399 (10342), 2227–2242. 10.1016/S0140-6736(22)00470-6. [DOI] [PubMed] [Google Scholar]
- Yang I. A.; Jenkins C. R.; Salvi S. S. Chronic obstructive pulmonary disease in never-smokers: risk factors, pathogenesis, and implications for prevention and treatment. Lancet Resp. Med. 2022, 10 (5), 497–511. 10.1016/S2213-2600(21)00506-3. [DOI] [PubMed] [Google Scholar]
- Seimetz M.; Sommer N.; Bednorz M.; Pak O.; Veith C.; Hadzic S.; Gredic M.; Parajuli N.; Kojonazarov B.; Kraut S.; et al. NADPH oxidase subunit NOXO1 is a target for emphysema treatment in COPD. Nat. Metab. 2020, 2 (6), 532–546. 10.1038/s42255-020-0215-8. [DOI] [PubMed] [Google Scholar]
- Martinez F. J.; Agusti A.; Celli B. R.; Han M. L. K.; Allinson J. P.; Bhatt S. P.; Calverley P.; Chotirmall S. H.; Chowdhury B.; Darken P.; et al. Treatment trials in young patients with chronic obstructive pulmonary disease and pre-chronic obstructive pulmonary disease patients time to move forward. Am. J. Respir. Crit. Care Med. 2022, 205 (3), 275–287. 10.1164/rccm.202107-1663SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbán C. J.; Han M. K.; Boes J. L.; Chughtai K. A.; Meyer C. R.; Johnson T. D.; Galbán S.; Rehemtulla A.; Kazerooni E. A.; Martinez F. J.; et al. Computed tomography-based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nat. Med. 2012, 18 (11), 1711–1715. 10.1038/nm.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratiu I. A.; Ligor T.; Bocos-Bintintan V.; Mayhew C. A.; Buszewski B. Volatile organic compounds in exhaled breath as fingerprints of lung cancer, asthma and COPD. J. Clin. Med. 2021, 10 (1), 32. 10.3390/jcm10010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin D. M. G.; Criner G. J.; Papi A.; Singh D.; Anzueto A.; Martinez F. J.; Agusti A. A.; Vogelmeier C. F.; Comm G. S. Global initiative for the diagnosis, management, and prevention of chronic obstructive lung disease the 2020 GOLD Science Committee report on COVID-19 and chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2021, 203 (1), 24–36. 10.1164/rccm.202009-3533SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmeier C. F.; Criner G. J.; Martinez F. J.; Anzueto A.; Barnes P. J.; Bourbeau J.; Celli B. R.; Chen R. C.; Decramer M.; Fabbri L. M.; et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report: GOLD executive summary. Eur. Respir. J. 2017, 49 (3), 1700214. 10.1183/13993003.00214-2017. [DOI] [PubMed] [Google Scholar]
- Sin D. D.; Hollander Z.; DeMarco M. L.; McManus B. M.; Ng R. T. Biomarker development for chronic obstructive pulmonary disease from discovery to clinical implementation. Am. J. Respir. Crit. Care Med. 2015, 192 (10), 1162–1170. 10.1164/rccm.201505-0871PP. [DOI] [PubMed] [Google Scholar]
- Shu Q.; Rajagopal M.; Fan J.; Zhan L.; Kong X.; He Y.; Rotcheewaphan S.; Lyon C. J.; Sha W.; Zelazny A. M.; et al. Peptidomic analysis of mycobacterial secreted proteins enables species identification. VIEW 2022, 3 (4), 20210019. 10.1002/VIW.20210019. [DOI] [Google Scholar]
- Song S.; Zhu L.; Wang C.; Yang Y. In vitro diagnostic technologies for the detection of extracellular vesicles: current status and future directions. VIEW 2023, 4 (2), 20220011. 10.1002/VIW.20220011. [DOI] [Google Scholar]
- Wang Z.; Locantore N.; Haldar K.; Ramsheh M. Y.; Beech A. S.; Ma W.; Brown J. R.; Tal-Singer R.; Barer M. R.; Bafadhel M.; et al. Inflammatory endotype-associated airway microbiome in chronic obstructive pulmonary disease clinical stability and exacerbations a multicohort longitudinal analysis. Am. J. Respir. Crit. Care Med. 2021, 203 (12), 1488–1502. 10.1164/rccm.202009-3448OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim W.; Wilde M. J.; Cordell R. L.; Richardson M.; Salman D.; Free R. C.; Zhao B.; Singapuri A.; Hargadon B.; Gaillard E. A.; et al. Visualization of exhaled breath metabolites reveals distinct diagnostic signatures for acute cardiorespiratory breathlessness. Sci. Transl. Med. 2022, 14 (671), eabl5849 10.1126/scitranslmed.abl5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Munoz J. M.; Rial M. J.; Sastre B.; Canas J. A.; Mahillo-Fernandez I.; Quirce S.; Sastre J.; Cosio B. G.; Del Pozo V. Circulating miRNAs as diagnostic tool for discrimination of respiratory disease: Asthma, asthma-chronic obstructive pulmonary disease (COPD) overlap and COPD. Allergy 2019, 74 (12), 2491–2494. 10.1111/all.13916. [DOI] [PubMed] [Google Scholar]
- Moll M.; Boueiz A.; Ghosh A. J.; Saferali A.; Lee S.; Xu Z. H.; Yun J. H.; Hobbs B. D.; Hersh C. P.; Sin D. D.; et al. Development of a blood-based transcriptional risk score for chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2022, 205 (2), 161–170. 10.1164/rccm.202107-1584OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander Z.; DeMarco M. L.; Sadatsafavi M.; McManus B. M.; Ng R. T.; Sin D. D. Biomarker development in COPD: moving from p values to products to impact patient care. Chest 2017, 151 (2), 455–467. 10.1016/j.chest.2016.09.012. [DOI] [PubMed] [Google Scholar]
- Butler C. C.; Gillespie D.; White P.; Bates J.; Lowe R.; Thomas-Jones E.; Wootton M.; Hood K.; Phillips R.; Melbye H.; et al. C-reactive protein testing to guide antibiotic prescribing for COPD exacerbations. N. Engl. J. Med. 2019, 381 (2), 111–120. 10.1056/NEJMoa1803185. [DOI] [PubMed] [Google Scholar]
- Huang Y. D.; Du S. Q.; Liu J.; Huang W. Y.; Liu W. S.; Zhang M. J.; Li N.; Wang R. M.; Wu J.; Chen W.; et al. Diagnosis and prognosis of breast cancer by high-performance serum metabolic fingerprints. Proc. Natl. Acad. Sci. U. S. A. 2022, 119 (12), e2122245119 10.1073/pnas.2122245119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtigall F. M.; Pereira A.; Trofymchuk O. S.; Santos L. S. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 2020, 38 (10), 1168–1173. 10.1038/s41587-020-0644-7. [DOI] [PubMed] [Google Scholar]
- Wang G.; Heijs B.; Kostidis S.; Mahfouz A.; Rietjens R. G. J.; Bijkerk R.; Koudijs A.; van der Pluijm L. A. K.; van den Berg C. W.; Dumas S. J.; et al. Analyzing cell-type-specific dynamics of metabolism in kidney repair. Nat. Metab. 2022, 4 (9), 1109–1118. 10.1038/s42255-022-00615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L.; Bu F.; Li X.; Zhang S.; Min L. Mass spectrometry-based extracellular vesicle micromolecule detection in cancer biomarker discovery: An overview of metabolomics and lipidomics. VIEW 2023, 4 (5), 20220086. 10.1002/VIW.20220086. [DOI] [Google Scholar]
- Su H. Y.; Li X. X.; Huang L.; Cao J.; Zhang M. J.; Vedarethinam V.; Di W.; Hu Z. Q.; Qian K. Plasmonic alloys reveal a distinct metabolic phenotype of early gastric cancer. Adv. Mater. 2021, 33 (17), 2007978. 10.1002/adma.202007978. [DOI] [PubMed] [Google Scholar]
- Chen Y. F.; Xu W.; Zhang W.; Tong R. Y.; Yuan A. C.; Li Z.; Jiang H. R.; Hu L. H.; Huang L.; Xu Y. D.; et al. Plasma metabolic fingerprints for large-scale screening and personalized risk stratification of metabolic syndrome. Cell Rep. Med. 2023, 4 (7), 101109. 10.1016/j.xcrm.2023.101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Shu W.; Zhao L.; Wan J. Advanced mass spectrometric and spectroscopic methods coupled with machine learning for in vitro diagnosis. VIEW 2023, 4 (1), 20220038. 10.1002/VIW.20220038. [DOI] [Google Scholar]
- Shao X.; Huang Y.; Wang G. Microfluidic devices for protein analysis using intact and top-down mass spectrometry. VIEW 2023, 4 (1), 20220032. 10.1002/VIW.20220032. [DOI] [Google Scholar]
- Wu J.; Liang C. M.; Wang X.; Huang Y. D.; Liu W. S.; Wang R. M.; Cao J.; Su X.; Yin T.; Wang X. L.; et al. Efficient metabolic fingerprinting of follicular fluid encodes ovarian reserve and fertility. Adv. Sci. 2023, 10 (23), 202302023. 10.1002/advs.202302023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. D.; Li B. C.; Stuart D. D.; Cheng Q. Three-dimensional printed microfluidic mixer/extractor for cell lysis and lipidomic profiling by matrix-assisted laser desorption/ionization mass spectrometry. VIEW 2023, 4 (1), 20220041. 10.1002/VIW.20220041. [DOI] [Google Scholar]
- Iakab S. A.; Rafols P.; Garcia-Altares M.; Yanes O.; Correig X. Silicon-based laser desorption ionization mass spectrometry for the analysis of biomolecules: a progress report. Adv. Funct. Mater. 2019, 29 (45), 1903609. 10.1002/adfm.201903609. [DOI] [Google Scholar]
- Mandal A.; Singha M.; Addy P. S.; Basak A. Laser desorption ionization mass spectrometry: Recent progress in matrix-free and label-assisted techniques. Mass Spectrom. Rev. 2019, 38 (1), 3–21. 10.1002/mas.21545. [DOI] [PubMed] [Google Scholar]
- Yang J.; Yin X.; Zhang L. Z.; Zhang X. W.; Lin Y.; Zhuang L. Z.; Liu W. S.; Zhang R.; Yan X. C.; Shi L.; et al. Defective Fe metal-organic frameworks enhance metabolic profiling for high-accuracy diagnosis of human cancers. Adv. Mater. 2022, 34 (26), 2201422. 10.1002/adma.202201422. [DOI] [PubMed] [Google Scholar]
- Wang R. M.; Gu Z. W.; Wang Y.; Yin X.; Liu W. S.; Chen W.; Huang Y. D.; Wu J.; Yang S. Z.; Feng L.; et al. A ″One-Stop Shop″ decision tree for diagnosing and phenotyping polycystic ovarian syndrome on serum metabolic fingerprints. Adv. Funct. Mater. 2022, 32 (45), 2206670. 10.1002/adfm.202206670. [DOI] [Google Scholar]
- Wei X.; Liu Z. H.; Jin X. L.; Huang L.; Gurav D. D.; Sun X. M.; Liu B. H.; Ye J.; Qian K. Plasmonic nanoshells enhanced laser desorption/ionization mass spectrometry for detection of serum metabolites. Anal. Chim. Acta 2017, 950, 147–155. 10.1016/j.aca.2016.11.017. [DOI] [PubMed] [Google Scholar]
- Chiang C.-K.; Chen W.-T.; Chang H.-T. Nanoparticle-based mass spectrometry for the analysis of biomolecules. Chem. Soc. Rev. 2011, 40 (3), 1269–1281. 10.1039/C0CS00050G. [DOI] [PubMed] [Google Scholar]
- Palermo A.; Forsberg E. M.; Warth B.; Aisporna A. E.; Billings E.; Kuang E.; Benton H. P.; Berry D.; Siuzdak G. Fluorinated gold nanoparticles for nanostructure imaging mass spectrometry. ACS Nano 2018, 12 (7), 6938–6948. 10.1021/acsnano.8b02376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelhamid H. N. Nanoparticle-based surface assisted laser desorption ionization mass spectrometry: a review. Microchim. Acta 2019, 186 (10), 682. 10.1007/s00604-019-3770-5. [DOI] [PubMed] [Google Scholar]
- Kim M.-J.; Yun T. G.; Noh J.-Y.; Song Z.; Kim H.-R.; Kang M.-J.; Pyun J.-C. Laser-induced surface reconstruction of nanoporous Au-modified TiO2 nanowires for in situ performance enhancement in desorption and ionization mass spectrometry. Adv. Funct. Mater. 2021, 31 (29), 2102475. 10.1002/adfm.202102475. [DOI] [Google Scholar]
- Yin X.; Yang J.; Zhang M. J.; Wang X. Y.; Xu W.; Price C. A. H.; Huang L.; Liu W. S.; Su H. Y.; Wang W. J.; et al. Serum metabolic fingerprints on bowl-shaped submicroreactor chip for chemotherapy monitoring. ACS Nano 2022, 16 (2), 2852–2865. 10.1021/acsnano.1c09864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.; Gurav D. D.; Wu S.; Xu W.; Vedarethinam V.; Yang J.; Su H. Y.; Wan X. Z.; Fang Y.; Shen B. Y.; et al. A multifunctional platinum nanoreactor for point-of-care metabolic analysis. Matter 2019, 1 (6), 1669–1680. 10.1016/j.matt.2019.08.014. [DOI] [Google Scholar]
- Chen W.; Yu H. J.; Hao Y.; Liu W. S.; Wang R. M.; Huang Y. D.; Wu J.; Feng L.; Guan Y. T.; Huang L.; et al. Comprehensive metabolic fingerprints characterize neuromyelitis optica spectrum disorder by nanoparticle-enhanced laser desorption/ionization mass spectrometry. ACS Nano 2023, 17 (20), 19779–19792. 10.1021/acsnano.3c03765. [DOI] [PubMed] [Google Scholar]
- Zhao T.; Chen L.; Liu M.; Lin R.; Cai W.; Hung C.-T.; Wang S.; Duan L.; Zhang F.; Elzatahry A.; et al. Emulsion-oriented assembly for Janus double-spherical mesoporous nanoparticles as biological logic gates. Nat. Chem. 2023, 15 (6), 832–840. 10.1038/s41557-023-01183-4. [DOI] [PubMed] [Google Scholar]
- Xu L.; Guo M.; Hung C.-T.; Shi X.-L.; Yuan Y.; Zhang X.; Jin R.-H.; Li W.; Dong Q.; Zhao D. Chiral skeletons of mesoporous silica nanospheres to mitigate Alzheimer’s β-amyloid aggregation. J. Am. Chem. Soc. 2023, 145 (14), 7810–7819. 10.1021/jacs.2c12214. [DOI] [PubMed] [Google Scholar]
- Huang L.; Zhou Y.; Zhu Y. X.; Su H. Y.; Yang S. Z.; Feng L.; Zhao L.; Liu S. R.; Qian K. Dual-modal nanoplatform integrated with smartphone for hierarchical diabetic detection. Biosens. Bioelectron. 2022, 210, 114254. 10.1016/j.bios.2022.114254. [DOI] [PubMed] [Google Scholar]
- Jiang B.; Li C. L.; Malgras V.; Imura M.; Tominakaa S.; Yamauchi Y. Mesoporous Pt nanospheres with designed pore surface as highly active electrocatalyst. Chem. Sci. 2016, 7 (2), 1575–1581. 10.1039/C5SC03779D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B.; Li C.; Imura M.; Tang J.; Yamauchi Y. Multimetallic mesoporous spheres through surfactant-directed synthesis. Adv. Sci. 2015, 2 (8), 1500112. 10.1002/advs.201500112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B.; Li C.; Henzie J.; Takei T.; Bando Y.; Yamauchi Y. Morphosynthesis of nanoporous pseudo Pd@Pt bimetallic particles with controlled electrocatalytic activity. J. Mater. Chem. A 2016, 4 (17), 6465–6471. 10.1039/C6TA00037A. [DOI] [Google Scholar]
- Huang X. Q.; Zhao Z. P.; Fan J. M.; Tan Y. M.; Zheng N. F. Amine-assisted synthesis of concave polyhedral platinum nanocrystals having {411} high-index facets. J. Am. Chem. Soc. 2011, 133 (13), 4718–4721. 10.1021/ja1117528. [DOI] [PubMed] [Google Scholar]
- Lim B.; Jiang M. J.; Camargo P. H. C.; Cho E. C.; Tao J.; Lu X. M.; Zhu Y. M.; Xia Y. N. Pd-Pt bimetallic nanodendrites with high activity for oxygen reduction. Science 2009, 324 (5932), 1302–1305. 10.1126/science.1170377. [DOI] [PubMed] [Google Scholar]
- Fan D. P.; Guo K.; Zhang Y.; Hao Q. Q.; Han M.; Xu D. D. Engineering high-entropy alloy nanowires network for alcohol electrooxidation. J. Colloid Interface Sci. 2022, 625, 1012–1021. 10.1016/j.jcis.2022.06.105. [DOI] [PubMed] [Google Scholar]
- Cao J.; Shi X. J.; Gurav D. D.; Huang L.; Su H. Y.; Li K. K.; Niu J. Y.; Zhang M. J.; Wang Q.; Jiang M. W.; et al. Metabolic fingerprinting on synthetic alloys for medulloblastoma diagnosis and radiotherapy evaluation. Adv. Mater. 2020, 32 (23), 2000906. 10.1002/adma.202000906. [DOI] [PubMed] [Google Scholar]
- Huang Y. D.; Yang H. J.; Li J. K.; Wang F. Q.; Liu W. S.; Liu Y. W.; Wang R. M.; Duan L. J.; Wu J.; Gao Z. W.; et al. Diagnosis of esophageal squamous cell carcinoma by high-performance serum metabolic fingerprints: A retrospective study. Small Methods 2023, 8, 202301046. 10.1002/smtd.202301046. [DOI] [PubMed] [Google Scholar]
- Pei C. C.; Wang Y.; Ding Y. J.; Li R. X.; Shu W. K.; Zeng Y.; Yin X.; Wan J. J. Designed concave octahedron heterostructures decode distinct metabolic patterns of epithelial ovarian tumors. Adv. Mater. 2023, 35 (18), 202209083. 10.1002/adma.202209083. [DOI] [PubMed] [Google Scholar]
- Lee J.; Lee J.; Chung T. D.; Yeo W.-S. Nanoengineered micro gold shells for LDI-TOF analysis of small molecules. Anal. Chim. Acta 2012, 736, 1–6. 10.1016/j.aca.2012.05.040. [DOI] [PubMed] [Google Scholar]
- Wu H.-P.; Su C.-L.; Chang H.-C.; Tseng W.-L. Sample-first preparation: A method for surface-assisted laser desorption/ionization time-of-flight mass spectrometry analysis of cyclic oligosaccharides. Anal. Chem. 2007, 79 (16), 6215–6221. 10.1021/ac070847e. [DOI] [PubMed] [Google Scholar]
- Liu K.; Bai Y. C.; Zhang L.; Yang Z. B.; Fan Q. K.; Zheng H. Q.; Yin Y. D.; Gao C. B. Porous Au-Ag nanospheres with high-density and highly accessible hotspots for SERS analysis. Nano Lett. 2016, 16 (6), 3675–3681. 10.1021/acs.nanolett.6b00868. [DOI] [PubMed] [Google Scholar]
- Stopka S. A.; Holmes X. A.; Korte A. R.; Compton L. R.; Retterer S. T.; Vertes A. Trace analysis and reaction monitoring by nanophotonic ionization mass spectrometry from elevated bowtie and silicon nanopost arrays. Adv. Funct. Mater. 2018, 28 (29), 1801730. 10.1002/adfm.201801730. [DOI] [Google Scholar]
- Li J. R.; Zhang G. N.; Wang J.; Maksymov I. S.; Greentree A. D.; Hu J. M.; Shen A. G.; Wang Y. L.; Trau M. Facile one-pot synthesis of nanodot-decorated gold-silver alloy nanoboxes for single-particle surface-enhanced Raman scattering activity. ACS Appl. Mater. Interfaces 2018, 10 (38), 32526–32535. 10.1021/acsami.8b10112. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Zhang Q.; Fu Z. W.; Qin D. Transformation of Ag nanocubes into Ag-Au hollow nanostructures with enriched Ag contents to improve SERS activity and chemical stability. ACS Appl. Mater. Interfaces 2014, 6 (5), 3750–3757. 10.1021/am500506j. [DOI] [PubMed] [Google Scholar]
- Li L. H.; Jiang R. T.; Shan B. B.; Lu Y. X.; Zheng C.; Li M. Near-infrared II plasmonic porous cubic nanoshells for in vivo noninvasive SERS visualization of sub-millimeter microtumors. Nat. Commun. 2022, 13 (1), 5249. 10.1038/s41467-022-32975-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka S. A.; Rong C.; Korte A. R.; Yadavilli S.; Nazarian J.; Razunguzwa T. T.; Morris N. J.; Vertes A. Molecular imaging of biological samples on nanophotonic laser desorption ionization platforms. Angew. Chem.-Int. Ed. 2016, 55 (14), 4482–4486. 10.1002/anie.201511691. [DOI] [PubMed] [Google Scholar]
- Yagnik G. B.; Hansen R. L.; Korte A. R.; Reichert M. D.; Vela J.; Lee Y. J. Large scale nanoparticle screening for small molecule analysis in laser desorption ionization mass spectrometry. Anal. Chem. 2016, 88 (18), 8926–8930. 10.1021/acs.analchem.6b02732. [DOI] [PubMed] [Google Scholar]
- Lai S. K. M.; Tang H. W.; Lau K. C.; Ng K. M. Nanosecond UV laser ablation of gold nanoparticles: enhancement of ion desorption by thermal-driven desorption, vaporization, or phase explosion. J. Phys. Chem. C 2016, 120 (36), 20368–20377. 10.1021/acs.jpcc.6b06261. [DOI] [Google Scholar]
- Brongersma M. L.; Halas N. J.; Nordlander P. Plasmon-induced hot carrier science and technology. Nat. Nanotechnol. 2015, 10 (1), 25–34. 10.1038/nnano.2014.311. [DOI] [PubMed] [Google Scholar]
- Linic S.; Chavez S.; Elias R. Flow and extraction of energy and charge carriers in hybrid plasmonic nanostructures. Nat. Mater. 2021, 20 (7), 916–924. 10.1038/s41563-020-00858-4. [DOI] [PubMed] [Google Scholar]
- Silina Y. E.; Koch M.; Volmer D. A. Influence of surface melting effects and availability of reagent ions on LDI-MS efficiency after UV laser irradiation of Pd nanostructures. J. Mass Spectrom. 2015, 50 (3), 578–585. 10.1002/jms.3564. [DOI] [PubMed] [Google Scholar]
- Chaleckis R.; Meister I.; Zhang P.; Wheelock C. E. Challenges, progress and promises of metabolite annotation for LC-MS-based metabolomics. Curr. Opin. Biotechnol. 2019, 55, 44–50. 10.1016/j.copbio.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Huang L.; Wan J. J.; Wei X.; Liu Y.; Huang J. Y.; Sun X. M.; Zhang R.; Gurav D. D.; Vedarethinam V.; Li Y.; et al. Plasmonic silver nanoshells for drug and metabolite detection. Nat. Commun. 2017, 8 (1), 220. 10.1038/s41467-017-00220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz D.; Mkorombindo T.; Schumann D. M.; Agusti A.; Ash S. Y.; Bafadhel M.; Bai C. X.; Chalmers J. D.; Gerard J. C.; Dharmage S. C.; et al. Towards the elimination of chronic obstructive pulmonary disease: a Lancet Commission. Lancet 2022, 400 (10356), 921–972. 10.1016/S0140-6736(22)01273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli B. R.; Wedzicha J. A. Update on clinical aspects of chronic obstructive pulmonary disease. N. Engl. J. Med. 2019, 381 (13), 1257–1266. 10.1056/NEJMra1900500. [DOI] [PubMed] [Google Scholar]
- Patil A. A.; Chiang C.-K.; Wen C.-H.; Peng W.-P. Forced dried droplet method for MALDI sample preparation. Anal. Chim. Acta 2018, 1031, 128–133. 10.1016/j.aca.2018.05.056. [DOI] [PubMed] [Google Scholar]
- Markley J. L.; Bruschweiler R.; Edison A. S.; Eghbalnia H. R.; Powers R.; Raftery D.; Wishart D. S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. 10.1016/j.copbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeloye D.; Song P. G.; Zhu Y. J.; Campbell H.; Sheikh A.; Rudan I.; Unit N. R. G. R. H. Global, regional, and national prevalence of, and risk factors for, chronic obstructive pulmonary disease (COPD) in 2019: a systematic review and modelling analysis. Lancet Resp. Med. 2022, 10 (5), 447–458. 10.1016/S2213-2600(21)00511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange P.; Halpin D. M.; O’Donnell D. E.; MacNee W. Diagnosis, assessment, and phenotyping of COPD: beyond FEV1. Int. J. Chron. Obstruct. Pulmon. Dis. 2016, 11, 3–12. 10.2147/copd.s85976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.; Chen Z.; Fang Z.; Zhao J.; Zhou Y.; Tang C. Multi-omics study on biomarker and pathway discovery of chronic obstructive pulmonary disease. J. Breath Res. 2021, 15 (4), 044001. 10.1088/1752-7163/ac15ea. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Covarrubias V.; Martínez-Martínez E.; del Bosque-Plata L. The potential of metabolomics in biomedical applications. Metabolites 2022, 12 (2), 194. 10.3390/metabo12020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmeier C. F.; Criner G. J.; Martinez F. J.; Anzueto A.; Barnes P. J.; Bourbeau J.; Celli B. R.; Chen R. C.; Decramer M.; Fabbri L. M.; et al. Global Strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. Am. J. Respir. Crit. Care Med. 2017, 195 (5), 557–582. 10.1164/rccm.201701-0218PP. [DOI] [PubMed] [Google Scholar]
- Rabe K. F.; Watz H. Chronic obstructive pulmonary disease. Lancet 2017, 389 (10082), 1931–1940. 10.1016/S0140-6736(17)31222-9. [DOI] [PubMed] [Google Scholar]
- Agustí A.; Celli B. R.; Criner G. J.; Halpin D.; Anzueto A.; Barnes P.; Bourbeau J.; Han M. K.; Martinez F. J.; de Oca M. M.; et al. Global initiative for chronic obstructive lung disease 2023 report: Gold executive summary. Am. J. Respir. Crit. Care Med. 2023, 207 (7), 819–837. 10.1164/rccm.202301-0106PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathioudakis A. G.; Janssens W.; Sivapalan P.; Singanayagam A.; Dransfield M. T.; Jensen J. U. S.; Vestbo J. Acute exacerbations of chronic obstructive pulmonary disease: in search of diagnostic biomarkers and treatable traits. Thorax 2020, 75 (6), 520–527. 10.1136/thoraxjnl-2019-214484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen M.; Ingebrigtsen T. S.; Marott J. L.; Dahl M.; Lange P.; Vestbo J.; Nordestgaard B. G. Inflammatory biomarkers and exacerbations in chronic obstructive pulmonary disease. JAMA-J. Am. Med. Assoc. 2013, 309 (22), 2353–2361. 10.1001/jama.2013.5732. [DOI] [PubMed] [Google Scholar]
- Hurst J. R.; Donaldson G. C.; Perera W. R.; Wilkinson T. M. A.; Bilello J. A.; Hagan G. W.; Vessey R. S.; Wedzicha J. A. Use of plasma biomarkers at exacerbation of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2006, 174 (8), 867–874. 10.1164/rccm.200604-506OC. [DOI] [PubMed] [Google Scholar]
- Durmuş U.; Dogan N. Ö.; Pekdemir M.; Yılmaz S.; Yaka E.; Karadaş A.; Güney Pınar S. The value of lactate clearance in admission decisions of patients with acute exacerbation of COPD. Am. J. Emerg. Med. 2018, 36 (6), 972–976. 10.1016/j.ajem.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Xu W.-F.; Upur H.; Wu Y.-H.; Mamtimin B.; Yang J.; Ga Y.-J.; You L. Metabolomic changes in patients with chronic obstructive pulmonary disease with abnormal Savda syndrome. Exp. Ther. Med. 2015, 9 (2), 425–431. 10.3892/etm.2014.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartziokas K.; Papaioannou A. I.; Loukides S.; Papadopoulos A.; Haniotou A.; Papiris S.; Kostikas K. Serum uric acid as a predictor of mortality and future exacerbations of COPD. Eur. Respir. J. 2014, 43 (1), 43–53. 10.1183/09031936.00209212. [DOI] [PubMed] [Google Scholar]
- Antus B.; Harnasi G.; Drozdovszky O.; Barta I. Monitoring oxidative stress during chronic obstructive pulmonary disease exacerbations using malondialdehyde. Respirology 2014, 19 (1), 74–79. 10.1111/resp.12155. [DOI] [PubMed] [Google Scholar]
- Marin-Corral J.; Minguella J.; Ramírez-Sarmiento A. L.; Hussain S. N. A.; Gea J.; Barreiro E. Oxidised proteins and superoxide anion production in the diaphragm of severe COPD patients. Eur. Respir. J. 2009, 33 (6), 1309–1319. 10.1183/09031936.00072008. [DOI] [PubMed] [Google Scholar]
- Ubhi B. K.; Riley J. H.; Shaw P. A.; Lomas D. A.; Tal-Singer R.; MacNeee W.; Griffin J. L.; Connor S. C. Metabolic profiling detects biomarkers of protein degradation in COPD patients. Eur. Respir. J. 2012, 40 (2), 345–355. 10.1183/09031936.00112411. [DOI] [PubMed] [Google Scholar]
- Cakmak A.; Nemutlu E.; Yabanoglu-Ciftci S.; Baysal I.; Kocaaga E.; Coplu L.; Inal-Ince D. Metabolomic, oxidative, and inflammatory responses to acute exercise in chronic obstructive pulmonary disease. Heart Lung 2023, 59, 52–60. 10.1016/j.hrtlng.2023.01.011. [DOI] [PubMed] [Google Scholar]
- Arshad H.; Siokis A.; Franke R.; Habib A.; Alfonso J. C. L.; Poliakova Y.; Lücke E.; Michaelis K.; Brönstrup M.; Meyer-Hermann M.; et al. Reprogramming of amino acid metabolism differs between community-acquired pneumonia and infection-associated exacerbation of chronic obstructive pulmonary disease. Cells 2022, 11 (15), 2283. 10.3390/cells11152283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. T.; Li Q. Y.; Liu C. Y.; Pang R. F.; Yin Y. X. Plasma metabolomics and lipidomics reveal perturbed metabolites in different disease stages of chronic obstructive pulmonary disease. Int. J. Chron. Obstruct. Pulmon. Dis. 2020, 15, 553–565. 10.2147/COPD.S229505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E.; Oka R.; Ishii T.; Korekane H.; Kurimoto A.; Kizuka Y.; Kitazume S.; Ariki S.; Takahashi M.; Kuroki Y.; et al. Fucosylated surfactant protein-D is a biomarker candidate for the development of chronic obstructive pulmonary disease. J. Proteomics 2015, 127, 386–394. 10.1016/j.jprot.2015.07.011. [DOI] [PubMed] [Google Scholar]
- Godbole S.; Labaki W. W.; Pratte K. A.; Hill A.; Moll M.; Hastie A. T.; Peters S. P.; Gregory A.; Ortega V. E.; DeMeo D.; et al. A metabolomic severity score for airflow obstruction and emphysema. Metabolites 2022, 12 (5), 368. 10.3390/metabo12050368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. Y.; Huang Y. D.; Hu C.; Du L. B.; Du Y. A.; Zhang Y. Q.; Qin J. J.; Liu W. S.; Wang R. M.; Yang S. Z.; et al. Efficient plasma metabolic fingerprinting as a novel tool for diagnosis and prognosis of gastric cancer: a large-scale, multicentre study. Gut 2023, 72 (11), 2051–2067. 10.1136/gutjnl-2023-330045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.