Abstract
Identification of replicable neuroimaging correlates of attention-deficit hyperactivity disorder (ADHD) has been hindered by small sample sizes, small effects, and heterogeneity of methods. Given evidence that ADHD is associated with alterations in widely distributed brain networks and the small effects of individual brain features, a whole-brain perspective focusing on cumulative effects is warranted. The use of large, multisite samples is crucial for improving reproducibility and clinical utility of brain-wide MRI association studies. To address this, a polyneuro risk score (PNRS) representing cumulative, brain-wide, ADHD-associated resting-state functional connectivity was constructed and validated using data from the Adolescent Brain Cognitive Development (ABCD, N = 5,543, 51.5% female) study, and was further tested in the independent Oregon-ADHD-1000 case–control cohort (N = 553, 37.4% female). The ADHD PNRS was significantly associated with ADHD symptoms in both cohorts after accounting for relevant covariates (p < 0.001). The most predictive PNRS involved all brain networks, though the strongest effects were concentrated among the default mode and cingulo-opercular networks. In the longitudinal Oregon-ADHD-1000, non-ADHD youth had significantly lower PNRS (Cohen's d = −0.318, robust p = 5.5 × 10−4) than those with persistent ADHD (age 7–19). The PNRS, however, did not mediate polygenic risk for ADHD. Brain-wide connectivity was robustly associated with ADHD symptoms in two independent cohorts, providing further evidence of widespread dysconnectivity in ADHD. Evaluation in enriched samples demonstrates the promise of the PNRS approach for improving reproducibility in neuroimaging studies and unraveling the complex relationships between brain connectivity and behavioral disorders.
Keywords: attention-deficit hyperactivity disorder, brain-wide association study, MRI, polyneuro score, polygenic score, resting-state functional connectivity
Significance Statement
Neuroimaging studies of ADHD have been hindered by small sample sizes, small effects, and differences among study methods. We demonstrate that an ADHD polyneuro risk score (PNRS), representing brain-wide connectivity patterns, was robustly associated with ADHD symptoms in two independent cohorts. The study used data from the Adolescent Brain Cognitive Development Study and the Oregon-ADHD-1000 cohort and provides further evidence of widespread dysconnectivity in ADHD. The findings highlight the promise of approaches examining cumulative, brain-wide effects, and the importance of using large samples for improving reproducibility of neuroimaging studies.
Introduction
Attention-deficit hyperactivity disorder (ADHD) is a common neurodevelopmental disorder, affecting approximately 5% of school-aged children (Song et al., 2019; Cordova et al., 2022). ADHD is characterized by age-inappropriate cognitive, behavioral, and emotional problems, though there is significant heterogeneity in both presentation and outcome (Sonuga-Barke, 2005; Fair et al., 2012; Posner et al., 2014; Luo et al., 2019; Nigg et al., 2020a). Substantial behavioral and quantitative genetic evidence suggests that ADHD symptoms exist in the population along a continuum and can be meaningfully studied as a dimensional trait (as we do here; Thapar, 2018).
Neuroimaging studies have aimed to elucidate the brain mechanisms and correlates of ADHD. Although these investigations have identified structural and functional brain features associated with ADHD (Albajara Sáenz et al., 2019; Hoogman et al., 2020; Mooney et al., 2021), replication has been disappointing. This is likely secondary to relatively small brain-behavior effect sizes (Smith and Nichols, 2018; Feczko et al., 2021; Pereira-Sanchez and Castellanos, 2021; Marek et al., 2022)—suggesting individual brain features are insufficient to meaningfully explain the ADHD phenotype. Indeed, considerable evidence supports the proposal that biological and physiological correlates of ADHD are not simply localized alterations in brain function but rather constitute widely distributed functional brain systems as evaluated by resting-state functional connectivity MRI (rs-fcMRI) studies. For example, although a systematic review of rs-fcMRI studies of ADHD through 2013 (Posner et al., 2014) found some consistent findings, such as altered connectivity within the default mode network (Castellanos et al., 2008; Fair et al., 2010; Qiu et al., 2011), an overall conclusion was that rs-fcMRI studies do not support a single neurocognitive deficit in ADHD. Rather, results suggest multiple brain networks are likely involved, providing support for a neurobiological basis for the heterogeneity observed in ADHD presentation (Saad et al., 2020).
Recent meta-analyses of rs-fcMRI studies have provided further evidence that connectivity in multiple brain networks is associated with ADHD (Gao et al., 2019; Sutcubasi et al., 2020). However, these studies have primarily focused on a few hypothesized networks, rather than examining effects brain-wide.
A large meta-analysis of whole-brain rs-fcMRI studies failed to find spatial convergence of ADHD-associated connectivity patterns (Cortese et al., 2021). This may be due to method variation across studies or to heterogeneity within ADHD. However, we hypothesize it may also be due to ADHD-associated dysconnectivity being broadly distributed across the entire brain, and it is the cumulative effect of many small connectivity alterations that correlates best with behavior.
These reviews highlight multiple linked challenges to brain-behavior associations with ADHD: (1) most brain-behavior associations for individual connections and/or brain regions show small effects (Smith and Nichols, 2018; Marek et al., 2022), (2) most studies are underpowered to detect small effects (Posner et al., 2014; Marek et al., 2022), (3) differences among study samples and methods continue to hinder reproducibility (Marek et al., 2022), (4) ADHD-related connectivity patterns may show interindividual differences (Feczko et al., 2019; Feczko and Fair, 2020; Nigg et al., 2020a), and (5) recent evidence suggests the involvement of multiple brain networks in ADHD (Gao et al., 2019; Sutcubasi et al., 2020). Collectively, these challenges point toward the importance of taking a whole-brain perspective, utilizing methods that can accommodate phenotypic heterogeneity, making use of large data sets for discovery, as well as enriched confirmation data sets to ensure relevance to clinical settings, and emphasizing validation/replication when studying the neural correlates of ADHD.
For instance, measures that represent the cumulative trait-associated effects of task-based fMRI measures across the entire brain were recently reported to explain a significantly greater proportion of trait variance than individual voxels (Zhao et al., 2019). In that study, a brain-wide association analysis was performed to identify voxels associated with performance on specific cognitive tasks. Resulting association measures were then used to construct a weighted sum of brain-wide activity, constituting a single fMRI-based summary measure of the trait.
This approach fits within a larger area of research focusing on multivariate modeling of behavioral traits using functional connectivity measures (Sui et al., 2020; Byington et al., 2023). These models range from relatively simple additive linear models (like the brain-wide scores we examined here), such as connectome-based predictive modeling (Rosenberg et al., 2016; Shen et al., 2017), to more complex, nonlinear multivariate models (Dadi et al., 2019; Pervaiz et al., 2020). These prior studies have demonstrated the utility of brain-wide connectivity scores, though conclusions about the best approaches have been limited by small sample sizes (Sui et al., 2020) and/or inadequate data processing to account for possible confounders (particularly regarding head motion; Ciric et al., 2017). There is some evidence, however, that linear models generalize better than more complex models in these types of applications (Traut et al., 2022). Similar methods have also been used with structural neuroimaging data (Kochunov et al., 2022).
Here, we build on prior work by generating scores representing brain-wide ADHD-associated resting-state connectivity using a large, diverse discovery data set, the Adolescent Brain Cognitive Development (ABCD) study (Casey et al., 2018; Feczko et al., 2021). These brain-wide scores, which we refer to as polyneuro risk scores [PNRS; Byington et al., 2023; akin to polyvertex scores (Zhao et al., 2019)], are somewhat analogous to polygenic scores (Choi et al., 2020; Li and He, 2021; Ronald et al., 2021) that represent cumulative genetic risk across the genome, although perhaps a more appropriate analogy is to polyepigenetic scores (Sugden et al., 2019; Suarez et al., 2020; Watkeys et al., 2020), given that PNRS can change over time in response to development and/or environmental influences. Nevertheless, PNRS, and other multivariate methods, address a number of limitations of neuroimaging studies, similar to the way polygenic risk scores (PRS) addressed the challenges of early genomic studies.
First, PNRS take advantage of large-scale consortium-level data sets (Miller et al., 2016; Casey et al., 2018; Feczko et al., 2021) to generate brain-wide effect estimates and allow testing of cumulative effects in smaller data sets that would be underpowered to detect small effects shown in meta-analyses (Pereira-Sanchez and Castellanos, 2021; Marek et al., 2022). Second, PNRS allow for heterogeneity among study subjects, because the same cumulative effect can result from varying effects of individual brain features. Third, cumulative brain-wide effects should provide significantly greater predictive power than individual brain features, if the trait of interest truly has a signal distributed across the brain. We test this hypothesis herein.
The increased predictive power (effect size) of the PNRS may also enable the examination of mediation effects, providing insight into the mechanisms of genetic or early environmental risk factors. Previous work has provided evidence for both structural and functional MRI measures partially mediating the effect of common genetic risk on ADHD diagnosis or symptoms (Hermosillo et al., 2020; Sudre et al., 2020; Mooney et al., 2021). A reasonable question is whether a brain-wide summary score will mediate a greater proportion of polygenic risk than individual brain features.
Our goals for this study were to (1) develop a PNRS associated with ADHD symptoms; (2) validate the predictive ability of the ADHD PNRS in a completely independent, enriched longitudinal ADHD case–control cohort; and (3) test whether the ADHD PNRS mediates common genetic risk for ADHD or whether effects are additive.
While the methodology for constructing PNRS is not novel, the use of two large, independent data sets with different ascertainment strategies for discovery and validation, including a case–control data set for validation to better reflect the situation in clinical prediction, along with the use of best practices for rs-fcMRI data processing represent important contributions to the field. This work stands to provide further evidence for the validity of brain-wide summary measures from neuroimaging data for the prediction of behavioral traits and to demonstrate how these scores can be used to provide insight into mechanistic and causal pathways.
Materials and Methods
Participants
The ABCD Study served as our discovery cohort (Casey et al., 2018). It is a 21-site, diverse cohort of over 11,000 children (age 9–10 at baseline). The children have been genotyped and are followed with extensive behavioral, cognitive, clinical, and MRI measures annually. The data set is publicly available on the NIMH Data Archive (NDA; http://dx.doi.org/10.15154/1504041; Feczko et al., 2021).
For internal replication, we split the ABCD cohort into two ABCD Reproducible Matched Samples (ARMS) matched on nine demographic factors thought to be involved with development: site, age, assigned sex at birth, race/ethnicity, grade, highest level of parental education, handedness, combined family income, and exposure to anesthesia (Feczko et al., 2020a,b).
The Oregon-ADHD-1000 cohort (Karalunas et al., 2017; Nigg et al., 2018, 2020b; Mooney et al., 2020, 2021) was used as an independent validation data set. It is a case–control cohort of ∼1,400 children, aged 7–11 years at baseline, that is enriched for psychopathology. Of these, 553 have clinical measures and MRI data at one or more time points. Genome-wide genotype data were available for 487 of these children (NDA Collection 2857).
rs-fcMRI data
Minimally processed, quality-controlled, baseline resting-state functional MRI data for the ABCD cohort were downloaded from the NDA, specifically the ABCD-BIDS Community Collection (ABCC; NDA Collection 3165). Details of the data access and processing can be found in the ABCC documentation (https://collection3165.readthedocs.io/en/stable/). Briefly, the data were processed using the ABCD-BIDS pipeline (Feczko et al., 2020a), which is a modified version of the Human Connectome Project pipeline (Glasser et al., 2013; Feczko et al., 2021). Processed ABCD data were further curated based on head motion, such that only frames that had a framewise displacement (FD) threshold of 0.2 mm were used. Of the surviving frames, additional frames were removed if they were detected as outliers (using the median absolute deviation) in the standard deviation of the bold signal across the whole brain. Trimming was done at random to limit bias in data selection. Only resting-state scans from participants trimmed to 10 min of usable data were kept for further analysis, resulting in a total of 5,543 participants. The final ABCD data set for our primary analyses consisted of N = 2,747 subjects (52.9% female) in ABCD ARMS-1 and N = 2,796 subjects (50.2% female) in ABCD ARMS-2.
Whole-brain functional connectivity was measured using the Gordon parcellation, an independent annotation derived from highly sampled individual data, which includes subcortical nuclei and the cerebellum (Gordon et al., 2016).
The Oregon cohort rs-fcMRI data was processed using the same pipeline as the ABCD cohort. After quality control (poor image quality, registration, or gray matter/white matter delineation) and exclusion of subjects with missing data in the Oregon cohort, a total of N = 553 subjects (37.4% female) were kept for longitudinal analyses and N = 494 for the baseline analysis (the smaller sample size here is due to missing covariates at the early time points).
ADHD composite symptom scores
ADHD symptom scores were created by averaging standardized (Z-scored) symptom measures across available parent-reported measures in both the ABCD and Oregon-ADHD-1000 cohorts. This parent-reported composite score was previously validated and replicated for ABCD in a confirmatory factor model with reproducible fit (Cordova et al., 2022) and was also previously validated for the Oregon-ADHD-1000 (Nigg et al., 2018). For ABCD, the measures used to create a composite ADHD symptom score were the attention problem scale from the Child Behavior Checklist (CBCL) and the inattentive and hyperactive symptom counts from the Kiddie Schedule for Affective Disorders and Schizophrenia (KSADS).
For the Oregon-ADHD-1000, the measures used to create a composite were the inattentive and hyperactive symptom counts from the ADHD Rating Scale, the KSADS, the Conners (third edition), and the Strengths and Weaknesses of Attention-Deficit/Hyperactivity Disorder Symptoms and Normal Behavior Scale (SWAN), as well as the hyperactivity raw score from the Strengths and Difficulties Questionnaire (Nigg et al., 2020b).
Reliability among the symptom measures in both cohorts was good to excellent (ABCD ARMS-1, Cronbach’s α = 0.87; ABCD ARMS-2, α = 0.86; Oregon-ADHD-1000, α = 0.96).
ADHD case identification in ABCD
Criteria for an ADHD diagnosis in ABCD were based on the Tier 4 criteria described by Cordova et al. (2022). That is, (1) cases that met the criteria on the computerized KSADS for School-Age Children interview as well as exceeding cutoffs of T-score > 65 on both parent and teacher normative ratings (using the Brief Problem Monitor) plus (2) participants who met diagnostic criteria for ADHD in the past (computerized KSADS), remained on ADHD medication despite not meeting diagnostic criteria on the interview currently, and had an elevated attention problem T-score from the teacher-reported Brief Problem Monitor (T-score > 65), suggesting that they were in fact true cases who were partially treated (partial remission), causing them not to meet full criteria. (Cordova et al. described this in their supplemental materials and noted in their discussion that adding these cases provides a viable alternative estimate.)
Brain-wide connectivity (polyneuro) scores
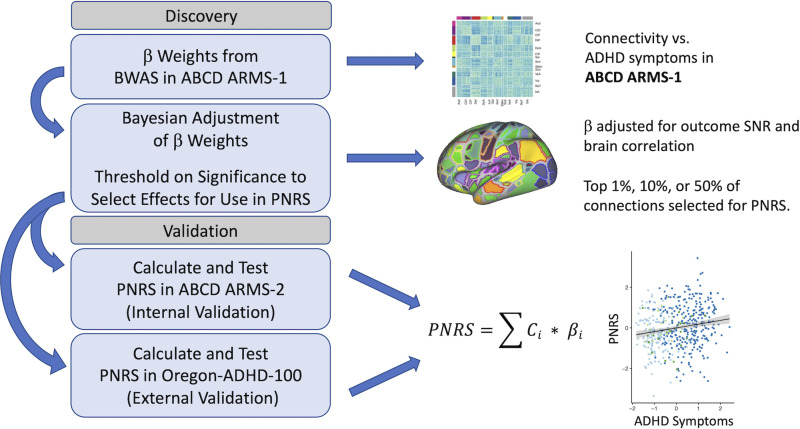
Polyneuro risk scores (PNRS), which represent brain-wide connectivity associated with ADHD symptoms, were created following a method (Fig. 1) similar to that described previously (Zhao et al., 2019). Effect estimates generated from a brain-wide association study (BWAS) performed on the ABCD discovery set (see below) were used to calculate PNRS for each subject in the validation data sets by multiplying the effect estimate for each connection, , by the subject’s connectivity measure, , and summing across all connections (Eq. 1). Scores were created using both unadjusted effect estimates, as well as effect estimates adjusted using a Bayesian procedure described previously (Zhao et al., 2019). The Bayesian adjustment accounts for both the observed correlation structure among all connections brain-wide, as well as the signal-to-noise ratio of the brain-behavior association. In addition, scores were created using only those connections that passed a particular significance threshold (determined in the discovery cohort): top 50%, top 10%, and top 1% most significant connections:
| (1) |
Figure 1.
Polyneuro risk score (PNRS) discovery and validation workflow. Note the thresholding done to select connections used in the PNRS is based on significance determined in the discovery cohort.
ADHD polygenic score
Details of the Oregon-ADHD-1000 genotyping procedure have been published previously (Nigg et al., 2018, 2020b; Mooney et al., 2021). For the ABCD cohort, saliva samples were collected at the baseline visit and sent to the Rutgers University Cell and DNA Repository for DNA isolation (Uban et al., 2018). Genotyping was performed using the Smokescreen array (Baurley et al., 2016). Processed genotypes were downloaded from the NDA (dx.doi.org/10.15154/1503209), and standard quality control checks were performed using the GWASTools R package (Gogarten et al., 2012). One batch of samples had a significantly lower call rate (∼85%) than all others (∼98%) and was removed (N = 126 samples). All single-nucleotide polymorphisms (SNPs) had an adequate call rate (>94%), and no significant batch effects were observed after examining allele frequency differences across batches.
For both cohorts, principal component analysis was conducted using the PC-Air method in the GENESIS Bioconductor package (Gogarten et al., 2019), and nongenotyped SNPs were imputed with IMPUTE2 (Howie et al., 2009) using the 1000 Genomes phase 3 reference panel. Only those SNPs imputed with high confidence (INFO > 0.8) were kept. Genotype probabilities were converted to best-guess genotypes, with the genotype set to missing if the probability is <0.8.
The PRS were constructed in both ABCD and Oregon-ADHD-1000 using the 2019 PGC + iPSYCH ADHD genome-wide association study (GWAS) meta-analysis (Demontis et al., 2019) as the discovery data set (20,183 ADHD cases; 35,191 controls). We used a method (LDpred) for calculating polygenic scores that has demonstrated good performance in a variety of data sets, as well as significantly improved performance compared with more traditional methods (Vilhjálmsson et al., 2015; Ni et al., 2021; Pain et al., 2021). Only SNPs with an INFO score of >0.8 in both the PGC + iPSYCH meta-analysis and the target data sets were considered. SNPs were further limited to the ∼1.2 million HapMap SNPs as suggested. Linkage disequilibrium was estimated using all unrelated individuals in the ABCD cohort, and PRS were created with the proportion of causal SNPs set to 0.3.
Experimental design and statistical analyses
Analyses testing the association between brain connectivity (i.e., each parcel-to-parcel brain correlation; we use the term “connection” throughout to refer to this measure of functional connectivity) and the ADHD composite symptom score in ABCD were carried out using the Sandwich Estimator for Neuroimaging Data (SEND; Feczko et al., 2020c, 2021). SEND constructs a marginal model (Guillaume et al., 2014) for brain-wide associations, where brain connectivity is the dependent variable, while covariates and independent variables are modeled as fixed effects. Standard errors are estimated with a sandwich estimator clustered for unique combinations of race, ethnicity, study site, and sex assigned at birth. Two covariates, subject age and highest parental education, and the independent variable ADHD composite symptom scores were modeled as fixed effects.
The association between the PNRS and ADHD composite symptom score was validated in both the ABCD cohort and the completely independent Oregon-ADHD-1000 cohort. First, ABCD ARMS-1 was used as the discovery cohort, and effect estimates were used to calculate PNRS in ABCD ARMS-2. The association between ADHD PNRS and ADHD symptoms was tested using a linear regression model, with age, sex, race/ethnicity, highest parental education, ABCD Study site, and the single most significant connection covaried. To assess whether family relatedness impacted the PNRS effect estimate, marginal models with family ID as the cluster variable were also fit. Marginal models were implemented with the gee R package (Carey, 2019). In addition, we tested the association between PNRS and symptoms in a matched case–control subsample of ABCD ARMS-2, consisting of 116 cases and 114 controls matched on age, sex assigned at birth, race/ethnicity, and study site. Given the low number of ADHD cases in the full ABCD cohort, this analysis allowed for a better comparison to the PNRS effect seen in the Oregon case–control cohort.
Second, the full ABCD cohort (ARMS-1 and ARMS-2 combined) was used as the discovery data set, and ADHD PNRS scores were calculated for all subjects in the Oregon-ADHD-1000 cohort. The association between ADHD PNRS and ADHD composite symptom score was validated in the Oregon cohort in three ways. (1) The earliest scan was chosen for each subject, and the association between PNRS and ADHD symptoms was tested in this baseline data using linear regression. Again, a marginal model with clusters defined by family ID was fit to verify effect estimates while accounting for the impact of siblings. (2) All available scans were used, and repeated measures were accounted for with a marginal model, again using the gee R package. (3) Finally, subjects were categorized as follows: persistent ADHD (ADHD at all time points), persistent control (control at all time points), remittent ADHD (ADHD at one or more early time points followed by non-ADHD status), and others. Again, a marginal model was fit to assess the association between these diagnostic categories and the ADHD PNRS over time. All models fit in the Oregon cohort including age, assigned sex at birth, race/ethnicity, highest parental education, puberty score, scan type, and the most significant individual connection as covariates. The most significant individual connection was included in the model to test whether the PNRS indeed explains more variation in symptoms than any single connection. Following best practices in the literature (Dennis et al., 2009; de Zeeuw et al., 2012), IQ was not included as a covariate in our models.
In both cohorts, we performed secondary analyses to determine if accounting for a history of ADHD medication usage impacted the association between PNRS and ADHD symptoms.
Finally, to examine whether the method used to construct the brain-wide summary score significantly impacted our results, we created an alternative PNRS using partial least squares regression (PLSR) and compared its performance to that of the PNRS constructed with a linear additive model as described above.
In all analyses examining the effect of ADHD PRS, the first three genomic principal components were covaried to adjust for potential population stratification. Analyses examining whether the effect of the PRS on ADHD symptoms is partially mediated by brain-wide connectivity (ADHD PNRS) were conducted using the mediation R package (Tingley et al., 2014).
Results
Overview of samples
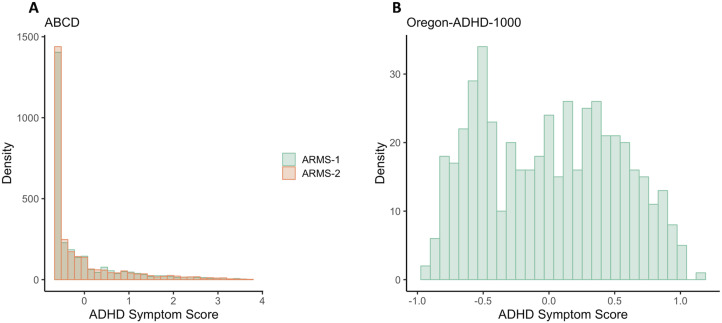
There were no significant differences in mean age, race/ethnicity, or ADHD prevalence between the two ABCD ARMS (all p-values > 0.1) but a slight difference in the proportion of females (chi-square p-value = 0.0463; Table 1). The distribution of symptom scores was not significantly different between ABCD ARMS-1 and ARMS-2 (p = 0.916).
Table 1.
Description of cohorts
| ABCD ARMS-1 | ABCD ARMS-2 | Oregon-ADHD-1000 | |
|---|---|---|---|
| Sample size | 2,747 | 2,796 | 553 |
| Mean age (SD) | 9.97 (0.63) | 9.98 (0.62) | 10.25 (1.56)a |
| % Female | 52.9 | 50.2 | 37.4 |
| % Caucasian | 65.0 | 65.5 | 79.7 |
| % ADHD Dx | 3.3 | 4.1 | 57.1a |
| Inattention symptoms‡ | 53.4 (5.7) | 53.3 (5.7) | 63.9 (16.8)a |
At earliest scan.
Inattention symptoms (T-scores) measured by parent-reported Child Behavior Checklist in ABCD and parent-reported Conners 3 in the Oregon-ADHD-1000.
By virtue of its case–control design, the Oregon-ADHD-1000 had a higher proportion of ADHD cases and, as a result, much greater variation in symptom scores compared with ABCD (Fig. 2; Mooney et al., 2023). The Oregon cohort also had a lower proportion of females and a greater proportion of Caucasians than the ABCD cohort (p-values <0.001; Table 1).
Figure 2.
Distribution of ADHD symptom scores in (A) both ARMS of ABCD (Mann–Whitney U test p-value = 0.916) and (B) the Oregon-ADHD-1000 case–control cohort. The ADHD composite symptom scores are the average of multiple standardized (mean = 0, SD = 1) symptom scales (see Materials and Methods).
Brain-wide association results
Brain-wide association analyses were performed in ABCD ARMS-1 and ARMS-2 separately, as well as in the full ABCD cohort. The correlation of effect estimates between ARMS-1 and ARMS-2 was significant, with Pearson’s correlation ranging between 0.46 for all connections and 0.79 for the top 1% of most significantly associated connections (Table 2).
Table 2.
Correlation of brain-wide effect estimates across ABCD ARMS
| T-stat quantile | Pearson’s correlation | p-value |
|---|---|---|
| All | 0.460 | 0 |
| Top 50% | 0.571 | 0 |
| Top 10% | 0.717 | 0 |
| Top 1% | 0.792 | 0 |
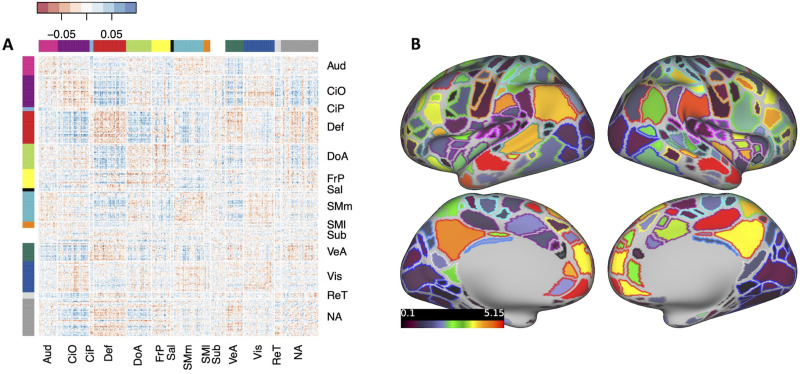
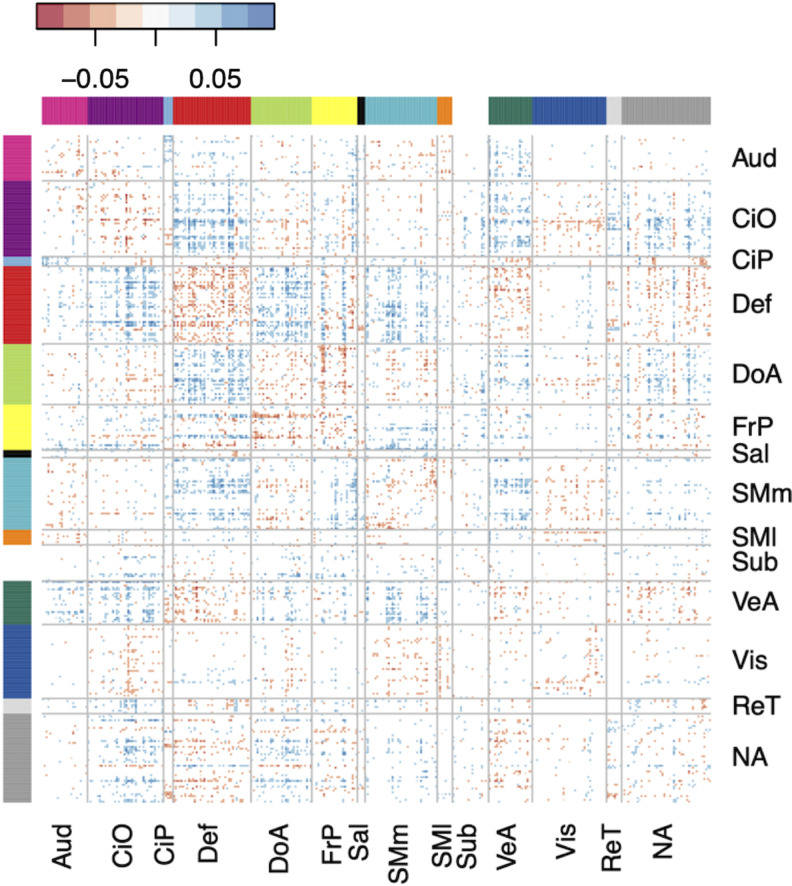
Effect estimates for individual connections were small, with standardized regression coefficients of <0.1. A matrix of regression coefficients for all connections brain-wide is shown in Figure 3A. The cumulative effect of connections involving each parcel (i.e., the sum of the absolute value of effect estimates across each row in the matrix), along with each parcel’s network assignment, is shown in Figure 3B.
Figure 3.
Brain-wide connectivity associated with ADHD symptoms in the ABCD Study cohort. A, The matrix of standardized regression coefficients showing the strength of association between all connections (organized by brain network) and ADHD symptoms. B, Gordon parcellation showing the relative contribution of each brain network to the ADHD PNRS. Only the top 10% most significant connections (representing the most predictive PNRS) are considered. The fill color represents the sum of the absolute value of β weights for all connections in which a parcel participates; the outline color represents network assignment. Aud, auditory; CiO, cingulo-opercular; CiP, cingulo-parietal; Def, default mode; DoA, dorsal attention; FrP, frontoparietal; Sal, salience; SMm, somatomotor medial; SMl, somatomotor lateral; Sub, subcortical; VeA, ventral attention; Vis, visual; ReT, retrosplenial temporal; NA, not assigned.
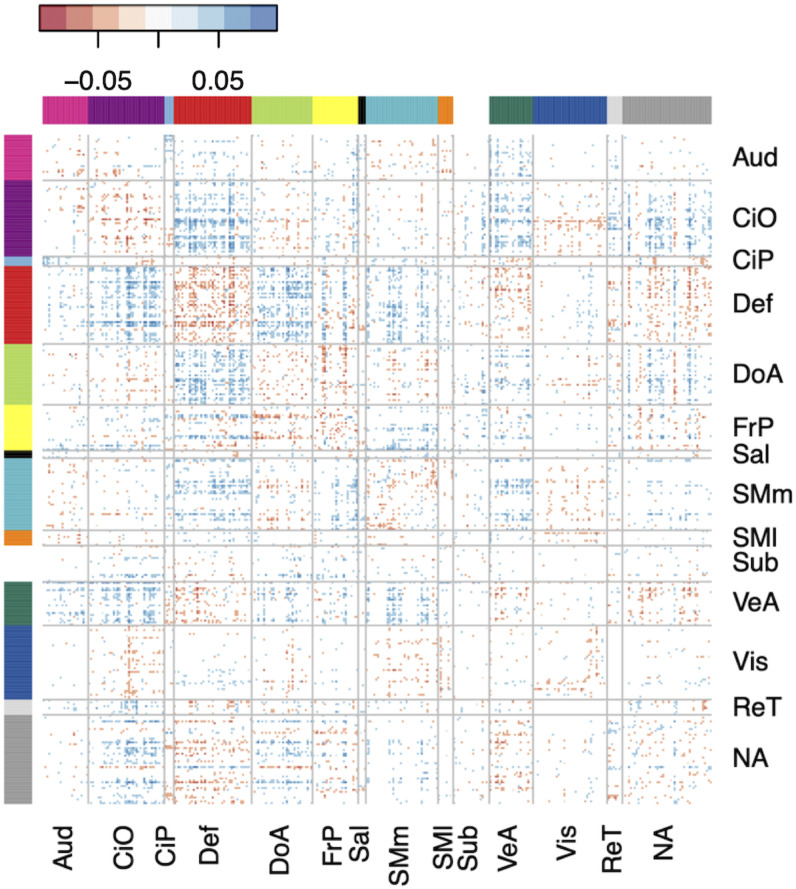
The top 10% most significant associations were spread across multiple brain networks but were particularly concentrated among connections involving the default mode and cingulo-opercular networks (Fig. 4).
Figure 4.
The matrix of standardized regression coefficients showing the strength of association between the top 10% most significant connections (organized by brain network) and ADHD symptoms. The cumulative effect of these connections comprised the most predictive PNRS, demonstrating the brain-wide, distributed nature of the ADHD PNRS. Aud, auditory; CiO, cingulo-opercular; CiP, cingulo-parietal; Def, default mode; DoA, dorsal attention; FrP, frontoparietal; Sal, salience; SMm, somatomotor medial; SMl, somatomotor lateral; Sub, subcortical; VeA, ventral attention; Vis, visual; ReT, retrosplenial temporal; NA, not assigned.
Polyneuro scores associated with ADHD symptoms
To assess whether the cumulative effect of brain-wide, ADHD-associated resting-state connectivity could predict ADHD symptoms, PNRS were constructed based on the effect estimates from BWAS in ABCD ARMS-1 and the full ABCD cohort.
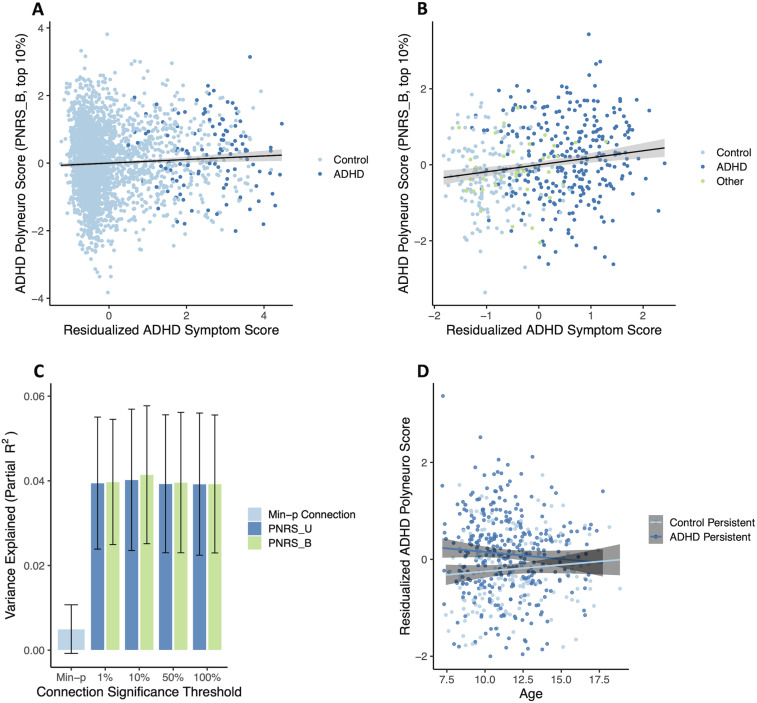
The ADHD PNRS (Bayesian-adjusted, top 10% of connections), based on effect estimates discovered in ABCD ARMS-1, was significantly associated with ADHD symptom scores in ABCD ARMS-2 [β = 0.078 (0.033, 0.123); p = 7.4 × 10−4, permutation p = 0.001; Fig. 5A], after adjusting for the single most significant connection. The PNRS explained a very small proportion of symptom variation (partial R2 = 0.004), which is not unexpected given the restricted range of symptoms in the ABCD cohort—the vast majority of ABCD participants have low symptom scores (Fig. 2). A larger proportion of symptom variation was explained by the PNRS when analyzing a matched case–control subsample (N = 116 cases, N = 114 controls) of ABCD ARMS-2 [β = 0.218 (0.034, 0.402); p = 0.021; partial R2 = 0.027]. Creating this subsample effectively downsampled the number of participants with low symptom scores, creating a symptom distribution much more similar to that in the Oregon-ADHD-1000 cohort.
Figure 5.
ADHD polyneuro score associated with ADHD symptoms. Polyneuro scores and residualized ADHD symptom scores, after adjusting for relevant covariates (see Materials and Methods), for all subjects in the (A) ABCD ARMS-2 (N = 2,796) and (B) Oregon-ADHD-1000 cohort, using each subject’s earliest scan (N = 494). C, The proportion of ADHD symptom score variance explained in the Oregon cohort, by the single most significantly associated connection (Min-p); polyneuro scores comprised of the top 1%, 10%, and 50% most significant connections; and all connections (bootstrapped standard errors are shown). D, Subjects in the Oregon-ADHD-1000 cohort with persistent ADHD showed higher ADHD PNRS than controls (p = 0.00142), though this difference decreases with age. PNRS_U, unadjusted polyneuro score; PNRS_B, Bayesian-adjusted polyneuro score.
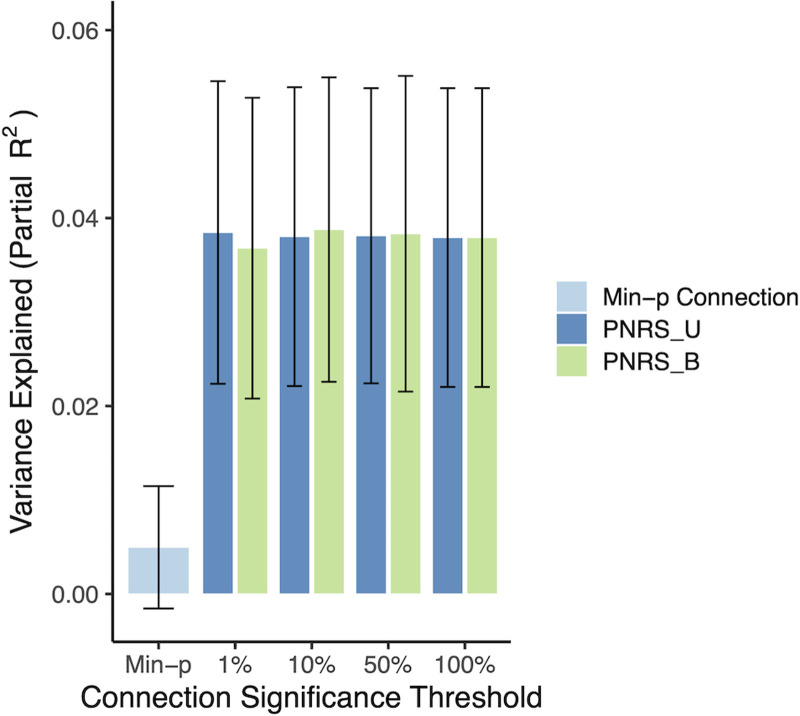
The full ABCD sample was used as the discovery cohort for validating the ADHD PNRS in the Oregon-ADHD-1000 cohort. When examining baseline data only (i.e., the earliest scan for each subject in the Oregon cohort; N = 494), the ADHD PNRS was significantly associated with ADHD symptoms, after accounting for relevant covariates (see Materials and Methods; Fig. 5B). All PNRS examined, regardless of the significance threshold used to select connections, were significantly associated with ADHD symptoms after adjusting for the single most significantly associated connection (all p-values < 0.001, Fig. 5C). The Bayesian-adjusted PNRS based on the top 10% most significant connections was the most predictive [β = 0.208 (0.113, 0.302); p = 2.1 × 10−5, permutation p = 2 × 10−4], explaining approximately 4.1% of the variation in the ADHD composite symptom score. The amount of variance explained by the PNRS was roughly eight times that explained by the single most significant connection (∼0.5%), demonstrating the benefit of the brain-wide score (Fig. 5C). Although the Bayesian-adjusted PNRS were slightly more predictive than the unadjusted PNRS, this difference was not significant (all p-values > 0.05); nor were there any significant differences among PNRS using different significance thresholds (top 1%, 10%, 50%, or all) to select connections (Fig. 5C). The effect size estimate from a marginal model, with clusters defined by family ID, was nearly identical (β = 0.208, robust p = 2.3 × 10−6), suggesting family structure had minimal influence on the associations observed.
Including all scans in the analysis (N = 553 subjects, N = 1,048 scans) and using a marginal model to account for repeated measures of the same individual resulted in a much reduced, but still significant, association between the PNRS and ADHD symptoms [β = 0.0626 (0.016, 0.110), robust p = 0.00903]. This smaller effect was consistent with the strength of association between the PNRS and ADHD symptoms decreasing with age.
Examining group effects among youth in the Oregon-ADHD-1000 with at least two scans (N = 320 subjects, N = 806 scans) showed that subjects with persistent ADHD had significantly higher PNRS than healthy controls across time [control group β = −0.331 (−0.518, −0.143), robust p = 5.5 × 10−4; Cohen’s d = −0.318; Fig. 5D]. There was a suggestive interaction between ADHD status and age at scan [interaction β = 0.0585 (−0.002, 0.119), robust p = 0.060], indicating the difference in PNRS between ADHD cases and controls may diminish somewhat with increasing age (Fig. 5D).
The results of sensitivity analyses to ensure findings were not significantly influenced by motion during scan acquisition, medication use, or sampling bias (including age-stratified analyses and assessment of test–retest reliability) are presented below.
ADHD polyneuro score relationship to ADHD polygenic risk score
In the ABCD ARMS-2, there was a weak but significant correlation between the PNRS and ADHD PRS (r = 0.177, p < 2 × 10−16). However, when included in the same regression model, both the ADHD PNRS [β = 0.071 (0.024, 0.118), p = 0.0032] and PRS [β = 0.178 (0.132, 0.225), p = 4.6 × 10−14] were significantly associated with symptoms, and there was no significant mediation of the genetic effect by the PNRS (bootstrapped p = 0.11).
In the Oregon cohort, similarly, both the ADHD PNRS [β = 0.217 (0.112, 0.322); p = 6.3 × 10−5] and the ADHD PRS [β = 0.215 (0.125, 0.306); p = 4.1 × 10−6] were significantly associated with ADHD symptoms when included in the same regression model. However, the two scores were not correlated (p = 0.857), and a mediation model showed no indication of the PNRS mediating genetic risk (bootstrapped p = 0.90).
Sensitivity analyses
Controlling for motion during scan acquisition
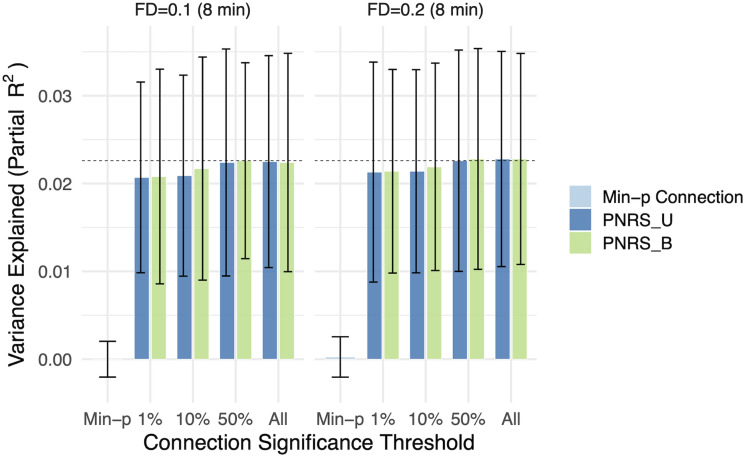
To ensure that findings were not significantly impacted by, or attributed to, motion during scan acquisition, we repeated analyses while accounting for motion in three ways. First, using a subsample of the ABCD discovery cohort (N = 2,863), BWAS were conducted using two FD thresholds (0.1 mm vs 0.2 mm; 8 min of data for both), and the results from each were used to construct PNRS in the Oregon cohort. The two resulting PNRS had nearly identical predictive power (Fig. 6), suggesting motion in the discovery cohort did not significantly impact the PNRS.
Figure 6.
ADHD polyneuro scores were robust to the motion threshold. The proportion of ADHD symptom score variance explained is nearly identical when analyzing the same set of subjects (N = 2,863) using a more stringent motion threshold (FD threshold of 0.1 mm vs 0.2 mm). Bootstrapped standard errors are shown. PNRS_U, unadjusted polyneuro score; PNRS_B, Bayesian-adjusted polyneuro score.
Second, an estimate of motion during each scan (mean FD) was included in regression models when testing the association between PNRS and ADHD symptoms in the Oregon cohort. This estimate of motion during scan acquisition was not significantly associated with ADHD symptoms (p = 0.690 for the baseline analysis). Although mean FD and the PNRS were significantly correlated (r = 0.31, p = 4.02 × 10−15), the strength of association between symptoms and the PNRS did not change meaningfully when including mean FD in the regression model (β = 0.202, after adjusting for mean FD in the baseline analysis).
Third, we repeated the BWAS in the full ABCD cohort (both ARMS combined), but this time including mean FD as a covariate (note: it was not possible to calculate mean FD for all participants; total N = 5,466 included in the FD-adjusted BWAS). Regression coefficients from this BWAS were highly correlated with those from the primary analysis (r > 0.99 across all functional connections; Figs. 4, 7), and PNRS based on this motion-adjusted BWAS performed very similarly. The Bayesian-adjusted PNRS based on the top 10% most significant connections was the most predictive again [β = 0.209 (0.110, 0.308); p = 4.15 × 10−5], explaining approximately 3.8% of the variation in the ADHD composite symptom score (Fig. 8).
Figure 7.
The matrix of standardized, motion-adjusted regression coefficients showing the strength of association between the top 10% most significant connections (organized by brain network) and ADHD symptoms. The regression coefficients shown here are from a BWAS that included mean FD as a covariate. Aud, auditory; CiO, cingulo-opercular; CiP, cingulo-parietal; Def, default mode; DoA, dorsal attention; FrP, frontoparietal; Sal, salience; SMm, somatomotor medial; SMl, somatomotor lateral; Sub, subcortical; VeA, ventral attention; Vis, visual; ReT, retrosplenial temporal; NA, not assigned.
Figure 8.
The proportion of ADHD symptom score variance explained in the Oregon cohort, by the single most significantly associated connection (Min-p); polyneuro scores comprised of the top 1%, 10%, and 50% most significant connections; and all connections. The results are shown for PNRS based on the BWAS that adjusted for mean FD. Bootstrapped standard errors are shown.
Polygenic risk score analyses
Because European-ancestry individuals are the largest ancestry group in both the ABCD and Oregon cohorts, we also examined an ADHD PRS constructed using the European-only PGC + iPSYCH meta-analysis (Demontis et al., 2019) as the discovery data set (19,099 ADHD cases and 34,194 controls). Other than the discovery data used, the methods for constructing the PRS were the same.
While we did see small differences in PRS effects between the two discovery sets, mediation results did not change meaningfully. In ABCD ARMS-2, the PRS effect was stronger for the score based on the European-ancestry discovery GWAS [β = 0.231 (0.168, 0.294), p = 6.7 × 10−13], and a very small, borderline significant mediation effect was observed (bootstrapped p = 0.052, proportion mediated = 0.016).
In the Oregon cohort, likewise, the PRS effect was slightly stronger when using the European-ancestry discovery data [β = 0.232 (0.135, 0.329); p = 3.7 × 10−6], but still no mediation was seen (bootstrapped p = 0.91).
Effect of ADHD medication usage
In the Oregon-ADHD-1000 cohort, a lifetime history of ADHD medication use was significantly associated with the PNRS (Cohen’s d = 0.22, p = 0.022), though not after adjusting for ADHD symptoms (p = 0.86). The PNRS remained significantly associated with ADHD symptoms after adjusting for a history of ADHD medication usage, although the effect was reduced slightly [β = 0.146 (0.061, 0.230); p = 7.8 × 10−4]. The same pattern held for the results in ABCD ARMS-2 [PNRS β = 0.062 (0.023, 0.102); p = 0.0018, after adjusting for ADHD medication use].
Effect of IQ
In the Oregon-ADHD-1000, IQ (full-scale IQ from the WISC-IV) measured at baseline was not significantly correlated with the PNRS (r = 0.029, p = 0.488). Adjusting for IQ had no meaningful impact on the association between the PNRS [β = 0.222 (0.121, 0.323)] and ADHD symptoms.
Test–retest reliability of the polyneuro risk score
To directly address the question of test–retest reliability in our data, we examined PNRS for the same subjects measured at multiple time points in the Oregon-ADHD-1000 longitudinal cohort. Of the 494 subjects included in our baseline analysis (earliest scan chosen), 55% had multiple scans available. When we repeated this cross-sectional analysis using the second available scan (approximately 1.8 years later on average) for those 55%, the strength of association between the PNRS and ADHD symptoms was nearly identical [β = 0.206 (0.110, 0.302)] to the baseline analysis [β = 0.208 (0.113, 0.302)].
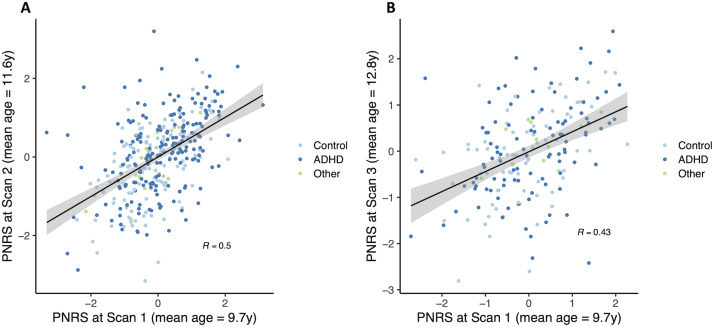
When specifically examining the relationship between scans performed at different times, we found that the PNRS was significantly correlated (r = 0.5, p = 4.5 × 10−23) when measured 1.83 years apart on average (Fig. 9). As expected, the correlation appeared to decrease with increasing time interval between MRI scans (r = 0.43, p = 3.8 × 10−10; for scans 3.10 years apart on average). Indeed, the amount of change in PNRS was significantly associated with the amount of time between the first and second scans (absolute value of PNRS change: β = 0.477, p = 0.0289).
Figure 9.
ADHD polyneuro scores measured in the same subject were significantly correlated when measured (A) approximately 2 years apart and (B) approximately 3 years apart.
Discovery data set sample size
Because the explanatory power of the PNRS depends on precise estimates of the effect of each individual brain feature (functional connections in this case), maximizing the size of the discovery data set is crucial. Because of this, we chose to use the full ABCD cohort (ARMS-1 and ARMS-2 combined) as the discovery set when validating the score in the Oregon-ADHD-1000. However, because internal validation in the ABCD cohort relied on only the ABCD ARMS-1 as the discovery cohort, we examined the impact of using this smaller discovery data set in the Oregon cohort as well. We found that the association between the PNRS and ADHD symptoms was slightly weaker [β = 0.179 (0.086, 0.272), p = 1.8 × 10−4, permutation p = 0.0006; partial R2 = 0.034] but comparable to the association when the full ABCD cohort was used for discovery [β = 0.208 (0.113, 0.302)] in the baseline analysis.
Bootstrapping to assess sampling bias
To assess whether sampling bias may have driven the observed small effects of the PNRS, bootstrapping analyses were conducted in both cohorts. First, the small effect seen in ABCD internal validation was confirmed through bootstrapped (N = 5,000 replications) resampling of the ABCD ARMS-2 [bootstrapped β = 0.078 (0.0329, 0.1226)]. Note: the higher number of replications (5,000 for ABCD vs 1,000 for Oregon below) was needed to accurately estimate the confidence interval due to the smaller effect size in ABCD.
In the Oregon cohort, we performed bootstrapping with N = 1,000 replications in each of two cross-sectional samples: one including the earliest scan available for all subjects (Cross-sectional-1) and another including the second available scan for the 55% of subjects with multiple scans (Cross-sectional-2). In both samples, the bootstrapped results confirm our findings: for the Cross-sectional-1 sample, bootstrapped β = 0.208 (0.119, 0.294), and for the Cross-sectional-2 sample, bootstrapped β = 0.206 (0.110, 0.309).
We also examined age-stratified bootstrap results in the Oregon cohort, by analyzing scans for participants aged <12 years old (mean age = 10.1 years) separately from scans for participants aged >12 years old (mean age = 13.3 years). For this analysis, we used only the Cross-sectional-2 sample, given the small number of “older” scans in Cross-sectional-1. The association between PNRS and ADHD symptoms was significant for scans done before age 12 [bootstrapped β = 0.231 (0.128, 0.319)], but not for those done after age 12 [bootstrapped β = 0.084 (−0.115, 0.294)], providing further evidence of decreased predictive power of the PNRS with increasing age.
PLSR
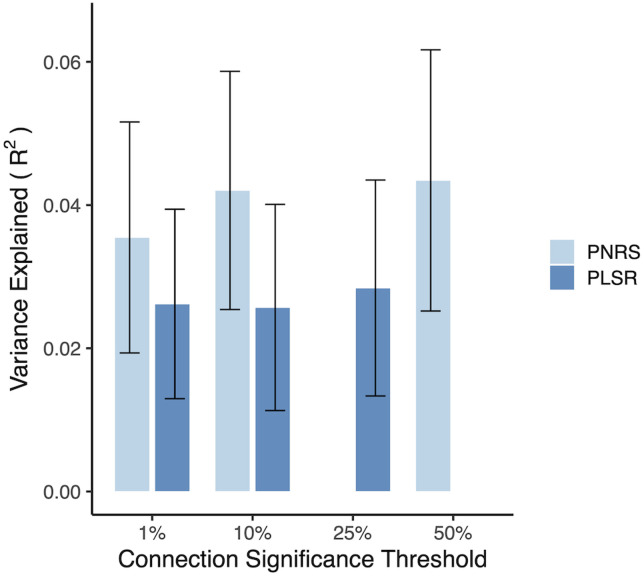
To determine whether the method used to construct the PNRS significantly impacted our results, we implemented an alternative PNRS using a commonly used multivariate method, PLSR. In this case, we used the same independent training (ABCD) and test (Oregon) samples as used for the primary PNRS. PLSR (Rosipal and Krämer, 2006) aims to predict dependent variables from a large set of independent variables through a latent variable (components) approach by maximizing the covariance between both the predictor and response variables. This method has been validated and used several times by our group (Rudolph et al., 2017, 2018; Kovacs-Balint et al., 2019; Miranda-Domínguez et al., 2020; Silva-Batista et al., 2021; Ragothaman et al., 2022; Byington et al., 2023). Here, we used functional connectivity as the independent variable and ADHD composite score as the dependent variable. For each threshold (top 1%, top 10%, and top 25% of connections), the connections included in the model were selected based on their univariate ability to predict within-sample outcomes. We explored the performance of PLSR using a different number of components. In this implementation, we use leave-one-out cross-validation to identify the optimal number of components able to predict the outcome. Once the optimal number of components was identified in the training sample, the entire ABCD data set was used to recalculate the weights. The resulting optimal models were applied to the independent Oregon cohort. The PLSR model showed predictive power (maximum proportion of variance explained in the Oregon cohort = 2.8%) slightly lower than, but comparable to, the primary PNRS (Fig. 10).
Figure 10.
The proportion of ADHD symptom score variance explained by both the PNRS and the PLSR model in the Oregon baseline cohort (without adjusting for covariates). Due to the computational complexity of the PLSR, that model was fit with a maximum of 25% of the most significant functional connections. Nevertheless, the two methods provide comparable predictive power across various connection inclusion thresholds, suggesting there is no meaningful benefit from the more complex PLSR model. Bootstrapped standard errors are shown.
Discussion
PNRS reveals a distributed view of the brain-wide association with ADHD symptoms
Our findings demonstrate a robust association between brain-wide connectivity patterns (PNRS) and ADHD symptoms in two independent cohorts with different recruitment procedures, supporting the generalizability of these findings. The findings demonstrate the importance of evaluating these types of measures in cohorts enriched for the trait of interest, given the larger effects seen in both the Oregon-ADHD-1000 case–control cohort and the ABCD matched case–control subsample. Importantly, the effect sizes seen in these enriched cohorts may better represent the effects seen in certain clinical populations.
PNRS that represent cumulative, brain-wide effects explained a significantly greater proportion of the variation in symptom scores than the most significant individual connections. Although the strongest contributions to the PNRS were concentrated among connections involving the default mode and cingulo-opercular networks, the most predictive PNRS incorporated connections involving all networks (Fig. 4) and explained ∼4% of the variation in symptoms—roughly eight times the variation explained by the single most significant connection. Though the PNRS incorporating the top 10% of most significant connections was most predictive in our data, there was no significant difference among the various significance thresholds tested, and the optimal threshold should be examined in future studies. Discovery set sample size and the distribution of effect sizes across the brain are likely to be important factors influencing the optimal inclusion threshold, as has been shown for polygenic scores (Dudbridge, 2013; Wray et al., 2014). In addition, other methods for modeling connectivity–behavior associations, such as alternate strategies for parcellation or estimating measures of connectivity (Dadi et al., 2019; Pervaiz et al., 2020), may improve the predictive power of PNRS and should be an area of future research.
In the enriched Oregon cohort, subjects with an ADHD PNRS in the highest 10% had 3.86 times greater odds of an ADHD diagnosis than those with a PNRS below the median (Fisher’s exact p = 1.2 × 10−5), demonstrating the potential predictive utility of the brain-wide summary measure. In ABCD ARMS-2, this odds ratio was 2.32 (p = 0.0023), demonstrating that even at the population level, extreme values of the PNRS may have utility for risk prediction when combined with other risk factors. The explanatory power of the PNRS examined here is comparable to PRS and will likely improve with larger discovery data sets. Like polygenic scores (in most cases), PNRS do not currently have utility for diagnostic decision-making; however, they can be useful in research settings for risk stratification and exploration of causal mechanisms (Yang et al., 2022).
The specific network contributions to the brain-wide score should be further evaluated and clarified with additional data. Our results are consistent with previous results implicating the default mode and cognitive control networks in ADHD but also demonstrate that the effects of individual networks do not fully capture connectivity patterns associated with the disorder. Our findings emphasize a distributed view of brain function, where complex cognitive behaviors or traits are best explained by cumulative effects across most brain networks (Rosenberg et al., 2016; Zhao et al., 2019; Marek et al., 2022).
Our results also exemplify the need to better understand the phenotypic heterogeneity among those with ADHD. Identification of PNRS specific to certain subgroups (e.g., ages, disorder subtypes, and patterns of comorbidity) may provide insight into the mechanisms driving this heterogeneity.
Sensitivity analyses suggest a robust association between brain-wide connectivity and ADHD symptoms
Sensitivity analyses were conducted to ensure that potential sources of bias (relatedness among subjects, history of ADHD medication usage, and motion during MRI acquisition) were accounted for, lending confidence to our results. In addition, while Bayesian-adjusted PNRS, which account for outcome signal-to-noise ratio and the correlation of effects across the brain, were slightly more predictive than unadjusted PNRS, the difference was not significant, contrary to previous work (Zhao et al., 2019). It is possible that our use of parcellated connectivity data, which already accounts for spatial correlation across the brain (by collapsing multiple highly correlated, spatially adjacent voxels into a single feature, or parcel), reduced the need for the adjustment. However, given the previously demonstrated benefit of this type of Bayesian adjustment (Vilhjálmsson et al., 2015; Zhao et al., 2019), we believe it to be good practice and its effect should be examined further in future studies of brain-wide summary scores, including those using other types of brain features (e.g., structural features).
In addition, an alternative method for constructing the PNRS (using PLSR) showed comparable results to the linear additive model used for the primary PNRS reported here. These results suggest that, for these types of brain-wide summary scores, the use of large sample sizes for discovery and appropriate quality control procedures is far more important than the method for combining/summarizing the brain-wide signal.
Lack of evidence for mediation of ADHD polygenic risk by the PNRS
Given the small effects of both ADHD PNRS and ADHD PRS, it is not surprising we did not observe evidence of the ADHD PNRS mediating common genetic risk for ADHD. However, it is important to note that larger sample sizes may allow for more precise scores and the detection of small but meaningful mediation effects. In addition, access to more diverse GWAS discovery cohorts for PRS is sorely needed to address the generalizability of PRS effects (Martin et al., 2019).
The lack of mediation effect, however, does not rule out the possibility of environmental factors influencing ADHD-related connectivity patterns. Low-to-moderate heritability for many brain features (Miranda-Dominguez et al., 2017; Sudre et al., 2017; Adhikari et al., 2018) and the growing recognition of the important role of environmental and developmental context in the etiology of ADHD suggest more work is needed to understand the interplay between genetic liability, environmental exposures, and brain endophenotypes in causal models of ADHD.
Longitudinal extension of PNRS models can assess specificity to childhood ADHD
Another important direction for future work should be the examination of how ADHD-associated connectivity changes with age (Kessler et al., 2016). The PNRS reported here were derived from data on children 9–10 years of age in the ABCD sample and showed at least suggestive evidence in the Oregon cohort of reduced predictive power in older subjects. This result raises the question of whether the scores examined here are specific to childhood ADHD or whether brain connectivity is simply less predictive of ADHD in older individuals. Subsequent waves of data released from the ABCD Study will allow further exploration of age effects.
We recognize that the stringent quality control procedures used here likely reduced the diversity of the discovery data set, given the known associations between rs-fcMRI data quality and several clinical and sociodemographic variables in ABCD (Cosgrove et al., 2022) and hence the generalizability of the PNRS. In addition to exploring the effect of age, the PNRS should be further evaluated in more diverse samples.
PNRS approach leverages small effect sizes to develop generalizable brain measures for clinical research
Overall, our findings provide strong evidence for the use of brain-wide summary measures of resting-state connectivity as a predictive measure of ADHD—and the importance of evaluating these measures in multiple large data sets, including those enriched for the trait of interest. These polyneuro scores have several advantages. First, because of the larger effect sizes for PNRS, they will increase the utility of small neuroimaging data sets (which are underpowered to detect associations with individual brain features) to test brain-behavior associations—similar to the way PRS have allowed testing for associations with cumulative genetic risk in relatively small samples. Second, PNRS partially alleviate issues of heterogeneity, given that the same cumulative risk can be the result of different individual risk factors. Third, PNRS may provide new ways to examine shared brain mechanisms across disorders (e.g., does an ADHD PNRS predict depression symptoms?) and provide insights into relationships between diagnostic categories and the risk of psychiatric comorbidities. Finally, further development of the PNRS approach and integration with other risk factors (e.g., polygenic and environmental measures) has the potential to provide clinically meaningful prediction of behavioral disorders and patient outcomes.
References
- Adhikari BM, et al. (2018) Heritability estimates on resting state fMRI data using ENIGMA analysis pipeline. Pac Symp Biocomput Pac Symp Biocomput 23:307–318. 10.1142/9789813235533_0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albajara Sáenz A, Villemonteix T, Massat I (2019) Structural and functional neuroimaging in attention-deficit/hyperactivity disorder. Dev Med Child Neurol 61:399–405. 10.1111/dmcn.14050 [DOI] [PubMed] [Google Scholar]
- Baurley JW, Edlund CK, Pardamean CI, Conti DV, Bergen AW (2016) Smokescreen: a targeted genotyping array for addiction research. BMC Genomics 17:145. 10.1186/s12864-016-2495-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byington N, et al. (2023) Polyneuro risk scores capture widely distributed connectivity patterns of cognition. Dev Cogn Neurosci 60:101231. 10.1016/j.dcn.2023.101231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey VJ (2019) gee: generalized estimation equation solver. Available at: https://CRAN.R-project.org/package=gee [Accessed September 15, 2021].
- Casey BJ, et al. (2018) The adolescent brain cognitive development (ABCD) study: imaging acquisition across 21 sites. Dev Cogn Neurosci 32:43–54. 10.1016/j.dcn.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos FX, et al. (2008) Cingulate-precuneus interactions: a new locus of dysfunction in adult attention-deficit/hyperactivity disorder. Biol Psychiatry 63:332–337. 10.1016/j.biopsych.2007.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Mak TS-H, O’Reilly PF (2020) Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc 15:2759–2772. 10.1038/s41596-020-0353-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciric R, et al. (2017) Benchmarking of participant-level confound regression strategies for the control of motion artifact in studies of functional connectivity. NeuroImage 154:174–187. 10.1016/j.neuroimage.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova MM, Antovich DM, Ryabinin P, Neighbor C, Mooney MA, Dieckmann NF, Miranda-Dominguez O, Nagel BJ, Fair DA (2022) Attention-deficit/hyperactivity disorder: restricted phenotypes prevalence, comorbidity, and polygenic risk sensitivity in the ABCD baseline cohort. J Am Acad Child Adolesc Psychiatry 61:1273–1284. 10.1016/j.jaac.2022.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese S, Aoki YY, Itahashi T, Castellanos FX, Eickhoff SB (2021) Systematic review and meta-analysis: resting-state functional magnetic resonance imaging studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 60:61–75. 10.1016/j.jaac.2020.08.014 [DOI] [PubMed] [Google Scholar]
- Cosgrove KT, McDermott TJ, White EJ, Mosconi MW, Thompson WK, Paulus MP, Cardenas-Iniguez C, Aupperle RL (2022) Limits to the generalizability of resting-state functional magnetic resonance imaging studies of youth: an examination of ABCD Study® baseline data. Brain Imaging Behav 16:1919–1925. 10.1007/s11682-022-00665-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadi K, Rahim M, Abraham A, Chyzhyk D, Milham M, Thirion B, Varoquaux G (2019) Benchmarking functional connectome-based predictive models for resting-state fMRI. NeuroImage 192:115–134. 10.1016/j.neuroimage.2019.02.062 [DOI] [PubMed] [Google Scholar]
- Demontis D, et al. (2019) Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 51:63. 10.1038/s41588-018-0269-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M, Francis DJ, Cirino PT, Schachar R, Barnes MA, Fletcher JM (2009) Why IQ is not a covariate in cognitive studies of neurodevelopmental disorders. J Int Neuropsychol Soc 15:331–343. 10.1017/S1355617709090481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zeeuw P, Schnack HG, van Belle J, Weusten J, van Dijk S, Langen M, Brouwer RM, van Engeland H, Durston S (2012) Differential brain development with low and high IQ in attention-deficit/hyperactivity disorder. PLoS ONE 7:e35770. 10.1371/journal.pone.0035770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F (2013) Power and predictive accuracy of polygenic risk scores. PLoS Genet 9:e1003348. 10.1371/journal.pgen.1003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Bathula D, Nikolas MA, Nigg JT (2012) Distinct neuropsychological subgroups in typically developing youth inform heterogeneity in children with ADHD. Proc Natl Acad Sci U S A 109:6769–6774. 10.1073/pnas.1115365109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Posner J, Nagel BJ, Bathula D, Dias TGC, Mills KL, Blythe MS, Giwa A, Schmitt CF, Nigg JT (2010) Atypical default network connectivity in youth with attention-deficit/hyperactivity disorder. Biol Psychiatry 68:1084–1091. 10.1016/j.biopsych.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feczko E, et al. (2021) Adolescent brain cognitive development (ABCD) community MRI collection and utilities. Available at: https://www.biorxiv.org/content/10.1101/2021.07.09.451638v1 [Accessed August 25, 2021].
- Feczko E, Earl E, Perrone A, Fair D (2020a) ABCD-BIDS community collection (ABCC). Available at: https://osf.io/psv5m/ [Accessed May 4, 2021].
- Feczko E, Earl E, Perrone A, Fair D (2020b) ABCD reproducible matched samples (ARMS) software. Available at: https://osf.io/7xn4f/ [Accessed May 4, 2021].
- Feczko E, Earl E, Perrone A, Fair D, Nichols T, Thompson WK (2020c) Sandwich estimator for neuroimaging dData (SEND). Available at: https://osf.io/rjse2/ [Accessed October 20, 2021].
- Feczko E, Fair DA (2020) Methods and challenges for assessing heterogeneity. Biol Psychiatry 88:9–17. 10.1016/j.biopsych.2020.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feczko E, Miranda-Dominguez O, Marr M, Graham AM, Nigg JT, Fair DA (2019) The heterogeneity problem: approaches to identify psychiatric subtypes. Trends Cogn Sci 23:584–601. 10.1016/j.tics.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, et al. (2019) Impairments of large-scale functional networks in attention-deficit/hyperactivity disorder: a meta-analysis of resting-state functional connectivity. Psychol Med 49:2475–2485. 10.1017/S003329171900237X [DOI] [PubMed] [Google Scholar]
- Glasser MF, et al. (2013) The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 80:105–124. 10.1016/j.neuroimage.2013.04.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten SM, et al. (2012) GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinforma Oxf Engl 28:3329–3331. 10.1093/bioinformatics/bts610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten SM, Sofer T, Chen H, Yu C, Brody JA, Thornton TA, Rice KM, Conomos MP (2019) Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics 35:5346–5348. 10.1093/bioinformatics/btz567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE (2016) Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb Cortex 26:288–303. 10.1093/cercor/bhu239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume B, Hua X, Thompson PM, Waldorp L, Nichols TE (2014) Fast and accurate modelling of longitudinal and repeated measures neuroimaging data. NeuroImage 94:287–302. 10.1016/j.neuroimage.2014.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermosillo RJM, et al. (2020) Polygenic risk score–derived subcortical connectivity mediates attention-deficit/hyperactivity disorder diagnosis. Biol Psychiatry Cogn Neurosci Neuroimaging 5:330–341. 10.1016/j.bpsc.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogman M, et al. (2020) Consortium neuroscience of attention deficit/hyperactivity disorder and autism spectrum disorder: the ENIGMA adventure. Hum Brain Mapp 43:37–55. 10.1002/hbm.25029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5:e1000529. 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalunas SL, Gustafsson HC, Dieckmann NF, Tipsord J, Mitchell SH, Nigg JT (2017) Heterogeneity in development of aspects of working memory predicts longitudinal attention deficit hyperactivity disorder symptom change. J Abnorm Psychol 126:774–792. 10.1037/abn0000292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D, Angstadt M, Sripada C (2016) Growth charting of brain connectivity networks and the identification of attention impairment in youth. JAMA Psychiatry 73:481–489. 10.1001/jamapsychiatry.2016.0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, et al. (2022) Translating ENIGMA schizophrenia findings using the regional vulnerability index: association with cognition, symptoms, and disease trajectory. Hum Brain Mapp 43:566–575. 10.1002/hbm.25045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs-Balint Z, et al. (2019) Early developmental trajectories of functional connectivity along the visual pathways in rhesus monkeys. Cereb COoRrTtex 29:3514–3526. 10.1093/cercor/bhy222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, He Q (2021) Polygenic scores for ADHD: a meta-analysis. Res Child Adolesc Psychopathol 49:297–310. 10.1007/s10802-021-00774-4 [DOI] [PubMed] [Google Scholar]
- Luo Y, Weibman D, Halperin JM, Li X (2019) A review of heterogeneity in attention deficit/hyperactivity disorder (ADHD). Front Hum Neurosci 13:42. 10.3389/fnhum.2019.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S, et al. (2022) Reproducible brain-wide association studies require thousands of individuals. Nature 603:654–660. 10.1038/s41586-022-04492-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ (2019) Current clinical use of polygenic scores will risk exacerbating health disparities. Nat Genet 51:584–591. 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KL, et al. (2016) Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci 19:1523–1536. 10.1038/nn.4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Feczko E, Grayson DS, Walum H, Nigg JT, Fair DA (2017) Heritability of the human connectome: a connectotyping study. Netw Neurosci 2:175–199. 10.1162/netn_a_00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Domínguez Ó, Ragothaman A, Hermosillo R, Feczko E, Morris R, Carlson-Kuhta P, Nutt JG, Mancini M, Fair D, Horak FB (2020) Lateralized connectivity between globus pallidus and motor cortex is associated with freezing of gait in Parkinson’s disease. Neuroscience 443:44–58. 10.1016/j.neuroscience.2020.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney MA, et al. (2023) Joint polygenic and environmental risks for childhood attention-deficit/hyperactivity disorder (ADHD) and ADHD symptom dimensions. JCPP Adv 3:e12152. 10.1002/jcv2.12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney MA, Bhatt P, Hermosillo RJM, Ryabinin P, Nikolas M, Faraone SV, Fair DA, Wilmot B, Nigg JT (2021) Smaller total brain volume but not subcortical structure volume related to common genetic risk for ADHD. Psychol Med 51:1279–1288. 10.1017/S0033291719004148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney MA, Ryabinin P, Wilmot B, Bhatt P, Mill J, Nigg JT (2020) Large epigenome-wide association study of childhood ADHD identifies peripheral DNA methylation associated with disease and polygenic risk burden. Transl Psychiatry 10:1–12. 10.1038/s41398-020-0710-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni G, et al. (2021) A comparison of ten polygenic score methods for psychiatric disorders applied across multiple cohorts. Biol Psychiatry 90:611–620. 10.1016/j.biopsych.2021.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg JT, Gustafsson HC, Karalunas SL, Ryabinin P, McWeeney SK, Faraone SV, Mooney MA, Fair DA, Wilmot B (2018) Working memory and vigilance as multivariate endophenotypes related to common genetic risk for attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 57:175–182. 10.1016/j.jaac.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg JT, Karalunas SL, Feczko E, Fair DA (2020a) Toward a revised nosology for attention-deficit/hyperactivity disorder heterogeneity. Biol Psychiatry Cogn Neurosci Neuroimaging 5:726–737. 10.1016/j.bpsc.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg JT, Karalunas SL, Gustafsson HC, Bhatt P, Ryabinin P, Mooney MA, Faraone SV, Fair DA, Wilmot B (2020b) Evaluating chronic emotional dysregulation and irritability in relation to ADHD and depression genetic risk in children with ADHD. J Child Psychol Psychiatry 61:205–214. 10.1111/jcpp.13132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain O, et al. (2021) Evaluation of polygenic prediction methodology within a reference-standardized framework. PLoS Genet 17:e1009021. 10.1371/journal.pgen.1009021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Sanchez V, Castellanos FX (2021) Neuroimaging in attention-deficit/hyperactivity disorder. Curr Opin Psychiatry 34:105–111. 10.1097/YCO.0000000000000669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervaiz U, Vidaurre D, Woolrich MW, Smith SM (2020) Optimising network modelling methods for fMRI. NeuroImage 211:116604. 10.1016/j.neuroimage.2020.116604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner J, Park C, Wang Z (2014) Connecting the dots: a review of resting connectivity MRI studies in attention-deficit/hyperactivity disorder. Neuropsychol Rev 24:3–15. 10.1007/s11065-014-9251-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M, Ye Z, Li Q, Liu G, Xie B, Wang J (2011) Changes of brain structure and function in ADHD children. Brain Topogr 24:243–252. 10.1007/s10548-010-0168-4 [DOI] [PubMed] [Google Scholar]
- Ragothaman A, Mancini M, Nutt JG, Fair DA, Miranda-Dominguez O, Horak FB (2022) Resting state functional networks predict different aspects of postural control in Parkinson’s disease. Gait Posture 97:122–129. 10.1016/j.gaitpost.2022.07.003 [DOI] [PubMed] [Google Scholar]
- Ronald A, de Bode N, Polderman TJC (2021) Systematic review: how the attention-deficit/hyperactivity disorder polygenic risk score adds to Our understanding of ADHD and associated traits. J Am Acad Child Adolesc Psychiatry 60:1234–1277. 10.1016/j.jaac.2021.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MD, Finn ES, Scheinost D, Papademetris X, Shen X, Constable RT, Chun MM (2016) A neuromarker of sustained attention from whole-brain functional connectivity. Nat Neurosci 19:165–171. 10.1038/nn.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosipal R, Krämer N (2006) Overview and recent advances in partial least squares. In: Subspace, latent structure and feature selection (Saunders C, Grobelnik M, Gunn S, Shawe-Taylor J, eds), pp 34–51. Lecture notes in computer science. Berlin, Heidelberg: Springer. [Google Scholar]
- Rudolph MD, et al. (2017) At risk of being risky: the relationship between “brain age” under emotional states and risk preference. Dev Cogn Neurosci 24:93–106. 10.1016/j.dcn.2017.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MD, Graham AM, Feczko E, Miranda-Dominguez O, Rasmussen JM, Nardos R, Entringer S, Wadhwa PD, Buss C, Fair DA (2018) Maternal IL-6 during pregnancy can be estimated from newborn brain connectivity and predicts future working memory in offspring. Nat Neurosci 21:765. 10.1038/s41593-018-0128-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JF, Griffiths KR, Korgaonkar MS (2020) A systematic review of imaging studies in the combined and inattentive subtypes of attention deficit hyperactivity disorder. Front Integr Neurosci 14:31. 10.3389/fnint.2020.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Finn ES, Scheinost D, Rosenberg MD, Chun MM, Papademetris X, Constable RT (2017) Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat Protoc 12:506–518. 10.1038/nprot.2016.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Batista C, Ragothaman A, Mancini M, Carlson-Kuhta P, Harker G, Jung SH, Nutt JG, Fair DA, Horak FB, Miranda-Domínguez O (2021) Cortical thickness as predictor of response to exercise in people with Parkinson’s disease. Hum Brain Mapp 42:139–153. 10.1002/hbm.25211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Nichols TE (2018) Statistical challenges in “big data” human neuroimaging. Neuron 97:263–268. 10.1016/j.neuron.2017.12.018 [DOI] [PubMed] [Google Scholar]
- Song M, Dieckmann NF, Nigg JT (2019) Addressing discrepancies between ADHD prevalence and case identification estimates among U.S. Children utilizing NSCH 2007–2012. J Atten Disord 23:1691–1702. 10.1177/1087054718799930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonuga-Barke EJS (2005) Causal models of attention-deficit/hyperactivity disorder: from common simple deficits to multiple developmental pathways. Biol Psychiatry 57:1231–1238. 10.1016/j.biopsych.2004.09.008 [DOI] [PubMed] [Google Scholar]
- Suarez A, et al. (2020) A polyepigenetic glucocorticoid exposure score at birth and childhood mental and behavioral disorders. Neurobiol Stress 13:100275. 10.1016/j.ynstr.2020.100275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudre G, Choudhuri S, Szekely E, Bonner T, Goduni E, Sharp W, Shaw P (2017) Estimating the heritability of structural and functional brain connectivity in families affected by attention-deficit/hyperactivity disorder. JAMA Psychiatry 74:76–84. 10.1001/jamapsychiatry.2016.3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudre G, Frederick J, Sharp W, Ishii-Takahashi A, Mangalmurti A, Choudhury S, Shaw P (2020) Mapping associations between polygenic risks for childhood neuropsychiatric disorders, symptoms of attention deficit hyperactivity disorder, cognition, and the brain. Mol Psychiatry 25:2482–2492. 10.1038/s41380-019-0350-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden K, et al. (2019) Establishing a generalized polyepigenetic biomarker for tobacco smoking. Transl Psychiatry 9:92. 10.1038/s41398-019-0430-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Jiang R, Bustillo J, Calhoun V (2020) Neuroimaging-based individualized prediction of cognition and behavior for mental disorders and health: methods and promises. Biol Psychiatry 88:818–828. 10.1016/j.biopsych.2020.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcubasi B, Metin B, Kurban MK, Metin ZE, Beser B, Sonuga-Barke E (2020) Resting-state network dysconnectivity in ADHD: a system-neuroscience-based meta-analysis. World J Biol Psychiatry 21:662–672. 10.1080/15622975.2020.1775889 [DOI] [PubMed] [Google Scholar]
- Thapar A (2018) Discoveries on the genetics of ADHD in the 21st century: new findings and their implications. Am J Psychiatry 175:943–950. 10.1176/appi.ajp.2018.18040383 [DOI] [PubMed] [Google Scholar]
- Tingley D, Yamamoto T, Hirose K, Keele L, Imai K (2014) Mediation: R package for causal mediation analysis. J Stat Softw 59:1–38. 10.18637/jss.v059.i0526917999 [DOI] [Google Scholar]
- Traut N, et al. (2022) Insights from an autism imaging biomarker challenge: promises and threats to biomarker discovery. NeuroImage 255:119171. 10.1016/j.neuroimage.2022.119171 [DOI] [PubMed] [Google Scholar]
- Uban KA, Horton MK, Jacobus J, Heyser C, Thompson WK, Tapert SF, Madden PAF, Sowell ER (2018) Biospecimens and the ABCD study: rationale, methods of collection, measurement and early data. Dev Cogn Neurosci 32:97–106. 10.1016/j.dcn.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhjálmsson BJ, et al. (2015) Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet 97:576–592. 10.1016/j.ajhg.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkeys OJ, Cohen-Woods S, Quidé Y, Cairns MJ, Overs B, Fullerton JM, Green MJ (2020) Derivation of poly-methylomic profile scores for schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry 101:109925. 10.1016/j.pnpbp.2020.109925 [DOI] [PubMed] [Google Scholar]
- Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, Middeldorp CM (2014) Research review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry 55:1068–1087. 10.1111/jcpp.12295 [DOI] [PubMed] [Google Scholar]
- Yang FN, Liu TT, Wang Z (2022) Functional connectome mediates the association between sleep disturbance and mental health in preadolescence: a longitudinal mediation study. Hum Brain Mapp 43:2041–2050. 10.1002/hbm.25772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Palmer CE, Thompson W, Jernigan TL, Dale AM, Fan CC (2019) The Bayesian polyvertex score (PVS-B): a whole-brain phenotypic prediction framework for neuroimaging studies. bioRxiv:813915.