Abstract
Background
Low-dose computed tomography screening can reduce lung cancer-related mortality. However, most screen-detected pulmonary abnormalities do not develop into cancer and it often remains challenging to identify malignant nodules, particularly among indeterminate nodules. We aimed to develop and assess prediction models based on radiological features to discriminate between benign and malignant pulmonary lesions detected on a baseline screen.
Methods
Using four international lung cancer screening studies, we extracted 2,060 radiomic features for each of 16,797 nodules (513 malignant) among 6,865 participants. After filtering out low-quality radiomic features, 642 radiomic and 9 epidemiologic features remained for model development. We used cross-validation and grid search to assess three machine learning (ML) models (XGBoost, Random Forest, LASSO) for their ability to accurately predict risk of malignancy for pulmonary nodules. We report model performance based on the area under the curve (AUC) and calibration metrics in the held-out test set.
Results
The LASSO model yielded the best predictive performance in cross-validation and was fit in the full training set based on optimized hyperparameters. Our radiomics model had a test-set AUC of 0.93 (95% CI: 0.90–0.96) and out-performed the established PanCan model (AUC=0.87, 95% CI: 0.85–0.89) for nodule assessment. Our model performed well among both solid (AUC=0.93, 95% CI: 0.89–0.97) and subsolid nodules (AUC=0.91, 95% CI: 0.85–0.95).
Conclusions
We developed highly-accurate machine learning models based on radiomic and epidemiologic features from four international lung cancer screening studies that may be suitable for assessing indeterminate screen-detected pulmonary nodules for risk of malignancy.
Introduction
Lung cancer is the leading cause of cancer mortality globally [1]. Only 10–20% of lung cancer patients live up to five years after diagnosis [2]. However, several large randomized screening trials have demonstrated that low-dose computed tomography (CT) screening can significantly reduce lung cancer mortality through early detection [3–6]. The National Lung Screening Trial (NLST) observed a 20% reduction in lung cancer-related mortality following CT screening [4], while the Dutch-Belgian trial (NELSON) observed a reduction in mortality of 24% in men and 33% in women [3].
Despite the promise of screening, the clinical management of screen-detected pulmonary nodules and the false-positive rate are important determinants for screening program efficacy. Across several studies, the average nodule detection rate was 20%, meanwhile, more than 90% of screen-detected nodules were benign [7]. Inaccurate assessment of indeterminate nodules may lead to unnecessary diagnostic workup, including: additional imaging studies (which confer higher radiation exposure); invasive procedures such as bronchoscopy, CT biopsy, or surgery; and may lead to overdiagnosis of indolent cancers [7]. Unnecessary follow-up carries significant healthcare costs, utilizes critical hospital and human resources, may lead to adverse events and complications, including premature death, and can cause anxiety and decreased quality of life for the screened participant.
Several guidelines have been developed to help inform screen-detected lung nodule management, however, there remains significant heterogeneity in these recommendations [8–22]. To address these issues, probability models have been developed to help identify high-risk lesions and guide clinical decision-making [23–25]. These models have traditionally been based on patient characteristics (e.g., age, smoking history, etc.) and clinically-collected nodule morphology and textural features (e.g., size, attenuation, etc.). These features characterize important aspects of the nodule and are routinely collected as part of the clinical management of pulmonary findings.
Nodule probability models based on routinely-collected patient and nodule information have shown good performance, however, there is growing interest in leveraging medical images directly to perform automated quantitative image analysis, enabling the quantification of hundreds or thousands of radiomic features that may capture important information otherwise imperceptible to the human eye. Radiomic features quantify aspects of the 3-dimensional (3D) morphology and grayscale distribution for a region-of-interest [26]. It is expected that radiomic features, in combination with patient-level information, will be able to accurately discriminate between benign and malignant pulmonary nodules beyond what has been achieved with traditional clinical features. However, it is currently unknown which features will be most important and whether they will generalize well to other screening cohorts. In addition, the use of deep learning for nodule malignancy assessment is growing in popularity and several studies have been developed models for this purpose [27,28]. However, the black-box nature of deep learning models often lack full transparency with open-source code, and high levels of model parameterization continue to hamper clinical implementation. Models based on extracted features have shown comparable performance while offering improved model interpretability and greater relative ease-of-implementation.
The goal of the current study was to perform quantitative image analysis and evaluate the predictive performance of high-dimensional radiomic features for pulmonary nodule malignancy assessment, and to develop and validate models using data from several large independent international lung cancer screening studies.
Methods
Lung Cancer Screening Studies
As part of the Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) program, we used data collected by four independent lung cancer screening studies for this analysis: 1. National Lung Screening Trial (NLST), 2. PanCanadian Early Detection of Lung Cancer (PanCan) Study, 3. International Early Lung Cancer Action Program (IELCAP-Toronto), and 4. Pittsburgh Lung Screening Study (PLuSS). Details of each study have been described previously [4,5,29–32]. We provide brief descriptions of each study in the following sections. Details about the study protocol used by each study are included in the Supplemental Materials.
National Lung Screening Trial (NLST)
NLST was a large randomized multi-center lung cancer screening study comparing low-dose helical CT to standard chest radiography (CXR) for screening adult heavy smokers [4,5]. Eligible participants were age 55 to 74 years, with 30 or more pack-years history of smoking, and former smokers quitting no more than 15 years prior. NLST enrolled 53,456 participants across 33 centers in the United States in 2002. We only used image data from the CT screening arm in the current study. Any non-calcified nodules (NCN) with a diameter of 4mm or greater was considered a positive screen-detected finding.
Pan-Canadian Early Detection of Lung Cancer Study (PanCan)
PanCan was a multi-center, single-arm prospective lung cancer screening study that included 2,537 participants [29]. Participants were recruited from eight sites across Canada. Eligible participants included those 50 to 75 years of age, without a self-reported history of lung cancer, current or former smokers, an estimated 6-year risk of lung cancer of at least 2% based on an earlier edition of the PLCOm2012 model [33], and an ECOG performance status of 0 or 1. Screening was performed with multi-detector row CT scanners. Each scan was reviewed by a trained radiologist and up to 10 lung nodules were identified and recorded.
International Early Lung Cancer Action Program (IELCAP-Toronto)
IELCAP was an international single-arm multi-centre study evaluating low-dose CT for lung cancer screening of high-risk individuals [30,31]. A common study protocol was adopted for screening regimen, however, each site was able to make decisions regarding enrollment criteria. The Toronto location (hereafter referred to as IELCAP-Toronto), was based out of Princess Margaret Cancer Centre and began in 2003. IELCAP-Toronto enrolled 4,782 adults age 50 or older who were ever-smokers with more than 10 pack-years history of smoking. Participants were screened with milt-detector-row CT scanners. Positive findings were considered as any NCN found on a baseline scan.
Pittsburgh Lung Screening Study (PLuSS)
PLuSS was a lung cancer screening study that recruited 3,642 eligible participants between January 2002 and April 2005 [32]. Eligible participants included those age 50 to 79 years, with no personal history of lung cancer, no concurrent participation in other lung screening studies, no chest CT within the preceding year, current or former smoker with 0.5 pack-years history of smoking for at least 25 years, no smoking cessation within 10 years of enrollment, and body weight less than 400 pounds. Participants underwent low-dose chest CT and any NCN was considered a positive finding.
Pulmonary Nodule Segmentation
We performed supervised, semi-automated segmentation of screen-detected pulmonary nodules using the open-source 3D Slicer software [34] and the Chest Imaging Platform extension [35,36]. Our radiologist (HAS) located and reviewed each pulmonary lesion. Upon locating the lesion, the radiologist placed a seed-point at the approximate centroid of the lesion; semi-automated segmentation was performed based on the single seed-point, and manual touch-ups were performed at the discretion of the radiologist to fix over- or under-segmentation. All nodules were reviewed using standard lung windows. Segmentations for PanCan were performed by the PanCan investigators using an automated segmentation algorithm based on a commercial software and images and masks were provided without further processing, except those relevant to the feature extraction, detailed in the following section. We also collected detailed nodule information, including: lung and lobe location, suspicion of nodule malignancy, a nodule-specific LungRADS score (based on LungRADS 1.1 [8]), and ratings for semantic nodule features including margin, sphericity, subtlety, spiculation, solidity, calcification, structure, and lobulation. Details on the ratings systems for semantic nodule features are described in Supplemental Table 1.
Radiomics Feature Extraction
We performed radiomic feature extraction for baseline screen-detected pulmonary nodules using PyRadiomics (version 3.0.1) [26]. Due to heterogeneity in image acquisition settings between and within screening studies, all images and masks were resampled and interpolated to have unit (1mm3) voxel spacing (i.e., isotropy). We used a linear interpolator for images and nearest-neighbours interpolator for masks (to preserve labels). Grayscale intensities were discretized into bins using a bin width of 25 for histogram-based features. Voxel intensities were right-shifted by 1000 units prior to feature extraction to avoid negative values during feature computations.
Feature classes and the number of features per class were: (1) first-order statistics [18 features], (2) shape-based [14 features], (3) gray level coocurrence matrix [24 features], (4) gray level run length matrix [16 features], (5) gray level size zone matrix [16 features], (6) neighbouring gray tone difference matrix [5 features], and (7) gray level dependence matrix [14 features]. The list of radiomic features for each class are provided in Supplemental Table 2. We extracted shape and intensity-based features using the original image. We also extracted intensity-based features from images after applying several transformations, including: wavelet, Laplacian of Gaussian (LoG), Square, SquareRoot, Logarithm, Exponential, Gradient, and LocalBinaryPattern3D. In total, we extracted 2,060 radiomic features per nodule.
Statistical Analysis
Epidemiologic covariates and outcomes
Epidemiologic data were harmonized across the four screening studies to establish a common set of patient-level covariates. After harmonization, age, sex, family history of lung cancer among a first-degree relative, history of COPD or emphysema, smoking status, smoking duration, smoking intensity, years since quitting, and body mass index were included. We combined epidemiologic and radiomic features from the four screening studies to form our candidate predictor set. Missing data were generally minimal in the harmonized epidemiologic data and patients were excluded if missing data were present (Supplemental Figure 1). Nodule-level malignancy status was the outcome of interest, with no minimum time-to-diagnosis. Nodule malignancy status was available for PanCan and IELCAP-Toronto, and for NLST and PLuSS it was determined from patient-level diagnoses and based on radiologist assessment of individual nodules. More details are provided in the Supplemental Methods.
Model Development
We used subject-level random sampling to split the data into training (80%) and testing (20%) sets, ensuring all nodules for a specific participant were in the same split. The training set was further split into five folds using subject-level random sampling to perform cross-validation (CV). We had 2,060 radiomic features to assess for their ability to classify benign and malignant pulmonary nodules. Many radiomic features have correspondences to established clinically-collected (i.e., semantic) nodules features, however, many features have an unknown predictive value. We performed an initial set of filtering steps to remove zero variance (n=78), low quality (n=11), and weakly predictive (FDR-adjusted P-value > 0.05 in univariate models, n=248), and highly-redundant features (pairwise correlation > 0.9, n=1,081), described in detail in the Supplemental Methods.
Using the 9 epidemiologic covariates and 647 radiomic features retained after filtering, we performed cross-validation to identify the top-performing ML model. All predictors were normalized prior to model fitting. We assessed the following ML models: penalized logistic regression (LASSO), Random Forest (RF), and Gradient Boosted Trees (XGBoost). We first performed grid-search over a set of hyperparameters chosen using a Latin hypercube space-filling design [37]. We then performed random grid search over a finer set of hyperparameters for the top-performing model. The optimal hyperparameter(s) were then fit to the full training set and model performance was evaluated in the hold-out test set (referred to as the INTEGRAL-Radiomics model). A schematic of the analytic approach used in this study is presented in Figure 1. All statistical analysis was performed using Python 3.7.10 and R 4.0.5 [38,39].
Figure 1.
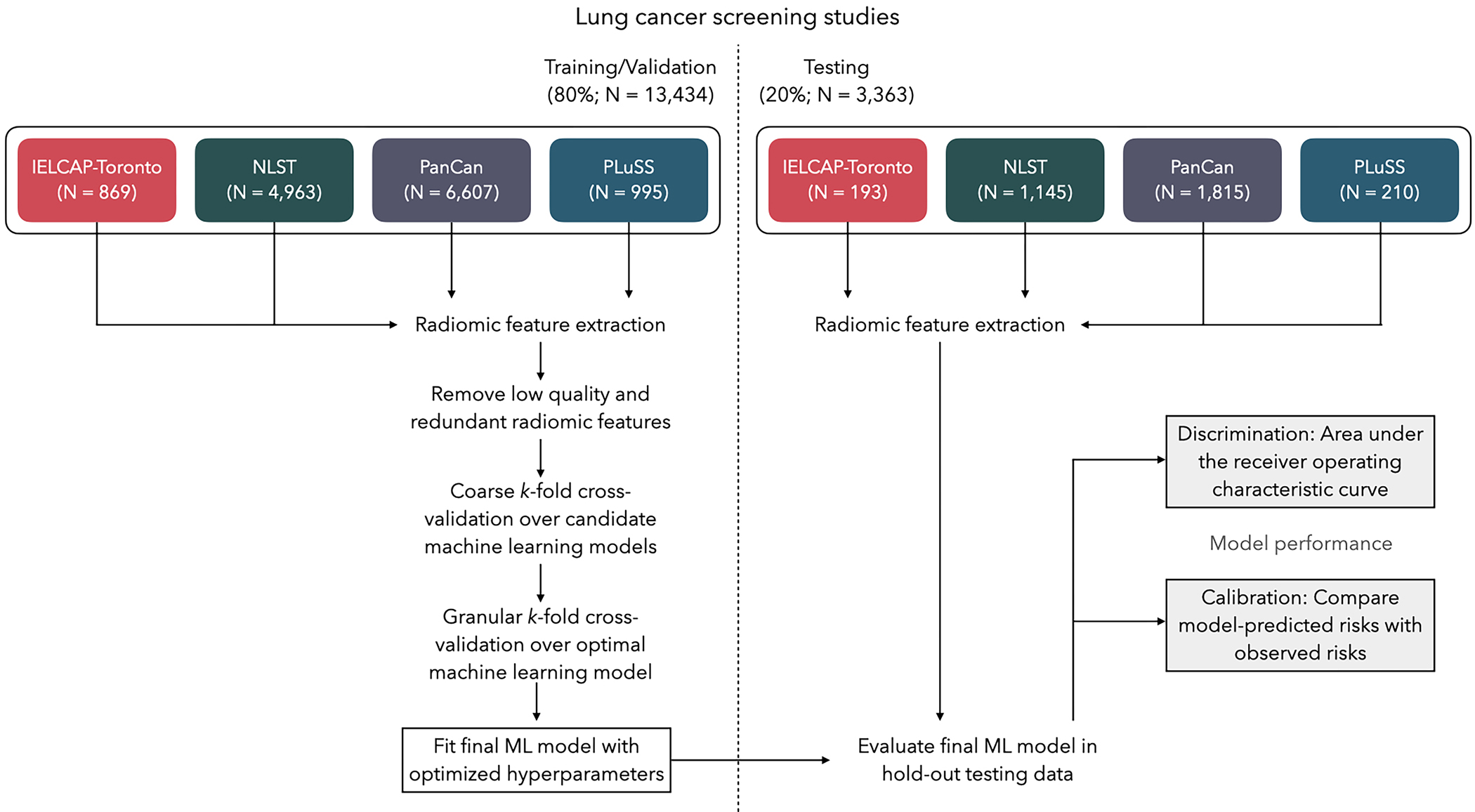
Schematic for the analytic framework used in this study. Data were partitioned into training/validation and testing splits using group-based random sampling to ensure all nodules for a participant were in a single set to avoid data leakage. Radiomic features were extracted and subject to filtering to exclude low-quality and highly-redundant features. K-fold cross-validation was performed to identify the optimal machine learning (ML) model and the optimal set of hyperparameters. The final ML model was fitted to the entire training data set and tested for out-of-sample performance in the hold-out test data; discrimination and calibration performance metrics are reported.
Model Performance
We evaluated model performance in two complementary ways: (1) area under the receiver operating characteristic curve (AUC) to assess a model’s ability to assign higher risks to malignant lesions than to benign lesions (i.e., discrimination), and (2) compare model-estimated risks to observed risks (i.e., calibration). For calibration, we compared predicted and observed risks within bins of predicted risks, and also assessed the ratio of expected to observed number of cancers and the difference between expected and observed number of cancers. We report the AUC and calibration metrics with percentile-based bootstrap confidence intervals. We compared our model performance with the established PanCan nodule malignancy model, previously reported in [40]. In brief, the PanCan model is a logistic regression model consisting of demographics, medical history, and nodule characteristics (Supplemental Methods).
Results
Basic demographics about the participants and nodules in the four lung cancer screening cohorts are presented in Table 1. Participants were similar in age between the cohorts. There were more males than females in NLST (57% vs. 43%), PanCan (53% vs. 47%), and PLuSS (51% vs. 49%), while IELCAP-Toronto (39% vs. 61%) had more women. The four cohorts had differing proportions of current and former smokers, and smoking histories (i.e., duration, intensity, and years since quitting) varied between studies. All four cohorts generally consisted of heavy current and former smokers. On average, PanCan had more nodules per participant, and also smaller nodules, compared to the other studies.
Table 1.
Patient-level and nodule-level descriptive statistics for each of the four screening cohorts included in this study. Means and standard deviations are reported for numeric variables and counts and proportions are reported for categorical variables.
| Total Participants (N = 6,865) | ||||
|---|---|---|---|---|
| IELCAP-Toronto (n = 502) |
NLST (n = 3,743) |
PanCan (n = 1,785) |
PLuSS (n = 835) |
|
| No. lung cancers (%) | 12 (2.4%) | 336 (9.0%) | 40 (2.2%) | 51 (6.1%) |
| Age (years) | 62.8 [7.5] | 62.4 [5.3] | 63.2 [6.0] | 60.5 [7.0] |
| Sex | ||||
| Male | 194 (38.6%) | 2,149 (57.4%) | 953 (53.4%) | 426 (51.0%) |
| Female | 308 (61.4%) | 1,594 (42.6%) | 832 (46.6%) | 309 (49.0%) |
| Body mass index (kg/m2) | 26.4 [4.4] | 27.5 [4.9] | 26.6 [4.5] | 28.1 [5.3] |
| Family history of lung cancer | ||||
| No | 388 (77.3%) | 2,894 (77.3%) | 1,288 (72.2%) | 190 (82.6%) |
| Yes | 114 (22.7%) | 849 (22.7%) | 497 (27.8%) | 145 (17.4%) |
| History of COPD or Emphysema | ||||
| No | 430 (85.7%) | 3,232 (86.3%) | 1,496 (83.8%) | 740 (88.6%) |
| Yes | 72 (14.3%) | 511 (13.7%) | 289 (16.2%) | 95(11.4%) |
| Smoking status | ||||
| Current | 66 (13.1%) | 1,845 (49.3%) | 1,133 (63.5%) | 580 (69.5%) |
| Former | 436 (86.9%) | 1,898 (50.7%) | 652 (36.5%) | 255 (30.5%) |
| Years smoked | 30.6 [10.7] | 40.9 [7.5] | 42.6 [8.8] | 40.9 [7.9] |
| Cigarettes per day | 21.3 [9.9] | 28.4 [11.4] | 24.7 [10.5] | 25.9 [9.8] |
| Years since cessation | 14.5 [10.6] | 19.9 [20.3] | 2.6 [5.7] | 2.0 [3.5] |
| Total Nodules (N = 16,797) | ||||
| IELCAP-Toronto (n = 1,062) |
NLST (n = 6,108) |
PanCan (n = 8,422) |
PLuSS (n = 1,205) |
|
| Nodules per participant | 3.2 [2.0] | 2.4 [1.7] | 8.0 [5.2] | 2.0 [1.4] |
| Nodule solidity | ||||
| Solid | 783 (74%) | 4,585 (79%) | 6,015 (80%) | 1,042 (86%) |
| Subsolid | 277 (26%) | 1,199 (21%) | 1,503 (20%) | 163 (14%) |
| Major axis length (mm) | 9.1 [5.2] | 11.0 [8.7] | 5.5 [4.5] | 12.5 [8.0] |
| Least axis length (mm) | 5.2 [2.7] | 5.8 [3.8] | 2.6 [2.4] | 6.6 [3.9] |
| Mesh Volume (mm3) | 446.5 [2,172.8] | 872.1 [5,202.0] | 168.4 [1,914.6] | 1,171.8 [6,489.3] |
| Sphericity | 0.76 [0.08] | 0.73 [0.10] | 0.79 [0.08] | 0.72 [0.09] |
Abbreviations: COPD, chronic obstructive pulmonary disease; IELCAP, International Early Lung Cancer Action Plan; mm, millimeter; NLST, National Lung Screening Trial; No., number; PanCan, PanCanadian Early Detection of Lung Cancer Study; PLuSS, Pittsburgh Lung Screening Study.
We excluded 1,284 nodules from our study due technical issues with feature extraction, 2,574 nodules not first-appearing on baseline scans, and another 2,103 nodules due to missing patient-level data for the harmonized set of epidemiologic covariates. In total, we had 16,797 baseline screen-detected nodules among 6,865 participants for our analytic sample. The median time-to-diagnosis for baseline-detected nodules was 134 days (IQR=59–452 days). A complete flow chart for nodule inclusion in the analytic sample is presented in Supplemental Figure 1. The distributions of patient-level and nodule-level traits in the training and testing data are shown in Supplemental Table 3 and Supplemental Table 4, respectively. Distributional measures for the radiomic features based on the original CT image are presented in Supplemental Table 5.
We started with 2,060 radiomics features for model development. We removed 78 features due to zero-variance and 11 features due to observed numerical instability (i.e., implausible values) for a large number of participants. Next, we fit univariate models for each feature in the training data, and retained features with a FDR-adjusted p-value less than 0.05 (n=248). Lastly, we evaluated all pairwise sets of predictors with correlation in the training set greater than 0.9 (in descending order) and removed the predictor with the larger p-value. We retained 642 radiomic features for model development. More details can be found in the Supplemental Materials and Supplemental Figure 2. The 642 radiomics features retained for model development are presented in Supplemental Table 6. We performed unsupervised clustering in the training data set using the 642 radiological features which revealed three distinct clusters of participants with similar radiomics profiles (see Supplemental Figure 3). We compared the three clusters based on their proportions of malignant pulmonary nodules and found statistically significant differences (PExact < 0.05).
We fit three different machine learning models (LASSO, XGBoost, Random Forest) using 5-fold cross-validation based on the 642 radiomics features and 9 epidemiologic covariates. We first fit a coarse grid of 50 sets of hyperparameters for each ML model. The results for this first-pass cross-validation are presented in Table 2 and Supplemental Figure 4 and 5. We selected the top performing model (LASSO) based on the combination of discrimination (AUC) and calibration (calibration ratio) and performed a final cross-validation and grid search over a finer grid of hyperparameters. The optimal penalty value for the LASSO, based on CV, was used to fit the final model based on the full training data set, and predictions were made on the held-out test set to evaluate model performance.
Table 2.
Area under the receiver operating characteristic curve (AUC) based on the K-fold cross-validation of three different machine learning classification models for nodule malignancy prediction based on epidemiologic and radiomic features. We present the cross-validated AUC and confidence intervals.
| ML Model | Optimal hyperparameters | CV-AUC (95% CI) |
|---|---|---|
| XGBoost | Num. of trees = 149 Tree depth = 11 Minimum node size = 15 Num. of predictors = 452 Learning rate = 0.0673 Loss reduction = 4.315 |
0.933 (0.923-0.944) |
| LASSO 1 | Penalty = 0.00044 | 0.930 (0.914-0.946) |
| Random Forest | Num. of trees = 147 Num. of predictors = 53 Minimum node size = 26 |
0.916 (0.904-0.929) |
Abbreviations: AUC, area under the curve; CI, confidence interval; LASSO, least absolute shrinkage and selection operator; ML, machine learning; Num, number; XGBoost, eXtreme Gradient Boosting.
The penalty parameter for the LASSO model was a L1 (i.e., LASSO) penalty.
The top ML submodels that yielded the highest cross-validated AUC were XGBoost (AUC=0.933, 95% CI: 0.923–0.944), LASSO (AUC=0.930, 95% CI: 0.914–0.946), and Random Forest (AUC=0.916, 95% CI: 0.904–0.929). However, calibration was superior for the LASSO model and was chosen as the top model (see Supplemental Figure 6). In total, 142 predictors were retained in the final LASSO model with non-zero coefficients (See Supplemental Figure 7). We assessed the relative stability of the features across CV folds for those retained in the final INTEGRAL-Radiomics model (Supplemental Figure 8). In general, these top features were selected often and consistently in different folds of CV. The top ML model based on epidemiologic variables only achieved a CV-AUC of 0.778 compared to 0.926 for radiomic features only.
We compared our model with the established PanCan Model and our radiomics model had better discrimination (P-value=0.0002), with a test-set AUC of 0.93 (95% CI: 0.90–0.96) compared to 0.87 (95% CI: 0.85–0.89) for the PanCan Model (see Figure 2). Our model performed well in both solid and subsolid (part-solid and non-solid) nodules with test-set AUC of 0.93 (95% CI: 0.89–0.97) and 0.91 (95% CI: 0.85–0.95), respectively. We present AUC according to other key factors (nodule size, sex, age, and smoking status) in Supplemental Table 7. Our model demonstrated excellent calibration when comparing observed risks with model-predicted (i.e., expected) risks, within bins of predicted risk. Our model had superior calibration compared to the PanCan Model (see Figure 3). We estimated the observed and expected number of malignant nodules (per 1,000 nodules) for the PanCan model and our INTEGRAL-Radiomics model. Our model had excellent calibration ratios (Exp / Obs) of 1.02 (95% CI: 0.89–1.18) and calibration differences (Exp - Obs) of 0.69 (95% CI: −4.0, 5.1), versus 1.25 (95% CI: 1.15–1.36) and 11.7 (95% CI: 7.7, 15.8) for the PanCan Model, respectively. We compare clinically-relevant metrics (e.g., sensitivity, specificity, etc.) between our model and the PanCan model in Table 3. At nearly every probability threshold, our model has higher sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy, while identifying fewer lesions as positive (i.e., suspicious), when compared to the PanCan model.
Figure 2.
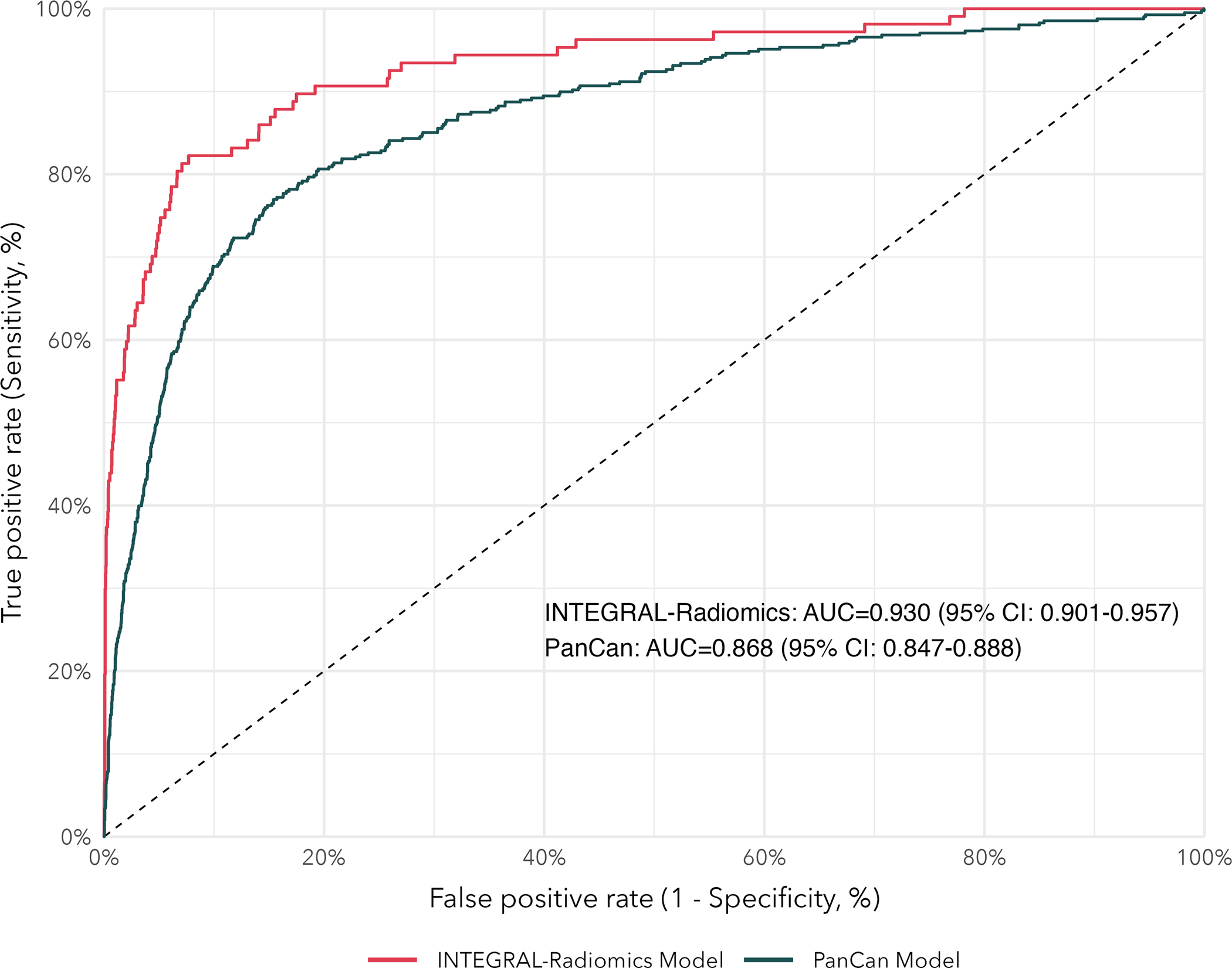
Receiver operating characteristic (ROC) curves for our INTEGRAL-Radiomics models and the established PanCan Model. Area under the curve (AUC) and 95% confidence intervals are reported.
Figure 3.
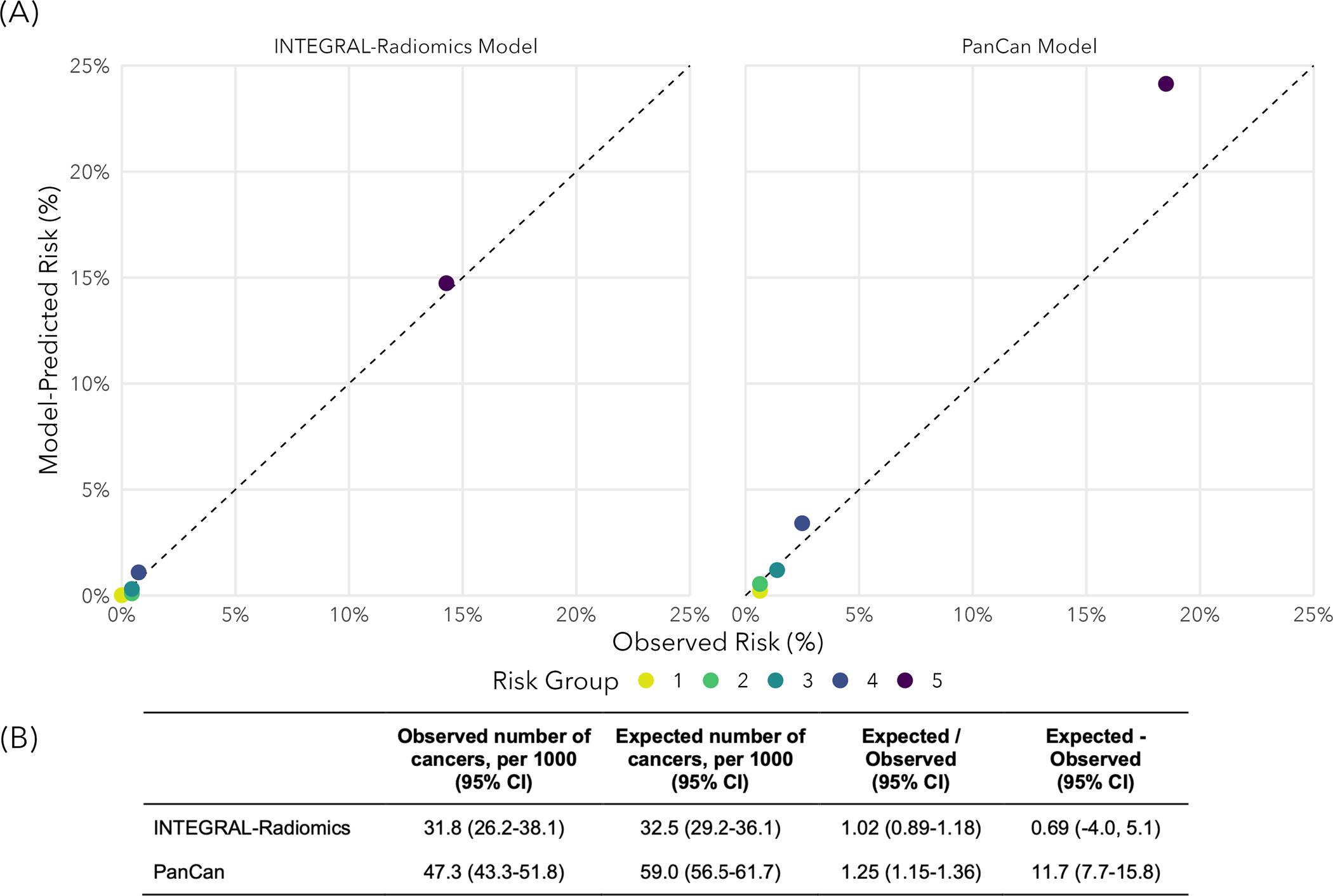
Calibration of our INTEGRAL-Radiomics model and the PanCan model (McWilliams et al., 2013) in hold-out test-data. (A) Model-predicted risks versus observed risks across quintiles of model-predicted risks. The diagonal dashed line indicates perfect calibration. (B) Observed and expected (model-predicted) number of malignant pulmonary nodules (per 1,000 nodules) including the calibration ratio (Exp / Obs) and difference (Exp - Obs). Calibration ratios less than 1 (or differences less than 0) indicate underestimation of risk and ratios greater than 1 (or differences greater than 0) indicate overestimation of risk.
Table 3.
Comparison of sensitivity, specificity, positive predictive value, negative predictive value, accuracy, and positive prevalence between our INTEGRAL-Radiomics model and the established PanCan Model. We provide point estimates and 95% percentile-based bootstrap confidence intervals for each statistic.
| Probability Threshold | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | Positive Prevalence (%) |
|---|---|---|---|---|---|---|
| INTEGRAL-Radiomics 1,2 | ||||||
| ≥2% | 89.7 (83.8-95.1) | 81.6 (80.3-82.9) | 13.8 (11.4-16.6) | 99.6 (99.4-99.8) | 81.9 (80.6-83.2) | 20.6 (19.4-22.0) |
| ≥5% | 82.2 (75.2-89.4) | 90.9 (89.9-91.8) | 23.0 (19.1-27.4) | 99.4 (99.1-99.6) | 90.7 (89.6-91.6) | 11.4 (10.4-12.5) |
| ≥10% | 74.8 (66.7-82.6) | 94.7 (94.0-95.5) | 31.9 (26.4-37.9) | 99.1 (98.8-99.5) | 94.1 (93.3-94.9) | 7.5 (6.6-8.4) |
| ≥15% | 64.5 (56.1-73.4) | 96.7 (96.1-97.3) | 39.2 (32.4-47.0) | 98.8 (98.4-99.2) | 95.7 (95.0-96.3) | 5.2 (4.5-6.0) |
| ≥20% | 61.7 (53.3-70.8) | 97.8 (97.2-98.3) | 47.5 (39.3-56.2) | 98.7 (98.4-99.1) | 96.6 (96.0-97.2) | 4.1 (3.4-4.8) |
| ≥25% | 55.1 (45.8-64.6) | 98.5 (98.0-98.9) | 54.6 (45.2-64.1) | 98.5 (98.1-98.9) | 97.1 (96.6-97.7) | 3.2 (2.6-3.8) |
| ≥30% | 55.1 (45.8-64.6) | 98.8 (98.5-99.2) | 60.8 (51.4-70.8) | 98.5 (98.1-98.9) | 97.4 (96.9-97.9) | 2.9 (2.3-3.5) |
| PanCan Model 3 | ||||||
| ≥2% | 87.7 (84.7-90.6) | 64.5 (63.5-65.5) | 10.9 (9.9-12.1) | 99.1 (98.8-99.3) | 65.6 (64.7-66.6) | 38.0 (37.0-39.0) |
| ≥5% | 80.6 (76.9-84.0) | 79.8 (78.9-80.7) | 16.6 (15.0-18.3) | 98.8 (98.5-99.0) | 79.9 (79.0-80.7) | 23.0 (22.2-23.9) |
| ≥10% | 72.3 (68.1-76.5) | 88.0 (87.3-88.7) | 23.0 (20.6-25.6) | 98.5 (98.2-98.7) | 87.3 (86.5-88.0) | 14.9 (14.1-15.6) |
| ≥15% | 65.0 (60.6-69.4) | 91.6 (91.0-92.2) | 27.7 (24.9-30.7) | 98.1 (97.8-98.4) | 90.3 (89.7-91.0) | 11.1 (10.4-11.7) |
| ≥20% | 58.3 (53.6-63.0) | 93.7 (93.2-94.2) | 31.5 (28.2-34.9) | 97.8 (97.5-98.2) | 92.0 (91.5-92.6) | 8.8 (8.1-9.3) |
| ≥25% | 51.7 (47.0-56.3) | 94.9 (94.5-95.4) | 33.7 (30.0-37.4) | 97.5 (97.2-97.9) | 92.9 (92.4-93.4) | 7.3 (6.7-7.8) |
| ≥30% | 43.9 (39.1-48.7) | 96.1 (95.6-96.5) | 35.6 (31.4-39.7) | 97.2 (96.8-97.5) | 93.6 (93.1-94.1) | 5.8 (5.3-6.3) |
Abbreviations: NPV, negative predictive value; PPV, positive predictive value.
Note: Sensitivity is the proportion of malignant nodules correctly identified as malignant. Specificity is the proportion of benign nodules correctly identified as benign. PPV is the proportion of positive predictions that are malignant nodules. NPV is the proportion of negative predictions that are benign nodules. Accuracy is the total number of correct predictions out of the total number of nodules. Positive prevalence is the proportion of positive predictions divided by the total number of predictions.
The radiomics model was evaluated in the 20% hold-out test data not used for model development (N = 3,363).
Our model was chosen based on K-fold cross-validations using the training sample, and performance metrics are reported based on the hold-out test sample. Our final model was a LASSO model with a penalty (lambda) value of 0.000442 and retained 142 predictors in the model.
The PanCan Model was evaluated in the entire eligible set of participants from IELCAP-Toronto, NLST, and PLuSS (N = 8,622).
Discussion
We developed and validated a pulmonary nodule malignancy assessment model based on radiomics and epidemiologic data from four large, international lung cancer screening cohorts using a machine learning approach. We found that the top-performing models were based on gradient boosted trees (XGBoost) and penalized logistic regression (LASSO), while the LASSO model provided the most optimal calibration. The use of quantitative imaging features (i.e., radiomics) showed improved performance compared to an established model based primarily on semantic nodule features. Radiomic features have demonstrated value for their ability to predict nodule malignancy risk and may improve the management of screen-detected pulmonary nodules by providing clinicians with supporting information for clinical decision-making.
Historically, the large quantity of medical images acquired during lung cancer screening have been under-utilized for extracting important information to inform nodule management. Traditionally, a modest set of semantic nodule traits are qualitatively assessed by expert radiologists to provide a high-level characterization of nodule morphology. High-throughput quantitative image analysis removes this layer of inter-reader subjectivity, while also collecting many more features that may further enhance our ability to characterize nodule morphology and intranodular textural heterogeneity [41]. Radiomic features can describe various aspects of the nodule morphology in ways that are imperceptible to the human eye (i.e., subtle intratumoral textural changes) [26]. The combination of radiomic features with known important patient-level features are expected to improve clinical management of nodules.
Previous studies demonstrated that quantitative image analysis can identify important prognostic signatures in head and neck cancer [41]. The feature extraction presented in the study by Aerts et al. [41] was formalized as a free and open-source software [26] and has enabled transparency and reproducibility for feature extraction, and contributed to the growing interest in quantitative image analysis in many areas of medical imaging, including lung cancer screening. To date, many of the radiomic studies for pulmonary nodule assessment have been performed based on relatively small data sets and with no ground-truth for nodule cancer status. Previous studies have shown that radiomic features may help identify lung cancer subtypes [42,43] and the presence of therapy-targetable somatic mutations (e.g., EGFR, KRAS) [44–48], though these findings require further validation in larger studies. The use of non-invasive image features is growing in popularity and will help improve lung cancer screening program efficiency. While there have been several studies that have used deep learning approaches for nodule malignancy risk assessment [49–52], including one that is commercially available for clinical use [49], our model shows comparable levels of performance while being simpler (i.e., fewer parameters) and with better interpretability.
To our knowledge, our radiomics study is the largest study to date to systematically investigate the importance of radiomics for pulmonary nodule assessment. We performed supervised, semi-automated segmentation of pulmonary nodules for three lung cancer screening studies using an open-source tool that is available for anyone to use. Our study was based on 16,797 nodules among 6,865 participants from four lung cancer screening cohorts. We used a systematic approach to develop a machine learning prediction model using radiomics features that were consistently predictive across each of these four independent screening cohorts. With increasing usage of computer-aided diagnostic (CAD) software, the segmentation process can be fully automated. The model presented here can be easily implemented without additional processing need for a large-amount of images with the added advantage of minimum inter-reader variability.
Our study has several limitations worth highlighting. First, ground-truth nodule-level malignancy status was unavailable for two of the screening studies (NLST, PLuSS). As such, we used a set of rules to assign nodule-level malignancy status for participants with a lung cancer diagnosis. Imperfect assignment will lead to missclassification errors that can bias the results of our study. However, we used a relatively conservative approach based on suspicion of malignancy determined by expert review of nodules by a radiologist who has extensive experience in lung CT assessment. For this reason, we believe the potential for missclassification bias is limited. There were feature extraction issues that excluded 5.7% of the candidate nodules. Nearly 80% of these issues were due to very small nodules with segmentation masks containing only a single voxel or were 1-dimensional after resampling and interpolation. These micronodules have a very low prior probability of being malignant and their exclusion are unlikely to bias our results. The training and testing data for this study were based on the random split of a combined data from four lung cancer screening studies. While these four studies are geographically distinct and represent different patient populations, our model may perform differently in patient populations not represented by these data and needs to be further validated on independent external data. Lastly, there was numerical instability for a small set of radiomic features when computing on derived images (i.e., after transformations). We minimized potential bias from these unstable features by excluding them for the filters where identifiable problems arose. All radiomic features appeared stable based on the original image.
In summary, developed a nodule assessment model based on quantitative imaging and patient-level features collected from four international lung cancer screening cohorts. We believe this study contributes important insights into the role that high-dimensional radiomic features can play in accurately assessing nodule malignancy risk and that these features generalize well to geo-temporally distinct screening cohorts. At present, there is emerging interest in analyzing medical images using deep learning computer vision approaches, although limited transparency in model development and lack of model interpretability can pose challenges for clinical implementation and widespread adoption [53,54]. In the future, our model may help to improve nodule malignancy assessment and provide supplemental information that can help guide decision-making for screen-detected nodule management.
Supplementary Material
Key Messages.
What is already known on this topic:
Indeterminate screen-detected pulmonary nodules are a challenge for lung cancer screening programs.
What this study adds:
In this study, we use data from four international lung cancer screening studies to develop machine learning models based on radiomic and epidemiologic features that accurately classify malignant nodules.
How this study might affect research, practice or policy:
Our radiomic model may be suitable to help assess screen-detected pulmonary nodules.
Funding
This work was supported by Canadian Institutes of Health Research (FDN 167273) and the National Institutes of Health (U19 CA203654).
Role of the funders
The funding sources had no role in the conception, design, implementation, analysis, or interpretation of the study, or writing of the report, or the decision to submit the report for publication.
Footnotes
Disclosures
The authors report no potential conflicts of interests.
Ethics Approval
Research ethics for this project is approved by the Mount Sinai Hospital (MSH) Research Ethics Board (REB) for the Integrative Analysis of Lung Cancer (INTEGRAL) project (MSH REB 17–0119-E).
Data Availability Statement
All data used in the present study may be made available upon reasonable request to the Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) program upon approval by the Data Access Committee. The model reported in the study and example code are publicly available on GitHub (https://github.com/mattwarkentin/INTEGRAL-Radiomics).
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2021; [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone A, Krapcho M, Miller D, Bishop K, Kosary C, Yu M, Ruhl J, Tatalovich Z, Mariotto A, others. SEER cancer statistics review, 1975–2014, national cancer institute. Bethesda, MD. 2017;1–12. [Google Scholar]
- 3.Koning HJ de, Aalst CM van der, Jong PA de, Scholten ET, Nackaerts, Heuvelmans MA, Lammers J-WJ, Weenink C, Yousaf-Khan U, Horeweg N, others. Reduced lung-cancer mortality with volume CT screening in a randomized trial. New England Journal of Medicine. 2020;382(6):503–13. [DOI] [PubMed] [Google Scholar]
- 4.Team NLSTR. Reduced lung-cancer mortality with low-dose computed tomographic screening. New England Journal of Medicine. 2011;365(5):395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Lung Screening Trial Research Team. Lung cancer incidence and mortality with extended follow-up in the national lung screening trial. Journal of Thoracic Oncology. 2019;14(10):1732–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastorino U, Silva M, Sestini S, Sabia F, Boeri M, Cantarutti A, Sverzellati N, Sozzi G, Corrao G, Marchianò A. Prolonged lung cancer screening reduced 10-year mortality in the MILD trial: New confirmation of lung cancer screening efficacy. Annals of Oncology. 2019;30(7):1162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bach PB, Mirkin JN, Oliver TK, Azzoli CG, Berry DA, Brawley OW, Byers T, Colditz GA, Gould MK, Jett JR, others. Benefits and harms of CT screening for lung cancer: A systematic review. Jama. 2012;307(22):2418–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.American College of Radiology Committee on Lung-RADS. Lung-RADS assessment categories version1.1. Available at https://www.acr.org/-/media/ACR/Files/RADS/Lung-RADS/LungRADSAssessmentCategoriesv1-1.pdf;
- 9.I-ELCAP protocol. Available at https://www.ielcap.org/sites/default/files/I-ELCAP-protocol-summary.pdf;
- 10.Xu DM, Gietema H, Koning H de, Vernhout R, Nackaerts K, Prokop M, Weenink C, Lammers J-W, Groen H, Oudkerk M, others. Nodule management protocol of the NELSON randomised lung cancer screening trial. Lung cancer. 2006;54(2):177–84. [DOI] [PubMed] [Google Scholar]
- 11.Horeweg N, Rosmalen J van, Heuvelmans MA, Aalst CM van der, Vliegenthart R, Scholten ET, Haaf K ten, Nackaerts K, Lammers J-WJ, Weenink C, others. Lung cancer probability in patients with CT-detected pulmonary nodules: A prespecified analysis of data from the NELSON trial of low-dose CT screening. The Lancet Oncology. 2014;15(12):1332–41. [DOI] [PubMed] [Google Scholar]
- 12.Oudkerk M, Devaraj A, Vliegenthart R, Henzler T, Prosch H, Heussel CP, Bastarrika G, Sverzellati N, Mascalchi M, Delorme S, others. European position statement on lung cancer screening. The Lancet Oncology. 2017;18(12):e754–66. [DOI] [PubMed] [Google Scholar]
- 13.Callister M, Baldwin D, Akram A, Barnard S, Cane P, Draffan J, Franks K, Gleeson F, Graham R, Malhotra P, others. British thoracic society guidelines for the investigation and management of pulmonary nodules: Accredited by NICE. Thorax. 2015;70(Suppl 2):ii1–54. [DOI] [PubMed] [Google Scholar]
- 14.Baldwin DR, Callister ME. The british thoracic society guidelines on the investigation and management of pulmonary nodules. Thorax. 2015;70(8):794–8. [DOI] [PubMed] [Google Scholar]
- 15.Yip R, Henschke CI, Yankelevitz DF, Smith JP. CT screening for lung cancer: Alternative definitions of positive test result based on the national lung screening trial and international early lung cancer action program databases. Radiology. 2014;273(2):591–6. [DOI] [PubMed] [Google Scholar]
- 16.NCCN practice guidelines in oncology lung cancer screening guideline version 4.2019. https://www.nccn.org/professionals/physician_gls/default.aspx;
- 17.Zhou Q, Fan Y, Wang Y, others. Guidelines for low-dose spiral CT screening of lung cancer in china (2018 edition). Zhongguo Fei Ai Za Zhi. 2018;21(2):67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bueno J, Landeras L, Chung JH. Updated fleischner society guidelines for managing incidental pulmonary nodules: Common questions and challenging scenarios. Radiographics. 2018;38(5):1337–50. [DOI] [PubMed] [Google Scholar]
- 19.MacMahon H, Naidich DP, Goo JM, Lee KS, Leung AN, Mayo JR, Mehta AC, Ohno Y, Powell CA, Prokop M, others. Guidelines for management of incidental pulmonary nodules detected on CT images: From the fleischner society 2017. Radiology. 2017;284(1):228–43. [DOI] [PubMed] [Google Scholar]
- 20.Tammemagi MC, Lam S. Screening for lung cancer using low dose computed tomography. Bmj. 2014;348. [DOI] [PubMed] [Google Scholar]
- 21.Lim KP, Marshall H, Tammemägi M, Brims F, McWilliams A, Stone E, Manser R, Canfell K, Weber M, Connelly L, others. Protocol and rationale for the international lung screening trial. Annals of the American Thoracic Society. 2020;17(4):503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kakinuma R, Ashizawa K, Kusunoki Y, Kobayashi T, Kondo T, Nakagawa T, Hatakeyama M, Y M. The pulmonary nodules management committee of the japanese society of CT screening. Guidelines for the management of pulmonary nodules detected by low-dose CT lung cancer screening version 3. [Google Scholar]
- 23.Toumazis I, Bastani M, Han SS, Plevritis SK. Risk-based lung cancer screening: A systematic review. Lung Cancer. 2020;147:154–86. [DOI] [PubMed] [Google Scholar]
- 24.Fox AH, Tanner NT. Approaches to lung nodule risk assessment: Clinician intuition versus prediction models. Journal of Thoracic Disease. 2020;12(6):3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loverdos K, Fotiadis A, Kontogianni C, Iliopoulou M, Gaga M. Lung nodules: A comprehensive review on current approach and management. Annals of Thoracic Medicine. 2019;14(4):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RG, Fillion-Robin J-C, Pieper S, Aerts HJ. Computational radiomics system to decode the radiographic phenotype. Cancer research. 2017;77(21):e104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJ. Artificial intelligence in radiology. Nature Reviews Cancer. 2018;18(8):500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kann BH, Hosny A, Aerts HJ. Artificial intelligence for clinical oncology. Cancer Cell. 2021;39(7):916–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tammemagi MC, Schmidt H, Martel S, McWilliams A, Goffin JR, Johnston MR, Nicholas G, Tremblay A, Bhatia R, Liu G, others. Participant selection for lung cancer screening by risk modelling (the pan-canadian early detection of lung cancer [PanCan] study): A single-arm, prospective study. The lancet oncology. 2017;18(11):1523–31. [DOI] [PubMed] [Google Scholar]
- 30.Roberts HC, Patsios D, Paul NS, McGregor M, Weisbrod G, Chung T, Herman S, Boerner S, Waddell T, Keshavjee S, others. Lung cancer screening with low-dose computed tomography: Canadian experience. Canadian Association of Radiologists Journal. 2007;58(4):225. [PubMed] [Google Scholar]
- 31.Menezes RJ, Roberts HC, Paul NS, McGregor M, Chung TB, Patsios D, Weisbrod G, Herman S, Pereira A, McGregor A, others. Lung cancer screening using low-dose computed tomography in at-risk individuals: The toronto experience. Lung Cancer. 2010;67(2):177–83. [DOI] [PubMed] [Google Scholar]
- 32.Wilson DO, Weissfeld JL, Fuhrman CR, Fisher SN, Balogh P, Landreneau RJ, Luketich JD, Siegfried JM. The pittsburgh lung screening study (PLuSS) outcomes within 3 years of a first computed tomography scan. American journal of respiratory and critical care medicine. 2008;178(9):956–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tammemägi MC, Katki HA, Hocking WG, Church TR, Caporaso N, Kvale PA, Chaturvedi AK, Silvestri GA, Riley TL, Commins J, others. Selection criteria for lung-cancer screening. New England Journal of Medicine. 2013;368(8):728–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, others. 3D slicer as an image computing platform for the quantitative imaging network. Magnetic resonance imaging. 2012;30(9):1323–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.San Jose Estepar R, Ross JC, Harmouche R, Onieva J, Diaz AA, Washko GR. Chest imaging platform: An open-source library and workstation for quantitative chest imaging. In: C66 Lung imaging II: New probes and emerging technologies. American Thoracic Society; 2015. p. A4975–5. [Google Scholar]
- 36.Krishnan K, Ibanez L, Turner WD, Jomier J, Avila RS. An open-source toolkit for the volumetric measurement of CT lung lesions. Optics Express. 2010;18(14):15256–66. [DOI] [PubMed] [Google Scholar]
- 37.McKay MD, Beckman RJ, Conover WJ. A comparison of three methods for selecting values of input variables in the analysis of output from a computer code. Technometrics [Internet]. 1979. [cited 2022 Apr 6];21(2):239–45. Available from: http://www.jstor.org/stable/1268522 [Google Scholar]
- 38.Van Rossum G, Drake FL. Python 3 reference manual. Scotts Valley, CA: CreateSpace; 2009. [Google Scholar]
- 39.R Core Team. R: A language and environment for statistical computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2021. Available from: https://www.R-project.org/ [Google Scholar]
- 40.McWilliams A, Tammemagi MC, Mayo JR, Roberts H, Liu G, Soghrati K, Yasufuku K, Martel S, Laberge F, Gingras M, others. Probability of cancer in pulmonary nodules detected on first screening CT. New England Journal of Medicine. 2013;369(10):910–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, others. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature communications. 2014;5(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Gao L, Ma H, Arefan D, He J, Wang J, Liu H. Radiomics-based features for prediction of histological subtypes in central lung cancer. Frontiers in Oncology. 2021;11:1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linning E, Lu L, Li L, Yang H, Schwartz LH, Zhao B. Radiomics for classifying histological subtypes of lung cancer based on multiphasic contrast-enhanced computed tomography. Journal of computer assisted tomography. 2019;43(2):300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu S, Shen G, Mao J, Gao B. CT radiomics in predicting EGFR mutation in non-small cell lung cancer: A single institutional study. Frontiers in Oncology. 2020;2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong D, Xu K, Zhang L, Wan X, Guo Y. Radiomics signature as a predictive factor for EGFR mutations in advanced lung adenocarcinoma. Frontiers in oncology. 2020;10:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia T-Y, Xiong J-F, Li X-Y, Yu W, Xu Z-Y, Cai X-W, Ma J-C, Ren Y-C, Larsson R, Zhang J, others. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. European radiology. 2019;29(9):4742–50. [DOI] [PubMed] [Google Scholar]
- 47.Velazquez ER, Parmar C, Liu Y, Coroller TP, Cruz G, Stringfield O, Ye Z, Makrigiorgos M, Fennessy F, Mak RH, others. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer research. 2017;77(14):3922–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Kim J, Balagurunathan Y, Li Q, Garcia AL, Stringfield O, Ye Z, Gillies RJ. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clinical lung cancer. 2016;17(5):441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baldwin DR, Gustafson J, Pickup L, Arteta C, Novotny P, Declerck J, Kadir T, Figueiras C, Sterba A, Exell A, others. External validation of a convolutional neural network artificial intelligence tool to predict malignancy in pulmonary nodules. Thorax. 2020;75(4):306–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venkadesh KV, Setio AA, Schreuder A, Scholten ET, Chung K, W. Wille MM, Saghir Z, Ginneken B van, Prokop M, Jacobs C. Deep learning for malignancy risk estimation of pulmonary nodules detected at low-dose screening CT. Radiology. 2021;300(2):438–47. [DOI] [PubMed] [Google Scholar]
- 51.Kim RY, Oke JL, Pickup LC, Munden RF, Dotson TL, Bellinger CR, Cohen A, Simoff MJ, Massion PP, Filippini C, others. Artificial intelligence tool for assessment of indeterminate pulmonary nodules detected with CT. Radiology. 2022;304(3):683–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, others. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nature medicine. 2019;25(6):954–61. [DOI] [PubMed] [Google Scholar]
- 53.Lam S, Bryant H, Donahoe L, Domingo A, Earle C, Finley C, Gonzalez AV, Hergott C, Hung RJ, Ireland AM, others. Management of screen-detected lung nodules: A canadian partnership against cancer guidance document. Canadian Journal of Respiratory, Critical Care, and Sleep Medicine. 2020;4(4):236–65. [Google Scholar]
- 54.Massion PP, Antic S, Ather S, Arteta C, Brabec J, Chen H, Declerck J, Dufek D, Hickes W, Kadir T, others. Assessing the accuracy of a deep learning method to risk stratify indeterminate pulmonary nodules. American journal of respiratory and critical care medicine. 2020;202(2):241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in the present study may be made available upon reasonable request to the Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) program upon approval by the Data Access Committee. The model reported in the study and example code are publicly available on GitHub (https://github.com/mattwarkentin/INTEGRAL-Radiomics).


