Abstract
The global loss of heterochromatin during aging has been observed in eukaryotes from yeast to humans, and this has been proposed to be one of the causes of aging. However, the cause of this age-associated loss of heterochromatin has remained enigmatic. Here we show that heterochromatin markers, including histone H3K9 di-/tri-methylation and HP1, decrease with age in murine muscle stem cells (MuSCs) as a consequence of the depletion of the methyl donor SAM. We find that restoration of intracellular SAM in aged MuSCs restores heterochromatin content to youthful levels and rejuvenates age-associated features including DNA damage accumulation, increased cell death, and defective muscle regeneration. SAM is not only a methyl group donor for transmethylation but it is also an aminopropyl donor for polyamine synthesis. Excessive consumption of SAM in polyamine synthesis may reduce its availability for transmethylation. Consistent with this premise, we observe that perturbation of increased polyamine synthesis by inhibiting spermidine synthase (Srm) restores the intracellular SAM content as well as heterochromatin formation, leading to improvements in aged MuSC function and regenerative capacity in male and female mice. Together, our studies demonstrate a direct causal link between polyamine metabolism and epigenetic dysregulation during MuSC aging.
INTRODUCTION
Alterations of chromatin architecture have been associated with cellular dysfunction during aging 1. One of the earliest models proposing the involvement of epigenetics in aging is the ‘heterochromatin loss model of aging’2. This model suggests that heterochromatin domains, established early in embryogenesis, decline during aging, causing the lack of proper chromatin packing and global changes in nuclear architecture. The lack of packed chromatin is predictive of genomic instability, including higher levels of DNA damage and the accumulation of aberrant transcripts3. Increased expression of transposable elements (TEs) due to the heterochromatin loss also leads to disruption of cellular homeostasis and DNA damage during aging4. The loss of heterochromatin during aging has been observed in eukaryotes from yeast to mammals5–7. Compared to young cells, aged cells show reduced levels of heterochromatin markers such as di- and tri-methylation of histone H3K9 (H3K9me2, H3K9me3) and HP1 proteins (HP1α/β/γ/)8, reflecting a more relaxed chromatin structure. Furthermore, studies have shown that the decline of heterochromatin markers is correlated with age-related impairment, while restoring heterochromatin levels extends the lifespan of yeast and flies5,6,9.
Previous work from our group and others have revealed a critical link between heterochromatin formation and stem cell function10–12. For example, a recent study showed that SIRT7 maintains the repressive state of heterochromatin by forming a complex with nuclear lamina proteins and heterochromatin proteins in human mesenchymal stem cells10. Accordingly, deficiency of SIRT7 results in loss of heterochromatin and accelerates cellular senescence10. A study from our group revealed that the Jumonji family member Hairless (Hr) regulates heterochromatin maintenance via its role as a H3K9 demethylase antagonist in quiescent MuSCs11. Hr-deficient MuSCs exhibit a loss in H3K9me3-marked heterochromatin and harbor an increased susceptibility to genotoxic stress and an impaired ability to contribute to muscle regeneration.
Recent work has highlighted an important link between metabolism and chromatin remodeling. Several metabolites, including S-adenosylmethionine (SAM), α-ketoglutarate (αKG), and acetyl-CoA not only play important roles in central carbon metabolism, but have also been shown to directly impact the modification of chromatin13,14. SAM is a major methyl donor for histone/DNA methylation, αKG is a cofactor for histone/DNA demethylases, and acetyl CoA is a major acyl donor for histone acetylation15–17. Recent evidence has suggested that alterations in the metabolic pathways that consume or produce these metabolites can change the epigenetic state and function of stem cells18–20. For instance, glutamine deprivation, and the subsequent depletion of αKG, can lead to an increase in tri-methylation of H3K9, H3K27, H3K26, and H4K20 in mouse embryonic stem cells (ESCs), which can be reversed through supplementation with cell permeable αKG18. In addition, in human ESCs, methionine deprivation, and the subsequent decline in cellular SAM levels, has been shown to result in a significant decrease in tri-methylation of H3K4 and a reduction in global DNA methylation19. These data highlight a direct link between changes in metabolism and the subsequent alterations in chromatin structure, which can lie upstream of cell fate and tissue function.
Aging is accompanied by a decline in the ability of tissues to regenerate and repair upon external stress, due largely to an age-related decline in the numbers and function of adult stem cells21–23. MuSCs are indispensable for muscle repair, and MuSC dysfunction with age results in impaired regeneration of aged muscle24–26. Our group and others have reported changes in epigenetic modifications during MuSC aging11,27,28. In particular, our group has previously reported an increase in H3K27me3 and a decrease in H3K4me3 in aged MuSCs28. However, the potential role of altered metabolism in driving these changes, remain largely unknown.
In this study, we show that heterochromatin declines during MuSC aging and that the depletion of the methyl donor SAM is associated with this decline. Restoring SAM levels increases the level of heterochromatin markers and alleviates age-associated features including DNA damage accumulation, increased cell death upon activation, and defective muscle regeneration. SAM is also an essential aminopropyl donor in the formation of the polyamines, spermidine and spermine. Our findings show that an age-related increase in spermidine synthesis and its utilization of SAM reduces availability of SAM for histone transmethylation. Restoration of SAM levels by the inhibition of spermidine synthesis improves aged MuSC function and regenerative capacity. Altogether, this report highlights a connection between altered polyamine metabolism and heterochromatin formation during aging via alterations in cellular SAM levels.
RESULTS
Heterochromatin loss occurs in MuSCs during aging
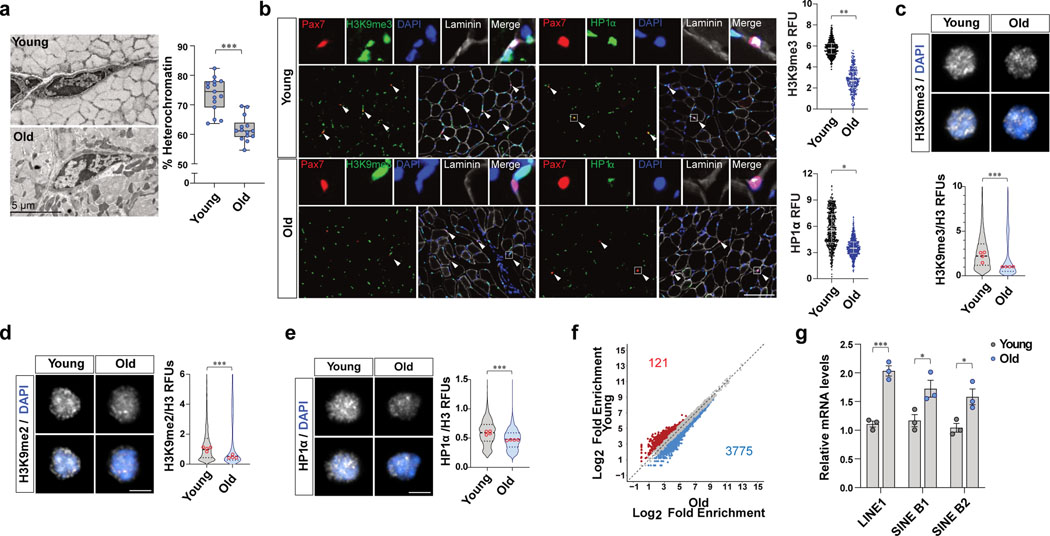
In order to directly assess whether heterochromatin loss occurs in MuSCs during aging, we used electron microscopy (EM) to quantify heterochromatin in MuSCs., as previously described11. We found a significant decrease in heterochromatin in MuSCs from old mice compared to young mice (Fig. 1a). In order to check the known heterochromatin markers including H3K9me3, H3K9me2, and HP1α, we quantified the markers in quiescent MuSCs in vivo by immunohistochemical staining of muscle cryosections as well as in MuSCs ex vivo which were isolated from young and old mice by fluorescence-activated cell sorting (FACS) (Fig. 1b–e and Extended Data Fig. 1a–d). These analyses revealed that old MuSCs have significantly lower levels of heterochromatin markers compared to young MuSCs (Fig. 1b–e and Extended Data Fig. 1b–d).
Figure 1. The loss of heterochromatin occurs in MuSC with age.
a, (Left) Representative transmission electron microscopy (TEM) images of MuSCs from young and old mice. MuSCs on EDL sections were imaged at 1500–3000x magnification. (Right) Quantification of heterochromatin content. The percentage of heterochromatin cross-sectional area over total cross-sectional area of nucleus is plotted1a (n=14 cells examined over 3 independent animals per group). The box represents the interquartile range (IQR), with the lower and upper hinges indicating the 25th and 75th percentiles, respectively. The horizontal line inside the box marks the median score. The whiskers extend to the minimum and maximum values. b, (Left) Representative tissue immunostaining images of young and old Pax7CreER;R26RRFP mice. (Right) Quantification of relative fluorescence units (RFUs) of H3K9me3 and HP1α in RFP-positive MuSCs. (Scale bar, 100 μm, n=4) c-e, Representative confocal immunofluorescence images of MuSCs of young and old mice. Freshly isolated MuSCs were plated and stained with antibodies specific to H3K9me2, H3K9me3, and HP1α (Scale bar, 5 μm). RFUs of H3K9me3 (c), H3K9me2 (d), or HP1α (e) were normalized to fluorescence intensities of total H3 (n=4). f, Differential chromatin accessibility between young and old MuSCs based on the ATAC-seq peaks with different number of reads that exhibit an over 50%-fold change in signals and the p-value less than 0.05. g, Relative mRNA expression of transposable elements (TEs). MuSCs isolated from young and old mice were subjected to qRT-PCR, and mRNA level of each TE was normalized to Gapdh (n=3). Data are shown as mean ± SD (g) and as median and quartiles (b-e). P values were calculated by two-sided unpaired Student’s t-tests (a-e, g). *P < 0.05; **P < 0.01; ***P < 0.001. Statistical details are provided in Source Data.
Since the loss of heterochromatin is associated with an enhanced chromatin accessibility29, we conducted Assay for Transposase Accessible Chromatin using sequencing (ATAC-seq) on MuSCs. The Pearson correlation assay revealed a clear difference between young and old MuSCs (Extended Data Fig. 1e). Comparison of the ATAC-seq peaks around the transcriptional start site (TSSs) across the genome showed more open peaks in old MuSCs (Extended Data Fig. 1f, g). From differential peak analysis of ATAC-seq peaks, we found more than 3,000 peaks showing increased accessibility in old MuSCs (Fig. 1f and Extended Data Fig. 1h). We then measured mRNA expression of TEs, including LINE, SINE B1, and SINE B2. There was a significant increase in the expression of TEs in old MuSCs (Fig. 1g). Taken together, these results are consistent with the premise that the loss of heterochromatin is accompanied by an increase in chromatin openness during aging of MuSCs.
Intracellular SAM is depleted in old MuSCs
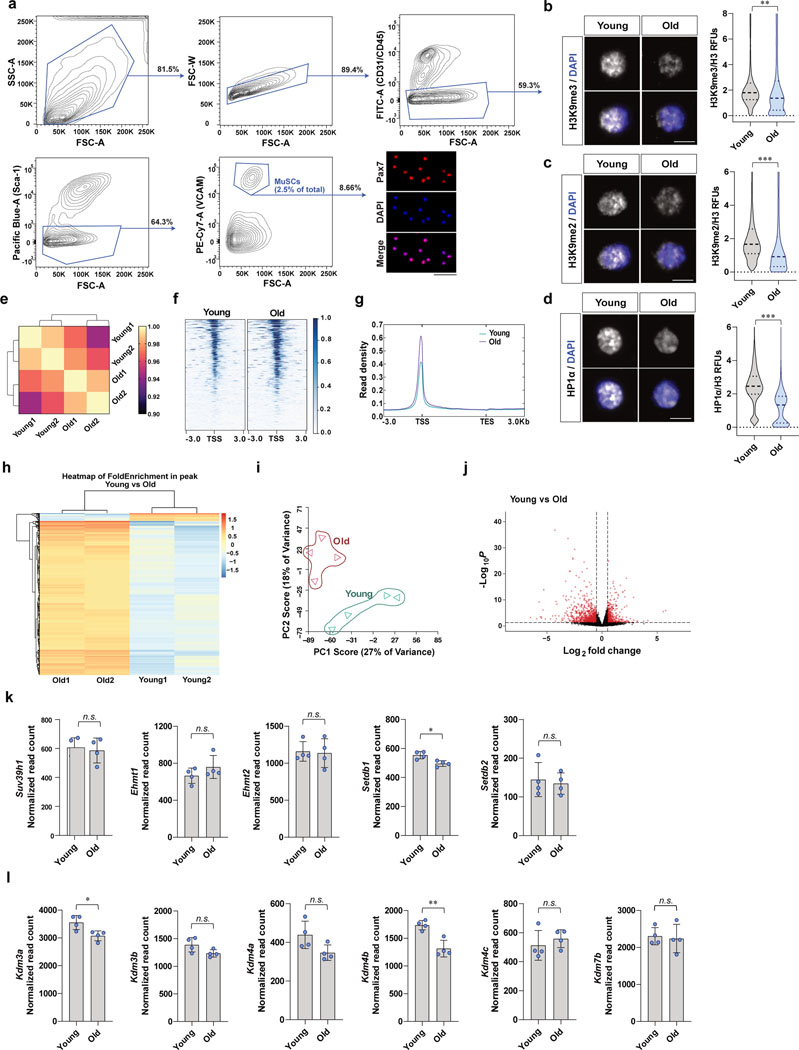
Next, we examined the cause of the decreased H3K9 methylation with age. We reasoned that the decreases in di- and tri-methylation of H3K9 might be due to the decreased expression of histone methyltransferases or the increased expression of histone demethylases. From our RNA sequencing data conducted with young and old MuSCs (Extended Data Fig. 1i, j), no significant change was observed in the expression levels of genes encoding H3K9 methyltransferases, including Suv39h1, Ehmt1/2, and Setdb1/2, or of genes encoding H3K9 demethylases, including Kdm3a/b, Kdm4a/b/c, and Kdm7b (Extended Data Fig. 1k,l).
We then asked whether the decrease in H3K9 methylation may be the result of a decreased abundance of the necessary substrate, S-adenosylmethionine (SAM), for these methylation reactions. Indeed, it has been recently shown that histone methylation can be affected by alterations in the cellular concentration of SAM19, which serves as the methyl donor for methyltransferases to catalyze the methylation reaction. Interestingly, based on an analysis of our untargeted metabolomic data recently published30, intracellular SAM levels are markedly lower in old MuSCs compared to young MuSCs (Fig. 2a). To confirm this, we quantified SAM levels in MuSCs using an ELISA assay. Consistent with our metabolomics data, the levels of intracellular SAM significantly decreased with age (Fig. 2b). In addition, we discovered that intracellular S-adenosylhomocysteine (SAH), which is formed upon SAM-dependent methylation, also declines with age (Fig. 2a).
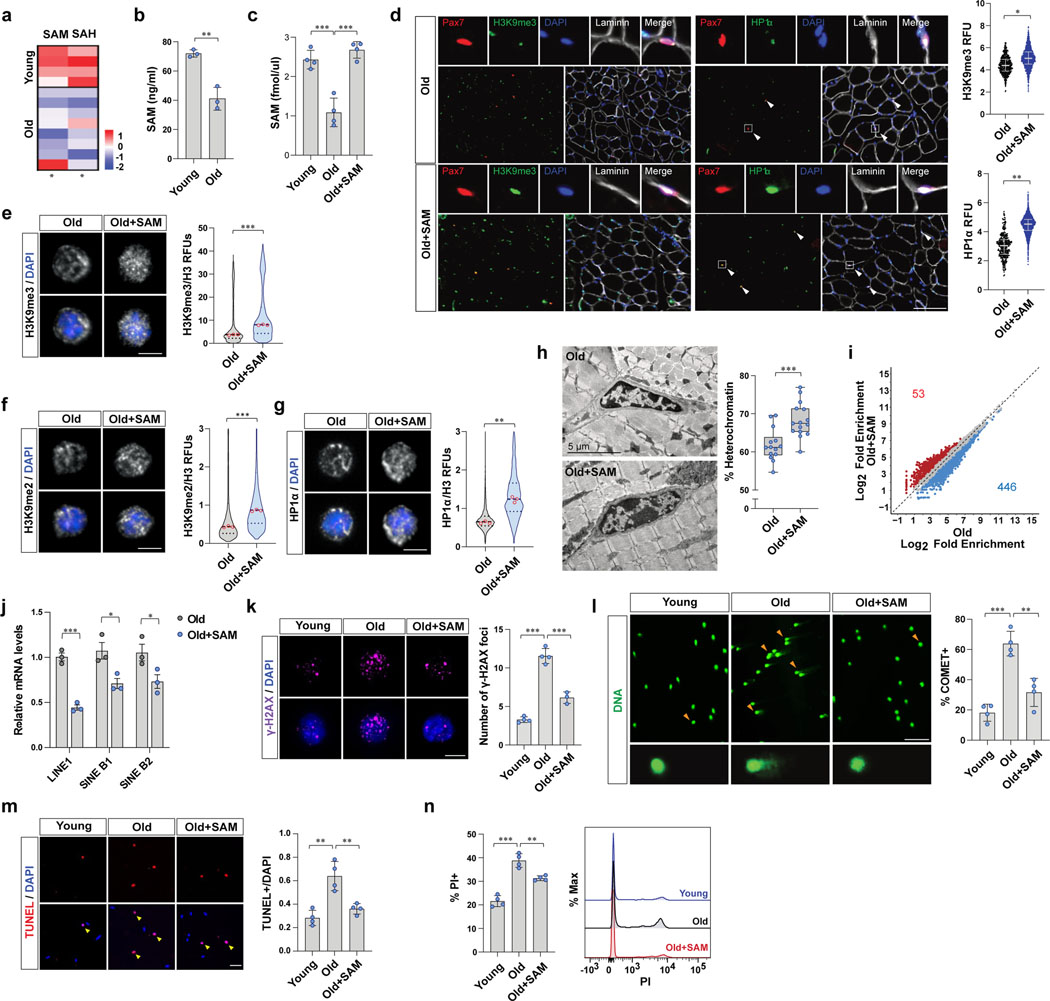
Figure 2. Restoration of SAM promotes heterochromatin formation and rescues dysfunctions of old MuSCs.
a, A heat map of relative levels of SAM and SAH detected by untargeted metabolomics based on LC-MS. b, SAM content of MuSCs measured by SAM ELISA (n=3). c, Quantitative LC-MS of SAM (n=4). d, (Left) Representative tissue immunostaining images of Pax7CreER;R26RRFP mice. (Right) RFUs of H3K9me3 and HP1α in RFP-positive MuSCs (Scale bar, 100 μm, n=4). e-g, (Left) Representative confocal immunofluorescence images (Scale bar, 1 μm). (Right) Violin plots showing RFUs of H3K9me3 (e), H3K9me2 (f), and HP1α (g), which were normalized to RFUs of total H3 (n=3). h, (Left) Representative TEM images (Scale bar, 5 μm). (Right) Quantification of heterochromatin cross-sectional area over total cross-sectional area of nucleus (n=14 cells examined over 3 independent animals per group). The box represents the IQR, with the lower and upper hinges indicating the 25th and 75th percentiles, respectively. The horizontal line inside the box marks the median. The whiskers extend to the minimum and maximum values. i, Differential ATAC-seq peaks were determined with different number of reads that exhibit an over 50%-fold change in signals (p-value< 0.05). j, The mRNA level of each TE was normalized to Gapdh (n=3). k, (Left) Representative confocal images (Scale bar, 5 μm). (Right) The number of γ-H2AX foci in a cell a day after irradiation (n=4 for young and old mice, n=3 for SAM-treated old mice). l, (Left) Representative images of cells after alkaline comet assay (Scale bar, 50 μm). (Right) The percentage of cells with DNA damage was quantified (n=4). m, A day after 7 Gy γ-irradiation, isolated MuSCs were cultured for 2 days. (Left) Representative images of TUNEL assay (Scale bar, 50 μm). (Right) The ratio of TUNEL positive MuSCs over total MuSCs (n=4). n, A day after 7 Gy γ-irradiation, isolated MuSCs were cultured for 2 days. (Left) The percentage of PI positive cells (n=4). (Right) Representative FACS plot. Data are shown as mean ± SD (b-c, j-n) and as median and quartiles (d-g). P values were calculated by two-sided unpaired Student’s t-tests (b-h, j-n). *P < 0.05; **P < 0.01; ***P < 0.001.
Restoration of SAM induces heterochromatin formation
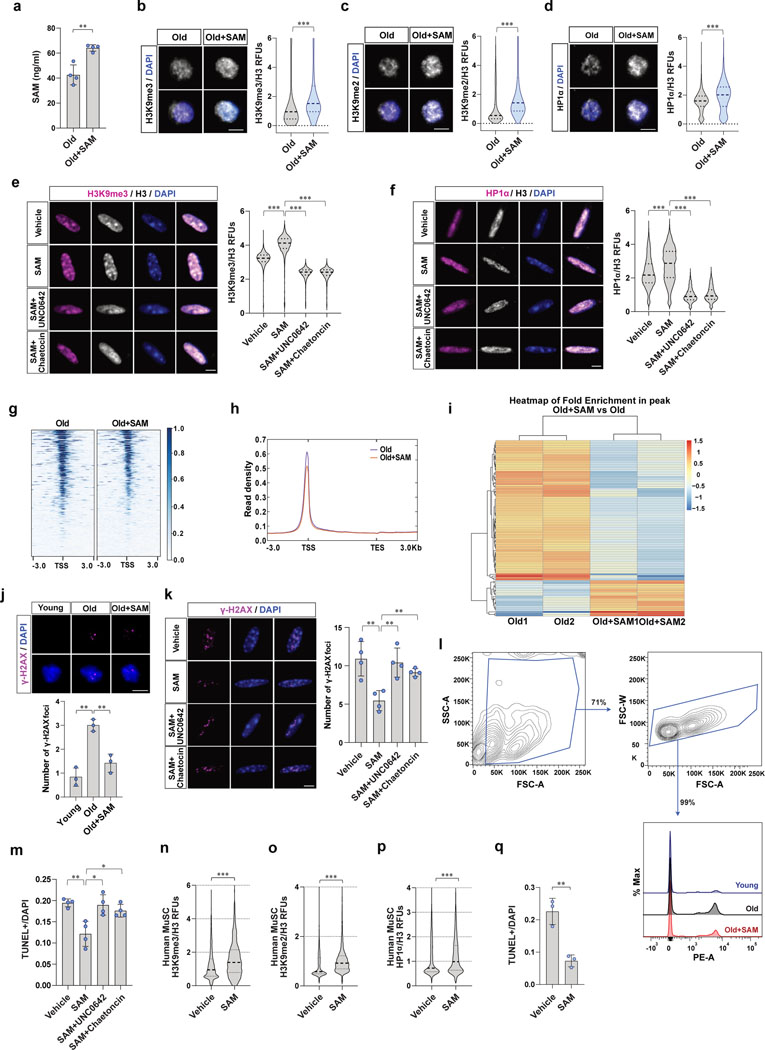
We next asked if restoring SAM back to young levels could rescue the reduction of H3K9 methylation in MuSCs with age. To test this, we injected SAM or vehicle intraperitoneally to aged mice and measured intracellular SAM levels of MuSCs. We found that the SAM levels in aged MuSCs were significantly increased upon in vivo SAM treatment (Fig. 2c and Extended Data Fig. 2a). We next tested whether restoration of SAM could promote heterochromatin formation in aged MuSCs. Indeed, we observed a significant upregulation of heterochromatin markers in MuSCs upon SAM treatment both in vivo and ex vivo (Fig. 2d–g and Extended Data Fig. 2b–d). In order to test if the changes in heterochromatin markers are dependent on the activity of histone H3K9-methyltransferases, we treated old MuSCs with SAM in the presence or absence of UNC0642 or Chaetocin. UNC0642 is a specific inhibitor of EHMT1/2, which are the enzymes that catalyze mono and di-methylation of H3K931. Chaetocin is an inhibitor of Suv39h1 and EHMT2, which are enzymes that catalyze tri-methylation of H3K932. We observed a significant decrease of heterochromatin markers upon UNC0642 or Chaetocin treatment of old MuSCs (Extended Data Fig. S2e, f). We also confirmed that aged MuSCs treated with SAM have expanded heterochromatin compared to MuSCs treated with vehicle (Fig. 2h). SAM treatment reduced ATAC-seq peak density around the TSSs and the number of accessible peaks compared to control old MuSCs (Fig. 2i and Extended Data Fig. 2g–i). In addition, the expression of TEs exhibited a significant decrease in SAM-treated old MuSCs compared to control old MuSCs (Fig. 2j).
Restoration of SAM reduces DNA damage and cell death
Previous work has shown that the loss of heterochromatin renders the genome more susceptible to DNA damage3. In order to test whether aged MuSCs have accumulated DNA damage and whether this can be rescued by restoration of SAM, we quantified the number of γ-H2AX foci in MuSCs. Young MuSCs harbor only a few γ-H2AX foci but the number of foci increases significantly with age, as we have previously shown25. Aged MuSCs isolated from SAM-treated mice contained significantly fewer γ-H2AX foci compared to cells from control mice (Extended Data Fig. 2j). We then tested whether aged MuSCs are more susceptible to genotoxic stress and whether SAM restoration can rescue this. In response to X-irradiation, aged MuSCs exhibited a significantly greater number of foci, an effect that was partially rescued by SAM treatment (Fig. 2k). Using Comet assays, we found that irradiation-induced DNA damage increased significantly with age, which was also partially rescued upon SAM treatment (Fig. 2l). The cell protective effect of SAM treatment was significantly ablated by inhibiting the activity of H3K9-methyltransferases (Extended Data Fig. 2k), suggesting the effect is due to H3K9 methylation and heterochromatin formation.
Since we have recently shown that aged MuSCs with a higher susceptibility to genotoxic stress have impaired ability to survive upon activation from quiescence11,25, we wanted to know if the restoration of SAM could rescue the impaired cell survival of old MuSCs following irradiation. We quantified the activation-induced cell death of MuSCs. The isolated MuSCs were activated ex vivo, and apoptotic cells were quantified. This assay revealed more than a two-fold increase in the number of aged MuSCs that were dying compared to young MuSCs. Furthermore, we found that SAM treatment could indeed rescue the survival impairments of aged MuSCs (Fig. 2m, n and Extended Data Fig. 2l). In addition, the beneficial effect of old MuSC survival was significantly ablated by inhibiting activity of H3K9-methyltransferases (Extended Data Fig. 2m). Similarly, directly treating isolated human MuSCs with SAM also increased heterochromatin markers and reduced cell death following genotoxic stress in culture (Extended Data Fig. 2n–q). Taken together, these results demonstrate that the restoration of intracellular SAM content can rescue the survival defects of aged MuSCs during exit from quiescence.
Inhibiting spermidine synthesis restores heterochromatin
When SAM is converted to decarboxylated S-adenosylmethionine (dcSAM) during polyamine synthesis it is used as an aminopropyl donor. This reaction results in the conversion of dcSAM to 5’-methylthioadenosine (MTA)33 (Fig. 3a). From our metabolomics data30, we discovered that dcSAM and MTA increased in MuSCs with age (Fig. 3b), suggesting that the utilization of SAM for polyamine biosynthesis increases with age. This could thus potentially explain the reduction of SAM levels in aged MuSCs.
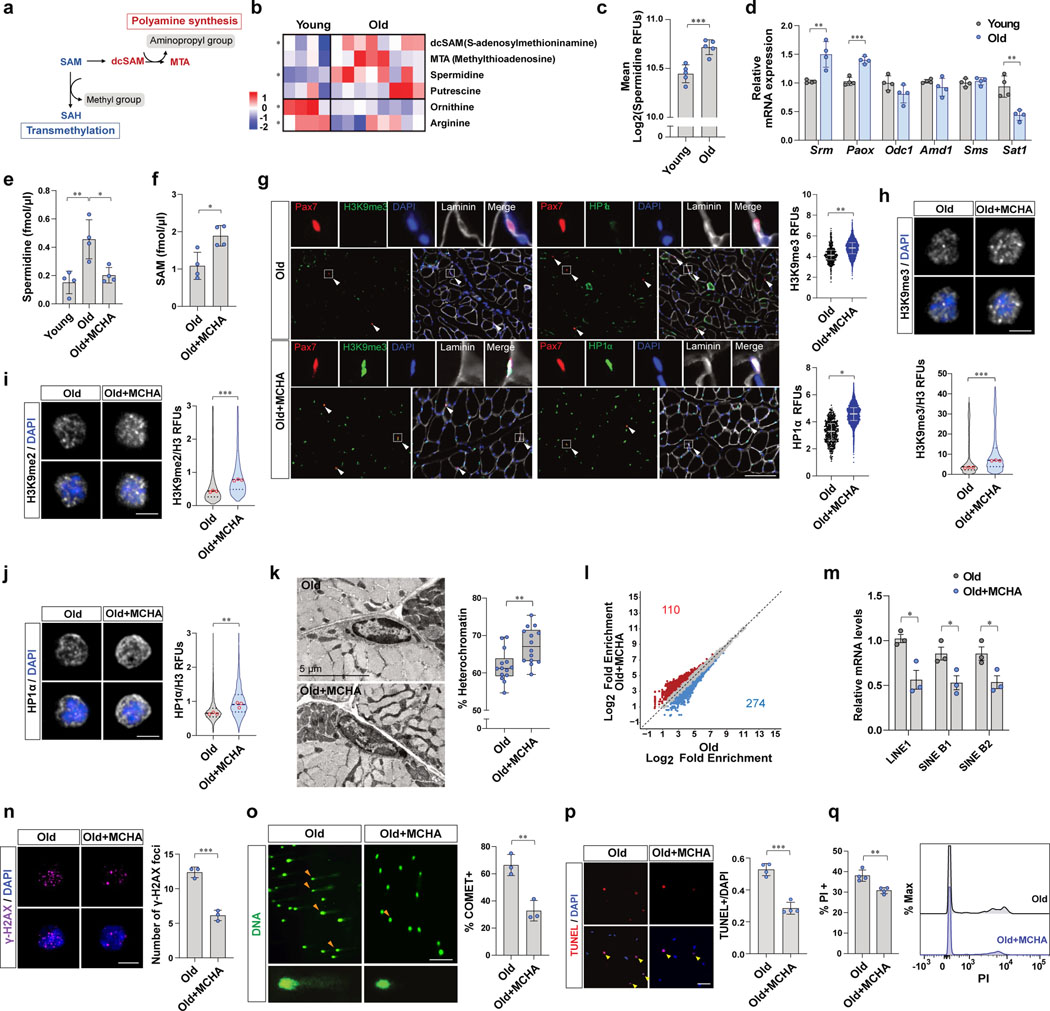
Figure 3. Excessive spermidine synthesis depletes SAM and elicits the heterochromatin loss in old MuSCs.
a, Schematic flow of SAM utilization. b, A heat map of relative levels of polyamine metabolism related metabolites detected by untargeted metabolomics based on LC-MS. c, Quantification of RFUs of spermidine in MuSCs (n=5). d, The mRNA expression of polyamine metabolism-related enzymes was normalized to Gapdh (n=4). e, Spermidine was quantified by LC-MS (n=4). f, SAM was quantified by LC-MS (n=4). g, (Left) Representative tissue immunostaining images of Pax7CreER;R26RRFP mice (Scale bar, 100 μm). (Right) Quantification of RFUs of H3K9me3 and HP1α in RFP-positive MuSCs (n=4). h-j, Representative confocal images (Scale bar, 5 μm). RFUs of H3K9me3 (h), H3K9me2 (i), and HP1α (j) were normalized to RFUs of total H3 (n=3). k, (Left) Representative TEM images. (Right) The percentage of heterochromatin was quantified (n=14 cells examined over 3 independent animals per group). The box represents the IQR, with the lower and upper hinges indicating the 25th and 75th percentiles, respectively. The horizontal line inside the box marks the median score. The whiskers extend to the minimum and maximum values. l, Differential ATAC-seq peaks were determined with different number of reads that exhibit an over 50%-fold change in signals (p-value<0.05). m, The mRNA level of each TE was normalized to Gapdh (n=3). n, (Left) Representative confocal images of γ-H2AX immunofluorescence (Scale bar, 5 μm). (Right) The number of γ-H2AX foci in a cell a day after irradiation (n=3). o, (Left) Representative comet images (Scale bar, 50 μm). (Right) The percentage of comet positive cells (n=3). p, A day after 7 Gy γ-irradiation, isolated MuSCs were cultured for 2 days. (Left) Representative images of TUNEL assay (Scale bar, 50 μm). (Right) The ratio of TUNEL positive MuSCs over total MuSCs (n=4). q, A day after 7 Gy γ-irradiation, isolated MuSCs were cultured for 2 days. (Left) The percentage of PI positive cells (n=4). (Right) A representative FACS plot. Data are shown as mean ± SD (c-f, m-q) and as median and quartiles (g-k). P values were calculated by two-sided unpaired Student’s t-tests (c-k, m-q). *P < 0.05; **P < 0.01; ***P < 0.001.
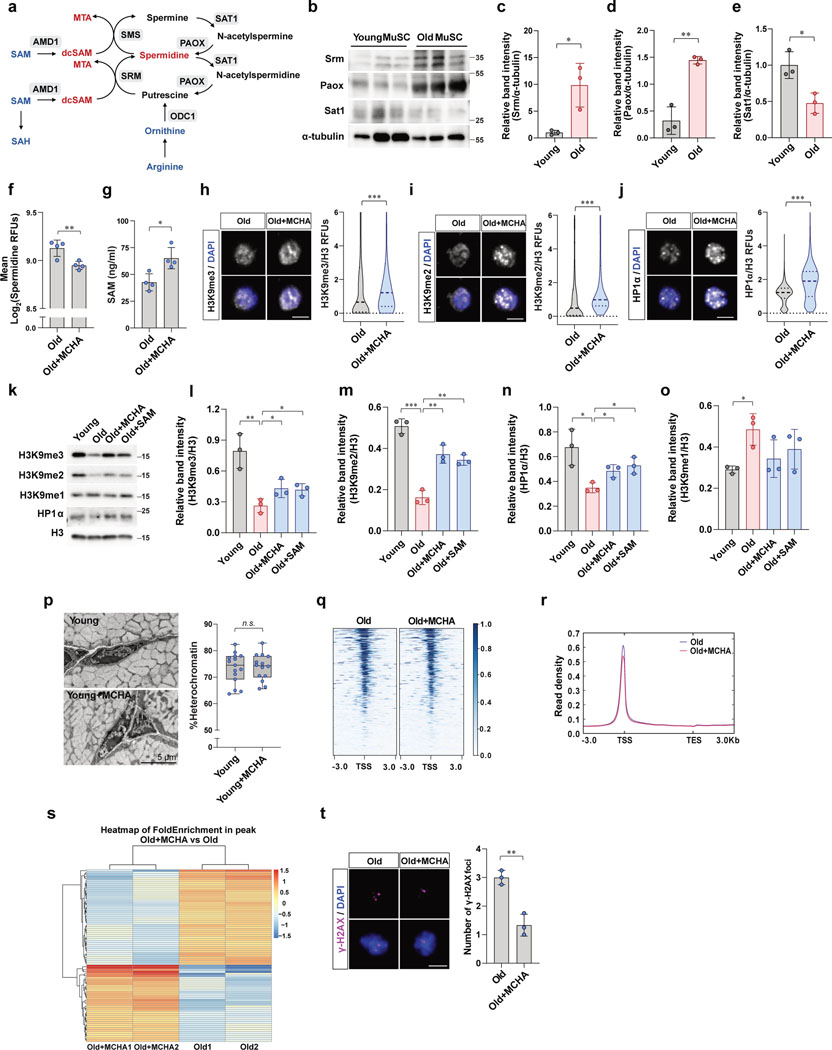
Consistent with this premise, we discovered that polyamines themselves, including spermidine and putrescine, increase with age, whereas the precursors of polyamine biosynthesis, namely ornithine and arginine, decline with age in MuSCs (Fig. 3b). We also quantified spermidine by staining MuSCs with an anti-spermidine antibody. Consistent with metabolomics data, we found that the intracellular spermidine levels increase with age (Fig. 3c). In order to understand why spermidine synthesis increases in aged MuSCs, we quantified the mRNA levels of enzymes related to polyamine metabolism, including spermidine synthase (Srm), spermine synthase (Sms), peroxisomal acetyl-oxidase (Paox), ornithine decarboxylase (Odc1), SAM decarboxylase (Amd1), and spermidine/spermine N1-acetyltransferase 1 (Sat1) (Extended Data Fig. 3a). This revealed that the expression of Srm and Paox increase, whereas that of Sat1, a gene encoding enzyme catalyzing degradation of spermidine and spermine, decrease with age (Fig. 3d). We confirmed that protein levels of SRM and PAOX are significantly higher whereas SAT1 is lower in old MuSCs than young MuSCs (Extended Data Fig. 3b–e). On the other hand, Odc1, Amd1, and Sms transcript levels were similar in young and old MuSCs (Fig. 3d).
As SRM is an enzyme that directly consumes dcSAM and increases with age, we hypothesized that a high level of SRM expression could lead to over-utilization of SAM in polyamine synthesis and result in SAM depletion in aged MuSCs. To address this, we tested whether inhibition of SRM could restore SAM content in the aged MuSCs. Aged mice were treated with MCHA, which is a specific inhibitor of SRM, and the intracellular SAM and spermidine levels were quantified. We found that inhibiting SRM not only decreased intracellular spermidine level but also significantly increased the intracellular SAM content (Fig. 3e, f and Extended Data Fig. 3f, g).
Given that MCHA is capable of restoring SAM content in aged MuSCs, we tested whether it could also increase heterochromatin formation. Indeed, treatment of aged mice with MCHA in vivo resulted in significant increases of heterochromatin markers (Fig. 3g–j and Extended Data Fig. 3h–n). In addition, we found that the percentage of heterochromatin in aged MuSCs was significantly increased following in vivo MCHA treatment (Fig. 3k), whereas no change was observed in the level of heterochromatin in response to MCHA treatment in young MuSCs (Extended Data Fig. 3p). Our ATAC-seq data also indicated increased heterochromatin in old MuSCs upon MCHA treatment by showing a decrease in ATAC-seq peak density and in the number of accessible peaks in old MuSCs treated with MCHA (Fig. 3l and Extended Data Fig. 3q–s). We also observed a significant decrease in the expression of TEs in MCHA-treated old MuSCs (Fig. 3m). On the other hand, there was a significant increase in level of H3K9me1with age and a slight decrease upon SAM or MCHA treatment (Extended Data Fig. 3k, o). This could be a consequence of loss of H3K9me2/3 or an adaptive mechanism to preserve heterochromatin stability as recently reported34.
In order to test whether inhibiting spermidine synthesis reduces accumulation of DNA damage in aged MuSCs, we isolated MuSCs from vehicle- or MCHA-treated aged mice and quantified the extent of DNA damage. We observed that inhibiting spermidine synthesis decreased the number of γ-H2AX foci in old MuSCs (Extended Data Fig. 3t). To quantify the extent of DNA damage upon genotoxic stress, MuSCs were isolated from aged mice that had been subjected to X-irradiation. We found that inhibiting spermidine synthesis decreased the number of γ-H2AX foci and Comet-positive cells from MCHA-treated old mice (Fig. 3n, o). In line with the reduced DNA damage accumulation, the MuSCs from MCHA-treated old mice exhibited decreased cell death upon activation from quiescence following X-irradiation (Fig. 3p, q).
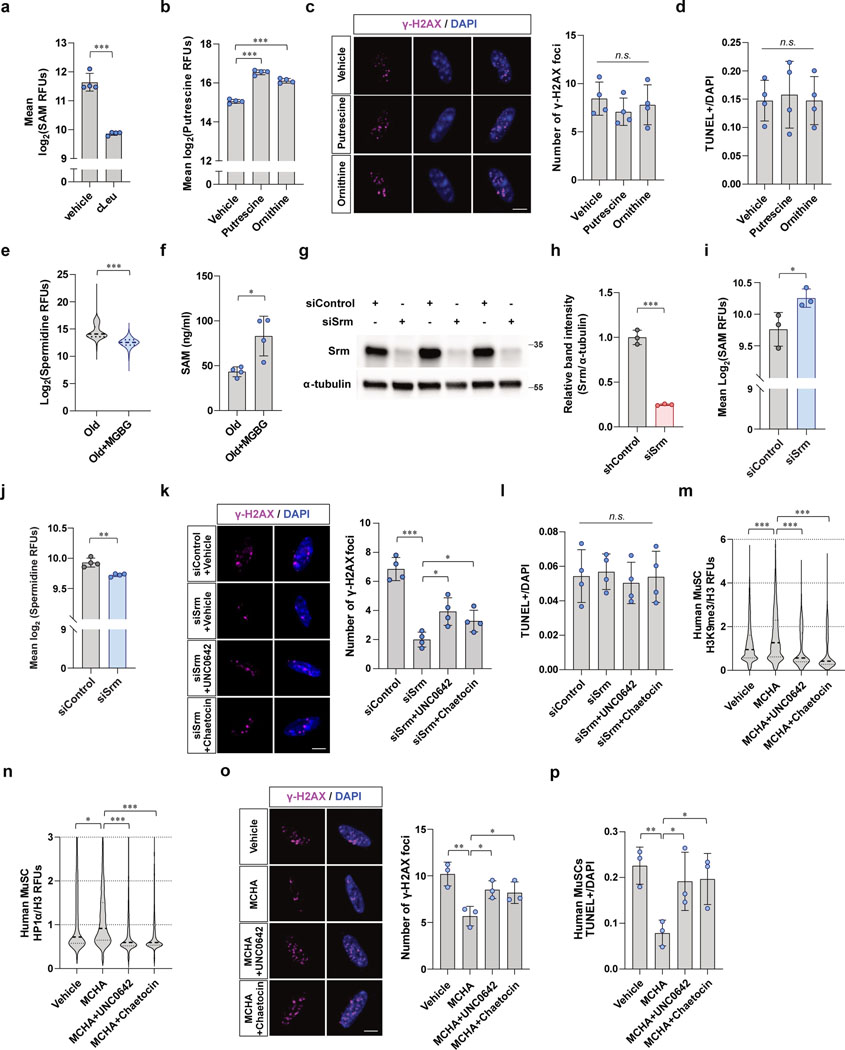
We wanted to test whether the beneficial effects we observed from MCHA-treated aged MuSCs are due to decreased spermidine levels or to the restoration of SAM by testing the effect of each independently. To test whether the beneficial effects of MCHA treatment were specifically due to the decrease in spermidine levels, we prevented the increase in SAM in MCHA-treated cells by treatment with cycloleucine (cLeu), an inhibitor of methionine adenosyltransferase (Fig. 4a). We first confirmed that treating cells with cLeu inhibited SAM synthesis in MuSCs (Extended Data Fig. 4a). Treatment of MCHA-treated cells with cLeu led to a decrease in spermidine levels without an increase in SAM levels (Fig. 4b, c). Conversely, to test whether the beneficial effects of MCHA treatment were specifically due to an increase in SAM levels, we prevented the decrease in spermidine in MCHA-treated cells by the addition of exogenous spermidine to the medium. Treating cells with spermidine concomitantly with MCHA has been shown to preserve intracellular spermidine levels in the face of SRM blockade35. Indeed, treatment of MCHA with exogenous spermidine led to an increase in SAM levels without a concurrent decrease in spermidine levels (Fig. 4b, c).
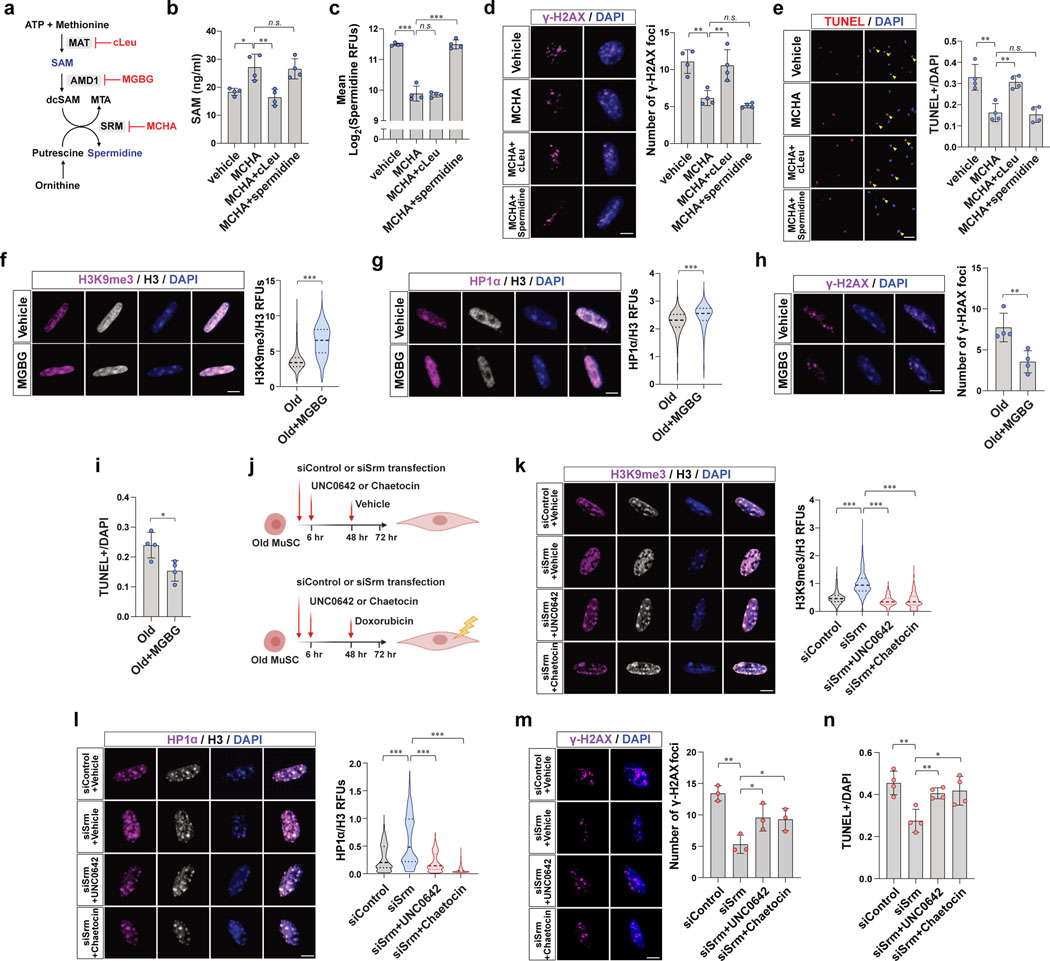
Figure 4. Heterochromatin re-formation by restoration of SAM ameliorates susceptibility to genotoxic stress and cell death in old MuSCs.
a, Diagram showing actions of cLeu, MGBG, and MCHA on the synthesis of SAM and spermidine. b, Old MuSCs were treated as indicated for 2 days. SAM was quantified by SAM ELISA (n=4). c, Mean RFUs of intracellular spermidine in old MuSCs treated as described in (b) (n=4). d, After treating old MuSCs as described in (b), 0.75 μM of doxorubicin was added to the media, and the cells were cultured for one more day. (Left) Representative confocal images of γ-H2AX foci (Scale bar, 5 μm). (Right) The ratio of γ-H2AX foci per cell (n=4). e, (Left) Representative TUNEL assay images of old MuSCs treated as indicated (Scale bar, 50 μm). (Right) The ratio of apoptotic cells over total cells was quantified (n=4). f-g, (Left) Representative confocal immunofluorescence images of old MuSCs treated with vehicle or MGBG for 48 hr (Scale bar, 5 μm). (Right) RFUs of H3K9me3 (f), or HP1α (g) were normalized to RFUs of total H3 (n=4). h, After treating old MuSCs with vehicle or MGBG, 0.75 μM of doxorubicin was added to the media, and the cells were cultured for one more day. (Left) Representative confocal images (Scale bar, 5 μm). (Right) The ratio of γ-H2AX foci per cell (n=4). i, The ratio of apoptotic cells over total cells (n=4). j, Timetable and procedure of ex vivo drug treatment of old MuSCs used in (k-n). k-l, (Left) Representative confocal images of MuSCs treated as described in (j) (Scale bar, 5 μm). (Right) RFUs of H3K9me3 (k) and HP1α (l) were normalized to RFUs of total H3 (n=4). m, (Left) Representative confocal images of γ-H2AX (Scale bar, 5 μm). (Right) Quantification of the number of damaged foci (n=3). n, The ratio of TUNEL positive cells over total cells was plotted (n=4). Data are shown as mean ± SD (b-e, h, i, m, n) and as median and quartiles (f, g, k, l). P values were calculated by two-sided unpaired Student’s t-tests (b-i, k-n). *P < 0.05; **P < 0.01; ***P < 0.001.
With the ability to independently assess the effects of increases in SAM levels and decreases in spermidine levels by MCHA, we tested which accounted for the benefits of SRM inhibition. We quantified the extent of DNA damage and cell death in populations of aged MuSCs under these different experimental conditions. Cells treated with both MCHA and cLeu exhibited greater DNA damage as well as increased cell death compared to controls (Fig. 4d, e), suggesting that the beneficial effects of SRM inhibition are due to the increases of SAM levels. By contrast, preservation of spermidine levels in MCHA-treated old MuSCs resulted in benefits of reduced DNA damage and cell death similar to what was observed with MCHA treatment alone (Fig. 4d, e), indicating that a decrease in spermidine does not account for the beneficial effects of SRM inhibition in old MuSCs.
Inhibiting spermidine synthesis by MCHA not only increases SAM and decreases spermidine but it also increases precursors of spermidine such as putrescine or ornithine36. As putrescine also has ability to stabilize DNA structure37, we tested if increased levels of putrescine could affect DNA damage and cell survival of aged MuSCs. We first measured intracellular putrescine levels in aged MuSCs treated with putrescine or its precursor, ornithine. Treatment with either putrescine or ornithine significantly increased intracellular putrescine levels in aged MuSCs (Extended Data Fig. 4b). However, we could not detect any significant beneficial effects on DNA damage or cell survival in putrescine- or ornithine-treated aged MuSCs (Extended Data Fig. 4c, d). These results support the conclusion that the beneficial effects of inhibition of spermidine synthesis are not due to the accumulation of polyamine precursors.
Inhibiting heterochromatin formation induces DNA damage
To further investigate whether the protective effect of SAM restoration is due to the reformation of heterochromatin, H3K9 methylation was inhibited by treating cells with UNC0642 or chaetocin (Fig. 4j). First, to induce the SAM restoration, we silenced SRM by using siRNA. Similar to MCHA-treated cells, we confirmed that knockdown of SRM significantly increased intracellular SAM and reduced spermidine of old MuSCs (Extended Data Fig. 4k–n). In addition, depletion of SRM significantly increased heterochromatin markers, which was prevented by methyltransferase inhibitor treatment (Fig. 4k, l). Knocking down SRM led to a dramatic reduction in DNA damage, an effect that was partially prevented by inhibiting H3K9 methylation (Extended Data Fig. 4o). Without extrinsic genotoxic stress, we did not observe a significant difference in cell death between groups (Extended Data Fig. 4p). To instigate DNA damage, we treated each group of cells with doxorubicin prior to quantifying DNA damage and cell death. As expected, we observed that SRM-deficient cells exhibited a lower DNA damage accumulation and a decreased propensity to undergo cell death, an effect that was again reversed by inhibiting H3K9 methylation (Fig. 4m, n). Similarly, we observed that treating human MuSCs with MCHA increased heterochromatin markers, which were decreased upon methyltransferases inhibitor treatment (Extended Data Fig. 4q, r). DNA damage and cell death were also quantified from human MuSCs with genotoxic stimuli in culture. As expected, we found that MCHA reduced DNA damage and cell death, effects that were prevented by additional treatment of methyltransferase inhibitors (Extended Data Fig. 4s, t). These findings suggested that inhibiting heterochromatin formation by reducing H3K9 methylation leads to an increase of DNA damage and survival impairments in SRM-deficient old MuSCs.
As AMD1 promotes SAM consumption in polyamine synthesis, we tested whether inhibiting AMD1 redirects SAM to transmethylation. As expected, treating aged MuSCs with MGBG, which is a specific inhibitor of AMD1, resulted in a decrease in spermidine and an increase in SAM (Extended Data Fig. 4e, f). We observed significantly increased heterochromatin markers, reduced DNA damage accumulation, and reduced cell death in MGBG-treated aged MuSCs (Fig. 4f–i). These data support our conclusion that restoration of SAM leads to protection against DNA damage in aged MuSCs.
Inhibiting spermidine synthesis does not alter autophagy of old MuSCs
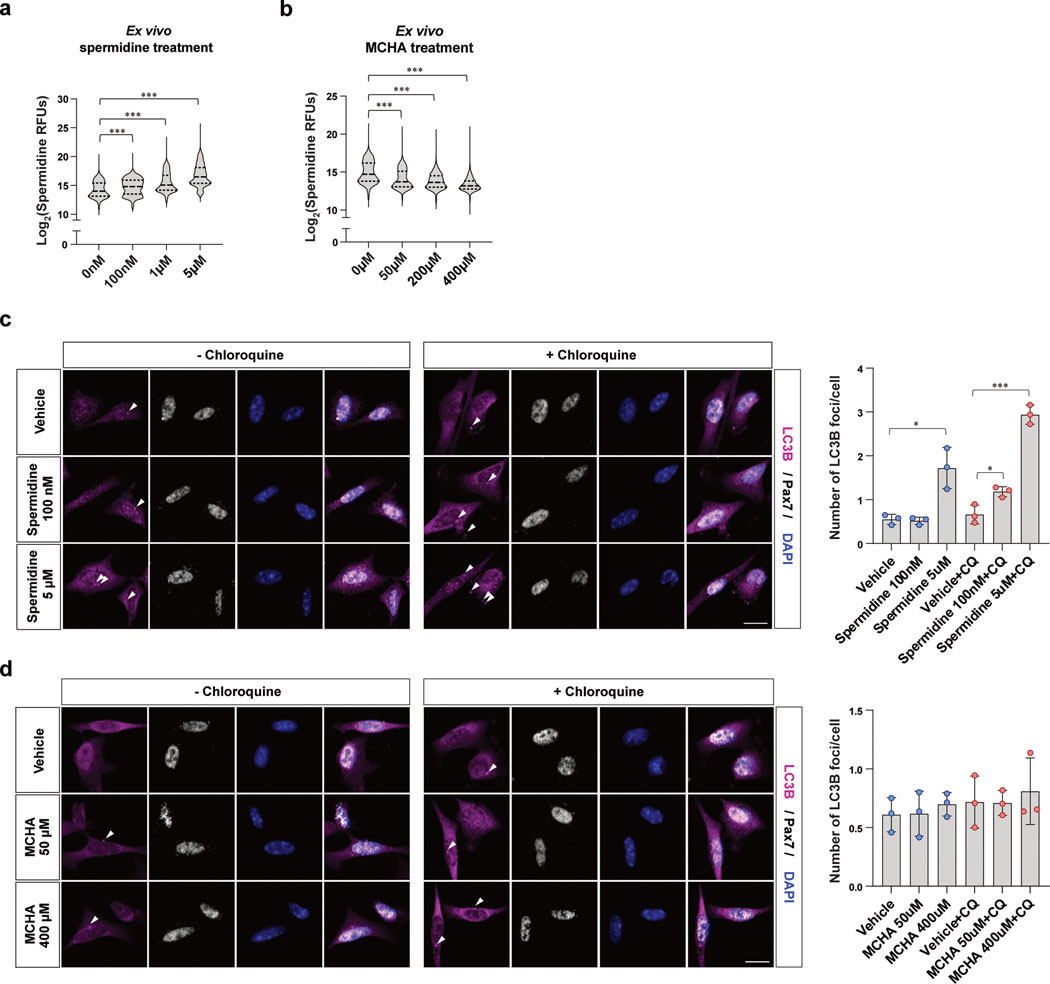
As spermidine is a well-known inducer of autophagy38, we tested whether inhibition of spermidine synthesis with MCHA impairs autophagy of aged MuSCs. We first measured spermidine levels in aged MuSCs treated with different doses of MCHA or spermidine. We observed a dose-dependent decrease in spermidine levels upon MCHA treatment and a dose-dependent increase in spermidine levels upon spermidine treatment (Extended Data Fig. 5a, b). Autophagic flux was significantly increased by both low and high dose of spermidine treatment in chloroquine-treated cells (Extended Data Fig. 5c). In contrast, aged MuSCs treated with MCHA did not show any significant change in autophagic flux (Extended Data Fig. 5d). Collectively, these data suggest that although an increase in intracellular spermidine promote autophagic flux, a decrease in intracellular spermidine does not significantly impair autophagy of aged MuSCs.
Restoration of SAM improves regenerative potency of old MuSC
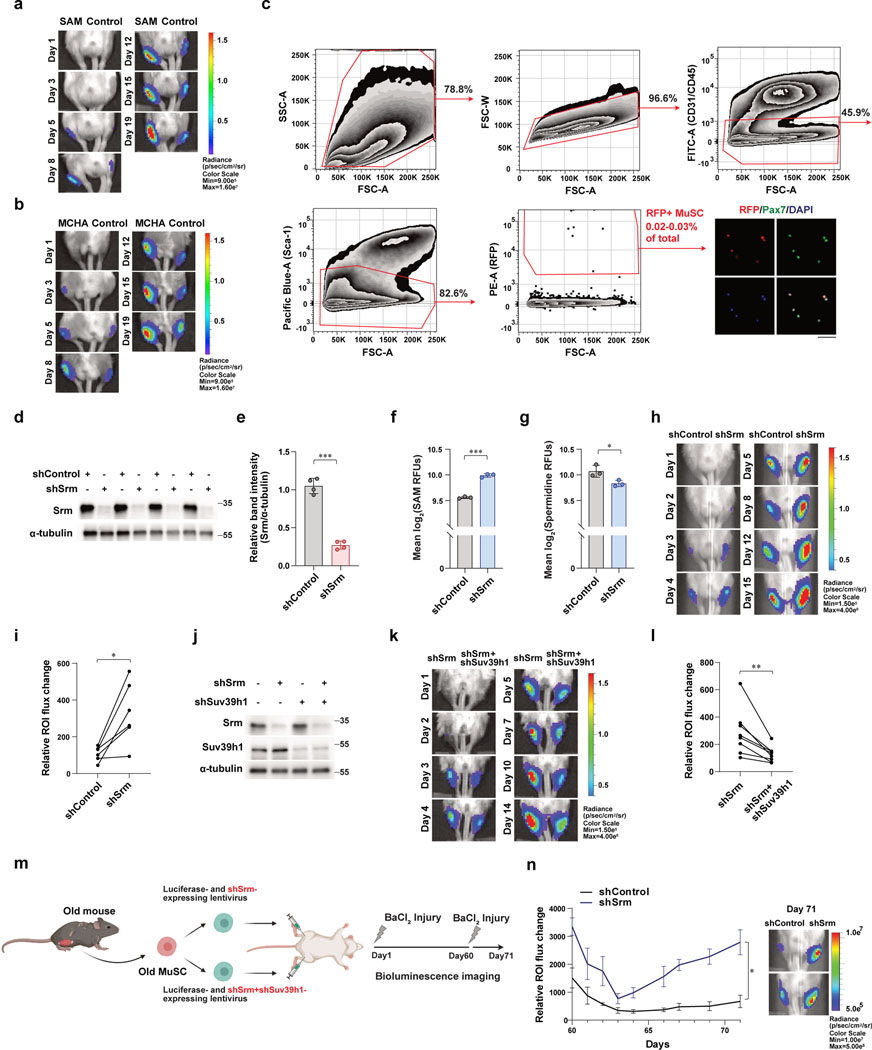
Given that restoration of SAM in old MuSCs produced a striking improvement in cell survival, we next tested the effect of SAM restoration on the in vivo potency of MuSCs to regenerate new muscle in a transplantation assay (Fig. 5a). We monitored transplanted MuSCs using bioluminescence imaging (BLI) and found that muscles transplanted with SAM- or MCHA-treated MuSCs showed higher signals from 3–5 days post-transplantation (Fig. 5b, c and Extended Data Fig. 6a, b), suggesting improved tissue regenerative potential. We tested if SAM- or MCHA-treated aged MuSCs are more resistant to cell death in the host muscle tissue under transplantation conditions. To track the fate of transplanted cells, we used MuSCs isolated from old Pax7creER;R26RRFP mice in which the administration of Tamoxifen induces the expression of RFP as a reporter specifically in MuSCs, treated with SAM or MCHA. RFP-positive MuSCs from different cohorts were transplanted, and muscles were collected at two days after transplantation, and RFP-labeled MuSCs were counted by FACS (Extended Data Fig. 6c). We found that a significantly higher number of RFP-positive MuSCs in muscles transplanted with SAM- or MCHA-treated MuSCs compared to those with vehicle-treated MuSCs (Fig. 5d, e). Given the short time point (2 days) of MuSC re-isolation after transplantation, the number of RFP-positive MuSCs is most likely the result of MuSC survival rather than proliferation. Taken together, these data demonstrate that the effect of SAM restoration on cell survival is associated with improved capacity of aged MuSCs to regenerate new muscle.
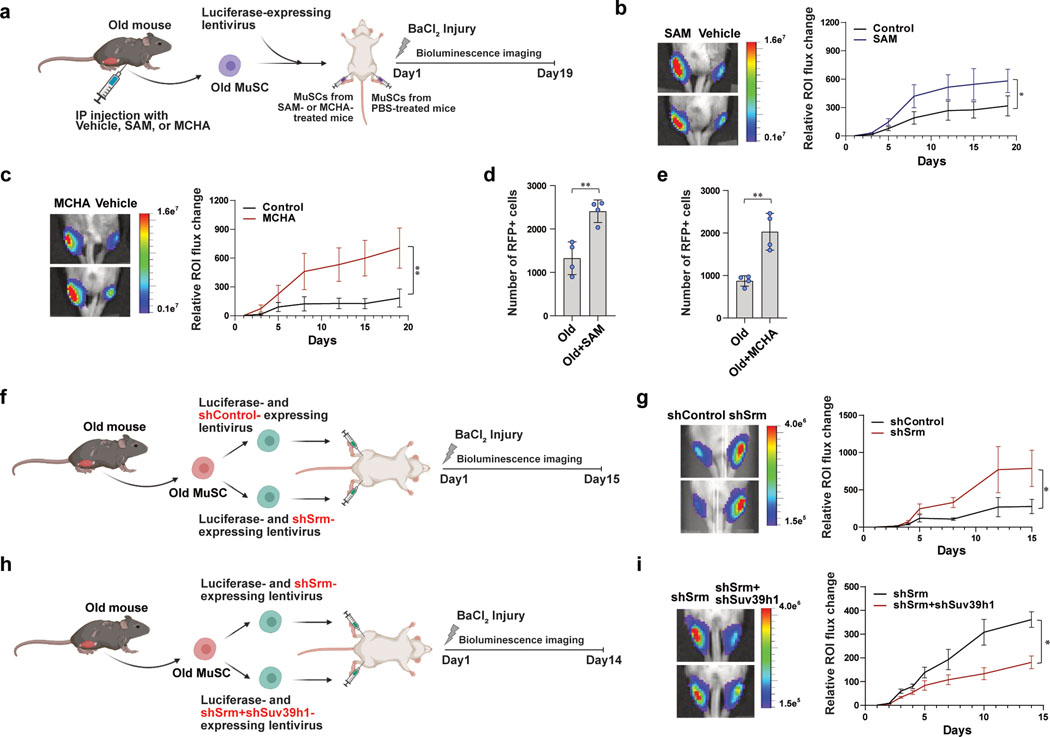
Figure 5. Restoration of heterochromatin improves in vivo potency of old MuSC to regenerate new muscle.
a, Schematic of MuSC transplantation. b, (Left) Representative images of BLI observed 19 days after transplantation. (Right) Quantified results of BLI measured at different time points up to 19 days following transplantation (n=7). c, (Left) Representative images of BLI observed 19 days after transplantation. (Right) Quantified results of BLI measured at different time points up to 19 days following transplantation (n=6). d-e, Quantification of number of RFP-positive MuSCs isolated from each transplanted TA muscle. The number of RFP-positive MuSCs isolated from SAM-treated (d) or MCHA-treated (e) mice was higher than that from vehicle-treated mice (n=4). f, Schematic of transplantation. Old MuSCs were transduced with sh-lentiviruses containing a control scrambled sequence (shControl) or a sequence targeting Srm (shSrm). TA muscles of NSG mice were injected with the MuSCs resuspended in BaCl2 and monitored for 15 days by BLI. g, (Left) Representative images of BLI captured 8 days after transplantation in TA muscles of NSG mice. (Right) Quantified results of BLI measured at different time points up to 15 days following transplantation (n=6). h, Schematic of transplantation. Old MuSCs were transduced with sh-lentiviruses containing a sequence targeting Srm (shSrm) or both Srm and Suv39h1 (shSrm+shSuv39h1). The cells were transplanted as indicated and monitored for 14 days by BLI. i, (Left) Representative images of bioluminescence captured 10 days after transplantation of MuSCs treated as described in (h). (Right) Quantified results of BLI measured at different time points up to 14 days following transplantation (n=8). ROI: Region Of Interest. Data are shown as mean ± SEM (b, c, g, i) and as mean ± SD (d, e). P values were calculated by one-sided student’s t test, pairwise between groups (b, c, g, i) and by two-sided unpaired Student’s t-tests (d, e). *P < 0.05; **P < 0.01; ***P < 0.001.
To further test whether the improved tissue regenerative potential is due to increased heterochromatin, we performed two additional transplantations using shRNA-expressinglentiviruses. First, old MuSCs were transduced with lentivirus expressing luciferase and either scrambled shRNA sequence or shRNA sequence targeting Srm, and the knock-down efficiency of shSrm was confirmed (Extended Data Fig. 6d, e). Depleting SRM significantly increased SAM and decreased spermidine in aged MuSCs (Extended Data Fig. 6f, g). Consistent with MCHA-treated cells, SRM-deficient cells contributed more robustly to the formation of new muscle fibers (Fig. 5f, g and Extended Data Fig. 6h, i). To assess whether formation of H3K9 tri-methylation leads to enhanced regenerative potency of SRM-deficient old MuSCs, old MuSCs were transduced with lentivirus expressing luciferase and shRNA sequence targeting either Srm alone or both Srm and Suv39h1. The knock-down efficiency of SRM and SUV39H1 was confirmed (Extended Data Fig. 6j). Interestingly, we found that muscles transplanted with both SRM and SUV39H1-deficient old MuSCs showed much lower signals compared to those with SRM-deficient old MuSCs (Fig. 5h, i and Extended Data Fig. 6k, l). Collectively, the data supports our previous finding that increasing H3K9 tri-methylation by inhibiting spermidine synthesis rescues impaired tissue regenerative capacity of old MuSCs.
Additionally, we performed repeated injuries following transplantation of shRNA-mediated SRM-deficient cells. Sixty days after the first injury, previously injured muscles were reinjured, and transplanted MuSCs were monitored for additional ten days (Extended Data Fig. 6m). SRM-deficient MuSCs again showed significantly higher signal compared to control cells (Extended Data Fig. 6n). This suggests that restoration of SAM can be targeted to stimulate long-term regeneration without depleting the stem cell pool.
Restoration of SAM improves muscle regeneration of aged mice
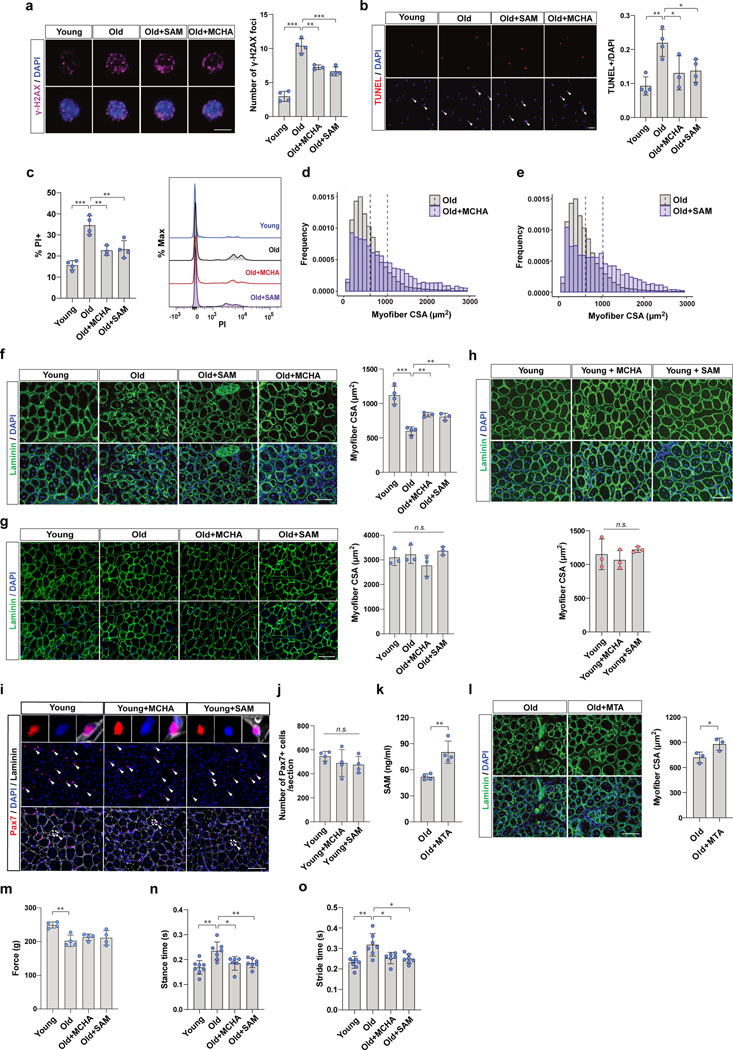
We next wanted to determine whether the reduced DNA damage and cell death in MuSCs from SAM- or MCHA-treated mice were associated with improved muscle regeneration capacity of aged mice. To address this question, we first tested whether SAM or MCHA treatment could reduce the DNA damage and cell death of old MuSCs upon in vivo activation by muscle injury (Fig. 6a). We found that aged mice possess a higher number of damaged MuSCs compared to young mice which was reversed by treatment with SAM or MCHA (Fig. 6b and Extended Data Fig 7a). We further sought to determine whether SAM- or MCHA-treated MuSCs are more resistant to cell death during in vivo activation. We observed a 25% decrease in the number of activated MuSCs that were TUNEL+ in SAM- or MCHA-treated aged mice (Fig. 6c and Extended Data Fig. 7b). In addition, we confirmed that the number of dead cells, which were labeled with PI, was reduced by approximately 40% from SAM- or MCHA-treated old MuSCs compared to vehicle-treated old MuSCs (Fig. 6d and Extended Data Fig. 7c).
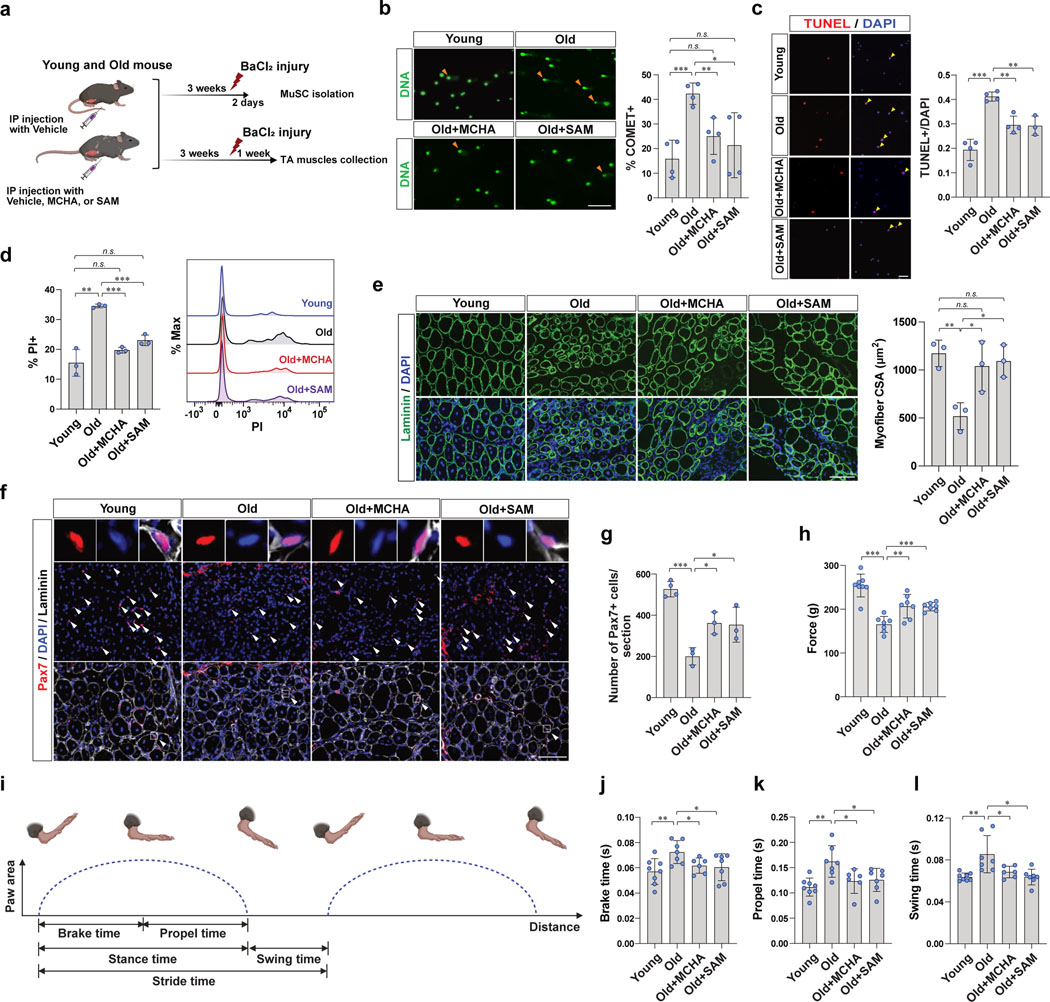
Figure 6. SAM restoration is associated with improved muscle regeneration of aged mice.
a, Schematic showing the experimental design of MuSC isolation (b-d) and tissue collection (e-g), after BaCl2 injury. In order to isolate activated MuSCs, MuSCs were isolated from injured the muscles at 2 days post injury (dpi). During the 2 days after injury, mice were continuously treated with vehicle, MCHA, or SAM. TA muscles were collected at 7 dpi. During the 7 days of after injury, mice were continuously treated with vehicle, MCHA, or SAM. b, (Left) Representative images of cells after alkaline comet assay (Scale bar, 50 μm). (Right) The percentage of comet positive cells (n=4). c, (Left) Representative images of TUNEL positive or negative MuSCs (Scale bar, 50 μm). (Right) Quantification of TUNEL positive MuSCs (n=4 for vehicle-treated young mice and vehicle- or MCHA-treated old mice, n=3 for SAM-treated old mice). d, (Left) The percentage of PI positive cells (n=3). (Right) The representative FACS plot. e, (Left) Representative immunofluorescence images of cross-sections of TA muscles (Scale bar, 100 μm). (Right) The cross-sectional areas (CSAs) of centrally nucleated myofibers of each tissue were quantified (n=3). f, Representative tissue immunostaining images of TA muscle sections (n=4 for young mice, n=3 for old mice per group, Scale bar, 100 μm). g, Quantification of the number of Pax7-positive MuSCs per each tissue section (n=4 for young mice, n=3 for old mice per group). h, Grip force measured from each group of mice at 16 dpi (n=8 for young mice, n=7 for old mice per group). i, Graphical depiction of parameters of a mouse’s gait. j-l, Gait measurements of each group of mice at 14 dpi. Mice were placed individually on a motor-driven treadmill at 10 cm/sec, and brake time (j), propel time (k), and swing time (l) of each mouse were measured (n=8 for young mice, n=7 for old mice treated with vehicle or SAM, n=6 for old mice treated with MCHA). Data are shown as mean ± SD (b-e, g, h, j-l). P values were calculated by two-sided unpaired Student’s t-tests (b-e, g, h, j-l). *P < 0.05; **P < 0.01; ***P < 0.001.
To determine whether the reduced DNA damage and cell death of SAM- or MCHA-treated MuSCs was associated with improved muscle regeneration capacity of aged mice, we collected muscles from each group of mice and examined regeneration histologically (Fig. 6a). We found that muscle regeneration efficacy, as measured by regenerating myofiber cross-sectional area (CSA), decreased significantly with age, but treatment of old mice with SAM or MCHA enhanced regeneration efficacy similar to that seen in young mice (Fig. 6e and Extended Data Fig. 7d–f). On the other hand, without muscle injury, SAM or MCHA did not affect CSA of myofibers in the homeostatic state (Extended Data Fig. 7g). In addition, SAM or MCHA treatment had no effect on the muscle regeneration efficacy of young mice (Extended Data Fig. 7h).
To test whether the improved muscle regeneration capacity was associated with increased MuSC survival within the injured muscle, we measured total Pax7-positive cells after muscle injury. We found a significant decrease in the number of MuSCs in old muscle compared to young muscle, and this was partially rescued by SAM or MCHA treatment (Fig. 6f, g). Consistent with the muscle regeneration results, there was no significant change in the number of Pax7-positive cells in young mice treated with SAM or MCHA compared to controls (Extended Data Fig. 7i, j). These data imply that impaired muscle regeneration in aging is associated with increased DNA damage and cell death, and this could be partially rescued by the restoration of SAM.
It has been noted that the efficiency of cellular uptake of SAM from the extracellular environment is low due to lack of a dedicated SAM transporter in the mammalian plasma membrane39. However, it has been well documented that extracellular SAM can enter the cell through the MTA and methionine salvage pathway40. SAM is unstable at physiological condition and rapidly degrades into MTA through a non-enzymatic cleavage reaction41, and MTA can be used to generate SAM intracellularly via the methionine salvage pathway42. To test whether extracellular MTA can be used to restore intracellular SAM levels in aged MuSCs and improve muscle regenerative capacity, we administered MTA to old mice and assessed muscle regeneration. We observed that MTA supplementation significantly increased intracellular SAM levels in aged MuSCs and increased muscle regenerative capacity in old mice (Extended Data Fig. 7k, l). These data provide additional support for the importance of SAM repletion, either via exogenous SAM or exogenous MTA treatment, for improving the regeneration of old muscle.
Lastly, to assess functional recovery from SAM restoration, we measured muscle force by grip test after muscle injury. We found that aged mice exhibited lower grip strength compared to young mice, and this was partially rescued by SAM or MCHA treatment (Fig. 6h). Consistent with the myofiber CSA results without muscle injury, SAM or MCHA did not affect grip strength of old mice (Extended Data Fig. 7m). In addition, we monitored gait after muscle injury and observed that aged mice showed significantly delayed average duration of one gait cycle compared to that of young mice, which was partially rescued by SAM- or MCHA-treatment (Fig. 6i–l and Extended Data Fig. 7n, o). Taken together, SAM- or MCHA-induced heterochromatin re-formation could improve muscle regenerative capacity and eventually improve functional recovery of the aged mice.
DISCUSSION
Our work aims to answer the fundamental question of why heterochromatin declines with age. We show that reduced intracellular SAM availability reduces H3K9 methylation as well as heterochromatin formation in old MuSCs. Although it has been previously documented that alterations in the levels of intracellular SAM can affect histone methylation43,44, our study provides a link between depletion of SAM with age and the age-associated decrease in histone H3K9 methylation. We further demonstrate that systemic restoration of SAM in old MuSCs to a youthful state induces heterochromatin re-formation and rescues numerous age-associated MuSC phenotypes, including DNA damage, susceptibility to genotoxic stress, and delayed muscle regeneration.
It has been well documented that altering SAM levels can potentially impact histone and DNA methylation43,44. Modulation of dietary methionine, which is the precursor for SAM, has a direct impact on both cellular SAM levels and H3K4/H3K9 methylation in mouse liver and human cancer cells45,46. In our study, we discovered that the intracellular levels of SAM decrease with age, and treatment with exogenous SAM can alter both intracellular SAM and H3K9 methylation in mouse MuSCs. Although we have shown that the regulation of SAM can directly alter a critical key histone methylation mark, it is noteworthy that there are multifactorial determinants of histone modifications47,48. For example, in our previous study, we showed higher levels of H3K27me3 in quiescent MuSC in old mice compared to young mice28. As the level of methylation of any histone lysine is a result of many factors, not just the activity of the relevant methyltransferase, an increase in H3K27me3 could occur even in the setting of a reduction of SAM levels in old MuSC.
Although SAM is widely known as the sole methyl donor for numerous methyl-transfer reactions, another major role of SAM is in polyamine biosynthesis. Here, SAM is first decarboxylated by AMD1 to form dcSAM, and the aminopropyl group of dcSAM is transferred to putrescine to generate spermidine and spermine. Polyamines, which include spermidine, spermine, and putrescin, are involved in a series of biological events, including autophagy induction, apoptosis, transcription, and DNA stability49–51. For example, spermidine was shown to be protective against age-related skeletal muscle atrophy by enhancing autophagy through the mediation of the AMPK-FOXO3a signaling pathway52. Multiple studies have demonstrated the role of polyamines in MuSC activation and differentiation53,54. Inhibition of polyamine biosynthesis decelerates MuSC activation54. In addition, spermidine is known as a source of hypusination, a unique post-translational modification55, and inhibition of hypusination on eukaryotic initiation factor 5A impaired MuSC differentiation53.
It has been reported that polyamine levels decrease with age in multiple tissues, including muscle, heart, and liver56–58. In skeletal muscle, spermidine and spermine were significantly decreased in aged mice with decreased expression levels of AMD1 and spermine oxidase which are enzymes involved in the synthesis of spermidine and spermine56. On the other hand, polyamines are generally elevated in cancer cells59,60. In cancer, as polyamines are essential for cell growth and proliferation, elevated polyamine levels due to dysregulated polyamine metabolism are necessary for transformation and tumor progression60. Intriguingly, polyamine levels have also been reported to be increased in the brains of patients with age-related neurodegenerative diseases61,62. Untargeted metabolomic analysis of human plasma and several different regions of brain revealed that polyamine levels were increased in patients with Alzheimer’s disease63,64.
A metabolomics study showed that the level of spermidine and spermine decreased by 50% with age in skeletal muscle56. However, we found that spermidine levels significantly increase with age specifically in MuSCs, which represent only a tiny fraction of the cytoplasmic volume of skeletal muscle tissue. The reason for this increase remains obscure and further study is required to fully elucidate the upstream cause. One possibility, however, is that an increase in spermidine with age is a compensatory mechanism initiated by old MuSCs to maintain maximum cellular functionality in the face of aging-associated functional decline. Indeed, increased spermidine synthesis in skeletal muscle has been reported in uncoupling protein 1 transgenic mice, which is a model for skeletal muscle-specific mitochondrial dysfunction65. As a compensatory cytoprotective response of mitochondrial distress, spermidine synthesis increased as evidenced by increased Srm gene expression and muscle levels of spermidine and spermine to maintain cellular stress tolerance and survival65. In addition, recent work from our lab has shown that old MuSCs engage the compensatory upregulation of glutathione biosynthetic metabolism to counteract the age-associated elevation in cellular reactive oxygen species30.
Our results provide strong evidence that maintaining cellular SAM levels by inhibiting increased spermidine synthesis confers a rejuvenating effect on aged MuSCs and promotes muscle regeneration, ultimately resulting in the alleviation of aging-associated muscle regenerative defects. So far, several studies have elucidated that rejuvenating aged MuSC through pharmacological interventions targeting such as STAT3 and p38 MAPK improves muscle regeneration66,67. However, our approaches to rejuvenate aged MuSC include not only pharmacological inhibition of Srm but also systemic supplementation of the metabolite SAM. To support the translational poential, we have shown that the beneficial effects of SAM restoration on MuSCs can be also extended to isolated human MuSCs. Taken together, our study provides insights that link SAM and polyamine metabolism and heterochromatin maintenance with aging of MuSCs and a potential way to improve muscle regeneration by rejuvenating aged MuSC.
METHODS
Animals
All animals were housed in the Veterinary Medical Unit at the Veterans Affairs Palo Alto Health Care System. Animal protocols were approved by the Institutional Animal Care and Use Committee of the Veterinary Medical Unit at the Veterans Affairs Palo Alto Health Care System. Young mice used in this study were 2–4 months old; old mice were 20–24 months old. Mice were maintained on a 12/12 hr light/dark cycle between 20–24°C with 40–60% humidity in cages that were individually ventilated. Mice were fed with rodent diets (Teklad global 18% protein, irradiated; #2918), and food and fresh water were provided ad libitum. Health checks were performed weekly on young mice and twice weekly for the aged mice. To minimize variability between individual animals used in analyses, mice used in this study were mostly male. Since the sex was not considered in study design, we performed key experiments in female mice in Extended Data Fig. 1b–d, 2b–d, 3h–j, 7a–c, and 7f. Young C57BL6 mice (#000664) and NSG mice (#005557) were purchased from The Jackson Laboratory. Old C57BL6 mice were obtained from Charles River Laboratories through the National Institute on Aging. Pax7CreER; ROSA26RFP mice were obtained by crossbreeding Pax7CreER mice (#017763) with ROSA26RFP (#007914) mice purchased from The Jackson Laboratory. The inducible CreER recombinase system was activated by injecting Tamoxifen (Sigma-Aldrich; # T5648) at a dose of 20 mg/mL in corn oil. Mice received 2 mg Tamoxifen intraperitoneally daily for five consecutive days. In certain experiments, mice were injected intraperitoneally with 200 μmol/kg of MCHA (Sigma Aldrich; #CDS019932) or 5 mg/kg of SAM (New England Biolabs; B9003S) which are dissolved in sterile PBS daily for 3 weeks. Mice were given 1 mg/mL of MTA (Sigma Aldrich; #D5011) in the drinking water for three weeks. For X-irradiation, mice were anesthetized with ketamine and protected in lead shields so that only the hindlimbs were exposed to the required dose in an X-RAD 320 γ-irradiator. Hindlimb muscles of mice received 7 Gy of γ-irradiation a day prior to sacrifice.
Mouse muscle injury
Before muscle injuries, mice were anesthetized with 2% isoflurane, and tibialis anterior (TA) muscles were disinfected with ethanol and iodopovidone. TA muscles were injected with 50 μl of 1.2% (w/v in sterile H2O) of BaCl2 (Sigma-Aldrich; # B0750) with 31-gauge insulin syringes. Mice received buprenorphine analgesia postoperatively and were monitored daily for the first two days and once every two days thereafter.
Human muscle specimen
The human muscle biopsy specimens were taken from three donors whose information is listed in Supplementary Table 1. The muscle biopsy samples were donated via Donor Network West (DNW). The biopsy samples were taken after patients gave informed consent, as part of a human studies research protocol which was approved by the Stanford University Institutional Review Board. The experiments were performed using fresh muscle specimens.
MuSC isolation and treatment
MuSCs were isolated from mouse hindlimbs as previously described68 using surface-antigen-based isolation (Extended Data Fig. 1a) using flow cytometry FACSDIVA 8+ (BD Bioscience). For fluorescence-activated cell sorting (FACS) isolation of mouse MuSC, disassociated muscle suspension was stained with FITC-anti-CD31, FITC-anti-CD45, Pacific blue-anti-Sca1, and PE/Cy7-anti-VCAM. For human MuSC isolation, muscle suspensions were stained with APC-anti-human CD31, FITC-anti-human CD45, PE-anti-human CD90, and PE/Cy7-anti-human CD82. The human MuSC population was obtained by FACS as previously described69. Freshly isolated MuSCs that we used in ex vivo analyses are the MuSCs that went through preparation steps from dissection of mouse to FACS-sorting which typically takes four to five hours. For analysis of cell death by flow cytometry, sorted cells were treated with propidium iodide (Thermo Scientific/Fisher; # P3566) immediately after sorting, and analyzed by flow cytometry. For culturing MuSCs, isolated MuSCs were plated in wells that were pre-coated with poly-D-lysine (0.1 mg/ml, EMD Millipore; # A-003-E) and ECM (25 mg/ml, Sigma-Aldrich; # E1270) and cultured in growth medium (GM: Ham’s F10, 10% horse serum, 100 U/ml penicillin, and 100 mg/ml streptomycin). For ex vivo drug treatment, 200 μM SAM (New England Biolabs; # B9003S), 50 μM MCHA (trans-4-Methylcyclohexylamine; Sigma-Aldrich; #CDS019932), 100 nM spermidine (Sigma-Aldrich; # S0266), 100 μM cycloleucine (Sigma-Aldrich; # A48105), 1 μM UNC0642 (Selleckchem; # S7230), 5 nM chaetocin (Selleckchem; # S8068), or 0.75 μM doxorubicin (Sigma-Aldrich; # 44583) was added in GM. For siRNA transfection, freshly isolated cells were incubated in GM and transfected with a freshly prepared mix of ON-TARGETplus siRNA (Dharmacon) and RNA iMax (Invitrogen; # 13778075) for 6 hr, and the cells were then maintained at least 2 days in fresh GM. For shRNA transduction, cells were incubated with concentrated virus and polybrene (Santa Cruz Biotechnology; # sc-134220), which were spun for 1 hr at 2,550g at 4°C. Sequences of oligonucleotide for siRNA and shRNA are listed in Supplementary Table 2.
Liquid chromatography-tandem mass spectrometry (LC-MS)
Authentic standards for spermidine and SAM were used for assay set up and preparation of calibration curve standards. Stable isotope labeled spermidine-d6 and SAM-d3 were used as internal standards (IS). MuSC pellets were homogenized in 80% methanol/1% acetic acid. Samples were diluted with 50% MeOH and spiked with IS. Separation of the analytes was performed on Acquity UHPLC I-class system (Waters) using a 100 × 2.1 mm, 2.7 μm Poroshell 120 HILIC-Z column (Agilent) after injection of sample at a 0.5 mL/min flow rate. Mobile phase A was 40 mM ammonium formate, pH 3.2, in water. Mobile phase B was 40 mM ammonium formate in 80% acetonitrile. The gradient program was as follows: 0–1 min – 95% B; 5–6.5 min – 15% B; 7–10 min 95% B. Observed retention times for spermidine and SAM were 4.5 min and 4.24 min, respectively. Xevo TQ-XS triple quadrupole mass spectrometer (Waters) was operated in positive electrospray ionization mode. The capillary voltage was set to 1.00 kV, and cone voltage was set to 50 V. Detection of the analytes was performed in selected reaction monitoring mode. Monitored transitions were 145.1 > 71.9 and 145.1 > 112.1 for spermidine, 399.1 > 250.1 and 399.1 > 97.0 for SAM, 152.1 > 72.0 and 152.1 > 118.1 for spermidine-d6, and 402.1 > 250.1 and 402.1 > 136.0 for SAM-d3. Eight level calibration curve prepared in 1:20 diluted charcoal stripped plasma was used to cover the range of 5 pM to 25 nM. The concentration of the IS in analyzed samples and calibration curve standards was 10 nM. Obtained data was analyzed using MassLynx and TargetLynx software.
SAM ELISA
For quantifying intracellular SAM content, a SAM ELISA kit (My Bio Source; MBS2603023) was used. MuSCs were lysed with RIPA buffer with 0.5 M NaCl2 at 4°C for 15 min, and the lysates were centrifuged at 13,000g for 15 min. Supernatants were used for ELISA assays. The cell lysate samples or different concentration of mouse SAM standards were added to corresponding wells and incubated at 37°C for 90 min. After incubation, ELISA plates were washed twice with wash buffer provided in the kit, and biotinylated mouse SAM antibody was added and incubated at 37°C for 60 min. After washing, enzyme-conjugate liquid was added to each well and incubated at 37°C for 30min. The plate was washed, and color reagent liquid was added to individual well. The plate was incubated in the dark at 37°C, until the color for the high concentration of standard became darker and the color gradient appeared. Lastly, color reagent C was added to individual wells, and optical density (OD) at 450 nm of each well was measured within 10 min. For calculations, the OD value of each sample was subtracted by that of a blank well, and the standard curve was drawn using CurveExpert Professional.
Electron microscopy
Extensor digitorum longus (EDL) muscles were fixed in 2% glutaraldehyde (EMS; #16000) and 4% paraformaldehyde (EMS; #15700) in 0.1 M sodium cacodylate (EMS; #12300) buffer (pH 7.4) for 1 hr at room temperature. The fixative was then replaced with cold 1% osmium tetroxide (EMS; #19100) and the muscles were allowed to warm to room temperature for 2 hr, rotating in a hood. Then muscles were then washed with ultrafiltered water and stained in 1% uranyl acetate for 2 hr while rotating. Muscle tissues were dehydrated in 50% and 70% of ethanol for 30 min each at room temperature and then incubated at 4°C overnight. They were placed in cold 95% EtOH and allowed to warm to room temperature, changed to 100% EtOH two times, then Propylene Oxide (PO) for 15 min. Samples were infiltrated with EMbed-812 resin (EMS; #14120), mixed 1:2, 1:1, and 2:1 with PO for 2 hr each, leaving samples in 2:1 resin to PO overnight, rotating in the hood. The samples were then placed into Embed-812 for 2 to 4 hr and then placed into molds with fresh resin at 65°C overnight. Sections were taken around 80 nm and picked up on formvar/Carbon coated 100 mesh Cu grids, stained for 40 sec in 3.5% uranyl acetate in 50% acetone, and then stained in Sato’s Lead Citrate for 2 min. The grids were observed in a JEOL JEM-1400 120kV transmission electron microscope. Images were taken using a Gatan Orius 832 4k x 2.6k digital camera with 9 μm pixel. The percentage of heterochromatin was quantified as the ratio between darkly stained areas filled with the dense heterochromatin the dark area and the total cross-sectional area of the nucleus of each MuSC using ImageJ.
Alkaline comet assay
Alkaline comet assays were performed by following the manufacturer’s instruction (Trevigen; #4250–050-K). Briefly, freshly isolated MuSCs were washed with PBS and mixed with low melt agarose which was pre-warmed at 37°C at a ratio of 1:10 (v/v). The mixture of cells and low melt agarose was immediately placed onto a comet slide. The slides were placed flat at 4°C in the dark until a 0.5 mm clear ring appeared at the edge of comet slide area. Slides were then immersed in 4°C lysis solution for 30 min, and then incubated in freshly prepared alkaline unwinding solution (pH>13) overnight at 4°C in the dark. The electrophoresis was run at 21V for 30 min. After electrophoresis, the slides were stained with diluted SYBR Gold at a ratio of 1:10,000 for 30 min at room temperature in the dark. Cell images were captured with an AxioObserver Z1 epifluorescence microscope (Carl Zeiss) equipped with a Hamamatsu Orca-R2 CCD camera, and the cells with comet tails were quantified using ImageJ.
TUNEL (Terminal deoxynucleotidyl transferase biotin–dUTP Nick End Labeling) assay
To detect single- and double-stranded DNA breaks, an In Situ Cell Death Detection Kit (Roche; #12156792910) was used according to the manufacturer’s protocol. In brief, fixed and permeabilized cells were incubated with the TUNEL reaction mixture that contains TdT and TMR-dUTP at 37°C for 1 hr. Cells with apoptotic DNA fragmentations which were labeled with TMR red were visualized on an ImageXpress Pico (Molecular devices) and quantified using CellReporterXpress.
RNA-sequencing
RNA was extracted from MuSCs using a miRNeasy kit (Qiagen; # 217084) according to the manufacturer’s protocol. The Smarter Ultra Low Input V4 RNAseq kit (Takara Bio; # 634848) was used to generate cDNA from 2 ng of total RNA. Amplified cDNA was purified using SPRI Ampure Beads (Beckman Coulter; # A63880). The quality and quantity of cDNA were assessed using a High Sensitivity DNA chip on an Agilent 2100 Bioanalyzer. Illumina Nextra-XT kit (# FC-131–1024) was used to generate libraries from 150 pg of amplified cDNA. Equal quantities of uniquely indexed libraries were pooled, and the libraries underwent paired-end 150 bp sequencing on a NovaSeq 2000 at the Stanford Genome Sequencing Service Center to a depth of 20–40 million reads. Raw reads were pre-processed with Trim Galore (v0.4.2) to remove adapters and improve the quality of the reads. Trimmed reads were then aligned to the mouse GRCm39 genome using STAR aligner, and Subread package (v1.5.1) with featurecounts module was used to generate raw count reads mapped on exonic regions. Raw counts analysis was done using RStudio software (v4.0.4). DESeq2 package was used for differential gene expression analysis. DESeq normalization was conducted by using median-of-ratio method as proposed by Anders and Huber70. ggplot2 package was used for data visualization.
ATAC-sequencing
ATAC-seq libraries were generated using an ATAC-seq kit (Active Motif; #53150). In brief, MuSCs were resuspended in cold lysis buffer and centrifuged immediately at 1600g for 10 min at 4°C to collect nuclei. For tagmentation reactions, the samples were incubated at 37°C for 30 min in a thermomixer set at 800 rpm. The tagmented DNA was purified using a DNA purification column, and the purified DNA was amplified in the presence of a different combination of i7/i5 indexed primers which are based on Illumina’s Nextera adapters. PCR products were then subjected to SPRI clean-up. PCR amplified libraries were analyzed to assess size distribution with a High Sensitivity DNA chip on an Agilent 2100 Bioanalyzer before sequencing. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using a TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on an Illumina Hiseq platform and 150 bp paired-end reads were generated at Novogene to a depth of 40 million reads. Nextera adaptor sequences were first trimmed from the reads using skewer (0.2.2). These reads were aligned to a reference genome using BWA, with standard parameters. These reads were then filtered for high quality (MAPQ ≥ 13), non-mitochondrial DNA, and properly paired reads (longer than 18 nt). All peak calling was performed with macs2 using ‘macs2 callpeak --nomodel --keepdup all --call-summits’. For simulations of peaks called per input read, aligned and de-duplicated BAM files were used without any additional filtering.
Western blotting
Lysates of MuSCs were electrophoresed on 4–15%, 8–16%, or 12% polyacrylamide gels (Bio-Rad) depending on the size of targeted protein, and then transferred to PVDF membranes. The membranes were blocked with blocking solution (Advansta; #R-03726-E10) for 1 hr at room temperature and probed with indicated antibodies diluted in the same blocking solution at 4°C overnight. Protein levels were visualized using an Enhanced Chemi-Luminescence detection system (GE Healthcare Life Sciences; #RPN2232). The intensities of protein bands were normalized by the intensities of α-tubulin using ImageJ densitometry.
Quantitative real-time PCR
Total RNA was extracted from MuSCs using the RNeasy Micro Plus Kit (Qiagen; # 74034) according to the manufacturer’s instructions. Isolated RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems; # 4368814). Quantitative real-time PCR was performed on a LightCycler 480 (Roche) with matching SYBR Green master mix. The mRNA values of targeted genes were calculated relative to Gapdh expression for each sample. All reactions were performed as triplicates. The nucleotide sequences of PCR primers are summarized in Supplementary Table 3.
Immunocytochemistry
For imaging freshly isolated MuSCs, cells were plated on either 8 well slides (Millicell EZ slide; Millipore/Fisher) or 96 well plates (Corning). To promote cell attachment, the slides or the plates were spun for 3 min at 2,550g at 4°C, and the cells were fixed with 4% PFA immediately after spinning. For imaging cultured MuSCs, cultured MuSCs were fixed and permeabilized with 0.5% Triton X-100 in PBS for 15 min at room temperature. The cells were blocked in blocking solution (10% FBS, 0.5% Triton X-100, 1% BSA, and 0.01% NaN3 in PBS) for 1 hr and incubated with primary antibodies diluted in the same blocking solution at 4°C overnight. After washing out primary antibodies with PBST (0.1% Tween-20 in PBS) three times for 10 min each, cells were then incubated with secondary antibodies and DAPI for 1 hr at room temperature in the dark. The stained cells on 8 well slides were mounted with FluoroSave (Calbiochem; # 345789) and imaged using a high-resolution confocal microscope (LSM-880 (Carl Zeiss)). Confocal images were taken as 8 to 15 z-stacks and deconvoluted using Huygens Essential. The stained cells on 96 well plates were imaged using an ImageXpress Pico (Molecular devices), and the relative fluorescence units (RFUs) of each cell were automatically quantified in CellReporterXpress. ImageJ was used for analysis and quantification.
Histology and immunohistochemistry
TA muscles were harvested and frozen in Optimal Cutting Temperature compound (Fisher Scientific; # 23–730-571) immediately after dissection and stored at −80°C until cryosectioning. Cryopreserved TA muscles were sectioned at the mid-belly of the muscle into 10 μm sections. Sections were fixed in 2% PFA in PBS at room temperature for 10 min, blocked in blocking solution (10% FBS, 0.5% Triton X-100, 1% BSA and 0.01% NaN3 in PBS) for 1 hr, and incubated with primary antibodies in the same blocking solution overnight at 4°C. After washing out primary antibodies with PBST (0.1% Tween-20 in PBS), sections were incubated with secondary antibodies for 45 min at room temperature in the dark. For visualization of Pax7-positive MuSCs on tissue sections, we either used endogenous RFP signals from Pax7CreER;R26RRFP mouse or Pax7 staining using Pax7 antibody (DHSB; Pax7-c) and M.O.M. (mouse on mouse) immunodetection kit (Vector laboratories; #BMK-2022). For the antigen retrieval, the sections were placed in double-boiled antigen retrieval buffer (10 mM Sodium citrate (pH 6.0), 0.05% Tween-20) for 10 min and blocked in blocking solution with M.O.M. solution for 1 hr at room temperature. Sections were then incubated with Pax7 antibody in the blocking solution overnight at 4°C. After washing out primary antibodies with PBST, sections were incubated with biotinylated anti-mouse IgG reagent (provided in the kit) in the blocking solution for 15 min at room temperature. Sections were then washed and incubated with secondary antibodies conjugated with streptavidin for 30 min at room temperature in the dark. Sections were washed with PBST and mounted with FluoroSave (Calbiochem; # 345789). Imaging was performed with an AxioObserver Z1 epifluorescence microscope (Carl Zeiss) equipped with a Hamamatsu Orca-R2 CCD camera. The cross-sectional area of centrally nucleated muscle fiber was quantified using the contour functions (findContours, contourArea, and arcLength) of the opensource Python package, OpenCV (https://pypi.org/project/opencv-python/).
MuSC Transplantation
Freshly isolated MuSCs from each donor group were transduced with luciferase reporters which was subcloned into a third generation HIV-1 lentiviral vector (CD51X DPS, SystemBio). For transduction, cells which were incubated with concentrated virus and polybrene (Santa Cruz Biotechnology; # sc-134220) in 6 well plates, which were spun for 1 hr at 2,550g at 4°C. Cells were then washed with fresh Ham’s F10 media three times, scraped from the plate in 1.2% BaCl2. Recipient NSG mice were injected with 10,000 cells resuspended in 50 μl of 1.2% BaCl2 under isoflurane anesthesia. To maximize the efficiency of transplantation of freshly isolated RFP-positive MuSCs, recipient NSG mice were prepared for transplantation with 1200 cGy of irradiation to the lower hindlimbs while under ketamine anesthesia. Then BaCl2 was injected into TA muscles, as described above, 3 days prior to transplantation. Freshly isolated RFP-positive MuSCs were pelleted and resuspended in PBS. Resuspended cells were injected into the TA muscles of recipient mice that were under isoflurane anesthesia. Two days after transplantation, RFP-positive MuSCs from the TA muscles were collected for FACS analysis.
Bioluminescence imaging
Bioluminescent imaging was performed using a Xenogen IVIS-Spectrum System (Caliper Life Sciences). Each mouse was intraperitoneally injected with 100 μl of sterile D-luciferin solution (50 mg/ml, Biosynth; #L-8220) under anesthesia with 2% isoflurane. Immediately after the injection, the mice were imaged with the IVIS Spectrum In Vivo Imaging System (Perkin Elmer). Imaging was taken for 1 min at maximum sensitivity at the highest resolution until the peak intensity of the bioluminescent signal began to diminish. Analysis of each image was performed using Living Image Software, version 4.0 (Caliper Life Sciences). The same sizes of manually generated circles were placed on top of the regions of interest (ROI) on the transplanted legs of recipient mouse. Similarly, background ROIs were placed on regions of mice outside the transplanted leg. Peak photon flux of bioluminescent in an ROI measured from each transplanted leg was normalized by photon flux in a background ROI measured at the same time point. The bioluminescence was monitored for 15–19 days as indicated. Every normalized ROI photon flux measured on different days was divided by a normalized ROI photon flux from day 1 post-transplantation.
Grip strength test
Grip strength was measured by using a grip test instrument (BIO-GS3; BIOSEB). Each mouse was allowed to grab the metal grid with four paws and gently pulled backward in the horizontal plane until the mouse releases the grasping tool. The force applied to the grid is recorded as the peak force just before the mouse loses the grip. Measurements were performed at least five times per mouse, and the values were averaged.
Gait analysis
A DigiGait apparatus (Mouse Specifics, Inc.) was used to assess gait parameters. Mice were placed individually on a motor-driven treadmill at 10 cm/s with a transparent treadmill belt in a small testing chamber. Mice that did not keep up with the treadmill speed were excluded from the analyses. A high-speed camera mounted below the belt captured digital video images of the underside of the mouse (80 frames/s). The frames were recorded using DigiGait Imager software (v. 12.3) and analyzed using the companion DigiGait Analysis program (v. 16.2).
Quantification and statistics
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications71. Sample size for each experiment is indicated in the corresponding figure legend. Data distribution was assumed to be normal but this was not formally tested. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. Any animals or data points were not excluded from the analyses for any reason. Statistical analyses were performed using Excel 2016 or GraphPad Prism 5.
Extended Data
Extended Data Fig. 1. Differential chromatin accessibility and gene expression in young and old MuSCs.
a, Gating strategy for fluorescence-activated cell sorting (FACS) isolation of muscle stem cells (MuSCs). Purity of isolated MuSCs is > 98% as assessed by staining for Pax7 of cells fixed one hour after plating. (Scale bar, 50 μm) b-d, Representative confocal immunofluorescence images of MuSCs of young and old female mice (Scale bar, 5 μm). RFUs of H3K9me3 (b), H3K9me2 (c), or HP1α (d) were normalized to fluorescence intensities of total H3 (n=4). e, Pearson correlation of ATAC-seq profiles from young and old MuSCs (n=2). A total of about 60,000 signal summits were found in each sample. f, Representative heat maps of ATAC-seq tag intensity 3kb around transcription start sites (TSSs) in young and old MuSCs. g, Read density within 3 kb upstream of TSSs and 3 kb downstream of TESs. h, Hierarchical clustering of peak enrichment patterns between young and old MuSCs. i, PCA plot of RNAseq data from freshly isolated young and old MuSCs. j, Volcano plots of differentially expressed genes (DEGs) between young and old MuSCs. Dashed lines indicate fold-change (log2FC>0.5) and p-value cut-offs (Padj < 0.05). Total 1,641 DEGs were found. Among the DEGs, 866 genes were highly expressed in old MuSCs, and 775 genes were highly expressed in young MuSCs. k, Normalized DESeq read counts of methyltransferases for H3K9 (Suv39h1, Ehmt1, Ehmt2, Setdb1, and Setdb2). (n=4) l, Normalized DESeq read counts of demethylases for H3K9 (Kdm3a, Kdm3b, Kdm4a, Kdm4b, Kdm4c, and Kdm7b). (n=4) DESeq normalization was conducted by using median-of-ratio method as described in the Methods. Data are shown as median and quartiles (b-d) and as mean SD (k, l). P values were calculated by two-sided unpaired Student’s t-tests (b-d, k, l). *P < 0.05; **P < 0.01; ***P < 0.001. Statistical details are provided in Source Data.
Extended Data Fig. 2. Restoration of intracellular SAM promotes heterochromatin formation and reduces susceptibility to DNA damage and cell death of old MuSCs.
a, Intracellular SAM content of MuSCs measured by SAM ELISA (n=4). b-d, Representative confocal immunofluorescence images of MuSCs (Scale bar, 5 μm). RFUs of H3K9me3 (b), H3K9me2 (c), or HP1α (d) were normalized to RFUs of total H3 (n=4). e-f, Representative confocal immunofluorescence images of old MuSCs treated as indicated (Scale bar, 5 μm). RFUs of H3K9me3 (e) or HP1α (f) were normalized to RFUs of total H3 (n=4). g, Representative heat maps of ATAC-seq tag intensity 3 kb around TSSs in vehicle- or SAM-treated old MuSCs. h, Read density within 3 kb upstream of TSSs and 3 kb downstream of the TESs. i, Hierarchical clustering of peak enrichment patterns. j, (Top) Representative confocal immunofluorescence images of γ-H2AX foci (Scale bar, 5 μm). (Bottom) Quantification of the number of γ-H2AX foci per cell (n=3). k, (Left) Representative confocal images of γ-H2AX foci. (Scale bar, 5 μm) (Right) The ratio of γ-H2AX foci per cell was quantified from old MuSCs treated as indicated (n=4). l, FACS analysis of propidium iodide (PI)-positive MuSCs. A day after the administration of 7 Gy γ-irradiation to the hind limbs, MuSCs were isolated from young, old, or old mice treated with SAM, and subsequently cultured for 2 days. m, The ratio of apoptotic cells over total cells was quantified by TUNEL assay and plotted (n=4). n-p, Relative fluorescence units (RFUs) of H3K9me3 (n), H3K9me2 (o), and HP1α (p) were normalized to RFUs of total H3 (n=3). q, Human MuSCs were treated with SAM for 2 days, and 0.75 μM of doxorubicin was added and incubated for another day. TUNEL positive cells were quantified (n=3). Data are shown as mean SD (a, j, k, m, q) and as median and quartiles (b-f, n-p). P values were calculated by two-sided unpaired Student’s t-tests (a-f, j, k, m-q). *P < 0.05; **P < 0.01; ***P < 0.001.
Extended Data Fig. 3. Inhibiting spermidine synthesis promotes heterochromatin re-formation and reduces DNA damage in old MuSCs.
a, Diagram of spermidine metabolism. The synthesis of spermidine and spermine requires dcSAM as aminopropyl donor, which is produced from SAM by AMD1. b, Representative Western Blots of SRM, PAOX, SAT1, and α-tubulin in MuSCs which were freshly isolated form young and old mice (n=3). c-e, Band intensities of SRM (c), PAOX (d), and SAT1 (e) were normalized to the level of α-tubulin (n=3). f, Quantification of RFUs of intracellular spermidine in MuSCs from vehicle- or MCHA-treated old mice (n=4). g, Intracellular SAM content of MuSCs measured by SAM ELISA (n=4). h-j, Representative confocal immunofluorescence images of MuSCs of old female mice treated with vehicle or MCHA (Scale bar, 5 μm). RFUs of H3K9me3 (h), H3K9me2 (i), or HP1α (j) were normalized to RFUs of total H3 (n=4). k, Representative western blots of H3K9me3, H3K9me2, H3K9me1, HP1α, and total H3 of MuSCs (n=3). l-o, Band intensities of H3K9me3 (l), H3K9me2 (m), HP1α (n), and H3K9me1 (o) were normalized to the band intensities of total H3 (n=3). p, (Left) Representative transmission electron microscopy images of MuSCs on EDL sections. (Right) The percentage of heterochromatin cross sectional area over total cross-sectional area of nucleus is quantified (n=15 cells examined over 3 independent young mice treated with vehicle, n=14 cells examined over 3 independent young mice treated with MCHA). The box represents the interquartile range, with the lower and upper hinges indicating the 25th and 75th percentiles, respectively. The horizontal line inside the box marks the median score. The whiskers extend to the minimum and maximum values. q, Representative heat maps of ATAC-seq tag intensity 3kb around TSSs. r, Read density within 3 kb upstream of TSSs and 3 kb downstream of the TESs. s, Hierarchical clustering of peak enrichment patterns. t, (Left) Representative confocal immunofluorescence images of γ-H2AX foci. (Right) Quantification of the number of γ-H2AX foci per cell (n=3). Data are shown as mean SD (c-g, l-o, t) and as median and quartiles (h-j, p). P values were calculated by two-sided unpaired Student’s t-tests (c-j, l-p, t). *P < 0.05; **P < 0.01; ***P < 0.001.
Extended Data Fig. 4. Heterochromatin re-formation induced by restoration of SAM reduces susceptibility to genotoxic stress and cell death of old MuSCs.
a, Quantification of RFUs of intracellular SAM (n=4). b, Quantification of RFUs of intracellular putrescine. Freshly isolated old MuSCs were treated with vehicle, putrescine, or ornithine for 48 hr (n=4). c, After treating old MuSCs as described in (b), doxorubicin was added to the media, and the cells were cultured for another day. (Left) Representative confocal images of γ-H2AX foci. (Scale bar, 5 μm) (Right) The ratio of γ-H2AX foci per cell (n=4). d, Old MuSCs were treated as described in (c). The ratio of apoptotic cells over total cells was quantified by TUNEL assay (n=4). e, Quantification of mean RFUs of intracellular spermidine (n=4). f, Quantification of intracellular SAM measured by SAM ELISA (n=4). g, Representative western blots of SRM and α-tubulin in old MuSCs (n=3). h, Band intensity of SRM was normalized to band intensity of α-tubulin (n=3). i-j, Old MuSCs transfected with siControl or siSrm were stained with antibodies against SAM (i) or spermidine (j). Mean RFUs of intracellular SAM or spermidine in each group of cells were quantified (n=3 for SAM staining, n=4 for spermidine staining). k, Old MuSCs were transfected with siControl or siSrm and treated with vehicle, UNC0642, or Chaetocin for 24 hr as indicated. (Left) Representative confocal immunofluorescence images of γ-H2AX foci (Scale bar, 5 μm). (Right) Quantification of the number of γ-H2AX foci per cell (n=4). l, Quantification of the ratio of TUNEL positive cells over total cells (n=4). m-n, Freshly isolated human MuSCs were treated as indicated for 48 hr. RFUs of H3K9me3 (m) or HP1α (n) were normalized to RFUs of total H3 (n=3). o, Human MuSCs treated as described in (m-n) were additionally treated with doxorubicin for 24 hr. (Left) Representative confocal images (Scale bar, 5 μm). (Right) Quantification of the number of γ-H2AX foci per cell (n=3). p, The human MuSCs treated as described in (o) were subjected to TUNEL assay. The ratio of TUNEL positive cells over total cells was quantified (n=3). Data are shown as mean SD (a-d, f, h-l, o, p) and as median and quartiles (e, m, n). P values were calculated by two-sided unpaired Student’s t-tests (a-f, h-p). *P < 0.05; **P < 0.01; ***P < 0.001.
Extended Data Fig. 5. Inhibiting spermidine synthesis does not impair autophagy of aged MuSCs.
a-b, Quantification of intracellular spermidine levels. Violin plots showing RFUs of spermidine in old MuSCs treated with each dose of spermidine (a) or MCHA (b) as indicated (n=3). c-d, (Left) Representative confocal immunofluorescence images of old MuSCs treated with vehicle or each dose of spermidine (c) or MCHA (d) for 48 hr. In each case, cells were treated with or without 50 μM of chloroquine (CQ) for the last 3 hr of incubation. (Right) Quantification of number of LC3B foci per cell. (Scale bar, 10 μM) Data are shown as median and quartiles (a, b) and mean ± SD (c, d). P values were calculated by two-sided unpaired Student’s t-tests (a-d). *P < 0.05; **P < 0.01; ***P < 0.001.
Extended Data Fig. 6. Restoration of SAM and H3K9 methylation improves in vivo potency of old MuSCs to regenerate new muscle.
a-b, Representative images of bioluminescence of vehicle- or SAM-treated MuSCs (a) and vehicle- or MCHA-treated old MuSCs (b) which had been transplanted into muscles of NSG mice. A day after transplantation is Day 1 (n=6). c, Representative FACS plots for analysis of RFP-positive MuSCs isolated from recipient NSG mice. Purity of isolated RFP-positive MuSCs is > 98% as assessed by staining for Pax7 of cells (Scale bar, 50 μm). d, Old MuSCs were transduced with shRNA containing lentiviruses as indicated. After 48 hr of culture, the cells were subjected to western blotting (n=4). e, Band intensity of Srm was normalized to the band intensity of α-tubulin (n=4). f-g, Old MuSCs transduced with shControl or shSrm were stained with antibodies against SAM (f) or spermidine (g). Mean RFUs of intracellular SAM or spermidine were quantified (n=3). h, Representative BLI images taken from Day 1 to Day 15. MuSCs transduced with shControl or shSrm were transplanted into muscles of NSG mice (n=6). i, Paired comparisons of relative bioluminescence flux measured from the right and left muscles of each mouse eight days after transplantation. j, Western blot analysis of SRM and SUV39H1 in MuSCs. Old MuSCs were transduced with shRNA containing lentiviruses as indicated. After 48 hr of transduction, the cells were subjected to western blotting (n=3). k, Representative BLI images of MuSCs transduced with shSrm or shSrm with shSuv39h1 which had been transplanted into muscles of NSG mice (n=8). l, Paired comparisons of relative bioluminescence flux measured from the right and left muscles of each mouse ten days after transplantation. m, Schematic of transplantation. n, (Left) Quantified results of BLI measured at different time points from 60 to 71 days following transplantation (n=6). (Right) Representative images of bioluminescence captured 71 days after transplantation. Data are shown as mean SD (e-g). P values were calculated by two-sided unpaired Student’s t-tests (e-g) and by one-sided student’s t test, pairwise between groups (i, l, n). *P < 0.05; **P < 0.01; ***P < 0.001.
Extended Data Fig. 7. Restoration of SAM is associated with improved muscle regeneration and function of aged mice.
a, MuSCs were isolated from the injured muscles of female mice at 2 days post injury (dpi). (Left) Representative confocal images. (Scale bar, 5 μm) (Right) The ratio of γ-H2AX foci per cell was quantified (n=4 for young mice, and old mice treated with vehicle or SAM, n=3 for old mice treated with MCHA). b, (Left) Representative TUNEL assay images of female MuSCs isolated at 2dpi (Scale bar, 50 μm). (Right) The ratio of apoptotic cells over total cells was quantified (n=4 for young mice, and old mice treated with vehicle or SAM, n=3 for old mice treated with MCHA). c, (Left) The percentage of PI positive female MuSCs isolated at 2dpi (n=4 for young mice, and old mice treated with vehicle or SAM, n=3 for old mice treated with MCHA). (Right) The representative FACS plot. d-e, Frequency histogram of cross-sectional areas (CSAs) of centrally nucleated myofibers in TA muscle sections. TA muscles were harvested from old mice treated with vehicle, MCHA (d) or SAM (e) at 7 dpi (n=3). f, (Left) Representative immunostaining of regenerating muscle fibers from female mice (Scale bar, 100 μm). (Right) Quantification of the mean CSAs of myofibers with centrally located nuclei in TA muscle sections (n=4 for young and old female mice treated with vehicle, n=3 for old female mice treated with MCHA or SAM). g, (Left) Representative immunostaining of non-regenerating muscle fibers (Scale bar, 100 μm). (Right) Quantification of the mean CSAs of myofibers in TA muscle sections (n=3). h, (Top) Representative immunostaining of regenerating muscle fibers (Scale bar, 100 μm). (Bottom) Quantification of the mean CSAs of myofibers with centrally located nuclei in TA muscle sections (n=3). i, Representative tissue immunostaining images of TA muscle sections from young mice collected at 7 dpi (n=4, Scale bar, 100 μm). j, Quantification of the number of Pax7-positive MuSCs per each tissue section (n=4). k, Old mice were given vehicle or MTA in drinking water, and MuSCs from each mouse were subjected to SAM ELISA (n=4). l, (Top) Representative immunostaining of regenerating muscle fibers from old mice treated with vehicle or MTA (Scale bar, 100 μm). (Bottom) Quantification of the mean CSAs of myofibers with centrally located nuclei in TA muscle sections (n=3). m, Grip force measured without injury (n=4). n-o, Stance time (n) and stride time (o) were measured at 14 dpi (n=8 for young mice, n=7 for old mice treated with vehicle or SAM, n=6 for old mice treated with MCHA). Data are shown as mean SD (a-c, f-h, j-o). P values were calculated by two-sided unpaired Student’s t-tests (e-g) and by one-sided student’s t test, pairwise between groups (a-c, f-h, j-o). *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1A6A3A03031423) to J.K., and by funding from the Glenn Foundation for Medical Research and the NIH (P01AG036695, R01 AG068667, R01 AR073248, and AG071783) to T.A.R. Work using TEM described here was supported, in part, by NIH S10 Award (1S10OD028536-01), titled “OneView 4kX4k sCMOS camera for transmission electron microscopy applications” from the Office of Research Infrastructure Programs (ORIP). The RNA sequencing data was generated with instrumentation purchased with NIH funds (S10OD025212 and 1S10OD021763). LC-MS based metabolite quantification was supported by the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry. It utilized the Waters Xevo TQ-XS mass spectrometer system (RRID:SCR_018510) that was purchased with funding from National Institutes of Health Shared Instrumentation grant S10OD026962. Human biopsy samples were obtained from Donor Network West (DNW). We hereby express our thanks for the cooperation of DNW and all of tissue donors and their families, for giving the gift of life and the gift of knowledge, by their generous donation. Schematic images in figures were created with BioRender.com.
Footnotes
CODE AVAILABILITY
• This paper does not report original code.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AVAILABILITY
RNA Sequencing data have been deposited in NCBI’s Gene Expression Omnibus (GEO) with accession number GSE229853.
ATAC Sequencing data have been deposited in NCBI’s Gene Expression Omnibus (GEO) with accession number GSE229851.
GRCm39 genome is available in NCBI RefSeq (GCF_000001635.27-RS_2023_04).
All related source data is made available to the readers as part of the manuscript files.
REFERENCES
- 1.Benayoun BA, Pollina EA & Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol 16, 593–610 (2015). 10.1038/nrm4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villeponteau B. The heterochromatin loss model of aging. Exp Gerontol 32, 383–394 (1997). 10.1016/s0531-5565(96)00155-6 [DOI] [PubMed] [Google Scholar]
- 3.Ljungman M. & Hanawalt PC Efficient protection against oxidative DNA damage in chromatin. Mol Carcinog 5, 264–269 (1992). 10.1002/mc.2940050406 [DOI] [PubMed] [Google Scholar]
- 4.Wood JG & Helfand SL Chromatin structure and transposable elements in organismal aging. Front Genet 4, 274 (2013). 10.3389/fgene.2013.00274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larson K. et al. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet 8, e1002473 (2012). 10.1371/journal.pgen.1002473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smeal T, Claus J, Kennedy B, Cole F. & Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84, 633–642 (1996). 10.1016/s0092-8674(00)81038-7 [DOI] [PubMed] [Google Scholar]
- 7.Wasserzug-Pash P. et al. Loss of heterochromatin and retrotransposon silencing as determinants in oocyte aging. Aging Cell 21, e13568 (2022). 10.1111/acel.13568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scaffidi P. & Misteli T. Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063 (2006). 10.1126/science.1127168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JH, Kim EW, Croteau DL & Bohr VA Heterochromatin: an epigenetic point of view in aging. Exp Mol Med 52, 1466–1474 (2020). 10.1038/s12276-020-00497-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi S. et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell 11, 483–504 (2020). 10.1007/s13238-020-00728-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L. et al. Hairless regulates heterochromatin maintenance and muscle stem cell function as a histone demethylase antagonist. Proc Natl Acad Sci U S A 118 (2021). 10.1073/pnas.2025281118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang C. et al. Stabilization of heterochromatin by CLOCK promotes stem cell rejuvenation and cartilage regeneration. Cell Res 31, 187–205 (2021). 10.1038/s41422-020-0385-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger SL & Sassone-Corsi P. Metabolic Signaling to Chromatin. Cold Spring Harb Perspect Biol 8 (2016). 10.1101/cshperspect.a019463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su X, Wellen KE & Rabinowitz JD Metabolic control of methylation and acetylation. Curr Opin Chem Biol 30, 52–60 (2016). 10.1016/j.cbpa.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang PK et al. S-Adenosylmethionine and methylation. FASEB J 10, 471–480 (1996). [PubMed] [Google Scholar]
- 16.Mews P. et al. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381–386 (2017). 10.1038/nature22405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukada Y. et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006). 10.1038/nature04433 [DOI] [PubMed] [Google Scholar]
- 18.Carey BW, Finley LW, Cross JR, Allis CD & Thompson CB Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416 (2015). 10.1038/nature13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiraki N. et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab 19, 780–794 (2014). 10.1016/j.cmet.2014.03.017 [DOI] [PubMed] [Google Scholar]
- 20.Yucel N. et al. Glucose Metabolism Drives Histone Acetylation Landscape Transitions that Dictate Muscle Stem Cell Function. Cell Rep 27, 3939–3955 e3936 (2019). 10.1016/j.celrep.2019.05.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunet A, Goodell MA & Rando TA Ageing and rejuvenation of tissue stem cells and their niches. Nat Rev Mol Cell Biol 24, 45–62 (2023). 10.1038/s41580-022-00510-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodell MA & Rando TA Stem cells and healthy aging. Science 350, 1199–1204 (2015). 10.1126/science.aab3388 [DOI] [PubMed] [Google Scholar]
- 23.Liu B, Qu J, Zhang W, Izpisua Belmonte JC & Liu GH A stem cell aging framework, from mechanisms to interventions. Cell Rep 41, 111451 (2022). 10.1016/j.celrep.2022.111451 [DOI] [PubMed] [Google Scholar]
- 24.Conboy IM & Rando TA Aging, stem cells and tissue regeneration: lessons from muscle. Cell Cycle 4, 407–410 (2005). 10.4161/cc.4.3.1518 [DOI] [PubMed] [Google Scholar]
- 25.Liu L. et al. Impaired Notch Signaling Leads to a Decrease in p53 Activity and Mitotic Catastrophe in Aged Muscle Stem Cells. Cell Stem Cell 23, 544–556 e544 (2018). 10.1016/j.stem.2018.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porpiglia E. et al. Elevated CD47 is a hallmark of dysfunctional aged muscle stem cells that can be targeted to augment regeneration. Cell Stem Cell 29, 1653–1668 e1658 (2022). 10.1016/j.stem.2022.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernando-Herraez I. et al. Ageing affects DNA methylation drift and transcriptional cell-to-cell variability in mouse muscle stem cells. Nat Commun 10, 4361 (2019). 10.1038/s41467-019-12293-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L. et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep 4, 189–204 (2013). 10.1016/j.celrep.2013.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X. et al. The loss of heterochromatin is associated with multiscale three-dimensional genome reorganization and aberrant transcription during cellular senescence. Genome Res 31, 1121–1135 (2021). 10.1101/gr.275235.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamin DI et al. Multiomics reveals glutathione metabolism as a driver of bimodality during stem cell aging. Cell Metab (2023). 10.1016/j.cmet.2023.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F. et al. Discovery of an in vivo chemical probe of the lysine methyltransferases G9a and GLP. J Med Chem 56, 8931–8942 (2013). 10.1021/jm401480r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greiner D, Bonaldi T, Eskeland R, Roemer E. & Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol 1, 143–145 (2005). 10.1038/nchembio721 [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi H. & Sato S. Fractionation of nonhistone proeins on a column of daunomycin-CH-Sepharose 4B. Biochim Biophys Acta 532, 113–121 (1978). 10.1016/0005-2795(78)90454-3 [DOI] [PubMed] [Google Scholar]
- 34.Haws SA et al. Methyl-Metabolite Depletion Elicits Adaptive Responses to Support Heterochromatin Stability and Epigenetic Persistence. Mol Cell 78, 210–223 e218 (2020). 10.1016/j.molcel.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtani M. et al. Spermidine regulates insulin synthesis and cytoplasmic Ca(2+) in mouse beta-TC6 insulinoma cells. Cell Struct Funct 34, 105–113 (2009). 10.1247/csf.09008 [DOI] [PubMed] [Google Scholar]
- 36.Massaro C, Thomas J. & Phanstiel Iv O. Investigation of Polyamine Metabolism and Homeostasis in Pancreatic Cancers. Med Sci (Basel) 5 (2017). 10.3390/medsci5040032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabor H. The protective effect of spermine and other polyamines against heat denaturation of deoxyribonucleic acid. Biochemistry 1, 496–501 (1962). 10.1021/bi00909a021 [DOI] [PubMed] [Google Scholar]
- 38.Eisenberg T. et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11, 1305–1314 (2009). 10.1038/ncb1975 [DOI] [PubMed] [Google Scholar]
- 39.Agrimi G. et al. Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem J 379, 183–190 (2004). 10.1042/BJ20031664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y. & Locasale JW Rethinking the bioavailability and cellular transport properties of S-adenosylmethionine. Cell Stress 6, 1–5 (2022). 10.15698/cst2022.01.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu SE, Huskey WP, Borchardt RT & Schowen RL Chiral instability at sulfur of S-adenosylmethionine. Biochemistry 22, 2828–2832 (1983). 10.1021/bi00281a009 [DOI] [PubMed] [Google Scholar]
- 42.Albers E. Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5’-methylthioadenosine. IUBMB Life 61, 1132–1142 (2009). 10.1002/iub.278 [DOI] [PubMed] [Google Scholar]
- 43.Mentch SJ et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab 22, 861–873 (2015). 10.1016/j.cmet.2015.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu W. et al. One-Carbon Metabolism Supports S-Adenosylmethionine and Histone Methylation to Drive Inflammatory Macrophages. Mol Cell 75, 1147–1160 e1145 (2019). 10.1016/j.molcel.2019.06.039 [DOI] [PubMed] [Google Scholar]
- 45.Dai Z, Mentch SJ, Gao X, Nichenametla SN & Locasale JW Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat Commun 9, 1955 (2018). 10.1038/s41467-018-04426-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadhu MJ et al. Nutritional control of epigenetic processes in yeast and human cells. Genetics 195, 831–844 (2013). 10.1534/genetics.113.153981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torano EG, Garcia MG, Fernandez-Morera JL, Nino-Garcia P. & Fernandez AF The Impact of External Factors on the Epigenome: In Utero and over Lifetime. Biomed Res Int 2016, 2568635 (2016). 10.1155/2016/2568635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hyun K, Jeon J, Park K. & Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med 49, e324 (2017). 10.1038/emm.2017.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan YH et al. The critical roles of polyamines in regulating ColE7 production and restricting ColE7 uptake of the colicin-producing Escherichia coli. J Biol Chem 281, 13083–13091 (2006). 10.1074/jbc.M511365200 [DOI] [PubMed] [Google Scholar]
- 50.Rato C, Amirova SR, Bates DG, Stansfield I. & Wallace HM Translational recoding as a feedback controller: systems approaches reveal polyamine-specific effects on the antizyme ribosomal frameshift. Nucleic Acids Res 39, 4587–4597 (2011). 10.1093/nar/gkq1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang C. et al. Defining the molecular requirements for the selective delivery of polyamine conjugates into cells containing active polyamine transporters. J Med Chem 46, 5129–5138 (2003). 10.1021/jm030223a [DOI] [PubMed] [Google Scholar]
- 52.Fan J. et al. Spermidine coupled with exercise rescues skeletal muscle atrophy from D-gal-induced aging rats through enhanced autophagy and reduced apoptosis via AMPK-FOXO3a signal pathway. Oncotarget 8, 17475–17490 (2017). 10.18632/oncotarget.15728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luchessi AD et al. Involvement of eukaryotic translation initiation factor 5A (eIF5A) in skeletal muscle stem cell differentiation. J Cell Physiol 218, 480–489 (2009). 10.1002/jcp.21619 [DOI] [PubMed] [Google Scholar]
- 54.Machado L. et al. Tissue damage induces a conserved stress response that initiates quiescent muscle stem cell activation. Cell Stem Cell 28, 1125–1135 e1127 (2021). 10.1016/j.stem.2021.01.017 [DOI] [PubMed] [Google Scholar]
- 55.Park MH The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J Biochem 139, 161–169 (2006). 10.1093/jb/mvj034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uchitomi R. et al. Metabolomic Analysis of Skeletal Muscle in Aged Mice. Sci Rep 9, 10425 (2019). 10.1038/s41598-019-46929-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishimura K, Shiina R, Kashiwagi K. & Igarashi K. Decrease in polyamines with aging and their ingestion from food and drink. J Biochem 139, 81–90 (2006). 10.1093/jb/mvj003 [DOI] [PubMed] [Google Scholar]
- 58.Minois N, Carmona-Gutierrez D. & Madeo F. Polyamines in aging and disease. Aging (Albany NY) 3, 716–732 (2011). 10.18632/aging.100361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casero RA Jr., Murray Stewart T. & Pegg AE Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer 18, 681–695 (2018). 10.1038/s41568-018-0050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murray-Stewart TR, Woster PM & Casero RA Jr. Targeting polyamine metabolism for cancer therapy and prevention. Biochem J 473, 2937–2953 (2016). 10.1042/BCJ20160383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewandowski NM et al. Polyamine pathway contributes to the pathogenesis of Parkinson disease. Proc Natl Acad Sci U S A 107, 16970–16975 (2010). 10.1073/pnas.1011751107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polis B, Karasik D. & Samson AO Alzheimer’s disease as a chronic maladaptive polyamine stress response. Aging (Albany NY) 13, 10770–10795 (2021). 10.18632/aging.202928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morrison LD, Becker L, Ang LC & Kish SJ Polyamines in human brain: regional distribution and influence of aging. J Neurochem 65, 636–642 (1995). 10.1046/j.1471-4159.1995.65020636.x [DOI] [PubMed] [Google Scholar]
- 64.Graham SF et al. Untargeted metabolomic analysis of human plasma indicates differentially affected polyamine and L-arginine metabolism in mild cognitive impairment subjects converting to Alzheimer’s disease. PLoS One 10, e0119452 (2015). 10.1371/journal.pone.0119452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ost M. et al. Muscle mitohormesis promotes cellular survival via serine/glycine pathway flux. FASEB J 29, 1314–1328 (2015). 10.1096/fj.14-261503 [DOI] [PubMed] [Google Scholar]
- 66.Cosgrove BD et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med 20, 255–264 (2014). 10.1038/nm.3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price FD et al. Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nat Med 20, 1174–1181 (2014). 10.1038/nm.3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu L, Cheung TH, Charville GW & Rando TA Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat Protoc 10, 1612–1624 (2015). 10.1038/nprot.2015.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charville GW et al. Ex Vivo Expansion and In Vivo Self-Renewal of Human Muscle Stem Cells. Stem Cell Reports 5, 621–632 (2015). 10.1016/j.stemcr.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brett JO et al. Exercise rejuvenates quiescent skeletal muscle stem cells in old mice through restoration of Cyclin D1. Nat Metab 2, 307–317 (2020). 10.1038/s42255-020-0190-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA Sequencing data have been deposited in NCBI’s Gene Expression Omnibus (GEO) with accession number GSE229853.
ATAC Sequencing data have been deposited in NCBI’s Gene Expression Omnibus (GEO) with accession number GSE229851.
GRCm39 genome is available in NCBI RefSeq (GCF_000001635.27-RS_2023_04).
All related source data is made available to the readers as part of the manuscript files.