Abstract
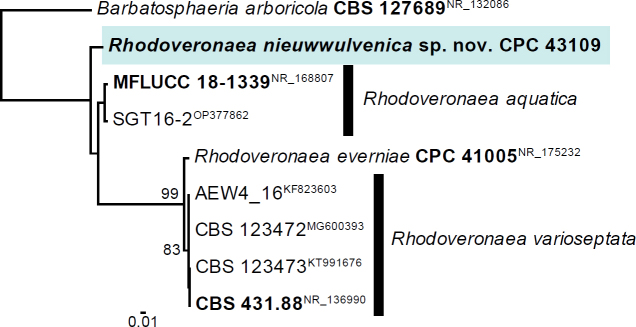
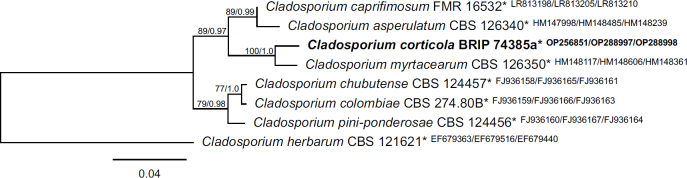
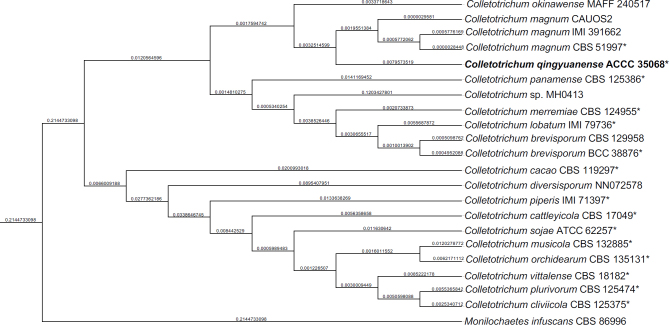
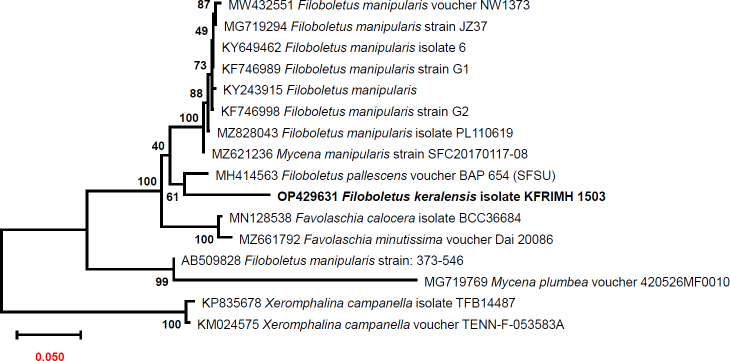
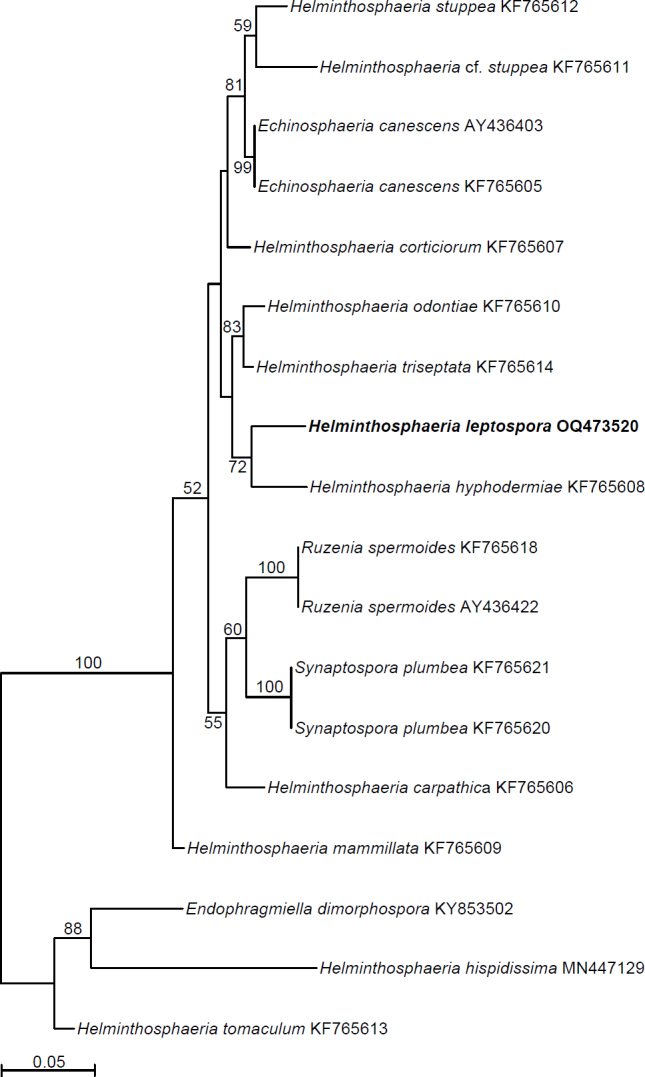
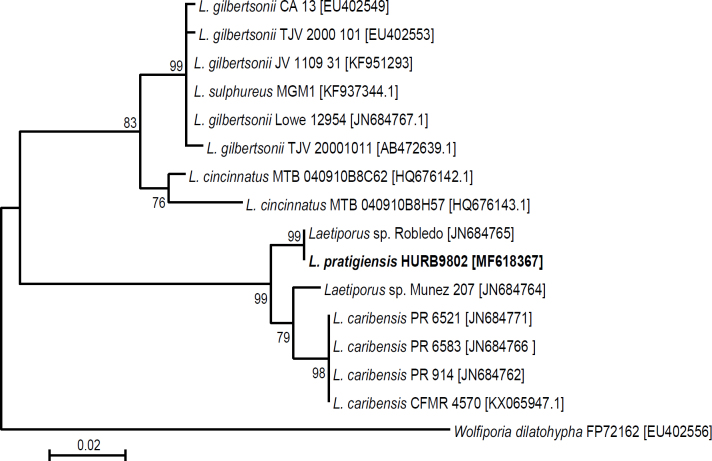
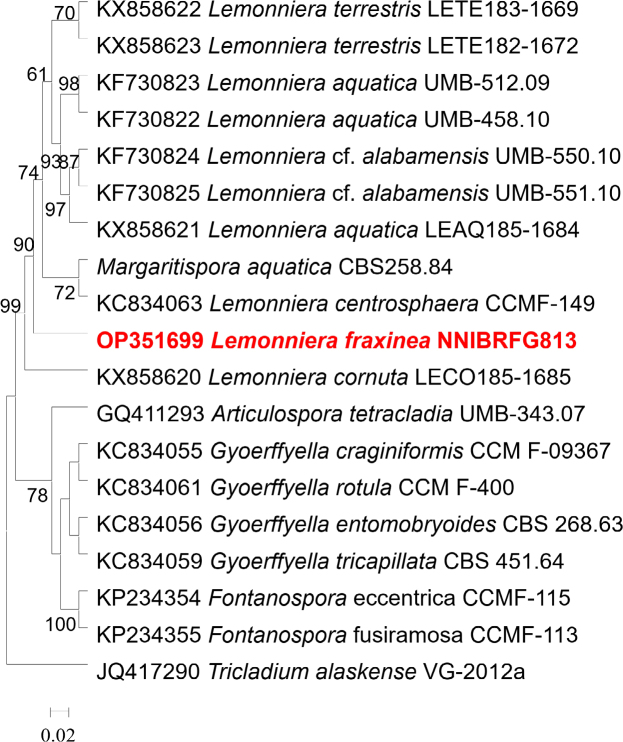
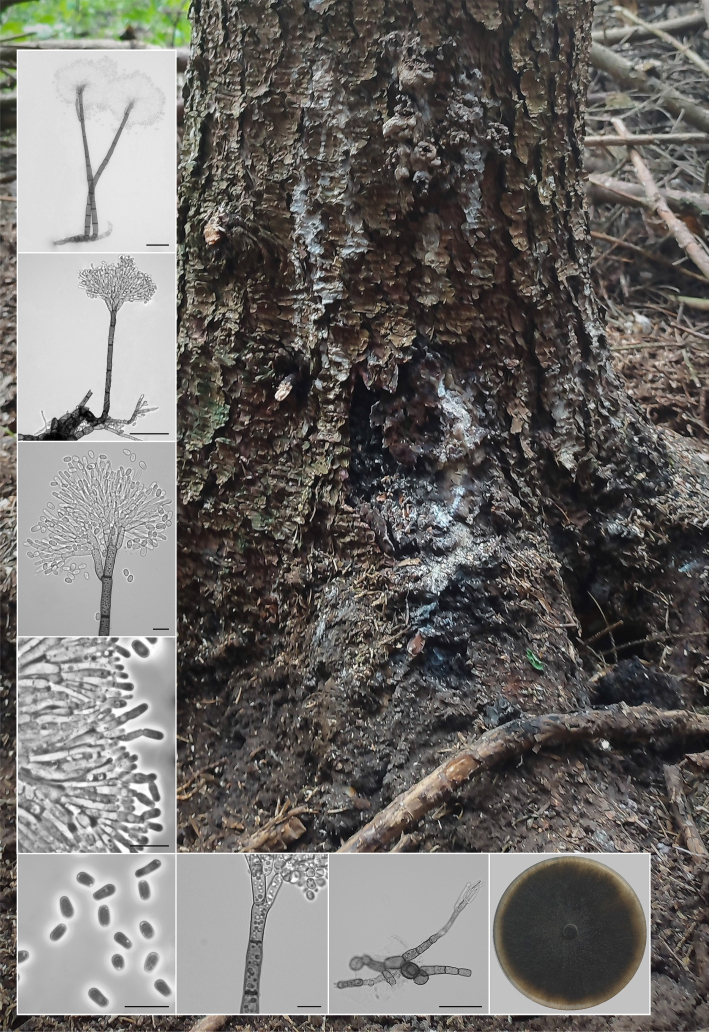
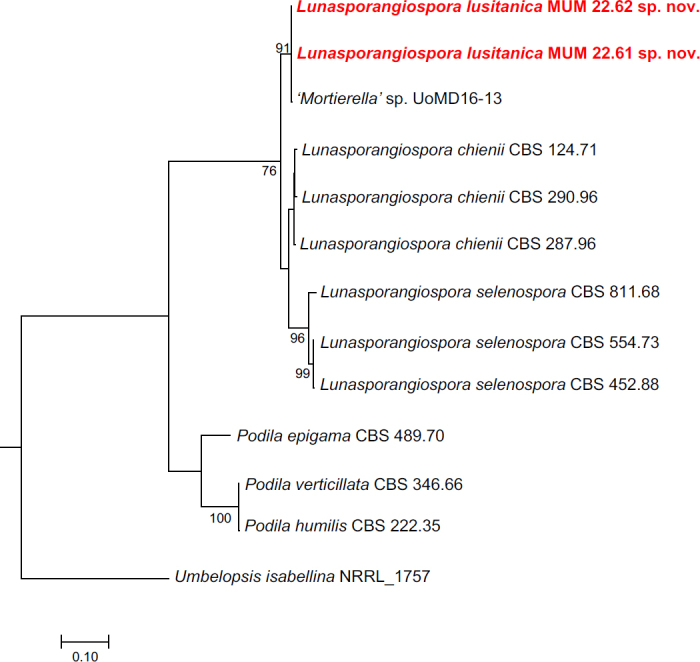
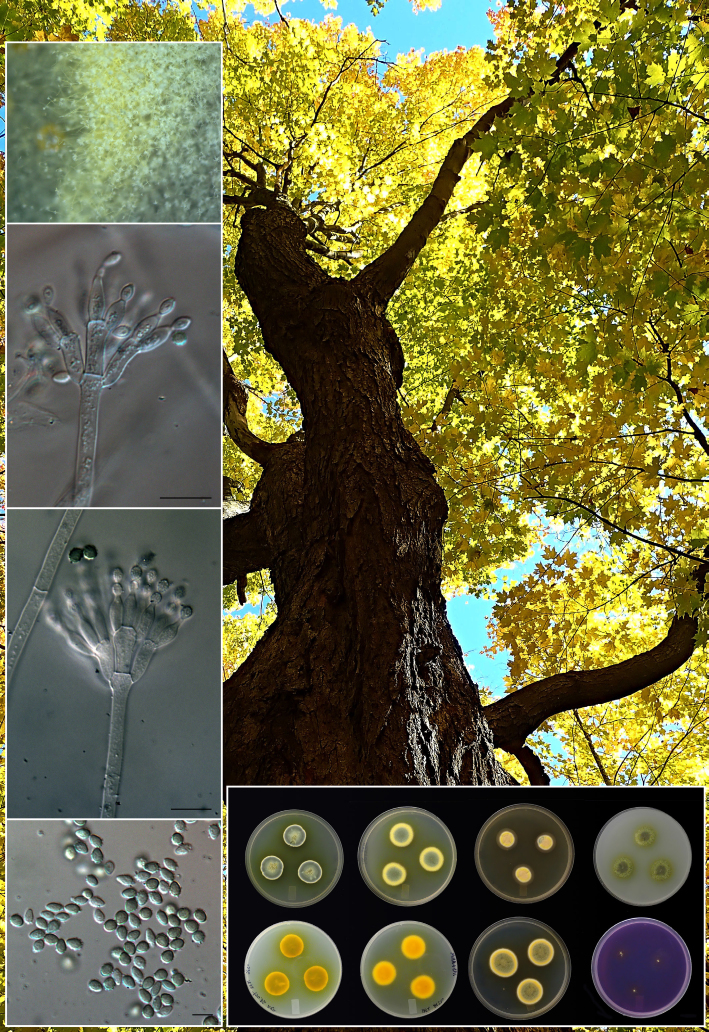
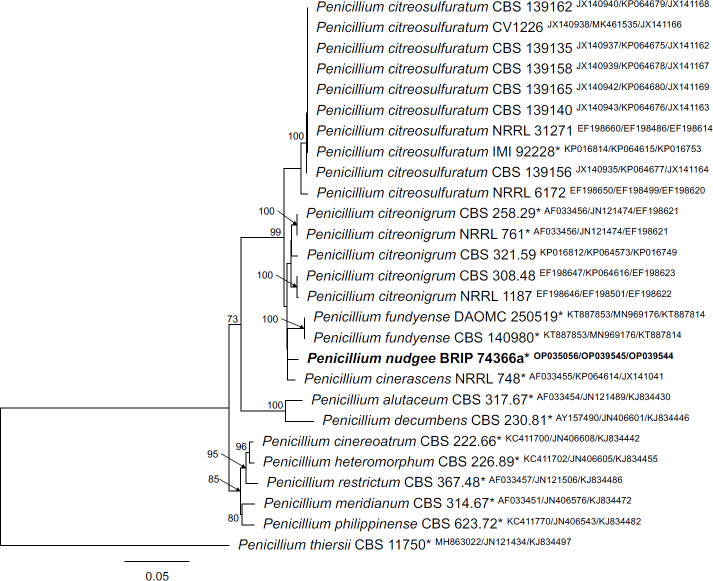
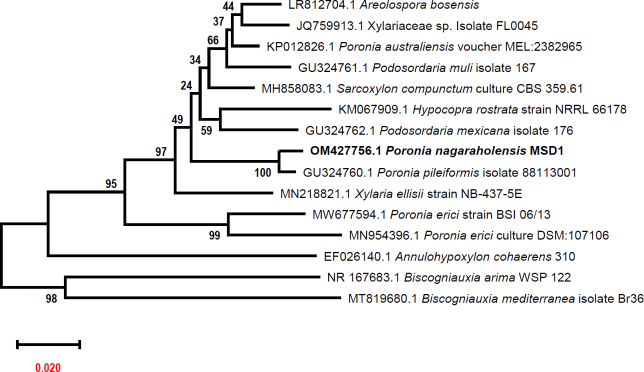
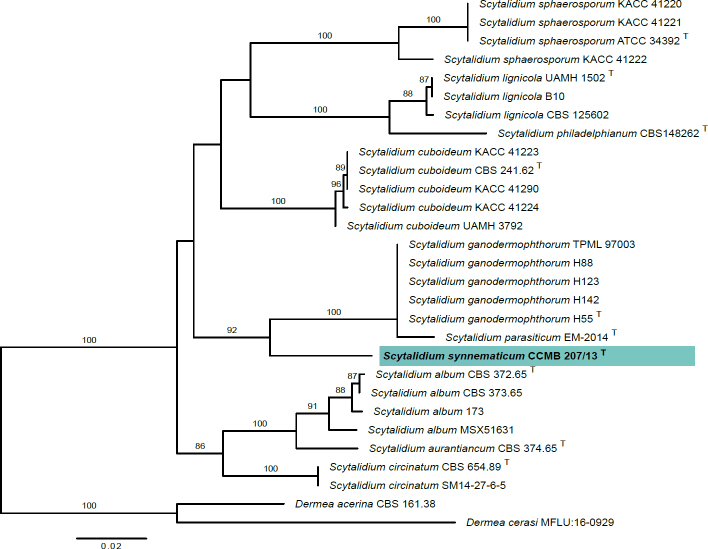
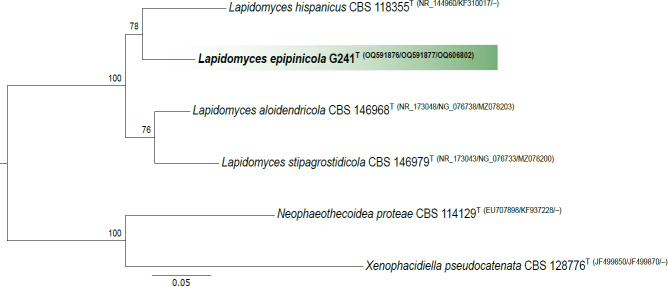
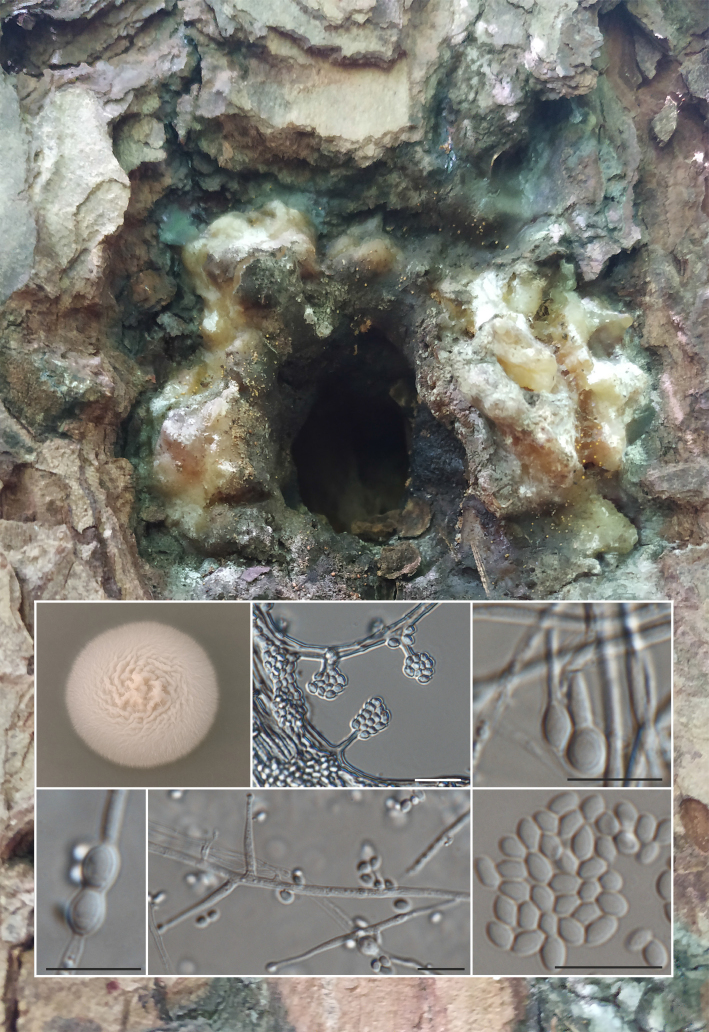
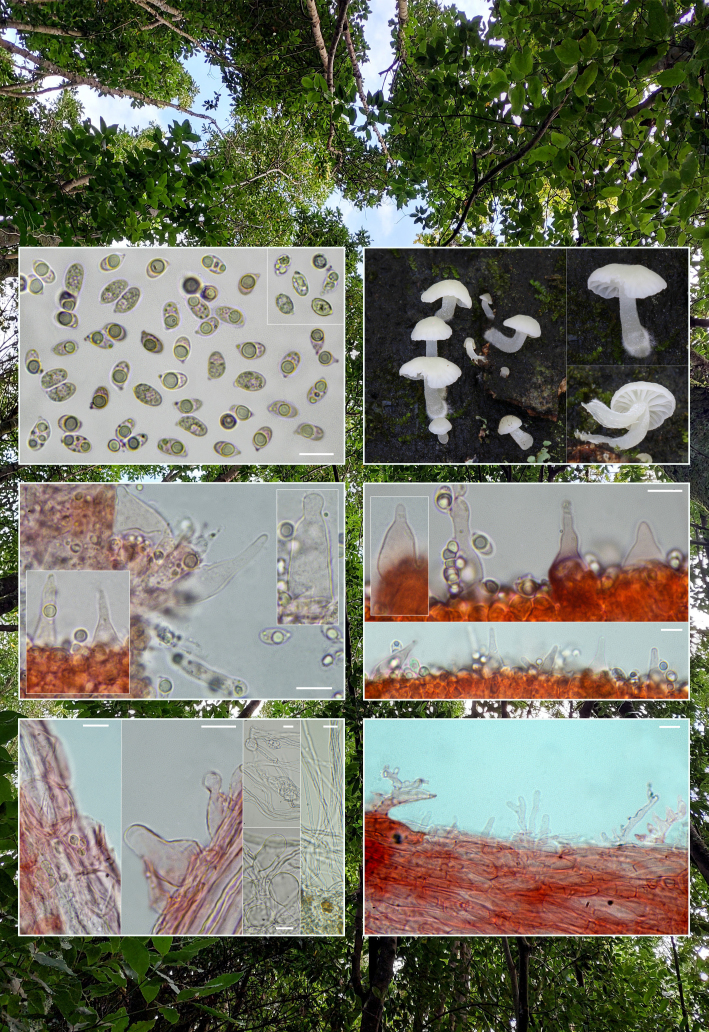
Novel species of fungi described in this study include those from various countries as follows: Australia, Aschersonia mackerrasiae on whitefly, Cladosporium corticola on bark of Melaleuca quinquenervia, Penicillium nudgee from soil under Melaleuca quinquenervia, Pseudocercospora blackwoodiae on leaf spot of Persoonia falcata, and Pseudocercospora dalyelliae on leaf spot of Senna alata. Bolivia, Aspicilia lutzoniana on fully submersed siliceous schist in high-mountain streams, and Niesslia parviseta on the lower part and apothecial discs of Erioderma barbellatum on a twig. Brazil, Cyathus bonsai on decaying wood, Geastrum albofibrosum from moist soil with leaf litter, Laetiporus pratigiensis on a trunk of a living unknown hardwood tree species, and Scytalidium synnematicum on dead twigs of unidentified plant. Bulgaria, Amanita abscondita on sandy soil in a plantation of Quercus suber. Canada, Penicillium acericola on dead bark of Acer saccharum, and Penicillium corticola on dead bark of Acer saccharum. China, Colletotrichum qingyuanense on fruit lesion of Capsicum annuum. Denmark, Helminthosphaeria leptospora on corticioid Neohypochnicium cremicolor. Ecuador (Galapagos), Phaeosphaeria scalesiae on Scalesia sp. Finland, Inocybe jacobssonii on calcareous soils in dry forests and park habitats. France, Cortinarius rufomyrrheus on sandy soil under Pinus pinaster, and Periconia neominutissima on leaves of Poaceae. India, Coprinopsis fragilis on decaying bark of logs, Filoboletus keralensis on unidentified woody substrate, Penicillium sankaranii from soil, Physisporinus tamilnaduensis on the trunk of Azadirachta indica, and Poronia nagaraholensis on elephant dung. Iran, Neosetophoma fici on infected leaves of Ficus elastica. Israel, Cnidariophoma eilatica (incl. Cnidariophoma gen. nov.) from Stylophora pistillata. Italy, Lyophyllum obscurum on acidic soil. Namibia, Aureobasidium faidherbiae on dead leaf of Faidherbia albida, and Aureobasidium welwitschiae on dead leaves of Welwitschia mirabilis. Netherlands, Gaeumannomycella caricigena on dead culms of Carex elongata, Houtenomyces caricicola (incl. Houtenomyces gen. nov.) on culms of Carex disticha, Neodacampia ulmea (incl. Neodacampia gen. nov.) on branch of Ulmus laevis, Niesslia phragmiticola on dead standing culms of Phragmites australis, Pseudopyricularia caricicola on culms of Carex disticha, and Rhodoveronaea nieuwwulvenica on dead bamboo sticks. Norway, Arrhenia similis half-buried and moss-covered pieces of rotting wood in grass-grown path. Pakistan, Mallocybe ahmadii on soil. Poland, Beskidomyces laricis (incl. Beskidomyces gen. nov.) from resin of Larix decidua ssp. polonica, Lapidomyces epipinicola from sooty mould community on Pinus nigra, and Leptographium granulatum from a gallery of Dendroctonus micans on Picea abies. Portugal, Geoglossum azoricum on mossy areas of laurel forest areas planted with Cryptomeria japonica, and Lunasporangiospora lusitanica from a biofilm covering a biodeteriorated limestone wall. Qatar, Alternaria halotolerans from hypersaline sea water, and Alternaria qatarensis from water sample collected from hypersaline lagoon. South Africa, Alfaria thamnochorti on culm of Thamnochortus fraternus, Knufia aloeicola on Aloe gariepensis, Muriseptatomyces restionacearum (incl. Muriseptatomyces gen. nov.) on culms of Restionaceae, Neocladosporium arctotis on nest of cases of bag worm moths (Lepidoptera, Psychidae) on Arctotis auriculata, Neodevriesia scadoxi on leaves of Scadoxus puniceus, Paraloratospora schoenoplecti on stems of Schoenoplectus lacustris, Tulasnella epidendrea from the roots of Epidendrum × obrienianum, and Xenoidriella cinnamomi (incl. Xenoidriella gen. nov.) on leaf of Cinnamomum camphora. South Korea, Lemonniera fraxinea on decaying leaves of Fraxinus sp. from pond. Spain, Atheniella lauri on the bark of fallen trees of Laurus nobilis, Halocryptovalsa endophytica from surface-sterilised, asymptomatic roots of Salicornia patula, Inocybe amygdaliolens on soil in mixed forest, Inocybe pityusarum on calcareous soil in mixed forest, Inocybe roseobulbipes on acidic soils, Neonectria borealis from roots of Vitis berlandieri × Vitis rupestris, Sympoventuria eucalyptorum on leaves of Eucalyptus sp., and Tuber conchae from soil. Sweden, Inocybe bidumensis on calcareous soil. Thailand, Cordyceps sandindaengensis on Lepidoptera pupa, buried in soil, Ophiocordyceps kuchinaraiensis on Coleoptera larva, buried in soil, and Samsoniella winandae on Lepidoptera pupa, buried in soil. Taiwan region (China), Neophaeosphaeria livistonae on dead leaf of Livistona rotundifolia. Türkiye, Melanogaster anatolicus on clay loamy soils. UK, Basingstokeomyces allii (incl. Basingstokeomyces gen. nov.) on leaves of Allium schoenoprasum. Ukraine, Xenosphaeropsis corni on recently dead stem of Cornus alba. USA, Nothotrichosporon aquaticum (incl. Nothotrichosporon gen. nov.) from water, and Periconia philadelphiana from swab of coil surface. Morphological and culture characteristics for these new taxa are supported by DNA barcodes.
Citation: Crous PW, Osieck ER, Shivas RG, et al. 2023. Fungal Planet description sheets: 1478–1549. Persoonia 50: 158– 310. https://doi.org/10.3767/persoonia.2023.50.05.
Keywords: ITS nrDNA barcodes, LSU, new taxa, systematics
Acknowledgments
The work of P.W. Crous and colleagues benefitted from funding by the European Union’s Horizon 2020 research and innovation program (RISE) under the Marie Skłodowska-Curie grant agreement No. 101008129, project acronym ‘Mycobiomics’, and the Dutch NWO Roadmap grant agreement No. 2020/ENW/00901156, project ‘Netherlands Infrastructure for Ecosystem and Biodiversity Analysis – Authoritative and Rapid Identification System for Essential biodiversity information’ (acronym NIEBAARISE). G. Gulden, B. Rian and I. Saar thank K. Bendiksen at the fungarium and G. Marthinsen at NorBol, both Natural History Museum, University of Oslo for valuable help with the collections, and the sequencing of our finds of A. similis from 2022. Sincere thanks to A. Voitk for assistance with the colour plate and review of the manuscript. I. Saar was supported by the Estonian Research Council (grant PRG1170). P. Rodriguez-Flakus and co-authors are greatly indebted to their colleagues and all staff of the Herbario Nacional de Bolivia, Instituto de Ecología, Universidad Mayor de SanAndrés, La Paz, for their generous long-term cooperation. Their research was financially supported by the National Science Centre (NCN) in Poland (grants numbers 2018/02/X/NZ8/02362 and 2021/43/B/NZ8/02902). Y.P. Tan and colleagues thank M.K. Schutze (Department of Agriculture and Fisheries, Queensland, Australia) for determining the identity of the insect hosts for Aschersonia mackerrasiae. The Australian Biological Resources Study funded the project that led to the discovery of Aschersonia mackerrasiae. K.G.G. Ganga acknowledges support from the University Grants Commission (UGC), India, in the form of a UGC research fellowship (Ref No. 20/12/2015(ii) EU-V), and the authorities of the University of Calicut for providing facilities to conduct this study. S. Mahadevakumar acknowledges the Director, KSCSTE - Kerala Forest Research Institute and Head of Office, Botanical Survey of India, Andaman and Nicobar Regional Centre, Port Blair for the necessary support and M. Madappa, Department of Studies in Botany, University of Mysore for technical assistance. A.R. Podile thanks the Department of Science and Technology, Govt. of India for the JC Bose Fellowship (Grant No. JCB/2017/000053) & MoE and IOE-Directorate-UOH for project (Grant No.UOH-IOE-RC3-21-065). Financial support was provided to R. de L. Oliveira and K.D. Barbosa by the Coordenação deAperfeiçoamento de Pessoal de Nível Superior - Brazil (CAPES) – Finance code 001, and to I.G. Baseia and M.P. Martín by the National Council for Scientific and Technological Development (CNPq) under CNPq-Universal 2016 (409960/2016-0) and CNPq-visiting researcher (407474/2013-7). E. Larsson acknowledges the Swedish Taxonomy Initiative, SLU Artdatabanken, Uppsala, Sweden. H.Y. Mun and J. Goh were supported by a grant from the Nakdonggang National Institute of Biological Resources (NNIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NNIBR202301106). J. Trovão and colleagues were financed by FEDER - Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020 - Operational Programme for Competitiveness and Internationalisation (POCI), and by Portuguese funds through FCT-Fundação para a Ciência e a Tecnologia in the framework of the project POCI-01-0145-FEDER-PTDC/EPH-PAT/3345/2014. Their research was carried out at the R & D Unit Centre for Functional Ecology – Science for People and the Planet (CFE), with reference UIDB/04004/2020, financed by FCT/MCTES through national funds (PIDDAC). João Trovão was supported by POCH - Programa Operacional Capital Humano (co-funding by the European Social Fund and national funding by MCTES), through a ‘FCT-Fundação para a Ciência e Tecnologia’ PhD research grant (SFRH/BD/132523/2017). O. Kaygusuz and colleagues thank the Research Fund of the Isparta University of Applied Sciences for their financial support under the project number 2021-ILK1-0155. They also thank N. Sánchez Biezma of the Department of Drawing and Scientific Photography at the Alcalá University for his help in the digital preparation of the photographs. The research of M. Spetik and co-authors was supported by project No. IGA-ZF/2021-SI1003. V. Darmostuk and colleagues acknowledge our colleagues and all staff of the Herbario Nacional de Bolivia, Instituto de Ecología, Universidad Mayor de San Andrés, La Paz, for their generous long-term cooperation. They would also like to thank the SERNAP (http://sernap.gob.bo), and all protected areas staff, for providing permits for scientific studies, as well as their assistance and logistical support during the field works. This research was financially supported by the National Science Centre (NCN) in Poland (grant number DEC-2013/11/D/NZ8/03274). M. Kaliyaperumal and co-authors thank the Centre of Advanced Studies in Botany, University of Madras for the laboratory facilities. M. Kaliyaperumal thanks the Extramural Research-SERB, DST (EMR/2016/003078), Government of India, for financial assistance. M. Kaliyaperumal and K. Kezo thanks RUSA 2.0 (Theme-1, Group-1/2021/49) for providing a grant. M. Shivannegowda and colleagues thank C.R. Santhosh, Department of Studies in Microbiology, University of Mysore, Manasagangotri, Mysuru for technical support. They also thank K.R. Sridhar, Mangalore University, Karnataka, India and S.S.N. Maharachchikumbura, University of Electronic Science and Technology of China, Chengdu for their support and helping with technical inputs. The study of G.G. Barreto and co-authors was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brazil (CAPES - Finance Code 001), and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq - Proc. 131503/2019-7; Proc. 312984/2018-9); the authors also thank to Programa de Pós-Graduação em Botânica – PPGBOT. L.F.P. Gusmão thanks to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for a research grant. T. Nkomo and colleagues thank the National Research Foundation of SouthAfrica for funding this study, with additional funding from the Forestry and Agricultural Biotechnology Institute and the University of Pretoria. G. Delgado is grateful to W. Colbert and S. Ward (Eurofins Built Environment) for continual encouragement and provision of laboratory facilities. J.G. Maciá-Vicente acknowledges support from the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) of the state of Hesse within the framework of the Cluster for Integrative Fungal Research (IPF) of Goethe University Frankfurt. F. Esteve-Raventós and colleagues acknowledge P. Juste and J.C. Campos for the loan of some collections for study and N. Subervielle and L. Hugot (Conservatoire Botanique National de Corse, Office de l’Environnement de la Corse, Corti) for their assistance. They also acknowledge the Balearic Mycology Group (FCB) for their permanent help in the search for collections in the Balearic Islands, and Y. Turégano for obtaining some of the sequences presented here, and L. Parra for his suggestions and help on nomenclatural issues. S. Mongkolsamrit and colleagues were financially supported by the Platform Technology Management Section, National Centre for Genetic Engineering and Biotechnology (BIOTEC), Project Grant No. P19-50231. S. De la Peña-Lastra and colleagues thank the Atlantic Islands National Maritime-Terrestrial Park authorities and guards. A. Mateos and co-authors would like to thank Secretaria Regional doAmbiente eAlterações Climáticas Açores for the permission granted for the sampling (Licença nº 16/2021/DRAAC). To the ECOTOX group for co-funding the trip. J. Mack & D.P. Overy were funded byAgriculture &Agri-Food Canada (Project ID#002272: Fungal and Bacterial Biosystematics-bridging taxonomy and “omics” technology in agricultural research and regulation) and are grateful for molecular sequencing support from the Molecular Technologies Laboratory (MTL) at the Ottawa Research & Development Centre of Agriculture & Agri-Food Canada. The study of P. Czachura was funded by the National Science Centre, Poland, under the project 2019/35/N/NZ9/04173. The study of M. Piątek and co-authors was funded by the National Science Centre, Poland, under the project 2017/27/B/NZ9/02902. O. Yarden and L. Granit were funded by the Israel Science Foundation (grant number 888/19). H. Taşkın and colleagues received support from the Bulgarian Academy of Sciences and the Scientific and Technological Research Council of Türkiye (Bilateral grant agreement between BAS and TÜBİTAK, project number 118Z640). The authors would also like to thank S. Şahin (İzmir, Türkiye) for conveying one of the localities of A. abscondita. Andrew Miller would like to thank the Roy J. Carver Biotechnology Center at the University of Illinois for Sanger sequencing. E.R. Osieck thanks Staatsbosbeheer for permission to collect fungi in Nieuw Wulven, in the Netherlands. P. van ‘t Hof and co-authors thank the Galapagos Genetic Barcode project supported by UK Research and Innovation, Global Challenges Research Fund, Newton Fund, University of Exeter, Galapagos Science Center, Universidad San Francisco de Quito, Galapagos Conservation Trust, and Biosecurity Agency of Galapagos (ABG).
REFERENCES
- Abdel-Wahab MA, Dayarathne MC, Suetrong SG. et al. 2017. New saprobic marine fungi and a new combination. Botanica Marina 60: 469–488. [Google Scholar]
- Adamčíková K, Juhásová G, Kobza M. 2011. The first report of Libertella spp. on Fagaceae in Slovakia. Mycoscience 52: 268–270. [Google Scholar]
- Ahmadi N, Arzanlou M, Narmani A. 2021. Molecular phylogeny and morphology differentiate a new Neosetophoma species from Iran. Nova Hedwigia 112: 383–397. [Google Scholar]
- Alvarado P, Cabero J, Moreno-Mateos D. et al. 2021. Phylogenetic relationships among false truffle genera of Paxillaceae – Alpova, Melanogaster, Neoalpova, and Paralpova, gen. nov. Mycologia 113: 828–841. [DOI] [PubMed] [Google Scholar]
- Alvarado P, Moreno G, Manjón JL. 2012. Comparison between Tuber gennadii and T. oligospermum lineages reveals the existence of the new species T. cistophilum (Tuberaceae, Pezizales). Mycologia 104: 894–910. [DOI] [PubMed] [Google Scholar]
- Antonín V, Noordeloos ME. 2004. A monograph of the genera Hemimycena, Delicatula, Fayodia, Gamundia, Myxomphalia, Resinomycena, Rickenella, and Xeromphalina (Tribus Mycenae sensu Singer, Mycena excluded) in Europe. IHW-Verlag. [Google Scholar]
- Aptroot A. 2006. Mycosphaerella and its anamorphs: 2. Conspectus of Mycosphaerella. CBS Biodiversity Series 5: 1–231. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. [Google Scholar]
- Arauzo S, Iglesias P. 2014. La familia Geoglossaceae ss. str. en la península Ibérica y la Macaronesia. Errotari 11: 166–259. [Google Scholar]
- ARSEF dataset. 2023. U.S. Department of Agriculture, Agricultural Research Service, Biological Integrated Pest Management Research Unit (2016). ARS Collection of Entomopathogenic Fungal Cultures (ARSEF). U.S. Department of Agriculture, Agricultural Research Service. 10.15482/USDA.ADC/1326695 (accessed 2023-02-17). [DOI] [Google Scholar]
- Arzanlou M, Groenewald JZ, Fullerton RA. et al. 2008. Multiple gene genealogies and phenotypic characters differentiate several novel species of Mycosphaerella and related anamorphs on banana. Persoonia: 20: 19–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W. et al. 2007. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied Genera. Studies in Mycology 58: 57–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzanlou M, Khodaei S. 2012. Aureobasidium iranianum, a new species on bamboo from Iran. Mycosphere 3: 404–408. [Google Scholar]
- Bandini D, Oertel B, Eberhardt U. 2021. A fresh outlook on the smooth-spored species of Inocybe: type studies and 18 new species. Mycological Progress 20: 1019–1114. [Google Scholar]
- Bandini D, Oertel B, Eberhardt U. 2022a. More smooth-spored species of Inocybe (Agaricales, Basidiomycota): type studies and 12 new species from Europe. Persoonia 48: 91–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandini D, Oertel B, Eberhardt U. 2022b. Noch mehr Risspilze (3): Einundzwanzig neue Arten der Familie Inocybaceae. Mycologia Bavarica 22: 31–138. [Google Scholar]
- Banik MT, Lindner DL, Ortiz-Santana B. et al. 2012. A new species of Laetiporus (Basidiomycota, Polyporales) from the Caribbean basin. Kurtziana 37: 15–21. [Google Scholar]
- Barr ME. 1992. Additions to and notes on the Phaeosphaeriaceae (Pleosporales, Loculoascomycetes). Mycotaxon 43: 371–400. [Google Scholar]
- Beker HJ, Eberhardt U, Vesterholt J. 2016. Hebeloma (Fr.) P. Kumm. Fungi Europaei 14: 1–1218. Ed. Tecnografica. [Google Scholar]
- Bellanger J-M, Bidaud A, Moreau P-A. 2021. Cortinarius subturibulosus. « Illumina »-tion d’un champion de l’adaptation. Journal des JEC 23: 3–15. [Google Scholar]
- Bellanger J-M, Moreau P-A, Corriol G. et al. 2015. Plunging hands into the mushroom jar: a phylogenetic framework for Lyophyllaceae (Agaricales, Basidiomycota). Genetica 143: 169–194. [DOI] [PubMed] [Google Scholar]
- Bensch K, Braun U, Groenewald JZ. et al. 2012. The genus Cladosporium. Studies in Mycology 72: 1–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch K, Groenewald JZ, Braun U. et al. 2015. Common but different: The expanding realm of Cladosporium. Studies in Mycology 82: 23–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bills GF, Menéndez VG, Platas G. 2012. Kabatiella bupleuri sp. nov. (Dothideales), a pleomorphic epiphyte and endophyte of the Mediterranean plant Bupleurum gibraltarium (Apiaceae). Mycologia 104: 962–973. [DOI] [PubMed] [Google Scholar]
- Boscaiu M, Ballesteros G, Naranjo MA. et al. 2011. Responses to salt stress in Juncus acutus and J. maritimus during seed germination and vegetative plant growth. Plant Biosystems 145: 770–777. [Google Scholar]
- Branco S, Ree RH. 2010. Serpentine soils do not limit mycorrhizal fungal diversity. PLoS ONE 5: e11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun U, Cunnington J, Priest MJ. et al. 2005. Annotated checklist of Ramularia species in Australia. Australasian Plant Pathology 34: 1–7. [Google Scholar]
- Bresadola G. 1902. Mycetes Lusitanici novi. Atti dell’ Imperiale Regia Accademia di Scienze. Lettere ed Arti Degli Agiati di Rovereto 3: 127–133. [Google Scholar]
- Brodie HJ. 1975. The Bird’s Nest Fungi. University of Toronto Press, Toronto, Canada. [Google Scholar]
- Cabral A, Groenewald JZ, Rego C. et al. 2012. Cylindrocarpon root rot: multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycological Progress 11: 655–688. [Google Scholar]
- Cailleux A. 1981. Code des couleurs des sols. Boubée, Paris. [Google Scholar]
- Calonge FD, Pasaban PM. 1993. Nuevos datos sobre los hongos hipogeos de España V. Registro de nueve citas nuevas. Boletín de la Sociedad Micológica de Madrid 18: 41–58. [Google Scholar]
- Calvelo S, Liberatore S. 2002. Catálogo de los líquenes de la Argentina. Kurtziana 29: 7–170. [Google Scholar]
- Câmara MP, Ramaley AW, Castlebury LA. 2003. Neophaeosphaeria and Phaeosphaeriopsis, segregates of Paraphaeosphaeria. Mycological Research 107: 516–522. [DOI] [PubMed] [Google Scholar]
- Capote N, Del Río MÁ, Herencia JF. et al. 2022. Molecular and pathogenic characterization of Cylindrocarpon-like anamorphs causing root and basal rot of almonds. Plants 11: 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalabuda TV. 1950. Species novae e genere Penicillium Link. Notulae systematicae e Sectione Cryptogamica Instituti Botanici nomine VL Komarovii Academiae Scientiarum URSS Botanicheskie materialy 6: 161–169. [Google Scholar]
- Chatin A. 1896. Truffes (Terfaz) de Grèce, Terfezia gennadii. Bulletin de la Société Botanique de France 43: 611–617. [Google Scholar]
- Chen W-H, Han Y-F, Liang J-D. et al. 2020. Morphological and phylogenetic characterisations reveal three new species of Samsoniella (Cordycipitaceae, Hypocreales) from Guizhou, China. MycoKeys 74: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomor O, Von Haeseler A, Quang Minh B. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65: 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CY. 1972. Mortierella umbellata, a new species from Georgia. Mycologia 34: 99–102. [Google Scholar]
- Clémençon H. 1982. Types studies and typifications in Lvophyllum (Agaricales). I. Staining species. Mycotaxon 15: 67–94. [Google Scholar]
- Consiglio G. 1998. Lyophyllum aemiliae, Rivista di Micologia 41: 99–104. [Google Scholar]
- Consiglio G, Contu M. 2002. Il Genere Lyophyllum P. Karst. Emend. Huhnér, in Italia. Rivista di Micologia II: 99–181. [Google Scholar]
- Consiglio G, Papetti C. 2001. Funghi d’Italia, ed. AMB 2: 613–619. [Google Scholar]
- Consiglio G, Papetti C. 2009. Funghi d’Italia, ed. AMB 3: 1090–1101. [Google Scholar]
- Cripps CL, Larsson E, Vauras J. 2020. Nodulose-spored Inocybe species from the Rocky Mountain alpine zone: molecularly linked to European type specimens. Mycologia 112: 133–153. [DOI] [PubMed] [Google Scholar]
- Crous PW, Begoude BAD, Boers J. et al. 2022a. New and interesting fungi. 5. Fungal Systematics and Evolution 10: 19–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Boers J, Holdom D. et al. 2022b. Fungal Planet description sheets: 1383-1435. Persoonia 48: 261–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kölling G. et al. 2020a. Fungal Planet description sheets: 1112-1181. Persoonia 45: 251–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kölling G. et al. 2021a. Fungal Planet description sheets: 1182-1283. Persoonia 46: 313–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hernández-Restrepo M, Schumacher RK. et al. 2021b. New and interesting fungi. 4. Fungal Systematics and Evolution 7: 255–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Mohammed C, Glen M. et al. 2007. Eucalyptus microfungi known from culture. 3. Eucasphaeria and Sympoventuria genera nova, and new species of Furcaspora, Harknessia, Heteroconium and Phacidiella. Fungal Diversity 25: 19–36. [Google Scholar]
- Crous PW, Osieck ER, Jurjević Ž. et al. 2021c. Fungal Planet description sheets: 1284-1382. Persoonia 47: 178–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Akulov A. et al. 2019a. New and Interesting Fungi. 2. Fungal Systematics and Evolution 3: 57–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W. et al. 2014. Fungal Planet description sheets: 214-280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI. et al. 2016. Fungal Planet description sheets: 469-557 Persoonia 37: 218–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI. et al. 2017a. Fungal Planet description sheets: 558-624. Persoonia 38: 240–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI. et al. 2017b. Fungal Planet description sheets: 625-715. Persoonia 39: 270–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J. et al. 2013. Fungal Planet description sheets: 154-213. Persoonia 31: 188–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Lombard L. et al. 2019b. Fungal Planet description sheets: 951-1041. Persoonia 43: 223–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK. et al. 2020b. New and interesting fungi 3. Fungal Systematics and Evolution 6: 157–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz RHSF, Baseia IG. 2014. Four new Cyathus species (Nidulariaceae, Basidiomycota, Fungi) from the semi-arid region of Brazil. Journal of the Torrey Botanical Society 141: 173–180. [Google Scholar]
- Dähncke RM, Contu M, Vizzini A. 2009. Some rare, critical, interesting taxa of the genus Lyophyllum s.l. (Basidiomycota, Agaricomycetes) from La Palma (Canary Islands, Spain). Österreichische Zeitschrift Pilzkunde 18: 129–139. [Google Scholar]
- Dähncke RM, Contu M, Vizzini A. 2011. Two new species of Lyophyllum s.l. (Basidiomycota, Agaricomycetes) from La Palma (Canary Islands, Spain). Mycotaxon 115: 63–71. [Google Scholar]
- Dai SJ, Dai YC. 2018. Morphological characters and molecular data reveal a new species of Physisporinus (Basidiomycota) from Southeast Asia. Mycosystema 37: 145–150. [Google Scholar]
- Damm U, Sato T, Alizadeh A. et al. 2018. The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Studies in Mycology 90: 71–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayarathne MC, Phookamsak R, Hyde KD. et al. 2016. Halodiatrype, a novel diatrypaceous genus from mangroves with H. salinicola and H. avicenniae spp. nov. Mycosphere 7: 612–627. [Google Scholar]
- Dayarathne MC, Wanasinghe D N, Devadatha B. et al. 2020. Modern taxonomic approaches to identifying diatrypaceous fungi from marine habitats, with a novel genus Halocryptovalsa Dayarathne & K.D. Hyde, gen. nov. Cryptogamie, Mycologie 41: 21–67. [Google Scholar]
- De Beer W, Procter M, Wingfield MJ. et al. 2022. Generic boundaries in the Ophiostomatales reconsidered and revised. Studies in Mycology 101: 57–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wildeman E. 1894. Notes mycologiques. Fascicle 3. Annual Society Belge Microscopy XVIII: 135–161. [Google Scholar]
- Dereeper A, Guignon V, Blanc G. et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. NucleicAcids Research 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederich P, Garnier-Delcourt M, Lücking R. et al. 2022. Flora of lichenicolous fungi. Vol. 1, Basidiomycota. National Museum of Natural History, Luxembourg. [Google Scholar]
- Dong W, Wang B, Hyde KD. et al. 2020. Freshwater Dothideomycetes. Fungal Diversity 105: 319–575. [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egidi E, De Hoog GS, Isola D. et al. 2014. Phylogeny and taxonomy of meristematic rock-inhabiting black fungi in the Dothideomycetes based on multi-locus phylogenies. Fungal Diversity 65: 127–165. [Google Scholar]
- Ellis JB, Everhart BM. 1888. New species of fungi from various localities. Journal of Mycology 4: 73–82. [Google Scholar]
- Ellis MB. 1971. Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew, Surrey, England. [Google Scholar]
- Ertz D, Diederich P, Lawrey JD. et al. 2015. Phylogenetic insights resolve Dacampiaceae (Pleosporales) as polyphyletic: Didymocyrtis (Pleosporales, Phaeosphaeriaceae) with Phoma-like anamorphs resurrected and segregated from Polycoccum (Trypetheliales, Polycoccaceae fam. nov.). Fungal Diversity 74: 53–89. [Google Scholar]
- Esteve-Raventós F, Pancorbo F, Larsson E. et al. 2022. Inocybe vaurasii (Agaricales, Inocybaceae), a new species of the I. xanthomelas group and similar European species with asteriform spores. Phytotaxa 566: 171–188. [Google Scholar]
- Etayo J. 2017. Hongos Liquenícolas de Ecuador. Opera Lilloana 50: 1–535. [Google Scholar]
- Etayo J, Flakus A, Kukwa M. 2013. Niesslia echinoides (Niessliaceae, Ascomycota), a new lichenicolous fungus on Erioderma from Bolivia. The Lichenologist 45: 21–24. [Google Scholar]
- Fang F, Chen JJ, Ji XH. et al. 2017. Phylogeny and diversity of the morphologically similar polypore genera Rigidoporus, Physisporinus, Oxyporus, and Leucophellinus. Mycologia 109: 749–765. [DOI] [PubMed] [Google Scholar]
- Favre J. 1955. Les champignons supérieurs de la zone alpine du Parc National Suisse. Ergebnisse der Wissensnschaftlichen Untersuchungen des Schweizerrischen National Parks 5: 1–212. [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Feuerer T, Ahti T, Vitikainen O. 1998. Lichenological investigations in Bolivia. In: Marcelli MP, Seaward MRD. (eds), Lichenology in LatinAmerica: History, current knowledge and applications. CETESB, Sao Paulo: 71–86. [Google Scholar]
- Finy P, Papp V, Knapp DG. et al. 2021. Geastrum dolomiticum, a new earthstar species from Central Europe. Plant Systematics and Evolution 307: 43. [Google Scholar]
- Flakus A, Sipman HJM, Bach K. et al. 2013. Contribution to the knowledge of the lichen biota of Bolivia. 5. Polish Botanical Journal 58: 697–733. [Google Scholar]
- Fletcher A, Purvis OW, Coppins BJ. 2009. Aspicilia A. Massal. In: Smith CW, Aptroot A, Coppins BJ. et al. (eds), The Lichens of Great Britain and Ireland. The British Lichen Society, London: 181–188. [Google Scholar]
- Fotedar R, Kolecka A, Boekhout T. et al. 2018. Fungal diversity of the hypersaline Inland Sea in Qatar. Botanica Marina 61: 595–609. [Google Scholar]
- Fryday AM, Wheeler TB, Etayo J. 2021. A new species of Aspicilia (Megasporaceae), with a new lichenicolous Sagediopsis (Adelococcaceae), from the Falkland Islands. The Lichenologist 53: 307–315. [Google Scholar]
- Fuckel L. 1860. Enumeratio Fungorum Nassoviae. Jahrbücher des Nassauischen Vereins für Naturkunde 15: 1–123. [Google Scholar]
- Galloway DJ, Quilhot W. 1998. Checklist of Chilean lichen-forming and lichenicolous fungi. Gayana Botanica 55: 111–185. [Google Scholar]
- Gams W. 1977. A key to the species of Mortierella. Persoonia 9: 381–391. [Google Scholar]
- Gams W, Stielow B, Gräfenhan T. et al. 2019. The ascomycete genus Niesslia and associated monocillium-like anamorphs. Mycological Progress 18: 1–72. [Google Scholar]
- Ge Y, Liu Z, Zeng H. et al. 2021. Updated description of Atheniella (Mycenaceae, Agaricales), including three new species with brightly coloured pilei from Yunnan Province, southwest China. MycoKeys 81: 139–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons AT, Idnurm A, Seiter M. et al. 2019. Amblypygid-fungal interactions: The whip spider exoskeleton as a substrate for fungal growth. Fungal Biology 123: 497–506. [DOI] [PubMed] [Google Scholar]
- Giraldo A, Crous PW. 2019. Inside Plectosphaerellaceae. Studies in Mycology 92: 227–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh YK, Goh TK, Marzuki NF. et al. 2015. Scytalidium parasiticum sp. nov., a new species parasitizing on Ganoderma boninense isolated from oil palm in Peninsular Malaysia. Mycobiology 43: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Góis JS, Cruz RHSF, Baseia IG. 2021. Taxonomic review and updates of the genus Cyathus (Agaricales, Basidiomycota) from Brazil. The Journal of the Torrey Botanical Society 148: 155–196. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V. et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Haji Moniri M, Gromakova AB, Lőkös L. et al. 2017. New members of the Megasporaceae (Pertusariales, lichen-forming Ascomycota): Megaspora iranica spec. nova and Oxneriaria gen. nova. Acta Botanica Hungarica 59: 343–370. [Google Scholar]
- Haridas S, Albert R, Binder M. et al. 2020. 101 Dothideomycetes genomes: A test case for predicting lifestyles and emergence of pathogens. Studies in Mycology 96: 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL. 1975. Notes on British lichenicolous fungi, I. Kew Bulletin 30: 183–203. [Google Scholar]
- Hernández-Restrepo M, Groenewald JZ, Crous PW. 2016a. Taxonomic and phylogenetic re-evaluation of Microdochium, Monographella and Idriella. Persoonia 36: 57–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Groenewald JZ, Elliott ML. et al. 2016b. Take-all or nothing. Studies in Mycology 83: 19–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirooka Y, Ichihara Y, Masuya H. et al. 2012. Seed rot, a new disease of beech tree caused by Neonectria ramulariae (anamorph: Cylindrocarpon obtusiusculum). Journal of Phytopathology 160: 504–506. [Google Scholar]
- Hoang DT, Chernomor O, Von Haeseler A. et al. 2018. UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner HT, Tiffany LH, Knaphus G. 1995. Oak-leaf-litter rhizomorphs from Iowa and Texas: Calcium oxalate producers. Mycologia 87: 34–40. [Google Scholar]
- Houbraken J, Kocsube S, Visagie CM. et al. 2020. Classification ofAspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Samson RA. 2011. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Visagie CM, Meijer M. et al. 2014. A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Studies in Mycology 78: 373–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JH, Yu WJ, Deng LS. et al. 2023. The detection of major clades and new species of Mallocybe (Inocybaceae, Agaricales) from China with elongate cheilocystidia. Mycological Progress 22: 15. [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hustad VP. 2015. A circumscription of the earth tongue fungi class Geoglossomycetes. Doctoral dissertation, University of Illinois at Urbana-Champaign. [Google Scholar]
- Hyde KD, Chaiwan N, Norphanphoun C. et al. 2018. Mycosphere notes 169–224. Mycosphere 9: 271–430. [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R. et al. 2020. Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 16: 1–273. [Google Scholar]
- Hyde KD, Hongsanan S, Jeewon R. et al. 2016. Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. [Google Scholar]
- Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B. et al. 2002. Decorospora, a new genus for the marine ascomycete Pleospora gaudefroyi. Mycologia 94: 651–659. [DOI] [PubMed] [Google Scholar]
- Ingold GT. 1942. Aquatic hyphomycetes of decaying alder leaves. Transactions of British Mycological Society 25: 339–417. [Google Scholar]
- Isola D, Prigione VP, Zucconi L. et al. 2022. Knufia obscura sp. nov. and Knufia victoriae sp. nov., two new species from extreme environments. International Journal of Systematic and Evolutionary Microbiology 72: 10. [DOI] [PubMed] [Google Scholar]
- Jacobsson S, Larsson E. 2012. Inocybe (Fr.) Fr. In: Knudsen H, Vesterholt J. (eds), Funga Nordica (Agaricoid, boletoid, clavarioid, cyphelloid and gastroid genera. Nordsvamp, Copenhagen: ): 981–1021. [Google Scholar]
- Jaklitsch W, Baral H-O, Lücking R. et al. 2016. Syllabus of plant families. Part 1/2 Ascomycota. Borntraeger, Stuttgart. [Google Scholar]
- Justo A, Morgenstern I, Hallen-Adams HE. et al. 2010. Convergent evolution of sequestrate forms in Amanita under Mediterranean climate conditions. Mycologia 102: 675–688. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF. et al. 2017. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Sigler L, Lee J. et al. 2010. Xylogone ganodermophthora sp. nov., an ascomycetous pathogen causing yellow rot on cultivated mushroom Ganoderma lucidum in Korea. Mycologia 102: 1167–1184. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple SequenceAlignment Software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygusuz O, Knudsen H, Bandini D. et al. 2022: Inocybe viscida (Inocybaceae: Agaricomycetes) a new species from Mediterranean forests of Turkey. Turkish Journal of Botany 46: 517–527. [Google Scholar]
- Kearse M, Moir R, Wilson A. et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kizlik S, Trescol F. 1991. Cortinarius subturibulosus sp. nov. Documents mycologiques XXI: 41–42. [Google Scholar]
- Klaysuban A, Sakayaroj J, Jones EBG. 2014. An additional marine fungal lineage in the Diatrypaceae, Xylariales: Pedumispora rhizophorae. Botanica Marina 57: 413–420. [Google Scholar]
- Kokkonen K. 2020. Diversity of boreal small species of Cortinarius subgenus Telamonia with Salix. Karstenia 58: 60–117. [Google Scholar]
- Kong HZ, Qi ZT. 1988. Three new species of Penicillium. Mycosystema 1: 107–114. [Google Scholar]
- Kornerup A, Wanscher JH. 1967. Methuen Handbook of Colour. 2nd edn. Methuen & Co Ltd, London, England. [Google Scholar]
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour. 3rd ed. Eyre Methuen, London. [Google Scholar]
- Kornerup A, Wanscher JH. 1981. Metheun’s Handbook of colours, 3rd ed. Methuen and Co. Ltd., London. [Google Scholar]
- Kosecka M, Kukwa M, Jabłońska A. et al. 2022. Phylogeny and ecology of Trebouxia photobionts from Bolivian lichens. Frontiers in Microbiology 13: 779784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T. et al. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhar F, Castiglia V, Papinutti L. 2013. Geastrum species of the La Rioja province. Mycotaxon 122: 145–156. [Google Scholar]
- Kühner R. 1955. Compléments à la “FloreAnalytique”. V. Inocybe léiosporés cystidiés. Bulletin de la Société des Naturalistes d’Oyonnax, supplement Mémoire hors série No. 2, 9: 3–95. [Google Scholar]
- Kühner R. 1988. Diagnoses de quelques nouveaux Inocybes récoltés en zone alpine de la Vanoise (Alpes françaises). Documents Mycologiques 19: 1–27. [Google Scholar]
- Kumar S, Stecher G, Li M. et al. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. 2017. Inocybe unicolor. Retrieved from the MushroomExpert.Com Web site: http://www.mushroomexpert.com/inocybe_unicolor.html.
- Kuyper TW. 1986. A revision of the genus Inocybe in Europe I. Subgenus Inosperma and the smooth-spored species of subgenus Inocybe. Persoonia supplement 3: 1–247. [Google Scholar]
- Kytövuori I, Nummela-Salo U. et al. 2005: Helttasienten ja tattien levinneisyystaulukko. Distribution table of agarics and boletes in Finland. In: Salo P, Niemelä T. et al. (eds), Suomen helttasienten ja tattien ekologia, levinneisyys ja uhanalaisuus. Suomen ympäristökeskus, Helsinki. Suomen ympäristö: 769: 109–224. [Google Scholar]
- La Spina L. 2021. Funghi di Sicilia, Tomo IV, ed. T. Italgrafica: 2193–2194. [Google Scholar]
- Landry J, Labbé R. 2023. Les champignons du Québec - Base de données de Mycoquébec. https://www.mycoquebec.org.
- Lanfear R, Calcott B, Ho SYW. et al. 2012. PartitionFinder: Combined Selection of Partitioning Schemes and Substitution Models for Phylogenetic Analyses. Molecular Biology and Evolution 29: 1695–1701. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM. et al. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP. et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Larsson E, Vauras J, Cripps CL. 2014. Inocybe leiocephala, a species with an intercontinental distribution range – disentangling the I. leiocephala – subbrunnea – catalaunica morphological species complex. Karsternia 54: 15–39. [Google Scholar]
- Lee BG, Shin HT, Hur JS. 2022. A new lichen-forming fungus, Aspicilia humida, from a forested wetland in South Korea, with a taxonomic key for aspicilioid species of Korea. Mycobiology 50: 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liimatainen K, Niskanen T, Dima B. et al. 2020. Mission impossible completed: unlocking the nomenclature of the largest and most complicated subgenus of Cortinarius, Telamonia. Fungal Diversity 104: 291–331. [Google Scholar]
- Linde CC, May TW, Phillips RD. et al. 2017. New species of Tulasnella associated with terrestrial orchids in Australia. IMA Fungus 8: 28–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ma ZY, Hou LW. et al. 2022. Updating species diversity of Colletotrichum, with a phylogenomic overview. Studies in Mycology 101: 1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Hyde KD, Jones EG. et al. 2015. Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–97. [Google Scholar]
- Liu XZ, Wang QM, Göker M. et al. 2015. Towards an integrated phylogenetic classification of the Tremellomycetes. Studies in Mycology 81: 85–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd CG. 1907. New notes on the Geasters. Mycological Writings 2: 309–317. [Google Scholar]
- Loizides M, Bellanger J-M, Yiangou Y. et al. 2018. Preliminary phylogenetic investigations into the genusAmanita (Agaricales) in Cyprus, with a review of previous records and poisoning incidents. Documents Mycologiques 37: 201–218. [Google Scholar]
- Lombard L, Houbraken J, Decock C. et al. 2016. Generic hyper-diversity in Stachybotriaceae. Persoonia 36: 156–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luangsa-ard JJ, Ridkaew R, Mongkolsamt S. et al. 2010. Ophiocordyceps barnesii and its relationship to other melolonthid pathogens with dark stromata. Fungal Biology 739–745. [DOI] [PubMed] [Google Scholar]
- Luangsa-ard JJ, Tasanathai K, Mongkolsamrit S. et al. 2007. Atlas of inverte-brate-pathogenic fungi of Thailand, vol 1. BIOTEC, National Science and Technology Development Agency, Pathumthani. [Google Scholar]
- Ludwig E. 2001. Pilzkompendium 1: 306. IHW-Verlag. [Google Scholar]
- Ludwig E. 2017. Pilzkompendium 4 (parts 1 & 2). Fungicon, Berlin. [Google Scholar]
- Lumbsch HT, Feige GB, Schmitz KE. 1994. Systematic studies in the Pertusariales I. Megasporaceae, a new family of lichenizedAscomycetes. The Journal of the Hattori Botanical Laboratory 75: 295–304. [Google Scholar]
- Luo ZL, Hyde KD, Liu JK. et al. 2019. Freshwater Sordariomycetes. Fungal Diversity 99: 451–660. [Google Scholar]
- Maas Geesteranus RA. 1992. Mycenas of the Northern Hemisphere. II. Conspectus of the Mycenas of the Northern Hemisphere. North-Holland, Amsterdam. [Google Scholar]
- Maciá-Vicente JG, Jansson HB, Abdullah SK. et al. 2008. Fungal root endophytes from natural vegetation in Mediterranean environments with special reference to Fusarium spp. FEMS Microbiology Ecology 64: 90–105. [DOI] [PubMed] [Google Scholar]
- Maciá-Vicente JG, Nau T, Piepenbring M. 2016. Low diversity and abundance of root endophytes prevail throughout the life cycle of an annual halophyte. Mycological Progress 15: 1303–1311. [Google Scholar]
- Magdama F, Sosa D, Espinoza F. et al. 2020. Guayaquilia gen. nov., typified by Idriella cubensis. Mycotaxon 135: 501–512. [Google Scholar]
- Malençon G, Bertault R. 1970. Flore des Champignons supérieurs du Maroc, Vol. 1. Institut Scientifique Chérifien et Faculté des Sciences, Rabat. [Google Scholar]
- Malloch D, Sigler L, Hambleton S. et al. 2016. Fungi associated with hibernating bats in New Brunswick caves: the genus Leuconeurospora. Botany 94: 1171–1181. [Google Scholar]
- Marin-Felix Y, Hernández-Restrepo M, Iturrieta-González I. et al. 2019. Genera of phytopathogenic fungi: GOPHY 3. Studies in Mycology 94: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny BP, Swenie RA. 2018. The Inocybe geophylla group in North America: a revision of the lilac species surrounding I. lilacina. Mycologia 110: 618–634. [DOI] [PubMed] [Google Scholar]
- Mattirolo O. 1900. Gli ipogei di Sardegna e di Sicilia. Malpighia 14:1–74 [Google Scholar]
- Melis M, Contu M. 2000. Una nuova specie di Lyophyllum sect. Lyophyllum dalla Sardegna meridionale: L. maleolens spec. nov. Micologia e Vegetazione Mediterranea 15: 101–105. [Google Scholar]
- Métrod G. 1956. Les inocybes leiosporés a cystides courtes. Bulletin trimestrale de la Société Mycologique de France 72: 122–131. [Google Scholar]
- Miller AN, Huhndorf SM, Fournier J. 2014. Phylogenetic relationships of five uncommon species of Lasiosphaeria and three new species in the Helminthosphaeriaceae (Sordariomycetes). Mycologia 106: 505–524. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA: 1–8. [Google Scholar]
- Miller OK, Horak E. 1992. Observations on the genus Torrendia and a new species from Australia. Mycologia 84: 64–71. [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O. et al. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsamrit S, Khonsanit A, Noisripoom W. et al. 2014. Aschersonia narathiwatensis sp. nov. from southern Thailand. Mycotaxon 139: 33–40. [Google Scholar]
- Mongkolsamrit S, Noisripoom W, Thanakitpipattana D. et al. 2018. Disentangling cryptic species with isaria-like morphs in Cordycipitaceae. Mycologia 110: 230–257. [DOI] [PubMed] [Google Scholar]
- Montecchi A, Sarasini M. 2000. Funghi ipogei d’Europa. Trento, Italy: AMB Fondazione Centro Studi Micologici. [Google Scholar]
- Moreau PA, Rochet J, Richard F. et al. 2011. Taxonomy of Alnus-associated hypogeous species of Alpova and Melanogaster (Basidiomycota, Paxillaceae) in Europe. Cryptogamie, Mycologie 32: 33–62. [Google Scholar]
- Munsell Color 1994. Soil Color Charts (revised edition). Macbeth Division of Kollmorgen Instruments Corporation, New Windsor, New York, USA. [Google Scholar]
- Neville P, Pourmat S. 2004. Amaniteae: Amanita, Limacella & Torrendia. Fungi Europaei 9. Candusso, Italy. [Google Scholar]
- Nguyen L-T, Schmidt HA, Von Haeseler A. et al. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin A, Savic S, Tibell L. 2010. Phylogeny and taxonomy of Aspicilia and Megasporaceae. Mycologia 102: 1339–1349. [DOI] [PubMed] [Google Scholar]
- Núñez M, Ryvarden L. 2001. East Asian polypores 2. Polyporaceae s. lato. Synopsis Fungorum 14: 170–522. [Google Scholar]
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden. [Google Scholar]
- Nylander W. 1861. Additamentum ad lichenographiam Andium Boliviensium. Annales des Sciences Naturelles, la Botanique 15: 365–382. [Google Scholar]
- Oppicelli N. 2020. Funghi in Italia, ed. Erredì Grafiche: 360–361. [Google Scholar]
- Örstadius L, Ryberg M, Larsson E. 2015. Molecular phylogenetics and taxonomy in Psathyrellaceae (Agaricales) with focus on psathyrelloid species: introduction of three new genera and 18 new species. Mycological Progress 14: 1–42. [Google Scholar]
- Owe-Larsson B, Nordin A, Tibell L. 2007. Aspicilia. In: Nash III TH, Ryan BD, Diederich P. et al. (eds), Lichen Flora of the Greater Sonoran Desert Region, Vol. 3: 61–108. Lichens Unlimited, Arizona State University, Tempe. [Google Scholar]
- Persoon CH. 1794. Neuer Versuch einer systematischen Einteilung der Schwämme. Neues Magazin für die Botanik 1: 63–128. [Google Scholar]
- Peterson SW, Manitchotpisit P, Leathers TD. 2013. Aureobasidium thailandense sp. nov. isolated from leaves and wooden surfaces. International Journal of Systematic and Evolutionary Microbiology 63: 790–795. [DOI] [PubMed] [Google Scholar]
- Pitt JI. 1980. The Genus Penicillium and Its Teleomorphic States Eupenicillium and Talaromyces. Academic Press, London. [Google Scholar]
- Pordel A, Khodaparast S, Mckenzie E. et al. 2017. Two new species of Pseudopyricularia from Iran. Mycological Progress 16: 729–736. [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ. et al. 2014. Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2016. FigTree v. 1.4.0. http://tree.bio.ed.ac.uk/software/figtree/.
- Ramirez C, Martinez T. 1980. Some new species of Penicillium recovered from the atmosphere in Madrid and from other substrata. Mycopathology 72181–191 [DOI] [PubMed] [Google Scholar]
- Raper KB, Thom C. 1949. Manual of the Penicillia. The Williams & Wilkins Company, Baltimore, United States. [Google Scholar]
- Rashmi M, Kushveer JS, Sarma VV. 2019. A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 10: 798–1079. [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. Commonwealth Mycological Institute & British Mycological Society, Kew, Richmond. [Google Scholar]
- Réblová M. 2009. Teleomorph of Rhodoveronaea (Sordariomycetidae) discovered and re-evolution of Pleurophragmium. Fungal Diversity 36: 129–139. [Google Scholar]
- Redhead SA. 2012. Nomenclatural novelties. Index Fungorum 14: 1. [Google Scholar]
- Rincón A, Santamaría BP, Ocaña L. et al. 2014. Structure and phylogenetic diversity of post-fire ectomycorrhizal communities of maritime pine. Mycorrhiza 24: 131–141. [DOI] [PubMed] [Google Scholar]
- Roberts P. 1999. Rhizoctonia-forming fungi: a taxonomic guid. Royal Botanic Gardens, Kew, Surrey, UK. [Google Scholar]
- Robich G. 2003. Mycena d’Europa. Associazione Micologica Bresadola, Trento, Italy. [Google Scholar]
- Rodriguez-Flakus P, Kukwa M, Etayo J. et al. 2016. Preliminary catalogue of lichens and lichenicolous fungi from Bolivia. https://bio.botany.pl/lichens-bolivia.
- Romagnesi H. 1987. Sur la tribu des Lyophylleae Kühner (Agaricales, Tricholomataceae). Beiträge zur Kenntnis der Pilze Mitteleuropas 3: 117–123. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P. et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman AY. 2021. In memoriam: Marie Leonore (“Lennie”) Farr, 6 September 1927–13 May 2014 First Woman President of MSA. Mycologia 113: 509–511. [Google Scholar]
- Rossman AY, Allen WC, Braun U. et al. 2016. Overlooked competing asexual and sexually typified generic names ofAscomycota with recommendations for their use or protection. IMA Fungus 7: 289–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Masson D, Bricaud O. et al. 2011. Flore et végétation des lichens et champignons lichénicoles de quatre réserves naturelles des Pyrénées-Orientales (France). Bulletin de la Société Linnéenne de Provence 14: 3–151. [Google Scholar]
- Roy M, Rochet J, Manzi S. et al. 2013. What determines Alnus-associated ectomycorrhizal community diversity and specificity? A comparison of host and habitat effects at a regional scale. New Phytologist 198: 1228–1238. [DOI] [PubMed] [Google Scholar]
- Royal Horticultural Society. 2015. Colour Chart, 6th ed.; Royal Horticultural Society: London, UK. [Google Scholar]
- Ryberg M, Nilsson RH, Kristiansson E. et al. 2008. Mining metadata from unidentified ITS sequences in GenBank: a case study in Inocybe (Basidiomycota). BMC Evolutionary Biology 8: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method:A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Samson RA. 1974. Paecilomyces and some allied hyphomycetes. Study in Mycology 6: 1–119. [Google Scholar]
- Samuels GJ, Candoussau F, Magni J-F. 1997. Fungicolous pyrenomycetes 1. Helminthosphaeria and the new family Helminthosphaeriaceae. Mycologia 89: 141–155. [Google Scholar]
- Sánchez-Gavilán I, Ramírez E, de la Fuente V. 2021. Bioactive compounds in Salicornia patula Duval-Jouve: A Mediterranean edible euhalophyte. Foods 10: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguy E. 1936. Encyclopedie Pratique du Naturaliste, 30. Paul Lechevalier, Paris. [Google Scholar]
- Sigler L, Carmichael JW. 1976. Taxonomy of Malbranchea and some other hyphomycetes with arthroconidia. Mycotaxon 4: 349–488. [Google Scholar]
- Sigler L, Lumley TC, Currah RS. 2000. New species and records of saprophytic ascomycetes (Myxotrichaceae) from decaying logs in the boreal forest. Mycoscience 41: 495–502. [Google Scholar]
- Simmons EG. 2007. Alternaria. An identification manual. CBS Biodiversity Series 6. Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands. [Google Scholar]
- Smith AH. 1947. North American species of Mycena. University of Michigan Press (and Oxford University Press). [Google Scholar]
- Soares F, Trovão J, Portugal A. 2022. Phototrophic and fungal communities inhabiting the Roman cryptoporticus of the national museum Machado de Castro (UNESCO site, Coimbra, Portugal). World Journal of Microbiology and Biotechnology 38: 157. [DOI] [PubMed] [Google Scholar]
- Sogonov MV, Schroers HJ, Gams W. et al. 2005. The hyphomycete Teberdinia hygrophila gen. nov., sp. nov. and related anamorphs of Pseudeurotium species. Mycologia 97: 695–709. [DOI] [PubMed] [Google Scholar]
- Sohrabi M, Leavitt SD, Rico VJ. et al. 2013. Teuvoa, a new lichen genus in Megasporaceae (Ascomycota: Pertusariales), including Teuvoa junipericola sp. nov. The Lichenologist 45: 347–360. [Google Scholar]
- Song J, Cui BK. 2017. Phylogeny, divergence time and historical biogeography of Laetiporus (Basidiomycota, Polyporales). BMC Evolutionary Biology 17: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML v. 8: A tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez JP, Weiß M, Abele A. et al. 2006. Diverse tulasnelloid fungi form mycorrhizas with epiphytic orchids in an Andean cloud forest. Mycological Research 110: 1257–1270. [DOI] [PubMed] [Google Scholar]
- Suárez-Santiago VN, Ortega A, Peintner U. et al. 2009. Study on Cortinarius subgenus Telamonia section Hydrocybe in Europe, with especial emphasis on Mediterranean taxa. Mycological Research 113: 1070–1090. [DOI] [PubMed] [Google Scholar]
- Sunhede S. 1989. Geastraceae (Basidiomycotina). Morphology, ecology and systematics with special emphasis on the North European species. Synopsis Fungorum 1: 1–534. [Google Scholar]
- Sutton BC. 1980. The Coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata. Commonwealth Mycological Institute, Kew. [Google Scholar]
- Swofford DL. 2003. PAUP* 4.0: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Tamura K, Peterson D, Peterson N. et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. 2021. MEGA 11: Molecular Evolutionary GeneticsAnalysis Version 11. Molecular Biology and Evolution 38: 3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YP, Bishop-Hurley SL, Shivas RG. et al. 2022. Fungal Planet description sheets: 1436-1477. Persoonia 49: 261–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Hirayama K, Yonezawa H. et al. 2015. Revision of the Massarineae (Pleosporales, Dothideomycetes). Studies in Mycology 82: 75–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanney JB, Seifert KA. 2020. Mollisiaceae: An overlooked lineage of diverse endophytes. Studies in Mycology 95: 293–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanakitpipattana D, Tasanathai K, Mongkolsamrit S. et al. 2019. Fungal pathogens occurring on Orthopterida in Thailand. Persoonia 44: 140–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JW. 1984. AmericanArctic Lichens. 2. The Microlichens. University of Wisconsin Press, Madison. [Google Scholar]
- Thüs H, Schultz M. 2009. Fungi 1. Lichens. In: Büdel B, Gärtner G, Krienitz L. et al. (eds), Freshwater Flora of Central Europe. Heidelberg: Spektrum Akademischer Verlag: 211–229. [Google Scholar]
- Trierveiler-Pereira L, Gomes-Silva AC, Baseia IG. 2009. Notes on gasteroid fungi in the Brazilian Amazon rainforest. Mycotaxon 110: 73–80. [Google Scholar]
- Trifinopoulos J, Nguyen L-T, von Haeseler A. et al. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44: W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulasne LR, Tulasne C. 1844. Recherches sur l’organisation et le mode de frutification des champignons de la tribu des Nidulariées, suivies d’um essai monographique. Annales des Sciences Naturalles series 3: 41–107. [Google Scholar]
- Van Ryckegem G. 2005. Fungi on common reed (Phragmites australis). Fungal diversity, community structure and decompositions processes. PhD thesis, Universiteit Gent, Belgium. [Google Scholar]
- Van Waveren EK. 1985. The Dutch, French and British species of Psathyrella. Persoonia supplement 2: 3–300. [Google Scholar]
- Vandepol N, Liber J, Desirò A. et al. 2020. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Diversity 104: 267–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauras J, Larsson E. 2016. Inocybe caprimulgi and I. lacunarum, two new nodulose-spored species from Fennoscandia. Karstenia 55: 1–18. [Google Scholar]
- Vellinga EC. 1988. Glossary. In: Bas C, Kuyper TW, Noordeloos ME. et al. (eds), Flora Agaricina Neerlandica vol. 1: 54–64. Balkema, Rotterdam, The Netherlands. [Google Scholar]
- Vesterholt J, Ludwig E. 2012. Lyophyllum. In: Knudsen H, Vesterholt J. (eds), Funga Nordica, 2nd edition. Nordsvamp, Copenhagen, 2 vols. [Google Scholar]
- Visagie CM, Renaud JB, Burgess KMN. et al. 2016b. Fifteen new species of Penicillium. Persoonia 36: 247–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Seifert KA, Houbraken J. et al. 2016a. A phylogenetic revision of Penicillium sect. Exilicaulis, including nine new species from fynbos in South Africa. IMA Fungus 7: 75–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Yilmaz N. 2022. Along the footpath of Penicillium discovery: Six new species from the Woodville Big Tree Forest Trail. Mycologia 28: 1–20. [DOI] [PubMed] [Google Scholar]
- Vittadini C. 1842. Monographia Lycoperdineorum. Ex Officina Regia, Italy. [Google Scholar]
- Voitk A, Saar I, Lücking R. et al. 2020. Surprising morphological, ecological and ITS sequence diversity in the Arrhenia acerosa complex (Basidiomycota: Agaricales: Hygrophoraceae). Sydowia 73: 133–162. [Google Scholar]
- Von Brackel W. 2014. Kommentierter Katalog der flechtenbewohnenden Pilze Bayerns. Bibliotheca Lichenologica 109: 1–476. [Google Scholar]
- Wächter D, Melzer A. 2020. Proposal for a subdivision of the family Psathyrellaceae based on a taxon-rich phylogenetic analysis with iterative multigene guide tree. Mycological Progress 19: 1151–1265. [Google Scholar]
- Wang B, Yu Y, Wang L. 2014. Penicillium fusisporum and P. zhuangii, two new monoverticillate species with apical-swelling stipes of section Aspergilloides isolated from plant leaves in China. PLoS One 9: e101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XC, Chen K, Zeng ZQ. et al. 2017. Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Scientific Reports 7: 8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YB, Wang Y, Fan Q. et al. 2020. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): New taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Diversity 103: 1–46. [Google Scholar]
- Wang Z, Wang Y, Dong Q. et al. 2022. Morphological and phylogenetic characterization reveals five new species of Samsoniella (Cordycipitaceae, Hypocreales). Journal of Fungi 8: 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watling R, Işıloğlu M. 1991. Torrendia pulchella Bres. A new and interesting record from Türkiye. Turkish Journal of Botany 15: 297–299. [Google Scholar]
- Wei TP, Zhang H, Zeng XY. et al. 2022. Re-evaluation of Sympoventuriaceae. Persoonia 48: 219–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield MJ, Harrington TC, Crous PW. 1994. Three new Leptographium species associated with conifer roots in the United States. Canadian Journal of Botany 72: 227–238. [Google Scholar]
- Woudenberg JH, Groenewald JZ, Binder M. et al. 2013. Alternaria redefined. Studies in Mycology 75: 171–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rogers J. 2004. A postharvest fruit rot in d’Anjou pears caused by Sphaeropsis pyriputrescens sp. nov. Plant Disease 88: 114–118. [DOI] [PubMed] [Google Scholar]
- Yang E-F, Phookamsak R, Jiang H-B. et al. 2022. Taxonomic reappraisal of Periconiaceae with the description of three new Periconia species from China. Journal of Fungi 8: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden. O. 2014. Fungal association with sessile marine invertebrates. Frontiers in Microbiology 5: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri Z, Divakar PK, Otte V. 2017. Taxonomy and phylogeny of Aspiciliella, a resurrected genus of Megasporaceae, including the new species A. portosantana. Herzogia 30: 166–176. [Google Scholar]
- Zamora JC, Calonge FD, Martín MP. 2015. Integrative taxonomy reveals an unexpected diversity in Geastrum section Geastrum (Geastrales, Basidiomycota). Persoonia 34: 130–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller SM. 1939. New and Noteworthy Gasteromycetes. Mycologia 31: 1–32. [Google Scholar]
- Zeller SM, Dodge CW. 1937. Melanogaster. Annals of the Missouri Botanical Garden 23: 639–655. [Google Scholar]
- Zhang Z, Dong C, Chen W. et al. 2020. The enigmatic Thelebolaceae (Thelebolales, Leotiomycetes): one new genus Solomyces and five new species. Frontiers in Microbiology 11: 572596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZY, Han YF, Chen WH. et al. 2023. Additions to Thelebolales (Leotiomycetes, Ascomycota): Pseudogeomyces lindneri gen. et sp. nov. and Pseudogymnoascus campensis sp. nov. MycoKeys 95: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZY, Shao QY, Li X. et al. 2021. Culturable fungi from urban soils in China I: description of 10 new taxa. Microbiology Spectrum 9: e00867–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Crous PW, Hou LW. et al. 2021. Fungi of quarantine concern for China I: Dothideomycetes. Persoonia 47: 45–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao RL, Jeewon R, Desjardin DE. et al. 2007. Ribosomal DNA phylogenies of Cyathus: Is the current infrageneric classification appropriate? Mycologia 99: 385–395. [DOI] [PubMed] [Google Scholar]
- Zhu H, Pan M, Wijayawardene NN. et al. 2021. The hidden diversity of Diatrypaceous fungi in China. Frontiers in Microbiology 12: 646262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurbenko MP, Pino Bodas R. 2017. A revision of lichenicolous fungi growing on Cladonia, mainly from the Northern Hemisphere, with a worldwide key to the known species. Opuscula Philolichenum 16: 188–266. [Google Scholar]