Abstract
Background
Ovarian cancer (OC) is the leading cause of death from gynecologic malignancies in the Western world. Contributing factors include a high frequency of late-stage diagnosis, the development of chemoresistance, and the evasion of host immune responses. Currently, debulking surgery and platinum-based chemotherapy are the treatment cornerstones, although recurrence is common. As the clinical efficacy of immune checkpoint blockade is low, new immunotherapeutic strategies are needed. Chimeric antigen receptor (CAR) T cell therapy empowers patients’ own T cells to fight and eradicate cancer, and has been tested against various targets in OC. A promising candidate is the MUC16 ectodomain. This ectodomain remains on the cell surface after cleavage of cancer antigen 125 (CA125), the domain distal from the membrane, which is currently used as a serum biomarker for OC. CA125 itself has not been tested as a possible CAR target. In this study, we examined the suitability of the CA125 as a target for CAR T cell therapy.
Methods
We tested a series of antibodies raised against the CA125 extracellular repeat domain of MUC16 and adapted them to the CAR format. Comparisons between these candidates, and against an existing CAR targeting the MUC16 ectodomain, identified K101 as having high potency and specificity. The K101CAR was subjected to further biochemical and functional tests, including examination of the effect of soluble CA125 on its activity. Finally, we used cell lines and advanced orthotopic patient-derived xenograft (PDX) models to validate, in vivo, the efficiency of our K101CAR construct.
Results
We observed a high efficacy of K101CAR T cells against cell lines and patient-derived tumors, in vitro and in vivo. We also demonstrated that K101CAR functionality was not impaired by the soluble antigen. Finally, in direct comparisons, K101CAR, which targets the CA125 extracellular repeat domains, was shown to have similar efficacy to the previously validated 4H11CAR, which targets the MUC16 ectodomain.
Conclusions
Our in vitro and in vivo results, including PDX studies, demonstrate that the CA125 domain of MUC16 represents an excellent target for treating MUC16-positive malignancies.
Keywords: receptors, chimeric antigen; immunotherapy; T-lymphocytes
WHAT IS ALREADY KNOWN ON THIS TOPIC.
Many chimeric antigen receptor (CAR) have been developed against ovarian cancer (OC) . To date, one anti-MUC16 CAR has been identified which targets a part of the extracellular domain that does not get cleaved. It was previously suggested that the cleaved part of MUC16, CA125, did not represent an efficient CAR target.
WHAT THIS STUDY ADDS
Our study demonstrates that CA125 is indeed an attractive target. The CAR derived from the anti-MUC16 antibody, K101, was efficient against OC models in vitro and in vivo. Furthermore, when CA125 was present at high levels, it did neither activate K101CAR T cell nor suppress tumor cell killing.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
MUC16 is an attractive target, but it was thought to be limited in targeting possibilities due to the CA125 cleavage which removes a large part of the extracellular domain. Here, we show that this huge CA125 domain can indeed be targeted, and this opens the possibility to develop additional CAR molecules.
Introduction
Ovarian cancer (OC) is the eighth most common cancer in women globally, for both incidence and deaths,1 with a 5-year survival rate of 49.7%.2 3 This is driven by a high frequency of late-stage diagnoses—when OC is particularly aggressive—the development of chemoresistance and evasion of host immune responses.4 Standard treatment consists of cytoreductive surgery combined with platinum-based chemotherapy. This may be followed by adjuvant maintenance treatment with anti-angiogenic agents and inhibitors of poly-ADP-ribose polymerase.4–6 Nevertheless, a majority of patients will relapse, and treatment will shift from curative to palliative care.4
High-grade serous tubo-ovarian carcinoma (HGSC) is classified into three different immunologic phenotypes—desert, excluded, and inflamed ovarian tumors—based on the degree of T-cell infiltration into the epithelial compartment.7 Immunotherapy in the form of immune checkpoint inhibitors performs poorly in OC.8 The general failure in T-cell priming observed in cold and excluded tumors reflects the urgent need for strategies that can deliver autologous or allogeneic effector cells into HGSC tumors. CAR T-cell therapy faces challenges similar to adoptive cell therapy in general,9 but induction of a strong antitumor response, plus the inclusion of additional factors in the engineered cells (eg, fourth-generation CARs) should help overcome these factors.10
Among the potential CAR targets for OC, MUC16/CA125 is one of the most promising in the field,11 being expressed by the majority of later-stage OC.12 The normal biological function of MUC16 is to provide a protective lubricating barrier at mucosal surfaces, but in cancer, it can facilitate peritoneal metastasis by promoting proliferation and inhibiting apoptosis.13 14 MUC16 is a single-pass integral membrane glycosylated mucin protein, with a large extracellular region comprising multiple tandem repeats. Cleavage and release of this extracellular domain results in the generation of the cancer antigen 125 (CA125).15 This cleaved CA125 has three main topographic domains, can be detected in the serum of patients with OC, and is used as a diagnostic marker for OCs.16 17
It is speculated that soluble CA125 may interfere with CAR reactivity by binding to the CAR receptor in the absence of target cells.18 These concerns are partially supported by the observations of interference caused by shedding of other CAR targets (eg, Sun et al 19). However, we note that not all antigens, when shed, result in interference (eg, Liu et al 20). In the case of B-cell maturation antigen, multiple studies found no effect of the soluble antigen on CAR function,21 22 while elsewhere it was found to inhibit CAR T-cell functionality.23 The ability of soluble ligands to dimerize CARs appears to be critical for generating this latter effect.24 For MUC16, there have been numerous efforts to target the extracellular ectodomain that remains after cleavage of CA125—sometimes referred to as MUC-CD or MUC16ecto—by various approaches, namely, antibodies,25 CAR T cells,18 26 and antibody-derived bispecific T-cell engagers.27–30
Soluble CA125 has been shown to reassociate with the remaining extracellular stub.31 Such reassociation is the basis of CA125 targeting in experimental immunologic approaches. Notably, administration of antibodies against CA125 restricts the growth of OVCAR3 tumors in mice.32 To our knowledge, interference of CAR function by soluble CA125 has not been definitively proven. Therefore, considering (1) the distribution of MUC16/CA125 on tumor cells and (2) the expected accessibility of CA125 epitopes (relative to the remaining stub, after reassociation), we wanted to examine the feasibility of targeting epitopes on the CA125 repeat domain for CAR T-cell therapy.
In the present study, we adapted antibodies from three hybridomas from our hospital’s collection33 to a CAR format. Of these, K101CAR induced the strongest response against MUC16-positive (MUC16pos) tumor cells in vitro and was selected for further testing. Soluble CA125 did not interfere with K101CAR T-cell cytotoxicity, and the functional activity of this CAR was superior to existing anti-MUC16ecto CAR in vitro and showed significant impairment of tumor growth in vivo. K101CAR represents a promising candidate for clinical CAR-based therapy.
Materials and methods
Analysis of RNA-Seq data
RNA-Seq data from The Cancer Genome Atlas (TCGA, released July 27, 2022) project for OC for 378 tumor samples was used. The analysis was performed in R (https://www.r-project.org/ (accessed in 2022)) using Bioconductor (https://www.bioconductor.org) packages. The corresponding OC clinical data as well as raw counts of gene expression for tumor-associated samples were downloaded through the TCGA biolinks package. The Harmonized database (https://portal.gdc.cancer.gov/), which is mapped to the reference genome GRCh38 (hg38), was used. Clinical staging in OC, using the FIGO (International Federation of Gynaecology and Obstetrics) system, categorized patients into eight distinct groups (I, II, IIIA, IIIB, IIIC, IVA, IVB and unknown stage). In this staging, three factors of tumor size, lymph node, and spread of metastasis were used to classify samples. To identify differential expression of genes on the TCGA cohort, the R package DESeq2 was used for normalization of raw counts. Prefiltering was applied to keep only genes that have at least 10 reads in total. Differential expression and design of the analysis were also performed with the DEseq2 package.34 The clinical stage of tumor samples was used in the design of the analysis. For false discovery rate (FDR) correction, adjusted p values <0.05 were selected. Differential expression of genes was considered to be statistically significant if their |log2 fold change|≥2 and FDR <0.05. Since the MUC16 gene is known to be highly expressed in OC tissues, we focused on a selection of genes (MUC16, MUC1, MSLN, FOLR1) in order to show the significant differential expression of MUC16 in the TCGA database.35 To visualize the results of differential expression analyses of selected genes (MUC16, MUC1, MSLN, FOLR1), pheatmap was used.
Cell culture and cell lines
Standard in vitro culture was at 37°C, with 5% CO2, in a humidified environment. Cell lines, OVCAR3 (HTB-161) and HeLa (CCL-2.2), were obtained from ATCC and cultured in RPMI-1640 (Biowest, Nuaillé, France) with 10% fetal calf serum (FCS) (Thermo Fisher Scientific, USA) and 10 µg/mL gentamycin (Thermo Fisher Scientific), except HEK-293 (CRL-1573) cells, also from ATCC, which were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Sigma, USA) with 10% HyClone FBS (GE Healthcare Life Sciences, USA) and 10 µg/mL gentamycin.
For regular passaging, adherent cells were washed twice with phosphate-buffered saline (PBS), then detached with trypsin/EDTA (Sigma-Aldrich, USA, T3924-100ML). Prior to co-culture experiments, target cells were detached with EDTA alone (5 mM in PBS, without trypsin).
Spheroids composed of tumor cells were generated using standard methods. Briefly, flat bottom 96-well plates were coated with 1% agarose dissolved in PBS. After 2 hours at room temperature, 2000 tumor cells in 200 µL of culture medium were added per well.
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood of healthy donors, under an approved institutional protocol, and were cultured in X-vivo 15 (Lonza, Switzerland), supplemented with 5% human serum (TCS Biosciences, Buckingham MK18 2LR, UK) and interleukin-2 (IL-2) (Clinigen, UK) at 100 U/mL. For activation, 24-well culture plates were precoated with 500 µL/well of PBS containing 0.5 µg anti-CD3 (functional grade OKT3, eBioscience, USA, #16-0037-85) and 0.5 µg anti-CD28 (functional grade CD28.6, eBioscience, #16-0288-85) antibodies for 2 hours at room temperature. This was removed, and PBMCs were added at 106 cells/mL per well. After 2–3 days, cells were counted and available for transduction.
Production of retroviral vectors, transduction, and protein production
Production of retroviral particles and transduction of T cells were performed as described previously.36 Briefly, human embryonic kidney (HEK) cells were transiently transfected with accessory plasmids and a retroviral vector bearing the CAR sequence. After 2 days, the supernatant was harvested and applied to J76-NFAT- green fluoresecent protein (GFP) cells, or activated PBMCs, in one or two rounds of transduction, respectively.
Flow cytometry
Testing of antibodies raised against CA125 was performed as follows. Target cell lines were detached with EDTA (see above) and then incubated with hybridoma supernatant diluted 1/20 for 15 min at room temperature (RT). As a positive control, we used an anti-CA125 antibody (clone X75, Thermo Fisher Scientific, #MA1-90039) at a dilution of 1/200. Cells were then washed twice with flow buffer and incubated with goat anti-mouse IgG secondary antibody (minimal cross-reactivity, BioLegend, USA, #405308) for 15 min at RT. Cells labeled with secondary antibody alone were used as negative controls. Cells were washed once more and resuspended in flow buffer for flow cytometry analysis.
Expression of the anti-MUC16CARs was detected by flow cytometry. T cells were labeled with a biotinylated anti-mFab antibody (1/200 dilution, Jackson ImmunoResearch, USA #115-066-072) followed by incubation with a Streptavidin-APC secondary antibody (1/200-1/400, BioLegend, #405207).
Cells were acquired on a FACSCanto 10 instrument (BD Biosciences, USA) and analyzed using FlowJo software V.10 (BD Biosciences).
2D and 3D cell cultures staining
For staining of two-dimensional (adherent monolayer, 2D) and three-dimensional (spheroids, 3D) cell cultures, the IncuCyte (IncuCyte S3, Sartorius Lab Instruments) was used to image seeded cells and quantify X75 staining. Briefly, HeLa and OVCAR3 cells were grown in flat bottom wells; for 2D, 20,000 cells were seeded per well. 3D cell culture was obtained by seeding 4000 cells, per well, in 1% agarose-coated flat bottom wells for spheroid formation. Cells were allowed to grow for 2 days and then stained with X75 antibody, or isotype controls. Total red integrated intensity was quantified with the IncuCyte software.
Sampling of detached CA125
For assessment of shedding by cell lines in culture, cells were allowed to remain in the media for at least 2 days and grown to at least 70% confluency. About 500 µL of culture media (“supernatant”) was removed and spun down (500×g for 5 min) to remove cellular material, and 400 µL of this was then frozen. Mouse serum samples were collected from whole blood via cardiac puncture or from the saphenous vein. Blood was allowed to coagulate for 4 hours at RT and then centrifuged to remove debris. Serum was then removed and transferred to a new tube. Sampling of the peritoneal fluid was via washing with PBS. After anesthesia, the skin was cut and retracted, and a small incision was made into the abdominal cavity. A 1 mL syringe (without needle) was used to introduce 500 µL of sterile PBS into the peritoneum. The abdomen was massaged briefly, and then fluid was removed with the same syringe and transferred to a clean tube. This was spun down (500×g for 5 min) to remove cellular material, and then 400 µL of this was then frozen. Samples were then submitted and processed as clinical samples. CA125 was measured using the Elecsys CA125 II assay on the Cobase601 platform (Roche Diagnostics, Mannheim, Germany).
Sequencing of hybridomas and generation of CAR constructs
CA125 contains three topographically distinct antigenic determinants, with most monoclonal antibodies grouped as OC125-like (group A) or M11-like (group B), while OV197 alone constitutes a third category, group C.37 The hybridomas producing the anti-CA125 antibodies used in this study were generated at the Department of Medical Biochemistry, Oslo University Hospital, Radium Hospital, Oslo, Norway. We selected hybridomas K93 and K95 (both group A) and K97 and K101 (both group B).38 Sequencing of hybridoma samples was performed as in the literature.39 Briefly, mRNA was extracted from frozen, pelleted hybridoma cells. This was used for 5′-RACE sequencing, and the resulting heavy and light chain sequences were codon-optimized and adapted as a single chain variable fragment (scFv) using the orientation of light chain-linker-heavy chain. The CAR constructs were finalized by fusing the scFv to a CD8 hinge, CD8 transmembrane domain, a 4-1BB costimulatory domain, and a CD3ζ intracellular domain, as described in the literature.40 The anti-CD19 scFv derived from FMC63 (a kind gift from Martin Pule, UCL Cancer Institute, London, UK) was incorporated in the same arrangement. The sequence of the anti- MUC16ecto clone 4H11 heavy and light chains was obtained from patent WO2011119979A2.
To generate luciferase-expressing cell lines for bioluminescent in vitro and in vivo assays, we adapted a construct incorporating the firefly luciferase-GFP (a kind gift from Rainer Loew, BioNTech IMFS, Idar-Oberstein, Germany)41 into the retroviral vector pMP71.36 Target cell lines were transduced with this vector, and GFP-positive cells were sorted by FACS.
The NFAT-GFP construct used in the reporter assays (pSIRV-NFAT-eGFP) was a gift from Peter Steinberger (Addgene plasmid #118031).42
Co-culture assays
For the reporter assays, Jurkat-76 cells (J76, a kind gift of Professor Mirjam Heemskerk) were transduced with the NFAT-GFP reporter construct42 to generate a J76-NFAT-GFP clone that expressed GFP in response to T-cell activation signals.43 This clone was subsequently transduced with the CAR vectors and then co-cultured with GFP-negative target cells at an effector:target (E:T) ratio of 1:2 overnight in RPMI with 10% FCS. CAR-mediated reactivity was assessed by flow cytometry of GFP expression.
CAR T-cell functionality was assessed by using CD107a as a degranulation marker. CAR-transduced T cells were co-cultured with target cells (E:T=1:2) for 5 hours in the presence of anti-CD107a (BD Biosciences, catalog number 555802) at the recommended concentration. Degranulation was assessed by flow cytometry. Here, effector and target cells were distinguished by GFP expression (GFP+target cells) or by prelabeling with Cell Trace Violet, as per the manufacturer’s methods (Thermo Fisher Scientific).
Bioluminescence-based cytotoxicity assays were performed as previously described.44 Briefly, luciferase-expressing tumor cells were co-cultured with CAR T cells in a 96-well plate in the presence of D-luciferin (75 µg/mL, Perkin Elmer, Norway) at the indicated E:T ratios. Cells were placed in an incubator at 37°C, 5% CO2, and bioluminescence was measured with a luminometer (VICTOR Multilabel Plate Reader, Perkin Elmer) at several time points up to 24 hours.
For killing assays using the IncuCyte (IncuCyte S3, Sartorius Lab Instruments) target cell lines, at 2000 cells/well, were seeded as spheroids for 5 days. T cells were added (10,000 cells/well), along with Annexin V red, at the recommended concentration (Sartorius, USA, catalog number 4641). Annexin V signal was recorded hourly, and the total red object integrated intensity (RCU×µm²/Image) was calculated.
For examination of cytokine secretion, CAR T cells were co-cultured with target cells (E:T=1:2) overnight in serum-free RPMI or X-vivo V.15. After centrifugation (500×g for 5 min), the supernatant was removed and immediately frozen at −80°C until use. Samples were prepared as per the manufacturer’s methods (Bio-Plex Pro Human Cytokine Group I panel, 17-plex kit, Bio-Rad Laboratories, USA).
Patient samples
Samples were obtained from patients undergoing primary debulking surgery at the Department of Gynaecological Oncology, Oslo University Hospital (n=4), or from frozen patient peritoneal effusions (n=2) from the Department of Pathology, Oslo University Hospital. The samples from debulking surgery were dissociated by physical and enzymatic means. Briefly, tumor samples were washed with RPMI with 10% FCS and reduced to 1–2 mm3 sized pieces by cutting with scalpels. This was centrifuged for 5 min at 100×g, and the supernatant was removed. Samples were resuspended in 10% RPMI, supplemented with collagenase II (Sigma Aldrich, C6885-500MG, at 4.7 mg/mL final) and DNase (Sigma Aldrich, DN25-100MG, at 0.57 mg/mL final), and incubated at 37°C with constant rotation for 1 hour, or longer if necessary. Samples were then passed successively through 100, 70, and 40 µm cell strainers. Samples were centrifuged at 500×g (5 min), then resuspended in 5 mL ACK lysis buffer (Lonza, #10-548E) for 30 s, before 5 mL of 10% RPMI was added. Samples were centrifuged at 400×g (10 min), then resuspended and frozen, or used fresh.
To test the reactivity of CAR T cells against OC samples, the samples were labeled with commercial anti-CA125 antibody (X75 at 1/200 dilution, Thermo Fisher Scientific, #MA1-90039) followed by a secondary antibody (APC goat anti-mouse IgG, BioLegend #405308, at 1/200 dilution), and labeling intensity was normalized against the same samples labeled with the secondary antibody only. For clarity, samples were grouped as CA125-low/negative, medium, or high. CAR T cells and patient samples were co-cultured overnight at an E:T ratio of 1:2 in the presence of anti-CD107a (BD Biosciences, catalog number 555802). CD107a labeling was compared with CAR T cells cultured under the same conditions but without patient cells present.
An established patient-derived xenograft (PDX) model derived from primary patient tumors, categorized as a stage IV HGSC, was used in this study. This PDX was generated from a treatment-naïve patient tumor, sampled from primary debulking surgery (Gynaecologic Cancer Biobank, Women’s Clinic, Haukeland University Hospital, Bergen, Norway). This tumor sample was cut into small tissue pieces of 1–2 mm3 in size. Tissue pieces were enzymatically dissociated with collagenase II (300 U/mL, catalog #17101015, Gibco, USA) and DNase (0.1 mg/mL, #07900, STEMCELL Technologies, UK) supplemented with calcium chloride (3 mM) for 2 hours with constant agitation (250 rpm) at 37°C. Digested tumor tissue was washed in PBS, strained through a 40 µm cell strainer and centrifuged. Cell viability was determined with trypan blue staining. Samples were cryopreserved or immediately injected orthotopically into the bursa of the ovary.45 After successful engraftment into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice, the passaged PDX material (passage 2) was cryopreserved before further use in in vivo preclinical MUC16 CAR T-cell treatment studies.
PET-CT imaging and analysis
For quantitation of the PDX model, PET-CT scans were acquired using the integrated nanoScan PC PET/CT (Mediso, Hungary) featuring spatial resolution of 800 and 300 µm of the positron emission tomography (PET) and CT detector systems, respectively. The field of view (FOV) of the stacked images was 9.6 axial×10 cm transaxial allowing whole-body 3D imaging of the mice. Animals were scanned using a dual mouse bed with integrated heating (37°C). Each PET scan was conducted over 20 min, 45 min after intravenous administration of 18F-fluorodeoxyglucose (18F-FDG; 5–12 MBq/mouse). Prior to PET acquisition, a whole-body CT scan (helical projections with tube energy of 50 kVp, exposure time 300 ms, 7.2 projections, max FOV, binning 1–4) was performed providing anatomic information.
PET images were reconstructed using the Nucline software and parameters were static normal, while CT images were reconstructed using a RamLak filter. The PET and CT images were coregistered automatically. Images were reconstructed with a voxel size of 0.25×0.25×0.25 mm3 for CT and 0.4×0.4×0.4 mm3 for PET. Data analyses were performed using InterView Fusion V.3.03.078.0000 (Mediso). Standard uptake value (SUV) was calculated using the equation: SUV=CPET(T)/(ID/BW), where CPET(T) was the measured activity in tissue, ID is the injected dose measured in kBq (corrected using https://www.cyclotron.nl/decay-calculator/), and BW is the mouse’s body weight in kg. For each scan, a signal above 2.5 SUV was regarded as a true tumor signal and a spherical volume of interest (VOI) was drawn semiautomatically for estimation and calculation of SUVmean and SUVmax.
Soluble CA125
To represent cleaved CA125 in our experiments, we used human calibration-grade CA125 protein (#abx060961, Abbexa, UK). Where this was used in co-culture assays (bioluminescence-based cytotoxicity and CD107a functional assays), it was added in solution, at the indicated concentrations.
Dot blots
Proteins of interest were applied to a nitrocellulose membrane and allowed to dry. The membrane was then blocked with 5% bovine serum albumin (BSA) in Tris-buffered saline (TBS), before incubation with the primary antibody (various). Next, the membrane was washed three times in TBS-Tween, then incubated with horseradish peroxidase-conjugated anti-mouse IgG secondary antibody (#62-6520, Thermo Fisher Scientific). After another three washes with TBS-Tween, the membrane was incubated for 1 min with enhanced chemiluminescent substrate, then the luminescent signal was read on an Amersham Imager 600 (G.E. Healthcare, USA). Semiquantitative analysis was performed using GelQuant software (BiochemLabSolutions.com).
In vivo assays
Female NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, OUS and UiB) and NXG (NOD-Prkdcscid-IL2rgTm1 /Rj, OUS) mice were maintained in pathogen-free conditions under the respective institutional animal care protocols. All animal experiments were conducted in compliance with the procedures from the Norwegian State Commission for Laboratory Animals and approved by the Norwegian Food Safety Authority.
For the cell line study, luciferase-expressing OVCAR3 or HeLa cells were trypsinized and washed twice in PBS. NXG mice 6–10 weeks old were injected intraperitoneally with 1×106 luciferase-expressing OVCAR3 or HeLa cells in 200 µL PBS. After 3 days, 200 µL of D-luciferin (20 mg/mL, #122799-5, Perkin Elmer, Waltham, USA) was injected intraperitoneally, and engraftment was confirmed by bioluminescence imaging (BLI) using the IVIS Spectrum in vivo imaging system (Perkin Elmer). Mice were allocated to treatment groups based on comparable tumor loads. The same day 5×106 T cells (Mock, K101CAR, or 4H11CAR) were injected intraperitoneally. This was followed by a second injection of the same cells, 8 days later for the OVCAR3 experiment, and 4 days later for the HeLa experiment.
For the PDX study, NSG mice 6–8 weeks old were injected with 1×105 PDX P2 cells orthotopically into the bursa of the right ovary.45 After 22 days, engrafted mice received 5×106 Mock (n=8) or K101CAR T cells (n=7) intraperitoneally, in 200 µL of serum-free RPMI. A second injection was delivered 2 days later. At 41 days (19 days after first T cell injection), mice were examined by PET-CT. This was repeated at 86 days (64 days after T cell injection). The humane endpoint was reached on day 276.
Statistical analysis
Statistical analyses, as indicated below (paired t-tests, one-way and two-way analyses of variance (ANOVAs), simple linear regression, Mann-Whitney, and log-rank tests), were calculated using GraphPad Prism V.9 (GraphPad Software, San Diego, California, USA).
Results
MUC16 is expressed in a large proportion of OC samples
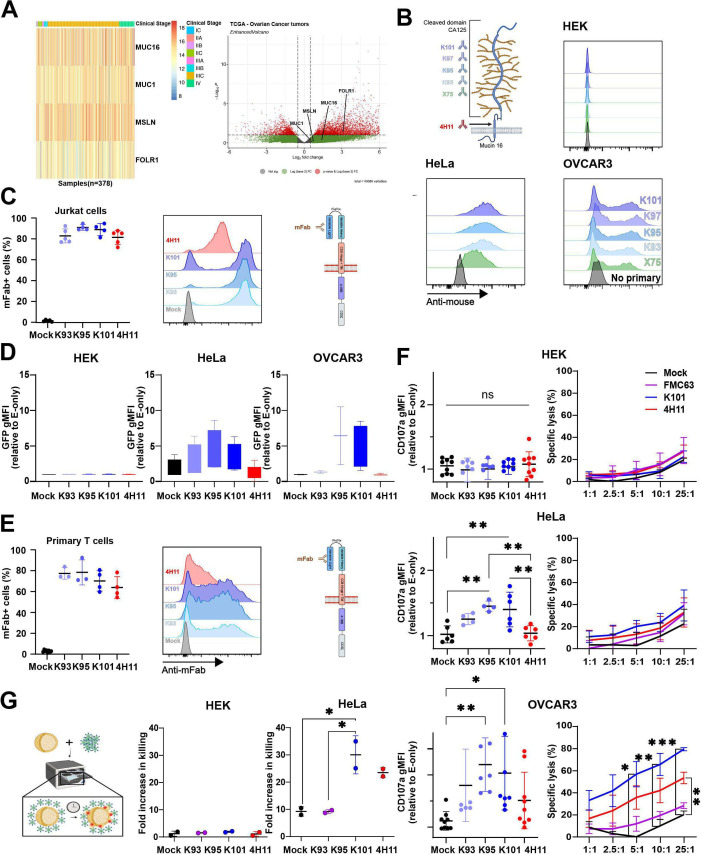
Although already recognized as a valid OC target, MUC16 distribution was analyzed from a TCGA cohort of 375 ovarian tumor samples.46 We compared MUC16 expression to that of other OC-validated targets (MSLN, FOLR1 and MUC1)47–49 and included the clinical stage information. We conducted DESeq2 analysis on the TCGA tumor samples, confirming that MUC16 was expressed in the majority of OC samples (figure 1A, left). We then used a volcano plot to visualize whether MUC16 expression was linked to clinical stage (figure 1A, right). MUC16 was compared with the three other targets but none of these genes were in the top 100 for RNA expression; however, they all show positive fold changes throughout the different clinical staging groups. These data confirm that MUC16 is a marker for OC and therefore a potential drug target for treating patients with advanced disease.
Figure 1.
MUC16 expression, CAR design and functionality in vitro. (A) Left—Gene expression of MUC16, MSLN, MUC1 and FOLR1 in patient samples across clinical stages. Data were taken from a TCGA OC cohort of 375 patients. The color scale varies from the low expression (blue) to high expression (red). The different clinical stages are shown in a graphical colorful legend as indicated. Right—Volcano plot where the x-axis is the fold change in gene expression and y-axis is the statistical significance (-log10 p value). Genes with a high degree of significance (p values <0.05) and substantial fold changes are in red and genes with lower significance or smaller fold changes are in green. Non-significant genes are in gray. (B) Representation of MUC16 protein at the plasma membrane and recognition sites of the antibodies used in this study. K-series hybridoma supernatants staining of HEK, HeLa, and OVCAR3 cell lines. The commercial anti-CA125 antibody, X75, was used as a positive control, and staining with the secondary only (anti-mouse) was used as a background control. Samples were then analyzed by flow cytometry. (C) Expression of MUC16CAR constructs in J76-NFAT-GFP assessed by staining with biotinylated anti-mFab antibody and fluorescent streptavidin. Samples were then analyzed by flow cytometry. Left—CAR expression in J76-NFAT-GFP as percent mFab-positive cells (N=4–5 independent transductions, expression of CARs, data are mean±SD). Right—Representative staining of J76-NFAT-GFP transduced or not (Mock) with the indicated MUC16CAR construct. (D) J76-NFAT-GFP cells expressing the indicated MUC16CAR construct were co-cultured (E:T=1:2) overnight with MUC16pos (HeLa, OVCAR3) or MUC16neg (HEK) cell lines, with effector-only (E-only) condition used as a baseline. CAR reactivity was assessed on the basis of GFP expression, N=3 independent experiments, data are mean±SD. (E) As in (C) using primary T cells from N=3–4 healthy donors. (F) Functional activity of primary T cells incubated with the indicated target cells (top: HEK, middle: HeLa, bottom: OVCAR3). Left— Degranulation activity: CAR-transduced T cells were co-cultured with target cells at E:T=1:2 overnight, in the presence of anti-CD107a fluorescent antibody. Target cells were gated out and CD107a was detected on T cells. Data are mean±SD, n=2–4 donors, N=4–6 independent experiments, one-way ANOVA, *p<0.05, **p<0.01. Right—Cytotoxic activity: CAR T cells were tested in bioluminescence-based citotoxicity co-cultures at different E:T ratios, as indicated. Anti-CD19 CAR, FMC63, was included as an irrelevant CAR control. The time point displayed is at 10 hours of co-culture. Data are mean±SD, N=3 independent experiments, two-way ANOVAs with Dunnett’s multiple comparisons, **p<0.01, ***p<0.001. (G) Cytotoxic assay in three-dimensional (3D) cultures (spheroids). Left—Representation of the assay. Right—HeLa and HEK cells were established as spheroids for 5 days. T cells were added, and killing was measured 24 hours later (day 6). Annexin V was recorded in IncuCyte live-cell imaging, and fold increase relative to target cells alone was calculated. Data are mean of normalized values±SD, N=2 different donors, one-way ANOVA, *p<0.05. ANOVA, analysis of variance; bioluminescence-based citotoxicity, bioluminescence-based cytotoxicity; CAR, chimeric antigen receptor; E:T, effector:target ratio; HEK, human embryonic kidney; OC, ovarian cancer; TCGA, The Cancer Genome Atlas.
Anti-CA125 scFv leads to functional CAR constructs
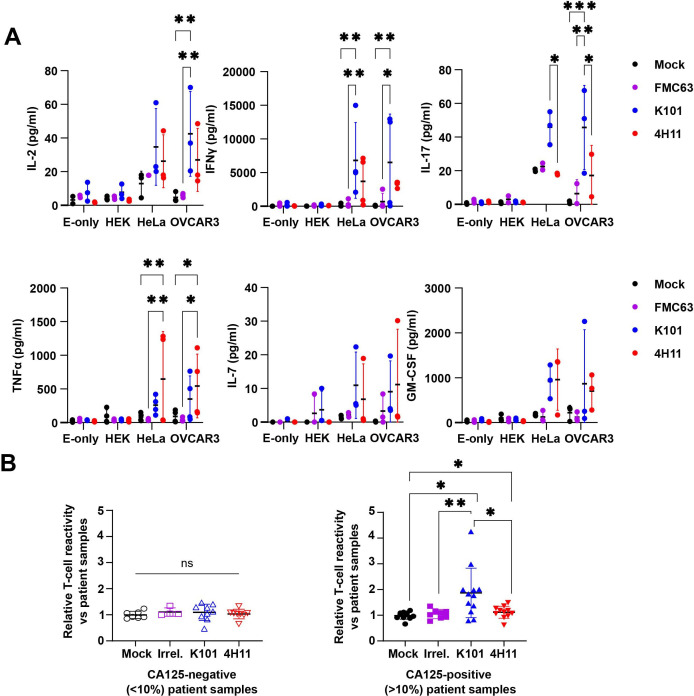
Supernatants from four anti-CA125 hybridomas (K-series33) were tested against CA125-positive and CA125-negative cell lines (figure 1B). The commercial anti-CA125 antibody, X75, was included for reference. As shown, labeling with each of the supernatants of the K-series closely matched the commercial X75 antibody signal. We next identified the heavy and light variable chain sequences from each of the K-hybridomas, except K97, and designed scFv (online supplemental figure 1A–F). In order to compare K-series CAR to an anti-MUC16 validated product, we also cloned the 4H11CAR,18 which is specific to the MUC16ecto. First, a J76-NFAT-GFP reporter cell clone43 50 was transduced with each of the CAR constructs, and their expression was confirmed by staining for mFab (figure 1C). These reporter cells were then co-cultured with different MUC16pos (HeLa, OVCAR3) or MUC16-negative (HEK) cell lines (figure 1D) and GFP signal was detected. As shown, only the MUC16pos cells induced GFP expression, suggesting that the CAR constructs were functional and specific. Then, primary T cells from healthy donors were transduced with the different CAR constructs (figure 1E), and degranulation (CD107a) was used as a marker of T cell activation (figure 1F, left panels). Among the K-series CARs, K95 and K101CARs demonstrated a superior activity toward OVCAR3 and HeLa cells and no reactivity against the MUC16-negative (MUC16neg) HEK cells (figure 1F). However, K95CAR T cells reacted against MUC16neg BL41 lymphoma cell line (online supplemental figure 2) and was therefore eliminated from the study. In addition, K93CAR showed weak reactivity in the different assays and further development was abandoned. On the other hand, 4H11CAR weakly activated J76 cells in the reporter assay (figure 1D, red) and demonstrated a non-significant primary T cell CD107a response against MUC16pos cells (figure 1F, red), although the CAR was well detected on effector cells (figure 1C,E, red), suggesting that MUC16ecto might not be well displayed in these experimental designs. Thus, in vitro studies of MUC16 CAR targeting are not trivial and even validated constructs might not properly respond. We then studied the cytotoxic function of K101 and 4H11CAR T cells by bioluminescence-based cytotoxicity assays at a range of E:T ratios (figure 1F, right panels). We included an irrelevant CAR as a control, anti-CD19 FMC63 (purple). As anticipated, none of the CAR T cells reacted against HEK cells, but K101CAR T cells demonstrated superior killing efficacy against OVCAR3, compared with the other constructs. Of note, 4H11CAR T cells were also able to efficiently kill OVCAR3 cells despite the low CD107a detection. Intriguingly, although CD107a was detected on K101CAR T cells incubated with HeLa cells, no killing was observed with any of the construct (figure 1F). We speculated that the 2D cell growth may influence MUC16 levels which might prevent proper killing. Staining of HeLa and OVCAR3 cells in 2D and as spheroids (3D) indicated that it did indeed affect MUC16 expression which increased in 3D for both cell lines (online supplemental figure 3). We thus tested CAR T cell cytotoxicity in a 3D-based live-cell imaging assay. We observed an increase in killing of HeLa spheroids with both anti-MUC16CARs, whereas no killing was observed against HEK spheroids (figure 1G). Thus, HeLa cells might change their MUC16 distribution following culture conditions. In order to complete the in vitro evaluation of K101CAR T cells, we examined their cytokine responses when co-cultured with target cells for 24 hours by Bioplex assay (figure 2A). We found that K101CAR T cells stimulated a strong release of IL-2, IL-17, and IFN-γ, whereas 4H11CAR T cells lead to TNF-α release. Together, these data support the specificity and effectiveness of K101CAR T cells against MUC16pos targets. In order to validate the robustness of K101CAR T cells, we tested them against a series of primary patient samples, including peritoneal effusions and samples derived from debulking surgery of solid tumors. MUC16 expression was categorized as positive or negative based on the percentage of cells labeled measured by anti-CA125 (X75) labeling (online supplemental figure 4). MUC16CAR T cell reactivity against positive or negative samples was assessed by degranulation and anti-CD107a staining. We observed that K101CAR T cells reactivity followed MUC16 presence, whereas 4H11CAR T cells reacted less than K101CAR T cells with the positive samples (figure 2B). From these data, K101CAR appears more potent than 4H11CAR to redirect T cells against MUC16pos targets.
Figure 2.
Cytokine production of MUC16CAR T cells and reactivity against primary patient samples. (A) Cytokine quantification. CAR T cells were co-cultured for 24 hours with target cells at an E:T ratio of 1:2, and the supernatant was collected for Bioplex cytokine assays. Selected cytokines are shown, data are mean±SD, N=3–4 independent measurements, two-way ANOVA. *p<0.05, **p<0.01. (B) Reactivity of CAR T cells against CA125-positive and CAR-negative samples. Tumor samples from patients with OC (effusions or material from debulking surgery) were labeled for CA125 (X75 antibody). Samples were categorized as CA125-positive (n>9), or CA125-negative (n>7), based on the percentage of cells labeling for CA125 (threshold 10%). These samples were co-cultured with CAR T cells overnight at E:T=1:2 in the presence of anti-CD107a. Effector cells were distinguished by prelabeling CAR T cells with Cell Trace Violet. CAR T cells cultured alone were used for normalization. Data are presented as mean±SD, one-way ANOVA. *p<0.05, **p<0.01. ANOVA, analysis of variance; CA125, cancer antigen 125; CAR, chimeric antigen receptor; E:T, effector:target ratio; HEK, human embryonic kidney; IL, interleukin.
jitc-2023-008179supp001.pdf (239.8KB, pdf)
jitc-2023-008179supp002.pdf (57.4KB, pdf)
jitc-2023-008179supp003.pdf (41KB, pdf)
jitc-2023-008179supp004.pdf (26.5KB, pdf)
jitc-2023-008179supp005.pdf (19.5KB, pdf)
Soluble CA125 does not alter K101CAR T cell activity
An important concern when targeting a cleaved antigen, such as CA125 which is released in the serum, is the risk of reducing the CAR activity. This can happen by competition mechanisms as we previously reported with an anti-IgΚCAR blocked by serum IgGs.51 We first confirmed K101 antibody binding to calibration-grade CA125 protein. Dot blot experiments were designed with top CA125 concentration fixed to 300 kU/L which would mimic a diagnostic situation for OC and other gynecologic cancers where the lower cut-off is set to 20 kU/L.52 53 For the staining, the commercial X75 antibody was used as a positive control and the background level was fixed using an irrelevant hybridoma supernatant, producing an anti-CEA antibody, CEA10. We then tested the indicated amounts or dilutions of antibodies and observed K101 binding to CA125 (online supplemental figure 5A,B). Thus, K101 also reacts with CA125 in a cell-free context, suggesting a risk of potential inhibition of K101CAR by serum CA125.
jitc-2023-008179supp006.pdf (202.4KB, pdf)
We verified shedding of CA125 by the cell lines used in this study with a clinical CA125 assay. CA125 was detectable in the culture media of OVCAR3 cells, and HeLa cells, but not of HEK cells (online supplemental figure 5C). Shedding by OVCAR3 cells was also tested in vivo, with mice receiving an injection of OVCAR3 cells intraperitoneally. After 5 weeks, shed CA125 could be detected in the serum of these mice. CA125 was also detected in the serum of mice with orthotopically engrafted PDX26 cells (online supplemental figure 5D). Finally, CA125 was also detected in peritoneal washes of the mice-bearing OVCAR3 cells (online supplemental figure 5E). These data show that CA125 was released at detectable levels in our different experimental formats and but did not affect CAR T cell activity in vitro (figure 1) and in vivo (see below).
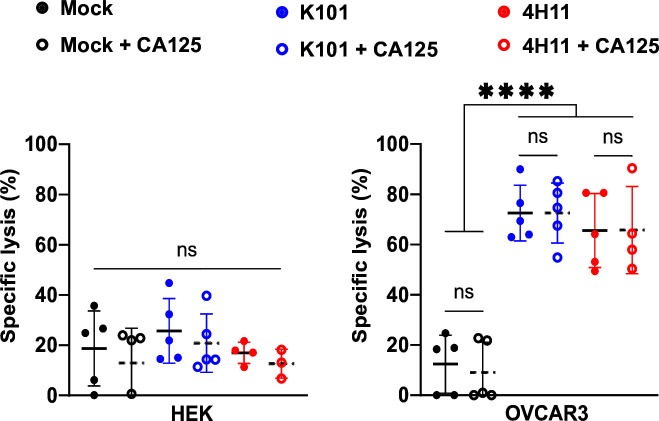
We next performed a co-culture assay of CAR T cells and target cells in the presence or not of 300 U/L calibration-grade CA125. We observed no difference between conditions for all MUC16CAR T cell killing efficacy against the MUC16pos OVCAR3 cells. As expected, no change with the MUC16neg control HEK was detected too and, due to its MUC16ecto specificity, 4H11CAR target recognition was not affected (figure 3). Thus, K101CAR T cells are functional even at a high concentration of CA125 and therefore not sensitive to the presence of soluble target.
Figure 3.
MUC16CAR T cell activity is unaffected by soluble antigen. Bioluminescence-based cytotoxicity assays in the presence of recombinant CA125 (300 kU/L) were performed at an E:T ratio of 10:1. Data are mean±SD, N=5 donors, one-way analysis of variance, with Šidák’s multiple comparisons test. ****p<0.0001. CA125, cancer antigen 125; CAR, chimeric antigen receptor; E:T, effector:target ratio; HEK, human embryonic kidney; ns, not significant.
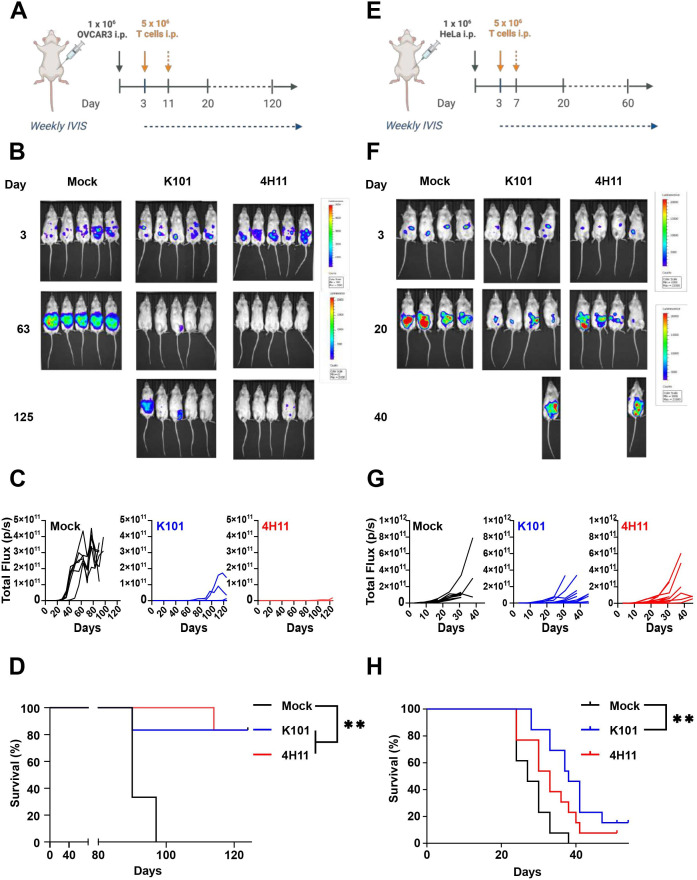
K101CAR T cells are efficient in vivo
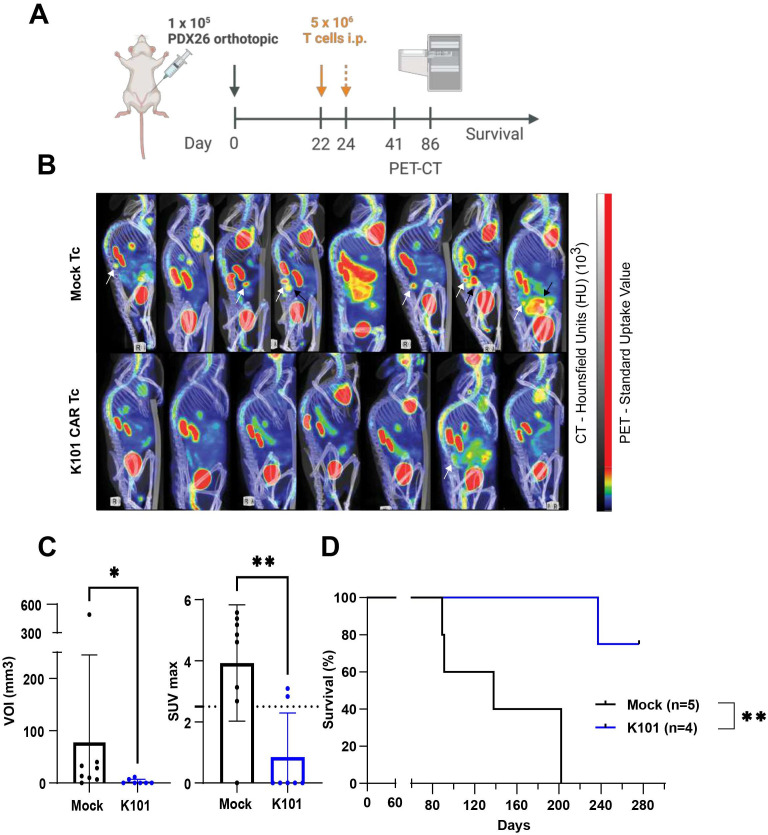
We tested the efficacy of anti-MUC16CAR T cells against OVCAR3 and HeLa cell lines in mouse xenograft models. T cells were activated, transduced, and then expanded in vitro, with cell proliferation and CAR expression monitored to ensure that CAR T cell populations were comparable (online supplemental figure 6). After intraperitoneal engraftment of the slow-growing OC cell line OVCAR3, immunodeficient mice received two injections (days 3 and 11) of CAR T cells or Mock T cell controls (figure 4). We observed that OVCAR3 form numerous small, solid tumors, with considerable expansion of ascites in the peritoneal space (figure 4B). The mice receiving Mock T cells developed a high tumor burden in approximately 30 days, whereas very little signal was detected in mice receiving the MUC16CAR T cells until around day 95 (figure 4C). Accordingly, we observed prolonged survival compared with the Mock T cell group (figure 4D). Ultimately, four out of five of the mice treated with MUC16CAR T cells were alive at the 4-month endpoint of the experiment. In order to assess the robustness of the MUC16CAR constructs, we tested the cervical cancer cell line HeLa because it expresses lower levels of MUC16 at the surface (figure 1B) and grow at a faster rate in vitro. As for OVCAR3 assay, mice were treated twice with T cell but the injections were performed closer in time (at days 3 and 7, figure 4E). As predicted, the HeLa tumors progressed rapidly (figure 4F–G) and were more resistant to CAR T cell control than OVCAR3. Nevertheless, K101CAR T cells result in a significant survival benefit versus Mock T cells in HeLa-engrafted mice, whereas survival with 4H11 T cells was not significantly different from the Mock control (figure 4H). Thus, K101CAR T cells are robust and can control a fast-growing cell line, with moderate levels of MUC16 expression. We finally wanted to evaluate how K101CAR T cells were performing in a complex model. We thus used patient-derived cells, PDX26, with a confirmed expression of MUC16 (online supplemental figure 7) matching the average of the primary samples tested (online supplemental figure 5) to orthotopically injected animals in the bursa. The mice were subsequently treated twice with either 5×106 K101CAR (n=7) or Mock T cells (n=8) intraperitoneally at days 22 and 24 post engraftment (figure 5A). Tumor development was monitored by PET-CT at days 41 and 86 after PDX engraftment (figure 5A,B). Injection of K101CAR T cells leads to a reduction in PDX tumor size and dissemination, where six out of seven K101CAR T cell-treated mice had no tumors observable by necropsy. While PET-CT at day 86 indicated high signal uptake in the ovary observed in six out of eight mock control mice, 18F-FDG uptake could only be observed in one of the treated mice (white arrow, figure 5B). In addition, three animals in the control group showed a large metastatic mass (black arrow, figure 5B) suggesting an efficient spreading of the tumor. Individual quantification supports the protective effect of K101CAR T cells and efficient engraftment of PDX26 in the mock control group. Finally, survival analysis demonstrates a clear protective effect of K101CAR T cells (figure 5C). Taken together, these data show that K101CAR T cells can control different types of MUC16pos tumor in vivo, even aggressive and disseminated models.
Figure 4.
MUC16CAR T cells control in vivo tumor growth. (A) Timeline of OVCAR3 in vivo experiment; 3 days after intraperitoneal injection of 1×106 OVCAR3 cells, engraftment was confirmed by bioluminescence-based cytotoxicity, mice were allocated to study groups, and the first injection of T cells was delivered (5×106, intraperitoneal) on the same day. A second injection of T cells was delivered 8 days later, and mice were observed weekly, including IVIS imaging. (B) IVIS imaging at the indicated time points. (C) Quantification of (B) bioluminescence signal intensity (pixel/second, p/s) over time (days). (D) Kaplan-Meier curve using log-rank test, n=5 mice per group. *p<0.05, **p<0.01. (E) HeLa cells (1×106) were injected intraperitoneally. After 3 days, engraftment was confirmed by BLI, mice were allocated to each group, and the first injection of T cells was delivered (5×106, intraperitoneal) on the same day. A second injection of T cells was delivered 4 days later, and mice were observed weekly, including IVIS imaging. (F) as in (B). (G) as in (C). (H) as in (D), n=4–5 mice per group, N=2 separated experiments, *p<0.05, **p<0.01. CAR, chimeric antigen receptor.
Figure 5.
K101CAR T cells control an orthotopic PDX model. (A) PDX26 cells (1×105) were engrafted orthotopically in the bursa of the ovary in immunodeficient mice. These were allowed to engraft for 22 days, prior to two injections (day 22 and day 24) of T cells (Mock or K101CAR). Tumor development was subsequently monitored with PET-CT using 18F-fluordesoxyglucose. (B) PET-CT images of the PDX-engrafted mice, treated with Mock or K101CAR T cells, taken on day 86 after engraftment. White arrows indicate primary tumor/ovarian tumor mass, black arrows indicate metastatic masses. (C) Spherical volume of interest (VOI (left)) and standard update value (SUV (right)) analyses of Mock and K101CAR T cell-treated mice at day 86. An SUV of 2.5 was considered a true tumor value. Columns are presented as mean±SD, n=7–8, Mann-Whitney test, *p<0.05, **p<0.01. (D) Kaplan-Meier curve of Mock and K101CAR T cell-treated mice (graft-versus-host disease (GvHD) events excluded), n=4–5, log-rank test, *p<0.05, **p<0.01. CAR, chimeric antigen receptor; PDX, patient-derived xenograft.
jitc-2023-008179supp007.pdf (21.6KB, pdf)
jitc-2023-008179supp008.pdf (214KB, pdf)
Discussion
OC is a common gynecologic cancer with high mortality rates. This is due in part to typically late diagnosis, limited treatment options, and high rates of recurrence.4 Conventional therapies are limited in efficacy and range, and new treatment modalities are needed. CAR therapy has the potential to direct immune cells to targets in immunologically cold tumor microenvironments. In this paper, opposite to what was previously predicted,18 we demonstrate that a CAR that targets CA125—the cleaved domain of MUC16—can efficiently recognize and kill MUC16pos tumors. This also confirms the reassociation of CA125 with tumor cells at levels sufficient to trigger a cytotoxic response.31 Another argument precluding the targeting of CA125 was the presence of the soluble antigen in the patient serum which could inhibit a CAR by competing with the cellular CA125. While the replication of “true” in situ conditions can be challenging, we also show that high levels of soluble CA125 in the immediate vicinity of the target cells did not affect CAR T cell activity in vitro. Furthermore, we detected CA125 in the serum and peritoneum of our in vivo tumor models, suggesting that anti-MUC16CAR T cells were not sensitive to high levels of CA125, confirming that the presence of this marker might not preclude K101CAR T cell activity in clinical settings.
Our work also questioned the value of in vitro assays alone in predicting CAR T functionality. OVCAR3 cells, which are often used as a validation model, do present all the expected features; they are slow growing and MUC16high, and, as expected, were easily killed by the anti-MUC16CARs. However, the cervical tumor-derived HeLa cells were markedly harder to control, especially in 2D culture. We suspect that this was due to (1) heterogeneity of MUC16 expression for which we confirmed the reports of previous authors,54 and/or (2) the influence of culture conditions on CA125 expression and distribution.55 Our microscopy study indeed supports the idea that CA125 presence is increased in 3D culture, and it is tempting to speculate that HeLa cells in 2D do not present detectable levels of MUC16 to stimulate K101CAR, whereas 3D becomes targetable. This could explain how K101CAR T cells kill HeLa cells grown in 3D and in vivo, these observations challenge the validity of the in vitro systems widely used by the CAR community, while supporting previous propositions that 3D systems are more representative or predictive of the in vivo context.56 57 We found that the advantage of our CAR carried over into testing on CA125-positive patient-derived samples; K101CARs typically showed stronger reactivity than did the 4H11CARs in vitro, although the nature of the samples prevented us from directly comparing cytotoxicity against them. Nevertheless, testing of the K101CAR in vivo in an orthotopic PDX model confirmed killing under these conditions. Interestingly, the PDX-26 did not demonstrate excessive presence of MUC16 at the surface, suggesting that the protein might be cycling, which might preclude clear correlation between staining and actual killing. To our knowledge, this is the first demonstration of CAR-mediated killing of patient-derived tubo-ovarian carcinomas in vivo. We plan in the future to use this orthotopic system to compare different CAR constructs.
Among CAR-based immunotherapeutics to treat OC, the 4H11CAR has been the main focus of CAR research, progressing to clinical trials in multiple formats.9 58 This focus on the targeting of the MUC16ecto 18 is evidently based on the assumption that CA125 is not associated (or reassociated) with the tumor cells at levels sufficient for targeting, and/or that the shed (soluble) CA125 interferes with CAR T cell functionality. These questions provided a context for the current study, and we have answered both. First, CA125 is detectable on tumor cells lines and primary tumor samples, at levels sufficient for targeting by CAR T cells both in vitro and in vivo. Second, we found no evidence that soluble CA125, even at high concentrations, had any effect on CAR T-cell cytotoxicity nor specificity. In comparison to the ectodomain-targeting 4H11-based CAR, the efficient killing of MUC16pos target cells by the K101CAR T cells indicates that the extracellular repeat domain is indeed an attractive target. Taken together, our results also open the path to the development of other anti-CA125 constructs38 and warrant further clinical development of the K101CAR.
jitc-2023-008179supp009.pdf (4.1MB, pdf)
Acknowledgments
We thank Sepehr Teimouri (Oslo University Hospital) who assisted with quantification of IVIS imaging and Åsmund Nybøen (Oslo University Hospital) who assisted with handling and processing of patient samples. LB, EMC and SW are grateful to OvaCure (Denmark) for their support in finding novel treatments for OC. We are grateful to the flow cytometry core facility of the Oslo University Hospital for excellent technical assistance. Images in figures 1, 4, 5 and 6 were created using Biorender.com.
Footnotes
@LabOslo
KK and CF contributed equally.
Contributors: NPC: Experiments, manuscript writing, first draft. KK: Experiments. CF: Experiments, manuscript writing. PFG: Experiments, manuscript writing. EB: Experiments. SJ: Experiments. BC: Experiments. CHG: Experiments. EGdJ: Experiments. FK: Experiments. DJW: Experiments, provided critical material. KL: Provided critical clinical material, manuscript writing. ER: Provided critical clinical material. BD: Analysis, essential material. MRM: Experiments. LB: Manuscript writing, funding, provided critical material. EMC: Funding, provided critical material. EMI: Supervision, manuscript writing, funding. SW: Supervision, experiments, manuscript writing, first draft, funding. SW was the guarantor.
Funding: NPC was a postdoctoral fellow funded by the South-Eastern Norway Regional Health Authority (# 2019062) and is presently supported by a Fight Kids Cancer “IMAGINE” (# 40085). CF is supported by a POLNOR-2019 fellowship of the research project “ALTERCAR” (# 0056/2019) fellowship, BC is a postdoctoral fellow of the Norwegian Cancer Society and the University of Oslo (Scientia Fellow II—Marie Skłodowska-Curie Actions # 801133), and SJ is supported by the Norwegian Research Council under the frame of the Era-Net EURONANOMED-3 European Research project “NAN-4-TUM” (# 310531). This project was also supported by grants from the Norwegian Cancer Society (# 6829007; #223171) to SW and from the Norwegian Research Council KSP-2021 “CellFit” grant (# 326811) to EMI. KK and LB were supported by Helse Vest RHF (# F-12183/4800003665). EMC and PG were supported by the Norwegian Cancer Society (# 223171 and 208012), the Research Council of Norway (# 326300 and 326811), and through its Centers of Excellence funding scheme (# 223250).
Competing interests: SW, EMI, DJW, NPC and EB have filled a patent application on chimeric antigen receptor for ovarian cancer. The other authors declare no conflict of interest.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
All investigations were carried out according to the 2013 revision of the Declaration of Helsinki. This research project has been conducted using material from The Cancer Biobank, Oslo University Hospital (REK ID S-04300). PBMCs were obtained from healthy donors according to the Regional Committee for Medical Research Ethics South-East (REK ID 60772). The PDX model was derived from a patient tumor sample from the Gynaecologic Cancer Biobank, Women’s Clinic, Haukeland University Hospital, Bergen, Norway. The biobanking and the present study are approved by REC West (REK ID: 2014/1907, 2015/548 and 2018/72). Where necessary, written informed consent was obtained from all women before the collection of fresh tumor tissues and clinicopathologic parameters were initiated. The animal study protocol was approved by the Institutional Review Board (or Ethics Committee) of Norwegian Food Safety Authority (OUS FOTS IDs 11118 and 26731. UiB FOTS ID 14128).
References
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Institute NC . Ovarian cancer — cancer STAT facts. 2019.
- 3. Chien J, Poole EM. Ovarian cancer prevention, screening, and early detection: report from the 11th biennial ovarian cancer research symposium. Int J Gynecol Cancer 2017;27:S20–2. 10.1097/IGC.0000000000001118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jayson GC, Kohn EC, Kitchener HC, et al. Ovarian cancer. Lancet 2014;384:1376–88. 10.1016/S0140-6736(13)62146-7 [DOI] [PubMed] [Google Scholar]
- 5. Ledermann JA, Raja FA, Fotopoulou C, et al. Newly diagnosed and Relapsed epithelial ovarian carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;24 Suppl 6:vi24–32. 10.1093/annonc/mdt333 [DOI] [PubMed] [Google Scholar]
- 6. Colombo N, Ledermann JA, ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org . Updated treatment recommendations for newly diagnosed epithelial ovarian carcinoma from the ESMO clinical practice guidelines. Ann Oncol 2021;32:1300–3. 10.1016/j.annonc.2021.07.004 [DOI] [PubMed] [Google Scholar]
- 7. Li Y, Tian R, Liu J, et al. Deciphering the immune landscape dominated by cancer-associated fibroblasts to investigate their potential in indicating prognosis and guiding therapeutic regimens in high grade Serous ovarian carcinoma. Front Immunol 2022;13. 10.3389/fimmu.2022.940801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bronger H. Immunology and immune Checkpoint inhibition in ovarian cancer – current aspects. Geburtshilfe Frauenheilkd 2021;81:1128–44. 10.1055/a-1475-4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu JWY, Dand S, Doig L, et al. T-cell receptor therapy in the treatment of ovarian cancer: A mini review. Front Immunol 2021;12:672502. 10.3389/fimmu.2021.672502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kandalaft LE, Dangaj Laniti D, Coukos G. Immunobiology of high-grade Serous ovarian cancer: lessons for clinical translation. Nat Rev Cancer 2022;22:640–56. 10.1038/s41568-022-00503-z [DOI] [PubMed] [Google Scholar]
- 11. Benard E, Casey NP, Inderberg EM, et al. SJI 2020 special issue: A catalogue of ovarian cancer targets for CAR therapy. Scand J Immunol 2020;92:e12917. 10.1111/sji.12917 [DOI] [PubMed] [Google Scholar]
- 12. Bast RC, Klug TL, St John E, et al. A Radioimmunoassay using a Monoclonal antibody to monitor the course of epithelial ovarian cancer. N Engl J Med 1983;309:883–7. 10.1056/NEJM198310133091503 [DOI] [PubMed] [Google Scholar]
- 13. Singh AP, Senapati S, Ponnusamy MP, et al. Clinical potential of Mucins in diagnosis, prognosis, and therapy of ovarian cancer. Lancet Oncol 2008;9:1076–85. 10.1016/S1470-2045(08)70277-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gubbels JAA, Belisle J, Onda M, et al. Mesothelin-Muc16 binding is a high affinity, N-Glycan dependent interaction that facilitates peritoneal metastasis of ovarian tumors. Mol Cancer 2006;5:50. 10.1186/1476-4598-5-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O’Brien TJ, Beard JB, Underwood LJ, et al. The CA 125 gene: an extracellular superstructure dominated by repeat sequences. Tumour Biol 2001;22:348–66. 10.1159/000050638 [DOI] [PubMed] [Google Scholar]
- 16. Duffy MJ, Bonfrer JM, Kulpa J, et al. Ca125 in ovarian cancer: European group on tumor markers guidelines for clinical use. Int J Gynecol Cancer 2005;15:679–91. 10.1111/j.1525-1438.2005.00130.x [DOI] [PubMed] [Google Scholar]
- 17. Bast RC, Badgwell D, Lu Z, et al. New tumor markers: Ca125 and beyond. Int J Gynecol Cancer 2005;15 Suppl 3:274–81. 10.1111/j.1525-1438.2005.00441.x [DOI] [PubMed] [Google Scholar]
- 18. Chekmasova AA, Rao TD, Nikhamin Y, et al. Successful eradication of established peritoneal ovarian tumors in SCID-beige mice following adoptive transfer of T cells genetically targeted to the Muc16 antigen. Clin Cancer Res 2010;16:3594–606. 10.1158/1078-0432.CCR-10-0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun L, Gao F, Gao Z, et al. Shed antigen-induced blocking effect on CAR-T cells targeting Glypican-3 in hepatocellular carcinoma. J Immunother Cancer 2021;9:e001875. 10.1136/jitc-2020-001875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu X, Onda M, Watson N, et al. Highly active CAR T cells that bind to a Juxtamembrane region of Mesothelin and are not blocked by shed Mesothelin. Proc Natl Acad Sci USA 2022;119:19. 10.1073/pnas.2202439119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedman KM, Garrett TE, Evans JW, et al. Effective targeting of multiple B-cell maturation antigen-expressing hematological Malignances by anti-B-cell maturation antigen Chimeric antigen receptor T cells. Hum Gene Ther 2018;29:585–601. 10.1089/hum.2018.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carpenter RO, Evbuomwan MO, Pittaluga S, et al. B-cell maturation antigen is a promising target for adoptive T-cell therapy of multiple myeloma. Clin Cancer Res 2013;19:2048–60. 10.1158/1078-0432.CCR-12-2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. García-Guerrero E, Sierro-Martínez B, Pérez-Simón JA. Overcoming Chimeric antigen receptor (CAR) modified T-cell therapy limitations in multiple myeloma. Front Immunol 2020;11:1128. 10.3389/fimmu.2020.01128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang ZL, Lorenzini MH, Chen X, et al. Rewiring T-cell responses to soluble factors with Chimeric antigen receptors. Nat Chem Biol 2018;14:317–24. 10.1038/nchembio.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aithal A, Rauth S, Kshirsagar P, et al. Muc16 as a novel target for cancer therapy. Expert Opin Ther Targets 2018;22:675–86. 10.1080/14728222.2018.1498845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koneru M, O’Cearbhaill R, Pendharkar S, et al. A phase I clinical trial of adoptive T cell therapy using IL-12 Secreting MUC-16(Ecto) directed Chimeric antigen receptors for recurrent ovarian cancer. J Transl Med 2015;13:102. 10.1186/s12967-015-0460-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crawford A, Haber L, Kelly MP, et al. A Mucin 16 Bispecific T cell-engaging antibody for the treatment of ovarian cancer. Sci Transl Med 2019;11:eaau7534. 10.1126/scitranslmed.aau7534 [DOI] [PubMed] [Google Scholar]
- 28. Winer IS, Shields AF, Yeku OO, et al. A phase I/II, multicenter, open-label study of Regn5668 (Mucin [MUC]16 X Cd28 Bispecific antibody [bsAb]) with Cemiplimab (programmed death [PD]-1 AB) or Regn4018 (Muc16 X Cd3 bsAb) in recurrent ovarian cancer (rOVCA). JCO 2021;39:TPS5602-TPS. 10.1200/JCO.2021.39.15_suppl.TPS5602 [DOI] [Google Scholar]
- 29. Frontiers . Bispecific T-cell engaging antibodies against Muc16 demonstrate efficacy against ovarian cancer in monotherapy and in combination with PD-1 and VEGF inhibition. 2023. [DOI] [PMC free article] [PubMed]
- 30. Wang Q, Ma X, Wu H, et al. Oncolytic adenovirus with Muc16-bite shows enhanced antitumor immune response by reversing the tumor Microenvironment in PDX model of ovarian cancer. OncoImmunology 2022;11. 10.1080/2162402X.2022.2096362 Available: https://doiorg/101080/2162402X20222096362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aithal A, Junker WM, Kshirsagar P, et al. Development and characterization of Carboxy-terminus specific Monoclonal antibodies for understanding Muc16 cleavage in human ovarian cancer. PLoS One 2018;13:e0193907. 10.1371/journal.pone.0193907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schultes BC, Zhang C, Xue LY, et al. Immunotherapy of human ovarian carcinoma with OVAREXTM MAB-B43.13 in a human-PBL-SCID/BG mouse model. Hybridoma 1999;18:47–55. 10.1089/hyb.1999.18.47 [DOI] [PubMed] [Google Scholar]
- 33. Warren DJ, Nustad K, Beard JB, et al. Expression and EPITOPE characterization of a recombinant CA 125 repeat: fourth report from the ISOBM TD-1 workshop. Tumor Biol 2009;30:51–60. 10.1159/000209988 [DOI] [PubMed] [Google Scholar]
- 34. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with Deseq2. Genome Biol 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Belisle JA, Horibata S, Jennifer GAA, et al. Identification of Siglec-9 as the receptor for Muc16 on human NK cells, B cells, and monocytes. Mol Cancer 2010;9:1–14.:118. 10.1186/1476-4598-9-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wälchli S, Løset GÅ, Kumari S, et al. A practical approach to T-cell receptor cloning and expression. PLoS One 2011;6:e27930. 10.1371/journal.pone.0027930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nustad K, Bast, Jr. RC, O’Brien TJ, et al. Specificity and affinity of 26 Monoclonal antibodies against the CA 125 antigen: first report from the ISOBM TD-1 workshop. Tumor Biol 1996;17:196–219. 10.1159/000217982 [DOI] [PubMed] [Google Scholar]
- 38. Nap M, Vitali A, Nustad K, et al. Immunohistochemical characterization of 22 Monoclonal antibodies against the Ca125 antigen: 2ND report from the ISOBM TD-1 workshop. Tumour Biol 1996;17:325–31. [PubMed] [Google Scholar]
- 39. Köksal H, Baken E, Warren DJ, et al. Chimeric antigen receptor preparation from Hybridoma to T-cell expression. Antib Ther 2019;2:56–63. 10.1093/abt/tbz007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Köksal H, Dillard P, Josefsson SE, et al. Preclinical development of Cd37Car T-cell therapy for treatment of B-cell lymphoma. Blood Adv 2019;3:1230–43. 10.1182/bloodadvances.2018029678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loew R, Heinz N, Hampf M, et al. Improved TET-responsive promoters with minimized background expression. BMC Biotechnol 2010;10:81. 10.1186/1472-6750-10-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jutz S, Leitner J, Schmetterer K, et al. Assessment of Costimulation and Coinhibition in a triple parameter T cell reporter line: simultaneous measurement of NF-kappaB, NFAT and AP-1. J Immunol Methods 2016;430:10–20. 10.1016/j.jim.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 43. Efficient Chimeric antigen receptor (CAR) targeting of a central EPITOPE of Cd22. J Biol Chem 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Walseng E, Köksal H, Sektioglu IM, et al. A TCR-based Chimeric antigen receptor. Sci Rep 2017;7:10713. 10.1038/s41598-017-11126-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Popa M, Fosse V, Kleinmanns K, et al. Xenograft models of ovarian cancer for therapy evaluation. Methods Mol Biol 2022;2424:275–93. 10.1007/978-1-0716-1956-8_18 [DOI] [PubMed] [Google Scholar]
- 46. Thériault C, Pinard M, Comamala M, et al. Muc16 (Ca125) regulates epithelial ovarian cancer cell growth, tumorigenesis and metastasis. Gynecol Oncol 2011;121:434–43. 10.1016/j.ygyno.2011.02.020 [DOI] [PubMed] [Google Scholar]
- 47. Frontiers . MSLN correlates with immune infiltration and Chemoresistance as a Prognostic biomarker in ovarian cancer. 2023. [DOI] [PMC free article] [PubMed]
- 48. Bax HJ, Chauhan J, Stavraka C, et al. Folate receptor alpha in ovarian cancer tissue and patient serum is associated with disease burden and treatment outcomes. Br J Cancer 2023;128:342–53. 10.1038/s41416-022-02031-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Engelstaedter V, Heublein S, Schumacher AL, et al. Mucin-1 and its relation to grade, stage and survival in ovarian carcinoma patients. BMC Cancer 2012;12:1–9. 10.1186/1471-2407-12-600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cerveira J, Begum J, Di Marco Barros R, et al. An imaging flow Cytometry-based approach to measuring the Spatiotemporal calcium Mobilisation in activated T cells. J Immunological Method 2015;423:120–30. 10.1016/j.jim.2015.04.030 [DOI] [PubMed] [Google Scholar]
- 51. Köksal H, Dillard P, Juzeniene A, et al. Combinatorial CAR design improves target restriction. J Biol Chem 2021;296:100116. 10.1074/jbc.RA120.016234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alagoz T, Buller RE, Berman M, et al. What is a normal Ca125 level Gynecol Oncol 1994;53:93–7. 10.1006/gyno.1994.1093 [DOI] [PubMed] [Google Scholar]
- 53. Altaras MM, Goldberg GL, Levin W, et al. What is a normal CA-125 level? Gynecol Oncol 1995;56:475–6. 10.1006/gyno.1995.1086 [DOI] [PubMed] [Google Scholar]
- 54. Garg G, Gibbs J, Belt B, et al. Novel treatment option for Muc16-positive malignancies with the targeted TRAIL-based fusion protein Meso-Tr3. BMC Cancer 2014;14:35. 10.1186/1471-2407-14-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Latifi A, Luwor RB, Bilandzic M, et al. Isolation and characterization of tumor cells from the Ascites of ovarian cancer patients: molecular phenotype of Chemoresistant ovarian tumors. PLoS ONE 2012;7:e46858. 10.1371/journal.pone.0046858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Frontiers | is it time to start Transitioning from 2d to 3d cell culture? 2023. [DOI] [PMC free article] [PubMed]
- 57. Kim YE, Jeon HJ, Kim D, et al. Quantitative Proteomic analysis of 2d and 3d cultured colorectal cancer cells: profiling of Tankyrase inhibitor Xav939-induced Proteome. Sci Rep 2018;8:13255. 10.1038/s41598-018-31564-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhu X, Cai H, Zhao L, et al. CAR-T cell therapy in ovarian cancer: from the bench to the bedside. Oncotarget 2017;8:64607–21. 10.18632/oncotarget.19929 Available: https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.v8i38 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-008179supp001.pdf (239.8KB, pdf)
jitc-2023-008179supp002.pdf (57.4KB, pdf)
jitc-2023-008179supp003.pdf (41KB, pdf)
jitc-2023-008179supp004.pdf (26.5KB, pdf)
jitc-2023-008179supp005.pdf (19.5KB, pdf)
jitc-2023-008179supp006.pdf (202.4KB, pdf)
jitc-2023-008179supp007.pdf (21.6KB, pdf)
jitc-2023-008179supp008.pdf (214KB, pdf)
jitc-2023-008179supp009.pdf (4.1MB, pdf)
Data Availability Statement
Data are available upon reasonable request.