Abstract
Discovery and optimization of monoclonal antibodies for therapeutic applications relies on large sequence libraries but is hindered by developability issues such as low solubility, high aggregation, and high immunogenicity. Generative language models, trained on millions of protein sequences, are a powerful tool for on-demand generation of realistic, diverse sequences. We present Immunoglobulin Language Model (IgLM), a deep generative language model for creating synthetic antibody libraries. Compared with prior methods that leverage unidirectional context for sequence generation, IgLM formulates antibody design based on text-infilling in natural language, allowing it to re-design variable-length spans within antibody sequences using bidirectional context. We trained IgLM on 558M antibody heavy- and light-chain variable sequences, conditioning on each sequence’s chain type and species-of-origin. We demonstrate that IgLM can generate full-length antibody sequences from a variety of species, and its infilling formulation allows it to generate infilled CDR loop libraries with improved in silico developability profiles. A record of this paper’s Transparent Peer Review process is included in the Supplemental Information.
Keywords: antibodies, deep learning, language modeling
eTOC Blurb
Synthetic antibody libraries are a powerful tool for therapeutic discovery, yet often produce sequences that are not human-like or developable. IgLM is a generative language model trained on 558M natural antibodies. IgLM generates full sequences, conditioned on species and chain type, and enables infilling of sequences for synthetic library design.
Introduction
Antibodies have become popular for therapeutics because of their diversity and ability to bind antigens with high specificity [46]. Traditionally, monoclonal antibodies (mAbs) have been obtained using hybridoma technology, which requires the immunization of animals [40], or transgenic animal systems, which involve integration of human immune loci into alternative species (e.g., mice) [47, 21]. In 1985, the development of phage display technology allowed for in vitro selection of specific, high-affinity mAbs from large antibody libraries [24, 42, 11]. Despite such advances, therapeutic mAbs derived from display technologies face issues with developability, such as poor expression, low solubility, low thermal stability, and high aggregation [48, 15]. Display technologies rely on a high-quality and diverse antibody library as a starting point to isolate high-affinity antibodies that are more developable [2]. Synthetic antibody libraries are prepared by introducing synthetic DNA into regions of the antibody sequences that define the complementarity-determining regions (CDRs), allowing for human-made antigen-binding sites. To discover antibodies with high affinity, massive synthetic libraries on the order of 1010–1011 variants must be constructed. However, the space of possible synthetic antibody sequences is very large (diversifying 10 positions of a CDR yields 2010 ≈ 1013 possible variants), meaning these approaches still vastly undersample the possible space of sequences. Further, sequences from randomized libraries often contain substantial fractions of non-functional antibodies [2, 40]. These liabilities could be reduced by restricting libraries to sequences that resemble natural antibodies, and are thus more likely to be viable therapeutics.
Recent work has leveraged natural language processing methods for unsupervised pre-training on massive databases of raw protein sequences for which structural data are unavailable [35, 8, 23]. These works have explored a variety of pre-training tasks and downstream model applications. For example, the ESM family of models (trained for masked language modeling) have been applied to representation learning [35], variant effect prediction [25], and protein structure prediction [20]. Masked language models have also shown promise for optimization and humanization of antibody sequences through suggestion of targeted mutations [13]. Autoregressive language modeling, an alternative paradigm for pre-training, has also been applied to protein sequence modeling. Such models have been shown to generate diverse protein sequences, which often adopt natural folds despite diverging considerably in residue makeup [10, 26]. In some cases, these generated sequences even retain enzymatic activity comparable to natural proteins [22]. Autoregressive language models have also been shown to be powerful zero-shot predictors of protein fitness, with performance in some cases continuing to improve with model scale [12, 26].
Another set of language models have been developed specifically for antibody-related tasks. The majority of prior work in this area has focused on masked language modeling of sequences in the Observed Antibody Space (OAS) database [16]. Prihoda et al. developed Sapiens, a pair of distinct models (each with 569K parameters) for heavy and light chain masked language modeling [29]. The Sapiens models were trained on 20M and 19M heavy and light chains respectively, and shown to be effective tools for antibody humanization. Similarly, likelihoods from antibody-specific masked language models have also been used as a proxy for immunogenic risk (or naturalness) [3]. Ruffolo et al. developed AntiBERTy, a single masked language model (26M parameters) trained on a corpus of 558M sequences, including both heavy and light chains [37]. AntiBERTy has been applied to representation learning for protein structure prediction [36]. Leem et al. developed AntiBERTa, a single masked language model (86M parameters) trained on a corpus of 67M antibody sequences (both heavy and light) [18]. Representations for AntiBERTa were used for paratope prediction. Olsen et al. developed AbLang, a pair of masked language models trained on 14M heavy chains and 187K light chains, for sequence restoration [27]. For sequence generation, autoregressive generative models have been trained on antibody sequences and used for library design [1, 39]. Akbar et al. [1] trained an LSTM for autoregressive generation of CDR H3 loops and conducted an in silico investigation of their potential for binding antigens. LSTMs have also been trained on phage display data to aid in discovery of optimized variants [38]. Towards a more general method for library generation, Shin et al. [39] experimentally validated a set of nanobody sequences with generated CDR3 loops and showed promising improvements to viability and binding discovery when compared to traditional approaches, despite the library being over 1000-fold smaller. However, because this generative model was unidirectional, it could not be used to directly re-design the CDR3 loop within the sequence, and instead had to be oversampled to produce sequences matching the residues following the loop.
Here, we present Immunoglobulin Language Model (IgLM), a generative language model that leverages bidirectional context for designing antibody sequence spans of varying lengths while training on a large-scale natural antibody dataset. We show that IgLM can generate full-length antibody sequences conditioned on chain type and species-of-origin. Furthermore, IgLM can diversify loops on an antibody to generate high-quality libraries that display favorable predicted biophysical properties while resembling human antibodies. IgLM should be a powerful tool for antibody discovery and optimization.
Results
Immunoglobulin language model
Our method for antibody sequence generation, IgLM, is trained on 558 million natural antibody sequences for both targeted infilling of residue spans, as well as full-length sequence generation. IgLM generates sequences conditioned on the species-of-interest and chain type (heavy or light), enabling controllable generation of antibody sequences.
Infilling language model
Design of antibody libraries typically focuses on diversification of the CDR loop sequences in order to facilitate binding to a diverse set of antigens. Through traditional diversification technologies, many putative antibody sequences can be produced and subjected to experimental screening, enabling the discovery or optimization of specific antibodies. However, such techniques typically produce large fractions on non-viable or poorly behaved sequences, as they are not constrained to the natural space of antibody sequences. Generative models of protein sequences, such as language models, offer an alternative means of efficiently sampling from the natural space of proteins to produce large libraries of sequences. However, existing approaches to protein sequence generation (including antibodies) typically adopt left-to-right decoding strategies [26, 10]. While these models have proven effective for generation of diverse and functional sequences, they are ill-equipped to re-design specific segments of interest within proteins. To address this limitation, we developed IgLM, an infilling language model for immunoglobulin sequences. IgLM uses a standard left-to-right decoder-only transformer architecture based on GPT-2, but it is trained for infilling through rearrangement of sequences. Specifically, we adopt the infilling language model formulation from natural language processing [6], wherein arbitrary-length sequence segments (spans) are masked during training and appended to the end of the sequence. By training on these rearranged sequences, models learn to predict the masked spans conditioned on the surrounding sequence context.
To train IgLM, we collected antibody sequences from the Observed Antibody Space (OAS) [16]. The OAS database contains natural antibody sequences from six species: human, mouse, rat, rabbit, rhesus, and camel. To investigate the impacts of model capacity, we trained two versions of the model: IgLM and IgLM-S, with 13M and 1.4M trainable parameters, respectively. Both IgLM models were trained on a set of 558M non-redundant sequences, clustered at 95% sequence identity. During training, we randomly masked spans of ten to twenty residues within the antibody sequence to enable diversification of arbitrary spans during inference. Additionally, we conditioned sequences on the chain type (heavy or light) and species-of-origin. Providing this context enables controllable generation of species-specific antibody sequences. An example of training data construction is illustrated in Figure 1A. Unless otherwise specified, we use the larger IgLM model for all experiments.
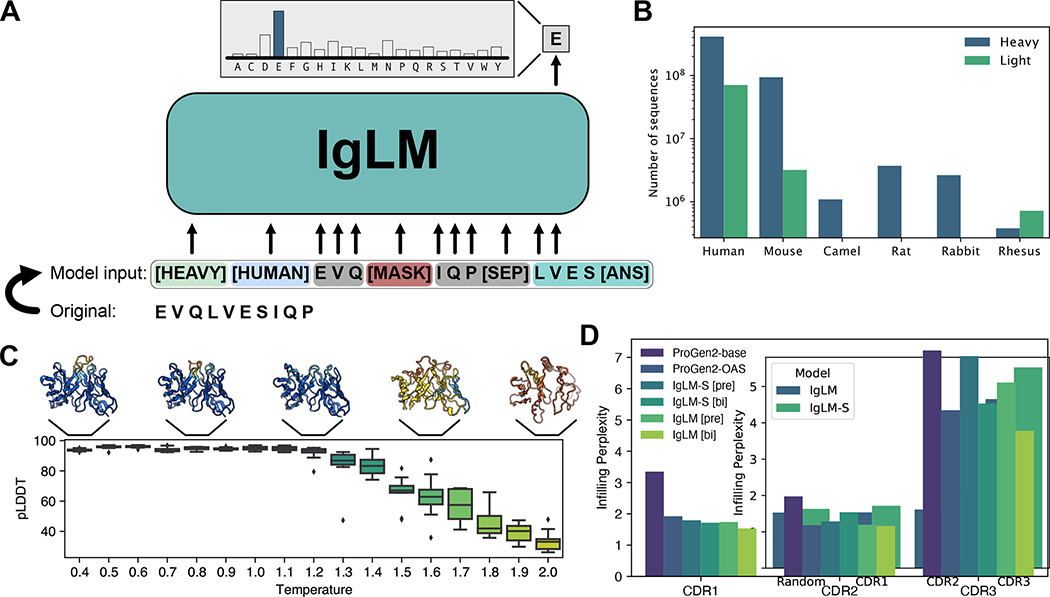
Figure 1.
Overview of IgLM model for antibody sequence generation. (A) IgLM is trained by autoregressive language modeling of reordered antibody sequence segments, conditioned on chain and species identifier tags. (B) Distribution of sequences in clustered OAS dataset for various species and chain types. (C) Effect of increased sampling temperature for full-length generation. Structures at each temperature are predicted by AlphaFold-Multimer and colored by prediction confidence (pLDDT), with blue being the most confident and orange being the least [n = 170]. (D) CDR loop infilling perplexity for IgLM and ProGen2 models on heldout test dataset of 30M sequences. IgLM models are evaluated with bidirectional infilling context ([bi]) and preceding context only ([pre]). Confidence intervals calculated from boostrapping (100 samples) had a width less than 0.01 and are therefore not shown.
IgLM generates foldable antibody sequences in silico
As an initial validation of the antibody sequence generation capabilities of IgLM, we conducted a small scale investigation of full-length generation (Methods). Specifically, we investigated the impacts of sampling temperature for tuning the diversity of generated sequences. Sampling temperature values above one effectively flatten the amino acid distribution at each step of generation, resulting in more diverse sequences, while temperature below one sharpens the distribution at each position, resembling a greedy decoding strategy. We generated a set of full-length sequences at temperatures ranging from 0.4 to 2.0, providing the model with human heavy and human light conditioning tags. Because IgLM was trained for sequence infilling, generated sequences contain discontinuous segments of sequence segments, which we simply reordered to produce full-length antibodies. Heavy and light chain sequences were generated independently of each other, as IgLM only considers single chains. Sequences were then paired according to sampling temperature and their structures predicted using AlphaFold-Multimer [9]. In general, IgLM generates sequences with correspondingly confident predicted structures at lower temperatures (up to 1.2), before beginning to degrade in quality at higher temperatures (Figure 1C). For subsequent experiments, we sampled with a maximum temperature of 1.2 to remain within foldable antibody space, and used the much faster IgFold model [36] for high-throughput structure predictions.
Language modeling evaluation
We evaluated IgLM as a language model by computing the per-token perplexity for infilled spans within an antibody, which we term the infilling perplexity. Because the infilled segment is located at the end of the sequences, computing the infilling perplexity is equivalent to taking the per-token perplexity after the [SEP] token (Methods). We compared the infilling perplexity of IgLM and IgLM-S given bidirectional context (IgLM [bi] and IgLM-S [bi]) and preceding context only (IgLM [pre] and IgLM-S [pre]) on a heldout test dataset of 30M sequences. We additionally computed infilling perplexity for ProGen2-base and ProGen2-OAS, which only use preceding context (Methods) [26]. Results are tabulated by CDR loop for each method (Figure 1D). As expected, the CDR3 loop, which is the longest and most diverse, has the highest infilling perplexity for all methods. For IgLM, providing bidirectional context yielded reduced perplexity, demonstrating that the sequence following CDR loops is important for determining their content. Both ProGen2 models evaluated have 764M parameters, substantially more than the 13M parameters of IgLM. However, with bidirectional context, IgLM is able to better fit the distribution of CDR loops than either model, demonstrating the importance of aligning the model pre-training objective with the downstream task.
The diversity of antibody sequences varies by species and chain type. For example, heavy chains introduce additional diversity into their CDR3 loops via D-genes, while some species (e.g., camels) tend to have longer loops. To investigate how these differences impact the performance of IgLM in different settings, we also tabulated the heldout set infilling perplexity by species and chain type. For CDR1 and CDR2 loop infilling, perplexity values are typically lower for human and mouse antibodies (Figure S1), which are disproportionately represented in the OAS database. In general, both models still perform better on these loops than the more challenging CDR3 loops, regardless of species. One exception is for rhesus CDR2 loops, on which IgLM-S performs considerably worse than the larger IgLM model. This appears to be due to poor fitting of rhesus CDR L2 loops, as reflected in the similarity high infilling average perplexity observed when tabulated by chain type (Figure S2). The highest infilling perplexity is observed for camel CDR3 loops, which tend to be longer than other species. Across all species and chain types, the larger IgLM model achieves lower infilling perplexity than IgLM-S, suggesting that further increasing model capacity would yield additional improvements.
Controllable generation of antibody sequences
Having demonstrated that IgLM can generate well-formed full-length sequences, we next considered the controllability of IgLM for generating antibody sequences with specific traits. Controllable generation uses conditioning tags to provide the model with additional context about the expected sequence.
Generating species- and chain-controlled sequences
To evaluate the controllability of IgLM, we generated a set of 220K full-length sequences using all viable combinations of conditioning tags, as well as a range of sampling temperatures (Figure 2A). For every species (except camel), we sampled with both heavy and light conditioning tags. For camel sequence generation, we only sampled heavy chains, as they do not produce light chains. To produce a diverse set of sequences for analysis, we sampled using a range of temperatures . Sampling under these conditions resulted in a diverse set of antibody sequences. However, we observed that the sequences frequently featured N-terminal truncations. These truncations are frequently observed in the OAS database used for training, with over 40% of sequences missing the first fifteen or more residues [27]. For heavy chains, these N-terminal deletions appeared as a left-shoulder in the sequence length distribution (Figure 2B, left) with lengths ranging from 100 to 110 residues. For light chains, we observed a population of truncated chains with lengths between 98 and 102 residues (Figure 2B, right). To address truncation in generated sequences, we used a prompting strategy, wherein we initialize each sequence with a three-residue motif corresponding to the species and chain type tags. The specific initialization sequences were selected according to germline sequences in the IMGT database [19] and are documented in Table S2. For light chains, we identified prompts corresponding to both lambda and kappa classes and divided the generation budget between the two. For both heavy and light chains, prompting with initial residues markedly reduced the population of truncated sequences (Figure 2B). For the following analysis, we consider only sequences generated with prompting.
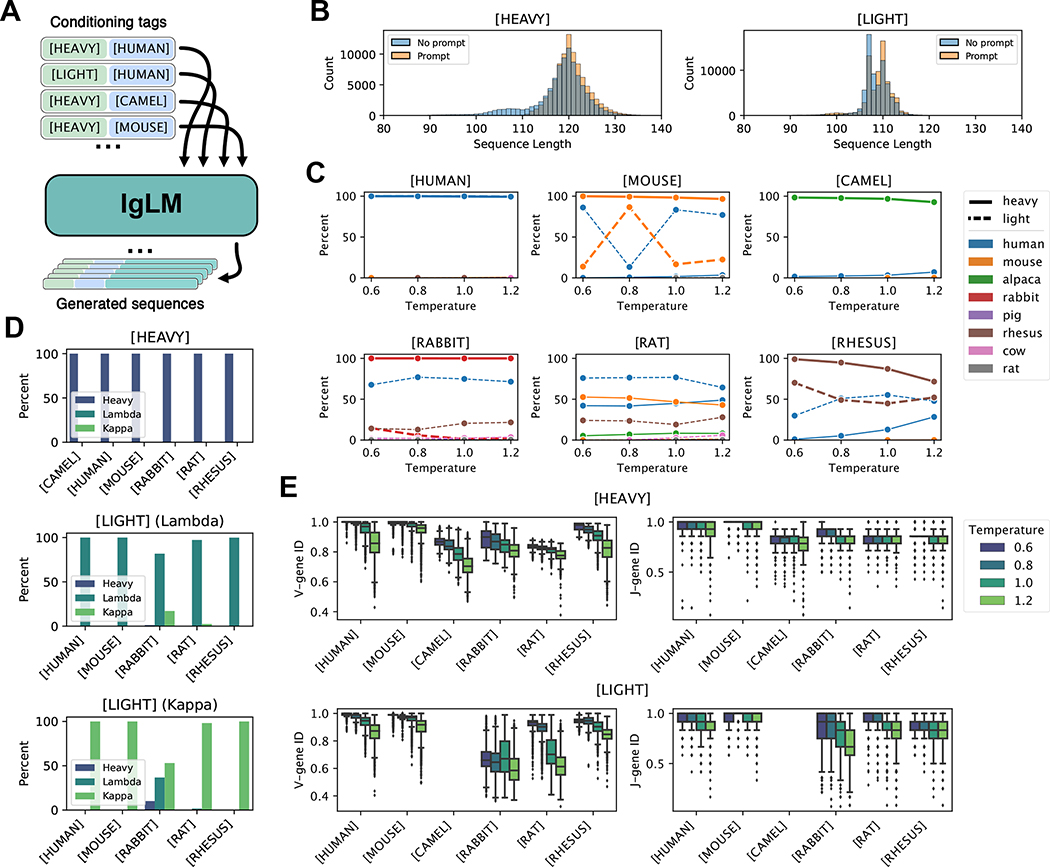
Figure 2.
Controllable antibody sequence generation. (A) Diagram of procedure for generating full-length antibody sequences given a desired species and chain type with IgLM. (B) Length of generated heavy and light with and without initial three residues provided (prompting). (C-E) Analysis of full-length generated sequences under different conditioning settings [n = 220,000]. (C) Adherence of generated sequences to species conditioning tags. Each plot shows the species classifications of antibody sequences generated with a particular species conditioning tag (indicated above plots). Solid and dashed lines correspond to sequences generated with heavy- and light-chain conditioning, respectively. (D) Adherence of generated sequences to chain conditioning tags. Top plot shows the percentage of heavy-chain-conditioned sequences classified as heavy chains, for each species conditioning tag. Lower plots show the percentage of light-chain-conditioned sequences, further divided by whether initial residues were characteristic of lambda or kappa chains, classified as lambda or kappa chains. (E) Effect of sampling temperature on germline identity for generated heavy and light chain sequences. As sampling temperature increases, generated sequences diverge from the closest germline V- and J-gene sequences.
Adherence to conditioning tags
To evaluate the effectiveness of controllable generation, we considered the agreement between the provided conditioning tags and the sequences produced by IgLM. For each generated sequence, we classified the species and chain type using ANARCI [7]. We note that the species classes provided by ANARCI diverge in some cases from those provided by the OAS database, but there was a suitable corresponding class for each conditioning token (e.g., alpaca for [CAMEL]). In Figure 2C, we show the makeup of sequences for each species conditioning tag, according to sampling temperature. In each plot, the percentage of heavy and light chain sequences classified as each species are indicated by solid and dashed lines, respectively. For most species (human, mouse, camel, rabbit, rhesus), IgLM is able to successfully generate heavy chain sequences at every temperature. The exception to this trend is rat sequences, for which we were unable to produce any sequences that ANARCI classified as belonging to the intended species.
The ability to generate sequences is not directly explained by prevalence in the training dataset, as the model is trained on an order of magnitude more rat heavy chain sequences than rhesus (Table S1). IgLM is generally less effective at generating light chain sequences for most species. With the exception of human light chains, all species have a large proportion of sequences classified as belonging to an unintended species (typically human). For mouse and rhesus light chains, IgLM generates the correct species in 34.89% and 88.14% of cases, respectively (Table S3). The disproportionately low recovery of mouse sequences may be due to inclusion of transgenic mice immune repertoires, which are harvested from mice but consist of human genetic material. For rabbit and rat light chains, IgLM was not exposed to any examples during training. Despite having seen no such sequences during training, IgLM is capable of generating sequences classified by ANARCI as rabbit light chains for 6.89% of samples (1,120 sequences). The majority of these sequences are cases where the model has instead generated a rabbit heavy chain. However, for 35 of these 1,120 cases, IgLM has produced rabbit light chain sequences. We further investigated the plausibility of these sequences by aligning to the nearest germline sequences assigned by ANARCI with Clustal-Omega [41]. The sequences appear to align well to rabbit germlines, though with considerable mutations to the framework regions (Figure S3). To investigate the structural viability of the generated rabbit light chain sequences, we predicted structures with IgFold [36]. All structures were predicted confidently in the framework residues, with the CDR loops being the most uncertain (Figure S4). Although rare (35 sequences out of 20,000 attempts), these results suggest that IgLM is capable of generating rabbit light chain sequences despite having never observed such sequences during training. This may be achieved by producing a consensus light chain, with some rabbit-likeness conferred from the heavy chain examples.
We next evaluated the adherence of IgLM-generated sequences to chain type conditioning tags. In Figure 2D, we show the percentage of sequences classified by ANARCI as heavy or light for each conditioning tag. Light chains are further divided into lambda and kappa classes. When conditioned towards heavy chain generation, IgLM effectively produces heavy chains for all species. For light chains, we observe a similar trend, with IgLM producing predominantly light chain sequences for all species. Only for rabbit sequences do we observe a population of heavy chains when conditioning for light chains. As noted above, these are cases where IgLM has instead produced a rabbit heavy chain. When generating light chain sequences, we provide initial residues characteristic of both lambda and kappa chains in equal proportion (Table S2). For most species (except rabbit), the generated sequences are aligned with light chain type indicated by the initial residues. However, as noted above, many of the light sequences for poorly represented species are human-like, rather than resembling the desired species. These results suggest that the chain type conditioning tag is a more effective prior for IgLM than species.
Sampling temperature controls mutational load
Increasing sampling temperature has the effect of flattening the probability distribution at each position during sampling, resulting in a greater diversity of sequences. We evaluated the effect of sampling temperature on the diversity of generated sequences by measuring the fractional identity to the closest germline sequences using ANARCI [7]. In Figure 2E, we show the germline identity for V- and J-genes for each species and chain type. At the lowest sampling temperature , IgLM frequently recapitulates germline sequences in their entirety for some species (human, mouse, rhesus). As temperature increases, sequences for every species begin to diverge from germline, effectively acquiring mutations. To evaluate whether these mutations emerge at biologically relevant positions, we calculated the positional entropy of generated sequences according to the Chothia numbering scheme (Methods). As expected, we observe markedly higher entropy in the CDR loops, with temperature further increasing the entropy at these positions (Figure S5). J-gene sequences typically acquire fewer mutations than V-genes for both heavy and light chains. This is likely a reflection of the concentration of CDR loops within the V-gene (CDR1 and CDR2). Only a portion of the CDR3 loop is contributed by the J-gene, with the remaining sequence being conserved framework residues.
Therapeutic antibody diversification
Diversification of antibody CDR loops is a common strategy for antibody discovery or optimization campaigns. Through infilling, IgLM is capable of replacing spans of amino acids within antibody sequences, conditioned on the surrounding context. To demonstrate this functionality, we generated infilled libraries for a set of therapeutic antibodies and evaluated several therapeutically relevant properties. Based on in silico measures of developability and humanness, we show that IgLM proposes libraries containing antibody sequences resembling natural antibodies with controllable diversity, which could then be experimentally screened to discover new high-affinity binders.
Infilled libraries for therapeutic antibodies
To evaluate the utility of infilling with IgLM for diversifying antibody sequences, we created infilled libraries for 49 therapeutic antibodies from Thera-SAbDab [33]. These antibodies were selected because they had experimentally determined structures and had been previously evaluated for developability screening [32]. For each antibody, we removed the CDR H3 loop (according to Chothia definitions [5]) and generated a library of infilled sequences using IgLM (Figure 3A). To produce diverse sequences, we used a combination of sampling temperatures and nucleus sampling probabilities . Nucleus sampling effectively clips the probability distribution at each position during sampling, such that only the most probable amino acids (summing to ) are considered. For each of the 49 therapeutic antibodies, we generated one thousand infilled sequences for each combination of and , totaling nine thousand variants per parent antibody. In Figure 3D, we show predicted structures (using IgFold [36]) for a subset of ten infilled loops derived from the trastuzumab antibody. The infilled loops vary in length and adopt distinct structural conformations. Across the infilled libraries, we see a variety of infilled CDR H3 loop lengths, dependent on the parent antibody’s surrounding sequence context (Figure 3B). The median length of infilled loops across antibodies ranges from 11 to 16 residues. IgLM occasionally generated very short CDR H3 loops (fewer than five residues), which were assigned correspondingly low log likelihoods by the model (Figure S6). We observe little impact on the length of infilled loops when varying the sampling temperature and nucleus probabilities (Figure 3C).
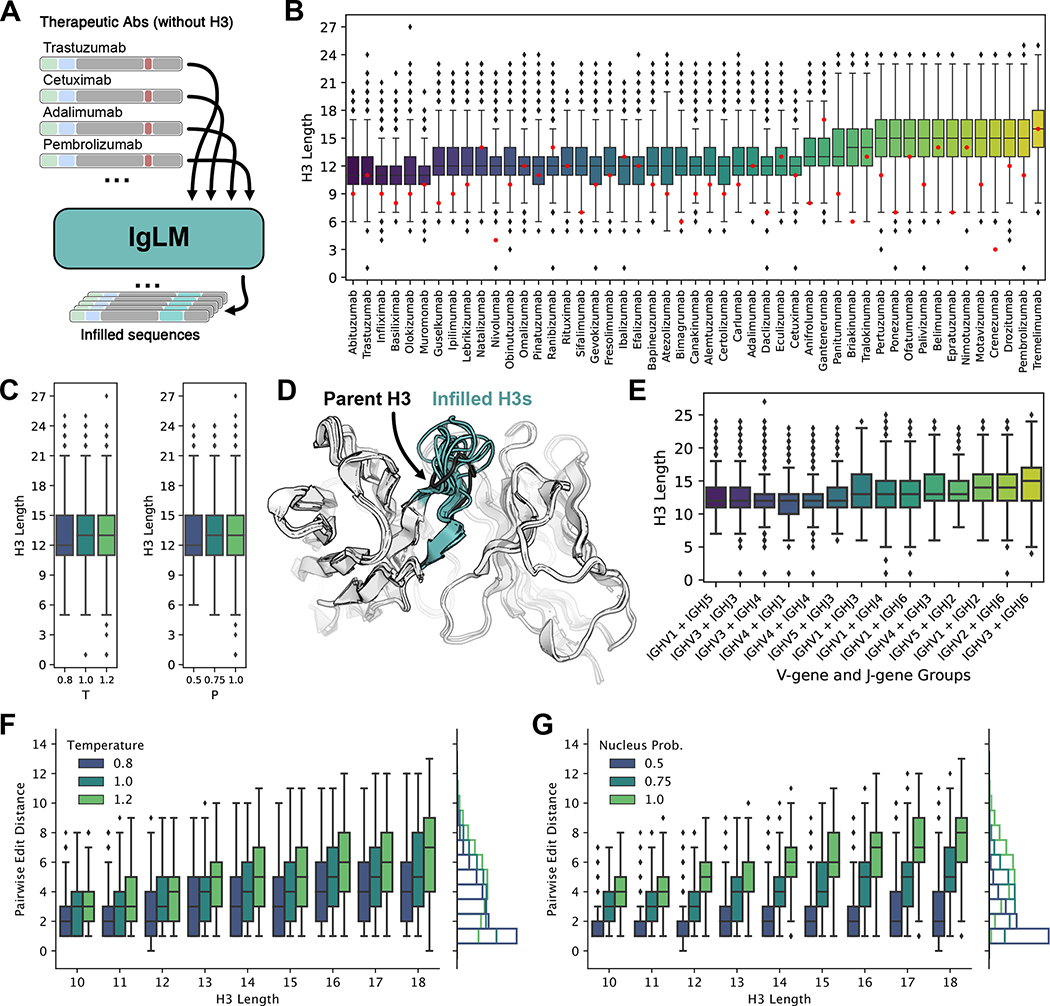
Figure 3.
Generation of infilled therapeutic antibody libraries. (A) Diagram of procedure for generating diverse antibody libraries by infilling the CDR H3 loops of therapeutic antibodies. (B) Distribution of infilled CDR H3 loop lengths for 49 therapeutic antibodies. Parent CDR H3 lengths are indicated in red. (C) Relationship between sampling temperature () and nucleus probability and length of infilled CDR H3 loops [n = 432,763]. (D) Infilled CDR H3 loops for trastuzumab therapeutic antibody adopt diverse lengths and conformations. Structures for infilled variants are predicted with IgFold [n = 432,763]. (E) Distribution of infilled CDR H3 loop lengths for therapeutic antibodies grouped by nearest germline gene groups [n = 432,763]. (F-G) Effect of sampling temperature () and nucleus probability (P) on diversity of infilled CDR H3 loops for lengths between 10 and 18 residues [n = 432,763]. Pairwise edit distance measures the minimum edits between each infilled loop to another in the same set of generated sequences (i.e., within the set of sequences produced with the same and parameters). For both parameters, less restrictive sampling produces greater infilled loop diversity.
The distributions of infilled loop lengths vary considerably over the 49 therapeutic antibodies. Because IgLM is trained on natural antibody sequences, we hypothesized that the model may be performing a sort of germline matching, wherein sequences with similar V- and J-genes lead to similar distributions of loop lengths. To test this, we identified the closest germline genes for each antibody with ANARCI [7]. We then group parent antibodies according to common V- and J-gene groups and compared the distributions of infilled loop lengths for each group (Figure 3E). While there may be some tendency for similar V- and J-genes to lead to similar distributions of infilled loop lengths, we observe considerable variation. This suggests that IgLM is not purely performing germline matching, but rather is considering other properties of the parent antibody.
Infilling generates diverse loop sequences
Diverse loop libraries are essential for discovering or optimizing sequences against an antigen target. To evaluate the diversity of infilled loops produced by IgLM, we measured the pairwise edit distance between each loop sequence and its closest neighbor amongst the sequences generated with the same sampling parameters. We then compared the diversity of sequences according to loop length and choice of sampling parameters (Figure 3F–G). Generally, we observe that generated loops are more diverse at longer lengths, as expected given the increased combinatorial complexity available as more residues are added. Increasing both sampling temperature and nucleus probability results in a greater diversity of sequences. However, these parameters affect the relationship between length and diversity in distinct ways. For a given loop length, increasing temperature produces more variance in the pairwise edit distance, while increases to nucleus probability provides a more consistent increase in diversity across loop lengths. Indeed, the marginal distribution of pairwise edit distance as nucleus probability is increased produces a much larger shift (Figure 3G, marginal) than that of temperature (Figure 3F, marginal). In practice, a combination of sampling parameters may be suitable for producing a balance of high-likelihood (low temperature and low nucleus probability) and diverse sequences.
Infilled loops display improved developability in silico
Developability encompasses a set of physiochemical properties – including aggregation propensity and solubility – that are critical for the success of a therapeutic antibody. Libraries for antibody discovery or optimization that are enriched for sequences with improved developability can alleviate the need for time-consuming post-hoc engineering. To evaluate the developability of sequences produced by IgLM, we used high-throughput computational tools to calculate the aggregation propensity (SAP score [4]) and solubility (CamSol Intrinsic [43]) of the infilled therapeutic libraries. As a precursor to calculation of aggregation propensity, we used IgFold [36] to predict the structures of the infilled antibodies (including the unchanged light chains). We then compared the predicted aggregation propensities and solubility values of the infilled sequences to those of the parent antibodies. For aggregation propensity, we observed a significant improvement (negative is better) by infilled sequences over the parent antibodies (Figure 4A, Figure S7). Similarly for solubility, infilled sequences tended to be predicted to be more soluble than their parent antibodies (Figure 4B, Figure S8). In both cases, the largest improvements tend to correspond to the shorter loops. Further, we observe a positive correlation between improvements to predicted aggregation propensity and solubility (Figure 4C, Figure S9). These results suggest that infilling can be used to generate libraries enriched for sequences with improved developability.
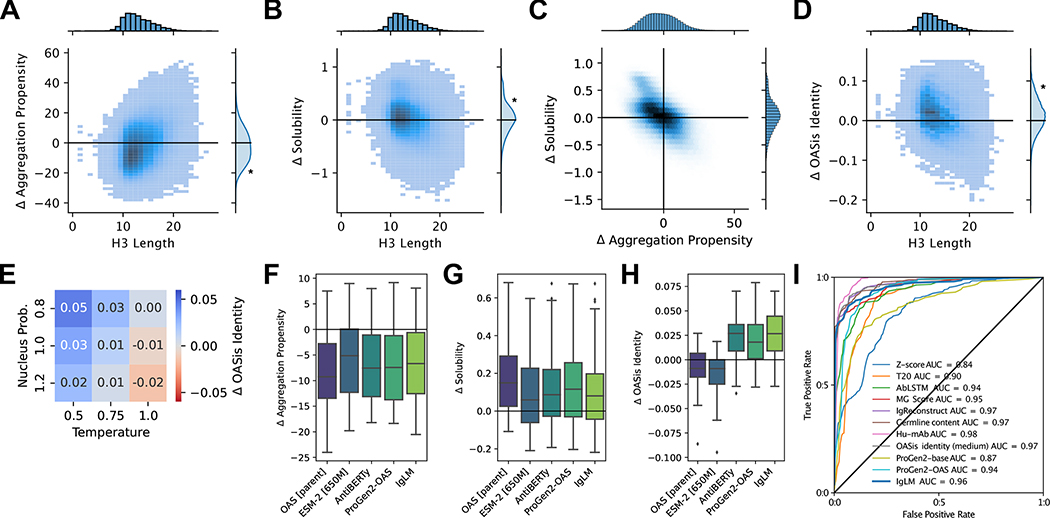
Figure 4.
Therapeutic properties of infilled antibody libraries. Asterisks indicate statistical significance (p ¡ 0.001) from a one-sample t-test (A, B, D) or a two-sample t-test (E). (A) Change in predicted aggregation propensity of infilled sequences relative to their parent antibodies. Infilled sequences display reduced aggregation propensity (negative is improved), particularly for shorter loops [n = 432,763]. (B) Change in predicted solubility of infilled sequences relative to their parent antibodies. Infilled sequences display increased solubility (positive is improved) [n = 432,763]. (C) Relationship between predicted changes in aggregation propensity and solubility for infilled sequence libraries [n = 432,763]. (D) Change in humanness of infilled sequences relative to their parent antibodies. Humanness is calculated as the OASis identity of the heavy chain sequence, with positive larger values being more human-like [n = 432,763]. (E) Relationship between sampling temperature () and nucleus probability () and change in human-likeness (OASis identity) of infilled heavy chains relative to their parent sequences [n = 432,763]. (F-G) Comparison of infilled library developability generated using alternative language models for loops with lengths between six and seventeen residues [n = 1,709,696]. (F) Change in predicted aggregation propensity for infilling methods. (G) Change in predicted solubility for infilling methods. (H) Change in humanness for infilling methods. (I) Receiver operating characteristic (ROC) curves for human sequence classification methods [n = 487]. The area under the curve (AUC) is shown for each method.
We next investigated whether choice of sampling parameters affects the developability of infilled sequences. When we compared the predicted aggregation propensity and solubility of infilled sequences according to the sampling temperature and nucleus sampling probability, we found marginal practical differences (Figure S10). This is likely explained by the relative consistency of infilled loop lengths across sampling parameters (Figure 3C). These results suggest that developability should not be a concern when tuning the diversity of a generated library.
Infilled loops are more human-like
Therapeutic antibodies must be human-like to avoid provoking an immune response and to be safe for use in humans. To evaluate the human-likeness of infilled sequences, we calculated the OASis identity (at medium stringency) [29]. OASis divides an antibody sequence into a set of 9-mers and calculates the fraction that have been observed in human repertoires. Thus, higher OASis identity indicates a sequence that is more similar to those produced by humans. When compared to their respective parent antibodies, sequences infilled by IgLM were typically more human-like (Figure 4D). This is expected, given that IgLM is trained on natural human antibodies, but not trivial as the parent sequences have been optimized and shown to be safe in humans. To achieve higher humanness, sequences from IgLM must better adhere to the natural distribution of human antibodies than the parent sequences. We also investigated the impact of sampling parameters on the human-likeness of infilled sequences. For both sampling temperature and nucleus probability, we find that less restrictive sampling tends to produce less human-like sequences (Figure 4E). For practical purposes, this suggests that sampling with lower temperature and nucleus probability may be more suitable when immunogenicity is a concern.
Libraries from alternative language models
To contextualize the properties of IgLM-generated infilled libraries, we conducted a benchmark using several alternative protein language models. The benchmark includes ESM-2, a masked language model trained on diverse sequences, AntiBERTy, an antibody-specific masked language model, and ProGen2-OAS, an autoregressive language model trained on antibody sequences [20, 37, 26]. We also compared with a baseline of sequences generated from the OAS data used to train IgLM. Sequences for the OAS baseline, OAS [parent], were generated by sampling from positional amino acid frequencies for loop lengths matching the parent sequence.
For all infilled libraries, we predicted structures with IgFold [36] and computed aggregation propensity [4], solubility [43], and humanness [29] for all sequences (Figure S11). To remove length-dependent biases from the evaluation, we compared the developability properties of only loops matching the parent CDR H3 loop length. In general, we found that all methods were able to generate infilled libraries predicted to have improved aggregation propensity and solubility relative to the parent sequences (Figure 4F–G). This illustrates the utility of drawing from informed sequence distributions (such as those derived from OAS or learned by language models), rather than randomly mutating sequences as is the norm for library construction. The OAS baseline performed particularly well, indicating that the natural makeup of CDR H3 loops are biophysically well-behaved. However, to produce human-like antibody libraries, we found that antibody-specific language models were substantially more effective than alternative approaches (Figure 4H). Among these models, IgLM produced slightly more human-like sequences than ProGen2-OAS, in accordance with the lower infilling perplexity demonstrated on the heldout set human sequences (Figure S1).
Sequence likelihood is an effective predictor of humanness
Likelihoods from autoregressive language models trained on proteins have been shown to be effective zero-shot predictors of protein fitness [12, 26]. Antibody-specific language models in particular have been used to measure the ”naturalness” of designed sequences [3], a measure related to humanness. To evaluate the effectiveness of IgLM for distinguishing human from non-human antibodies, we used the model’s likelihood to classify sequences from the IMGT mAb DB [28]. Sequences in this set span a variety of species (human and mouse) and engineering strategies (e.g., humanized, chimeric, felinized). We considered all sequences not specifically labeled as human to be non-human, and calculated a likelihood (conditioned on human species) for each. All sequences had both a heavy and light chain, for which we calculated separate likelihoods and then multiplied.
We compared the performance of IgLM to that of a number of other methods previously benchmarked by Prihoda et al. [29] using a receiver operating characteristic (ROC) curve (Figure 4I). The results here for alternative methods are adapted from those presented by Prihoda et al., but with several redundant entries removed to avoid double-counting. We additionally evaluated model likelihoods from ProGen2-base and ProGen2-OAS [26], which are models similar to IgLM that contain substantially more parameters (764M). ProGen2-base is trained on a diverse set of protein sequences, while ProGen2-OAS is trained on a dataset similar to IgLM (OAS clustered at 85% sequence identity). We find that IgLM is competitive with state-of-the-art methods designed for human sequence classification, though not the best. IgLM outperforms ProGen2-OAS (ROC AUC of 0.96 for IgLM vs. 0.94 for ProGen2-OAS), despite having fewer parameters (13M vs. 764M). This may result from the different strategies for constructing training datasets from OAS. By filtering at a less stringent 95% sequence identity, IgLM is likely exposed to a greater proportion of human antibody sequences, which dominate the OAS database. These distinctions highlight the importance of aligning training datasets with the intended application and suggest that training on only human sequences may further improve performance for human sequence classification.
Discussion
Antibody libraries are a powerful tool for discovery and optimization of therapeutics. However, they are hindered by large fractions of non-viable sequences, poor developability, and immmunogenic risks. Generative language models offer a promising alternative to overcome these challenges through on-demand generation of high-quality sequences. However, previous work has focused entirely on contiguous sequence decoding (N-to-C or C-to-N) [26, 39]. While useful, such models are not well-suited for generating antibody libraries, which vary in well-defined regions within the sequence, and for which changes may be undesirable in other positions. In this work, we presented IgLM, an antibody-specific language model for generation of full-length sequences and infilling of targeted residue spans. IgLM was trained for sequence infilling on 558M natural antibody sequences from six species. During training, we provide the model with conditioning tags that indicate the antibody’s chain type and species-of-origin, enabling controllable generation of desired types of sequences.
Concurrent work on autoregressive language models for antibody sequence generation have been trained on similar sets of natural antibody sequences and explored larger model sizes [26]. However, models like ProGen2-OAS are limited in utility for antibody generation and design, as they are difficult to guide towards generation of specific types of sequences (e.g., species or chain types). Both this work and the ProGen2-OAS paper have used prompting strategies to guide model generation towards full-length sequences. While these strategies may help in some cases (particularly to overcome dataset limitations), substantially more residues may need to be provided to guide the model towards a specific sequence type (e.g., human vs rhesus heavy chain). In contrast, by including conditioning information for species and chain type in the model’s training, IgLM is able to generate sequences of the desired type without additional prompting. Still, as shown in this work, increasing the capacity of models like IgLM may lead to better performance for sequence infilling (lower perplexity) and scoring (better likelihood estimation), a promising direction for future work.
Antibody-specific language models have recently proliferated [29, 37, 18, 27], showing promise for a broad range of traditional antibody engineering tasks [36]. Such models are typically trained on the Observed Antibody Space database [16], which comes with a particular set of biases that are reflected in the behavior of such models. For example, sampling from IgLM with higher temperatures largely corresponds to increased mutational distance from germline sequences, reflecting the nature of immune repertoire datasets. In other work, antibody-specifc language models have been found to underperform universal protein models on antibody fitness prediction tasks [26] – including binding affinity, thermal stability, and expression – despite being trained on considerably more antibody sequences. These findings suggest that we must carefully consider the utility of language models trained on immune repertoire datasets based on the particular task at hand. For generative tasks, training on immune repertoire data may be an intuitive and necessary way to produce large numbers of natural antibody sequences. Meanwhile, for fitness prediction tasks in protein engineering workflows, universal models may better capture the critical developability properties of interest that are divergent from the selective pressures on the immune system.
IgLM’s primary innovation is the ability to generate infilled residue spans at specified positions within the antibody sequence. In contrast to traditional generative language models that only consider preceding the residues, this enables IgLM to generate within the full context of the region to be infilled. IgLM therefore acts as a tool for developing synthetic libraries for large-scale experimental screening by diversifying regions of an existing antibody. Because IgLM is trained on a massive dataset of natural antibodies, it proposes sequences that more efficiently explore the sequence space of natural antibodies, which can reduce the fraction of non-functional antibodies in IgLM-designed libraries compared with randomized synthetic libraries. We demonstrate the utility of infilling by generating libraries 49 therapeutic antibodies. We found that IgLM was capable of generating diverse CDR H3 loop sequences, and that diversity was largely tunable by choice of sampling parameters. Further, as measured by in silico tools, the infilled libraries possessed desirable developability traits (aggregation propensity, solubility) while being more human-like on average than their parent sequences. Notably, IgLM achieves these improvements over antibodies that are already highly optimized, as all of the parent sequences have been engineered for mass-production and use in humans. Although we focused on antibody loop infilling in this work, similar strategies may be useful for proteins generally. For example, a universal protein sequence infilling model may be applicable to re-design of contiguous protein active sites or for generating linkers between separate domains for protein engineering.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jeffrey Gray (jgray@jhu.edu).
Materials Availability
This study did not generate new unique reagents.
Code and Data Availability
Generated sequences and developability metrics have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table.
All original code has been deposited at https://github.com/Graylab/IgLM and Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Infilled therapeutic antibody sequences and developability metrics for IgLM and alternative methods | This paper | 10.5281/zenodo.8248326 |
| Software and algorithms | ||
| Code for IgLM | This paper | 10.5281/zenodo.8248335 |
METHOD DETAILS
Infilling formulation
Designing spans of amino acids within an antibody sequence can be formulated as an infilling task, similar to text-infilling in natural language. We denote an antibody sequence , where represents the amino acid at position of the antibody sequence. To design a span of length starting at position along the sequence, we first replace the span of amino acids with a single [MASK] token to form a sequence . To generate reasonable variable-length spans to replace given , we seek to learn a distribution .
We draw inspiration from the Infilling by Language Modeling (ILM) framework proposed for natural language infilling [6] to learn . For assembling the model input, we first choose a span and concatenate , [SEP],, and [ANS]. We additionally prepend conditioning tags and to specific the chain type (heavy or light) and species-of-origin (e.g., human, mouse, etc.) of the antibody sequence. The fully formed sequence of tokens for IgLM is:
| (1) |
We then train a generative model with parameters to maximize , which can be decomposed into a product of conditional probabilities:
| (2) |
Model implementation
The IgLM model uses a Transformer decoder architecture based on a modified version of the GPT-2 Transformer [30] as implemented in the HuggingFace Transformers library [? ]. We trained two models, IgLM and IgLM-S, for sequence infilling. Hyperparameter details are provided in Table 1.
Table 1.
IgLM model hyperparameters.
|
| ||
|---|---|---|
| IgLM | IgLM-S | |
|
| ||
| Number of layers | 4 | 3 |
| Embedding dimension | 512 | 192 |
| Hidden dimension | 512 | 192 |
| Attention heads | 8 | 6 |
| Feed-forward dimension | 2048 | 768 |
|
| ||
| Total parameters | 12,889,600 | 1,439,616 |
|
| ||
Antibody sequence dataset
To train IgLM, we collected unpaired antibody sequences from the Observed Antibody Space (OAS) [16]. OAS is a curated set of over one billion unique antibody sequences compiled from over eighty immune repertoire sequencing studies. After removing sequences indicated to have potential sequencing errors, we were left with 809M unique antibody sequences. We then clustered these sequences using LinClust [44] at 95% sequence identity, leaving 588M non-redundant sequences. The distribution of sequences corresponding to each species and chain type are documented in Figure 1B and Table S1. The dataset is heavily skewed towards human antibodies, particularly heavy chains, which make up 70% of all sequences.
The highly conserved nature of antibody sequences, which are recombined and mutated from a common of set germline components, makes construction of distinct training and validation sets challenging, as overly aggressive splitting may result in exclusion of entire germline lineages from training. For this work, we held out a random 5% of the clustered sequences as a test set to evaluate model performance. Of the remaining sequences, we randomly selected 558M sequences for training and 1M for validation. This splitting criteria ensures that the model is exposed to all of the available conserved regions of antibody sequences, but can be evaluated on how well it captures mutations to those sequences.
Model training
During training, for each sequence we chose a mask length uniformly at random from [10, 20] and a starting position uniformly at random from . We prepended two conditioning tags and denoting the chain type and species-of-origin of each sequence as annotated in the OAS database. Models were trained with a batch size of 512 and 2 gradient accumulation steps using DeepSpeed [31, 34]. Training required approximately 3 days when distributed across 4 NVIDIA A100 GPUs.
Infilling perplexity
Language models are commonly evaluated using perplexity, which computes the exponentiated average negative log-likelihood across tokens in a dataset. For a dataset with total tokens across sequences, this corresponds to computing:
| (3) |
where indexes over the sequences in the dataset. Since IgLM re-designs antibodies by infilling spans conditioned on the surrounding context, rather than evaluating model likelihood on all tokens in a sequence, we define an infilling perplexity metric to evaluate model likelihood only on tokens within infilled spans. For a dataset with sequences masked by our infilling formulation procedure above, we compute infilled perplexity with IgLM as:
| (4) |
where indexes over the sequences in the dataset, represents the span with the [ANS] token appended to it, and represents the total length of all across the dataset. In other words, infilling perplexity is equivalent to taking the per-token perplexity after the [SEP] token.
In Figure 1D, we also compared IgLM infilling perplexity to methods using only preceding context (IgLM [pre], IgLM-S [pre], ProGen2-base, ProGen2-OAS). For these methods, rather than compute perplexity using our infilling formulation procedure, we instead provide only the amino acid sequence context preceding the span to be predicted. We additionally prepend the appropriate conditioning tokens for each model (i.e. the chain type and species-of-origin tokens for IgLM, and the 1 character token for the ProGen2 models) prior to inference. We then compute per-token perplexity over the predicted span and the first residue following the span, where the first residue following the span acts as a proxy for the [ANS] token. In this way, we compute infilling perplexity over the same number of tokens with these methods while only providing the preceding amino acid sequence context.
Full-length antibody generation
Given a chain type and species-of-origin, IgLM samples full-length antibodies by autoregressively sampling from until the [ANS] is sampled, where is the chain token , and is the species token . Because IgLM is trained with the infilling formulation, the model will generate a [MASK], [SEP], and [ANS] token within the sampled sequence . To form the full-length antibody sequence, we replaced the [MASK] token with the span between [SEP] and [ANS] and removed all non-amino acid tokens. Any sampled sequences without [MASK], [SEP], and [ANS] in the correct order were discarded.
Because the OAS database we used for training frequently features sequences with N-terminal truncations, we used a prompting strategy: in addition to providing a chain species token, we provided an initial three-residue motif based on the species and chain type tags. Specific initialization sequences are documented in Table S2.
Positional entropy of full-length sequences
To observe whether mutations from germline tend to occur at biologically relevant positions in generated full-length sequences, we computed the positional entropy of sequences according to Chothia numbering. Specifically, we first selected all sequences for which the species and chain type classifications from ANARCI matched the full-length sequence generation parameters specified in Table S2. For each chain type, species, and temperature setting, we aligned the remaining sequences and aggregated residues at insertion points in the numbering scheme with the prior non-insertion residue. Then, we computed the entropy at each position as:
| (5) |
where indexes over the Chothia position of the aligned sequences, represents the set of all 20 residues, and denotes the proportion of residues at position that correspond to residue .
Sequence infilling
Because IgLM is trained under the infilling framework, the model can re-design spans within a given sequence. To re-design a span of length starting at position within an antibody sequence , we conditioned on . To generate a span , we sequentially sampled until the [ANS] token was sampled. To form our designed sequences, we replaced [MASK] in with and removed all non-amino acid tokens.
Sampling parameters
As we sampled sequences under the model, we applied temperature sampling to shape the probability distribution for each token. Applying temperature corresponds to scaling the logits from the last layer before applying softmax:
| (6) |
where denotes the probability assigned during sampling to token out of possible tokens in the vocabulary. Intuitively, sampling with higher temperatures results in more diverse sequences, with the probability distribution across tokens becoming nearly uniform when is large.
In addition to applying temperature, we also applied nucleus sampling to vary the diversity of sequences generated by IgLM [14]. In nucleus sampling with probability , the probability distribution during sampling is clipped such that only the smallest set of tokens whose cumulative probability exceeds are considered during sampling. Intuitively, a lower restricts sampling to highly probable tokens, which decreases the diversity of sequences while increasing confidence.
Therapeutic antibody diversification benchmarks
To highlight the advantage of IgLM’s infilling framework for CDR H3 loop diversification, we benchmarked against randomized baselines, as well as other protein and antibody language models.
We generated randomized baselines by sampling from position-wise amino acid frequencies for CDR H3s from the OAS database. Specifically, for each CDR H3 loop length, we computed position-wise amino acid frequencies across all CDR H3s of that length from sequences in the training dataset, resulting in a position frequency matrix (PFM) for each CDR H3 loop length. For a given therapeutic antibody, we generated two libraries of 1000 sequences: a fixed-length library and a variable-length library. In the fixed-length library, we sampled from the PFM corresponding to the native CDR H3 loop length to obtain 1000 sequences of the same length. In the variable-length library, for each sequence, we first sampled a loop length from the distribution of CDR H3 loop lengths among the training set before sampling from the PFM corresponding to the sampled loop length.
AntiBERTy is a 26M parameter antibody-specific language model trained with a masked language modeling objective on the same dataset that IgLM uses for training [37]. For a given therapeutic antibody, to generate a library with diversified CDR H3 loops using AntiBERTy, we replaced all residues of the CDR H3 with [MASK] tokens and repeatedly sampled from the model to autoregressively fill in [MASK] tokens from left to right.
ESM-2 is a large protein language model trained with a masked language modeling objective on sequences from UniRef50 [45, 20]. In our benchmarks, due to computational limitations, we used the 650M parameter ESM-2 model, which is the third largest publicly available ESM-2 model behind the 3B parameter and 15B parameter models. For a given therapeutic antibody, to generate a library with diversified CDR H3 loops using ESM-2, we replaced all residues of the CDR H3 with ⟨mask⟩ tokens and repeatedly sampled from the model to autoregressively fill in ⟨mask⟩ tokens from left to right.
ProGen2-OAS is a 764M parameter language model trained with a next-token prediction learning objective on 554M OAS sequences clustered at 85% sequence identity [26]. For a given therapeutic antibody, to generate a library with diversified CDR H3 loops using ProGen2-OAS, we provided a 1 character token followed by the sequence context preceding the CDR H3 to the model. We then sampled until the 2 character token (the end of sequence token) was generated. After sampling, we annotated the CDR H3 loop of the generated sequence using Chothia definitions [5] and replaced the CDR H3 in the parent antibody sequence with the CDR H3 generated by ProGen2-OAS.
For all baselines, we sampled to obtain a set of 1000 unique sequences. For all language model baselines, we sampled sequences using a combination of sampling temperatures and nucleus sampling probabilities . For certain combinations of and , low generated sequence diversity yielded redundant sequences. In these cases, we could not obtain 1000 unique sequences and instead used all unique sequences found among 10000 sampling attempts.
Classification of species and chain type
To evaluate the adherence of IgLM-generated sequences to provided species and chain type conditioning tags, we used the ANARCI software [7]. ANARCI uses a set of antibody-specific HMMs to compare a given antibody to a database of germline sequences across several species and chain types. To classify the chain type and species for generated sequences, we used the corresponding species and chain type for the top V-gene match returned by ANARCI.
Evaluation of sequence properties
To assess the developability and humanness of infilled therapeutic antibody sequences, we used a set of in silico tools previously developed for antibodies. Aggregation propensity was calculated based on the predicted structures for each antibody using the Rosetta [17] implementation of the spatial aggregation propensity (SAP) score [4]. Solubility was calculated based on sequence alone, using the public CamSol-Intrinsic web server [43]. To measure humanness (a proxy for immunogenicity), we used the BioPhi OASis identity [29]. OASis identity measures the fraction of 9-mers for a given sequence that have been observed in human repertoires in the OAS database [16].
Supplementary Material
Highlights.
IgLM is a generative language model trained on 558M natural antibody sequences.
IgLM generates full-length antibody sequences conditioned on species and chain type.
Infilled CDR H3 loops libraries generated by IgLM display improved developability.
Acknowledgements
We thank Dr. Sai Pooja Mahajan and Dr. Rahel Frick for insightful discussions and advice. This work was supported by the National Science Foundation grant DBI-1950697 (R.W.S.) and National Institutes of Health grants R01-GM078221 (J.A.R.) and R35-GM141881 (J.A.R.) J.A.R. was supported as a Johns Hopkins-AstraZeneca Fellow. Computation was performed using the Advanced Research Computing at Hopkins (ARCH) core facility.
Footnotes
Declaration of Interests
R.W.S., J.A.R., and J.J.G are inventors of the IgLM technology developed in this study. The Johns Hopkins University has filed international patent application PCT/US2022/052178 Generative Language Models And Related Aspects For Peptide And Protein Sequence Design, which relates to the IgLM technology. R.W.S., J.A.R., and J.J.G may be entitled to a portion of revenue received from commercial licensing of the IgLM technology and any intellectual property therein. J.J.G. is an unpaid member of the Executive Board of the Rosetta Commons. Under an institutional participation agreement between the University of Washington, acting on behalf of the Rosetta Commons, and the Johns Hopkins University (JHU), JHU may be entitled to a portion of revenue received on licensing of Rosetta software used in this paper. J.J.G. has a financial interest in Cyrus Biotechnology. Cyrus Biotechnology distributes the Rosetta software, which may include methods used in this paper. These arrangements have been reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Akbar R, Robert PA, Weber CR, Widrich M, Frank R, Pavlović M, Scheffer L, Chernigovskaya M, Snapkov I, Slabodkin A et al. (2022). In silico proof of principle of machine learning-based antibody design at unconstrained scale. Mabs 14, 2031482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Almagro JC, Pedraza-Escalona M, Arrieta HI and Pérez-Tapia SM (2019). Phage display libraries for antibody therapeutic discovery and development. Antibodies 8, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bachas S, Rakocevic G, Spencer D, Sastry AV, Haile R, Sutton JM, Kasun G, Stachyra A, Gutierrez JM, Yassine E et al. (2022). Antibody optimization enabled by artificial intelligence predictions of binding affinity and naturalness. bioRxiv, 2022–08. [Google Scholar]
- [4].Chennamsetty N, Voynov V, Kayser V, Helk B and Trout BL (2010). Prediction of aggregation prone regions of therapeutic proteins. The Journal of Physical Chemistry B 114, 6614–6624. [DOI] [PubMed] [Google Scholar]
- [5].Chothia C and Lesk AM (1987). Canonical structures for the hypervariable regions of immunoglobulins. Journal of molecular biology 196, 901–917. [DOI] [PubMed] [Google Scholar]
- [6].Donahue C, Lee M and Liang P (2020). Enabling language models to fill in the blanks. arXiv preprint arXiv:2005.05339. [Google Scholar]
- [7].Dunbar J and Deane CM (2016). ANARCI: antigen receptor numbering and receptor classification. Bioinformatics 32, 298–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Elnaggar A, Heinzinger M, Dallago C, Rehawi G, Wang Y, Jones L, Gibbs T, Feher T, Angerer C, Steinegger M et al. (2021). Prottrans: Toward understanding the language of life through self-supervised learning. IEEE transactions on pattern analysis and machine intelligence 44, 7112–7127. [DOI] [PubMed] [Google Scholar]
- [9].Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J et al. (2021). Protein complex prediction with AlphaFold-Multimer. biorxiv, 2021–10. [Google Scholar]
- [10].Ferruz N, Schmidt S and Höcker B (2022). ProtGPT2 is a deep unsupervised language model for protein design. Nature communications 13, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Griffiths AD, Williams SC, Hartley O, Tomlinson I, Waterhouse P, Crosby WL, Kontermann R, Jones P, Low N and Allison T. a. (1994). Isolation of high affinity human antibodies directly from large synthetic repertoires. The EMBO journal 13, 3245–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hesslow D, Zanichelli N, Notin P, Poli I and Marks D (2022). RITA: a Study on Scaling Up Generative Protein Sequence Models. arXiv preprint arXiv:2205.05789. [Google Scholar]
- [13].Hie BL, Shanker VR, Xu D, Bruun TU, Weidenbacher PA, Tang S, Wu W, Pak JE and Kim PS (2023). Efficient evolution of human antibodies from general protein language models. Nature Biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Holtzman A, Buys J, Du L, Forbes M and Choi Y (201. 9). The curious case of neural text degeneration. arXiv preprint arXiv:1904.09751. [Google Scholar]
- [15].Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y et al. (2017). Biophysical properties of the clinical-stage antibody landscape. Proceedings of the National Academy of Sciences 114, 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kovaltsuk A, Leem J, Kelm S, Snowden J, Deane CM and Krawczyk K (2018). Observed antibody space: a resource for data mining next-generation sequencing of antibody repertoires. The Journal of Immunology 201, 2502–2509. [DOI] [PubMed] [Google Scholar]
- [17].Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman KW, Renfrew PD, Smith CA, Sheffler W et al. (2011). ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. In Methods in enzymology vol. 487, pp. 545–574. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Leem J, Mitchell LS, Farmery JH, Barton J and Galson JD (2022). Deciphering the language of antibodies using self-supervised learning. Patterns 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lefranc M-P, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J et al. (2009). IMGT®, the international ImMunoGeneTics information system®. Nucleic acids research 37, D1006–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lin Z, Akin H, Rao R, Hie B, Zhu Z, Lu W, Smetanin N, Verkuil R, Kabeli O, Shmueli Y et al. (2023). Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130. [DOI] [PubMed] [Google Scholar]
- [21].Lonberg N (2005). Human antibodies from transgenic animals. Nature biotechnology 23, 1117–1125. [DOI] [PubMed] [Google Scholar]
- [22].Madani A, Krause B, Greene ER, Subramanian S, Mohr BP, Holton JM, Olmos JL Jr, Xiong C, Sun ZZ, Socher R et al. (2023). Large language models generate functional protein sequences across diverse families. Nature Biotechnology, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Madani A, McCann B, Naik N, Keskar NS, Anand N, Eguchi RR, Huang P-S and Socher R (2020). Progen: Language modeling for protein generation. arXiv preprint arXiv:2004.03497. [Google Scholar]
- [24].McCafferty J, Griffiths AD, Winter G and Chiswell DJ (1990). Phage antibodies: filamentous phage displaying antibody variable domains. nature 348, 552–554. [DOI] [PubMed] [Google Scholar]
- [25].Meier J, Rao R, Verkuil R, Liu J, Sercu T and Rives A (2021). Language models enable zero-shot prediction of the effects of mutations on protein function. Advances in Neural Information Processing Systems 34, 29287–29303. [Google Scholar]
- [26].Nijkamp E, Ruffolo J, Weinstein EN, Naik N and Madani A (2022). ProGen2: exploring the boundaries of protein language models. arXiv preprint arXiv:2206.13517. [DOI] [PubMed] [Google Scholar]
- [27].Olsen TH, Moal IH and Deane CM (2022). AbLang: an antibody language model for completing antibody sequences. Bioinformatics Advances 2, vbac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Poiron C, Wu Y, Ginestoux C, Ehrenmann F, Duroux P and Lefranc M (2010). IMGT/mAb-DB: the IMGT® database for therapeutic monoclonal antibodies. Poster no101 11. [Google Scholar]
- [29].Prihoda D, Maamary J, Waight A, Juan V, Fayadat-Dilman L, Svozil D and Bitton DA (2022). BioPhi: a platform for antibody design, humanization, and humanness evaluation based on natural antibody repertoires and deep learning. MAbs 14, 2020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Radford A, Wu J, Child R, Luan D, Amodei D, Sutskever I et al. (2019). Language models are unsupervised multitask learners. OpenAI blog 1, 9. [Google Scholar]
- [31].Rajbhandari S, Rasley J, Ruwase O and He Y (2020). Zero: Memory optimizations toward training trillion parameter models. pp. 1–16, IEEE. [Google Scholar]
- [32].Raybould MI, Marks C, Krawczyk K, Taddese B, Nowak J, Lewis AP, Bujotzek A, Shi J and Deane CM (2019). Five computational developability guidelines for therapeutic antibody profiling. Proceedings of the National Academy of Sciences 116, 4025–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Raybould MI, Marks C, Lewis AP, Shi J, Bujotzek A, Taddese B and Deane CM (2020). Thera-SAbDab: the therapeutic structural antibody database. Nucleic acids research 48, D383–D388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ren J, Rajbhandari S, Aminabadi RY, Ruwase O, Yang S, Zhang M, Li D and He Y (2021). Zero-offload: Democratizing billion-scale model training. arXiv preprint arXiv:2101.06840. [Google Scholar]
- [35].Rives A, Meier J, Sercu T, Goyal S, Lin Z, Liu J, Guo D, Ott M, Zitnick CL, Ma J et al. (2021). Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proceedings of the National Academy of Sciences 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ruffolo JA, Chu L-S, Mahajan SP and Gray JJ (2023). Fast, accurate antibody structure prediction from deep learning on massive set of natural antibodies. Nature communications 14, 2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ruffolo JA, Gray JJ and Sulam J (2021). Deciphering antibody affinity maturation with language models and weakly supervised learning. arXiv preprint arXiv:2112.07782. [Google Scholar]
- [38].Saka K, Kakuzaki T, Metsugi S, Kashiwagi D, Yoshida K, Wada M, Tsunoda H and Teramoto R (2021). Antibody design using LSTM based deep generative model from phage display library for affinity maturation. Scientific reports 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shin J-E, Riesselman AJ, Kollasch AW, McMahon C, Simon E, Sander C, Manglik A, Kruse AC and Marks DS (2021). Protein design and variant prediction using autoregressive generative models. Nature communications 12, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sidhu SS and Fellouse FA (2006). Synthetic therapeutic antibodies. Nature chemical biology 2, 682–688. [DOI] [PubMed] [Google Scholar]
- [41].Sievers F and Higgins DG (2014). Clustal Omega, accurate alignment of very large numbers of sequences. In Multiple sequence alignment methods pp. 105–116. Springer. [DOI] [PubMed] [Google Scholar]
- [42].Smith GP (1985). Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317. [DOI] [PubMed] [Google Scholar]
- [43].Sormanni P, Aprile FA and Vendruscolo M (2015). The CamSol method of rational design of protein mutants with enhanced solubility. Journal of molecular biology 427, 478–490. [DOI] [PubMed] [Google Scholar]
- [44].Steinegger M and Söding J (2018). Clustering huge protein sequence sets in linear time. Nature communications 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH and Consortium U (2015). UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Suzuki M, Kato C and Kato A (2015). Therapeutic antibodies: their mechanisms of action and the pathological findings they induce in toxicity studies. Journal of toxicologic pathology 28, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Taylor LD, Carmack CE, Schramm SR, Mashayekh R, Higgins KM, Kuo C-C, Woodhouse C, Kay RM and Lonberg N (1992). A transgenic mouse that expresses a diversity of human sequence heavy and light chain immunoglobulins. Nucleic acids research 20, 6287–6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wolf Pérez A-M, Sormanni P, Andersen JS, Sakhnini LI, Rodriguez-Leon I, Bjelke JR, Gajhede AJ, De Maria L, Otzen DE, Vendruscolo M et al. (2019). In vitro and in silico assessment of the developability of a designed monoclonal antibody library. MAbs 11, 388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Generated sequences and developability metrics have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table.
All original code has been deposited at https://github.com/Graylab/IgLM and Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Infilled therapeutic antibody sequences and developability metrics for IgLM and alternative methods | This paper | 10.5281/zenodo.8248326 |
| Software and algorithms | ||
| Code for IgLM | This paper | 10.5281/zenodo.8248335 |