Abstract
This article presents the results of an ongoing inventory of Ascomycota in Yunnan, China, carried out as part of the research project series “Exploring ascomycete diversity in Yunnan”. From over 100 samples collected from diverse host substrates, microfungi have been isolated, identified and are currently being documented. The primary objective of this research is to promote the discovery of novel taxa and explore the ascomycete diversity in the region, utilising a morphology-phylogeny approach. This article represents the second series of species descriptions for the project and introduces three undocumented species found in the families Bambusicolaceae, Dictyosporiaceae and Periconiaceae, belonging to the suborder Massarineae (Pleosporales, Dothideomycetes). These novel taxa exhibit typical morphological characteristics of Bambusicola, Periconia and Trichobotrys, leading to their designation as Bambusicolahongheensis, Periconiakunmingensis and Trichobotryssinensis. Comprehensive multigene phylogenetic analyses were conducted to validate the novelty of these species. The results revealed well-defined clades that are clearly distinct from other related species, providing robust support for their placement within their respective families. Notably, this study unveils the phylogenetic affinity of Trichobotrys within Dictyosporiaceae for the first time. Additionally, the synanamorphism for the genus Trichobotrys is also reported for the first time. Detailed descriptions, illustrations and updated phylogenies of the novel species are provided, and thus presenting a valuable resource for researchers and mycologists interested in the diversity of ascomycetes in Yunnan. By enhancing our understanding of the Ascomycota diversity in this region, this research contributes to the broader field of fungal taxonomy and their phylogenetic understanding.
Key words: Ascomycota, Bambusicola , Periconia , phylogeny, polyphasic approach, taxonomy, the Greater Mekong Subregion, Trichobotrys
Introduction
Pleosporales is the largest order of Dothideomycetes, comprising two main suborders (viz. Massarineae and Pleosporineae), 91 families, 653 genera (including Pleosporales genera incertae sedis) and a quarter of all Dothideomycetes species (Hongsanan et al. 2020; Wijayawardene et al. 2022b). The order was invalidly introduced by Luttrell (1955) and later validated by Barr (1987) and is characterised by perithecial ascomata with typically a papillate ostiole, bitunicate, fissitunicate asci and hyaline to pigmented, variedly shaped, mostly septate ascospores. The asexual morph is represented by both coelomycetes and hyphomycetes (Zhang et al. 2012; Hyde et al. 2013; Hongsanan et al. 2020). Members of Pleosporales are ecologically and morphologically diverse and also shown to be polyphyletic in various groups, as well as contained within species complexes still waiting to be resolved (Zhang et al. 2012; Hyde et al. 2013; Jaklitsch et al. 2016a; Hongsanan et al. 2020). Pleosporalean species are cosmopolitan and ubiquitous in diverse ecological niches. Their life modes include epiphytes, endophytes or parasites on living organisms, hyperparasites on fungi or insects, saprobes, pathogens and lichenised fungi (Zhang et al. 2012; Hyde et al. 2013; Tanaka et al. 2015; Jaklitsch et al. 2016a; Hongsanan et al. 2020). Of these, several genera, such as Alternaria, Bipolaris, Didymella, Leptospharia, Parastagonospora, Phaeosphaeria and Pyrenophora, have been reported as plant pathogens causing severe diseases on economic crops (Quaedvlieg et al. 2013; Woudenberg et al. 2013, 2014, 2015; Manamgoda et al. 2014; Ariyawansa et al. 2015a, b; Chen et al. 2015, 2017; Tanaka et al. 2015; El-Demerdash 2018; Khiralla et al. 2019; Bhunjun et al. 2020; Hongsanan et al. 2020; Backes et al. 2021; Bartosiak et al. 2021; Li et al. 2023).
A comprehensive study of the genera in Pleosporales was carried out by Zhang et al. (2012), based on morphological studies of the type specimens coupled with phylogenetic analyses. Consequently, the taxonomic treatment of numerous Pleosporales was updated by various authors, based on polyphasic taxonomic approaches, mainly using morphology-phylogeny-based taxonomy (Ariyawansa et al. 2014, 2015a, b; Phookamsak et al. 2014, 2015; Tanaka et al. 2015; Thambugala et al. 2015; Boonmee et al. 2016; Jaklitsch and Voglmayr 2016; Jaklitsch et al. 2016a, b, 2018; Su et al. 2016; Chen et al. 2017; Hashimoto et al. 2017; Wanasinghe et al. 2017a, b). Even though novel taxa of Pleosporales have been dramatically increasing over the last ten years after the taxonomic circumscription provided by Zhang et al. (2012) and Hyde et al. (2013), there is still over a quarter of the total known species lacking molecular data and/or reliable phylogenetic markers for clarifying the placements in Pleosporales.
Yunnan is known as one part of the 36 global biodiversity hotspots where over 17,000 species of vascular plants are known, including highly endemic species (Feng and Yang 2018; Cai et al. 2019). Highly diverse environments and geographical distribution, as well as flourishing vegetation, have shown the Province to be one of the richest sources of fungi, covering over 40% of the known species in China (Feng and Yang 2018; Liu et al. 2018). Feng and Yang (2018) estimated a species number of fungi existing in Yunnan Province, based on the ratio of local vascular plants and fungi (1:6) following the suggestion of Hawksworth (2001). With this estimation, Yunnan may harbour over 104,000 fungal species; of which only 6000 described species have been reported from the Province, including approximately 3000 species of Ascomycota and Basidiomycota (Feng and Yang 2018).
Since Feng and Yang (2018) updated the status of fungal diversity in this region, the taxonomic study of ascomycetes has steadily increased and over 300 novel species have been discovered in the last five years (Luo et al. 2019; Phookamsak et al. 2019; Dong et al. 2020; Hyde et al. 2020a, b; Wanasinghe et al. 2020, 2022; Wang et al. 2020; Mortimer et al. 2021; Wijayawardene et al. 2021b, 2022a; Gu et al. 2022; Jiang et al. 2022; Yang et al. 2022a, b; Si et al. 2023). However, most studies were restricted to certain groups of ascomycetes, such as bambusicolous fungi (Jiang et al. 2019, 2021b; Dai et al. 2022; Phookamsak et al. 2022), cordycipitoid fungi (Wang et al. 2020; Fan et al. 2021; Dong et al. 2022; Tang et al. 2023), endolichenic fungi (Si et al. 2021, 2023), lignicolous freshwater fungi (Luo et al. 2018a, b, 2019; Su et al. 2018; Dong et al. 2020; Shen et al. 2022), nematode-trapping fungi (Zhang et al. 2020, 2022a, b, c, 2023; Yang et al. 2023b) and woody litter-inhabiting fungi (Mortimer et al. 2021; Wanasinghe et al. 2022), as well as fungi associated with specific host plants (e.g. Camellia, Coffea, Magnolia, Mangifera and Rhododendron) (Wanasinghe et al. 2020; Gu et al. 2022; Lu et al. 2022; Tibpromma et al. 2022; Wijayawardene et al. 2022a; Yang et al. 2022a, b, 2023a). Comparable with the total estimated number of species that may be found in this region, these fungal inventories are still only representing a small number of extant ascomycetes in Yunnan.
The present study aims to introduce three novel pleosporalean species from Yunnan, based on morphological characteristics and phylogenetic evidence coupled with the differences in nucleotide pairwise comparison amongst closely-related species.
Materials and methods
Sample collection, isolation, morphological examination and preservation
Samples were collected from Yunnan Province, China during 2016–2021 at three different collecting sites: Honghe (rice terraces), Kunming (botanical garden) and Xishuangbanna (secondary forest). Specimens were collected during the rainy (September) and dry seasons (January and April) and brought to the laboratory in sealed plastic Ziploc bags for further observation and examination. The samples were observed and axenic cultures, via single spore isolation, were obtained within 1–2 weeks after collection. Single spore isolation was performed using the spore suspension technique (Senanayake et al. 2020). Two sets (five spores per set) of the germinated spores were placed separately on to freshly sterilised potato dextrose agar (PDA) medium and incubated under normal day/night light conditions at room temperature (15–25 °C depending on the rainy and dry seasons). Culture characteristics, growth and sporulation in vitro were observed and recorded after one and four-week intervals.
Macro-morphological features, such as ascomata and fungal colonies visualised on host substrates, were observed using an Olympus SZ61 series stereomicroscope and photo-captured by a digital camera. Micro-morphological features were examined by differential interference contrast (DIC) microscopy using a Nikon ECLIPSE Ni-U compound microscope and images captured with a Nikon DS-Ri2 camera. The mucilaginous sheath that covered the ascospores was checked by staining with India Ink and the fungal centrum was stained using Congo red for checking the clearity of conidiophores and conidiogenous cells. Lactoglycerol was added to preserve important morphological features on permanent slides. All morphological features were measured using Tarosoft (R) Image FrameWork version 0.9.7. and photographic plates were edited and combined using Adobe Photoshop CS6 software (Adobe Systems Inc., San Jose, CA, USA).
Axenic living cultures were preserved in PDA and sterilised double-distilled water (ddH2O) at 4 °C for short-term storage and long-term glycerol storage at -20 °C and -80 °C, respectively. Ex-type living cultures were deposited at the collection of Rungtiwa Phookamsak housed at Honghe Center for Mountain Futures (RPC) and duplicated in the Culture Collection of the Herbarium of Cryptogams Kunming Institute of Botany, Academia Sinica (KUNCC), Kunming, China Mae Fah Luang University Culture Collection (MFLUCC), Chiang Rai, Thailand. The type specimens were preserved with silica gel and deposited in the Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS), China. Index Fungorum numbers (http://www.indexfungorum.org; accessed on 25 May 2023) were obtained for the newly-described taxa.
DNA extraction, PCR amplification and sequencing
Fungal genomic DNA was extracted from fresh mycelia using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux, Hangzhou, China) following the procedure from the manufacturer. The genomic DNA was also extracted from ascomata using a Forensic DNA Kit (Omega, Norcross, GA, USA) in case the fungus could not be obtained from the pure culture. Amplicons were generated by polymerase chain reaction (PCR) using five phylogenetic markers, including the internal transcribed spacers region of ribosomal DNA (ITS; ITS1-5.8S-ITS2), the partial 28S large subunit nuclear ribosomal DNA (LSU), the partial 18S small subunit rDNA (SSU), the partial RNA polymerase II second largest subunit (rpb2) and the partial translation elongation factor 1-alpha (tef1-α). The ITS region was amplified with the primer pair ITS4 and ITS5 (White et al. 1990), the LSU region with LR0R and LR5 (Vilgalys and Hester 1990), the SSU region with NS1 and NS4 (White et al. 1990), the rpb2 region with fRPB2-5F and fRPB2-7cR (Liu et al. 1999) and the tef1-α region with EF1-983F and EF1-2218R (Rehner and Buckley 2005). The component of PCR reaction was performed in a total volume of 25 μl, containing 2 μl DNA template (30–50 ng/μl), 1 μl of each forward and reverse primer (10 μM), 12.5 μl Master Mix (mixture of EasyTaqTM DNA Polymerase, dNTPs and optimised buffer; Beijing TransGen Biotech Co., Ltd., Chaoyang District, Beijing, China) and 8.5 µl of double-distilled water (ddH2O). The thermal cycle of PCR amplification for ITS, LSU, SSU, rpb2 and tef1-α was set up following Phookamsak et al. (2014, 2023). PCR products were purified and sequenced by using PCR primers at TsingKe Biological Technology (Kunming City, Yunnan Province, China). The quality of raw sequence data was checked and trimmed of low-quality segments with BioEdit 7.1.3.0 (Hall 1999). The consensus sequences of the newly-generated strains were assembled using SeqMan Pro version 11.1.0 (DNASTAR, Inc. Madison, WI, USA) and submitted to the GenBank database to further encourage accession within the scientific community.
Sequence alignments and phylogenetic analyses
The newly-generated sequences were subjected to the nucleotide BLAST search tool on the NCBI website for checking the correctness of species identification and searching for closely-related taxa that were further included in the sequence alignment dataset. Reference sequences from relevant publications and BLAST results of the closely-related species were downloaded from GenBank to supplement the datasets (Tables 1–3). Three datasets were prepared to construct the phylogenetic trees for clarifying phylogenetic relationships of the novel taxa in Bambusicolaceae (Table 1), Dictyosporiaceae (Table 2) and Periconiaceae (Table 3). The individual gene dataset was aligned using MAFFT v.7 (Katoh et al. 2019) and improved manually where necessary in Bioedit 7.1.3.0 (Hall 1999). The alignments of individual gene datasets were prior analysed by Maximum Likelihood (ML) for checking the congruence of tree topologies and further combined into a multigene dataset. Phylogenetic analyses were performed, based on ML and Bayesian Inference (BI) analyses.
Table 1.
Species details and GenBank accession numbers used in phylogenetic analysis of Bambusicola species (Bambusicolaceae, Pleosporales). The new sequences are indicated in bold and the ex-type strains are indicated by superscript “T”. Missing sequences are indicated by “–”.
| Species name | Strain/specimen no. | GenBank accession numbers | ||||
|---|---|---|---|---|---|---|
| ITS | LSU | rpb2 | SSU | tef1-α | ||
| Bambusicolaaquatica T | MFLUCC 18-1031 | MT627729 | MN913710 | MT878462 | MT864293 | MT954392 |
| Bambusicolaautumnalis T | CGMCC 3.24280 | OQ427824 | OQ427825 | OQ507621 | OQ427823 | OQ507622 |
| Bambusicolaautumnalis | UESTCC 23.0001 | OQ609612 | OQ550210 | OQ556791 | OQ550209 | OQ556792 |
| Bambusicolabambusae T | MFLUCC 11-0614 | JX442031 | JX442035 | KP761718 | JX442039 | KP761722 |
| Bambusicoladidymospora T | MFLUCC 10-0557 | KU940116 | KU863105 | KU940163 | KU872110 | KU940188 |
| Bambusicoladimorpha T | MFLUCC 13-0282 | KY026582 | KY000661 | KY056663 | KY038354 | – |
| Bambusicolaficuum T | MFLUCC 17-0872 | – | MT215580 | – | MT215581 | MT199326 |
| Bambusicolafusispora T | MFLUCC 20-0149 | MW076532 | MW076531 | MW034589 | MW076529 | – |
| Bambusicolaguttulata T | CGMCC 3.20935 | ON332909 | ON332927 | ON383985 | ON332919 | ON381177 |
| Bambusicolahongheensis T | BN06/ KUN-HKAS 129042 | OR233600 | OR335804 | OR540736 | OR501419 | – |
| Bambusicolairregulispora T | MFLUCC 11-0437 | JX442032 | JX442036 | KP761719 | JX442040 | KP761723 |
| Bambusicolaloculata T | MFLUCC 13-0856 | KP761732 | KP761729 | KP761715 | KP761735 | KP761724 |
| Bambusicolamassarinia T | MFLUCC 11-0389 | JX442033 | JX442037 | KP761716 | JX442041 | KP761725 |
| Bambusicolapustulata T | MFLUCC 15-0190 | KU940118 | KU863107 | KU940165 | KU872112 | KU940190 |
| Bambusicolananensis T | MFLUCC 21-0063 | NR_176767 | NG_081535 | – | – | – |
| Bambusicolasichuanensis T | SICAUCC 16-0002 | MK253473 | MK253532 | MK262830 | MK253528 | MK262828 |
| Bambusicolasplendida T | MFLUCC 11-0439 | JX442034 | JX442038 | KP761717 | JX442042 | KP761726 |
| Bambusicolasubthailandica T | SICAU 16-0005 | MK253474 | MK253533 | MK262831 | MK253529 | MK262829 |
| Bambusicolathailandica T | MFLUCC 11-0147 | KU940119 | KU863108 | KU940166 | – | KU940191 |
| Bambusicolatriseptatispora T | MFLUCC 11-0166 | KU940120 | KU863109 | KU940167 | – | – |
| Corylicolaitalica | MFLU 19-0500 | MT554925 | MT554926 | MT590776 | MT554923 | – |
| Corylicolaitalica T | MFLUCC 20-0111 | MT633085 | MT626713 | MT635596 | MT633084 | MT590777 |
| Leucaenicolaaseptata T | MFLUCC 17-2423 | MK347746 | MK347963 | MK434891 | MK347853 | MK360059 |
| Leucaenicolacamelliae T | NTUCC 18-093-4 | MT112302 | MT071278 | MT743283 | MT071229 | MT374091 |
| Leucaenicolaphraeana T | MFLUCC 18-0472 | MK347785 | MK348003 | MK434867 | MK347892 | MK360060 |
| Occultibambusabambusae T | MFLUCC 13-0855 | KU940123 | KU863112 | KU940170 | KU872116 | KU940193 |
| Occultibambusakunmingensis T | KUN-HKAS 102151 | MT627716 | MN913733 | MT878453 | MT864342 | MT954407 |
| Occultibambusasichuanensis T | CGMCC 3.20938 | ON332913 | ON332931 | ON383989 | – | ON381181 |
| Palmiascomagregariascomum T | MFLUCC 11-0175 | KP744452 | KP744495 | KP998466 | KP753958 | – |
| Palmiascomaqujingense T | KUMCC 19-0201 | MT477183 | MT477185 | MT495782 | MT477186 | – |
| Pseudotetraploabambusicola T | CGMCC 3.20939 | ON332915 | ON332933 | ON383991 | ON332923 | ON381183 |
| Pseudotetraploacurviappendiculata T | JCM 12852 | AB524792 | AB524608 | – | AB524467 | – |
| Seriascomabambusae T | KUMCC 21-0021 | MZ329039 | MZ329035 | MZ325470 | MZ329031 | MZ325468 |
| Seriascomadidymosporum T | MFLUCC 11-0179 | KU940127 | KU863116 | KU940173 | KU872119 | KU940196 |
| Seriascomayunnanense T | MFLU 19-0690 | – | MN174695 | MN210324 | MN174694 | MN381858 |
| Versicolorisporiumtriseptatum T | JCM 14775 | AB365596 | AB330081 | – | AB524501 | – |
| Versicolorisporiumtriseptatum | NMX1222 | OL741378 | OL741318 | – | OL741381 | – |
Table 3.
Species details and GenBank accession numbers used in phylogenetic analysis of Periconia species (Periconiaceae, Pleosporales). The new sequences are indicated in bold and the ex-type strains are indicated by superscript “T”. Missing sequences are indicated by “–”.
| Species | Strain No. | GenBank accession numbers | |||
|---|---|---|---|---|---|
| ITS | LSU | SSU | tef1-α | ||
| Flavomycesfulophazae | CBS 135664 | KP184000 | KP184039 | KP184081 | – |
| Flavomycesfulophazae T | CBS 135761 | NR_137960 | NG_058131 | NG_061191 | – |
| Lentitheciumaquaticum T | CBS 123099 | NR_160229 | NG_064211 | NG_016507 | GU349068 |
| Lentitheciumclioninum T | KT 1149A | LC014566 | AB807540 | AB797250 | AB808515 |
| Lentitheciumclioninum | KT 1220 | LC014567 | AB807541 | AB797251 | AB808516 |
| Massarinacisti T | CBS 266.62 | – | AB807539 | AB797249 | AB808514 |
| Massarinaeburnea | CBS 473.64 | – | GU301840 | GU296170 | GU349040 |
| Morosphaeriaramunculicola | KH 220 | – | AB807554 | AB797264 | AB808530 |
| Morosphaeriavelatispora | KH 221 | LC014572 | AB807556 | AB797266 | AB808532 |
| Periconiaalgeriana T | CBS 321.79 | MH861212 | MH872979 | – | – |
| Periconiaalishanica T | MFLUCC 19-0145 | MW063165 | MW063229 | – | MW183790 |
| Periconiaaquatica T | MFLUCC 16-0912 | KY794701 | KY794705 | – | KY814760 |
| Periconiaartemisiae T | KUMCC 20-0265 | MW448657 | MW448571 | MW448658 | MW460898 |
| Periconiaartemisiae | G1782 | MK247789 | – | – | – |
| Periconiaatropurpurea | CBS 381.55 | MH857524 | MH869061 | – | – |
| Periconiabanksiae T | CBS 129526 | JF951147 | NG_064279 | – | – |
| Periconiabyssoides | KUMCC 20-0264 | MW444854 | MW444855 | MW444856 | MW460895 |
| MAFF 243869 | LC014582 | AB807569 | AB797279 | AB808545 | |
| MFLUCC 17-2292 | MK347751 | MK347968 | MK347858 | MK360069 | |
| MFLUCC 18-1553 | MK347806 | MK348025 | MK347914 | MK360068 | |
| MFLUCC 20-0172 | MW063162 | MW063226 | – | – | |
| NCYUCC 19-0314 | MW063163 | MW063227 | – | – | |
| Periconiacaespitosa T | LAMIC 110 16 | MH051906 | MH051907 | – | – |
| Periconiachengduensis T | CGMCC 3.23930 | OP955987 | OP956012 | OP956056 | OP961453 |
| Periconiachengduensis | UESTCC 22.0140 | OP955977 | OP956002 | OP956046 | OP961443 |
| Periconiachimonanthi T | KUMCC 20-0266 | MW448660 | MW448572 | MW448656 | MW460897 |
| Periconiacircinata | CBS 263.37 | MW810265 | MH867413 | – | MW735660 |
| Periconiacitlaltepetlensis T | ENCB 140251 = IOM 325319.1 | MH890645 | MT625978 | – | – |
| Periconiacitlaltepetlensis | IOM 325319.2 | MT649221 | MT649216 | – | – |
| Periconiacookei | MFLUCC 17-1399 | MG333490 | MG333493 | – | MG438279 |
| MFLUCC 17-1679 | – | MG333492 | – | MG438278 | |
| UESTCC 22.013 | OP955968 | OP955993 | OP956037 | – | |
| Periconiacortaderiae T | MFLUCC 15-0457 | KX965732 | KX954401 | KX986345 | KY310703 |
| Periconiacynodontis T | CGMCC 3.23927 | OP909925 | OP909921 | OP909920 | OP961434 |
| Periconiacyperacearum T | CPC 32138 | NR_160357 | NG_064549 | – | – |
| Periconiadelonicis T | MFLUCC 17-2584 | – | NG_068611 | NG_065770 | MK360071 |
| Periconiadidymosporum T | MFLU 15-0058 | KP761734 | KP761731 | KP761738 | KP761728 |
| Periconiadigitata | CBS 510.77 | LC014584 | AB807561 | AB797271 | AB808537 |
| Periconiaelaeidis T | MFLUCC 17-0087 | MG742713 | MH108552 | MH108551 | – |
| Periconiaepilithographicola | MFLUCC 21–0153 | OL753687 | OL606155 | OL606144 | OL912948 |
| Periconiaepilithographicola T | CBS 144017 | NR_157477 | – | – | – |
| Periconiafestucae T | CGMCC 3.23929 | OP955973 | OP955998 | OP956042 | OP961439 |
| Periconiagenistae T | CBS 322.79 | MH861213 | MH872980 | – | – |
| Periconiahomothallica T | CBS 139698/ KT916 | AB809645 | AB807565 | AB797275 | AB808541 |
| Periconiaigniaria | CBS 379.86 | LC014585 | AB807566 | AB797276 | AB808542 |
| Periconiaimperatae T | CGMCC 3.23931 | OP955984 | OP956009 | OP956053 | OP961450 |
| Periconiaimperatae | UESTCC 22.0145 | OP955979 | OP956004 | OP956048 | OP961445 |
| Periconiakunmingensis T | KUMCC 18-0173/ RPC 15-017 | MH892346 | MH892399 | OR225814 | MH908963 |
| Periconialateralis | CBS 292.36 | MH855804 | MH867311 | – | – |
| Periconiamacrospinosa | CBS 135663 | KP183999 | KP184038 | KP184080 | – |
| REF144 | JN859364 | JN859484 | – | – | |
| Periconiaminutissima | MFLUCC 15-0245 | KY794703 | KY794707 | – | – |
| MUT 2887 | MG813227 | – | – | – | |
| Periconianeobrittanica T | CPC 37903 | NR_166344 | NG_068342 | – | – |
| Periconiapalmicola T | MFLUCC 14-0400 | – | NG_068917 | MN648319 | MN821070 |
| Periconiapenniseti T | CGMCC 3.23928 | OP955971 | OP955996 | OP956040 | OP961437 |
| Periconiaprolifica T | CBS 209.64 | MH858422 | MH870050 | – | – |
| Periconiapseudobyssoides | KUMCC 20-0263 | MW444851 | MW444852 | MW444853 | MW460894 |
| Periconiapseudodigitata | KT 644 | MW444852 | AB807562 | AB797272 | AB808538 |
| Periconiapseudodigitata T | KT 1395 | MW444853 | NG_059396 | NG_064850 | AB808540 |
| Periconiasahariana T | CBS 320.79 | MW444854 | MH872978 | – | – |
| Periconiasalina T | GJ374/ MFLU 19–1235 | MW444855 | MN017846 | MN017912 | – |
| Periconiaspodiopogonis T | CGMCC 3.23932 | MW444856 | OP955988 | OP956032 | OP961429 |
| Periconiasubmersa T | MFLUCC 16-1098 | MW444857 | KY794706 | – | KY814761 |
| Periconiathailandica T | MFLUCC 17-0065 | MW444858 | KY753888 | KY753889 | – |
| Periconiathysanolaenae T | KUMCC 20-0262 | MW444859 | MW444850 | MW448659 | MW460896 |
| Periconiavariicolor T | SACCR-64 | MW444860 | – | – | – |
| Periconiaverrucosa T | MFLUCC 17-2158 | MT310617 | MT214572 | MT226686 | MT394631 |
| Periconiaverrucosa | UESTCC 22.0136 | OP955966 | OP955991 | OP956035 | OP961432 |
| KT 1825 | – | AB807573 | AB797283 | AB808549 | |
| KT 1820A | – | AB807572 | AB797282 | AB808548 | |
Table 2.
Species details and GenBank accession numbers used in phylogenetic analysis of taxa in Dictyosporiaceae (Pleosporales). The new sequences are indicated in bold and the ex-type strains are indicated by superscript “T”. Missing sequences are indicated by “–”.
| Species name | Strain/ specimen no. | GenBank accession numbers | |||
|---|---|---|---|---|---|
| ITS | LSU | SSU | tef1-α | ||
| Anthosulcatisporasubglobosa T | MFLUCC 17-2065/ MFLU 17-1473 | MT310636 | NG_073851 | MT226705 | MT394649 |
| Aquadictyosporalignicola T | MFLUCC 17-1318 | MF948621 | MF948629 | – | MF953164 |
| Aquaticheirosporalignicola T | RK-2006a/ HKUCC10304 | AY864770 | AY736378 | AY736377 | – |
| Cheirosporiumtriseriale T | HMAS 180703 | EU413953 | EU413954 | – | – |
| Chromolaenicolananensis T | MFLUCC 17-1473 | MN325015 | NG_070942 | MN325009 | MN335648 |
| Darksideaalpha T | CBS 135650 | NR_137619 | KP184019 | KP184049 | KP184166 |
| Dendryphiellafasciculata T | MFLUCC 17-1074 | NR_154044 | NG_059177 | – | – |
| Dendryphiellavariabilis T | CBS 584.96 | LT963453 | LT963454 | – | – |
| Dictyocheirosporabannica T | KH 332 | LC014543 | AB807513 | AB797223 | AB808489 |
| Dictyocheirosporarotunda T | MFLUCC 14-0293b | KU179099 | KU179100 | – | – |
| Dictyosporiumbulbosum | yone 221 | LC014544 | AB807511 | AB797221 | AB808487 |
| Dictyosporiumelegans T | NBRC 32502 | DQ018087 | DQ018100 | DQ018079 | – |
| Didymosphaeriarubi-ulmifolii T | MFLUCC 14-0023 | – | KJ436586 | KJ436588 | – |
| Digitodesmiumbambusicola T | CBS 110279 | DQ018091 | DQ018103 | – | – |
| Falciformisporasenegalensis T | CBS 196.79 | MH861195 | NG_057981 | NG_062928 | KF015687 |
| Fuscosphaeriahungarica T | DSE883, CBS 147250 | MW209054 | MW209059 | MW209065 | MW238843 |
| Gregaritheciumcurvisporum T | HHUF 30134 | NR_154049 | NG_059394 | NG_061002 | AB808523 |
| Gregaritheciumcurvisporum | MS224 | LC482117 | – | – | – |
| DCR17 | MZ047572 | – | – | – | |
| Helicascuselaterascus | KT 2673/ MAFF 243867 | AB809626 | AB807533 | AB797243 | AB808508 |
| Immotthiabambusae T | KUN-HKAS 112012AI | MW489455 | MW489450 | MW489461 | MW504646 |
| KUN-HKAS 112012B | MW489457 | MW489452 | – | – | |
| Jalapriyapulchra T | MFLUCC 15-0348 | KU179108 | KU179109 | KU179110 | – |
| Jalapriyatoruloides T | CBS 209.65 | DQ018093 | DQ018104 | DQ018081 | – |
| Katumotoabambusicola T | KT1517a | LC014560 | AB524595 | AB524454 | AB539108 |
| Lentitheciumclioninum T | KT1149A/ HHUF:28199 | NR_154137 | NG_059391 | NG_064845 | AB808515 |
| Lentitheciumpseudoclioninum T | HHUF 29055 | AB809633 | NG_059392 | NG_064847 | AB808521 |
| Loculosulcatisporathailandica T | KUMCC 20-0159 | MT376742 | MT383964 | MT383968 | MT380476 |
| Magnicamarosporiumiriomotense T | HHUF 30125/ KT 2822 | NR_153445 | NG_059389 | NG_060999 | AB808485 |
| Montagnulacirsii T | MFLUCC 13-0680 | KX274242 | KX274249 | KX274255 | KX284707 |
| Morosphaeriamuthupetensis T | NFCCI4219 | MF614795 | MF614796 | MF614797 | MF614798 |
| Murilentitheciumclematidis T | MFLUCC 14-0562 | KM408757 | KM408759 | KM408761 | KM454445 |
| Neodendryphiellamali T | CBS 139.95 | LT906655 | LT906657 | EF204511 | – |
| Neodendryphiellamichoacanensis T | FMR 16098 | NR_160583 | LT906658 | – | – |
| Neohelicascusaquaticus | MFLUCC 10-0918/ KT 1544 | AB809627 | AB807532 | AB797242 | AB808507 |
| Paradictyocheirosporatectonae T | NFCCI 4878/ AMH 10301 | MW854646 | MW854647 | – | MW854832 |
| Phaeosphaeriaoryzae T | CBS 110110 | KF251186 | KF251689 | GQ387530 | – |
| Phaeosphaeriopsisglaucopunctata T | MFLUCC 13-0265 | KJ522473 | KJ522477 | KJ522481 | MG520918 |
| Pseudocoleophomabauhiniae T | MFLUCC 17–2586 | MK347736 | MK347953 | MK347844 | MK360076 |
| Pseudocoleophomacalamagrostidis T | KT 3284/ HHUF 30450 | LC014592 | LC014609 | LC014604 | LC014614 |
| Pseudoconiothyriumbroussonetiae T | CBS:145036/ CPC:33570 | NR_163377 | NG_066331 | – | MK442709 |
| Pseudoconiothyriumtyphicola T | MFLUCC 16-0123 | KX576655 | KX576656 | – | – |
| Pseudocyclothyriellaclematidis T | MFLUCC 17-2177A | MT310595 | MT214548 | – | MT394730 |
| Pseudocyclothyriellaclematidis | MFLU 16-0280 | MT310596 | MT214549 | – | – |
| PseudodictyosporiumelegansT (=Cheiromoniliophoraelegans) | CBS 688.93 | DQ018099 | DQ018106 | DQ018084 | – |
| Pseudodictyosporiumthailandica T | MFLUCC 16-0029 | NR_154347 | NG_059688 | NG_063611 | KX259526 |
| Sajamaeamycophila T | APA-2999 | MK795715 | MK795718 | – | – |
| Sulcatisporaacerina T | KT 2982 | LC014597 | LC014610 | LC014605 | LC014615 |
| Tingoldiagograminicola T | KH68 | LC014598 | AB521743 | AB521726 | AB808561 |
| Trichobotryseffusus | 1179 | KJ630313 | – | – | – |
| HNNUZCJ-94 | OM281094 | – | – | – | |
| FS524 | MN545626 | – | – | – | |
| SYSU-MS4729 | MH050972 | – | – | – | |
| DFFSCS021 | JX156367 | – | – | – | |
| Trichobotryssinensis T | RPC 21-007/ KUNCC 23-14554 | OR233595 | OR335805 | OR501420 | OR547995 |
| Trichobotrys sp. [as Gregarithecium sp.] | MFLUCC 13-0853 | KX364281 | KX364282 | KX364283 | – |
| GMB1217 | – | – | OM836759 | – | |
| Trematosphaeriapertusa T | CBS 122368 | NR_132040 | NG_057809 | FJ201991 | KF015701 |
| Verrucoccumcoppinsii T | E 00814291 | MT918785 | MT918770 | NG_081399 | – |
| Verrucoccumspribillei T | SPO 1154 | MT918781 | MT918764 | MT918772 | – |
| Vikalpaaustraliense | HKUCC 8797 | DQ018092 | – | – | – |
Maximum Likelihood (ML) implemented by the Randomised Axelerated Maximum Likelihood (RAxML), was performed in RAxML-HPC v.8 on the XSEDE (8.2.12) tool via the online web portal CIPRES Science Gateway v. 3.3 (Miller et al. 2010) using default settings, but adjusted with 1000 bootstrap replicates and a gamma-distributed rate variation of a general time reversible model (GTR) was applied. The BI analyses were conducted by MrBayes on XSEDE v. 3.2.7a via the same web portal as in ML, with two parallel runs. The best-fit model of nucleotide substitution was determined by MrModelTest v. 2.3 (Nylander et al. 2008). Six simultaneous Markov chains were run for 1–5 million generations, but stopped automatically when the critical value for the topological convergence diagnostic reached 0.01. Trees were sampled every 100th generation. The initial 10% of sample trees were treated as burn-in (estimated by Tracer v. 1.7; Rambaut et al. (2018)) and discarded. The remaining trees were used to calculate the posterior probabilities in the majority rule consensus tree. The phylograms were visualised using Figtree v. 1.4.0 (Rambaut and Drummond 2012) and backbone trees were laid out and edited in Adobe Illustrator version 20.0.0. software (Adobe Systems Inc., San Jose, CA, USA).
Results
Phylogenetic analyses
In this study, three phylogenetic analyses were conducted to clarify the phylogenetic placements of our new taxa within the Bambusicolaceae (Analysis 1), Dictyosporiaceae (Analysis 2) and Periconiaceae (Analysis 3), as follows:
Analysis 1
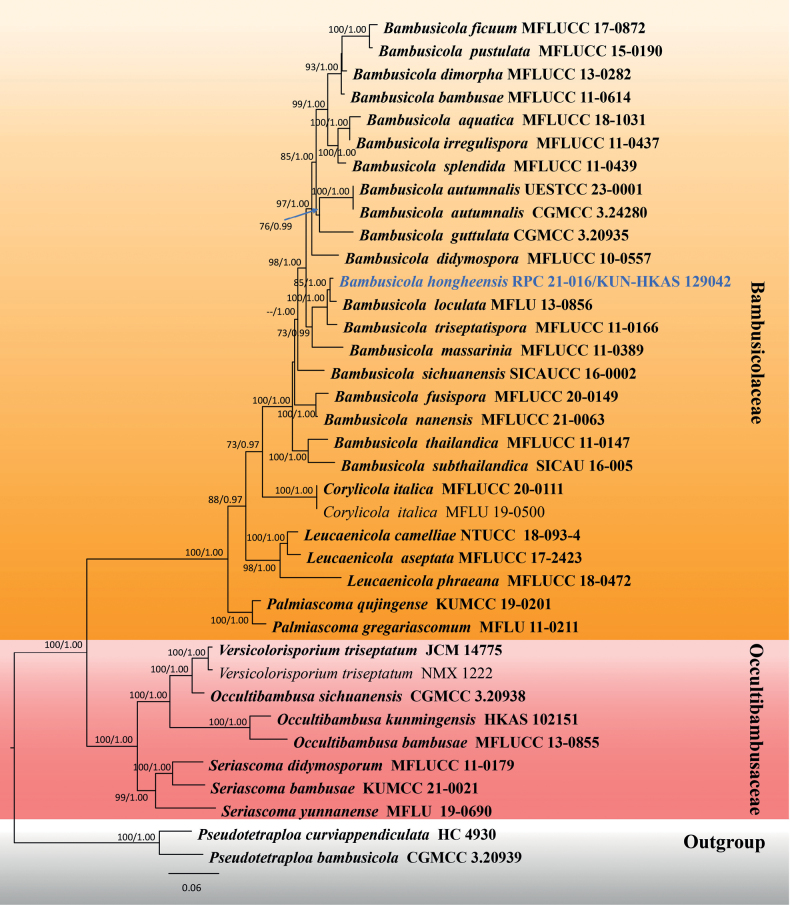
The Bambusicola species tree was constructed using a sequence dataset of the concatenated ITS, LSU, rpb2, SSU and tef1-α of all Bambusicola species, as well as representatives of other related genera. A total of 37 strains were included, with two strains of Pseudotetraploabambusicola (CGMCC 3.20939) and P.curviappendiculata (JCM 12852) as the outgroup. Primarily, phylogenetic analysis of the concatenated LSU, SSU and ITS sequence dataset was conducted, based on ML and compared with the multigene phylogenetic analysis (the concatenated ITS, LSU, rpb2, SSU and tef1-α sequence dataset). Phylogenetic analysis, based on the concatenated LSU, SSU and ITS gene regions, showed a similar topology with the concatenated ITS, LSU, rpb2, SSU and tef1-α gene regions and were not significantly different (data not shown). Hence, multigene phylogenetic analysis of the concatenated ITS, LSU, rpb2, SSU and tef1-α gene regions was selected to represent the phylogenetic relationships of the new species with other closely-related species in Bambusicolaceae. The aligned dataset contained 4929 characters, including gaps. Phylogenetic relationships were inferred by conducting analyses using both ML and BI methods. The best-scoring RAxML tree was selected to represent the relationships amongst taxa, with a final likelihood value of -29592.797597 (Fig. 1). The matrix contained 1905 distinct alignment patterns, with a 22.83% proportion of gaps and completely undetermined characters. The estimated base frequencies of A = 0.243583, C = 0.258293, G = 0.271748, T = 0.226375; substitution rates AC = 1.393909, AG = 2.806593, AT = 1.064133, CG = 1.193703, CT = 6.412290, GT = 1.000000; gamma distribution shape parameter α = 0.589535; Tree-Length = 1.823129. For BI analysis, GTR + I + G was selected as the best-fit model by AIC in MrModelTest for each gene (ITS, LSU, rpb2, SSU and tef1-α). Six simultaneous Markov chains were set to run for 1,000,000 generations, but stopped at 25,000 generations because the convergence diagnostic hit the stop value, resulting in 251 total trees. The first 10% of trees were discarded as the burn-in phase of the analyses and the remaining trees were used for calculating posterior probabilities in the majority rule consensus tree, of which the final average standard deviation of split frequencies at the end of total MCMC generations was 0.005298.
Figure 1.
Phylogram of the best-scoring ML consensus tree of taxa in Bambusicolaceae and Occultibambusaceae. The new isolate is indicated in blue. Isolates from type materials are in bold. The ML ultrafast bootstrap and Bayesian PP values greater than 60% and 0.90 are shown at the nodes.
Multigene phylogenetic analyses demonstrated that all genera of Bambusicolaceae formed well-resolved clades (up to 98% ML, 1.00 PP; Fig. 1) in the present study. The new species, Bambusicolahongheensis (KUN-HKAS 129042), clustered with the clade containing B.loculata (MFLUCC 13-0856) (85% ML, 1.00 PP) and B.triseptatispora (MFLUCC 11-0166) with high statistical support (100% ML, 1.00 PP). These three species have close relationships with B.massarinia (MFLUCC 11-0389) (73% ML, 0.99 PP), the type genus of Bambusicola.
Analysis 2
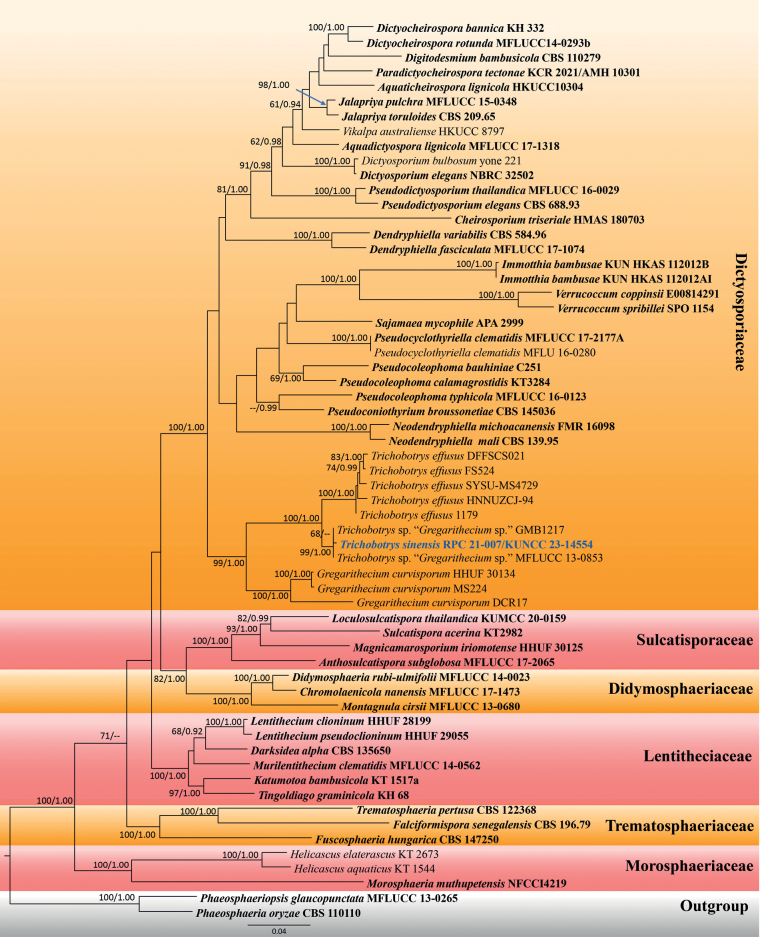
The Trichobotrys tree was constructed using sequence data from ITS, LSU, SSU and tef1-α. A total of 61 strains of taxa in Dictyosporiaceae and closely-related families (viz. Didymosphaeriaceae, Lentitheciaceae, Morosphaeriaceae, Sulcatisporaceae and Trematosphaeriaceae) were included, with Phaeosphaeriaoryzae (CBS 110110) and Phaeosphaeriopsisglaucopunctata (MFLUCC 13-0265) (Phaeosphaeriaceae) as the outgroup. Primarily, phylogenetic analysis of the concatenated LSU, SSU and ITS sequence dataset was conducted, based on ML and compared with phylogenetic analysis of the concatenated ITS, LSU, SSU and tef1-α sequence dataset. Phylogenetic analysis, based on the concatenated LSU, SSU and ITS sequence dataset, showed a similar topology with the concatenated ITS, LSU, SSU and tef1-α sequence dataset and were not significantly different (data not shown). Hence, multigene phylogenetic analysis of the concatenated ITS, LSU, SSU and tef1-α gene regions was selected to represent the phylogenetic relationships of Trichobotryssinensis sp. nov. with other closely-related species in Dictyosporiaceae. The aligned dataset contained 3729 characters, including gaps. Phylogenetic relationships were inferred by conducting analyses using both ML and BI methods. The best-scoring RAxML tree was selected to represent the relationships amongst taxa, with a final likelihood value of -28366.415110 (Fig. 2). The matrix contained 1566 distinct alignment patterns, with a 39.19% proportion of gaps and completely undetermined characters. The estimated base frequencies of A = 0.239629, C = 0.244575, G = 0.269426, T = 0.246371; substitution rates AC = 1.123110, AG = 2.634717, AT = 1.787337, CG = 0.836519, CT = 6.160493, GT = 1.000000; gamma distribution shape parameter α = 0.461486; Tree-Length = 3.107341. For BI analysis, GTR + I + G was selected as the best-fit model by AIC in MrModelTest for each gene (ITS, LSU, SSU and tef1-α). Six simultaneous Markov chains were run for 4,085,000 generations, resulting in 40,851 total trees. The first 10% of trees were discarded as the burn-in phase of the analyses and the remaining trees were used for calculating posterior probabilities in the majority rule consensus tree, of which the final average standard deviation of split frequencies at the end of total MCMC generations was 0.009998.
Figure 2.
Phylogram of the best-scoring ML consensus tree of Trichobotrys species in Dictyosporiaceae and closely-related families viz. Didymosphaeriaceae, Lentitheciaceae, Morosphaeriaceae, Sulcatisporaceae and Trematosphaeriaceae. The new isolate is indicated in blue. Isolates from type materials are in bold. The ML ultrafast bootstrap and Bayesian PP values greater than 70% and 0.95 are shown at the nodes.
Multigene phylogenetic analyses of the concatenated ITS, LSU, SSU and tef1-α demonstrated that all representative families formed well-resolved clades in the present study. Our new isolate grouped with two unnamed Gregarithecium sp. (strains GMB1217 and MFLUCC 13-0853), with high support in ML and BI analyses (99% ML, 100 PP; Fig. 2) and clustered with Trichobotryseffusus (strains 1179, HNNUZCJ-94, FS524, SYSU-MS4729 and DFFSCS021) with high support (100% ML, 1.00 PP; Fig. 2) in Dictyosporiaceae. Gregarithecium sp. (strains GMB1217 and MFLUCC 13-0853) is unpublished and showed to be conspecific with our new isolate. Therefore, our new isolate is introduced as Trichobotryssinensis, based on phylogenetic evidence coupled with morphological characteristics. Trichobotrys formed a highly-supported subclade with Gregarithecium (99% ML, 1.00 PP; Fig. 2) in the present study. However, these two genera are represented by different morphs. Therefore, the congeneric status of these two genera is doubtful in the study pending future study.
Analysis 3
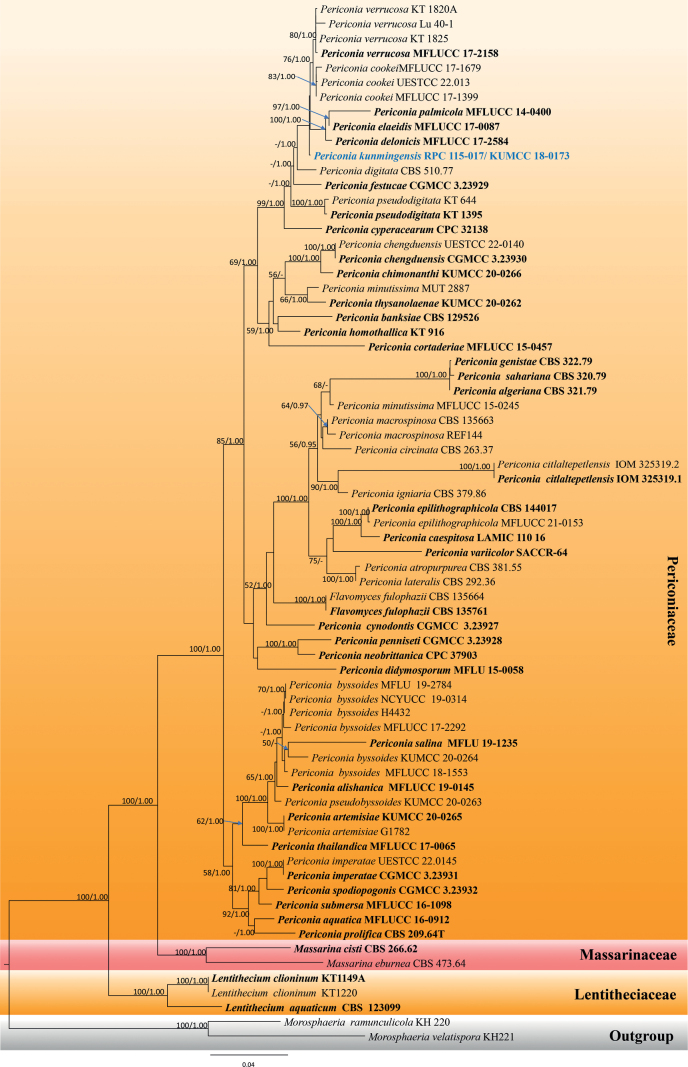
The Periconia species tree was constructed using sequence data from ITS, LSU, SSU and tef1-α of all taxa in Periconiaceae and other related families (viz. Lentitheciaceae, and Massarinaceae). A total of 71 strains were included, with Morosphaeriaramunculicola (KH 220) and M.velatispora (KH 221) as the outgroup. The aligned dataset contained 3646 characters, including gaps. The best-scoring RAxML tree was selected to represent the relationships amongst taxa, with a final likelihood value of -19141.848334 (Fig. 3). The matrix contained 1265 distinct alignment patterns, with a 32.87% proportion of gaps and completely undetermined characters. The estimated base frequencies of A = 0.239678, C = 0.253426, G = 0.268914, T = 0.237981; substitution rates AC = 1.751555, AG = 3.051838, AT = 1.900841, CG = 1.359429, CT = 9.411951, GT = 1.000000; gamma distribution shape parameter α = 0.505775; Tree-Length = 1.483987. For BI analysis, GTR + I + G was selected as the best-fit model by AIC in MrModelTest for each gene (ITS, LSU, SSU and tef1-α). Six simultaneous Markov chains were run for 555,000 generations, resulting in 5551 total trees. The first 10% of trees were discarded as the burn-in phase of the analyses and the remaining trees were used for calculating posterior probabilities in the majority rule consensus tree, of which the final average standard deviation of split frequencies at the end of total MCMC generations was 0.009941.
Figure 3.
Phylogram of the best-scoring ML consensus tree of taxa in Periconiaceae and the closely-related families Lentitheciaceae and Massarinaceae. The new isolate is indicated in blue. Isolates from type materials are in bold. The ML ultrafast bootstrap and Bayesian PP values greater than 50% and 0.95 are shown at the nodes.
Multigene phylogenetic analyses demonstrated that the new species Periconiakunmingensis (KUMCC 18-0173) formed a distinct lineage and clustered with the clade containing P.cookei (MFLUCC 17-1679, MFLUCC 17-1399 and UESTCC 22.013), P.delonicis, (MFLUCC 17-2584), P.elaeidis (MFLUCC 17-0087), P.palmicola (MFLUCC 14-0400) and P.verrucosa (MFLUCC 17-2158, Lu40-1, KT1820A and KT1825), with strong statistical support (100% ML, 1.00 PP; Fig. 3).
Taxonomy
. Bambusicolaceae
D.Q. Dai & K.D. Hyde, Fungal Diversity 63: 49 (2013)
896D3838-ABD8-5F94-B86C-28C0447E2920
Index Fungorum: IF804293
Notes.
Bambusicolaceae was first introduced by Hyde et al. (2013) to accommodate Bambusicola with B.massarinia being the type species. Subsequently, another three genera were accommodated in this family viz. Corylicola (Wijesinghe et al. 2020), Leucaenicola (Jayasiri et al. 2019) and Palmiascoma (Liu et al. 2015). Species of these genera have been reported from various hosts, such as Camellia, Corylus, Eucalyptus, Fagaceae sp., Leucaena, Osmanthus and palm and so far, found distributed in China (Sichuan and Yunnan), Italy and Thailand (Liu et al. 2015; Jayasiri et al. 2019; Ariyawansa et al. 2020a, b; Hongsanan et al. 2020; Wijesinghe et al. 2020; Monkai et al. 2021). Members of Bambusicolaceae are mainly saprobes; however, Ariyawansa et al. (2020a, b) reported that species of Leucaenicola associated with leaf spot diseases on Camellia and Osmanthus in Taiwan (China). Bambusicolaceae is a well-studied family, based on morphological characteristics of sexual-asexual morphs and multigene phylogenetic evidence. Recent taxonomic treatment carried out by Hongsanan et al. (2020) revealed that the family belongs to the suborder Massarineae, order Pleosporales of Dothideomycetes, comprising four genera and 25 species (http://www.indexfungorum.org; accessed on 25 May 2023).
. Bambusicola
D.Q. Dai & K.D. Hyde, Cryptog. Mycol. 33(3): 367 (2012)
42DF2EDA-6DEB-58CF-9B43-8CDD5E37978A
Index Fungorum: IF801041
Notes.
Bambusicola was introduced by Dai et al. (2012) to accommodate four saprobic species associated with bamboo, namely B.bambusae, B.irregulispora, B.massarinia and B.splendida. Subsequently, many species were included in the genus which were mainly known as saprobes on different bamboos in terrestrial habitats (Dai et al. 2012, 2015, 2017; Thambugala et al. 2017; Monkai et al. 2021; Phukhamsakda et al. 2022; Yu et al. 2022). However, B.sichuanensis and B.subthailandica were reported as parasites on Phyllostachysheteroclada (Yang et al. 2019). While B.aquatica was reported as a saprobe submerged in freshwater (Dong et al. 2020) and B.ficuum was reported on dead twigs of Ficus (Brahmanage et al. 2020). Bambusicola is morphologically well-studied and appear pleomorphic. Besides, its phylogenetic affinities have been well-clarified, based on multigene phylogenetic evidence (e.g. B.didymospora, B.massarinia, B.triseptatispora) (Dai et al. 2012, 2017). Currently, there are 17 species accepted in the genus, mostly distributed in the Sichuan and Yunnan Provinces of China and Thailand (http://www.indexfungorum.org; accessed on 25 May 2023). In the present study, we introduce a novel species B.hongheensis which was collected from dead bamboo culms in Yunnan, China.
. Bambusicola hongheensis
Phookamsak, Bhat & Hongsanan sp. nov.
EBE6B320-314F-5BB9-9DA5-839369AD3EE3
Index Fungorum: IF900830
Figure 4.
Bambusicolahongheensis (KUN-HKAS 129042, holotype) A the appearance of ascomata on the host surface B vertical section of an ascoma C, D peridia E pseudoparaphyses F, G asci embedded in pseudoparaphyses H–K ascospores L, M ascospores stained in India Ink show a thin mucilaginous sheath surrounding ascospores. Scale bars: 100 μm (B); 20 μm (C–G); 10 μm (H–M).
Etymology.
The specific epithet “hongheensis” refers to the locality, Honghe Hani and Yi Autonomous Prefecture (Yunnan, China), where the holotype was collected.
Holotype.
KUN-HKAS 129042.
Description.
Saprobic on dead culm of bamboo in terrestrial habitats, visible as black, shiny, gnarled on the host surface. Sexual morph: Ascomata 225–350 μm high, 340–590 μm diam., scattered, sometimes forming stroma with a clustered 1–3 ascomata, gregarious, semi-immersed, raised, becoming superficial, dark brown, dome-shaped to subconical or subglobose, glabrous, coriaceous, ostiolate with inconspicuous papilla. Peridium 40–80(–130) μm wide at sides towards the apex, 10–25 μm wide at the base, composed of several layers of small, dark brown pseudoparenchymatous cells, outer layer fused with host cells, arranged in textura angularis to textura globulosa, inner layer composed of 1–3 strata of flattened cells, of textura globulosa to textura prismatica, with thick, palisade-like cells at the sides. Hamathecium composed of 1–3 μm wide, filiform, dense, septate, branched, pseudoparaphyses, anastomosed between and above the asci, embedded in a gelatinous matrix. Asci (58–)70–90(–105)(–119) × 12–15(–17) μm (x̄ = 80.5 × 13.5 μm, SD = ± 13.2 × 1.8, n = 25), 8-spored, bitunicate, fissitunicate, cylindrical-clavate, shortly pedicellate, apically rounded with well-developed ocular chamber. Ascospores 22–26(–30) × 4.5–7 μm (x̄ = 24.6 × 5.4 μm, SD = ± 2.3 × 0.5, n = 30), overlapping 1–3-seriate, hyaline, fusiform, slightly curved, 1-septate, occasionally 2–3-septate, slightly constricted at the septum, the upper cell slightly larger than the lower cell, smooth-walled, surrounded by a thin, indistinct, mucilaginous sheath. Asexual morph: Undetermined.
Distribution.
China (Yunnan).
Specimen examined.
China. Yunnan Province: Honghe Hani and Yi Autonomous Prefecture, Honghe County, rice terraces, on dead culm of bamboo, 26 Jan 2021, R. Phookamsak BN06 (KUN-HKAS 129042, holotype). Notes: As the axenic culture is not active, the sequences of SSU and rpb2 were obtained from genomic DNA extracted from ascomata and dried culture.
Notes.
Based on the NCBI nucleotide BLAST search of ITS sequence, Bambusicolahongheensis (KUN-HKAS 129042) has the closest match with B.triseptatispora (MFLUCC 11-0166, ex-type strain) with 98.71% similarity (Identities = 535/542 with no gap) and is similar to B.loculata (MFLU 15-0056, ex-type strain) with 98.69% similarity (Identities = 528/535 with 1 gap) and B.splendida (MFLUCC 11-0611) with 98.25% similarity (Identities = 392/399 with no gap). The NCBI nucleotide BLAST search of LSU sequence indicated that B.hongheensis has the closest match with B.triseptatispora (MFLUCC 11-0166, ex-type strain) and B.didymospora (MFLUCC 10-0557, ex-type strain) with 100% similarity (Identities = 802/802 with no gap) and is similar to B.loculata (MFLU 15-0056, ex-type strain) with 99.75% similarity (Identities = 813/815 with 2 gaps) and B.nanensis (MFLUCC 21-0063, ex-type strain) with 99.49% similarity (Identities = 785/789 with no gap). The NCBI nucleotide BLAST search of rpb2 sequence indicated that B.hongheensis has the closest match with B.loculata (MFLU 15-0056, ex-type strain) with 99.90% similarity (Identities = 1042/1043 with no gaps) and is also similar to B.triseptatispora (MFLUCC 11-0166, ex-type strain) with 97.92% similarity (Identities = 990/1011 with no gap) and B.massarinia (voucher MFLU 11-0389) with 93.57% similarity (Identities = 975/1042 with 4 gaps).
Phylogenetic analyses of a concatenated ITS, LSU, rpb2, SSU and tef1-α sequence dataset demonstrated that Bambusicolahongheensis formed a separate branch (85% ML, 1.00 PP; Fig. 1), and clustered with B.loculata and B.triseptatispora with high support (100% ML, 1.00 PP; Fig. 1) and also clustered with the generic type of Bambusicola, B.massarinia with significant support (73% ML, 0.99 PP; Fig. 1). A nucleotide pairwise comparison of ITS sequence indicated that B.hongheensis differs from B.triseptatispora in 35/600 bp (5.83%), differs from B.loculata in 16/547 bp (2.92%) and differs from B.massarinia in 72/608 bp (11.84%). Whereas the nucleotide pairwise comparison of LSU sequence indicated that B.hongheensis is consistent with B.triseptatispora (0/802 bp) and B.loculata (1/816 bp), but differs from B.massarinia in 7/803 bp (0.87%). Furthermore, the nucleotide pairwise comparison of rpb2 sequence indicated B.hongheensis is not significantly different from B.loculata (1/1043 bp), but differs from B.triseptatispora in 21/1012 bp (2.07%) and differs from B.massarinia in 68/1042 bp (6.52%).
Morphologically, Bambusicolahongheensis resembles B.loculata and B.triseptatispora in terms of the size range of ascomata, asci and ascospores. However, B.hongheensis has comparatively smaller ascomata (340–590 μm diam. of B.hongheensis vs. 350–600 μm diam. of B.loculata vs. 470–730 μm diam. of B.triseptatispora), shorter and wider asci ((58–)70–90(–105)(–119) × 12–15(–17) μm vs. 80–105 × 8–13 μm vs. (78–)80–100(−110) × 10–12(−14) μm, respectively) and sharing the size range of ascospores (22–26(–30) × 4.5–7 μm vs. 22–26.5 × 5–6 μm vs. (25–)26–30(−31) × 4–6 μm, respectively). The ascospores of B.hongheensis are typically hyaline, 1-septate, whereas B.triseptatispora has hyaline to pale brown and 3-septate ascospores (Dai et al. 2017). Distinguishing B.loculata from B.hongheensis, based on morphological characteristics alone is challenging, but B.loculata can be differentiated by its larger ascomata and asci (Dai et al. 2015). However, a clear differentiation is achieved through phylogenetic evidence (Fig. 2) and nucleotide pairwise comparison of ITS gene region (2.92% difference).
. Dictyosporiaceae
Boonmee & K.D. Hyde, in Boonmee et al., Fungal Diversity: 10.1007/s13225-016-0363-z, [7] (2016)
DEEF0297-BCD6-5176-A27C-B85343343124
Index Fungorum: IF551574
Notes.
Dictyosporiaceae was introduced by Boonmee et al. (2016) to initially accommodate ten genera that were mainly represented by the hyphomycetous asexual morph, forming cheiroid, digitate, palmate and/or dictyosporous conidia. The sexual morph is scarcely known for this family, of which species of genera Dictyosporium, Gregarithecium, Immotthia, Pseudocoleophoma, Sajamaea and Verrucoccum have been represented as the sexual morph (Boonmee et al. 2016; Piątek et al. 2020; Atienza et al. 2021; Jiang et al. 2021a). Members of Dictyosporiaceae are morphologically diverse in various ecological niches, commonly known as saprobes on plant litter in terrestrial and freshwater habitats (Tanaka et al. 2015; Boonmee et al. 2016; Li et al. 2017; Crous et al. 2019; Rajeshkumar et al. 2021; Tian et al. 2022; Tennakoon et al. 2023). Besides, some genera were known as fungicolous (hyperparasites and mycoparasites) and lichenicolous fungi as well as inhabiting soil and herbivore dung (Iturrieta-González et al. 2018; Piątek et al. 2020; Atienza et al. 2021; Jiang et al. 2021a). An updated taxonomic description of Dictyosporiaceae was provided by Hongsanan et al. (2020) who listed 15 genera in this family, while Wijayawardene et al. (2022b) listed 17 genera in Dictyosporiaceae. Tennakoon et al. (2023) provided a backbone tree of Dictyosporiaceae and currently listed 20 genera in this family, namely Aquadictyospora, Aquaticheirospora, Cheirosporium, Dendryphiella, Dictyocheirospora, Dictyopalmispora, Dictyosporium, Digitodesmium, Gregarithecium, Immotthia, Jalapriya, Neodendryphiella, Neodigitodesmium, Pseudocoleophoma, Pseudoconiothyrium, Pseudocyclothyriella, Pseudodictyosporium, Sajamaea, Verrucoccum and Vikalpa.
. Trichobotrys
Penz. & Sacc., Malpighia 15(7–9): 245 (1902) [1901]
2FA25B85-ED64-579E-8EA3-600AA01538B2
Index Fungorum: IF10275
Notes.
Trichobotrys was introduced by Penzig and Saccardo (1902) to accommodate the type species T.pannosus [as ‘pannosa’]. The genus is scarcely known and only five species are available in Index Fungorum (http://www.indexfungorum.org; accessed on 25 May 2023), of which only T.effusus [as ‘effusa’] has molecular data available in GenBank. The genus is known as only a hyphomycetous asexual morph and is characterised by dark brown to black, effuse to velvety colonies, partly immersed to superficial mycelium, non-stromatic, macronematous, mononematous, dark brown to reddish-brown, verruculose or echinulate conidiophores, bearing short, smooth, fertile, often unciform lateral branches, with sterile, setiform apex, polyblastic, integrated, terminal or discrete, determinate, ellipsoidal, spherical or subspherical conidiogenous cells and catenated, in branched acropetal chains, spherical, brown, aseptate, verruculose or minutely echinulate conidia (Ellis 1971; D’Souza and Bhat 2001). The taxonomic classification of the genus is doubtful due to the lack of molecular phylogeny. Recently, Wijayawardene et al. (2022b) treated Trichobotrys as Ascomycota genus incertae sedis, pending future study. In the present study, the novel species, T.sinensis is introduced and the phylogenetic analyses demonstrated the genus affinity in Dictyosporiaceae.
. Trichobotrys sinensis
Phookamsak, Bhat & Hongsanan sp. nov.
49C20070-18C2-5650-87C4-419C204D1167
Index Fungorum: IF900832
Figure 5.
Trichobotryssinensis (KUN-HKAS 129041, holotype) A, B the appearance of colonies on the host surface C mycelium D–H conidiophores bearing conidiogenous cells and conidia I conidia in a short acropetal chain J–N conidia O culture characteristics on PDAP conidioma forming on PDA after eight weeks Q pycnidial wall R–T conidiogenous cells (note: T = stained in Congo red) U conidia. Scale bars: 100 μm (P); 50 μm (C); 10 μm (D–H, Q–U); 5 μm (J–N).
Etymology.
The specific epithet “sinensis” refers to the country, China, where the holotype was collected.
Holotype.
KUN-HKAS 129041.
Description.
Saprobic on dead culm of Brachiariamutica, submerged in a small stream. Sexual morph: Undetermined. Asexual morph: Colonies dull, black, effuse, visible as hairy fluffy on the host. Mycelia up to 1 mm long, 2–4 µm wide, superficial, composed of brown to dark brown, branched, septate, thick-walled, echinulate hyphae. Conidiophores (9–)15–40(–70) × 2–4 µm (x̄ = 26.9 × 3.3 μm, n = 30), sometimes reduced to conidiogenous cells, macronematous, mononematous, straight or flexuous, brown to dark brown, septate, verruculose or echinulate, bearing short, lateral, unciform, fertile branches, with setiform apex. Conidiogenous cells 1–3.5 × 2.5–5 µm (x̄ = 2.1 × 2.5 μm, n = 30), polyblastic, subhyaline to pale brown, ellipsoidal or hemispherical (2.5–5 × 3.5–6 µm), intercalary or terminal, integrated or discrete, sometimes denticulate on branches. Conidia 7–11 × 8–12 µm (x̄ = 10 × 10 μm, n = 30) simple, solitary, brown to dark brown, spherical, aseptate, verruculose; sometimes in short acropetal chains. In vitroConidiomata 280–470 µm high, 280–570 µm diam., black, pycnidial, solitary or clustered in a small group (2–4-loculate), scattered to gregarious, globose to subglobose, glabrous, covered by brown to dark brown mycelium, becoming a packed pycnidial wall, ostiolate, with inconspicuous, minute papilla. Pycnidial wall 20–35 µm wide, thick-walled of unequal thickness, thicker at the base, composed of multi-layered, dark brown to black pseudoparenchymatous cells, outer layers composed of textura intricata, inner layers composed of flattened cells of textura angularis to textura prismatica. Conidiophores reduced to conidiogenous cells. Conidiogenous cells (6.5–)10–16(–25) × 2–4.5 µm (x̄ = 13.4 × 3.2 μm, n = 30), holoblastic to phialidic, hyaline, cylindrical to subcylindrical, terminal or intercalary, septate, smooth-walled, with distinct collarette. Conidia 2–3 × 1.5–2.5 µm (x̄ = 2.8 × 2 μm, n = 30) hyaline, ellipsoidal to ovoid, aseptate, smooth-walled, with a guttulate.
Culture characteristics.
Colonies on PDA reaching 25–28 mm diam. after two weeks at room temperature (20–27 °C), medium dense, circular, surface smooth with an entire edge, flattened, slightly raised, fairly fluffy to feathery; from above, initially white, with cream conidial masses, becoming white to cream at the margin, pale yellowish towards the centre with age; from below, white at the margin, dark grey to black towards the centre; pigmentation not produced in PDA. Sporulation in PDA after two weeks, initially visible as cream conidial masses, later forming black conidiomata with hyaline to cream conidial masses on colonies.
Distribution.
China (Yunnan).
Specimen examined.
China. Yunnan Province: Xishuangbanna Dai Autonomous Prefecture, Mengla County, Bubeng, 21°36'30.13"N, 101°35'52.54"E, 664 + 5 m a.s.l., on culms of Brachiariamutica submerged in a freshwater stream, 27 Apr 2021, R. Phookamsak BB21-007 (KUN-HKAS 129041, holotype), ex-type living culture, RPC 21-007 = KUNCC 23-14554.
Notes.
Based on NCBI nucleotide BLAST search of ITS sequence, the closest hit of Trichobotryssinensis (RPC 21-007/ KUNCC 23-14554) is Gregarithecium sp. DQD-2016a strain MFLUCC 13-0853 with 99.03% similarity (Identities = 508/513 with 2 gaps) and is similar to Trichobotryseffusus [as ‘effusa’] isolate 1179 (93.51% similarity, Identities = 504/539 with 13 gaps), T.effusus [as ‘effusa’] strain FS522 (93.35% similarity, Identities = 477/511 with 12 gaps) and T.effusus [as ‘effusa’] isolate HNNUZCJ-94 (93.08% similarity, Identities = 471/506 with 16 gaps). In LSU nucleotide BLAST search, the closest hit of T.sinensis (RPC 21-007/ KUNCC 23-14554) is Gregarithecium sp. DQD-2016a strain MFLUCC 13-0853 with 99.88% similarity (Identities = 848/849 with 1 gap) and is similar to Gregarithecium sp. isolate L13E (99.40% similarity, Identities = 830/835 with 3 gaps) and G.curvisporum HHUF 30134 (97.74% similarity, Identities = 822/841 with 5 gaps).
Multigene phylogenetic analyses of a concatenated ITS, LSU, SSU and tef1-α sequence dataset demonstrated that Trichobotryssinensis (RPC 21-007/ KUNCC 23-14554) shared a branch length with Gregarithecium sp. DQD-2016a strain MFLUCC 13-0853 and Gregarithecium sp. isolate GMB 1217 and clustered with the clade of T.effusus (Fig. 2). However, Gregarithecium sp. DQD-2016a strain MFLUCC 13-0853 and Gregarithecium sp. isolate GMB 1217 are unpublished strains. Hence, Trichobotryssinensis (RPC 21-007/ KUNCC 23-14554) is introduced herein as a new species and Gregarithecium sp. (strains MFLUCC 13-0853 and isolate GMB 1217) is re-identified as T.sinensis to avoid misidentification. Morphologically, T.sinensis (RPC 21-007/ KUNCC 23-14554) is typical of Trichobotrys, but can be distinguished from T.effusus, T.pannosus, T.ramosus and T.trechisporus in having larger conidia (2 µm diam. of T.effusus vs. 4 µm diam. of T.pannosus vs. 3–5 µm diam. of T.ramosus vs. 5 × 3 µm or 4 µm diam. of T.trechisporus) (Berkeley and Broome 1873; Penzig and Saccardo 1902; Petch 1917; Ellis 1971; D’Souza and Bhat 2001).
. Periconiaceae
Nann., Repert. mic. uomo: 482 (1934)
D0747C5C-7962-5E6C-8223-63898767E14E
Index Fungorum: IF81124
Notes.
Periconiaceae was resurrected by Tanaka et al. (2015) who provided an updated taxonomic treatment and placed the family in the suborder Massarineae (Pleosporales). Tanaka et al. (2015) accepted four genera namely, Periconia (Tode 1791), Noosia (Crous et al. 2011), Bambusistroma (Adamčík et al. 2015) and Flavomyces (Knapp et al. 2015), as well as included Sporidesmiumtengii in the Periconiaceae. Yang et al. (2022b) re-circumscribed genera Bambusistroma, Noosia and Periconia, based on type studies compared with their new findings. Hence, Yang et al. (2022b) treated Bambusistroma and Noosia as synonyms of Periconia due to morphological resemblances and phylogenetic evidence, while the generic status of Flavomyces is doubted pending further studies.
. Periconia
Tode, Fung. mecklenb. sel. (Lüneburg) 2: 2 (1791)
900300E7-F98F-5EFA-AF57-08869540F86F
Index Fungorum: IF9263
Notes.
Periconia was established by Tode (1791) to accommodate dematiaceous hyphomycetes that were unique in forming macronematous, mononematous, branched, septate, pigmented conidiophores, bearing spherical conidial heads that produced globose to ellipsoidal, aseptate, verruculose to echinulate, pigmented conidia (Tanaka et al. 2015; Hongsanan et al. 2020; Yang et al. 2022b). Species of Periconia are typically known by their asexual morph; only a few species have been reported with their sexual morph (Tanaka et al. 2015; Hongsanan et al. 2020; Yang et al. 2022b). Periconia species have been commonly reported as saprobes occurring on various host substrates in terrestrial and aquatic habitats worldwide. However, some species have been reported as endophytes, plant pathogens (e.g. P.circinata, P.digitata and P.macrospinosa) and human pathogens, as well as producing economically-important bioactive compounds (Sarkar et al. 2019; Gunasekaran et al. 2021; Hongsanan et al. 2020; Samarakoon et al. 2021; Azhari and Supratman 2021; Yang et al. 2022b; Su et al. 2023). Even though over 200 species of Periconia were listed in Index Fungorum (http://www.indexfungorum.org; accessed on 25 May 2023), less than half of a quarter have molecular data to clarify phylogenetic placement. Of these, the type species of Periconia, P.lichenoides, also lacks molecular data. This suggests that there is a huge research gap in the taxonomic classification of the genus Periconia. In the present study, we follow the latest taxonomic treatment of Yang et al. (2022b) and Su et al. (2023) and the new species Periconiakunmingensis occurring on fern, is introduced.
. Periconia kunmingensis
Phookamsak & Hongsanan sp. nov.
4E5988EE-BB87-5946-B836-46DC18DBD7C7
Index Fungorum: IF900833
Figure 6.
Periconiakunmingensis (KUN-HKAS102239, holotype) A, B the appearance of fungal colonies on host substrate C–E conidiophores F, G closed-up conidiophores with spherical heads H, I conidiogenous cells bearing conidia J conidia catenate in acropetal short chain K–P conidia. Scale bars: 500 µm (A, B); 50 µm (C–E); 20 µm (F, G); 10 µm (J); 5 µm (H, I, K–P).
Etymology.
The specific epithet “kunmingensis” refers to the Kunming Institute of Botany, Kunming, Yunnan, China, where the holotype was collected.
Holotype.
KUN-HKAS 102239.
Description.
Saprobic on dead, standing rachis of a fern. Sexual morph: Undetermined. Asexual morph: Colonies on the substrates superficial, numerous, effuse, brown to dark brown, floccose. Mycelia 6–7 μm wide, partly superficial, composed of septate, branched, dark brown hyphae. Conidiophores 100–260 μm long, 7–12 μm diam., macronematous, mononematous, solitary, dark brown, 3–5-septate, unbranched below, branched only at the apex, erect, straight or slightly flexuous, sometimes swollen near the base, with 1–2 spherical guttules in each cell, forming a spherical head at the tip. Conidiogenous cells (4–)5–8(–10) × 2.5–5(–6) μm (x̄ = 6.4 × 4 μm, n = 30) mono- to polyblastic, terminal, discrete, subspherical to fusiform, subhyaline to pale brown, verruculose. Conidia 4.5–7(–9) × 4–7(–8) μm (x̄ = 6 × 5.9 μm, n = 50), solitary to catenate, in acropetal short chains, subglobose to globose, subhyaline to pale brown, aseptate, minutely verruculose to short-spinulose.
Culture characteristics.
Colonies on PDA reaching 23–25 mm diam. after two weeks at room temperature (20–30 °C). Colony dense, circular, flattened, slightly raised, surface smooth, edge fimbriate, velvety, with fairly fluffy at the margin; colony from above, white to white-grey, separated from the centre by greenish-grey radiating near the margin; colony from below, pale yellowish to cream at the margin, deep green near the margin, with dark green concentric ring, separating the margin from greenish-grey to dark green centre; slightly produced light yellowish pigment tinted agar.
Distribution.
China (Yunnan).
Specimen examined.
China. Yunnan Province: Kunming, Kunming Institute of Botany, on dead, standing rachis of a fern, 23 Sep 2016, R. Phookamsak KIB004 (KUN-HKAS 102239, holotype), ex-type living culture RPC 15-017 = KUMCC 18-0173 = MFLUCC 18-0679. Addition GenBank no: rpb2 = OR547996.
Notes.
Based on the NCBI nucleotide BLAST search of ITS sequence, the closest hits of Periconiakunmingensis are Periconia sp. strain 8R5B1-3 and Periconia sp. isolate LS77 with 99.80% similarity (Identities = 507/508 and 498/499 with no gap, respectively) and is similar to P.verrucosa isolate HNNU0545 with 99.60% similarity (Identities = 502/504 with 1 gap), Periconia sp. strain MFLUCC 17-0087 with 99.59% similarity (Identities = 482/484 with 1 gap) and P.elaeidis isolate PT49 with 99.57% similarity (Identities = 464/466 with 1 gap). In the LSU nucleotide BLAST search, P.kunmingensis is similar to P.verrucosa isolate MFLUCC 17-2158 (Identities = 847/847 with no gap), Periconia sp. KT 1825 (Identities = 843/843 with no gap), P.elaeidis strain GZCC19-0435 (Identities = 842/842 with no gap), P.cookei strain IHEM:28143 (Identities = 826/826 with no gap), Pleosporales sp. A1039 (Identities = 815/815 with no gap) and P.verrucosa isolate w232_2 (Identities = 812/812 with no gap), isolate Lu53_1 (Identities = 807/807 with no gap) and isolate Lu98_2 (Identities = 796/796 with no gap), with 100% similarities. In the tef1-α nucleotide BLAST search, the closest hit of P.kunmingensis is Periconia sp. KT 1820A (Identities = 745/747 with no gap) and P.delonicis voucher MFLU 20-0696 (Identities = 736/738 with no gap) with 99.73% similarity. Periconiakunmingensis is also similar to P.delonicis strain MFLUCC 17-2584 and P.verrucosa isolate MFLUCC 17-2158 with 99.60% similarity (Identities = 744/747 with no gap).
Phylogenetic analyses of the concatenated ITS, LSU, SSU and tef1-α sequence data showed that Periconiakunmingensis formed a distinct branch basally to P.verrucosa, P.cookei, P.palmicola, P.elaeidis and P.delonicis, respectively (Fig. 3). The ITS nucleotide pairwise comparison indicated that P.kunmingensis differs from P.verrucosa (MFLUCC 17–2158, ex-type strain) in 3/512 bp (0.59%), differs from P.cookei in 2/465 bp (0.43%) of MFLUCC 17–1399 and 3/465 bp (0.65%) of UESTCC 22.0134 and differs from P.elaeidis (MFLUCC 17–0087, ex-type strain) in 14/518 bp (2.70%). The rpb2 nucleotide pairwise comparison indicated that P.kunmingensis differs from P.verrucosa (UESTCC 22.0136) in 35/849 bp (4.12%), differs from P.cookei (UESTCC 22.0134) in 30/819 bp (3.66%) and differs from P.delonicis (MFLUCC 17–2584, ex-type strain) in 54/1073 bp (5.03%). The tef1-α nucleotide pairwise comparison indicated that P.kunmingensis differs from P.verrucosa (MFLUCC 17–2158, ex-type strain) in 108/929 bp (11.63%), differs from P.cookei in 4/736 bp (0.54%) of MFLUCC 17-1399 and 107/906 bp (11.81%) of MFLUCC 17-1679, differs from P.palmicola (MFLUCC 14-0400, ex-type strain) in 19/991 bp (1.92%) and differs from P.delonicis (MFLUCC 17–2584, ex-type strain) in 105/987 bp (10.64%).
Distinguishing Periconiakunmingensis from other Periconia species, based on morphological features alone, presents challenges. However, differentiation can be achieved by considering variations in the sizes of conidiophores, conidiogenous cells and conidia, as well as the number of conidiophores originating from the stromatic, swollen part of the conidiophores, septation characteristics and the occurrence and origin of the host. A comprehensive morphological comparison is provided in Table 4.
Table 4.
Morphological comparison of Periconiakunmingensis with other related species. A novel species is indicated by black bold.
| Species | Conidiophores | Conidiogenous cells | Conidia | Host occurrence | Origin | Reference |
|---|---|---|---|---|---|---|
| Periconiacookei | 360–800 µm high, singly or in groups (up to six), 2–6-septate, swollen at the apex, dark brown at the lower part, pale brown at the upper part | 7–11 µm diam., spherical, ovoid or pyriform, initially hyaline, smooth-walled, becoming brown, verrucose on age | 13–16 µm diam., with the wall up to 2 µm thick, spherical, brown, verrucose, singly or in short chains of 2–3 on conidiogenous cells | On stems of Heracleumsphondylium | Great Britain | Mason and Ellis (1953) |
| (IMI 16174, holotype) | ||||||
| Periconiadelonicis | 360−420 μm high, singly, septate, greyish-brown to dark brown, unbranched, smooth to minutely verruculose | Monoblastic, proliferating, ovoid to globose, hyaline | 5.5−7 μm diam., subglobose to globose, subhyaline to pale brown, verruculose, singly or in short chains | On pods of Delonixregia | Thailand | Jayasiri et al. (2019) |
| (MFLU 18−2100, holotype) | ||||||
| Periconiaelaeidis | 200−400 μm high, singly, 4−7-septate, grayish-brown to dark brown, unbranched, smooth to minutely verruculose | Polyblastic, proliferating, ovoid to globose, pale brown, smooth | 4.5−6.5 μm diam., subglobose to globose, subhyaline to pale brown, verruculose, solitary | On dead leaves of oil palm | Thailand | Hyde et al. (2018) |
| (MFLU 18−0626, holotype) | ||||||
| Periconiakunmingensis (KUN-HKAS 102239, holotype) | 100–260 μm high, solitary, 3–5-septate, dark brown, unbranched below, branched only at the apex, sometimes swollen near the base | (4–)5–8(–10) × 2.5–5(–6) μm, mono- to polyblastic, subspherical to fusiform, subhyaline to pale brown, verruculose | 4.5–7(–9) × 4–7(–8) μm, subglobose to globose, subhyaline to pale brown, minutely verruculose to short-spinulose, solitary to catenate, in acropetal short chains | On dead standing rachis of a fern | Yunnan, China | This study |
| Periconiapalmicola (MFLU 14-0198, holotype) | 151–188 μm high, singly or in groups, septate, dark brown to black, branched at the apex | 3–3.5 × 3–4.8 μm, mono- to polyblastic, globose, hyaline to subhyaline | 5.1–7.4 × 4.8–6.1 μm, subglobose to globose, light brown to brown, verruculose, solitary to catenate, in acropetal short chains | On dead, fallen leaves of unidentified palm | Thailand | Hyde et al. (2020) |
| Periconiaverrucosa (MFLU 17–1516, holotype) | 170–296 µm high, singly, 2–4-septate, dark brown, with 3–4 short branches at the apex | 11–26 × 6–14 μm, mono- to polyblastic, retrogressive, oblong, pale brown | 7–15 μm diam., globose, dark brown to reddish-brown, verrucose, acrogenous in branched chains | On dead stems of Clematisviticella | Belgium | Phukhamsakda et al. (2020) |
Discussion
This paper, in the series “Exploring ascomycete diversity in Yunnan”, presents three novel taxa in the suborder Massarineae (Pleosporales), viz. Bambusicolahongheensis (Bambusicolaceae), Periconiakunmingensis (Periconiaceae) and Trichobotryssinensis (Dictyosporiaceae). The novelties of these taxa were well-justified, based on morphological characteristics and phylogenetic evidence, as well as the differences in nucleotide pairwise comparison of reliable genes amongst closely-related taxa. This provides a better fundamental knowledge of the taxonomic framework of ascomycetes in this region.
Bambusicolahongheensis is justified, based on multigene phylogeny and the differences in nucleotide pairwise comparison of the ITS region with closely-related species. Monkai et al. (2021) mentioned that many Bambusicola species have similar morphology, but these species can be distinguished, based on multigene phylogeny and they also recommended the use of the rpb2 gene for delineating species level of Bambusicola. Unfortunately, the rpb2 sequence did not distinguish B.hongheensis from B.loculata in the present study; however, the ITS region of B.hongheensis showed > 1.5% nucleotide differences amongst the closely-related species viz. B.loculata, B.massarinia and B.triseptatispora. This provides adequate justification for the species’ novelty following the recommendation of Jeewon and Hyde (2016).
Although many Bambusicola species are morphologically somewhat similar, it is notable that they can also be distinguished by their represented asexual morphs that are easily sporulated in vitro as well as on natural substrates. For instance, coelomycetous asexual morphs of B.massarinia and B.triseptatispora sporulated in vitro; of which B.massarinia can be distinguished from B.triseptatispora in having pale brown, 1-septate, cylindrical conidia (Dai et al. 2012). Whereas conidia of B.triseptatispora are light brown, 3-septate, cylindrical to cylindrical-clavate (Dai et al. 2017). Unfortunately, the asexual morphs of B.hongheensis and B.loculata have not yet been determined. Hence, further studies on their asexual morphs sporulated in vitro should be carried out for a better understanding through their sexual-asexual reproduction, as well as gaining criteria of species delineation.
Trichobotryssinensis is morphologically typical of Trichobotrys. Trichobotrys was previously classified into Ascomycota genus incertae sedis (Wijayawardene et al. 2022b). Although the sequence data of the type species of Trichobotrys is currently unavailable, the inclusion of available sequence data along with our new species that morphologically align well with Trichobotrys in the phylogenetic analyses, provides compelling evidence supporting the placement of Trichobotrys within the Dictyosporiaceae. This information contributes to our understanding of taxonomic relationships and highlights the need for further studies to explore the molecular characteristics and genetic diversity of Trichobotrys species within the Dictyosporiaceae.
Synanamorph is the term of use for fungal taxa producing two or more different asexual morphs which were often linked by the sporulation in culture (Wijayawardene et al. 2021a, 2022c). Many fungal taxa have been reported for their synanamorphism, such as Botryosphaeria with dichomera-like in vitro and Neofusicoccum (as Fusicoccum) (Barber et al. 2005), Barbatosphaeriafagi (≡ Calosphaeriafagi) with ramichloridium-like and sporothrix-like asexual morphs (Réblová et al. 2015) and Synnemasporella with sporodochial and pycnidial asexual morphs on natural hosts (Fan et al. 2018). The formation of two or more different morphs in a single species has led to misidentification and the distinct morphs have been somehow counted as different species (Wijayawardene et al. 2021a, 2022c). It has further caused problems in the dual nomenclature of pleomorphic fungi that proposed one name for one fungus (McNeill and Turland 2012; Rossman et al. 2015). Interestingly, Trichobotryssinensis formed two different asexual morphs, one in nature (as Trichobotrys) and another in vitro (pycnidial coelomycetous asexual morph) which is the first report of the synanamorphism for the genus Trichobotrys. This new finding provides insight into pleomorphism which is essential in further revision of taxonomic boundaries and easing of existing complications. It is noteworthy that Trichobotrys formed a well-resolved clade with Gregarithecium in the present phylogenetic analyses. Unfortunately, the sexual morph of Trichobotrys has not yet been determined. Similarly, the asexual morph of Gregarithecium has also not yet been reported. Hence, the sexual-asexual connection between Gregarithecium and Trichobotrys is doubtful pending future study.
Periconiakunmingensis is introduced in this paper, based on its morphology and phylogeny. Morphologically, P.kunmingensis fits well with the generic concept of Periconia and its phylogenetic affinity is also well-clarified within Periconiaceae. It is noteworthy that the ITS region could not be used to separate P.kunmingensis from other closely-related species, including P.cookei and P.verrucosa, based on the nucleotide pairwise comparison. Whereas, the ITS sequences of P.delonicis, P.elaeidis and P.palmicola are unavailable. The interspecific variation amongst these species may be questionable. However, the rpb2, and tef1-α gene regions which have sufficient genetic variation can be used to distinguish these species. Nevertheless, the rpb2 gene of most Periconia species is unavailable. Therefore, the sequences of protein-coding genes (e.g. rpb2 and tef1-α) are acquired to offer reliable phylogenetic markers for species delineation.
Over the past five years, the number of newly-described fungal species has been rapidly increasing in Yunnan. Several novel and interesting ascomycetes were described and illustrated from various host plants and on different substrates and habitats. Many studies of ascomycetous taxonomy on specific host substrates have become essential and challenging for mycologists across the region. For instance, D.N. Wanasinghe and his colleagues (2018–2022) carried out research studies on fungal biogeography and published over 40 novel taxa of wood-inhabiting fungi, as well as other substrates in this region (Bao et al. 2019; Wanasinghe et al. 2020, 2021; Yasanthika et al. 2020; Mortimer et al. 2021; Ren et al. 2021; Wijayawardene et al. 2022a; Maharachchikumbura et al. 2022). Simultaneously, S. Tibpromma and her colleagues (2018–2022) have also carried out research studies of fungal taxonomy and diversity on various host plants, such as agarwood, coffee, Pandanus, para rubber and tea plants. They introduced 20 novel taxa from Pandanus (Tibpromma et al. 2018), while taxonomic studies on the other plants (approximately 45 novel species on agarwood, coffee and para rubber) are pending (S. Tibpromma, personal data information). A comprehensive study of freshwater Sordariomycetes in Yunnan has been carried out by Luo et al. (2018a, b, 2019) who introduced more than 50 novel taxa and reported more than 75 freshwater Sordariomycetes species in Yunnan. Even though these studies unravelled a substantial number of ascomycetes in Yunnan, there is still a huge gap of knowledge in hitherto undescribed novel taxa in this region. If considering only the plant and fungal ratio, many of the so far fungal taxonomic studies on land plants have underestimated these in Yunnan, especially on those economic and horticulture plants. Hence, the inventory of ascomycetes on these land plants will be interesting in further research studies.
Conclusion
In conclusion, this study introduces three novel species in the suborder Massarineae (Pleosporales): Bambusicolahongheensis, Periconiakunmingensis and Trichobotryssinensis. These species were found as saprobes in different habitats, with B.hongheensis and P.kunmingensis occurring in terrestrial environments, while T.sinensis was discovered in a freshwater stream. Notably, the presence of Trichobotrys in a freshwater habitat is a significant finding, as it aligns with other aquatic lignicolous species within the family Dictyosporiaceae. The novelty of B.hongheensis is supported by multigene phylogeny and nucleotide pairwise comparison, although further genetic analysis is recommended. Differentiation between Bambusicola species can also be achieved through the examination of their asexual morphs. Trichobotryssinensis, morphologically typical of Trichobotrys, is phylogenetically placed within Dictyosporiaceae and highlights the need for additional studies on molecular characteristics and genetic diversity within the genus. The observation of synanamorphism in T.sinensis adds complexity to its morphological identification and taxonomic boundaries. The introduction of Periconiakunmingensis is supported by its morphology and phylogenetic affinity within the family Periconiaceae, although the use of protein-coding genes is recommended for reliable species delineation. This study contributes to our understanding of ascomycete diversity in Yunnan and emphasises the importance of such investigations to enhance our knowledge of newly-discovered taxa.
Supplementary Material
Acknowledgements
We express our sincere thanks to the Biology Experimental Center, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences for providing the facilities of the molecular work. We are also grateful to Dr. Shaun Pennycook at Manaaki Whenua – Landcare Research for his kind advice in naming the new fungal species. Chun-Fang Liao and Diana Sandamali at Mae Fah Luang University, Thailand are also thanked for helping with fungal isolation and molecular analyses. Dr. Hongbo Jiang and Qinxian Li at Kunming Institute of Botany, Chinese Academy of Sciences are thanked for their general assistance. Sinang Hongsanan would like to thank Chiang Mai University, Thailand and Shenzhen University, China, for supporting the works. Rungtiwa Phookamsak thanks Chiang Mai University and the Yunnan Revitalization Talent Support Program “Young Talent” Project (grant no. YNWR-QNBJ-2020-120) for financial research support. D. Jayarama Bhat and Turki M. Dawoud gratefully acknowledge the financial support provided under the Distinguished Scientist Fellowship Programme (DSFP), at King Saud University, Riyadh, Saudi Arabia.
Citation
Phookamsak R, Hongsanan S, Bhat DJ, Wanasinghe DN, Promputtha I, Suwannarach N, Kumla J, Xie N, Dawoud TM, Mortimer PE, Xu J, Lumyong S (2024) Exploring ascomycete diversity in Yunnan II: Introducing three novel species in the suborder Massarineae (Dothideomycetes, Pleosporales) from fern and grasses. In: Wijayawardene N, Karunarathna S, Fan X-L, Li Q-R (Eds) Taxonomy and secondary metabolites of wood-associated fungi. MycoKeys 104: 9–50. https://doi.org/10.3897/mycokeys.104.112149
Funding Statement
Chiang Mai University (Post–Doctoral Fellowship 2022; grant no. R000031743) The Project on Key Technology for Ecological Restoration and Green Development in Tropical Dry-Hot Valley, under the Yunnan Department of Sciences and Technology of China (grant no: 202202AE090091).
Contributor Information
Jianchu Xu, Email: jxu@mail.kib.ac.cn.
Saisamorn Lumyong, Email: scboi009@gmail.com.
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
The research was supported by Post–Doctoral Fellowship 2022 for Chiang Mai University (grant no. R000031743). This research study is also supported by the Project on Key Technology for Ecological Restoration and Green Development in Tropical Dry-Hot Valley, under the Yunnan Department of Sciences and Technology of China (grant no: 202302AE090023) and Smart Yunnan Project (Young Scientists) under project code E13K281261.
Author contributions
Conceptualisation: RP, SH. Data curation: RP. Formal analysis: DNW, SH. Funding acquisition: IP, JX, NX, SL, TMD. Investigation: DJB, DNW, RP, SH. Methodology: DNW, RP, SH. Project administration: RP, NS, JK. Supervision: SL, JX, IP, NX, PEM.: Writing – original draft: RP, SH, DNW. Writing – review and editing: DJB, NS, IP, JK, JX, NX, PEM, TMD.
Author ORCIDs
Rungtiwa Phookamsak https://orcid.org/0000-0002-6321-8416
Sinang Hongsanan https://orcid.org/0000-0003-0550-3152
Darbhe Jayarama Bhat http://orcid.org/0000-0002-3800-5910
Dhanushka N. Wanasinghe https://orcid.org/0000-0003-1759-3933
Itthayakorn Promputtha https://orcid.org/0000-0003-3376-4376
Nakarin Suwannarach https://orcid.org/0000-0002-2653-1913
Jaturong Kumla https://orcid.org/0000-0002-3673-6541
Ning Xie https://orcid.org/0000-0002-5866-8535
Turki M. Dawoud http://orchid.org/0000-0002-1444-4185
Peter E. Mortimer https://orcid.org/0000-0002-8507-7407
Jianchu Xu https://orcid.org/0000-0002-2485-2254
Saisamorn Lumyong https://orcid.org/0000-0002-6485-414X
Data availability
All of the data that support the findings of this study are available in the main text.
References
- Adamčík S, Cai L, Chakraborty D, Chen XH, Cotter HVT, Dai DQ, Dai YC, Das K, Deng C, Ghobad-Nejhad M, Hyde KD, Langer E, Latha KPD, Liu F, Liu SL, Liu T, Wei LV, Shu-Xia LV, Machado AR, Pinho DB, Pereira OL, Prasher IB, Rosado AWC, Qin J, Qin WM, Verma RK, Wang Q, Yang ZL, Yu XD, Zhou LW, Buyck B. (2015) Fungal biodiversity profiles 1–10. Cryptogamie. Mycologie 36(2): 121–166. 10.7872/crym/v36.iss2.2015.121 [DOI] [Google Scholar]
- Ariyawansa HA, Tanaka K, Thambugala KM, Phookamsak R, Tian Q, Camporesi E, Hongsanan S, Monkai J, Wanasinghe DN, Mapook A, Chukeatirote E, Kang JC, Xu JC, McKenzie EHC, Jones EBG, Hyde KD. (2014) A molecular phylogenetic reappraisal of the Didymosphaeriaceae (=Montagnulaceae). Fungal Diversity 68(1): 69–104. 10.1007/s13225-014-0305-6 [DOI] [Google Scholar]
- Ariyawansa HA, Phukhamsakda C, Thambugala KM, Bulgakov TS, Wanasinghe DN, Perera RH, Mapook A, Camporesi E, Kang JC, Jones EBG, Bahkali AH, Jayasiri SC, Hyde KD, Liu ZY, Bhat DJ. (2015a) Revision and phylogeny of Leptosphaeriaceae. Fungal Diversity 74(1): 19–51. 10.1007/s13225-015-0349-2 [DOI] [Google Scholar]
- Ariyawansa HA, Thambugala KM, Manamgoda DS, Jayawardena RS, Camporesi E, Boonmee S, Wanasinghe DN, Phookamsak R, Hongsanan S, Singtripop C, Chukeatirote E, Kang JC, Jones EBG, Hyde KD. (2015b) Towards a natural classification and backbone tree for Pleosporaceae. Fungal Diversity 71(1): 85–139. 10.1007/s13225-015-0323-z [DOI] [Google Scholar]
- Ariyawansa HA, Tsai I, Hozzein WN, Thambugala KM. (2020a) Leucaenicolaosmanthi sp. nov. (Bambusicolaceae, Pleosporales), causing leaf spot of Osmanthusfragrans in Taiwan. Phytotaxa 437: 23–31. 10.11646/phytotaxa.437.1.3 [DOI] [Google Scholar]
- Ariyawansa HA, Tsai I, Thambugala KM, Chuang WY, Lin SR, Hozzein WN, Cheewangkoon R. (2020b) Species diversity of Pleosporalean taxa associated with Camelliasinensis (L.) Kuntze in Taiwan. Scientific Reports 10(1): e12762. 10.1038/s41598-020-69718-0 [DOI] [PMC free article] [PubMed]
- Atienza V, Hawksworth DL, Pérez-Ortega S. (2021) Verrucoccum (Dothideomycetes, Dictyosporiaceae), a new genus of lichenicolous fungi on Lobarias. lat. for the Dothideahymeniicola species complex. Mycologia 113: 1233–1252. 10.1080/00275514.2021.1966281 [DOI] [PubMed] [Google Scholar]
- Azhari A, Supratman U. (2021) The chemistry and pharmacology of fungal genus Periconia: A review. Scientia Pharmaceutica 89(3): 1–34. 10.3390/scipharm89030034 [DOI] [Google Scholar]
- Backes A, Guerriero G, Ait Barka E, Jacquard C. (2021) Pyrenophorateres: Taxonomy, morphology, interaction with barley, and mode of control. Frontiers in Plant Science 12: e614951. 10.3389/fpls.2021.614951 [DOI] [PMC free article] [PubMed]
- Bao DF, Wanasinghe DN, Luo ZL, Mortimer PE, Kumar V, Su HY, Hyde KD. (2019) Murisporaaquatica sp. nov. and Murisporafagicola, a new record from freshwater habitat in China. Phytotaxa 416(1): 1–13. 10.11646/phytotaxa.416.1.1 [DOI] [Google Scholar]
- Barber PA, Burgess TJ, Hardy G, Slippers B, Keane PJ, Wingfield MJ. (2005) Botryosphaeria species from Eucalyptus in Australia are pleoanamorphic, producing Dichomera synanamorphs in culture. Mycological Research 109: 1347–1363. 10.1017/S0953756205003989 [DOI] [PubMed] [Google Scholar]
- Barr ME. (1987) Prodromus to Class Loculoascomycetes. University of Massachusetts, Amherst, 67 pp. [Google Scholar]
- Bartosiak SF, Arseniuk E, Szechyńska-Hebda M, Bartosiak E. (2021) Monitoring of natural occurrence and severity of leaf and glume blotch diseases of winter wheat and winter triticale incited by necrotrophic fungi Parastagonospora spp. and Zymoseptoriatritici. Agronomy 11(5): e967. 10.3390/agronomy11050967 [DOI]
- Berkeley MJ, Broome CE. (1873) Enumeration of the Fungi of Ceylon. Part II., containing the remainder of the Hymenomycetes, with the remaining established tribes of Fungi (Continued.). The Journal of the Linnean Society. Botany 14(74): 65–140. 10.1111/j.1095-8339.1873.tb00302.x [DOI] [Google Scholar]
- Bhunjun CS, Dong Y, Jayawardena RS, Jeewon R, Phukhamsakda C, Bundhun D, Hyde KD, Sheng J. (2020) A polyphasic approach to delineate species in Bipolaris. Fungal Diversity 102(1): 225–256. 10.1007/s13225-020-00446-6 [DOI] [Google Scholar]
- Boonmee S, D’souza MJ, Luo Z, Pinruan U, Tanaka K, Su H, Bhat JD, McKenzie EHC, Jones EBG, Taylor JE, Phillips AJL, Hirayama K, Eungwanichayapant PD, Hyde KD. (2016) Dictyosporiaceae fam. nov. Fungal Diversity 80(1): 457–482. 10.1007/s13225-016-0363-z [DOI] [Google Scholar]
- Brahmanage RS, Dayarathne MC, Wanasinghe DN, Thambugala KM, Jeewon R, Chethana KWT, Samarakoon MC, Tennakoon DS, De Silva NI, Camporesi E, Raza M, Yan JY, Hyde KD. (2020) Taxonomic novelties of saprobic Pleosporales from selected dicotyledons and grasses. Mycosphere 11(1): 2481–2541. 10.5943/mycosphere/11/1/15 [DOI] [Google Scholar]
- Cai J, Yu WB, Zhang T, Wang H, Li DZ. (2019) China’s biodiversity hotspots revisited: A treasure chest for plants. PhytoKeys 130: 1–24. 10.3897/phytokeys.130.38417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Jiang JR, Zhang GZ, Cai L, Crous PW. (2015) Resolving the Phomaenigma. Studies in Mycology 82(1): 137–217. 10.1016/j.simyco.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Hou LW, Duan WJ, Crous PW, Cai L. (2017) Didymellaceae revisited. Studies in Mycology 87(1): 105–159. 10.1016/j.simyco.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG, Edwards J, Seifert KA, Alfenas AC, Alfenas RF, Burgess TI, Carnegie AJ, Hardy GESt J, Hiscock N, Hüberli D, Jung T, Louis-Seize G, Okada G, Pereira OL, Stukely MJC, Wang W, White GP, Young AJ, McTaggart AR, Pascoe IG, Porter IJ, Quaedvlieg W. (2011) Fungal Planet description sheets: 69–91. Persoonia – Molecular Phylogeny and Evolution of Fungi 26: 108–156. 10.3767/003158511X581723 [DOI] [Google Scholar]
- Crous PW, Schumacher RK, Akulov A, Thangavel R, Hernández-Restrepo M, Carnegie AJ, Cheewangkoon R, Wingfield MJ, Summerell BA, Quaedvlieg W, Coutinho TA, Roux J, Wood AR, Giraldo A, Groenewald JZ. (2019) New and Interesting fungi. 2. Fungal Systematics and Evolution 3(1): 57–134. 10.3114/fuse.2019.03.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza MA, Bhat DJ. (2001) A new species of Trichobotrys from the Western Ghat forests, India. Mycotaxon 80: 105–108. [Google Scholar]
- Dai DQ, Bhat DJ, Liu JK, Chukeatirote E, Zhao RL, Hyde KD. (2012) Bambusicola, a new genus from bamboo with asexual and sexual morphs. Cryptogamie. Mycologie 33(3): 363–379. 10.7872/crym.v33.iss3.2012.363 [DOI] [Google Scholar]
- Dai DQ, Bahkali AH, Li WJ, Bhat DJ, Zhao RL, Hyde KD. (2015) Bambusicolaloculata sp. nov. (Bambusicolaceae) from bamboo. Phytotaxa 213(2): 122–130. 10.11646/phytotaxa.213.2.5 [DOI] [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2017) Bambusicolous fungi. Fungal Diversity 82(1): 1–105. 10.1007/s13225-016-0367-8 [DOI] [Google Scholar]
- Dai DQ, Wijayawardene NN, Dayarathne MC, Kumla J, Han LS, Zhang GQ, Zhang X, Zhang TT, Chen HH. (2022) Taxonomic and phylogenetic characterizations reveal four new species, two new asexual morph reports, and six new country records of bambusicolous Roussoella from China. Journal of Fungi 8(5): e532. 10.3390/jof8050532 [DOI] [PMC free article] [PubMed]
- Dong W, Wang B, Hyde KD, McKenzie EHC, Raja HA, Tanaka K, Abdel-Wahab MA, Abdel-Aziz FA, Doilom M, Phookamsak R, Hongsanan S, Wanasinghe DN, Yu XD, Wang GN, Yang H, Yang J, Thambugala KM, Tian Q, Luo ZL, Yang JB, Miller AN, Fournier J, Boonmee S, Hu DM, Nalumpang S, Zhang H. (2020) Freshwater Dothideomycetes. Fungal Diversity 105(1): 319–575. 10.1007/s13225-020-00463-5 [DOI] [Google Scholar]
- Dong QY, Wang Y, Wang ZQ, Tang DX, Zhao ZY, Wu HJ, Yu H. (2022) Morphology and phylogeny reveal five novel species in the genus Cordyceps (Cordycipitaceae, Hypocreales) from Yunnan, China. Frontiers in Microbiology 13: e846909. 10.3389/fmicb.2022.846909 [DOI] [PMC free article] [PubMed]
- El-Demerdash A. (2018) Chemical diversity and biological activities of Phaeosphaeria fungi genus: A systematic review. Journal of Fungi 4(4): e130. 10.3390/jof4040130 [DOI] [PMC free article] [PubMed]
- Ellis MB. (1971) Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Key Surrey, Kew, 608 pp. [Google Scholar]
- Fan XL, Bezerra JD, Tian CM, Crous PW. (2018) Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Persoonia – Molecular Phylogeny and Evolution of Fungi 40: 119–134. 10.3767/persoonia.2018.40.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Wang YB, Zhang GD, Tang DX, Yu H. (2021) Multigene phylogeny and morphology of Ophiocordycepsalboperitheciata sp. nov., a new entomopathogenic fungus attacking Lepidopteran larva from Yunnan, China. Mycobiology 49(2): 133–141. 10.1080/12298093.2021.1903130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Yang Z. (2018) Studies on diversity of higher fungi in Yunnan, southwestern China: A review. Plant Diversity 40(4): 165–171. 10.1016/j.pld.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu R, Bao DF, Shen HW, Su XJ, Li YX, Luo ZL. (2022) Endophytic Pestalotiopsis species associated with Rhododendron in Cangshan Mountain, Yunnan Province, China. Frontiers in Microbiology 13: e1016782. 10.3389/fmicb.2022.1016782 [DOI] [PMC free article] [PubMed]
- Gunasekaran R, Janakiraman D, Rajapandian SGK, Appavu SP, Venkatesh PN, Prajna L. (2021) Periconia species-An unusual fungal pathogen causing mycotic keratitis. Indian Journal of Medical Microbiology 39: 36–40. 10.1016/j.ijmmb.2020.10.006 [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: A user–friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hashimoto A, Matsumura M, Hirayama K, Tanaka K. (2017) Revision of Lophiotremataceae (Pleosporales, Dothideomycetes): Aquasubmersaceae, Cryptocoryneaceae, and Hermatomycetaceae fam. nov. Persoonia – Molecular Phylogeny and Evolution of Fungi 39: 51–73. 10.3767/persoonia.2017.39.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL. (2001) The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycological Research 105(12): 1422–1432. 10.1017/S0953756201004725 [DOI] [Google Scholar]
- Hongsanan S, Hyde KD, Phookamsak R, Wanasinghe DN, McKenzie EHC, Sarma VV, Boonmee S, Lücking R, Pem D, Bhat DJ, Liu NG, Tennakoon DS, Anuruddha Karunarathna A, Jiang SH, Jones EBG, Phillips AJL, Manawasinghe IS, Tibpromma S, Jayasiri SC, Sandamali D, Jayawardena RS, Wijayawardene NN, Ekanayaka AH, Jeewon R, Lu YZ, Dissanayake AJ, Zeng XY, Luo ZL, Tian Q, Phukhamsakda C, Thambugala KM, Dai DQ, Chethana KWT, Ertz D, Doilom M, Liu JK. (2020) Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 11(1): 1553–2107. 10.5943/mycosphere/11/1/13 [DOI] [Google Scholar]
- Hyde KD, Jones EBG, Liu JK, Ariyawansa HA, Boehm E, Boonmee S, Braun U, Chomnunti P, Crous PW, Dai DQ, Diederich P, Dissanayake A, Doilom M, Doveri F, Hongsanan S, Jayawardena R, Lawrey JD, Li YM, Liu YX, Lücking R, Monkai J, Muggia L, Nelsen MP, Pang KL, Phookamsak R, Senanayake IC, Shearer CA, Suetrong S, Tanaka K, Thambugala KM, Wijayawardene NN, Wikee S, Wu HX, Zhang Y, Aguirre-Hudson B, Alias SA, Aptroot A, Bahkali AH, Bezerra JL, Bhat DJ, Camporesi E, Chukeatirote E, Gueidan C, Hawksworth DL, Hirayama K, De Hoog S, Kang JC, Knudsen K, Li WJ, Li XH, Liu ZY, Mapook A, McKenzie EHC, Miller AN, Mortimer PE, Phillips AJL, Raja HA, Scheuer C, Schumm F, Taylor JE, Tian Q, Tibpromma S, Wanasinghe DN, Wang Y, Xu JC, Yacharoen S, Yan JY, Zhang M. (2013) Families of Dothideomycetes. Fungal Diversity 63(1): 1–313. 10.1007/s13225-013-0263-4 [DOI] [Google Scholar]
- Hyde KD, Chaiwan N, Norphanphoun C, Boonmee S, Camporesi E, Chethana KWT, Dayarathne MC, de Silva NI, Dissanayake AJ, Ekanayaka AH, Hongsanan S, Huang SK, Jayasiri SC, Jayawardena RS, Jiang HB, Karunarathna A, Lin CG, Liu JK, Liu NG, Lu YZ, Luo ZL, Maharachchimbura SSN, Manawasinghe IS, Pem D, Perera RH, Phukhamsakda C, Samarakoon MC, Senwanna C, Shang Q, Tennakoon DS, Thambugala KM, Tibpromma S, Wanasinghe DN, Xiao YP, Yang J, Zeng XY, Zhang JF, Zhang SN, Bulgakov TS, Bhat DJ, Cheewangkoon R, Goh TK, Jones EBG, Kang JC, Jeewon R, Liu ZY, Lumyong S, Kuo CH, McKenzie EHC, Wen TC, Yan JY, Zhao Q. (2018) Mycosphere notes 169–224. Mycosphere 9(2): 271–430. 10.5943/mycosphere/9/2/8 [DOI] [Google Scholar]
- Hyde KD, de Silva NI, Jeewon R, Bhat DJ, Phookamsak R, Doilom M, Boonmee S, Jayawardena RS, Maharachchikumbura SSN, Senanayake IC, Manawasinghe IS, Liu NG, Abeywickrama PD, Chaiwan N, Karunarathna A, Pem D, Lin CG, Sysouphanthong P, Luo ZL, Wei DP, Wanasinghe DN, Norphanphoun C, Tennakoon DS, Samarakoon MC, Jayasiri SC, Jiang HB, Zeng XY, Li JF, Wijesinghe SN, Devadatha B, Goonasekara ID, Brahmanage RS, Yang EF, Aluthmuhandiram JVS, Dayarathne MC, Marasinghe DS, Li WJ, Dissanayake LS, Dong W, Huanraluek N, Lumyong S, Liu JK, Karunarathna SC, Jones EBG, Al-Sadi AM, Xu JC, Harishchandra D, Sarma VV. (2020a) AJOM new records and collections of fungi: 1–100. Asian Journal of Mycology 3(1): 22–294. 10.5943/ajom/3/1/3 [DOI] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Bhat DJ, Jones EBG, Liu NG, Abeywickrama PD, Mapook A, Wei DP, Perera RH, Manawasinghe IS, Pem D, Bundhun D, Karunarathna A, Ekanayaka AH, Bao DF, Li JF, Samarakoon MC, Chaiwan N, Lin CG, Phutthacharoen K, Zhang SN, Senanayake IC, Goonasekara ID, Thambugala KM, Phukhamsakda C, Tennakoon DS, Jiang HB, Yang J, Zeng M, Huanraluek N, Liu JK, Wijesinghe SN, Tian Q, Tibpromma S, Brahmanage RS, Boonmee S, Huang S-K, Thiyagaraja V, Lu Y-Z, Jayawardena RS, Dong W, Yang E-F, Singh SK, Singh SM, Rana S, Lad SS, Anand G, Devadatha B, Niranjan M, Sarma VV, Liimatainen K, Aguirre-Hudson B, Niskanen T, Overall A, Alvarenga RLM, Gibertoni TB, Pfliegler WP, Horváth E, Imre A, Alves AL, da Silva Santos AC, Tiago PV, Bulgakov TS, Wanasinghe DN, Bahkali AH, Doilom M, Elgorban AM, Maharachchikumbura SSN, Rajeshkumar KC, Haelewaters D, Mortimer PE, Zhao Q, Lumyong S, Xu J, Sheng J. (2020b) Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100(1): 5–277. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Iturrieta-González I, Gené J, Guarro J, Castañeda-Ruiz RF, García D. (2018) Neodendryphiella, a novel genus of the Dictyosporiaceae (Pleosporales). MycoKeys 37: 19–38. 10.3897/mycokeys.37.27275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Voglmayr H. (2016) Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Studies in Mycology 85(1): 35–64. 10.1016/j.simyco.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch W, Baral HO, Lücking R, Lumbsch HT. (2016a) Ascomycota. In: Frey W (Ed.) Syllabus of Plant Families – Adolf Engler’s Syllabus der Pflanzenfamilien (Part 1/2, 13th edn.). Borntraeger Science Publishers, Stuttgart, 322 pp. [Google Scholar]
- Jaklitsch WM, Olariaga I, Voglmayr H. (2016b) Teichospora and the Teichosporaceae. Mycological Progress 15(3): 1–20. 10.1007/s11557-016-1171-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Checa J, Blanco MN, Olariaga I, Tello S, Voglmayr H. (2018) A preliminary account of the Cucurbitariaceae. Studies in Mycology 90(1): 71–118. 10.1016/j.simyco.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasiri SC, Hyde KD, Jones EBG, McKenzie EHC, Jeewon R, Phillips AJL, Bhat DJ, Wanasinghe DN, Liu JK, Lu YZ, Kang JC, Xu J, Karunarathna SC. (2019) Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10(1): 1–186. 10.5943/mycosphere/10/1/1 [DOI] [Google Scholar]
- Jeewon R, Hyde KD. (2016) Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 7(11): 1669–1677. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Jiang HB, Hyde KD, Jayawardena RS, Doilom M, Xu JC, Phookamsak R. (2019) Taxonomic and phylogenetic characterizations reveal two new species and two new records of Roussoella (Roussoellaceae, Pleosporales) from Yunnan, China. Mycological Progress 18(4): 577–591. 10.1007/s11557-019-01471-9 [DOI] [Google Scholar]
- Jiang HB, Jeewon R, Karunarathna SC, Phukhamsakda C, Doilom M, Kakumyan P, Suwannarach N, Phookamsak R, Lumyong S. (2021a) Reappraisal of Immotthia in Dictyosporiaceae, Pleosporales: Introducing Immotthiabambusae sp. nov. and Pseudocyclothyriellaclematidis comb. et gen. nov. based on morphology and phylogeny. Frontiers in Microbiology 12: e656235. 10.3389/fmicb.2021.656235 [DOI] [PMC free article] [PubMed]
- Jiang HB, Phookamsak R, Hyde KD, Mortimer PE, Xu JC, Kakumyan P, Karunarathna SC, Kumla J. (2021b) A taxonomic appraisal of bambusicolous fungi in Occultibambusaceae (Pleosporales, Dothideomycetes) with new collections from Yunnan Province, China. Life 11(9): e932. 10.3390/life11090932 [DOI] [PMC free article] [PubMed]
- Jiang HB, Phookamsak R, Hongsanan S, Bhat DJ, Mortimer PE, Suwannarach N, Kakumyan P, Xu JC. (2022) A review of bambusicolous Ascomycota in China with an emphasis on species richness in southwest China. Studies in Fungi 7(1): 1–33. 10.48130/SIF-2022-0020 [DOI] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khiralla A, Spina R, Saliba S, Laurain-Mattar D. (2019) Diversity of natural products of the genera Curvularia and Bipolaris. Fungal Biology Reviews 33(2): 101–122. 10.1016/j.fbr.2018.09.002 [DOI] [Google Scholar]
- Knapp DG, Kovács GM, Zajta E, Groenewald JZ, Crous PW. (2015) Dark septate endophytic pleosporalean genera from semiarid areas. Persoonia – Molecular Phylogeny and Evolution of Fungi 35: 87–100. 10.3767/003158515X687669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WL, Luo ZL, Liu JK, Bhat D, Bao DF, Su HY, Hyde KD. (2017) Lignicolous freshwater fungi from China I: Aquadictyosporalignicola gen. et sp. nov. and new record of Pseudodictyosporiumwauense from northwestern Yunnan Province. Mycosphere 8(10): 1587–1597. 10.5943/mycosphere/8/10/1 [DOI] [Google Scholar]
- Li JF, Jiang HB, Jeewon R, Hongsanan S, Bhat DJ, Tang SM, Lumyong S, Mortimer PE, Xu JC, Camporesi E, Bulgakov TS, Zhao GJ, Suwannarach N, Phookamsak R. (2023) Alternaria: Update on species limits, evolution, multi-locus phylogeny, and classification. Studies in Fungi 8(1): 1–61. 10.48130/SIF-2023-0001 [DOI] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16(12): 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Liu JK, Hyde KD, Jones EBG, Ariyawansa HA, Bhat DJ, Boonmee S, Maharachchikumbura SSN, McKenzie EHC, Phookamsak R, Phukhamsakda C, Shenoy BD, Abdel-Wahab MA, Buyck B, Chen J, Chethana KWT, Singtripop C, Dai DQ, Dai YC, Daranagama DA, Dissanayake AJ, Doilom M, D’souza MJ, Fan XL, Goonasekara ID, Hirayama K, Hongsanan S, Jayasiri SC, Jayawardena RS, Karunarathna SC, Li WJ, Mapook A, Norphanphoun C, Pang KL, Perera RH, Peršoh D, Pinruan U, Senanayake IC, Somrithipol S, Suetrong S, Tanaka K, Thambugala KM, Tian Q, Tibpromma S, Udayanga D, Wijayawardene NN, Wanasinghe DN, Wisitrassameewong K, Zeng XY, Abdel-Aziz FA, Adamčík S, Bahkali AH, Boonyuen N, Bulgakov T, Callac P, Chomnunti P, Greiner K, Hashimoto A, Hofstetter V, Kang JC, Lewis D, Li XH, Liu XZ, Liu ZY, Matsumura M, Mortimer PE, Rambold G, Randrianjohany E, Sato G, Sri-Indrasutdhi V, Tian CM, Verbeken A, von Brackel W, Wang Y, Wen TC, Xu JC, Yan JY, Zhao RL, Camporesi E. (2015) Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72(1): 1–197. 10.1007/s13225-015-0324-y [DOI] [Google Scholar]
- Liu D, Cheng H, Bussmann RW, Guo Z, Liu B, Long C. (2018) An ethnobotanical survey of edible fungi in Chuxiong City, Yunnan, China. Journal of Ethnobiology and Ethnomedicine 14(1): 1–42. 10.1186/s13002-018-0239-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Karunarathna SC, Dai DQ, Xiong YR, Suwannarach N, Stephenson SL, Elgorban AM, Al-Rejaie S, Jayawardena RS, Tibpromma S. (2022) Description of rour novel species in Pleosporales associated with coffee in Yunnan, China. Journal of Fungi 8(10): e1113. 10.3390/jof8101113 [DOI] [PMC free article] [PubMed]
- Luo ZL, Hyde KD, Bhat DJ, Jeewon R, Maharachchikumbura SSN, Bao DF, Li WL, Su XJ, Yang XY, Su HY. (2018a) Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycological Progress 17(5): 511–530. 10.1007/s11557-018-1377-6 [DOI] [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, Bhat DJ, Bao DF, Li WL, Su HY. (2018b) Lignicolous freshwater fungi from China II: Novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 9(3): 444–461. 10.5943/mycosphere/9/3/2 [DOI] [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, Maharachchikumbura SSN, Jeewon R, Bao DF, Bhat DJ, Lin CG, Li WL, Yang J, Liu NG, Lu YZ, Jayawardena RS, Li JF, Su HY. (2019) Freshwater Sordariomycetes. Fungal Diversity 99(1): 451–660. 10.1007/s13225-019-00438-1 [DOI] [Google Scholar]
- Luttrell ES. (1955) The ascostromatic Ascomycetes. Mycologia 47(4): 511–532. 10.1080/00275514.1955.12024473 [DOI] [Google Scholar]
- Maharachchikumbura SSN, Wanasinghe DN, Elgorban AM, Al-Rejaie SS, Kazerooni EA, Cheewangkoon R. (2022) Brunneosporopsisyunnanensis gen. et sp. nov. and Allocryptovalsaxishuangbanica sp. nov., new terrestrial Sordariomycetes from Southwest China. Life 12(5): e635. 10.3390/life12050635 [DOI] [PMC free article] [PubMed]
- Manamgoda DS, Rossman AY, Castlebury LA, Crous PW, Madrid H, Chukeatirote E, Hyde KD. (2014) The genus Bipolaris. Studies in Mycology 79(1): 221–288. 10.1016/j.simyco.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason EW, Ellis MB. (1953) British species of Periconia. Mycological Papers 56: 1–126. http://www.cybertruffle.org.uk/cyberliber [Google Scholar]
- McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Marhold K, Prado J, Prud’homme Van Reine WF, Smith GF, Wiersema JH, Turland NJ [Eds] (2012) International Code of Nomenclature for algae, fungi and plants (Melbourne Code) adopted by the Eighteen International Botanical Congress Melbourne, Australia, July 2011. Koeltz Scientific Books, Königstein, 40 pp. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), November 14, 2010, New Orleans, Louisiana, 8 pp. 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Monkai J, Wanasinghe DN, Jeewon R, Promputtha I, Phookamsak R. (2021) Morphological and phylogenetic characterization of fungi within Bambusicolaceae: Introducing two new species from the Greater Mekong Subregion. Mycological Progress 20(5): 721–732. 10.1007/s11557-021-01694-9 [DOI] [Google Scholar]
- Mortimer PE, Jeewon R, Xu JC, Lumyong S, Wanasinghe DN. (2021) Morpho-phylo taxonomy of novel dothideomycetous fungi associated with dead woody twigs in Yunnan Province, China. Frontiers in Microbiology 12: e654683. 10.3389/fmicb.2021.654683 [DOI] [PMC free article] [PubMed]
- Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL. (2008) AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24(4): 581–583. 10.1093/bioinformatics/btm388 [DOI] [PubMed] [Google Scholar]
- Penzig AGO, Saccardo PA. (1902) Diagnoses fungorum novorum in insula Java collectorum. Series III. Malpighia 15: 201–260. [Google Scholar]
- Petch T. (1917) Additions to Ceylon fungi. Annals of the Royal Botanic Gardens, Peradeniya 6: 195–256. 10.2307/4111564 [DOI] [Google Scholar]
- Phookamsak R, Liu JK, McKenzie EHC, Manamgoda DS, Ariyawansa HA, Thambugala KM, Dai DQ, Camporesi E, Chukeatirote E, Wijayawardene NN, Bahkali AH, Mortimer PE, Xu JC, Hyde KD. (2014) Revision of Phaeosphaeriaceae. Fungal Diversity 68(1): 159–238. 10.1007/s13225-014-0308-3 [DOI] [Google Scholar]
- Phookamsak R, Norphanphoun C, Tanaka K, Dai DQ, Luo ZL, Liu JK, Su HY, Bhat DJ, Bahkali AH, Mortimer PE, Xu JC, Hyde KD. (2015) Towards a natural classification of Astrosphaeriella-like species; introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov and Astrosphaeriellopsis, gen. nov. Fungal Diversity 74(1): 143–197. 10.1007/s13225-015-0352-7 [DOI] [Google Scholar]
- Phookamsak R, Hyde KD, Jeewon R, Bhat DJ, Jones EBG, Maharachchikumbura SSN, Raspé O, Karunarathna SC, Wanasinghe DN, Hongsanan S, Doilom M, Tennakoon DS, Machado AR, Firmino AL, Ghosh A, Karunarathna A, Mešić A, Dutta AK, Thongbai B, Devadatha B, Norphanphoun C, Senwanna C, Wei D, Pem D, Ackah FK, Wang GN, Jiang HB, Madrid H, Lee HB, Goonasekara ID, Manawasinghe IS, Kušan I, Josep Cano J, Gené J, Li JF, Das K, Acharya K, Raj KNA, Latha KPD, Chethana KWT, He MQ, Dueñas M, Jadan M, Martín MP, Samarakoon MC, Dayarathne MC, Raza M, Park MS, Telleria MT, Chaiwan N, Matočec N, de Silva NI, Pereira OL, Singh PN, Manimohan P, Uniyal P, Shang QJ, Bhatt RP, Perera RH, Alvarenga RLM, Nogal-Prata S, Singh SK, Vadthanarat S, Oh SY, Huang SK, Rana S, Konta S, Paloi S, Jayasiri SC, Jeon SJ, Mehmood T, Gibertoni TB, Nguyen TTT, Singh U, Thiyagaraja V, Sarma VV, Dong W, Yu XD, Lu YZ, Lim YW, Chen Y, Tkalčec Z, Zhang ZF, Luo ZL, Daranagama DA, Thambugala KM, Tibpromma S, Camporesi E, Bulgakov TS, Dissanayake AJ, Senanayake IC, Dai DQ, Tang LZ, Khan S, Zhang H, Promputtha I, Cai L, Chomnunti P, Zhao RL, Lumyong S, Boonmee S, Wen TC, Mortimer PE, Xu JC. (2019) Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Diversity 95(1): 1–273. 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- Phookamsak R, Jiang HB, Suwannarach N, Lumyong S, Xu JC, Xu S, Liao CF, Chomnunti P. (2022) Bambusicolous fungi in Pleosporales: Introducing four novel taxa and a new habitat record for Anastomitrabeculiadidymospora. Journal of Fungi 8(6): e630. 10.3390/jof8060630 [DOI] [PMC free article] [PubMed]
- Phookamsak R, Hongsanan S, Bhat DJ, Xu JC, Mortimer PE, Suwannarach N, Kumla J, Dawoud TM, Lumyong S. (2023) Scolecohyalosporiumthailandense sp. nov. (Parabambusicolaceae, Pleosporales) collected on Imperata sp. (Poaceae) in northern Thailand. Phytotaxa 594(4): 267–282. 10.11646/phytotaxa.594.4.4 [DOI] [Google Scholar]
- Phukhamsakda C, McKenzie EHC, Phillips AJL, Jones EBG, Bhat DJ, Stadler M, Bhunjun CS, Wanasinghe DN, Thongbai B, Camporesi E, Ertz D, Jayawardena RS, Perera RH, Ekanayake AH, Tibpromma S, Doilom M, Xu JC, Kevin D, Hyde KD. (2020) Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Diversity 102(1): 1–203. 10.1007/s13225-020-00448-4 [DOI] [Google Scholar]
- Phukhamsakda C, Nilsson RH, Bhunjun CS, de Farias ARG, Sun YR, Wijesinghe SN, Raza M, Bao DF, Lu L, Tibpromma S, Dong W, Tennakoon DS, Tian XG, Xiong YR, Karunarathna SC, Cai L, Luo ZL, Wang Y, Manawasinghe IS, Camporesi E, Kirk PM, Promputtha I, Kuo C-H, Su H-Y, Doilom M, Li Y, Fu Y-P, Hyde KD. (2022) The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Diversity 114(1): 327–386. 10.1007/s13225-022-00502-3 [DOI] [Google Scholar]
- Piątek M, Rodriguez-Flakus P, Domic A, Palabral-Aguilera AN, Gómez MI, Flakus A. (2020) Phylogenetic placement of Leptosphaeriapolylepidis, a pathogen of Andean endemic Polylepistarapacana, and its newly discovered mycoparasite Sajamaeamycophila gen. et sp. nov. Mycological Progress 19(1): 1–14. 10.1007/s11557-019-01535-w [DOI] [Google Scholar]
- Quaedvlieg W, Verkley GJM, Shin H-D, Barreto RW, Alfenas AC, Swart WJ, Groenewald JZ, Crous PW. (2013) Sizing up Septoria. Studies in Mycology 75: 307–390. 10.3114/sim0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeshkumar KC, Verma RK, Boonmee S, Chandrasiri S, Hyde KD, Ashtekar NIKHIL, Lad S, Wijayawardene NN. (2021) Paradictyocheirosporatectonae, a novel genus in the family Dictyosporiaceae from India. Phytotaxa 509(3): 259–271. 10.11646/phytotaxa.509.3.1 [DOI] [Google Scholar]
- Rambaut A, Drummond AJ. (2012) FigTree: Tree Figure Drawing Tool. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh.
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67(5): syy032. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed]
- Réblová M, Réblová K, Štěpánek V. (2015) Molecular systematics of Barbatosphaeria (Sordariomycetes): multigene phylogeny and secondary ITS structure. Persoonia – Molecular Phylogeny and Evolution of Fungi 35: 21–38. 10.3767/003158515X687434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner SA, Buckley E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97(1): 84–98. 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- Ren GC, Wanasinghe DN, Monkai J, Mortimer PE, Hyde KD, Xu JC, Pang A, Gui H. (2021) Novel saprobic Hermatomyces species (Hermatomycetaceae, Pleosporales) from China (Yunnan Province) and Thailand. MycoKeys 82: 57–79. 10.3897/mycokeys.82.67973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman AY, Crous PW, Hyde KD, Hawksworth DL, Aptroot A, Bezerra JL, Bhat JD, Boehm E, Braun U, Boonmee S, Camporesi E, Chomnunti P, Dai DQ, D’souza MJ, Dissanayake A, Jones EBG, Groenewald JZ, Hernández-Restrepo M, Hongsanan S, Jaklitsch WM, Jayawardena R, Li WJ, Kirk PM, Lawrey J, Mapook A, McKenzie EHC, Monkai J, Phillips AJL, Phookamsak R, Raja HA, Seifert KA, Senanayake IC, Slippers B, Suetrong S, Taylor JE, Thambugala KM, Tian Q, Tibpromma S, Wanasinghe DN, Wijayawardene NN, Wikee S, Woudenberg JHC, Wu HX, Yan J, Yang T, Zhang Y. (2015) Recommended names for pleomorphic genera in Dothideomycetes. IMA Fungus 6(2): 507–523. 10.5598/imafungus.2015.06.02.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakoon BC, Phookamsak R, Karunarathna SC, Jeewon R, Chomnunti P, Xu JC, Li YJ. (2021) New host and geographic records of five pleosporalean hyphomycetes associated with Musa spp. (Banana). Studies in Fungi 6(1): 92–115. 10.5943/sif/6/1/5 [DOI] [Google Scholar]
- Sarkar T, Chakraborty P, Karmakar A, Saha A, Saha D. (2019) First report of Periconiamacrospinosa causing leaf necrosis of pointed gourd in India. Journal of Plant Pathology 101(4): e1281. 10.1007/s42161-019-00348-w [DOI]
- Senanayake IC, Rathnayaka AR, Marasinghe DS, Calabon MS, Gentekaki E, Lee HB, Hurdeal VG, Pem D, Dissanayake LS, Wijesinghe SN, Bundhun D, Nguyen TT, Goonasekara ID, Abeywickrama PD, Bhunjun CS, Jayawardena RS, Wanasinghe DN, Jeewon R, Bhat DJ, Xiang MM. (2020) Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 11(1): 2678–2754. 10.5943/mycosphere/11/1/20 [DOI] [Google Scholar]
- Shen HW, Bao DF, Bhat DJ, Su HY, Luo ZL. (2022) Lignicolous freshwater fungi in Yunnan Province, China: An overview. Mycology 13(2): 119–132. 10.1080/21501203.2022.2058638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si HL, Su YM, Zheng XX, Ding MY, Bose T, Chang RL. (2021) Phylogenetic and morphological analyses of Coniochaeta isolates recovered from Inner Mongolia and Yunnan revealed three new endolichenic fungal species. MycoKeys 83: 105–121. 10.3897/mycokeys.83.71140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si H, Wang Y, Liu Y, Li S, Bose T, Chang R. (2023) Fungal diversity associated with thirty-eight lichen species revealed a wew genus of endolichenic fungi, Intumescentia gen. nov. (Teratosphaeriaceae). Journal of Fungi 9(4): e423. 10.3390/jof9040423 [DOI] [PMC free article] [PubMed]
- Su HY, Hyde KD, Maharachchikumbura SSN, Ariyawansa HA, Luo ZL, Promputtha I, Tian Q, Lin C, Shang Q, Zhao YC, Chai HM, Liu XY, Bahkali AH, Bhat JD, McKenzie EHC, Zhou DQ. (2016) The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Diversity 80(1): 375–409. 10.1007/s13225-016-0362-0 [DOI] [Google Scholar]
- Su XJ, Luo ZL, Jeewon R, Bhat DJ, Bao DF, Li WL, Hao YE, Su HY, Hyde KD. (2018) Morphology and multigene phylogeny reveal new genus and species of Torulaceae from freshwater habitats in northwestern Yunnan, China. Mycological Progress 17(5): 531–545. 10.1007/s11557-018-1388-3 [DOI] [Google Scholar]
- Su P, Lu Z, Tian W, Chen Y, Maharachchikumbura SSN. (2023) Six additions to the genus Periconia (Dothideomycetes: Periconiaceae) from graminaceous plants in China. Journal of Fungi 9(3): e300. 10.3390/jof9030300 [DOI] [PMC free article] [PubMed]
- Tanaka K, Hirayama K, Yonezawa H, Sato G, Toriyabe A, Kudo H, Hashimoto A, Matsumura M, Harada Y, Kurihara Y, Shirouzu T, Hosoya T. (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes). Studies in Mycology 82(1): 75–136. 10.1016/j.simyco.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Huang O, Zou W, Wang Y, Wang Y, Dong Q, Sun T, Yang G, Yu H. (2023) Six new species of zombie-ant fungi from Yunnan in China. IMA Fungus 14(1): 1–9. 10.1186/s43008-023-00114-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennakoon DS, de Silva NI, Maharachchikumbura SSN, Bhat DJ, Kumla J, Suwannarach N, Lumyong S. (2023) Exploring more on Dictyosporiaceae: The species geographical distribution and intriguing novel additions from plant litter. Diversity 15(3): e410. 10.3390/d15030410 [DOI]
- Thambugala KM, Hyde KD, Tanaka K, Tian Q, Wanasinghe DN, Ariyawansa HA, Jayasiri SC, Boonmee S, Camporesi E, Hashimoto A, Hirayama K, Schumacher RK, Promputtha I, Liu ZY. (2015) Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Diversity 74(1): 199–266. 10.1007/s13225-015-0348-3 [DOI] [Google Scholar]
- Thambugala KM, Wanasinghe DN, Phillips AJL, Camporesi E, Bulgakov TS, Phukhamsakda C, Ariyawansa HA, Goonasekara ID, Phookamsak R, Dissanayake AJ, Tennakoon DS, Tibpromma S, Chen YY, Liu ZY, Hyde KD. (2017) Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 8(4): 697–796. 10.5943/mycosphere/8/4/13 [DOI] [Google Scholar]
- Tian W, Chen Y, Maharachchikumbura SSN. (2022) Neodigitodesmium, a novel genus of family Dictyosporiaceae from Sichuan Province, China. Phytotaxa 559(2): 176–184. 10.11646/phytotaxa.559.2.6 [DOI] [Google Scholar]
- Tibpromma S, Hyde KD, McKenzie EHC, Bhat DJ, Phillips AJL, Wanasinghe DN, Samarakoon MC, Jayawardena RS, Dissanayake AJ, Tennakoon DS, Doilom M, Phookamsak R, Tang AMC, Xu JC, Mortimer PE, Promputtha I, Maharachchikumbura SSN, Khan S, Karunarathna SCK. (2018) Fungal diversity notes 840–928: Microfungi associated with Pandanaceae. Fungal Diversity 92(1): 1–160. 10.1007/s13225-018-0408-6 [DOI] [Google Scholar]
- Tibpromma S, Karunarathna SC, Bhat JD, Suwannarach N, Stephenson SL, Elgorban AM, Al-Rejaie S, Xu JC, Mortimer PE. (2022) Using culture-dependent and molecular techniques to identify endophytic fungi associated with tea leaves (Camellia spp.) in Yunnan Province, China. Diversity 14(4): e287. 10.3390/d14040287 [DOI]
- Tode HJ. (1791) Fungi Mecklenburgenses selecti. Fasc. II, Generum Novorum Appendicem. Lüneburg, Germany, 67 pp. 10.5962/bhl.title.148599 [DOI] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Hyde KD, Jeewon R, Crous PW, Wijayawardene NN, Jones EBG, Bhat DJ, Phillips AJL, Groenewald JZ, Dayarathne MC, Phukhamsakda C, Thambugala KM, Bulgakov TS, Camporesi E, Gafforov YS, Mortimer PE, Karunarathna SC. (2017a) Phylogenetic revision of Camarosporium (Pleosporineae, Dothideomycetes) and allied genera. Studies in Mycology 87(1): 207–256. 10.1016/j.simyco.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Phookamsak R, Jeewon R, Li WJ, Hyde KD, Jones EBG, Camporesi E, Promputtha I. (2017b) A family level rDNA based phylogeny of Cucurbitariaceae and Fenestellaceae with descriptions of new Fenestella species and Neocucurbitaria gen. nov. Mycosphere 8(4): 397–414. 10.5943/mycosphere/8/4/2 [DOI] [Google Scholar]
- Wanasinghe DN, Wijayawardene NN, Xu JC, Cheewangkoon R, Mortimer PE. (2020) Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China). PLoS ONE 15(7): e0235855. 10.1371/journal.pone.0235855 [DOI] [PMC free article] [PubMed]
- Wanasinghe DN, Mortimer PE, Xu JC. (2021) Insight into the systematics of microfungi colonizing dead woody twigs of Dodonaeaviscosa in Honghe (China). Journal of Fungi 7(3): e180. 10.3390/jof7030180 [DOI] [PMC free article] [PubMed]
- Wanasinghe DN, Ren GC, Xu JC, Cheewangkoon R, Mortimer PE. (2022) Insight into the taxonomic resolution of the Pleosporalean epecies associated with dead woody litter in Natural Forests from Yunnan, China. Journal of Fungi 8(4): e375. 10.3390/jof8040375 [DOI] [PMC free article] [PubMed]
- Wang YB, Wang Y, Fan Q, Duan DE, Zhang GD, Dai RQ, Dai YD, Zeng WB, Chen ZH, Li DD, Tang DX, Xu ZH, Sun T, Nguyen TT, Tran N-L, Dao V-M, Zhang CM, Huang LD, Liu YJ, Zhang XM, Yang DR, Sanjuan T, Liu XZ, Yang ZL, Yu H. (2020) Multigene phylogeny of the family Cordycipitaceae (Hypocreales): New taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyceshepiali. Fungal Diversity 103(1): 1–46. 10.1007/s13225-020-00457-3 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee SJWT, Taylor JL. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR Protocols: A Guide to Methods and Applications.Academic Press, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Bahram M, Sánchez-Castro I, Dai DQ, Ariyawansa KGSU, Jayalal U, Suwannarach N, Tedersoo L. (2021a) Current insight into culture-dependent and culture-independent methods in discovering Ascomycetous Taxa. Journal of Fungi 7(9): e703. 10.3390/jof7090703 [DOI] [PMC free article] [PubMed]
- Wijayawardene NN, Dissanayake LS, Li QR, Dai DQ, Xiao Y, Wen TC, Karunarathna SC, Wu HX, Zhang H, Tibpromma S, Kang JC, Wang Y, Shen XC, Tang LZ, Deng CY, Liu YX, Kang YQ. (2021b) Yunnan–Guizhou Plateau: A mycological hotspot. Phytotaxa 523(1): 1–31. 10.11646/phytotaxa.523.1.1 [DOI] [Google Scholar]
- Wijayawardene NN, Dai DQ, Zhu ML, Wanasinghe DN, Kumla J, Zhang GQ, Zhang TT, Han LS, Tibpromma S, Chen HH. (2022a) Fungi associated with dead branches of Magnoliagrandiflora: A case study from Qujing, China. Frontiers in Microbiology 13: e954680. 10.3389/fmicb.2022.954680 [DOI] [PMC free article] [PubMed]
- Wijayawardene NN, Hyde KD, Dai DQ, Sánchez-García M, Goto BT, Saxena RK, Erdoğdu M, Selçuk F, Rajeshkumar KC, Aptroot A, Błaszkowski J, Boonyuen N, da Silva GA, de Souza FA, Dong W, Ertz D, Haelewaters D, Jones EBG, Karunarathna SC, Kirk PM, Kukwa M, Kumla J, Leontyev DV, Lumbsch HT, Maharachchikumbura SSN, Marguno F, Martínez-Rodríguez P, Mešić A, Monteiro JS, Oehl F, Pawłowska J, Pem D, Pfliegler WP, Phillips AJL, Pošta A, He MQ, Li JX, Raza M, Sruthi OP, Suetrong S, Suwannarach N, Tedersoo L, Thiyagaraja V, Tibpromma S, Tkalčec Z, Tokarev YS, Wanasinghe DN, Wijesundara DSA, Wimalaseana SDMK, Madrid H, Zhang GQ, Gao Y, Sánchez-Castro I, Tang LZ, Stadler M, Yurkov A, Thines M. (2022b) Outline of Fungi and fungus-like taxa-2021. Mycosphere 13(1): 53–453. 10.5943/mycosphere/13/1/2 [DOI] [Google Scholar]
- Wijayawardene NN, Phillips AJL, Pereira DS, Dai DQ, Aptroot A, Monteiro JS, Druzhinina IS, Cai F, Fan X, Selbmann L, Coleine C, Castañeda-Ruiz RF, Kukwa M, Flakus A, Fiuza PO, Kirk PM, Rajesh Kumar KC, Arachchi ISL, Suwannarach N, Tang LZ, Boekhout T, Tan CS, Jayasinghe RPPK, Thines M. (2022c) Forecasting the number of species of asexually reproducing fungi (Ascomycota and Basidiomycota). Fungal Diversity 114(1): 463–490. 10.1007/s13225-022-00500-5 [DOI] [Google Scholar]
- Wijesinghe SN, Wang Y, Camporesi E, Wanasinghe DN, Boonmee S, Hyde KD. (2020) A new genus of Bambusicolaceae (Pleosporales) on Corylusavellana (Fagales) from Italy. Biodiversity Data Journal 8: e55957. 10.3897/BDJ.8.e55957 [DOI] [PMC free article] [PubMed]
- Woudenberg JHC, Groenewald JZ, Binder M, Crous PW. (2013) Alternaria redefined. Studies in Mycology 75: 171–212. 10.3114/sim0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudenberg JHC, Truter M, Groenewald JZ, Crous PW. (2014) Large-spored Alternaria pathogens in section Porri disentangled. Studies in Mycology 79(1): 1–47. 10.1016/j.simyco.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudenberg JHC, Seidl MF, Groenewald JZ, De Vries M, Stielow JB, Thomma BPHJ, Crous PW. (2015) AlternariasectionAlternaria: Species, formae speciales or pathotypes? Studies in Mycology 82(1): 1–21. 10.1016/j.simyco.2015.07.001 [DOI] [PMC free article] [PubMed]
- Yang CL, Xu XL, Liu YG. (2019) Two new species of Bambusicola (Bambusicolaceae, Pleosporales) on Phyllostachysheteroclada from Sichuan, China. Nova Hedwigia 108(3–4): 527–545. 10.1127/nova_hedwigia/2019/0526 [DOI] [Google Scholar]
- Yang EF, Karunarathna SC, Dai DQ, Stephenson SL, Elgorban AM, Al-Rejaie S, Xiong YR, Promputtha I, Samarakoon MC, Tibpromma S. (2022a) Taxonomy and phylogeny of fungi associated with Mangiferaindica from Yunnan, China. Journal of Fungi 8(12): e1249. 10.3390/jof8121249 [DOI] [PMC free article] [PubMed]
- Yang EF, Phookamsak R, Jiang HB, Tibpromma S, Bhat DJ, Karunarathna SC, Dai DQ, Xu JC, Promputtha I. (2022b) Taxonomic reappraisal of Periconiaceae with the description of three new Periconia species from China. Journal of Fungi 8(3): e243. 10.3390/jof8030243 [DOI] [PMC free article] [PubMed]
- Yang EF, Tibpromma S, Karunarathna SC, Phookamsak R, Xu JC, Zhao ZX, Karunanayake C, Promputtha I. (2022c) Taxonomy and phylogeny of novel and extant taxa in Pleosporales associated with Mangiferaindica from Yunnan, China (Series I). Journal of Fungi 8(2): e152. 10.3390/jof8020152 [DOI] [PMC free article] [PubMed]
- Yang EF, Dai DQ, Bhat JD, Dawoud TM, Promputtha I, Adikaram N, Stephenson SL, Karunarathna SC, Tibpromma S. (2023a) Taxonomic and phylogenetic studies of saprobic fungi associated with Mangiferaindica in Yunnan, China. Journal of Fungi 9(6): e680. 10.3390/jof9060680 [DOI] [PMC free article] [PubMed]
- Yang YQ, Zhang F, Li ZQ, Zhou FP, Yang XY, Xiao W. (2023b) Morphological and multigene phylogenetic analyses reveal two new nematode-trapping fungi (Arthrobotrys, Orbiliaceae) from Yunnan, China. Phytotaxa 591(4): 263–272. 10.11646/phytotaxa.591.4.3 [DOI] [Google Scholar]
- Yasanthika E, Dissanayake LS, Wanasinghe DN, Karunarathna SC, Mortimer PE, Samarakoon BC, Monkai J, Hyde KD. (2020) Lonicericolafuyuanensis (Parabambusicolaceae) a new terrestrial pleosporalean ascomycete from Yunnan Province, China. Phytotaxa 446: 103–113. 10.11646/phytotaxa.446.2.3 [DOI] [Google Scholar]
- Yu XD, Zhang SN, Liu JK. (2022) Morpho-phylogenetic evidence reveals novel Pleosporalean taxa from Sichuan Province, China. Journal of Fungi 8(7): e720. 10.3390/jof8070720 [DOI] [PMC free article] [PubMed]
- Zhang Y, Crous PW, Schoch CL, Hyde KD. (2012) Pleosporales. Fungal Diversity 53(1): 1–221. 10.1007/s13225-011-0117-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Zhou XJ, Monkai J, Li FT, Liu SR, Yang XY, Wen X, Hyde KD. (2020) Two new species of nematode-trapping fungi (Dactylellina, Orbiliaceae) from burned forest in Yunnan, China. Phytotaxa 452(1): 65–74. 10.11646/phytotaxa.452.1.6 [DOI] [Google Scholar]
- Zhang F, Boonmee S, Bhat JD, Xiao W, Yang XY. (2022a) New Arthrobotrys nematode-trapping species (Orbiliaceae) from terrestrial soils and freshwater sediments in China. Journal of Fungi 8(7): e671. 10.3390/jof8070671 [DOI] [PMC free article] [PubMed]
- Zhang F, Boonmee S, Monkai J, Yang XY, Xiao W. (2022b) Drechslerelladaliensis and D.xiaguanensis (Orbiliales, Orbiliaceae), two new nematode-trapping fungi from Yunnan, China. Biodiversity Data Journal 10: e96642. 10.3897/BDJ.10.e96642 [DOI] [PMC free article] [PubMed]
- Zhang X, Zhang F, Jiang L, Yang YQ, Yang XY, Xiao W. (2022c) Two new nematode-trapping fungi (Arthrobotrys, Orbiliaceae) from Yunnan, China. Phytotaxa 568(3): 255–266. 10.11646/phytotaxa.568.3.2 [DOI] [Google Scholar]
- Zhang F, Yang YQ, Zhou FP, Xiao W, Boonmee S, Yang XY. (2023) Morphological and phylogenetic characterization of five novel nematode-trapping fungi (Orbiliomycetes) from Yunnan, China. Journal of Fungi 9(7): e735. 10.3390/jof9070735 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.