Abstract
Sulfur is an essential macroelement in plant and animal nutrition. Plants assimilate inorganic sulfate into two sulfur-containing amino acids, cysteine and methionine. Low supply of sulfate leads to decreased sulfur pools within plant tissues. As sulfur-related metabolites represent an integral part of plant metabolism with multiple interactions, sulfur deficiency stress induces a number of adaptive responses, which must be coordinated. To reveal the coordinating network of adaptations to sulfur deficiency, metabolite profiling of Arabidopsis has been undertaken. Gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry techniques revealed the response patterns of 6,023 peaks of nonredundant ion traces and relative concentration levels of 134 nonredundant compounds of known chemical structure. Here, we provide a catalogue of the detected metabolic changes and reconstruct the coordinating network of their mutual influences. The observed decrease in biomass, as well as in levels of proteins, chlorophylls, and total RNA, gives evidence for a general reduction of metabolic activity under conditions of depleted sulfur supply. This is achieved by a systemic adjustment of metabolism involving the major metabolic pathways. Sulfur/carbon/nitrogen are partitioned by accumulation of metabolites along the pathway O-acetylserine to serine to glycine, and are further channeled together with the nitrogen-rich compound glutamine into allantoin. Mutual influences between sulfur assimilation, nitrogen imbalance, lipid breakdown, purine metabolism, and enhanced photorespiration associated with sulfur-deficiency stress are revealed in this study. These responses may be assembled into a global scheme of metabolic regulation induced by sulfur nutritional stress, which optimizes resources for seed production.
For living organisms with a complex hierarchical organization, such as plants, the necessity for close coordination of various elements requires systemic organization at the level of the whole organism. Additional complexity is imposed on the system through environmental variability. Due to the inability to escape unfavorable environmental conditions, plants have evolved complex mechanisms to sense and transmit external signals to the internal decision points to trigger the adaptive response program for homeostatic maintenance. Such adaptive programs are accomplished at multiple organizational levels, e.g. gene and enzymatic activities, being finally manifested in altered metabolite concentrations. This study describes the metabolic component of the whole-system response to sulfur deficiency, as an environmental perturbation, with special emphasis on the mechanisms of the response coordination.
Despite the difficulties in measuring metabolites due to their dynamic behavior and complex chemistry, new methods allow profiling of low molecular weight compounds, with gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS) being the most robust (Stitt and Fernie, 2003). In this study, plant material was analyzed with different approaches to measure as many as possible metabolites in plant extracts, including GC-MS and LC-MS as profiling methods. This allowed an unprecedented cataloging of the metabolic changes occurring in plants in responses to insufficient sulfur supply.
Sulfur in higher plants is an essential component for the synthesis of the nutritionally important amino acids Cys and Met, as well as for a wide range of sulfur-containing metabolites (Hesse and Hoefgen, 2003; Saito, 2004). A useful approach to study the regulation of sulfur metabolism is to subject plants to sulfur starvation, followed by the analysis of changes in metabolite concentrations. This study extends previous observations on individual metabolic responses of Arabidopsis (Arabidopsis thaliana) plants to sulfur starvation (Kutz et al., 2002; Hirai et al., 2003; Nikiforova et al., 2003) through analysis of the coordination in the metabolite component of the response cascade. Revelation of the chains of successive changes in metabolite concentrations in this cascade can be achieved by simultaneous metabolite profiling of different nutritional states. Initial studies on metabolite profiling of sulfur deficiency were accompanied by integrated expression profiling data on the same experimental material (Hirai et al., 2004; Nikiforova et al., 2004). In these studies, general distribution of sulfur-induced responses between transcriptome and metabolome was examined (Nikiforova et al., 2004), and specific metabolic pathways, such as glucosinolate metabolism, were characterized (Hirai et al., 2004). Here, we provide the first catalog of the simultaneous metabolic changes occurring under sulfur starvation, as determined by high-throughput metabolite profiling, and explore coordination inside the metabolome. Analysis of these data allowed the assembly of the global scheme for the metabolic responses to sulfur nutritional stress.
RESULTS
To evoke metabolic responses to nutrient perturbation, seedlings of Arabidopsis plants were subjected to constitutive (experiment 1) and induced (experiment 2) sulfur starvation, as described by Nikiforova et al. (2003). For both experiments, two data points were chosen for sampling: before (time point 1) and after (time point 2) appearance of the first visible phenotypic changes. In total, from this experimental design, we obtained eight experimental points (four sulfur-starvation conditions with four corresponding sulfur-sufficient controls).
Fading of General Biosynthetic Activity under Sulfur Starvation
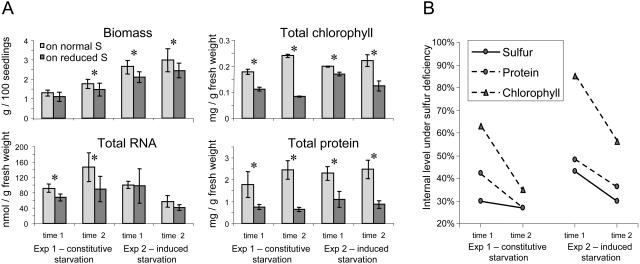
At the time points of sampling, the total biomass of plants under sulfur-deficiency stress had declined, and the total protein and total chlorophyll contents had decreased significantly in all experimental points. Notably, there were no visible changes in phenotype at time point 1. Total RNA content decreased significantly under constitutive starvation and showed a downward trend under induced starvation (Fig. 1A). The decline in the total protein levels (Fig. 1B) was stronger than in chlorophyll levels, while the rate of the decline (the incline) was faster for chlorophyll than for total protein.
Figure 1.
Analysis of general biosynthetic activity in control and sulfur-starved plants. Exp, Experiment. A, Values ± sd characterize the average of five independent repetitions; asterisks indicate pairs of significantly different values with P < 0.05, as determined by Student's t test. B, The decline in the internal protein and chlorophyll levels, depicted as a percentage to the level in sulfur-sufficient conditions, which was assigned to 100%. Slopes depict the rate of the decline. The decline of the internal sulfur level was reported earlier (Nikiforova et al., 2003) and is given here for comparison.
Combined Metabolite Profiles of Sulfur Deficiency
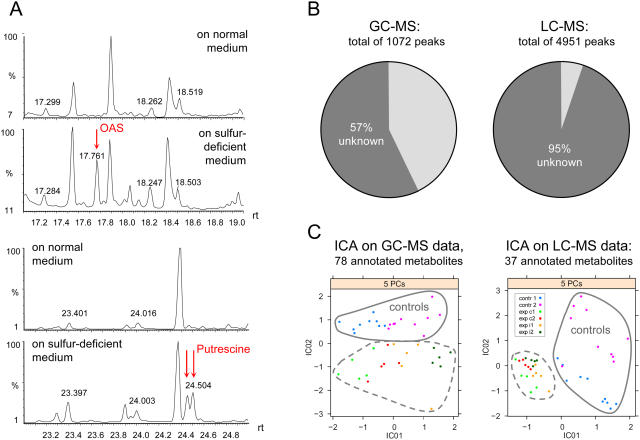
Metabolites were extracted from plant samples in five replicate pools of plants for each of eight experimental points. To obtain profiles of polar (hydrophilic) metabolites of low molecular masses (up to about 600), the methoxyaminated and trimethylsilylated derivatives were analyzed by GC-MS (1,072 ion peaks listed in Supplemental Table I; exemplary fragments are shown in Fig. 2A); nonderivatized metabolites of higher molecular masses (200 to optimum 2,000, maximum 4,000) were analyzed by LC-MS (4,951 ion peaks); the thiol-containing metabolites Cys and glutathione were analyzed by HPLC (Nikiforova et al., 2003); chemical elements were measured by inductively coupled plasma-atomic emission spectroscopy (ICP-AES); and anthocyanins and total chlorophylls were measured using spectrophotometry. In total, 134 nonredundant compounds of known chemical structure (78 by GC-MS, 37 by LC-MS, 15 by ICP-AES, 2 by spectrophotometer, and 2 by HPLC), belonging to different chemical classes, were profiled. As a general rule for MS analyses, the number of resolved ion peaks is higher than the number of resolved compounds. Each metabolite is represented by, on average, three to five peaks. This redundancy originates from several derivatives per compound (for GC-MS) and several quantitative ion traces per spectrum (for GC-MS and LC-MS) being analyzed. The high portion of nonidentified peaks in metabolite profiles (Fig. 2B) remains a major obstacle for these types of analysis. However, Independent Component Analysis (Scholz et al., 2004; an algorithm incorporated into the MetaGeneAlyse Web tool; Daub et al., 2003) applied for two subsets of annotated metabolites obtained by GC-MS and LC-MS allowed clear separation between control and treated samples, as well as between first and second time points and between constitutive and induced starvation (Fig. 2C).
Figure 2.
A, Chromatogram fragments derived from GC-MS analysis containing OAS and putrescine peaks. B, Portion of ion peaks belonging to nonidentified metabolites. C, Independent Component Analysis (ICA) applied for annotated metabolites (exp, experiment; c1 and c2, constitutive starvation time points 1 and 2; i1 and i2, induced starvation time points 1 and 2).
The changes between relative concentrations in plants grown on normal medium as a control and plants grown on sulfur-deficient medium were analyzed by calculating the response ratio (R) of the average relative concentration from five repetitions on sulfur-deficient medium to the average relative concentration from five repetitions on control sulfur-sufficient medium for each analyte. Statistical significance of the differences in relative concentrations was analyzed with the t test. Relative concentrations of 7.1% (after 6 d) and 11.5% (after 13 d) of the analytes were significantly changed (R > 2.5 or <0.4 with P < 0.05) under sulfur starvation in at least one of four experimental points (GC-MS profiles; Table I), which is a higher proportion than the induced changes at the transcript level (Nikiforova et al., 2003).
Table I.
Comparative analysis of changes in transcript and metabolic levels in response to sulfur deficiency
R, Ratio of average level at depleted sulfur (−S)/average level at normal S; P, probability value of the t test.
| Experiment ID
|
Sulfur-Responding Analytes, R > 2.5 or <0.4 with P < 0.05, in % to Total No. of Analyzed Analytes
|
|
|---|---|---|
| Expressed Sequence Tag Clonesa (R > 2.5/R < 0.4) | Metabolites (R > 2.5/R < 0.4) | |
| Experiment 2.1, 6 d at −S | 2.0 (1.0/0.9) | 7.1 (5.9/1.2) |
| Experiment 2.2, 10 d at −S | 2.5 (1.9/0.6) | 9.3 (7.0/2.3) |
| Experiment 1.1, 10 d at −S | 3.4 (2.3/1.1) | 11.4 (8.6/2.8) |
| Experiment 1.2, 13 d at −S | 4.9 (1.3/3.6) | 11.5 (8.6/2.9) |
Numbers are calculated from the transcript data reported earlier (Nikiforova et al., 2003).
The changes detected in metabolite levels are cataloged in Table II. Of 15 measured chemical elements, internal levels of none except sulfur changed significantly in sulfur-starved plants (data not shown). Those metabolites, where significantly altered concentrations could be determined in sulfur-starved plants (P value of the t test is less than 0.05; ratios in bold in Table II), were regarded as sulfur-responding metabolites if R was more than 2.5 or less than 0.4. Among them, concentrations of sulfur-containing metabolites decreased as expected with the decreasing levels of total internal sulfur: Cys, glutathione, sulfolipids, and glucosinolates of all three classes (aliphatic, indolyl, and aralkyl) are included in this group. Lipids and chlorophylls are the other major groups of metabolites whose concentrations decreased under conditions of sulfur deficiency. Proportional changes in different lipid levels are shown in Figure 3A. Within the chemical classes of amino acids, organic acids, sugars, and sugar alcohols, the response was not uniform; some metabolites of these classes were induced and some were reduced. Flavonoids of different chemical subclasses accumulated in sulfur-starved plants.
Table II.
Metabolic profiles of sulfur deficiency
R, Ratioa of average relative concentration at depleted sulfur (−S) to average relative concentration at normal S; c1 and c2, constitutive starvation time points 1 and 2; i1 and i2, induced starvation time points 1 and 2; SpPh, spectrophotometry; DH-, dehydro-; GI, glucosinolate; PE, phosphatidylethanolamine; alc, alcohol.
| Compound | Method | Class | R c1 | R c2 | R i1 | R i2 |
|---|---|---|---|---|---|---|
| Ala | GC-MS | Amino acid | 1.28 | 2.14 | 1.05 | 2.18 |
| Arg | GC-MS | Amino acid | 0.19 | 0.15 | 0.63 | 0.23 |
| Asn | GC-MS | Amino acid | 3.04 | 3.65 | 2.77 | 2.28 |
| Aspartic | GC-MS | Amino acid | 0.62 | 0.89 | 1.46 | 0.59 |
| β-Ala | GC-MS | Amino acid | 3.56 | 3.87 | 4.29 | 3.28 |
| 4-Aminobutyric | GC-MS | Amino acid | 3.03 | 1.80 | 2.34 | 1.94 |
| Glutamic | GC-MS | Amino acid | 1.26 | 0.81 | 1.02 | 1.13 |
| Gln | GC-MS | Amino acid | 5.20 | 6.75 | 3.10 | 4.18 |
| Gly | GC-MS | Amino acid | 2.63 | 1.21 | 8.92 | 1.20 |
| Homoserine | GC-MS | Amino acid | 1.41 | 1.09 | 1.99 | 1.38 |
| Ile | GC-MS | Amino acid | 1.80 | 1.33 | 1.99 | 2.27 |
| Lys | GC-MS | Amino acid | 1.28 | 1.10 | 1.40 | 1.00 |
| Met | GC-MS | Amino acid | 0.81 | 0.75 | 1.43 | 0.85 |
| N-Acetylserine | GC-MS | Amino acid | 9.21 | 10.60 | 2.63 | 2.22 |
| OASb | GC-MS | Amino acid | 24.22 | 14.48 | 8.53 | 3.71 |
| Orn | GC-MS | Amino acid | 0.06 | 0.27 | 0.57 | 0.06 |
| Phe | GC-MS | Amino acid | 1.99 | 1.01 | 1.92 | 1.16 |
| Pro | LC-MS | Amino acid | 0.93 | 0.74 | 1.19 | 0.74 |
| Oxoproline | LC-MS | Amino acid | 0.87 | 3.72 | 0.37 | 0.71 |
| t-4-HO-Pro | GC-MS | Amino acid | 3.45 | 3.58 | 2.64 | 2.35 |
| Pyroglutamic | GC-MS | Amino acid | 1.44 | 0.94 | 0.95 | 1.30 |
| Serb | GC-MS | Amino acid | 2.60 | 1.99 | 2.51 | 2.25 |
| Thr | GC-MS | Amino acid | 1.86 | 1.35 | 2.05 | 1.47 |
| Trpb | GC-MS | Amino acid | 28.31 | 7.06 | 16.60 | 5.67 |
| Val | GC-MS | Amino acid | 1.96 | 1.41 | 2.01 | 1.76 |
| Aconitic | GC-MS | Organic acid | 2.04 | 1.05 | 1.03 | 0.68 |
| α-Ketoglutaric | GC-MS | Organic acid | 1.43 | 0.98 | 1.84 | 1.40 |
| Allantoin | GC-MS | Organic acid | 6.12 | 4.77 | 15.09 | 5.57 |
| Benzoic | GC-MS | Organic acid | 1.00 | 0.84 | 2.22 | 0.82 |
| Citric | GC-MS | Organic acid | 0.76 | 1.00 | 1.41 | 0.60 |
| c-Sinapic | GC-MS | Organic acid | 1.34 | 0.85 | 1.44 | 1.04 |
| DH-ascorbic | GC-MS | Organic acid | 0.75 | 0.85 | 1.02 | 0.61 |
| Fumaric | GC-MS | Organic acid | 0.79 | 0.82 | 2.27 | 1.12 |
| Galactonic | GC-MS | Organic acid | 0.78 | 0.89 | 1.30 | 0.64 |
| Gluconic | GC-MS | Organic acid | 0.78 | 1.05 | 2.17 | 0.60 |
| Glucuronic | GC-MS | Organic acid | 1.03 | 0.95 | 1.09 | 0.90 |
| Glutaric | GC-MS | Organic acid | 1.12 | 0.87 | 0.22 | 0.82 |
| Glyceric | GC-MS | Organic acid | 1.31 | 0.56 | 3.12 | 1.39 |
| Gulonic | GC-MS | Organic acid | 1.28 | 1.02 | 1.26 | 1.08 |
| Maleic | GC-MS | Organic acid | 0.39 | 0.66 | 1.24 | 0.21 |
| Malic | GC-MS | Organic acid | 1.41 | 0.64 | 2.04 | 1.14 |
| Nicotinic | GC-MS | Organic acid | 1.73 | 1.01 | 1.65 | 0.75 |
| Phosphoric | GC-MS | Organic acid | 2.05 | 1.68 | 1.32 | 1.73 |
| Phosphoric methyl ester | GC-MS | Organic acid | 1.53 | 1.61 | 1.38 | 0.66 |
| Shikimic | GC-MS | Organic acid | 0.81 | 1.30 | 1.08 | 1.05 |
| Succinic | GC-MS | Organic acid | 1.03 | 0.63 | 1.53 | 0.86 |
| t-Ferulic | GC-MS | Organic acid | 1.33 | 0.85 | 1.50 | 0.91 |
| Threonic | GC-MS | Organic acid | 0.89 | 0.83 | 2.00 | 1.32 |
| t-Sinapic | GC-MS | Organic acid | 1.35 | 0.79 | 1.54 | 0.92 |
| Uric | GC-MS | Organic acid | 10.60 | 1.05 | 5.81 | 1.05 |
| Urea | GC-MS | Urea | 2.42 | 2.03 | 2.85 | 2.17 |
| GI_437 | LC-MS | GI aliphatic | 1.48 | 0.001 | 0.25 | 0.001 |
| GI_478 | LC-MS | GI indolyl | 0.16 | 0.04 | 0.27 | 0.03 |
| GI_493 | LC-MS | GI aralkyl | 0.05 | 0.003 | 0.07 | 0.001 |
| Hydroxylamine | GC-MS | Amine | 0.06 | 0.88 | 0.57 | 0.05 |
| Noradrenalin | GC-MS | Amine | 2.47 | 2.40 | 4.10 | 3.18 |
| Norvaline | GC-MS | Amine | 0.98 | 0.39 | 0.83 | 1.10 |
| Putrescine | GC-MS | Amine | 5.15 | 8.75 | 8.87 | 2.61 |
| Tyramine | GC-MS | Amine | 1.14 | 4.50 | 1.19 | 0.95 |
| SAH (AdoHcy) | LC-MS | Nucleoside | 1.03 | 1.04 | 1.11 | 1.23 |
| SAM (AdoMet) | LC-MS | Nucleoside | 0.16 | 0.03 | 0.29 | 0.03 |
| Sulfurb | ICP-AES | Chemical element | 0.30 | 0.27 | 0.43 | 0.30 |
| Glutathioneb | HPLC | Peptide | 0.10 | 0.06 | 0.14 | 0.11 |
| Chlorophyll a | LC-MS | Chlorophyll | 0.52 | 0.26 | 0.49 | 0.50 |
| Chlorophyll b | LC-MS | Chlorophyll | 0.64 | 0.43 | 0.57 | 0.58 |
| Chlorophyll total | SpPh | Chlorophyll | 0.63 | 0.35 | 0.85 | 0.56 |
| Hydroxypheo_a | LC-MS | Chlorophyll | 0.45 | 0.52 | 0.56 | 0.68 |
| Pheophytin | LC-MS | Chlorophyll | 1.36 | 1.27 | 0.68 | 0.96 |
| Pheophytin_a | LC-MS | Chlorophyll | 0.23 | 0.96 | 0.64 | 0.84 |
| Anthocyanins | SpPh | Flavonoid | 3.31 | 4.26 | 2.19 | 2.25 |
| Flavonoid624 | LC-MS | Flavonoid | 1.07 | 1.65 | 1.22 | 0.92 |
| Hesperidin | LC-MS | Flavonoid | 1.31 | 2.64 | 0.82 | 1.98 |
| Kaempferol578 | LC-MS | Flavonoid | 1.54 | 2.11 | 1.41 | 0.81 |
| Kaempferol594 | LC-MS | Flavonoid | 1.02 | 1.53 | 1.41 | 0.95 |
| Kaempferol740 | LC-MS | Flavonoid | 1.51 | 1.66 | 1.43 | 1.01 |
| Kaempferol756 | LC-MS | Flavonoid | 1.33 | 1.71 | 1.66 | 1.27 |
| DAG608 | LC-MS | Lipid | 0.76 | 0.89 | 1.19 | 0.52 |
| DAG610 | LC-MS | Lipid | 0.73 | 0.93 | 1.18 | 0.60 |
| DAG632 | LC-MS | Lipid | 0.70 | 0.74 | 1.12 | 0.59 |
| DGDG932 | LC-MS | Lipid | 0.40 | 0.33 | 0.71 | 0.44 |
| DGDG954 | LC-MS | Lipid | 0.36 | 0.34 | 0.75 | 0.31 |
| DGDG956 | LC-MS | Lipid | 0.31 | 0.35 | 0.69 | 0.42 |
| MGDG746a | LC-MS | Lipid | 0.61 | 0.40 | 0.84 | 0.26 |
| MGDG764 | LC-MS | Lipid | 0.29 | 0.26 | 1.00 | 0.15 |
| MGDG792 | LC-MS | Lipid | 0.33 | 0.32 | 0.84 | 0.21 |
| MGDG794 | LC-MS | Lipid | 0.74 | 0.59 | 0.75 | 0.33 |
| PE766 | LC-MS | Lipid | 0.39 | 0.34 | 1.16 | 0.18 |
| PG742 | LC-MS | Lipid | 0.36 | 0.16 | 0.42 | 0.28 |
| PG762 | LC-MS | Lipid | 0.76 | 0.50 | 1.17 | 0.52 |
| PG768 | LC-MS | Lipid | 0.50 | 0.41 | 1.06 | 0.24 |
| Sulfolipid834 | LC-MS | Lipid | 0.12 | 0.08 | 0.31 | 0.17 |
| Sulfolipid856 | LC-MS | Lipid | 0.07 | 0.03 | 0.20 | 0.07 |
| Sulfolipid858 | LC-MS | Lipid | 0.05 | 0.04 | 0.15 | 0.07 |
| Arabinose | GC-MS | Sugar | 1.09 | 0.88 | 1.21 | 0.92 |
| Fructose | GC-MS | Sugar | 1.34 | 1.35 | 0.81 | 1.17 |
| Fructose-6-P | GC-MS | Sugar | 2.14 | 1.95 | 1.58 | 2.23 |
| Galactose | GC-MS | Sugar | 0.91 | 1.44 | 1.50 | 0.93 |
| Galactose-6-P | GC-MS | Sugar | 2.00 | 3.65 | 1.23 | 2.35 |
| Glucose | GC-MS | Sugar | 1.02 | 1.63 | 1.14 | 1.02 |
| Glucose-6-P | GC-MS | Sugar | 1.52 | 1.97 | 1.23 | 1.12 |
| Maltose | GC-MS | Sugar | 0.72 | 0.46 | 0.70 | 0.52 |
| Mannose | GC-MS | Sugar | 2.11 | 2.34 | 1.42 | 1.09 |
| Mannose-6-P | GC-MS | Sugar | 1.61 | 2.04 | 1.28 | 1.25 |
| Melibiose | GC-MS | Sugar | 1.36 | 2.06 | 2.40 | 2.23 |
| Raffinose | GC-MS | Sugar | 42.80 | 3.00 | 24.25 | 2.65 |
| Ribose | GC-MS | Sugar | 1.35 | 1.25 | 1.51 | 0.94 |
| Sucrose | GC-MS | Sugar | 1.00 | 0.83 | 0.90 | 1.02 |
| Trehalose | GC-MS | Sugar | 0.26 | 0.18 | 0.23 | 0.06 |
| Xylose | GC-MS | Sugar | 0.86 | 0.87 | 1.20 | 0.66 |
| Erythritol | GC-MS | Sugar alc | 0.97 | 0.95 | 1.49 | 1.31 |
| Glycerol | GC-MS | Sugar alc | 0.79 | 0.78 | 1.11 | 0.62 |
| Glycerol-1-P | GC-MS | Sugar alc | 3.04 | 4.26 | 3.81 | 2.57 |
| Hex2Glycerol | LC-MS | Sugar alc | 0.32 | 0.31 | 0.34 | 0.87 |
| Maltitol | GC-MS | Sugar alc | 0.72 | 1.40 | 1.27 | 0.87 |
| myo-Inositol | GC-MS | Sugar alc | 0.20 | 1.29 | 1.40 | 0.16 |
| myo-Ino-2-P-1 | GC-MS | Sugar alc | 1.96 | 1.38 | 1.33 | 1.75 |
| Ononitol | GC-MS | Sugar alc | 0.38 | 0.65 | 1.20 | 0.56 |
| Sorbitol | GC-MS | Sugar alc | 1.68 | 1.93 | 1.40 | 1.28 |
Ratios in bold are statistically significant with P values less than 0.05.
Ratios are calculated from concentration levels measured earlier (Nikiforova et al., 2003).
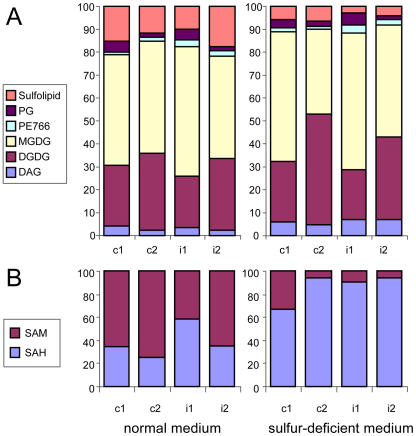
Figure 3.
Proportional changes in the levels of some metabolites under sulfur deficiency, measured by LC-MS, depicted as percentage of the sum of the metabolites in the respective chemical class (c1 and c2, constitutive starvation time points 1 and 2; i1 and i2, induced starvation time points 1 and 2). A, Lipids (PG, PE, MGDG, DGDG, and DAG). B, SAM and SAH.
For the analysis of photorespiratory fluxes, the levels of the key intermediates of one-carbon metabolism S-adenosylmethionine (AdoMet, or SAM) and S-adenosylhomocysteine (AdoHcy, or SAH) are important but traditionally difficult to measure (Edwards, 1995; Wise and Fullerton, 1995; Hanson and Roje, 2001). To estimate the impact of changes in photorespiration as part of the response to sulfur deficiency, we determined relative SAM and SAH levels (Table II) using the improved method of metabolite extraction followed by the LC-MS technique. As SAM decreased (3–30-fold) under sulfur deficiency, whereas SAH remained unchanged, the SAM/SAH ratio decreased accordingly (Fig. 3B).
Metabolic responses to constitutive and induced sulfur starvation were quite similar. Only small quantitative differences could be observed in some cases, e.g. accumulation of O-acetylserine (OAS), Trp, and uric acid was stronger under constitutive starvation (Table II).
Previously, in various studies, the individual levels of 19 metabolites have been reported for various plant species in sulfur-starvation experiments, including thiols, Ser, Trp, OAS, anthocyanin, and glucosinolates determined in Arabidopsis (references collected in Table III). These metabolites are included in the present metabolic profiles and corroborate previous findings (compare Tables II and III), with one exception: in this study, levels of Arg were significantly reduced under sulfur starvation.
Table III.
Individual metabolite responses to sulfur starvation in different plant species determined so fara
References to earlier experiments on sulfur starvation regarding the reaction of several nitrogen-containing compounds were collected by Rabe (1990); there accumulation of Arg, Asn, Gln, citrulline, Ser, Gly, and agmatine in different plant species (excluding Arabidopsis) was reported.
In the GC-MS and in the LC-MS profiles, some of the unknown analytes showed prominent changes caused by sulfur deficiency. The metabolites of unknown chemical structure are not considered further in this study. Along with the genes of unknown function identified in the transcript profiles, these analytes constitute a promising source of data for future consideration, requiring the determination of the corresponding chemical structures and genomic functions.
Shift of Metabolic Pathways in Response to Sulfur Deficiency
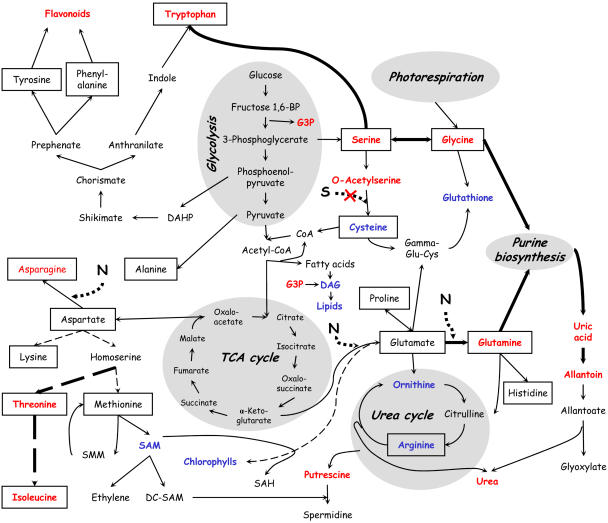
To evaluate coordination in metabolic changes under sulfur-deficiency stress, the measured metabolite concentrations were mapped on to plant biosynthetic pathways (Fig. 4). With a limited input of sulfur, levels of sulfur-containing metabolites such as Cys and glutathione decreased. Although levels of Met, the next sulfur-containing amino acid in the sulfur assimilation pathway, remained almost unchanged, the downstream product, SAM, decreased. Furthermore, the metabolic pathway via the Met precursor, homoserine, was redirected along the competing pathway branch, resulting in higher levels of Thr and Ile under sulfur deficiency. Another consequence of this redirection was the accumulation of putrescine, as the conversion to its downstream polyamine, spermidine, was presumably blocked due to reduced SAM availability. Similarly, accumulation of OAS, as the Cys precursor, and increasing levels of its upstream metabolites, Ser and Gly, as well as Trp reflect rechanneling of assimilated carbon.
Figure 4.
Mapping of measured metabolite concentrations onto plant biosynthetic pathways, with those showing increased levels under sulfur deficiency depicted in red and decreased levels in blue; standard amino acids are framed.
Assimilated nitrogen accumulated in Gln, Asn, and, more strongly, allantoin, while the levels of Arg and Orn, the other nitrogen-rich compounds, were significantly decreased (Table II, data from GC-MS). These results obtained for Arabidopsis differ from data reported earlier, where sulfur/nitrogen imbalance under sulfur deficiency resulted in the accumulation of Arg in various plant species: clover (Trifolium pratense), tomato (Lycopersicon esculentum), flax (Linum usitatissimum), pea (Pisum sativum), alfalfa (Medicago sativa), mint (Mentha sp.; Rabe, 1990), spinach (Spinacea oleracea), sugar beet (Beta vulgaris), and tobacco (Nicotiana tabacum; Table III). To confirm the present findings, Arg levels were measured using LC-MS after pooling 20 extracts (4 time points in 5 repetitions each) from control plants and 20 extracts from sulfur-starved plants. The level of Arg measured this way was 5-fold reduced in sulfur-starved plants, while Gln was 5-fold increased. These results show that for Arabidopsis, at least in these experimental conditions, the problem of sulfur/nitrogen imbalance under sulfur starvation is resolved differently from other plant species examined to date. Here, excess nitrogen together with excess carbon is channeled via Gln into allantoin, instead of Arg. Detected decrease in Arg under sulfur deficiency corroborates the increased transcript level of Arg decarboxylase (SPE2 gene; Hirai et al., 2003), the enzyme that converts Arg to agmatine on the polyamine biosynthetic pathway leading to putrescine, and corresponds to the accumulation of putrescine, as well as to the induction of putative arginase (At4g08870) in a background of the SULTR1;2 sulfate transporter mutant (Maruyama-Nakashita et al., 2003).
Integrated Analysis of Transcript and Metabolic Profiles
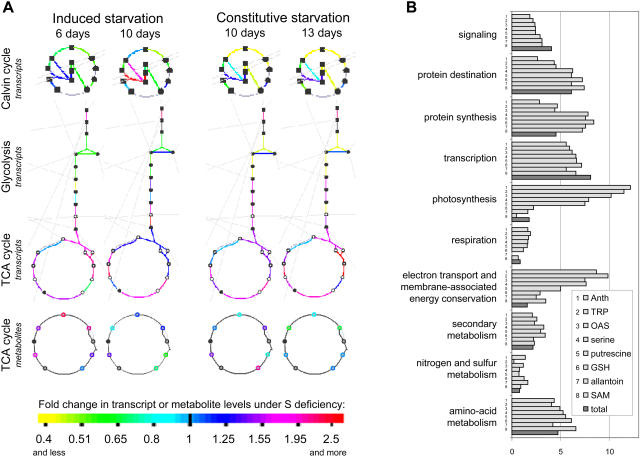
Combining transcript and metabolic data further enables the precise delineation of the extent of the coordinated response. The high number of metabolites altered in their concentrations (Table I) is indicative of the importance of metabolic control in the hypo-sulfur stress response. Using an integrated analysis of transcript and metabolite profiles, the responses to sulfur deficiency of those parts of metabolism, which are controlled mainly either at the transcript or metabolite level, were investigated. To define these parts, metabolic and transcriptional (a set of expression data available for the same plant material: Nikiforova et al., 2003) changes in sulfur-starved plants were mapped to the predesigned scheme of Arabidopsis metabolism, using the tool Omics Viewer. This tool is available in AraCyc, the first plant-specific database of biochemical pathways, developed for Arabidopsis (http://www.arabidopsis.org/tools/aracyc/; Mueller et al., 2003). The tool can be used to map either absolute transcript/metabolite levels or differences (ratios) between two treatments. Using the tool, transcript and metabolite ratios (sulfur-starved plants to control plants) were mapped for four analyzed experimental points (Supplemental Fig. 1). This allowed identification of coherent changes along biochemical pathways. The largest perturbations were found in the expression of genes encoding enzymes of the Calvin cycle, glycolysis, and TCA cycle (zoomed fragments containing these pathways are shown in Fig. 5A). These findings together with the demonstrated general decrease of metabolic activity in conditions of sulfur deficiency reflect a strong involvement of central metabolism in sulfur stress responses. In addition, it is clear that part of the adaptation is regulated at the transcriptional level.
Figure 5.
Integrated analysis of transcriptome and metabolome. A, Mapping of the transcript and metabolite ratios (sulfur-starved plants to control plants) on metabolic pathways from the AraCyc database, using an Omics Viewer tool. Those pathways are depicted, which show the strongest and the most consistent changes on transcript level, and thus are considered to be mostly regulated transcriptionally. B, Functional categorization of the genes, correlating significantly (P < 0.05) to the sulfur-responding metabolites anthocyanins (Anth), tryptophan (TRP), OAS, Ser, putrescine, glutathione (GSH), allantoin, and SAM, depicted as percentage of genes from the corresponding functional category among the whole set of genes, significantly correlating to the corresponding metabolite. These results are put together in comparison to the percent representation of the corresponding functional category in the total set of annotated genes (dark columns).
To characterize the possible roles of sulfur-responding metabolites in the development of the response to sulfur depletion, the representation of different functional categories as a percentage in a set of genes, significantly correlating (P < 0.05) to the metabolite, was calculated (Fig. 5B). The genes from the sets of significantly correlating ones were assigned to different functional categories automatically using the Munich Information Center for Protein Sequences Arabidopsis database (MATDB; http://mips.gsf.de/proj/thal/db). Representation of a functional category in the total set of analyzed genes is depicted by a dark column and used as a control. The most prominent enrichments in correlations revealed by this method were found between metabolites 4 to 8 (Ser, putrescine, glutathione, allantoin, and SAM) and the genes of protein synthesis, and between metabolites 1 to 5 (anthocyanins, Trp, OAS, Ser, and putrescine) and the genes of photosynthesis/energy assimilation, while correlations to genes involved in signaling and transcription were not stronger than average for any of the considered metabolites.
DISCUSSION
Analysis of plants under sulfur stress provides one of the first case studies, in which the systems response is characterized by high-throughput methods at different organizational levels. Previously, transcript profiling of the sulfur-deficiency stress response was performed on Arabidopsis plants from 24 h to 13 d of sulfur starvation (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003), which resulted in a detailed listing of the changes at the transcript level. Several specific pathways in steady-state sulfur-deficient metabolome have been described by Hirai et al. (2004). With this study, the analysis is complemented by adding metabolome data on the time course resulting from metabolite profiling of plants exposed to sulfur deficiency.
Systems Priorities in the Adjustment of Metabolism
The necessity to maintain viability in conditions where sulfur, a crucially important macronutrient, is deficient results in a systemic internal rebalancing of plant metabolism. This is reflected by decreased or increased levels of distinct metabolites in sulfur-deficient plants. Analysis of these changes reveals the priorities of the system in the rebalancing process aimed at economizing resources for survival and, eventually, seed production. These priorities are met by the earlier flowering of plants concurrently retarded in growth (Nikiforova et al., 2004) and accompanied by a general fading of metabolic activities, reflected by decreased levels of total proteins and chlorophyll and reduced biomass and RNA levels (Fig. 1), as well as by an assumed reduction of photosynthetic activity (Hirai et al., 2004). Decrease of chlorophyll under sulfur deficiency was described in wheat (Triticum aestivum; Gilbert et al., 1997), spinach (Warrilow and Hawkesford, 1998), oilseed rape (Brassica napus; Lencioni et al., 1997; Blake-Kalff et al., 1998), sugar beet (Thomas et al., 2000), and rice (Oryza sativa; Resurreccion et al., 2002). For spinach, a decrease in total protein content has been described (Warrilow and Hawkesford, 1998), and a decline of total RNA abundance under sulfur deficiency has been reported (Prosser et al., 1997). Decreased total protein levels in sulfur-starved plants are not surprising, considering that about one-half of all internal sulfur is allocated to proteins, as was shown for oilseed rape, a plant from the same family as Arabidopsis (Blake-Kalff et al., 1998). Insufficient sulfur as a direct cause of decreased protein is supported by the rate of its decline, which parallels that of the internal sulfur (metabolic control, Fig. 1B), and by a strong correlation between the genes encoding protein synthesis and the sulfur-responding metabolites Ser, putrescine, glutathione, and allantoin (transcriptional control, Fig. 5B).
Evidence for Direct Metabolic Control of Systems Adjustment
Within the main metabolic pathways, there are several branch-points, where adjustments of fluxes may be triggered by metabolic changes, for example, the accumulation of OAS as Cys precursor and Ser as a substrate for OAS synthesis in conditions of reduced Cys production under depleted sulfur; the accumulation of putrescine in conditions of reduced SAM, as its decarboxylated form is necessary to produce spermidine from putrescine; shift of the metabolic pathway from Asp through the branch-point homoserine toward accumulating Thr and Ile, due to the assumingly reduced metabolic flow along sulfur assimilation pathway; and decrease in glucosinolates (shown also by Blake-Kalff et al., 1998; Hirai et al., 2004), as glucosinolate biosynthesis requires a sulfur donor for the thiol sulfur, and in glucosinolate catabolism sulfate is released.
Several changes in metabolite profiles of sulfur-starved plants indicate possible metabolic and/or regulatory ways for the decline in chlorophylls. The sulfur-containing metabolite SAM decreases 3.5- to 37-fold under sulfur deficiency (Table II). In addition to many other methylation reactions, SAM is required in a late step of chlorophyll biosynthesis, and its decreased availability may result in a bottleneck in chlorophyll formation. In barley (Hordeum vulgare), blocking of SAM-methyltransferase led to inhibition of chlorophyll synthesis (Vothknecht et al., 1995).
One of the most dramatic changes in sulfur-starved plants was the overall decline in lipid content under sulfur deficiency (Table II). The assembly of glycerolipids includes the sequential transfer of synthesized fatty acids to glycerol-3-P (G3P) with the formation of diacylglycerol (DAG), followed by the addition of the head group with the formation of glycolipids such as monogalactosyl diacylglycerol (MGDG), digalactosyl diacylglycerol (DGDG), and sulfolipid, or phospholipids. When compared to control plants, G3P levels were 3- to 4-fold increased, while DAG and all measured glyco- and phospholipids were significantly reduced (Table II), indicating a block on metabolic flux from G3P into DAG. From these findings, fatty acid synthesis appears to be the limiting step in lipid formation under sulfur-limiting conditions. In the biosynthesis of fatty acids, two sulfur-containing molecules are involved, acetyl-CoA and acyl carrier protein. Acetyl-CoA is used as the central building block for assembly of the carbon backbone of long chains, and all the subsequent steps of plant fatty acid synthesis require acyl carrier protein (Andrews and Ohlrogge, 1990; Somerville et al., 2000). Thus, lipid break down may also be explained, at least partially, by limitations in sulfur-containing molecules.
Altered Purine Metabolism Contributes to the Shift of Metabolic Pathways for Carbon/Sulfur/Nitrogen Balancing
Several results indicate an increased catabolism of purine and pyrimidine bases in sulfur-deficient plants. The general decrease of metabolic activities and the decline in total RNA content cause an accumulation of β-Ala (Table II) as a product of pyrimidine catabolism (Katahira and Ashihara, 2002) and an accumulation of ureides (uric acid and allantoin, Table II), released from purine catabolism. A 37-fold reduction of SAM (Table II), which contains the purine base adenine, may further increase purine catabolism and accumulation of nitrogen-rich ureides. In the methylation cycle, SAM is converted to SAH and further hydrolyzed to homocysteine and adenosine, which is forwarded to nucleotide salvage reactions. As a strong competitive inhibitor of transmethylation reactions, SAH must be effectively hydrolyzed, and for this, efficient removal of adenosine is critical (Hanson and Roje, 2001). A gene encoding adenosylhomocysteinase (At4g13940), an enzyme that catalyzes this recycling step, was 2-fold induced in sulfur-starved plants (Nikiforova et al., 2003). This observation supports the impact of sulfur starvation on increased purine catabolism, which may have a role in keeping the SAM methylation cycle functional.
The other aspect of the sulfur stress response/purine metabolism interaction is the possibility to channel excess nitrogen to ureides via the purine metabolism pathway. For some plant families, ureides are the dominant forms of stored nitrogen (Reinbothe and Mothes, 1962), and the purine synthesis pathway is dominant for primary nitrogen metabolism in nitrogen-fixing nodules of tropical legumes (Atkins and Smith, 2000). Here, we show that sulfur-starved Arabidopsis utilizes this pathway to store nitrogen, which is sensed as being in excess under sulfur/nitrogen imbalance. The enhanced photorespiration under sulfur-deficient conditions (see below) followed by the release of additional ammonia could contribute to the total carbon:sulfur:nitrogen imbalance. Thus, rechanneling toward purine metabolism, which merges the products of photorespiration and nitrogen assimilation (Fig. 4), seems to be an efficient mechanism for detoxification of ammonium and carbon/sulfur/nitrogen equilibration under sulfur deficiency.
Assembling the Global Scheme of Metabolic Regulation of Sulfur Nutritional Stress
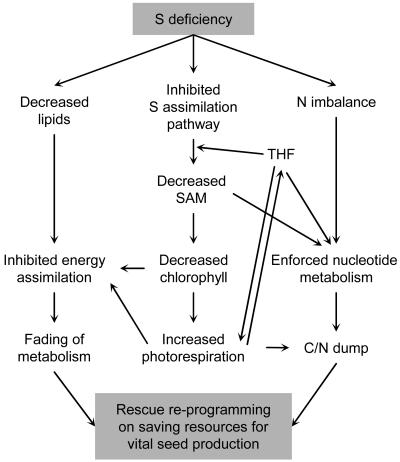
By summarizing the information on the coordination between different metabolic changes, a network of mutual cross-influences in the sulfur stress response may be assembled (Fig. 6).
Figure 6.
Summary scheme of the revealed mutual influences of physiological processes in sulfur-starved plants that lead to the rebalancing of the system.
To hasten seed production through minimization of biomass, general metabolic activities are damped, with the process being regulated mainly at the transcript level (Fig. 5A). A major part of metabolic regulation is promoted through decreased SAM, which influences the cessation of photosynthesis. This is connected with a strong decline in lipid content. A possible effect of altered contents of sulfolipids on the light assimilation capacity was discussed previously regarding hypo-sulfur stress, under which genes encoding accessory proteins of electron transport and membrane-associated energy conservation are significantly down-regulated (Nikiforova et al., 2003). Here, measurements of relative concentrations of membrane lipids in sulfur-starved plants showed a decline for all of them (Table II), including both chloroplast anionic lipids, phospholipid phosphatidylglycerol (PG) and sulfur-containing glycolipid (further termed as sulfolipid). Anionic lipids are required for normal chloroplast structure and function and are kept in a constant proportion even under adverse conditions such as phosphorus starvation (Yu and Benning, 2003). However, under sulfur deficiency, the proportion of anionic lipids decreases about 2-fold (Fig. 3A). Together with the impaired chlorophyll biosynthesis (see above), this may be assumed to affect photosynthesis.
Down-regulation of the Rubisco-encoding genes under sulfur deficiency (Nikiforova et al., 2003) further aggravates problems with energy assimilation in sulfur-starved plants. Low catalytic activity of Rubisco severely limits photosynthesis and leads to photorespiration (Spreitzer and Salvucci, 2002; Ogren, 2003; Dhingra et al., 2004), which in turn further increases the energy costs associated with photosynthesis (Siedow and Day, 2000). The effect of sulfur stress on photorespiration is further supported by the observed accumulation of Gly (Table II), which has been shown to be increased under enhanced photorespiration (Novitskaya et al., 2002), and by 5- to 9-fold heightened transcript levels of the Gly/Ser hydroxymethyltransferase gene (At1g36370; Nikiforova et al., 2003), which may encode the enzyme for these reactions. Additional ammonia, which is released in the mitochondria during photorespiration by the Gly decarboxylation reaction, may contribute to the sulfur/nitrogen imbalance under sulfur deficiency, resulting in active conversion to ureides via the purine metabolism pathway and leading to urate, allantoin, and urea accumulation.
One-carbon transfer reactions represent another link of mutual influences between photorespiration, sulfur assimilation (Met biosynthesis), and purine metabolism. A central cofactor in C1 metabolism is tetrahydrofolate (THF). Specific C1 derivatives of THF are generated from Ser/Gly metabolism and are used, among other reactions, to convert homocysteine to Met (the largest folate-dependent C1 flux) or to synthesize purines (Hanson and Roje, 2001). Moreover, C1 transfers are central for photorespiratory fluxes, in which Ser and Gly are interconverted via the action of Ser hydroxymethyltransferase and THF as a cofactor (Douce and Neuburger, 1999; Wingler et al., 2000). The strongly decreased ratio between SAM and SAH (Fig. 3B), the key intermediates of one-carbon metabolism, may further influence metabolic pathway cross-disturbance, due to increased SAH-mediated inhibition of SAM-dependent transmethylation reactions. Furthermore, as THF is not used for SAM biosynthesis, it may accumulate and act in a feedback manner on Ser/Gly metabolism and photorespiration. In turn, altered Ser levels may be involved in the regulation of the expression levels of the genes, which encode enzymes for photosynthesis and energy-related processes, as indicated by their cross-correlation (Fig. 5B).
Thus, mutual influences between the revealed changes in sulfur-starved plants form a network of coordination (Fig. 6), aimed at saving resources for seed production. Combination of all these changes results in a new metabolic balance of the system under conditions of sulfur deficiency.
Assumed Mechanisms of Adaptation to Sulfur-Deficiency Stress
Metabolite profiling of Arabidopsis revealed adaptive mechanisms of the system indicating a shift to a new sulfur-deficient homeostasis. As expected, alterations of sulfur-containing molecules (SAM, Cys, glutathione) occurred. These changes provoked a propagation of events along the main biosynthetic pathways, either upstream resulting in the accumulation of precursors (OAS, Ser, putrescine), or downstream by causing concentrational changes due to limited substrate or cofactor availability for further metabolic reactions (lipids, chlorophylls). Hence, the accumulating products lead to the activation of alternative metabolic branches (Thr, Trp, ureides). Each of these metabolic changes may further influence the expression of genes that encode the catalytical enzymes of the pathway reactions. In turn, expressional changes may contribute further to enhanced or inhibited biochemical reactions via the altered levels of activity of the corresponding enzymes. Thus, mutual influences between metabolic levels and gene expression cause coherent changes, propagating to the response endpoints. The number of system elements involved in the response development is high, with more than 2,000 genes significantly altered in expression by sulfur deficiency (Nikiforova et al., 2003), and at the metabolic level, the fraction of sulfur-responding analytes being even greater (Table I). The mutual influences form a dense network, interconnecting the elements into a coordinated systems response. As both transcript and, now, metabolite profiles are becoming available, the whole-system response network of mutual influences is amenable to reconstruction. However, in the future this analysis must go beyond mapping of the elements on just the known pathways in order to determine the full extent of the systems involved.
MATERIALS AND METHODS
Plant Material; Physiological Experiment on Sulfur Depletion
Arabidopsis (Arabidopsis thaliana) genotype Col-0 G1 (Torjek et al., 2003) plants were grown on a solidified agarose medium in sterile petri dishes (half-normal Murashige and Skoog salts for control sulfur-sufficient medium, 89% less sulfur for sulfur-depleted medium). Sulfur depletion was applied in four conditions: as constitutive (10 and 13 d, experiment 1) and induced (6 and 10 d, experiment 2) stress; seedlings grown on sulfur-sufficient medium were used as 4 corresponding controls. Material for each of 8 experimental points was collected as whole seedlings, in 5 repetitive pools, containing 500 to 600 plants in each pool. The experiment was described in detail previously (Nikiforova et al., 2003). The same plant material was used for transcript profiling (as reported earlier; Nikiforova et al., 2003), RNA isolation, and chlorophyll, anthocyanin, and metabolite extraction.
Metabolite Profiling 1: GC-MS
For GC-MS analysis, polar metabolite fractions were extracted from 60 mg of frozen plant material, ground to a fine powder, with hot MeOH/CHCl3. The fraction of polar metabolites was prepared by liquid partitioning into water as described earlier (Roessner et al., 2000; Wagner et al., 2003). Metabolite samples were derivatized by methoxyamination, using a 20 mg/mL solution of methoxyamine hydrochloride in pyridine, and subsequent trimethylsilylation, with N-methyl-N-(trimethylsilyl)-trifluoroacetamide (Fiehn et al., 2000; Roessner et al., 2000). A C12, C15, C19, C22, C28, C32, and C36 n-alkane mixture was used for the determination of retention time indices (Wagner et al., 2003). Ribitol and deuterated Ala were added for internal standardization. Samples were injected in splitless mode (1 μL/sample) and analyzed using a quadrupole-type GC-MS system (MD800; ThermoQuest, Manchester, UK). For 8 samples in 5 repetitions, 40 chromatograms were obtained. The chromatograms and mass spectra were evaluated using the MASSLAB software (ThermoQuest, Manchester, UK).
Metabolite Profiling 2: LC-MS
Prior to LC-MS analysis, a two-step extraction procedure was developed to get as many metabolites extracted as possible. The following combinations of the extractants were tested: 80% methanol/100% methanol, 80% methanol/100% acetone, 80% acetone/100% acetone, and 80% methanol/100% isopropanol; of them, the last was chosen as giving the best chromatography, allowing better coverage of components in a sample. The whole procedure was performed at 4°C. In the first step, 150 μL of 80% methanol (precooled at −20°C) was added to 100 mg frozen in liquid nitrogen and ground plant material, followed by vortexing, 5-min extraction, 5-min centrifugation at maximal speed, and collecting the supernatant. In the second step, 150 μL of isopropanol (precooled at −20°C) was added to the pellet, followed by vortexing, 5-min extraction, 5-min centrifugation at maximal speed, and collecting the supernatant, which was further combined with the supernatant from the first step. The collected extracts were analyzed by LC-MS, using hydrophilic interaction liquid chromatography coupled to electrospray ionization MS for polar compounds (Tolstikov and Fiehn, 2002), and monolithic reversed-phase LC coupled to electrospray mass spectrometry for nonpolar compounds (Tolstikov et al., 2003), giving altogether 80 chromatograms for 8 samples in 5 repetitions. Each chromatogram was manually analyzed in both positive and negative modes. Mass spectrometry settings were used as given by Tolstikov and Fiehn (2002) and Tolstikov et al. (2003), with continuous switching between positive and negative electrospray modes. For each repetition series, a representative chromatogram was then manually investigated for finding new peaks. These peaks were compiled as search list into the Xcalibur peak-finding and quantification software (version 1.3; ThermoFinnigan, San Jose, CA), combining both unidentified and known compounds. SAH (m/z 385) could be resolved by both reversed-phase and hydrophilic interaction liquid chromatography; SAM (m/z 399) was detectable and resolved well by the reversed-phase LC-MS. To confirm compound identity, spiking experiments have been performed using pure SAH (Sigma and Fluka; St. Louis) and SAM (Fluka) as standard compounds added to reference Arabidopsis leaf extracts. For quantification, raw peak areas were normalized to the sum of all detected compounds for each chromatogram. Such normalized peaks are called relative metabolite levels and were used for statistical analysis of relative responses under stressed and unstressed environmental conditions.
Other Biochemical Measurements
Total RNA content was detected after the appropriate extraction with RNeasy plant kit (Qiagen GmbH, Hilden, Germany). Concentrations of isolated RNA were measured on electropherograms by a 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA).
Total soluble protein content was determined as described by Bradford (1976).
Chlorophyll was extracted from 60 mg of seedlings frozen and ground in pools of 500 to 600 plants in each, with 1 mL of dimethylformamide. Absorbance values at 647 nm (A647) and 664.5 nm (A664.5) were measured on a UVIKON 942 spectrophotometer (KONTRON Instruments, Milan) in 1-cm cuvettes. The total chlorophyll (mg/g fresh weight) was calculated as (17.90A647 + 8.08A664.5)/60 (Inskeep and Bloom, 1985).
Anthocyanin levels were determined spectrophotometrically, based on a common method summarized by Bariola et al. (1999), with some variations: anthocyanins were extracted from 50 mg of frozen and ground seedlings with 750 μL of acidified (1% HCl, v/v) methanol for 24 h at 4°C, then 500 μL of water and 700 μL of chloroform were added, and the mixture was vortexed; after separating the phases by centrifugation for 3 min at 6,000g, 1 mL of the upper aqueous/methanol phase was assayed at 530 nm (for anthocyanins), followed by assaying at 657 nm (to compensate for the absorbance of chlorophyll); and difference A530 minus A657 was used as a measure of anthocyanin content.
Elemental content was determined by ICP-AES (Applied Research Laboratories, Accuris, Ecublens, Switzerland).
Cross-Correlation Analysis of Produced Transcript and Metabolite Datasets
For calculations on Student's t test and Pearson product moment correlation coefficient r, an algorithm, incorporated into the Microsoft Excel 2000 software program, was used. In all t test analyses, the difference was considered statistically significant with a probability of P < 0.05. To evaluate the significance of correlations, calculated with the use of Pearson correlation coefficient, depending on a number of considered experimental points, the following formula, incorporated into the SAS 8.1 statistical software package (SAS Institute, Cary, NC) was used:
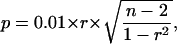 |
where P is a significance level, r is a Pearson correlation coefficient, and n is a number of experimental points. For 8 experimental points (in 5 repetitions for each; each repetition is a pool of 500–600 seedlings), the significance limit for Pearson correlation coefficient r with probability value P < 0.05 was determined as |r| > 0.898.
Supplementary Material
Acknowledgments
We thank Claudia Birkemeyer for kind assistance in the analysis of GC-MS chromatograms. We thank also Dr. Peter Dörmann and Dr. Rita Zrenner for helpful discussions of the lipid and nucleotide parts of the study.
This work was supported by the European Union commission through funding of FP5 project QLRT–2000–00103 and by the Max-Planck-Society. Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.053793.
References
- Andrews J, Ohlrogge J (1990) Fatty acid and lipid biosynthesis and degradation. In DT Dennis, DH Turpin, eds, Plant Physiology, Biochemistry and Molecular Biology. Longman Scientific & Technical, Essex, UK, pp 339–352
- Atkins CA, Smith PMC (2000) Ureide synthesis in legume nodules. In EJ Triplett, ed, Procaryotic Nitrogen Fixation: A Model System for the Analysis of a Biological Process. Horizon Scientific Press, Wymondham, Norfolk, UK, pp 559–587
- Awazuhara M, Hirai MY, Hayashi H, Chino M, Naito S, Fujiwara T (2000) Effects of sulfur and nitrogen nutrition on O-acetyl-L-serine contents in Arabidopsis thaliana. In C Brunold, J-C Davidian, L De Kok, H Rennenberg, I Stulen, eds, Sulfur Nutrition and Sulfur Assimilation in Higher Plants. Paul Haupt Publishers, Bern, Switzerland, pp 331–333
- Bariola PA, MacIntosh GC, Green PJ (1999) Regulation of S-like ribonuclease levels in Arabidopsis. Antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiol 119: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake-Kalff MMA, Harrison KR, Hawkesford MJ, Zhao FJ, McGrath SP (1998) Distribution of sulfur within oilseed rape leaves in response to sulfur deficiency during vegetative growth. Plant Physiol 118: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Daub CO, Kloska S, Selbig J (2003) MetaGeneAlyse: analysis of integrated transcriptional and metabolite data. Bioinformatics 19: 2332–2333 [DOI] [PubMed] [Google Scholar]
- Dhingra A, Portis AR, Daniell H (2004) Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proc Natl Acad Sci USA 101: 6315–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R, Neuburger M (1999) Biochemical dissection of photorespiration. Curr Opin Plant Biol 2: 214–222 [DOI] [PubMed] [Google Scholar]
- Edwards R (1995) Determination of S-adenosyl-L-methionine and S-adenosyl-L-homocysteine in plants. Phytochem Anal 6: 25–30 [Google Scholar]
- Fiehn O, Kopka J, Trethewey RN, Willmitzer L (2000) Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal Chem 72: 3573–3580 [DOI] [PubMed] [Google Scholar]
- Gilbert S, Clarkson DT, Cambridge M, Lambers H, Hawkesford MJ (1997) Sulphate-deprivation has an early effect on the content of ribulose 1,5-bisphosphate carboxylase/oxygenase and photosynthesis in young leaves of wheat. Plant Physiol 115: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Roje S (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52: 119–137 [DOI] [PubMed] [Google Scholar]
- Hesse H, Hoefgen R (2003) Molecular aspects of methionine biosynthesis. Trends Plant Sci 8: 259–262 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-L-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33: 651–663 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inskeep WP, Bloom PR (1985) Extinction coefficients of chlorophyll-a and chlorophyll-b in N,N-dimethylformamide and 80-percent acetone. Plant Physiol 77: 483–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmoker JL, Clarkson DT, Saker LR, Rooney JM, Purves JV (1991) Sulphate deprivation depresses the transport of nitrogen to the xylem and the hydraulic conductivity of barley (Hordeum vulgare L.) roots. Planta 185: 269–278 [DOI] [PubMed] [Google Scholar]
- Katahira R, Ashihara H (2002) Profiles of pyrimidine biosynthesis, salvage and degradation in disks of potato (Solanum tuberosum L.) tubers. Planta 215: 821–828 [DOI] [PubMed] [Google Scholar]
- Kim H, Hirai MY, Hayashi H, Chino M, Naito S, Fujiwara T (1999) Role of O-acetyl-L-serine in the coordinated regulation of the expression of a soybean seed storage-protein gene by sulfur and nitrogen nutrition. Planta 209: 282–289 [DOI] [PubMed] [Google Scholar]
- Kutz A, Müller A, Hennig P, Kaiser WM, Piotrowski M, Weiler EW (2002) A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J 30: 95–106 [DOI] [PubMed] [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass ADM, Touraine B (1999) Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. Plant J 18: 89–95 [DOI] [PubMed] [Google Scholar]
- Lencioni L, Ranieri A, Fergola S, Soldatini GF (1997) Photosynthesis and metabolic changes in leaves of rapeseed grown under long-term sulfate deprivation. J Plant Nutr 20: 405–415 [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yarnaya T, Takahashi H (2003) Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol 132: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum JA, Pither-Joyce M, Shaw M (2002) Sulfur deprivation and genotype affect gene expression and metabolism of onion roots. J Am Soc Hortic Sci 127: 583–589 [Google Scholar]
- Migge A, Bork C, Hell R, Becker TW (2000) Negative regulation of nitrate reductase gene expression by glutamine or asparagine accumulating in leaves of sulfur-deprived tobacco. Planta 211: 587–595 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Zhang PF, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Gakière B, Kempa S, Adamik M, Willmitzer L, Hesse H, Hoefgen R (2004) Towards dissecting nutrient metabolism in plants: a systems biology case study on sulfur metabolism. J Exp Bot 55: 1861–1870 [DOI] [PubMed] [Google Scholar]
- Novitskaya L, Trevanion SJ, Driscoll S, Foyer CH, Noctor G (2002) How does photorespiration modulate leaf amino acid contents? A dual approach through modelling and metabolite analysis. Plant Cell Environ 25: 821–835 [Google Scholar]
- Ogren WL (2003) Affixing the O to Rubisco: discovering the source of photorespiratory glycolate and its regulation. Photosynth Res 76: 53–63 [DOI] [PubMed] [Google Scholar]
- Prosser IM, Purves JV, Saker LR, Clarkson DT (2001) Rapid disruption of nitrogen metabolism and nitrate transport in spinach plants deprived of sulphate. J Exp Bot 52: 113–121 [PubMed] [Google Scholar]
- Prosser IM, Schneider A, Hawkesford MJ, Clarkson DT (1997) Changes in nutrient composition, metabolite concentrations and enzyme activities in spinach in the early stages of S-deprivation. In WJ Cram, LJ De Kok, I Stulen, C Brunold, H Rennenberg, eds, Sulphur Metabolism in Higher Plants. Backhuys Publishers, Leiden, The Netherlands, pp 339–342
- Rabe E (1990) Stress physiology: the functional significance of the accumulation of nitrogen-containing compounds. J Hortic Sci 65: 231–243 [Google Scholar]
- Reinbothe H, Mothes K (1962) Urea, ureides, and guanidines in plants. Annu Rev Plant Physiol 13: 129–151 [Google Scholar]
- Resurreccion AP, Makino A, Bennett J, Mae T (2002) Effect of light intensity on the growth and photosynthesis of rice under different sulfur concentrations. Soil Sci Plant Nutr 48: 71–77 [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Saito K (2004) Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol 136: 2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz M, Gatzek S, Sterling A, Fiehn O, Selbig J (2004) Metabolite fingerprinting: detecting biological features by Independent Component Analysis. Bioinformatics 20: 2447–2454 [DOI] [PubMed] [Google Scholar]
- Siedow JN, Day DA (2000) Respiration and photorespiration. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 676–728
- Somerville C, Browse J, Jaworski JG, Ohlrogge J (2000) Lipids. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 456–527
- Spreitzer RJ, Salvucci ME (2002) Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol 53: 449–475 [DOI] [PubMed] [Google Scholar]
- Stitt M, Fernie AR (2003) From measurements of metabolites to metabolomics: an ‘on the fly’ perspective illustrated by recent studies of carbon-nitrogen interactions. Curr Opin Biotechnol 14: 136–144 [DOI] [PubMed] [Google Scholar]
- Thomas SG, Bilsborrow PE, Hocking TJ, Bennett J (2000) Effect of sulphur deficiency on the growth and metabolism of sugar beet (Beta vulgaris cv Druid). J Sci Food Agric 80: 2057–2062 [Google Scholar]
- Thomas SG, Hocking TJ, Bilsborrow PE (2003) Effect of sulphur fertilisation on the growth and metabolism of sugar beet grown on soils of differing sulphur status. Field Crops Res 83: 223–235 [Google Scholar]
- Tolstikov VV, Fiehn O (2002) Analysis of highly polar compounds of plant origin: combination of hydrophilic interaction chromatography and electrospray ion trap mass spectrometry. Anal Biochem 301: 298–307 [DOI] [PubMed] [Google Scholar]
- Tolstikov VV, Lommen A, Nakanishi K, Tanaka N, Fiehn O (2003) Monolithic silica-based capillary reversed-phase liquid chromatography/electrospray mass spectrometry for plant metabolomics. Anal Chem 75: 6737–6740 [DOI] [PubMed] [Google Scholar]
- Torjek O, Berger D, Meyer RC, Mussig C, Schmid KJ, Sorensen TR, Weisshaar B, Mitchell-Olds T, Altmann T (2003) Establishment of a high-efficiency SNP-based framework marker set for Arabidopsis. Plant J 36: 122–140 [DOI] [PubMed] [Google Scholar]
- Vidmar JJ, Schjoerring JK, Touraine B, Glass ADM (1999) Regulation of the hvst1 gene encoding a high-affinity sulfate transporter from Hordeum vulgare. Plant Mol Biol 40: 883–892 [DOI] [PubMed] [Google Scholar]
- Vothknecht UC, Willows RD, Kannangara CG (1995) Sinefungin inhibits chlorophyll synthesis by blocking the S-adenosyl-methionine—MG-protoporphyrin IX O-methyltransferase in greening barley leaves. Plant Physiol Biochem 33: 759–763 [Google Scholar]
- Wagner C, Sefkow M, Kopka J (2003) Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62: 887–900 [DOI] [PubMed] [Google Scholar]
- Warrilow AGS, Hawkesford MJ (1998) Separation, subcellular location and influence of sulphur nutrition on isoforms of cysteine synthase in spinach. J Exp Bot 49: 1625–1636 [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC (2000) Photorespiration: metabolic pathways and their role in stress protection. Philos Trans R Soc Lond B Biol Sci 355: 1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise C, Fullerton F (1995) Analytical procedure for determination of S-adenosylmethionine, S-adenosylhomocysteine, and S-adenosylethionine in same isocratic HPLC run, with a procedure for preparation and analysis of the analog S-adenosylhomocysteine sulfoxide. J Liq Chromatogr 18: 2005–2017 [Google Scholar]
- Yu B, Benning C (2003) Anionic lipids are required for chloroplast structure and function in Arabidopsis. Plant J 36: 762–770 [DOI] [PubMed] [Google Scholar]
- Zhao F, Evans EJ, Bilsborrow PE, Syers JK (1994) Influence of nitrogen and sulphur on the glucosinolate profile of rapeseed (Brassica napus L.). J Sci Food Agric 64: 295–304 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.