Abstract
Phosphorylation of cytosolic pre-S domains of the duck hepatitis B virus (DHBV) large envelope protein (L) was identified as a regulatory modification involved in intracellular signaling. By using biochemical and mass spectrometric analyses of phosphopeptides obtained from metabolically radiolabeled L protein, a single phosphorylation site was identified at serine 118 as part of a PX(S/T)P motif, which is strongly preferred by ERK-type mitogen-activated protein kinases (MAP kinases). ERK2 specifically phosphorylated L at serine 118 in vitro, and L phosphorylation was inhibited by a coexpressed MAP kinase-specific phosphatase. Furthermore, L phosphorylation and ERK activation were shown to be induced in parallel by various stimuli. Functional analysis with transfected cells showed that DHBV L possesses the ability to activate gene expression in trans and, by using mutations eliminating (S→A) or mimicking (S→D) serine phosphorylation, that this function correlates with L phosphorylation. These mutations had, however, no major effects on virus production in cell culture and in vivo, indicating that L phosphorylation and transactivation are not essential for hepadnavirus replication and morphogenesis. Together, these data suggest a role of the L protein in intracellular host-virus cross talk by varying the levels of pre-S phosphorylation in response to the state of the cell.
Hepatitis B viruses (HBVs), or hepadnaviruses, are small enveloped DNA viruses which replicate in the livers of their respective hosts via reverse transcription of an RNA pregenome (18, 37). Although causing liver damage by the host immune response, hepadnavirus infections are per se noncytopathic, indicating that these viruses have optimized a strategy aimed at establishing inapparent, productive, long-term persistent infections. This is particularly well illustrated in case of the avian viruses, such as duck HBV (DHBV), which normally circumvent host defense mechanisms by vertical transmission. With the mammalian viruses, the nonpathogenic carrier state is probably best illustrated by HBV-transgenic mice, which produce virus titers comparable to those of chronic HBV patients without any indication of liver disease (21).
These and other indications of a well-balanced replication strategy which avoids major pathogenic effects suggest the existence of an intimate cross talk between virus and host. Evidence for such mechanisms has become apparent in a number of recent reports demonstrating a variation in hepadnavirus gene expression as a result of changes in the intracellular state of the host hepatocyte in response to extracellular signals. Perhaps most notable is the cytokine-mediated down regulation of HBV expression in HBV-transgenic mice upon infection with various animal viruses (8). Another example is the rapid termination of virus production which is observed upon plating of hepatocytes from HBV-transgenic mice, with a similar effect also being seen in freshly plated DHBV-infected primary duck hepatocytes (PDHs) (41). Furthermore, establishment of DHBV infection has been shown to be sensitive to drugs that raise the intracellular cyclic AMP level (23). Finally, it has long been known that DHBV covalently closed circular DNA is vastly overamplified in aging PDHs (56), suggesting the existence of regulatory mechanisms compensating for the reduced viral gene expression.
The mechanisms underlying these responses are presently only poorly understood, and equally little is known about the potential role of hepadnavirus gene products in these processes. Circumstantial experimental evidence suggests that hepadnaviruses encode proteins with regulatory functions which help to establish and maintain infection through direct or indirect modulation of cellular targets. The mammalian viruses, such as human HBV or woodchuck HBV, express low levels of the nonstructural X protein, which has been shown to activate gene expression from a great variety of cellular and viral promoters (46). Consistent with a regulatory function, the HBx gene product is not required for virus production from transfected HBV DNA genomes (60); it appears to be essential, however, for the establishment of infection in vivo (61). No homolog to the HBx gene, or any other open reading frame for nonstructural proteins, is present in the genomes of the avian hepadnaviruses, which nevertheless respond to changes in the state of the cell. This raises the possibility that structural proteins may be used for regulatory functions, at least in the avian hepadnaviruses.
Several observations suggest that this may indeed be the case for products of the pre-S/S gene, which encodes the viral envelope proteins. The largest of these proteins (the large envelope protein [L protein]) carries, in addition to the S domain, which constitutes the small envelope protein (S protein), an N-terminal extension, the pre-S domain, which has important functions at different stages of the viral life cycle; it mediates the receptor interaction during cellular virus uptake (30, 57) and, in an alternate topology with a cytosolic pre-S domain, presumably interacts, in a matrix-like function, with the nucleocapsid during virus formation (6, 18). The cytosolic pre-S domain has also been implicated in regulating the level of intracellular genome replication. Mutational analysis with DHBV-infected cells had indicated that mature nucleocapsids were not exported in the absence of L protein but instead were redirected into the nuclear reimport pathway, enhancing the level of nuclear DNA templates (33, 54). In these studies, not all the effects of certain pre-S mutations could be explained by this simple stoichiometric feedback model. For example, N-terminal truncations of the L protein caused both elevated levels of covalently closed circular DNA and enhanced virus production (54), suggesting the existence of an additional, nonstoichiometric regulatory role of cytosolic DHBV pre-S domains. That the DHBV L protein may carry a regulatory function is also suggested by the observation that the protein is phosphorylated to variable extents in cytosolic pre-S domains (19), as is often observed in regulatory proteins. Finally, despite its multiple functions, the pre-S region (and the underlying spacer region in the polymerase gene) shows by far the highest sequence variation between the various members of the hepadnavirus family (53); it could therefore represent a genome segment especially apt to evolve accessory regulatory functions.
While potential regulatory pre-S functions cannot be analyzed in experimental HBV infections, a transactivating activity with characteristics similar to the HBx gene product has been identified by transfection analysis of HBV pre-S variants. Initially detected in C-terminally truncated middle envelope proteins with inverted topologies (25, 26), this activity has more recently also been observed in the HBV L protein (the full-length pre-S/S gene product) and correlated with a cytosolic pre-S disposition (24). However, evidence for an involvement of this transactivation function in virus-controlled regulatory processes is lacking.
Based on the above-described observations, and in view of the limitations of the mammalian systems, we decided to study a possible regulatory role of pre-S phosphorylation in the DHBV animal model, which allows functional studies in cell culture and in vivo. After mapping of the L phosphorylation site and subsequent functional analysis with respect to virus replication, we have obtained evidence for a possible interactive role of L phosphorylation in cellular regulatory circuits. We report a DHBV L transactivating activity and its correlation with L phosphorylation by an ERK-type mitogen-activated protein kinase (MAP kinase) in response to extracellular stimuli, and we propose a model where L phosphorylation modulates viral replication in response to the host cell status.
MATERIALS AND METHODS
Nomenclature.
By assigning the designations p36 to the phosphorylated form of DHBV L (molecular mass, 36.2 kDa) and p35 to the nonphosphorylated form, we follow Grgacic and Anderson (19) and deviate from earlier publications using p37/p35 or p37/p36, respectively (15, 42, 49).
Plasmids and mutations.
Plasmids for expression of DHBV L in tissue culture were based on a construct, pMT-DL, which contains nucleotides (nt) 727 to 2815 of the DHBV subtype 16 (DHBV16) genome (34), encoding the pre-S/S gene downstream of the human metallothionine IIA promoter (55). L transcripts from this promoter are predicted to be very similar to the pre-S transcripts produced in vivo, which start at nt 732 and terminate at around nt 2800 (39). To facilitate introduction of mutations at the serine 118 codon (nt 1152 to 1154), two unique restriction sites (not affecting the pre-S open reading frame and making only serine-to-threonine exchanges in the polymerase) were introduced by point mutations, T1157A (BstXI) and A1124T (BspEI). Synthetic oligonucleotide duplexes between these sites were then used to change codon 118 from TCC to GCC (coding for alanine) in plasmid pMD-L-S118A and from TCC to GAC (coding for aspartate) in plasmid pMD-L-S118D. pMD-LΔ116 expresses L protein with a deletion of amino acids 11 to 126 (55).
Plasmid pSG5-CL100myc for the expression of myc-tagged CL100 (MAP kinase phosphatase [2]) was kindly provided by S. M. Keyse.
For production of DHBV particles from transfected LMH cells, we used pMT-D3, a plasmid containing a 10% overlength DHBV3 genome (53) driven by the metallothionine IIA promoter to produce genomic DHBV transcripts, in analogy to the cytomegalovirus promoter-driven plasmid pCD0 (39). Codon 118 in the L gene (TCT) was changed to GCT (for alanine) or to GAT (aspartate) by site-directed mutagenesis. In the polymerase open reading frame, these mutations change isoleucine 328 to serine or arginine, respectively. As isoleucine 328 is in the variable spacer region of the polymerase gene, these mutations are not expected to affect the functionality of the enzyme (44).
32P metabolic labeling of DHBV-infected PDHs and preparation of L protein.
Primary hepatocytes from DHBV3-infected ducks were prepared and maintained as described previously (45, 56). At day 7 to 14 postplating, 1.2 × 107 cells in a 10-cm-diameter plate were phosphate starved for 1 h in phosphate-free Dulbecco’s modified Eagle’s medium (ICN) and incubated with 4 to 6 mCi of 32Pi in 3.5 ml of the same medium. Incubation was for 5 h at 37°C with gentle rocking. Cells were washed three times with phosphate-buffered saline, scraped from the plate in 1 ml of TNE (10 mM Tris, 100 mM NaCl, 1 mM EDTA, pH 8.0), and broken by three cycles of freeze-thawing. Crude membrane fractions were prepared as described previously (19) by centrifugation for 2 min at 10,000 rpm and 5 min at 6,500 rpm (Eppendorf Microfuge), resuspension of the membrane pellet in TNE–1% Nonidet P-40 (NP-40), and centrifugation again for 2 min at 10,000 rpm and 5 min at 6,500 rpm to remove the nuclei. L protein was immunoprecipitated from the resulting supernatant with the monoclonal antibody (MAb) 4F8 (recognizing pre-S amino acids 100 to 105) covalently coupled to 20 μl of protein A-Sepharose beads (CL-4B; Pharmacia) with dimethylpimelimidate. After repeated washing with TNE–1% NP-40, the beads were boiled in 20 μl of protein sample buffer (200 mM Tris-HCl [pH 8.8], 10% sucrose, 3% sodium dodecyl sulfate [SDS], 5 mM EDTA, 2% 2-mercaptoethanol, 0.1% bromophenol blue), and the extract was applied to a 10% gel. By this method, 6,000 to 20,000 cpm of 32P-labeled L protein (detected as a p36 band) was obtained. For preparing large amounts of unlabeled L protein, the procedure was upscaled by using freshly prepared hepatocytes from a perfused DHBV-infected liver (a 30-g liver yielding approximately 10 g of hepatocytes). Crude membrane fractions (30 to 40 ml from 10 g of hepatocytes) were cleared by centrifugation for 20 min at 17,000 × g. Samples were preabsorbed with protein A-Sepharose beads for 2 h to reduce background binding and then immunoprecipitated for 3 h at 4°C with approximately 1 mg of MAb 4F8 covalently coupled to 200 μl of protein A-Sepharose beads. After repeated washing, L protein was recovered from the beads by boiling in 100 μl of protein sample buffer. The beads were reextracted twice with 100 μl of water, and pooled extracts were concentrated to 50 μl by evaporation in a Speed-Vac and loaded onto an SDS–10% polyacrylamide gel. The starting material for analysis of AspN peptides (see Fig. 1C and Table 1) was p36 protein isolated from 30 g of DHBV-infected hepatocytes supplemented with 5,000 cpm of 32P-labeled p36.
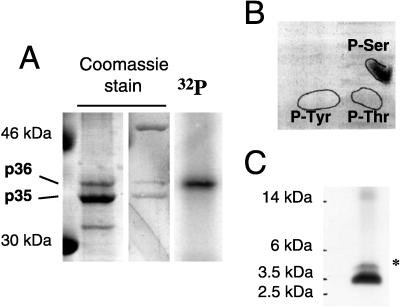
FIG. 1.
DHBV L contains a single major phosphopeptide with serine phosphorylation. (A) Preparative purification of phosphorylated DHBV L protein. Membrane-associated L protein from DHBV-infected hepatocytes was enriched by immunoadsorption and purified by SDS–10% PAGE (lane 1) (starting material, approximately 5 g of hepatocytes). In parallel, L proteins from 32P-labeled hepatocytes were separated. The single 32P-labeled band (lane 3) coincided with p36, as visualized by Coomassie blue staining (lane 2). (B) Phosphoamino acid analysis of 32P-labeled p36. After acid hydrolysis, the constituent amino acids were separated by two-dimensional thin-layer chromatography. The positions of phosphoamino acid standards detected by ninhydrin staining are indicated by solid lines on the autoradiogram. (C) Separation of 32P-labeled p36 AspN peptides. After SDS-PAGE on a 16.5% Tris-Tricine gel (48), radiolabeled peptides were detected by autoradiography. ∗, minor AspN peptide.
TABLE 1.
Mass spectrometric analysis of DHBV L phosphopeptides
In-gel protease digestion and extraction of peptides from gel slices.
For in-gel protease digestion, a Coomassie blue-stained L protein band (approximately 8 by 2 mm) was excised from an SDS–10% polyacrylamide gel and cut into small cubes (approximately 2 by 2 mm). Gel pieces were destained four times with 100 μl of acetonitrile-water (1:1) and suspended in 50 μl of acetonitrile, resulting in shrinking of the gel cubes. The supernatant was discarded, and 50 μl of 100 mM NH4HCO3 was added. After 15 min of swelling, the gel pieces were again shrunk in 50 μl of acetonitrile and dried briefly in a Speed-Vac. The dried pieces were then submerged in 100 μl of protease digestion buffer containing 12.5 μg of trypsin or AspN (Boehringer Mannheim, sequencing grade) per ml; digestion buffer for trypsin contained 50 mM NH4HCO3, 5 mM CaCl2, and 10% acetonitrile, and digestion buffer for AspN contained 50 mM phosphate buffer (pH 8.0) and 10% acetonitrile. After soaking of the gel cubes by incubation for 30 min on ice, surplus digestion buffer was replaced by buffer without protease so that the gel pieces were just covered, and samples were then incubated at 37°C overnight for trypsin digestion or for 6 h for AspN digestion. The supernatants were saved and pooled with the peptide-containing extracts described below. Peptides were extracted from the gel pieces twice with 50 μl of 25 mM NH4HCO3 and then three times with 50 μl of 5% formic acid (or two times with 70% trifluoracetic acid–water and two times with trifluoracetic acid-acetonitrile [1:1]), with a 50-μl of acetonitrile shrinking after every step. All extraction steps were carried out with gentle shaking at 30 to 37°C. Volumes were kept as small as possible to reduce the final volume of the peptide pool. All extracts (including the acetonitrile from shrinking steps) were pooled and dried down in a Speed-Vac. For high-pressure liquid chromatography (HPLC) analysis, tryptic peptides (extracted with trifluoracetic acid) were resuspended in 0.1% trifluoracetic acid. For analysis on a protein gel, AspN peptides were resuspended in 20 μl of sample buffer (50 mM Tris-HCl [pH 6.8], 12% [wt/vol] glycerol, 4% SDS, 2% 2-mercaptoethanol, 0.01% bromophenol blue). The described elution procedure generally resulted in recovery of about 80% of 32P-labeled peptides.
For high resolution of AspN peptides, separation was performed on a 16.5% Tris-Tricine gel (48). Peptide bands (approximately 8 by 2 mm) were excised, cut into very small cubes (1 by 1 mm), washed for 15 min in 40 μl of 50% methanol–10% acetic acid and then for 10 min in 40 μl of 10% methanol–7.5% acetic acid, and shrunk with 20 μl of acetonitrile. Washes were discarded, and peptides were extracted by incubating the gel pieces for 1 h each in 30 μl of 100 mM NH4HCO3, 30 μl of 25 mM NH4HCO3, and 30 μl of 25 mM NH4HCO3–acetonitrile (1:1), with shrinking with 20 μl of acetonitrile after every step. Extractions were carried out with gentle shaking at 37°C in 200-μl tubes with a tiny hole in the bottom so that liquid could be removed into a collection tube by a short spin. Pooled extracts were concentrated to 2 to 5 μl in a Speed-Vac and directly analyzed by mass spectrometry.
Mass spectrometry.
Matrix-assisted laser desorption-ionization (MALDI) mass spectra were recorded with α-cyano-4-hydroxycinnamic acid as the matrix on a Reflex II time-of-flight instrument (Bruker-Franzen, Bremen, Germany) equipped with a SCOUT multiprobe inlet and a 337-nm nitrogen laser. Tryptic peptides were analyzed in the positive-ion reflector mode with an ion acceleration voltage of 28.5 kV and a reflector voltage of 30 kV. AspN peptides were recorded in the positive-ion reflector mode with delayed extraction. The ion acceleration voltage was set to 28.5 kV, the reflector voltage was 30 kV, and the first extraction plate was set to 18.5 kV. Mass spectra were obtained by averaging 50 to 100 individual laser shots. Calibration of the spectra was performed externally by a two-point linear fit with angiotensin I and insulin.
Phosphoamino acid analysis.
The procedures used here are basically as described by Boyle et al. (5). For total acid hydrolysis, tryptic 32P-labeled peptides (containing 1,500 cpm) were incubated in 300 μl of 6 M HCl with 0.1% phenol for 4 h at 110°C under nitrogen. The sample was repeatedly frozen, lyophilized, and resuspended in water to result in a clear pellet. Two microliters of pH 3.5 running buffer (pyridine-glacial acetic acid-water, 1:10:189 [vol/vol/vol]), 2 μl of unlabeled phosphoaminoacid standard (phosphoserine-phosphothreonine-phosphotyrosine, each 10 μg/μl), and 1 μl of xylene cyanol (1 mg/ml) were added. The sample was applied to a Merck 20- by 20-cm thin-layer cellulose plate (premoistened with pH 3.5 running buffer) and separated by two-dimensional electrophoresis in a flat-bed electrophoresis unit. Electrophoresis in the first dimension was performed with the pH 3.5 buffer for 1.5 h at 850 V (13 W). The plate was dried, moistened with pH 1.9 buffer (88% formic acid–glacial acetic acid–water, 25:78:897 [vol/vol/vol]), and electrophoresis in the second dimension was performed with pH 1.9 buffer for 2.5 h at 1,000 V (15 W). After drying, standard phosphoamino acids were detected by ninhydrin staining (0.2% in ethanol; Sigma), and labeled phosphoamino acids were detected by exposure on a PhosphorImager.
Transient expression and detection of L protein.
LMH cells (11) or COS7 cells were seeded at a density of 5 × 106 to 6 × 106 cells per 10-cm-diameter dish and transfected 24 h later with 10 μg of pMT-DL by the calcium phosphate method. Cells were lysed at 2 to 4 days posttransfection by incubation for 10 min at 4°C in 1 ml of TNE–1% NP-40. Cell debris was removed from the lysate by centrifugation at 13,000 rpm for 10 min, and L protein was immunoprecipitated from the supernatant with polyclonal antiserum (D084) directed against the N-terminal two-thirds of DHBV pre-S. Typically, immunoprecipitates from one or half of a 10-cm-diameter dish (COS7 or LMH cells, respectively) were analyzed for L protein by Western blotting with MAb 4F8 (recognizing amino acids 100 to 105 in pre-S). In cotransfection experiments, 10 μg of pMT-DL and 15 μg of pSG5-CL100myc were used per 10-cm-diameter dish. CL100myc was immunoprecipitated and detected in Western blots with a myc MAb (9E10; Invitrogen). For λ-phosphatase digestion of L protein, the immunoprecipitated L protein was digested directly on beads with 400 U of λ-phosphatase (New England Biolabs) for 30 min at 30°C in a final volume of 50 μl of reaction buffer containing 0.5% NP-40.
Hepatocyte lysates and cell stimulation.
To determine the phosphorylation state of L protein in lysates of DHBV-infected liver, small pieces of liver tissue (or pelleted PDHs) were weighed and Dounce homogenized in a small volume of protein sample buffer, and volumes were adjusted with protein sample buffer to 25 mg of tissue per ml. Plated PDHs were lysed by applying 400 μl of protein sample buffer per six-well plate (2 × 106 cells), resulting in lysates with approximately 25 mg of protein per ml. In general, for the gels shown in Fig. 5, 250 μg of protein was applied per lane. Comparable protein contents of the samples were confirmed on a Coomassie blue-stained gel in parallel with the Western blot for L protein (MAb 4F8). Antisera for detection of total ERK1/2 and active ERK1/2 (anti-ACTIVE-MAPK) in hepatocyte lysates were obtained from Santa Cruz Biotechnology and Promega, respectively.
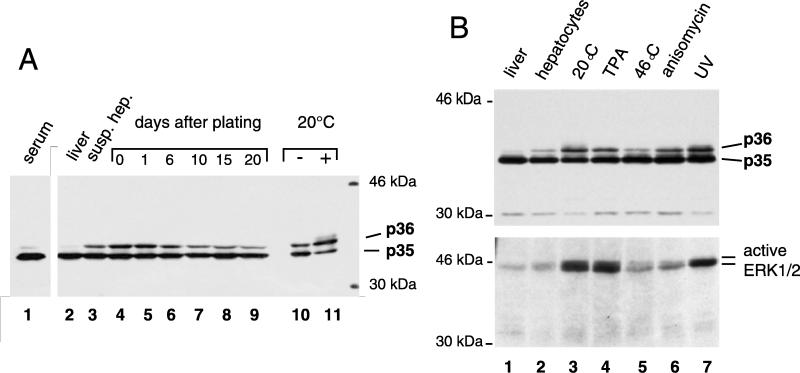
FIG. 5.
L phosphorylation induced by extracellular stimuli correlates with ERK activation. (A) Time course of preparation and cell culture of hepatocytes from a DHBV-infected duck. Total proteins from hepatocyte lysates (lanes 2 to 11) were separated by SDS–10% PAGE, and L protein was detected by Western blotting. In lane 6, 1.3 times more lysate was applied to compensate for a reduction of DHBV proteins reproducibly observed at around day 6 postplating. Cold treatment (lanes 10 and 11) was performed for 12 h starting at 6 h postplating. Two microliters of a DHBV-positive serum, containing approximately 5 × 1010 subviral particles, was applied as a reference (lane 1). susp. hep., suspended hepatocytes. (B) Effects of various stimuli on PDHs from a second DHBV-infected duck. Hepatocyte lysates were separated by SDS–10% PAGE, and L protein (upper panel) and active ERK1/2 (lower panel) were detected by Western blotting.
PDHs from DHBV-infected ducks were subjected to different stimuli starting 4 to 6 h postplating: incubation at 20°C for 14 to 16 h or at 46°C for 15 min, UV irradiation at 30 to 80 J/m2 for 45 min, or treatment with 100 nM 12-O-tetradecanoylphorbol-13-acetate (TPA) for 15 min or with 10 μg of anisomycin per ml for 30 min.
In vitro kinase assays with L protein from subviral particles.
DHBV particles were purified from sera of wild-type DHBV- or S118A-DHBV-infected ducklings essentially as described previously (30). Briefly, 30 ml of serum, containing ca. 5 × 109 viral particles/ml and ca. 5 × 1012 subviral particles/ml, was layered on a sucrose gradient and sedimented onto a cushion of 70% sucrose in 1× phosphate-buffered saline–1 mM EDTA. Subviral particle-containing fractions were pooled, concentrated by polyethylene glycol precipitation (see below), and further purified by centrifugation into a second sucrose step gradient (70 to 20% sucrose in 5% steps, 1 ml each) at 218,000 × g for 4 h at 20°C. Subviral particle-containing fractions were directly used for in vitro kinase assays: 5 μl (containing 100 to 200 ng of L protein) was incubated for 20 min at 30°C with 100 ng of activated mouse glutathione S-transferase (GST)-tagged ERK2 (GST-p42MAPK; UBI) in 20 μl of kinase reaction buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 0.1 mM EDTA, 0.5% NP-40, 2 mM dithiothreitol, and 10 mM β-glycerophosphate) containing 5 μCi of [γ-32P]ATP. L protein was immunoprecipitated with anti-pre-S antiserum D084 and subjected to SDS–10% polyacrylamide gel electrophoresis (SDS–10% PAGE). 32P-labeled proteins were detected in dried gels with a PhosphorImager.
CAT assays.
HepG2, LMH, or 293 cells were seeded at a density of 3 × 106 to 5 × 106 cells per 6-cm-diameter dish and transfected 24 h later with 2 μg of the chloramphenicol acetyltransferase (CAT) reporter construct p3xAP1-CAT (27) and either 6 to 8 μg of different L expression plasmids or a control plasmid without L insert by the Lipofectin method. At 2 days posttransfection, cells were lysed in 250 mM Tris-HCl, pH 8.0. After centrifugation to remove nuclei and cell debris, the cytosolic fraction was used to determine CAT activities as previously described (25).
Infection of PDHs with virus from cloned DHBV DNA.
Wild-type DHBV and S118A- and S118D-DHBV were harvested from the media at days 5 and 8 following transfection of LMH cells with 10 μg of pMT-D3-derived plasmid per 10-cm-diameter dish. To determine the yield of enveloped virions, an aliquot of the sample was centrifuged into a CsCl step gradient (1.4, 1.3, and 1.2 g/ml and 20% sucrose), and DHBV DNA in each fraction was quantified relative to a standard by dot blot hybridization as previously described (39). If necessary, virus was concentrated to titers of up to 3 × 109 DNA-containing virus particles per ml by precipitation with polyethylene glycol (final concentration, 6.5%) in 350 mM NaCl. Concentrated virus stocks were kept in phosphate-buffered saline containing 10% glycerol at −20°C.
PDHs (2 × 106 cells in one six-well plate, 3 days postplating) were infected overnight with 108 virus particles. Infection efficiency and virus replication were monitored by determining the DHBV DNA by DNA dot blotting of the culture supernatants from days 1 to 5, 5 to 9, 9 to 13. Cell lysates (lysis buffer was 0.1 M Tris-HCl [pH 8.8], 1% SDS, 10 mM EDTA, and 0.1% 2-mercaptoethanol) were digested with proteinase K and extracted with phenol-chloroform to increase the sensitivity of viral DNA detection by dot blot hybridization.
RESULTS
DHBV L protein is phosphorylated at serine 118.
The nonphosphorylated and the phosphorylated forms of the L protein (p35 and p36, respectively) were efficiently separated by preparative SDS-PAGE (Fig. 1A, lane 1) of L immunoabsorbed from crude membrane preparations of DHBV3-infected PDHs. In a parallel preparation from metabolically 32P-labeled cells (Fig. 1A, lanes 2 and 3), p36 was the only phospholabeled L protein species, as reported previously (19). For further analysis, the bands containing either p36 or p35 were excised from the gel, and proteins were digested with endoproteinase AspN or with trypsin as described in Materials and Methods. The resulting mixture of peptide fragments was either used for phosphoamino acid analysis or further resolved for identification of individual phosphopeptides by mass spectrometry.
Phosphoamino acid analysis of 32P-labeled p36, performed with the tryptic digest eluted from the gel, showed that the radiolabel was linked exclusively to serine residues, with no radiolabel being detected at the positions of phosphothreonine or phosphotyrosine (Fig. 1B). Separation of 32P-labeled AspN peptides on a 16.5% Tris-Tricine gel (Fig. 1C) revealed that 90% of the radioactivity migrated as a single band with an apparent molecular mass of 3.5 kDa, a result demonstrating the presence of a single major phosphopeptide in DHBV L. After elution from the gel, a mass of 4,221.8 Da was determined for this peptide. This value corresponds closely to the mass calculated for a singly phosphorylated AspN cleavage product, comprising amino acid residues 93 to 127 in the pre-S domain of DHBV L (4,220.6 Da) (Table 1 and Fig. 2), whereas values for any other phosphoserine-containing peptide, possibly arising from complete or partial AspN cleavage of DHBV L, were calculated to be at least 372 Da lower or 92 Da higher. In DHBV3, the subtype used in this experiment, the peptide from amino acid 93 to 127 contains only a single serine residue at position 118 (Fig. 2); we therefore concluded that this serine residue was the site for DHBV L phosphorylation. This interpretation was further supported by the good correlation of the mass values determined and calculated for the corresponding AspN peptide from the unphosphorylated L protein species, p35, which were 4,140.0 and 4,140.6 Da, respectively (data not shown). Moreover, a second, minor 32P-labeled AspN phosphopeptide, representing the remaining 10% of the radioactivity and migrating at approximately 4.5 kDa (marked by an asterisk in Fig. 1C), was found to possess a mass of 4,282.2 Da, which is only 60.4 Da higher than the one determined for the major AspN phosphopeptide (Table 1). As outlined above, this value does not correspond to any other singly or doubly phosphorylated peptide resulting from AspN cleavage of L protein. Furthermore, the relative fraction of this minor peptide, by 32P content, varied severalfold between experiments (not shown); it therefore most likely represents the product of a modification, apparently caused by unspecified variations of the experimental conditions during isolation, adding 60 Da to the major phosphopeptide.
FIG. 2.
DHBV L protein is phosphorylated at serine 118. Phosphopeptides matching the mass values obtained by mass spectrometry (see Table 1 and text) are shown in the context of the DHBV3 L protein amino acid sequence. As phosphoserine was the only phosphoamino acid detected in p36 (Fig. 1B), serine 118 must be the DHBV L phosphorylation site.
Tryptic peptides from the p36 protein were characterized by reverse-phase HPLC of 32P-labeled digestion products. Peptides from three major peak fractions which contained similar amounts of radiolabel (40, 34, and 26%, respectively) were analyzed further. Mass spectrometry revealed that the first two peptides, which were present in two directly adjacent fractions, contained two closely related peptides with mass values determined to be 2,583.6 and 2,569.5 Da (peptide 1 and peptide 2, respectively [Table 1]). Further mass spectrometric analysis in the reflectron mode (3), showed a metastable fragmentation of phosphoric acid, indicating that both peptides contained only a single phosphate group (data not shown). The mass determined for tryptic peptide 2 corresponds well to the mass of 2,566.7 Da predicted for the tryptic cleavage product encompassing phosphorylated serine 118 (amino acids 103 to 123) (Table 1 and Fig. 2), whereas mass values for other phosphoserine-containing tryptic pre-S peptides are predicted to be at least 100 Da higher or 1,000 Da lower. The apparent mass increase of 14 Da in peptide 1 is therefore best explained by the addition of a single oxygen atom (16 Da) to peptide 2, possibly as the result of proline or tryptophan hydroxylation (31). Mass spectrometric analysis of the third HPLC fraction, containing 26% of the radiolabel, did not give conclusive results. Conceivably, this peptide may have also contained a derivative of the tryptic phosphopeptide from amino acid 103 to 123, possibly carrying the modification adding 60 Da that was observed with the phosphopeptides from the AspN digest. Thus, the majority of the phosphorylated tryptic peptides recovered from metabolically labeled p36 was found to correspond to phosphopeptides containing serine 118, a result confirming the more complete mapping of the phosphorylation site by analysis of the AspN peptides presented above.
Confirmation of DHBV L phosphorylation at serine 118 by mutational analysis.
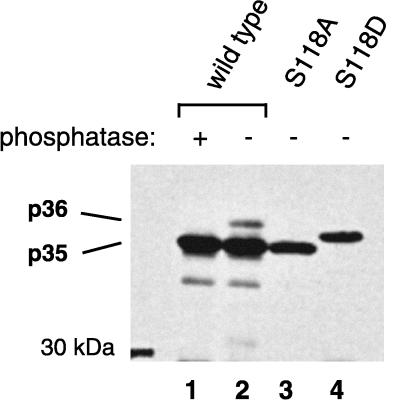
As phosphorylation results in reduced migration of DHBV L in SDS-PAGE, mutations changing serine 118 are expected to influence the electrophoretic mobility of the L protein. To test this prediction, serine 118 was replaced either by alanine, thereby eliminating phosphorylation at this site, or by aspartate, which is commonly used to functionally mimic protein phosphorylation. Wild-type or mutant DHBV16 L proteins were expressed in transfected LMH cells (a chicken hepatoma cell line [11]) and analyzed by immunoblotting for changes in migration during SDS-PAGE. As expected, replacement by alanine (S118A) resulted in the complete disappearance of the slower-migrating p36 species (Fig. 3, compare lanes 2 and 3), as was observed after incubation of phosphorylated wild-type protein with λ-phosphatase (lane 1). Replacement of serine 118 by aspartate (S118D) resulted in a single L protein species migrating between p35 and p36 (Fig. 3, lane 4), an observation demonstrating that the negative charge introduced with the aspartate residue at position 118 was sufficient to reduce the electrophoretic mobility of DHBV L, although not to the same degree as the doubly charged phosphate. Taken together, these results demonstrate that the reduced mobility of the p36 L protein species in SDS-polyacrylamide gels correlates with its phosphorylation at serine 118; they thus prove the conclusions drawn from the biochemical mapping of the phosphorylation site. Moreover, as biochemical mapping was performed with L protein from DHBV3 and mutational analysis was performed with DHBV16, these data demonstrate that pre-S phosphorylation at serine 118 is independent of the DHBV subtype.
FIG. 3.
Mutational analysis confirming that phosphorylation at serine 118 causes the electrophoretic mobility shift leading to the p36 L species. LMH cells were transfected with DHBV 16 L expression constructs coding for wild-type (lanes 1 and 2) or mutated (S118A for removing the phosphorylation site [lane 3] and S118D for mimicking phosphoserine [lane 4]) L proteins. L proteins in cell lysates at day 8 posttransfection were immunoprecipitated, separated on an SDS–10% polyacrylamide gel, and detected by anti-pre-S Western blotting.
DHBV L protein is phosphorylated by an ERK-type MAP kinase.
The extracellular signal-regulated protein kinases ERK1 and ERK2 (isoforms) are members of the MAP kinase family (7). In the DHBV L protein sequence, serine 118 is part of a PXSP sequence (Fig. 2), a motif that is recognized by ERKs in preference to other SP sites (12, 51). There are no basic residues in its vicinity, which argues against the possibility that phosphorylation may alternatively involve protein kinase C, protein kinase A, or a member of the cyclin-dependent kinase family, the other group of SP- or TP-targeted kinases (28, 51).
In order to test whether ERK was indeed participating in L phosphorylation, L protein from detergent-disrupted subviral particles was used as a substrate in an in vitro phosphorylation assay with GST-tagged ERK2. As shown in Fig. 4A, ERK2 was indeed capable of phosphorylating DHBV L, resulting in a strongly radiolabeled p36 band detected by SDS-PAGE (lane 1). Phosphorylation was specific for serine 118, as mutant L protein carrying the S118A mutation was not converted into p36 (Fig. 4A, lane 2), even though comparable amounts of L protein had been present in the reaction mixture (as demonstrated by Coomassie blue staining [not shown]). Weak radioactive signals were seen slightly above the position of p35 with the wild-type and the mutant proteins. These were probably due to minor L phosphorylation of (S/T)P sites outside serine 118, which do not cause a major mobility shift in SDS-PAGE (20). In summary, these data demonstrate that highly purified ERK2 can strongly and specifically phosphorylate the DHBV L protein at serine 118, the site shown to be the target of phosphorylation in vivo.
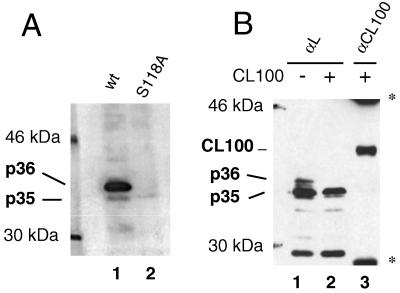
FIG. 4.
DHBV L is phosphorylated by an ERK-type MAP kinase. (A) In vitro kinase assay. Wild-type (wt) or S118A-DHBV particles were incubated with GST-tagged ERK2 and [γ32P]ATP in a reaction buffer which contained 0.5% NP-40 to disrupt the particles. L proteins were immunoprecipitated and separated by electrophoresis on an SDS–10% polyacrylamide gel, and radiolabeled proteins were detected by autoradiography. (B) Inhibition of phosphorylation at serine 118 by CL100, a phosphatase which specifically inactivates MAP kinases. COS7 cells were transfected with an L expression plasmid (pMT-DL) and a myc-tagged CL100 expression construct (pSG5-CL100myc) as indicated. Cells were lysed at day 2 posttransfection, and L proteins were immunoprecipitated and detected by Western blotting after separation on an SDS–10% polyacrylamide gel (lanes 1 and 2). To check for this presence, CL100myc protein was also immunoprecipitated from the lysate of cells cotransfected with L and CL100 and detected on the same Western blot (lane 3) (double stained with anti-pre-S and anti-myc). ∗, signals from mouse immunoglobulin G used for precipitating CL100myc.
To confirm an involvement of a MAP kinase in DHBV L phosphorylation, we investigated whether CL100, a phosphatase specifically inactivating MAP kinases by returning them to the dephosphorylated inactive state (reviewed in reference 29), had an inhibitory effect on L phosphorylation in transfected COS7 cells. As shown in Fig. 4B, L protein phosphorylation (lane 1) was indeed reduced strongly upon cotransfection of a construct expression CL100 (lane 2), confirming that an activated MAP kinase was most likely responsible for L phosphorylation. Direct dephosphorylation of L by CL100 is improbable because of the high specificity of this phosphatase for MAP kinases (2). Taken together, the results from kinase assays performed in vitro and with transfected cells strongly suggest that an ERK-type MAP kinase is responsible for L protein phosphorylation at serine 118.
L protein phosphorylation induced by extracellular stimuli correlates with activation of ERK 1/2.
In our initial experiments, major variations in the extent of L protein phosphorylation were observed between DHBV-infected duck livers and PDH cultures that had been maintained for different time periods. In a more systematic follow-up, liver samples and cultured hepatocytes from the same DHBV-infected donor duck were analyzed before and during the course of liver perfusion, as well as over extended time periods after plating. Changes in L protein phosphorylation, indicated by the p35/p36 ratio, were monitored by anti-pre-S Western blots (Fig. 5A, lanes 2 to 11), with L protein from serum DHBV particles (known to be only marginally phosphorylated) being included as a reference (Fig. 5A, lane 1). As shown in Fig. 5A (lane 2), the phosphorylated form of the L protein (p36) was barely detectable in the liver. It increased progressively, however, as the liver cells were liberated by perfusion and suspended in the maintenance medium, as is demonstrated in Fig. 5A (lane 3) by L protein obtained from suspended cells taken 30 min after the start of perfusion. Samples taken at earlier time points during collagenase treatment showed intermediate p36 levels which increased with time. In PDH cell cultures, p36/p35 ratios initially stayed constant and decreased slowly after day 6 (Fig. 5A, lanes 7 to 9). It should be noted that the increase in L phosphorylation in response to perfusion and plating varied between PDH cultures from different test animals (compare Fig. 5A, lanes 2 and 4, to Fig. 5B, upper panel, lanes 1 and 2), as did the decrease of p36 levels after day 6 (data not shown).
The levels of phosphorylated L protein could be enhanced further, particularly in freshly plated PDHs, in response to various extracellular stimuli. Strong increases were observed after exposure to low temperature (overnight at 20°C [Fig. 5A, lane 11, and B upper panel, lane 3]), UV irradiation (Fig. 5B, upper panel, lane 7), or a mitogenic phorbolester (TPA) (Fig. 5B, upper panel, lane 4). A stimulatory response was also obtained with anisomycin, an inhibitor of translation (Fig. 5B, upper panel, lane 6), while no change in p36 levels was observed after heat shock at 46°C (lane 5) (the avian body temperature is 42°C). Except for the effect of anisomycin, which is known to stimulate stress-activated MAP kinases and not ERKs, these observations are again in keeping with an involvement in L phosphorylation of mitogen-activated ERK-type MAP kinases. In contrast, there was little correlation with the activation profile of stress-activated MAP kinases, such as JNK and p38, which are typically stimulated by heat shock (32). UV irradiation, which enhanced L protein phosphorylation, is known to stimulate primarily JNKs but also ERKs (43); the strong activation in response to cold treatment observed here has, to our knowledge, not yet been noticed in any other system.
Taken together, the data presented so far suggested that L phosphorylation correlated with ERK activation. To test whether this correlation was also observed during the changes in L phosphorylation in freshly plated hepatocytes, Western blots of PDH lysates from the above-described experiments were probed with an antiserum recognizing only the active form of ERK1/2 (Fig. 5B, lower panel) and, as a control, with an antiserum recognizing both inactive and active ERK1/2 (not shown). In this analysis, the amounts of activated ERK varied in close correlation with induction of L phosphorylation (Fig. 5B, compare upper and lower panels), while the total ERK protein content was found to be comparable in all samples. These observations again support the hypothesis that an ERK-type MAP kinase is responsible for phosphorylation of the DHBV L protein.
Phosphorylation at serine 118 mediates activation of gene expression in trans.
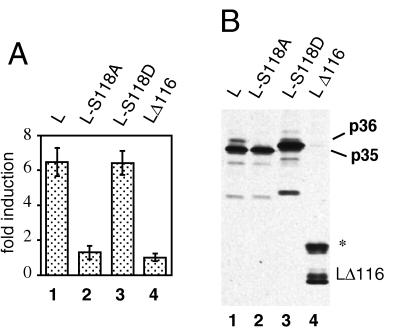
Pre-S transactivation has been extensively studied in HBV, and a segment essentially corresponding to the pre-S2 subdomain has been identified in transient-transfection experiments as the minimal transactivating unit (24, 25). To test whether the L protein of DHBV carried a similar transactivator activity, HepG2 cells, a human hepatoma cell line, were transfected with plasmids expressing different L protein variants (Fig. 6A) or a control plasmid without L insert, together with a CAT reporter construct used previously in transactivation assays with the HBV L protein (p3xAP1-CAT [24]). At 2 days posttransfection, cells were lysed and the cytosolic fraction was used to determine CAT activities. Wild-type DHBV L protein was found to activate expression of the reporter gene up to sevenfold (Fig. 6A, lane 1) relative to a DHBV L deletion mutant lacking two-thirds of the pre-S domain (LΔ116) (Fig. 6A, lane 4), indicating that an intact pre-S domain is a prerequisite for a transactivation function of DHBV L. In other CAT assays with transfected LMH cells, up to 10-fold transactivation was observed relative to the vector plasmid without DHBV insert (not shown). To examine whether L transactivation was linked to a cytosolic orientation of the pre-S domain, we included a construct which results in cotranslational translocation of the pre-S domain into the lumen of the endoplasmic reticulum through an N-terminally fused signal sequence (sigL1 [55]). As expected, the lack of cytosolic pre-S domains resulted in complete loss of transactivating activity (data not shown).
FIG. 6.
Serine 118 is essential for transactivating function of the DHBV L protein. (A) CAT assays. HepG2 cells were cotransfected with expression plasmids for DHBV16 L proteins and a 3xAP 1-driven CAT reporter construct. CAT activity was measured in cell lysates prepared at 2 days posttransfection. Bars represent mean values with standard deviations from four transfections. Fold inductions were calculated relative to the value for the pre-S deletion mutant LΔ116. (B) Comparable expression of the different L proteins was confirmed by Western blotting of immunoprecipitated L proteins. LΔ116, 22 kDa; ∗, glycosylated LΔ116 protein (55).
We next examined whether the L transactivation function was related to phosphorylation at serine 118 by testing L protein mutations abolishing and functionally mimicking phosphorylation (S118A and S118D, respectively) in the transactivation assay. Transactivation was completely lost with the S118A mutant (Fig. 6A, lane 2), whereas the transactivation potential of L was conserved after replacement of serine 118 with aspartate (Fig. 6A, lane 3).
To exclude the possibility that the changes observed were related to potential differences in L protein expression and/or protein stability, the L proteins were visualized by Western blotting and were found to be expressed at comparable levels (Fig. 6B). The transactivating activity of L thus appears to depend on the presence of a negative charge at position 118 (provided by either phosphoserine or the substituting aspartate). The low level of phospho-L in the wild-type protein (Fig. 6B, lane 1, p36) does not contradict this interpretation if we assume that a lower transactivation potential of L-S118D was compensated for by the higher number of transactivating protein molecules. This hypothesis is supported by observations with other phosphoproteins (for example, MAP kinase kinase-1 [1]) indicating that comparable substitutions may only partially mimic the activating effect of a phosphorylated amino acid. Furthermore, the amounts of transactivating proteins and the levels of their transactivating activities do not necessarily correlate directly (14). Nevertheless, although not quantitatively interpretable, the apparent equivalence of transactivation by wild-type DHBV L and L-S118D is an important complement to the loss of transactivation function by the alanine substitution.
Mutations in DHBV L that eliminate or mimic phosphorylation at serine 118 do not significantly influence viral replication.
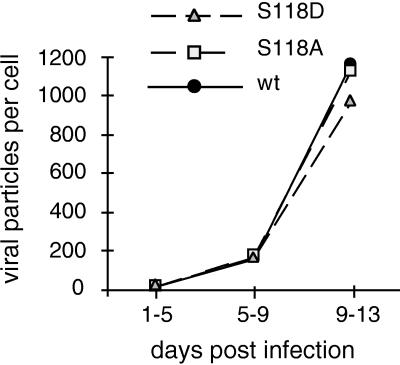
To test whether mutational changes at serine 118 which affect L protein phosphorylation and transactivation corresponded to a phenotype in virus replication, we introduced the S118A and the S118D mutations into a cloned replication-competent, overlength DHBV genome. These constructs were then transfected into LMH cells to produce DHBV particles, which were subsequently assayed for their competence to initiate the complete viral replication cycle in PDHs or in infected ducklings. In these experiments, transfected LMH cells produced virions from S118A-DHBV or from S118D-DHBV with yields comparable to those obtained with wild-type DHBV, thus demonstrating that phosphorylation was not of central importance for virus formation and maturation or any other late step in the viral life cycle. In DHBV-infected PDHs, again no differences in infection competence were observed between wild-type virus and either of the mutant viruses as judged from the amounts of DHBV DNA secreted into the tissue culture medium (Fig. 7) or by virus cell-to-cell spread as visualized by immunofluorescence staining at day 4 or 9 post infection (data not shown). Replication kinetics were also tested by quantifying total intracellular virus DNA in PDH lysates (between days 1 and 17 post infection), and again no differences exceeding a factor of 2 were observed (data not shown). We therefore conclude that mutations affecting phosphorylation at serine 118 have no significant effects on viral infection and replication in the cell culture system.
FIG. 7.
Mutations at serine 118 do not significantly influence viral replication. Primary duck hepatocytes were infected with wild-type (wt) DHBV or variant viruses (carrying L-S118A or L-S118D) produced from LMH cells. The viral DNA contents in tissue culture supernatants collected at the indicated time intervals were determined by dot blot analysis relative to a DHBV DNA standard by using a PhosphorImager. Each point is the average of values from two wells.
To test the situation in vivo, ducklings were infected 1 day after hatching with wild-type or mutant virus produced from LMH cells. As judged from serum DHBV DNA titers determined before and at several time points after infection, all animals developed titers of between 2 × 109 and 5 × 1010 DNA-containing virus particles per ml at 1 to 2 weeks after infection. Serum viremia varied substantially between individual test animals, but these differences in virus titer and kinetics were not reproducibly related to the genotype of the infecting virus. These results, like those from the infection studies with PDHs presented above, thus indicate that phosphorylation at serine 118 is not of critical importance for viral replication.
DISCUSSION
In this study we show that a hepadnavirus envelope protein with functions in virus morphogenesis and cellular entry can serve an additional regulatory function through phosphorylation-mediated cross talk influencing gene expression. The results reported here for the DHBV L protein support and extend earlier observations with HBV envelope proteins indicating that cytosolically exposed pre-S domains possessed the potential to activate gene expression from cellular promoter elements in trans (24, 25); this further suggests that this unconventional property is evolutionarily conserved despite the fact that there is only fragmentary amino acid sequence identity between the pre-S domains of avian and mammalian hepadnaviruses (53).
Other known examples of transactivation activities of structural viral proteins are the influenza virus hemagglutinin (40) and the large hepatitis delta antigen (58). In contrast to DHBV L, however, there is no evidence that these transactivating activities can be modulated in response to extracellular stimuli. Our studies on DHBV L further indicate that this modulation is mediated by phosphorylation at a unique serine residue: L protein with the mutation S118A completely lost transactivation activity, whereas the protein with the S118D mutation transactivated an AP1 promoter-linked indicator gene comparably to the (partly phosphorylated) wild-type protein (Fig. 6). These results strongly suggest that a negative charge in position 118 (provided by either phosphoserine or the substituting aspartate) is essential for the transactivating activity of DHBV pre-S.
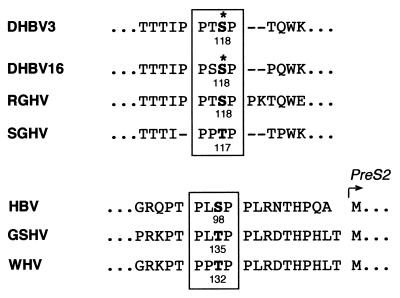
Phosphorylation at serine 118 results in altered migration in SDS-PAGE (Fig. 3). As noted by others, this was not the case for several additional, minor pre-S phosphorylation sites, which were detectable only after 33P labeling of mutants and two-dimensional gel electrophoresis (20). Therefore, serine 118 appears to be situated at a structurally distinguished position in the DHBV pre-S sequence. A high density of proline residues, as well as a substantial variability (including amino acid insertions and deletions in closely related avian viruses [Fig. 8] and, particularly, a 7-amino-acid insertion in the more distantly related heron hepatitis B virus [52]), is characteristic of the amino acid sequence surrounding this site in the L proteins of avian hepadnaviruses. This suggests that serine 118 may be part of a hinge element separating the receptor-binding domain (mapped to pre-S amino acids 30 to 115 [57]) from the C-terminal part of DHBV pre-S, for which mutational analysis and peptide scanning data indicate that it is required for interaction with the cytosolic nucleocapsid during morphogenesis (22, 33). Thus, phosphorylation at serine 118 may conceivably trigger structural alterations which directly or indirectly modulate structural and nonstructural pre-S functions (such as transactivation), possibly through homomeric or heteromeric pre-S interactions, as is known to be the case in other regulatory proteins.
FIG. 8.
DHBV L phosphorylation occurs at a MAP kinase recognition motif which is conserved in the pre-S domains of avian and mammalian hepadnaviruses. The motif is boxed. Phosphorylation target sites within the PX(S/T)P motif are indicated in boldface. The experimentally determined phosphorylation site of DHBV L is marked with an asterisk. RGHV, Ross’s goose hepatitis B virus (38); SGHV, snow goose hepatitis B virus (59); GSHV, ground squirrel hepatitis B virus (50); WHV, woodchuck hepatitis B virus (16). The HBV sequence shown is derived from subtype ayw (17). The MAP kinase motif is not conserved in the homologous sequence of heron hepatitis B virus, another avian hepadnavirus (52).
Of particular interest is the novel finding that L phosphorylation, and therefore most likely also L-mediated transactivation, is modulated in response to intra- or extracellular signals. The levels of the phosphorylated species, while barely detectable under physiological conditions in the liver, increased rapidly when the hepatocytes were liberated during liver perfusion, and L phosphorylation was further enhanced by a variety of specific stimuli (Fig. 5). Several lines of evidence indicate that this modulation of L phosphorylation, and of phospho-L-mediated signaling, involves MAP kinases of the ERK-type (7, 35): (i) the phosphorylation site, serine 118, is part of the ERK consensus target sequence PX(S/T)P (Fig. 8); (ii) recombinant ERK2 phosphorylated wild-type L protein but not the mutant S118A in vitro; (iii) L protein phosphorylation was inhibited in transfected cells by coexpression of CL100, a phosphatase specifically inactivating MAP kinases; (iv) the induction pattern observed for phospho-L (p36) in DHBV-infected PDHs matched that of ERK-type MAP kinases with respect to the positive response to proliferative stimuli such as treatment with TPA or liberation of the hepatocytes from the liver tissue; and (v) induction of L phosphorylation was closely correlated with ERK activation in DHBV-infected PDHs (Fig. 5B). Although predominantly in the cell nucleus, activated ERK protein kinases are initially cytosolic (10), and therefore, even membrane-associated pre-S should be accessible for this type of kinase. Our data thus support a model in which L-mediated modulation of gene expression varies in response to specific extracellular stimuli through serine 118 phosphorylation by ERK-type MAP kinases.
The enhancement of gene expression observed in cotransfection with an AP-1-driven reporter construct (up to 10-fold induction) was in a range similar to that observed in analogous experiments characterizing the transactivation activity of the HBV L protein (24) or of the HBV X protein (46). It is thus conceivable that DHBV L initiates signaling via the Ras-Raf-MAP kinase cascade from a cytoplasmic location, as has been proposed for activation of gene expression by HBV X and HBV L (see, e.g., references 13 and 24). Two other mechanisms also discussed for the HBV transactivators appear to be less likely: nuclear localization and direct promoter activation, as proposed for the X protein (9, 13), would require the release of the L transactivator protein from its membrane-anchored state. Alternatively, a mechanism involving oxidative stress due to accumulation of overexpressed viral membrane proteins in the endoplasmic reticulum of transfected cells, as postulated to explain the transactivation potential of truncated HBV envelope proteins and of the influenza virus hemagglutinin (36, 40), is highly unlikely for DHBV pre-S in view of our finding that transactivation was abolished by a single amino acid exchange without L protein expression levels being affected.
In the context of this discussion, it may be recalled that there is, despite extensive efforts, still no conclusive model reconciling rather conflicting data on the mechanisms involved in HBV X transactivation and on the primary targets of this apparently pleiotropically acting protein. In addition to experimental difficulties, this probably relates to the fact that research on HBV X (as well as HBV L) function has so far been mainly aimed at finding a potential link to HBV-associated carcinogenesis, while no major studies have been undertaken to elucidate the role by which the HBV transactivators contribute to the establishment and/or maintenance of infection by mammalian hepadnaviruses. As exemplified by this study, DHBV offers better opportunities to investigate hepadnavirus transactivator function also in infection experiments, and furthermore, it encodes, to our present knowledge, only a single transactivator protein. Information gained from this avian system may therefore also contribute to a better understanding of analogous mechanisms in HBV: for example, it will be of interest to determine whether the HBV L protein is phosphorylated in the pre-S domain at a MAP kinase recognition motif analogous to the one analyzed here for DHBV and conserved between mammalian hepadnaviruses; this predicted target site (serine 98 in the HBV ayw subtype [Fig. 8]), however, lies outside the HBV pre-S region determined previously to specify transactivation (essentially the pre-S2 subdomain starting at amino acid residue 109 [25]).
L phosphorylation and the associated transactivation function are apparently not essential for the basic viral life cycle. Cell culture and in vivo experiments showed no significant influence on virus replication of the mutation inactivating the phosphorylation site (S118A) (Fig. 7), in agreement with data from two other studies identifying this site by mutational analysis (4, 20). Furthermore, no revertants to the wild-type pre-S sequence were detected upon serial passage in test animals. Likewise, no significant effects of the S118D mutation on virus replication were observed, except for a selective disadvantage of S118D-DHBV in direct competition with wild-type virus in mixed infections in vivo (47). This lack of major effects with respect to virus replication is, however, in contrast to the appearance of a distinct pathogenic phenotype with severe growth retardation and pathologic liver histology in S118D-DHBV-infected animals (47). Given the low level of L phosphorylation in the steady-state liver, such a change to a pathogenic phenotype caused by a mutation mimicking constitutive phosphorylation supports the hypothesis that L phosphorylation and transactivation provide a subtle regulatory device mediating intracellular host-virus cross talk in response to an unbalanced state of the cell.
ACKNOWLEDGMENTS
We thank Bärbel Glass for preparation of PDHs and virus stocks, Armin Bosserhoff and Rainer Frank for advice on peptide analysis, Hans Will for providing unpublished sequence information from new isolates of avian hepadnaviruses, Elizabeth Grgacic and David Anderson for the exchange of unpublished data, Karin Coutinho for expert editorial assistance, and Elisabeth Grgacic for critically reading the manuscript during revision.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 229) and by Fonds der Chemischen Industrie.
REFERENCES
- 1.Alessi D R, Saito Y, Campbell D G, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall C J, Cowley S. Identification of the sites in MAP kinase kinase-phosphorylated by p74raf-1. EMBO J. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alessi D R, Smythe C, Keyse S M. The human CL100 gene encodes a Tyr/Thr-protein phosphatase which potently and specifically inactivates MAP kinase and suppresses its activation by oncogenic ras in Xenopus oocyte extracts. Oncogene. 1993;8:2015–2020. [PubMed] [Google Scholar]
- 3.Annan R S, Carr S A. Phosphopeptide analysis by matrix-assisted laser desorption time-of-flight mass spectrometry. Anal Chem. 1996;68:3413–3421. doi: 10.1021/ac960221g. [DOI] [PubMed] [Google Scholar]
- 4.Borel C, Sunyach C, Hantz O, Trepo C, Kay A. Phosphorylation of DHBV pre-S: identification of the major site of phosphorylation and effects of mutations on the virus life cycle. Virology. 1998;242:90–98. doi: 10.1006/viro.1997.9004. [DOI] [PubMed] [Google Scholar]
- 5.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 6.Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350–9357. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cano E, Mahadevan L C. Parallel signal processing among mammalian MAPKs. Trends Biochem Sci. 1995;20:117–122. doi: 10.1016/s0968-0004(00)88978-1. [DOI] [PubMed] [Google Scholar]
- 8.Cavanaugh V J, Guidotti L G, Chisari V C. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infection in transgenic mice. J Virol. 1998;72:2630–2637. doi: 10.1128/jvi.72.4.2630-2637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheong J H, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen P. The search for physiological substrates of MAP and SAP kinases in mammalian cells. Trends Cell Biol. 1997;7:353–361. doi: 10.1016/S0962-8924(97)01105-7. [DOI] [PubMed] [Google Scholar]
- 11.Condreay L D, Aldrich C E, Coates L, Mason W S, Wu T T. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J Virol. 1990;64:3249–3258. doi: 10.1128/jvi.64.7.3249-3258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis R J. The mitogen-activated protein kinase signal transduction pathway. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 13.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faktor O, Shaul Y. The identification of hepatitis B virus X gene responsive elements reveals functional similarity of X and HTLV-I tax. Oncogene. 1990;5:867–872. [PubMed] [Google Scholar]
- 15.Fernholz D, Wildner G, Will H. Minor envelope proteins of duck hepatitis B virus are initiated at internal pre-S AUG codons but are not essential for infectivity. Virology. 1993;197:64–73. doi: 10.1006/viro.1993.1567. [DOI] [PubMed] [Google Scholar]
- 16.Galibert F, Chen T N, Mandart E. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: comparison with the hepatitis B virus sequence. J Virol. 1982;41:51–65. doi: 10.1128/jvi.41.1.51-65.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 18.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Field’s virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 19.Grgacic E V, Anderson D A. The large surface protein of duck hepatitis B virus is phosphorylated in the pre-S domain. J Virol. 1994;68:7344–7350. doi: 10.1128/jvi.68.11.7344-7350.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grgacic E V L, Lin B, Gazina E V, Snooks M J, Anderson D A. Normal phosphorylation of duck hepatitis B virus L protein is dispensable for infectivity. J Gen Virol. 1998;79:2743–2751. doi: 10.1099/0022-1317-79-11-2743. [DOI] [PubMed] [Google Scholar]
- 21.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hild M. Zelluläre Funktionen während der frühen und späten Schritte im Infektionszyklus des Enten Hepatitis B Virus. Ph.D. thesis. Heidelberg, Germany: University of Heidelberg; 1997. [Google Scholar]
- 23.Hild M, Weber O, Schaller H. Glucagon treatment interferes with an early step of duck hepatitis B infection. J Virol. 1998;72:2600–2606. doi: 10.1128/jvi.72.4.2600-2606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildt E, Saher G, Bruss V, Hofschneider P H. The hepatitis B virus large surface protein (LHBs) is a transcriptional activator. Virology. 1996;225:235–239. doi: 10.1006/viro.1996.0594. [DOI] [PubMed] [Google Scholar]
- 25.Hildt E, Urban S, Hofschneider P H. Characterization of essential domains for the functionality of the MHBst transcriptional activator and identification of a minimal MHBst activator. Oncogene. 1995;11:2055–2066. [PubMed] [Google Scholar]
- 26.Kekule A S, Lauer U, Meyer M, Caselmann W H, Hofschneider P H, Koshy R. The preS2/S region of integrated hepatitis B virus DNA encodes a transcriptional transactivator. Nature. 1990;343:457–461. doi: 10.1038/343457a0. [DOI] [PubMed] [Google Scholar]
- 27.Kekule A S, Lauer U, Weiss L, Luber B, Hofschneider P H. Hepatitis B virus transactivator HBx uses a tumour promoter signalling pathway. Nature. 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 28.Kemp B E, Pearson R B. Protein kinase recognition sequence motifs. Trends Biochem Sci. 1990;15:342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- 29.Keyse S M. An emerging family of dual specificity MAP kinase phosphatases. Biochim Biophys Acta. 1995;1265:152–160. doi: 10.1016/0167-4889(94)00211-v. [DOI] [PubMed] [Google Scholar]
- 30.Klingmüller U, Schaller H. Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J Virol. 1993;67:7414–7422. doi: 10.1128/jvi.67.12.7414-7422.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishna R G, Wold F. Post-translational modifications of proteins. In: Imahori K, Sakiyma F, editors. Methods in protein sequence analysis. New York, N.Y: Plenum Press; 1993. pp. 167–172. [Google Scholar]
- 32.Kyriakis J M, Avruch J. Sounding the alarm: protein kinase cascades activated by stress and inflammation. J Biol Chem. 1996;271:24313–24316. doi: 10.1074/jbc.271.40.24313. [DOI] [PubMed] [Google Scholar]
- 33.Lenhoff R J, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandart E, Kay A, Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984;49:782–7892. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marais R, Marshall C J. Control of the ERK MAP kinase cascade by Ras and Raf. Cancer Surv. 1996;27:101–125. [PubMed] [Google Scholar]
- 36.Meyer M, Caselmann W H, Schluter V, Schreck R, Hofschneider P H, Baeuerle P A. Hepatitis B virus transactivator MHBst: activation of NF-kappa B, selective inhibition by antioxidants and integral membrane localization. EMBO J. 1992;11:2991–3001. doi: 10.1002/j.1460-2075.1992.tb05369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hep. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 38.Newbold, J. E., et al. GenBank accession number M95589.
- 39.Obert S, Zachmann Brand B, Deindl E, Tucker W, Bartenschlager R, Schaller H. A spliced hepadnavirus RNA that is essential for virus replication. EMBO J. 1996;15:2565–2574. [PMC free article] [PubMed] [Google Scholar]
- 40.Pahl H L, Baeuerle P A. Expression of influenza virus hemagglutinin activates transcription factor NF-kappa B. J Virol. 1995;69:1480–1484. doi: 10.1128/jvi.69.3.1480-1484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Protzer, U., U. Klöcker, and H. Schaller. Unpublished data.
- 42.Pugh J C, Sninsky J J, Summers J W, Schaeffer E. Characterization of a pre-S polypeptide on the surfaces of infectious avian hepadnavirus particles. J Virol. 1987;61:1384–1390. doi: 10.1128/jvi.61.5.1384-1390.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radler-Pohl A, Sachsenmaier C, Gebel S, Auer H P, Bruder J T, Rapp U, Angel P, Rahmsdorf H J, Herrlich P. UV-induced activation of AP-1 involves obligatory extranuclear steps including Raf-1 kinase. EMBO J. 1993;12:1005–1012. doi: 10.1002/j.1460-2075.1993.tb05741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rigg R J, Challer H S. Duck hepatitis B virus infection of hepatocytes is not dependent on low pH. J Virol. 1992;66:2829–2836. doi: 10.1128/jvi.66.5.2829-2836.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossner M T. Hepatitis B virus X-gene product: a promiscuous transcriptional activator. J Med Virol. 1992;36:101–117. doi: 10.1002/jmv.1890360207. [DOI] [PubMed] [Google Scholar]
- 47.Rothmann, K., W. J. Hofmann, B. Glass, and H. Schaller. 1998. Unpublished data.
- 48.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 49.Schlicht H J, Kuhn C, Guhr B, Mattaliano R J, Schaller H. Biochemical and immunological characterization of the duck hepatitis B virus envelope proteins. J Virol. 1987;61:2280–2285. doi: 10.1128/jvi.61.7.2280-2285.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seeger C, Ganem D, Varmus H E. Nucleotide sequence of an infectious molecularly cloned genome of ground squirrel hepatitis virus. J Virol. 1984;51:367–375. doi: 10.1128/jvi.51.2.367-375.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Songyang Z, Lu K P, Kwon Y T, Tsai L H, Filhol O, Cochet C, Brickey D A, Soderling T R, Bartleson C, Graves D J, DeMaggio A J, Hoekstra M F, Blenis J, Hutner T, Cantley L C. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sprengel R, Kaleta E F, Will H. Isolation and characterization of a hepatitis B virus endemic in herons. J Virol. 1988;62:3832–3839. doi: 10.1128/jvi.62.10.3832-3839.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sprengel R, Kuhn C, Will H, Schaller H. Comparative sequence analysis of duck and human hepatitis B virus genomes. J Med Virol. 1985;15:323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- 54.Summers J, Smith P M, Huang M J, Yu M S. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swameye I, Schaller H. Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation. J Virol. 1997;71:9434–9441. doi: 10.1128/jvi.71.12.9434-9441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuttleman J S, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 57.Urban S, Breiner K M, Fehler F, Klingmüller U, Schaller H. Avian hepatitis B virus infection is initiated by the interaction of a distinct pre-S subdomain with the cellular receptor gp180. J Virol. 1998;72:8089–8097. doi: 10.1128/jvi.72.10.8089-8097.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei Y, Ganem D. Activation of heterologous gene expression by the large isoform of the hepatitis delta antigen. J Virol. 1998;72:2089–2096. doi: 10.1128/jvi.72.3.2089-2096.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Will, H., H. J. Netter, and S. F. Chang. Personal communication.
- 60.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]