Abstract
High-grade glioma (HGG) is the most common cause of cancer death in children and the most common primary central nervous system tumor in adults. While pediatric HGG was once thought to be biologically similar to the adult form of disease, research has shown these malignancies to be significantly molecularly distinct, necessitating distinct approaches to their clinical management. However, emerging data have shown shared molecular events in pediatric and adult HGG including the histone H3K27M mutation. This somatic missense mutation occurs in genes encoding one of two isoforms of the Histone H3 protein, H3F3A (H3.3), or HIST1H3B (H3.1), and is detected in up to 80% of pediatric diffuse midline gliomas and in up to 60% of adult diffuse gliomas. Importantly, the H3K27M mutation is associated with poorer overall survival and response to therapy compared to patients with H3 wild-type tumors. Here, we review the clinical features and biological underpinnings of pediatric and adult H3K27M mutant glioma, offering a groundwork for understanding current research and clinical approaches for the care of patients suffering with this challenging disease.
Keywords: epigenetics, glioma, histone, H3K27M, mutant
Graphical Abstract
Graphical Abstract.
Key Points.
Shared molecular events in pediatric and adult high-grade glioma include the histone H3K27M mutation.
The H3K27M mutation is associated with distinct tumor molecular biology and clinical outcomes.
The H3K27M mutation results in an altered 3D chromatin structure and transcription regulation.
H3K27M mutant glioma cell lineages gather additional mutations.
H3K27M Mutation in Glioma
Historically, intracranial diffuse midline gliomas (DMGs) arising in the brainstem were diagnosed radiographically alone, given their pathognomonic appearance on MRI scans and the relative risk of surgical biopsy for histologic analysis (Figure 1).1 This led to a paucity of tumor tissue for molecular testing, impeding progress in understanding disease biology, and hence in identifying effective therapies. As a result, DMGs persist as the number one cause of pediatric cancer death. Unfortunately, pediatric high-grade gliomas (HGGs) in the brainstem represent approximately 10% of all pediatric brain tumors, with 80% of these located in the pons, and are associated with a dismal prognosis: median survival is just 9 months, while virtually all patients die within 2 years of diagnosis.2,3 Standard clinical management for pontine glioma consists of radiation therapy, which results in a brief period of symptom relief (2–4 months), however, does not increase the overall survival of these patients. Fortunately, recent advancements in neurosurgical techniques, including the widespread application of image-guided stereotactic biopsy capability, have led to increased access to tumor tissue for molecular analysis. Indeed, biopsy of thalamic and brainstem lesions can now be performed with a less than 5% risk of transient neurologic deficit, and a less than 1% risk of permanent disability.4,5
Figure 1.
T2 Sagittal (left) and axial (right) MR images of the brain, revealing a diffuse midline glioma (DMG) in the pons, also known as diffuse intrinsic pontine glioma (DIPG, white arrows).
Analysis of these newfound tumor tissues led to the discovery of the H3K27M mutation, first reported in DMGs in 2012 in 2 independent publications.6,7 This somatic missense mutation results in a Lys27 to Met substitution on the N-terminal tail of the Histone H3 protein. The H3K27 residue plays a central and important role in cellular epigenomic regulation. The H3K27 residue is typically acetylated (Ac) or trimethylated (me3), which is generally associated with transcription expression and repression, respectively. Further, the H3K27M mutant histone functions as a dominant negative inhibitor of the polycomb repressor complex 2 (PRC2), which deposits methylation marks on the H3K27 residue. PRC2 inhibition results in a global loss of H3K27me3 and corresponding upregulation of many genes that may be responsible for activation of oncogenic pathways.8–10 Subsequent research over the past 10 years has revealed that H3K27M mutant glioma is clinically and molecularly distinct from the H3K27 wild-type (H3K27WT) gliomas. For example, multiple reports demonstrate distinct protein, gene, and DNA methylation profiles, as well as unique epigenetic characteristics and chromatin 3D structure in the presence of the H3K27M mutation.11,12 In addition, H3K27M mutant tumors tend to harbor additional somatic mutations and exhibit a unique immunologic phenotype, as detailed in the sections below.
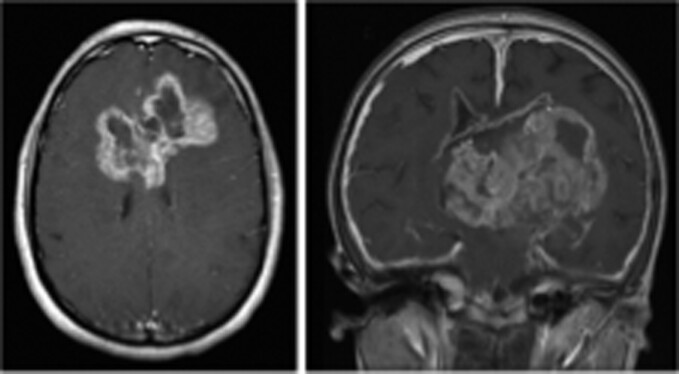
As with the pediatric disease, adults with H3K27M mutant glioma exhibit higher mortality than those harboring H3K27WT tumors. Further, while the majority of H3K27M mutant gliomas are diffuse midline lesions, the H3K27M mutation is occasionally found in circumscribed gliomas and in tumors in nonmidline locations (ie, cerebral hemispheres, Figure 2). As in diffuse and midline gliomas, the H3K27M mutation in hemispheric and circumscribed gliomas is associated with a poorer prognosis; however, circumscribed glioma patients exhibit longer overall survival relative to those with diffuse lesions, likely due to feasibility (ie, extent) of surgical resection.13 The presence of H3K27M mutation in both adult and pediatric glioma is associated with an inferior prognosis, regardless of the histological grade of the lesion.14 As a result, H3K27M mutant tumors are now conferred a Grade IV assignment in the 2021 World Health Organization Classification of CNS tumors, irrespective of tissue histological features.15,16 In turn, clinical H3K27me3 testing via tissue immunohistochemistry, or next-generation sequencing for H3K27M, has become routine for diffuse and histologically HGGs at advanced medical centers across the world. Additionally, H3K27WT gliomas frequently overexpress enhancer of zeste interacting protein (EZHIP), a PRC2 inhibitor, to recapitulate the loss of H3K27me3 and gain of H3K27ac observed in H3K27M mutant tumors.17–21 Together, these are defined as “H3K27-altered” gliomas. Here, however, we focus on the epigenetic, immunological, and genetic features specifically observed in H3K27M mutant gliomas, introducing their downstream mechanisms to contextualize emerging findings in this more common disease entity.
Figure 2.
Postcontrast T1 axial (right) and coronal (left) MR images of the brain, revealing a circumscribed high-grade glioma spanning bilateral cerebral hemispheres (white arrows).
Epigenetic Features of H3K27M Mutant Glioma
H3K27M mutant gliomas are characterized by several distinct epigenetic features, including decreased H3K27me3 and increased H3K27ac, which facilitate a unique transcriptional profile (Figure 3). The mechanisms by which this effect is mediated underscores the complex effects of the H3K27M mutation on chromatin structure and function. For example, polycomb repressor complex 2 (PRC2), a principal writer of facultative heterochromatin, is responsible for catalyzation of H3K27 methylation and transcription repression. PRC2 is composed of enhancer of zeste 1/2 (EZH1/2), embryonic ectoderm development, suppressor of zeste 12 (SUZ12), and retinoblastoma-binding protein p48.22 PRC2 is recruited to CpG islands in both healthy and H3K27M mutant cells, but fails to spread H3K27me3 in the presence of the H3K27M mutant protein, potentially defined by boundaries of elevated H3K36me2 deposition.23–27 EZH1/2 is the catalytic domain of PRC2, blocked by the methionine of H3K27M oncohistones, and likely remains inhibited even after dissociation with H3K27M.24,26,28
Figure 3.
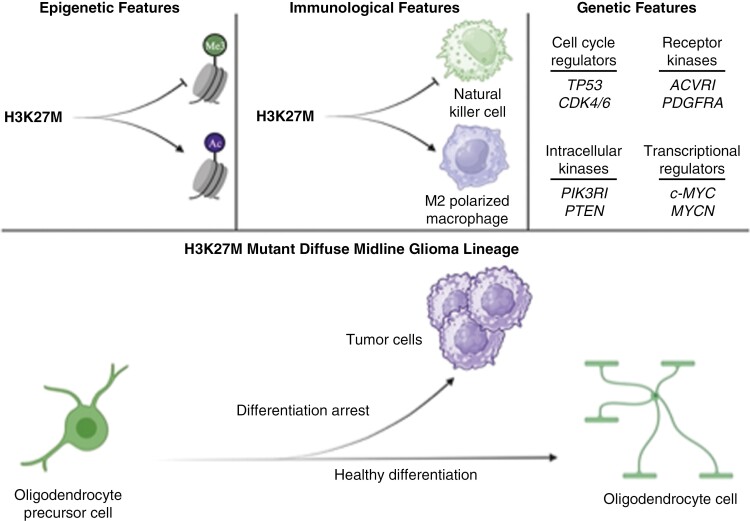
Major biological underpinnings and cell of origin in H3K27M mutant diffuse midline glioma. Unique epigenetic, immunological, and genetic alterations are observed in this disease entity, with their cumulative effect being an arrest of oligodendrocyte precursor cell differentiation to facilitate tumor cell formation, maintenance, and progression. Created with BioRender.com.
As mentioned, global loss of H3K27me3 in H3K27M mutant glioma is associated with an increase in H3K27ac, a transcriptional activating mark.29–32 Genomic enrichment with H3K27ac further increases chromatin accessibility to transcriptional machinery, including RNA Polymerase II (RNAPII) via formation of H3K27M-K27ac heterotypic nucleosomes, which recruit the histone acetyl group readers bromodomain-containing proteins 2 and 4 (BRD2/4).9,32,33 Further, H3K27M and BRD2/4 are preferentially colocalized into super-enhancer and promoter regions, facilitating transcription.32 One H3K27M-driven promoter is that of AFF4, the scaffolding subunit of the super-enhancer complex, which employs cyclin-dependent kinase 9 (CDK9) to catalyze RNAPII phosphorylation.34 This releases RNAPII from promoter-proximal pausing to increase transcription of several oncogenes, including MYC and MCL-1.35
While global de-repression of PRC2 target loci is observed in H3K27M mutant glioma to both directly and indirectly promote transcription, there are notable exceptions to this rule, with certain preexisting polycomb target sites actually receiving more PRC2-mediated transcription repression.29,36 For example, the CDKN2A locus is a likely target of this residual PRC2 activity, with observed increased H3K27me3 enrichment and decreased transcription.37–39 This is similarly true for many neuronal differentiation genes, thereby promoting a de-differentiated tumor phenotype in the presence of the H3K27M mutant protein.32,40–42
Indeed, DMGs are thought to arise from the oligodendrocyte precursor cell (OPC), most commonly during the ventral pontine expansion characteristic of middle childhood (Figure 3).43–48 Specifically, the presence of the H3K27M mutant protein promotes a neurodevelopmentally defined transcriptional profile that arrests OPCs’ differentiation into oligodendrocytes (OLs).11,49 OLs are the mature, myelinating, postmitotic, and postmigratory cells of the central nervous system (CNS).50 This offers an important explanation into the cycling and migratory capacities of the DMG lineage as well as a potential modality of differentiation therapy. Indeed, agents that re-initiate differentiation reprogram this tumor lineage’s transcriptional profile toward one more characteristic of OLs or astrocytes, and it remains an exciting avenue for further investigation.32,34,51–57
Partner mutations then arise that are secondary to this founding histone mutation and augment these transcriptional changes to accelerate gliomagenesis (Figure 3).29,58–64 For example, an H3.1K27M cosegregating gain-of-function mutation in ACVR1 drives SMAD-mediated transcription to enhance proliferation and migration; similarly, an H3.3K27M cosegregating amplification in PDGFRA drives ERK-mediated transcription for the same phenotypic outcome.65,66 In addition to these epi(genetic) alterations observed in H3K27M mutant glioma, notable efforts have been recently made to further stratify OPC-like DMG cells based on tumor location, with less differentiated lineages observed more frequently in pontine tumors and more differentiated lineages observed in thalamic tumors.45 This important finding encourages an evaluation of both molecular and spatial characteristics in the diagnostic workflow.
Immunological Features of H3K27M Mutant Glioma
Oncoimmunology is an expanding field that promises a novel therapeutic strategy for achieving more effective clinical outcomes in cancer by utilizing the body’s innate immune response to target tumorigenesis. Meanwhile, advances in multiple sclerosis research have driven discoveries on immune modulation of the OL lineage, suggesting an applicability of this modality to OL-derived HGGs such as DMG.67 Along with multiple other solid tumors, HGGs, including those harboring the H3K27M mutation, exhibit a unique inflammatory expression profile that researchers have characterized in recent years and is the basis for a multitude of clinical trials. H3K27M mutant gliomas are characterized by distinct immunological features, including decreased natural killer (NK) cells and potentially increased macrophage 2 (M2) polarization, which engender a principally “immunocold” state, though tumor location appears to play a role in their oncoimmunologic profile (Figure 3). In general, diffuse midline H3K27M mutant gliomas, including diffuse intrinsic pontine gliomas (DIPGs), have a less inflammatory expression profile than hemispheric HGGs, due to low immune infiltration and immune checkpoint molecule expression.68,69
Myelin-associated CD4+ T cells harboring NK receptors target OLs in vitro.70 Indeed, NK cells lyse DIPG cell cultures but are decreased in patients with DIPG.71,72 Although hemispheric HGGs have increased macrophage infiltration, DIPGs do not.71,73 H3K27M mutant glioma cells do not show repolarization of macrophages, which is observed in H3K27WT HGG.71 In addition, only one-quarter of H3K27M mutant glioma cells express programmed death-ligand 1 (PD-L1), a checkpoint immunosuppressor expressed in OLs that serves as a biomarker of immunotherapeutic efficacy.74,75 Another study corroborated with this finding, concluding that DIPG cells do not show overexpression of PD-L1 nor transforming growth factor-β1 (TGF-β1), another regulator of inflammatory responses.71 Moreover, the CD45+ leukocyte compartment of DIPGs contains more CD11b+ macrophages than CD3+ T-lymphocytes, an anti-inflammatory M2 phenotypic signature that could lead to immune suppression and tumor progression.68,76,77 Therapeutic strategies targeting these characteristics are now the basis of multiple clinical trials. However, M2-derived activin-A drives OPC differentiation in CNS remyelination, potentially offering a vulnerability of the tumor to an immunologically based differentiation therapy.78 Looking into the future, there may be a nonsynergistic relationship between immune-activating immunotherapies and this new differentiation therapy, cautioning their combination or at least necessitating their carefully timed application through the clinical course.
Genetic Features in H3K27M Mutant Glioma
Although H3K27M mutations are common in DMG and have a critical role in regulating gene expression and maintaining chromosomal stability, additional associated mutations unique to each patient contribute to tumor development, resulting in varying degrees of poor prognosis.79,80 The mutation patterns associated with H3.3K27M and H3.1K27M tumors differ. For example, TP53 gene mutations are more commonly observed in H3.3K27M tumors, and they are believed to promote tumor development and progression by disrupting p53 protein function.81 In contrast, H3.1K27M tumors frequently partner with ACVR1 gene mutations, characterized by a substitution of arginine for glycine at position 34 (G34R) in the ACVR1 protein. These mutations are associated with enhanced activation of the bone morphogenic protein (BMP) signaling pathway and increased tumor growth.
To develop effective treatments for H3K27M mutant glioma, it is crucial to understand the genetic landscape of the disease, including recurring somatic mutations and related pathways, as well as the influence of subclonal populations on tumor biology and response to therapy. Advanced technologies, such as single-cell sequencing and spatial transcriptomics, are being used to study somatic heterogeneity at a higher resolution, potentially leading to the discovery of new therapeutic targets.
Tumor Protein p53
The p53 protein encoded by the TP53 gene is a key tumor suppressor protein that helps to maintain the genomic stability of cells.82 Mutations in several phosphorylation sites of p53 can lead to loss of both DNA-binding capacity and protein stability. The loss of p53 function due to mutations in the TP53 gene can lead to genomic instability, uncontrolled cell division, and decreased apoptosis. This can result in the accumulation of mutant p53 protein and a decrease in the activity of WT p53, leading to tumorigenesis and cancer progression. Moreover, p53 is a necessary mediator of OPC differentiation in healthy systems and exerts a tumor-suppressive effect in glioma by inhibiting OPC proliferation, a principle step in differentiation.83–85TP53 mutations are prevalent in DMG, with rates ranging from 60% to 80%, making it the most common mutation after H3 mutations (Figure 3).80 These mutations co-occur with both the H3.3K27M and H3K27WT subgroups of DMG, and to a lesser extent with the H3.1K27M subgroup (less than 15%). The mutations are often found alongside amplification of PDGFRA. Patients with both H3.3K27M mutation and TP53 mutations tend to have increased radiation therapy resistance, more aggressive tumors, and a poorer prognosis.5,79 These data suggest that loss of TP53 might allow for continued cell cycling and, in collaboration with developmental gene transcriptional dysregulation engendered by the H3K27M mutation, subsequently arrest OPC differentiation to drive DMG formation (Figure 3).
Cyclin-Dependent Kinases
Other alterations in genes controlling the cell cycle have been implicated in DMG, including amplifications of cyclin D family members, such as CCND1, CCND2, and CCND3, as well as CDK4 and CDK6 (Figure 3).80,86 These factors specifically regulate the G1-to-S-phase transition, a period during which temporal regulatory processes of the OL lineage, including p27 accumulation, cause cell cycle exit to initiate differentiation.87,88 The CDK4/CDK6 complex with cyclins CCND1, CCND2, or CCND3 phosphorylates and inactivates retinoblastoma-associated protein, a tumor suppressor protein expressed in G1-phase.65 This interaction allows for the activation of E2F1 target genes, inducing transcription of cell-cycle genes to facilitate progression through the G1-to-S-phase transition. These data further suggest defects in cell-cycle regulation underlie a cycling OPC state that resists differentiation to promote DMG.
Receptor Tyrosine Kinases
Receptor tyrosine kinases (RTKs) are receptors located on cell membranes that have an important role in various signaling pathways related to cell growth, differentiation, and survival. Pertinent RTKs include EGFR and PDGFRA. EGFR characterizes pre-OPCs as they are specified from outer radial glial cells, and later, PDGFRA marks early OPCs before they begin exhibiting morphological features of more mature OPC derivatives.79,89–91 Amplifications and mutations in PDGF and PDGFRA are commonly observed in DMG, play a crucial role in facilitating phosphorylation at different phosphotyrosine domains, which ultimately activate PI3K/AKT/mTOR and Ras/Raf/MEK/ERK downstream signaling pathways (Figure 3).65 Amplification of PDGFRA is observed in approximately 30% of DMG tumors, and PDGFRA mutation is seen in almost 15% of DMG patients.80PDGFRA alterations are found primarily with the H3.3K27M mutation and exhibit an proneural gene expression profile, which is linked with clinical aggressiveness irrespective of histological categorization. Alongside retinoic acid treatment, withdrawal of PDGFA induces OPC differentiation in vitro; moreover, suppression of PDFRA expression is observed in healthy OPC differentiation.92–94 Therefore, it is likely that PDFRA signaling maintains OPCs in a cycling state. Indeed, activation of PDGFRA triggers dimerization and autophosphorylation of its tyrosine kinase domain, which attracts downstream effectors and phosphorylates downstream targets such as MYC and JUN, both of which are members of the PI3K pathway.80 In turn, this drives the activity of MYC and JUN, creating a positive-feedback loop that increases the expression of multiple other RTKs, leading to oncogene addiction. Unfortunately, PDGFRA heterogeneity and co-occurring amplifications in other RTKs limit the therapeutic value of targeting PDGFRA.65 Amplification of PDGFRA plays a significant role in the initiation of gliomagenesis in vivo, indicating that it is an early event in tumorigenesis. This leads to the development of a clinically aggressive glioma that is unresponsive to tyrosine kinase inhibitors.80
Activin A Receptor Type 1
ACVR1, which encodes ALK2, is a receptor in the BMP signaling pathway of the TGF-β superfamily.65,80 The BMP pathway is tumor-suppressive and promotes maturity in H3.3K27M DMG, yet it has an opposing function in H3.1K27M mutant DMG, resulting instead in OPC differentiation arrest and population expansion in one valuable murine model.60,95 Indeed, although activating mutations in ACVR1 are observed in over 30% of DMG tumors, almost 90% co-occur with the H3.1K27M mutation (Figure 3).
These mutations are clustered around the serine/threonine kinase domain of ACVR1 at arginine 258 (R258G), glycine 328 and 356 (G328E, G328V, G328W, G356D), and in the glycine-serine (GS) rich domain (R206H), causing a conformational change in the kinase and leading to the constitutive activation of the BMP pathway.65,86 ALK2 phosphorylates SMAD transcription factors, which drive expression of DNA-binding protein inhibitors ID1 and ID2, promoting tumor initiation, angiogenesis, and repressing differentiation. ACVR1 mutations are more frequent in younger children, but have been linked to improved median overall survival.
Intracellular Kinases
The PI3K/Akt signaling pathway drives OPC differentiation and OL myelination, but activating mutations in its components, such as PIK3CA and PIK3R1, have been observed in DMGs (Figure 3).65,96 Surprisingly, these mutations result in cellular immaturity, requiring further investigation; nonetheless, they contribute to tumor growth and progression. Mutations in PIK3CA are found in approximately 12% of DMG cases, while those in PIK3R1 are present in 18% of cases.80PIK3R1 mutations are an obligatory partner to H3.3K27M and are found in clonal populations of DIPGs, while PIK3CA mutations are considered accessory drivers and have been observed in subclonal populations.
The inactivation of PTEN, a tumor suppressor and negative regulator of the PI3K pathway, occurs in a smaller percentage of H3K27M mutant glioma cases, with 4% of H3.1K27M and 6% of H3.3K27M cases showing this mutation (Figure 3).63,80
Transcriptional Regulators
MYC alterations are present in 20% of H3.3K27M mutant gliomas, while MYCN alterations are observed in 8% of overall DMGs (Figure 3).80 These factors regulate growth signal transduction pathways, metabolic processes, and cell maturity. For example, c-Myc (Myc) drives expression of LIN28B, an oncofetal RNA-binding protein which increases proto-oncogene expression to suppress differentiation across developmental lineages and promotes H3K27M mutant glioma proliferation and migration.97 Meanwhile, loss of H3K27me3 at Myc target genes results in their increased transcription.98 The H3K27M mutation also activates the RAS pathway, and in turn, the RAS pathway component ERK5 stabilizes Myc to increase its activity.16–19 Altogether, MYC alterations have been demonstrated to boost gene expression in DMG, leading to the activation of key oncogenic pathways that drive tumor growth and progression.86 Amplification and overexpression of the transcription factor MYCN and its protein target PVT1 are also linked to tumor initiation, progression, and recurrence, with the MYCN molecular subtype of DMG notable for hypermethylation and chromosomal rearrangement, including aneuploidy, leading to recurrent MYCN amplification and increased histological grade.79
Conclusion
H3K27M mutant glioma is a high-grade CNS tumor with distinct molecular biology and poor clinical outcomes. While much has been learned about H3K27M mutant glioma in recent years, ongoing work is needed in order to translate this increased understanding of tumor biology to improved clinical outcomes. Indeed, exciting lines of inquiry into the biological underpinnings of this disease have increasingly revealed an inextricable relationship between the biology of H3K27M mutant glioma and that of its cell of origin, the OPC. Here, we have introduced several epigenetic, immunological, and genetic aspects of H3K27M mutant gliomas that in part explain its ontogeny as a disease of OPC differentiation. These insights are now laying the groundwork for novel therapeutic approaches, including differentiation therapy, with the promise for improved clinical care for patients with this devastating disease.
Contributor Information
Amanda M Saratsis, Advocate Aurora Healthcare, Chicago, Illinois, USA.
Truman Knowles, Claremont McKenna College, Claremont, California, USA.
Antonela Petrovic, DMG Research Center, Department of Oncology, University Children’s Hospital, University of Zürich, Zürich, Switzerland.
Javad Nazarian, Research Center for Genetic Medicine, Children’s National Health System, Washington, District of Columbia, USA; Brain Tumor Institute, Children’s National Health System, Washington, District of Columbia, USA; DMG Research Center, Department of Pediatrics, University Children’s Hospital, University of Zurich, Zürich, Switzerland.
Conflict of interest statement
A.S. is an independent contractor, clinical development, for Chimerix Inc. The remaining authors have no conflicts of interest to declare.
Supplement sponsorship
This article appears as part of the supplement “H3 K27M-mutant Glioma: Disease State Overview,” sponsored by Chimerix, Inc.
Funding
The authors have no funding sources to report for this review.
References
- 1. Packer RJ, Nicholson HS, Vezina LG, Johnson DL.. Brainstem gliomas. Neurosurg Clin N Am. 1992;3(4):863–879. [PubMed] [Google Scholar]
- 2. Hoffman LM, Veldhuijzen van Zanten SEM, Colditz N, et al. Clinical, radiologic, pathologic, and molecular characteristics of long-term survivors of diffuse intrinsic pontine glioma (DIPG): a collaborative report from the International and European Society for Pediatric Oncology DIPG Registries. J Clin Orthod. 2018;36(19):1963–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vitanza NA, Monje M.. Diffuse intrinsic pontine glioma: from diagnosis to next-generation clinical trials. Curr Treat Options Neurol. 2019;21(8):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Puget S, Beccaria K, Blauwblomme T, et al. Biopsy in a series of 130 pediatric diffuse intrinsic Pontine gliomas. Childs Nerv Syst. 2015;31(10):1773–1780. [DOI] [PubMed] [Google Scholar]
- 5. Kline C, Jain P, Kilburn L, et al. Upfront biology-guided therapy in diffuse intrinsic pontine glioma: therapeutic, molecular, and biomarker outcomes from PNOC003. Clin Cancer Res. 2022;28(18):3965–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khuong-Quang DA, Buczkowicz P, Rakopoulos P, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124(3):439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 8. Bender S, Tang Y, Lindroth AM, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–672. [DOI] [PubMed] [Google Scholar]
- 9. Lewis PW, Müller MM, Koletsky MS, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan KM, Fang D, Gan H, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27(9):985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J, Huang TYT, Hou Y, et al. Epigenomic landscape and 3D genome structure in pediatric high-grade glioma. Sci Adv. 2021;7(23):eabg4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saratsis AM, Kambhampati M, Snyder K, et al. Comparative multidimensional molecular analyses of pediatric diffuse intrinsic pontine glioma reveals distinct molecular subtypes. Acta Neuropathol. 2014;127(6):881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pratt D, Natarajan SK, Banda A, et al. Circumscribed/non-diffuse histology confers a better prognosis in H3K27M-mutant gliomas. Acta Neuropathol. 2018;135(2):299–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buczkowicz P, Hoeman C, Rakopoulos P, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014;46(5):451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mosaab A, El-Ayadi M, Khorshed EN, et al. Histone H3K27M mutation overrides histological grading in pediatric gliomas. Sci Rep. 2020;10(1):8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jain SU, Rashoff AQ, Krabbenhoft SD, et al. H3 K27M and EZHIP impede H3K27-methylation spreading by inhibiting allosterically stimulated PRC2. Mol Cell. 2020;80(4):726–735.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jain SU, Do TJ, Lund PJ, et al. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat Commun. 2019;10(1):2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pratt D, Quezado M, Abdullaev Z, et al. Diffuse intrinsic pontine glioma-like tumor with EZHIP expression and molecular features of PFA ependymoma. Acta Neuropathol Commun. 2020;8(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Castel D, Kergrohen T, Tauziède-Espariat A, et al. Histone H3 wild-type DIPG/DMG overexpressing EZHIP extend the spectrum diffuse midline gliomas with PRC2 inhibition beyond H3-K27M mutation. Acta Neuropathol. 2020;139(6):1109–1113. [DOI] [PubMed] [Google Scholar]
- 21. Sievers P, Sill M, Schrimpf D, et al. A subset of pediatric-type thalamic gliomas share a distinct DNA methylation profile, H3K27me3 loss and frequent alteration of EGFR. Neuro Oncol. 2021;23(1):34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simon JA, Kingston RE.. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. [DOI] [PubMed] [Google Scholar]
- 23. An S, Camarillo JM, Huang TYT, et al. Histone tail analysis reveals H3K36me2 and H4K16ac as epigenetic signatures of diffuse intrinsic pontine glioma. J Exp Clin Cancer Res. 2020;39(1):261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harutyunyan AS, Krug B, Chen H, et al. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat Commun. 2019;10(1):1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oksuz O, Narendra V, Lee CH, et al. Capturing the onset of PRC2-mediated repressive domain formation. Mol Cell. 2018;70(6):1149–1162.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stafford JM, Lee CH, Voigt P, et al. Multiple modes of PRC2 inhibition elicit global chromatin alterations in H3K27M pediatric glioma. Sci Adv. 2018;4(10):eaau5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu JR, LeRoy G, Bready D, et al. The H3K36me2 writer-reader dependency in H3K27M-DIPG. Sci Adv. 2021;7(29):eabg7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee CH, Yu JR, Granat J, et al. Automethylation of PRC2 promotes H3K27 methylation and is impaired in H3K27M pediatric glioma. Genes Dev. 2019;33(19-20):1428–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brien GL, Bressan RB, Monger C, et al. Simultaneous disruption of PRC2 and enhancer function underlies histone H3.3-K27M oncogenic activity in human hindbrain neural stem cells. Nat Genet. 2021;53(8):1221–1232. [DOI] [PubMed] [Google Scholar]
- 30. Krug B, De Jay N, Harutyunyan AS, et al. Pervasive H3K27 acetylation leads to ERV expression and a therapeutic vulnerability in H3K27M gliomas. Cancer Cell. 2019;35(5):782–797.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pasini D, Malatesta M, Jung HR, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of polycomb group target genes. Nucleic Acids Res. 2010;38(15):4958–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Piunti A, Hashizume R, Morgan MA, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med. 2017;23(4):493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herz HM, Morgan M, Gao X, et al. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014;345(6200):1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dahl NA, Danis E, Balakrishnan I, et al. Super elongation complex as a targetable dependency in diffuse midline glioma. Cell Rep. 2020;31(1):107485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anshabo AT, Milne R, Wang S, Albrecht H.. CDK9: a comprehensive review of its biology, and its role as a potential target for anti-cancer agents. Front Oncol. 2021; 11(May 10):678559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fang D, Gan H, Cheng L, et al. H3.3K27M mutant proteins reprogram epigenome by sequestering the PRC2 complex to poised enhancers. eLife. 2018;7(June 22):e36696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bracken AP, Kleine-Kohlbrecher D, Dietrich N, et al. The polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21(5):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cordero FJ, Huang Z, Grenier C, et al. Histone H3.3K27M Represses p16 to accelerate gliomagenesis in a murine model of DIPG. Mol Cancer Res. 2017;15(9):1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mohammad F, Weissmann S, Leblanc B, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017;23(4):483–492. [DOI] [PubMed] [Google Scholar]
- 40. Funato K, Major T, Lewis PW, Allis CD, Tabar V.. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346(6216):1529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kfoury-Beaumont N, Prakasam R, Pondugula S, et al. The H3K27M mutation alters stem cell growth, epigenetic regulation, and differentiation potential. BMC Biol. 2022;20(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sanders LM, Cheney A, Seninge L, et al. Identification of a differentiation stall in epithelial mesenchymal transition in histone H3-mutant diffuse midline glioma. GigaScience. 2020;9(12):giaa136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Filbin MG, Tirosh I, Hovestadt V, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018;360(6386):331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lindquist RA, Guinto CD, Rodas-Rodriguez JL, et al. Identification of proliferative progenitors associated with prominent postnatal growth of the pons. Nat Commun. 2016;7(1):11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu I, Jiang L, Samuelsson ER, et al. The landscape of tumor cell states and spatial organization in H3-K27M mutant diffuse midline glioma across age and location. Nat Genet. 2022;54(12):1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Monje M, Mitra SS, Freret ME, et al. Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. Proc Natl Acad Sci U S A. 2011;108(11):4453–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nagaraja S, Vitanza NA, Woo PJ, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 2017;31(5):635–652.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tate MC, Lindquist RA, Nguyen T, et al. Postnatal growth of the human pons: a morphometric and immunohistochemical analysis: human postnatal pons development. J Comp Neurol. 2015;523(3):449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lewis NA, Klein RH, Kelly C, Yee J, Knoepfler PS.. Histone H3.3 K27M chromatin functions implicate a network of neurodevelopmental factors including ASCL1 and NEUROD1 in DIPG. Epigenetics Chromatin. 2022;15(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barateiro A, Fernandes A.. Temporal oligodendrocyte lineage progression: in vitro models of proliferation, differentiation and myelination. Biochim Biophys Acta Mol Cell Res. 2014;1843(9):1917–1929. [DOI] [PubMed] [Google Scholar]
- 51. Anastas JN, Zee BM, Kalin JH, et al. Re-programing chromatin with a bifunctional LSD1/HDAC inhibitor induces therapeutic differentiation in DIPG. Cancer Cell. 2019;36(5):528–544.e10. [DOI] [PubMed] [Google Scholar]
- 52. Balakrishnan I, Danis E, Pierce A, et al. Senescence induced by BMI1 inhibition is a therapeutic vulnerability in H3K27M-mutant DIPG. Cell Rep. 2020;33(3):108286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ehteda A, Simon S, Franshaw L, et al. Dual targeting of the epigenome via FACT complex and histone deacetylase is a potent treatment strategy for DIPG. Cell Rep. 2021;35(2):108994. [DOI] [PubMed] [Google Scholar]
- 54. Katagi H, Takata N, Aoi Y, et al. Therapeutic targeting of transcriptional elongation in diffuse intrinsic pontine glioma. Neuro Oncol. 2021;23(8):1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Przystal JM, Cianciolo Cosentino C, Yadavilli S, et al. Imipridones affect tumor bioenergetics and promote cell lineage differentiation in diffuse midline gliomas. Neuro Oncol. 2022;24(9):1438–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Silveira AB, Kasper LH, Fan Y, et al. H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol. 2019;137(4):637–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Surowiec RK, Ferris SF, Apfelbaum A, et al. Transcriptomic analysis of diffuse intrinsic pontine glioma (DIPG) Identifies a targetable ALDH-positive subset of highly tumorigenic cancer stem-like cells. Mol Cancer Res. 2021;19(2):223–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Castel D, Philippe C, Calmon R, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130(6):815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Castel D, Philippe C, Kergrohen T, et al. Transcriptomic and epigenetic profiling of ‘diffuse midline gliomas, H3 K27M-mutant’ discriminate two subgroups based on the type of histone H3 mutated and not supratentorial or infratentorial location. Acta Neuropathol Commun. 2018;6(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fortin J, Tian R, Zarrabi I, et al. Mutant ACVR1 arrests glial cell differentiation to drive tumorigenesis in pediatric gliomas. Cancer Cell. 2020;37(3):308–323.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Larson JD, Kasper LH, Paugh BS, et al. Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell. 2019;35(1):140–155.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nagaraja S, Quezada MA, Gillespie SM, et al. Histone variant and cell context determine H3K27M reprogramming of the enhancer landscape and oncogenic state. Mol Cell. 2019;76(6):965–980.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nikbakht H, Panditharatna E, Mikael LG, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun. 2016;7(1):11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sarthy JF, Meers MP, Janssens DH, et al. Histone deposition pathways determine the chromatin landscapes of H3.1 and H3.3 K27M oncohistones. eLife. 2020;9(Sept 9):e61090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Duchatel RJ, Jackson ER, Alvaro F, et al. Signal transduction in diffuse intrinsic pontine glioma. Proteomics. 2019;19(21–22):1800479. [DOI] [PubMed] [Google Scholar]
- 66. Peeters SM, Muftuoglu Y, Na B, Daniels DJ, Wang AC.. Pediatric gliomas. Neurosurg Clin N Am. 2021;32(2):181–190. [DOI] [PubMed] [Google Scholar]
- 67. Harrington EP, Bergles DE, Calabresi PA.. Immune cell modulation of oligodendrocyte lineage cells. Neurosci Lett. 2020;715(Jan 10):134601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin GL, Nagaraja S, Filbin MG, et al. Non-inflammatory tumor microenvironment of diffuse intrinsic pontine glioma. Acta Neuropathol Commun. 2018;6(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang Z, Guo X, Gao L, et al. Classification of pediatric gliomas based on immunological profiling: implications for immunotherapy strategies. Mol Ther Oncolytics. 2021;20(Dec 25):34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zaguia F, Saikali P, Ludwin S, et al. Cytotoxic NKG2C+ CD4 T cells target oligodendrocytes in multiple sclerosis. J Immunol. 2013;190(6):2510–2518. [DOI] [PubMed] [Google Scholar]
- 71. Lieberman NAP, DeGolier K, Kovar HM, et al. Characterization of the immune microenvironment of diffuse intrinsic pontine glioma: implications for development of immunotherapy. Neuro Oncol. 2019;21(1):83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang L, Yu H, Xue Y, Liu Y.. Decreased natural killer cells in diffuse intrinsic pontine glioma patients. Childs Nerv Syst. 2020;36(7):1345–1346. [DOI] [PubMed] [Google Scholar]
- 73. Ross JL, Chen Z, Herting CJ, et al. Platelet-derived growth factor beta is a potent inflammatory driver in paediatric high-grade glioma. Brain. 2021;144(1):53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jha P, Manjunath N, Singh J, et al. Analysis of PD-L1 expression and T cell infiltration in different molecular subgroups of diffuse midline gliomas. Neuropathology. 2019;39(6):413–424. [DOI] [PubMed] [Google Scholar]
- 75. Chauhan P, Lokensgard J.. Glial Cell Expression of PD-L1. Iran J Med Sci. 2019;20(7):1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hayden E, Holliday H, Lehmann R, et al. Therapeutic targets in diffuse midline gliomas—an emerging landscape. Cancers. 2021;13(24):6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mackay A, Burford A, Molinari V, et al. Molecular, pathological, radiological, and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY phase II randomized trial. Cancer Cell. 2018;33(5):829–842.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Miron VE, Boyd A, Zhao JW, et al. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat Neurosci. 2013;16(9):1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Damodharan S, Lara-Velazquez M, Williamsen BC, Helgager J, Dey M.. Diffuse intrinsic pontine glioma: molecular landscape, evolving treatment strategies and emerging clinical trials. J Prenat Med. 2022;12(5):840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Findlay IJ, De Iuliis GN, Duchatel RJ, et al. Pharmaco-proteogenomic profiling of pediatric diffuse midline glioma to inform future treatment strategies. Oncogene. 2022;41(4):461–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Aziz-Bose R, Monje M.. Diffuse intrinsic pontine glioma: molecular landscape and emerging therapeutic targets. Curr Opin Oncol. 2019;31(6):522–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Aubrey BJ, Strasser A, Kelly GL.. Tumor-suppressor functions of the TP53 pathway. Cold Spring Harb Perspect Med. 2016;6(5):a026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tokumoto YM, Tang DG, Raff MC.. Two molecularly distinct intracellular pathways to oligodendrocyte differentiation: role of a p53 family protein. EMBO J. 2001;20(18):5261–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Billon N, Terrinoni A, Jolicoeur C, et al. Roles for p53 and p73 during oligodendrocyte development. Development. 2004;131(6):1211–1220. [DOI] [PubMed] [Google Scholar]
- 85. Gonzalez PP, Kim J, Galvao RP, et al. p53 and NF 1 loss plays distinct but complementary roles in glioma initiation and progression. Glia. 2018;66(5):999–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lapin DH, Tsoli M, Ziegler DS.. Genomic insights into diffuse intrinsic pontine glioma. Front Oncol. 2017;7(Mar 28):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zezula J, Casaccia-Bonnefil P, Ezhevsky SA, et al. p21 cip1 is required for the differentiation of oligodendrocytes independently of cell cycle withdrawal. EMBO Rep. 2001;2(1):27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Durand B, Raff M.. A cell-intrinsic timer that operates during oligodendrocyte development. Bioessays. 2000;22(1):64–71. [DOI] [PubMed] [Google Scholar]
- 89. Galichet C, Clayton RW, Lovell-Badge R.. Novel tools and investigative approaches for the study of oligodendrocyte precursor cells (NG2-Glia) in CNS development and disease. Front Cell Neurosci. 2021;15(Apr 29):673132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Huang W, Bhaduri A, Velmeshev D, et al. Origins and proliferative states of human oligodendrocyte precursor cells. Cell. 2020;182(3):594–608.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tiane A, Schepers M, Rombaut B, et al. From OPC to oligodendrocyte: an epigenetic journey. Cells. 2019;8(10):1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Barres BA, Lazar MA, Raff MC.. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120(5):1097–1108. [DOI] [PubMed] [Google Scholar]
- 93. Zhou L, Shao CY, Xie YJ, et al. Gab1 mediates PDGF signaling and is essential to oligodendrocyte differentiation and CNS myelination. eLife. 2020;9(Jan 16):e52056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fang N, Cheng J, Zhang C, et al. Sirt2 epigenetically down-regulates PDGFRα expression and promotes CG4 cell differentiation. Cell Cycle. 2019;18(10):1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sun Y, Yan K, Wang Y, et al. Context-dependent tumor-suppressive BMP signaling in diffuse intrinsic pontine glioma regulates stemness through epigenetic regulation of CXXC5. Nat Cancer. 2022;3(9):1105–1122. [DOI] [PubMed] [Google Scholar]
- 96. Ishii A, Furusho M, Macklin W, Bansal R.. Independent and cooperative roles of the Mek/ERK1/2-MAPK and PI3K/Akt/mTOR pathways during developmental myelination and in adulthood. Glia. 2019;67(7):1277–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Knowles T, Huang T, Qi J, et al. LIN28B and let-7 in diffuse midline glioma: a review. Cancers. 2023;15(12):3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pajovic S, Siddaway R, Bridge T, et al. Epigenetic activation of a RAS/MYC axis in H3.3K27M-driven cancer. Nat Commun. 2020;11(1):6216. [DOI] [PMC free article] [PubMed] [Google Scholar]