Abstract
The cochlea is an important sensory organ for both balance and sound perception, and the formation of the cochlea is a complex developmental process. The development of the mouse cochlea begins on embryonic day (E)9 and continues until postnatal day (P)21 when the hearing system is considered mature. Small extracellular vesicles (sEVs), with a diameter ranging from 30 to 200 nm, have been considered a significant medium for information communication in both physiological and pathological processes. However, there are no studies exploring the role of sEVs in the development of the cochlea. Here, we isolated tissue-derived sEVs from the cochleae of FVB mice at P3, P7, P14, and P21 by ultracentrifugation. These sEVs were first characterized by transmission electron microscopy, nanoparticle tracking analysis, and western blotting. Next, we used small RNA-seq and mass spectrometry to characterize the microRNA transcriptomes and proteomes of cochlear sEVs from mice at different ages. Many microRNAs and proteins were discovered to be related to inner ear development, anatomical structure development, and auditory nervous system development. These results all suggest that sEVs exist in the cochlea and are likely to be essential for the normal development of the auditory system. Our findings provide many sEV microRNA and protein targets for future studies of the roles of cochlear sEVs.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04164-x.
Keywords: Small extracellular vesicles, Cochlea, Development, MicroRNAs, Proteins, Proteomics
Introduction
The cochlea in the inner ear is an important auditory signal transduction organ that develops from embryonic day (E)9 through postnatal day (P)21 [1]. The detection of sound waves and transmission of sound information to the brain are both dependent on cochlear hair cell (HCs) [2]. The first cochlear HCs develop at E11, and ultimately three rows of outer hair cells (OHCs), one row of inner hair cells (IHCs), and supporting cells (SCs) beneath the HCs are formed [3]. By P3, the total number of HCs peaks and will remain basically unchanged, while the morphology of the HCs will change as the HCs mature from P3 to P21 [4, 5].
HC maturation involves many complex developmental processes, such as the formation of hair bundles, synapses, and mechanical transduction channels (METs) [6–8]. Hearing formation requires the establishment of proper innervation, and the afferent nerves of the inner ear gradually form an outer spiral bundle of OHCs from P0 to P3 [9]. In the first 7 days after birth, hair bundles and METs develop gradually, and mature innervation patterns emerge gradually between P14 and P21 [9, 10]. At P7, HCs have mature mechanical transduction abilities, which is the most important aspect of formation of the auditory system [11]. Hearing onset and HC synapses gradually mature at P12–14, during which time mice gradually obtain the initial ability to hear [12]. Adult ranges of cochlear potentials can be initially tested at P11, but adult ranges and sensitivities of cochlear potential do not mature until P14 [13]. Mature cochlear potentials are essential for the formation of hearing. At P21, the morphology and function of the cochlea are mature, and hearing function can be measured by auditory brainstem response. During the process of HC maturation, the characteristics of SCs, especially inner ear progenitors, also change dramatically. SCs have been reported to act as inner ear stem cells and to transdifferentiate into HCs by induction of Wnt signaling or inhibition of Notch signaling in newborn mice [14, 15]. However, the stemness of SCs deteriorates with age, and their capacity to divide is completely lost by P14 [16].
It has been reported that many important transcription factors and signaling pathways are associated with the development of the cochlea, such as Sox2, Atoh1 [17], and the Wnt, Notch, and FGF signaling pathways [18, 19]. In addition, many microRNAs (miRNAs), such as miR182, miR183, and miR124, are also reported to regulate inner ear tissue differentiation and to maintain cell differentiation and proliferation [20, 21]. However, the cochlea’s development is a complicated process, and many regulatory processes and the factors that are involved remain to be elucidated.
Small extracellular vesicles (sEVs) have become a research hotspot in recent years and are reported to be involved in intercellular signal transmission during many important pathological and physiological processes [22–24]. sEVs have sizes from 30 to 200 nm and can be generated by various cells [25]. The contents of sEVs include numerous proteins and nucleic acids that are protected by a phospholipid bilayer structure from being digested by extracellular substances, and these materials can be delivered to recipient cells and thus contribute to cellular communication and signal transmission [26, 27]. sEVs participate in cell proliferation and differentiation in both pathological and healthy situations through signaling pathways mediated by miRNAs [28–30], and sEVs are involved in intercellular signal transmission during the development of brain neural circuits and in regulating growth patterns during embryonic development [31, 32].
Although sEVs have been extensively studied in cancer and other diseases, limited studies have been performed on the role of sEVs in the cochlea. This may be because as the mice age the otic vesicle outside the cochlea gradually becomes ossified and becomes rigid, especially after P10, which makes it difficult to obtain the substances inside the cochlea. However, it is known that in the utricle SC-derived exosomes can protect HCs against neomycin-induced ototoxicity [33] and that inner ear stem cell-derived exosomes can reduce ototoxic drug damage by transferring miR-182-5p to HEI-OC1 cells [34, 35]. At present, the research on inner ear-derived sEVs is based on in vitro models, and there is no research on sEVs in intact inner ear tissues.
In this study, we extracted cochlear tissue-derived sEVs from mice at different ages after birth and systematically analyzed and characterized their protein and miRNA contents for the first time. We used transmission electron microscopy (TEM), western blotting, and nanoparticle tracking analysis (NTA) to quantify the characteristics of sEVs and then performed proteomics and small RNA-seq to analyze the differentially expressed proteins and miRNAs and to predict the functions of these proteins and miRNAs. These results are expected to provide important information for the subsequent functional analysis of sEVs in the cochlea.
Materials and methods
Isolation of cochlear tissue-derived sEVs
The cochleae were obtained from P3, P7, P14, and P21 FVB mice. sEVs were isolated from 45 mouse cochleae according to the ultracentrifugation method as previously reported [36, 37]. Briefly, the cochleae were dissected, placed in a centrifuge tube with PBS buffer, and then ground for 1 min at 40 Hz in a grinder (Jingxin, Shanghai, China). The sample was filtered with a pore size of 0.22 µm after differential centrifugation to eliminate cell debris and microvesicles (600 × g for 10 min, 2000×g for 15 min, and 12,000×g for 50 min, all at 4 °C). The filtered samples were concentrated to 1–1.5 ml in a 50 ml 100 kDa molecular weight cutoff (MWCO) ultrafiltration centrifuge tube (Millipore) at 3000×g for 15 min at 4 °C. The samples were then ultracentrifuged at 110,000×g for 2 h at 4 °C to obtain sEVs. After discarding the supernatant, the sEV pellets were resuspended, washed with PBS once, and ultracentrifuged a second time at 110,000×g for 2 h at 4 °C. The sEVs were finally resuspended in 400–500 μl PBS for the following experiments.
Transmission electron microscopy
A total of 10 µl of sEV sample was dropped on an electron microscope copper grid and dried at room temperature and then negatively stained with 1.5% phosphotungstic acid (pH 7.4) at room temperature for 5 min. The images of cochlear sEVs were obtained by TEM (JEM-2100, Hitachi, Tokyo, Japan) with an accelerating voltage of 200 kV.
Nanoparticle tracking analysis
NTA (NS300, Malvern, United Kingdom) was used to identify the size and concentration of sEVs. A total of five 60-s videos were obtained for each sample, and the dispersed light signal of the sEVs was gathered using an optical microscope. According to Brownian motion of particles, the sizes and concentrations of the sEVs were averaged from the five videos.
Immunofluorescent staining
Immunofluorescent staining was performed according to a previous study [38]. In brief, cochleae were decalcified with 0.5 M ethylene diamine tetraacetic acid (EDTA) after being fixed in 4% (v/v) paraformaldehyde. The cochleae were then blocked and incubated with primary antibodies. Fluorescence-conjugated secondary antibodies were then added and bound to primary antibodies. A Zeiss LSM 700 confocal microscope was used to capture fluorescent images of the cochleae. The primary antibodies included anti-myosin7a (Proteus Bioscience, #25-6790, 1:1000 dilution), anti-Sox2 (R&D systems AF2018-SP, 1:1,000 dilution), anti-CD63 (ab217345, 1:1,000 dilution), and anti-CD9 (ab92726, 1:1500 dilution). Alexa Fluor 647 donkey anti-goat IgG (Invitrogen, A-21447, 1:400 dilution), Alexa Fluor 555 donkey anti-rabbit IgG (Invitrogen, A-31572, 1:400 dilution), and Alexa Fluor 488 donkey anti-mouse IgG (Invitrogen, A-21202, 1:400 dilution) were used as secondary antibodies.
Western blotting
Small extracellular vesicles were freeze-dried and then lysed in 200 μl RIPA lysis buffer (Beyotime) with 1 × protease cocktail (Roche) for 30 min at 4 °C. The protein quality was assessed using a BCA kit (Beyotime). The samples were boiled for 15 min at 95 °C in 5 × sodium dodecyl sulfate (SDS) loading buffer. SDS polyacrylamide gel electrophoresis was utilized to isolate the sEV proteins, which were then transferred onto a polyvinylidene difluoride membrane at 275 mA for 90 min. The membrane was blocked with 5% BSA (5% (v/v) bovine serum albumin in 0.1% (v/v) Tween-20 in phosphate buffered solution (PBS)) for 1 h at room temperature and then incubated with primary antibody overnight at 4 °C. The second day, the membrane was incubated with HRP-conjugated secondary antibody (Abclonal, 1:2,000 dilution). SuperSignal West Pico Plus chemiluminescent substrate (Thermo Scientific) was employed for visualizing the target bands on a Tanon-5200 automatic chemical imaging system. The primary antibodies were anti-CD63 (ab217345, 1:1000 dilution), anti-CD9 (ab92726, 1:1500 dilution), anti-Tsg101 (ab125011, 1:2000 dilution), anti-mouse EEA1 (Santa Cruz Biotechnology, 1:100 dilution), anti-rabbit Rab7 (Cell Signaling Technology, 1:1,000 dilution), anti-GAPDH (Kangchen, KC-5G4, 1:2000 dilution), anti-Synapsin-1 (Cell Signaling Technology 5297T, 1:1000), and anti-VGLUT3 (Synaptic Systems 135203, 1:1000).
RNA extraction and quantitative real-time PCR
Small extracellular vesicle samples were mixed with 1 ml Trizol (Invitrogen, 15596-026) on ice for 5 min, then centrifuged at 17,970×g for 5 min at 4 °C. The sample was mixed with 200 μl chloroform, vortexed to mix well, and then placed on ice for 10 min. After centrifugation at 17,970×g for 15 min at 4 °C, the supernatant was mixed with an equal amount of isopropanol, incubated for 10 min, and centrifuged at 17,970×g for 10 min at 4 °C. The RNA pellet was washed with 70% ethanol after removing the supernatant then dissolved in 25 μl RNase-free water.
Total RNA from sEVs was reverse transcribed to cDNA using an miRNA first-strand cDNA synthesis kit (Vazyme #MR101) following the manufacturer’s directions. Real-time PCR was done using an Applied Biosystems real-time PCR instrument with miRNA Universal SYBR qPCR Master Mix (Vazyme, #MR101-01) to quantify the miRNA expression levels. All primer sequences are listed in the supplemental table. The levels of miRNAs were compared using two-tailed, unpaired Student's t tests after being standardized to small nuclear RNA U6.
Small RNA sequencing and analysis
For the small RNA-seq library, a minimum of 2 μg RNA single sample (n = 3) was used as the starting material. Following the manufacturer's protocol, sequencing libraries were created using the NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, USA), and miRNA data were evaluated by FASTQC (v 0.11.5). Sequences were aligned to the reference genome derived from MirBase v22.1 (http://www.mirbase.org/) using Bowtie2 (v 2.2.5). The miRNA expression level in each sample was determined by featureCounts (v 2.0.0) and then normalized with the CPM (counts-per-million) algorithm, and differential expression analysis was performed in edgeR (v 3.30.3) using |log2FoldChange|> 2.0 and p < 0.05 as the threshold. Short Time-Series Expression Miner (STEM) (v 1.3.13) software was used for expression trend analysis. To avoid too many false positives, only miRNA-targeted genes in the Tarbase v7.0 database [39], which were identified experimentally, were selected.
DIANA-279 miRPath v.3 was used to assess miRNA enrichment pathways [40], and the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were employed to investigate functional annotation and pathway enrichment. The cumulative effects of the specified miRNAs were evaluated using the "genes-Union" algorithm. The Fisher accurate test with a microT threshold of 0.8, false discovery rate (FDR) correction, and a p value threshold of 0.05 was used for enrichment analysis.
Protein digestion
The freeze-dried sEVs were dissolved in buffer consisting of phosphatase inhibitor cocktails, 10 mM TCEP, 40 mM 2-chloroacetamide, 12 mM sodium deoxycholate, 50 mM Tris–HCl, and 12 mM sodium lauroyl sarcosinate (pH 8.5) (Sigma-Aldrich) by boiling for 10 min at 95 °C. After that, the samples were diluted fivefold with 50 mM triethylammonium bicarbonate and digested for 3 h at 37 °C with Lys-C (Wako) at 1:100 (w/w). To further degrade the peptides, the samples were treated overnight at 37 °C with trypsin at a ratio of 1:50 (w/w). To acidify the sample with a concentration of 1% TFA, ethyl acetate solution and 10% trifluoroacetic acid (TFA) were adjusted in a 1:1 ratio according to the aforementioned combination. The sample solution was vortexed before being centrifuged at 15,000×g for 3 min. The organic phase on the top was discarded, and the aqueous phase at the base was harvested and freeze-dried by refrigerated vacuum centrifuge (Laconco CentriVap). The desalting was performed on an 8 mm extraction disk as directed by the manufacturer (3 M Empore 2240-SDB-XC). All samples were stored at – 80 °C.
LC–MS/MS and quantitative data analysis
LC–MS/MS was performed as previously described [41]. Briefly, the peptides were solubilized in 10 µL 0.1% formic acid (FA), and 2 µL of the nanoElute was used for proteomics analysis. All peptides could be separated in a 25 cm internal packed column in the mobile phase with a fluid velocity of 300 nl/min. The timsTOF Pro mass spectrometer (Bruker) was connected to the nanoElute in real time, and the data settings were adjusted to full scan (m/z 100–1700) by the mass spectrometer.
Using the PEAKS Studio X+ program (Bioinformatics Solutions Inc), the raw files were explicitly compared with the UniProt database to obtain clean data. There were no duplicate entries in the identification of proteins and peptides, but special peptides and proteins were found. To examine differential proteins, markers of exosomes, and isolated inner ear proteins in the various samples, the intensities of the peptides were quantified using a label-free approach. The Perseus software was utilized to investigate the differential expression of sEV proteins of the cochlea based on these data. DAVID (https://david.ncifcrf.gov/) was conducted to identify biological process terms from GO and KEGG pathway analyses, and the protein–protein interaction network was obtained by STRING database (http://string-db.org/).
Statistical analysis
All data in this study are shown as the mean ± SD, and all analyses were performed using GraphPad Prism 7 software. When analyzing the different groups, we performed a two-tailed, unpaired Student’s t tests to evaluate statistical significance. Statistical significance was defined as a value of p < 0.05.
Results
Isolation and characterization of cochlear tissue-derived sEVs
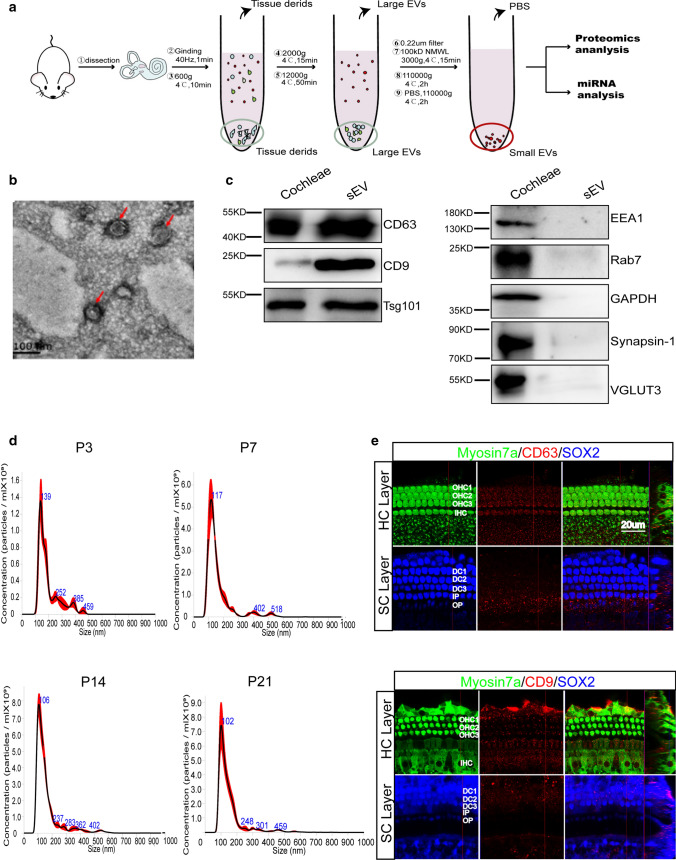
Small extracellular vesicles were isolated from the cochlear tissue of mice at P3, P7, P14, and P21 by ultracentrifugation as described previously [36, 37] (Fig. 1a). Considering that the cochlea is surrounded by the rigid otic vesicle, we dissected the cochleae and ground them in a grinder at 40 Hz as gently as possible so as not to break open the cells. The samples were centrifuged at low speed (600×g and 2000×g) to remove cell debris and then at high speed (12,000×g) to remove large EVs. After passing through a 0.22 µm filter, the samples were concentrated by ultrafiltration with a 100 kDa MWCO ultrafilter. Finally, sEVs were isolated by ultracentrifugation at 110,000×g. The RNAs and proteins extracted from sEVs were used for miRNA sequencing and proteomics analysis, respectively. TEM by negative staining indicated the oval shape of sEVs (Fig. 1b), and we characterized the size and number of sEVs from mice of different ages by NTA and confirmed that the diameter of the sEVs was 30–200 nm (Fig. 1d). Typical sEV marker proteins—such as the tetraspanins CD63 and CD9—and the composition of ESCRT-Ι complex Tsg101 were detected in cochlear tissue-derived sEVs by western blotting (Fig. 1c). Marker proteins for other vesicles, including EEA1 (endosome marker), Rab7 (lysosome marker), GAPDH, and Synapsin-1 and VGLUT3 (synaptic vesicles markers) were used as negative markers of sEVs and were not detected in the sEV samples (Fig. 1c). We also used immunofluorescent staining to confirm the presence of CD63 and CD9 in HCs and SCs (Fig. 1e). We further characterized the morphology and protein markers of sEVs and other vesicle markers in the four age groups by TEM and western blotting (Supplementary Fig. S1), which all showed a cup-like shape and typical sEV marker expression. Together, these results suggest that this isolation method of cochlear tissue-derived sEVs is feasible and can yield relatively pure sEVs.
Fig. 1.
Isolation and characterization of cochlear tissue-derived sEVs. a The workflow for isolating cochlear tissue-derived sEVs by ultracentrifugation. b TEM of cochlear sEVs. Scale bar = 100 nm. c Western blotting of cochlear tissue lysate (TL) and sEV samples. CD63, CD9, and Tsg101 were used as sEV markers, and EEA1, Rab7, GAPDH, Synapsin-1, and VGLUT3 from other organelles were used as negative markers. d NTA of cochlear sEVs from P3, P7, P14, and P21 mice. e Immunofluorescent staining of CD63 and CD9 (red) in the P3 cochlea. Myo7a (green) and Sox2 (blue) were used as HC and SC markers, respectively. OHC, outer hair cell. IHC, inner hair cell. DC, Deiters’ cell. IPC, inner pillar cell. OPC, outer pillar cell. IPhC, inner phalangeal cell. Scale bar = 20 μm
Micro analysis of cochlear tissue-derived sEVs from mice of different ages
Small extracellular vesicles contain a variety of RNAs, especially miRNAs, which play important roles in gene regulation and thus mediate numerous biological processes [42, 43]. Because the role of sEV miRNA in the cochlea is poorly understood, we employed small RNA-seq to evaluate the cochlear tissue-derived sEVs from P3, P7, P14, and P21 mice and to identify differentially expressed miRNA during the development of the cochlea.
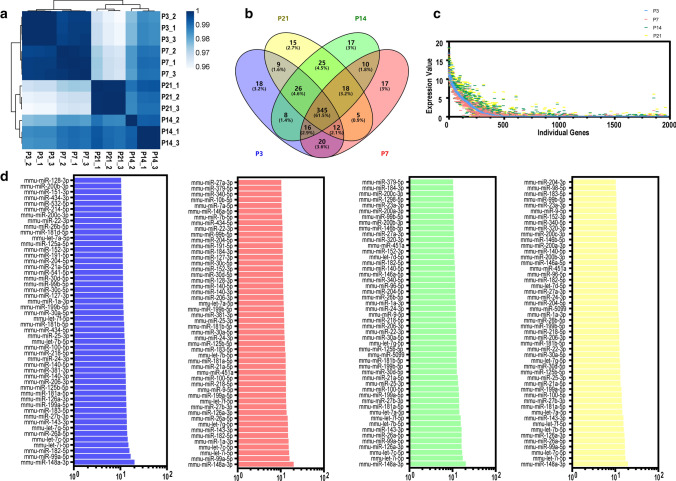
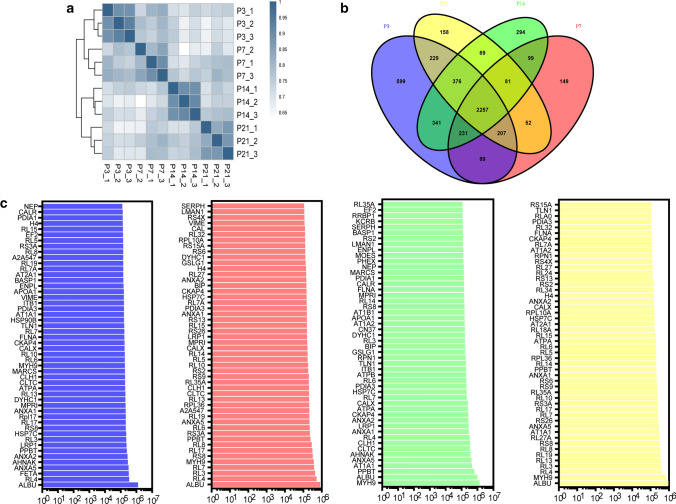
The correlations of the samples were tested by hierarchical clustering analysis, and the P3, P7, P14, and P21 groups were well-separated according to their Spearman correlation coefficients (Fig. 2a). We detected a total of 561 miRNAs, including 454, 453, 465, and 455 miRNAs from cochlear tissue-derived sEVs from P3, P7, P14, and P21 mice, respectively (Fig. 2b). Furthermore, there were 18, 17, 17, and 15 miRNAs that were uniquely expressed at P3, P7, P14, and P21, respectively (Fig. 2b). The expression levels of all miRNAs at P3, P7, P14, and P21 are shown in Fig. 2c. We compared the differentially expressed miRNAs between each of the age groups pairwise (Supplementary Fig. S2), and the top 50 most-abundant miRNAs in the four age groups are shown in Fig. 2d.
Fig. 2.
Transcriptome analysis of cochlear sEV miRNAs from P3, P7, P14, and P21 mice. a Cluster analysis of cochlear sEV miRNA sequencing data. b The Venn diagram of the miRNA sequencing data. c All differentially expressed miRNAs in the four samples. P3 data were used as the control as indicated by the blue line. d The top 50 highly expressed cochlear sEV miRNAs from P3 (blue), P7 (red), P14 (green), and P21 (yellow) mice
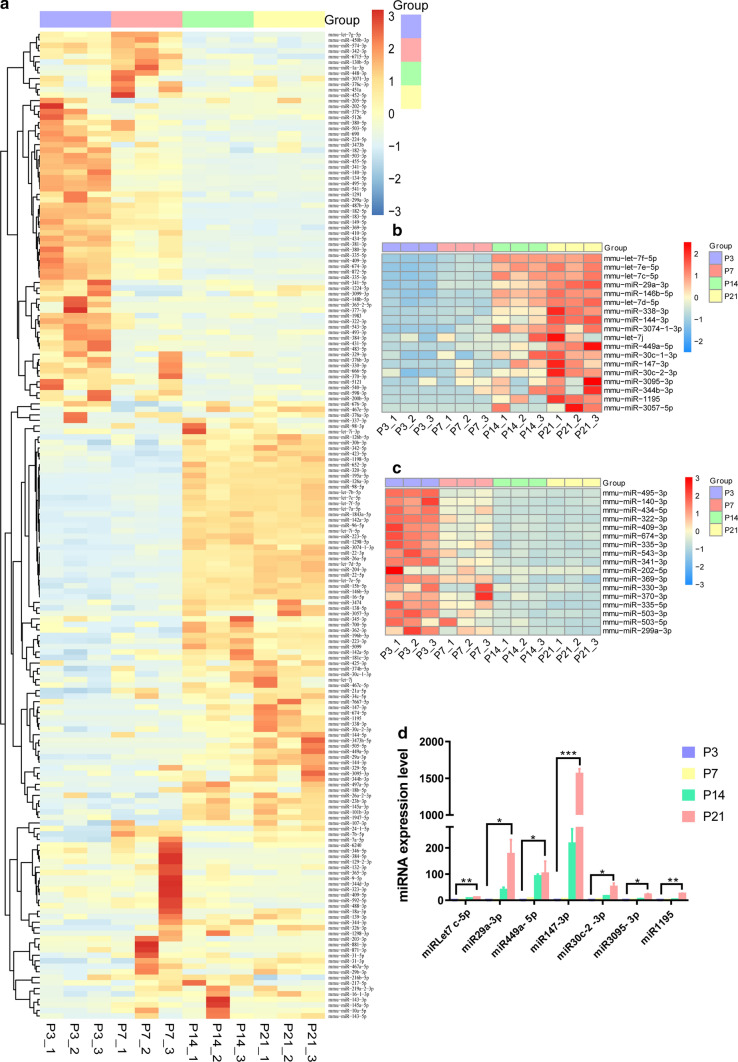
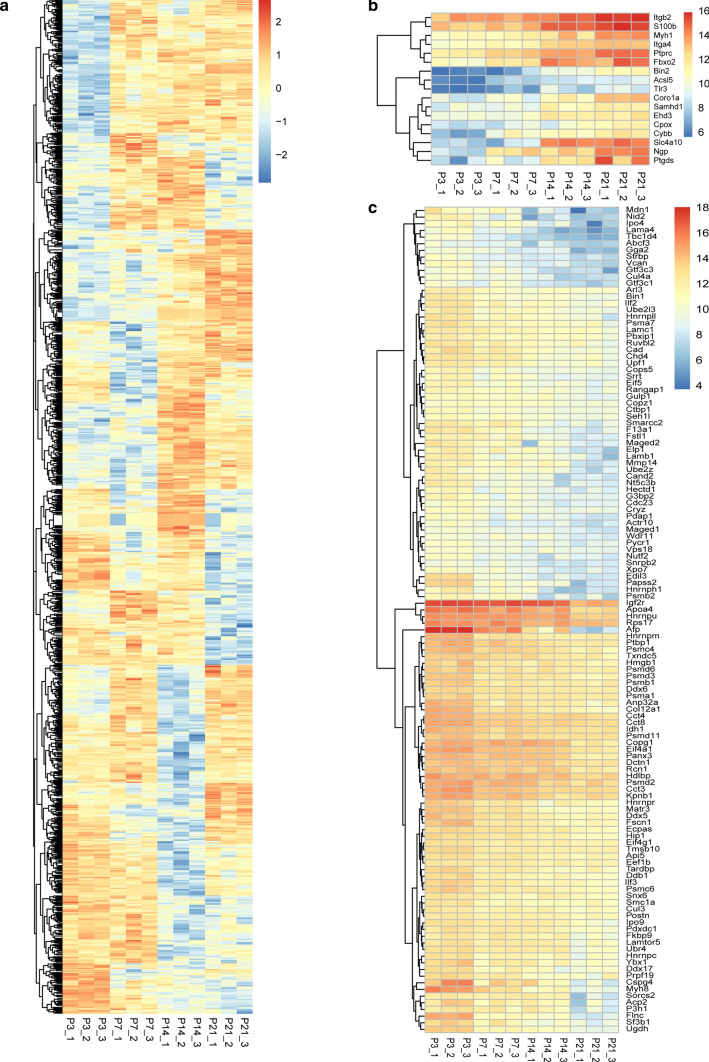
We found 179 miRNAs that were differentially expressed across the four age groups by pairwise comparison, including 57, 33, 29, and 60 miRNAs that were highly expressed at P3, P7, P14, and P21, respectively (Fig. 3a, p < 0.05, fold change > 2). Further analysis of the 179 differentially expressed miRNAs showed that 18 of these miRNAs became more prevalent in sEVs with age, while 17 miRNAs decreased with age (Fig. 3b, c). Of the increased miRNAs, miRLet-7f-5p [44], miRLet-7e-5p [45], miRLet-7c-5p [46], miR29a-3p [47], miR146b-5p [48], miRLet-7d-5p [49], miR338-3p [50], miR144-3p [51], miRLet-7j [52], miR449a-5p [53], miR30c-1-3p [54], miR147-3p [55], miR30c-2-3p [56], and miR1195 [57] have been attributed to a range of biological processes including cellular proliferation, cellular differentiation, and cellular signaling and communication. miR3074-1-3p, miR3095-3p, miR344b-3p, and miR3057-5p have no reported biological functions. For the decreased miRNAs, miR495-3p [58], miR140-3p [59], miR434-5p [60], miR322-3p [61], miR409-3p [62], miR674-3p [63], miR335-3p [61], miR543-3p [64], miR341-3p [65], miR202-5p [66], miR369-3p [67], miR330-3p [68], miR370-3p [69], miR335-5p [70], miR503-3p [71], and miR503-5p [72] have been reported to be related to biological processes, and only miR299a-3p has no reported function. We verified the 18 increased miRNAs by qPCR, and Fig. 3d shows that the expression of seven miRNAs (miRLet7c-5p, miR29a-3p, miR449a-5p, miR147-3p, miR30c-2-3p, miR3095-3p, and miR1195) was matched to the results of the bioinformatics analysis. The specific information for these seven miRNAs is listed in Table 1, including gene ID, number of predicted target genes, functional description, and references.
Fig. 3.
The differentially expressed cochlear sEV miRNAs from P3, P7, P14, and P21 mice. a Heatmap of the differentially expressed miRNAs. b Heatmap of the up-regulated miRNAs as mice age. c Heatmap of the down-regulated miRNAs as mice age. d qPCR verification of some differentially expressed miRNAs. Values lower and higher than the mean are shown by blue and red scales, respectively. *p < 0.05, **p < 0.01, ***p < 0.001, n = 3
Table 1.
Validated miRNA candidates of age-related cochlear tissue-derived sEVs with the numbers of target genes and functional categories
| miRNA | Gene ID | Number of target genes | Functional description | Reference |
|---|---|---|---|---|
| mmu-miRLet7c-5p | 387246 | 558 | Cell differentiation, anatomical structure development, ion binding, cell death, growth, developmental maturation, cell cycle, endocytosis, axon guidance, TNF signaling pathway, AMPK signaling pathway, TGF-beta signaling pathway, mTOR signaling pathway, MAPK signaling pathway, MAPK signaling pathway | Chen et al. [45], Morgado et al. [131], Yao et al. [132], Zhang et al. [133] and Chen et al. [134] |
| mmu-miR29a-3p | 387222 | 451 | Cell differentiation, anatomical structure development, ion binding, cell cycle, cell death, anatomical structure formation involved in morphogenesis, cell division, growth, developmental maturation, positive regulation of apoptotic process, axon guidance, TNF signaling pathway, AMPK signaling pathway, TGF-beta signaling pathway, mTOR signaling pathway, MAPK signaling pathway, Hippo signaling pathway | Qu et al. [47], You et al. [135], Volpicelli et al. [136], Wang et al. [137] and Tumaneng et al. [138] |
| mmu-miR449a-5p | 723868 | 469 | Cell differentiation, anatomical structure development, ion binding, cell differentiation, anatomical structure development, ion binding, cell motility, cytoplasmic membrane-bounded vesicle, cell junction, endocytosis, axon guidance, AMPK signaling pathway, TGF-beta signaling pathway, mTOR signaling pathway, MAPK signaling pathway, Hippo signaling pathway | Shu et al. [16], Ni et al. [53], Wu et al. [138] and Bou Kheir et al. [139] |
| mmu-miR30c-2-3p | 723964 | 209 | Anatomical structure development, regulation of epidermal cell differentiation | Tang et al. [56], Hu et al. [140], Hand et al. [141] and Liang et al. [ 142] |
| mmu-miR3095-3p | 100526502 | 156 | MicroRNA metabolic process, tissue homeostasis, inner cell mass cell differentiation, positive regulation of cAMP-mediated signaling, post-embryonic development, post-embryonic body morphogenesis, cell proliferation in the forebrain, regulation of canonical Wnt signaling pathway, auditory receptor cell fate commitment, positive regulation of transcription of Notch receptor target, regulation of timing of cell differentiation, long-term depression, mTOR signaling pathway | Chiang et al. [143] and Kozomara et al. [144] |
| mmu-miR147-3p | 387165 | 13 | Wnt and MAPK signaling, inflammatory responses | Tang et al. [55] and Liu et al. [108] |
| mmu-miR1195 | 100316676 | 79 | Cell differentiation, apoptosis | Zhang et al. [145] and Tagne et al. [57] |
Functional analysis of differentially expressed miRNAs in cochlear sEVs
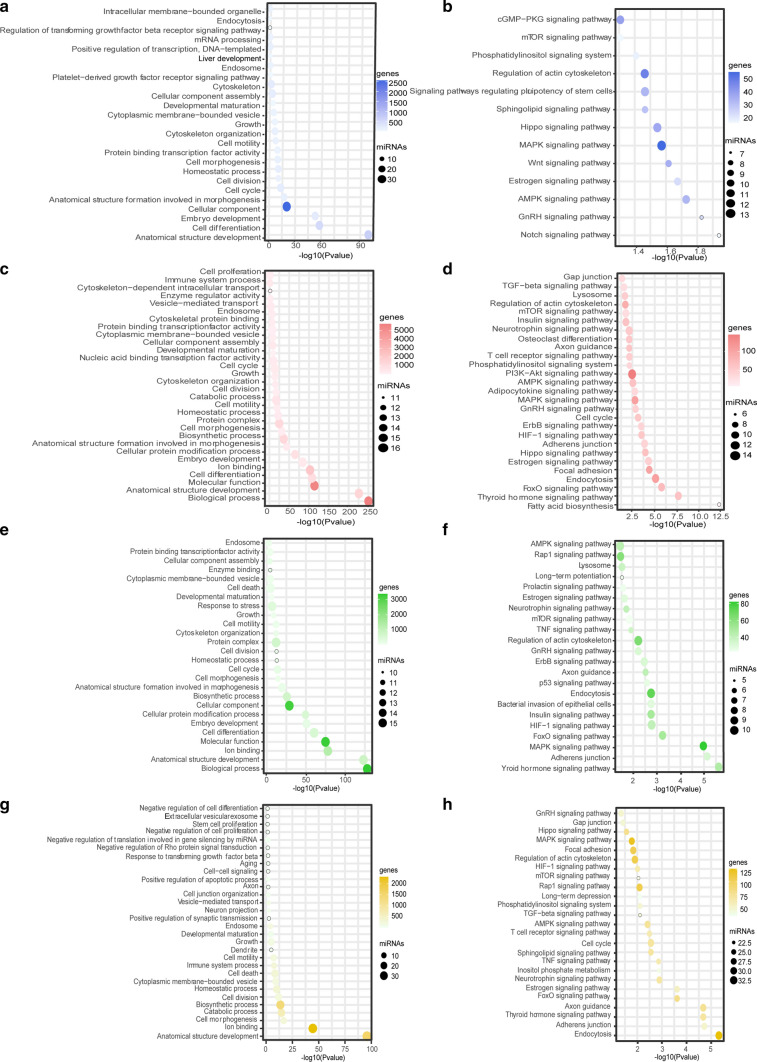
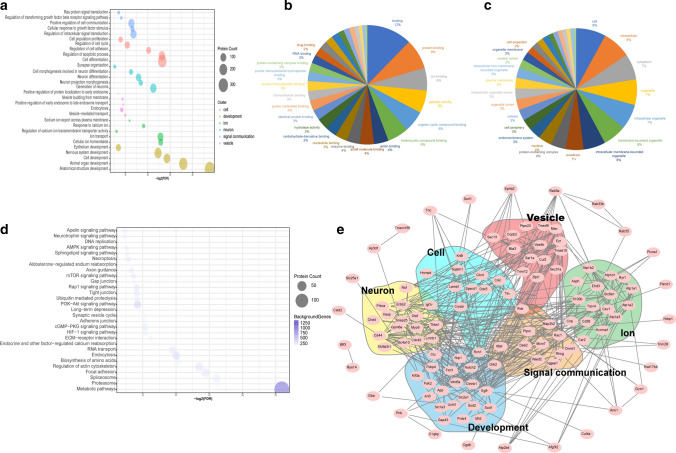
The GO and KEGG pathway analyses of the highly expressed miRNAs at P3, P7, and P14 were performed with DIANA-mirPath v.3 (http://snf-515788.vm.okeanos.grnet.gr/) using the target genes in the Tarbase v7.0 database (http://www.microrna.gr/tarbase). These miRNAs in cochlear sEVs at different ages have different biological functions (Fig. 4). Notably, the GO analysis showed that these miRNAs are mainly involved in anatomical structure development, cell differentiation, developmental maturation, growth, cell cycle, and vesicle-mediated transport (Fig. 4a, c, e, g). Figure 4b, d, f, h shows that the highly expressed miRNAs at P3, P7, P14, P21 are involved in the mTOR, PI3K-Akt, TGF-β, Wnt, Hippo, Notch, and cGMP-PKG signaling pathways. These findings suggest that these pathways are likely to be involved in the development of the cochlea and the formation of the auditory system. Among them Wnt, Notch, TGF-β, and Hippo signaling have been implicated in progenitor cell proliferation and differentiation as well as cell plane polarity during inner ear development [73, 74].
Fig. 4.
The GO and KEGG pathway analysis of differentially expressed cochlear sEV miRNAs from P3, P7, P14, and P21 mice. GO analysis of miRNAs of P3 (a), P7 (c), P14 (e), and P21 (g) mouse cochlear sEVs. KEGG enrichment pathways analysis of miRNAs of P3 (b), P7 (d), P14 (f), and P21 (h) mouse cochlear sEVs. The size of the bubble indicates the number of miRNAs, and the intensity of the color shows the number of genes targeted by the miRNA in all figures. P3 (blue), P7 (red), P14 (green), P21 (yellow)
Label-free quantitative proteomics analysis of cochlear tissue-derived sEVs from mice of different ages
Considering that proteins in sEVs also play important roles as biomarkers and in multiple biological processes [75, 76], the protein contents of the sEV sample from the cochleae of P3, P7, P14, and P21 mice was analyzed utilizing label-free quantitative proteomics. Each group included three biological replicates, and the samples clustered well with no outliers (Fig. 5a). A total of 5231 proteins were identified, and 2257 of these were present in all four groups (Fig. 5b). sEV marker proteins (Tsg101, CD63, CD9, CD81, and Flotillin-1) were also found among these proteins by mass spectrometry (Table 2). Figure 5c shows the top 50 most-abundant proteins in the P3, P7, P14, and P21 sEV samples. In addition, we compared all identified proteins with the Exocarta and Vesiclepedia databases and found that 978 proteins overlapped with Exocarta and 115 proteins overlapped with Vesiclepedia (Supplementary Fig. S3a). Figure S3b shows that 8, 6, 8, and 7 proteins of the top 100 proteins in P3, P7, P14, and P21 cochlea-derived sEVs were reported among the top 100 proteins in the Exocarta and Vesiclepedia databases. These results suggest that many sEV proteins in the Exocarta and Vesiclepedia databases were also found in our sEV samples and that there were sEV proteins in our samples that were not in the EV databases and thus might be newly identified EV proteins in cochlear tissue-derived sEVs.
Fig. 5.
Proteomics analysis of cochlear sEV proteins from P3, P7, P14, and P21 mice. a Cluster analysis of cochlear sEV proteomics data. b The Venn diagram of cochlear sEV proteomics data. c The top 50 most highly expressed proteins of cochlear sEVs from P3 (blue), P7 (red), P14 (green), and P21 (yellow) mice
Table 2.
Mass spectrometry analysis identified typical sEV proteins
| Name | Accession | − 10lgP | Intensity | |||
|---|---|---|---|---|---|---|
| P3 | P7 | P14 | P21 | |||
| TSG101 | Q61187 | 214.54 | 4.11E+03 | 1.57E+03 | 2.75E+03 | 1.79E+03 |
| CD9 | P40240 | 293.38 | 4.24E+04 | 1.99E+04 | 5.30E+04 | 3.44E+04 |
| CD63 | P41731 | 195.28 | 7.86E+03 | 2.46E+03 | 1.52E+04 | 4.64E+03 |
| CD81 | P35762 | 248.37 | 1.46E+04 | 5.28E+03 | 2.05E+04 | 5.33E+03 |
| Flotillin-1 | O08917 | 376.55 | 1.03E+04 | 3.66E+03 | 1.31E+04 | 4.46E+03 |
We performed quantitative analysis of cochlear sEV proteins and found many differentially expressed proteins between the different age groups (Supplementary Fig. S4), suggesting that the expression of many sEV proteins changes with the development of the cochlea. We found 3,120 proteins that are differentially expressed across the four age groups (Fig. 6a), and among them the expression level of 17 proteins increased with age (Fig. 6b), while the expression level of 124 proteins decreased with age (Fig. 6c). These results suggest that the expression patterns of proteins in the cochlear tissue-derived sEVs are correlated with age and may play significant roles in the formation of the inner ear system.
Fig. 6.
The differentially expressed cochlear sEV proteins from P3, P7, P14, and P21 mice. a Heatmap of the differentially expressed cochlear sEV proteins. b Heatmap of the up-regulated proteins as mice age. c Heatmap of the down-regulated proteins as mice age. Values lower and higher than the mean are shown by blue and red scales, respectively
Functional analysis of differentially expressed cochlear sEV proteins
We performed GO analysis to identify the biological processes, molecular functions, and cell membrane components of the differentially expressed proteins (Fig. 7a–c). For the enriched biological processes, GO annotations indicated that these proteins are involved in cell, development, ion, neuron, signal communication, and vesicle processes. We mapped the top 20 molecular functions and cell membrane components of these proteins, and this showed that these proteins are mostly involved in ion binding, catalytic activity, protein binding, and RNA binding. In addition, the cellular components analysis revealed that these proteins are mostly found in the cytoplasm, the endomembrane system, and the plasma membrane.
Fig. 7.
GO and KEGG pathway analysis of differentially expressed cochlear sEV proteins from P3, P7, P14, and P21 mice. Differentially expressed cochlear sEV proteins were identified by GO analysis as part of biological processes (a), molecular components (b), and cell membranes (c). d KEGG pathway analysis showing the significantly enriched pathways of differentially expressed cochlear sEV proteins in the four age groups. The size of the bubble shows the amount of protein, and the intensity of the color indicates the number of genes targeted by the protein in a and d. e The STRING network analysis for cochlear sEV proteins
We next conducted KEGG pathway analysis of the differentially expressed proteins, which showed that these proteins are mostly involved in the neurotrophin, AMPK, mTOR, PI3K-Akt, and cGMP-PKG signaling pathways and in endocytosis (Fig. 7d). According to GO, KEGG analysis and previous reports, we screened out 20 proteins from 141 age-related proteins (17 increased and 124 decreased with age as we showed above) that are reported to play roles in the inner ear and/or to be involved in the processes of development, cell differentiation, and proliferation. Table 3 lists these 20 top candidate proteins, including their UniProt accession numbers, functional descriptions, and references, which should be studied in the future to elucidate their roles in cochlear sEVs. These results suggest that cochlear sEVs may act as mediators in intercellular communications. Finally, to analyze the interactions between differentially expressed sEV proteins, we created a STRING protein interaction network (Fig. 7e).
Table 3.
Protein candidates of age-related cochlear tissue-derived sEVs with functional categories
| Protein | Accession | Functional description | Expression trends with age | Reference |
|---|---|---|---|---|
| Fbxo2 | Q80UW2 | Biological regulation, regulation of cell population proliferation, regulator for age-related hearing loss | ↑ | Atkin et al. [99] and Nelson et al. [100] |
| Tlr3 | Q99MB1 |
Biological regulation, anatomical structure development, immune system process, regulation of apoptotic process, positive regulation of cell communication, animal organ development, signal transduction |
↑ | Yamada et al. [101], Gollmann-Tepeköylü et al. [102] and Krisher et al. [146] |
| Slc4a10 | Q5DTL9 | Ion transport, system development, anatomical structure development, nervous system development, cell differentiation, generation of neurons, sodium ion transmembrane transport, neuron differentiation, multicellular organism growth | ↑ | Sinning et al. [98], Huebner et al. [103] and Christensen et al. [147] |
| S100b | P50114 | Metabolic process, regulation of cellular component organization, response to stimulus, system development, anatomical structure development, multicellular organism development, regulation of cell death, ion homeostasis, cell differentiation, regulation of apoptotic process, nervous system development, neurogenesis, regulation of intracellular signal transduction, positive regulation of cell communication | ↑ | Raponi et al. [148], Zeng et al. [149], Nishiyama et al. [150] and Riuzzi et al. [151] |
| Ddx5 | Q61656 | Regulation of localization, regulation of transport, positive regulation of biological process, metabolic process, cell proliferation and differentiation | ↓ | Legrand et al. [152], Dardenne et al. [153] and Arun et al. [154] |
| Chd4 | Q6PDQ2 | Cellular metabolic process, transmembrane transport, cellular localization, response to oxidative stress, neuronal differentiation | ↓ | Xu et al. [111], Tomofuji et al. [155] and Sun et al. [156] |
| Sorcs2 | Q9EPR5 | Maintain ciliary morphology, apoptosis, neuroprotection | ↓ | Forge et al. [118], Leloup et al. [157] and Malik et al. [158] |
| Idh1 | O88844 | Cellular metabolic process, multicellular organism development, anatomical structure development, system development, system development | ↓ | Zhang et al. [54], Kim et al. [121], Itsumi et al. [159] and Rohle et al. [160] |
| Panx3 | Q8CEG0 | Establishment of localization, cellular component organization or biogenesis, transport | ↓ | Abitbol et al. [119, 120] and Ishikawa et al. [161] |
| Hnrnp | P49312 | Cellular localization, metabolic process, regulation of cellular protein localization, mRNA processing, multicellular organism development, anatomical structure development, system development, cellular developmental process, cell differentiation, positive regulation of cell cycle process, cell division, animal organ development | ↓ | Chen et al. [105] and Ruan et al. [162] |
| Ptbp1 | P17225 | Cellular metabolic process, RNA processing, regulation of protein localization, multicellular organism development, anatomical structure development, system development, generation of neurons, nervous system development, regulation of cell development, cell differentiation, regulation of neuron projection development, regulation of neuron differentiation | ↓ | Mokabber et al. [112], Linares et al. [163], Georgilis et al. [164] and Vuong et al. [165] |
| Lama4 | P97927 | Regulation of localization, cell adhesion, multicellular organism development, system development, cellular developmental process, cell differentiation, regulation of multicellular organismal development, regulation of cell migration | ↓ | Suzuki et al. [49], Kim et al. [116] and Gorfu et al. [166] |
| Ilf3 | Q9Z1X4 | Negative regulation of transcription, cell proliferation, cell apoptosis | ↓ | Jia et al. [107] and Larcher et al. [175] |
| Lamtor5 | Q9D1L9 | Cellular localization, regulation of metabolic process, regulation of cell death, regulation of apoptotic process, regulation of intracellular signal transduction, positive regulation of cell communication | ↓ | Zhang et al. [88] and Liu et al. [108] |
| Psmd2 | Q8VDM4 | Cellular catabolic process, regulation of biological process, cell proliferation, cell cycle | ↓ | Li et al. [109] |
| Ddb1 | Q3U1J4 | Metabolic process, response to stimulus, organelle organization, regulation of cell death, homeostatic process, regulation of apoptotic process, regulation of cell cycle, signal transduction | ↓ | Zheng et al. [110], Zhao et al. [126], Yang et al. [167] and Gao et al. [168] |
| Ruvbl2 | Q3TXT7 | Establishment of localization, metabolic process, positive regulation of biological process, cellular component assembly, cellular response to stress, regulation of gene expression, regulation of signaling, regulation of cell communication, regulation of signal transduction | ↓ | Zhang et al. [42] and Hong et al. [114] |
| Cul4a | Q3TCH7 | Metabolic process, regulation of biological process, developmental process, anatomical structure development, animal organ development, regulation of response to stimulus, regulation of cell differentiation, immune system process, regulation of multicellular organismal development, regulation of cell cycle, regulation of cell population proliferation | ↓ | Liu et al. [115], Pan et al. [169], Kopanja et al. [170] and Waning et al. [171] |
| P3h1 | Q3V1T4 | Skeletal development, hearing loss | ↓ | Fratzl-Zelman et al. [172], Pokidysheva et al. [173] and Vranka et al. [174] |
Discussion
Small extracellular vesicles are important mediators in cellular communication and signal transmission, and they also can be used as naturally occurring carriers for drugs and biomarkers in clinical trials. At present, most researchers extract sEVs from in vitro culture systems, and previous research on inner ear sEVs has also relied on in vitro culture systems [33, 77]. However, the in vitro culture environment cannot truly replicate the in vivo environment, and sEVs derived from inner ear tissues can more accurately depict sEV functions in the inner ear. Therefore, we extracted sEVs from cochlear tissue for the first time and studied the miRNA transcriptomes and proteomics of the cochlear tissue-derived sEVs. We found that typical sEVs could be isolated from the cochlea by ultracentrifugation, and we identified 561 miRNAs and 5,231 proteins in cochlear tissue-derived sEVs that are engaged in multiple biological functions, including cellular communication, development, and vesicle production.
The cochlea is surrounded by the otic vesicle that gradually ossifies and becomes rigid as the mouse ages, especially after P10, and this makes it difficult to dissect the basilar membrane for extracting cochlear tissue-derived sEVs. Some recent studies have used enzyme digestion for the purpose of maintaining the integrity of the cells as much as possible to extract EVs from fat, brain, and tumor tissues [36, 78–81], while other studies have ground the tissues as a necessary step for extracting EVs [80, 82–84]. Crescitelli et al. showed that the digestive enzymes in the existing tissue extraction methods are ineffective for bone tissue, and the methods for this type of tissue need further optimization [85]. Considering the above factors, we improved the extraction method based on the scheme of Crewe et al. [36]. We used low-frequency grinding of the cochlear tissue to avoid breaking open the cells, and we increased the centrifugal force (12,000×g) for removing large vesicles and for isolating sEVs. TEM and NTA showed that the cochlear tissue-derived sEVs we extracted had typical sEV shapes and sizes. The western blotting also showed that the typical sEV markers—CD63, CD9, and Tsg101—could be detected in the sEV samples, while contaminating proteins Rab7, EEA1, Synapsin-1 and VGLUT3 from other vesicles and the intracellular protein GAPDH were not detected, which further confirmed the integrity and relative purity of the sEVs extracted by our method.
One of the important contents of sEVs is nucleic acids, which include miRNAs, lncRNAs, tRNAs, mtDNAs, and ssDNA [86]. Among them, miRNAs are reported to have a role in numerous biological processes including organ development and maturation and cell communication [87, 88]. In addition, miR-318 from mesenchymal stem cell-derived sEVs promotes chondrogenesis by suppressing TAOK1 [43], and miR135a derived from epithelial exosomes accelerates the mesenchymal production of dentin matrix proteins by triggering the Wnt/β-catenin signaling pathway [37]. Therefore, small RNA-seq was performed to characterize the miRNAs in cochlear tissue-derived sEVs and to elucidate their possible roles in the cochlea. We identified 561 miRNAs in cochlear sEVs, including 179 differentially expressed miRNAs, and we found that the expression of 18 miRNAs increased and 17 miRNAs decreased as the mice aged. We used qPCR to verify the expression of miRNAs and found that 7 miRNAs (miRLet7c-5p, miR29a-3p, miR449a-5p, miR147-3p, miR30c-2-3p, miR3095-3p, and miR1195) were consistent with the RNA-seq analysis results. Overexpression of miRLet7c-5p can inhibit laryngeal squamous cell carcinoma cell proliferation and can regulate microglial activation during the repair of brain injury [46, 89], and upregulation of miR30c-2-3p suppresses gastric cancer and the proliferation of renal cell carcinomas [56, 90]. miR29a-3p, miR449a-5p, and miR147-3p have been reported to be accumulated in exosomes derived from oral squamous cells, macrophages, and bronchoalveolar lavage fluid [53, 55, 91, 92]. In addition, upregulation of miR29a-3p rescues bronchopulmonary dysplasia and has a negative regulatory effect on the Smad, NFκB, and canonical Wnt signaling pathways [93–95]. Importantly, miR29a-3p directly targets the Wnt-related genes DVL3 (Dishevelled 3), CSNK2A2 (casein kinase 2 alpha 2 polypeptide), FZD3 (Frizzled family receptor 3), and FZD5 (Frizzled family receptor 5) [95]. These seven miRNAs whose expression increases as mice age are listed in Table 1, and they may be involved in the development of the cochlea after birth and may act as new targets to be further studied in the future to elucidate the detailed mechanisms behind cochlear development.
We also performed GO and KEGG analysis on the highly expressed miRNAs. GO analysis showed that these miRNAs are important for growth, development, maturation, anatomical structure development, ion binding, cell differentiation, and cell proliferation, all of which are relevant to cochlear development events. The enriched miRNAs in the sEVs are involved in the Hippo, MAPK, Wnt, Notch, TGF-β, and PI3K-Akt signaling pathways, most of which are essential to the development of the cochlea and in regulating the pluripotency of stem cells. These results showed that miRNAs enriched in cochlear tissue-derived sEVs may be essential for cell communication during inner ear development.
Proteins are another major component of sEVs and play significant roles in cell communication, mediation of immune responses, and proliferation of cancer cells and as markers for disease diagnosis [96, 97]. We performed proteomics analysis of the cochlear tissue-derived sEV proteins and identified 5,231 proteins, including 3,120 differentially expressed proteins, in the four age groups. We also found the sEV marker proteins CD63, CD9, CD81, and Tsg101 in the proteomics data, which again verified the purity of our isolated cochlear sEVs. We identified 1,051 proteins in the cochlear sEVs that overlapped with proteins in the Vesiclepedia and Exocarta databases.
Among the differentially expressed sEV proteins, we found that the expression of 17 proteins increased and the expression 124 proteins decreased as the mice aged. For the 17 increased proteins, Slc4a10, Fbxo2, and S100b are related to the process of neurodevelopment and in regulating the differentiation and excitability of neurons [98, 99], which suggests that these three proteins may be involved in the innervation of the cochlea that is required for hearing function. Fbxo2 is enriched in the inner ear and is a key regulator of age-related hearing loss [100]. Tlr3 is also present in the inner ear and regulates immune responses [101, 102], and Tlr4 acts as a mediator in protecting HCs from damage by exosomes secreted by SCs [33]. This suggests that Tlr3 might also have a protective role on inner ear development. In addition, Slc4a10 is important for maintaining ion homeostasis in the inner ear, and the absence of Slc4a10 can lead to hearing loss [103, 104]. Among the proteins that decrease with age, Hnrnp [105], Ddx5 [106], Ilf3 [107], Lamtor5 [108], Psmd2 [109], Ddb1 [110], and Chd4 [111] are reported to be related to cell proliferation and differentiation. Ptbp1 [112], Chd4 [113], Ruvbl2 [114], Cul4a [115], and Lama4 [116] are required for early developmental processes and neuronal differentiation, and some proteins are also present in the inner ear, such as P3h1 [117], Sorcs2 [118], Panx3 [119, 120], Idh1 [121], and Lamb1 [122]. P3h1 knockout mice showed dysplasia of middle ear bones and hearing impairment [117]. Sorcs2 regulates HC development by maintaining the shape of the cilia [118], while Idh1 is a protein found in the cochlea and may play a role in age-related hearing loss by acting as an antioxidant [121, 123]. Panx3 is a pannexin channel protein and is mainly present in the cochlear bone structure and is essential for the maintenance of cochlear morphology [119, 120], and the expression of Panx3 is regulated during development and reaches its peak at P8 [119]. The GO and KEGG analysis of 17 increased and 124 decreased proteins and previous reports of these proteins, some of which are expressed in the inner ear, showed that they affect the ion balance of the inner ear and mediate inner ear immunity and cilia formation [98, 99, 101, 103, 119]. In addition, some proteins are involved in various developmental processes of the skeletal and nervous systems [111, 115, 117] and regulate cell proliferation, differentiation, apoptosis, and other cellular communication processes [110, 111]. Table 3 lists the 20 top candidate proteins among them. These cochlear sEV proteins may play important roles and may be used as new targets for studying the development of the cochlea in the future.
We conducted GO and KEGG analysis of the differentially expressed proteins. The GO analysis revealed that cochlear tissue-derived sEV proteins play significant roles in various biological processes such as Ras protein signal transduction, cell proliferation, cell differentiation, neuron differentiation, endocytosis, cellular ion homeostasis, nervous system development, and organ development and that these proteins are involved in many molecular functions, including ion binding, protein binding, and RNA binding. The cellular components analysis showed that sEVs can be secreted from the cell, cytoplasm, and endomembrane system. According to the KEGG pathway analysis, these proteins are involved in axon guidance, the synaptic vesicle cycle, the AMPK signaling pathway, the mTOR signaling pathway, the PI3K-Akt signaling pathway, and endocytosis. Synapses on HCs are connected to spiral neurons for transmitting signals to the brain, and this activity is essential for hearing function [8, 124, 125]. In addition, these pathways have also been reported to be critical for the biological functions of the inner ear. Down-regulating the AMPK signaling pathway can reduce noise-induced damage to HCs and can prevent age-related hearing loss [126, 127]. The mTOR signaling pathway is involved in reprograming Myc/NICD to promote HC regeneration [16], and age-related hearing loss and HC damage can be relieved by inhibiting the mTOR signaling pathway [128, 129]. Balancing the AMPK and mTOR signaling pathways can further protect HCs from damage by ototoxic drugs [130]. We also created a STRING protein-interaction network investigating the interactions between differentially expressed sEV proteins, and this showed that sEV proteins involved in vesicles, development, neurons, signal communication, cellular processes, and ion homeostasis have close interactions with each other and with other differentially expressed cochlear sEV proteins. These results indicate that sEVs may be critical for the development of the cochlear nervous system, as well as for the protection and regeneration of HCs during development.
In summary, we isolated cochlear tissue-derived sEVs from mice of different ages after birth by ultracentrifugation and characterized the miRNA transcriptomes and proteomics of these sEVs to elucidate their possible roles. We found 561 miRNAs and 5,231 proteins in the cochlear sEVs, and among them 179 miRNAs and 3,120 proteins were differentially expressed at different ages. We further analyzed these differentially expressed miRNAs and proteins and found that the expression of many miRNAs and proteins may be relevant to the maturation of HCs, to changes in SC characteristics, to neural development, and to the protection of HCs from P3 to P21. These miRNAs and proteins might be used as new targets for further studying the detailed mechanism of cochlear development after birth.
Based on our results, we speculate that sEVs play a regulatory role in the maturation of HCs, HC regeneration from inner ear stem cells, and neural development in the inner ear after birth, and this should be further confirmed in future studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Thanks to all colleagues who contributed to this research.
Funding
This work was supported by grants from the National Key RD Program of China (nos. 2021YFA1101300, 2020YFA0112503), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA16010303), the National Natural Science Foundation of China (nos. 82171149, 81970892, 82030029, 81970882), the Natural Science Foundation of Jiangsu Province (nos. BK20190062 and BE2019711), the Science and Technology Department of Sichuan Province (no. 2021YFS0371), the Shenzhen Fundamental Research Program (no. JCYJ20190814093401920, JCYJ20210324125608022), Open Research Fund of State Key Laboratory of Genetic Engineering, Fudan University (No. SKLGE-2109), and the Fundamental Research Funds for the Central Universities for the Support Program of Zhishan Youth Scholars of Southeast University (no. 2242021R41136).
Availability of data and material
All the data and material analyzed in this research are included in this article and its supplementary files.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval and consent to participate
All animal studies followed the authorized guidelines of Southeast University's Animal Care and Use Committee and the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The number of animals was kept to a minimum, and all efforts were made to reduce their suffering. Written informed consent was obtained from individuals or their guardians.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pei Jiang, Xiangyu Ma, Shanying Han and Leyao Ma contributed equally to this work.
Contributor Information
W. Andy Tao, Email: watao@purdue.edu.
Shasha Zhang, Email: zhangss5576@163.com.
Renjie Chai, Email: renjiec@seu.edu.cn.
References
- 1.Wright TJ, et al. Expression of mouse fibroblast growth factor and fibroblast growth factor receptor genes during early inner ear development. Dev Dyn. 2003;228(2):267–272. doi: 10.1002/dvdy.10362. [DOI] [PubMed] [Google Scholar]
- 2.LeMasurier M, Gillespie PG. Hair-cell mechanotransduction and cochlear amplification. Neuron. 2005;48(3):403–415. doi: 10.1016/j.neuron.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson PJ, et al. Sensory hair cell development and regeneration: similarities and differences. Development. 2015;142(9):1561–1571. doi: 10.1242/dev.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns JC, et al. In vivo proliferative regeneration of balance hair cells in newborn mice. J Neurosci. 2012;32(19):6570–6577. doi: 10.1523/JNEUROSCI.6274-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raft S, et al. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134(24):4405–4415. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- 6.Goutman JD, Elgoyhen AB, Gómez-Casati ME. Cochlear hair cells: the sound-sensing machines. FEBS Lett. 2015;589(22):3354–3361. doi: 10.1016/j.febslet.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim KX, Fettiplace R. Developmental changes in the cochlear hair cell mechanotransducer channel and their regulation by transmembrane channel-like proteins. J Gen Physiol. 2013;141(1):141–148. doi: 10.1085/jgp.201210913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fettiplace R. Hair cell transduction, tuning, and synaptic transmission in the mammalian cochlea. Compr Physiol. 2017;7(4):1197–1227. doi: 10.1002/cphy.c160049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang LC, et al. Spatiotemporal definition of neurite outgrowth, refinement and retraction in the developing mouse cochlea. Development. 2007;134(16):2925–2933. doi: 10.1242/dev.001925. [DOI] [PubMed] [Google Scholar]
- 10.Fettiplace R, Kim KX. The physiology of mechanoelectrical transduction channels in hearing. Physiol Rev. 2014;94(3):951–986. doi: 10.1152/physrev.00038.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maoiléidigh DÓ, Ricci AJ. A bundle of mechanisms: inner-ear hair-cell mechanotransduction. Trends Neurosci. 2019;42(3):221–236. doi: 10.1016/j.tins.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun S, et al. Hair cell mechanotransduction regulates spontaneous activity and spiral ganglion subtype specification in the auditory system. Cell. 2018;174(5):1247–1263.e15. doi: 10.1016/j.cell.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikaelian D, Ruben RJ. Development of hearing in the normal Cba-J mouse: correlation of physiological observations with behavioral responses and with cochlear anatomy. Acta Otolaryngol. 1965;59(2–6):451–461. doi: 10.1001/archotol.1964.00750040430011. [DOI] [PubMed] [Google Scholar]
- 14.Samarajeewa A, et al. Transcriptional response to Wnt activation regulates the regenerative capacity of the mammalian cochlea. Development. 2018 doi: 10.1242/dev.166579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang T, et al. Lgr5+ cells regenerate hair cells via proliferation and direct transdifferentiation in damaged neonatal mouse utricle. Nat Commun. 2015;6:6613. doi: 10.1038/ncomms7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu Y, et al. Renewed proliferation in adult mouse cochlea and regeneration of hair cells. Nat Commun. 2019;10(1):5530–5530. doi: 10.1038/s41467-019-13157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7(11):837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, et al. Co-regulation of the Notch and Wnt signaling pathways promotes supporting cell proliferation and hair cell regeneration in mouse utricles. Sci Rep. 2016;6:29418. doi: 10.1038/srep29418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiernan AE. Notch signaling during cell fate determination in the inner ear. Semin Cell Dev Biol. 2013;24(5):470–479. doi: 10.1016/j.semcdb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang D, et al. miR-124 promotes the neuronal differentiation of mouse inner ear neural stem cells. Int J Mol Med. 2016;38(5):1367–1376. doi: 10.3892/ijmm.2016.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010;30(9):3254–3263. doi: 10.1523/JNEUROSCI.4948-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borghesan M, et al. Small extracellular vesicles are key regulators of non-cell autonomous intercellular communication in senescence via the interferon protein IFITM3. Cell Rep. 2019;27(13):3956–3971.e6. doi: 10.1016/j.celrep.2019.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, et al. Mesenchymal stromal cell-derived small extracellular vesicles induce ischemic neuroprotection by modulating leukocytes and specifically neutrophils. Stroke. 2020;51(6):1825–1834. doi: 10.1161/STROKEAHA.119.028012. [DOI] [PubMed] [Google Scholar]
- 24.Loyer X, et al. Intra-cardiac release of extracellular vesicles shapes inflammation following myocardial infarction. Circ Res. 2018;123(1):100–106. doi: 10.1161/CIRCRESAHA.117.311326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 26.Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Chen IH, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci USA. 2017;114(12):3175–3180. doi: 10.1073/pnas.1618088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, et al. Exosome-mediated miR-200b promotes colorectal cancer proliferation upon TGF-β1 exposure. Biomed Pharmacother. 2018;106:1135–1143. doi: 10.1016/j.biopha.2018.07.042. [DOI] [PubMed] [Google Scholar]
- 29.Liao FL, et al. Hematopoietic stem cell-derived exosomes promote hematopoietic differentiation of mouse embryonic stem cells in vitro via inhibiting the miR126/Notch1 pathway. Acta Pharmacol Sin. 2018;39(4):552–560. doi: 10.1038/aps.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo L, et al. Breast cancer cell-derived exosomal miR-20a-5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med. 2019;8(12):5687–5701. doi: 10.1002/cam4.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma P, Schiapparelli L, Cline HT. Exosomes function in cell–cell communication during brain circuit development. Curr Opin Neurobiol. 2013;23(6):997–1004. doi: 10.1016/j.conb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGough IJ, Vincent JP. Exosomes in developmental signalling. Development. 2016;143(14):2482–2493. doi: 10.1242/dev.126516. [DOI] [PubMed] [Google Scholar]
- 33.Breglio AM, et al. Exosomes mediate sensory hair cell protection in the inner ear. J Clin Investig. 2020;130(5):2657–2672. doi: 10.1172/JCI128867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Men Y, et al. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat Commun. 2019;10(1):4136. doi: 10.1038/s41467-019-11534-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai R, et al. Exosomes derived from mouse inner ear stem cells attenuate gentamicin-induced ototoxicity in vitro through the miR-182-5p/FOXO3 axis. J Tissue Eng Regen Med. 2020;14(8):1149–1156. doi: 10.1002/term.3089. [DOI] [PubMed] [Google Scholar]
- 36.Crewe C, et al. An endothelial-to-adipocyte extracellular vesicle axis governed by metabolic state. Cell. 2018;175(3):695–708.e13. doi: 10.1016/j.cell.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang N, et al. Exosomes mediate epithelium-mesenchyme crosstalk in organ development. ACS Nano. 2017;11(8):7736–7746. doi: 10.1021/acsnano.7b01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, et al. Knockdown of Foxg1 in supporting cells increases the trans-differentiation of supporting cells into hair cells in the neonatal mouse cochlea. Cell Mol Life Sci. 2020;77(7):1401–1419. doi: 10.1007/s00018-019-03291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlachos IS, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43(Database issue):D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlachos IS, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, et al. Synergistically bifunctional paramagnetic separation enables efficient isolation of urine extracellular vesicles and downstream phosphoproteomic analysis. ACS Appl Mater Interfaces. 2021;13(3):3622–3630. doi: 10.1021/acsami.0c19400. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Small extracellular vesicles ameliorate peripheral neuropathy and enhance chemotherapy of oxaliplatin on ovarian cancer. J Extracell Vesicles. 2021;10(5):e12073. doi: 10.1002/jev2.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jing H, et al. miR-381-abundant small extracellular vesicles derived from kartogenin-preconditioned mesenchymal stem cells promote chondrogenesis of MSCs by targeting TAOK1. Biomaterials. 2020;231:119682. doi: 10.1016/j.biomaterials.2019.119682. [DOI] [PubMed] [Google Scholar]
- 44.Chen G, et al. Hypoxia-induced let-7f-5p/TARBP2 feedback loop regulates osteosarcoma cell proliferation and invasion by inhibiting the Wnt signaling pathway. Aging (Albany NY) 2020;12(8):6891–6903. doi: 10.18632/aging.103049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Handgraaf S et al (2020) Let-7e-5p regulates GLP-1 content and basal release from enteroendocrine L cells from DIO male mice. Endocrinology 161(2) [DOI] [PubMed]
- 46.Wu Y, et al. Circular RNA circCORO1C promotes laryngeal squamous cell carcinoma progression by modulating the let-7c-5p/PBX3 axis. Mol Cancer. 2020;19(1):99. doi: 10.1186/s12943-020-01215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu F, et al. LncRNA HOXA-AS3 promotes gastric cancer progression by regulating miR-29a-3p/LTβR and activating NF-κB signaling. Cancer Cell Int. 2021;21(1):118. doi: 10.1186/s12935-021-01827-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu Z, et al. Loss of miR-146b-5p promotes T cell acute lymphoblastic leukemia migration and invasion via the IL-17A pathway. J Cell Biochem. 2019;120(4):5936–5948. doi: 10.1002/jcb.27882. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki A, et al. MicroRNA-124-3p suppresses mouse lip mesenchymal cell proliferation through the regulation of genes associated with cleft lip in the mouse. BMC Genom. 2019;20(1):852. doi: 10.1186/s12864-019-6238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang R, et al. MicroRNA-338-3p suppresses ovarian cancer cells growth and metastasis: implication of Wnt/catenin beta and MEK/ERK signaling pathways. J Exp Clin Cancer Res. 2019;38(1):494. doi: 10.1186/s13046-019-1494-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Hou G, et al. LncRNA GAS6-AS2 promotes non-small-cell lung cancer cell proliferation via regulating miR-144-3p/ MAPK6 axis. Cell Cycle. 2021;20(2):179–193. doi: 10.1080/15384101.2020.1867782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Wang N, Xu A. miR-10b-3p, miR-8112 and let-7j as potential biomarkers for autoimmune inner ear diseases. Mol Med Rep. 2019;20(1):171–181. doi: 10.3892/mmr.2019.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ni Z, et al. The exosome-like vesicles from osteoarthritic chondrocyte enhanced mature IL-1β production of macrophages and aggravated synovitis in osteoarthritis. Cell Death Dis. 2019;10(7):522. doi: 10.1038/s41419-019-1739-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang LS, et al. Identification of altered microRNAs in retinas of mice with oxygen-induced retinopathy. Int J Ophthalmol. 2019;12(5):739–745. doi: 10.18240/ijo.2019.05.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang B, et al. Small RNA sequencing reveals exosomal miRNAs involved in the treatment of asthma by scorpio and centipede. Biomed Res Int. 2020;2020:1061407. doi: 10.1155/2020/1061407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang CT, et al. RAB31 targeted by MiR-30c-2-3p regulates the GLI1 signaling pathway, affecting gastric cancer cell proliferation and apoptosis. Front Oncol. 2018;8:554. doi: 10.3389/fonc.2018.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tagne JB, et al. Transcription factor and microRNA interactions in lung cells: an inhibitory link between NK2 homeobox 1, miR-200c and the developmental and oncogenic factors Nfib and Myb. Respir Res. 2015;16(1):22. doi: 10.1186/s12931-015-0186-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia Y, et al. lncRNA NEAT1 facilitates melanoma cell proliferation, migration, and invasion via regulating miR-495-3p and E2F3. J Cell Physiol. 2019;234(11):19592–19601. doi: 10.1002/jcp.28559. [DOI] [PubMed] [Google Scholar]
- 59.Dou D, et al. Circ_0008039 supports breast cancer cell proliferation, migration, invasion, and glycolysis by regulating the miR-140-3p/SKA2 axis. Mol Oncol. 2021;15(2):697–709. doi: 10.1002/1878-0261.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu WI, et al. Effect of Xiaoai Jiedu recipe on mIRNA expression profiles in H22 tumor-bearing mice. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2016;36(9):1112–1118. [PubMed] [Google Scholar]
- 61.Meyer SU, et al. Integrative analysis of microRNA and mRNA data reveals an orchestrated function of microRNAs in skeletal myocyte differentiation in response to TNF-α or IGF1. PLoS ONE. 2015;10(8):e0135284. doi: 10.1371/journal.pone.0135284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X, et al. MiR-409-3p and MiR-1896 co-operatively participate in IL-17-induced inflammatory cytokine production in astrocytes and pathogenesis of EAE mice via targeting SOCS3/STAT3 signaling. Glia. 2019;67(1):101–112. doi: 10.1002/glia.23530. [DOI] [PubMed] [Google Scholar]
- 63.Eom TY, et al. Schizophrenia-related microdeletion causes defective ciliary motility and brain ventricle enlargement via microRNA-dependent mechanisms in mice. Nat Commun. 2020;11(1):912. doi: 10.1038/s41467-020-14628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu X, et al. Regulatory mechanism of miR-543-3p on GLT-1 in a mouse model of Parkinson's disease. ACS Chem Neurosci. 2019;10(3):1791–1800. doi: 10.1021/acschemneuro.8b00683. [DOI] [PubMed] [Google Scholar]
- 65.Gässler A, et al. Overexpression of Gjb4 impairs cell proliferation and insulin secretion in primary islet cells. Mol Metab. 2020;41:101042. doi: 10.1016/j.molmet.2020.101042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu T, Guo J, Zhang X. MiR-202-5p/PTEN mediates doxorubicin-resistance of breast cancer cells via PI3K/Akt signaling pathway. Cancer Biol Ther. 2019;20(7):989–998. doi: 10.1080/15384047.2019.1591674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galleggiante V, et al. Quercetin-induced miR-369-3p suppresses chronic inflammatory response targeting C/EBP-β. Mol Nutr Food Res. 2019;63(19):e1801390. doi: 10.1002/mnfr.201801390. [DOI] [PubMed] [Google Scholar]
- 68.Li Q, et al. Circular RNA circ-0016068 promotes the growth, migration, and invasion of prostate cancer cells by regulating the miR-330-3p/BMI-1 axis as a competing endogenous RNA. Front Cell Dev Biol. 2020;8:827. doi: 10.3389/fcell.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao X, et al. Pulmonary silicosis alters microRNA expression in rat lung and miR-411-3p exerts anti-fibrotic effects by inhibiting MRTF-A/SRF signaling. Mol Ther Nucleic Acids. 2020;20:851–865. doi: 10.1016/j.omtn.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu X, Yao X, Liu D. Up-regulation of microRNA-335-5p reduces inflammation via negative regulation of the TPX2-mediated AKT/GSK3β signaling pathway in a chronic rhinosinusitis mouse model. Cell Signal. 2020;70:109596. doi: 10.1016/j.cellsig.2020.109596. [DOI] [PubMed] [Google Scholar]
- 71.Kim KS, et al. ELK3 expressed in lymphatic endothelial cells promotes breast cancer progression and metastasis through exosomal miRNAs. Sci Rep. 2019;9(1):8418. doi: 10.1038/s41598-019-44828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jee YH, et al. mir-374-5p, mir-379-5p, and mir-503-5p regulate proliferation and hypertrophic differentiation of growth plate chondrocytes in male rats. Endocrinology. 2018;159(3):1469–1478. doi: 10.1210/en.2017-00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Munnamalai V, Fekete DM. Wnt signaling during cochlear development. Semin Cell Dev Biol. 2013;24(5):480–489. doi: 10.1016/j.semcdb.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riccomagno MM, Takada S, Epstein DJ. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005;19(13):1612–1623. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eguchi T et al (2020) Cell stress induced stressome release including damaged membrane vesicles and extracellular HSP90 by prostate cancer cells. Cells 9(3) [DOI] [PMC free article] [PubMed]
- 76.Štok U, et al. Characterization of plasma-derived small extracellular vesicles indicates ongoing endothelial and platelet activation in patients with thrombotic antiphospholipid syndrome. Cells. 2020 doi: 10.3390/cells9051211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wong EHC, et al. Inner ear exosomes and their potential use as biomarkers. PLoS ONE. 2018;13(6):e0198029. doi: 10.1371/journal.pone.0198029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crescitelli R, et al. Subpopulations of extracellular vesicles from human metastatic melanoma tissue identified by quantitative proteomics after optimized isolation. J Extracell Vesicles. 2020;9(1):1722433–1722433. doi: 10.1080/20013078.2020.1722433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang Y, et al. Influence of species and processing parameters on recovery and content of brain tissue-derived extracellular vesicles. J Extracell Vesicles. 2020;9(1):1785746–1785746. doi: 10.1080/20013078.2020.1785746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vella LJ, et al. A rigorous method to enrich for exosomes from brain tissue. J Extracell Vesicles. 2017;6(1):1348885. doi: 10.1080/20013078.2017.1348885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muraoka S, et al. Proteomic and biological profiling of extracellular vesicles from Alzheimer's disease human brain tissues. Alzheimers Dement. 2020;16(6):896–907. doi: 10.1002/alz.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gallart-Palau X, Serra A, Sze SK. Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR. Mol Neurodegener. 2016;11(1):41. doi: 10.1186/s13024-016-0108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perez-Gonzalez R, et al. The exosome secretory pathway transports amyloid precursor protein carboxyl-terminal fragments from the cell into the brain extracellular space. J Biol Chem. 2012;287(51):43108–43115. doi: 10.1074/jbc.M112.404467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wan S, et al. CD8α(+)CD11c(+) extracellular vesicles in the lungs control immune homeostasis of the respiratory tract via TGF-β1 and IL-10. J Immunol. 2018;200(5):1651–1660. doi: 10.4049/jimmunol.1701447. [DOI] [PubMed] [Google Scholar]
- 85.Crescitelli R, Lässer C, Lötvall J. Isolation and characterization of extracellular vesicle subpopulations from tissues. Nat Protoc. 2021;16(3):1548–1580. doi: 10.1038/s41596-020-00466-1. [DOI] [PubMed] [Google Scholar]
- 86.Kalluri R, LeBleu VS (2020) The biology, function, and biomedical applications of exosomes. Science 367(6478) [DOI] [PMC free article] [PubMed]
- 87.Mensà E, et al. Small extracellular vesicles deliver miR-21 and miR-217 as pro-senescence effectors to endothelial cells. J Extracell Vesicles. 2020;9(1):1725285. doi: 10.1080/20013078.2020.1725285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang H, et al. BMMSC-sEV-derived miR-328a-3p promotes ECM remodeling of damaged urethral sphincters via the Sirt7/TGFβ signaling pathway. Stem Cell Res Ther. 2020;11(1):286. doi: 10.1186/s13287-020-01808-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lv J, et al. MicroRNA let-7c-5p improves neurological outcomes in a murine model of traumatic brain injury by suppressing neuroinflammation and regulating microglial activation. Brain Res. 2018;1685:91–104. doi: 10.1016/j.brainres.2018.01.032. [DOI] [PubMed] [Google Scholar]
- 90.Mathew LK, et al. Restricted expression of miR-30c-2-3p and miR-30a-3p in clear cell renal cell carcinomas enhances HIF2α activity. Cancer Discov. 2014;4(1):53–60. doi: 10.1158/2159-8290.CD-13-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cai J, et al. Oral squamous cell carcinoma-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am J Physiol Cell Physiol. 2019;316(5):C731–c740. doi: 10.1152/ajpcell.00366.2018. [DOI] [PubMed] [Google Scholar]
- 92.Zhou J, et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res. 2018;6(12):1578–1592. doi: 10.1158/2326-6066.CIR-17-0479. [DOI] [PubMed] [Google Scholar]
- 93.Zhong Q, et al. Long non-coding RNA TUG1 modulates expression of elastin to relieve bronchopulmonary dysplasia via sponging miR-29a-3p. Front Pediatr. 2020;8:573099. doi: 10.3389/fped.2020.573099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Song Q, et al. Long non-coding RNA LINC00473 acts as a microRNA-29a-3p sponge to promote hepatocellular carcinoma development by activating Robo1-dependent PI3K/AKT/mTOR signaling pathway. Ther Adv Med Oncol. 2020;12:1758835920937890. doi: 10.1177/1758835920937890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Le LT, et al. The microRNA-29 family in cartilage homeostasis and osteoarthritis. J Mol Med (Berl) 2016;94(5):583–596. doi: 10.1007/s00109-015-1374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Servage KA, et al. Proteomic profiling of small extracellular vesicles secreted by human pancreatic cancer cells implicated in cellular transformation. Sci Rep. 2020;10(1):7713. doi: 10.1038/s41598-020-64718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vinik Y et al (2020) Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response. Sci Adv 6(40) [DOI] [PMC free article] [PubMed]
- 98.Sinning A, Liebmann L, Hübner CA. Disruption of Slc4a10 augments neuronal excitability and modulates synaptic short-term plasticity. Front Cell Neurosci. 2015;9:223. doi: 10.3389/fncel.2015.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Atkin G, et al. Loss of F-box only protein 2 (Fbxo2) disrupts levels and localization of select NMDA receptor subunits, and promotes aberrant synaptic connectivity. J Neurosci. 2015;35(15):6165–6178. doi: 10.1523/JNEUROSCI.3013-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nelson RF, et al. Selective cochlear degeneration in mice lacking the F-box protein, Fbx2, a glycoprotein-specific ubiquitin ligase subunit. J Neurosci. 2007;27(19):5163–5171. doi: 10.1523/JNEUROSCI.0206-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamada T, et al. Toll-like receptor ligands induce cytokine and chemokine production in human inner ear endolymphatic sac fibroblasts. Auris Nasus Larynx. 2017;44(4):398–403. doi: 10.1016/j.anl.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 102.Gollmann-Tepeköylü C et al (2020) Shock waves promote spinal cord repair via TLR3. JCI Insight 5(15) [DOI] [PMC free article] [PubMed]
- 103.Huebner AK, et al. Early hearing loss upon disruption of Slc4a10 in C57BL/6 mice. J Assoc Res Otolaryngol. 2019;20(3):233–245. doi: 10.1007/s10162-019-00719-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sun S, et al. Solute carrier family 4 member 1 might participate in the pathogenesis of Meniere's disease in a murine endolymphatic hydrop model. Acta Otolaryngol. 2019;139(11):966–976. doi: 10.1080/00016489.2019.1663365. [DOI] [PubMed] [Google Scholar]
- 105.Chen EB, et al. HnRNPR-CCNB1/CENPF axis contributes to gastric cancer proliferation and metastasis. Aging (Albany NY) 2019;11(18):7473–7491. doi: 10.18632/aging.102254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Du C, et al. DDX5 promotes gastric cancer cell proliferation in vitro and in vivo through mTOR signaling pathway. Sci Rep. 2017;7:42876. doi: 10.1038/srep42876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jia R, et al. Oncogenic splicing factor SRSF3 regulates ILF3 alternative splicing to promote cancer cell proliferation and transformation. RNA. 2019;25(5):630–644. doi: 10.1261/rna.068619.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu BW, et al. Oncoprotein HBXIP induces PKM2 via transcription factor E2F1 to promote cell proliferation in ER-positive breast cancer. Acta Pharmacol Sin. 2019;40(4):530–538. doi: 10.1038/s41401-018-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li Y, et al. PSMD2 regulates breast cancer cell proliferation and cell cycle progression by modulating p21 and p27 proteasomal degradation. Cancer Lett. 2018;430:109–122. doi: 10.1016/j.canlet.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 110.Zheng W et al (2019) DDB1 regulates sertoli cell proliferation and testis cord remodeling by TGFβ pathway. Genes (Basel) 10(12) [DOI] [PMC free article] [PubMed]
- 111.Xu N, et al. CHD4 mediates proliferation and migration of non-small cell lung cancer via the RhoA/ROCK pathway by regulating PHF5A. BMC Cancer. 2020;20(1):262. doi: 10.1186/s12885-020-06762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mokabber H, Najafzadeh N, Mohammadzadeh Vardin M. miR-124 promotes neural differentiation in mouse bulge stem cells by repressing Ptbp1 and Sox9. J Cell Physiol. 2019;234(6):8941–8950. doi: 10.1002/jcp.27563. [DOI] [PubMed] [Google Scholar]
- 113.Hirota A, et al. The nucleosome remodeling and deacetylase complex protein CHD4 regulates neural differentiation of mouse embryonic stem cells by down-regulating p53. J Biol Chem. 2019;294(1):195–209. doi: 10.1074/jbc.RA118.004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hong S, et al. RuvB-like protein 2 (Ruvbl2) has a role in directing the neuroectodermal differentiation of mouse embryonic stem cells. Stem Cells Dev. 2016;25(18):1376–1385. doi: 10.1089/scd.2016.0076. [DOI] [PubMed] [Google Scholar]
- 115.Liu L, et al. Essential role of the CUL4B ubiquitin ligase in extra-embryonic tissue development during mouse embryogenesis. Cell Res. 2012;22(8):1258–1269. doi: 10.1038/cr.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim IM, et al. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem. 2005;280(23):22278–22286. doi: 10.1074/jbc.M500936200. [DOI] [PubMed] [Google Scholar]
- 117.Pokidysheva E, et al. Prolyl 3-hydroxylase-1 null mice exhibit hearing impairment and abnormal morphology of the middle ear bone joints. Matrix Biol. 2013;32(1):39–44. doi: 10.1016/j.matbio.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Forge A, et al. Disruption of SorCS2 reveals differences in the regulation of stereociliary bundle formation between hair cell types in the inner ear. PLoS Genet. 2017;13(3):e1006692. doi: 10.1371/journal.pgen.1006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Abitbol JM, et al. Differential effects of pannexins on noise-induced hearing loss. Biochem J. 2016;473(24):4665–4680. doi: 10.1042/BCJ20160668. [DOI] [PubMed] [Google Scholar]
- 120.Abitbol JM, et al. Double deletion of Panx1 and Panx3 affects skin and bone but not hearing. J Mol Med (Berl) 2019;97(5):723–736. doi: 10.1007/s00109-019-01779-9. [DOI] [PubMed] [Google Scholar]
- 121.Kim YR, et al. Expression patterns of members of the isocitrate dehydrogenase gene family in murine inner ear. Biotechnol Histochem. 2017;92(7):536–544. doi: 10.1080/10520295.2017.1367034. [DOI] [PubMed] [Google Scholar]
- 122.Meyerzum Gottesberge AM. Felix H, Abnormal basement membrane in the inner ear and the kidney of the Mpv17−/− mouse strain: ultrastructural and immunohistochemical investigations. Histochem Cell Biol. 2005;124(6):507–516. doi: 10.1007/s00418-005-0027-7. [DOI] [PubMed] [Google Scholar]
- 123.Tadros SF, et al. Gene expression changes for antioxidants pathways in the mouse cochlea: relations to age-related hearing deficits. PLoS ONE. 2014;9(2):e90279. doi: 10.1371/journal.pone.0090279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kearney G, et al. Developmental synaptic changes at the transient olivocochlear-inner hair cell synapse. J Neurosci. 2019;39(18):3360–3375. doi: 10.1523/JNEUROSCI.2746-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Michanski S, et al. Mapping developmental maturation of inner hair cell ribbon synapses in the apical mouse cochlea. Proc Natl Acad Sci USA. 2019;116(13):6415–6424. doi: 10.1073/pnas.1812029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao J, et al. Down-regulation of AMPK signaling pathway rescues hearing loss in TFB1 transgenic mice and delays age-related hearing loss. Aging (Albany NY) 2020;12(7):5590–5611. doi: 10.18632/aging.102977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nagashima R, et al. Acoustic overstimulation activates 5'-AMP-activated protein kinase through a temporary decrease in ATP level in the cochlear spiral ligament prior to permanent hearing loss in mice. Neurochem Int. 2011;59(6):812–820. doi: 10.1016/j.neuint.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 128.Fu X, et al. Tuberous sclerosis complex-mediated mTORC1 overactivation promotes age-related hearing loss. J Clin Investig. 2018;128(11):4938–4955. doi: 10.1172/JCI98058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Leitmeyer K, et al. Inhibition of mTOR by rapamycin results in auditory hair cell damage and decreased spiral ganglion neuron outgrowth and neurite formation in vitro. Biomed Res Int. 2015;2015:925890. doi: 10.1155/2015/925890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bodmer D, Levano-Huaman S. Sesn2/AMPK/mTOR signaling mediates balance between survival and apoptosis in sensory hair cells under stress. Cell Death Dis. 2017;8(10):e3068–e3068. doi: 10.1038/cddis.2017.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yao K, Qiu S, Tian L, Snider WD, Flannery JG, Schaffer DV, Chen B. Wnt regulates proliferation and neurogenic potential of Müller glial cells via a Lin28/let-7 miRNA-dependent pathway in adult mammalian retinas. Cell Rep. 2016;17(1):165–178. doi: 10.1016/j.celrep.2016.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang W, Liu H, Liu W, Liu Y, Xu J. Polycomb-mediated loss of microRNA let-7c determines inflammatory macrophage polarization via PAK1-dependent NF-κB pathway. Cell Death Differ. 2015;22(2):287–297. doi: 10.1038/cdd.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen PY, Qin L, Barnes C, Charisse K, Yi T, Zhang X, Ali R, Medina PP, Yu J, Slack FJ, Anderson DG. FGF regulates TGF-β signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep. 2012;2(6):1684–1696. doi: 10.1016/j.celrep.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.You L, Chen H, Xu L, Li X. Overexpression of miR-29a-3p suppresses proliferation, migration, and invasion of vascular smooth muscle cells in atherosclerosis via targeting TNFRSF1A. BioMed Res Int. 2020 doi: 10.1155/2020/9627974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Volpicelli F, Speranza L, Pulcrano S, De Gregorio R, Crispino M, De Sanctis C, Leopoldo M, Lacivita E, di Porzio U, Bellenchi GC, Perrone-Capano C. The microRNA-29a modulates serotonin 5-HT7 receptor expression and its effects on hippocampal neuronal morphology. Mol Neurobiol. 2019;56(12):8617–8627. doi: 10.1007/s12035-019-01690-x. [DOI] [PubMed] [Google Scholar]
- 136.Wang Y, Zhang D, Tang Z, Zhang Y, Gao H, Ni N, Shen B, Sun H, Gu P. REST regulated by RA through miR-29a and the proteasome pathway plays a crucial role in RPC proliferation and differentiation. Cell Death Dis. 2018 doi: 10.1038/s41419-018-0473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, Fitamant J, Bardeesy N, Camargo FD, Guan KL. YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nature Cell Biol. 2012;14(12):1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, Mastick GS, Xu C, Yan W. Two miRNA clusters miR-34b/c and miR-449 are essential for normal brain development motile ciliogenesis and spermatogenesis. Proceed Nat Acad Sci. 2014;111(28):E2851–E2857. doi: 10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bou Kheir T, Futoma-Kazmierczak E, Jacobsen A, Krogh A, Bardram L, Hother C, Grønbæk K, Federspiel B, Lund AH, Friis-Hansen L. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Molecular Cancer. 2011 doi: 10.1186/1476-4598-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hu F, Wang M, Xiao T, Yin B, He L, Meng W, Dong M, Liu F. miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes. 2015;64(6):2056–2068. doi: 10.2337/db14-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hand NJ, Master ZR, Eauclaire SF, Weinblatt DE, Matthews RP, Friedman JR. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterol. 2009;136(3):1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liang T, Zhou B, Shi L, Wang H, Chu Q, Xu F, Li Y, Chen R, Shen C, Schinckel AP. IncRNA AK017368 promotes proliferation and suppresses differentiation of myoblasts in skeletal muscle development by attenuating the function of miR-30c. FASEB J. 2018;32(1):377–389. doi: 10.1096/fj.201700560rr. [DOI] [PubMed] [Google Scholar]
- 143.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, Blelloch R. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Develop. 2010;24(10):992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database):D152–D157:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang Q, Liu H, McGee J, Walsh EJ, Soukup GA, He DZ. Identifying microRNAs involved in degeneration of the organ of corti during age-related hearing loss. PLoS ONE. 2013;8(4):e62786. doi: 10.1371/journal.pone.0062786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Krisher T, Bar-Shavit Z. Regulation of osteoclastogenesis by integrated signals from toll-like receptors. J Cell Biochem. 2014;115(12):2146–2154. doi: 10.1002/jcb.24891. [DOI] [PubMed] [Google Scholar]
- 147.Christensen IB, Wu Q, Bohlbro AS, Skals MG, Damkier HH, Hübner CA, Fenton RA, Praetorius J. Genetic disruption of slc4a10 alters the capacity for cellular metabolism and vectorial ion transport in the choroid plexus epithelium. Fluids Barriers CNS. 2020 doi: 10.1186/s12987-019-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Raponi E, Agenes F, Delphin C, Assard N, Baudier J, Legraverend C, Deloulme JC. S100B expression defines a state in which GFAP-expressing cells lose their neural stem cell potential and acquire a more mature developmental stage. Glia. 2007;55(2):165–177. doi: 10.1002/glia.20445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zeng X, Ye M, Resch JM, Jedrychowski MP, Hu B, Lowell BB, Ginty DD, Spiegelman BM. Innervation of thermogenic adipose tissue via a calsyntenin 3β–S100b axis. Nature. 2019;569(7755):229–235. doi: 10.1038/s41586-019-1156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nishiyama H, Knöpfel T, Endo S, Itohara S. Glial protein S100B modulates long-term neuronal synaptic plasticity. Proceed Nat Acad Sci. 2002;99(6):4037–4042. doi: 10.1073/pnas.052020999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Riuzzi F, Sorci G, Beccafico S, Donato R. S100B engages RAGE or bFGF/FGFR1 in myoblasts depending on its own concentration and myoblast density. Implications for Muscle Regeneration. PLoS ONE. 2012;7(1):e28700. doi: 10.1371/journal.pone.0028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Legrand J, Chan AL, La HM, Rossello FJ, Änkö ML, Fuller-Pace FV, Hobbs RM. DDX5 plays essential transcriptional and post-transcriptional roles in the maintenance and function of spermatogonia. Nature Commun. 2019 doi: 10.1038/s41467-019-09972-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dardenne E, Espinoza MP, Fattet L, Germann S, Lambert MP, Neil H, Zonta E, Mortada H, Gratadou L, Deygas M, Chakrama FZ. RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 2014;7(6):1900–1913. doi: 10.1016/j.celrep.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 154.Arun G, Akhade VS, Donakonda S, Rao MR. mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Mol Cell Biol. 2012;32(15):3140–3152. doi: 10.1128/MCB.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tomofuji Y, Takaba H, Suzuki HI, Benlaribi R, Martinez CD, Abe Y, Morishita Y, Okamura T, Taguchi A, Kodama T, Takayanagi H. Chd4 choreographs self-antigen expression for central immune tolerance. Nature Immunol. 2020;21(8):892–901. doi: 10.1038/s41590-020-0717-2. [DOI] [PubMed] [Google Scholar]
- 156.Sun F, Yang Q, Weng W, Zhang Y, Yu Y, Hong A, Ji Y, Pan Q. Chd4 and associated proteins function as corepressors of Sox9 expression during BMP-2-induced chondrogenesis. J Bone Mineral Res. 2013;28(9):1950–1961. doi: 10.1002/jbmr.1932. [DOI] [PubMed] [Google Scholar]
- 157.Leloup N, Chataigner LM, Janssen BJ. Structural insights into SorCS2–Nerve Growth Factor complex formation. Nature Commun. 2018 doi: 10.1038/s41467-018-05405-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Malik AR, Szydlowska K, Nizinska K, Asaro A, van Vliet EA, Popp O, Dittmar G, Fritsche-Guenther R, Kirwan JA, Nykjaer A, Lukasiuk K, Aronica E, Willnow TE. SorCS2 controls functional expression of amino acid transporter EAAT3 and protects neurons from oxidative stress and epilepsy-induced pathology. Cell Rep. 2019;26(10):2792–2804.e6. doi: 10.1016/j.celrep.2019.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Itsumi M, Inoue S, Elia AJ, Murakami K, Sasaki M, Lind EF, Brenner D, Harris IS, Chio II, Afzal S, Cairns RA, Cescon DW, Elford AR, Ye J, Lang PA, Li WY, Wakeham A, Duncan GS, Haight J, You-Ten A, Snow B, Yamamoto K, Ohashi PS, Mak TW. Idh1 protects murine hepatocytes from endotoxin-induced oxidative stress by regulating the intracellular NADP+/NADPH ratio. Cell Death Differ. 2015;22(11):1837–1845. doi: 10.1038/cdd.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, Kunii K, Pedraza A, Schalm S, Silverman L, Miller A, Wang F, Yang H, Chen Y, Kernytsky A, Rosenblum MK, Liu W, Biller SA, Su SM, Brennan CW, Chan TA, Graeber TG, Yen KE, Mellinghoff IK. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ishikawa M, Iwamoto T, Fukumoto S, Yamada Y. Pannexin 3 inhibits proliferation of osteoprogenitor cells by regulating Wnt and p21 signaling. J Biol Chemistry. 2014;289(5):2839–285. doi: 10.1074/jbc.M113.523241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ruan QT, Yazdani N, Blum BC, Beierle JA, Lin W, Coelho MA, Fultz EK, Healy AF, Shahin JR, Kandola AK, Luttik KP, Zheng K, Smith NJ, Cheung J, Mortazavi F, Apicco DJ, Varman DR, Ramamoorthy S, Ash PEA, Rosene DL, Emili A, Wolozin B, Szumlinski KK, Bryant CD. A mutation in Hnrnph1 that decreases methamphetamine-induced reinforcement, reward, and dopamine release and increases synaptosomal hnRNP H and mitochondrial proteins. J Neurosci. 2020;40(1):107–130. doi: 10.1523/JNEUROSCI.1808-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Linares AJ, Lin CH, Damianov A, Adams KL, Novitch BG, Black DL. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. eLife. 2015 doi: 10.7554/eLife.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Georgilis A, Klotz S, Hanley CJ, Herranz N, Weirich B, Morancho B, Leote AC, D'Artista L, Gallage S, Seehawer M, Carroll T, Dharmalingam G, Wee KB, Mellone M, Pombo J, Heide D, Guccione E, Arribas J, Barbosa-Morais NL, Heikenwalder M, Thomas GJ, Zender L, Gil J. PTBP1-mediated alternative splicing regulates the inflammatory secretome and the pro-tumorigenic effects of senescent cells. Cancer Cell. 2018;34(1):85–102.e9. doi: 10.1016/j.ccell.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vuong JK, Lin CH, Zhang M, Chen L, Black DL, Zheng S. PTBP1 and PTBP2 serve both specific and redundant functions in neuronal pre-mRNA splicing. Cell Rep. 2016;17(10):2766–2775. doi: 10.1016/j.celrep.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gorfu G, Virtanen I, Hukkanen M, Lehto VP, Rousselle P, Kenne E, Lindbom L, Kramer R, Tryggvason K, Patarroyo M. Laminin isoforms of lymph nodes and predominant role of α5-laminin(s) in adhesion and migration of blood lymphocytes. J Leukoc Biol. 2008;84(3):701–712. doi: 10.1189/jlb.0108048. [DOI] [PubMed] [Google Scholar]
- 167.Yang L, Chen W, Li L, Xiao Y, Fan S, Zhang Q, Xia T, Li M, Hong Y, Zhao T, Li Q, Liu W-H, Xiao N. Ddb1 Is essential for the expansion of CD4+ helper T cells by regulating cell cycle progression and cell death. Frontiers Immunol. 2021 doi: 10.3389/fimmu.2021.722273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gao J, Buckley SM, Cimmino L, Guillamot M, Strikoudis A, Cang Y, Goff SP, Aifantis I. The CUL4-DDB1 ubiquitin ligase complex controls adult and embryonic stem cell differentiation and homeostasis. eLife. 2015 doi: 10.7554/eLife.07539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pan Y, Wang B, Yang X, Bai F, Xu Q, Li X, Gao L, Ma C, Liang X. CUL4A facilitates hepatocarcinogenesis by promoting cell cycle progression and epithelial-mesenchymal transition. Sci Rep. 2015 doi: 10.1038/srep17006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kopanja D, Stoyanova T, Okur MN, Huang E, Bagchi S, Raychaudhuri P. Proliferation defects and genome instability in cells lacking Cul4A. Oncogene. 2009;28(26):2456–2465. doi: 10.1038/onc.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Waning DL, Li B, Jia N, Naaldijk Y, Goebel WS, HogenEsch H, Chun KT. Cul4A is required for hematopoietic cell viability and its deficiency leads to apoptosis. Blood. 2008;112(2):320–329. doi: 10.1182/blood-2007-11-126300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fratzl-Zelman N, Bächinger HP, Vranka JA, Roschger P, Klaushofer K, Rauch F. Bone matrix hypermineralization in prolyl-3 hydroxylase 1 deficient mice. Bone. 2016;85:15–22. doi: 10.1016/j.bone.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 173.Pokidysheva E, Tufa S, Bresee C, Brigande JV, Bächinger HP. Prolyl 3-hydroxylase-1 null mice exhibit hearing impairment and abnormal morphology of the middle ear bone joints. Matrix Biol. 2013;32(1):39–44. doi: 10.1016/j.matbio.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vranka JA, Pokidysheva E, Hayashi L, Zientek K, Mizuno K, Ishikawa Y, Maddox K, Tufa S, Keene DR, Klein R, Bächinger HP. Prolyl 3-hydroxylase 1 null mice display abnormalities in fibrillar collagen-rich tissues such as tendons, skin, and bones. J Biol Chem. 2010;285(22):17253–17262. doi: 10.1074/jbc.M110.102228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Larcher JC, Gasmi L, Viranaïcken W, Eddé B, Bernard R, Ginzburg I, Denoulet P. Ilf3 and NF90 associate with the axonal targeting element of Tau mRNA. FASEB J. 2004;18(14):1761–1763. doi: 10.1096/fj.04-1763fje. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data and material analyzed in this research are included in this article and its supplementary files.