Abstract
Endothelial damage is the initial and crucial factor in the occurrence and development of vascular complications in diabetic patients, contributing to morbidity and mortality. Although hyperglycemia has been identified as a damaging effector, the detailed mechanisms remain elusive. In this study, identified by ATAC-seq and RNA-seq, JunB reverses the inhibition of proliferation and the promotion of apoptosis in human umbilical vein endothelial cells treated with high glucose, mainly through the cell cycle and p53 signaling pathways. Furthermore, JunB undergoes phase separation in the nucleus and in vitro, mediated by its intrinsic disordered region and DNA-binding domain. Nuclear localization and condensation behaviors are required for JunB-mediated proliferation and apoptosis. Thus, our study uncovers the roles of JunB and its coacervation in repairing vascular endothelial damage caused by high glucose, elucidating the involvement of phase separation in diabetes and diabetic endothelial dysfunction.
Keywords: hyperglycemia, JunB, phase separation, endothelial damage
Introduction
Diabetes mellitus (DM) is a chronic disease attributed to insufficient insulin secretion or utilization (or both) and can be divided into type 1 and type 2 (Ahmad et al., 2022). Micro- and macro-vascular complications contribute to the morbidity and mortality of patients with DM, including neuropathy, nephropathy, retinopathy, coronary heart disease, cerebrovascular disease, and peripheral arterial disease (Chen et al., 2017). Evidence suggests that endothelial damage and dysfunction not only initiate vascular complications in DM, but also play an essential role in its development (Lorenzi and Cagliero, 1991; Cohen, 1993; Piqueras et al., 2007; Higashi et al., 2009; Gimbrone and Garcia-Cardena, 2016; Vanhoutte et al., 2017). The endothelium, composed of a single-cell layer, acts as the barrier between the blood and the vascular wall. Moreover, it plays both endocrine and paracrine roles in the body, producing vasodilators such as nitric oxide and vasoconstrictors such as endothelin-1 (Sumpio et al., 2002; Vane et al., 1990; Rafii et al., 2016; Yang and Cao, 2022). Endothelial cells (ECs) are involved in cell adhesion, angiogenesis, hemostasis, and blood flow (Feletou and Vanhoutte, 2006). Once the endothelium gets damaged, surrounding healthy ECs with a rapid proliferation rate replace the damaged cells to prevent the exposure of the vascular cavity surface (Hansson et al., 1985; Itoh et al., 2010). In addition, enhancing apoptosis also evokes the proliferation of ECs (Wright, 1968; Schwartz and Benditt, 1977; Kaiser et al., 1997; Sakao et al., 2005; Foteinos et al., 2008; Chiu and Chien, 2011).
Hyperglycemia has been consistently reported as the major risk factor for vascular complications in early-stage DM (Ruderman et al., 1992; Diabetes et al., 1993; Susztak et al., 2006). Exposure to high glucose inhibits the proliferation of vascular ECs and prompts apoptosis, leading to endothelial damage and dysfunction (Lorenzi et al., 1985; Risso et al., 2001; McGinn et al., 2003; Quagliaro et al., 2003; Varma et al., 2005; Piqueras et al., 2007; Bhatt et al., 2013; Liu et al., 2014; Chen et al., 2017). This is primarily mediated by the PI3K/Akt, ERK1/2, JNK, NF-κB, and p38 pathways (Bach et al., 1997; Brouard et al., 2000; Bubici et al., 2006; Curtis et al., 2009; Chaudhury et al., 2010; Zhang et al., 2011; Charreau, 2012; Abeyrathna and Su, 2015). High glucose induces endothelial damage through diverse mechanisms, including stimulation of the polyol pathway, elevation of advanced glycation end-products, activation of protein kinase C (PKC), activation of the hexosamine biosynthetic pathway, and induction of oxidative stress (Quyyumi et al., 1995; Forstermann and Li, 2011). Nevertheless, the underlying mechanisms of endothelial dysfunction caused by hyperglycemia are still vague. Investigating the detailed processes of endothelial dysfunction induced by high glucose has critical clinical significance and may improve the prevention and treatment of vascular complications in diabetes.
Biological liquid‒liquid phase separation (LLPS) is a process by which multiple molecules are concentrated into a liquid-like compartment that coexists with the surrounding environment. Protein LLPS is mainly triggered by polyvalent interactions and mediated by proteins carrying intrinsic disordered regions (IDRs) (Bergeron-Sandoval et al., 2016; Banani et al., 2017; Shin and Brangwynne, 2017). Furthermore, it is also modulated by environmental changes such as media composition, concentration, temperature, pH, and salt concentration (Banani et al., 2017). LLPS has been reported to be involved in a myriad of physiological activities, including autophagy, immunity, stress response, tight junctions, cell division, cell signaling, and aging (Ganassi et al., 2016; Banani et al., 2017; Shin and Brangwynne, 2017; Boeynaems et al., 2018; Franzmann et al., 2018; Hofweber et al., 2018; Zhang et al., 2018a; Beutel et al., 2019; Wolozin and Ivanov, 2019; Bhattacharya and Behrends, 2020; Fujioka et al., 2020; Huang et al., 2021). However, whether and how phase separation contributes to diabetic endothelial dysfunction remain unknown.
JunB belongs to the AP-1 transcription factor (TF) family, participating in lymphatic vascular morphogenesis, differentiation of erythroid cells and naive T helper cells, regeneration of tadpole tails through positive regulation of cell proliferation, modulation of lipotoxic β-cell death, the transition from S to G2/M phase by activating cyclin A, the proliferation of embryonic fibroblasts, inhibition of NF-κB-dependent inflammation, overproduction of type I collagen, development of dermatofibrosis, and cell proliferation regulation (Bakiri et al., 2000; Andrecht et al., 2002; Jacobs-Helber et al., 2002; Cunha et al., 2014; Kiesow et al., 2015; Ponticos et al., 2015; Zhang et al., 2015; Jia et al., 2016; Carr et al., 2017; Hasan et al., 2017; Yamazaki et al., 2017; Nakamura et al., 2020; Katagiri et al., 2021). JunB is also identified to affect the functions of ECs. Some reports imply that JunB inhibition affects the migration but not proliferation of ECs (Bakiri et al., 2000; Jia et al., 2016), while others report that JunB affects cell morphology in human primary microvascular ECs (Yoshitomi et al., 2017). In this study, we characterized the landscape and relationship between chromatin accessibility and transcript expression in high glucose-treated human umbilical vein endothelial cells (HUVECs) by performing the assay for transposase-accessible chromatin using sequencing (ATAC-seq) and RNA sequencing (RNA-seq) analyses, identifying that JunB displays corresponding declines in both motif accessibility and expression level. The data show that JunB and its condensation promote proliferation and inhibit apoptosis in vascular ECs under hyperglycemic conditions and thus indicate a new connection between LLPS and diabetic endothelial damage and recovery.
Results
JunB exhibits reductions in both motif accessibility and gene expression level under high-glucose condition
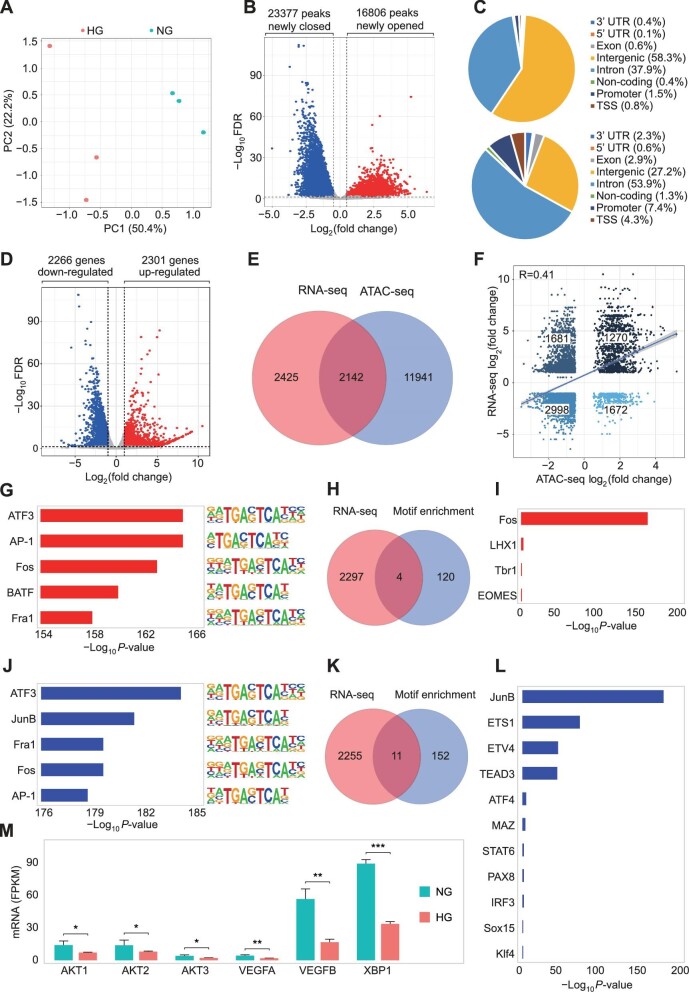
We aimed to investigate genome-wide changes in chromatin accessibility, gene expression, and transcriptional regulation during vascular endothelial dysfunction caused by hyperglycemia. ATAC-seq and RNA-seq were used and combined to obtain comprehensive information in HUVECs treated with high or normal glucose. In previous studies, a wide range of glucose concentrations from 30 to 120 mM were used in HUVECs to mimic hyperglycemic conditions and simulate high-glucose damage (Chao et al., 2016; Rezabakhsh et al., 2017; Yu et al., 2017; Tousian et al., 2020). At the beginning of our study, we tried different glucose concentrations to find the optimal dose and discovered that the glucose concentration of 60 mM showed more obvious and stable effects on the cell number of HUVECs (Supplementary Figure S1A). Therefore, we used the glucose dose of 60 mM in our following study. By performing ATAC-seq, we found that nucleosome-free, mononucleosome, and dinucleosome regions were present in the fragment length distribution of all samples, displaying typical ATAC-seq nucleosome banding patterns (Supplementary Figure S1B). The ATAC-seq data obtained from libraries were sufficient, represented by >260000 peaks (Supplementary Figure S1C) and >35 million non-duplicate, non-mitochondrial aligned reads (Supplementary Figure S1D). The alignment rates were >97%, showing that the libraries were not contaminated by other chromatin (Supplementary Figure S1E). Non redundant fraction and polymerase chain reaction (PCR) bottlenecking coefficient 1 (PBC1) values for each library were >0.8 (Supplementary Figure S1F and G), and the number of PBC2 exceeded 4 (Supplementary Figure S1H). These results indicated that the library complexity was sufficient for subsequent deep analyses. The transcription start site enrichment scores ranged from 10 to 14 (Supplementary Figure S1I), and the fraction of reads in the called peak regions (FRiP) scores were >0.2 (Supplementary Figure S1J), implying that our libraries presented the high signal-to-noise ratio and quality.
The principal component analysis (PCA) of peak read counts showed significant segregation of samples treated with high glucose or normal glucose (Figure 1A). By analyzing differential chromatin accessibility between the two conditions, we found a total of 40183 peaks with differential accessibility (absolute log2(fold change) >0.5 and −log10FDR < 0.01), among which 16806 peaks showed increased accessibility and 23377 peaks showed decreased accessibility (Supplementary Figure S1K and L; Figure 1B). Peak annotation showed that major accessibility-increased peaks (58.3%) were located in intergenic regions and major accessibility-decreased peaks were in introns (53.9%) (Figure 1C). Gene Ontology (GO) analysis showed that regions with increased accessibility were related to biological processes including axon development and genesis, molecular functions, including metal ion transmembrane transporter and cation channel activity, and cellular components including synaptic membrane and postsynaptic membrane, while regions with decreased accessibility were associated with actin filament organization, regulation of supramolecular fiber organization, phosphatidylinositol (PI) binding and serine/threonine kinase (STK) activity, and cellular components such as cell-substrate junction and focal adhesion molecules (Supplementary Figure S1M and N). Coincidentally, the molecular functions, including PI binding and STK activity, enriched in our GO analysis have been reported to mediate the proliferation and apoptosis of high glucose-treated HUVECs, further leading to vascular endothelial dysfunction (Baron et al., 1991; Natali et al., 1997; Kim et al., 2006). In addition, high glucose might induce cell death or vascular endothelial dysfunction through the caspase pathway and autophagic programmed cell death (Kothakota et al., 1997; Ohsumi, 2001; Gozuacik and Kimchi, 2004; Elmore, 2007).
Figure 1.
JunB shows decreased motif accessibility and gene expression in HUVECs treated with high glucose. (A) PCA of counts across all ATAC-seq regions in HUVECs treated with 5.5 mM glucose (NG, blue) or 60 mM glucose (HG, red). (B) Volcano plot of ATAC-seq peaks comparing HG to NG. Peaks with differential chromatin accessibility (FDR < 0.01 and |log2(fold change)| > 0.5) are highlighted. The numbers of peaks with open (red) and closed (blue) chromatin accessibility are shown. (C) Percentages of peak annotation types of ATAC-seq peaks with differential accessibility (FDR < 0.01 and |log2(fold change)| > 0.5). (D) Volcano plot of RNA-seq genes comparing HG to NG. Genes differentially expressed (FDR < 0.05 and |log2(fold change)| > 1) are highlighted. The numbers of genes with increased (red) and decreased (blue) expression are shown. (E) Venn diagram illustrating the overlap between differentially expressed genes (red circle) and the nearest genes of differentially accessible regions (blue circle). (F) Correlation analysis between differentially accessible regions and their nearest differentially expressed genes. Each dot represents a region with significantly differential accessibility and associated with differential expression. Pearson's correlation coefficient (R) and the number of dots in different quadrants are shown. (G and J) Motif enrichment analysis of increased (G) or decreased (J) accessible peaks comparing HG to NG via Homer findMotifsGenome.pl. The top 5 significantly enriched TF motifs (with least P-values) are plotted. (H and K) Venn diagram showing overlap between the enriched TF motifs in increased accessible regions and up-regulated genes (H) or between the enriched TF motifs in decreased accessible regions and down-regulated genes (K). Only the enriched TF motifs with a P-value < 0.01 were included. (I and L) Column chart showing the identified up-regulated (I) or down-regulated (L) TFs. The length of each column represents the P-value for the enriched motif. (M) The mRNA expression levels of JunB downstream genes. Error bars represent mean ± SEM, n = 3 independent experiments. Significance was determined using two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate how high glucose affects the gene expression of HUVECs, RNA-seq was performed with three biological replicates of HUVECs treated with high glucose or normal glucose. We observed 2301 up-regulated genes and 2266 down-regulated genes in HUVECs treated with high glucose compared with normal glucose (absolute log2(fold change) >1 and −log10FDR < 0.05) (Figure 1D). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that the up-regulated genes were mainly related to the cell cycle, the TGF-β signaling pathway, deoxyribonucleic acid (DNA) replication, and mismatch repair, whereas the down-regulated genes were primarily involved in lysosome, the HIF-1 signaling pathway, and fructose and mannose metabolism (Supplementary Figure S1O and P). All these pathways were connected with cell proliferation and apoptosis in previous studies (Li, 1999; Goda et al., 2003; Groth-Pedersen and Jaattela, 2013; Zhang et al., 2017; Jiang et al., 2021; Xiao et al., 2021). Through comprehensive analyses of ATAC-seq and RNA-seq data, we identified 2142 overlaps between the differentially expressed genes and the peaks with differential accessibility (Figure 1E). The differential gene accessibility positively correlated with the differential expression in a correlation analysis (Pearson correlation coefficient R = 0.41), suggesting that alterations in chromatin accessibility contributed to the concomitant differential messenger RNA (mRNA) expression in HUVECs treated with high glucose (Figure 1F). Then, HOMER motif analysis was utilized to identify TF motifs enriched in the regions with differential accessibility. We discovered 124 enriched TF motifs (ATF3, AP-1, Fos, BATF, and Fra1 motifs displaying the highest enrichments) in the regions with increased accessibility (Figure 1G and H) and 163 enriched TF motifs (ATF3, JunB, Fra1, Fos, and AP-1 displaying the highest enrichments) in the regions with decreased accessibility (Figure 1J and K). By combining RNA-seq and HOMER motif analyses, we identified four up-regulated TFs, Fos, LHX1, Tbr1, and EOMES (Figure 1H and I) and 11 down-regulated TFs, JunB, ETS1, ETV4, TEAD3, ATF4, MAZ, STAT6, PAX8, IRF3, Sox15, and Klf4, among which, JunB exhibited the highest degree of motif enrichment in the regions with decreased accessibility (Figure 1K and L). Furthermore, the downstream transcriptional targets of JunB, including AKT1, AKT2, AKT3, VEGFA, VEGFB, and XBP1, all showed decreased mRNA levels under high-glucose condition (Figure 1M; Supplementary Figure S1Q), confirming the reduced motif accessibility and expression level of JunB.
JunB reverses the effects of high glucose on HUVEC proliferation and apoptosis
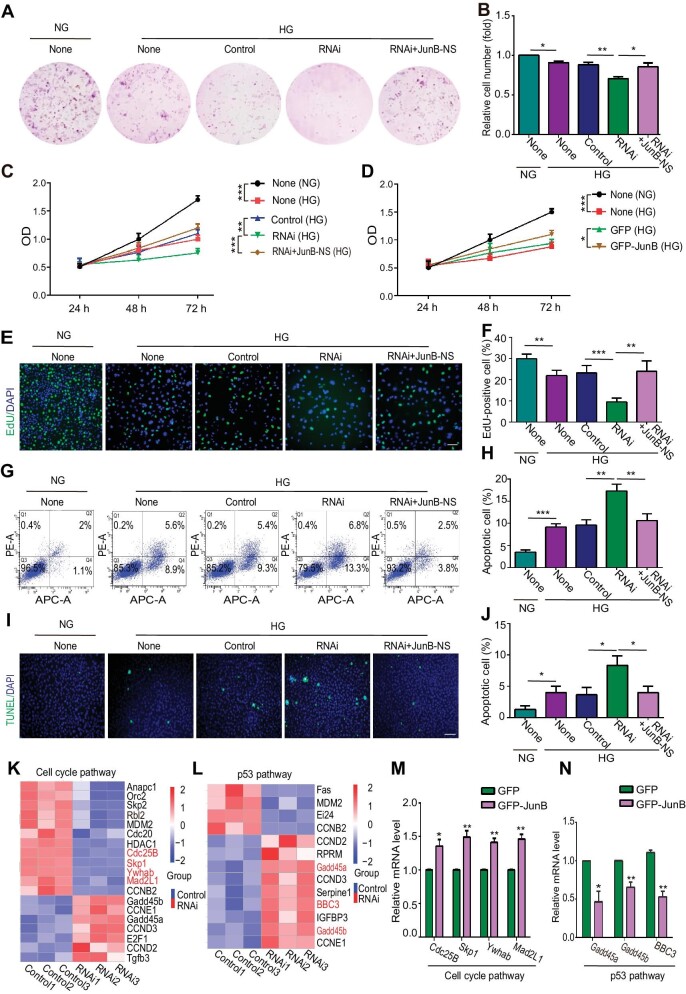
To confirm the reduction of JunB expression level in HUVECs under high glucose, reverse transcription–quantitative PCR (RT–qPCR) and western blotting analyses were performed. We confirmed that JunB mRNA and protein levels were both down-regulated (Supplementary Figure S2A–C). By cell cloning and counting assays, we found that JunB depletion by interference RNA (RNAi) amplified high glucose-induced decrease in HUVEC number, while overexpressing a JunB non-sense mutant (JunB-NS) resistant to JunB RNAi rescued the decreased cell number (Figure 2A and B; Supplementary Figure S2B).
Figure 2.
JunB reverses the effects of high glucose on EC proliferation and apoptosis. HUVECs were treated as indicated: NG, 5.5 mM glucose; HG, 60 mM glucose; None, without transfection reagents or oligonucleotides; Control, only with transfection reagents; RNAi, with both transfection reagents and oligonucleotides; RNAi+JunB-NS, infected by lentivirus after adding transfection reagents and oligonucleotides. (A and B) The colony forming ability of cells. Representative colony formation pictures (A) and the ratio of cell number relative to the None (NG) group (B) (n = 5). (C and D) The optical density (OD) of cells with JunB depletion (C) or GFP-JunB overexpression (D) was determined by the CCK8 assay. (E and F) Cell proliferation was determined by the EdU assay. Scale bar, 75 μm. (G and H) Cell apoptosis was determined by flow cytometry with Annexin V (abscis) and PI (ordinate) staining. Apoptotic cell percentage was the sum of Q2 and Q4 percentages. (I and J) Cell apoptosis was determined by the TUNEL assay. Scale bar, 250 μm. (K and L) RNA-seq performed with control or JunB-depleted HUVECs under high glucose determined the differential expression of genes in the cell cycle pathway (K) or p53 pathway (L). The genes verified positive by RT‒qPCR are indicatedin red. (M and N) RT‒qPCR showed up-regulation of Cdc25B, Skp1, Ywhab, and Mad2L1 and down-regulation of Gadd45a, Gadd45b, and BBC3 in HUVECs overexpressing GFP-JunB. Error bars represent mean ± SEM, n = 3 independent experiments if not stated. Significance was determined using two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
To further explore how JunB regulates the EC number under high glucose, we examined both cell proliferation and apoptosis. We found that JunB RNAi suppressed whereas JunB overexpression promoted the proliferation of HUVECs treated with high glucose, as evidenced by CCK8 evaluation, EdU, and phosphohistone H3 (pH3) staining (Figure 2C–F; Supplementary Figure S2C–G). In addition, bidirectional deviations of JunB modulated the activation of apoptosis under high-glucose condition, as indicated by flow cytometry and terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assays (Figure 2G–J; Supplementary Figure S2H). Importantly, the effects of JunB RNAi on HUVEC proliferation and apoptosis under high glucose were reversed by overexpressing JunB-NS (Figure 2C and E–J; Supplementary Figure S2B, D, and E). Thus, JunB promoted cell proliferation and suppressed cell apoptosis in HUVECs under high-glucose condition.
To further uncover the underlying mechanisms of JunB-mediated cell proliferation and apoptosis during diabetic vascular endothelial injury, we compared genes expressed in the HUVECs treated with control and JunB RNAi under high-glucose condition using RNA-seq. Clusters of up-regulated and down-regulated genes were identified in JunB-depleted HUVECs (Supplementary Figure S2I and J). The KEGG enrichment analysis showed that the down-regulated genes were mainly involved in the mTOR signaling, tight junction, and cell cycle pathways, contributing to cell proliferation (Gonzalez-Mariscal et al., 2012; Lu et al., 2021; Pan et al., 2021), while genes associated with the JAK/STAT signaling pathway, p53 signaling pathway, and cell adhesion molecules that mediate cell apoptosis (Zhang et al., 2018b; Jiang et al., 2019; Carracedo et al., 2001; Meyer et al., 2020; Yang et al., 2021) were enriched in the up-regulated gene cluster (Supplementary Figure S2K and L). Upon JunB depletion, cell cycle pathway genes, including Cdc25B, Skp1, Ywhab, and Mad2L1, were down-regulated in our RNA-seq data, whereas p53 signaling pathway genes, including Gadd45a, Gadd45b, and BBC3, were upregulated (Figure 2K and L). Meanwhile, JunB overexpression increased the mRNA levels of Cdc25B, Skp1, Ywhab, and Mad2L1 and decreased that of Gadd45a, Gadd45b, and BBC3 (Figure 2M and N). Overall, JunB controlled the proliferation and apoptosis of HUVECs under hyperglycemic conditions through the cell cycle and p53 signaling pathways, respectively.
JunB undergoes LLPS in nuclei and in vitro mediated by IDR and DNA-binding domain
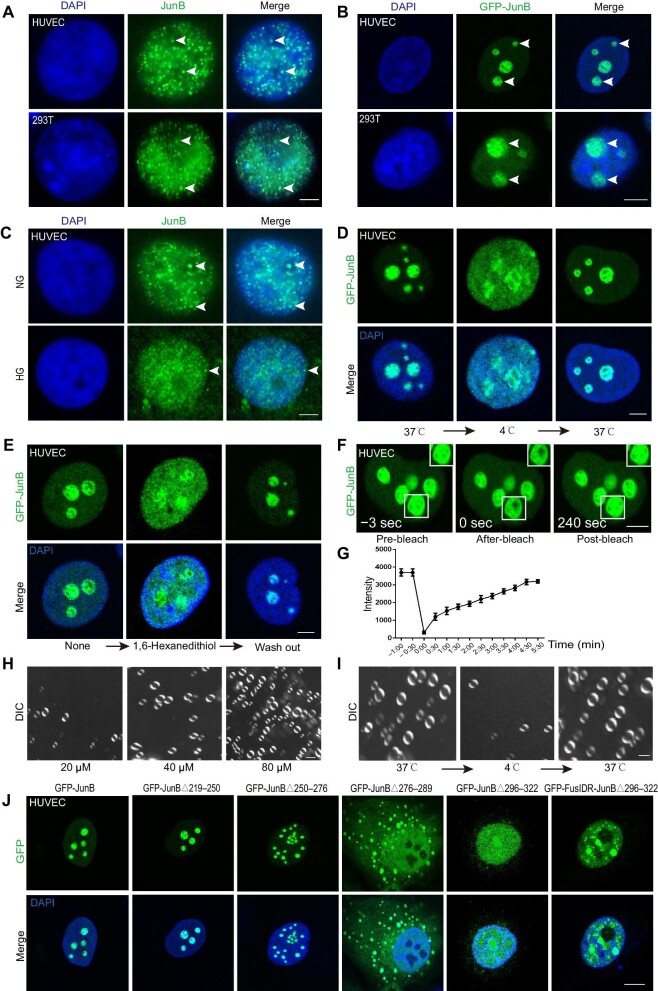
The IDRs were distributed among the majority of JunB proteins, inferring that JunB might exert LLPS behaviors (Supplementary Figure S3A). Consistently, we observed by immunofluorescence that endogenous JunB and exogenous GFP-JunB formed multiple puncta in the nuclei of HUVECs and 293T cells (Figure 3A and B). Interestingly, high glucose not only reduced JunB protein level but also impaired its phase separation property (Figure 3C). Previous studies indicated that protein LLPS was modulated by the temperature shift, which was reversible within a certain temperature range (Jiang et al., 2015; Cinar et al., 2019). We exposed HUVECs to different temperatures to further verify whether JunB puncta displayed LLPS properties. We discovered that the puncta formed by endogenous or exogenous JunB diffused when the cells were transferred from 37°C to 4°C and re-formed when transferred back to 37°C (Figure 3D; Supplementary Figure S3B). By incubating HUVECs with alcohol 1,6-hexanediol, which is widely used to destroy LLPS condensates (Duster et al., 2021; Shi et al., 2021, 2022), we revealed that droplets of endogenous or exogenous JunB were dissolved, and drug wash out reversed this effect (Figure 3E; Supplementary Figure S3C). Thus, the temperature shift and 1,6-hexanediol treatment assays elucidated that endogenous or exogenous JunB formed reversible puncta in the nucleus. To further assess JunB condensate fluidity in the nucleus, we performed a fluorescence recovery after photobleaching (FRAP) assay in HUVECs overexpressing GFP-JunB. We found that the fluorescence-quenched region of GFP-JunB droplet was restored in ∼4 min, indicating the highly dynamic diffusion coefficient of the JunB condensate (Figure 3F and G). Then, we purified His-JunB protein in vitro and demonstrated that the formation of JunB droplets was regulated by protein concentration, temperature, and salt ion concentration (Figure 3H and I; Supplementary Figure S3D and F). Moreover, the addition of a TRE-repeat DNA fragment (3×TRE or 7×TRE) that specifically binds to JunB (Chiu et al., 1989; Yukimasa et al., 1999) accelerated in vitro condensation (Supplementary Figure S3G).
Figure 3.
JunB undergoes LLPS in the nucleus and in vitro. (A and B) Representative images of JunB immunofluorescence (A) or overexpressed GFP-JunB (B) showing the formation of multiple puncta in the nuclei (visualized by DAPI) of HUVECs and 293T cells. Scale bar, 5 μm. (C) High glucose diminished JunB signal and puncta formation in the nuclei of HUVECs. Scale bar, 5 μm. (D) Temperature-dependent GFP-JunB puncta diffusion and re-formation in the nuclei of HUVECs. Temperature ramp, 4°C–37°C. Scale bar, 5 μm. (E) 1,6-hexanediol incubation and wash out led to GFP-JunB puncta diffusion and re-formation in the nuclei of HUVECs. Scale bar, 5 μm. (F) Representative images showing fluorescence recovery of GFP-JunB condensates in the nuclei of HUVECs after photobleaching. The photobleached area is indicated by the white box. Scale bar, 5 μm. (G) Fluorescence recovery curves after photobleaching for GFP-JunB condensates. n = 5 granules. (H and I) The JunB protein concentration (H) and temperature (I) modulated the formation of JunB droplets in vitro. Scale bar, 2 μm. (J) Representative images showing the formation of puncta in HUVECs overexpressing different GFP-JunB mutants. Scale bar, 10 μm.
To understand the mechanism of JunB coacervation, we analyzed the structure of JunB protein and found a major IDR located at 38–322 aa (Supplementary Figure S3A). We truncated the entire IDR (38–322 aa) into four different regions and generated mutants by deleting each region: JunBΔ38–90, JunBΔ91–140, JunBΔ141–218, and JunBΔ219–322 (Supplementary Figure S3E). Compared with wild-type JunB (JunB1–347), JunBΔ219–322, but not JunBΔ38–90, JunBΔ91–140, or JunBΔ141–218, destroyed droplet formation in HUVECs (Supplementary Figure S3H). As the 219–322 aa region of JunB contains the DNA-binding (250–276 aa), nuclear localization signal (NLS) (276–289 aa), and leucine zipper (296–317 aa) domains (Chen, 2003), we constructed four other mutants: JunBΔ219–250, JunBΔ250–276, JunBΔ276–289, and JunBΔ296–322 (Supplementary Figure S3E). As shown in Figure 3J, JunBΔ219–250 formed similar droplets as wild-type JunB. JunBΔ250–276 deficient in interacting with DNA showed attenuated droplet formation, constituting smaller condensates. JunBΔ276–289 without the NLS domain also constituted smaller droplets in the cytoplasm but not in the nucleus, further revealing the requirement of DNA binding for JunB coacervation. Coincidentally, no visible droplets were formed when only JunBΔ296–322 was overexpressed in HUVECs, indicating that the 296–322 aa region mainly contributed to JunB condensation (Figure 3J). Thus, JunB underwent phase separation in vivo and in vitro, mediated by its IDR region (aa 296–322) and DNA-binding property.
JunB nuclear localization and condensation control its functions in EC proliferation and apoptosis induced by high glucose
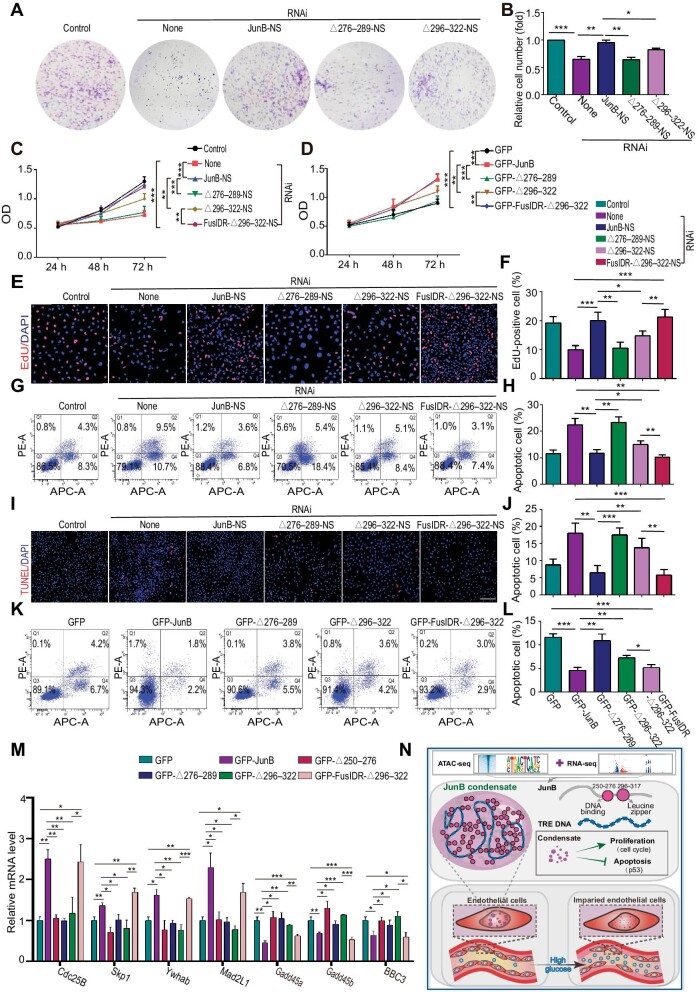
To study the role of JunB condensation and nuclear localization in HUVEC proliferation and apoptosis stimulated by high glucose, we overexpressed JunB mutants JunBΔ276–289-NS, JunBΔ296–322-NS, or JunB-NS in JunB-depleted HUVECs treated with high glucose. Since the 296–322 aa region of JunB comprises a leucine zipper structure that usually forms heterodimers with other AP-1 family members (Deng and Karin, 1993; Basbous et al., 2003; Ghosh et al., 2005), we additionally utilized another JunBΔ296–322 construct fused with the N-terminus of the fused in sarcoma (FUS) protein (FusIDR-JunBΔ296–322) to discriminate the heterodimer formation from the condensation property of JunB in the following functional rescue assays (Supplementary Figure S3E). In previous studies, the intrinsically disordered N-terminal region of FUS required for its phase separation was usually leveraged to replace the IDR of other coacervating proteins to confirm and study their LLPS behaviors (Lin et al., 2017; King and Petry, 2020; Rawat et al., 2021). Accordingly, FusIDR-JunBΔ296–322 re-formed visible puncta in the nuclei of HUVECs, compared with JunBΔ296–322 (Figure 3J). We found that both JunBΔ276–289-NS and JunBΔ296–322-NS exhibited weaker reversing effects than JunB-NS on cell number reduction (Figure 4A and B), proliferation inhibition (Figure 4C, E, and F), and apoptosis induction (Figure 4G–J; Supplementary Figure S4A and B) caused by JunB depletion under high glucose condition, but FusIDR-JunBΔ296–322-NS displayed stronger reversing effects than JunBΔ296–322-NS that were comparable to the effects of JunB-NS. Meanwhile, JunBΔ276–289 or JunBΔ296–322 showed weaker induction of cell proliferation (Figure 4D; Supplementary Figure S4C–F) and suppression of cell apoptosis (Figure 4K and L) under high-glucose condition compared with wild-type JunB-WT, while FusIDR-JunBΔ296–322 exerted more significant effects than JunBΔ296–322. To further study the function and mechanism of JunB condensation and transcription behaviors in the proliferation and apoptosis of HUVECs under high glucose, we verified the mRNA levels of JunB-regulated targets in the cell cycle and p53 signaling pathways by qPCR. The results showed that overexpressing JunBΔ250–276, JunBΔ276–289, or JunBΔ296–322 in HUVECs treated with high glucose did not significantly affect the mRNA levels of Cdc25B, Skp1, Ywhab, and Mad2L1 in the cell cycle signaling pathway and Gadd45a, Gadd45b, and BBC3 in the p53 signaling pathway (Figure 4M). FusIDR-JunBΔ296–322 exerted similar effects as wild-type JunB on up-regulating cell cycle-related genes and down-regulating p53-related genes (Figure 4M). Thus, our data proposed that JunB underwent phase separation in the nucleus and in vitro, modulated by its IDR at 296–322 aa (leucine zipper domain) and DNA-binding property. JunB condensation and transcription behaviors regulated the proliferation and apoptosis of vascular ECs induced by high glucose via the cell cycle and p53 signaling pathway, respectively (Figure 4N). As the leucine zipper domain generally mediates dimerization (Angel and Karin, 1991; Chinenov and Kerppola, 2001), JunB dimerization might be necessary for its phase separation, and the FusIDR-JunBΔ296–322 chimera might restore the phase separation propensity under the modulation of the FUS IDR-mediated polymerization.
Figure 4.
The functions of JunB nuclear localization and condensation in EC proliferation and apoptosis induced by high glucose. (A–M) HUVECs were treated as indicated. (A and B) The colony forming ability of cells. Representative colony formation photos (A) and the ratio of cell number relative to the Control group (B) (n = 5). (C and D) The ODof cells was determined by the CCK8 assay. (E and F) Cell proliferation was determined by the EdU assay. Scale bar, 75 μm. (G and H, K and L) Cell apoptosis was determined by flow cytometry with Annexin V (abscis) and PI (ordinate) staining. Apoptotic cell percentage was the sum of Q2 and Q4 percentages. (I and J) Cell apoptosis was determined by the TUNEL assay. Scale bar, 250 μm. (M) mRNA levels of Cdc25B, Skp1, Ywhab, Mad2L1, Gadd45a, Gadd45b, and BBC3 were determined by RT‒qPCR. (N) An illustration depicting how JunB was identified and that high glucose can affect the proliferation and apoptosis of ECs by affecting the protein level and phase separation of JunB, thus leading to endothelial damage. Error bars represent mean ± SEM, n = 3 independent experiments if not stated. Significance was determined using two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
We further analyzed the interaction between JunB and downstream transcriptional targets by chromatin immunoprecipitation (ChIP)‒qPCR. Since VEGFB and Ywhab emerged top in the target list from our ATAC-seq and RNA-seq analyses, they might act as downstream targets of JunB in regulating cell proliferation (Figures 1M and 2K). ChIP‒qPCR results demonstrated that high glucose attenuated the interaction between JunB and the promoter of VEGFB or Ywhab, which was rescued by overexpression of GFP-JunB or FusIDR-JunBΔ296–322, but not JunBΔ296–322 (Supplementary Figure S4G). In addition, we isolated vascular ECs from C57BL/6 wild-type and db/db mice by using fluorescence-activated cell sorting (FACS) with the vascular EC marker CD31 and analyzed the interaction between JunB and the promoter of VEGFB or Ywhab by ChIP‒qPCR. The interaction between JunB and the promoter of VEGFB or Ywhab was decreased in db/db mice compared with C57BL/6 wild-type mice (Supplementary Figure S4H), validating the reduced JunB target gene accessibility under clinically relevant hyperglycamic condition.
Discussion
In this study, we investigated genome-wide landscape changes in chromatin accessibility and the transcriptome of vascular ECs treated with high glucose. Comprehensive analyses showed that JunB expression level and motif accessibility both significantly decline upon high glucose-induced damage, inferring that JunB may adjust cell responses during endothelial damage under high-glucose conditions. Then, we found that JunB promotes cell proliferation through the cell cycle signaling pathway and attenuates cell apoptosis via the p53 signaling pathway in HUVECs treated with high glucose. Additionally, we uncovered that JunB undergoes LLPS in vitro and in the nucleus, modulated by its IDR at 296–322aa and DNA-binding domain, which contribute to its impact on the proliferation and apoptosis of HUVECs treated by high glucose and effects on EC recovery under high-glucose damage. Thus, our data reveal the potential involvement of LLPS in diabetic endothelial dysfunction, inferring an additional candidate and strategy to attenuate vascular endothelial damage under hyperglycemic condition from the angle of LLPS.
Previous studies showed that JunB promotes the migration of ECs and knocking down JunB in ECs reduces MMP13 expression, which ultimately inhibits EC migration (Licht et al., 2006). Vascular endothelial growth factor (VEGF), upon binding to its EC-specific receptors VEGF-R1 and VEGF-R2, can induce HUVEC migration and increase JunB mRNA levels; conversely, JunB RNAi impaired the migration of HUVECs induced by VEGF (Jia et al., 2016). Furthermore, in human retinal microvascular ECs, VEGFA-induced PKCθ phosphorylation amplifies JunB expression, which further promotes VEGFR3 expression and the activation of STAT3 to control cell migration (Kumar et al., 2020). In our study, we validated that LLPS mediates JunB activities in endothelial recovery upon high-glucose damage, as well as its transcriptional activities to regulate the mRNA levels of its downstream genes involved in cell proliferation and apoptosis. Therefore, JunB might also function in endothelial migration via its condensation property.
As a TF, JunB is implicated in the regulation of the VEGF signaling pathway, the differentiation of Th2 and Th17 cells, and the expression of human papillomavirus e6 and e7 oncoproteins (Rengarajan et al., 2002; Henderson et al., 2004; Schmidt et al., 2007; He and Luo, 2012; Carr et al., 2017; Hasan et al., 2017; Yoshitomi et al., 2017; Xu et al., 2020). It has been reported that individual or multiple TFs form intracellular mono- or multi-element condensates to regulate the corresponding transcriptional activities (Smith et al., 2021). For instance, HSF1, acting as the transcriptional regulator of heat shock protein (HSP), relies on the coacervation behavior upon stress to adjust the transcription of HSP and HSP-mediated cell protection (Biamonti and Vourc'h, 2010; Gaglia et al., 2020). As another example, OCT4 and GCN4 form phase-separated droplets with Mediator in vitro and in vivo, promoting gene activation (Boija et al., 2018; Sabari et al., 2018). The AP-1 family comprises four subfamilies, i.e. Jun (c-Jun, JunB, and JunD), c-Fos (c-Fos, FosB, Fra1, and Fra2), Maf (c-Maf, MafA, MafB, Maff, Mafg, Mafk, and Nrl), and ATF (ATF2, ATF3, ATF4, ATFa, BATF, JDP1, and JDP2) (Karin et al., 1997; Leonard et al., 1997). AP-1 family members form heterodimers or homodimers to exert their transcription effects via the leucine zipper motif (Leonard et al., 1997). Regarding this ability, previous studies proposed that the heterodimer Jun/Fos shows higher DNA affinity and transcriptional activity than the homodimer Jun/Jun (Hai et al., 1989; Eferl and Wagner, 2003). Thus, we infer that other AP-1 family members may participate in the phase separation of JunB in vivo to modulate its transcriptional activities and functions.
Diverse small-molecule compounds have been reported to prevent the RNA-dependent condensation of RNA-binding proteins associated with ALS, TDP-43, FUS, and HNRNPA2B1 (Fang et al., 2019). 4,4′-dianilino-1,1′-diphthyl-5,5′-diabetic acid (bis ANS) and silicon compounds, ATP, flavonoid myricetin, and others have been shown to modulate LLPS of related proteins (Hayes et al., 2018; Kang et al., 2018, 2019; Babinchak et al., 2020; Dai et al., 2021; Song, 2021; Xu et al., 2022). In addition, our data showed that high glucose decreases JunB expression levels to disrupt its phase separation and attenuate endothelial recovery upon damage. Accordingly, further research on whether high glucose influences JunB LLPS behavior through direct physical binding is needed.
Materials and methods
Cell culture
HUVECs and HEK293 cells (ATCC) were cultured at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (Gibco) containing 10% fetal bovine serum (Gibco). For experiments, cells were treated with normal glucose (NG, 5.5 mM glucose) or high glucose (HG, 60 mM glucose) for 96 h.
Construction of different vectors
The cDNA encoding JunB was cloned into pET-32a to express the His-JunB fusion protein in bacteria. The cDNA encoding JunB was cloned into pLVX-AcGFP1-C1 or pLVX-mCherry-C1 as GFP-JunB or mCherry-JunB, respectively, for packaging virus. JunB mutants (JunB-NS, JunBΔ38–90, JunBΔ91–140, JunBΔ141–218, JunBΔ219–322, JunBΔ219–250, JunBΔ250–276, JunBΔ276–289, and JunBΔ296–322) were constructed on pLVX-AcGFP1-C1-JunB plasmids using the Q5 Site-Directed Mutagenesis Kit (NEB). The cDNA encoding FusIDR was cloned into JunBΔ296–322 as FusIDR-JunBΔ296–322. JunBΔ250–276-NS, JunBΔ276–289-NS, and JunBΔ296–322-NS were built on JunB-NS using the Q5 Site-Directed Mutagenesis Kit. FusIDR-JunBΔ296–322-NS was built on FusIDR-JunBΔ296–322 using the Q5 Site-Directed Mutagenesis Kit. The primer sequences and restriction sites used for the constructs are listed in Supplementary Table S1.
Knockdown of the JunB gene in HUVECs
We used the JunB siRNA oligonucleotides (5′-CGACUACAAACUCCUGAAATT-3′ and 3′-UUUCAGGAGUUUGUAGUCGTT-5′) designed by Shanghai GenePharma for JunB knockdown. When HUVECs were grown to 70% confluence, 40 pM of each siRNA oligonucleotide was transfected into cells using Lipofectamine RNAiMAX (Invitrogen) with gentle shaking to ensure mixing. The efficiency was detected after 72 h.
Lentivirus packaging and infection of HUVECs
293T cells (80% confluence) were cultured in a 10-cm petri dish. pLVX-AcGFP1-C1 and its helper plasmids pCMV-dR8.91 and pMD2.G were transfected into 293T cells with Lipo8000 (Beyotime). The fluid was changed at 8 h, and the virus supernatant was collected at 48 and 72 h. The virus was concentrated with polyethylene glycol (PEG) and collected by centrifugation at 24 h later for virus titer detection. Lentivirus-infected HUVECs were added to the medium containing 1:1000 polybrene (Solarbio). The efficiency of virus infection was examined under a fluorescence microscope, and the cell infection rate was >90%.
RT–qPCR
Total RNA was extracted from HUVECs with TRIzol and quantified using an ND-2000C NanoDrop spectrophotometer (NanoDrop Technologies). First-strand cDNA was synthesized from 1 μg of total RNA with HiScript III RT SuperMix for qPCR (Vazyme). qPCR was performed with ChamQ Universal SYBR qPCR Master Mix (Vazyme) on a C1000 Touch Thermal Cycler (Bio-Rad). All the primers are shown in Supplementary Table S2. β-Actin served as the reference gene. The 2−ΔΔCT method was used for data calculation.
CCK8 assay
Cells were seeded into 96-well plates (1000 cells/well in 100 μl medium), and 10% CCK8 was added at 24, 48, and 72 h, respectively. After incubation for 1.5 h, the absorbance at 450 nm was measured by a microplate reader.
EdU assay
Cells were seeded into 96-well plates, incubated with 10 μM EdU (Abbkine) for 2 h at 37°C, and fixed in 4% paraformaldehyde (PFA) for 20 min. Each well was washed three times with 0.1 ml BSA Wash Solution, and cells were permeabilized in 0.2% Triton X-100 (Biofroxx) in phosphate-buffered saline (PBST) for 30 min. Each sample was incubated with 100 μl of Click-iT reaction mixture for 30 min at room temperature in the dark. Then, to remove the reaction mixture, each well was washed three times with 0.1 ml BSA Wash Solution. Nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI) solution (Solarbio).
Immunofluorescence
Cells seeded on coverslips (Thermo Fisher Scientific) were washed twice in PBS, fixed in 4% PFA for 20 min, and permeabilized in PBST for 30 min. After incubation with anti-JunB antibody (1:200, Abcam) or anti-pH3 antibody (1:1000, Millipore) at 4°C overnight and washed three times with PBST for 10 min, the cells were incubated with goat anti-rabbit IgG (H+L) highly cross-adsorbed secondary antibody (1:1000, Thermo Fisher Scientific) at room temperature for 1 h and labeled with DAPI solution to visualize nuclei. Images were taken with Leica fluorescence stereo microscopes or Leica TCS-SP8 confocal microscope.
Colony formation assay and cell counting
Cells were counted after trypsin digestion, plated into 12-well plates (100 cells per well), and incubated for 7–14 days to assay for colony forming ability. Colonies were stained with 0.5% crystal violet in 6% glutaraldehyde solution.
Cells were washed twice in PBS, fixed in 4% PFA for 20 min, and permeabilized in PBST for 30 min. DAPI staining was followed by fluorescence microscope sampling and counting.
Analysis of cell apoptosis by flow cytometry
Cells were digested with trypsin, washed three times with PBS, and stained. Cells were resuspended in binding buffer at the density of 1 × 106–5 × 106/ml and incubated for 5 min in the dark with 5 μl of Annexin V/Alexa Fluor 647, followed by 10 μl of 20 μg/ml propidium iodide (PI) and 400 μl of PBS for immediate detection. Flow cytometry was performed by using a FACS Aria II sorter (BD Biosciences).
TUNEL staining
Cells seeded on coverslips (Thermo Fisher Scientific) were washed twice in PBS, fixed in 4% PFA for 20 min, and permeabilized in PBST for 5 min. TUNEL staining was then performed to detect apoptotic cells using the DeadEnd™ Fluorometric TUNEL System (Promega) or TUNEL BrightRed Apoptosis Detection Kit (Vazyme).
1,6-hexanediol assay and temperature change assay of phase separation
For phase separation in vivo, 1,6-hexanediol (Sigma) was added to the medium until the concentration of 1,6-hexanediol was 5%. Cells seeded on coverslips were incubated for 7 min in the medium containing 1,6-hexanediol (as the 1,6-hexanediol incubation group). Some of these cells were then incubated in the medium without 1,6-hexanediol (as the wash out group). MG-132 (Selleck) was added to the medium until the concentration of MG-132 was 5 μM. Cells seeded on coverslips were incubated at 4°C in the medium containing MG-132 for 5 h (as the 4°C group). Some of these cells were then incubated at 37°C for 12 h (as the 37°C group).
For phase separation in vitro, the His-JunB protein was added to the flow cell on the slide, followed by incubation at 37°C for 2 min, then at 4°C for 30 min, and then at 37°C for 2 min. The His-JunB protein was processed at different temperature conditions, and the images were collected.
Droplet experiment and FRAP
JunB protein was diluted in ice-cold PBS buffer containing 5% PEG. The solution was incubated at 37°C for 2 min and then loaded into a flow cell made with two pieces of double sticky scotch taps for immediately imaging with an upright microscope (Leica) under differential interference contrast (DIC). The DIC images were taken using Scan-DIC with a sequential scan. For DIC imaging, the light angle was set to be the same for all samples imaged based on the shadows around the edge of the droplets.
HUVECs were overexpressed with GFP-JunB to form protein puncta, and the FRAP assay was performed on protein puncta by the bleaching mode on a structure illumination microscope (Nikon). The region of protein puncta was selected with a circular pattern, and each protein puncta was bleached with a 488-nm laser at 50% power for ∼1 sec. The fluorescence recovery was calculated using the CellSens software (OLYMPUS).
ATAC-seq
The ATAC-seq libraries were prepared as previously described (Corces et al., 2017). Briefly, 50000 cells per sample were suspended in ice-cold PBS. Cells were pelleted, resuspended in lysis buffer (10 mM Tris–HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% NP40, and 0.1% Tween-20), and washed once with 1 ml of cold lysis buffer not containing NP40. Following a 10-min centrifugation at 4°C, nucleic extracts were resuspended in Tn5 transposition reaction mix (Vazyme) for 30 min at 37°C. Transposed DNA was purified using the MinElute PCR Purification Kit (Qiagen). PCR was performed to amplify the library for 12 cycles. Libraries were cleaned up twice with VAHTS DNA Clean Beads (Vazyme), the first using 0.55× volume to remove large DNA fragments (>1 kb), and the second using 1× volume to clean up the remaining small fragments. Libraries were sequenced on the Illumina HiSeq 500 with 150-bp paired-end reads.
The raw data were processed with the ENCODE ATAC-seq pipeline (https://github.com/ENCODE-DCC/atac-seq-pipeline). In short, the reads were trimmed, filtered, and aligned against hg38 using Bowtie2. PCR duplicates and reads mapped to the mitochondrial chromosome or repeated regions were removed. To correct for the Tn5 transposase insertion, mapped reads were shifted +4/−5. Peak calling was performed using MACS2, with P-value <0.01 as the cutoff. Then, the peak merging script (https://github.com/corceslab/ATAC_IterativeOverlapPeakMerging) was used to create the union peak set. We used featucounts to calculate within-peak read counts, which were analyzed for differential chromatin accessibility via DESeq2. Homer annotatepeaks were used to annotate MACS2 enrichments based on their genomic locations. Binary alignment map format (BAM) files used for peak calling were reads per kilobase per million mapped reads (RPKM)-normalized using deeptools bamCoverage and a binSize of 10. For heatmap visualization, RPKM-normalized bigwig files from biological replicates of each genotype were merged using bigWigMerge, which was then used as the input for deeptools computeMatrix to obtain the ATAC-seq signal that is lined up at the center of peaks. RPKM-normalized bigwig files were used for visualization in IGV.
RNA-seq
Total amount and integrity of RNA were assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system. The mRNA sequencing libraries were constructed using the Illumina TruSeq Stranded mRNA Library Prep kit with 200 ng of total RNA following the manufacturer's specifications. Library quantity and quality were measured using Qubit2.0 Fluorometer and Agilent 2100 bioanalyzer, respectively. Libraries were sequenced by the Illumina NovaSeq 6000. The end reading of the 150-bp pairing is generated.
Raw reads were first processed through the fastp software. Clean reads were obtained by removing reads containing adapters, reads containing N bases, and low-quality reads from the raw data. The index of the reference genome was built using HISAT2 (v2.0.5), and paired-end clean reads were aligned to the reference genome hg38 using HISAT2. featureCounts was used to count the number of reads mapped to each gene. Differential expression analysis of the two groups was performed using the DESeq2 R package. The resulting P-values were adjusted using Benjamini and Hochberg's approach for controlling the false discovery rate (FDR).
Quantification and statistical analysis
Statistical significance was determined by unpaired two-tailed t-test or one-way analysis of variance (ANOVA) with Tukey‒Kramer's post hoc test for normal distribution and equal variance, unless indicated otherwise. Sample sizes, experimental replicates, and specific statistical tests used are described in figure legends. Data were plotted using GraphPad Prism 9 software as the mean values. Error bars represent standard error of the mean (SEM).
Data and material availability
The RNA-seq and ATAC-seq data that support the findings of this study have been deposited in the Sequence Read Archive under BioProject ID: PRJNA925697 (corresponding to data in Figures 1 and 2; Supplementary Figures S1 and S2).
The data sets generated and/or analyzed during this study are available with no restrictions from the corresponding author upon reasonable request. Custom codes are available from the corresponding author upon request. All key reagents used in this study are listed in Supplementary Table S3, which are available from the corresponding author with a completed Materials Transfer Agreement.
Supplementary Material
Contributor Information
Xuxia Ren, Laboratory for Aging and Cancer Research, Frontiers Science Center Disease-related Molecular Network, State Key Laboratory of Respiratory Health and Multimorbidity and National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu 610041, China.
Zexu Cui, Laboratory for Aging and Cancer Research, Frontiers Science Center Disease-related Molecular Network, State Key Laboratory of Respiratory Health and Multimorbidity and National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu 610041, China.
Qiaoqiao Zhang, Key Laboratory of Gene Engineering of the Ministry of Education, State Key Laboratory of Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China.
Zhiguang Su, Molecular Medicine Research Center, West China Hospital, Sichuan University, Chengdu 610041, China.
Wei Xu, Laboratory for Aging and Cancer Research, Frontiers Science Center Disease-related Molecular Network, State Key Laboratory of Respiratory Health and Multimorbidity and National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu 610041, China.
Jinhui Wu, Center of Geriatrics and Gerontology, National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu 610041, China.
Hao Jiang, Laboratory for Aging and Cancer Research, Frontiers Science Center Disease-related Molecular Network, State Key Laboratory of Respiratory Health and Multimorbidity and National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu 610041, China.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32090043 and 31771505), the National Key Research and Development Program of China (2017YFC0840101 and 2018YFA0108302), Sichuan Science and Technology Program, the Central Government Guides Local Science and Technology Development Projects (2022ZYD0078), Sichuan Science and Technology Program (2023YFQ0008), Science and Technology Department of Tibet, the Central Government Guides Local Science and Technology Development Projects (XZ202102YD0026C), the 1.3.5 Project for Disciplines of Excellence (ZYYC20001, ZYGD20010, and ZY2017201), the Project of Max Cynader Academy of Brain Workstation, WCHSCU (HXYS19005), and the National Clinical Research Center for Geriatrics (Z20191011, Z20201009, and Z2023YY003).
Conflict of interest: none declared.
Author contributions: H.J.: conceptualization, writing original draft, writing review and editing, funding acquisition, resources, and supervision; J.W.: writing review and editing, funding acquisition, resources, and supervision; X.R.: conceptualization, methodology, investigation, writing original draft, and writing review and editing; Z.C.: methodology, investigation, writing original draft, and writing review and editing; Q.Z.: investigation; Z.S.: investigation; W.X.: methodology.
References
- Abeyrathna P., Su Y. (2015). The critical role of Akt in cardiovascular function. Vasc. Pharmacol. 74, 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad E., Lim S., Lamptey R. et al. (2022). Type 2 diabetes. Lancet North Am. Ed. 400, 1803–1820. [DOI] [PubMed] [Google Scholar]
- Andrecht S., Kolbus A., Hartenstein B. et al. (2002). Cell cycle promoting activity of JunB through cyclin A activation. J. Biol. Chem. 277, 35961–35968. [DOI] [PubMed] [Google Scholar]
- Angel P., Karin M. (1991). The role of Jun, Fos and the Ap-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072, 129–157. [DOI] [PubMed] [Google Scholar]
- Babinchak W.M., Dumm B.K., Venus S. et al. (2020). Small molecules as potent biphasic modulators of protein liquid‒liquid phase separation. Nat. Commun. 11, 5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach F.H., Hancock W.W., Ferran C. (1997). Protective genes expressed in endothelial cells: a regulatory response to injury. Immunol. Today 18, 483–486. [DOI] [PubMed] [Google Scholar]
- Bakiri L., Lallemand D., Bossy-Wetzel E. et al. (2000). Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 19, 2056–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S.F., Lee H.O., Hyman A.A. et al. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron A.D., Laakso M., Brechtel G. et al. (1991). Mechanism of insulin resistance in insulin-dependent diabetes mellitus: a major role for reduced skeletal muscle blood flow. J. Clin. Endocrinol. Metab. 73, 637–643. [DOI] [PubMed] [Google Scholar]
- Basbous J., Arpin C., Gaudray G. et al. (2003). The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 278, 43620–43627. [DOI] [PubMed] [Google Scholar]
- Bergeron-Sandoval L.P., Safaee N., Michnick S.W. (2016). Mechanisms and consequences of macromolecular phase separation. Cell 165, 1067–1079. [DOI] [PubMed] [Google Scholar]
- Beutel O., Maraspini R., Pombo-Garcia K. et al. (2019). Phase separation of zonula occludens proteins drives formation of tight junctions. Cell 179, 923–936.e11. [DOI] [PubMed] [Google Scholar]
- Bhatt M.P., Lim Y.C., Kim Y.M. et al. (2013). C-peptide activates AMPKα and prevents ROS-mediated mitochondrial fission and endothelial apoptosis in diabetes. Diabetes 62, 3851–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D., Behrends C. (2020). When separation causes activation: biophysical control of bulk autophagy initiation. Mol. Cell 78, 379–381. [DOI] [PubMed] [Google Scholar]
- Biamonti G., Vourc'h C. (2010). Nuclear stress bodies. Cold Spring Harb. Perspect. Biol. 2, a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S., Alberti S., Fawzi N.L. et al. (2018). Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A., Klein I.A., Sabari B.R. et al. (2018). Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouard S., Otterbein L.E., Anrather J. et al. (2000). Carbon monoxide generated by heme oxygenase 1 suppresses endothelial cell apoptosis. J. Exp. Med. 192, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubici C., Papa S., Pham C.G. et al. (2006). The NF-κB-mediated control of ROS and JNK signaling. Histol. Histopathol. 21, 69–80. [DOI] [PubMed] [Google Scholar]
- Carr T.M., Wheaton J.D., Houtz G.M. et al. (2017). JunB promotes Th17 cell identity and restrains alternative CD4+ T-cell programs during inflammation. Nat. Commun. 8, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo J., Ramirez R., Martin-Malo A. et al. (2001). Role of adhesion molecules in mononuclear cell apoptosis induced by cuprophan hemodialysis membranes. Nephron 89, 186–193. [DOI] [PubMed] [Google Scholar]
- Chao A.C., Lee T.C., Juo S.H.H. et al. (2016). Hyperglycemia increases the production of amyloid beta-peptide leading to decreased endothelial tight junction. CNS Neurosci. Ther. 22, 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charreau B. (2012). Signaling of endothelial cytoprotection in transplantation. Hum. Immunol. 73, 1245–1252. [DOI] [PubMed] [Google Scholar]
- Chaudhury H., Zakkar M., Boyle J. et al. (2010). c-Jun N-terminal kinase primes endothelial cells at atheroprone sites for apoptosis. Arterioscler. Thromb. Vasc. Biol. 30, 546–553. [DOI] [PubMed] [Google Scholar]
- Chen F. (2003). JUNB (jun B proto-oncogene). Atlas Genet. Cytogenet. Oncol. Haematol., http://atlasgeneticsoncology.org/gene/178/junb-(jun-b-proto-oncogene) [Google Scholar]
- Chen X., Duong M.N., Psaltis P.J. et al. (2017). High-density lipoproteins attenuate high glucose-impaired endothelial cell signaling and functions: potential implications for improved vascular repair in diabetes. Cardiovasc. Diabetol. 16, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y., Kerppola T.K. (2001). Close encounters of many kinds: Fos–Jun interactions that mediate transcription regulatory specificity. Oncogene 20, 2438–2452. [DOI] [PubMed] [Google Scholar]
- Chiu J.J., Chien S. (2011). Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives. Physiol. Rev. 91, 327–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R., Angel P., Karin M. (1989). Jun-B differs in its biological properties from and is a negative regulator of, c-Jun. Cell 59, 979–986. [DOI] [PubMed] [Google Scholar]
- Cinar H., Fetahaj Z., Cinar S. et al. (2019). Temperature, hydrostatic pressure, and osmolyte effects on liquid‒liquid phase separation in protein condensates: physical chemistry and biological implications. Chemistry 25, 13049–13069. [DOI] [PubMed] [Google Scholar]
- Cohen R.A. (1993). Dysfunction of vascular endothelium in diabetes-mellitus. Circulation 87, 67–76. [Google Scholar]
- Corces M.R., Trevino A.E., Hamilton E.G. et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha D.A., Gurzov E.N., Naamane N. et al. (2014). JunB protects β-cells from lipotoxicity via the XBP1–AKT pathway. Cell Death Differ. 21, 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis A.M., Wilkinson P.F., Gui M. et al. (2009). p38 mitogen-activated protein kinase targets the production of proinflammatory endothelial microparticles. J. Thromb. Haemost. 7, 701–709. [DOI] [PubMed] [Google Scholar]
- Dai B., Zhong T., Chen Z.X. et al. (2021). Myricetin slows liquid‒liquid phase separation of Tau and activates ATG5-dependent autophagy to suppress Tau toxicity. J. Biol. Chem. 297, 101222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T., Karin M. (1993). JunB differs from c-Jun in its DNA-binding and dimerization domains and represses c-Jun by formation of inactive heterodimers. Genes Dev. 7, 479–490. [DOI] [PubMed] [Google Scholar]
- Diabetes C., Complications Trial Research G., Nathan D.M. et al. (1993). The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N. Engl. J. Med. 329, 977–986. [DOI] [PubMed] [Google Scholar]
- Duster R., Kaltheuner I.H., Schmitz M. et al. (2021). 1,6-Hexanediol, commonly used to dissolve liquid‒liquid phase separated condensates, directly impairs kinase and phosphatase activities. J. Biol. Chem. 296, 100260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R., Wagner E.F. (2003). AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868. [DOI] [PubMed] [Google Scholar]
- Elmore S. (2007). Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35, 495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M.Y., Markmiller S., Vu A.Q. et al. (2019). Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron 103, 802–819.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feletou M., Vanhoutte P.M. (2006). Endothelial dysfunction: a multifaceted disorder (The Wiggers Award Lecture). Am. J. Physiol. Heart Circ. Physiol. 291, H985–H1002. [DOI] [PubMed] [Google Scholar]
- Forstermann U., Li H. (2011). Therapeutic effect of enhancing endothelial nitric oxide synthase (eNOS) expression and preventing eNOS uncoupling. Br. J. Pharmacol. 164, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foteinos G., Hu Y., Xiao Q. et al. (2008). Rapid endothelial turnover in atherosclerosis-prone areas coincides with stem cell repair in apolipoprotein E-deficient mice. Circulation 117, 1856–1863. [DOI] [PubMed] [Google Scholar]
- Franzmann T.M., Jahnel M., Pozniakovsky A. et al. (2018). Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654. [DOI] [PubMed] [Google Scholar]
- Fujioka Y., Alam J.M., Noshiro D. et al. (2020). Phase separation organizes the site of autophagosome formation. Nature 578, 301–305. [DOI] [PubMed] [Google Scholar]
- Gaglia G., Rashid R., Yapp C. et al. (2020). HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat. Cell Biol. 22, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganassi M., Mateju D., Bigi I. et al. (2016). A surveillance function of the HSPB8–BAG3–HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol. Cell 63, 796–810. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Wu Y., Li R. et al. (2005). Jun proteins modulate the ovary-specific promoter of aromatase gene in ovarian granulosa cells via a cAMP-responsive element. Oncogene 24, 2236–2246. [DOI] [PubMed] [Google Scholar]
- Gimbrone M.A. Jr, Garcia-Cardena G. (2016). Endothelial cell dysfunction and the pathophysiology of atherosclerosis. Circ. Res. 118, 620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda N., Dozier S.J., Johnson R.S. (2003). HIF-1 in cell cycle regulation, apoptosis, and tumor progression. Antioxid. Redox Signal. 5, 467–473. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L., Bautista P., Lechuga S. et al. (2012). ZO-2, a tight junction scaffold protein involved in the regulation of cell proliferation and apoptosis. Ann. NY Acad. Sci. 1257, 133–141. [DOI] [PubMed] [Google Scholar]
- Gozuacik D., Kimchi A. (2004). Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23, 2891–2906. [DOI] [PubMed] [Google Scholar]
- Groth-Pedersen L., Jaattela M. (2013). Combating apoptosis and multidrug resistant cancers by targeting lysosomes. Cancer Lett. 332, 265–274. [DOI] [PubMed] [Google Scholar]
- Hai T.W., Liu F., Coukos W.J. et al. (1989). Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 3, 2083–2090. [DOI] [PubMed] [Google Scholar]
- Hansson G.K., Chao S., Schwartz S.M. et al. (1985). Aortic endothelial cell death and replication in normal and lipopolysaccharide-treated rats. Am. J. Pathol. 121, 123–127. [PMC free article] [PubMed] [Google Scholar]
- Hasan Z., Koizumi S.I., Sasaki D. et al. (2017). JunB is essential for IL-23-dependent pathogenicity of Th17 cells. Nat. Commun. 8, 15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M.H., Peuchen E.H., Dovichi N.J. et al. (2018). Dual roles for ATP in the regulation of phase separated protein aggregates in Xenopus oocyte nucleoli. eLife 7, e35224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Luo Y. (2012). Brg1 regulates the transcription of human papillomavirus type 18 E6 and E7 genes. Cell Cycle 11, 617–627. [DOI] [PubMed] [Google Scholar]
- Henderson A., Holloway A., Reeves R. et al. (2004). Recruitment of SWI/SNF to the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 24, 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y., Noma K., Yoshizumi M. et al. (2009). Endothelial function and oxidative stress in cardiovascular diseases. Circ. J. 73, 411–418. [DOI] [PubMed] [Google Scholar]
- Hofweber M., Hutten S., Bourgeois B. et al. (2018). Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13. [DOI] [PubMed] [Google Scholar]
- Huang S., Zhu S., Kumar P. et al. (2021). A phase-separated nuclear GBPL circuit controls immunity in plants. Nature 594, 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y., Toriumi H., Yamada S. et al. (2010). Resident endothelial cells surrounding damaged arterial endothelium reendothelialize the lesion. Arterioscler. Thromb. Vasc. Biol. 30, 1725–1732. [DOI] [PubMed] [Google Scholar]
- Jacobs-Helber S.M., Abutin R.M., Tian C. et al. (2002). Role of JunB in erythroid differentiation. J. Biol. Chem. 277, 4859–4866. [DOI] [PubMed] [Google Scholar]
- Jia J., Ye T., Cui P. et al. (2016). AP-1 transcription factor mediates VEGF-induced endothelial cell migration and proliferation. Microvasc. Res. 105, 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Sun M., Li S. et al. (2021). Long non-coding RNA DICER1-AS1-low expression in arsenic-treated A549 cells inhibits cell proliferation by regulating the cell cycle pathway. Environ. Toxicol. Pharmacol. 84, 103617. [DOI] [PubMed] [Google Scholar]
- Jiang H., Wang S., Huang Y. et al. (2015). Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 163, 108–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zhao X.H., Mao Y.L. et al. (2019). Long non-coding RNA RP11-468E2.5 curtails colorectal cancer cell proliferation and stimulates apoptosis via the JAK/STAT signaling pathway by targeting STAT5 and STAT6. J. Exp. Clin. Cancer Res. 38, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D., Freyberg M.A., Friedl P. (1997). Lack of hemodynamic forces triggers apoptosis in vascular endothelial cells. Biochem. Biophys. Res. Commun. 231, 586–590. [DOI] [PubMed] [Google Scholar]
- Kang J., Lim L., Lu Y. et al. (2019). A unified mechanism for LLPS of ALS/FTLD-causing FUS as well as its modulation by ATP and oligonucleic acids. PLoS Biol. 17, e3000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Lim L., Song J. (2018). ATP enhances at low concentrations but dissolves at high concentrations liquid‒liquid phase separation (LLPS) of ALS/FTD-causing FUS. Biochem. Biophys. Res. Commun. 504, 545–551. [DOI] [PubMed] [Google Scholar]
- Karin M., Liu Z., Zandi E. (1997). AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246. [DOI] [PubMed] [Google Scholar]
- Katagiri T., Kameda H., Nakano H. et al. (2021). Regulation of T cell differentiation by the AP-1 transcription factor JunB. Immunol. Med. 44, 197–203. [DOI] [PubMed] [Google Scholar]
- Kiesow K., Bennewitz K., Miranda L.G. et al. (2015). Junb controls lymphatic vascular development in zebrafish via miR-182. Sci. Rep. 5, 15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.A., Montagnani M., Koh K.K. et al. (2006). Reciprocal relationships between insulin resistance and endothelial dysfunction: molecular and pathophysiological mechanisms. Circulation 113, 1888–1904. [DOI] [PubMed] [Google Scholar]
- King M.R., Petry S. (2020). Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat. Commun. 11, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothakota S., Azuma T., Reinhard C. et al. (1997). Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science 278, 294–298. [DOI] [PubMed] [Google Scholar]
- Kumar R., Mani A.M., Singh N.K. et al. (2020). PKCθ–JunB axis via upregulation of VEGFR3 expression mediates hypoxia-induced pathological retinal neovascularization. Cell Death Dis. 11, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard D.A., Rajaram N., Kerppola T.K. (1997). Structural basis of DNA bending and oriented heterodimer binding by the basic leucine zipper domains of Fos and Jun. Proc. Natl Acad. Sci. USA 94, 4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.M. (1999). The role of mismatch repair in DNA damage-induced apoptosis. Oncol. Res. 11, 393–400. [PubMed] [Google Scholar]
- Licht A.H., Pein O.T., Florin L. et al. (2006). JunB is required for endothelial cell morphogenesis by regulating core-binding factor β. J. Cell Biol. 175, 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Currie S.L., Rosen M.K. (2017). Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 292, 19110–19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.Y., Yao J., Li X.M. et al. (2014). Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 5, e1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi M., Cagliero E. (1991). Pathobiology of endothelial and other vascular cells in diabetes mellitus. Call for data. Diabetes 40, 653–659. [DOI] [PubMed] [Google Scholar]
- Lorenzi M., Cagliero E., Toledo S. (1985). Glucose toxicity for human endothelial cells in culture. Delayed replication, disturbed cell cycle, and accelerated death. Diabetes 34, 621–627. [DOI] [PubMed] [Google Scholar]
- Lu Z.N., Song J., Sun T.H. et al. (2021). UBE2C affects breast cancer proliferation through the AKT/mTOR signaling pathway. Chin. Med. J. 134, 2465–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn S., Saad S., Poronnik P. et al. (2003). High glucose-mediated effects on endothelial cell proliferation occur via p38 MAP kinase. Am. J. Physiol. Endocrinol. Metab. 285, E708–E717. [DOI] [PubMed] [Google Scholar]
- Meyer L.K., Verbist K.C., Albeituni S. et al. (2020). JAK/STAT pathway inhibition sensitizes CD8 T cells to dexamethasone-induced apoptosis in hyperinflammation. Blood 136, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Yoshida H., Takahashi E. et al. (2020). The AP-1 transcription factor JunB functions in Xenopus tail regeneration by positively regulating cell proliferation. Biochem. Biophys. Res. Commun. 522, 990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natali A., Taddei S., Quinones Galvan A. et al. (1997). Insulin sensitivity, vascular reactivity, and clamp-induced vasodilatation in essential hypertension. Circulation 96, 849–855. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. (2001). Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2, 211–216. [DOI] [PubMed] [Google Scholar]
- Pan L., Feng F., Wu J. et al. (2021). Diosmetin inhibits cell growth and proliferation by regulating the cell cycle and lipid metabolism pathway in hepatocellular carcinoma. Food Funct. 12, 12036–12046. [DOI] [PubMed] [Google Scholar]
- Piqueras L., Reynolds A.R., Hodivala-Dilke K.M. et al. (2007). Activation of PPARβ/δ induces endothelial cell proliferation and angiogenesis. Arterioscler. Thromb. Vasc. Biol. 27, 63–69. [DOI] [PubMed] [Google Scholar]
- Ponticos M., Papaioannou I., Xu S. et al. (2015). Failed degradation of JunB contributes to overproduction of type I collagen and development of dermal fibrosis in patients with systemic sclerosis. Arthritis Rheumatol. 67, 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quagliaro L., Piconi L., Assaloni R. et al. (2003). Intermittent high glucose enhances apoptosis related to oxidative stress in human umbilical vein endothelial cells: the role of protein kinase C and NAD(P)H-oxidase activation. Diabetes 52, 2795–2804. [DOI] [PubMed] [Google Scholar]
- Quyyumi A.A., Dakak N., Andrews N.P. et al. (1995). Nitric oxide activity in the human coronary circulation. Impact of risk factors for coronary atherosclerosis. J. Clin. Invest. 95, 1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii S., Butler J.M., Ding B.S. (2016). Angiocrine functions of organ-specific endothelial cells. Nature 529, 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat P., Boehning M., Hummel B. et al. (2021). Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol. Cell 81, 1013–1026.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J., Mowen K.A., McBride K.D. et al. (2002). Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezabakhsh A., Ahmadi M., Khaksar M. et al. (2017). Rapamycin inhibits oxidative/nitrosative stress and enhances angiogenesis in high glucose-treated human umbilical vein endothelial cells: role of autophagy. Biomed. Pharmacother. 93, 885–894. [DOI] [PubMed] [Google Scholar]
- Risso A., Mercuri F., Quagliaro L. et al. (2001). Intermittent high glucose enhances apoptosis in human umbilical vein endothelial cells in culture. Am. J. Physiol. Endocrinol. Metab. 281, E924–E930. [DOI] [PubMed] [Google Scholar]
- Ruderman N.B., Williamson J.R., Brownlee M. (1992). Glucose and diabetic vascular disease. FASEB J. 6, 2905–2914. [DOI] [PubMed] [Google Scholar]
- Sabari B.R., Dall'Agnese A., Boija A. et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakao S., Taraseviciene-Stewart L., Lee J.D. et al. (2005). Initial apoptosis is followed by increased proliferation of apoptosis-resistant endothelial cells. FASEB J. 19, 1178–1180. [DOI] [PubMed] [Google Scholar]
- Schmidt D., Textor B., Pein O.T. et al. (2007). Critical role for NF-κB-induced JunB in VEGF regulation and tumor angiogenesis. EMBO J. 26, 710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S.M., Benditt E.P. (1977). Aortic endothelial cell replication. I. Effects of age and hypertension in the rat. Circ. Res. 41, 248–255. [DOI] [PubMed] [Google Scholar]
- Shi J., Chen S.Y., Shen X.T. et al. (2022). NOP53 undergoes liquid‒liquid phase separation and promotes tumor radio-resistance. Cell Death Discov. 8, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., You K., Chen T. et al. (2021). Quantifying the phase separation property of chromatin-associated proteins under physiological conditions using an anti-1,6-hexanediol index. Genome Biol. 22, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Brangwynne C.P. (2017). Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- Smith N.C., Kuravsky M., Shammas S.L. et al. (2021). Binding and folding in transcriptional complexes. Curr. Opin. Struct. Biol. 66, 156–162. [DOI] [PubMed] [Google Scholar]
- Song J. (2021). Adenosine triphosphate energy-independently controls protein homeostasis with unique structure and diverse mechanisms. Protein Sci. 30, 1277–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpio B.E., Riley J.T., Dardik A. (2002). Cells in focus: endothelial cell. Int. J. Biochem. Cell Biol. 34, 1508–1512. [DOI] [PubMed] [Google Scholar]
- Susztak K., Raff A.C., Schiffer M. et al. (2006). Glucose-induced reactive oxygen species cause apoptosis of podocytes and podocyte depletion at the onset of diabetic nephropathy. Diabetes 55, 225–233. [PubMed] [Google Scholar]
- Tousian H., Razavi B.M., Hosseinzadeh H. (2020). Alpha-mangostin decreased cellular senescence in human umbilical vein endothelial cells. DARU J. Pharm. Sci. 28, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vane J.R., Anggard E.E., Botting R.M. (1990). Regulatory functions of the vascular endothelium. N. Engl. J. Med. 323, 27–36. [DOI] [PubMed] [Google Scholar]
- Vanhoutte P.M., Shimokawa H., Feletou M. et al. (2017). Endothelial dysfunction and vascular disease—a 30th anniversary update. Acta Physiol. 219, 22–96. [DOI] [PubMed] [Google Scholar]
- Varma S., Lal B.K., Zheng R. et al. (2005). Hyperglycemia alters PI3k and Akt signaling and leads to endothelial cell proliferative dysfunction. Am. J. Physiol. Heart Circ. Physiol. 289, H1744–H1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B., Ivanov P. (2019). Stress granules and neurodegeneration. Nat. Rev. Neurosci. 20, 649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright H.P. (1968). Endothelial mitosis around aortic branches in normal guinea pigs. Nature 220, 78–79. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Lai Y., Yu Y. et al. (2021). The exocrine differentiation and proliferation factor (EXDPF) gene promotes ovarian cancer tumorigenesis by up-regulating DNA replication pathway. Front. Oncol. 11, 669603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Mo X., Chen J. et al. (2022). Myricetin inhibits α-synuclein amyloid aggregation by delaying the liquid-to-solid phase transition. Chembiochem 23, e202200216. [DOI] [PubMed] [Google Scholar]
- Xu M., Cao L., Zhang X. et al. (2020). miR-3133 inhibits proliferation and angiogenesis by targeting the JUNB/VEGF pathway in human umbilical vein endothelial cells. Oncol. Rep. 44, 1699–1708. [DOI] [PubMed] [Google Scholar]
- Yamazaki S., Tanaka Y., Araki H. et al. (2017). The AP-1 transcription factor JunB is required for Th17 cell differentiation. Sci. Rep. 7, 17402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wang C., Shu J. et al. (2021). Porcine epidemic diarrhea virus induces vero cell apoptosis via the p53–PUMA signaling pathway. Viruses 13, 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.L., Cao Y.H. (2022). The impact of VEGF on cancer metastasis and systemic disease. Semin. Cancer Biol. 86, 251–261. [DOI] [PubMed] [Google Scholar]
- Yoshitomi Y., Ikeda T., Saito H. et al. (2017). JunB regulates angiogenesis and neurovascular parallel alignment in mouse embryonic skin. J. Cell Sci. 130, 916–926. [DOI] [PubMed] [Google Scholar]
- Yu C.H., Suriguga G.,M., Liu W.J. et al. (2017). High glucose induced endothelial to mesenchymal transition in human umbilical vein endothelial cell. Exp. Mol. Pathol. 102, 377–383. [DOI] [PubMed] [Google Scholar]
- Yukimasa N., Isobe K., Nagai H. et al. (1999). Successive occupancy by immediate early transcriptional factors of the tyrosine hydroxylase gene TRE and CRE sites in PACAP-stimulated PC12 pheochromocytoma cells. Neuropeptides 33, 475–482. [DOI] [PubMed] [Google Scholar]
- Zhang G., Wang Z., Du Z. et al. (2018). mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell 174, 1492–1506.e22. [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang X., Li X. et al. (2019). Effect of CCNB1 silencing on cell cycle, senescence, and apoptosis through the p53 signaling pathway in pancreatic cancer. J. Cell. Physiol. 234, 619–631. [DOI] [PubMed] [Google Scholar]
- Zhang X., Jin J.Y., Wu J. et al. (2015). RNA-Seq and ChIP-Seq reveal SQSTM1/p62 as a key mediator of JunB suppression of NF-κB-dependent inflammation. J. Invest. Dermatol. 135, 1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Tang N., Hadden T.J. et al. (2011). Akt, FoxO and regulation of apoptosis. Biochim. Biophys. Acta 1813, 1978–1986. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Alexander P.B., Wang X.F. (2017). TGF-β family signaling in the control of cell proliferation and survival. Cold Spring Harb. Perspect. Biol. 9, a022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.