Abstract
Expression of genes of the plasminogen activator (PA) system declines at the G0/G1-S-phase boundary of the cell cycle. We found that overexpression of E2F1-3, which acts mainly in late G1, inhibits promoter activity and endogenous expression of the urokinase-type PA (uPA) and PA inhibitor 1 (PAI-1) genes. This effect is dose dependent and conserved in evolution. Mutation analysis indicated that both the DNA-binding and transactivation domains of E2F1 are necessary for this regulation. Interestingly, an E2F1 mutant lacking the pRB-binding region strongly repressed the uPA and PAI-1 promoters. An E2F-mediated negative effect was also observed in pRB and p107/p130 knockout cell lines. This is the first report that E2F can act as a repressor independently of pocket proteins. Mutation of AP-1 elements in the uPA promoter abrogated E2F-mediated transcriptional inhibition, suggesting the involvement of AP-1 in this regulation. Results shown here identify E2F as an important component of transcriptional control of the PA system and thus provide new insights into mechanisms of cellular proliferation.
Extracellular proteolysis, especially that mediated by the plasminogen activator (PA) system, plays an important role in various physiological and pathological processes, such as angiogenesis, wound healing, inflammation, and tumor metastasis (1, 27). PAs, urokinase-type PA (uPA) and tissue-type PA (tPA), are secreted serine proteases that convert the ubiquitous zymogen plasminogen to plasmin. This trypsin-like protease degrades a wide range of substrates, including various extracellular matrix proteins, such as fibronectin, vitronectin, and fibrin. Of the two PAs, uPA is considered to be engaged more in cell-associated proteolysis due to the presence of a cell surface-associated uPA receptor (uPAR). The activities of both uPA and tPA are negatively regulated by the binding of PA inhibitor 1 (PAI-1) and PAI-2, which are members of the serine protease inhibitor superfamily. Interestingly, both uPA and PAI-1 are highly expressed in various metastatic tumors, suggesting that controlled proteolysis is important for metastasis (1).
The PA system may also have a significant role in cell cycle progression, where cells undergo detachment from neighboring cells and the extracellular matrix. It has been reported that PAI-1 and uPA mRNAs are rapidly induced soon after exposure of growth-arrested cells to serum-containing medium and that this expression declines prior to DNA synthesis in the G1-to-S transition phase (21, 55, 66). This pattern of expression appears also in the second cell cycle of synchronized cells, suggesting that the regulation is cell cycle dependent. Induction of the transcription of these genes in early-to-mid-G1 phase is thought to be mediated through AP-1- and c-myc-responsive elements present in their promoters (33, 55). However, the suppression mechanism acting on the transcription of these genes in late G1 has not been elucidated.
One of the key regulators of cell cycle events at the boundary of the G0/G1 and S phases is the E2F transcription factor (38, 63). This factor regulates the transcription of several genes required for DNA replication and cell cycle progression (23, 35, 49). E2F acts as a transcription activator or repressor, depending on the promoter context. Active E2F is a heterodimer of an E2F family member (E2F1-6) and a DP family member (DP1 or DP2) (65). The E2F protein is composed of functionally distinct domains responsible for binding to DNA, dimerization with a DP partner, and transactivation. The latter domain contains sites for interaction with a pocket-binding protein (pRB, p107, or p130) and other cofactors such as CBP (CREB-binding protein) (61), MDM2 (40), and TRRAP (transformation-transcription domain-associated protein) (41). Binding of a pocket protein to E2F suppresses its transactivation activity (14, 51) or converts it to an active repressor (24, 67) that exerts its inhibitory effect partly by recruiting histone deacetylase (9) or by interaction with general transcription factors (64). E2F6 shares homology with other E2F family members in the DNA-binding and dimerization domains but lacks transactivation and pocket protein-binding domains. Thus, it acts as a negative regulator countering the activity of other E2F members (19, 60).
In the present work, we investigated the role of the PA system in cell cycle regulation by examining the control of uPA and PAI-1 gene expression by the E2F transcription factor. We found that overexpression of E2F1, E2F2, and E2F3 can repress transcription of the uPA and PAI-1 genes. We provide evidence that active repression by E2F is independent of pocket protein partners. We demonstrated the importance of the AP-1-responsive element in E2F transcriptional regulation of the uPA promoter.
MATERIALS AND METHODS
Cell culture.
LLC-PK1 pig epithelial cells, 293 human kidney epithelial cells, mouse embryo fibroblasts (MEF), and U2OS and SAOS-2 human osteosarcoma cells were cultured in Dulbecco's modified Eagle's medium (GIBCO-BRL) supplemented with 10% (vol/vol) fetal calf serum (AMIMED), streptomycin at 0.2 mg/ml, and penicillin at 50 U/ml at 37°C in a humidified CO2 (5%) incubator. WI-38 human lung fibroblasts were cultured in minimum essential medium (GIBCO-BRL) supplemented with 15% fetal calf serum.
Plasmids.
The wild-type and mutant E2F1 expression vectors pRcCMV-E2F1, pRcCMV-E132, pRcCMV-del24, and pRcCMV-del5 have been previously described (24, 29, 31). The uPA-CAT 5′ deletion constructs were provided by D. von der Ahe (32), pBL-PAI-1-CAT was from A. Riccio (52), −1452 tPA-CAT (42) and −1.1 PAI-2-CAT (15) were from R. Medcalf, and the PAI-1–luciferase construct p800 was from D. Loskutoff (62). AP-1 binding site mutant forms of the uPA promoter were constructed by site-directed mutagenesis on the basis of a −2.5 uPA chloramphenicol acetyltransferase (CAT) reporter plasmid. tPA(+enh).CAT was constructed by inserting the uPA enhancer fragment (−1968 to −1870) covering two AP-1 sites but void of Ets and NFκ sites immediately upstream of the minimal tPA promoter. CAT reporter genes for the pig uPA promoter (36) and the mouse uPA promoter (12) were described previously. The E2F1-responsive reporter construct p(−3407)19ARF.CAT (53) was provided by K. D. Robertson, and pRSV-c-Jun was provided by P. Angel (2). The p3×AP1-tk-Luc construct was described previously (6).
Transient-transfection assays.
LLC-PK1 cells (0.3 × 106/well) were plated in six-well (35-mm-diameter) tissue culture plates and transfected by the calcium phosphate precipitation method (Pharmacia Biotech Inc.). Amounts of transfected DNA are indicated in the figure legends. Cell extracts were assayed for luciferase activity as described previously (6) using a luminometer (Autolumat LB 953; Berthold) and for CAT activity using a CAT enzyme-linked immunosorbent assay kit (Boehringer Mannheim).
Passage 5 to 7 MEF cells (6 × 105/well) were plated in 24-well tissue culture plates and transfected with Lipofectamine (Life Technologies). Each well contained 400 ng of the reporter gene, 40 ng of the expression vector, and 2 μl of Lipofectamine. Cells were incubated with the DNA-liposome mixture for 2 h, washed twice with 1× phosphate-buffered saline, and incubated in medium containing 10% serum. After 24 h, cells were harvested and assayed for luciferase activity as described above.
Viruses.
Adenovirus (Ad)-cytomegalovirus (CMV), Ad-E2F1, Ad-E2F2, Ad-E2F3, Ad-E2F4, Ad-E2F5, and Ad-DP1 recombinant viruses were provided by J. R. Nevins (16), and viral stocks were created as previously described (57). Titers of viral stocks were determined by a plaque assay on 293 cells and defined as PFU per milliliter. Quiescent or randomly growing WI-38 cells were infected, at an input multiplicity of 1 PFU/cell (except Ad-E2F3 [2 PFU/cell]), by adding viral stocks directly to the culture medium. For analysis, cells were harvested 16 h following infection.
RNA isolation and Northern blot analysis.
Total RNA (10 μg), prepared with the acid guanidinium thiocyanate-phenol-chloroform method (13), was resolved by gel electrophoresis under denaturing conditions and transferred to a nylon membrane as previously described (68). rRNA was stained on the filters with methylene blue (25) to assess RNA loading and transfer. Hybridization was performed as previously described (68). The cDNA clones for human uPA, human PAI-1, and human uPAR were provided by F. Asselberg, D. Loskutoff, and E. K. Kruithof, respectively. The DNA inserts from each plasmid were labeled with [α-32P] dATP using the random oligonucleotide-primed reaction (18). The mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was obtained from Ambion. Levels of specific RNA were measured in a Molecular Dynamics PhosphorImager.
Western blot analysis.
Cells from a 10-cm-diameter dish were lysed for 15 min on ice in 800 μl of a lysis buffer consisting of 50 mM Tris (pH 7.5), 250 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, 1 mM dithiothreitol, aprotinin at 10 μl/ml, and leupeptin at 10 μl/ml. Cell extracts (10 μg of protein) were fractionated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis, blotted onto polyvinylidene difluoride transfer membranes (Millipore), and analyzed using antibodies against different E2F members. Monoclonal antibodies against E2F2(C-20), E2F3(C-18), E2F4(C-108), and E2F5(C-20) were obtained from Santa Cruz Biotechnology, and a monoclonal antibody against E2F1 was obtained from Upstate Biotechnology. Signals were detected by ECL (Amersham).
Immunofluorescence analysis.
WI-38 cells were plated on glass coverslips, starved, and virus infected. Immunofluorescence analysis was performed as previously described (43). The following antibodies were used for detection of E2Fs: rabbit polyclonal anti-E2F1 (C-20) and anti-E2F4 (C-20) antibody (Santa Cruz Biotechnology), followed by Cy3-conjugated donkey anti-rabbit immunoglobulin G antibody (Milan Analytica AG). The nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI; Roche). The cellular distribution of E2F was examined using a Leica IRBE inverted microscope with a 40 × 1.25 NA lens.
RESULTS
E2F1 inhibits uPA, PAI-1, and PAI-2 promoter activity.
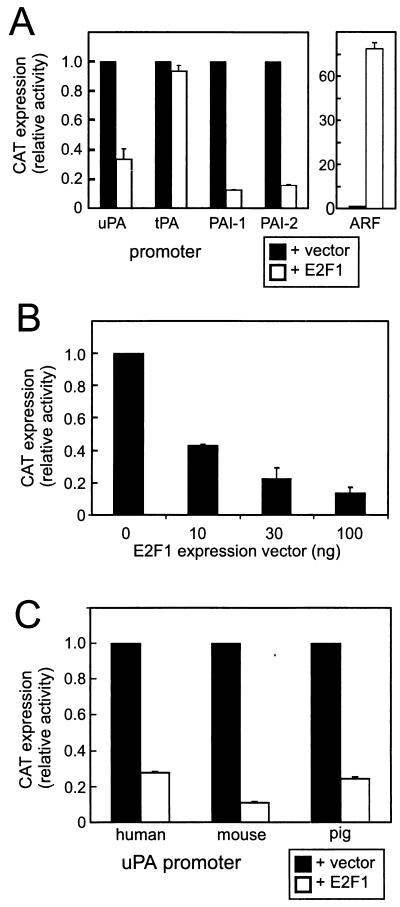
The effects of E2F1 on different genes of the PA system were analyzed by transient-cotransfection assays using CAT reporter genes in LLC-PK1 nontransformed epithelial cells. As shown in Fig. 1A, E2F1 overexpression reduced CAT expression from the human uPA, PAI-1, and PAI-2 promoters to 30, 10, and 17%, respectively, of the control values. The tPA promoter was not affected. In contrast, the alternate reading frame (ARF) cell cycle regulatory gene promoter (53), which we examined as a positive control for the E2F effect, was strongly enhanced. The inhibitory effect of the E2F1 expression vector on the PAI-1 promoter was dose dependent (Fig. 1B). The human, pig, and mouse uPA promoters were all strongly suppressed by E2F1 overexpression, indicating that this negative regulation of the uPA gene has been conserved during evolution (Fig. 1C). Taken together, these data suggest that downregulation of the uPA, PAI-1, and PAI-2 genes is a promoter-specific effect of E2F1.
FIG. 1.
E2F1 inhibits transcription from uPA, PAI-1, and PAI-2 promoters. LLC-PK1 kidney epithelial cells were transfected with different CAT reporter genes (2 μg/assay) together with an empty vector (pRcCMV) or a vector encoding wild-type E2F1 (200 ng/assay). After 24 h, cells were harvested and assayed for CAT expression. (A) Comparison of human uPA (−2.5 kb), tPA (−1.4 kb), PAI-1 (−0.8 kb), PAI-2 (−1.1 kb), and alternate reading frame (ARF) (−3.5 kb) promoters. (B) Dose dependency of the effect of the E2F1 expression vector on the PAI-1 promoter. (C) Comparison of human uPA (−2.5 kb), mouse uPA (−8.2 kb), and pig uPA (−4.6 kb) promoters. The results shown are averages of two independent experiments. Assays were done in duplicate, and mean values are shown with error bars. The results were normalized with luciferase expression from a cotransfected minimal tPA promoter (0.5 kb)-luciferase reporter gene that did not respond to E2F1.
We focused further analysis mainly on the uPA and PAI-1 genes, because they are known to be involved in a cooperative manner in various biological processes (for a review, see reference 27). The following analysis also included uPAR gene expression because this gene plays an important role in the interplay of uPA and PAI-1 in various cellular activities.
The DNA-binding and transactivation domains, but not the pRB- and cyclin A-binding domains, of E2F1 are involved in repression of the uPA and PAI-1 promoters.
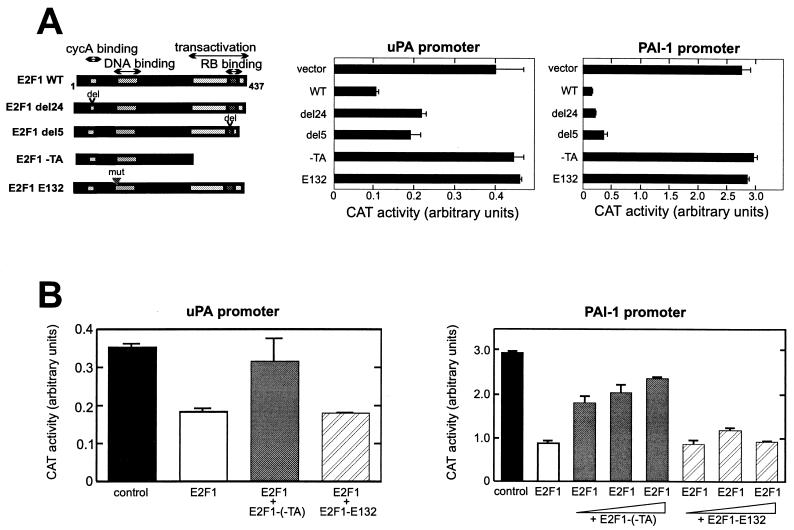
To understand the molecular mechanism of E2F-mediated inhibition, we examined the effects of different E2F1 mutants on the uPA and PAI-1 promoters in LLC-PK1 cells (Fig. 2A). Mutation of the DNA-binding domain (E132), which abolished the DNA-binding activity of E2F1, completely abrogated the inhibitory effect of E2F1 on both promoters. This indicates that DNA binding is essential for E2F1 suppression of these promoters. A deletion in the amino-terminal CycA-cdk2 binding domain (del24) did not affect E2F-mediated inhibition, which excludes the involvement of the CycA-cdk2 complex in this process. Deletion of the transactivation domain (−TA) of E2F1 abolished its inhibitory effect. Interestingly, an E2F1 mutant defective in pRB binding (del5) also exhibited strong inhibitory activity on the uPA and PAI-1 promoters. Taken together, these results indicate that the transactivation domain is essential for the E2F1 inhibitory effect, possibly through interaction with transcription coactivator proteins other than pRB. This suggests a novel mechanism for E2F-mediated repression which is pocket protein independent.
FIG. 2.
Effects of E2F1 mutations on negative regulation of the uPA and PAI-1 promoters. (A) The wild type (WT; 200 ng) and different mutant forms of the pRcCMV-E2F1 expression vector (200 ng) were cotransfected with uPA CAT (−2.5 kb; 2 μg) or PAI-1 CAT (−0.8 kb; 2 μg) in LLC-PK1 cells. RB, pRB; del, deletion; mut, mutation. (B) Competition between wild-type E2F1 and (−TA) and E132 mutants. Cells were transfected with the reporter gene and wild-type E2F1 (60 ng) together with increasing amounts of (−TA) or E132 expression vectors (100 ng for uPA promoter analysis and 100, 150, and 300 ng for PAI-1 promoter analysis). CAT expression was measured as described in the legend to Fig. 1.
The dependency of E2F1 negative regulation on DNA binding was further tested by a competition experiment between the wild type and a transactivation domain-negative (−TA) or DNA binding-deficient (E132) E2F1 mutant. The −TA mutation interfered with the negative effect of wild-type E2F1 on both the uPA and PAI-1 promoters. In contrast, the E132 mutation did not overcome the negative regulation by wild-type E2F1 (Fig. 2B). These results suggest that E2F1 must bind to the uPA and PAI-1 promoters to exert its inhibitory effect.
Pocket protein family members are not involved in the negative regulation of the uPA and PAI-1 promoter by E2F1.
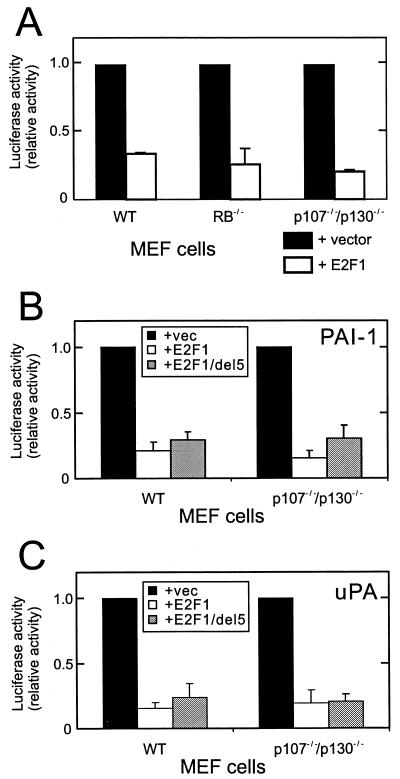
Although it was shown previously that transcriptional repression mediated by E2F involves its association with a pocket protein partner (14, 51, 67), the above mutation analysis suggests a new mechanism for the negative regulation of the uPA and PAI-1 promoters by E2F1 that is independent of pRB. To test this hypothesis, we examined the effect of E2F1 overexpression on the PAI-1 promoter in cells lacking functional pRB or p107 and p130. As shown in Fig. 3A, PAI-1 promoter activity in pRB-deficient MEF cells was reduced by E2F1 overexpression as strongly as in wild-type MEF cells. Likewise, overexpression of E2F1 in p107/p130 double-knockout MEF cells suppressed PAI-1 promoter activity. Similar results were obtained for the uPA promoter (data not shown). Negative regulation by E2F overexpression in MEF cells was specific for the PAI-1 and uPA promoters. The control ARF gene promoter was highly induced by E2F overexpression in these cells (data not shown). Both knockout cell lines, however, still carry at least one active pocket protein. To exclude the possibility that one of the remaining pocket proteins functionally replaces deleted pocket proteins, we transfected p107/p130 (−/−) MEF with an E2F1 mutant that is incapable of pRB binding. This mutant suppressed uPA and PAI-1 promoter activity to an extent similar to that of wild-type E2F1 (Fig. 3B). Taken together, these results support the hypothesis that E2F1-mediated inhibition of the PAI-1 and uPA promoters does not require pRB, p107, or p130.
FIG. 3.
Repression of the PAI-1 promoter is pocket protein independent. (A) The reporter construct of PAI-1 luciferase (400 ng/assay) was transfected together with either a control expression vector or a vector encoding E2F1 (40 ng/assay) into wild-type (WT), pRB−/−, or p107−/−/p130−/− MEF cells. RB, pRB. (B and C) The PAI-1 luciferase (B) or uPA-CAT (C) (400 ng/assay) reporter construct was transfected together with either an empty control vector (vec) or a vector encoding wild-type E2F1 or mutant E2F1 that does not bind pRB, E2F1d5 (40 ng/assay) into wild-type or p107−/−/p130−/− MEF cells. After 24 h, cells were harvested and assayed for luciferase activity and CAT expression. The results shown are averages of at least two independent experiments and were normalized with CAT expression or luciferase activity from a cotransfected tPA-luc or tPA-CAT reporter gene, respectively.
Differential ability of E2F family members to regulate uPA, PAI-1, and uPAR gene expression.
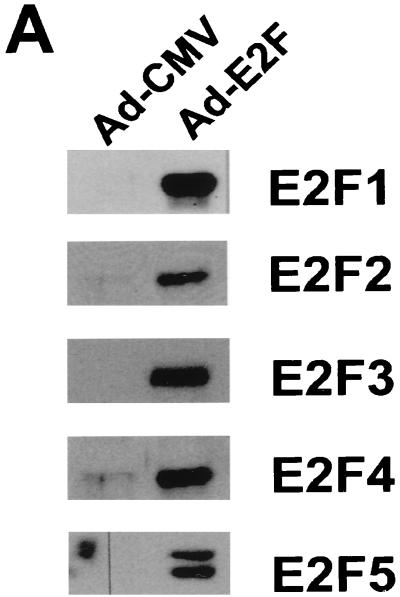
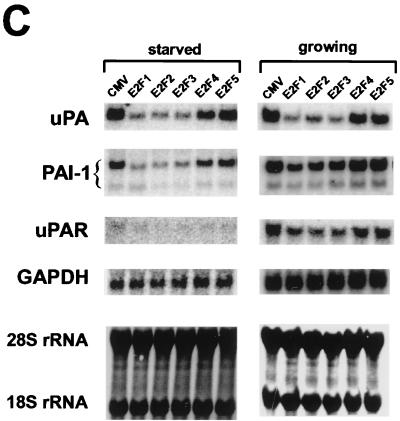
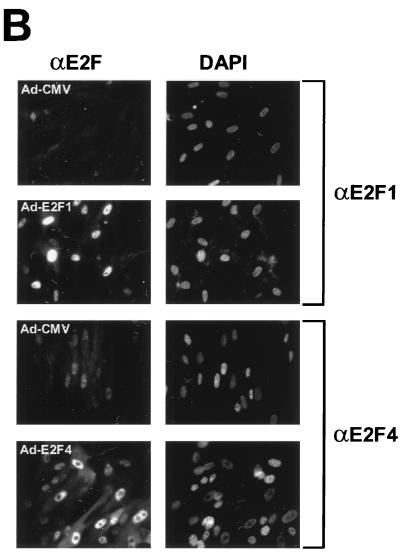
The E2F family has six members, which can be divided into three subgroups, E2F1-3, E2F4/5, and E2F6, based on structural and functional features. The members of subgroup E2F1-3 share a conserved amino-terminal domain containing binding sites for cyclin A-cdk2 (31) and Sp1 (30), which are absent in subgroups E2F4/5 and E2F6. E2F1-3 proteins bind preferentially to pRB and have very low affinity for p107 and p130 (37). These two members of the pocket protein family bind mainly to the carboxy terminus of E2F4 and E2F5 (56). E2F6 lacks the transactivation domain and acts as a negative regulator competing with other E2F members (19, 60). To compare the abilities of E2F family members to regulate genes of the PA system, we used recombinant Ad-based vectors expressing different E2F proteins. These proteins were overexpressed together with DP1, which is a binding partner of E2F, in nontransformed, low-passage WI-38 human primary fibroblasts. Coexpression of DP1 is known to enhance the level of E2F activity, especially of E2F3, E2F4, and E2F5 (16). Starved WI-38 primary cells expressed high levels of the corresponding proteins after Ad infection (Fig. 4A) and thus provide an appropriate platform for studying transcriptional regulation of E2F-targeted genes. Immunostaining analysis showed that E2F proteins were efficiently expressed and translocated to the nucleus after infection of serum-starved cells (Fig. 4B).
FIG. 4.
Differential effects of E2F family members on genes of the PA system. (A) Overexpression of each of the E2F family members in WI-38 human primary fibroblast cells (passage 20). WI-38 cells were deprived of serum for 24 h and then infected either with the control recombinant Ad (Ad-CMV) or with a recombinant expressing the indicated E2F family member (E2F1, E2F2, E2F3, E2F4, or E2F5) at a multiplicity of infection of 1 PFU/cell for viruses Ad-CMV, Ad-E2F1, Ad-E2F2, Ad-E2F4, and Ad-E2F5 and 2 PFU/cell for Ad-E2F3. A virus expressing the DP1 protein, Ad-DP1, was coinfected (multiplicity of 1 PFU/cell) with each E2F-expressing vector. After 16 h, cells were harvested for Western blot analysis using the corresponding monoclonal antibody against each E2F family member. (B) Analysis of infection efficiency by immunostaining. WI-38 cells were treated as described above. Immunofluorescence analysis was performed by staining of infected cells with polyclonal antibodies against E2F1 and E2F4. The nuclei were costained with DAPI. (C) Effects of E2F family members on serum-starved or randomly growing cells. Serum-starved WI-38 cells (left) were infected as described above. After 16 h, cells were harvested for Northern blot analysis using the probes indicated. Equivalent loading of RNA samples was confirmed by probing with GAPDH cDNA and methylene blue staining of rRNAs. Effects of E2F family members on randomly growing cells are shown on the right. WI-38 cells grown in the presence of 10% serum were infected with the indicated recombinant viruses. After 16 h, cells were harvested and processed for Northern blot analysis as described above.
The results in Fig. 4C show that endogenous mRNAs encoding uPA and PAI-1 were reduced by E2F1-3 overexpression but not by E2F4 or E2F5 overexpression. In serum-starved cells, uPAR mRNA levels were rather low and therefore the effects of E2F were not obvious. None of the E2F family members significantly affected the expression of the control GAPDH gene.
As the E2F1-3 proteins can promote cell progress into S phase (16), their inhibitory effects on the uPA, PAI-1, and uPAR genes may be indirect, i.e., associated with cell cycle events. Therefore, we measured cell cycle distribution by fluorescence-activated cell sorter analysis of cells infected with an E2F1-expressing virus. At the time when mRNA levels were examined (16 h postinfection), the majority of cells remained in the G0/G1 phase when they were infected with either a CMV control (78%) or an E2F1-expressing virus (68%) (data not shown). We also examined the effects of E2F expression in randomly growing cells. Under these conditions, uPA, PAI-1, and uPAR mRNA expression levels were again reduced by E2F1-3 but not by E2F4 or E2F5 (Fig. 4D). These results suggest the direct suppression of uPA, PAI-1, and uPAR mRNA levels by E2F1-3 expression and not via a change in cell cycle phase.
The −2 kb enhancer of the uPA promoter is critical for E2F1-mediated inhibition.
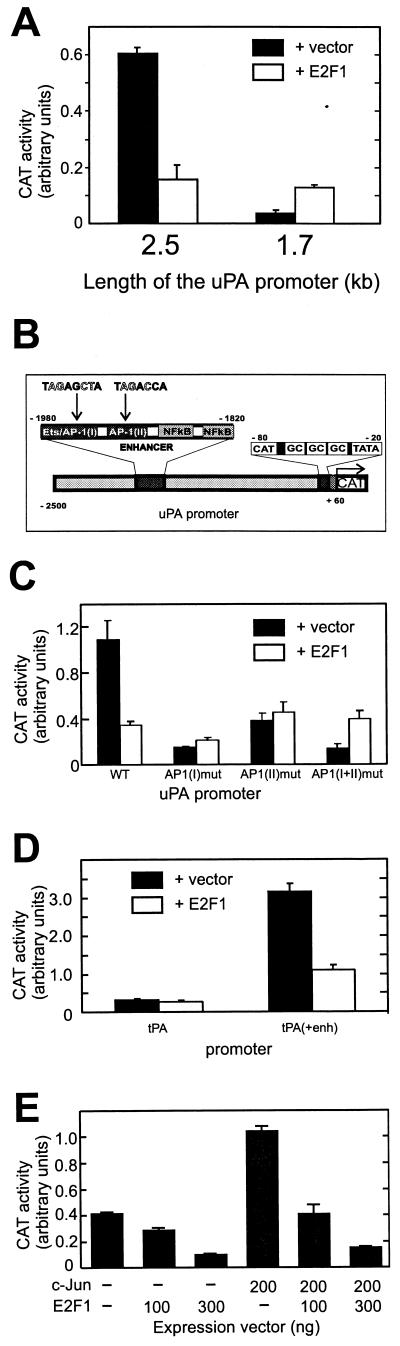
To gain insight into the molecular mechanism of negative E2F regulation, we characterized the uPA promoter. Regulation of this promoter by various signals has been extensively studied by our group and others (reviewed in reference 7). A 5′ deletion analysis of the uPA promoter showed that the −2.5-kb construct responded negatively to E2F1, whereas the −1.7-kb construct responded positively (Fig. 5A). This suggests that the region between −2.5 and −1.7 kb contains a site(s) responsible for the E2F1 negative effect and that the uPA promoter can respond positively to E2F under certain conditions when the negative regulatory site(s) is absent.
FIG. 5.
Role of the −2-kb enhancer in the uPA promoter in E2F1-mediated negative regulation. (A) LLC-PK1 cells were transfected with different 5′ deletion mutant forms (−2.5 or −1.7 kb) of the uPA-CAT reporter genes together with a control vector or an E2F1-expressing vector and harvested for CAT assay. CAT expression was measured and normalized as described in the legend to Fig. 1. (B) Schematic representation of uPA promoter reporter constructs. White letters indicate nucleotide changes in mutated constructs. (C) Effects of AP-1 site mutations on E2F negative regulation. LLC-PK1 cells were transfected with the wild type (WT) or AP-1 site-mutated uPA-CAT reporter genes together with a control vector or an E2F1-expressing vector. CAT expression was measured and normalized as described above. (D) Involvement of AP-1 elements in E2F1-mediated repression. Cells were transfected with −1.4 tPA-CAT or tPA(+enh)-CAT (2 μg) together with a control vector or an E2F1-expressing vector (200 ng). CAT expression was measured and normalized as described above. (E) Abrogation of the transcription-enhancing effect of AP-1 by E2F1. Cells were transfected with −2.5 uPA-CAT (2 μg) together with pRSV-c-Jun (0 or 200 ng) and pRcCMV-E2F1 (0, 100, or 300 ng). CAT expression was measured as described above. Empty vectors, pRSV and pRcCMV, were included if necessary to normalize the amount of DNA.
Region −2.5 to −1.7 kb of the uPA promoter contains a well-characterized enhancer that is a composite of cis-acting elements for the AP-1, Ets, and NFκB factors (Fig. 5B). The enhancer is highly conserved among humans, pigs, and mice (48), and the importance of this site for uPA gene regulation has been confirmed for all three organisms (for a review, see reference 7). To determine whether the same enhancer is involved in E2F1-mediated negative regulation of the promoter, we introduced mutations into AP-1 sites. Mutation of the major AP-1 site immediately 3′ of the Ets site reduced basal template activity and, at the same time, abrogated the E2F1 inhibitory effect (Fig. 5C). Mutation of another downstream AP-1 site also reduced both basal template activity and E2F1-mediated inhibition. The double mutant with changes at both AP-1-binding sites also completely abrogated E2F1 repression. These results suggest that the two AP-1 sites are cooperatively involved in transcription suppression. Mutation of the NFκB site within the uPA enhancer reduced enhancer activity by 30% but did not change the E2F1 inhibitory effect. Thus, this enhancer element is not the target of E2F1 negative regulation (data not shown). To see whether E2F-mediated repression of transcription via the AP-1 sites is promoter independent, we examined these AP-1 sites in the background of the tPA promoter, which by itself is insensitive to E2F1. Insertion of the uPA enhancer covering the two AP-1 sites but void of Ets and NFκB sites upstream of the tPA promoter enhanced basal template activity and, at the same time, rendered the promoter susceptible to E2F1 negative regulation (Fig. 5D). In accordance with this, overexpression of c-Jun, which is part of the AP-1 complex, upregulated a wild-type uPA promoter which was suppressed dose dependently by E2F1 (Fig. 5E).
DISCUSSION
In the present work, we demonstrated that E2F1 specifically inhibits the uPA and PAI-1 promoters and also reduces endogenous mRNA levels of these genes, as well as that of uPAR. This effect was dose dependent, conserved in evolution, and not cell type specific. Our findings are in accord with a previous report that transcription of these genes declines during the G0/G1-to-S-phase transition (8, 55, 66), when E2F1 and E2F3 levels are increased (38). Therefore, we propose that E2F plays a role in inhibiting expression of these genes during the G1-S-phase transition of the cell cycle.
As to how E2F suppresses uPA and PAI-1 gene expression, an obvious possibility is that involving pRB. Depending on the promoter context, E2F in association with a pocket protein partner has been shown to bind to E2F recognition elements in the promoter and actively suppress transcription. This mechanism is operative under certain conditions, as on the B-myb (34), E2F1 (28), cyclin A (26), and cyclin E (9) promoters. However, this mechanism for negative regulation appears unlikely in our studies. E2F1 mutants lacking the pRB-binding domain still exerted a suppressing effect on the uPA and PAI-1 promoters (Fig. 2). Wild-type E2F1, as well as a pRB binding-defective mutant, also strongly inhibited the transcription of these genes in MEF cells lacking the pRB gene or the p107/p130 genes (Fig. 3). In particular, the pRB-E2F4, p107-E2F4, and p130-E2F4 complexes have so far been implicated in transcriptional repression (39, 44). However, in our studies, E2F1, E2F2, and E2F3 but not E2F4 and E2F5, inhibited expression of the uPA and PAI-1 genes. These data provide strong evidence that E2F can act as a transcription repressor independently of interaction with pocket protein partners. E2F6 has also been shown to be a negative regulator in a pocket protein-independent manner. However, this protein lacks a transactivation domain and acts by countering the activity of other E2F family members (19, 60).
The carboxy-terminal transactivation domain was essential for E2F1 suppression of the uPA and PAI-1 promoters (Fig. 2). Several proteins are known to bind to this region and stimulate transactivation activity of E2F, such as MDM2 (40), CBP (CREB-binding protein) (61), and TRRAP (41). The transactivation domain also interacts with the basal transcription machinery by binding TATA box-binding protein (TBP) and transcription factor IIH (TFIIH) (50). It has been proposed that competition between pRB and both TBP and TFIIH for binding to the E2F1 transactivation domain is a mechanism by which pRB can inhibit activation by E2F1 (22). We excluded the possibility that pocket-binding proteins are responsible for E2F-mediated negative regulation of uPA and PAI-1 gene expression. Thus, we speculate that E2F1 binds to an unidentified protein to repress the PAI-1 and uPA promoters.
The inhibition of uPA promoter activity by E2F1 requires the enhancer element at −2.0 kb. Mutation analysis suggested that two AP-1-binding sites within this region act cooperatively for high basal promoter activity and that E2F1 inhibits the promoter by suppressing the activity of this enhancer. These elements are highly conserved among humans, mice, and pigs (48). We reported previously that the AP-1 elements in the enhancer are bound by c-Jun family members (17). We have shown that upregulation of the uPA promoter by overexpression of c-Jun could be titrated out by E2F1 overexpression. Furthermore, we also observed that E2F1 suppressed a reporter gene containing AP-1 sites of the uPA enhancer (Fig. 5D) or three copies of the AP-1-binding element from the collagenase promoter (data not shown). However, the uPA enhancer sequences do not possess E2F1 consensus binding sites. E2F1 regulation via nonconsensus sites in the promoter has only been shown for the herpes simplex virus thymidine kinase promoter, where E2F1 can bind to GC-rich elements (58). Using nuclear extracts from U2OS cells in a gel shift assay, we were unable to detect changes in patterns of protein binding to the uPA enhancer upon addition of insect cell-expressed E2F1/DP1 (data not shown). It may have been that E2F1 binding to the uPA enhancer was not stable enough to be detected in this assay or that some additional promoter elements were required. The latter, however, is not likely because the uPA enhancer containing only the two AP-1 sites rendered an otherwise insensitive tPA promoter susceptible to E2F1 negative regulation when located upstream of this promoter (Fig. 5D). The adapter CBP or its relative p300 is implicated in transcription activation by AP-1 (4, 5). Since deletion of the transactivation domain of the E2F1 protein, which binds CBP or p300 (61), abolishes its inhibitory effect on the uPA promoter, it may be that E2F1 exerts its effect on the uPA promoter by interacting with CBP and disrupting the ability of CBP to activate transcription. This is similar to the known mechanism of p53-mediated repression of AP-1-regulated promoters, which involves recruitment of p300 or CBP by p53 (3). We propose the model in which E2F represses uPA gene expression by inactivating the activity of an enhancer located at −2.0 kb from the transcription initiation site in a manner which interferes with the interaction between AP-1 proteins bound to the enhancer and the basic transcriptional machinery or adapter molecules like CBP. Details of this model at the molecular level remain to be elucidated. Characterization of the PAI-1 promoter is currently under way.
Among the five E2F family members tested, only subset E2F1-3 had an inhibitory effect on transcription. Given the fact that this group induces S-phase entry into quiescent fibroblasts while E2F4 and E2F5 do so only weakly (16), it could be argued that the transcription repression is due to a change in cell cycle phase. However, infection of WI-38 cells with E2F1-expressing Ad did not shift cells to S phase after 16 h, the time when transcript levels were measured. We obtained the same inhibitory effect in randomly growing cells, which suggests that transcription repression is not caused by a change in cell cycle stage but by direct action of E2F. Indeed, it is not surprising that proteins sharing structural and functional features and classified into one group of the E2F family had the same effect on uPA and PAI-1 gene expression. Common to their protein structure is the amino-terminal cyclin A-cdk2 (31)-binding domain, which is absent in E2F4 and E2F5. However, as we showed in the E2F mutant studies, deletion of the cyclin A-binding domain does not abolish E2F1-mediated transcription inhibition and, thus, this interaction is not important for the negative effect.
Because repression of the uPA and PAI-1 promoters was observed under conditions in which E2F was ectopically expressed, it may be asked whether our observation reflects the physiological situation. Our data do reflect cell cycle events. Transcripts of both uPA and PAI-1 genes decrease in the G1/G0-to-S1 transition phase (21, 55, 66), when the levels of pocket protein-free, and therefore transcriptionally active, E2F1 increase (38, 63). Also, our recent observation supports a role for E2F1 as a negative regulator of the PAI-1 gene in vivo. As active pRB binds E2F1 and reduces the levels of free E2F1, we attempted to sequester active E2F1 from the uPA and PAI-1 promoters and derepress these genes in mutant SAOS2 cells expressing temperature-sensitive pRB (59). As predicted, we found that PAI-1 mRNA was induced in mutant cells by shifting the culture to permissive temperature, while it was not affected in wild-type SAOS2 cells (data not shown). uPA mRNA was not detected in these cells.
The transcription regulation described in this paper concerns a very important event during cell cycle progression. Ordered expression of genes of the PA system may help cells during the cell cycle to go through the process of detachment from or attachment to the extracellular matrix and neighboring cells. We are also aware of the presence of other genes involved in extracellular proteolysis, such as collagenase and stromelysin. Their functions may overlap, to some extent, those of the PA system.
Passage through the cell cycle may not be the only situation in which E2F regulates PAI-1 gene expression. PAI-1 is highly expressed in senescent cells and is a good marker for this cell state (46), although its physiological significance is unclear. Upon serum activation of quiescent fibroblasts, the PAI-1 mRNA level declines in late G1 prior to entry into S phase when cells are in early passages, whereas the PAI-1 mRNA level remains high when cells are in late passages (47). This difference is not attributable to a change in mRNA stability with increasing cell passages. In further independent work, it was shown that the E2F1 protein level is significantly reduced in senescent fibroblasts (20). All such results suggest a causal relationship between high E2F expression and low PAI-1 gene expression. A further situation in which E2F may regulate the PA system is wound healing. The PA system plays an important role in this process (11, 54). Vascular injury induces smooth muscle cells to migrate and proliferate to form a neointima layer inside a vein. This process was pathologically accelerated in PAI-1-deficient mice and could be reduced by intravenous injection of an Ad vector expressing PAI-1 (10). Transfection of a double-stranded DNA oligonucleotide with high-affinity binding sites for E2F, which sequesters E2F from active transcription, inhibited hyperplasia formation after vascular injury of animals (45). Thus, high levels of E2F1 and low levels of PAI-1 are associated with increased vascular neointima. These observations combine to suggest that E2F1 has a function(s) other than its well-established role in G0/S control.
ACKNOWLEDGMENTS
We thank Michael Berman and Patrick King for critical reading of the manuscript. We are grateful to David Cobrinik and Philip W. Hinds for generously providing us with the MEF and SAOS2 cell lines, respectively, and to those mentioned in the text for various plasmids and viruses. We also thank Ulrich Müller for introducing us to the Ad expression vector system.
REFERENCES
- 1.Andreasen P A, Kjoller L, Christensen L, Duffy M J. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72:1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 2.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 3.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 6.Besser D, Presta M, Nagamine Y. Elucidation of a signaling pathway induced by FGF-2 leading to uPA gene expression in NIH 3T3 fibroblasts. Cell Growth Differ. 1995;6:1009–1017. [PubMed] [Google Scholar]
- 7.Besser D, Verde P, Nagamine Y, Blasi F. Signal transduction and the u-PA/u-PAR system. Fibrinolysis. 1996;10:215–237. [Google Scholar]
- 8.Boehm J R, Kutz S M, Sage E H, Staiano-Coico L, Higgins P J. Growth state-dependent regulation of plasminogen activator inhibitor type-1 gene expression during epithelial cell stimulation by serum and transforming growth factor-beta1. J Cell Physiol. 1999;181:96–106. doi: 10.1002/(SICI)1097-4652(199910)181:1<96::AID-JCP10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 9.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 10.Carmeliet P, Moons L, Lijnen R, Janssens S, Lupu F, Collen D, Gerard R D. Inhibitory role of plasminogen activator inhibitor-1 in arterial wound healing and neointima formation: a gene targeting and gene transfer study in mice. Circulation. 1997;96:3180–3191. doi: 10.1161/01.cir.96.9.3180. [DOI] [PubMed] [Google Scholar]
- 11.Carmeliet P, Schoonjans L, Kieckens L, Ream B, Degen J, Bronson R, De Vos R, van den Oord J J, Collen D, Mulligan R C. Physiological consequences of loss of plasminogen activator gene function in mice. Nature. 1994;368:419–424. doi: 10.1038/368419a0. [DOI] [PubMed] [Google Scholar]
- 12.Cassady A I, Stacey K J, Nimmo K A, Murphy K M, von der Ahe D, Pearson D, Botteri F M, Nagamine Y, Hume D A. Constitutive expression of the urokinase plasminogen activator gene in murine RAW264 macrophages involves distal and 5′ non-coding sequences that are conserved between mouse and pig. Nucleic Acids Res. 1991;19:6839–6847. doi: 10.1093/nar/19.24.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 14.Cobrinik D, Whyte P, Peeper D S, Jacks T, Weinberg R A. Cell cycle-specific association of E2F with the p130 E1A-binding protein. Genes Dev. 1993;7:2392–2404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- 15.Cousin E, Medcalf R L, Bergonzelli G E, Kruithof E K. Regulatory elements involved in constitutive and phorbol ester- inducible expression of the plasminogen activator inhibitor type 2 gene promoter. Nucleic Acids Res. 1991;19:3881–3886. doi: 10.1093/nar/19.14.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Orazio D, Besser D, Marksitzer R, Kunz C, Hume D A, Kiefer B, Nagamine Y. Cooperation of two PEA3/AP1 sites in uPA gene induction by TPA and FGF-2. Gene. 1997;201:179–187. doi: 10.1016/s0378-1119(97)00445-9. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 19.Gaubatz S, Wood J G, Livingston D M. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc Natl Acad Sci USA. 1998;95:9190–9195. doi: 10.1073/pnas.95.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Good L, Dimri G P, Campisi J, Chen K Y. Regulation of dihydrofolate reductase gene expression and E2F components in human diploid fibroblasts during growth and senescence. J Cell Physiol. 1996;168:580–588. doi: 10.1002/(SICI)1097-4652(199609)168:3<580::AID-JCP10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Grimaldi G, Di Fiore P, Locatelli E K, Falco J, Blasi F. Modulation of urokinase plasminogen activator gene expression during the transition from quiescent to proliferative state in normal mouse cells. EMBO J. 1986;5:855–861. doi: 10.1002/j.1460-2075.1986.tb04295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 24.Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrin D L, Schmidt G W. Rapid, reversible staining of Northern blots prior to hybridization. BioTechniques. 1988;6:196–200. [PubMed] [Google Scholar]
- 26.Huet X, Rech J, Plet A, Vie A, Blanchard J M. Cyclin A expression is under negative transcriptional control during the cell cycle. Mol Cell Biol. 1996;16:3789–3798. doi: 10.1128/mcb.16.7.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irigoyen J P, Munoz-Canoves P, Montero L, Koziczak M, Nagamine Y. The plasminogen activator system: biology and regulation. Cell Mol Life Sci. 1999;56:104–132. doi: 10.1007/PL00000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 29.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 30.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Spl with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W G, Jr, Livingston D M. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 32.Kunz C, Pebler S, Otte J, von der Ahe D. Differential regulation of plasminogen activator and inhibitor gene transcription by the tumor suppressor p53. Nucleic Acids Res. 1995;23:3710–3717. doi: 10.1093/nar/23.18.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutz S M, Nickey S A, White L A, Higgins P J. Induced PAI-1 mRNA expression and targeted protein accumulation are early G1 events in serum-stimulated rat kidney cells. J Cell Physiol. 1997;170:8–18. doi: 10.1002/(SICI)1097-4652(199701)170:1<8::AID-JCP2>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 34.Lam E W, Watson R J. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavia P, Jansen-Dürr P. E2F target genes and cell-cycle checkpoint control. Bioessays. 1999;21:221–230. doi: 10.1002/(SICI)1521-1878(199903)21:3<221::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 36.Lee J S, von der Ahe D, Kiefer B, Nagamine Y. Cytoskeletal reorganization and TPA differently modify AP-1 to induce the urokinase-type plasminogen activator gene in LLC-PK1 cells. Nucleic Acids Res. 1993;21:3365–3372. doi: 10.1093/nar/21.15.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lees J A, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindeman G J, Gaubatz S, Livingston D M, Ginsberg D. The subcellular localization of E2F-4 is cell-cycle dependent. Proc Natl Acad Sci USA. 1997;94:5095–5100. doi: 10.1073/pnas.94.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin K, Trouche D, Hagemeier C, Sorensen T S, La Thangue N B, Kouzarides T. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 41.McMahon S B, Van Buskirk H A, Dugan K A, Copeland T D, Cole M D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 42.Medcalf R L, Ruegg M, Schleuning W D. A DNA motif related to the cAMP-responsive element and an exon-located activator protein-2 binding site in the human tissue-type plasminogen activator gene promoter cooperate in basal expression and convey activation by phorbol ester and cAMP. J Biol Chem. 1990;265:14618–14626. [PubMed] [Google Scholar]
- 43.Messerschmitt A, Disela C, Dilworth S, Marti A G, Ballmer-Hofer K. Polyomavirus middle-T antigen lacking a membrane anchor sequence accumulates in the nucleus. J Gen Virol. 1996;77:17–26. doi: 10.1099/0022-1317-77-1-17. [DOI] [PubMed] [Google Scholar]
- 44.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morishita R, Gibbons G H, Horiuchi M, Ellison K E, Nakama M, Zhang L, Kaneda Y, Ogihara T, Dzau V J. A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc Natl Acad Sci USA. 1995;92:5855–5859. doi: 10.1073/pnas.92.13.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mu X C, Higgins P J. Differential growth state-dependent regulation of plasminogen activator inhibitor type-1 expression in senescent IMR-90 human diploid fibroblasts. J Cell Physiol. 1995;165:647–657. doi: 10.1002/jcp.1041650324. [DOI] [PubMed] [Google Scholar]
- 47.Mu X C, Staiano-Coico L, Higgins P J. Increased transcription and modified growth state-dependent expression of the plasminogen activator inhibitor type-1 gene characterize the senescent phenotype in human diploid fibroblasts. J Cell Physiol. 1998;174:90–98. doi: 10.1002/(SICI)1097-4652(199801)174:1<90::AID-JCP10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 48.Nagamine Y, Lee J S, Menoud P-A, Nanbu R. Structure and function of the urokinase-type plasminogen activator gene. In: Glas-Grenwalt P, editor. Fibrinolysis in disease. Boca Raton, Fla: CRC Press; 1995. pp. 10–20. [Google Scholar]
- 49.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 50.Pearson A, Greenblatt J. Modular organization of the E2F1 activation domain and its interaction with general transcription factors TBP and TFIIH. Oncogene. 1997;15:2643–2658. doi: 10.1038/sj.onc.1201451. [DOI] [PubMed] [Google Scholar]
- 51.Qin X Q, Livingston D M, Ewen M, Sellers W R, Arany Z, Kaelin W G., Jr The transcription factor E2F-1 is a downstream target of RB action. Mol Cell Biol. 1995;15:742–755. doi: 10.1128/mcb.15.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riccio A, Lund L R, Sartorio R, Lania A, Andreasen P A, Dano K, Blasi F. The regulatory region of the human plasminogen activator inhibitor type-1 (PAI-1) gene. Nucleic Acids Res. 1988;16:2805–2824. doi: 10.1093/nar/16.7.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robertson K D, Jones P A. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romer J, Bugge T H, Pyke C, Lund L R, Flick M J, Degen J L, Dano K. Impaired wound healing in mice with a disrupted plasminogen gene. Nat Med. 1996;2:287–292. doi: 10.1038/nm0396-287. [DOI] [PubMed] [Google Scholar]
- 55.Ryan M P, Kutz S M, Higgins P J. Complex regulation of plasminogen activator inhibitor type-1 (PAI-1) gene expression by serum and substrate adhesion. Biochem J. 1996;314:1041–1046. doi: 10.1042/bj3141041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg R A. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwarz J K, Bassing C H, Kovesdi I, Datto M B, Blazing M, George S, Wang X F, Nevins J R. Expression of the E2F1 transcription factor overcomes type beta transforming growth factor-mediated growth suppression. Proc Natl Acad Sci USA. 1995;92:483–487. doi: 10.1073/pnas.92.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin E K, Tevosian S G, Yee A S. The N-terminal region of E2F-1 is required for transcriptional activation of a new class of target promoter. J Biol Chem. 1996;271:12261–12268. doi: 10.1074/jbc.271.21.12261. [DOI] [PubMed] [Google Scholar]
- 59.Tiemann F, Hinds P W. Induction of DNA synthesis and apoptosis by regulated inactivation of a temperature-sensitive retinoblastoma protein. EMBO J. 1998;17:1040–1052. doi: 10.1093/emboj/17.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trimarchi J M, Fairchild B, Verona R, Moberg K, Andon N, Lees J A. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci USA. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trouche D, Kouzarides T. E2F1 and E1A(12S) have a homologous activation domain regulated by RB and CBP. Proc Natl Acad Sci USA. 1996;93:1439–1442. doi: 10.1073/pnas.93.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Zonneveld A J, Curriden S A, Loskutoff D J. Type 1 plasminogen activator inhibitor gene: functional analysis and glucocorticoid regulation of its promoter. Proc Natl Acad Sci USA. 1988;85:5525–5529. doi: 10.1073/pnas.85.15.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z M, Yang H, Livingston D M. Endogenous E2F-1 promotes timely G0 exit of resting mouse embryo fibroblasts. Proc Natl Acad Sci USA. 1998;95:15583–15586. doi: 10.1073/pnas.95.26.15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 65.Wu C L, Zukerberg L R, Ngwu C, Harlow E, Lees J A. In vivo association of E2F and DP family proteins. Mol Cell Biol. 1995;15:2536–2546. doi: 10.1128/mcb.15.5.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zavizion B, White J H, Bramley A J. Cell cycle-dependent fluctuation of urokinase-type plasminogen activator, its receptor, and inhibitors in cultured bovine mammary epithelial and myoepithelial cells. Biochim Biophys Acta. 1998;1403:141–150. doi: 10.1016/s0167-4889(98)00035-4. [DOI] [PubMed] [Google Scholar]
- 67.Zhang H S, Postigo A A, Dean D C. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta, and contact inhibition. Cell. 1999;97:53–61. doi: 10.1016/s0092-8674(00)80714-x. [DOI] [PubMed] [Google Scholar]
- 68.Ziegler A, Knesel J, Fabbro D, Nagamine Y. Protein kinase C down-regulation enhances cAMP-mediated induction of urokinase-type plasminogen activator mRNA in LLC-PK1 cells. J Biol Chem. 1991;266:21067–21074. [PubMed] [Google Scholar]