Abstract
Invasive graft biopsies assess the efficacy of immunosuppression through lagging indicators of transplant rejection. We report on a microporous scaffold implant as a minimally invasive immunological niche to assay rejection before graft injury. Adoptive transfer of T cells into Rag2−/− mice with mismatched allografts induced acute cellular allograft rejection (ACAR), with subsequent validation in wild-type animals. Following murine heart or skin transplantation, scaffold implants accumulate predominantly innate immune cells. The scaffold enables frequent biopsy, and gene expression analyses identified biomarkers of ACAR before clinical signs of graft injury. This gene signature distinguishes ACAR and immunodeficient respiratory infection before injury onset, indicating the specificity of the biomarkers to differentiate ACAR from other inflammatory insult. Overall, this implantable scaffold enables remote evaluation of the early risk of rejection, which could potentially be used to reduce the frequency of routine graft biopsy, reduce toxicities by personalizing immunosuppression, and prolong transplant life.
Engineered tissue implants surveil for transplant rejection before graft injury onset and distinguish rejection from infection.
INTRODUCTION
Solid organ transplant recipients receive life-long immune suppression that protects grafts from rejection but substantially increases the risk of opportunistic infection (1), malignancy (2), and renal impairment (3). This risk of rejection and immunosuppression drug toxicity is particularly impactful for pediatric or young adult transplant recipients who must undergo decades of immunosuppression burden (4–6). Physicians typically apply aggressive prophylactic immunosuppression in a blanket approach (7) as there are no methods to determine which allografts will be rejected, and symptoms may only present late in the progression of graft rejection or injury (8). An assay to continuously monitor transplant health could enable precision immunosuppression while maintaining graft acceptance. Strategies for monitoring graft rejection, including acute cellular allograft rejection (ACAR), are limited (9). Endomyocardial biopsy (EMB) remains the gold standard for heart transplant (HTx) surveillance and ACAR diagnosis (7, 10, 11). However, biopsy is a severe stressor on HTx recipients (7, 12) and has consequential sampling error and interobserver variability (13, 14). In addition, this histological evidence of rejection inherently lags behind molecular signs of rejection (15) and does not assess parenchymal injury as well as molecular assessment (16). Last, no consensus has emerged as to the actionable importance of mild rejection identified through EMB, although it may cause persistent injury after the rejection is treated or subsides (3, 13, 17).
The limitations of graft biopsy have motivated the development of alternative diagnostic assays to surveil graft health. Imaging techniques, including advances in echocardiography, can monitor cardiac graft function (7), but graft dysfunction is a late indicator of ACAR (18). The alternative to analyzing the graft itself is to analyze the activity of systemic immune responses for indications of graft injury. Cell-free donor-derived DNA assays, such as the Allosure test, assess graft cell death (8, 19, 20) but lack positive predictive power for allograft rejection while also only being approved for use several months posttransplantation (21). Gene expression profiling of blood (e.g., the Allomap test) is being applied as an assay for T cell proliferation, yet this strategy also lacks positive predictive power for allograft rejection (22–24). In general, these HTx surveillance modalities are limited to late indicators of tissue injury and dysfunction which cannot predict the onset of rejection. More recently, an emerging technology to assess immune function involves a microporous scaffold that is implanted as an immunological niche (25), which captures tissue phenotypic immune responses, unlike liquid biopsy, to facilitate early disease detection (26). When implanted subcutaneously, these scaffolds become vascularized and offer a distinct immunological niche for circulating immune cells that exhibit tissue specific phenotypes upon extravasation (25). The scaffold implants can be biopsied more easily than the transplanted organ and have been reported to recapitulate immune aspects of disease progression in murine metastatic cancer and multiple sclerosis (27–29).
In this work, we investigated the potential of analyzing gene expression from the implanted scaffolds to predict allograft rejection. Scaffolds were implanted subcutaneously in murine skin transplant (STx) and heterotopic HTx recipients (30, 31). We performed initial studies in mice deficient in mature T cells and induced ACAR by adoptive T cell transfer, which enabled a focus on alloimmune injury responses and reduced immune modifications from ischemia-reperfusion injury (IRI) and wound healing dynamics. We biopsied scaffolds at various stages of ACAR to determine captured immune cell populations. RNA sequencing (RNA-seq) of the scaffold biopsies revealed pathway and gene differentiators between syngeneic graft recipients, allogeneic recipients with ACAR before graft injury onset, and recipients with ACAR during graft injury. In addition, we assessed the capacity of these scaffold-derived gene biomarkers to predict the onset of graft injury in ACAR and distinguish ACAR from an alternative inflammatory insult in immunodepleted mice, such as respiratory infection. Last, we validated the predictive capacity of these scaffold-derived biomarkers in wild-type mice with ACAR following immune suppression. A minimally invasive implant that forecasts ACAR by capturing immune responses that precede graft injury could ultimately enable personalized immunosuppression to minimize rejection episodes and susceptibility to iatrogenic toxicities.
RESULTS
Scaffold implants capture innate and adaptive immune cells without accelerating rejection
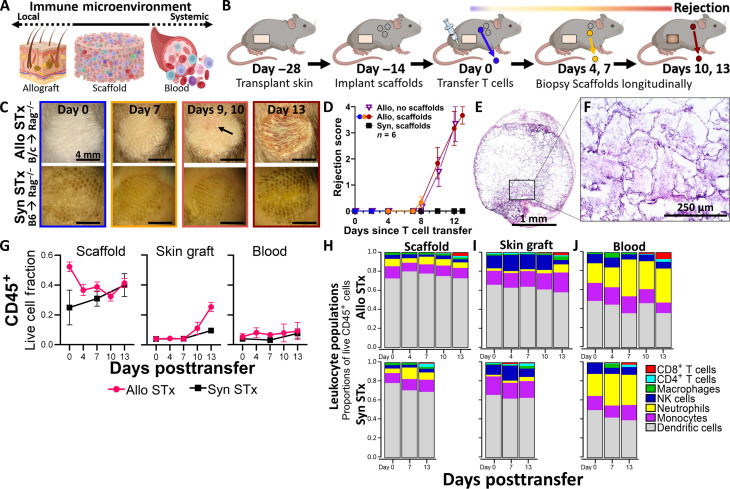
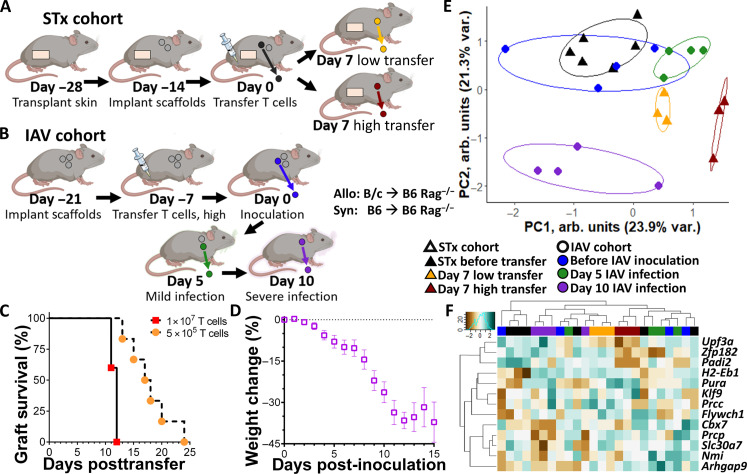
We performed initial studies with the scaffold implant as an allograft immunological niche in full major histocompatibility mismatch STx (Fig. 1, A and B). Rag2−/− immunodeficient mice, on a C57BL/6 background, received tail skin grafts onto the dorsum as allogenic grafts (BALB/c onto Rag2−/−) or syngeneic grafts (C57BL/6 onto Rag2−/−). Grafts healed over 28 days, with three to four polycaprolactone (PCL) scaffolds implanted subcutaneously after 14 days. At 14 days postscaffold implantation, we adoptively transferred 1 × 107 T cells from naïve C57BL/6 (B6) donors to initiate ACAR (Fig. 1, C and D, and fig. S1). For allograft recipients, ACAR developed over approximately 13 days, with graft injury (i.e., wound formation) visible beginning at day 9 or 10 following T cell transfer and complete graft wounding observed on day 13 post–T cell transfer (Fig. 1, C and D). We considered day 7 a pre-injury time point for ACAR. Implanted subcutaneous scaffolds and their longitudinal explants after adoptive T cell transfer did not alter the tempo of ACAR in allograft recipients (Fig. 1D). Syngeneic grafts integrated well with the host and did not show signs of rejection following T cell transfer. Histological analysis of retrieved implants indicated cell infiltration and vascularization throughout the scaffold (Fig. 1, E and F).
Fig. 1. Scaffold implants capture predominantly innate immune cell populations during acute skin allograft rejection.
(A) Schematic representation of the immunological niche scaffolds as a bridge between the local immune responses in the allograft and the systemic immune responses present in the blood and hematopoietic tissues. (B) Schematic representation of scaffold implants applied to monitor acute cellular rejection of murine STx after adoptive transfer of syngeneic T cells. (C) Representative images of skin grafts on dorsum of C57BL/6 recipients, with N = 6 independent graft recipients as biological replicates per group. Top row: BALB/c tail skin grafts on C57BL/6 Rag2−/− recipients (Allo STx). Arrow indicates early signs of rejection at graft on day 9 following adoptive T cell transfer. Bottom row: C57BL/6 tail skin grafts on C57BL/6 Rag2−/− recipients (Syn STx). Each graft is approximately 8 mm in diameter. Scale bars, 4 mm. (D) Skin grafts scored following adoptive T cell transfer by severity of rejection. Mean ± SEM; N = 6 independent graft recipients per group. Plot of allograft recipients with scaffold implants colored according to stage of allograft rejection. (E) Cross-sectional images of scaffold explant from an allogeneic STx recipient before T cell transfer (day 0), stained by hematoxylin and eosin (H&E). Images representative of scaffolds from N = 3 independent mice. Scale bar, 1 mm. (F) Insert at ×20 magnification. Scale bar, 250 μm. (G) CD45+ cells as a fraction of all live cells found in the scaffold implant (left), the skin graft (center), and the blood (right) before and at various time points after adoptive T cell transfer; mean ± SEM with N = 4 independent graft recipients per group. (H to J) Proportions of immune cell types in allogeneic and syngeneic STx recipients in the (H) scaffold implants, (I) the skin graft, and (J) the blood. Statistics performed by two-tailed Student’s t test. NK cells, natural killer cells.
We subsequently recovered cells from the scaffold, skin grafts, and blood, which were isolated before T cell transfer and at days 4, 7, 10, and 13 after transfer, for assessment of leukocyte populations (Fig. 1, G to J, and fig. S2). Before visual evidence of graft injury (before day 10), scaffolds captured an approximately threefold greater proportion of leukocytes compared to the blood and in the graft prior (Fig. 1G). Skin allografts at days 10 and 13 had an influx of CD45+ cells (Fig. 1G) that corresponded with the onset of visible ACAR graft injury (Fig. 1, C and D). This influx was absent in syngeneic graft recipients. These results identify day 7 and earlier as pre-alloimmune injury time points of ACAR at which it is unlikely that rejection risk would be identified through standard approaches such as biopsy assessment for lymphocyte infiltration (18, 32) or noninvasive assays of T cell proliferation or donor cell death (20).
Flow cytometric analysis revealed that scaffolds contained predominantly innate immune cells (Fig. 1H), with a greater proportion of dendritic cells (DCs) than was found at the skin graft or in the blood (Fig. 1, I and J). Neutrophil proportion, by contrast, was diminished in the scaffold (Fig. 1H) in comparison to the blood (Fig. 1J). Leukocyte proportions changed little, however, within each tissue during ACAR progression or compared to nonrejecting graft recipients (Fig. 1, H to J), until the graft was infiltrated by CD4+ and CD8+ T cells on day 13 (Fig. 1I), corresponding to late ACAR and extensive graft injury (Fig. 1, C and D). Beyond the graft during late-stage rejection, this T cell expansion was also observed in the scaffold and, in even greater proportion, in the circulation (Fig. 1, H and J). The similarity in leukocyte proportions and ratios (fig. S3) before ACAR graft injury indicates that population dynamics cannot predict rejection, which supported subsequent studies to assess the specific phenotype and gene expression profile of these immune cells. This result also highlights the limitations of analyzing T cell responses to predict rejection, as the proliferation and invasion of these cells are not observed until late in the process of rejection when extensive injury has occurred.
Scaffolds identify a biomarker panel of early ACAR in STx
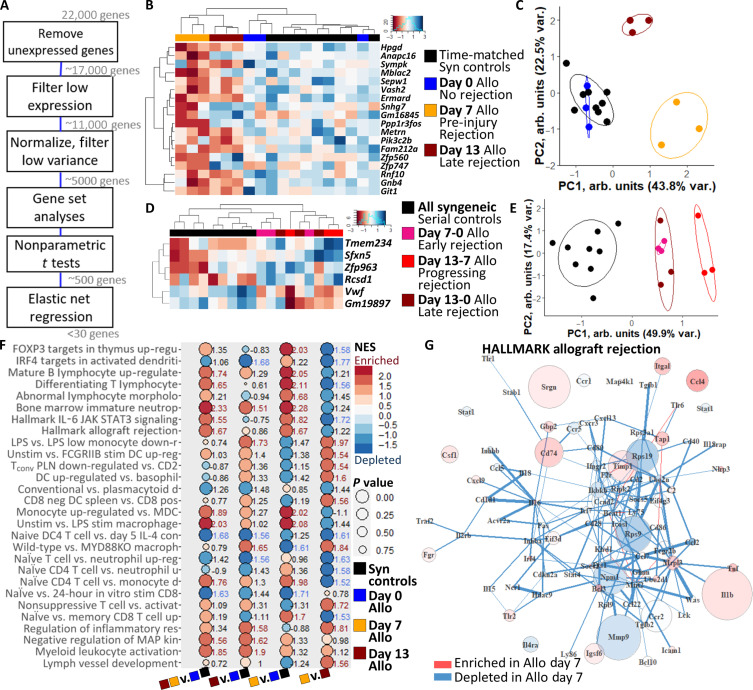
We then analyzed gene expression of the scaffold-captured cells to identify biomarkers of ACAR. RNA was extracted from longitudinal scaffold explants and sequenced (RNA-seq), with expression filtered to remove unexpressed, lowly expressed, and uniformly expressed genes (Fig. 2A and fig. S4). From the transcriptomic profiles of the scaffolds, we identified differential gene expression (Fig. 2, B to E) and pathway enrichment (Fig. 2, F and G) between mid-injury rejecting, pre-injury rejecting, and nonrejecting graft recipients. We derived 136 highly differentially expressed genes at the scaffold during ACAR (fig. S5A and table S1). To create a sparse diagnostic biomarker panel of ACAR in STx with the fewest necessary indicators, we developed a pathway-agnostic elastic net regression which identified 18 differentially expressed biomarkers (Fig. 2B and table S1). Eleven of these 18 genes were down-regulated at the scaffold during pre-injury ACAR and late ACAR compared to nonrejecting grafts, while 7 genes were down-regulated in pre-injury ACAR but up-regulated in late ACAR. Unsupervised clustering of these 18 genes distinguished mice with syngeneic or allogeneic grafts before adoptive T cell transfer (i.e., both nonrejecting time points), pre-injury ACAR (allogeneic day 7), and late ACAR (allogeneic day 13) (Fig. 2C). Principal component loadings of this 18-gene panel in unsupervised clustering identified Gnb4, Sepw1, Gm16845, Rnf10, and Git1 as the top differentiators of healthy and rejecting skin allografts (fig. S5, B and C).
Fig. 2. Gene expression at the scaffold immunological niche distinguishes stages of acute cellular skin allograft rejection.
(A) Representation of gene expression RNA-seq analysis for identifying sparse gene panels and enriched pathways of ACAR at the scaffold immunological niche. (B) Clustered heatmap of 18-gene panel distinguishing skin graft rejection stages, with expression normalized by row. Columns indicate individual samples; N = 3 biological replicates for each time point. (C) Principal components clustering of scaffolds based on 18-gene panel. Points indicate individual scaffold samples. All ellipses = 70% confidence interval (CI). (D) Clustered heatmap of six-gene panel distinguishing skin graft rejection stage transitions. Columns indicate individual samples. (E) Principal components clustering of scaffolds based on six-gene panel. Points indicate individual scaffold samples. All ellipses = 70% CI. (F) Most highly enriched immune pathways in pairwise comparisons of pre-injury ACAR (Allo day 7), late ACAR (Allo day 13), and nonrejecting graft recipients (Allo day 0 and time-matched Syn) at the scaffold. NES displayed, where red text indicates NES > 1.5 and blue text indicates NES < −1.5. Pathways selected by greatest sum of NES values and lowest sum of P value. Shortened pathway names, see table S1 for full names. IRF4, interferon regulatory factor 4; JAK, Janus kinase; STAT, signal transducers and activators of transcription; LPS, lipopolysaccharide; MDC, macrophage-derived chemokine. (G) Differential network of the Hallmark allograft rejection pathway between scaffolds during pre-injury ACAR and nonrejecting graft recipients at the scaffold. Gene expression displayed for pathway constituents where enrichment or depletion is defined as a differential connectivity P value < 0.01. PC, Principal component; FOXP3, forkhead box P3 protein; Tconv, Resting conventional T cells; PLN, pancreatic draining lymph node; MYD88KO, myeloid differentiation primary response 88 know out.
The scaffold provides the opportunity for frequent, longitudinal surveillance of intragraft immune events and a distinct measure of subclinical ACAR to minimize graft damage. As the scaffold can be easily biopsied, longitudinal gene expression can be individualized by monitoring the change across ACAR development for each recipient. With this individualized gene expression change, ACAR—both pre-injury and late injury—can be distinguished from time-matched syngeneic graft recipients with an elastic net–derived six-gene panel (Fig. 2, D and E, and table S1). Tmem234, Sfxn5, Zfp963, and Rcsd1 increased in expression over time as ACAR progressed compared to healthy recipients; whereas Vwf and Gm19897 decreased in expression during rejection development. Notably, this six-gene panel differentiates the progression from pre-injury to late injury ACAR as strongly as it differentiates these two stages from the time-matched syngeneic recipients. Collectively, these data indicate that gene expression analyses at the scaffold can both identify ACAR and distinguish between stages of ACAR and that longitudinal analyses enabled by the distally implanted scaffold can increase computational rigor and individualize graft surveillance.
The biology underlying this shift from the pre-injury to injury stage of acute rejection was next analyzed using gene set enrichment analysis (GSEA). We identified 28 differentially enriched pathways by normalized enrichment score (NES) (NES > |1.5| and NES sum > |3.5|) at the scaffold with a low false discovery rate (FDR < 0.1) between pairwise comparisons of the three groups: nonrejecting grafts, pre-injury ACAR, and mid-injury ACAR (Fig. 2F and table S1). Overall, comparing both stages of ACAR to nonrejecting graft recipients highlights myeloid cell activation (Fig. 2F, first column; cumulative |NES| = 37.9), including macrophages and monocytes and neutrophil maturity as highly enriched in ACAR. Between the two stages of ACAR, enrichment of these pathways at the scaffold in late injury ACAR (Allo day 13; Fig. 2F, second column) is more similar to nonrejecting graft recipients (NES = 36.8) than to pre-injury ACAR (Allo day 7; Fig. 2F, third column; NES = 43.1). Numerous differentially regulated T cell pathways associated with activation and differentiation were readily identified in pre-injury ACAR (Fig. 2F, third column), despite few T cells present at the scaffold at day 7 (Fig. 1H). Along with the enrichment of these T cell processes, the scaffold had enriched myeloid cell–related activation and activity during pre-injury ACAR compared to either nonrejecting recipients or late injury ACAR (Fig. 2F, third and fourth columns). These myeloid pathways included monocytes, stimulated DCs and plasmacytoid DCs, and stimulated macrophages. While both stages of ACAR displayed enriched T cell pathways at the scaffold, these T cell pathways were more strongly enriched during late ACAR (Fig. 2B, fourth column) reflective of systemic and graft-localized T cell dynamics (Fig. 1, H to J).
Myeloid cell proportions were largely unchanged as ACAR progressed (Fig. 1, H to J, and fig. S3), yet processes enriched and depleted at the scaffold demonstrate the differential expression and activity of these myeloid cells and their possible participation in T cell signaling preceding graft injury (33). Mapping these pathways highlights genes and gene connections likely playing key roles during the early stages or rejection (Fig. 2G and fig. S6). Notably, the Hallmark database pathway of allograft rejection (34) was enriched at the scaffold during pre-injury ACAR (Fig. 2, F and G), with up-regulation of Ccl4, Cd74, and Tnf, down-regulation of Mmp9 and Rps9, and depleted relationships between Cd1d1-Il16, Cd28-Stat4, and Cd80-Cxcr3 (Fig. 2G), among others. Ccl4 and Tnf are mediators shared in both CD8 T cell stimulation in ACAR and CD16a natural killer cell stimulation in antibody-mediated rejection (35). Cd74 regulates DC migration and T lymphocyte homing, proliferation, and cytokine secretion (36).
To assess pathway specificity for ACAR, we identified the pathways that were most strongly correlated with ACAR using gene set variation analysis (GSVA) applied to the RNA-seq data and then an elastic net regression to specify the fewest pathways needed to distinguish ACAR from nonrejecting grafts at the scaffold (fig. S7 and table S1). These differentiating pathways include processes associated with epigenetic regulation of cell death, apoptosis, and cytomegalovirus infection. Viral infections limit the power of noninvasive assays at predicting ACAR, as many viral infections similarly activate T cell responses (37) and even induce prorejection inflammation (38, 39). As these immunological pathways corresponding to ACAR may overlap with those of other inflammatory insults, we probed further to deduce correlative biomarkers with higher specificity for ACAR over other inflammatory insults at the scaffold environment.
Scaffolds predict early risk of ACAR in HTx recipients
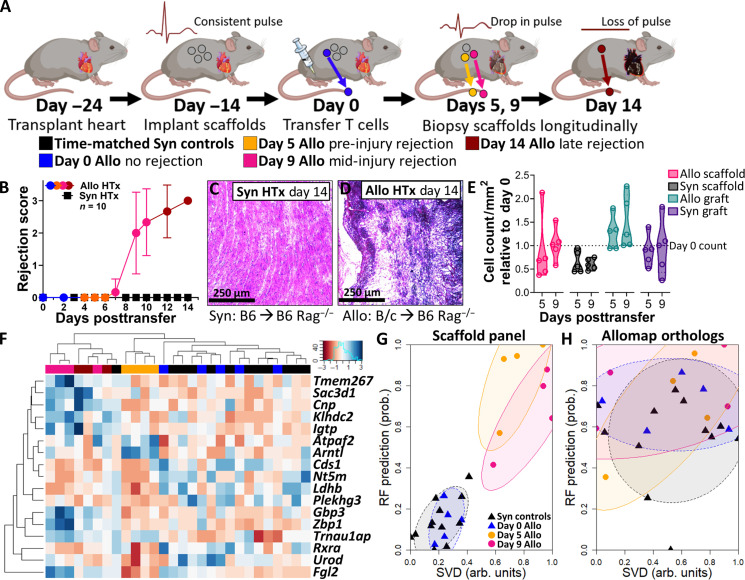
We next investigated biomarkers of ACAR in murine heterotopic HTx using the immunological niche of the scaffold (Fig. 3A). We performed syngeneic (B6 to B6) and allogeneic (B/c to B6) HTx, similarly followed by scaffold implantation and adoptive transfer of 2 × 107 syngeneic T cells from naïve B6 donors. Syngeneic HTx recipients remained free of ACAR (Fig. 3, B and C). For allograft recipients, however, ACAR occurred over 14 days (Fig. 3, B and D, and fig. S8, A and B), with indications of graft dysfunction (i.e., edema and weakened pulse) beginning at day 8 or 9 and complete graft failure on day 13 or 14. Scaffolds were serially explanted from HTx recipients corresponding to ACAR state, with an earlier pre-injury time point included at day 5, a mid-rejection point on day 9, and a late rejection day 14 explant. Cell count per area as an indicator of immune cell infiltration indicates no difference in allogeneic or syngeneic grafts at day 5 or 9 following T cell transfer (Fig. 3E), as similarly seen in the scaffold (Fig. 3E and fig. S8, C and D).
Fig. 3. Gene expression at the scaffold immunological niche distinguishes stages of acute cellular heart allograft rejection.
(A) Schematic representation of scaffold implants applied to monitor acute cellular rejection of murine heart allografts after adoptive transfer of T cells. (B) Heart grafts scored following adoptive T cell transfer by severity of rejection, N = 10 independent graft recipients per group as biological replicates. Plot of allograft recipients with scaffold implants colored according to stage of allograft rejection. Mean ± SEM. Plot of allograft recipient scores colored according to stage of allograft rejection. (C and D) Cross-sectional images of (C) syngeneic and (D) allogeneic heart grafts 14 days after adoptive T cell transfer, stained by H&E. Images representative of N = 3 heart grafts from independent graft recipients. Scale bars, 250 μm. (E) Histological analysis of the scaffold implant and HTx graft across the rejection cascade before graft failure (0 to 9 days post–T cell transfer), with cell count analyses performed from three averaged representative slides of scaffolds or heart grafts from N = 5 independent mice and normalized to day 0 values. Statistical analysis performed by unpaired, nonparametric t test (Mann-Whitney) between allogeneic and syngeneic at each time point and tissue type. (F) Clustered heatmap of elastic net–derived 17-gene panel at the scaffold for distinguishing stages of cardiac ACAR. (G) Unsupervised singular value decomposition (SVD) and supervised random forest (RF) scoring of samples based on 17-gene panel derived at the scaffold. (H) Unsupervised SVD and supervised RF scoring of the murine orthologs of the blood-based Allomap test, applied to the scaffold. Points indicate individual scaffold samples. All ellipses = 70% CI.
We identified 107 differentially expressed genes in HTx ACAR at the scaffold (fig. S9A and table S2), and 17 differentially expressed genes were further identified as a sparse biomarker panel of HTx ACAR via elastic net regression (Fig. 3E; fig. S9, B to E; and table S2), with no shared genes with the sparse STx panel. Cds1, Ntrm, and Ldhb were moderately down-regulated at the scaffold during all stages of ACAR. Atpaf2 and Arntl, by contrast, were strongly up-regulated in pre-injury ACAR compared to both nonrejecting recipients and mid-injury ACAR, while Rxra, Urod, and Plekhg3 were down-regulated in pre-injury ACAR only. Last, the remaining eight genes were strongly up-regulated in mid-injury ACAR only. While this sparse gene panel distinguished pre-injury and mid-injury ACAR from nonrejecting recipients, they were also readily distinguished from one another (fig. S9D). Late-stage ACAR, however, was not well-distinguished (fig. S9D), highlighting both the distinct stage of ACAR pregraft injury and the resolving nature of late ACAR response in a heterotopic HTx model where the graft is not functionally necessary for survival.
We then developed a robust scoring system to distinguish between nonrejecting grafts or ACAR. Two approaches were used to identify rejection, singular value decomposition (SVD) for unsupervised scoring and a random forest bagged tree algorithm for supervised scoring of samples with leave-one-out validation (Fig. 3F). The scaffold-derived panel distinguishes pre-injury and mid-injury ACAR from nonrejecting graft recipients (Fig. 3F). Mouse orthologs of the blood-derived Allomap test (40), conversely, were unable to distinguish scaffolds from mice with nonrejecting HTx and ACAR (Fig. 3G). Fgl2, Arntl, and Cnp exhibit the greatest variable importance in this two-metric system for identifying ACAR (fig. S9E). Most notably, Fgl2 is down-regulated in pre-injury ACAR but up-regulated in mid-injury ACAR relative to both late ACAR and nonrejecting graft recipients. Fgl2 plays a key role in DC maturation (41) and regulatory T cell–based regulation of T cell proliferation (42) and can be inhibited to ameliorate graft rejection (43). This early identification at the scaffold could create a therapeutic window for immune suppression interventions to abrogate ACAR before overt graft injury.
Scaffolds identify conserved ACAR pathways and biomarkers among graft types
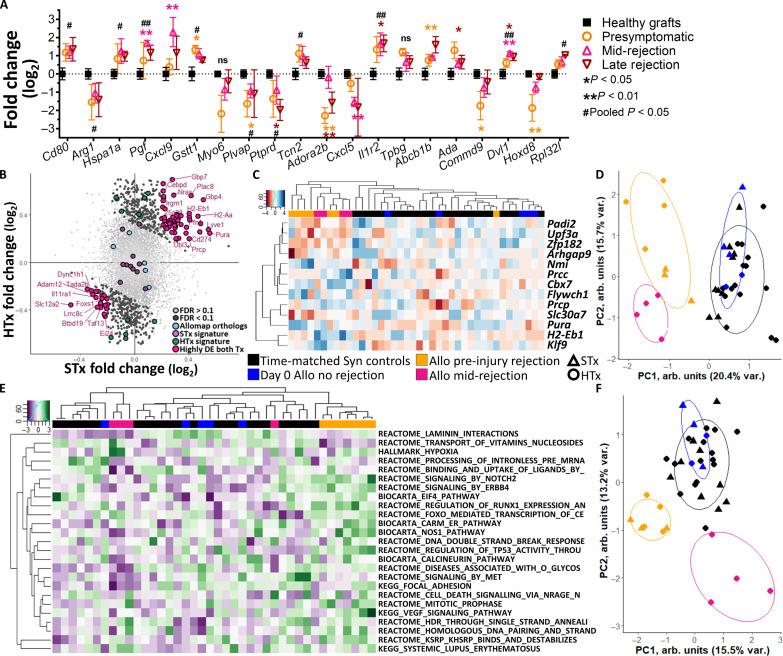
Graft biopsy is the cornerstone for investigating rejection and injury and their relationships to patient outcomes. We probed the scaffold for gene expression markers identified as canonical biopsy markers of clinical graft rejection to determine whether local aspects of graft rejection are recapitulated at the scaffold. Because some injury responses are shared across organs (44) and ACAR superimposed on tissue injury can be defined by conserved alloimmune states (45, 46), we assessed known canonical markers of rejection identified in human kidney transplantation due to greater patient numbers and rejection characterization (45, 47, 48). Although kidney transplant recipients received a combination of a calcineurin or mammalian target of rapamycin (mTOR) inhibitor, prednisone, and antimetabolite (45) and may have a degree of convoluting nonrejection graft dysfunction (49), many of these human kidney graft biopsy markers associated with clinical T cell–mediated rejection were differentially expressed at the scaffold during murine HTx rejection (Fig. 4A). Cd80, Arg1, Hspa1a, Gstt1, Plvap, Tcn2, and Il1r2 were expressed at consistent levels during all the stages of ACAR. The scaffold provides, however, a uniquely nuanced assessment of these canonical markers, as some, such as Adora2b, Ada, Commd9, and Hoxd8 are most divergent in expression before ACAR injury onset. Others still, such as Pgf, Cxcl9, Cxcl5, or Dvl1, differ from expression in nonrejecting recipients most during mid-injury rejection. Expression of these canonical markers was not significantly altered during late ACAR, however, when graft dysfunction is most readily observable. Sampling of the tissue-like immune responses at the scaffold identified biomarkers of acute T cell–mediated transplant rejection that may be conserved across allograft types at a remote site to the graft.
Fig. 4. Scaffolds identify conserved biomarkers and pathways of ACAR across skin and heart allografts.
(A) Gene expression of mouse orthologs of human kidney biopsy canonical markers of graft rejection expressed at the scaffold during stages of HTx ACAR, as represented by log2 fold change from the scaffold in healthy graft recipients. *P < 0.05, **P < 0.01, #Pooled ACAR samples P < 0.05, ##Pooled ACAR samples P < 0.01. Mean ± SEM with scaffold explants from N = 4 independent graft recipients per group and time point. ns, not significant. (B) Log2 fold change of gene expression at the scaffold comparing ACAR to healthy recipients in STx and HTx where genes with an FDR above 0.1 were excluded, where 43 differentially expressed (DE) genes with the greatest summed fold change are labeled. (C) Clustered heatmap of elastic net–derived 13-gene panel for distinguishing stages of ACAR conserved across both allograft types; with expression normalized by row. Columns indicate individual samples; N = 3 (STx) or 4 (HTx) biological replicates for each time point. (D) Principal components clustering of scaffolds based on 13-gene panel; points indicate individual scaffold samples, and all ellipses = 70% CI. (E) Twenty-five pathways identified by elastic net regression of GSVA-converted gene expression which sparsely distinguish between stages of ACAR; with expression normalized by row. Columns indicate individual samples; N = 3 (STx) or 4 (HTx) biological replicates for each time point. Shortened pathway names, for full names see table S3. (F) Principal components clustering of scaffolds based on 25-pathway set; points indicate individual scaffold samples, and all ellipses = 70% CI.
We then aggregated the data from skin and HTx recipients (fig. S10) with the goal of identifying a tissue-independent panel for ACAR injury onset, which may provide greater translation in solid organ transplantation and improved specificity to distinguish rejection from other inflammatory insults such as viral infection. Genes with a low FDR (<0.1) were identified by fold change in expression between nonrejecting recipients and ACAR (Fig. 4B). A total of 43 genes with high differential expression in each transplant type were identified between ACAR recipients and nonrejecting recipients (table S3). This group of potential biomarkers does not include the genes identified in the tissue-specific derived STx or HTx signatures. Elastic net regression reduced this group of strong differentiators to 13 genes (Fig. 4C and table S3) as a minimal panel for distinguishing ACAR in both STx and HTx recipients (Fig. 4D and fig. S11, A to C), in which late-stage ACAR was excluded from the derivation of the gene panel to emphasize predicting and identifying graft injury onset. Four of these genes, Padi2, Upf3a, Zfp182, and Slc30a7, were down-regulated in all stages of ACAR, while the other nine genes were up-regulated compared to nonrejecting graft recipient scaffolds. From this 13-gene panel, Arhgap9, Zfp182, Prcc, and Klf9 had the greatest variable importance in the two-metric system for identifying ACAR (fig. S11, D and E), where the 13-gene panel distinguished both pre-injury and mid-injury ACAR from nonrejecting recipients.
These aggregated expression data were analyzed at the pathway level (fig. S12) by conversion to gene set variation scores (50) using the Molecular Signatures Database (MSigDB) collections (51). Elastic net regression derived 25 pathways as a sparse, indicative ACAR panel (Fig. 4E and table S3). These 25 pathways distinguish both pre-injury and mid-injury ACAR from nonrejecting HTx recipients, with divergent clustering (Fig. 4F). We focused our analyses on pathways in HTx rather than STx due to available clinical data for comparison (45) and as the clinical consequences of a developing HTx rejection episode are severe compared with other solid organ transplants (SOT). This dissimilar pathway representation suggests a transition in ACAR state corresponding with the onset of graft injury. Among this sparse 25-pathway set, pathways associated with calcineurin and Vegf signaling are most differently represented by the two stages of ACAR (fig. S12), consistent with intragraft clinical and experimental findings (52–54). Vegf signaling, along with RunX1 regulation, ErbB4 signaling, and TP53 activity, is enriched in pre-injury ACAR compared to nonrejecting graft recipients and mid-injury ACAR. Conversely, the calcineurin pathway is depleted in mid-injury ACAR compared to pre-injury ACAR and nonrejecting graft recipients. Focusing on the differences in these two stages, we identified an additional 31 pathways with highly differential enrichment between pre-injury and mid-injury ACAR (fig. S13, A and B, and table S3). The most enriched pathways in mid-injury ACAR were degradation of β-catenin by the destruction complex and the interleukin-6 (IL-6) pathway, while leukocyte transendothelial migration was most enriched in pre-injury ACAR (fig. S13, C and D). Overall, the pathway differences between pre-injury and mid-injury ACAR indicate that the scaffold can discriminate between distinct biological processes taking place as ACAR transitions to a graft injury stage with T cell infiltration.
Scaffolds distinguish pre-injury ACAR from respiratory infection in immunodeficient mice
We next investigated whether the gene signature could distinguish the immune activation resulting from ACAR from that associated with a nonrejection inflammatory insult, such as viral infection. Viral infections limit the power of noninvasive assays at predicting ACAR (38, 39). Suppressing the adaptive immune system can result in infections such as influenza A virus (IAV) (1), and we thus performed STx (Fig. 5A) or infected Rag2−/− mice with IAV (Fig. 5B). Adoptive transfer of 1 × 107 T cells again led to complete rejection of the STx graft within 13 days (Fig. 5C), with graft wounding first observed on day 10 or 11, as previously (Fig. 1, C and D). At a lower adoptive transfer of T cells (5 × 105), STx recipients had a more varied onset of wounding ranging from day 13 to day 24 after adoptive transfer (Fig. 5A). We also explanted scaffolds from Rag2−/− mice with an IAV infection (55, 56) in which syngeneic T cell transfer (1 × 107) occurred 7 days before viral inoculation. Weight loss was tracked following influenza inoculation as a relative measure of infection severity, with scaffolds explanted from the inoculated mice 3 days before inoculation and 5 and 10 days post-inoculation. A moderate infection was apparent at 5 days post-inoculation, while significant weight loss 10 days post-inoculation indicated severe infection (Fig. 5D) leading to euthanasia of all infected mice by day 15. Analysis of the 13-gene tissue-independent signature at these scaffold explants (Fig. 5, E and F) identified STx recipients, 3 to 4 days before onset of graft injury, as a distinct group relative to both scaffolds from STx recipients before T cell transfer and STx recipient >5 days before graft injury onset. Scaffolds explanted both before and during moderate influenza infection were indistinguishable from the nonrejecting STx scaffold explants. Conversely, scaffolds explanted during severe infection exhibited highly differential panel expression compared to all other groups, although still distinct from pre-injury ACAR. These gene expression results demonstrate the potential of a scaffold-derived biomarker panel in identifying ACAR before graft injury onset, beginning to approximate timing of oncoming graft injury, and distinguishing ACAR from a viral respiratory infection as an example of nonrejection inflammatory insult.
Fig. 5. Early and specific detection of pre-injury ACAR in Rag2−/− skin graft rejection via the scaffold.
(A) Schematic representation of scaffold implants in STx ACAR after high (1 × 107) or low (5 × 105) adoptive transfer of T cells. (B) Schematic representation of scaffold implants in IAV infection after high adoptive transfer of T cells. (C) Graft survival of Rag2−/− STx recipients following high or low T cell adoptive transfer to initiate ACAR. Nonsurvival events defined as the first day at which graft wounding is identified at ×10 magnification; N = 6 independent graft recipients per group as biological replicates. (D) Weight loss as a measure of infection severity for mice inoculated with influenza virus. Mean ± SEM with N = 7 inoculated mice per group as biological replicates. Plot of IAV infected mice with scaffold implants colored according to severity of weight loss. Symbol represents animal euthanized due to >35% weight loss. (E) Principal components clustering of scaffolds based on previously derived 13-gene panel for distinguishing STx allograft recipient mice with pre-injury ACAR from mice with influenza infection. N = 3 or 4 scaffolds from independent mice per group as biological replicates. Points indicate individual scaffold samples, and all ellipses = 70% CI. (F) Clustered heatmap of elastic net–derived 13-gene panel for distinguishing allograft-conserved ACAR from mice with influenza infection; with expression normalized by row. Columns indicate individual samples; N = 3 or 4 scaffolds from independent mice per group as biological replicates.
Scaffolds predict onset of ACAR graft injury in immune suppressed wild-type mice
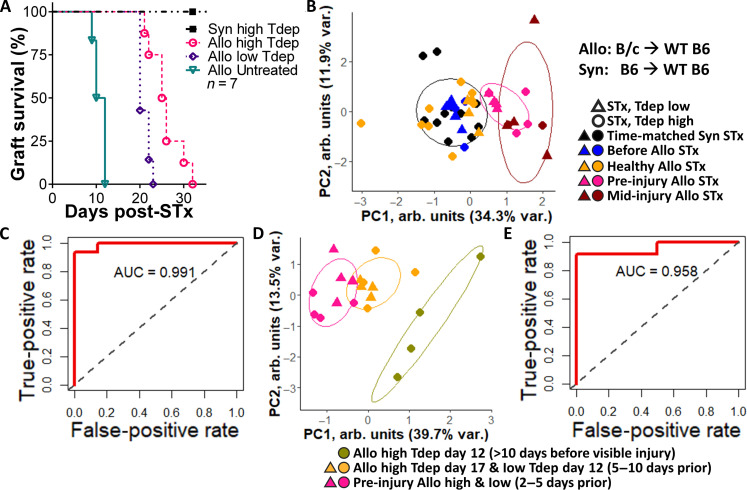
Last, we investigated the ability to detect ACAR at the scaffold before injury onset in wild-type C57BL/6 mice undergoing transient T cell depletion (57–59) by monoclonal anti-CD4 and anti-CD8 antibodies as a more clinically relevant model. The artificial Rag2−/− model used to identify a tissue-independent signature of ACAR (Figs. 1 to 5) enabled graft integration and allowed a focus on rejection without potential interference from wound healing dynamics yet also has altered endogenous T cell processes (e.g., central tolerance, proliferation, migration, and, possibly, activation) (60, 61) and avoided the IRI period immediately following transplantation, a period of great clinical need for surveilling early rejection (8, 54) and distinguishing nonrejection graft dysfunction (49, 62) (e.g., cardiomyopathies). In these wild-type STx with varied doses of transient T cell depletion (Fig. 6A) by αCD4 and αCD8 monoclonal antibodies, the delayed onset of ACAR progressed over different kinetics. Wild-type STx recipients given low T cell depletion (αCD4 and αCD8) first exhibited graft injury on days 19 to 22 following transplant (Fig. 6A), approximately 10 days following untreated wild-type recipients. High T cell depletion (αCD4 and αCD8) further delayed the onset of ACAR graft injury by 2 to 10 days. Principal components clustering of the 13-gene panel expression (Fig. 6B) indicates that, independent of the T cell depletion dose, the scaffold-derived signature separates recipients based on the number of days before graft injury onset at which the scaffolds were explanted. Performance characteristics of the 13-gene panel to identify pre-injury ACAR were computed (Fig. 6, C and D). The receiver operator characteristic area under the curve (AUC) was 0.991 for identifying 2 to 10 days pre-injury ACAR (Fig. 6C), and the AUC was 0.958 when identifying <5 days pre-injury ACAR (Fig. 6D). These results highlight that the 13-gene panel captures aspects of the immune responses preceding graft injury and their relative severity.
Fig. 6. Early detection of STx acute cellular rejection in transient T cell–depleted wild-type mice via the scaffold.
(A) Graft survival of C57BL/6 STx recipients following high or low doses of T cell depleting antibodies (Tdep) to heterogeneously delay ACAR. Nonsurvival events are defined as the first day at which graft injury is identified. N = 4 or 5. (B) Principal components clustering of scaffolds based on previously derived 13-gene panel for identifying wild-type (WT) STx recipient mice before or after transplant and receiving varied doses of transient T cell depletion. Points indicate individual scaffold samples. (C) Receiver operator characteristic curve for sensitivity and specificity of identifying pre-injury ACAR using the scaffold-derived 13-gene panel. N = 3 or 4. (D) Principal components clustering of scaffolds based on previously derived 13-gene panel for identifying wild-type STx allograft recipient mice with pre-injury ACAR after receiving varied doses of transient T cell depletion. Points indicate individual scaffold samples. (E) Receiver operator characteristic curve for sensitivity and specificity of identifying pre-injury ACAR using the scaffold-derived 13-gene panel. N = 3 or 4 scaffolds from independent transplant recipients per group and time point as biological replicates.
DISCUSSION
Organ transplants are monitored for rejection through biopsy, coupled with blood-based assays, but these surveillance modalities identify graft injury as a lagging indicator of rejection and cannot predict onset of rejection (9). EMB—the gold standard—identifies graft injury as a retrospective indicator of allograft immunity through histological assessment of lymphocyte infiltration, myocardial necrosis, and edema (18, 32). However, patient discomfort and potential complications limit EMB frequency to “snapshot” clinical assessments (63), challenging early detection of rejection (44). Blood gene expression profiling and plasma donor–derived cell-free DNA have reduced pressure to perform procedural biopsies. While these blood-based assays may rule out the need for graft biopsy by identifying patients at low risk for rejection, EMB has remained a vital window to the states of both rejection and parenchymal injury (44). The engineered immunological implant reported herein functions as a remote, lymphatic niche that captures immune cell phenotypes that can predict acute allograft injury for more frequent longitudinal monitoring than is possible with graft biopsy, creating the opportunity for precision immune suppression.
Analysis of gene expression from scaffolds identified biomarkers indicative of immune activation leading to graft rejection, yet at a stage that is pre-injury. While injury-induced changes are strong molecular correlates of dysfunction and graft loss (64), mapping molecular injury changes for transplant surveillance is precarious as injury due to rejection has been shown to persist after rejection is treated or subsides (64). For these reasons, scaffold-derived pre-injury biomarkers of ACAR could be greatly beneficial in transplant surveillance. In heart and skin transplantation, rejection responses assessed at the scaffold through a gene expression signature could be identified before lymphocyte infiltration or graft injury. Signature genes identified with the scaffold differ significantly from transcripts that are associated with EMB, which likely results from our signature being early in the rejection response and involving more innate cells whereas EMB is later in rejection and tends to be T cell centric. Previous reports have indicated that gene expression by immune cells in circulation does not correlate with gene expression of cells within the scaffold (26, 29), likely due to a combination of selective recruitment and phenotype changes at the scaffold. Collectively, biopsy of this scaffold implant may provide an opportunity to identify rejection responses that involves less risk relative to invasive graft biopsy while providing more specificity about immune activation than is now available with liquid biopsy. A minimally invasive assay that can predict acute allograft rejection, particularly before evidence of graft injury, could create a therapeutic window to preserve graft function.
The remotely implanted scaffold captures markers of early rejection predominantly through innate immune cells (Fig. 1, G and H) and can thus identify immune responses that precede graft insult (33). Innate immune cell participation has been underemphasized in contributing to allograft rejection as immune suppressive drugs largely target T cell maturation or responses, yet these responses are highly localized (65) and, as shown in Fig. 1 (H to J), expand in circulation, the scaffold, and the graft only late in the rejection cascade. Alloimmune graft injury involving non-antigen–specific inflammatory pathways can be mediated by innate immune cell populations, specifically myeloid-derived monocytes, macrophages, and DCs (66). The high proportion of DCs, monocytes, and macrophages which home to the scaffold due to the foreign body response (67) may be favorable for identifying early allorecognition (68). DCs have been implicated in allorecognition and greatly contribute to rejection (69, 70). Additional work is necessary to fully characterize the recruitment and subsequent phenotypic changes of these cells at the scaffold implant. Here, we observed Igtp, an endoplasmic reticulum–related guanosine triphosphatase identified in the HTx ACAR signature (Fig. 3E) which has been shown to control cross-presentation in DCs (71). In addition, early down-regulation of Fgl2, as identified preceding graft injury at the scaffold (Fig. 3E), may allow antigen presenting cell maturation. Similarly, macrophages play an important role in promoting allograft rejection (72) and tolerance (73). Git1 has been shown to regulate macrophage inflammatory responses (74). H2-Eb1 has been associated with antigen-presenting macrophages in murine cardiac allografts (75). Also, Zbp1 regulates innate immune-mediated inflammatory cell death, particularly through macrophages (76). Myd88 expression in macrophage pathways is differentially enriched in scaffold implants during ACAR and has been shown to be critical to both toll-like receptor functions in allograft rejection (77) and calcineurin inhibitor–induced acute kidney injury (78). As calcineurin inhibition is the primary clinical strategy for maintaining immune quiescence posttransplant, the scaffold implant should be investigated further as a tool to monitor suppression effectiveness. In addition, awareness is growing that the pathogenesis of donor-specific antibodies in organ transplantation also depends on the activation of innate pathways (79). Capturing innate immune cell responses may be a potential advantage of the scaffold, as the innate responses are systemic and precede substantial T cell accumulation at the graft.
T cells, the primary mediators of rejection, are not observed to increase in numbers until late stages of rejection, yet the innate cells captured at the scaffold before alloimmune graft injury participate in early T cell signaling processes for activation, migration, and activation (Fig. 2F). The active pathways identified from sequencing indicate that the scaffold recapitulates pathways associated with allograft injury and T cell signaling, including IL-6 Janus kinase–signal transducers and activators of transcription signaling, mitogen-activated protein (MAP) kinase, nuclear factor κB (NF-κB) activity, vascular endothelial growth factor (VEGF) signaling, calcineurin regulation, Erb-B2 Receptor Tyrosine Kinase 4 signaling, Tp53 activity, runt-related transcription factor expression, inflammation from antigen stimuli, β-catenin degradation (80), and numerous T cell and myeloid cell activation pathways (Figs. 2F and 4E). MAP kinase and NF-κB activity are involved in corticosteroid signaling pathways in transplant immunosuppression (81) and immune suppression–induced nephrotoxicity (82, 83), giving further credence to investigation of the scaffold implant as a means to monitor common immunosuppression regimes consisting of calcineurin inhibitors and corticosteroids. Secreted fibrinogen-like protein 2 (FGL2) by CD4 T cells potentially mediates acute graft rejection and is induced by tumor necrosis factor–α or interferon-γ stimulation through MAP kinase signaling (84). VEGF signaling has been linked to acute and chronic rejection (54, 85), and the high differential regulation of both VEGF signaling and Vwf expression during pre-injury rejection highlights that while this work has remain largely focused on the scaffold-recruited leukocytes, endothelial cell and endothelial-immune interactions enriched at the scaffold may also have prognostic and mechanistic relevance for ACAR. Tp53 is pivotal to cell cycling and DNA damage repair pathways in renal grafts during acute rejection (86, 87). RUNX1 has been identified as an important transcription factor of differentially expressed genes in lung transplant biopsies (88). IL-6 production in DCs, which is an enriched pathway at the scaffold in pre-injury ACAR, has been shown to promote allograft rejection by interaction with CD4 T cells (89, 90). Calcineurin inhibitors have revolutionized immunosuppression, which regulates pathways that are also depleted from pre-injury to mid-injury ACAR in the scaffold. In addition, several of the sparse gene indicators of ACAR derived at the scaffold, including Ppp1r3fos, Metrn, Rxra, and Flywch1, have been implicated in calcineurin inhibitor nephrotoxicity (91). These pathway enrichments further corroborate that T cells do not need to be abundant at the scaffold to monitor T cell signaling and immunosuppression targets preceding graft injury onset. The remotely implanted scaffold also captures genes regulating T cells. Arntl, identified in HTx acute rejection at the scaffold, regulates mitochondrial metabolism in the heart (92, 93). Differential Vwf expression was identified at the scaffold during graft rejection, with increased Vwf expression identified in early adverse events (94, 95), yet plasma levels have not been found to correlate with primary graft dysfunction (96).
A substantial challenge in monitoring grafts is distinguishing rejection from other immune activity, and we demonstrated that the scaffold could distinguish rejection from viral respiratory infection in mice with a partially reconstituted lymphocyte compartment. Many innate and adaptive immunity responses share features with viral infection and other inflammation, creating diagnostic challenges (38). Several of the tissue-specific, sparse biomarkers we identified in the scaffold for skin or heart graft ACAR are modulated during viral infection. Thus, we used a signature that was based on conserved markers of acute rejection across both HTx and STx, which enabled a distinction from IAV infection in these mice. The biomarkers associated with viral infection included Zbp1, which is activated by viral infection such as human cytomegalovirus (HCMV) binding (97). FGL2 transcription has been found to be initiated in two separate pathways by alloimmunity and viral proteins (98). During HCMV infection of fibroblasts, NF-κB activation appears to follow a specific sequence in which the pathway is active early in infection (97). The scaffold-derived 13-gene signature of tissue-independent ACAR differentiated this IAV infection from rejection, with severe influenza infection distinctly activating these 13 genes relative to their activation induced by rejection.
The scaffold-derived gene signature of acute rejection identifies early rejection responses even in the presence of IRI. Innate immune responses have been largely considered in the context of IRI response following transplantation. The Rag2−/− model used identified a tissue-independent signature of ACAR without potential interference from wound healing dynamics and IRI following transplantation. In wild-type STx with transient T cell depletion, independent of depletion dose, the scaffold-derived signature separates recipients based on the number of days before graft injury onset at which the scaffolds were explanted. The 13-gene panel derived at the scaffold and monitored through the scaffold captures aspects of the immune responses preceding clinical graft injury and their relative severity. Current immunosuppressive drugs suppress T cell responses, leaving innate immune cells programmed during IRI to inflict graft damage in acute graft rejection or stimulate mild T cell responses over time (79). The scaffold environment reflects these critical early allograft responses even in the presence of background IRI.
As SOT recipients undergo chronic immune suppression, the clinical safety and sustained efficacy of the scaffold device are critical to translation as a minimally invasive assay of the state of the graft. Implant site infection and foreign body response including fibrous encapsulation are important considerations for any implant. In the clinic, various implants have been found safe and effective in chronically suppressed SOT recipients, including cochlear implants (99, 100), various joint replacement surgeries (101–104), mesh hernia repair (105), breast reconstruction (106), permanent pacemakers (107), dental implants (108), and both subcutaneous contraceptive implants (109) and intrauterine devices (110–112). Translation of the scaffold implant to the clinic will likewise involve preclinical assessment of the implant parameters, including surface topography (113), to coordinate prevention of fibrous encapsulation with timing of scaffold residence and biopsy for immunosurveillance in extended tempos of rejection relative to the murine models used in this study.
In conclusion, we report a subcutaneous implant that recruits immune cells from circulation and whose gene expression can be used to predict the onset of acute T cell–mediated graft rejection. The gene expression patterns reflect that the cell phenotypes, not simply the cell numbers or types, are significantly altered before alloimmune injury. Scaffold-derived gene expression captures both tissue-specific and tissue-independent molecular responses of alloimmunity and can distinguish T cell–mediated transplant rejection from viral respiratory infection in immunodeficiency. Clinical translation of the scaffold will involve identifying pre-injury rejection events during chronic immunosuppression (e.g., tacrolimus). Within the context of transplant rejection, identifying immune activation before tissue injury could enable personalized immune suppression to prolong graft life while minimizing risks associated with sustained immune suppression. Clinical transplant endpoints incorporate multilevel, complementary surveillance, including systemic immune quiescence and drug toxicity through liquid biopsy, graft injury via histology, and graft function via imaging. The immunological niche developed at the scaffold may provide a bridge between the two ends of the rejection continuum, namely, transplanted organ and peripheral blood cells (114). The scaffold device could further refine the paradigm of rejection to include immune responses before graft insult for identifying more than simply immune quiescence through the circulation.
MATERIALS AND METHODS
Study design
The aim of this study was to develop an implant for remotely surveilling allograft health. We performed murine skin and heterotopic HTx in mature T cell–deficient mice in which we implanted microporous polymeric scaffolds and then initiated acute cellular rejection through adoptive T cell transfer. Using the Rag2−/− mouse model, we evaluated the scaffold by flow cytometry, bulk RNA-seq gene expression, and gene set variation for differential expression in stages of acute allograft rejection. Using an elastic net and random forest bagged tree algorithm, we identified gene signatures of STx rejection, HTx rejection, and a tissue-independent signature of rejection. We then validated the rejection specificity of this signature via quantitative polymerase chain reaction (qPCR) against influenza infection and in wild-type transplant recipients with transient T cell depletion. All animal care and procedures were performed in accordance with standards from the Guide for the Care and Use of Laboratory Animals and were carried out in compliance with protocols approved by the Institutional Animal Care and Use Committee (protocol nos. 00009777 and 00010704) at the University of Michigan (UM). Sample sizes were calculated on the basis of previous experience by the investigators. The investigators were not blinded to allocation during experiments or analyses. No outlier values were excluded.
Materials
Ester terminated poly(ε-caprolactone) (Evonik Corp., Birmingham, AL) was purchased for generating microporous scaffolds. Suture (Ethicon, Raritan, NJ), carprofen, and punch biopsies (Henry Schein Inc., Melville, NY) were purchased to perform scaffold implants, skin or HTx, and scaffold biopsies in mice. A hematoxylin and eosin (H&E) staining kit (Abcam, Cambridge, UK) was purchased to stain histological samples, and a direct-zol RNA Miniprep Plus (Zymo Research, Irvine, CA) was purchased to isolate RNA from tissue and scaffold samples. A naive pan mouse T cell isolation kit and LS columns were purchased from Miltenyi Biotec (Bergisch Gladbach, North Rhine-Westphalia, Germany). Stocks of the VR-95 influenza A/PR8/34 H1N1 were purchased from American Type Culture Collection (Manassas, VA). The original stock was aliquoted and stored at −80°C until use. Anti-mouse CD4 (clone GK1.5) and anti-mouse CD8α (clone 2.43) monoclonal antibodies were purchased from Bio X Cell (Lebanon, NH). TaqMan gene probes, gene expression mastermix, and all other reagents were purchased from Thermo Fisher Scientific (Waltham, MA) unless otherwise stated. To maintain integrity, materials were maintained at room temperature, 4°C, or −20°C according to the manufacturer’s instructions. Water from a Millipore filtration system (Darmstadt, Germany) with 18.2 M ohm cm resistivity was used unless otherwise stated.
Microporous scaffold generation
Microporous PCL scaffolds with 250- to 425-μm pores were generated as described previously (27, 29, 115). Briefly, microporous PCL scaffolds were generated by first mixing 99 g of sieved NaCl (250 to 425 μm) with 3 g of PCL pellets at 85°C for 1 hour. A total of 77.5 mg of the PCL-NaCl melt dispersion was pressed into 5- or 6-mm-diameter scaffolds. Scaffolds were heated at 65°C for 5 min on each side to anneal the PCL. Upon cooling, the scaffolds were immersed in water for 1 hour three times to dissolve the NaCl porogen. The scaffolds were then sanitized by ethanol immersion and washed with sterile water in preparation for murine implantation.
STx and heterotopic HTx models
Male and female C57BL/6 (B6), BALB/c (B/c), and B6.Cg-Rag2tm1.1Cgn (Rag2−/−) mice were purchased from the Jackson Laboratory (Bar Harbor, ME) at 8 to 12 weeks old. Mice were housed in a pathogen-free environment under a 12-hour light-dark cycle. Rag2−/− mice were bred, and offspring were used as transplant recipients at 8 to 12 weeks of age. Transplants were performed male-to-male and female-to-female in approximately equal numbers. Four transplant cohorts were performed: STx training cohort, heterotopic HTx training cohort, STx validation cohort, and STx wild-type immunosuppression cohort. Murine heterotopic HTx or STx were performed as described previously (116). In murine STx, donor adult mice were euthanized as approved, and the tails were cleansed with 70% ethanol. The tails were transected at the base, and a longitudinal incision to deglove the tail skin was made. Tail skin was then trimmed into circles of 8-mm diameter with a punch biopsy tool. Recipient adult mice were anesthetized as approved and received subcutaneous carprofen (5 mg/kg) before surgery. The dorsal skin of the recipient was depilated with ointment, rinsed with water, and then scrubbed with ethanol and betadine. A 6-mm-diameter skin portion was excised via punch biopsy. The donor graft was placed on the recipient site and attached to the graft bed by 8-0 suture. The graft area was then impregnated with triple antibiotic ointment and wrapped with gauze. After 24 hours, another dose of carprofen was given. Dressings were removed on postoperative days 5 to 7, and sutures were removed on day 10. On the basis of a previous experience, the STx recipient mice were then rested for an additional 18 days to allow for skin graft healing (28 total days posttransplant before adoptive T cell transfer). For STx recipients in which the graft failed within 5 days of transplant, these failures were considered surgical error. No graft failures took place after 5 days but before adoptive T cell transfer. Following adoptive T cell transfer, STx recipients were monitored for graft loss and euthanized at the time of graft rejection for analysis of pathological changes within the grafts. STx was performed as follows: B6 graft to Rag2−/− recipient (syngeneic), B/c graft to Rag2−/− recipient (allogeneic), B6 graft to B6 recipient (syngeneic), or B/c graft to B6 recipient (allogeneic).
In murine HTx, grafts were collected from adult donor mice. The heterotopic transplant was maintained in the recipient neck for monitoring the viability of the transplanted heart by inspection and palpation. HTx was performed as B6 graft to Rag2−/− recipient (syngeneic) or B/c graft to Rag2−/− recipient (allogeneic). The donor heart ascending aorta and pulmonary artery were anastomosed to the recipient common carotid artery and external jugular vein, respectively. A prophylactic dose of antibiotics was administered to reduce the risk of infection. Analgesic carprofen was administered preemptively and every day for 48 hours and then as needed. During the initial 72 hours post-operation, animals were monitored to confirm activity and health. On the basis of a previous experience, the HTx recipient mice were then rested for an additional 21 days to allow for heart graft healing (24 total days posttransplant before adoptive T cell transfer). For HTx recipients in which the graft failed within 72 hours of transplant, these failures were considered surgical error. No graft failures took place after 72 hours but before adoptive T cell transfer. Following adoptive T cell transfer, all failures were considered immunological rejection. Graft failure was determined by the lack of a palpable heartbeat.
Scaffold implants in transplant recipients
Mice were anesthetized with 2% (v/v) isoflurane for subcutaneous implantation of scaffolds and serial scaffold explants. Mice received carprofen immediately before surgery and 24 hours after surgery. Four to six scaffolds were implanted subcutaneously 14 days before inducing graft rejection in Rag2−/− mice or before transplant in immunocompetent B6 mice. The subcutaneous space was selected for minimal invasiveness (27, 29, 115), and below the dorsal skin was selected for convenience when implanting in mice. After vascularization and cell in-growth, scaffolds were then explanted before and during graft rejection. With the implant of numerous 5- to 6-mm scaffolds in the dorsal subcutaneous space, scaffolds were explanted no more frequently than 4 days apart with a minimal surgical incision of 2 to 4 mm to allow for incision healing and limit local inflammation at the scaffold explant site. Time-matched syngeneic recipient scaffold explants were used in part to control for the local inflammation encountered due to scaffold explant. Scaffold explants were carefully trimmed of any surrounding fibrous tissue, and resultant explants were approximately the same height and diameter as when implanted. Cardiac drawn blood and primary grafts (terminal points) were also collected.
Inducing graft rejection
Transplant recipient Rag2−/− or B6 mice were allowed 10 or 14 days for heart or skin grafts to heal, respectively, with another 14 days for implanted scaffolds to become vascularized and additional graft healing. For adoptive transfer induction of ACAR, B6 mice were euthanized, and splenic pan naïve B6 T cells were collected via magnetic-activated cell sorting. These syngeneic B6 T cells were counted with an automated cell counter (Countess 3 FL, Invitrogen, Waltham, MA) and injected intraperitoneally into transplant recipient Rag2−/− mice. For STx, 5 × 105 or 1 × 107 cells were transferred per mouse; while for HTx, 2 × 107 cells were transferred per mouse to induce ACAR. For these adoptive transfer studies, the day of T cell transfer was considered day 0, and graft recipients were monitored for graft rejection and euthanized at differing stages of rejection for various analyses. Heart graft rejection was scored on a 0 to 3 scale: 0 = strong pulse, no swelling; 1 = weak pulse or swelling, 2 = weak pulse and swelling, 3 = no pulse. Skin graft rejection was scored as percentage of the graft area that was visibly inflamed. STx grafts were considered fully rejected at the time of sloughing or upon complete conversion to a hard avascular eschar. Grafts were imaged and scored daily following T cell transfer (117).
Tissue and nucleic acid isolation
To collect serial scaffold explants, mice were anesthetized with isoflurane and shaved closely near the implant site. A small dorsal incision made aside the surface of the implanted scaffold was opened with sterile forceps, with the implanted scaffold and encapsulating tissue gently pulled through and excised. The incision was then sutured closed. Implants for RNA isolation or histology were flash frozen in isopentane, kept on dry ice, and stored at −80°C until further processing and analysis. Implants used for flow cytometry were immediately placed into phosphate-buffered saline (PBS) kept on ice. To collect blood and primary grafts, mice were anesthetized, and blood was terminally drawn via intracardiac puncture with EDTA to prevent clotting. The mice were then euthanized as approved, and skin or heart grafts were carefully excised from surrounding tissue. All blood samples were resuspended in Ammonium-Chloride-Potassium lysing buffer to lyse red blood cells, washed in PBS, and then resuspended in TRIzol reagent and stored at −80°C until further processing and analysis. Frozen tissues were homogenized in TRIzol reagent, and RNA was isolated with a direct-zol RNA Miniprep Plus kit following the manufacturer’s instructions.
Histology
For histological analyses, skin, heart, and scaffold samples were each embedded into Optimal cutting temperature compound (OCT)–30% sucrose and sectioned onto slides using a Cryostat (Microm HM 525, Thermo Fisher Scientific, Waltham, MA) at 14-μm thickness. The slides were stained with H&E following the standard procedures. Images were taken at 20× within the thickness of the scaffold or heart graft. The H&E-stained histological slides were imaged and processed using QuPath software (version 0.4.4), as indicated by example shown in fig. S8 (C and D). Cases in which the slides were not suitable for analysis (image obscured by slide markings or out of focus or image scanned below ×20 magnification) were excluded. The parameter adjusted for cell detection was minimum area of immune cell nuclei, which with a default at 5 μm2. If cell detection was suboptimal (due to cell clusters misidentified), then the minimum nuclei area was adjusted (range: 4 to 7 μm2), to adequately separate cells.
Flow cytometry
Biopsied scaffolds or grafts were prepared for flow cytometry by mechanical dissection and enzymatic incubation (28, 115). Briefly, samples were minced, incubated for 20 min in Liberase TL (Roche, Basel, Switzerland) at 37°C, then mashed through a 70-μm filter, which was washed extensively with fluorescence-activated cell sorting buffer: PBS with 0.5% bovine serum albumin (Sigma-Aldrich, St. Louis, MO) and 2 mM EDTA. Single-cell suspensions from blood samples were similarly prepared. Cells were equally divided to enable staining and analysis of innate and adaptive immune cells from the same sample (table S4) and then blocked with anti-CD16/32 (1:50; clone 93, eBioscience). Each tube was stained with Live/Dead Fixable Red (Life Technologies) and Alexa Fluor 700 anti-CD45 (1:125; clone 30-F11, BioLegend). The adaptive immune panel was also stained with fluorescein isothiocyanate (FITC) anti-CD8 (1:25; clone 53-6.7, BioLegend), phycoerythrin (PE)–Cy7 anti-CD49b (1:30; clone DX5, BioLegend), and V500 anti-CD4 (1:100; clone RM4-5, BD Biosciences). The innate immune panel was also stained with APC anti-CD11c (1:80; clone N418, BioLegend), FITC anti-Ly6C (1:100; clone HK.14, BioLegend), Pacific Blue anti–Ly-6G/Ly-6C (Gr-1) (1:70; clone RB6-8C5, BioLegend), PE-Cy7 anti-F4/80 (1:80; clone BM8, BioLegend), and V500 anti-CD11b (1:100; clone M1/70, BD Biosciences). Samples were analyzed on a Cytoflex Cell Analyzer, and single-color controls and fluorescence-minus-one (FMO) controls were used to aid with gating and compensation (fig. S2), with data analysis on FlowJo v.10 software.
RNA sequencing
For high-throughput gene expression analysis, bulk RNA-seq analysis and quality control were performed on a fee-for-service basis by the UM Advanced Genomics Core, with a NextSeq HO 150 cycle and the QuantStudio 12k Flex system (Thermo Fisher Scientific). A total of n = 3 or N = 4 samples per transplant type (allogeneic or syngeneic) were analyzed from each time point. Each sample was from an independent mouse, and samples were alternated between healthy and rejecting on the plate to minimize spatial bias. Samples were analyzed to filter genes based on low expression and expression variance to reduce noise. Raw counts were then batch-corrected and normalized as needed via the DESeq2 package (118).
Differential expression and selecting genes of interest
Genes of interest were selected by log2 fold change > 1.5 and Padj < 0.1, attempting to include both genes that increased and decreased in expression during rejection to make the model more robust. Gene expression was then analyzed by elastic net regularization (27, 30) for high-dimensional differential expression across mice, allogeneic or syngeneic graft donor, and time points in the HTx and STx training cohorts to generate a biomarker panel of approximately 10 to 24 genes which categorize mice by graft health or stage of ACAR. Through SVD and supervised machine learning (random forest) (119), to derive single-metric scores, we constructed a two-metric scoring system based on biomarker panel expression for the likelihood that a mouse will reject the transplanted graft.
Comparing genes with public datasets and pathway analyses
Genes identified as significant via nonparametric t test between healthy and rejecting transplant recipients were converted to their human orthologs using biomaRt (120). Preranked gene lists were assembled for significant genes (adjusted P value <0.05) using the logarithmic fold change between healthy and rejecting transplant recipients as the ranking variable. The corresponding human ortholog genes underwent GSEA (51, 121) using a preranked gene list to identify significantly enriched pathways. Gene sets were sampled from the HALLMARK, BIOCARTA, Kyoto Encyclopedia of Genes and Genomes (KEGG), and REACTOME sets obtained from the MSigDB collections (34, 51). GSEA was performed using the GSEA v4.3.2 software (Broad Institute), computing gene set overlaps between canonical pathways and Gene Ontology biological process. Differentially enriched pathways were defined by |NES| > 1.5 and |NES sum| > 3.5. GSVA was also performed to identify sparse pathway lists classifying ACAR distinctly from healthy graft recipients (50). Last, the DNApath package was used to visual gene-gene connectivity in pathways of interest by generating differential network analyses (122), with differential connectivity score P values < 0.01.
Influenza inoculation
Rag2−/− mice received scaffold implants as previously, and 1 × 107 T cells were adoptively transferred 14 days later. After another 7 days to allow for syngeneic T cell proliferation, mice were lightly anesthetized with isoflurane and given 400 plaque-forming units of IAV in 40 μl of PBS intranasally (123), which was considered day 0 for subsequent analyses. Mice were monitored daily following infection for weight loss and morbidity and were euthanized after loss of 30% of their preinfection body weight. As described for transplant recipients, implanted scaffolds were explanted from infected mice on day −1, day 5, and day 10 relative to initiating infection. The scaffolds from N = 4 randomly selected mice were selected for gene panel validation via qPCR.
Wild-type T cell depletion study
Wild-type B6 mice received scaffold implants 14 days before STx. Anti-CD4 and anti-CD8 were given in combination on days −3 and −1 before skin graft at 100 or 200 μg per monoclonal antibody per injection (124, 125). Following allogeneic (B/c to T cell depleted B6) or syngeneic (B6 to T cell depleted B6) STx, wild-type graft recipients were then monitored for acute cellular rejection as previously. Implanted scaffolds were explanted from wild-type mice on days −3, 7, 12, 17, 22, and 28 relative to transplant. The scaffolds from N = 3 randomly selected mice were selected for gene panel validation via qPCR.
Panel validation via RT-qPCR
Once the 13 genes of interest were identified as described above, validation and prediction studies were conducted via reverse transcription qPCR (RT-qPCR) analysis in 384-well plates. Three housekeeping genes were also selected by analyzing each of the 16 OpenArray reference genes for the lowest variance overall and across transplant type (fig. S14) to select the three most stable reference genes (Gapdh, Ppia, and Trib3) to be used as reference in RT-qPCR. Single-strand cDNA was synthesized from isolated RNA with the SuperScript VILO cDNA Synthesis Kit according to the manufacturer’s instructions using a thermocycler (Mastercycler gradient, Eppendorf, Hamburg, Germany). RT was performed with RNA concentrations of 200 ng/μl. TaqMan probes of the genes of interest were ordered, and cDNA samples were placed in randomly assigned order on the 384-well plate. TaqMan Gene Expression Mastermix was used, and a final volume of 9 μl was reached for each well of the 384-well plate. RT-qPCR was performed on the QuantStudio ViiA 7 system (Applied Biosystems, Waltham, MA), and Cq values were determined by the accompanying software. ΔCq values were calculated for each gene from the average of the reference genes for that sample.
Statistical analyses and reproducibility
Statistical analyses for validation assays were performed using R and GraphPad Prism 9 software (GraphPad) when N > 2 with unpaired Student’s t test or one- or two-way with Tukey’s multiple comparisons test. P values lower than 0.05 were considered statistically significant. For two-group comparisons, Student’s t tests were used to analyze flow cytometry data as comparisons were made between healthy and rejecting within each time point. All data are presented as means ± SEM with the number of samples provided [distinct samples or mice (N) or images within a tissue (N); not repeated measures], unless otherwise indicated. The details of specific statistical tests used for each experiment and probability values are detailed in the figure legends, and raw data and details of statistical analyses are provided in data S1 to S7. For histological images, at least three samples were acquired, and representative images were displayed for each transplant group and time point. For multiple group analysis and when comparing pooled control recipients versus rejecting recipient mice, two-way analysis of variance (ANOVA) was used with a post hoc Bonferroni correction to determine significance. Log-rank test (Mantel-Cox) test was used for survival comparison. P values below 0.05 were considered statistically significant.
Acknowledgments
We thank lab managers R. Maramraju and S. Wood for excellent technical assistance, the UM Advanced Genomic Core for sample analysis, the UM Flow Cytometry Core for training and technical assistance, and F. Lakkis and D. Rothstein for critical review of the proof of concept.
Funding: This work was supported by funds and/or resources from the Falk Medical Trust Catalyst Award #N029877 (R.R.U., D.R.G., and L.D.S.), the Falk Medical Trust Transformational Award #N033434 (R.R.U., D.R.G., and L.D.S.), the National Institutes of Health R01 #AI138347 (D.R.G.), the UM Undergraduate Research Opportunity Program (C.X.), the National Institutes of Health F31 #HL158003 (J.C.), and the National Institutes of Health K99 EB028840 (A.M.).
Author contributions: Conceptualization: R.R.U., A.M., D.R.G., and L.D.S. Methodology: R.R.U., D.F., A.M., D.R.G., and L.D.S. Murine transplantation: R.R.U., D.F., and J.F. Investigation: R.R.U., A.M., E.H., C.X., J.C., E.L., J.Z.L., D.R.G., and L.D.S. Visualization: R.R.U., E.H., C.X., D.R.G., and L.D.S. Funding acquisition: R.R.U., D.R.G., and L.D.S. Project administration: R.R.U., D.R.G., and L.D.S. Supervision: D.R.G. and L.D.S. Writing—original draft: R.R.U., E.H., D.R.G., and L.D.S. Writing—review and editing: R.R.U., A.M., E.H., J.Z.L., D.R.G., and L.D.S.
Competing interests: R.R.U., A.M., D.R.G., and L.D.S. are inventors on a patent application related to this work filed by The Regents of the UM (US Provisional Patent Application no. 63/310,201, filed 14 February 2023). The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Tables S1 to S4
Legends for data S1 to S7
Other Supplementary Material for this manuscript includes the following:
Data S1 to S7
REFERENCES AND NOTES
- 1.Fishman J. A., Infections in immunocompromised hosts and organ transplant recipients: Essentials. Liver Transpl. 17, S34–S37 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Adami J., Gäbel H., Lindelöf B., Ekström K., Rydh B., Glimelius B., Ekbom A., Adami H.-O., Granath F., Cancer risk following organ transplantation: A nationwide cohort study in Sweden. Br. J. Cancer 89, 1221–1227 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J. M., Canter C. E., Hsu D. T., Kindel S. J., Law Y. M., McKeever J. E., Pahl E., Schumacher K. R., Current topics and controversies in pediatric heart transplantation: Proceedings of the Pediatric Heart Transplantation Summit 2017. World J. Pediatr. Congenit. Heart Surg. 9, 575–581 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Blondet N. M., Healey P. J., Hsu E., Immunosuppression in the pediatric transplant recipient. Semin. Pediatr. Surg. 26, 193–198 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Knackstedt E. D., Danziger-Isakov L., Infections in pediatric solid-organ transplant recipients. Semin. Pediatr. Surg. 26, 199–205 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Yanik E. L., Smith J. M., Shiels M. S., Clarke C. A., Lynch C. F., Kahn A. R., Koch L., Pawlish K. S., Engels E. A., Cancer risk after pediatric solid organ transplantation. Pediatrics 139, e20163893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller C. A., Fildes J. E., Ray S. G., Doran H., Yonan N., Williams S. G., Schmitt M., Non-invasive approaches for the diagnosis of acute cardiac allograft rejection. Heart 99, 445–453 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Khush K. K., Patel J., Pinney S., Kao A., Alharethi R., DePasquale E., Ewald G., Berman P., Kanwar M., Hiller D., Yee J. P., Woodward R. N., Hall S., Kobashigawa J., Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: A prospective multicenter study. Am. J. Transplant. 19, 2889–2899 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crespo-Leiro M. G., Barge-Caballero G., Couto-Mallon D., Noninvasive monitoring of acute and chronic rejection in heart transplantation. Curr. Opin. Cardiol. 32, 308–315 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Chi N.-H., Chou N.-K., Tsao C.-I., Huang S.-C., Wu I.-H., Yu H.-Y., Chen Y.-S., Wang S.-S., Endomyocardial biopsy in heart transplantation: Schedule or event? Transplant. Proc. 44, 894–896 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Vlaminck I. D., Valantine H. A., Snyder T. M., Strehl C., Cohen G., Luikart H., Neff N. F., Okamoto J., Bernstein D., Weisshaar D., Quake S. R., Khush K. K., Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci. Transl. Med. 6, 241ra77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grady K. L., Wang E., White-Williams C., Naftel D. C., Myers S., Kirklin J. K., Rybarczyk B., Young J. B., Pelegrin D., Kobashigawa J., Higgins R., Heroux A., Factors associated with stress and coping at 5 and 10 years after heart transplantation. J. Heart Lung Transplant. 32, 437–446 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H.-M., Lai C. K., Gjertson D. W., Baruch-Oren T., Ra S. H., Watts W., Wallace W. D., Shintaku P., Kobashigawa J. A., Fishbein M. C., Has the 2004 revision of the International Society of Heart and Lung Transplantation grading system improved the reproducibility of the diagnosis and grading of cardiac transplant rejection? Cardiovasc. Pathol. 18, 198–204 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Crespo-Leiro M. G., Zuckermann A., Bara C., Mohacsi P., Schulz U., Boyle A., Ross H. J., Parameshwar J., Zakliczynski M., Fiocchi R., Stypmann J., Hoefer D., Lehmkuhl H., Deng M. C., Leprince P., Berry G., Marboe C. C., Stewart S., Tazelaar H. D., Baron H. M., Coleman I.-C., Vanhaecke J., Concordance among pathologists in the second cardiac allograft rejection gene expression observational study (CARGO II). Transplantation 94, 1172–1177 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Snyder T. M., Khush K. K., Valantine H. A., Quake S. R., Universal noninvasive detection of solid organ transplant rejection. Proc. Natl. Acad. Sci. U.S.A. 108, 6229–6234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Famulski K. S., de Freitas D. G., Kreepala C., Chang J., Sellares J., Sis B., Einecke G., Mengel M., Reeve J., Halloran P. F., Molecular phenotypes of acute kidney injury in kidney transplants. J. Am. Soc. Nephrol. 23, 948–958 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winters G. L., Loh E., Schoen F. J., Natural history of focal moderate cardiac allograft rejection. Circulation 91, 1975–1980 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Clemmensen T. S., Firooznia N., Olawi F. M., Løgstrup B. B., Poulsen S. H., Eiskjær H., Assessment of acute rejection by global longitudinal strain and cardiac biomarkers in heart-transplanted patients. Front. Immunol. 13, 841849 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander M. M., Teuteberg J., Detecting cardiac allograft rejection in the era of personalized medicine: A review of current genomic surveillance techniques. Curr. Transpl. Rep. 3, 367–374 (2016). [Google Scholar]
- 20.Verhoeven J. G. H. P., Boer K., Van Schaik R. H. N., Manintveld O. C., Huibers M. M. H., Baan C. C., Hesselink D. A., Liquid biopsies to monitor solid organ transplant function: A review of new biomarkers. Ther. Drug Monit. 40, 515–525 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Agbor-Enoh S., Shah P., Tunc I., Hsu S., Russell S., Feller E., Shah K., Rodrigo M. E., Najjar S. S., Kong H., Pirooznia M., Fideli U., Bikineyeva A., Marishta A., Bhatti K., Yang Y., Mutebi C., Yu K., Kyoo Jang M., Marboe C., Berry G. J., Valantine H. A., Cell-free DNA to detect heart Allograft acute rejection. Circulation 143, 1184–1197 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M.-C. Parent, B. A. Clarke, K. K. Khush, “Noninvasive Tools for Monitoring Acute Cardiac Allograft Rejection: State of the Art” in The Pathology of Cardiac Transplantation: A Clinical and Pathological Perspective, 265–277 (2016).
- 23.Crespo-Leiro M. G., Stypmann J., Schulz U., Zuckermann A., Mohacsi P., Bara C., Ross H., Parameshwar J., Zakliczyński M., Fiocchi R., Hoefer D., Colvin M., Deng M. C., Leprince P., Elashoff B., Yee J. P., Vanhaecke J., Clinical usefulness of gene-expression profile to rule out acute rejection after heart transplantation: CARGO II. Eur. Heart J. 37, 2591–2601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobashigawa J., Patel J., Azarbal B., Kittleson M., Chang D., Czer L., Daun T., Luu M., Trento A., Cheng R., Esmailian F., Randomized pilot trial of gene expression profiling versus heart biopsy in the first year after heart transplant. Circ. Heart Fail. 8, 557–564 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Vegas A. J., Veiseh O., Doloff J. C., Ma M., Tam H. H., Bratlie K., Li J., Bader A. R., Langan E., Olejnik K., Fenton P., Kang J. W., Hollister-Locke J., Bochenek M. A., Chiu A., Siebert S., Tang K., Jhunjhunwala S., Aresta-Dasilva S., Dholakia N., Thakrar R., Vietti T., Chen M., Cohen J., Siniakowicz K., Qi M., McGarrigle J., Graham A. C., Lyle S., Harlan D. M., Greiner D. L., Oberholzer J., Weir G. C., Langer R., Anderson D. G., Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates. Nat. Biotechnol. 34, 345–352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris A. H., Hughes K. R., Oakes R. S., Cai M. M., Miller S. D., Irani D. N., Shea L. D., Engineered immunological niches to monitor disease activity and treatment efficacy in relapsing multiple sclerosis. Nat. Commun. 11, 3871 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bushnell G. G., Hardas T. P., Hartfield R. M., Zhang Y., Oakes R. S., Ronquist S., Chen H., Rajapakse I., Wicha M. S., Jeruss J. S., Biomaterial scaffolds recruit an aggressive population of metastatic tumor Cells In vivo. Cancer Res. 79, 2042–2053 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguado B. A., Hartfield R. M., Bushnell G. G., Decker J. T., Azarin S. M., Nanavati D., Schipma M. J., Rao S. S., Oakes R. S., Zhang Y., Biomaterial scaffolds as pre-metastatic niche mimics systemically alter the primary tumor and tumor microenvironment. Adv. Healthc. Mater. 7, e1700903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oakes R. S., Bushnell G. G., Orbach S. M., Kandagatla P., Zhang Y., Morris A. H., Hall M. S., LaFaire P., Decker J. T., Hartfield R. M., Brooks M. D., Wicha M. S., Jeruss J. S., Shea L. D., Metastatic conditioning of myeloid cells at a subcutaneous synthetic niche reflects disease progression and predicts therapeutic outcomes. Cancer Res., 602–612 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corry R. J., Winn H. J., Russell P. S., Primarily vascularized allografts of hearts in mice. Transplantation 16, 343–350 (1973). [DOI] [PubMed] [Google Scholar]
- 31.Westhofen S., Jelinek M., Dreher L., Biermann D., Martin J., Vitzhum H., Reichenspurner H., Ehmke H., Schwoerer A. P., The heterotopic heart transplantation in mice as a small animal model to study mechanical unloading – Establishment of the procedure, perioperative management and postoperative scoring. PLOS ONE 14, e0214513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M. S., Tadwalkar R. V., Fearon W. F., Kirtane A. J., Patel A. J., Patel C. B., Ali Z., Rao S. V., Cardiac allograft vasculopathy: A review. Catheter. Cardiovasc. Interv. 92, E527–E536 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Ochando J., Ordikhani F., Boros P., Jordan S., The innate immune response to allotransplants: Mechanisms and therapeutic potentials. Cell. Mol. Immunol. 16, 350–356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P., The molecular signatures database hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkes M. D., Halloran P. F., Hidalgo L. G., Mechanistic sharing between NK cells in ABMR and effector T cells in TCMR. Am. J. Transplant. 18, 63–73 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Wang H.-X., Zhang Q., Zhang J., Luan R., Liang Z., Tan L., Xu Y., Zhang P., Zheng L., Zhao Y., Qiu Y.-R., CD74 regulates cellularity and maturation of medullary thymic epithelial cells partially by activating the canonical NF-κB signaling pathway. FASEB J. 35, e21535 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Higdon L. E., Tan J. C., Maltzman J. S., Infection, rejection, and the connection. Transplantation 107, 584–595 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. F. Henao-Martínez, J. G. Montoya, Infections in Heart, Lung, and Heart-Lung Transplantation. Principles and Practice of Transplant Infectious Diseases 21–39 (2018).
- 39.Beus J. M., Hashmi S. S., Selvaraj S. A., Duan D., Stempora L. L., Monday S. A., Cheeseman J. A., Hamby K. M., Speck S. H., Larsen C. P., Kirk A. D., Kean L. S., Heterologous immunity triggered by a single, latent virus in Mus musculus: Combined costimulation- and adhesion- blockade decrease rejection. PLOS ONE 8, e71221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng M. C., The AlloMap™ genomic biomarker story: 10 years after. Clin. Transplant. 31, e12900 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Liu H., Shalev I., Manuel J., He W., Leung E., Crookshank J., Liu M. F., Diao J., Cattral M., Clark D. A., Isenman D. E., Gorczynski R. M., Grant D. R., Zhang L., Phillips M. J., Cybulsky M. I., Levy G. A., The FGL2-FcγRIIB pathway: A novel mechanism leading to immunosuppression. Eur. J. Immunol. 38, 3114–3126 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Shalev I., Liu H., Koscik C., Bartczak A., Javadi M., Wong K. M., Maknojia A., He W., Liu M. F., Diao J., Winter E., Manuel J., McCarthy D., Cattral M., Gommerman J., Clark D. A., Phillips M. J., Gorczynski R. R., Zhang L., Downey G., Grant D., Cybulsky M. I., Levy G., Targeted deletion of fgl2 leads to impaired regulatory T cell activity and development of autoimmune glomerulonephritis. J. Immunol. 180, 249–260 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Ning Q., Sun Y., Han M., Zhang L., Zhu C., Zhang W., Guo H., Li J., Yan W., Gong F., Chen Z., He W., Koscik C., Smith R., Gorczynski R., Levy G., Luo X., Role of fibrinogen-like protein 2 prothrombinase/fibroleukin in experimental and human allograft rejection. J. Immunol. 174, 7403–7411 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Halloran P. F., Madill-Thomsen K. S., The molecular microscope diagnostic system: Assessment of rejection and injury in heart transplant biopsies. Transplantation 107, 27–44 (2023). [DOI] [PubMed] [Google Scholar]
- 45.Halloran P. F., Venner J. M., Madill-Thomsen K. S., Einecke G., Parkes M. D., Hidalgo L. G., Famulski K. S., Review: The transcripts associated with organ allograft rejection. Am. J. Transplant. 18, 785–795 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Harden J. T., Wang X., Toh J., Sang A. X., Brown R. A., Esquivel C. O., Martinez O. M., Krams S. M., High-resolution phenotyping of early acute rejection reveals a conserved alloimmune signature. Cell Rep. 34, 108806 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Halloran P. F., Venner J. M., Famulski K. S., Comprehensive analysis of transcript changes associated with allograft rejection: Combining universal and selective features. Am. J. Transplant. 17, 1754–1769 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Reeve J., Sellarés J., Mengel M., Sis B., Skene A., Hidalgo L., de Freitas D. G., Famulski K. S., Halloran P. F., Molecular diagnosis of t cell-mediated rejection in human kidney transplant biopsies. Am. J. Transplant. 13, 645–655 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Devarajan P., Update on mechanisms of ischemic acute kidney injury. J. Am. Soc. Nephrol. 17, 1503–1520 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Hänzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong B. W., Lymphatic vessels in solid organ transplantation and immunobiology. Am. J. Transplant. 20, 1992–2000 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Nykänen A. I., Sandelin H., Krebs R., Keränen M. A. I., Tuuminen R., Kärpänen T., Wu Y., Pytowski B., Koskinen P. K., Ylä-Herttuala S., Alitalo K., Lemström K. B., Targeting lymphatic vessel activation and CCL21 production by vascular endothelial growth factor receptor-3 inhibition has novel immunomodulatory and antiarteriosclerotic effects in cardiac allografts. Circulation 121, 1413–1422 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Dashkevich A., Raissadati A., Syrjälä S. O., Zarkada G., Keränen M. A. I., Tuuminen R., Krebs R., Anisimov A., Jeltsch M., Leppänen V.-M., Alitalo K., Nykänen A. I., Lemström K. B., Ischemia–reperfusion injury enhances lymphatic endothelial VEGFR3 and rejection in cardiac allografts. Am. J. Transplant. 16, 1160–1172 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Kingsley C. I., Nadig S. N., Wood K. J., Transplantation tolerance: Lessons from experimental rodent models. Transpl. Int. 20, 828–841 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larsen C. P., Elwood E. T., Alexander D. Z., Ritchie S. C., Hendrix R., Tucker-Burden C., Cho H. R., Aruffo A., Hollenbaugh D., Linsley P. S., Winn K. J., Pearson T. C., Long-term acceptance of skin and cardiac allografts after blocking CD40 and CD28 pathways. Nature 381, 434–438 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Wood K. J., Bushell A., Jones N. D., Immunologic unresponsiveness to alloantigen in vivo: A role for regulatory T cells. Immunol. Rev. 241, 119–132 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Sykes M., Immune tolerance: Mechanisms and application in clinical transplantation. J. Intern. Med. 262, 288–310 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Monaco A. P., Immunosuppression and tolerance for clinical organ allografts. Curr. Opin. Immunol. 1, 1174–1177 (1989). [DOI] [PubMed] [Google Scholar]
- 60.L. Gunaratnam, A. M. Jevnikar, R. B. Mannon, “Small Animal Models of Transplantation” in Textbook of Organ Transplantation, 158–184 (2014).
- 61.Belizário J. E., Immunodeficient Mouse Models: An Overview. Open J. Immunol. 2, 79–85 (2009). [Google Scholar]
- 62.Lee M. S., Zimmer R., Kobashigawa J., Long-term outcomes of orthotopic heart transplantation for hypertrophic cardiomyopathy. Transplant. Proc. 46, 1502–1505 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Naranjo M., Agrawal A., Goyal A., Rangaswami J., Neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio predict acute cellular rejection in the kidney allograft. Ann. Transplant. 23, 467–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madill-Thomsen K. S., Reeve J., Aliabadi-Zuckermann A., Cadeiras M., Crespo-Leiro M. G., Depasquale E. C., Deng M., Goekler J., Kim D. H., Kobashigawa J., Macdonald P., Potena L., Shah K., Stehlik J., Zuckermann A., Halloran P. F., Assessing the relationship between molecular rejection and parenchymal injury in heart transplant biopsies. Transplantation 106, 2205–2216 (2022). [DOI] [PubMed] [Google Scholar]
- 65.Celli S., Albert M. L., Bousso P., Visualizing the innate and adaptive immune responses underlying allograft rejection by two-photon microscopy. Nat. Med. 17, 744–749 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Lakkis F. G., Li X. C., Innate allorecognition by monocytic cells and its role in graft rejection. Am. J. Transplant. 18, 289–292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aguado B. A., Bushnell G. G., Rao S. S., Jeruss J. S., Shea L. D., Engineering the pre-metastatic niche. Nat. Biomed. Eng. 1, 0077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schroth S., Glinton K., Luo X., Thorp E. B.et al. , Front. Immunol. 11, 869 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morelli A. E., Dendritic cells of myeloid lineage. Curr. Opin. Organ Transplant. 19, 20–27 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Zhuang Q., Lakkis F. G., Dendritic cells and innate immunity in kidney transplantation. Kidney Int. 87, 712–718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amigorena S., Savina A., Intracellular mechanisms of antigen cross presentation in dendritic cells. Curr. Opin. Immunol. 22, 109–117 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Braza M. S., van Leent M. M. T., Lameijer M., Sanchez-Gaytan B. L., Arts R. J. W., Pérez-Medina C., Conde P., Garcia M. R., Gonzalez-Perez M., Brahmachary M., Fay F., Kluza E., Kossatz S., Dress R. J., Salem F., Rialdi A., Reiner T., Boros P., Strijkers G. J., Calcagno C. C., Ginhoux F., Marazzi I., Lutgens E., Nicolaes G. A. F., Weber C., Swirski F. K., Nahrendorf M., Fisher E. A., Duivenvoorden R., Fayad Z. A., Netea M. G., Mulder W. J. M., Ochando J., Inhibiting inflammation with myeloid cell-specific nanobiologics promotes organ transplant acceptance. Immunity 49, 819–828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conde P., Rodriguez M., van der Touw W., Jimenez A., Burns M., Miller J., Brahmachary M., Chen H., Boros P., Rausell-Palamos F., Yun T. J., Riquelme P., Rastrojo A., Aguado B., Stein-Streilein J., Tanaka M., Zhou L., Zhang J., Lowary T. L., Ginhoux F., Park C. G., Cheong C., Brody J., Turley S. J., Lira S. A., Bronte V., Gordon S., Heeger P. S., Merad M., Hutchinson J., Chen S.-H., Ochando J., DC-SIGN+ macrophages control the induction of transplantation tolerance. Immunity 42, 1143–1158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao S.-J., Liu H., Chen J., Qian D.-F., Kong F.-Q., Jie J., Yin G.-Y., Li Q.-Q., Fan J., Macrophage GIT1 contributes to bone regeneration by regulating inflammatory responses in an ERK/NRF2-dependent way. J. Bone Miner. Res. 35, 2015–2031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang C., Xu H., Li Y., Zhang X., Cui J., Zou Y., Yu J., Wu J., Xia J., Single-cell RNA sequencing reveals immune cell dynamics and local intercellular communication in acute murine cardiac allograft rejection. Theranostics 12, 6242–6257 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karki R., Kanneganti T.-D., ADAR1 and ZBP1 in innate immunity, cell death, and disease. Trends Immunol. 44, 201–216 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goldstein D. R., Tesar B. M., Akira S., Lakkis F. G., Critical role of the Toll-like receptor signal adaptor protein MyD88 in acute allograft rejection. J. Clin. Invest. 111, 1571–1578 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang Q., Wang X., Li H., Yin X., Liu H., Hu W., Qing Y., Ding L., Yang L., Li Z., Sun H., Integrative analysis of renal microRNA and mRNA to identify hub genes and pivotal pathways associated with cyclosporine-induced acute kidney injury in mice. Hum. Exp. Toxicol. 42, 9603271231215499 (2023). [DOI] [PubMed] [Google Scholar]
- 79.Wang G., Kong G., Li X. C., Adaptive features of innate immune cells and their relevance to graft rejection. Curr. Opin. Organ Transplant. 24, 664–669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Staal F. J. T., Arens R., Wnt signaling as master regulator of T-lymphocyte responses. Transplantation 100, 2584–2592 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Rekers N. V., de Fijter J. W., Claas F. H. J., Eikmans M., Mechanisms and risk assessment of steroid resistance in acute kidney transplant rejection. Transpl. Immunol. 38, 3–14 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Bonezi V., Genvigir F. D. V., de Cássia Salgado P., Felipe C. R., Tedesco-Silva H. Jr., Medina-Pestana J. O., Cerda A., Doi S. Q., Hirata M. H., Hirata R. D. C., Differential expression of genes related to calcineurin and mTOR signaling and regulatory miRNAs in peripheral blood from kidney recipients under tacrolimus-based therapy. Ann. Transl. Med. 8, 1051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yen N. T. H., Phat N. K., Oh J.-H., Park S.-M., Moon K.-S., Thu V. T. A., Cho Y.-S., Shin J.-G., Long N. P., Kim D. H., Pathway-level multi-omics analysis of the molecular mechanisms underlying the toxicity of long-term tacrolimus exposure. Toxicol. Appl. Pharmacol. 473, 116597 (2023). [DOI] [PubMed] [Google Scholar]
- 84.Zhao Z., Wang L., Yang C., Zhao T., Li L., Hu L., Wu D., Rong R., Xu M., Zhu T., Soluble FGL2 induced by tumor necrosis factor-α and interferon-γ in CD4+ T cells through MAPK pathway in human renal allograft acute rejection. J. Surg. Res. 184, 1114–1122 (2013). [DOI] [PubMed] [Google Scholar]
- 85.Dormond O., Dufour M., Seto T., Bruneau S., Briscoe D. M., Targeting the intragraft microenvironment and the development of chronic allograft rejection. Hum. Immunol. 73, 1261–1268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vitalone M., Sigdel T. K., Salomonis N., Sarwal R. D., Hsieh S.-C., Sarwal M. M., Transcriptional perturbations in graft rejection. Transplantation 99, 1882–1893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Curci C., Sallustio F., Serino G., De Palma G., Trpevski M., Fiorentino M., Rossini M., Quaglia M., Valente M., Furian L., Toscano A., Mazzucco G., Barreca A., Bussolino S., Gesualdo L., Stratta P., Rigotti P., Citterio F., Biancone L., Schena F. P., Potential role of effector memory T cells in chronic T cell-mediated kidney graft rejection. Nephrol. Dial. Transplant. 31, 2131–2142 (2016). [DOI] [PubMed] [Google Scholar]
- 88.Xiu M., Liu Z., Tang J., Screening and identification of key regulatory connections and immune cell infiltration characteristics for lung transplant rejection using mucosal biopsies. Int. Immunopharmacol. 87, 106827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Booth A. J., Grabauskiene S., Wood S. C., Lu G., Burrell B. E., Bishop D. K., IL-6 Promotes Cardiac Graft Rejection Mediated by CD4+ Cells. J. Immunol. 187, 5764–5771 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wan S., Xia C., Morel L., IL-6 produced by dendritic cells from lupus-prone mice inhibits CD4+CD25+ T cell regulatory functions. J. Immunol. 178, 271–279 (2007). [DOI] [PubMed] [Google Scholar]
- 91.Benway C. J., Iacomini J., Defining a microRNA-mRNA interaction map for calcineurin inhibitor induced nephrotoxicity. Am. J. Transplant. 18, 796–809 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Vasudevan P., Wolfien M., Lemcke H., Lang C. I., Skorska A., Gaebel R., Koczan D., Lindner T., Engelmann R., Vollmar B., Krause B. J., Wolkenhauer O., Lang H., Steinhoff G., David R., Cardiomyocyte transplantation after myocardial infarction alters the immune response in the heart. Cells 9, 1825 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tarazón E., Pérez-Carrillo L., García-Bolufer P., Triviño J. C., Feijóo-Bandín S., Lago F., González-Juanatey J. R., Martínez-Dolz L., Portolés M., Roselló-Lletí E., Circulating mitochondrial genes detect acute cardiac allograft rejection: Role of the mitochondrial calcium uniporter complex. Am. J. Transplant. 21, 2056–2066 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pereboom I. T. A., Adelmeijer J., Van Leeuwen Y., Hendriks H. G. D., Porte R. J., Lisman T., Development of a severe von Willebrand factor/ADAMTS13 dysbalance during orthotopic liver transplantation. Am. J. Transplant. 9, 1189–1196 (2009). [DOI] [PubMed] [Google Scholar]
- 95.Ko S., Okano E., Kanehiro H., Matsumoto M., Ishizashi H., Uemura M., Fujimura Y., Tanaka K., Nakajima Y., Plasma ADAMTS13 activity may predict early adverse events in living donor liver transplantation: Observations in 3 cases. Liver Transpl. 12, 859–869 (2006). [DOI] [PubMed] [Google Scholar]
- 96.Covarrubias M., Ware L. B., Kawut S. M., De Andrade J., Milstone A., Weinacker A., Orens J., Lama V., Wille K., Bellamy S., Shah C., Demissie E., Christie J. D.; Lung Transplant Outcomes Group , Plasma intercellular adhesion molecule-1 and von Willebrand factor in primary graft dysfunction after lung transplantation. Am. J. Transplant. 7, 2573–2578 (2007). [DOI] [PubMed] [Google Scholar]
- 97.Goodwin C. M., Ciesla J. H., Munger J., Who’s driving? Human cytomegalovirus, interferon, and NFκB signaling. Viruses 10, 447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu J., Yan J., Rao G., Latha K., Overwijk W. W., Heimberger A. B., Li S., The duality of Fgl2 - secreted immune checkpoint regulator versus membrane-associated procoagulant: Therapeutic potential and implications. Int. Rev. Immunol. 35, 325–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Patterson D. M., Telischi F. F., Connell S. S., Ulubil S. A., Hodges A. V., Eshraghi A. A., Balkany T. J., Cochlear implantation in organ transplantation. Laryngoscope 118, 116–119 (2008). [DOI] [PubMed] [Google Scholar]
- 100.Ayub B., Young N. M., Cochlear implantation of solid organ transplant patients receiving immunosuppressive therapy. Int. J. Pediatr. Otorhinolaryngol. 91, 19–22 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Testa G., Goldstein R. M., Toughanipour A., Abbasoglu O., Jeyarajah D. R., Levy M. F., Husberg B. S., Gonwa T. A., Klintmalm G. B., Guidelines for surgical procedures after liver transplantation. Ann. Surg. 227, 590–599 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bucci J. R., Oglesby R. J., Agodoa L. Y., Abbott K. C., Hospitalizations for total hip arthroplasty after renal transplantation in the united states. Am. J. Transplant. 2, 999–1004 (2002). [DOI] [PubMed] [Google Scholar]
- 103.Chalmers B. P., Ledford C. K., Statz J. M., Perry K. I., Mabry T. M., Hanssen A. D., Abdel M. P., Survivorship after primary total hip arthroplasty in solid-organ transplant patients. J. Arthroplasty 31, 2525–2529 (2016). [DOI] [PubMed] [Google Scholar]
- 104.Sayed-Noor A. S., Joint arthroplasties other than the hip in solid organ transplant recipients. Open Orthop. J. 3, 27–31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Varga M., Matia I., Kucera M., Oliverius M., Adamec M., Polypropylene mesh repair of incisional hernia after kidney transplantation: Single-center experience and review of the literature. Ann. Transplant. 16, 121–125 (2011). [DOI] [PubMed] [Google Scholar]
- 106.Koonce S. L., Giles B., McLaughlin S. A., Perdikis G., Waldorf J., Lemaine V., TerKonda S., Breast reconstruction after solid organ transplant. Ann. Plast. Surg. 75, 343–347 (2015). [DOI] [PubMed] [Google Scholar]
- 107.Jones D. G., Mortsell D. H., Rajaruthnam D., Hamour I., Hussain W., Markides V., Banner N. R., Wong T., Permanent pacemaker implantation early and late after heart transplantation: Clinical indication, risk factors and prognostic implications. J. Heart Lung Transplant. 30, 1257–1265 (2011). [DOI] [PubMed] [Google Scholar]
- 108.Paredes V., López-Pintor R. M., Torres J., de Vicente J. C., Sanz M., Hernández G., Implant treatment in pharmacologically immunosuppressed liver transplant patients: A prospective-controlled study. Clin. Oral Implants Res. 29, 28–35 (2018). [DOI] [PubMed] [Google Scholar]
- 109.Lew J., Sheeder J., Lazorwitz A., Etonogestrel contraceptive implant uptake and safety among solid organ transplant recipients. Contraception 104, 556–560 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McIntosh T., Puerzer P., Li M. T., Malat G., Sammons C., Norris M., Fallah T., Trofe-Clark J., DuBois J. M., Iltis A., Mohan S., Sawinski D., A survey of solid organ transplant recipient attitudes and concerns regarding contraception and pregnancy. Clin. Transplant. 37, e14948 (2023). [DOI] [PubMed] [Google Scholar]
- 111.E. Schmidt, S. L. Pachtman, J. T. Diedrich, “Contraception in Chronic Kidney Disease and Renal Transplantation” in Obstetric and Gynecologic Nephrology: Women’s Health Issues in the Patient With Kidney Disease, 225–243 (2020).
- 112.Agarwal K. A., Pavlakis M., Sexuality, contraception, and pregnancy in kidney transplantation. Kidney Med. 3, 837–847 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Capuani S., Malgir G., Chua C. Y. X., Grattoni A., Advanced strategies to thwart foreign body response to implantable devices. Bioeng Transl. Med. 7, e10300 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meng H., Liang Y., Hao J., Lu J., Comparison of rejection-specific genes in peripheral blood and allograft biopsy from kidney transplant. Transplant. Proc. 50, 115–123 (2018). [DOI] [PubMed] [Google Scholar]
- 115.Rao S. S., Bushnell G. G., Azarin S. M., Spicer G., Aguado B. A., Stoehr J. R., Jiang E. J., Backman V., Shea L. D., Jeruss J. S., Enhanced survival with implantable scaffolds that capture metastatic breast cancer cells in vivo. Cancer Res. 76, 5209–5218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shen H., Heuzey E., Mori D. N., Wong C. K., Colangelo C. M., Chung L. M., Bruce C., Slizovskiy I. B., Booth C. J., Kreisel D., Goldstein D. R., Haptoglobin enhances cardiac transplant rejection. Circ. Res. 116, 1670–1679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schwoebel F., Barsig J., Wendel A., Hamacher J., Quantitative assessment of mouse skin transplant rejection using digital photography. Lab. Anim. 39, 209–214 (2005). [DOI] [PubMed] [Google Scholar]
- 118.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Breiman L., Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 120.Durinck S., Spellman P. T., Birney E., Huber W., Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mootha V. K., Lindgren C. M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C., PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 122.Grimes T., Potter S. S., Datta S., Integrating gene regulatory pathways into differential network analysis of gene expression data. Sci. Rep. 9, 5479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen J., Deng J. C., Zemans R. L., Bahmed K., Kosmider B., Zhang M., Peters-Golden M., Goldstein D. R., Age-induced prostaglandin E2 impairs mitochondrial fitness and increases mortality to influenza infection. Nat. Commun. 13, 6759 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.De Fazio S. R., Masli S., Gozzo J. J., Effect of monoclonal anti-cd4 and anti-cd8 on skin allograft survival in mice treated with donor bone marrow cells. Transplantation 61, 104–110 (1996). [DOI] [PubMed] [Google Scholar]
- 125.Mai H.-L., Boeffard F., Longis J., Danger R., Martinet B., Haspot F., Vanhove B., Brouard S., Soulillou J.-P., IL-7 receptor blockade following T cell depletion promotes long-term allograft survival. J. Clin. Invest. 124, 1723–1733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Tables S1 to S4
Legends for data S1 to S7
Data S1 to S7