Abstract
Technological breakthroughs in genomics have had a significant impact on clinical therapy for human diseases, allowing us to use patient genetic differences to guide medical care. The “synthetic lethal approach” leverages on cancer-specific genetic rewiring to deliver a therapeutic regimen that preferentially targets malignant cells while sparing normal cells. The utility of this system is evident in several recent studies, particularly in poor prognosis cancers with loss-of-function mutations that become “treatable” when two otherwise discrete and unrelated genes are targeted simultaneously. This review focuses on the chemotherapeutic targeting of epigenetic alterations in cancer cells and consolidates a network that outlines the interplay between epigenetic and genetic regulators in DNA damage repair. This network consists of numerous synergistically acting relationships that are druggable, even in recalcitrant triple-negative breast cancer. This collective knowledge points to the dawn of a new era of personalized medicine.
Keywords: Synthetic lethality, Epigenetics, Precision medicine, Cancers, Gene network
Introduction
Anti-cancer chemotherapy reached a critical juncture in recent years, with the realization that subtle genetic variations could be leveraged to create better and more targeted therapies, with improved patient care and fewer adverse effects. The advent of powerful genetic tools, such as next-generation sequencing, allowed for correlations between chemotherapeutic responses and specific genetic backgrounds. Chemotherapeutic agents to this point—developed more than 50 years ago—were first-generation drugs that preferentially targeted actively proliferating cells, which were presumed to be cancerous. This presumption was based on the knowledge that most normal somatic cells are predominantly quiescent, with the exception of progenitor cells at sites subjected to constant abrasion, such as cells in the skin, hair follicles, bone marrow, and digestive tracts, which require continual replacement. Consequently, these normal, highly proliferative regions are also targeted by anti-cancer drugs, leading to significant side effects for the patient.
This awareness, along with the knowledge that the “one-size-fits-all” approach to cancer treatment was not universally beneficial [1, 2], led to more recent objectives toward personalized or precision medicine. This coincided with the advent of powerful genetic tools, such as next-generation sequencing, which provided opportunities to correlate chemotherapeutic responses with specific patient genetic backgrounds. Indeed, the identification of specific genetic polymorphisms in cancers were proposed to serve as not only prognostic or diagnostic markers, but as targets for cancer treatment regimens [3, 4]. Recognizing the power of this approach, various medical establishments initiated consorted efforts to streamline genomic acquisition toward precision medicine for chemotherapeutic success [5, 6].
One of the most significant discoveries in personalized medicine arose from the findings of oncogenic addiction. First proposed almost two decades ago [7], oncogenic addiction describes how tumors rely on cancer-specific, oncogenic proteins that arise from genetic instability events for their survival and growth. For example, in most patients with chronic myelogenous leukemia (CML; [8]), chromosomal translocation between chromosomes 9 and 22 leads to the formation of a new chromosome 22 (Philadelphia chromosome), which contains the BCR-ABL fusion gene [9]. Chromosomal translocations occur in response to numerous DNA double strand breaks (DSBs) that are misjoined rather than repaired, resulting in the formation of aberrant chromosomes. The later development of the Imatinib tyrosine kinase inhibitor against the BCR-ABL fusion product revolutionized the field, with significant improvement in patient responses and survival rates, raising the hope that protein kinase inhibitors would also act as “magic bullets” for other cancer types. However, aberrations deriving from chromosomal changes account for only a small fraction of malignancies [10–12], and alternative approaches are required for most patients.
The synthetic lethality approach in personalized medicine
About 100 years ago, the study of gene–gene interactions in budding yeasts and fruit flies revealed a phenomenon referred to as “synthetic lethality” [13], which described a loss of cell viability following the concurrent disruption of synergistically acting genes, but not either gene alone [13, 14]. The concept of synthetic lethality has since found applications in cancer therapy, taking advantage of the changes in cellular rewiring that occur in response to altered or mutated gene expression to invoke new vulnerabilities. Conceptually, the same addiction/dependency unique to cancer cells that presents through genetic rewiring can be leveraged to kill the cancer cells, posing minimal side effects to the normal cells that lack these rewired networks or mutations. This is best exemplified in BRCA1/BRCA2-deficient cells in ovarian and breast cancers. These BRCA-deficient cells are defective in homologous recombination (HR) and must rely on the less-stringent non-homologous end joining (NHEJ) for the repair of DNA DSBs. This dependency renders the cells susceptible to inhibitors against poly (ADP-ribose) polymerase (PARP), a nuclear enzyme that aids in the detection of DNA damage and is involved in end-joining, including NHEJ [15, 16]. The loss of both genes results in an accumulation of replication defects and causes cell death. The successes that ensued from PARPi in BRCA-deficient cancer cells led to the search for other targeted approaches against a range of gain-of-function and loss-of-function mutations hitherto considered to be undruggable [14].
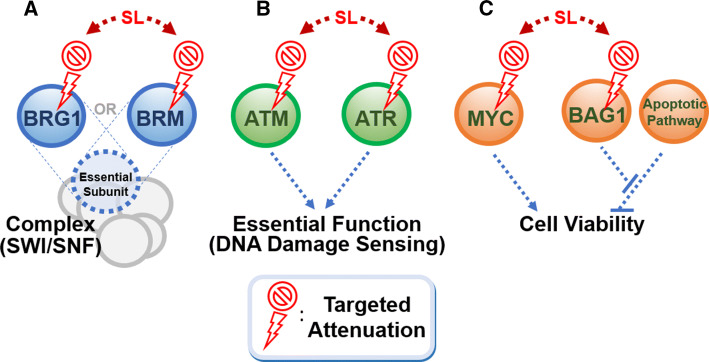
There are three key scenarios where the synthetic lethal approach can be leveraged (Fig. 1): (1) where two genes are exclusive, key members of an essential protein complex. For example, BRG1 and BRM are two mutually exclusive catalytic subunits of the chromatin remodeling switch/sucrose non-fermentable (SWI/SNF) complex, which is increasingly implicated in cell survival in several cancer types [17, 18]. BRM and BRG1 are frequently inactivated in kidney, ovarian, and lung cancers, and a recent study shows that targeting BRG1 in BRM-deficient cells in lung cancer is synthetic lethal, reminiscent of the BRCA/PARP scenario in breast and ovarian cancers (Fig. 1a). (2) Where there is a dependency on a specific pathway for survival following an inactivating mutation that occurs in a parallel regulatory mechanism (Fig. 1b). For example, ataxia telangiectasia (ATM) and ataxia telangiectasia and Rad3-related (ATR) both transmit DNA damage signals to activate a checkpoint kinase [19, 20]. Loss-of-function mutations in ATM are commonly found in cancers, and predispose cells to uncontrolled growth [21]. Treatment of ATM-deficient cells with ATR inhibitors can lead to synthetic lethality in lung adenocarcinoma, gastric cancer and mantel cell lymphoma [22–24]. (3) Finally, where a repressor protein keeps an anti-survival pathway in check. For example, BAG1 keeps in check the route to MYC-induced apoptosis. Thus, its downregulation in conjunction with MYC overexpression would induce apoptosis (Fig. 1c) [25].
Fig. 1.
Genetic interactions that can contribute to a synthetic lethal relationship. The concurrent inactivation of two factors in the genetic relationship can result in cumulative functional inactivation or cell death. a BRG1 and BRM are mutually exclusive subunits of a complex. b ATR and ATM act synergistically to sense DNA damage, an important process that maintains genomic stability in the cell. c The repressor protein BAG1 inhibits an anti-proliferative apoptotic mechanism that counteracts proliferation induced by the Myc oncoprotein
Yet, identifying which targets can be used in synthetic lethal combinations is not straightforward. There has been much effort invested into performing synthetic lethal screens using siRNA libraries to search for targetable factors. One example is TAK1/MAP3K7 kinase, which was identified through a siRNA-mediated screen for factors that, when downregulated, enhanced the potency of the topoisomerase I inhibitor, camptothecin. The downregulation of TAK1 resulted in breast cancer cell death in conjunction with the LMP-400 Top1 inhibitor [26]. However, screens are costly and can be technically challenging. Model organisms such as yeast are also used as an alternative first-line screening option [27, 28]. Indeed, using the fission yeast model organism, we previously showed that vacuolar ATPase acts alongside the ABC drug-transporter multidrug resistance protein 1 (MDR1; also known as p-glycoprotein) to sensitize cells to doxorubicin, a topoisomerase II inhibitor [29, 30]. Doxorubicin also induces cell death when delivered with a histone deacetylase inhibitor in fission yeast [28]. Although DNA damage response pathways remain one of the most—if not the most—useful pathways for inducing synthetic lethality in cancer cells, recent findings point to leveraging the co-operation between epigenetic regulators of chromatin architecture and canonical cancer-related signaling mechanisms to induce cell death.
Epigenetic dysregulation in cancer
Epigenetic regulation involves genomic alterations that are independent of changes in DNA sequences [31, 32]. The genome can be epigenetically regulated through chemical modifications to the scaffolding histone proteins [33] and DNA nucleotide bases [34]; through altered nucleosomal spacing [35, 36]; and via post-translational regulation of transcribed templates, mediated by RNA interference (RNAi) mechanisms with non-coding small RNAs (e.g., microRNAs [miRNAs] or small interference-RNA [siRNA]) [37, 38]. Long non-coding RNA (LncRNA) can also affect the localization and activity of chromatin enzymatic complexes in conjunction with histone and DNA modifications [39–41]. These epigenetic regulations, in turn, affect DNA metabolic pathways such as gene transcription, chromosomal segregation mechanisms, and DNA replication, recombination, and the damage detection/repair [31, 33, 35].
Except when mutated, DNA sequences remain unchanged. In contrast, the epigenetic status can be remodeled in accordance with environmental cues and growth signals [42, 43], which are stably and faithfully maintained across cell generations [44–46]. This level of plasticity makes epigenetic regulation ideal to maintain developmental fate, as observed in dosage compensation, X-inactivation, and genomic imprinting [31]. Epigenetic dysregulation is thus often associated with or drives the development of human disease, particularly cancer, with different stages of oncogenesis liable to epigenetic control [47, 48]. Epigenetic abnormalities may, therefore, underlie cancer-specific phenotypes and represent targetable, molecular vulnerabilities for cancer therapy using the synthetic lethality approach.
Epigenetic aberrations caused by gene fusion (chromosomal translocation) can give rise to tumor-specific fusion products and resemble cancers with gain-of-function mutations that lead to oncogenic addiction. For instance, fusion between the transcriptional activation domain of NUP98 with the methylated histone H3 lysine 4 (H3K4)-binding domain of the H3K4 demethylase JARID1 underlies a subset of acute myeloid leukemia [49, 50], causing aberrant transcription of the homeobox genes that maintain stemness in bone marrow cells. Like many other fusion products and gain-of-function mutations, fusion events between epigenetic regulators are rare, and this has discouraged the pursuit of inhibitors for therapies against such aberrations. Yet, recurrent loss-of-function mutations in epigenetic regulators, particularly in histone modifiers and chromatin remodeling factors [51–57], have been useful in the stratification of tumors [58, 59] and employed in synthetic lethality approaches to target cancer cell viability. Below, we will explore how chromatin remodeling and histone modification pathways have been targeted in the treatment of cancer using the synthetic lethality approach.
Epigenetic dysregulation as a basis for synthetic lethality
Chromatin remodeling
The chromatin structure—repeating nucleosome units of DNA wound around histone proteins—represents the first-line of defense against agents that threaten to undermine the integrity of DNA. Indeed, the loss of nucleosomal integrity leads to a substantial increase in chromosomal breaks [60, 61]. Yet, DNA sequences must be readily accessible to interact with protein machineries during replication, repair and recombination.
DNA damage can be caused by a range of exogenous (UV exposure, ionizing radiation, chemical exposure and cytotoxic drugs) and endogenous (errors in replication, spontaneous deamination, oxidation and methylation changes) factors, and the cell employs various processes to circumvent or repair damage. Chromatin remodeling complexes, for example, hydrolyze ATP to overcome the energy barrier to slide, reposition via disassembly, and replace histone octamers on the DNA template [35, 36]. The SWI/SNF complex is one of several conserved chromatin-modifying complexes that uses ATP hydrolysis to mobilize nucleosomes and remodel chromatin. Mutations in the SWI/SNF complex have been found in genomic studies in multiple cancers [62, 63]. First identified in budding yeast and subsequently shown to be conserved in human cells, the SWI/SNF complex hosts two mutually exclusive DNA-dependent ATPases: BRG1/SMARCA4 and Brahma/BRM/SMARCA2 [35, 36]. The loss of BRM or BRG1 is commonly found in kidney, ovarian, and lung cancers [63–66]. Recent ChIP-seq efforts revealed colocalization of BRM and BRG1 on overlapping set of genes in the TNFα–NFκB pathway. These genes are transcriptionally co-regulated by SWI/SNF factors [67], and are targeted by hypoxia-induced transcription factor [68] and growth factors [69]. BRM and BRG1 can also differentially interact with RB and p53, checkpoint proteins that regulate progression within the cell cycle [70, 71].
A recent unbiased shRNA screen in > 50 cancer cell lines showed that BRG1/SMARCA4-mutant cancer cells are highly sensitive to BRM/SMARCA2 depletion [18]. Indeed, in BRG1-deficient non-small cell lung cancer (NSCLC) cells, BRM depletion can attenuate cell growth [17]. BRG1 and BRM thus likely constitute mutually exclusive catalytic subunits of different sub-populations of the SWI/SNF complex required for essential cellular processes, such as transcription [67, 72] (Fig. 2).
Fig. 2.
Mutually exclusive ‘sibling’ subunits; for example, within SWI/SNF chromatin remodeling and polycomb repressive complex, PRC2. a SWI/SNF complex contains either one of the ATPase counterparts BRG1 or BRM; or b ARID1A or ARID1B. c PRC2 complex contains either one of the two enhancer of zeste homologues EZH1 or EZH2 catalytic subunits. Concurrent downregulation of both ‘sibling’ factors renders the complex inactive and results in synthetic lethality (SL)
The SWI/SNF complex also contains the AT-rich interactive domain 1A and 1B (ARID1A and ARID1B) subunit pair, which are also implicated in cancer. Like BRM and BRG1, ARID1A and ARID1B also seem to be alternatively expressed and mutually exclusive [73]. ARID1A is frequently mutated in cancers, with ~ 57% of ovarian clear cell carcinomas (OCCC) associated with ARID1A mutations [63, 74–76]. ARID1B, on the other hand, is associated with only minor perturbations to chromatin accessibility following its knockdown in colorectal cancer cells [77], and mutations have been detected in liver cancer, neuroblastoma, and melanoma [55, 78, 79]. There is also a report of the co-occurrence of ARID1A and ARID1B mutations in ovarian cancer [80]. Although ARID1B plays a less significant role in the SWI/SNF complex, it compensates for the absence of ARID1A function, and thus presents a specific vulnerability in cancers with ARID1A mutations [81]. Knocking down ARID1B in ARID1A-deficient cells increases chromatin accessibility, especially at enhancer sequences, to control the binding of transacting factors [77]. The mutually exclusive occurrence of ARID1A and ARID1B, therefore, results in specific “subtypes” of the SWI/SNF complex that control nucleosomal spacing, which is essential for the control of cellular events, such as the transcription of important genes; these subtypes of SWI/SNF complexes (containing different ARID1 subunits) could, therefore, be leveraged for synthetic lethal targeting (Fig. 2).
Modifying enzymes of histones
Histone H3 lysine 27 methylation
Enhancer of zeste homolog 2 (EZH2) is the catalytic subunit of the polycomb repressive complex 2 (PRC2), a histone-lysine N-methyl transferase that primarily trimethylates histone H3 at lysine 27 (H3 K27me) to silence developmental genes in metazoans [82]. Mutations in EZH2 have been detected in multiple cancers and are associated with poor prognosis, whereas mutations in the core components of the PRC2 (EED and SUZ12) are found in nerve sheath tumors [83–92]. Previous work has shown that inhibition of EZH2 activity and downregulation of EED and SUZ12 can counter tumor growth; this strongly suggests an oncogenic driver role for the PRC2 complex [93, 94], and EZH2 as a potential therapeutic target [93, 95–98]. Furthermore, the dual inhibition of EZH1 and EZH2—as mutually exclusive catalytic subunits of PRC2—offers greater anti-tumorigenicity than inhibiting EZH2 alone [99] (Fig. 2).
OCCC is an aggressive form of ovarian cancer that shows poor prognosis and is refractive to the canonical cisplatin-based chemotherapeutic regimens [100, 101]. Sequencing of OCCC revealed that up to 57% of tumors bear ARID1A mutations, and a shRNA-based screen further showed that inhibition of EZH2 can destabilize ARID1A-deficient OCCC by suppressing the PI3K-AKT pathway to reduce cell growth and induce apoptosis [76]. This synthetic lethal strategy advances the therapeutic hope surrounding this largely incurable cancer. The same approach of targeting EZH2 can be used in cancers that are deficient in other subunits of the SWI/SNF complex [102] (Fig. 2). For example, EZH2 inhibition can significantly increase the susceptibility of BRG1-deficient lung cancer cells to a topoisomerase II inhibitor [103].
H3K27me recruits a PRC1 complex comprising BMI1 (B cell specific, Moloney murine leukemia virus integration site 1). This subunit is implicated in stem cell renewal and may act as a cancer-initiating factor because of its telomerase-activating and senescence-suppressing activities. Recent work [104, 105] notes that the concurrent downregulation of BMI1 and EZH2 can be used against glioblastoma tumors, suggesting that PRC1 and PRC2 may not simply act sequentially in the same epistatic pathway, but are involved in some non-overlapping roles as part of a much more complicated network.
Histone H3 lysine 36 methylation
Histone H3 lysine 36 methylation (H3K36me) facilitates a wide range of cellular processes, including transcription and splicing, and recent studies have focused on the connection between this histone modification and the detection and repair of DNA damage [106, 107]. The histone methyltransferase SETD2, which trimethylates H3K36, is commonly mutated in cancer [57, 108–110], suggesting a tumor suppressor role for the protein [107]. It also has potential prognostic value in cancers like gastric cancer, renal cancer, and leukemia [111–113]. Targeting other loss-of-function mutations in association with a SETD2 mutation, while challenging, could be approached using synthetic lethality. Pfister et al. reported that H3K36me-deficient cancer cells and SETD2-attenuated xenografts show preferential susceptibility toward an inhibitor of the Wee1 kinase, which suppresses cyclin-dependent kinase (CDK). SETD2 disruption downregulates the RRM2 ribonucleotide reductase (RNR) subunit, which, when combined with Wee1 inhibition (activating CDK and repressing RRM2), causes cells to enter into and arrest in S-phase. Cells that linger in S-phase are induced to undergo apoptosis [114] (Fig. 3).
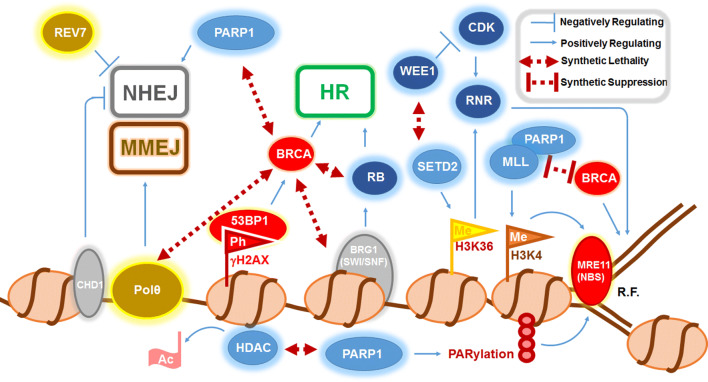
Fig. 3.
The genetic–epigenetic interplay involved in cell cycle regulation and DNA damage repair. Cell cycle regulators are in dark blue (WEE1, CDK, RNR, RB), DNA repair factors in red (BRCA, 53BP1, MRE11, NBS); chromatin remodeling factors in grey (CHD1, BRG1), DNA polymerase-linked factors in yellow (Polθ and REV7), nucleosomes in beige, and chromatin-modifying proteins in light blue (HDAC, PARP1, SETD2, MLL) Flags indicate histone modifications that include phosphorylation (Ph) of histone H2AX (γH2AX), methylation (Me) of histone H3 lysine 4 (H3K4) and histone H3 lysine 36 (H3K36). Many regulators of homologous recombination repair (HR) and non-homologous end joining (NHEJ) and microhomology end joining (MMEJ) show synthetic lethal relationship with BRCA proteins. Abbreviations: R.F. replication fork; Ac acetyl group; RNR ribonucleotide reductase; CDK cyclin-dependent kinase; RB retinoblastoma protein; HDAC histone deacetylase; PARP1 poly (ADP) ribose polymerase 1; MLL mixed lineage leukemia; NBS Nijmegen Breakage Syndrome; Polθ polymerase θ. Double-headed maroon lines indicate interconnecting factors that showed synthetic lethality or suppression upon downregulation. Blue lines represent an induction (positively regulating) and repression (negatively regulating)
SETD2 also methylates other targets that affect cancer development; for example, the oncogenic signaling factor STAT1 [115] and tubulin for cytoskeleton remodeling [116, 117]. These findings add more complexity—but also opportunities—to exploit SETD2 inhibition in the synthetic lethal targeting of cancers.
Histone H2AX phosphorylation and other chromatin modulations in DNA damage response—PARP and BRCA at center stage
The formation of DNA DSBs is the most detrimental type of DNA damage that can threaten genomic stability and cell viability. Upon detection of a break, cells activate signal transduction via the phosphorylation of histone H2AX (γH2AX) primarily by two DNA damage checkpoint signaling kinases—ATM and ATR. γH2AX is detectable over a long stretch of chromatin encompassing the break site, and this is thought to amplify the damage signal and recruit DNA damage repair factors [118]. ATM and ATR exist synergistically, and their relationship can be exploited to induce synthetic lethality when following attenuation of other DNA damage repair pathway regulators, such as topoisomerase I and DNA polymerase δ [119–121].
Through chemical screens for γH2AX-interacting factors, several studies have identified that both 53BP1, a DSB repair factor, and MDC1, a signaling kinase substrate of ATM, interact with γH2AX via their BRCT domains [122–124]. These factors direct the recruitment of downstream DNA repair factors—BRCA1, Rad51 and the NBS component, Mre11—to regulate HR repair [125–128]. 53BP1, however, is also associated with NHEJ at specific chromosomal loci, and it appears that stabilization of 53BP1 is associated with an increase in NHEJ, which is observed when the chromatin remodeler CHD1 is downregulated in prostate cancer cells [129, 130] (Fig. 3).
Even though direct targeting of γH2AX is not commonly employed in cancer strategies, the loss of its downstream effector, BRCA, predisposes cells to become susceptible to PARPi, and this understanding exposes the prominent “Achilles’ heel” in ovarian and breast cancers that could similarly be exploited for other cancers. Indeed, a similar synergistic relationship is apparent for Rad51C with PARPi [15, 16, 131]. Thus, the use of PARPi in situations with HR attenuation is currently actively translated for clinical treatments in combination with conventional DNA damaging chemotherapeutics [132–134].
Unfortunately, despite the success of PARPi in BRCA-deficient cells, numerous parallel signaling pathways in cancer cells can confer chemoresistance toward PARPi. For example, activation of a backup DNA end resection pathway can bypass the early step of Rad51 recruitment coordinated by BRCA proteins [135, 136]. One of these is the repression of Rad51 accumulation by REV7, a translesion synthesis (TLS) polymerase ζ component, which is recruited by γH2AX independently of polζ, but requires a physical interaction with 53BP1. Enhancing the role of REV7 downplays HR and, in combination with PARPi, results in synthetic lethality thus proposing the usefulness of an agonist of REV7 to effect synthetic lethal targeting in cancer cells [137, 138] (Fig. 3).
HR repair also acts in parallel with the error-prone microhomology-mediated end joining (MMEJ) or alternative NHEJ (alt-NHEJ, or alternative end joining [alt-EJ]) catalyzed by polymerase θ (Polθ), which is encoded by the POLQ gene [134, 139–144]. Polθ downregulation results in a heavier reliance on HR activity through the release of RAD51 protein, which can be sequestered upon physical binding to a RAD51-binding motif on Polθ [142]. Consistently, the loss of Polθ function in HR-deficient epithelial ovarian cancer cells and BRCA−/− mouse embryonic fibroblasts results in synthetic lethality [141, 142]. Translocation of Polθ on chromatin can also facilitate the removal of single-stranded DNA-stabilizing Replication Protein A (RPA) complex from resected DSBs to expose stretches of homology for annealing and subsequent joining by MMEJ [145]. Thus, in cells with altered HR activity, MMEJ can overcome the effect of PARPi. However, the additional use of a Polθ inhibitor could potentially prevent this route of chemoresistance (Fig. 3).
Unexpectedly, the simultaneous loss of PARP and BRCA function can lead to synergistic viability [146, 147]. A considerable proportion of the damage found in BRCA-deficient cells is due to disrupted replication fork stability, which can be strengthened by preventing the recruitment of MRE11 to degrade nascent DNA at the fork. This is sufficient to restore viability to BRCA-deficient cells even in the presence of PARPi and platinum-based agents. However, MRE11 recruitment relies on several histone modifications at the replication fork, including H3K4me and poly ADP-ribosylation (PARylation) [148, 149]. Therefore, the downregulation of MLL3/4 H3K4 methyltransferase complex combined with PARPi treatment can suppress the growth defect in BRCA-deficient cells [146, 147] (Fig. 3). As a result, the sequence of inactivation of PARP and BRCA function is important, rendering either a synthetic lethal (BRCA inactivation before PARP inactivation) or rescue phenotype [147]. This observation profoundly impacts the chemotherapeutic application of PARPi. Although the inhibitor would be efficacious in a total loss-of-function BRCA−/− background, most BRCA mutations do not result in total loss of function. Thus, the pharmacokinetics of PARPi and the administration sequence must be carefully considered for the efficacy of this approach and, in some cases, BRCA function must be downregulated before that of PARP. Furthermore, the extent of functional impairment to BRCA genes is conceptually important, as a deletion that removes the bulk of its BRCT and DNA binding domains still retains HR competency. Thus, the type of BRCA mutation in patients may reliably predict the efficacy of PAPRi-based regimens [150].
Histone acetylation will also affect the efficacy of PARPi. In vitro experiments have shown that PARylation can occur preferentially on acetylated histones [149]. Histone acetyltransferases (HAT) and deacetylases (HDAC) play essential roles not only in gene transcription (e.g., in the transcription of the chief oncogene, MYC [151]), but also in DNA damage repair, and inhibitors of these enzymes can induce cancer cell death. HDAC inhibitors (HDACi), such as suberoylanilide hydroxamic acid (SAHA), are approved for chemotherapy against cutaneous lymphoma [152–154]. HDACi sensitize cancer cells toward many DNA damaging chemotherapeutic drugs, presumably by inducing an opened chromatin conformation to facilitate DNA access or by impeding cell cycle progression by disrupting DNA replication integrity. Thus, HDACi can act in a synthetic lethal manner with other DNA aberrations (drug-induced or otherwise) to induce apoptosis [28, 152, 155, 156]. Unexpectedly, HDACs were recently shown to positively regulate several key HR genes, and HDAC inhibition was found to dampen HR and enhance the susceptibility of triple-negative breast cancer cells towards PARPi [157] (Fig. 3). This insight and others highlight the need to fine-tune therapeutic applications of HDACi for specific patients and in other types of cancers.
There is a pressing need for a more comprehensive understanding of the interactive network between PARPi and BRCA genes and their role in anti-cancer therapy [158]. The links between epigenetic and genetic factors in governing pathway choices is complex and likely enriched by synergistically acting factors (Fig. 3). Collectively, these studies have highlighted the options for targeted therapy and offer potential treatment regimens for cancers, including triple-negative breast cancers, which have been problematic to treat up to now.
Concluding remarks
Technology that permits whole-genome interrogation brings forth the dawn of precision medicine, allowing links to be made between previously disconnected factors within the global gene network. This also excites the possibility of targeting the epigenetic mechanisms that affect the entire genome. First-generation epigenetic drugs tend to be toxic and associated with significant side effects. However, a better understanding of the networks involved should help in the selection of more applicable drugs and sidestep the off-target effects seen with drugs that targets cells based on proliferation. Furthermore, experimental successes in cultured cells or animal models must be further scrutinized in primary human normal and cancerous cells; these largely intractable models await further gene-editing breakthroughs. However, with the success of the synthetic lethality approach in targeting once-intractable cancers, there is a high hope of customized and personalized treatment strategies for patients, and it will be exciting to see the development in this field in the coming years.
Acknowledgements
I apologize to those authors whose work could not be cited due to space limitations. I thank Rebecca Jackson for editing a draft of this manuscript. This work was supported by a Singapore Ministry of Education Academic Research Fund (MOE2016-T2-2-063).
Abbreviations
- HR
Homologous recombination
- NHEJ
Non-homologous end joining
- MMEJ
Microhomology-mediated end joining
- HDAC
Histone deacetylase
- HDAC
Histone deacetylase inhibitor
- DSB
Double-stranded DNA break
- PARP
Poly (ADP-ribose) polymerase
- PARPi
Poly (ADP-ribose) polymerase inhibitor
- PRC
Polycomb repressive complex
- miRNA
Micro-RNA
- siRNA
Small interference-RNA
- LncRNA
Long non-coding RNA
- RNAi
RNA interference
- H3K4
Histone H3 lysine 4
- H3K4me
Methylated histone H3 lysine 4
- H3K27me
Methylated histone H3 lysine 27
- H3K36me
Methylated histone H3 lysine 36
- SAHA
Suberoylanilide hydroxamic acid
- PARylation
Poly ADP ribosylation
Compliance with ethical standards
Conflict of interest
The author declares that he has no conflict of interest.
References
- 1.De Iuliis F, Salerno G, Taglieri L, Scarpa S. Are pharmacogenomic biomarkers an effective tool to predict taxane toxicity and outcome in breast cancer patients? Cancer Chemother Pharmacol. 2015;76:679–690. doi: 10.1007/s00280-015-2818-4. [DOI] [PubMed] [Google Scholar]
- 2.Roscilli G, et al. Human lung adenocarcinoma cell cultures derived from malignant pleural effusions as model system to predict patients chemosensitivity. J Transl Med. 2016;14:61. doi: 10.1186/s12967-016-0816-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tremblay J, Hamet P. Role of genomics on the path to personalized medicine. Metabolism. 2013;62:S2–S5. doi: 10.1016/j.metabol.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 4.Lionetti M, Neri A. Utilizing next-generation sequencing in the management of multiple myeloma. Expert Rev Mol Diagn. 2017;17:653–663. doi: 10.1080/14737159.2017.1332996. [DOI] [PubMed] [Google Scholar]
- 5.Weitzel KW, et al. IGNITE network. The IGNITE network: a model for genomic medicine implementation and research. BMC Med Genomics. 2016;9:1. doi: 10.1186/s12920-015-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inaba H, Azzato EM, Mullighan CG. Integration of next-generation sequencing to treat acute lymphoblastic leukemia with targeted lesions: the St. Jude Children’s Research Hospital Approach. Front Pediatr. 2017;5:258. doi: 10.3389/fped.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinstein IB. Disorders in cell circuitry during multistage carcinogenesis: the role of homeostasis. Carcinogenesis. 2000;21:857–864. doi: 10.1093/carcin/21.5.857. [DOI] [PubMed] [Google Scholar]
- 8.Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–303. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 9.Hannah AL. Kinases drug discovery targets in hematologic malignancies. Curr Mol Med. 2005;5:625–642. doi: 10.2174/156652405774641106. [DOI] [PubMed] [Google Scholar]
- 10.Tolbert VP, Coggins GE, Maris JM. Genetic susceptibility to neuroblastoma. Curr Opin Genet Dev. 2017;42:81–90. doi: 10.1016/j.gde.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlic H, Schlögl EE, Nowotny H, Grüner H, Valent A, Auer H, Heinz R. Rare occurrence of Philadelphia chromosome negative (but BCR-ABL positive) CML in a central European population. Am J Hematol. 1994;47:253–254. doi: 10.1002/ajh.2830470331. [DOI] [PubMed] [Google Scholar]
- 12.Sukov WR, Hodge JC, Lohse CM, Akre MK, Leibovich BC, Thompson RH, Cheville JC. ALK alterations in adult renal cell carcinoma: frequency, clinicopathologic features and outcome in a large series of consecutively treated patients. Mod Pathol. 2012;25:1516–1525. doi: 10.1038/modpathol.2012.107. [DOI] [PubMed] [Google Scholar]
- 13.Nijman SMB. Synthetic lethality: general principles, utility and detection using genetic screens in human cells. FEBS Lett. 2011;585:1–6. doi: 10.1016/j.febslet.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson RA, Chen ES. Synthetic lethal approaches for assessing combinatorial efficacy of chemotherapeutic drugs. Pharmacol Ther. 2016;162:69–85. doi: 10.1016/j.pharmathera.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 16.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 17.Oike T, et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 2013;73:5508–5518. doi: 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman GR, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci USA. 2014;111:3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, Golec JM, Pollard JR. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 20.Menezes DL, et al. A synthetic lethal screen reveals enhanced sensitivity to ATR inhibitor treatment in mantle cell lymphoma with ATM loss-of-function. Mol Cancer Res. 2015;13:120–129. doi: 10.1158/1541-7786.MCR-14-0240. [DOI] [PubMed] [Google Scholar]
- 21.Manic G, Obrist F, Sistigu A, Vitale I. Trial watch: targeting ATM-CHK2 and ATR-CHK1 pathways for anticancer therapy. Mol Cell Oncol. 2015;2:e1012976. doi: 10.1080/23723556.2015.1012976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min A, et al. AZD6738, a novel inhibitor of ATR induces synthetic lethality with ATM deficiency in gastric cancer cells. Mol Cancer Ther. 2017;16:566–577. doi: 10.1158/1535-7163.MCT-16-0378. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt A, et al. ATM deficiency is associated with sensitivity to PARP1- and ATR inhibitors in lung adenocarcinoma. Cancer Res. 2017;77:3040–3056. doi: 10.1158/0008-5472.CAN-16-3398. [DOI] [PubMed] [Google Scholar]
- 24.Menezes DL, et al. A synthetic lethal screen reveals enhanced sensitivity to ATR inhibitor treatment in mantle cell lymphoma with ATM loss-of-function. Mol Cancer Res. 2015;13:120–129. doi: 10.1158/1541-7786.MCR-14-0240. [DOI] [PubMed] [Google Scholar]
- 25.Zhang XY, et al. Inhibition of the single downstream target BAG1 activates the latent apoptotic potential of MYC. Mol Cell Biol. 2011;31:5037–5045. doi: 10.1128/MCB.06297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin SE, Wu ZH, Gehlhaus K, Jones TL, Zhang YW, Guha R, Miyamoto S, Pommier Y, Caplen NJ. RNAi screening identifies TAK1 as a potential target for the enhanced efficacy of topoisomerase inhibitors. Curr Cancer Drug Targets. 2011;11:976–986. doi: 10.2174/156800911797264734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Pel DM, Stirling PC, Minaker SW, Sipahimalani P, Hieter P. Saccharomyces cerevisiae genetics predicts candidate therapeutic genetic interactions at the mammalian replication fork. G3 (Bethesda) 2013;G3:273–282. doi: 10.1534/g3.112.004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen TT, et al. Predicting chemotherapeutic drug combinations through gene network profiling. Sci Rep. 2016;6:18658. doi: 10.1038/srep18658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tay Z, et al. P-glycoprotein and vacuolar ATPase synergistically confer anthracycline resistance to fission yeast and human cells. Curr Med Chem. 2014;21:251–260. doi: 10.2174/09298673113206660269. [DOI] [PubMed] [Google Scholar]
- 30.Tay Z, Eng RJ, Sajiki K, Lim KK, Tang MY, Yanagida M, Chen ES. Cellular robustness conferred by genetic crosstalk underlies resistance against chemotherapeutic drug doxorubicin in fission yeast. PLoS One. 2013;8:e55041. doi: 10.1371/journal.pone.0055041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allis CD, Jenuwein T, Reinberg D, Caparros ML, editors. Epigenetics. Woodbury, New York: Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- 32.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 33.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39:310–318. doi: 10.1016/j.tibs.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Zhou CY, Johnson SL, Gamarra NI, Narlikar GJ. Mechanisms of ATP-dependent chromatin remodelling motors. Annu Rev Biophys. 2016;45:153–181. doi: 10.1146/annurev-biophys-051013-022819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 37.Morris KV. RNA-mediated transcriptional gene silencing in human cells. Curr Top Microbiol Immunol. 2008;320:211–224. doi: 10.1007/978-3-540-75157-1_10. [DOI] [PubMed] [Google Scholar]
- 38.Chitwood DH, Timmermans MC. Small RNAs are on the move. Nature. 2010;467:415–419. doi: 10.1038/nature09351. [DOI] [PubMed] [Google Scholar]
- 39.Sun M, et al. LncRNA HOXA11-AS Promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 40.O’Leary VB, Hain S, Maugg D, Smida J, Azimzadeh O, Tapio S, Ovsepian SV, Atkinson MJ. Long non-coding RNA PARTICLE bridges histone and DNA methylation. Sci Rep. 2017;7:1790. doi: 10.1038/s41598-017-01875-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C, Wang L, Ding Y, Lu X, Zhang G, Yang J, Zheng H, Wang H, Jiang Y, Xu L. LncRNA structural characteristics in epigenetic regulation. Int J Mol Sci. 2017 doi: 10.3390/ijms18122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choudhuri S, Cui Y, Klaassen CD. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol. 2010;245:378–393. doi: 10.1016/j.taap.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 44.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 45.Williams BP, Gehring M. Stable transgenerational epigenetic inheritance requires a DNA methylation-sensing circuit. Nat Commun. 2017;8:2124. doi: 10.1038/s41467-017-02219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunner AM, Nanni P, Mansuy IM. Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics Chromatin. 2014;7:2. doi: 10.1186/1756-8935-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 48.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang CC, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chi P, Allis CD, Wang GG. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anjanappan M, et al. A system for detecting high impact-low frequency mutations in primary tumors and metastases. Oncogene. 2018;37:185–196. doi: 10.1038/onc.2017.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan L, et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature. 2017;543:265–269. doi: 10.1038/nature21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujimoto A, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 56.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johann PD, et al. Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell. 2016;29:379–393. doi: 10.1016/j.ccell.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamane K, Mizugichi T, Cui B, Zofall M, Noma K, Grewal SI. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell. 2011;41:56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- 62.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oike T, Ogiwara H, Amornwichet N, Nakano T, Kohno T. Chromatin-regulating proteins as targets for cancer therapy. J Radiat Res. 2014;55:613–628. doi: 10.1093/jrr/rrt227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsubara D, et al. Lung cancer with loss of BRG1/BRM, shows epithelial mesenchymal transition phenotype and distinct histologic and genetic features. Cancer Sci. 2013;104:266–273. doi: 10.1111/cas.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rao Q, Xia QY, Shen Q, Shi SS, Tu P, Shi QL, Zhou XJ. Coexistent loss of INI1 and BRG1 expression in a rhabdoid renal cell carcinoma (RCC): implications for a possible role of SWI/SNF complex in the pathogenesis of RCC. Int J Clin Exp Pathol. 2014;7:1782–1787. [PMC free article] [PubMed] [Google Scholar]
- 66.Karnezis AN, et al. Dual loss of the SWI/SNF complex ATPase SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J Pathol. 2016;238:389–400. doi: 10.1002/path.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raab JR, Runge JS, Spear CC, Magnuson T. Co-regulation of transcription by BRG1 and BRM, two mutually exclusive SWI/SNF ATPase subunits. Epigenetics Chromatin. 2017;10:62. doi: 10.1186/s13072-017-0167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sena JA, Wang L, Hu CJ. BRG1 and BRM chromatin-remodeling complexes regulate the hypoxia response by acting as coactivators for a subset of hypoxia-inducible transcription factor target genes. Mol Cell Biol. 2013;33:3849–3863. doi: 10.1128/MCB.00731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lan J, Li H, Luo X, Hu J, Wang G. BRG1 promotes VEGF-A expression and angiogenesis in human colorectal cancer cells. Exp Cell Res. 2017;360:236–242. doi: 10.1016/j.yexcr.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 70.Marquez-Vilendrer SB, Rai SK, Gramling SJ, Lu L, Reisman DN. BRG1 and BRM loss selectively impacts RB and P53, respectively: BRG1 and BRM have differential functions in vivo. Oncoscience. 2016;3:337–350. doi: 10.18632/oncoscience.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vélez-Cruz R, Manickavinayaham S, Biswas AK, Clary RW, Premkumar T, Cole F, Johnson DG. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes Dev. 2016;30:2500–2512. doi: 10.1101/gad.288282.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vangamudi B, et al. The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF-mutant cancers: insights from cDNA rescue and PFI-3 inhibitor studies. Cancer Res. 2015;75:3865–3878. doi: 10.1158/0008-5472.CAN-14-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, Nagl NG, Wilsker D, Van Scoy M, Pacchione S, Yaciuk P, Dallas PB, Moran E. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem J. 2004;383:319–325. doi: 10.1042/BJ20040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bitler BG, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kelso TWR, Porter DK, Amaral ML, Shokhirev MN, Benner C, Hargreaves DC. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers. Elife. 2017 doi: 10.7554/elife.30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sausen M, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coatham M, et al. Concurrent ARID1A and ARID1B inactivation in endometrial and ovarian dedifferentiated carcinomas. Mod Pathol. 2016;29:1586–1593. doi: 10.1038/modpathol.2016.156. [DOI] [PubMed] [Google Scholar]
- 81.Helming KC, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med. 2014;20:251–254. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 83.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 84.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.He LJ, Cai MY, Xu GL, Li JJ, Weng ZJ, Xu DZ, Luo GY, Zhu SL, Xie D. Prognostic significance of overexpression of EZH2 and H3k27me3 proteins in gastric cancer. Asian Pac J Cancer Prev. 2012;13:3173–3178. doi: 10.7314/APJCP.2012.13.7.3173. [DOI] [PubMed] [Google Scholar]
- 86.Behrens C, et al. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin Cancer Res. 2013;19:6556–6565. doi: 10.1158/1078-0432.CCR-12-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu C, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toll AD, Dasgupta A, Potoczek M, Yeo CJ, Kleer CG, Brody JR, Witkiewicz AK. Implications of enhancer of zeste homologue 2 expression in pancreatic ductal adenocarcinoma. Hum Pathol. 2010;41:1205–1209. doi: 10.1016/j.humpath.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 89.Wagener N, Macher-Goeppinger S, Pritsch M, Hüsing J, Hoppe-Seyler K, Schirmacher P, Pfitzenmaier J, Haferkamp A, Hoppe-Seyler F, Hohenfellner M. Enhancer of zeste homolog 2 (EZH2) expression is an independent prognostic factor in renal cell carcinoma. BMC Cancer. 2010;10:524. doi: 10.1186/1471-2407-10-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cao W, Feng Z, Cui Z, Zhang C, Sun Z, Mao L, Chen W. Up-regulation of enhancer of zeste homolog 2 is associated positively with cyclin D1 overexpression and poor clinical out- come in head and neck squamous cell carcinoma. Cancer. 2012;118:2858–2871. doi: 10.1002/cncr.26575. [DOI] [PubMed] [Google Scholar]
- 91.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 92.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 93.Knutson SK, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci USA. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, Orkin SH. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 96.Knutson SK, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 97.Qi W, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci USA. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gehling VS, et al. Discovery, design, and synthesis of indole-based EZH2 inhibitors. Bioorg Med Chem Lett. 2015;25:3644–3649. doi: 10.1016/j.bmcl.2015.06.056. [DOI] [PubMed] [Google Scholar]
- 99.Honma D, et al. Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci. 2017;108:2069–2078. doi: 10.1111/cas.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takano M, Tsuda H, Sugiyama T. Clear cell carcinoma of the ovary: is there a role of histology-specific treatment? J Exp Clin Cancer Res. 2012;31:53. doi: 10.1186/1756-9966-31-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fujiwara K, Shintani D, Nishikawa T. Clear-cell carcinoma of the ovary. Ann Oncol. 2016;27(Suppl 1):i50–i52. doi: 10.1093/annonc/mdw086. [DOI] [PubMed] [Google Scholar]
- 102.Kim KH, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med. 2015;21:1491–1496. doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fillmore CM, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520:239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells. 2012;30:372–378. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 105.Jin X, et al. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat Med. 2017;23:1352–1361. doi: 10.1038/nm.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wagner E, Carpenter PB. Understanding the language of Lys 36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li J, Duns G, Westers H, Sijmons R, van den Berg A, Kok K. SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget. 2016;7:50719–50734. doi: 10.18632/oncotarget.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Piva F, et al. BAP1, PBRM1 and SETD2 in clear-cell renal cell carcinoma: molecular diagnostics and possible targets for personalized therapies. Expert Rev Mol Diagn. 2015;15:1201–1210. doi: 10.1586/14737159.2015.1068122. [DOI] [PubMed] [Google Scholar]
- 109.Parker H, et al. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia. 2016;30:2179–2186. doi: 10.1038/leu.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McKinney M, et al. The genetic basis of hepatosplenic T-cell lymphoma. Cancer Discov. 2017;7:369–379. doi: 10.1158/2159-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen Z, Raghoonundun C, Chen W, Zhang Y, Tang W, Fan X, Shi X. SETD2 indicates favourable prognosis in gastric cancer and suppresses cancer cell proliferation, migration, and invasion. Biochem Biophys Res Commun. 2018;498:579–585. doi: 10.1016/j.bbrc.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 112.Liu L, Guo R, Zhang X, Liang Y, Kong F, Wang J, Xu Z. Loss of SETD2, but not K3K36me3, correlates with aggressive clinicopathological features of clear cell renal cell carcinoma patients. Biosci Trends. 2017;11:214–220. doi: 10.5582/bst.2016.01228. [DOI] [PubMed] [Google Scholar]
- 113.Martinelli G, et al. SETD2 and histone H3 lysine 36 methylation deficiency in advanced systemic mastocytosis. Leukemia. 2018;32:139–148. doi: 10.1038/leu.2017.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pfister SX, et al. WEE1 selectively kills histone H3K36me3-deficient cancers by dNTP starvation. Cancer Cell. 2015;28:557–568. doi: 10.1016/j.ccell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen K, Liu J, Liu S, Xia M, Zhang X, Han D, Jiang Y, Wang C, Cao X. Methyltransferase SETD2-mediated methylation of STAT1 is critical for interferon antiviral activity. Cell. 2017;170:492–506. doi: 10.1016/j.cell.2017.06.042. [DOI] [PubMed] [Google Scholar]
- 116.Park IY, Chowdhury P, Tripathi DN, Powell RT, Dere R, Terzo EA, Rathmell WK, Walker CL. Methylated α-tubulin antibodies recognize a new microtubule modification on mitotic microtubules. MAbs. 2016;8:1590–1597. doi: 10.1080/19420862.2016.1228505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park IY, et al. Dual chromatin and cytoskeletal remodeling by SETD2. Cell. 2016;166:950–962. doi: 10.1016/j.cell.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Georgoulis A, Vorgias CE, Chrousos GP, Rogakou EP. Genome instability and γH2AX. Int J Mol Sci. 2017 doi: 10.3390/ijms18091979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Min A, et al. AZD6738, a novel inhibitor of ATR, induces synthetic lethality with ATM deficiency in gastric cancer cells. Mol Cancer Ther. 2017;16:566–577. doi: 10.1158/1535-7163.MCT-16-0378. [DOI] [PubMed] [Google Scholar]
- 120.Jossé R, et al. ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase I inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 2014;74:6968–6979. doi: 10.1158/0008-5472.CAN-13-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hocke S, et al. A synthetic lethal screen identifies ATR-inhibition as a novel therapeutic approach for POLD1-deficient cancers. Oncotarget. 2016;7:7080–7095. doi: 10.18632/oncotarget.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated H2AX to regulate cellular responses to DNA double-stranded breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 123.Lee MS, Edwards RA, Thede GL, Glover JN. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the gamma-H2AX histone tail. J Biol Chem. 2005;280:32053–32056. doi: 10.1074/jbc.C500273200. [DOI] [PubMed] [Google Scholar]
- 124.Kleiner RE, Verma P, Molloy KR, Chait BT, Kapoor TM. Chemical proteomics reveals gammaH2AX-53BP1 interaction in the DNA damage response. Nat Chem Biol. 2015;11:807–814. doi: 10.1038/nchembio.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Goldberg M, Stucki M, Falck J, D’Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 126.Stewart GS, Wang B, Bignell CR, Tylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 127.Zhang J, Ma Z, Treszezamsky A, Powell SN. MDC1 interacts with Rad51 and facilitates homologous recombination. Nat Struct Mol Biol. 2005;12:902–909. doi: 10.1038/nsmb991. [DOI] [PubMed] [Google Scholar]
- 128.Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sennoy TR, et al. CHD1 loss sensitizes prostate cancer to DNA damaging therapy by promoting error-prone double-strand break repair. Ann Oncol. 2017;28:1495–1507. doi: 10.1093/annonc/mdx165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lottersberger F, Bothmer A, Robbiani DF, Nussenzweig MC, de Lange T. Role of 53BP1 oligomerization in regulating double-stranded break repair. Proc Natl Acad Sci USA. 2013;110:2146–2151. doi: 10.1073/pnas.1222617110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Somyajit K, Mishra A, Jameei A, Nagaraju G. Enhanced non-homologous end joining contributes toward synthetic lethality of pathological RAD51C mutants with poly (ADP-ribose) polymerase. Carcinogenesis. 2015;36:13–24. doi: 10.1093/carcin/bgu211. [DOI] [PubMed] [Google Scholar]
- 132.Gray HJ, Bell-McGuinn K, Fleming GF, Cristea M, Xiong H, Sullivan D, Luo Y, McKee MD, Munasinghe W, Martin LP. Phase I combination study of the PARP inhibitor veliparib plus carboplatin and gemcitabine in patients with advanced ovarian cancer and other solid malignancies. Gynecol Oncol. 2018;148:507–514. doi: 10.1016/j.ygyno.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 133.Ledermann J, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 134.Bhattacharjee S, Nandi S. Synthetic lethality in DNA repair network: a novel avenue in targeted cancer therapy and combination therapeutics. IUBMB Life. 2017;69:929–937. doi: 10.1002/iub.1696. [DOI] [PubMed] [Google Scholar]
- 135.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 136.Zhao W, et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature. 2017;550:360–365. doi: 10.1038/nature24060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu G, et al. REV7 counteracts DNA double-stranded break resection and impacts PARP inhibition. Nature. 2015;521:541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bhattacharjee S, Nandi S. DNA damage response and cancer therapeutics through the lens of the Fanconi Anemia DNA repair pathway. Cell Commun Signal. 2017;15:41. doi: 10.1186/s12964-017-0195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38:5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human RNA polymerase θ. Nat Struct Mol Biol. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ceccaldi R, et al. Homologous-recombination-deficient tumors are dependent on Polθ-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wyatt DW, Feng W, Conlin MP, Yousefzaden MJ, Roberts SA, Mieczkowski P, Wood RD, Gupta GP, Ramsden DA. Essential roles of polymerase θ-mediated end joining in the repair of chromosome breaks. Mol Cell. 2016;63:662–673. doi: 10.1016/j.molcel.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bhattacharjee S, Nandi S. Choices have consequences: the nexus between DNA repair pathways and genomic instability in cancer. Clin Transl Med. 2016;5:45. doi: 10.1186/s40169-016-0128-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mateos-Gomez PA, Kent T, Deng SK, McDevitt S, Kashkina E, Hoang TM, Pomerantz RT, Sfeir A. The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat Struct Mol Biol. 2017;24:1116–1123. doi: 10.1038/nsmb.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ray Chaudhuri A, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ding X, et al. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat Commun. 2016;7:12425. doi: 10.1038/ncomms12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nicolae CM, Aho ER, Choe KN, Constantin D, Hu HJ, Lee D, Myung K, Moldovan GL. A novel role for the mono-ADP-ribosyltransferase PARP14/ARTD8 in promoting homologous recombination and protecting against replication stress. Nucleic Acids Res. 2015;43:3143–3153. doi: 10.1093/nar/gkv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Boulikas T. Poly(ADP-ribosylated) histones in chromatin replication. J Biol Chem. 1990;265:14638–14647. [PubMed] [Google Scholar]
- 150.Davies H, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, Kohno T. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov. 2016;6:430–445. doi: 10.1158/2159-8290. [DOI] [PubMed] [Google Scholar]
- 152.Ronnekleiv-Kelly SM, Sharma A, Ahuja N. Epigenetic therapy and chemosensitization in solid malignancy. Cancer Ther Rev. 2017;55:200–208. doi: 10.1016/j.ctrv.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 153.Hess-Stumpp H. Histone deacetylase inhibitors and cancer: from cell biology to the clinic. Eur J Cell Biol. 2005;84:109–121. doi: 10.1016/j.ejcb.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 154.Marks PA, Xu WS. Histone deacetylase inhibitors: potential in cancer therapy. J Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Conti C, Leo E, Eichler GS, Sordet O, Martin MM, Fan A, Aladjem MI, Pommier Y. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70:4470–4480. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Seah KS, Loh JY, Nguyen TT, Tan HL, Hutchinson PE, Lim KK, Dymock BW, Long YC, Lee EJD, Shen HM, et al. SAHA and cisplatin sensitize gastric cancer cells to doxorubicin by induction of DNA damage, apoptosis and perturbation of AMPK-mTOR signalling. Exp Cell Res. 2018 doi: 10.1016/j.yexcr.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 157.Wiegmans AP, Yap P, Ward A, Lim YC, Khanna KK. Differences in expression of key DNA damage repair genes after epigenetic-induced BRCAness dictate synthetic lethality with PARP1 inhibition. Mol Cancer Ther. 2015;14:2321–2331. doi: 10.1158/1535-7163.MCT-15-0374. [DOI] [PubMed] [Google Scholar]
- 158.Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]