Abstract
The recent impact of cancer immunotherapies has firmly established the ability and importance of the immune system to fight malignancies. However, the intimate interaction between the highly dynamic tumor and immune cells leads to a selection process driven by genetic and epigenetic processes. As the molecular pathways of cancer resistance mechanisms to immunotherapy become increasingly known, novel therapeutic targets are being tested in combination with immune-stimulating approaches. We here review recent insights into the molecular mechanisms of tumor resistance with particular emphasis on epigenetic processes and place these in the context of previous models.
Keywords: EZH2, EED, PRC2, Tumor escape, Immunogenicity, Checkpoint inhibitor, PD-1, CTLA-4, Cytokine, IL-2
Introduction
In 1891, William Coley injected cancer patients with live Streptococcus pyogenes and observed tumor shrinkage, but also lethal systemic infections. Thus, he modified his treatment regimen and since 1893 he began to study intratumoral injections of heat-inactivated S. pyogenes and Serratia marcescens, later called Coley’s toxins, which in some patients resulted in complete remission of sarcomas [1, 2]. Since that time, a large number of research studies contributed to the understanding of the steps needed to get an effective anti-tumor immune response, starting with innate immune recognition and antigen presentation in the secondary lymphoid organs, inducing effector T cell responses able to recognize cancer cells and overcome the immunosuppressive tumor microenvironment. A successful cancer immunotherapy ideally fulfills each of these steps.
Several immunotherapeutic strategies have been developed and validated for the treatment of aggressive cancers. These include cancer vaccines, adoptive transfer of ex vivo activated T-cells, recombinant cytokines with the aim of stimulating the immune system, and monoclonal antibodies blocking immune checkpoint pathways such as cytotoxic T lymphocyte-associated protein 4 (CTLA-4) and programmed cell death protein 1 (PD-1). The approval and use of immune checkpoint inhibitors in patients with advanced cancers led to durable clinical responses revitalizing the field of cancer immunotherapy [3]. However, a percentage of patients do not benefit from this therapy and others relapse after initial response [4–7]. Elucidating the mechanisms of primary and acquired resistance to immunotherapy is essential to improve the outcome of cancer immunotherapy and find complementary and alternative strategies, which has been expertly reviewed previously by Sharma et al. [8].
Numerous factors can render cancer cells resistant to the effector mechanisms of the immune system, either in the first instance (the so-called primary resistance) or following initially successful immunotherapy (termed secondary or adaptive resistance). Thus, T-cells can fail to recognize cancer cells due to the absence of high-affinity T-cell receptors (TCR) or the lack of expression of tumor antigens on cancer cells may be due to defects in the antigen presentation machinery [9–12]. Moreover, modification or silencing of several tumor intrinsic signaling pathways can promote resistance. Relevant examples are the Wnt/β-catenin, PTEN/phosphoinositide 3-kinase (PI3K) and Janus kinase (JAK) 1 and JAK2 pathways, which lead to dendritic cell (DC) dysfunction and loss of interferon-γ (IFN-γ) response [13–18]. Of note, the prototypic T helper (Th) 1 cytokine IFN-γ exerts crucial roles in anti-tumor immune responses as well as immunity to intracellular pathogens. Moreover, the Wnt/β-catenin pathway affects T-cell trafficking to the tumor site by curbing the production of the T-cell-attractant chemokines CXCL9 and CXCL10, as evidenced in β-catenin-positive tumors [19]. Furthermore, recent studies have shown that genetic mutations in certain signaling pathways cause constitutive expression of programmed death-ligand 1 (PD-L1) inhibiting T-cell-mediated anti-tumor effects [20, 21]. In consonance with this, persistent antigen stimulation negatively regulates the activity of T-cells thus leading to the expression of inhibitory receptors and resulting in exhaustion and dysfunction of effector T-cells [22]. The administration of antibodies blocking the CTLA-4 or PD-1–PD-L1 inhibitory axes can lead to the upregulation of other inhibitory signaling pathways, such as lymphocyte-activation gene 3 (LAG-3), T-cell immunoglobulin and mucin domain 3 (TIM-3), T-cell immunoreceptor with Ig and ITIM domains (TIGIT), and V-domain Ig suppressor of T-cell activation (VISTA). Upregulation of these latter inhibitory molecules has been associated to the activation of various pathways such as the PI3K–AKT and interferon signaling pathways [23–26]. In this context, metabolic reprogramming in the tumor microenvironment associated with acute hypoxia, high concentration of tumor-derived lactate and glucose deprivation can restrain proper T-cell effector functions [27]. T-cell infiltration and activity can also be altered by the chemokine milieu and the relative accumulation of immune cells playing an inhibitory role within the tumor microenvironment. Well-characterized immunosuppressive cell subsets are CD4+ T-regulatory (Treg) cells, myeloid-derived suppressor cells (MDSCs) and alternatively activated M2 macrophages [28–36].
Additional factors influencing the anti-tumor immune response include angiogenesis, the extracellular matrix, and the microbiome, to name a few. Angiogenesis is a key process for the growth and metastasis of tumors. Increased levels of angiogenic activators such as vascular endothelial growth factor (VEGF) allow the extravasation of immunosuppressive Treg cells while decreasing the levels of effector T-cells [37]. Moreover, T-cells can encounter biological barriers such as low levels of enzymes needed to degrade the extracellular matrix to reach target cells [38]. Another relevant parameter to tumor resistance is the association between the microbiome diversity and the lack of response or relapse of certain patients to cancer immunotherapy. A representative example is a study showing that the anti-cancer properties of the approved anti-CTLA-4 antibody are influenced by the microbiota composition [39].
Within the scope of this review we will focus on the ability of the immune system to contribute to cancer resistance mechanisms by engaging epigenetic pathways, which altogether can result in tumor immune escape. We will first revisit the published data on the effects of epigenetic regulation contributing to tumor resistance mechanisms. Subsequently, we will focus on enhancer of zeste homolog 2 (EZH2), as this histone methyltransferase has been an intensively studied epigenetic modulator in cancer biology, including its contribution to adaptive cancer resistance.
Epigenetic regulation
Epigenetic regulation of gene expression
The contribution of genomic instability to tumor resistance has been extensively investigated but classic genetics cannot fully explain how tumor cells are able to escape from immune control, especially in situations of secondary resistance. Epigenetics is the term used to describe the molecular pathways modulating the expression of genes without altering the DNA sequence and has become a key area of research for cancer development and progression. The mechanisms involved in epigenetic gene regulation include methylation of cytosines in the DNA and enzymatic modifications of histones. DNA methylation is catalyzed by DNA methyltransferases (DNMT) and is associated with the maintenance of chromatin in a silent state. Aberrant levels of DNA methylation have been linked with enhanced expression of essential proteins involved in metastasis development or inactivation of tumor suppressor genes [40]. Histone modifications, such as acetylation, methylation and phosphorylation, affect their interaction with other proteins, thus modulating the activity of the associated DNA. The acetylation of lysine residues (K) of histones (H) depends on the activity of histone acetyl transferases and histone deacetylases (HDAC). The effect of histone methylation, catalyzed by histone methyl transferases (HMT), depends on the residue and the site of methylation. For example, methylation of H3 at K4 is linked to transcriptional activation, whereas methylation of H3 at K9 or K27 leads to transcriptional repression [41]. Unlike genetic alterations, epigenetic modifications are usually reversible with pharmacologic agents inhibiting DNA methylation, histone methylation and deacetylation. The first DNMT inhibitors developed were nucleoside analogs of cytidine including 5-azacytidine (AZA) and 5-aza-2′-deoxycytidine (5-AZA-2). This was followed by the development of HDAC inhibitors and drugs targeting HMTs [42]. Accordingly, various in vitro studies investigating the role of epigenetic modifications on cancer cells have been performed using this broad range of inhibitors. These studies, described more in detail below, revealed strategies to overcome epigenetic modifications to improve anti-tumor immune responses by inducing tumor antigen expression, processing and presentation together with sensitization of tumors to apoptotic signals.
Epigenetic control of tumor antigen expression
Cancer cells express on their cell surface tumor-associated antigens (TAA) presented on major histocompatibility complex (MHC) class-I molecules (MHC-I). MAGE-type antigens are a class of TAAs with normal expression confined to male germ cells and trophoblastic cells, both of which usually do not express MHC-I [43]. The expression of MAGE-type antigens in normal and cancerous tissue is regulated by epigenetic mechanisms (Table 1). MAGE-type antigens have been detected in a variety of malignancies including bladder, lung, ovary, pancreas, and head and neck cancer, as well as melanoma and multiple myeloma [43, 44]. The restricted expression profile of these antigens make them an ideal target for T-cells, although minimal central tolerance toward germline antigens has been reported [45]. Endogenous cytotoxic T-cell activity against a number of MAGE-type antigens have been examined, and MAGE-A1-specific T-cells were initially identified in melanoma followed by MAGE-A3, NY-ESO-1 and SSX-2 [46]. Downregulation of this class of antigens upon hypermethylation has been observed in several tumors [47–49].
Table 1.
Epigenetic mechanisms and their targeting agents impacting anti-tumor immune responses
| Resistance development | Epigenetic mechanism | Affected proteins | Targeting agents | References |
|---|---|---|---|---|
| Downregulation of tumor-associated antigen expression | DNA methylation |
MAGE-A1 SSX |
5-AZA-2 AZA |
[50–53, 55] |
| Histone methylation |
MAGE-A1 MAGE-A3 NY-ESO-1 |
DZNep | [54] | |
| EZH2 |
Pmel TYRP1 |
GSK503 | [122, 125] | |
| Downregulation of antigen presentation | DNA methylation |
MHC-I MHC-II |
AZA | [58] |
| Histone deacetylation |
MHC-I MHC-II TAP-1 CD40 |
IFN-γ Vorinostat Trichostatin A |
[59–64] | |
| EZH2 |
MHC-I TAP-1 TAP-2 |
GSK503 | [122] | |
| Modulation of apoptotic pathways | DNA methylation | Caspase-8 | 5-AZA-2 | [68–70] |
| Histone deacetylation |
FasL Fas TRAIL |
VPA NaBt MS-275 |
[71–73] |
AZA azacytidine, DZNep 3-deazaneplanocin A, FasL Fas ligand, MAGE-A melanoma-associated antigen-A, MHC major histcompatibility complex, NaBt sodium butyrate, NY-ESO-1 New York esophageal squamous cell carcinoma-1, Pmel premelanosome protein, SSX synovial sarcoma X, TAP transporter associated with antigen processing, TRIAL TNF-related apoptosis-inducing ligand, VPA valproic acid
Treatment with 5-AZA-2 has been shown to upregulate expression of various MAGE-type antigens in tumor cell lines leading to the recognition and consequent elimination by antigen-specific cytotoxic T-cells [50–53]. Inhibition of histone methylation has also been shown to contribute to MAGE-type antigen upregulation in lung cancer cells [54]. Pursuant to these studies, in a phase II clinical trial for multiple myeloma a similar DNA methylation inhibitor, AZA, was used in combination with lenalidomide. The combination treatment led to increased expression of MAGE-type antigens in the bone marrow compared to samples before treatment. Furthermore, antigen-specific T-cell response was detected in peripheral blood mononuclear cells (PBMCs) of patients [55].
Epigenetic regulation of tumor antigen presentation
DNA methylation levels can regulate the expression of molecules essential for the recognition of cancer cells by the immune system such as the components of the antigen presentation machinery [44, 56]. One mechanism described in melanoma cells was the hypermethylation of MHC genes leading to low levels of transcription [57]. Furthermore, low dose AZA treatment was shown to induce expression of the antigen processing and presentation molecules together with IFN-γ signaling in human breast, colorectal and ovarian cancer cells [58]. Downregulation of the transporter associated with antigen processing (TAP)-1 due to loss of histone acetylation was demonstrated in metastatic murine cell lines. TAP-1 expression and consequent MHC-I presentation was restored by IFN-γ exposure [59]. Moreover, HDAC inhibitors were shown to increase the expression of TAP-1 and TAP-2, leading to better tumor control [60, 61].
Few studies suggested epigenetic mechanisms regulating the development and function of DCs, which prime antitumor T-cell responses through the presentation and/or cross-presentation of TAAs and the provision of costimulatory signals. Deacetylation suppresses the expression of MHC class-II (MHC-II) and T-cell costimulatory genes CD40, CD80 and CD86, which can be restored by treatment with the HDAC inhibitor trichostatin A [62–66]. Moreover, histone acetylation regulates interleukin (IL)-6 production by DCs affecting antigen-specific T-cell responses [67].
Epigenetic control of tumor cell killing
The final goal of the immune response during cancer immunotherapy is the elimination of the cancer cells. Cytotoxic T-cells and natural killer (NK) cells are the main players in this process. Although the activation process of T and NK cells is quite different, they share effector mechanisms, mainly granule exocytosis and the death ligand–death receptor system. Some studies have demonstrated that tumor cells are able to evade cell lysis through epigenetic modulation of several apoptotic pathways. Expression of various cell extrinsic or intrinsic apoptosis inducers have been shown to be downregulated by DNA methylation in cancers [68]. One such example is hypermethylation of the caspase-8 promoter, which can be rescued by 5-AZA-2 treatment [69]. Similarly, DNMT inhibitors were shown to overcome the resistance to tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) therapy in a mouse model of glioblastoma [70]. Factors that are frequently methylated in tumors include, but are not limited to, death-associated protein kinase (DAPK), Ras-association domain family 1A (RASSF1A), and apoptotic protease-activating factor 1 (APAF-1) [68].
In addition to DNA methylation, histone modification can contribute to epigenetic escape of apoptosis. The anti-tumorigenic effect of HDAC inhibitors observed in leukemia depends on activation of TRAIL and Fas (CD95) signaling pathways. Upstream mediators such as Fas ligand (FasL, also known as CD95L), Fas and TRAIL were upregulated by HDAC inhibitors, which led to increased caspase activity [71, 72]. These findings were also translated to pancreatic cancer cell lines, where treatment with the HDAC inhibitor sodium butyrate (NaBt) sensitized the cells to both Fas- mediated and mitochondria-triggered apoptosis [73].
Epigenetic regulation of T-cell response
An important role of DNA methylation has been shown for T-cell development and function. Epigenetic modulation of the CD8 co-receptor affects the activation status of T-cells upon encountering the MHC-I–TAA complex and controls the transcriptional activation of IFN-γ [74–76]. As previously introduced, persistent antigen stimulation and inflammatory microenvironment observed in chronic infection and cancer can lead to a dysfunctional T-cell state, termed T-cell exhaustion. The exhausted T-cell state is characterized by co-expression of high levels of immune checkpoint molecules, such as PD-1, CTLA-4, TIM-3 and LAG-3, and loss of secretion of effector cytokines such as IL-2, TNF and IFN-γ. Exhausted T-cells also manifest a low proliferation rate, altered metabolism and distinct transcription factor expression and activity.
The expression levels of the transcription factors T-bet and Eomesodermin (Eomes) demonstrated heterogeneity within the exhausted T-cells. While the T-bethigh PD-1mid cell subset is responsive to reinvigoration by checkpoint inhibitors, Eomeshigh PD-1high cells are terminally differentiated [77]. T-cell exhaustion restricts the effector functions of T-cells, preventing control of infection or tumor growth. Targeting this dysfunctional T-cell state with blocking antibodies to PD-1 (or its ligand PD-L1) and CTLA-4 led to increased anti-tumor T-cell responses and showed considerable clinical benefits in metastatic cancer patients.
Virus-specific CD8+ T-cells generated in response to persistent chronic infection kept their exhausted phenotype, characterized by high PD-1 expression and decreased cytokine responses, when transferred into naïve mice [78]. Surprisingly, these cells were able to proliferate and mediated cytotoxic responses to eliminate viral infections upon re-challenge while maintaining their exhausted phenotype. These results revealed a heritable alteration within exhausted T-cells. In line with this study, demethylation of the Pdcd1 locus (encoding PD-1) in antigen-specific CD8+ T-cells was identified in chronic viral infections in the effector stage. While memory T-cells were able to remethylate the region, the demethylation state was maintained in exhausted T-cells even if the target antigen was absent [79–81]. It would be interesting to investigate whether similar mechanisms are operational in cancer.
Further studies using transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) have demonstrated global epigenetic remodeling during T-cell activation and differentiation [82], which includes the transition of naïve T-cells to effectors with many of these -cells undergoing apoptosis, while a few survive as memory T-cells and other effector T-cells may become exhausted if the antigen persists [83]. Chromatin accessibility of enhancer regions were altered during T-cell differentiation, positively regulating gene expression of distinct programs corresponding with the acquired phenotype [82]. In a similar study, the effect of inhibition of PD-1–PD-L1 axis on exhausted T-cells was investigated. In terms of gene expression, PD-L1 blockade favored an effector T-cell phenotype rather than a memory T-cell phenotype [84]. This rejuvenation was lost with time upon sustained antigen exposure, suggesting re-exhaustion of T-cells. The analysis of the epigenetic landscape revealed that PD-L1 blockade only led to minimal alterations in accessible chromatin regions. This suggests that without altering the underlying epigenetic program, it is very challenging to restore the activity and cytotoxic capacity of CD8+ T-cells. In a recent study, a combination of HDAC and DNMT inhibitors in non-small cell lung cancer cells correlated with increased IFN-α/β signaling, upregulation of the antigen presentation machinery and depletion of MYC signaling [85]. This led to enhanced tumor control correlating with increased T-cell infiltration and reversion of T-cell exhaustion, thereby skewing the system toward memory and effector T-cell responses. Capitalizing on these findings, Dnmt3a-c-mediated de novo DNA methylation was shown to be crucial for exhausted T-cell reprogramming in chronic inflammation and cancer [86]. Dnmt3a-c-deficient T-cells were selected against their wild-type counterparts as they retained the capacity to proliferate, produce effector cytokines and maintain a T-bethigh Eomeslow expression profile. 5-AZA-2 treatment synergized with PD-1 blockade in tumor control, increasing the fitness of both antigen-specific and polyclonal tumor-infiltrating CD8+ T-cells. By contrast, tumor cell intrinsic demethylation can also cause the upregulation of inhibitory pathways leading to T-cell exhaustion. In vitro, non-small cell lung cancer cell lines upregulate PD-L1 upon treatment with AZA [87]. Moreover, higher levels of the inhibitory molecules PD-L1, PD-L2, PD-1 and CTLA-4 were detected in a cohort of myelodysplastic syndrome, chronic myelomonocytic leukemia and acute myeloid leukemia patients treated with 5-AZA-2 [88].
In summary, the described epigenetic alterations are associated with mechanisms developed by the tumor cells to evade immune control (Fig. 1). Furthermore, recent studies have depicted a key role of altered histone methylation in the development of cancer, especially through the activity of EZH2.
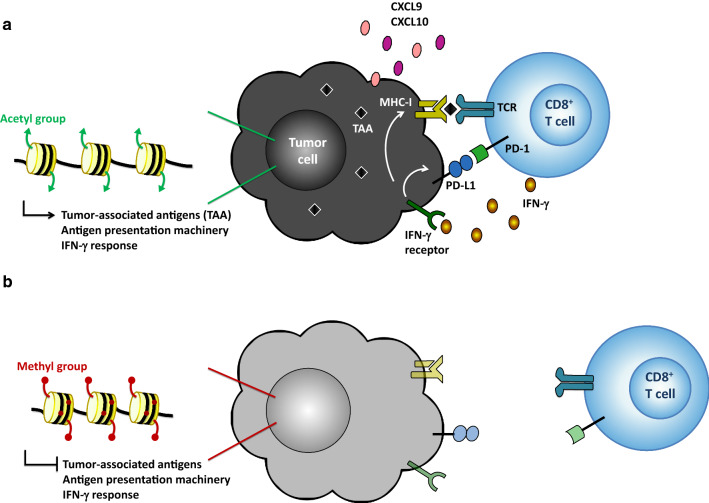
Fig. 1.
Epigenetic mechanisms leading to loss of immunogenicity. a Tumor cell (black) maintains expression of tumor-associated antigens (TAA) and immune-related processes, such as antigen presentation machinery, interferon (IFN)-γ receptor, and chemokines CXCL9 and CXCL10. This leads to the accumulation of CD8+ T-cells (blue) in the tumor microenvironment and subsequent recognition of tumors via the major histocompatibility complex class-I (MHC-I)—TAA—T-cell receptor (TCR) complex. Infiltrating CD8+ and CD4+ T-cells (not shown) produce IFN-γ, which further promotes antigen presentation and expression of programmed death-ligand 1 (PD-L1) on tumor cells. Persistent antigen stimulation, interaction of programmed cell death protein-1 (PD-1) and PD-L1 and expression of other inhibitory signals lead to exhaustion of CD8+ T-cells. b Tumor cell (gray) loses the expression of the above-mentioned genes due to DNA and histone methylation. Lack of T-cell-attracting chemokines and antigen presentation inhibits T-cell infiltration and tumor recognition
Epigenetic regulation of suppressor-type immune cells
A major tumor extrinsic factor mediating resistance to anti-cancer immune responses is the presence of immunosuppressive cells in the tumor microenvironment, including Treg cells and MDSCs. Treg cells are a subset of CD4+ T-cells, commonly characterized by expression of the transcription factor forkhead box P3 (FoxP3) and high surface levels of IL-2 receptor α (also known as CD25), although there also exist other Treg cell subsets with variable expression of these molecules. Treg cells inhibit effector T and NK cell function by secretion of immunosuppressive cytokines such as IL-10, IL-35 and TGF-β, expression of inhibitory molecules, and induction of cytolysis. Moreover, Treg cells depend on, consume and scavenge IL-2, an essential cytokine for the proliferation and development of effector and memory T-cells [89]. Also, Treg cells are able to kill or modulate antigen-presenting cells by endo- or trogocytosis of costimulatory molecules [90, 91].
FoxP3 is the major transcription factor mediating Treg differentiation and function. Its expression in Treg cells is very stable during their lifespan, maintained by hypomethylation of the conserved non-coding sequence 2 (CNS2) at the 5′ untranslated region of the Foxp3 gene [92]. DNA hypomethylation is also observed for other Treg markers including CD25, CTLA-4 and Tnfrsf18 (also known as GITR) [93, 94]. It has been shown that inhibition of DNA methylation promotes and stabilizes FoxP3 expression [95, 96]. Based on this data, it could be speculated that use of DNA methyltransferase inhibitors in cancer treatment could support Treg function or even promote their differentiation in the tumor microenvironment.
A second cell type exerting immunosuppressive effects in cancer constitute MDSCs. These cells develop under chronic inflammatory conditions from immature myeloid cells. MDSCs are a distinct cell subset with the ability to suppress various T-cell functions. The epigenetic mechanisms regulating MDSC function in the tumor microenvironment remain still mostly unknown. One study revealed that HDAC11 regulates MDSCs function in vivo. MDSCs from HDAC11-deficient mice were highly suppressive leading to more aggressive tumors compared to wild-type controls [97]. On the other hand, another study using checkpoint inhibitors in poorly immunogenic tumor models showed that combination treatment of mice with entinostat, a class-I HDAC inhibitor, and anti-PD-1 or anti-CTLA-4 enhanced tumor control and inhibited metastasis development correlating with low numbers of MDSCs in the tumor [98]. Hence, cytotoxic T-cells obtained upon immune checkpoint blockade were not fully functional unless immune suppressor cells were reduced by treatment with epigenetic modulators. Such studies show the complexity behind epigenetic regulation having opposite effects on the function and migration of immunosuppressive cells. Youn et al. demonstrated that HDAC2 promoted differentiation of monocytic MDSCs into polymorphonuclear MDSC, the latter of which are the major MDSC subtype accumulating in the tumor [99]. These two MDSC subtypes use different suppressive mechanisms. The impact for cancer immunotherapy of this transition from one MDSC subtype to the other, needs to be elucidated.
There are two groups of macrophages within the tumor, referred to as tumor-associated macrophages (TAMs). Type I macrophages (M1) represent potent effector cells, mediating tumor cell killing and the production of proinflammatory cytokines. In contrast, type II or alternatively activated macrophages (M2) suppress inflammatory responses, promote angiogenesis, and enhance tumor invasion [100–102]. Even though some studies suggested that macrophage polarization was epigenetically regulated, the molecular mechanisms behind the suppressive functions of M2 need to be elucidated [36, 103]. Ishii et al. reported that the expression of M2 signature genes was regulated by histone methylation, i.e., H3K4 versus H3K27 [104]. Furthermore, IL-4 was shown to reduce H3K27 methylation in the promoter regions of M2 genes by concomitant activation of H3K27 demethylase Jumonji domain containing 3 (JMJD3). This was mediated through the signal transducer and activator of transcription (STAT) 6 [104], the latter being downstream of the IL-4 receptor signaling pathway [105]. One of the major features of M2 is the down-regulation of MHC-II. In line with this, it has been shown that decoy receptor 3, a member of the TNF receptor superfamily, when overexpressed in tumors leads to the upregulation of genes characteristic of M2 and inhibits the expression of MHC-II on macrophages via histone deacetylation [106].
EZH2-specific regulation
EZH2, a prominent epigenetic regulator
The EZH2 catalytic subunit of the polycomb repressive complex 2 (PRC2) is a highly conserved histone methyltransferase. It mediates the tri-methylation of lysine 27 on histone 3 (H3K27me3), thus inducing chromatin compaction and transcriptional repression of target genes (Fig. 2) [107, 108]. Several studies have shown an essential role of EZH2 for cancer development and progression. High levels of EZH2 have been associated with more aggressive forms of melanoma, renal cell carcinoma, prostate, breast, bladder, endometrial and gastric cancer. Overexpression of EZH2 in many cancers is due to either a gain-of-function mutation in tyrosine 641 or a loss-of-function mutation in EZH2 antagonists, including ubiquitously transcribed tetratricopeptide repeat gene on X chromosome (UTX) and BRG1-/BRM-associated factor (BAF), the latter of which is the human analog of switch/sucrose non-fermentable-A (SWI/SNF-A) [109, 110]. Interestingly, EZH2 overexpression has been linked to inefficient T-cell response in uveal melanoma [111]. Because of its role in cancer, EZH2 has been validated as a promising target for the development of small molecule inhibitors such as 3-deazaneplanocin A (DZNep), GSK126, GSK343, and GSK503 (see also Table 2) [112, 113].
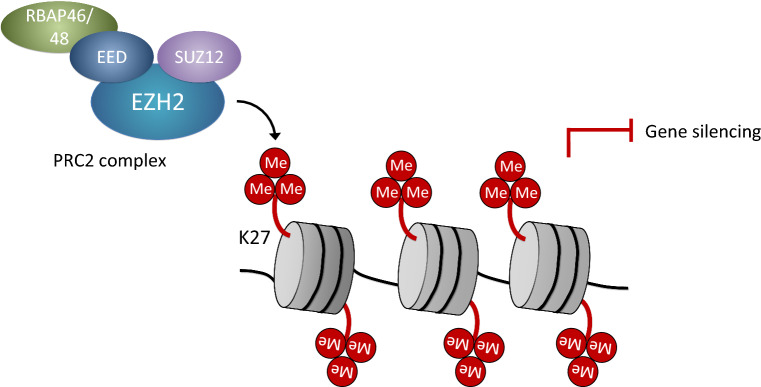
Fig. 2.
Polycomb repressive complex 2-mediated transcriptional repression. The polycomb repressive complex 2 (PRC2) is made of the core components enhancer of zeste homolog 2 (EZH2), embryonic ectoderm development (EED), and suppressor of zeste 12 (SUZ12). These associate with other subunits, such as RBAP46 (also termed RBBP7), RBAP48 (RBBP4), Jumonji and AT-rich interaction domain containing 2 (JARID2; not shown), AE binding protein 2 (AEBP2; not shown) and polycomb-like proteins (PCL; not shown). The PRC2 complex binds to unmethylated cytosin-guanine dinucleotide (CpG) islands and, via tri-methylation of lysine 27 in histone 3 (H3K27m3) of adjacent nucleosomes, mediates silencing of repressed genes
Table 2.
PRC2-targeting agents in clinical development.
Source: Clinicaltrials.gov; accessed 8 Aug 2018
| PRC2-targeting agent | Type of malignancy | Status |
|---|---|---|
| EZH2 inhibitors | ||
| Tazemetostat (also known as EPZ-6438 and E7438) (Epizyme, Inc.) |
B-cell non-Hodgkin lymphoma Diffuse large B-cell lymphoma Follicular lymphoma Malignant mesothelioma Malignant rhabdoid tumors of different organs (kidney, ovary) Atypical teratoid rhabdoid tumors Synovial sarcoma Epitheliod sarcoma Mesothelioma Poorly differentiated chordoma Renal medullary carcinoma Advanced solid tumors |
Phase I/II |
| CPI-1205 (Constellation Pharmaceuticals) |
B-cell lymphoma Advanced solid tumors (including metastatic castration-resistant prostate cancer) |
Phase Ib/II |
| GSK2816126 (also known as GSK126) (GlaxoSmithKline) |
Diffuse large B-cell lymphoma Transformed follicular lymphoma Non-Hodgkin lymphoma Multiple myeloma Advanced solid tumors (including castration-resistant prostate cancer) |
Phase I |
| PF-06821497 (Pfizer) |
Diffuse large B-cell lymphoma Follicular lymphoma Small cell lung cancer Castration-resistant prostate cancer |
Phase I |
| SHR2554 (Jiangsu HengRui Medicine Co., Ltd.) | Mature lymphoid neoplasms | Phase I |
| EED inhibitor | ||
| MAK683 (Novartis Pharmaceuticals) | Diffuse large B-cell lymphoma | Phase I/II |
EZH2-mediated regulation of cancer cells
Although the involvement of EZH2 on cancer development and progression is supported by several studies, the molecular mechanisms linking EZH2 to tumor immune escape are beginning to emerge. There is evidence that EZH2 induces dedifferentiation and proliferation of cancer cells, which is in line with its biological role in maintaining stem cell function [114–116]. Furthermore, EZH2 can suppress DNA repair pathways leading to accumulation of driver mutations and promote epithelial-to-mesenchymal transition (EMT) [117, 118]. In addition to promoting tumorigenesis, traits such as invasion, dedifferentiation, stemness and EMT have been linked to immune evasion.
Recent studies suggested a role of EZH2 on T-cell immunity. It has been well-established that infiltration of CD8+ T-cells to the tumor microenvironment correlates with better prognosis of cancer patients. As previously mentioned, T-cell migration to the site of cancer depends mainly on the chemokines CXCL9 and CXCL10 [119]. Inhibition of EZH2 leads to increased CXCL9 and CXCL10 expression in human ovarian and colon cancer cells, as well as in B16-F10 mouse melanoma in vivo, suggesting that EZH2 alters the anti-tumor immune response by interfering with T-cell trafficking [120–122]. A similar effect on chemokine expression was observed upon inhibition of DNMT-1 in ovarian cancer cells. In line with these results, a combination of GSK126 and 5-AZA-2, synergized with adoptive cell therapy in a mouse model of ovarian cancer, lead to increased T-cell infiltration and improved tumor control [120].
We have recently investigated the role of EZH2 in acquired resistance to cancer immunotherapy. Using three different mouse models of melanoma, our studies revealed that treatment with the checkpoint inhibitor anti-CTLA-4 or use of a CD122-biased IL-2 immunotherapy (more precisely, IL-2 complexes [123, 124]) promoted EZH2 upregulation in cancer cells leading to loss of tumor control [122]. Tumor intrinsic activation of EZH2 led to methylation of H3K27 and consequent suppression of essential immune-related genes, including those involved in melanocyte lineage, MHC-I, antigen processing and presentation machinery, immunoproteasome, and T-cell-attractant chemokines. Interestingly, EZH2 upregulation by melanoma cells was dependent on T-cell infiltration into the tumor and T-cell-dependent TNF production. Upon EZH2 inhibition using GSK503 or tumor cell-specific RNA interference, tumor immunogenicity and T-cell infiltration was restored, resulting in improved tumor control upon immunotherapy [122]. These findings provide a mechanistic insight into the upstream and downstream effects of EZH2 activation, which overall results in adaptive cancer resistance to immunotherapy. Furthermore, these data nicely fit and extend the report by Tüting et al. who described reversible dedifferentiation of melanoma cells in response to TNF exposure [125].
A recent study has identified another key chromatin regulator of tumor immune resistance. Using a genome-scale CRISPR-Cas9 screen the investigators of that study were able to identify key genes involved in resistance of tumor cells to T-cell-mediated lysis. The study revealed that inactivation of any of the three unique genes ARID2, PBRM1 and BRD7 of the polybromo and BRG1-associated factors (PBAF) complex sensitized melanoma cells to cytotoxic T-cells via an enhanced response to IFN-γ [126]. PBAF played a central role in epigenetic regulatory mechanisms by the regulation of chromatin accessibility for transcription factors. PBAF-deficient tumor cell lines produced higher amounts of CXCL9 and CXCL10, therefore allowing better T-cell infiltration to the tumor site. Importantly, inactivating mutations of these genes in patients with metastatic cancers were associated with improved clinical responses to checkpoint inhibitors.
Regulation of effector-type immune cells by EZH2
EZH2 expression is not restricted to cancer or stem cells but is also present in immune cells. It has been shown that EZH2 is important for differentiation of CD4+ T-cells. Upon in vitro polarization into Th1, Th2 and Th17 cells, EZH2-deficient CD4+ T-cells produced higher concentrations of all signature cytokines compared to their wild-type counterparts [127, 128]. This was most notable for the Th1 cytokine IFN-γ, which was synthesized at increased levels even by non-polarized EZH2-deficient CD4+ T-cells. However, when challenged with the intracellular pathogen T. gondii, CD4cre EZH2flox mice with a CD4+ T-cell-restricted deficiency in EZH2 produced lower amounts of IFN-γ and failed to control the infection. This was due to premature senescence of CD4+ T-cells, illustrating the requirement of EZH2 for optimal effector CD4+ T-cell differentiation and function [128]. In line with this, conditional deletion of EZH2 from CD4+ T-cells reduced numbers of pathogenic Th1 cells in a mouse model of aplastic anemia [129].
In addition, a function of EZH2 on CD8+ T-cells has been illustrated. EZH2 expression has been detected in human CD8+ T-cells in various tissues, including in ovarian cancer. EZH2+ CD8+ T-cells did not express markers associated with exhaustion and were able to produce effector cytokines [130]. Further analysis revealed that EZH2 promoted Notch signaling leading to increased survival and effector function of CD8+ T-cells. In the tumor microenvironment, the percentage of EZH2+ CD8+ T-cells were lower compared to peripheral blood and non-malignant tissues, suggesting that the tumor microenvironment interfered with EZH2 expression of T-cells. Indeed, cancer cells restricted T-cell-mediated immunity via inhibition of their EZH2 expression thereby limiting their glycolysis pathway. Thus, EZH2 expression can lead to the advantageous survival of CD8+ T-cells in the tumor microenvironment. Likewise, EZH2 expression by T-cells correlated with better survival and prognosis in patients. Similarly, another study revealed that EZH2 was crucial for the development and maintenance of T-cell memory precursors, correlating with enhanced tumor control [131]. Furthermore, Gunawan et al. showed that the infiltration of EZH2-deficient DCs into inflamed tissues was impaired, leading to poor T-cell responses [132].
These studies suggested that EZH2 can play opposite roles on T-cells and tumor cells revealing effects on immune cells that should be taken into consideration with the systemic use of epigenetic therapies. However, the role of EZH2 on immune cells might depend on the tumor type and tumor microenvironment, as systemic inhibition of EZH2 did not affect T-cell proliferation and effector functions in mouse models of melanoma [122].
NK cells are able to generate rapid responses against foreign, infected or cancerous cells in the absence of antigen presentation by MHC-I. Several studies have described the contribution of epigenetic mechanisms to the development and fate of NK cells. In vitro studies showed that inhibition of histone deacetylation promotes apoptosis, decreases cytotoxicity and chemokine secretion by NK cells [133]. Moreover, the expression of essential genes for NK cell diversity and maturity has been shown to be regulated by DNA methylation and histone acetylation [134, 135]. Furthermore, a role for the PRC2 complex in NK cell development and differentiation has been initially described through the regulation of homeobox genes HOXA9 and HOXA10 [136]. A subsequent study showed that EZH2 inactivation correlated with an increased number of NK cell precursors [137]. In line with this, EZH2 blockade promoted NK cell development and led to higher expression of the IL-2 receptor β (also termed CD122), NK cell-activating receptor NKG2D, Toll-like receptors, and granzymes in NK cells. This correlated with the proliferation, activation and cytotoxic activity of NK cells, providing new insights into systemic effect of EZH2 interference.
EZH2-mediated regulation of Treg cells
EZH2 has also been shown to be required for Treg cell function. Thus, although they did not display a significant phenotypic difference in vivo, EZH2-deficient mature Treg cells were unable to establish tolerance or suppress experimental colitis [128]. Moreover, EZH2-deficient Treg cells were unable to maintain a FoxP3 differentiation program and thus were unstable and dysfunctional upon activation [128, 138]. A recent study has demonstrated a selective upregulation of EZH2 in tumor-infiltrating Treg cells compared to effector T-cells or Treg cells in peripheral blood. Specific disruption of the EZH2 gene in Treg cells resulted in an increased anti-cancer immune response correlating with improved tumor control [139]. In line with these findings, systemic treatment with the EZH2 inhibitor GSK503 in combination with IL-2 immunotherapy correlated with lower intratumoral IL-10 and TGF-β expression [122]. Collectively, Treg cells appear to require an EZH2-mediated epigenetic program for their suppressive activity and maintenance.
Conclusion
The discovery of immune checkpoint inhibitors led to the development of novel and efficient treatments for patients with metastatic malignancies. However, a major limitation of single or combined immunotherapies is the development of secondary or acquired resistance. To improve immunotherapy, efforts taken in studying the different mechanisms of intrinsic and extrinsic resistance development need to be intensified. Epigenetic reprogramming is involved in a variety of mechanisms used by neoplastic cells to escape from immune control. The inherent reversibility of epigenetic modifications makes this mechanism an attractive target for the development of inhibitors able to modulate the activity of DNA methyltransferases, histone deacetylases and histone methyltransferases (Tables 1, 2). The study of these epigenetic drugs in vitro and in various pre-clinical models showed their capacity to regulate different pathways and molecules involved in the interaction of the immune system with cancer cells. These efforts are currently translating to clinical trials on epigenetic-modifying drugs in combination with immune checkpoint inhibitors, such as NCT03179943 and NCT02664181.
However, other studies performed using epigenetic drugs for cancer therapy showed that they can play a Janus-faced role in the tumor microenvironment, challenging their clinical application. One example is the histone methyltransferase EZH2, the activity of which can lead to opposite effects on cancer and T-cells. EZH2 upregulation on cancer cells contributes to tumor immune escape through the downregulation of tumor antigen presentation and chemokine expression resulting in reduced T-cell infiltration, thereby contributing to tumor cell intrinsic and extrinsic mechanisms of immune resistance [120–122]. Thus, inhibition of EZH2 can result in improved tumor control through a variety of mechanisms (Fig. 3). By contrast, EZH2 might convey to T-cells’ survival advantages in the tumor microenvironment [130], which however appear to depend on the tumor type and the treatment used [122].
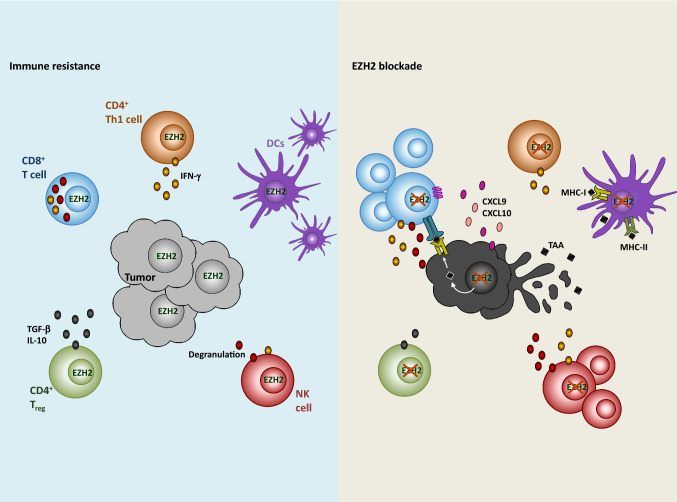
Fig. 3.
EZH2-mediated immune resistance in anti-tumor immune responses. EZH2 expression (left) mediates downregulation of T-cell-attracting chemokines resulting in decreased T-cell infiltration to the tumor site. The lack of antigen presentation limits T-cell effector functions. Upon EZH2 blockade (right), expression of CXCL9 and CXCL10 increases and promotes CD8+ T-cell (blue) infiltration. The recognition of cancer cells by CD8+ T cell through the MHC-I–TAA–TCR complex leads to the secretion of effector molecules, such as perforin and granzymes (dark red). CD4+ T cells (orange) require EZH2 for their differentiation and maintenance. Thus, the function of Th1 cells and regulatory T (Treg) cells can be altered upon EZH2 blockade, including production of IFN-γ, interleukin-10 (IL-10) and transforming growth factor-β (TGF-β). EZH2 is important for infiltration of dendritic cells (DCs; purple) to the inflammatory microenvironment. However, EZH2-dependent downregulation of TAA affects DC-mediated priming of T-cells. Natural killer (NK) cells (red) mature and show enhanced lytic activity upon EZH2 inhibition. In summary, although EZH2 inhibition can affect DC migration and Th1 cell function, different mechanisms involved in tumor resistance can be efficiently reversed, thus improving tumor control
In conclusion, the recent data clearly highlight the role of EZH2 inhibitors in reversing, at least, certain features of adaptive resistance of tumors to immunotherapy. Future directions of research and development should focus on which immunotherapies best combine with EZH2 inhibition. Thus, IL-2 immunotherapy as well as anti-CTLA-4 antibody treatment combined well with EZH2 inhibition, whereas PD-1 blockade did not show any significant synergy in the tested melanoma models [122]. Such effects might also depend on the cancer type, the stage of cancer, or the EZH2-targeting agent [120, 122]. Moreover, it will be important to determine whether EZH2 inhibitors will be equally efficacious in malignancies with wild-type EZH2 versus gain-of-function EZH2 mutations. In certain malignancies, the systemic use of EZH2 inhibitors could lead to antagonistic effects. Hence, the development of tumor-targeted EZH2 inhibition could be beneficial. Alternatively, the different effects of EZH2 blockade might require different doses of EZH2 inhibition. Furthermore, the analysis of tumor samples from treatment responders, non-responders and those showing a long-term disease stabilization following immunotherapy could provide valuable information on the epigenetic changes in cancer cells and the tumor microenvironment contributing to treatment response versus development of resistance. Such analysis might also provide insight into the contribution of epigenetic factors in tumor, stromal and immune cells. Additionally, such studies might help in the discovery of biomarkers, including those predicting a favorable response to EZH2 blockers as well as prognosis and response to combination immunotherapy.
Acknowledgements
We thank Catherine Crowley-Kühn for critical reading of the manuscript. This work was funded by the Sassella Foundation (to N. A. R.), Swiss National Science Foundation Grant 310030-172978 (to O. B.), Swiss Cancer Research Grant KFS-4136-02-2017 (to O. B.), and Hochspezialisierte Medizin Schwerpunkt Immunologie (HSM-2-Immunologie; to O. B.).
Footnotes
Natalia Arenas-Ramirez and Dilara Sahin contributed equally.
Contributor Information
Natalia Arenas-Ramirez, Phone: +41 44 255 2069, Email: natalia.venetzarenas@uzh.ch.
Onur Boyman, Phone: +41 44 255 2069, Email: onur.boyman@uzh.ch.
References
- 1.Coley WB. The treatment of malignant tumors by repeated inoculations of erysipelas: with a report of ten original cases. Am J Med Sci. 1893;105:487–511. [PubMed] [Google Scholar]
- 2.McCarthy EF. The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop J. 2006;26:154–158. [PMC free article] [PubMed] [Google Scholar]
- 3.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garon EB, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 5.Larkin J, Hodi FS, Wolchok JD. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373(13):1270–1271. doi: 10.1056/NEJMc1509660. [DOI] [PubMed] [Google Scholar]
- 6.Ribas A, et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016;315(15):1600–1609. doi: 10.1001/jama.2016.4059. [DOI] [PubMed] [Google Scholar]
- 7.Robert C, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372(26):2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 8.Sharma P, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anagnostou V, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7(3):264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482(7385):400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 12.Verdegaal EM, et al. Neoantigen landscape dynamics during human melanoma–T cell interactions. Nature. 2016;536(7614):91–95. doi: 10.1038/nature18945. [DOI] [PubMed] [Google Scholar]
- 13.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 14.Peng W, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6(2):202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin DS, et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2017;7(2):188–201. doi: 10.1158/2159-8290.CD-16-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaretsky JM, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375(9):819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, et al. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167(2):397 e9–404 e9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel SJ, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548(7669):537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spranger S, et al. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 2017;31(5):711.e4–723.e4. doi: 10.1016/j.ccell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casey SC, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352(6282):227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lastwika KJ, et al. Control of PD-L1 expression by oncogenic activation of the AKT–mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76(2):227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 22.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spranger S, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5(200):200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benci JL, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540.e12–1554.e12. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shayan G, et al. Adaptive resistance to anti-PD1 therapy by Tim-3 upregulation is mediated by the PI3K–Akt pathway in head and neck cancer. Oncoimmunology. 2017;6(1):e1261779. doi: 10.1080/2162402X.2016.1261779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med. 2017;23(5):551–555. doi: 10.1038/nm.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brand A, et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 2016;24(5):657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 28.Kiss Mate, Van Gassen Sofie, Movahedi Kiavash, Saeys Yvan, Laoui Damya. Myeloid cell heterogeneity in cancer: not a single cell alike. Cellular Immunology. 2018;330:188–201. doi: 10.1016/j.cellimm.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Lin YC, et al. Activated but not resting regulatory T cells accumulated in tumor microenvironment and correlated with tumor progression in patients with colorectal cancer. Int J Cancer. 2013;132(6):1341–1350. doi: 10.1002/ijc.27784. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13(1):23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solito S, et al. A human promyelocytic-like population is responsible for the immune suppression mediated by myeloid-derived suppressor cells. Blood. 2011;118(8):2254–2265. doi: 10.1182/blood-2010-12-325753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L, et al. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell. 2004;6(4):409–421. doi: 10.1016/j.ccr.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 33.Kiss M, et al. Myeloid cell heterogeneity in cancer: not a single cell alike. Cell Immunol. 2018 doi: 10.1016/j.cellimm.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13(10):739–752. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis CE, Harney AS, Pollard JW. The multifaceted role of perivascular macrophages in tumors. Cancer Cell. 2016;30(2):365. doi: 10.1016/j.ccell.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Mantovani A, et al. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motz GT, et al. Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat Med. 2014;20(6):607–615. doi: 10.1038/nm.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caruana I, et al. Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes. Nat Med. 2015;21(5):524–529. doi: 10.1038/nm.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vetizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maio M, et al. Molecular pathways: at the crossroads of cancer epigenetics and immunotherapy. Clin Cancer Res. 2015;21(18):4040–4047. doi: 10.1158/1078-0432.CCR-14-2914. [DOI] [PubMed] [Google Scholar]
- 41.Kanwal R, Gupta K, Gupta S. Cancer epigenetics: an introduction. Methods Mol Biol. 2015;1238:3–25. doi: 10.1007/978-1-4939-1804-1_1. [DOI] [PubMed] [Google Scholar]
- 42.Nebbioso A, et al. Trials with ‘epigenetic’ drugs: an update. Mol Oncol. 2012;6(6):657–682. doi: 10.1016/j.molonc.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coulie PG, et al. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14(2):135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 44.Heninger E, Krueger TE, Lang JM. Augmenting antitumor immune responses with epigenetic modifying agents. Front Immunol. 2015;6:29. doi: 10.3389/fimmu.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huijbers IJ, et al. Minimal tolerance to a tumor antigen encoded by a cancer-germline gene. J Immunol. 2012;188(1):111–121. doi: 10.4049/jimmunol.1002612. [DOI] [PubMed] [Google Scholar]
- 46.Scanlan MJ, et al. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 47.James SR, Link PA, Karpf AR. Epigenetic regulation of X-linked cancer/germline antigen genes by DNMT1 and DNMT3b. Oncogene. 2006;25(52):6975–6985. doi: 10.1038/sj.onc.1209678. [DOI] [PubMed] [Google Scholar]
- 48.Yu J, et al. Methylation profiles of thirty four promoter-CpG islands and concordant methylation behaviours of sixteen genes that may contribute to carcinogenesis of astrocytoma. BMC Cancer. 2004;4:65. doi: 10.1186/1471-2407-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu J, et al. Methylation profiling of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. BMC Cancer. 2002;2:29. doi: 10.1186/1471-2407-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith HA, et al. Expression and immunotherapeutic targeting of the SSX family of cancer-testis antigens in prostate cancer. Cancer Res. 2011;71(21):6785–6795. doi: 10.1158/0008-5472.CAN-11-2127. [DOI] [PubMed] [Google Scholar]
- 51.Weber J, et al. Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2′-deoxycytidine. Cancer Res. 1994;54(7):1766–1771. [PubMed] [Google Scholar]
- 52.Dubovsky JA, McNeel DG. Inducible expression of a prostate cancer-testis antigen, SSX-2, following treatment with a DNA methylation inhibitor. Prostate. 2007;67(16):1781–1790. doi: 10.1002/pros.20665. [DOI] [PubMed] [Google Scholar]
- 53.Goodyear O, et al. Induction of a CD8+ T-cell response to the MAGE cancer testis antigen by combined treatment with azacitidine and sodium valproate in patients with acute myeloid leukemia and myelodysplasia. Blood. 2010;116(11):1908–1918. doi: 10.1182/blood-2009-11-249474. [DOI] [PubMed] [Google Scholar]
- 54.Rao M, et al. Inhibition of histone lysine methylation enhances cancer-testis antigen expression in lung cancer cells: implications for adoptive immunotherapy of cancer. Cancer Res. 2011;71(12):4192–4204. doi: 10.1158/0008-5472.CAN-10-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toor AA, et al. Epigenetic induction of adaptive immune response in multiple myeloma: sequential azacitidine and lenalidomide generate cancer testis antigen-specific cellular immunity. Br J Haematol. 2012;158(6):700–711. doi: 10.1111/j.1365-2141.2012.09225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia-Lora A, et al. MHC class I-deficient metastatic tumor variants immunoselected by T lymphocytes originate from the coordinated downregulation of APM components. Int J Cancer. 2003;106(4):521–527. doi: 10.1002/ijc.11241. [DOI] [PubMed] [Google Scholar]
- 57.Serrano A, et al. Rexpression of HLA class I antigens and restoration of antigen-specific CTL response in melanoma cells following 5-aza-2′-deoxycytidine treatment. Int J Cancer. 2001;94(2):243–251. doi: 10.1002/ijc.1452. [DOI] [PubMed] [Google Scholar]
- 58.Li H, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5(3):587–598. doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Setiadi AF, et al. Epigenetic control of the immune escape mechanisms in malignant carcinomas. Mol Cell Biol. 2007;27(22):7886–7894. doi: 10.1128/MCB.01547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Setiadi AF, et al. Epigenetic enhancement of antigen processing and presentation promotes immune recognition of tumors. Cancer Res. 2008;68(23):9601–9607. doi: 10.1158/0008-5472.CAN-07-5270. [DOI] [PubMed] [Google Scholar]
- 61.Ritter C, et al. Epigenetic priming restores the HLA class-I antigen processing machinery expression in Merkel cell carcinoma. Sci Rep. 2017;7(1):2290. doi: 10.1038/s41598-017-02608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chou SD, et al. Histone acetylation regulates the cell type specific CIITA promoters, MHC class II expression and antigen presentation in tumor cells. Int Immunol. 2005;17(11):1483–1494. doi: 10.1093/intimm/dxh326. [DOI] [PubMed] [Google Scholar]
- 63.Khan AN, Magner WJ, Tomasi TB. An epigenetically altered tumor cell vaccine. Cancer Immunol Immunother. 2004;53(8):748–754. doi: 10.1007/s00262-004-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Magner WJ, et al. Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J Immunol. 2000;165(12):7017–7024. doi: 10.4049/jimmunol.165.12.7017. [DOI] [PubMed] [Google Scholar]
- 65.Maeda T, et al. Up-regulation of costimulatory/adhesion molecules by histone deacetylase inhibitors in acute myeloid leukemia cells. Blood. 2000;96(12):3847–3856. [PubMed] [Google Scholar]
- 66.Nencioni A, et al. Histone deacetylase inhibitors affect dendritic cell differentiation and immunogenicity. Clin Cancer Res. 2007;13(13):3933–3941. doi: 10.1158/1078-0432.CCR-06-2903. [DOI] [PubMed] [Google Scholar]
- 67.Rosenzweig JM, et al. KLF4 modulates expression of IL-6 in dendritic cells via both promoter activation and epigenetic modification. J Biol Chem. 2013;288(33):23868–23874. doi: 10.1074/jbc.M113.479576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hervouet E, et al. DNA methylation and apoptosis resistance in cancer cells. Cells. 2013;2(3):545–573. doi: 10.3390/cells2030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hopkins-Donaldson S, et al. Silencing of death receptor and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ. 2003;10(3):356–364. doi: 10.1038/sj.cdd.4401157. [DOI] [PubMed] [Google Scholar]
- 70.Eramo A, et al. Inhibition of DNA methylation sensitizes glioblastoma for tumor necrosis factor-related apoptosis-inducing ligand-mediated destruction. Cancer Res. 2005;65(24):11469–11477. doi: 10.1158/0008-5472.CAN-05-1724. [DOI] [PubMed] [Google Scholar]
- 71.Lucas DM, et al. The histone deacetylase inhibitor MS-275 induces caspase-dependent apoptosis in B-cell chronic lymphocytic leukemia cells. Leukemia. 2004;18(7):1207–1214. doi: 10.1038/sj.leu.2403388. [DOI] [PubMed] [Google Scholar]
- 72.Insinga A, et al. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med. 2005;11(1):71–76. doi: 10.1038/nm1160. [DOI] [PubMed] [Google Scholar]
- 73.Natoni F, et al. Sodium butyrate sensitises human pancreatic cancer cells to both the intrinsic and the extrinsic apoptotic pathways. Biochim Biophys Acta. 2005;1745(3):318–329. doi: 10.1016/j.bbamcr.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 74.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 75.de Araújo-Souza PS, Hanschke SCH, Viola JPB. Epigenetic control of interferon-gamma expression in CD8 T cells. J Immunol Res. 2015;2015:849573. doi: 10.1155/2015/849573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harland KL, et al. Epigenetic plasticity of Cd8a locus during CD8(+) T-cell development and effector differentiation and reprogramming. Nat Commun. 2014;5:3547. doi: 10.1038/ncomms4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Utzschneider DT, et al. T cells maintain an exhausted phenotype after antigen withdrawal and population reexpansion. Nat Immunol. 2013;14(6):603–610. doi: 10.1038/ni.2606. [DOI] [PubMed] [Google Scholar]
- 79.Ahn E, et al. Demethylation of the PD-1 promoter is imprinted during the effector phase of CD8 T cell exhaustion. J Virol. 2016;90(19):8934–8946. doi: 10.1128/JVI.00798-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Youngblood B, et al. Cutting edge: prolonged exposure to HIV reinforces a poised epigenetic program for PD-1 expression in virus-specific CD8 T cells. J Immunol. 2013;191(2):540–544. doi: 10.4049/jimmunol.1203161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Youngblood B, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35(3):400–412. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sen DR, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354(6316):1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raeber ME, et al. The role of cytokines in T-cell memory in health and disease. Immunol Rev. 2018;283(1):176–193. doi: 10.1111/imr.12644. [DOI] [PubMed] [Google Scholar]
- 84.Pauken KE, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354(6316):1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Topper MJ, et al. Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell. 2017;171(6):1284.e21–1300.e21. doi: 10.1016/j.cell.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ghoneim HE, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170(1):142 e19–157 e19. doi: 10.1016/j.cell.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wrangle J, et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget. 2013;4(11):2067–2079. doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang H, et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia. 2014;28(6):1280–1288. doi: 10.1038/leu.2013.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12(3):180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 90.Wing K, Yamaguchi T, Sakaguchi S. Cell-autonomous and -non-autonomous roles of CTLA-4 in immune regulation. Trends Immunol. 2011;32(9):428–433. doi: 10.1016/j.it.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 91.Letourneau S, et al. IL-2- and CD25-dependent immunoregulatory mechanisms in the homeostasis of T-cell subsets. J Allergy Clin Immunol. 2009;123(4):758–762. doi: 10.1016/j.jaci.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 92.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38(3):414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 93.Overacre AE, Vignali DA. T(reg) stability: to be or not to be. Curr Opin Immunol. 2016;39:39–43. doi: 10.1016/j.coi.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37(5):785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 95.Lal G, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182(1):259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Polansky JK, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38(6):1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 97.Sahakian E, et al. Histone deacetylase 11: a novel epigenetic regulator of myeloid derived suppressor cell expansion and function. Mol Immunol. 2015;63(2):579–585. doi: 10.1016/j.molimm.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim K, et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc Natl Acad Sci USA. 2014;111(32):11774–11779. doi: 10.1073/pnas.1410626111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Youn JI, et al. Epigenetic silencing of retinoblastoma gene regulates pathologic differentiation of myeloid cells in cancer. Nat Immunol. 2013;14(3):211–220. doi: 10.1038/ni.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4(1):71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 102.Sica A, et al. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer. 2006;42(6):717–727. doi: 10.1016/j.ejca.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 103.Ivashkiv LB. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 2013;34(5):216–223. doi: 10.1016/j.it.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ishii M, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114(15):3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heeb LE, et al. Regulation of neutrophils in type 2 immune responses. Curr Opin Immunol. 2018;54:115–122. doi: 10.1016/j.coi.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 106.Chang YC, et al. Epigenetic control of MHC class II expression in tumor-associated macrophages by decoy receptor 3. Blood. 2008;111(10):5054–5063. doi: 10.1182/blood-2007-12-130609. [DOI] [PubMed] [Google Scholar]
- 107.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Comet I, et al. Maintaining cell identity: pRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16(12):803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 109.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22(2):128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Christofides A, et al. Epigenetic regulation of cancer biology and anti-tumor immunity by EZH2. Oncotarget. 2016;7(51):85624–85640. doi: 10.18632/oncotarget.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holling TM, et al. A role for EZH2 in silencing of IFN-gamma inducible MHC2TA transcription in uveal melanoma. J Immunol. 2007;179(8):5317–5325. doi: 10.4049/jimmunol.179.8.5317. [DOI] [PubMed] [Google Scholar]
- 112.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 113.Verma SK, et al. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med Chem Lett. 2012;3(12):1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van Vlerken LE, et al. EZH2 is required for breast and pancreatic cancer stem cell maintenance and can be used as a functional cancer stem cell reporter. Stem Cells Transl Med. 2013;2(1):43–52. doi: 10.5966/sctm.2012-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Adhikary G, et al. Survival of skin cancer stem cells requires the Ezh2 polycomb group protein. Carcinogenesis. 2015;36(7):800–810. doi: 10.1093/carcin/bgv064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zingg D, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 117.Cao Q, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27(58):7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ma DN, et al. MicroRNA-26a suppresses epithelial-mesenchymal transition in human hepatocellular carcinoma by repressing enhancer of zeste homolog 2. J Hematol Oncol. 2016;9:1. doi: 10.1186/s13045-015-0229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mikucki ME, et al. Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints. Nat Commun. 2015;6:7458. doi: 10.1038/ncomms8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Peng D, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527(7577):249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nagarsheth N, et al. PRC2 epigenetically silences Th1-type chemokines to suppress effector T-cell trafficking in colon cancer. Cancer Res. 2016;76(2):275–282. doi: 10.1158/0008-5472.CAN-15-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zingg D, et al. The histone methyltransferase Ezh2 controls mechanisms of adaptive resistance to tumor immunotherapy. Cell Rep. 2017;20(4):854–867. doi: 10.1016/j.celrep.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 123.Arenas-Ramirez N, Woytschak J, Boyman O. Interleukin-2: biology, design and application. Trends Immunol. 2015;36(12):763–777. doi: 10.1016/j.it.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 124.Arenas-Ramirez N, et al. Improved cancer immunotherapy by a CD25-mimobody conferring selectivity to human interleukin-2. Sci Transl Med. 2016;8(367):367ra166. doi: 10.1126/scitranslmed.aag3187. [DOI] [PubMed] [Google Scholar]
- 125.Landsberg J, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490(7420):412–416. doi: 10.1038/nature11538. [DOI] [PubMed] [Google Scholar]
- 126.Pan D, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018;359(6377):770–775. doi: 10.1126/science.aao1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tumes DJ, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 2013;39(5):819–832. doi: 10.1016/j.immuni.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 128.Yang XP, et al. EZH2 is crucial for both differentiation of regulatory T cells and T effector cell expansion. Sci Rep. 2015;5:10643. doi: 10.1038/srep10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tong Q, et al. Ezh2 regulates transcriptional and posttranslational expression of T-bet and promotes Th1 cell responses mediating aplastic anemia in mice. J Immunol. 2014;192(11):5012–5022. doi: 10.4049/jimmunol.1302943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao E, et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat Immunol. 2016;17(1):95–103. doi: 10.1038/ni.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.He S, et al. Ezh2 phosphorylation state determines its capacity to maintain CD8(+) T memory precursors for antitumor immunity. Nat Commun. 2017;8(1):2125. doi: 10.1038/s41467-017-02187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gunawan M, et al. The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat Immunol. 2015;16(5):505–516. doi: 10.1038/ni.3125. [DOI] [PubMed] [Google Scholar]
- 133.Shi X, et al. Epigenetic suppression of the antitumor cytotoxicity of NK cells by histone deacetylase inhibitor valproic acid. Am J Cancer Res. 2016;6(3):600–614. [PMC free article] [PubMed] [Google Scholar]
- 134.Santourlidis S, et al. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169(8):4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 135.Santourlidis S, et al. Lineage-specific transition of histone signatures in the killer cell Ig-like receptor locus from hematopoietic progenitor to NK cells. J Immunol. 2008;180(1):418–425. doi: 10.4049/jimmunol.180.1.418. [DOI] [PubMed] [Google Scholar]
- 136.Nagel S, et al. Polycomb repressor complex 2 regulates HOXA9 and HOXA10, activating ID2 in NK/T-cell lines. Mol Cancer. 2010;9:151. doi: 10.1186/1476-4598-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yin J, et al. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc Natl Acad Sci USA. 2015;112(52):15988–15993. doi: 10.1073/pnas.1521740112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.DuPage M, et al. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity. 2015;42(2):227–238. doi: 10.1016/j.immuni.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang D, et al. Targeting EZH2 reprograms intratumoral regulatory T cells to enhance cancer immunity. Cell Rep. 2018;23(11):3262–3274. doi: 10.1016/j.celrep.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]