Abstract
Exit from mitosis and completion of cytokinesis require the inactivation of mitotic cyclin-dependent kinase (Cdk) activity. In budding yeast, Cdc14 phosphatase is a key mitotic regulator that is activated in anaphase to counteract Cdk activity. In metaphase, Cdc14 is kept inactive in the nucleolus, where it is sequestered by its inhibitor, Net1. At anaphase onset, downregulation of PP2ACdc55 phosphatase by separase and Zds1 protein promotes Net1 phosphorylation and, consequently, Cdc14 release from the nucleolus. The mechanism by which PP2ACdc55 activity is downregulated during anaphase remains to be elucidated. Here, we demonstrate that Cdc55 regulatory subunit is phosphorylated in anaphase in a Cdk1–Clb2-dependent manner. Interestingly, cdc55-ED phosphomimetic mutant inactivates PP2ACdc55 phosphatase activity towards Net1 and promotes Cdc14 activation. Separase and Zds1 facilitate Cdk-dependent Net1 phosphorylation and Cdc14 release from the nucleolus by modulating PP2ACdc55 activity via Cdc55 phosphorylation. In addition, human Cdk1–CyclinB1 phosphorylates human B55, indicating that the mechanism is conserved in higher eukaryotes.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03086-5) contains supplementary material, which is available to authorized users.
Keywords: Cell cycle, Phosphatases, Mitosis, Mitotic exit, PP2A–Cdc55, Zds1, Separase, FEAR
Introduction
Mitotic exit encompasses an ordered series of events that occur between chromosome segregation during anaphase and the completion of cell division by cytokinesis (reviewed in Refs. [1, 2]). The budding yeast phosphatase, Cdc14, is a major regulator of these events. Cdc14 is the major Cdk-counteracting phosphatase, which promotes cyclin destruction, inducing the accumulation of the Cdk1 inhibitor Sic1 and dephosphorylating Cdk1 targets [3, 4]. Cdc14 is kept inactive in the nucleolus by binding to its nucleolar inhibitor, Net1 (also called Cfi1), for most of the cell cycle [5, 6]. Release of Cdc14 from the nucleolus depends on two regulatory networks: the Cdc-Fourteen Early Anaphase Release (FEAR) network, which initiates Cdc14 release, and the mitotic exit network (MEN, also known as the Hippo pathway in higher eukaryotes) that maintains Cdc14 activity following release. Numerous proteins, including separase, Cdk1–Clb2, PP2ACdc55, Zds1, Slk19, polo-like kinase Cdc5, Spo12, Fob1 and Bns1, have been implicated in FEAR-Cdc14 release (reviewed in Refs. [7–9]). The MEN is a G-protein signaling cascade (reviewed in Ref. [10, 11]) that is associated with the spindle pole body (SPBs, yeast centrosomes) and this localization is crucial for their function in mitotic exit [12–16].
In metaphase, upon activation of the anaphase-promoting complex (APC) by its co-activator Cdc20, APCCdc20, securin is ubiquitinated, thereby activating separase. The release of separase from its inhibitor securin is crucial to Cdc14 activation [17–19]. FEAR-dependent Cdc14 release requires Net1 phosphorylation at Cdk consensus sites [20]. Net1 is maintained in an under-phosphorylated state in metaphase by the phosphatase PP2ACdc55 [17]. Separase and Zds1/2-dependent PP2ACdc55 downregulation specifically initiates Cdk1-dependent phosphorylation of Net1 in anaphase [21, 22]. Cdc14 release from the nucleolus is also regulated by Cdc5-dependent Net1 phosphorylation [23, 24]. The first wave of Cdc14 release induced by FEAR eventually activates MEN by dephosphorylating the PAK kinase Cdc15, which further activates the downstream effectors Dbf2–Mob1 and Cdc14 [18, 25–28]. Downregulation of PP2ACdc55 phosphatase also regulates MEN activity through its action on Bfa1 and Mob1 [29]. However, the mechanism by which PP2ACdc55 activity is inactivated during anaphase has yet to be elucidated.
It has been proposed that Zds1 acts as a PP2ACdc55 regulator by recruiting Cdc55 to the nucleolus [22] via their physical interaction through the Zds1-C motif [22, 30]. Nevertheless, PP2ACdc55 cannot be modulated solely as a function of its localization; an additional regulatory step may be required. Here, we demonstrate that Cdc55 phosphorylation is increased upon PP2ACdc55 down-regulation at anaphase onset. Remarkably, Cdc55 phosphorylation depends on Cdk1–Clb2, regulates Cdc14 activation and directly controls PP2ACdc55 phosphatase activity towards Net1. Moreover, Cdc55 phosphorylation requires active separase and functional Zds1 protein. Excitingly, down-regulation of PP2ACdc55 is achieved by increasing the PP2ACdc55 and Net1 physical interaction acting as a substrate trap and could serve as a new paradigm for how PP2A may regulate it substrates. Strikingly, human PP2A-B55 is also phosphorylated by Cdk1–CyclinB1, suggesting that the mechanism is conserved in mammals.
Methods
Yeast strains, plasmids and cell-cycle synchronization procedures
All yeast strains used in this study were derivatives of W303 and are summarized in Table 1. Epitope tagging of endogenous genes and gene deletions was performed by gene targeting using polymerase chain reaction (PCR) products. Cell synchronization using α-factor and metaphase arrest by Cdc20 depletion and entry into synchronous anaphase by Cdc20 re-introduction were also performed as previously described [31]. The plasmids containing Cdc55 mutations were obtained by site-directed mutagenesis using the plasmids E362 (promCDC55-HA3-CDC55-3′utrCDC55) and E427 (promCDC55-Pk3-CDC55-3′utrCDC55): E367 (promCDC55-HA3-CDC55-T174E), E368 (promCDC55-HA3-CDC55-S301D), E377 (promCDC55-HA3-CDC55-T174D-S301D), E378 (promCDC55-HA3-CDC55-T174A), E379 (promCDC55-HA3-CDC55-S301A), E380 (promCDC55-HA3-CDC55-T174A-S301A), E430 (promCDC55-Pk3-CDC55-T174A-S301A) and E463 (promCDC55-Pk3-CDC55-T174E-S301D). All experiments were done at least three times.
Table 1.
List of strains used in this study
| Strain | Genotype | Origin |
|---|---|---|
| Y610 | MATα pph22::HA-PPH21 TRP1::GAL1-FLAG3-ZDS1 CDC14-MYC18 cdc20::MET-CDC20 | This study |
| Y888 | MATa cdc55::HA3-CDC55 TRP1::GAL1-FLAG3-ZDS1 CDC14-Pk9 cdc20::MET-CDC20 | This study |
| Y769 | Matα cdc20::MET-CDC20 TRP1::GAL1-FLAG3-ZDS1 cdc55::HA3-CDC55 | This study |
| Y770 | Matα cdc20::MET-CDC20 cdc55::HA3-CDC55 TRP1::GAL1-FLAG3-zds1Δ803-916 | This study |
| Y1015 | Mata cdc20::MET-CDC20 TRP1::GAL1-FLAG3-ZDS1 cdc55::HA3-CDC55 CDC14-Pk9 NET1-GFP | This study |
| Y2541 | Mata cdc20::GAL1-CDC20 cdc55::HA3-CDC55 | This study |
| Y492 | Mata cdc20::GAL1-CDC20 cdc55::HA3-CDC55 cdc28::cdc28-as1 | This study |
| Y537 | MATa HA3-CDC55 TRP1::GAL1-FLAG3-ZDS1 cdc55::HA3-CDC55 CDC14-Pk9 cdc20::MET-CDC20 cdc28::cdc28-as1 | This study |
| Y1125 | Mata cdc20::MET-CDC20 clb2Δ TRP1::GAL1-FLAG3-ZDS1 cdc55::HA3-CDC55 CDC14-Pk9 | This study |
| Y569 | MATa cdc20::MET-CDC20 cdc55::HA3-CDC55 | This study |
| Y480 | MATa cdc20::MET-CDC20 cdc55::HA3-CDC55 cdc28::cdc28-as1 | This study |
| Y1281 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55 NET1-MYC18 | This study |
| Y1284 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-2A NET1-MYC18 | This study |
| Y1081 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55 GAL1-FLAG3-ZDS1 | This study |
| Y1087 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-2A GAL1-FLAG3-ZDS1 | This study |
| Y1211 | Mata cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55 CDC14-Pk9 | This study |
| Y1215 | Mata cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-T174A CDC14-Pk9 | This study |
| Y1216 | Mata cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55 -S301A CDC14-Pk9 | This study |
| Y1217 | Mata cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55 -2A CDC14-Pk9 | This study |
| Y3034 | MATa cdc20::MET-CDC20 zds1∆ CDC14-Pk9 | Our lab |
| Y564 | MATa cdc20::MET-CDC20 cdc55::HA3-CDC55 | This study |
| Y879 | Mata cdc20::MET-CDC20 cdc55Δ CDC14-Pk9 | Our lab |
| Y1212 | Mata cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-T174E CDC14-Pk9 | This study |
| Y1213 | Mata cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-S301D CDC14-Pk9 | This study |
| Y1214 | Mata cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-ED CDC14-Pk9 | This study |
| Y500 | Mata cdc20::GAL1-CDC20 cdc55::Pk3-CDC55 | This study |
| Y2312 | MATa cdc20::GAL1-CDC20 CDC14-HA6 CLB2-Pk3 | Our lab |
| Y1049 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55 | This study |
| Y1077 | MATa cdc20::MET-CDC20 cdc55Δ cdc55::HA3-CDC55 | This study |
| Y1080 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-2A | This study |
| Y824 | MATα cdc20::MET-CDC20 | Our lab |
| Y1294 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-ED NET1-MYC18 | This study |
| Y1285 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55 ZDS1-PK6 | This study |
| Y639 | MATa cdc20::MET-CDC20 NET1-MYC18 CDC14-Pk9 | Our lab |
| Y1118 | MATa cdc20::MET-CDC20 zds1∆zds2∆ cdc55::HA3-CDC55 YCplac22 | This study |
| Y1192 | MATa cdc20::MET-CDC20 zds1∆zds2∆ cdc55::HA3-CDC55 YCplac22-Zds1Promoter-FLAG3-ZDS1_full length | This study |
| Y1119 | MATa cdc20::MET-CDC20 zds1∆zds2∆ cdc55::HA3-CDC55 YCplac22-Zds1Promoter-FLAG3-zds1∆461-916 | This study |
| Y1122 | MATa cdc20::MET-CDC20 zds1∆zds2∆ cdc55::HA3-CDC55 YCplac22-Zds1Promoter-FLAG3-zds1∆1-802 | This study |
| Y1117 | MATa cdc20::MET-CDC20 zds1∆zds2∆ cdc55::HA3-CDC55 YCplac22-Zds1Promoter-FLAG3-zds1∆74-802 | This study |
| Y1120 | MATa cdc20::MET-CDC20 zds1∆zds2∆ cdc55::HA3-CDC55 YCplac22-Zds1Promoter-FLAG3-zds1∆461-802 | This study |
| Y1288 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-ED ZDS1-PK6 | This study |
| Y1291 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-2A ZDS1-PK6 | This study |
| Y1597 | MATα cdc20::MET-CDC20 zds1Δ zds2Δ cdc28::cdc28Y19F cdc55Δ CDC14-HA6 pRS316 | This study |
| Y1598 | MATα cdc20::MET-CDC20 zds1Δ zds2Δ cdc28::cdc28Y19F cdc55Δ CDC14-HA6 pRS316-Pk3-CDC55 | This study |
| Y1600 | MATα cdc20::MET-CDC20 zds1Δ zds2Δ cdc28::cdc28Y19F cdc55Δ CDC14-HA6 pRS316-Pk3-CDC55-ED | This study |
| Y536 | MATa cdc55::HA3-CDC55 | This study |
| Y567 | MATa cdc55::HA3-CDC55 esp1::esp1-2 | This study |
| Y568 | MATa cdc55::HA3-CDC55 esp1::esp1-2 | This study |
| Y565 | MATa cdc20::MET-CDC20 cdc55::HA3-CDC55 esp1::esp1-2 | This study |
| Y888 | MATa cdc20::MET-CDC20 TRP1::GAL1-FLAG3-ZDS1 cd55::HA3-CDC55 CDC14-Pk9 | This study |
| Y524 | MATa cdc20::MET-CDC20 esp1::esp1-2 TRP1::GAL1-FLAG3-ZDS1 cdc55::HA3-CDC55 CDC14-Pk9 | This study |
| Y2782 | MATα cdc20::MET-CDC20 esp1::esp1-2 cdc55Δ CDC14-HA6 pRS316 | This study |
| Y1561 | MATα cdc20::MET-CDC20 esp1::esp1-2 cdc55Δ CDC14-HA6 pRS316-Pk3-CDC55 | This study |
| Y1481 | MATα cdc20::MET-CDC20 esp1::esp1-2 cdc55Δ CDC14-HA6 pRS316-Pk3-CDC55-ED | This study |
| Y755 | MATa cdc55::HA3-CDC55 ZDS1-FLAG3 | This study |
| Y986 | MATa cdc20::MET-CDC20 cdc55Δ | Our lab |
| Y616 | Mata cdc20::MET-CDC20 pph22::PPH21-HA | This study |
| Y1649 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55 | This study |
| Y1651 | MATa cdc20::MET-CDC20 cdc55Δ URA3::HA3-CDC55-2A | This study |
| Y3227 | MATa cdc20::GAL1-CDC20 CDC14-Pk9 | Our lab |
| Y3034 | Mata cdc20::MET-CDC20 zds1Δ CDC14-Pk9 | Our lab |
Immunoprecipitation, phosphatase assay, and kinase assays
The co-immunoprecipitation assays were performed as described [22, 24]. Protein extracts were prepared by mechanical lysis using glass beads. The clarified extracts were incubated with antibody, and the immunocomplexes were adsorbed onto magnetic protein-A Dynabeads® (Life Technologies). The beads were washed in extraction buffer and the protein-bound fraction eluted with SDS-PAGE loading buffer. The antibody used for immunoprecipitation was the α-Pk clone SV5-Pk1 (Serotec), α-Clb2 (Santa Cruz Biotechnologies) and α-myc 9E10 (Covance). The images were obtained using the Amersham Imager 600 (GE Healthcare) and quantified using ImageQuant software (GE Healthcare). For the quantification of the western blots, all data points from a replicate were first normalized to the control and later mean and SEM were calculated.
The radioactive Cdk1–Clb2 kinase assays were done as described [17]. The non-radioactive kinase assays were done using Pro-Q Diamond to detect phosphorylation [24]. Pk epitope-tagged Clb2 or Clb2 protein were immunopurified on protein-A Dynabeads, as above. The beads were washed two times more with kinase buffer (50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, 5 mM β-glycerolphosphate, 150 μM ATP) and incubated with 20 μL kinase reaction mix containing bacteria-purified GST-Cdc55(1–193) or GST-Cdc55(1–193)-T174A for 30 min at 20 °C. Reactions were terminated by adding 2 × SDS-PAGE loading buffer. Phosphorylation was detected by Pro-Q Diamond staining (Life Technologies) following the manufacturer’s instructions, and images were acquired using Typhoon FLA 9500 (GE Healthcare). Clb2 recovered from the beads and the GST-Cdc55 were visualized using the Amersham Imager 600 (GE Healthcare) and quantified using ImageQuant software (GE Healthcare). For the quantification of the specific kinase activity, the radioactive signal was first normalized by the Clb2 kinase and GST-Cdc55 levels for each point. Then, all data points from a replicate were normalized to the control, mean and SEM were calculated and the analysis of the unpaired two-tailed t test was performed to assess statistical significance.
For the PP2A–Cdc55 phosphatase assays, bacteria-purified Net1(1–600) was previously phosphorylated with Cdk1–Clb2, as before, but after the kinase reaction the phosphorylated-Net1 was recovered from the supernatant and reserved for the phosphatase assays. Pk epitope-tagged Cdc55 was immunopurified on protein-A Dynabeads, as before, and the beads were washed two times more with phosphatase buffer (50 mM Tris–HCl, pH 7.5, 1 mM EGTA, 0.1% β-mercaptoethanol). Reaction mix (phosphatase buffer containing P-Net1) was added to the beads and incubated at 30 °C for 15 min. Reactions were terminated by adding 2 × SDS-PAGE loading buffer. Remaining Net1 phosphorylation was detected by Pro-Q Diamond staining and images were acquired using Typhoon FLA 9500 (GE Healthcare). Cdc55 recovered on the beads was quantified using the Amersham Imager 600 (GE Healthcare) and ImageQuant software (GE Healthcare). For the quantification of the remaining Net1 phosphorylation, the Pro-Q diamond signal was first normalized by the Cdc55 and Net1(1–600)-His levels for each point. Then, all data points from a replicate were first normalized to the control and, mean values and SEM were calculated.
For the kinase assays of human proteins (Fig. 7a), commercially available purified recombinant Cdk1 and Cyclin B1 co-expressed in baculovirus Sf9 insect cells (SRP5009, Sigma-Aldrich) and recombinant human PPP2R2A (B55 alpha subunit) purified from wheat germ (ab159172, Abcam) were purchased and the kinase assays were performed following the manufacturer’s instructions. Briefly, a mix of 10 μL Kinase solution (0.05 μg/μL), 3 μL of substrate solution (0.05 μg/μL), 5 μL of ATP (at final concentrations of 20, 80, and 160 μM) and 5 μL of cold water per sample were prepared, incubated at 30 °C for 15 min and reaction was terminated by adding 2 × SDS-PAGE loading buffer. Phosphorylation was detected by Pro-Q Diamond staining (Life Technologies) following the manufacturer’s instructions, and images were acquired using Typhoon FLA 9500 (GE Healthcare).
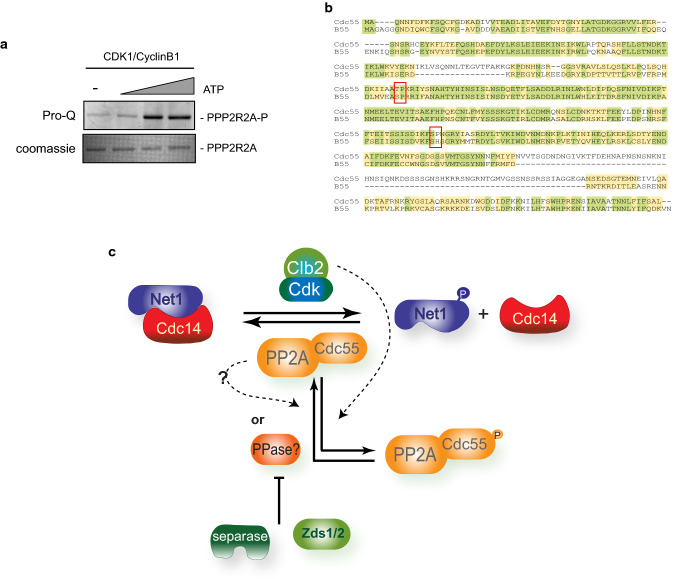
Fig. 7.
Phosphorylation of B55 by Cdk1 is conserved in humans. a Cdk1–CyclinB1 phosphorylates human B55. Kinase assays using recombinant human Cdk1–Cyclin B1 and B55 were performed. A representative image of one of the experiments is depicted. b Alignment of the budding yeast Cdc55 and human B55. Red boxes mark the conserved T174 and S301 in Cdc55. c Model of the Net1 phosphorylation and Cdc14 release regulated by downregulation of the PP2ACdc55 via phosphorylation of the regulatory subunit Cdc55. During metaphase, futile cycles of Cdk1–Clb2 phosphorylation and PP2ACdc55 dephosphorylation maintain low levels of phosphorylated-Net1. During anaphase, Cdc55 phosphorylation promotes the inactivation of the phosphatase activity of the PP2ACdc55 (indicated as black dashed lines) and, as a consequence, Net1 phosphorylation is increased and Cdc14 is released from the nucleolus
Identification of Cdc55 phosphorylation by mass-spectrometry
The immunoprecipitation of Cdc55 was performed as before but using 2 × 109 cells (1 L cultures). Purified Cdc55 were resolved in 4–15% SDS-PAGE gels (Bio-rad) and stained using EZ Blue Coomassie (Sigma). The bands were then excised and in-gel digested with trypsin for LC–MS/MS analysis. Before the LC–MS/MS analysis the peptides were purified with a C18 reverse-phase column to remove any contamination and traces of acrylamide. Peptides were analyzed by LC–MS/MS using an LTQ-Orbitrap Lumos mass spectrometer (Thermo Scientific). Eluted peptides were subjected to electrospray ionization in an emitter needle (PicoTipTM, New Objective, Scientific Instrument Services, Ringoes, NJ, USA) with an applied voltage of 2000 V. Peptides were fragmented using CID (collision-induced dissociation) in the linear ion trap using helium as collision gas with 38% normalized collision energy. Multistage activation was enabled to favor the detection of phosphopeptides. Peptides were identified using Thermo Xcalibur software (v.2.2) (Termo Fisher Scientific). The data analysis was performed in the MaxQuant software (v.1.6.2.6a) using the Andromeda Search Engine with the Saccharomyces cerevisiae SwissProt database (July 2018). The searches were filtered to get a 1% FDR at both protein and peptide level.
rDNA chromatin immunoprecipitation assays
The ChIP-qPCR experiments were performed as previously described [32]. The antibody used for immunoprecipitation was the α-Pk clone SV5-Pk1 (Serotec). Oligonucleotide sequences for quantitative real-time PCR were described in Ref. [33].
Molecular interactions by surface plasmon resonance experiments
The surface plasmon resonance experiments were performed using a BIACORE T200 (GE Healthcare) equipped with a research-grade CM5 sensor chip. The Net1(1–600)-His ligand was immobilized using amine-coupling chemistry. The ligand at a concentration of 0.5 μM in 10 mM sodium acetate, pH 5.0, was immobilized at a density of 500 RU on flow cell 2 and 4; flow cell 1 and 3 were left blank to serve as a reference surface. To collect kinetic binding data, the GST(1–193)-Cdc55 and GST-(1–193)-Cdc55-P analytes in 10 mM HEPES, 500 mM NaCl, 0.005% P-20, pH 7.4, were injected at concentrations of 0.35–5 μM at a flow rate of 30 μL/min and at 25 °C. The complex was allowed to associate and dissociate for 120 and 300 s, respectively. The surfaces were regenerated with a 30 s injection of 50 mM NaOH. Duplicate injections of each analyte were flowed over the surfaces. The data were fit to a simple 1:1 kinetics interaction model using the global data analysis option available or to a steady-state affinity curve within BiaEvaluation 3.1 software.
Other techniques
Protein extracts for western blots were obtained by NaOH or TCA protein extraction. For the phostag gels, 20 μM Phostag (Wako) 6% gels containing 100 μM MnCl2 were used. Protein gels were washed with 1 mM EDTA before protein transfer to remove the manganese. For the quantification of the Cdc55 phosphorylation, the ratio of the quantified upper and lower bands for each point was calculated. Then, all data points from a replicate were normalized to the control and, mean and SEM were calculated.
Antibodies used for western blots and immunofluorescence were α-HA clone 12CA5 (Roche), α-phosphoserine/threonine antibody clone 22A (BD Biosciences), α-Cdc14 (yE-17) sc-12045 (Santa Cruz Biotechnology), α-myc 9E10 (Covance), α-FLAG clone M2 (Sigma), α-Pk clone SV5-Pk1 (Serotec), α-Clb2 (y-180) sc-907 (Santa Cruz Biotechnology), α-tubulin clone YOL1/34 (Serotec), α-GST (GE Healthcare), α-his ab9108 (Abcam), α-phosphoCdc28Y19 (phosphoCdc2-Tyr15) clone 9111 (Cell Signaling Technology), α-Cdc28 sc-53 (Santa Cruz Biotechnology) α-Tpd3 (Innovagen) and α-phosphoglycerate kinase (Life Technologies). The secondary antibodies were Cy3-labeled α-mouse (GE Healthcare), fluorescein-conjugated α-rat (Millipore) and Cy3-labeled α-goat (Jackson ImmunoResearch).
Statistical analysis
Statistical significance was tested by analysis of the unpaired two-tailed Student’s t test (GraphPad Prism). Values of p < 0.05 were consider statistically significant, p < 0.001***, p > 0.001**; p > 0.01*.
Results
The PP2A regulatory subunit Cdc55 is phosphorylated at anaphase onset
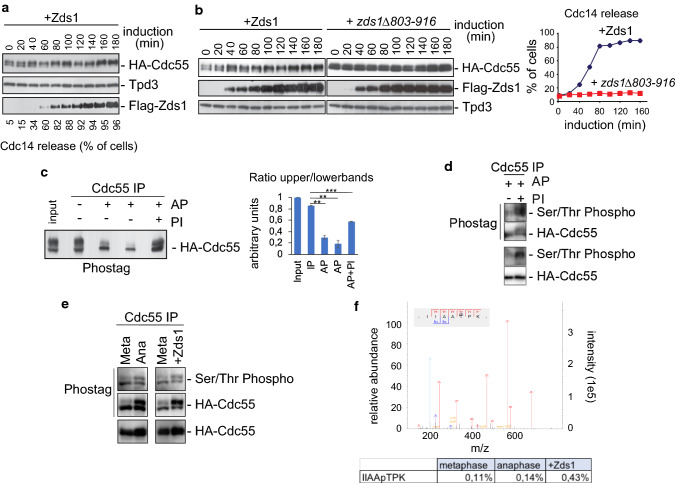
Ectopic Zds1 expression induces the downregulation of PP2ACdc55 and promotes Net1 phosphorylation [21], the Zds_C motif being sufficient to achieve this [22]. As a first approach to studying the detailed mechanism underlying PP2ACdc55 downregulation at anaphase onset, we visualized the three PP2ACdc55 subunits (Pph21, Tpd3 and Cdc55) by western blot after ectopic Zds1 expression. Two isoforms were detected for Cdc55 and Pph21 (Fig. 1a and Fig. S1). We observed that the overexpression of Zds1 in metaphase-arrested cells by Cdc20 depletion promotes the accumulation of the slower migrating form of Cdc55 simultaneously with the nucleolar release of Cdc14 (Fig. 1a). This slowly migrating Cdc55 form is not accumulated when a truncated Zds1 protein lacking the Zds1_C motif is induced. This protein cannot promote the nucleolar release of Cdc14, and is therefore incapable of downregulating PP2ACdc55 [22] (Fig. 1b). These results suggest that Cdc55 suffers a post-translational modification upon Zds1 ectopic expression.
Fig. 1.
PP2A regulatory subunit Cdc55 is phosphorylated upon PP2ACdc55 downregulation. a PP2ACdc55 downregulation induces the accumulation of a Cdc55 post-translational modification. Strain Y888 was arrested in metaphase by Cdc20 depletion, and Zds1 ectopic expression was induced by galactose addition. Cdc55, Tpd3 and Zds1 proteins were analyzed by western blot. Cdc14 release from the nucleolus was visualized by immunofluorescence. b Ectopic expression of a non-functional version of Zds1 cannot promote the Cdc55 post-translational modification. Strains Y769 and Y770 were arrested in metaphase by Cdc20 depletion, and Zds1 ectopic expression was induced. Cdc55, Zds1 and Tpd3 proteins were analyzed by western blot (left). Tpd3 was used as a loading control. Cdc14 release from the nucleolus was visualized by immunofluorescence (right). c Cdc55 experiences phosphorylation modifications. Strain Y1015 was arrested in metaphase by Cdc20 depletion, and Zds1 ectopic expression was induced. After 180 min of Zds1 induction, protein extracts were prepared, Cdc55 was immunoprecipitated, and alkaline phosphatase (AP) treatment was performed. 2x PhosSTOP (Roche) was used as an alkaline phosphatase inhibitor (PI). A representative experiment of one of the assays is shown (left). A quantification of the slower and faster Cdc55 migration isoforms is depicted (right). Mean values and SEM are shown. d Cdc55 is phosphorylated in Ser/Thr residues. Cdc55 immunopurified as in c were detected using an anti-phosphoserine/threonine antibody and anti-HA as a control. One independent experiment is shown. e Phosphorylation of Cdc55 in Ser/Thr residues is enhanced in anaphase and upon Zds1 induction. Protein extracts from strain Y2541 arrested in metaphase (Meta) by Cdc20 depletion, and 20 min (Ana) after released into synchronous anaphase were prepared. Cell extract from Y1015 after 180 min of Zds1 induction was also prepared. Cdc55 was immunopurified, resolved in phostag gels and Ser/Thr phosphorylation was detected using anti-phosphoserine/threonine antibody. Purified HA–Cdc55 protein was also detected by western blot in phostag and normal gels as controls. f In vivo phosphorylation of Cdc55 at T174. Cdc55 immunopurified as in e were submitted to mass-spectrometry. The best MS/MS spectrum corresponding to the anaphase sample is shown. The ratios of modified/unmodified peptide are depicted
To ascertain the nature of the Cdc55 post-translational modification we purified Cdc55 after Zds1 ectopic expression in metaphase-arrested cells and treated it with alkaline phosphatase (AP) (Fig. 1c). In the presence of alkaline phosphatase, the slower migrating form of Cdc55 was greatly reduced but was partially recovered when the alkaline phosphatase was combined with phosphatase inhibitors (PI). Therefore, these results suggest that Cdc55 is phosphorylated upon Zds1 induction.
Next, we wondered whether the post-translational modification corresponds to a Ser/Thr phosphorylation. First, we repeat the alkaline phosphatase treatment in immunopurified-Cdc55 as in (c) and incubate the immunoblots with an anti-phosphoSer/Thr antibody (Fig. 1d). In the presence of AP the Ser/Thr phosphorylation signal was greatly reduced, suggesting that Cdc55 phosphorylation is almost abrogated upon AP treatment. On the contrary, upon inhibition of the AP by PI addition, the Ser/Thr phosphorylation signal was recovered. This suggests that the anti-phosphoSer/Thr antibody does not cross-react with the non-phosphorylated Cdc55 isoforms. Interestingly, these data indicate that the two Cdc55 migrating isoforms contain a mix of phosphorylated and non-phosphorylated Cdc55. Second, we purified Cdc55 in synchronized cells at the metaphase-to-anaphase transition by Cdc20 depletion and release into anaphase by Cdc20 re-addition (Fig. 1e). Purified-Cdc55 was resolved in Phostag gels to improve the visualization of the phosphorylated forms. Two Cdc55 isoforms were resolved in the Phostag gels. The anti-Ser/Thr antibody recognized the lower and faster migration Cdc55 isoforms, suggesting that both bands contained Cdc55-phosphorylated isoforms. Interestingly, the signal of the slower migration band increased in anaphase and upon Zds1 induction. These results indicate that Cdc55 is phosphorylated at Ser/Thr residues and that the phosphorylation intensifies during anaphase and upon Zds1 induction. Finally, to unequivocally identify the Cdc55 phosphorylation we submitted the purified-Cdc55 to mass-spectrometry analysis (Fig. 1f). The phosphopeptide IIAApTPK corresponding to the Thr174 of Cdc55 was identified in metaphase, anaphase and upon Zds1 induction. More interestingly, the relative amount of the phosphopeptide increased 27% at anaphase compare to metaphase cells. This demonstrates that the Cdc55 phosphorylation at Thr174 enhances in anaphase and upon Zds1 induction as suggested by the western blot.
We next wanted to establish whether Cdc55 phosphorylation is cell cycle-dependent. This prompted us to analyze the Cdc55 phospho-isoforms in cell-cycle progression after synchronous release from G1 (Fig. S2). To improve the visualization of the phosphorylated forms we used Phostag gels. Two slower migrating Cdc55 isoforms appeared by 60 min that was coincident with the peak of Clb2 levels, suggesting that Cdc55 was hyperphosphorylated during mitosis. To investigate the Cdc55 phosphorylation further we synchronized cells at the metaphase-to-anaphase transition by Cdc20 depletion and release into anaphase by Cdc20 re-addition (Fig. 2a, WT panel). An accumulation of the slower migration form of Cdc55 was detected during anaphase (after 15–30 min). These results suggest that Cdc55 is hyperphosphorylated during mitosis, specifically during anaphase, coinciding with the PP2ACdc55 downregulation required for Cdc14 activation. All these findings lead us to propose that Cdc55 phosphorylation at anaphase might be the mechanism controlling PP2ACdc55 activity.
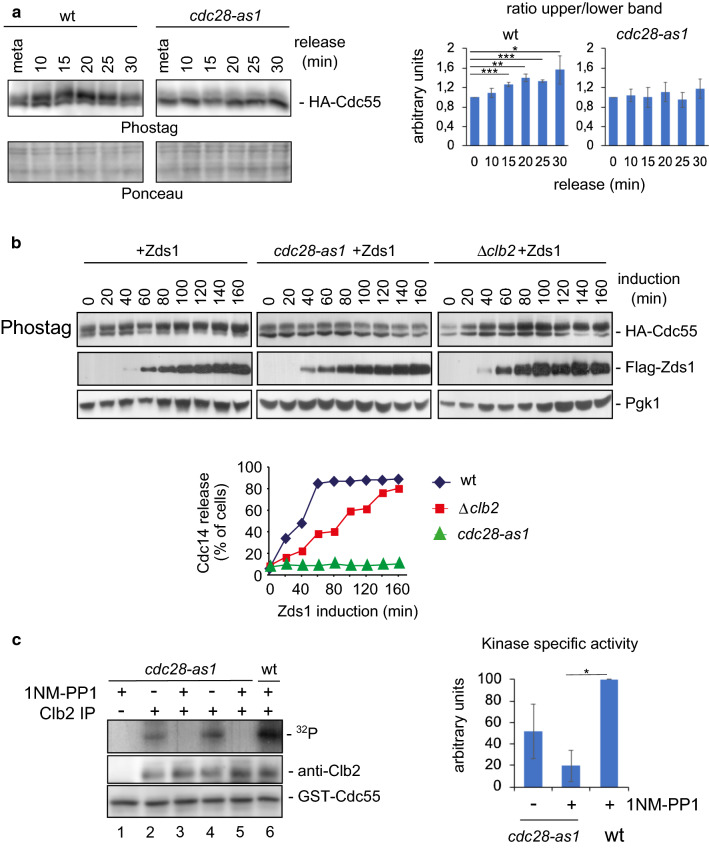
Fig. 2.
Cdk1–Clb2 complex phosphorylates Cdc55. a Cdk1 is required for Cdc55 phosphorylation. Strains Y2541 and Y492 were arrested in metaphase by depletion of Cdc20, the cdc28-as1 allele was inactivated by addition of 1 μM 1NM-PP1 20 min before release into anaphase by Cdc20 re-addition. Cdc55 protein phosphorylation was analyzed by western blot. Ponceau staining is presented as loading control. A representative experiment of one of the assays is shown (left). Quantifications of the slower and faster Cdc55 migration isoforms are depicted (right). Mean values and SEM are illustrated. b The Cdk1–Clb2 complex is necessary for proper Zds1-induced Cdc55 phosphorylation. Strains Y1015, Y537, and Y1125 were arrested in metaphase, the cdc28-as1 allele was inhibited by the addition of 1NM-PP1 during 20 min, and Zds1 expression was induced. Cdc14 release from the nucleolus was monitored by in situ immunofluorescence (bottom) and the Cdc55 phosphorylation and Zds1 levels were analyzed by western blot. Pgk1 was used as loading control. c The Cdk1–Clb2 complex phosphorylates Cdc55 in vitro. Strains Y564 and Y480 were arrested in metaphase by Cdc20 depletion and the cdc28-as1 allele was inactivated using 1 μM 1NM-PP1. Clb2 was purified by immunoprecipitation, and the Cdc28–Clb2 kinase assay was performed using bacteria-purified GST-Cdc55(1–193) as the substrate. An immunoprecipitation using rabbit IgG, instead of the Clb2 antibody, was used as a negative control for the kinase assay. A representative image of one of the experiments is depicted (left). A quantification of four independent immunoprecipitations was performed (right). Mean values and SEM are shown
Cdc28–Clb2 participates in Cdc55 phosphorylation
Cdc14 activation in early anaphase depends on Cdk1-dependent Net1 phosphorylation [17, 20]. In order to test whether Cdc55 phosphorylation is also regulated by Cdk1, we used Phostag gels to check the Cdc55 isoforms in cells carrying the ATP analog-sensitive allele cdc28-as1 [34]. Cells were arrested in metaphase by Cdc20 depletion, then cdc28-as1 was inhibited with 1NM-PP1 and Cdc20 was re-induced. Control cells containing wild-type Cdc28 showed two Cdc55 isoforms at metaphase and the slower migrating isoform accumulated during anaphase (Fig. 2a). In contrast, the Cdc55 slower migrating isoform was greatly reduced in the cdc28-as1 cells in metaphase and anaphase cells. This result suggests that Cdk1 is responsible for Cdc55 phosphorylation. Nevertheless, since the Cdc55 slower migrating band in the cdc28-as1 cells is not completely abolished, we cannot rule out the possibility that another kinase also contributes to Cdc55 phosphorylation.
To investigate further the role of Cdk1–Clb2 in regulating the Cdc55 phosphorylation, we checked Cdc55 phosphorylation upon Zds1 ectopic expression in cells lacking Cdk1 activity. Cells carrying the cdc28-as1 allele or a clb2 deletion were arrested in metaphase by Cdc20 depletion and Zds1 expression was induced (Fig. 2b). 1NM-PP1 and galactose to induce Zds1 were added simultaneously in cdc28-as1 cells. Cells containing wild-type Cdc28 accumulated the slower migrating form of Cdc55, as expected. But inhibition of Cdc28-as1 by 1NM-PP1 avoided further phosphorylation of Cdc55 upon Zds1 induction. Accumulation of the Cdc55 phosphorylated form was delayed in clb2Δ cells concomitantly with a delay in Cdc14 release from the nucleolus. These data suggest that Cdk1–Clb2 participates in Cdc55 phosphorylation.
To explicitly test Cdk1-dependent Cdc55 phosphorylation we measured Cdk1–Clb2 in vitro activity against Cdc55 after Clb2 purification from metaphase-arrested cells, when Cdk1–Clb2 is more active (Fig. 2c). The substrate was a recombinant fragment of Cdc55 spanning amino acids 1–193, containing the phosphorylated residue detected by mass spectrometry: Thr174. Cdc55 phosphorylation was observed in the presence of Cdk1–Clb2: wild-type cells (lane 6) and cdc28-as1 strain without drug (lanes 2, 4). Remarkably, the phosphorylation was impaired in the cdc28-as1 allele inhibited by 1NM-PP1 (lanes 3, 5). Therefore, Clb2-associated Cdk1 activity against GST-Cdc55 was determined, indicating that Cdc55 is an in vitro substrate of Cdk1–Clb2.
Cdc55 phosphorylation regulates nucleolar release of Cdc14
The results reported before indicate that Cdk1–Clb2 participates in Cdc55 phosphorylation during anaphase. Within Cdc55 we identified one residue, Thr174, with a full consensus for Cdk1 (S/TxP-x-K/R), and a second residue, Ser301, containing the Cdk1 minimal consensus site (S/TxP). We prepared strains containing these residues mutated to alanine (non-phosphorylable mutants) and to aspartic/glutamic (phosphorylated mimetic mutants) in a cdc55Δ background. The growth and the mitotic kinetics of the control HA–Cdc55 cdc55Δ were equivalent to the control strain with endogenous Cdc55 epitope tagging (data not shown). The control HA–Cdc55 and the mutant alleles cdc55-T174A-S301A (hereafter, cdc55-2A), cdc55-T174E-S301D (hereafter, cdc55-ED) showed equivalent growth rates, rescued the cold-sensitivity of the cdc55Δ mutant (Fig. S3a), suppressed the elongated bud phenotypes of the cdc55Δ mutant (Fig. S3b), physically interact with the PP2A catalytic subunit Pph21 (Fig. S3c) and presented similar amounts of inhibitory Cdk1 phosphorylation at Y19 (Fig. S3d). All these results indicate that the mutant Cdc55 alleles behave similar to the control Cdc55 outside mitosis.
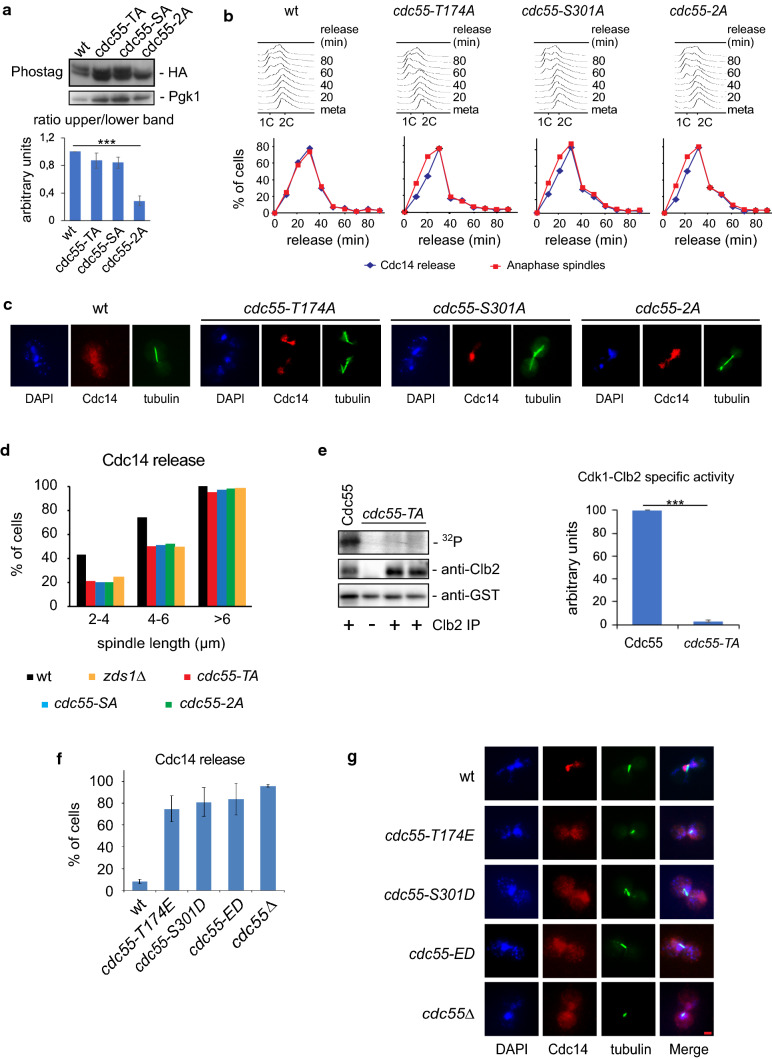
To determine whether either of these residues is required for Cdc55 phosphorylation in vivo, first, we analyzed Cdc55 phosphorylation in the non-phosphorylable mutants (Fig. 3a) in metaphase cells. Reduced Cdc55 phosphorylation was observed in the cdc55-2A mutant allele compared with levels in control Cdc55 and in the single mutants cdc55-T174A and cdc55-S301A. These data indicate that the slower migrating form of Cdc55 corresponds to the phosphorylation in both residues (T174 and S301) and suggests that the two residues are phosphorylated in vivo. Second, we investigated the mitotic progression and Cdc14 release from the nucleolus in anaphase-synchronized cells by Cdc20 depletion in the non-phosphorylable mutants (Fig. 3b, c). Control cells released Cdc14 from the nucleolus at anaphase onset, as expected. In contrast, in cdc55-T174A, cdc55-S301A and cdc55-2A, Cdc14 release from the nucleolus was retarded by 10 min relative to the onset of spindle elongation, a delay similar to that observed in FEAR mutants [21]. These results suggest that Cdc55 phosphorylation is required for proper Cdc14 release from the nucleolus in anaphase.
Fig. 3.
Cdc55 phosphorylation alters Cdc14 release from the nucleolus. a Cdc55 phosphorylation is impaired in the non-phosphorylable cdc55-2A mutant. Protein extracts from strains Y1211, Y1215, Y1216 and Y1217 arrested in metaphase by Cdc20 depletion were prepared. Cdc55 phosphorylation was analyzed by western blot. Pgk1 protein levels are shown as loading control. A representative experiment of one of the assays is presented (top). Quantifications of the slower and faster Cdc55 migration isoforms are depicted (bottom). Mean values and SEM are shown. b Cdc14 release is impaired during early anaphase in non-phosphorylable Cdc55 mutants. Strains Y1211, Y1215, Y1216, and Y1217 were arrested in metaphase by Cdc20 depletion and released into synchronous anaphase by Cdc20 re-introduction. Mitosis progression was monitored by FACS analysis of DNA content (up). Cdc14 release from the nucleolus and anaphase spindles were monitored by immunofluorescence (middle). Representative images of cells in early anaphase with a spindle length of 3–5 μm are depicted in c. Scale bar, 2 μm. d Cdc14 nucleolar release from b was analyzed as a function of spindle length. The strain Y3034 was used as a FEAR mutant control. At least 50 cells were scored for each spindle length. e Mutation of the T174A within Cdc55 impaired Cdk1–Clb2 phosphorylation in vitro. Strain Y564 was arrested in metaphase by Cdc20 depletion, Clb2 was purified by immunoprecipitation, and the Cdc28–Clb2 kinase assay was performed using bacteria-purified GST-Cdc55(1–193) and GST-Cdc55(1–193)-T174A as substrates. A representative experiment is depicted (left). Mean values and SEM of four independent immunoprecipitations are shown (right). f Cdc14 is prematurely released during metaphase in Cdc55 phospho-mimetic mutants. Strains Y879, Y1211, Y1212, Y1213, and Y1214 expressing Cdc55 phospho-mimetic mutants of the Cdk1 consensus sites within Cdc55 were arrested in metaphase by Cdc20 depletion. Cdc14 release was checked by immunofluorescence. At least 100 cells were scored for each strain. Mean values and SD of three independent experiments are shown. Photographs are of cells in early anaphase with a spindle length of 3–5 μm (g). Scale bar, 2 μm
To study the delay in Cdc14 activation relative to an internal marker for mitotic progression further, we measured Cdc14 release from the nucleolus with respect to the spindle length (Fig. 3d). Control cells started to release Cdc14 at spindle lengths of > 2 μm. In contrast, the non-phosphorylable mutants released Cdc14 when the spindle length was 4–6 μm, similar to what occurs in zds1Δ cells [21]. Similar results were obtained synchronizing cells from G1 (Fig. S3). Therefore, Cdc55 phosphorylation is required for the proper release of Cdc14 from the nucleolus in early anaphase. Thus, phosphorylation of both residues, T174A and S301A, contributes similarly to timely Cdc14 activation in early anaphase.
Next, we examined whether these Cdc55 phosphorylation sites are responsible for the Cdk1–Clb2 phosphorylation of Cdc55 observed before in vitro. We were not able to purify the Cdc55 full-length recombinant protein, so we could only test the T174 residue by changing the residue to alanine, GST-Cdc55-T174A. Cdk1–Clb2 activity against control GST-Cdc55 was again detected as before (Figs. 2c, 3e); in contrast, GST-Cdc55-T174A phosphorylation was greatly reduced, indicating that T174 is responsible for the Cdk1–Clb2-dependent phosphorylation of Cdc55 in vitro.
To determine whether Cdc55 phosphorylation is sufficient to bring about PP2ACdc55 downregulation we prepared phospho-mimetic mutants by changing the threonine into a glutamic residue and the serine to an aspartic residue. If Cdc55 phosphorylation induces PP2ACdc55 inactivation, the phospho-mimetic mutants should lead to a premature Cdc14 release from the nucleolus, reminiscent of the cdc55Δ deletion mutant [17]. We constructed the single and double phospho-mimetic mutants, cdc55-T174E, cdc55-S301D and cdc55-ED and measured the Cdc14 release in metaphase-arrested cells by Cdc20 depletion (Fig. 3f, g). In controls cells, Cdc14 was sequestered in the nucleoli of metaphase cells, as expected. Conversely, Cdc14 was prematurely released from the nucleolus in all the phospho-mimetic mutants, similar to the phenotype of the cdc55Δ mutant. All these results suggest that PP2ACdc55 inactivation by Cdc55 phosphorylation regulates timely Cdc14 activation.
Cdc55 phosphorylation modulates PP2ACdc55 phosphatase activity
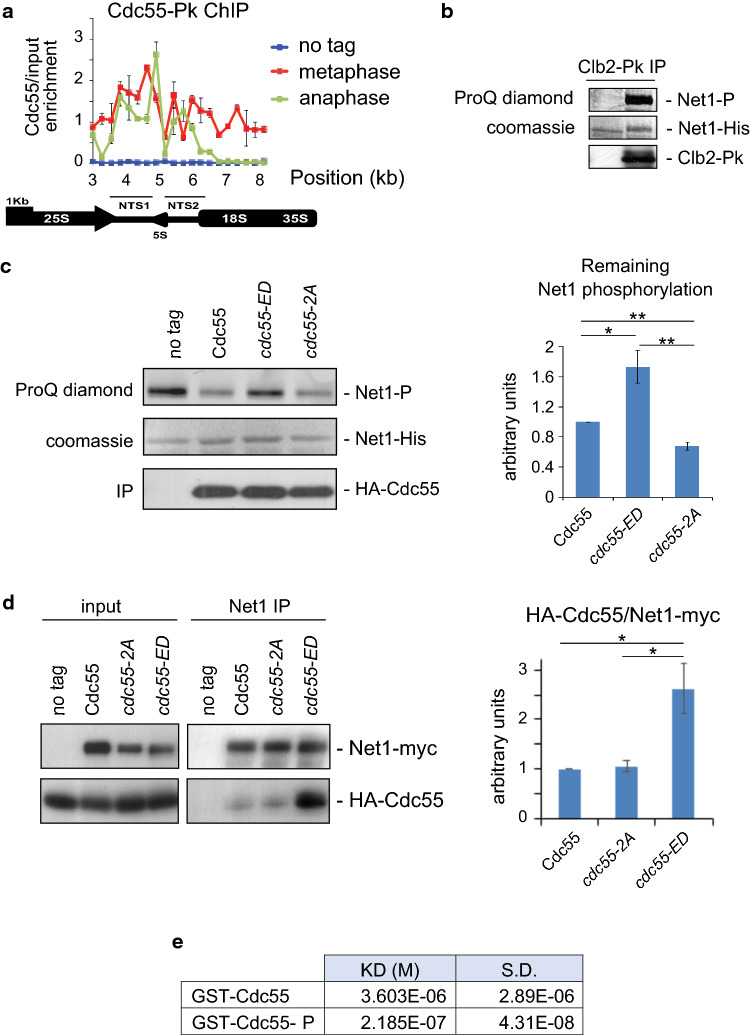
It has been previously reported that Cdc55 and its substrate, Net1, are located at the nucleolus and that this location does not change throughout mitosis [17, 22]. By contrast, a reduction of Cdc55 signal during mitosis was also proposed [35]. To find out whether nucleolar Cdc55 localization is regulated during anaphase, we used a non-subjective and quantitative technique: ChIP-qPCR. This assay allows nucleolar localization to be evaluated by quantifying the enrichment of a given protein at the ribosomal DNA (rDNA) region [32], and it has often been used to detect Net1 and Cdc14 nucleolar localization. To test whether we can detect Cdc14 redistribution throughout the entire cell during anaphase using this assay, we performed Cdc14-Pk ChIP-qPCR in metaphase- and anaphase-synchronized cells by Cdc20 depletion and reintroduction (Fig. S5). Cdc14 was enriched at the NTS1 and NTS2 region at metaphase, and this enrichment was greatly reduced during anaphase, concomitantly with the Cdc14 release. Therefore, we can use this technique to detect nucleolar localization of Cdc55.
Next, we repeated the ChIP-qPCR assays for Pk-Cdc55 at metaphase and anaphase cells (Fig. 4a). Cdc55 was enriched at the NTS1 and NTS2 region in metaphase cells, indicating that Cdc55 is bound to the nucleolus (rDNA). In anaphase cells, Cdc55 enrichment at the NTS1 region and most of the NTS2 was similar to that in metaphase cells; nevertheless, enrichment at the end of the NTS2 region and downstream towards the 18S region was reduced in anaphase cells. This differential Cdc55 enrichment along the rDNA during anaphase might explain the controversial results previously reported using other localization approaches. Therefore, nucleolar Cdc55 localization might be mildly altered during anaphase and could contribute to the regulation of PP2ACdc55 activity. We do not fully understand what the reduction at the NTS2-18S region means, but overall, our experiments suggest that Cdc55 is still bound to the rDNA in anaphase cells (at least at the NTS1 region and partially at the NTS2 region) and changes in Cdc55 localization alone cannot explain the PP2ACdc55 inactivation during anaphase.
Fig. 4.
Cdc55 phosphorylation regulates PP2ACdc55 phosphatase activity. a Cdc55 nucleoli localization downstream from the NTS2 is reduced in anaphase. Strain Y500 was synchronized at the metaphase-to-anaphase transition. Binding to the NTS1 and NTS2 regions of the rDNA was determined by Pk-Cdc55 chromatin immunoprecipitation experiments. Mean values and SEM of three independent experiments are shown. b Net1 phosphorylation by Cdk1–Clb2. Strain Y2312 was arrested in metaphase by Cdc20 depletion, Clb2-Pk was purified by immunoprecipitation and Cdc28–Clb2 kinase assay was performed using bacteria-purified Net1(1–600)-his as substrate. Strain Y2541 without the Clb2-Pk epitope was used as negative control for the kinase assay. c PP2ACdc55 phosphatase activity in the Cdc55 phosphorylation mutants. Strains Y1049, Y1077 and Y1080 were arrested in metaphase by Cdc20 depletion. Phosphatase activity of Cdc55-immunopurified complexes was measured as described in the “Methods”. Strain Y824 without HA–Cdc55 epitope was used as a negative control for the phosphatase assay. A representative image of one of the experiments is depicted (left). Mean values and SEM of remaining Net1 phosphorylation in three independent experiments are presented (right). d The Cdc55 and Net1 interaction is less dynamic in the Cdc55 phospho-mimetic mutants. Cell extracts were prepared from strains Y1281, Y1284 and Y1294 arrested in metaphase and co-immunoprecipitation of Cdc55 with Net1 was determined. Strain Y1285 without Net1-myc epitope was used as a negative control. Immunoblots of one representative experiment are illustrated (left). Mean values and SEM of three independent experiments are shown (right). e Cdc55 phosphorylation increases Net1–Cdc55 interaction. Bacteria-purified Net1(1–600)-his was immobilized in CM5 chips and GST-Cdc55(1–193) and GST-Cdc55(1–193)-P (phosphorylated by Cdk1–Clb2) were used as analytes in surface plasmon resonance experiments. The mean KD and the SD are depicted
Downregulation of PP2ACdc55 phosphatase-specific activity occurs at anaphase onset [17] and upon Zds1 ectopic expression [21]. To establish whether Cdc55 phosphorylation modulates PP2ACdc55 phosphatase activity, we measured the in vitro phosphatase activity of the non-phosphorylable and the phospho-mimetic mutants of Cdc55. As a substrate we prepared in vitro-phosphorylated Net1(1–600)-his by Cdk1–Clb2 (Fig. 4b). Cdc55 immunoprecipitates from metaphase-arrested cells which were incubated with this substrate and the remaining Net1 phosphorylation was quantified using Pro-Q Diamond-stained gels (Fig. 4c). Cells containing wild-type Cdc55 dephosphorylated Net1, as expected. The non-phosphorylable mutant cdc55-2A presented a mild increase in phosphatase activity compared to control Cdc55 cells. The quantification of the Net1 remaining phosphorylation (Fig. 4c right panel) showed a 30% reduction in the Net1 phosphorylation, suggesting that the cdc55-2A allele is hyperactive. In the phospho-mimetic mutant cdc55-ED the Net1 phosphorylation detected was similar to the levels in negative control cells in which Cdc55 was not tagged. Quantification of the phosphatase-specific activity was consistent with PP2ACdc55 activity being inactivated in the cdc55-ED mutant (Fig. 4c right panel). We conclude that the phosphorylation of Cdc55 regulates PP2ACdc55 phosphatase activity.
PP2A regulatory subunits provide substrate specificity. Co-immunoprecipitation experiments showed that Cdc55 interacts with Net1 in metaphase-arrested cells (Fig. 4d). To establish whether Cdc55 phosphorylation affects substrate recognition we examined whether Cdc55 phospho-isoforms physically interact with Net1 substrate.
An interaction was also detected between cdc55-2A and Net1, similar to what is observed in control cells. Strikingly, the cdc55-ED and Net1 interaction was stronger (Fig. 4d right panel). To further confirm the Net1–Cdc55-binding dynamics we studied the molecular interactions using the surface plasmon resonance Biacore system (Fig. 4e). Net1(1–600)-his was purified from E. coli and immobilized in CM5 chips. Bacteria-purified GST-Cdc55 was phosphorylated in vitro by Cdk1–Clb2 and phosphorylated and non-phosphorylated GST-Cdc55 were used as analytes. The phosphorylated GST-Cdc55-P presented a lower KD 2.18E−07 compared to the non-phosphorylated GST-Cdc55 (KD 3.6E−06) one, indicating a higher affinity towards Net1. These results indicate that phosphorylation of Cdc55 modulates PP2ACdc55 activity by altering enzyme–substrate-binding dynamics. In this case, the cdc55-ED mutant acts as a “substrate trapping” mutant [36] that retains a high binding affinity for substrates but blocks catalysis, making the interaction between the enzyme and its substrate less dynamic.
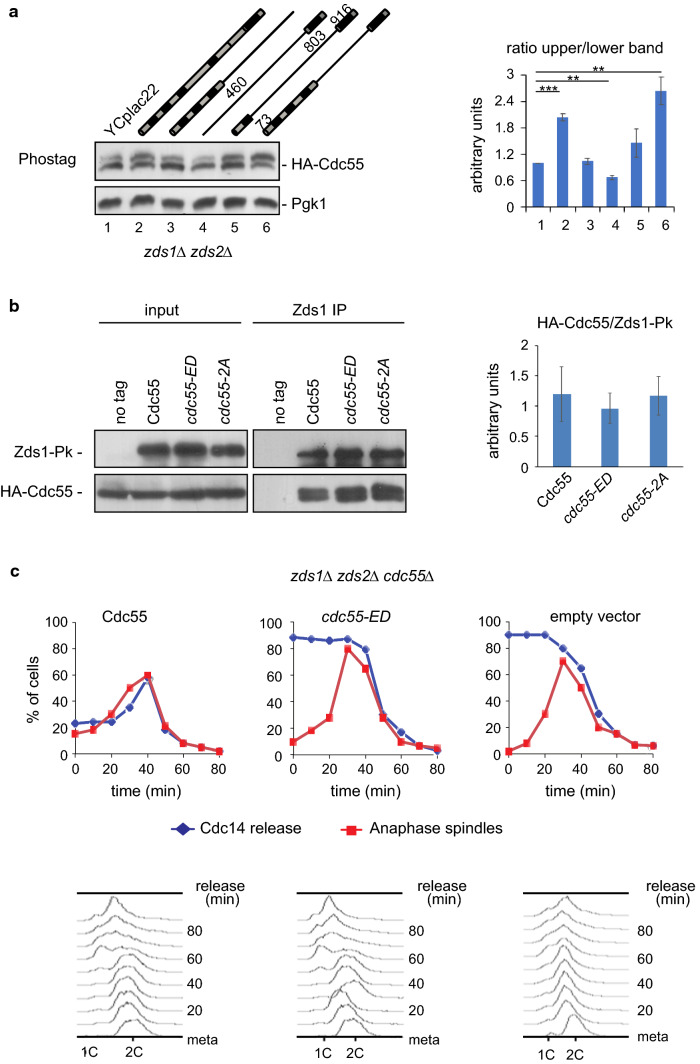
Functional Zds1 is required for proper Cdc55 phosphorylation
As reported above, Zds1 ectopic expression promotes the accumulation of the Cdc55 phosphorylation. Conversely, Cdc55 phosphorylation should be altered in absence of Zds1. Cdc55 phosphorylation was checked in the double mutant zds1Δzds2Δ in cycling cells (Fig. 5a, lane 1). Phosphorylation levels of Cdc55 were lower in the absence of Zds1/2 (compare lane 1 and 2), indicating that the presence of Zds1/2 is required to properly maintain Cdc55 phosphorylation. To test which domain of the Zds1 protein is required for the Cdc55 phosphorylation, we introduced truncated versions of Zds1 in the zds1Δ zds2Δ double mutant (lanes 3-6). We previously described that the truncated zds1Δ461-802 protein (lane 6) is the only functional to release Cdc14 from the nucleolus [22]. Only the truncated version containing the functional protein zds1Δ461-802 (lane 6) was able to restore the Cdc55 phosphorylation to comparable levels of the full length Zds1 (lane 2). Thus, not only Zds1’s presence, but its functional form is also required to promote Cdc14 activation for proper Cdc55 phosphorylation.
Fig. 5.
Zds1 is required for proper Cdc55 phosphorylation. a Cdc55 phosphorylation is impaired in the absence of Zds proteins. Protein cell extracts were prepared from strains Y1118, Y1192, Y1119, Y1122, Y1117 and Y1120 in cycling cells. Cdc55 phosphorylation was analyzed by western blot. Pgk1 protein levels are shown as loading control. A representative image of one of the experiments is depicted (left). Mean values and SEM of three independent experiments are presented (right). b Cdc55 and Zds1’s physical interaction is not affected by Cdc55 phosphorylation. Protein cell extracts of strains Y1285, Y1288 and Y1291 arrested in metaphase were prepared, and co-immunoprecipitation of Cdc55 with Zds1 was determined (left). The strain Y1281 without the Zds1-Pk epitope was used as a negative control. Mean values and SEM of two independent experiments are shown (right). c The inactive cdc55-ED phospho-mimetic mutant promotes premature Cdc14 release from the nucleolus in the zds1Δ zds2Δ mutant. Strains Y1597, Y1598 and Y1600 were arrested in metaphase by Cdc20 depletion and release into anaphase by Cdc20 re-introduction. Cdc14 release from the nucleolus and anaphase spindles was determined by immunofluorescence. Mitosis progression was monitored by FACS analysis of DNA content
Zds1 physically interacts with PP2ACdc55 via its regulatory subunit Cdc55 [22, 30]. Therefore, we examined whether the Zds1 and Cdc55 physical interaction is affected by Cdc55 phosphorylation. To test this possibility, co-immunoprecipitation experiments were performed using the Cdc55 phospho-mutants. In Zds1 immunoprecipitates from metaphase-arrested cells, we detected similar levels of co-purified Cdc55, cdc55-ED and cdc55-2A by western blot (Fig. 5b). These results indicate that Zds1 and Cdc55 interact independent of Cdc55 phosphorylation status.
If the defects in the Cdc14 release from the nucleolus observed in zds mutants are due to the impaired Cdc55 phosphorylation, the introduction of the cdc55-ED phospho-mimetic allele should rescue the Cdc14 release. To study in greater depth whether Zds1 promotes Cdc14 activation by regulating Cdc55 phosphorylation, we introduced the cdc55-ED phospho-mimetic mutant into a zds1Δzds2Δ cdc55Δ CDC28Y19F background. The CDC28Y19F is refractory to inhibition by phosphorylation at Cdc28-Y19 and rescues the slow-growing phenotype and the elongated morphology of the zds1Δzds2Δ double mutant, allowing us to synchronize cells. We previously showed that the defective-Cdc14 release was equivalent in zds1Δzds2Δ and zds1Δzds2Δ CDC28Y19F mutants [22]. We arrested cells at the metaphase-to-anaphase transition by Cdc20 depletion and analyzed the Cdc14 localization. When we introduced the wild-type Cdc55, the Cdc14 release was delayed as expected for zds mutants (Fig. 5c). On the other hand, in the absence of Cdc55 (empty vector), Cdc14 was released from the nucleolus prematurely in most of the metaphase-arrested cells (Fig. 5c, and [21]). The inactive cdc55-ED allele also promoted the premature release of Cdc14 from the nucleolus in the zds1Δzds2Δ background. Therefore, inactivation of Cdc55 by phosphorylation also promotes Cdc14 activation in the absence of Zds1. This is consistent with Zds1 being involved in the regulation of Cdc55 phosphorylation.
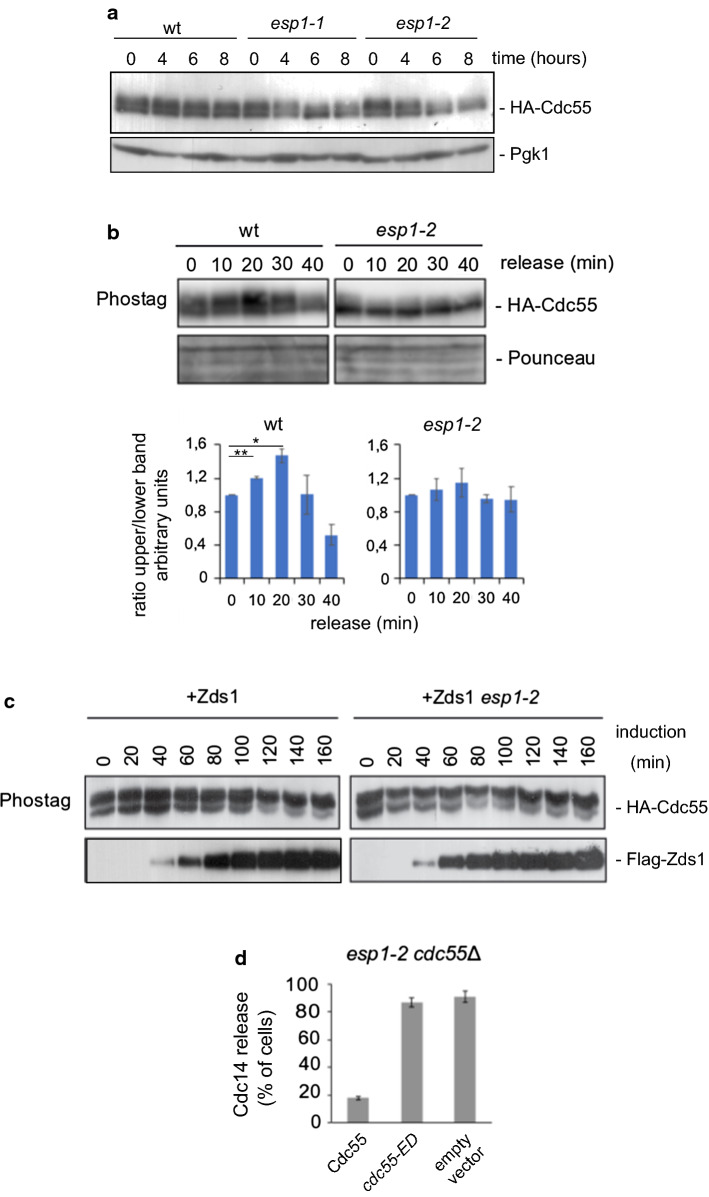
Separase is needed to maintain Cdc55 phosphorylation
Separase activation is essential for Cdc14 nucleolar release, and downregulation of PP2ACdc55 at anaphase onset depends on separase [17]. To discover how separase regulates PP2ACdc55 we studied Cdc55 phosphorylation in separase thermo-sensitive mutants. In cycling cells, after 6–8 h at the restrictive temperature when the separase alleles esp1-1 and esp1-2 are inactivated, the slower migrating form of Cdc55 was reduced (Fig. 6a), suggesting that separase inactivation impaired Cdc55 phosphorylation. Later on, we synchronized cells at the metaphase-to-anaphase transition by Cdc20 depletion in wild-type and esp1-2 mutant cells (Fig. 6b). Upon inactivation of the separase allele, esp1-2, the slower migrating form of Cdc55 was reduced at the restrictive temperature already at metaphase cells (time 0). In addition, the accumulation of the slower migration form of Cdc55 was not detected during anaphase in esp1-2 cells, contrary to control cells. This indicates that separase activation is required to maintain Cdc55 phosphorylation.
Fig. 6.
Separase is necessary for proper Cdc55 phosphorylation. a Cdc55 phosphorylation is defective in separase mutants. Protein cell extracts were prepared in cycling cells from strains Y536, Y568 and Y567 incubated at 32 °C for the indicated times. Cdc55 phosphorylation was analyzed by western blot. Pgk1 is shown as loading control. b Impaired Cdc55 phosphorylation in anaphase in separase mutants. Strains Y564 and Y565 were arrested in metaphase, incubated at 32 °C for 2 h to inactivate the esp1-2 allele and release into anaphase. Cdc55 phosphorylation was analyzed by western blot. Ponceau staining is shown as loading control. A representative image of one of the experiments is depicted (top). Mean values and SEM of three independent experiments are presented (bottom). c Separase is not required for Zds1-induced Cdc55 phosphorylation. Strains Y888 and Y524 were arrested in metaphase by Cdc20 depletion, incubated at 32 °C for 2 h, and Zds1 ectopic expression was induced. d Premature Cdc14 release in separase mutants upon expression of the Cdc55 phospho-mimetic mutant, cdc55-ED. Strains Y2782, Y1561 and Y1481 were arrested in metaphase. Cdc14 release was analyzed by immunofluorescence. Mean values and SEM of three independent experiments are shown
Zds1 is required for separase-induced Cdc14 activation while Zds1 can release Cdc14 in absence of separase activity, suggesting that Zds1 acts in concert with separase, or downstream of it [21]. To analyze the relationship of these proteins, we examined Cdc55 phosphorylation upon Zds1 ectopic expression in the esp1-2 cells in metaphase-arrested cells by Cdc20 depletion at the restrictive temperature (Fig. 6c). Zds1 expression in metaphase-arrested wild-type or separase mutant esp1-2 cells led to Cdc55 phosphorylation with similar kinetics. These results are consistent with Zds1 acting in parallel or downstream of separase regulating PP2ACdc55 activity, as previously described.
To confirm the influence of separase on Cdc55 phosphorylation, we introduced the cdc55-ED phospho-mimetic mutant into the esp1-2 mutant allele. We arrested cells in metaphase by Cdc20 depletion and incubated the cells at the restrictive temperature to inactivate the separase. Upon introduction of the control Cdc55 into the esp1-2 cdc55Δ mutant (Fig. 6d), Cdc14 was sequestered in the nucleolus, as previously described [17]. By contrast, the inactive cdc55-ED allele promoted the release of Cdc14 from the nucleolus, similar to what occurs in the absence of Cdc55. This is consistent with separase-facilitating Cdk-dependent Net1 phosphorylation and Cdc14 release from the nucleolus by modulating PP2ACdc55 activity via Cdc55 phosphorylation.
Cdk1–CyclinB1 phosphorylates PP2A-B55 in higher eukaryotes
PP2A phosphatases regulate almost every stage of the cell cycle by forming diverse heterotrimeric holoenzymes (reviewed in Refs. [37, 38]). Many aspects and components of the mitotic exit are conserved in higher eukaryotes. Human PP2A-B55 has several roles in mitosis. Before mitosis initiation, PP2A-B55 dephosphorylates Wee1 and Greatwall kinase, thereby inactivating them [39–41]. CDC25 dephosphorylation by PP2A-B55 subsequently promotes mitotic exit [42, 43]. On the other hand, Cdk1 phosphorylates Greatwall and in turn Greatwall-ENSA inhibits PP2A-B55 [41, 44–47]. Therefore, we considered it worthwhile checking whether PP2ACdc55 regulation by Cdk1 phosphorylation is conserved. For this purpose, we investigated whether the human PP2A-B55 was phosphorylated by Cdk1–CyclinB1 in vitro. Recombinant human PP2A-B55 was incubated with a greater amount of human recombinant Cdk1–CyclinB1 at 30 °C for 30 min. PP2A-B55 phosphorylation levels were visualized by Pro-Q Diamond staining (Fig. 7a). PP2A-B55 was phosphorylated in vitro in the presence of Cdk1–CyclinB1. This result suggests that Cdk1-dependent phosphorylation of the B regulatory subunit is conserved in human cells. Strikingly, the yeast Thr174 is a conserved residue corresponding to human Ser167, which is a minimal Cdk1 consensus site SP (Fig. 7b). The yeast Ser301 is also conserved in human B55, which corresponds to Ser294, although in this case it does not match with any Cdk1 consensus site. Therefore, we propose that phosphorylation at T174 (or S167 in human) might be involved in the conserved mechanism of Cdk1-dependent phosphorylation of Cdc55 (human B55). Although some of the effectors may differ in mammalian cells, studying the molecular mechanism governing yeast mitotic exit may yield insights into mitotic progression in other contexts and organisms.
Discussion
Cdk1-dependent Cdc55 phosphorylation regulates timely Cdc14 activation
In S. cerevisiae Cdc14 phosphatase is a key mitotic component whose essential function is to counteract Cdk1–Clb2 activity during anaphase, allowing cells to exit from mitosis [4]. Initial FEAR-Cdc14 activation at anaphase onset depends on downregulation of PP2ACdc55, promoting Net1 phosphorylation and Cdc14 release from the nucleolus [17]. Anaphase-specific inactivation of PP2ACdc55 phosphatase activity arises from the phosphorylation of its regulatory B subunit, Cdc55, by the Cdk1–Clb2 kinase. Cdk1-dependent Cdc55 phosphorylation does not provoke a change in substrate recognition; instead, Cdc55 phosphorylation affects the dynamic interaction between Net1 and Cdc55. Notably, active separase and functional Zds1 are required to sustain Cdc55 phosphorylation and a constitutive inactive version of Cdc55, cdc55-ED, can compensate for the defects of Cdc14 release from the nucleolus in the absence of Zds1 and separase activities. Therefore, separase and Zds1 control Cdc14 activation by modulating Cdc55 phosphorylation or PP2ACdc55 phosphatase activity.
Phosphorylation of Cdc55 at the Cdk1 consensus sites within T174 and Ser301 is equally required for Cdc14 activation since no additive effect was observed in the double mutants cdc55-2A and cdc55-ED compared to the single phospho-mutants. It has recently been reported that PP2ACdc55 imposes ordered cell-cycle phosphorylation by opposing threonine phosphorylation [48] and PP2A-B55 preference for threonine residues is conserved in mammals [49]. Therefore, Cdk1–Clb2 can phosphorylate both residues during mitosis, but the fine-tuning of Cdc55 phosphorylation might be regulated by preferential (auto)-dephosphorylation of T174 by PP2ACdc55.
The different subunits of PP2ACdc55 are known to suffer post-translational modifications, such as phosphorylation and carboxy-methylations [50–55]. The Leu309 carboxy-methylation of Pph21 is involved in the assembly of the holoenzymes [55] and the Pph21/22 phosphorylation at Tyr307 destabilized the heterotrimeric PP2ACdc55 complex. Although we cannot rule out the possibility that some other post-translational modification might also affect PP2ACdc55 phosphatase activity, we did not detect any anaphase-specific modification in Pph21 and Tpd3 by western blot or mass-spectrometry. In addition, in global yeast phosphoproteome studies, several phosphorylations for Cdc55 were described in the S124, S155 and S440 residues in asynchronous cells [56–59]. In our mass-spectrometry analysis of Cdc55 phosphorylation we identified the T174 phosphorylation. We were not able to detect any peptide (modified or unmodified) containing the S301. Although, we detected phosphorylation at (non-Cdk1) residues T133 and S403 during mitosis. Therefore, we cannot discount the possibility that Cdc55 undergoes other non-Cdk1-dependent phosphorylation or even other post-translational modifications, since in our experiments in the absence of Cdk1 and with the alkaline phosphatase treatment, a residual slower migrating form of Cdc55 is still detectable.
Zds1 and separase are involved in maintaining proper Cdc55 phosphorylation
Our results demonstrate that functional Zds1 is required for proper Cdc55 phosphorylation, and that the same functional domains of Zds1 regulating Cdc14 activation [22] are also involved in maintaining proper Cdc55 phosphorylation (Fig. 5a). In addition, introduction of the inactive cdc55-ED allele promotes the premature Cdc14 release from the nucleolus in the absence of Zds1 (Fig. 5c). In the same way, the need for active separase to maintain proper Cdc55 phosphorylation and Cdc14 activation (Fig. 6a–c) is compensated by a constitutive Cdc55 phosphorylation (cdc55-ED, Fig. 6d). All these findings prompted us to propose a model in which separase and Zds1 control the activity of the proteins regulating Cdc55 phosphorylation (Fig. 7c).
Proposed model of the regulation of PP2ACdc55 and Cdc14 during mitosis
Protein phosphorylation and dephosphorylation are regulated by the interplay of protein kinases and phosphatases. Reversible phosphorylation of protein substrates commonly involved futile cycles correlated with an exponential signal-response curve [17, 60]. Nevertheless, coherent feed-forward loops reduce futile cycling, ensuring that the kinase and phosphatase are not fully active at the same time and generate a sigmoidal signal–response curve helping to make more definitive decisions. In this sense, our proposed model (Fig. 7c) fits perfectly with a coherent feed-forward loop in which the kinase, Cdk1–Clb2, phosphorylates the Net1 substrate and helps inactivate the phosphatase, PP2ACdc55, by phosphorylating it (the new mechanism described in this work, indicated as black dashed lines). In this model, the active form of the phosphatase PP2ACdc55 can promote its own activation by auto-dephosphorylation or a second phosphatase (marked as PPase) might contribute to PP2ACdc55 dephosphorylation (marked with a question mark since this step is not proven).
In the proposed model, some protein effectors of the system might regulate the kinase, Cdk1–Clb2 or the phosphatase, PP2ACdc55. Zds1 and separase could therefore be involved in regulating Cdk1–Clb2 or PP2ACdc55. Our results favor a model in which separase and Zds1 act to regulate PP2ACdc55 and/or the second phosphatase (PPase). Further work is required to elucidate the mechanism by which Zds1 and separase control the activity of the phosphatases. PP1 (Glc7 in budding yeast) is known to have different roles in yeast during mitosis [61–66], Xenopus [67–69] and human cells [70–72]. Therefore, PP1 is a candidate phosphatase to regulate PP2ACdc55. In addition, PP1 and PP2A physically interact and are mutually regulated [66], suggesting that both phosphatases may be closely regulated in different organisms.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Joanjo Bech-Serra and Carolina De LaTorre for helpful discussion and advice about the mass-spectrometry assays; and Silvia Barceló-Batllori for advice and discussion on the surface plasmon resonance (Biacore) experiments. We also thank all the members of our laboratory for discussing the work and for their critical reading of the manuscript. We thank CERCA Program/Generalitat de Catalunya for institutional support. Our laboratory is funded by the Spanish Ministry of Economy, Industry and Competitiveness (MINECO), which is part of the State Agency, through the projects BFU2013-43132-P and BFU2016-77975-R, (co-funded by the European Regional Development Fund, ERDF, a way to build Europe). The proteomics analyses were performed in the IDIBELL Clinical Proteomics Unit which is part of Proteored, PRB3 and is supported by Grant PT17/0019, of the PE I+D+i 2013-2016, funded by ISCIII and ERDF.
Author contributions
SJ, IC, YMR, PG and EQ performed the experiments. SJ, IC and EQ designed the experiments and interpreted the data. EQ wrote the manuscript. All authors read and discussed the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Morgan DO. The cell cycle, principles of control. London: New Science Press; 2007. [Google Scholar]
- 2.Meadows JC, Millar JBA. Sharpening the anaphase switch. Biochem Soc Trans. 2015;43:19–22. doi: 10.1042/BST20140250. [DOI] [PubMed] [Google Scholar]
- 3.Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/S0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 4.Visintin R, Craig K, Hwang ES, et al. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/S1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 5.Shou W, Seol JH, Shevchenko A, et al. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/S0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 6.Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 7.Queralt E, Uhlmann F. Cdk-counteracting phosphatases unlock mitotic exit. Curr Opin Cell Biol. 2008;20:661–668. doi: 10.1016/j.ceb.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rock JM, Amon A. The FEAR network. Curr Biol. 2009;19:R1063–R1068. doi: 10.1016/j.cub.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mocciaro A, Schiebel E. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J Cell Sci. 2010;123:2867–2876. doi: 10.1242/jcs.074815. [DOI] [PubMed] [Google Scholar]
- 10.Baro B, Queralt E, Monje-Casas F. Regulation of mitotic exit in Saccharomyces cerevisiae. Methods Mol Biol. 2017;1505:3–17. doi: 10.1007/978-1-4939-6502-1_1. [DOI] [PubMed] [Google Scholar]
- 11.Hotz M, Barral Y. The mitotic exit network: new turns on old pathways. Trends Cell Biol. 2014;24:145–152. doi: 10.1016/j.tcb.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Pereira G, Schiebel E. The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr Opin Cell Biol. 2001;13:762–769. doi: 10.1016/S0955-0674(00)00281-7. [DOI] [PubMed] [Google Scholar]
- 13.Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/S0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 14.Pereira G, Hofken T, Grindlay J, et al. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. doi: 10.1016/S1097-2765(05)00017-1. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Luo G, Bahk YY, Song K. Cdc5-dependent asymmetric localization of bfa1 fine-tunes timely mitotic exit. PLoS Genet. 2012;8:e1002450. doi: 10.1371/journal.pgen.1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monje-Casas F, Amon A. Cell polarity determinants establish asymmetry in MEN signaling. Dev Cell. 2009;16:132–145. doi: 10.1016/j.devcel.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Queralt E, Lehane C, Novak B, Uhlmann F. Downregulation of PP2ACdc55 phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 18.Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/S0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan M, Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat Cell Biol. 2003;5:249–254. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azzam R, Chen SL, Shou W, et al. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science (80-) 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- 21.Queralt E, Uhlmann F. Separase cooperates with Zds1 and Zds2 to activate Cdc14 phosphatase in early anaphase. J Cell Biol. 2008;182:873–883. doi: 10.1083/jcb.200801054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calabria I, Baro B, Rodriguez-Rodriguez J-AA, et al. Zds1 regulates PP2A(Cdc55) activity and Cdc14 activation during mitotic exit through its Zds_C motif. J Cell Sci. 2012;125:2875–2884. doi: 10.1242/jcs.097865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shou W, Azzam R, Chen SL, et al. Cdc5 influences phosphorylation of Net1 and disassembly of the RENT complex. BMC Mol Biol. 2002;3:3. doi: 10.1186/1471-2199-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Rodriguez J-AJ-A, Moyano Y, Játiva S, Queralt E. Mitotic exit function of polo-like kinase Cdc5 is dependent on sequential activation by Cdk1. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.04.079. [DOI] [PubMed] [Google Scholar]
- 25.Menssen R, Neutzner A, Seufert W. Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr Biol. 2001;11:345–350. doi: 10.1016/S0960-9822(01)00095-1. [DOI] [PubMed] [Google Scholar]
- 26.Jaspersen SL, Morgan DO. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr Biol. 2000;10:615–618. doi: 10.1016/S0960-9822(00)00491-7. [DOI] [PubMed] [Google Scholar]
- 27.Pereira G, Manson C, Grindlay J, Schiebel E. Regulation of the Bfa1p–Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J Cell Biol. 2002;157:367–379. doi: 10.1083/jcb.200112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rock JM, Lim D, Stach L, et al. Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes. Science. 2013;340:871–875. doi: 10.1126/science.1235822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baro B, Rodriguez-Rodriguez J-A, Calabria I, et al. Dual regulation of the mitotic exit network (MEN) by PP2A–Cdc55 phosphatase. PLoS Genet. 2013 doi: 10.1371/journal.pgen.1003966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasutis K, Vignali M, Ryder M, et al. Zds2p regulates Swe1p-dependent polarized cell growth in Saccharomyces cerevisiae via a novel Cdc55p interaction domain. Mol Biol Cell. 2010;21:4373–4386. doi: 10.1091/mbc.e10-04-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monje-Casas F, Queralt E. The mitotic exit network. New York: Humana Press; 2017. [Google Scholar]
- 32.Huang J, Iglesias N, Moazed D. Evaluation of the nucleolar localization of the RENT complex to ribosomal DNA by chromatin immunoprecipitation assays. In: Monje-Casas F, Queralt E, editors. The mitotic exit network: methods and protocols. New York: Springer; 2017. pp. 195–213. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bishop AC, Ubersax JA, Petsch DT, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 35.Rossio V, Yoshida S. Spatial regulation of Cdc55–PP2A by Zds1/Zds2 controls mitotic entry and mitotic exit in budding yeast. J Cell Biol. 2011;193:445–454. doi: 10.1083/jcb.201101134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powers BL, Hall H, Charbonneau H, Hall MC. A substrate trapping method for identification of direct Cdc14 phosphatase targets. Methods Mol Biol. 2017;1505:119–132. doi: 10.1007/978-1-4939-6502-1_10. [DOI] [PubMed] [Google Scholar]
- 37.Hunt T. On the regulation of protein phosphatase 2A and its role in controlling entry into and exit from mitosis. Adv Biol Regul. 2013;53:173–178. doi: 10.1016/j.jbior.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Wlodarchak N, Xing Y. PP2A as a master regulator of the cell cycle. Crit Rev Biochem Mol Biol. 2016;51:162–184. doi: 10.3109/10409238.2016.1143913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harvey SL, Enciso G, Dephoure N, et al. A phosphatase threshold sets the level of Cdk1 activity in early mitosis in budding yeast. Mol Biol Cell. 2011;22:3595–3608. doi: 10.1091/mbc.e11-04-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hégarat N, Vesely C, Vinod PK, et al. PP2A/B55 and Fcp1 regulate greatwall and ensa dephosphorylation during mitotic exit. PLoS Genet. 2014;10:e1004004. doi: 10.1371/journal.pgen.1004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P, Larouche M, Normandin K, et al. Spatial regulation of greatwall by Cdk1 and PP2A-Tws in the cell cycle. Cell Cycle. 2016;15:528–539. doi: 10.1080/15384101.2015.1127476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forester CM, Maddox J, Louis JV, et al. Control of mitotic exit by PP2A regulation of Cdc25C and Cdk1. Proc Natl Acad Sci. 2007;104:19867–19872. doi: 10.1073/pnas.0709879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson ES, Kornbluth S. Phosphatases driving mitosis. Prog Mol Biol Transl Sci. 2012;106:327–341. doi: 10.1016/B978-0-12-396456-4.00008-0. [DOI] [PubMed] [Google Scholar]
- 44.Gharbi-Ayachi A, Labbe J-C, Burgess A, et al. The substrate of greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science (80-) 2010;330:1673–1677. doi: 10.1126/science.1197048. [DOI] [PubMed] [Google Scholar]
- 45.Mochida S, Maslen SL, Skehel M, Hunt T. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science (80-) 2010;330:1670–1673. doi: 10.1126/science.1195689. [DOI] [PubMed] [Google Scholar]
- 46.Mochida S, Rata S, Hino H, et al. Two bistable switches govern M phase entry. Curr Biol. 2016;26:3361–3367. doi: 10.1016/j.cub.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Juanes MA, Khoueiry R, Kupka T, et al. Budding yeast greatwall and endosulfines control activity and spatial regulation of PP2ACdc55 for timely mitotic progression. PLoS Genet. 2013 doi: 10.1371/journal.pgen.1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Godfrey M, Touati SA, Kataria M, et al. PP2A(Cdc55) phosphatase imposes ordered cell-cycle phosphorylation by opposing threonine phosphorylation. Mol Cell. 2017;65:393.e3–402.e3. doi: 10.1016/j.molcel.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cundell MJ, Hutter LH, Bastos RN, et al. A PP2A-B55 recognition signal controls substrate dephosphorylation kinetics during mitotic exit. J Cell Biol. 2016;214:539–554. doi: 10.1083/jcb.201606033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J, Martin BL, Brautigan DL. Regulation of protein serine-threonine phosphatase type-2A by tyrosine phosphorylation. Science (80-) 1992;257:1261–1264. doi: 10.1126/science.1325671. [DOI] [PubMed] [Google Scholar]
- 51.Guo H, Damuni Z. Autophosphorylation-activated protein kinase phosphorylates and inactivates protein phosphatase 2A. Proc Natl Acad Sci USA. 1993;90:2500–2504. doi: 10.1073/pnas.90.6.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei H, Ashby DG, Moreno CS, et al. Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. J Biol Chem. 2001;276:1570–1577. doi: 10.1074/jbc.M008694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hombauer H, Weismann D, Mudrak I, et al. Generation of active protein phosphatase 2A Is coupled to holoenzyme assembly. PLoS Biol. 2007;5:e155. doi: 10.1371/journal.pbio.0050155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail) Trends Biochem Sci. 2008;33:113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 55.Löw C, Quistgaard EM, Kovermann M, Anandapadamanaban M, Balbach JNP. Structural basis for PTPA interaction with the invariant C-terminal tail of PP2A. Biol Chem. 2014;395:881. doi: 10.1515/hsz-2014-0106. [DOI] [PubMed] [Google Scholar]
- 56.Albuquerque CP, Smolka MB, Payne SH, et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteom. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holt LJ, Tuch BB, Villén J, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soulard A, Cremonesi A, Moes S, et al. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell. 2010;21:3475–3486. doi: 10.1091/mbc.e10-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swaney DL, Beltrao P, Starita L, et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novak B, Kapuy O, Domingo-Sananes MR, Tyson JJ. Regulated protein kinases and phosphatases in cell cycle decisions. Curr Opin Cell Biol. 2010;22:801–808. doi: 10.1016/j.ceb.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hisamoto N, Sugimoto K, Matsumoto K. The Glc7 type 1 protein phosphatase of Saccharomyces cerevisiae is required for cell cycle progression in G2/M. Mol Cell Biol. 1994;14:3158–3165. doi: 10.1128/MCB.14.5.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinsky BA, Nelson CR, Biggins S. Protein phosphatase I regulates exit from the spindle checkpoint in budding yeast. Curr Biol. 2009;19:1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr Biol. 2011;21:942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marquina M, Queralt E, Casamayor A, Ariño J. Lack of the Glc7 phosphatase regulatory subunit Ypi1 activates the morphogenetic checkpoint. Int J Biochem Cell Biol. 2012;44:1862–1871. doi: 10.1016/j.biocel.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 65.Böhm S, Buchberger A. The budding yeast Cdc48(Shp1) complex promotes cell cycle progression by positive regulation of protein phosphatase 1 (Glc7) PLoS One. 2013;8:e56486. doi: 10.1371/journal.pone.0056486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grallert A, Boke E, Hagting A, et al. A PP1–PP2A phosphatase relay controls mitotic progression. Nature. 2014;517:94–98. doi: 10.1038/nature14019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu JQ, Guo JY, Tang W, et al. PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat Cell Biol. 2009;11:644–651. doi: 10.1038/ncb1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heim A, Konietzny A, Mayer TU. Protein phosphatase 1 is essential for greatwall inactivation at mitotic exit. EMBO Rep. 2015;16:1501–1510. doi: 10.15252/embr.201540876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma S, Vigneron S, Robert P, et al. Greatwall dephosphorylation and inactivation upon mitotic exit is triggered by PP1. J Cell Sci. 2016;129:1329 LP–1339 LP. doi: 10.1242/jcs.178855. [DOI] [PubMed] [Google Scholar]
- 70.Bollen M, Gerlich DW, Lesage B. Mitotic phosphatases: from entry guards to exit guides. Trends Cell Biol. 2009;19:531–541. doi: 10.1016/j.tcb.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 71.Sivakumar S, Janczyk PŁ, Qu Q, et al. The human SKA complex drives the metaphase-anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. Elife. 2016;5:12902. doi: 10.7554/eLife.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Della Monica R, Visconti R, Cervone N, et al. Fcp1 phosphatase controls greatwall kinase to promote PP2A-B55 activation and mitotic progression. Elife. 2015;4:e10399. doi: 10.7554/eLife.10399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.