Abstract
Background
The ADAM10-mediated cleavage of transmembrane proteins regulates cellular processes such as proliferation or migration. Substrate cleavage by ADAM10 has also been implicated in pathological situations such as cancer or Morbus Alzheimer. Therefore, identifying endogenous molecules, which modulate the amount and consequently the activity of ADAM10, might contribute to a deeper understanding of the enzyme’s role in both, physiology and pathology.
Method
To elucidate the underlying cellular mechanism of the TBX2-mediated repression of ADAM10 gene expression, we performed overexpression, RNAi-mediated knockdown and pharmacological inhibition studies in the human neuroblastoma cell line SH-SY5Y. Expression analysis was conducted by e.g. real-time RT-PCR or western blot techniques. To identify the binding region of TBX2 within the ADAM10 promoter, we used luciferase reporter assay on deletion constructs and EMSA/WEMSA experiments. In addition, we analyzed a TBX2 loss-of-function Drosophila model regarding the expression of ADAM10 orthologs by qPCR. Furthermore, we quantified the mRNA level of TBX2 in post-mortem brain tissue of AD patients.
Results
Here, we report TBX2 as a transcriptional repressor of ADAM10 gene expression: both, the DNA-binding domain and the repression domain of TBX2 were necessary to effect transcriptional repression of ADAM10 in neuronal SH-SY5Y cells. This regulatory mechanism required HDAC1 as a co-factor of TBX2. Transcriptional repression was mediated by two functional TBX2 binding sites within the core promoter sequence (− 315 to − 286 bp). Analysis of a TBX2 loss-of-function Drosophila model revealed that kuzbanian and kuzbanian-like, orthologs of ADAM10, were derepressed compared to wild type. Vice versa, analysis of cortical brain samples of AD-patients, which showed reduced ADAM10 mRNA levels, revealed a 2.5-fold elevation of TBX2, while TBX3 and TBX21 levels were not affected.
Conclusion
Our results characterize TBX2 as a repressor of ADAM10 gene expression and suggest that this regulatory interaction is conserved across tissues and species.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2998-2) contains supplementary material, which is available to authorized users.
Keywords: ADAM10, Alpha-secretase, APP-processing, Kuzbanian, Omb, TBX2, Transcriptional regulation
Background
Cleavage of type I integral transmembrane proteins, a process designated as ectodomain shedding, represents a pivotal regulatory component of physiological and pathophysiological cellular processes. Particularly, the ADAM (a disintegrin and metalloproteinase)-mediated shedding of cell surface proteins controls important biological functions such as cell fate determination, migration and proliferation [1]. The human ADAM family encodes type I transmembrane proteinases and consists of 21 members [2]. Thirteen of them have been described as being catalytically active by containing a typical zinc binding motif. Within the secretory pathway, the prodomain of these enzymes, which serves as an intrinsic enzyme inhibitor, is removed by proprotein convertases [3]. One of these catalytically competent enzymes—ADAM10—has been investigated intensively due to its wide range of substrates [4, 5] and involvement in neuronal activity [6, 7]. It is expressed in all mammalian species; orthologs are found in invertebrates such as C. elegans (SUP-17) and Drosophila melanogaster [kuzbanian (kuz) and kuzbanian-like (kul)] [8–11]. Knock-out of ADAM10 in mice leads to early embryonic lethality [12], which for a long time hindered investigations regarding its role in development and consequently emphasizes the high relevance of this enzyme in the cell.
Activity of ADAM10 has been associated with e.g. the formation of the brain cortex [13]: mice with a conditional knock-out of ADAM10 in neural progenitor cells die perinatal due to a severe disturbance in neocortex development. This has been shown to be associated with an impaired shedding of ADAM10-dependent targets such as Notch-1 or the amyloid precursor protein (APP). Besides the above mentioned substrates, ADAM10 is known to cleave a variety of type I transmembrane proteins and also GPI-anchored substrates such as the metastasis-associated protein C4.4A [4, 14]. More recently, the investigation of neuroligin-1 (NL-1) and nerve-glia antigen 2 (NG2) as physiological substrates of this protease revealed that ADAM10-mediated shedding is associated with excitatory activity within neuronal networks [6, 7]. In addition to its physiological function, ADAM10 is involved in pathological processes: expression is increased in several types of cancer and increased shedding of cell adhesion molecules leads to development of a motile, invasive phenotype of tumor cells, consequently resulting in a higher risk for metastasis [15–19]. In contrast, ADAM10 has been discussed as a protective protease in the context of Alzheimer’s disease (AD) [20–23]. In this regard, ADAM10-mediated cleavage of APP prevents the generation of neurotoxic A-beta peptides, the key players in AD-pathogenesis, and moreover gives rise to the neuroprotective and neurotrophic fragment APPs-alpha [13, 24–28].
Because of its implication in physiological as well as pathophysiological processes in the cell, it is of great interest to analyze, how the amount of ADAM10 is controlled at the molecular level. An important contribution is probably made by transcription factors (TFs) binding to the ADAM10 promoter. The human ADAM10 promoter region has been described to be located from – 2179 to − 1 bp upstream of the translation initiation start site [29]. Bioinformatical characterization of this sequence revealed several potential TF binding sites for factors such as MZF1, Sp1, and USF, of which the last two were found functional by shift assay analysis [29]. Additionally, retinoic acid receptors have been described to induce ADAM10 expression [23, 29, 30], a finding which already led to conceptualization of a novel therapeutic approach in AD [22]. Additionally, PPAR-alpha, another type of nuclear receptor, has been reported to increase ADAM10 gene expression in neurons via a PPAR/RXR response element within the ADAM10-promoter sequence [31].
We conducted and reported a systematic approach to identify transcriptional modulators of ADAM10 expression [32]: analysis of a luciferase-based screening of 704 human TFs for their potential to modulate ADAM10-promoter activity revealed known and also novel transcriptional activators of ADAM10 such as the unfolded protein response mediator XBP-1. We also identified, for the first time to our knowledge, a transcriptional repressor of ADAM10 expression: the T-box family member TBX2, which is implicated in cell cycle control and several developmental processes such as heart, limb, and brain development [33–36]. The novel TBX2-dependent regulation may play an important role in different functional contexts by altering the amount of ADAM10.
Here we describe that TBX2 decreased expression of ADAM10 in neuronal cells and consequently reduced alpha-secretase-dependent shedding of APP, while the amount of neurotoxic A-beta peptides was increased. To assess the general nature of the observed regulatory interaction, we quantified expression of Drosophila ADAM10 homologues in flies lacking omb function (the Drosophila ortholog of TBX2) and Adam10 amount in murine fibroblasts with elevated Tbx2 expression. Finally, TBX2 mRNA levels in cortical tissue samples of AD-patients were measured to determine whether regulation of ADAM10 by TBX2 is potentially relevant in a human neurodegenerative disease.
Results
Identification of TBX2 as a modulator of the ratio of ADAM10- to BACE-1-promoter activity
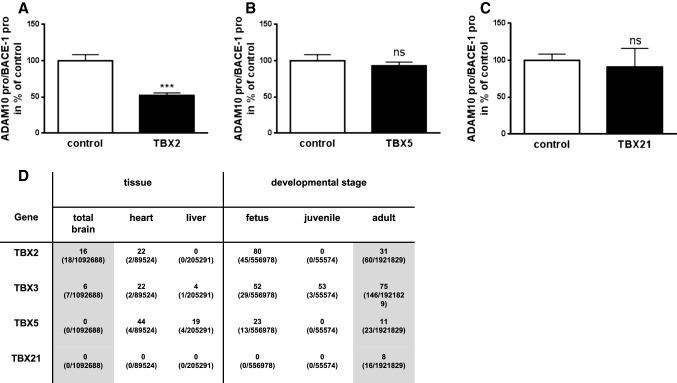
We previously reported a screening of 704 human transcription factors (TFs) for transcriptional regulation of AD-relevant proteases ADAM10 and BACE-1 expression in human SH-SY5Y cells [32]. BACE-1 is the major A-beta generating beta-secretase in humans [37]. TBX2 was found to significantly reduce the ratio of ADAM10- to BACE-1-promoter activity to 52% as compared to empty vector transfected cells (Fig. 1a, S1), while overexpression of the T-box family members TBX5 and TBX21 revealed no significant influence (93% and 91%, Fig. 1b, c, S1). TBX2 belongs to the T-box gene family containing a highly conserved DNA-binding domain—the T-box [38, 39]. In Fig. 1d we provide information on the expression levels of TBX2 and T-box genes which we used as controls. They were ranked according to their relative expression level in different tissues or developmental stages using EST profile analysis. In addition to the three TFs included in the screening approach (TBX2, 5 and 21) we also included TBX3, which has been described as highly homologous to TBX2 with overlapping expression patterns during organogenesis [39–42]. Nonetheless, TBX2 and TBX3 are functionally not equivalent [43, 44].
Fig. 1.
Influence of selected T-box family members (TBX2, TBX5 and TBX21) on ADAM10- and BACE-1-promoter activity. a–c Data were obtained from a screening approach of 704 human TFs described previously [32]. Bars display mean ± SD from three independent experiments (ns p > 0.05, ***p < 0.001, Unpaired Student’s t test). d EST profile analysis of selected human T-box family members. Indicated TFs were ranked according to relative expression level in the brain as compared to other tissue and in adult compared to other developmental stages using NCBI database information. Values represent transcripts per million (Gene EST/Total EST in pool). Respective values for total brain and adult are marked grey
TBX2 has been described in the literature as a mostly developmental factor [45], which is corroborated by a high relative expression level of TBX2 in human fetus [80 transcripts per million (TPM)] as compared to other developmental stages such as juvenile (Fig. 1d). However, TBX2 is strongly re-expressed in adults (31 TPM), which hints at a potential involvement of TBX2 in non-developmental processes. Consistent with a previously reported role for TBX2 in the regulation of heart development [35], we obtained a high TBX2 expression level of 22 TPM in human heart tissue from database analysis (Fig. 1d). From all selected TBX family members only TBX2 and TBX3 displayed expression in the brain based on EST analysis with TBX2 revealing the highest expression level in human brain tissue (16 TPM, Fig. 1d). This might indicate a potential role in brain function or in pathological conditions of neuronal tissue.
TBX2 regulates ADAM10 gene expression in neuronal cells
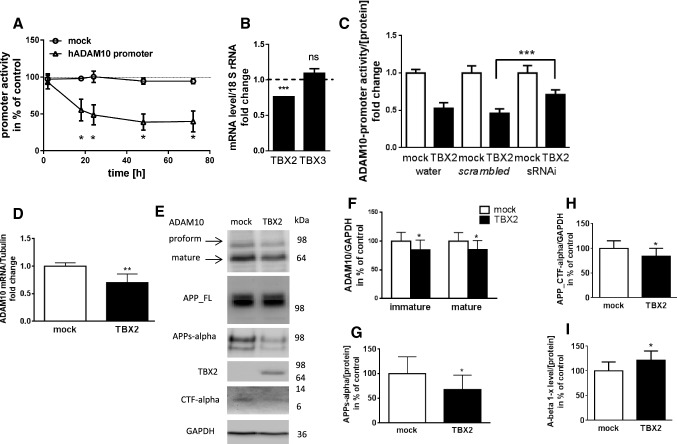
To further characterize TBX2 regulation of ADAM10 transcription, we followed ADAM10-promoter activity over time using a secreted Gaussia luciferase reporter. Human neuroblastoma SH-SY5Y cells were transiently transfected with the reporter constructs together with TBX2 expression plasmid or empty vector and luminescence was recorded from supernatants at indicated time points. TBX2 expression caused a significant decrease of ADAM10-promoter activity to 55% as compared to control cells already 18 h after transfection (Fig. 2a) and inhibition persisted during the entire observation period (40% compared to control at 72 h post transfection).
Fig. 2.
Characterization of TBX2-mediated repression of ADAM10 gene expression. a Time-resolved measurement of ADAM10-promoter activity. SH-SY5Y cells were transiently transfected with human ADAM10-promoter reporter construct or respective promoter-less control vector (mock) together with TBX2 expression plasmid or empty vector. Promoter activity was recorded by luminescence derived from secreted Gaussia princeps luciferase. Values obtained for empty vector transfected cells were set to 100% for each time point (indicated as red dashed line). Statistical analysis was performed by using multiple t test (*p < 0.05). b TBX2 knock-down was verified by real-time RT-PCR. Detection of TBX3 mRNA levels was used for specificity control. Exemplary results are shown from one experiment performed in duplicate (Unpaired Student’s t test was performed against the respective control; (ns p > 0.05, ***p < 0.001). c Test of TBX2-knock-down specificity. TBX2 expression plasmid or empty vector were transfected together with ADAM10-promoter reporter construct in combination with TBX2 specific sRNAi, scrambled control sRNAi or water (solvent control) in SH-SY5Y cells. Cell lysates obtained after 48 h total incubation period were analyzed via luciferase-based measurement. Values were normalized to protein content of lysates. The experiments were performed three times independently in triplicate and values denote mean ± SD. d Effect of TBX2 overexpression on ADAM10 mRNA levels. Total RNA from TBX2 expression plasmid or empty vector transfected SH-SY5Y cells was extracted and subjected to real-time RT-PCR. Obtained values for ADAM10 mRNA were normalized to those of Tubulin. Bars represent mean ± SD from three independent experiments performed in duplicate. e Analysis of the effect of TBX2 overexpression on ADAM10 and full-length APP protein level. SH-SY5Y cells were transfected with TBX2 expression vector or empty vector and cell lysates (20 µg of protein per lane) were subjected to western blot method using specific antibodies for the human proteins. GAPDH was used as a loading control and TBX2 overexpression demonstrated by a TBX2 detecting antibody. e, f–i Quantitation of ADAM10 and APP/APP cleavage product protein levels in TBX2 overexpressing cells. Exemplary blots are shown in e. Obtained values for both, the proform and mature ADAM10 (f), as well as for APP CTF_alpha (h) were normalized to the values obtained for GAPDH and ratios for empty vector transfected cells were set to 100%. Obtained values for secreted APPs-alpha were normalized to total protein amount of respective cell lysate (g). Bars represent mean ± SD of four independent experiments conducted in triplicate. i Determination of A-beta levels. Conditioned medium from TBX2 overexpressing cells was subjected to ELISA measurement and protein content of cell lysates served as normalization. Bars represent mean ± SD from two independent experiments performed in quadruplicate. (ns p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, Unpaired Student’s t test)
In order to test whether ADAM10-promoter activity also responded to a decrease of TBX2 expression, we performed knock-down experiments using TBX2-specific sRNAi. TBX2 mRNA levels were reduced by about 25% in SH-SY5Y cells, while those of TBX3 were not affected (Fig. 2b). Overexpression of TBX2 together with an unspecific scrambled control sRNAi led to a decrease of human ADAM10-promoter activity of about 50% (Fig. 2c). The effect was significantly attenuated (71% as compared to controls), when TBX2 was knocked down using TBX2-specific sRNAi. The results obtained by reporter gene assays were verified by mRNA and protein analysis: SH-SY5Y cells were transiently transfected with TBX2 expression plasmid or empty vector and RNA samples subjected to quantitative RT-PCR. ADAM10 mRNA level was decreased in TBX2 overexpressing cells to 70% as compared to control transfected cells 48 h after transfection (Fig. 2d). This 30% reduction of endogenous ADAM10 mRNA level is weaker than the effect seen in the ADAM10-promoter assay (decrease of about 50%). Such a difference between promoter activity and endogenous mRNA level is consistent with our previous report [31]. Protein analysis using western blot method revealed that both, the proform and the mature ADAM10 were decreased to 85% due to TBX2 overexpression (Fig. 2e, f). This was followed by a reduced ADAM10-dependent cleavage of APP: secretion of APPs-alpha into the cell culture supernatant was decreased about 30% (Fig. 2e, g). In addition, the alpha-cleavage-derived C-terminal fragments of APP (APP_CTF-alpha) were significantly reduced in cell lysates of TBX2 overexpressing SH-SY5Y cells (Fig. 2e, h). However, the expression of full-length APP (APP_FL) was not changed under these experimental conditions (100 ± 32% versus 107 ± 24%, p = 0.5; data not shown). To asses if TBX2 overexpression also affects the generation of neurotoxic A-beta peptides, we analyzed conditioned medium of TBX2 overexpressing cells using ELISA. A-beta 1 − x levels were significantly increased to 122% as compared to values obtained from empty vector transfected cells (Fig. 2i).
Contribution of TBX2 functional domains to the regulation of ADAM10 transcription
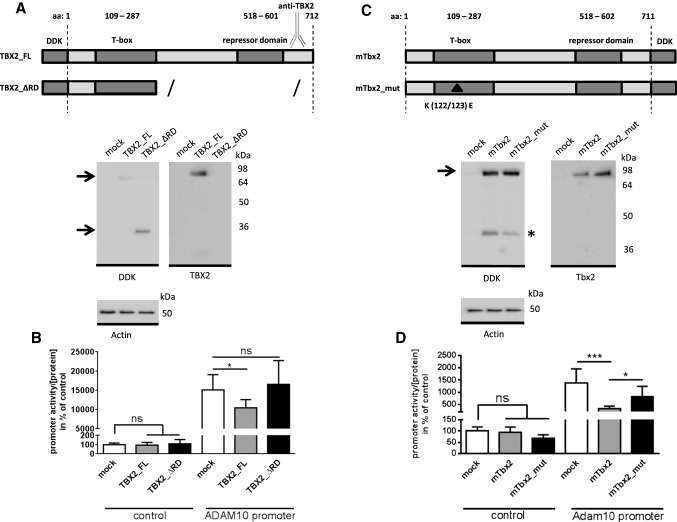
To investigate the contribution of functional domains within the TBX2 protein to the repression of ADAM10 transcription, we generated a TBX2 mutant lacking the C-terminal part of the protein previously identified as a repressor domain (aa 518–aa 601, Fig. 3a, [34, 46, 47]). Expression of the mutant TBX2 (Δ_RD, 33 kDa) as well as the full-length (FL, 75 kDa) protein was verified by western blot using antibodies against the C-terminus of TBX2 and the N-terminal DDK (Flag) epitope (Fig. 3a). Overexpression of the tagged FL-TBX2 in neuronal cells decreased ADAM10-promoter activity to 70% (Fig. 3b), whereas TBX2_ΔRD had no effect on ADAM10 transcription, even though it was more strongly expressed than the wild type protein (Fig. 3a, DDK-specific western blot). To analyze, whether the TBX2-mediated effect on ADAM10 transcriptional level is conserved among species, we transfected murine Neuro2a (N2a) cells with murine wild type Tbx2 expression vector (overexpression demonstrated by western blot, Fig. 3c) together with a murine Adam10-promoter reporter vector. Expression of murine Tbx2 reduced Adam10-promoter activity by 76% as compared to empty vector transfected cells. This demonstrates that repression of ADAM10-promoter is conserved between human and mouse (Fig. 3b, d). In addition, we analyzed a point mutated (R122/123E) murine Tbx2 variant. The missense mutations are located within the highly conserved DNA binding domain (the T-box [48]) but allow expression of a protein of about 75 kDa (Fig. 3c). The mutant Tbx2 with reduced DNA binding [46, 49] caused attenuated repression of Adam10 transcription compared to wild type Tbx2 (reduction of promoter activity only by 40% compared to mock transfected cells, Fig. 3d). This is consistent with previous reports, stating that such a mutant Tbx2 is less efficient than the corresponding wild type protein in repression of the E-cadherin promoter, but not fully defective [50, 51].
Fig. 3.
Effect of TBX2 mutants on ADAM10 transcriptional activity. a Analysis of TBX2 repressor domain (RD) deletion mutant: schematic view of applied human TBX2 protein variants: DDK-tagged full-length TBX2 and respective protein sequence lacking the C-terminal part including the repressor domain (ΔRD). Expression of the TBX2 variants was verified by western blot method using both, TBX2 and DDK antibodies. Actin was detected for loading control. b TBX2 expression constructs were transfected together with ADAM10-promoter luciferase construct or promoter-less control plasmid in human SH-SY5Y cells. Luminescence values were normalized to protein content of respective cell lysate and values obtained for empty vector transfected cells set to 100%. c Effect of DNA-binding attenuated mutant of Tbx2 on ADAM10-promoter activity: schematic view of DDK-tagged murine full-length Tbx2 and the Tbx2 R122/123E DNA-binding impaired mutant (mTbx2_mut, mutation is indicated with a black triangle, both with a C-terminal DDK-Tag). Overexpression in murine N2a cells was analyzed by western blot and Actin served as loading control. A low molecular weight band occurring in Tbx2 overexpressing cells might represent a degradation product (band is marked with *). d N2a cells were transfected with respective Tbx2 expression plasmids together with murine Adam10-promoter reporter plasmid or promoter-less control vector. Values were normalized to protein content and values for control vector transfected cells were set to 100%. b, d Bars represent mean ± SD of three independent experiments conducted in triplicate (ns p > 0.05, *p < 0.05, One-way ANOVA, Bonferroni post-test)
Identification of functional TBX2 binding sites within the human ADAM10-promoter
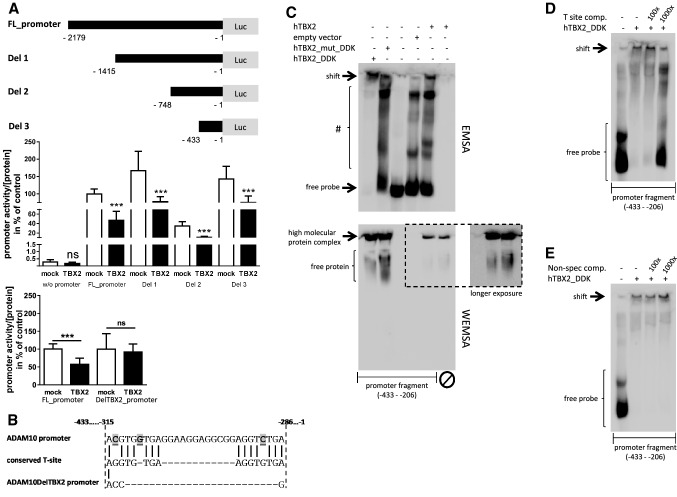
To further substantiate the finding that the DNA binding domain of TBX2 is required for repression of ADAM10 transcription (Fig. 3d), we analyzed the human promoter region for potential TBX2 binding sites. We used luciferase reporter vectors with progressive truncations of the human ADAM10-promoter (Fig. 4a) to approximately identify the TBX2-sensitive promoter area. TBX2 expression in neuronal SH-SY5Y cells significantly decreased ADAM10-promoter activity of all four tested constructs. Under the same conditions, a promoter-less control vector showed no influence when TBX2 was overexpressed (Fig. 4a). Therefore, we concluded that potential TBX2 binding sites might be located within the first 433 bp upstream of the translation initiation start site, which constitutes the major part of the core promoter region [29]. In general, TBX2 mediates transcriptional repression via specific DNA binding elements, the T-sites, comprising the conserved sequence AGGTGTGA [52]. In silico analysis of the respective promoter region revealed two tandem sequences (− 286 to – 294 and – 306 to − 315 bp), which had strong similarity to the consensus T-site (Fig. 4b). When these potential binding sites were eliminated by deletion mutation (sequence information is given in Fig. 4b: ADAM10DelTBX2 promoter), the ADAM10 promoter no longer responded to overexpression of TBX2 within luciferase reporter assays (Fig. 4a, lower graph).
Fig. 4.
Identification of TBX2 binding sites in the human ADAM10-promoter. a Schematic overview of ADAM10-promoter constructs with sequential sequence deletion. Effect of TBX2 overexpression on ADAM10 deletion mutants and on a mutant lacking the TBX2 binding sites (DelTBX2_promoter) was analyzed in comparison to the full-length promoter sequence (FL_promoter) by luciferase-based reporter gene assay. Values represent mean ± SD from three independent experiments performed in triplicate. (ns p > 0.05, ***p < 0.001, Unpaired Student’s t test). b A putative TBX2 binding sequence within the human ADAM10-promoter (− 315 to − 286 bp upstream of the translation initiation site) consisting of two partially conserved T-sites. Sequence discrepancy between the ADAM10-promoter and consensus T-site sequence is highlighted. Additionally, the sequence of the mutant lacking the binding sites is shown. c Electrophoretic mobility shift assay (EMSA) for the identified region of the ADAM10-promoter sequence comprising a putative TBX2 binding motif. The biotinylated promoter fragment (− 433 to − 206) was incubated with nuclear fractions from HEK 293 cells either overexpressing DDK-tagged TBX2, DDK-tagged mutant TBX2 (G121A/R122S), untagged TBX2 or fractions from empty vector transfected cells. Protein-DNA complexes were separated from the free DNA probe by electrophoresis in a native polyacrylamide gel and chemiluminescent signals were visualized by HRP-coupled streptavidin conjugate. Free probe and high molecular weight, TBX2 containing complexes (indicated as shift) are marked. Formed DNA–protein complexes with lower molecular weight are indicated with #. In addition, TBX2 proteins were visualized by western blot-EMSA (WEMSA) using a TBX2-specific antibody under the same conditions as for EMSA analysis. d Nuclear extracts from human TBX2_DDK expressing cells were incubated with the promoter fragment and separated as described in c with or without the presence of an unlabeled competitive oligonucleotide comprising a fully conserved palindromic T-site ([54], see “Methods” section) or an unrelated oligonucleotide, lacking any binding potency towards TBX2, as a specificity control (e). Molar excess of unlabeled positive oligonucleotide is indicated
To test for direct binding of TBX2 to the presumptive T-sites, we performed an electrophoretic shift assay. When the amplified promoter region from – 433 to − 206 bp, including both potential T-sites, was incubated with nuclear extracts of TBX2 overexpressing HEK293 cells, a shift due to DNA–protein binding was observed (Fig. 4c, lane 1). This was strongly attenuated in favor of the free probe when nuclear extracts from cells expressing a DNA-binding impaired mutant of human TBX2 was used (G121A/R122S, hTBX2_mut_DDK, Fig. 4c, lane 2). In addition to the high molecular shift, we observed multiple shift bands with lower molecular weight (indicated by #, Fig. 4c). This may be caused by the use of a large EMSA probe and by using nuclear extracts containing an endogenous pool of nuclear proteins besides the overexpressed TBX2 variants. Therefore, we conclude that these complexes occur due to the binding of nuclear proteins to the sequence or by interaction of DNA stabilizing agents such as histones, which are also present in the nuclear fractions. However, the occurrence of multiple band shifts is consistent with a previous report using nuclear extracts from TBX2 overexpressing HEK293 cells [53]. A high molecular shift complex was also detected using nuclear extracts of untagged TBX2 (hTBX2) but not with nuclear extracts obtained from cells transfected with empty vector (Fig. 4c, lane 4 and 5). Under the same conditions, no signal was obtained by only applying nuclear extracts without DNA (lane 6, Fig. 4c). In parallel, the same samples were analyzed for protein via western blot-EMSA (WEMSA, Fig. 4c, lower part of the picture). A high molecular complex containing TBX2 was observed in all samples with TBX2 overexpression (Fig. 4c, lane 1, 2, 5 and 6). However, TBX2 was replaced from this complex in favor of the unbound protein in the binding reaction, when the mutant of TBX2 with impaired DNA-binding was used and also in the sample completely lacking the DNA component (Fig. 4c, lower part, lane 2 and 6). Formation of the high molecular TBX2-dependent shift-complex was attenuated when reaction was co-incubated with a 1000-fold molar excess of an oligonucleotide comprising a perfect conserved T-site (Fig. 4d, [54]). Competition with the consensus T-site oligonucleotide also strongly increased the free EMSA probe. Under the same conditions, the shift of the high molecular DNA–protein complex was not affected when competing with an unrelated oligonucleotide (non-specific competitor, Fig. 4e). Apparently, TBX2 along with other proteins in the nuclear extract can form an unusually large and stable complex with the analyzed ADAM10-promoter fragment containing the identified T-sites within – 315 to − 286 bp of the translation initiation start site.
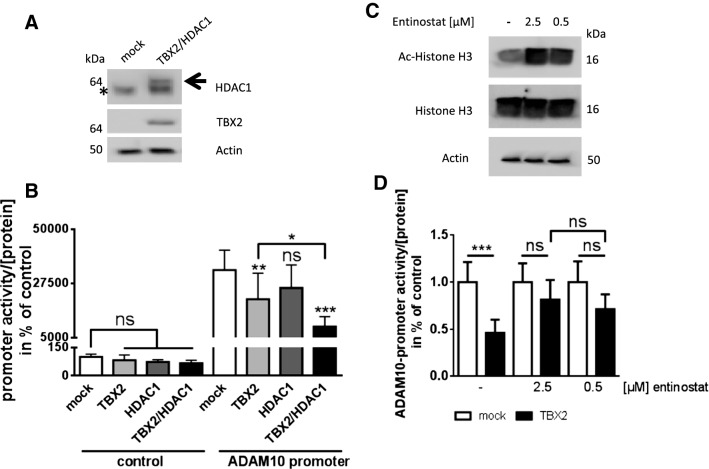
The co-factor histone deacetylase 1 (HDAC1) contributes to the TBX2-mediated repression of ADAM10 expression
In previous reports it has been described, that recruitment of the class I histone deacetylase 1 (HDAC1) is essential for the transcriptional repression of TBX2-target genes such as p21. Thereby, formation of a repressive chromatin structure results in the silencing of respective target gene promoters [36, 55]. To investigate the role of HDAC1 as a potential co-factor for the TBX2-based decrease of ADAM10 transcriptional activity, we performed co-expression studies: successful overexpression of TBX2 and HDAC1 was monitored by western blotting (Fig. 5a). Overexpression of TBX2 revealed the expected decrease in ADAM10-promoter activity (63% of control transfected cells, Fig. 5b), whereas overexpression of HDAC1 alone had no significant influence on ADAM10 gene expression and on protein level (for protein level analysis please see Figure S3). Combination of TBX2 and HDAC1 led to a strong decrease of promoter activity to 29% compared to control, indicating a synergistic effect of TBX2 and HDAC1. To further substantiate our results, we analyzed the effect of TBX2 overexpression combined with pharmacological inhibition of HDAC activity: the inhibitor entinostat has been reported to inhibit selectively HDAC1 and HDAC3 at low µM concentrations, while other members of the HDAC family such as HDAC4, 6, 8 or 10 are not affected (IC50 > 100 µM) [56]. 0.5 µM entinostat has been demonstrated to specifically act on HDAC1 while not affecting other class I HDACs [57]. Effectivity of the inhibitor was monitored by detection of acetylated histone H3 (Lys9) in western blots (Fig. 5c). Overexpression of TBX2 in cells treated with solvent control decreased ADAM10 gene expression to about 50% as observed before (Fig. 5d). When in addition to TBX2 overexpression, HDAC1 and 3 (2.5 µM) or specifically HDAC1 (0.5 µM) were inhibited by entinostat, the TBX2-mediated repression of ADAM10 transcription was significantly attenuated (only 82 and 72% of solvent-treated cells, Fig. 5d). In addition, we observed no statistically significant difference comparing both substance concentrations in TBX2 overexpressing cells indicating an HDAC1 specific effect.
Fig. 5.
Contribution of the co-factor HDAC1 to the TBX2 mediated repression of ADAM10 gene expression. a Overexpression of HDAC1 and TBX2 in SH-SY5Y cells was verified by western blot. A non-specific band in the HDAC1 detecting blot part is marked with *. Actin was detected as loading control. b TBX2 and HDAC1 expression plasmids or empty vector were transiently transfected together with ADAM10-promoter luciferase plasmid. Relative light units (RLU) obtained from cell lysates were normalized to protein amount and values for controls set to 100%. Values are given as mean ± SD from three independently conducted experiments in triplicate (ns p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, One-way ANOVA, Bonferroni post-test). c Inhibition of HDACs was achieved by pharmacological intervention using 2.5 µM or 0.5 µM entinostat respectively. Functionality of inhibitors was demonstrated by detection of acetylated histone H3 (Lys9) as compared to respective total histone or Actin via western blot (20 µg of proteins per lane). DMSO served as solvent control. d Cells were transiently transfected with TBX2 expression vector or empty vector (mock) together with ADAM10-promoter reporter construct and subsequently incubated with entinostat in indicated concentrations or solvent (DMSO) for 24 h. Obtained luminescence values were normalized to protein content of cell lysates and set in relation to empty vector transfected cells. Results are represented as mean ± SD from three independent experiments performed in triplicate. (ns p > 0.05, ***p < 0.001, One-way ANOVA, Bonferroni post-test)
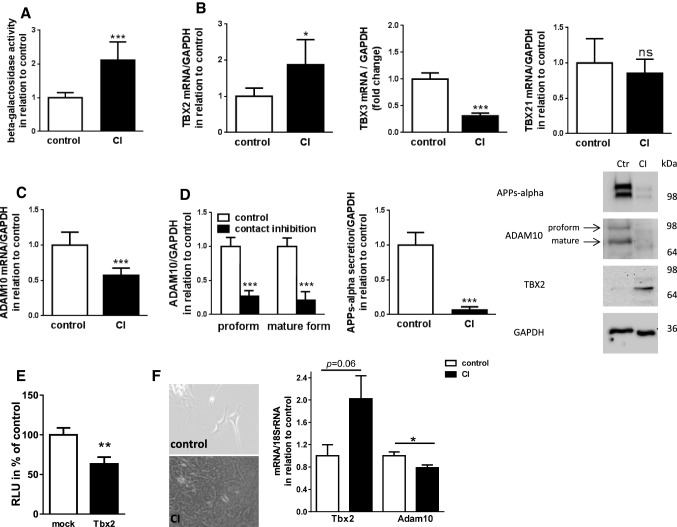
Expression analysis of TBX2 and ADAM10 in contact inhibited cells
TBX2 has been previously described as an immortalizing factor overcoming cell cycle arrest by transcriptional repression of p21 [34, 36]. To investigate the TBX2-mediated repression of ADAM10 transcription in the context of cell cycle control, we cultured SH-SY5Y cells under a contact inhibition (CI) paradigm. In contrast to other cancer cells, SH-SY5Y cells have been reported to be able to undergo growth arrest at least pharmacologically induced [58, 59]. To demonstrate growth inhibition in the CI situation, we measured activity of the senescence-associated beta-galactosidase (SA beta-Gal, [60, 61]), which was about 2-fold increased at high cell density compared to non-contact inhibited controls (Fig. 6a). Under these conditions, the endogenous mRNA level of TBX2 was significantly increased 1.9-fold, whereas the mRNA level of the T-box family member TBX21 was not affected (Fig. 6b). It has been described that TGF-beta-induced expression of TBX3 negatively regulates the TBX2-mediated repression of p21 during cell cycle control [62]. Under contact inhibition we observed significantly decreased TBX3 mRNA levels (Fig. 6b), which might contribute to the increase of TBX2. The elevated TBX2 expression level was paralleled by a decreased ADAM10 expression of about 43% (Fig. 6c). We confirmed these effects at the protein level: TBX2 was strongly increased under CI-conditions (Fig. 6d), while both, the proform and the mature ADAM10 were reduced to 27% and 21%. Moreover, secretion of APPs-alpha, derived by alpha-secretase cleavage, was significantly decreased by 93% in the cell culture supernatant of contact inhibited cells (Fig. 6d). To analyze the regulation of ADAM10 in a different cell type, we transfected murine primary dermal fibroblasts with murine Tbx2-expression vector and again were able to observe a nearly 50% reduction of Adam10 mRNA level (Fig. 6e). Additionally, in contact inhibited fibroblasts (Fig. 6f), Tbx2 was increased twofold (p = 0.06) and Adam10 significantly decreased.
Fig. 6.
Expression and activity of ADAM10 in contact inhibited SH-SY5Y cells and murine dermal fibroblasts. a Senescence was induced in SH-SY5Y cells by contact inhibition (CI) and detected by fluorescence-based measurement of senescence-associated beta-galactosidase activity. mRNA levels of ADAM10 (c), TBX2 and T-box family members TBX3 and TBX21 (b) were analyzed by real-time RT-PCR. Obtained values were normalized to those of GAPDH and set in relation to control cells (low plating density). d APPs-alpha, ADAM10, and TBX2 protein level were determined by western blot using 20 µg of protein from cell lysates and the respective antibodies. Both, the proform and mature ADAM10 were analyzed separately and set in relation to values from control cells. Secreted proteins of respective supernatants were subjected to western blot analysis and ADAM10-dependent soluble fragment of APP, APPs-alpha, was detected with a specific N-terminal antibody. GAPDH was used as loading control. Results are displayed as mean ± SD of two independent experiments performed in triplicate (ns p > 0.05, *p < 0.05, ***p < 0.001, Unpaired Student’s t test). e Primary murine dermal fibroblasts from four independent donor mice were co-transfected with Tbx2-expression vector and murine Adam10 promoter reporter. Luciferase-activity as a reporter for Adam10 transcriptional activity was measured and normalized to empty control vector transfected cells (set to 100%) (**p < 0.01, Unpaired Student’s t test). f Murine dermal fibroblasts were grown in two different densities (control and contact inhibited) and Adam10 as well as Tbx2 mRNA level quantified via real time RT PCR. 18SrRNA levels served for normalization and control values were set to 1 (*p < 0.05, Unpaired Student’s t test)
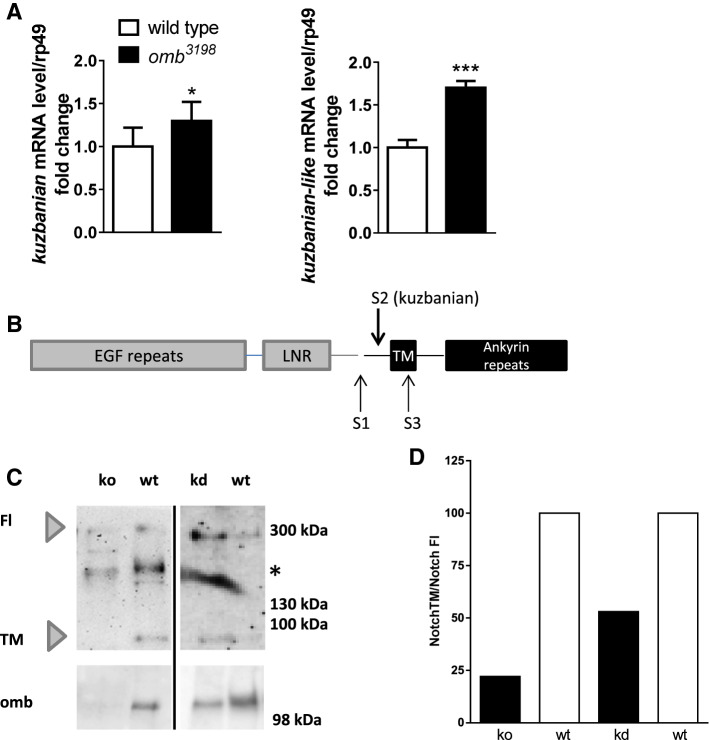
Analysis of kuzbanian and kuzbanian-like expression in omb mutant Drosophila tissue
To extend our results regarding TBX2-mediated regulation of ADAM10 expression to an in vivo situation, we analyzed the mRNA expression level of kuzbanian and kuzbanian-like, the Drosophila homologues of ADAM10 [10, 11], in Drosophila wing imaginal discs. Most cells of this tissue express optomotor-blind (omb), the TBX2 ortholog of the fly [63, 64]. Expression of kuz and kul was compared between wild type and l(1)omb3198 mutant tissue. l(1)omb3198 is a null mutant, encoding a truncated, DNA binding deficient protein [65]. Kuz as well as kul mRNA level were significantly increased about 1.3- and 1.7-fold respectively in mutant wing imaginal discs as compared to wild type larvae (kuz: p = 0.0398; kul: p < 0.0001, Fig. 7a). In Drosophila, Notch (N) is a target for Kuz processing [66]. Unlike the situation in mammals, Drosophila N may not be processed at cleavage site S1 by a furin protease in the absence of signaling, hence existing in a full-length form (Fig. 7b; [67]). We investigated the cleavage products of Notch in the Drosophila wing imaginal disc either lacking (l(1)omb3198) or with reduced omb expression (knock-down). We found in both cases a reduced amount of the protein serving as an ADAM10 substrate in relation to the full length protein (TM; Fig. 7c, d) which might indicate an increased turnover by enhanced kuz/kul expression.
Fig. 7.
Evaluation of the physiological relevance for the TBX2-homologue omb-mediated repression of ADAM10 gene expression in Drosophila. a Expression analysis of Drosophila ADAM10 orthologs kuzbanian and kuzbanian-like in omb-mutant Drosophila larval wing imaginal discs using real-time RT-PCR. Obtained values were normalized to those of ribosomal protein 49 (rp49) and set in relation to the respective wild type samples (*p < 0.05, ***p < 0.001, Unpaired Student’s t test). b Schematic view on Notch processing. Upon cleavage by pro-protein convertases (S1), mature Notch can be cleaved by ADAM10 (S2) cleavage which subsequently allows S3-cleavage (mediated by gamma-secrtase). c Protein lysates of wing imaginal disc obtained by 10 individual larvae with lack of (ko) or lowered (kd) omb expression were subjected to 14% SDS polyacrylamide gels and transferred to nitrocellulose membrane. Omb and Notch protein fragments were detected with the appropriate antibodies. d Amount of full-length Notch (FL) and S2-cleavage educt (TM) were densitometrically determined and are indicated as a ratio of both
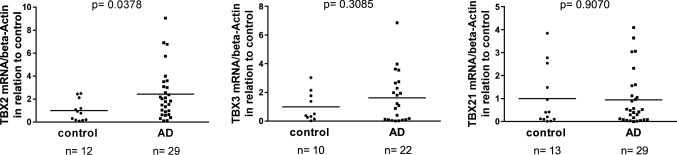
Expression of TBX2 is affected in Alzheimer’s disease (AD) post-mortem brain tissue
To analyze TBX2 and ADAM10 in a pathophysiological context, we assessed TBX2 expression in AD post-mortem brain tissue. The characterization of both sample groups (healthy controls and AD) has been previously described [32]: samples were matched according to age, gender and post-mortem delay and disease state was assessed via CERAD score and Braak stages (Suppl. Table 1A). We have demonstrated earlier, that mRNA levels of ADAM10 were significantly reduced in post-mortem frontal cortex tissue of AD patients as compared to non-demented controls (Suppl. Table 1B) [32]. Analyzing the same set of brain samples for TBX2 expression, we observed a statistically significant increase of TBX2 mRNA level in the AD group compared to healthy controls (2.5-fold, p = 0.0378, Fig. 8) while mRNA levels of the gene family members TBX3 and TBX21 showed no significant differential regulation among groups (p = 0.3085 and p = 0.9070, respectively).
Fig. 8.
TBX2-expression in samples from Alzheimer’s disease-affected patients. Expression of selected T-box family members in AD post-mortem cortical brain tissue. Analysis of TBX2, TBX3 and TBX21 mRNA level in frontal cortex tissue of AD-patients compared to healthy, age-matched controls using real-time RT-PCR. For characterization of samples see [32] and Suppl. Table 1 (p values are indicated, Unpaired Student’s t test)
Discussion
By a luciferase-based screening approach of 704 human transcription factors we identified transcriptional modulators of ADAM10 [32], an enzyme with a protective role in Alzheimer’s disease. By using SH-SY5Y cells, we chose a widely accepted in vitro model for neurodegenerative diseases that is easy to propagate and to transfect. Nevertheless, these cells do not represent mature neurons without being differentiated and are of cancer origin [68]; we therefore included also other in vitro and in vivo models such as primary mouse fibroblasts and Drosophila larvae to substantiate our finding that TBX2 acts as one of the first described transcriptional repressors of ADAM10 gene expression.
In general, members of the T-box family are primarily involved in developmental processes [38]. For example, TBX2 is expressed in specific cardiac structures such as the atrioventricular channel, regulating chamber formation by repressing critical target genes such as Connexin43 [69, 70]. Databank information on EST profiles confirmed these findings from the literature: TBX2 is relatively highly expressed in the heart and also in fetal tissue. ADAM10 has also been reported to be involved in heart development: ADAM10 enzymatic activity mediates cleavage of the Notch-1 receptor and consequently regulates the Notch-1 dependent formation of the atrioventricular channel in mice [71, 72]. Interestingly, our analysis of EST profiles revealed TBX2 expression in total brain tissue of adults, which might indicate a role for TBX2 in regulating cerebral ADAM10 level. Therefore, the regulatory mechanism described here might be of general relevance for cellular processes throughout the entire lifespan of the respective organism and for different organ systems.
In the context of TBX2-mediated transcriptional repression of ADAM10, we identified two well conserved T-sites located between – 315 to − 286 bp upstream of the translation initiation start of ADAM10. A previous report identified the region from – 508 to − 300 bp as the core promoter sequence of ADAM10, by which transcription of this gene is mainly influenced due to the occurrence of several functional binding sites [29]. The two newly identified TBX2 binding elements are within or in close proximity to the described core promoter region, which emphasizes the high relevance of these binding sites in regulating ADAM10 transcription. For TBX2, binding to the consensus sequence AGGTGTGA has been described previously [52, 53]. For example, within the p21 promoter region this binding sequence has been found to be 100% conserved in the human, mouse and rat gene, and has been characterized as functional by band shift assays using in vitro transcribed/translated TBX2 [36]. The putative T-box binding elements (TBEs) in the ADAM10-promoter differ from the consensus in one or two positions. The consensus originally was obtained from a set of sequences bound by T-box proteins in vitro with maximum affinity (e.g. [73, 74]). However, TBEs often differ from the consensus sequence and tend to occur in direct or inverted repeats separated by short spacers. For example, the TBX2 TBE of the well conserved mammalian Connexin43 promoter consists of two tandemly repeated 8 bp elements separated by an 11 bp spacer [75]; the TBX2 TBE of the human p14ARF is made up of an inverted repeat separated by a 1 bp spacer [46]. In these cases, cooperative binding apparently compensates for the lower affinity of the individual variant sites. In the presumptive ADAM10 TBE, two 8 bp elements are tandemly repeated and separated by 12 bp (Fig. 4b). Functionality of these sites regarding TBX2 binding capability was confirmed by EMSA experiments using nuclear extracts of TBX2 overexpressing cells.
It has been described previously, that TBX2 interacts with a variety of binding partners such as RB1 or HDAC1 [55, 54, 76, 77]. HDAC1, but also other members of this HDAC family, were recognized as co-repressors of TBX2 [55, 78, 79] but also of other TBX proteins [80, 81]. For example, HDAC1 directly interacts with the repressor domain of TBX2 and consequently contributes to transcriptional regulation of p21 expression in B16 melanoma cells [55]. Our analysis using a neuronal cell line demonstrates that HDAC1 is required for the TBX2-mediated transcriptional repression of ADAM10 expression in a neuronal context.
Previously, HDAC1 has also been correlated with a physiological role in the developing zebrafish model by regulating Notch-1 signaling [82, 83]. Interestingly, function of TBX2 has also been associated with developmental processes regarding the regulation by Notch-1 downstream targets [35, 84]. Notch-1 is, in regard to developmental processes, an important substrate of ADAM10 in mammals [12, 85]. In addition, in Drosophila the ADAM10 homologue, kuzbanian (kuz), mediates proteolytic processing of Notch, thereby regulating lateral inhibition during Drosophila neurogenesis [86, 66]. Moreover, the Drosophila Notch ligand Delta is shedded by a second ADAM10 ortholog kuzbanian-like (kul) [87]. This ensures unidirectional Notch signaling within the fly cell and for instance drives wing imaginal disc development [87, 88]. Analyzing wing imaginal discs of animals lacking omb (the TBX2 homologue in flies), revealed increased expression of both, kuzbanian and kuzbanian-like. Our findings therefore indicate that TBX2-mediated regulation of ADAM10 expression might play an important role in the context of Notch-dependent larval development, consistent with altered cleavage ratios of Notch observed here. In addition to our analysis using a human experimental cell model and our results derived from Drosophila, we confirmed the conservation of the effect in a rodent-derived model: overexpression of the murine Tbx2 protein in murine neuronal cells decreased activity of the murine Adam10 promoter and murine dermal fibroblasts also revealed reduced Adam10 expression when Tbx2 was overexpressed. Therefore, we conclude that this regulation is a conserved mechanism at least in human, mouse and fly.
There is emerging evidence that TBX2 is an essential modulator of cell cycle control by down regulating p21 expression [55]. In the context of cell cycle regulation, we analyzed a neuronal cellular model, in which the cell overcomes experimentally induced growth arrest by elevating the endogenous level of TBX2. This was accompanied by a decrease of ADAM10 expression in SH-SY5Y cells and in murine primary fibroblasts. In this regard it is of note, that the cell cycle mechanism has been reported to be impaired in AD: hippocampal neurons from AD-patients seem to be able to re-enter the cell cycle [89]. This attempt by neurons to re-enter mitosis leads to a defective cell cycle and consequently to neuronal degeneration [90]. Additionally, the regulator of cell cycle (RGCC) has been found to be upregulated in the frontal cortex of MCI and AD patients compared to healthy controls and to correlate on the protein level with poorer ante-mortem cognitive performance [91]. To test whether the TBX2-mediated repression of ADAM10 might be involved in this neurodegenerative disease, we analyzed cortical samples derived from AD-patients. We observed a 2.5-fold increased expression of TBX2, while the family members TBX3 and TBX21 remained unaffected. We have demonstrated previously, that ADAM10 expression is decreased by about 30% in the same tissue samples as investigated here [32]. Therefore we conclude that elevated levels of TBX2 might in part contribute to the decreased expression of ADAM10 in the brain of patients suffering from AD.
Conclusion
In summary, we describe that the transcription factor TBX2 is able to restrain ADAM10 gene expression and that this mechanism might play a role in regulating cellular processes in health, development and disease.
Methods
Luciferase encoding reporter vectors
Human ADAM10-promoter luciferase-reporter vector and dual ADAM10/BACE-1 luciferase-reporter (based on pGL4.76; Promega, Madison, WI, USA) were described previously [32]. The murine Adam10-promoter reporter construct has been described in [23].
Promoter reporter vectors coding for secreted Gaussia princeps luciferase (pGL4.76_hADAM10-promoter_GLuc and pGL4.76_GLuc)
Gaussia princeps luciferase cDNA sequence was amplified from pCMV_GLuc (New England Biolabs, Ipswich, MA, USA) using FailSafe PCR system (Epicentre, Madison, WI, USA) with the following oligonucleotides: GLuc_for: 5′-AGATCTGATCCAGCCACCATGGGAG-3′ and GLuc_rev: 5′-GGATCCAGCATGCCTGCTATTGCAG-3′ (introduced restriction sites for BglII or BamHI are underlined; the start codon of luciferase cDNA is marked bold and underlined). The amplicon was subcloned into the EcoRV site of a modified pUC19 vector (New England Biolabs, Ipswich, MA, USA) via T/A cloning method. Gaussia princeps luciferase coding sequence was removed by BglII and BamHI restriction and used for replacement of Renilla reniformis luciferase cDNA within the pGL4.76_hADAM10-promoter_Renilla_Luc reporter plasmid [32].
Truncated ADAM10-promoter containing reporter constructs (Del1, Del2 and Del3)
For the identification of a TBX2-sensitive area within the human ADAM10-promoter, we constructed luciferase reporter vectors with truncated promoter sequences. Subfragments of the full-length human ADAM10-promoter (described in [29]) were amplified using the FailSafe PCR system (Epicentre, Madison, WI, USA) and the following oligonucleotides: Del1 (− 1415 to − 1 bp) Del1_for: 5′-ACTGCTAGCCCAGCACTTGTGGGAGGC-3′, Del 2 (− 748 to − 1 bp) Del2_for 5′-ACTGCTAGCCATCACTGGGGCAGCTGC-3′ and pGL4.76_rev (for both deletion constructs) 5′-GAGGCCCAGTGATCATGCG-3′ (NheI restriction sites are underlined). The obtained amplicons were digested with BglII and NheI and subsequently inserted as a replacement of the BglII and NheI site flanked promoter sequence of the pGL 4.76 ADAM10-promoter reporter vector. For the Del3 deletion mutant (− 433 to − 1 bp), the Del1 construct (− 1415 to − 1 bp) was digested with StuI, producing a fragment comprising 433 bp of the ADAM10-promoter region upstream of the translation initiation start site together with Renilla luciferase coding sequence. The second, vector DNA including fragment, was subsequently digested with NheI to remove the remaining ADAM10-promoter sequence from – 1415 to − 434 bp. Blunting of 5′ overhangs was performed by incubation with T4 DNA polymerase (New England Biolabs, Ipswich, MA, USA) and obtained fragments were ligated to generate the Del3 construct. To delete the potential TBX2 binding sites within the human ADAM10 promoter, a part of the promoter (starting downstream of the binding sites and ending downstream of the BglII site of the vector) was amplified using the oligonucleotides TBXDel_for: 5′CGGTTTCGAAGGAGG3′ and TBXDel_Rev: 5′ AGTACCGGATTGC3′. The amplified region was used to replace the respective sequence of the full length promoter by digestion with PmaCI (upstream of the binding sites) and BglII. Positive clones containing the mutated vector were identified by the newly inserted AgeI restriction site (ACCGGT).
Expression vectors
Expression vectors for HDAC1 (pCMV-Entry_Myc_DDK_HDAC1), TBX2 (pCMV6_TBX2), TBX5 (pCMV6_TBX5), TBX21 (pCMV6_TBX21) and respective empty vector were obtained from Origene (Rockville, MD, USA).
Construction of the TBX2 repressor domain deletion mutant (TBX2_delta RD)
To obtain a mutant TBX2 protein with deleted transcriptional repressor domain, we generated a plasmid encoding for a tagged (His, DDK) TBX2 mutant lacking the C-terminal part of the protein (300–712 aa). For control studies, we also constructed a corresponding vector expressing the full-length (FL) DDK-tagged TBX2 (see Fig. 3a). Briefly, sequences encoding TBX2_FL (1–712 aa) or TBX2_delta RD (1–299 aa) were amplified from pCMV6_TBX2 construct (Origene, Rockville, MD, USA) with oligonucleotides as follows: TBX2_for: 5′-GCGATCG CCATGAGAGAGCCGGCGCTG-3′ (SgfI site is underlined, start codon is marked bold and underlined), TBX2_FL_rev: 5′-ACGCGTTCACTTGGGCGACTC CCG-3′ and TBX2_delta RD_rev: 5′-ACGCGTCAGCTGCTGCTTCCTTTTCTC-3′ (MluI sites are underlined, stop codons are marked bold and underlined). Obtained amplicons were inserted into pUC19Tmod (pUC19 vector (New England Biolabs, Ipswich, MA, USA) with an introduced EcoRV restriction site and thymidine overhang) by T/A cloning and subsequently cloned as a SgfI-MluI fragment into pCMV6_AN_His_DDK vector (Origene, Rockville, MD, USA).
Generation of human TBX2 expression vectors
The pT-REx-DEST30/hTBX2_DDK expression plasmid has been described previously [92]. The vector pT-REx-DEST30/TBX2 (G121A/R122S) was constructed as described for the analogous TBX3 mutant [50], which encodes a DNA-binding deficient double mutant protein.
Generation of murine Tbx2 expression vectors
Expression vectors for murine wild type Tbx2 (p3xFlagCMV14_mTbx2) and respective point mutated sequence (R122/123E) leading to impaired DNA-binding (p3xFlagCMV14_mTbx2_mut) were kindly provided by C. Goding (University of Oxford, United Kingdom) and described previously [46, 51, 93].
All restriction enzymes used in this study were from New England Biolabs (Ipswich, MA, USA) or Thermo Scientific (Karlsruhe, Germany). All inserted/mutated sequences were verified by sequencing (Eurofins Genomics, Ebersberg, Germany) and/or restriction analysis. DNA for transfection experiments was prepared endotoxin-free by Nucleo Bond Xtra Midi Plus EF kit (Macherey–Nagel, Düren, Germany).
Electrophoretic mobility Shift Assay (EMSA)
For electrophoretic mobility shift assay (EMSA) the LightShift Chemiluminecent EMSA Kit (Thermo Scientific, Waltham, MA, USA) was used as recommended by the manufacturer. Oligonucleotides were biotinylated using the Biotin 3′ End DNA Labeling Kit (Thermo Scientific, Waltham, MA, USA). ADAM10-promoter sequence (− 433 to − 206 bp) used for initial band shift analysis was amplified from the ADAM10-promoter deletion construct Del3 (described above) using Perfect Taq polymerase kit (5Prime, Hilden, Germany) with oligonucleotides as follows: Del3_for: 5′-CCTAGCAGCACGGGAAC-3′ and Del3_rev: 5′-CCCCTCGTTCGTTTCCTT C-3′. Double stranded oligonucleotides with the conserved T-site were generated by annealing of T-site_for: 5′-CTAGAGGGAATTTCACACCTAGGTGTGAAATTCCCT-3′ and T-site_ rev: 5′-AGGGAATTTCACACCTAGGTGTGAAATTCCCTCTAG-3′ (potential TBX2 binding sites are underlined). Oligonucleotides for the unspecific competitor were: Comp_for: 5′-GCCTCCCAAGCCCAA TCCCAGCTCTCCGCCG-3′ and Comp_rev: 5′-CGGCGGAGAGCTGGGATTGGGCTTGG GAGGC-3′. Nuclear protein extracts from HEK293 cells transiently transfected with TBX2 encoding plasmids or empty vector were obtained using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Waltham, MA, USA). We used HEK293 cells, because they have been found suitable as a model system with high expression rates after transient transfection (e.g. [94]). Successful overexpression of either TBX2 species was verified by western blot method (Figure S2 A). Conservation of the TBX2-mediated effect on ADAM10 transcription in HEK293 cells was confirmed via luciferase reporter gene assay (Figure S2 B).
Protein-DNA complexes were separated on native 6% polyacrylamide gels containing 0.5 × TBE buffer (50 mM Tris, 45 mM boric acid, 0.5 mM Na2-EDTA, pH 8.3). After electrophoresis, nucleic acids and proteins were blotted onto Biodyne Precut Nylon Membranes (Thermo Scientific, Waltham, MA, USA) and fixed by UV irradiation using a Stratalinker (Stratagene, La Jolla, CA, USA). Chemiluminescent signals of biotinylated molecules were captured using a CCD-camera imaging system (Raytest, Straubenhardt, Germany).
For western blot EMSA method (WEMSA, [95]), proteins were transferred to a nitrocellulose membrane after electrophoresis, essentially using the same conditions as for EMSA analyses (described above). The following procedure regarding e.g. antibody incubation was identical to that described in the respective western blotting section.
Cell culture
Cell lines were maintained at humidified air (95%), 5% CO2 and 37 °C. The human neuroblastoma cell line SH-SY5Y was cultured in DMEM/F12 (Life Technologies, Darmstadt, Germany) supplemented with 10% FCS and 1% glutamine (both GE Healthcare, Piscataway, NJ, USA). The human embryonic kidney 293 cells (HEK 293) and the murine neuroblastoma cell line N2a were cultured in DMEM medium (GE Healthcare, Piscataway, NJ, USA) supplemented with 10% FCS, 1% glutamine and 1% sodium-pyruvate. SH-SY5Y cells were passaged twice a week 1:2–1:3, HEK 293 and N2a cells were passaged 1:10–1:12 twice a week. For inhibition of Histone deacetylases (HDACs), SH-SY5Y cells were treated with 2.5 μM (specific for HDAC1 and 3) or 0.5 µM (HDAC1 specific) entinostat (Selleckchem, Houston, TX, USA) for 24 h. DMSO served as solvent control.
Murine dermal fibroblasts were obtained from ear biopsy material of wild type C57B6/J mice (aged 2–3 months). Briefly, tissue was disinfected with Braunol, transferred to 70% ethanol and then to sterile PBS containing 1% Penicillin/Streptomycin. After 30 min digestion with Collagenase/Dispase mixture (4 mg/ml in DMEM, Roche, Rotkreuz Risch, CH) and manual disruption by a sterile scalpel, cells were cultivated in DMEM medium (GE Healthcare, Piscataway, NJ, USA) supplemented with 10% FCS, 1% Penicillin/Streptomycin (Sigma Aldrich, Steinheim, Germany). Cell debris and dead cells from preparation were aspirated with regular medium exchange and cells were used for experiments at 8–10 DIV.
Transfection and luciferase assay
Co-transfections of TBX2 expression plasmids together with reporter constructs were performed as described previously [32]. Luciferase activity was measured with Renilla luciferase assay kit (Promega, Madison, WI, USA) following the manufacturer’s protocol. Relative light units (RLUs) were normalized to protein content in the cell lysate, quantified by Bradford Nanoquant reagent (Roth, Karlsruhe, Germany). For time-resolved measurements of promoter activities either ADAM10-promoter reporter plasmid or promoter-less Gaussia luciferase plasmid (mock) together with TBX2 expression vector were transfected in SH-SY5Y cells. After 5 h, transfection medium was replaced by fresh culture medium and 10 µl aliquots of the supernatant were collected at indicated time points, stored at − 20 °C and luminescence signal derived from secreted Gaussia luciferase was measured and normalized as described above.
Transfection of murine dermal fibroblasts was conducted using the NeonTransfection System combined with the 10 µl size kit components (Life Technologies, Darmstadt, Germany). We followed instructions of the vendor despite adapting the parameters of electroporation due to pre-tests considering viability and transfection efficacy optimization for murine dermal fibroblasts (1300 V, 20 s pulse, 3 pulses).
sRNAi transfection
SH-SY5Y cells were transfected with 50 ng TBX2 expression plasmid or empty vector together with 50 ng luciferase reporter vector. This was combined with 1 pmol TBX2 specific sRNAi (HSS110509, Life Technologies, Darmstadt, Germany), scrambled control sRNAi with medium GC content (Life Technologies, Darmstadt, Germany) or water for specificity control. Cells were transfected using 0.5 µl Lipofectamine2000 (Life Technologies, Darmstadt, Germany) per 96-well, harvested 48 h post transfection, and promoter activities were subsequently analyzed via luciferase-based assay as described above.
RNA isolation from cultured cells and quantitative real-time RT-PCR
Total RNA from SH-SY5Y cells or dermal fibroblast grown on 6-well plates was extracted using the RNA II Kit (Macherey&Nagel, Düren, Germany) following the manufacturer’s protocol. Samples were stored at − 80 °C and freshly diluted for application. 100 ng of RNA were subjected to real-time RT-PCR analysis, performed with the thermocycler StepOne Plus (Life Technologies, Darmstadt, Germany) and QuantiTect SYBR Green RT PCR Kit (Qiagen, Hilden, Germany). QuantiTect Primer Assays (Qiagen, Hilden, Germany) were used for detection of the following genes: ADAM10 (Hs_ADAM10_1 _SG), TBX2 (Hs_TBX2_1_SG), TBX3 (Hs_TBX3_1_SG), TBX21 (Hs_TBX21_1_SG), Tubulin (Hs_TUBB_1_SG), 18 S rRNA (Hs_RRN18S_1_SG) and GAPDH (Hs_GAPDH_2_SG), murine 18 S rRNA (Mm_Rn18s_3_SG), murine Tbx2 (Mm_Tbx2_1_SG). Obtained mRNA quantities of each reaction were calculated from calibration curve and normalized to those of the respective housekeeping gene (Tubulin, 18 S rRNA or GAPDH). Normalized data were set in relation to the corresponding control-treated cell population.
Quantitation of kuzbanian and kuzbanian-like expression levels in omb3198 mutant flies
For analysis, Drosophila stock l(1)omb3198 and wild type controls were used [96]. The mutation introduces a stop codon within the omb coding sequence, leading to a truncated form of the protein with impaired DNA-binding properties [97, 98]. For expression analysis of the ADAM10 Drosophila homologues kuzbanian (kuz) and kuzbanian-like (kul) male hemizygous larvae were selected because of omb being localized on the X-chromosome. Total RNA from wing imaginal discs of wild type or omb-mutant larvae (70 wing discs per group) was extracted using the RNA II kit (Macherey&Nagel, Düren, Germany) following company instructions. 13 ng RNA of each sample (technical replicates n = 6) was subsequently subjected to real-time RT-PCR analysis using a StepOne Plus cycler (Life Technologies, Darmstadt, Germany) and the QuantiTect SYBR Green RT PCR kit (Qiagen, Hilden, Germany). The sequences of the used oligonucleotides were as follows: kuz_for: 5′-CGGCCTGTCGCAATTCC-3′ and kuz_rev: 5′-AAATCTCTGTAGGTGAAC ACGTAAGC-3′; rp49_for: 5′-TACAGGCCCAAGATCGTGAA-3′ and rp49_rev: 5′-TCTCCTTGCGCTTCTTGGA-3′; QuantiTect Primer Assay (Qiagen, Hilden, Germany) was used for detection of kuzbanian-like (Dm_Kul_1 _SG). mRNA level obtained for kuz and kul were quantified using the 2−(ΔΔCt) method and data were normalized to rp49 mRNA levels.
Evaluation of the expression level of T-box family members in AD post-mortem brains
Total RNA of frozen frontal cortex tissue (BrainNet, Munich, Germany) was extracted using Trizol (Life Technologies, Darmstadt, Germany) following the manufacturer’s protocol. 2 µg RNA were reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Darmstadt, Germany) and subsequently subjected to qPCR analysis using the 7500 Fast real-time PCR cycler (Life Technologies, Darmstadt, Germany) and Fast SYBR Green Master Mix (Life Technologies, Darmstadt, Germany). Oligonucleotides for the house keeping gene beta-Actin were as described [99, 93]. Oligonucleotides for TBX-gene family members were as follows: TBX2_for: 5′-TCCTGAAGCTGCCTTACAGC-3′ and TBX2_rev: 5′-CTT CAGCTGTGTGATCTTGTCA-3′, TBX3_for: 5′-TGGGACCTCTGATGAGTCCT-3′ and TBX3_rev: 5′-CGCTGGGACATAAATCTTTGA-3′ and TBX21_for: 5′-TTGAGTCCATGT ACACATCTGTTG-3′ and TBX21_rev: 5′-TGGTTGGGTAGGAGAGGAGA-3′. The mRNA levels of the respective gene were quantified using the 2−(ΔΔCt) method [100] and obtained values were normalized to beta-Actin mRNA levels. For sample characterization see (Suppl. Table 1 and [32]).
Preparation of whole cell lysates and western blotting
SH-SY5Y cells were seeded on 12-well plates at a density of 5*105 cells per well. Cells were transiently transfected with expression plasmids or empty vector or incubated with histone deacetylase inhibitors as described above. After the respective incubation time, cells were lysed in LDS sample buffer (Life Technologies, Darmstadt, Germany) containing 10% DTT (v/v, 1 M) and 1x protease inhibitor cocktail (Roche, Mannheim, Germany). Samples were incubated for 10 min at 95 °C and stored at − 20 °C. Lysates were assayed for protein content using the Nanoquant reagent (Roth, Karlsruhe, Germany). 20 µg protein of whole cell lysate were separated on 10% SDS–polyacrylamide gels and blotted onto nitrocellulose membrane at 100 V for 2 h. Detection of the signals was carried out by blocking the membranes for 1 h in blocking solution and incubation overnight at 4 °C with the appropriate primary antibody at a dilution of 1:1000. Antibodies were as follows: TBX2 mouse monoclonal antibody (ab58249, Abcam, Cambridge, UK), histone deacetylase 1 (HDAC1) rabbit polyclonal antibody (PA1-860, Thermo Scientific, Karlsruhe, Germany), Actin rabbit polyclonal antibody (A2066, Sigma Aldrich, Steinheim, Germany), ADAM10 rabbit polyclonal antibody (Merck, Darmstadt, Germany), GAPDH rabbit monoclonal antibody (14C10, Cell Signaling, Danvers, MA, USA), DYKDDDDK Tag mouse monoclonal antibody (9A3, Cell Signaling, Danvers, MA, USA), APP N-terminal mouse monoclonal antibody (6E10, Covance, Madison, WI, USA) or APP C-terminal antibody (described previously [101]), Acetyl-Histone H3 (Lys9, C5B11) or total-Histone H3 (D1H2) rabbit monoclonal antibodies (both Cell Signaling, Danvers, MA, USA), anti-dNotch-1 antibody (1:500; C17.9C6, Developmental Studies Hybridoma Bank (DSHB), University of Iowa; [102]), anti-Omb antibody [97]. Blots were incubated with secondary antibody coupled with horseradish peroxidase (Thermo Scientific, Karlsruhe, Germany). Signals obtained by applying SuperSignal West Femto chemiluminescent substrate (Thermo Scientific, Karlsruhe, Germany) were captured using a CCD-camera imaging system (Raytest, Straubenhardt, Germany). Quantitative analysis was carried out with AIDA image analyzer 4.26 software (Raytest, Straubenhardt, Germany).
The effect of omb knock-down on N processing was tested in the following genotypes: w[1118] P{w[+ mW.hs] = GawB}Bx[MS1096]/w[1118]; P{w[+ mC] = UAS-Dcr-2.D}2/II; UAS-omb-RNAi[C4]/III (experimental) and w[1118] P{w[+ mW.hs] = GawB}Bx[MS1096]/w[1118]; P{w[+ mC] = UAS-Dcr-2.D}2/II (control), based on the Bloomington stock 25706. Wing discs were dissected from female third instar larvae.
ELISA measurement
SH-SY5Y cells were transfected with TBX2 expression plasmid or empty vector for 32 h as described above. Medium was exchanged followed by a secretion period of 16 h and conditioned medium was collected and centrifuged for 5 min at 1000 rpm. 200 µl of cell supernatant was applied to human A-beta 1 − x ELISA Kit (IBL, Hamburg, Germany) as recommended by the manufacturer. Values obtained for A-beta peptide concentration were normalized to the protein content of the respective cell lysate.
Senescence analysis
Growth arrest was induced in SH-SY5Y cells by contact inhibition. Cells were seeded at a density of 2.5 × 105 cells/ml for control cell population and 7.5 × 105 cells/ml for contact inhibition conditions. Induction of senescence was assessed by detection of senescence-associated beta-galactosidase (SA beta-Gal) activity using a fluorescence-based assay kit (cellbiolabs, San Diego, CA, USA) as recommended by the manufacturer. mRNA and protein analysis were carried out as described in the method section (quantitative real-time RT-PCR and western blotting).
EST-profile analysis of T-box family members
Selected members of the T-box gene-family (TBX2, TBX3, TBX5 and TBX21) were ranked according to their relative expression levels in different tissues (brain, heart and liver) and additionally, to various developmental stages (fetus, juvenile and adult) using databank information on EST profiles (http://www.ncbi.nlm.nih.gov/unigene). Values indicate transcripts per million (TPM) and gene EST per total EST in pool.
Statistical analysis
Statistical significance was tested with One-way ANOVA followed by Bonferroni post-test or by unpaired Student’s t test when appropriate (GraphPad Prism version 6, San Diego, CA, USA). Values of p < 0.05 were considered statistically significant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Federal Ministry of Education and Research (BMBF) in the framework of the National Genome Research Network (NGFN), FKZ01GS08130, 01GS08125 and 01GS08129-5 and by the Alfons Geib Stiftung. We thank K. Hilger, A. Bruns, and S. Schneider (all University Medical Center Mainz, Germany) for technical assistance; we also are grateful to Inka Hoffmann, Fred Eichinger, and Melanie Heyde (University of Mainz, Germany) for preparation of Drosophila wing imaginal discs and to Sven Grösgen (Saarland University, Germany) for experiments regarding human samples; we want to thank Colin Goding (University of Oxford, UK) for murine TBX2 expression constructs and Christian Haass (LMU Munich, Germany) for APP C-terminal antibody 6687. Stock 25706 obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) was used in this study.
Abbreviations
- AD
Alzheimer’s disease
- ADAM10
A disintegrin and metalloproteinase 10
- A-beta
Amyloid-beta
- APP
Amyloid precursor protein
- BACE-1
Beta site APP cleaving enzyme-1
- CERAD
Consortium to establish a registry for Alzheimer’s disease
- EST
Expressed sequence tag
- EMSA
Electrophoretic mobility shift assay
- FL
Full-length
- HDAC1
Histone deacetylase 1
- kuz
Kuzbanian
- kul
Kuzbanian-like
- NG2
Nerve glia antigen 2
- NL-1
Neuroligin-1
- Omb
Optomotor-blind
- PPAR-alpha
Peroxisome proliferator activated receptor-alpha
- RB1
Retinoblastoma 1
- Rp49
Ribosomal protein 49
- SA beta-GAL
Senescence associated beta-galactosidase
- TF
Transcription factor
- TPM
Transcripts per million
- XBP-1
X-box binding protein-1
Author contributions
KE conceived and coordinated the study, drafted the manuscript, performed experiments with dermal fibroblasts and generated the TBX2 binding site deletion mutant. SR carried out the molecular biology and biochemistry studies, performed analysis on EST profiles, conducted the statistical analysis, and helped to draft the manuscript. FS constructed secreted luciferase reporter vectors and helped to revise the manuscript. NS performed overexpression experiments of HDAC1 to assess effects on ADAM10 and luciferase reporter gene assays for analyzing the TBX2 binding site deletion mutant. MG and TH designed and coordinated studies regarding human brain tissue and revised the manuscript. GP provided expression constructs for human DDK-tagged TBX2 and Drosophila larval samples. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the German Federal Ministry of Education and Research (BMBF) in the framework of the National Genome Research Network (NGFN) and FKZ01GS08130 and by the Alfons-Geib Stiftung.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Compliance with ethical standards
Ethical approval and consent to participate
All experiments were conducted in accordance to the official regulations for the care and use of laboratory animals and approved by local authorities (University of Mainz, Germany).
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Saftig P, Reiss K. The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: novel drug targets with therapeutic potential? Eur J Cell Biol. 2011;90:527–535. doi: 10.1016/j.ejcb.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 2001;15:1837–1839. doi: 10.1096/fj.01-0007fje. [DOI] [PubMed] [Google Scholar]
- 4.Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol. 2009;20:164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Endres K, Deller T. Regulation of alpha-secretase ADAM10 in vitro and in vivo: genetic, epigenetic, and protein-based mechanisms. Front Mol Neurosci. 2017;10:56. doi: 10.3389/fnmol.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakry D, Neitz A, Singh J, Frischknecht R, Marongiu D, Biname F, Perera SS, Endres K, Lutz B, Radyushkin K, et al. Oligodendrocyte precursor cells modulate the neuronal network by activity-dependent ectodomain cleavage of glial NG2. PLoS Biol. 2014;12:e1001993. doi: 10.1371/journal.pbio.1001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki K, Hayashi Y, Nakahara S, Kumazaki H, Prox J, Horiuchi K, Zeng M, Tanimura S, Nishiyama Y, Osawa S, et al. Activity-dependent proteolytic cleavage of neuroligin-1. Neuron. 2012;76:410–422. doi: 10.1016/j.neuron.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Jarriault S, Greenwald I. Evidence for functional redundancy between C. elegans ADAM proteins SUP-17/Kuzbanian and ADM-4/TACE. Dev Biol. 2005;287:1–10. doi: 10.1016/j.ydbio.2005.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riddle DL, Blumenthal T, Meyer BJ, Priess JR. Introduction to C. elegans. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans I. New York: Cold Spring Harbor; 1997. [Google Scholar]
- 10.Rooke J, Pan D, Xu T, Rubin GM. KUZ, a conserved metalloprotease-disintegrin protein with two roles in Drosophila neurogenesis. Science. 1996;273:1227–1231. doi: 10.1126/science.273.5279.1227. [DOI] [PubMed] [Google Scholar]
- 11.Wei S, Whittaker CA, Xu G, Bridges LC, Shah A, White JM, Desimone DW. Conservation and divergence of ADAM family proteins in the Xenopus genome. BMC Evol Biol. 2010;10:211. doi: 10.1186/1471-2148-10-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, Saftig P. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- 13.Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, Snellinx A, Craessaerts K, Thathiah A, Tesseur I, et al. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci. 2010;30:4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esselens CW, Malapeira J, Colome N, Moss M, Canals F, Arribas J. Metastasis-associated C4.4A, a GPI-anchored protein cleaved by ADAM10 and ADAM17. Biol Chem. 2008;389:1075–1084. doi: 10.1515/BC.2008.121. [DOI] [PubMed] [Google Scholar]
- 15.Gavert N, Conacci-Sorrell M, Gast D, Schneider A, Altevogt P, Brabletz T, Ben-Ze’ev A. L1, a novel target of beta-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J Cell Biol. 2005;168:633–642. doi: 10.1083/jcb.200408051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohutek ZA, diPierro CG, Redpath GT, Hussaini IM. ADAM-10-mediated N-cadherin cleavage is protein kinase C-alpha dependent and promotes glioblastoma cell migration. J Neurosci. 2009;29:4605–4615. doi: 10.1523/JNEUROSCI.5126-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCulloch DR, Akl P, Samaratunga H, Herington AC, Odorico DM. Expression of the disintegrin metalloprotease, ADAM-10, in prostate cancer and its regulation by dihydrotestosterone, insulin-like growth factor I, and epidermal growth factor in the prostate cancer cell model LNCaP. Clin Cancer Res. 2004;10:314–323. doi: 10.1158/1078-0432.CCR-0846-3. [DOI] [PubMed] [Google Scholar]
- 18.Moss ML, Stoeck A, Yan W, Dempsey PJ. ADAM10 as a target for anti-cancer therapy. Curr Pharm Biotechnol. 2008;9:2–8. doi: 10.2174/138920108783497613. [DOI] [PubMed] [Google Scholar]
- 19.Wu E, Croucher PI, McKie N. Expression of members of the novel membrane linked metalloproteinase family ADAM in cells derived from a range of haematological malignancies. Biochem Biophys Res Commun. 1997;235:437–442. doi: 10.1006/bbrc.1997.6714. [DOI] [PubMed] [Google Scholar]
- 20.Endres K, Fahrenholz F. Upregulation of the alpha-secretase ADAM10–risk or reason for hope? FEBS J. 2010;277:1585–1596. doi: 10.1111/j.1742-4658.2010.07566.x. [DOI] [PubMed] [Google Scholar]
- 21.Endres K, Fahrenholz F. Regulation of alpha-secretase ADAM10 expression and activity. Exp Brain Res. 2012;217:343–352. doi: 10.1007/s00221-011-2885-7. [DOI] [PubMed] [Google Scholar]
- 22.Endres K, Fahrenholz F, Lotz J, Hiemke C, Teipel S, Lieb K, Tuscher O, Fellgiebel A. Increased CSF APPs-alpha levels in patients with Alzheimer disease treated with acitretin. Neurology. 2014;83:1930–1935. doi: 10.1212/WNL.0000000000001017. [DOI] [PubMed] [Google Scholar]
- 23.Tippmann F, Hundt J, Schneider A, Endres K, Fahrenholz F. Up-regulation of the alpha-secretase ADAM10 by retinoic acid receptors and acitretin. FASEB J. 2009;23:1643–1654. doi: 10.1096/fj.08-121392. [DOI] [PubMed] [Google Scholar]
- 24.Corrigan F, Vink R, Blumbergs PC, Masters CL, Cappai R, van den Heuvel C. sAPPalpha rescues deficits in amyloid precursor protein knockout mice following focal traumatic brain injury. J Neurochem. 2012;122:208–220. doi: 10.1111/j.1471-4159.2012.07761.x. [DOI] [PubMed] [Google Scholar]
- 25.Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29:3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milosch N, Tanriover G, Kundu A, Rami A, Francois JC, Baumkotter F, Weyer SW, Samanta A, Jaschke A, Brod F, et al. Holo-APP and G-protein-mediated signaling are required for sAPPalpha-induced activation of the Akt survival pathway. Cell Death Dis. 2014;5:e1391. doi: 10.1038/cddis.2014.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postina R, Schroeder A, Dewachter I, Bohl J, Schmitt U, Kojro E, Prinzen C, Endres K, Hiemke C, Blessing M, et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J Clin Invest. 2004;113:1456–1464. doi: 10.1172/JCI20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thornton E, Vink R, Blumbergs PC, Van Den Heuvel C. Soluble amyloid precursor protein alpha reduces neuronal injury and improves functional outcome following diffuse traumatic brain injury in rats. Brain Res. 2006;1094:38–46. doi: 10.1016/j.brainres.2006.03.107. [DOI] [PubMed] [Google Scholar]
- 29.Prinzen C, Muller U, Endres K, Fahrenholz F, Postina R. Genomic structure and functional characterization of the human ADAM10 promoter. FASEB J. 2005;19:1522–1524. doi: 10.1096/fj.04-3619fje. [DOI] [PubMed] [Google Scholar]
- 30.Endres K, Postina R, Schroeder A, Mueller U, Fahrenholz F. Shedding of the amyloid precursor protein-like protein APLP2 by disintegrin-metalloproteinases. FEBS J. 2005;272:5808–5820. doi: 10.1111/j.1742-4658.2005.04976.x. [DOI] [PubMed] [Google Scholar]
- 31.Corbett GT, Gonzalez FJ, Pahan K. Activation of peroxisome proliferator-activated receptor alpha stimulates ADAM10-mediated proteolysis of APP. Proc Natl Acad Sci USA. 2015;112:8445–8450. doi: 10.1073/pnas.1504890112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinhardt S, Schuck F, Grosgen S, Riemenschneider M, Hartmann T, Postina R, Grimm M, Endres K. Unfolded protein response signaling by transcription factor XBP-1 regulates ADAM10 and is affected in Alzheimer’s disease. FASEB J. 2014;28:978–997. doi: 10.1096/fj.13-234864. [DOI] [PubMed] [Google Scholar]
- 33.Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, Silver LM, Papaioannou VE. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131:5041–5052. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- 34.Jacobs JJ, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, van Lohuizen M. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 35.Kokubo H, Tomita-Miyagawa S, Hamada Y, Saga Y. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development. 2007;134:747–755. doi: 10.1242/dev.02777. [DOI] [PubMed] [Google Scholar]
- 36.Prince S, Carreira S, Vance KW, Abrahams A, Goding CR. Tbx2 directly represses the expression of the p21(WAF1) cyclin-dependent kinase inhibitor. Cancer Res. 2004;64:1669–1674. doi: 10.1158/0008-5472.CAN-03-3286. [DOI] [PubMed] [Google Scholar]
- 37.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 38.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- 39.Papaioannou VE, Silver LM. The T-box gene family. BioEssays. 1998;20:9–19. doi: 10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 40.Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 41.Gibson-Brown JJ, Agulnik SI, Silver LM, Niswander L, Papaioannou VE. Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development. 1998;125:2499–2509. doi: 10.1242/dev.125.13.2499. [DOI] [PubMed] [Google Scholar]
- 42.Takabatake Y, Takabatake T, Takeshima K. Conserved and divergent expression of T-box genes Tbx2-Tbx5 in Xenopus. Mech Dev. 2000;91:433–437. doi: 10.1016/S0925-4773(99)00329-9. [DOI] [PubMed] [Google Scholar]
- 43.Douglas NC, Papaioannou VE. The T-box transcription factors TBX2 and TBX3 in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2013;18:143–147. doi: 10.1007/s10911-013-9282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peres J, Davis E, Mowla S, Bennett DC, Li JA, Wansleben S, Prince S. The highly homologous T-box transcription factors, TBX2 and TBX3, have distinct roles in the oncogenic process. Genes Cancer. 2010;1:272–282. doi: 10.1177/1947601910365160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowley M, Grothey E, Couch FJ. The role of Tbx2 and Tbx3 in mammary development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2004;9:109–118. doi: 10.1023/B:JOMG.0000037156.64331.3f. [DOI] [PubMed] [Google Scholar]
- 46.Lingbeek ME, Jacobs JJ, van Lohuizen M. The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator. J Biol Chem. 2002;277:26120–26127. doi: 10.1074/jbc.M200403200. [DOI] [PubMed] [Google Scholar]
- 47.He M, Wen L, Campbell CE, Wu JY, Rao Y. Transcription repression by Xenopus ET and its human ortholog TBX3, a gene involved in ulnar-mammary syndrome. Proc Natl Acad Sci USA. 1999;96:10212–10217. doi: 10.1073/pnas.96.18.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demay F, Bilican B, Rodriguez M, Carreira S, Pontecorvi M, Ling Y, Goding CR. T-box factors: targeting to chromatin and interaction with the histone H3 N-terminal tail. Pigment Cell Res. 2007;20:279–287. doi: 10.1111/j.1600-0749.2007.00389.x. [DOI] [PubMed] [Google Scholar]
- 50.Fischer K, Pflugfelder GO. Putative breast cancer driver mutations in TBX3 cause impaired transcriptional repression. Front Oncol. 2015;5:244. doi: 10.3389/fonc.2015.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez M, Aladowicz E, Lanfrancone L, Goding CR. Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer Res. 2008;68:7872–7881. doi: 10.1158/0008-5472.CAN-08-0301. [DOI] [PubMed] [Google Scholar]
- 52.Macindoe I, Glockner L, Vukasin P, Stennard FA, Costa MW, Harvey RP, Mackay JP, Sunde M. Conformational stability and DNA binding specificity of the cardiac T-box transcription factor Tbx20. J Mol Biol. 2009;389:606–618. doi: 10.1016/j.jmb.2009.04.056. [DOI] [PubMed] [Google Scholar]
- 53.Sinha S, Abraham S, Gronostajski RM, Campbell CE. Differential DNA binding and transcription modulation by three T-box proteins, T, TBX1 and TBX2. Gene. 2000;258:15–29. doi: 10.1016/S0378-1119(00)00417-0. [DOI] [PubMed] [Google Scholar]
- 54.Vance KW, Shaw HM, Rodriguez M, Ott S, Goding CR. The retinoblastoma protein modulates Tbx2 functional specificity. Mol Biol Cell. 2010;21:2770–2779. doi: 10.1091/mbc.e09-12-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 2005;65:2260–2268. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- 56.Saito A, Yamashita T, Mariko Y, Nosaka Y, Tsuchiya K, Ando T, Suzuki T, Tsuruo T, Nakanishi O. A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc Natl Acad Sci USA. 1999;96:4592–4597. doi: 10.1073/pnas.96.8.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu E, Dul E, Sung CM, Chen Z, Kirkpatrick R, Zhang GF, Johanson K, Liu R, Lago A, Hofmann G, et al. Identification of novel isoform-selective inhibitors within class I histone deacetylases. J Pharmacol Exp Ther. 2003;307:720–728. doi: 10.1124/jpet.103.055541. [DOI] [PubMed] [Google Scholar]
- 58.Narath R, Ambros IM, Kowalska A, Bozsaky E, Boukamp P, Ambros PF. Induction of senescence in MYCN amplified neuroblastoma cell lines by hydroxyurea. Genes Chromosomes Cancer. 2007;46:130–142. doi: 10.1002/gcc.20393. [DOI] [PubMed] [Google Scholar]
- 59.Yu X, Li X, Jiang G, Wang X, Chang HC, Hsu WH, Li Q. Isradipine prevents rotenone-induced intracellular calcium rise that accelerates senescence in human neuroblastoma SH-SY5Y cells. Neuroscience. 2013;246:243–253. doi: 10.1016/j.neuroscience.2013.04.062. [DOI] [PubMed] [Google Scholar]
- 60.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang NC, Hu ML. The limitations and validities of senescence associated-beta-galactosidase activity as an aging marker for human foreskin fibroblast Hs68 cells. Exp Gerontol. 2005;40:813–819. doi: 10.1016/j.exger.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 62.Li J, Ballim D, Rodriguez M, Cui R, Goding CR, Teng H, Prince S. The anti-proliferative function of the TGF-beta1 signaling pathway involves the repression of the oncogenic TBX2 by its homologue TBX3. J Biol Chem. 2014;289:35633–35643. doi: 10.1074/jbc.M114.596411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grimm S, Pflugfelder GO. Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science. 1996;271:1601–1604. doi: 10.1126/science.271.5255.1601. [DOI] [PubMed] [Google Scholar]
- 64.Pflugfelder GO, Eichinger F, Shen J. T-box genes in drosophila limb development. Curr Top Dev Biol. 2017;122:313–354. doi: 10.1016/bs.ctdb.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 65.Poeck B, Hofbauer A, Pflugfelder GO. Expression of the Drosophila optomotor-blind gene transcript in neuronal and glial cells of the developing nervous system. Development. 1993;117:1017–1029. doi: 10.1242/dev.117.3.1017. [DOI] [PubMed] [Google Scholar]
- 66.Lieber T, Kidd S, Young MW. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kidd S, Lieber T. Furin cleavage is not a requirement for Drosophila Notch function. Mech Dev. 2002;115:41–51. doi: 10.1016/S0925-4773(02)00120-X. [DOI] [PubMed] [Google Scholar]
- 68.Kovalevich J, Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol Biol. 2013;1078:9–21. doi: 10.1007/978-1-62703-640-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boogerd KJ, Wong LY, Christoffels VM, Klarenbeek M, Ruijter JM, Moorman AF, Barnett P. Msx1 and Msx2 are functional interacting partners of T-box factors in the regulation of Connexin43. Cardiovasc Res. 2008;78:485–493. doi: 10.1093/cvr/cvn049. [DOI] [PubMed] [Google Scholar]
- 70.Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn. 2004;229:763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- 71.Weber S, Niessen MT, Prox J, Lullmann-Rauch R, Schmitz A, Schwanbeck R, Blobel CP, Jorissen E, de Strooper B, Niessen CM, Saftig P. The disintegrin/metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development. 2011;138:495–505. doi: 10.1242/dev.055210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang C, Tian L, Chi C, Wu X, Yang X, Han M, Xu T, Zhuang Y, Deng K. Adam10 is essential for early embryonic cardiovascular development. Dev Dyn. 2010;239:2594–2602. doi: 10.1002/dvdy.22391. [DOI] [PubMed] [Google Scholar]
- 73.Kispert A, Hermann BG. The Brachyury gene encodes a novel DNA binding protein. EMBO J. 1993;12:4898–4899. doi: 10.1002/j.1460-2075.1993.tb06179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sen A, Grimm S, Hofmeyer K, Pflugfelder GO. Optomotor-blind in the development of the Drosophila HS and VS lobula plate tangential cells. J Neurogenet. 2014;28:250–263. doi: 10.3109/01677063.2014.917645. [DOI] [PubMed] [Google Scholar]
- 75.Chen JR, Chatterjee B, Meyer R, Yu JC, Borke JL, Isales CM, Kirby ML, Lo CW, Bollag RJ. Tbx2 represses expression of Connexin43 in osteoblastic-like cells. Calcif Tissue Int. 2004;74:561–573. doi: 10.1007/s00223-003-0106-5. [DOI] [PubMed] [Google Scholar]
- 76.Abrahams A, Parker MI, Prince S. The T-box transcription factor Tbx2: its role in development and possible implication in cancer. IUBMB Life. 2010;62:92–102. doi: 10.1002/iub.275. [DOI] [PubMed] [Google Scholar]
- 77.Paxton C, Zhao H, Chin Y, Langner K, Reecy J. Murine Tbx2 contains domains that activate and repress gene transcription. Gene. 2002;283:117–124. doi: 10.1016/S0378-1119(01)00878-2. [DOI] [PubMed] [Google Scholar]
- 78.Zhu B, Zhang M, Byrum SD, Tackett AJ, Davie JK. TBX2 blocks myogenesis and promotes proliferation in rhabdomyosarcoma cells. Int J Cancer. 2014;135:785–797. doi: 10.1002/ijc.28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu B, Zhang M, Williams EM, Keller C, Mansoor A, Davie JK. TBX2 represses PTEN in rhabdomyosarcoma and skeletal muscle. Oncogene. 2016;35:4212–4224. doi: 10.1038/onc.2015.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaltenbrun E, Greco TM, Slagle CE, Kennedy LM, Li T, Cristea IM, Conlon FL. A Gro/TLE-NuRD corepressor complex facilitates Tbx20-dependent transcriptional repression. J Proteome Res. 2013;12:5395–5409. doi: 10.1021/pr400818c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lewandowski SL, Janardhan HP, Smee KM, Bachman M, Sun Z, Lazar MA, Trivedi CM. Histone deacetylase 3 modulates Tbx5 activity to regulate early cardiogenesis. Hum Mol Genet. 2014;23:3801–3809. doi: 10.1093/hmg/ddu093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cunliffe VT. Histone deacetylase 1 is required to repress Notch target gene expression during zebrafish neurogenesis and to maintain the production of motoneurones in response to hedgehog signalling. Development. 2004;131:2983–2995. doi: 10.1242/dev.01166. [DOI] [PubMed] [Google Scholar]
- 83.Lightman EG, Harrison MR, Cunliffe VT. Opposing actions of histone deacetylase 1 and Notch signalling restrict expression of erm and fgf20a to hindbrain rhombomere centres during zebrafish neurogenesis. Int J Dev Biol. 2011;55:597–602. doi: 10.1387/ijdb.113315el. [DOI] [PubMed] [Google Scholar]
- 84.Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 86.Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/S0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 87.Sapir A, Assa-Kunik E, Tsruya R, Schejter E, Shilo BZ. Unidirectional Notch signaling depends on continuous cleavage of Delta. Development. 2005;132:123–132. doi: 10.1242/dev.01546. [DOI] [PubMed] [Google Scholar]
- 88.Sotillos S, Roch F, Campuzano S. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development. 1997;124:4769–4779. doi: 10.1242/dev.124.23.4769. [DOI] [PubMed] [Google Scholar]
- 89.Currais A, Hortobagyi T, Soriano S. The neuronal cell cycle as a mechanism of pathogenesis in Alzheimer’s disease. Aging (Albany NY) 2009;1:363–371. doi: 10.18632/aging.100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McShea A, Lee HG, Petersen RB, Casadesus G, Vincent I, Linford NJ, Funk JO, Shapiro RA, Smith MA. Neuronal cell cycle re-entry mediates Alzheimer disease-type changes. Biochim Biophys Acta. 2007;1772:467–472. doi: 10.1016/j.bbadis.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 91.Counts SE, Mufson EJ. Regulator of cell cycle (RGCC) expression during the progression of Alzheimer’s disease. Cell Transplant. 2017;26:693–702. doi: 10.3727/096368916X694184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schneider MA, Scheffer KD, Bund T, Boukhallouk F, Lambert C, Cotarelo C, Pflugfelder GO, Florin L, Spoden GA. The transcription factors TBX2 and TBX3 interact with human papillomavirus 16 (HPV16) L2 and repress the long control region of HPVs. J Virol. 2013;87:4461–4474. doi: 10.1128/JVI.01803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thomas P, Smart TG. HEK293 cell line: a vehicle for the expression of recombinant proteins. J Pharmacol Toxicol Methods. 2005;51:187–200. doi: 10.1016/j.vascn.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 95.Moeenrezakhanlou A, Nandan D, Reiner NE. Identification of a calcitriol-regulated Sp-1 site in the promoter of human CD14 using a combined western blotting electrophoresis mobility shift assay (WEMSA) Biol Proced Online. 2008;10:29–35. doi: 10.1251/bpo140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Poeck B, Balles J, Pflugfelder GO. Transcript identification in the optomotor-blind locus of Drosophila melanogaster by intragenic recombination mapping and PCR-aided sequence analysis of lethal point mutations. Mol Gen Genet. 1993;238:325–332. doi: 10.1007/BF00291990. [DOI] [PubMed] [Google Scholar]
- 97.Pflugfelder GO, Roth H, Poeck B, Kerscher S, Schwarz H, Jonschker B, Heisenberg M. The lethal(1)optomotor-blind gene of Drosophila melanogaster is a major organizer of optic lobe development: isolation and characterization of the gene. Proc Natl Acad Sci USA. 1992;89:1199–1203. doi: 10.1073/pnas.89.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sen A, Gadomski C, Balles J, Abassi Y, Dorner C, Pflugfelder GO. Null mutations in Drosophila Optomotor-blind affect T-domain residues conserved in all Tbx proteins. Mol Genet Genom. 2010;283:147–156. doi: 10.1007/s00438-009-0505-z. [DOI] [PubMed] [Google Scholar]
- 99.Grimm MO, Grosgen S, Rothhaar TL, Burg VK, Hundsdorfer B, Haupenthal VJ, Friess P, Muller U, Fassbender K, Riemenschneider M, et al. Intracellular APP domain regulates serine-palmitoyl-CoA transferase expression and is affected in alzheimer’s disease. Int J Alzheimers Dis. 2011;2011:695413. doi: 10.4061/2011/695413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 101.Steiner H, Kostka M, Romig H, Basset G, Pesold B, Hardy J, Capell A, Meyn L, Grim ML, Baumeister R, et al. Glycine 384 is required for presenilin-1 function and is conserved in bacterial polytopic aspartyl proteases. Nat Cell Biol. 2000;2:848–851. doi: 10.1038/35041097. [DOI] [PubMed] [Google Scholar]
- 102.Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MA, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-L. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.