Abstract
The P3H1/CRTAP/PPIB complex is essential for prolyl 3-hydroxylation and folding of procollagens in the endoplasmic reticulum (ER). Deficiency in any component of this ternary complex is associated with the misfolding of collagen and the onset of osteogenesis imperfecta. However, little structure information is available about how this ternary complex is assembled and retained in the ER. Here, we assessed the role of the KDEL sequence of P3H1 and probed the spatial interactions of PPIB in the complex. We show that the KDEL sequence is essential for retaining the P3H1 complex in the ER. Its removal resulted in co-secretion of P3H1 and CRTAP out of the cell, which was mediated by the binding of P3H1 N-terminal domain with CRTAP. The secreted P3H1/CRTAP can readily bind PPIB with their C-termini close to PPIB in the ternary complex. Cysteine modification, crosslinking, and mass spectrometry experiments identified PPIB surface residues involved in the complex formation, and showed that the surface of PPIB is extensively covered by the binding of P3H1 and CRTAP. Most importantly, we demonstrated that one disease-associated pathological PPIB mutation on the binding interface did not affect the PPIB prolyl-isomerase activity, but disrupted the formation of P3H1/CRTAP/PPIB ternary complex. This suggests that defects in the integrity of the P3H1 ternary complex are associated with pathological collagen misfolding. Taken together, these results provide novel structural information on how PPIB interacts with other components of the P3H1 complex and indicate that the integrity of P3H1 complex is required for proper collagen formation.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03102-8) contains supplementary material, which is available to authorized users.
Keywords: Osteogenesis imperfecta, Sulfo-GMBS, Hyperelastosis cutis, Hydroxylase, Chaperone
Introduction
Type I collagen is the most abundant extracellular matrix protein in humans, and has important roles in building the structural framework of tissues such as bone, skin, tendon, and cartilage. It is characterized by a unique repeating Gly-X-Y motif that is required for the formation of its triple-helical tertiary structure [1, 2]. Its precursor procollagen is synthesized and folded into a heterotrimer with two α1 and one α2 chains through multiple steps involving many different proteins in the rough endoplasmic reticulum (rER) [3–5].
Briefly, after translocation of a growing polypeptide chain of procollagen into the rER, proline residues become 4-hydroxylated by prolyl 4-hydroxylase. 4-Hydroxylation of proline residues increases the stability of the triple helix and is a key element in its folding [6–8]. The chain selection and association for triple helix formation are determined by carboxyl-terminal propeptides in fibrillar collagens [9, 10]. Premature association between procollagen chains is thought to be prevented by chaperones such as protein-disulfide isomerase, BiP/GRP78, GRP94, HSP47, and FKBP65, and by collagen-modifying enzymes until the biosynthesis of the individual chain is complete [11]. Additional modifications include the 3-hydroxylation of proline residues by the P3H1/CRTAP/PPIB complex [12], the hydroxylation of lysine residues by lysyl hydroxylases [13], and glycosylation. The chains are then selected, and trimers are formed by association of the carboxyl-terminal propeptides with disulfide bonds between the chains. Triple helix formation proceeds from the carboxyl-terminal end toward the amino-terminal end in a zipper-like fashion [10]. The folded procollagen is subsequently shipped to the Golgi apparatus for packing and secretion. The rate-limiting step in this process is believed to be the cis–trans isomerization of peptide bonds catalyzed by peptidyl–prolyl cis–trans isomerase PPIB [14]. Overall, these modification enzymes, cofactors, and chaperones coordinately function together as “folding machines” to allow proper procollagen formation.
Mutations in the COL1A1 or COL1A2 genes that encode the two type I procollagen alpha chains often result in defects in procollagen formation and the subsequent onset of osteogenesis imperfecta (OI) [15–17]. Individuals with OI are characterized by fragile bones with high susceptibility to fracture. However, over the last decade, deficiencies in the molecular chaperones or modification enzymes of procollagen synthesis such as FKBP10 [18], HSP47 [19, 20], and the collagen prolyl 3-hydroxylation complex P3H1/CRTAP/PPIB have also been associated with OI in humans [21]. Many knock-out mouse models have also confirmed the importance of these proteins for proper procollagen formation [22–24].
The P3H1/CRTAP/PPIB ternary complex was first purified from chicken embryos by gelatin affinity columns [12]. Laser light scattering and velocity sedimentation analysis have shown that the three proteins form a 1:1:1 complex. P3H1, which is known to hydroxylate a single residue (Pro986) in type I collagen chains, consists of two major domains: a carboxyl-terminal dioxygenase domain that contains the enzymatic activity of the complex and a unique amino-terminal domain containing four-cysteine (CXXXC) repeats. CRTAP is believed to be the helper protein of the complex and shares homology with P3H1, but it does not contain the common dioxygenase domain [25, 26]. It has been suggested that CRTAP and the N-terminal domain of P3H1 may function as a disulfide isomerase in the ER [27]. Null mutations in either P3H1 or CRTAP gene result in a substantial decrease of both proteins but no reduction in the amount of PPIB. Therefore, it has been suggested that P3H1 and CRTAP are mutually stabilized within the ER, with the absence of either protein resulting in the absence of both proteins [28]. PPIB is the third component of the complex and is an abundant ER-resident peptidyl–prolyl cis–trans isomerase that catalyzes the rate-limiting step in procollagen folding [14]. Interestingly, amongst all three components of the ternary complex, only P3H1 has a KDEL ER-retrieval sequence at its C-terminus. It has been shown that truncation of the C-terminal 18 amino acids of P3H1, including this KDEL sequence, results in the loss of the P3H1 complex in the ER and is associated with non-lethal OI [29].
As it is not well understood how this ternary complex is retained within the ER, and little structural information is available about how PPIB binds P3H1 and CRTAP, we investigated the role of the P3H1 KDEL sequence by generating various P3H1 truncations and probed the interactions of PPIB within the ternary complex through cysteine modification and crosslinking experiments. Our results allow us to build an initial model showing the spatial interrelationship of the three components of P3H1 complex. More importantly, we assessed the effect of pathological PPIB mutations on the formation of the P3H1 ternary complex and showed the integrity of P3H1 ternary complex which is required for collagen formation.
Materials and methods
Materials
The full-length cDNA of human P3H1 and CRTAP was amplified from the total RNA of human control fibroblasts as described previously [30], and the full-length cDNA of PPIB was purchased from Addgene. The primary antibodies used in this study were rabbit anti-GAPDH (Abcam Catalog: ab9485), rabbit anti-Lepre1 (Abcam Catalog: ab154799), rabbit anti-CRTAP (Abcam Catalog: ab183720), rabbit anti-PPIB (Proteintech Catalog: 11607-1-AP), and anti-6 × His-tag (Proteintech Catalog: HRP-66005). The chemical crosslinker Sulfo-GMBS was purchased from Thermo Fisher, and mPEG-2000 for cysteine modifications was purchased from Iris Biotech GmbH. All columns for protein purification were purchased from GE Healthcare.
Cloning, mutagenesis, expression, and purification of recombinant proteins
The cDNAs of human P3H1, human CRTAP, and human PPIB were sub-cloned into the mammalian expression vector pCEP4 (Invitrogen, Karlsruhe, Germany). All variants (Fig. 1) were constructed by PCR mutagenesis using the KOD Plus mutagenesis kit (TYOBO). Notably, a P3H1 variant with its KDEL sequence at C-terminus truncated was named as PD4. His-tags were added at the C-terminal tails of various P3H1 or CRTAP or PPIB variants (Fig. 1). HEK 293E cells were cultured in DMEM supplemented with 10% FBS at 37 °C and 5% CO2. The day before transfection, 20 mL of HEK 293E cells at 6 × 105 cells/mL were plated in a 150 mm dish. Aliquots (15 μg) of expression construct DNA were diluted in 1 mL of serum-free medium, as were 45 μg aliquots of Polyethylenimine (PEI) with molecular weight ~ 40,000 Da (Polyscience). Diluted DNA was combined with the diluted PEI and incubated at room temperature (RT) for 30 min. The DNA–PEI complexes were subsequently added to each dish. After 4 h of incubation, the transfection medium was replaced by the Freestyle-293 medium (Thermo Fisher), and the culture medium was harvested after 72 h. PD4/CRTAP, PD4-His/CRTAP, and PD4/CRTAP-His complex in the culture medium was partially purified by a 5-mL Q FF column and eluted with buffer congaing 0.5 M NaCl and 10 mM Tris–HCl, pH 7.4. Samples were dialyzed against PBS (Phosphate-Buffered Saline) overnight and then concentrated before use. The purity of the partially purified PD4/CRTAP complex was assessed to be approximately 80% pure by SDS-PAGE and the concentrations were estimated by the Nanodrop 2000C spectrophotometer using a calculated coefficient according to the amino acid sequences of PD4 and CRTAP.
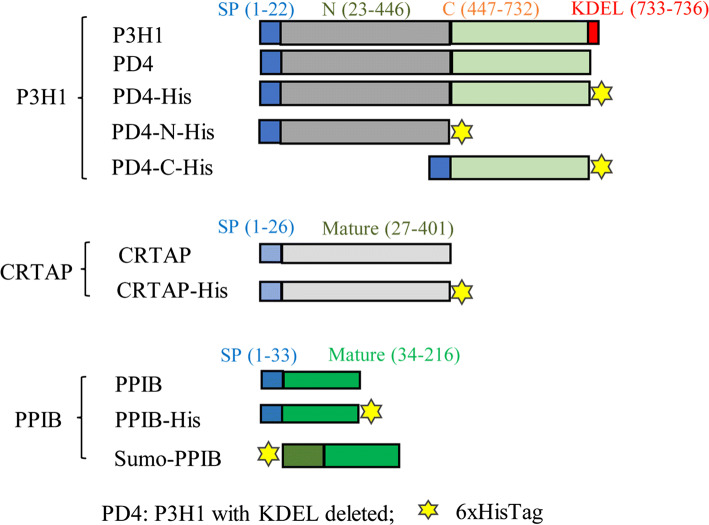
Fig. 1.
Schematic representation of P3H1, CRTAP, and PPIB variants. Various constructs were prepared to explore the interactions among P3H1/CRTAP/PPIB complex members. Blue squares represent the signal peptides of each construct, red squares represent the KDEL sequence of P3H1, and the yellow stars represent the His6 sequence. P3H1 with the KDEL sequence removed from the tail was named PD4. The N-terminal segment of P3H1 (1–446) was named PD4-N-His, and the C-terminal dioxygenase segment (447–732) was named PD4-C-His. Sumo-PPIB variant was expressed in E. coli, and all other P3H1, CRTAP, and PPIB variants were prepared from HEK 293E cells
The PPIB variants were sub-cloned into the E. coli expression vector pE-Sumo3, where the PPIB was expressed as a Sumo-fusion protein with a His-tag at the N-termini of Sumo. The expression constructs were transformed into BL21 (DE3) cells and grown in 2 × TY at 37 °C until the optical density at 600 nm reached 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.5 mM, and the culture was further incubated in a shaker at 25 °C for another 12 h. The cells were collected by centrifugation, re-suspended in ice-cold buffer A (20 mM Tris–HCl, pH 7.4, 0.5 M NaCl, and 20 mM imidazole), and disrupted by a high-pressure cell breaker. The supernatant of the cell lysate was loaded onto a 5-mL HiTrap FF column and eluted by a 0.02–0.2 M imidazole gradient. The peak fractions were then collected. To remove the Sumo-tag, the fusion proteins were digested by His-tagged protease SENP2 and dialyzed against buffer A overnight. Then, the proteins were reloaded onto a 5-mL HisTrap FF column to remove the Sumo-tag and protease, and the recombinant PPIB was concentrated and stored at − 80 °C before use. PPIB variants where the surface-exposed PPIB residues K35, T49, D67, Q71, T81, S85, K113, Q119, D131, H134, E145, T155, and E184 based on PDB 1CYN were mutated to cysteines individually were prepared similarly. All PPIB variants used for the protein crosslinking and modification experiments were based on the PPIB (C170S) backbone.
Western analysis
For western blot analysis, cells were cultured in 6-well plates and transfected with various combinations of P3H1, PD4, CRTAP, and PPIB expression plasmids (Fig. 2) using lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. As the expression vector pCEP4 is 10.4 kb long, all the expression constructs are of similar sizes (pCEP4-P3H1, 12.5 kb; pECP4-CRTAP, 11.6 kb, pCEP4-PPIB, 11 kb). The day before transfection, 2 mL of HEK 293E cells at 5 × 105 cells/mL were plated in each well of the 6-well plate. Aliquots of 2.4 μg total expression construct DNA (e.g., Fig. 2 lane 1: 0.8 μg pCEP4-P3H1 DNA and 1.6 μg pCEP4 vector; lane 5: 0.8 μg pCEP4-P3H1, 0.8 μg pCEP4-CRTAP, and 0.8 μg pCEP4 vector; lane 10: 0.8 μg pCEP4-P3H1, 0.8 μg pCEP4-CRTAP, and 0.8 μg pCEP4-PPIB) were diluted in 100 μL of serum-free medium, as were 10 μL lipofectamine 2000 reagent (Invitrogen). The diluted DNA was then combined with the diluted lipofectamine 2000 and incubated at room temperature for 15 min. The DNA–lipofectamine 2000 complexes were subsequently added to each well. Freestyle-293 medium was applied after transfection and the culture medium were collected after 36 h. We measured the transfection efficiency in our study using pCEP4-GFP as a control plasmid. The efficiency is about 70% when assessed by GFP fluorescence measurement (Supplemental Fig. 1a). Cells were collected with cold PBS and lysed in radio-immunoprecipitation assay (RIPA) buffer consisting of 50 mM Tris–HCl, pH 7.4, 0.5% Nonidet P-40, 0.25% Nadeoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, and protease inhibitor cocktail (Sigma). The samples of culture medium and cell lysate were prepared in reducing SDS-loading buffer and the loadings were normalized according to cell numbers and corresponding culture medium. It was estimated that proteins from about 10,000 HEK 293E cells or corresponding culture medium were loaded on each lane of SDS gel for western blot analysis. The primary antibodies used for the western blot analysis were rabbit anti-GAPDH (1:5000), rabbit anti-P3H1 (1:5000), rabbit anti-CRTAP (1:5000), rabbit anti-PPIB (1:2000), and anti-6 × His-tag (1:10,000) antibody. The bands from western blot were quantified by densitometry with the Image J System.
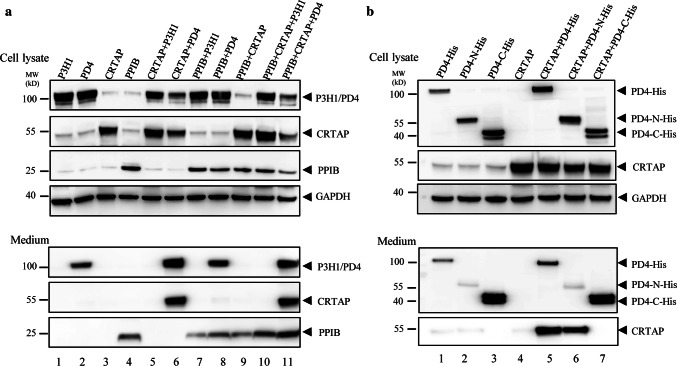
Fig. 2.
The role of KDEL in co-translocation of P3H1/CRTAP. a Transfection of various combinations among P3H1, PD4, CRTAP, and PPIB into cells, followed by the analysis of the media and cell lysates by western blot. Wild-type P3H1 and CRTAP were almost all retained in the cells in HEK 293E cell lines (lanes 1, 3, 5, and 10). Substantial amount of the PD4 was secreted into the medium (lane 2), while large amount of CRTAP was secreted along with PD4 (lanes 6 and 11). This shows the importance of the KDEL sequence in the retention of both P3H1 and CRTAP in ER. GAPDH in the cells was blotted as a loading control. b The full-length of PD4-His, the N-terminal segment of PD4 (PD4-N-His), and the C-terminal dioxygenase domain of PD4 (PD4-C-His) were transfected alone or with CRTAP. The samples were analyzed by SDS-PAGE and western blot. Both domains of P3H1 could be secreted into the culture medium, and CRTAP was mostly retained in the cells; however, when co-expressed with PD4-His (lane 5) or PD4-N-His (lane 6), substantial amount of CRTAP was secreted into the medium. Little CRTAP was detected in the medium when it was co-expressed with PD4-C-His (lane 7). All experiments were performed ≥ 3 times independently
Gel filtration analysis
For assessing the complex formation, a mixture containing 0.5 μM PD4/CRTAP or PD4-His/CRTAP or PD4/CRTAP-His with 1.5 μM Sumo-PPIB in 500 μL PBS was loaded onto a Superpose 6 gel filtration column (25 mL bed volume), and chromatographed in 10 mM Tris pH 7.4 and 100 mM NaCl at a flow rate of 0.5 mL/min. Fractions were collected and analyzed by 10% SDS-PAGE and western blot. The gel filtration column was calibrated using a mixture of thyroglobulin (669 kDa), ferritin (440 kDa), BSA (67 kDa), and ribonuclease A (13.7 kDa) according to the manufacturer’s instructions.
Modification of the free thiol group by mPEG
For the thiol group modification experiment, mPEG (Meo-PEG-mal-2000, Iris Biotech GmbH) stock solutions were prepared at 1 mM in water. The single-cysteine PPIB variants based on the PPIB C170S backbone were first reduced with 1 mM dithiothreitol (DTT) and buffer exchanged by a desalting column (1.5 mL) to remove excess DTT. The variants at 200 nM were then incubated alone or with 1 μM PD4/CRTAP complex on ice for 30 min in 100 μL PBS. The mixture was subsequently treated with 50 μM mPEG at room temperature for 30 min. The reaction was quenched by adding 2 mM DTT, and the samples were analyzed by SDS-PAGE and Western blot.
Chemical crosslinking
The surface-exposed free thiols of the PD4/CRTAP complex were first blocked by incubation with 10 mM iodoacetamide in PBS, and the samples were buffer exchanged by a desalting column (1.5 mL) to remove excess iodoacetamide. For the crosslinking experiment, 2 μM PD4/CRTAP complex was first incubated with 250 μM N-maleimidobutylody sulfosuccinimide ester (Sulfo-GMBS, Thermo Fisher, 7.3 Å spacer arm) in 1 mL PBS for 30 min at RT, and then, excess reagent was removed using a desalting column. Subsequently, 5 μM of PPIB variants with thiol group activated by DTT as above were mixed with 1 μM Sulfo-GMBS-modified PD4/CRTAP in 100 μL PBS and incubated for 2 min at RT. The reaction was stopped by the addition of 5 μL of 1 M DTT. The samples were analyzed by reducing SDS-PAGE and Western blot where 2.5% BSA was used to block the nitrocellulose membrane.
LC–MS/MS analysis
To identify the crosslinked peptides derived from the P3H1 complex, the crosslinked samples were digested as previously described [31]. The peptides were lyophilized using a SpeedVac (ThermoSavant) and re-suspended in 10 μL 0.3% formic acid/5% acetonitrile. All mass spectrometric experiments were performed on an Orbitrap LUMOS mass spectrometer connected to an Easy-nLC 1200 via an Easy Spray (Thermo Fisher Scientific). The peptide mixtures were loaded onto a 15 cm by 0.075 mm inner diameter column packed with C18 2-μm Reversed Phase resins (PepMap RSLC) and eluted within 60 min using a linear gradient from 95% solvent A (0.1% formic acid/2% acetonitrile/98% water) to 35% solvent B (0.1% formic acid/100% acetonitrile) at a flow rate of 300 nL/min. The spray voltage was set to 2 kV and the temperature of the ion transfer capillary was 275 °C. One full MS scan from 350 to 1500 m/z was acquired at high resolution = 120,000 (defined at m/z = 400), and then, the 20 most abundant multiply charged ions by filter dynamic exclusion were followed by EthCD fragmentation at resolution = 15,000 (defined at m/z = 400).
A database with lysine containing peptides derived from P3H1 and CRTAP linked with D67C of PPIB peptides by Sulfo-GMBS was generated and the MS/MS ion spectra were analyzed using the database by pLink 2.3.1 [32] to identify the crosslinked peptides. The pLink search parameters were as follows: precursor mass tolerance 10 p.p.m., fragment mass tolerance 20 p.p.m.; crosslinker composition: C(8)H(7)N(1)O(3), and monocomposition: C(8)H(9)N(1)O(3) (crosslinking alpha sites C and beta sites K, linker mass 165.043, monolink mass 167.058), fixed modification carbamidomethyl (C), variable modification oxidation (M), deamidated (Q), and deamidated (R), peptide length minimum 6 amino acids and maximum 60 amino acids per chain, peptide mass minimum 600 and maximum 6000 Da per chain, and enzyme trypsin three missed cleavage sites per chain. The result filter tolerance was set to 10 p.p.m., and separate FDR < 1% at the spectral level.
Prolyl-isomerase activity assay
The prolyl-isomerase activities were measured by a chymotrypsin-coupled PPIase assay [33, 34], which is rate-limited by the cis–trans isomerization of the Ala-Pro peptide bond of the synthetic Suc-AAPF-pNA substrate. The assay mixture contained 50 mM HEPES buffer pH 8.0, 100 mM NaCl, 50 μg a-chymotrypsin (Sigma-Aldrich), 25 μM Suc-AAPF-pNA (5 mM stock dissolved in trifluoroethanol supplemented with 0.45 M LiCl), and appropriate amounts of enzyme. The assay buffer was mixed with chymotrypsin and subsequently with cyclophilin. The reaction was initiated inside a cuvette with the addition of the peptide. The increase in absorbance at 390 nm was monitored at 8 °C using a spectrophotometer. Under these conditions, the cis–trans isomerization of the prolyl bond was a single exponential process, and its rate constant was determined using GraFit software.
Pull-down assay
To assess the binding between PPIB variants and PD4/CRTAP, 500 μL of 40 nM SUMO-PPIB variants in pull-down buffer (50 mM sodium-phosphate, pH 7.4, 300 mM NaCl, 0.01% Tween-20) were loaded onto to 2 μL Ni magnetic beads and the beads were washed three times with 300 μL pull-down buffer, to remove excess Sumo-PPIB. Then, 50 nM of PD4/CRTAP in 500 μL pull-down buffer was incubated with the beads for 30 min RT. The beads were then washed three times as above and the bound proteins were eluted with 50 μL pull-down buffer containing 300 mM imidazole. The samples were analyzed by SDS-PAGE and western blot using corresponding antibodies.
Results
The KDEL sequence of P3H1 is essential for intracellular localization of P3H1 and CRTAP
P3H1, originally isolated as the matrix proteoglycan leprecan, could be purified from chicken embryos as a P3H1/CRTAP/PPIB ternary complex [12]. Here, we confirmed that endogenous P3H1, CRTAP, and PPIB protein in human dermal fibroblasts-adult (HDF-a, Sciencell, USA) cell lysates formed complexes as all three proteins could be immunoprecipitated with anti-CRTAP antibody (Supplemental Fig. 1b), which is consistent with the previous observations [22]. P3H1 is the only component in the complex with a KDEL sequence. To examine the role of this KDEL sequence on the distribution of the complex, we generated a P3H1 mutant without the KDEL sequence, named PD4 (Fig. 1). Various combinations of P3H1, PD4, CRTAP, and PPIB were then co-transfected into HEK 293E cells. Proteins secreted in the medium or retained in the cells were analyzed by SDS-PAGE and western blot respectively (Fig. 2). To more accurately quantify the ratio of protein present in the medium and the cells, the cell lysate and corresponding amount of culture medium were also analyzed on a same gel by western blot (Supplemental Fig. 2a). The results showed that the wild-type P3H1 was retained within the cell as expected, while about 20% of total PD4 was detected in the culture medium (Fig. 2a, lanes 1, 2; Supplemental Fig. 2a). When co-expressed with CRTAP, about 60% of total PD4 was detected in the medium (lane 6). Even more (about 70%) PD4 was detected in the medium when it was co-expressed with CRTAP and PPIB. This suggests that complex formation stabilizes P3H1 during secretion, which is in line with the previous findings that P3H1 and CRTAP are mutually stabilized in the ER [28].
CRTAP has a signal peptide sequence and has no KDEL sequence, but it is predominantly retained in the cells when expressed alone (Fig. 2a, lane 3) and only trace amount of CRTAP could be detected in the medium (Supplemental Fig. 2c). It is also mostly retained in cells when co-expressed with P3H1 or PPIB (Fig. 2a, lane 5 and 9). However, when co-expressed with PD4 for 36 h, about 60% CRTAP was present in the medium (Fig. 2a, lane 6; Supplemental Fig. 2a). This indicates that PD4 binds to CRTAP and brings CRTAP out of the cell. As the expression levels of endogenous P3H1 complex proteins in HEK 293E are extremely low when compared with the overexpressed proteins (Supplemental Fig. 1c), and also the ration of the overexpressed P3H1 complex is similar to that of endogenous proteins (Supplemental Fig. 1d). Therefore, it is unlikely that the endogenous forms would play a significant role in the distribution of overexpressed P3H1 complex proteins.
It has been shown that PPIB is an abundant protein in the ER and secretory pathways, and is released in biological fluids [35]. Here, we show that overexpressed PPIB was also present in both cells and the medium (Fig. 2a, lane 4), and its distribution was only slightly affected by the co-expression of P3H1 or CRTAP. However, significantly more PPIB was present in the medium when it was co-expressed with PD4 and CRTAP (Fig. 2a, lane 11, Supplemental Fig. 2a).
Overall, these data indicate that the KDEL sequence keeps P3H1 within the cell and CRTAP is inherently an ER retention protein, even though it does not have a KDEL sequence. The removal of P3H KDEL sequence resulted in co-secretion of overexpressed P3H1 and CRTAP into the medium, which suggests a direct physical interaction between P3H1 and CRTAP, independent of PPIB.
P3H1 binds CRTAP through its N-terminal domain
It has been suggested that P3H1 and CRTAP are mutually stabilized in the ER [28], but little is known about how they bind to each other. Since PD4, when co-expressed with CRTAP, could bring CRTAP out of the cell and into the culture medium, we assessed which part of PD4 was responsible for the co-translocation of PD4 and CRTAP. PD4 was split into two parts: the N-terminal domain (residue 1–446) with an added His-tag, termed PD4-N-His, which shares a high similarity with CRTAP, and the C-terminal dioxygenase domain (447–732) with an added His-tag termed PD4-C-His (Fig. 1). The full-length PD4 and its truncations were then expressed alone or co-expressed with CRTAP in cells. The culture medium and cells were harvested after 36 h and analyzed by western blot as above. The results showed that PD4-His and PD4-N-His were secreted into the medium and most PD4-C-His was secreted (Fig. 2b, lanes 1–3). The doublet bands of PD4-C-His are due to heterogeneity in N-glycosylation of residues N470 and N546 of P3H1, and replacement of these residues with glutamines resulted a single band of this domain (Supplemental Fig. 3). CRTAP was predominantly retained in cells when expressed alone or co-expressed with PD4-C-His (Fig. 2b, lanes 4, 7); however, large amounts of CRTAP were detected in the culture medium when it was co-transfected with PD4 or PD4-N-His (Fig. 2b, lanes 5, 6). This indicates that the secretion of CRTAP observed here is mediated by the N-terminal domain of P3H1. Therefore, P3H1 most likely binds CRTAP through its N-terminal domain.
A His-tag added at the carboxyl terminus of P3H1 or CRTAP impedes the binding of PPIB
As PD4 and CRTAP could be secreted into the medium as a complex, this provides an attractive method for preparing recombinant P3H1 complex for further study. We first tested if the secreted PD4 and CRTAP can form a ternary complex with recombinant PPIB in vitro. PD4/CRTAP complex was purified by ionic exchange column from the expression medium of HEK 293E cells transfected with PD4 and CRTAP, and then mixed with recombinant PPIB fusion protein Sumo-PPIB (with a His-tag at the N-terminus of Sumo), which was purified from an E. coli expression system. The mixture was loaded onto a gel filtration chromatography column, where higher molecular weight species were eluted off from the column earlier. As a result, PD4 and CRTAP were co-eluted with Sumo-PPIB in a peak corresponding to the molecular weight of the ternary complex (Fig. 3a). The later peak corresponded to that of free Sumo-PPIB when sumo-PPIB was analyzed on the same column (Supplemental Fig. 4b). Therefore, PD4 and CRTAP secreted in the medium by HEK 293E cells can readily form a ternary complex with PPIB and the P3H1 complex could be co-purified by this approach.
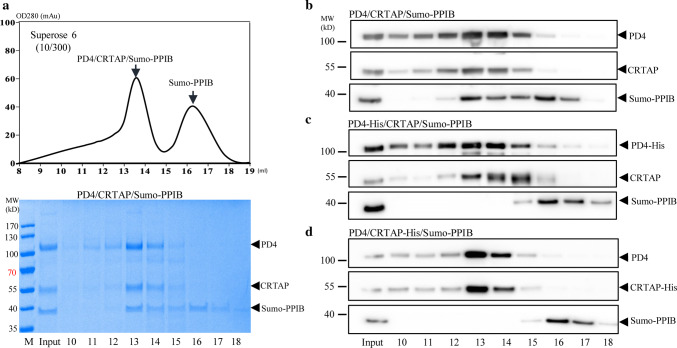
Fig. 3.
The role of the C-termini of P3H1 and CRTAP in forming a complex with PPIB. a Partially purified PD4/CRTAP was mixed with Sumo-PPIB and then analyzed by a Superose 6 gel filtration column with eluted fractions analyzed by coomassie-stained SDS-PAGE. This shows that PPIB can readily form ternary complex with the secreted PD4/CRTAP and was eluted in the early peak with PD4/CRTAP. b–d The partially purified PD4/CRTAP or PD4-His/CRTAP or PD4/CRTAP-His were mixed with Sumo-PPIB and then directly analyzed by gel filtration. The elution positions of PD4, CRTAP, and Sumo-PPIB were analyzed by SDS-PAGE and western blot. Complex formation in fraction 13 and 14 was observed from PD4/CRTAP/Sumo-PPIB in b, as seen in a, but not from the mixture of Sumo-PPIB with PD4-His/CRTAP (c) or PD4/CRTAP-His (d). All experiments were performed ≥ 3 times independently
To fine-tune the purification procedure, we also tested whether other constructs with His-tags directly added at the C-termini of P3H1 or CRTAP could yield the ternary complex in a similar way. To our surprise, neither of these constructs could be purified by the same procedure. To confirm this, the partially purified co-expressing PD4/CRTAP, PD4-His/CRTAP, or PD4/CRTAP-His (Supplemental Fig. 4a) were mixed with Sumo-PPIB, respectively, and then analyzed directly by gel filtration with the elution positions of P3H1, CRTAP, and PPIB assessed by western blot. As shown in Fig. 3b, about 50% of Sumo-PPIB was eluted in the early fractions corresponding to the ternary complex when mixed with PD4 and CRTAP. This is consistent with the above gel (Fig. 3a). However, no such early peak of Sumo-PPIB was observed with PD4-His/CRTAP (Fig. 3c) or PD4/CRTAP-His (Fig. 3d). This suggests that the His-tags added at the C-termini of P3H1 or CRTAP prevent the ternary complex formation. Therefore, it is likely that the C-termini of P3H1 and CRTAP are in close proximity to PPIB in the ternary complex, and the extension of these C-termini thus blocks PPIB binding. As PD4-His (where the KDEL sequence was replaced by six histidines) is only two residues longer than wild-type P3H1, this further emphasizes the closeness of the KDEL sequence with PPIB in the ternary complex. Nevertheless, the binding of PD4 on CRTAP/PPIB does not exclude the possibility that the KDEL sequence could form certain stabilizing interactions with CRTAP and/or PPIB in the normal P3H1 complex.
Mapping the surface area of PPIB that interacts with P3H1/CRTAP
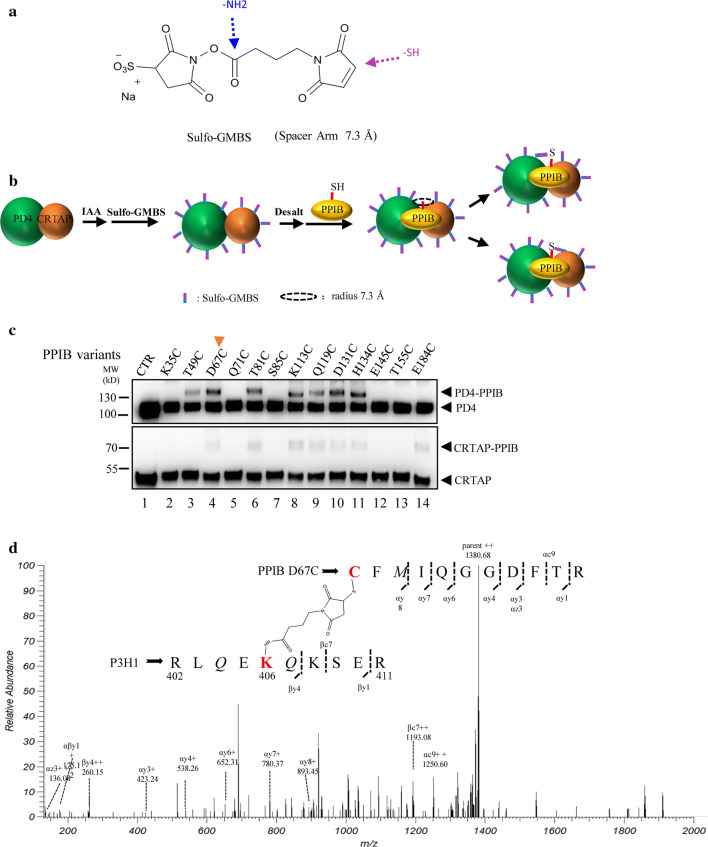
PPIB is a peptidyl–prolyl cis–trans isomerase involved in the folding of the procollagen triple helix, and its crystal structure is known. However, little structural information exists about P3H1 and CRTAP. As PPIB readily formed a ternary complex with PD4 and CRTAP in vitro, we assessed which part of the PPIB surface is involved in binding to P3H1 or CRTAP. First, based on the crystal structure of PPIB complexed with a peptide inhibitor (PDB 1CYN) [36], we randomly selected ten surface-exposed hydrophilic residues (Asp, Glu, Lys, Ser, and Thr) across the whole PPIB surface and three residues (Q71, Q119, and H134) nearby the substrate-binding pocket of PPIB, and mutated them individually to cysteines. We expected that replacement of these solvent accessible residues would not disturb the overall fold of the protein. These PPIB mutants were purified and then assessed for the accessibility of these cysteines for specific modification by mPEG in the presence or absence of PD4/CRTAP. Any cysteine exposed on the protein surface should be readily modified by mPEG, with each modification resulting in an increase in the molecular weight of 2000 Da. Thirteen single-cysteine PPIB variants were tested and analyzed by western blot (Fig. 4a). The results showed that the cysteines in different positions of PPIB, except T155C, were exposed on the surface and could be modified by mPEG as expected. In the presence of PD4 and CRTAP, cysteines of eight PPIB variants, including K35C, D67C, T81C, K113C, D131C, E145C, and E184C, were not modified by mPEG, indicating that these residues are covered by the binding of PD4 and CRTAP (Fig. 4b). Only about 50% of T49C could be modified by mPEG. This indicates that T49 is only partially exposed in the ternary complex. Other residues such as S85C, Q71C, Q119C, and H134C seemed unaffected by the binding of PD4 and CRTAP. As Q71, Q119, and H134 are located in the substrate-binding pocket of PPIB, this indicates that PPIB in the ternary complex retains its ability to bind its substrate, consistent with the prolyl-isomerase function of P3H1 complex.
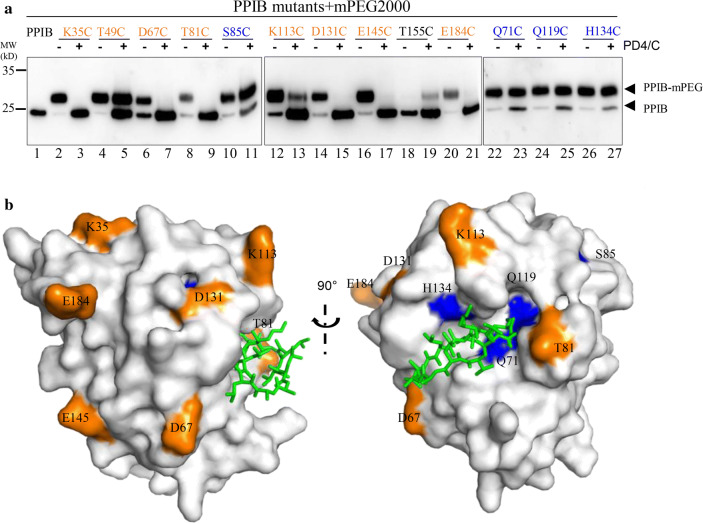
Fig. 4.
Probing the surface area of PPIB that interacts with the PD4/CRTAP complex. a PPIB variants with residues Lys35, Thr49, Asp67, Gln71, Thr81, Ser85, Lys113, Gln119, Asp131, His134, Glu145, Thr155, and Glu184 mutated to cysteine individually, were mixed with mPEG in the absence or presence of the co-expressed PD4/CRTAP. The samples were then analyzed by western blot. mPEG is a cysteine-specific alkylation reagent with each modification of PPIB (PPIB-mPEG), resulting in an increase in the molecular weight of 2000 Da. PPIB variants (S85C, Q71C, Q119C, and H134C), where their modification is not affected by the presence of PD4/CRTAP, are labeled in blue. PPIB variants (K35C, D67C, T81C, K113C, D131C, E145C, and E184C), where their modification is inhibited by the presence of PD4/CRTAP, are labeled in orange. T49C and K113C could be partially modified by mPEG in the presence of PD4 and CRTAP. T155C is resistant to the mPEG modification. b The positions of selected residues on the surface of PPIB structure are colored according to their accessibility in the P3H1 ternary complex. PPIB inhibitor CsA binds PPIB in the substrate-binding pocket and is shown in sticks (green). T49C is located in the backside of the current viewpoint
Since the T155C variant by itself could not be modified by mPEG, we reexamined the crystal structure of PPIB and found that T155 is involved in crystal packing. This part of the PPIB molecule is stabilized in this particular exposed conformation by a neighboring PPIB molecule in the crystal lattice. Therefore, it is likely that T155 is not surface exposed in solution, as seen in the crystal structure. Interestingly, in the presence of PD4 and CRTAP, it became partially accessible for modification (Fig. 4a, lane 19), indicating certain conformational changes around this area of PPIB upon ternary complex formation.
Overall, the cysteine mapping experiment showed that the PPIB surface is extensively covered by the binding of P3H1 and CRTAP, while the PPIB substrate-binding pocket in the ternary complex is available for its prolyl cis–trans isomerase function (Fig. 4b).
Chemical crosslinking of PPIB variants to the PD4/CRTAP complex
To gain further insight into the detailed interaction between PPIB and PD4/CRTAP, we carried out a crosslinking experiment utilizing the single-cysteine PPIB variants prepared above and the dual-specific crosslinking reagent Sulfo-GMBS, which modifies amine groups and thiol groups simultaneously (Fig. 5a). Here, the partially purified PD4/CRTAP from the culture medium (Supplemental Fig. 4a) was first treated with iodoacetic amine to block any free thiol groups of these two proteins and then mixed with Sulfo-GMBS. This allowed specific modifications of amine groups of the surface lysine residues of PD4/CRTAP complex. Subsequently, the modified PD4/CRTAP complex was desalted to remove excess Sulfo-GMBS and then incubated with PPIB variants. Once the ternary complex was formed, amine-linked GMBS on PD4 or CRTAP could specifically react with the free thiols of PPIB within a 7.3 Å radius (Fig. 5b). The reactions were terminated after 30 min, and samples were analyzed by western blot using P3H1 and CRTAP antibodies, respectively. This two-step crosslinking strategy with removal of excess crosslinking reagents would avoid non-specific crosslinking amongst the complex components. Also we confirmed that pre-modified PD4/CRTAP complex retains the ability to form ternary complexes with PPIB (Supplemental Fig. 5).
Fig. 5.
Chemical crosslinking of PPIB variants to the PD4/CRTAP complex. a Sulfo-GMBS is a water-soluble amine-to-sulfhydryl crosslinker with a short spacer arm (7.3 Å). b Sulfo-GMBS conjugates with the free amines of PD4 and CRTAP through its N-hydroxysuccinimide ester. Once PPIB is added, the maleimide group of sulfo-GMBS will form a covalent linkage with a nearby cysteine residue of PPIB variant. c The PD4/CRTAP were incubated with Sulfo-GMBS, and then mixed with the PPIB variants according to the procedure in b, and samples were analyzed by western blot. The high-molecular-weight species represent the crosslinked PD4–PPIB or CRTAP–PPIB complex. The crosslinking data are summarized in Fig. 6a according to the relative density of the bands. d The crosslinked sample of PD4/CRTAP with PPIB D67C was digested with trypsin and analyzed by LC-mass spectrometry with Sulfo-GMBS containing peptides identified. The PPIB D67C was found to crosslink with K406 of P3H1 or K363 of PD4 or K120 of CRTAP (Supplemental Fig. 6)
As the crosslinked PD4 or CRTAP have higher molecular weights, it was clear from the gel that PPIB D67C, T81C, K113C, D131C, and H134C could be readily crosslinked to PD4, and other PPIB variants such as T49C and Q119C could also be linked, but to a lesser extent (Fig. 5c). Similarly, crosslinking also occurred between CRTAP and PPIB variants such as D67C, T81C, K113C, Q119C, D131C, H134C, and E184C. It seemed that K35C, Q71C, S85C, E145C, and T155C could not be crosslinked to either PD4 or CRTAP, while T49C could only be linked to PD4 and E184C could only be linked to CRTAP (Fig. 5c). Most interestingly, Q119C and H134C, two of the residues in the PPIB substrate-binding pocket, could be crosslinked to both PD4 and CRTAP. This indicates that both P3H1 and CRTAP bind closely to the rims of the PPIB substrate-binding pocket.
To identify the direct linkage between the residues of PPIB and PD4/CRTAP, we performed tryptic digests and liquid chromatography (LC)–mass spectrometry analysis of the crosslinked products from PPIB D67C and T81C, respectively. A database of potential crosslinked peptides by GMBS between the cysteines of PPIB and lysines of PD4 or CRTAP was generated and scanned for fragments across the mass spectrometry peaks. Although no crosslinked peptides from PPIB T81C could be found, crosslinked peptides from PPIB D67C were identified. The cysteine from peptide CFMIQGGDFTR (67–77) of PPIB was linked with K406 of the P3H1 peptide RLQEKQKSER (402–411) (Fig. 5d), K363 of the P3H1 peptide ESAKEYRQR (360–368) (Supplemental Fig. 6a), or K120 of CRTAP peptide RAHCLKR (115–121) (Supplemental Fig. 6b), with corresponding ions highlighted. This indicates that D67 of PPIB is within 7.3 Å distance from K363 and K406 of P3H1 and K120 of CRTAP in the ternary complex. As both K363 and K406 are located in the N-terminal domain of P3H1, this further indicates the involvement of the P3H1 N-terminal domain in the binding of both CRTAP and PPIB in the ternary complex.
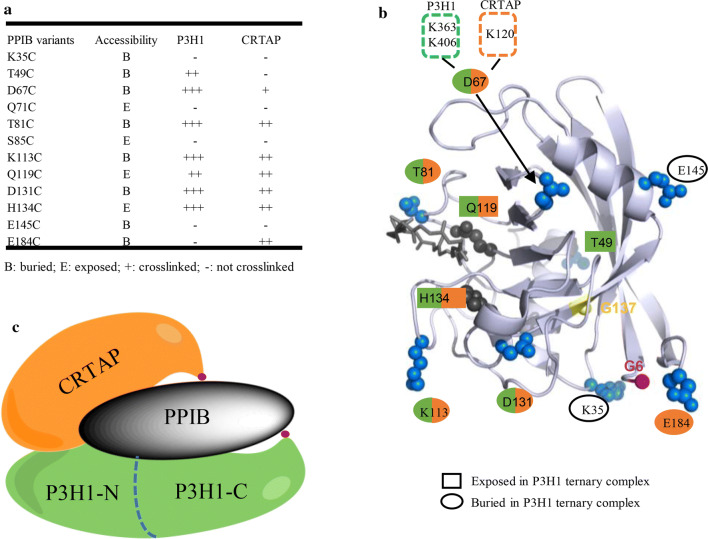
These results are summarized in Fig. 6 with an illustration showing the relative positions of the three components of the P3H1 complex. The PPIB residues involved in binding to P3H1 and CRTAP are also mapped onto the surface of the PPIB structure (Fig. 6b).
Fig. 6.
Summarized PPIB intramolecular interactions in the P3H1 ternary complex. a Accessibility of PPIB surface residues in the P3H1 ternary complex was derived from Fig. 4, and the crosslinking information was derived from Fig. 5c. b The key residues of PPIB tested here are highlighted on the surface of PPIB structure showing their accessibility and crosslinking property. The buried PPIB residues upon ternary complex formation are shown in blue spheres. The residues crosslinked to P3H1 (T49, D67, T81, K113, Q119, D131, and H134) are marked in green, and the residues crosslinked to CRTAP (D67, T81, K113, Q119, D131, H134, and E184) are marked in orange. D67 was identified to be close to K363 and K406 (green dotted box) of P3H1, and the K120 (orange dotted box) of CRTAP. c A working model showing relative spatial positions of each component in the P3H1/CRTAP/PPIB complex. The C-termini of P3H1 and CRTAP are labeled as red dots
Pathological mutations of PPIB impede P3H1 complex formation
It has been well documented that mutations in the PPIB gene are associated with OI. Most of these mutations result in the truncation of PPIB with the subsequent loss of the PPIB protein. However, two pathological point-mutations of PPIB, Gly6Arg and Gly137Asp, have been reported recently. The first mutation is associated with hyperelastosis cutis (HC) in the American quarter horse, resulting in a delay in the folding and secretion of procollagen [37]. The second mutation was identified from a Chinese family with autosomal recessive OI [38]. Both residues are located around the PPIB-binding surface in the ternary complex, and Gly6 in particular is a surface-exposed residue located at the N-terminal strand close to residue Glu184 in the PPIB C-terminus (Fig. 6b). It was not very clear on how these two mutations would cause the defect in the folding of procollagen, so we prepared these two PPIB mutants and assessed their peptidyl–prolyl cis–trans isomerase activity and their ability to form a ternary complex with P3H1 and CRTAP.
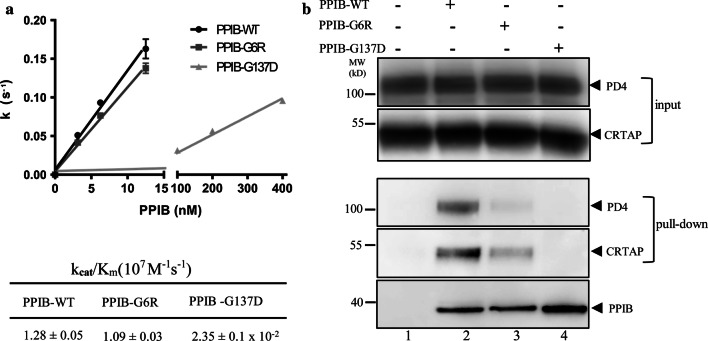
PPIB isomerization activity assays showed that PPIB-G6R has a similar activity as PPIB-WT with a kcat/Km of 1.09 × 107M-1s-1, consistent with a previous report [39]. However, PPIB-G137D has a kcat/Km of 0.024 × 107M-1s-1, which is about 45-fold lower (Fig. 7a). Subsequently, we assessed the ability of these PPIB variants to form ternary complex with P3H1 and CRTAP. Ni-chelating beads loaded with Sumo-PPIB variants were mixed with the partially purified PD4/CRTAP. After incubation, the beads were extensively washed, and the bound proteins were analyzed by SDS-PAGE and western blot. As shown in Fig. 7b, PPIB-WT could readily pull down PD4 and CRTAP (lane 2) which is consistent with gel filtration results that secreted PD4/CRTAP can readily form ternary complexes with PPIB (Fig. 3b). However, the PPIB-G6R mutant pulled down a significantly lesser amount of PD4 and CRTAP (lane 3) and no PD4 or CRTAP could be pulled down PPIB-G137D mutant (lane 4). These data suggest that these two pathological mutations largely abolished the ability of PPIB to form a ternary complex with P3H1 and CRTAP, and this defect of PPIB mutants likely plays a key role in the onset of pathological collagen misfolding-related diseases.
Fig. 7.
Characterization of pathological PPIB mutants. a The kinetics of PPIB variants in cis/trans isomerization of the Suc-AAPF-pNA substrate were measured by following the absorbance at 390 nm in a spectrophotometer at 8 °C with spectra shown (Supplemental Fig. 7). The rate of cis–trans transition in the substrate was plotted against the concentrations of PPIB variants with the calculated kcat/Km values listed at the bottom. The activity of PPIB-G137D is very low and little activity could be detected when its concentration is below 20 nM. The break in the x-axis is between 15 and 100 nM. PPIB-G6R has similar catalyzing activity as the wild-type PPIB; however, the PPIB-G137D mutant is about 45 times less active. b The binding of PPIB variants towards P3H1 and CRTAP was assessed by a pull-down assay. PPIB variants immobilized on Ni magnetic beads were incubated with partially purified PD4/CRTAP. The bound proteins were then analyzed by SDS-PAGE and western blot using antibodies against P3H1 and CRTAP, respectively. Lane 1, immobilized Sumo protein as a control; lane 2, Sumo-PPIB-WT; lane 3, Sumo-PPIB G6R; lane 4, Sumo-PPIB G137D. The two pathological mutations G6R and G137D largely abolished the ability of PPIB to form ternary complexes with P3H1 and CRTAP (lane 3, 4). All experiments were performed ≥ 3 times independently. Error bars represent mean ± SEM
Discussion
Recent investigations have revealed that the “brittle bone” phenotype in OI is caused not only by dominant mutations in collagen type I genes, but also by recessively inherited mutations in genes responsible for the post-translational processing of type I procollagen as well as for bone formation [22, 40–43]. Deficiencies in genes that encode proteins of P3H1 ternary complex are associated with diminished Pro986 hydroxylation and misfolding of procollagen. As P3H1/CRTAP/PPIB complex can function as a molecular chaperone, a proline 3-hydroxylase, and a disulfide isomerase [27, 44], it is not clear which function(s) of the P3H1 complex or other unknown functions of the ternary complex, are indispensable during procollagen folding. Many questions regarding this complex remain. For example, how is this ternary complex retained within the ER where only P3H1 has a KDEL sequence? Why does it preferentially hydroxylate the single Pro986 of procollagen? Which function of this ternary complex is most important during the folding of procollagen? How do these proteins bind to each other to form the ternary complex? Is the integrity of this complex required for its functions? Here, we have addressed some of these questions.
Retention of the P3H1 complex within the ER
Recently, in a unique non-lethal OI case, a mutation in P3H1 was found to result in the truncation of the C-terminal 18 amino acids of P3H1, including the KDEL ER-retrieval sequence. Although it appeared that this truncation resulted in the loss of the P3H1 complex in the cell [29], it is unclear if this is solely because of the loss of these four residues (KDEL). Since P3H1 is the only component of the P3H1 complex with a KDEL sequence, we, therefore, assessed the role of this sequence in the retention of the complex in the cell through mutagenesis studies. Our data show that most P3H1 was expressed in the intracellular compartments, while the P3H1 mutant PD4 with KDEL deleted could be secreted into the culture medium, suggesting that the KDEL sequence is essential for P3H1 intracellular localization.
Furthermore, we found that large amounts of CRTAP were also secreted out of cells when co-expressed with PD4, while most CRTAP is retained in the ER when expressed alone or co-expressed with normal P3H1. Trace amount of CRTAP could be detected in the culture medium of HEK293E cells overexpressing CRTAP. Similarly, only trace amount of CRTAP could be detected in the medium from HEK293E and fibroblast-derived HT1080 cells (data not shown). This is in line with the previous observation with osteoblasts and osteoclasts [22] where trace amount of CRTAP could be detected in the medium. It is also somewhat consistent with a previous report that small amount (~ 10%) of CRTAP could be detected in the culture medium of fibroblast cells [28]. The difference in the amount of secreted CRTAP in the medium is unclear and could be due to differences in the cell lines. Nevertheless, these data indicate that CRTAP is mainly an ER-resident protein, even though it does not have a KDEL sequence and its signal peptide is not very efficient in directing CRTAP secretion. In addition, we confirm that the ER-retrieval sequence at the carboxyl terminus of P3H1 is indispensable for P3H1/CRTAP intracellular localization.
Intermolecular interactions of the P3H1 complex
Although it has been reported previously that P3H1 and CRTAP are mutually stabilized in the prolyl 3-hydroxylation complex, and that the absence of either one results in the degradation of the other; the specific binding areas between these two proteins have been unclear. Here, based on the co-secretion of P3H1 with CRTAP after the removal of the KDEL sequence, we discovered that this co-secretion is mediated by the N-terminal domain of P3H1 (Fig. 2b). As this domain has a four-cysteine repeat (CXXC), homologous to that of CRTAP, this suggests that the CXXC motifs of both P3H1 and CRTAP may not only function as a disulfide isomerase, but also play an important role in their special interdependent relationship.
As the secreted P3H1/CRTAP can readily form ternary complex with PPIB in vitro, this allows us to map the binding surface of PPIB in the ternary complex. Cysteine modification and crosslinking experiments showed that the PPIB surface is largely covered by bound P3H1/CRTAP but with the substrate-binding site vacant. This agrees with a previous finding that cyclosporine A (CsA) can bind to and inhibit the activity of PPIB, yet CsA binding on PPIB does not interfere with the interaction between P3H1 and PPIB [24]. The delineated physical interactions of PPIB with P3H1 and CRTAP through crosslinking and mass spectrometry experiments showed that the residue D67 of PPIB is close to K363 and K406 of P3H1, as well as K120 of CRTAP. As both K363 and K406 of P3H1 are located in the N-terminal domain of P3H1, this further implies the importance of this CXXC domain in P3H1 complex formation. In addition, by serendipity, we found that a slight extension at the C-termini of P3H1 or CRTAP blocked the formation of the complex with PPIB, indicating potential interactions of PPIB with the C-terminal parts of P3H1 and CRTAP. This provides novel spatial information on how these three proteins bind each other and allows us to build an initial model showing the assembly of the P3H1 ternary complex (Fig. 6).
Implication of pathological mutations on P3H1 ternary complex function
It is well documented that deficiencies in any component of the P3H1 ternary complex are associated with pathological dysregulation of collagen formation; however, the relative contribution of losing this complex’s 3-hydroxylation, PPIase, or collagen chaperone activities to the phenotype of recessive OI is not clear. None of the pathological mutations on either P3H1 or CRTAP identified so far are informative in distinguishing the roles of the P3H1 complex during procollagen formation, as the absence of one protein results in the absence of the other, ultimately leading to the loss of the whole complex in the ER. The lack of hydroxylation of a single site, Pro986, in the triple-helical domain of the α1 chain was originally thought to be part of the causative disease mechanism, but a recent knock-in mouse model with an inactive P3H1 mutant showed that the bone development of these mice is largely unaffected, even though they lack hydroxylation at Pro986 in procollagen [45]. It is also consistent with some PPIB-deficient patients with normal hydroxylation of Pro986 [46]. This seems to indicate that Pro986 hydroxylation is dispensable during procollagen formation, and that its absence is unlikely to be the major disease-causing factor. Thus, other functions of P3H1 and CRTAP or the ternary complex, apart from hydroxylation, are essential for the proper folding of procollagen.
The effect of pathological PPIB mutations on collagen formation are rather complex. Mutations in PPIB often lead to a marked reduction in the amount of stable PPIB protein produced, but they have different effects on the levels of P3H1 and CRTAP. This leads to variation in the amount of Pro986 hydroxylation in the α1 chain by skin fibroblasts [24, 42, 46, 47]. The underlying mechanisms seem to vary, as well. In some cases, PPIB deficiencies are associated with a delay in the type I procollagen chain association and result in perinatal lethal-to-moderate OI phenotypes, while, in other cases, a PPIB deficiency has a limited effect on the folding of procollagen and cartilage formation [46, 47]. Nevertheless, these phenotypes in patients can be attributed to the loss of PPIB isomerase activity and/or to the loss of other functions of PPIB, such as interactions with other binding partners, including P3H1 and CRTAP as described here.
One natural PPIB mutant PPIB-G137D associated with OI is characterized here, and it shows impaired isomerase activity and an abolished binding affinity towards the P3H1/CRTAP complex. This is not so surprising as Gly137 is located in the hydrophobic core of PPIB protein, replacement of this residue with a polar aspartic acid is expected to affect the packing of this mutant and ultimately affects its function (Fig. 6b). Again, both defects of PPIB-G137D in isomerase activity and in the formation of a complex with P3H1 and CRTAP likely contribute to the phenotype of PPIB-G137D observed, and it is hard to tell which defect plays a major role. Most informatively, however, the other pathological PPIB mutant (PPIB-G6R) examined here has normal isomerase activity, but an impaired ability to form a ternary complex with P3H1 and CRTAP. This mutant is associated with a delay in procollagen folding and secretion [37]. Clearly, the phenotype observed most likely arises from defects in PPIB functions other than as isomerase. Although it was suggested that this procollagen folding defects might be caused by its impaired binding towards P domain of calreticulin, the binding affinity measurements did not give conclusive support [37]. Notably, the interaction between P domain of calreticulin and PPIB is relatively weak with a Kd about 10 μM [48], while the interaction between P3H1/CRTAP and PPIB is much stronger with a Kd in the nanomolar range according to our pull-down assay (Fig. 7). Given the overwhelming evidences showing the key role of P3H1 complex in collagen formation and the finding here that PPIB-G6R mutation significantly disrupted the interaction between PPIB and P3H1/CRTAP, we believe that it is the loss of the integrity of the P3H1 ternary complex, not the loss of PPIB isomerase activity or the loss of calreticulin P domain binding of PPIB-G6R, that caused the collagen folding defects in HC horses. The normal functions of P3H1/CRTAP as proline hydroxylase together with normal activity of PPIB as isomerase in HC horses are insufficient for the proper folding of procollagen and it is the intact ternary complex that plays a critical role during procollagen formation. Although how three components of the ternary complex synchronize their functions during the procollagen folding needs to be further defined, it has been proposed that the complex functions as a chaperone or by positioning PPIB onto the carboxyl end of the collagen helix, where folding is initiated [46]. Clearly, further investigation is required to unravel the underlying mechanisms of this molecular machinery.
Overall, the data presented here provide novel spatial information about the binding interactions of PPIB in the P3H1 ternary complex, and the detailed characterization of pathological PPIB mutants indicates that the integrity of the P3H1 ternary complex is critical for proper folding of procollagen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (PPTX 26481 kb)
Acknowledgements
This work was supported in part by Grants from the National Natural Science Foundation of China (81870309, 31570824, and 81572090), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, Shanghai PuJiang Program, and Innovation Program of Shanghai Municipal Education Commission (no. 12ZZ113).
Abbreviations
- ER
Endoplasmic reticulum
- P3H1
Prolyl 3-hydroxylase 1
- CRTAP
Cartilage-associated protein
- PPIB
Peptidyl–prolyl cis–trans isomerase B
- OI
Osteogenesis imperfecta
- mPEG
NEM-PEG2000
- MW
Molecular weight
- HC
Hyperelastosis cutis
- WT
Wild type
- IPTG
Isopropyl-β-d-thiogalactopyranoside
- DTT
Dithiothreitol
Author contributions
JW and WZ prepared all the P3H1/CRTAP/PPIB mutants. WZ, ZS, JZ, and LF performed the in vitro experiment about the interaction of P3H1 and CRTAP. J.W carried out the PPIB cysteine modification and crosslinking experiment. XL helped to establish the mass spectrometry method. JW and AZ wrote the paper. WY, NZ, and AZ conceived and designed all the experiments.
Funding
This work was supported in part by Grants from the National Natural Science Foundation of China (81870309, 31570824, and 81572090), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, Shanghai PuJiang Program, and Innovation Program of Shanghai Municipal Education Commission (no. 12ZZ113).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiawei Wu, Wenting Zhang, and Li Xia have contributed equally to this work.
Contributor Information
Wei Ye, Email: jyyewei@163.com.
Naiyan Zeng, Email: zengny@shsmu.edu.cn.
Aiwu Zhou, Email: awz20@shsmu.edu.cn.
References
- 1.Bächinger HP, Mizuno K, Vranka JA, Boudko SP. Collagen formation and structure. In: Mander L, Liu HW, editors. Comprehensive natural products II: chemistry and biology. Oxford: Elsevier; 2010. pp. 469–530. [Google Scholar]
- 2.Hamaia S, Farndale RW. Integrin recognition motifs in the human collagens. Adv Exp Med Biol. 2014;819:127–142. doi: 10.1007/978-94-017-9153-3_9. [DOI] [PubMed] [Google Scholar]
- 3.Myllyharju J. Intracellular post-translational modifications of collagens. Top Curr Chem. 2005;247:115–147. doi: 10.1007/b103821. [DOI] [Google Scholar]
- 4.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20(1):33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Lamande SR, Bateman JF. Procollagen folding and assembly: the role of endoplasmic reticulum enzymes and molecular chaperones. Semin Cell Dev Biol. 1999;10(5):455–464. doi: 10.1006/scdb.1999.0317. [DOI] [PubMed] [Google Scholar]
- 6.Jimenez S, Harsch M, Rosenbloom J. Hydroxyproline stabilizes the triple helix of chick tendon collagen. Biochem Biophys Res Commun. 1973;52(1):106. doi: 10.1016/0006-291X(73)90960-1. [DOI] [PubMed] [Google Scholar]
- 7.Berg RA, Prockop DJ. The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple-helix of collagen. Biochem Biophys Res Commun. 1973;52(1):115. doi: 10.1016/0006-291X(73)90961-3. [DOI] [PubMed] [Google Scholar]
- 8.Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22(1):15–24. doi: 10.1016/S0945-053X(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 9.Lees JF, Tasab M, Bulleid NJ. Identification of the molecular recognition sequence which determines the type-specific assembly of procollagen. EMBO J. 1997;16(5):908. doi: 10.1093/emboj/16.5.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doege KJ, Fessler JH. Folding of carboxyl domain and assembly of procollagen I. J Biol Chem. 1986;261(19):8924–8935. [PubMed] [Google Scholar]
- 11.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7(4):523. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 12.Vranka JA, Sakai LY, Bachinger HP. Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J Biol Chem. 2004;279(22):23615–23621. doi: 10.1074/jbc.M312807200. [DOI] [PubMed] [Google Scholar]
- 13.Kellokumpu S, Sormunen R, Heikkinen J, Myllylã R. Lysyl hydroxylase, a collagen processing enzyme, exemplifies a novel class of luminally-oriented peripheral membrane proteins in the endoplasmic reticulum. J Biol Chem. 1994;269(48):30524–30529. [PubMed] [Google Scholar]
- 14.Steinmann B, Bruckner P, Superti-Furga A. Cyclosporin A slows collagen triple-helix formation in vivo: indirect evidence for a physiologic role of peptidyl-prolyl cis–trans-isomerase. J Biol Chem. 1991;266(2):1299–1303. [PubMed] [Google Scholar]
- 15.Forlino A, Cabral WA, Barnes AM, Marini JC. New perspectives on osteogenesis imperfecta. Nat Rev Endocrinol. 2011;7(9):540–557. doi: 10.1038/nrendo.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marini JC, Forlino A, Bächinger HP, Bishop NJ, Byers PH, Paepe AD, Fassier F, Fratzl-Zelman N, Kozloff KM, Krakow D, Montpetit K, Semler O. Osteogenesis imperfecta. Nat Rev Dis Prim. 2017;3:17052. doi: 10.1038/nrdp.2017.52. [DOI] [PubMed] [Google Scholar]
- 17.Forlino A, Marini JC. Osteogenesis imperfecta. Lancet. 2016;387(10028):1657–1671. doi: 10.1016/S0140-6736(15)00728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alanay Y, Avaygan H, Camacho N, Utine GE, Boduroglu K, Aktas D, Alikasifoglu M, Tuncbilek E, Orhan D, Bakar FT. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86(4):551–559. doi: 10.1016/j.ajhg.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christiansen HE, Schwarze U, Pyott SM, Alswaid A, Balwi MA, Alrasheed S, Pepin MG, Weis MA, Eyre DR, Byers PH. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86(3):389–398. doi: 10.1016/j.ajhg.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito S, Nagata K. Biology of Hsp47 (Serpin H1), a collagen-specific molecular chaperone. Semin Cell Dev Biol. 2017;62:142–151. doi: 10.1016/j.semcdb.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Marini JC, Reich A, Smith SM. Osteogenesis imperfecta due to mutations in non-collagenous genes: lessons in the biology of bone formation. Curr Opin Pediatr. 2014;26(4):500–507. doi: 10.1097/MOP.0000000000000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morello R, Bertin TK, Chen Y, Hicks J, Tonachini L, Monticone M, Castagnola P, Rauch F, Glorieux FH, Vranka J, Bachinger HP, Pace JM, Schwarze U, Byers PH, Weis M, Fernandes RJ, Eyre DR, Yao Z, Boyce BF, Lee B. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127(2):291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 23.Vranka JA, Pokidysheva E, Hayashi L, Zientek K, Mizuno K, Ishikawa Y, Maddox K, Tufa S, Keene DR, Klein R. Prolyl 3-hydroxylase 1 null mice display abnormalities in fibrillar collagen-rich tissues such as tendons, skin and bones. J Biol Chem JBC. 2010;M110:102228. doi: 10.1074/jbc.M110.102228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi JW, Sutor SL, Lindquist L, Evans GL, Madden BJ, Bergen HR, 3rd, Hefferan TE, Yaszemski MJ, Bram RJ. Severe osteogenesis imperfecta in cyclophilin B-deficient mice. PLoS Genet. 2009;5(12):e1000750. doi: 10.1371/journal.pgen.1000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marini JC, Cabral WA, Barnes AM, Chang W. Components of the collagen prolyl 3-hydroxylation complex are crucial for normal bone development. Cell Cycle. 2007;6(14):1675–1681. doi: 10.4161/cc.6.14.4474. [DOI] [PubMed] [Google Scholar]
- 26.Tonachini L, Morello R, Monticone M, Skaug J, Scherer SW, Cancedda R, Castagnola P. cDNA cloning, characterization and chromosome mapping of the gene encoding human cartilage associated protein (CRTAP) Cytogenet Genome Res. 1999;87(3–4):191. doi: 10.1159/000015463. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa Y, Bachinger HP. An additional function of the rough endoplasmic reticulum protein complex prolyl 3-hydroxylase 1. Cartilage-associated protein–cyclophilin B: the CXXXC motif reveals disulfide isomerase activity in vitro. J Biol Chem. 2013;288(44):31437–31446. doi: 10.1074/jbc.M113.498063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang W, Barnes AM, Cabral WA, Bodurtha JN, Marini JC. Prolyl 3-hydroxylase 1 and CRTAP are mutually stabilizing in the endoplasmic reticulum collagen prolyl 3-hydroxylation complex. Hum Mol Genet. 2010;19(2):223–234. doi: 10.1093/hmg/ddp481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takagi M, Ishii T, Barnes AM, Weis M, Amano N, Tanaka M, Fukuzawa R, Nishimura G, Eyre DR, Marini JC, Hasegawa T. A novel mutation in LEPRE1 that eliminates only the KDEL ER- retrieval sequence causes non-lethal osteogenesis imperfecta. PLoS One. 2012;7(5):e36809. doi: 10.1371/journal.pone.0036809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbirato C, Trancozo M, Almeida MG, Almeida LS, Santos TO, Duarte JC, Reboucas MR, Sipolatti V, Nunes VR, Paula F. Mutational characterization of the P3H1/CRTAP/CypB complex in recessive osteogenesis imperfecta. Genet Mol Res. 2015;14(4):15848–15858. doi: 10.4238/2015.December.1.36. [DOI] [PubMed] [Google Scholar]
- 31.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 32.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9(9):904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 33.Kofron JL, Kuzmic P, Kishore V, Colonbonilla E, Rich DH. Determination of kinetic constants for peptidyl prolyl cis–trans isomerases by an improved spectrophotometric assay. Biochemistry. 1991;30(25):6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- 34.Dimou M, Venieraki A, Liakopoulos G, Kouri ED, Tampakaki A, Katinakis P. Gene expression and biochemical characterization of Azotobacter vinelandii cyclophilins and protein interaction studies of the cytoplasmic isoform with dnaK and lpxH. J Mol Microbiol Biotechnol. 2011;20(3):176–190. doi: 10.1159/000329486. [DOI] [PubMed] [Google Scholar]
- 35.Galat A, Bouet F. Cyclophilin-B is an abundant protein whose conformation is similar to cyclophilin-A. FEBS Lett. 1994;347(1):31. doi: 10.1016/0014-5793(94)00501-X. [DOI] [PubMed] [Google Scholar]
- 36.Mikol V, Kallen J, Walkinshaw MD. X-Ray structure of a cyclophilin B/cyclosporin complex: comparison with cyclophilin A and delineation of its calcineurin-binding domain. Proc Natl Acad Sci USA. 1994;91(11):5183–5186. doi: 10.1073/pnas.91.11.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishikawa Y, Vranka JA, Boudko SP, Pokidysheva E, Mizuno K, Zientek K, Keene DR, Rashmirraven AM, Nagata K, Winand NJ. Mutation in cyclophilin B that causes hyperelastosis cutis in american quarter horse does not affect peptidylprolyl cis–trans isomerase activity but shows altered cyclophilin B–protein interactions and affects collagen folding. J Biol Chem. 2012;287(26):22253–22265. doi: 10.1074/jbc.M111.333336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, Pan J, Guo D, Zhang W, Xie J, Fang Z, Guo C, Fang Q, Jiang W, Guo Y. Two novel mutations in the PPIB gene cause a rare pedigree of osteogenesis imperfecta type IX. Clinica Chimica Acta. 2017;469:111–118. doi: 10.1016/j.cca.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 39.Price ER, Zydowsky LD, Jin MJ, Baker CH, Mckeon FD, Walsh CT. Human cyclophilin B: a second cyclophilin gene encodes a peptidyl-prolyl isomerase with a signal sequence. Proc Natl Acad Sci USA. 1991;88(5):1903–1907. doi: 10.1073/pnas.88.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnes AM, Chang W, Morello R, Cabral WA, Weis M, Eyre DR, Leikin S, Makareeva E, Kuznetsova N, Uveges TE. Deficiency of cartilage-associated protein in recessive lethal osteogenesis imperfecta. N Engl J Med. 2006;355(26):2757–2764. doi: 10.1056/NEJMoa063804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cabral WA, Chang W, Barnes AM, Weis M, Scott MA, Leikin S, Makareeva E, Kuznetsova NV, Rosenbaum KN, Tifft CJ, Bulas DI, Kozma C, Smith PA, Eyre DR, Marini JC. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet. 2007;39(3):359–365. doi: 10.1038/ng1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dijk FSV, Nesbitt IM, Zwikstra EH, Nikkels PGJ, Piersma SR, Fratantoni SA, Jimenez CR, Huizer M, Morsman AC, Cobben JM. PPIB mutations cause severe osteogenesis imperfecta. Am J Hum Genet. 2009;85(4):521–527. doi: 10.1016/j.ajhg.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baldridge D, Schwarze U, Morello R, Lennington J, Bertin TK, Pace JM, Pepin MG, Weis M, Eyre DR, Walsh J, Lambert D, Green A, Robinson H, Michelson M, Houge G, Lindman C, Martin J, Ward J, Lemyre E, Mitchell JJ, Krakow D, Rimoin DL, Cohn DH, Byers PH, Lee B. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum Mutat. 2008;29(12):1435–1442. doi: 10.1002/humu.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishikawa Y, Wirz J, Vranka JA, Nagata K, Bachinger HP. Biochemical characterization of the prolyl 3-hydroxylase 1. Cartilage-associated protein–cyclophilin B complex. J Biol Chem. 2009;284(26):17641–17647. doi: 10.1074/jbc.M109.007070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Homan EP, Lietman C, Grafe I, Lennington J, Morello R, Napierala D, Jiang MM, Munivez EM, Dawson B, Bertin TK, Chen Y, Lua R, Lichtarge O, Hicks J, Weis MA, Eyre D, Lee BH. Differential effects of collagen prolyl 3-hydroxylation on skeletal tissues. PLoS Genet. 2014;10(1):e1004121. doi: 10.1371/journal.pgen.1004121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnes AM, Carter EM, Cabral WA, Weis M, Chang W, Makareeva E, Leikin S, Rotimi CN, Eyre DR, Raggio CL, Marini JC. Lack of cyclophilin B in osteogenesis imperfecta with normal collagen folding. N Engl J Med. 2010;362(6):521–528. doi: 10.1056/NEJMoa0907705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pyott SM, Schwarze U, Christiansen HE, Pepin MG, Leistritz DF, Dineen R, Harris C, Burton BK, Angle B, Kim K, Sussman MD, Weis M, Eyre DR, Russell DW, McCarthy KJ, Steiner RD, Byers PH. Mutations in PPIB (cyclophilin B) delay type I procollagen chain association and result in perinatal lethal to moderate osteogenesis imperfecta phenotypes. Hum Mol Genet. 2011;20(8):1595–1609. doi: 10.1093/hmg/ddr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozlov G, Bastos-Aristizabal S, Maattanen P, Rosenauer A, Zheng F, Killikelly A, Trempe JF, Thomas DY, Gehring K. Structural basis of cyclophilin B binding by the calnexin/calreticulin P-domain. J Biol Chem. 2010;285(46):35551–35557. doi: 10.1074/jbc.M110.160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (PPTX 26481 kb)