Abstract
Low-density lipoprotein receptor-related protein 4 (LRP4) is a multi-functional protein implicated in bone, kidney and neurological diseases including Cenani-Lenz syndactyly (CLS), sclerosteosis, osteoporosis, congenital myasthenic syndrome and myasthenia gravis. Why different LRP4 mutation alleles cause distinct and even contrasting disease phenotypes remain unclear. Herein, we utilized the zebrafish model to search for pathways affected by a deficiency of LRP4. The lrp4 knockdown in zebrafish embryos exhibits cyst formations at fin structures and the caudal vein plexus, malformed pectoral fins, defective bone formation and compromised kidney morphogenesis; which partially phenocopied the human LRP4 mutations and were reminiscent of phenotypes resulting form a perturbed Notch signaling pathway. We discovered that the Lrp4-deficient zebrafish manifested increased Notch outputs in addition to enhanced Wnt signaling, with the expression of Notch ligand jagged1b being significantly elevated at the fin structures. To examine conservatism of signaling mechanisms, the effect of LRP4 missense mutations and siRNA knockdowns, including a novel missense mutation c.1117C > T (p.R373W) of LRP4, were tested in mammalian kidney and osteoblast cells. The results showed that LRP4 suppressed both Wnt/β-Catenin and Notch signaling pathways, and these activities were perturbed either by LRP4 missense mutations or by a knockdown of LRP4. Our finding underscore that LRP4 is required for limiting Jagged–Notch signaling throughout the fin/limb and kidney development, whose perturbation representing a novel mechanism for LRP4-related diseases. Moreover, our study reveals an evolutionarily conserved relationship between LRP4 and Jagged–Notch signaling, which may shed light on how the Notch signaling is fine-tuned during fin/limb development.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2928-3) contains supplementary material, which is available to authorized users.
Keywords: Skeletogenesis, Bone disorders, Pronephros, Morphant, Phenocopy, EGF-like domain, wt1b, HES1
Introduction
The genes underlying the human genetic syndromes manifesting limb and digit malformations are well conserved in evolution, and some perform the same roles in the teleostean fins [1–3]. Using the zebrafish, the signaling pathways of fin development and regeneration can be well dissected. For example, the zebrafish has been developed as a model of Fraser Syndrome (FS), a recessive polygenic, multisystem congenital human disorder characterized by syndactyly of the soft tissue of digits, cryptophthalmos and renal agenesis. In the FS zebrafish mutant, the blister underneath the basement membrane of the fin epidermis is formed, which represents the limb malformation in FS patients [4]. The zebrafish model has also emerged as a powerful tool for dissecting molecular mechanisms that govern development, disease and regeneration of multiple organs or tissues such as the blood vasculature and the kidney [5, 6].
Low-density lipoprotein receptor-related protein 4 (LRP4) is a type I single transmembrane protein of the low-density lipoprotein receptor (LDLR) family. LRP4 mutations have been identified in Cenani-Lenz syndrome (CLS; MIM#212780) [7], congenital myasthenic syndrome 17 (CMS17; MIM 616304) [8] and Sclerosteosis 2 (MIM 614305) [9]. Different mutations in the LRP4 locus result in distinct disease phenotypes: CLS is a rare autosomal recessive congenital disorder characterized by highly variable degree of syndactyly and/or oligodactyly of fingers and toes, which are associated with kidney agenesis or hypoplasia, nail aplasia or dysplasia, teeth anomalies and mild facial dysmorphisms; CMS17 patients present with neuromuscular disorder, but no syndactyly and kidney defects; Sclerosteosis 2 is a severe sclerosing bone dysplasia characterized by progressive skeletal overgrowth, with a variable manifestation of syndactyly. In contrast, the association between LRP4 and osteoporosis is revealed by genome-wide studies [10]. Autoantibodies against LRP4 has emerged as a diagnostic marker in Acetylcholine receptor (AChR)- and MuSK-negative myasthenia gravis [11]. The spectrum of disease phenotypes caused by various LRP4 mutant alleles suggests a complicated role for LRP4 in development and homeostasis. Diverse functions of LRP4 have been further revealed by a series of mouse mutants, such as Lrp4ECD/ECD, Lrp4mte/mte, Lrp4mitt/mitt and Lrp4mdig/mdig [12–14]. The mutant mice present with syndactyly, characterized by the fusion and duplication of digits at both fore and hind limbs, some involve the malformation of carpal bones and entire limbs. Though renal anomalies are associated with some, but not all of the human LRP4 mutations, they have been identified in the Lrp4 mutant mice [15]. The Lrp4 mutant mice also exhibit malformations of neuromuscular junctions, mammary glands and teeth. However, in patients with LRP4 mutation-associated bone diseases, no consistent neuromuscular dysfunctions have been described [16, 17]. Why different LRP4 mutation alleles lead to distinct and even contrasting disease phenotypes is still unclear, and the answer may lie in the possible network of signaling pathways modulated by LRP4 protein.
LRP4 is localized exclusively to the plasma membrane of epithelial cells and demonstrates an antagonistic effect on LRP5/6-mediated activation of Wnt/β-catenin signaling, possibly due to a lack of an Axin-binding domain [12, 14]. However, the phenotypes of Lrp4 mutant mice are more severe than what observed in Lrp5 and Lrp6 mutants [18, 19]. Moreover, novel ligands of LRP4 have been identified, including Wise, DKK1, Sclerostin, Gremlin1, Agrin, APP, MuSK, ApoE, Wnt9a and Wnt11 [16, 20–26]; which implicates multiple signaling mechanisms regulated by LRP4. The extracellular domain of LRP4 can interact with DKK1 and Sclerostin in osteoblasts, and with Wise in skin appendage and tooth development, suggesting that the Wnt inhibitory activity of LRP4 may depend on its interaction with the Wnt antagonists [22, 23, 26, 27]. However, LRP4 could also modulate Wnt/β-catenin signaling in tooth development through interacting with Wnt ligands in the absence of Wise [27]. LRP4 participates in the development of neuromuscular junction by interacting with Agrin, MuSK and APP [20, 21, 25]. In addition, LRP4 binds the BMP4 antagonist Gremlin1 in vitro [24], and is the receptor for ApoE in the rat brain [28]. These studies support the idea that LRP4 interacts with different ligands in a context-dependent manner. However, whether and how LRP4 serves to integrate multiple signaling pathways during dynamic processes of development and physiology remains to be investigated.
To explore how LRP4 regulates signaling pathways in different tissues, we have set out to identify and characterize the zebrafish ortholog of lrp4. A knockdown of zygotic RNA and protein expressions of the lrp4 led to blistering of fin structures and caudal vein plexus, defective skeletogenesis and kidney phenotypes that partially phenocopied human LRP4 mutations, with an activation of Wnt/β-Catenin signaling pathway. Notably, we found that the Jagged–Notch signaling was activated in the fin structures upon a knockdown of lrp4 in fish. In human embryonic kidney cells, LRP4 was confirmed to repress the Notch signaling activity, and this repressing activity was compromised by a novel missense mutation c.1117C > T (p.R373W) and other missense alleles of Lrp4 that cause the CLS phenotype, a severe form of the LRP4-related diseases. Knockdown of LRP4 expression in human osteoblasts induced upregulated expressions of Notch signaling components. Consistently, overexpression of Lrp4 in mouse osteoblasts suppressed the expressions of Notch signaling components. The loss-of-function and overexpression results together implicate that abnormal activation of the Jagged–Notch pathway is a novel mechanism underlying the pathology of LRP4-related bone disorders. On the other hand, our study suggests that the negative regulation of Jagged–Notch pathway by LRP4 is a conserved mechanism shared between fin and limb development; possibly implicated in how Notch signaling controls the balance between self-renewal and differentiation.
Materials and method
Zebrafish husbandry
Zebrafish (Danio rerio) were raised according to standard protocols [29]. Embryos were obtained by natural crosses of wild-type and transgenic fish, and staged as previously described [30]. Embryos for histological analysis were treated with 0.03% phenylthiourea (Sigma) from 12-h post-fertilization (hpf) onwards to inhibit pigmentation. The following lines were used: Tg(wt1b:GFP) (line1) (from Christoph Englert, Fritz-Lipmann Institute, Jena, Germany); AB (ZIRC, Eugene, Oregon). All experimental procedures on zebrafish were approved by the Institutional Animal Care and Use Committee of Tunghai University (IRB Approval NO. 104-24) and carried out in accordance with the approved guidelines.
Cloning of zebrafish lrp4
lrp4 5′RACE primers were designed and given in Table S1. 5′RACE was carried out using ExactSTART™ Eukaryotic mRNA 5′- & 3′-RACE Kit (Epicentre,USA). Twenty embryos at 6 hpf were collected and lysed by TRIzol Reagent (Ambion, USA). 10 μg total RNA was treated by alkaline phosphatase. The 5′ m7GPPPN structure of RNA was changed to 5′ single phosphoric acid by tobacco acid pyrophosphatase (TAP). 5′ adapters were ligated with 5′ single phosphoric acid RNA by T4 RNA ligase. First-strand cDNAs were synthesized by reverse transcriptase. After PCR amplification, target fragments were purified and TA cloned for screening. Full-length of lrp4 was finally cloned to pCS2+ vector.
In situ hybridization (ISH), immunohistochemistry (IHC), 3β-Hsd activity staining and Acridine Orange staining
Whole-mount ISH assays were performed as described in [31, 32] with modifications. Digoxigenin (DIG)-labeled riboprobes were synthesized from linearized plasmids containing lrp4, fras1, pax2a, wnt5b, dkk1b and jag1b, and DIG-labeled probes were detected with alkaline phosphatase conjugated anti-DIG antibody (Roche). The expressions were detected with BM purple (Roche). Stained embryos from the ISH assay were cleared in 50% glycerol in PBS, and photographed under Nomarski optics on an Olympus BX51 and Nikon SMZ25 microscope systems.
To detect the Lrp4 protein, Tg(wt1b:GFP) embryos were fixed and embedded in 4% NuSieve GTG low-melting agarose (Lonza), cut into 100 μm sections with a VT1000M vibratome (Leica), and permeabilized with PBS containing 1% Triton X-100 before incubation with mouse anti-LRP4 monoclonal antibody [S207-27] at 1:200 (Abcam). Dylight 650-conjugated anti-mouse IgG (Abcam) were used as secondary antibodies at 1:200. Images were captured with an LSM510 confocal microscope with version 3.5 software (Zeiss).
To quantify the distances between various tissue structures, images of embryos in each group were taken with identical magnification using an Axioskop 2 Plus microscope equipped with AxioVision 3.0 software (Carl Zeiss). Lengths were measured using Image Gauge Program, version 4.0 (Fuji Photo Film).
Histochemical staining for 3β-Hsd enzymatic activity was performed on whole embryos according to the described protocol [33]. For simultaneous analysis of interrenal steroidogenic activity and GFP fluorescence, 3β-Hsd staining signals were captured using transmitted light, while the fluorescent signals were captured with Argon 488-nm laser connected to the confocal microscope. Image processing and analysis was performed using the LSM 510 version 3.5 software.
Acridine orange staining to detect apoptotic cell death in living embryos was performed according to [34]. Live embryos at 2 days post-fertilization (dpf) were incubated in 2 μg/mL acridine orange (Sigma Aldrich) for 30 min followed by several rinses in the embryo medium, and were photographed using an LSM510 confocal microscope with version 3.5 software (Zeiss).
Microinjection of morpholinos (MOs) and mRNA
MOs were synthesized at Genetools, LLC. The nucleotide sequences of the MOs were: lrp4-sd MO, 5′-ATC AGT GTT GTT GAT CTG ACC TGC A-3′; lrp4-atgMO, 5′-ACA AAG CAC CGC AGC GAG CCT CAT C-3′; tp53MO, 5′- GCG CCA TTG CTT TGC AAG AAT TG-3′ [35], STD-MO, 5′-CCT CTT ACC TCA GTT ACA ATT TAT A-3′ [36]. A 2 mM stock solution was prepared by dissolving lyophilized MO powder in 1 × Danieau solution before further dilution into the required concentrations. Full-length of mouse Lrp4 cDNA was cloned into the pCS2+MT plasmid, and the capped mouse Lrp4 mRNA was made by mMESSAGE mMACHINE SP6 Transcription kit (Thermo Fisher Scientific). MO- and mRNA-containing solutions were injected into one- to two-cell stage embryos using a Nanoject (Drummond Scientific). tp53MO, STD-MO and mouse Lrp4 mRNA were injected at the dosage of 0.5 pmol, 1.2 pmol and 100 pg per embryo, respectively.
Patients and clinical assessment
The patient was initially diagnosed at the National Center for Diabetes, Endocrinology and Genetics in Jordan. Genomic DNA from saliva samples from the four members of the kindred were obtained after parents gave their informed consent forms and the local ethics commission gave its approval. All human studies were approved by the Review Board of Northwest University.
Mutation analysis
Direct candidate gene sequencing was performed. Primer pairs for each exon and the flanking intron regions of LRP4 were designed with Primer 3, and the primer sequences were given in Table S1. Sequence analysis was done with BigDye Terminator cycle sequencing kit and PCR products were run on ABI PRISM 3730 Analyzer (Applied Biosystems).
Plasmids
Mouse Lrp4 was subcloned into pcDNA3.1-3XFLAG from Lrp4-pcDNA3.1/V5-His-TOPO (kindly provided by Wollnik B. Laboratory). R373W, C160Y and D449N Lrp4 mutant constructs were generated by site-directed mutagenesis. Mouse Lrp6 was cloned into pcDNA3.1-3XFLAG. Mouse pUSE-Wnt1 was kindly provided by Virshup D. Laboratory. For construction of Lrp4 overexpression stable lines, mouse Lrp4 was subcloned into pCDH-CMV-MCS-EF1-Puro vector. pLP1, pLP2 and pLP-VSVG plasmids were purchase from System Biosciences (USA). Full-length zebrafish lrp4 was cloned by RACE, and in-fusion to pCS2+vector. Zebrafish fras1 plasmid was kindly provided by Carney TJ. Laboratory.
Cell culture and transfections
HEK293T cells and MC3T3-E1 cells were maintained in DMEM (Gibco, USA) supplemented with 10% FBS (Gibco, USA), streptomycin and penicillin (Gibco, USA) at 37 °C. hFOB cells were obtained from ATCC (USA) and maintained in Ham’s F12 DMEM (Gibco, USA) supplemented with 10% FBS (Gibco, USA) and 0.3 mg/ml G418 (Gibco, USA) at 33.5 °C. Cells were transfected with the use of Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions.
Luciferase assays
HEK293T cells were seeded at 700,000 cells/well in 6-well plates. To detect canonical Wnt signal pathway, transfections were performed in triplicate with the use of the TOP-Flash reporter system and the indicated expression plasmids with the following concentrations: 1000 ng wild-type (wt) Lrp4 or 1000 ng mutants, 500 ng Lrp6, 500 ng Wnt1, 200 ng Topflash Vector, 10 ng Renilla (p-RL-TK). To detect Notch signaling pathway, transfections were performed in triplicate with the use of the HES1-pGL4.10 reporter system and the indicated expression plasmids with the following concentrations: 2000 ng wild-type (wt) Lrp4 or 2000 ng mutants, 200 ng HES1-pGL4.10 Vector, 10 ng Renilla (p-RL-TK). Transfections were performed via Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. Two days after transfection, cells were lysed and luciferase activity was measured with the use of the Dual Luciferase Reporter Assay Kit (Promega, USA) and luminometer (Molecular Devices, USA). Each transfection was also measured in triplicates.
RNA interference and stable line generation
hFOB cells were seeded at 700,000 cells/well in 6-well plates and transfected via Lipofectamine 3000 (Invitrogen,USA) with two siRNAs specific for human LRP4 (100 pmol of each) or standard siRNA as a negative control. The siRNAs were purchased from GenePharma (China), and their sequences are given in Table S1. For SYBR green real-time PCR, cells were assayed 48 h after transfection. For stable line generation, HEK293T cells were seeded at 1000,000 cells per 100 mm dish, co-transfected with pLP1, pLP2, pLP-VSVG and pCDH-CMV-MCS-EF1-Puro-Lrp4-FLAG, after 72 h of transfection, the lentiviral particles were collected and concentrated. MC3T3-E1 cells were transfected with lentiviral particles, containing 5 µg/mL puromycin (Gibco, USA). Individual puromycin-resistant clone was isolated using separation rings, transferred, and sub-cultured.
Reverse transcription PCR (RT-PCR) analysis
hFOB cells, MC3T3-E1 cells and zebrafish embryos were subject to total RNA extraction using TRIzol™ reagent (Ambion, USA). 1 μg of total RNA isolated from each sample was subject to cDNA synthesis by SuperScript™ system (Invitrogen). Semi-quantitative PCR was performed using Phusion Hot Start II DNA Polymerase (Thermo Fisher Scientific) and GeneAmp PCR System 2700 (Applied Biosystems), and ImageJ software was used for densitometric analysis. Quantitative PCR was performed with SYBR Green PCR Master Mix (Kapa Biosystems, USA) and ViiA 7 Real-Time PCR System (Applied Biosystems). The quantified values were generated from the average results of three independent experiments, and the value of each experiment was from the average of triplicate PCR runs. Primers sequences are given in Table S1.
Statistical analysis
Each experiment was repeated at least three times, and the data were expressed as mean ± SE. The Student’s t test was performed for comparing whether mean values were significantly different between two sample groups, whereas analysis of variance (ANOVA) followed by Duncan’s new multiple range test (Duncan’s multiple test) was used for multiple comparisons among mean values from more than two sample groups. P < 0.05 was considered statistically significant.
Results
Identification of the lrp4 gene in the zebrafish
To investigate the LRP4 functions in vivo, we have isolated the complete cDNA sequence of lrp4 in the zebrafish. Partial genomic and cDNA sequences of the zebrafish lrp4 gene were obtained from Ensembl, and the full-length cDNA sequence was determined through a 5′-RACE method. Comparisons of the deduced amino acid sequence of the zebrafish Lrp4 to the human and mouse LRP4 sequences displayed a 77% overall amino acid identity (Fig. S1). Homology modeling of human, mouse and zebrafish proteins using SWISS-MODEL and PyMOL reveals that the structures of LDLa repeats, β-propeller domains, and EGF-like domains; where most of the known LRP4 mutations are located, are all highly similar among the three species (Fig. S2). The high amino acid sequence homology and the conserved 3D structures lead to a hypothesis that the lrp4 gene isolated from the zebrafish is a functional ortholog of the mammalian LRP4, and the biochemical properties of LRP4 are highly conserved among vertebrates.
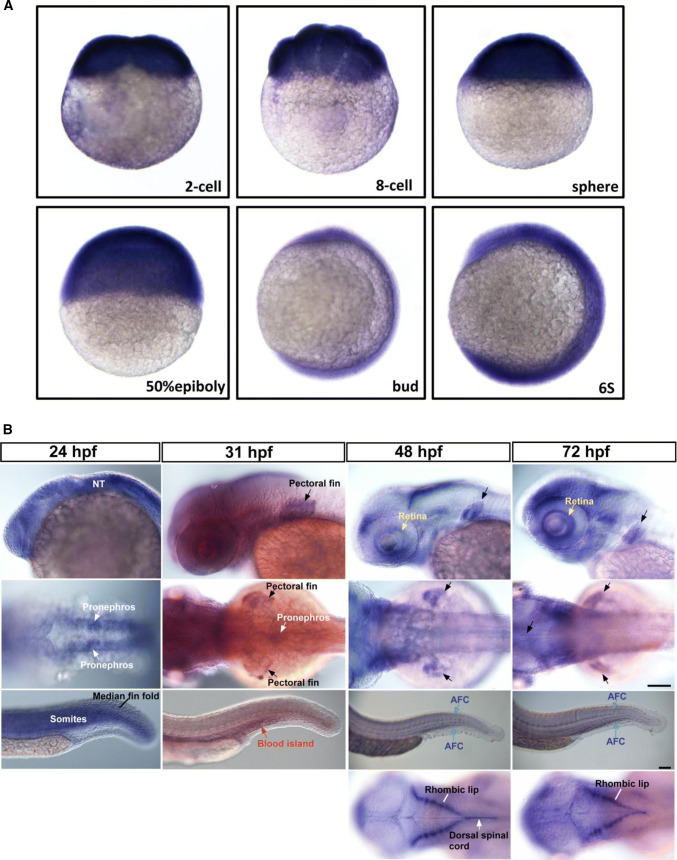
ISH analysis revealed that the expression of lrp4 mRNA was ubiquitous during stages of cleavage, gastrula and early somitogenesis (Fig. 1a), yet became restrictive from 24 hpf onwards (Fig. 1b). The lrp4 mRNA was broadly expressed at the head region at 24 hpf, and was specifically distributed at the retina and the rhombic lips from 48 to 72 hpf. An evident expression of lrp4 was detected at part of the dorsal spinal chord at 48 but not 72 hpf. At the pronephric kidney, the lrp4 mRNA was clearly detected at 24 and 31 hpf and yet undetectable from 2 dpf onwards. As the zebrafish nephron is differentiated and functionally assembled at around 2 dpf, the results in Fig. 1b indicate that lrp4 mRNA expression is activated at the kidney before the nephron differentiation. Meanwhile, the lrp4 expression was abundant at pectoral fins from 31 hpf onwards. In the posterior trunk and tail regions, the lrp4 gene showed a dynamic pattern of expression. The expression of lrp4 at somites was strong at 24 hpf and weak during later stages. lrp4 transcripts were detected at median fin fold at 24 hpf, at the posterior blood island (forming caudal vein plexus) at 31 hpf, and at the actinotrichia forming cells (AFC; essential for fin skeleton formation) from 48 to 72 hpf. The expression of lrp4 in the developing zebrafish implicates its multiple roles for tissue formation and organogenesis, and is generally consistent with those in humans and in mice.
Fig. 1.
The mRNA expression pattern of lrp4 in the developing zebrafish. a The ubiquitous expression of lrp4 mRNA during cleavage and gastrula stages. b The dynamic expression of lrp4 mRNA at multiple tissues including the neural tube (NT) and the retina at the head (first row; lateral view with anterior towards the left); pectoral fins and pronephros at the midtrunk (second row; dorsal view with anterior towards the left); the median fin fold, somites, the (posterior) blood island, and actinotrichia forming cells (AFC) in the posterior trunk and tail regions (third row; lateral view with anterior towards the left). Restrictive expression of lrp4 mRNA at rhombic lips and the dorsal spinal chord is evident from the dorsal view, starting from about 48 hpf onwards (last row). The ISH results shown are representative of at least 30 embryos for each stage
Fin and kidney phenotypes in the zebrafish lrp4 morphant
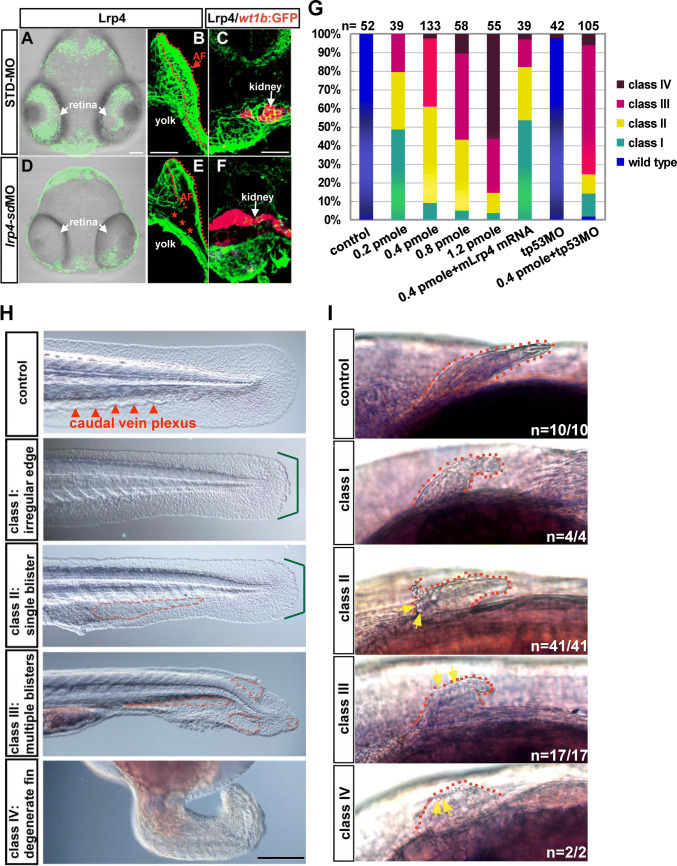
To model the disorders caused by a loss of LRP4 function, we performed lrp4 MO injections in developing zebrafish embryos. The antisense MO was designed against the exon 15-intron 15 junction of the lrp4 pre-mRNA. Injections of the lrp4 splice-donor MO (lrp4-sdMO) into embryos specifically blocked the pre-mRNA splicing of the lrp4 gene (Fig. S3A). Increasing dosages of lrp4-sdMO led to dose-dependent decrease in wild-type lrp4 mRNA levels, as estimated by semi-quantitative RT-PCR analysis (Fig. S3b). On the other hand, quantitative RT-PCR (qRT-PCR) analysis using the intron-15 specific primer to detect expression levels of the mis-spliced product displayed an increase of mis-splicing from 0.2 to 0.4 pmol/embryo, yet essentially no significant difference among the dosage groups of 0.4, 0.6 and 1.2 pmol/embryo (Fig. S3C). In STD-MO but not lrp4-sdMO injected embryos, IHC analysis revealed the localization of Lrp4 protein in the retina, the inside of apical folds and the proximal region of the pectoral fin, as well as the kidney (Fig. 2a–f). Although a decrease of lrp4 mRNA was evident in the lrp4-sd morphant as early as 10 hpf (Fig. S3a, b), no noticeable phenotype could be detected until 24 hpf. At 24 hpf, all of the lrp4-sd morphants displayed very mild dysmorphogenesis at the median fin fold and the caudal vein plexus (Fig. S4a). For the dosages higher than 0.2 pmol/embryo, the majority of lrp4-sd morphants displayed either single or multiple blisters at the median fin fold at 2 dpf. Therefore, we classified the lrp4-sd morphants into four categories according to the severity of median fin fold phenotypes (Fig. 2h). Moreover, the lrp4-sdMO caused a dose-dependent effect on the median fin fold, ranging from irregular edge to severe degeneration, which could be alleviated by a co-injection of mouse Lrp4 mRNA (Fig. 2g, h). A co-injection of tp53MO did not rescue the blistering phenotypes (Fig. 2g), and no apoptotic cells were detected at the median fin fold of the lrp4-sd morphant (Fig. S4c), testifying that the blistering phenotype was not due to an off-targeting effect mediated through p53 activation. An analysis of live lrp4-sd morphants revealed that the single blister phenotype (class II) was focused on the caudal vein plexus where the proliferation of hematopoietic stem cells occurs. Blisters in the class III embryos were distributed at both the caudal vein plexus and the posterior median fin fold (Fig. S4B). The results in Fig. 2g, S3b, S3c and S4b suggest that the dose-dependent effect of the lrp4-sdMO on posterior trunk and tail phenotypes are closely correlated with the extent of lrp4 mRNA knockdown. In addition, all of the lrp4-sd morphants examined displayed dysmorphogenesis of the pectoral fin (Fig. 2i), and pectoral fin blistering were exhibited by class II-IV morphants.
Fig. 2.
Phenotypes of the lrp4-sd morphant at the median fin fold and the pectoral fin. The expression of Lrp4 protein at the retina (a, d), pectoral fins (b, e) and the kidney (c, f) was compared between the STD-MO and lrp4-sdMO injected embryos. IHC analysis was performed on vibratome sections of 50 hpf Tg(wt1b:GFP) embryos injected with STD-MO (a–c) or lrp4-sdMO (0.4 pmol/embryo) (d–e) to detect the Lrp4 expression (green fluorescence; pseudo-color), while the wt1b:GFP was shown as red fluorescence (pseudo-color). The apical fold (AF) of the pectoral fin is highlighted by red broken lines. The absence of Lrp4 protein in the proximal region of the pectoral fin in the lrp4-sd morphant is indicated by red stars. The images of STD-MO and lrp4-sdMO injected embryos are representative of 4 samples for each treatment. glrp4-sdMO led to a dose-dependent effect in the posterior trunk and tail regions, and the morphant phenotype was rescued by co-injected mouse Lrp4 mRNA but not by tp53MO. h Classification of the lrp4-sd morphants is based on the phenotype of irregular fin edges (bracketed in blue) and formation of blisters (highlighted by broken red lines) as observed by Nomarski microscopy. i All classes of lrp4-sd morphants displayed small pectoral fins with irregular edges, and blisters (yellow arrows) were detected at the edge of pectoral fins of class II-IV morphants. Scale bar: 100 μm
To further verify the results of the lrp4-sdMO knockdown, an ATG morpholino (lrp4-atgMO) was utilized for phenotypic analysis. The results in Fig. S5a showed that the lrp4-atgMO caused essentially the same blistering phenotypes at the caudal vein plexus and the median fin fold, albeit with lower penetrance and expressivity.
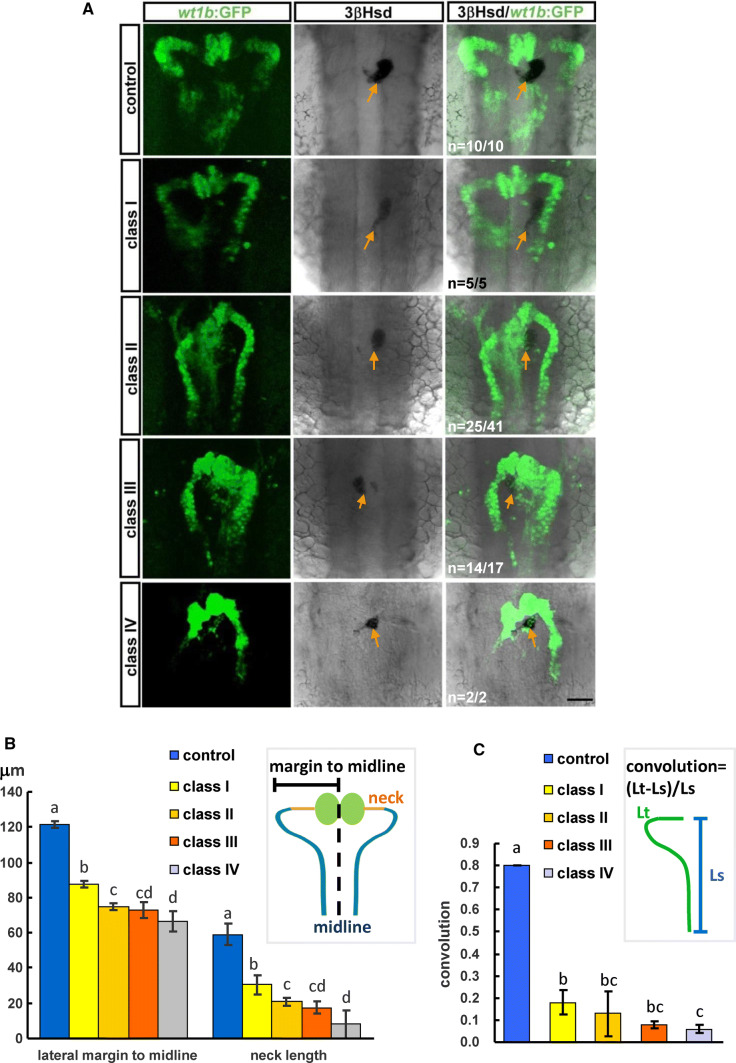
To know whether a loss of lrp4 function in the zebrafish affects the pronephric kidney development, we analyzed the kidney morphologies in lrp4-sd morphants which were classified into classes I to IV according to caudal fin phenotypes as described in Fig. 2h (Fig. 3a). lrp4-sdMO injections were performed on Tg(wt1b:GFP) embryos where developing kidney cells were marked by GFP, and the morphants were subjected to 3β-Hydroxysteroid dehydrogenase (3β-Hsd) activity staining for co-labeling steroidogenic interrenal tissues, the teleostean counterpart of the mammalian adrenal cortex. The kidney and interrenal morphologies were analyzed at 50 hpf when the pronephric nephron starts to function. The nephron morphology, as assessed by the distance from the lateral margin of proximal convoluted tubule to the midline, the length of pronephric neck segment, as well as the degree of convolution, was significantly compromised in all classes of lrp4-sd morphants (Fig. 3b, c). While kidney and interrenal cells were well segregated in all control embryos, the wt1b:GFP was detected in the steroidogenic interrenal tissues in 61.0% (n = 41) of lrp4-sd class I morphants and 82.4% of class II morphants (n = 17), indicating a defective segregation between kidney and steroidogenic interrenal cells (Fig. 3a).
Fig. 3.
The phenotype of kidney and interrenal tissue in the lrp4-sd morphant. a The kidney morphology was delineated by the wt1b:GFP, and the steroidogenic interrenal cells were detected by 3β-Hsd activity staining (orange arrows). b The pronephric structure was quantified by measuring the average distance from bilateral margins to the midline, and the average length of bilateral pronephric necks. c The pronephric convolution was arbitrarily defined as the ratio of the length of the proximal pronephric tubules marked by the wt1b:GFP (Lt) minus the length of a straight line connecting the ends of this segment (Ls) divided by the length of this straight line. Histograms with different letters above them are significantly different (ANOVA and Duncan’s multiple test, P<0.05)
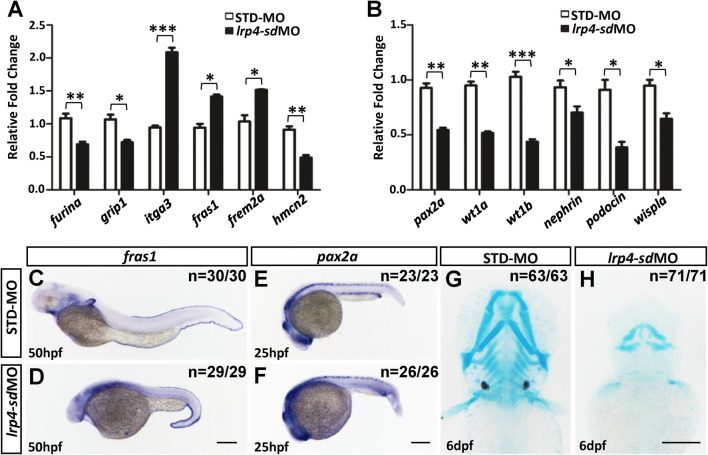
Furthermore, the expressions of fin and kidney markers were significantly changed as detected by real-time qRT-PCR analysis in lrp4 morphants (Fig. 4a, b). By 50 hpf, the expression of fras1, the zebrafish orthologue of the human Fraser syndrome gene FRAS1 whose mutation leads to similar epidermal blistering, was increased in the median fin fold in lrp4-sd morphants as compared to control embryos (Fig. 4c, d). On the other hand, pax2a expression in pronephric duct and proctodeum was reduced in lrp4-sd morphants, which indicated a kidney malformation by loss of lrp4 function in zebrafish (Fig. 4e, f). By 6 dpf, lrp4-sd morphants exhibited major skeletal anomalies by Alcian blue staining (Fig. 4g, h). Thus, the fin, kidney and skeletal phenotypes observed upon knockdown of lrp4 in zebrafish partially phenocopy the human LRP4 mutations, revealing a conserved role of LRP4 in fin/limb development, kidney morphogenesis and skeletogenesis.
Fig. 4.
lrp4 knockdown in zebrafish embryos affects gene expressions in the skeletal and kidney development. The expressions of fin (a) and kidney (b) markers, as determined by qRT-PCR, were significantly changed in lrp4-sd morphants at 2 dpf. The results of qRT-PCR were the average of triplicate experiments using 10 embryos in each treatment group for one RT reaction. c STD-MO injected control and dlrp4-sd morphant embryos at 50 hpf stained for the fin marker fras1. Note the enhanced expression in the median fin fold of lrp4-sd morphants. e STD-MO injected control and flrp4-sd morphant embryos at 25 hpf stained for the kidney marker pax2a. Note the reduced expression in the pronephric duct and the proctodeum of lrp4-sd morphants. Alcian blue staining showed severe skeletal anomalies in lrp4-sd morphants (h) compared with STD-MO-injected control (g) embryos at 6 dpf. Scale bar: 200 μm. Values in a and b represent mean ± SE of data from three independent experiments, *P < 0.05, **P < 0.01, and ***P < 0.0001 (Student’s t test)
Loss of Lrp4 function leads to upregulated Wnt and Notch signalings in the zebrafish
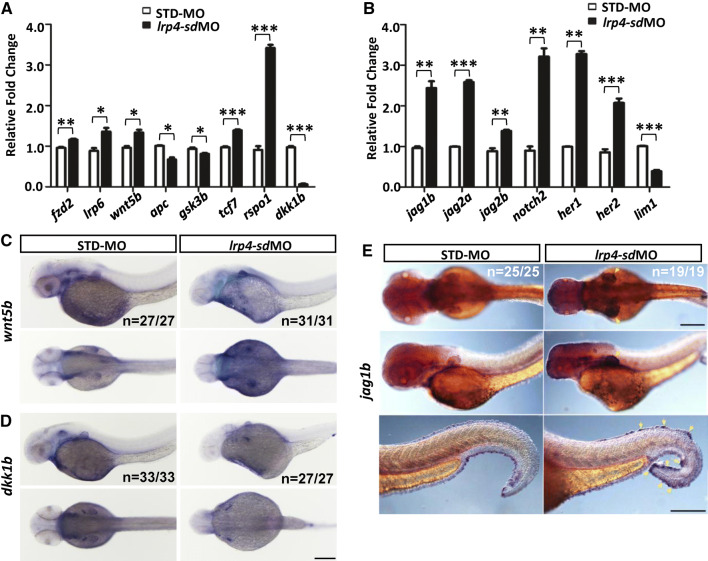
To check whether Lrp4 modulates Wnt/β-catenin signaling in the zebrafish, we analyzed the expression of Wnt/β-catenin pathway components by qRT-PCR (Fig. 5a). The expressions of many Wnt pathway components were increased in lrp4-sd morphants, remarkably labeled by Wnt receptor fzd2, co-receptor lrp6, ligand wnt5b, transcription factor tcf7 and regulator rspo1. In contrast, the expression of Wnt inhibitor dkk1b was dramatically decreased. By 50 hpf, ISH analysis presented that the expression of wnt5b was increased in pectoral fin buds in lrp4-sd morphants (Fig. 5c). Meanwhile, dkk1b expression was visibly dampened in pectoral fin buds in lrp4-sd morphants compared to those of control embryos (Fig. 5d).
Fig. 5.
The activation of Wnt and Notch pathways in lrp4-sd morphants. The expression of genes which are components of Wnt pathway (a) or Notch pathway (b) was quantitatively analyzed by qRT-PCR for lrp4 morphants (black bar) and STD-MO injected control embryos (white bar) at 2 dpf. The results of qRT-PCR were the average of triplicate experiments using 10 embryos in each treatment group for one RT reaction. ISH analysis of wnt5b (c), dkk1b (d) and jag1b (e) expressions was performed on the lrp4-sd morphant and STD-MO injected control embryos. Scale bar: 200 μm. Values in a and b represent mean ± SE of data from three independent experiments, *P < 0.05, **P < 0.01, and ***P < 0.0001 (Student’s t test)
The loss of lrp4 function in the zebrafish led to a kidney phenotype reminiscent of those resulted from defective Jagged–Notch signaling (Fig. 3a) [37]. A disruption of Jagged and Notch molecules has been shown to cause defective limb development in human and mice [38–40]. It is, therefore, of interest to examine whether the Jagged–Notch pathway is affected by a loss of lrp4. We discovered by ISH that the expression of jag1b demonstrates an evident and tissue-specific increase at the pectoral fin and the median fin fold (Fig. 5e, yellow arrows). Concomitant with the upregulated jag1b expression detected by ISH, qRT-PCR analysis of lrp4-sd morphants also exhibited significantly increased expression levels of Notch ligands jag1b, jag2a and jag2b; Notch receptor notch2; as well as Notch effectors her1 and her2, indicating upregulated Notch signaling upon Lrp4 deficiency (Fig. 5b). In contrast, the expression of lim1, a target of Notch signaling whose expression is repressed by the Notch pathway, demonstrated an evident decrease in the lrp4-sd morphant and further validated the activated Notch signaling. These results suggest that the Notch activity is negatively regulated by Lrp4 in the zebrafish. Expression levels of jag1b, jag2a and her1 were also significantly elevated in the lrp4-atg morphant, however, with a less marked increase (Fig. S5B). The expression of jag2b, notch2 and her2 was activated by the lrp4-sdMO but not by the lrp4-atgMO, and the limb1 expression was not suppressed by lrp4-atgMO. A comparison between the effects of lrp4-sdMO and lr4-atgMO implicates that the extent of Notch pathway activation may be correlated with the severity of lrp4 knockdown phenotype.
A novel missense mutation in the 1st EGF-like domain of LRP4 represses canonical Wnt signaling activity
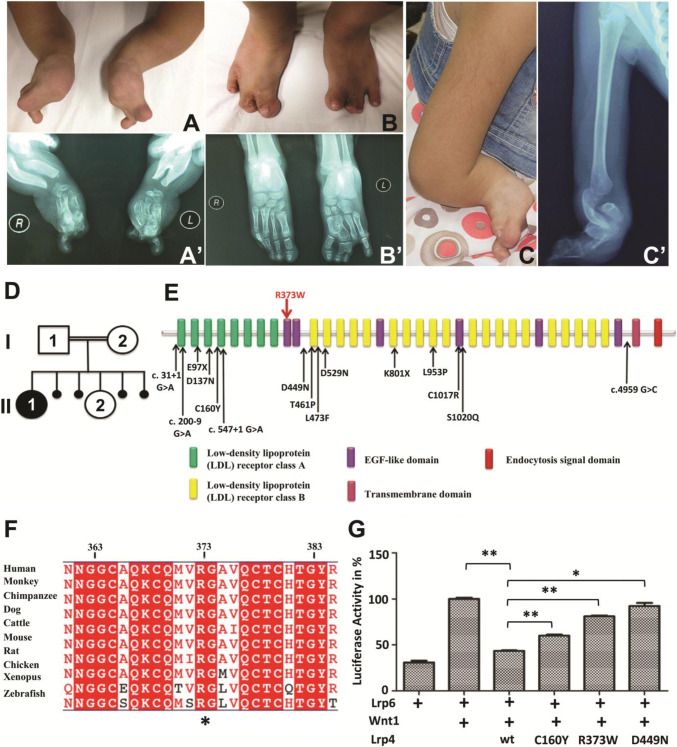
While the Jagged–Notch signaling pathway was found to be antagonized by Lrp4 in the zebrafish, we sought to investigate whether the mechanism is relevant to the human LRP4 mutation. The patient from a consanguineous Jordanian family was examined and presented with a typical expression of CLS clinical symptoms, mainly including short forearms, radius-ulnar synostosis, syndactyly of hands and feet, disorganized metacarpal and phalangeal bones, nail aplasia, as well as vertebral anomalies. The lower extremities were less severely affected than upper limbs (Fig. 6a–c′). Renal agenesis, presenting with absence of right kidney and hypertrophy of left kidney, was also observed (Data not shown). The index patient was subjected to Sanger sequencing of LRP4, and a novel missense mutation c.1117C > T (p.R373W) was detected. This variant was observed in a homozygous state in the index patient while heterozygous in her parents and the unaffected sibling (Fig. 6d). The variant was not found after being further screened in at least 100 healthy control individuals. This missense mutation is located in the 1st extracellular EGF-like domain of LRP4 (Fig. 6e), highly conserved across various species (Fig. 6f).
Fig. 6.
Phenotypic characteristics of investigated patient with typical Cenani-Lenz syndactyly syndrome caused by a point mutation in LRP4. The picture (a) and X-ray radiograph (a′) of patient’s hands, the picture (b) and X-ray radiograph (b′) of patient’s feet, showing syndactyly, disorganized metacarpal and phalangeal bones. The picture (c) and X-ray radiograph (c′) of patient’s right upper limb, showing short forearms and radius-ulnar synostosis. d The patient, female (II:1), was born to first cousin parents (I:1 and I:2) with one unaffected child (II:2) and four abortions. e Schematic view of LRP4 domains and localization of identified CLS mutations, where the novel mutation R373W is labeled by the red arrow and other known mutations labeled by black arrows. f LRP4 is highly conserved across different vertebrate species; the conserved R373 is highlighted with a star (*). g The effect of R373W mutation and LRP4 knockdown on the Wnt signaling in vitro. TOP-Flash luciferase assay was performed to quantify the Wnt activity in HEK293T cells co-transfected with Lrp6, Wnt1, and the indicated Lrp4 cDNA. Values represent mean ± SE of data from three independent experiments, *P < 0.05, **P < 0.01. (Student’s t test)
To examine whether the R373W mutation leads to loss of LRP4 function, we performed TOP-Flash luciferase assay to analyze the effect of R373W mutation on the activation of canonical Wnt signaling. Similar to C160Y and D449N missense mutations [16], R373W abolished the antagonistic LRP4 effect on LRP6-mediated activation of Wnt/β-catenin signaling (Fig. 6g), which could be due to the failure of mutant LRP4 to be efficiently transported to the plasma membrane. The effect of R373W missense mutation, along with the siRNA knockdown, was then utilized for studying how loss of LRP4 function affects Jagged–Notch signaling.
Jagged–Notch signaling is activated by loss of LRP4 function and repressed by LRP4 overexpression
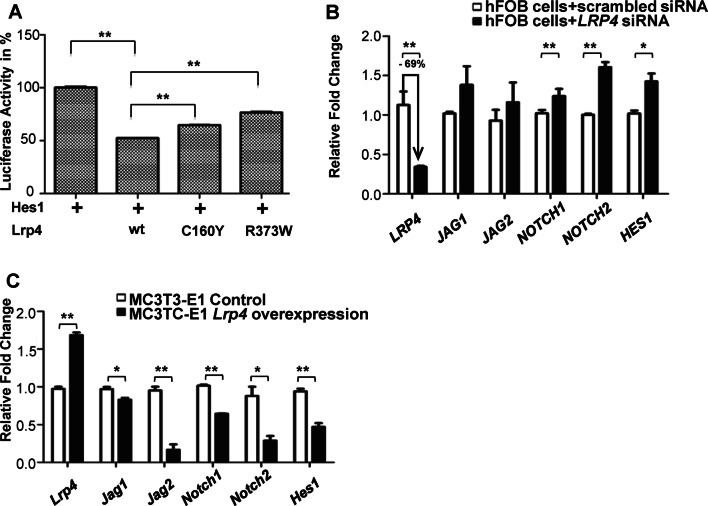
To investigate whether the Notch activity was negatively regulated by the LRP4-mediated signaling in mammalian cells, we examined how wild-type and mutant forms of mouse Lrp4 affected the Notch pathway by a dual luciferase reporter assay in HEK293T cells. We found that Lrp4 was able to antagonize the Notch activation. In contrast, coexpression of Lrp4 C160Y or R373W mutant with the reporter of Notch effector Hes1 abolished the antagonistic effect of Lrp4 on Notch signaling (Fig. 7a). Analysis of siRNA knockdown of human LRP4 in hFOB cells exhibited markedly increased NOTCH signaling by the expression of NOTCH1, NOTCH2 and HES1 (Fig. 7b). Consistent with the upregulation of Notch signaling by LRP4 knockdown in hFOB cells, the expression of Notch signaling pathway components was significantly reduced in mouse Lrp4-overexpressing stable MC3T3-E1 cells (Fig. 7c). Taken together, our results from zebrafish, human and mouse cells show that both Wnt/β-catenin and Notch pathways were activated upon the LRP4 deficiency, suggesting a cross talk between the two pathways.
Fig. 7.
Loss of LRP4 results in activation of Notch signaling. a Dual luciferase reporter assay to quantify the Notch signaling activity as Hes1 was coexpressed with the indicated Lrp4 cDNA in HEK293T cells. bLRP4 siRNA in hFOB cells achieved 69% knockdown with upregulated gene expressions of NOTCH signaling components. c Gene expressions of Notch signaling components were downregulated when Lrp4 was overexpressed stably in MC3T3-E1 cells. Values in a–c represent mean ± SE of data from three independent experiments, *P < 0.05, **P < 0.01. (Student’s t test)
Discussion
The analysis of sequence, expression and phenotype of the zebrafish lrp4 gene in this study supports that the structure and function of LRP4 are conserved between fish and mammals. The knockdown of lrp4 mRNA in the lrp4-sd morphant (Fig. S3A,B) resembles the severe yet incomplete reduction of Lrp4 mRNA in the Lrp4dan/dan mouse mutant [13]. However, Lrp4dan/dan exhibits syndactyly but no noticeable kidney agenesis. The major mis-spliced product in the lrp4-sd morphant (Fig. S3a, c), if successfully translated, is expected to result in one substitution at the end of 1st β-propeller domain (G701Q) followed by 26 incorrect amino acids, leading to a premature stop codon. It cannot be ruled out that the putative truncated protein resulted from the mis-spliced product may also play a role for the lrp4-sd morphant phenotype. The mouse Lrp4 mitt allele that contains an early stop mutation at the 1st EGF-like domain (Q377 stop) demonstrates phenotypes including syndactyly, kidney agenesis and early death [14]; which are more severe than those caused by the Lrp4 dan allele. In human, a homozygous nonsense mutation at the very beginning of the large extracellular domain (p.E97X) leads to severe CLS phenotypes including renal hypoplasia/agenesis [41]. Compound heterozygosity of a nonsense allele (p.K801X) and a frameshift allele (p.S1020Qfs*27) results in a prenatal lethal form of CLS [42]. These studies support that the truncating mutations of LRP4 cause more severe phenotypes. It is possible that the fin and kidney phenotypes in the lrp4-sd morphant are resulted from combinatorial effects of lrp4 mRNA reduction and mis-spliced RNA product, although the effect of the mis-spliced RNA product may not be MO dose-dependent (Fig. 3c). Characterization of the lrp4 gene in the zebrafish led us to discover that lrp4 acted as an antagonist of Jagged–Notch signaling during development. This finding in fish has further been confirmed by the studies of human missense LRP4 mutations, knockdown of LRP4 by siRNAs, and overexpression of Lrp4. The Notch activity, quantified by the HES1 reporter assay in embryonic kidney cells, was repressed by LRP4, and the repressing effect was partially abrogated by R373W and C160Y mutations. Through siRNA knockdown and lentivirus-mediated stable expression of Lrp4 in osteoblasts, both Wnt/β-Catenin and Jagged–Notch signaling pathways were found to be activated by a deficiency of LRP4 and suppressed upon the overexpression of Lrp4. The experiments in human and mouse osteoblast cells, as well as those in human embryonic kidney cells, therefore, testified the inhibitory action of LRP4 upon Jagged–Notch signaling activity in mammals.
While this study suggests that LRP4 is required for negatively regulating the Jagged–Notch pathway, it is intriguing to ask the biological importance of this mechanism during fin/limb development. In mammalian osteoblastogenesis and chondrogenesis, the Notch signaling activity serves to maintain stemness of progenitors by inhibiting osteoblast or chondrogenic cell proliferation [40, 43]. Consistently, in the regenerating adult zebrafish fins, the Notch signaling activity promotes proliferation of undifferentiated precursor cells in the blastema and prevents osteoblast differentiation. Therefore, in the skeletal development of mammals and fish, the Notch signaling activity is critical for maintaining a balance between self-renewal and differentiation. In this regard, the function of LRP4 may serve to ensure osteoblast differentiation in the developing fins/limbs. Interestingly, the upregulation of Jagged–Notch signaling in the lrp4-sd morphant is reminiscent of the knockdown phenotype of chsy1, a zebrafish orthologue of human Temtamy preaxial brachydactyly syndrome (MIM 605282) [3], implicating that aberrantly upregulated Jagged–Notch signaling is an important pathogenic mechanism for limb/fin morphogenetic disorders.
It is possible that different mutation alleles of LRP4 exert differential effects on the balance of Jagged–Notch signaling, thus leading to distinct bone phenotypes including syndactyly, sclerosteosis, and osteoporosis. Syndactyly, sclerosteosis and osteoposis are all known to be associated with dysregulations of Notch signaling whose strength and activity levels are critical for skeletal development and bone remodeling. For example, Notch gain of function in the osteoblasts of transgenic mice leads to increased number of osteoblasts and massive sclerosteosis [44]. Osteoporosis is manifested in Hajdu-Cheney Syndrome due to a sustained activation of NOTCH2 activity [45], and JAG1 polymorphisms are correlated with risk of osteoporotic fracture in several populations [46]. Mutations of Jag2 cause syndactyly in mice [39]. It remains unknown how and through which domain LRP4 modulates the Jagged–Notch activity, and whether relative abundance of Notch pathway components in various tissues could partially account for the distinct phenotypes caused by LRP4 mutations. As the expression of Jag1b was significantly increased in the zebrafish lrp4-sd morphant (Fig. 5e), it is possible that the Jagged–Notch signaling may function downstream of the canonical Wnt pathway in the zebrafish embryo. However, it may not be ruled out that the Notch pathway and Canonical Wnt pathways could also be modulated in a parallel manner, either directly or indirectly, by LRP4. It will be of interest to explore whether different LRP4 alleles, that cause distinct disease phenotypes and implicated in different pathways, would differentially affect the gene expression and function of the Jagged–Notch pathway.
We also provide evidence to support that abnormally increased activity of the Jagged–Notch signaling may, at least partially, explain the often association between limb and kidney disorders in the human LRP4 mutation. Upregulated expressions of Jagged–Notch signaling pathway components in either kidney podocytes or tubules have been implicated in diabetic nephropathy, folic acid-induced kidney injury and focal segmental glomerulosclerosis [47, 48]. While Jagged2 (Serrate2) mutant mice exhibit syndactylism due to defects in limb-bud patterning and interdigital apoptosis [38, 39], knockdown of either jagged2 or notch3 in the zebrafish disrupts development of the pronephric duct [49, 50]. These reports and our study together point out that precise control of Jagged–Notch signaling is essential to prevent limb defects and kidney anomalies.
The dysmorphogenesis of fin and caudal vein plexus in the lrp4 zebrafish morphant highly resembles that in the reported zebrafish mutations of human Fraser syndrome (MIM 219000) [4], indicating a link between lrp4 and Fraser syndrome genes for the extracellular matrix patterning. Mutations of large basement membrane protein-encoding genes including FRAS1 (MIM 607830), FREM1 (MIM 608944) or FREM2 (MIM 608945) lead to the Fraser syndrome in humans, and a blebbed phenotype in mice; both are characterized by syndactyly and bilateral renal agenesis [51, 52]. Mutations of Fraser genes in the zebrafish result in fin blistering which is due to a disruption of basement membranes [4]. The high phenotypic similarity between lrp4 morphants and Fraser syndrome gene mutations in zebrafish, therefore, suggests that basement membrane formation and function during fin/limb and kidney development might be regulated by Wnt/β-Catenin and/or Jagged–Notch signaling pathways. Although the expression of Fraser syndrome genes fras1 and frem2 were mildly upregulated upon the knockdown of lrp4 in zebrafish (Fig. 4a), it remains unclear whether there is any direct regulatory relationship between Wnt/Notch signaling and Fraser syndrome gene expressions. Unexpectedly, the swelling and abnormal blood accumulation at the caudal vein plexus was found in lrp4 morphants (Fig. S4B), suggesting the importance of lrp4 during blood vessel formation that is difficult to be observed in mammals. It remains to be explored whether the blood vasculature phenotype was due to defective regulation of Wnt/β-catenin or Jagged–Notch signaling.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1. Alignment of human, mouse and zebrafish LRP4 amino acid sequences. The zebrafish sequence lacks 39 amino acid residues at the C-terminal region. (PDF 982 kb)
Figure S2. Homology modeling of human, mouse and zebrafish LRP4 proteins. Template-based structure prediction of LRP4 protein structures was performed by SWISS-MODEL, and the predicted 3D structures from three species were superimposed by PyMOL. The templates used for modeling each region are: Region I, Low-density lipoprotein receptor (1n7d.1.A); Region II and III, Low-density lipoprotein receptor-related protein 6 chain A (5gje.1.A). AA, amino acids. (PDF 551 kb)
Figure S3. The effect of lrp4-sdMO on the RNA splicing of lrp4. (A) RT-PCR analysis was used to examine whether the lrp4-sdMO affects the splicing of intron 15 of lrp4 pre-mRNA. The primer set was designed to amplify the cDNA sequence spanning through the junction between exons 15 and 16. The exon–intron organization was predicted by BLASTing the lrp4 cDNA sequence to GRCz10 (Ensembl). Retention of partial intron 15 of the lrp4 pre-mRNA sequence (#2 band of RT-PCR products) was detected in the lrp4 morphant at 10 hpf, which is predicted to cause an in-frame early stop codon (highlighted in light blue). (B) Increasing doses of lrp4-sdMO led to reductions of lrp4 mRNA expression as analyzed by semi-quantitative RT-PCR. The gel picture shown is a representative of triplicate experiments. The intensity of #1 band from the gel picture was subject to densitometric analysis (lower panel) for semi-quantitative measurements of wild-type lrp4 mRNA expressions. The average of three separate experiments shows a significant dose-dependent decrease of #1 band product from 0.2 to 1.2 pmol of lrp4-sdMO injections. Apart from the #2 band of mis-spliced product, one more mis-spliced product was present in trace amounts (indicated by yellow star). (C) Quantitative analysis of mis-spliced products by qRT-PCR using the intron-15 specific primer. A representative gel picture of the mis-spliced product and its housekeeping control (eelf1a) from qRT-PCR is shown in the lower panel. The results of semi-quantitative RT-PCR in (B) and qRT-PCR in (C) were the average of triplicate experiments using 10 embryos in each treatment group for one RT reaction. *P<0.05. (Student’s t test) (PDF 579 kb)
Figure S4. The posterior trunk and tail phenotypes of lrp4-sd morphants. (A) Injections of lrp4-sd MO (0.4 pmol/embryo), tp53MO (0.5 pmol/embryo) and lrp4-sdMO/tp53MO led to mild dysmorphogenesis in the posterior trunk and tail regions at 24 hpf. (B) Abnormal accumulation of blood cells (red arrows) at the caudal vein plexus in the class I to III of lrp4-sd morphants was captured by live imaging. (C) Acridine orange staining revealed apoptotic cells (orange arrows) in the tail region of the control embryo, which were apparently reduced in the tp53 morphant; whereas no apoptotic cells were detected in either lrp4-sd morphants or lrp4-sd/tp53 double morphants. Scale bar: 100 μm. (PDF 373 kb)
Figure S5. The effect of lrp4-atgMO on the median fin fold phenotype and the Notch pathway. (A) lrp4-sdMO led to blistering phenotype in the caudal vein plexus and the median fin fold. Phenotypic classification is according to what is described in Fig. 2H. (B) The expression of genes which are components of Notch pathway was analyzed by qRT-PCR for lrp4-atg morphants and STD-MO injected control embryos at 2 dpf. The results were the average of triplicate experiments using 10 embryos in each treatment group for one RT reaction. ***P<0.0001 (Student’s t test). (PDF 212 kb)
Acknowledgements
We are indebted to the family for kindly partaking in this study. We are grateful to Prof. Bernd Wollnik, Dr. Thomas J. Carney and Prof. David Virshup for the kind provision of plasmids. We also thank Prof. Baojie Li for the kind gift of MC3T3-E1 cell line; Prof. Christoph Englert for the kind gift of the Tg(wt1b:GFP)(line 1) zebrafish strain; Dr. Xingang Wang and Ms. Pang Zhan for assistance on gene cloning; Mr. Kuan-Chieh Wang for statistical help; Ms. Wei-Ru (Lydia) Hsiao and Chia-Yu Chang for aquarium care; the Taiwan Zebrafish Core Facility at NHRI (TZCF@NHRI), the Taiwan Zebrafish Core Facility at Academia Sinica (TZCAS) and Northwest University Zebrafish Core Facility for assistance with fish culture.
Authors’ contributions
JT and Y-WL conceived the study, designed the experiments and prepared the manuscript. JS, CL, H-YH, C-WC, GL, YK, Y-HC, M-JC, ZL, W-LC, Y-FC and Y-HS prepared the samples, performed the experiments and analyzed the data. MS, ME-K, and OQS diagnosed the patient.
Funding
This work was supported by Natural Science Foundation of Shaanxi Province, China (2016JM3018); Opening Foundation of State Key Laboratory of Freshwater Ecology and Biotechnology, China (2018FB10); Ministry of Science and Technology, Taiwan (MOST) 106-2313-B-029-002-MY3, 105-2313-B-029-002 and 102-2628-B-029-002-MY3.
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interests.
Contributor Information
Jing Tian, Phone: 86-29-88302339, Email: tianjing@nwu.edu.cn.
Yi-Wen Liu, Phone: 886-4-2359-0121, Email: dlslys@thu.edu.tw.
References
- 1.Nakamura T, Gehrke AR, Lemberg J, Zymaszek JS, Shubin NH. Digits and fin rays share common developmental histories. Nature. 2016;537:225. doi: 10.1038/nature19322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grandel H, Schulte-Merker S. The development of the paired fins in the zebrafish (Danio rerio) Mech Dev. 1998;79:99–120. doi: 10.1016/S0925-4773(98)00176-2. [DOI] [PubMed] [Google Scholar]
- 3.Tian J, Ling L, Shboul M, et al. Loss of CHSY1, a secreted FRINGE enzyme, causes syndromic brachydactyly in humans via increased NOTCH Signaling. Am J Hum Genet. 2010;87:768–778. doi: 10.1016/j.ajhg.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carney TJ, Feitosa NM, Sonntag C, et al. Genetic analysis of fin development in zebrafish identifies furin and hemicentin 1 as potential novel fraser syndrome disease genes. PLoS Genet. 2010;6:e1000907. doi: 10.1371/journal.pgen.1000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 6.Gerlach GF, Wingert RA. Kidney organogenesis in the zebrafish: insights into vertebrate nephrogenesis and regeneration. Wiley Interdiscip Rev Dev Biol. 2013;2:559–585. doi: 10.1002/wdev.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cenani A, Lenz W. Total syndactylia and total radioulnar synostosis in 2 brothers. A contribution on the genetics of syndactylia. Z Kinderheilkd. 1967;101:181–190. doi: 10.1007/BF00438491. [DOI] [PubMed] [Google Scholar]
- 8.Leupin O, Piters E, Halleux C, et al. Bone overgrowth-associated mutations in the LRP4 gene impair sclerostin facilitator function. J Biol Chem. 2011;286:19489–19500. doi: 10.1074/jbc.M110.190330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohkawara B, Cabrera-Serrano M, Nakata T, et al. LRP4 third beta-propeller domain mutations cause novel congenital myasthenia by compromising agrin-mediated MuSK signaling in a position-specific manner. Hum Mol Genet. 2014;23:1856–1868. doi: 10.1093/hmg/ddt578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivadeneira F, Styrkarsdottir U, Estrada K, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199-U1158. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pevzner A, Schoser B, Peters K, et al. Anti-LRP4 autoantibodies in AChR- and MuSK-antibody-negative myasthenia gravis. J Neurol. 2012;259:427–435. doi: 10.1007/s00415-011-6194-7. [DOI] [PubMed] [Google Scholar]
- 12.Johnson EB, Hammer RE, Herz J. Abnormal development of the apical ectodermal ridge and polysyndactyly in Megf7-deficient mice. Hum Mol Genet. 2005;14:3523–3538. doi: 10.1093/hmg/ddi381. [DOI] [PubMed] [Google Scholar]
- 13.Simon-Chazottes D, Tutois S, Kuehn M, et al. Mutations in the gene encoding the low-density lipoprotein receptor LRP4 cause abnormal limb development in the mouse. Genomics. 2006;87:673–677. doi: 10.1016/j.ygeno.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Weatherbee SD, Anderson KV, Niswander LA. LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development. 2006;133:4993–5000. doi: 10.1242/dev.02696. [DOI] [PubMed] [Google Scholar]
- 15.Tanahashi H, Tian QB, Hara Y, Sakagami H, Endo S, Suzuki T. Polyhydramnios in Lrp4 knockout mice with bilateral kidney agenesis: defects in the pathways of amniotic fluid clearance. Sci Rep. 2016;6:20241. doi: 10.1038/srep20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Pawlik B, Elcioglu N, et al. LRP4 mutations alter Wnt/beta-catenin signaling and cause limb and kidney malformations in Cenani-Lenz syndrome. Am J Hum Genet. 2010;86:696–706. doi: 10.1016/j.ajhg.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan TN, Klar J, Ali Z, Khan F, Baig SM, Dahl N. Cenani-Lenz syndrome restricted to limb and kidney anomalies associated with a novel LRP4 missense mutation. Eur J Med Genet. 2013;56:371–374. doi: 10.1016/j.ejmg.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Tamai K, Semenov M, Kato Y, et al. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 19.Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 20.Kim N, Stiegler AL, Cameron TO, et al. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell. 2008;135:334–342. doi: 10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Luo SW, Wang Q, Suzuki T, Xiong WC, Mei L. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60:285–297. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi HY, Dieckmann M, Herz J, Niemeier A. Lrp4, a novel receptor for Dickkopf 1 and Sclerostin, Is expressed by osteoblasts and regulates bone growth and turnover In vivo. PLoS One. 2009;4:e7930. doi: 10.1371/journal.pone.0007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn Y, Sims C, Logue JM, Weatherbee SD, Krumlauf R. Lrp4 and Wise interplay controls the formation and patterning of mammary and other skin appendage placodes by modulating Wnt signaling. Development. 2013;140:583–593. doi: 10.1242/dev.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karner CM, Dietrich MF, Johnson EB, et al. Lrp4 regulates initiation of ureteric budding and is crucial for kidney formation—a mouse model for Cenani-Lenz syndrome. PLoS One. 2010;5:e10418. doi: 10.1371/journal.pone.0010418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi HY, Liu Y, Tennert C, et al. APP interacts with LRP4 and agrin to coordinate the development of the neuromuscular junction in mice. Elife. 2013;2:e00220. doi: 10.7554/eLife.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohazama A, Johnson EB, Ota MS, et al. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One. 2008;3:e4092. doi: 10.1371/journal.pone.0004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn Y, Sims C, Murray MJ, et al. Multiple modes of Lrp4 function in modulation of Wnt/beta-catenin signaling during tooth development. Development. 2017;144:2824–2836. doi: 10.1242/dev.150680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, Tian QB, Endo S, Suzuki T. A role for LRP4 in neuronal cell viability is related to apoE-binding. Brain Res. 2007;1177:19–28. doi: 10.1016/j.brainres.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 29.Westerfield M. The zebrafish book: guide for the laboratory use of zebrafish (Danio rerio) Eugene: Univ. of Oregon Press; 2000. [Google Scholar]
- 30.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dynam. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 31.Chou CW, Zhuo YL, Jiang ZY, Liu YW. The hemodynamically-regulated vascular microenvironment promotes migration of the steroidogenic tissue during its interaction with chromaffin cells in the zebrafish embryo. PLoS One. 2014;9:e107997. doi: 10.1371/journal.pone.0107997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian J, Yam C, Balasundaram G, Wang H, Gore A, Sampath K. A temperature-sensitive mutation in the nodal-related gene cyclops reveals that the floor plate is induced during gastrulation in zebrafish. Development. 2003;130:3331–3342. doi: 10.1242/dev.00544. [DOI] [PubMed] [Google Scholar]
- 33.Chou CW, Lin J, Hou HY, Liu YW. Visualizing the interrenal steroidogenic tissue and its vascular microenvironment in zebrafish. J Vis Exp. 2016;118:e54820. doi: 10.3791/54820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tucker B, Lardelli M. A rapid apoptosis assay measuring relative acridine orange fluorescence in zebrafish embryos. Zebrafish. 2007;4:113–116. doi: 10.1089/zeb.2007.0508. [DOI] [PubMed] [Google Scholar]
- 35.Louw JJ, Bastos RN, Chen XW, et al. Compound heterozygous loss-of-function mutations in KIF20A are associated with a novel lethal congenital cardiomyopathy in two siblings. PLoS Genet. 2018;14:e1007138. doi: 10.1371/journal.pgen.1007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu YW, Guo L. Endothelium is required for the promotion of interrenal morphogenetic movement during early zebrafish development. Dev Biol. 2006;297:44–58. doi: 10.1016/j.ydbio.2006.04.464. [DOI] [PubMed] [Google Scholar]
- 37.Chou CW, Lin J, Jiang YJ, Liu YW. Aberrant global and Jagged-mediated Notch signaling disrupts segregation between wt1-expressing and steroidogenic tissues in zebrafish. Endocrinology. 2017;158:4206–4217. doi: 10.1210/en.2017-00548. [DOI] [PubMed] [Google Scholar]
- 38.Pan YH, Liu ZY, Shen J, Kopan R. Notch 1 and 2 cooperate in limb ectoderm to receive an early Jagged2 signal regulating interdigital apoptosis. Dev Biol. 2005;286:472–482. doi: 10.1016/j.ydbio.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 39.Sidow A, Bulotsky MS, Kerrebrock AW, et al. Serrate2 is disrupted in the mouse limb-development mutant syndactylism. Nature. 1997;389:722–725. doi: 10.1038/39587. [DOI] [PubMed] [Google Scholar]
- 40.Hilton MJ, Tu XL, Wu XM, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kariminejad A, Stollfuss B, Li Y, et al. Severe Cenani-Lenz syndrome caused by loss of LRP4 function. Am J Med Genet A. 2013;161A:1475–1479. doi: 10.1002/ajmg.a.35920. [DOI] [PubMed] [Google Scholar]
- 42.Lindy AS, Bupp CP, McGee SJ, et al. Truncating mutations in LRP4 lead to a prenatal lethal form of Cenani-Lenz syndrome. Am J Med Genet A. 2014;164:2391–2397. doi: 10.1002/ajmg.a.36647. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe N, Tezuka Y, Matsuno K, et al. Suppression of differentiation and proliferation of early chondrogenic cells by Notch. J Bone Miner Metab. 2003;21:344–352. doi: 10.1007/s00774-003-0428-4. [DOI] [PubMed] [Google Scholar]
- 44.Tao JN, Chen S, Yang T, et al. Osteosclerosis Owing to Notch Gain of Function Is Solely Rbpj-Dependent. J Bone Miner Res. 2010;25:2175–2183. doi: 10.1002/jbmr.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simpson MA, Irving MD, Asilmaz E, et al. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat Genet. 2011;43:303–305. doi: 10.1038/ng.779. [DOI] [PubMed] [Google Scholar]
- 46.Kung AWC, Xiao SM, Cherny S, et al. Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am J Hum Genet. 2010;86:229–239. doi: 10.1016/j.ajhg.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murea M, Park JK, Sharma S, et al. Expression of Notch pathway proteins correlates with albuminuria, glomerulosclerosis, and renal function. Kidney Int. 2010;78:514–522. doi: 10.1038/ki.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niranjan T, Bielesz B, Gruenwald A, et al. The Notch pathway in podocytes plays a role in the development of glomerular disease. Nat Med. 2008;14:290–298. doi: 10.1038/nm1731. [DOI] [PubMed] [Google Scholar]
- 49.Ma M, Jiang Y-J. Jagged2a-notch signaling mediates cell fate choice in the zebrafish pronephric duct. PLoS Genet. 2007;3:e18. doi: 10.1371/journal.pgen.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Pathak N, Kramer-Zucker A, Drummond IA. Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development. 2007;134:1111–1122. doi: 10.1242/dev.02806. [DOI] [PubMed] [Google Scholar]
- 51.McGregor L, Makela V, Darling SM, et al. Fraser syndrome and mouse blebbed phenotype caused by mutations in FRAS1/Fras1 encoding a putative extracellular matrix protein. Nat Genet. 2003;34:203–208. doi: 10.1038/ng1142. [DOI] [PubMed] [Google Scholar]
- 52.Slavotinek AM, Tifft CJ. Fraser syndrome and cryptophthalmos: review of the diagnostic criteria and evidence for phenotypic modules in complex malformation syndromes. J Med Genet. 2002;39:623–633. doi: 10.1136/jmg.39.9.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Alignment of human, mouse and zebrafish LRP4 amino acid sequences. The zebrafish sequence lacks 39 amino acid residues at the C-terminal region. (PDF 982 kb)
Figure S2. Homology modeling of human, mouse and zebrafish LRP4 proteins. Template-based structure prediction of LRP4 protein structures was performed by SWISS-MODEL, and the predicted 3D structures from three species were superimposed by PyMOL. The templates used for modeling each region are: Region I, Low-density lipoprotein receptor (1n7d.1.A); Region II and III, Low-density lipoprotein receptor-related protein 6 chain A (5gje.1.A). AA, amino acids. (PDF 551 kb)
Figure S3. The effect of lrp4-sdMO on the RNA splicing of lrp4. (A) RT-PCR analysis was used to examine whether the lrp4-sdMO affects the splicing of intron 15 of lrp4 pre-mRNA. The primer set was designed to amplify the cDNA sequence spanning through the junction between exons 15 and 16. The exon–intron organization was predicted by BLASTing the lrp4 cDNA sequence to GRCz10 (Ensembl). Retention of partial intron 15 of the lrp4 pre-mRNA sequence (#2 band of RT-PCR products) was detected in the lrp4 morphant at 10 hpf, which is predicted to cause an in-frame early stop codon (highlighted in light blue). (B) Increasing doses of lrp4-sdMO led to reductions of lrp4 mRNA expression as analyzed by semi-quantitative RT-PCR. The gel picture shown is a representative of triplicate experiments. The intensity of #1 band from the gel picture was subject to densitometric analysis (lower panel) for semi-quantitative measurements of wild-type lrp4 mRNA expressions. The average of three separate experiments shows a significant dose-dependent decrease of #1 band product from 0.2 to 1.2 pmol of lrp4-sdMO injections. Apart from the #2 band of mis-spliced product, one more mis-spliced product was present in trace amounts (indicated by yellow star). (C) Quantitative analysis of mis-spliced products by qRT-PCR using the intron-15 specific primer. A representative gel picture of the mis-spliced product and its housekeeping control (eelf1a) from qRT-PCR is shown in the lower panel. The results of semi-quantitative RT-PCR in (B) and qRT-PCR in (C) were the average of triplicate experiments using 10 embryos in each treatment group for one RT reaction. *P<0.05. (Student’s t test) (PDF 579 kb)
Figure S4. The posterior trunk and tail phenotypes of lrp4-sd morphants. (A) Injections of lrp4-sd MO (0.4 pmol/embryo), tp53MO (0.5 pmol/embryo) and lrp4-sdMO/tp53MO led to mild dysmorphogenesis in the posterior trunk and tail regions at 24 hpf. (B) Abnormal accumulation of blood cells (red arrows) at the caudal vein plexus in the class I to III of lrp4-sd morphants was captured by live imaging. (C) Acridine orange staining revealed apoptotic cells (orange arrows) in the tail region of the control embryo, which were apparently reduced in the tp53 morphant; whereas no apoptotic cells were detected in either lrp4-sd morphants or lrp4-sd/tp53 double morphants. Scale bar: 100 μm. (PDF 373 kb)
Figure S5. The effect of lrp4-atgMO on the median fin fold phenotype and the Notch pathway. (A) lrp4-sdMO led to blistering phenotype in the caudal vein plexus and the median fin fold. Phenotypic classification is according to what is described in Fig. 2H. (B) The expression of genes which are components of Notch pathway was analyzed by qRT-PCR for lrp4-atg morphants and STD-MO injected control embryos at 2 dpf. The results were the average of triplicate experiments using 10 embryos in each treatment group for one RT reaction. ***P<0.0001 (Student’s t test). (PDF 212 kb)