Abstract
The molecular dysfunction in X-linked dystonia-parkinsonism is not completely understood. Thus far, only noncoding alterations have been found in genetic analyses, located in or nearby the TATA-box binding protein-associated factor 1 (TAF1) gene. Given that this gene is ubiquitously expressed and is a critical component of the cellular transcription machinery, we sought to study differential gene expression in peripheral models by performing microarray-based expression profiling in blood and fibroblasts, and comparing gene expression in affected individuals vs. ethnically matched controls. Validation was performed via quantitative polymerase chain reaction in discovery and independent replication sets. We observed consistent downregulation of common TAF1 transcripts in samples from affected individuals in gene-level and high-throughput experiments. This signal was accompanied by a downstream effect in the microarray, reflected by the dysregulation of 307 genes in the disease group. Gene Ontology and network analyses revealed enrichment of genes involved in RNA polymerase II-dependent transcription, a pathway relevant to TAF1 function. Thus, the results converge on TAF1 dysfunction in peripheral models of X-linked dystonia-parkinsonism, and provide evidence of altered expression of a canonical gene in this disease. Furthermore, our study illustrates a link between the previously described genetic alterations and TAF1 dysfunction at the transcriptome level.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2159-4) contains supplementary material, which is available to authorized users.
Keywords: Microarray, Expression profiling, Transcriptomics, Transcriptional dysregulation, Neurodegeneration
Introduction
X-linked dystonia-parkinsonism (XDP, DYT3, “Lubag Disease”, OMIM #314250) is a neurodegenerative disorder currently found exclusively in Filipinos due to a genetic founder effect [1, 2]. The condition is characterized by adult-onset dystonia, followed in later years by parkinsonism; premature death occurs due to aspiration pneumonia or suicide [3]. A study of brains has revealed neuronal loss in the striatum, with a pattern similar to Huntington’s disease [4, 5].
The molecular genetic mechanisms surrounding XDP are complex. Seven different DNA sequence variants segregate with disease, lying in the vicinity of the TAF1 (TATA-box binding protein-associated factor 1) gene. They include five single-nucleotide substitutions, one 48-bp deletion, and one 2627-bp SINE-VNTR-Alu retrotransposon insertion [6, 7]. The variants occur in complete linkage disequilibrium, and define a disease-specific “XDP haplotype” (Fig. 1a) [2]. Nevertheless, their relevance to TAF1 function—and consequently to disease causation—is undetermined, given that none are found in protein-coding transcripts [7, 8]. Furthermore, while TAF1 is known to participate extensively in the activation of gene transcription in most cell types in eukaryotes [9], it is not understood if, how, or why the variants cause an exclusively neurologic phenotype.
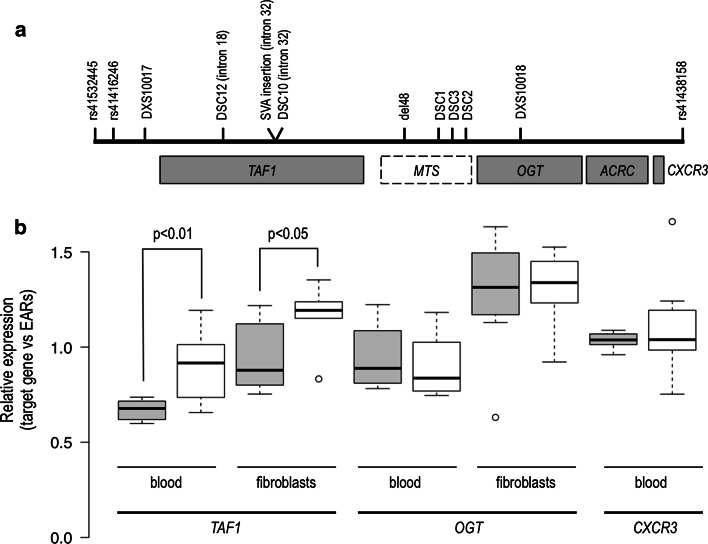
Fig. 1.
a Linked region in Xq13.1 showing XDP-associated genetic changes (“XDP haplotype”: DSC12, SVA retrotransposon insertion, DSC10, del48, DSC1, DSC2, DSC3), located either in introns of TAF1 or within an untranslated region (“Multiple Transcript System”, MTS) distal to the gene. b qPCR analysis showing significant downregulation of TAF1 in the XDP group (shaded boxes) in both blood and fibroblast-derived cDNA, and no difference in two other genes in the linked region. TAF1 and OGT are both expressed in blood and fibroblasts, while CXCR3 is expressed only in blood. Boxplot whiskers extend to data points that are within ×1.5 the interquartal ranges. EARs expressed Alu repeats
One proposed mechanism is the reduced expression of a neuron-specific, alternatively spliced isoform of TAF1 (nTAF1), an alteration that is thought to result in dysregulation of neuronal TAF1 targets [7]. Currently, however, no definite causal link has been established between the genetic variants associated with the disease and this derangement. Another study suggests the existence of a “Multiple Transcript System” (MTS) distal to TAF1, encoding five additional exons [6, 10]. Combinations of these exons can be transcribed independently or spliced to TAF1 mRNA, and one genetic alteration within the XDP haplotype—the only variant that is transcribed [10]—is located in one such noncanonical exon [6]. Importantly, nonconventional MTS transcripts have not been shown to be abundant [11], and thus are also of questionable relevance.
Alternatively, any one or a combination of the genetic alterations within the XDP haplotype could impact regulation of canonical TAF1 mRNA expression, i.e., affecting quantitative rather than qualitative levels of transcription and translation. In addition, altered regulation of a ubiquitously expressed gene does not necessarily have to occur only where the main disease phenotype manifests, but may also be present in more easily accessible peripheral tissue [12–14]. Studying expression changes in blood or in primary fibroblast cultures derived from skin biopsies could thus lead to important insights.
Given this, we hypothesized dysregulated TAF1 expression in peripheral models of XDP, i.e., (1) blood, and (2) fibroblasts, the latter representing an endogenous cell model with the same embryonic origin as neurons. We investigated concomitant changes in global gene expression through genome-wide expression profiling, followed by validation using gene-level quantification in the discovery and in replication sets. Our study reveals foremost reduced expression of conventional TAF1 transcripts in blood and in fibroblasts in XDP, thus linking the disease-associated genetic alterations to the dysfunction of a canonical gene.
Materials and methods
Subjects and samples
Affected individuals (all male) were recruited from multimodal studies on XDP. All were in the combined dystonic-parkinsonian stage of disease. Age-, sex-, and ethnicity-matched healthy controls were also recruited. After informed consent, blood samples from 9 affected individuals and 13 controls were collected in PAXGene tubes (Qiagen). From these, RNA was extracted using the manufacturer’s protocols. Skin biopsies were obtained from 14 affected individuals (including the 9 patients who also provided a blood sample), and from 8 controls. This study was approved by the local ethics committee of the University of Lübeck.
Primary fibroblast cultures were grown on Dulbecco’s modified Eagle medium (Gibco) supplemented with 10 % fetal bovine serum and 1 % penicillin/streptomycin, passaged when confluent, and at passages 7–9, harvested via Accutase (Gibco) treatment. RNA was extracted from a defined amount of cells and after DNaseI treatment (Qiagen). Prior to sending samples for microarray expression profiling, genotypes were determined via Sanger sequencing and PCR-based analyses (methods described in Ref. [2]).
Microarray expression profiling
Ten blood-derived RNA samples from affected individuals and 12 from controls were subjected to microarray-based expression profiling (see Supplementary Figure for study workflow). For the analysis in fibroblasts, 15 samples from affected individuals and 10 from controls were submitted as a separate group (thus, 1–2 technical replicates were included in each group). Expression profiling was performed by OakLabs GmbH (Hennigsdorf, Germany) on an Agilent Sureprint G3 Human Gene Expression 8x60K v2 oligonucleotide microarray (Agilent Technologies). Importantly, all samples belonging to a tissue group were handled and hybridized onto the array in a single batch, eliminating the possibility of batch effects [15]. Also, all RNAs were determined to have RIN values >8.0 on the Bioanalyzer 2100 (Agilent Technologies).
Quality control and differential expression analysis
The following groups were compared: (1) 9 blood-derived RNA samples from affected individuals vs. 13 from controls, (2) 15 fibroblast-derived samples from affected individuals vs. 10 from controls. After quantile normalization, probes with intensities less than the median value in all samples were removed. Expression values in probes that were mapped to the same Entrez genes were averaged, resulting in 12,351 genes in the fibroblast microarray experiment and 12,940 in the experiment performed on blood RNA. Differential expression analyses (XDP vs. controls, blood and fibroblast-derived data treated independently) was then performed in R, using (1) Wilcoxon rank-sum tests, and (2) the limma statistic with SAM (Statistical Analysis of Microarrays) correction, both at 0.10 FDR (False Discovery Rate)-corrected cut-offs [16]. The list of differentially expressed genes (DEGs) was then ranked according to SAM q value and Wilcoxon score (Supplementary Table); genes for qPCR analysis were selected from this list by hand, using biological function and expression in the brain as criteria for selection. In particular, we probed the freely available data of the UK Brain Expression Consortium (UKBEC, http://www.braineac.org/) to determine genes that are expressed in the human brain based on microarray experiments performed in control individuals.
qPCR analysis
Quantitative PCR (qPCR) analysis was performed on selected DEGs. First, RNA from 8 randomly selected affected individuals and 8 controls, and from the same set of fibroblast RNAs that was used in the microarray (discovery set) was reverse transcribed into cDNA (Thermo Scientific). Real-time qPCR was then performed using SYBR Green chemistry (Fermentas) on a Roche 480 Light Cycler, and using primer sequences obtained from a publicly available database (http://primerdepot.nci.nih.gov). Expression levels were computed using the Advanced Relative Quantification method, determined by the Roche LightCycler software (release 1.5.0). This automated quantification method approximates the expression level calculated using the well-accepted delta–delta-CT method [17], but corrects for differences in efficiency between target and reference genes. The expression levels of target genes were normalized against the expression of Expressed Alu Repeats (EARs) [18]. Fold-change was determined by obtaining the ratio of the average expression value in affected individuals vs. the average in controls. When this value was below 1, the inverse was obtained. The p value was obtained by performing a two-tailed Student’s t test between expression values in the XDP group vs. controls. Significance level was set at p < 0.10.
Replication experiments
For the replication analyses, fibroblasts from 5 affected individuals and 6 controls were regrown in culture (technical replication set). In addition, fibroblasts from 5 new affected individuals and 2 new controls were obtained (independent replication set). When the replication experiments were repeated, we added 3 technical replicates for the XDP group and 2 controls (total 8 each group). RNA was extracted, reverse transcribed, and qPCR analyses of target genes were performed as in the discovery set.
Aside from group-level analyses using normalization against EARs, sample-level analyses were performed from data obtained from the replication experiments in fibroblasts. Here, we normalized against reference genes. First, the Genevestigator platform (https://genevestigator.com/gv/) was used to determine the usual expression profile in primary fibroblasts/skin cells of the target genes. The RefGenes [19] tool of the platform was then used to determine stable, suitable reference genes (in this case, FLAD1 and ZNF672). Fold change of expression difference was then computed by obtaining the ratio of normalized expression of each target gene (computed for using the Relative Quantification method, as above) in each sample against the average normalized expression in 8 controls (6 from technical replicate group, and 2 independently collected). For the sample-level analysis in blood, we used the discovery set of RNAs, and reference genes (FPGS, UBC) that have been shown to be stable in previous studies performed in peripheral blood [20].
Enrichment and network analyses
From the list of DEGs in fibroblasts, enrichment analysis was performed in EXPANDER (Expression Analysis and Displayer), a java-based tool designed for gene expression analysis, and in particular, enrichment analysis of gene lists (http://acgt.cs.tau.ac.il/expander/) [21, 22]. Search for enrichment of Gene Ontology (GO) terms was performed using the TANGO procedure [23], for pathway enrichment using the KEGG and Wikipathways features, and for miRNA enrichment using FAME [24]. Enriched transcription factor motifs and putative transcriptional regulators were determined using PRIMA [25]. We performed enrichment analysis of the upregulated and downregulated gene sets separately in order to characterize more precisely the biological functions and processes possibly involved. Additional GO/network analysis was performed using GSEA (Gene Set Enrichment Analysis) [26] and GeneMania (http://www.genemania.org) [27].
Results
Preliminary qPCR experiments
Preiliminary qPCR analysis of common TAF1 transcripts in blood- and fibroblast-derived cDNA revealed statistically significant reduced expression of TAF1 in the XDP group (p value in blood <0.01, in fibroblasts <0.05), albeit with low magnitude, i.e., fold change (fc) <1.5 in both (Fig. 1b). We failed to detect sufficient amounts of nTAF1 or of the MTS in either tissue to allow for calculation of differences in expression of nonconventional transcripts. The expression of two other genes (OGT, CXCR3) in the disease locus on Xq13.1 was similar in the XDP and control groups, i.e., TAF1 was the only gene in the disease-linked region with differential gene expression (Fig. 1a, b). While expanding our fibroblast cultures, we observed generally faster growth in the XDP group (data not shown).
Microarray expression profiling
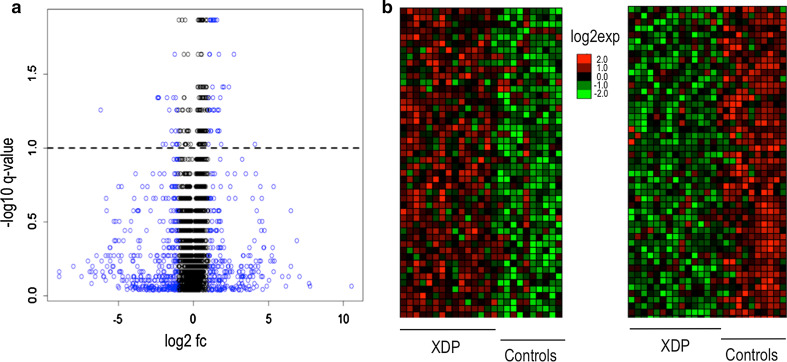
307 DEGs were identified in the microarray experiment performed on fibroblasts using a cut-off q value of <0.10 in the SAM test, representing 2.4 % of the total number of transcripts analyzed; 250 of these transcripts were upregulated and 57 were downregulated in the XDP group (Fig. 2). In contrast, no DEGs were identified in the blood microarray experiment, even when the q value cut-off was relaxed to 0.20. Of note, with the exception of a handful, all DEGs discovered in the fibroblast experiment had small fold changes, i.e., between 1.1 and 2.0. Consistent with the results of the preliminary qPCR analysis, TAF1 was one of the downregulated DEGs in the fibroblast microarray experiment; two probes targeting common TAF1 transcripts (Table 1) showed lower expression in the XDP group compared to controls (fc = 1.2, averaged q value = 0.03). ACRC, the only other gene in the linked region that was not investigated in preliminary qPCR experiments, was not differentially expressed in the microarray experiment (q value = 0.27). Microarray data are available at the NCBI’s Gene Expression Omnibus (GEO) through GEO Series accession number GSE74068.
Fig. 2.
a Volcano plot showing differentially expressed genes (DEGs) in the fibroblast microarray experiment. Blue dots represent genes with log2fc >2.0, dotted line represents cut-off -log10 q value of <0.10 (SAM test). Among DEGs with q values below this cut-off, there were only a few with log2fc >2.0. b Expression heat maps of two gene groups from the fibroblast microarray experiment, showing good separation of up- and downregulated genes. The Supplementary Table to this manuscript lists all DEGs
Table 1.
TAF1 probes, genomic locations and sequences
| Experiment | Probe | Transcript/s | Loc | Sequence |
|---|---|---|---|---|
| qPCR analysis | TAF1_SYBR_Forward | NM_001286074 | Exon 32 | AGAGTCGGGAGAGCTTTCTG |
| NM_004606 | ||||
| TAF1_SYBR_Reverse | NM_138923 | Exon 33 | CACAATCTCCTGGGCAGTCT | |
| Microarray | A_23_P11237 | NM_001286074 | Exon 38 | GCATGCTTCAGGAGAACACAAGGATGGACATGGAAAATGAAGAAAGCATGATGTCCTATG |
| NM_004606 | Exon 37 | |||
| NM_138923 | Exon 37 | |||
| Microarray | A_32_P192615 | NM_001286074 | Exon 39 (UTR) | GGCTGAGATGAGACTGAAAGATGGGCAGGAAGTATATCATCACAAGCTTTGTGTTTGATG |
| NM_138923 | Exon 38 (UTR) | |||
| Microarray | A_33_P3353111a | ENST00000373775 (nonsense mediated decay) | Exon 15 (UTR) | TCTTTGTGACTGGTTACTTCATCCAGGGGACCAATTTCTCTCTGGAATATTAGTCCCGCA |
| ENST00000462588 (untranslated) | Exon 11 |
NM_001286074, NM_004606, and NM_138923 are the three full-length and most abundant transcripts of TAF1
aTwo examples of alternatively spliced untranslated transcripts interrogated by A_33_P3353111 are given; this probe was eventually filtered out of the analysis due to low expression
qPCR analysis in discovery set
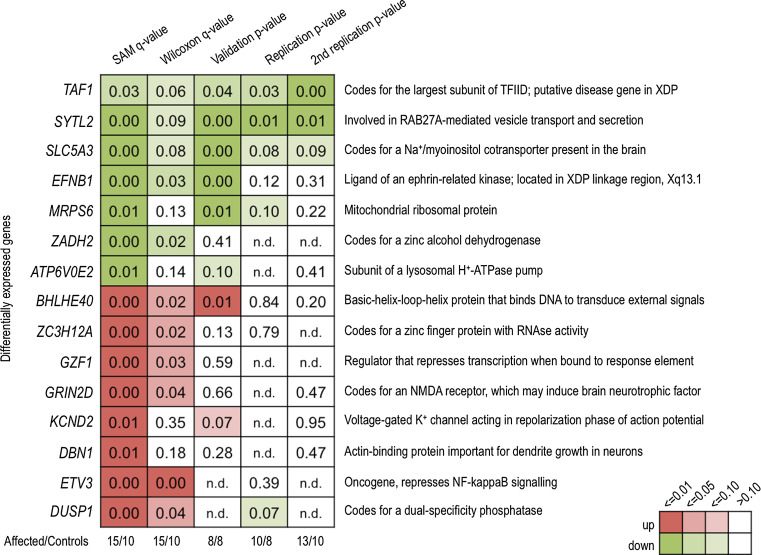
Twelve genes were chosen for qPCR analysis, using their biological function (Fig. 3) and expression in the brain (Supplementary Figure 2) as criteria for selection. qPCR analysis of these 12 DEGs (6 downregulated and 6 upregulated genes) confirmed significant differential expression of 7 genes in fibroblast-derived RNA (p value <0.10: SYTL2, EFNB1, SLC5A3, MRPS6, ATP6V0E2, BHLHE40, KCND2). Of note, five of seven genes would still be differentially expressed at a significance level p < 0.05 (Fig. 3). The expression of other genes chosen for qPCR analysis was not significantly different between the XDP group and controls.
Fig. 3.
Heat map showing the performance of different genes chosen for follow-up, using microarray (SAM q value, Wilcoxon q value) and qPCR analysis. Data is from fibroblast-derived RNA. One criteria for selection of genes to follow-up was biological function (right-hand labels). Amongst differentially expressed genes, there were strikingly consistent signals in TAF1, SYTL2, and SLC5A3, and less consistently so, in EFNB1 and MRPS6. SAM significance analysis of microarrays
Replication experiments
DEGs that were validated in the preceding experiment, and TAF1, were then further investigated using technical and independent replicates. Fresh fibroblast-derived RNA from these replicates also enabled validation of two more DEGs (ETV3 and DUSP1). qPCR analysis in cDNA from these sets revealed consistently lower expression of TAF1 in the XDP group (p value = 0.03), although again with a modest magnitude (fc = 1.3) (Fig. 3). Repeating the experiment in a larger number of samples from affected individuals and controls revealed the same fc (1.3) but with higher statistical significance (p value = 2.1 × 10−6). Consistent with the microarray and validation experiments, significant downregulation of SYTL2 (p value = 0.01), SLC5A3 (p value = 0.08), EFNB1 (p value = 0.12), and MRPS6 (p value = 0.10) in the XDP group was observed. No other genes tested were significantly differentially expressed in the XDP group.
Sample-level analysis
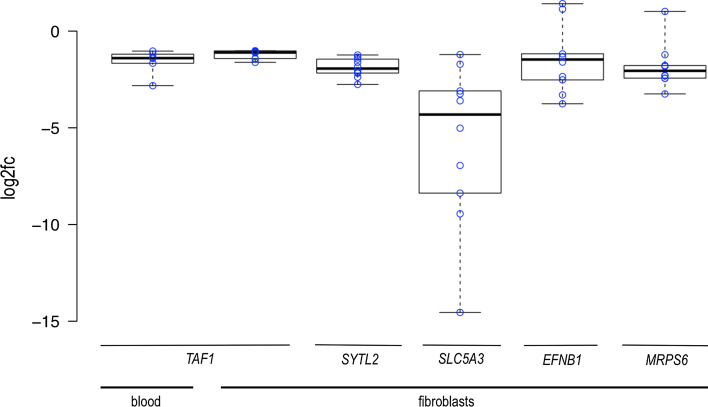
Reduced expression of TAF1, SYTL2, and SLC5A3 was observed in every fibroblast-derived sample obtained from an affected individual when treated individually and compared to average expression in controls (fc ranges: TAF1: 1.0–1.6, SYTL2: 1.2–2.75, SLC5A3: 1.2–14.5; Fig. 4). We went back to the qPCR data in blood, and also observed consistently lower expression of TAF1 (fc range 1.0–2.8) in every XDP sample in this tissue. Although our study was not powered to detect a possible correlation between clinical status and changes in expression, we noted greatest downregulation of TAF1 (in blood and fibroblasts) and EFNB1 (in fibroblasts) in samples that came from affected individuals with longer disease duration.
Fig. 4.
Sample level analysis in blood- and fibroblast-derived cDNA, target vs. reference genes. Blue circles represent log2 relative expression in individual samples from affected individuals vs. average expression in controls. There were no samples from affected individuals with overexpression of TAF1, SYTL2, and SLC5A3. Boxplot whiskers extend to minimum and maximum values
Enrichment and network analysis
Significantly enriched GO terms seen in upregulated DEGs from the fibroblast experiment included: “vasculature development” (q value = 6 × 10−4), “regulation of cell differentiation” (q value = 0.0052), and “regulation of transcription from RNA polymerase II promoter” (q value = 0.012). Upregulated DEGs were enriched for targets of the transcription factors SRF and PRDM1. There were no enrichments seen in downregulated DEGs upon GO and transcription factor analysis. There were no significantly enriched KEGG pathways/Wikipathways or miRNA enrichments seen. Gene set enrichment analysis (GSEA) revealed only one enriched gene set at 0.2 FDR: “cell cycle” (q value = 0.03).
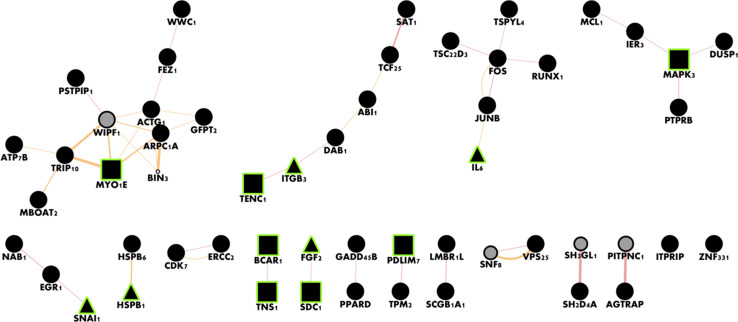
Network analysis using GeneMania [27] revealed enriched GO terms only in upregulated DEGs. These enrichments were similar to the ones seen in the initial GO analysis, and included some redundant terms, i.e., “DNA-dependent transcription” (q value = 0.035) and “negative regulation of sequence-specific DNA-binding transcription factors” (q value = 0.045); “response to oxidative stress” (q value = 0.0035) and “response to reactive oxygen species” (q value = 0.045); and “actin binding” (q value = 0.014) and “actin cytoskeleton” (p value = 0.047) (Table 2). Overexpressed genes were enriched for tissue-specific genes related to the growth and maturation of fibroblasts (Fig. 5).
Table 2.
Gene ontology (GO) term enrichments with q value <0.05
| q valuea | GO annotation/s |
|---|---|
| 1.5 × 10−7 | Adherens junction, anchoring junction, focal adhesions |
| 0.0026 | Positive regulation of cell migration/motility/locomotion |
| 0.0035 | Response to oxidative stress |
| 0.0086 | Blood vessel development, blood vessel morphogenesis |
| 0.0140 | Actin binding, actin cytoskeleton organization |
| 0.0170 | Muscle organ development, muscle structure development |
| 0.0340 | DNA-dependent transcription, initiation |
| 0.0340 | Positive regulation of angiogenesis, vasculature development |
| 0.0340 | Transmembrane receptor protein serine/threonine kinase signalling |
| 0.0450 | Negative regulation of sequence-specific DNA binding transcription factors |
| 0.0450 | Angiogenesis |
| 0.0450 | Response to reactive oxygen species |
Aside from GO terms related to RNA polymerase II-based transcription, terms related to the tissue derivation (cultured fibroblasts) are enriched, i.e., “adhesion”, “vascular development”, “actin binding and organization”, reflecting tissue-specific effects
aOverlapping terms collapsed into single rows; lowest p value noted
Fig. 5.
GeneMANIA analysis of overexpressed genes in the microarray reveals active connected components. Black nodes denote differentially expressed genes and lines denote physical interactions. Gray nodes were added by the GeneMANIA analysis. Rectangular nodes are related to adherens junction (q = 0.0041), and triangular nodes are related to positive regulation of cell migration (q = 0.013). This network analysis reveals tissue-specific effects, while also serving as a sanity check on the fibroblast microarray experiment
Discussion
TAF1 codes for the largest component of the TFIID transcription factor complex, a general transcription factor intrinsic to RNA polymerase II-dependent transcription of most, if not all protein-coding genes [9, 28]. TAF1 is also the putative disease gene in XDP, mainly by virtue of association—the seven genetic alterations specific to XDP lie either in introns of TAF1 or in the region directly distal to the gene. As no protein-coding mutation linked to the disease has been discovered so far, our hypothesis was that alteration of TAF1 function in XDP occurs at the expression level, influencing the role of the gene in the transcriptional machinery.
We observed consistent downregulation of common TAF1 transcripts in the XDP group in our qPCR- and microarray-based experiments. Although the degree of downregulation was small, the reliability of the signal was striking. TAF1 expression was reduced in the XDP group in both tissues studied, i.e., blood and fibroblasts, and TAF1 was the only gene in the disease-linked region showing expression difference. Heterogeneity of global expression profiles did not allow TAF1 dysregulation to reach significance in the microarray profiling experiment performed in blood. Nevertheless, taking our experiments as a whole, we provide evidence of downregulation of common TAF1 transcripts in XDP.
In this study, we used p and q values, as statistics that incorporate variability, to allow us to infer the robustness of our comparisons. While fold change reflects effect size, the measure inadequately accounts for variance, and an arbitrary cutoff (such as >2.0, as in most expression studies) limits the discovery to genes that vary wildly between groups [29, 30], without accounting for biological relevance, consistency, or a previous hypothesis. As we had a standing hypothesis of TAF1 dysfunction that was already supported by preliminary qPCR experiments, we followed-up the signal in TAF1 despite the small fold change.
Notably, all other DEGs in the fibroblast microarray experiment—except for a handful of genes with low expression at baseline—had small fold change as well; this is likely related to the endogenous nature of the model that we used (i.e., not an overexpressed system) and to the homogeneity of expression profiles in fibroblasts. Furthermore, globally reduced expression variation in cultured cells is a recognized phenomenon [31] and is due to the controlled environment and absence of influences introducing noise. Because of this, the signal obtained from the transcriptome in this experiment can be inferred to be secondary mainly to the mutation status.
The probes used in the qPCR and microarray analyses were located in different exons of the gene and interrogate the full-length and most abundant protein-coding transcripts of TAF1. While we could have also investigated the differential expression of non-canonical TAF1 transcripts, i.e., nTAF1 and transcripts of the MTS, we found the amounts of these isoforms in blood and fibroblast-derived RNA to be too small to allow precise measurements of expression changes. Using brain-derived RNA, previous studies report the relative expression of these isoforms to be as much as 3- to 10-fold lower in comparison to common TAF1 transcripts [7, 8]. Our objective was rather to investigate gene expression changes occurring at endogenous levels, hence we avoided the use of overexpression models and limited our experiments to well-expressed TAF1 transcript variants.
We also hypothesized that changes in gene expression occur genome-wide corollary to TAF1 dysfunction, given its role as a transcriptional activator. Despite the low magnitude of downregulation in TAF1, we saw an extensive signal composed of 307 DEGs in the microarray analysis, representing a downstream effect that implies indeed dysregulation of a critical component of the cell’s transcriptional machinery. Furthermore, in network and GO analyses, there was a clear and consistent enrichment of genes involved in RNA polymerase II-based transcriptional processes.
Importantly, XDP is frequently cited as a transcriptional dysregulation syndrome, as is Huntington’s disease (HD) [32], with which XDP shares similarities in brain pathology [4, 5]. Previous work also demonstrated that TAF7, which codes for a transcriptional coactivator that interacts closely with TAF1 as part of TFIID complex [33], is dysregulated in models and patients with HD [12, 34]. Thus, enrichments of genes involved in transcriptional processes in microarray data from both diseases could denote changes in global gene expression (1) as a direct downstream effect, or (2) as a result of genome-wide alterations in transcription factor activity, to compensate for the downregulation of the disease-related gene. Similarly, altered interaction of TATA-box binding protein (TBP) to TFIIB has been shown as the molecular mechanism underlying neurodegeneration in transgenic spinocerebellar ataxia 17 (SCA17) mice; the enhanced interaction between the activator and the transcription factor results in dysregulated target genes [35, 36].
Amongst discovered DEGs, consistent signatures were seen in SYTL2, SLC5A3, and EFNB1. SYTL2- encoded Synaptotagmin-like 2 is a RAB27A interactor that participates in membrane trafficking in peripheral secretory cells [37, 38]. Expression profiling in d3–d4-overexpressed cells in an overexpression model of XDP—if it is to be assumed that d3–d4 TAF1 isoforms contribute to the molecular mechanism of the disease—also identified altered vesicle trafficking and synaptogenesis as a significant signature using pathway analysis [8]. Furthermore, disruption of endosomal membrane trafficking is a cellular process that is disrupted in many neurodegenerative diseases [39, 40], including genetic forms of Parkinson’s disease [41]. SLC5A3 meanwhile codes for a sodium/myoinositol transporter that regulates brain osmoregulation [42]. Inositol metabolism has recently been associated with neural dysfunction in HD, observed as loss of the inositol polyphosphate multikinase enzyme in the striatum in humans, and in cellular and murine models of the disease [43]. Mutations in EFNB1 cause a neurodevelopmental phenotype (craniofrontonasal syndrome, OMIM #304110) [44]. Interestingly, as this manuscript was being prepared, exonic and splicing TAF1 mutations were found to underlie a group of neurodevelopmental syndromes with characteristic facial dysmorphic features [45]. EFNB1 is located (as is TAF1) on Xq13.1, the chromosomal region associated with XDP. Although outside the disease-linked haplotype containing all known XDP-associated mutations to date [2], long-range intrachromosomal DNA interactions affecting the gene’s expression may provide a mechanism for the alteration observed here [46]. Lastly, DNAJC6, mutations in which cause juvenile parkinsonism due to impaired synaptic vesicle recycling [47, 48], is also one of the overexpressed genes in the microarray, as is GRIN2D, a gene coding for a glutamate receptor that was recently identified to be differentially expressed in Parkinson’s disease patients vs. healthy controls [49]. The contributions of these target genes to the molecular mechanisms surrounding XDP should be explored in further studies.
We did not intend in this study to search for biomarkers or to correlate disease status with expression changes in our peripheral models. XDP is characterized by large variations in age of onset, duration, and clinical findings [1], such that a biomarker study would have required more samples from affected individuals in different, well-delineated disease stages. Despite this limitation, we observed more striking expression changes in TAF1 and EFNB1 in affected individuals with longer disease duration. XDP is a progressive, degenerative disease; neuroimaging and neuropathologic studies also report more severe involvement of brain regions and greater neuronal loss in patients in later stages [4, 5]. Thus, the relevance of increasing downregulation of TAF1 and EFNB1 with longer disease duration, and hypothetically, greater neuronal loss, is intriguing. In relation to this, the network analysis we performed also revealed enrichment of genes related to the capacity of cells to deal with oxidative stress, a common molecular feature of age-related neurodegenerative diseases [50, 51]. Thus, a plausible hypothesis is age-related increase in TAF1 dysfunction in mutation carriers, accompanied by increasing oxidative damage as the disease progresses. Such temporally defined derangements may be related to the adult-onset nature of the disease, to the evolution of the phenotype from dystonia to degenerative parkinsonism, and to the progressive imaging and pathologic findings.
Lastly, in the set of upregulated genes, we observed an enrichment of genes and processes related to positive growth and differentiation. This mainly serves as a sanity check on the results of our microarray experiment, and on the analyses performed in silico, given that we observed faster growth in vitro in fibroblast cultures obtained from XDP-affected individuals. Notably, BHLHE40, a transcription factor and an upregulated DEG, interacts with the TFIID complex (with which TAF1 is involved) to promote differentiation [52], while also mediating p53-dependent premature senescence [53]—another intriguing link between the molecular signature observed in this study and the neurodegenerative phenotype of XDP.
In summary, we report (1) consistent dysregulation of common TAF1 transcripts in peripheral models of XDP, accompanied by (2) altered gene expression genome-wide, and (3) enrichment of genes and networks related to transcriptional processes. These findings corroborate the notion that XDP is a transcriptional dysregulation syndrome, while also linking the genetic alterations specific to XDP to dysfunction of a canonical gene. We infer that TAF1 dysregulation likewise drives changes in gene expression in the brains of XDP-affected individuals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1. Study workflow showing the high-throughput (microarray-based, left-side) and gene-level (qPCR-based, right-side) experiments and analyses. One lane on the microarray had a defect that was undetected during hybridization but was later made obvious by quality control assessments. The blood-derived sample from an affected individual was not included in further analyses. (PDF 98 kb)
Supplementary Fig. 2. Expression in various parts of the brain of the different genes chosen for follow-up. Data is derived from the UK Brain Expression Consortium (http://www.braineac.org/). From top left, clockwise: transcript-level expression of SYTL2, SLC5A3, EFNB1, MRPS6, BHLHE40, KCND2, ATP6V0E2, TAF1. (PDF 296 kb)
Acknowledgments
This research was funded by a Thyssen Foundation research grant, and by a Jake’s Ride for Dystonia research grant (through the Bachmann-Strauss Dystonia and Parkinson Foundation) to Ana Westenberger. Aloysius Domingo is supported by the German Academic Exchange Service (DAAD). David Amar is supported by the Azrieli Foundation and the Edmond J. Safra Center for Bioinformatics at Tel Aviv University. Ron Shamir is supported by the Raymond and Beverly Sackler Chair in Bioinformatics. Christine Klein is supported by the Hermann and Lilly Schilling Foundation.
Gene names
- ACRC
Acid repeat-containing gene
- ATP6V0E2
ATPase, H+ transporting, lysosomal, 9-kD, V0 subunit E2
- BHLHE40
Basic helix-loop-helix family, member E40
- CXCR3
Chemokine, CXC motif, receptor 3
- DBN1
Drebrin E
- DUSP1
Dual-specificity phosphatase 1
- EFNB1
Ephrin B1
- ETV3
ETS variant gene 3
- GRIN2D
Glutamate receptor, ionotropic, N-methyl-d-aspartate, subunit 2D
- GZF1
GDNF-inducible zinc finger protein 1
- KCND2
Potassium voltage-gated channel, Shal-related subfamily, member 2
- MRPS6
Mitochondrial ribosomal protein S6
- OGT
O-linked N-acetylglucosamine transferase
- PRDM1
PR-domain containing protein 1
- SLC5A3
Solute carrier family 5 (inositol transporter), member 3
- SRF
Serum response factor
- SYTL2
Synaptotagmin-like 2
- TAF1
TATA-box binding protein-associated factor 1
- TBP
TATA-box binding protein
- ZADH2
Zinc alcohol dehydrogenase
- ZC3H12A
Zinc finger CCCH domain-containing protein 12A
Others
- DEG
Differentially expressed gene
- DSC
Disease-specific single-nucleotide change
- GO
Gene ontology
- HD
Huntington’s disease
- MTS
Multiple transcript system
- SAM
Significance analysis of microarrays
- XDP
X-linked dystonia-parkinsonism
Compliance with ethical standards
Conflict of interest
Authors report no potential conflicts of interest.
References
- 1.Lee LV, Rivera C, Teleg R, et al. The unique phenomenology of X-linked dystonia-parkinsonism (XDP, DYT3, “Lubag”) Int J Neurosci. 2011;121(Suppl):3–11. doi: 10.3109/00207454.2010.526728. [DOI] [PubMed] [Google Scholar]
- 2.Domingo A, Westenberger A, Lee LV, et al. New insights into the genetics of X-linked dystonia-parkinsonism (XDP, DYT3) Eur J Hum Genet. 2015;23:1334–1340. doi: 10.1038/ejhg.2014.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosales RL. X-linked dystonia parkinsonism: clinical phenotype, genetics and therapeutics. J Mov Disord. 2010;3:32–38. doi: 10.14802/jmd.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goto S, Lee LV, Munoz EL, et al. Functional anatomy of the basal ganglia in X-linked recessive dystonia-parkinsonism. Ann Neurol. 2005;58:7–17. doi: 10.1002/ana.20513. [DOI] [PubMed] [Google Scholar]
- 5.Pasco PMD, Ison CV, Munoz EL, et al. Understanding XDP through imaging, pathology, and genetics. Int J Neurosci. 2011;121:12–17. doi: 10.3109/00207454.2010.526729. [DOI] [PubMed] [Google Scholar]
- 6.Nolte D, Niemann S, Müller U. Specific sequence changes in multiple transcript system DYT3 are associated with X-linked dystonia parkinsonism. Proc Natl Acad Sci USA. 2003;100:10347–10352. doi: 10.1073/pnas.1831949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makino S, Kaji R, Ando S, et al. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am J Hum Genet. 2007;80:393–406. doi: 10.1086/512129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herzfeld T, Nolte D, Grznarova M, et al. X-linked dystonia parkinsonism syndrome (XDP, lubag): disease-specific sequence change DSC3 in TAF1/DYT3 affects genes in vesicular transport and dopamine metabolism. Hum Mol Genet. 2013;22:941–951. doi: 10.1093/hmg/dds499. [DOI] [PubMed] [Google Scholar]
- 9.Thomas MC, Chiang C-M. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 10.Herzfeld T, Nolte D, Müller U. Structural and functional analysis of the human TAF1/DYT3 multiple transcript system. Mamm Genome. 2007;18:787–795. doi: 10.1007/s00335-007-9063-z. [DOI] [PubMed] [Google Scholar]
- 11.Müller U, Herzfeld T, Nolte D, et al. Letters to the editor. Am J Hum Genet. 2007;81:414–421. doi: 10.1086/519528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borovecki F, Lovrecic L, Zhou J, et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington’s disease. Proc Natl Acad Sci USA. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y, Schapiro MB, Franz DN, et al. Blood expression profiles for tuberous sclerosis complex 2, neurofibromatosis type 1, and Down’s syndrome. Ann Neurol. 2004;56:808–814. doi: 10.1002/ana.20291. [DOI] [PubMed] [Google Scholar]
- 14.Strand AD, Aragaki AK, Shaw D, et al. Gene expression in Huntington’s disease skeletal muscle: a potential biomarker. Hum Mol Genet. 2005;14:1863–1876. doi: 10.1093/hmg/ddi192. [DOI] [PubMed] [Google Scholar]
- 15.Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Marullo M, Zuccato C, Mariotti C, et al. Expressed Alu repeats as a novel, reliable tool for normalization of real-time quantitative RT-PCR data. Genome Biol. 2010;11:R9. doi: 10.1186/gb-2010-11-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hruz T, Wyss M, Docquier M, et al. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genom. 2011;12:156. doi: 10.1186/1471-2164-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamova BS, Apperson M, Walker WL, et al. Identification and validation of suitable endogenous reference genes for gene expression studies in human peripheral blood. BMC Med Genom. 2009;2:49. doi: 10.1186/1755-8794-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shamir R, Maron-Katz A, Tanay A, et al. EXPANDER–an integrative program suite for microarray data analysis. BMC Bioinform. 2005;6:232. doi: 10.1186/1471-2105-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulitsky I, Maron-Katz A, Shavit S, et al. Expander: from expression microarrays to networks and functions. Nat Protoc. 2010;5:303–322. doi: 10.1038/nprot.2009.230. [DOI] [PubMed] [Google Scholar]
- 23.Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Syst Biol. 2007;1:8. doi: 10.1186/1752-0509-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulitsky I, Laurent LC, Shamir R. Towards computational prediction of microRNA function and activity. Nucleic Acids Res. 2010;38:e160-e160. doi: 10.1093/nar/gkq570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elkon R, Linhart C, Sharan R, et al. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Subramanian A, Tamayo P, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim TH, Barrera LO, Zheng M, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalman MR, Deeter A, Nimishakavi G, Duan Z-H. Fold change and p value cutoffs significantly alter microarray interpretations. BMC Bioinformatics. 2012;13:S11. doi: 10.1186/1471-2105-13-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 31.White MP, Rufaihah AJ, Liu L, et al. Limited gene expression variation in human embryonic stem cell and induced pluripotent stem cell-derived endothelial cells. Stem Cells. 2013;31:92–103. doi: 10.1002/stem.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bithell A, Johnson R, Buckley NJ. Transcriptional dysregulation of coding and non-coding genes in cellular models of Huntington’s disease. Biochem Soc Trans. 2009;37:1270–1275. doi: 10.1042/BST0371270. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharya S, Lou X, Hwang P, et al. Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D. Proc Natl Acad Sci USA. 2014;111:9103–9108. doi: 10.1073/pnas.1408293111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunah AW, Jeong H, Griffin A, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 35.Friedman MJ, Shah AG, Fang Z-H, et al. Polyglutamine domain modulates the TBP-TFIIB interaction: implications for its normal function and neurodegeneration. Nat Neurosci. 2007;10:1519–1528. doi: 10.1038/nn2011. [DOI] [PubMed] [Google Scholar]
- 36.Shah AG, Friedman MJ, Huang S, et al. Transcriptional dysregulation of TrkA associates with neurodegeneration in spinocerebellar ataxia type 17. Hum Mol Genet. 2009;18:4141–4152. doi: 10.1093/hmg/ddp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuroda TS, Fukuda M, Ariga H, Mikoshiba K. The Slp homology domain of synaptotagmin-like proteins 1–4 and Slac2 functions as a novel Rab27A binding domain. J Biol Chem. 2002;277:9212–9218. doi: 10.1074/jbc.M112414200. [DOI] [PubMed] [Google Scholar]
- 38.Fukuda M. Versatile role of Rab27 in membrane trafficking: focus on the Rab27 effector families. J Biochem. 2005;137:9–16. doi: 10.1093/jb/mvi002. [DOI] [PubMed] [Google Scholar]
- 39.Schreij AMA, Fon EA, McPherson PS. Endocytic membrane trafficking and neurodegenerative disease. Cell Mol Life Sci. 2015 doi: 10.1007/s00018-015-2105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perrett RM, Alexopoulou Z, Tofaris GK. The endosomal pathway in Parkinson’s disease. Mol Cell Neurosci. 2015;66:21–28. doi: 10.1016/j.mcn.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Munsie LN, Milnerwood AJ, Seibler P, et al. Retromer-dependent neurotransmitter receptor trafficking to synapses is altered by the Parkinson’s disease VPS35 mutation p. D620N. Hum Mol Genet. 2015;24:1691–1703. doi: 10.1093/hmg/ddu582. [DOI] [PubMed] [Google Scholar]
- 42.Fenili D, Weng Y-Q, Aubert I, et al. Sodium/myo-Inositol transporters: substrate transport requirements and regional brain expression in the TgCRND8 mouse model of amyloid pathology. PLoS ONE. 2011;6:e24032. doi: 10.1371/journal.pone.0024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed I, Sbodio JI, Harraz MM, et al. Huntington’s disease: neural dysfunction linked to inositol polyphosphate multikinase. Proc Natl Acad Sci USA. 2015;112:9751–9756. doi: 10.1073/pnas.1511810112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wieland I, Jakubiczka S, Muschke P, et al. Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome. Am J Hum Genet. 2004;74:1209–1215. doi: 10.1086/421532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Rawe JA, Wu Y, Dörfel MJ, et al. TAF1 variants are associated with dysmorphic features, intellectual disability, and neurological manifestations. Am J Hum Genet. 2015;97:922–932. doi: 10.1016/j.ajhg.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartkuhn M, Renkawitz R. Long range chromatin interactions involved in gene regulation. Biochim Biophys Acta. 2008;1783:2161–2166. doi: 10.1016/j.bbamcr.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Edvardson S, Cinnamon Y, Ta-Shma A, et al. A deleterious mutation in DNAJC6 encoding the neuronal-specific clathrin-uncoating Co-chaperone auxilin, is associated with juvenile parkinsonism. PLoS ONE. 2012;7:4–8. doi: 10.1371/journal.pone.0036458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koroglu C, Baysal L, Cetinkaya M, et al. DNAJC6 is responsible for juvenile parkinsonism with phenotypic variability. Parkinsonism Relat Disord. 2013;19:320–324. doi: 10.1016/j.parkreldis.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Liu S, Zhang Y, Bian H, Li X. Gene expression profiling predicts pathways and genes associated with Parkinson’s disease. Neurol Sci Off J Ital Neurol Soc Ital Soc Clin Neurophysiol. 2016;37:73–79. doi: 10.1007/s10072-015-2360-5. [DOI] [PubMed] [Google Scholar]
- 50.Coppede F, Migliore L. DNA damage in neurodegenerative diseases. Mutat Res. 2015;776:84–97. doi: 10.1016/j.mrfmmm.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 51.Schapira AHV, Olanow CW, Greenamyre JT, Bezard E. Slowing of neurodegeneration in Parkinson’s disease and Huntington’s disease: future therapeutic perspectives. Lancet (London, England) 2014;384:545–555. doi: 10.1016/S0140-6736(14)61010-2. [DOI] [PubMed] [Google Scholar]
- 52.Shen M. Basic helix-loop-helix protein DEC1 promotes chondrocyte differentiation at the early and terminal stages. J Biol Chem. 2002;277:50112–50120. doi: 10.1074/jbc.M206771200. [DOI] [PubMed] [Google Scholar]
- 53.Qian Y, Zhang J, Yan B, Chen X. DEC1, a basic helix-loop-helix transcription factor and a novel target gene of the p53 family, mediates p53-dependent premature senescence. J Biol Chem. 2008;283:2896–2905. doi: 10.1074/jbc.M708624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Study workflow showing the high-throughput (microarray-based, left-side) and gene-level (qPCR-based, right-side) experiments and analyses. One lane on the microarray had a defect that was undetected during hybridization but was later made obvious by quality control assessments. The blood-derived sample from an affected individual was not included in further analyses. (PDF 98 kb)
Supplementary Fig. 2. Expression in various parts of the brain of the different genes chosen for follow-up. Data is derived from the UK Brain Expression Consortium (http://www.braineac.org/). From top left, clockwise: transcript-level expression of SYTL2, SLC5A3, EFNB1, MRPS6, BHLHE40, KCND2, ATP6V0E2, TAF1. (PDF 296 kb)