Abstract
Histones are the primary protein component of chromatin, the mixture of DNA and proteins that packages the genetic material in eukaryotes. Large amounts of histones are required during the S phase of the cell cycle when genome replication occurs. However, ectopic expression of histones during other cell cycle phases is toxic; thus, histone expression is restricted to the S phase and is tightly regulated at multiple levels, including transcriptional, post-transcriptional, translational, and post-translational. In this review, we discuss mechanisms of regulation of histone gene expression with emphasis on the transcriptional regulation of the replication-dependent histone genes in the model yeast Saccharomyces cerevisiae.
Keywords: Yeast, Transcription, Hir1, Regulation of histone gene expression, Rtt106
Introduction
Chromatin refers to the complex mixture of DNA and proteins that together constitute the eukaryotic chromosome. The nucleosome, the fundamental repeating unit of chromatin, is composed of a histone octamer around which a 147-bp stretch of DNA is wrapped. Each octamer is composed of two H3–H4 histone dimers bridged together as a stable tetramer that is flanked on either side by two separate H2A–H2B dimers [1]. The four core histones (H2A, H2B, H3, and H4) are among the most conserved eukaryotic proteins in primary amino acid sequence and are uniformly small in size (<20 kD), highly enriched in basic amino acids (>20 % lysine/arginine residues), and contain the conserved histone fold domain that interacts non-specifically with the surrounding DNA and mediates histone–histone interactions [1, 2].
Histone proteins are generally divided into two classes. The first class is composed of replication-dependent (RD) histones whose expression is induced right before and during DNA replication (see below). This class includes the core set of histones that compose the nucleosome, H2A, H2B, H3, H4, as well as the linker histones that bind the linker DNA located between nucleosomes. Linker histones are thought to coordinate the packing of nucleosomes into higher order chromatin structure. The second class of histone proteins is composed of histone variants that are generally expressed at a relatively low level through all stages of the cell cycle, and are therefore classified as replication-independent (RI). RI histone variants have specialized, distinct functions. For example, the variants of the core histones H2A and H3, named H2A.Z and H3.3, respectively, function specifically in transcription [3], whereas the H2A variant, H2A.X, is linked to maintenance of genome stability [4]. We will focus on a review of mechanisms that regulate the expression of the set of RD core histone genes encoding H2A, H2B, H3, and H4.
A detailed understanding of the regulation of RD histone expression is of considerable general interest for several reasons. First, expression of RD core histone gene expression is subject to exquisite regulation in eukaryotes. When a eukaryotic cell replicates its DNA in the synthetic or S phase of the cell cycle, the genome doubles in size, demanding a corresponding doubling of histones to package the newly synthesized genome. Therefore, a burst of RD histone expression is needed to produce histones for quick and efficient packaging of the newly synthesized DNA. In fact, histone expression in yeast starts prior to S phase—during late G1 phase—to ensure that adequate histones are present as DNA is synthesized [5]. Second, DNA replication in the absence of new histone synthesis in the model yeast Saccharomyces cerevisiae (Sc) is lethal to the cell [6]. Additionally, if RD core histones are present in excess and accumulate as free soluble proteins, cell fitness is compromised and cells become sensitive to DNA-damaging agents [7]. Therefore, RD core histone synthesis must be repressed outside of S phase—during G1, G2 mitosis (M phase)—and when DNA damage forces the cell to abruptly stop replicating its DNA in order to repair it. The achievement of RD histone expression homeostasis requires complex regulation, which occurs at different levels ranging from transcriptional to post-translational. Here, we review the available literature on the mechanisms underlying the regulation of histone expression in Sc with emphasis on transcriptional regulation.
RD core histones in Yeast
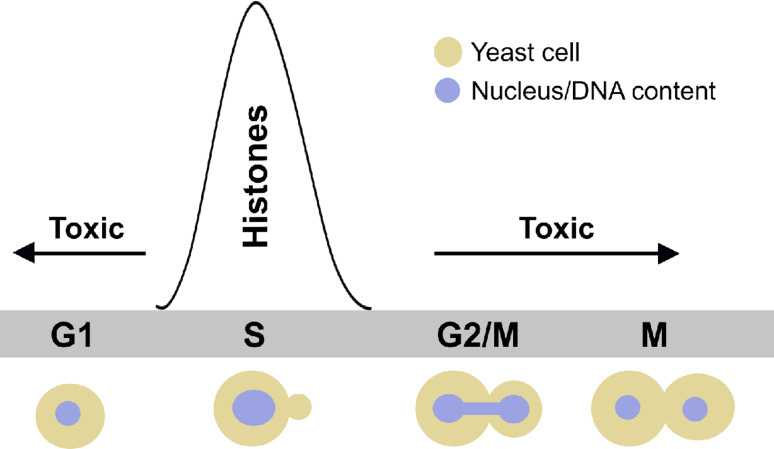
All known eukaryotes possess more than one gene encoding each RD core histone protein. For example, the human and mouse genomes encode 14 copies of the RD histone H4 gene alone [8]. Consequently, molecular genetic analysis of core histone genes can be technically challenging. The most experimentally manageable eukaryote with respect to genetic analysis of RD histones is Sc, which arguably remains the most useful model to understand the mechanisms regulating core histone gene expression. The Sc genome contains two copies of each RD histone gene, each arranged in opposite orientation to a gene encoding its partner within the nucleosome: HHT1–HHF1 and HHT2–HHF2, the two gene pairs encoding core histones H3 and H4 [9], and HTA1–HTB1 and HTA2–HTB2, the two gene pairs encoding H2A and H2B [10]. Each pair of genes is regulated by centrally located, divergent promoter elements [11]. All eight genes share a common expression pattern that is cell cycle-dependent (Fig. 1): very low expression occurs outside of S phase except in late G1 (immediately prior to initiation of DNA replication) when histone gene transcripts begin to accumulate before peaking in mid-S phase, after which expression levels decline until cells exit S phase and enter into the G2- and M-phases [5].
Fig. 1.
Core histone expression during the yeast cell cycle. RD core histone genes are expressed exclusively in late G1 and S phase of the cell cycle, in parallel with the replication of DNA. Outside of the S phase, histone expression is toxic; therefore, cells tightly restrict histone production to the S phase. See text for details
Intuitively, it makes sense that RD histone expression should occur primarily in S phase, but what is the evidence that restricted histone expression is important for proper cell division and fitness? Several dramatic phenotypes are associated with inappropriate histone gene expression: (1) if levels of core histones are inadequate yet DNA replication proceeds, yeast cells exhibit dramatic genome instability or mitotic arrest [12, 13], (2) a failure to repress histone expression following DNA replication and RD chromatin assembly—for example, in cells with over- or ectopic expression of core histones—is highly toxic to Sc and other eukaryotic cells (Fig. 1), due to failure to correctly segregate chromosomes at mitosis [14], and (3) the accumulation of soluble histones results in sensitivity to DNA-damaging chemicals which is exacerbated by deletion of genes whose wild-type (WT) role is to negatively regulate expression of RD histones [7]. For example, a loss of function mutation in the LSM1 gene, which results in increased histone gene mRNA outside of S phase (see section below on post-transcriptional regulation), causes a genome instability phenotype manifested experimentally by sensitivity to hydroxyurea (HU), a chemical that depletes deoxyribonuleotides and causes DNA replication fork disruption, thus triggering a DNA damage checkpoint which arrests cells within S phase, allowing them to repair their damaged DNA. The HU-sensitive phenotype of an lsm1Δ strain can be suppressed by lowering histone dosage through deletion of one of the two respective H2A–H2B or H3–H4 encoding gene pairs [15]. The deleterious effects of histone over-expression are not exclusive to yeast/fungi. For example, in Drosophila melanogaster, a loss of function mutation in the abnormal oocyte (ABO) gene, which encodes a protein that negatively regulates core histone gene expression during embryogenesis, is maternal-lethal suggesting that WT expression levels of genes encoding core histones is critical for normal embryogenesis [16].
Replication-dependent (RD) histone gene expression must also be repressed sometime within, in addition to outside, S phase. Upon genotoxic stress, DNA replication may be abruptly blocked or disrupted. If the replication fork encounters DNA damage, a DNA damage checkpoint is triggered, and soluble RD histones may accumulate to toxic levels if expression continued unabated. To prevent this, eukaryotic cells possess mechanisms that tightly link histone expression to DNA replication progression so that histone transcription may be repressed quickly within S phase once a delay or blockage in DNA replication has occurred. In Sc, this phenomenon was initially observed when histone expression was assessed in the DNA replication (temperature-sensitive) mutant cdc8 [5], which encodes thymidylate kinase, an enzyme important for S phase progression [17]. When DNA synthesis is arrested by shifting cdc8 ts cells to the non-permissive temperature, RD histone expression is quickly halted as well [5]. Further analysis revealed that a rapid arrest of RD histone gene expression also occurs in Sc in response to HU [18].
As outlined above, expression of RD histone genes is regulated in a cell cycle-specific manner and in response to DNA damage. In many eukaryotic cells, regulation of histone levels involves transcriptional, post-transcriptional, translational, and post-translational mechanisms. Although in Sc regulation of core histone genes is best understood at the transcriptional level, increasing evidence points to regulation at other levels as well. Below, we focus mainly of the regulation of core histone gene transcription in Sc with reference to other regulatory mechanisms and models where appropriate.
Cis- and Trans-acting factors regulating transcription of RD core histone genes in Sc
Cis-acting sequences regulating histone gene transcription in Sc
The Sc HTA1–HTB1 locus has provided an excellent model for understanding mechanisms involved in the transcriptional regulation of RD histone genes. The first model to explain the mechanism of S phase-specific expression of histone genes proposed a direct coupling of transcription to the DNA replication apparatus, and was based on physical linkage of the HTB1 gene to an autonomously replicating sequence (ARS) [11]. However, subsequent experiments revealed no effect of the ARS on transcription, but instead uncovered the existence of two separate and important classes of cis-acting sequences located in the ~800-bp intergenic regulatory region between HTA1 and HTB1 [11]. The first cis-acting sequence is referred to as a histone upstream activating sequence (UAS) and is responsible for S phase-specific activation of transcription. In HTA1–HTB1, several copies of the 16-bp UAS sequence (GCGAAAAANTNNGAAC) are situated ~200–300 bp upstream of each transcriptional start site and each copy has a positive and additive effect on the S phase-specific periodicity of HTA1 and HTB1 mRNA levels [11]. The UASs are found in the intergenic regions of each of the four sets of RD core histone genes in Sc [19], and are sufficient to confer cell cycle-regulated expression of a constitutively expressed reporter gene [11].
The second class of cis-acting sequence is the cell cycle control region (CCR), which was identified through analysis of a LacZ reporter gene linked to the HTA1–HTB1 regulatory region. Deletion of the CCR in this context causes constitutive expression reporter gene expression, suggesting that CCR negatively influences transcription of RD histone genes [11]. In addition, deletion of the CCR region (also referred to as NEG) causes a loss of cell cycle control of expression of HTA1–LacZ despite the continued presence of the UAS that confers cell cycle-dependent expression in the presence of NEG. Importantly, NEG is also required for the rapid arrest of RD histone expression when Sc is treated with HU which triggers the intra-S checkpoint as described above [18]. A single copy of the NEG sequence is found in the intergenic regions of three of the four divergent histone gene pairs with the exception being the HTA2–HTB2 regulatory region. Thus, there are similarities (copies of histone UASs upstream of all eight RD histone genes) and differences (presence of NEG in three of the four histone gene pairs) in transcriptional regulation of the four Sc histone gene pairs. A major focus in the field has been to elucidate how these two classes of cis-acting sequences collaborate to coordinate the periodic transcription of the core histone genes.
Trans-acting factors with roles in repression of histone gene transcription
HIR complex
HIR1, HIR2, and HIR3 (histone regulation) were the first genes identified as regulators of RD histone transcription, in a screen for mutants with defective HTA1–lacZ reporter gene expression [20]. The so-called Hir− phenotype of the three mutants results from a failure to repress expression of both the lacZ reporter, and the actual HTA1 locus is de-repressed as well. Failure to repress reporter/HTA1 expression in any of the hir mutants is also observed when an intra-S DNA damage checkpoint is triggered with HU [20]. If Hir2 is artificially tethered to a HTA1–lacZ reporter gene, it represses transcription in a manner requiring both Hir1 and Hir3 [21], consistent with a role for the Hir proteins as histone gene transcriptional repressors. A fourth gene showing a mutant Hir− phenotype, histone promoter control (HPC2), was identified in a separate genetic screen [22]. Interestingly, the Hir− phenotype of the Hir/HPC mutant strains phenocopies NEG deletion, and repression of a NEG-containing HTA1–LACZ reporter is lost in a hir mutant background [20], suggesting that HIR/HPC2 gene products act through NEG. Also, deletions of each of the HIR/HPC genes affects expression of only three of the four histone gene pairs, the exception being HTA2–HTB2, which lacks NEG. Biochemical experiments support a direct regulatory relationship between the Hir/HPC proteins: all four of these gene products co-purify as a protein complex [23, 24] from yeast extracts and individual deletion of any of the four genes compromises integrity of the complex [23]. In addition, all four Hir/Hpc subunits specifically localize to chromatin at the HTA1–HTB1 region that contains NEG [23, 25] as assessed using chromatin immuno-precipitation (ChIP). Insight into the mechanism of HIR-mediated repression at NEG-dependent histone loci derived from the demonstration that the HIR complex functions as a RI histone H3–H4 chaperone [23, 24]. Correspondingly, individual deletion mutants of members of the HIR complex exhibit a significant decrease in histone density in the regulatory region of NEG/HIR-regulated histone gene pairs [25]. Thus, an intact HIR complex localizes to chromatin at the CCR/NEG region and is necessary for repression of three of the four histone loci (HTA1–HTB1, HTT1–HHF1, and HTT2–HFF2). HIR-mediated histone gene repression is a consequence of its activity as a histone chaperone, likely because the Hir complex helps assemble histones that occlude RNA polymerase II (RNAPII) recruitment onto promoter sequences (see below).
How is HIR recruited to NEG? Consistent with specific localization of Hir proteins to chromatin, gel shift assays showed that purified HIR complex binds both nucleosomes and DNA [24]. However, the DNA-binding activity of the HIR complex appears non-specific since HIR binds equally well to NEG and non-NEG containing double-stranded DNA oligonucleotides [24]. Although HIR does not appear to directly bind NEG, the site seems necessary for recruitment of HIR to chromatin in the region. Thus, the precise function of NEG remains enigmatic. Is NEG sufficient or is an additional trans-acting factor required to recruit HIR to CCR/NEG region? It has been speculated that there is likely an as yet unidentified sequence-specific DNA-binding factor(s) whose function is to recruit HIR to the CCR/NEG region. The identification of a short peptide sequence within Hpc2, that when deleted prevents HIR recruitment to histone gene promoters but does not disrupt the integrity of the complex suggests, that one function of Hpc2 within HIR may be to physically interact with this hypothetical trans-acting factor [26]. Because the histone UASs flank CCR/NEG, it will be interesting to determine the effect of deleting SPT10, a UAS-binding histone gene activator (see below), on HIR recruitment to the CCR/NEG region.
The universality of HIR function in the regulation of histone gene expression remains to be determined. The genes encoding all four HIR components of Sc are conserved within the crown eukaryotes (plants, fungi, animal) with the caveat that most have a single protein, HIRA, that is the equivalent of a fusion between the N-terminus of Sc Hir1 and the C-terminus of Sc Hir2 [27]. In support of a universal role in the regulation of histone gene transcription, ectopic expression of HIRA in human U2OS cells represses transcription of histone genes leading to a block in DNA synthesis [28]. Recently, it was demonstrated that phosphorylation by the WEE1 kinase of H2B at tyrosine 37 upstream of the Hist1 gene cluster of RD histone genes [8] in human cells is required to recruit HIRA in order to repress transcription of these genes [29]. Mutation of the corresponding tyrosine on H2B in Sc (H2BY40A) increases expression of all four histone gene pairs [29] and is dependent on the WEE1-related cell cycle kinase, Swe1. It is not yet known whether H2B phosphorylation disrupts HIR binding in Sc, although other mechanisms must also be in play since both NEG-regulated and non-NEG-regulated histone gene loci are affected in the H2B phosphorylation mutant [29]. Besides HIRA, the roles of other putative HIR members in the regulation of histone gene expression (UBN1 similar to Hpc2 [30], CABIN1 similar to Hir3 [31]) have not been tested outside of Sc. Nonetheless, in human cells, HIRA, UBN1, and Asf1a localize preferentially to nucleosome-free regions in transcription start sites in gene promoters [32]. Taken together, these data suggest that important aspects of HIR repression of histone gene expression are conserved throughout eukaryote evolution.
Asf1 and Rtt106
HIR-mediated repression of RD histone transcription also involves two additional H3-H4 histone chaperones, Asf1 and Rtt106. Several lines of evidence, including yeast two hybrid assays and affinity purification, have demonstrated that Asf1 interacts with the HIR complex via direct physical interaction with Hir1 [23, 33], an interaction that appears evolutionarily conserved. Consistent with their physical association, deletion of ASF1 results in a Hir− phenotype [33] with respect to RD histone expression and Asf1 localizes to NEG-containing chromatin [34]. Deletion of ASF1 does not disrupt integrity of the HIR complex or localization of HIR subunits to CCR/NEG-containing chromatin [23, 25], but it does cause a decrease in histone density at the CCR/NEG region, similar to that observed for individual deletion of HIR subunits [25]. Together, these results suggest a role for Asf1 in RD histone gene regulation downstream of HIR.
Regulator of Ty1 Transposition (RTT106) was originally identified as a gene whose deletion causes an increase in transposition of the Ty1 retroelement [35]. A later large-scale screen revealed that deletion of RTT106 also enhances the defective gene silencing of a pol30-879 strain which is mutated for PCNA, an essential DNA replication factor [36]. Subsequent genetic analysis revealed that the RTT106 function in silencing is linked to ASF1 and HIR1, but independent of CAF-1, another chromatin assembly factor involved with histone assembly on replicating DNA [36]. More recently, a role for Rtt106 in RD histone gene repression was discovered using a novel dual reporter-based synthetic genetic array (R-SGA) screen [25]. In the R-SGA, array-based yeast genetics was used to engineer yeast cells containing a plasmid with the HTA1 upstream regulatory sequences directing cell cycle-specific synthesis of a green fluorescent protein (GFP) reporter, and also expressing an integrated control reporter composed with a constitutive promoter driving expression of tdTomato [37] (RFP). The two reporters were introduced into WT cells and the so-called non-essential deletion array, composed of strains individually deleted for each of the ~5,000 non-essential genes in the yeast genome [38]. As its name implies, the R-SGA method exploits SGA technology [39] that automates yeast genetics, enabling rapid introduction of marked genetic elements of interest into the yeast deletion mutant and other arrayed collections through several replica-pinning steps [40]. In R-SGA, the deletion strains carrying the two reporter genes are monitored for relative GFP:RFP fluorescence levels and compared to that of WT Sc. In the HTA1–GFP screen, decreased GFP:RFP identified potential HTA1 activators, while higher GFP:RFP identified putative repressors. As expected for a repressor of HTA1, deletion of ASF1 or any one of the four HIR genes resulted in increased GFP:RFP [25]. Interestingly, increased fluorescence was also observed in the strain containing a deletion of RTT106, implicating Rtt106 in repression of HTA1 [25]. Subsequent experiments showed that Rtt106 specifically cross-links to CCR/NEG-containing chromatin in an ASF1- and HIR-dependent fashion [25]. Also, deletion of RTT106, like that of ASF1 and HIR1, causes a decrease in histone density in the regulatory region of the NEG-dependent gene pairs [25]. Rtt106 functions by binding histones H3 and H4 and has in vitro nucleosome assembly activity, consistent with a histone chaperone role [36]. Rtt106 directly recruits the RSC chromatin remodeling complex (see below) to NEG-containing chromatin outside of S phase [41, 42].
Yta7
The Yeast tat-binding analog 7 (Yta7) protein also functions in HIR/Asf1/Rtt106-mediated repression of the NEG-regulated histone gene pairs in Sc. YTA7 encodes a protein that possesses an AAA-ATPase domain and a bromodomain (BD)-like region [43]. BDs are well characterized acetyl-lysine binding domains, but elucidating the function of Yta7’s non-canonical BD has not been straightforward. Deletion studies indicate that, although the Yta7 BD might not bind acetylated histones as do canonical BDs, it is required for specific Yta7 distribution along chromosomes [44]. Yta7 functions as a boundary protein, acting as a physical barrier between regions of silent and active chromatin around the HMR locus, a role that requires the presence of the Yta7 BD [43–45]. Deletion of YTA7 results in the spreading of silent chromatin from the HMR locus causing repression of a flanking reporter gene [43].
Yta7 was initially implicated in regulation of histone gene expression because deletion of yta7 causes a genetic interaction with deletion of HIR1 [46]. Subsequent ChIP analysis revealed that Yta7 localizes specifically to regulatory regions of the HIR-regulated histone loci [25, 34, 46]. Consistent with these experiments, the R-SGA screen discussed above identified YTA7 as a putative activator of HTA1, since a yta7Δ strain had reduced GFP:RFP fluorescence relative to a WT strain [25]. Also, HTA1 mRNA levels are reduced in yeast cells deleted for YTA7 [25, 34]. ChIP experiments from a yta7Δ strain showed significant mis-localization of Rtt106, but not Hir1 or Asf1, across the HTA1 locus, with spreading of Rtt106 from the NEG region through the HTA1 ORF where it is not normally found [25, 34]. Additionally, the Yta7 AAA-ATPase domain is required to prevent Rtt106 spreading, since a yta7-K460A point mutation which is predicted to disrupt AAA-ATPase activity does not stop Yta7 localization to chromatin but is unable to prevent Rtt106 spreading [47]. Thus, one function of Yta7 in histone gene repression appears to be the restriction of HIR- and Asf1-recruited Rtt106 to the regulatory region of NEG-regulated histone gene pairs. Rtt106 spreading into the HTA1 coding region is therefore postulated to result in repression of transcription, presumably by inhibiting transcriptional elongation by RNAPII, explaining the identification of Yta7 as an activator of HTA1–GFP in an R-SGA assay [25]. Like HIRA and Asf1, Yta7 is conserved in higher order eukaryotes [48]. The ATAD2 protein of humans is highly similar to Yta7 and its expression has been correlated with the proliferative growth of several aggressive cancers; however, its precise function and any relationship with RD core histone gene expression remains to be determined [49–51]. Although unrelated at the sequence level, Yta7 appears to function similarly to mammalian CCCTC-binding factor (CTCF), which binds insulator elements to act as a barrier protein in vertebrates, and also regulates gene expression by binding to the regulatory region of imprinted genes [52].
Model for histone gene repression
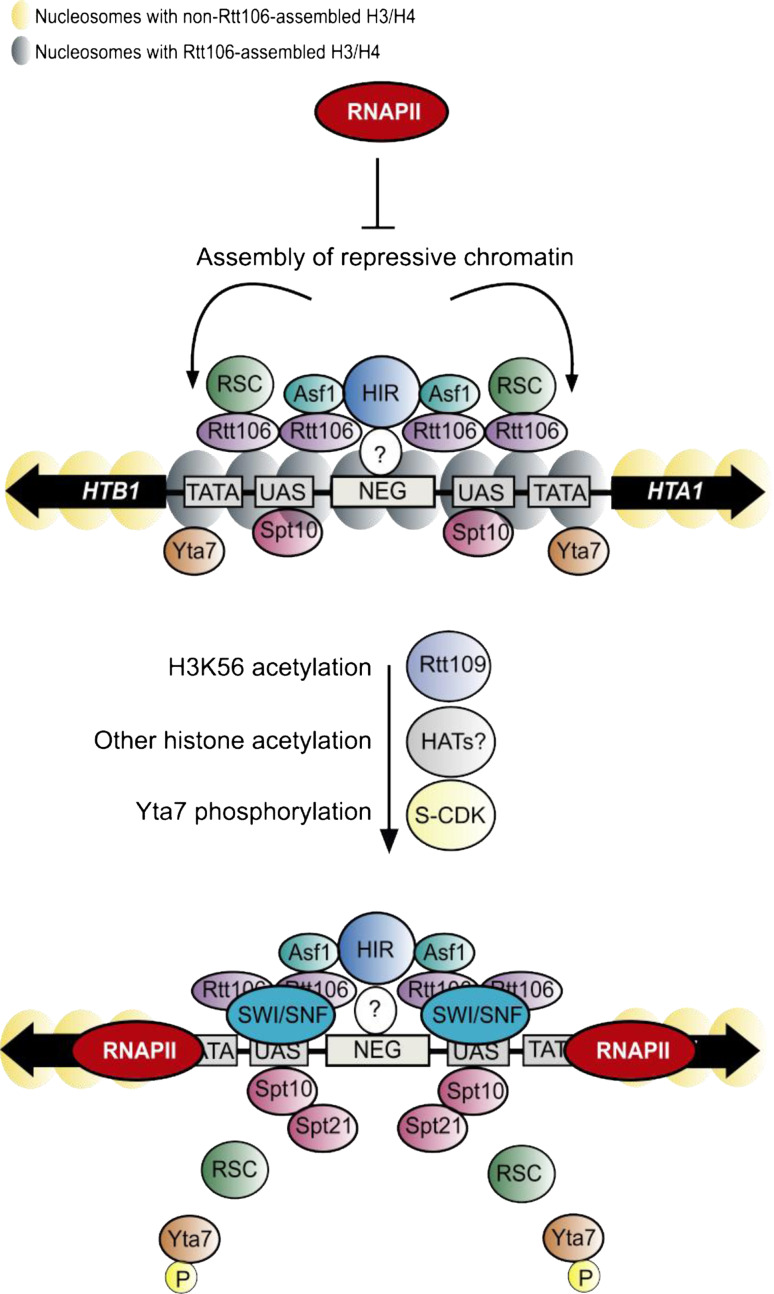
How do the HIR, Asf1, and Rtt106 histone H3–H4 chaperones collaborate with boundary protein Yta7 to repress transcription of HTA1–HTB1, HHT1–HHF1, and HHT2–HHF2? Although HIR and Asf1 interact directly, Rtt106 interaction with Asf1 (or HIR) appears to be indirect and mediated through histones H3–H4. Mutations that disrupt/reduce the histone-binding ability of Asf1 and/or Rtt106 result in reduced or abolished recuitment of Rtt106 to the histone loci as well as increased transcription of HTA1 [34, 53, 54]. Rtt106’s function in repression appears linked to another protein complex previously implicated in repression of CCR/NEG-regulated histone loci, the ATP-dependent chromatin remodeling complex remodels structure of chromatin (RSC). A genome-wide analysis of RSC localization revealed HIR-dependent RSC association with CCR/NEG-containing chromatin regions [42]. RSC recruitment to histone loci coincides with periods of histone gene repression [42], and is dependent on Asf1 and Rtt106 [41]. Consistent with a functional linkage to Rtt106, the RSC component Rsc8 spreads along with Rtt106 in a yta7 mutant background [47]. Although the presence of RSC at histone regulatory regions is tighly linked to that of Rtt106, its exact contribution to repression remains unclear. Whether NEG-regulated histone gene repression depends on expression of any RSC complex components remains to be tested. A simple model explaining repression of NEG-dependent RD core histone genes outside of S phase is diagramed in Fig. 2. In this model, Rtt106 collaborates with RSC to assemble H3–H4 onto chromatin. Chromatin assembly occludes promoter sequences, which prevents the recruitment of the general transcription apparatus and RNAPII [47].
Fig. 2.
Repression/activation model for the regulation of RD histone gene transcription in yeast. Upper panel a repressed HTA1–HTB1 gene promoter. Here, HIR recruits Rtt106, RSC, Asf1, and Yta7 in order to generate a repressive chromatin structure (nucleosomes with Rtt106-assembled H3/H4) at promoters. NEG also CCR (cell cycle control region), UAS upstream activating sequence and TATA (TATA-box). A repressive chromatin structure prevents the recruitment of the basal transcription machinery and RNAPII. See text for details. Lower panel the HTA1–HTB1 promoter in an active state. Rtt109-dependent incorporation of H3 acetylated at K56 (H3K56ac) enables recruitment of SWI/SNF, which generates a nucleosome-depleted promoter, enabling RNAPII recruitment. Other unidentified HATs are very likely involved in this process as well (see text for details). S phase forms of CDK1 (S-CDK) then phosphorylate Yta7 causing its eviction from promoters (which is tracked by the RSC), which is important for efficient promoter escape and transcript-elongation by RNAPII. Spt10 and Spt21 have important roles in these processes as well, but their molecular functions remain to be revealed
What is the signal for repression of histone transcription as the S phase draws to a close? The negative feedback model [34, 55, 56] proposes that soluble histone levels are monitored by HIR/Asf1/Rtt106. According to this hypothesis, when DNA replication is almost finished there are fewer available genomic locations for assembly of newly synthesized histones. Therefore, at this time, all histone chaperones including HIR, Asf1, and Rtt106 become fully charged with their histone substrate, which in turn facilitates their assembly onto NEG-containing chromatin with concomitant promoter occlusion and reduced recruitment of RNAPII. This model is supported by analysis of yeast strains deleted for genes encoding the chromatin assembly factors/histone chaperones Spt4, Spt5, and Spt6, which exhibit reduced transcription of HTA1–lacZ in a manner dependent on NEG [57]; in several cases, the defective activation phenotype is suppressed by H3–H4 over-expression [57]. These observations are consistent with the negative feedback model: deletion of genes encoding other histone chaperones like Spt4, Spt5, and Spt6 may decrease the histone-binding ‘capacity’ in the cell, promoting loading/charging of other chaperones like HIR/Asf1/Rtt106 earlier than normal; this may cause premature repression of histone genes transcription and thus less histone mRNA. In this scenario, NEG-dependence is a simple consequence of HIR/Asf1/Rtt106-mediated assembly or repressive chromatin—without NEG, there can be no HIR-mediated repression.
Trans-acting factors with roles in activation of histone gene transcription
SWI/SNF
The first activator identified to function in histone gene expression was the SWI/SNF ATP-dependent chromatin remodeling complex [58]. SWI/SNF subunits Swi2 and Snf5 specifically cross-link to NEG region chromatin in a cell cycle-regulated fashion, with peak association during late G1/S when histone genes are transcribed [41, 58]. Deletion of SNF5 strongly reduces histone gene expression but only in the presence of intact NEG or HIR, suggesting that NEG-dependent, HIR-mediated repression (also including Asf1 and Rtt106, see above) creates a transcriptional block that must be overcome by ATP-dependent chromatin remodeling [58]. Snf5 ChIP assays revealed that recruitment of SWI/SNF to the HTA1 locus requires the putative lysine acetyltransferase SPT10 (see below) and is abolished in a H3K56R mutant [59]. Specific dependence of SWI/SNF recruitment on the H3K56-specific histone acetyl-transferase (HAT) RTT109 (see below) remains to be established.
In addition, deletion of genes encoding any of the four HIR subunits, Asf1, or Rtt106 prevents recruitment of SWI/SNF subunits to histone genes [15, 58]. Rtt106 is likely to directly recruit SWI/SNF through a physical interaction that has demonstrated both in vivo and in vitro [41].
Spt10 and Spt21
SPT10 (suppressor of Ty) and SPT21 were originally identified in a genetic screen for mutations that suppress a transcriptional defect linked to insertion of a Ty1 transposon, permitting transcription of a flanking reporter to initiate from the 3′ long terminal repeat (LTR) of Ty1 elements [60, 61]. SPT10 is required for transcription of all core histone genes including the non-NEG-containing HTA2–HTB2 loci [59, 62]. SPT10 encodes a DNA-binding protein that binds with high affinity, and in a cooperative manner, to pairs of the histone UAS elements [63], four of which are found in the regulatory regions of the four histone gene pairs (see above). In addition to its DNA-binding domain, Spt10 possesses a histone acetyl-transferase (HAT) domain of the Gcn5-related N-acetyltransferases (GNAT) family [64]. Consistent with a role in histone acetylation, SPT10 is required for the S phase-dependent accumulation of H3K9ac, H3K18ac, and H3K56ac in the regulatory region of all four histone gene loci [59]. Mutation of a putative catalytic residue in the HAT domain of Spt10 [62] results in a loss-of-function phenotype but HAT activity for Spt10 has never been directly demonstrated. Also, the catalytic-site mutant of Spt10 is very unstable in vivo [64], making it difficult to link the phenotype to a loss of putative HAT activity.
The Spt21 protein is functionally linked to Spt10 [65]. Spt21 is required for S phase binding of Spt10 to the regulatory region of all four histone gene pairs [66]. Deletion of SPT21 results in significant reduction of transcription of the HTA2–HTB2 [67] gene pair that does not contain a CCR/NEG region. In addition, Spt10 and Spt21 physically interact in vitro and, like Spt10, Spt21 cross-links to the regulatory region of all four histone gene pairs [66]. Spt10 and Spt21 are also functionally linked to transcriptional silencing [68] and affect repression of the three NEG-dependent histone loci [69]. The precise roles of Spt10 and Spt21 in regulating histone gene transcription and whether HAT activity is involved remain to be elucidated.
MBF and SBF
MBF and SBF are activators of G1/S-expressed genes [70] and were functionally linked to the histone gene activation using global ChIP analyses [71, 72]. SBF and MBF share a common regulatory subunit called Swi6, and have dedicated DNA binding subunits, Swi4 and Mbp1, in SBF and MBF, respectively [73]. Consistent with a role in activation of histone gene expression, deletion of either SWI4 or MBP1 reduces levels of histone mRNA, and either mutant is lethal when combined with a mutation of SPT10 [66]. This synthetic lethality makes sense in that consensus binding sites for Swi4 and Mbp1 overlap with the histone UAS, suggesting that SBF/MBP1 and Spt10 function in separate activation pathways [74]. In fact, Spt10 and SBF together can recapitulate the WT cell-cycle dependence of HTA1–HTB1 transcription with SBF responsible for a small early peak of transcription (late G1) and Spt10 for the remainder (S phase) [74]. The mechanism by which SBF and MBP1 activate histone gene transcription remains to be determined. Given what is known about SBF/MBF activation of G1-S genes, possible mechanisms include antagonism of HIR/Rtt106-assembled chromatin or recruitment of classic co-activators such as SAGA or Mediator.
Rtt109–Vps75
As well as identifying new repressors of histone genes (see above), R-SGA analysis of HTA1–GFP also identified RTT109 and VPS75 as genes encoding potential activators of histone gene expression [25]. Expression of HTA1 is decreased in cells deleted for RTT109 consistent with a role in activation [25]. RTT109 encodes a HAT that, in conjunction with Asf1 [75], catalyzes H3K56ac [76–78], a modification associated with RD chromatin assembly [79]. During S phase, Rtt109–Asf1-catalyzed H3K56ac enhances RD chromatin assembly by increasing the interaction between H3–H4 dimers and Rtt106 and CAF-1 histone chaperones [80]. Additionally, H3K56ac leads to chromatin disassembly during transcription activation of PHO5 [81]. During this process, Rtt109-acetylated H3K56 on Asf1-bound H3–H4 is exchanged with unacetylated histones on chromatin leading to disassembly in conjunction with ATP-dependent chromatin remodeling. R-SGA also implicated VPS75 in regulation of HTA1 [25]. In vivo, Rtt109 forms a stable protein complex with Vps75 which is also an H3–H4 histone chaperone [82, 83]. Rtt109–Vps75 acetylates H3K9ac in vivo in a pathway that requires ASF1 [84].
Model for histone gene activation
We previously proposed that activation of S phase-specific transcription of NEG-regulated histone genes occurs by overcoming constitutive HIR/Rtt106 repressive chromatin [25]. Specifically, we suggest that Rtt109 enhances transcription of the HTA1 gene (and likely other histone genes) by encouraging chromatin disassembly at the locus through deposition of H3K56ac–H4 dimers, promoting a more “open” nucleosome conformation [79, 85]. When incorporated into chromatin at the NEG-region, H3K56ac promotes SWI/SNF-dependent chromatin disassembly and subsequent activation of transcription. SWI/SNF is possibly recruited initially to the NEG-regulated histone loci via its demonstrated physical interaction with Rtt106 (Fig. 2). Evidence for this model includes the observation that SWI/SNF recruitment to HTA1 is abolished in a H3K56R strain [59], and the fact that histone promoters are nucleosome-free in asf1Δ, hir1Δ, and rtt106Δ strains [25], suggesting that proper NEG-based repression is a function of chromatin assembly. Further tests of the model ought to include assessment of SWI/SNF recruitment in RTT109 (and VPS75) mutants, and analysis of the interplay between SWI/SNF, Rtt106, and H3K56ac in promoting proper histone gene expression.
A seeming paradox exists in this model in that we propose Rtt109 activates histone gene expression but Asf1, its cellular collaborator in H3 acetylation, is a repressor. This paradox is potentially resolvable in a scenario in which Asf1 both activates and represses core histone transcription, an idea that has been previously suggested by Sutton et al. [33]. Thus, we propose that, like Rtt106, HIR and Asf1 are also tethered to histone promoters through the cell cycle [25]. Continuous HIR/Asf1-based recruitment of Rtt106 would ensure repressive chromatin is always assembled outside of S phase. However, in late G1 and S phase when core histone genes are transcribed, Rtt109 works via the tethered Asf1 to acetylate H3 which is then deposited into chromatin via Rtt106, leading to activation.
The model for histone gene activation we summarize above raises many questions that remain to be addressed:
What directs Rtt109’s acetylation activity to histone promoters? We suggest that UAS-recruited Spt10, either by itself or with Spt21, directs the assembly of Rtt109-acetylated H3–H4 (H3K56ac) into chromatin at histone promoters in late G1/S. Vps75 may also have a role since SPT10 is also required for accumulation of H3K9ac (59), which is catalyzed by Rtt109–Vps75 in an Asf1-dependent pathway [84].
Is there redundancy in histone acetylation at histone gene promoters during transcriptional activation? To date, Rtt109 has only been shown to activate HTA1. Interestingly, Gcn5, the other H3K9 HAT in yeast, acetylates H3K18 and has been implicated in RD chromatin assembly of H3 and H4. In vivo, SPT10 is required for accumulation of H3K9ac and H3K18ac in addition to H3K56ac in the coding region of HTA2 [59]. It will be interesting to determine if Gcn5 functions in a partially redundant pathway with Rtt109 in histone gene expression downstream of Spt10. Interestingly, H3K56ac is also enriched in chromatin at histone promoters in human cells [86]. The specific HAT(s) responsible for H3K56ac at histone promoters and identity of any downstream proteins recruited following histone modification remain to be determined.
How are Spt10/21 activated in S phase? In the related ascomycete yeast S. pombe, S phase activation of core histone transcription is directed by the Ams2 protein [87], a member of the GATA-type transcription factor family [88]. Activation by Ams2 appears to be restricted to S phase by different E3 ligases that promote its ubiquitination and degradation pre- (G1) and post-S phase (G2/M) [89, 90]. Although any evolutionary relationship between Ams2 and the Spt10 or Spt21 is unclear, it will be interesting to determine if they are subject to similar post-translational regulation.
Regulation of transcriptional elongation at RD histone loci
Cell cycle control of histone gene expression is not restricted to regulation of RNAPII recruitment. Transcriptional elongation at the RD histone loci is also subject to cell cycle control. Outside of the S phase, the AAA-ATPase domain of Yta7 is important for boundary function, as defined by correct positioning of RSC and Rtt106 [47]. During S phase, the N-terminus of Yta7 (where the AAA-ATPase domain is located) is hyper-phosphorylated by both the S phase-specific forms of cyclin-dependent kinase 1 (Cdk1) and casein kinase 2 (CK2). Cdk1 is the major cell cycle kinase in yeast [91] while CK2 has general roles in regulating transcription [92]. Phosphorylation of Yta7 promotes its dissociation from HTA1 chromatin during S phase, which in turn is essential for promoter escape and elongation by RNAPII [47], probably through histone eviction [93]. Notably, Yta7 regulation by Cdk1 represents the first concrete mechanistic link between the cell cycle machinery and to histone gene transcription. In human cells, Cdk colocalizes with the NPAT proteins to histone gene clusters in subnuclear Cajal bodies [94, 95] and phosphorylation of NPAT by cyclin E-CDK2 activates histone gene transcription [94–96]. The underlying molecular mechanisms of NPAT regulation, however, remain elusive.
Spt4, Spt5, Spt6, and the FACT complex, composed of Pob3 and Spt16, are regulators of transcriptional elongation, and all three can be cross-linked specifically to the coding region of the constitutively transcribed PMA1 gene [25]. Spt16 also associates with HTA1 chromatin during the time in the cell cycle when HTA1 is repressed [47], implicating FACT in repression of histone gene transcription. Spt6 and Rtt106 are known to function in parallel to suppress cryptic initiation at an internal promoter within the FLO8 gene and both associate with the coding region of constitutively transcribed genes ([25], see above, [97]). However, at the HTA1–HTB1 locus there exists a physical separation of Rtt106 (and HIR/Asf1), which crosslinks to the regulatory region, from Spt4, Spt5, Spt6, and FACT, which cross-link to the coding region [25]. Thus, although one function of Yta7 may be to restrict Rtt106 to the regulatory region, an additional function may be to ensure exclusive regulation by HIR/Asf1/Rtt106 by preventing FACT and/or Spt4, Spt5, and Spt6 from accessing the region.
Post-transcriptional regulation of histone mRNAs
Clear mechanisms exist to regulate histone mRNAs post-transcriptionally. In metazoans, RD core histone genes are not polyadenylated, but contain a unique 3′ end structure which consists of an unusual highly conserved stem loop structure that forms in the 3′-untranslated region (UTR) of the mRNA. The conserved stem-loop binding protein (SLBP) interacts with this structure and regulates all downstream events including proper termination, mRNA export, translation, and regulated mRNA degradation post-S phase [98]. Most lower eukaryotes do not share this unique form of regulation although some protozoans appear to encode proteins with similarity to SLBP [99]. In yeast, histone mRNAs are polyadenylated, albeit possessing shorter polyA tails than most yeast genes [100, 101]. Despite the absences in Sc of both a stem loop in 3′-UTR of RD core histone mRNA and the SLBP protein, 3′-UTR-dependent post-transcriptional regulation is a significant contributor to their characteristic cell cycle expression pattern [18]. Fusion of the 3′-UTR of HTB1 to a reporter gene under the control of the GAL1 promoter is sufficient to cause S phase-specific transcript accumulation in the presence of galactose [102]. Thus, it could be argued that mechanisms exist throughout the cell cycle to degrade histone mRNAs, and that this mechanism is antagonized specifically in S phase. Consistent with this hypothesis, a protein that specifically binds the HTB1 3′-UTR has been seen using a gel shift assay with S phase-specific whole cell extracts [103]. The identity of this protein remains unknown.
What are the mechanisms in Sc that monitor histone mRNA accumulation? Regulated mRNA degradation generally occurs in either the 3′–5′ or 5′–3′ direction. Generally, the first step in degradation of a polyadenylated mRNA is deadenylation, which produces an oligoadenylated mRNA. Degradation of oligoadenylated mRNA then proceeds via either of two riboexonucleolytic pathways: the exosome complex degrades mRNA from the 3′–5′ direction, while Xrn1 degrades messages from the 5′–3′ direction. For the Xrn1 exoribonuclease to function, de-capping of the mRNA must first occur (in Sc by Dcp1/Dcp2). The Lsm1-7–Pat1 complex is composed of seven Sm-like proteins (Lsm1 through Lsm7 as well Pat1) and functions to stimulate de-capping by binding to oligoadenylated mRNAs. Binding of Lsm1-7–Pat1 to deadenylated mRNA is enhanced on mRNAs that also carry U-rich sequences at their 3′ end [104]. There is evidence that RD core histone accumulation is monitored post-transcriptionally by both the exosome and Xrn1. [15]. For example, deletion of exosome component RRP6 results in continued accumulation of HTB1 mRNA after S phase [105], while deletion of LSM1 causes a G1-specific increase in histone mRNA levels and a synthetic growth defect in combination with a HIR1 deletion [15]. Consistent with an important interplay of transcriptional post-transcriptional regulatory mechanisms for ensuring the appropriate histone, gene deletion of both HIR1 and LSM1 causes a dramatic fitness defect. Also, combined deletion of LSM1 and the exosome component SKI2 causes greater accumulation of all histone mRNAs than seen in either single deletion strain, indicating that RD histone mRNA abundance is regulated by both 3′–5′ or 5′–3′ degradation pathways [15].
What targets Lsm1-7-Pat1 to histone mRNA? Post-S phase, the metazoan Lsm1-7-Pat1 complex is recruited to histone mRNA as a consequence of SLBP-stimulated oligouridylation of histone mRNA, due to recruitment of terminal uridylyl transferases (TUTases) [106]. The core histone genes do not appear to contain an obvious U-rich sequence in their 3′-UTR and no TUTase has been characterized in yeast. One candidate worth testing are the non-canonical poly A-polymerase Trf4 and Trf5, members of the TRAMP complex previously implicated in the regulation of histone mRNA abundance in Sc [107]. Deletion of TRF5 is strongly synthetic sick/lethal with rad53Δ and hir1Δ, and trf5Δ mutants have elevated levels of histone mRNA [107]. It has been suggested that Trf4/5 may function upstream of the exosome [107], but whether it functions upstream of Lsm1-7–Pat1 and has TUTase activity remains to be tested.
The data we summarize above shows that core histone mRNA accumulation is monitored by the major exoribonucleic activities of the cell. An important regulatory question is how S phase histone mRNAs avoid the action of exoribonucleic pathways to enable S phase-specific transcript accumulation. The answer probably lies in the 3′ UTR of RD core histone mRNA. As mentioned above, a previous study found an unidentified S phase protein that can specifically associate with HTB1 mRNA in vitro [103]. This protein—or another regulator—may function to prevent association of the Lsm1-7–Pat1 complex and encourage guidance of histone mRNAs to the ribosome for productive translation. In addition, histone mRNAs are subject to regulation at the level of polyA tail length. As noted above, histone mRNAs have very short polyA tails during S phase, and relatively longer polyA tails in G1 phase [101]. SEN1 encodes an essential helicase-like protein that binds Nab3 and Nrd1, which together function in transcription termination, and interacts with the exosome via Nrd1 [108]. One model proposes that Sen1 coordinates transcription termination with the addition of a longer polyA tail to histone mRNAs outside of S phase, which ultimately results in exosome-mediated transcript degradation [101]. An untested prediction of this model is that Sen1 should cross-link to RD core histone genes outside of S phase. A more speculative extension to this model proposes that Yta7, which does not interact with the NEG region during S phase, coordinates Sen1-based termination of transcription so that longer polyA tails are produced, which are then shunted to the nuclear exosome for degradation. A genetic study suggest the alternate possibility that Yta7 functions in the same pathway as Mex67, a poly(A) RNA binding protein involved in regulated nuclear mRNA export, and a component of the nuclear pore complex [109]. A derivative of Mex67 encoded by the mex67-5 conditional allele remains in the cytoplasm at the restrictive temperature, a phenotype that is suppressed by deletion of YTA7 [109]. Since Yta7 localizes to chromatin when histone expression is not required, an additional function of Yta7 may be to antagonize the nuclear export of histone mRNA. As noted earlier, a Yta7-13A phospho-mutant remains on chromatin through S phase and has a severe defect in S phase histone expression [47], and HTB1 mRNA is retained in the nucleus during G1 [101]. It will be interesting to determine if the yta7-13A mutant shows nuclear retention of HTB1 mRNA in S phase.
Translational and post-translational regulation of histone mRNAs
In the slime mold Physarum polycephalum and protist Leishmania infantum, regulation of RD core histone synthesis occurs at least in part at the level of translation [110, 111]. In metazoans, SLBP coordinates translation by interacting with SLIP1, which interacts with translation initiation factor eIF4G [112]. In human cells, the La protein, which contains the RNA binding La motif, has been reported to stabilize S phase histone mRNA, promoting their translation [113]. Whether or not RD core histone translation is subject to cell cycle regulation in Sc remains unclear, but the La-related proteins Sro9 and Slf1 co-purify with RD core histone mRNA [114]. Sro9 shuttles between the nucleus and cytoplasm and specifically cross-links to actively transcribed genes [115]. Further work is required to determine whether Sro9 and/or Slf1 function to escort S phase histone mRNA to the ribosome, antagonizing mRNA degradation pathways.
Histone expression is also regulated at post-translational level in Sc. For example, the negative feedback model described above proposes that histone protein levels are monitored and a feedback mechanism induces repression of transcription. In addition, a Rad53-dependent mechanism exists to degrade excess soluble histone proteins in Sc [7, 116]. The Rad53 checkpoint kinase phosphorylates soluble histones not bound to a histone chaperone leading to their ubiquitination and subsequent degradation by the proteasome [117].
Summary/future perspectives
In this review, we have summarized work exploring many facets of histone gene regulation, many of which impinge in some way on the chromatin at histone loci regulatory regions. Nucleosome occupancy at histone loci has been assessed in two ways. First, cross-linking of histone H3 or H2B to the HTA1–HTB1 promoter revealed that H3 and H2B levels are significantly reduced in chromatin isolated from asf1∆, hir1∆, or rtt106∆ mutant backgrounds [25]. Second, nucleosome occupancy at the NEG-regulated histone gene pairs was assessed using MNase to specifically purify nucleosome-bound DNA which was then hybridized to tiling microarrays with 4-bp resolution [25, 118]. This experiment revealed that the NEG-containing regulatory region including the promoter sequence is nucleosome-free in hir1∆ and rtt106∆ strains [25]. These experiments were performed with chromatin isolated from asynchronously growing cells. It will be useful to assay nucleosome occupancy at histone loci using yeast chromatin harvested from cells proceeding synchronously through the cell cycle. The model in Fig. 2 predicts that histone promoter regions will be nucleosome-free during time cell cycle periods corresponding to histone gene expression (late G1 and during S phase). In contrast, at all other times of the cell cycle when histone expression is repressed, nucleosomes should be positioned over the promoters as a result of recruitment of the HIR/Asf1/Rtt106 histone chaperones, which in turn assemble histones into a repressive chromatin structure that spreads towards the promoter, inhibiting transcription. The same repressive nucleosome pattern should be seen along the histone gene regulatory regions upon DNA damage, in a manner dependent on HIR1 and RTT106. These types of experiments will provide a view of the dynamics of nucleosome assembly at histone gene regulatory regions in response to genomic stress and cell cycle regulatory signals.
Several interesting questions concerning HIR-, Asf1-, and Rtt106-mediated chromatin assembly remain to be addressed. Asf1, HIR, and Rtt106 all appear to be individually capable of depositing histones so why do they function in an apparent linear pathway? Which of these three histone H3–H4 chaperones deposit histones onto DNA and what are their roles as chaperones of parental versus newly synthesized histones [119, 120]? Does HIR function at NEG-regulated histone gene pairs simply to recruit Rtt106 via Asf1, or is HIR involved in a more complicated histone transfer pathway? Genetic evidence supports a more complex role for the HIR complex: the defect in HTA1 transcription in an rtt106Δ strain is not as severe as that seen in a HIR1 deletion mutant [25]. This phenotype may reflect dual roles for Rtt106 in both activation (via SWI/SNF recruitment) and repression (via chromatin assembly and recruitment of RSC), or may point to an additional as yet uncharacterized histone chaperone pathway leading to HIR-dependent repression. The mechanism of Rtt106 spreading in the absence of the chromatin boundary protein Yta7 [25] deserves further investigation. In particular, it will be important to assess whether Rtt106 spreading is dependent on Rtt106 homo-oligomerization [121], or its preference for H3K56ac–H4 dimers. Interestingly, despite the absence of Yta7 in S phase, Rtt106 remains restricted to the CCR/NEG region of chromatin at HTA1 throughout the entire cell cycle [25]. This and other observations hint at the existence of a previously unknown and potentially novel mechanism of chromatin remodeling and transcriptional regulation.
Although it is clear that the H3K56 HAT Rtt109 and the putative HAT Spt10 are involved in histone gene regulation, their mechanisms of action remain obscure. It will be useful to ask whether H3K56ac levels at histone gene promoters are abolished in cells deleted for RTT109, consistent with a role upstream of Rtt109 in H3K56ac at histone gene promoters. Also, the role of Rtt109-binding partner and H3–H4 histone chaperone Vps75 needs be clarified. Possible Vps75 functions include stabilization of Rtt109 or Rtt109–Vps75-based acetylation of H3K9 or K56 [84, 122]. Furthermore, the precise relationship between H3K56ac, Rtt109, and Spt10 remains unclear, and acetylation targets of Spt10 are unknown.
Many other questions remain concerning the activation of RD core histone expression in Sc. For example, it is unclear how efficient DNA replication through the histone gene loci is coordinated with the high levels of S phase transcription of RD core histone genes. R-SGA of HTA1–GFP in an RRM3 deletion strain revealed a phenotype consistent with a role in HTA1 activation [25]. RRM3 (also called RTT104 [35]) encodes a DNA helicase that promotes progression of the replication fork through obstacles in the DNA, as might occur during active transcription. Further analysis will be required to understand how Rrm3 may coordinate these two processes to promote RD core histone transcription.
Although it is clear that histone transcription is linked to CDKs (see discussion of Yta7 above), very few direct targets of CDKs are known to regulate histone transcription. For example, mitotic B-type cyclins are involved in the alleviation of HIR-mediated repression at the G1 to S transition [26], but the target of this regulation and the underlying mechanisms are unknown. Kinase targets involved in the repression of histone genes following genotoxic stress also remain elusive. Checkpoint kinases directly regulate the inhibition of DNA replication by phosphorylation of key factors [123]. As DNA-replication and histone transcription are tightly linked, it is very likely that checkpoint kinases also act on histone regulators to repress histone gene transcription in DNA damaging conditions.
Much of what is known about the regulation of histone gene expression in Sc applies to the well-studied CCR/NEG containing histone gene pairs. By contrast, little is known concerning mechanisms underlying the regulation of the HTA2–HTB2 gene pair that contains a histone UAS but not NEG in its regulatory region. Why one of the core histone gene pairs, HTA2–HTB2, is regulated by a HIR-independent mechanism remains unclear, and this apparently distinct pathway deserves further exploration. We do know that all four of the histone pairs are subject to histone UAS-mediated cell cycle activation by Spt10/Spt21 and SBF. However, what is the basis of activation? Are a different set of proteins involved in overcoming repressive chromatin at HTA2–HTB2? Functional genomic approaches such as R-SGA using an HTA2–GFP reporter may reveal specific regulators that act at the HTA2–HTB2 locus. In addition, the R-SGA approach [25] can be extended to explore other arrayed mutant collections, such as the collection of strains carrying temperature-sensitive alleles of essential genes and arrays of strains expressing over-expression alleles [124, 125]. Finally, R-SGA of a GFP reporter linked not to the promoter but to the 3′-UTR of HTA1 has the potential to yield new information about post-transcriptional regulation of RD histone expression.
Acknowledgments
Work on histone gene regulation in the Fillingham and Andrews laboratories is supported by an NSERC Discovery Grant to J.S.F. (grant number: 386646-2010) and a grant from the Canadian Institutes of Health Research (CIHR) to B.J.A. and Tim Hughes (BMB-210972). C.F.K. was supported by an EMBO long-term fellowship.
Contributor Information
Brenda Andrews, Email: brenda.andrews@utoronto.ca.
Jeffrey Fillingham, Email: jeffrey.fillingham@ryerson.ca.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billon P, Cote J. Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochim Biophys Acta. 2012;1819:290–302. doi: 10.1016/j.bbagrm.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Fillingham J, Keogh MC, Krogan NJ. GammaH2AX and its role in DNA double-strand break repair. Biochem Cell Biol. 2006;84:568–577. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- 5.Hereford LM, Osley MA, Ludwig TR, 2nd, McLaughlin CS. Cell-cycle regulation of yeast histone mRNA. Cell. 1981;24:367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- 6.Kim UJ, Han M, Kayne P, Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae . EMBO J. 1988;7:2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae . Cell. 2003;115:537–549. doi: 10.1016/S0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 8.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. doi: 10.1006/geno.2002.6850. [DOI] [PubMed] [Google Scholar]
- 9.Smith MM, Murray K. Yeast H3 and H4 histone messenger RNAs are transcribed from two non-allelic gene sets. J Mol Biol. 1983;169:641–661. doi: 10.1016/S0022-2836(83)80163-6. [DOI] [PubMed] [Google Scholar]
- 10.Hereford L, Fahrner K, Woolford J, Jr, Rosbash M, Kaback DB. Isolation of yeast histone genes H2A and H2B. Cell. 1979;18:1261–1271. doi: 10.1016/0092-8674(79)90237-X. [DOI] [PubMed] [Google Scholar]
- 11.Osley MA, Gould J, Kim S, Kane MY, Hereford L. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986;45:537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- 12.Prado F, Aguilera A. Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Mol Cell Biol. 2005;25:1526–1536. doi: 10.1128/MCB.25.4.1526-1536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han M, Chang M, Kim UJ, Grunstein M. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 1987;48:589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- 14.Meeks-Wagner D, Hartwell LH. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 15.Herrero AB, Moreno S. Lsm1 promotes genomic stability by controlling histone mRNA decay. EMBO J. 2011;30:2008–2018. doi: 10.1038/emboj.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berloco M, Fanti L, Breiling A, Orlando V, Pimpinelli S. The maternal effect gene, abnormal oocyte (abo), of Drosophila melanogaster encodes a specific negative regulator of histones. Proc Natl Acad Sci USA. 2001;98:12126–12131. doi: 10.1073/pnas.211428798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jong AY, Kuo CL, Campbell JL. The CDC8 gene of yeast encodes thymidylate kinase. J Biol Chem. 1984;259:11052–11059. [PubMed] [Google Scholar]
- 18.Lycan DE, Osley MA, Hereford LM. Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae . Mol Cell Biol. 1987;7:614–621. doi: 10.1128/mcb.7.2.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breeden L. Cell cycle-regulated promoters in budding yeast. Trends Genet. 1988;4:249–253. doi: 10.1016/0168-9525(88)90031-5. [DOI] [PubMed] [Google Scholar]
- 20.Osley MA, Lycan D. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol Cell Biol. 1987;7:4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spector MS, Raff A, DeSilva H, Lee K, Osley MA. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, Kim UJ, Schuster T, Grunstein M. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae . Mol Cell Biol. 1992;12:5249–5259. doi: 10.1128/mcb.12.11.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, 3rd, Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 2005;19:2534–2539. doi: 10.1101/gad.1341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fillingham J, Kainth P, Lambert JP, van Bakel H, Tsui K, Pena-Castillo L, Nislow C, Figeys D, Hughes TR, Greenblatt J, et al. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol Cell. 2009;35:340–351. doi: 10.1016/j.molcel.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Amin AD, Dimova DK, Ferreira ME, Vishnoi N, Hancock LC, Osley MA, Prochasson P. The mitotic Clb cyclins are required to alleviate HIR-mediated repression of the yeast histone genes at the G1/S transition. Biochim Biophys Acta. 2012;1819:16–27. doi: 10.1016/j.bbagrm.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balaji S, Iyer LM, Aravind L. HPC2 and ubinuclein define a novel family of histone chaperones conserved throughout eukaryotes. Mol Biosyst. 2009;5:269–275. doi: 10.1039/b816424j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson DM, Ye X, Hall C, Santos H, Ma T, Kao GD, Yen TJ, Harper JW, Adams PD. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol Cell Biol. 2002;22:7459–7472. doi: 10.1128/MCB.22.21.7459-7472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahajan K, Fang B, Koomen JM, Mahajan NP. H2B Tyr37 phosphorylation suppresses expression of replication-dependent core histone genes. Nat Struct Mol Biol. 2012;19:930–937. doi: 10.1038/nsmb.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banumathy G, Somaiah N, Zhang R, Tang Y, Hoffmann J, Andrake M, Ceulemans H, Schultz D, Marmorstein R, Adams PD. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol Cell Biol. 2009;29:758–770. doi: 10.1128/MCB.01047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rai TS, Puri A, McBryan T, Hoffman J, Tang Y, Pchelintsev NA, van Tuyn J, Marmorstein R, Schultz DC, Adams PD. Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex. Mol Cell Biol. 2011;31:4107–4118. doi: 10.1128/MCB.05546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pchelintsev NA, McBryan T, Rai TS, van Tuyn J, Ray-Gallet D, Almouzni G, Adams PD. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 2013;3:1012–1019. doi: 10.1016/j.celrep.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton A, Bucaria J, Osley MA, Sternglanz R. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zunder RM, Rine J. Direct interplay among histones, histone chaperones, and a chromatin boundary protein in the control of histone gene expression. Mol Cell Biol. 2012;32:4337–4349. doi: 10.1128/MCB.00871-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang S, Zhou H, Katzmann D, Hochstrasser M, Atanasova E, Zhang Z. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc Natl Acad Sci USA. 2005;102:13410–13415. doi: 10.1073/pnas.0506176102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 38.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 39.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 40.Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, et al. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- 41.Ferreira ME, Flaherty K, Prochasson P. The Saccharomyces cerevisiae histone chaperone Rtt106 mediates the cell cycle recruitment of SWI/SNF and RSC to the HIR-dependent histone genes. Plos ONE. 2011;6:e21113. doi: 10.1371/journal.pone.0021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tackett AJ, Dilworth DJ, Davey MJ, O’Donnell M, Aitchison JD, Rout MP, Chait BT. Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gradolatto A, Smart SK, Byrum S, Blair LP, Rogers RS, Kolar EA, Lavender H, Larson SK, Aitchison JD, Taverna SD, et al. A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity. Mol Cell Biol. 2009;29:4604–4611. doi: 10.1128/MCB.00160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jambunathan N, Martinez AW, Robert EC, Agochukwu NB, Ibos ME, Dugas SL, Donze D. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics. 2005;171:913–922. doi: 10.1534/genetics.105.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gradolatto A, Rogers RS, Lavender H, Taverna SD, Allis CD, Aitchison JD, Tackett AJ. Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics. 2008;179:291–304. doi: 10.1534/genetics.107.086520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurat CF, Lambert JP, van Dyk D, Tsui K, van Bakel H, Kaluarachchi S, Friesen H, Kainth P, Nislow C, Figeys D, et al. Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein. Genes Dev. 2011;25:2489–2501. doi: 10.1101/gad.173427.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leachman NT, Brellier F, Ferralli J, Chiquet-Ehrismann R, Tucker RP. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev Growth Differ. 2010;52:747–755. doi: 10.1111/j.1440-169X.2010.01211.x. [DOI] [PubMed] [Google Scholar]
- 49.Raeder MB, Birkeland E, Trovik J, Krakstad C, Shehata S, Schumacher S, Zack TI, Krohn A, Werner HM, Moody SE, et al. Integrated genomic analysis of the 8q24 amplification in endometrial cancers identifies ATAD2 as Essential to MYC-dependent cancers. Plos ONE. 2013;8:e54873. doi: 10.1371/journal.pone.0054873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fouret R, Laffaire J, Hofman P, Beau-Faller M, Mazieres J, Validire P, Girard P, Camilleri-Broet S, Vaylet F, Leroy-Ladurie F, et al. A comparative and integrative approach identifies ATPase family, AAA domain containing 2 as a likely driver of cell proliferation in lung adenocarcinoma. Clin Cancer Res. 2012;18:5606–5616. doi: 10.1158/1078-0432.CCR-12-0505. [DOI] [PubMed] [Google Scholar]
- 51.Caron C, Lestrat C, Marsal S, Escoffier E, Curtet S, Virolle V, Barbry P, Debernardi A, Brambilla C, Brambilla E, et al. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene. 2010;29:5171–5181. doi: 10.1038/onc.2010.259. [DOI] [PubMed] [Google Scholar]
- 52.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lambert JP, Fillingham J, Siahbazi M, Greenblatt J, Baetz K, Figeys D. Defining the budding yeast chromatin-associated interactome. Mol Syst Biol. 2010;6:448. doi: 10.1038/msb.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silva AC, Xu X, Kim HS, Fillingham J, Kislinger T, Mennella TA, Keogh MC. The replication-independent histone H3–H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J Biol Chem. 2012;287:1709–1718. doi: 10.1074/jbc.M111.316489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eriksson PR, Ganguli D, Nagarajavel V, Clark DJ. Regulation of histone gene expression in budding yeast. Genetics. 2012;191:7–20. doi: 10.1534/genetics.112.140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moran L, Norris D, Osley MA. A yeast H2A–H2B promoter can be regulated by changes in histone gene copy number. Genes Dev. 1990;4:752–763. doi: 10.1101/gad.4.5.752. [DOI] [PubMed] [Google Scholar]
- 57.Compagnone-Post PA, Osley MA. Mutations in the SPT4, SPT5, and SPT6 genes alter transcription of a subset of histone genes in Saccharomyces cerevisiae . Genetics. 1996;143:1543–1554. doi: 10.1093/genetics/143.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell. 1999;4:75–83. doi: 10.1016/S1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- 59.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 60.Fassler JS, Winston F. Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae . Genetics. 1988;118:203–212. doi: 10.1093/genetics/118.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Natsoulis G, Dollard C, Winston F, Boeke JD. The products of the SPT10 and SPT21 genes of Saccharomyces cerevisiae increase the amplitude of transcriptional regulation at a large number of unlinked loci. New Biol. 1991;3:1249–1259. [PubMed] [Google Scholar]
- 62.Hess D, Liu B, Roan NR, Sternglanz R, Winston F. Spt10-dependent transcriptional activation in Saccharomyces cerevisiae requires both the Spt10 acetyltransferase domain and Spt21. Mol Cell Biol. 2004;24:135–143. doi: 10.1128/MCB.24.1.135-143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mendiratta G, Eriksson PR, Shen CH, Clark DJ. The DNA-binding domain of the yeast Spt10p activator includes a zinc finger that is homologous to foamy virus integrase. J Biol Chem. 2006;281:7040–7048. doi: 10.1074/jbc.M511416200. [DOI] [PubMed] [Google Scholar]
- 64.Neuwald AF, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/S0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 65.Natsoulis G, Winston F, Boeke JD. The SPT10 and SPT21 genes of Saccharomyces cerevisiae . Genetics. 1994;136:93–105. doi: 10.1093/genetics/136.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hess D, Winston F. Evidence that Spt10 and Spt21 of Saccharomyces cerevisiae play distinct roles in vivo and functionally interact with MCB-binding factor, SCB-binding factor and Snf1. Genetics. 2005;170:87–94. doi: 10.1534/genetics.104.039214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dollard C, Ricupero-Hovasse SL, Natsoulis G, Boeke JD, Winston F. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae . Mol Cell Biol. 1994;14:5223–5228. doi: 10.1128/mcb.14.8.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang JS, Winston F. Spt10 and Spt21 are required for transcriptional silencing in Saccharomyces cerevisiae . Eukaryot Cell. 2011;10:118–129. doi: 10.1128/EC.00246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sherwood PW, Osley MA. Histone regulatory (hir) mutations suppress delta insertion alleles in Saccharomyces cerevisiae . Genetics. 1991;128:729–738. doi: 10.1093/genetics/128.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cooper K. Rb, whi it’s not just for metazoans anymore. Oncogene. 2006;25:5228–5232. doi: 10.1038/sj.onc.1209630. [DOI] [PubMed] [Google Scholar]
- 71.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 72.Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/S0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- 73.Macpherson N, Measday V, Moore L, Andrews B. A yeast taf17 mutant requires the Swi6 transcriptional activator for viability and shows defects in cell cycle-regulated transcription. Genetics. 2000;154:1561–1576. doi: 10.1093/genetics/154.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eriksson PR, Ganguli D, Clark DJ. Spt10 and Swi4 control the timing of histone H2A/H2B gene activation in budding yeast. Mol Cell Biol. 2011;31:557–572. doi: 10.1128/MCB.00909-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 77.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 79.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 80.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci USA. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae . Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 83.Selth L, Svejstrup JQ. Vps75, a new yeast member of the NAP histone chaperone family. J Biol Chem. 2007;282:12358–12362. doi: 10.1074/jbc.C700012200. [DOI] [PubMed] [Google Scholar]
- 84.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell. 2009;33:417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takayama Y, Takahashi K. Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res. 2007;35:3223–3237. doi: 10.1093/nar/gkm213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen ES, Saitoh S, Yanagida M, Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell. 2003;11:175–187. doi: 10.1016/S1097-2765(03)00011-X. [DOI] [PubMed] [Google Scholar]
- 89.Takayama Y, Mamnun YM, Trickey M, Dhut S, Masuda F, Yamano H, Toda T, Saitoh S. Hsk1- and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Dev Cell. 2010;18:385–396. doi: 10.1016/j.devcel.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trickey M, Fujimitsu K, Yamano H. Anaphase-promoting complex/cyclosome-mediated proteolysis of Ams2 in the G1 phase ensures the coupling of histone gene expression to DNA replication in fission Yeast. J Biol Chemi. 2013;288:928–937. doi: 10.1074/jbc.M112.410241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Enserink JM, Kolodner RD. An overview of Cdk1-controlled targets and processes. Cell Div. 2010;5:11. doi: 10.1186/1747-1028-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 93.Lombardi LM, Ellahi A, Rine J. Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc Natl Acad Sci USA. 2011;108:E1302–E1311. doi: 10.1073/pnas.1116819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. doi: 10.1101/gad.827700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ye X, Wei Y, Nalepa G, Harper JW. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Imbeault D, Gamar L, Rufiange A, Paquet E, Nourani A. The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast. J Biol Chem. 2008;283:27350–27354. doi: 10.1074/jbc.C800147200. [DOI] [PubMed] [Google Scholar]
- 98.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davila Lopez M, Samuelsson T. Early evolution of histone mRNA 3′ end processing. RNA. 2008;14:1–10. doi: 10.1261/rna.782308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fahrner K, Yarger J, Hereford L. Yeast histone mRNA is polyadenylated. Nucleic Acids Res. 1980;8:5725–5737. doi: 10.1093/nar/8.23.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beggs S, James TC, Bond U. The PolyA tail length of yeast histone mRNAs varies during the cell cycle and is influenced by Sen1p and Rrp6p. Nucleic Acids Res. 2012;40:2700–2711. doi: 10.1093/nar/gkr1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu HX, Johnson L, Grunstein M. Coding and noncoding sequences at the 3′ end of yeast histone H2B mRNA confer cell cycle regulation. Mol Cell Biol. 1990;10:2687–2694. doi: 10.1128/mcb.10.6.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Campbell SG, Li Del Olmo M, Beglan P, Bond U. A sequence element downstream of the yeast HTB1 gene contributes to mRNA 3′ processing and cell cycle regulation. Mol Cell Biol. 2002;22:8415–8425. doi: 10.1128/MCB.22.24.8415-8425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Canavan R, Bond U. Deletion of the nuclear exosome component RRP6 leads to continued accumulation of the histone mRNA HTB1 in S-phase of the cell cycle in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:6268–6279. doi: 10.1093/nar/gkm691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reis CC, Campbell JL. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics. 2007;175:993–1010. doi: 10.1534/genetics.106.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 109.Estruch F, Peiro-Chova L, Gomez-Navarro N, Durban J, Hodge C, Del Olmo M, Cole CN. A genetic screen in Saccharomyces cerevisiae identifies new genes that interact with me67–5, a temperature-sensitive allele of the gene encoding the mRNA export receptor. Mol Genet Genomics. 2009;281:125–134. doi: 10.1007/s00438-008-0402-x. [DOI] [PubMed] [Google Scholar]
- 110.Soto M, Iborra S, Quijada L, Folgueira C, Alonso C, Requena JM. Cell-cycle-dependent translation of histone mRNAs is the key control point for regulation of histone biosynthesis in Leishmania infantum . Biochem J. 2004;379:617–625. doi: 10.1042/BJ20031522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Laffler TG, Carrino J. Cell-cycle-regulated translation of histone mRNA in Physarum plasmodia . J Bacteriol. 1987;169:2291–2293. doi: 10.1128/jb.169.5.2291-2293.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol Cell Biol. 2008;28:1182–1194. doi: 10.1128/MCB.01500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McLaren RS, Caruccio N, Ross J. Human La protein: a stabilizer of histone mRNA. Mol Cell Biol. 1997;17:3028–3036. doi: 10.1128/mcb.17.6.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schenk L, Meinel DM, Strasser K, Gerber AP. La-motif-dependent mRNA association with Slf1 promotes copper detoxification in yeast. RNA. 2012;18:449–461. doi: 10.1261/rna.028506.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rother S, Burkert C, Brunger KM, Mayer A, Kieser A, Strasser K. Nucleocytoplasmic shuttling of the La motif-containing protein Sro9 might link its nuclear and cytoplasmic functions. RNA. 2010;16:1393–1401. doi: 10.1261/rna.2089110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gunjan A, Paik J, Verreault A. The emergence of regulated histone proteolysis. Curr Opin Genet Dev. 2006;16:112–118. doi: 10.1016/j.gde.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 117.Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 119.Verzijlbergen KF, van Welsem T, Sie D, Lenstra TL, Turner DJ, Holstege FC, Kerkhoven RM, van Leeuwen F. A barcode screen for epigenetic regulators reveals a role for the NuB4/HAT-B histone acetyltransferase complex in histone turnover. Plos Genet. 2011;7:e1002284. doi: 10.1371/journal.pgen.1002284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim HJ, Seol JH, Han JW, Youn HD, Cho EJ. Histone chaperones regulate histone exchange during transcription. EMBO J. 2007;26:4467–4474. doi: 10.1038/sj.emboj.7601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fazly A, Li Q, Hu Q, Mer G, Horazdovsky B, Zhang Z. Histone chaperone Rtt106 promotes nucleosome formation using (H3-H4)2 tetramers. J Biol Chem. 2012;287:10753–10760. doi: 10.1074/jbc.M112.347450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Radovani E, Cadorin M, Shams T, El-Rass S, Karsou AR, Kim HS, Kurat CF, Keogh MC, Greenblatt J, Fillingham JS. The carboxyl terminus of Rtt109 functions in chaperone control of histone acetylation. Eukaryot Cell. 2013;12:654–664. doi: 10.1128/EC.00291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zegerman P, Diffley JF. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature. 2010;467:474–478. doi: 10.1038/nature09373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, Vizeacoumar FS, Min R, Vandersluis B, Bellay J, Devit M, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]