Abstract
Topoisomerases are vital enzymes specialized in controlling DNA topology, in particular supercoiling and decatenation, to properly handle nucleic acid packing and cell dynamics. The type IIA enzymes act by cleaving both strands of a double helix and having another strand from the same or another molecule cross the DNA gate before a re-sealing event completes the catalytic cycle. Here, we will consider the two types of IIA prokaryotic topoisomerases, DNA Gyrase and Topoisomerase IV, as crucial regulators of bacterial cell cycle progression. Their synergistic action allows control of chromosome packing and grants occurrence of functional transcription and replication processes. In addition to displaying a fascinating molecular mechanism of action, which transduces chemical energy into mechanical energy by means of large conformational changes, these enzymes represent attractive pharmacological targets for antibacterial chemotherapy.
Keywords: DNA-topology, Topoisomerases, DNA Gyrase, Topoisomerase IV, Prokaryotic enzymes, Antibacterials
Introduction
When we think of DNA, our mind springs to the inspiring image of a double helix. Although duplex structure represents a hallmark in modern biology, at first glance base stacking and braided architecture lend it unusual stiffness, so it might appear as a biological device more suited to safe genetic information storage than to dynamic action in living systems. Rather, the nucleic acid behaves as a flexible molecule and shows a remarkable plasticity when performing essential functions such as replication and transcription. In fact, we should take into account that DNA is organized in large loops both in eukaryotic and prokaryotic cells, which essentially render it a closed circular system. As a consequence, it may adopt several different configurations (relaxed, positively or negatively supercoiled, knotted, catenanes) (Fig. 1). They refer to otherwise identical molecules differing in their spatial arrangement only, called topological isomers or topoisomers. These forms are dynamically generated and interconvert during DNA processing. For example, replication, which demands local opening of the double helix, produces positive supercoiling ahead of the replication fork and interwinding of daughter chromosomes (precatenanes) behind it, an event that manifestly requires topological intervention. The same is true for transcription, which leads to the production of positive supercoiling ahead of the transcription machinery and negative supercoiling behind it. More importantly, it is now clear that DNA topology plays an active role in regulating DNA processing [1–3]: basic biological processes, including control of gene transcription, faithful DNA replication and segregation, maintenance of the genome and cellular differentiation are largely directed by the energetics of DNA structure in addition to the regulation provided by the sequence itself [4, 5]. If topology were not taken care of appropriately, these fundamental processes could not occur. Hence, to dynamically modulate its activity and grant physiologic occurrence of essential cellular events, (circular/constrained) DNA must be efficiently and timely converted from one topological state to another [6].
Fig. 1.
Different topological states of covalently closed circular DNA
Topoisomerases
In all organisms, the DNA topology control is carried out by an essential family of enzymes, the topoisomerases. Variation in the degree of supercoiling of a circular duplex as well as separation of catenated circles require phosphodiester bond cleavage followed by DNA strand passage and, finally, resealing of the original linkage. Therefore, the mechanism underlying topoisomerase action cannot be simple. Indeed, it foresees the combination of functions characteristic of nucleases and ligases, besides the possibility of large conformational changes to cope with a strand-passing process. Moreover, in most instances, these functionalities are coupled to energy-producing ATPase activities [7]. In fact, topoisomerases consist of several domains. They can be divided into type I enzymes, which operate by introducing DNA single-strand breaks only and type II enzymes, which produce double-strand breaks. In turn, depending upon amino-acid sequence, structural, mechanistic, and evolutionary differences, type I can be subdivided into the A, B, and C subclasses, while type II consists of the A and B subclasses. All these subclasses reflect differences in the enzyme structure and mechanism of action.
Several recent reviews describe in detail the structural and biological properties of topoisomerases [8–15]. Here, we will focus on bacterial topoisomerases belonging to the type IIA subclass, with special attention to the most recent knowledge rendered available in the field.
Bacterial type IIA topoisomerases: DNA Gyrase (Gyr) and topoisomerase IV (Topo IV)
In general, every organism requires at least one type I and one type II enzyme to fully deal with maintaining appropriate levels of DNA supercoiling and removing chromosomal entanglements. In addition, most bacteria express two distinct topoisomerases of type IIA, referred to as DNA Gyrase (Gyr) and Topoisomerase IV (Topo IV). As notable exceptions, a few bacteria such as Treponema pallidum, Deinococcus radiodurans, Helicobacter pylori, Campylobacter jejuni together with the actinobacteria Mycobacterium tuberculosis, Mycobacterium leprae and Corynebacterium glutamicum are reported to express one topo II (Gyr) only [16–20].
Gyr activity was discovered over 30 years ago [21]. The E. coli functional enzyme was soon after shown to consist of a tetramer (A2B2, MW 400 kDa) formed by two pairs of identical subunits (GyrA and GyB) having molecular weights of about 97 and 90 kDa respectively [22]. About 14 years later, a new bacterial topoisomerase II, named topoisomerase IV, was discovered in E. coli and shown to be assembled in a way similar to Gyr, with subunit ParC (MW 83.7 kDa) corresponding to GyrA and ParE (MW 70.2 kDa) to GyrB [23, 24]. The above subunits are encoded by the GyrA, GyrB, ParC and ParE genes, respectively [25, 26].
Enzymatic cycle of bacterial type II topoisomerases
Gyr and Topo IV share mechanistic features common to all type II enzymes described to date, spanning from archeabacterial to eukaryotic species. In fact, a transphosphorylation process generates a transient covalent linkage between the protein and the nucleic acid, resulting in dsDNA breakage (cleavage complex) [21, 27].
Global catalytic reaction is performed by two Tyr residues conveniently located at the active site, which form phosphotyrosine linkages at the 5′ side of the attacked phosphodiester bond. Topoisomerase II-mediated breaks are not produced at the level of the two residues belonging to the same base pair, but are staggered 4 base pairs apart from each other [28, 29].
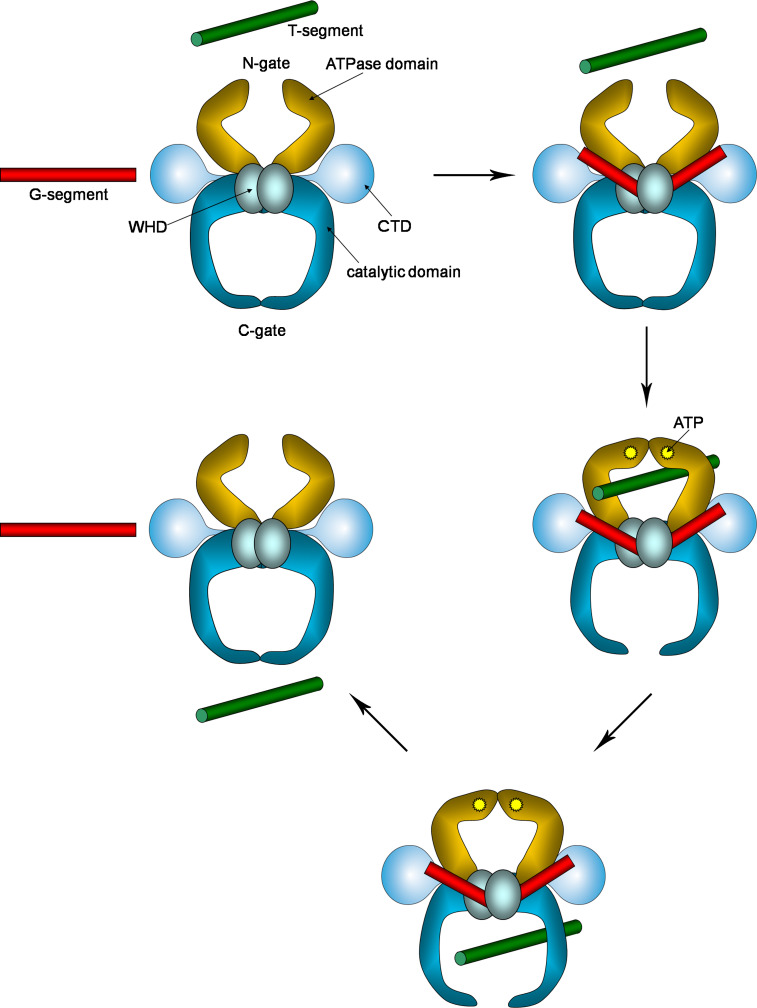
A generally accepted mechanism for the DNA topological changes mediated by type II topoisomerases, the so-called “two-gate mechanism”, was proposed long ago [30, 31]. Based upon this, the DNA segment being transported to produce strand passing (T) can exit from the interior of the enzyme through a gate (C) located on the opposite side of the entrance gate (N) (Fig. 2). The DNA double-stranded breakage forms an additional temporary gate (DNA gate) through which the T DNA double helix strand-passage occurs, which actually leads to a “three-gate mechanism”. The concerted gates opening/closure grants one-way strand passage of the T segment: indeed each gate can be opened only when the others are closed according to a double-lock rule [32, 33]. This allows a powerful unidirectional action.
Fig. 2.
Relevant steps of the two-gate mechanism for type IIA topoisomerases. CTD C-terminal domain, WHD winged-helix domain, containing the catalytic Tyr residue. The G-segment is engaged at the GyrA/GyrB or ParC/ParE domain interface, where it is sharply bent, T-segment is recruited in the ATP-clamp (N-gate), which is closed when ATP binds. Then, the G-gate opens, forming the cleavage complex and allows T-segment transport. Finally, T-segment exits through the C-gate and the enzyme is reset for a subsequent catalytic cycle
ATP hydrolysis and catalytic metal ions are the two main cofactors required by type IIA topoisomerases for successful DNA processing [21].
At the moment we do not know precisely which step makes use of the energy of ATP hydrolysis, but this might be needed (1) to securely hold the two G segments while DNA is cleaved to provide the gate for transport and/or (2) to favor the conformational changes (N gate closure) needed to push the T segment through the G segment into the enzyme central cavity. Additionally, the enzyme is functionally reset only through hydrolysis of a second ATP molecule. As a consequence, all type II enzymes require two ATP molecules to undergo subsequent rounds of catalysis [34].
Concomitantly, a divalent metal ion co-factor (usually Mg2+) is required to catalyse the DNA cleavage/rejoining chemical process [21]. Mg2+ is physiologically present in the bacterial intracellular environment and is expected to assist the transphosphorylation process in several ways. In particular, it can stabilize the incoming negative tyrosinate species, neutralize the penta-coordinated negatively charged transition state and/or assist in the stabilization of the negatively charged leaving group. The information presently available suggests participation of two ions per DNA strand cleavage event [35].
Clearly, the requisites of tetramer assembly, ATP hydrolysis and metal ion binding confer a remarkable specificity to the system, which can be activated/deactivated under individually modulated environmental conditions during the cell cycle and localized at selected loci as appropriate [36]. In addition, topoisomerases of different organisms exhibit different co-factor dependencies of their catalytic activity to optimally respond to individual needs [37, 38].
Domain organization
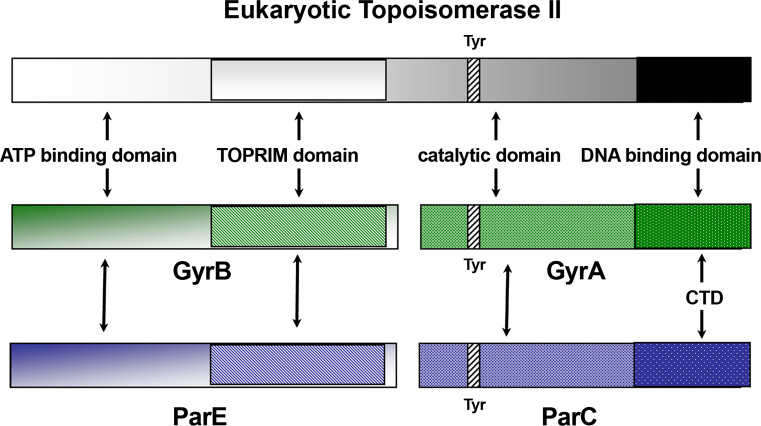
Since the DNA cleavage-resealing process performed by type IIA topoisomerases proceeds parallel on the two DNA strands, a symmetrical arrangement of the functional enzyme is expected [29]. In fact, the double-strand cutting process is handled by the formation of a protein tetramer as mentioned above [22]. Analytical ultracentrifugation and X-ray scattering studies enabled the overall shapes of Gyr subunits in solution to be defined [39, 40]. GyrA forms a dimeric heart-like core accompanied by two pear-shaped lateral densities (the carboxyl terminal domains), closely flanking the core and lying in the lower part of the protein, far from the cleavage site [39]. GyrB as such is monomeric and shows a tadpole-like structure, suggestive of a modular organization of the protein domains thus revealing potential structural flexibility of the various subdomains [40]. Dimerization of this subunit is then triggered by the presence of GyrA (and the DNA substrate). In addition, the results indicate that GyrB should be located on the top of GyrA in the active tetrameric assembly [40].
The domain organization of the various subunits is schematically reported in Fig. 3.
Fig. 3.
Schematic organization of functional domains in type IIA topoisomerases. Location of catalytic tyrosine (Tyr) is indicated
Interestingly, Gyr and Topo IV share a common distribution of functional domains.
The catalytic Tyr is located in the N-terminal domain of GyrA/ParC, whereas DNA binding occurs at the C-terminal domain of these proteins. In turn, the ATP-binding region is positioned at the N-terminal domain of GyrB/ParE, while their C-terminal domains contain the TOPRIM region, spanning about 100 residues, involved in catalytic Mg2+ coordination. Interestingly, the latter region is shared by all type IA and type II topoisomerases as well as by several other nucleases [41].
Although the various functional domains of the bacterial enzymes are situated in distinct subunits, a close similarity to the eukaryotic counterpart(s) is apparent. Additionally, Gyr and Topo IV subunits from various sources (including both Gram-negative and Gram-positive bacteria) share similar functional and structural arrangements [15], in spite of their relatively modest sequence homology (Table 1).
Table 1.
Optimal sequence similarity scores for selected DNA Gyrase and Topoisomerase IV subunits from Gram-negative and Gram-positive bacteria
| GyrA | ParC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | B. subtilis | S. aureus | S. pneum. | B. burg. | M. tub. | E. coli | B. subtilis | S. aureus | S. pneum. | B. burg. | |
| No. of amino acids | 875 | 821 | 889 | 822 | 810 | 838 | 752 | 806 | 800 | 823 | 599 |
| GyrA | |||||||||||
| E.coli | 53 | 48 | 49 | 41 | 45 | 33 | 40 | 37 | 36 | 20 | |
| B. subtilis | 64 | 59 | 42 | 49 | 32 | 42 | 39 | 35 | 24 | ||
| S. aureus | 59 | 41 | 46 | 34 | 39 | 38 | 33 | 24 | |||
| S. pneum. | 42 | 46 | 31 | 40 | 38 | 39 | 23 | ||||
| B. burg. | 39 | 29 | 33 | 35 | 32 | 23 | |||||
| M. tub. | 31 | 36 | 36 | 33 | 22 | ||||||
| ParC | |||||||||||
| E. coli | 28 | 29 | 27 | 19 | |||||||
| B. subtilis | 58 | 53 | 23 | ||||||||
| S. aureus | 52 | 24 | |||||||||
| S. pneum. | 23 | ||||||||||
| GyrB | ParE | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | B. subtilis | S. aureus | S. pneum. | B. burg. | M. tub. | E. coli | B. subtilis | S. aureus | S. pneum. | B. burg. | |
| No. of amino acids | 804 | 638 | 644 | 648 | 639 | 675 | 630 | 655 | 663 | 647 | 626 |
| GyrB | |||||||||||
| E.coli | 44 | 54 | 53 | 52 | 48 | 38 | 49 | 48 | 49 | 32 | |
| B. subtilis | 68 | 66 | 56 | 55 | 39 | 56 | 55 | 55 | 36 | ||
| S. aureus | 62 | 52 | 54 | 38 | 52 | 52 | 50 | 35 | |||
| S. pneum. | 53 | 51 | 38 | 51 | 50 | 49 | 34 | ||||
| B. burg. | 49 | 38 | 48 | 48 | 48 | 35 | |||||
| M. tub. | 39 | 49 | 46 | 48 | 32 | ||||||
| ParE | |||||||||||
| E. coli | 37 | 36 | 39 | 29 | |||||||
| B. subtilis | 70 | 69 | 34 | ||||||||
| S. aureus | 68 | 35 | |||||||||
| S. pneum. | 31 | ||||||||||
Since the crystal structure of the full-length Gyr/Topo IV subunits is not available as of yet, domain similarity has been used to combine crystallographic data from several topoisomerases fragments to yield very valuable structure–function relationships. Although the conformation examined in a crystal structure represents but one of the (several) possible stable conformations adopted by the enzyme (fragment) during the catalytic cycle, different conditions used to obtain the crystals may give insight into the conformational changes the fragment under investigation can undergo. In fact, studies on different topoisomerase II DNA-binding and cleavage domains show that, in addition to substantial conformational changes in the DNA-opening region, a dramatic reorganization of accessory domains may occur during catalysis as well [42]. Also, ion and nucleotide co-factors play a remarkable role in dictating the three-dimensional architecture of the protein.
The DNA cleavage–rejoining process
The functional domains needed for bacterial topoisomerases to catalyse DNA cleavage-rejoining are located in both enzyme subunits, GyrA/ParC containing the catalytic Tyr residue and GyrB/ParE enclosing the TOPRIM region.
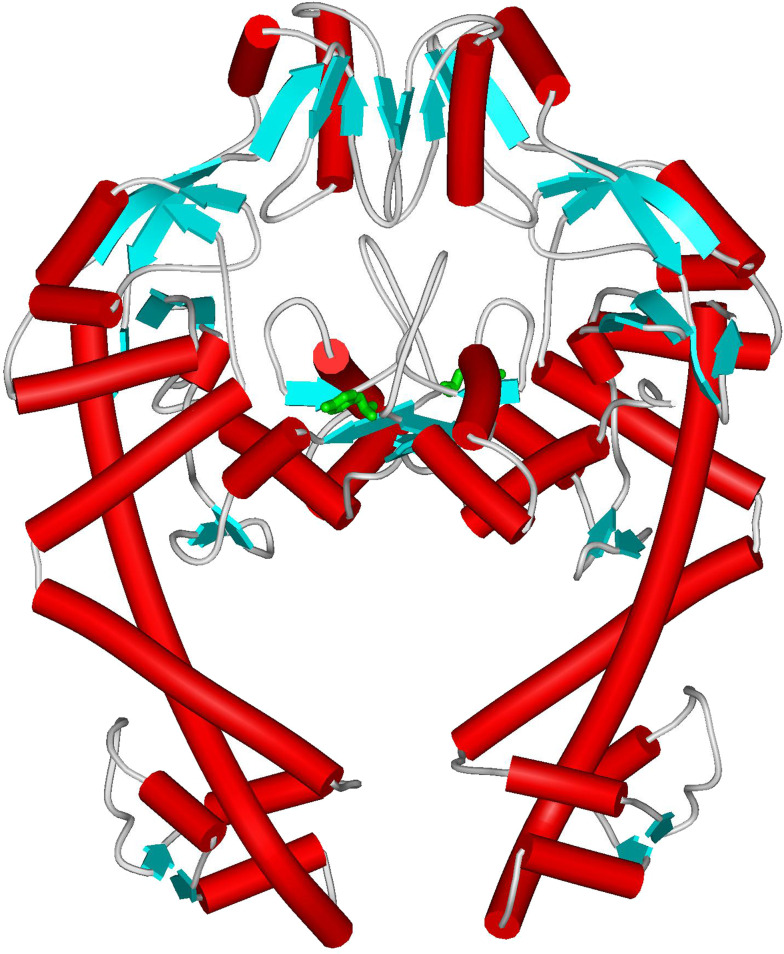
The GyrA 59-kDa N-terminal domain, GyrA59, entails the breakage–reunion machinery, including the winged-helix domain (WHD) with the active-site Tyr, and the ‘tower’ domain that stands on it and participates in DNA binding. It is catalytically functional in the presence of GyrB. Its crystal structure is dimeric, as expected by the symmetric features of enzyme action (Fig. 4) [43]. At the top of the interface, the ‘recognition’ helices make a head-to-head antiparallel dimer contact. Dimer formation generates a large concave surface on the top comprising the upper surface of the WHD domains and the sides of the two ‘tower’ domains. The active-site tyrosines are located in loops at either extreme of the dimer interface, 30 Å apart from each other, a distance very similar to that observed in yeast topoisomerase II [44]. They are positioned at the ends of strongly basic grooves created by the dimer-related monomers, ideally suited to accommodate a G segment. A 30-Å-wide central cavity generated by dimerization is adequate to let a T segment translocate through the cleaved G-segment. Two long helices produce the “tail” encompassing the primary dimerization interface. The latter can open to generate the C-gate during catalytic cycle progression. As it approaches its target, the G segment will dramatically change the local electrostatic potential and definitely promote further conformational changes to optimize the protein–DNA complex [43].
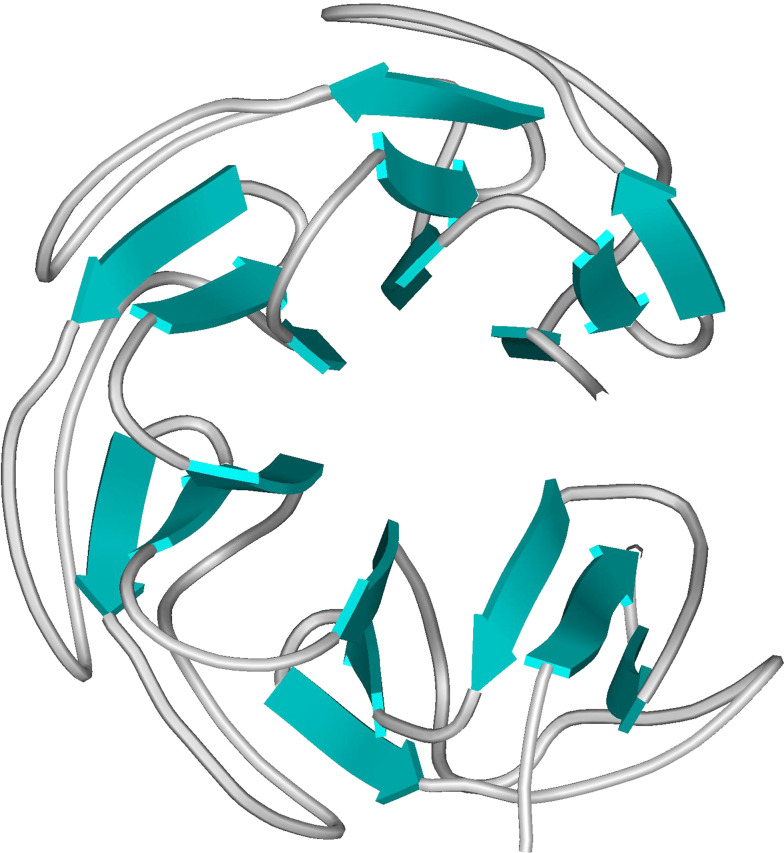
Fig. 4.
Crystal structure of E. coli GyrA59 dimer (PDB ID 1AB4). Red tubes refer to α-helix regions, blue arrows to β-sheet. The catalytic Tyr is highlighted in green (ball-and-stick model)
The structural features of ParC fragments from E. coli and S. pneumoniae, containing WHD and tower regions (the S. pneumoniae domain also in complex with DNA), have been characterized as well [45, 46]. The 55-kDa N-terminal breakage-reunion domain of Topo IV Par C from S. pneumoniae reveals a “closed” dimer folded like E. coli GyrA, but divergent from the “open-gate” structure of E. coli ParC [46, 47]. These differences probably reflect structural changes the proteins undergo to process DNA. In addition, while the G-segment binding surface in GyrA exhibits a large positive charge density, the DNA-binding site of ParC reveals a distinct pattern with alternating regions of opposite electron distribution matching the grooves and phosphate backbone of DNA [46]. Given the non-negligible distances between the catalytic tyrosine(s) and the fitted DNA phosphate backbone, a local re-shaping is required to allow their closing up in the cleavage-reunion process. Possible key steps in DNA binding and recognition involve interactions with the two active-site helices and the flexible loop between amino-acids 100 and 122. This rearrangement is indeed observed in the ParC-DNA covalent complex, although the presence of a drug molecule at the cleavage site might locally remodel the structure [48].
As far as the TOPRIM domain is concerned, the crystal structure of the 485–714 sequence of GyrB from M. tuberculosis has been solved very recently [49]. TOPRIM spans about 120 residues (486–604) and is characterized by the presence of a Glu residue and an Asp-x-Asp-x-Asp (DxDxD) motif in the IIA enzymes [41]. It includes a Rossmann-like fold containing a central β-sheet sandwiched by two α-helical bundles, with a tertiary scaffold nearly identical to other topoisomerases. The glutamate is located in a sharp turn at a β-α connection, a structurally analogous arrangement occurring at the level of the DxDxD element. As a consequence of TOPRIM folding, the above acidic residues are close in space and become available for concerted catalytic assistance through Mg2+ coordination [35].
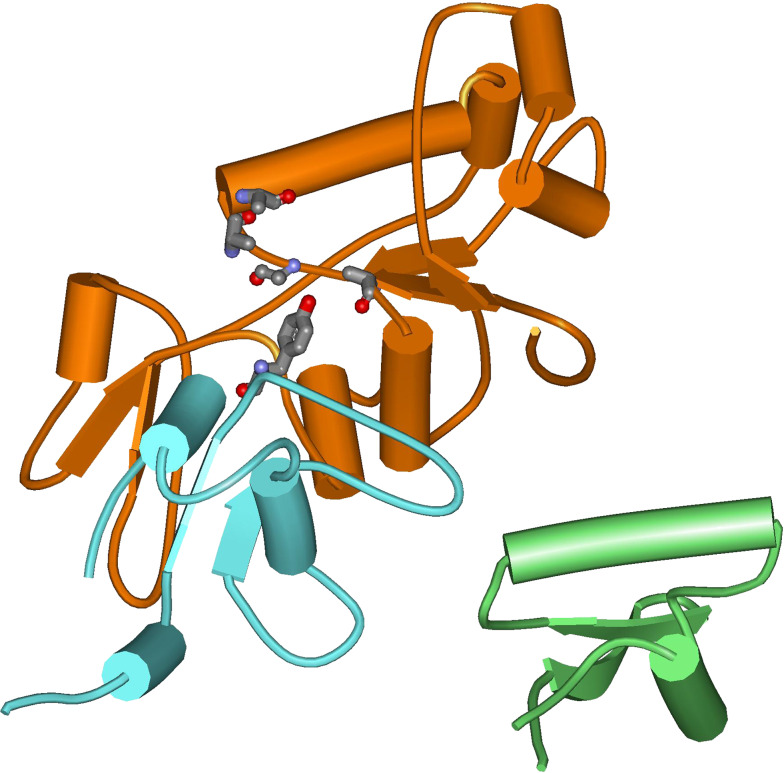
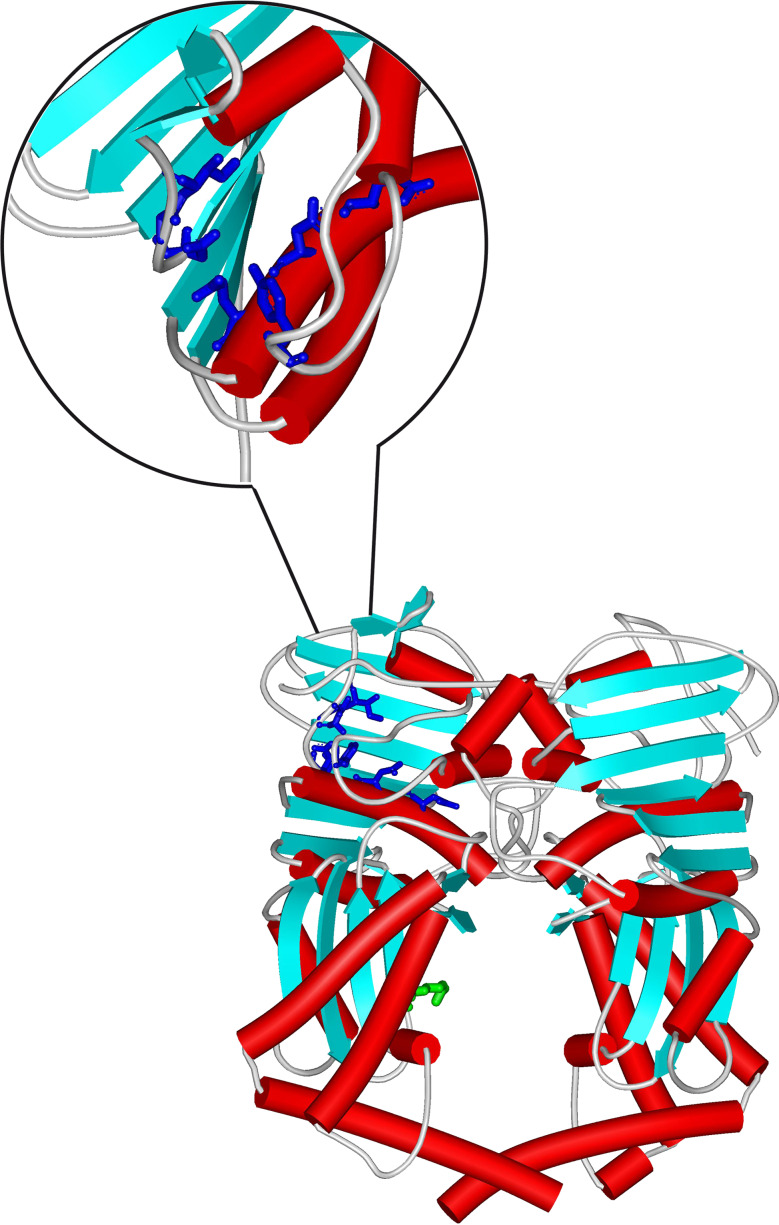
Valuable direct information about the cleavable complex structure in bacteria stems from the successful co-crystallization of the S. pneumoniae ParC cleavage-reunion domain with the ParE TOPRIM domain and a DNA substrate (Fig. 5) [48]. Both the proteins and the nucleic acid are profoundly affected by mutual interaction, which is consistent with the results obtained for yeast topoisomerase II [44]. The protein provides a large positively charged surface for nucleic acid binding and the effective positioning of tyrosine and ion co-factors is guaranteed by the closing up of the Mg2+ coordination tetrad to the phosphodiester bond to be processed. In line with the M. tuberculosis and yeast structures, the TOPRIM core exhibits four parallel β-sheets surrounded by α-helices [44, 49].
Fig. 5.
Crystal structure of the S. pneumoniae ParC breakage-reunion domain (ParC55) and ParE TOPRIM domain (ParE30) in the cleavage complex with DNA in the presence of the quinolone moxifloxacin (PDB ID 3FOF). Red tubes refer to α-helix regions, blue arrows to β-sheet. The DNA portion is in green sticks
The relative structural organization of the WHD, tower, and TOPRIM regions to form the Topo IV cleavage complex are shown in Fig. 6.
Fig. 6.
Relative location of the WHD (cyan), tower (green), and TOPRIM (brown) domains in the catalytic DNA cleavage-resealing pocket as observed in the crystal structure of the S. pneumoniae ParC breakage-reunion domain (ParC55) in combination with the ParE TOPRIM domain (ParE30) in the cleavage complex with DNA(PDB ID 3FOF). The catalytic Tyr residue in WHD and the four acidic residues involved in Mg2+ binding in TOPRIM are highlighted (ball-and-stick model). Tubes represent α-helix regions, arrows β-sheets
The DNA G-segment, similar to the yeast enzyme, is sharply bent by the protein (Fig. 5) [44]. Waiting for the experimental proof from a crystal structure, a significantly curved G segment structure can be reasonably expected to characterize the Gyr-DNA complex as well.
Biochemical role of bacterial topoisomerases II
The above mechanistic and structural similarities notwithstanding, an impressive amount of biological and biochemical evidence shows that Gyr and Topo IV are in fact specialized in performing distinct functions in bacterial cells [14, 50–52].
In particular, DNA Gyrase is suited for:
introducing negative supercoils into covalently closed, positively supercoiled or relaxed, double-stranded circular DNA molecules;
catenating and decatenating DNA rings;
relaxing negative supercoils in the absence of ATP.
Topoisomerase IV activities include:
catenation and decatenation of double-stranded circular DNA molecules;
relaxation of both positive and negative supercoils.
Considering these main functions, Gyr is effective in producing intramolecular DNA strand passing processes, in particular supercoiling, whereas Topo IV is fit for intermolecular events, such as catenation/decatenation.
The above individual features allow maintaining the supercoiling level of the bacterial chromosome as appropriate and dealing with the topological constraints that arise during DNA replication and transcription. To explain how a common general mechanism can be tuned to favor different biological pathways, DNA supercoiling and catenation must be more closely examined.
DNA supercoiling/relaxation
Both Gyr and Topo IV can relax supercoiled DNA but only gyrase is able to introduce negative supercoiling. Biological and structural studies indicate that Gyr can support DNA supercoiling by allowing DNA wrapping at its exterior [53, 54]. This process produces a single positive supercoil with a left-handed superhelical crossing: after passing through the open G segment, the supercoil handedness is converted from positive to negative. Footprinting experiments and electron microscopy studies show that Gyr is able to bind a sequence of about 130–140 bp, which allows processing G and T segments close to each other [55, 56]. In turn, Topo IV covers a small region of 34 bp centered about the cleavage site and, as a result, it bends DNA to an extent that is not sufficient to generate a T segment flanking a G segment [47, 57]. Hence, Topo IV processes DNA segments at a distance and is unable to introduce negative supercoils thus exhibiting a mechanism reminiscent of eukaryotic topoisomerases, rather than of Gyr [33].
The GyrA/ParC C-terminal domains are apparently responsible for DNA wrapping versus bending [58].
It is worth recalling that the prokaryotic CTDs, in particular GyrA CTD, are structurally distinct from the corresponding domains in eukaryotes, which strengthens the idea that they perform special functions in bacteria [47, 59]. Several three-dimensional structures are now available to assess their mechanistic significance. The GyrA CTD, a pear-shaped domain located aside the catalytic core according to X-ray scattering, adopts an unusual fold, first seen in the B. burgdorferi domain, namely a β-pinwheel, that is globally reminiscent of a β-propeller but is made up of six blades with a peculiar topology (Fig. 7) [39, 60]. A large, conserved basic amino acid sequence on the outer edge of this domain represents a likely site for binding and wrapping/bending DNA. FRET assays show that the GyrA CTD can sharply bend DNA over a 40-bp region to generate a T-segment in cis [60]. Such a combination fixes the positive crossover which, after strand passing, introduces a negative supercoil into circular DNA [53, 54].
Fig. 7.
The β-pinwheel structure of the E. coli GyrA CTD (PDB ID 1ZI0). Blue arrows refer to β- sheet tracts
The CTDs shape can vary from species to species. In fact, whereas the E. coli GyrA-CTD is spiral, the corresponding B. burgdorferi domain is flat [60, 61]. However, this difference is probably related to the peculiar character of CTD in B. burgdorferi, the only one expressed as an independent gene product [62]. Site-directed mutagenesis of M. tuberculosis GyrA suggests a key role played by one Tyr and two Arg residues in the third CTD blade as their mutation produced loss of enzymatic activities, without affecting ATPase activity [63].
Another distinctive feature of Gyr CTD is the so-called GyrA box characterized by the heptapeptide sequence Gln-Arg-Arg-Gly-Gly-Lys-Gly located at the first amino-terminal blade of the GyrA CTD [64]. Mutation/deletion of this box inhibits DNA wrapping around itself [65].
In addition, GyrA CTDs are characterized by a flexible connection to the catalytic core [39], which may help them interchange between at least two positional states: one located close to the N gate to grant DNA-wrapping prior to T segment engagement and the second, close to the C gate, possibly representing a resting conformation after strand passage. Sequence alignment and secondary structure analysis evidenced two notable differences between GyrA and ParC CTDs in E. coli. In fact, (1) there are six blades in the β-pinwheel fold for GyrA and five for ParC and (2) the GyrA box is missing in ParC [44, 65]. The superhelical spiral motif is conserved among different species as it occurs also in the B. stearothermophilus ParC CTD, which is structurally similar to the corresponding region in GyrA, but still lacks the GyrA-box [66]. Apparently, the Gyr domain is better suited to wrap DNA around itself as compared to the Topo IV domain [47]. In addition, the mobility of GyrA CTD is not observed in ParC CTD, which binds more tightly to the catalytic portion of the protein and cannot assist T fragment translocation [39, 47].
To summarize, the unique supercoiling activity exhibited by Gyr is explainable by the combination of CTD features: GyrA box (perhaps synergistically with the GyrB tail domain [40]) is possibly capable of electrostatically directing the G segment from the catalytic core to the CTD, where it can be appropriately wrapped to generate the T segment. The flexible connection of the CTD to the core domain would additionally help in transferring the T segment through the DNA gate by shifting from the “upper” to the “lower” position further facilitating DNA supercoiling [39].
In turn, supercoiled and entangled regions of DNA are expected to be locally bent, which renders them ideal substrates for recognition and processing by Topo IV [67].
This picture is fully consistent with the biochemical and structural data thus far available.
DNA catenation/decatenation
The use of isogenic E. coli strains that allowed selective in vivo inhibition of Gyr, Topo IV, or both, unequivocally clarified that Topo IV, not Gyr, is responsible for the decatenation process [68]. In fact, lack of Topo IV essentially leaves all newly synthesized plasmid DNA in the catenated form, Gyr exhibiting at least a hundred-fold less effective decatenase activity [69, 70]. Topo IV decatenates linked cyclic DNAs 10–40 times faster than it relaxes supercoiled DNA and unlinks supercoiled catenanes at an even faster rate [71]. Thus, supercoiling seems to favor (rather than inhibit) decatenation [72]. Additionally, Topo IV binds preferentially to positively supercoiled DNA possibly in a different conformation as compared to other topoisomers, including negatively supercoiled DNA [57, 73]. Further studies on the interplay between supercoiling, catenation, and knotting in E. coli cells with impaired Topo IV activity indicate that catenation and negative supercoiling compete with each other in replication intermediates formed in vivo, high catenation levels being accompanied by low supercoiling and, conversely, high supercoiling levels corresponding to low catenation as segregation proceeds [74].
This confirms that effective decatenation by Topo IV relies on its ability to recognize the type and level of DNA supercoiling, thus indicating that the topological state of the genome plays a dynamic role during progressive in vivo decatenation of sister chromosomes [74].
Together with catenation, knotting should be kept at as low a level as possible (see below) since it interferes with the normal processing of DNA [75]. Like catenanes, knot crossings are preferentially removed by Topo IV [70], although it should be considered that DNA supercoiling renders elimination of intramolecular knotting energetically favored [76].
Besides helping to rationalize different individual activity profiles, the above findings foreshadow the need for concerted actions of the two bacterial topoisomerases II (and, obviously, of the type I enzyme) and the need for a finely tuned equilibrium between time and location of enzyme action/expression. However, in bacteria not expressing Topo IV, Gyr is probably engineered to perform decatenation in addition to supercoiling. Indeed, Gyr from Mycobacterium smegmatis behaves as an efficient decatenase [77].
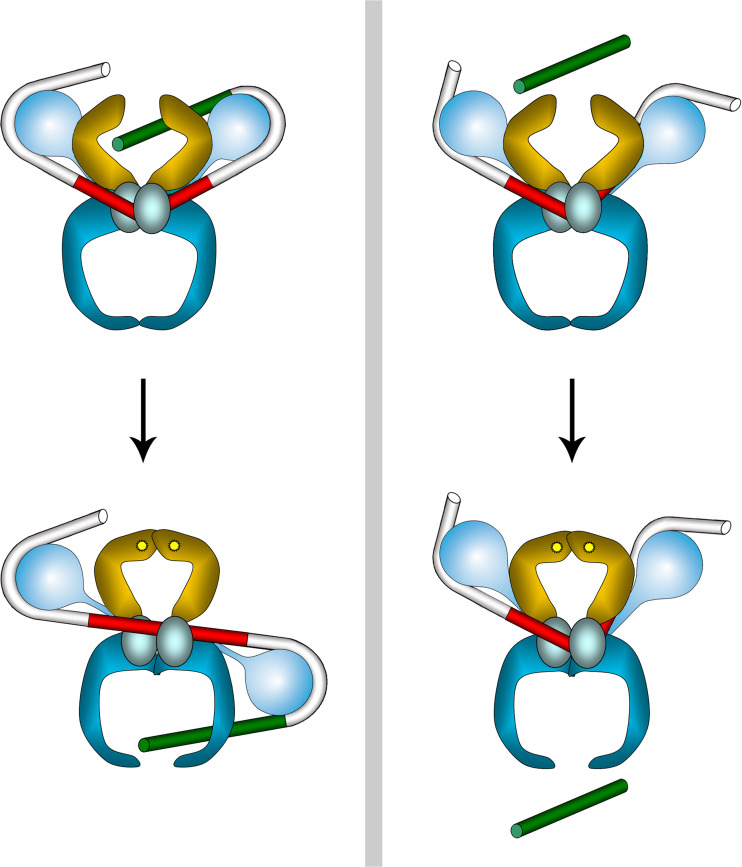
A cartoon model for the DNA-processing mechanisms exhibited by Gyr and Topo IV is presented in Fig. 8.
Fig. 8.
Proposed model for diverse recruitment of G (red) and T (green) segments by DNA Gyrase (left panel) and Topoisomerase IV (right panel). Labeling as in Fig. 2
DNA topological simplification
Unexpectedly, the steady-state fractions of relaxed and decatenated products generated by topoisomerase II action in vivo were found to be substantially higher than the corresponding equilibrium fractions [78]. Hence, the enzymes not only assist the reaching of equilibrium but also force it towards topological simplification. Moreover, the topoisomer distribution appears to be narrower than expected [78]. Though this fact is clearly rational from a biological point of view, it is not apparent how small enzymes could selectively affect the topology of very large DNA molecules sensing their complexity, as this corresponds to a global property of the assembly and cannot be determined by a spatially confined DNA–protein interaction. A three-binding-sites model was originally proposed, according to which binding of topoisomerase to a DNA crossover divides the circular DNA into two smaller cyclic domains [78]. This increases the probability of G-T segment collision in catenated, knotted, and supercoiled systems. As a corollary, sliding of the protein clamp along the DNA can trap a catenane or knot node in a small loop, thereby facilitating removal of topological links from DNA. Other models were afterwards suggested to account for the experimental topology simplification. One foresees kinetic proofreading by the enzyme which binds to entangled DNA in two successive binding events, the first of which activates the enzyme for subsequent strand passage [79]. A simpler explanation was next proposed considering a sharp bending of the G segment into a hairpin-like structure when bound to the enzyme [67]. As a result, the doorway for the T segment is placed inside the hairpin. Thus, the T segment can move across the G segment only from the interior to the exterior of the bend. This would represent a specific tracking element, allowing the strand passing reaction to occur monodirectionally relative to the curved G segment. Further statistical mechanic simulations considered a G segment oriented in a left-handed helical conformation, characterized by a winding angle and a height parameter [80]. The results show that the winding angle plays an important role in directing the chiral preference of the topoisomerase II for relaxation of positive versus negative supercoils, favoring the former when it exceeds 180°, as in the case of Topo IV [80, 81].
The use of tightly bound mutant EcoRI complexes on a plasmid allows ruling out a mechanism of the sliding of a protein clamp along DNA to trap supercoiled regions. In addition, the simplification process appears to be independent of circle size, which also disfavors tracking mechanisms [34, 82]. Moreover, the amount of free energy obtainable from ATP hydrolysis does not affect the extent of topological simplification, suggesting an energy facilitated geometric (or kinetic) selection of T segments for unidirectional strand passage [34, 82]. Apparently, the “hairpin” model is more adequate to explain the experimental results of enhanced relaxation and decatenation levels. No straightforward rationale can explain how the suggested “hairpin DNA” mechanism produces simplification of DNA topology; however a computational analysis shows that this is indeed the case [67]. Topoisomerase preference for hooked juxtapositions, where the nucleic acid strands are curved toward each other, might also be invoked to explain selectivity [83]. However, simultaneous binding of the enzyme to two juxtapositions is an unlikely event that would dramatically slow down disentanglement [49, 75]. In addition, the CTD regions principally responsible for DNA bending do not appear to contribute to DNA structure simplification [82]. Indeed, CTDs would provide an ideal physical location for bent T-segment selection to effectively recognize hooked crossovers.
An important issue to be further considered is the real significance of equilibrium information given the continuous topological changes occurring in the bacterial chromosome [84], which make disentanglement a process that is more kinetically than thermodynamically controlled.
A final concern deals with the substantially different simplification efficiency produced by enzymes of the topoisomerase II family, Topo IV being most effective in this regard [82]. This means that individual enzymes react differently to entanglement, shifting the equilibrium fraction of simplified structure to different extents. Hence, the fine details of entanglement sensing and resolution, although based on general rules, are dictated by (modest) adjustments in G and T segment recognition.
ATPase activity
The ATP requirement for topoisomerase II enzymes has been thoroughly discussed in recent reviews [7].
ATP hydrolysis by Gyrase follows a non-Michaelis–Menten kinetics and is consistent with dimerization of the protein in line with the symmetric assembly of the functional subunit [85]. Accordingly, the activity of the reconstituted enzyme prepared with varying ratios of the inactive mutant at Lys103 and wild-type GyrB suggest an interdependent action of both B subunits in ATP binding and hydrolysis to produce DNA supercoiling [86].
Available crystallographic data indicate that the 43-kDa N-terminal GyrB fragment accommodates a nonhydrolysable ATP analogue in a series of β-pleated sheets, containing both antiparallel and parallel motifs [87]. The protein undergoes dimer formation as a result of a polypeptide exchange involving 14 N-terminal amino acids (Fig. 9). The hydrophobic Ile10 residue and the nucleotide-protein contact mediated by the Tyr5 side chain appear to be particularly relevant for dimerization [88]. Moreover, according to ATPase activities of mutant proteins dimerization enhances the ATP-hydrolysis turnover [88]. In the complex, the Tyr109 residue provides a scaffold for the ATP-hydrolysis center. A channel formed at the dimer interface allows reactive water molecules to access and process the ATP γ-phosphate. Besides neutralizing the negative local charge density, a catalytically required Mg2+ ion provides appropriate orientation of the triphosphate group inside the reactive pocket [89]. A flexible GHKL loop, having conserved Gly residues allows accommodation of the phosphates, their negative charge being additionally counterbalanced by the conserved Lys103. In addition, a negatively charged Asp residue accounts for ATP over GTP specificity [87].
Fig. 9.
Crystal structure of the 43-kDa E. coli GyrB dimer in complex with ADPNP, a nonhydrolyzable analogue of ATP (PDB ID 1E11). Red tubes refer to α-helix regions, blue arrows to β-sheet. Residues Glu42, Asn46, Glu50, Asp73, Arg76, Gly77, Ile78 and Tyr109 in the ATP hydrolysis region are highlighted in dark blue (ball-and-stick models, see also close-up), Asp286 in the DNA binding region in green (see text for details)
Besides the ATPase domain, a large surface containing basic residues in the hole generated by dimerization was identified as a possible DNA-binding region [87]. Such a transducer region mediates signal transduction and coordinates the subsequent cascade of catalytic events. In human topoisomerase IIα, it is thought that a specific motif (QTK loop) mediates communication between ATPase and DNA cleavage regions [89]. Interestingly, the same motif is observed in E. coli and S. aureus GyrB as well as in E. coli and S. pneumoniae ParE transducer region at positions 335–337, 346–348, 332–334, and 333–335, respectively [90].
Computer-modeling studies using the dimeric structure of GyrB43 suggest that the putative DNA-binding cavity contains a constriction produced by Arg286 [91]. In a Gln mutant, unable to produce supercoiling, ATPase activity was not stimulated by DNA, which implies that nucleic acid binding to GyrB is a prerequisite for stimulation of the ATPase activity [91]. Similar studies on other GyrB mutants (Glu42, Asn46, Glu50, Asp73, Arg76, Gly77, and Ile78) suggest that all the above residues are directly involved in ATP hydrolysis, while Pro79 and Lys103 mutants support ATPase function, but not supercoiling. In addition, nucleotide hydrolysis at only one GyrB subunit is sufficient to promote supercoiling [86, 92, 93].
DNA gate opening represents a crucial step to trigger DNA-stimulated ATP hydrolysis, as indicated by GyrB cross-linking studies [94]. Moreover, blocking the ATP-clamp in the closed form by means of ADPNP, a non-hydrolysable ATP analogue, is compatible with reduced DNA binding and cleaving activity, but not with supercoiling [95]. These results suggest that a T segment may reach equilibrium across the DNA gate by entering through the ATP-operated clamp or through the exit gate [95].
The mechanism emerging from the available information envisages an ATP-operated clamp, which segregates a T segment prior to DNA-gate opening. The conformational changes required for effective DNA processing catalysis can be triggered by T-segment trapping.
The ATP-binding region ParE43 from E. coli, examined by X-ray techniques, is structurally very similar to its Gyr counterpart in all essential amino acids [45]. Also, ADPNP binding occurs in a strictly analogous fashion. This essentially overlapping architecture, which could not be anticipated from the subunits’ moderate sequence similarity, suggests a substantial conservation of the mechanism and effects of ATP binding and hydrolysis. Interestingly, a single amino-acid mutation, Met74Ile (Ile is found at the corresponding position in GyrB) produces remarkable changes in the enzyme’s substrate affinity and catalytic efficiency, showing fine tuning of biochemical response produced by minor changes in the sequence of homologous enzymes [45].
The above similarities notwithstanding, a certain heterogeneity is apparent when examining the ATPase process in related species. In fact, Gyr from B. subtilis, exhibits an activity profile comparable to the E. coli enzyme [96]. However, subtle differences are evidentiated by the fact that the rate-limiting step in the nucleotide cycle of B. subtilis GyrB is ATP hydrolysis, not product dissociation or a related conformational change. Moreover, no cooperativity phenomena connect DNA and ATP binding in B. subtilis with single steps varying significantly even when comparing closely related enzymes [96].
Another interesting issue refers to the sequential hydrolysis of the two ATP molecules reported for yeast topoisomerase II, one prior to and the other after the strand passage event [97]. While ATP (but also ADP) binding grants N-gate closure, the first hydrolytic process possibly allows T-segment pushing for strand passage, the second ATPase event, responsible for enzyme resetting, is not molecularly defined. So far, it is not clear whether this mechanism could also apply to the bacterial topoisomerases, in particular to the supercoiling process mediated by Gyr though the experimental results presented above are compatible with such a view.
However, it is less simple to rationalize the linkage between energy input and the energetically favored relaxation, decatenation and unknotting processes, since they would not require ATPase activity [7]. The simplest explanation rests in the energy requirements for the observed reduction of supercoiling or catenation below equilibrium. Apparently, this process requires less energy than provided by nucleotide hydrolysis (0.2 versus 150 kJ/mol), nor is simplification sensitive to the ATP:ADP ratio [82]. Hence, in terms of energy transduction, it would represent poorly efficient machinery. Perhaps, a proofreading process could account for selective T segment testing followed by removal of those not properly oriented towards successful topological simplification [7].
Single molecule studies
Deeper insight into critical issues related to topoisomerases mechanism of action at the molecular level can be gained by monitoring progression of the catalytic cycle “on line”. Recently, this was rendered feasible by applying single molecule techniques [98]. Torsional strain can be built up in the molecule when the DNA duplex ends are firmly bound one to a suitable surface and the other to a magnetic bead that could be rotated in a magnetic field. Under tension, as turns are added, the duplex extension remains essentially unchanged until a critical amount of torque accumulates and the molecule finally buckles, exchanging twist with writhe to form supercoiled structures [99]. This reduces the apparent DNA extension as the number of superhelical turns increases. Such a single molecule device can be useful to investigate enzyme action on differently stressed DNA as well as on DNA–protein assemblies like chromatin [100].
Topoisomerases are natural candidates for single-molecule studies. In fact, the activity of Gyr was monitored on a stretched DNA molecule [101]. Bursts of rotation were observed in the presence of ATP, which implies a processive introduction of negative supercoils in multiples of two. Increasing applied tension did not affect supercoiling kinetics, but reduced processivity. In view of that, processivity is explained in terms of kinetic competition between dissociation of the DNA–enzyme complex and tension-sensitive DNA wrapping [101]. Some features of this model were later criticized in view of discrepancies with biochemical data concerning hydrolysis product like the rate-limiting step and variation in the efficiency of coupling between ATP binding and strand passage [102].
Three distinct modes of Gyr activity were subsequently monitored as a function of force and torque [103, 104]. Under low mechanical stress, Gyr introduces negative supercoils by wrapping DNA. Instead, elevated tension or positive torque stimulates a mode of activity typical of the other topoisomerase II enzymes producing relaxation of left-handed braids and positive supercoils. A third mode accounts for the ATP-independent relaxation of negative supercoils [94]. It is interesting to note that applied tensile strength represents a key element to preferentially activate a specific mechanism of action.
Topo IV was shown to preferentially relax positively supercoiled DNA, probably by discriminating the supercoiling geometry of the left-handed versus right-handed arrangements [81]. DNA binding and single-molecule braiding experiments confirmed that Topo IV recognizes positive supercoils by a mechanism related to the chiral geometry of G- and T-segment crossings [105]. The enzyme is found to bind cooperatively to supercoiled DNA, suggesting that the enzyme subunits assemble efficiently upon binding to the nucleic acid [106]. Like Gyr, but unlike the eukaryotic homologues, Topo IV bends DNA changing local twist and/or writhe by 0.16 turns per complex unit. The enzyme/DNA complex changes between two states, one weakly bound and the other strongly bound, possibly reflecting different enzyme conformations upon T or G segments binding [106].
Subsequent studies on the relaxation rates of single and multiple DNA crossings by Topo IV showed that the enzyme processes DNA strands oriented almost perpendicularly [107]. Moreover, relaxation of positively supercoiled DNA is highly processive, whereas relaxation of negatively supercoiled DNA is purely distributive, which may account for the chiral substrate discrimination observed in Topo IV. These results show the importance of enzymatic processivity as compared to the direction of the crossing angle in chiral discrimination by Topo IV [107]. Interestingly, the eukaryotic type II enzyme from Drosophila melanogaster relaxes left-handed and right-handed braids at essentially the same rate. In addition, the prokaryotic and eukaryotic enzyme exhibit substantially different rate-limiting steps [108]. The remarkably different efficiencies exhibited by enzymes performing equivalent functions possibly reflect the different need to resolve decatenation in bacterial versus mammalian chromosomes.
Although the results from magnetic micromanipulation are quite informative and may give quantitative information on the biochemical processes at the single molecule level, the biological significance of experimental outcome remains still somewhat controversial, especially when the technique calls for application of high tensile strength onto DNA, which might represent a non-physiological state, which hence induces enzyme responses not directly transferable to real situations.
A further mechanistic issue addressed by a single-molecule smFRET approach refers to topoisomerase gate opening/closure timing during catalysis [109]. The experimental results bear evidence that DNA binding, cleavage, and gate opening are mechanistically distinct events. Additionally, DNA gate was shown to be prevalently in a closed form. This addresses concerns about the possible chromosomal damage generated by high content and long persistence of cleaved DNA intermediates produced by the action of topoisomerases [110].
At this point, a few mechanistic remarks are warranted:
DNA bending appears to represent a key determinant for bacterial topoisomerases functions in vivo, as strongly indicated by available information on the molecular mechanisms of enzyme action. The level of bending is sufficient to address DNA cleavage intra- or inter-molecularly with quite different biological consequences; bending is invoked to account for monodirectionality of the topological event, T segment passing through the interior of the curved G segment; bending is deemed responsible for sensing catenane handedness and resolving entanglement. It would be interesting to examine DNA hot spots for topoisomerase binding to assess their intrinsic tendency to produce bent conformations, hence to act as favored enzyme substrates.
Most mechanistic implications were obtained by combining data from crystal structures of topoisomerase fragments from different sources. Hence, relevant information for a detailed description of the interplay among domains either located in the same subunit or getting close in space as a result of functional tetramer assembly is still elusive. For example, the poorly understood functional role of the GyrB “tail” domain (residues 605–648), ubiquitous among type IIA topoisomerases, is now being clarified starting from the very recent crystal structure of the GyrB sequence 485–714, including the TOPRIM and the “tail” domains, and a C-terminal segment [49].
Finally, the significance of selective changes occurring in catalytically and structurally crucial residues in topoisomerases from different species needs to be analyzed much more deeply to explain important modifications in substrate recognition and processing on a case-by-case basis.
Cellular localization of Gyr and Topo IV
Progression of the replicative fork generates a condition where positive supercoils accumulate in front of it while precatenanes (strongly similar to catenanes) are generated behind [111]. This uneven topological distribution drives differential recruitment of the two enzymes along the genome being processed and, possibly, requires different timing of expression [51, 112].
This is confirmed by topoisomerases mutations that lead to a dramatic reduction in replication fork progression and production of double-stranded breaks into the nucleic acid [113]. DNA microarray experiments further suggest that in E. coli, both Gyr and Topo IV promote replication fork progression, which contributes to the elongation rate [114]. The same effect is present in transcription elongation as a solution to the positive supercoiling generated during this process. Hence, Gyr should be especially effective in front of the replication fork and Topo IV behind it, while they could cooperate in reducing positive superhelicity during transcription.
In addition to specific abilities of Gyr and Topo IV in processing DNA in vivo, an important issue refers to their cellular distribution as it can further differentiate the physiological role of the two enzymes [115, 116]. In living B. subtilis cells Topo IV and Gyr are reported to differentially localize on the nucleoids. Topo IV is apparently uniformly distributed, whereas Gyr forms discrete accumulations on the nucleoids in a large fraction of the cells, which showed highly dynamic movements. Three-dimensional time-lapse microscopy indicated that Gyr builds up and vanishes within a 1-min time scale. In addition, it is frequently co-localized with the central DNA replication machinery, confirming a major role for Gyr at the level of this process.
We should also recall that the supercoiling level changes locally due to compartmentation of DNA sequences during selected cell-cycle dependent functions, which may recruit different actions at defined genomic regions.
Altogether, the data thus far presented show how modest structural changes may produce specialized biological functions. Indeed, Topo IV might derive from Gyr by progressive loss or modification of structural characters of the CTD region, in particular deletion of the GyrA-box. This step would be sufficient to cause loss of Gyr supercoiling activity, while preserving relaxation and promoting decatenation abilities.
Bacterial IIA topoisomerases as drug targets
Since both Gyr and Topo IV are vital enzymes, any agent able to interfere with their catalytic cycle can be in principle exploited for antibacterial chemotherapy [50]. To grant effective pharmacological action and avoid toxic effects, a drug molecule should be able to inhibit the prokaryotic topoisomerases’ activities without impairing the host topoisomerases’ functions.
Two principal targets for effective drug intervention can be envisaged:
impairment of the DNA cleaving-rejoining functions;
inhibition of ATPase activity.
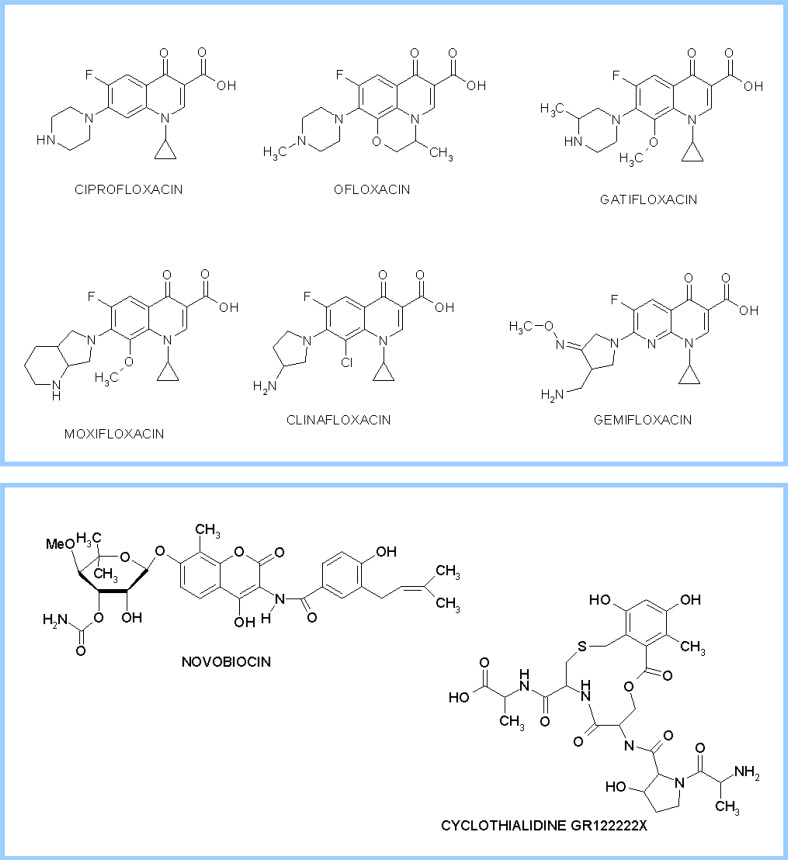
Representative drugs acting by the above mechanisms are reported in Fig. 10.
Fig. 10.
Representative bacterial topoisomerase II inhibitors. Chemical structure of cleavage complex poisons (upper panel) and ATPase inhibitors (lower panel)
Bacterial topoisomerase II poisons
The first target exploits the topoisomerase action, transforming a physiological process into a lethal event. In fact, an intermediate stage of the strand-passing event corresponds to the formation of a covalent DNA–enzyme complex in which duplex DNA is cut and its 3′-ends are temporarily uncapped. If the steady-state level of this “damaged” DNA is kept low by a physiological procedure of re-sealing, damage is transient and will soon be repaired without consequences. On the contrary, if the damage level is increased by the presence of an interfering drug molecule that stacks in the cleavage site and changes its local conformation, the phosphotyrosine group will no longer be located sufficiently close to the 3′-OH to engage it in the resealing transphosphorylation [117]. In this case, the steady-state level of cleaved DNA will increase and the cut DNA will trigger at least two types of events, one dependent and the other not upon protein synthesis. In particular, (1) the replication machinery is arrested and (2) oxidative damage is activated via peroxide/hydroxyl radical production [118, 119]. As a consequence, the bacterial chromosome becomes lethally fragmented and the SOS response is activated, finally leading to bacterial cell death [120]. Hence, antimicrobial action is the result of “poisoning” the topoisomerase II enzyme, which actively participates in producing bacterial cell death [121, 122]. Indeed, this topoisomerase-mediated mechanism has successfully led to clinically effective antibacterial agents. A similar mechanism is exploited by eukaryotic topoisomerase poisons which are currently used in anticancer chemotherapy.
Fluoroquinolones
The principal family of prokaryotic topoisomerases II-targeted drugs is represented by fluoroquinolones [123–125]. They consist of a 6-fluoro-quinoline ring system, with a keto-carboxylic function at position 3 and 4, with several possible substituents at positions N-1 and C-7. Successful representatives of this family are shown in Fig. 10, in the upper panel.
At the molecular level, the interactions of quinolones were found to involve mainly subunit A of Gyrase and ParC of Topo IV. In fact, a region spatially close to the active tyrosine called Quinolone Resistance Determining Region (QRDR) has been identified. Amino acid mutations in this region greatly reduce the affinity of quinolones for the enzyme–DNA complex. The most relevant positions in E. coli are Ser83 and Asp87 in GyrA and the corresponding Ser79 and Asp83 in ParC [126, 127]. Mutations in GyrB and ParE, although implicated in the fluoroquinolone resistance, are less frequently observed. The current hypothesis is that they occur at the interface between the two subunits inducing subtle structural changes in the overall protein architecture.
The precise mechanism of topoisomerase II inhibition by quinolones has long been an object of debate, specific issues dealing on mode and cooperativity of interaction with DNA and participation of metal-ion co-factors [128–130]. Very recently, the structural features of a quinolone bound to a Topo IV cleavage complex (S. pneumoniae ParC breakage-reunion, ParE TOPRIM and G-DNA) have been elucidated by crystallographic studies (Fig. 5) [48]. The quinolones employed were moxifloxacin and clinafloxacin, two very effective antimicrobial agents.
Moxifloxacin complex is formed from a ParC55 heart-shaped homodimer flanked on either side by ParE30 monomers. Structurally, the ParC55 dimer closely resembles the unliganded protein in the absence of ParE30 as well as GyrA59, with the active site Arg117 and Tyr118 on the 100–122 loop (see above). The C-gate is organized in the closed form and TOPRIM domain is correctly located to interact with DNA along with the WHD domain (Fig. 6). The nucleic acid participates in the cleavage complex with the active site tyrosines covalently bound to the 5′-phosphate residue. In this region, DNA produces notable local changes, closing up the protein domains, so that the inner part of the 100–122 loop leans toward the DNA backbone. A moxifloxacin residue is inserted into the cleft at each side of the 4-bp staggered cleavage site. The structure of the drug molecule intercalated between the bases immediately preceding (−1) or following (+1) the cleaved bond is reported in Fig. 11. The quinolone stacks against a purine belonging to the residue immediately adjacent to the cleaved (+1) phosphodiester link. The cyclopropane ring points in the direction of Ser79 and Asp83, which are known to mutate in quinolone-resistant bacterial strains. Apparently, the reason for resistance is not correlated to a direct interaction, but it might rest in changes in local conformation when a hydrophilic amino acid is eventually replaced by an aromatic residue (Phe, Tyr) in the mutants. The C7 substituent gets close to the DNA base on the opposite DNA strand and is easily accommodated inside a large, solvent-accessible cavity generated by cleavage. No particular interaction is evidenced by the 6-fluorine atom, which possibly optimizes the interaction through its negative charge density.
Fig. 11.
Close-up view of the quinolone moxifloxacin (stick model) bound to the S. pneumoniae ParC55-ParE30-DNA cleavage complex (line model). Source: PDB ID 3FOF
Surprisingly, the essential keto-carboxylic substituents are separated from the metal ion-binding TOPRIM residues. Since the supposed function of the drug substituents at positions 3 and 4 was to chelate the active site Mg2+ ion, the present data lead to the conclusion that this is not the case. However, given the fact that some domains of the full-length enzyme are missing and considering that the catalytic TOPRIM region must get close to the phosphotyrosine linkage (and the cleaved 3′-OH group) to complete the catalytic cycle, there might be a further conformational change needed to reseal the gap, which could account for “later” sequestering of the metal ion by the quinolone. This is, of course, a speculative matter. In any event, inspection of Fig. 11 shows the possibility of quinolone and phosphotyrosine oxygens’ engagement in coordination to a divalent ion. The DNA-base preferences observed (+1 G, +1 A) possibly account for the more efficient stacking of the planar portion of the quinolone onto a purine rather than a pyrimidine base.
The clinafloxacin analogue yielded essentially the same picture, which would suggest a common mode of quinolone recognition of the cleavage complex. Information at the molecular level will surely help in the future rational design of effective inhibitors.
New drugs are in fact badly needed, as the high efficiency and broad spectrum of activity shown by quinolones are now seriously challenged by the selection of resistant strains, even for naturally highly susceptible species like E. coli [126, 131–134]. Thus, great effort is presently devoted to overcoming resistance phenomena that are emerging as the most relevant complication in antibacterial therapy. Resistance to quinolones is generally acquired through mutation. In fact, as mentioned above, quinolone resistance is seen in selected GyrA and/or ParC mutants [135]. However, plasmid-mediated genes encoding quinolone resistance were recently discovered [136–138]. Some of them (QnrA1, QnrB, QnrS; QnrC) protect Gyr/Topo IV from drug attack, others (QepA) code for enzymes able to modify quinolones or for efflux pump. The promotion of resistance by these genes, their horizontally spreading abilities and the combined selection of resistance patterns raise serious concern on the practical use of quinolones.
New inhibitors
Antibacterials of the fluoroquinolone family were first utilized almost 40 years ago. Since then, well over 10,000 analogues were reported. Solid SAR (structure activity relationships) studies are available in the literature, which allow the exploitation of a large number of substituents in different positions of the quinolone ring [139, 140]. Besides positions 3 and 4, particularly significant in the modulation of activity are positions 1, 7, and 8. Bioisosteric substitutions as well as modifications in the ring structures were additionally performed. Recent studies suggest the potential usefulness of gatifloxacin and ofloxacin derivatives, quinolines, pyrazoles, isothiazolopyridones, and aminoquinazolinediones, some of which are especially suited to counteract the onset of resistance [141–148].
Dual inhibitors
When tested against Gyr and Topo IV, many quinolones inhibited both enzymes. This is not surprising, given the close mechanistic similarities exhibited by the latter. Nonetheless, traditional quinolones active against Gram-negative bacteria are more effective against Gyr than Topo IV, whereas the reverse is true for quinolones aimed at Gram-positive bacteria. However, this is not a general rule as preferential enzyme inhibition is species specific. Compounds exhibiting dual targeting are desirable since mutations must be introduced in both topoisomerases to produce resistant strains, hence the onset of resistance is less likely [149, 150]. Among quinolones showing dual targeting clinafloxacin, gemifloxacin, moxifloxacin, besifloxacin, and amminopyrrolidyl analogues deserve to be mentioned [151, 152]. Recently, isothiazolone derivatives and a ciprofloxacin analogue with a methyl substituent at position C-8 and a 4-hydroxy-3-methyl-1-piperidinyl moiety at C-7 have been reported to behave as effective dual inhibitors [153–155].
As an extension of dual inhibition, hybrid antibiotics targeting bacterial topoisomerases II together with DNA- or RNA-polymerase were also investigated and shown to develop resistance at quite low rates [156, 157].
Finally, a quinoline derivative linked to a piperidine carboxylic acid, exhibited potent activity against Gram-positive bacteria resistant to inhibition by fluoroquinolones [158]. Although it interferes with Gyr and Topo IV activities, its molecular mechanism of action seems to impair formation of the cleavage complex, rather than stabilizing it.
Inhibition of ATPase activity
Although until now no antibacterial drug presently in clinical use acts by inhibiting ATPase, the absolute requirement for ATP of the bacterial topoisomerases prompts significant efforts to design new derivatives that impair nucleotide hydrolysis. In fact, naturally occurring cyclic peptides of the coumarin family (novobiocin) and the cyclothialidines, have been known for a long time to represent effective Gyr inhibitors acting on the N-terminal GyrB subunit (Fig. 10, lower panel). The structural features of cyclothialidine recognition of the ATP site when complexed to a short (24-kDa) GyrB fragment in a monomeric assembly include a major contribution from the resorcinol group and several hydrogen bonds mediated by water molecules [159]. Novobiocin, in turn, shows a large network of hydrogen bonds, involving primarily the novobiose sugar. An interesting notion emerging from these studies is that the various inhibitors only partially overlap the ATP-binding site, which shows how different drugs may use different target-recognition strategies to reach the same goal. More informative is an investigation dealing with the structure of a dimeric GyrA43 fragment form T. thermophilus in complex with novobiocin. This antibiotic produces large conformational changes of the subdomains within the dimer. In particular, loop 98–118, closing the active site through dimeric contacts, generates an “open” conformation of the N gate as opposed to the “closed” conformation adopted by the competitor ATP [160]. More recent studies on the equivalent ParC fragment explain that novobiocin is less potent against Topo IV than against Gyr as a result of an unfavorable contact of the drug with a Met residue, replaced by Ile in Gyr [45]. Unfortunately, members of the above-mentioned natural families, although exhibiting activities in the nanomolar range, were not useful as drugs due to their inadequate pharmacological profile. However, their structures and the biochemical knowledge stemming from their use can be valuably employed to devise new potential inhibitors.
New ATPase inhibitors
Since the ATP-binding site of GyrB possesses a remarkable level of flexibility, consensus functionality maps were employed to identify functional groups able to recognize the ATPase region [161]. The calculations show that some yet unexplored binding sites could be considered for drug design and suggest the potential usefulness of several new chemical scaffolds. Moreover, since the crystal structure of a nonhydrolyzable ATP analogue bound to GyrB43 is available [87], a generally employed approach is to mimic key interactions occurring between the adenosine functional groups and protein invariant residues, in particular Asp73, Gly77, and Thr165 (Fig. 9). Another way to select new active compounds, unrelated to the previous ones, is the use of virtual screening techniques [162]. Although these techniques give valuable initial directions for drug discovery, their indication should be considered as preliminary. High-throughput screening of existing libraries represents another reasonable strategy for lead discovery.
Novel inhibitors thus far tested include indolinones, catechins, azoles, benzopyranones, imidazolo- and triazolo-pyridines, benzimidazoles and chiral barbituric acid derivatives [163–170]. Among these compounds, members of the latter two families were endowed with the ability to inhibit both GyrB and ParE ATPases, hence behaving as especially valuable dual inhibitors.
Natural antibacterial compounds
A number of natural compounds aimed at bacterial topoisomerases II have been discovered and investigated. The interest in these compounds stems from the fact that they efficiently recognize their targets, so that they behave as remarkably specific inhibitors. In addition, starting from the natural structure, a pharmacophoric pattern can be identified and subsequently used for drug design. Among the derivatives recently reported the toxins CcdB, Microcin B17, albicidin and the small protein YacG deserve attention [171–175].
A paradigmatic, well-studied, example is CcdB. The toxin (T) CcdB is encoded by the ccd operon on plasmid F of E. coli together with its antitoxin (A) CcdA. Protein TA systems are utilized by plasmids to ensure that only the daughter cells that inherit the plasmid survive after cell division [176]. CcdB has two effects on Gyr. In CcdB overproducing cells, the enzyme (A subunit) is bound to CcdB and loses its ability to bind and supercoil DNA. As a second mode of action, CcdB converts Gyr into a cytotoxic agent with a mechanism similar to that of quinolones. In fact, CcdB, as a dimer, can trap Gyr in a special conformation during strand passage, with the opening of the tower and catalytic domain, when the G segment is cleaved but crossing of the T segment has not yet occurred [175, 177]. In fact, the crystal structure of CcdB bound to the dimerization domain of the A subunit of Gyr (GyrA14) reveals an “open” conformation for the catalytic core when in complex with the toxin. This represents a novel mechanism, which freezes the cleavage complex before re-sealing, hence in a toxic arrangement [178]. This mechanism could be profitably exploited to develop new effective drug molecules. As a matter of fact, peptides based on the CcdB structure showed inhibitory properties towards Gyr-mediated DNA supercoiling [179].
Recently, a new antibiotic, simocyclinone D8, was isolated from S. antibioticus Tű 6040 [180, 181]. It shows activity against Gram-positive bacteria, as well as cytostatic effects against human tumor cell lines, while it is apparently ineffective against Gram-negative species. The interest for this compound arises from the fact that, notwithstanding the presence of a 3-amino-4,7-dihydroxycoumarin function, reminiscent of the coumarin antibiotics, it does not interfere with ATPase functions. Instead, it is a potent inhibitor of Gyr, acting by a novel mechanism possibly related to Gyr-DNA-binding impairment [182, 183], likely occurring through effective drug interactions with the GyrA subunit [184]. Once this mechanism is fully elucidated, new drug-development strategies on the identified target(s) could be envisaged.
Finally, a biotechnological approach has been employed to down-regulate Gyr A gene expression in E. coli and P. falciparum through antisense ribozymes and it shows remarkable effects on cell viability [185, 186].
Conclusions
The more we learn, the more we see nature’s amazing ability to create remarkable biological functionalities. In the case of topoisomerases, chemical energy is transduced into mechanical energy to grant effective and safe means for the cell to deal with DNA topology. Powerful methods such as X-ray crystallography and single-molecule techniques enable us to “see” such huge proteins at various stages of their action and to dissect the mechanisms by which they perform DNA strand passage to produce supercoiling or decatenation. By the additional contribution of other biochemical, biophysical, and biotechnological methods our understanding of the biological pathways of cell function is further and further enriched. Quite interestingly, evolution from one topoisomerase species to the other has allowed acquisition of very specialized functions. Gyr and Topo IV are a clear example of how modest structural rearrangements can make them solve specific topological problems generated by essential cellular actions like DNA replication and transcription. Not only do they act individually, they are synergistically operating to maintain the proper structural organization of the genome. In addition, DNA topology plays a more general role in the expression of virulence genes in pathogenic bacteria at different phases of the host–pathogen relationship. In fact, supercoiling levels control the gene regulatory network of the cell hierarchically, influencing the activities of a large number of promoters simultaneously [187].
Learning how essential enzymes work allows us to rationally devise new tools for interfering with their action not only to dissect known functions or unravel new ones but also to develop appropriate therapeutic strategies, for example when dealing with bacterial infections. Also, different targets in the same enzyme can be exploited, improving the chances for successful therapeutic intervention. In this connection, it would be interesting to identify drug leads able to interfere with the bacterial CTD functions, not present in the human enzymes therefore maximally reducing the concern that compounds acting against bacterial topoisomerases might exhibit minor albeit non-negligible effects also on the corresponding eukaryotic homologues, thus producing undesired toxic side-effects [188].
As a final remark, a programmed manipulation of the genes involved in the biosynthetic pathways of antibiotic-producing microorganisms could represent a valuable approach to yield combinatorial libraries of re-designed antibiotics [189, 190] to be successfully exploited against the so-called life-threatening “superbugs”.
Acknowledgments
This work was supported by AIRC, Associazione Italiana per la Ricerca sul Cancro (MP) (Grant #. 5826) and University of Padova (CS) (Grant # CPDA078422/07). The authors thank Susan Paris for expert proofreading of the manuscript.
References
- 1.Kalkbrenner T, Arnold A, Tans SJ. Internal dynamics of supercoiled DNA molecules. Biophys J. 2009;96:4951–4955. doi: 10.1016/j.bpj.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z, Deibler RW, Chan HS, Zechiedrich L. The why and how of DNA unlinking. Nucleic Acids Res. 2009;37:661–671. doi: 10.1093/nar/gkp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wasserman SA, Cozzarelli NR. Biochemical topology: applications to DNA recombination and replication. Science. 1986;232:951–960. doi: 10.1126/science.3010458. [DOI] [PubMed] [Google Scholar]
- 4.Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 5.Ohniwa RL, Morikawa K, Kim J, Ohta T, Ishihama A, Wada C, Takeyasu K. Dynamic state of DNA topology is essential for genome condensation in bacteria. EMBO J. 2006;25:5591–5602. doi: 10.1038/sj.emboj.7601414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bates AD, Maxwell A. Energy coupling in type II topoisomerases: why do they hydrolyze ATP? Biochemistry. 2007;46:7929–7941. doi: 10.1021/bi700789g. [DOI] [PubMed] [Google Scholar]
- 8.Baker NM, Rajan R, Mondragon A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009;37:693–701. doi: 10.1093/nar/gkn1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Tse-Dinh YC. Exploring DNA topoisomerases as targets of novel therapeutic agents in the treatment of infectious diseases. Infect Disord Drug Targets. 2007;7:3–9. doi: 10.2174/187152607780090748. [DOI] [PubMed] [Google Scholar]
- 11.Deweese JE, Osheroff MA, Osheroff N. DNA topology and topoisomerases: teaching a “Knotty” subject. Biochem Mol Biol Educ. 2008;37:2–10. doi: 10.1002/bmb.20244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–499. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 16.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori . Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 17.Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 18.Schmutz E, Muhlenweg A, Li SM, Heide L. Resistance genes of aminocoumarin producers: two type II topoisomerase genes confer resistance against coumermycin A1 and clorobiocin. Antimicrob Agents Chemother. 2003;47:869–877. doi: 10.1128/AAC.47.3.869-877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 20.Aubry A, Fisher LM, Jarlier V, Cambau E. First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis . Biochem Biophys Res Commun. 2006;348:158–165. doi: 10.1016/j.bbrc.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 21.Gellert M, Mizuuchi K, O’Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins NP, Peebles CL, Sugino A, Cozzarelli NR. Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc Natl Acad Sci USA. 1978;75:1773–1777. doi: 10.1073/pnas.75.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli . Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-B. [DOI] [PubMed] [Google Scholar]
- 24.Peng H, Marians KJ. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 25.Mizuuchi K, Mizuuchi M, O’Dea MH, Gellert M. Cloning and simplified purification of Escherichia coli DNA gyrase A and B proteins. J Biol Chem. 1984;259:9199–9201. [PubMed] [Google Scholar]
- 26.Pan XS, Fisher LM. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuuchi K, Fisher LM, O’Dea MH, Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc Natl Acad Sci USA. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JC (1987) DNA topoisomerases: from a laboratory curiosity to a subject in cancer chemotherapy. NCI Monogr 3–6 [PubMed]
- 29.Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- 30.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 31.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc Natl Acad Sci USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roca J. The path of the DNA along the dimer interface of topoisomerase II. J Biol Chem. 2004;279:25783–25788. doi: 10.1074/jbc.M402555200. [DOI] [PubMed] [Google Scholar]
- 33.Roca J. Topoisomerase II: a fitted mechanism for the chromatin landscape. Nucleic Acids Res. 2009;37:721–730. doi: 10.1093/nar/gkn994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maxwell A, Costenaro L, Mitelheiser S, Bates AD. Coupling ATP hydrolysis to DNA strand passage in type IIA DNA topoisomerases. Biochem Soc Trans. 2005;33:1460–1464. doi: 10.1042/BST20051460. [DOI] [PubMed] [Google Scholar]
- 35.Sissi C, Palumbo M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009;37:702–711. doi: 10.1093/nar/gkp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 37.Simon H, Roth M, Zimmer C. Biochemical complementation studies in vitro of gyrase subunits from different species. FEBS Lett. 1995;373:88–92. doi: 10.1016/0014-5793(95)01007-2. [DOI] [PubMed] [Google Scholar]
- 38.Huang WM. Bacterial diversity based on type II DNA topoisomerase genes. Annu Rev Genet. 1996;30:79–107. doi: 10.1146/annurev.genet.30.1.79. [DOI] [PubMed] [Google Scholar]
- 39.Costenaro L, Grossmann JG, Ebel C, Maxwell A. Small-angle X-ray scattering reveals the solution structure of the full-length DNA gyrase a subunit. Structure. 2005;13:287–296. doi: 10.1016/j.str.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Costenaro L, Grossmann JG, Ebel C, Maxwell A. Modular structure of the full-length DNA gyrase B subunit revealed by small-angle X-ray scattering. Structure. 2007;15:329–339. doi: 10.1016/j.str.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 41.Aravind L, Leipe DD, Koonin EV. Toprim–a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fass D, Bogden CE, Berger JM. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat Struct Biol. 1999;6:322–326. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 43.Morais Cabral JH, Jackson AP, Smith CV, Shikotra N, Maxwell A, Liddington RC. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 44.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 45.Bellon S, Parsons JD, Wei Y, Hayakawa K, Swenson LL, Charifson PS, Lippke JA, Aldape R, Gross CH. Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase. Antimicrob Agents Chemother. 2004;48:1856–1864. doi: 10.1128/AAC.48.5.1856-1864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laponogov I, Veselkov DA, Sohi MK, Pan XS, Achari A, Yang C, Ferrara JD, Fisher LM, Sanderson MR. Breakage-reunion domain of Streptococcus pneumoniae topoisomerase IV: crystal structure of a Gram-positive quinolone target. PLoS One. 2007;2:e301. doi: 10.1371/journal.pone.0000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J Mol Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 48.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thomson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat Struct Mol Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 49.Fu G, Wu J, Liu W, Zhu D, Hu Y, Deng J, Zhang XE, Bi L, Wang DC. Crystal structure of DNA gyrase B’ domain sheds lights on the mechanism for T-segment navigation. Nucleic Acids Res. 2009;37:5908–5916. doi: 10.1093/nar/gkp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levine C, Hiasa H, Marians KJ. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 52.Schoeffler AJ, Berger JM. Recent advances in understanding structure–function relationships in the type II topoisomerase mechanism. Biochem Soc Trans. 2005;33:1465–1470. doi: 10.1042/BST20051465. [DOI] [PubMed] [Google Scholar]
- 53.Kirkegaard K, Wang JC. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981;23:721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- 54.Liu LF, Wang JC. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978;15:979–984. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- 55.Fisher LM, Mizuuchi K, O’Dea MH, Ohmori H, Gellert M. Site-specific interaction of DNA gyrase with DNA. Proc Natl Acad Sci USA. 1981;78:4165–4169. doi: 10.1073/pnas.78.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison A, Cozzarelli NR. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc Natl Acad Sci USA. 1981;78:1416–1420. doi: 10.1073/pnas.78.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng H, Marians KJ. The interaction of Escherichia coli topoisomerase IV with DNA. J Biol Chem. 1995;270:25286–25290. doi: 10.1074/jbc.270.42.25286. [DOI] [PubMed] [Google Scholar]
- 58.Kampranis SC, Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc Natl Acad Sci USA. 1996;93:14416–14421. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sengupta T, Mukherjee M, Mandal C, Das A, Majumder HK. Functional dissection of the C-terminal domain of type II DNA topoisomerase from the kinetoplastid hemoflagellate Leishmania donovani . Nucleic Acids Res. 2003;31:5305–5316. doi: 10.1093/nar/gkg727. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc Natl Acad Sci USA. 2004;101:7293–7298. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruthenburg AJ, Graybosch DM, Huetsch JC, Verdine GL. A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J Biol Chem. 2005;280:26177–26184. doi: 10.1074/jbc.M502838200. [DOI] [PubMed] [Google Scholar]
- 62.Knight SW, Samuels DS. Natural synthesis of a DNA-binding protein from the C-terminal domain of DNA gyrase A in Borrelia burgdorferi . EMBO J. 1999;18:4875–4881. doi: 10.1093/emboj/18.17.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang YY, Deng JY, Gu J, Zhang ZP, Maxwell A, Bi LJ, Chen YY, Zhou YF, Yu ZN, Zhang XE. The key DNA-binding residues in the C-terminal domain of Mycobacterium tuberculosis DNA gyrase A subunit (GyrA) Nucleic Acids Res. 2006;34:5650–5659. doi: 10.1093/nar/gkl695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ward D, Newton A. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus . Mol Microbiol. 1997;26:897–910. doi: 10.1046/j.1365-2958.1997.6242005.x. [DOI] [PubMed] [Google Scholar]
- 65.Kramlinger VM, Hiasa H. The “GyrA-box” is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J Biol Chem. 2006;281:3738–3742. doi: 10.1074/jbc.M511160200. [DOI] [PubMed] [Google Scholar]
- 66.Hsieh TJ, Farh L, Huang WM, Chan NL. Structure of the topoisomerase IV C-terminal domain: a broken beta-propeller implies a role as geometry facilitator in catalysis. J Biol Chem. 2004;279:55587–55593. doi: 10.1074/jbc.M408934200. [DOI] [PubMed] [Google Scholar]
- 67.Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerases. Proc Natl Acad Sci USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zechiedrich EL, Khodursky AB, Cozzarelli NR. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli . Genes Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zechiedrich EL, Cozzarelli NR. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli . Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 70.Deibler RW, Rahmati S, Zechiedrich EL. Topoisomerase IV, alone, unknots DNA in E. coli . Genes Dev. 2001;15:748–761. doi: 10.1101/gad.872301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Belotserkovskii BP, Arimondo PB, Cozzarelli NR. Topoisomerase action on short DNA duplexes reveals requirements for gate and transfer DNA segments. J Biol Chem. 2006;281:25407–25415. doi: 10.1074/jbc.M603977200. [DOI] [PubMed] [Google Scholar]
- 72.Witz G, Stasiak A (2009) DNA supercoiling and its role in DNA decatenation and unknotting. Nucleic Acids Res doi:10.1093/nar/gkp1161 [DOI] [PMC free article] [PubMed]
- 73.Crisona NJ, Cozzarelli NR. Alteration of Escherichia coli topoisomerase IV conformation upon enzyme binding to positively supercoiled DNA. J Biol Chem. 2006;281:18927–18932. doi: 10.1074/jbc.M603068200. [DOI] [PubMed] [Google Scholar]
- 74.Martinez-Robles ML, Witz G, Hernandez P, Schvartzman JB, Stasiak A, Krimer DB. Interplay of DNA supercoiling and catenation during the segregation of sister duplexes. Nucleic Acids Res. 2009;37:5126–5137. doi: 10.1093/nar/gkp530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vologodskii A. Theoretical models of DNA topology simplification by type IIA DNA topoisomerases. Nucleic Acids Res. 2009;37:3125–3133. doi: 10.1093/nar/gkp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burnier Y, Dorier J, Stasiak A. DNA supercoiling inhibits DNA knotting. Nucleic Acids Res. 2008;36:4956–4963. doi: 10.1093/nar/gkn467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manjunatha UH, Dalal M, Chatterji M, Radha DR, Visweswariah SS, Nagaraja V. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 2002;30:2144–2153. doi: 10.1093/nar/30.10.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 79.Yan J, Magnasco MO, Marko JF. A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature. 1999;401:932–935. doi: 10.1038/44872. [DOI] [PubMed] [Google Scholar]
- 80.Klenin K, Langowski J, Vologodskii A. Computational analysis of the chiral action of type II DNA topoisomerases. J Mol Biol. 2002;320:359–367. doi: 10.1016/S0022-2836(02)00447-3. [DOI] [PubMed] [Google Scholar]
- 81.Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stuchinskaya T, Mitchenall LA, Schoeffler AJ, Corbett KD, Berger JM, Bates AD, Maxwell A. How do type II topoisomerases use ATP hydrolysis to simplify DNA topology beyond equilibrium? Investigating the relaxation reaction of nonsupercoiling type II topoisomerases. J Mol Biol. 2009;385:1397–1408. doi: 10.1016/j.jmb.2008.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buck GR, Zechiedrich EL. DNA disentangling by type-2 topoisomerases. J Mol Biol. 2004;340:933–939. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 84.Rui S, Tse-Dinh YC. Topoisomerase function during bacterial responses to environmental challenge. Front Biosci. 2003;8:d256–d263. doi: 10.2741/984. [DOI] [PubMed] [Google Scholar]
- 85.Ali JA, Jackson AP, Howells AJ, Maxwell A. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry. 1993;32:2717–2724. doi: 10.1021/bi00061a033. [DOI] [PubMed] [Google Scholar]
- 86.O’Dea MH, Tamura JK, Gellert M. Mutations in the B subunit of Escherichia coli DNA gyrase that affect ATP-dependent reactions. J Biol Chem. 1996;271:9723–9729. doi: 10.1074/jbc.271.16.9723. [DOI] [PubMed] [Google Scholar]
- 87.Wigley DB, Davies GJ, Dodson EJ, Maxwell A, Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 88.Brino L, Urzhumtsev A, Mousli M, Bronner C, Mitschler A, Oudet P, Moras D. Dimerization of Escherichia coli DNA gyrase B provides a structural mechanism for activating the ATPase catalytic center. J Biol Chem. 2000;275:9468–9475. doi: 10.1074/jbc.275.13.9468. [DOI] [PubMed] [Google Scholar]
- 89.Bendsen S, Oestergaard VH, Skouboe C, Brinch M, Knudsen BR, Andersen AH. The QTK loop is essential for the communication between the N-terminal ATPase domain and the central cleavage–ligation region in human topoisomerase IIalpha. Biochemistry. 2009;48:6508–6515. doi: 10.1021/bi9005978. [DOI] [PubMed] [Google Scholar]
- 90.data from: Protein knowledge base (UniproKb). www.uniprot.org
- 91.Tingey AP, Maxwell A. Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res. 1996;24:4868–4873. doi: 10.1093/nar/24.24.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kampranis SC, Maxwell A. Conformational changes in DNA gyrase revealed by limited proteolysis. J Biol Chem. 1998;273:22606–22614. doi: 10.1074/jbc.273.35.22606. [DOI] [PubMed] [Google Scholar]
- 93.Gross CH, Parsons JD, Grossman TH, Charifson PS, Bellon S, Jernee J, Dwyer M, Chambers SP, Markland W, Botfield M, Raybuck SA. Active-site residues of Escherichia coli DNA gyrase required in coupling ATP hydrolysis to DNA supercoiling and amino acid substitutions leading to novobiocin resistance. Antimicrob Agents Chemother. 2003;47:1037–1046. doi: 10.1128/AAC.47.3.1037-1046.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Williams NL, Maxwell A. Locking the DNA gate of DNA gyrase: investigating the effects on DNA cleavage and ATP hydrolysis. Biochemistry. 1999;38:14157–14164. doi: 10.1021/bi991478m. [DOI] [PubMed] [Google Scholar]
- 95.Williams NL, Howells AJ, Maxwell A. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J Mol Biol. 2001;306:969–984. doi: 10.1006/jmbi.2001.4468. [DOI] [PubMed] [Google Scholar]
- 96.Gottler T, Klostermeier D. Dissection of the nucleotide cycle of B. subtilis DNA gyrase and its modulation by DNA. J Mol Biol. 2007;367:1392–1404. doi: 10.1016/j.jmb.2007.01.055. [DOI] [PubMed] [Google Scholar]
- 97.Baird CL, Harkins TT, Morris SK, Lindsley JE. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc Natl Acad Sci USA. 1999;96:13685–13690. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cornish PV, Ha T. A survey of single-molecule techniques in chemical biology. ACS Chem Biol. 2007;2:53–61. doi: 10.1021/cb600342a. [DOI] [PubMed] [Google Scholar]
- 99.Bryant Z, Stone MD, Gore J, Smith SB, Cozzarelli NR, Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 100.Meglio A, Praly E, Ding F, Allemand JF, Bensimon D, Croquette V. Single DNA/protein studies with magnetic traps. Curr Opin Struct Biol. 2009;19:615–622. doi: 10.1016/j.sbi.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 101.Gore J, Bryant Z, Stone MD, Nollmann M, Cozzarelli NR, Bustamante C. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–104. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bates AD. DNA topoisomerases: single gyrase caught in the act. Curr Biol. 2006;16:R204–R206. doi: 10.1016/j.cub.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 103.Nollmann M, Stone MD, Bryant Z, Gore J, Crisona NJ, Hong SC, Mitelheiser S, Maxwell A, Bustamante C, Cozzarelli NR. Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat Struct Mol Biol. 2007;14:264–271. doi: 10.1038/nsmb1213. [DOI] [PubMed] [Google Scholar]
- 104.Higgins NP. Under DNA stress, gyrase makes the sign of the cross. Nat Struct Mol Biol. 2007;14:256–258. doi: 10.1038/nsmb0407-256. [DOI] [PubMed] [Google Scholar]
- 105.Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamante C, Cozzarelli NR. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc Natl Acad Sci USA. 2003;100:8654–8659. doi: 10.1073/pnas.1133178100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Charvin G, Strick TR, Bensimon D, Croquette V. Topoisomerase IV bends and overtwists DNA upon binding. Biophys J. 2005;89:384–392. doi: 10.1529/biophysj.105.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Neuman KC, Charvin G, Bensimon D, Croquette V. Mechanisms of chiral discrimination by topoisomerase IV. Proc Natl Acad Sci USA. 2009;106:6986–6991. doi: 10.1073/pnas.0900574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Charvin G, Bensimon D, Croquette V. Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc Natl Acad Sci USA. 2003;100:9820–9825. doi: 10.1073/pnas.1631550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gubaev A, Hilbert M, Klostermeier D. The DNA-gate of Bacillus subtilis gyrase is predominantly in the closed conformation during the DNA supercoiling reaction. Proc Natl Acad Sci USA. 2009;106:13278–13283. doi: 10.1073/pnas.0902493106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Malik M, Nitiss KC, Enriquez-Rios V, Nitiss JL. Roles of nonhomologous end-joining pathways in surviving topoisomerase II-mediated DNA damage. Mol Cancer Ther. 2006;5:1405–1414. doi: 10.1158/1535-7163.MCT-05-0263. [DOI] [PubMed] [Google Scholar]
- 111.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94:819–827. doi: 10.1016/S0092-8674(00)81740-7. [DOI] [PubMed] [Google Scholar]
- 112.Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli . J Biol Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- 113.Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Khodursky AB, Peter BJ, Schmid MB, DeRisi J, Botstein D, Brown PO, Cozzarelli NR. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc Natl Acad Sci USA. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tadesse S, Graumann PL. Differential and dynamic localization of topoisomerases in Bacillus subtilis . J Bacteriol. 2006;188:3002–3011. doi: 10.1128/JB.188.8.3002-3011.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hsu YH, Chung MW, Li TK. Distribution of gyrase and topoisomerase IV on bacterial nucleoid: implications for nucleoid organization. Nucleic Acids Res. 2006;34:3128–3138. doi: 10.1093/nar/gkl392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen CR, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- 118.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli . Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pohlhaus JR, Kreuzer KN. Norfloxacin-induced DNA gyrase cleavage complexes block Escherichia coli replication forks, causing double-stranded breaks in vivo. Mol Microbiol. 2005;56:1416–1429. doi: 10.1111/j.1365-2958.2005.04638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Malik M, Zhao X, Drlica K. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol Microbiol. 2006;61:810–825. doi: 10.1111/j.1365-2958.2006.05275.x. [DOI] [PubMed] [Google Scholar]
- 121.Abbanat D, Morrow B, Bush K. New agents in development for the treatment of bacterial infections. Curr Opin Pharmacol. 2008;8:582–592. doi: 10.1016/j.coph.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 122.Bradbury BJ, Pucci MJ. Recent advances in bacterial topoisomerase inhibitors. Curr Opin Pharmacol. 2008;8:574–581. doi: 10.1016/j.coph.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 123.Shea ME, Hiasa H. Distinct effects of the UvrD helicase on topoisomerase-quinolone-DNA ternary complexes. J Biol Chem. 2000;275:14649–14658. doi: 10.1074/jbc.275.19.14649. [DOI] [PubMed] [Google Scholar]
- 124.Wentzell LM, Maxwell A. The complex of DNA gyrase and quinolone drugs on DNA forms a barrier to the T7 DNA polymerase replication complex. J Mol Biol. 2000;304:779–791. doi: 10.1006/jmbi.2000.4266. [DOI] [PubMed] [Google Scholar]
- 125.Willmott CJ, Critchlow SE, Eperon IC, Maxwell A. The complex of DNA gyrase and quinolone drugs with DNA forms a barrier to transcription by RNA polymerase. J Mol Biol. 1994;242:351–363. doi: 10.1006/jmbi.1994.1586. [DOI] [PubMed] [Google Scholar]
- 126.Piddock LJ. Mechanisms of fluoroquinolone resistance: an update 1994–1998. Drugs 58 Suppl. 1999;2:11–8. doi: 10.2165/00003495-199958002-00003. [DOI] [PubMed] [Google Scholar]
- 127.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli . Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Critchlow SE, Maxwell A. DNA cleavage is not required for the binding of quinolone drugs to the DNA gyrase-DNA complex. Biochemistry. 1996;35:7387–7393. doi: 10.1021/bi9603175. [DOI] [PubMed] [Google Scholar]
- 129.Heddle JG, Barnard FM, Wentzell LM, Maxwell A. The interaction of drugs with DNA gyrase: a model for the molecular basis of quinolone action. Nucleosides Nucleotides Nucleic Acids. 2000;19:1249–1264. doi: 10.1080/15257770008033048. [DOI] [PubMed] [Google Scholar]
- 130.Palu G, Valisena S, Ciarrocchi G, Gatto B, Palumbo M. Quinolone binding to DNA is mediated by magnesium ions. Proc Natl Acad Sci USA. 1992;89:9671–9675. doi: 10.1073/pnas.89.20.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dutta S, Kawamura Y, Ezaki T, Nair GB, Iida K, Yoshida S. Alteration in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in Quinolone-resistant Shigella dysenteriae serotype 1 clinical isolates from Kolkata, India. Antimicrob Agents Chemother. 2005;49:1660–1661. doi: 10.1128/AAC.49.4.1660-1661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hu LF, Li JB, Ye Y, Li X. Mutations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in clinical strains of fluoroquinolone-resistant Shigella in Anhui, China. J Microbiol. 2007;45:168–170. [PubMed] [Google Scholar]
- 133.Madurga S, Sanchez-Cespedes J, Belda I, Vila J, Giralt E. Mechanism of binding of fluoroquinolones to the quinolone resistance-determining region of DNA gyrase: towards an understanding of the molecular basis of quinolone resistance. Chembiochem. 2008;9:2081–2086. doi: 10.1002/cbic.200800041. [DOI] [PubMed] [Google Scholar]
- 134.Rafii F, Park M, Novak JS. Alterations in DNA gyrase and topoisomerase IV in resistant mutants of Clostridium perfringens found after in vitro treatment with fluoroquinolones. Antimicrob Agents Chemother. 2005;49:488–492. doi: 10.1128/AAC.49.2.488-492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee JK, Lee YS, Park YK, Kim BS. Alterations in the GyrA and GyrB subunits of topoisomerase II and the ParC and ParE subunits of topoisomerase IV in ciprofloxacin-resistant clinical isolates of Pseudomonas aeruginosa . Int J Antimicrob Agents. 2005;25:290–295. doi: 10.1016/j.ijantimicag.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 136.Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance in Gram-negative bacterial species: an update. Curr Med Chem. 2009;16:1028–1046. doi: 10.2174/092986709787581879. [DOI] [PubMed] [Google Scholar]
- 137.Poirel L, Cattoir V, Nordmann P. Is plasmid-mediated quinolone resistance a clinically significant problem? Clin Microbiol Infect. 2008;14:295–297. doi: 10.1111/j.1469-0691.2007.01930.x. [DOI] [PubMed] [Google Scholar]
- 138.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 139.Peterson LR. Quinolone molecular structure–activity relationships: what we have learned about improving antimicrobial activity. Clin Infect Dis. 2001;33:S180–186. doi: 10.1086/321846. [DOI] [PubMed] [Google Scholar]
- 140.Mitscher LA. Bacterial topoisomerase inhibitors: quinolone and pyridone antibacterial agents. Chem Rev. 2005;105:559–592. doi: 10.1021/cr030101q. [DOI] [PubMed] [Google Scholar]
- 141.Dinakaran M, Senthilkumar P, Yogeeswari P, China A, Nagaraja V, Sriram D. Novel ofloxacin derivatives: synthesis, antimycobacterial and toxicological evaluation. Bioorg Med Chem Lett. 2008;18:1229–1236. doi: 10.1016/j.bmcl.2007.11.110. [DOI] [PubMed] [Google Scholar]
- 142.Ellsworth EL, Tran TP, Showalter HD, Sanchez JP, Watson BM, Stier MA, Domagala JM, Gracheck SJ, Joannides ET, Shapiro MA, Dunham SA, Hanna DL, Huband MD, Gage JW, Bronstein JC, Liu JY, Nguyen DQ, Singh R. 3-aminoquinazolinediones as a new class of antibacterial agents demonstrating excellent antibacterial activity against wild-type and multidrug resistant organisms. J Med Chem. 2006;49:6435–6438. doi: 10.1021/jm060505l. [DOI] [PubMed] [Google Scholar]
- 143.Gomez L, Hack MD, Wu J, Wiener JJ, Venkatesan H, Santillan A, Jr, Pippel DJ, Mani N, Morrow BJ, Motley ST, Shaw KJ, Wolin R, Grice CA, Jones TK. Novel pyrazole derivatives as potent inhibitors of type II topoisomerases. Part 1: synthesis and preliminary SAR analysis. Bioorg Med Chem Lett. 2007;17:2723–2727. doi: 10.1016/j.bmcl.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 144.Pan XS, Gould KA, Fisher LM. Probing the differential interaction of quinazolinedione PD 0305970 and quinolones with gyrase and topoisomerase IV. Antimicrob Agents Chemother. 2009;53:3822–3831. doi: 10.1128/AAC.00113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ramesh E, Manian RD, Raghunathan R, Sainath S, Raghunathan M. Synthesis and antibacterial property of quinolines with potent DNA gyrase activity. Bioorg Med Chem. 2009;17:660–666. doi: 10.1016/j.bmc.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 146.Sriram D, Aubry A, Yogeeswari P, Fisher LM. Gatifloxacin derivatives: synthesis, antimycobacterial activities, and inhibition of Mycobacterium tuberculosis DNA gyrase. Bioorg Med Chem Lett. 2006;16:2982–2985. doi: 10.1016/j.bmcl.2006.02.065. [DOI] [PubMed] [Google Scholar]
- 147.Tran TP, Ellsworth EL, Sanchez JP, Watson BM, Stier MA, Showalter HD, Domagala JM, Shapiro MA, Joannides ET, Gracheck SJ, Nguyen DQ, Bird P, Yip J, Sharadendu A, Ha C, Ramezani S, Wu X, Singh R. Structure–activity relationships of 3-aminoquinazolinediones, a new class of bacterial type-2 topoisomerase (DNA gyrase and topo IV) inhibitors. Bioorg Med Chem Lett. 2007;17:1312–1320. doi: 10.1016/j.bmcl.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 148.Wiener JJ, Gomez L, Venkatesan H, Santillan A, Jr, Allison BD, Schwarz KL, Shinde S, Tang L, Hack MD, Morrow BJ, Motley ST, Goldschmidt RM, Shaw KJ, Jones TK, Grice CA. Tetrahydroindazole inhibitors of bacterial type II topoisomerases. Part 2: SAR development and potency against multidrug-resistant strains. Bioorg Med Chem Lett. 2007;17:2718–2722. doi: 10.1016/j.bmcl.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 149.Fisher LM, Heaton VJ. Dual activity of fluoroquinolones against Streptococcus pneumoniae. J Antimicrob Chemother. 2003;51:463–464. doi: 10.1093/jac/dkg059. [DOI] [PubMed] [Google Scholar]
- 150.Strahilevitz J, Hooper DC. Dual targeting of topoisomerase IV and gyrase to reduce mutant selection: direct testing of the paradigm by using WCK-1734, a new fluoroquinolone, and ciprofloxacin. Antimicrob Agents Chemother. 2005;49:1949–1956. doi: 10.1128/AAC.49.5.1949-1956.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cambau E, Matrat S, Pan XS, Roth Dit Bettoni R, Corbel C, Aubry A, Lascols C, Driot JY, Fisher LM. Target specificity of the new fluoroquinolone besifloxacin in Streptococcus pneumoniae, Staphylococcus aureus and Escherichia coli . J Antimicrob Chemother. 2009;63:443–450. doi: 10.1093/jac/dkn528. [DOI] [PubMed] [Google Scholar]
- 152.Okumura R, Hirata T, Onodera Y, Hoshino K, Otani T, Yamamoto T. Dual-targeting properties of the 3-aminopyrrolidyl quinolones, DC-159a and sitafloxacin, against DNA gyrase and topoisomerase IV: contribution to reducing in vitro emergence of quinolone-resistant Streptococcus pneumoniae . J Antimicrob Chemother. 2008;62:98–104. doi: 10.1093/jac/dkn136. [DOI] [PubMed] [Google Scholar]
- 153.Charifson PS, Grillot AL, Grossman TH, Parsons JD, Badia M, Bellon S, Deininger DD, Drumm JE, Gross CH, LeTiran A, Liao Y, Mani N, Nicolau DP, Perola E, Ronkin S, Shannon D, Swenson LL, Tang Q, Tessier PR, Tian SK, Trudeau M, Wang T, Wei Y, Zhang H, Stamos D. Novel dual-targeting benzimidazole urea inhibitors of DNA gyrase and topoisomerase IV possessing potent antibacterial activity: intelligent design and evolution through the judicious use of structure-guided design and structure–activity relationships. J Med Chem. 2008;51:5243–5263. doi: 10.1021/jm800318d. [DOI] [PubMed] [Google Scholar]
- 154.Cheng J, Thanassi JA, Thoma CL, Bradbury BJ, Deshpande M, Pucci MJ. Dual targeting of DNA gyrase and topoisomerase IV: target interactions of heteroaryl isothiazolones in Staphylococcus aureus . Antimicrob Agents Chemother. 2007;51:2445–2453. doi: 10.1128/AAC.00158-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.East SP, White CB, Barker O, Barker S, Bennett J, Brown D, Boyd EA, Brennan C, Chowdhury C, Collins I, Convers-Reignier E, Dymock BW, Fletcher R, Haydon DJ, Gardiner M, Hatcher S, Ingram P, Lancett P, Mortenson P, Papadopoulos K, Smee C, Thomaides-Brears HB, Tye H, Workman J, Czaplewski LG. DNA gyrase (GyrB)/topoisomerase IV (ParE) inhibitors: synthesis and antibacterial activity. Bioorg Med Chem Lett. 2009;19:894–899. doi: 10.1016/j.bmcl.2008.11.102. [DOI] [PubMed] [Google Scholar]
- 156.Robertson GT, Bonventre EJ, Doyle TB, Du Q, Duncan L, Morris TW, Roche ED, Yan D, Lynch AS. In vitro evaluation of CBR-2092, a novel rifamycin-quinolone hybrid antibiotic: studies of the mode of action in Staphylococcus aureus . Antimicrob Agents Chemother. 2008;52:2313–2323. doi: 10.1128/AAC.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhi C, Long ZY, Manikowski A, Comstock J, Xu WC, Brown NC, Tarantino PM, Jr, Holm KA, Dix EJ, Wright GE, Barnes MH, Butler MM, Foster KA, LaMarr WA, Bachand B, Bethell R, Cadilhac C, Charron S, Lamothe S, Motorina I, Storer R. Hybrid antibacterials. DNA polymerase-topoisomerase inhibitors. J Med Chem. 2006;49:1455–1465. doi: 10.1021/jm0510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Black MT, Stachyra T, Platel D, Girard AM, Claudon M, Bruneau JM, Miossec C. Mechanism of action of the antibiotic NXL101, a novel nonfluoroquinolone inhibitor of bacterial type II topoisomerases. Antimicrob Agents Chemother. 2008;52:3339–3349. doi: 10.1128/AAC.00496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lewis RJ, Singh OM, Smith CV, Skarzynski T, Maxwell A, Wonacott AJ, Wigley DB. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 1996;15:1412–1420. [PMC free article] [PubMed] [Google Scholar]
- 160.Lamour V, Hoermann L, Jeltsch JM, Oudet P, Moras D. An open conformation of the Thermus thermophilus gyrase B ATP-binding domain. J Biol Chem. 2002;277:18947–18953. doi: 10.1074/jbc.M111740200. [DOI] [PubMed] [Google Scholar]
- 161.Schechner M, Sirockin F, Stote RH, Dejaegere AP. Functionality maps of the ATP binding site of DNA gyrase B: generation of a consensus model of ligand binding. J Med Chem. 2004;47:4373–4390. doi: 10.1021/jm0311184. [DOI] [PubMed] [Google Scholar]
- 162.Ostrov DA, Hernandez Prada JA, Corsino PE, Finton KA, Le N, Rowe TC. Discovery of novel DNA gyrase inhibitors by high-throughput virtual screening. Antimicrob Agents Chemother. 2007;51:3688–3698. doi: 10.1128/AAC.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gradisar H, Pristovsek P, Plaper A, Jerala R. Green tea catechins inhibit bacterial DNA gyrase by interaction with its ATP binding site. J Med Chem. 2007;50:264–271. doi: 10.1021/jm060817o. [DOI] [PubMed] [Google Scholar]
- 164.Grossman TH, Bartels DJ, Mullin S, Gross CH, Parsons JD, Liao Y, Grillot AL, Stamos D, Olson ER, Charifson PS, Mani N. Dual targeting of GyrB and ParE by a novel aminobenzimidazole class of antibacterial compounds. Antimicrob Agents Chemother. 2007;51:657–666. doi: 10.1128/AAC.00596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hassan GS, Farag NA, Hegazy GH, Arafa RK. Design and synthesis of novel benzopyran-2-one derivatives of expected antimicrobial activity through DNA gyrase-B inhibition. Arch Pharm (Weinheim) 2008;341:725–733. doi: 10.1002/ardp.200700266. [DOI] [PubMed] [Google Scholar]
- 166.Mani N, Gross CH, Parsons JD, Hanzelka B, Muh U, Mullin S, Liao Y, Grillot AL, Stamos D, Charifson PS, Grossman TH. In vitro characterization of the antibacterial spectrum of novel bacterial type II topoisomerase inhibitors of the aminobenzimidazole class. Antimicrob Agents Chemother. 2006;50:1228–1237. doi: 10.1128/AAC.50.4.1228-1237.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Miller AA, Bundy GL, Mott JE, Skepner JE, Boyle TP, Harris DW, Hromockyj AE, Marotti KR, Zurenko GE, Munzner JB, Sweeney MT, Bammert GF, Hamel JC, Ford CW, Zhong WZ, Graber DR, Martin GE, Han F, Dolak LA, Seest EP, Ruble JC, Kamilar GM, Palmer JR, Banitt LS, Hurd AR, Barbachyn MR. Discovery and characterization of QPT-1, the progenitor of a new class of bacterial topoisomerase inhibitors. Antimicrob Agents Chemother. 2008;52:2806–2812. doi: 10.1128/AAC.00247-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Oblak M, Grdadolnik SG, Kotnik M, Poterszman A, Atkinson RA, Nierengarten H, Desplancq D, Moras D, Solmajer T. Biophysical characterization of an indolinone inhibitor in the ATP-binding site of DNA gyrase. Biochem Biophys Res Commun. 2006;349:1206–1213. doi: 10.1016/j.bbrc.2006.08.172. [DOI] [PubMed] [Google Scholar]
- 169.Oblak M, Kotnik M, Solmajer T. Discovery and development of ATPase inhibitors of DNA gyrase as antibacterial agents. Curr Med Chem. 2007;14:2033–2047. doi: 10.2174/092986707781368414. [DOI] [PubMed] [Google Scholar]
- 170.Tanitame A, Oyamada Y, Ofuji K, Fujimoto M, Suzuki K, Ueda T, Terauchi H, Kawasaki M, Nagai K, Wachi M, Yamagishi J. Synthesis and antibacterial activity of novel and potent DNA gyrase inhibitors with azole ring. Bioorg Med Chem. 2004;12:5515–5524. doi: 10.1016/j.bmc.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 171.Hashimi SM, Wall MK, Smith AB, Maxwell A, Birch RG. The phytotoxin albicidin is a novel inhibitor of DNA gyrase. Antimicrob Agents Chemother. 2007;51:181–187. doi: 10.1128/AAC.00918-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Parks WM, Bottrill AR, Pierrat OA, Durrant MC, Maxwell A. The action of the bacterial toxin, microcin B17, on DNA gyrase. Biochimie. 2007;89:500–507. doi: 10.1016/j.biochi.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 173.Pierrat OA, Maxwell A. Evidence for the role of DNA strand passage in the mechanism of action of microcin B17 on DNA gyrase. Biochemistry. 2005;44:4204–4215. doi: 10.1021/bi0478751. [DOI] [PubMed] [Google Scholar]
- 174.Sengupta S, Nagaraja V. YacG from Escherichia coli is a specific endogenous inhibitor of DNA gyrase. Nucleic Acids Res. 2008;36:4310–4316. doi: 10.1093/nar/gkn355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Simic M, De Jonge N, Loris R, Vesnaver G, Lah J. Driving forces of gyrase recognition by the addiction toxin CcdB. J Biol Chem. 2009;284:20002–20010. doi: 10.1074/jbc.M109.014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Smith AB, Maxwell A. A strand-passage conformation of DNA gyrase is required to allow the bacterial toxin, CcdB, to access its binding site. Nucleic Acids Res. 2006;34:4667–4676. doi: 10.1093/nar/gkl636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.De Jonge N, Buts L, Vangelooven J, Mine N, Van Melderen L, Wyns L, Loris R. Purification and crystallization of Vibrio fischeri CcdB and its complexes with fragments of gyrase and CcdA. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:356–360. doi: 10.1107/S1744309107012092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Trovatti E, Cotrim CA, Garrido SS, Barros RS, Marchetto R. Peptides based on CcdB protein as novel inhibitors of bacterial topoisomerases. Bioorg Med Chem Lett. 2008;18:6161–6164. doi: 10.1016/j.bmcl.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 180.Oppegard LM, Hamann BL, Streck KR, Ellis KC, Fiedler HP, Khodursky AB, Hiasa H. In vivo and in vitro patterns of the activity of simocyclinone D8, an angucyclinone antibiotic from Streptomyces antibioticus . Antimicrob Agents Chemother. 2009;53:2110–2119. doi: 10.1128/AAC.01440-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sadiq AA, Patel MR, Jacobson BA, Escobedo M, Ellis K, Oppegard LM, Hiasa H, Kratzke RA. Anti-proliferative effects of simocyclinone D8 (SD8), a novel catalytic inhibitor of topoisomerase II. Invest New Drugs. 2010;28:20–25. doi: 10.1007/s10637-008-9209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Flatman RH, Howells AJ, Heide L, Fiedler HP, Maxwell A. Simocyclinone D8, an inhibitor of DNA gyrase with a novel mode of action. Antimicrob Agents Chemother. 2005;49:1093–1100. doi: 10.1128/AAC.49.3.1093-1100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sissi C, Vazquez E, Chemello A, Mitchenall LA, Maxwell A, Palumbo M. Mapping simocyclinone D8 interaction with DNA gyrase: evidence for a new binding site on GyrB. Antimicrob Agents Chemother. 2010;54:213–220. doi: 10.1128/AAC.00972-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Edwards MJ, Flatman RH, Mitchenall LA, Stevenson CE, Le TB, Clarke TA, McKay AR, Fiedler HP, Buttner MJ, Lawson DM, Maxwell A. A crystal structure of the bifunctional antibiotic simocyclinone D8, bound to DNA gyrase. Science. 2009;326:1415–1418. doi: 10.1126/science.1179123. [DOI] [PubMed] [Google Scholar]
- 185.Ahmed A, Sharma YD. Ribozyme cleavage of Plasmodium falciparum gyrase A gene transcript affects the parasite growth. Parasitol Res. 2008;103:751–763. doi: 10.1007/s00436-008-1036-y. [DOI] [PubMed] [Google Scholar]
- 186.Rao SS, Savithri HS, Raghunathan M. Down regulation of gyrase A gene expression in E. coli by antisense ribozymes using RT-PCR. Mol Biol Rep. 2008;35:575–578. doi: 10.1007/s11033-007-9126-y. [DOI] [PubMed] [Google Scholar]
- 187.Dorman CJ, Corcoran CP. Bacterial DNA topology and infectious disease. Nucleic Acids Res. 2009;37:672–678. doi: 10.1093/nar/gkn996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Smart DJ. Genotoxicity of topoisomerase II inhibitors: an anti-infective perspective. Toxicology. 2008;254:192–198. doi: 10.1016/j.tox.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 189.Walsh CT. Combinatorial biosynthesis of antibiotics: challenges and opportunities. Chembiochem. 2002;3:125–134. doi: 10.1002/1439-7633(20020301)3:2/3<124::AID-CBIC124>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 190.Heide L. Genetic engineering of antibiotic biosynthesis for the generation of new aminocoumarins. Biotechnol Adv. 2009;27:1006–1014. doi: 10.1016/j.biotechadv.2009.05.017. [DOI] [PubMed] [Google Scholar]