Significance
SR proteins are essential for pre-mRNA splicing, but the molecular details underlying their mechanism of action within the spliceosome remain unclear. We used proximity labeling to identify many proteins that are proximal to SRSF1 in HeLa cells. Among them, we uncovered and characterized a direct interaction between SRSF1 and the spliceosomal RNA helicase DDX23/PRP28. This interaction has potential functional implications for a discrete stage of spliceosome assembly. Detailed knowledge about molecular interactions between key components of the splicing machinery may yield unique approaches to target the spliceosome in pathologies that involve dysregulation of pre-mRNA splicing.
Keywords: SRSF1, DDX23, pre-mRNA splicing, proximity labeling, SR proteins
Abstract
SRSF1 is the founding member of the SR protein family. It is required—interchangeably with other SR proteins—for pre-mRNA splicing in vitro, and it regulates various alternative splicing events. Dysregulation of SRSF1 expression contributes to cancer and other pathologies. Here, we characterized SRSF1’s interactome using proximity labeling and mass spectrometry. This approach yielded 190 proteins enriched in the SRSF1 samples, independently of the N- or C-terminal location of the biotin-labeling domain. The detected proteins reflect established functions of SRSF1 in pre-mRNA splicing and reveal additional connections to spliceosome proteins, in addition to other recently identified functions. We validated a robust interaction with the spliceosomal RNA helicase DDX23/PRP28 using bimolecular fluorescence complementation and in vitro binding assays. The interaction is mediated by the N-terminal RS-like domain of DDX23 and both RRM1 and the RS domain of SRSF1. During pre-mRNA splicing, DDX23’s ATPase activity is essential for the pre-B to B spliceosome complex transition and for release of U1 snRNP from the 5′ splice site. We show that the RS-like region of DDX23’s N-terminal domain is important for spliceosome incorporation, while larger deletions in this domain alter subnuclear localization. We discuss how the identified interaction of DDX23 with SRSF1 and other SR proteins may be involved in the regulation of these processes.
Human SRSF1 was the first member of the SR protein family to be discovered in the early 1990s, when its activities in the complementation of pre-messenger RNA (pre-mRNA) splicing in vitro and in the regulation of 5′-splice-site selection were described (1–3). SRSF1 is a 248-residue protein with two RNA-Recognition Motifs (RRMs) at its amino-terminal end, followed by an RS domain with multiple consecutive RS or SR dipeptides—a defining feature of the SR protein family (4). The RRMs are responsible for specific RNA binding, but they can also engage in key interactions with SRSF1 functional partners and with the RS domain (5–7). The SRSF1 RS domain consists of 51 residues, with 37% arginine (R) and 39% serine (S) content, organized as blocks of consecutive dipeptide repeats. Many of the serine residues in the RS domain are phosphorylated by kinases that regulate SR protein localization and recruitment to nascent transcripts during splicing (8). At steady state, the SRSF1 RS domain is highly phosphorylated (9). In general, RS domains are responsible for protein–protein interactions, though unphosphorylated RS domains have also been proposed to interact with RNA (10, 11).
During early spliceosome assembly, SRSF1 engages in protein–protein interactions with a U1 small nucler Ribonucleoprotein (snRNP) polypeptide (U1-70K) at the 5′ splice site (5′ss) and with the U2AF1/U2AF2 heterodimer at the 3′ splice site (12), promoting the commitment of the pre-mRNA to splicing (13). SRSF1 also specifically binds U1 snRNA stem-loop 3 and can be recruited to the pre-mRNA via U1 snRNA, to participate in exon definition (14). Recent studies found additional connections of SRSF1 with later-stage spliceosome and associated components, including the SF3 complex (15) and the exon-junction complex (16–18). SRSF1’s RRM2 was recently crosslinked and modeled into a cryo-EM structure of the pre-Bact1 spliceosome complex, proximal to the 5′ end of the upstream exon, near the cap-binding complex and additional B-complex-specific proteins, including TCERG1, MFAP1, and UBL5 (19).
In this work, we used a proximity-labeling method (20) to characterize SRSF1’s interactome, and we then validated a strong direct interaction between SRSF1 and DDX23 (also known as PRP28 or U5-100kD). DDX23 is an 820-residue protein that belongs to the DEAD-box family of ATP-dependent RNA helicases and is involved in the displacement of U1 snRNP from the 5′ss during the pre-B to B spliceosome-complex transition (21, 22). Mapping of the regions required for the interaction revealed that both RRM1 and the RS domain of SRSF1 contact the N-terminal (Nt) RS-like domain of DDX23. Deletions within this DDX23 domain disrupted the interaction with SRSF1. Phosphorylation of the RS repeats in a minimal DDX23 construct also inhibited the interaction. Furthermore, deletion of the first 327 amino acids of DDX23 resulted in altered subnuclear localization, and a 37-residue deletion within the RS-like domain showed decreased spliceosome recruitment during in vitro splicing. Therefore, the Nt domain of DDX23 is involved in its subnuclear targeting, including incorporation into the spliceosome, and its interaction with SRSF1 likely facilitates or regulates these processes.
Results
SRSF1 Interactome Analysis by Proximity Labeling.
We designed two SRSF1 cDNA constructs fused to BioID2—a small, promiscuous biotin ligase developed for proximity labeling (23); in these constructs, the biotin-ligase domain is located at the N-terminus (BS) or at the C-terminus (SB), respectively (Fig. 1A). We designed an additional construct to express BioID2-EGFP-3xNLS (BG) as a nuclear nonspecifically biotinylating control (Fig. 1A). Retroviral transduction of HeLa cells followed by puromycin selection yielded three cell lines that express the proteins of interest. The expression level attained for the SRSF1 fusion constructs was markedly lower than the endogenous SRSF1 level, so as to limit unspecific biotinylation due to overexpression (Fig. 1B). Immunofluorescence staining demonstrated that the addition of the BioID2 domain at the N- or C-terminus of SRSF1 did not alter the predominantly nuclear localization pattern of SRSF1 (SI Appendix, Fig S1A). Moreover, fusion of the BioID2 domain to SRSF1 did not affect its dose-dependent ability to regulate the expression of endogenous SRSF1 (24) or alternative splicing of the endogenous target PEA15 (25), suggesting that the fusions do not hinder SRSF1’s splicing-regulatory activity (SI Appendix, Fig S1 B and C).
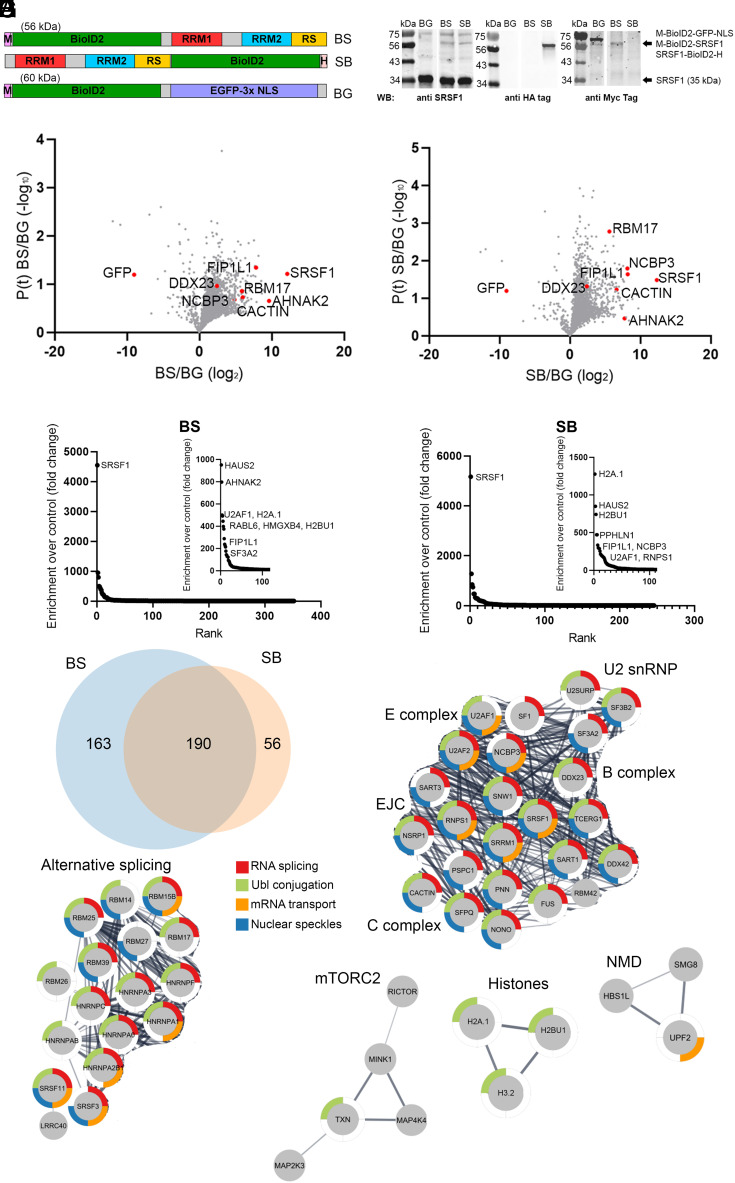
Fig. 1.
Proximity labeling highlights connections of SRSF1 to the spliceosome in addition to recently identified functions. (A) Constructs used for proximity labeling. M: Myc tag, H: HA tag. (B) Western blot analysis of lysates from HeLa cells transduced with the indicated retroviruses, after puromycin selection. Arrows point to the specific immunoreactive bands. (C) Volcano plot showing protein enrichment in the BS sample, compared to the BioID2-GFP control. Selected proteins are highlighted. (D) Volcano plot showing protein enrichment in the SB sample, compared to the BioID2-GFP control. Selected proteins are highlighted. (E) Representation of 353 proteins enriched in the BioID2-SRSF1 dataset. (F) Representation of 246 proteins enriched in the SRSF1-BioID2 dataset. (G) Venn diagram of proteins detected in the BS and SB datasets. (H) Protein-network visualization generated with Cytoscape and STRING database, derived from the 190 proteins in common between BS and SB datasets. Colors represent the indicated GO terms (RNA splicing, Ubl conjugation, mRNA transport, nuclear speckles). Proteins were clustered according to their functions. The thickness of the connecting lines represents the degree of confidence for protein–protein functional interaction, obtained from the STRING database.
We used these HeLa cell line derivatives (BG, BS, SB) to study SRSF1’s interactome by proximity labeling. After a 24 h biotin pulse in live cells and streptavidin purification of the biotinylated proteins, we analyzed triplicate samples for each condition by Liquid Chromatography with tandem Mass Spectrometry (LC/MS-MS) (Dataset S1). We compared the samples expressing the SRSF1 fusions to the nuclear EGFP control. We observed a general enrichment of proteins in the samples expressing SRSF1-BioID2 fusions (Fig. 1 C and D). SRSF1 was the most enriched protein in both samples, with an enrichment value of more than 4,000-fold (Fig. 1 E and F). A total of 353 proteins were enriched ≥5 times in the BS dataset with respect to the BG control, whereas 246 proteins were similarly enriched in the SB dataset. Of these proteins, 190 were present in both datasets, highlighting the strong overlap between the two lists, and suggesting that the retrieved candidates represent functionally relevant proteins proximal to SRSF1 (Fig. 1G, Dataset S1, and SI Appendix, Table S1).
Gene Ontology (GO)-term analysis of the overlap list revealed an enrichment for proteins related to RNA splicing (GO:0008380, FDR: 1.97E-36), which confirms that SRSF1 is still able to engage in pre-mRNA splicing in the presence of the BioID2 fusions. In addition, enrichment of proteins related to nuclear speckles (GO:0016607, FDR: 2.32E-20) suggests that the retrieved proteins colocalize with SRSF1 in its predominant subcellular compartment. The significant enrichment of proteins susceptible to ubiquitin-like conjugation (KW:0832, FDR: 7.97E-24) may reflect the reported role of SRSF1 in the SUMO conjugation pathway (26, 27), whereas the enrichment of proteins related to mRNA transport (GO:0051028, FDR:2.01E-6) is consistent with the ability of SRSF1 to promote mRNA export, and its known association with postsplicing events like translation and nonsense-mediated mRNA decay (NMD) (17, 18, 28–32) (Fig. 1H).
Among the proteins with the highest enrichment score, we found well-characterized SRSF1 interactors like U2AF1 (BS: 500× enrichment; SB: 68×) and core spliceosome proteins like SF3A2 (BS: 125×; SB: 156×), SF3B2 (BS: 13×; SB: 59×), TCERG1 (BS: 28×; SB: 39×), and CACTIN (BS: 127×; SB: 175×), proteins associated with the EJC like RNPS1 (BS:19×; SB: 66×) and PNN (BS: 32×; SB: 28×), and other splicing-associated proteins like NONO (BS: 32×; SB: 24×). In addition to splicing-related proteins, other functional aspects of SRSF1 were reflected. For instance, several histone proteins including H2A.1 (BS: 490×; SB: 1276×), Histone H2B type 3-B (H2BU1) (BS: 444×; SB: 741×), H3.2 (BS: 18×; SB: 54×), and H3.3 (BS: 24×; SB: 3×) were specifically enriched. These interactions are likely relevant, as SRSF1 was reported to be associated with chromatin, specifically with the histone H3 tail during interphase (33). Strong enrichment of proteins related to other RNA-processing events, like the alternative cap-binding protein NCBP3 (BS: 29×; SB: 287×) and the mRNA 3′-end processing protein FIP1L1 (BS: 226×; SB: 297×), suggests that coordination between splicing and these processing events could be mediated by SRSF1.
The proximity-labeling approach led to strong enrichment of several proteins that were not previously known to physically or functionally interact with SRSF1, including AHNAK2 (BS: 796×; SB: 217×), HAUS2 (BS: 950×; SB: 849×), RABL6 (BS: 374×; SB: 40×), HMGXB4 (BS: 397×; SB: 41×), and PPHLN1 (BS: 26×; SB: 471×). AHNAK2 is a 616-kDa PDZ-domain scaffold protein that interacts with many proteins (34) and was computationally inferred, using the Phylogenetic Annotation and Inference Tool (PAINT), to have a role in splicing regulation (35). HAUS2 is involved in mitotic-spindle assembly, maintenance of centrosome integrity, and cytokinesis (36). SRSF1 was reported to promote translation of mitosis-associated genes (28, 37), so a potential functional connection with HAUS2 during the events leading to mitosis warrants further exploration. Finally, connections between SRSF1 and the GTPase RABL6, the epigenetic repressor PPHLN1, and the transcription regulator HMGXB4 were not previously described, but their high enrichment scores suggest possible functional connections that merit further study.
The overlap list reflected all stages of spliceosome assembly, including E-complex proteins (U2AF1, U2AF2, and SF1), A-complex proteins (SF3A2, SF3B2, U2SURP), B/C-complex proteins (DDX23, TCERG1, SART1, CACTIN, SRRM1, SNW1, RBM42), and components of the EJC (RNPS1, PNN). This observation is consistent with spliceosome-composition studies that found that SRSF1 is associated with the spliceosome from the earliest stages of assembly and remains bound to the mature mRNA after splicing is completed (38). In addition, we detected several proteins that participate in alternative splicing, including other SR proteins (SRSF3 and SRSF11) and hnRNP-superfamily members (HNRNPC, HNRNPA3, HNRNPF, HNRNPA1, HNRNPA0, HNRNPAB, HNRNPA2B1). Proximity to these proteins likely reflects a shared functional environment, which is expected for RNA-splicing regulators (Fig. 1H). It is possible that some of these interactions take place when these proteins are bound to the same RNAs in close proximity.
Finally, two additional cytoplasmic functions of SRSF1 were reflected in the overlapping protein list, namely its proposed connection with mTOR kinase during translation (29) and the effect of SRSF1 on NMD (32). In particular, we detected enrichment of the mTOR-pathway components RICTOR (BS: 10×; SB: 11×) (specific to mTOR complex 2) and mTOR (BS: 17×; SB: 4×), as well as of the NMD-pathway components UPF2 (BS: 6×; SB: 6×) and SMG8 (BS: 11×; SB: 9×).
Analysis of the individual datasets separately highlights additional interactors, especially in the case of the BS dataset, which comprises a larger number of proteins (SI Appendix, Fig. S2). For example, TNPO3, which functions in the nuclear import of SR proteins, was specifically enriched in the BS dataset (BS: 9×; SB: 3×). In addition, the identification of SUMO2 (BS: 8×; SB: 0.5×) suggests a preference for this modification over other SUMO types. Finally, the detection of AHNAK (BS: 7×; SB: 2×) (related to AHNAK2) in the BS dataset suggests a common mode of interaction with proteins from the AHNAK family. The differences in enrichment between the two datasets may reflect the geometries of interaction with these proteins, which could be further studied using structural approaches.
Validation of Interaction Partners by Bimolecular Fluorescence Complementation (BiFC) in Living Cells.
To verify that selected proteins identified by proximity labeling can physically interact with SRSF1, we used an orthogonal approach. We mostly focused on spliceosomal proteins, in an attempt to identify unique SRSF1 interactors at various stages of spliceosome assembly and catalysis. We used BiFC to study the interaction between SRSF1 and 21 candidate protein partners identified above. We cloned SRSF1 cDNA in a fusion-protein vector, downstream of the first 173 Nt amino acids of mVenus. On the same vector, we cloned cDNAs for the selected candidates to express fusion proteins upstream of the last 82 C-terminal (Ct) amino acids of mVenus. For each construct, we also tested the fused candidate together with the isolated 173 Nt mVenus construct, as a control for background reassembly of the two mVenus protein halves (Fig. 2A and SI Appendix, Fig. S3).
Fig. 2.
BiFC validates interactions of SRSF1 with selected spliceosomal proteins. (A) Schematic representation of the BiFC approach. SRSF1 is shown fused to mVenus 173-Nt. mVenus-82-Ct is shown fused to a circle that represents the various interactor candidates tested. The interaction between the candidate and SRSF1 brings the two mVenus halves closer together, resulting in stronger green fluorescence. (B) BiFC results for DDX23 visualized by fluorescence microscopy. BF, bright field. Scale bar represents 130 μm in 10× images and 15 μm in 40× images. (C–F) BiFC results for the indicated proteins, plotted as Forward scatter (linear scale) vs. Green fluorescence (logarithmic scale). The vertical line indicates the threshold for cells that were considered mVenus positive. The bar graphs on the right of each panel represent % mVenus-positive cells and mean green fluorescence, respectively, for Interactor-mVCt coexpressed with Nt-mVenus vs. Interactor-mVCt coexpressed with SRSF1-Nt-mVenus. An unpaired t test was performed to compare the %mV+ cells and MGF values between the experimental conditions and the controls. ****P ≤ 0.0001. Error bars represent SD.
BiFC relies on the ability of the split mVenus to reassemble upon proximity of the two halves and emit fluorescence (39). For each protein pair, we then compared the percentage of mVenus-positive cells (%mV+) and the mean green fluorescence of the samples (MGF), between the construct that coexpresses SRSF1 with the potential interactor and the construct that coexpresses the interactor with the unfused Nt half of mVenus (Fig. 2 and SI Appendix, Figs. S3 and S4). This approach was successful in recapitulating known interactions of SRSF1, including with U2AF1 and TNPO3 (SI Appendix, Fig. S4 B and C). However, a limitation of this assay is the difficulty in detecting interactions between proteins that preferentially localize in different cellular compartments. Such was the case for the known SRSF1 interaction with SRPK1, a kinase that mainly phosphorylates SRSF1 in the cytoplasm (40). After phosphorylation, SRSF1 preferentially localizes in the nucleus, whereas SRPK1 tends to remain in the cytoplasm. Therefore, even though we did observe a positive trend, the assay was not able to properly recapitulate this known transient interaction (SI Appendix, Fig. S4D).
Using BiFC, we successfully validated the proximity of SRSF1 to proteins that were recently modeled near SRSF1 in a cryo-EM structure of the pre-Bact1 spliceosome, including CBP20 and MFAP1 (SI Appendix, Fig. S4). Proximity to additional spliceosomal proteins also present in the BioID2 dataset was confirmed for STEEP1, SNW1, DDX23, and SF3A2. Finally, we confirmed the proximity to SRSF1 of the EJC proteins MAGOH, Y14, RNPS1, and PNN, as well as of the splicing-related protein NONO (Fig. 2 B–F and SI Appendix, Fig. S4). In several cases, BiFC did not recapitulate interactions revealed by the BioID2 datasets (SART1, PRPF4B, and SFPQ). Subcellular localization, protein stability, or steric hindrance due to the fusions could in some cases interfere with the interactions. Therefore, the failure to detect proximity by BiFC should not be interpreted as proof of the absence of interaction, but only as an indication that a potential interaction cannot be observed by this method.
SRSF1 Interacts with the Nt of DDX23 via Its RRM1 and RS Domain.
Among the potential interactors that were positive both by proximity labeling and by BiFC, we further characterized in detail the interaction between SRSF1 and DDX23. DDX23 is an integral component of the U5 snRNP and the U4/U6.U5 tri-snRNP in humans, and its ATPase activity is crucial for progression from the pre-B complex to the B complex and for U1 snRNP release (41). DDX23 has a conserved DEAD-box helicase Ct domain that in the human protein is preceded by an Nt domain with a low-complexity RS-like region (Fig. 3 A and B). When human DDX23 was first identified, it was anticipated that its potential interaction with SR proteins via the RS-like region might stabilize its association with the tri-snRNP and the spliceosome (21). In addition, the same study proposed the idea that interactions mediated by DDX23’s RS-like domain might contribute to its ATPase/helicase activity. The presence of DDX23 in both our BioID (SI Appendix, Fig. S5) and BiFC datasets together with its RS-like region potentially serving as an interaction domain prompted us to further study the SRSF1-DDX23 interaction in vitro.
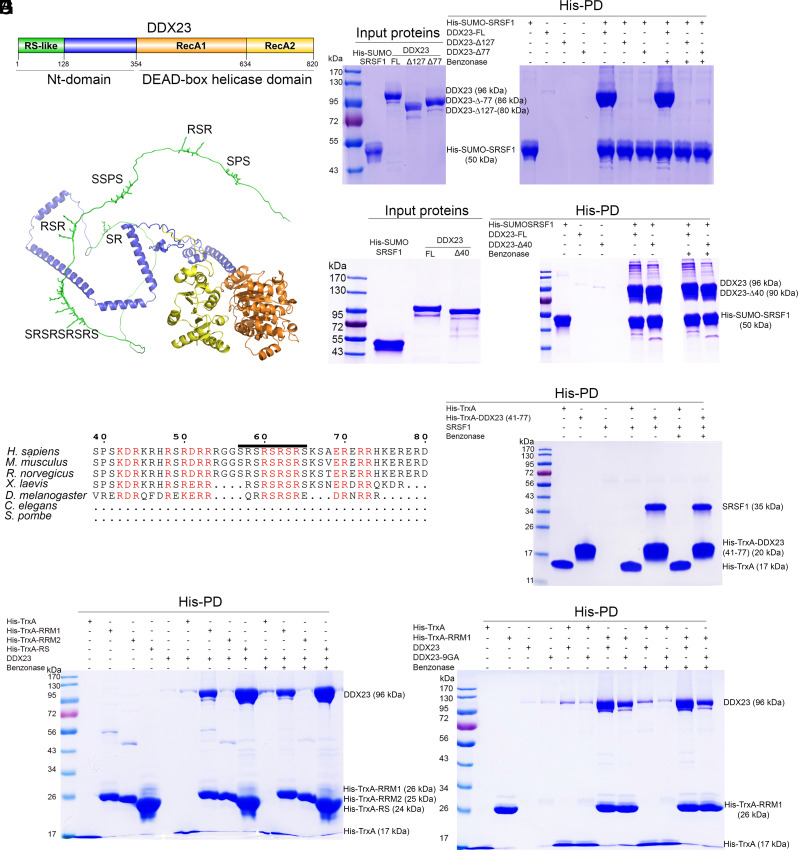
Fig. 3.
A direct interaction between SRSF1 and DDX23 can be recapitulated using recombinant proteins in vitro. (A) Schematic representation of domains of DDX23. (B) AlphaFold prediction of the structure of DDX23 (AF-Q9BUQ8-F1). RS/SP residues present in the RS-like domain are highlighted as sticks. RecA1 domain is shown in orange and RecA2 domain in yellow. (C) Representative His Pulldown assay using His-SUMO-SRSF1 and untagged DDX23, DDX23-Δ77, DDX23-Δ127, with or without benzonase treatment. (D) Representative His Pulldown assay using His-SUMO-SRSF1 and the indicated DDX23 constructs. (E) Sequence alignment of the SRSF1-interacting fragment of the RS-like domain of human DDX23 and its orthologs. This region is absent in the proteins from C. elegans and S. pombe. The location of the 9 SR residues is marked by a horizontal line. Charged residues conserved in the top five species are in red font. Numbers represent residue position in the human protein. (F) Representative His Pulldown assay using His-TrxA or His-TrxA-DDX23 (S41-R77) and untagged SRSF1. (G) Representative His Pulldown assay using the indicated truncated versions of SRSF1 or His-TrxA and FL-DDX23. (H) Representative His Pulldown assay using His-TrxA-RRM1, or His-TrxA and WT or 9GA-mutant DDX23.
We expressed recombinant SRSF1 and DDX23 in Escherichia coli Rosetta cells and achieved extensive phosphorylation of SRSF1’s RS domain by coexpression of CLK1 kinase (42). We purified a His-SUMO-tagged version of SRSF1 and performed a His-pulldown (PD) assay to test its interaction with untagged DDX23. Full-length (FL) DDX23 was strongly pulled down by His-SUMO SRSF1, and the interaction was benzonase-resistant. Partial or complete removal of the Nt RS-like domain of DDX23 (deletion mutants Δ127 and Δ77) completely abolished the interaction (Fig. 3C). In contrast, removal of the first 40 residues of DDX23 did not affect the interaction, suggesting that the region of DDX23 that contacts SRSF1 involves at least 37 residues, from S41 to R77 (Fig. 3D). To further test whether these 37 residues are sufficient for the interaction with SRSF1, we expressed TrxA (E. coli Thioredoxin 1) fused at its Ct to these residues and tested the interaction with untagged SRSF1. The PD analysis confirms that these DDX23 residues interact with SRSF1 (Fig. 3F). This region is conserved in higher eukaryotes (but is missing in Caenorhabditis elegans and Schizosaccharomyces pombe) and includes motifs with SK/KD/RK/ER/RR and RS consecutive repeats (Fig. 3E). The presence of these negatively and positively charged sequence features suggests potential electrostatic interactions with SRSF1.
To further delineate the interaction region between SRSF1 and DDX23, we generated a construct [DDX23-Δ(S41-R77)] that lacks the S41-R77 region that on its own binds SRSF1. Notably, this construct retained the ability to bind SRSF1, suggesting that redundant regions within the Nt-domain of DDX23 can still interact with SRSF1 in the absence of the S41-R77 region (SI Appendix, Fig. S6). Indeed, the first 40-residue peptide (M1-P40) also comprises many charged residues (35% K+R, 27.5% D+E), including 10 consecutive alternating-charge residues, and is conserved in higher eukaryotes (SI Appendix, Fig. S7). Our results are consistent with a flexible interaction between the unstructured Nt-region of DDX23 (M1-R77) and SRSF1, which may involve internally redundant segments of the former.
Deletion of the RS domain of SRSF1 did not abolish the interaction with FL-DDX23 (SI Appendix, Fig. S8A). We also tested isolated domains of SRSF1—fused to the Ct of His-TrxA—for interaction with DDX23, and both TrxA-RRM1 and the phosphorylated TrxA-RS domain—generated by coexpression with CLK1—successfully pulled down DDX23. In contrast, the isolated TrxA-RRM2 did not show detectable binding to DDX23 (Fig. 3G).
The presence of RS-like regions in spliceosomal proteins from higher eukaryotes is usually correlated with the evolutionary emergence of SR proteins (43). Therefore, it has been proposed that RS-like regions could bind to the RS domains of SR proteins and that these interactions could help regulate splicing in these organisms. The DDX23 RS-like domain has a conserved nine-residue stretch of RS repeats (S57 to S65), whereas the remaining RS repeats are shorter and dispersed (Fig. 3B). To test the relevance of the long RS-repeat stretch for the interaction, we generated a FL-DDX23 mutant with the RS repeats replaced by GA repeats (9GA mutant). We tested the interaction of SRSF1 with this mutant and found that the interaction was not affected (SI Appendix, Fig. S8B), presumably because of the above-mentioned redundancy with charged segments closer to the Nt of DDX23. Indeed, when we analyzed the interaction of the isolated TrxA-RRM1 with DDX23-9GA, the PD efficiency markedly decreased, suggesting that the long stretch of RS repeats contributes to the interaction with RRM1 (Fig. 3H). Additional PD assays confirmed that both TrxA-RRM1 and TrxA-RS domain of SRSF1 separately interact with FL-DDX23 and DDX23-Δ40 and that the S57-S65 RS region of DDX23 is important for interaction with TrxA-RRM1, but not with the TrxA-RS domain (SI Appendix, Fig. S8C).
Phosphorylation of the RS Repeats in DDX23 (S41-R77) Inhibits SRSF1 Binding.
DDX23 was previously shown to undergo phosphorylation of its RS repeats, with one of the kinases responsible for this phosphorylation being SRPK2 (44). Though the precise residues that undergo phosphorylation were not mapped, DDX23 phosphorylation was shown to be required for B-complex formation and for stable association of DDX23 with the tri-snRNP (44). Considering that the SRSF1-interaction region we mapped harbors several RS repeats, including the above-mentioned nine-residue stretch, we tested whether phosphorylation of these residues affects the interaction with SRSF1. We coexpressed His-TrxA-DDX23 (S41-R77) with CLK1 kinase in bacteria, to obtain a phosphorylated version of the protein. CLK1 was previously shown to phosphorylate DDX23 in vitro (21), and we determined the phosphorylation pattern by MS analysis (SI Appendix, Fig. S9). In addition, we generated a mutant version of the minimal protein fused to His-TrxA, with the 9RS motif (S57 to S65) substituted by GA repeats (9GA). Phosphorylation of the RS repeats in the minimal DDX23 construct markedly reduced the efficiency of the PD with SRSF1. In the context of this minimal version of DDX23, the 9GA mutant had no detectable interaction with SRSF1 (Fig. 4A). We conclude that the phosphorylation state of the conserved RS repeats affects the interaction with SRSF1 mediated by the positively charged arginines. Moreover, the fact that the 9GA mutant in this context impairs binding to SRSF1, whereas it has no effect in the context of FL-DDX23, is consistent with other charged regions within the DDX23 Nt domain helping to stabilize the interaction with SRSF1.
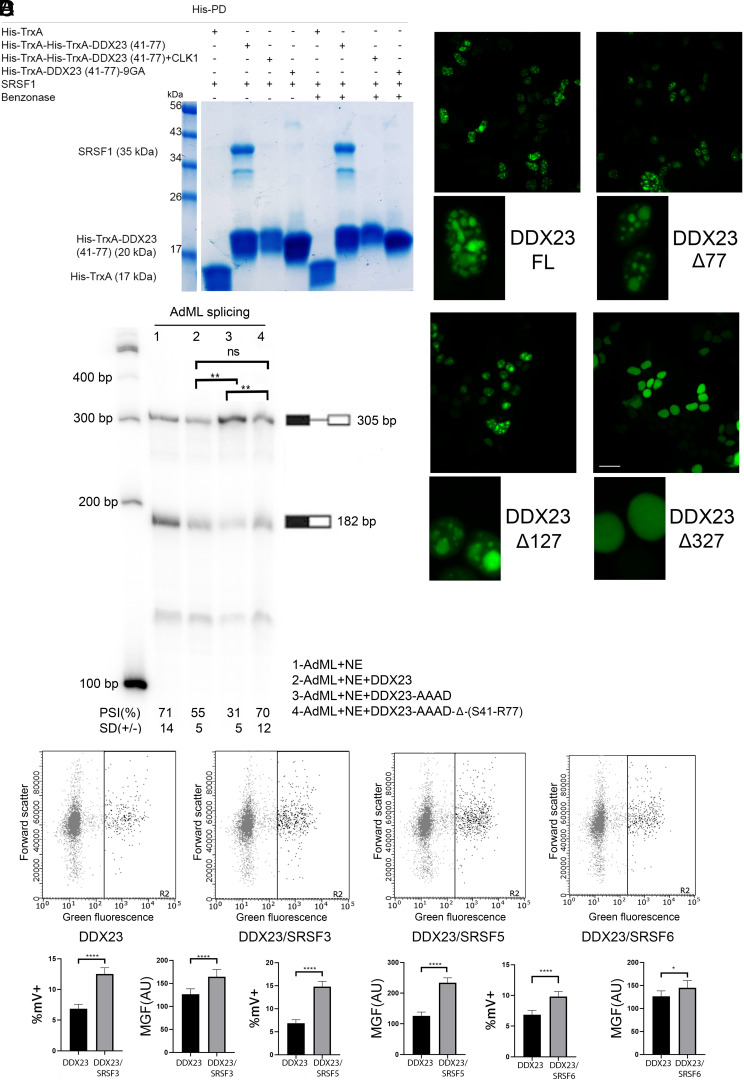
Fig. 4.
Possible functional roles of DDX23’s Nt-domain and conservation of the interaction with other SR protein family members. (A) Representative His PD assay using the indicated versions of His-TrxA-DDX23 (41 to 77). (B) In vitro splicing reactions using nuclear extract supplemented with the indicated versions of recombinant DDX23 protein. Calculated PSI values are reported for each condition, with their indicated SD. Triplicate samples were used for unpaired t tests. **P ≤ 0.01, ns = not significant. (C) Live fluorescence microscopy imaging of HEK293T cells transfected with the indicated mVenus-fused DDX23 constructs. Magnification was 40× (Top images). Scale bar represents ~15 μm. Cropped images (Bottom) are focused on one or two nuclei, so that the nuclear distribution of DDX23 can be observed more clearly. (D) BiFC results for the indicated proteins, plotted as Forward scatter (linear scale) vs. Green fluorescence (logarithmic scale). The vertical line indicates the threshold for cells that were considered mVenus-positive. The bar graphs below each panel represent %mVenus-positive cells and mean green fluorescence, respectively, for DDX23-mVCt coexpressed with Nt-mVenus vs. DDX23-mVCt coexpressed with SRSF3/5/6-Nt-mVenus. An unpaired t test was performed to compare the %mV+ cells and MGF values between the experimental conditions and the controls. *P ≤ 0.05; ****P ≤ 0.0001. Error bars represent SD.
DDX23’s (S41-R77) Region Is Important for Recruitment to the Spliceosome.
To better understand the possible functional consequences of the DDX23-SRSF1 interaction, we characterized the role of the Nt-domain of DDX23 in spliceosome assembly. A previous study demonstrated that an ATPase catalytically inactive mutant of DDX23 (DEAD to AAAD mutation) results in stalling of the spliceosome at the pre-B stage; therefore, the addition of recombinant DDX23-AAAD to in vitro splicing reactions results in splicing inhibition (41). We expressed and purified recombinant DDX23-AAAD and DDX23-AAAD-Δ(S41-R77)—a version of the protein that is both catalytically dead and lacks the (S41-R77) region. We performed in vitro splicing reactions using HeLa cell nuclear extract preincubated with recombinant DDX23-wt, DDX23-AAAD, or DDX23-AAAD-Δ(S41-R77). Splicing reactions with added DDX23-wt proceeded normally, whereas incubation with the DDX23-AAAD mutant resulted in splicing inhibition, as expected, reflecting competition of the AAAD mutant with endogenous DDX23-wt for spliceosome incorporation. In contrast, incubation with the DDX23-AAAD-Δ(S41-R77) mutant did not inhibit splicing. This result is consistent with the DDX23-AAAD-Δ(S41-R77) mutant failing to incorporate into the spliceosome (Fig. 4B). Given that SRSF1 interacts with this region of DDX23, it is possible that SR protein binding has a role in regulating DDX23’s recruitment to the spliceosome.
The Conserved DDX23 Nt Domain Is Required for Proper Localization.
To further characterize the function of DDX23’s Nt RS-like domain, we generated a series of mutants that lack the first 77, 127, or 327 residues of DDX23 and are fused at their N-terminus to mVenus. We used these constructs to transfect 293T cells and track the localization pattern of the DDX23 deletion mutants. Full-length DDX23 had an exclusively nuclear localization, with a nuclear speckled pattern, which we also observed with the smaller deletions (Δ77, Δ127). In contrast, deletion of the first 327 residues resulted in a diffused nuclear localization, lacking the characteristic speckled pattern, and with occasional cytoplasmic localization (Fig. 4C). These localization patterns indicate a role for the Nt domain of DDX23 in proper subnuclear targeting, which might involve interaction with SRSF1 and/or other SR proteins.
DDX23 Interacts with Additional SR Proteins.
SR proteins have related and, in some cases, overlapping functions, so we tested whether DDX23 can also interact with other family members. We tested seven additional SR proteins using BiFC and found that DDX23 interacted with SRSF3, SRSF4, SRSF5, SRSF6, SRSF7, SRSF9, and SRSF10 (Fig. 4D and SI Appendix, Fig. S10). Therefore, there may be a conserved function(s) among members of the SR protein family that involves direct interaction with DDX23. These results are consistent with our observation that RRM1 and the phosphorylated RS domain of SRSF1 can each efficiently pull down DDX23 and point to other SR proteins harboring these domains as candidates for interaction with DDX23.
Discussion
SRSF1 is an essential splicing factor that has been implicated in cancer, autoimmune diseases, and tautopathies. Therefore, further investigation of SRSF1 functions and its potential interactors can lead to unique insights into SRSF1 dysregulation in disease (45). Our previous study used IP-MS and computational approaches to study the SRSF1 interactome (15). However, many of the interactors we detected using BioID2 were not previously observed using IP-MS. Several advantages of proximity labeling may account for this observation. First, the labeling takes place in live cells, capturing interactions in the intact cellular compartments. Second, proximity labeling allows the detection of dynamic and transient interactions, such as those that occur during spliceosome assembly and splicing catalysis, which may be hard to capture in a copurification assay. Third, the protein isolation for proximity labeling is done under highly denaturing conditions, improving the solubility and recovery of labeled proteins and eliminating interference from bound nucleic acids. However, the proximity-labeling interactome reflects adjacency (up to 10 Å), so not all recovered proteins are necessarily part of a complex with SRSF1 (46). This is why complementary methods, such as BiFC and recombinant-protein interaction assays, can help to validate and map relevant protein–protein interactions.
Our datasets reflect SRSF1’s varied and dynamic subcellular localization, as well as SRSF1’s roles in splicing and in additional pathways like NMD and enhancement of translation (45). The detection of spliceosomal proteins involved in all stages of splicing reinforces the idea that SRSF1 remains associated with the spliceosome and later accompanies the spliced mRNA to the cytoplasm. Further characterization of these dynamic interactions should help clarify the molecular mechanisms underlying SR protein functions in pre-mRNA splicing and additional cellular functions of SRSF1.
Previous studies suggested that SR proteins play a role during the prespliceosome to spliceosome transition, helping to recruit the tri-snRNP to the spliceosome (47). We provide experimental evidence of a direct physical interaction between SRSF1 and DDX23, an essential protein that drives the pre-B to B complex transition, where stabilization of the tri-snRNP occurs. We detected the DDX23-SRSF1 interaction in live cells by two different approaches, as well as in vitro between purified recombinant proteins. The interaction involves several charged, conserved segments of the Nt-domain of DDX23 that function as alternative contact surfaces with SRSF1.
In SRSF1, the interaction involves two domains: RRM1 and the phosphorylated RS domain. The RS domain was previously characterized as a protein-interaction domain (10). Regarding RRM1, several studies likewise identified this domain as a mediator of protein–protein interactions, in addition to binding RNA (48–51).
In the Nt domain of DDX23, most of the RS repeats are relatively dispersed, except for a nine-residue stretch of RS repeats (S57-S65). A recent study of SRSF1 identified a negatively charged surface of RRM1 as a potential mediator of interactions with RS-repeat-containing peptides that is conserved in all SR protein family members (7). In the present study, mutation of the DDX23 RS repeats to GA (9GA mutant) decreased the interaction with RRM1, confirming that the long RS-repeat region contributes to SRSF1 binding through interactions with RRM1. However, binding was not completely abolished by the mutations, suggesting that other regions of the FL-DDX23 protein also contribute.
In contrast, in the context of the minimal DDX23 version (S41-R77), the 9GA mutation completely abolished binding to FL-SRSF1, whereas in the context of FL-DDX23, it had no effect. These results suggest that the flexibility of the disordered Nt domain of DDX23 permits multiple interactions with SRSF1 in the context of the whole protein. This notion is also confirmed by our results with the DDX23-Δ(S41-R77) mutant, which still binds SRSF1. We suggest that in the absence of the conserved (S57-S65) RS repeats, other charged repeats within the Nt domain of FL-DDX23 can interact instead, reflecting the intrinsic flexibility of the Nt domain.
Phosphorylation of the RS repeats of DDX23 was previously reported and proposed to be important for DDX23’s association with the tri-snRNP and for spliceosome B complex formation. It is not clear whether DDX23 is already phosphorylated when it associates with U5 snRNP or whether it becomes phosphorylated after tri-snRNP assembly (44). In addition, it seems plausible that DDX23 needs to be dephosphorylated to dissociate from the tri-snRNP after U1’s departure from the spliceosome. Our results suggest that the interaction with SRSF1 mediated by the RS repeats in DDX23 is inhibited by phosphorylation. Therefore, it is possible that regulation of the interaction with SRSF1 is mediated by the phosphorylation/dephosphorylation dynamics of DDX23, which would control the recruitment of DDX23 and its subsequent exit during spliceosome remodeling.
Several regions within the DDX23 Nt domain interact with spliceosome components, including a PRP8-SNU114 binding region (Y286 to H356), and a helical region (E158-L255) that connects BRR2 with PRP8. The Nt region also comprises the so-called 5′ exon mimic (D269-S283), a disordered loop that occupies the position of the 5′ exon in the pre-B spliceosome and is displaced by the 5′ exon upon DDX23’s departure (52) (Fig. 5A). Our results indicate that the S41-R77 region of DDX23 is important for spliceosome incorporation and that the M1-R77 region can interact with SRSF1 in multiple ways. This region also undergoes serine phosphorylation, which can affect the interactions with components of the tri-snRNP and with SRSF1.
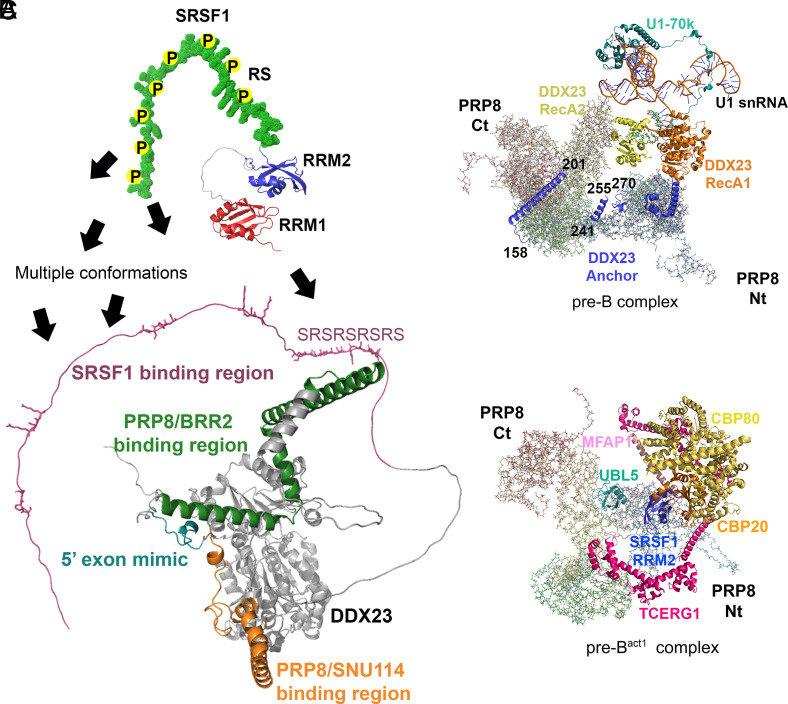
Fig. 5.
Structural predictions of SRSF1 and DDX23, and cryo-EM structures of spliceosomal complexes that contain DDX23 or SRSF1. (A) Structural representation of DDX23 N-terminal region (AF-Q9BUQ8-F1), which interacts with several spliceosome components, including PRP8, BRR2, SNU114, and SRSF1. The DDX23 5′ exon mimic, which occupies the same location as the 5′ exon in the B complex, is also depicted. A structural representation of SRSF1 (AF-J3KTL2-F1) is shown at the Top, with arrows denoting the interactions of the phosphorylated RS domain and RRM1 with the mapped interacting region in DDX23. (B) Cryo-EM spliceosome structure of the pre-B complex (PDB 6QX9). PRP8 is shown in the background, and the domains of DDX23 are highlighted, including fragments of the Nt-domain (numbers indicate residues); U1 snRNA and U1-70K (SRSF1 interactors) are also highlighted. (C) Cryo-EM spliceosome structure of the pre-Bact1 complex (7ABG). PRP8 is shown in the background; RRM2 of SRSF1 is highlighted (blue), together with CBP80/CBP20 (yellow/orange), TCERG1, UBL5, and MFAP1.
Larger deletions of the DDX23 Nt domain resulted in striking changes in the nuclear localization of DDX23 (Δ327 mutant). These changes may reflect the loss of protein–protein interactions that allow DDX23 to properly engage with spliceosome components, or to participate in liquid–liquid phase separation. A recent report on DDX23 from C. elegans showed that—similar to our observations with the human protein—the Nt domain is involved in the formation of nuclear foci, which involves liquid–liquid phase separation (53). Therefore, in higher eukaryotes, the presence of the extended Nt domain could play a role in several aspects of DDX23 regulation, from localization and phase separation, to phosphorylation and spliceosome incorporation. Dissecting the protein interactions involving this domain can therefore reveal key regulatory mechanisms that may be exclusive to higher eukaryotes.
Both human DDX23 and yeast Prp28p have very low RNA-dependent ATPase activity in vitro, and their presumed RNA-helicase activity has never been detected. Therefore, it is believed that key interactions between this enzyme and other spliceosome components are essential to achieve proper activation of DDX23 (22, 54). A recent study identified an interaction between Prp28p and phosphorylated Npl3p (P-S411) that is important for Prp28p’s ATPase activation and recruitment to the budding yeast spliceosome (55). Given that SR proteins are evolutionarily related to Npl3p, our results suggest that the reported contact observed in yeast is conserved in higher eukaryotes as a contact between DDX23 and SR proteins. We performed preliminary assays to investigate whether SRSF1 could stimulate DDX23’s very low basal RNA-dependent ATPase activity, but we did not observe activation under the conditions tested. However, one key difference between yeast Prp28p and human DDX23 is the lack of the RS-like Nt domain in the yeast protein. In addition, yeast Npl3p has RRMs with clear homology with those in SR proteins, but it lacks an RS domain (56), which in SRSF1 enhances the interaction with DDX23, as shown in the present study. Therefore, notwithstanding the conserved interaction between Prp28p-Npl3p and DDX23-SRSF1, the underlying interaction mechanisms are different, and so the functional consequences may also differ, such that additional protein interactions with human DDX23 may be required to stimulate its ATPase activity.
In addition to its crucial role in the pre-B to B spliceosomal-complex transition, yeast Prp28p was implicated in the earliest stage of spliceosome assembly (57). As SRSF1 also participates in early spliceosome assembly (58), it seems possible that the initial interaction with DDX23 could take place at an early stage. However, information derived from the cryo-EM structures of the human pre-B and pre-Bact1 spliceosomes places DDX23 in the pre-B complex and SRSF1’s RRM2 in the pre-Bact1 complex in the same vicinity, proximal to the 5’ss (19, 52) (Fig. 5 B and C). In addition, the fact that SRSF1 binds U1 snRNA and is involved in U1 snRNP recruitment, whereas DDX23 promotes U1 snRNP release, functionally links these two proteins and suggests that their physical interaction could help coordinate these sequential events, either by recruiting DDX23 or affecting the conformation of its Nt domain. Further studies, particularly structural approaches and analysis of DDX23 regulation by phosphorylation, will be necessary to fully understand the functional implications of this interaction.
Materials and Methods
Proximity Labeling and Streptavidin Pulldown.
Details about the generation of cell lines used for proximity labeling are in SI Appendix, Supplementary Methods section. For proximity labeling, we performed a 50-μM biotin pulse for 24 h, followed by streptavidin PD under denaturing conditions (Lysis buffer: 50 mM Tris-HCl pH 7.5, 8 M urea, 1 mM DTT, 1x protease inhibitor cocktail (Roche)). We subjected the washed beads corresponding to triplicate samples for each condition to on-bead trypsin digestion and analyzed the resulting peptides by LC/MS, followed by label-free quantification. We detected a total of 2,706 proteins, and for each one, we extracted the peptide ion chromatogram (XIC) and calculated the area under the curve ratio between SRSF1 samples and the GFP control. We filtered the proteins by their combined abundance across samples and set a lower threshold of 6 × 107, yielding 1,699 candidates for further analysis. We calculated the abundance ratios for each detected protein in the BS and SB samples, with respect to BG. We set an enrichment ratio of 5.0 as a minimal threshold to identify proteins enriched in the BS and SB datasets. Application of this threshold yielded 353 proteins enriched in the BS dataset and 246 proteins in the SB dataset, with respect to BG. We input this protein list into Cytoscape (59) to visualize the connections among the proteins and to perform a GO term analysis.
BiFC.
We seeded HEK-293 cells in 6-well plates (5 × 105 cells/well) and transfected them with the BiFC vectors at 70% confluency. For transfection, we mixed 100 μL of opti-MEM with X-treme gene 9 and the corresponding BiFC plasmid. We normalized the amount of plasmid to use by the size of the plasmid, as follows ng plasmid = 0.059 x length of plasmid (bp), and added 3 μL of X-treme gene 9 per μg of plasmid. After 18 h, we imaged live cells by fluorescence microscopy, collected them using 0.25% trypsin (500 μL), and resuspended them in 13 mL of DMEM. We transferred 200 μL of cells into 96-well round-bottom plates and analyzed them by flow cytometry (Guava). We analyzed eight technical replicates per transfection in parallel and compared them to an EGFP transfection control. We repeated each transfection at least 3 times. We analyzed the flow cytometry data using Guava Soft 4.5, with InCyte or Express pro. We gated the cells from a Forward versus Side scatter plot and obtained the percentage of mVenus-positive cells and mean green fluorescence values, which we plotted on Graphpad Prism.
His Pulldown Assay.
Details of expression and purification of recombinant proteins are in SI Appendix, Supplementary Methods section. We incubated with gentle rotation 10 μg of His-tagged protein for 1 h at 4 °C with IMAC beads (Chelating Sepharose Fast Flow beads charged with Ni2+) equilibrated in Pulldown buffer (50 mM Tris-HCl pH 7.5, 200 mM NaCl, 2 mM MgCl2, 0.5% CHAPs, 20 mM imidazole), in the presence of 10 μg BSA and, where indicated, 10 units of benzonase nuclease (Millipore Sigma). We then added 20 μg of untagged protein and incubated the suspension for an extra 2 h at 4 °C. Next, we spun the beads for 5 min at 500 × g and washed them three times with 500 μL of Pulldown buffer for 3 min, with rotation at 4 °C. After the last wash, we removed all the remaining buffer and resuspended the washed beads in 20 μL of Laemmli buffer with 5 mM EDTA. We heated the samples at 95 °C for 10 min and analyzed the proteins by SDS-PAGE and Coomassie Blue staining.
Sequence Alignment.
We retrieved the protein sequences of human DDX23 and its orthologs from the Uniprot database, visualized them in Seaview, and aligned them using Muscle (60). We generated a structural sequence alignment using ESPript 3.0 (61).
In Vitro Pre-mRNA Splicing Assay.
We transcribed and gel-purified the AdML pre-mRNA from BamHI-digested AdML-M3 plasmid (Addgene #11244) as described in SI Appendix, Supplementary Methods section. Pre-mRNA splicing was carried out with HeLa cell nuclear extract (Ipracell). For each version of DDX23 [DDX23-wt, DDX23-AAAD, or DDX23-AAAD-Δ(S41-R77)], we preincubated 1.2 μg of purified recombinant protein (stock concentration: 1.0 mg/mL) with 15 μL of nuclear extract at 30 °C for 30 min. After pre-incubation, we added reagents to a final concentration of 2 mM ATP, 20 mM creatine phosphate, 65 mM KCl, 3 mM MgCl2, and radioactively labeled pre-mRNA substrate, in a final reaction volume of 30 μL, such that the final recombinant protein concentration was 39 ng/μL. We incubated the splicing reactions at 30 °C for 1 h and quenched them by adding 200 μL of splicing stop solution (0.3 M sodium acetate pH 5.2, 0.1% SDS, 62.5 μg/mL tRNA). We extracted the RNA using 200 μL of acid phenol-chloroform, pH 4.5 and precipitated it with 500 μL of 100% EtOH. We resuspended the RNA pellets in 2x RNA sample buffer (90% formamide, 0.5 mM EDTA, 0.1% XC, 0.1% BB), heated the samples at 68 °C for 10 min, and analyzed them by denaturing-PAGE. We visualized RNA bands using a Typhoon biomolecular imager and performed the quantification using FIJI software.
Live Imaging of HEK-293T Cells.
We imaged the transfected cells under the GFP channel of a 488-nm laser, at 12.8% laser power, with 85 ms exposure time, using a spinning-disk laser-scanning confocal microscope (UltravVIEW Vox, PerkinElmer) with a Nikon Plan Apo 40x/0.95 NA objective lens. The platform incorporates a 488-nm diode laser (Vortran VersaLaseTM, Biovision Technology) with an ORCA-Fusion BT sCMOS camera (Hamamatsu, Japan). The platform is integrated with Volocity image-acquisition software (version 7.0.0, Quorum Technologies).
Graphics.
We generated protein-structure images with Pymol 1.7.4.0, and protein-domain graphics with IBS illustrator (62).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We acknowledge Dr. Peter Cherepanov from The Francis Crick Institute for kindly providing the His-SUMO-SRSF1 and CLK1 bacterial expression vectors, Dr. Maria Antonelli for her assistance with phosphorylation detection by MS, and members of the Krainer and Joshua-Tor labs for helpful discussions. This work was funded by National Institues of Health (NIH) grant GM42699 to A.R.K. Cold Spring Harbor Laboratory Shared Resources used in this work were funded in part by National Cancer Institute (NCI) Cancer Center Support Grant 5P30CA045508.
Author contributions
D.S. and A.R.K. designed research; D.S., D.W.A., N.H., P.S.T., T.-L.W., and P.C. performed research; L.J.-T. and A.R.K. contributed new reagents/analytic tools; D.S. analyzed data; and D.S. and A.R.K. wrote the paper.
Competing interests
A.R.K. is a co-founder, Director, and shareholder of Stoke Therapeutics, a member of the SABs of Skyhawk Therapeutics, Envisagenics, and Autoimmunity BioSolutions, and a consultant for Biogen. These relationships are unrelated to the present work.
Footnotes
Reviewers: G.G., University of California, San Diego; and J.V., Centre de Regulacio Genomica.
Data, Materials, and Software Availability
Interactome data have been deposited in ProteomeXchange, PRIDE database (Project accession: PXD048180, Project DOI: 10.6019/PXD048180) and is included as Dataset S1 (63). All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Krainer A. R., Conway G. C., Kozak D., Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 4, 1158–1171 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Krainer A. R., Conway G. C., Kozak D., The essential pre-mRNA splicing factor SF2 influences 5’ splice site selection by activating proximal sites. Cell 62, 35–42 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Ge H., Manley J. L., A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell 62, 25–34 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Manley J. L., Krainer A. R., A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24, 1073–1074 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clery A., et al. , Structure of SRSF1 RRM1 bound to RNA reveals an unexpected bimodal mode of interaction and explains its involvement in SMN1 exon7 splicing. Nat. Commun. 12, 428 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clery A., et al. , Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition. Proc. Natl. Acad. Sci. U.S.A. 110, E2802–E2811 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fargason T., et al. , Peptides that Mimic RS repeats modulate phase separation of SRSF1, revealing a reliance on combined stacking and electrostatic interactions. Elife 12, e84412 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannakouros T., Nikolakaki E., Mylonis I., Georgatsou E., Serine-arginine protein kinases: A small protein kinase family with a large cellular presence. FEBS J. 278, 570–586 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Hanamura A., Caceres J. F., Mayeda A., Franza B. R. Jr., Krainer A. R., Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4, 430–444 (1998). [PMC free article] [PubMed] [Google Scholar]
- 10.Howard J. M., Sanford J. R., The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip. Rev. RNA 6, 93–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen H., Kan J. L., Green M. R., Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell 13, 367–376 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Wu J. Y., Maniatis T., Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75, 1061–1070 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Fu X. D., Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature 365, 82–85 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Jobbins A. M., et al. , Exon-independent recruitment of SRSF1 is mediated by U1 snRNP stem-loop 3. EMBO J. 41, e107640 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akerman M., et al. , Differential connectivity of splicing activators and repressors to the human spliceosome. Genome Biol. 16, 119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh G., et al. , The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151, 915–916 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Aznarez I., et al. , Mechanism of nonsense-mediated mRNA decay stimulation by splicing factor SRSF1. Cell Rep. 23, 2186–2198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman M. A., Lin K. T., Bradley R. K., Abdel-Wahab O., Krainer A. R., Recurrent SRSF2 mutations in MDS affect both splicing and NMD. Genes Dev. 34, 413–427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Townsend C., et al. , Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation. Science 370, eabc3753 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teigelkamp S., Mundt C., Achsel T., Will C. L., Luhrmann R., The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA 3, 1313–1326 (1997). [PMC free article] [PubMed] [Google Scholar]
- 22.Mohlmann S., et al. , Structural and functional analysis of the human spliceosomal DEAD-box helicase Prp28. Acta Crystallogr. D Biol. Crystallogr. 70, 1622–1630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D. I., et al. , An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 27, 1188–1196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun S., Zhang Z., Sinha R., Karni R., Krainer A. R., SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat. Struct. Mol. Biol. 17, 306–312 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anczukow O., et al. , SRSF1-regulated alternative splicing in breast cancer. Mol. Cell 60, 105–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pozzi B., et al. , SUMO conjugation to spliceosomal proteins is required for efficient pre-mRNA splicing. Nucleic Acids Res. 45, 6729–6745 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelisch F., et al. , The serine/arginine-rich protein SF2/ASF regulates protein sumoylation. Proc. Natl. Acad. Sci. U.S.A. 107, 16119–16124 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maslon M. M., Heras S. R., Bellora N., Eyras E., Caceres J. F., The translational landscape of the splicing factor SRSF1 and its role in mitosis. Elife, 10.7554/eLife.02028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michlewski G., Sanford J. R., Caceres J. F., The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell 30, 179–189 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Sanford J. R., Gray N. K., Beckmann K., Caceres J. F., A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18, 755–768 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caceres J. F., Screaton G. R., Krainer A. R., A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12, 55–66 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z., Krainer A. R., Involvement of SR proteins in mRNA surveillance. Mol. Cell 16, 597–607 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Loomis R. J., et al. , Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol. Cell 33, 450–461 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komuro A., et al. , The AHNAKs are a class of giant propeller-like proteins that associate with calcium channel proteins of cardiomyocytes and other cells. Proc. Natl. Acad. Sci. U.S.A. 101, 4053–4058 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaudet P., Livstone M. S., Lewis S. E., Thomas P. D., Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 12, 449–462 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawo S., et al. , HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19, 816–826 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Haward F., et al. , Nucleo-cytoplasmic shuttling of splicing factor SRSF1 is required for development and cilia function. Elife 10, e65104 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agafonov D. E., et al. , Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol. Cell Biol. 31, 2667–2682 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerppola T. K., Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding J. H., et al. , Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol. Biol. Cell 17, 876–885 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boesler C., et al. , A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat. Commun. 7, 11997 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maertens G. N., et al. , Structural basis for nuclear import of splicing factors by human Transportin 3. Proc. Natl. Acad. Sci. U.S.A. 111, 2728–2733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Busch A., Hertel K. J., Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA 3, 1–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathew R., et al. , Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat. Struct. Mol. Biol. 15, 435–443 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Li D., Yu W., Lai M., Towards understandings of serine/arginine-rich splicing factors. Acta Pharm. Sin. B 13, 3181–3207 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bosch J. A., Chen C. L., Perrimon N., Proximity-dependent labeling methods for proteomic profiling in living cells: An update. Wiley Interdiscip. Rev. Dev. Biol. 10, e392 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roscigno R. F., Garcia-Blanco M. A., SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA 1, 692–706 (1995). [PMC free article] [PubMed] [Google Scholar]
- 48.Aubol B. E., Serrano P., Fattet L., Wuthrich K., Adams J. A., Molecular interactions connecting the function of the serine-arginine-rich protein SRSF1 to protein phosphatase 1. J. Biol. Chem. 293, 16751–16760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho S., et al. , Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1–70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. U.S.A. 108, 8233–8238 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagopian J. C., et al. , Adaptable molecular interactions guide phosphorylation of the SR protein ASF/SF2 by SRPK1. J. Mol. Biol. 382, 894–909 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fregoso O. I., Das S., Akerman M., Krainer A. R., Splicing-factor oncoprotein SRSF1 stabilizes p53 via RPL5 and induces cellular senescence. Mol. Cell 50, 56–66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Charenton C., Wilkinson M. E., Nagai K., Mechanism of 5’ splice site transfer for human spliceosome activation. Science 364, 362–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doi A., Suarez G. D., Droste R., Horvitz H. R., A DEAD-box helicase drives the partitioning of a pro-differentiation NAB protein into nuclear foci. Nat. Commun. 14, 6593 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacewicz A., Schwer B., Smith P., Shuman S., Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28. Nucleic Acids Res. 42, 12885–12898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeh F. L., et al. , Activation of Prp28 ATPase by phosphorylated Npl3 at a critical step of spliceosome remodeling. Nat. Commun. 12, 3082 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Birney E., Kumar S., Krainer A. R., Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21, 5803–5816 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price A. M., Gornemann J., Guthrie C., Brow D. A., An unanticipated early function of DEAD-box ATPase Prp28 during commitment to splicing is modulated by U5 snRNP protein Prp8. RNA 20, 46–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staknis D., Reed R., SR proteins promote the first specific recognition of Pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol. Cell Biol. 14, 7670–7682 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gouy M., Guindon S., Gascuel O., SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu W., et al. , IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Segovia D., Adams D. W., Hoffman N., Safaric Tepes P., Wee T.-L., et al. , SRSF1 interactome determined by proximity labeling reveals direct interaction with spliceosomal RNA helicase DDX23. PRIDE. http://www.proteomexchange.org/PXD048180. Deposited 30 December 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
Interactome data have been deposited in ProteomeXchange, PRIDE database (Project accession: PXD048180, Project DOI: 10.6019/PXD048180) and is included as Dataset S1 (63). All other data are included in the manuscript and/or supporting information.