Summary
Background
The field of precision medicine endeavors to transform the healthcare industry by advancing individualised strategies for diagnosis, treatment modalities, and predictive assessments. This is achieved by utilizing extensive multidimensional biological datasets encompassing diverse components, such as an individual's genetic makeup, functional attributes, and environmental influences. Artificial intelligence (AI) systems, namely machine learning (ML) and deep learning (DL), have exhibited remarkable efficacy in predicting the potential occurrence of specific cancers and cardiovascular diseases (CVD).
Methods
We conducted a comprehensive scoping review guided by the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) framework. Our search strategy involved combining key terms related to CVD and AI using the Boolean operator AND. In August 2023, we conducted an extensive search across reputable scholarly databases including Google Scholar, PubMed, IEEE Xplore, ScienceDirect, Web of Science, and arXiv to gather relevant academic literature on personalised medicine for CVD. Subsequently, in January 2024, we extended our search to include internet search engines such as Google and various CVD websites. These searches were further updated in March 2024. Additionally, we reviewed the reference lists of the final selected research articles to identify any additional relevant literature.
Findings
A total of 2307 records were identified during the process of conducting the study, consisting of 564 entries from external sites like arXiv and 1743 records found through database searching. After 430 duplicate articles were eliminated, 1877 items that remained were screened for relevancy. In this stage, 1241 articles remained for additional review after 158 irrelevant articles and 478 articles with insufficient data were removed. 355 articles were eliminated for being inaccessible, 726 for being written in a language other than English, and 281 for not having undergone peer review. Consequently, 121 studies were deemed suitable for inclusion in the qualitative synthesis. At the intersection of CVD, AI, and precision medicine, we found important scientific findings in our scoping review. Intricate pattern extraction from large, complicated genetic datasets is a skill that AI algorithms excel at, allowing for accurate disease diagnosis and CVD risk prediction. Furthermore, these investigations have uncovered unique genetic biomarkers linked to CVD, providing insight into the workings of the disease and possible treatment avenues. The construction of more precise predictive models and personalised treatment plans based on the genetic profiles of individual patients has been made possible by the revolutionary advancement of CVD risk assessment through the integration of AI and genomics.
Interpretation
The systematic methodology employed ensured the thorough examination of available literature and the inclusion of relevant studies, contributing to the robustness and reliability of the study's findings. Our analysis stresses a crucial point in terms of the adaptability and versatility of AI solutions. AI algorithms designed in non-CVD domains such as in oncology, often include ideas and tactics that might be modified to address cardiovascular problems.
Funding
No funding received.
Keywords: Artificial intelligence, Cardiovascular diseases, Deep learning, Personalised medicine, Precision medicine
Research in context.
Evidence before this study
Prior to initiating this study, there were limited existing research and programme and programme descriptions concerning the integration of personalised medicine (PM) and AI in managing CVD. Despite several study projects, the scholarly literature lacked a comprehensive synthesis of this information. This gap underscored the significance of our scoping assessment in consolidating and appraising the existing body of knowledge in this domain.
Added value of this study
Our study represents the first of its kind in exploring the integration of AI and PM in CVD management. Through a systematic scoping review, we have provided unique insights into the application of AI algorithms for tasks such as risk prediction, disease classification, and the identification of genetic biomarkers associated with CVD. These findings underscore the potential of AI to revolutionise cardiovascular risk assessment, enable early diagnosis, and optimise personalised treatment strategies.
Implications of all the available evidence
Our study underscores the importance of harnessing AI-driven approaches to enhance the accuracy of cardiovascular risk prediction and advance personalised medicine in managing CVD. Our findings provide valuable insights for healthcare providers and administrators to guide the development and implementation of AI-driven solutions aimed at optimising clinical care, reducing adverse outcomes, and enhancing patient outcomes in cardiovascular medicine. By consolidating and analysing the existing knowledge in this field, our study lays the foundation for further research and innovation to tackle the evolving challenges and opportunities in CVD management through AI-driven personalised medicine approaches.
Introduction
Personalised medicine (PM) is an approach to healthcare that tailors medical decisions and treatments to individual patients based on their unique characteristics, including their genetic makeup, lifestyle, environment, and other pertinent factors.1,2 The primary goal of PM is to provide more effective and precise treatments, reducing the likelihood of adverse reactions and optimizing patient outcomes.3 This approach recognises that not all patients with the same condition will respond similarly to a particular treatment. Hence, it aims to deliver targeted therapies that align with each patient's needs. It enables doctors to predict how patients will respond to a particular treatment, helping them make more informed decisions and avoid ineffective or potentially harmful therapies.4
PM offers numerous advantages. First, customizing treatments to a patient's unique genetic and clinical characteristics ensures that interventions, medications, and therapies are directed precisely where they are most likely to give positive results. This targeted approach enhances patient care and minimises the waste of valuable resources on treatments that may prove ineffective or less beneficial for certain individuals. Second, it can expedite drug discovery by screening for potentially useful compounds much more efficiently than a human medicinal chemist.5 This streamlines the research process, shortens drug development timelines, and brings potentially life-saving medications to market sooner. Third, it can help identify patients more likely to experience adverse reactions to certain drugs. This information allows for more precise prescribing and monitoring, reducing the risk of harmful side effects. Fourth, it also plays a role in cardiovascular intervention. With minimally invasive techniques, the interventional branch aims to diagnose and treat a wide range of cardiovascular problems. Procedures like percutaneous coronary intervention (PCI), which uses balloons and stents to open narrowed or blocked coronary arteries and structural heart interventions like transcatheter aortic valve replacement (TAVR), which corrects structural heart abnormalities, are essential to interventional cardiology.6, 7, 8, 9, 10 Lastly, it offers hope for patients with rare diseases or conditions that have been historically challenging to treat effectively.
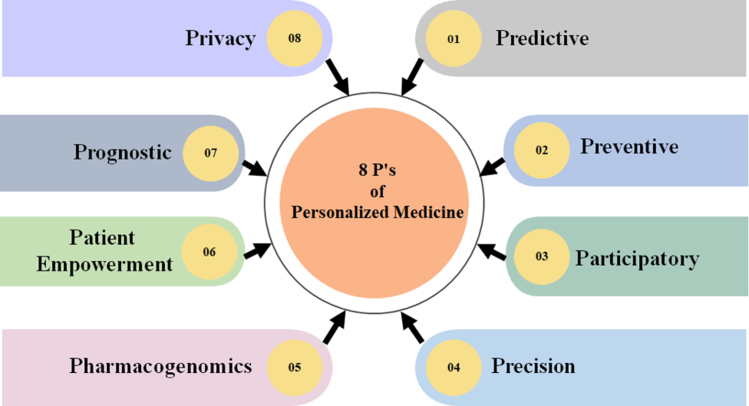
Nowadays, PM is applied in various areas, such as cancer therapy, where specific genetic mutations are targeted with precision medicines.11 It is making advancements in areas like genetic testing for disease risk assessment,12,13 personalised vaccines,14 and regenerative medicine.15 Fig. 1 depicts the 8 P's of PM that serve as the fundamental basis of this pioneering approach to healthcare, building on the longstanding intellectual legacy of the 4 P concept in medical science—predictive, preventive, precise, and participatory.27,28 These guiding concepts, predictive, preventive, participatory, precision, pharmacogenomics, patient empowerment, prognostic, and privacy, collectively shape a comprehensive framework for PM-based CVD.
Fig. 1.
The 8 P's of PM. Each “P” stands for the fundamental aspect that has contributed to developing and advancing the ground-breaking field of PM. The predictive aspect harnesses data and AI to forecast patients' disease risks and treatment responses.16,17 Preventive strategies focus on proactive interventions to mitigate disease risks, while participatory engagement empowers patients to be active partners in their healthcare decisions.18,19 Precision modifies the treatments to meet patients' needs and preferences.16,18,20, 21, 22, 23, 24, 25 Pharmacogenomics focuses on understanding how an individual's genetic makeup affects their drug response, leading to optimised drug selection and dosages.4,26 Patient empowerment provides patients with the knowledge, skills, and resources necessary to make informed decisions about their health. Prognostic models use AI to predict disease progression and patient outcomes, guiding personalised insights into a patient's health status. Finally, ensuring privacy safeguards patient data confidentiality, fostering trust in the PM journey.
It also faces several challenges that must be addressed to integrate into healthcare successfully. One significant challenge is the complexity of interpreting and integrating vast amounts of diverse data, the so-called “big” data,29 as PM harnesses a wealth of different data types, including genetic information, molecular profiling, electronic health records (EHRs), laboratory tests, patient histories, lifestyle data, and environmental factors motivating for a composite design.30 However, the seamless extraction and harmonization of these multifaceted data originating from multiple sources in a meaningful and standardised way poses technical and logistical challenges. Gathering patient data is the first step in orchestrating PM, such as genetic data from deoxyribonucleic acid (DNA) sequencing techniques, biomarker data from blood tests, and lifestyle data from wearables and health monitoring devices.31 Additionally, EHRs capture patients’ medical histories and treatment responses. All these data sources must be synergised for purposeful comprehension and precise predictions.
AI-driven advances in hardware have sparked revolutionary shifts in the personalised treatment of CVD. Data processing capabilities have been transformed by the increase of computational capacity, through devices such as multi-core CPUs, specialised GPUs, and Tensor Processing Units (TPUs).32,33 Improved diagnosis, risk assessment, and tailored therapy are made possible by these improvements that allow for the integration and analysis of many data modalities, such as genetic data, medical imaging, and electronic health records. AI-driven medical technologies are also becoming more widely available due to the availability and affordability of high-performance computer resources as well as the growth of cloud computing services. Furthermore, the effective processing of huge datasets (or big data) necessary for tailored CVD management is ensured by advancements in data management and storage, such as scalable cloud storage and high-performance solid-state drives (SSDs).29,34, 35, 36 As specialised hardware continues to advance, it promises to further enhance the efficacy and scalability of AI-driven approaches, ultimately optimizing patient care and outcomes in cardiovascular medicine.
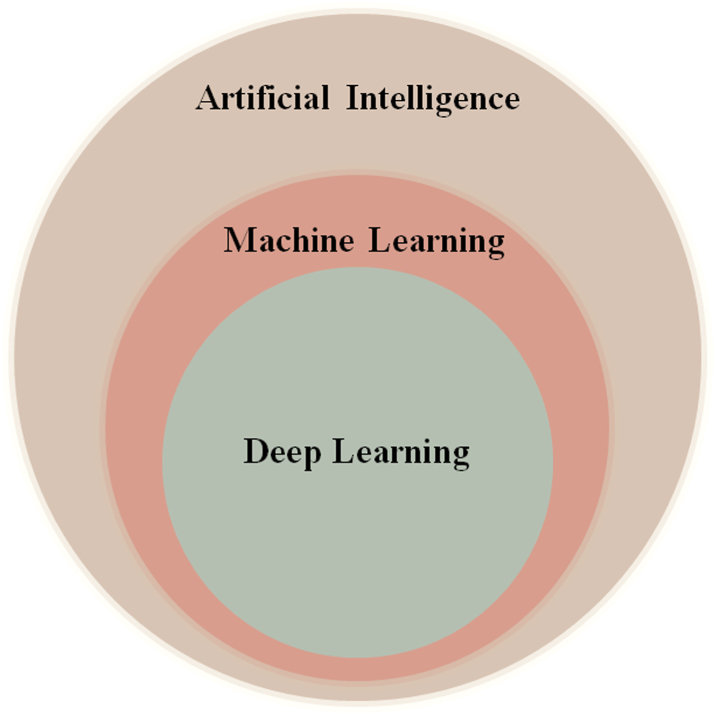
AI uses a diverse array of techniques, with supervised and unsupervised learning being two fundamental approaches.37 Supervised learning trains a model on labeled data, mapping input variables or risk predictors to corresponding output labels.38,39 For example, in cardiovascular risk prediction, patient demographics, clinical variables, and miRNA expression profiles are used to predict the presence or absence of cardiovascular diseases, so-called risk stratification. Unsupervised learning, on the other hand, does not need labeled data.40 Rather, it investigates data structures to find hidden correlations and patterns. In cardiology, techniques like clustering and dimensionality reduction identify patient subgroups or extract features from high-dimensional datasets. For instance, unsupervised learning can analyze miRNA expression data to identify molecular subtypes or novel biomarkers of cardiovascular disease progression. The key distinction lies in labeled data: supervised learning uses labels for training, while unsupervised learning relies solely on data structure.41 Both methods complement each other in cardiology, with supervised learning for predictive modeling and unsupervised learning for understanding data structure and complexity. Big data presents unique challenges in disease prediction.42 As datasets become more extensive and complex, traditional linear models may not be sufficient to capture all the nuances and interactions within the data. In such scenarios, non-linear relationships between risk factors and the gold standard or between the risk factors themselves can exist, further complicating the accurate prediction of outcomes.43 This is where the pivotal role of AI in PM becomes evident.44 Fig. 2 depicts the relationship between AI, ML, and DL. AI is the umbrella term, embracing computer systems designed to do activities that generally need human-like intellect. ML is a subset of AI dedicated to developing algorithms and statistical models that allow computers to learn from data and make informed predictions or decisions. ML techniques include supervised,37 unsupervised,45 and reinforcement learning.46 DL, a subset of ML inspired by the human brain's structure, uses artificial neural networks (ANN) with numerous layers that process and learn from data hierarchically.47
Fig. 2.
Venn diagram depicting AI, ML, and DL hierarchy.
It is a specialised and more advanced form of ML that has gained significant attention in PM due to its ability to handle large and complex datasets effectively because of its distributed computing capabilities and scalability.48 It automatically identifies essential elements from raw data, allowing it to capture intricate correlations. In PM, DL has been utilised in various tasks, such as image analysis,49 genomics,50 and EHR processing.51 These models have shown remarkable success in detecting anomalies and classifying medical images, enabling more accurate diagnoses.23,52 They have been instrumental in predicting disease risk and treatment responses by incorporating multi-modal data, such as genomic and imaging data, to improve predictive accuracy. However, the development, implementation, and use of AI technologies in cardiovascular care must be governed by a strong legal framework that addresses issues of patient consent, data privacy, liability, and regulatory compliance. Comprehensive steps are also required to guarantee the ethical and fair application of AI-driven solutions in clinical practice, including issues with algorithmic bias, accountability, and transparency.6,25,53, 54, 55
Methods
Literature sources
The authors MS and JSS conducted an extensive search up to March 2024 to identify relevant literature. Core search terms for CVD included: “CVD”, “AI in CVD”, “Genomics in CVD”, “Personalised Medicine in CVD”, “Personalised Risk Assessment in CVD”, “Deep Learning in CVD”, “Deep Learning in Personalised Medicine”, “Composite CVD Risk Assessment in Personalised Medicine”, “Personalised Medicine and CVD”, “Personalised Medicine-based biomarkers for CVD”, “AI and CVD”, “AI, CVD for Personalised Medicine”. We searched the following databases on August 10, 2023: Google Scholar, PubMed, IEEE Xplore, ScienceDirect, Web of Science and arXiv and then updated the searches on March 20, 2024. Within these databases, we searched for research articles, book chapters, and conference proceedings.
Literature selection
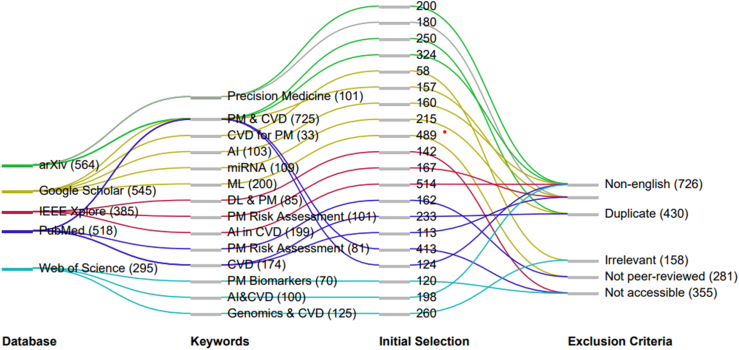
We conducted a two-stage screening process without restrictions on study design, location, time, or sex or gender differences. In the first stage, two reviewers (MS and JSS) independently screened titles and abstracts in duplicate. Documents were excluded if they did not pertain to CVD, PM, or AI. Any title/abstract deemed relevant by either reviewer advanced to the second stage. If a title seemed relevant but lacked an abstract, the article was also advanced to the second stage. During the second stage, two reviewers (MS and JSS) independently reviewed the full-text documents selected from stage one in duplicate. Fig. 3 represents the Sankey diagram illustrating the methodical journey taken in the review process from the initial identification stage across various databases to the application of exclusion criteria. The width of the lines in the diagram corresponds to the number of papers passing through each step, illustrating the filtering process. Initially, a sizable pool of studies was retrieved, reflecting the breadth of literature available in the field. The inclusion and exclusion criteria for full text review included.
Fig. 3.
Sankey diagram depicting the keywords used per database for initial paper identification and subsequent exclusion criteria.
Inclusion criteria
Studies were included if they focused on the topics of cardiovascular disease, personalised medicine, or artificial intelligence. There were no restrictions on study design, location, or gender.
Exclusion criteria
Studies were excluded if they did not focus on CVD, personalized medicine, or AI. Additionally, duplicate records, non-English language publications, non-peer-reviewed works, studies utilizing non-standard AI techniques, and studies with low-quality design were excluded.
Role of funding source
There was no exclusive funding for this project. Further, the development, design, and undertaking of this scoping review was implemented by co-authors itself. All authors had access to the data and were responsible for the decision to submit this scoping review for publication.
Results
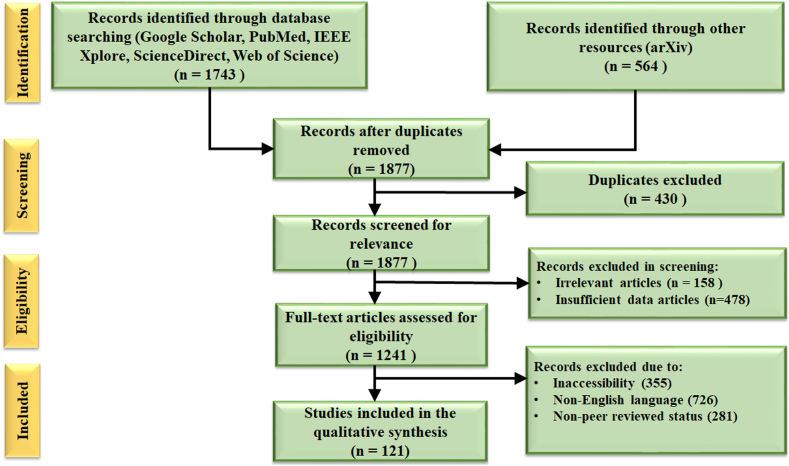
The search from the five databases identified 1743 records. The search from other sources (e.g., websites and arXiv) identified an additional 564 records. A total of 121 documents met our final inclusion criteria and were included in this review (Fig. 4).
Fig. 4.
PRISMA flowchart for systematic paper selection and quality assessment.
Personalised medicine and artificial intelligence
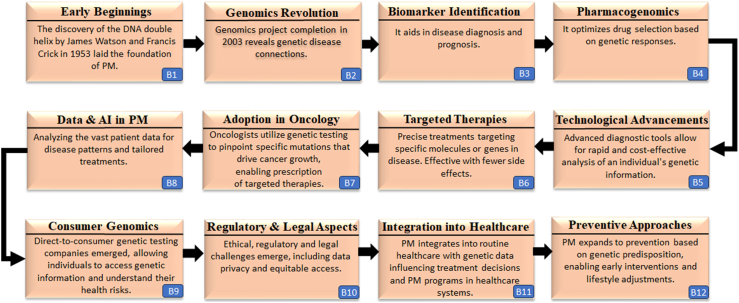
The journey of PM has been marked by significant milestones that have shaped the landscape of healthcare (Fig. 5). It all began with the ground-breaking discovery of DNA, the genetic material and molecular basis for heredity.56 Its structure laid the foundation for understanding genetic variations and heredity. Completing the Human Genome Project propelled the evolution of genomics, providing a comprehensive map of the human genome and advancing genomic research.57,58 Advancements in omics technologies enabled the identification of disease-related biomarkers, revolutionizing early diagnosis and opening doors to personalised treatments.59,60 The advent of consumer genomics services empowered individuals to access their genetic information, granting them greater control over their health decisions.61,62 The increasing acceptance of PM into mainstream clinical practice necessitates rethinking resilient and robust frameworks to address medical ethics and data privacy.63,64 While the landscape is evolving, PM continues to make strides in healthcare, seamlessly integrating into established clinical practices and transforming patient care.
Fig. 5.
Milestones in the journey of PM.
PM, also known as precision medicine,48 relies on various cutting-edge technologies, including high-performance computing (HPC) and vast biological datasets, to guide tailored diagnosis and treatment for individual patients.65 These advanced tools are essential for implementing a precise approach that can revolutionise healthcare outcomes. At the core of this strategy are sophisticated computer algorithms capable of identifying patterns in complex multidimensional datasets.66, 67, 68, 69, 70, 71, 72 These algorithms employ various feature or pattern extraction methods,73 which can then be used for classification of binary or multiclass scenarios based on gold standard labels.74 Note that the pattern recognition is a precursor for extraction of features while classification or risk stratification is a specific task performed by the classifier model to predict the risk or bin in the stratified risk. The trained classifier is actually designed using the training features with known gold standard labels. The training-based classifiers can be linear and non-linear methods.75,76 Examples of non-linear classifier models include Support Vector Machines, Random Forest, Decision Tree, and Artificial Neural Networks.16,17,77 These algorithms leverage knowledge from similar patient data to predict or optimise treatments for new individuals. While both neural network (NN) and random forest (RF) are non-linear, a NN's large parameter space greatly enhances its non-linear performance when compared to a RF. A NN's architecture enables it to learn hierarchical features from data, resulting in extremely complicated and non-linear mappings, whereas a RF depends on an ensemble of decision trees, each of which contributes to a non-linear decision boundary. For example, AI generated 3D models and simulations can be used by surgeons to rehearse operations in a safe setting. This method optimises operations based on each patient's distinct anatomy, improving surgical competence.78
Cardiovascular diseases
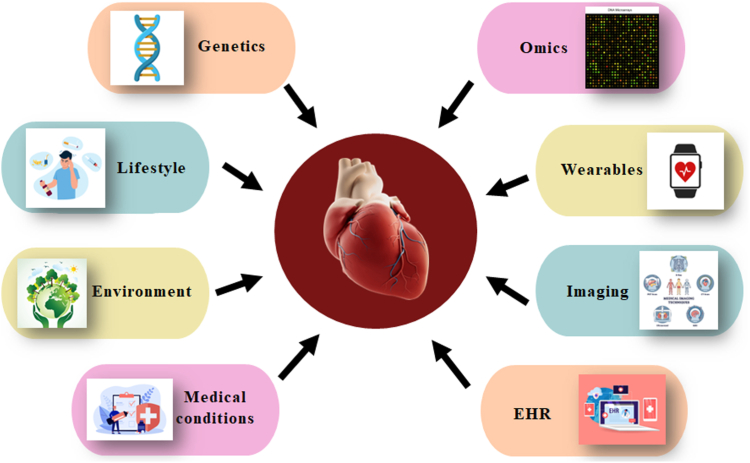
CVD pose a significant global health challenge, accounting for a substantial proportion of morbidity and mortality worldwide. According to statistics, CVD is the leading cause of death globally, responsible for more deaths than any other disease category.79 It includes coronary artery disease (CAD), heart failure (HF), stroke, and peripheral vascular disease (PVD). The burden of CVD affects both developed and developing countries, with an increasing prevalence in low- and middle-income nations due to changes in lifestyle, urbanization, and population aging.80 According to the World Health Organization (WHO), over 17.9 million people die yearly due to CVD, accounting for approximately 31% of all global deaths.81 Alarmingly, this number is projected to rise in the coming decades,82 posing significant challenges to healthcare systems and economies. Tackling this growing burden requires comprehensive prevention strategies, early detection, effective management, and continued research to address the complex interplay of risk factors contributing to CVD development on a global scale. Fig. 6 depicts the major risk components associated with CVD and the methods of data acquisition used in risk assessment. The depicted risk factors encompass both traditional elements, such as lifestyle factors (smoking status, alcohol, and fast-food consumption), as well as newly uncovered factors like genetic information and medical conditions. Moreover, some risk models also take environmental factors into consideration, further deepening the scope of risk assessment. This comprehensive integration of diverse risk factors, gathered through EHRs, medical imaging, omics data, wearables, and genetic testing, plays a crucial role in accurately and holistically assessing an individual's risk for developing CVD.
Fig. 6.
CVD risk factors and data acquisition.
The potential of DL in PM is further increased by assimilating the analysis of carotid vessel morphology since it offers vital insights into vascular health and contributes to a thorough understanding of cardiovascular problems. As the global burden of CVD grew, a paradigm shift emerged in the field of CVD risk assessment, embracing the integration of genomics and AI. Genomics offered new insights into the genetic basis of CVD risk, unveiling susceptibility and molecular mechanisms underlying the disease.83,84 Genetic testing can identify specific mutations or variants underlying certain cardiovascular disorders, aiding in accurate disease classification. It can provide crucial diagnostic clarity in ambiguous clinical presentations and facilitate appropriate management decisions. Identifying essential genes, pathways, and biological processes associated with CVD has deepened our knowledge of disease pathophysiology. This fundamental understanding lay the groundwork for precision medicine in CVD. Simultaneously, AI harnesses the power of big data analytics, allowing for the thorough analysis of diverse datasets to identify novel risk indicators and refine predictions.85
As one of the most advanced biomarkers for CVD, microRNAs (miRNAs) have many benefits that make them effective diagnostic instruments.86 These tiny, non-coding RNAs boast remarkable stability in circulation and tissue-specific expression patterns. Their early detection potential of CVD is made possible by the fact that they are in the bloodstream even before symptoms emerge. This enables rapid diagnosis and intervention. Because miRNAs amplify only at specific sequences, they are particularly good at detecting disorders related to the cardiovascular system with high specificity and sensitivity. For instance, increased levels of miR-21 are connected to inflammation and fibrosis, making it a significant biomarker for the development of heart failure. Its elevated expression is associated with the pathogenic mechanisms that underlie heart failure, suggesting that it may be used as a therapeutic target and prognostic marker to manage this condition.87 Conversely, miR-126, which is recognised for its function in controlling endothelial function, shows potential in forecasting the probability of atherosclerosis.88 One of the main factors to atherosclerosis is endothelial dysfunction, and miR-126 is essential for preserving endothelial integrity and function. It has been observed that dysregulation of miR-126 levels occurs in those who have atherosclerosis, indicating the potential use of this biomarker to guide preventative measures and assess cardiovascular risk. Furthermore, patient comfort is increased, and the danger associated with invasive procedures is decreased due to the minimally invasive nature of miRNA testing, which normally just takes a blood sample. Their revolutionary potential in early identification, risk assessment, and personalised treatment options is highlighted by the strong correlation observed between miRNA signatures and a range of cardiovascular diseases. Harnessing the therapeutic and diagnostic potential of miRNAs promises to usher in a new era of precision cardiovascular medicine, ultimately improving patient care and outcomes, as our understanding of miRNA biology continues to grow.
Table 1 provides an exhaustive overview of the use of AI and genomics in the context of CVD and non-CVD disorders. They utilise ML algorithms and techniques to identify biomarkers, classify different types of cardiomyopathies and diseases, develop prediction models for CVD risk, and explore novel genetic biomarkers associated with specific diseases. For example, in CVD studies, Phan et al.89 aimed to identify biomarkers for CVD using genomic datasets and applied various ML algorithms. The study achieved an accuracy ranging from 55% to an impressive 97%. Biomarker detection using AI and genomics has the potential to aid in early detection and management of CVD greatly. Alimadadi et al.90 used RNA-seq data from seven datasets to classify different forms of cardiomyopathies. Random forest (RF) outperformed others with 78%–84% accuracy among the algorithms employed. This shows the efficacy of AI algorithms in accurately distinguishing between various cardiomyopathy types. Steinfeldt et al.91 developed a prediction model for the 10-year risk of major adverse cardiac events (MACE) using NeuralCVD-DSM. The model showed an improved C-index and net reclassification index (NRI), providing better risk stratification for individuals with low to intermediate clinical risk. This demonstrates the potential of AI in enhancing cardiovascular risk prediction, aiding in better preventive measures. Using CNN-GWAS, Kwon et al.92 classified atrial fibrillation (AF) vs. non-AF. The model achieved moderate accuracy in predicting AF phenotype based on genetic information, with an area under the curve (AUC) ranging from 0.74 to 0.82. This highlights the potential of AI algorithms in leveraging genetic data for improved disease classification. Venkat et al.93 identified CVD-associated genes and predicted CVD diseases using a RF approach. The model achieved high accuracy, particularly in predicting HF, AF, and other CVD diseases, ranging from 90.9% to 95.9%. Evidence like this demonstrates the promise of AI in areas like identifying genes and disease prediction, opening avenues for progress in PM. For non-CVD studies, Khalifa et al.94 classified five types of cancer across four organs using BPSO-DT and CNN. The study achieved a high accuracy of 96.9% in classifying cancer types, surpassing the accuracy achieved by other related works. This emphasises the significance of AI in cancer classification, potentially aiding in precision oncology. Peng et al.95 identified high-risk individuals for various diseases, including Alzheimer's Disease (AD), Inflammatory Bowel Disease (IBD), Type 2 Diabetes (T2D), and Breast Cancer (BRCA) using Bidirectional Long Short Term Memory (BiLSTM). The model achieved promising performance with AUC values ranging from 0.66 to 0.86, outperforming traditional methods. This indicates the potential of AI in identifying individuals at risk for specific diseases. Li et al.96 classified AD patients and healthy controls using GWAS and ResNet, achieving accuracies of 71.38% for AD classification and 92.65% for healthy control classification. The study also discovered novel genetic biomarkers for AD. This showcases the potential of AI in genomics research and its role in uncovering new insights into complex diseases. Zekavat et al.97 calculated fractal dimension (FD) and vascular density (VD) using the U-Net-based Ensemble, identifying seven novel loci associated with FD and 13 with VD. The model achieved an outstanding accuracy of 95.6% in this task. This demonstrates the potential of AI in analyzing complex patterns in medical imaging and uncovering novel disease-related markers. Hahn et al.98 integrated genetic information and metabolite profiles to predict T2D risk using LR and RF. The RF-based model outperformed the logistic regression-based model, achieving an accuracy of 85.4%. This indicates the potential of AI in combining diverse data sources for improved disease risk prediction.
Table 1.
AI-genomics CVD/non-CVD.
| SN | Author & year | Disease type | Objective | Models | Dataset | Data Size | Input | Performance metrics | Conclusion |
|---|---|---|---|---|---|---|---|---|---|
| CVD | |||||||||
| 1 | Phan et al.89 (2012) | CVD | To describe the pipeline for biomarker identification for CVD and exemplify it by analyzing 4 genomic datasets. | KNN, linear SVM, LR, Bayesian | Blood Gene Exp. CAD, Baseline Macrophages Atherosclerosis, Monocytes FH, Monocytes Atherosclerosis. | 370 samples in 4 datasets | Gene expression data | ACC: 61%, 87%, 55%, 97% | A systematic pipeline for biomarker identification in CVD can be constructed using high-throughput genomic data and bioinformatics methods. |
| 2 | Alimadadi et al.90 (2020) | CM | To classify different types of cardiomyopathies. | svmRadial, pcaNNet, DT, ENet, RF | 7 datasets in the GEO database | 137 samples in 7 datasets | RNA-seq data | aACC: 80%, 83%, 78%, 84%, 82.66% | RF outperformed the others as it was the only one to show an increase in all the metrics. |
| 3 | Steinfeldt et al.91 (2022) | CVD | To develop and validate the prediction model for the 10-year risk of MACE. | NeuralCVD- DSM | UK Biobank | 395,713 CVD-free participants | 29 clinical predictors and 6 PGS | ΔC-index: 0.006, 95% CI: 0.005–0.007 NRI: 0.0116, 95% CI: 0.0066–0.0159 |
When additional high polygenic risk was present, those with low to moderate clinical risk and ages less than 50 years experienced a substantial rise in overall risk. |
| 4 | Kwon et al.92 (2022) | AF | To classify AF vs. non-AF. | CNN-GWAS | Yonsei AF cohort, Korea AF Network, KoGES, Korean Genome Rural cohort, 3-independent ethnic-specific GWAS | 6358 subjects selected from 4 cohorts | SNPs | AUC: 0.74–0.82 | CNN-GWAS algorithms predict AF phenotype moderately accurately using only genetic information, capturing cumulative gene effects and interactions. |
| 5 | Venkat et al.93 (2023) | HF, AF, other CVD disease | To identify genes associated with CVD diseases and predict the disease. | RF | Self-made dataset | 61 CVD patients | Gene expression data and clinical data | ACC: 90.9%, 95%,95.9% | Predicted the association of highly significant HF, AF, and other CVD genes with demographic variables. |
| Non-CVD | |||||||||
| 6 | Khalifa et al.94 (2020) | KIRC, BRCA, LUSC, LUAD, UCEC |
To classify 5 different types of cancer. | BPSO-DT, CNN | Tumor gene expression dataset | 2086 samples | RNA-Seq gene expression data | ACC: 96.90% | The present work exceeds prior relevant work regarding testing accuracy for 5 tumor types. |
| 7 | Peng et al.95 (2021) | AD, IBD, T2D, BRCA | To identify the high-risk individuals by calculating the PRS. | BiLSTM | UK biobank | 351,022 participants | SNPs, clinical features | AUC: 0.8624, 0.6585, 0.7316, 0.6660 | DeepPRS outperforms traditional techniques in terms of performance. |
| 8 | Li et al.96 (2021) | AD | To classify AD patients and HC. | GWAS, ResNet | ADNI database | 988 subjects | SNP genotype data | ACC: 71.38%, 92.65% | DLG model performs better than the traditional GWAS model. Also, they discovered novel genetic biomarkers of AD. |
| 9 | Zekavat et al.97 (2022) | Multiple | To calculate FD & VD. | U-Net Ensemble | UK biobank cohort | 54,813 participants | Retinal fundus photographs | ACC: 95.6% | GWAS identified 7 new loci related to FD and 13 with VD. PheWAS discovered systemic and ocular phenotypes associated with retinal microvasculature. |
| 10 | Hahn et al.98 (2022) | T2D | To integrate genetic information and metabolite profiles to predict T2D risk. | LR, RF | KoGES Ansan Ansung cohort | 1425 participants | Demo. +gPRS + clinical features+ metabolites |
ACC: 81.2%, 85.4% | RF-based model using clinical factors, gPRS, and metabolites predicted T2D risk more accurately than the LR-based model. |
CVD: Cardiovascular disease, CM: Cardiomypathy, AF: Atrial fibrillation, HF: Heart failure, MACE: Major adverse cardiac event, KNN: K-nearest neighbor, SVM: Support vector machine, LR: Logistic regression, svmRadial: Support vector machine with radial kernel, pcaNNet: Principal component analysis neural networks, DT: Decision tree, ENet: Elastic net, RF: Random forest, DSM: Deep survival machine, CNN: Convolutional neural network, GWAS: Genome-wide association studies, CAD: Coronary artery disease, FH: Familial hypercholesterolemia, GEO: Gene expression omnibus, KoGES: Korean genome Epidemiology Study, RNA: Ribonucleic acid, PGS: Polygenic score, ACC: Accuracy, CI: Confidence interval, C-index: Concordance index, NRI: Net reclassification index, AUC: Area under curve, KIRC: Kidney renal clear cell carcinoma, BRCA: Breast Cancer, LUSC: Lung squamous cell carcinoma, LUAD: Lung adenocarcinoma, UCEC: Uterine corpus endometrial carcinoma, AD: Alzheimer's disease, IBD: Inflammatory Bowel Disease, T2D: Type 2 Diabetes, BPSO-DT: Binary particle swarm optimization with decision tree, BiLSTM: Bidirectional Long Short Term Memory, ADNI: Alzheimer's Disease Neuroimaging Initiative, Demo.: Demographics, gPRS: genome-wide polygenic risk score, DLG: Deep learning genomics, FD: Fractal dimension, VD: Vascular density, PheWAS: Phenome-wide association study.
ACC: These are the average accuracies for the 5 algorithms in classifying different types of cardiomyopathies.
These studies demonstrate the power of AI algorithms in extracting meaningful patterns from complex genomic data, facilitating disease diagnosis, risk prediction, and the discovery of novel genetic biomarkers. This personalised approach to risk assessment signifies a promising step towards improving the precision of cardiovascular risk prediction and advancing precision medicine in the battle against CVD. By embracing such innovative approaches and continuing to explore the potential of genomics and AI, the future holds promise for a more proactive, personalised approach to CVD management, ultimately improving patient outcomes worldwide.
Discussion
In cardiovascular medicine, AI has distinct applications, ranging from the lab to the bedside and beyond. It has accelerated data analysis, identified genetic patterns, and predicted disease based on genetic information. Computational modelling leverages computers to simulate the behaviour of the human body, enabling personalised heart simulations and optimizing therapies tailored to individual patients. The construction of a personalised “digital twin” is a major goal of the synergistic integration of numerous cutting-edge technologies.99 This ground-breaking technology aims to seamlessly combine the distinct clinical data of each patient, which may come from various sources. Its goal is to provide clinicians with a never-before-seen level of individualised knowledge of a patient's overall cardiovascular health. This can completely transform how CVD are detected, treated, and managed. Notably, one of the distinct benefits of the “digital twin” idea is the capability to pretest cardiovascular therapies inside the patient's digital doppelganger. This cutting-edge method enables a complete and quantitative evaluation of possible outcomes without putting the actual patient through damaging operations.100 Synthetic data generation is valuable for tracking disease progression in large-scale clinical studies and addressing privacy and cost concerns.101 Meanwhile, integrated with AI analytics, mobile health devices and wearable technologies gather copious amounts of individual-level data, enabling detailed phenotyping of diseases and PM.
AI can automate diagnostic pathways, streamline patient triaging, and facilitate the identification of patients requiring further evaluation or monitoring for CVD, akin to other medical tests. This, in turn, can reduce patient waiting times and optimise clinical care, making the healthcare system more efficient. In the context of CVD, AI methods find utility in areas such as drug therapy,48 HF management,102 and risk stratification.103
Table 2 showcases the diverse applications of AI in CVD. The studies cover a wide range of applications, including identifying patients with acute decompensated heart failure (ADHF),104 predicting 30-day readmissions of HF patients,105 developing ML risk calculators for atherosclerotic cardiovascular disease (ASCVD),106 and predicting modified Rankin Scale (mRS) scores for stroke patients.107 Other applications include diagnosing asymptomatic left ventricular dysfunction (ALVD),108 predicting myocardial infarction (MI) from non-enhanced MRI,110 and assessing the risk of CVD mortality and hospitalization in hypertensive patients.113
Table 2.
Applications of AI in CVD.
| SN | Author & Year |
Disease Type |
Aim | Method | Dataset | N | Feature Types |
Performance Metrics |
Conclusion |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Blecker et al.104 (2018) | ADHF | To test 4 algorithms to identify patients with ADHF. | LR | Single center | 37,229 | EHR and clinical features | A1: Sen (0.98), PPV (0.14) A2: AUC (0.96) A3&A4: AUC (0.99) |
ML with the unstructured data had the best performance for identifying ADHF. |
| 2 | Golas et al.105 (2018) | HF | To develop this EMR-based prediction model for 30-day readmissions in HF patients. | LR, GBA, Maxout networks, DUNs | Hospitals in Partners Healthcare System | 11,510 | EHR | AUC: 0.664, 0.650, 0.695, 0.705 | At the classification threshold, the DUNs model had an accuracy of 76.4%, corresponding to the largest cost savings to the hospital. |
| 3 | Kakadiaris et al.106 (2018) | ASCVD | To develop a ML Risk Calculator. | SVM | MESA | 6459 | CRF | Sen:86%, Spe:95%, AUC:0.92 |
The FLEMENGHO-validated ML Risk Calculator outperformed the ACC/AHA Risk Calculator. |
| 4 | Xie et al.107 (2019) | Stroke | To predict mRS scores at 90 days. | XGB, GBM | Single center | 512 | CT Head and clinical parameters | AUC: 0.884, 0.877 | With a high AUC, decision tree-based GBMs may predict the recovery prognosis of stroke patients at admission. |
| 5 | Attia et al.108 (2019) | ALVD | Role of AI-based learning algorithms to diagnose asymptomatic left ventricular dysfunction. | CNN | Single center | 625,326 | ECG data | AUC: 0.93, Sen: 86.3%, Spe:85.7%, ACC: 85.7% | Patients without a positive AI test were four times more likely to acquire future ventricular dysfunction than those with a negative screen. |
| 6 | Lancaster et al.109 (2019) | CVD | Clustering for grouping directly observed diastolic function parameters and identifying patient phenotypes with similar behaviour to predict future adverse events. | Hierarchical clustering | Single center | 866 | Echocardiographic variables | Kappa: 0.41 (p < 0.001), 0.619 (p < 0.001) |
Using echocardiographic characteristics recommended by guidelines for assessing LVDD, hierarchical clustering produced groupings that distinguish patient prognosis better than standards-based categorization. |
| 7 | Zhang et al.110 (2019) | MI | To predict MI from non-enhanced MRI. | LSTM, Optical flow |
Single center | 299 | MRI images | AUC: 0.94, Sen: 89.8%, Spe: 99.1% |
Using nonenhanced cine MRI, the model could detect chronic MI (validated by LGE). |
| 8 | Jamthikar et al.19 (2020) | CVD/Stroke | To predict the risk of CVD/stroke on retrospective data while using EEGS as the surrogate endpoints. | RF | Single center | 202 | CRF and CUSIP | AUC: 0.99 | The AtheroRisk-integrated system outperforms the AtheroRisk-conventional system. |
| 9 | Chao et al.111 (2021) | CVD | CVD screening and risk prediction. | Deep CNN | NLST | 10,395 | LDCT | AUC: 0.871, 0.768 | DL can transform LDCT for lung cancer screening into a dual-screening quantitative tool for CVD risk estimate in high-risk individuals. |
| 10 | Krittanawong et al.112 (2021) | SCAD | To predict in-hospital mortality in patients with SCAD. | DL, AdaBoost, SVM, KNN, XGB, DT, LR, RF | Single center | 30,425 | EHR data | AUC: 0.98 (Best performing DL-algorithm) | DL outperformed both regression and ML models. |
| 11 | Lee et al.113 (2022) | CVD | Assess the risk of CVD mortality and hospitalization within a year in patients with hypertension. | LR and DNN | KNHIS | 2,037,027 | Clinical parameters | ACC: 92.5%, 78%, 86.3%, 65.5% | The DNN-based model showed higher performance than the LR in all the datasets. |
| 12 | Absi et al.114 (2022) | CVD | Distinguishing the CVD group from the non-CVD using DXA and retinal images. | CNN | QBB | 500 | Retinal images and DXA data | ACC: 78.3% | Integrating multi-modal data and DL techniques provides a fast and relatively non-invasive method for diagnosing CVD. |
ADHF: Acute decompensated heart failure, HF: heart failure, ASCVD: Atherosclerotic cardiovascular disease, ALVD: Asymptomatic left ventricular dysfunction, MI: Myocardial infarction, CVD: Cardiovascular disease, SCAD: Spontaneous coronary artery dissection, EMR: Electronic medical record, ML: Machine learning, AI: Artificial intelligence, mRS: modified Rankin Scale, MRI: Magnetic Resonance Imaging, EEGS: Event-equivalence gold standard, DXA: Dual-energy X-ray absorptiometry, LR: Logistic regression, GBA: Gradient boosting algorithm, DUNs: Deep unified networks, SVM: Support vector machine, XGB: Extreme gradient boosting, GBM: Gradient boosting machine, CNN: Convolutional neural network, LSTM: Long short term memory, RF: Random forest, KNN: K-nearest neighbor, DT: Decision tree, DNN: Deep neural network, MESA: Multi-Ethnic Study of Atherosclerosis, NLST: National Lung Screening Trial, KNHIS: Korean National Health Insurance Service, QBB: Qatar biobank, EHR: Electronic Health Record, CRF: Conventional risk factors, CT: Computed tomography, ECG: Electrocardiogram, CUSIP: Carotid ultrasound image pheno types, LDCT: Low dose computed tomography, Sen: Sensitivity, Spe: Specificity, AUC: Area under curve, PPV: Positive predictive value, ACC: Accuracy, DL: Deep learning, ACC/AHA: American College of Cardiology/American Heart Association, FLEMENGHO: Flemish Study of Environment, Genes and Health Outcomes, LVDD: Left ventricular diastolic dysfunction, LGE: Late gadolinium enhancement.
AI algorithms can diagnose heart murmurs,115 detect left ventricular dysfunction,116 and even identify HF patients through histological interpretation of biopsy slides.117 Moreover, blood flow dynamics within the aorta are simulated by AI-based computer models, which let physicians choose the best course of action based on the anatomy and features of each patient's aneurysm. AI algorithms can monitor alterations in the morphology of the aneurysm sac and detect patterns that indicate the effectiveness or failure of treatment by examining clinical factors and longitudinal imaging data. Personalised monitoring and intervention techniques can be made possible using AI-driven predictive models to stratify patients according to their risk of aneurysm rupture or progression.118 These strides in diagnostic precision carry substantial ramifications for clinical choices and the well-being of patients. PM in CVD is witnessing the convergence of genomics and clinical data with the revolutionary capabilities of DL for prognostic prediction.119 This approach is well-suited to address the complexity of heterogeneous multi-scale genomic profiles, commonly called multi-omics data.120 Precision cardiology is leading the way in this domain, which harnesses the vast potential of post-genomics advancements.
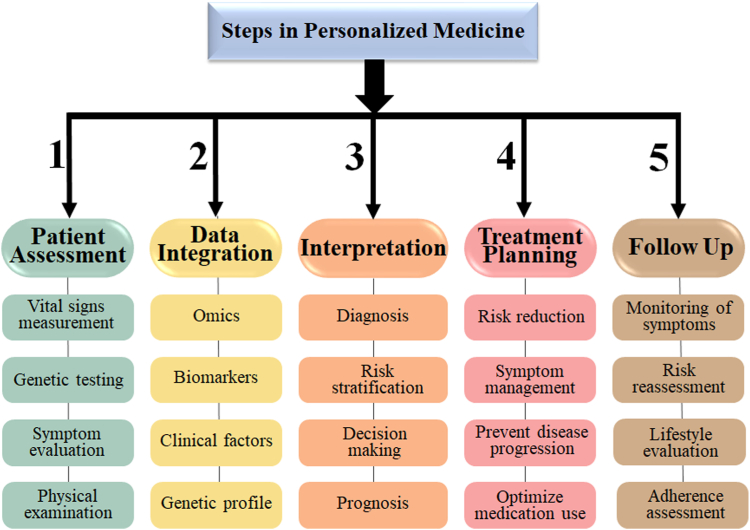
The personalised treatment for CVD follows a systematic approach (Fig. 7), beginning with comprehensive patient assessment, which includes vital signs measurement, genetic testing, symptom evaluation, physical examination, and history-taking to identify familial predispositions to cardiac disease. Data integration combines a variety of parameters such as omics, biomarkers, clinical aspects, and genetic profiles. The interpretation step includes diagnosis, risk stratification, decision-making, and prognosis. Based on this assessment, a treatment plan is developed, which includes risk reduction, symptom management, disease progression prevention, and drug optimization. Follow-up involves monitoring symptoms, analyzing risks, reviewing lifestyle changes, and maintaining drug adherence for successful management and improved cardiovascular health. Notably, a crucial feedback loop would be set up to allow the AI-driven PM algorithm to learn and enhance its effectiveness over time continuously. This iterative procedure allows the treatment plan to be dynamically adjusted in response to changes in the patient's state and the algorithm's evolving understanding, progressively refining the algorithm's ability to maintain the patient's health over time.
Fig. 7.
Five-step personalized treatment for CVDs.
Our study also has several limitations that should be acknowledged. First, the search strategy employed in this scoping review aimed to comprehensively capture relevant literature; however, there is a possibility that some relevant studies may have been overlooked. Despite efforts to employ systematic search techniques, the vast and diverse nature of the literature pertaining to PM and AI in CVD framework have led to the omission of certain relevant articles. Secondly, the inclusion criteria for this review focused on studies and literature specifically related to the intersection of PM, AI, and CVD. While this ensured relevance to our research objectives, it may have resulted in the exclusion of potentially valuable literature that intersects with related but distinct domains. Additionally, the synthesis of findings from diverse sources, including research articles, guidelines, conference proceedings, and dissertations, poses challenges in terms of data integration and interpretation. The heterogeneity in study designs, methodologies, and outcome measures across the included literature further complicates the synthesis and generalization of findings. Finally, our review primarily focused on summarizing existing literature and identifying gaps in knowledge. While we have highlighted implications for research, clinical practice, administration, and education, the translation of these implications into actionable strategies requires further investigation and validation. Future research should aim to address these limitations and build upon our findings to advance the understanding and application of PM, AI, and CVD in healthcare practice.
The healthcare field has already witnessed substantial progress due to AI, and there is optimistic anticipation that the accomplishments attained in cancer and other non-CVDs will likewise be mirrored in CVD. Integrating AI with PM is driving advancements in healthcare, enabling medical professionals and patients to access highly accurate medical diagnostic and treatment information. The confluence of these two factors contributes to the early identification and prevention of CVD in individuals, potentially decreasing the total burden of CVD and mitigating healthcare costs associated with preventive measures. The integration of AI algorithms facilitates the examination of extensive datasets encompassing genomes, proteomics, and clinical information, thereby enabling precise risk evaluations and personalised approaches to treatment. By adopting this synergistic strategy, CVD management is poised to witness significant advancements, resulting in enhanced cardiovascular health outcomes and a more robust population.
Contributors
M.S., J.S.S., V.R., and Z.R., did the conceptualization of the paper. N.N.K., J.R.L., and A.N., contributed to the analysis and interpretation of information. PRISMA was done by M.S., and J.S.S. The investigation was done by M.S., G.F., A.M.J., and L.E.M. J.F.E.F., J.S.T., and N.S. developed the methodology. M.M.F., R.S., and A.S., contributed to data verification. M.S., G.K., V.R., D.G. and I.M.S. gathered the required resources. L.S., A.F.L, D.L.B., D.G. and J.S.S. did the supervision. The figures were made by M.S. and A.K. The tables were made by M.S. The original draft was written by M.S. and J.S.S. K.T., M.A.M, E.R.I., S.C., K.I.P., D.P.M., V.V., M.K.K., reviewed and edited the entire paper. All authors agree to be fully accountable for ensuring the integrity and accuracy of the work and read and approved the final manuscript. The corresponding author had full access to all data in the study and assumed final responsibility for the decision to submit the manuscript for publication.
Data sharing statement
The data used in this scoping review is existing literature and publicly available.
Declaration of interests
The authors have no conflicts of interest to declare.
References
- 1.Vargas A.J., Harris C.C. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer. 2016;16(8):525–537. doi: 10.1038/nrc.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subramanian M., Wojtusciszyn A., Favre L., et al. Precision medicine in the era of artificial intelligence: implications in chronic disease management. J Transl Med. 2020;18:1–12. doi: 10.1186/s12967-020-02658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozomaro U., Wahlestedt C., Nemeroff C.B. Personalised medicine in psychiatry: problems and promises. BMC Med. 2013;11(1):1–35. doi: 10.1186/1741-7015-11-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogenberg F.R., Barash C.I., Pursel M. Personalised medicine: part 1: evolution and development into theranostics. P T. 2010;35(10):560. [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco-Gonzalez A., Cabezon A., Seco-Gonzalez A., et al. The role of ai in drug discovery: challenges, opportunities, and strategies. Pharmaceuticals. 2023;16(6):891. doi: 10.3390/ph16060891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumari V., Kumar N., Kumar K.S., et al. Deep learning paradigm and its bias for coronary artery wall segmentation in intravascular ultrasound scans: a closer look. J Cardiovasc Dev Dis. 2023;10(12):485. doi: 10.3390/jcdd10120485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boi A., Jamthikar A.D., Saba L., et al. A survey on coronary atherosclerotic plaque tissue characterization in intravascular optical coherence tomography. Curr Atheroscl Rep. 2018;20:1–17. doi: 10.1007/s11883-018-0736-8. [DOI] [PubMed] [Google Scholar]
- 8.Banchhor S.K., Araki T., Londhe N.D., et al. Five multiresolution-based calcium volume measurement techniques from coronary IVUS videos: a comparative approach. Comp Method Program Biomed. 2016;134:237–258. doi: 10.1016/j.cmpb.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Benjamin M.M., Rabbat M.G. Artificial intelligence in transcatheter aortic valve replacement: its current role and ongoing challenges. Diagnostics. 2024;14(3):261. doi: 10.3390/diagnostics14030261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho Y., Park S., Hwang S.H., et al. Aortic annulus detection based on deep learning for transcatheter aortic valve replacement using cardiac computed tomography. J Korean Med Sci. 2023;38(37) doi: 10.3346/jkms.2023.38.e306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jo S.D., Ku S.H., Won Y.-Y., Kim S.H., Kwon I.C. Targeted nanotheranostics for future personalized medicine: recent progress in cancer therapy. Theranostics. 2016;6(9):1362. doi: 10.7150/thno.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abul-Husn N.S., Owusu Obeng A., Sanderson S.C., Gottesman O., Scott S.A. Implementation and utilization of genetic testing in personalized medicine. Pharmacogenomics Pers Med. 2014:227–240. doi: 10.2147/PGPM.S48887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jena B., Saxena S., Nayak G.K., et al. Brain tumor characterization using radiogenomics in artificial intelligence framework. Cancers. 2022;14(16):4052. doi: 10.3390/cancers14164052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z., Ott P.A., Wu C.J. Towards personalised, tumour-specific, therapeutic vaccines for cancer. Nat Rev Immunol. 2018;18(3):168–182. doi: 10.1038/nri.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyles S.P., Hayden R.E., Meyer F.B., Terzic A. Regenerative medicine curriculum for next-generation physicians. NPJ Regen Med. 2019;4(1):3. doi: 10.1038/s41536-019-0065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxena S., Jena B., Mohapatra B., et al. Fused deep learning paradigm for the prediction of o6-methylguanine-DNA methyltransferase genotype in glioblastoma patients: a neuro-oncological investigation. Comput Biol Med. 2023;153:106492. doi: 10.1016/j.compbiomed.2022.106492. [DOI] [PubMed] [Google Scholar]
- 17.Maniruzzaman M., Rahman M.J., Ahammed B., et al. Statistical characterization and classification of colon microarray gene expression data using multiple machine learning paradigms. Comput Methods Program Biomed. 2019;176:173–193. doi: 10.1016/j.cmpb.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Al-Maini M., Maindarkar M., Kitas G.D., et al. Artificial intelligence-based preventive, personalized and precision medicine for cardiovascular disease/stroke risk assessment in rheumatoid arthritis patients: a narrative review. Rheumatol Intern. 2023;43(11):1965–1982. doi: 10.1007/s00296-023-05415-1. [DOI] [PubMed] [Google Scholar]
- 19.Jamthikar A., Gupta D., Khanna N.N., Saba L., Laird J.R., Suri J.S. Cardiovascular/stroke risk prevention: a new machine learning framework integrating carotid ultrasound image-based phenotypes and its harmonics with conventional risk factors. Ind Heart J. 2020;72(4):258–264. doi: 10.1016/j.ihj.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khanna N.N., Singh M., Maindarkar M., et al. Polygenic risk score for cardiovascular diseases in artificial intelligence paradigm: a review. J Korean Med Sci. 2023;38(46) doi: 10.3346/jkms.2023.38.e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saba L., Maindarkar M., Khanna N.N., et al. A pharmaceutical paradigm for cardiovascular composite risk assessment using novel radiogenomics risk predictors in precision explainable artificial intelligence framework. Clin Trial Tool Front Biosci. 2023;28(10) doi: 10.31083/j.fbl2810248. [DOI] [PubMed] [Google Scholar]
- 22.Bhagawati M., Paul S., Agarwal S., et al. Cardiovascular disease/stroke risk stratification in deep learning framework: a review. Cardiovasc Diagn Ther. 2023;13(3):557. doi: 10.21037/cdt-22-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain P.K., Dubey A., Saba L., et al. Attention-based UNet Deep Learning model for Plaque segmentation in carotid ultrasound for stroke risk stratification: an artificial Intelligence paradigm. J Cardiovasc Dev Dis. 2022;9(10):326. doi: 10.3390/jcdd9100326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxena S., Jena B., Gupta N., et al. Role of artificial intelligence in radiogenomics for cancers in the era of precision medicine. Cancers. 2022;14(12):2860. doi: 10.3390/cancers14122860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suri J.S., Paul S., Maindarkar M.A., et al. Cardiovascular/stroke risk stratification in Parkinson’s disease patients using atherosclerosis pathway and artificial intelligence paradigm: a systematic review. Metabolites. 2022;12(4):312. doi: 10.3390/metabo12040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancinelli L., Cronin M., Sadée W. Pharmacogenomics: the promise of personalised medicine. AAPS PharmSci. 2000;2:29–41. doi: 10.1208/ps020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hood L., Heath J.R., Phelps M.E., Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306(5696):640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 28.Flores M., Glusman G., Brogaard K., Price N.D., Hood L. P4 medicine: how systems medicine will transform the healthcare sector and society. Personal Med. 2013;10(6):565–576. doi: 10.2217/PME.13.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banchhor S.K., Londhe N.D., Araki T., et al. Calcium detection, its quantification, and grayscale morphology-based risk stratification using machine learning in multimodality big data coronary and carotid scans: a review. Comput Biol Med. 2018;101:184–198. doi: 10.1016/j.compbiomed.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Jamthikar A., Gupta D., Johri A.M., Mantella L.E., Saba L., Suri J.S. A machine learning framework for risk prediction of multi-label cardiovascular events based on focused carotid plaque B-Mode ultrasound: a Canadian study. Comput Biol Med. 2022;140:105102. doi: 10.1016/j.compbiomed.2021.105102. [DOI] [PubMed] [Google Scholar]
- 31.Tolani P., Gupta S., Yadav K., Aggarwal S., Yadav A.K. Big data, integrative omics and network biology. Adv Protein Chem Struct Biol. 2021;127:127–160. doi: 10.1016/bs.apcsb.2021.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Shen F., Narayanan R., Suri J.S. 2008 30th annual international conference of the IEEE Engineering in Medicine and Biology Society. IEEE; 2008. Rapid motion compensation for prostate biopsy using GPU. [DOI] [PubMed] [Google Scholar]
- 33.Narayanan R., Werahera P., Barqawi A., et al. Adaptation of a 3D prostate cancer atlas for transrectal ultrasound guided target-specific biopsy. Phys Med Biol. 2008;53(20):N397. doi: 10.1088/0031-9155/53/20/N03. [DOI] [PubMed] [Google Scholar]
- 34.Munjral S., Maindarkar M., Ahluwalia P., et al. Cardiovascular risk stratification in diabetic retinopathy via atherosclerotic pathway in COVID-19/non-COVID-19 frameworks using artificial intelligence paradigm: a narrative review. Diagnostics. 2022;12(5):1234. doi: 10.3390/diagnostics12051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paul S., Maindarkar M., Saxena S., et al. Bias investigation in artificial intelligence systems for early detection of Parkinson’s disease: a narrative review. Diagnostics. 2022;12(1):166. doi: 10.3390/diagnostics12010166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Baz A., Suri J.S. CRC Press; 2019. Big data in multimodal medical imaging. [Google Scholar]
- 37.Choudhary R., Gianey H.K. 2017 international conference on machine learning and data science (MLDS) IEEE; 2017. Comprehensive review on supervised machine learning algorithms. [Google Scholar]
- 38.Shrivastava V.K., Londhe N.D., Sonawane R.S., Suri J.S. A novel and robust Bayesian approach for segmentation of psoriasis lesions and its risk stratification. Comput Meth Program Biomed. 2017;150:9–22. doi: 10.1016/j.cmpb.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Araki T., Ikeda N., Shukla D., et al. PCA-based polling strategy in machine learning framework for coronary artery disease risk assessment in intravascular ultrasound: a link between carotid and coronary grayscale plaque morphology. Comput Method Program Biomed. 2016;128:137–158. doi: 10.1016/j.cmpb.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Saba L., Tiwari A., Biswas M., et al. Wilson’s disease: a new perspective review on its genetics, diagnosis and treatment. Front Biosci Elite. 2019;11(1):166–185. doi: 10.2741/E854. [DOI] [PubMed] [Google Scholar]
- 41.Johnson K.W., Torres Soto J., Glicksberg B.S., et al. Artificial intelligence in cardiology. J Am Coll Cardiol. 2018;71(23):2668–2679. doi: 10.1016/j.jacc.2018.03.521. [DOI] [PubMed] [Google Scholar]
- 42.Fan J., Han F., Liu H. Challenges of big data analysis. Natl Sci Rev. 2014;1(2):293–314. doi: 10.1093/nsr/nwt032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khanna N.N., Jamthikar A.D., Araki T., et al. Nonlinear model for the carotid artery disease 10-year risk prediction by fusing conventional cardiovascular factors to carotid ultrasound image phenotypes: a Japanese diabetes cohort study. Echocardiography. 2019;36(2):345–361. doi: 10.1111/echo.14242. [DOI] [PubMed] [Google Scholar]
- 44.Johnson K.B., Wei W.Q., Weeraratne D., et al. Precision medicine, AI, and the future of personalized health care. Clin Transl Sci. 2021;14(1):86–93. doi: 10.1111/cts.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckhardt C.M., Madjarova S.J., Williams R.J., et al. Unsupervised machine learning methods and emerging applications in healthcare. Knee Surgery. Sport Traumatol Arthrosc. 2023;31(2):376–381. doi: 10.1007/s00167-022-07233-7. [DOI] [PubMed] [Google Scholar]
- 46.Mahmud M., Kaiser M.S., Hussain A., Vassanelli S. Applications of deep learning and reinforcement learning to biological data. IEEE Transact Neural Networks Learn Syst. 2018;29(6):2063–2079. doi: 10.1109/TNNLS.2018.2790388. [DOI] [PubMed] [Google Scholar]
- 47.Saba L., Biswas M., Kuppili V., et al. The present and future of deep learning in radiology. Eur J Radiol. 2019;114:14–24. doi: 10.1016/j.ejrad.2019.02.038. [DOI] [PubMed] [Google Scholar]
- 48.Zhang S., Bamakan S.M.H., Qu Q., Li S. Learning for personalized medicine: a comprehensive review from a deep learning perspective. IEEE Review Biomed Eng. 2018;12:194–208. doi: 10.1109/RBME.2018.2864254. [DOI] [PubMed] [Google Scholar]
- 49.Janowczyk A., Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inf. 2016;7(1):29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alharbi W.S., Rashid M. A review of deep learning applications in human genomics using next-generation sequencing data. Hum Genom. 2022;16(1):1–20. doi: 10.1186/s40246-022-00396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solares J.R.A., Raimondi F.E.D., Zhu Y., et al. Deep learning for electronic health records: a comparative review of multiple deep neural architectures. J Biomed Informat. 2020;101:103337. doi: 10.1016/j.jbi.2019.103337. [DOI] [PubMed] [Google Scholar]
- 52.Tandel G.S., Tiwari A., Kakde O.G., Gupta N., Saba L., Suri J.S. Role of ensemble deep learning for brain tumor classification in multiple magnetic resonance imaging sequence data. Diagnostics. 2023;13(3):481. doi: 10.3390/diagnostics13030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khanna N.N., Maindarkar M.A., Viswanathan V. et al. Economics of artificial intelligence in healthcare: diagnosis vs. treatment. 2022 doi: 10.3390/healthcare10122493. Healthcare: MDPI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das S., Nayak G.K., Saba L., Kalra M., Suri J.S., Saxena S. An artificial intelligence framework and its bias for brain tumor segmentation: a narrative review. Comput Biol Med. 2022;143:105273. doi: 10.1016/j.compbiomed.2022.105273. [DOI] [PubMed] [Google Scholar]
- 55.Suri J.S., Bhagawati M., Paul S., et al. Understanding the bias in machine learning systems for cardiovascular disease risk assessment: the first of its kind review. Comput Biol Med. 2022;142:105204. doi: 10.1016/j.compbiomed.2021.105204. [DOI] [PubMed] [Google Scholar]
- 56.Pray L. Discovery of DNA structure and function: watson and crick. Nature Educ. 2008;1(1):100. [Google Scholar]
- 57.Gibbs R.A. The human genome project changed everything. Nat Rev Genet. 2020;21(10):575–576. doi: 10.1038/s41576-020-0275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collins F.S., Morgan M., Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300(5617):286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 59.Kim D.H., Kim Y.S., Son N.I., Kang C.K., Kim A.R. Recent omics technologies and their emerging applications for personalised medicine. IET Syst Biol. 2017;11(3):87–98. doi: 10.1049/iet-syb.2016.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ibrahim R., Pasic M., Yousef G.M. Omics for personalised medicine: defining the current we swim in. Expert Rev Mol Diagn. 2016;16(7):719–722. doi: 10.1586/14737159.2016.1164601. [DOI] [PubMed] [Google Scholar]
- 61.Foster M.W., Sharp R.R. Out of sequence: how consumer genomics could displace clinical genetics. Nat Rev Genet. 2008;9(6):419. doi: 10.1038/nrg2374. [DOI] [PubMed] [Google Scholar]
- 62.Khan R., Mittelman D. Consumer genomics will change your life, whether you get tested or not. Genome Biol. 2018;19(1):1–4. doi: 10.1186/s13059-018-1506-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gonzalez-Angulo A.M., Hennessy B.T., Mills G.B. Future of personalised medicine in oncology: a systems biology approach. J Clin Oncol. 2010;28(16):2777. doi: 10.1200/JCO.2009.27.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knowles L., Luth W., Bubela T. Paving the road to personalised medicine: recommendations on regulatory, intellectual property and reimbursement challenges. J Law Biosci. 2017;4(3):453–506. doi: 10.1093/jlb/lsx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uddin M., Wang Y., Woodbury-Smith M. Artificial intelligence for precision medicine in neurodevelopmental disorders. NPJ Digit Med. 2019;2(1):112. doi: 10.1038/s41746-019-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moon H., Ahn H., Kodell R.L., Baek S., Lin C.-J., Chen J.J. Ensemble methods for classification of patients for personalized medicine with high-dimensional data. Artif Intell Med. 2007;41(3):197–207. doi: 10.1016/j.artmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 67.Lin S., Schorpp K., Rothenaigner I., Hadian K. Image-based high-content screening in drug discovery. Drug Discov Today. 2020;25(8):1348–1361. doi: 10.1016/j.drudis.2020.06.001. [DOI] [PubMed] [Google Scholar]
- 68.Hamamoto R., Komatsu M., Takasawa K., Asada K., Kaneko S. Epigenetics analysis and integrated analysis of multiomics data, including epigenetic data, using artificial intelligence in the era of precision medicine. Biomolecules. 2019;10(1):62. doi: 10.3390/biom10010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bellazzi R., Zupan B. Predictive data mining in clinical medicine: current issues and guidelines. Int J Med Inf. 2008;77(2):81–97. doi: 10.1016/j.ijmedinf.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 70.Suri J.S., Rangayyan R.M. SPIE; Bellingham, WA, USA: 2006. Breast imaging, mammography, and computer-aided diagnosis of Breast cancer. [Google Scholar]
- 71.Saba L., Nardi V., Cau R., et al. Carotid artery plaque calcifications: lessons from histopathology to diagnostic imaging. Stroke. 2022;53(1):290–297. doi: 10.1161/STROKEAHA.121.035692. [DOI] [PubMed] [Google Scholar]
- 72.Cau R., Falaschi Z., Paschè A., et al. CT findings of COVID-19 pneumonia in ICU-patients. J Public Health Res. 2021;10(3) doi: 10.4081/jphr.2021.2270. jphr.2021.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nayak D.S.K., Mahapatra S., Routray S.P., et al. aiGeneR 1.0: An Artificial Intelligence Technique for the Revelation of Informative and Antibiotic Resistant Genes in Escherichia coli. Front Biosci Landmark. 2024;29(2):82. doi: 10.31083/j.fbl2902082. [DOI] [PubMed] [Google Scholar]
- 74.Acharya U.R., Mookiah M.R.K., Sree S.V., et al. Evolutionary algorithm-based classifier parameter tuning for automatic ovarian cancer tissue characterization and classification. Ultraschall in der Medizin-European. J Ultrasound. 2014;35(03):237–245. doi: 10.1055/s-0032-1330336. [DOI] [PubMed] [Google Scholar]
- 75.Singh J., Khanna N.N., Rout R.K., et al. GeneAI 3.0: powerful, novel, generalized hybrid and ensemble deep learning frameworks for miRNA species classification of stationary patterns from nucleotides. Sci Rep. 2024;14(1):7154. doi: 10.1038/s41598-024-56786-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ouyang F.-s., Guo B.-l., Ouyang L.-z., et al. Comparison between linear and nonlinear machine-learning algorithms for the classification of thyroid nodules. Eur J Radiol. 2019;113:251–257. doi: 10.1016/j.ejrad.2019.02.029. [DOI] [PubMed] [Google Scholar]
- 77.Biswas M., Kuppili V., Araki T., et al. Deep learning strategy for accurate carotid intima-media thickness measurement: an ultrasound study on Japanese diabetic cohort. Comput Biol Med. 2018;98:100–117. doi: 10.1016/j.compbiomed.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 78.Kato R.M., Parizotto Jd.O.L., Oliveira Ph.Jd., Gonçalves J.R. Artificial intelligence in orthognathic surgery–a narrative review of surgical digital tools and 3D orthognathic surgical planning. J Calif Dental Assoc. 2023;51(1):2202444. [Google Scholar]
- 79.Murray C.J., Lopez A.D. Alternative projections of mortality and disability by cause 1990–2020: global burden of disease study. Lancet. 1997;349(9064):1498–1504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- 80.Bowry A.D., Lewey J., Dugani S.B., Choudhry N.K. The burden of cardiovascular disease in low-and middle-income countries: epidemiology and management. Can J Cardiol. 2015;31(9):1151–1159. doi: 10.1016/j.cjca.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 81.Roth G.A., Abate D., Abate K.H., et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1736–1788. doi: 10.1016/S0140-6736(18)32203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amini M., Zayeri F., Salehi M. Trend analysis of cardiovascular disease mortality, incidence, and mortality-to-incidence ratio: results from global burden of disease study 2017. BMC Publ Health. 2021;21(1):1–12. doi: 10.1186/s12889-021-10429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O'Donnell C.J., Nabel E.G. Genomics of cardiovascular disease. N Engl J Med. 2011;365(22):2098–2109. doi: 10.1056/NEJMra1105239. [DOI] [PubMed] [Google Scholar]
- 84.Arnett D.K., Baird A.E., Barkley R.A., et al. Relevance of genetics and genomics for prevention and treatment of cardiovascular disease: a scientific statement from the American Heart Association Council on Epidemiology and Prevention, the Stroke Council, and the Functional Genomics and Translational Biology Interdisciplinary Working Group. Circulation. 2007;115(22):2878–2901. doi: 10.1161/CIRCULATIONAHA.107.183679. [DOI] [PubMed] [Google Scholar]
- 85.Rumsfeld J.S., Joynt K.E., Maddox T.M. Big data analytics to improve cardiovascular care: promise and challenges. Nat Rev Cardiol. 2016;13(6):350–359. doi: 10.1038/nrcardio.2016.42. [DOI] [PubMed] [Google Scholar]
- 86.Zhou S.-s., Jin J.-p., Wang J.-q., et al. miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacologica Sinica. 2018;39(7):1073–1084. doi: 10.1038/aps.2018.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Holland A., Enrick M., Diaz A., Yin L. Is miR-21 A therapeutic target in cardiovascular disease? Int J Drug Discov Pharmacol. 2023;2(1):26. doi: 10.53941/ijddp.0201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chistiakov D.A., Orekhov A.N., Bobryshev Y.V. The role of miR-126 in embryonic angiogenesis, adult vascular homeostasis, and vascular repair and its alterations in atherosclerotic disease. J Mol Cell Cardiol. 2016;97:47–55. doi: 10.1016/j.yjmcc.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 89.Phan J.H., Quo C.F., Wang M.D. Cardiovascular genomics: a biomarker identification pipeline. IEEE Trans Inf Technol Biomed. 2012;16(5):809–822. doi: 10.1109/TITB.2012.2199570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alimadadi A., Manandhar I., Aryal S., Munroe P.B., Joe B., Cheng X. Machine learning-based classification and diagnosis of clinical cardiomyopathies. Physiol Genomics. 2020;52(9):391–400. doi: 10.1152/physiolgenomics.00063.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Steinfeldt J., Buergel T., Loock L., et al. Neural network-based integration of polygenic and clinical information: development and validation of a prediction model for 10-year risk of major adverse cardiac events in the UK Biobank cohort. Lancet Digit Health. 2022;4(2):e84–e94. doi: 10.1016/S2589-7500(21)00249-1. [DOI] [PubMed] [Google Scholar]
- 92.Kwon O.-S., Hong M., Kim T.-H., et al. Genome-wide association study-based prediction of atrial fibrillation using artificial intelligence. Open Heart. 2022;9(1) doi: 10.1136/openhrt-2021-001898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Venkat V., Abdelhalim H., DeGroat W., Zeeshan S., Ahmed Z. Investigating genes associated with heart failure, atrial fibrillation, and other cardiovascular diseases, and predicting disease using machine learning techniques for translational research and precision medicine. Genomics. 2023;115(2):110584. doi: 10.1016/j.ygeno.2023.110584. [DOI] [PubMed] [Google Scholar]
- 94.Khalifa N.E.M., Taha M.H.N., Ali D.E., Slowik A., Hassanien A.E. Artificial intelligence technique for gene expression by tumor RNA-Seq data: a novel optimized deep learning approach. IEEE Access. 2020;8:22874–22883. [Google Scholar]
- 95.Peng J., Li J., Han R., et al. A deep learning-based genome-wide polygenic risk score for common diseases identifies individuals with risk. medRxiv. 2021:2021.11.. 17.21265352. [Google Scholar]
- 96.Li L., Huang Y., Han Y. In: Use of deep learning genomics to discriminate Alzheimer’s disease and healthy controls. 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) Jiang J., editor. IEEE; 2021. [DOI] [PubMed] [Google Scholar]
- 97.Zekavat S.M., Raghu V.K., Trinder M., et al. Deep learning of the retina enables phenome-and genome-wide analyses of the microvasculature. Circulation. 2022;145(2):134–150. doi: 10.1161/CIRCULATIONAHA.121.057709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hahn S.-J., Kim S., Choi Y.S., Lee J., Kang J. Prediction of type 2 diabetes using genome-wide polygenic risk score and metabolic profiles: a machine learning analysis of population-based 10-year prospective cohort study. EBioMedicine. 2022:86. doi: 10.1016/j.ebiom.2022.104383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Corral-Acero J., Margara F., Marciniak M., et al. The ‘Digital Twin’to enable the vision of precision cardiology. Eur Heart J. 2020;41(48):4556–4564. doi: 10.1093/eurheartj/ehaa159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Coorey G., Figtree G.A., Fletcher D.F., et al. The health digital twin to tackle cardiovascular disease—a review of an emerging interdisciplinary field. NPJ Digit Med. 2022;5(1):126. doi: 10.1038/s41746-022-00640-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jacobs F., D’Amico S., Benvenuti C., et al. Opportunities and challenges of synthetic data generation in oncology. JCO Clin Cancer Informat. 2023;7 doi: 10.1200/CCI.23.00045. [DOI] [PubMed] [Google Scholar]
- 102.Bui A.L., Fonarow G.C. Home monitoring for heart failure management. J Am Coll Cardiol. 2012;59(2):97–104. doi: 10.1016/j.jacc.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Peters S.A., Den Ruijter H.M., Bots M.L., Moons K.G. Improvements in risk stratification for the occurrence of cardiovascular disease by imaging subclinical atherosclerosis: a systematic review. Heart. 2012;98(3):177–184. doi: 10.1136/heartjnl-2011-300747. [DOI] [PubMed] [Google Scholar]
- 104.Blecker S., Sontag D., Horwitz L.I., et al. Early identification of patients with acute decompensated heart failure. J Cardiac Fail. 2018;24(6):357–362. doi: 10.1016/j.cardfail.2017.08.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Golas S.B., Shibahara T., Agboola S., et al. A machine learning model to predict the risk of 30-day readmissions in patients with heart failure: a retrospective analysis of electronic medical records data. BMC Med Informat Decision Making. 2018;18(1):1–17. doi: 10.1186/s12911-018-0620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kakadiaris I.A., Vrigkas M., Yen A.A., Kuznetsova T., Budoff M., Naghavi M. Machine learning outperforms ACC/AHA CVD risk calculator in MESA. J Am Heart Assoc. 2018;7(22) doi: 10.1161/JAHA.118.009476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie Y., Jiang B., Gong E., et al. Use of gradient boosting machine learning to predict patient outcome in acute ischemic stroke on the basis of imaging, demographic, and clinical information. Am J Roentgenol. 2019;212(1):44–51. doi: 10.2214/AJR.18.20260. [DOI] [PubMed] [Google Scholar]
- 108.Attia Z.I., Kapa S., Lopez-Jimenez F., et al. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nature Med. 2019;25(1):70–74. doi: 10.1038/s41591-018-0240-2. [DOI] [PubMed] [Google Scholar]
- 109.Lancaster M.C., Salem Omar A.M., Narula S., Kulkarni H., Narula J., Sengupta P.P. Phenotypic clustering of left ventricular diastolic function parameters: patterns and prognostic relevance. JACC Cardiovasc Imag. 2019;12(7 Part 1):1149–1161. doi: 10.1016/j.jcmg.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 110.Zhang N., Yang G., Gao Z., et al. Deep learning for diagnosis of chronic myocardial infarction on nonenhanced cardiac cine MRI. Radiology. 2019;291(3):606–617. doi: 10.1148/radiol.2019182304. [DOI] [PubMed] [Google Scholar]
- 111.Chao H., Shan H., Homayounieh F., et al. Deep learning predicts cardiovascular disease risks from lung cancer screening low dose computed tomography. Nature Commun. 2021;12(1):2963. doi: 10.1038/s41467-021-23235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Krittanawong C., Virk H.U.H., Kumar A., et al. Machine learning and deep learning to predict mortality in patients with spontaneous coronary artery dissection. Sci Rep. 2021;11(1):8992. doi: 10.1038/s41598-021-88172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee S.-J., Lee S.-H., Choi H.-I., et al. Deep Learning Improves Prediction of Cardiovascular Disease-Related Mortality and Admission in Patients with Hypertension: Analysis of the Korean National Health Information Database. J Clin Med. 2022;11(22):6677. doi: 10.3390/jcm11226677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Al-Absi H.R., Islam M.T., Refaee M.A., Chowdhury M.E., Alam T. Cardiovascular disease diagnosis from DXA scan and retinal images using deep learning. Sensors. 2022;22(12):4310. doi: 10.3390/s22124310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dominguez-Morales J.P., Jimenez-Fernandez A.F., Dominguez-Morales M.J., Jimenez-Moreno G. Deep neural networks for the recognition and classification of heart murmurs using neuromorphic auditory sensors. IEEE Transact Biomed Circuit Syst. 2017;12(1):24–34. doi: 10.1109/TBCAS.2017.2751545. [DOI] [PubMed] [Google Scholar]
- 116.Vaid A., Johnson K.W., Badgeley M.A., et al. Using deep-learning algorithms to simultaneously identify right and left ventricular dysfunction from the electrocardiogram. Cardiovasc Imag. 2022;15(3):395–410. doi: 10.1016/j.jcmg.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cooper L.T., Baughman K.L., Feldman A.M., et al. The role of endomyocardial biopsy in the management of cardiovascular disease: a scientific statement from the American Heart Association, the American College of Cardiology, and the European Society of Cardiology. Circulation. 2007;116(19):2216–2233. doi: 10.1161/CIRCULATIONAHA.107.186093. [DOI] [PubMed] [Google Scholar]
- 118.Pietrzak K., Maciąg R., Terlecki P. Prevention and management of type II endoleaks after endovascular aneurysm repair. Acta Angiol. 2023;29(4):141–148. [Google Scholar]
- 119.Lin J., Ngiam K.Y. How data science and AI-based technologies impact genomics. Singap Med J. 2023;64(1):59. doi: 10.4103/singaporemedj.SMJ-2021-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reilly M.P., Bornfeldt K.E. Integrative Multiomics approaches for discovery of new drug targets for cardiovascular disease. Am Heart Assoc. 2021:2471–2474. doi: 10.1161/CIRCULATIONAHA.121.054900. [DOI] [PMC free article] [PubMed] [Google Scholar]