Abstract
Many Gram-positive spore-forming rhizobacteria of the genus Bacillus show potential as biocontrol biopesticides that promise improved sustainability and ecological safety in agriculture. Here, we present a draft-quality genome sequence for Bacillus velezensis EU07, which shows growth-promotion in tomato plants and biocontrol against Fusarium head blight. We found that the genome of EU07 is almost identical to that of the commercially used strain QST713, but identified 46 single-nucleotide differences that distinguish these strains from each other. The availability of this genome sequence will facilitate future efforts to unravel the genetic and molecular basis for EU07's beneficial properties.
Keywords: Bacillus velezensis, biological control, genome sequence, plant-growth promoting
Data Summary
In this study, we generated genome sequence data, which has been deposited in public databases:
National Center for Biotechnology Information (NCBI) BioProject accession number PRJNA743875 – https://www.ncbi.nlm.nih.gov/bioproject/743875
Assembly NCBI GenBank accession number GCA_019997305.2 – https://www.ncbi.nlm.nih.gov/nuccore/JAIFZJ000000000
NCBI RefSeq accession number GCF_019997305.2
NCBI Sequence Read Archive (SRA) accession number SRR27184279.
Introduction
Many Gram-positive spore-forming rhizobacteria of the genus Bacillus show potential as biocontrol biopesticides that promise improved sustainability and ecological safety in agriculture [1,3]. Here, we present genomic sequencing data for Bacillus strain Egem-Utku 07, hereafter known as EU07. This strain was previously isolated from the rhizosphere of diseased tomato plants [4] in an effort to collect strains that could inhibit the soilborne pathogen Fusarium oxysporum f. sp. radicis-lycopersici [4], which causes crown rot in tomato. We demonstrated that EU07 inhibits this pathogen in vitro [4]. Furthermore, EU07 promotes growth and inhibits fusarium head blight in planta [5]. We previously established that EU07 is a member of the genus Bacillus, but its precise species identity was ambiguous. Furthermore, in the absence of sequence data, little was known about the potential molecular mechanisms for its beneficial properties. Here, we present a draft-quality genome sequence assembly and genomic sequence reads from strain EU07. This dataset will help in better understanding EU07’s phylogeny and taxonomy, and provide a resource to assist elucidation of the molecular mechanisms of EU07’s beneficial traits.
Methods
Bacterial strain and isolation of genomic DNA
We isolated genomic DNA from bacterial strain EU07 from fresh liquid culture grown for 24 h in nutrient broth pH 7.2. We note that this medium provides a laboratory environment quite different from the bacterium’s normal soil environment. The liquid culture was inoculated from a single colony and, therefore, was assumed to be clonal. We used the ISOLATE II genomic DNA kit (Bioline), following the manufacturer’s instructions. The quality and concentration of the genomic DNA were assessed using a NanoDrop 2000c spectrophotometer (ThermoFisher Scientific).
DNA sequencing
Genomic DNA was sent to the University of Exeter’s Sequencing Facility (https://biosciences.exeter.ac.uk/sequencing/) for Illumina Nextera XT library preparation and sequencing on the Illumina MiSeq platform to generate 748 528 pairs of 300 bp reads with a mean insert size of approximately 400 bp.
Genome sequence assembly
We performed adapter trimming and quality filtering on the MiSeq reads using Trim Galore version 0.6.7 [6], which incorporates Cutadapt version 3.5 [7]. The -q parameter was set to 30 and we used the --paired option. The resulting cleaned read-pairs served as input for de novo assembly using SPAdes version 3.13.1 [8] with the --careful option. The resulting scaffolds and contigs were re-ordered against the reference genome of strain FZB42 with the Mauve Contig Mover [9]. Annotation was added by the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline version 6.6 [10] after submission of the genome assembly. The command lines are documented in GitHub at https://github.com/davidjstudholme/bacillus_EU07/tree/main/assembly and in the Zenodo repository (https://doi.org/10.5281/zenodo.10968102) [11].
Assessment of genome-assembly quality
We calculated assembly statistics using quast version 5.2.0 [12]. We checked read coverage of the genome assembly by aligning the EU07 reads against the EU07 assembly and calculating alignment statistics with Qualimap version 2.3 [13]. The alignment was performed using bwa-mem version 0.7.17 [14]; then, we reformatted and sorted the output using SAMtools version 1.13 [15]. The full details of the command lines are documented at https://github.com/davidjstudholme/bacillus_EU07/blob/main/assemblyQC/README.md and in the Zenodo repository [11].
Average nucleotide identity (ANI)
We used fastANI [16] to calculate ANI between the genome of EU07 and each of the Bacillus amyloliquefaciens group (taxonomy ID: 1938374) genome assemblies retrieved from GenBank [17,18]. The exact command lines are documented in GitHub at https://github.com/davidjstudholme/bacillus_EU07/ and in the Zenodo repository [11].
Phylogenomics
To generate a maximum-likelihood phylogenetic tree based on genome-wide SNPs, we used PhaME [19] with FastTree [20]. The exact command lines used are documented at https://github.com/davidjstudholme/bacillus_EU07/ and in the Zenodo repository [11]. The resulting tree was rendered using the Interactive Tree of Life (iTOL) 6.8.1 [21].
Whole-genome alignment
Genome sequences were aligned using progressiveMauve version 2.4.0 [22] after first re-ordering the contigs against the reference genome of strain KNU-28 [23] with the Mauve Contig Mover [9]. The resulting alignment was visualized using Mauve snapshot_2015-02-25 [24]. The exact command lines used are documented at https://github.com/davidjstudholme/bacillus_EU07/ and in the Zenodo repository [11].
Further whole-genome analyses
We used the Proksee web server [25] to perform several analyses of the assembled EU07 genome. This included blastn searches against 888 related genomes, annotation of horizontally acquired genomic regions with Alien Hunter [26], and identification of bacteriophage sequences using VirSorter [27,28] and Phigaro [29]. Variant-calling was performed using the Parsnp tool in Harvest [30].
Results and discussion
Genome sequencing and assembly
We generated 748 528 pairs of 300 bp Illumina MiSeq sequencing reads from EU07 genomic DNA. This represents approximately 100× coverage of the 4.2 Mbp genome. Trimming and filtering with Trim Galore left 715 442 pairs of reads, with lengths ranging from 20 to 300 bp. De novo assembly with SPAdes yielded 266 contigs with a total length of 4.2 Mbp and N50 length of 52.8 kb. This was deposited in GenBank via the NCBI Submission Portal under accession number GCA_019997305.2. The NCBI’s contamination filtering removed 5 contigs, leaving 261. The NCBI PGAP annotation system predicted 4 273 genes, of which 4 081 encode putative proteins. The results of NCBI’s quality check with CheckM v1.2.2 [31,32] revealed a completeness of 98.16 % (85th percentile) and 0.47 % contamination.
Alignment of sequencing reads against the genome assembly and analysis with Qualimap revealed a mean coverage of 93.25× and standard deviation of 89.87. Almost all of the genome assembly (99.96 %) had at least 1× coverage, and 97.59 % of the assembly had at least 10× coverage. The full Qualimap report and output files are available in the Zenodo repository (https://doi.org/10.5281/zenodo.10968102) [11], allowing users of this data to take coverage into account when performing analyses. We note that the contig with least coverage is JAIFZJ020000237.1, having only 1.04× coverage. Nevertheless, blast searches reveal that this contig shows very high levels of sequence similarity to genomes of other Bacillus velezensis strains, increasing confidence in its validity.
EU07 belongs to the species B. velezensis
Previously, the phylogenetic and taxonomic position of strain EU07 had been ambiguous and we previously referred to it as ‘B. sp.’ and ‘B. subtilis’ [4,5]. To identify the species to which strain EU07 belongs, we uploaded the genome assembly to the Type Strain Genome Server (TYGS) [33]. This classified EU07 to the species B. amyloliquefaciens. Among the sequenced type strains in TYGS, the most similar to EU07 was FZB42 [34], which is the type strain of B. amyloliquefaciens subsp. plantarum [35]. However, this taxon is now considered to be synonymous with B. velezensis and distinct from B. amyloliquefaciens [36]. Hereafter, we refer to our strain as B. velezensis EU07.
EU07 belongs to a clade of plant-associated strains of B. velezensis
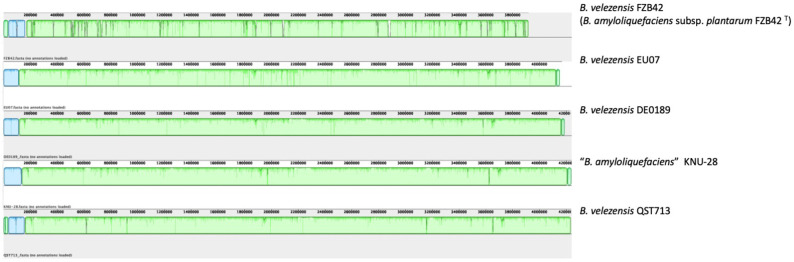
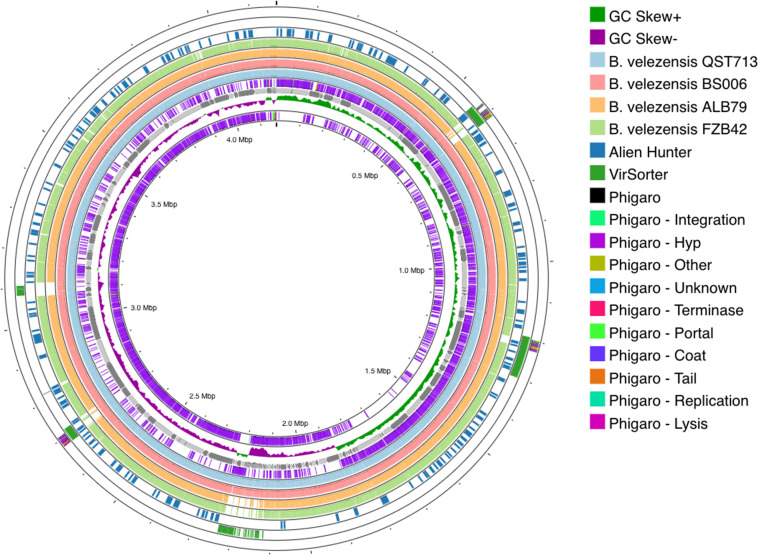
To identify previously sequenced similar genomes, we calculated ANI between B. velezensis EU07 and all 888 genome assemblies available in GenBank for the B. amyloliquefaciens group. This revealed that EU07 shares more than 99.9 % ANI with 13 previously sequenced genomes. Table 1 lists the genomes showing the highest levels of ANI to that of B. velezensis EU07. This includes strains that previously have been classified variously as B. amyloliquefaciens or B. velezensis. However, they all fall within the B. velezensis clade [36,38] and should be considered as belonging to that species. To further elucidate the evolutionary relationships of EU07, we generated a phylogenomic tree including these closely related strains and the relevant type strains; this is presented in Fig. 1. Consistent with the ANI results, strain EU07 falls within a clade that includes the same 13 strains that showed greatest ANI with EU07. Alignment of these genomes with Mauve (Fig. 2) reveals extensive conservation and co-linearity of the chromosome sequence among these strains. Comparison of the EU07 chromosome versus the genome sequences of related strains, as shown in Fig. 3, revealed that most of the presence–absence polymorphism was associated with loci predicted to originate from bacteriophage genomes.
Table 1. Genomes that share more than 99 % ANI with B. velezensis EU07.
| GenBank accession no. | Reference | Strain | ANI (%) |
| GCA_004421045.1 | [47] | ‘B. amyloliquefaciens’ FS1092 | 99.99 |
| GCA_021228895.1 | [48] | B. velezensis A4P130 | 99.99 |
| GCA_003986895.1 | – | B. velezensis BE2 | 99.99 |
| GCA_007678125.1 | [49] | B. velezensis DE0189 | 99.99 |
| GCA_003073255.1 | [37] | B. velezensis QST713 | 99.99 |
| GCA_026156445.1 | [50] | B. velezensis CHBv2 | 99.98 |
| GCA_001709055.1 | – | B. velezensis CFSAN034339 | 99.98 |
| GCA_019093835.1 | – | ‘B. amyloliquefaciens’ BK | 99.98 |
| GCA_014791945.1 | – | ‘B. amyloliquefaciens’ INH2-4b | 99.98 |
| GCA_028609625.1 | [42] | B. velezensis DMW1 | 99.98 |
| GCA_003149795.1 | [40] | ‘B. amyloliquefaciens’ ALB79 | 99.95 |
| GCA_024300805.1 | [23] | ‘B. amyloliquefaciens’ KNU-28 | 99.95 |
| GCA_001278635.1 | [39] | ‘B. amyloliquefaciens’ BS006 | 99.94 |
| GCA_024134605.1 | – | B. velezensis 2987tsa1 | 99.12 |
| GCA_000817575.1 | [51] | ‘B. amyloliquefaciens’ TF28 | 99.10 |
| GCA_034060585.1 | – | B. velezensis Y-4 | 99.07 |
| GCA_010671715.1 | [52] | B. velezensis HU-91 | 99.07 |
| GCA_009193045.1 | [53] | B. velezensis BPC6 | 99.07 |
| GCA_034061945.1 | – | B. velezensis YN-2A | 99.05 |
| GCA_026786545.1 | – | B. velezensis NRRL B-59289 | 99.04 |
| GCA_024138555.1 | [54] | ‘B. amyloliquefaciens’ TPS17 | 99.04 |
| GCA_029866505.1 | [55] | ‘B. amyloliquefaciens’ MN-13 | 99.03 |
| GCA_000341875.1 | [56] | B. velezensis UCMB5036 | 99.02 |
| GCA_009789615.1 | [57] | B. velezensis BA-26 | 99.02 |
| GCA_029910295.1 | – | B. velezensis PT4 | 99.01 |
| GCA_009738165.1 | [58] | B. velezensis HN-Q-8 | 99.01 |
| GCA_021559715.1 | [59] | B. velezensis CF57 | 99.01 |
| GCA_012647845.1 | [60] | B. velezensis UCMB5140 | 99.01 |
Fig. 1. Phylogenetic position of B. velezensis EU07 within the B. amyloliquefaciens group. The phylogenomic maximum-likelihood tree was generated using PhaME and FastTree. The black star highlights the position of strain EU07, whose genome sequence is presented in this study. The configuration file and the tree files are deposited in GitHub at https://github.com/davidjstudholme/bacillus_EU07. Accession numbers for the genome assemblies can be found in Tables 3Table 2Table 3. The tree can be viewed interactively at https://itol.embl.de/tree/14417323152242691702474608.
Fig. 2. Whole-genome-sequence alignment between B. velezensis EU07 and closely related strains. Genome sequences were re-ordered, aligned and visualized using Mauve. Accession numbers for the genome assemblies can be found in Table 3. Green blocks in each genome are homologous to green blocks in all the other genomes. Blue blocks are homologous to the blue blocks in all the other genomes.
Fig. 3. Overview of the genome of B. velezensis EU07 and comparison with closely related genomes. The circular plot of the EU07 chromosome was generated using Proksee. Data are arranged in nine concentric circular tracks as follows: (1) G+C skew, (2) EU07 contigs, (3) blastn hits against the QST713 genome, (4) blastn hits against the BS006 genome, (5) blastn hits against the ALB79 genome, (6) blastn hits against the FZB542 genome, (7) predicted horizontally acquired regions predicted by Alien Hunter, (8) phage loci predicted by VirSorter and (9) phage loci predicted by Phigaro.
Among the strains closely related to EU07 are several that previously have been described as having growth-promoting and/or pathogen-inhibitory properties. For example, strain BS006 was isolated from roots of Physalis peruviana in Colombia and promotes growth in banana [39]. Strain KNU-28 was isolated from peach leaves in Korea [23]. Strain ALB79 was isolated from grapes in northern California and shown to inhibit the growth of Listeria monocytogenes in vitro [40], while strain QST713 is used commercially (Serenade; Bayer) to protect mushroom crops against green mould disease and promotes growth in banana [37,41], among other applications. The endophytic Bacillus strain DMW1 was isolated from the inner tissues of potato tubers and exhibited strong biocontrol activity [42]. The near-identity of these genome sequences, independently isolated from plants in diverse geographical locations, suggests that EU07 is a member of a widely disseminated lineage of B. velezensis with biocontrol and growth-promoting properties. The molecular mechanisms and genetic determinants of these properties have been extensively reviewed elsewhere [43,45], and include gene clusters for secondary metabolites such as bacilysin, fengycin and macrolactin, which are conserved in the B. velezensis lineage that includes BS006 and EU07 [38].
Since our previous phenotypic comparisons between strains EU07 and QST713 revealed differences in their abilities to suppress fungal growth, we compared their genome sequences to identify possible genetic determinants of the observed differences. Their genomes are almost identical, with no detectable differences in their gene contents. However, we identified 46 single-nucleotide differences, which are listed in Table 2. These differences appear to be non-uniformly distributed across the genome. For example, 20 of the 46 SNPs occur within a single gene that encodes the beta subunit of a class-1b ribonucleoside-diphosphate reductase [46] (RefSeq WP_108702400.1; locus tag BVQ_RS09140). This suggests that these differences might be explained by recombination events associated with horizontal genetic transfer rather than point mutations. We also identified some sequence differences between EU07 and QST713 in the intergenic regions between several tRNA genes (GenBank accession no. JAIFZJ010000168.1). These genetic differences may explain the previously observed differences observed between the DNA fingerprints of these two strains when previously assayed using RAPDs [4].
Table 2. Forty-six SNPs between B. velezensis strains EU07 and QST713.
| Position in CP025079.1 | Nucleotide in QST713 | Nucleotide in EU07 | Amino acid change | Predicted gene product |
| 21 222 | A | G | K→E | BVQ_RS00080: serine-tRNA ligase |
| 230 096 | A | C | E→A | BVQ_RS21890: non-ribosomal peptide synthetase |
| 230 098 | A | C | K→Q | BVQ_RS21890: non-ribosomal peptide synthetase |
| 230 111 | C | A | A→E | BVQ_RS21890: non-ribosomal peptide synthetase |
| 530 737 | T | G | Y→STOP | BVQ_RS02595: hypothetical protein |
| 530 789 | T | G | L→V | BVQ_RS02595: hypothetical protein |
| 530 811 | T | G | I→>S | BVQ_RS02595: hypothetical protein |
| 531 288 | T | G | I→S | BVQ_RS02595: hypothetical protein |
| 705 298 | A | C | F→V | BVQ_RS03655: GNAT family N-acetyltransferase |
| 855 165 | A | C | Non-coding | |
| 1 168 486 | A | C | Non-coding | |
| 1 215 136 | A | C | F→C | BVQ_RS06330: contact-dependent growth inhibition system immunity protein |
| 1 851 920 | T | G | F→L | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 923 | A | T | G→G (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 925 | C | A | T→K | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 929 | G | T | K→N | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 932 | A | G | E→E (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 935 | A | G | Q→Q (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 938 | C | T | D→D (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 941 | T | G | T→T (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 944 | T | C | Y→Y (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 950 | A | G | K→K (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 953 | T | G | V→V (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 954 | T | C | L→L (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 956 | A | C | L→F | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 959 | T | C | A→A (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 962 | A | C | G→G (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 965 | T | G | L→L (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 969 | T | C | L→L (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 971 | A | G | L→L (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 972 | T | C | L→L (synonymous) | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 851 974 | G | T | L→F | BVQ_RS09140: class 1b ribonucleoside-diphosphate reductase subunit beta |
| 1 878 004 | T | G | Non-coding | |
| 2 191 740 | T | C | D→G | BVQ_RS10680: cysteine hydrolase family protein |
| 2 415 378 | C | A | Non-coding | |
| 2 415 381 | C | A | Non-coding | |
| 2 415 440 | C | A | Non-coding | |
| 2 722 225 | G | T | Non-coding | |
| 2 722 243 | T | G | Non-coding | |
| 3 268 938 | G | T | A→E | BVQ_RS16510: class 1 isoprenoid biosynthesis enzyme |
| 3 269 022 | T | G | N→T | BVQ_RS16510: class 1 isoprenoid biosynthesis enzyme |
| 3 467 035 | A | C | Non-coding | |
| 3 489 562 | A | G | F→F (synonymous) | BVQ_RS17685: lantibiotic immunity ABC transporter MutG family permease subunit |
| 3 490 697 | T | A | I→I (synonymous) | BVQ_RS17690: lantibiotic immunity ABC transporter MutE/EpiE family permease subunit |
| 3 573 178 | T | A | Non-coding | |
| 4 000 822 | T | G | Non-coding |
Table 3. Genome sequences included in the phylogenomic analysis.
| GenBank accession no. | Taxon | Reference |
| GCA_003149795.1 | ‘B. amyloliquefaciens’ ALB79 | [40] |
| GCA_019093835.1 | ‘B. amyloliquefaciens’ BK | – |
| GCA_001278635.1 | ‘B. amyloliquefaciens’ BS006 | [39] |
| GCA_000196735.1 | B. amyloliquefaciens DSM7T | [34] |
| GCA_004421045.1 | ‘B. amyloliquefaciens’ FS1092 | [47] |
| GCA_014791945.1 | ‘B. amyloliquefaciens’ INH2-4b | – |
| GCA_024300805.1 | ‘B. amyloliquefaciens’ KNU-28 | [23] |
| GCA_029866505.1 | ‘B. amyloliquefaciens’ MN-13 | [55] |
| GCA_000817575.1 | ‘B. amyloliquefaciens’ TF28 | [51] |
| GCA_024138555.1 | ‘B. amyloliquefaciens’ TPS17 | [54] |
| GCA_000262045.1 | B. siamensis KCTC 13613T | [61] |
| GCA_024134605.1 | B. velezensis 2987tsa1 | – |
| GCA_021228895.1 | B. velezensis A4P130 | [48] |
| GCA_001647965.1 | B. velezensis AP194 | [62] |
| GCA_009789615.1 | B. velezensis BA-26 | [57] |
| GCA_003986895.1 | B. velezensis BE2 | – |
| GCA_009193045.1 | B. velezensis BPC6 | [53] |
| GCA_003431885.1 | B. velezensis (B. methylotrophicus) CBMB205T | [63] |
| GCA_021559715.1 | B. velezensis CF57 | [59] |
| GCA_001709055.1 | B. velezensis CFSAN034339 | – |
| GCA_026156445.1 | B. velezensis CHBv2 | [50] |
| GCA_007678125.1 | B. velezensis DE0189 | [49] |
| GCA_028609625.1 | B. velezensis DMW1 | [42] |
| GCA_000015785.2 | B. velezensis (B. amyloliquefaciens subsp. plantarum) FZB42T | [34] |
| GCA_009738165.1 | B. velezensis HN-Q-8 | [58] |
| GCA_010671715.1 | B. velezensis HU-91 | [52] |
| GCA_001461835.1 | B. velezensis (='B. oryzicola') KACC 18228T | [64] |
| GCA_001267695.1 | B. velezensis KCTC 13012 | [65] |
| GCA_001461825.1 | B. velezensis NRRL B-41580T | [36] |
| GCA_026786545.1 | B. velezensis NRRL B-59289 | – |
| GCA_026787705.1 | B. velezensis NRRL BD-154 | – |
| GCA_029910295.1 | B. velezensis PT4 | – |
| GCA_003073255.1 | B. velezensis QST713 | [37] |
| GCA_000341875.1 | B. velezensis UCMB5036 | [56] |
| GCA_012647845.1 | B. velezensis UCMB5140 | [60] |
| GCA_034060585.1 | B. velezensis Y-4 | – |
| GCA_034061945.1 | B. velezensis YN-2A | – |
| GCA_019997305.1 | B. velezensis EU07 | This study |
Conclusion
Genome sequencing of potential biocontrol strain EU07 revealed that it belongs to the species B. velezensis, a species often closely associated with plant roots, and well known for promoting plant growth and biocontrol. The EU07 strain is genetically almost identical to the commercially used strain QST713 (Serenade) and several other previously sequenced and characterized strains; however, we identified several genes containing single-nucleotide differences that can distinguish between EU07 and QST713. Strain EU07 is more distantly related to the commercially used B. velezensis strain FZB24 (TAEGRO), previously known as the type-strain of B. amyloliquefaciens subsp. plantarum. The availability of this genome sequence will facilitate future efforts to unravel the genetic and molecular basis for the strains beneficial properties.
Acknowledgements
We acknowledge the University of Exeter’s DNA Sequencing Facility for performing library preparation and sequencing, and the University of Exeter’s Research IT team for provision of advanced research computing infrastructure used for sequence analysis.
Abbreviations
- ANI
average nucleotide identity
- NCBI
National Center for Biotechnology Information
Footnotes
Funding: Support for C. Jimenez-Quiros from the University of Worcester is gratefully acknowledged. This project utilized equipment funded by the Wellcome Trust Institutional Strategic Support Fund (WT097835MF), a Wellcome Trust Multi-User Equipment Award (WT101650MA) and a Biotechnology and Biological Sciences Research Council (BBSRC) LOLA award (BB/K003240/1).
Accession No: The genome sequence data generated in this work have been deposited in public databases: BioProject accession number PRJNA743875, https://www.ncbi.nlm.nih.gov/bioproject/743875; GenBank
assembly accession number GCA_019997305.2, https://www.ncbi.nlm.nih.gov/nuccore/JAIFZJ000000000; NCBI RefSeq accession number GCF_019997305.2; Sequence Read Archive (SRA) accession number SRX22864526.
Author contributions: Conceptualization: O.B. and M.T. Data curation: all authors. Formal analysis: all authors. Investigation: all authors. Writing – original draft: O.B., D.J.S. and M.T. Writing – review and editing: all authors.
Contributor Information
David J. Studholme, Email: d.j.studholme@exeter.ac.uk.
Catherine Jimenez-Quiros, Email: catherinejq@gmail.com.
Mahmut Tör, Email: m.tor@worc.ac.uk.
References
- 1.Saxena AK, Kumar M, Chakdar H, Anuroopa N, Bagyaraj DJ. Bacillus species in soil as a natural resource for plant health and nutrition. J Appl Microbiol. 2020;128:1583–1594. doi: 10.1111/jam.14506. [DOI] [PubMed] [Google Scholar]
- 2.Aloo BN, Makumba BA, Mbega ER. The potential of bacilli rhizobacteria for sustainable crop production and environmental sustainability. Microbiol Res. 2019;219:26–39. doi: 10.1016/j.micres.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Hashem A, Tabassum B, Fathi Abd Allah E. Bacillus subtilis: a plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J Biol Sci. 2019;26:1291–1297. doi: 10.1016/j.sjbs.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baysal Ö, Çalışkan M, Yeşilova Ö. An inhibitory effect of a new Bacillus subtilis strain (EU07) against Fusarium oxysporum f. sp. radicis-lycopersici. Physiol Mol Plant Pathol. 2008;73:25–32. doi: 10.1016/j.pmpp.2008.11.002. [DOI] [Google Scholar]
- 5.Jimenez-Quiros C, Okechukwu EC, Hong Y, Baysal Ö, Tör M. Comparison of antifungal activity of Bacillus strains against Fusarium graminearum in vitro and in planta. Plants. 2022;11:1999. doi: 10.3390/plants11151999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krueger F. Trim Galore! Babraham Bioinformatics. [14-July-2020]; http://www.bioinformatics.babraham.ac.uk/projects/trim_galore accessed.
- 7.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 8.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, et al. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Studholme DJ. 2024. davidjstudholme/bacillus_EU07: v1.0. Zenoob . [DOI]
- 12.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013:arXiv.1303.3997. doi: 10.48550/arXiv.1303.3997. [DOI] [Google Scholar]
- 15.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson DA. GenBank. Nucleic Acids Research. 2004;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, et al. GenBank. Nucleic Acids Res. 2019;47:D94–D99. doi: 10.1093/nar/gky989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shakya M, Ahmed SA, Davenport KW, Flynn MC, Lo C-C, et al. Standardized phylogenetic and molecular evolutionary analysis applied to species across the microbial tree of life. Sci Rep. 2020;10:1723. doi: 10.1038/s41598-020-58356-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darling ACE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M-J, Park T, Jeong M, Lee G, Jung D-R, et al. Complete genome sequence of Bacillus amyloliquefaciens KNU-28 isolated from peach leaves (Prunus persica [L.] Batsch) Microbiol Resour Announc. 2022;11:e0073422. doi: 10.1128/mra.00734-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant JR, Enns E, Marinier E, Mandal A, Herman EK, et al. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023;51:W484–W492. doi: 10.1093/nar/gkad326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vernikos GS, Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- 27.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9:37. doi: 10.1186/s40168-020-00990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starikova EV, Tikhonova PO, Prianichnikov NA, Rands CM, Zdobnov EM, et al. Phigaro: high-throughput prophage sequence annotation. Bioinformatics. 2020;36:3882–3884. doi: 10.1093/bioinformatics/btaa250. [DOI] [PubMed] [Google Scholar]
- 30.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chklovski A, Parks DH, Woodcroft BJ, Tyson GW. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat Methods. 2023;20:1203–1212. doi: 10.1038/s41592-023-01940-w. [DOI] [PubMed] [Google Scholar]
- 33.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rückert C, Blom J, Chen X, Reva O, Borriss R. Genome sequence of B. amyloliquefaciens type strain DSM7(T) reveals differences to plant-associated B. amyloliquefaciens FZB42. J Biotechnol. 2011;155:78–85. doi: 10.1016/j.jbiotec.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Borriss R, Chen X-H, Rueckert C, Blom J, Becker A, et al. Relationship of Bacillus amyloliquefaciens clades associated with strains DSM 7T and FZB42T: a proposal for Bacillus amyloliquefaciens subsp. amyloliquefaciens subsp. nov. and Bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on complete genome sequence comparisons. Int J Syst Evol Microbiol. 2011;61:1786–1801. doi: 10.1099/ijs.0.023267-0. [DOI] [PubMed] [Google Scholar]
- 36.Dunlap CA, Kim S-J, Kwon S-W, Rooney AP. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int J Syst Evol Microbiol. 2016;66:1212–1217. doi: 10.1099/ijsem.0.000858. [DOI] [PubMed] [Google Scholar]
- 37.Pandin C, Le Coq D, Deschamps J, Védie R, Rousseau T, et al. Complete genome sequence of Bacillus velezensis QST713: a biocontrol agent that protects Agaricus bisporus crops against the green mould disease. J Biotechnol. 2018;278:10–19. doi: 10.1016/j.jbiotec.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 38.Palazzini JM, Dunlap CA, Bowman MJ, Chulze SN. Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: genome sequencing and secondary metabolite cluster profiles. Microbiol Res. 2016;192:30–36. doi: 10.1016/j.micres.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Gamez RM, Rodríguez F, Bernal JF, Agarwala R, Landsman D, et al. Genome sequence of the banana plant growth-promoting rhizobacterium Bacillus amyloliquefaciens BS006. Genome Announc. 2015;3:e01391-15. doi: 10.1128/genomeA.01391-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran TD, Huynh S, Parker CT, Hnasko R, Gorski L, et al. Complete genome sequences of three Bacillus amyloliquefaciens strains that inhibit the growth of Listeria monocytogenes in vitro. Genome Announc. 2018;6:e00579-18. doi: 10.1128/genomeA.00579-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian L, Zhang W, Zhou G-D, Li S, Wang Y, et al. A biological product of Bacillus amyloliquefaciens QST713 strain for promoting banana plant growth and modifying rhizosphere soil microbial diversity and community composition. Front Microbiol. 2023;14:1216018. doi: 10.3389/fmicb.2023.1216018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu C, Chen H, Zhu L, Song Y, Jiang Q, et al. Profiling of antimicrobial metabolites synthesized by the endophytic and genetically amenable biocontrol strain Bacillus velezensis DMW1. Microbiol Spectr. 2023;11:e0003823. doi: 10.1128/spectrum.00038-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chowdhury SP, Hartmann A, Gao X, Borriss R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 – a review. Front Microbiol. 2015;6:780. doi: 10.3389/fmicb.2015.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, et al. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol. 2009;140:27–37. doi: 10.1016/j.jbiotec.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Magno-Pérez-Bryan MC, Martínez-García PM, Hierrezuelo J, Rodríguez-Palenzuela P, Arrebola E, et al. Comparative genomics within the Bacillus genus reveal the singularities of two robust Bacillus amyloliquefaciens biocontrol strains. Mol Plant Microbe Interact. 2015;28:1102–1116. doi: 10.1094/MPMI-02-15-0023-R. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Stubbe J. Bacillus subtilis class Ib ribonucleotide reductase is a dimanganese(III)-tyrosyl radical enzyme. Biochemistry. 2011;50:5615–5623. doi: 10.1021/bi200348q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu G, Gonzalez-Escalona N, Zheng J, Bolten S, Luo Y, et al. Genome sequences of Brevundimonas naejangsanensis strain FS1091 and Bacillus amyloliquefaciens strain FS1092, isolated from a fresh-cut-produce-processing plant. Microbiol Resour Announc. 2020;9:e01448-19. doi: 10.1128/MRA.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Becker R, Ulrich K, Behrendt U, Schneck V, Ulrich A. Genomic characterization of Aureimonas altamirensis C2P003 – a specific member of the microbiome of Fraxinus excelsior trees tolerant to ash dieback. Plants. 2022;11:3487. doi: 10.3390/plants11243487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C, Song W, Ma HR, Peng X, Anderson DJ, et al. Temporal encoding of bacterial identity and traits in growth dynamics. Proc Natl Acad Sci USA. 2020;117:20202–20210. doi: 10.1073/pnas.2008807117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nøhr-Meldgaard K, Struve C, Ingmer H, Agersø Y. Intrinsic tet(L) sub-class in Bacillus velezensis and Bacillus amyloliquefaciens is associated with a reduced susceptibility toward tetracycline. Front Microbiol. 2022;13:966016. doi: 10.3389/fmicb.2022.966016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang S, Jiang W, Li J, Meng L, Cao X, et al. Whole genome shotgun sequence of Bacillus amyloliquefaciens TF28, a biocontrol entophytic bacterium. Stand Genomic Sci. 2016;11:73. doi: 10.1186/s40793-016-0182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wash P, Yasmin H, Ullah H, Haider W, Khan N, et al. Deciphering the genetics of antagonism and antimicrobial resistance in Bacillus velezensis HU-91 by whole genome analysis. J King Saud Univ Sci. 2023;35:102954. doi: 10.1016/j.jksus.2023.102954. [DOI] [Google Scholar]
- 53.Sun W, Yan L, Chen C, Tian Y, Li X, et al. Identification and biocontrol effect of antagonistic bacterium Bacillus velezensis Bpc6 against soft rot and Sclerotinia rot diseases on lettuce. Chin J Biol. 2020;36:231. [Google Scholar]
- 54.Kuebutornye FKA, Lu Y, Abarike ED, Wang Z, Li Y, et al. In vitro assessment of the probiotic characteristics of three Bacillus species from the gut of Nile Tilapia, Oreochromis niloticus. Probiotics Antimicrob Proteins. 2020;12:412–424. doi: 10.1007/s12602-019-09562-5. [DOI] [PubMed] [Google Scholar]
- 55.Yang J, Gao MY, Li M, Li ZZ, Li H, et al. Bacillus amyloliquefaciens CotA degradation of the lignin model compound guaiacylglycerol-β-guaiacyl ether. Lett Appl Microbiol. 2018;67:491–496. doi: 10.1111/lam.13060. [DOI] [PubMed] [Google Scholar]
- 56.Manzoor S, Niazi A, Bejai S, Meijer J, Bongcam-Rudloff E. Genome sequence of a plant-associated bacterium, Bacillus amyloliquefaciens strain UCMB5036. Genome Announc. 2013;1:e0011113. doi: 10.1128/genomeA.00111-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang B, Liu C, Yang X, Wang Y, Zhang F, et al. Genomics-guided isolation and identification of active secondary metabolites of Bacillus velezensis BA-26. Biotechnol Biotechnol Equip. 2021;35:895–904. doi: 10.1080/13102818.2021.1934540. [DOI] [Google Scholar]
- 58.Zhao J, Zhou Z, Bai X, Zhang D, Zhang L, et al. A novel of new class II bacteriocin from Bacillus velezensis HN-Q-8 and its antibacterial activity on Streptomyces scabies. Front Microbiol. 2022;13:943232. doi: 10.3389/fmicb.2022.943232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang C, Chen L, Si H, Gao W, Liu P, et al. Study on the characteristics and mechanisms of nicosulfuron biodegradation by Bacillus velezensis CF57. J Basic Microbiol. 2020;60:649–658. doi: 10.1002/jobm.202000039. [DOI] [PubMed] [Google Scholar]
- 60.Reva ON, Larisa SA, Mwakilili AD, Tibuhwa D, Lyantagaye S, et al. Complete genome sequence and epigenetic profile of Bacillus velezensis UCMB5140 used for plant and crop protection in comparison with other plant-associated Bacillus strains. Appl Microbiol Biotechnol. 2020;104:7643–7656. doi: 10.1007/s00253-020-10767-w. [DOI] [PubMed] [Google Scholar]
- 61.Jeong H, Jeong D-E, Kim SH, Song GC, Park S-Y, et al. Draft genome sequence of the plant growth-promoting bacterium Bacillus siamensis KCTC 13613T. J Bacteriol. 2012;194:4148–4149. doi: 10.1128/JB.00805-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hassan MK. In vitro pectate lyase activity and carbon uptake assays and whole genome sequencing of Bacillus amyloliquefaciens subsp. plantarum strains for a pectin defective pathway. bioRxiv. 2021:2021.01.03.425148. doi: 10.1101/2021.01.03.425148. [DOI] [Google Scholar]
- 63.Hwangbo K, Um Y, Kim KY, Madhaiyan M, Sa TM, et al. Complete genome sequence of Bacillus velezensis CBMB205, a phosphate-solubilizing bacterium isolated from the rhizoplane of rice in the Republic of Korea. Genome Announc. 2016;4:e00654-16. doi: 10.1128/genomeA.00654-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chung EJ, Hossain MT, Khan A, Kim KH, Jeon CO, et al. Bacillus oryzicola sp. nov., an endophytic bacterium isolated from the roots of rice with antimicrobial, plant growth promoting, and systemic resistance inducing activities in rice. Plant Pathol J. 2015;31:152–164. doi: 10.5423/PPJ.OA.12.2014.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeong H, Park S-H, Choi S-K. Genome sequence of antibiotic-producing Bacillus amyloliquefaciens strain KCTC 13012. Genome Announc. 2015;3:e01121-15. doi: 10.1128/genomeA.01121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Studholme DJ. 2024. davidjstudholme/bacillus_EU07: v1.0. Zenoob . [DOI]