Abstract
Polymorphic basic residues near the C terminus of the prion protein (PrP) in humans and sheep appear to protect against prion disease. In heterozygotes, inhibition of prion formation appears to be dominant negative and has been simulated in cultured cells persistently infected with scrapie prions. The results of nuclear magnetic resonance and mutagenesis studies indicate that specific substitutions at the C-terminal residues 167, 171, 214, and 218 of PrPC act as dominant-negative, inhibitors of PrPSc formation (K. Kaneko et al., Proc. Natl. Acad. Sci. USA 94:10069–10074, 1997). Trafficking of substituted PrPC to caveaola-like domains or rafts by the glycolipid anchor was required for the dominant-negative phenotype; interestingly, amino acid replacements at multiple sites were less effective than single-residue substitutions. To elucidate which domains of PrPC are responsible for dominant-negative inhibition of PrPSc formation, we analyzed whether N-terminally truncated PrP(Q218K) molecules exhibited dominant-negative effects in the conversion of full-length PrPC to PrPSc. We found that the C-terminal domain of PrP is not sufficient to impede the conversion of the full-length PrPC molecule and that N-terminally truncated molecules (with residues 23 to 88 and 23 to 120 deleted) have reduced dominant-negative activity. Whether the N-terminal region of PrP acts by stabilizing the C-terminal domain of the molecule or by modulating the binding of PrPC to an auxiliary molecule that participates in PrPSc formation remains to be established.
The prion diseases are a group of neurodegenerative disorders that include kuru, Creutzfeldt-Jakob disease (CJD), Gerstmann-Sträussler-Scheinker disease, and fatal insomnia in humans, scrapie in sheep, and bovine spongiform encephalopathy in cattle. Prion diseases are unique in that they present as sporadic, inherited, and infectious disorders (36, 42). Additionally, the possible transmission of bovine spongiform encephalopathy prions to humans has aroused considerable concern (63, 64).
Prions consist largely, if not entirely, of an abnormal, disease-causing prion protein (PrP) isoform designated PrPSc. A posttranslational process modifies the conformation of the cellular PrP isoform (PrPC) to form PrPSc, which is rich in β-sheets (3, 8, 32). In all natural and experimental prion diseases, PrPSc or other abnormal PrP isoforms accumulate in the central nervous system, resulting in neurologic dysfunction (5, 10, 16).
PrPSc formation seems to occur in rafts or caveola-like domains (CLDs) on the external surfaces of cells (15, 23, 30, 57, 60). The results of transgenic (Tg) mouse studies in which PrP is expressed under the control of ectopic promoters suggest that a limited number of cell types are capable of converting PrPC into PrPSc (41). This result is consistent with those of earlier investigations using both Tg mice and cultured cells contending that an auxiliary factor participates in PrPSc formation (24, 59).
Transmission of human prions to Tg mice expressing either a chimeric human PrP (HuPrP)-mouse PrP (MoPrP) denoted MHu2M or HuPrPC on a null (Prnp0/0) background, but not to mice expressing both MoPrPC and HuPrPC, led us to conclude that MoPrPC inhibits HuPrPSc formation (59). In contrast, MoPrPC was a weak inhibitor of MHu2M PrPSc formation, suggesting that mouse residues at the N or C terminus of PrP compete effectively to reduce the inhibition manifested by wild-type (wt) MoPrPC. Since earlier studies suggested that the N terminus of PrP is not required to initiate or sustain PrPSc formation (38, 46), we concluded that the C-terminal region of PrPC must be the site where binding to another molecule must occur (59). Those results suggested either that MoPrPC binds to HuPrPSc in the inoculum, preventing its interaction with transgene-encoded HuPrPC, or that MoPrPC binds more tightly than does HuPrPC to a mouse factor that participates in PrPSc formation.
The first possibility, that MoPrPC binds to HuPrPSc and prevents its interaction with HuPrPC, suggested that MoPrPC has a higher affinity for HuPrPSc than does HuPrPC and led us to postulate that MoPrPC has a greater affinity for heterologous PrPSc than for homologous PrPC. However, results from earlier studies show that homologous PrPC and PrPSc interact much more readily than do the heterologous isoforms (39).
The second possibility, that MoPrPC binds more tightly to a mouse factor that participates in PrPSc formation than does HuPrPC, led to the hypothesis that MoPrPC binds to homologous PrPSc with a greater affinity than does heterologous PrPC. Additional evidence favoring this interpretation was provided by studies with chronically infected mouse neuroblastoma (ScN2a) cultured cells (24). In ScN2a cells, we found that a chimeric HuPrP-Mo PrP containing the seven C-terminal HuPrP-specific residues was not converted into PrPSc; moreover, this chimeric PrP did not inhibit the conversion of MoPrPC into PrPSc (24). In light of the results with ScN2a cells, we had argued that MoPrPC has a higher affinity for MoPrPSc than for HuPrPC.
The foregoing findings are more readily explained in terms of PrPC binding to an auxiliary molecule that participates in the transformation of PrPC into PrPSc. We presume that this auxiliary molecule is a protein based upon its exquisite specificity for PrPC and provisionally designated it protein X (24, 59). Although a variety of proteins have been reported to bind to PrP and hence are candidates for protein X, none have been shown to participate in PrPSc formation (12, 28, 31, 43, 65).
Using site-directed mutagenesis in conjunction with the nuclear magnetic resonance structure of recombinant Syrian hamster (SHa) PrP, the binding site on PrPC for protein X was defined (24). The side chains of four PrP residues that are sequentially distant but spatially quite proximal bind to protein X. Substitution of one or more of these residues with a basic amino acid inhibits PrPSc formation, apparently by binding to protein X. The binding of PrPC bearing basic residues at the protein X binding site seems to explain dominant-negative inhibition of PrPSc formation. In support of this concept, polymorphisms in humans and sheep that render these mammals resistant to prion disease (21, 52, 62) are the result of basic residue substitutions in PrP at the protein X binding site.
To define those regions of PrPC that have a role in the dominant-negative inhibition of PrPSc formation, we constructed a variety of truncated PrP molecules carrying the basic substitution Q218K. To distinguish recombinant PrPs from endogenous MoPrP in ScN2a cells, we substituted two residues found in SHa and HuPrP, but not MoPrP, which form an epitope for the 3F4 monoclonal antibody (MAb) (26). The resulting mutant PrP carrying the 3F4 epitope and the Q218K substitution was designated MHM2(Q218K) or MHM2 PrP(Q218K) (24, 49).
As reported here, we found that full-length MHM2 PrP(Q218K) completely abolished formation of MHM2 PrPSc in ScN2a cells. N-terminal truncations of PrP were less effective inhibitors of PrPSc formation than full-length PrP. Although an N-terminally truncated PrP(90-231,Q218K) was a reasonably good inhibitor of PrPSc formation, addition of residues from the extreme N terminus enhanced the inhibition. The residues in PrP(90-231) correspond to those found in PrP 27-30, which is the protease-resistant core of PrPSc that retains infectivity. As few as six residues from the N terminus of PrP (23KKRPKP29) fused to PrP(90-231,Q218K) substantially increased dominant-negative inhibition of PrPSc formation.
MATERIALS AND METHODS
Cultured cells.
Mouse neuroblastoma N2a cells were obtained from the American Tissue Culture Collection (Rockville, Md.). Scrapie-infected N2a cells (ScN2a cells) are chronically infected with scrapie prions and produce both MoPrPC and MoPrPSc as described previously (6). All cells were grown and maintained at 37°C in minimal essential medium supplemented with 10% fetal bovine serum. In some cases, cells were treated with phosphatidyl-inositol phospholipase C (PIPLC) by incubation for 4 h at 37°C in OptiMEM with 0.5 U of PIPLC (Boehringer Mannheim) per ml.
Plasmid constructions.
All plasmids used in this study were derived from the MHM2 PrP gene construct cloned into the pSPOX-neo vector as described previously (49). Introduction of the Q218K substitution was made by PCR using the oligonucleotides described previously (24) or by cloning the BstEII-XhoI fragment from MHM2(Q218K) (24) in the appropriate truncated constructs.
Deletions of residues 23 to 88 and residues 23 to 88 108 to 121 were described previously (29). Similarly, PrPC(Δ107-120) was obtained using the following primers: 5′ CCATAATCAGTGGAACAAGCCCAGCAAACCAAAACCGTGGG 3′ and 5′ GCCCCCCACGGTTTTGGTTTGCTGGGCTTGTTCCACTGATTATGGGGTAC 3′. The oligonucleotides that carry the restricted KpnI and EcoO109I sites were annealed. The double-stranded DNA fragment was treated with kinase, purified on a Sephadex G25 column, and ligated into a derivative of pSP72. The pSP72 derivative vector was constructed as follows: pSP72 was restricted with EcoO109I and treated with Klenow fragment to generate a vector lacking the EcoO109I site. To this vector, the MHM2 gene was ligated between the BglII and XhoI sites to generate the derivative vector. Ligation of the double-stranded DNA generated a plasmid that produced PrP with 13 amino acids within the N-terminal domain of PrPC deleted. The BglII-XhoI fragments were then inserted into pSPOX-neo vector between the BamHI and XhoI sites.
PrPC(Δ23-120) was constructed by using the T7 primer 5′ AAATTAATACGACTCACTAT 3′ and the oligonucleotide 5′ GTTCCACCCACCAGGCTTTGGCCGAAGGCCCCCCACGCAGAGGCCGAC 3′. The product obtained by PCR was restricted with BglII and EcoO109I and cloned into the pSP72-derived vector lacking EcoO109I and containing the entire MHM2 gene. Restriction with EcoO109I and ligation resulted in a vector that expressed PrP proteins with 97 amino acid residues missing from the N-terminal region of PrP, designated PrPC(Δ23-120). Restricted fragments BglII-XhoI, with or without the Q218K mutation, were subcloned into the expression vector pSPOX-neo.
PrPC(Δ112-145) was obtained by annealing the oligonucleotides 5′ GTCCACATGCT TGAGGT TGGT TT TTGGT TTGCTGGGCT TGT TCCACTGAT TATGGGTAC 3′ and 5′ CCATAATCAGTGGAACAAGCCCAGCAAACCAAAAACCAACCTCAAGCATGTG 3′. The double-stranded DNA fragment was cloned into pSP72 containing the MHM2 DNA construct between the KpnI and AvaII sites.
MHM2(V214I,Q218K) was obtained after PCR using the primer 5′ TAATAGGCCTGGGACTCCTTTTTGTACTGGGTGATGCACATCTG 3′ and the BstEII primer described previously (24). After restriction with BstEII and StuI, the DNA fragment was cloned into the MHM2 gene and inserted in the pSP72 vector. The BglII-XhoI fragment that contained the entire mutated gene was then subcloned into the pSPOX vector restricted with BamHI and XhoI. From the same construct, the BstEII-XhoI fragment was also cloned in the pSP72 vector that expressed MHM2(Q171R). Again, the BglII-XhoI-restricted fragment was subcloned into the pSPOX-neo vector to express the triply mutated PrP protein (Q171R, V214I, and Q218K).
PrPC(Δ23-111) was obtained using PCR with the following oligonucleotides, the T7 primer 5′ AAATTAATACGACTCACTAT 3′ and the primer 5′ GTTCCACCCACCAGGC T T TGGCCGAAGGCCCCCCACCACGGCCCC TGCCGCAGCAGCGCCGGCCATGCAGAGGCCGAC 3′. The double-stranded DNA fragment, restricted with BglII and EcoO109I, was cloned into the vector pSP72 mutated to eliminate the EcoO109I site. This vector contains the entire MHM2 gene between the BglII and XhoI sites. Again, BglII-XhoI-restricted fragments were ligated into the expression vector pSPOX-neo.
PrPC(Q218K,Δ231-253) was generated using PCR with the T7 primer and the oligonucleotide 5′ T TAAACGTACCCTCGAGTCACGATCGTCGGCCGTCGTAATAGGCCTGGGA 3′ on the pSP72 vector that contains the MHM2(Q218K) construct. The double-stranded DNA fragment was restricted with BglII and XhoI and cloned into the pSPOX-neo expression vector.
PrPC(Δ32-80) was generated using PCR with the T7 primer and the primer 5′ CCACTGATTGTGGGTACCCCCTCCTTGGCCCCACCCTCGAGGCTTTGGCCCGC 3′. The PCR fragment was then restricted with BglII and KpnI and cloned into the pSP72 vector that contains the entire MHM2 sequence. After restriction map verification and sequence, the BglII-XhoI fragments were ligated into the expression vector pSPOX-neo.
Similarly, PrP(Δ30-91) was obtained using PCR with the primers T7 and 5′ CCACTGATTGTGGGTACCCCCTCCAGGCTTTGGCCGCTTTTTGCGAGGCC 3′.
PrP(Δ52-91) and PrP(OR+6) were previously described (46).
Introduction of the substitution Q218K was made as described previously by cloning the BstEII-XhoI fragment of the MHM2(Q218K) construct (24).
Transfection, limited proteolysis, and Western blotting.
N2a and ScN2a cells were transiently transfected with appropriate DNA constructs (5 or 10 μg) using the DOTAP DNA transfection kit (Boehringer Mannheim). Cotransfections of the ScN2a cells with two different DNA constructs were performed as described previously (17, 24, 25). Cells were lysed in lysis buffer T (10 mM Tris-HCl [pH 8.00], 0.5% deoxycholate, 0.5% Nonidet P-40, 150 mM NaCl) as described previously (49). Proteinase K (PK) (Boehringer Mannheim) was added to the cell lysate to attain a final concentration of 20 μg/ml, and the samples were incubated at 37°C for 60 min. The reaction was stopped by adding phenylmethylsulfonyl fluoride at a final concentration of 1 mM. Insoluble PK-resistant PrPSc was recovered after centrifugation at 100,000 × g for 60 min, and the pellet was resolubilized in lysis buffer T and Laemmli sample buffer (27). In some cases, 30 μl of cell lysate was incubated overnight at 37°C with 1 U of PNGase (Boehringer-Mannheim).
After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were transferred to nitrocellulose and Western blots were performed as previously described (49). PrP was detected with the anti-PrP 3F4 MAb as well as with RO73, a polyclonal antibody. Anti-PrP 3F4 is a mouse MAb raised against SHaPrP 27-30 (26) that recognizes the Met 109-Met 112 epitope (45) and does not bind to MoPrP. RO73 is a rabbit antiserum raised against SHaPrP 27-30 (50) that recognizes MoPrP.
Indirect immunofluorescence.
Cells were grown on coverslips prior to transfection. When PIPLC treatment was performed, cells were treated with the enzyme for 4 h before indirect fluorescence was examined.
The cells were washed twice with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 30 min at room temperature, and then rinsed three times. To block nonspecific binding, cells were treated with PBS containing 5% dry milk and 1% bovine serum albumin for 1 h. In order to visualize the intracellular protein expression, permeabilization of the cells was performed in blocking solution containing 0.2% saponin. The MAb 3F4 (diluted 1:100) was incubated with the cells for 1 h at room temperature in the blocking solution. The cells were then washed five times with PBS and incubated for 30 min at room temperature with fluorescein isothiocyanate-conjugated rabbit anti-mouse immunoglobulin G (Boehringer) diluted 1:100 in PBS containing 1% bovine serum albumin. After the cells were washed five times with PBS, 5 μl of mounting medium (VECTASHIELD) containing diaminopimelic acid (VECTOR) was added to each glass slide, the coverslips were placed on the slides, and indirect immunofluorescence was visualized with a fluorescence microscope (LEICA) using a 100× oil objective.
RESULTS
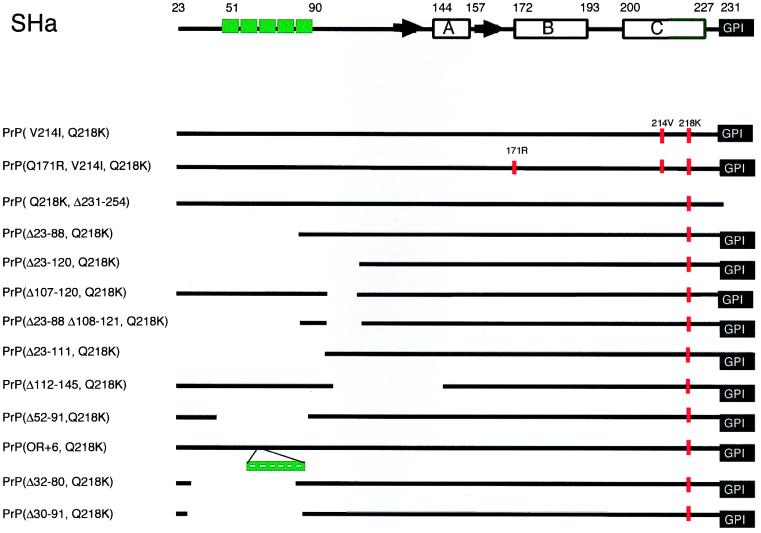
To identify those regions of PrP that participate in dominant-negative inhibition of PrPSc formation, we carried out a deletion mutagenesis study. Before making deletions in PrP, we asked if inhibition by the Q218K substitution could be augmented by additional substitutions of basic residues. Upon finding that additional basic amino acids diminished the inhibitory effect of the Q218K mutation, all subsequent deletion and truncation studies were restricted to constructs carrying only the Q218K substitution (Fig. 1). Recombinant PrPs were assessed by measuring their effect on PrPSc formation in ScN2a cells. Both mutant MHM2 PrP(Q218K) and wild-type (wt) MHM2 PrP were transfected into ScN2a cells, and the formation of MHM2 PrPSc was measured by Western blotting using the 3F4 MAb.
FIG. 1.
Schematic representations of the truncated PrP mutant proteins used in this study. At the top is a schematic representation of SHaPrP(23-231). The secondary structures of recombinant SHaPrP(90-231) are denoted as follows: horizontal boxes for the helical structures and black arrows for the β-strands. Deletions are shown as gaps, and the amino acid positions bordering deletions are shown in parentheses in the construct designations. The protein X binding domain is located at the C-terminal region of PrPC where substitutions at residues 167, 171, 214, and 218 are significant for the dominant-negative phenotype. Octarepeat sequences are depicted as green boxes, and amino acid substitutions are depicted as vertical red bars.
Multiple basic residues diminish dominant-negative inhibition.
The results of mutagenesis studies identified four residues, 167, 171, 214, and 218, that form the epitope for the binding of PrP to protein X (24). Many of the mutations at these residues abolished PrPSc formation, and some of these mutants exhibited dominant-negative inhibition (24). Because dominant-negative phenotypes are typically due to the sequestration of a rate-limiting factor in a metabolic process, we asked whether multiple mutations might be synergistic.
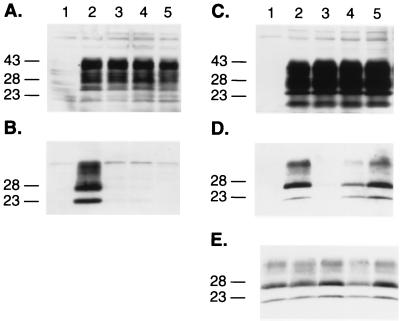
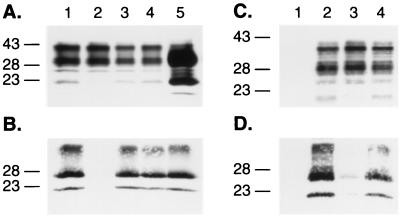
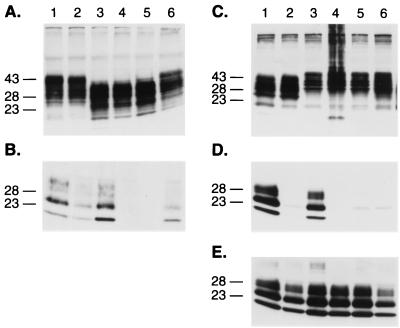
First, PrPs carrying multiple mutations were assessed for conversion into PrPSc upon transient transfection into scrapie-infected mouse neuroblastoma cells (ScN2a). Neither PrP(V214I,Q218K) nor PrP(Q171R,V214I,Q218K) was converted into PrPSc, as judged by the absence of PK-resistant PrP in ScN2a lysates (Fig. 1 and 2A and B). Second, the effects of the double and triple mutations on binding of PrP to protein X were analyzed by measuring conversion of the wt MHM2 PrPC into PrPSc; neither the double nor triple PrP mutant prevented the conversion of wt MHM2 PrPC into PrPSc (Fig. 2C to E). In contrast, the single PrP mutants, i.e., PrP(Q171R), PrP(V214I), or PrP(Q218K), did exhibit dominant-negative inhibition of conversion of wt MHM2 PrPC into PrPSc (24).
FIG. 2.
Combined effects of the substitutions at positions 171, 214, and 218 within the protein X binding site at the C-terminal domain of the PrP protein. (A) Western blot analysis of the expression of PrP mutant proteins in ScN2a cells and (B) conversion of individual MHM2 constructions into PrPSc after treatment with PK (20 μg/ml). Lanes: 1, mock transfection; 2, MHM2; 3, MHM2(Q218K); 4, MHM2(V214I,Q218K); 5, MHM2(Q171R,V214I,Q218K). (C to E) Influence of the PrP mutants in preventing the conversion of the wt MHM2 protein. Coexpression of the mutant proteins with MHM2 in the orientation described above for panels A and B prior (C) or after (D and E) PK treatment. ScN2a cells were transfected with the two DNA constructs, and the conversion of MHM2 PrPSc was specifically monitored using the MAb 3F4. Lanes: 1, mock transfection; 2, MHM2; 3, coexpression of MHM2 and MHM2(Q218K); 4, MHM2(V214I,Q218K); 5, MHM2(Q171R,V214I,Q218K). The gels were stained with anti-PrP 3F4 MAb (A to D) or with RO73 antibody (E), which recognizes endogenous MoPrPSc as well as chimeric constructs. Apparent molecular masses (in kilodaltons) based on migration of protein standards are given to the left of the gels.
Taken together, our results indicate that the effects of the C-terminal PrP mutations Q171R, V214I, and Q218K are not additive. We suspect that the combination of these mutations destabilizes the protein X binding site either through unfavorable charge-charge interactions between R171 and K218 or owing to the influence of the β-branched I214 on the stability of the α-helical backbone. Against this background, we proceeded to analyze the impact of mutations in other regions of the PrP(Q218K) molecule on the dominant-negative phenotype.
Dominant-negative phenotype requires targeting PrP to the cell surface.
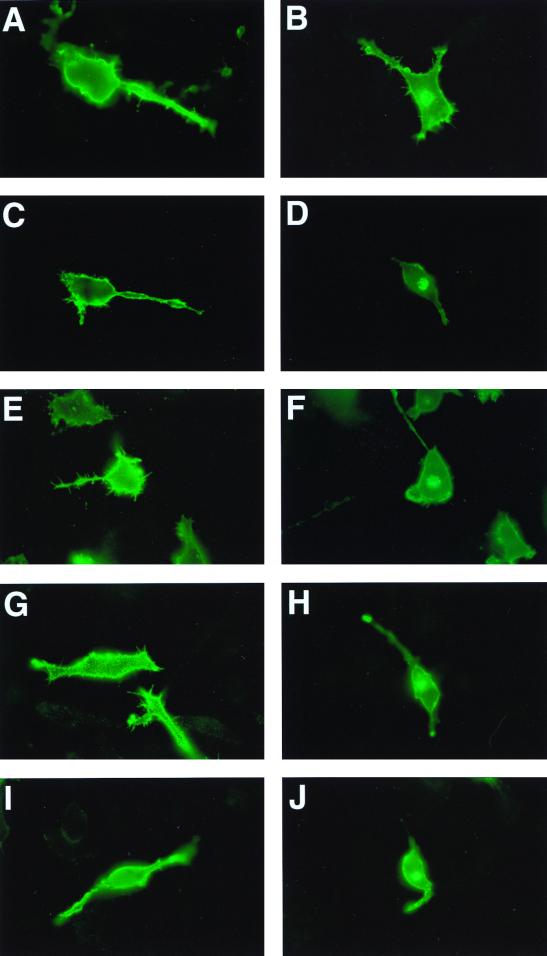
The mutant MHM2 PrP(Q218K) substitution was correctly targeted to the cell surfaces of neuroblastoma (N2a) cells as demonstrated by immunofluorescence microscopy (Fig. 3). N2a cells were transiently transfected with MHM2 PrP(Q218K) and treated with PIPLC. This PIPLC treatment released the mutant PrP from the cell surface, as demonstrated by immunoblotting (data not shown).
FIG. 3.
Cell surface localization of MHM2 and various PrP mutant proteins expressed in N2a cells as measured by indirect immunofluorescence microscopy. Cell surface expression (A, C, E, G, and I) and internal localization in permeabilized cells (B, D, F, H, and J) of proteins MHM2 (A and B), MHM2(Q218K) (C and D), MHM2(Δ23-88,Q218K) (E and F), MHM2(Δ52-91,Q218K) (G and H), and MHM2(Δ32-80,Q218K) (I and J) are shown. All the N2a cells were incubated with anti-PrP 3F4 MAb as described in Materials and Methods.
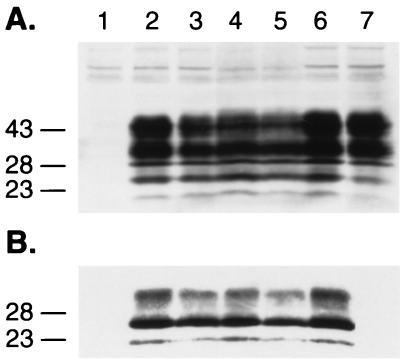
Mutant MHM2 PrP(Q218K,Δ231-254) lacking the glycosylphosphatidyl inositol (GPI) anchor signal sequence was transfected into ScN2a cells to determine if it could be converted into PrPSc as judged by the acquisition of protease resistance. As expected, no mutant PrPSc was found (Fig. 4A and B, lanes 4). Next, MHM2 PrP(Q218K,Δ231-254) was examined for inhibition of conversion of wt MHM2 PrPC into PrPSc. It is well documented that the cell surface localization of PrP is essential for PrPSc replication (23, 57). MHM2 PrPC was converted into PrPSc in cotransfected cells that expressed both MHM2 PrP(Q218K,Δ231-254) and wt MHM2 (Fig. 4A and B, lanes 6). These results indicate that the dominant-negative phenotype is manifested only if mutant PrP(Q218K) is correctly targeted to the cell surface.
FIG. 4.
Requirement of the GPI anchor signal sequence for efficient dominant-negative phenotype of the MHM2(Q218K) chimera. Expression of the chimeric PrP and PrP(Q218K) lacking the GPI anchor (A) and conversion of MHM2 PrPC into PrPSc (B and C). Lanes: 1, mock transfection; 2, MHM2; 3, coexpression of MHM2 and MHM2(Q218K); 4, MHM2(Δ231-254); 5, MHM2(Q218K,Δ231-254); 6, coexpression of MHM2 and MHM2(Q218K,Δ231-254). The PrP proteins were stained with the anti-PrP 3F4 MAb (A and B) or with the polyclonal RO73 antibody (C). Apparent molecular masses (in kilodaltons) based on migration of protein standards are given to the left of the gels.
N-terminal PrP deletions and protein X.
In ScN2a cells, MHM2 PrP(Δ23-88) was earlier reported to acquire PK resistance (46). To determine if MHM2 PrP(Δ23-88) binds to protein X, we cotransfected ScN2a cells with MHM2 PrP(Δ23-88,Q218K) and either MHM2 PrP or MHM2 PrP(Δ23-88). Immunofluorescence microscopy of N2a cells revealed that MHM2 PrP(Δ23-88,Q218K), like full-length MHM2 PrP(Q218K), is expressed on the external surface (Fig. 3C). In ScN2a cells, MHM2 PrP(Δ23-88,Q218K) was not converted into PrPSc (data not shown).
When MHM2 PrP(Δ23-88,Q218K) and full-length MHM2 PrP were coexpressed in ScN2a cells, the formation of MHM2 PrPSc was not impeded (Fig. 5A and B, lanes 5). In contrast, when MHM2 PrP(Δ23-88,Q218K) and MHM2 PrP(Δ23-88) were coexpressed in ScN2a cells, the formation of MHM2 PrPSc(Δ23-88) was inhibited (data not shown). These findings suggest that the avidity of MHM2 PrP(Δ23-88,Q218K) for protein X is less than that of full-length PrP(Q218K).
FIG. 5.
Reduced avidity of N-terminally truncated MHM2(Q218K) molecules for protein X. (A) Western blot analysis of the coexpression of mutant proteins with MHM2 and (B) conversion of MHM2 PrPC into PrPSc, monitored with 3F4 MAb, after treatment with PK (20 μg/ml). MHM2 alone (lane 1) and coexpressed with MHM2(Q218K) (lane 2), MHM2(Δ107-120,Q218K) (lane 3), MHM2(Δ23-120,Q218K) (lane 4), and MHM2(Δ23-88,Q218K) (lane 5). (C) Western blot analysis of PrP mutant protein that conserved the hydrophobic core coexpressed with MHM2 and (D) conversion of MHM2 into PrPSc when coexpressed with MHM2(Δ23-111,Q218K) derivatives. Lanes: 1, mock transfection; 2, MHM2 alone; 3, MHM2 and MHM2(Q218K); 4, MHM2 and MHM2(Δ23-111,Q218K). Apparent molecular masses (in kilodaltons) based on migration of protein standards are given to the left of the gels.
As MHM2 PrP(Δ23-88,Q218K) was found to bind to protein X but with a reduced degree of affinity, we enlarged this deletion to residue 120. Earlier studies reported that PrP(121-231) is an autonomously folding unit of PrP (18). Neither MHM2 PrP(Δ23-120) nor MHM2 PrP(Δ23-120,Q218K) was converted into PrPSc in ScN2a cells, as judged by the acquisition of protease resistance (data not shown). In cotransfection experiments, MHM2 PrP(Δ23-120,Q218K) did not inhibit the conversion of either full-length MHM2 PrP (Fig. 5A and B, lanes 4) or MHM2 PrP(Δ23-88) into PrPSc. Two additional deletion mutations were also investigated with respect to inhibition of PrPSc formation. Neither MHM2 PrP(Δ107-120,Q218K) (Fig. 5A and B, lanes 3) nor MHM2 PrP(Δ23-88,Δ108-121,Q218K) inhibited the conversion of MHM2 PrP into PrPSc in ScN2a cells.
Because dominant-negative inhibition was not observed with any of the mutants described above, we asked if preservation of the hydrophobic core (PrP residues 112 to 125) (22) could restore binding to protein X. To explore this issue, we constructed a truncated PrP molecule lacking residues 23 to 111 but preserving the hydrophobic core. The substitution Q218K was introduced. Cotransfection of PrP(Δ23-111,Q218K) and MHM2 PrP into ScN2a cells did not inhibit the formation of PrPSc (Fig. 5C and D, lanes 4). In view of these results, we restored residues 23 to 111 and deleted residues 112 to 145, which retained the region of recombinant PrP known to be highly structured, i.e., residues 146 to 231. We found that neither PrP(Δ112-145) nor PrP(Δ112-145,Q218K) was converted into PrPSc in ScN2a cells (data not shown); moreover, neither PrP(Δ112-145) (Fig. 6, lanes 3 and 4) nor PrP(Δ112-145,Q218K) inhibited the conversion of wt MHM2 PrPC into PrPSc (Fig. 6, lanes 5 and 6).
FIG. 6.
The highly structured region (residues 146 to 231) of PrP is insufficient for high-affinity binding to protein X. (A) Western blot analysis of the coexpression of MHM2(Δ112-145) and MHM2(Δ112-145,Q218K) with MHM2 in ScN2a cells and (B) conversion of MHM2 PrPC into PrPSc when coexpressed with mutant proteins after digestion with PK (20 μg/ml). Lanes: 1, mock transfection; 2, MHM2; 3, MHM2 and MHM2(Δ112-145) at 10 μg; 4, MHM2 and MHM2(Δ112-145) at 20 μg; 5, MHM2 and MHM2(Δ112-145,Q218K) at 5 μg; 6, MHM2 and MHM2(Δ112-145,Q218K) at 20 μg; 7, MHM2 and MHM2(Q218K). Apparent molecular masses (in kilodaltons) based on migration of protein standards are given to the left of the gels.
From the foregoing data, we concluded that the highly structured region of recombinant PrP consisting of helices A, B, and C (11, 22, 44) was insufficient for high-affinity binding to protein X. We also found that the addition of a variety of additional residues failed to produce mutant PrP molecules that bind to protein X except for PrP(Δ23-88), which is also called PrP(89-231).
Octarepeats and dominant-negative phenotype.
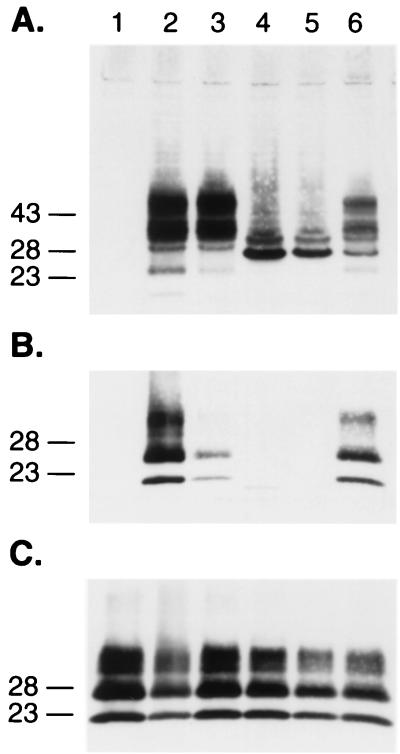
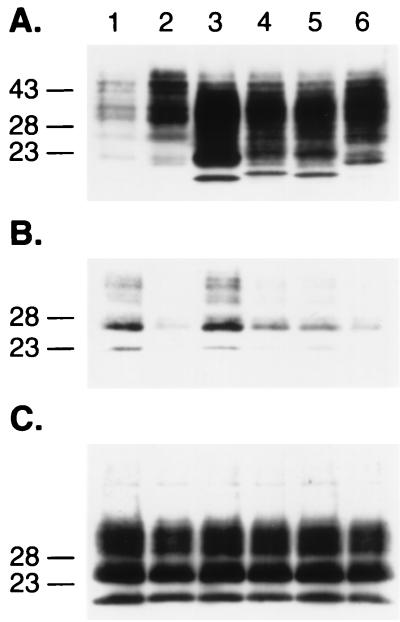
Having determined that PrP(Δ23-88) exhibits dominant-negative inhibition but at a diminished level when assayed against full-length PrPC, we explored the roles of residues between 23 and 88. We began by deleting the five octarepeats resulting in PrP(Δ52-91). The octarepeats are of considerable interest, since additional octarepeats cause inherited prion disease (14, 34) and the His residues of these repeats bind Cu2+ ions (4, 19, 33, 54, 55, 61). Immunofluorescence microscopy showed that both PrP(Δ52-91) and PrP(Δ52-91,Q218K) were expressed on the surfaces of N2a cells (Fig. 3D). In addition, both mutant PrPs were released from the cell surface by PIPLC digestion. Although PrP(Δ52-91) is efficiently converted into PrPSc in ScN2a cells (Fig. 1 and 7A and B) as previously reported (46), PrPC (Δ52-91,Q218K) was not converted into PrPSc. To investigate further whether the octarepeat region influences the binding of PrP to protein X, we cotransfected ScN2a cells with MHM2 PrP(Δ52-91,Q218K) and wt MHM2 PrP. We found that PrP(Δ52-91,Q218K) impeded the conversion of both MHM2 PrP(Δ52-91) and full-length MHM2 PrP into PrPSc in ScN2a cells.
FIG. 7.
Octarepeats of PrP and dominant-negative phenotype. (A and B) Avidity of PrP(Δ52-91,Q218K) for protein X is as high as full-length PrP(Q218K). Expression (A) and conversion (B) of PrPC(Δ52-91) into PrPSc after PK treatment. Lanes: 1, MHM2; 2, coexpression of MHM2 and MHM2(Q218K); 3, MHM2(Δ52-91); 4, MHM2(Δ52-91,Q218); 5, coexpression of MHM2(Δ52-91) and MHM2(Δ52-91,Q218K); 6, coexpression of MHM2(Δ52-91) and MHM2(Q218K). (C to E) Additional octarepeats (OR +6) impede the conversion of both full-length MHM2 and MHM2(OR+6). Coexpression of MHM2 or MHM2(OR+6) with mutant proteins (C) and after PK treatment (D and E). Lanes: 1, MHM2; 2, MHM2 and MHM2(Q218K); 3, MHM2 and MHM2(OR+6); 4, MHM2 and MHM2(OR+6,Q218K); 5, MHM2(OR+6) and MHM2(OR+6,Q218K); 6, MHM2(OR+6) and MHM2(Q218K). The gels were stained with anti-PrP 3F4 MAb (A to D) or with anti-PrP RO73 antibody (E). Apparent molecular masses (in kilodaltons) based on migration of protein standards are given to the left of the gels.
Since deletion of the octarepeats did not alter the dominant-negative phenotype, we asked if additional octarepeats might appreciably increase the inhibition displayed by PrP(Q218K). If this were the case, then additional octarepeats would provide a mechanism whereby mutant PrPs cause inherited human prion diseases. Earlier studies showed that PrP(OR+6) containing additional six octarepeats was readily expressed in N2a cells (35) and converted into PrPSc in ScN2a cells (46). In contrast to PrP(OR+6), PrP(OR+6,Q218K) was not converted into PrPSc in ScN2a cells (Fig. 1 and 7C to E). We also found that PrP(OR+6,Q218K) impeded the conversion of both MHM2 PrP(OR+6) and full-length MHM2 PrP into PrPSc in ScN2a cells.
The dominant-negative phenotypes of PrP(Δ52-91,Q218K) and PrP(OR+6,Q218K) seem to be similar to that of full-length PrP(Q218K). From these findings, we conclude that the avidity of PrP(Δ52-91,Q218K) and PrP(OR+6,Q218K) for protein X is indistinguishable from that of full-length PrP(Q218K) when assayed in ScN2a cells converting full-length MHM2 PrPC into PrPSc.
N-terminal positive charges modulate dominant-negative inhibition mediated by C-terminal residues.
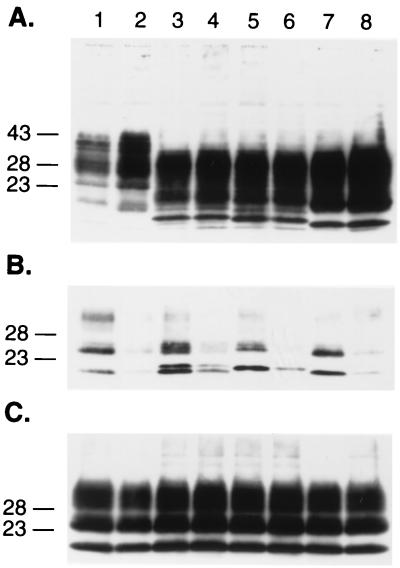
From the foregoing results comparing the dominant-negative phenotypes of PrP(Δ23-88,Q218K) and PrP(Δ52-91,Q218K), we concluded that residues 23 to 51 influence the affinity of PrP for protein X. To investigate the roles of the extreme N-terminal residues, two truncated PrPs were produced: PrP(Δ32-80) and PrP(Δ30-91). Both PrP(Δ32-80) and PrP(Δ30-91) were efficiently converted into PrPSc (Fig. 1 and 8). Conversion of these mutant PrPs was abolished in cotransfections with either truncated PrP carrying the Q218K mutation. Immunofluorescence analysis of PrP(Δ32-80,Q218K) expressed in N2a cells showed that it transports to the cell surface (Fig. 3E). Treatment with PIPLC released PrP(Δ32-80,Q218K) from the external surfaces of the cultured cells. Cotransfection studies using ScN2a cells indicated that either PrP(Δ32-80,Q218K) or PrP(Δ30-91,Q218K) inhibited conversion of PrP(Δ32-80), PrP(Δ30-91), and full-length MHM2 PrP into PrPSc (Fig. 1 and 9). Both PrP(Δ32-80,Q218K) and PrP(Δ30-91,Q218K) seem to exhibit a greater degree of dominant-negative inhibition than PrP(Δ23-88,Q218K). These findings argue that the extreme N-terminal sequence (23KKRPKP29) enhances the dominant-negative phenotype. This basic sequence is highly conserved in all species studied to date (1).
FIG. 8.
N-terminal positive charges are required for the dominant-negative phenotype. (A) Western blot analysis on the expression (A) and conversion of truncated PrP molecules into PrPSc after PK treatment (B and C). Lanes: 1, MHM2; 2, coexpression of MHM2 and MHM2(Q218K); 3, MHM2(Δ32-80); 4, coexpression of MHM2(Δ32-80) and MHM2(Δ32-80,Q218K); 5, MHM2(Δ30-91); 6, coexpression of MHM2(Δ30-91) and MHM2(Δ30-91,Q218K); 7, PrP(Δ23-88); 8, PrP(Δ23-88) and PrP(Δ23-88,Q218K). The gels were stained with anti-PrP 3F4 MAb (A and B) or with anti-PrP RO73 antibody (C). Apparent molecular masses (in kilodaltons) based on migration of protein standards are given to the left of the gels.
FIG. 9.
Western blot analysis on the dominant-negative inhibition of truncated PrP mutants on the conversion of the full-length MHM2 PrP protein. Expression of the different proteins in ScN2a cells (A) and conversion of MHM2 into PrPSc after PK digestion in the presence of truncated proteins (Q218K) that lack the N-terminal domain (B and C). The conversion of MHM2 was specifically monitored with MAb 3F4. Lanes: 1, MHM2; 2, MHM2 and MHM2(Q218K); 3, MHM2 and MHM2(Δ23-88,Q218K); 4, MHM2 and MHM2(Δ32-80,Q218K); 5, MHM2 and MHM2(Δ30-91,Q218K); 6, MHM2 and MHM2(Δ52-91,Q218K). The gels were stained with anti-PrP 3F4 MAb (A and B) or with anti-PrP RO73 antibody (C). Apparent molecular masses (in kilodaltons) based on migration of protein standards are given to the left of the gels.
DISCUSSION
The discovery that dominant-negative inhibition of PrPSc formation is governed by a discontinuous epitope in the C-terminal domain of PrPC has opened several new approaches to studies of prions. First, this discovery explains the apparent protection from CJD in people with a single allele encoding PrP(K219) and from scrapie in sheep with a single allele encoding PrP(R171) (21, 52, 62). Second, the discovery of this epitope in conjunction with the results described here defines the optimal ligands for both affinity purification and assay of protein X. Third, the dominant-negative phenotype argues that rational drug design should be targeted to the site of interaction between PrPC and protein X (V. Perrier, A. Wallace, K. Kaneko, S. B. Prusiner, and F. E. Cohen, unpublished data).
While all of the studies reported here were performed in ScN2a cells and thus demand a note of caution, preliminary findings with Tg mice are encouraging. Tg(MoPrP,Q167R)FVB/Prnp0/0 mice remain well for more than 250 days after inoculation with RML prions (V. Perrier, K. Kaneko, F. E. Cohen, and S. B. Prusiner, unpublished data). The level of transgene expression is similar to that of non-Tg FVB mice which have incubation times of ∼150 days with RML prions (7). The Tg(MoPrP,Q167R)FVB/Prnp0/0 mice are currently being crossed with FVB mice to produce Tg(MoPrP,Q167R)FVB/Prnp+/+ mice in order to determine if they display dominant-negative inhibition of prion replication. Similar experiments with mice expressing MoPrP(Q218K) transgenes are also in progress.
Binding of PrPC to protein X.
The binding of PrPC to protein X is likely to be accompanied by a conformational change in PrP. We have designated this intermediate as PrP* (9). The PrP*-protein X complex might represent the functionally active form of PrP. Considerable data argue that PrP is a copper binding protein (4, 19, 33, 54, 55, 61). Whether PrP binds copper in the extracellular milieu and protein X participates in this process remains to be established.
Several lines of evidence presented here and in earlier studies suggest that the PrP*-protein X complex is the substrate for binding to PrPSc. The PrPSc-PrP*-protein X complex is converted through an as yet undefined process by which PrP* is converted into PrPSc and protein X is released (40). How PrPSc acts as a template in directing the refolding of PrP* into a nascent PrPSc molecule remains to be established. Considerable evidence now argues that each prion strain represents a different conformation of PrPSc (2, 37, 47, 48, 58).
The results of combined mutagenesis and structural studies indicate that a discontinuous epitope in the C-terminal PrP domain binds to protein X (22, 24). This epitope includes residues 214 and 218 from one side of helix C, and a loop structure defined by residues 161 to 172. Selective substitutions at positions 167, 171, 214, and 218 prevented the conversion of the mutant proteins. Substitution of basic residues at position 167 or 218 produced mutant PrPs that act as dominant-negative inhibitors by diminishing the conversion of wt PrPC into PrPSc. The same substitutions in sheep and humans render them resistant to scrapie and CJD, respectively (20, 51, 62).
In the study reported here, we present evidence that the dominant-negative effects of substitutions at positions 171, 214, and 218 are not additive and that modifications of charges in three out of the four residues involved in the binding to protein X diminished or abolished PrPC-protein X complex formation. Therefore, we focused our studies on the peculiarity of the 218K substitution alone to define more precisely the domains of PrP that are required to bind to protein X.
Allosteric effects of N-terminal deletions.
The results reported here demonstrate that N-terminal deletions profoundly alter the binding of PrP to protein X. Unexpectedly, truncated PrPC molecules containing the Q218K substitution with deletion of residues 23 to 120, 107 to 120, or 23 to 111 did not exhibit dominant-negative inhibition of PrPSc formation.
In a series of deletion mutagenesis studies, we found that the N-terminal domain of PrP plays an important role in maintaining the integrity of the protein X binding site. Whether the N-terminal PrP domain modifies the binding of PrP to protein X by altering the structure of the binding epitope or whether it binds directly to protein X remains to be established. Sequence analysis revealed that the N-terminal region of PrP is highly conserved between species, which suggests an important structural role (1). Although PrP(Δ23-88) was converted into PrPSc molecules in ScN2a cells and PrP 27-30 generated by limited proteolysis of the N-terminal 67 amino acids of PrPSc is infectious, PrP(Δ23-88) was not readily converted into PrPSc in Tg(MHM2PrP,Δ23-88)Prnp0/0 mice inoculated with RML prions composed of full-length MoPrPSc (56). However, Tg(MHM2PrP,Δ23-88)Prnp+/0 mice hemizygous for Prnpa were susceptible to RML prions with incubation times much shorter than those seen in Prnp+/0 mice. An additional PrP deletion (of residues 141 to 176) rendered Tg(PrP106) mice expressing PrP(Δ23-88,Δ141-176), composed of 106 amino acids, susceptible to RML prions. Because half of the protein X binding site in PrP106 has been deleted, we considered the possibility that PrP106 may not bind protein X and that it assumes a PrP*-like conformation without the aid of protein X (56).
In contrast to our results with Tg mice, other investigators have reported that Tg(PrP,Δ32-88) and Tg(PrP,Δ32-93) mice are susceptible to RML prions (13, 53). In these Tg mice, the nine N-terminal amino acids (KKRPKPGGW) of PrP were preserved. Intrigued by the differences between our N-terminal PrP deletion mice and these Tg mice, we examined a series of N-terminal deletion mutants for conversion into PrPSc in ScN2a cells and for dominant-negative inhibition of wt PrPSc formation. Interestingly, deletion of the octarepeat sequences (residues 52 to 91) did not alter PrPSc formation and PrP(Δ52-91,Q218K) exhibited dominant-negative inhibition of wt PrPSc formation. PrP(Δ52-91,Q218K) was a much more potent inhibitor of wt PrPSc formation than PrP(Δ23-88,Q218K) (Table 1). We also found that an additional 22 residues could be deleted from the N terminus with only a slight diminution of the dominant-negative inhibition of PrPSc formation. That PrP(Δ30-91,Q218K) inhibited PrPSc formation argues that the extreme N-terminal PrP residues KKRPKPG modify the binding of PrP to protein X.
TABLE 1.
Dominant-negative inhibition of PrPSc formation
| Substitution in MHM2 PrP(Q218K) | Relative inhibition of PrPSc formationa
|
|
|---|---|---|
| PrP(Δ23-88) | PrP | |
| None | 10 | 10 |
| V214I | ND | 2 |
| Q171R,V214I | ND | 2 |
| Δ23-88 | 10 | 2 |
| Δ52-91 | 10 | 10 |
| Δ32-80 | 10 | 8 |
| Δ30-91 | 10 | 8 |
| OR+6 | ND | 10 |
| Δ112-145 | ND | 0 |
| Δ23-88,Δ108-121 | ND | 0 |
| Δ107-120 | ND | 0 |
| Δ23-120 | 0 | 0 |
| Δ23-111 | ND | 0 |
Relative inhibition of PrPSc formation by mutant PrPs carrying the Q218K polymorphism was assessed in ScN2a cells. Data on the dominant-negative inhibition of the conversion of PrP(Δ23-88) and full-length PrP into PrPSc are summarized. Full-length PrP(Q218K) displayed maximal dominant-negative inhibition and was assigned an inhibition score of 10. PrP(Δ23-88,Q218K) displayed minimal dominant-negative inhibition when full-length PrP was the substrate and was assigned an inhibition score of 2. No inhibition was scored as 0. ND, not determined.
Prion replication.
The discovery that dominant-negative inhibition of PrPSc formation is governed by a discontinuous epitope in the C-terminal domain suggested that the rate-limiting step in PrPSc formation is generally the binding of PrPC to protein X (40). Although protein X has not been isolated, the work presented here provide additional evidence for the existence of this protein. Defining the requirements for maximal binding of PrP to protein X should greatly facilitate efforts to identify this protein. Interestingly, the four positively charged residues (in bold type) in the sequence KKRPKP at the extreme N terminus of PrP seem to be responsible for the profound effect that the N terminus of PrP has on the binding of PrP to protein X. Determining the process by which these basic residues modify PrP binding to protein X is likely to be of considerable importance in elucidating the mechanism of PrPSc formation.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institutes of Health (NS14069, AG08967, AG02132, and AG10770), the American Health Assistance Foundation, and the French Foundation as well as by a gift from the G. Harold and Leila Y. Mathers Foundation. L.Z. was supported by a postdoctoral fellowship from the International Human Frontier Science Program.
REFERENCES
- 1.Bamborough P, Wille H, Telling G C, Yehiely F, Prusiner S B, Cohen F E. Prion protein structure and scrapie replication: theoretical, spectroscopic and genetic investigations. Cold Spring Harbor Symp Quant Biol. 1996;61:495–509. [PubMed] [Google Scholar]
- 2.Bessen R A, Marsh R F. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borchelt D R, Scott M, Taraboulos A, Stahl N, Prusiner S B. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J Cell Biol. 1990;110:743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown D R, Qin K, Herms J W, Madlung A, Manson J, Strome R, Fraser P E, Kruck T, von Bohlen A, Schulz-Schaeffer W, Giese A, Westaway D, Kretzschmar H. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 5.Bruce M E, McBride P A, Farquhar C F. Precise targeting of the pathology of the sialoglycoprotein, PrP, and vacuolar degeneration in mouse scrapie. Neurosci Lett. 1989;102:1–6. doi: 10.1016/0304-3940(89)90298-x. [DOI] [PubMed] [Google Scholar]
- 6.Butler D A, Scott M R D, Bockman J M, Borchelt D R, Taraboulos A, Hsiao K K, Kingsbury D T, Prusiner S B. Scrapie-infected murine neuroblastoma cells produce protease-resistant prion proteins. J Virol. 1988;62:1558–1564. doi: 10.1128/jvi.62.5.1558-1564.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson G A, Ebeling C, Yang S-L, Telling G, Torchia M, Groth D, Westaway D, DeArmond S J, Prusiner S B. Prion isolate specified allotypic interactions between the cellular and scrapie prion proteins in congenic and transgenic mice. Proc Natl Acad Sci USA. 1994;91:5690–5694. doi: 10.1073/pnas.91.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caughey B, Raymond G J. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- 9.Cohen F E, Prusiner S B. Pathologic conformations of prion proteins. Annu Rev Biochem. 1998;67:793–819. doi: 10.1146/annurev.biochem.67.1.793. [DOI] [PubMed] [Google Scholar]
- 10.DeArmond S J, Mobley W C, DeMott D L, Barry R A, Beckstead J H, Prusiner S B. Changes in the localization of brain prion proteins during scrapie infection. Neurology. 1987;37:1271–1280. doi: 10.1212/wnl.37.8.1271. [DOI] [PubMed] [Google Scholar]
- 11.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Structure of the recombinant full-length hamster prion protein PrP(29-231): the N terminus is highly flexible. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edenhofer F, Rieger R, Famulok M, Wendler W, Weiss S, Winnacker E-L. Prion protein PrPC interacts with molecular chaperones of the Hsp60 family. J Virol. 1996;70:4724–4728. doi: 10.1128/jvi.70.7.4724-4728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 14.Goldfarb L G, Brown P, McCombie W R, Goldgaber D, Swergold G D, Wills P R, Cervenakova L, Baron H, Gibbs C J J, Gajdusek D C. Transmissible familial Creutzfeldt-Jakob disease associated with five, seven, and eight extra octapeptide coding repeats in the PRNP gene. Proc Natl Acad Sci USA. 1991;88:10926–10930. doi: 10.1073/pnas.88.23.10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorodinsky A, Harris D A. Glycolipid-anchored proteins in neuroblastoma cells form detergent-resistant complexes without caveolin. J Cell Biol. 1995;129:619–627. doi: 10.1083/jcb.129.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegde R S, Mastrianni J A, Scott M R, DeFea K A, Tremblay P, Torchia M, DeArmond S J, Prusiner S B, Lingappa V R. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 17.Holt C E, Garlick N, Cornel E. Lipofection of cDNAs in the embryonic vertebrate central nervous system. Neuron. 1990;4:203–214. doi: 10.1016/0896-6273(90)90095-w. [DOI] [PubMed] [Google Scholar]
- 18.Hornemann S, Glockshuber R. Autonomous and reversible folding of a soluble amino-terminally truncated segment of the mouse prion protein. J Mol Biol. 1996;262:614–619. doi: 10.1006/jmbi.1996.0487. [DOI] [PubMed] [Google Scholar]
- 19.Hornshaw M P, McDermott J R, Candy J M, Lakey J H. Copper binding to the N-terminal tandem repeat region of mammalian and avian prion protein: structural studies using synthetic peptides. Biochem Biophys Res Commun. 1995;214:993–999. doi: 10.1006/bbrc.1995.2384. [DOI] [PubMed] [Google Scholar]
- 20.Hunter N, Cairns D, Foster J D, Smith G, Goldmann W, Donnelly K. Is scrapie solely a genetic disease? Nature. 1997;386:137. doi: 10.1038/386137a0. [DOI] [PubMed] [Google Scholar]
- 21.Hunter N, Moore L, Hosie B D, Dingwall W S, Greig A. Association between natural scrapie and PrP genotype in a flock of Suffolk sheep in Scotland. Vet Rec. 1997;140:59–63. doi: 10.1136/vr.140.3.59. [DOI] [PubMed] [Google Scholar]
- 22.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko K, Vey M, Scott M, Pilkuhn S, Cohen F E, Prusiner S B. COOH-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scrapie isoform. Proc Natl Acad Sci USA. 1997;94:2333–2338. doi: 10.1073/pnas.94.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kariya K, Karns L R, Simpson P C. Expression of a constitutively activated mutant of the β-isozyme of protein kinase C in cardiac myocytes stimulates the promoter of the β-myosin heavy chain isogene. J Biol Chem. 1991;266:10023–10026. [PubMed] [Google Scholar]
- 26.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R I, Wisniewski H M, Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Martins V R, Graner E, Garcia-Abreu J, de Souza S J, Mercadante A F, Veiga S S, Zanata S M, Neto V M, Brentani R R. Complementary hydropathy identifies a cellular prion protein receptor. Nat Med. 1997;3:1376–1382. doi: 10.1038/nm1297-1376. [DOI] [PubMed] [Google Scholar]
- 29.Muramoto T, Scott M, Cohen F E, Prusiner S B. Recombinant scrapie-like prion protein of 106 amino acids is soluble. Proc Natl Acad Sci USA. 1996;93:15457–15462. doi: 10.1073/pnas.93.26.15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naslavsky N, Stein R, Yanai A, Friedlander G, Taraboulos A. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J Biol Chem. 1997;272:6324–6331. doi: 10.1074/jbc.272.10.6324. [DOI] [PubMed] [Google Scholar]
- 31.Oesch B, Teplow D B, Stahl N, Serban D, Hood L E, Prusiner S B. Identification of cellular proteins binding to the scrapie prion protein. Biochemistry. 1990;29:5848–5855. doi: 10.1021/bi00476a029. [DOI] [PubMed] [Google Scholar]
- 32.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan K-M, Stahl N, Prusiner S B. Purification and properties of the cellular prion protein from Syrian hamster brain. Protein Sci. 1992;1:1343–1352. doi: 10.1002/pro.5560011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poulter M, Baker H F, Frith C D, Leach M, Lofthouse R, Ridley R M, Shah T, Owen F, Collinge J, Brown G, Hardy J, Mullan M J, Harding A E, Bennett C, Doshi R, Crow T J. Inherited prion disease with 144 base pair gene insertion. 1. Genealogical and molecular studies. Brain. 1992;115:675–685. doi: 10.1093/brain/115.3.675. [DOI] [PubMed] [Google Scholar]
- 35.Priola S A, Chesebro B. Abnormal properties of prion protein with insertional mutations in different cell types. J Biol Chem. 1998;273:11980–11985. doi: 10.1074/jbc.273.19.11980. [DOI] [PubMed] [Google Scholar]
- 36.Prusiner S B. Creutzfeldt-Jakob disease and scrapie prions. Alzheimer Dis Assoc Disord. 1989;3:52–78. doi: 10.1097/00002093-198903010-00007. [DOI] [PubMed] [Google Scholar]
- 37.Prusiner S B. Molecular biology of prion diseases. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 38.Prusiner S B, Bolton D F D C, Kent S B, Hood L E. Purification and structural studies of a major scrapie prion protein. Cell. 1984;38:127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
- 39.Prusiner S B, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson G A, Hoppe P C, Westaway D, DeArmond S J. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 40.Prusiner S B, Scott M R, DeArmond S J, Cohen F E. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 41.Raeber A J, Sailer A, Hegyi I, Klein M A, Rulike T, Fischer M, Brandner S, Aguzzi A, Weissmann C. Ectopic expression of prion protein (PrP) in T lymphocytes or hepatocytes of PrP knockout mice is insufficient to sustain prion replication. Proc Natl Acad Sci USA. 1999;96:3987–3992. doi: 10.1073/pnas.96.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ridley R M, Baker H F, Crow T J. Transmissible and nontransmissible neurodegenerative disease: similarities in age of onset and genetics in relation to aetiology. Psychol Med. 1986;16:199–207. doi: 10.1017/s0033291700002634. [DOI] [PubMed] [Google Scholar]
- 43.Rieger R, Edenhofer F, Lasmézas C I, Weiss S. The human 37-kDa laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nat Med. 1997;3:1383–1388. doi: 10.1038/nm1297-1383. [DOI] [PubMed] [Google Scholar]
- 44.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. NMR structure of the mouse prion protein domain PrP(121-231) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 45.Rogers M, Serban D, Gyuris T, Scott M, Torchia T, Prusiner S B. Epitope mapping of the Syrian hamster prion protein utilizing chimeric and mutant genes in a vaccinia virus expression system. J Immunol. 1991;147:3568–3574. [PubMed] [Google Scholar]
- 46.Rogers M, Yehiely F, Scott M, Prusiner S B. Conversion of truncated and elongated prion proteins into the scrapie isoform in cultured cells. Proc Natl Acad Sci USA. 1993;90:3182–3186. doi: 10.1073/pnas.90.8.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen F E, Prusiner S B. Eight prion strains have PrPSc molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 48.Scott M R, Groth D, Tatzelt J, Torchia M, Tremblay P, DeArmond S J, Prusiner S B. Propagation of prion strains through specific conformers of the prion protein. J Virol. 1997;71:9032–9044. doi: 10.1128/jvi.71.12.9032-9044.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott M R, Köhler R, Foster D, Prusiner S B. Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serban D, Taraboulos A, DeArmond S J, Prusiner S B. Rapid detection of Creutzfeldt-Jakob disease and scrapie prion proteins. Neurology. 1990;40:110–117. doi: 10.1212/wnl.40.1.110. [DOI] [PubMed] [Google Scholar]
- 51.Shibuya S, Higuchi J, Shin R-W, Tateishi J, Kitamoto T. Codon 219 Lys allele of PRNP is not found in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 1998;43:826–828. doi: 10.1002/ana.410430618. [DOI] [PubMed] [Google Scholar]
- 52.Shibuya S, Higuchi J, Shin R-W, Tateishi J, Kitamoto T. Protective prion protein polymorphisms against sporadic Creutzfeldt-Jakob disease. Lancet. 1998;351:419. doi: 10.1016/S0140-6736(05)78358-6. [DOI] [PubMed] [Google Scholar]
- 53.Shmerling D, Hegyi I, Fischer M, Blattler T, Brandner S, Gotz J, Rulicke T, Flechsig E, Cozzio A, von Mering C, Hangartner C, Aguzzi A, Weissmann C. Expression of amino-terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell. 1998;93:203–214. doi: 10.1016/s0092-8674(00)81572-x. [DOI] [PubMed] [Google Scholar]
- 54.Stöckel J, Safar J, Wallace A C, Cohen F E, Prusiner S B. Prion protein selectively binds copper (II) ions. Biochemistry. 1998;37:7185–7193. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 55.Sulkowski E. The saga of IMAC and MIT. Bioessays. 1989;10:170–175. doi: 10.1002/bies.950100508. [DOI] [PubMed] [Google Scholar]
- 56.Supattapone S, Bosque P, Muramoto T, Wille H, Aagaard C, Peretz D, Nguyen H-O B, Heinrich C, Torchia M, Safar J, Cohen F E, DeArmond S J, Prusiner S B, Scott M. Prion protein of 106 residues creates an artificial transmission barrier for prion replication in transgenic mice. Cell. 1999;96:869–878. doi: 10.1016/s0092-8674(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 57.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszio L, Prusiner S B. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibits formation of the scrapie isoform. J Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Telling G C, Parchi P, DeArmond S J, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner S B. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 59.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 60.Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond S J, Smart E J, Anderson R G, Taraboulos A, Prusiner S B. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci USA. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Viles J H, Cohen F E, Prusiner S B, Goodin D B, Wright P E, Dyson H J. Copper binding to the prion protein: structural implications of four identical cooperative binding sites. Proc Natl Acad Sci USA. 1999;96:2042–2047. doi: 10.1073/pnas.96.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westaway D, Zuliani V, Cooper C M, Da Costa M, Neuman S, Jenny A L, Detwiler L, Prusiner S B. Homozygosity for prion protein alleles encoding glutamine-171 renders sheep susceptible to natural scrapie. Genes Dev. 1994;8:959–969. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]
- 63.Will R G, Cousens S N, Farrington C P, Smith P G, Knight R S G, Ironside J W. Deaths from variant Creutzfeldt-Jakob disease. Lancet. 1999;353:979. doi: 10.1016/s0140-6736(99)01160-5. [DOI] [PubMed] [Google Scholar]
- 64.Will R G, Ironside J W, Zeidler M, Cousens S N, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith P G. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet. 1996;347:921–925. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 65.Yehiely F, Bamborough P, Da Costa M, Perry B J, Thinakaran G, Cohen F E, Carlson G A, Prusiner S B. Identification of candidate proteins binding to prion protein. Neurobiol Dis. 1997;3:339–355. doi: 10.1006/nbdi.1997.0130. [DOI] [PubMed] [Google Scholar]