Abstract
We have previously described an approach that employs retroviral receptor-ligand bridge proteins to target retroviral vectors to specific cell types. To determine whether targeted retroviral entry can also be achieved using a retroviral receptor–single-chain antibody bridge protein, the TVA-MR1 fusion protein was generated. TVA-MR1 is comprised of the extracellular domain of the TVA receptor for subgroup A avian leukosis viruses (ALV-A), fused to the MR1 single-chain antibody that binds specifically to EGFRvIII, a tumor-specific form of the epidermal growth factor receptor. We show that TVA-MR1 binds specifically to a murine version of EGFRvIII and promotes ALV-A entry selectively into cells that express this cell surface marker. These studies demonstrate that it is possible to target retroviral vectors to specific cell types through the use of a retroviral receptor–single-chain antibody fusion protein.
Several different strategies have been developed for targeting infection of specific cell types by retroviral vectors. The most common approach has employed recombinant viral envelope (Env) proteins that contain either cell type-specific ligands or single-chain antibodies that bind to specific cell surface molecules (1, 4, 7–12, 15, 18–20, 24–28, 31–35, 38, 39, 41, 42, 44, 46, 48, 51–54, 57). This approach requires that the specific alterations made to Env do not affect the biosynthesis, virion assembly, or fusogenic function of the viral glycoprotein (12, 54, 57).
An alternative approach for targeting retroviral entry that employs soluble retroviral receptor-ligand bridge proteins was recently developed (5, 47). These bridge proteins are bifunctional reagents: the ligand moiety binds to specific cell surface receptors and the retroviral receptor moiety binds to Env, activating viral entry. This technique does not require making alterations to the viral glycoprotein but instead relies on “wild-type” Env-receptor interactions to target viral entry.
To demonstrate the feasibility of this approach, the TVA-EGF and TVB-EGF fusion proteins were generated. These bridge proteins contained human epidermal growth factor (EGF) fused to the extracellular domains of either the TVA receptor for subgroup A avian leukosis viruses (ALV-A) or the TVB receptor for ALV-B and ALV-D, respectively. Each of these bridge proteins promoted specific retroviral entry into cells that express the EGF receptor (EGFR) when bound to cell surfaces before viral challenge (5, 47). Furthermore, murine leukemia virus (MLV) pseudotypes bearing ALV-B Env (EnvB) and preloaded with TVB-EGF were targeted specifically to cells that express EGFR (5). These data demonstrated that retroviral vectors could be targeted to specific cell types by binding retroviral receptor-ligand bridge proteins to virions or to cell surfaces before viral challenge.
To extend the utility of this approach, we have now asked whether targeted retroviral entry into cells can also be achieved using TVA-MR1, a bridge protein that contains the extracellular domain of TVA fused to the MR1 single-chain antibody. This antibody binds specifically to the extracellular region of EGFRvIII, a variant form of the EGFR that is expressed at the surface of human tumor cells, including those derived from lung and breast carcinomas and glioblastomas (14, 17, 30, 37, 40, 49, 55, 56). EGFRvIII lacks a substantial portion of the extracellular domain of the wild-type receptor as a consequence of a deletion or rearrangement that commonly occurs when the EGFR gene is amplified during tumor biogenesis. These alterations result in constitutive, ligand-independent activation of EGFRvIII (22), which, in turn, confers a transformed phenotype upon various cell lines (22, 40).
The MR1 antibody binds to a novel polypeptide sequence that is formed at the site of the deletion or rearrangement that gives rise to EGFRvIII (29). The combination of the tumor-restricted expression of EGFRvIII and the binding specificity of the MR1 antibody makes this an attractive model system to test whether retroviral receptor–single-chain antibody bridge proteins can mediate targeted viral entry into cells. In this report, we demonstrate that TVA-MR1 can support efficient and specific ALV entry into mammalian cells that have been engineered to express a murine form of EGFRvIII.
MATERIALS AND METHODS
Viruses and immunoadhesins.
The SUA-rIgG immunoadhesin was described elsewhere and is comprised of the surface protein of the Schmidt-Ruppin A strain of Rous sarcoma virus fused to a rabbit Fc chain (58). ALV-A-specific vectors encoding the enhanced green fluorescent protein (EGFP; Clontech) were generated by transfection of DF1 cells (45) with the RCASBP(A)-EGFP plasmid (provided by Mark Federspiel and Matt van Brocklin). The transfected cells were propagated until 100% of the population expressed EGFP as determined by fluorescence microscopy. Cells were then seeded in 1,700-cm2 roller bottles, and virions were harvested every 12 h in 50 ml of medium that was equilibrated with 5% CO2. This medium was pooled and filtered through 0.45-μm-pore-size filters and stored at −80°C. Before use, the virus-containing supernatants were thawed and subjected to centrifugation at 109,000 × g for 1.5 h at 4°C. The viral pellets were then resuspended overnight at 4°C in 1/100 of the original volume of TNE buffer (5).
Replication-defective MLV vectors encoding a synthetic transmembrane form of TVA (TVAsyn) (3) or a murine form of EGFRvIII were generated. The MLV vector pMMP.TVAsyn was generated by first preparing a DNA fragment that encodes TVAsyn by PCR amplification using pDW1 plasmid template DNA (D. Wenzke and J. A. T. Young, unpublished data) and the following two primers: 5′-GCATAGCGTACCATGGCTAGATTGCTTCCTGCATTGC-3′ and 5′-CG ATCGACATGCATCCGGAACTAATCGATCTGAGCAGCGTAATCTGG-3′. The resultant DNA fragment was digested with NcoI and BspEI restriction enzymes and was subcloned into the NcoI and BspEI sites of the pMMP.EGFP plasmid (K. Bradley and J. A. T. Young, unpublished data), generating the pMMP.TVAsyn plasmid. The MLV vector pSFG.EGFRvIII contains a gene encoding a murine form of EGFRvIII (R. Carter and R. C. Mulligan, unpublished data) located between the NcoI and BamHI restriction enzyme sites of the pSFG vector.
Stocks of MLV vectors pseudotyped with the VSV-G protein were prepared from human 293T cells by using a transient transfection system essentially as described previously (5) with 5 μg of the pMD.G plasmid encoding VSV-G, 15 μg of the pMD.old.gagpol plasmid encoding MLV Gag and Pol, and 15 μg of either plasmid pSFG.EGFRvIII or plasmid pMMP.TVAsyn. Virions were harvested at 48 and 72 h postinfection, and the supernatants containing MLV-EGFRvIII(VSV-G) and MLV-TVAsyn(VSV-G) viruses were separately pooled and filtered through a 0.45-μm-pore-size filter and then stored at −80°C.
Cell lines.
Human 293T cells were propagated in Dulbecco modified Eagle medium containing 5% fetal bovine serum. 293T-EGFRvIII cells were generated by transducing 293T cells with MLV-EGFRvIII(VSV-G). Approximately 72 h after viral challenge, the transduced cell population (107 cells) was incubated for 30 min at 4°C with 5 ml of extracellular supernatant containing TVA-MR1 and then with 5 ml of extracellular supernatant containing SUA-rIgG. These cells were then incubated for 10 min at 4°C with a fluorescein isothiocyanate (FITC)-conjugated swine anti-rabbit antibody (DAKO Corp.) diluted 1:100 in medium. The cells were washed with ice-cold medium, and those expressing the EGFRvIII protein were isolated by flow cytometric sorting using a Coulter Epics Elite cell sorter. 293T-TVAsyn cells were generated in a similar manner, except that 293T cells were transduced with MLV-TVAsyn(VSV-G). Cells expressing the TVAsyn receptor were isolated by flow cytometric sorting after binding SUA-rIgG to the cells for 30 min at 4°C, followed by binding the FITC-conjugated anti-rabbit antibody; 25% of these cells with the highest level of cell surface TVAsyn were then isolated by subjecting the population to an additional round of flow cytometric sorting.
Construction and expression of TVA-MR1.
A DNA fragment encoding the MR1 single-chain antibody (29) was generated by PCR amplification, using as template DNA the plasmid pCMMP.MR1, which contains a gene encoding MR1 located between the XbaI and HindIII restriction enzyme sites of the pCMMP vector (T. Niederman et al., unpublished data), and the following two oligonucleotide primers: 5′-GAACTCCTAGGGGGACCGCAGGTACAACTCCAGCAGTCCGGGGG-3′ and 5′-GAGGGGCCCTCTAGATTATAGAGCTTTTTCAAGCTTGGTGCCATCACCG-3′. The resultant DNA fragment was digested with ApaI, end repaired with the Klenow fragment of DNA polymerase, and digested with AvrII to generate a 758-bp DNA fragment encoding MR1. This fragment was then ligated with plasmid pSS8 that had been cut with Asp718, end repaired, and then digested with AvrII. Plasmid pSS8 contains the gene encoding a TVA-heregulinβ1 fusion protein (S. Snitkovsky and J. Young, unpublished data) located between the EcoRI and HpaI restriction enzyme sites of the pCI expression vector (Promega). The resultant plasmid pSS11 encodes the TVA-MR1 fusion protein, and the authenticity of this open reading frame was confirmed by DNA sequence analysis (performed by the core DNA sequencing facility in the Department of Microbiology and Molecular Genetics at Harvard Medical School).
To generate the TVA-MR1 fusion protein, plasmid pSS11 was transfected into human 293 cells as described previously (47). Aliquots of 40 μl of extracellular supernatant taken from transfected and nontransfected human 293 cells were subjected to electrophoresis on 10% polyacrylamide gel containing sodium dodecyl sulfate under nonreducing conditions. The proteins were then transferred to a nitrocellulose membrane, and TVA-MR1 was detected by immunoblotting with 10 ml of extracellular supernatant containing SUA-rIgG and then with a horseradish peroxidase-conjugated antibody specific for rabbit immunoglobulins (Amersham), followed by enhanced chemiluminescence.
TVA-MR1 binding studies.
A suspension of 3.5 × 105 293T-EGFRvIII cells was incubated for 1 h at 4°C with increasing amounts of TVA-MR1-containing extracellular supernatant. These samples were made up to a total volume of 500 μl with 293 cell-conditioned medium (medium that was obtained from a confluent monolayer of human 293 cells). A suspension of 3.5 × 105 293T cells was incubated for 1 h at 4°C with 500 μl of extracellular supernatant that either lacked or contained TVA-MR1. Both cell populations were then washed with 2 ml of ice-cold phosphate-buffered saline (PBS) and incubated for 1 h at 4°C with 500 μl of extracellular supernatant containing SUA-rIgG. The cells were washed again with ice-cold PBS and incubated for 30 min at 4°C with the FITC-conjugated swine anti-rabbit antibody (diluted 1:100) in medium. The cells were then washed again and resuspended in 500 μl of ice-cold PBS containing 1% formaldehyde, and the bound TVA-MR1 fusion proteins were detected by flow cytometry using a Coulter Epics Ex-L instrument.
The peptide competition binding experiments were performed by incubating 500 μl of extracellular supernatant containing TVA-MR1 for 1 h at 4°C with a final 100 nM concentration of either the MR1 epitope-containing peptide (LEEKKGNYVVTDH) (29) or a scrambled (control) version of this peptide (YKELGVEVDNKHT). These samples were then incubated with 293T-EGFRvIII cells for 1 h at 4°C. TVA-MR1 proteins that were bound to the cells were then detected using SUA-rIgG and the FITC-conjugated antibody as described above. For control purposes, 500 μl of 293 cell supernatants containing a 100 nM concentration of either the MR1 epitope-containing peptide or of the control peptide was placed on ice for 1 h and then incubated with 293T-TVAsyn cells for an additional hour. The cells were then washed with ice-cold PBS, and cell surface TVAsyn proteins were detected by flow cytometry as described above.
TVA-MR1-mediated infection.
Each of the following steps was performed at 4°C, with a suspension of approximately 5 × 105 cells of each cell type, and each experiment was performed in triplicate. 293T cells were rocked together for 1 h with 500 μl of 293 cell-conditioned medium that either lacked or contained TVA-MR1. 293T-EGFRvIII cells were similarly incubated with 500 μl of extracellular supernatant containing TVA-MR1. 293T-TVAsyn cells were incubated with 500 μl of 293 cell-conditioned medium for 1 h.
Following each of these incubations, the cells were washed and resuspended in 500 μl of medium with or without 5 μl of 100-fold-concentrated RCASBP(A)-EGFP (0.5 μl of 100-fold-concentrated virus for infection of 293T-TVAsyn cells). The cells were then rocked with virus for 1 h at 4°C before plating and incubation at 37°C. The medium was replaced 20 h later; 72 h after viral challenge, the cells were resuspended in Ca2+- and Mg2+-free PBS containing 1 mM EDTA and 7 μM propidium iodide and were analyzed for EGFP expression using a Coulter Epics Ex-L flow cytometer. Dead cells that had taken up propidium iodide were excluded from this analysis by electronic gating. The viral titer was then measured by first calculating the multiplicity of infection (MOI) observed from the input virus: MOI = −ln[1 − (number of EGFP fluorescent cells/total number of cells analyzed)]. The actual titers (per microliter of 100-fold-concentrated virus) were then calculated using the following equation: (MOI × total number of cells that were challenged/5 (or divided by 0.5 in the case of 293T-TVAsyn cells). The number of fluorescent cells seen in the absence of virus was subtracted from each of the calculations.
Peptide competition experiments were performed by incubating 500 μl of extracellular supernatant containing TVA-MR1 at 4°C for 1 h with a 100 nM concentration of either the MR1 epitope-containing peptide or of the control peptide. These samples were then incubated with 293T-EGFRvIII cells for another hour at 4°C. The cells were then washed and incubated for 1 h with 500 μl of medium containing 5 μl of 100-fold-concentrated RCASBP(A)-EGFP before plating and incubation at 37°C for 18 h. The medium was then replaced, and 72 h after viral challenge, the cells were analyzed by flow cytometry as described above.
RESULTS
Production of the TVA-MR1 protein.
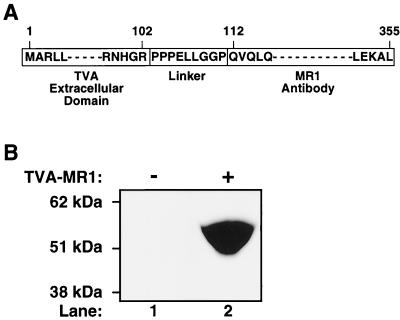
A gene that encodes the recombinant TVA-MR1 bridge protein was constructed in plasmid pSS11 (described in Materials and Methods). TVA-MR1 is comprised of the extracellular portion of the TVA receptor (2) fused to the N terminus of the MR1 single-chain antibody, which recognizes a unique sequence in the extracellular domain of EGFRvIII (29) (Fig. 1A). A proline-rich polypeptide linker that was described previously (47) was inserted between each of the two domains of TVA-MR1 (Fig. 1A) in order to maximize the probability that each domain would fold and function independently. TVA-MR1 was produced as a secreted protein, approximately 54 kDa in size, in the extracellular supernatants taken from human 293 cells transfected with plasmid pSS11 (Fig. 1B, compare lanes 1 and 2).
FIG. 1.
Production of TVA-MR1. (A) TVA-MR1 was comprised of the extracellular domain of the TVA receptor for ALV-A fused, via a proline-rich linker region, to the N-terminal end of the MR1 single-chain antibody that binds specifically to EGFRvIII. (B) Extracellular supernatants collected from transfected 293 cells that expressed TVA-MR1 (lane 2) or did not express the bridge protein (lane 1) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis under nonreducing conditions. The protein samples were then subjected to immunoblotting with SUA-rIgG (58) and a horseradish peroxidase-conjugated secondary antibody, and the TVA-MR1 protein was detected by enhanced chemiluminescence.
TVA-MR1 specifically binds to cells expressing an EGFRvIII protein.
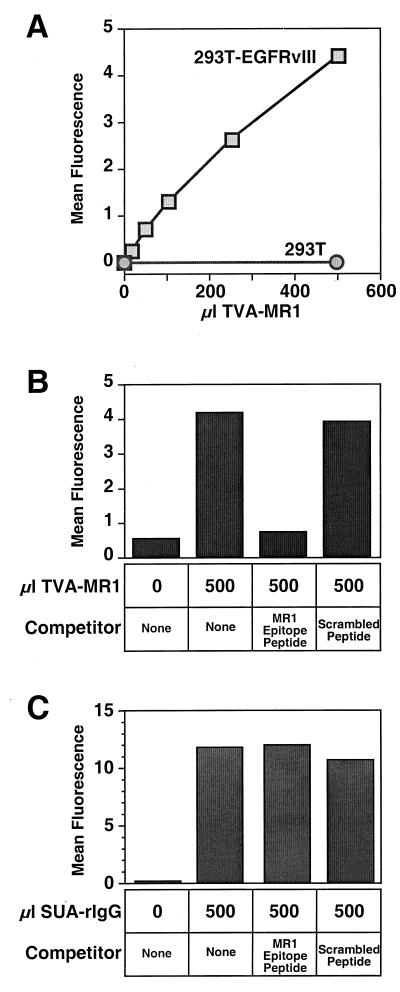
To determine whether the Env-binding and antigen-binding domains of TVA-MR1 can each function independently, we investigated whether this bridge protein can bind both to the ALV-A surface envelope (ALV-A SU) and to a cell surface EGFRvIII protein. Transfected human 293T cells expressing a murine form of EGFRvIII (293T-EGFRvIII cells) were incubated with increasing amounts of an extracellular supernatant containing TVA-MR1. For control purposes, the parental 293T cells were also incubated with TVA-MR1-containing supernatant. The bridge proteins that were bound to these cells were then detected by flow cytometry after the cells had been incubated with SUA-rIgG, an immunoadhesin composed of the ALV-A surface envelope fused to a rabbit Fc chain (58), and with an FITC-conjugated secondary antibody. TVA-MR1 bound specifically and in a dose-dependent manner to 293T-EGFRvIII cells (Fig. 2A), indicating that this bridge protein can bind simultaneously to both ALV-A SU and to cell surface EGFRvIII.
FIG. 2.
TVA-MR1 binds specifically to the EGFRvIII protein. (A) 293T cells and 293T-EGFRvIII cells were incubated with extracellular supernatants that either lacked or contained TVA-MR1. The bound TVA-MR1 proteins were then detected by flow cytometry using SUA-rIgG and an FITC-conjugated secondary antibody. (B) Extracellular supernatants containing TVA-MR1 were preincubated with either no peptide (none), the MR1 epitope-containing peptide, or a scrambled version of this peptide. These samples were then incubated with 293T-EGFRvIII cells, and the bound TVA-MR1 proteins were then detected by flow cytometry as shown in panel A. The results obtained were compared with the background levels of fluorescence obtained with cells incubated in the absence of TVA-MR1. (C) 293T-TVAsyn cells were incubated without SUA-rIgG or with SUA-rIgG and with either no peptide (none), the MR1 epitope-containing peptide, or the scrambled peptide. The bound immunoadhesin was then detected by flow cytometry using an FITC-conjugated secondary antibody. Results of a representative experiment are shown (A through C).
To obtain direct evidence that TVA-MR1 binds to the EGFRvIII protein, we examined whether the interaction of this bridge protein with 293T-EGFRvIII cells could be blocked by a synthetic peptide that contains the target epitope for the MR1 antibody (29). Indeed, this peptide blocked the binding of TVA-MR1 to these cells (Fig. 2B). By contrast, a control peptide (with the same amino acids scrambled in a different order) did not affect TVA-MR1 binding to these cells (Fig. 2B). To rule out the possibility that the MR1 epitope-containing peptide interfered nonspecifically with ALV-A SU–TVA interactions, this peptide was also tested for its effect on the binding of SUA-rIgG to a stably transfected line of human 293T cells (designated as 293T-TVAsyn) that express a transmembrane form of the TVA receptor. The MR1 epitope-containing peptide did not block the binding of SUA-rIgG to these cells (Fig. 2C), confirming that this peptide specifically blocks the interaction between TVA-MR1 and the EGFRvIII protein.
TVA-MR1 mediates specific ALV-A entry into 293T-EGFRvIII cells.
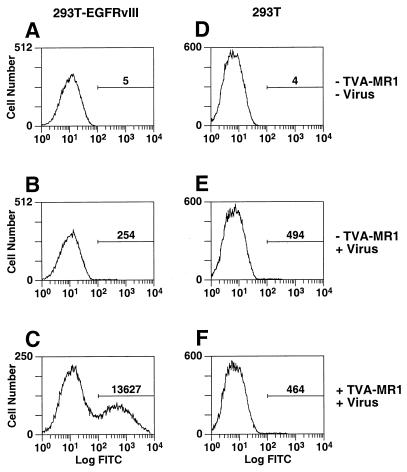
To determine whether TVA-MR1 can facilitate ALV-A infection of 293T-EGFRvIII cells, these cells and, for control purposes, 293T cells, were incubated with this bridge protein and then challenged with an ALV-A vector [RCASBP(A)-EGFP], encoding EGFP. The addition of TVA-MR1 led to a significant enhancement of ALV-A entry into 293T-EGFRvIII cells (Table 1) (Fig. 3, compare panels B and C). Indeed, TVA-MR1 acted in a dose-dependent manner to increase the susceptibility of these cells to ALV-A infection (data not shown). In these experiments, the level of TVA-MR1-mediated viral entry into 293T-EGFRvIII cells was 8.5% of the level seen with 293T-TVAsyn cells, and titers of approximately 108 infectious units/ml of 100-fold-concentrated virus were achieved (Table 1). In contrast, the addition of this bridge protein had no effect upon the susceptibility of the parental 293T cells to viral infection (Table 1) (Fig. 3, compare panels E and F).
TABLE 1.
Infection of human 293T cell lines by the ALV-A vector RCASBP(A)-EGFP
| Cell type | Viral titera
|
|
|---|---|---|
| − TVA-MR1 | + TVA-MR1 | |
| 293T | (5.96 ± 0.58) × 102 | (5.36 ± 0.10) × 102 |
| 293T-EGFRvIII | (5.94 ± 0.28) × 102 | (1.09 ± 0.01) × 105 |
| 293T-TVAsyn | (1.29 ± 0.02) × 106 | Not done |
Titers are expressed per microliter of 100-fold-concentrated virus.
FIG. 3.
TVA-MR1 promotes ALV-A infection of 293T-EGFRvIII cells. 293T-EGFRvIII cells (A through C) and 293T cells (D through F) were incubated with extracellular supernatant that contained (+) or lacked (−) TVA-MR1 as indicated. These cells were incubated in the absence or presence of the ALV-A vector RCASBP(A)-EGFP, and then aliquots of each cell type (5 × 104 293T-EGFRvIII cells and 105 293T cells) were analyzed by flow cytometry.
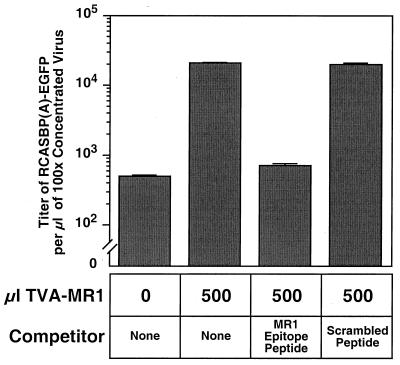
To confirm that TVA-MR1 had to be bound to the EGFRvIII protein in order to mediate enhanced levels of viral entry into 293T-EGFRvIII cells, we determined whether this effect could be blocked by the MR1 epitope-containing peptide. The MR1 epitope-containing peptide, but not the control peptide, blocked this bridge protein-dependent viral infection (Fig. 4). Therefore, TVA-MR1 is capable of facilitating targeted ALV-A entry into mammalian cells when bound to the EGFRvIII protein.
FIG. 4.
The TVA-MR1–EGFRvIII interaction is required for targeted viral entry. Extracellular supernatant containing TVA-MR1 was incubated with no peptide (none), the MR1 epitope-containing peptide, or the scrambled peptide before addition to 293T-EGFRvIII cells. The cells were then challenged with RCASBP(A)-EGFP and analyzed by flow cytometry 72 h later. For control purposes, viral infection of 293T-EGFRvIII cells was also assessed in the absence of any TVA-MR1.
DISCUSSION
In this report we have demonstrated the feasibility of using a soluble retroviral receptor–single-chain antibody bridge protein for targeting retroviral infection to a specific cell type. Cell-binding studies and peptide competition experiments confirmed that TVA-MR1 is a bifunctional reagent that can bind to a cell surface EGFRvIII protein expressed on 293T cells and to ALV-A SU. Furthermore, the binding of TVA-MR1 to the EGFRvIII protein allowed specific viral entry at a level that was approximately 7% (ranging from 1.8 to 14% in separate experiments) of that found in ALV-A infection of 293T-TVAsyn cells (Table 1 and data not shown). In contrast, TVA-MR1 did not bind to or promote infection of the parental 293T cells, which are known to express cell surface EGFRs (50). Indeed, based upon previous results that demonstrated the unique specificity of the MR1 antibody for EGFRvIII, we fully expected that TVA-MR1 would allow only ALV-A infection of cells expressing mutant, but not wild-type, EGFRs. For example, Lorimer et al. (29) have already shown that this single-chain antibody, engineered in the context of an MR1-toxin fusion protein, directs the potent killing of cells that express 400,000 EGFRvIII molecules (at a concentration of 7 to 10 ng/ml), whereas this recombinant toxin is unable to kill cells expressing 200,000 wild-type EGFR molecules even when added at concentrations as high as 1,000 ng/ml. Taken together, our studies have demonstrated that TVA-MR1 can be an efficient and specific facilitator of viral entry into cells when bound to EGFRvIII.
During these experiments we found that ALV-A is capable of infecting human 293T cells and 293T-EGFRvIII cells at a low “background” level that was 1/2,000 to 1/5,000 of the level seen with 293T-TVAsyn cells (Table 1 and data not shown). The addition of TVA-MR1 led to an 180-fold to 200-fold increase in the susceptibility of 293T-EGFRvIII cells to ALV-A infection but did not affect the “background” level of infection seen with 293T cells (Table 1). The “background” level of ALV-A infection seen with human 293T cells is similar to that seen with some other human cell lines, e.g., U250 glioma cells, but it is approximately 100-fold higher than that seen with other mammalian cell lines (6). It is important to understand why certain mammalian cell types are more susceptible to ALV infection than are others, since this information may help us eliminate such infections and thus optimize the use of retroviral receptor-ligand and retroviral receptor–single-chain antibody bridge proteins for viral targeting.
The results presented in this report suggest that it might be possible both in vivo as well as in vitro, to use retroviral receptor–single-chain antibody bridge proteins as tools to deliver retroviral vectors to specific cell types that express cognate cell surface antigens. With regard to in vivo applications, it is not yet clear whether the background level of ALV-A infection that was observed in cultured human 293T cells will represent a significant hurdle for cell type-specific viral targeting: viral targeting studies, performed with transgenic lines of mice that express a transmembrane form of TVA in specific cell types, indicate that the “background” level of ALV-A infection seen with cells that lack this viral receptor is extremely low (16). Furthermore, in considering the potential utility of this system for delivering viral vectors to tumor cells, the use of ALV-based or MLV-based vectors for gene delivery affords another level of specificity since these viruses only establish proviral DNA in dividing cell types (36, 43). Indeed, several groups have already shown that it is possible to use this feature of MLV vectors to deliver genes specifically to tumor cells, even in the absence of a selective Env-targeting system (13, 23). However, since the retroviral vectors used in these studies have a broad host range, there exists the potential for infecting other dividing cell types in addition to the target tumor cells. The use of retroviral receptor-ligand and retroviral receptor–single-chain antibody bridge proteins for viral delivery may lead to more specific viral targeting in vivo. Indeed, it will be interesting to determine the efficiency and specificity of in vivo viral targeting that can be achieved with TVA-MR1 in mouse models of human cancer that employ tumor cells expressing EGFRvIII (21). An added advantage of using retroviral receptor–single-chain antibody fusion proteins is that such bridge proteins might be useful for targeting retroviral vector infection to cells expressing a number of different cell surface factors, including those with no known ligands.
ACKNOWLEDGMENTS
We acknowledge John Daly at the Dana-Farber Cancer Institute for expert assistance with flow cytometry and members of the core DNA sequencing facility in the Department of Microbiology and Molecular Genetics at Harvard Medical School for help with DNA sequencing. We thank members of the Young laboratory for comments, suggestions, and helpful discussions and Nathan Astrof and Walther Mothes for critical reading of the manuscript. We also thank John Naughton for help preparing the final figures and Mark Federspiel and Matt von Brocklin for kindly providing viral vectors.
This work was supported by NIH grant CA 70810 from the National Cancer Institute and by grant DAMD17-98-1-8488 from the Department of the Army.
REFERENCES
- 1.Ager S, Nilson B H, Morling F J, Peng K W, Cosset F L, Russell S J. Retroviral display of antibody fragments; interdomain spacing strongly influences vector infectivity. Hum Gene Ther. 1996;7:2157–2164. doi: 10.1089/hum.1996.7.17-2157. [DOI] [PubMed] [Google Scholar]
- 2.Bates P, Young J A T, Varmus H E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 3.Bélanger C, Zingler K, Young J A T. Importance of cysteines in the LDLR-related domain of the subgroup A avian leukosis and sarcoma virus receptor for viral entry. J Virol. 1995;69:1019–1024. doi: 10.1128/jvi.69.2.1019-1024.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benedict C A, Tun R Y, Rubinstein D B, Guillaume T, Cannon P M, Anderson W F. Targeting retroviral vectors to CD34-expressing cells: binding to CD34 does not catalyze virus-cell fusion. Hum Gene Ther. 1999;10:545–557. doi: 10.1089/10430349950018625. [DOI] [PubMed] [Google Scholar]
- 5.Boerger A L, Snitkovsky S, Young J A T. Retroviral vectors preloaded with a viral receptor-ligand bridge protein are targeted to specific cell types. Proc Natl Acad Sci USA. 1999;96:9867–9872. doi: 10.1073/pnas.96.17.9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bova-Hill C, Olsen J C, Swanstrom R. Genetic analysis of the Rous sarcoma virus subgroup D env gene: mammal tropism correlates with temperature sensitivity of gp85. J Virol. 1991;65:2073–2080. doi: 10.1128/jvi.65.4.2073-2080.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chadwick M P, Morling F J, Cosset F L, Russell S J. Modification of retroviral tropism by display of IGF-I. J Mol Biol. 1999;285:485–494. doi: 10.1006/jmbi.1998.2350. [DOI] [PubMed] [Google Scholar]
- 8.Chu T H, Dornburg R. Retroviral vector particles displaying the antigen-binding site of an antibody enable cell-type-specific gene transfer. J Virol. 1995;69:2659–2663. doi: 10.1128/jvi.69.4.2659-2663.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu T H, Dornburg R. Toward highly efficient cell-type-specific gene transfer with retroviral vectors displaying single-chain antibodies. J Virol. 1997;71:720–725. doi: 10.1128/jvi.71.1.720-725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu T H, Martinez I, Sheay W C, Dornburg R. Cell targeting with retroviral vector particles containing antibody-envelope fusion proteins. Gene Ther. 1994;1:292–299. [PubMed] [Google Scholar]
- 11.Cosset F L, Morling F J, Takeuchi Y, Weiss R A, Collins M K, Russell S J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosset F L, Russell S J. Targeting retrovirus entry. Gene Ther. 1996;3:946–956. [PubMed] [Google Scholar]
- 13.Culver K W, Ram Z, Wallbridge S, Ishii H, Oldfield E H, Blaese R M. In vivo gene transfer with retroviral vector-producer cells for treatment of experimental brain tumors. Science. 1992;256:1550–1552. doi: 10.1126/science.1317968. [DOI] [PubMed] [Google Scholar]
- 14.Ekstrand A J, Sugawa N, James C D, Collins V P. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci USA. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fielding A K, Maurice M, Morling F J, Cosset F L, Russell S J. Inverse targeting of retroviral vectors: selective gene transfer in a mixed population of hematopoietic and nonhematopoietic cells. Blood. 1998;91:1802–1809. [PubMed] [Google Scholar]
- 16.Fisher G H, Orsulic S, Holland E, Hively W P, Li Y, Lewis B C, Williams B O, Varmus H E. Development of a flexible and specific gene delivery system for production of murine tumor models. Oncogene. 1999;18:5253–5260. doi: 10.1038/sj.onc.1203087. [DOI] [PubMed] [Google Scholar]
- 17.Garcia de Palazzo I E, Adams G P, Sundareshan P, Wong A J, Testa J R, Bigner D D, Weiner L M. Expression of mutated epidermal growth factor receptor by non-small cell lung carcinomas. Cancer Res. 1993;53:3217–3220. [PubMed] [Google Scholar]
- 18.Hall F L, Gordon E M, Wu L, Zhu N L, Skotzko M J, Starnes V A, Anderson W F. Targeting retroviral vectors to vascular lesions by genetic engineering of the MoMLV gp70 envelope protein. Hum Gene Ther. 1997;8:2183–2192. doi: 10.1089/hum.1997.8.18-2183. [DOI] [PubMed] [Google Scholar]
- 19.Han X, Kasahara N, Kan Y W. Ligand-directed retroviral targeting of human breast cancer cells. Proc Natl Acad Sci USA. 1995;92:9747–9751. doi: 10.1073/pnas.92.21.9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatziioannou T, Delahaye E, Martin F, Russell S J, Cosset F L. Retroviral display of functional binding domains fused to the amino terminus of influenza hemagglutinin. Hum Gene Ther. 1999;10:1533–1544. doi: 10.1089/10430349950017860. [DOI] [PubMed] [Google Scholar]
- 21.Holland E C, Hively W P, DePinho R A, Varmus H E. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H S, Nagane M, Klingbeil C K, Lin H, Nishikawa R, Ji X D, Huang C M, Gill G N, Wiley H S, Cavenee W K. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 23.Hurford R K, Dranoff G, Mulligan R C, Tepper R I. Gene therapy of metastatic cancer by in vivo retroviral gene targeting. Nat Genet. 1995;10:430–435. doi: 10.1038/ng0895-430. [DOI] [PubMed] [Google Scholar]
- 24.Jiang A, Chu T H, Nocken F, Cichutek K, Dornburg R. Cell-type-specific gene transfer into human cells with retroviral vectors that display single-chain antibodies. J Virol. 1998;72:10148–10156. doi: 10.1128/jvi.72.12.10148-10156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang A, Dornburg R. In vivo cell type-specific gene delivery with retroviral vectors that display single chain antibodies. Gene Ther. 1999;6:1982–1987. doi: 10.1038/sj.gt.3301043. [DOI] [PubMed] [Google Scholar]
- 26.Jiang A, Fisher H, Pomerantz R J, Dornburg R. A genetically engineered spleen necrosis virus-derived retroviral vector that displays the HIV type 1 glycoprotein 120 envelope peptide. Hum Gene Ther. 1999;10:2627–2636. doi: 10.1089/10430349950016663. [DOI] [PubMed] [Google Scholar]
- 27.Kasahara N, Dozy A M, Kan Y W. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science. 1994;266:1373–1376. doi: 10.1126/science.7973726. [DOI] [PubMed] [Google Scholar]
- 28.Konishi H, Ochiya T, Chester K A, Begent R H, Muto T, Sugimura T, Terada M, Begent R H. Targeting strategy for gene delivery to carcinoembryonic antigen-producing cancer cells by retrovirus displaying a single-chain variable fragment antibody. Hum Gene Ther. 1998;9:235–248. doi: 10.1089/hum.1998.9.2-235. [DOI] [PubMed] [Google Scholar]
- 29.Lorimer I A, Keppler-Hafkemeyer A, Beers R A, Pegram C N, Bigner D D, Pastan I. Recombinant immunotoxins specific for a mutant epidermal growth factor receptor: targeting with a single chain antibody variable domain isolated by phage display. Proc Natl Acad Sci USA. 1996;93:14815–14820. doi: 10.1073/pnas.93.25.14815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malden L T, Novak U, Kaye A H, Burgess A W. Selective amplification of the cytoplasmic domain of the epidermal growth factor receptor gene in glioblastoma multiforme. Cancer Res. 1988;48:2711–2714. [PubMed] [Google Scholar]
- 31.Marin M, Noel D, Valsesia-Wittman S, Brockly F, Etienne-Julan M, Russell S, Cosset F L, Piechaczyk M. Targeted infection of human cells via major histocompatibility complex class I molecules by Moloney murine leukemia virus-derived viruses displaying single-chain antibody fragment-envelope fusion proteins. J Virol. 1996;70:2957–2962. doi: 10.1128/jvi.70.5.2957-2962.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin F, Kupsch J, Takeuchi Y, Russell S, Cosset F L, Collins M. Retroviral vector targeting to melanoma cells by single-chain antibody incorporation in envelope. Hum Gene Ther. 1998;9:737–746. doi: 10.1089/hum.1998.9.5-737. [DOI] [PubMed] [Google Scholar]
- 33.Martin F, Neil S, Kupsch J, Maurice M, Cosset F L, Collins M. Retrovirus targeting by tropism restriction to melanoma cells. J Virol. 1999;73:6923–6929. doi: 10.1128/jvi.73.8.6923-6929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matano T, Odawara T, Iwamoto A, Yoshikura H. Targeted infection of a retrovirus bearing a CD4-Env chimera into human cells expressing human immunodeficiency virus type 1. J Gen Virol. 1995;76:3165–3169. doi: 10.1099/0022-1317-76-12-3165. [DOI] [PubMed] [Google Scholar]
- 35.Maurice M, Mazur S, Bullough F J, Salvetti A, Collins M K, Russell S J, Cosset F L. Efficient gene delivery to quiescent interleukin-2 (IL-2)-dependent cells by murine leukemia virus-derived vectors harboring IL-2 chimeric envelope glycoproteins. Blood. 1999;94:401–410. [PubMed] [Google Scholar]
- 36.Miller D G, Adam M A, Miller A D. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 1990;10:4239–4242. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moscatello D K, Holgado-Madruga M, Godwin A K, Ramirez G, Gunn G, Zoltick P W, Biegel J A, Hayes R L, Wong A J. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res. 1995;55:5536–5539. [PubMed] [Google Scholar]
- 38.Nguyen T H, Pages J C, Farge D, Briand P, Weber A. Amphotropic retroviral vectors displaying hepatocyte growth factor-envelope fusion proteins improve transduction efficiency of primary hepatocytes. Hum Gene Ther. 1998;9:2469–2479. doi: 10.1089/hum.1998.9.17-2469. [DOI] [PubMed] [Google Scholar]
- 39.Nilson B H, Morling F J, Cosset F L, Russell S J. Targeting of retroviral vectors through protease-substrate interactions. Gene Ther. 1996;3:280–286. [PubMed] [Google Scholar]
- 40.Nishikawa R, Ji X D, Harmon R C, Lazar C S, Gill G N, Cavenee W K, Huang H J. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng K W, Morling F J, Cosset F L, Murphy G, Russell S J. A gene delivery system activatable by disease-associated matrix metalloproteinases. Hum Gene Ther. 1997;8:729–738. doi: 10.1089/hum.1997.8.6-729. [DOI] [PubMed] [Google Scholar]
- 42.Peng K W, Vile R G, Cosset F L, Russell S J. Selective transduction of protease-rich tumors by matrix-metalloproteinase-targeted retroviral vectors. Gene Ther. 1999;6:1552–1557. doi: 10.1038/sj.gt.3300982. [DOI] [PubMed] [Google Scholar]
- 43.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell S J, Hawkins R E, Winter G. Retroviral vectors displaying functional antibody fragments. Nucleic Acids Res. 1993;21:1081–1085. doi: 10.1093/nar/21.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaefer-Klein J, Givol I, Barsov E V, Whitcomb J M, Van Brocklin M, Foster D N, Federspiel M J, Hughes S H. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology. 1998;248:305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 46.Schnierle B S, Moritz D, Jeschke M, Groner B. Expression of chimeric envelope proteins in helper cell lines and integration into Moloney murine leukemia virus particles. Gene Ther. 1996;3:334–342. [PubMed] [Google Scholar]
- 47.Snitkovsky S, Young J A T. Cell-specific viral targeting mediated by a soluble retroviral receptor-ligand fusion protein. Proc Natl Acad Sci USA. 1998;95:7063–7068. doi: 10.1073/pnas.95.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somia N V, Zoppe M, Verma I M. Generation of targeted retroviral vectors by using single-chain variable fragment: an approach to in vivo gene delivery. Proc Natl Acad Sci USA. 1995;92:7570–7574. doi: 10.1073/pnas.92.16.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugawa N, Ekstrand A J, James C D, Collins V P. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci USA. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas D, Bradshaw R A. Differential utilization of SHCA tyrosine residues and functional domains in the transduction of epidermal growth factor-induced mitigen-activated protein kinase activation in 293T cells and nerve growth factor-induced neurite outgrowth in PC12 cells—identification of a new GRB2-center-DOT-SOS1 binding site. J Biol Chem. 1997;272:22293–22299. doi: 10.1074/jbc.272.35.22293. [DOI] [PubMed] [Google Scholar]
- 51.Valsesia-Wittmann S, Drynda A, Deleage G, Aumailley M, Heard J M, Danos O, Verdier G, Cosset F L. Modifications in the binding domain of avian retrovirus envelope protein to redirect the host range of retroviral vectors. J Virol. 1994;68:4609–4619. doi: 10.1128/jvi.68.7.4609-4619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valsesia-Wittmann S, Morling F J, Hatziioannou T, Russell S J, Cosset F L. Receptor co-operation in retrovirus entry: recruitment of an auxiliary entry mechanism after retargeted binding. EMBO J. 1997;16:1214–1223. doi: 10.1093/emboj/16.6.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valsesia-Wittmann S, Morling F J, Nilson B H, Takeuchi Y, Russell S J, Cosset F L. Improvement of retroviral retargeting by using amino acid spacers between an additional binding domain and the N terminus of Moloney murine leukemia virus SU. J Virol. 1996;70:2059–2064. doi: 10.1128/jvi.70.3.2059-2064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verma I M, Somia N. Gene therapy—promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 55.Wong A J, Ruppert J M, Bigner S H, Grzeschik C H, Humphrey P A, Bigner D S, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamazaki H, Fukui Y, Ueyama Y, Tamaoki N, Kawamoto T, Taniguchi S, Shibuya M. Amplification of the structurally and functionally altered epidermal growth factor receptor gene (c-erbB) in human brain tumors. Mol Cell Biol. 1988;8:1816–1820. doi: 10.1128/mcb.8.4.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y, Zhu L, Lee S, Li L, Chang E, Soong N W, Douer D, Anderson W F. Identification of the block in targeted retroviral-mediated gene transfer. Proc Natl Acad Sci USA. 1999;96:4005–4010. doi: 10.1073/pnas.96.7.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zingler K, Young J A T. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J Virol. 1996;70:7510–7516. doi: 10.1128/jvi.70.11.7510-7516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]