Abstract
Pulmonary infections pose formidable challenges in clinical settings with high mortality rates across all age groups worldwide. Accurate diagnosis and early intervention are crucial to improve patient outcomes. Artificial intelligence (AI) has the capability to mine imaging features specific to different pathogens and fuse multimodal features to reach a synergistic diagnosis, enabling more precise investigation and individualized clinical management. In this study, we successfully developed a multimodal integration (MMI) pipeline to differentiate among bacterial, fungal, and viral pneumonia and pulmonary tuberculosis based on a real-world dataset of 24,107 patients. The area under the curve (AUC) of the MMI system comprising clinical text and computed tomography (CT) image scans yielded 0.910 (95% confidence interval [CI]: 0.904–0.916) and 0.887 (95% CI: 0.867–0.909) in the internal and external testing datasets respectively, which were comparable to those of experienced physicians. Furthermore, the MMI system was utilized to rapidly differentiate between viral subtypes with a mean AUC of 0.822 (95% CI: 0.805–0.837) and bacterial subtypes with a mean AUC of 0.803 (95% CI: 0.775–0.830). Here, the MMI system harbors the potential to guide tailored medication recommendations, thus mitigating the risk of antibiotic misuse. Additionally, the integration of multimodal factors in the AI-driven system also provided an evident advantage in predicting risks of developing critical illness, contributing to more informed clinical decision-making. To revolutionize medical care, embracing multimodal AI tools in pulmonary infections will pave the way to further facilitate early intervention and precise management in the foreseeable future.
Graphical abstract
Public summary
-
•
Multimodal integration pipeline could diagnose pulmonary infections based on clinical characteristics, CT images, and laboratory test results.
-
•
The pipeline achieved diagnostic performance comparable with senior physicians.
-
•
In addition to assessing pulmonary infections, the automated system could also predict progression to critical illness and facilitate early intervention.
Introduction
Lower respiratory tract infections are one of the leading causes of death worldwide and contribute to a significant disease burden, which impacted 344 million people and resulted in 2.18 million deaths according to the 2021 Global Burden of Diseases study.1,2 Pneumonia—attributable to bacteria, viruses, and fungi—is recognized as the most pervasive infectious disease globally, further aggravated by the COVID-19 pandemic.3 In addition, Mycobacterium tuberculosis also poses a significant lung infection risk, with 6.2 million people diagnosed with pulmonary tuberculosis (PTB) worldwide.4,5 There are variations in the treatment strategies for different pathogens, but a staggering 62% of patients lack a definitive pathogen diagnosis.6,7,8 The traditional diagnostic gold standard requires obtaining respiratory samples for microbial smears or cultures, which is plagued by insensitivity and protracted processing time.3,9 Once pulmonary infections patients receive delayed treatment and develop severe disease, their mortality rate soars to 50%.10 Hence, a rapid and accurate method for pulmonary infection diagnosis, pathogen detection, and severe disease prediction is urgently needed in clinical scenarios.
The diagnosis of pulmonary infections calls for a multifaceted approach encompassing clinical assessments, laboratory tests, and other indicators.3,11 The predominant symptoms, cough and expectoration, are common clinical manifestations. Additionally, peripheral blood leukopenia has significance in bacterial infection, signaling critical illness and an unfavorable prognosis. Chest computed tomography (CT) also acts as a crucial supportive tool for diagnosing pulmonary infections, offering three-dimensional representations of pulmonary structures.12 Discrepancies exist in the imaging manifestations of different types of pathogenic infections. For example, the imaging of bacterial pneumonia typically reveals alveolar infiltrates or solid changes, whereas the imaging of viral pneumonia is characterized mostly by ground-glass opacities (GGOs) alongside solid shadows.11 In cases of severe pneumonia, the percentage of lesion-involved areas in the chest CT images of patients frequently exceeds 50%.13 However, the radiographic manifestations of pneumonia exhibit an overlap across diverse pathogens, and the imaging signs of specific pathogenic infections can vary among individuals. This phenomenon complicates the accurate diagnosis of pulmonary infections for physicians and renders the non-invasive identification of the pathogen even more challenging.
The rapid development of artificial intelligence (AI) technology has catalyzed innovation regarding medical tasks.14,15,16,17,18 AI models have garnered remarkable results in skin cancer subtype classification, diabetic retinopathy diagnosis, respiratory disease diagnosis, and prognosis assessment.19,20,21,22,23,24 With the evolution of AI technology, a trend of multimodal fusion has emerged, providing a more comprehensive approach that resonates with real-world clinical applications.25,26,27,28 In terms of pulmonary infections, a surge of intelligent diagnostic models has been noted in recent years.29,30,31,32,33,34,35,36,37 For instance, advanced diagnostic architectures employing the DenseNet-121 deep learning network have been established to identify viral, non-viral, and COVID-19 pneumonia based on chest X-ray (CXR) images, demonstrating impressive discrimination capabilities (area under the curve [AUC]: 0.867–0.966).29 Another convolutional neural network model using CXR images was constructed to diagnose PTB with an AUC of 0.992 (95% confidence interval [CI]: 0.961–1.000), and the performance of the pre-trained network was 11% higher than that of the untrained network.30 However, CXR is a two-dimensional imaging technique with inherent limitations regarding diagnostic utility. Furthermore, an AI model based on the quantitative analysis of CT images was constructed to automatically localize typical foci of pneumonia, such as GGOs and solid changes, accurately distinguishing pneumonia with remarkable preformance (AUC: 0.975).31 Additionally, investigations have integrated CT images with clinical information from diverse cohorts to rapidly diagnose pulmonary infections patients, outperforming the single-modality model (AUC: 0.92 vs. 0.80).32 However, the majority of studies focus narrowly on individual pathogen but ignore the complexity of multiple infection types. Moreover, merely utilizing unimodal imaging data for detecting pneumonia have curtailed the ability of these AI models to make a more detailed and accurate diagnosis compared to the use of multimodal information, which integrates data from various dimensions.
In this study, we developed a multimodal integration (MMI) pipeline to assist with the precise diagnosis of bacterial, fungal, and viral pneumonia as well as PTB and to facilitate pathogen prediction (Figure 1). In addition, crucial features were filtered to predict the probability of severe pneumonia. The MMI system holds the potential to assist physicians with improving clinical efficiency and personalizing the fine management of patients with pulmonary infections.
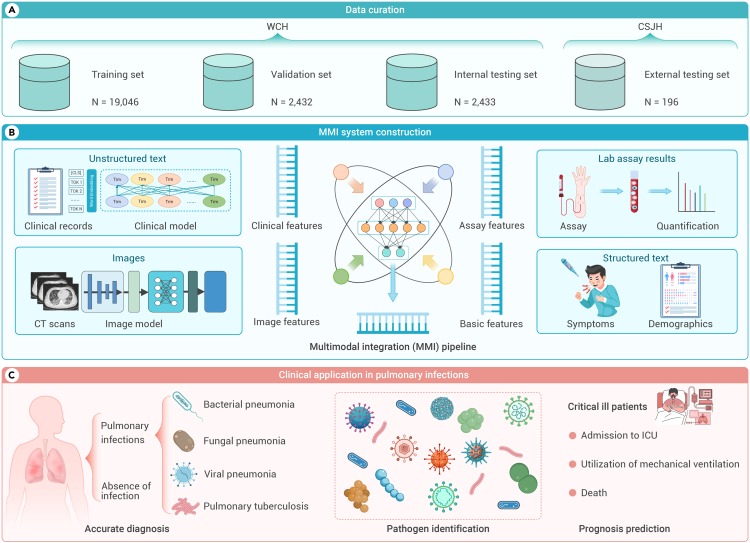
Figure 1.
Overview of the multimodal integration (MMI) pipeline for pulmonary infection management
(A) Patients were identified and divided into training (WCH, N = 19,046), validation (WCH, N = 2,432), internal (WCH, N = 2,433), and external testing (CSJH; N = 196) datasets for the MMI system.
(B) Development of the MMI system through the integration of multimodal data. It integrated multimodal information, including unstructured text, images, lab assay results, and structured text to construct the system.
(C) Application of the MMI system in clinical settings for the diagnosis of infectious diseases, identification of pathogens, and risk prediction of critical patients, showcasing the actual utility of the AI model in managing pulmonary infections. CSJH, Chengdu ShangJin Nanfu Hospital; ICU, intensive care unit; WCH, West China Hospital.
Results
Participant characteristics
An extensive large dataset of 161,258 chest CT scans from 54,581 patients with associated clinical information and laboratory examinations was assembled from West China Hospital (WCH) of Sichuan University and Chengdu ShangJin Nanfu Hospital (CSJH). The data from WCH were employed for training, validation, and internal testing, while the data from CSJH served as an external testing set to ensure the robustness of the model. The study population (subset 1, primary prediction) consisted of 24,107 patients to discriminate pulmonary infections (Table S1).
To advance the retrospective diagnosis of infection subtypes and prognosticate severe acute respiratory failure, we developed the multimodal system to predict four infections, specific infectious pathogens, and severe pneumonia utilizing distinct subsets (Figure S1). Subset 2 (infection classification, N = 13,361) was utilized to construct and validate the performance of the MMI system in predicting primary infections. The infections were categorized into four types: bacterial pneumonia (BP), fungal pneumonia (FP), viral pneumonia (VP), and PTB. Additionally, subset 3 (virus subtypes prediction, N = 2,520) and subset 4 (bacteria subtypes prediction, N = 535) were employed to identify the specific infectious pathogens associated with the viral and bacterial pneumonia. Furthermore, subset 5, consisting of 2,672 patients with severe pneumonia, was thoroughly examined. Severe pneumonia was defined based on criteria including clinical need for transfer to the intensive care unit (ICU), the requirement for mechanical ventilation, or death. These patients were identified based on their progression to critical outcomes after admission. This subset was specifically selected to evaluate the performance of our system in predicting critical outcomes that occurred 14 days after admission, specifically targeting patients at a higher risk of developing acute respiratory failure.
MMI system development
The proposed disease recognition system incorporated clinical medical records, laboratory test results, and CT images to expedite the identification of pulmonary infections. To effectively extract information from medical records text, we leveraged bidirectional encoder representations from transformers (BERT), an advanced network-based pre-training technique for natural language processing.38,39 Additionally, to capture the spatial features of the CT image findings, we utilized image-based backbone networks known as Swin-transformer structures.40,41 The Swin-transformer is an architecture based on transformer that hierarchically process input tokens. This hierarchical processing allows for the incorporation of larger patch sizes and deeper models, thus enhancing performance in image recognition tasks while maintaining computational efficiency.
To further bolster the performance and exploit the complementary information from textual and image features, this multimodal system integrated basic demographic details, chief complaints, laboratory test results, and CT scans to assess infectious diseases. Employing an attention architecture, we amalgamated the unimodal feature spaces learned from clinical and image data into a unified representation, forming the multimodal features. By extracting discriminative features from the clinical and image data, this system captured the specific information characteristics of each modality. These features were then integrated into a shared feature space, allowing the capture of intricate relationships and leveraging complementary information across different modalities. This integration augmented disease identification accuracy, empowering the MMI system to integrate multiple sources of information and effectively make precise predictions.
Accurate diagnosis of pulmonary infections
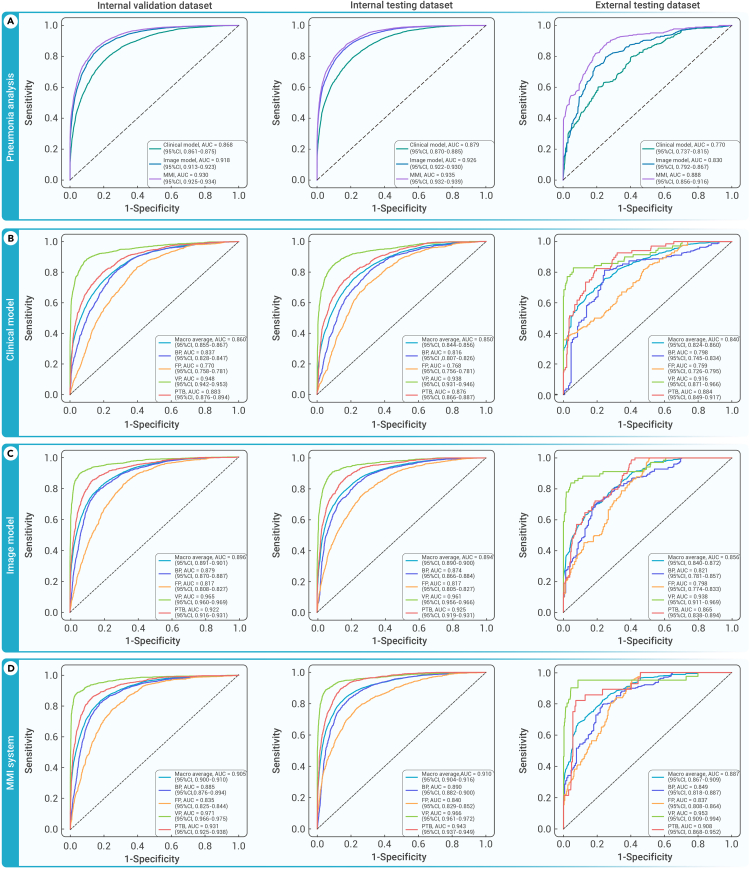
In this primary prediction task, our findings demonstrated that both clinical features and image features alone exhibited significant discriminative power in distinguishing infections from cases without infections, thus achieving a satisfactory baseline performance (Figure 2A; Table S2). Specifically, the clinical model achieved an AUC of 0.879 (95% CI: 0.870–0.885), while the image model achieved an AUC of 0.926 (95% CI: 0.922–0.930) on the internal testing cohort. These results underscore the potential of both clinical and image features as valuable discriminators for infections amid lower respiratory tract conditions. Nevertheless, integrating these features resulted in a substantial improvement in the performance of the MMI system. When combined, the accuracy of MMI system increased significantly to 0.849 (95% CI: 0.844–0.855), with a sensitivity of 0.866 (95% CI: 0.857–0.874), specificity of 0.838 (95% CI: 0.829–0.848), and AUC of 0.935 (95% CI: 0.932–0.939) (Table S2). These findings emphasized the critical role of multi-information integration and the effectiveness of fusion strategies in enhancing the accuracy of infection identification. The combination of clinical and image features furnished a more comprehensive and more robust framework to accurately discriminate pulmonary infections from other diseases with heightened accuracy.
Figure 2.
Performance of the MMI system in identifying patients with pulmonary infections
(A) Comparison of the receiver operating characteristic (ROC) curves of the clinical model, image model, and MMI system for detecting pulmonary infections in the validation, internal testing, and external testing datasets.
(B–D), The ROCs for predicting four pulmonary infection types (bacterial pneumonia, fungal pneumonia, viral pneumonia, and PTB) utilizing the clinical model (B), image model (C), and MMI system (D) in the validation, internal testing, and external testing datasets. BP, bacterial pneumonia; FP, fungal pneumonia; PTB, pulmonary tuberculosis; VP, viral pneumonia.
Remarkably, the multimodal system demonstrated exemplary performance in identifying infections within the external testing set. It achieved a mean accuracy of 0.919 (95% CI: 0.898–0.937), signifying a remarkably high level of overall correctness (Table S2). Additionally, the AUC exhibited a significant improvement, increasing from 0.830 (95% CI: 0.792–0.867) of the image model to 0.888 (95% CI: 0.856–0.916) of the MMI system. This notable increase further corroborated superior generalization ability of the MMI system in handling unknown data distributions and highlights its robustness in accurately distinguishing between infected and non-infected cases.
Performance of MMI system in single and mixed infections
It is notable that the current AI models tend to focus on single infection, often neglecting mixed infections. This narrow focus will potentially jeopardize their efficacy and impede their practical applications. To solve this problem, we utilized multilabel system to distinguish between single infection type and more than one infection type. This approach enabled us to assess the performance of the model in a more diverse and clinically relevant setting.
When comparing the diagnosis of single-infection patients to that of mixed infections, the latter posed greater challenges, as indicated in Table S3. Remarkably, the performance of the single infection model yielded an impressive AUC of 0.949 (95% CI: 0.943–0.954), while the mixed infections cases still achieved a respectable AUC of 0.876 (95% CI: 0.861–0.890). These results highlighted the capability of our multilabel multimodal model to deliver superior performance in both the relatively simpler cases of single infection and the more complex cases of mixed infections.
Precise classification of infectious diseases
Pulmonary infections encompass various subtypes, such as bacterial, fungal, and viral pneumonia and PTB. Nevertheless, accurate differentiation of respiratory tract infections presents a formidable challenge, even for experienced physicians in tertiary hospitals. When considering clinical features alone, the model achieved an accuracy of 0.788 (95% CI: 0.777–0.798) and an average mean AUC of 0.850 (95% CI: 0.844–0.856) in the internal testing dataset (Figures 2B; Table 1). In a similar vein, exclusively employing image features, the model achieved an accuracy of 0.821 (95% CI: 0.812–0.830) and an average mean AUC of 0.894 (95% CI: 0.890–0.900) (Figure 2C). Significantly, amplification of the model’s efficacy by supplementing information and purging noisy or incomplete data from these modalities culminated in peak performance. The MMI system realized an accuracy of 0.848 (95% CI: 0.838–0.856), a sensitivity of 0.846 (95% CI: 0.830–0.860), a specificity of 0.847 (95% CI: 0.837–0.858), and an AUC of 0.910 (95% CI: 0.904–0.916) in the internal testing dataset (Figure 2D). In external testing dataset, the AUC of the MMI system yielded 0.887 (95% CI: 0.867–0.909). These results demonstrated the iterative improvements achieved by optimizing the model and incorporating relevant information. By refining the fusion of multiple modalities, we obtained optimal performance in accurately identifying and differentiating the subtypes within the four-class task. A delineation of performance breakdown for the individual four-class task is presented in Table 1 and Figure S2.
Table 1.
Performance of the MMI system in identifying BP, FP, VP, and PTB
| Dataset | Sensitivity (95% CI) | Specificity (95% CI) | Accuracy (95% CI) | AUC (95% CI) | |
|---|---|---|---|---|---|
| Clinical model | Validation | 0.830 (0.816–0.847) | 0.760 (0.747–0.772) | 0.786 (0.777–0.795) | 0.860 (0.855–0.867) |
| Internal testing | 0.785 (0.764–0.804) | 0.779 (0.767–0.792) | 0.788 (0.777–0.798) | 0.850 (0.844–0.856) | |
| External testing | 0.706 (0.644–0.765) | 0.875 (0.846–0.904) | 0.788 (0.755–0.821) | 0.840 (0.824–0.860) | |
| Image model | Validation | 0.838 (0.822–0.854) | 0.832 (0.820–0.843) | 0.832 (0.824–0.842) | 0.896 (0.891–0.901) |
| Internal testing | 0.847 (0.829–0.861) | 0.806 (0.795–0.819) | 0.821 (0.812–0.830) | 0.894 (0.890–0.900) | |
| External testing | 0.883 (0.853–0.920) | 0.711 (0.674–0.749) | 0.755 (0.727–0.788) | 0.856 (0.840–0.872) | |
| MMI system | Validation | 0.836 (0.820–0.853) | 0.850 (0.840–0.861) | 0.846 (0.838–0.855) | 0.905 (0.900–0.910) |
| Internal testing | 0.846 (0.830–0.860) | 0.847 (0.837–0.858) | 0.848 (0.838–0.856) | 0.910 (0.904–0.916) | |
| External testing | 0.880 (0.819–0.944) | 0.800 (0.766–0.839) | 0.835 (0.807–0.865) | 0.887 (0.867–0.909) |
Multimodal data interconnection
The primary objective of the multimodal system was to elucidate associations and shared information across diverse modalities. Then we aimed to gain new insights into the AI-based diagnosis of pulmonary infections and potentially identify new biological signatures conducive to this process. To accomplish this, a spectrum of various fusion approaches was employed, including early fusion, intermediate fusion, and late fusion, and we conducted extensive experiments to evaluate their effectiveness (Figure S3).
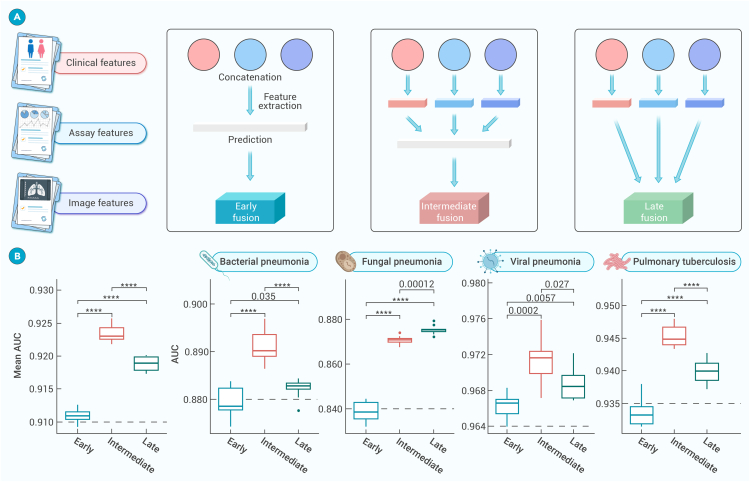
In these experiments, cross-attention fusion methods were employed to amalgamate the image and clinical results into a single layer, thereby engendering multi-information, patient-level result features (Figure 3). The classic early fusion model achieved a mean AUC of 0.910 (95% CI: 0.904–0.916) coupled with 0.848 (95% CI: 0.838–0.856) accuracy in the internal testing dataset. It is noteworthy that the intermediate fusion approach yielded a preeminent AUC of 0.923 (95% CI: 0.919–0.927) and an accuracy of 0.870 (95% CI: 0.862–0.879). Similarly, the late fusion approach garnered an AUC of 0.917 (95% CI: 0.913–0.923) and an accuracy of 0.859 (95% CI: 0.852–0.868) (Figure S4; Table S4). Overall, the intermediate approach evinced superior performance compared with the classic early fusion and late fusion models in both validation cohorts for the infections identification tasks. These results highlighted the effectiveness of the intermediate fusion approach in integrating multimodal data and elevating the accuracy for infection identification.
Figure 3.
Performance of the MMI system using different fusion approaches in identifying pulmonary infections types
(A) Structure diagram of different fusion strategies: early fusion, intermediate fusion, and late fusion. Further details were provided in Figure S4.
(B) Evaluation of the three fusion approaches to predict pneumonia types (average performance, bacterial pneumonia, fungal pneumonia, viral pneumonia, and pulmonary tuberculosis) in the internal testing dataset. The performance measures were visualized through boxplots representing the statistical distribution of AUCs, including maximum, minimum, median, and upper and lower quartiles of a set of data (first/third quartile) as well as outliers. A t test was used to calculate the p value between approaches: ∗∗∗∗p < 0.0001.
Comparison of the MMI system against physicians
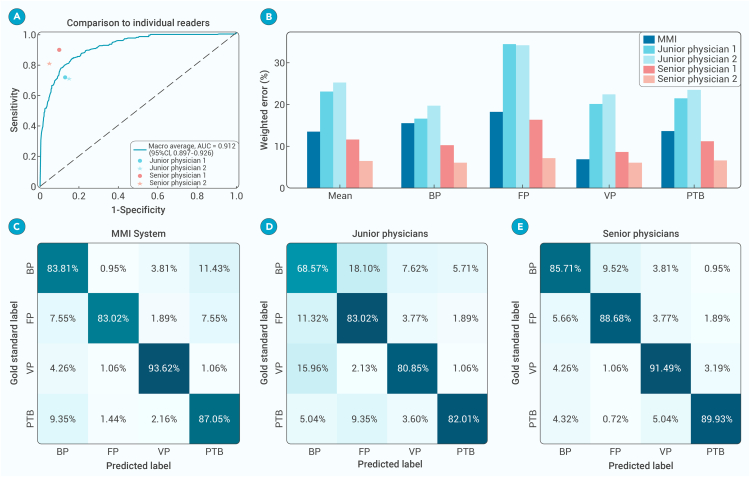
We conducted a comparative study involving four board-certified physicians divided into two groups based on their experience (the junior group, consisting of physicians with less than 10 years of professional experience, and the senior group, consisting of physicians with over 10 years of experience). In order to ensure a fair comparison, all physicians were provided with a uniform set of information encompassing CT scans, demographic details, chief complaints, and laboratory assay results. The dataset comprised 400 cases including 94 cases of viral pneumonia, 55 cases of fungal pneumonia, 108 cases of bacterial pneumonia, and 143 cases of PTB. Each physician was charged with the independent assessment of whether patients had been diagnosed with one of the four distinct infectious diseases. Subsequently, their performance was juxtaposed with that of the MMI system. The results indicated that the AI system surpassed each junior physician and demonstrated performance comparable to the senior physicians (Figure 4). Furthermore, the superiority of the AI system’s performance was evident numerically as well. When comparing the weighted errors in patient diagnosis between the AI system and the physicians, the AI system yielded 13.52%. Conversely, the range of weighted errors by expert physicians varied from 6.45% to 25.17%, with a mean of 8.98% for the senior group and 24.10% for the junior group (Table S5).
Figure 4.
Benchmarking the MMI system vs. physicians in diagnosing pulmonary infections
(A) The diagnostic performance of the MMI system and physicians for BP, FP, VP, and PTB.
(B) Weighted error results based on penalty scores.
(C–E), The confusion matrix of the MMI system (C), junior physicians (D), and senior physicians (E) to differentiate among various types of pulmonary infections.
Pathogen identification of pulmonary infections
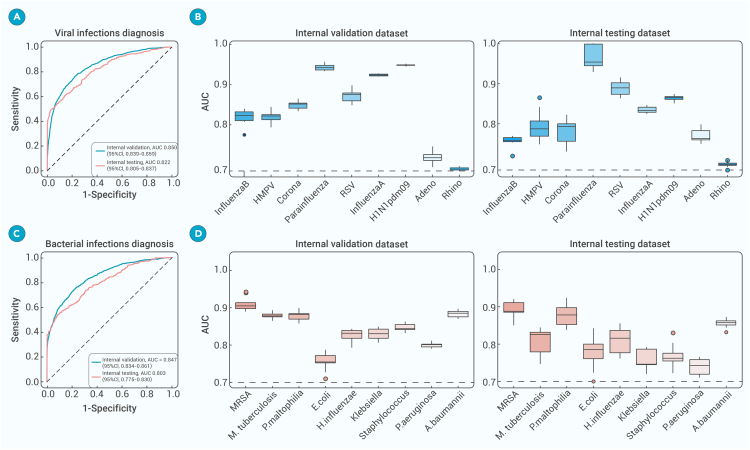
Furthermore, the MMI system also had the capability to play a pivotal role in pinpointing specific pathogens to aid precision treatment decisions. Subset 3 covered a total of 11,375 CT scans from 2,520 patients, all of whom underwent molecular testing for a spectrum of 9 common respiratory viruses: influenza B virus, human metapneumovirus (HMPV), coronavirus, parainfluenza virus, respiratory syncytial virus (RSV), influenza A virus, H1N1pdm09, adenovirus, and rhinovirus. Subset 4 comprised 2,278 CT scans from 535 patients who had been diagnosed with 9 bacterial subtypes: methicillin-resistant Staphylococcus aureus (MRSA), Mycobacterium tuberculosis, Pseudomonas maltophilia, Escherichia coli, Haemophilus influenzae, Klebsiella, Staphylococcus, Pseudomonas aeruginosa, and Acinetobacter baumannii. This comprehensive testing approach ensured the accurate identification of pathogens. For the challenging task of discerning viruses and bacteria, the system utilized transfer learning alongside a pre-trained transformer architecture initially trained on different tasks involving four types of infections and then meticulously fine tuned on specific datasets to recognize the distinctive features and patterns indicative of pathogens from CT images and clinical records.
The performance of the MMI system in accurately identifying viral infections from CT images was exceptional, as evidenced by its mean AUC score of 0.850 (95% CI: 0.839–0.859) on the validation set and 0.822 (95% CI: 0.805–0.837) on the testing set for the overall nine-way classification (Figures 5A and 5B). This remarkable performance underscored the high accuracy and reliability of the system in correctly identifying cases of viral pneumonia with minimal false positives. To gauge the discriminatory power of the model, we calculated the AUC for each viral subtype class, which quantified the ability of the MMI system to differentiate between viral pneumonia cases and other individuals, with AUC values spanning from 0.700 to 0.949 across the nine subgroups, which showcased its excellent capability to differentiate among the various virus subtypes.
Figure 5.
Performance of our AI system in identifying different subtypes of viruses and bacterial subtypes
(A) Mean ROCs demonstrating MMI system accuracy in identifying viral infections within the internal validation and testing cohorts.
(B) Boxplots showing MMI system performance for classifying nine viral subtypes.
(C) Mean ROCs highlighting MMI system accuracy in identifying bacterial infections within the internal validation and testing cohorts.
(D) Boxplots presenting MMI system performance for classifying nine bacterial subtypes. A.baumannii, Acinetobacter baumannii; E.coli, Escherichia coli; HMPV, human metapneumovirus; H.influenzae, Haemophilus influenzae; MRSA, Methicillin-resistant Staphylococcus aureus; MMI, multimodal integration;M.tuberculosis, Mycobacterium tuberculosis; P.maltophilia, Pseudomonas maltophilia; P.aeruginosa, Pseudomonasaeruginosa; RSV, respiratory syncytial virus.
By leveraging our multimodal AI system, we processed the CT images and generated predictions for the specific pathogen subtypes in each scan (Figures 5C and 5D). To evaluate the performance of our system across different bacterial subtypes, we utilized boxplot visualizations to juxtapose the AUC scores obtained from the system on both the validation cohort and testing cohort. The specific bacteria diagnostic system achieved an AUC of 0.847 (95% CI: 0.834–0.861) and 0.803 (95% CI: 0.775–0.830) on the validation and testing cohorts. The MMI system detected MRSA with an AUC exceeding 0.900. The MMI system could accurately distinguish among different bacteria subtypes based on CT images and clinical records, holding promise for aiding healthcare workers to make prompt and accurate diagnoses.
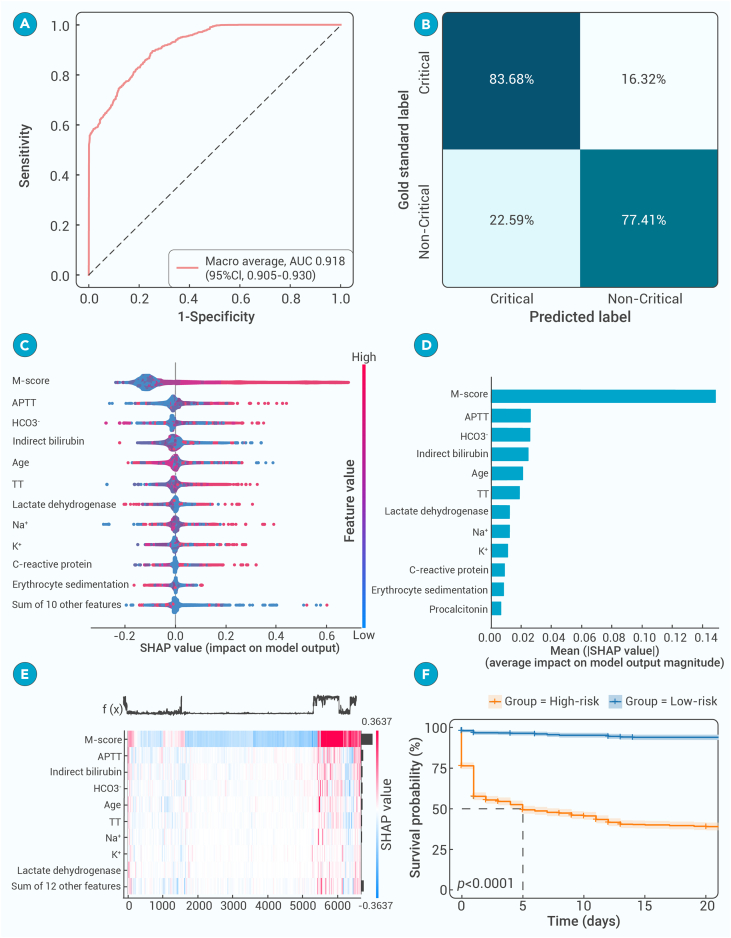
Prognosis prediction for critical illness
Regarding more actual clinical deployment, the MMI system demonstrated extraordinary performance in identifying patients with severe pneumonia. The multimodal model exhibited a robust ability to differential among varying degrees of disease severity, with an AUC of 0.918 (95% CI: 0.905–0.930) in the internal testing dataset (Figures 6A and 6B), paving the way for a deeper understanding of the critical factors influencing disease prognosis. Then, through comprehensive analysis, we estimated the most influential factors that played a crucial role in the progression toward critical illness, emphasizing the necessity of considering multiple data dimensions when assessing patient outcomes (Figures 6C and 6D). Notably, non-quantized multimodal features score (M-score) played a significant role in predicting the progression to critical illness, aligning with existing research identifying them as determinants of disease outcomes. Liver biochemical markers, including albumin, serum lactate dehydrogenase (LDH), and indirect bilirubin, emerged as important prognostic indicators, elucidating the association between liver dysfunction and adverse clinical outcomes.42 Coagulation markers, including thrombin time (TT), activated partial thromboplastin time (APTT), and platelet count, were also prognostic factors, reflecting disturbances in the coagulation system observed in critical illness.43 Furthermore, markers related to electrolyte and acid-base balance, alongside inflammation markers including C-reactive protein, lymphocyte count, and neutrophil count, contributed significantly to predicting clinical prognosis, emphasizing their importance in assessing disease severity and predicting outcomes, and were consistent with previous research.44
Figure 6.
Risk factors and clinical prognosis analysis for critical illness
(A) ROC for a binary classification of progression to critical illness in the internal testing dataset.
(B) Diagnostic accuracy for critical illness progression prediction represented by a confusion matrix.
(C and D) Predictive features for critical illness progression through Shapley Additive exPlanation (SHAP) value analysis.
(E) Correlative analysis of predictive features influencing progression to critical illness.
(F) When patients were stratified into high-risk and low-risk groups, the Kaplan-Meier curves demonstrated a significant difference in survival probabilities.
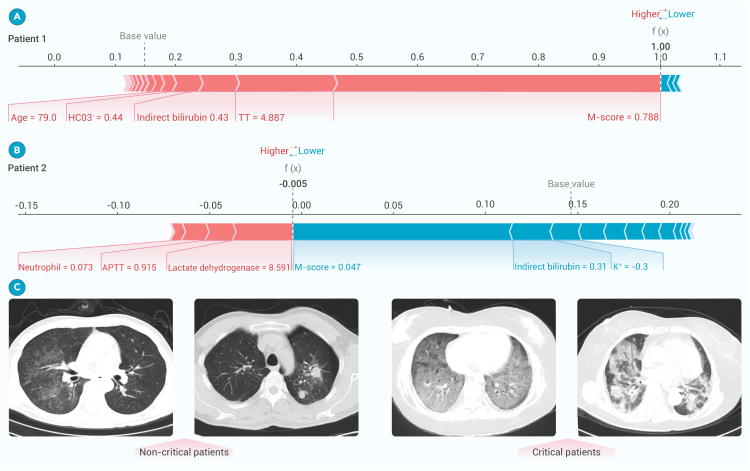
These analyses revealed that clinical parameters and coagulation markers played complementary roles in prognosis, without significant correlations observed among the leading prognostic indicators (Figure 6E). In a specific clinical case, elevated levels of TT and indirect bilirubin levels were found to be associated with more severe outcomes. Notably, LDH and APTT factors were particularly valuable for predicting non-critical disease. The high-risk group, characterized by these prognostic markers, exhibited significantly lower survival probability than the low-risk group, with a statistically significant p value of less than 0.001 based on the log-rank test (Figure 6F). These findings not only reaffirmed the expansive capability of the multimodal model in predicting critical illness but also pinpointed the specific markers pivotal for an accurate prognostic assessment using real-world examples (Figure 7). Although CT images in critically ill patients had shown specific manifestations, AI system recognized unique features that were previously unreadable to the human eye. These results indicated that accurate prognostic prediction required an integration of clinical data with lung lesion characteristics. This synergy enhanced our understanding and aided in the development of more accurate prognostic predictive tools.
Figure 7.
Illustration of the AI system for prognosis estimation of pneumonia patients
(A and B) Real-world cases for prognostic evaluation. One patient with critical illness (patient 1) and the other with non-critical disease (patient 2), to demonstrate how lung lesion characteristics and clinical features serve as input variables to predict prognosis. The influence of these inputs on risk assessment was depicted, with pink indicators increasing the projected risk on the right and blue indicators decreasing it on the left.
(C) Comparative CT scans of non-critical and critical pulmonary infection cases.
Discussion
In this study, the MMI pipeline was constructed to facilitate accurate diagnosis of pulmonary infections based on a large-scale dataset and tested on an external cohort. Integrating radiological, clinical, and laboratory multimodal features, the MMI system enabled discriminative diagnosis of bacterial, fungal, and viral pneumonia and PTB. Meanwhile, a critical illness prediction model incorporating deep learning features and multidimensional parameters was established to provide timely intervention for high-risk patients. This pipeline is poised to significantly expedite the diagnostic process for pulmonary infectious diseases and mitigate the overall disease burden.
Pulmonary infections constitute a substantial global disease burden, epitomized by the high morbidity and mortality rates. Obstacles in ascertaining the pathogens lead to the misuse of antibiotics, thereby exacerbating the proliferation of drug-resistant bacteria.45 Existing studies mostly focus on the diagnosis of specific pathogens, which is crucial for decision-making regarding specific types of pneumonia in pandemics.46,47,48,49 A deep learning-based automated detection algorithm developed using 60,989 CXR scans was used to identify active PTB and performed better than human experts.50 Another AI model based on a multicountry chest CT dataset automatically located the parietal pleura and lung parenchyma and classified COVID-19 pneumonia patients with an accuracy of 90.8%.51 However, there are still frequent types of pneumonia that require accurate and rapid diagnosis. A model based on 432 CT scans was developed to distinguish active PTB from community-acquired pneumonia.52 Recently, the Pneumonia-Plus model based on a dataset of 2,763 patients was leveraged to identify bacterial, fungal, and viral pneumonia with a mean AUC of 0.822.53 By contrast, our study has advanced the field by developing an AI model that not only broadens the spectrum of detectable diseases to include bacterial, fungal, and viral pneumonia and PTB but also achieves superior diagnostic precision with an elevated AUC of 0.910. Moreover, the MMI system precisely targeted frequent pneumonia pathogens, thus optimizing therapeutic interventions. Despite the constrained dataset of patients with partially pathogen-positive infections resulting decent preformance for pathogen prediction, these insights can still function as a critical reference for clinicians, guiding the decision-making for pathogen-specific treatments to enhance antibiotic efficacy.
Meanwhile, accurate disease diagnosis necessitates integrated multimodal data analysis, encompassing clinical features, imaging, and laboratory tests.25,54,55 However, previous studies were often limited by the relatively small number of labeled samples.30,32 Here, our approach entailed the construction of a multimodal fusion AI model, leveraging a substantial dataset, which yielded performance on par with advanced physicians. The MMI system architecture was refined based on IRENE, a transformer-based representation learning model with unified processing of multimodal input for clinical diagnostics.27 Unprocessed laboratory test indicators, clinical complaints, demographic information (such as age and sex), and chest images were automatically coded and embedded. It demonstrated superior diagnostic accuracy for pulmonary diseases compared with the single-dimensional model (AUC: 0.887 vs. 0.840). In this study, this normalized multimodal fusion architecture was employed to diagnose pulmonary infections, which also surpassed the performance of single-dimension models. The development of AI technologies, including large language models, heralds a future where medical AI models will incorporate a broader array of clinically significant multimodal data, such as breath sounds, thereby facilitating precise medical interventions.56,57,58
Then we compared three feature fusion strategies: early, late, and intermediate fusion. Different fusion architectures could affect the diagnostic efficiency of the model.59,60,61 Early fusion utilized global information from different sources, providing a holistic feature representation. However, it may also introduce redundancy or inconsistency, given the crucial disparities among information sources, potentially overlooking unique information specific to each source. Late fusion preserved the independence of each information source, but it lacked the capacity to share global information during the feature extraction phase, potentially hindering the exploitation of local advantages from one source to another. In contrast, intermediate fusion sought to merge features from multiple sources during the feature extraction process, generating a unified feature set for the final classification. This approach enhanced information propagation among sources and interactively optimized the feature extraction process.60,61 Moreover, a multistage fusion strategy was implemented in the proposed intermediate fusion architecture, allowing multimodal features to interact throughout the entire feature extraction process.
In addition, early warning of severe illness remains another tough challenge in the management of pulmonary infections. Previous studies have confirmed that radiomics could quantify the severity of pneumonia, and multiomics biomarkers, including clinical parameters, microbiome signatures, and radiomics, have been identified as prognostic indicators for patients with pneumonia.62,63,64,65,66,67 For example, a regression model constructed with age, BMI, CD4+ T lymphocytes, and serum interleukin-6 levels enabled effective prediction of the risk of developing severe disease in patients with viral pneumonia.62 The presence of Staphylococcus, Ralstonia, and Enterococcus was strongly associated with acute respiratory distress syndrome by microbiome investigations.63 Additionally, a neural network model based on CT images was utilized to assess disease severity in patients with viral pneumonia (AUC: 0.75).67 The present study has pinpointed multiple risk factors from multimodal characteristics as potential predictors of severe pneumonia. The integration of diverse data sources will allow for a comprehensive approach to predicting critical illness, paving the way for effective risk stratification of patients and preemptive medical interventions.
Notably, co-infection with multiple pathogens increases the public health burden.68 The present study provides an initial exploration of predicting co-infection, but the intricate mechanism by which mixed infections affect imaging performance remains unclear. The advent of single-cell technology makes it possible to unravel the dynamic interplays in the disease microenvironment, which may be a promising opportunity.69,70,71,72,73 Several studies have elucidated that T cells and natural killer cells in patients with bacterial pneumonia complicated by sepsis exhibited activation and depletion characteristics, respectively, and that the proportion of plasma cells is significantly increased.71 In contrast, cell subsets such as DC_c4-LILRA4, B_c05-MZB1-XBP1, and Neu_c3-CST7 in viral pneumonia are closely associated with disease severity.72 Furthermore, gene expression profiles have revealed distinct patterns: CCL7, CCL8, CCL13, and IFIT2 are highly expressed in viral pneumonia, while BAG3, HIF1A and IL1B are upregulated in other types of pneumonia (including bacterial infections, etc.), and CCL2, RNASE1, and MAFB genes were highly expressed in different types of pneumonia.73 These specific molecular events forge new insights for the deep excavation of biological mechanisms underpinning macroscopic imaging observations, providing a pivotal foundation for more nuanced diagnostic and therapeutic decision-making.
This study had several limitations that should be acknowledged. First, mixed infections present as a prevalent yet intricate challenge in clinical scenarios. We constructed a preliminary prediction model, but quantitative evaluation of mixed infections needs to be explored in conjunction with high-throughput molecular detection methods. Second, the practical application of AI models necessitates enhancements of both usability and accuracy. The integration of a predictive AI model with a clinical workflow must be seamless to achieve optimal results.74 The existing models still require further optimization to meet the demands of medical applications. Last but not the least, future research should focus on validating these findings using a broader range of central datasets, in this way reinforcing the generalizability of the AI system.
In summary, this study pioneered a multitask model based on multimodal clinical data from over 20,000 patients. The MMI system fused clinical, imaging, and laboratory assay features to diagnose various types of pulmonary infections and pathogens. It also incorporated key indicators to warn critically ill patients. This comprehensive approach could not only enhance the accuracy of diagnostics, but also guide the accurate use of medication in clinical settings. It holds the potential to evolve into a robust tool for precise interventions among patients with pulmonary infections, propelling the advancement into a new epoch of clinical care.
Materials and methods
For materials and methods, see the supplemental information.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (82341083, 82100119), the Science and Technology Project of Sichuan (2020YFG0473, 2022ZDZX0018), the Beijing Municipal Science and Technology Planning Project (Z211100003521009), Hong Kong Research Grants Council through General Research Fund (Grant 17207722), the Sichuan University from “0” to “1” Innovation Project (2023SCUH0051), and the 1.3.5 Project for Disciplines Excellence of West China Hospital, Sichuan University (ZYYC23027).
Author contributions
C.W. conceived the idea and designed the experiments. J.S., J.M., Y.Y., S.Z., and W.L. implemented and performed the experiments. J.S., J.M., W.W., and C.W. analyzed the data and experimental results. J.S., J.M., and C.W. wrote the paper. All authors commented on the paper.
Declaration of interests
The authors declare no competing interests.
Published Online: May 22, 2024
Footnotes
It can be found online at https://doi.org/10.1016/j.xinn.2024.100648.
Contributor Information
Weimin Li, Email: weimi003@scu.edu.cn.
Chengdi Wang, Email: chengdi_wang@scu.edu.cn.
Lead contact website
Supplemental information
References
- 1.GBD 2021 Causes of Death Collaborators Global burden of 288 causes of death and life expectancy decomposition in 204 countries and territories and 811 subnational locations, 1990-2021: a systematic analysis for the Global Burden of Disease Study 2021. Lancet. 2024;403:2100–2132. doi: 10.1016/s0140-6736(24)00367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD 2021 Diseases and Injuries Collaborators Global incidence, prevalence, years lived with disability (YLDs), disability-adjusted life-years (DALYs), and healthy life expectancy (HALE) for 371 diseases and injuries in 204 countries and territories and 811 subnational locations, 1990-2021: a systematic analysis for the Global Burden of Disease Study 2021. Lancet. 2024;403:2133–2161. doi: 10.1016/s0140-6736(24)00757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres A., Cilloniz C., Niederman M.S., et al. Pneumonia. Nat. Rev. Dis. Primers. 2021;7(1):25. doi: 10.1038/s41572-021-00259-0. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization . 2022. Global Tuberculosis Report 2022. [Google Scholar]
- 5.Furin J., Cox H., Pai M. Tuberculosis. Lancet. 2019;393(10181):1642–1656. doi: 10.1016/s0140-6736(19)30308-3. [DOI] [PubMed] [Google Scholar]
- 6.Jain S., Self W.H., Wunderink R.G., et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N. Engl. J. Med. 2015;373(5):415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H., Lu S. COVID-19 and tuberculosis. J. Transl. Int. Med. 2020;8(2):59–65. doi: 10.2478/jtim-2020-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Hu T.Y. Strategies for advanced personalized tuberculosis diagnosis: current technologies and clinical approaches. Precis. Clin. Med. 2021;4(1):35–44. doi: 10.1093/pcmedi/pbaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aliberti S., Dela Cruz C.S., Amati F., et al. Community-acquired pneumonia. Lancet. 2021;398(10303):906–919. doi: 10.1016/s0140-6736(21)00630-9. [DOI] [PubMed] [Google Scholar]
- 10.Cillóniz C., Torres A., Niederman M.S. Management of pneumonia in critically ill patients. BMJ. 2021;375 doi: 10.1136/bmj-2021-065871. [DOI] [PubMed] [Google Scholar]
- 11.Musher D.M., Thorner A.R. Community-acquired pneumonia. N. Engl. J. Med. 2014;371(17):1619–1628. doi: 10.1056/NEJMra1312885. [DOI] [PubMed] [Google Scholar]
- 12.Claessens Y.E., Debray M.P., Tubach F., et al. Early chest computed tomography scan to assist diagnosis and guide treatment decision for suspected community-acquired pneumonia. Am. J. Respir. Crit. Care Med. 2015;192(8):974–982. doi: 10.1164/rccm.201501-0017OC. [DOI] [PubMed] [Google Scholar]
- 13.Péju E., Belicard F., Silva S., et al. Management and outcomes of pregnant women admitted to intensive care unit for severe pneumonia related to SARS-CoV-2 infection: the multicenter and international COVIDPREG study. Intensive Care Med. 2022;48(9):1185–1196. doi: 10.1007/s00134-022-06833-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singhal K., Azizi S., Tu T., et al. Large language models encode clinical knowledge. Nature. 2023;620(7972):172–180. doi: 10.1038/s41586-023-06291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bera K., Braman N., Gupta A., et al. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat. Rev. Clin. Oncol. 2022;19(2):132–146. doi: 10.1038/s41571-021-00560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong F., de la Fuente-Nunez C., Collins J.J. Leveraging artificial intelligence in the fight against infectious diseases. Science. 2023;381(6654):164–170. doi: 10.1126/science.adh1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theodosiou A.A., Read R.C. Artificial intelligence, machine learning and deep learning: Potential resources for the infection clinician. J. Infect. 2023;87(4):287–294. doi: 10.1016/j.jinf.2023.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Shao J., Feng J., Li J., et al. Novel tools for early diagnosis and precision treatment based on artificial intelligence. Chin. Med. J. Pulm. Crit. Care Med. 2023;1(3):148–160. doi: 10.1016/j.pccm.2023.05.001. [DOI] [Google Scholar]
- 19.Esteva A., Kuprel B., Novoa R.A., et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kermany D.S., Goldbaum M., Cai W., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Wang C., Ma J., Zhang S., et al. Development and validation of an abnormality-derived deep-learning diagnostic system for major respiratory diseases. NPJ Digit. Med. 2022;5(1):124. doi: 10.1038/s41746-022-00648-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S., Liu X., Zhou L., et al. Intelligent prognosis evaluation system for stage I-III resected non-small-cell lung cancer patients on CT images: a multi-center study. eClinicalMedicine. 2023;65 doi: 10.1016/j.eclinm.2023.102270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C., Shao J., Lv J., et al. Deep learning for predicting subtype classification and survival of lung adenocarcinoma on computed tomography. Transl. Oncol. 2021;14(8) doi: 10.1016/j.tranon.2021.101141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Q., Wang C., Guo J., et al. The gap in the thickness: estimating effectiveness of pulmonary nodule detection in thick- and thin-section CT images with 3D deep neural networks. Comput. Methods Programs Biomed. 2023;229 doi: 10.1016/j.cmpb.2022.107290. [DOI] [PubMed] [Google Scholar]
- 25.Boehm K.M., Khosravi P., Vanguri R., et al. Harnessing multimodal data integration to advance precision oncology. Nat. Rev. Cancer. 2022;22(2):114–126. doi: 10.1038/s41568-021-00408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao J., Ma J., Zhang Q., et al. Predicting gene mutation status via artificial intelligence technologies based on multimodal integration (MMI) to advance precision oncology. Semin. Cancer Biol. 2023;91:1–15. doi: 10.1016/j.semcancer.2023.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Zhou H.Y., Yu Y., Wang C., et al. A transformer-based representation-learning model with unified processing of multimodal input for clinical diagnostics. Nat. Biomed. Eng. 2023;7(6):743–755. doi: 10.1038/s41551-023-01045-x. [DOI] [PubMed] [Google Scholar]
- 28.Li J., Zhou Y., Ma J., et al. The long-term health outcomes, pathophysiological mechanisms and multidisciplinary management of long COVID. Signal Transduct. Target. Ther. 2023;8(1):416. doi: 10.1038/s41392-023-01640-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G., Liu X., Shen J., et al. A deep-learning pipeline for the diagnosis and discrimination of viral, non-viral and COVID-19 pneumonia from chest X-ray images. Nat. Biomed. Eng. 2021;5(6):509–521. doi: 10.1038/s41551-021-00704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakhani P., Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 31.Zhang K., Liu X., Shen J., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433.e11. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mei X., Lee H.C., Diao K.Y., et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26(8):1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javaheri T., Homayounfar M., Amoozgar Z., et al. CovidCTNet: an open-source deep learning approach to diagnose covid-19 using small cohort of CT images. npj Digit. Med. 2021;4(1):29. doi: 10.1038/s41746-021-00399-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lessmann N., Sánchez C.I., Beenen L., et al. Automated assessment of COVID-19 reporting and data system and chest CT severity scores in patients suspected of having COVID-19 using artificial intelligence. Radiology. 2021;298(1):18–28. doi: 10.1148/radiol.2020202439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dayan I., Roth H.R., Zhong A., et al. Federated learning for predicting clinical outcomes in patients with COVID-19. Nat. Med. 2021;27(10):1735–1743. doi: 10.1038/s41591-021-01506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C., Wang Z., Wang G., et al. COVID-19 in early 2021: current status and looking forward. Signal Transduct. Target. Ther. 2021;6(1):114. doi: 10.1038/s41392-021-00527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhan Y., Wang Y., Zhang W., et al. Diagnostic accuracy of the artificial intelligence methods in medical imaging for pulmonary tuberculosis: a systematic review and meta-analysis. J. Clin. Med. 2022;12(1) doi: 10.3390/jcm12010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devlin J., Chang M.W., Lee K., et al. BERT: pre-training of deep bidirectional transformers for language understanding. arXiv. 2018 doi: 10.48550/arXiv.1810.04805. Preprint at. [DOI] [Google Scholar]
- 39.Lan Z., Chen M., Goodman S., et al. ALBERT: a lite BERT for self-supervised learning of language representations. arXiv. 2019 doi: 10.48550/arXiv.1909.11942. Preprint at. [DOI] [Google Scholar]
- 40.He K., Zhang X., Ren S., et al. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. Deep residual learning for image recognition. [Google Scholar]
- 41.Liu Z., Lin Y., Cao Y., et al. 2021 IEEE/CVF International Conference on Computer Vision. ICCV; 2021. Swin Transformer: Hierarchical vision Transformer using shifted windows. [Google Scholar]
- 42.Wong A.K.H., Woodhouse I., Schneider F., et al. Broad auto-reactive IgM responses are common in critically ill patients, including those with COVID-19. Cell Rep. Med. 2021;2(6) doi: 10.1016/j.xcrm.2021.100321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terpos E., Ntanasis-Stathopoulos I., Elalamy I., et al. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020;95(7):834–847. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Huang X., Li F., et al. Serum-integrated omics reveal the host response landscape for severe pediatric community-acquired pneumonia. Crit. Care. 2023;27(1):79. doi: 10.1186/s13054-023-04378-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/s0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Y., Qi Q., Qi S., et al. Classification of COVID-19 from community-acquired pneumonia: Boosting the performance with capsule network and maximum intensity projection image of CT scans. Comput. Biol. Med. 2023;154 doi: 10.1016/j.compbiomed.2023.106567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng F., Kottlors J., Shahzad R., et al. AI support for accurate and fast radiological diagnosis of COVID-19: an international multicenter, multivendor CT study. Eur. Radiol. 2023;33(6):4280–4291. doi: 10.1007/s00330-022-09335-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loubet P., Tubiana S., Claessens Y.E., et al. Community-acquired pneumonia in the emergency department: an algorithm to facilitate diagnosis and guide chest CT scan indication. Clin. Microbiol. Infect. 2020;26(3):382.e1–382.e7. doi: 10.1016/j.cmi.2019.06.026. [DOI] [PubMed] [Google Scholar]
- 49.Wang M., Xia C., Huang L., et al. Deep learning-based triage and analysis of lesion burden for COVID-19: a retrospective study with external validation. Lancet Digit. Health. 2020;2(10):506–515. doi: 10.1016/s2589-7500(20)30199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang E.J., Park S., Jin K.N., et al. Development and validation of a deep learning-based automatic detection algorithm for active pulmonary tuberculosis on chest radiographs. Clin. Infect. Dis. 2019;69(5):739–747. doi: 10.1093/cid/ciy967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harmon S.A., Sanford T.H., Xu S., et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;11(1):4080. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han D., Chen Y., Li X., et al. Development and validation of a 3D-convolutional neural network model based on chest CT for differentiating active pulmonary tuberculosis from community-acquired pneumonia. Radiol. Med. 2023;128(1):68–80. doi: 10.1007/s11547-022-01580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang F., Li X., Wen R., et al. Pneumonia-Plus: a deep learning model for the classification of bacterial, fungal, and viral pneumonia based on CT tomography. Eur. Radiol. 2023;33(12):8869–8878. doi: 10.1007/s00330-023-09833-4. [DOI] [PubMed] [Google Scholar]
- 54.Lipkova J., Chen R.J., Chen B., et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell. 2022;40(10):1095–1110. doi: 10.1016/j.ccell.2022.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sammut S.J., Crispin-Ortuzar M., Chin S.F., et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature. 2022;601(7894):623–629. doi: 10.1038/s41586-021-04278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang L.Y., Liu X.C., Nejatian N.P., et al. Health system-scale language models are all-purpose prediction engines. Nature. 2023;619(7969):357–362. doi: 10.1038/s41586-023-06160-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heitmann J., Glangetas A., Doenz J., et al. DeepBreath-automated detection of respiratory pathology from lung auscultation in 572 pediatric outpatients across 5 countries. NPJ Digit. Med. 2023;6(1):104. doi: 10.1038/s41746-023-00838-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moor M., Banerjee O., Abad Z.S.H., et al. Foundation models for generalist medical artificial intelligence. Nature. 2023;616(7956):259–265. doi: 10.1038/s41586-023-05881-4. [DOI] [PubMed] [Google Scholar]
- 59.Baltrusaitis T., Ahuja C., Morency L.P. Multimodal machine learning: a survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 2019;41(2):423–443. doi: 10.1109/tpami.2018.2798607. [DOI] [PubMed] [Google Scholar]
- 60.Zitnik M., Nguyen F., Wang B., et al. Machine learning for integrating data in biology and medicine: principles, practice, and opportunities. Inf. Fusion. 2019;50:71–91. doi: 10.1016/j.inffus.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang T., Xu H., Wang H., et al. Artificial intelligence for medicine: progress, challenges, and perspectives. Innovat. Med. 2023;1(2) doi: 10.59717/j.xinn-med.2023.100030. [DOI] [Google Scholar]
- 62.Chen C., Wang H., Liang Z., et al. Predicting illness severity and short-term outcomes of COVID-19: a retrospective cohort study in China. Innovation. 2020;1(1) doi: 10.1016/j.xinn.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Montassier E., Kitsios G.D., Radder J.E., et al. Robust airway microbiome signatures in acute respiratory failure and hospital-acquired pneumonia. Nat. Med. 2023;29(11):2793–2804. doi: 10.1038/s41591-023-02617-9. [DOI] [PubMed] [Google Scholar]
- 64.Chouchane O., Schuurman A.R., Reijnders T.D.Y., et al. The plasma lipidomic landscape in patients with sepsis due to community-acquired pneumonia. Am. J. Respir. Crit. Care. Med. 2024;209(8):973–986. doi: 10.1164/rccm.202308-1321OC. [DOI] [PubMed] [Google Scholar]
- 65.Ning W., Lei S., Yang J., et al. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat. Biomed. Eng. 2020;4(12):1197–1207. doi: 10.1038/s41551-020-00633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Byeon S.K., Madugundu A.K., Garapati K., et al. Development of a multiomics model for identification of predictive biomarkers for COVID-19 severity: a retrospective cohort study. Lancet Digit. Health. 2022;4(9):632–645. doi: 10.1016/s2589-7500(22)00112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lassau N., Ammari S., Chouzenoux E., et al. Integrating deep learning CT-scan model, biological and clinical variables to predict severity of COVID-19 patients. Nat. Commun. 2021;12(1):634. doi: 10.1038/s41467-020-20657-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang J., Gong Y., Zhang C., et al. Co-existence and co-infection of influenza A viruses and coronaviruses: public health challenges. Innovation. 2022;3(5) doi: 10.1016/j.xinn.2022.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silvin A., Chapuis N., Dunsmore G., et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182(6):1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Melms J.C., Biermann J., Huang H., et al. A molecular single-cell lung atlas of lethal COVID-19. Nature. 2021;595(7865):114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang T., Zhang X., Liu Z., et al. Single-cell RNA sequencing reveals the sustained immune cell dysfunction in the pathogenesis of sepsis secondary to bacterial pneumonia. Genomics. 2021;113(3):1219–1233. doi: 10.1016/j.ygeno.2021.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ren X., Wen W., Fan X., et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184(23):5838. doi: 10.1016/j.cell.2021.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grant R.A., Morales-Nebreda L., Markov N.S., et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590(7847):635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dvijotham K.D., Winkens J., Barsbey M., et al. Enhancing the reliability and accuracy of AI-enabled diagnosis via complementarity-driven deferral to clinicians. Nat. Med. 2023;29(7):1814–1820. doi: 10.1038/s41591-023-02437-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.